- 1Graduate School of Science and Engineering, University of the Ryukyus, Okinawa, Japan
- 2Department of Chemistry, Biology and Marine Science, Faculty of Science, University of the Ryukyus, Okinawa, Japan
Stony corals possess major components of the circadian system, which oscillate in response to light-dark cycles in aquatic environments. However, the extent to which the circadian system influences physiological processes remains unknown. This study investigated the role of circadian genes (cry1, cry2, cry3, clock, cycle, and slmb) in modulating the transcription of photoreceptor (opsin1, opsin2, and opsin3), calcification/metabolism-related (ca, pmca, sglt, and ppp1r), and homeostasis/stress-related (hif1α, egln, sod, and hsp70) genes in Acropora digitifera, a stony coral inhabiting shallow water. Nubbins of A. digitifera were reared under light-dark (LD) and constant darkness (DD) conditions and sampled at 4-h intervals. Quantitative PCR analysis revealed that cry1, cry2, cry3, and clock expression increased during the daytime under LD conditions and attenuated during the subjective daytime under DD conditions, suggesting that these genes are light-responsive. In contrast, cycle and slmb exhibited similar expression profiles with increases during the daytime/subjective daytime under both LD and DD conditions, implying robust roles in the circadian system. The abundance of opsin1 showed minimal change under LD and DD conditions, whereas the abundances of opsin2 and opsin3 increased during daytime/subjective daytime under both conditions, indicating circadian regulation. Some genes tested by qPCR significantly fluctuated with light (pmca, sglt, ppp1r, egln, sod, and hsp70) and time (ca, pmca, sglt, ppp1r, hif1α, egln, sod, and sod). Principal component analysis revealed significant correlations of circadian genes (cycle or slmb) with calcification/metabolism-related (pmca), oxygen homeostasis (hif1α), and stress indicator (sod) genes under both LD and DD conditions. Therefore, some physiological responses in A. digitifera exhibit daily changes and are partially regulated by the circadian system.
1 Introduction
Corals exhibit diurnal rhythmicity in their behavioral and physiological responses, including calcification and skeleton production (Moya et al., 2006; Sorek et al., 2014; Gutner-Hoch et al., 2016; Neder et al., 2022), tentacle movement (Levy et al., 2001, 2006b), metabolism (Bertucci et al., 2015; Hemond and Vollmer, 2015; Ruiz-Jones and Palumbi, 2015), and immunity (Rinsky et al., 2022; Gong et al., 2023). Rhythmic changes in light likely play a major role in oscillating and entraining these responses. Thus far, the involvement of the circadian system in these responses remains unclear, although researchers have identified some circadian genes in corals (Vize, 2009; Brady et al., 2011).
The circadian network in mammals comprises a central oscillator in the suprachiasmatic nucleus (SCN) of the hypothalamus and peripheral oscillators in all tissues. Photoreceptors in the retina perceive changes in light, and signals are conveyed to the SCN and then to the peripheral tissues via neural and endocrine pathways to synchronize and entrain the physiological responses in the peripheral tissues (Mazzoccoli et al., 2012; Nakao et al., 2015). A negative feedback loop regulates the circadian system: clock and bmal1 heterodimers activate transcription of per and cry; PER and CRY proteins then inhibit their own expression by repressing clock/bmal1 activity (Zhang and Kay, 2010; Takahashi, 2017). This loop, in conjunction with post-transcriptional modifications (Torres et al., 2018), generates approximately 24-h oscillations of clock protein levels and activity (Nakao et al., 2015).
Studies have examined the circadian system of stony corals (Vize, 2009; Brady et al., 2011; Hoadley et al., 2011; Levy et al., 2011): circadian oscillation was observed in 24 genes, including transcription factors, nuclear translocators, membrane receptors, kinases, cAMP pathway components, and ubiquitin protein-ligase in Acropora millepora (Vize, 2009); rhythmic expression patterns were detected in nine circadian genes including clock, cry2, cycle, and eyes-absent in A. millepora (Brady et al., 2011); and the expression profiles of cry1, cry2, clock, and cycle in Favia fragum were reported (Hoadley et al., 2011). Therefore, corals likely possess the general components of the circadian system with daily oscillations (Levy et al., 2007, 2011; Vize, 2009; Reitzel et al., 2010; Brady et al., 2011; Hoadley et al., 2011; Sorek and Levy, 2012; Sorek et al., 2014). However, the functional mechanisms by which these components entrain physiological responses in corals remain unclear.
This study investigated the integrated roles of the daily and circadian system in regulating physiological processes in A. digitifera, a stony coral inhabiting shallow waters. We examined circadian genes, including five transcription factors (cry1, cry2, cry3, clock, and cycle) and one ubiquitin-proteasome (slmb, Vize, 2009). We also assessed the daily fluctuations of three membrane photoreceptors (opsin1, opsin2, and opsin3), which belong to the G-protein coupled receptor (GPCR) family (Nathans and Hogness, 1983; Porter et al., 2011). Additionally, we examined daily and circadian fluctuations of two calcification-related genes (ca and pmca), two sugar metabolism genes (sglt and ppp1r), two oxygen homeostasis genes (hif1α and egln), and two stress indicator genes (sod and hsp70). These genes were selected based on a previous transcriptomic survey of day-night differences in gene expression (Ruiz-Jones and Palumbi, 2015). Nubbins of A. digitifera were exposed to natural light-dark (LD) and constant darkness (DD) conditions. The daily and circadian changes in transcript levels of the aforementioned genes were measured by real-time quantitative PCR, and interrelationships among the genes were statistically compared.
2 Materials and methods
2.1 Coral collection
The collection of coral was approved by the government of Okinawa Prefecture (approval nos. 27–76 and 28–84). Ten A. digitifera colonies were collected from reefs (average depth 2–4 m) along south Sesoko Island, Okinawa, Japan (26°38′13.964″N, 127°51′56.112″E) on October 20, 2023. After collection, the colonies were transported to Sesoko Station, the Tropical Biosphere Research Center, University of the Ryukyus, Okinawa, Japan, and then divided into nubbins (1.5–3.0 cm branch lengths). Nubbins from the colonies were randomly placed into two 800-L outdoor tanks with running seawater, each containing ten sub-tank replicates. The nubbins acclimated for 5 days under ambient temperatures of 26–27°C and natural light intensity, recorded using a HOBO logger (Onset, Bourne, MA, USA). The day after acclimation, the nubbins in one tank remained under a natural photoperiod (LD; sunrise 06h33, sunset 17h53), whereas the other tank was covered with black sheets and maintained in constant darkness (DD) (Supplementary Figure 1). Nubbins (10 samples per time point, 1.1 ± 0.3 g) from both tanks were sampled at 4-h intervals starting at 08h00, flash-frozen in liquid nitrogen, and stored at –80°C. Samples in the LD tank were subjected to natural environmental light, whereas samples in the DD tank were collected during photophase in a black chamber, with strict avoidance of any environmental light. Samples were collected during scotophase under dim red light (~2.4 lux).
2.2 RNA extraction and cDNA synthesis
Total RNA was extracted from the nubbins using TriPure isolation reagent (Sigma-Aldrich, St. Louis, MO, USA) and purified using the RNeasy Mini Kit (QIAGEN, Hilden, Germany), in accordance with the manufacturers’ protocols. After the quality and quantity of the RNA samples had been assessed with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), reverse transcription was performed to synthesize cDNA from total RNA (500 ng) using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Kusatsu, Japan), in accordance with the manufacturer’s protocol.
2.3 Cloning and sequencing
The selected genes were cloned according to the method of Izumi et al. (2023), using primers (Supplementary Table 1) designed based on highly conserved sequence regions obtained from the A. digitifera genome database (http://marinegenomics.oist.jp). After cloning, plasmid samples (n = 5) were sent to Macrogen Japan (Kyoto, Japan) for DNA sequence confirmation using a 3730xl DNA Analyzer (Applied Biosystems, Waltham, MA, USA).
The open-reading frame (ORF) of each cloned gene was identified and translated into amino acid sequences using a web-based ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). To confirm their characteristics, the obtained ORFs were analyzed with BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences were aligned with related sequences from other vertebrates and invertebrates using ClustalW (Thompson et al., 1994). Subsequently, a phylogenic tree was constructed using the neighbor-joining method with the Jones–Taylor–Thornton model and 1000 bootstrap replications. Sequence alignment and phylogenetic construction were performed in MEGA X (Kumar et al., 2018).
2.4 Real-time quantitative PCR
The transcript levels of the selected genes were analyzed using a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) and the TB Green premix PCR kit (Takara Bio). Forward and reverse primers of each gene were designed based on the sequence results (Supplementary Table S1). Specific primer assays were performed using serial dilutions of A. digitifera cDNA, and amplification efficiencies of approximately 100% were obtained. Each PCR assay was conducted in a final volume of 10 μL containing 5 μL TB Premix Ex Taq II (Tli RNaseH Plus), 0.3 μL of 10 pM forward and reverse primers, 2.4 μL nuclease-free water, and 2 μL of cDNA template. Each sample had two technical replicates. The PCR protocol consisted of an initial 2 min at 95°C, followed by 39 cycles of 5 s at 95°C and 30 s at 60°C. Melting curve analysis was performed subsequently to ensure single amplicon amplification. The mRNA levels of target genes in each sample were normalized to the amounts of β-actin and ef1α (internal controls). Both internal control genes were selected based on stability and minimal variation across the tested samples.
2.5 Statistics
Statistical analyses were conducted using RStudio (v4.3.1; Posit Team, 2024). Two-way ANOVA was applied to evaluate the significance of mRNA expression changes induced by light and time, followed by Dunnett’s test to assess the significance at each time point. All relative mRNA expression data were log-transformed to meet the assumption of normal distribution. Data are presented as the mean ± standard error of the mean (SEM). Principal component analysis (PCA) was utilized to reveal correlations among the 17 genes under LD and DD conditions, using the FactoMine R package (Lê et al., 2008). Missing values were handled using the multiple imputation method with the MIPCA function (Li et al., 2015). Pearson correlations were calculated to assess the linear relationship between each pair of tested genes, using the ggcorrplot package (Kassambara, 2023).
3 Results
3.1 Identification of circadian genes
The genes cry1, cry2, and cry3 had ORFs of 717, 984, and 819 bp, respectively, corresponding to 238, 327, and 272 amino acid residues (Supplementary Figures 2-4); clock, cycle, and slmb had ORFs of 444, 774, and 801 bp, respectively, corresponding to 147, 257, and 266 amino acid residues (Supplementary Figures 5-7). In the phylogenetic analyses, these six circadian genes exclusively clustered with their orthologs (Supplementary Figures 8-11).
3.2 Diurnal expression patterns
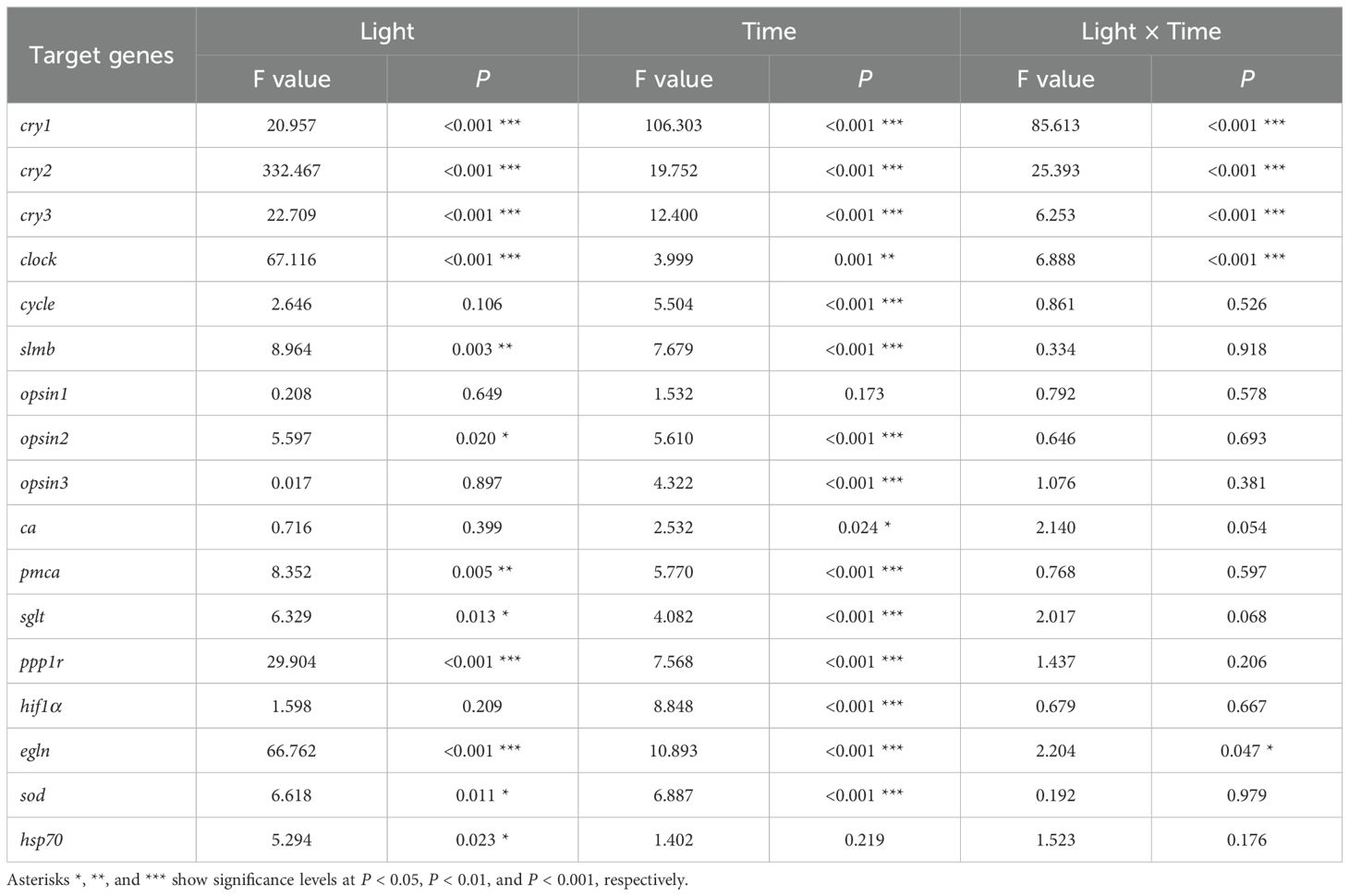
When A. digitifera nubbins were reared under LD conditions, the transcript levels of cry1, cry2, cry3, and clock increased during the daytime and decreased at night. The cry1 transcripts peaked at 12h00 and were lowest at 24h00, whereas those of cry2, cry3, and clock peaked at 16h00. Under DD conditions, there were minimal increases in their expression during the subjective day (Figures 1A–D). Significant increases in cycle and slim under LD and DD conditions occurred only at 08h00* the next morning local time (Figures 1E, F). The changes in circadian genes were significantly affected by light (Plight < 0.001) and time (Ptime < 0.001; Table 1). Significant effects were observed in the expression of cry1-3 and clock (Plight × time < 0.001), cycle (Ptime < 0.001), and slmb (Plight = 0.003; Ptime < 0.001).
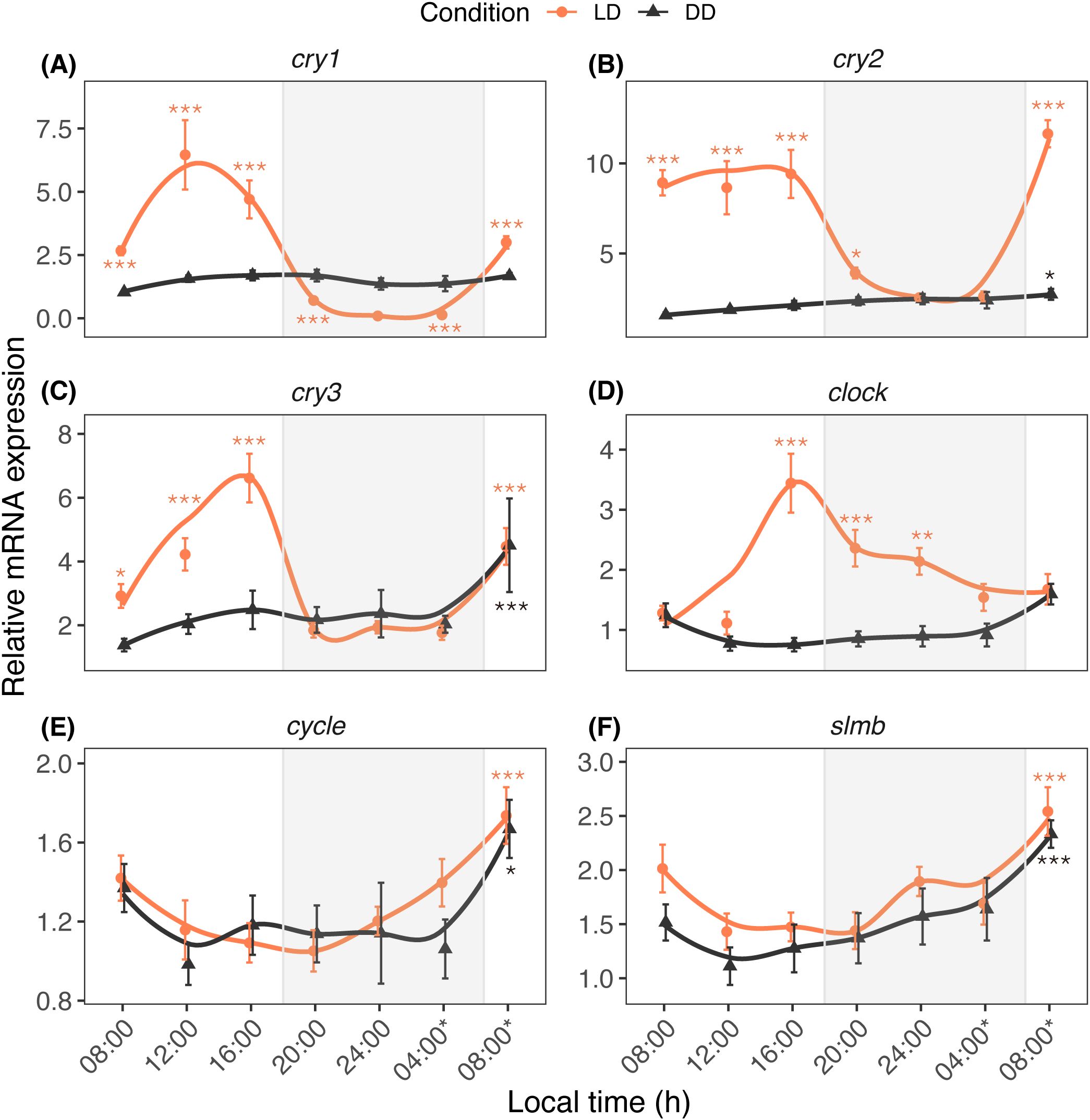
Figure 1. Diurnal expression profiles of circadian genes cry1 (A), cry2 (B), cry3 (C), clock (D), cycle (E) and slmb (F) in A. digitifera. The nubbins were exposed to natural light and dark (LD, orange line) and constant darkness (DD, black line) conditions, and sampled at an interval of 4 hours. Shaded areas denote periods of darkness during the night under LD. Relative mRNA expression values (mean ± SE, n = 10) were analyzed using two-way ANOVA followed by a Dunnett’s test. Time points with asterisks are significantly different, * means P ≤ 0.05; ** means P ≤ 0.01; *** means P ≤ 0.001.
Figure 2 shows the daily profiles of opsin1–3 transcripts in the nubbins reared under LD and DD conditions. The daily fluctuation of opsin1 was not significantly affected by light (Plight = 0.649) or time (Ptime= 0.173). Significant fluctuation was observed in the levels of opsin2 (Plight = 0.02; Ptime < 0.001) and opsin3 (Ptime < 0.001) transcripts; opsin2 mRNA increased at 08h00* under LD and DD conditions, whereas opsin3 mRNA increased at 24h00, 04h00, and 08h00* under LD conditions.
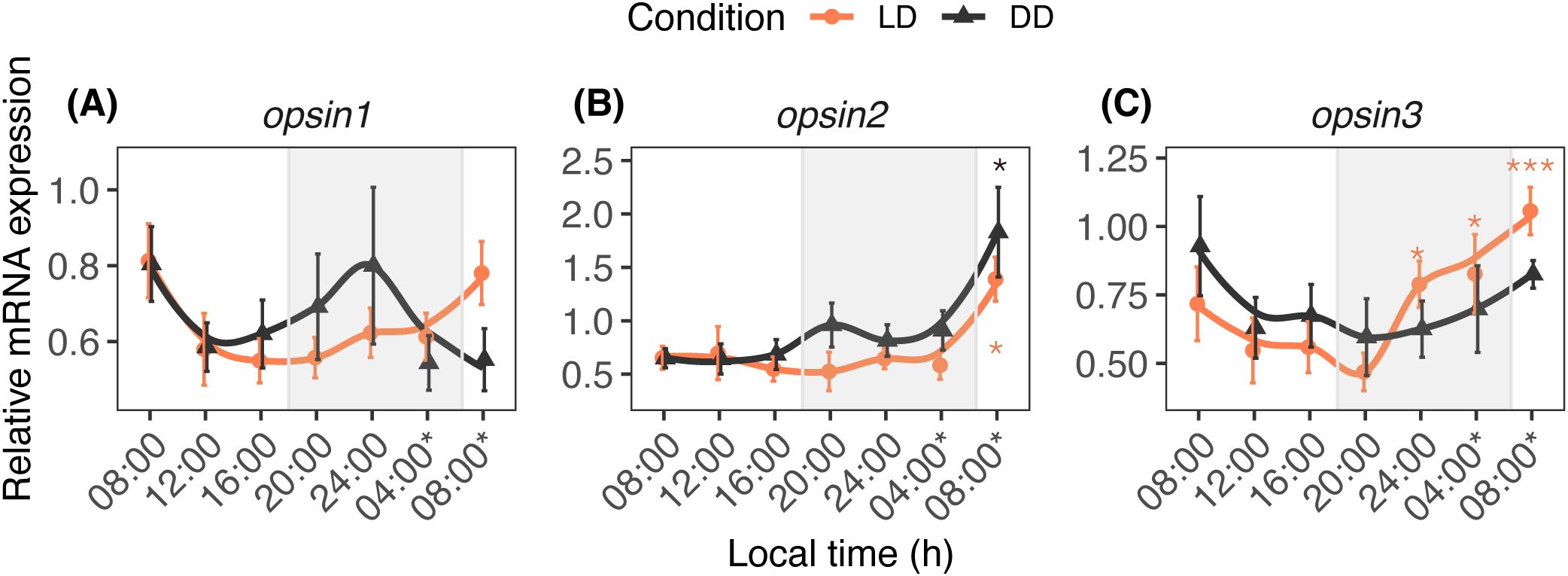
Figure 2. Diurnal expression profiles of opsin1 (A), opsin2 (B), and opsin3 (C) in A. digitifera. The nubbins were exposed to natural light and dark (LD, orange line) and constant darkness (DD, black line) conditions, and sampled at an interval of 4 hours. Shaded areas denote periods of darkness during the night under LD. Relative mRNA expression values (mean ± SE, n = 10) were analyzed using two-way ANOVA followed by a Dunnett’s test. Time points with asterisks are significantly different, * means P ≤ 0.05; *** means P ≤ 0.001.
The study examined the daily and circadian fluctuations of calcification/metabolism-related (ca, pmca, sglt, and ppp1r) (Figure 3) and homeostasis/stress-related (hif1α, egln, sod, and hsp70) (Figure 4) genes. The time factor altered the ca transcription pattern (Ptime = 0.024), whereas the pmca (Plight = 0.005; Ptime < 0.001), sglt (Plight = 0.013; Ptime < 0.001), and ppp1r (Plight < 0.001; Ptime < 0.001) transcription patterns significantly fluctuated according to light and time. Light and time had significant effects on egln (Plight < 0.001; Ptime < 0.001), sod (Plight < 0.001; Ptime < 0.001), hsp70 (Plight = 0.023), and hif1α (Ptime < 0.001).
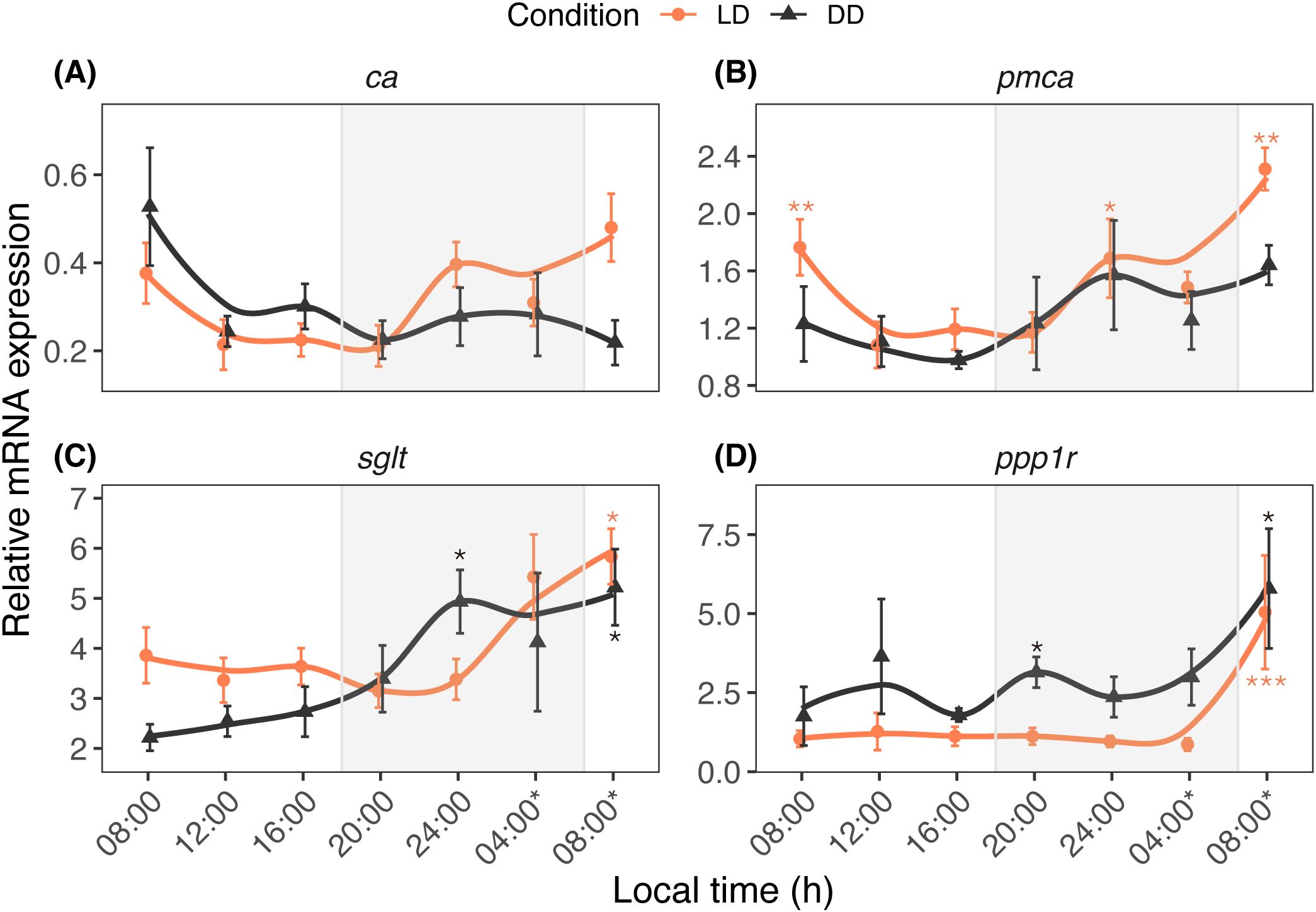
Figure 3. Diurnal expression profiles of ca (A), pmca (B), sglt (C), and ppp1r (D) in A. digitifera. The ca and pmca genes are associated with calcification processes, while sglt and ppp1r are involved in sugar transport and metabolism. The nubbins were exposed to natural light and dark (LD, orange line) and constant darkness (DD, black line) conditions, and sampled at an interval of 4 hours. Shaded areas denote periods of darkness during the night under LD. Relative mRNA expression values (mean ± SE, n = 10) were analyzed using two-way ANOVA followed by a Dunnett’s test. Time points with asterisks are significantly different, * means P ≤ 0.05; ** means P ≤ 0.01; *** means P ≤ 0.001.
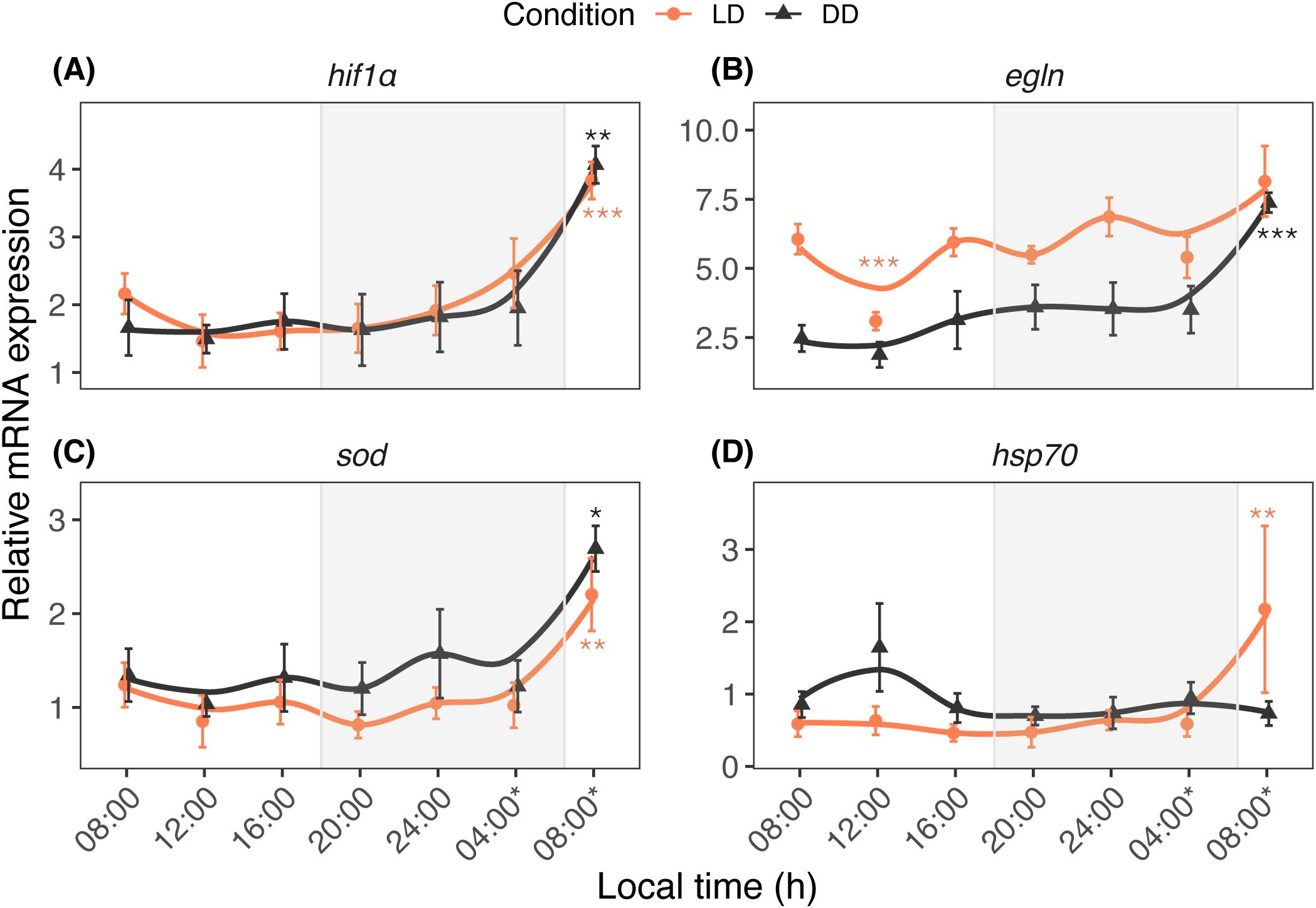
Figure 4. Diurnal expression profiles of hif1α (A), egln (B), sod (C), and hsp70 (D) in A. digitifera. The hif1α and egln genes are critical for oxygen homeostasis, while sod and hsp70 are expressed under stress conditions. The nubbins were exposed to natural light and dark (LD, orange line) and constant darkness (DD, black line) conditions, and sampled at an interval of 4 hours. Shaded areas denote periods of darkness during the night only for the LD condition, DD condition was kept in 24-hour dark. Relative mRNA expression values (mean ± SE, n = 10) were analyzed using two-way ANOVA followed by a Dunnett’s test. Time points with asterisks are significantly different, * means P ≤ 0.05; ** means P ≤ 0.01; *** means P ≤ 0.001.
3.3 PCA and correlations
PCA was used to elucidate interrelationships of circadian genes with other genes under LD and DD conditions (Figure 5). The cluster analysis based on time points revealed two distinct clusters for LD, observed at 16h00 (light green) and 12h00 (deep blue) (Figure 5A). The top five genes contributing to daily variation—cry1, sod, hif1α, cry2, and cry3—collectively explained 83.7% of the variance (Figure 5C).
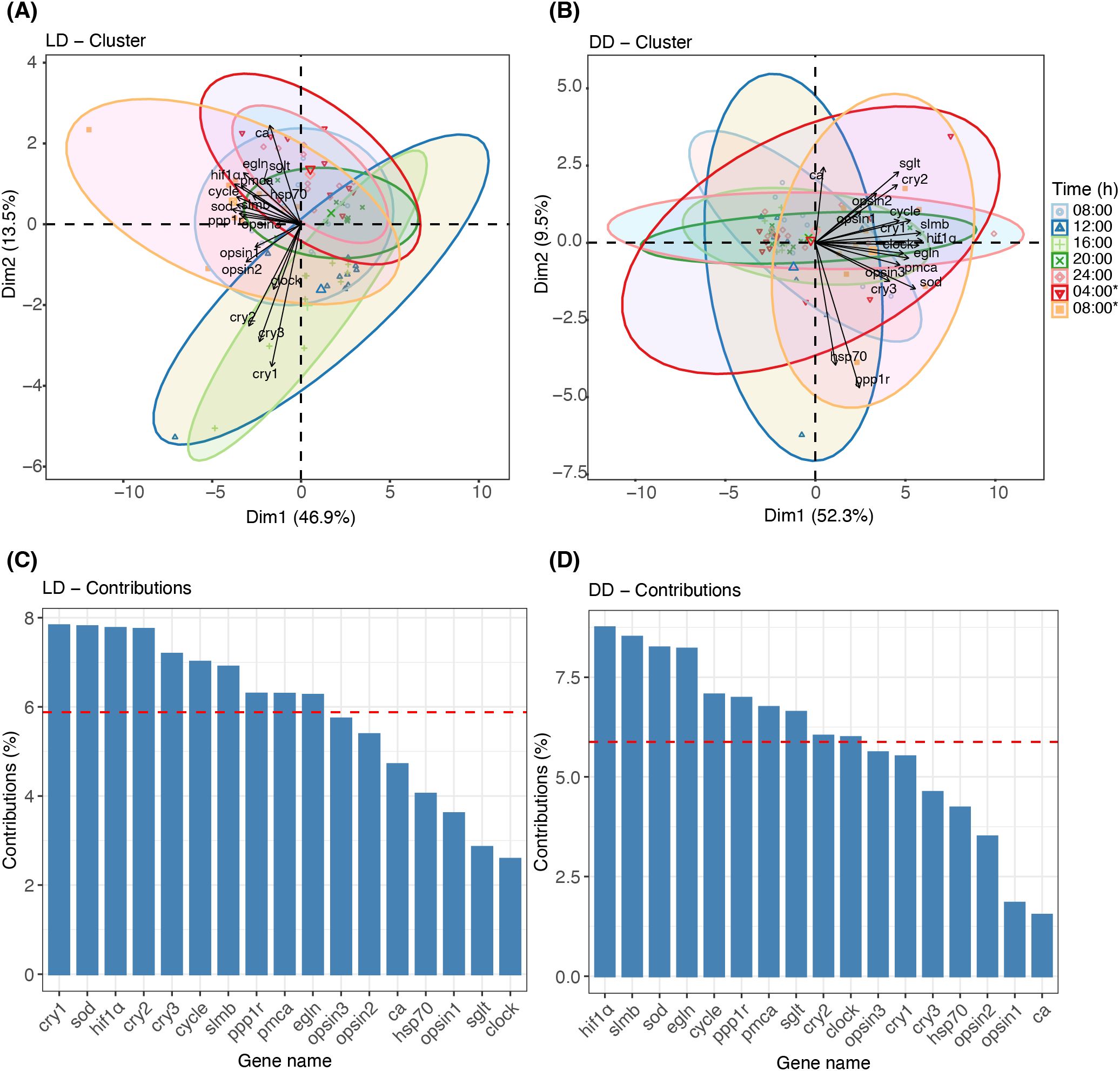
Figure 5. Principal Component Analysis (PCA) of gene expression under LD and DD conditions. (A, B) Biplots depicting the contributions of tested genes to dimensions 1 (Dim 1) and 2 (Dim 2) under LD and DD conditions, respectively. Data are clustered by seven sampling time points, with colored ellipses indicating the corresponding time points. (C, D) Bar plots showing the percentage contribution of each tested gene to the combined Dim 1 and Dim 2 under LD and DD conditions, respectively. The red line highlights the top 10 genes contributing most to the variations.
In the correlation analyses, a score over 0.7 was considered significant (Figure 6). There were robust interaction correlations among the cry genes (cry1 vs. cry2, r = 0.8; cry1 vs. cry3, r = 0.75; cry2 vs. cry3, r = 0.76; Figure 6A). All genes except ca and sglt were positively correlated with cycle, which had strong correlations with hif1α (r = 0.72) and sod (r = 0.74). Similarly, slmb exhibited strong correlations with opsin3 (r = 0.77), pmca (r = 0.77), and hif1α (r = 0.79).
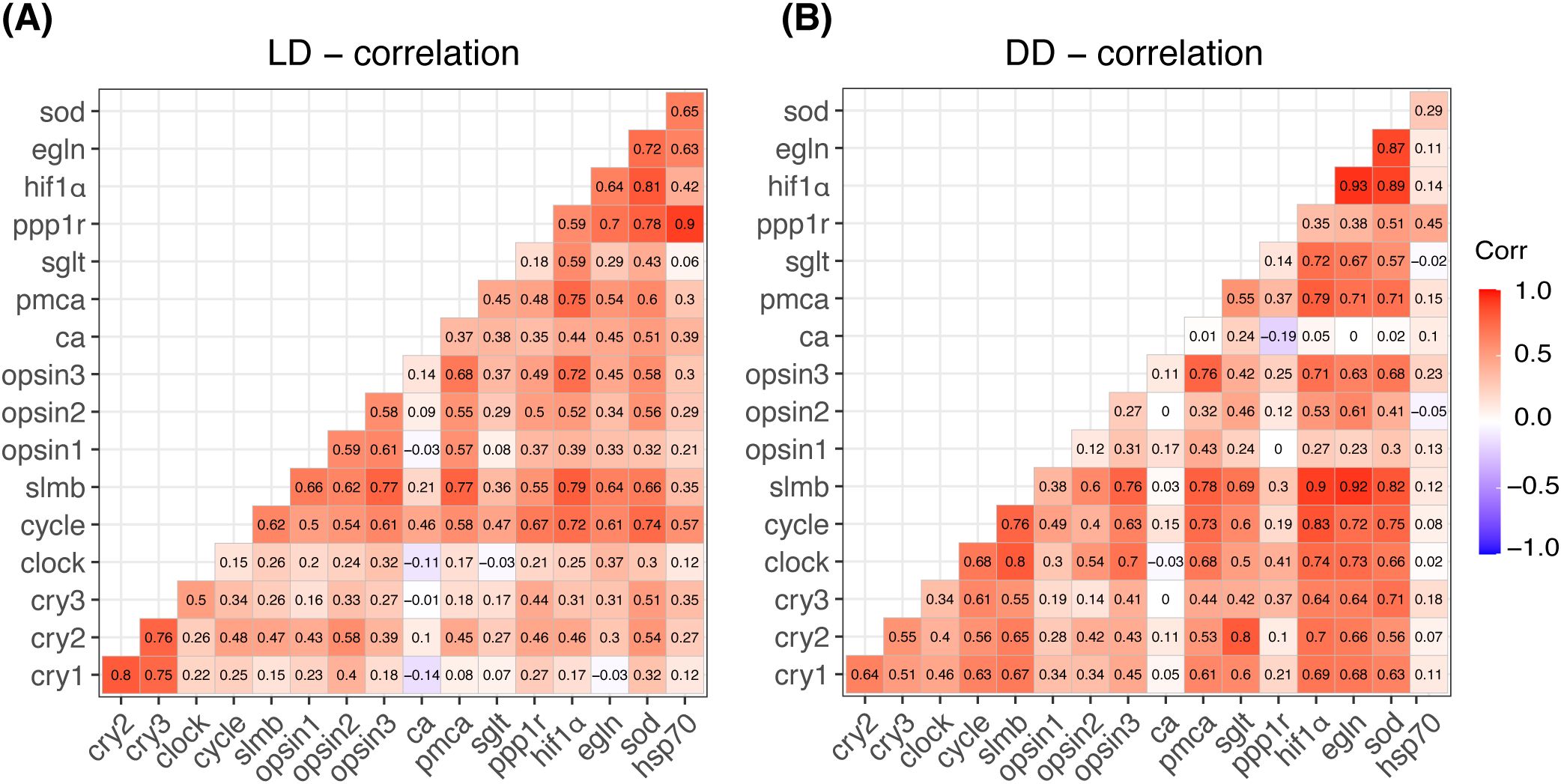
Figure 6. Heatmap displaying Pearson’s correlation coefficients between each pair of tested genes under LD (A) and DD (B) conditions. The color scale ranges from red (indicating positive correlation) to blue (indicating negative correlation), with values closer to 1 signifying a higher correlation. The numbers within each grid represent the exact correlation scores calculated from normalized data.
Figures 5B, D show the PCA results under DD conditions. The cluster analysis demonstrated a high degree of overlap among different time points (Figure 5B). The predominant explanatory genes—hif1α, slmb, sod, egln, and cycle—explained 81.4% of the variance (Figure 5D). High circadian gene scores (> 0.7) were observed between clock and slmb (r = 0.8, Figure 6B) and between cycle and slmb (r = 0.76). Positive correlations between circadian genes and other tested genes were also detected for cry2 (sglt, r = 0.8), cry3 (sod, r = 0.71), clock (opsin3, r = 0.7; hif1α, r = 0.74; and egln, r = 0.73), cycle (pmca, r = 0.73; hif1α, r = 0.83; egln, r = 0.72; and sod, r = 0.75), and slmb (opsin3, r = 0.75; pmca, r = 0.78; hif1α, r = 0.9; egln, r = 0.92; and sod, r = 0.82).
4 Discussion
This study revealed that the cry1, cry2, cry3, and clock transcripts fluctuated daily with an increase during the daytime and decrease at night when A. digitifera nubbins were reared under LD conditions. This suggests that these circadian genes fluctuate daily in response to day-night conditions in their habitat. Rhythmic profiles of these genes have been studied in stony corals under LD conditions: cry1 and cry2 increased at 08h00 and 12h00, respectively, in A. millepora (Levy et al., 2007); cry1, cry2, and clock peaked at 10h00, 18h00, and 18h00, respectively, in Favia fragum (Hoadley et al., 2011). Our findings indicate that the increases in these genes during subjective daytime were attenuated in the nubbins reared under DD conditions. Therefore, they likely function as light-responsive genes in A. digitifera. However, different cry2 and clock profiles were observed in A. millepora larvae, with robust rhythmicity in cry2 and clock under LD and DD conditions (Brady et al., 2011). This difference may be partly attributed to the developmental stages studied (larvae or adults) and seasonal variation in each study; seasonal expression of circadian genes has been reported in some fish species (Davie et al., 2009; Herrero and Lepesant, 2014). The circadian system appears to be predominantly involved in the expression of cycle because its rhythmicity was observed under both LD and DD conditions. Similar rhythmicity of this circadian gene under LD and DD conditions was noted in A. millepora (Brady et al., 2011) and F. fragum (Hoadley et al., 2011). Although slmb is a member of the F-box/WD40 protein family and an essential circadian component in Drosophila (Grima et al., 2002), few studies of this gene have been conducted in corals. The slmb expression does not vary between LD and DD conditions, suggesting that ubiquitin-mediated protein degradation is regulated, at least in part, by circadian mechanisms in corals. Overall, the regulation of circadian genes such as cycle and slmb likely differs from that of light-responsive genes such as cry1, cry2, cry3, and clock.
In our study, qPCR analysis revealed a rhythmical circadian pattern in the expression profiles of opsin2 and opsin3. Correlation analysis also indicated that light-responsive circadian genes had a weak interrelationship with opsin2 and opsin3, whereas slmb exhibited a strong interrelationship with these photoreceptor genes. The daily variation in these photoreceptor genes appears to be strongly regulated in a circadian manner. The regulation of photoreceptor genes by a circadian oscillator has been observed in the retinas of the toad Bufo marinus and fish Haplochromis burtoni, in which the rod opsins showed daily variation with an increase during subjective daytime under DD conditions (Korenbrot and Fernald, 1989). In the zebrafish Danio rerio, clock influences the circadian expression of long-wavelength cone opsin mRNA, as inhibition of clock translation reduced the circadian rhythms of this opsin mRNA (Lê et al., 2008). In the present study, clock likely influenced the opsin3 transcript levels in A. digitifera because the correlation analysis showed a positive interrelationship between clock and opsin3 under DD conditions. The GloSenor assay demonstrated that acropsin 2 and acropsin 3 in A. millepora failed to increase light-induced intracellular cAMP levels in opsin-expressing HEK293S cells, although a chimeral mutant of acropsin 2 exhibited changes in intracellular cAMP levels after light exposure (Mason et al., 2023). Considering that light induced a significant difference in the daily profile of opsin2, this gene may fluctuate in a light-dependent manner. However, there was minimal variation in the opsin1 expression in the present study, which differed from previous reports: intercellular signal transducer (cAMP) levels increased in cultured cells expressing acropsin1 after light exposure in A. millepora (Mason et al., 2023); opsin1 increased at 12h00 and decreased at 24h00 under natural light, and it was upregulated after LED light exposure (Shi et al., 2024). Moreover, acropsin 1 activated coral G protein in a light-dependent manner in Acropora palmata (Mason et al., 2012). The failure to observe light-responsiveness of opsin1 in the present study may be partially attributed to variation in the moon phase under natural conditions. In this context, opsin-dependent signaling in the marine bristle worm Platynereis dumerilii was influenced by a blue light receptor similar to cry, which can perceive changes in moonlight (Zurl et al., 2022). Because our day–night experiment was conducted under natural light, the possibility that opsin1 oscillation was masked by other photoreceptors cannot be excluded.
The present study examined daily and circadian expression patterns of calcification/metabolism-related (ca, pmca, sglt, and ppp1r) and homeostasis/stress-related (hif1α, egln, sod, and hsp70) genes. Based on qPCR results and correlation analysis, the tested genes can be categorized into at least two distinct patterns. The first pattern includes genes regulated by both light and circadian oscillation, such as pmca and sod. The PCA results indicate that these genes, alongside cry genes, contributed significantly to the observed variation under LD conditions. This suggests that cry genes may play a role in coordinating essential functions, including calcification and stress response, with daily light cycles. In coral calicoblasts, pmca functions as a Ca2+ pump, contributing to calcification processes (Zoccola et al., 2004; Krebs, 2022). In the giant clam Tridacna squamosa, both the transcripts of pmca and the protein abundance of PMCA in the inner mantle increased after light exposure, suggesting light-enhanced calcification (Ip et al., 2017). In addition to its modulation by light, calcification is regulated by the circadian system (Gutner-Hoch et al., 2016; Neder et al., 2022). In contrast, sod functions as an antioxidant, neutralizing harmful oxygen free radicals and hydrogen peroxide to prevent cellular damage in living tissue (McCord and Fridovich, 1969; Levy et al., 2006). A previous study revealed that the SOD and CAT activities in corals such as Favia favus, Plerogyra sinuosa, and Goniopora lobata exhibited a daily pattern, increasing from morning (08h00) to midday (10h00–14h00) and decreasing from afternoon (16h00) until night (05h00), suggesting that light intensity activates these enzymes (Levy et al., 2006a). Our results imply that the circadian system also plays a role in the diurnal regulation of this free-radical-scavenger enzyme activity in A. digitifera. The interplay between external light cues and internal circadian regulation on the daily oscillation of sod suggests an active light-defense system in shallow water corals. This provides a molecular basis for the adaptation of shallow water species to high-light conditions (Levy et al., 2006a), and implies a potential mechanism for coping with elevated light or temperature conditions under future climate change scenarios.
The second pattern involves genes with expression patterns primarily regulated by circadian rhythms. Hif1α likely falls into this category because the qPCR results indicated a significant effect of time (but not light) on its expression. Correlation analysis also showed positive correlations of hif1α with cycle and slmb under LD and DD conditions. Hif1α plays a crucial role in regulating oxygen levels under hypoxic conditions (Semenza, 2000). There are interactions between daily oxygen rhythms and the circadian clock (Adamovich et al., 2017), and hypoxia affects the expression of circadian genes in different tissues (Manella et al., 2020). Therefore, a functional link might exist between oxygen and circadian rhythm sensing (Kvitt et al., 2022). Our findings suggest that the expression of hif1α is regulated in a circadian manner. Given the importance of daily oxygen fluctuations in coral survival, the oscillatory expression of hif1α likely helps maintain oxygen homeostasis, even under DD conditions. This mechanism may support coral resilience by adapting to environmental oxygen changes independent of light exposure.
In conclusion, daily fluctuations in some genes in A. digitifera, including photoreceptor, calcification/metabolism-related, and homeostasis/stress-related genes, are regulated by light or the circadian system. The present study could not exclude the involvement of the circadian system in the expression patterns of other tested genes because we established a relatively high threshold for demonstrating the circadian system. Additionally, the constant darkness (DD) condition used in experiment does not reflect the natural light corals received in their environment, which may influence the physiological responses observed. Further studies are necessary to clarify the involvement of circadian genes in the photophysiology of corals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the studied coral is a low invertebrate animal.
Author contributions
ZS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Visualization, Writing – original draft. AT: Conceptualization, Investigation, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by University of the Ryukyus Suzuki Syohe Funding (2023-No.61).
Acknowledgments
We gratefully thank staff of Sesoko Station, Tropical Biosphere Research Center, University of the Ryukyus, Okinawa, Japan, for use of facilities. We also greatly appreciate the kind assistance of manuscript preparation by Dr. T. Singh, Dr. Tan, Dr. H. Takekata, Dr. K. Fukunaga, and Dr. S. Udagawa as well as of sample collection by X. Yin, N. Fujihara, K. Kimura, and R. Gomez.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1494597/full#supplementary-material
Abbreviations
cry1, cryptochrome1; cry2, cryptochrome2; cry3, chrptochrome3; clock, circadian locomotor output cycles kaput; cycle, cycle, also known as aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL), or brain and muscle ARNT-like 1 (BMAL1); slmb, supernumerary limbs; ca, carbonic anhydrase; pmca, plasma membrane calcium transporting ATPase 2; hif1α, hypoxia inducible factor 1 alpha; egln, egg-laying defective nine homolog 1, also known as HIF prolyl hydroxylase domain (PHD); sglt, sodium glucose cotransporter; ppp1r, protein phosphate 1 regulator; sod, superoxide dismutase; hsp70, heat shock protein 70.
References
Adamovich Y., Ladeuix B., Golik M., Koeners M. P., Asher G. (2017). Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell. Metab. 25, 93–101. doi: 10.1016/j.cmet.2016.09.014
Bertucci A., Forêt S., Ball E. E., Miller D. J. (2015). Transcriptomic differences between day and night in Acropora millepora provide new insights into metabolite exchange and light-enhanced calcification in corals. Mol. Ecol. 24, 4489–4504. doi: 10.1111/mec.13328
Brady A. K., Snyder K. A., Vize P. D. (2011). Circadian cycles of gene expression in the coral, Acropora millepora. PloS One 6, e25072. doi: 10.1371/journal.pone.0025072
Davie A., Minghetti M., Migaud H. (2009). Seasonal variations in clock-gene expression in atlantic salmon (Salmo Salar). Chronobiol. Int. 26, 379–395. doi: 10.1080/07420520902820947
Gong S., Li G., Liang J., Xu L., Tan Y., Jin X., et al. (2023). Day-night cycle as a key environmental factor affecting coral-Symbiodiniaceae symbiosis. Ecol. Indic. 146, 1–10. doi: 10.1016/j.ecolind.2023.109890
Grima B., Lamouroux A., Chélot E., Papin C., Limbourg-Bouchon B., Rouyer F. (2002). The F-box protein Slimb controls the levels of clock proteins Period and Timeless. Nature 420, 173–178. doi: 10.1038/nature01171
Gutner-Hoch E., Schneider K., Stolarski J., Domart-Coulon I., Yam R., Meibom A., et al. (2016). Evidence for rhythmicity pacemaker in the calcification process of scleractinian coral. Sci. Rep. 6, 1–8. doi: 10.1038/srep20191
Hemond E. M., Vollmer S. V. (2015). Diurnal and nocturnal transcriptomic variation in the Caribbean staghorn coral, Acropora cervicornis. Mol. Ecol. 24, 4460–4473. doi: 10.1111/mec.13320
Herrero M. J., Lepesant J. M. J. (2014). Daily and seasonal expression of clock genes in the pituitary of the European sea bass (Dicentrarchus labrax). Gen. Comp. Endocrinol. 208, 30–38. doi: 10.1016/j.ygcen.2014.08.002
Hoadley K. D., Szmant A. M., Pyott S. J. (2011). Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PloS One 6, e19755. doi: 10.1371/journal.pone.0019755
Ip Y. K., Hiong K. C., Goh E. J. K., Boo M. V., Choo C. Y. L., Ching B., et al. (2017). The whitish inner mantle of the giant clam, Tridacna squamosa, Expresses an apical plasma membrane Ca2+-ATPase (PMCA) which displays light-dependent gene and protein expressions. Front. Physiol. 8. doi: 10.3389/fphys.2017.00781
Izumi R., Tan E. S., Higa H., Shi Z., Takeuchi Y., Isomura N., et al. (2023). Effects of light intensity and spectral composition on the growth and physiological adaptation of Acroporid corals. Coral. Reefs. 42, 385–398. doi: 10.1007/s00338-023-02348-w
Kassambara A. (2023). Package “ggcorrplot”. Available online at: http://www.sthda.com/english/wiki/ggcorrplot-visualization-of-a-correlation-matrix-using-ggplot2 (Accessed July 9, 2024).
Korenbrot J. I., Fernald R. D. (1989). Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature 337, 454–457. doi: 10.1038/337454a0
Krebs J. (2022). Structure, function and regulation of the plasma membrane calcium pump in health and disease. Int. J. Mol. Sci. 23, 2–16. doi: 10.3390/ijms23031027
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kvitt H., Malik A., Ben-Tabou de-Leon S., Shemesh E., Lalzar M., Gruber D. F., et al. (2022). Transcriptional responses indicate acclimation to prolonged deoxygenation in the coral Stylophora pistillata. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.999558
Lê S., Josse J., Husson F. (2008). FactoMineR: An R Package for multivariate analysis. J. Stat. Software 25, 1–18. doi: 10.18637/jss.v025.i01
Levy O., Achituv Y., Yacobi Y. Z., Dubinsky Z., Stambler N. (2006a). Diel “tuning” of coral metabolism: Physiological responses to light cues. J. Exp. Biol. 209, 273–283. doi: 10.1242/jeb.01983
Levy O., Appelbaum L., Leggat W., Gothlif W., Hayward D. C., Miller D. J., et al. (2007). Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318, 463–467. doi: 10.1126/science.1144642
Levy O., Dubinsky Z., Achituv Y., Erez J. (2006b). Diurnal polyp expansion behavior in stony corals may enhance carbon availability for symbionts photosynthesis. J. Exp. Mar. Biol. Ecol. 333, 1–11. doi: 10.1016/j.jembe.2005.11.016
Levy O., Kaniewska P., Alon S., Eisenberg E., Karako-Lampert S., Bay L. K., et al. (2011). Complex diel cycles of gene expression in coral-algal symbiosis. Science 331, 175. doi: 10.1126/science.1196419
Levy O., Mizrahi L., Chadwick-Furman N. E., Achituv Y. (2001). Factors controlling the expansion behavior of Favia favus (Cnidaria: Scleractinia): Effects of light, flow, and planktonic prey. Biol. Bull. 200, 118–126. doi: 10.2307/1543305
Li P., Stuart E. A., Allison D. B. (2015). Multiple imputation: A flexible tool for handling missing data. J. Am. Med. Assoc. 314, 1966–1967. doi: 10.1001/jama.2015.15281
Manella G., Aviram R., Bolshette N., Muvkadi S., Golik M., Smith D. F., et al. (2020). Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. PNAS 117, 779–786. doi: 10.1073/pnas.1914112117
Mason B. M., Koyanagi M., Sugihara T., Iwasaki M., Slepak V., Miller D. J., et al. (2023). Multiple opsins in a reef-building coral, Acropora millepora. Sci. Rep. 13, 1–8. doi: 10.1038/s41598-023-28476-5
Mason B., Schmale M., Gibbs P., Miller M. W., Wang Q., Levay K., et al. (2012). Evidence for multiple phototransduction pathways in a reef-building coral. PloS One 7, e50371. doi: 10.1371/journal.pone.0050371
Mazzoccoli G., Pazienza V., Vinciguerra M. (2012). Clock genes and clock-controlled genes in the regulation of metabolic rhythms. Chronobiol. Int. 29, 227–251. doi: 10.3109/07420528.2012.658127
McCord J. M., Fridovich I. (1969). Superoxide dismutase - An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055. doi: 10.1016/S0021-9258(18)63504-5
Moya A., Tambutté S., Tambutté E., Zoccola D., Caminiti N., Allemand D. (2006). Study of calcification during a daily cycle of the coral Stylophora pistillata: Implications for “light-enhanced calcification. J. Exp. Biol. 209, 3413–3419. doi: 10.1242/jeb.02382
Nakao A., Nakamura Y., Shibata S. (2015). The circadian clock functions as a potent regulator of allergic reaction. Allergy 70, 467–473. doi: 10.1111/all.12596
Nathans J., Hogness D. S. (1983). Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell 34, 807–814. doi: 10.1016/0092-8674(83)90537-8
Neder M., Saar R., Malik A., Antler G., Mass T. (2022). New insights on the diurnal mechanism of calcification in the stony coral, Stylophora pistillata. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.745171
Porter M. L., Blasic J. R., Bok M. J., Cameron E. G., Pringle T., Cronin T. W., et al. (2011). Shedding new light on opsin evolution. Proc. R. Soc B. 279, 3–14. doi: 10.1098/rspb.2011.1819
Posit Team. (2024). RStudio: Integrated Development Environment for R. Posit Software, PBC, Boston, MA. Available at: https://www.posit.co/.
Reitzel A. M., Behrendt L., Tarrant A. M. (2010). Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: The evolution of the animal circadian clock. PloS One 5, 1–9. doi: 10.1371/journal.pone.0012805
Rinsky M., Weizman E., Waldman Ben-Asher H., Eyal G., Zhu B., Levy O. (2022). Temporal gene expression patterns in the coral Euphyllia paradivisa reveal the complexity of biological clocks in the cnidarian-algal symbiosis. Sci. Adv. 8, 1–15. doi: 10.1126/sciadv.abo6467
Ruiz-Jones L. J., Palumbi S. R. (2015). Transcriptome-wide changes in coral gene expression at noon and midnight under field conditions. Biol. Bull. 228, 227–241. doi: 10.1086/BBLv228n3p227
Semenza G. L. (2000). Expression of hypoxia-inducible factor 1: Mechanisms and consequences. Biochem. Pharmacol. 59, 47–53. doi: 10.1016/s0006-2952(99)00292-0
Shi Z., Tan E. S., Takemura A. (2024). Day–night expression patterns of opsin genes in the coral Acropora digitifera under natural and LED light conditions. Coral. Reefs. 43, 1535–1547. doi: 10.1007/s00338-024-02558-w
Sorek M., Díaz-Almeyda E. M., Medina M., Levy O. (2014). Circadian clocks in symbiotic corals: The duet between Symbiodinium algae and their coral host. Mar. Genomics 14, 47–57. doi: 10.1016/j.margen.2014.01.003
Sorek M., Levy O. (2012). Influence of the quantity and quality of light on photosynthetic periodicity in coral endosymbiotic algae. PloS One 7, e43264. doi: 10.1371/journal.pone.0043264
Takahashi J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi: 10.1038/nrg.2016.150
Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Torres M., Becquet D., Franc J. L., François-Bellan A. M. (2018). Circadian processes in the RNA life cycle. Wiley. Interdiscip. Rev. RNA. 9, e1467. doi: 10.1002/wrna.1467
Vize P. D. (2009). Transcriptome analysis of the circadian regulatory network in the coral Acropora millepora. Biol. Bull. 216, 131–137. doi: 10.1086/BBLv216n2p131
Zhang E. E., Kay S. A. (2010). Clocks not winding down: Unravelling circadian networks. Nat. Rev. Mol. Cell. Biol. 11, 764–776. doi: 10.1038/nrm2995
Zoccola D., Tambutté E., Kulhanek E., Puverel S., Scimeca J. C., Allemand D., et al. (2004). Molecular cloning and localization of a PMCA P-type calcium ATPase from the coral Stylophora pistillata. Biochim. Biophys. Acta Biomembr. 1663, 117–126. doi: 10.1016/j.bbamem.2004.02.010
Keywords: circadian, coral, opsin, calcification, homeostasis, metabolism, photophysiology
Citation: Shi Z and Takemura A (2025) Interplay between light and circadian rhythms in the regulation of photoreception and physiological processes in the stony coral Acropora digitifera. Front. Mar. Sci. 11:1494597. doi: 10.3389/fmars.2024.1494597
Received: 11 September 2024; Accepted: 11 December 2024;
Published: 06 January 2025.
Edited by:
Juan José Chiesa, National University of Quilmes, ArgentinaCopyright © 2025 Shi and Takemura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akihiro Takemura, dGFrZW11cmFAc2NpLnUtcnl1a3l1LmFjLmpw
 Zongyan Shi
Zongyan Shi Akihiro Takemura
Akihiro Takemura