- 1Chinese Sturgeon Research Institute, China Three Gorges Corporation, Yichang, Hubei, China
- 2Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes, Yichang, Hubei, China
- 3National Pathogen Collection Center for Aquatic Animals, Shanghai Ocean University, Shanghai, China
- 4School of Marine Science and Engineering, Qingdao Agricultural University, Qingdao, Shandong, China
- 5China Three Gorges Corporation, Wuhan, Hubei, China
Introduction: Our understanding of the regulatory mechanisms that govern the development of sexual characteristics in sturgeon, particularly the Chinese sturgeon (Acipenser sinensis), is limited. Foxl2, a member of the forkhead/HNF-3-related transcription factor family, is known for its role in precise gene regulation within specific tissues and its control over ovarian aromatase, a key enzyme in ovarian development and maturation. This study aimed to investigate the sexually dimorphic expression of foxl2 during the molecular and morphological sex differentiation stages in Chinese sturgeon.
Methods: We successfully isolated the full-length cDNAs of foxl2 in Chinese sturgeon. We then examined the expression profiles of this gene across various somatic tissues in both female and male specimens.
Results: Foxl2 was predominantly expressed in the ovulatory organs of two-year-old Chinese sturgeon, with significantly lower expression in other somatic tissues. We observed sexual dimorphism in foxl2 expression within gonads that had achieved morphological differentiation.
Discussion: The results suggest that foxl2 expression is intricately linked to the process of ovarian differentiation in Chinese sturgeon. Furthermore, we propose that the mRNA quantities of foxl2 could serve as an accurate biomarker for sex determination in sturgeon during their early growth stages.
1 Introduction
Chinese sturgeon (Acipenser sinensis) belongs to the order Acipenseriformes, a cartilaginous fish with a history of evolution spanning over 184 million years (Peng et al., 2007). The Chinese sturgeon is an infrequent anadromous migratory waterborne creature, designated as critically endangered by the International Union for Conservation of Nature (IUCN) and classified as a Class I protected animal within China. This migratory species primarily inhabits the continental shelves of East China Sea, with spawning occurring in the upstream areas of the Yangtze River in China. As one of the most at-risk aquatic organisms globally, the Chinese sturgeon exhibits a complex life history that encompasses both marine and freshwater habitats. The Chinese sturgeon’s life cycle starts in freshwater habitats, where the eggs are fertilized and the embryos mature within the safety of the female’s body. Following the hatching process, the offspring, referred to as alevins, remain in the breeding grounds typically situated in the higher sections of the Yangtze River. During this stage, they rely on their yolk sac for nourishment. Once the yolk sac is absorbed, the juveniles start to feed on plankton and small aquatic invertebrates. As they grow, they gradually move downstream, eventually entering the estuarine and marine environments. This period marks a significant change in their morphology, preparing them for life in the ocean. Chinese sturgeon juveniles enter the East China Sea, where they spend most of their lives. In the marine environment, they continue to grow and mature, feeding on a variety of benthic invertebrates, including crustaceans and mollusks. The length of their stay in the marine environment can span several years to more than a decade. Sexual maturity in Chinese sturgeon is achieved after approximately 9 to 12 years. When ready to spawn, these fish undertake a remarkable anadromous migration, returning to the freshwater rivers where they were born. This migration, prompted by environmental and physiological cues, can cover vast distances and is a critical part of their life cycle.
The number of wild Chinese sturgeon has dropped considerably as a result of excessive fishing (Zhuang et al., 2002). Since 1983, in order to avoid the Chinese sturgeon extinction, researchers began to try this species artificial breeding (Chen et al., 2006). However, this species is an extremely late-maturing and asynchronous species (A range of 14 to 26 years for females and 8 to 18 years for males) (Cao et al., 2009). Furtherly, only the limited number of females cultivated in a regulated environment can successfully overpass the developmental threshold of the ovary from phase II to III, involving the deposition of yolk, making conservation and culture costly and time consuming (Wei et al., 2013). Nonetheless, the fundamental genetic aspects of gonadal differentiation and maturation in sturgeon remain elusive.
Principal genes that dictate sex have been recognized in a number of vertebrates: DMRT1 (doublesex and Mab-3 related transcription factor 1) in chickens (Smith et al., 2009), DM-W (W-linked paralogue of DMRT1) in the African clawed frog (Yoshimoto et al., 2008), Sry (sex-determining region of the Y chromosome) in placental mammals (Koopman et al., 1991; Sinclair et al., 1990), and dmy (duplicated copy of DMRT1) in medaka fish (Matsuda et al., 2002; Nanda et al., 2002). The system for sex determination seems to be preserved, featuring Sry in mammals and DMRT1 in avian species. However, in fish, there is significant diversity among sex determination candidate genes, such as sdY in rainbow trout (Oncorhynchus mykiss) (Yano et al., 2012), which diverges from the immune system, GSDF in Oryzias luzonensis (Myosho et al., 2012), amhy (duplicated copy of amh) in Patagonian pejerrey (Odontesthes hatcheri) (Hattori et al., 2012), and amhr2 in fugu (Takifugu rubripes) (Kamiya et al., 2012).
Identifying sturgeon sex at early developmental stages is crucial for producing valuable caviar from their ovaries. Nevertheless, sturgeons do not display outward sexual differences, and up until now, no regions or sex chromosomes determining sex have been pinpointed in these species. Various investigations have tried to identify gender-specific DNA sequences or genes using DNA profiling techniques for determining sex, but were unsuccessful (Wuertz et al., 2006; McCormick et al., 2008). Current investigations are concentrating on locating pivotal genes that play a role in sex determination and development, grounded on preserved molecular processes seen across diverse vertebrate taxa. Nevertheless, the results have been inconsistent across different sturgeon species.
In Amur sturgeon (A. schrenckii), the expression of sox9 and sox2 are higher in the testis compared to the ovary (Chen et al., 2006), whereas in lake sturgeon (A. fulvescens), no dimorphic sox genes have been identified (McCormick et al., 2008). For wild shovelnose sturgeon (Scaphirhynchus platorynchus), there have been no noteworthy differences in DMRT1 expression between the sexes (Amberg et al., 2010), while in Siberian sturgeon (A. baerii) (Berbejillo et al., 2012) and lake sturgeon (Hale et al., 2010a), DMRT1 expression is significantly different between sexes. Investigating the genetic sequences within cDNA libraries of male and female gonads in Adriatic sturgeon (A. naccarii) revealed that five genes (wt1, lhx1, cyp19a1, fhl3, and fem1a) and two genes (ar and emx2) are sex-specific, though PCR validation was not conducted (Vidotto et al., 2013). In Chinese sturgeon, lhx1 is detected solely in the testis, accompanied by IGF-1 and sox11, while foxl2, cyp19a1a, nanos3b, and GNRHR are uniquely present in the ovarian gene expression profile (Ye et al., 2015). Sequencing the gonadal genetic transcripts at various growth stages in Russian sturgeon (A. gueldenstaedtii) (9-month-old intersex, 2-year-old, and 5-year-old immature individuals) demonstrated differential expression of sox9 and foxl2 between sexes (Chen et al., 2016). In the case of sterlet at two years of age, hsd11b2 and DMRT1 expression is elevated from the incipient phase of spermatogenesis in males, while cyp19a1 and foxl2 show higher expression throughout all differentiated ovarian stages in females (Wang et al., 2017). Analyzing the expression patterns of DMRT1 and foxl2 in differentiated gonads indicated that only foxl2 shows gender-specific expression patterns (Okada et al., 2017). A group of scientists have proposed that the principal sex-regulating gene may not be expressed in the gonads of young adult sturgeons or adolescent, but rather during an earlier developmental stage (Hale et al., 2010b). Consequently, Hagihara et al. (2014) tried to find suitable gender-differentiating markers for sturgeons by examining the RNA sequences of undifferentiated gonads in Russian sturgeon. The proposition was made that GSDF, foxl2, hsd17b1, and cyp19a1 might act as suitable sexing indicators for undifferentiated Russian sturgeon, though PCR confirmation was not carried out. The variability in these results across sturgeon species may be attributed to differences during the phases of gonad development (Hagihara et al., 2014). Hence, comprehensive data on sex-associated genes and their expression trajectories across various phases of gonadal maturation, starting from gonad differentiation, is necessitated.
Foxl2, a component of the Forkhead (FKH) domain-containing Fox protein subset, is recognized as the initial definitive indicator of ovarian development in mammals (Klugmann et al., 2009). Its predominant expression in the ovaries of certain hermaphroditic fish has been verified, and its essential function in the development of ovaries has been shown in Nile tilapia (Oreochromis niloticus) (Zhang et al., 2017), rainbow trout (Oncorhynchus mykiss) (Bertho et al., 2018), and zebrafish (Danio rerio) (Yang et al., 2017). Even though foxl2 is a universally retained transcription factor across metazoans, its developmental history among osteichthyans is intricate (Bertho et al., 2016). Prior to the morphological and sexual differentiation in Nile tilapia, Cyp19A1A (cytochrome P450 family 19 subfamily A polypeptide 1A) and foxl2 initially exhibit specific expression within the undifferentiated reproductive glands of genetically female organisms at 5 days post-hatching (DAH); DMRT1, a gender-determining and Mab-3 related transcription factor, exhibits predominant male expression in the immature gonads of males specified by genetics at 6 DAH (Ijiri et al., 2008). The stage preceding the initiation of physical sex differentiation is referred to as molecular sex differentiation. Typically, foxl2 plays a crucial role in estrogen production and the development of ovaries in vertebrates, exhibiting greater transcription in the immature gonads of genetically female individuals compared to males, as also observed in rainbow trout (Oncorhynchus mykiss) and medaka (Oryzias latipes) (Vizziano et al., 2007; Nakamoto et al., 2006b). At present, the molecular processes underpinning sex differentiation in sturgeon are not fully comprehended, and the available molecular data is scant. A segment of foxl2 cDNA was isolated from wild paddlefish (Scaphirhynchus platorynchus), with higher transcript abundance in the ovary than in the testis (Amberg et al., 2010). Recently, next-generation sequencing technology has been used for RNA-seq analysis of the undifferentiated gonads of Russian sturgeon. revealing that the expression of foxl2 was more elevated in the immature gonads than in the prospective testis at 9 MAH (Hagihara et al., 2014). In this research, we discovered a foxl2 gene in the Chinese sturgeon (Asfoxl2) and explored its mRNA expression profiles across a variety of somatic tissues in both female and male specimens. The objective of this study is to expand our fundamental comprehension of foxl2 in the Chinese sturgeon.
2 Materials and methods
This study’s fish handling and experimental methodologies were sanctioned by the Chinese Sturgeon Research Institute, the China Three Gorges Corporation. The research was executed adhering to the ARRIVE guidelines, and all procedures were conducted in line with pertinent guidelines and regulations. The authors affirm that the research was carried out in a manner that is both ethical and responsible.
2.1 Experimental fish, sampling and sex identification
The specimens of Chinese sturgeon used in this study were procured from the Chinese Sturgeon Research Institute (Hubei Province, China). These samples originated from induced spawning practices and were incubated under conditions of 18.0 ± 0.5°C. After a 24-hour period of abstinence from food, we sampled three healthy Chinese sturgeon individuals, both male and female, aged two years (with an average full length of 70.00 ± 4.83 cm and a body mass of 1.29 ± 0.06 kg). The gender of the Chinese sturgeon was determined using sex-linked markers (F: CAGGAAAGGCAAGGATGT; R: TAAAGGGAGACGGCAGAT), developed in our laboratory (Hu et al., 2021). Intensive anesthesia was realized through the use of a 0.05% concentration of ethyl 3-aminobenzoate mesylate-222. (MS-222, Sigma). Multiple organs, comprising the hypothalamus, pituitary, liver, brain, intestine, heart, testicles, stomach, muscle, and ovaries, were carefully sectioned and immediately immersed in liquid nitrogen to facilitate subsequent gene cloning and studies on tissue distribution.
2.2 Isolation of RNA and synthesis of complementary DNA
Total RNA was isolated employing Trizol lysate buffer followed by purification with an RNA simple kit (Takara, Dalian, China) according to the manufacturer’s protocols. The quality and integrity of RNA were assessed via agarose gel electrophoresis and absorbance spectroscopy, respectively. The SMARTTM cDNA kit (Clonetech, Shanghai, China) was used for cDNA cloning and qPCR analysis was performed With the aid of the PrimeScript quantitative PCR assay kit (Takara, Dalian, China). The cDNA was then stored at -80°C as a template for qPCR.
2.3 Molecular cloning procedures for Asfoxl2 and an in-depth bioinformatics analysis
The primer of foxl2-3 showed in Table 1, was used to obtain the open reading frame of Asfoxl2, which was based on the sequence tags derived from Asfoxl2 transcripts of our lab’s transcriptome data sequencing. Subsequently, cDNA fragments of Chinese sturgeon’s foxl2 were procured via PCR amplification. The PCR amplification was performed with a total volume of 50 μL, which included 4 μL of a 10 mmol/L dNTP mixture, 2 μL cDNA, 1 μL of each primer solution (refer to Table 1), 5 μL reaction buffer, 36.6 μL of nuclease-free water and 0.4 μL of Taq polymerase (Takara), The initial denaturation was executed at 94°C for 3 minutes, followed by 35 amplification cycles comprising 30 seconds at 94°C, 30 seconds of annealing at 60°C, and a 30-second extension at 72°C. A final extension was performed at 72°C for 10 minutes. The PCR outputs were subsequently analyzed by gel electrophoresis using a 1.5% agarose matrix and made visible through ethidium bromide treatment. The presumed gene sequences were inserted into the PMD18-T cloning vector (Takara, Japan) following PCR product purification and sequenced using an ABI3730XL sequencer (Applied Biosystems, Foster City, CA).
The nucleotide sequence was deciphered into protein sequence, and the invariant domains of Asfoxl2 proteins were investigated with DNAMAN version 7 and NCBI’s CDD utilities (http://www.ncbi.nlm.nih.gov/), respectively. We employed the ExPASy Compute pI/Mw utility to determine the molecular weight and theoretical isoelectric point of the predicted Asfoxl2 protein. The SignalP-5.0 tool was used to forecast the signal peptide cleavage positions in largemouth bass lamps. The SMART platform was used to forecast probable protein domain characteristics. The TMHMM server (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) was employed for the prediction of possible transmembrane domains. The identification of potential N-linked and O-linked glycosylation sites was carried out using NetNGlyc-1.0 and NetOGlyc-4.0, respectively. The NetPhos-3.1 server was employed for the prediction of potential phosphorylation locations. The subcellular localization was predicted using the CELLO online predictor (cello.life.nctu.edu.tw) with default parameters. The outcomes of gene functional categorization were derived from the PredictProtein web-based tool The prediction of Asfoxl2 protein’s secondary structure and three-dimensional conformation was accomplished using PSIPRED and SWISS-MODEL, respectively, with default parameters.
The Asfoxl2 sequence and its amino acid were analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) for sequence and phylogenetic analysis utilizing online bioinformatics tools. The MEGA5.0 program was employed for the alignment of amino acids and conducting phylogenetic studies with the bootstrapped neighbor-joining technique. Aligned sequences of Asfoxl2 and corresponding orthologs from Salmo salar (XP_014018845.1), Acipenser schrenckii (BAZ96608.1), Pelodiscus sinensis (KP734210.1), Mauremys reevesii (KU821114.1), Mauremys mutica (XM_045029577.1), Homo sapiens (NM_023067.4), Oreochromis niloticus (NM_001279778.1), Oryzias latipes (NM_001104888.1), Gallus (NM_001012612.1), Acipenser ruthenus (MN627262.1), Polyodon spathula (XP_041125750.1).
2.4 Cellular Localization of the Asfoxl2 protein
The full-length open reading frame (ORF) of Asfoxl2, with the termination codon, was enlarged by employing the primers detailed in Table 1. Afterwards, it was ligated into the XhoI-cleaved plasmid vector pCDNA3.1-EGFP using the pEASY-Basic Seamless Cloning and Assembly Kit (TransGen, Beijing). HEK293t cells were cultivated in Dulbecco’s Modified Eagle Medium (HyClone), enriched with 10% Fetal Bovine Serum (Gibco) and 1% Penicillin-Streptomycin mixture, at 37°C in a sterile incubation chamber with a 5% CO2 atmosphere. The modified expression vector was transfected into HEK293t cells using the EZ Trans transfection reagent from Liji Biotechnology Co., Ltd., Shanghai. Following a 24-hour cultivation period, the cells underwent fixation with a 4% paraformaldehyde solution, were dyed with DAPI—a stain specific for cell nuclei—and their distribution was examined under a fluorescence microscope model Eclipse 80i from Nikon. Furthermore, the open reading frame (ORF) of Lamin A/B was replicated from HEK293t cells and linked into the pCDNA3.1-RFP vector, adhering to the methodology delineated earlier. Then, the vectors pcDNA3.1-SIRT1-EGFP and pcDNA3.1-Lamin-RFP were co-delivered into HEK293t cells to ascertain the positioning of Asfoxl2, and the Nikon Eclipse 80i fluorescence microscope was utilized for observation.
2.5 Quantitative real-time PCR
Polymerase chain reaction (PCR) experiments were carried out using a 96-well plate on an ABI PRISM 7500 Real-Time PCR instrument, with each reaction having a total volume of 20 μL. The primer of foxl2-rt4 showed in Table 1, was used for the Real-Time PCR, and the size of the amplicon for foxl2-rt4 is 190bp, with β-actin serving as the internal control gene. Every assay consisted of 0.2 μL of each primer (10 μM), 10 μL of 2×SYBR Green Real-Time PCR Master Mix (ABI, cat no. 4367659), 9.1 μL of H2O, and 0.5 μL of 5-fold diluted cDNA template. The procedure for amplification was established in the following manner: 2 minutes at 50°C, 5 minutes at 95°C, then 40 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, and elongation at 72°C for 30 seconds, ending with an analysis of the dissociation curve. Sterile deionized water was utilized in place of the cDNA template to create negative controls. Each sample was subjected to triplicate trials to ensure technical replication. Data from assorted assay plates targeting the same gene were standardized using a consistent threshold within the acceptable error range for amplification efficiency (R), before the utilization of external calibration curves for the analysis.
2.6 Statistical analyses
Statistical analyses were performed using SPSS Statistics. Concentrations of sex steroids and quantitative PCR results were displayed as the average ± standard error of the mean (SEM). All data were subjected to assessments for normal distribution and equal variance using the Shapiro-Wilk test and Levene’s test, respectively. After verifying these prerequisites, the data were analyzed using a single-factor analysis of variance (ANOVA), subsequently followed by the Tukey post-hoc test. Information failing to fulfill the prerequisites were modified (such as applying log10, sine, or square root transformations) and subsequently reassessed for adherence to normal distribution and uniformity of variance. If the data failed to meet the prerequisites even with data transformation, the Kruskal-Wallis test, a non-parametric approach, was utilized for all between-group comparisons. A P-value below 0.05 was considered indicative of statistical significance.
3 Results
3.1 Characterization of Asfoxl2
The complete open reading frame (ORF) of Asfoxl2 (GenBank No. PP895200) was successfully amplified through PCR techniques. Subsequent sequence analysis revealed that the Asfoxl2 gene comprises 918 nucleotides, which translate into 306 amino acids (Figure 1A). An NCBI-CDD search confirmed the presence of a foxl2 domain at the C-terminal end of the amino acid chain (Figure 1B). The calculated molecular weight of the resultant protein was approximately 34.5 kDa, and its isoelectric point was determined to be 9.13. The analysis also identified no signaling peptides or transmembrane regions, but it did find 15 predicted O-glycosylation sites, 4 potential N-glycosylation sites, and 68 predicted phosphorylation sites within Asfoxl2. Gene ontology analysis predicted that Asfoxl2’s primary molecular function is as a DNA-binding transcription factor, participating in the control of gene expression and metabolic routes. Predictions using the CELLO subcellular localization tool indicated a nuclear localization for Asfoxl2. Further subcellular localization studies in HEK 293 t cells confirmed predominant nuclear localization of the Asfoxl2 protein (Figure 2).
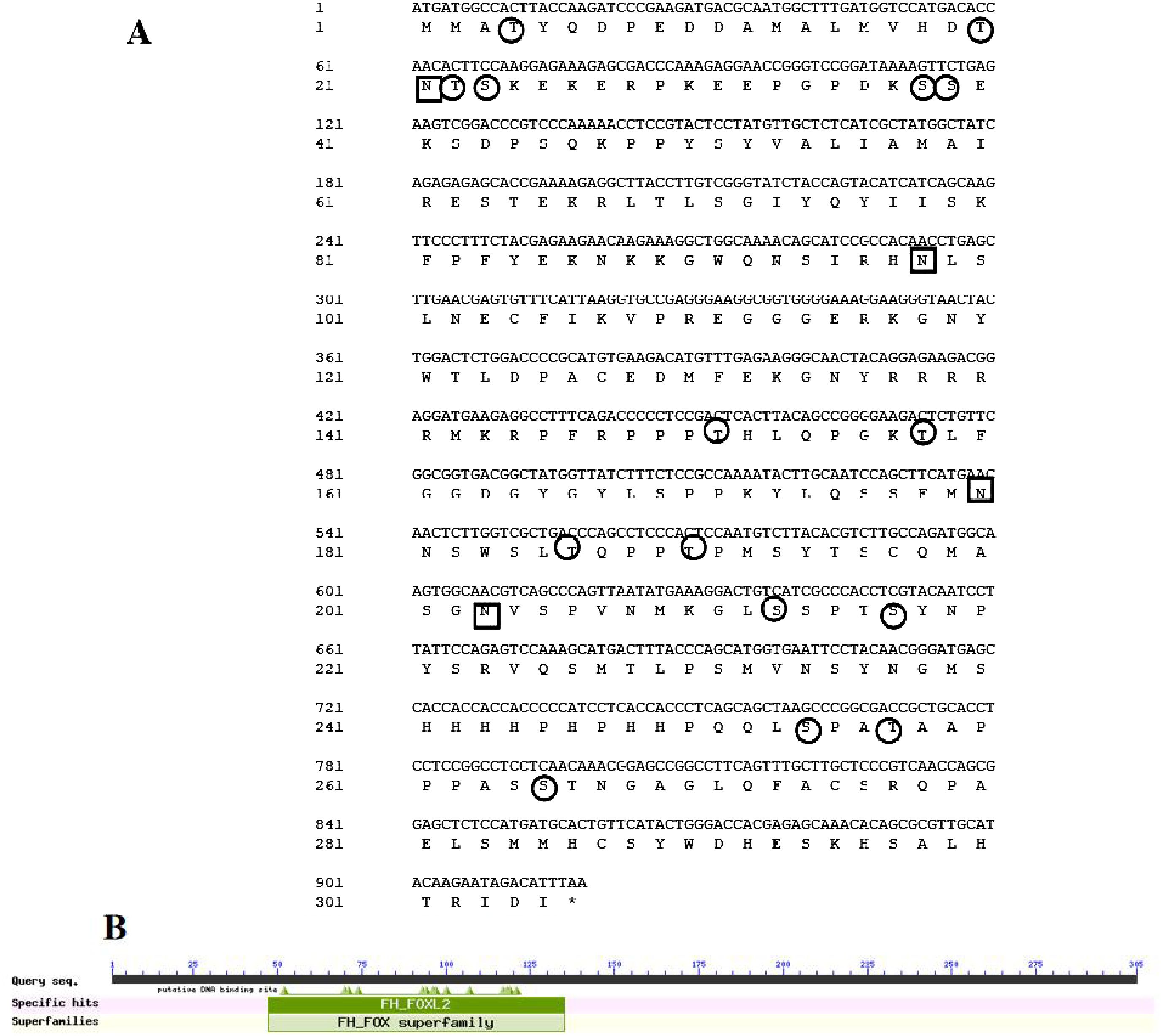
Figure 1. Examination of Asfoxl2’s structure and sequence. (A) The CDS sequence and the inferred protein sequence of Asfoxl2. (B) CDD examination revealing the conserved domain within Asfoxl2 proteins. The positions of nucleotides and amino acids are denoted to the left. The termination codon is indicated with a black asterisk (*). Potential N-glycosylation locations are indicated by squares, while potential O-glycosylation sites are indicated with circles.
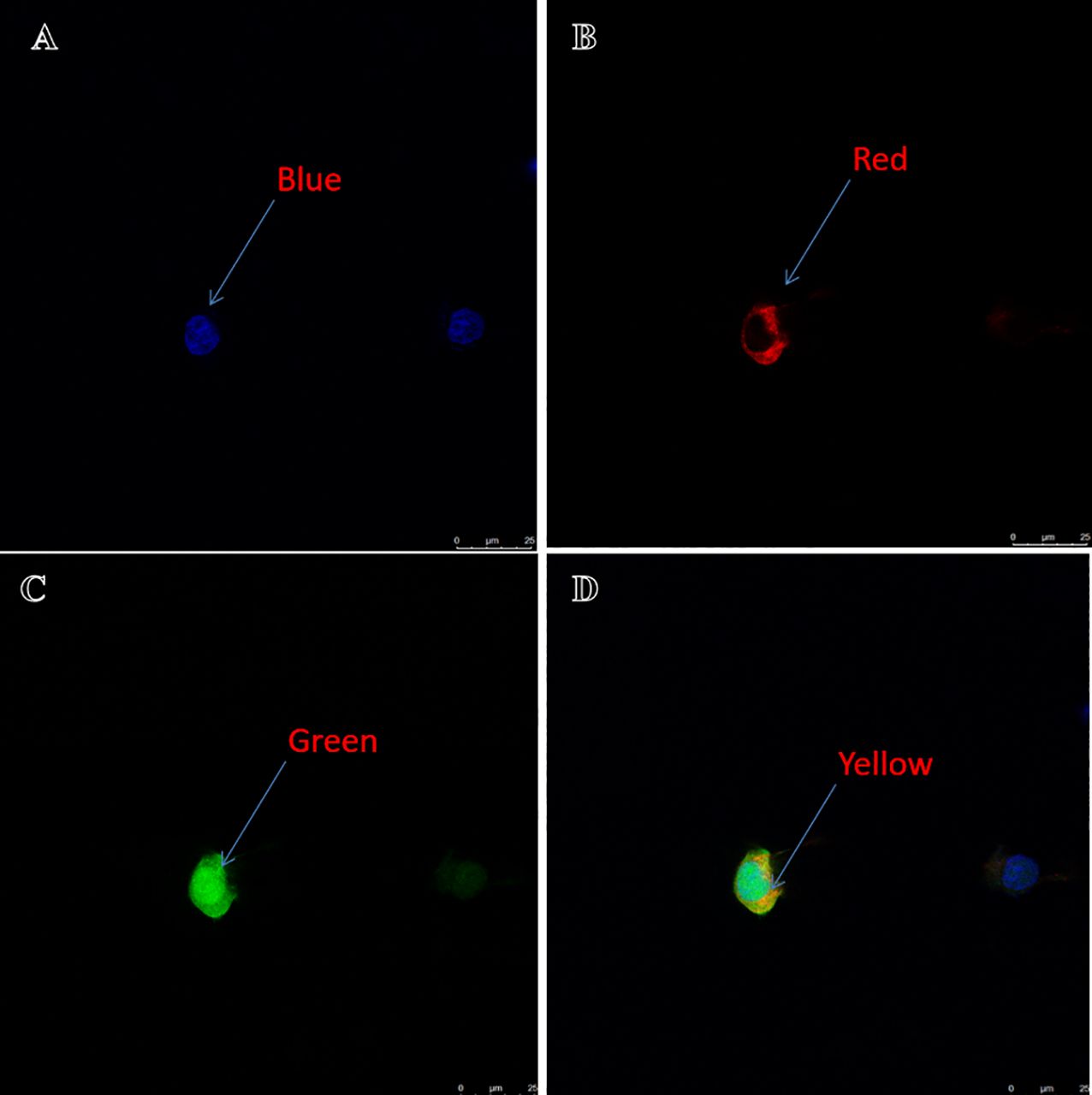
Figure 2. Localization of Asfoxl2 in HEK293t cells. (A) Blue signifies the nuclear location of the stained cell. (B) Red indicates the membrane location of the stained cell. (C) Green denotes the location of target gene protein expression. (D) Yellow represents the overlap of green and red. The yellow hue indicates details regarding the precise site of the target gene’s protein expression.
The outcomes of the secondary structure examination revealed that the protein Asfoxl2 consisted of 10 helices and a pair of β-strands, as identified through web-based protein structure prediction (Figure 3A). By utilizing SWISS MODEL, a three-dimensional molecular representation of Asfoxl2 was produced. The results showed that the sequence similarity between Asfoxl2 and the reference model (Forkhead box protein L2, AlphaFold DB entry Q1JPV2_DANRE with ID Q1JPV2.1.A) was 85.62%. This implies that the three-dimensional model of the Asfoxl2 protein was logically sound and is classified under the foxl2 family (Figure 3B).
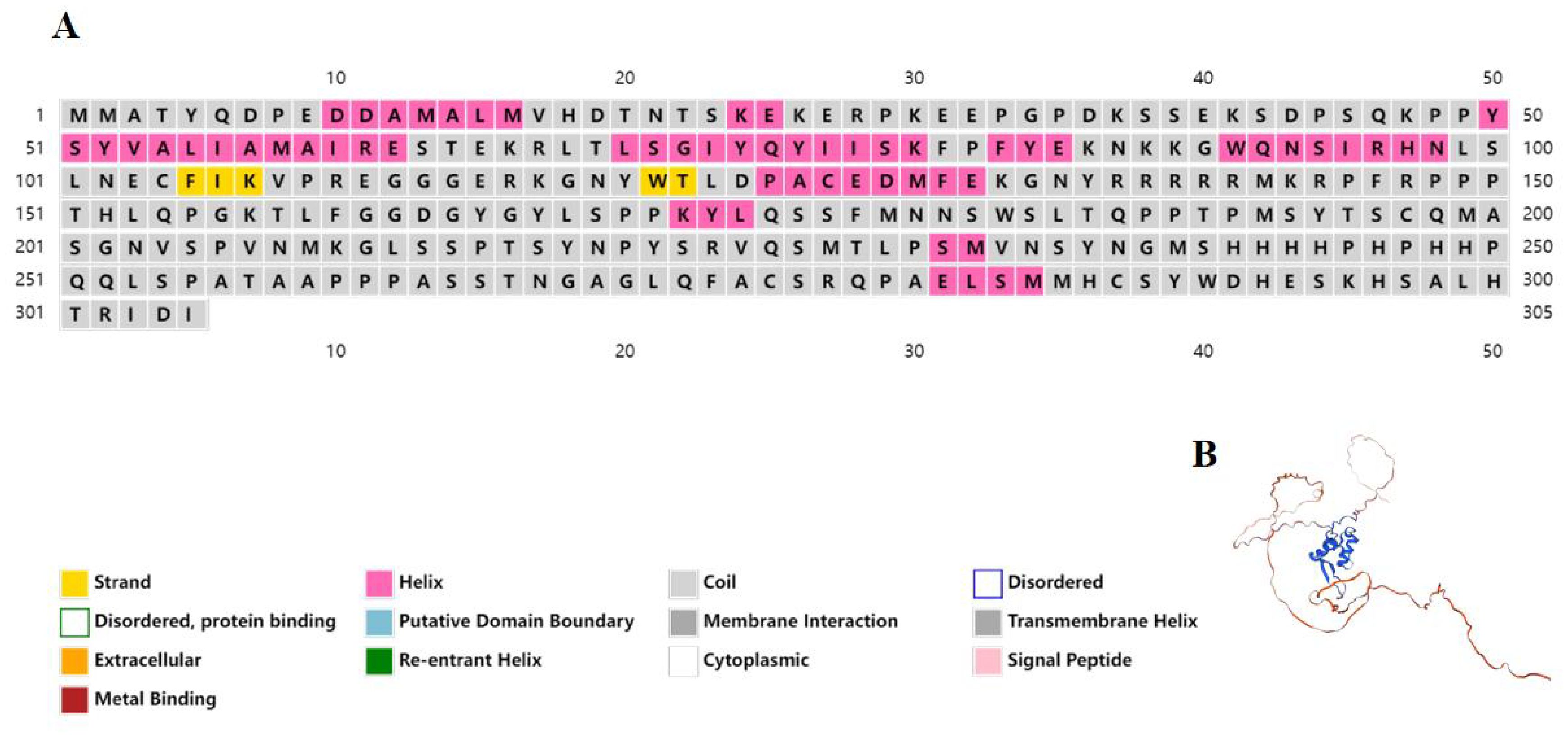
Figure 3. Structural analysis of asfoxl2. (A) The forecasted secondary structure of Asfoxl2 protein indicated a composition of 10 alpha-helices and 2 beta-strands, as ascertained by an internet-based protein conformation predictor. (B) A 3D model of the Asfoxl2 protein was generated using SWISS MODEL.
To enhance our comprehension of genetic similarities, the foxl2 gene from the Chinese sturgeon was analyzed in comparison with those from other vertebrates through the use of the NCBI database. The DNA sequence of the foxl2 gene found in Chinese sturgeon exhibited a pronounced resemblance to those found in certain fish species, notably Acipenser. Our analyses involving multiple sequence alignments highlighted that the foxl2 gene in the Chinese sturgeon exhibits substantial sequence identity with the foxl2 genes from various species (Figure 4). In order to explore the evolutionary links among the foxl2 genes across different species, a phylogenetic diagram was assembled (Figure 5). Information from the genetic lineage analysis suggest that the foxl2 gene of the Chinese sturgeon is most closely aligned with that of the Acipenser species. These findings align with the known taxonomic and evolutionary positions of these organisms.
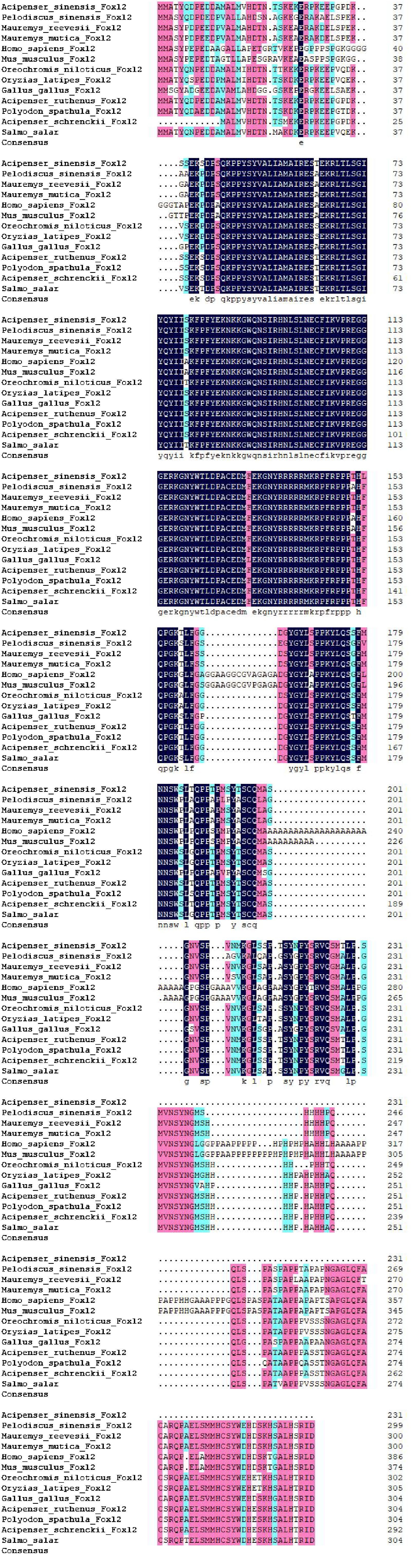
Figure 4. Cross-species sequence comparison of Asfoxl2. Utilizing the NCBI database, we performed a comparative study of the foxl2 gene sequence between Chinese sturgeon and other vertebrates.
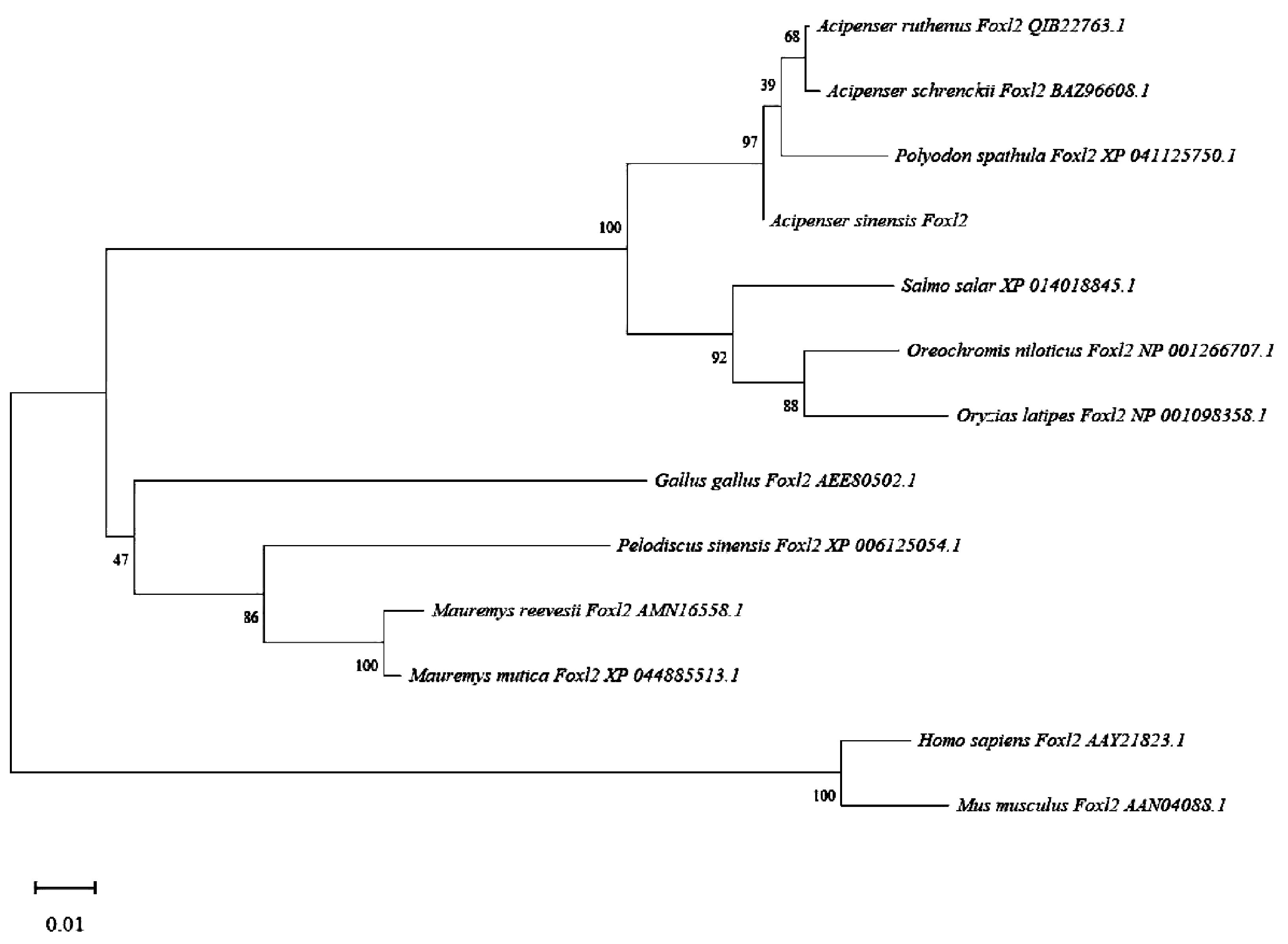
Figure 5. Phylogenetic analysis of asfoxl2. To elucidate the evolutionary linkages between the anticipated foxl2 of the Chinese sturgeon and foxl2 from other species, we generated a phylogenetic tree.
3.2 Expression patterns of foxl2 in somatic tissues of Chinese sturgeon
To ascertain the distribution of Asfoxl2 across various somatic tissues, an RT-PCR analysis was conducted on a selection of 10 tissues: these included the liver, pituitary, heart, brain, stomach, hypothalamus, testicles, muscle, intestine, and ovary (Figure 6). Transcripts of the Asfoxl2 gene were identified in both male and female specimens in this investigation. The transcription of the foxl2 gene was predominantly found in the ovaries of two-year-old Chinese sturgeon, with significantly lower levels of expression detected in multiple bodily tissues. The expression of Asfoxl2 exhibited sexual dimorphism in morphologically differentiated gonads. The gene showed significant levels of expression in the ovary, with moderate expression in the pituitary and hypothalamus. A segment of the β-actin gene was increased in quantity in every sample. This investigation revealed that female specimens exhibited significantly elevated Asfoxl2 mRNA levels in comparison to their male counterparts (P < 0.05).
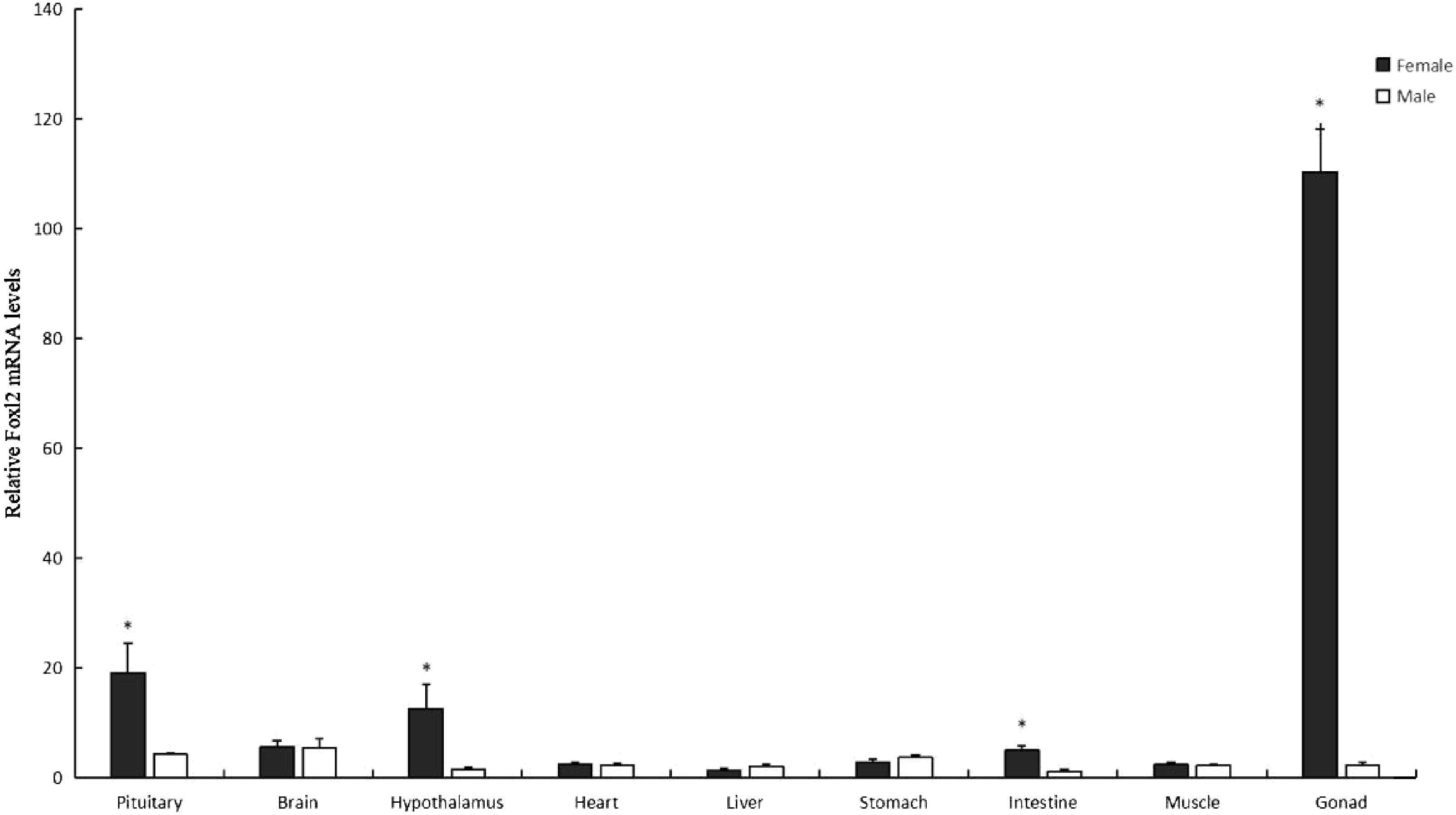
Figure 6. The expression profile of Asfoxl2, as established through qPCR analysis, is presented. Data indicated by a black asterisk showed a statistically significant difference (p < 0.05).
4 Discussion
The expression patterns of genes associated with sex differentiation could aid in the early determination of sex, particularly if these genes demonstrate expression that is specific to or predominant in one sex. For instance, before the onset of visible sex differentiation in Nile tilapia, the foxl2 gene and the cyp19a1a are uniquely transcribed in the immature gonads of genetically female individuals commencing at 5 dph, whereas the DMRT1 exhibits predominant expression in the undifferentiated gonads of genetically male individuals from 6 dph (Ijiri et al., 2008). The phase before the physical distinction of sexes is commonly known as molecular sex differentiation. Typically, the foxl2 and cyp19a1a genes play a pivotal role in the production of estrogen and the development of ovaries in vertebrates, exhibiting higher expression levels in the immature gonads of genetically female individuals than in those of genetically male ones, a pattern seen in rainbow trout (Vizziano et al., 2007) and medaka (Nakamoto et al., 2006a). DMRT1, belonging to the DM family, plays a crucial role as a transcription factor in testicular differentiation in numerous organisms, encompassing teleosts. Considering the data presented, it seems reasonable to deduce that these genes could display gender-specific or male/female predominant expression in the immature gonads of sturgeon before the onset of visible sex differentiation.
The genetic mechanisms controlling sex differentiation in sturgeon are not yet completely understood, given the limited molecular information available. Within the gonads of mature wild shovelnose sturgeon, a segment of the foxl2 complementary DNA was extracted, exhibiting greater transcript levels in the ovary than in the testis (Amberg et al., 2010). Lately, research on the RNA sequencing of immature gonads in Russian sturgeon has been carried out using advanced sequencing techniques. The study disclosed that the quantity of foxl2 mRNA was higher in immature gonads on the brink of becoming ovaries compared to those that would develop into testes at 9 MAH, whereas DMRT1 showed no gender-specific expression pattern (Hagihara et al., 2014). As highlighted by earlier studies, limited information is available about the genes DMRT1 and foxl2 in sturgeon, and their expression profiles during molecular and morphological sex differentiation are largely uncharted territories.
In summary, this investigation presents the expression patterns of foxl2 in Chinese sturgeon throughout the process of molecular and morphological sex differentiation. Foxl2 was predominantly found in the ovaries of two-year-old Chinese sturgeon, with significantly lower expression levels in various somatic tissues. It is proposed that the mRNA levels of Asfoxl2 could effectively act as a sex identification marker in sturgeon during the initial phases of development. Future research will focus on enhancing our comprehension of sex differentiation in sturgeon by investigating additional genes related to sex determination.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Chinese Sturgeon Research Institute, the China Three Gorges Corporation, and the Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. RT: Writing – original draft. YL: Writing – original draft, Writing – review & editing. TS: Writing – original draft. GY: Data curation, Project administration, Writing – review & editing. ZC: Data curation, Project administration, Writing – review & editing. HW: Investigation, Software, Writing – original draft. XL: Data curation, Methodology, Writing – original draft. XZ: Project administration, Validation, Writing – original draft. BW: Formal analysis, Software, Writing – original draft. BG: Data curation, Formal analysis, Writing – original draft. RW: Formal analysis, Validation, Writing – original draft. HQ: Investigation, Writing – original draft. HS: Writing – original draft. DZ: Writing – original draft. HD: Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. ZL: Writing – review & editing. JC: Writing – review & editing. DW: Formal analysis, Funding acquisition, Project administration, Resources, Validation, Visualization, Writing – review & editing. DX: Data curation, Formal analysis, Methodology, Project administration, Resources, Visualization, Writing – review & editing. WJ: Funding acquisition, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. QL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YY: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors declare that this study received funding from the Hubei Provincial Natural Science Foundation of China (no, 2023AFB989, 2022CFB803, and 2022CFB738) and Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes (no. 2022001- ZHX), China Three Gorges Corporation (no. WWKY-2021-0351), the National Natural Science Foundation of China (32403023, and 32202967), Hubei Provincial Chenguang Project for Young Scientific and Technological Talents (Grant No. 202350). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of interest
Authors YH, YL, TS, XL, XZ, BW, BG, RW, HQ, HS, DZ, HD, ZL, JC, DW, WJ, and YY were employed by China Three Gorges Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amberg J. J., Goforth R., Stefanavage T., Sepulveda M. S. (2010). Sexually dimorphic gene expression in the gonad and liver of shovelnose sturgeon (Scaphirhynchus platorynchus). Fish Physiol. Biochem. 36, 923–932. doi: 10.1007/s10695-009-9369-8
Berbejillo J., Martinez-Bengochea A., Bedo G., Brunet F., Volff J. N., Vizziano-Cantonnet D. (2012). Expression and phylogeny of candidate genes for sex differentiation in a primitive fish species, the Siberian sturgeon, Acipenser baerii. Mol. Reprod. Dev. 79, 504–516. doi: 10.1002/mrd.22053
Bertho S., Herpin A., Branthonne A., Jouanno E., Yano A., Nicol B., et al. (2018). The unusual rainbow trout sex determination gene hijacked the canonical vertebrate gonadal differentiation pathway. Proc. Natl. Acad. Sci. U S A. 115, 12781–12786. doi: 10.1073/pnas.1803826115
Bertho S., Pasquier J., Pan Q., Le Trionnaire G., Bobe J., Postlethwait J. H., et al. (2016). Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex Dev. 10, 111–129. doi: 10.1159/000447611
Cao H., Zhou L., Zhang Y. Z., Wei Q. W., Chen X. H., Gui J. F. (2009). Molecular characterization of Chinese sturgeon gonadotropins and cellular distribution in pituitaries of mature and immature individuals. Mol. Cell Endocrinol. 303, 34e42. doi: 10.1016/j.mce.2009.01.015
Chen J., Yuan H., Sun D., Liang B., Zhang S. (2006). Sequence and expression of three members of the Sox gene in Amur sturgeon (Acipenser schrenckii). J. Appl. Ichthyol. 22, 77–81. doi: 10.1111/j.1439-0426.2007.00764.x
Chen X., Wei Q., Yang D., Zhu Y. (2006). Observations on the formation and development of the primary germinal tissue of cultured Chinese sturgeon, Acipenser sinensis. J. Appl. Ichthyol 22, 358. doi: 10.1111/j.1439-0426.2007.00984.x
Chen Y., Xia Y., Shao C., Han L., Chen X., Yu M., et al. (2016). Discovery and identification of candidate sex-related genes based on transcriptome sequencing of Russian sturgeon (Acipenser gueldenstaedtii) gonads. Physiol. Genomics 48, 464–476. doi: 10.1152/physiolgenomics.00113.2015
Hagihara S., Yamashita R., Yamamoto S., Ishihara M., Abe T., Ijiri S., et al. (2014). Identification of genes involved in gonadal sex differentiation and the dimorphic expression pattern in undifferentiated gonads of Russian sturgeon Acipenser gueldenstaedtii Brandt &Ratzeburg 1833. J. Appl. Ichthyol. 30, 1557–1564. doi: 10.1111/jai.12588
Hale M. C., Jackson J. R., DeWoody J. A. (2010a). Discovery and evaluation of candidate sex-determining genes and xenobiotics in the gonads of lake sturgeon (Acipenser fulvescens). Genetica. 138, 745–756. doi: 10.1007/s10709-010-9455-y
Hale M. C., Jackson J. R., DeWoody J. A. (2010b). Discovery and evaluation of candidate sex-determining genes and xenobiotics in the gonads of lake sturgeon (Acipenser fulvescens). Genetica. 138, 745–756. doi: 10.1007/s10709-010-9455-y
Hattori R. S., Murai Y., Oura M., Masuda S., Majhi S. K., Sakamoto T., et al. (2012). A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U.S.A. 109, 2955–2959. doi: 10.1073/pnas.1018392109
Hu Y., Wang B., Du H., Liu X., Xiao K., Zeng Q., et al. (2021). A rapid method for identifying the genetic sex of the Chinese sturgeon using a genomic sequence fragment ZHXF-1 and its application CN113136437B.
Ijiri S., Kaneko H., Kobayashi T., Wang D. S., Sakai F., Paul-Prasanth B., et al. (2008). Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 78, 333–341. doi: 10.1095/biolreprod.107.064246
Kamiya T., Kai W., Tasumi S., Oka A., Matsunaga T., Mizuno N., et al. (2012). A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PloS Genet. 8, 470–471. doi: 10.1371/journal.pgen.1002798
Koopman P., Gubbay J., Vivian N., Goodfellow P., Lovell-Badge R. (1991). Male development of chromosomally female mice transgenic for Sry. Nature. 351, 117–121. doi: 10.1038/351117a0
Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., et al. (2002). DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 417, 559–563. doi: 10.1038/nature751
McCormick C., Bos D., DeWoody J. (2008). Multiple molecular approaches yield no evidence for sex-determining genes in lake sturgeon. J. Appl. Ichthyol. 1, 643–645. doi: 10.1111/j.1439-0426.2008.01156.x
Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y., Fujiyama A., et al. (2012). Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics. 191, 163–170. doi: 10.1534/genetics.111.137497
Nakamoto M., Matsuda M., Wang D. S., Nagahama Y., Shibata N. (2006a). Molecularcloning and analysis of gonadal expression of Foxl2 in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 344, 353–361. doi: 10.1016/j.bbrc.2006.03.137
Nakamoto M., Matsuda M., Wang D. S., Nagahama Y., Shibata N. (2006b). Molecular cloning and analysis of gonadal expression of Foxl2 in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 344, 353–361. doi: 10.1016/j.bbrc.2006.03.137
Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C., Shimizu A., et al. (2002). A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. U.S.A. 99, 11778–11783. doi: 10.1073/pnas.182314699
Okada H., Hagihara S., Yamashita K., Ijiri S., Adachi S. (2017). Expression pattern of foxl2 and dmrt1 in gonad of Amur sturgeon Acipenser schrenckii in relation to sex differentiation. Aquaculture. 479, 712–720. doi: 10.1016/j.aquaculture.2017.07.020
Peng Z., Ludwig A., Wang D., Diogo R., Wei Q., He S. (2007). Age and biogeography of major clades in sturgeons and paddlefishes (Pisces: Acipenseriformes). Mol. Phylogenet. Evol. 42, 854–862. doi: 10.1016/j.ympev.2006.09.008
Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 346, 240–244. doi: 10.1038/346240a0
Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., Doran T. J., et al. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 461, 267–271. doi: 10.1038/nature08298
Vidotto M., Grapputo A., Boscari E., Barbisan F., Coppe A., Grandi G., et al. (2013). Transcriptome sequencing and de novo annotation of the critically endangered Adriatic sturgeon. BMC Genomics 14, 407–423. doi: 10.1186/1471-2164-14-407
Vizziano D., Randuineau G., Baron D., Cauty C., Guiguen Y. (2007). Characterization of early molecular sex differentiation in rainbow trout, Oncorhynchus mykiss. Dev. Dyn. 236, 2198–2206. doi: 10.1002/dvdy.v236:8
Wang W., Zhu H., Dong Y., Tian Z., Dong T., Hu H., et al. (2017). Dimorphic expression of sex-related genes in different gonadal development stages of sterlet, Acipenser ruthenus, a primitive fish species. Fish Physiol. Biochem. 43, 1557–1569. doi: 10.1007/s10695-017-0392-x
Wei Q., Li L., Du H., Zhang X., Xiong W., Zhang H., et al. (2013). Research on technology for controlled propagation of cultured Chinese sturgeon (Acipenser sinensis). J. Fish Sci. China 20, 1–11. doi: 10.3724/SP.J.1118.2013.00001
Wuertz S., Gaillard S., Barbisan F., Carle S., Congiu L., Forlani A., et al. (2006). Extensive screening of sturgeon genomes by random screening techniques revealed no sex-specific marker. Aquaculture. 258, 685–688. doi: 10.1016/j.aquaculture.2006.03.042
Yang Y. J., Wang Y., Li Z., Zhou L., Gui J. F. (2017). Sequential, divergent, and cooperative requirements of foxl2a and foxl2b in ovary development and maintenance of zebrafish. Genetics 205, 1551–1572. doi: 10.1534/genetics.116.199133
Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., Klopp C., et al. (2012). An Immune-Related Gene Evolved into the Master Sex-Determining Gene in Rainbow Trout, Oncorhynchus mykiss. Curr. Biol. 22, 1423–1428. doi: 10.1016/j.cub.2012.05.045
Ye H., Li C. J., Yue H. M., Yang X. G., Wei Q. W. (2015). Differential expression of fertility genes boule and dazl in Chinese sturgeon (Acipenser sinensis), a basal fish. Cell Tissue Res. 1-13, 413–425. doi: 10.1007/s00441-014-2095-2
Yoshimoto S., Okada E., Umemoto H., Tamura K., Uno Y., Nishida-Umehara C., et al. (2008). A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 105, 2469–2474. doi: 10.1073/pnas.0712244105
Zhang X., Li M., Ma H., Liu X., Shi H., Li M., et al. (2017). Mutation of foxl2 or cyp19a1a results in female to male sex reversal in XX Nile Tilapia. Endocrinology 158, 2634–2647. doi: 10.1210/en.2017-00127
Keywords: Chinese sturgeon, FOXL2, sex differentiation, gonad, endangered fish
Citation: Hu Y, Tan R, Li Y, Shu T, Yang G, Chu Z, Wang H, Liu X, Zhu X, Wang B, Guo B, Wang R, Qu H, Song H, Zhang D, Du H, Li Z, Chang J, Wang D, Xu D, Jiang W, Li Q and Yang Y (2024) Molecular cloning, expression and functional analysis of foxl2 from Chinese sturgeon (Acipenser sinensis) in relation to sex differentiation. Front. Mar. Sci. 11:1506932. doi: 10.3389/fmars.2024.1506932
Received: 06 October 2024; Accepted: 21 November 2024;
Published: 16 December 2024.
Edited by:
Fabio Carneiro Sterzelecki, Federal Rural University of the Amazon, BrazilReviewed by:
Bruno José Corecha Fernandes Eiras, Federal Rural University of the Amazon, BrazilIgor Hamoy, Federal Rural University of the Amazon, Brazil
Copyright © 2024 Hu, Tan, Li, Shu, Yang, Chu, Wang, Liu, Zhu, Wang, Guo, Wang, Qu, Song, Zhang, Du, Li, Chang, Wang, Xu, Jiang, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanjin Yang, eWFuZ195dWFuamluQGN0Zy5jb20uY24=; Qingfei Li, bGlxaW5nZmVpQHFhdS5lZHUuY24=; Wei Jiang, amlhbmdfd2VpNkBjdGcuY29tLmNu; Dan Xu, ZHh1QHNob3UuZWR1LmNu; Dianchang Wang, d2FuZ19kaWFuY2hhbmdAY3RnLmNvbS5jbg==
†These authors have contributed equally to this work and share first authorship
 Yacheng Hu
Yacheng Hu Ruihua Tan
Ruihua Tan Yang Li1,2†
Yang Li1,2† Tingting Shu
Tingting Shu Binzhong Wang
Binzhong Wang Jianbo Chang
Jianbo Chang Dan Xu
Dan Xu Qingfei Li
Qingfei Li