- 1Fishery College, Zhejiang Ocean University, Zhoushan, Zhejiang, China
- 2Zhoushan Marine Workstation, East China Sea Branch of State Oceanic Administration, Zhoushan, Zhejiang, China
Global climate change has caused rapid temperature changes in marine environments. Understanding how marine organisms respond to temperature changes can help predict their richness of future biodiversity. In this study, we examined the gene expression levels and the difference in the pathways that are responsive to acute temperature stress in low- and high-latitude populations of the shore swimming crab, Charybdis japonica. The two populations of C. japonica were exposed to low- and high-temperature stresses (15°C and 28°C) and used for transcriptome sequencing. Genetic regulatory ability changes were compared to determine the diverse response of the two crab populations to temperature change. The gene expression levels and functional enrichment analysis showed that the low-latitude crab regulated more genes (938) that were mainly enriched in DNA replication and metabolic pathways, whereas the high-latitude crab regulated less genes (309) that were mainly enriched in genetic information processing at low-temperature stress. Furthermore, the low-latitude crab regulated less genes (33) that were mainly enriched in genetic information processing, whereas the high-latitude crab regulated more genes (280) that were mainly enriched in signal transduction and cellular process at high-temperature stress. These results implied that the low-latitude population was more resilient to high-temperature stress, while the high-latitude population was more resilient to low-temperature stress. This study enhances our understanding of how different geographic C. japonica populations respond to varying temperature environments in their living zone, which could be helpful for predicting future biodiversity trends of intertidal crustaceans under global climate change.
1 Introduction
With the global climate change, land temperatures have been exhibiting significant year-to-year fluctuations (Solomon et al., 2007; Bopp et al., 2013). As an important environmental factor, temperature variation has a remarkable influence on the intrinsic physiological state of organisms (Pörtner and Farrell, 2008; Hofmann and Todgham, 2010; Zhang et al., 2020). However, the ocean as another habitat for living organisms, its environmental temperature also has a significant effect to the marine organisms (Dissanayake and Ishimatsu, 2011; Johansen et al., 2014; Greenberg and Palen, 2021). As we all known, although the specific heat capacity of sea water is higher than terrenes, the temperature in it also variation caused by many factors such as latitude, tides and circadian rhythms (Edwards and Richardson, 2004; Perry et al., 2005). Thus, the survival and sustainable development of marine organisms are under serious threat by extreme variations in water temperature (Bopp et al., 2013). To date, many studies have shown that the temperature fluctuations of the marine environment affect the reproductive, swimming, feeding, and migration behavior and distribution of marine organisms (Green and Fisher, 2004; Saucedo et al., 2004; Scott and Johnston, 2012; García Molinos et al., 2016; Poloczanska et al., 2016; Boyd et al., 2018). To deal with variations in water temperature, marine organisms can alleviate the negative effect of temperature stress through physiological adaptability (Yang et al., 2022; Li et al., 2023). Thus, understanding the underlying molecular mechanism of physiological adaptation of marine organisms to temperature stress may help reveal their genetic adaptation to temperature variation and protect marine biological germplasm resources.
Gene expression regulation in marine organisms were thought to play a crucial role in the process of physiological adaptability (Oulhen et al., 2007). Previous studies demonstrated that many marine organisms, like fishes, mollusks, and crustaceans, have their own gene regulatory network in response to environmental temperature changes (Qian and Xue, 2016; Lyu et al., 2018; Ning et al., 2021; Shrestha et al., 2021; Jahan et al., 2022; Yang et al., 2022). In fish, the zebrafish Danio rerio can alleviate apoptosis induced by low-temperature stress through mitochondrial β-oxidation of hepatocytes to improve their frost resistance (Sun et al., 2021). In mollusk, Crassostrea gigas, Mytilus galloprovincialis, and Katelysia rhytiphora can improve the activity of antioxidant enzymes in hemocytes to activate the immune response under high-temperature stress (Rahman et al., 2019). In crustaceans, the porcelain crab Petrolisthes cinctipes can improve the expression of heat shock protein to adapt to thermal stress (Stillman and Tagmount, 2009). As a result, these organisms can produce different reactions to temperature changes. Furthermore, to adapt to temperature changes, different geographical groups of marine species may have varying coping strategies because the heterogeneity of long-term environmental temperature directly leads to the evolution of temperature adaptability among different biological groups (Thiyagarajan et al., 2003; Yannic et al., 2014). Previous studies have shown that two different populations of Japanese mantis shrimp Oratosquilla oratoria have dissimilar molecular stress responses under thermal stress, which the low-latitude mantis shrimps regulated the genes of metabolic pathways, whereas the high-latitude mantis shrimps regulated genes of genetic information processing, immune and environmental information processing (Lou et al., 2019). Furthermore, two different populations of manila clam Ruditapes philippinarum also have dissimilar molecular stress responses under acute temperature changes, which the low-latitude clams regulated the genes of xenobiotic metabolism pathway under both heat and cold stress, whereas the high-latitude clams regulated genes of encoding peroxisomes under heat stress (Jahan et al., 2022). Thus, studies on the differences between different geographical groups of organisms under varied temperature conditions are crucial.
The shore swimming crab, Charybdis japonica, belongs to the family Portunidae. It is a commercial marine crab in China that has high nutritional value (Yu et al., 2004). As an intertidal crustacean, C. japonica has a wide adaptation range to temperature (5-30 °C) and salinity (6.5-45.5‰) (Yu et al., 2004; Lou et al., 2022). Therefore, it is an ideal material for studying the response of organisms to temperature stress. Although a previous study has demonstrated that C. japonica can activate more genes under heat stress and these genes were mainly enriched in the endoplasmic reticulum pathway, whereas activate less genes under cold stress and these genes were mainly enriched in the insect hormone biosynthesis pathway (Lou et al., 2022), its gene regulatory ability of physiological adaptation to acute temperature changes in different geographic populations is unknown. In addition, verifying molecular regulatory mechanisms to acute temperature stress in different geographic populations would help us thoroughly understand the temperature adaptation of C. japonica populations and improve the resource management and protection of C. japonica under global climate change.
In this study, the low- and high-latitude populations of C. japonica were exposed to low- and high-temperature conditions. After acute temperature stress treatment, the muscle tissues of C. japonica were collected and sequenced. Subsequently, the differentially expressed genes (DEGs) and molecular pathways of related functional genes were analyzed to determine the molecular regulation mechanisms of different geographical groups of C. japonica to temperature stress. This study was the first to identify how the different geographical groups of C. japonica respond to short-term changes in ambient temperature. Our results can provide significant referential value for other crustaceans.
2 Materials and methods
2.1 C. japonica collection
Twenty-five healthy C. japonica individuals were collected in November 2022 from the coastal area of Zhoushan, Zhejiang Province (low-latitude group) and Dalian, Liaoning Province (high-latitude group). The crabs were transported to the laboratory at Zhejiang Ocean University. Before the experiment, the crab individuals of two different populations were distributed into three plastic aquariums (length × width × height: 55 cm × 40 cm × 35 cm) and acclimated in sand-filtered seawater with temperature of 20°C and salinity of 25‰ for 48 h. During the temporary maintenance period, all crabs were not fed and oxygenated continuously to prevent any factors on subsequent experiments. The water of the plastic aquariums was also changed daily to ensure that the water was clean.
2.2 Temperature treatment and tissue sampling
In this study, two crab populations were randomly divided into low- and high-temperature stress groups and the control group. A total of six groups were set, namely, two control groups (ZSC and DLC) in which the water temperature was maintained at 20°C, two low-temperature stress groups (ZSL and DLL) in which the water temperature was maintained at 15°C, and two high-temperature stress groups (ZSH and DLH) in which the water temperature was maintained at 28°C. After the temperature stress treatment for 12 h, the crabs in the six groups were anesthetized using the MS-22 Tranquillizer (Henan Nanhua Qianmu biological technology Co., Ltd.), and the muscle tissue of the crabs was dissected immediately. There were three replicates for each group, and each replicate contained three crabs. Finally, the samples were frozen in liquid nitrogen and stored at −80°C for the subsequent experiments.
2.3 Total RNA extraction and transcriptome sequencing
Total RNA of 18 tissue samples was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The RNA samples were then subjected to strict quality control. The integrity of RNA was verified using 1% agarose gel electrophoresis. The concentration and quality of RNA were determined using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, San Jose, CA, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Eligible RNA samples were then used for subsequent transcriptome sequencing.
Approximately 1.5 μg of total RNA per sample was treated with the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following the manufacturer’s protocols to establish the pair-end RNA-Seq library. First, mRNA with poly A tail was enriched by Oligo (dT) magnetic beads, and the purified mRNA was randomly interrupted with divalent cations in NEB Fragmentation Buffer. Second, the first-strand cDNA and second-strand cDNA were synthesized under the M-MuLV reverse transcriptase system and DNA polymerase I system, respectively. Third, double-stranded cDNA was purified and repaired, and the poly A-tail was added. The 250–300bp cDNAs were screened using AMPure XP beads (Beckman Coulter, Beverly, USA), and PCR amplification was performed to obtain the final library. All RNA-seq libraries were quantified using Qubit 2.0 Fluorometer, diluted to 1.5 ng/µL, and purified using the Agilent Bioanalyzer 2100 system. Finally, the 18 cDNA libraries were used for de novo transcriptome sequencing on an Illumina HiSeq 2000 sequencer (Illumina, San Diego, CA, USA).
2.4 Assembly and annotation of transcriptome
The raw reads were trimmed using Trimmomatic-0.36 (Bolger et al., 2014), and the clean reads were subjected to quality control using FastQC (Andrews, 2010). After the low-quality reads (Phred score ≤ 5), non-polyA reads, unknown nucleotides (N ratio > 10%), and reads with adapters were removed, the high-quality clean RNA-Seq reads were aligned to the reference genome of C. japonica (NCBI BioProject database under BioProjectID PRJNA766329) using Hisat2 v2.0.5, and novel transcripts were generated using StingTie software (Kim et al., 2015). Furthermore, novel transcripts from each sample were integrated using Cuffmerge software and compared with the reference genome of C. japonica (Han et al., 2022). The nonredundant sequences were annotated in the Pfam database, SUPERFAMILY database, GO database, and KEGG database using HMMER 3.0 package and Blast2GO software.
2.5 Differential expression analysis
The gene expression levels of the C. japonica samples were calculated using RSEM software (Li and Dewey, 2011; Kim et al., 2015). FPKM (fragments per kilobase of the exon model per million mapped fragments) value was used to describe the gene expression level. The DEGs between the groups ZSC versus ZSH, ZSC versus ZSL, DLC versus DLH, and DLC versus DLL were identified using the DEG method (Robinson et al., 2010). The regulation of gene expression in each temperature treatment of C. japonica was determined using the Bioconductor package DEGSeq2 (Love et al., 2014), and |log2 fold change (FC)| > 1 and false discovery rate (FDR) < 0.05 were used as the threshold criteria for significant DEGs. Finally, a volcano plot was used to present the different numbers of DEGs of each group. We used GOseq and KOBAS to perform the GO and KEGG enrichment analyses of DEGs (Mao et al., 2005; Young et al., 2010).
2.6 Validation of transcriptomic data by quantitative real-time PCR
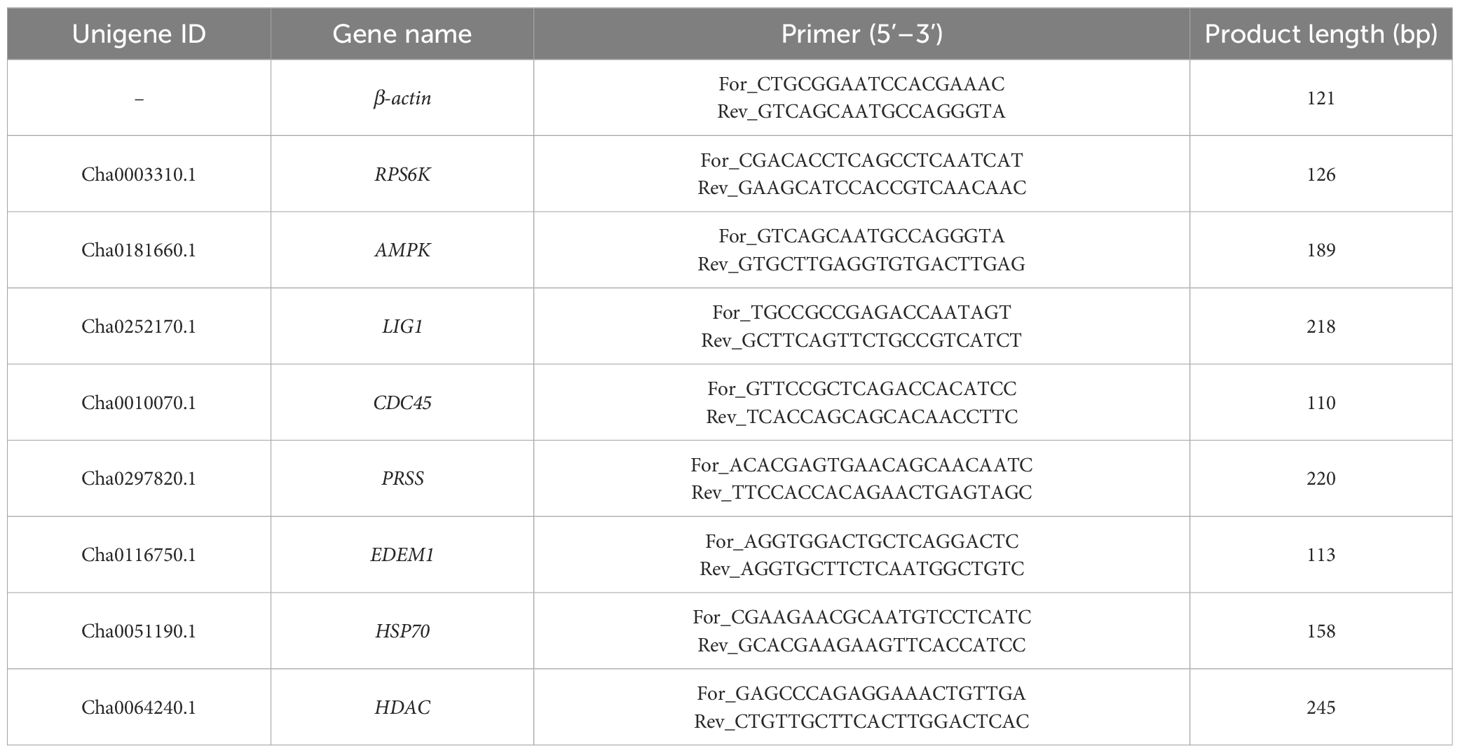
To validate the expression level of DEGs in RNA-seq data, we selected eight DEGs from the high- and low-temperature treatment groups for qRT-PCR analysis. The RNA samples of each treatment group were used to synthesize the cDNA using the Prime Script™ RT reagent Kit with gDNA Eraser (TaKaRa). The primers of the internal reference gene (β-actin) and eight DEGs were designed using Primer Premier 6.0 and are listed in Table 1. The reaction system of qRT-PCR was as follows: 10 μL of SYBR Green qPCR master mix, 0.5 μL of forward primer, 0.5 μL of reverse primer, 1 μL of cDNA, and 8 μL of RNase Free ddH2O. The reaction conditions were as follows: 95°C for 3 min, 40 cycles at 95°C for 10 s, and 58°C for 30 s. Three technical replicates were performed on each biological sample. Finally, the relative expression levels of each genes were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
3 Results
3.1 Sequencing data, assembly, and annotation of transcriptome
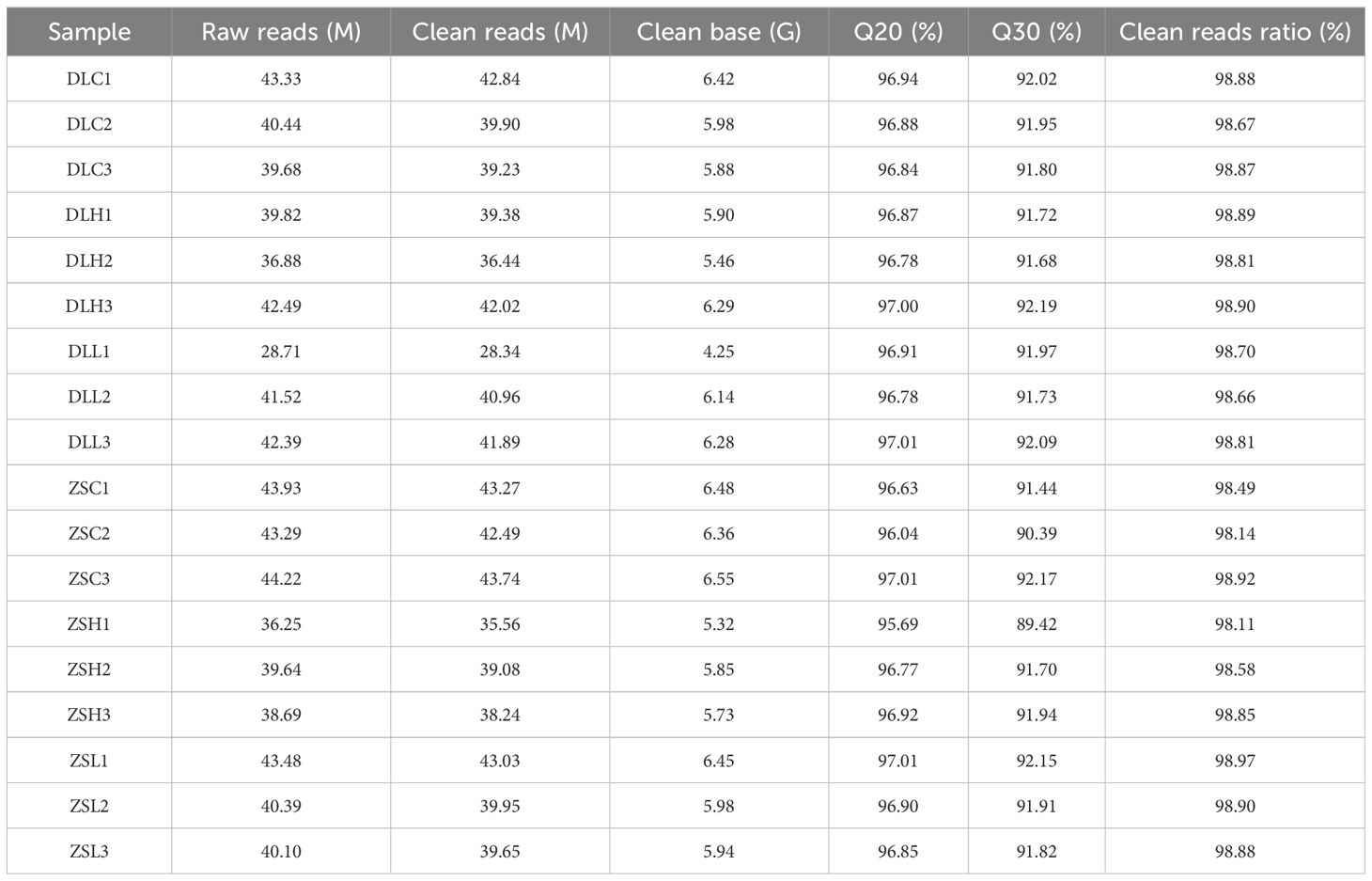
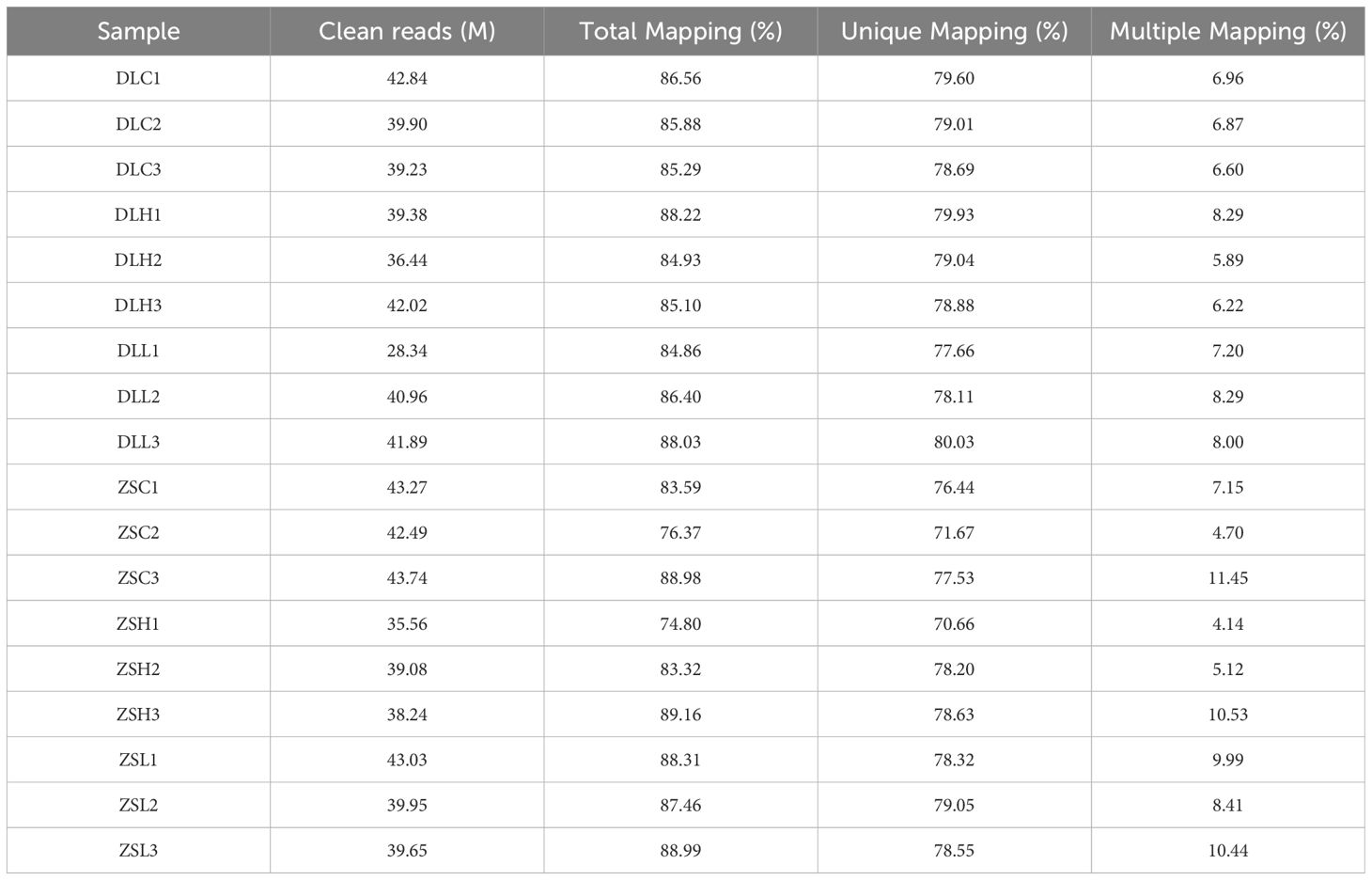
In this study, we obtained 107.26 Gb clean reads from 18 cDNA libraries after the quality control of the raw data. The Q30 (percentage of Phred quality score > 30) ratio of the clean reads ranged from 89.42% to 92.19%. A summary of sequencing data is shown in Table 2. After we aligned the clean reads to the reference genome of C. japonica, the mapping ratio of each sample ranged from 74.80% to 89.16% (Table 3). A total of 45,952 novel transcripts for annotated genes were obtained, including 21,453 coding transcripts and 24,499 noncoding transcripts (Table 4).
3.2 Quantitative analysis of DEGs
In both populations, the high-temperature treatment group (ZSH and DLH) and low-temperature treatment group (ZSL and DLL) were compared with the control group (ZSC and DLC). The DEGs in each population group were statistically analyzed. Under high-temperature stress, the results showed that 33 DEGs with 28 upregulated genes and 5 downregulated genes were found in the low-latitude population, and 280 DEGs with 84 upregulated genes and 196 downregulated genes were found in the high-latitude population (Figures 1A, C). Under low-temperature stress, the results showed that 938 DEGs with 68 upregulated genes and 870 downregulated genes were found in the low-latitude population, and 309 DEGs with 182 upregulated genes and 127 downregulated genes were found in the high-latitude population (Figures 1B, D). Venn-diagram analysis of the four comparisons showed that 28 DEGs were shared between two temperature treatments in Dalian population, seven DEGs were shared between two temperature treatments in Zhoushan population, 47 DEGs were shared between two populations under cold stress, and none DEGs were shared between two populations under heat stress (Figure 2; Supplementary Tables S1–S3).
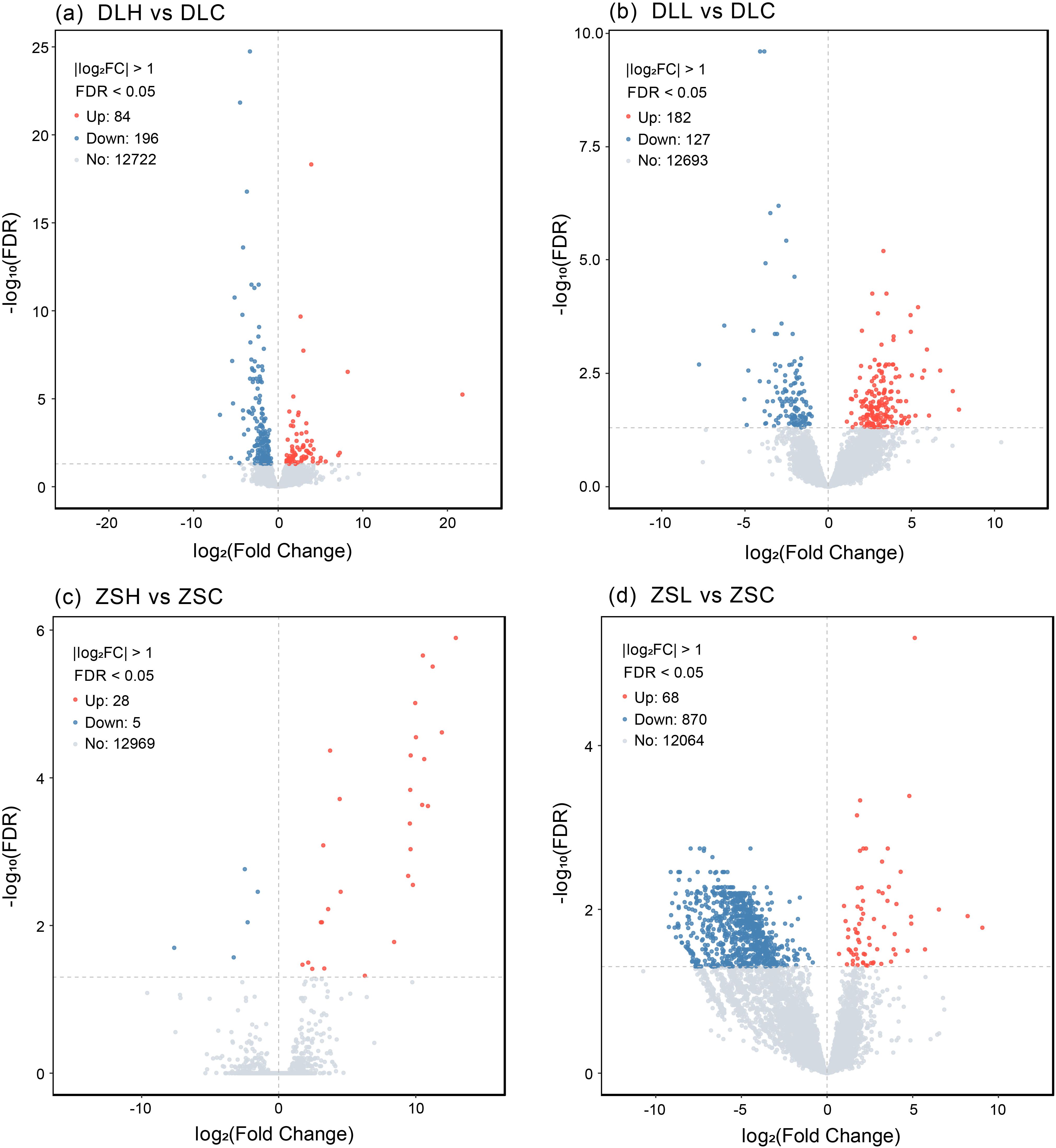
Figure 1. Volcanic maps of differentially expressed genes in different temperature groups of Zhoushan population and Dalian population. (A)Represent the DEGs between 20 and 28 °C in Dalian population; (B)represent the DEGs between 20 and 15 °C in Dalian population; (C)represent the DEGs between 20 and 28 °C in Zhoushan population; (D)represent the DEGs between 20 and 15 °C in Zhoushan population. DLH, Dalian population under high temperature stress; DLC, Dalian population control group; DLL, Dalian population under low temperature stress; ZSH, Zhoushan population under high temperature stress; ZSC, Zhoushan population control group; ZSL, Zhoushan population under low temperature stress; Up, upregulated genes; Down, downregulated genes; No, no significant difference.
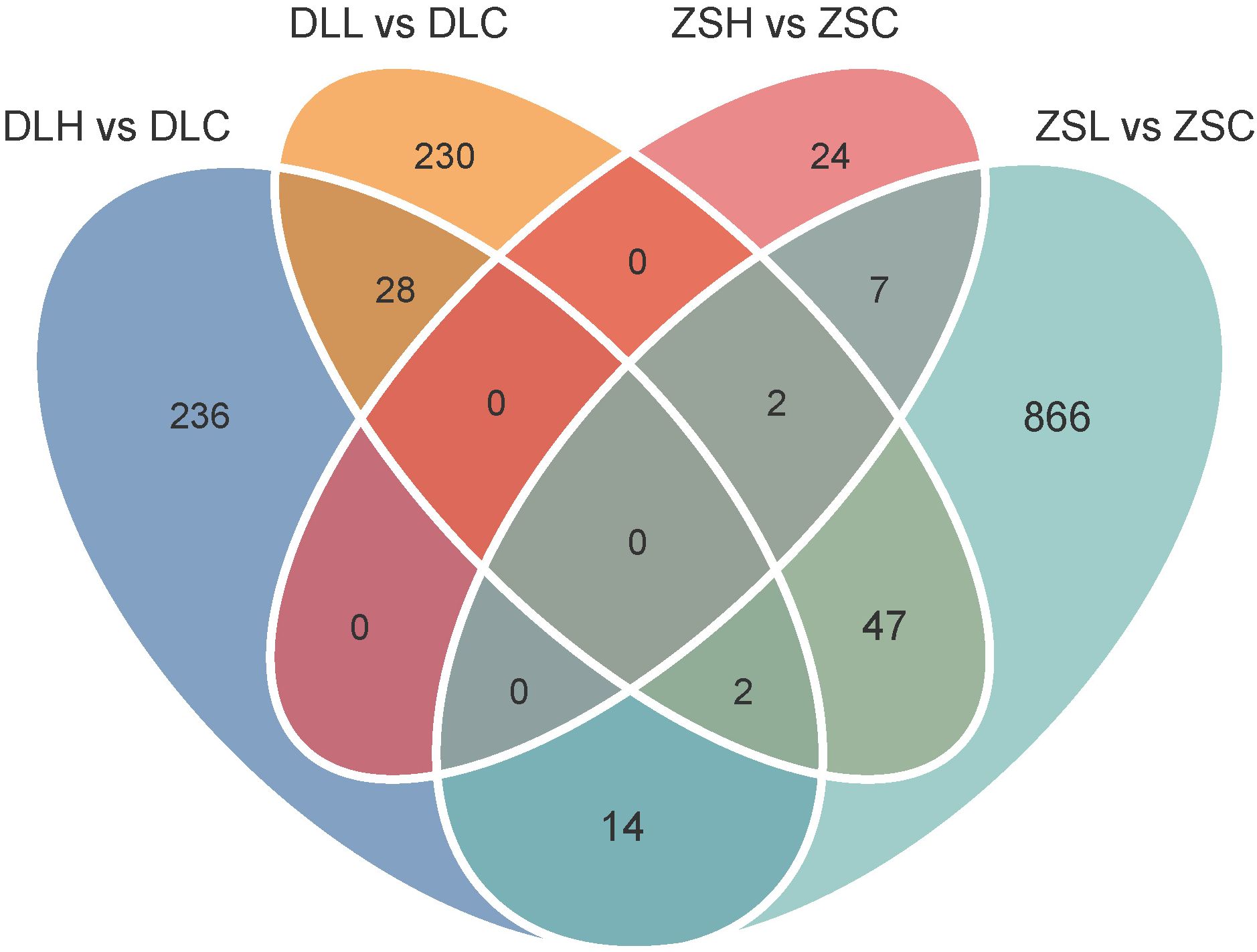
Figure 2. Venn diagrams showing unigenes significantly differentially expressed in all populations. Different colors indicate different comparison groups.
3.3 GO enrichment analysis of DEGs in C. japonica under different temperature stresses
The main biological functions of DEGs and their properties were determined by GO functional enrichment analysis (Figure 3). In the low-latitude population, compared with the ZSC group, the DEGs in the ZSH group were mainly annotated with the structural constituent of cuticle (GO:0042302) and DNA replication initiation (GO:0006270; Supplementary Table S4). The DEGs in the ZSL group were mainly annotated with the nucleosome (GO:0000786) and nucleus (GO:0005634; Supplementary Table S5). In the high-latitude population, compared with the DLC group, the DEGs in the DLH group were mainly annotated in protein kinase activity (GO:0004672), protein phosphorylation (GO:0006468), and steroid hormone receptor activity (GO:0003707; Supplementary Table S6). The DEGs in the DLL group were mainly annotated with extracellular space (GO:0005615), peptidyl-dipeptidase activity (GO:0008241), and sequence-specific DNA binding (GO:0043565; Supplementary Table S7).
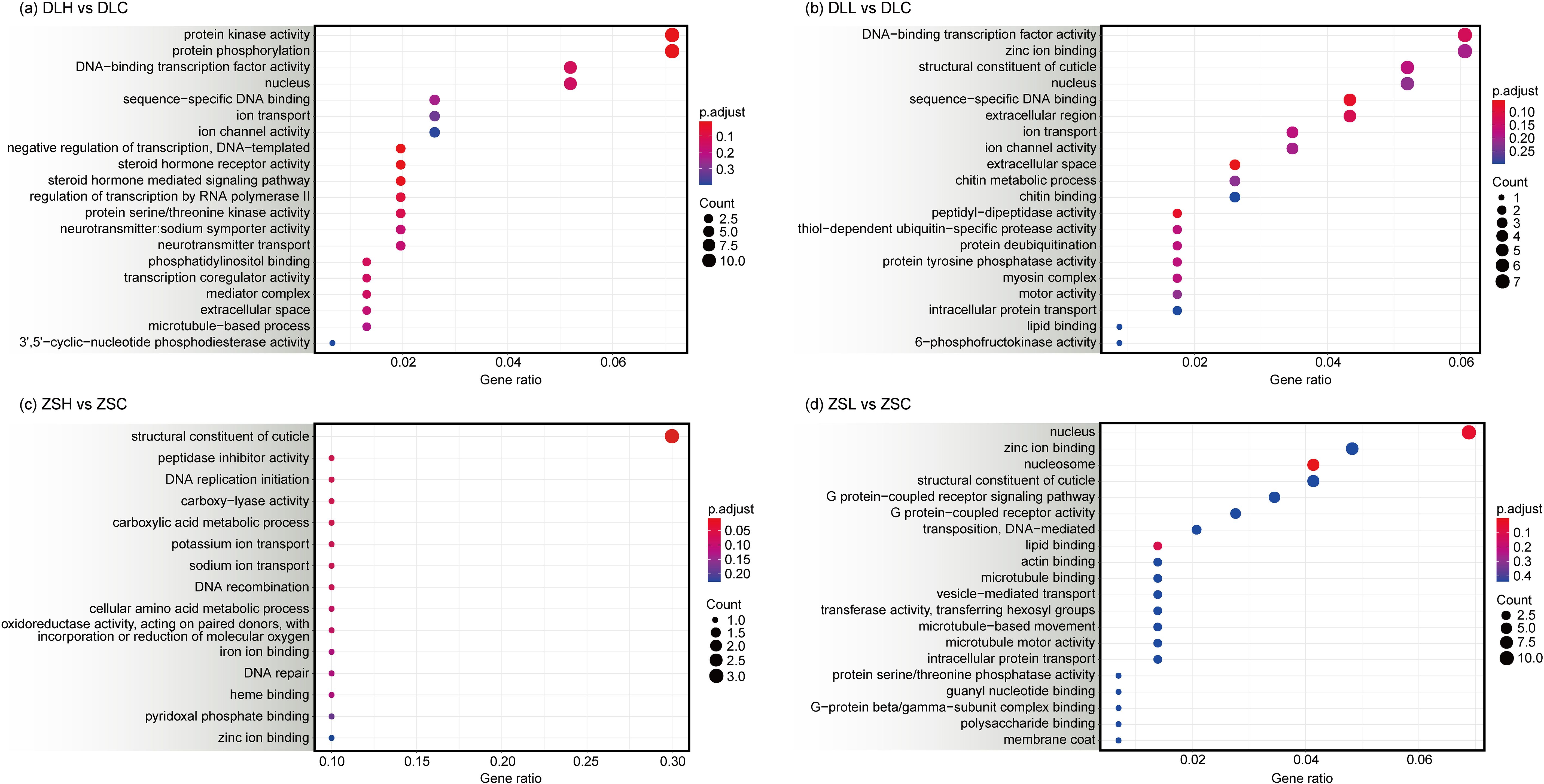
Figure 3. GO categorization (top 20) of unigenes in the transcriptome of C. japonica between DLH vs DLC (A), DLL vs DLC (B), ZSH vs ZSC (C) and ZSL vs ZSC (D). Count indicates the number of differential genes annotated to the GO number. The colour bar in p.adjust enrichment analysis represents the significance of enrichment. The red dot size indicates highly enriched unigenes.
3.4 KEGG enrichment analysis of DEGs in C. japonica under different temperature stresses
The DEGs were analyzed by KEGG enrichment to identify the biological regulatory pathways in the two crab populations under different temperature stress conditions (Figure 4). At 28°C, compared with the control group, the DEGs in the ZSH group were enriched into seven pathways, and the most significant pathway was DNA replication (Supplementary Table S8). The DEGs in the DLH group were enriched into 152 pathways and mainly included the apelin signaling pathway and autophagy (Supplementary Table S9). At 15°C, compared with the control group, the DEGs in the ZSL group were enriched into 88 pathways, including alcoholism and systemic lupus erythematosus (Supplementary Table S10). The DEGs in the DLL group were enriched into 100 pathways, including influenza A, protein processing in endoplasmic reticulum, and cholinergic synapse (Supplementary Table S11).
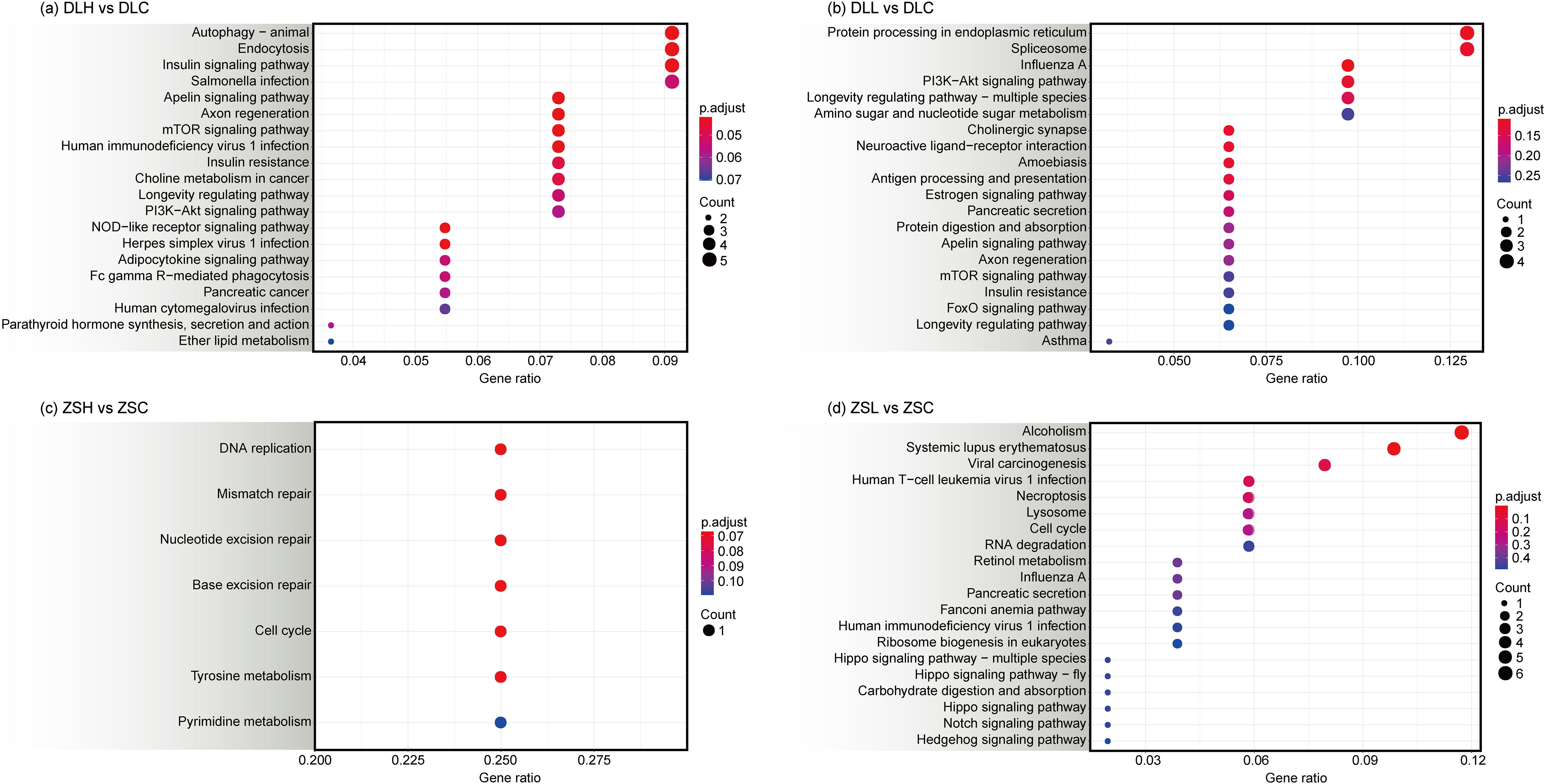
Figure 4. KEGG assignment (top 20) of unigenes in the transcriptome of C. japonica between DLH vs DLC (A), DLL vs DLC (B), ZSH vs ZSC (C) and ZSL vs ZSC (D). Count indicates the number of differential genes annotated to the KEGG number. The colour bar in p.adjust enrichment analysis represents the significance of enrichment. The red dot size indicates highly enriched unigenes.
3.5 Transcriptomic data validation
A total of eight DEGs were chosen for qRT-PCR analysis. The relative expression levels of these eight randomly selected genes based on qRT-PCR and transcriptome analysis were consistent (Figure 5), which meant that the RNA-seq data were reliable. Of these eight DEGs, RPS6K and AMPK were downregulated in the high-latitude population, whereas LIG1 was upregulated and CDC45 was downregulated in the low-latitude population under high-temperature stress. In addition, PRSS and EDEM1 were upregulated and HSP70 was downregulated in the high-latitude population, whereas HDAC was downregulated in the low-latitude population under low-temperature stress.
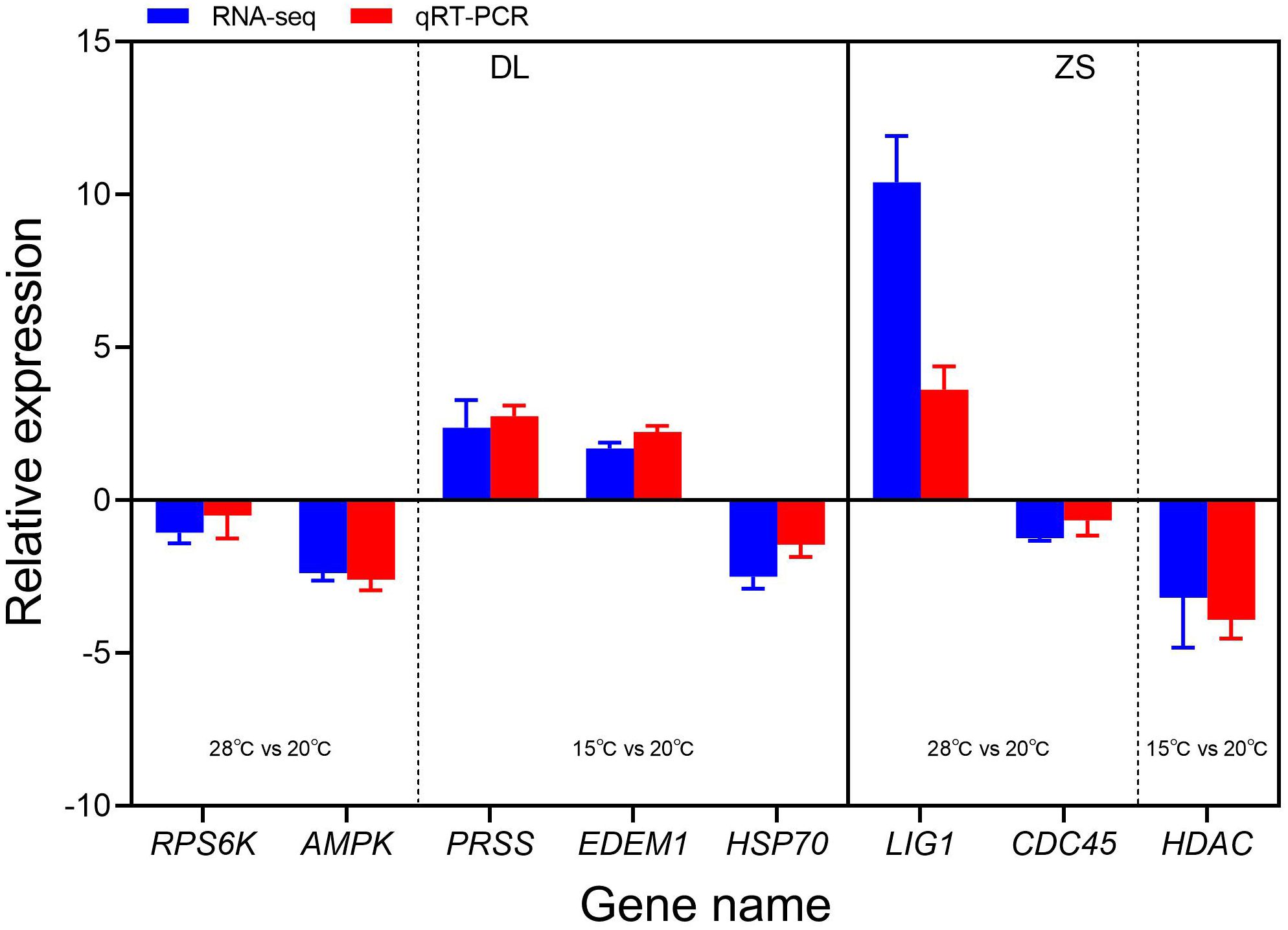
Figure 5. Relative expression levels of eight randomly selected genes obtained based on RNA-seq and qRT-PCR.
4 Discussion
Global climate change is already causing a number of adverse impacts on marine ecosystems and biodiversity. As a crucial element in climate variability, ocean temperature not only affects the growth, reproduction, and metabolism of marine organisms but also influences their spatial distribution, habitat preferences, and migration patterns (Saucedo et al., 2004; Scott and Johnston, 2012; García Molinos et al., 2016; Poloczanska et al., 2016; Boyd et al., 2018; Zhang et al., 2020). As poikilotherms, crustaceans are highly sensitive to temperature changes (Stillman and Somero, 2000; Somero, 2010; Soofi et al., 2014; Legrand et al., 2018). Previous studies have shown that some crustaceans can adapt to environmental temperature changes by regulating related gene expression levels (Stillman and Tagmount, 2009; Lou et al., 2022). Furthermore, the adaptive capacity of crustaceans in different geographical groups varies under ocean temperature changes (Cheng et al., 2024). However, the differences in molecular regulation of crustaceans to temperature stresses from different populations need to be elucidated. C. japonica, an intertidal crustacean, is widely distributed along the Chinese coast, ranging from the Bohai Sea to the East China Sea and covering a wide range of ambient temperatures (Lou et al., 2022). To adjust to variations in environmental temperature, different populations of C. japonica may develop various survival strategies or unique temperature adaptability to cope with temperature stress. In this study, we showed, for the first time, how C. japonica from two populations regulated their functional gene expression levels to adapt to changes in environmental temperature.
The differential gene expression levels and gene function enrichment were compared between the two crab populations at 15°C and 28°C. The results showed that the number of upregulated genes of the high-latitude population at 15°C and 28°C was higher than that of the low-latitude population. These results further confirmed that the species belonging to different geographical groups may exhibit varying physiological adaptations in response to changes in environmental temperature (Archambault et al., 2018; Pfeiffer et al., 2018). Furthermore, as the temperature increased, the number of differentially expressed genes in the two populations decreased. Compared with the control group (DLC and ZSC), a total of 309 and 938 DEGs were found in the DLH and ZSH groups at 15°C and 280 and 33 DEGs in the DLL and ZSL groups at 28°C. The gene regulatory trend of C. japonica to temperature stress was similar to that of O. oratoria in previous studies (Lou et al., 2019). The shared 28 DEGs between two temperature treatments in Dalian population were mainly corresponded to zinc finger protein of which plays an important role under temperature stress in many organisms (Sugano et al., 2003; Yang et al., 2020), whereas the shared seven DEGs between two temperature treatments in Zhoushan population were mainly corresponded to cuticle protein of which plays an important role in environmental adaptation in arthropods (Zheng et al., 2023). Furthermore, the shared 47 DEGs between two populations under cold stress were mainly corresponded to metabolism related genes of which plays an important role under cold stress in fish (Sun et al., 2021).
Functional enrichment analysis showed that the DEGs of the high-temperature group and low-temperature group of both populations were mainly enriched in GO terms such as protein kinase activity, protein phosphorylation, hemoglobin binding, structural constituent of cuticle, and extracellular space. Previous studies have shown that phosphorylation is an important cellular regulatory mechanism, and protein kinases play important roles in it (Johnson, 2009). When an organism is under temperature stress, temperature signals are primarily transduced to downstream targets by protein kinases, activating the expression of related genes (Jost et al., 2015; Chen et al., 2021). Furthermore, under environmental pressure, the distribution of hemoglobin transport protein genes differs among various populations, and gene inactivation and horizontal transfer may play important roles in the occurrence of intra-species genetic diversity (Lemos and Osorio, 2007). Moreover, the DEGs of both populations were enriched in cell structure-related GO terms, such as the structural constituent of cuticle and extracellular space. The cell function and structure of C. japonica changed under temperature stress, and some functional enzymes and structural proteins were activated to help C. japonica resist external stresses (Pinney et al., 2021). These cellular stress responses are considered a defensive response of organisms to environmental stress, and they usually lead to the deformation or damage of DNA, proteins, or other basic macromolecules (Kültz, 2020). In summary, C. japonica of both populations could regulate related genes to cope with temperature stress. However, disparities exist in the regulatory ability of the two groups to handle temperature stress.
Most of the enriched KEGG pathways are related to signal transduction, genetic information processing, cellular process, DNA replication, and metabolic process. Under high-temperature stress, the DEGs (280) of the high-latitude population were remarkably enriched in signal transduction and cellular process. In the apelin signaling pathway and autophagy animal pathway, we found that ribosomal protein S6 kinase (RPS6K) and AMP-activated protein kinase (AMPK) showed a downregulation trend. Previous studies have shown that the phosphorylation of RPS6K can promote body growth by improving protein synthesis (Jastrzebski et al., 2007; Chauvin et al., 2014), so we speculated that the body growth of C. japonica is inhibited when the environment is too hot. In addition, AMPK is a major cellular regulator of metabolic process in organisms (Jiang et al., 2023). In general, the activation of AMPK weakens the related metabolic process of consuming ATP, which preserves ATP for acute cell survival processes and replenishes ATP through catabolic processes (Li et al., 2019). Thus, we speculated that the ATP consumption of C. japonica increased by downregulating AMPK under high-temperature stress to support their growth in this study. By contrast, we noted less DEGs (33) in the low-latitude population at 28 °C, and the DEGs were mainly enriched in genetic information processing, among which the most significant pathway was DNA replication. We found that DNA ligase 1 (LIG1) showed an upregulation trend, whereas cell division control protein 45 (CDC45) showed a downregulation trend under this treatment. Previous studies have shown that LIG1 and CDC45 are involved in cell proliferation, especially in regulating the initiation and elongation stages of eukaryotic chromosome DNA replication (Srinivasan et al., 2013; Liddiard et al., 2019). Therefore, we speculated that C. japonica of the low-latitude population increased the expression of LIG1 and reduced the expression of CDC45 to inhibit cell proliferation to adapt to heat stimulation. These results demonstrated that C. japonica of the low- and high-latitude populations showed different genetic regulatory mechanisms under high temperature, and the low-latitude population was more resilient to high-temperature stress than the high-latitude population. This result was similar to that of Cynoscion nebulosus in previous studies (Song and McDowell, 2021).
Under low-temperature stress, the DEGs (309) of the high-latitude population were significantly enriched in genetic information processing. The expression of trypsin (PRSS), which is involved in physiological processes such as digestion, absorption, immune defense, cell growth, cell death, and apoptosis (Kaur and Singh, 2022), is significantly upregulated in the influenza A pathway. The antifreeze protein (AFGP) gene originates from the trypsin gene, and the chimera of AFGP-trypsin as the antifreeze glycoprotein can withstand cold stress in many fishes and other organisms (Cheng and Chen, 1999). Thus, the increased expression of trypsin may improve the cold resistance of C. japonica in high latitude. In addition, endoplasmic reticulum degradation enhancer, mannosidase alpha-like 1 (EDEM1), shows an upregulation trend, whereas heat shock 70 kDa protein (HSP70) shows a downregulation trend in the endoplasmic reticulum protein processing pathway. EDEM1 is induced by endoplasmic reticulum stress (Hosokawa et al., 2001). When the temperature environment changes, the increase in misfolded proteins disrupts the homeostasis of the endoplasmic reticulum, and upregulated EDEM1 can degrade the misfold proteins (Zuber et al., 2007). However, HSP70, which can fold and assemble proteins, regulate protein activity, and proofread protein structures, is downregulated (Ryan and Pfanner, 2001; Mayer and Bukau, 2005). Therefore, we speculated that upregulated EDEM1 can offset the downregulated HSP70 in high-latitude C. japonica under low-temperature pressure. By contrast, there were more DEGs (938) in the low-latitude population than in the high-latitude population at 15 °C, and the DEGs were mainly enriched in metabolic pathways. Histone deacetylase (HDAC) is significantly decreased in the alcoholism pathway. HDAC has been shown to regulate the activity and function of proteins, and it can activate gene transcription (Cui et al., 2023). In addition, HDAC has been found to mediate thermal plasticity in zebrafish (Danio rerio), and HDAC inhibition promotes regenerative neurogenesis in D. rerio (Seebacher and Simmonds, 2019; Kiyooka et al., 2020). Therefore, we speculated that the downregulation of HDAC is the adaptation of low-latitude C. japonica to cold stimuli. The above results revealed that low- and high-latitude C. japonica showed different genetic regulatory mechanisms under low-temperature stress. The high-latitude population regulated less genes than the low-latitude population, which indicated that the former was more resilient to low-temperature stress than the latter.
5 Conclusions
Temperature is an environmental factor that significantly impacts the behavior and physiology of intertidal crustaceans. This study analyzed the transcriptome of two populations of C. japonica at low- and high-temperature stress to identify differences in the genetic regulation. Results showed that C. japonica from the low-latitude population regulated more genes at low-temperature stress, and C. japonica from the high-latitude population regulated more genes at high-temperature stress. In addition, the physiological processes differed with temperature fluctuations between the two populations. The disparity in gene expression levels and the distinct pathways that respond to temperature stress in the two populations of C. japonica highlight the importance of genetic regulatory adaptations in response to fluctuations in environmental temperature. The results of this study provide valuable information on the sensitivity of low- and high-latitude C. japonica populations to temperature-related environmental stress. Our results offer valuable insights into the molecular regulatory mechanisms of temperature fluctuations in other crustaceans.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by The Animal Care and Use Committee of Zhejiang Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SS: Data curation, Funding acquisition, Investigation, Writing – original draft. ZHe: Data curation, Formal analysis, Investigation, Writing – original draft. FZ: Formal analysis, Investigation, Writing – review & editing. ZHa: Conceptualization, Funding acquisition, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32070513), the Project Supported by Scientific Research Fund of Zhejiang Provincial Education Department (Y202353931), and the Fundamental Research Funds for Zhejiang Provincial Universities and Research Institutes (2024J004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1491685/full#supplementary-material
References
Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data (Cambridge, United Kingdom: Babraham Bioinformatics, Babraham Institute).
Archambault J. M., Cope W. G., Kwak T. J. (2018). Chasing a changing climate: Reproductive and dispersal traits predict how sessile species respond to global warming. Divers. Distrib. 24, 880–891. doi: 10.1111/ddi.12740
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bopp L., Resplandy L., Orr J. C., Doney S. C., Dunne J. P., Gehlen M., et al. (2013). Multiple stressors of ocean ecosystems in the 21st century: projections with CMIP5 models. Biogeosciences 10, 6225–6245. doi: 10.5194/bg-10-6225-2013
Boyd P. W., Collins S., Dupont S., Fabricius K., Gattuso J. P., Havenhand J., et al. (2018). Experimental strategies to assess the biological ramifications of multiple drivers of global ocean change—a review. Glob. Change Biol. 24, 2239–2261. doi: 10.1111/gcb.14102
Chauvin C., Koka V., Nouschi A., Mieulet V., Hoareau-Aveilla C., Dreazen A., et al. (2014). Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33, 474–483. doi: 10.1038/onc.2012.606
Chen X., Ding Y., Yang Y., Song C., Wang B., Yang S., et al. (2021). Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 63, 53–78. doi: 10.1111/jipb.13061
Cheng C. H. C., Chen L. (1999). Evolution of an antifreeze glycoprotein. Nature 401, 443–444. doi: 10.1038/46721
Cheng J., Zhang Z., Li Y., Zhang L., Hui M., Sha Z. (2024). Rolling with the punches: Organism-environment interactions shape spatial pattern of adaptive differentiation in the widespread mantis shrimp Oratosquilla oratoria. Sci. Total Environ. 917, 170244. doi: 10.1016/j.scitotenv.2024.170244
Cui X., Dard A., Reichheld J. P., Zhou D. X. (2023). Multifaceted functions of histone deacetylases in stress response. Trends Plant Sci. 28, 1245–1256. doi: 10.1016/j.tplants.2023.06.006
Dissanayake A., Ishimatsu A. (2011). Synergistic effects of elevated CO2 and temperature on the metabolic scope and activity in a shallow-water coastal decapod (Metapenaeus joyneri; Crustacea: Penaeidae). ICES J. Mar. Sci. 68, 1147–1154. doi: 10.1093/icesjms/fsq188
Edwards M., Richardson A. J. (2004). Impact of climate change on marine pelagic phenology and trophic mismatch. Nature 430, 881–884. doi: 10.1038/nature02808
García Molinos J., Halpern B. S., Schoeman D. S., Brown C. J., Kiessling W., Moore P. J., et al. (2016). Climate velocity and the future global redistribution of marine biodiversity. Nat. Clim. Change 6, 83–88. doi: 10.1038/nclimate2769
Green B. S., Fisher R. (2004). Temperature influences swimming speed, growth and larval duration in coral reef fish larvae. J. Exp. Mar. Biol. Ecol. 299, 115–132. doi: 10.1016/j.jembe.2003.09.001
Greenberg D. A., Palen W. J. (2021). Hydrothermal physiology and climate vulnerability in amphibians. Proc. Biol. Sci. 288, 20202273. doi: 10.1098/rspb.2020.2273
Han Z. Q., Gao T. X., Lou F. R., Liu Q. (2022). Charybdis japonica genome provides insights into desiccation adaptation and sex-determining region. Zool. Res. 43, 927–930. doi: 10.24272/j.issn.2095-8137.2022.155
Hofmann G. E., Todgham A. E. (2010). Living in the now: physiological mechanisms to tolerate a rapidly changing environment. Annu. Rev. Physiol. 72, 127–145. doi: 10.1146/annurev-physiol-021909-135900
Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., et al. (2001). A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415–422. doi: 10.1093/embo-reports/kve084
Jahan K., Nie H., Yin Z., Zhang Y., Li N., Yan X. (2022). Comparative transcriptome analysis to reveal the genes and pathways associated with adaptation strategies in two different populations of Manila clam (Ruditapes philippinarum) under acute temperature challenge. Aquaculture 552, 737999. doi: 10.1016/j.aquaculture.2022.737999
Jastrzebski K., Hannan K. M., Tchoubrieva E. B., Hannan R. D., Pearson R. B. (2007). Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Fact. 25, 209–226. doi: 10.1080/08977190701779101
Jiang D., Yang C., Wang X., Ma X., He Z., Wang L., et al. (2023). The involvement of AMP-activated protein kinase α in regulating glycolysis in Yesso scallop Patinopecten yessoensis under high temperature stress. Fish Shellfish Immunol. 140, 108998. doi: 10.1016/j.fsi.2023.108998
Johansen J., Messmer V., Coker D., Hoey A., Pratchett M. (2014). Increasing ocean temperatures reduce activity patterns of a large commercially important coral reef fish. Glob. Change Biol. 20, 1067–1074. doi: 10.1111/gcb.12452
Johnson L. N. (2009). The regulation of protein phosphorylation. Biochem. Soc Trans. 37, 627–641. doi: 10.1042/bst0370627
Jost J. A., Keshwani S. S., Abou-Hanna J. J. (2015). Activation of AMP-activated protein kinase in response to temperature elevation shows seasonal variation in the zebra mussel, Dreissena polymorpha. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 182, 75–83. doi: 10.1016/j.cbpa.2014.11.025
Kaur J., Singh P. K. (2022). Trypsin detection strategies: a review. Crit. Rev. Anal. Chem. 52, 949–967. doi: 10.1080/10408347.2020.1846490
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kiyooka M., Shimizu Y., Ohshima T. (2020). Histone deacetylase inhibition promotes regenerative neurogenesis after stab wound injury in the adult zebrafish optic tectum. Biochem. Biophys. Res. Commun. 529, 366–371. doi: 10.1016/j.bbrc.2020.06.025
Kültz D. (2020). Evolution of cellular stress response mechanisms. J. Exp. Zool. A Ecol. Integr. Physiol. 333, 359–378. doi: 10.1002/jez.2347
Legrand E., Riera P., Pouliquen L., Bohner O., Cariou T., Martin S. (2018). Ecological characterization of intertidal rockpools: Seasonal and diurnal monitoring of physico-chemical parameters. Reg. Stud. Mar. Sci. 17, 1–10. doi: 10.1016/j.rsma.2017.11.003
Lemos M. L., Osorio C. R. (2007). Heme, an iron supply for vibrios pathogenic for fish. Biometals 20, 615–626. doi: 10.1007/s10534-006-9053-8
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 1–16. doi: 10.1186/1471-2105-12-323
Li Y., Sun R., Zou J., Ying Y., Luo Z. (2019). Dual roles of the AMP-activated protein kinase pathway in angiogenesis. Cells 8, 752. doi: 10.3390/cells8070752
Li Z., Zhao Z., Luo L., Wang S., Zhang R., Guo K., et al. (2023). Immune and intestinal microbiota responses to heat stress in Chinese mitten crab (Eriocheir sinensis). Aquaculture 563, 738965. doi: 10.1016/j.aquaculture.2022.738965
Liddiard K., Ruis B., Kan Y., Cleal K., Ashelford K. E., Hendrickson E. A., et al. (2019). DNA Ligase 1 is an essential mediator of sister chromatid telomere fusions in G2 cell cycle phase. Nucleic Acids Res. 47, 2402–2424. doi: 10.1093/nar/gky1279
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lou F., Han Z., Gao T. (2019). Transcriptomic responses of two ecologically divergent populations of Japanese mantis shrimp (Oratosquilla oratoria) under thermal stress. Animals 9, 399. doi: 10.3390/ani9070399
Lou F., Wang Y., Han Z., Shui B. (2022). Comparative transcriptome reveals the molecular regulation mechanism of Charybdis japonica to high-and low-temperature stresses. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.849485
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Lyu L., Wen H., Li Y., Li J., Zhao J., Zhang S., et al. (2018). Deep transcriptomic analysis of black rockfish (Sebastes schlegelii) provides new insights on responses to acute temperature stress. Sci. Rep. 8, 9113. doi: 10.1038/s41598-018-27013-z
Mao X., Cai T., Olyarchuk J. G., Wei L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Mayer M. P., Bukau B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Ning J., Zou D., Lu X., Cao W., Chen M., Liu B., et al. (2021). Transcriptomic analyses provide insights into the adaptive responses to heat stress in the ark shells, Scapharca subcrenata. Comp. Biochem. Physiol. Part D Genomics Proteomics 38, 100813. doi: 10.1016/j.cbd.2021.100813
Oulhen N., Morales J., Cosson B., Mulner-Lorillon O., Bellé R., Cormier P. (2007). Gene expression regulation at the translational level: contribution of marine organisms. J. Soc Biol. 201, 297–306. doi: 10.1051/jbio:2007028
Perry A. L., Low P. J., Ellis J. R., Reynolds J. D. (2005). Climate change and distribution shifts in marine fishes. Science 308, 1912–1915. doi: 10.1126/science.1111322
Pfeiffer J. M., Sharpe A. E., Johnson N. A., Emery K. F., Page L. M. (2018). Molecular phylogeny of the Nearctic and Mesoamerican freshwater mussel genus Megalonaias. Hydrobiologia 811, 139–151. doi: 10.1007/s10750-017-3441-7
Pinney M. M., Mokhtari D. A., Akiva E., Yabukarski F., Sanchez D. M., Liang R., et al. (2021). Parallel molecular mechanisms for enzyme temperature adaptation. Science 371, eaay2784. doi: 10.1126/science.aay2784
Poloczanska E. S., Burrows M. T., Brown C. J., García Molinos J., Halpern B. S., Hoegh-Guldberg O., et al. (2016). Responses of marine organisms to climate change across oceans. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00062
Pörtner H. O., Farrell A. P. (2008). Physiology and climate change. Science 322, 690–692. doi: 10.1126/science.1163156
Qian B., Xue L. (2016). Liver transcriptome sequencing and de novo annotation of the large yellow croaker (Larimichthy crocea) under heat and cold stress. Mar. Genomics 25, 95–102. doi: 10.1016/j.margen.2015.12.001
Rahman M. A., Henderson S., Miller-Ezzy P., Li X. X., Qin J. G. (2019). Immune response to temperature stress in three bivalve species: Pacific oyster Crassostrea gigas, Mediterranean mussel Mytilus galloprovincialis and mud cockle Katelysia rhytiphora. Fish Shellfish Immunol. 86, 868–874. doi: 10.1016/j.fsi.2018.12.017
Robinson M. D., Mccarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Ryan M. T., Pfanner N. (2001). Hsp70 proteins in protein translocation. Adv. Protein Chem. 59, 223–242. doi: 10.1016/S0065-3233(01)59007-5
Saucedo P. E., Ocampo L. A., Monteforte M., Bervera H. (2004). Effect of temperature on oxygen consumption and ammonia excretion in the Calafia mother-of-pearl oyster, Pinctada mazatlanica (HanleHanley 1856). Aquaculture 229, 377–387. doi: 10.1016/s0044-8486(03)00327-2
Scott G. R., Johnston I. A. (2012). Temperature during embryonic development has persistent effects on thermal acclimation capacity in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 109, 14247–14252. doi: 10.1073/pnas.1205012109
Seebacher F., Simmonds A. I. M. (2019). Histone deacetylase activity mediates thermal plasticity in zebrafish (Danio rerio). Sci. Rep. 9, 8216. doi: 10.1038/s41598-019-44726-x
Shrestha A. M. S., Lilagan C. A. I., Guiao J. E. B., Romana-Eguia M. R. R., Ablan Lagman M. C. (2021). Comparative transcriptome profiling of heat stress response of the mangrove crab Scylla serrata across sites of varying climate profiles. BMC Genomics 22, 1–14. doi: 10.1186/s12864-021-07891-w
Solomon S., Manning M., Marquis M., Qin D. (2007). Climate change 2007-the physical science basis: Working group I contribution to the fourth assessment report of the IPCC. Cambridge university press, Cambridge.
Somero G. N. (2010). The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J. Exp. Biol. 213, 912–920. doi: 10.1242/jeb.037473
Song J., McDowell J. R. (2021). Comparative transcriptomics of spotted seatrout (Cynoscion nebulosus) populations to cold and heat stress. Ecol. Evol. 11, 1352–1367. doi: 10.1002/ece3.7138
Soofi W., Goeritz M. L., Kispersky T. J., Prinz A. A., Marder E., Stein W. (2014). Phase maintenance in a rhythmic motor pattern during temperature changes in vivo. J. Neurophysiol. 111, 2603–2613. doi: 10.1152/jn.00906.2013
Srinivasan S. V., Dominguez-Sola D., Wang L. C., Hyrien O., Gautier J. (2013). Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep. 3, 1629–1639. doi: 10.1016/j.celrep.2013.04.002
Stillman J. H., Somero G. N. (2000). A comparative analysis of the upper thermal tolerance limits of eastern Pacific porcelain crabs, genus Petrolisthes: influences of latitude, vertical zonation, acclimation, and phylogeny. Physiol. Biochem. Zool. 73, 200–208. doi: 10.1086/316738
Stillman J. H., Tagmount A. (2009). Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol. Ecol. 18, 4206–4226. doi: 10.1111/j.1365-294X.2009.04354.x
Sugano S., Kaminaka H., Rybka Z., Catala R., Salinas J., Matsui K., et al. (2003). Stress-responsive zinc finger gene ZPT2-3 plays a role in drought tolerance in petunia. Plant J. 36, 830–841. doi: 10.1046/j.1365-313X.2003.01924.x
Sun S., Cao X., Gao J. (2021). C24: 0 avoids cold exposure-induced oxidative stress and fatty acid β-oxidation damage. iScience 24, 103409. doi: 10.2139/ssrn.3881356
Thiyagarajan V., Harder T., Qian P. Y. (2003). Combined effects of temperature and salinity on larval development and attachment of the subtidal barnacle Balanus trigonus Darwin. J. Exp. Mar. Biol. Ecol. 287, 223–236. doi: 10.1016/s0022-0981(02)00570-1
Yang K., Huang Z., Ma A., Liu X., Yang S. (2020). Transcriptome study of kidney of turbot under high-temperature stress. Prog. Fish. Sci. 41, 86–95. doi: 10.19663/j.issn2095-9869.20181211001
Yang Y., Xu W., Jiang Q., Ye Y., Tian J., Huang Y., et al. (2022). Effects of low temperature on antioxidant and heat shock protein expression profiles and transcriptomic responses in crayfish (Cherax destructor). Antioxidants 11, 1779. doi: 10.3390/antiox11091779
Yannic G., Pellissier L., Ortego J., Lecomte N., Couturier S., Cuyler C., et al. (2014). Genetic diversity in caribou linked to past and future climate change. Nat. Clim. Change 4, 132–137. doi: 10.1038/nclimate2074
Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, 1–12. doi: 10.1186/gb-2010-11-2-r14
Yu C. G., Song H. T., Yao G. Z. (2004). Assessment of the crab stock biomass in the continental shelf waters of the East China Sea. J. Fish. China 28, 41–46. doi: 10.3321/j.issn:1000-0615.2004.01.007
Zhang W. Y., Storey K. B., Dong Y. W. (2020). Adaptations to the mudflat: Insights from physiological and transcriptional responses to thermal stress in a burrowing bivalve Sinonovacula constricta. Sci. Total Environ. 710, 136280. doi: 10.1016/j.scitotenv.2019.136280
Zheng Y., Liu C., Wang S., Qian K., Feng Y., Yu F., et al. (2023). Genome-wide analysis of cuticle protein family genes in rice stem borer Chilo suppressalis: Insights into their role in environmental adaptation and insecticidal stress response. Int. J. Biol. Macromol. 242, 124989. doi: 10.1016/j.ijbiomac.2023.124989
Keywords: Charybdis japonica, temperature stress, Illumina sequencing, gene expression level, adaption
Citation: Sun S, He Z, Zhang F and Han Z (2024) Comparative transcriptome analysis of low- and high-latitude populations of Charybdis japonica under temperature stress. Front. Mar. Sci. 11:1491685. doi: 10.3389/fmars.2024.1491685
Received: 05 September 2024; Accepted: 17 October 2024;
Published: 04 November 2024.
Edited by:
Yu Zhang, Shanghai Jiao Tong University, ChinaReviewed by:
Vittoria Roncalli, Anton Dohrn Zoological Station Naples, ItalyFangrui Lou, Yantai University, China
Copyright © 2024 Sun, He, Zhang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Han, ZDYzMzkxMjRAMTYzLmNvbQ==
 Shaolei Sun
Shaolei Sun Zhiqi He1
Zhiqi He1 Zhiqiang Han
Zhiqiang Han