- 1Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 2State Key Laboratory of Marine Pollution, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 3Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
- 4Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
- 5Liaoning Key Laboratory of Marine Animal Immunology, Dalian Ocean University, Dalian, China
Editorial on the Research Topic
Applications of environmental DNA in the aquatic ecosystem management of East Asia
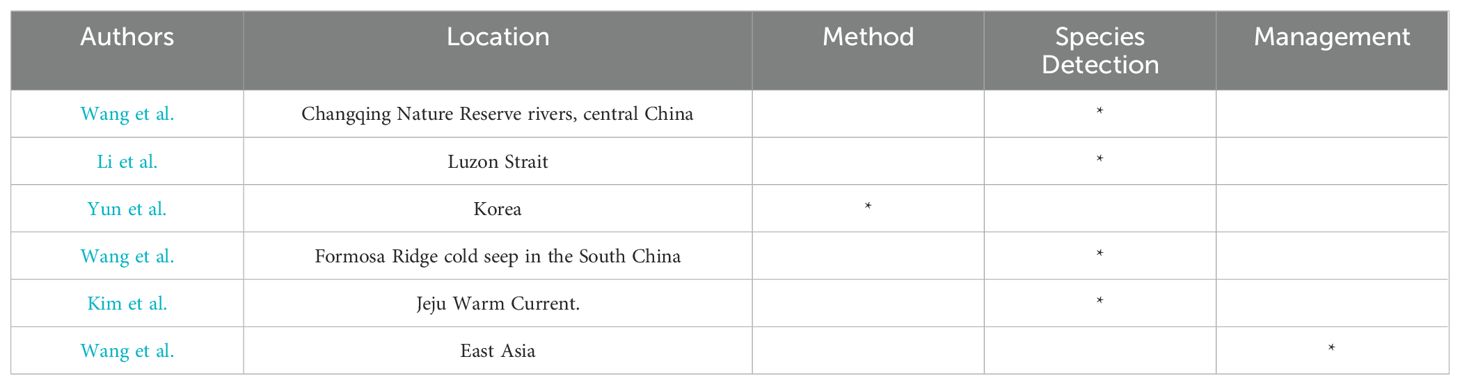
Environmental DNA (eDNA) is the extra-organismal genetic material from the target species, which allows us to detect the presence or recent presence of species without direct observation or capturing whole organisms. The idea of using eDNA and molecular analysis to infer organism presence has repeatedly and independently occurred in aquatic environments. The advances in eDNA-based identification are expected to promote the sensitivity, accuracy and cost-effectiveness of monitoring and the subsequent assessment. Thus, eDNA analysis has rapidly developed into a promising tool across a large variety of research fields including biodiversity assessment, alien invasive species monitoring, rare species conservation, etc. Furthermore, the fast pace of development and improvements in next-generation sequencing (NGS) technologies can reflect broader and more robust applications in eDNA research. In Europe, North America and South America, eDNA are widely used as near-stand-alone biodiversity assessments and beginning to expand from water samples into air and soil environmental samples (Jerde, 2021). In East Asia, including countries like China, Japan, and South Korea, the applications of eDNA are also becoming increasingly widespread. This includes its use in biodiversity monitoring, invasive alien species detection, technological innovation, and integration with management and policy. This Research Topic includes six papers on eDNA research in the East Asian region. While they may not cover all aspects, it is hoped that they can provide valuable reference points for further studies (Table 1).
Wang et al. compared fish diversity and distribution across 29 sampling points in the Changqing Nature Reserve rivers, central China, using both eDNA and traditional sampling methods. They identified 46 unique fish species: eDNA detected 34 species, while traditional methods found 22. Notably, eDNA revealed more species within individual points, identifying 24 species not found by traditional methods, which in turn identified 12 unique species. Although eDNA showed a broader range of species, traditional methods often provided higher Shannon diversity index values. Using β-diversity indices (Bray-Curtis and Jaccard), along with NMDS and PCoA, differences were explored between the methods. Despite distinct capabilities in capturing biodiversity, no significant statistical differences were found in overall biodiversity measurement. Therefore, the integrated methodology enriches species detection and unveils nuanced understanding of species distribution and relative abundance across different locales.
Li et al. used DNA barcoding to assess larval fish diversity in the Luzon Strait and adjacent waters. From 15 stations, 385 larval fish were collected, with 354 successfully barcoded, identifying 147 species from 93 genera, 44 families, and 22 orders. Interspecific K2P divergence was significantly higher than intraspecific divergence, confirming DNA barcoding’s feasibility. Significant variations in species diversity and community composition were observed, with greater diversity in western regions influenced by the Kuroshio current. Economically valuable species were identified, highlighting their ecological significance. Establishing a local DNA sequence database will enhance species identification in eDNA applications, supporting sustainable fisheries management and conservation.
Yun et al. developed a novel method for selectively detecting intracellular DNA (iDNA) from specific species in eDNA samples by applying PMA treatment to Alexandrium spp., preventing non-selective extracellular DNA (exDNA) signals. Optimizing filter size in sampling allowed selective collection and analysis of iDNA, particularly from Alexandrium spp. cysts in sediment. The combined use of PMA treatment and filter size optimization significantly enhanced selective detection of iDNA. These results demonstrate the practical applicability of this method for eDNA monitoring, advancing environmental conservation, monitoring, and ecological research.
Wang et al. used 16S rRNA amplicons to analyze bacterial communities in six deep-sea sponge species from the Formosa Ridge cold seep in the South China Sea. Bacterial communities in these closely related sponge species were dominated by Proteobacteria (mainly Gammaproteobacteria and Alphaproteobacteria) but varied in diversity and composition. Most samples were dominated by the SUP05 clade (Thioglobaceae), with diverse SUP05 operational taxonomic units (OTU) phylotypes indicating significant divergence. OTUs from the Methylomonadaceae family also showed substantial genetic distance. These findings support the hypothesis of host-species specificity in sponge-associated bacterial communities. Dominant functional microbes, such as sulfur- and methanol-oxidizing bacteria, likely play crucial roles as chemosynthetic symbionts in adapting sponges to the cold seep environment. This study enhances our understanding of bacterial diversity in deep-sea sponges and the significance of functional microbes in cold seep ecosystems.
Kim et al. used 18S rRNA amplicon sequencing and light microscopy at nine stations to analyze protistan community structures. The Jeju Warm Current created a thermohaline front, dividing the region into two areas with similar planktonic biomass but distinct protistan communities. Dinoflagellates were more prevalent in warm, saline waters (stations E35, E44, E45), while diatoms and picochlorophytes were more common in cooler waters (stations E32, E42). Higher species richness and Shannon Diversity Index values were observed in the warm waters, suggesting increased protistan diversity due to the Jeju Warm Current. Harmful algal bloom species were found in the warm Kuroshio-originating waters, indicating potential risks for the Yellow Sea and Korea Strait. The study highlights that changes in regional current systems could significantly impact protistan community structure.
Wang et al. reviewed major aquatic invasive species in East Asian countries and the application of eDNA technology in their detection. Using China as a case study, it discussed integrating the 4E strategy (Early detection, Eradication, Education, and Enforcement) with eDNA technology for monitoring biological invasions. The paper explored the potential of eDNA technology in species diversity management and policy development, providing guidance for establishing effective monitoring systems. The integration of eDNA with the 4E strategy offered significant promises for improving policies related to aquatic biological invasions and biodiversity management.
This Research Topic aims to stimulate interdisciplinary research on the application of eDNA methods or results in aquatic environmental assessment, especially the river, lake or bay area in east Asia. The bays and riversides of East Asia are densely populated areas. In recent years, with the increasing population activities, the aquatic environment has undergone dramatic changes. The problems of biological invasions, biodiversity and rare animal conservation have gradually received more and more attention. In the field of eDNA research in East Asia, we also face challenges related to data formatting and sharing. To address this issue, we advocate for researchers to actively participate in the “Making eDNA FAIR (Findable, Accessible, Interoperable, Reusable)” project (Takahashi and Berry, 2023). By doing so, researchers can help ensure that eDNA data is more easily discoverable, accessible, interoperable, and reusable. This will not only improve research efficiency but also foster interdisciplinary and cross-regional collaboration, ultimately advancing the field of eDNA research. The application of eDNA methods may play an important role in the assessment of aquatic environments.
Author contributions
CL: Writing – review & editing, Writing – original draft. MY: Writing – review & editing. WX: Writing – review & editing. NL: Writing – review & editing. LG: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. CL acknowledges support from the National Key R&D Program of China (2022YFC2601500 & 2022YFC2601504).
Acknowledgments
The editors would like to thank the authors, reviewers, and the Frontiers in Marine Science development team, whose efforts have led to the success of this Research Topic.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Jerde C. (2021). Can we manage fisheries with the inherent uncertainty from eDNA. J. Fish Biol. 8, 341–353. doi: 10.1111/jfb.14218
Keywords: environmental DNA, aquatic environment, biodiversity, invasive species, rare wild species
Citation: Liu C, Yan M, Xiong W, Li N and Gao L (2024) Editorial: Applications of environmental DNA in the aquatic ecosystem management of East Asia. Front. Mar. Sci. 11:1473463. doi: 10.3389/fmars.2024.1473463
Received: 31 July 2024; Accepted: 02 September 2024;
Published: 18 September 2024.
Edited and Reviewed by:
Christopher L. Jerde, University of California, Santa Barbara, United StatesCopyright © 2024 Liu, Yan, Xiong, Li and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Conghui Liu, bGl1Y29uZ2h1aUBjYWFzLmNu
†Present address: Ning Li, State Key Laboratory of Mariculture Breeding, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
 Conghui Liu
Conghui Liu Meng Yan
Meng Yan Wei Xiong
Wei Xiong Ning Li
Ning Li Lei Gao
Lei Gao