Abstract
Schizothorax lissolabiatus and Schizothorax griseus are native Chinese fish species with significant ecological and economic importance. We wanted to support sustainable aquaculture practices by exploring the differences in metabolic, histological, and microbiota between the two Schizothorax species. This study analyzed and compared the digestive enzyme activities, metabolic indicators, gut histology, and microbiota composition of S. lissolabiatus and S. griseus under identical farming conditions. Our comparative analysis reveals both shared characteristics and species-specific differences. While a-amylase and trypsin activities showed no significant differences between species, a consistent trend was observed, with enzyme activity highest in the hindgut, followed by the midgut, foregut, esophagus, and liver. In terms of specific metabolic markers, S. griseus shows higher lipase activity in the liver and foregut, as well as higher aspartate aminotransferase and catalase levels in the liver, along with an increased serum glucose content compared to S. lissolabiatus. Serum metabolomics analysis revealed 21 differentially abundant metabolites linked to glycerophospholipid metabolism, autophagy, purine metabolism, and necroptosis, reflecting distinct metabolic adaptations and nutritional needs for each species. These metabolic differences provide a basis for optimizing feed composition and aquaculture practices tailored to each species. Notably, S. lissolabiatus displays a significantly higher goblet cell count in the hindgut compared to S. griseus. Additionally, both species exhibit greater villi number, length, width, crypt depth, and muscle thickness in the esophagus than in the foregut, midgut, and hindgut. S. griseus also has a higher number of operational taxonomic units (OTUs) and greater microbial diversity in its intestinal microbiota, which may enhance its nutrient utilization capabilities. While both species’ dominant microbial phyla include Fusobacteria, Firmicutes, Proteobacteria, and Bacteroidetes, S. griseus demonstrated superior lipid digestion capabilities. Furthermore, the midgut and hindgut in both species display higher starch and protein digestive enzyme activities than other digestive tissues. These findings highlight the physiological differences between the two Schizothorax species, suggesting targeted strategies to improve health, growth, and sustainability in aquaculture.
1 Introduction
Schizothorax lissolabiatus and Schizothorax griseus are two native fish species of considerable ecological and economic importance, especially in Chinese aquaculture. These species belong to the order Osteichthyes, suborder Cypriniformes, family Cyprinidae, subfamily Schizothorax, and genus Schizothorax. They exhibit distinct geographical distributions: S. lissolabiatus mainly inhabits the upper reaches of the Lancang, Yuan, and Nanpan rivers (Chen, 2013; Le, 2000), while S. griseus is found in the Jinsha, upper Nanpan, Longchuan, Daying, and Wujiang rivers (Kang et al., 2015). Schizothorax species play a unique ecological role in Chinese fish diversity, akin to salmon in Western ecosystems.
In northwest China, the Schizothorax species have become economically valuable in aquaculture due to their high nutritional value and strong market demand. These fish play a key role in supporting local economies (Deng et al., 2013; Jiang et al., 2018; Lu et al., 2024). However, factors such as late sexual maturity, low fertility, slow growth rates, and high environmental requirements—coupled with habitat degradation and increased human interference—have led to a significant reduction in their habitat range and population numbers (Gao, 2019; Tang et al., 2013; Yang et al., 2021). The economic value of these species in aquaculture further underscores the need for conservation efforts.
Both S. lissolabiatus and S. griseus have been successfully domesticated and artificially bred in locations such as Kunming and Dali in Yunnan Province, China (Jiang et al., 2018; Yang et al., 2021). Enhancement and release programs in the Jinsha River aim to restore wild populations annually. Previous research has focused on their nutritional quality (Jin et al., 2024; Wang et al., 2018), reproductive biology (Jiang et al., 2018; Gao, 2019), and genetic diversity (Yang et al., 2021), providing valuable insights for aquaculture. However, studies on digestive tract morphology, digestive physiology, and physiological metabolism remain limited, which are crucial areas for optimizing feeding strategies and enhancing nutrient absorption. Understanding these aspects can support the development of nutritionally balanced, artificial feeds, thereby promoting sustainable aquaculture practices.
The digestive tract is the primary site for nutrient digestion and absorption, directly influencing fish growth, development, and reproductive success (Kumar et al., 2005). Recent studies on other Schizothorax species, such as Schizothorax kozlovi (Hu et al., 2016), Schizothorax grahami (Rong et al., 2023), Schizothorax biddulphi (Ren et al., 2020), Schizothorax prenanti (Xiang et al., 2012), and Schizothorax wangchiachii (Liang et al., 2020), have investigated digestive enzyme activities, intestinal tissue morphology, and intestinal microbial communities. These findings demonstrate that digestive tract structure, enzyme activities, and microbial composition significantly influence nutrient digestion and metabolic function in Schizothorax species.
Intestinal microbial communities, comprising thousands of microorganisms, are integral to fish health, growth, metabolism, and immune function (Xu et al., 2009). For instance, these microbes can produce enzymes that aid in food digestion, such as lipases and proteases (Ghanbari et al., 2015; Ray et al., 2012). Metagenomic analyses of these microbial communities provide insights into nutrient utilization and overall fish health (Fadrosh et al., 2014).
Metabolomics, the study of small-molecule metabolites within an organism, reveals interactions between metabolites and physiological or environmental factors (Jiye et al., 2015; Long et al., 2017). This approach has been applied in fish nutrition, reproductive physiology, and environmental toxicology, with promising results (Rigaud et al., 2020; Roques et al., 2020; Xu et al., 2016). However, there are limited studies on the physiological metabolic indicators and microbiome composition in Schizothorax species under artificial breeding conditions.
This study aims to compare the digestive enzyme activities, physiological metabolic indicators, serum metabolome, digestive tract histology, and intestinal microbiota composition of 2-year-old S. lissolabiatus and S. griseus reared under identical farming conditions. The results will not only enhance our understanding of digestive physiology and microbiota in Schizothorax species but also provide foundational knowledge for developing species-specific feeds and improving aquaculture sustainability.
2 Materials and methods
The Animal Welfare and Ethical Committee of Dali University approved all experimental protocols and guidelines (No. 2020-PZ-46). Fish were maintained in well-aerated water, anesthetized with eugenol prior to sampling, and their viscera were extracted following the Guidelines for the Care and Use of Laboratory Animals in China.
2.1 Experimental fish
Schizothorax lissolabiatus and Schizothorax griseus were obtained from the Dali Plateau Aquatic Resources Breeding Farm. Both species were co-cultivated in a circular concrete pool with an area of 30–40 m², a depth of 1.5 m, and water levels maintained between 0.8 and 1.1 m. The breeding water was sourced from Cangshan Creek, with transparency exceeding 120 cm, a pH of 7.6, and an annual temperature range of 8°C–23°C. During the breeding period, fish were fed size-specific diets tailored to their growth stages. Fry measuring 4–11 cm received a feed particle diameter of 0.5 mm, juveniles of 12–30 cm were fed with a 1.0-mm diameter, and adult fish over 30 cm were given a 2.0-mm diameter feed. Details of the dietary composition and nutritional levels for each growth stage are outlined in Table 1. Fish were fed twice daily, at 8:00 a.m. and 5:00 p.m., with a daily feed amount of approximately 3% of their body mass.
Table 1
| Items | Different culture sizes of fish | ||
|---|---|---|---|
| 4–11 cm | 12–30 cm | > 30 cm | |
| Ingredients (%) | |||
| Fish meal | 12 | 12 | 10 |
| Soybean meal | 30 | 30 | 30 |
| Rapeseed meal | 15 | 15 | 14 |
| Cottonseed meal | 13 | 13 | 10 |
| Wheat bran | 10 | 10 | 11 |
| Wheat flour | 12 | 12 | 18 |
| Fish oil | 2 | 2 | 2 |
| Soybean oil | 2 | 2 | 1 |
| Vitamin premixa | 1 | 1 | 1 |
| Mineral premixb | 1 | 1 | 1 |
| Calcium dihydrogen phosphate | 2 | 2 | 2 |
| Feed diameter (mm) | 0.5 | 1 | 2 |
| Proximate composition (%) | |||
| Moisture | 10 | 10 | 10 |
| Crude protein | 36 | 36 | 34 |
| Total lipids | 6 | 6 | 5 |
| Ash | 8 | 8 | 8 |
Formulations and nutritional components of feed.
Vitamin premix (per kg diet): vitamin A, 40000 IU, vitamin C, 300 mg, vitamin D3, 18000 IU, vitamin E, 1.0 g, vitamin K3, 60 mg, vitamin B1, 100 mg, vitamin B2, 100 mg, vitamin B6, 150 mg, vitamin B12, 0.30 mg, biotin, 6 mg, D-calcium pantothenate, 200 mg, folic acid, 20 mg, inositol, 200 mg.
Mineral premix (per kg diet): FeSO4.H2O, 180 mg, CuSO4.5H2O, 60 mg, ZnSO4.H2O, 200 mg, MnSO4.H2O, 120 mg, MgSO4.H2O, 180 mg, KH2PO4, 3.0 g, NaH2PO4, 200 mg, KI, 6 mg, CoCl2.6H2O, 6 mg, Na2SeO3, 2 mg.
2.2 Sample collection
Five 2-year-old individuals of S. lissolabiatus and five S. griseus with similar body weights (254.75 ± 29.71 g and 215.27 ± 52.97 g, respectively) were selected to ensure homogeneity in experimental conditions. This sample size was chosen to balance resource limitations and ensure sufficient statistical power for the study. Random sampling was used to avoid selection bias, and all fish were kept under identical farming conditions before sampling.
Each fish was anesthetized with eugenol (30 mg/L) for approximately 1 min. To prevent contamination, surface moisture was carefully removed before blood sampling. Blood was drawn from the caudal vein using a 1.0-mL syringe and immediately transferred to 1.5-mL tubes. The blood was then centrifuged at 10,000g for 5 min at 4°C to separate the serum. The serum was divided into three aliquots: two aliquots were stored at −20°C for analysis of physiological metabolism, antioxidant capacity, and immune indicators, while the third aliquot was frozen in liquid nitrogen and stored at −80°C for metabolomics analysis.
Fish were dissected post-anesthesia, with livers excised, weighed, and stored at −80°C to determine digestive enzyme activities, metabolism, antioxidant, and immune indices. Approximately 200 mg of hindgut contents was collected, frozen in liquid nitrogen, and stored at −80°C for analysis of intestinal microbial communities.
The gut was divided into three segments—foregut, midgut, and hindgut—based on the method of Ren et al. (2020). Approximately 0.5 cm of each segment was sampled, and after the intestinal contents and surrounding fat tissue were removed, samples were rinsed with sterile saline and fixed in 4% paraformaldehyde for histological analysis. The esophagus and gut samples were rinsed, contents expelled, and then stored at −40°C for digestive enzyme activity analysis.
2.3 Biochemical indices analysis
Approximately 0.2 g of liver or intestinal tissue was mixed with 1 mL of ice-cold physiological saline, homogenized with an IKA homogenizer (T10B, IKA Co., Germany), and centrifuged at 10,000g for 10 min at 4°C. The supernatant was collected for biochemical analysis. Additionally, 0.2 g of liver tissue was homogenized with 1 mL of methanol and centrifuged under the same conditions to analyze total cholesterol (TC) and triglyceride (TG) levels.
Levels of total soluble protein (TP), trypsin, lipase, α-amylase, total cholesterol (TC), triglyceride (TG), alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), glucose (GLU), total antioxidant capacity (T-AOC), catalase (CAT), total superoxide dismutase (T-SOD), malondialdehyde (MDA), lysozyme (LZM), acid phosphatase (ACP), and alkaline phosphatase (ALP) in tissue homogenates and serum were measured using a spectrophotometer (T6 New Century, Beijing Purkinje General Instrument Co., Ltd., Beijing, China) and commercial kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Each biochemical index was analyzed separately to ensure accuracy and consistency.
2.4 Serum metabolomics analysis
Serum samples stored at −80°C were thawed at room temperature. A 100-µL aliquot of serum was added to a 2-mL test tube with 10 µL of 0.3 mg/mL L-2-chlorophenylalanine in methanol as an internal standard. The mixture was vortexed for 10 s, followed by the addition of 300 µL methanol-acetonitrile (2:1, v/v) solution, vortexed for 1 min, sonicated in an ice-water bath for 10 min, and kept at −20°C for 30 min. After centrifugation at 10,000g for 10 min at 4°C, 300 µL of supernatant was transferred, evaporated under nitrogen, reconstituted, and further processed for metabolomics analysis using an AB Exion LC system coupled with an AB Triple TOF 6600 plus mass spectrometer. Chromatographic and mass spectrometry conditions are detailed in Supplementary Tables S1, S2.
Data processing was conducted using Progenesis QI v2.3, and compounds were identified through the EMDB database. Differential metabolites were selected based on a variable importance in projection (VIP) value > 1.0 and a t-test with p < 0.05. These metabolites were subjected to Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis.
2.5 Digestive tract histology analysis
The esophagus, foregut, midgut, and hindgut were fixed in 4% paraformaldehyde, rinsed, dehydrated, cleared with xylene, embedded in paraffin, and sectioned at 5 μm. Hematoxylin-eosin (HE) staining was performed, and sections were sealed with neutral gum. An Olympus microscope was used to observe and photograph crypt depth, villus length, and muscle thickness in tissue sections, which were analyzed with K-Viewer v1.5.3.
2.6 Intestinal microbial community
Total DNA from hindgut contents was extracted with the E.Z.N.A. DNA kit and confirmed through 1% agarose gel electrophoresis. Primers targeting the V3 and V4 regions of the bacterial 16S rRNA gene (343F/798R) were used. PCR amplification conditions included an initial denaturation at 94°C, 35 cycles of denaturation, annealing, extension, and a final extension. PCR products were confirmed, purified, quantified, and sequenced on the Illumina NovaSeq platform. Alpha and beta diversity were calculated with QIIME2, and bacterial phenotypes were predicted using the BugBase database.
2.7 Statistical analysis
Statistical analysis was conducted using SPSS 17.0, with results expressed as mean ± standard error (SE). An independent sample t-test was used to compare S. lissolabiatus and S. griseus, with p < 0.01 and p < 0.05 indicating highly significant and significant differences, respectively. For indices across different tissues, a one-way analysis of variance was followed by Duncan’s multiple range test for post hoc comparisons when significant differences were detected (p < 0.05). Assumptions of normality and homogeneity of variance were tested to ensure statistical rigor.
3 Results
3.1 Digestive enzyme activities in the liver and gut
The digestive enzyme activities in different regions of the digestive tract—esophagus, foregut, midgut, and hindgut—are shown in Table 2. S. griseus exhibited significantly higher lipase activity in the liver and foregut compared to S. lissolabiatus (p < 0.05), while no significant differences were found in the esophagus, midgut, or hindgut between the two species. Additionally, no significant differences in α-amylase and trypsin activities were observed in these regions, although S. lissolabiatus showed slightly higher activity overall.
Table 2
| Items | S. lissolabiatus | S. griseus |
|---|---|---|
| Lipase (U/g protein) | ||
| Liver | 8.54 ± 0.53bD | 16.53 ± 1.16aB |
| Esophagus | 27.59 ± 2.08A | 22.51 ± 1.47A |
| Foregut | 15.68 ± 0.68bC | 21.71 ± 1.74aA |
| Midgut | 18.85 ± 1.61BC | 15.15 ± 1.09B |
| Hindgut | 22.68 ± 1.97B | 16.77 ± 2.17B |
| α-Amylase (U/mg protein) | ||
| Liver | 0.32 ± 0.08D | 0.43 ± 0.05D |
| Esophagus | 0.69 ± 0.10CD | 0.49 ± 0.05D |
| Foregut | 1.06 ± 0.10BC | 0.82 ± 0.04C |
| Midgut | 1.55 ± 0.23B | 1.10 ± 0.06B |
| Hindgut | 2.22 ± 0.31A | 1.66 ± 0.16A |
| Trypsin (× 103 U/mg protein) | ||
| Liver | 0.79 ± 0.01C | 0.76 ± 0.02D |
| Esophagus | 6.36 ± 0.62B | 4.78 ± 0.55C |
| Foregut | 7.40 ± 0.32B | 6.56 ± 0.33BC |
| Midgut | 7.75 ± 0.71B | 7.10 ± 0.44B |
| Hindgut | 17.40 ± 2.21A | 12.75 ± 1.22A |
Digestive enzymes activities of S. lissolabiatus and S. griseus.
Data are presented as mean ± SE (N = 5). Values in the same row with different lowercase letters are significantly different (p < 0.05). Values in the same column with different uppercase letters are significantly different (p < 0.05).
In S. lissolabiatus, lipase activity varied across digestive regions, ranking highest in the esophagus, followed by the hindgut, midgut, foregut, and liver. For α-amylase, activity was ordered as hindgut > midgut > foregut > esophagus > liver, and for trypsin, the ranking was hindgut > midgut > foregut > esophagus > liver. S. griseus displayed a similar but distinct pattern, with the highest lipase activity in the esophagus, followed by the foregut, hindgut, liver, and midgut. Both α-amylase and trypsin activities followed similar trends in S. griseus.
In summary, both species exhibited distinct patterns of digestive enzyme distribution across gut regions, suggesting species-specific adaptations in nutrient processing. These enzyme activity differences may reflect physiological adaptations related to each species’ unique digestive and nutritional needs.
3.2 Metabolic indices
The metabolic indicators of both Schizothorax species are shown in Table 3. No significant differences were observed in total protein (TP), TC, TGs, ALT, BUN, or GLU levels in the liver between S. lissolabiatus and S. griseus (p > 0.05). However, AST activity was significantly higher in the liver of S. griseus than in S. lissolabiatus (p < 0.05). Serum GLU levels were also significantly higher in S. griseus (p < 0.05).
Table 3
| Items | S. lissolabiatus | S. griseus |
|---|---|---|
| Liver | ||
| TP (mg/g tissue) | 81.35 ± 2.28 | 76.16 ± 1.85 |
| TC (mmol/100g tissue) | 0.29 ± 0.03 | 0.28 ± 0.08 |
| TG (mmol/100g tissue) | 0.61 ± 0.07 | 0.65 ± 0.15 |
| AST (U/mg protein) | 64.09 ± 13.58b | 105.03 ± 2.42a |
| ALT (U/mg protein) | 57.25 ± 13.89 | 26.91 ± 5.72 |
| Urea (mmol/g protein) | 0.30 ± 0.04 | 0.50 ± 0.10 |
| Glucose (mg/g tissue) | 48.23 ± 4.22 | 45.65 ± 3.89 |
| Serum | ||
| TP (g/L) | 15.25 ± 0.84 | 16.30 ± 0.74 |
| TC (mmol/L) | 9.05 ± 1.66 | 11.99 ± 0.90 |
| TG (mmol/L) | 3.53 ± 0.31 | 3.64 ± 0.35 |
| AST (U/mL) | 114.00 ± 9.07 | 140.90 ± 17.56 |
| ALT (U/mL) | 3.21 ± 0.91 | 1.28 ± 1.79 |
| Urea (mmol/L) | 5.83 ± 0.76 | 7.19 ± 1.09 |
| Glucose (mg/dL) | 117.54 ± 9.10b | 156.56 ± 12.91a |
Metabolic indices of liver and serum between S. lissolabiatus and S. griseus.
Data are presented as mean ± SE (N = 5). Values in the same row with different lowercase letters are significantly different (p < 0.05). TP, total soluble protein; TC, total cholesterol; TG, triglycerides; AST, aspartate aminotransferase; ALT, alanine aminotransferase.
Overall, while most metabolic indicators did not differ significantly, elevated AST and GLU levels in S. griseus may suggest species-specific differences in liver function and energy metabolism, potentially indicating distinct metabolic adaptations between the species.
3.3 Antioxidant capacity and immunity indices in the liver and serum
The antioxidant and immune indices of S. lissolabiatus and S. griseus are presented in Table 4. S. griseus showed significantly higher CAT activity and lower MDA levels in the liver compared to S. lissolabiatus (p < 0.05). ACP activity was higher in S. lissolabiatus. No significant differences were observed in T-AOC, T-SOD, LZM, ACP, or ALP levels between the species.
Table 4
| Items | S. lissolabiatus | S. griseus |
|---|---|---|
| Liver | ||
| T-AOC (U/g protein) | 1.04 ± 0.19 | 1.08 ± 0.04 |
| CAT (U/g protein) | 2.44 ± 0.04b | 2.71 ± 0.05a |
| T-SOD (U/mg protein) | 10.35 ± 1.43 | 13.09 ± 0.37 |
| MDA (nmol/mg protein) | 2.21 ± 0.51a | 0.85 ± 0.15b |
| LZM (μg/mg protein) | 0.22 ± 0.04 | 0.12 ± 0.02 |
| ALP (U/g protein) | 5.86 ± 1.41 | 7.46 ± 0.98 |
| ACP (U/g protein) | 190.60 ± 29.28 | 271.67 ± 22.47 |
| Serum | ||
| T-AOC (U/mL) | 10.33 ± 0.83 | 8.63 ± 0.53 |
| CAT (U/mL) | 3.12 ± 0.87 | 4.67 ± 0.51 |
| T-SOD (U/mL) | 55.46 ± 7.96 | 70.04 ± 2.68 |
| MDA (nmol/mL) | 3.52 ± 0.39 | 3.49 ± 0.16 |
| LZM (μg/mL) | 1.40 ± 0.36 | 1.42 ± 0.21 |
| ALP (U/mL) | 2.50 ± 0.10 | 2.56 ± 0.26 |
| ACP (U/mL) | 18.19 ± 1.54 | 26.61 ± 7.70 |
Antioxidant and immune indices of liver and serum of S. lissolabiatus and S. griseus.
Data are presented as mean ± SE (N = 5). Values in the same row with different lowercase letters are significantly different (p < 0.05). T-AOC, total antioxidant capacity; CAT, catalase; T-SOD, total superoxide dismutase; MDA, malondialdehyde; LZM, lysozyme; ALP, alkaline phosphatase; ACP, acid phosphatase.
In summary, the increased CAT activity and lower MDA levels in S. griseus suggest a potentially stronger antioxidant capacity compared to S. lissolabiatus. However, immune indices were generally similar, suggesting limited differences in immune function between the species.
3.4 Serum metabolomics
The partial least squares discriminant analysis (PLS-DA) score plot of serum samples showed clear separation between S. lissolabiatus and S. griseus (Figure 1), confirming the model’s reliability with an R²Y value of 0.99 and a Q² value of 0.802 (Figure 2). Differentially abundant metabolites, identified with a VIP score > 1.0 and p < 0.05, included 21 metabolites linked to various metabolic pathways (e.g., glycerophospholipid metabolism, autophagy, and necroptosis), as shown in Table 5, Supplementary Table S3; Figure 3.
Figure 1

Partial least squares discriminant analysis (PLS-DA) score plot based on the metabolites in the serum of S. lissolabiatus and S. griseus from same culture conditions. R2X [1] = 0.325, R2X [2] = 0.139, Ellipse: Hotelling’s T2 (95%).
Figure 2
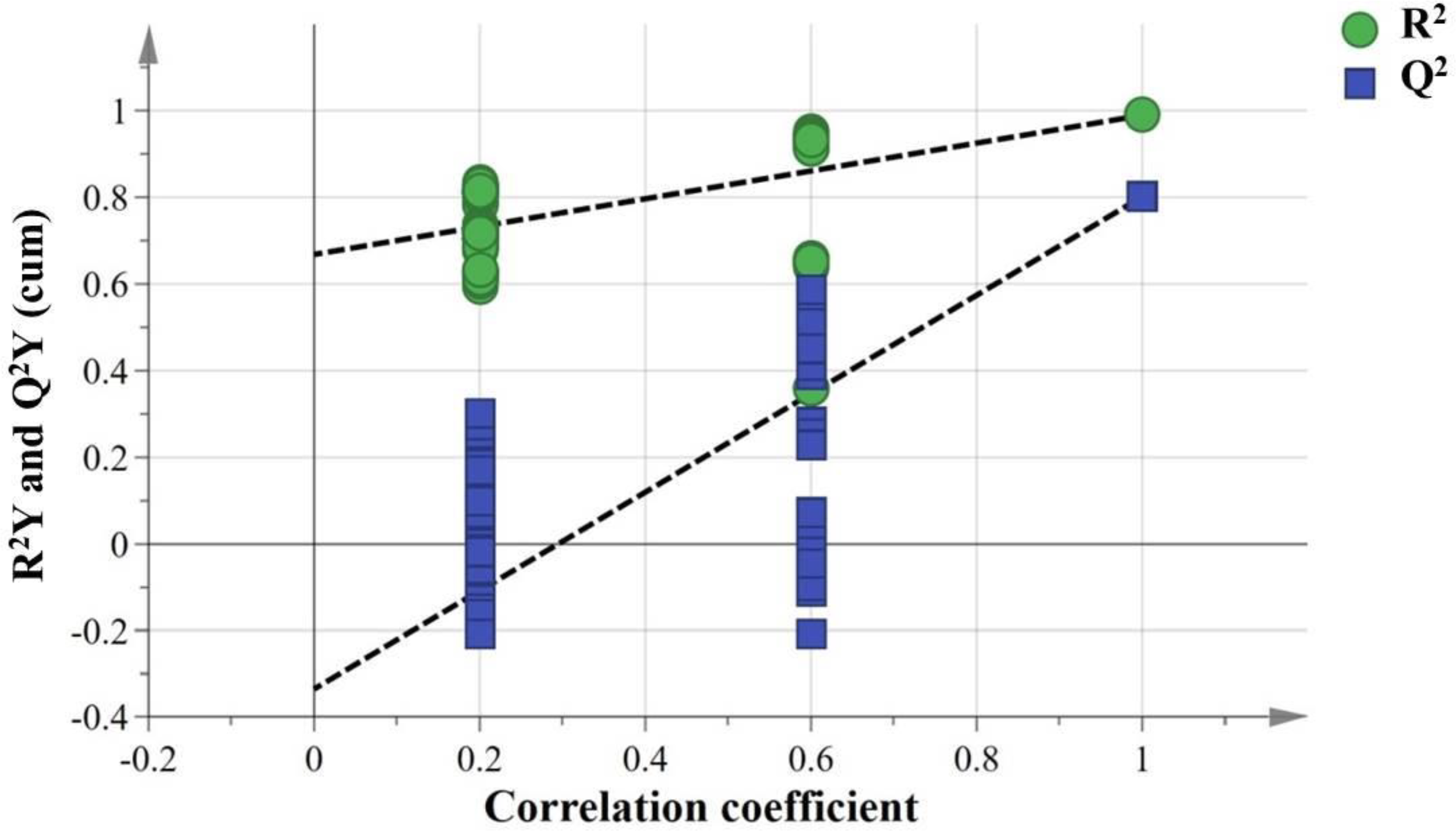
Permutation test of the serum. R2: green circle, Q2: blue square. The green line represents the regression line for R2 and the blue line for Q2. The intercept: R2 = 0.668, Q2 = −0.336.
Table 5
| Differential metabolites | VIPa | P-valueb | FCc | Level d |
|---|---|---|---|---|
| PI(18:1(9Z)/20:3(8Z,11Z,14Z)) | 13.95 | 3.61 × 10−3 | 15.67 | Up |
| Aminocaproic acid | 3.20 | 2.27 × 10−2 | 2.22 | Up |
| D-Glucuronic acid | 1.44 | 3.41 × 10−3 | 25.79 | Up |
| Norvaline | 1.33 | 1.32 × 10−2 | 1.85 | Up |
| PC(20:3(8Z,11Z,14Z)/18:2(9Z,12Z)) | 44.26 | 9.10 × 10−4 | 0.02 | Down |
| PC(16:0/16:0) | 22.15 | 2.81 × 10−2 | 0.03 | Down |
| PC(20:3(5Z,8Z,11Z)/20:4(8Z,11Z,14Z,17Z)) | 18.38 | 4.35 × 10−3 | 0.08 | Down |
| PC(20:5(5Z,8Z,11Z,14Z,17Z)/20:3(5Z,8Z,11Z)) | 11.18 | 2.12 × 10−2 | 0.19 | Down |
| SM(d18:0/16:1(9Z)) | 9.79 | 2.41 × 10−2 | 0.03 | Down |
| PE(20:3(8Z,11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | 7.23 | 1.57 × 10−2 | 0.26 | Down |
| PC(16:0/20:5(5Z,8Z,11Z,14Z,17Z)) | 6.88 | 4.12 × 10−2 | 0.30 | Down |
| PC(20:3(8Z,11Z,14Z)/22:4(7Z,10Z,13Z,16Z)) | 6.07 | 1.65 × 10−2 | 0.00 | Down |
| PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/P-16:0) | 5.13 | 4.22 × 10−2 | 0.16 | Down |
| PC(18:1(9Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | 5.13 | 4.76 × 10−2 | 0.41 | Down |
| PC(18:4(6Z,9Z,12Z,15Z)/16:1(9Z)) | 4.39 | 2.35 × 10−2 | 0.33 | Down |
| PC(16:0/14:1(9Z)) | 4.25 | 6.32 × 10−3 | 0.00 | Down |
| PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/18:4(6Z,9Z,12Z,15Z)) | 3.62 | 1.22 × 10−2 | 0.19 | Down |
| PS(18:0/22:5(7Z,10Z,13Z,16Z,19Z)) | 2.67 | 1.13 × 10−2 | 0.45 | Down |
| Uric acid | 1.69 | 6.27 × 10−3 | 0.40 | Down |
| Deoxyinosine | 1.54 | 3.25 × 10−3 | 0.32 | Down |
| PC(18:4(6Z,9Z,12Z,15Z)/16:0) | 1.52 | 3.93 × 10−2 | 0.14 | Down |
The differential metabolites in the serum between S. lissolabiatus and S. griseus.
Variable importance in the projection (VIP) was acquired from the PLS-DA model with a threshold of 1.0.
P-values were calculated from two-tailed Student t-test.
FC, Fold change, mean value of peak area obtained from S. griseus/mean value of peak area obtained from S. lissolabiatus. If the FC value is > 1, it means that metabolites in S. griseus are more than in S. lissolabiatus.
Up: an increased in the S. griseus, down: a decrease in the S. griseus.
Figure 3
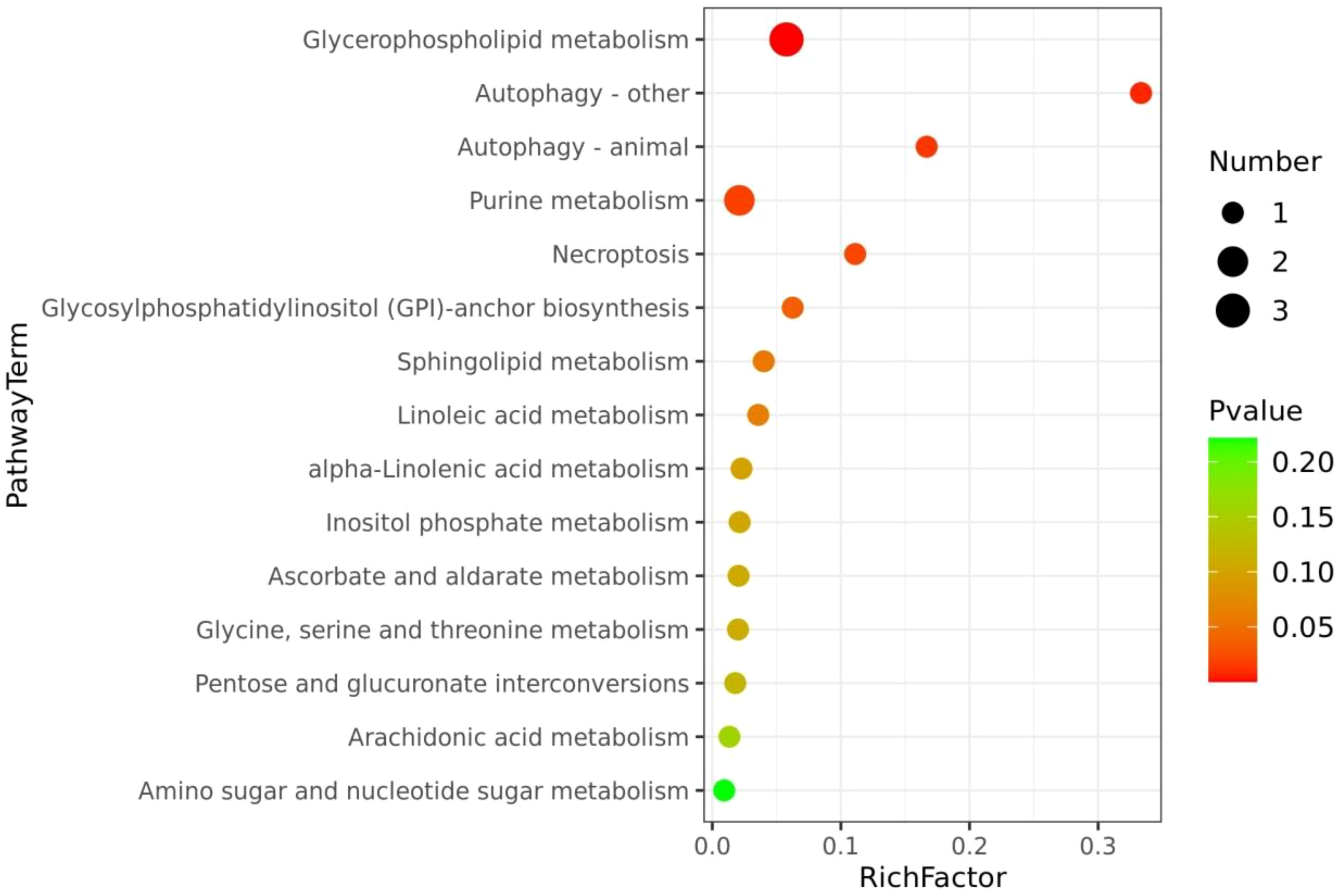
The metabolome view map of significant metabolic pathways characterized in the serum between the S. lissolabiatus and S. griseus. This figure illustrates significantly changed pathways based on enrichment and topology analysis. The x-axis represents pathway enrichment, and the y-axis represents pathway impact. Larger sizes and darker colors represent greater pathway enrichment and higher pathway impact values, respectively.
In summary, the PLS-DA analysis indicated significant metabolic differentiation between the two species, with specific metabolite profiles suggesting possible physiological and ecological adaptations.
3.5 Digestive tract histology
Table 6 and Figure 4 present the histological indices of the digestive tracts in S. lissolabiatus and S. griseus. Both species exhibited similar histology structures across most regions, although S. lissolabiatus had a significantly higher goblet cell count in the hindgut (p < 0.05). This increase may enhance mucosal protection and digestion in the hindgut, suggesting functional adaptations.
Table 6
| Items | S. lissolabiatus | S. griseus |
|---|---|---|
| Villus number | ||
| Esophagus | 41.33 ± 4.62A | 41.00 ± 2.16A |
| Foregut | 30.50 ± 1.19B | 33.25 ± 2.17AB |
| Midgut | 26.25 ± 1.89B | 29.50 ± 3.07B |
| Hindgut | 27.25 ± 1.65B | 28.67 ± 1.89B |
| Villus length (μm) | ||
| Esophagus | 990.36 ± 100.91A | 1236.61 ± 103.59A |
| Foregut | 744.99 ± 69.06AB | 805.62 ± 76.81B |
| Midgut | 662.16 ± 43.59B | 650.09 ± 50.61B |
| Hindgut | 737.32 ± 106.01AB | 771.76 ± 28.16B |
| Villus width (μm) | ||
| Esophagus | 184.07 ± 14.00A | 193.65 ± 18.70 |
| Foregut | 167.58 ± 4.89AB | 158.02 ± 9.22 |
| Midgut | 154.00 ± 5.97B | 162.75 ± 15.03 |
| Hindgut | 179.64 ± 4.43AB | 160.70 ± 10.87 |
| Crypt depth (μm) | ||
| Esophagus | 935.77 ± 97.55A | 1196.02 ± 81.96A |
| Foregut | 701.68 ± 73.70AB | 746.43 ± 65.11B |
| Midgut | 521.98 ± 28.81B | 524.33 ± 64.36C |
| Hindgut | 649.61 ± 101.53B | 675.08 ± 30.08BC |
| Muscle thickness (μm) | ||
| Esophagus | 220.88 ± 33.03A | 260.67 ± 20.48A |
| Foregut | 157.57 ± 7.00AB | 142.09 ± 15.56B |
| Midgut | 128.86 ± 6.14B | 142.01 ± 10.70B |
| Hindgut | 172.46 ± 29.55AB | 137.02 ± 1.70B |
| Goblet cell number (per villus) | ||
| Esophagus | 23.44 ± 2.02 | 21.72 ± 2.33 |
| Foregut | 26.88 ± 1.74 | 25.47 ± 3.58 |
| Midgut | 20.78 ± 1.77 | 21.47 ± 1.18 |
| Hindgut | 23.03 ± 0.55a | 18.67 ± 0.91b |
Histology of the digestive tract of S. lissolabiatus and S. griseus.
Data are presented as mean ± SE (N = 5). Values in the same row with different lowercase letters are significantly different (p < 0.05). Values in the same column with different uppercase letters are significantly different (p < 0.05).
Figure 4
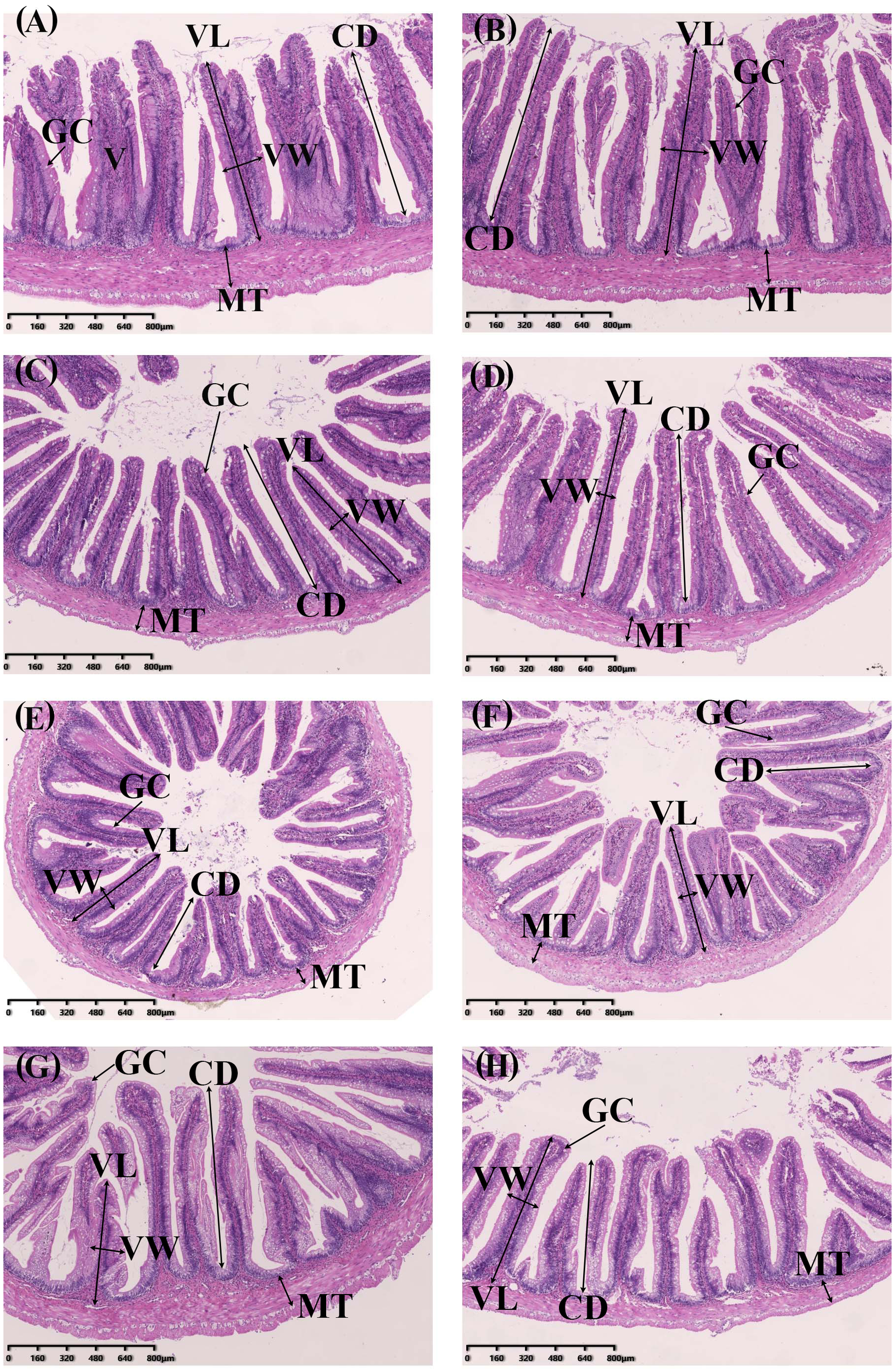
The intestinal morphology of the S. lissolabiatus and S. griseus. (A) esophagus of S. lissolabiatus, (B) esophagus of S. griseus, (C) foregut of S. lissolabiatus, (D) foregut of S. griseus, (E) midgut of S. lissolabiatus, (F) midgut of S. griseus, (G) hindgut of S. lissolabiatus, and (H) hindgut of S. griseus. GC, gobelt cell; MT, muscle thickness; V, villi; VL, villus length; VW, villus width; CD, crypt depth.
3.6 Intestinal microbial community
A total of 656 operational taxonomic units (OTUs) were identified, with S. griseus exhibiting a higher OTU count (Figure 5) and α-diversity (Table 7), indicating a more diverse intestinal microbiota. Despite this diversity, n-diversity analysis (Figures 6A, B) showed that both species shared similar microbial community structures, dominated by phyla such as Fusobacteria, Firmicutes, Proteobacteria, and Bacteroidetes (Figures 7, 8). In summary, the higher microbial diversity in S. griseus may support enhanced nutrient utilization, while the similar microbial composition across both species indicates shared functional adaptations in the gut microbiota.
Figure 5
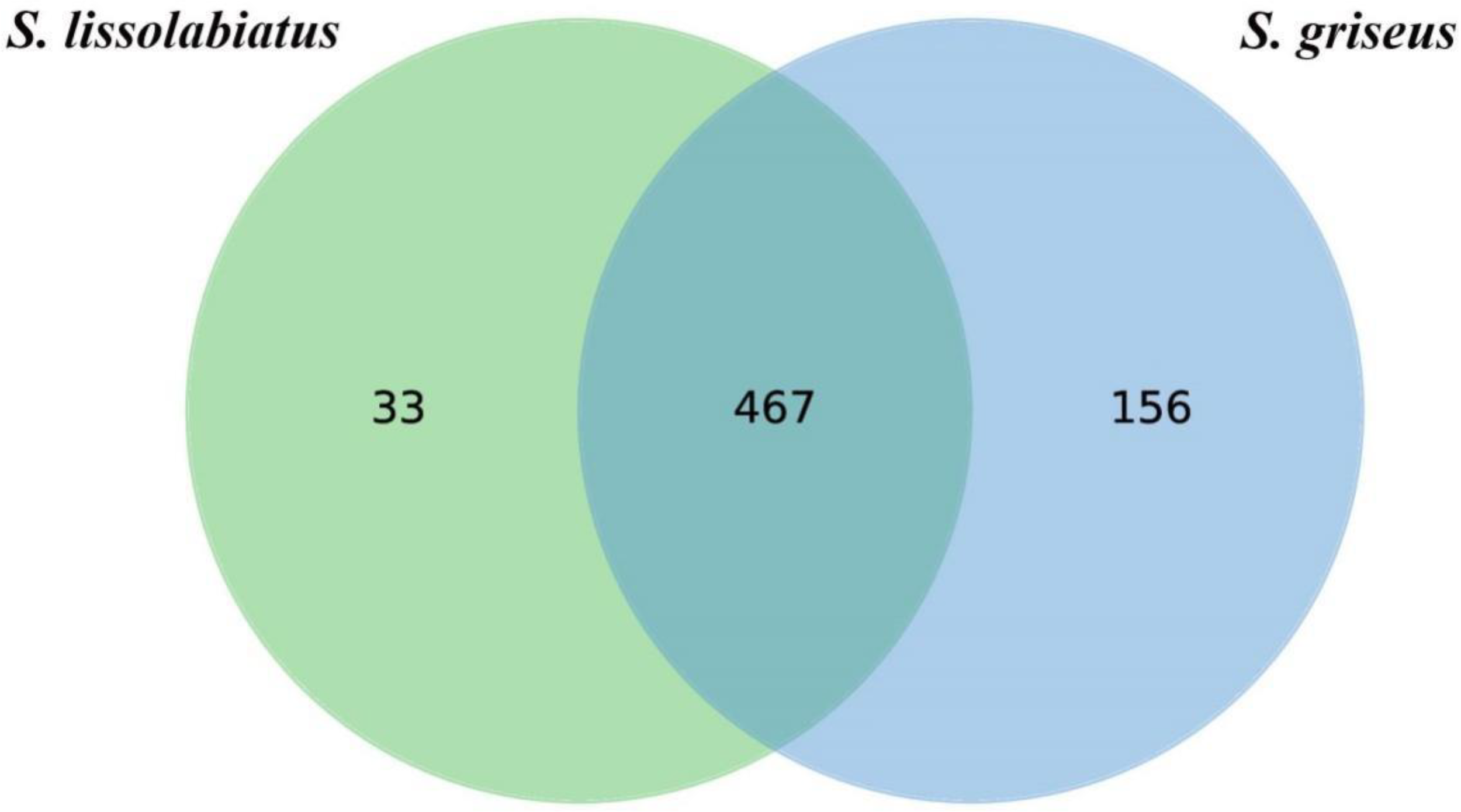
OTU Venn diagram of intestinal tract microorganisms of S. lissolabiatus and S. griseus.
Table 7
| Items | S. lissolabiatus | S. griseus |
|---|---|---|
| Chao1 | 420.69 ± 22.38 | 456.02 ± 14.73 |
| Shannon | 1.64 ± 0.14 | 1.72 ± 0.12 |
| Observed_species | 334.13 ± 15.76b | 410.05 ± 16.20a |
| Simpson | 0.48 ± 0.05 | 0.45 ± 0.04 |
| PD_whole_tree | 14.57 ± 0.72b | 17.56 ± 0.74a |
| Goods_coverage | 0.9986 ± 0.0001 | 0.9987 ± 0.0001 |
Microbial alpha diversity indexes of intestinal contents of S. lissolabiatus and S. griseus.
Data are presented as mean ± SE (N = 5). Values in the same row with different lowercase letters are significantly different (p < 0.05).
Figure 6
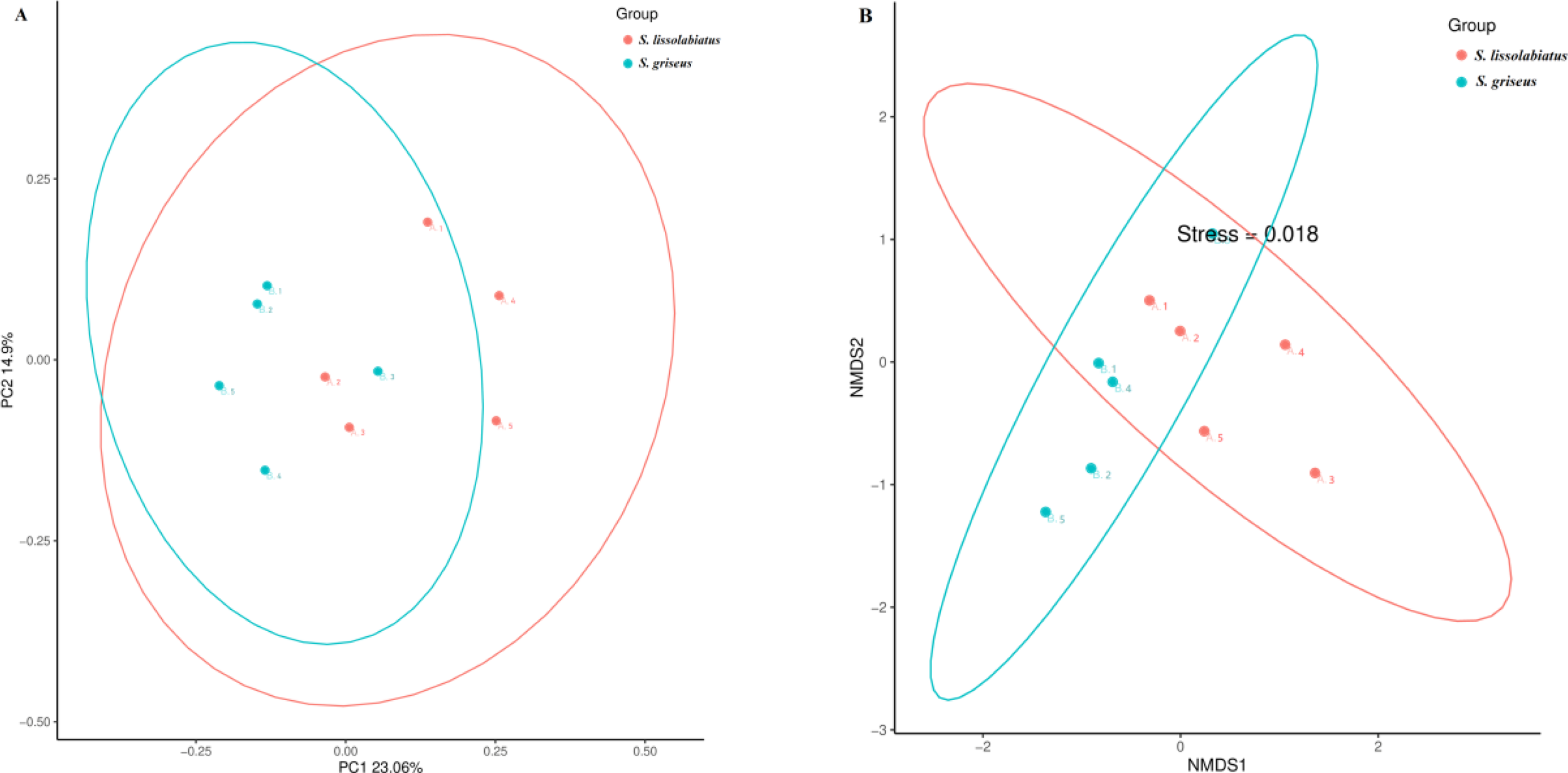
The principal coordinate analysis (A) and non-metric multidimensional scaling analysis (B) of intestinal microbial community composition of S. lissolabiatus and S. griseus..
Figure 7
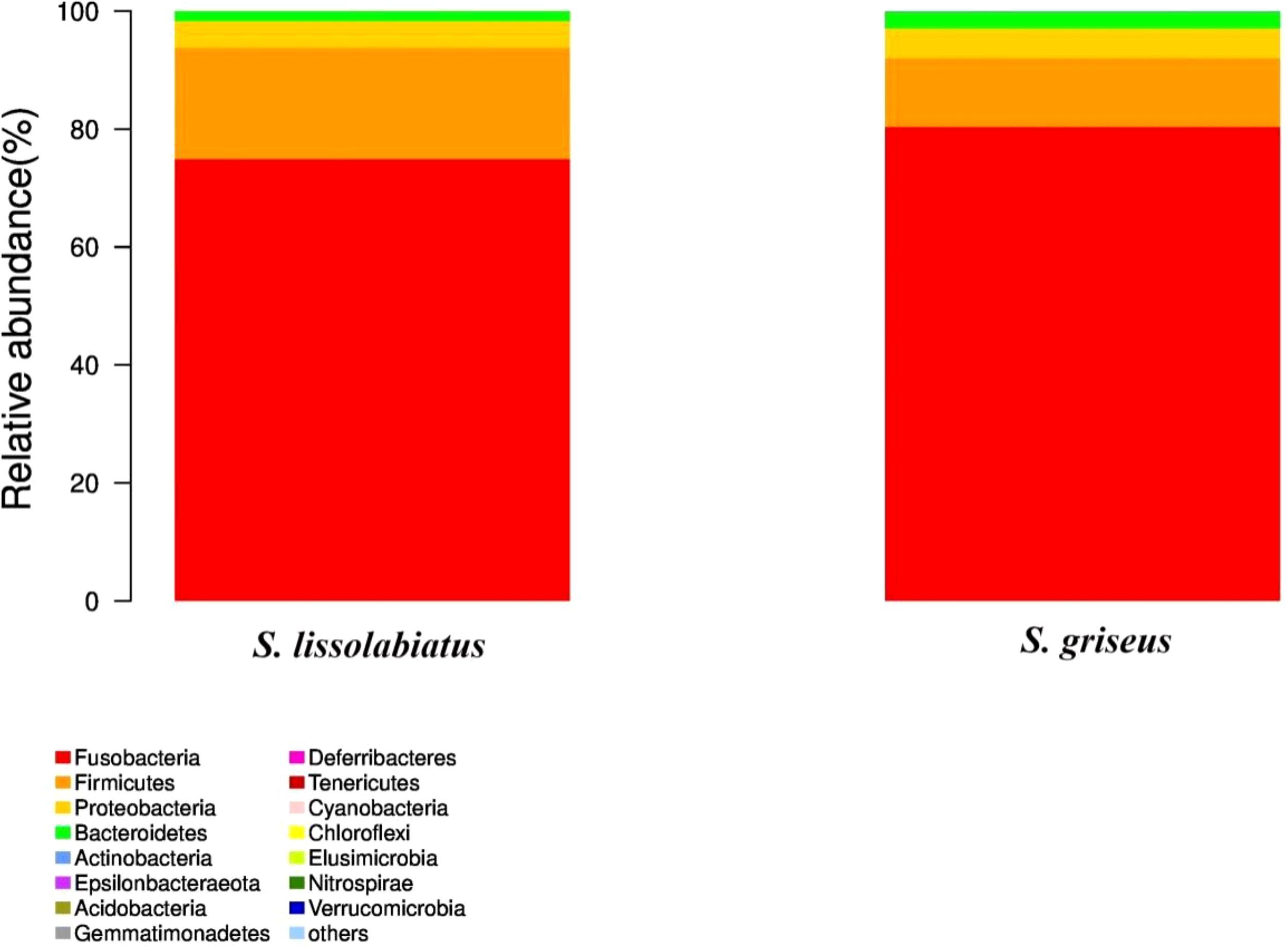
Intestinal microbiota structure and relative abundance of structure and relative abundance of intestinal microbiota of S. lissolabiatus and S. griseus at the phylum level.
Figure 8
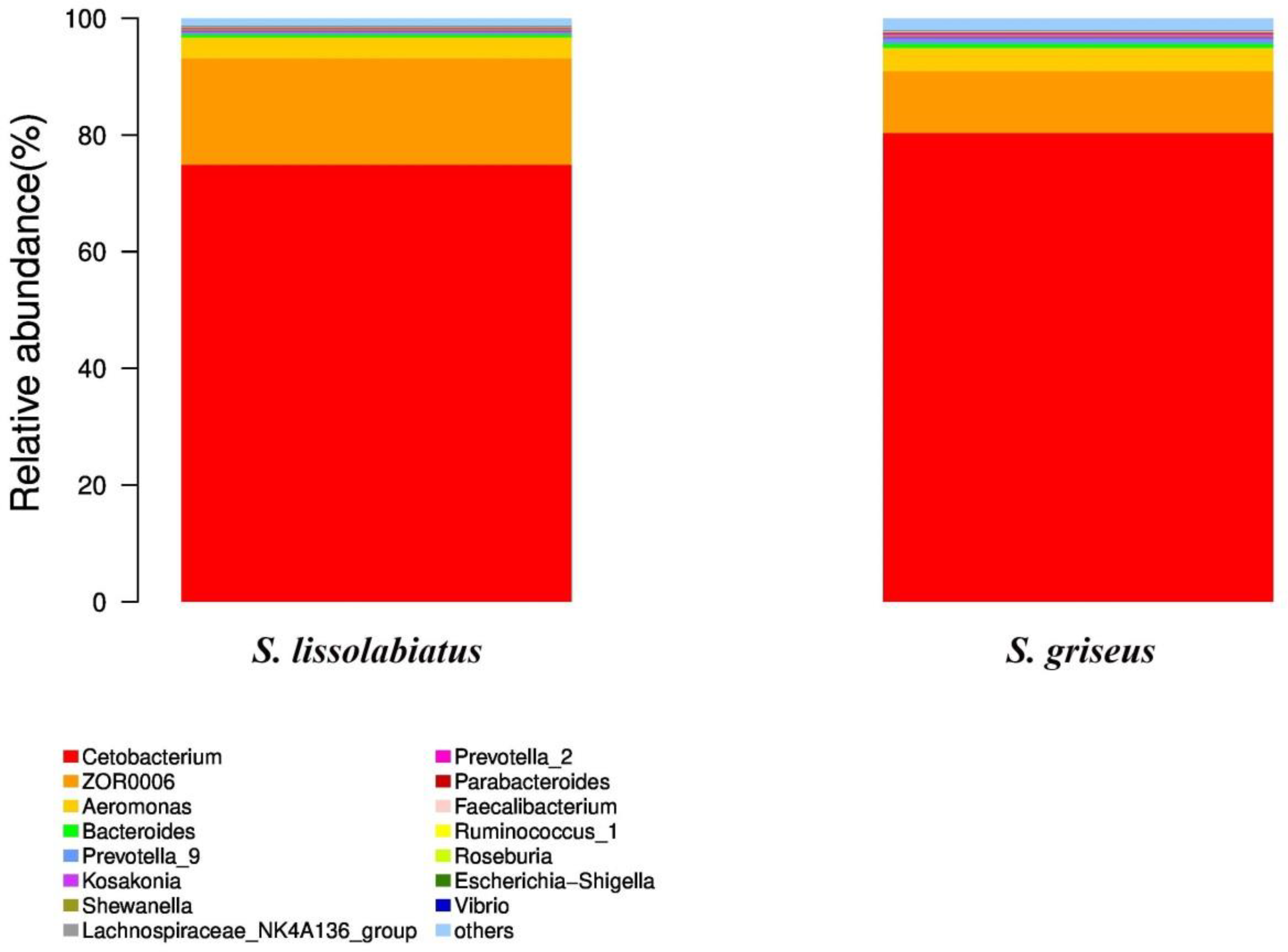
Intestinal microbiota structure and relative abundance of structure and relative abundance of intestinal microbiota of S. lissolabiatus and S. griseus at the genus level.
4 Discussion
4.1 Enzyme activity
Digestive enzyme activity is a critical indicator of the nutritional and physiological status of fish. Variations in enzyme activity reveal potential adaptations related to nutrient processing and ecological niches (Baragi and Lovell, 1986; Martinez et al., 1999). Distinct lipase activity patterns were observed with S. griseus showing higher activity in the foregut and liver, indicating adaptations for lipid digestion. Such differences suggest that S. griseus may have evolved a more robust lipid digestion capacity, potentially reflecting adaptations to diet or habitat. This observation aligns with findings in Schizothorax kozlovi, where the esophagus was also identified as a primary site of lipase activity (Hu et al., 2011).
Both species showed comparable α-amylase and trypsin activities across gut regions, likely due to the shared habitat and controlled feeding conditions used in this study. Similar enzyme activity across digestive regions has been documented in Ctenopharyngodon idella and Danio rerio, highlighting the potential for convergent digestive strategies among fish with similar ecological requirements (Chakrabarti et al., 1995; López-Vásquez et al., 2009). Future research could explore whether such similarities persist across varied environmental conditions or diet formulations, as this would provide insights into the environmental plasticity of these enzymatic patterns.
4.2 Metabolic differences
The observed differences in ALT and AST levels between the species provide insights into potential variations in protein metabolism and hepatic function. Elevated AST in S. griseus may indicate a higher dietary protein requirement or a greater reliance on endogenous amino acid synthesis pathways (Vijayavel and Balasubramanian, 2006). Elevated ALT in S. lissolabiatus, however, suggests a more active protein synthesis capacity, potentially supporting greater tissue growth or repair.
This study also revealed species-specific differences in blood GLU levels, with higher GLU observed in S. griseus. This difference, despite identical feeding conditions, implies inherent species-level metabolic differences that may be linked to variations in energy utilization. In practical aquaculture, these findings could inform dietary formulations tailored to meet the specific metabolic needs of each species, potentially optimizing growth and health outcomes. These metabolic differences between species may influence their adaptation to farming environments and dietary requirements.
4.3 Antioxidant and immune responses
Differences in CAT activity and MDA levels suggest that S. griseus may possess stronger antioxidant defenses, potentially increasing its resilience to oxidative stress in high-density aquaculture environments (Cabiscol et al., 2000). The elevated CAT activity in S. griseus might reflect an evolutionary adaptation to its natural habitat, where environmental stressors may select for enhanced oxidative defenses. This antioxidant capacity could be an important consideration in aquaculture management, where environmental factors can induce oxidative stress. The study’s findings on nonspecific immune markers, such as ACP and ALP, further support that both species exhibit similar baseline immune capabilities, indicating that health management practices may be equally applicable across both species.
4.4 Metabolomics and functional implications
Serum metabolomics analysis highlighted distinct metabolic profiles between the species, with S. griseus showing higher levels of D-glucuronic acid, an important metabolite involved in ascorbate and aldarate metabolism. This pathway is associated with cellular redox balance, inflammation reduction, and immune responses (Marselos et al., 1975; Hiroko et al., 2015). Higher D-glucuronic acid levels in S. griseus suggest a stronger anti-inflammatory potential, which could confer a competitive advantage in aquaculture settings by improving resilience to disease and other environmental stressors.
Differences in phospholipid metabolism, especially in phosphatidylcholine and phosphatidylethanolamine, were also noted. S. lissolabiatus exhibited higher levels of phosphatidylcholine, which is essential for cell membrane structure and lipid metabolism, and is associated with increased antioxidant capacity. Enhanced phosphatidylcholine levels may support greater cellular repair and growth in S. lissolabiatus, potentially making this species more robust under high-energy dietary regimens commonly used in aquaculture (Fokina et al., 2014). The elevated phosphatidylethanolamine levels in S. lissolabiatus also suggest a role in autophagy, which is crucial for cellular maintenance and could indicate an adaptation to handle cellular stress effectively (Xia et al., 2019).
4.5 Microbial diversity
Intestinal microbiota plays vital roles in digestion, nutrient absorption, and immunity in fish (Valdes et al., 2018; Tran et al., 2018). In this study, S. griseus exhibited a higher OTU count and α-diversity index, indicating a richer microbial community than S. lissolabiatus. Higher microbial diversity has been linked to improved health and nutrient utilization in fish, potentially giving S. griseus an advantage in aquaculture settings where diverse microbiota support resilience against pathogens and better nutrient absorption (Sullam et al., 2015).
The microbiota of both species was dominated by phyla such as Fusobacteria, Firmicutes, Proteobacteria, and Bacteroidetes, consistent with findings in other fish species (Ghanbari et al., 2015). The role of these phyla in supporting metabolic functions is well documented; for instance, Firmicutes are known to aid in energy balance, while Proteobacteria are often associated with host immune responses (Semova et al., 2012; Blandford et al., 2018). No significant differences in major microbial genera were found between the two species, suggesting that while microbial diversity differs, the core functional microbiota may provide similar benefits across species.
4.6 Implications for aquaculture practices
The findings have practical implications for aquaculture practices, particularly in dietary formulation and health management. The higher lipase activity and lipid metabolism in S. griseus suggest that diets enriched in lipids could enhance growth and energy efficiency for this species. In contrast, the higher phospholipid levels in S. lissolabiatus indicate a potential benefit from diets with balanced fatty acid compositions, supporting cellular health and oxidative resilience.
The distinct antioxidant profiles and immune capabilities also suggest that specific supplementation strategies, such as antioxidants or immune-supportive prebiotics, might optimize the health outcomes of each species. Additionally, the differences in gut microbiota diversity indicate that incorporating probiotics or prebiotics into the diet could further enhance microbial diversity, especially for S. lissolabiatus, supporting its digestive efficiency and disease resistance.
By recognizing these interspecies differences, aquaculture practices can be adapted to support optimal growth, health, and resilience in Schizothorax species, contributing to sustainable fish farming in the long term. Future research should focus on tailored feeding strategies to further enhance aquaculture sustainability for each species.
5 Conclusion
This study presents a comparative analysis of the physiological metabolism, intestinal morphology, and microbial composition of two artificially cultured Schizothorax species, S. lissolabiatus and S. griseus, raised under identical conditions. This study reveals distinct nutrient utilization patterns with S. griseus exhibiting traits advantageous for large-scale aquaculture. Specifically, S. griseus exhibited more efficient growth rates and better feed conversion efficiency, factors that contribute to its resilience under environmental stress. The enrichment of metabolic pathways associated with robust immune responses, suggested by serum metabolite analysis, was observed in both species. However, S. griseus stood out with a significantly higher number of OTUs and greater microbial diversity, as revealed by high-throughput 16S rRNA sequencing and alpha diversity analysis. This enriched microbial community structure potentially offers additional benefits for nutrient utilization and health maintenance. Despite similarities in dominant gut microbial phyla—Fusobacteria, Firmicutes, Proteobacteria, and Bacteroidetes—S. griseus’s enhanced microbial diversity emphasizes its potential as a promising candidate for aquaculture development. This not only contributes to the economic growth of Schizothorax farming but also aligns with ecological and sustainability goals by fostering healthy microbial communities in aquaculture systems. These insights reinforce the importance of selecting species like S. griseus to advance sustainable aquaculture practices.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
This study was approved by the Animal Welfare and Ethics Committee of Dali University (Approval Number: 2020-PZ-46). All experiments were conducted in strict accordance with the Guidelines for the Care and Use of Laboratory Animals in China.
Author contributions
XL: Conceptualization, Funding acquisition, Project administration, Writing – review & editing. CZ: Data curation, Investigation, Writing – original draft. LC: Data curation, Investigation, Writing – original draft. ZH: Data curation, Investigation, Writing – review & editing. YH: Formal analysis, Investigation, Writing – review & editing. YJ: Investigation, Methodology, Writing – review & editing. JZ: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Dali University Doctoral Research Startup Fee Project (No. KYBS2021064), the Yunnan Province ‘Xingdian Talent Support Plan’ Young Talents Special Project (No. 230212524080), the Yunnan Fundamental Research Projects (No. 202101AT070028), and the Foundation of Yunnan Province Science and Technology Department (No. 202305AM070003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1510780/full#supplementary-material
Supplementary Table 1Elution gradient of mobile phase.
Supplementary Table 2Parameters of ion scanning mass spectrometry.
Supplementary Table 3Metabolic pathways identified from the significantly different metabolites in the serum between S. lissolabiatus and S. griseus.
References
1
Baragi V. M. Lovell R. T. (1986). Digestive enzyme activities in striped bass from first feeding through larva development. Trans. Am. Fish. Soc115, 478–484. doi: 10.1577/1548-8659(1986)115<478:DEAISB>2.0.CO;2
2
Blandford M. I. Taylor B. A. Schlacher T. A. Nowak B. Polkinghorne A. (2018). Epitheliocystis in fish: An emerging aquaculture disease with a global impact. Transbound Emerg. Dis.65, 1436–1446. doi: 10.1111/tbed.12908
3
Cabiscol E. Tamarit J. Ros J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol.3, 3–8. doi: 10.2307/41125178
4
Chakrabarti I. Gani A. Chaki K. Sur R. Misra K. (1995). Digestive enzymes of 11 freshwater teleost fish species in relation to food habit and niche segregation. Comp. Biochem. Physiol.A112, 162–177. doi: 10.1016/0300-9629(95)00072-F
5
Chen X. (2013). Checklist of fishes in yunnan. Zool. Res.34, 281–337. doi: 10.11813/j.issn.0254-5853.2013.4.0281
6
Deng J. M. Zhang X. Long X. W. Kang B. Ding D. H. Bi B. L. (2013). Analysis and evaluation of nutritional composition in muscle of three Schizothorax species. Acta Nutrimenta Sin.35, 391–393. doi: 10.13325/j.cnki.acta.nutr.sin.2013.04.003
7
Fadrosh D. W. Ma B. Gajer P. Sengamalay N. Ott S. Brotman R. M. et al . (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeg platform. Microbiome2, 1–7. doi: 10.1186/2049-2618-2-6
8
Fokina N. N. Bakhmet I. N. Shklyarevich G. A. Nemova N. N. (2014). Effect of seawater desalination and oil pollution on the lipid composition of blue mussels Mytilus edulis L. from the White Sea. Ecotox. Environ. Safe.110, 103–109. doi: 10.1016/j.ecoenv.2014.08.010
9
Gao H. (2019). Study on key techniques of wildization and rleasing of Schizothorax lissolabiatus tsao artificial breeding fry in Lancang river. Mod. Agric. Sci. Technol.739, 197–200. doi: 10.3969/j.issn.1007-5739.2019.05.111
10
Ghanbari M. Kneifel W. Domig K. J. (2015). A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture448, 464–475. doi: 10.1016/j.aquaculture.2015.06.033
11
Hiroko F. Hitoshi I. Shigehisa E. Keiko N. Yasukazu Y. Yoshihisa H. et al . (2015). Ascorbic acid attenuates acute pulmonary oxidative stress and inflammation caused by zinc oxide nanoparticles. J. Occup. Health57, 118–125. doi: 10.1539/joh.14-0161-oa
12
Hu S. Yu Z. Chen Y. Zhao H. (2011). Research on activity of lipase in the digestive system of Schizothorax kozlovi nikolsky. J. Bijie Univ29, 94–97. doi: 10.13557/j.cnki.issn1002-2813.2016.03.011
13
Hu S. Zeng C. Yang S. Zhao H. Chen Y. (2016). Digestive enzyme activity research of Schizothorax kozlovi nikolsky. Feed Res.3, 41–44. doi: 10.13557/j.cnki.issn1002-2813.2016.03.011
14
Jiang Y. Zhao S. Xiao W. Huang Z. (2018). Study on artificial propagation technique of Schizothorax griseus. J. Dali Univ.3, 79–81. doi: 10.3969/j.issn.2096-2266.2018.12.017
15
Jin F. Zuo P. Wu J. Zhao J. Wang Z. Leng Y. (2024). Comparison of nutrient components and nutritional value evaluation of Schizothorax lissolabiatus Tsao musele under different culture modes. Heilongjiang Anim. Sci. Vet. Med.8, 110–116. doi: 10.13881/j.cnki.hljxmsy.2023.04.0065
16
Jiye A. Trygg J. Gullberg J. Johansson A. I. Jonsson P. Antti H. et al . (2015). Extraction and GC/MS analysis of the Human blood plasma metabolome. Anal. Chem.77, 8086–8094. doi: 10.1021/ac051211v
17
Kang Z. Yang D. Huang J. Li H. (2015). New record of fish species in Hunan Province: Schizothorax griseus. Sichuan J. Zool.34, 434. doi: CNKI:SUN:SCDW.0.2015-03-026
18
Kumar S. Sahu N. P. Pal A. K. Choudhury D. Yengkokpam S. Mukherjee S. C. (2005). Effect of dietary carbohydrate on hematology, respiratory burst activity and histological changes in L. rohita juveniles. Fish Shellfish Immunol.19, 331–344. doi: 10.1016/j.fsi.2005.03.001
19
Le P. Q. (2000). Fauna Sinica: Osteichthyes, Cypriniformes: Volume II (Beijing: Science Press), 273–290.
20
Liang M. Wei K. J. Zhu X. Y. Ma B. S. Xu B. Xu J. (2020). Effects on fish body composition, digestive enzyme activity and RNA/DNA of Schizothorax wangchiachii juveniles: starvation and refeeding. Chin. Agric. Sci. Bulletin.36, 151–160. doi: 10.11924/j.issn.1000-6850.casb20190800508
21
Long T. Hicks M. Yu H. C. Biggs W. H. Kirkness E. F. Menni C. et al . (2017). Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet.49, 568–578. doi: 10.1038/ng.3809
22
López-Vásquez K. Castro-Pérez C. A. Val A. L. (2009). Digestive enzymes of eight Amazonian teleosts with different feeding habits. J.Fish Biol.74, 1620–1628. doi: 10.1111/j.1095-8649.2009.02196.x
23
Lu K. Ma B. Huo B. Chu Z. Xu B. Zhu X. (2024). Analysis on the difference of nutrient composition in muscle of schizothorax wangchiachii with different sizes. China Anim. Husbandry Vet. Med.51, 1094–1102. doi: 10.16431/j.cnki.1671-7236.2024.03.021
24
Marselos M. Rantanen T. Puhakainen E. (1975). Comparison of the effect of phenobarbital on the d-glucuronic acid pathway in euthyroid and hypothyroid rats. Acta Pharmacol. Toxicol.37, 415–424. doi: 10.1111/j.1600-0773.1975.tb00860.x
25
Martinez I. Moyano F. J. Fernandez-diaz C. Yufera M. (1999). Digestive enzyme activity during larval development of the Senegal sole (Solea Senegalensis). Fish Physiol. Biochem.21, 317–323. doi: 10.1023/A:1007802708459
26
Ray A. K. Ghosh K. Ringo E. (2012). Enzyme-producing bacteria isolated from fish gut: A review. Aquac. Nutr.18, 465–492. doi: 10.1111/j.1365-2095.2012.00943.x
27
Ren Y. Nie Z. Yang Z. Zhang S. Wei J. (2020). Morphology and histology studies of the digestive system of Schizothorax biddulphi. Progr. Fish. Sci.41, 49–57. doi: 10.19663/j.issn2095-9869.20191225001
28
Rigaud C. Eriksson A. Rokka A. Skaugen M. Lihavainen J. Keinänen M. et al . (2020). Retene, pyrene and phenanthrene cause distinct molecular-level changes in the cardiac tissue of rainbow trout (Oncorhynchus mykiss) larvae, part 2-Proteomics and metabolomics. Sci. Total Environ.746, 141161. doi: 10.1016/j.scitotenv.2020.141161
29
Rong H. Zhang L. Wang X. Wu X. Wang J. Hu Q. et al . (2023). Comparative study on digestive enzyme activity and intestinal tissue morphology of four fishes with different feeding habits. Freshw. Fish.53, 29–35. doi: 10.13721/j.cnki.dsyy.2023.02.008
30
Roques S. Deborde C. Guimas L. Marchand Y. Richard N. Jacob D. et al . (2020). Integrative metabolomics for assessing the effect of insect (Hermetia illucens) protein extract on rainbow trout metabolism. Metabolites10, 83–100. doi: 10.3390/metabo10030083
31
Semova I. Carten J. D. Stombaugh J. Mackey L. C. Knight R. Farber S. A. et al . (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe12, 277–288. doi: 10.1016/j.chom.2012.08.003
32
Sullam K. E. Rubin B. E. Dalton C. M. Kilham S. S. Flecker A. S. Russell J. A. (2015). Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of trinidadian guppies. Isme J.9, 1508. doi: 10.1038/ismej.2014.231
33
Tang J. Ye S. Li W. Jia S. (2013). Status and historical changes in the fish community in Erhai Lake. Chin. J. Oceanol. Limnol.31, 712–723. doi: 10.1007/s00343-013-2324-7
34
Tran N. T. Zhang J. Xiong F. Wang G. T. Li W. X. Wu S. G. (2018). Altered gut microbiota associated with intestinal disease in grass carp (Ctenopharyngodon idellus). World J. Microb. Biot.34, 1–9. doi: 10.1007/s11274-018-2447-2
35
Valdes A. M. Walter J. Segal E. Spector T. D. (2018). Role of the gut microbiota in nutrition and health. Brit. Med. J.361, 17–22. doi: 10.1136/bmj.k2179
36
Vijayavel K. Balasubramanian M. P. (2006). Fluctuations of biochemical constituents and marker enzymes as a consequence of naphthalene toxicity in the edible estuarine crab scylla serrata. Ecotoxicol Environ. Saf.63, 141–147. doi: 10.1016/j.ecoenv.2005.02.004
37
Wang S. Zheng Y. Tang H. Yin J. Li J. Zhao S. (2018). Analysis and evaluation for the nutrition components of Schizothorax griseus muscles. Freshw. Fish.48, 80–86. doi: 10.13721/j.cnki.dsyy.2018.02.012
38
Xia X. Wang X. Qin W. Jiang J. Cheng L. (2019). Emerging regulatory mechanisms and functions of autophagy in fish. Aquaculture511, 734212–734220. doi: 10.1016/j.aquaculture.2019.734212
39
Xiang X. Zhou X. Chen J. Li D. Wang W. Zhou X. (2012). Effects of dietary protein and animal protein levels on growth, body composition and digestive enzyme activities of juvenile prenant’s Schizothoracin (Schizothorax prenanti). J. Chin. Cereals Oils Assoc.27, 74–80. doi: 10.3969/j.issn.1003-0174.2012.05.016
40
Xu G. Chen X. Du J. Mu Z. (2009). Fish digestive system: It’s structure, function and the distributions and characteristics of digestive enzymes. Chin. J. Fish.22, 49–55. doi: 10.3969/j.issn.1005-3832.2009.04.013
41
Xu G. Du F. Nie Z. Li Y. Xu P. (2016). Integrated application of transcriptomics and metabolomics yields insights into population-asynchronous ovary development in Coilia nasus. Sci. Rep.6, 31835–31845. doi: 10.1038/srep31835
42
Yang W. Dai Y. An D. Han H. (2021). genetic diversity of genome DNA in schizothorax lissolabiatus population from the Beipan river based on AFLP analysis. Mar. Fish.43, 385–394. doi: 10.3969/j.issn.1004-2490.2021.04.001
Summary
Keywords
gut histology, gut microbiota, metabolomics, physiological metabolism, Schizothorax lissolabiatus , Schizothorax griseus
Citation
Zhong C, Chen L, Huang Z, Hu Y, Jiang Y, Zhou J and Long X (2024) Comparison of metabolism, gut histology, and microbiota between Schizothorax lissolabiatus and Schizothorax griseus under identical farming conditions. Front. Mar. Sci. 11:1510780. doi: 10.3389/fmars.2024.1510780
Received
13 October 2024
Accepted
11 November 2024
Published
05 December 2024
Volume
11 - 2024
Edited by
Yafei Duan, South China Sea Fisheries Research Institute, China
Reviewed by
D. K. Meena, Central Inland Fisheries Research Institute (ICAR), India
Sofia Priyadarsani Das, National Taiwan Ocean University, Taiwan
Updates
Copyright
© 2024 Zhong, Chen, Huang, Hu, Jiang, Zhou and Long.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowen Long, 15900771883@163.com; Jun Zhou, junz2003@126.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.