- 1Key Laboratory of Marine Environment and Ecology, Ministry of Education of China, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Ecology and Environmental Science, Laoshan Laboratory, Qingdao, China
Dust deposition can supply nutrients to the upper ocean, and subsequently affect primary production and biodiversity in planktonic ecosystem, but the differential response among taxa and their interactions are not fully understood. Here, we performed 7-day onboard incubation experiment amended with different dust loadings (0, 0.2 and 2 mg L-1) in the Kuroshio-Oyashio transition region of the Northwest Pacific Ocean and characterized the community structure and microbial network of a planktonic microbiome in response to dust addition. Chlorophyll a and nutrient analysis indicated that dust-derived nitrogen promoted the growth of phytoplankton (165.8–293.6%) and phytoplankton size structure shifted towards larger cells (>3 μm). Metabarcoding sequencing, targeting prokaryotic and eukaryotic microbes, revealed the changes in community structure and co-occurrence network in response to dust addition. Dust induced a shift from dinoflagellate dominance toward diatom dominance in phytoplankton, and favored members of Cercozoa, Labyrinthulomycetes and Saprospirae, which showed positive correlation with diatom. Temporal response patterns among taxa were categorized into five clusters, and collectively pointed to a more sensitive microeukaryotic community than prokaryotic one in response to dust. The community turnover during the incubation was dominated by moderate taxa with 55.71-62.26% moderate OTUs transitioning to abundant or rare taxa, and dust addition stimulated the transitions of rare taxa. Moreover, biotic factors shaped planktonic microbiome more than abiotic factors, particularly the cross-domain interaction significantly affected microeukaryotic community. Notably, dust addition enhanced the co-occurrence network complexity, with the number of keystone taxa increased, suggesting more interspecies interactions were induced by dust. With integrated analysis, our findings highlight the differential sensitivity of planktonic microbiome to dust deposition and the effects could pass on other organisms through interspecies interaction.
1 Introduction
Atmospheric dust deposition serves as a unique source of external nutrients to the upper ocean (Duce et al., 1991; Jickells et al., 2005; Chien et al., 2016), providing relatively substantial level of nitrogen (N) and iron (Fe) with lower levels of phosphorus (P; Moreno and Martiny, 2017; Chu et al., 2018). The external nutrient input from dust plays a role in modulating nutrient structure, primary production and the biogeochemical cycles in marine ecosystems. For example, on a segment of the Kuroshio Current near the shelf break of the East China Sea, the continuous dust eruption process caused the increase of surface N in the sea area (Chung et al., 2011). Similarly, the high N:P ratio (N:P>40) of nutrient input through African dust also contributed to P limitation in the western tropical Atlantic Ocean off Barbados (Chien et al., 2016). A well-recognized response of marine ecosystem to the dust deposition includes the stimulation of phytoplankton biomass as demonstrated in observational, modelling and microcosm studies (Li et al., 2021; Ma et al., 2021). However, due to the difference in the trophic state of the ambient waters and the nutrient composition in dust, the fertilizing effects of dust-derived N are not uniform across marine ecosystems: three main categories, relief, supplementation and enhancement of the N demand, have been characterized in low-nutrient low-chlorophyll zone (LNLC), high-nutrient low-chlorophyll zone (HNLC) and euphotic zone, respectively (Zhang et al., 2019a).
The differential response of phytoplankton to nutrient fertilization from the dust deposition is size-dependent (Guo et al., 2012; Liu et al., 2013). For example, dust enrichment shifted the dominant phytoplankton from pico-sized to nano- or micro-sized cells in the Yellow Sea (Liu et al., 2013; Zhang et al., 2018), while it stimulated pico-sized phytoplankton in the Mediterranean Sea and the Western Tropical Atlantic Ocean, and favored all size fractions in some cases (Giovagnetti et al., 2013; Chien et al., 2016). The relationship between the size-dependent contribution to total chlorophyll a (Chl a) and the available nutrient has been quantified based on integrative analysis of multiple regions (Zhang et al., 2022). The species- or lineage-specific physiology and behavior also contribute to the succession of dominant phytoplankton, such as the different nutrient demand and uptake efficiency between diatom and dinoflagellate (Maranon et al., 2015). Dust deposition could also directly affect other players in planktonic ecosystem including heterotrophic bacteria, heterotrophic and mixotrophic protist, or through trophic interactions (Guo et al., 2014; Chen et al., 2019). Both abundance and activity of plankton could be affected, as shown in heterotrophic bacteria (Chen et al., 2020). The changes in size, community and trophic structure, as well as biodiversity will further drive ecosystem function and stability.
Asian dust is the second largest contributor to global dust aerosols (Tegen and Schepanski, 2009; Chen et al., 2017), with annual dust emissions ranging from 230 to 540 Tg (Yumimoto and Takemura, 2015). Driven by strong westerly winds, Asian dust transports large amounts of nutrients to downwind areas, including Chinese marginal seas and open oceans in the Northwest Pacific Ocean (NWPO; Shao et al., 2011). The Kuroshio-Oyashio transition region is one of the typical locations in NWPO for exploring the response of plankton community to Asian dust deposition. It is located between the Kuroshio extension and the Subarctic front and is influenced by the southwestern Kuroshio and the northern Oyashio currents (Long et al., 2019). The surface seawater of this region shows HNLC characteristics. Dust deposition as well as haze particles supplements the N demand of phytoplankton and shifts the phytoplankton community size structure towards larger cells, as evidenced by size-fractionated Chl a and microscopic analysis in several independent onboard incubation experiments conducted in this region (Zhang et al., 2018, 2019a; b; Zhang et al., 2020). However, how the diversity of non-phytoplankton microbes changes with dust deposition in this region has received much less attention.
Metabarcoding methods have been widely used to assess the composition and diversity of planktonic microbiomes in global oceans (e.g. de Vargas et al., 2015; Tapolczai et al., 2019), and also their response to atmospheric deposition (e.g. Meng et al., 2016; Chen et al., 2020; Li et al., 2021; Maki et al., 2021; Wang et al., 2022). However, prokaryotes and eukaryotes are examined separately in the majority of these studies. Despite the technical requirement to prepare amplicon libraries separately for prokaryotes and eukaryotes due to their unique nucleotide characters, prokaryotic and eukaryotic microbes collectively constitute the natural microbiome and collaboratively contribute to ecological processes. Recently, there has been a growing focus on the combined examination of microbial prokaryotic and eukaryotic plankton (e.g. Santi et al., 2019; Sun et al., 2022). This has unveiled that the distribution patterns and environmental sensitivity differ significantly between the two domains, and that interactions across these domains are crucial in shaping the community, sometimes even taking precedence over abiotic factors. Also, such joint analysis allows to better understand the multitrophic diversity from bacteria to invertebrate in the aquatic ecosystem under environmental changes (Li et al., 2020). In addition, there is growing evidence from high-throughput metabarcoding studies that non-abundant taxa (i.e. rare and moderate) are the major players in driving ecosystem multifunctionality and maintaining stability (Sun et al., 2017; Ziegler et al., 2018; Xiong et al., 2020). Several investigations have revealed that aquatic protistan assemblage can rapidly and regularly reconstruct themselves to yield unique combination of dominant taxa. A highly dynamic and resilient microbial subcommunity, often rare and moderate taxa respond swiftly to changing environments and contribute to the community turnover (Vigil et al., 2009; Lynch and Neufeld, 2015; Newton and Shade, 2016; Sun et al., 2017). However, to our best knowledge, previous studies using metabarcoding approach to investigate the effect of atmospheric deposition only looked at either one domain (prokaryotes/eukaryotes) or target phytoplankton using lineage-specific primers. Little is known on the plankton microbiome dynamics as a whole, how cross-domain interaction and non-abundant biosphere contribute to the community shift in response to dust deposition.
Here, we conducted onboard incubation amended with low and high loadings of Asian dust using seawater sampled from the Kuroshio-Oyashio transition region in the NWPO. We profiled biodiversity from bacterial to microeukaryotic plankton in the incubated seawater using metabarcoding approach, and investigated the effects of dust deposition on the community composition, dynamics and molecular ecological network. As the first attempt, we jointly analyzed prokaryotic and eukaryotic microbes, and characterized the subcommunities of different abundance under the effect of dust. Meanwhile, total and size-fractioned Chl a, and nutrients of incubated seawater were also determined. It is hoped that this study will contribute to a deeper understanding on the response of planktonic microbiome to dust deposition and its driving factors.
2 Materials and methods
2.1 Preparation of Asian dust
The dust used in this study originated from a source of Asian dust events crossing over the NWPO (Shao et al., 2011; Tan et al., 2013), Mu Us Desert (37.92°N, 107.11°E). The soil samples were collected and artificially modified following Zhang et al. (2018) to simulate the aging process of dust particles in the atmosphere. Briefly, soil particles smaller than 20 µm were selected and mixed with synthetic cloud water. The treated dust was weighted and stored dry at -20°C.
2.2 Study area and experimental design
The 7-day microcosm experiment was performed from April 26 to May 3, 2015, during the spring cruise in NWPO onboard the R/V DongFangHong 2. In the Kuroshio-Oyashio transition region, surface seawater was collected at B1 station (37.93°N, 146.98°E) from a depth of 5 m, using the water sampler mounted on CTD (SeaBird SBE 911). For the ambient baseline condition (hereafter ‘day 0’), after prefiltered through 200-μm mesh to remove large zooplankton, seawater was immediately collected for size-fractionated Chl a, DNA and ancillary measurements as described below. For the microcosm experiment, prefiltered seawater was aliquoted into nine 20-L acid-washed polycarbonate carboys (round and clear plastic container for large volume culture, Nalgene). Three groups were prepared in triplicate: i) ‘control’, no dust addition; ii) ‘low’, 0.2 mg L-1 of dust particles in incubated seawater; iii) ‘high’, 2 mg L-1 of dust. We added such amount of dust, aiming to simulate the deposition flux to the surface of the NWPO in mild (2 g m-2) and strong (20 g m-2) dust events, respectively (Iwamoto et al., 2011; Shi et al., 2012). The carboys were placed in an on-deck incubator with circulating seawater to keep near-ambient surface temperature. Neutral density filters were used to cover the carboys, reducing light intensity to ~40% of the incident. Temperature and light intensity were monitored three times per day. Size-fractionated Chl a and nutrient concentrations were measured on daily basis, while seawater samples for morphological and metabarcoding analysis were collected on day 0, 2, 4 and 7.
2.3 Chl a
For each carboy, 300 mL incubated seawater was sequentially filtered through 3 µm (Φ47 mm, Millipore) and 0.2 µm polycarbonate filters. Filters were wrapped in aluminum foil and stored at -20 °C. After 24-h extraction by 90% acetone (v/v) at -20°C in dark, the pigment content on each filter were quantified by measuring the absorbance of supernatant using a Trilogy fluorometer (Turner Designs). The total Chl a concentration was the sum of the two size fractions.
2.4 Inorganic nutrients
The leachable inorganic nutrients in the artificially modified dust were determined in the laboratory before the cruise. Briefly, the dust (10 mg L-1) was added into prefiltered (0.45 μm) oligotrophic seawater, and placed in ultrasonic bath at 0 °C for 30 min. The leaching solution was then filtered through acid-washed cellulose acetate membranes and the filtrate was immediately stored in acid-washed high-density polyethylene bottles at -20°C for nutrient analysis. In the field, 200 mL ambient seawater and 200 mL incubated seawater from each carboy were collected following the same sampling protocol. NO3-, NO2-, PO43- and SiO32- concentrations were measured by Auto Analyzer3 continuous-flow analyser (SEAL Analytical).
2.5 Microscopic analysis of phytoplankton
Subsample (500 mL) from the ambient seawater (day 0) and incubated seawater (day 2, 4 and 7) were fixed with 1% neutral Utermöhl’s solutions and stored at 4 °C in dark. Before microscopic examinations, fixed samples were settled for 48 h in dark and then concentrated to 10~50 mL. Phytoplankton were identified under microscope (YS100, Nikon) and cell count was obtained using Sedgwick-Rafter chamber.
2.6 DNA extraction, PCR amplification and amplicon sequencing
On day 0, day 2, day 4 and day 7, 1 L sample water were sequentially filtered through 3 µm and 0.2 µm polycarbonate membrane (Millipore, Φ47 mm). The filters were immediately stored in 1 mL 100 mM EDTA, and then frozen in liquid nitrogen until DNA extraction.
Membrane filters of 3 μm and 0.2 μm from the same incubated samples were pooled together for DNA extraction. DNA extraction followed the method described by Yuan et al. (2015). Briefly, membrane filters were cut with sterile scissors and homogenized using microbeads in sample homogenizer (Bioprep-24, Allsheng). Homogenate in 2 mL DNA extraction buffer (100 mM EDTA, 1% SDS, and 10 mg mL-1 proteinase K) was incubated at 56°C for 24 h. DNA was extracted using the CTAB method, purified with the DNA Clean & Concentrator kit (Zymo Research), and eluted in 25 μL 10 mM Tris-HCl. DNA concentrations and purities were examined on a NanoDrop 2000 Spectrophotometer (Thermo Scientific). DNA integrity was determined using 1% agarose gel electrophoresis.
The V4 region of the eukaryotic 18S rRNA gene was amplified with the primer pair 18SNCF3 (5’- CGCGHAAATTRCCCAATCY -3’) and 18SV4R2 (5’- CTWACTTTCGTTCTTGATYAA -3’). Thermal cycling was as follows: 95°C for 5 min, followed by 5 cycles of 95°C for 10 s, 56°C for 30 s, and 72°C for 30 s, and then 25 cycles of 95°C for 10 s, 52°C for 30 s, and 72°C for 30 s with a final extension at 72°C for 10 min. The V4 region of prokaryotic 16S rRNA gene was amplified with primer pair 515F (5’-GTGYCAGCMGCCGCGGTAA-3’) and 806R (5’- CAAYAAATCYRAGAATTTCACCTCT-3’) (Caporaso et al., 2011), following the thermal cycling: 95°C for 5 min, followed by 25 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 40 s, with a final extension at 72°C for 10 min. PCR reactions were performed in a total volume of 25 µL using Ex Taq (Takara), with 20~300 ng template gDNA. All amplicons were purified and paired-end sequenced using Illumina MiSeq PE300 platform (GENEWIZ).
2.7 Sequence analysis
Raw reads were subjected to quality filtration by removing reads of low quality, adaptor contamination, high N content and low complexity to obtain clean reads with Cutadapt (v1.9.1; Martin, 2011), Vsearch (v1.9.6; Rognes et al., 2016) and Qiime (v1.9.1; Bolyen et al., 2019). The remaining high-quality paired-end reads were assembled into metabarcodes, and non-singleton ones subsequently clustered into OTUs (Operational Taxonomy Units) with 97% similarity using USEARCH (v7.0.1090; Edgar, 2013). For taxonomic assignment, the representative OTU sequences of 18S rDNA were classified using sintax algorithm (Edgar, 2016) based on PR2 database (Guillou et al., 2012), with a confidence threshold set at 0.8. The representative OTU sequences of 16S rDNA were classified using the RDP classifier (Ribosomal Database Program, v2.2; Wang et al., 2007) based on the Greengene database (DeSantis et al., 2006). The ecological functions of bacterial communities were further annotated using FAPROTAX (Louca et al., 2016).
In addition to the total community, abundant, moderate and rare subcommunities were analyzed separately. OTUs with a relative abundance greater than 0.1% of the overall sequences in a single library (i.e. sample) were considered as ‘abundant’ OTUs, those with a relative abundance less than 0.01% were considered as ‘rare’ OTUs, and the rest were considered as ‘moderate’ OTUs (Campbell et al., 2011). Moreover, we further classified all OTUs into six categories (Xue et al., 2018): Define OTUs that were abundant OTU in all samples as ‘always abundant’ OTUs; OTUs that were rare OTU in all samples as ‘always rare’ OTUs; OTUs that were always moderate OTU in all samples as ‘always moderate’ OTUs; OTUs that were abundant or moderate OTU in all samples as ‘conditional abundant’ OTUs; OTUs that were rare or moderate OTU in all samples as ‘conditional rare’ OTUs; and the rest were considered as ‘conditional abundant and rare’ OTUs.
2.8 Net nitrogen conversion index
Net nitrogen conversion index (NCEI, μg μmol-1) was calculated using the equation below (Zhang et al., 2018):
where t represented the incubation duration (7 days), Chl ai represented the concentration of Chl a at the ith day (μg L-1), Chl a0 represented the initial concentration of Chl a on the 0th day in the control (μg L-1), ΔN represented consumption value of dissolved inorganic nitrogen (DIN) during incubation (μmol L-1).
2.9 Diversity and statistical analyses
For alpha-diversity, the ‘vegan’ package in R (v4.2.2) was used to calculate diversity indices including Chao 1, Pielou’ evenness and Shannon-Wiener index (Oksanen et al., 2013). Beta-diversity was visualized in Non-Metric Multidimensional Scaling (NMDS) with Bray-Curtis dissimilarity distance calculated in PRIMER 6 (Clarke and Gorley, 2009), and the significance of differences among groups was tested using ANOSIM. Beta diversity was partitioned into turnover and nestedness components by applying the function beta.pair within the R package (‘betapart’) following Jiao et al. (2017).
To characterize the differential response of different taxa, the fold change of relative abundance between treatment and control were calculated. Heatmap with hierarchical clustering was generated by ‘heatmap’ and ‘ggplot’ package in R. Complete linkage and Euclidean distance were used in hierarchical clustering. To further grouping the time series pattern, soft clustering was performed in Mfuzz.
2.10 Molecular ecological network construction
To ensure that the network analysis was supported by sufficient data, control and low dust treatment were merged into ‘Con-Low’ group to characterize planktonic microbiome that were not or weakly affected by dust additions. Low and high dust treatments were merged into the ‘Low-High’ group to characterize the community affected or strongly affected by dust additions. Spearman’s correlations between the relative abundance of OTUs were calculated using the ‘rcorr’ function in the ‘Hmisc’ package, and co-occurrence networks were constructed based on these correlations. OTUs detected in over 30% of all samples with robust correlation (|spearman rho| > 0.8 and p < 0.01) were selected for network analysis. Networks were visualized in Gephi (v0.9.4; Bastian et al., 2009) and topological parameters of networks were calculated in the ‘igraph’ R package (Csardi and Nepusz, 2006). Spearman’s correlation between topological parameters and environmental factors were calculated and visualized in circus plot using the ‘circlize’ package in R. Nodes featured with high degree (>120) and low betweenness centrality (<2000) were considered to be keystone species in the co-occurrence network.
3 Results
3.1 Variation in nutrients and Chl a
The baseline conditions at B1 were featured as HNLC, with a N:P ratio (14:1) close to the Redfield ratio but low Chl a concentration 0.78 g L-1 (Supplementary Table 1). The >3 μm size fraction only accounted for 26.1% of the total Chl a.
Prior to the field experiments, we determined the leachable nutrient contents in the dust. The NO3-+NO2 content reached 989.23 μmol g-1, while the PO43- (4.53 μmol g-1) and SiO32- (6.60 μmol g-1) contents were relatively low (Figure 1A). By summing the baseline values and dust-derived ones, we calculated the amended nutrient concentrations in the dust treatments, yielding 3.9% and 39.0% increase of NO3-+NO2 in the low and high dust treatments, respectively (Figure 1B). But the increases of PO43- and SiO32- concentrations were negligible, even in the high dust treatments, which were only 2.5% and 0.2%, respectively.
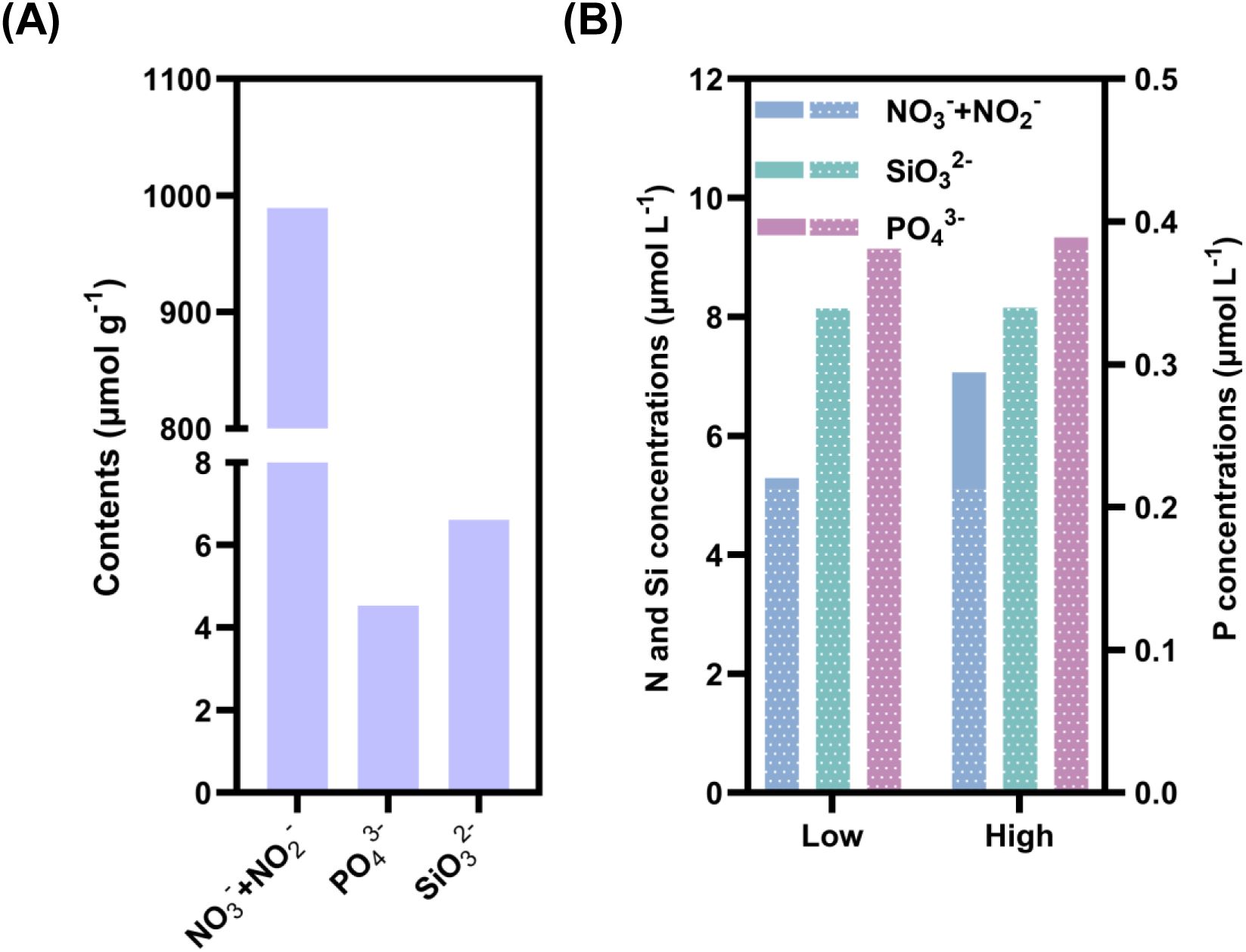
Figure 1. (A) Nutrient content in dust and (B) changes in incubated seawater after the addition of dust.
During the on-board incubation, nutrients and size-fractionated Chl a were monitored on a daily basis. Overall, nutrients showed a decline trend over the time in all groups (Figures 2A–C), with the greatest decrease found in the high dust treatment (13.6~43.0%). In spite of the decrease, the NO3-+NO2 concentrations in the high dust treatment remained higher than those in control and low treatment, reaching a comparable concentration on day 7. The PO43- and SiO32- concentrations were roughly similar among groups in the first 4 days, but the differences were more pronounced towards the end of the incubation. N:P ratios in the control and low dust treatment varied in the range of 11-17, while those in the high treatment (16-18) maintained above Redfield ratio with a slight decrease detected on day 6 and 7. Although the N:Si ratios were elevated by dust-derived N, ratios remained below one regardless of treatments (Figure 2E).
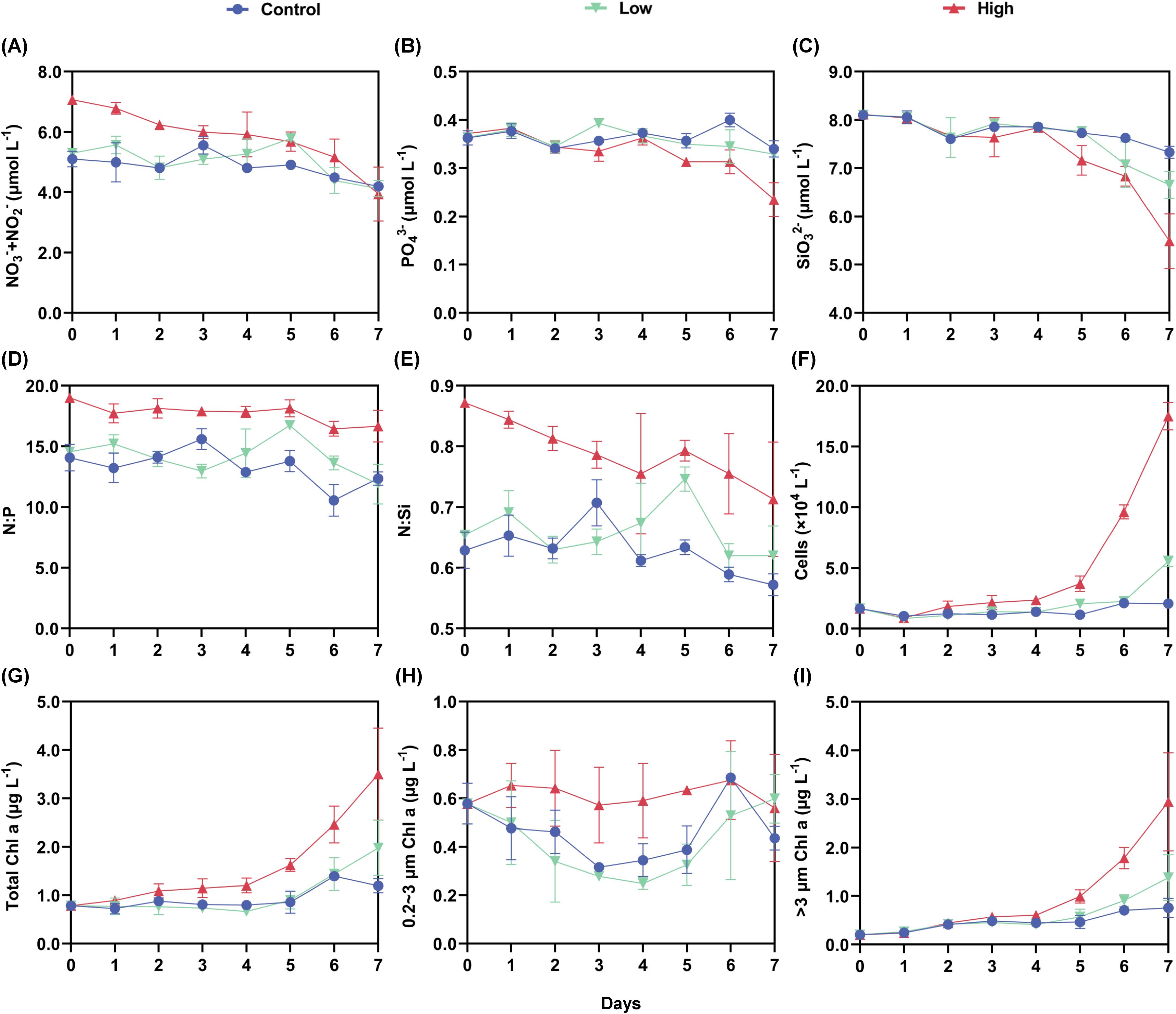
Figure 2. Variation in macronutrient concentrations, stoichiometric ratios and Chl a concentrations during the incubation experiment. (A) NO3-+NO2-. (B) PO43-. (C) SiO32-. (D) N:P. (E) N:Si. (F) Cell density of phytoplankton. (G) Total Chl a. (H) 0.2~3 μm fraction Chl a. (I) 3~200 μm fraction Chl a.
The consumption of nutrients came with the increase of Chl a concentration and phytoplankton abundance (Figures 2G–I). The high dust addition promoted the total Chl a concentration throughout the incubation with the significant increase detected after day 4 (p<0.05), and the value peaked on day 7 being 293.6% of the control. But the enhancing effect of low dust addition on total Chl a was not significant until day 7 (p>0.05). The Chl a increase was mainly contributed by >3 µm size fraction, taking up 69.7% and 84.0% of the total Chl a in low and high dust treatment on day 7, respectively. The specific growth rate of larger phytoplankton (using either >3 µm size fraction Chl a or phytoplankton abundance) was positively correlated with available N in the incubation (R2 = 0.2134, p<0.05, Supplementary Figure 3). However, such correlation was not observed in 0.2~3 µm size fraction. NCEI value further indicated the positive effect of the added N on phytoplankton growth, which was more pronounced in high dust treatment and >3 µm size fraction (Supplementary Figure 4).
3.2 Taxonomic composition
Microeukaryotic and prokaryotic plankton communities were examined parallelly, yielding 903,119 high-quality 18S rDNA metabarcodes and 2,510 OTUs, 464,927 high-quality 16S rDNA metabarcodes and 838 OTUs, respectively. The taxonomic composition was uneven across samples showing both temporal and intergroup variation (Figure 3). The microeukaryotic communities were further analyzed in three categories, phytoplankton, heterotrophic protist and metazoan. the relative abundance of phytoplankton reads in dust treatments peaked on day 7, which was 2.05-2.16 folds of the control (Supplementary Figure 5A), in concert with the trend of Chl a concentration and cell density (Supplementary Figure 7). Compared to the baseline condition, a shift from dinoflagellate dominance toward diatom dominance was observed in all groups, but dust addition particularly enhanced the relative abundance of diatom, which was increased by 120.5~137.7% relative to the control on day 7. Such increase was mainly contributed by genera Thalassiosira, Chaetoceros, Pseudo-nitzschia, and Navicula (Supplementary Figure 8), accounting for 80.28~87.88% of the total diatom reads in dust treatments. The relative abundance of reads and cell density were positively correlated in diatom (R2 = 0.6348, p<0.01) but not observed in dinoflagellate (Supplementary Figure 7).
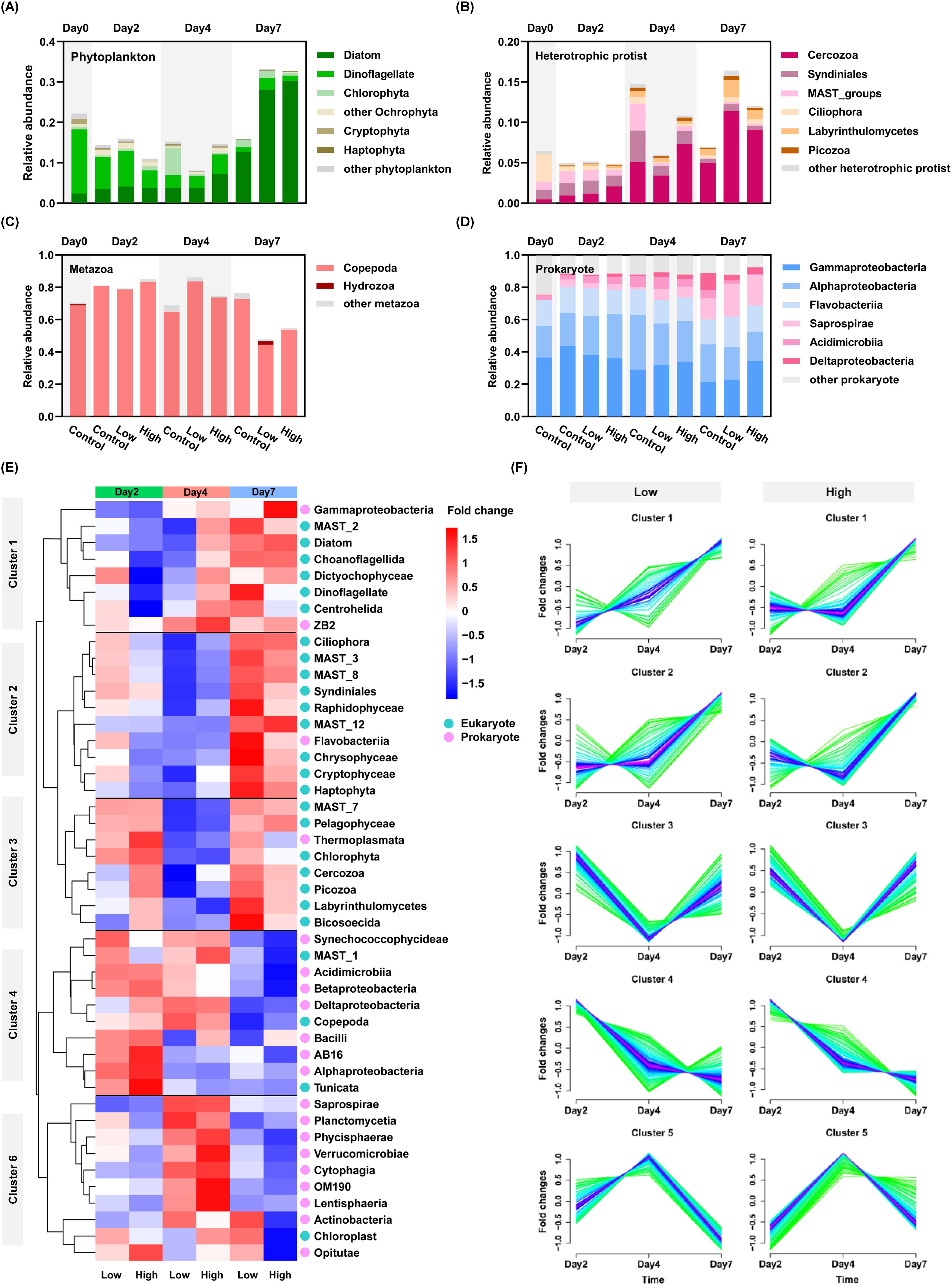
Figure 3. Community composition and variation pattern of plankton microbiome during the incubation experiment. (A) Phytoplankton, (B) Heterotrophic protist, (C) Metazoan, (D) Prokaryote, (E) Heatmap showing the ratio of relative abundance between the control and dust treatments. The clustering used Euclidean distances to calculate distances between data, a complete method to determine similarity, and the relative abundance ratios were scaled by row (z-core). (F) Mfuzz analysis showing temporal dynamics in the ratio of relative abundance between dust treatments and Control.
The relative abundance of heterotrophic protist in the dust treatments generally showed an increasing trend over time, while that in the control peaked on day 4 and fell back to a comparable level to the baseline (Figure 3B; Supplementary Figure 5B). The relative abundance of Cercozoa in the low and high dust treatments was 2.34 and 1.71 folds of that in the control, respectively. This was mainly due to the increase of Cercozoa, in which filose taxa (subphylum Filosa) were predominant, taking up 29.2~39.7% of the total Cercozoa reads in dust treatments. The relative abundance of Cercozoa was positively correlated with diatom (R2 = 0.7366, p<0.01, Supplementary Figure 9A). Moreover, despite lower abundance, Labyrinthulomycetes was also stimulated by dust addition, being 148.14~277.78% of the control on day 7. Majority (>93.1%) of the metazoan reads were affiliated with copepod, followed by Urochodata (tunicate) and Hydrozoa. Slight increases were observed in all groups on day 2 and 4, but pronounced decrease occurred in dust treatments on day 7 with the relative abundance falling to 62.5~71.5% of that in the control. The variation in taxonomic composition of bacterioplankton was not as strong as that of microeukaryotes (Figure 3D). Gammaproteobacteria, Alphaproteobacteria and Flavobacteriia dominated across all samples (60.09~80.25%). The most remarkable change was observed in Saprospirae, of which the relative abundance on day 7 increased 5.2-8.3 folds relative to the baseline condition, and that in the low and high dust treatments was 156.74% and 144.01% of that in the control. The relative abundance of Saprospirae was positively correlated with that of diatoms (R2 = 0.9066, p<0.001, Supplementary Figure 9B). The ecological function cellulolysis detected in bacterioplankton was exclusively affiliated with Saprospirae (Supplementary Figure 10).
To better characterize the differential response among taxa to dust addition and describe the temporal dynamics, we performed cluster analyses based on the fold change between the relative abundance of certain taxa in the dust treatments and that in the control. With the effect of dust, the variability of taxonomic profile in eukaryotes was significantly greater than that in prokaryotes, but functional profile was relatively stable in terms of prokaryotes, except for the function cellulolysis (Figure 3E; Supplementary Figure 11). Both lineage- and OTU-level clustering yielded 5 clusters (Figures 3E, F). Generally, cluster 1-3 pointed to the promotional effects of dust which was characterized by the higher relative abundance in the dust treatments on day 7, while cluster 4-5 showed the opposite trend. When considering temporal variations, more complex response was observed among lineages. Except for the metazoan, the majority of eukaryotic taxa fell into cluster 1-3. The dominant prokaryotic taxa, Gammaproteobacteria, Flavobacteriia and Saprospirae, were also found in cluster 1-3, while the rest of prokaryotes in cluster 4-5.
3.3 Dynamics of abundant, moderate and rare subcommunities
To investigate the dynamics of different subcommunities throughout the incubation, we categorized OTUs by their relative abundance in each sample. Both microeukaryotic and prokaryotic plankton communities were dominated by few abundant taxa, as 8.0~24.7% of OTUs per sample were classified as abundant taxa and accounted for 49.3~87.8% of reads (Figure 4A). Non-abundant taxa (i.e. moderate and rare taxa) contributed to the majority of community richness, representing 75.3~92.0% of OTUs per sample. Remarkably, the contribution of moderate taxa was significantly higher in prokaryotic community relative to that in eukaryotic one, taking up averagely 35.67% of the richness and 41.32% of the reads (Supplementary Figure 12). Moreover, most of the control- or dust treatment-specific OTUs belonged to rare taxa (Supplementary Figure 13).
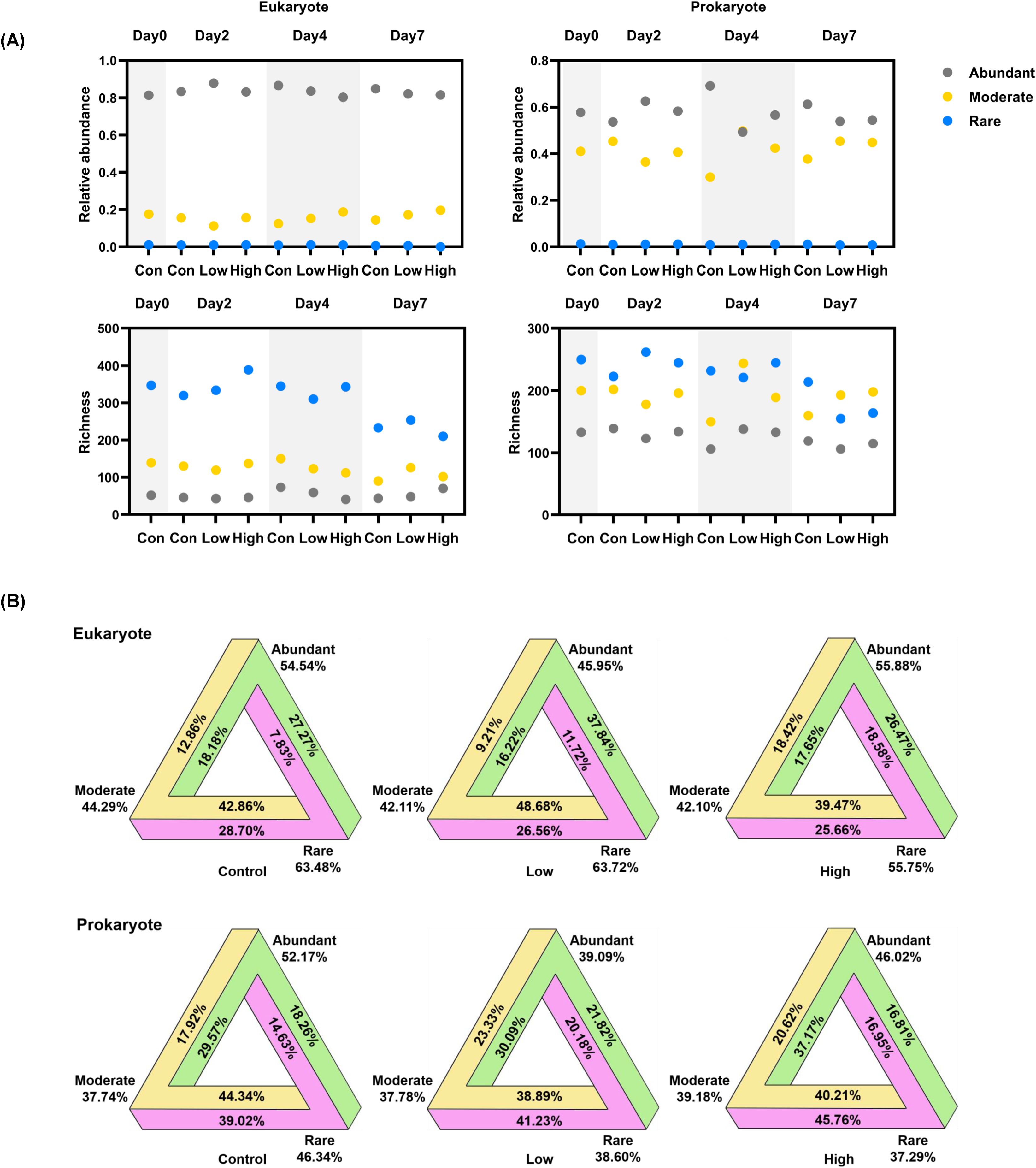
Figure 4. Dynamics of abundant, moderate and rare subcommunities. (A) Relative abundance and richness of abundant, moderate and rare subcommunities; (B) The proportion of each subcommunity transit to other subcommunities during the incubation (from day 0 to day 7).
To address the possible transitions among abundant, moderate and rare taxa, we further categorized OTUs by their relative abundance across all samples (see methods). A total of 617 18S rDNA OTUs (59.91%) representing 62.52% of reads, and 596 16S rDNA OTUs (70.87%) representing 70.85% of reads, were stable in terms of their relative abundance level throughout the incubation (Supplementary Table 2). The rest OTUs were assigned as oscillating taxa (conditionally to be rare and/or abundant taxa) indicating 29.13~37.48% of the community had been undergone transition. Further quantification revealed that all three subcommunities were dynamic with the moderate taxa having the highest proportion of OTUs (55.71-62.26%) that transitioned to abundant or rare taxa (Figure 4B). Moreover, dust addition stimulated the transitions of rare taxa. Particularly, the proportion of eukaryotic rare taxa that transitioned to abundant taxa increased with dust addition. Similar trend was also observed in prokaryotic rare taxa that transitioned to moderate taxa.
3.4 Patterns and drivers of diversity
Temporal and intergroup variation were also observed in alpha diversity but eukaryotic and prokaryotic communities showed different patterns. For eukaryotic communities, dust addition decreased alpha diversity from day 0 to day 4, followed by a pronounced increase of Pielou’s evenness and Shannon diversity towards day 7 (Figures 5C–E). For prokaryotic communities, dust addition promoted the alpha diversity (Chao 1, Pielou’s evenness and Shannon diversity) and functional richness from day 0 to day 4, followed by a decrease towards day 7 (Figures 5F–H). Although relative to the control, higher functional evenness and diversity with lower functional redundancy was observed in high dust treatment on day 4, the general trend over time was similar among control and dust treatments (Supplementary Figure 12).
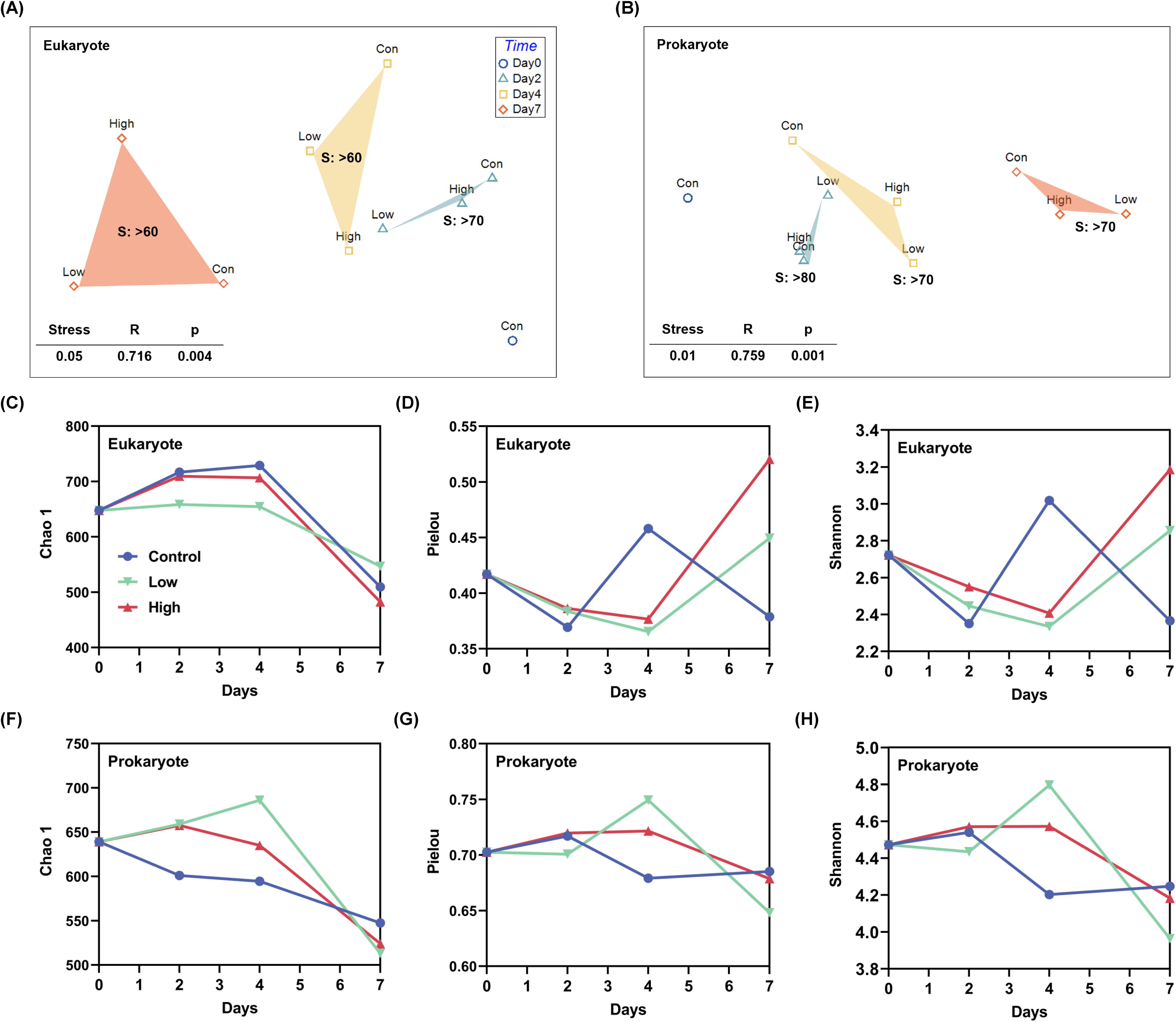
Figure 5. Community structuring of eukaryotic and prokaryotic plankton during the incubation experiment. (A, B) Non-metric multidimensional scaling (NMDS) ordination of plankton communities based on Bray–Curtis distances. Con, control group with no addition; Low, low group with 0.2 mg L-1 dust addition; High, high group with 2 mg L-1 dust addition; S, similarity. Variation in the α diversity (Chao 1, Pielou’ evenness and Shannon-Wiener index) of (C-E) eukaryotic and (F-H) prokaryotic microplankton communities.
NMDS ordinations and ANOSIM analysis revealed the distinct separation of plankton communities by time (Figures 5A, B). On the same sampling day, the community similarity between dust treatments and control decreased (Figures 5A, B), indicating that dust also played a role in community divergence, which was greater in microeukaryotic plankton relative to that in the prokaryotes. This is also the case when analyzing the functional structure of prokaryotic community with much weaker community divergence (similarity >80%, Supplementary Figure 4A). We further partitioned total beta diversity into turnover and nestedness, revealing that species replacement (turnover) played a major role in driving the community succession, and the percentage of turnover in eukaryotic communities was significantly higher than that in prokaryotic ones (Supplementary Figure 15). All these results indicated that the sensitivity of microeukaryotic and prokaryotic plankton to dust addition was different.
Correlation analysis suggested that the relative abundance of some lineages was significantly correlated with inorganic nutrient conditions (Supplementary Figure 16). Remarkably, the relative abundance of most eukaryotic lineages (18 out of 20) was significantly correlated with prokaryotic community, while 7 out of 20 prokaryotic lineages showed significantly correlation with eukaryotes. This pointed to that microeukaryotic plankton was more sensitive to cross-domain biotic factors, which was also revealed by Mantel test at community level (Supplementary Table 3).
3.5 Co-occurrence network of planktonic microbiome
To further explore the potential biotic interaction in the planktonic microbiome affected by dust addition, we conducted a co-occurrence network analysis (Figure 6). Compared to the Con-Low network, the Low-High network was more complex with 4% more nodes, 13% more positive edges and 30% more negative edges (Supplementary Table 5). Although positive edges dominated both networks (>60%), the proportions of negative edges slightly increased (30%) due to dust addition. Among the increased edges, 8.4%, 75.2% and 16.4% were contributed by the increased eukaryote-prokaryote, eukaryote-eukaryote and prokaryote-prokaryote connections, respectively. Phytoplankton-bacteria edges dominated the eukaryote-prokaryote connections (>45.06%), and the number of connections between diatoms and their correlated taxa including Cercozoa, Labyrinthulomycetes, Copepoda and Saprospirae were higher in Low-High network (Supplementary Table 6). We compared the unique node-level topological characteristics of the two networks and found that the degree and closeness centrality of the Low-High network were significantly higher than that of the Con-Low network (p<0.05), while betweenness centrality did not show significant difference (Figures 6C–E). Our results implied that microbial communities display more complex networks when exposed to increased dust addition treatments. A total of 44 OTUs belonging to 14 taxa in the Con-Low network and 87 OTUs belonging to 22 taxa in the Low-High network were identified as keystone taxa (Figure 6F). Only 5 keystone taxa were shared between two networks, belonging to Alphaproteobacteria (2 OTUs), Gammaproteobacteria (2 OTUs) and Syndiniales (1 OTU). Five OTUs belonging to diatoms were identified as keystone taxa.
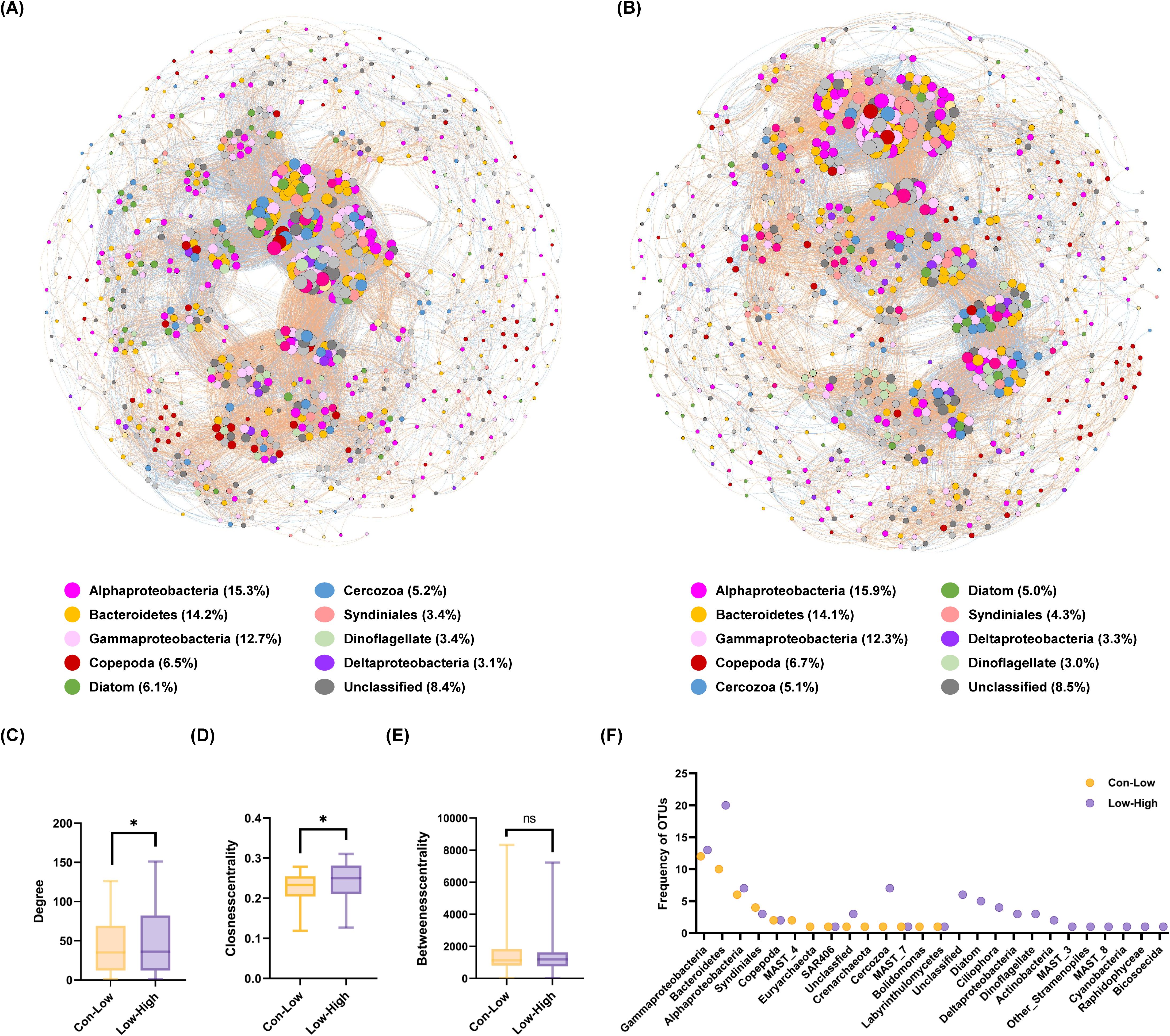
Figure 6. Co-occurrence networks of planktonic microbiome affected by dust addition. To ensure that the network analysis was supported by sufficient data, the Control and Low groups were merged into ‘Con-Low’ group to characterize planktonic microbial communities that were not affected or weakly affected by dust additions (A), and the Low and High groups were merged into the ‘Low-High’ group to characterize planktonic microbial communities that were affected or strongly affected by dust additions (B). The nodes of the network are colored according to taxa. The red and blue lines represent positive and negative association (edge), respectively. The sizes of the nodes are scaled to the degree of connection. (C-E) Comparison of node-level topological features (degree, closeness centrality and betweenness centrality) between two co-occurrence networks. Asterisk indicates the significant level at P < 0.05 level determined by nonparametric Mann–Whitney U test. (F) Comparison of the keystone taxa frequency between two co-occurrence networks.
4 Discussion
4.1 Fertilization by dust-derived nitrogen
The N:P ratio at B1 station was close to Redfield ratio, indicating that N was not the primary limiting factor (Figure 2D). Despite this, dust addition still stimulated the growth of phytoplankton by supplying a considerable amount of N and the promotion effect showed a dust loading-dependent pattern as evidenced by both microscopic analysis and Chl a concentration (Figures 2F–I). Our results are in concert with previous studies in Kuroshio-Oyashio transition region (Zhang et al., 2018, 2019a; b; Zhang et al., 2020). Dust-derived N has been recognized as a supplementation of the N demand for phytoplankton in this HNLC zone, when compared with the response of phytoplankton in LNLC and eutrophic zones (Zhang et al., 2019). We further used NCEI to quantify the N-utilization efficiency among groups (Supplementary Figure 4). A higher NCEI value indicates that the positive effect of added N on phytoplankton growth is stronger. Although Chl a increased with dust loading, the NCEI value is supposed to be independent of N loading. However, we found that the NCEI value in high dust treatment was higher than that in low dust treatment, implying that other substances leached from dust may influence the N-utilization efficiency in phytoplankton. Phytoplankton at B1 is possibly limited by Fe and other trace metals, such as Cu and Zn (Yang et al., 2019), which could be alleviated by dust deposition and in turn facilitates the N uptake as shown in high dust treatment. Moreover, the Asian dust provides considerable amount of N but negligible P, making the N:P ratios in high dust treatment (16-18) maintained above Redfield ratio, which potentially caused depletion of dissolved inorganic phosphorus (DIP). In our results, the enhanced DIN uptake by phytoplankton suggested the utilization of a supplementary P source, such as dissolved organic phosphorus (DOP), which is more abundant relative to DIP in the oceans. A recent study by Jin et al. (2024) provides a possible explanation: atmospheric deposition can stimulate the biological utilization of DOP by depleting DIP due to increasing N. Under DIP deficiency, phytoplankton most commonly use alkaline phosphatase but also other hydrolases to convert DOP to DIP for cellular uptake (Lin et al., 2016) and the activity of alkaline phosphatase is induced by low DIP concentrations. Based on microcosm experiments and global ocean analysis, Jin et al. (2024) proposed a threshold ratio of Chl a to DIP [Log10 (Chl a/DIP) > 1.20] to determine whether DOP is an important P source. The value of [Log10 (Chl a/DIP)] at B1 is 2.31, pointing to that DOP utilization might play a role in enhancing DIN uptake by phytoplankton.
Although dust input induced significant phytoplankton biomass accumulation, the degree of impact was varied between size fractions. Size as one of the most important fitness-related traits, its distribution can be used as indicator of biological transitions, and even regime shift in ecosystem (Spanbauer et al., 2016). In response to dust input, phytoplankton size structure shifted towards larger cells over incubation time (Figures 2G–I). Correlation between size-fractioned Chl a and N supply indicates that dust addition favored the growth of >3 μm-sized phytoplankton, which can be partially explained by the higher nutrient uptake rate of larger phytoplankton under nutrient-replete conditions (Maranon, 2015). Although dust addition slightly promoted the growth of picoplankton, no time-dependent increase was observed (Figure 2H). Trophic interaction may provide a possible explanation: increased abundance of protist grazers and their efficient grazing on picophytoplankton under atmospheric deposition keep the picophytoplankton biomass under control, as found in the microcosm experiments carried out in South China Sea (Guo et al., 2012; 2014). Contrastingly, a previous study in the Southern Yellow Sea showed that the abundance of heterotrophic and mixotrophic protists were inhibited by Asian dust deposition (Chen et al., 2019). Thus, trophic structure should be considered when evaluating the effects of atmospheric deposition on marine planktonic ecosystems. In addition, usually mesozooplankton (>200 μm) is excluded in microcosm experiments, which may technically lower the grazing pressure for micro- and nanophytoplankton.
4.2 Community dynamics and differential response of planktonic taxa to dust deposition
Besides size-dependent response of phytoplankton to dust deposition, we further profiled the differential response at a fine taxonomic resolution covering both microeukaryotic and prokaryotic plankton. Communities were distinctly separated by time, and dust exerted stronger effect on community divergence towards the end of incubation (Figures 5A, B), indicating that both magnitude and duration of the deposition event played a role in modulating planktonic microbiome. Numerous incubation experiments in the NWPO find that the Chl a generally started to increase with 0-5 days after dust deposition (e.g. Zhang et al., 2019a, 2020), and subarctic NWPO showed the longest response time (up to 40 days) to high dust deposition events, compared to other regions (Meng et al., 2022). We have detected the change of community composition and structure since day 2 and characterized the temporal dynamics by clustering analysis (Figures 3E, F). Time- and lineage-dependent responses were complex within 7-day incubation.
4.2.1 Positive response of diatom and its associated effects
When taking a closer look at the taxonomic profile, we found that the majority of eukaryotic taxa including phytoplankton and heterotrophic protist, positively responded to dust addition based relative to control (cluster 1-3 in Figures 3E, F). But fluctuations were also observed on day 4 (cluster 2&3). Among phytoplankton, dust addition particularly enhanced the relative abundance of diatom by over 120% relative to the control, shifting the dominance from dinoflagellate to diatom (Figure 3A). As evidenced by observational study, incubation experiments and modelling, the changes in nutrient regime significantly affected diatom-dinoflagellate competition (Xiao et al., 2018; Bi et al., 2021; Cheung et al., 2021), which is also the case for nutrient supplied by dust (Zhang et al., 2019a). At high nutrient concentrations, particularly with external N-supply, diatoms show a competitive superiority over dinoflagellate with higher nitrate reductase activity, more NO3- transporters and a higher NO3- use efficiency in diatoms relative to dinoflagellates (Glibert et al., 2016). While the versatile nutrient utilization and mixotrophy make dinoflagellates less sensitive to nutrient input (Dagenais-Bellefeuille and Morse, 2013; Edwards, 2019). However, with the combined effect of temperature and N-P imbalance, dinoflagellates tend to prevail in the warming and eutrophic ocean (Wu et al., 2018; Wei et al., 2024). Studies in coastal regions also found that dinoflagellates benefit from increased N:P ratios and phosphorus limitation (Zhang et al., 2024). However, for the dinoflagellates at B1 station, such benefit derived from dust addition seems very limited. The discrepancy can be attributed to that the physiological difference between coastal and oceanic algal species (strain), or the effects of nutrients other than DIN and DIP leached from dust.
Among heterotrophic protists, Cercozoa dominated by filose taxa were particularly stimulated by dust addition, and their relative abundance was positively correlated with diatoms (Supplementary Figure 9A). The Cercozoa-diatom association evidenced by metabarcodes was also found in previous studies: Cercozoa were present when diatoms developed in an oligomesotrophic lake in France where (Lepère et al., 2006), and high abundance of parasitic Cercozoa coincided with a decline of diatom bloom in the surface water of easter North Pacific, where (Berdjeb et al., 2018). These parasitic cercozoan were hardly detectable when the abundance of diatoms was low (Berdjeb et al., 2018), as we saw in the ambient water at B1, Cercozoa only took up 0.47% of the total reads, but proliferated with diatom after dust addition. Cercozoa is known to feed on bacteria, fungi and even some eukaryotes, and parasitic cercozoan were only described from diatoms (Scholz et al., 2016). The correlation between Cercozoa and diatoms in our study suggests the potential diatom infection by parasitic cercozoan. Increased predation on bacteria may also contribute to the cercozoan proliferation, as dust addition can improve bacteria abundance in oligotrophic NWPO (Chen et al., 2020). Moreover, Labyrinthulomycets were also induced by dust addition, which are prevalent and associated with living marine algae, sea grasses and animals as parasites, commensals or mutualists (Raghukumar, 2002). Remarkably, the parasitic Labyrinthulomycets were also mainly described from diatom hosts (Scholz et al., 2016; Pereboev and Bubnova, 2023).
In terms of metazoan, although seawater was prefiltered through 200-μm mesh to remove large zooplankton before incubation experiment, small-sized zooplankton and their young life stages could still remain. Pronounced decrease of relative abundance in copepod was induced by dust addition on day 7, when the relative abundance of diatom peaked. As a means of defense against predators, some diatoms generate compounds like aldehydes, oxylipins and domoic acid, which could cause detrimental impacts on copepod reproduction and development (Ban et al., 1997). Evidence from the field showed that after feeding on a diatom-dominated bloom, the hatching success of wild copepods decreased, and embryonic development was hindered (Miralto et al., 1999). Laboratory study showed the negative effects of these chemicals on nauplii development (Brugnano et al., 2016). These findings provide a possible explanation to the opposite pattern of copepod and diatom observed in the incubation experiment.
4.2.2 Differential sensitivity of microeukaryotes and prokaryotes to dust
Despite the variation in microeukaryotes composition, the bacterial communities over the time of incubation were consistently dominated by three classes, including Gammaproteobacteria, Alphaproteobacteria and Flavobacteriia (Figure 3D). The members of these classes are typically the most dominant bacteria in phytoplankton bloom, as identified by 16S rDNA surveys (reviewed in Buchan et al., 2014). The dust-induced phytoplankton growth in our study can be regarded as an experimental phytoplankton bloom. Organic matter produced by phytoplankton offers a broad spectrum of nutritional resources that can be utilized by associated heterotrophic bacterial community. Thus, the abundance of three dominant bacteria classes often correlates with the succession patterns of phytoplankton populations in the field (Riemann et al., 2000). However, in our study, the variation of bacterial taxonomic profile was weaker than that of phytoplankton (Supplementary Figure 15), which might attribute to the magnitude of phytoplankton bloom, or relatively small temporal scale. The most noticeable change was found in the family Saprospiraceae (class Saprospirae), of which the relative abundance was promoted by dust (44.01~56.74% increase relative to control), and positively correlated with that of diatoms (Supplementary Figure 9B). Many members of the family Saprospiraceae have algicidal or predatory properties, capable of utilizing complex carbon sources (McIlroy and Nielsen, 2014). Saprospiraceae can actively prey on diatom by capturing and lysing diatom through cell-to-cell contact (Furusawa et al., 2003). Several field studies evidenced the Saprospiraceae-phytoplankton association: it was the dominant bacterial family during a dinoflagellate-diatom in coastal waters of China (Shao et al., 2020), and was found to be enriched in particle-attached bacteria community during a spring phytoplankton bloom in southern North Sea (Wang et al., 2024). As algicidal bacteria can play an active role in controlling phytoplankton bloom (Mayali and Azam, 2004), we speculate that the increase of Saprospiraceae may also control the diatom proliferation induced by dust addition. But 7-day incubation is not long enough to observe a collapse of diatoms population caused by protist and bacterial predation.
As discussed above, our results found that Asian dust deposition likely impacts every link of the marine planktonic food chain in NWPO, which is also suggested by a study on microbial food web dynamics in response to a Saharan dust event (Pulido-Villena et al., 2014). Differential response to dust addition existed among taxa, and collectively led to the difference between eukaryotic and prokaryotic communities. There is increasing recognition that the sensitivity of eukaryotic and prokaryotic microbes to environmental filtering is different, as shown in soil and aquatic ecosystems facing different stressors such as glacial retreat, physical mixing and damming (Jiang et al., 2018; Lu et al., 2022; Sun et al., 2022). In our study, multiple lines of evidence support that microeukaryotic plankton is more sensitive to dust addition than prokaryotic counterpart: i) the community divergence between control and dust treatments over time was greater in microeukaryotic plankton (Figures 5A, B); ii) the percentage of species replacement (turnover) in driving the succession of microeukaryotic community was higher (Supplementary Figure 15); iii) the variability of taxonomic profile in eukaryotes was greater (Figure 3E; Supplementary Figure 11). This is consistent with previous researches on protist and bacteria in East China Sea’s euphotic zone (Wu et al., 2018) and in damming river (Lu et al., 2022), but contradict with that in global surface waters (Logares et al., 2020), upwelling system (Sun et al., 2022) and blue hole (Chen et al., 2022). Such inconsistency may be explained by the combined effect of habitat type, community composition and stressor gradient.
4.2.3 Non-abundant taxa dominate the community turnover
Besides the taxonomic variability, we also characterize the dynamics of abundant, moderate and rare subcommunities (Figure 4). A skewed abundance and richness distribution was also detected in the studied plankton microbiome, as normally found across habitats (reviewed in Xue et al., 2018): a few abundant taxa dominated the community, and non-abundant taxa contributed to the majority of community richness. But it is surprising that the contribution of moderate taxa was significantly higher in prokaryotic community (averagely 41.32% abundance) relative to that in eukaryotic one (15.81%). This is also higher than the contribution of moderate taxa observed in previous studies, including microeukaryotes in surface coastal waters of Europe (up to 30%, Logares et al., 2014), microeukaryotes (13%) and prokaryotes (17%) in blue hole (Chen et al., 2022), and estuarine ciliates (up to 20%; Sun et al., 2017). We speculated that high abundance and richness of moderate taxa in prokaryotes increased the evenness among three subcommunities, leading to a lower environmental sensitivity to dust addition. Moreover, moderate taxa had the highest proportion in transition irrespective to treatments, i.e. moderate taxa were most dynamic or responsive during incubation (Figure 4B). A highly dynamic turnover of moderate taxa was also found in estuarine ciliate communities, suggesting their roles in maintaining community stability and function under changing environments (Sun et al., 2017). Remarkably, dust addition stimulated the transitions of rare taxa. Rare taxa are recognized as a microbial seed bank with a pool of ecological potential (reviewed in Lynch and Neufeld, 2015). They can become dominant under favorable conditions. The recruitment of rare taxa is important in maintaining ecosystem processes after environmental disturbance (Sjöstedt et al., 2012).
4.3 Dust addition enhanced microbial network complexity
Mantel test revealed that biotic factor shaped both bacterial and microeukaryotic community more than abiotic factors, particularly the cross-domain interaction significantly affected microeukaryotic community (Supplementary Table 3). Dust deposition, as a disturbance to the surface ocean, may first affect the most sensitive lineages and its effects could pass on other organisms through interspecies interaction, and other components of the ecosystems (Battisti et al., 2016). Here, as the first attempt, we used co-occurrence network to portrait the biotic interactions responding to dust deposition. Ecological complexities can be depicted through networks, where species are represented as nodes and their interactions as connecting edges. This representation is essential for understanding the dynamics of species interactions within an ecosystem (Berry and Widder, 2014). Dust addition altered the network topology by increasing the number of edges, closeness centrality and the number of keystone taxa, resulting in a more densely linked network (Figure 6). Network complexity and connectivity are considered to be associated with the sensitivity of microbes to environmental disturbances, and strongly correlated with stability (Zhou et al., 2010). Some insights have been revealed into the impact of environmental stressors, such as climate warming and salinity stress, on the microbial ecological network (Yuan et al., 2021; Li et al., 2023). But the induced changes in complexity and stability of network are different, depending on the type of stressor and temporal scale. Remarkably, our co-occurrence network analysis revealed that the number of diatom-related edges were increased by dust addition, suggesting that diatoms were actively interconnected with their symbionts, competitors, predators and other associated microbes. This confirms that dust deposition does not only induce a succession of phytoplankton in NWPO, which has drawn most of the attention, but also a progression of heterotrophic protists, metazoan, bacteria and other players in the planktonic ecosystem.
Although positive association dominated both networks, more negative associations were induced by dust addition (Figure 6; Supplementary Table 4). Positive and negative associations primarily represent cooperation and competition, respectively (Yuan et al., 2021). Dai et al. (2022) studied the effects of nutrient supply on network association in marine bacterial communities and found that positive associations are more dominant in nutrient-scarce pelagic regions, while the proportion of negative associations are higher in nutrient-rich coastal sediment. High nutrient concentrations favor the actively growing bacteria which may inhibit the growth of competitors by producing metabolites (Ratzke et al., 2020), or cause competition for other essential nutrients. Based on this, we speculate that the slight increase of negative associations in dust-affected network could be partly explained by the nutrient supply from dust, and diatom is one of the strong competitors. Previous findings on negative association induced by nutrient supply are solely based on soil and marine bacterial studies (Ratzke et al., 2020; Dai et al., 2022), but whether this can be applied to interactions among microeukaryotes and cross-domain interactions remain elusive.
Dust-affected network also had greater number and more diverse keystone taxa. Keystone taxa regulate the network stability, which could be significantly reduced when keystone taxa were removed (Liu et al., 2022). Additionally, only 5 keystone taxa were shared between two networks, and several OTUs belonging to diatoms and their associated microbes became keystone taxa in the dust-affected network. This implies that dust addition may alter the microbial network by displacing keystone nodes, as they are associated with topological feature in co-occurrence network (Berry and Widder, 2014).
5 Limitation and perspective
Onboard microcosm has been widely used to evaluate the impact of atmospheric deposition on marine communities, with sufficient replicates to assess a range of treatments. However, the features of natural environment cannot be fully reproduced and limitations such as bottle effect may exist. Mesocosm, the large-scale incubation, is considered closer to representing natural ecosystems and has been also used to study the dust affect (e.g. Ridame et al., 2014). Despite the different incubation volume, mesocosm and microcosm yield similar phytoplankton response to dust addition, such as the relieved nitrogen limitation, rapid and long-lasting increase of Chl a. Moreover, it is believed that incubation and sampling time is critical to reduce the bottle effect, and the first rising period of Chl a during the incubation can best reflect the nutrient state of the phytoplankton (Zhang et al., 2019a). In this study, following previous microcosms investigating the response of NWPO surface water to Asian dust, we monitored the planktonic microbiome for 7 days until all groups reached the maximum Chl a concentration. The duration is also within the range of the dust residence time in mixed layer (Croot et al., 2004). However, the response time of subarctic NWPO surface seawater to some high dust deposition events may be up to 40 days (Meng et al., 2022). Thus, long-term impacts of dust on planktonic microbiome merit further investigation, particularly in terms of resilience of communities facing disturbance. There are also limitations in the use of metabarcoding. A prevalent issue is the discrepancy in the number of rDNA gene copies across various species, especially in microbial eukaryotes, which complicates the direct conversion of rDNA read counts into the actual number of organisms present (Burki et al., 2021). The presence of larger organisms or those with unique genome architecture (e.g. ciliate and dinoflagellate) is likely to complicate the rDNA data interpretation, since the number of rDNA copies within a genome is positively linked to the cell size and biovolume (de Vargas et al., 2015). Despite the fact that metabarcoding analysis derived only relative or semi-quantitative data, it is assumed that these molecular biases remain consistent across samples, enabling the comparison of different ecological states (Pawlowski et al., 2016). As shown in our results, metabarcoding, microscopic and Chl a analysis collectively evidenced the promotion effect of dust on phytoplankton growth and particularly favored diatom growth. Moreover, we found 30.51~46.93% of OTUs were not detected in the ambient water (day 0), and these rare OTUs transitioned to moderate or abundant taxa during the incubation, contributing to the diversity increase and community turnover. Whether a taxon can be detected by metabarcoding approach is related to the choice of barcode region and primers, as well as the number of cells (Marinchel et al., 2023). It is also argued that due to the variations in rDNA copy numbers in certain lineages, the ecological interactions inferred from metabarcodes-derived OTUs might be overestimated (Lima-Mendez et al., 2015). However, large-scale batch analysis by high-throughput sequencing uncovers a wide range of potential interactions. This is particularly valuable when traditional methods of morphological examination and physiological testing are not feasible, particularly in natural environments. In the future, profiling the metabolic activity and machinery in plankton may unveil the strategy underlying the biointeraction in response to dust deposition.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://db.cngb.org/, CNP0005306.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
YW: Data curation, Formal Analysis, Visualization, Writing – original draft, Writing – review & editing, Investigation. YZ: Data curation, Formal Analysis, Funding acquisition, Project administration, Writing – review & editing, Writing – original draft, Conceptualization, Investigation, Resources. SW: Formal Analysis, Visualization, Writing – original draft, Investigation. HC: Investigation, Writing – review & editing, Conceptualization. WW: Investigation, Writing – review & editing. CZ: Investigation, Writing – review & editing, Resources. HG: Project administration, Supervision, Writing – review & editing, Conceptualization, Resources. GL: Supervision, Writing – review & editing, Resources.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No.42276134, No.42376150) and the Fundamental Research Funds for the Central Universities (No. 202261073).
Acknowledgments
We are grateful to Yousong Huang for his help in sampling. We also thank Huan Zhang for technical assistance in DNA sample processing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1468739/full#supplementary-material
References
Ban S., Burns C. W., Castel J., Chaudron Y., Christou E. D., Escribano R., et al. (1997). The paradox of diatom-copepod interactions*. Mar. Ecol. Prog. Ser. 157, 287–293. doi: 10.3354/MEPS157287
Bastian M., Heymann S., Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. Proc. Third Int. ICWSM Conf (ICWSM) 8, 361–362. doi: 10.13140/2.1.1341.1520
Battisti C., Poeta G., Fanelli G. (2016). The concept of disturbance. Environ. Sci. Eng. 7–12 doi: 10.1007/978-3-319-32476-0_2
Berdjeb L., Parada A., Needham D. M., Fuhrman J. A. (2018). Short-term dynamics and interactions of marine protist communities during the spring–summer transition. ISME J. 12, 1907–1917. doi: 10.1038/s41396-018-0097-x
Berry D., Widder S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00219
Bi R., Cao Z., Ismar-Rebitz S. M. H., Sommer U., Zhang H., Ding Y., et al. (2021). Responses of Marine diatom-dinoflagellate competition to multiple environmental drivers: abundance, elemental, and biochemical aspects. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.731786
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 1091. doi: 10.1038/s41587-019-0252-6
Brugnano C., Granata A., Guglielmo L., Minutoli R., Zagami G., Ianora A. (2016). The deleterious effect of diatoms on the biomass and growth of early stages of their copepod grazers. J. Exp. Mar. Biol. Ecol. 476, 41–49. doi: 10.1016/j.jembe.2015.11.015
Buchan A., LeCleir G. R., Gulvik C. A., Gonzalez J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Burki F., Sandin M. M., Jamy M. (2021). Diversity and ecology of protists revealed by metabarcoding. Curr. Biol. 31, R1267–R1280. doi: 10.1016/j.cub.2021.07.066
Campbell B. J., Yu L., Heidelberg J. F., Kirchman D. L. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. United States America 108, 12776–12781. doi: 10.1073/pnas.1101405108
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chen S., Huang J., Kang L., Wang H., Ma X., He Y., et al. (2017). Emission, transport, and radiative effects of mineral dust from the Taklimakan and Gobi deserts: comparison of measurements and model results, Atmos. Chem. Phys. 17, 2401–2421. doi: 10.5194/acp-17-2401-2017
Chen X., Liu G., Huang X., Chen H., Zhang C., Zhao Y. (2019). ). Effects of asian dust and phosphorus input on abundance and trophic structure of protists in the southern yellow sea. Water 11, 1188. doi: 10.3390/w11061188
Chen X., Zhang X., Zhao Y., Liu G., Zhang C., Gao H. (2020). Response of heterotrophic bacteria abundance and community structure to asian dust addition in the oligotrophic northwest pacific ocean. J. Ocean Univ. China. 19, 722–728. doi: 10.1007/s11802-020-4126-9
Chen T., Zhuang Y., Chen C., Mao X., Ge R., Chen H., et al. (2022). Metabarcoding reveals the differential sensitivity of planktonic microbiome to environmental filtering and biointeraction in Sansha Yongle blue hole. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.1046808
Cheung Y. Y., Cheung S., Mak J., Liu K., Liu H. (2021). Distinct interaction effects of warming and anthropogenic input on diatoms and dinoflagellates in an urbanized estuarine ecosystem. Global Change Biol. 27, 3463–3473. doi: 10.1111/gcb.15667
Chien C. T., Mackey K. R., Dutkiewicz S., Mahowald N. M., Prospero J. M., Paytan A. (2016). Effects of African dust deposition on phytoplankton in the western tropical Atlantic Ocean off Barbados. Global Biogeochemical Cycles 30, 716–734. doi: 10.1002/2015GB005334
Chu Q., Liu Y., Shi J., Zhang C., Gong X., Yao X., et al. (2018). Promotion effect of Asian dust on phytoplankton growth and potential dissolved organic phosphorus utilization in the South China Sea. J. Geophys Res.-Biogeo. 123, 1101–1116. doi: 10.1002/2017JG004088
Chung C. C., Chang J., Gong G. C., Hsu S. C., Chiang K. P., Liao C. W. (2011). Effects of asian dust storms on synechococcus populations in the subtropical Kuroshio current. Mar. Biotechnol. 13, 751–763. doi: 10.1007/s10126-010-9336-5
Croot P. L., Streu P., Baker A. R. (2004). Short residence time for iron in surface seawater impacted by atmospheric dry deposition from Saharan dust events. Geophys. Res. Lett. 31, L23S08. doi: 10.1029/2004GL020153
Csardi G., Nepusz T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695.
Dagenais-Bellefeuille S., Morse D. (2013). Putting the N in dinoflagellates. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00369
Dai T., Wen D., Bates C. T., Wu L., Gao X., Liu S., et al. (2022). Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 13, 175. doi: 10.1038/s41467-021-27857-6
DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. Jul. 72, 5069–5072. doi: 10.1128/AEM.03006-05
de Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348, 29223618–29223625. doi: 10.1126/science.1261605
Duce R. A., Liss P. S., Merrill J. T., Atlas E. L., Buat-Menard P., Hicks B. B., et al. (1991). The atmospheric input of trace species to the world ocean. Glob. Biogeochem. Cycles. 5, 193–259. doi: 10.1029/91GB01778
Edgar R. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. (2016). SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv. doi: 10.1101/074161
Edwards K. F. (2019). Mixotrophy in nanoflagellates across environmental gradients in the ocean. Proc. Natl. Acad. Sci. 116, 6211–6220. doi: 10.1073/pnas.1814860116
Furusawa G., Yoshikawa T., Yasuda A., Sakata T. (2003). Algicidal activity and gliding motility of Saprospira sp. Canadian J. Microbiol. 49 (2), 92–100. doi: 10.1139/w03-017
Giovagnetti V., Brunet C., Conversano F., Tramontano F., Obernosterer I., Ridame C., et al. (2013). Assessing the role of dust deposition on phytoplankton ecophysiology and succession in a low-nutrient low-chlorophyll ecosystem: a mesocosm experiment in the Mediterranean Sea. Biogeosciences 10, 2973–2991. doi: 10.5194/BG-10-2973-2013
Glibert P. M., Wilkerson F. P., Dugdale R. C., Raven J. A., Dupont C. L., Leavitt P. R., et al. (2016). Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnology Oceanography 61, 165–197. doi: 10.1002/lno.10203
Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L., et al. (2012). Protist ribosomal reference database (PR2): A catalog ofunicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Guo C., Liu H., Yu J., Zhang S., Wu C. (2014). Role of microzooplankton grazing in regulating phytoplankton biomass and community structure in response to atmospheric aerosol input. Mar. Ecol. Prog. Ser. 507, 69–79. doi: 10.3354/meps10809
Guo C., Yu J., Ho T. Y., Wang L., Song S., Kong L., et al. (2012). Dynamics of phytoplankton community structure in the South China Sea in response to the East Asian aerosol input. Biogeosciences 9, 1519. doi: 10.5194/bg-9-1519-2012
Iwamoto Y., Yumimoto K., Toratani M., Tsuda A., Miura K., Uno I., et al. (2011). Biogeochemical implications of increased mineral particle concentrations in surface waters of the northwestern North Pacific during an Asian dust event. Geophys. Res.Lett. 38. doi: 10.1029/2010GL045906
Jiang Y., Lei Y., Yang Y., Korpelainen H., Niinemets U., Li C. (2018). Divergent assemblage patterns and driving forces for bacterial and fungal communities along a glacier forefield chronosequence. Soil Biol. Biochem. 118, 207–216. doi: 10.1016/j.soilbio.2017.12.019
Jiao S., Chen W., Wei G. (2017). Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol. Ecol. 26, 5305–5317. doi: 10.1111/mec.14218
Jickells T. D., An Z. S., Andersen K. K., Baker A. R., Bergametti G., Brooks N., et al. (2005). Global iron connections between desert dust, ocean biogeochemistry, and climate. Science 308, 67–71. doi: 10.1126/science.1105959
Jin H., Zhang C., Meng S., Wang Q., Ding X., Meng L., et al. (2024). Atmospheric deposition and river runoff stimulate the utilization of dissolved organic phosphorus in coastal seas. Nat. Commun. 15, 658. doi: 10.1038/s41467-024-44838-7
Lepère C., Boucher D., Jardillier L., Domaizon I., Debroas D. (2006). Succession and regulation factors of small eukaryote community. composition in a lacustrine ecosystem (Lake pavin). Appl. Environ. Microbiol. 72, 2971–2981. doi: 10.1128/AEM.72.4.2971-2981.2006
Li F., Altermatt F., Yang J., An S., Li A., Zhang X. (2020). Human activities’ fingerprint on multitrophic biodiversity and ecosystem functions across a major river catchment in China. Glob. Change Biol. 26, 6867–6879. doi: 10.1111/gcb.15357
Li H., Chen Y., Zhou S., Wang F., Yang T., Zhu Y., et al. (2021). Change of dominant phytoplankton groups in the eutrophic coastal sea due to atmospheric deposition. Sci. Total Environ. 753, 141961. doi: 10.1126/science.1262073
Li C., Jin L., Zhang C., Li S., Zhou T., Hua Z., et al. (2023). Destabilized microbialNetworks with distinct performances of abundant and rareBiospheres in maintaining networks under increasing salinityStress. iMeta 2, e79. doi: 10.1002/imt2.795
Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F., et al. (2015). Determinants of community structure in the global plankton interactome. Science 348, 1262073. doi: 10.1126/science.1262073
Lin S., Litaker R. W., Sunda W. (2016). Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol. 52, 10–36. doi: 10.1111/jpy.12365
Liu S. W., Yu H., Yu Y., Huang J., Zhou Z., Zeng J., et al. (2022). Ecological stability of microbial communities in Lake Donghu regulated by keystone taxa. Ecol. Indicators. 136, 108695. doi: 10.1016/j.ecolind.2022.108695
Liu Y., Zhang T., Shi J., Gao H., Yao X. (2013). Responses of chlorophyll a to added nutrients, Asian dust, and rainwater in an oli-gotrophic zone of the Yellow Sea: Implications for promotion and inhibition effects in an incubation experiment. J. Geophysical Research: Biogeosciences 118, 1763–1772. doi: 10.1002/2013JG002329
Logares R., Audic S., Bass D., Bittner L., Boutte C., Christen R., et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr. Biol. 24, 813–821. doi: 10.1016/j.cub.2014.02.050
Logares R., Deutschmann I. M., Junger P. C., Giner C. R., Krabberød A. K., Schmidt T. S. B., et al. (2020). Disentangling the mechanisms shaping the surface ocean microbiota. Microbiome 8, 55. doi: 10.1186/s40168-020-00827-8
Long Y., Zhu X.-H., Guo X. (2019). The Oyashio nutrient stream and its nutrient transport to the mixed water region. Geophysical Res. Lett. 46, 1513–1520. doi: 10.1029/2018GL081497
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lu L., Tang Q., Li H., Li Z. (2022). Damming river shapes distinct patterns and processes of planktonic bacterial and microeukaryotic communities. Environ. Microbiol. 24, 1760–1774. doi: 10.1111/1462-2920.15872
Lynch M., Neufeld J. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Ma Q., Chen Y., Wang F., Li H. (2021). Responses of primary productivity and phytoplankton community to the atmospheric nutrient deposition in the East China sea. Atmosphere 12, 210. doi: 10.3390/atmos12020210
Maki T., Lee K. C., Pointing S. B., Watanabe K., Aoki K., Archer S. D. J., et al. (2021). Desert and anthropogenic mixing dust deposition influences microbial communities in surface waters of the western Pacific Ocean. Sci. Total Environ. 791, 148026. doi: 10.1016/j.scitotenv.2021.148026
Maranon E. (2015). Cell size as a key determinant ofphytoplankton metabolism and community structure. Annu. Rev. Mar. Sci. 7, 241–264. doi: 10.1002/lno.10138
Maranon E., Cermeno P., Latasa M., Tadonleke R. D. (2015). Resource supply alone explains the variability of marine phytoplankton size structure. Limnol. Oceanogr. 60, 1848–1854. doi: 10.1002/lno.10138
Marinchel N., Marchesini A., Nardi D., Girardi M., Casabianca S., Vernesi C., et al. (2023). Mock community experiments can inform on the reliability of eDNA metabarcoding data: a case study on marine phytoplankton. Sci. Rep. 13, 20164. doi: 10.1038/s41598-023-47462-5
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBNet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mayali X., Azam F. (2004). Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryotic Microbiol. 51 2, 125–262. doi: 10.1111/j.1550-7408.2004.tb00538.x
McIlroy S. J., Nielsen P. H. (2014). “The family saprospiraceae,” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Springer Berlin Heidelberg, Berlin; Heidelberg), 863–889. doi: 10.1007/978-3-642-38954-2_138
Meng X., Chen Y., Wang B., Ma Q., Wang F. (2016). Responses of phytoplankton community to the input of different aerosols in the East China Sea. Geophys. Res. Lett. 43, 7081–7088. doi: 10.1002/2016GL069068
Meng X., Yao F., Zhang J., Liu Q., Liu Q., Shi L., et al. (2022). Impact of dust deposition on phytoplankton biomass in the Northwestern Pacific: A long-term study from 1998 to 2020. Sci. Total Environ. 813, 152536. doi: 10.1016/j.scitotenv.2021.152536
Miralto A., Barone G., Romano G., Poulet S. A., Ianora A., Russo G. L., et al. (1999). The insidious effect of diatoms on copepod reproduction. Nature 402, 173–176. doi: 10.1038/46023
Moreno A. R., Martiny A. C. (2017). Ecological stoichiometry of ocean plankton. Annu. Rev. Mar. Sci. 10, 43–69. doi: 10.1146/annurev-marine-121916-063126
Newton R. J., Shade A. (2016). Lifestyles of rarity: understanding heterotrophic strategies to inform the ecology of the microbial rare biosphere. Micro. Ecol. 78, 51–63. doi: 10.3354/ame01801
Oksanen J., Blanchet F., Kindt R., Legendre P., Minchin P., O’Hara R., et al. (2013). vegan: Community Ecology Package, version 2.0–7. R package.
Pawlowski J., Lejzerowicz F., Apotheloz-Perret-Gentil L., Visco J., Esling P. (2016). Protist metabarcoding and environmental biomonitoring: time for change. Eur. J. Protisto 55, 12–25. doi: 10.1016/j.ejop.2016.02.003
Pereboev D. D., Bubnova E. N. (2023). Marine labyrinthulomycetes. Russ J. Mar. Biol. 49, 241–250. doi: 10.1134/S1063074023040107
Pulido-Villena E., Baudoux A., Obernosterer I., Landa M., Capparros J., Catala P., et al. (2014). Microbial food web dynamics in response to a Saharan dust event: Results from a mesocosm study in the oligo- trophic Mediterranean Sea. Biogeosciences Discuss 11, 5607–5619. doi: 10.5194/bg-11-5607-2014
Raghukumar S. (2002). Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur. J. Protistology 38, 127–145. doi: 10.1078/0932-4739-00832
Ratzke C., Barrere J., Gore J. (2020). Strength of species interactions determines biodiversity and stability in microbial communities. Nat. Ecol. Evolution. 4, 376–383. doi: 10.1038/s41559-020-1099-4
Ridame C., Dekaezemacker J., Guieu C., Bonnet S., L’Helguen S., Malien F. (2014). Contrasted Saharan dust events in LNLC environments: Impact on nutrient dynamics and primary production. Biogeosciences 11, 4783–4800. doi: 10.5194/bg-11-4783-2014
Riemann L., Steward G. F., Azam F. (2000). Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66, 578–587. doi: 10.1128/AEM.66.2.578-587.2000
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Santi I., Tsiola A., Dimitriou P. D., Fodelianakis S., Kasapidis P., Papageorgiou N., et al. (2019). Prokaryotic and eukaryotic microbial community responses to N and P nutrient addition in oligotrophic Mediterranean coastal waters: Novel insights from DNA metabarcoding and network analysis. Mar. Environ. Res. 150, 104752. doi: 10.1016/j.marenvres.2019.104752
Scholz B., Guillou L., Marano A., Neuhauser S., Sullivan B., Karsten U., et al. (2016). Zoosporic parasites infecting marine diatoms e A black box that needs to be opened Corresponding editor. Fungal Ecol. 19, 59–76. doi: 10.1016/j.funeco.2015.09.002
Shao Q., Lin Z., Zhou C., Zhu P., Yan X. (2020). Succession of bacterioplankton communities over complete Gymnodinium-diatom bloom cycles. Sci. Total Environ. 709, 135951. doi: 10.1016/j.scitotenv.2019.135951
Shao Y., Wyrwoll K. H., Chappell A., Huang J., Lin Z., McTainsh G. H., et al. (2011). Dust cycle: An emerging core theme in Earth system science. Aeolian Res. 2, 181–204. doi: 10.1016/j.aeolia.2011.02.001
Shi J., Gao H., Zhang J., Tan S., Ren J., Liu C., et al. (2012). Examination of causative link between a spring bloom and dry/wet deposition of Asian dust in the Yellow Sea, China. J. Geophys. Res.-Atmos. 117. doi: 10.1029/2012JD017983
Sjöstedt J., Koch-Schmidt P., Pontarp M., Canbäck B., Tunlid A., Lundberg P., et al. (2012). Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl. Environ. Microbiol. 78. doi: 10.1128/AEM.05542-11
Spanbauer T. L., Allen C. R., Angeler D. G., Eason T., Fritz S. C., Garmestani A. S., et al. (2016). Body size distributions signal a regime shift in a lake ecosystem. Proc. R. Soc B 283, 20160249. doi: 10.1098/rspb.2016.0249
Sun P., Huang L., Xu D., Huang B., Chen N., Warren A. (2017). Marked seasonality and high spatial variation in estuarine ciliates are driven by exchanges between the ‘abundant’ and ‘intermediate’ biospheres. Sci. Rep. 7, 9494. doi: 10.1038/s41598-017-10308-y
Sun P., Wang Y., Huang X., Huang B., Wang L. (2022). Water masses and their associated temperature and cross-domain biotic factors co-shape upwelling microbial communities. Water Res. 215, 118274. doi: 10.1016/j.watres.2022.118274
Tan S., Yao X., Gao H., Shi G., Yue X. (2013). Variability in the correlation between Asian dust storms and chlorophyll a concentration from the north to equatorial Pacific. PloS One 8, e57656. doi: 10.1371/journal.pone.0057656
Tapolczai K., Keck F., Bouchez A., Rimet F., Kahlert M., Vasselon V. (2019). Diatom dna metabarcoding for biomonitoring: strategies to avoid major taxonomical and bioinformatical biases limiting molecular indices capacities. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00409
Tegen I., Schepanski K. (2009). The global distribution of mineral dust. IOP Conf. Ser. Earth Environ. Sci. 7, 12001. doi: 10.1088/1755-1307/7/1/012001
Vigil P., Countway P. D., Rose J., Lonsdale D. J., Gobler C. J., Caron D. A. (2009). Rapid shifts in dominant taxa among microbial eukaryotes in estuarine ecosystems. Aquat Microb. Ecol. 54, 83–100. doi: 10.3354/ame01252
Wang F. Q., Bartosik D., Sidhu C., Siebers R., Lu D. C., Trautwein-Schultet A., et al. (2024). Particle-attached bacteria act as gatekeepers in the decomposition of complex phytoplankton polysaccharides. Microbiome 12, 32. doi: 10.1186/s40168-024-01757-5
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang Q., Zhang C., Jin H., Chen Y., Yao X., Gao H. (2022). Effect of anthropogenic aerosol addition on phytoplankton growth in coastal waters: role of enhanced phosphorus bioavailability. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.915255
Wei Y., Luan Q., Shan X., Cui H., Qu K., Cui Z., et al. (2024). Temperature and nutrients drive distinct successions between diatoms and dinoflagellates over the past 40 years: Implications for climate warming and eutrophication. Sci. Total Environ. 931, 172997. doi: 10.1016/j.scitotenv.2024.172997
Wu W., Lu H. P., Sastri A., Yeh Y., Gong G., Chou W., et al. (2018). Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 12, 485–494. doi: 10.1038/ismej.2017.183
Xiao W., Liu X., Irwin A., Laws E. A., Wang L., Chen B., et al. (2018). Warming and eutrophication combine to restructure diatoms and dinoflagellates. Water Res. 128, 206–216. doi: 10.1016/j.watres.2017.10.051
Xiong C., He J., Singh B., Zhu Y., Zhang L. (2020). Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 23, 1907–1924. doi: 10.1111/1462-2920.15262
Xue Y., Chen H., Yang J., Liu M., Huang B., Yang J. (2018). Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 12, 2263–2277. doi: 10.1038/s41396-018-0159-0
Yang T., Chen Y., Zhou S., Li H. (2019). Impacts of aerosol copper on marine phytoplankton: A review. Atmosphere 10, 414. doi: 10.3390/atmos10070414
Yuan M. M., Guo X., Wu L., Wu L., Zhang Y., Xiao N., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348. doi: 10.1038/s41558-021-00989-9
Yuan J., Li M., Lin S. (2015). An improved DNA extraction method for efficient and quantitative recovery of phytoplankton diversity in natural assemblages. PloS One 10, e0133060–e0133060. doi: 10.1371/journal.pone.0133060
Yumimoto K., Takemura T. (2015). Long-term inverse modeling of asian dust: interannual variations of its emission, transport, deposition, and radiative forcing. J. Geophys. Res. Atmos. 120, 1582–1607. doi: 10.1002/2014JD022390
Zhang C., Chu Q., Mu Y., Yao X., Gao H. (2022). Weakened fertilization impact of anthropogenic aerosols on marine phytoplankton—A comparative analysis of dust and haze particles. Ecotoxicology Environ. Saf. 230, 113162. doi: 10.1016/j.ecoenv.2022.113162
Zhang C., Gao H., Yao X., Shi Z., Shi J., Yang Y., et al. (2018). Phytoplankton growth response to Asian dust addition in the northwest Pacific Ocean versus the Yellow Sea. Biogeosciences. 15, 749. doi: 10.5194/bg-15749-2018
Zhang C., He J., Yao X., Mu Y., Guo X., Ding X., et al. (2020). Dynamics of phytoplankton and nutrient uptake following dust additions in the northwest Pacific. Sci. Total Environ. 739, 139999. doi: 10.1016/j.scitotenv.2020.139999
Zhang C., Ito A., Shi Z., Aita M. N., Yao X., Chu Q., et al. (2019b). Fertilization of the Northwest Pacific Ocean by East Asia air pollutants. Global Biogeochemical Cycles 33, 690–702. doi: 10.1029/2018GB006146
Zhang C., Yao X., Chen Y., Chu Q., Yu Y., Shi J., et al. (2019a). Variations in the phytoplankton community due to dust additions in eutrophication, LNLC and HNLC oceanic zones. Sci. Total Environ. 669, 282–293. doi: 10.1016/j.scitotenv.2019.02.068
Zhang X., Yu K., Li M., Jiang H., Gao W., Zhao J., et al. (2024). Diatom-dinoflagellate succession in the Bohai Sea: The role of N/P ratios and dissolved organic nitrogen components. Water Res. 251, 121150. doi: 10.1016/j.watres.2024.121150
Zhou J., Deng Y., Luo F., He Z., Tu Q., Zhi X. (2010). Functional molecular ecological networks. Mbio 1, e00169–e00110. doi: 10.1128/mbio.00169-10
Keywords: dust deposition, metabarcoding, planktonic microbiome, microbial network, community structure
Citation: Wang Y, Zhuang Y, Wang S, Chen H, Wang W, Zhang C, Gao H and Liu G (2024) Dust deposition drives shifts in community structure and microbial network complexity of a planktonic microbiome in the Northwest Pacific Ocean. Front. Mar. Sci. 11:1468739. doi: 10.3389/fmars.2024.1468739
Received: 22 July 2024; Accepted: 26 August 2024;
Published: 12 September 2024.
Edited by:
Zhang-Xian Xie, Xiamen University, ChinaCopyright © 2024 Wang, Zhuang, Wang, Chen, Wang, Zhang, Gao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunyun Zhuang, eXVueXVuLnpodWFuZ0BvdWMuZWR1LmNu
 Yingxia Wang
Yingxia Wang Yunyun Zhuang
Yunyun Zhuang Shanshan Wang1
Shanshan Wang1 Hongju Chen
Hongju Chen Weimin Wang
Weimin Wang Huiwang Gao
Huiwang Gao Guangxing Liu
Guangxing Liu