- 1Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, China
- 2State Key Laboratory Breeding Base of Marine Genetic Resources, Third Institute of Oceanography, Xiamen, China
- 3Fujian Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, Xiamen, China
Introduction: Alcanivorax, a typical alkane-degrading bacterium, has demonstrated the ability to utilize inorganic electron donor in some reports. However, a comprehensive analysis of its potentiality to utilize inorganic electron donor is still lacking.
Methods: In this study, genomic and phylogenetic analyzes were used to explore the potential oxidative capacity of inorganic compounds in Alcanivorax. And its functions were verified through physiological experiments.
Results: The sulfur oxidation-related genes sqr and tsdA are prevalent and have various evolutionary origins. Potential genes for CO oxidation were present in 39 strains, whereas genes associated with iron, hydrogen, and ammonia oxidation were either rare or absent. The physiological functions of Sqr and TsdA were confirmed in six representative strains under heterotrophic conditions. Adding thiosulfate enhanced Alcanivorax growth. However, Alcanivorax bacteria perform sulfide detoxification through Sqr rather than by gaining energy via sulfide oxidation Although no strain was confirmed to be chemoautotrophs, we discovered that the two clades, A. xenomutans and A. profundimaris, can grow under conditions with very low organic matter.
Discussion: The ability to utilize inorganic compounds as a supplementary energy source and adapt to carbon oligotrophic growth may contribute to the prevalence of Alcanivorax in marine ecosystems.
Introduction
Marine microorganisms play a crucial role in marine ecosystems, playing a significant role in maintaining the balance of marine ecology and biogeochemical cycles. Generally, marine heterotrophs primarily utilize carbon from sinking organic particles (Engel et al., 2022), microbial remnants (Cerro-Gálvez et al., 2021; Katayama et al., 2024), dissolved organic matter (DOM) (Baltar et al., 2021; Quigley et al., 2019), and alkanes from the crust (Dong et al., 2022; Love et al., 2021). Interestingly, some studies have revealed that in areas where the concentration of organic matter is limited but the concentrations of reductive inorganic substances are high, certain groups of heterotrophic microorganisms, including Alcanivorax, are abundant (Dede et al., 2023; Chernikova et al., 2020; Kesava et al., 2020; Bhattacharya et al., 2020). When using chemoautotrophic culture media for enrichment, these heterotrophic groups are sometimes enriched (Dede et al., 2022; Wang et al., 2020; Wei et al., 2022; Hedrich et al., 2011). A new perspective suggests that Alcanivorax may be able to utilize these reductive inorganic substances in specific environments to achieve chemolithotrophic growth. Even fix inorganic carbon as the primary carbon source like chemoautotrophs (Wang et al., 2020). The main difference between chemolithotrophs and chemoautotrophs is where they get carbon. chemolithotrophs can get carbon from organic or inorganic sources and energy from inorganic sources. chemoautotrophs get both carbon and energy from inorganic sources.
Alcanivorax is a prominent genus that frequently emerges in studies of marine microorganisms (Wei et al., 2022; Wang et al., 2020). It is ubiquitous in oceanic environments, ranging from coastal areas to the deep sea, and even in hydrothermal vents (Yang et al., 2018; Dede et al., 2023; Chernikova et al., 2020; Wei et al., 2022; Wang et al., 2020). Its prevalence is notable, constituting approximately 0.1% of the microbial community in open ocean settings and exceeding 1% in coastal areas (Supplementary Figure 1, data from IMNGS (Lagkouvardos et al., 2016)).
The Alcanivorax genus is an important group within the class Gammaproteobacteria. The first strain of Alcanivorax was identified and named Yakimov in 1998, which initially gained significant attention due to its remarkable capacity to degrade alkanes (Yakimov et al., 1998). In past decades, an increasing number of Alcanivorax species have been isolated and identified, with a total of 29 species reported by 2024. However, according to the latest report, the phylotaxogenomic analysis showed that Alcanivorax species formed three clades. The original Alcanivorax genus has been further divided into Alloalcanivorax, Isoalcanivorax, and the narrow Alcanivorax (Rai et al., 2023).
Alcanivorax species are regarded as a group of traditionally heterotrophic microorganisms (Liu and Shao, 2005; Yang et al., 2018; Zhu et al., 2021). However, our recent report indicated that Alcanivorax bacteria can grow through iron oxidation (Wei et al., 2022). In hydrothermal environments, where alkanes and other organic compounds are not as abundant as reduced inorganic compounds (such as sulfide), their relative abundance can reach a significant level of up to 40% (Dede et al., 2023). These findings suggest that Alcanivorax may have special metabolic strategies for adapting to oligotrophic environments (Lv et al., 2024; Wang et al., 2020).
However, to date, there is no comprehensive investigation detailing the ability of Alcanivorax to oxidize inorganic reducing compounds. It is still unclear what types of inorganic energy it might use and whether it can fix inorganic carbon to achieve chemoautotrophic growth. In this study, we explored the aforementioned issues by analyzing the genomes of 167 Alcanivorax strains covering all 29 species and performing experimental validations on representative strains.
Materials and methods
Representative strains and cultural conditions
Six representative strains were selected base on phylogenetic tree and their genomes, which encode TsdA, Sqr and Sdo proteins, for the following experiments. These strains included A. hongdengensisT MCCC1A01496, Alcanivorax sp. MCCC1A02275 (most similar to A. borkumensis), Alcanivorax sp. MCCC1A04522 (most similar to A. modilis), A. venustensis MCCC1A04970, A. profundimarisT MCCC1A7714, and A. xenomutans MCCC1A05661. Strains A. hongdengensisT MCCC1A01496, Alcanivorax sp. MCCC1A02275, A. venustensis MCCC1A04970, and A. profundimarisT MCCC1A7714 were isolated from upper seawater samples. A. xenomutans MCCC1A05661 was isolated from deep seawater sample, and Alcanivorax sp. MCCC1A04522 was specifically isolated from hydrothermal sediment sample. All strains were cultured on 2216E agar plates. Subsequently, the culture was thoroughly washed with phosphate buffered saline (PBS) three times before being inoculated into different media.
The composition of the heterotrophic sulfide (thiosulfate) oxidation media are as follows (L-1): 2 g of acetate, 0.2 g of yeast, 25 g of NaCl, 0.5 g of KCl, 0.84 g of NaNO3, 0.14 g of K2HPO4, 0.2 g of NaHCO3, 1 mL of vitamins, 1 mL of trace elements (Balch et al., 1979), 1 mM Na2S·9H2O (10 mM Na2S2O4). Furthermore, heterotrophic sulfide oxidation was tested under an anaerobic environment to avoid spontaneous sulfide oxidation in a short time. Another anaerobic heterotrophic growth experiment without sulfide was designed as a blank group to survey strains’ growth ability under an anaerobic environment. One percent hexadecane was used to replace 2 g of acetate and 0.2 g of yeast was used for another heterotrophic thiosulfate oxidation experiment.
The composition of the chemoautotrophic thiosulfate oxidate media was as follows (L-1): 25 g of NaCl, 0.5 g of KCl, 0.84 g of NaNO3, 0.14 g of K2HPO4, 0.84 g of NaHCO3, 1 mL of vitamins, 1 mL of trace elements and 20 mM Na2S2O4.
Genomes annotation
Over the past two decades, 167 Alcanivorax genomes have been successfully sequenced by the Marine Culture Collection of China (MCCC). For translation and preliminary annotation analysis, Prokka (v1.13) was used, while EggNOG-mapper (v2.0) was employed for a deeper exploration of metabolic pathways.
Key metabolic genes prediction
To identify functional genes responsible for reducing inorganic compound oxidation, the genes encoding proteins involved in sulfur oxidation (Sulfide: quinone reductase, Sqr; Flavocytochrome c sulfide dehydrogenase, FCSD; reverse dissimilatory sulfite reduction, Dsr; heterodisulide reductase-like, Hdr; sulfur oxygenase/reductase, Sor; sulfur dioxygenase, Sdo; thiosulfate oxidation complex, SoxABCDXYZ; adenylylsulfate reductase, Apr; sulfate adenylyltransferase, Sat; thiosulfate dehydrogenase, Tsd) (Hutt, 2017; Du et al., 2022; Zhang et al., 2020; Powell et al., 2017; Landry et al., 2021; Dahl et al., 2005), ammonia oxidation (ammonia monooxygenase, Amo; hydroxylamine oxidoreductase, Hao) (Kuypers et al., 2018; Stein, 2019), carbon monoxide oxidation (carbon monoxide dehydrogenase, CoxMSL) (Stein, 2019), and hydrogen oxidation (hydrogenase, Hya) (Abdellatif et al., 2017; Petersen et al., 2011) were identified by a local BLASTP protein similarity search with an amino acid similarity cutoff of 40%, coverage >80%, and an e-value cutoff of 1E-5. Additionally, the nitrogen-reducing genes were also surveyed to predict their ability to use nitrate as an electron acceptor via the same method. FeGenie was used to search for iron oxidation functional genes (Garber et al., 2020). The results obtained from both BLASTP and FeGenie were further checked against conserved domain search (CD-search) results (Lu et al., 2020).
Phylogenetic analysis
A genome-based phylogenetic tree encompassing 92 core genes was constructed by UBCG (v2) (bcg files were supported in Supplementary Material 1) (Na et al., 2018). The amino acid sequences of functional proteins, including Sqr and TsdA, were subjected to multiple sequence alignments with reference sequences obtained from the NCBI and UniProt databases (all sequences are listed in Supplementary Materials 2) via MAFFT (v7) (Katoh and Standley, 2013). Subsequently, these aligned sequences were employed in MEGA11, utilizing the JTT model and 1000 replicates, to construct individual neighbor-joining (NJ) phylogenetic trees for the respective functional genes (Tamura et al., 2021).
Verification of carbon fixation through δ13C isotope analysis
In the 13C isotopic tracer experiments for carbon fixation, the same medium used for chemoautotrophic thiosulfate utilization was used, and NaHCO3 was replaced with NaH13CO3 as the sole source of the 13C isotope. The cells were harvested from the heterotrophic medium (2216E) at 4500 rpm for 30 min, and then thoroughly washed three times with sterile chemoautotrophic medium to remove any residual organic matter. After completing the washing process, the concentration of the organisms was adjusted to OD600 = 0.6. Subsequently, the cells were inoculated into the chemoautotrophic medium at a concentration of 5% inoculum. The cells were then collected after 3 hours and 7 days of incubation. After centrifuging the cells at 8000 rpm for 20 min, the supernatant was discarded. To remove any residual NaH13CO3 adhering to the cells, they were washed by chemoautotrophic medium without isotopes and then centrifuged again for collection.
The 13C isotopes of the cells were accurately determined with a stable isotope ratio mass spectrometer (IRMS) (measuring accuracy = ± 0.2; DeltaVAdvantage, Bremen, Germany).
Detection of total organic carbon
Total organic carbon (TOC) detection was conducted in a similar way to isotope experiments (but without the use of isotope-labeled media), and samples were individually collected on days 0, 7, and 21. The samples were acidified with hydrochloric acid (pH<2) before testing, and the acidification process was extended beyond 2 days to guarantee thorough elimination of inorganic carbon. TOC detection was then performed using a 900 Laboratory TOC Analyzer. To ensure the reliability of the results, three parallel samples were used for both the isotope test and the TOC measurement.
Detection of sulfur compounds
Sulfide was detected through the methylene blue method (Trüper and Schlegel, 1964), while thiosulfate, sulfite, and tetrathionate were detected through the spectrophotometric iodometric method, which relies on measuring the optical density at OD350. When thiosulfate and sulfite are both present in a mixture, sulfite can be masked by formaldehyde. In the event of sufficient alkalinity, 2 mol of tetrathionate will yield 3 mol of thiosulfate and 2 mol of sulfite. Given that 2 μM of thiosulfate and 1 μM of sulfite both result in an identical decrease in OD350, the sulfite and tetrathionate concentrations can be accurately calculated after the addition of formaldehyde and sufficient alkaline, respectively. The sulfate content in the cultures was determined by a turbidimetric method (Kolmert et al., 2000).
Results and discussion
Basic genomes and phylogenomic analysis
A total of 167 genomes were counted, with sizes ranging from 3.05 to 5.09 Mb. Among them, the 108 genomes included 17 species, mainly A. borkumensis, A. marinus, A. profundimaris, A. venustensis, A. xenomutansand, and 12 other species. An additional 59 isolates have not yet been definitively classified into a definite species. These isolates were collected from diverse environments, including deep-sea water, deep-sea sediments, hydrothermal sediments, and upper seawater (Figure 1, with further details provided in Supplementary Table 1). A phylogenomic tree encompassing 92 core genes was constructed by UBCG (v2). Based on the phylogenetic tree results and the original strain names, numerous clades were delineated (Figure 1).
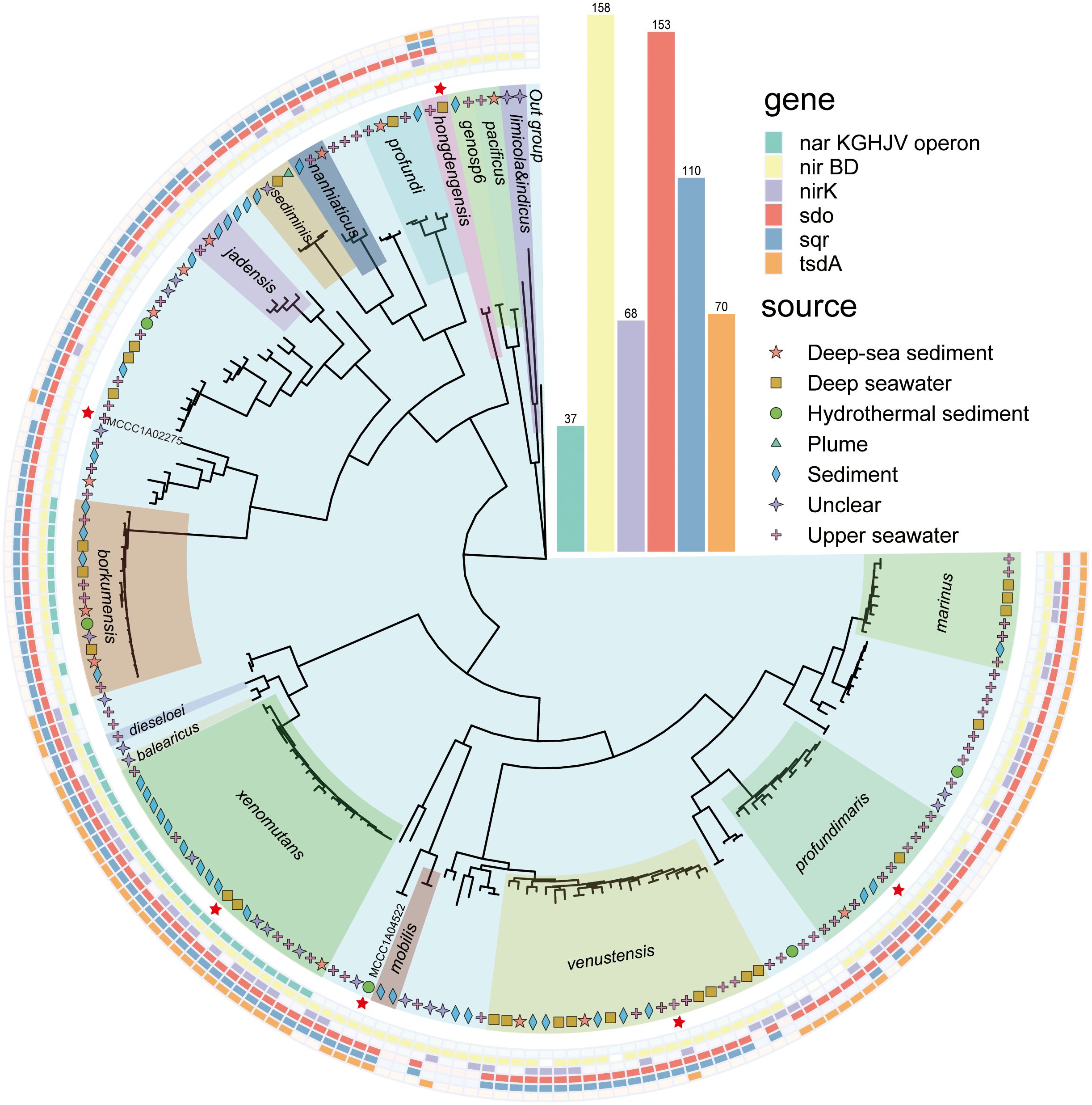
Figure 1. Phylogenetic tree of the genus Alcanivorax constructed using UBCG(v2). The different colors inside represent different species of Alcanivorax, which were divided into 17 clades, and unnamed strains not showing names in this tree without a color block. The inner symbol indicates the strain source. The red stars are representative strains used for the following experiments. The outside ranges with different colors show the results of functional genes identified in their genomes. The inside out indicates the nar operon, nirBD, nirK, sdo, sqr, and tsdA.
Identification of potential genes involved in chemolithotrophic growth
Using local BLASTP, Sqr was identified in 110 of 167 genomes, while Sdo was detected in 153 genomes. Sqr is predominant present in A. hongdengesis, A. nanhaiticus, A. sediminis, A. jadensis, A. borkumensis, A. venustensis, A. xenomutans, A. balearicus, and A. profundimaris, but is mainly absent in A. limicola, A. pacificus, and A.profundi. Sdo was prevalent in most genomes except for those of A. limicola and A. pacificus. Notably, all strains harboring the sqr gene also exhibited the presence of the sdo gene upstream, indicating a potential strategy for efficient sulfide oxidation in environments with high sulfide concentrations (Ran et al., 2022; Xia et al., 2017; Wang et al., 2014; Wu et al., 2017; Cuevasanta et al., 2017). TsdA was identified in a total of 70 genomes, including those of A. xenomutans, A. profundimaris, A. hongdengesis, A. gelatinphageus, A. marinus, and A. mibilis. This finding aligns with the results obtained from Prokka analysis.
Despite attempts at local BLASTP and a 40% identity threshold, certain genes crucial for sulfur oxidation, such as the sox complex, apr, and dsrAB, remained undetected. Similarly, genes associated with nitrogen and hydrogen oxidation were also elusive, in line with the results obtained from direct annotation using Prokka. However, a potential coxLMS operon was identified in species such as A. profundimaris, A. gelatinphageus, A. marinus, and several unnamed strains closely related to A. marinus, with a total of 39 sequences encoding carbon monoxide dehydrogenase (further details are provided in Supplementary Material 3). Notably, despite BLASTP identification falling below the 40% identity threshold, these sequences exhibit functional domains similar to those of CoxLMS. Moreover, attempts to identify the iron oxidase gene using Fegenie revealed the presence of Cyc1 in two strains, Alcanivorax sp. MCCC1A05192 and MCCC1A04750. Genes commonly associated with iron oxidation, including Cyc2, the Fox operon, Sulfocyanin, PioABC, and MotAB, as reviewed by Garber, were conspicuously absent in the genomes (Garber et al., 2020).
Over one-third of the strains showed the presence of the tsdA gene, while more than two-thirds contained the sqr gene, which is responsible for sulfide oxidation. Additionally, sdo genes were nearly ubiquitous across all strains. The analysis of iron oxidation genes in individual bacterial species aligned with previous reports (Wei et al., 2022), indicating the potential for iron oxidation among these strains. These findings suggest that Alcanivorax bacteria may have a diverse range of available inorganic energy sources. Traditionally, Alcanivorax strains have been regarded as typical heterotrophic bacteria capable of utilizing various carbon sources, including some recalcitrant compounds. However, a recent breakthrough in 2022 revealed that a strain of Alcanivorax exhibits remarkable iron oxidation capabilities and potential for carbon fixation (Wei et al., 2022), thereby challenging the long-held perception of Alcanivorax as purely heterotrophs. This discovery suggested that they may belong to a group of chemolithotrophs.
Identification of nitrogen-reducing genes
There is no doubt that Alcanivorax strains can use oxygen as an electron acceptor (Liu and Shao, 2005; Zhu et al., 2021), but it is still unclear whether they can use nitrate as an electron acceptor under microaerobic or anaerobic conditions. Therefore, the nitrogen-reducing genes of Alcanivorax were also surveyed.
The ability to perform complete assimilative nitrate reduction is a feature found across all Alcanivorax genomes. However, the capacity for dissimilatory nitrate reduction is restricted to certain clades. Notably, the narKGHJV operon, which is essential for converting nitrate to nitrite (Kuypers et al., 2018), is unique to the A. barkumensis and A. xenomutans clades. In contrast, the NirBD system, which is responsible for the conversion of nitrite to ammonium, is nearly universally present across nearly all genomes. Furthermore, within the denitrification pathway, NirK, which catalyzes nitrite to nitric oxide, was identified in 68 genomes, predominantly within the A. xenomutans, A. venustensis, A. profundimaris, and A. mariuis clades.
Nitrate reduction genes are widespread among Alcanivorax species, indicating their potential to use nitrate as an electron acceptor, thereby facilitating electron transfer within respiratory chains (Kuypers et al., 2018). This capability is particularly relevant in environments where thiosulfate or sulfide accumulates under microaerobic or anaerobic conditions. Alcanivorax, a commonly encountered and abundant bacterial group in marine environments (Dede et al., 2023), possesses a high proportion of genes related to sulfur oxidation and nitrate reduction, suggesting that it plays a significant role in the inorganic nitrogen and sulfur cycles in marine environments.
Potential carbon fixation pathway analysis
To date, a total of nine distinct pathways for microbial fixation of inorganic carbon have been reported, including six classical natural pathways and three alternate pathways Mall et al., 2018; Sánchez-Andrea et al., 2020; Steffens et al., 2021; Correa et al., 2023). However, despite thorough investigations, neither the entire suite of microbial inorganic carbon fixation pathways nor their crucial genes have been comprehensively identified in Alcanivorax (Supplementary Material 4). Nevertheless, the reverse Tricarboxylic Acid cycle (rTCA) and Calvin-Benson-Bassham cycle (CBB) pathways share relatively high integrity in Alcanivorax. Unfortunately, the key genes of the rTCA and CBB pathways were all absent.
In the rTCA pathway, most of the genes can be found in genomes. Although a few genes are missing in some genomes, they are not absent in special species. For instance, Citryl-CoA lyase (ccl) is present in the genomes of A. xenomuant and A. profundimaris. However, the key genes ATP-citrate lyase (acl) and citryl-CoA synthetase (ccs), which are considered essential as key genes, are absent from all the genomes.
In the CBB pathway, the crucial functional gene ribulose-bisphosphate carboxylase is conspicuously absent from the genomes investigated. Only a few genes encoding fructose-bisphosphate aldolase (Fba) and phosphoglycerate kinase (Pgk) were identified. Based on the finding that most of the genes in the CBB pathway are absent, it seems improbable that carbon fixation would occur through the CBB pathway in these organisms.
Phylogenetic analysis of Sqr and TsdA
The phylogenetic tree of Sqr of Alcanivorax exhibited similarity to the reported classification (Marcia et al., 2010), with a clear division into FCSDs and six types of Sqr. All Sqr homologs of Alcanivorax belonged to type II, except for a single copy of Alcanivorax sp. MCCC1A05192, which was categorized as type I (Figure 2). They were categorized into three distinct clusters. Except for the single copy of Alcanivorax sp. MCCC1A05192, all others exhibited proximity to the neighboring branches of Pseudomonas aeruginosa and other related genera of Gammaproteobacteria. The Sqr close to Alcanivorax predominantly was origin from heterotrophic bacteria and some eukaryotes, whereas the reference sequences located in the more distant cluster within type II Sqr are mostly derived from chemoautotrophs, such as Sulfurimonas. In addition, the type I Sqr of Alcanivorax sp. MCCC1A05192 exhibits the highest similarity to the Sqr of Acidithiobacillus ferrooxidans, a strain that is evolutionarily remote from Alcanivorax.
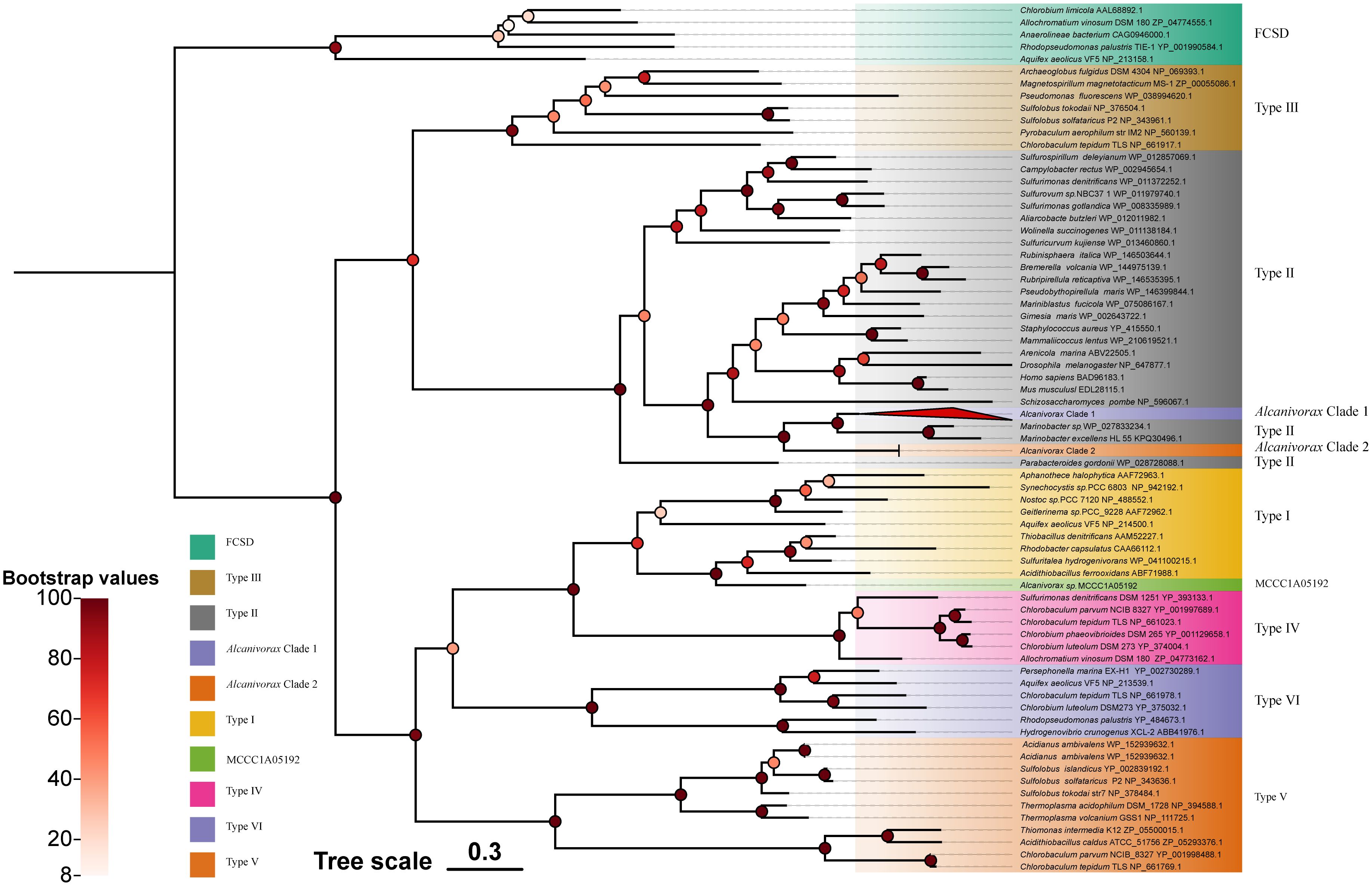
Figure 2. Phylogenetic tree of Alcanivorax and reference Sqr sequences. The different square blocks represent the FCSD, six-type Sqr, and Alcnivorax clades or strains. The red triangle block (Alcanivorax clade I) contains most of the Sqr sequence branches of Alcanivorax, counting 107.
Phylogenetic analyses revealed that the Sqr branches of Alcanivorax clustered, characterized by a high degree of Sqr similarity within the genus, indicating minimal variability and pronounced homology with Sqr originating from neighboring species.
Among the type II Sqr, the Sqr from chemoautotrophs and heterotrophic bacteria are clearly divided into two distinct branches. This separation suggests that type II Sqr enzymes may have evolved from two distinct common ancestors. Additionally, the Sqr of Alcanivorax originated from heterotrophic ancestors. Type II Sqr primarily perform detoxification functions in organisms, as has been demonstrated in mitochondria and diverse bacterial species (Marcia et al., 2009; Powell et al., 2017; Szabo, 2018). Although type II Sqr are also present in chemoautotrophic sulfide bacteria, their genomes are also characterized by other types of Sqr, such as type IV-V Sqr, which have been confirmed to support chemoautotrophic growth when sulfide serves as the sole electron donor (Han and Perner, 2015). Hence, it is difficult to determine whether type II Sqr provides sufficient energy to support the growth of Alcanivorax, which needs to be verified by wet experiments as follows.
The phylogenetic tree of TsdA shows that the TsdA genes of Alcanivorax are grouped into two distinct clades (Figure 3). Clade I, comprising 35 members, includes A. marinus, A. gelattinphagus, A. profundimaris, A. hongdengsis, and several unnamed strains. Clade II, with 36 members, encompasses A. xenomutans, A. mobilis, A. balearicus, and a subset of A. profundimaris. Notably, the most recent branches within clade I TsdA exclusively comprise chemoautotrophs that have been documented to be phototrophs (Kurth et al., 2016). However, it has been reported that the neighboring branches of the two clades are predominantly composed of TsdA originating from common heterotrophic sulfur-oxidizing taxa, which are distributed in distinct clusters compared to the TsdA of chemoautotrophs. After a thorough analysis of the protein cluster arrangements of TsdA in both classes, it became evident that the TsdA of these two classes exhibited significant differences in their genomic organization. Specifically, TsdB was observed upstream of TsdA in clade II, whereas its counterpart was conspicuously absent in clade I. Even though TsdA does not rely on TsdB or cytochrome c4 as an electron acceptor, some microorganisms can still function independently (Denkmann et al., 2012). The absence of TsdB in clade I attracted our attention, given that the TsdA of clade I are significantly longer (by 220 amino acid residues, Supplementary Materials 5) than the TsdA of clade II and the reported TsdA. Furthermore, the three neighboring branches harbor taxa in which TsdBA fusions have been documented (Kurth et al., 2016). A subsequent CD-search analysis of the conserved structures within certain sequences revealed that the initial approximately 200 amino acid residues of clade I belong to the Cyt553 superfamily, the same protein superfamily as TsdB (Lu et al., 2020). In conclusion, type I TsdA fused with TsdB, analogous to the phototrophic groups Thiocystis violascens, Thioflavicoccus mobilis, and Marichromatium purpuratum (Kurth et al., 2016; Yu, 2022). This marks the first discovery of two TsdA types within a single genus and even within a single strain (A. profundimaris).
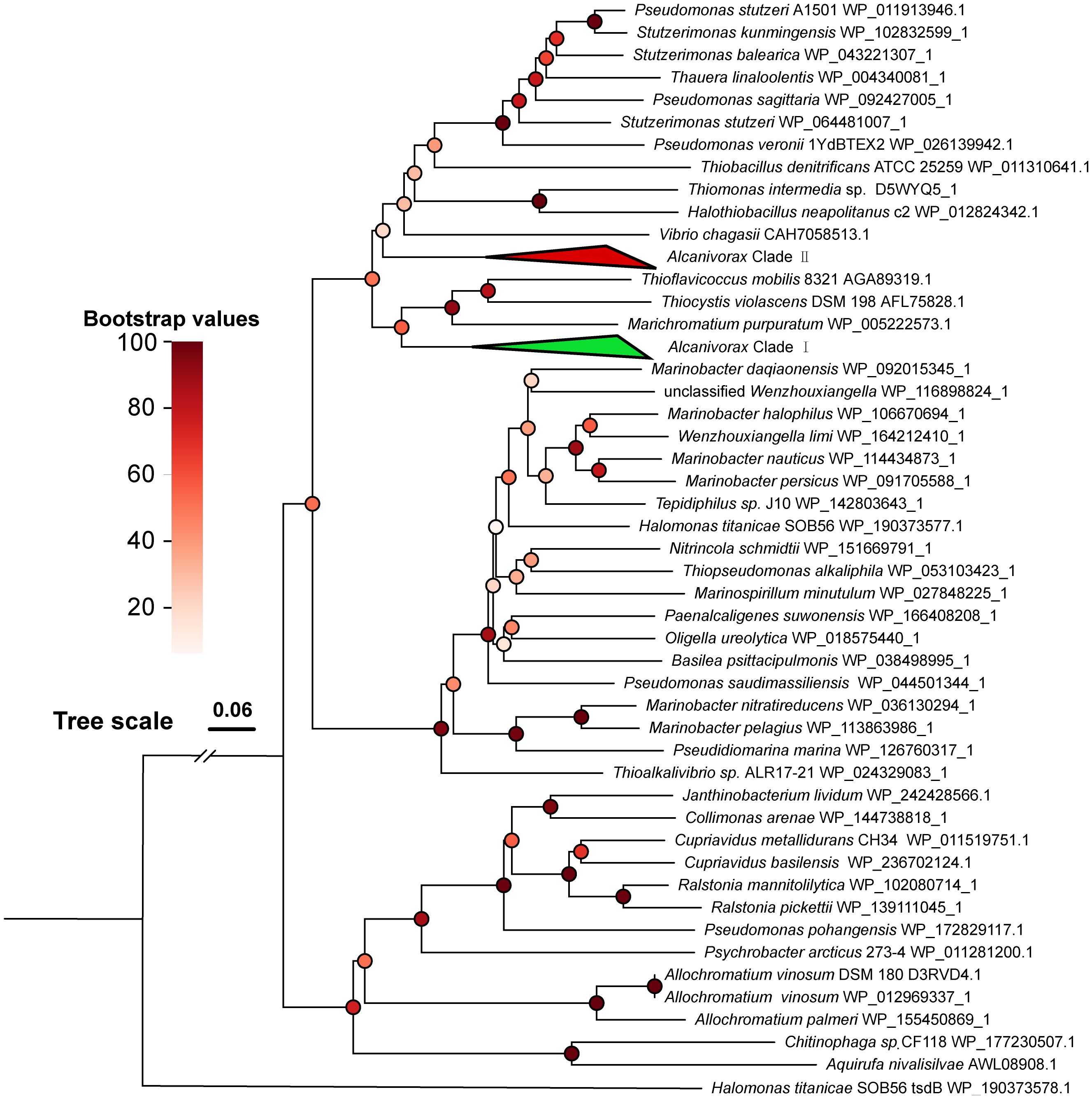
Figure 3. Phylogenetic tree of Alcanivorax and reference TsdA sequences. The green triangle block (Alcanivorax clade I) contains 35 branches of Alcanivorax, including TsdA of A. marinus, A. gelattinphagus, A. profundimaris, and A. hongdengsis. The red triangle block (Alcanivorax Clade II) contains 36 branches of Alcanivorax, including Tsd(B)A of A. xenomutans, A. mobilis, A. balearicus, and a subset of A. profundimaris.
Despite the seemingly diverse origins of the two types of TsdA found in Alcanivorax, they exhibit remarkable structural similarity and perform analogous functions with minor variations in their enzymatic characteristics (Brito et al., 2015; Kurth et al., 2016; Jenner et al., 2019). When Tsd(B)A oxidates thiosulfate, the two electrons that are produced are typically presumed to be relayed to TsdB or HiPIP proteins, subsequently conveyed to cytochrome cbb3, and ultimately integrated into the respiratory chain. The electron transfer process of the fusion variant of Tsd(B)A not only involves direct transport to cytochrome cbb3 but also has the potential to integrate into the electron chain of the photosynthetic reaction (Kurth et al., 2016). Despite these diligent searches, no photosynthetic pigment genes have been discovered in Alcanivorax, thus eliminating the prospect of its involvement in photosynthetic reactions for electron transfer. As a result, the electron transfer mechanism for both types of Tsd(B)A in Alcanivorax is expected to be analogous, ultimately culminating in their integration into the respiratory chain to generate reducing power, which then provides energy for growth. In M. purpuratum, studies have demonstrated that the fused type of Tsd(B)A exhibits a heightened affinity for substrates (Kurth et al., 2016), suggesting that the presence of various Tsd(B)A types may be essential for accommodating different thiosulfate concentrations.
Heterotrophic sulfur oxidation assays by representative strains
Six representative strains of Alcanivarax were selected to confirm the physiologic functions of Sqr and TsdA in media including artificial seawater, sulfide (or thiosulfate), and organic matter (acetic and yeast, or hexadecane). The sulfide oxidation experiment showed that all six strains were capable of promoting sulfide oxidation in an aerobic environment. Moreover, the efficiency of sulfide oxidation was comparable among all six strains, as sulfide was reduced from 1 mM to approximately 0.3 mM by each strain, while the noninoculated group still retained 0.5 mM sulfide within 60 minutes. After 180 minutes, 1 mM sulfide was completely consumed by the strains, whereas the noninoculated group still contained 0.1 mM residual sulfide (Figure 4).
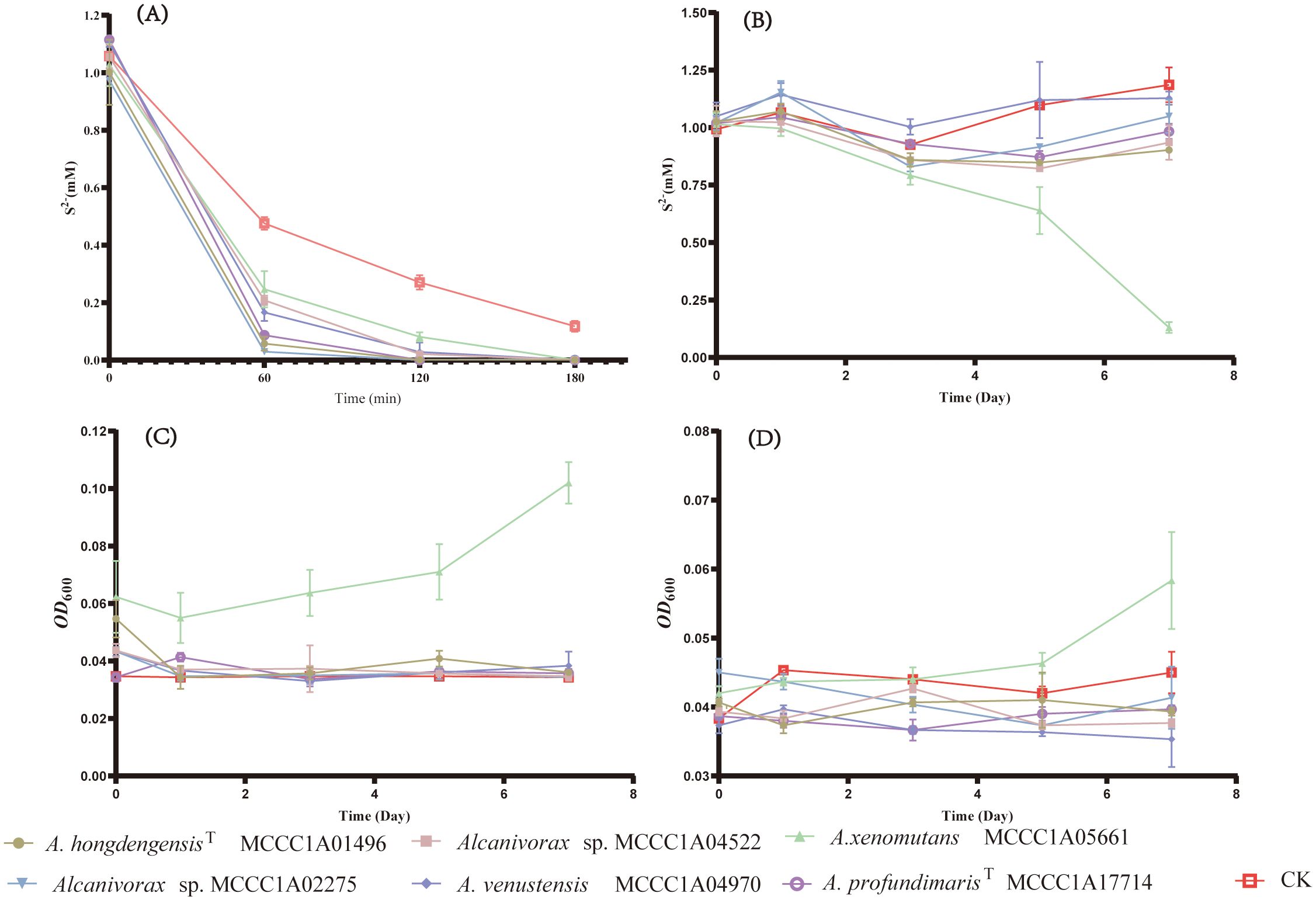
Figure 4. The growth and sulfide oxidation of representative strains under aerobic and anaerobic conditions. (A) Changes in the concentration of aerobic heterotrophic sulfide, (B) Changes in the concentration of anaerobic heterotrophic sulfide, (C) Changes in the anaerobic heterotrophic OD600 without sulfide, (D) Changes in the anaerobic heterotrophic OD600 with 1mM sulfide.
Under heterotrophic anaerobic conditions, A. xenomutans MCCC1A05661 was the sole strain exhibiting growth and sulfide oxidation capabilities. For seven days, the sulfide concentration markedly decreased by 0.88 mM, accompanied by a corresponding increase in biomass, as indicated by the increase in the OD600, which increased from 0.04 to 0.06. Conversely, the OD600 of the control group, which did not receive any sulfide, exhibited a more significant increase, reaching 0.1 of the OD600. After seven days, the sulfide concentration and OD600 values of the other five strains, as well as those of the blank control group, remained almost unchanged. Based on the quantification results of A. xenomutans MCCC1A05661, we observed a substantial inhibitory impact on the growth of A. xenomutans MCCC1A05661 (Supplementary Figure 2). This suggests that sulfide exerts a specific toxic effect during the growth phase of Alcanivorax. While sulfide may release a certain quantity of energy through the oxidation process by Sqr, its harmful impact on cellular toxicity significantly outweighs its marginal positive influence on cell growth.
Therefore, the primary role of Sqr in Alcanivorax is to facilitate the detoxification of sulfide rather than contributing significantly to the growth of sulfur-oxidizing bacteria such as Beggiatoa, which are capable of utilizing sulfide as an energy source (Berg et al., 2014). Furthermore, the inability of the remaining five strains to grow is likely attributed to the absence of the Nar operon, which is essential for effectively utilizing nitrate as an electron acceptor (Stein, 2019). Given the experimental conditions outlined in this study, it is highly probable that these five strains lack suitable electron acceptors.
The oxidative ability of thiosulfate was observed under both heterotrophic and chemoautotrophic conditions. Among the heterotrophic cultures with sodium acetate and yeast, A. xenomutans MCCC1A05661, A. venstensis MCCC1A04970, Alcanivorax sp. MCCC1A02275 and A. profundimarisT MCCC1A7714 were able to oxidize thiosulfate, whereas A. hongdengensisT MCCC1A01496 and Alcanivorax sp. MCCC1A04522 exhibited no such activity. Notably, A. xenomutans MCCC1A05661 showed the lowest ability to oxidize thiosulfate under heterotrophic (acetate and yeast) conditions. Within 48 hours, A. xenomutans MCCC1A05661 consumed 1 mM thiosulfate and produced 0.5 mM tetrathionate. In contrast, the other species consumed 5 to 8 mM thiosulfate and generated 2 to 4 mM tetrathionate within the same period (Figures 5A–C). However, when hexadecane served as the sole source of carbon and energy, all strains exhibited the ability to oxidize thiosulfate. They consumed 5 mM thiosulfate and produced 2 to 3 mM tetrathionate within 2 days (Figures 5D–F). Notably, under heterotrophic conditions, the addition of thiosulfate significantly enhanced the growth of the bacteria, regardless of whether sodium acetate with yeast or hexadecane was the primary carbon source (Supplementary Figures 3, 4).
In heterotrophic sulfur oxidation experiments, the introduction of inorganic energy sources has been shown to stimulate bacterial growth. This finding implies that Alcanivorax may belong to a neglected class of chemolithotrophs rather than heterotrophs. The utilization of sulfur oxidation as an auxiliary energy source could explain the prevalent abundance of Alcanivorax in sulfur-rich environments, such as hydrothermal fluids (Salazar et al., 2016; Wang et al., 2020; Dede et al., 2023);. Although it has been postulated that TsdA-mediated oxidation of thiosulfate results in an increase in pH, thereby indirectly fostering bacterial growth, we cannot discount this potential effect. However, it is noteworthy that the electrons generated during this oxidation process ultimately feed into the respiratory chain, supplying a considerable amount of energy. The impact of this energy derived from thiosulfate oxidation, along with changes in culture conditions such as pH, varies among different bacterial strains (Dlu, 2003; Hutt, 2017). It is now generally accepted that the energy derived from TsdA-mediated thiosulfate oxidation, in the absence of soxB, is insufficient to sustain bacterial growth and is considered a supplementary energy source (Denkmann et al., 2012). This finding aligns with the results observed under the culture conditions established in the current study. However, following the silencing of the soxB gene in Paracoccus thiocyanatus, which naturally harbors both the TsdA and SoxB oxidation systems, the strain showed typical features of TsdA-mediated thiosulfate oxidation. Notably, the cell density initially increased from approximately 5×106 cells/mL to 5×107 cells/mL but subsequently decreased to 1×107 cells/mL upon complete consumption of thiosulfate (Rameez et al., 2020). Hence, despite the previous assumption that the energy produced by TsdA oxidation of thiosulfate solely functions as a supplementary energy source, the potential for this process to support bacterial growth remains a topic of discussion (Denkmann et al., 2012; Hutt, 2017).
Oligotrophic growth and inorganic carbon fixation in Alcanivorax
By exploring the potential for Alcanivorax to utilize thiosulfate for chemoautotrophic growth, we discovered that the A. xenomuants MCCC1A05661 and A. profundmarisT MCCC1A7714 displayed thiosulfate oxidation ability, but others did not exhibit significant alterations in thiosulfate or tetrathionate levels. A. xenomuants MCCC1A05661 consumed approximately 8 mM thiosulfate and converted it to 4 mM tetrathionate, and A. profundmarisT MCCC1A7714 exhibited a similar pattern, utilizing 15 mM thiosulfate to produce 7 mM tetrathionate in 10 days (Figures 5H, I). The cells underwent a remarkable increase in cell density during 10 days of chemoautotrophic culture, growing from approximately 1.0×107 cells/L to approximately 5.0×108 cells/L. However, others did not show any notable changes in cell numbers (Figure 5G). It seems that the A. xenomuants MCCC1A05661 and A. profundmarisT MCCC1A7714 utilize thiosulfate for chemoautotrophic growth. However, the results of 13C isotope analysis and total organic carbon (TOC) measurements fail to substantiate the possibility of chemoautotrophic growth via thiosulfate oxidation. Despite the isotope experiments indicating that a fraction of inorganic carbon entered the cytosol, the assimilation rate increased only marginally from 1.1% to 1.3% (Figure 6A). This percentage is even lower than the assimilation rate of inorganic carbon by Bacillus subtilis under heterotrophic conditions, which is 5% (Braun et al., 2021). Hence, the assimilation of inorganic carbon likely occurs through a replenishment pathway, which usually occurs in heterotrophs. The TOC results showed that after 21 days of culture, the organic matter concentration increased from 2 ppm to 15 ppm in the CK group (Figure 6B), suggesting that these two strains were oligotrophs. These observations suggest that the carbon source sustaining the growth of A. xenomuants MCCC1A05661 and A. profundmarisT MCCC1A7714 under the specified chemoautotrophic conditions is not primarily inorganic carbon, most likely due to air pollution.
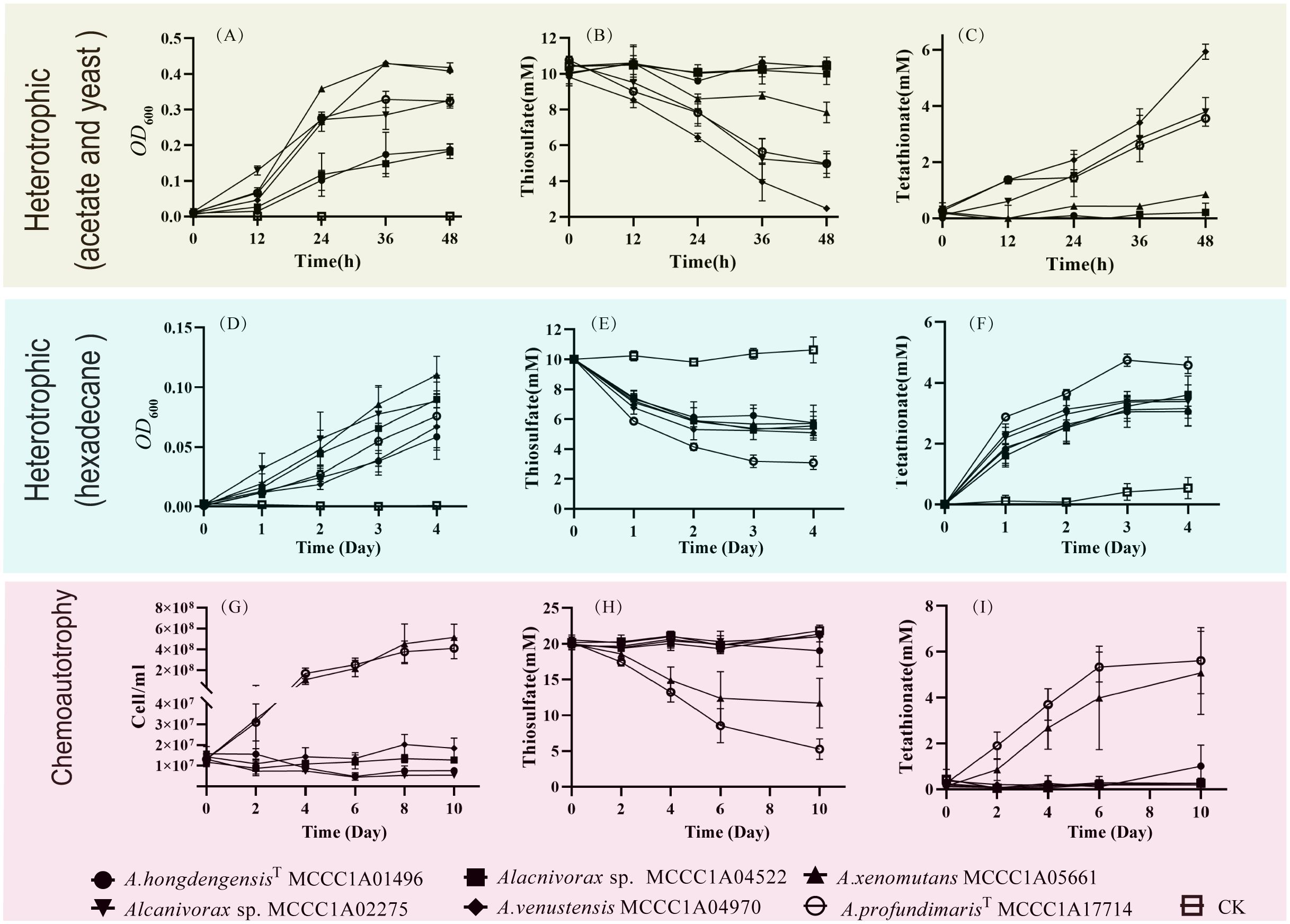
Figure 5. The growth and thiosulfate oxidation of Alcanivorax under different conditions. (A–C) Changes in OD600, thiosulfate, and tetrathionate in acetate and yeast heterotrophic sulfur-oxidizing media; (D–F) Changes in OD600, thiosulfate, and tetrathionate under hexadecane heterotrophic conditions; (G–I) Changes in cell concentration, thiosulfate and tetrathionate in chemoautotrophic media.
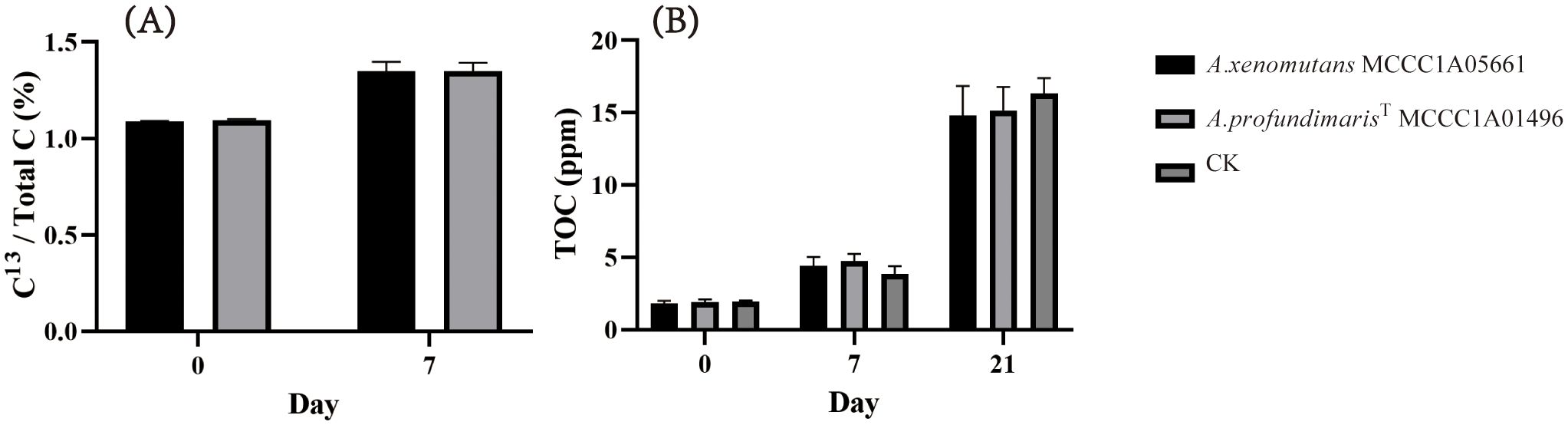
Figure 6. Carbon fixation ability of A xenomuants MCCC1A05661 and A profundmarisT MCCC1A7714. (A) δ13C assimilation, (B) TOC changes under chemoautotrophic conditions.
Although the A. xenomuants MCCC1A05661 and A. profundmarisT MCCC1A7714 cannot efficiently fix inorganic carbon, they exhibit remarkable resilience, demonstrating their capacity to thrive in extremely oligotrophic conditions, even at concentrations of organic matter as low as 5 ppm or below. Notably, similar results were obtained for other strains of A. xenomuants and A. profundmaris (data not show). However, the other strains lacked similar adaptability. This suggests that the capacity to proliferate under extreme oligotrophic conditions may not be a universal trait among Alcanivorax species; rather, it may indicate species or substrate specificity. Consequently, the presence of distinct trace organics in various regions may significantly contribute to the prevalence of specific Alcanivorax types in the pelagic environment.
Summary and conclusions
This study investigated the metabolic characteristics of Alcanivorax species in marine environments, particularly their ability to utilize inorganic compounds. The genes associated with sulfur oxidation, sqr and tsdA, were prevalent, and their distribution was closely tied to species evolution rather than habitat.
Experimental validations confirmed that the addition of thiosulfate can enhance Alcanivorax growth, suggesting its potential as a supplementary energy source. However, Alcanivorax bacteria primarily use Sqr for sulfide detoxification rather than for energy acquisition through sulfide oxidation. Although no strains were confirmed as chemoautotrophs, certain lineages, A. xenomutans and A. profundimaris, can grow under conditions with very low organic matter. The potential for chemolithotrophy and carbon oligotrophy likely drives the widespread distribution of Alcanivorax. Further research is needed to fully understand the mechanisms underlying this adaptation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZC: Writing – review & editing, Writing – original draft, Software, Methodology. SX: Writing – review & editing, Data curation. YL: Writing – original draft. QL: Writing – review & editing. CD: Writing – review & editing. JL: Writing – review & editing. GL: Writing – review & editing, Writing – original draft, Methodology, Funding acquisition. ZS: Writing – review & editing, Writing – original draft, Methodology, Data curation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (Nos. 42030412 and 32200045), Scientific Research Foundation of Third Institute of Oceanography, MNR (No. 2023006), the Natural Science Foundation of Xiamen (No.3502Z20227089), National Key R&D Program of China (2022YFC2804003).
Acknowledgments
Thanks MCCC for bacterial preservation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1491690/full#supplementary-material
References
Abdellatif L., Ben-Mahmoud O. M., Yang C., Hanson K. G., Gan Y., Hamel C. (2017). The H2-oxidizing Rhizobacteria associated with field-grown lentil promote the growth of lentil Inoculated with Hup+ Rhizobium through multiple modes of action. J. Plant Growth Regul. 36, 348–361. doi: 10.1007/s00344-016-9645-7
Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. (1979). Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43, 260–296. doi: 10.1128/mr.43.2.260-296.1979
Baltar F., Alvarez-Salgado X. A., Arístegui J., Benner R., Hansell D. A., Herndl G. J., et al. (2021). What is refractory organic matter in the ocean? Frot Mar. Sci. 8. doi: 10.3389/fmars.2021.642637
Berg J. S., Schwedt A., Kreutzmann A. C., Kuypers M. M., Milucka J. (2014). Polysulfides as intermediates in the oxidation of sulfide to sulfate by Beggiatoa spp. Appl. Environ. Microbiol. 80, 629–636. doi: 10.1128/AEM.02852-13
Bhattacharya S., Roy C., Mandal S., Sarkar J., Rameez M. J., Mondal N., et al. (2020). Aerobic microbial communities in the sediments of a marine oxygen minimum zone. FEMS Microbiol. Lett. 367, fnaa157. doi: 10.1093/femsle/fnaa157
Braun A., Spona-Friedl M., Avramov M., Elsner M., Baltar F., Reinthaler T., et al. (2021). Reviews and syntheses: Heterotrophic fixation of inorganic carbon – significant but invisible flux in global carbon cycling. Biogeosciences 18, 3689–3700. doi: 10.5194/bg-2020-465
Brito J. A., Denkmann K., Pereira I. A., Archer M., Dahl C. (2015). Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290, 9222–9238. doi: 10.1074/jbc.M114.623397
Cerro-Gálvez E., Dachs J., Lundin D., Fernández-Pinos M.-C., Sebastián M., Vila-Costa M. (2021). Responses of coastal marine microbiomes exposed to anthropogenic dissolved organic carbon. Environ. Sci. Ecotechnol 55, 9609–9621. doi: 10.1021/acs.est.0c07262
Chernikova T. N., Bargiela R., Toshchakov S. V., Shivaraman V., Lunev E. A., Yakimov M. M., et al. (2020). Hydrocarbon-degrading bacteria Alcanivorax and Marinobacter associated with microalgae Pavlova lutheri and Nannochloropsis oculata. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.572931
Correa S. S., Schultz J., Lauersen K. J., Rosado A. S. (2023). Natural carbon fixation and advances in synthetic engineering for redesigning and creating new fixation pathways. J. Adv. Res. 47, 75–92. doi: 10.1016/j.jare.2022.07.011
Cuevasanta E., Möller M. N., Alvarez B. (2017). Biological chemistry of hydrogen sulfide and persulfides. Arch. Biochem. Biophys. 617, 9–25. doi: 10.1016/j.abb.2016.09.018
Dahl C., Engels S., Pott-Sperling A. S., Schulte A., Sander J., Lübbe Y., et al. (2005). Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol 187, 1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005
Dede B., Hansen C. T., Neuholz R., Schnetger B., Kleint C., Walker S., et al. (2022). Niche differentiation of sulfur-oxidizing bacteria (SUP05) in submarine hydrothermal plumes. ISME J. 16, 1479–1490. doi: 10.1038/s41396-022-01195-x
Dede B., Priest T., Bach W., Walter M., Amann R., Meyerdierks A. (2023). High abundance of hydrocarbon-degrading Alcanivorax in plumes of hydrothermally active volcanoes in the South Pacific Ocean. ISME J. 17, 600–610. doi: 10.1038/s41396-023-01366-4
Denkmann K., Grein F., Zigann R., Siemen A., Bergmann J., Van Helmont S., et al. (2012). Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x
Dlu S. (2003). Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72, 641–653. doi: 10.1023/B:MICI.0000008363.24128.e5
Dong X., Zhang C., Peng Y., Zhang H. X., Shi L. D., Wei G., et al. (2022). Phylogenetically and catabolically diverse diazotrophs reside in deep-sea cold seep sediments. Nat. Commun. 13, 4885. doi: 10.1038/s41467-022-32503-w
Du R., Di G., Wang Y., Liu L., Cheng J., Liu J., et al. (2022). Heterotrophic sulfur oxidation of Halomonas titanicae SOB56 and its habitat adaptation to the hydrothermal environment. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.888833
Engel A., Kiko R., Dengler M. (2022). Organic matter supply and utilization in oxygen minimum aones. Ann. Rev. Mar. Sci. 14, 355–378. doi: 10.1146/annurev-marine-041921-090849
Garber A. I., Nealson K. H., Okamoto A., McAllister S. M., Chan C. S., Barco R. A., et al. (2020). FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00037
Han Y., Perner M. (2015). The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00989
Hedrich S., Schlömann M., Johnson D. B. (2011). The iron-oxidizing proteobacteria. Microbiology 157, 1551–1564. doi: 10.1099/mic.0.045344-0
Hutt L. P. (2017). Taxonomy, physiology and biochemistry of the sulfur bacteria (Plumoutn, UK: University of Plymouth). Doctoral dissertation.
Jenner L. P., Kurth J. M., Van Helmont S., Sokol K. P., Reisner E., Dahl C., et al. (2019). Heme ligation and redox chemistry in two bacterial thiosulfate dehydrogenase (TsdA) enzymes. J. Biol. Chem. 294, 18002–18014. doi: 10.1074/jbc.RA119.010084
Katayama T., Nobu M. K., Imachi H., Hosogi N., Meng X. Y., Morinaga K., et al. (2024). A marine group a isolate relies on other growing bacteria for cell wall formation. Nat. Microbiol. 9, 1954–1963. doi: 10.1038/s41564-024-01717-7
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kesava P. R., Raju R., Inga L., Chellandi M., Rathinam A. J. (2020). Genomic features and copper biosorption potential of a new Alcanivorax sp. VBW004 isolated from the shallow hydrothermal vent (Azores, Portugal). Genomics 115, 3268–3273. doi: 10.1016/j.ygeno.2020.06.015
Kolmert A., Wikström P., Hallberg K. B. (2000). A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J. Microbiol. Meth 41, 179–184. doi: 10.1016/s0167-7012(00)00154-8
Kurth J. M., Brito J. A., Reuter J., Flegler A., Koch T., Franke T., et al. (2016). Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. J. Biol. Chem. 291, 24804–24818. doi: 10.1074/jbc.M116.753863
Kuypers M. M. M., Marchant H. K., Kartal B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Lagkouvardos I., Joseph D., Kapfhammer M., Giritli S., Horn M., Haller D., et al. (2016). IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci. Rep. 23, 33721. doi: 10.1038/srep33721
Landry A. P., Ballou D. P., Banerjee R. (2021). Hydrogen sulfide oxidation by sulfide quinone oxidoreductase. Chembiochem 22, 949–960. doi: 10.1002/cbic.202000661
Liu C., Shao Z. (2005). Alcanivorax dieselolei sp. nov., a novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int. J. Syst. Evol. Microbiol. 55, 1181–1186. doi: 10.1099/ijs.0.63443-0
Love C. R., Arrington E. C., Gosselin K. M., Reddy C. M., Van Mooy B. A. S., Nelson R. K., et al. (2021). Microbial production and consumption of hydrocarbons in the global ocean. Nat. Microbiol. 6, 489–498. doi: 10.1038/s41564-020-00859-8
Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, 265–268. doi: 10.1093/nar/gkz991
Lv S., Cui K., Zhao S., Li Y., Liu R., Hu R., et al. (2024). Continuous generation and release of microplastics and nanoplastics from polystyrene by plastic-degrading marine bacteria. J. Hazard Mater Adv. 465, 133339. doi: 10.1016/j.jhazmat.2023.133339
Mall A., Sobotta J., Huber C., Tschirner C., Kowarschik S., Bačnik K., et al. (2018). Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359, 563–567. doi: 10.1126/science.aao2410
Marcia M., Ermler U., Peng G., Michel H. (2009). The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. PNAS 106, 9625–9630. doi: 10.1073/pnas.0904165106
Marcia M., Ermler U., Peng G., Michel H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78, 1073–1083. doi: 10.1002/prot.22665
Na S. I., Kim Y. O., Yoon S. H., Ha S. M., Baek I., Chun J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. doi: 10.1007/s12275-018-8014-6
Petersen J. M., Zielinski F. U., Pape T., Seifert R., Moraru C., Amann R., et al. (2011). Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476, 176–180. doi: 10.1038/nature10325
Powell C. R., Dillon K. M., Matson J. B. (2017). A review of hydrogen sulfide (H2S) donors: Chemistry and potential therapeutic applications. Biochem. Pharmacol. 149, 110–123. doi: 10.1016/j.bcp.2017.11.014
Quigley L. N. M., Edwards A., Steen A. D., Buchan A. (2019). Characterization of the interactive effects of labile and recalcitrant organic matter on microbial growth and metabolism. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00493
Rai A., Suresh G., Ria B., PK S., Ipsita S., Sasikala C., et al. (2023). Phylogenomic analysis of the genus Alcanivorax: Proposal for division of this genus into the emended genus Alcanivorax and two novel genera Alloalcanivorax gen. nov. and Isoalcanivorax gen. nov. Int. J. Syst. Evol. Microbiol. 73, 5672. doi: 10.1099/ijsem.0.005672
Rameez M. J., Pyne P., Mandal S., Chatterjee S., Alam M., Bhattacharya S., et al. (2020). Two pathways for thiosulfate oxidation in the alphaproteobacterial chemolithotroph Paracoccus thiocyanatus SST. Microbiol. Res. 230, 126345. doi: 10.1016/j.micres.2019.126345
Ran M., Li Q., Xin Y., Ma S., Zhao R., Wang M., et al. (2022). Rhodaneses minimize the accumulation of cellular sulfane sulfur to avoid disulfide stress during sulfide oxidation in bacteria. Redox Biol. 53, 102345. doi: 10.1016/j.redox.2022.102345
Salazar G., Cornejo-Castillo F. M., Benítez-Barrios V., Fraile-Nuez E., Álvarez-Salgado X. A., Duarte C. M., et al. (2016). Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 10, 596–608. doi: 10.1038/ismej.2015.137
Sánchez-Andrea I., Guedes I. A., Hornung B., Boeren S., Lawson C. E., Sousa D. Z., et al. (2020). The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat. Commun. 11, 5090. doi: 10.1038/s41467-020-18906-7
Steffens L., Pettinato E., Steiner T. M., Mall A., König S., Eisenreich W., et al. (2021). High CO2 levels drive the TCA cycle backwards towards autotrophy. Nature 592, 784–788. doi: 10.1038/s41586-021-03456-9
Stein L. Y. (2019). Insights into the physiology of ammonia-oxidizing microorganisms. Curr. Opin. Chrm Biol. 49, 9–15. doi: 10.1016/j.cbpa.2018.09.003
Szabo C. (2018). A timeline of hydrogen sulfide (H2S) research: From environmental toxin to biological mediator. Biochem. Pharmacol. 149, 5–19. doi: 10.1016/j.bcp.2017.09.010
Tamura K., Stecher G., Kumar S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Trüper H. G., Schlegel H. G. (1964). Sulphur metabolism in thiorhodaceae I. quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 30, 225–238. doi: 10.1007/BF02046728
Wang W., Li Z., Zeng L., Dong C., Shao Z. (2020). The oxidation of hydrocarbons by diverse heterotrophic and mixotrophic bacteria that inhabit deep-sea hydrothermal ecosystems. ISME J. 14, 1994–2006. doi: 10.1038/s41396-020-0662-y
Wang H., Liu S., Liu X., Li X., Wen Q., Lin J. (2014). Identification and characterization of an ETHE1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl. Microbiol. Biot 98, 7511–7522. doi: 10.1007/s00253-014-5830-4
Wei M., Zeng X., Han X., Shao Z., Xie Q., Dong C., et al. (2022). Potential autotrophic carbon-fixer and Fe(II)-oxidizer Alcanivorax sp. MM125-6 isolated from Wocan hydrothermal field. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.930601
Wu W., Pang X., Lin J., Liu X., Wang R., Lin J., et al. (2017). Discovery of a new subgroup of sulfur dioxygenases and characterization of sulfur dioxygenases in the sulfur metabolic network of Acidithiobacillus caldus. PloS One 12, e0183668. doi: 10.1371/journal.pone.0183668
Xia Y., Lü C., Hou N., Xin Y., Liu J., Liu H., et al. (2017). Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 11, 2754–2766. doi: 10.1038/ismej.2017.125
Yakimov M. M., Golyshin P. N., Lang S., Moore E. R. B., Abraham W. R., Lünsdorf H., et al. (1998). Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 48, 339–348. doi: 10.1099/00207713-48-2-339
Yang S., Li M., Lai Q., Li G., Shao Z. (2018). Alcanivorax mobilis sp. nov., a new hydrocarbon-degrading bacterium isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 68, 1639–1643. doi: 10.1099/ijsem.0.002612
Yu Q. (2022). Studies on the thiosulfate oxidation pathway in sulfur-reducing Shewanella oneidensis (Zhejiang, China: Zhejiang University). Doctoral dissertation.
Zhang J., Liu R., Xi S., Cai R., Zhang X., Sun C. (2020). A novel bacterial thiosulfate oxidation pathway provides a new clue about the formation of zero-valent sulfur in deep sea. ISME J. 14, 2261–2274. doi: 10.1038/s41396-020-0684-5
Keywords: Alcanivorax, chemolithotrophic, carbon oligotrophic, sulfur oxidation, TsdA, Sqr
Citation: Chen Z, Xiang S, Lu Y, Lai Q, Dong C, Li J, Li G and Shao Z (2024) Adaptation mechanisms of Alcanivorax facilitating its predominance in marine environments. Front. Mar. Sci. 11:1491690. doi: 10.3389/fmars.2024.1491690
Received: 05 September 2024; Accepted: 28 October 2024;
Published: 15 November 2024.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Aparna Chakkamadathil Rajeev, Kerala Forest Research Institute, IndiaSuping Yang, Huaqiao University, China
Copyright © 2024 Chen, Xiang, Lu, Lai, Dong, Li, Li and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guizhen Li, bGlndWl6aGVuLm9rQDE2My5jb20=; Zongze Shao, c2hhb3p6QDE2My5jb20=
 Zhen Chen
Zhen Chen Shizheng Xiang
Shizheng Xiang Yao Lu
Yao Lu Qiliang Lai
Qiliang Lai Chunming Dong
Chunming Dong Jianyang Li
Jianyang Li Guizhen Li
Guizhen Li Zongze Shao
Zongze Shao