- 1Departament de Biologia Marina i Oceanografia, Institut de Ciències del Mar (ICM-CSIC), Barcelona, Catalonia, Spain
- 2Limnology laboratory, INIBIOMA-CONICET, Universidad Nacional del Comahue, San Carlos de Bariloche, Argentina
- 3Marine and Freshwater Solutions Unit, Finnish Environment Institute (Syke), Helsinki, Finland
Microplastics are ubiquitous in marine ecosystems and are suitable matrices for bacterial attachment and growth. Studies on the microbes growing on plastics are mainly done using flow cytometry and massive sequencing, which do not allow for the quantification of specific groups and their activity. Here we present the results from a mesocosm experiment, designed to compare the effects of biodegradable and conventional microplastics on planktonic communities of the Baltic Sea. Our specific aim was to study the effects on bacterial activity and abundance using epifluorescence microscopy techniques. Specifically, we applied BONCAT-FISH which simultaneously allows for phylogenetic identification and the detection of the activity of individual bacterial cells. In our experiment, mesocosms were filled with Baltic brackish seawater and amended with 20 microplastic beads·ml-1 in triplicates for several treatments: (i) None (control), (ii) PS, (iii) PLGA and (iv) PS + PLGA. Our results show a low impact of the presence and quality of microplastics on marine bacterial communities during the first 11 days of exposure, with only weak differences in the activity of bacterial communities growing with biodegradable or conventional microplastics additions.
1 Introduction
The term Plastic Age has been used to describe the current period of human history (Thompson et al., 2009). The intense consumption and rapid disposal of plastic products are leading to a visible accumulation of plastic debris (Barnes et al., 2009), with the global load of plastic on the open ocean surface estimated to be in the order of tens of thousands of tons (Cózar et al., 2014; Van Sebille et al., 2015), causing rising concern. When released into the environment, plastic litter undergoes fragmentation through various physical and chemical processes. Physical processes, such as UV radiation, wave action, and abrasion from sand or other particles, contribute to breaking down larger plastic items into smaller fragments (Andrady, 2011; Gewert et al., 2015). Additionally, chemical processes, including oxidation and hydrolysis, further degrade plastics by altering their molecular structure and increasing brittleness (Fotopoulou and Karapanagioti, 2012). Together, these processes lead to the formation of small plastic particles, commonly referred to as microplastics (MPs) (<5 mm) (Barnes et al., 2009; Dussud et al., 2018). These fragments, when smaller than 1 mm in size, are easily ingested by zooplankton and subsequently excreted within fecal pellets (Gunaalan et al., 2023). These pellets act as means of vertical transportation, in addition to providing a source of food for marine organisms. Biofouled plastics also transport non-native species (Urbanek et al., 2018) and, as a result, microorganisms that naturally occur in one part of the world can be transported by marine debris into new distinct habitats, potentially leading to adverse effects on marine ecosystems (Zettler et al., 2013; De Tender et al., 2015; Debroas et al., 2017).
Polyethylene (PE) prevails in the composition of plastic waste at sea surface, followed by polypropylene (PP) and polystyrene (PS) (Auta et al., 2017). PS is a long-lasting thermoplastic that is generally thought to be nonbiodegradable. Actually, biodegradation of PS does occur but at a very slow rate in natural environments, therefore PS can persist for long periods of time as solid waste (Ho et al., 2018; Vesamäki et al., 2022). Kaplan et al. (1979) stated that in cultivated soils containing a wide range of fungi, microbes, and invertebrates, degradation of PS is less than 1% after 90 days with no significant increase in degradation rate beyond this point. In contrast, Otake et al. (1995) reported that a sheet of PS buried in soil for 32 years showed no signs of degradation. As an alternative for these conventional plastics, the interest in biodegradable plastics has risen, although their degradation rates are very slow in nature (Wang et al., 2021) unless they are under certain industrial conditions (Haider et al., 2019). PolyLactic co-Glycolic Acid (PLGA) is a biodegradable plastic that showed a 100% degradation rate after 270 days in seawater (Wang et al., 2021).
In recent years, the environmental impacts of microplastics (MPs) have garnered increased attention, particularly regarding their prevalence and potential effects on marine ecosystems (Da Costa et al., 2020). Studies highlight that MPs are not only pervasive but may pose significant ecological risks due to their persistence and interaction with various marine organisms (Reichelt and Gorokhova, 2020). Recent research underscores the role of MPs as vectors for pollutant transport and potential disruptors of microbial community dynamics in the Baltic Sea and other marine environments (Schernewski et al., 2020). Additionally, findings from the Warnow Estuary in the southwestern Baltic Sea emphasize the diverse sources of MPs in urbanized estuaries, illustrating the complex pathways by which these pollutants enter marine systems and accumulate (Piehl et al., 2021).
Bacteria play a key role in the degradation of organic matter in the ocean as well as of anthropogenic materials such as microplastics (Torena et al., 2021; Miri et al., 2022; Thakur et al., 2023). To date, research on the MP-bacteria interaction has focused on three main areas: (a) the establishment of plastic-specific biofilms (the so-called plastisphere); (b) the enrichment of pathogenic bacteria, particularly members of the genus Vibrio, coupled to a vector function of MPs; and (c) the microbial degradation of MPs in the marine environment (Oberbeckmann and Labrenz, 2019). Although some authors have observed stimulation of bacterial activity after MP addition (Romera-Castillo et al., 2022), others found a decrease in extracellular enzyme activity (Caruso et al., 2018). However, fewer studies have attempted to identify the specific bacterial groups that exhibit increased activity (Birnstiel et al., 2022). New advances in culture-independent methods for measuring cellular activity, such as BioOrthogonal NonCanonical Amino acid Tagging (BONCAT) (Hatzenpichler et al., 2014) coupled with FISH (CAtalysed Reporter Deposition Fluorescence In Situ Hybridization) (Pernthaler et al., 2004), show promise in answering questions related to substrate preference, relative activity levels, and phylogenetic identity of active members of marine communities (Buongiorno, 2018).
The persistence of both conventional and biodegradable plastics in marine environments presents complex ecological challenges, as they may alter microbial communities and biogeochemical processes (Eronen-Rasimus et al., 2022). Recent findings indicate that degradation rates and microbial colonization vary significantly among bioplastic materials, suggesting that specific environmental conditions, like those in the brackish Baltic Sea, may influence microbial responses and degradation dynamics. This highlights the need to evaluate the environmental fate of bioplastics across diverse marine ecosystems to inform sustainable material choices and pollution mitigation efforts. Therefore, the aim of our work was to elucidate the stimulation of bacterial growth of α-, β- and γ-proteobacteria and Bacteroidetes in response to the addition of conventional (PS) and biodegradable (PLGA) microplastics, as well as a mixture of both (MIX). We hypothesized that PLGA, due to its higher lability, would show a stimulation of certain primary bacterial colonizer groups, such as γ-proteobacteria, after the 10th day of incubation.
2 Materials and methods
2.1 Experimental setup
Coastal Baltic seawater (salinity 5.34‰, temperature 14°C) was pumped from 5 m depth in the middle of the Gulf of Finland (depth of the station 70m) on the research vessel Aranda into 4 m3 tanks on 15th of August, 2019. The water was pre-filtered through a 100 µm mesh into twelve mesocosms (320 l). Ammonium (~3 µmol·l-1) and phosphate (0.6-0.8 µmol·l-1) were present in the water and oxygen was 5.75 ml·l-1.
Mesocosms were amended with 20 MP beads·ml-1 (10 µm in diameter) in triplicates of each treatment group: None (K, control), conventional polystyrene (PS), biodegradable polylactic co-glycolic acid (PLGA), and a combination of both PS and PLGA (MIX). PS (Thermo Fisher) was fluorescent under green light (orange) and PLGA (Sigma-Aldrich) was fluorescent under blue light (green).
The experiment was conducted at a controlled temperature of 14°C and light/dark cycle of 12 h/12 h from August 26th to September 6th at Syke Marine Research Laboratory, Finland. The mesocosms were sampled every two days for bacterial production, and for BONCAT, FISH and DAPI on the last day. Immediately following each sampling event, the samples underwent filtration and fixation, and filters were then transported frozen from Helsinki to the ICM to follow the hybridization, fluorescence and counting procedures.
2.2 Fluorescent techniques
We used three different fluorescent approaches for our study. In all instances, we filtered 5 ml of sampled water through a 0.2 µm pore-size polycarbonate filter (Whatman® Cyclopore®) for free-living bacteria on the 11th day and they were observed under the microscope using: DAPI (4′,6-diamidino-2-phenylindole), which stains all bacteria; CARD-FISH, which highlights specific targeted groups of bacteria; and BONCAT, which shows translationally active cells.
2.2.1 Bacterial abundance
DAPI staining was used to estimate bacterial abundance, as well as to double-check cells analyzed with other fluorescent techniques such as CARD-FISH or BONCAT, following a modified protocol from Porter and Feig (1980). Samples (4.5 ml) were fixed with 0.5 ml of 10% glutaraldehyde and incubated for at least one hour (but no more than 24 h) at 4°C in the dark. The samples were stained by adding 50 µl of DAPI (0.5 mg·ml-1) to 5 ml of fixed sample and filtered through 0.2 µm polycarbonate black filters.
2.2.2 BONCAT-FISH
Single-cell activity was estimated following a modification to the Hatzenpichler and Orphan (2015) protocol by applying a triple labelling with DAPI, CARD-FISH and BONCAT to assess the in situ translational activity of all cells and specific groups of bacteria.
This technique is based on the in vivo incorporation of artificial amino acids that carry modifiable chemical tags into newly synthesized proteins. After the chemical fixation of cells, surrogate-containing proteins can be detected by whole-cell fluorescence staining using azide-alkyne click chemistry.
For this experiment, 4.5 ml samples were collected from each treatment group and incubated with L-Homopropargylglycine (HPG; final concentration 1 μM) for 2 h. Incubations were performed in triplicate. Samples were fixed with 0.2 μm-filtered paraformaldehyde (PFA; final concentration 4% [v/v]) overnight at 4°C, filtered onto a 0.2 μm pore size white polycarbonate filter, and stored at −80°C until further processing. After thawing, the filters were dipped in a previously boiled 0.1% (w/w) low-gelling-point agarose solution to ensure cell attachment to the filter and to prevent cell loss during permeabilization and downstream procedures. These were then dried at 37°C and dehydrated with 95% ethanol. Bacterial cells were permeabilized with lysozyme (10 mg·ml−1; 0.05 M EDTA, 0.1 M Tris-HCL, 1 h) and achromopeptidase (2 μL·ml−1; 0.01 M NaCl, 0.01 M Tris-HCl, pH 8 30 min) at 37°C.
For CARD-FISH, we followed a modified version of Pernthaler et al. (2004) protocol as previously outlined by Sebastián et al. (2019). Filter portions were hybridized with the following horseradish peroxidase (HRP)-labelled (fluorescent marker) probes which bind by complementarity to specific rRNA sequences: ALF968 which targets α-proteobacteria (Neef, 1997), BET42a which targets β-proteobacteria (Manz et al., 1992), GAM42a which targets γ-proteobacteria (Manz et al., 1992) and CF968 which targets Bacteroidetes (Acinas et al., 2014). Specific hybridization conditions were established by addition of formamide to the hybridization buffers (45% formamide for the ALF968 probe and 55% for the other probes). Hybridization was performed overnight at 35°C. For amplification, a tyramide labelled with Alexa488 was used.
Cu(I)-catalysed click chemistry was later conducted following a method by Leizeaga et al. (2017). For the click reaction, the dye-premix was prepared by mixing 12.5 μL of copper sulphate (CuSO4(H2O)5; 20mM), 10 μL of Alexa594 azide dye and 25 μL of Tris[(1-hydroxypropyl-1H-1,2,3-triazol-4-yl)methyl]amine (THPTA; 50 mM), and left to react at room temperature for 3 minutes in absence of light. Meanwhile, 125 µL of a freshly prepared 10 mM solution of sodium ascorbate (in phosphate buffer saline, PBS) and 125 µL of a freshly prepared 10 mM solution of aminoguanidine hydrochloride (in PBS) were added to 2.21 ml of PBS solution. In order to prepare the click reaction mixture, the dye-premix was added to the PBS-ascorbate-aminoguanidine mix and the tube was inverted twice. The portions of filters were then placed in an empty 1.5 ml Eppendorf tube and the reaction mix was added up to the cap, to further avoid production of air bubbles and to maintain reducing conditions. The click reaction was incubated at room temperature for 30 minutes in the dark and the filters were then washed three times for 3 minutes each in PBS-filled petri dishes. A final 1 h wash with a 1:1 PBS: Ethanol solution was performed to reduce the fluorescence background.
Filters portions were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; final concentration 0.5 μg·ml−1) in order to quantify the relative abundance of each targeted phylogenetic group and their single-cell activity.
Black and white images were obtained using an Axio Imager.Z2m epifluorescence microscope connected to a Zeiss camera (AxioCam MRm, Carl Zeiss MicroImaging, S.L., Barcelona, Spain) at 630x magnification, through use of the Axiovision software. The coordinates of the samples were obtained using the SamLoc software and subsequently analyzed using ACMEtool. A Colibri LED light source (Carl Zeiss) with three light-emitting diodes was used and all images (between 10 and 48 images per filter) were acquired using 30-60 ms exposure time for DAPI, 100 - 150 ms exposure time for CARD-FISH and 150 - 350 ms exposure time for BONCAT (A594). The exposure times of each bacteria group were calculated for each treatment at each time point. The percentage of FISH+ and BONCAT+ cells was calculated in relation to the DAPI counts. ACMEtool was used for the analysis of the images, as described in Leizeaga et al. (2017), and for subtracting the background of the samples. To address background noise, the signal-to-background ratio of the cells was adjusted.
2.3 Bacterial production
Bacterial production was measured by determining the incorporation of 14C-leucine (Leu) for protein synthesis (BPL). At all sampling dates, five subsamples of 1 ml were taken from each mesocosm unit. Two of the subsamples were fixed with formaldehyde (final concentration 1.85%) and served as a blank of non-biological adsorption of the radio isotope. All samples were spiked with [14C (U)]-leucine (PerkinElmer) at a final saturating concentration of 150 nM. The samples were incubated at the experimental temperature (14°C) for 1 h in darkness. The incubations were stopped by adding formaldehyde to a final concentration of 1.85%. The unincorporated radioactive substrate was removed, using the standard cold trichloroacetic acid (TCA) extraction method (Fuhrman and Azam, 1982). The samples (1 ml) were centrifuged (Smith and Azam, 1992) and the pellets were dissolved in an Insta-Gel Plus scintillation cocktail (PerkinElmer), and the radioactivity was determined with a Wallac WinSpectral 1414 counter.
2.4 Statistical analysis
To assess variability among the treatments set in the mesocosm experiments, statistical analyses were performed on our key response variables: bacterial abundance, activity, community composition and bacterial production. A one-way ANOVA was used for single-factor analyses, while multifactorial two-way ANOVA with interaction terms was employed to explore the combined effects of the treatments and incubation time on the response variables. Tukey’s post-hoc tests were conducted following ANOVA to identify significant differences between treatment levels. Normality and homoscedasticity of residuals were assessed using the Shapiro-Wilk test for normality, and homoscedasticity was tested with the Brown-Forsythe test for one-way ANOVA or Spearman’s test for two-way ANOVA. All statistical analyses were conducted in GraphPad Prism 9, with significance defined as p < 0.05.
3 Results
Bacterial concentrations on the last day of the experiment varied between 4.45 x 106 and 7.19 x 106 cells·ml-1 (Figure 1), with no statistical differences observed among treatments (one-way ANOVA test, p = 0.2401).
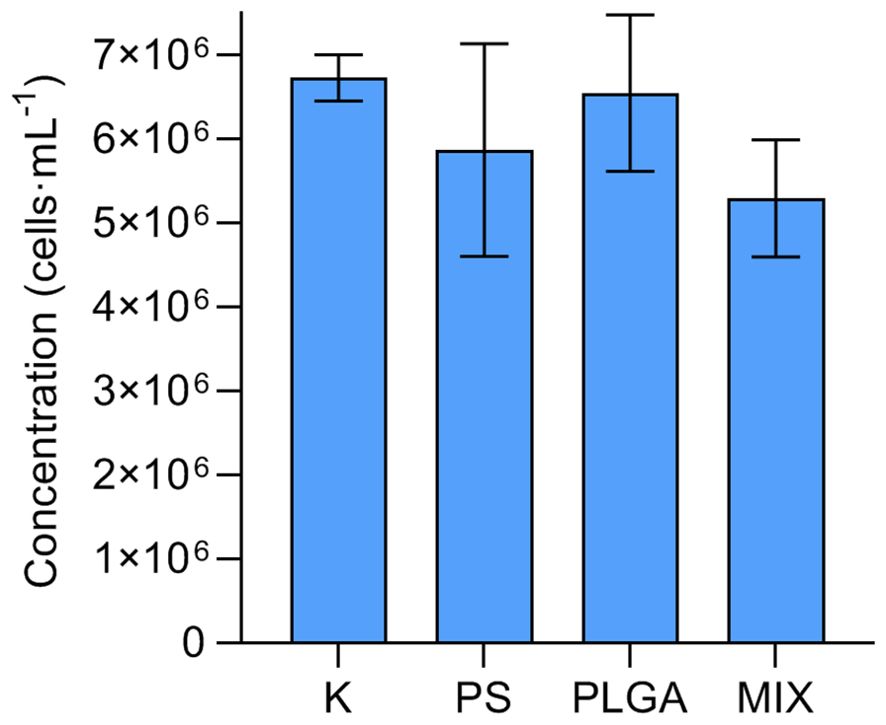
Figure 1. Bacterial concentration (mean ± standard deviation, n=3) at the end of the experiment (day 11) in the treatments. K: control; PS (polystyrene); PLGA (biodegradable MP); MIX (mixture of PS+PLGA).
The percentage of BONCAT+, i.e. active bacteria (Figure 2), was significantly higher (one-way ANOVA test, p = 0.0010) in all treatments amended with MPs compared to the control K (53%): PS (67%, p = 0.0063**); PLGA (62%, p = 0.0275*) MIX (68%, p = 0.0007***). No differences were found between the MP-amended treatments (Tukey p > 0.05).
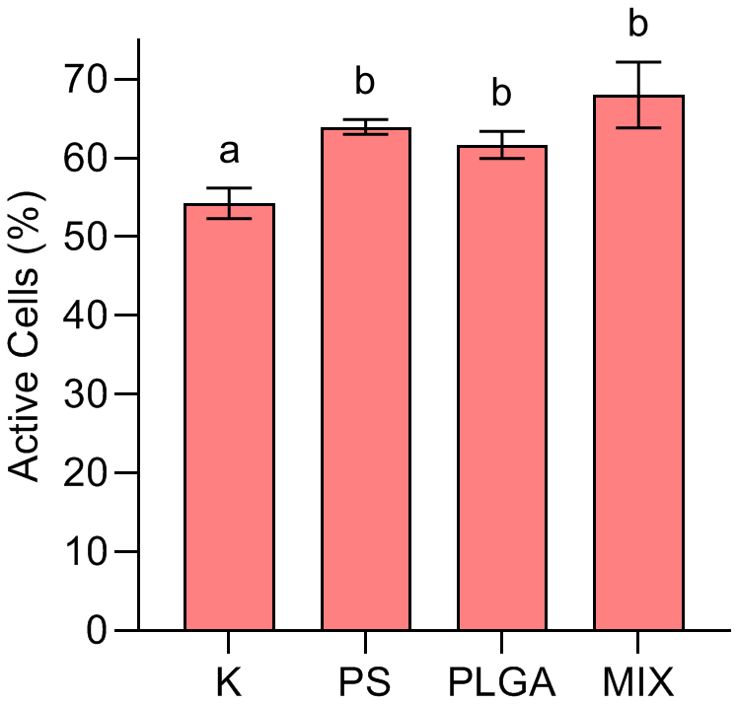
Figure 2. Percentage of BONCAT+ bacteria at the end of the experiment (day 11) in the treatments. K: control; PS (polystyrene); PLGA (biodegradable MP); MIX (mixture of PS+PLGA). Different letters indicate significant differences among treatments [ANOVA followed by a post hoc Tukey’s test (P < 0.05)].
The four CARD-FISH probes chosen for our experiment (α-proteobacteria, β-proteobacteria, γ-proteobacteria and Bacteroidetes) covered between 27% and 37% of all bacteria present in the experiment. For better clarity, results are presented based on the known part of the community. The most abundant group was found to be β-proteobacteria, accounting for between 50% and 56%, while the least abundant group was Bacteroidetes, accounting for 8%-11% (Figure 3). No significant differences in bacterial composition among treatments were detected (two-way ANOVA p = 0.3602) nor in the interaction between treatments and phylogenetic groups (p = 0.9962), leaving phylogenetic groups as the only significant source of variation (p < 0.0001****). These probes targeted an average of 30% of the total bacteria detected, with a higher detection in the K treatment and similar detection rates among treatment groups amended with MPs (K: 37%; PS: 28%; PLGA: 27%; MIX: 27%).
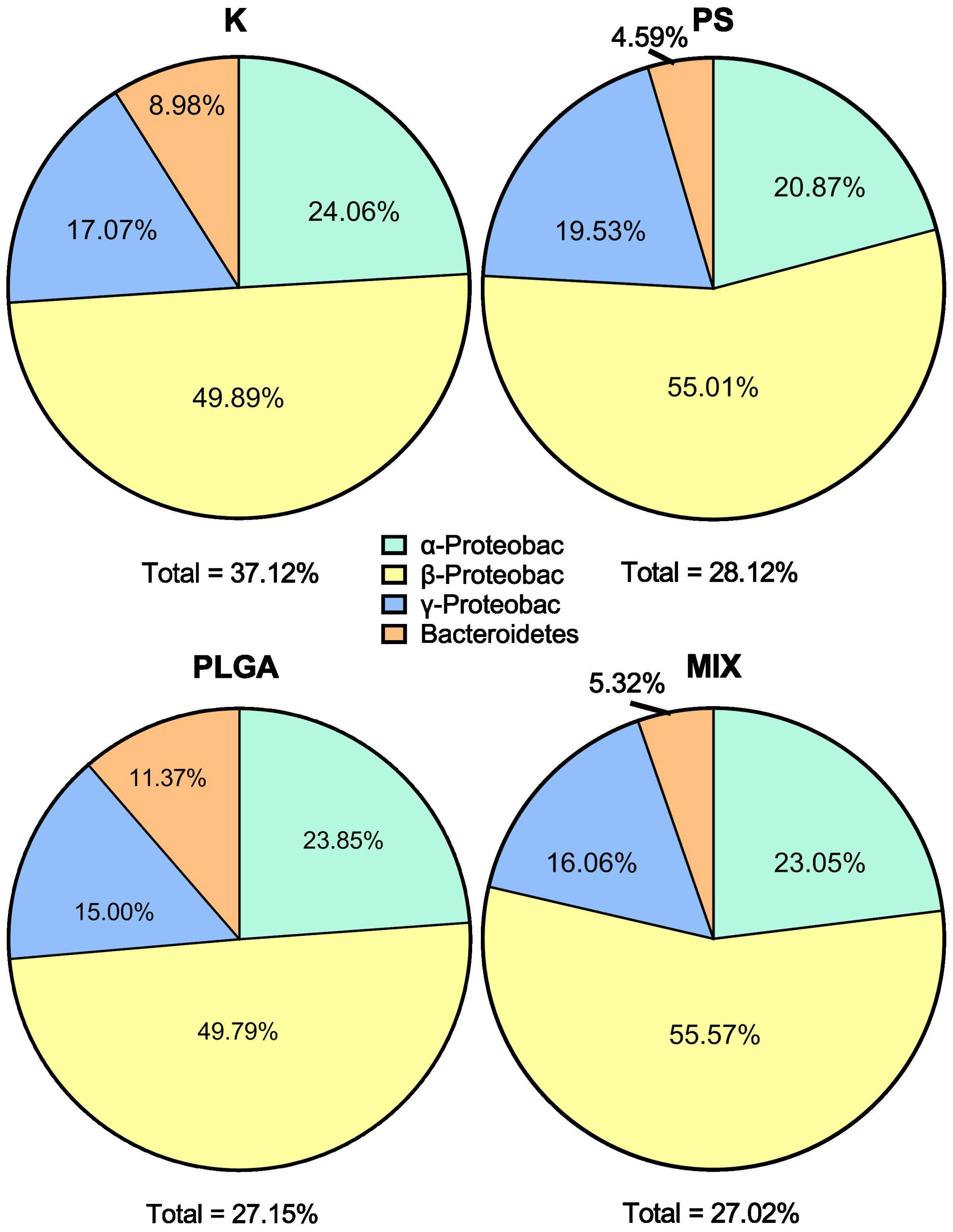
Figure 3. Bacterial community composition (%) at the end of the experiment (day 11) in the treatments. K: control; PS (polystyrene); PLGA (biodegradable MP); MIX (mixture of PS+PLGA).
The combination of the three fluorescence techniques (BONCAT, CARD-FISH and DAPI) allowed for observation of active bacteria within each phylogenetic group (Figure 4). No significant differences among treatments were found (two-way ANOVA test, p = 0.3879) nor interaction between treatments and phylogenetic groups (p = 0.2250), leaving phylogenetic groups as the only significant source of variation (p < 0.0001****).
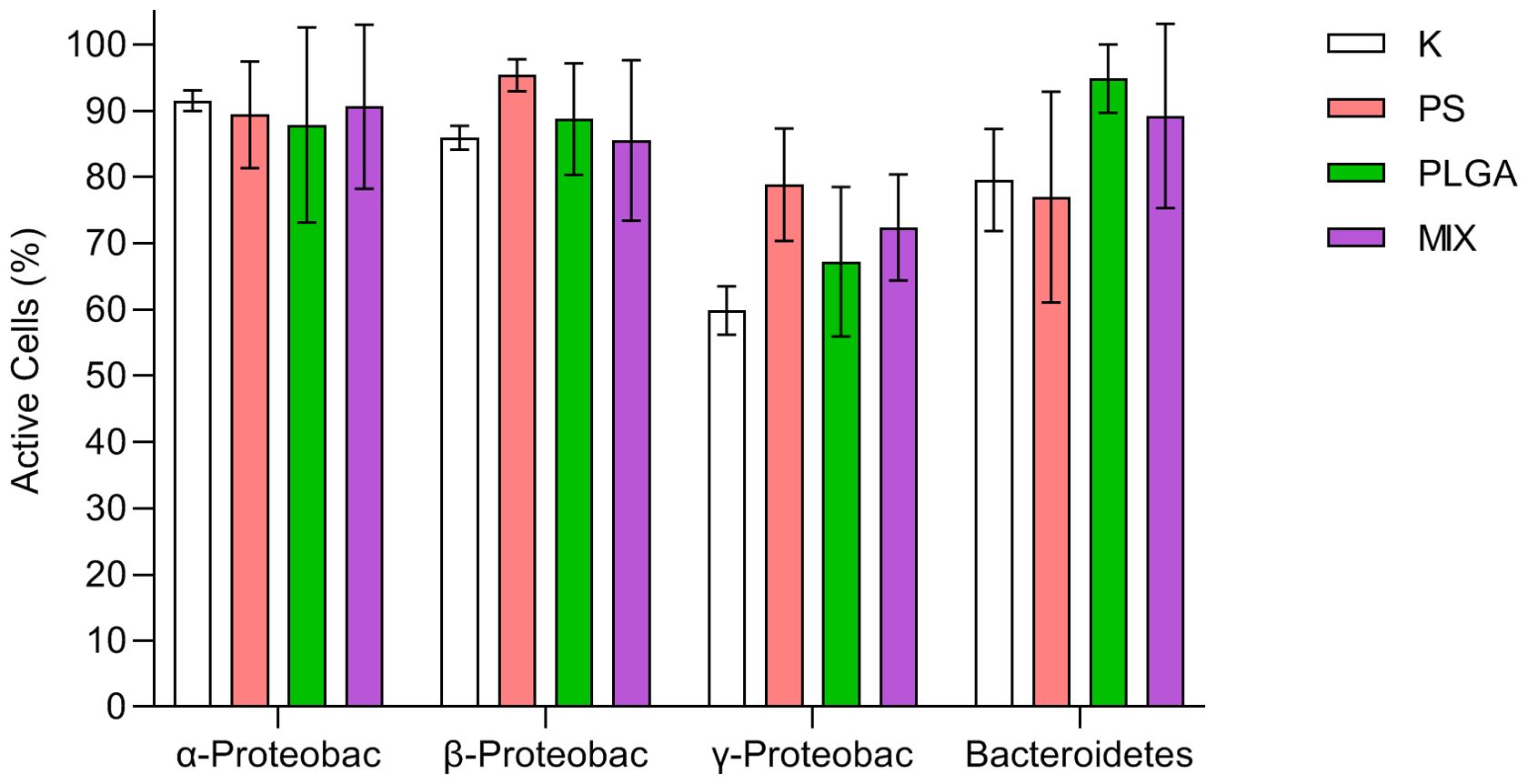
Figure 4. Percentage of active bacteria (mean ± standard deviation, n=3) at the end of the experiment (day 11) in the treatments. K: control; PS (polystyrene); PLGA (biodegradable MP); MIX (mixture of PS+PLGA).
Bacterial production, measured through leucine incorporation, followed the same trend in all treatments with no significant treatment effect (two-way ANOVA test p = 0.4323), increasing from 1.73 ± 0.13 nmol·l-1·h-1 on day 1 to 6.43 ± 1.26 nmol·l-1·h-1 on day 5 and then decreasing to a level 5.33 ± 0.77 nmol·l-1·h-1 for the rest of the experiment (Figure 5).
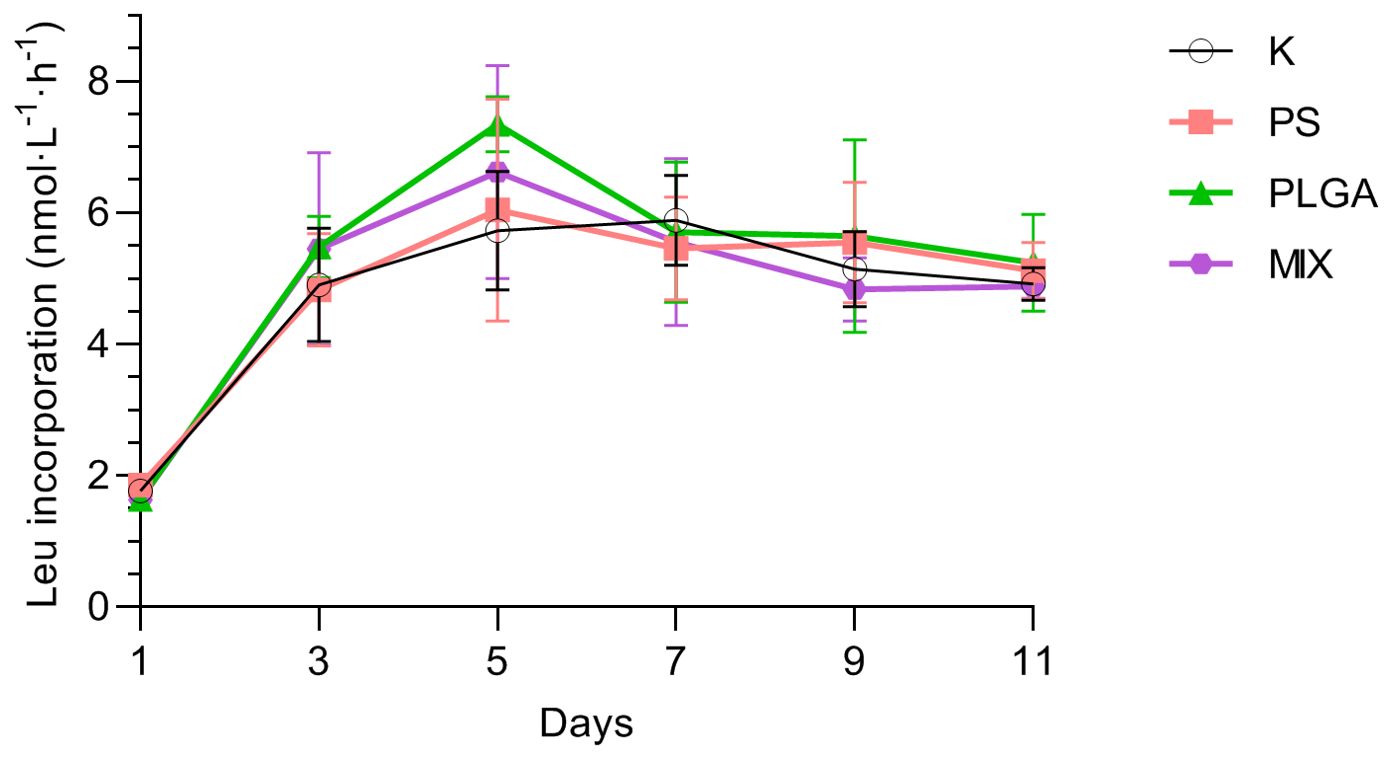
Figure 5. Total leucine incorporation (mean ± standard deviation, n=3) during the experiment. K: control; PS (polystyrene); PLGA (biodegradable MP); MIX (mixture of PS+PLGA).
4 Discussion
Microplastics are present in the Baltic Sea, with average concentrations of 0.21 ± 0.15 particles·m-3 in water (Beer et al., 2018) and 1.34 particles per individual coastal fish (Sainio et al., 2021). However, no increase in MP concentration has been proven from 1987 to 2015 (Beer et al., 2018). Other studies have found up to 10 times higher concentrations (Lusher et al., 2014) or even up to 501 particles·m-3 (Enders et al., 2015), underscoring the need for standardized methods for the determination of microplastics in water to facilitate better comparison across studies. We amended our experiment with a lower concentration of MPs than previous studies (Grigorakis and Drouillard, 2018) which may be a more suitable approach to assess the impacts of higher concentrations of MPs in seawater in the future. By the time of the experiment, we did not know the concentration of 10 µm particles in natural waters but they have been detected later in the marine environment (Ripken et al., 2021). The concentration of 20 MP beads·ml-1 is higher compared to natural concentration of 350 μm – 5.7 mm particles in seawater accounting for < 1 - 7.73·m-3 (Gewert et al., 2017) in the Baltic Sea or 0.2 - 0.9·m-3 for particles > 500μm globally (Everaert et al., 2018). MPs ranged between 3 – 20 μm in size were found in concentrations up to 16800 particles·m-3 in Los Angeles river and up to 11950 particles·m-3 on its coastline (Wiggin and Holland, 2019). Since we used particles close to the nanoplastics range, which are more abundant due to fragmentation, we assumed their concentration would be a few orders of magnitude higher. Our concentrations, although still higher than in nature in order to trigger significant effects, were markedly lower than those added in similar experiments.
The results of our comparison of the effects of conventional and biodegradable microplastics on bacteria in the Baltic Sea show a general increase in the percentage of active bacteria with the addition of MPs. However, there were no differences in bacterial abundance, composition, or leucine incorporation. This minimal impact of both conventional and biodegradable microplastics (MPs) suggests that microbial communities in the Baltic Sea may have inherent resilience to short-term MP exposure. The weak microbial response could be influenced by the availability of other organic matter sources within the mesocosms, which may reduce the bacteria’s dependence on MPs as carbon or nutrient sources. This is consistent with findings by Romera-Castillo et al. (2018), who observed that microbial activity related to MP-derived organic matter tends to be more evident in environments where MPs are one of the main carbon sources. Similar observations were made by Marchant et al. (2023), who found that even under nutrient enrichment, microplastic exposure did not significantly affect freshwater community composition or ecosystem function in semi-natural mesocosms. This aligns with findings suggesting that moderate MP levels may not disrupt microbial communities in complex environments with multiple resource inputs (Stanković et al., 2022, Yıldız et al., 2022). The Baltic Sea’s specific environmental conditions, including lower temperatures, could also constrain microbial growth, preventing a pronounced response to MPs in the short term.
In comparison with previous studies, our results align with those that have observed weak or negligible effects of MPs on microbial communities (e.g., Romera-Castillo et al., 2022). While some studies report shifts in bacterial community composition following MP exposure (e.g., Caruso et al., 2018; Gong et al., 2023), such effects were generally observed over longer durations or at higher MP concentrations. This study, with lower MP concentrations and a shorter exposure time, reflects conditions closer to those in natural settings and supports the hypothesis that moderate MP levels may not significantly disrupt microbial communities in the early stages. Contrastingly, experiments with prolonged MP exposure, such as those by Eich et al. (2015), reveal potential longer-term shifts in bacterial diversity and function, particularly under nutrient-enriched or closed-system conditions. Throughout the experiment, no microorganisms were observed to attach to the MPs. This finding is consistent with the results of a shorter, 6-day experiment conducted by Romera-Castillo et al. (2022). However, microplastics not only provide a surface for bacterial growth (Jacquin et al., 2018), but can also leach carbon, enhancing bacterial growth (Galgani et al., 2018; Romera-Castillo et al., 2018) or decreasing extracellular enzyme activity (Caruso et al., 2018). In environments where plastics are the sole carbon source, it is likely that bacteria utilize this carbon for growth as suggested by Ho et al. (2018). Nonetheless, in environments where other carbon sources are readily available, the rate of MP biodegradation may be significantly diminished. In contrast with our initial hypothesis, PLGA failed to stimulate growth nor promote changes in bacterial composition when compared to PS, despite its accelerated biodegradability (Wang et al., 2021). We expected to observe significant changes in bacterial concentrations and activity in the first 11 days of the experiment given that MPs leached carbon that could be used by bacterioplankton in a 6-day experiment (Romera-Castillo et al., 2022). Previous studies have demonstrated that biodegradable MPs enhance the uptake of exogenous carbohydrates and amino acids by bacteria after 60 days in soil experiments (Sun et al., 2022) as well as in sediment experiments amended with conventional plastics (Yao et al., 2023). This comparison underscores the need for studies across different temporal and spatial scales to accurately predict MP impacts on marine microbial ecosystems.
No clear differences in the abundance of the four phylogenetic groups were observed between the MP-amended treatments. Furthermore, the proportion of active cells in all groups was similar. The CARD-FISH probes selected for this study were chosen based on their suitability for detecting the most abundant bacterial groups in the Baltic Sea. Although the Baltic Sea is predominantly populated by Bacteroidetes, it is significantly influenced by typical freshwater phylogenetic groups, such as β-proteobacteria, whose abundance is observed to increase during the summer months (Riemann et al., 2008). However, we must consider that the four CARD-FISH probes used in this experiment targeted, on average, a 30% of the total bacteria present in the experiment, a lower percentage than what is typically recovered in the Mediterranean. The absence of differences in the bacterial composition and the number of active cells in our experiment may hide the existence of a fast-responding group of bacteria which may not have been detected with our probes.
CARD-FISH offers a robust approach for quantifying diverse bacterial groups, although it has several limitations. First, the technique is dependent on the number and specificity of probes used, which are limited to previously identified lineages. Additionally, CARD-FISH may exhibit biases in both coverage and specificity due to the pronounced seasonality of microbial communities (Alonso-Sáez et al., 2014). As a result, the taxonomic composition of targeted groups may fluctuate across seasons, potentially altering probe specificity and impacting abundance estimates. Furthermore, the accuracy of CARD-FISH can be affected by several factors, including incomplete probe coverage and specificity, variations in the amount and activity of endogenous peroxidases across different phylogenetic groups and environmental samples, challenges in detecting low-abundance or inactive community members, and difficulties in counting aggregated cells (Pernthaler et al., 2004; Amann and Fuchs, 2008).
The bacterial groups studied in our experiment have been found to be dominant in previous studies. In soil experiments with conventional MPs, Proteobacteria and Bacteroidetes were the dominant bacterial groups, with their relative abundances increasing after 90 days of incubation (Li et al., 2022). Koskinen et al. (2010) found that γ-proteobacteria were the most prevalent group, representing 53% of all PCR reads. Generally, γ-proteobacteria and α-proteobacteria have been identified as key players in primary colonization whereas Bacteroidetes stand out as a secondary colonizer (Oberbeckmann et al., 2015; Díaz-Mendoza et al., 2020). However, with time, members of the Bacteroidetes phylum have also become increasingly abundant (Lee et al., 2008). Moreover, most bacteria that are able to degrade MPs belong to the phylum Proteobacteria, and have been studied in most biodegradation experiments (Matjašič et al., 2021) as well as Firmicutes, particularly the genus Bacillus (Muthukumar and Veerappapillai, 2015).
Gong et al. (2023) found that Proteobacteria (15.3–24.1%) were dominant in soil amended with 5 μm polystyrene beads, significantly increasing its abundance compared to a control. A study conducted in coastal sediments revealed that, after 14 days, communities were dominated by the genera Arcobacter and Colwellia (γ-proteobacteria) which collectively accounted for 84%–93% of sequences in the presence of polyethylene (Harrison et al., 2014).
BONCAT-FISH did not provide any significant differences in abundance or composition of the bacterial community, nor in the activity within those bacteria groups in the presence of biodegradable or conventional MPs. Nonetheless, significant differences were observed in the colonization over extended periods (6 weeks) of non-biodegradable and biodegradable plastics in terms of marine bacterial abundance, diversity and activity (Eich et al., 2015; Dussud et al., 2018). Similar to our findings, the presence of MPs did not have effects on the composition and abundance of the bacterial composition. However, it did influence the activity of these bacteria, as evidenced by a reduction in microbial extracellular enzymatic activities (Caruso et al., 2018), specifically those which are responsible for polymer degradation (Sala and Güde, 2004).
It is noteworthy to mention that biodegradable plastics can only be considered so under specific conditions, as demonstrated in both controlled and uncontrolled environments (Kim et al., 2023). Biodegradable MPs have been observed not only to increase bacterial diversity but also to enrich environmentally friendly taxa (Hu et al., 2022), while conventional MPs have been shown to reduce biodiversity (Ogonowski et al., 2018; Xiang et al., 2024) and likely promote the growth of harmful taxa while hindering the recovery of community richness (Li et al., 2022; Li and Xiao, 2023). Additionally, the concentration of conventional MPs also seems to play an important role in diversity, with high concentrations enriching dominant bacteria and decreasing diversity indexes, whilst low MP concentrations seemed to have the opposite effect (Yuan et al., 2023).
Our findings hold important implications for environmental management and plastic pollution policies. The weak microbial response to MPs observed here suggests that, in the short term, the current levels of MPs in the Baltic Sea may not pose a substantial risk to microbial communities. However, as highlighted by Piehl et al. (2021) and Schernewski et al. (2020), the accumulation of MPs over time, particularly in enclosed or semi-enclosed seas like the Baltic, could alter microbial functions critical for ecosystem health. Such changes may affect processes like organic matter decomposition and nutrient cycling, which are essential for maintaining marine ecosystem balance. Given the regulatory challenges associated with microplastic pollution, Da Costa et al. (2020) emphasize the importance of effective legislative frameworks that focus not only on plastic waste reduction but also on the mitigation of its long-term ecological impacts. Understanding the microbial interactions with MPs is therefore essential to inform strategies for plastic waste management and support the development of policies aimed at minimizing plastic pollution in marine ecosystems.
5 Conclusions
The addition of microplastics showed a weak increase in the percentage of active cells compared to the control. This could lead to an increase in the concentration of those bacterial groups that are able to degrade plastics. However, this was not reflected by an increase in the activity of the specific groups targeted with the CARD-FISH probes used in this experiment. No significant differences were observed between the composition nor activity of the bacterial community in the presence of conventional or biodegradable MPs. The weak effect of MPs observed might be due to the low concentration of the additions made (20 MP beads·ml-1) and the short duration of the experiment. This also suggests that the low MP concentrations that are found in nature do not have any clear effects in the first stages of MP addition. Many studies are carried out with unrealistically high MP concentrations, so more experiments with MP additions closer to natural conditions are needed.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
UL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. LS: Writing – review & editing. IF: Methodology, Writing – review & editing. FP: Writing – review & editing. JP: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. HK: Data curation, Formal analysis, Methodology, Writing – review & editing. ML: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. MS: Conceptualization, Data curation, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project (PLASMIC: Effects of conventional and biodegradable microPLAStics on marine MICrobial communities; PIs: U. Lora and M.M. Sala) was supported by the EU H2020-INFRAIA project [grant number 731065] AQUACOSM: Network of Leading European AQUAtic MesoCOSM Facilities Connecting Mountains to Oceans from the Arctic to the Mediterranean - funded by the European Commission. Additional funding was partly provided by the EU HORIZON-CL6-2023-ZEROPOLLUTION-01 project ONE-BLUE (Ref No. 101134929)" and by the European Union - NextGeneration EU - as part of the MITECO program for the Spanish Recovery, Transformation and Resilience Plan (Recovery and Resilience Facility of the European Union established by the Regulation (EU) 2020/2094), and was entrusted to CSIC, AZTI, SOCIB, and the universities of Vigo and Cadiz.
Acknowledgments
We thank Marta Sebastián for her contributions and suggestions on writing this paper. We also thank all the researchers in Syke that gave a helping hand in sampling and technical assistance making this project possible. And thanks to Nicole Peto for her selfless advice in helping to write and check this work. The study utilized Syke Marine Research Laboratory infrastructures as a part of the national FINMARI consortium.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acinas S. G., Ferrera I., Sarmento H., Díez-Vives C., Forn I., Ruiz-González C., et al. (2014). Validation of a new catalysed reporter deposition-fluorescence in situ hybridization probe for the accurate quantification of marine Bacteroidetes populations. Environ. Microbiol. 17, 3557–3569. doi: 10.1111/1462-2920.12517
Alonso-Sáez L., Zeder M., Harding T., Pernthaler J., Lovejoy C., Bertilsson S., et al. (2014). Winter bloom of a rare betaproteobacterium in the Arctic Ocean. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00425
Amann R., Fuchs B. M. (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6, 339–348. doi: 10.1038/nrmicro1888
Andrady A. L. (2011). Microplastics in the marine environment. Mar. pollut. Bull. 62, 1596–1605. doi: 10.1016/j.marpolbul.2011.05.030
Auta H. S., Emenike C. U., Fauziah S. H. (2017). Distribution and importance of microplastics in the marine environment a review of the sources, fate, effects, and potential solutions. Environ. Int. 102, 165–176. doi: 10.1016/j.envint.2017.02.013
Barnes D. K. A., Galgani F., Thompson R. C., Barlaz M. (2009). Accumulation and fragmentation of plastic debris in global environments. Phil. Trans. R. Soc B364, 1985–1998. doi: 10.1098/rstb.2008.0205
Beer S., Garm A., Huwer B., Dierking J., Nielsen T. G. (2018). No increase in marine microplastic concentration over the last three decades – A case study from the Baltic Sea. Sci. Total Environ. 621, 1272–1279. doi: 10.1016/j.scitotenv.2017.10.101
Birnstiel S., Sebastián M., Romera-Castillo C. (2022). Structure and activity of marine bacterial communities responding to plastic leachates. Sci. Total Environ. 834, 155264. doi: 10.1016/j.scitotenv.2022.155264
Buongiorno J. (2018). Microbial communities and biogeochemistry in marine sediments of the Baltic Sea and the high Arctic, Svalbard (University of Tennessee). Available online at: https://trace.tennessee.edu/utk_graddiss/5268/.
Caruso G., Pedà C., Cappello S., Leonardi M., La Ferla R., Lo Giudice A., et al. (2018). Effects of microplastics on trophic parameters, abundance and metabolic activities of seawater and fish gut bacteria in mesocosm conditions. Environ. Sci. pollut. Res. 25, 30067–30083. doi: 10.1007/s11356-018-2926-x
Cózar A., Echevarria F., Gonzalez-Gordillo J. I., Irigoien X., Ubeda B., Hernández-León S., et al. (2014). Plastic debris in the open ocean. PNAS 111, 10239–10244. doi: 10.1073/pnas.1314705111
Da Costa J. P., Mouneyrac C., Costa M., Duarte A. C., Rocha-Santos T. (2020). The role of legislation, regulatory initiatives and guidelines on the control of plastic pollution. Front. Environ. Sci. 8. doi: 10.3389/fenvs.2020.00104
Debroas D., Mone A., Ter Halle A. (2017). Plastics in the North Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci. Total Environ. 599-600, 1222–1232. doi: 10.1016/j.scitotenv.2017.05.059
De Tender C. A., Devriese L. I., Haegeman A., Maes S., Ruttink T., Dawyndt P. (2015). Bacterial community profiling of plastic litter in the Belgian part of the North Sea. Environ. Sci. Technol. 49, 9629–9638. doi: 10.1021/acs.est.5b01093
Díaz-Mendoza C., Mouthon-Bello J., Pérez-Herrera N. L., Escobar-Díaz S. M. (2020). Plastics and microplastics, effects on marine coastal areas: a review. Environ. Sci. pollut. Res. 27, 39913–39922. doi: 10.1007/s11356-020-10394-y
Dussud C., Hudec C., George M., Fabre P., Higgs P., Bruzaud S., et al. (2018). Colonization of non-biodegradable and biodegradable plastics by marine microorganisms. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01571
Eich A., Mildenberger T., Laforsch C., Weber M. (2015). Biofilm and diatom succession on polyethylene (pe) and biodegradable plastic bags in two marine habitats: early signs of degradation in the pelagic and benthic zone? PloS One 10, e0137201. doi: 10.1371/journal.pone.0137201
Enders K., Lenz R., Stedmon C. A., Nielsen T. G. (2015). Abundance, size and polymer composition of marine microplastics ≥ 10 μm in the Atlantic Ocean and their modelled vertical distribution. Mar. pollut. Bull. 100, 70–81. doi: 10.1016/j.marpolbul.2015.09.027
Eronen-Rasimus E. L., Nakki P. P., Kaartokallio H. P. (2022). Degradation rates and bacterial community compositions vary among commonly used bioplastic materials in a brackish marine environment. Enviro. Sci. Technol. 56, 15760–15769. doi: 10.1021/acs.est.2c06280
Everaert G., Van Cauwenberghe L., De Rijcke M., Koelmans A. A., Mees J., Vandegehuchte M., et al. (2018). Risk assessment of microplastics in the ocean: Modelling approach and first conclusions. Environ. pollut 242, 1930-1938. doi: 10.1016/j.envpol.2018.07.069
Fotopoulou K. N., Karapanagioti H. K. (2012). Surface properties of beached plastic pellets. Mar. Environ. Res. 81, 70–77. doi: 10.1016/j.marenvres.2012.08.010
Fuhrman J., Azam F. (1982). Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66, 109–120. doi: 10.1007/BF00397184
Galgani L., Engel A., Rossi C., Donati A., Loiselle S. A. (2018). Polystyrene microplastics increase microbial release of marine chromophoric dissolved organic matter in microcosm experiments. Sci. Rep. 8, 14635. doi: 10.1038/s41598-018-32805-4
Gewert B., Ogonowski M., Barth A., MacLeod M. (2017). Abundance and composition of near surface microplastics and plastic debris in the Stockholm Archipelago, Baltic Sea. Mar. pollut. Bull. 120, 292–302. doi: 10.1016/j.marpolbul.2017.04.062
Gewert B., Plassmann M. M., MacLeod M. (2015). Pathways for degradation of plastic polymers floating in the marine environment. Environ. Sci.: processes impacts 17, 1513–1521. doi: 10.1039/C5EM00207A
Gong X., Ge Z., Ma Z., Li Y., Huang D., Zhang J. (2023). Effect of different size microplastic particles on the construction of algal-bacterial biofilms and microbial communities. J. Environ. Manage. 343, 118246. doi: 10.1016/j.jenvman.2023.118246
Grigorakis S., Drouillard K. G. (2018). Effect of microplastic amendment to food on diet assimilation efficiencies of PCBs by fish. Environ. Sci. Technol. 52, 10796–10802. doi: 10.1021/acs.est.8b02497
Gunaalan K., Nielsen T. G., Rodríguez Torres R., Lorenz C., Vianello A., Andersen C. A., et al. (2023). Is zooplankton an entry point of microplastics into the marine food web? Environ. Sci. Technol.; 57, 11643–11655. doi: 10.1021/acs.est.3c02575
Haider T. P., Völker C., Kramm J., Landfester K., Wurm F. R. (2019). Plastics of the future? the impact of biodegradable polymers on the environment and on society. Angew. Chem. Int. Ed. 58, 50. doi: 10.1002/anie.201805766
Harrison J. P., Schratzberger M., Sapp M., Osborn A. M. (2014). Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 14 (1), 232. doi: 10.1186/s12866-014-0232-4
Hatzenpichler R., Scheller S., Tavormina P. L., Babin B. M., Tirrell D. A., Orphan V. J. (2014). In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ. microbiol.. 16 (8), 2568-2590. doi: 10.1111/1462-2920.12436
Hatzenpichler R., Orphan V. J. (2015). “Detection of protein-synthesizing microorganisms in the environment via bioorthogonal noncanonical amino acid tagging (BONCAT),” in Hydrocarbon and Lipid Microbiology Protocols. Springer Protocols Handbooks. Eds. McGenity T. J., Timmis K. N., Nogales B. (Springer, Berlin, Heidelberg). doi: 10.1007/8623_2015_61
Ho B. T., Roberts T. K., Lucas S. (2018). An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Crit. Rev. Biotechnol. 38, 308–320. doi: 10.1080/07388551.2017.1355293
Hu X., Gu H., Wang Y., Liu J., Yu Z., Li Y., et al. (2022). Succession of soil bacterial communities and network patterns in response to conventional and biodegradable microplastics: A microcosmic study in Mollisol. J. Hazard. Mater. 436, 129218. doi: 10.1016/j.jhazmat.2022.129218
Jacquin J., Cheng J., Odobel C., Conan P., Pujo-pay M., Meistertzheim A., et al. (2018). Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the ‘plastisphere’. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00865
Kaplan D. L., Hartenstein R., Sutter J. (1979). Biodegradation of polystyrene, poly(methyl methacrylate), and phenol formaldehyde. Appl. Environ. Microbiol. 38, 551–553. doi: 10.1128/aem.38.3.551-553.1979
Kim M. S., Chang H., Zheng L., Yan Q., Pfleger B. F., Klier J., et al. (2023). A review of biodegradable plastics: chemistry, applications, properties, and future research needs. Chem. Rev. 123, 9915–9939. doi: 10.1021/acs.chemrev.2c00876
Koskinen K., Hultman J., Paulin L., Auvinen P., Kankaanpää H. (2010). Spatially differing bacterial communities in water columns of the northern Baltic Sea. FEMS Microbiol. Ecol. 75, 99–110. doi: 10.1111/j.1574-6941.2010.00987.x
Lee J.-W., Nam J.-H., Kim Y.-H., Lee K.-H., Lee D.-H. (2008). Bacterial communities in the initial stage of marine biofilm formation on artificial surfaces. J. Microbiol. 46, 174–182. doi: 10.1007/s12275-008-0032-3
Leizeaga A., Estrany M., Forn I., Sebastián M. (2017). Using click-chemistry for visualizing in situ changes of translational activity in planktonic marine bacteria. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02360
Li W., Wang Z., Li W., Li Z. (2022). Impacts of microplastics addition on sediment environmental properties, enzymatic activities and bacterial diversity. Chemosphere 307, 135836. doi: 10.1016/j.chemosphere.2022.135836
Li W., Xiao Y. (2023). Microplastics increase soil microbial network complexity and trigger diversity-driven community assembly. Environ. pollut. 333, 122095. doi: 10.1016/j.envpol.2023.122095
Lusher A. L., Burke A., O’Connor I., Officer R. (2014). Microplastic pollution in the Northeast Atlantic Ocean: validated and opportunistic sampling. Mar. pollut. Bull. 88, 325–333. doi: 10.1016/j.marpolbul.2014.08.023
Manz W., Amann R., Ludwig W., Wagner M., Schleifer K. H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15, 593–600. doi: 10.1016/S0723-2020(11)80121-9
Marchant D. J., Rodríguez A. M., Francelle P., Jones J. I., Kratina P. (2023). Contrasting the effects of microplastic types, concentrations and nutrient enrichment on freshwater communities and ecosystem functioning. Ecotoxicol. Environ. Saf. 255, 114834. doi: 10.1016/j.ecoenv.2023.114834
Matjašič T., Simčič T., Medvešček N., Bajt O., Dreo T., Mori N. (2021). Critical evaluation of biodegradation studies on synthetic plastics through a systematic literature review. Sci. Total Environ. 752, 141959. doi: 10.1016/j.scitotenv.2020.141959
Miri S., Saini R., Davoodi S. M., Pulicharla R., Brar S. K., Magdouli S. (2022). Biodegradation of microplastics: Better late than never. Chemosphere 286, 131670. doi: 10.1016/j.chemosphere.2021.131670
Muthukumar A., Veerappapillai S. (2015). Biodegradation of plastics – A brief review. Int. J. Pharm. Sci. Res. 31, 204–209. Available at: https://www.researchgate.net/publication/282050386_Biodegradation_of_plastics_-_A_brief_review. (accessed June 20, 2024)
Neef A. (1997). Anwendung der in situ-einzelzell-identifizierung von bakterien zur populationsanalyse in komplexen mikrobiellen biozönosen. Munich, Germany: Univ. Munich.
Oberbeckmann S., Labrenz M. (2019). Marine microbial assemblages on microplastics: diversity, adaptation, and role in degradation. Annu. Rev. Mar. Sci. 12 (1), 209-232. doi: 10.1146/annurev-marine-010419-010633
Oberbeckmann S., Loeder M. G. J., Labrenz M. (2015). Marine microplastic associated biofilms - a review. Environ. Chem. 12, 551–562. doi: 10.1071/EN15069
Ogonowski M., Motiei A., Ininbergs K., Hell E., Gerdes Z., Udekwu K. I., et al. (2018). Evidence for selective bacterial community structuring on microplastics. Environ. Microbiol. 20, 2796–2808. doi: 10.1111/1462-2920.14120
Otake Y., Kobayashi T., Asabe H., Murakami N., Ono K. (1995). Biodegradation of low-density polyethylene, polystyrene, polyvinyl chloride, and urea formaldehyde resin buried under soil for over 32 years. J. Appl. Polym. Sci. 56, 1789–1796. doi: 10.1002/app.1995.070561309
Pernthaler A., Pernthaler J., Amann R. (2004). Sensitive multi-color fluorescence in situ hibridization for the identification of environmental microorganisms. Mol. Microb. Ecol. Man. 711–726. doi: 10.1128/aem.68.6.3094-3101.2002
Piehl S., Hauk R., Robbe E., Richter B., Kachholz F., Schilling J., et al. (2021). Combined approaches to predict microplastic emissions within an urbanized estuary (Warnow, Southwestern Baltic Sea). Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.616765
Porter K. G., Feig Y. S. (1980). The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25, 943–948. doi: 10.4319/lo.1980.25.5.0943
Reichelt S., Gorokhova E. (2020). Micro-and nanoplastic exposure effects in microalgae: a meta-analysis of standard growth inhibition tests. Front. Environ. Sci. 8. doi: 10.3389/fenvs.2020.00131
Riemann L., Leitet C., Pommier T., Simu K., Holmfeldt K., Larsson U., et al. (2008). The native bacterioplankton community in the central Baltic Sea is influenced by freshwater bacterial species. Appl. Environ. Microbiol. 74, 503–515. doi: 10.1128/AEM.01983-07
Ripken C., Kotsifaki D. G., Nic Chormaic S. (2021). Analysis of small microplastics in coastal surface water samples of the subtropical island of Okinawa, Japan. Sci. Total Environ. 760, 143927. doi: 10.1016/j.scitotenv.2020.143927
Romera-Castillo C., Birnstiel S., Álvarez-Salgado X. A., Sebastián M. (2022). Aged plastic leaching of dissolved organic matter is two orders of magnitude higher than virgin plastic leading to a strong uplift in marine microbial activity. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.861557
Romera-Castillo C., Pinto M., Langer T. M., Álvarez-Salgado X. A., Herndl G. J. (2018). Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat. Commun. 9, 1430. doi: 10.1038/s41467-018-03798-5
Sainio E., Lehtiniemi M., Setälä O. (2021). Microplastic ingestion by small coastal fish in the northern Baltic Sea, Finland. Mar. pollut. Bull. 172, 112814. doi: 10.1016/j.marpolbul.2021.112814
Sala M. M., Güde H. (2004). Ectoenzymatic activities and heterotrophic bacteria decomposing detritus. Arch. Hydrobiol. 160, 289–304. doi: 10.1127/0003-9136/2004/0160-0289
Schernewski G., Radtke H., Hauk R., Baresel C., Olshammar M., Osinski R., et al. (2020). Transport and behavior of microplastics emissions from urban sources in the Baltic Sea. Front. Environ. Sci. 8. doi: 10.3389/fenvs.2020.579361
Sebastián M., Estrany M., Ruiz-González C., Forn I., Sala M. M., Gasol J. M., et al. (2019). High growth potential of long-term starved deep ocean opportunistic heterotrophic bacteria. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00760
Smith D. C., Azam F. (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6, 107–114.
Stanković J., Milošević D., Jovanović B., Savić-Zdravković D., Petrović A., Raković M., et al. (2022). In situ effects of a microplastic mixture on the community structure of benthic macroinvertebrates in a freshwater pond. Environ. Toxicol. Chem. 41, 888–895. doi: 10.1002/etc.5119
Sun Y., Duan C., Cao N., Ding C., Huang Y., Wang J. (2022). Biodegradable and conventional microplastics exhibit distinct microbiome, functionality, and metabolome changes in soil. J. Hazard. Mater. 424, 127282. doi: 10.1016/j.jhazmat.2021.127282
Thakur B., Singh J., Singh J., Angmo D., Vig A. P. (2023). Biodegradation of different types of microplastics: Molecular mechanism and degradation efficiency. Sci. Total Environ. 877, 162912. doi: 10.1016/j.scitotenv.2023.162912
Thompson R. C., Swan S. H., Moore C. J., vom Saal F. S. (2009). Our plastic age. Philos. Trans. R. Soc Lond. B. Biol. Sci. 364, 1973–1976. doi: 10.1098/rstb.2009.0054
Torena P., Alvarez-Cuenca M., Reza M. (2021). Biodegradation of polyethylene terephthalate microplastics by bacterial communities from activated sludge. Can. J. Chem. Eng.; 99, S69–S82. doi: 10.1002/cjce.24015
Urbanek A. K., Rymowicz W., Mirończuk A. M. (2018). Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 102, 7669–7678. doi: 10.1007/s00253-018-9195-y
Van Sebille E., Wilcox C., Lebreton L., Maximenko N., Hardesty B. D., van Franeker J. A., et al. (2015). A global inventory of small floating plastic debris. Environ. Res. Lett. 10, 124006. doi: 10.1088/1748-9326/10/12/124006
Vesamäki J. S., Nissinen R., Kainz M. J., Pilecky M., Tiirola M., Taipale S. J. (2022). Decomposition rate and biochemical fate of carbon from natural polymers and microplastics in boreal lakes. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1041242
Wang G.-X., Huang D., Ji J.-H., Völker C., Wurm F. R. (2021). Seawater-degradable polymers—fighting the marine plastic pollution. Adv. Sci. 8, 2001121. doi: 10.1002/advs.202001121
Wiggin K. J., Holland E. B. (2019). Validation and application of cost and time effective methods for the detection of 3–500 μm sized microplastics in the urban marine and estuarine environments surrounding Long Beach, California. Mar. pollut. Bull. 143, 152–162. doi: 10.1016/j.marpolbul.2019.03.060
Xiang P., Liao W., Xiong Z., Xiao W., Luo Y., Peng L., et al. (2024). Effects of polystyrene microplastics on the agronomic traits and rhizosphere soil microbial community of highland barley. Sci. Total Environ. 907, 167986. doi: 10.1016/j.scitotenv.2023.167986
Yao Y., Zhao J., Adyel T. M., Liu Y., Liu J., Miao L. (2023). Sediment bacterial and fungal communities exhibit distinct responses to microplastic types and sizes in Taihu lake. Environ. pollut. 320, 121092. doi: 10.1016/j.envpol.2023.121092
Yıldız D., Yalçın G., Jovanović B., Boukal D. S., Vebrová L., Riha D., et al. (2022). Effects of a microplastic mixture differ across trophic levels and taxa in a freshwater food web: In situ mesocosm experiment. Sci. Total. Environ. 836, 155407. doi: 10.1016/j.scitotenv.2022.155407
Yuan Y., Zu M., Li R., Zuo J., Tao J. (2023). Soil properties, microbial diversity, and changes in the functionality of saline-alkali soil are driven by microplastics. J. Hazard. Mater. 446, 130712. doi: 10.1016/j.jhazmat.2022.130712
Keywords: microplastics, CARD-FISH, BONCAT, mesocosm experiment, conventional plastic, biodegradable plastic
Citation: Lora U, Schenone L, Forn I, Peters F, Piiparinen J, Kaartokallio H, Lehtiniemi M and Sala MM (2024) Weak effects of conventional and biodegradable microplastics on marine microbial communities. Front. Mar. Sci. 11:1502825. doi: 10.3389/fmars.2024.1502825
Received: 27 September 2024; Accepted: 25 November 2024;
Published: 13 December 2024.
Edited by:
Gordon T. Taylor, Stony Brook University, United StatesReviewed by:
Sedat Gundogdu, Çukurova University, TürkiyeAditi Pandit, University of South Bohemia in České Budějovice, Czechia
Copyright © 2024 Lora, Schenone, Forn, Peters, Piiparinen, Kaartokallio, Lehtiniemi and Sala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulises Lora, dWxvcmFAaWNtLmNzaWMuZXM=
 Ulises Lora
Ulises Lora Luca Schenone
Luca Schenone Irene Forn
Irene Forn Francesc Peters
Francesc Peters Jonna Piiparinen
Jonna Piiparinen Hermanni Kaartokallio
Hermanni Kaartokallio Maiju Lehtiniemi
Maiju Lehtiniemi Maria Montserrat Sala
Maria Montserrat Sala