Spiralia is arguably the most diverse of the three superphyla that include extant bilaterally symmetrical animal species. In addition to their remarkable morphological disparity, the approximately 14 phyla (Figure 1, Liao et al., 2023) that constitute the Spiralia play a crucial role in various ecosystems and are of significant cultural and economic importance to humans globally.
Figure 1
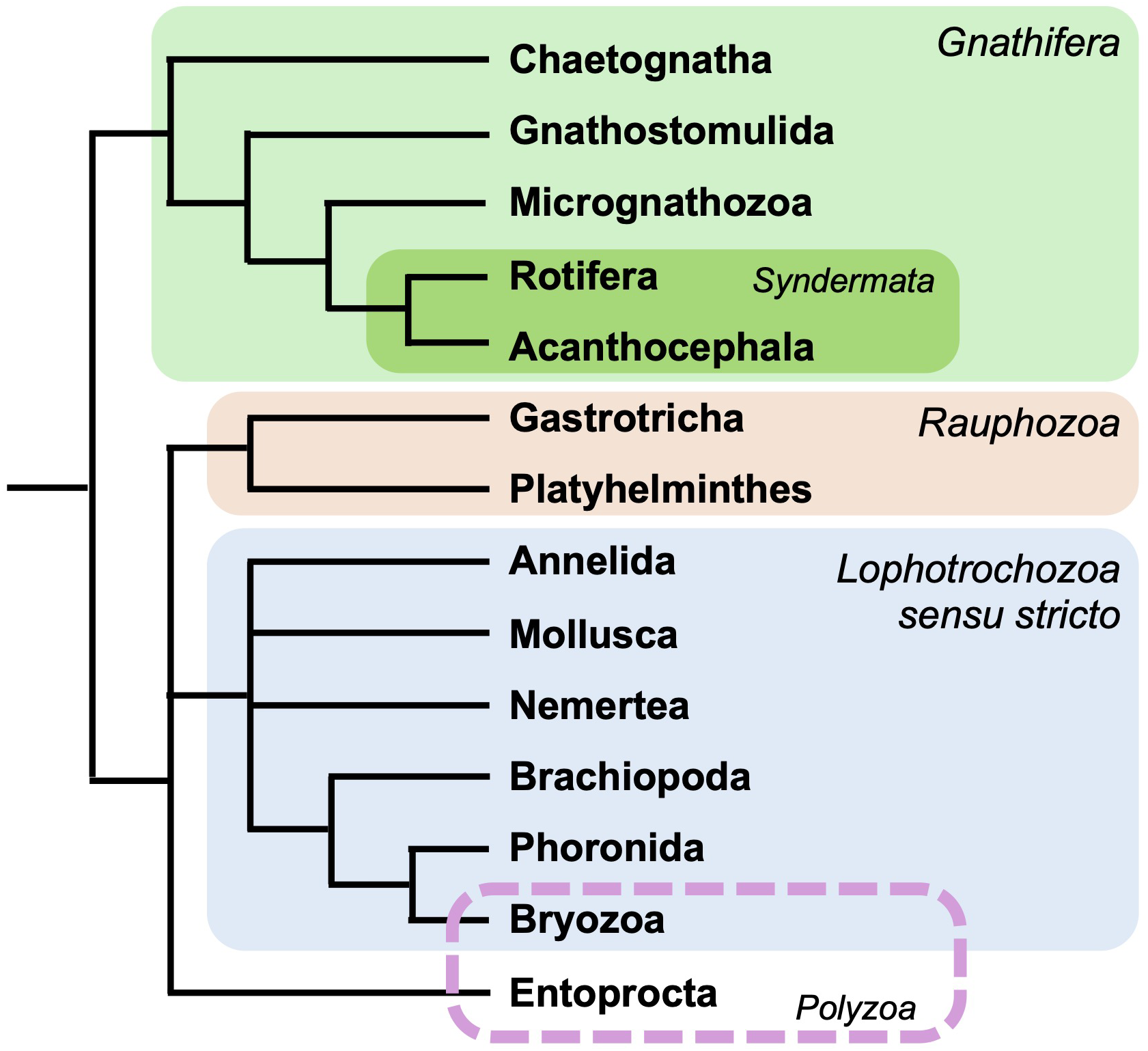
Cladogram depicting a consensus of spiralian interrelationships, based on Liao et al., 2023. Three phyla are excluded (Orthonectida, Dicyemida, and Cyclophora).
Despite their ubiquity and significance, the study of spiralians has lagged behind that of other phyla, particularly those containing well-established model organisms. This has led to significant gaps in our understanding of their biology across various levels. Fundamental resources, such as genome sequences, are sparse within this clade, and spiralians are under-investigated compared to vertebrates and insects. Coupled with the genetic and morphological novelties common among spiralians, our ability to fully comprehend the origins of their intriguing traits is still developing.
The multi-faceted challenges posed by climate change highlight the growing problem of our limited understanding of spiralian biology. Within Spiralia, numerous species play crucial ecological roles and are now experiencing rapid climatic shifts. Many of these species rely on finely tuned chemical processes, such as those involved in biomineralization (e.g., shell formation), making them particularly vulnerable to temperature-related impacts. Additionally, issues like ocean acidification have especially severe effects on spiralians – consequences of which we are currently ill-prepared to address.
Fortunately, the emergence of third-generation sequencing platforms, such as Oxford Nanopore and PacBio systems, has transformed our ability to generate genomic data. This technological leap has significantly accelerated data production by consortia like the Earth BioGenome Project (Lewin et al., 2022), but also enabled smaller laboratories to investigate their model species of interest. Coupled with the growing accessibility and decreasing cost of transcriptomic sequencing techniques, both in the wet lab and at the computational level, studying the impact of climate change on spiralian organisms had never been easier.
This Research Topic highlights several global efforts where laboratories have successfully generated high-quality genomic and transcriptomic datasets to understand the biology of spiralian species. For instance, Daniels et al., combined PacBio and Oxford Nanopore long-read technologies with Illumina short-read sequencing to create a highly contiguous and well-annotated genomic resource for the coastal gastropod, Kelletia kelletii. This species plays a crucial role as a fisheries and ecological resource. The genomic and transcriptomic resources presented here will enable future studies on its distribution and response to climatic pressures.
These new tools can also be applied to study individual traits relevant to climate change. Achilleos et al. investigated the molecular mechanisms underlying biomineralization in the bryozoan species, Cellaria immersa. This is a crucial but under-explored feature of many spiralian organisms, which could be directly affected by shifts in temperature, pH, and ocean chemistry. Such changes could lead to the loss of the three-dimensional structure of benthic ecosystems, with cascading effects on food webs. This is an example of how transcriptomic tools provide rapid insights into the molecular basis of biomineralization in non-model spiralian organisms, potentially aiding in mitigating of climate change impacts.
In Zúñiga-Soto et al., similar transcriptomic tools, combined with histology, were used to investigate biomineralization in the oyster Crassostrea gigas under low pH conditions. This study offers a direct glimpse into the environments organisms will soon face globally and their potential responses to these changes. Differences in the mantle secretome and morphology observed under these conditions suggest that organisms may adapt to low pH, illustrating how these changes can be studied. However, moving forward, we must recognize that these changes in adaptations may be suboptimal, potentially leading to downstream issues stemming from alterations in the biomineralization processes.
In addition to genomic and transcriptomic tools, the availability of new, comprehensive resources enables the study of gene family evolution through comparative genomics. Edsinger and Moroz utilized diverse resources to investigate lineage-specific gene family expansions, focusing on heat-sensing TRPM channels in molluscs. Their findings suggest that local segmental duplications may drive adaptation to challenging environments, offering potential targets for species management. This work demonstrates how recent advances in sequencing technologies have generated new hypotheses about the mechanisms underlying adaptation in spiralians and provided a robust framework for testing these hypotheses.
Altogether, the latest sequencing technologies are a powerful tool in bridging gaps in our understanding of spiralian biology and the potential impacts of climate change. We hope that the “Spiralian Genomics in a Changing World” Research Topic serves as a springboard for a broader range of investigations into this fascinating clade, offering valuable tools to address some of the most pressing challenges that face our planet.
Statements
Author contributions
NJK: Writing – original draft, Writing – review & editing. FA: Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Lewin H. A. Richards S. Lieberman Aiden E. Allende M. L. Archibald J. M. Bálint M. et al . (2022). The earth bioGenome project 2020: starting the clock. Proc. Natl. Acad. Sci. United States America119, e2115635118. doi: 10.1073/pnas.2115635118
2
Liao I. J. Lu T. M. Chen M. E. Luo Y. J. (2023). Spiralian genomics and the evolution of animal genome architecture. Briefings Funct. Genomics22, 498–508. doi: 10.1093/bfgp/elad029
Summary
Keywords
spiralians, lophotrochozoa, genomics, transcriptomics, climate change, ecology, development, mitigation
Citation
Kenny NJ and Aguilera F (2024) Editorial: Spiralian genomics in a changing world. Front. Mar. Sci. 11:1501345. doi: 10.3389/fmars.2024.1501345
Received
24 September 2024
Accepted
03 October 2024
Published
17 October 2024
Volume
11 - 2024
Edited and reviewed by
Ciro Rico, Spanish National Research Council (CSIC), Spain
Updates
Copyright
© 2024 Kenny and Aguilera.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathan J. Kenny, nathan.kenny@otago.ac.nz; Felipe Aguilera, faguilera@udec.cl
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.