- 1Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, Ministry of Natural Resources of China; Fujian Key Laboratory of Marine Genetic Resources, Xiamen, China
- 2School of Environment, Harbin Institute of Technology, Harbin, China
- 3Second institute of oceanography, Ministry of Natural Resources, Hangzhou, China
- 4Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai, China
Microplastics are widespread pollutants in aquatic environments, posing a significant threat to the health of marine ecosystems. However, little is known about the impact of plastics on deep-sea microbial communities. In this paper, we investigated the effects of polystyrene (PS) microplastics with three particle sizes (60 nm, 600 nm and 1 µm) and three concentrations (10, 50, 150 mg/L) as well as PS films (1 × 1 cm) on the deep-sea microbial community inoculated with water of 3370 m water depth from Pacific Ocean by using reactive oxygen species (ROS) detection, growth rate, scanning electron microscope (SEM) and high-throughput sequencing. Microplastics surface rotting (600 nm and 1 µm) and further fragmentation (60 nm) were observed caused by plastic-degrading microbial erosion after 50 days’ incubation. Similarly, deformation of PS film, including formation of obvious wrinkles and deep pits and the generation of microplastics and nanoplastics were also observed. Microplastics from commercial and plastic films could stimulate the bacterial community to secrete extracellular polymeric substance (EPS), favouring biofilm formation and resistance to external stress. Compared with larger microplastics, 60 nm microplastics and plastic films significantly inhibited the growth of bacterial communities with enhanced ROS production. The abundance of Moraxellaceae dominated in all enriched samples with the addition of microplastics, while the abundance of Alcanivoracaceae also increased in the 60 nm and plastic film enrichments, in contrast to dominant bacteria of Colwelliaceae, Marinobacteraceae, Rhodobacteraceae and Alcanivoracaceae the deep seawater in situ. Correspondingly, the functional changes of the communities were observed via functional prediction by 16S rRNA gene based on their alterations in bacterial community structure. The study provides insights into the effects of microplastics and nanoplastics on deep-sea microbial communities.
1 Introduction
Plastic pollution has become one of the major environmental problems affecting the oceans, with millions of tonnes entering the ocean each year (Jones et al., 2024). Larger plastics in the ocean are often formed into small plastic particles (i.e., microplastics,< 5 mm; nanoplastics,< 1 µm) by sunlight, mechanical abrasion and microbial degradation (Lv et al., 2024). Microplastics can affect more than 690 marine species due to their high specific surface area and high pollutant adsorption capacity (Carbery et al., 2018; Li et al., 2023b). Polystyrene (PS) is a plastic polymer widely used in the packaging, toy and construction industries and so on (Goncalves et al., 2023). In particular, expanded polystyrene (EPS) plastics are heavily used in marine aquaculture (pontoons, buoys and floats) which has resulted in PS debris being found throughout the world’s oceans (Xayachak et al., 2023). Therefore, a comprehensive understanding on the impacts of PS microplastic pollution in marine ecosystems is essential.
Microplastics in the environment have been reported to alter the composition of microbial communities. Campylobacterota was significantly enriched in mangrove soil due to the addition of polyethylene (PE) microplastics (200 - 500 µm) (Zhou et al., 2024). The relative abundance of phyla Patescibacteria and Bacteroidetes was increased in wheat rhizosphere soil when PE microplastics (200 µm) were added (Zhu et al., 2022). Further, changes in bacterial community structure may affect C and N cycling. For example, accumulation of microplastics could promote CO2 emissions, ammonia and nitrite oxidation, and nitrification and denitrification (Chen et al., 2020; Seeley et al., 2020; Chen et al., 2023). The alteration of these functional properties could lead to potential impacts on carbon, nitrogen and phosphorus cycling, which could have significant ecological consequences for biogeochemical processes. However, most of studies have focused on plastics with micro- or greater particle sizes, the effects of nanoplastics on the structure and function of biological communities in seawater need to be further investigated.
It has been demonstrated that smaller microplastic particles can negatively impact the growth of microorganisms. Such as, nano-polystyrene (30 nm) increased ROS level and contributed to DNA damage within E.coli (Ning et al., 2022). Streptomyces M145 exposed to nanoplastics could produce excess ROS, leading to cell membrane breakdown and cell death (Liu et al., 2021). Polylactic acid (PLA) microplastics (3-4 mm, flake shape) could cause oxidative damage to the cell membrane, disrupt cell wall components and inhibit cell growth of Bacillus amyloliquefaciens for 3 days (Li et al., 2023a). However, current research on microbial toxicity of microplastics was limited to pure bacterial strains that cannot degrade microplastics. The community structure in nature is complex and further exploration is needed regarding the effects of microplastic toxicity on communities.
Traces of microplastics have now been found in deep-sea areas. For example, Microplastic counts ranged from 0 to 13.6 fragments per g dry sediment in deep-sea sediments from the Great Australian Bight (1655 - 3062 m water depth) (Barrett et al., 2020). The investigation was carried out in the abyssal plain of Northwest Pacific (5705 m water depth) where it was found that microplastic abundance could be as high as 341.0 × 104 pieces/m2 (Tsuchiya et al., 2024). It is evident that the deep sea microplastics pollution is very serious and it may become a repository for microplastics in the future. Marine microorganisms are important participants in biogechemical processes, which make it crucial to understand the impact of microplastics on marine communities.
In this study, we explored the effects of nanoplastics on the structure and function of microbial communities in deep seawater with PS, a petroleum-based plastic commonly found in the ocean. To assess the effects of PS on microbial communities in the long term, microplastics of different sizes (60 nm, 600 nm and 1 µm) and concentrations (10 mg/L, 50 mg/L and 150 mg/L), and plastic films were added to microbial communities and incubated for 50 days. ROS detection, growth rate, SEM and high-throughput sequencing of enriched samples were used to observe the changes in microbial community structure and function caused by different conditions of plastics, providing a basis for the study of the effect of plastics/microplastics on the microbial communities.
2 Materials and methods
2.1 Collection and enrichment of samples
Seawater was collected in the Pacific Ocean during the DY79 cruise of COMRA in December 2023, at water depths of 3370 m, located at 19.92°N, 173.33°E (Supplementary Figure S1). Deep-sea seawater was added to MMC medium containing polystyrene microspheres of different particle sizes (60 nm, 600 nm and 1 µm) (Tianjin BaseLine ChromTech Research Centre) and different concentrations (10 mg/L, 50 mg/L and 150 mg/L), and PS plastic pieces (1 × 1 cm) (Sigma−Aldrich, USA), respectively. Subsequently, continuous static incubation was carried out for 50 days at 28 °C. MMC is an artificial seawater medium with PS as the sole carbon and energy source, the formulation of which is described in the Text S1.
2.2 Strains isolated from enriched samples with verified PS degradation activity
The enrichment samples with gradient dilutions of 10-6, 10-7, 10-8 and 10-9 was spread on marine broth 2216 (BD, DIFCO, USA) agar plates. The plates were incubated in the dark at 28 °C under aerobic conditions for 5 days. All resultant colonies were purified by subcultivation in the same medium. The genomic DNA of the isolated pure culture was extracted as per manufacturer’s instructions using the genome extraction kit (Shanghai SBS Genetech Co., Ltd). Then 16S rRNA gene was amplified by the universal primers: 27 F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492 R (5′-TAC GGC TAC CTT GTT ACG ACT T-3′). The PCR procedure (30 cycles of 45 s at 94 °C, 45 s at 55 °C, and 90 s at 72 °C) was performed using a thermal cycler (Biometra TProfessional Standard gradient 96, Germany) (Liu et al., 2023). Then, the PCR products were sequenced (Shanghai Sangon Biotech Co., Ltd) and the sequence reads were used as queries to search against the EzBioCloud database (http://www.ezbioclound.net). To test the ability of pure strains to utilize PS, cells from 2216 agar plates were collected and washed four times using MMC medium at 3500 rpm for 8 min. Subsequently, a single isolate was cultured in MMC medium with PS and incubated for 30 days at 28 °C, 150 rpm/min. Bacterial growth was observed by light microscope (Nikon 80i, Japan).
2.3 The morphology of microplastics interacting with microorganisms
Morphological observations were carried out on the bacterial communities exposed to 150 mg/L different particle sizes of microplastics and microplastics released from PS films. Briefly, it was fixed with a 1:2 (v/v) ratio of bacterial culture and 2.5% glutaraldehyde for 2 h. The fixative was removed at 9000 rpm for 10 min using the high-speed freezing centrifuge (Eppendorf Centrifuge 5804R, Germany), and the excess fixative was washed twice with PBS. Subsequently, the cells were dehydrated with ethanol at a gradient of 50%, 60%, 70%, 80% and 90% while they were finally stored in anhydrous ethanol. A little of the liquid was taken and placed on silicon wafer during the experiment and waited for the ethanol to evaporate before spraying platinum. Samples were analysed using SEM (Hitachi SU8600, Japan).
2.4 Extraction and analysis of microplastics
The enriched samples were collected to remove the organic components by digestion. A ratio of 1:10 (v/v) between the sample and 10% KOH was added to 250 mL conical flasks. The reaction was carried out at 60 °C for 24 h until the microorganisms were completely digested. The solution obtained after digestion was density-separated with saturated NaCl and allowed to stand overnight. The supernatant was then filtered through a PC membrane (0.22 µm, 47 mm). Microplastics and nanoplastics were transferred to ultrapure water for following analyses by shaking the PC membrane for 30 s. Morphological changes on the microplastic surface were observed by SEM (Hitachi SU8600, Japan).
2.5 Cellular reactive oxygen speciesassay
The ROS levels in bacterial cells after different treatments were detected using the 2’,7’-dichlorofluorescein diacetate (DCFH-DA) ROS assay kit (UElandy, China). After diluting the bacteria to the same cell concentration (5 × 105/mL), 100 µL bacterial cells were centrifuged and washed three times with 2% NaCl. Then, the bacterial cells were resuspended with 100 µL (10 µmol/L) DCFH-DA and incubated for 30min in the dark at 28 °C. The cells were washed three times with 2% NaCl to remove the excess DCFH-DA. The fluorescence intensity values were measured using a microplate reader (Varioskan LUX, USA) at excitation and emission wavelengths of 504 and 529 nm, respectively. The intracellular ROS levels were expressed as the ratio of fluorescence intensity of the exposed group over the control group (Ning et al., 2022).
2.6 Dissolved organic carbon analysis
The samples enriched for 50 days were filtered through 50 nm PCTE membranes (Whatman, UK) to remove the bacterial community and microplastic particles. DOC was assessed by a total organic carbon (TOC) analyser (Sievers InnovOx, USA) using wet chemical oxidation.
2.7 Growth rate
Cells were counted at 40 × light microscope (Nikon 80i, Japan) using a Neubauer-improved haemocytometer (Supplementary Figure S2). The control group (CK) was MMC medium containing sodium acetate (2 g/L) without plastic films/microplastics. The growth rate was calculated as follow.
where C was the cell density of control group and T was the cell density of the treated group.
2.8 Extracellular polymeric substance extraction analysis
Samples were collected on 50th day to determine the protein and polysaccharide contents of EPS. The samples pretreatment was as follows: the samples were mixed with phosphate buffered saline (PBS) to make the same cell concentration and subjected to ultrasonic treatment for 3 min using an ultrasonic cleaner (KQ-800DE, China). Finally, the samples were centrifuged with 12000 g (4 °C) for 20 min (Eppendorf Centrifuge 5804R, Germany), and the supernatant was collected for EPS composition analysis. The polysaccharide content of EPS was determined by the phenol-sulphuric acid method and the protein content was determined by BCA protein assay kit (Thermo Fisher Scientific 23225, USA).
2.9 Seawater DNA extraction, high-throughput sequencing, and data analysis
Seawater bacterial communities were detected using high-throughput sequencing technology. Bacterial DNA was extracted using the Power Soil DNA Isolation Kit (MoBio, USA) according to the manufacturer’s instructions. The V3 – V4 region of the 16S rRNA genes was amplified using forward primer 338 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse primer 806 R (5′-GGACTACHVGGGTWTCTAAT-3′). DNA sequencing was performed by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Functional and metabolic pathway predictions were analyzed using PICRUSt2, BugBase and FAPROTAX.
3 Results and discussion
3.1 Biodegradation of microplastics and plastic films
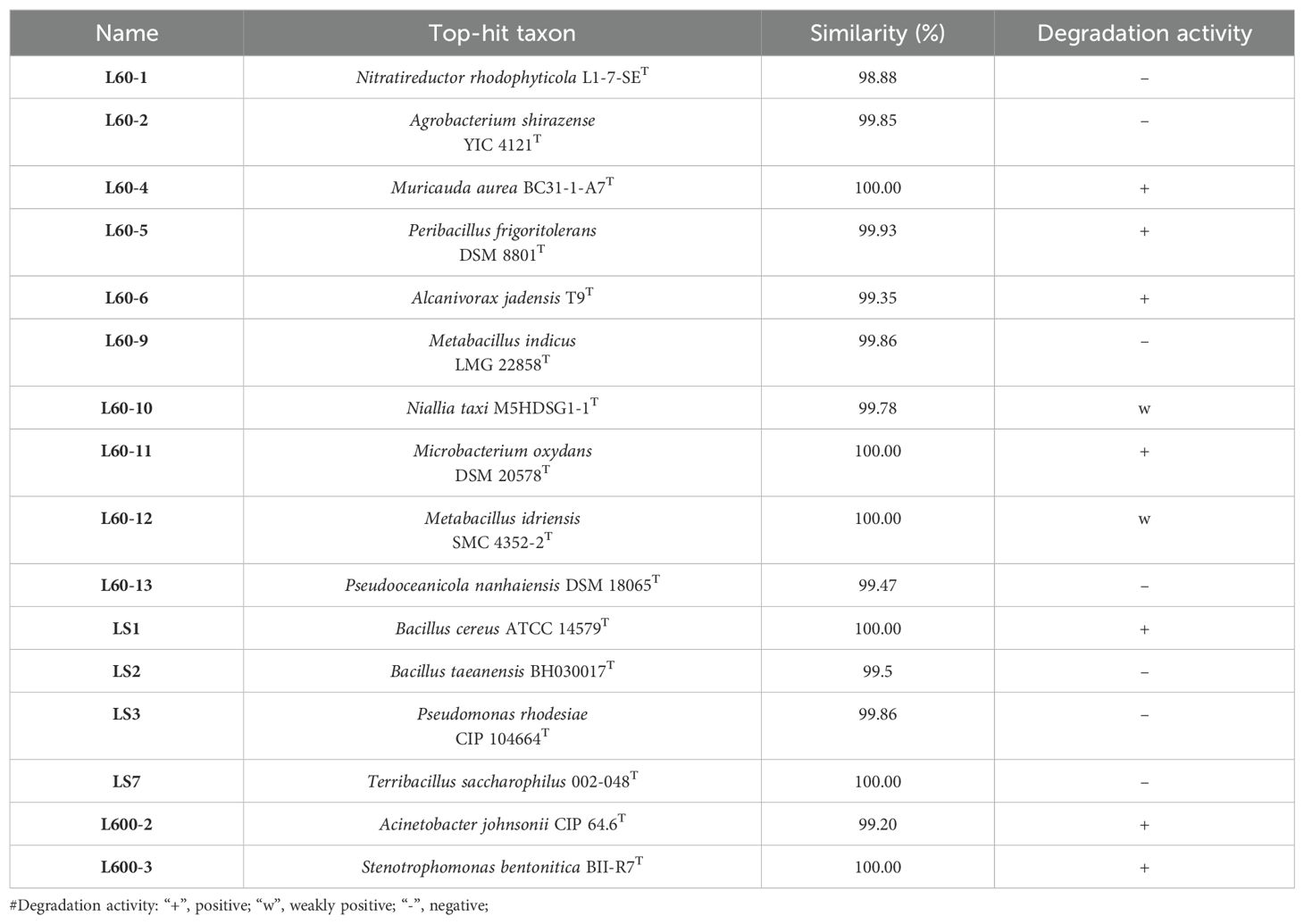
In order to analyse the action of the bacterial community on the plastic, the plastic surface was observed using SEM. As shown in Figure 1, the 60 nm microplastics was broken up, where smaller particles were found on the surface. Compared with the flat and smooth surface of PS microplastics (600 nm and 1 µm) (Supplementary Figure S3), the surface was uneven after 50 days of incubation with the bacterial community. This was consistent that the phenomenon was observed on Rhodococcus ruber C208 degradation of nano-sized PS (Eom et al., 2023). Wrinkles and deep pits were observed on the plastic films surface. The change of microplastic and plastic films surface morphology implied that there are plastic-degrading bacteria in the microbial community. Further to prove this, the samples were diluted and spread on MB agar plates. After several rounds of purification by plate streaking, a total of 16 colonies were isolated. Different colonies were selected for 16S rRNA gene sequencing and uploaded to the EzBioCloud (http://www.ezbiocloud.net) for comparison. The isolated pure strains were cultured in MMC medium with PS as the sole carbon and energy source, and their growth was observed to judge the degradation ability (Supplementary Figure S4). Seven strains were initially found to be able to grow well with plastic (Table 1), where Bacillus cereus and Acinetobacter johnsonii have been reported that they could degrade PS plastics (Yuan et al., 2022; Zhao et al., 2024). Some bacteria were isolated that were unable to degrade plastics, and they may have survived on metabolites of other bacteria. In addition, smaller irregular nanoplastic were found agglomerated together in the medium containing PS films, suggesting that the films could release microplastics and nanoplastics in the medium which could have an effect on the bacterial community.
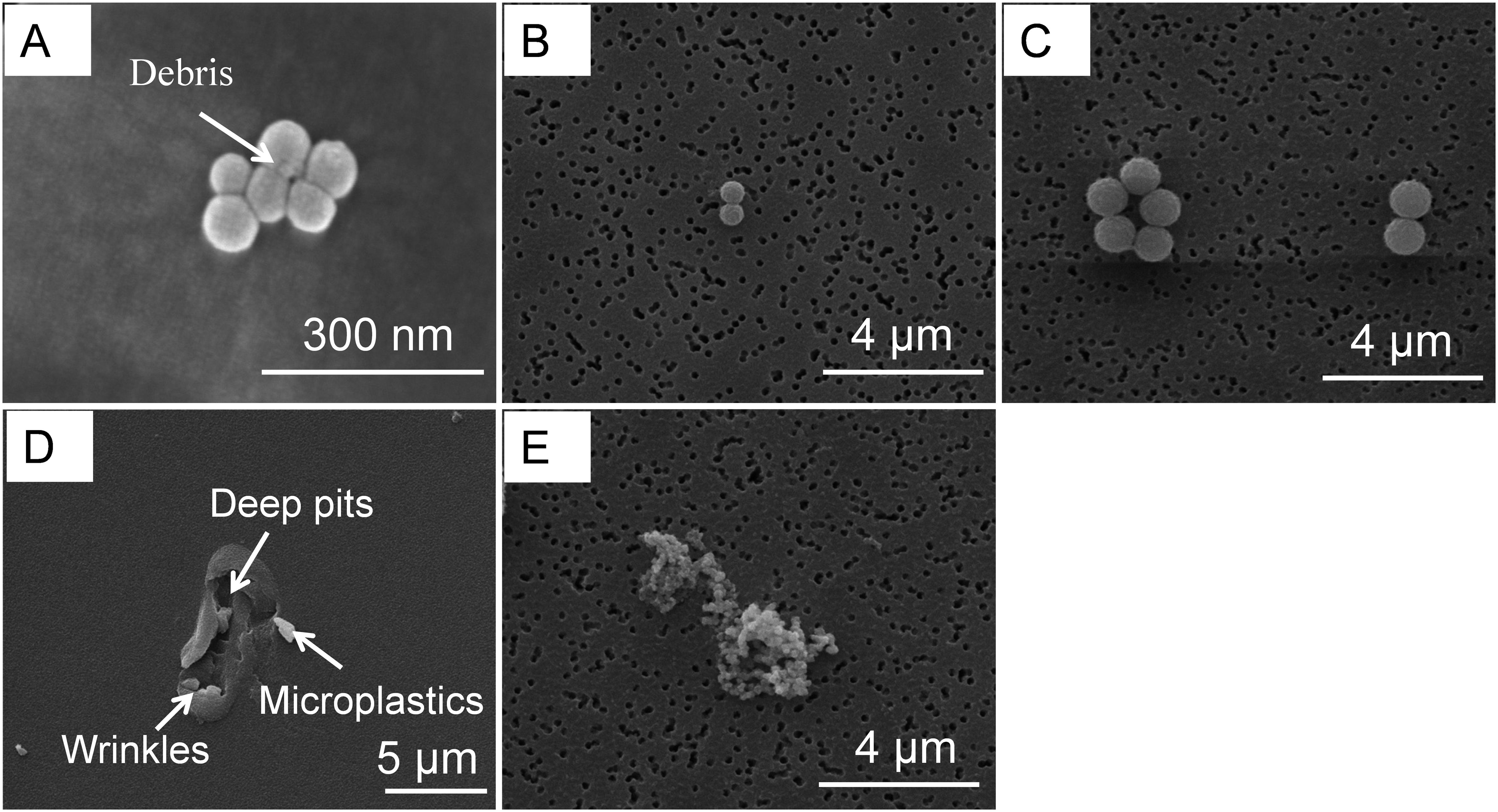
Figure 1. The change of plastic morphology with bacterial communities cultured for 50 days at 28 °C. (A) 60 nm. (B) 600 nm. (C) 1 µm. (D) plastic films surface. (E) microplastic released from plastic films.
3.2 Interaction of bacterial community with plastics
To determine the interaction between microplastics and microbial communities, the phenomenon was observed by SEM (Figure 2). In these images, PS nanoplastics (60 nm) form aggregates that were tightly clustered around the cells (Figure 2A). The agglomeration phenomenon can cause direct physical damage to the cells, such as alteration of the membrane structure or disruption of the cell wall (Zhao et al., 2020; Gong et al., 2023). In contrast, the plastic particles of 600 nm and 1 µm microplastics were relatively spread out without a significant aggregation phenomenon (Figure 2B, C). It was also observed that EPS, which consists mainly of protein and polysaccharide, was secreated by microorganisms. EPS plays a key role in the formation of microbial biofilm. Firstly, it helped microorganisms in the environment to be colonized easily, forming a small ecological niche known as the “Plastisphere” (He et al., 2022). Secondly, EPS could protect the microorganisms from toxic external stresses (Gong et al., 2023; Zhao et al., 2023). They could actively adjust their EPS release and composition on response to unfavourable stress conditions when subjected to excessive external stress (Supplementary Figure S5). Large pieces of microplastic were also found in the samples enriched for bacterial communities using plastic films, which had fragmented into nanoplastics and be attached by microorganisms (Figure 2D). This phenomenon suggested that microplastics were released by the plastic films into the culture medium, which may be utilized by the bacterial community or affect the growth of the microorganisms.
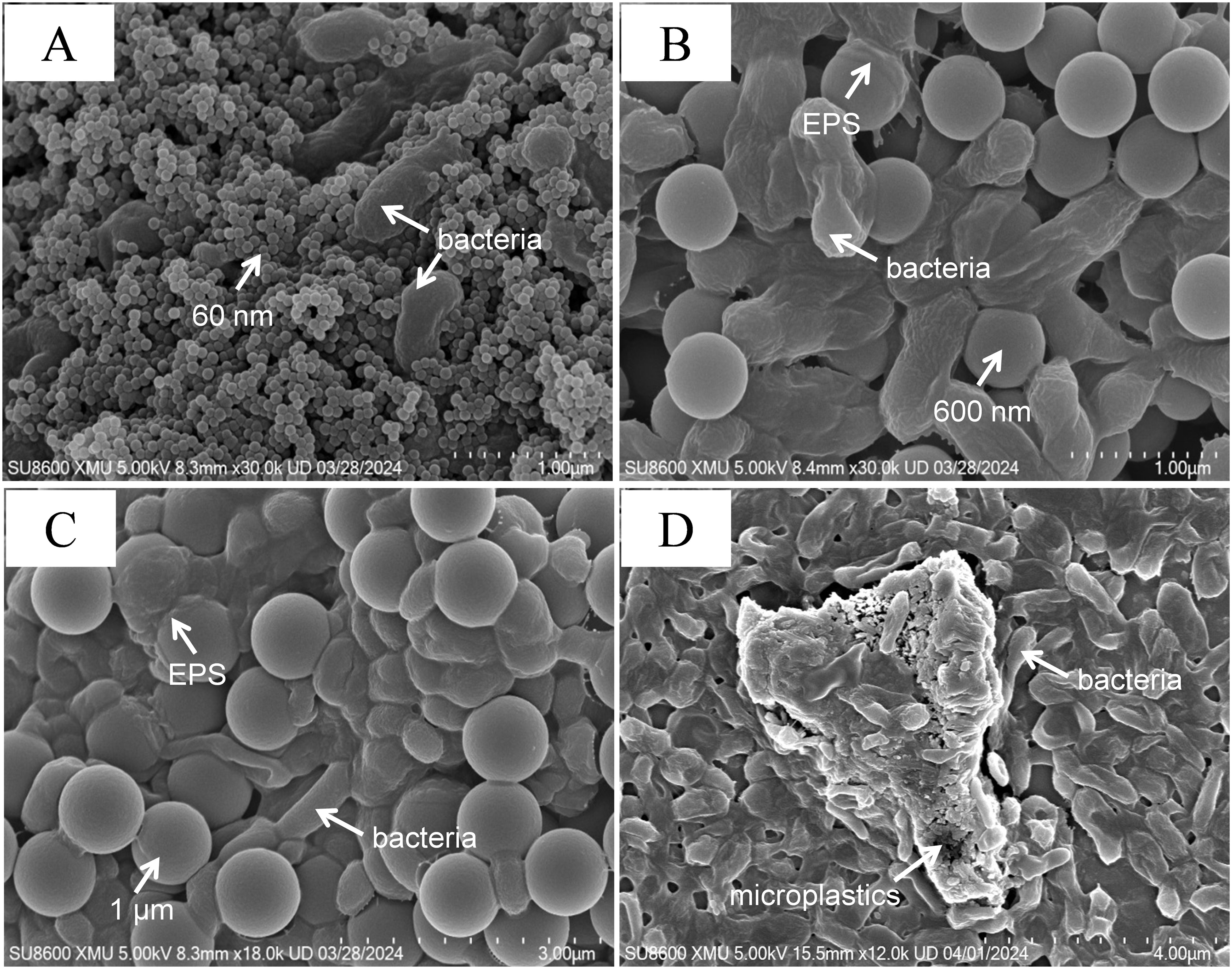
Figure 2. SEM images of bacterial communities interacting with PS. (A) 60 nm. (B) 600 nm. (C) 1 µm. (D) microplastics released from PS films.
In order to observe the effects of microplastics and films on the growth of deep-sea bacterial communities, the growth rates of bacterial communities in the enriched samples were examined (Figure 3A). In all samples, the growth of the bacterial community was inhibited. The strong size effect resulted in the lowest growth of the bacterial community exposed to 60 nm microplastics, which did not exceed 1%. And there was no significant difference between different concentrations (p > 0.05). Exposed to 600 nm and 1 µm microplastics, the growth rate of the bacterial community increased significantly with increasing microplstic concentration (p< 0.05). This may be due to the presence of DOC in the medium as an additional carbon source for the growth of heterotrophic microbes. The source of DOC in the samples was attributed to two reasons, one is that PS can be degraded by plastic-degrading bacteria of communities to produce water-soluble low molecular weight oligomers. Secondly, plastics could leach DOC in seawater, accompanied by an increase in plastic-derived DOC with the addition of plastic (Romera-Castillo et al., 2018; Chen et al., 2023). Further, plastic biodegradation could lead to aging, which released more DOC than new plastics. For instance, the amount of DOC leached from aging plastics (PE and PP) was up to two orders of magnitude higher than that of virgin plastics (Romera-Castillo et al., 2022). The levels of DOC in all samples were determined during the experiment, as predicted (Supplementary Figure S6). The little growth rate of microorganisms was observed in the medium containing the plastic films, which was considered to be inhibited by nanoplastics released from the films. This was confirmed by SEM of microplastics in the medium within the incubation process (Figure 2).
Figure 3. Toxic effects of PS on bacterial community after 50 days of incubation. (A) Growth rate. (B) ROS. Different letters above the bars indicate significant differences (p< 0.05) among samples. CK: untreated bacterial community; B1: 60 nm 10 mg/L; B2: 60 nm 50 mg/L; B3: 60 nm 150 mg/L; B4: 600 nm 10 mg/L; B5: 600 nm 50 mg/L; B6: 600 nm 150 mg/L; B7: 1 µm 10 mg/L; B8: 1 µm 50 mg/L; B9: 1 µm 150 mg/L; B10: plastic films treatment.
Oxidative damage caused by ROS generation was usually considered as one of the main toxicity mechanisms of nanoparticles (Ning et al., 2022). In this study, intracellular ROS generation of bacterial communities exposed to microplastics and plastic films was measured at the same cell density. As shown in Figure 3B, ROS level was significantly enhanced in all samples when compared with the control group (p< 0.05). Lower levels of ROS were produced by the bacterial community exposed to 600 nm and 1 µm microplastics for 50 days and there was no significant difference between the different concentrations (p > 0.05). Bacterial community in the medium containing 60 nm microplastic and plastic films produced a higher amount of ROS. Exposure to 60 nm led to more ROS production with increase concentration of nanoplastics (p< 0.05). The possible reason for this was that the external environmental stress generated by the higher concentrations of microplastics resulted in increased level of ROS (Qiang and Cheng, 2021). The ROS was also produced in the medium containing plastic film (B10), which was supposed to be the result of oxidative stress in the microorganisms caused by the small particle size microplastics released from the film. This was consistent with the results of other studies. For example, Escherichia coli and Bacillus sp. produced more ROS exposed to 60 nm for 8-11 h than the larger PS particles (220, 430, 700, 1040, 1700 and 2260 nm) (Kim et al., 2022). The high level of ROS production may result in tolerant microorganisms remaining alive during long-term enrichment of the bacterial community.
3.3 Effect of mocroplastics on bacterial community diversity
The rarefaction curves based on the indices showed smoothness under different conditions, indicating that the sequencing volume was sufficient to cover the microbial community in all samples (Supplementary Figure S7). The specific values are listed in Supplementary Table S1. The Sobs index was used to indicate community richness. Compared with the control, the addition of PS particles or films resulted in a decrease in the abundance of the biological communities in varying degrees. The lower Sobs values in the 600 nm and 1 µm treatments indicated that larger particle sizes of microplastics had a greater impact on the richness of the microbial communities. The Shannon index was used to indicate the diversity of the biological community. Compared with the control, the diversity of the 60 nm group remained almost unchanged or increased slightly. The diversity of the 600 nm group and 1µm group decreased, implying that the bacterial community was effectively enriched. This further demonstrated that microplastics with larger particle sizes may be better utilized by microorganisms as the sole source of carbon and energy. The growth rate of microorganisms verified this result (Figure 3A). It was also worth our attention that the Sobs and Shannon values in the samples enriched by the addition of PS films were in the middle of the values of the 60 nm and 600 nm experimental groups, which may be due to the production of smaller particle size microplastics from PS films during the microbial degradation process. It has been shown that the average particle size of microplastics produced was 614 nm and 496 nm when PS films was biodegraded by Alcanivorax xenomutans and Halomonas titanicae for 30 days, respectively (Lv et al., 2024). PCoA analysis based on OUT levels showed that the community composition of bacteria was significantly separated under the influence of plastic microspheres and films (Supplementary Figure S8).
3.4 Effect of microplastics on composition and richness of the microbial community structure
Microplastics have a selective effect on microbial communities, while microbial community structural characteristics and compositional types play a crucial role in plastics degradation. Overall, Proteobacteria were dominant in all samples. Microbial populations in all samples were relatively similar at the phylum level, but differences in abundance were observed. Compared with the control, the relative abundance of Proteobacteria decreased and the relative abundance of Bacteroidota increased in all samples with plastic as the sole carbon source. The relative abundance of Actinobacteriota (B1, B2, B5, B6, B7, B10) and WPS-2 (B1) increased (Supplementary Figure S9).
As the level of family (Figure 4), Moraxellaceae have the highest relative abundance in the enriched samples, accounting for 48.2%-96.0% of species. It was followed by Alcanivoracaceae (0.1%-21.8%), Rhizobiaceae (0.1%-12.1%). There was a decrease in the abundance of Moraxellaceae and an increase in the abundance of Alcanivoracaceae in the samples exposed to 60 nm and plastic films than that in the samples exposed to 600 nm and 1 µm microplastics. It was evident that the growth of Moraxellaceae would be inhibited due to its relative sensitivity to 60 nm nanoplastics. And the reason for this phenomenon in plastic films is that smaller nanoplastics were released during biodegradation. The increase in the abundance of Alcanivoracaceae may have a certain degree of tolerance to small particle size microplastics. Among the available reports, Alcanivoracaceae occupied considerable abundance in polluted areas containing polycyclic aromatic hydrocarbons (PAHs), oil, etc., which was seen to be adaptive to environmental changes (Chikere et al., 2019; Gosai et al., 2022). In summary, microplastics and nanoplastics may be detrimental to the growth of certain species of microorganisms. Also, microplastics as a carbon source may promote the development of rather complex microbial communities.
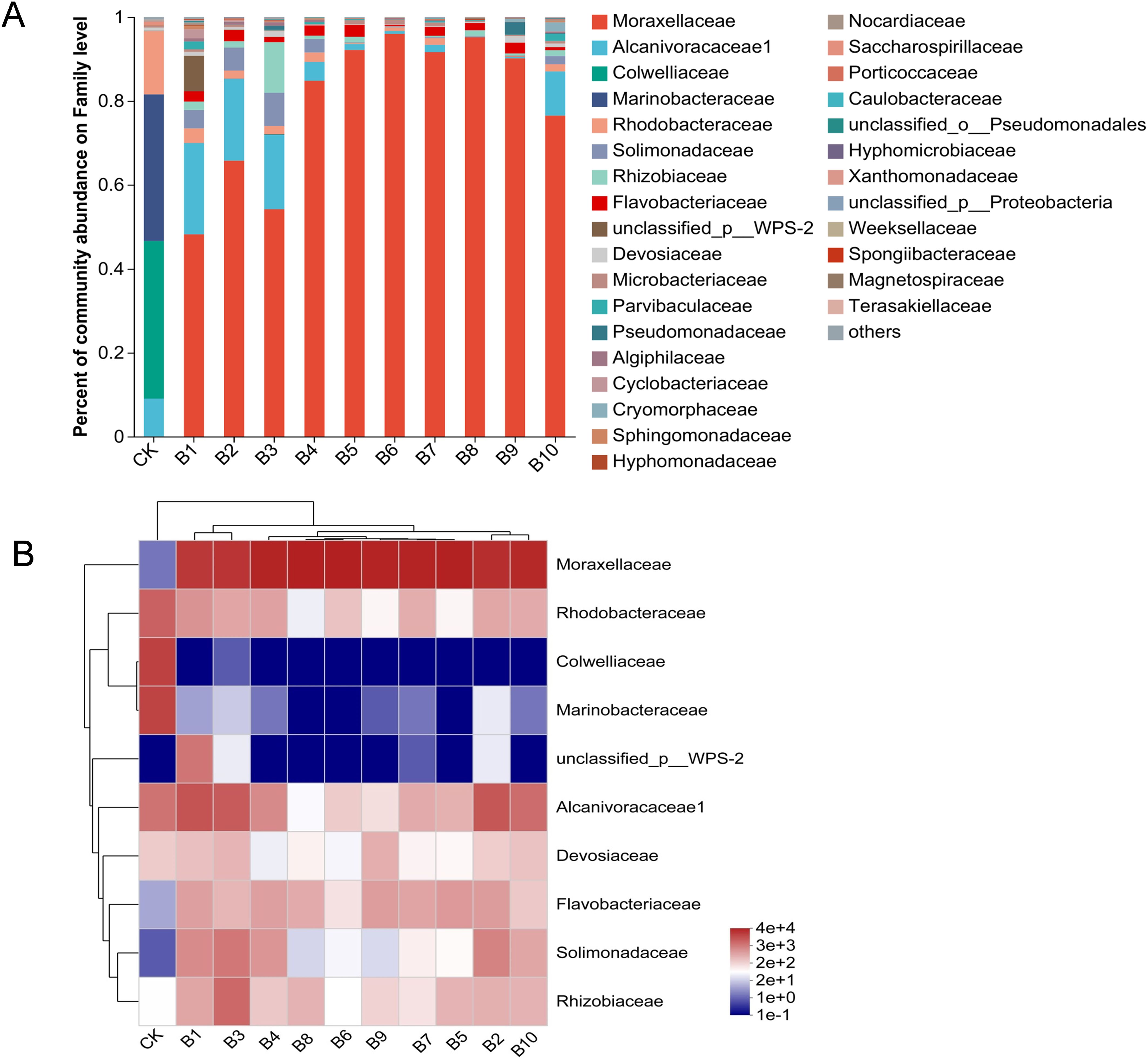
Figure 4. Bacterial community diversity analyses after 50 days enrichment of deep-sea seawater with microplastics and films. (A) Relative abundance of bacteria at the family level of the sample enriched with microplastics and films. (B) Heatmap and hierarchical cluster analysis of the major families. CK: untreated bacterial community; B1: 60 nm 10 mg/L; B2: 60 nm 50 mg/L; B3: 60 nm 150 mg/L; B4: 600 nm 10 mg/L; B5: 600 nm 50 mg/L; B6: 600 nm 150 mg/L; B7: 1 µm 10 mg/L; B8: 1 µm 50 mg/L; B9: 1 µm 150 mg/L; B10: plastic films treatment.
3.5 Analysis of microbial community function
To further investigate the functional characteristics of the microplastics or films enriched samples, the KEGG database was used to analyse the metabolic pathways. As shown in Figure 5A, the KEGG level 1 pathways included six pathways. The order of relative abundance was metabolism > genetic information processing > environmental information processing > human diseases > cellular processes > organismal systems. KEGG level 2 analysis revealed a total of 46 secondary functional genes detected (Figure 5B). The largest proportions of functional abundance were found with global and overview maps, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, and energy metabolism. Compared with the control, amino acid metabolism and carbohydrate metabolism were slightly decreased by the addition of microplastics and films. This may be caused by two factors, one is that ROS produced by microorganisms are constantly stimulated and scavenged due to the prolonged presence of microplastic, further leading to genotoxicity affecting the metabolic functions of microorganisms (Zhang et al., 2023). The second is that some microorganisms in the community can grow using the products resulting from the plastic biodegradation by other microorganisms as a carbon source (Sun et al., 2022a). In contrast, energy metabolism pathways increased, which was probably due to the fact that microorganisms require more energy to sustain growth in a relatively oligotrophic environment and stress from microplastics (Sun et al., 2022a; Wu et al., 2023). Membrane transport is a protein system that governs unfavorable changes in the bacterial environment and plays an essential part in informing environmental risk assessment (Hamlin et al., 2015). Regarding membrane transport and signal transduction genes, functional abundance was decreased due to the addition of microplastics and films. This result showed that both commercial microplastics and secondary microplastics released from plastic films could impair the ability of biofilms to handle environmental information, inhibiting them from rapidly adapting to environmental changes. Higher cell motility protects against toxic and harmful substances (Gong et al., 2023). The abundance of cell motility decreased when microplastics and films were added, especially exposed to 600 nm and 1 µm microplastics, suggesting that larger particle size microplastics could lead to a decrease in the ability of microorganisms to avoid toxic and harmful substances.
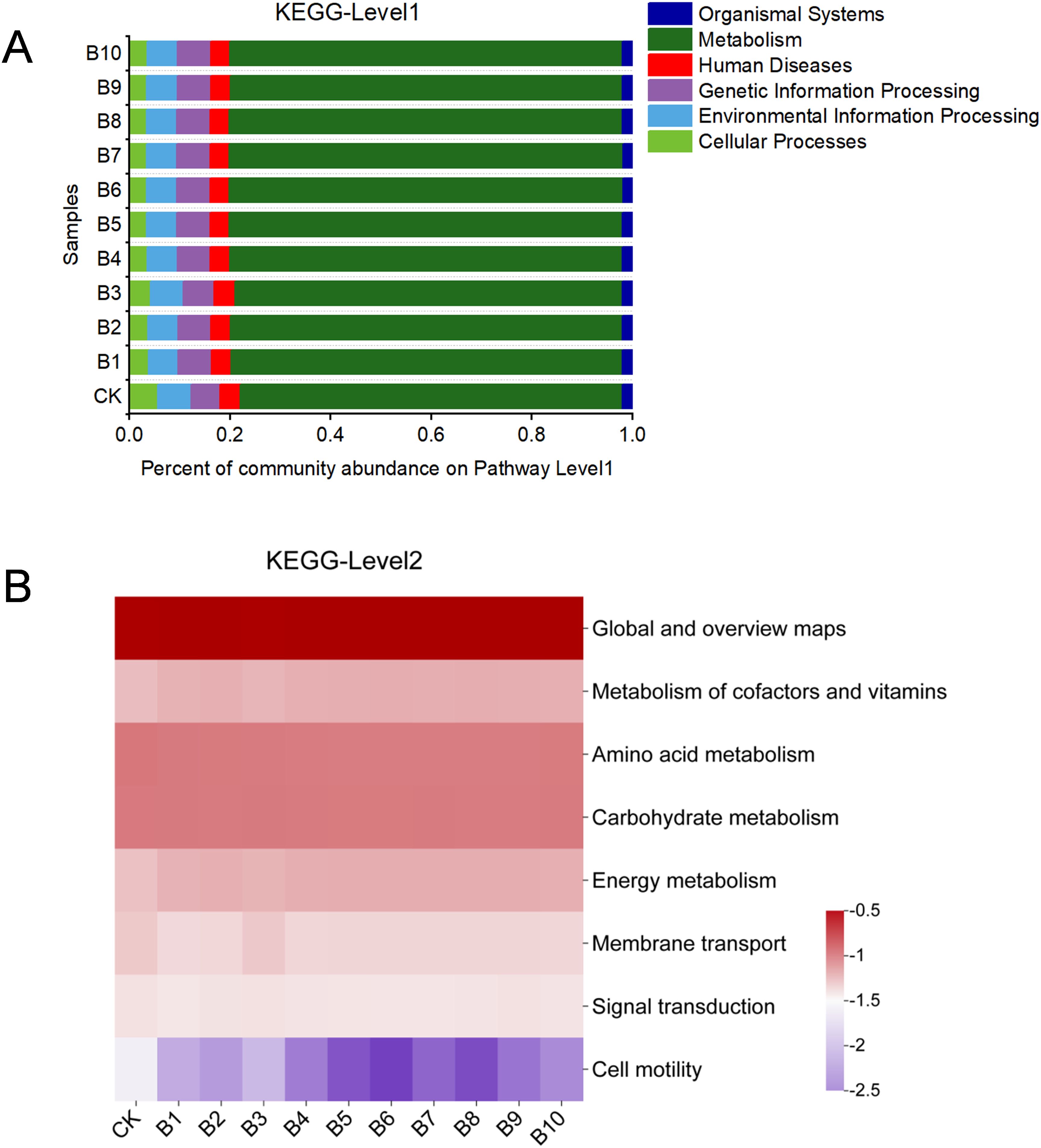
Figure 5. Barplot of KEGG pathway level1 (A) and Heatmap of level2 (B) of the deep-sea bacterial communities with microplastics and films. CK: untreated bacterial community; B1: 60 nm 10 mg/L; B2: 60 nm 50 mg/L; B3: 60 nm 150 mg/L; B4: 600 nm 10 mg/L; B5: 600 nm 50 mg/L; B6: 600 nm 150 mg/L; B7: 1 µm 10 mg/L; B8: 1 µm 50 mg/L; B9: 1 µm 150 mg/L; B10: plastic films treatment.
The phenotypic changes of the bacterial community were analysed using BugBase. As shown in Figure 6A, form-biofilm relative abundance phenotypes increased in the enriched samples after microplastic and film exposure, which is consistent with the enrichment of bacterial communities in soil by microplastics (PS, PVC, PE) (Zhu et al., 2022). Exposure to microplastics and plastic films reduced the relative abundance of potentially pathogenic within 50d. This is not consistent with current results that pathogenic bacteria could be enriched by microplastic. The possible reason for this is that pathogenic bacteria can be enriched in the short term and unfavourable under long-term stress, and the results of bacterial communities enriched by PS microplastics in freshwater sediments confirmed our suspicion (He et al., 2024).
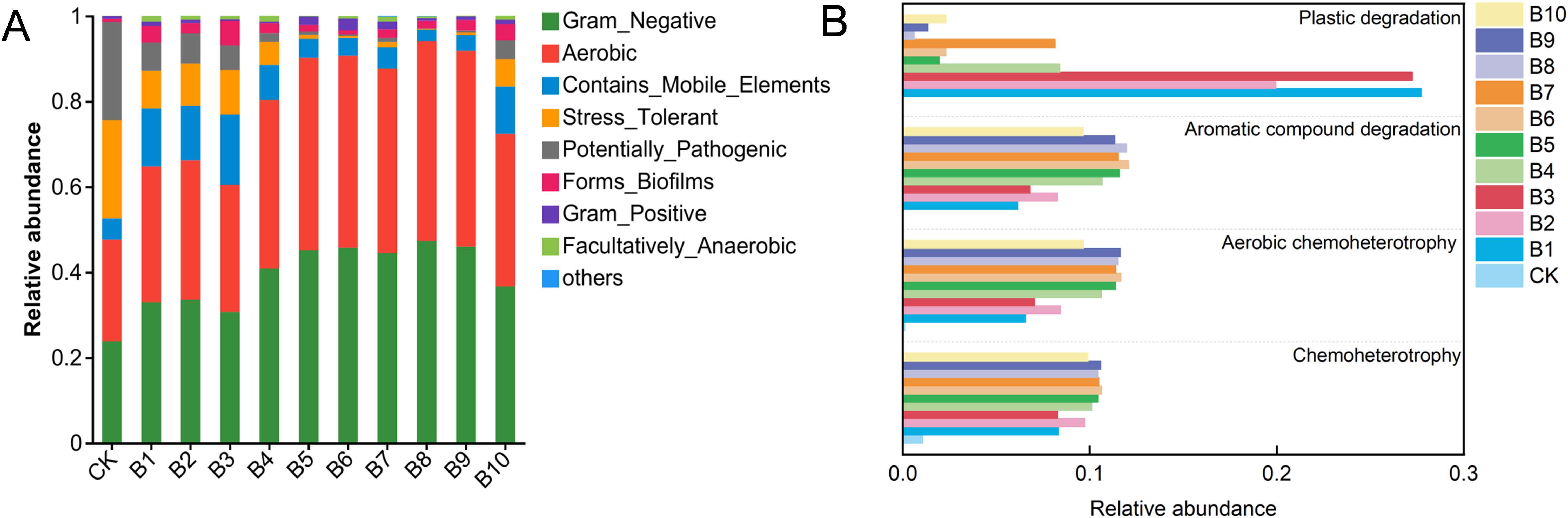
Figure 6. Bacterial community phenotypes (A) and functional gene predictions of microbial communities based on FAPROTAX (B) in different treatment samples. CK: untreated bacterial community; B1: 60 nm 10 mg/L; B2: 60 nm 50 mg/L; B3: 60 nm 150 mg/L; B4: 600 nm 10 mg/L; B5: 600 nm 50 mg/L; B6: 600 nm 150 mg/L; B7: 1 µm 10 mg/L; B8: 1 µm 50 mg/L; B9: 1 µm 150 mg/L; B10: plastic films treatment.
FAPROTAX analysis was used to map the 16S rRNA sequencing data to metabolic or other ecologically relevant functions (Sun et al., 2022b). As shown in Figure 6B, it was highly enriched in all samples on chemoheterotrophy, aerobic chemoheterotrophy and aromatic compound degradation. The relative abundance of chemoheterotrophy and aerobic chemoheterotrophy increased suggesting that the bacteria obtained carbon and energy for their growth mainly by oxidizing organic compounds (Zhang et al., 2018; Sun et al., 2022b). PS is a single-use thermoplastic composed of aromatic styrene monomers (Bhanot et al., 2024). The increase on aromatic compound degradation genes indicated that the samples were effectively enriched. Compared with 60 nm and plastic films, 600 nm and 1 µm resulted in higher enrichment of aromatic compound degradation genes. It was believed that larger particles of microplastics would be more susceptible to microbial growth as a carbon and energy source. Notably, the 60 nm microplastics were significantly enriched for plastic degradation gene, although they severely inhibited the growth of microbial communities. This may be due to the fact that most of the plastic-degrading bacteria in the environment can adapt to the toxic chemical microenvironment of microplastics. There is no much enrichment of plastic degradation gene in the medium containing plastic films even though smaller microplastics could be produced. It may be related to its concentration, shape and other factors, which need to be further verified. The functional analysis of the bacterial community in this study was based on 16S rRNA gene, and further metagenomic analyses are required to fully understand the function of the bacterial community in the enriched samples.
4 Conclusion
This study compared the effects of three particle sizes of PS microplastics (60 nm, 600 nm and 1 µm) in concentration rang of 10 to 150 mg/L on deep-sea microbial communities in control with PS plastic films. The microplastics could be degraded by the bacterial community leading to plastic surface roughness and fragmentation. Microplastics from commercial and plastic films could stimulate the bacterial community to secrete EPS, favouring biofilm formation and resistance to external stress. Larger microplastics stimulated specific bacterial groups with the highest relative abundances of Moraxellaceae. Compared with the larger microplastics, the 60 nm and nanoplastics from plastic films could significantly inhibit the growth of microorganisms and increased ROS production. Functional prediction analysis based on 16S rRNA indicated that the addition of plastics promoted metabolic function and enrichment of functional genes (plastic degradation and aromatic compound degradation) of the bacterial community. However, the effects of microplastics on deep-sea bacteria in situ remain to be elucidated in future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1142753.
Author contributions
SL: Writing – original draft, Writing – review & editing, Visualization, Formal analysis, Data curation. YuL: Writing – review & editing, Visualization, Data curation. QY: Writing – review & editing, Visualization, Data curation. YaL: Writing – review & editing, Data curation. YY: Writing – review & editing, Data curation. YZ: Writing – review & editing, Resources. RL: Writing – review & editing, Resources. SZ: Writing – review & editing, Resources. JX: Writing – review & editing, Resources. LZ: Writing – review & editing, Resources. ZS: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Projects of the National Science Foundation of China (No. 42030412, 92251302 and 91851203), Scientific Research Foundation of Third Institute of Oceanography (No. 2019021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1479919/full#supplementary-material
References
Barrett J., Chase Z., Zhang J., Holl M. M. B., Willis K., Williams A., et al. (2020). Microplastic pollution in deep-sea sediments from the great Australian bight. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.576170
Bhanot V., Mamta, Gupta S., Panwar J. (2024). Phylloplane fungus Curvularia dactyloctenicola VJP08 effectively degrades commercially available PS product. J. Environ. Manage. 351, 119920. doi: 10.1016/j.jenvman.2023.119920
Carbery M., O’Connor W., Thavamani P. (2018). Trophic transfer of microplastics and mixed contaminants in the marine food web and implications for human health. Environ. Int. 115, 400–409. doi: 10.1016/j.envint.2018.03.007
Chen C., Deng Y., Zhou H., Jiang L., Deng Z., Chen J., et al. (2023). Revealing the response of microbial communities to polyethylene micro (nano)plastics exposure in cold seep sediment. Sci. Total Environ. 881, 163366. doi: 10.1016/j.scitotenv.2023.163366
Chen X., Chen X., Zhao Y., Zhou H., Xiong X., Wu C. (2020). Effects of microplastic biofilms on nutrient cycling in simulated freshwater systems. Sci. Total Environ. 719, 137276. doi: 10.1016/j.scitotenv.2020.137276
Chikere C. B., Mordi I. J., Chikere B. O., Selvarajan R., Ashafa T. O., Obieze C. C. (2019). Comparative metagenomics and functional profiling of crude oil-polluted soils in Bodo West Community, Ogoni, with other sites of varying pollution history. Ann. Microbiol. 69, 495–513. doi: 10.1007/s13213-019-1438-3
Eom J., Sarwar A., Lee E. C., Im G.-B., Kim S.-W., Nguyen L. T., et al. (2023). Cytotoxic effect and mechanism of nano-sized polystyrene degraded by Rhodococcus ruber C208. Chem. Eng. J. 473, 145094. doi: 10.1016/j.cej.2023.145094
Goncalves J. M., Benedetti M., d’Errico G., Regoli F., Bebianno M. J. (2023). Polystyrene nanoplastics in the marine mussel Mytilus galloprovincialis.*. Environ. pollut. 333, 122104. doi: 10.1016/j.envpol.2023.122104
Gong X., Ge Z., Ma Z., Li Y., Huang D., Zhang J. (2023). Effect of different size microplastic particles on the construction of algal-bacterial biofilms and microbial communities. J. Environ. Manage. 343, 118246. doi: 10.1016/j.jenvman.2023.118246
Gosai H. B., Panseriya H. Z., Patel P. G., Patel A. C., Varjani S., Shankar A., et al. (2022). Exploring bacterial communities through metagenomics during bioremediation of polycyclic aromatic hydrocarbons from contaminated sediments. Sci. Total Environ. 842, 156794. doi: 10.1016/j.scitotenv.2022.156794
Hamlin H. J., Marciano K., Downs C. A. (2015). Migration of nonylphenol from food-grade plastic is toxic to the coral reef fish species Pseudochromis fridmani. Chemosphere 139, 223–228. doi: 10.1016/j.chemosphere.2015.06.032
He W., Huang J., Liu S., Yu H., Li E., Zhang W., et al. (2024). Effects of microplastics on sedimentary geochemical properties and microbial ecosystems combined with hydraulic disturbance. Sci. Total Environ. 923, 171350. doi: 10.1016/j.scitotenv.2024.171350
He S., Jia M., Xiang Y., Song B., Xiong W., Cao J., et al. (2022). Biofilm on microplastics in aqueous environment: Physicochemical properties and environmental implications. J. Hazard. Mater. 424, 127286. doi: 10.1016/j.jhazmat.2021.127286
Jones D. M., Potts J., Hale M. S. (2024). The sampling and analysis of coastal microplastic and mesoplastic: Development of a citizen science approach. J. Coast. Conserv. 28 (1), 14. doi: 10.1007/s11852-023-01023-0
Kim S. Y., Kim Y. J., Lee S.-W., Lee E.-H. (2022). Interactions between bacteria and nano (micro)-sized polystyrene particles by bacterial responses and microscopy. Chemosphere 306, 135584. doi: 10.1016/j.chemosphere.2022.135584
Li Y., Lu Q., Yang J., Xing Y., Ling W., Liu K., et al. (2023b). The fate of microplastic pollution in the Changjiang River estuary: A review. J. Clean. Prod. 425, 138970. doi: 10.1016/j.jclepro.2023.138970
Li R., Tao J., Huang D., Zhou W., Gao L., Wang X., et al. (2023a). Investigating the effects of biodegradable microplastics and copper ions on probiotic (Bacillus amyloliquefaciens): Toxicity and application. J. Hazard. Mater. 443, 130081. doi: 10.1016/j.jhazmat.2022.130081
Liu X., Ma J., Yang C., Wang L., Tang J. (2021). The toxicity effects of nano/microplastics on an antibiotic producing strain - Streptomyces coelicolor M145. Sci. Total Environ. 764, 142804. doi: 10.1016/j.scitotenv.2020.142804
Liu R., Zhao S., Zhang B., Li G., Fu X., Yan P., et al. (2023). Biodegradation of polystyrene (PS) by marine bacteria in mangrove ecosystem. J. Hazard. Mater. 442, 130056. doi: 10.1016/j.jhazmat.2022.130056
Lv S., Cui K., Zhao S., Li Y., Liu R., Hu R., et al. (2024). Continuous generation and release of microplastics and nanoplastics from polystyrene by plastic-degrading marine bacteria. J. Hazard. Mater. 465, 133339. doi: 10.1016/j.jhazmat.2023.133339
Ning Q., Wang D., An J., Ding Q., Huang Z., Zou Y., et al. (2022). Combined effects of nanosized polystyrene and erythromycin on bacterial growth and resistance mutations in Escherichia coli. J. Hazard. Mater. 422, 126858. doi: 10.1016/j.jhazmat.2021.126858
Qiang L., Cheng J. (2021). Exposure to polystyrene microplastics impairs gonads of zebrafish (Danio rerio). Chemosphere 263, 128161. doi: 10.1016/j.chemosphere.2020.128161
Romera-Castillo C., Birnstiel S., Alvarez-Salgado X. A., Sebastian M. (2022). Aged plastic leaching of dissolved organic matter is two orders of magnitude higher than virgin plastic leading to a strong uplift in marine microbial activity. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.861557
Romera-Castillo C., Pinto M., Langer T. M., Alvarez-Salgado X. A., Herndl G. J. (2018). Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat. Commun. 9, 1430. doi: 10.1038/s41467-018-03798-5
Seeley M. E., Song B., Passie R., Hale R. C. (2020). Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat. Commun. 11 (1), 2372. doi: 10.1038/s41467-020-16235-3
Sun Y., Duan C., Cao N., Ding C., Huang Y., Wang J. (2022a). Biodegradable and conventional microplastics exhibit distinct microbiome, functionality, and metabolome changes in soil. J. Hazard. Mater. 424, 127282. doi: 10.1016/j.jhazmat.2021.127282
Sun Y., Duan C., Cao N., Li X., Li X., Chen Y., et al. (2022b). Effects of microplastics on soil microbiome: The impacts of polymer type, shape, and concentration. Sci. Total Environ. 806, 150516. doi: 10.1016/j.scitotenv.2021.150516
Tsuchiya M., Kitahashi T., Nakajima R., Oguri K., Kawamura K., Nakamura A., et al. (2024). Distribution of microplastics in bathyal- to hadal-depth sediments and transport process along the deep-sea canyon and the Kuroshio Extension in the Northwest Pacific. Mar. pollut. Bull. 199, 115466. doi: 10.1016/j.marpolbul.2023.115466
Wu Y. Y., Tian W. F., Cheng C. X., Yang L., Ye Q. Q., Li W. H., et al. (2023). Effects of cadmium exposure on metabolism, antioxidant defense, immune function, and the hepatopancreas transcriptome of Cipangopaludina cathayensis. Ecotoxicol. Environ. Saf. 264, 115416. doi: 10.1016/j.ecoenv.2023.115416
Xayachak T., Haque N., Lau D., Emami N., Hood L., Tait H., et al. (2023). White spill: Life cycle assessment approach to managing marine EPS litter from flood-released pontoons. Chemosphere 337, 139400–139400. doi: 10.1016/j.chemosphere.2023.139400
Yuan J., Cao J., Yu F., Ma J. (2022). Microbial degradation of polystyrene microplastics by a novel isolated bacterium in aquatic ecosystem. Sustain. Chem. Pharm. 30, 100873. doi: 10.1016/j.scp.2022.100873
Zhang X., Hu B. X., Ren H., Zhang J. (2018). Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Sci. Total Environ. 633, 518–528. doi: 10.1016/j.scitotenv.2018.03.158
Zhang X., Li Y., Lei J., Li Z., Tan Q., Xie L., et al. (2023). Time-dependent effects of microplastics on soil bacteriome. J. Hazard. Mater. 447, 130762. doi: 10.1016/j.jhazmat.2023.130762
Zhao S., Liu R., Lv S., Zhang B., Wang J., Shao Z. (2024). Polystyrene-degrading bacteria in the gut microbiome of marine benthic polychaetes support enhanced digestion of plastic fragments. Commun. Earth Environ. 5, 162. doi: 10.1038/s43247-024-01318-6
Zhao T., Tan L., Zhu X., Huang W., Wang J. (2020). Size-dependent oxidative stress effect of nano/micro-scaled polystyrene on Karenia mikimotoi. Mar. pollut. Bull. 154, 111074. doi: 10.1016/j.marpolbul.2020.111074
Zhao Z., Zheng X., Han Z., Yang S., Zhang H., Lin T., et al. (2023). Response mechanisms of Chlorella sorokiniana to microplastics and PFOA stress: Photosynthesis, oxidative stress, extracellular polymeric substances and antioxidant system. Chemosphere 323, 138256. doi: 10.1016/j.chemosphere.2023.138256
Zhou X., Xiao C., Zhang B., Chen T., Yang X. (2024). Effects of microplastics on carbon release and microbial community in mangrove soil systems. J. Hazard. Mater. 465, 133152. doi: 10.1016/j.jhazmat.2023.133152
Keywords: microplastics, deep-sea microbial community, biodegradation, oxidative stress, 16S rRNA
Citation: Lv S, Li Y, Yuan Q, Lu Y, Ye Y, Zhong Y, Liu R, Zhao S, Xia J, Zeng L and Shao Z (2024) Influence of microplastics on the structure and function of deep-sea communities during long-term enrichment processes. Front. Mar. Sci. 11:1479919. doi: 10.3389/fmars.2024.1479919
Received: 13 August 2024; Accepted: 16 October 2024;
Published: 07 November 2024.
Edited by:
Hans Uwe Dahms, Kaohsiung Medical University, TaiwanReviewed by:
Naga Radha Srinivas Tanuku, Council of Scientific and Industrial Research (CSIR), IndiaDongdong Zhang, Zhejiang University, China
Copyright © 2024 Lv, Li, Yuan, Lu, Ye, Zhong, Liu, Zhao, Xia, Zeng and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongze Shao, c2hhb3p6QDE2My5jb20=
 Shiwei Lv
Shiwei Lv YuFei Li1
YuFei Li1 Yonglian Ye
Yonglian Ye Zongze Shao
Zongze Shao