
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 26 August 2024
Sec. Marine Biotechnology and Bioproducts
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1462825
The discovery of natural products derived from marine sources has demonstrated a consistent upward trajectory for the decade of 2011-2020, holding significant promise for the development of novel drugs and many other marine bioproducts. In recent years, the spotlight has shifted away from marine sponges (Porifera) towards marine microorganisms as the primary source of discovery. Despite reports of marine sponges spanning 20 different orders and being the subject of 769 papers between 2011 and 2020, they only contributed to 19.29% of all new compounds discovered, in contrast to 51.94% by marine microorganisms and phytoplankton. 563 new compounds were reported from marine sponge-associated microbes, more than doubling the number for the previous decade (2001-2010). It heralds a positive outlook for a sustainable resource strategy as the extraction of bioactive compounds produced by pure cultures of sponge-associated microbes could overcome supply challenges that arise with isolation from host sponges for the same compound. However, the application of novel marine natural products (MNPs) remains challenging due to the limited yield of compounds from large amounts of sponges. This review covers the literature published between 2011 and 2020, focusing on MNPs isolated from marine sponges. A total of 2603 new compounds are documented, detailing their chemical classification, biological activities, source country or geographic locations, and the taxonomic information of the source organisms, including order, family, genus, and species.
During the last decade 2011 to 2020, marine natural product discovery has shifted the primary focus from marine sponges to marine microorganisms, especially marine-derived fungi and bacteria (Blunt et al., 2012). However, sponges were still the second largest source of new marine natural products, along with the growing realization that microbial symbionts could be the real producers of “sponge” specialized metabolites. This new information highlights the need for more detailed metagenomic and biosynthetic analyses of sponge matrices (Reveillaud et al., 2014); and indicates that some of these microorganisms engage in symbiotic relationships with their host sponges, while others interact with various organisms within the sponge habitat (Harvey et al., 2015). Ultimately, both sponges and sponge-associated microorganisms contribute to the production of bioactive compounds in the marine environment (Mehbub et al., 2014; Brinkmann et al., 2017). However, in recent decades sponge-associated fungi were also found to be a rich source of bioactive metabolites (Jin et al., 2016). In the present review, it was discovered that sponge-associated microbes, primarily fungi such as Aspergillus, Penicillium and actinomycetes such as Streptomyces and Verrucosispora, contribute up to 563 new compounds. Altogether sponges and their associated microbes are responsible for producing over 3166 new compounds. In contrast, marine microorganisms and phytoplankton, which are mostly available in sediments, marine organisms and mangroves (Debbab et al., 2010; Carroll et al., 2023), generate a total of 7009 compounds. Nevertheless, sponges and their associated microbes together constitute the second largest sources of new marine-derived natural compounds (23.46%), accounting for almost half the number compared to those produced by marine microorganisms and phytoplankton (47.77%).
The production of bioactive compounds in sponges is a complex process that is regulated by a variety of factors, including environmental cues, nutrient availability, and genetic factors (Atanasov et al., 2021). Specialized cells in sponge called choanocytes and sclerocytes were reported to be responsible for producing and secreting bioactive secondary metabolites (Varijakzhan et al., 2021). Some of these compounds play a role in the sponge defense against predators, while others may help the sponge compete for resources in its environment (Helber et al., 2018).
Depth can influence the production of bioactive compounds in sponges in the ocean (Steffen et al., 2022). Studies have shown that the abundance and diversity of bioactive compounds produced by sponges can vary with depth due to differences in environmental conditions, such as light availability, temperature, pressure, and nutrient availability (Wulff, 2012). Lower light levels can affect the photosynthetic activity of symbiotic algae associated with some sponges and may also influence the synthesis of bioactive compounds by the sponge itself. Additionally, pressure and temperature can affect the stability and activity of the enzymes involved in the biosynthesis of secondary metabolites, leading to differences in bioactive compound production with depth (Parte et al., 2017). Some studies have shown that sponges in deeper waters may produce a greater diversity and quantity of bioactive compounds than those in shallower waters (Skropeta and Wei, 2014).
The color of the sponge can be an important factor in the production of bioactive compounds in sponges due to its relationship with light and its role in regulating the sponge’s photosynthesis and metabolism (Kim and Dewapriya, 2012; Muñoz-Miranda and Iñiguez-Moreno, 2023). Some species of sponges particularly colored sponges contain photosynthetic symbionts, such as algae or cyanobacteria, which are important sources of energy for the sponge (Erwin and Thacker, 2008; Gao et al., 2017) and may contribute to the sponge metabolic pathways and chemical diversity (Pineda et al., 2016) as a means of deterring potential predators or competitors (Taylor et al., 2007). In sponges, the colors come from tiny pigment granules inside special cells called amoebocytes (Bergquist, 1978). Encrusting sponge Acanthancora cyanocrypta is usually cobalt blue due to a symbiotic blue-green alga but an orange color morph occurs when the alga is absent (Bakus and Abbott, 1980). Some of the colorful sponges are Monanchora sp., Antho sp. (red), Stylissa flabelliformis (pale orange), Psammoclema sp. (bright orange), Chondropsis sp. (greenish), Batzella sp. (pale yellow) (Petek et al., 2017).
Additionally, light wavelengths (blue and red) have been shown to have different effects on the growth and metabolic activity of photosynthetic organisms, and this can also impact the production of bioactive compounds (Curdt et al., 2022; Zhang et al., 2023). However, the specific role of color will depend on the species and environmental conditions (Brusca and Brusca, 2002).
Further research is required to better understand the factors that influence the production of bioactive compounds in sponges and to identify the optimal conditions for their production.
Over the past five decades, the number of new compounds discovered from all marine sources has increased almost linearly (Figure 1). However, in the most recent decade from 2011 to 2020, the rate of increase in new compounds has surged significantly, resulting in a total of 13,492 new compounds. This trend is illustrated in the last 4 years of the previous decade (2001-2010) where more than 960 new compounds were discovered compared to over 1400 in the comparable period of the last decade (2011-2020) (Supplementary Figure S1). The escalating trend presents abundant opportunities for developing new drugs to combat various deadly diseases and a myriad of novel marine bioproducts for many other applications.
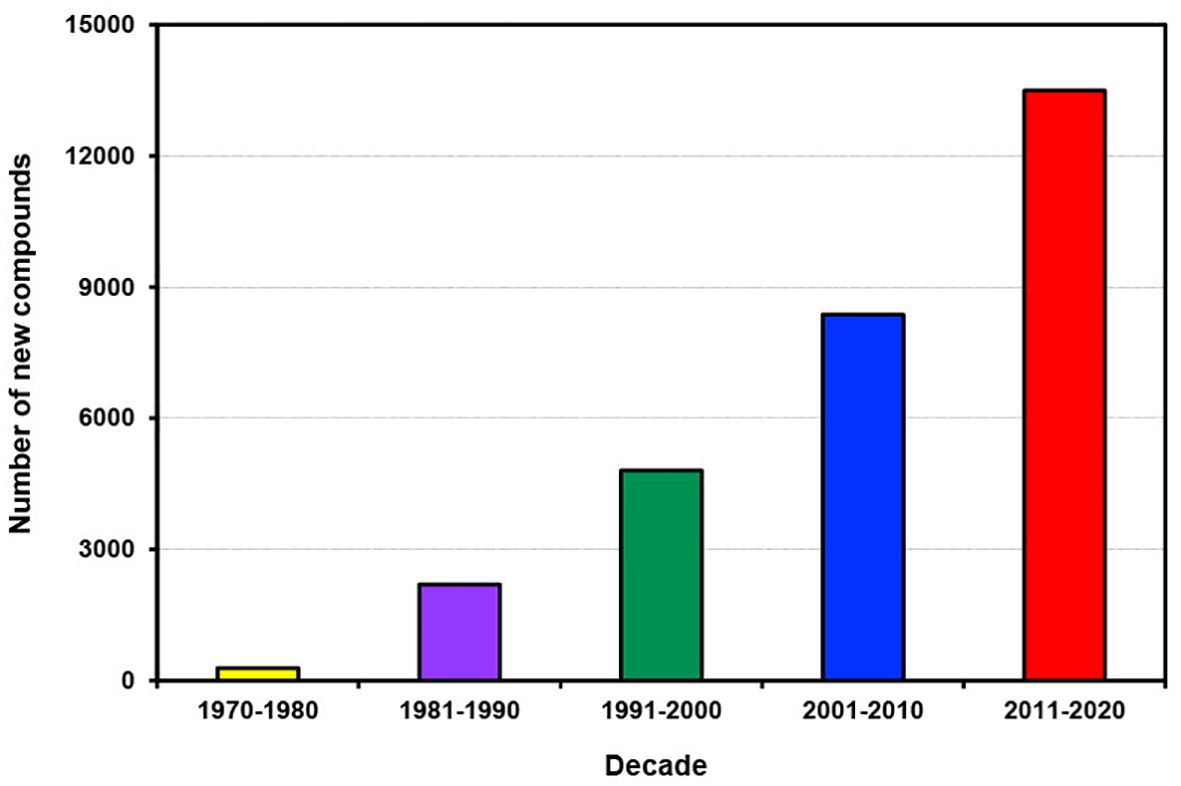
Figure 1. Number of new compounds discovered from different marine sources per decade from 1970 to 2020.
According to Mehbub et al. (2015) between 2000 and 2012, a total of 269 novel compounds were isolated from sponge-associated microbes. Of these, 186 compounds were derived from 27 genera of fungi, including two compounds from a single yeast genus. In contrast, only 56 new compounds were isolated from seven actinobacterial genera, and 27 new compounds were obtained from seven bacterial genera. This shift towards fungal sources is further supported by the number of publications on fungal metabolites, which totaled 69, compared to 20 publications on actinobacteria and 15 on bacteria during the review period (Mehbub et al., 2015).
With the advancement of new technologies, scientists have become increasingly interested in isolating novel compounds from sponge-associated microbes. Between 2011 and 2020, a total of 563 new compounds were discovered. Specifically, 473 compounds were isolated from 46 different genera of fungi, 74 compounds from 10 different genera of actinomycetes, and 16 compounds from other bacteria such as Bacillus sp. In fact, the discovery rate of new compounds from sponge-associated microbes in the recent decade doubled compared to the previous decade, highlighting significant progress in this field. More details about these microbes is beyond the scope of this paper.
In a study by Pita et al. (2018) the concept of holobionts was explored, describing them as dynamic ecosystems that interact across multiple scales and exhibit responses to environmental changes. The authors argued that sponges exemplify the notion of nested ecosystems, thereby providing a novel framework for comprehending holobionts within marine environments (Pita et al., 2018). This perspective underscores the complexity of interactions between the sponge host and its associated microorganisms, which extend beyond simple commensal or mutualistic relationships. Competition for space and nutrients may cause marine bacteria associated with sponges to produce more antibiotic substances (Bibi et al., 2017). Sponge symbionts reside both within cells and in extracellular spaces, with each symbiotic microorganism occupying a distinct niche in the host (Lee et al., 2001). This symbiotic relationship offers advantages to the associated microbes, including nutrient acquisition, stabilization of the sponge’s structure, elimination of metabolic waste, and synthesis of secondary metabolites (Thomas et al., 2010). The primary bacteria responsible for producing secondary metabolites in sponges come from various phyla, including Proteobacteria, Firmicutes, Actinobacteria, and Cyanobacteria (S, 2003). Marine sponge-associated actinomycetes are a valuable resource for drug discovery due to their production of diverse and potent bioactive compounds. Streptomyces species from sponges like Halichondria and Dendrilla nigra produce mayamycin and nocapyrones with strong antibacterial and anticancer properties (Selvin et al., 2009; Schneemann et al., 2010a). Actinomycetes such as Nocardiopsis and Micromonospora yield compounds effective against tuberculosis, HIV, and malaria (Hill et al., 2004; Schneemann et al., 2010b). Many isolated actinomycetes contain genes for secondary metabolite production, highlighting their therapeutic potential which include the anti-MRSA kocurin and new antimicrobials from sponge-derived actinomycetes (Palomo et al., 2013). Proteobacteria are a treasure trove of bioactive metabolites from sponges, second only to Actinobacteria. Within this phylum, the α-Proteobacteria and γ-Proteobacteria classes stand out for their prolific production of compounds with impressive antimicrobial, antitumor, and anticancer properties (Bibi et al., 2017). Alphaproteobacteria share several genetic traits with dominant sponge symbionts that are currently challenging to cultivate. These traits potentially enhance host fitness by detoxifying substance such as heavy metals and metabolic waste, breaking down aromatic compounds, and synthesizing essential vitamins like B6 and B12. They also contribute to nutrient cycling, particularly involving organic sulfur and nitrogen (Karimi et al., 2019). Moreover, they play a role in chemical defense by producing compounds like polyketides and terpenoids (Karimi et al., 2019). For instance, a Pseudomonas species from the family of sponge Aplysinellidae produces quinolones with potent antibacterial effects (Debitus et al., 1998). Pseudoalteromonas piscicida, found in the sponge Hymeniacidon perleve, generates the powerful antimicrobial compound norharman, effective against a variety of pathogens (Zheng et al., 2005). Additionally, Proteobacteria strains associated with sponges are known for their anti-inflammatory, cytotoxic, and anti-angiogenic activities, making them a fascinating focus for drug discovery and therapeutic development (Thakur et al., 2005). Alphaproteobacteria associated with sponges were reported widely to exhibit the ability to produce polyketides, terpenes and bacteriocins (Naughton et al., 2017; Versluis et al., 2018) which suggest a pivotal role of this bacterial group in shaping the chemical complexity, diversity of natural product biosynthesis and taxonomic composition of the marine sponge microbiome (Karimi et al., 2019). Firmicutes, especially Bacillus strains from sponges, are notable for producing potent secondary metabolites. Bacillus cereus from Halichondria japonica synthesizes antibiotics and thiopeptides effective against drug-resistant bacteria (Nagai et al., 2003). Bacillus subtilis A184 from Aplysina aerophoba produces diverse antimicrobial lipopeptides like iturins and fengycins, active against various pathogens (Pabel et al., 2003; Thomas et al., 2010). Other Bacillus species associated with sponges yield compounds such as Apteniol A, which shows strong antimicrobial activity (Devi et al., 2010). Bacteria found in sponge Halichondria panacea, particularly from the Cytophaga genus, are notable for producing agarase enzymes (Imhoff and Stöhr, 2003). In another instance, Arthrobacter species associated with sponge Spirastrella sp. produce acetylcholinesterase, highlighting their enzymatic capabilities (Mohapatra and Bapuji, 1998). Across different marine sponges, bacteria produce enzymes like proteases, amylases, and cellulases, with significant applications in biotechnology (Mohapatra et al., 2003). These microbes also contribute enzymes such as dehalogenases, chitinases, urethanases, and phospholipase A2 (PLA2), showcasing their role in both industry and environmental processes (Selvin et al., 2010).
Furthermore, understanding the dynamics of sponge-microbe interactions has broader implications for marine ecology and conservation. Sponges are essential components of marine ecosystems, contributing to nutrient cycling, sediment stabilization, and biodiversity maintenance (Maldonado et al., 2012). The role of microbial symbionts in sponge physiology and ecology underscores the need for integrated approaches to marine ecosystem management that consider the intricate relationships between microorganisms and sponges (Mehbub, 2015).
The supply of chemical compounds from sponges is still limited for several reasons. The major constraints are the low levels of compound produced per unit weight of sponge tissue, the slow growth of sponge, the need for sustainability and continuous production prevent large scale collections. Sponges are not found in high densities in all areas of the ocean, and some species are rare, making it difficult to obtain sufficient quantities of bioactive compounds for commercial purposes. Harvesting sponges from their natural habitats can be challenging, and many species are protected from over-collection due to their vulnerable populations and the slow rate of growth and reproduction. In most cases the compound is purified from a small amount of sponge sample and its structure is elucidated from milligrams of pure compound leaving enough for a bioassay. This leaves a number of compounds being tested out of their potency range and in a non-optimized manner. Understanding the biosynthetic mechanisms of chemical production and how to stimulate it in sponges is still a major challenge (Mehbub et al., 2016; Mehbub et al., 2018). The cost of producing bioactive compounds from sponges can be high due to the challenges of harvesting, processing, and purifying the compounds. In addition, the chemical synthetic production of these natural compounds with often complex structures can be academically feasible but may be expensive and difficult for industry synthesis. These challenges restrict the chemical synthesis and subsequent supply of bioactive compounds from sponges, hindering the scalability of production for commercial purposes. However, ongoing research aimed at enhancing our understanding of sponge biology and chemistry could lead to more efficient methods for harvesting, processing, and producing these bioactive compounds by synthetic biotechnology.
One way to mitigate supply issues is to develop sponge farming techniques that allow for the sustainable harvest of sponges (Maslin et al., 2021). This would provide a reliable and sustainable source of bioactive compounds and could reduce the pressure on wild populations of sponges. Another way is to develop improved collection techniques that allow for a more efficient and sustainable collection of sponges from the wild (Schiefenhövel and Kunzmann, 2012). This includes the use of remote operated vehicles (ROVs) and other underwater technologies to access deeper and more remote habitats, as well as the development of more selective and sustainable collection methods. Another potential solution to the supply issue is to use techniques to stimulate the production of bioactive compounds in sponge in an aquarium. The bioactive compounds and other organic compounds produced are captured by a hydrophobic resin (Hill et al., 2004) and placed in a porous bag in the sponge tank. The resin keeps the sponge alive by removing any toxic materials, as well as capturing the compounds produced by the stimulation and can be replaced with fresh resin before the next stimulation. Meanwhile, the resin is eluted to keep the ‘milking’ process going. This would allow for the large-scale production of bioactive compounds without the need for wild-collected sponges (Mehbub et al., 2016; Carroll et al., 2022).
The new compounds isolated during the last decade 2011-2020 are based on the data reviewed by Blunt et al. and Carroll et al. from Natural Product Reports of 2012 to 2022 (Blunt et al., 2012; Blunt et al., 2013; Blunt et al., 2014; Blunt et al., 2015; Blunt et al., 2016; Blunt et al., 2017; Blunt et al., 2018; Carroll et al., 2019; Carroll et al., 2020; Carroll et al., 2022). The World Porifera Database and The World Register of Marine Species (WoRMS) database (Bernot et al., 2024) were used to cross-check detailed taxonomical information (order and family) for each surveyed species and to validate and/or update their scientific names.
In recent years, there has been a noticeable shift in the field of marine natural product discovery. Instead of focusing primarily on compounds derived from sponges, it consistently shows an increasing trend of discovery for marine microorganisms and phytoplankton over the last decade (2011 to 2020), as depicted in Figure 2. While there has been a decreasing trend for sponge-derived natural products (Supplementary Figure S2). This shift reflects a growing understanding of the complex symbiotic relationships within marine ecosystems and the potential wealth of bioactive compounds they contain. Furthermore, advances in metagenomic and metabolomic techniques have revolutionized our ability to explore the complex interactions between sponges and their microbial partners (Hentschel et al., 2012; Taylor et al., 2012), facilitating the identification and characterization of bioactive molecules previously hidden within these symbiotic networks (Aguiar-Pulido et al., 2016; El-Samak et al., 2023). This shift towards sponge-associated microbes as a prolific source of natural compounds underscores the importance of understanding and preserving these delicate ecosystems in the face of environmental challenges and anthropogenic impacts (Mehbub et al., 2016; Mehbub et al., 2018; Li et al., 2023).
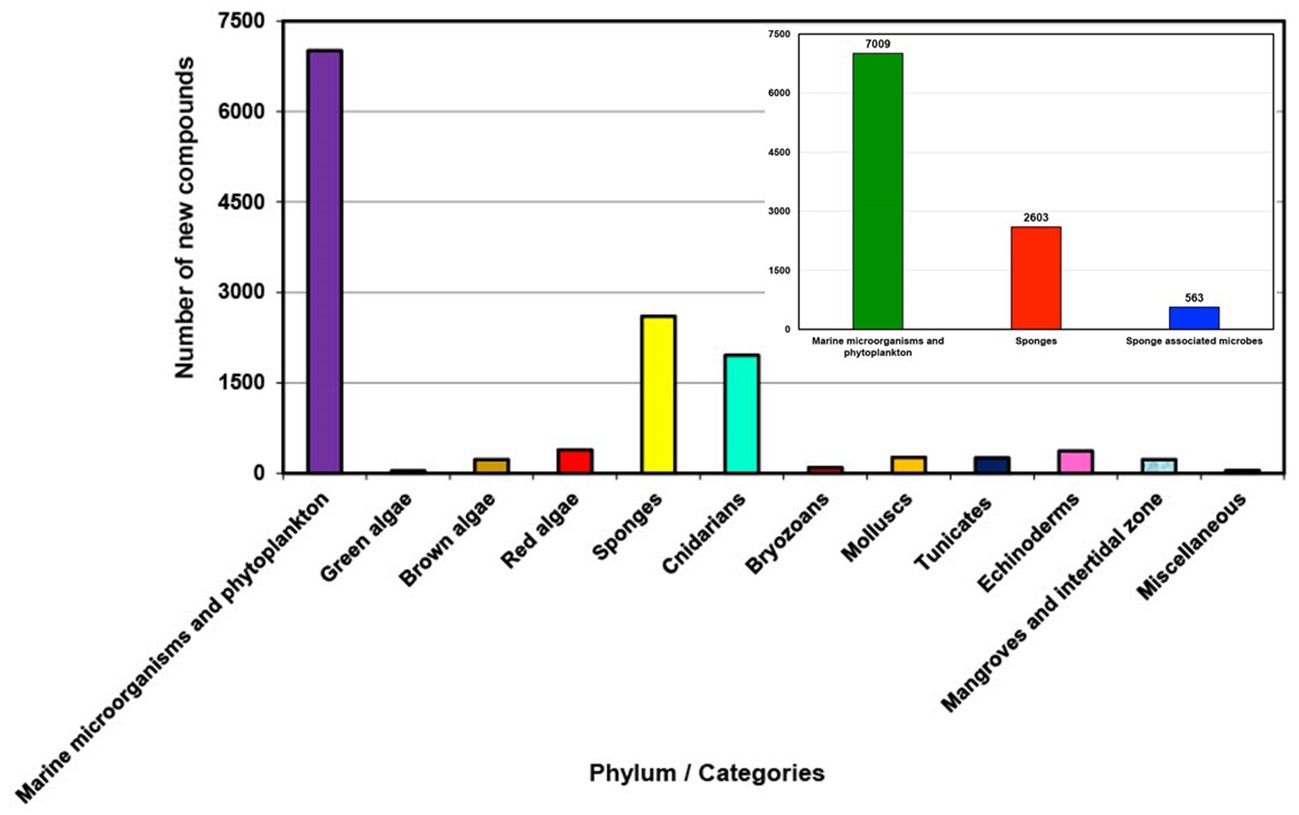
Figure 2. Total number of new compounds isolated from different groups of marine organisms/resources between 2011 and 2020.
Marine-sourced fungi and bacteria emerged as the primary producers of novel compounds contributed by marine microorganisms and phytoplankton during the last decade spanning from 2011 to 2020, as depicted in Figure 3.
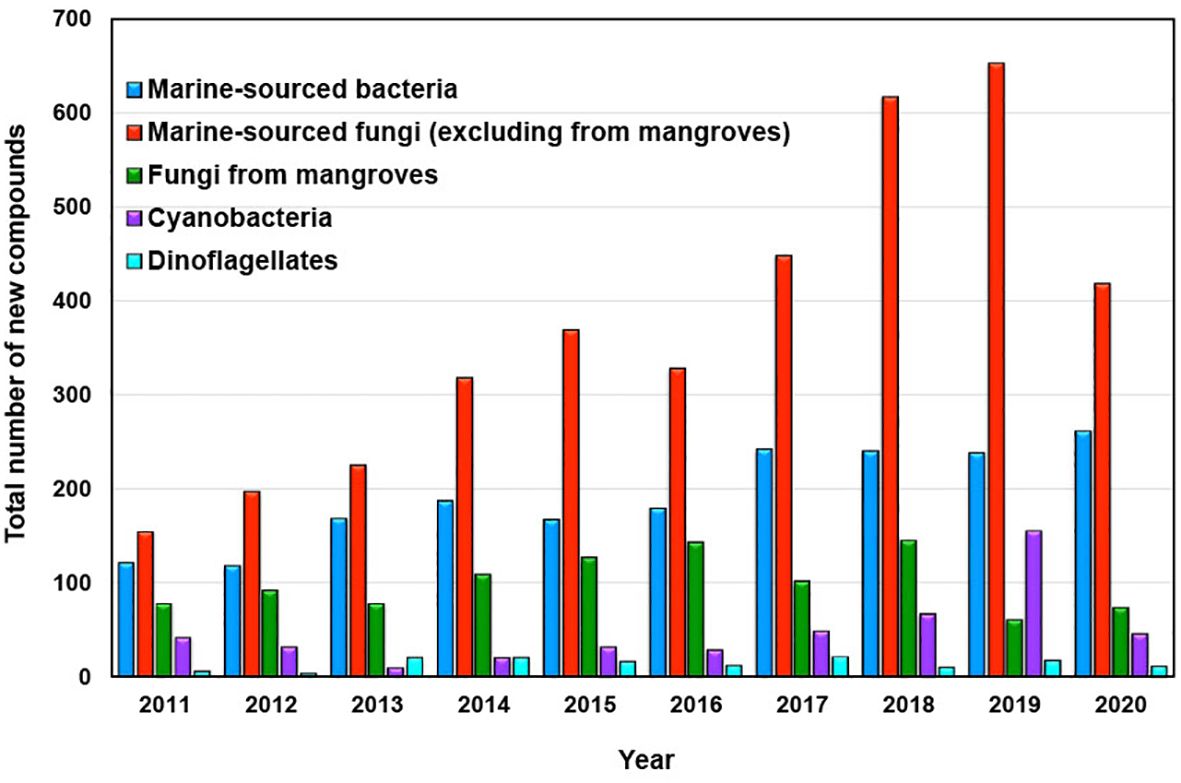
Figure 3. New compounds isolated from marine microorganisms and phytoplankton between 2011 and 2020.
The discovery of potent bioactive compounds from sponges, such as the anticancer agent ara-A from Tethya crypta in the 1950s, marked the beginning of a new era in natural product research (Bergmann and Feeney, 1950; Bergmann and Feeney, 1951).Throughout the latter decades of the 20th century and into the early 2000s, sponges reigned supreme as a prolific source of novel natural compounds (Sipkema et al., 2005). This intensive exploration effort yielded a wealth of bioactive compounds with diverse pharmacological properties, including antiviral, antibacterial, antifungal, and anticancer activities (Gunasekera et al., 1990; Blunt et al., 2009; Blunt et al., 2012; Blunt et al., 2014; Blunt et al., 2016; Blunt et al., 2018; Carroll et al., 2020; Carroll et al., 2021; Carroll et al., 2022). Over the last decade, 2011 to 2020, more than 2600 novel compounds have been discovered from 311 families, 487 genera, and 677 species of sponges. A comprehensive compilation of this data drew upon 769 references.
The identification of sponge species has progressed significantly, with estimates suggesting that up to 15,000 species have been characterized to date (Han et al., 2019). From 2011 to 2020 the distribution of new compounds (2603) came from 20 different orders of sponges. One of the driving factors behind the sustained interest in sponges is their intricate associations with microorganisms, encompassing bacteria and fungi (Zhang et al., 2017; Buttachon et al., 2018; Kiran et al., 2018). The interplay between sponges and microorganisms is highly complex, as sponge-derived metabolites can selectively enrich microbial populations (Sathiyanarayanan et al., 2017).
During the last decade, the sponges studied can be classified as 19 known orders and two sponges of unknown identity (Figure 4). These included Agelasida, Astrophorida, Axinellida, Biemnida, Bubarida, Clathrinida, Clionaida, Dendroceratida, Dictyoceratida, Halichondrida, Haplosclerida, Homosclerophorida, Lyssacinosida, Poecilosclerida, Polymastiida, Scopalinida, Suberitida, Tetractinellida, and Verongiida whereas in 2001 to 2010 sponges were collected from the orders that had changed their nomenclature (Mehbub et al., 2014) as the taxonomy of sponges was amended in 2015 (Morrow and Cárdenas, 2015).
The distribution of new natural products from sponges is quite diverse and extensive as shown in Figure 4; Table 1, mainly discovered from Dictyoceratida (25.04%), Haplosclerida (15.98%), Poecilosclerida (12.86%), Tetractinellida (10.52), Verongiida (7.10%), Homsclerophorida (5.91%) and Suberitida (5.91%) (Supplementary Figure S3). In contrast, when we analyzed the new compounds isolated from sponge-associated microorganisms (Figure 5), most of the new compounds discovered were from Haplosclerida, Unknown sponges, Dictyoceratida, Axinellida and Suberitiida (Figure 5).
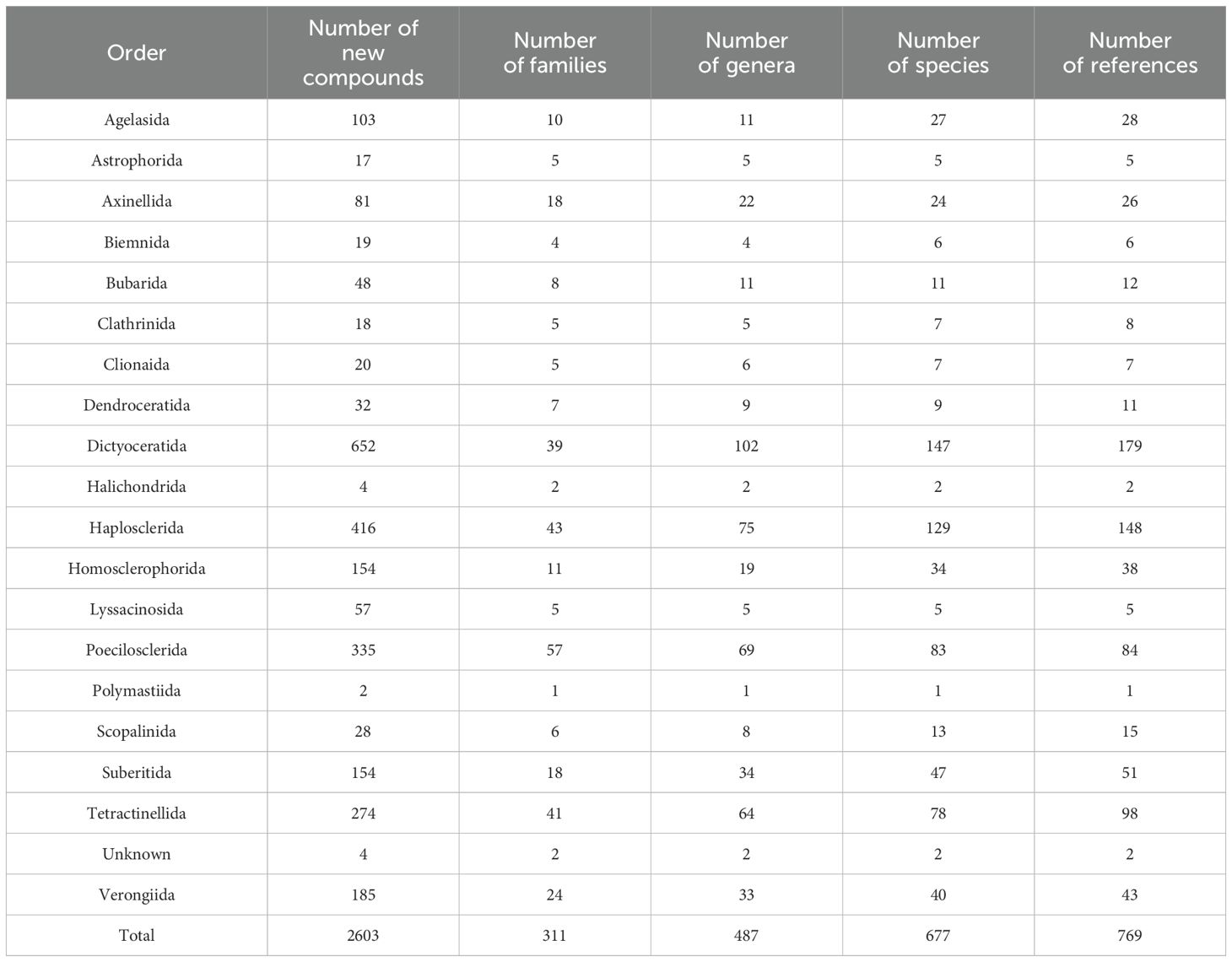
Table 1. Total number of new compounds isolated from different orders of marine sponges with number of families, genera, species and references published from 2011 to 2020.
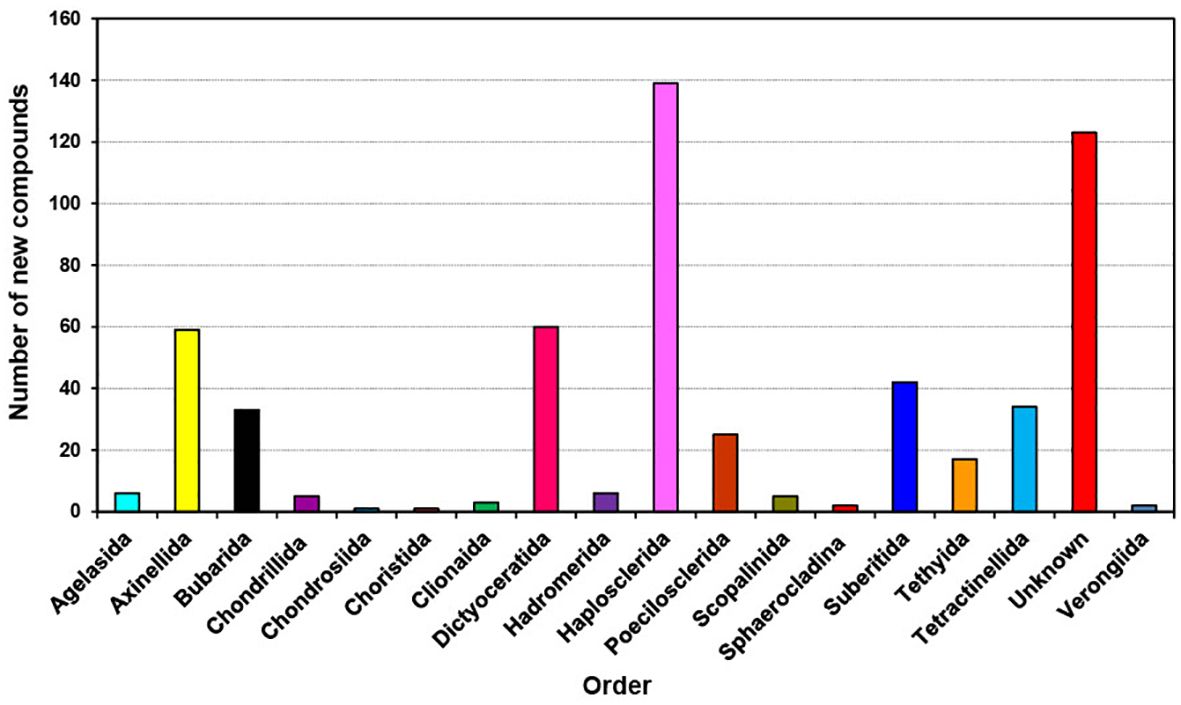
Figure 5. Total number of new compounds isolated from marine microbes associated with different orders of marine sponges from 2011 to 2020.
On average, during the last decade Dictyoceratida contributed an average of 25.9% of new compounds each year, with a minimum of 9.3% and a maximum of 36.4% (Supplementary Figure S3). Haplosclerida contributed an average of 15.8% of the new compounds each year with the highest number of new compounds in the 2011, at 26.7%. Poecilosclerida was the third highest producing order which ranged between 4.9 to 23.1%. Although these three orders showed a similar trend when compared with the previous decade (Mehbub et al., 2014), the contribution of the 4th and 5th highest orders changed in the recent decade. Tetractinellida and Verongiida contributed more from 2011 to 2020 than Halichondrida and Astrophorida (2001 to 2010).
The sponge order Haplosclerida is known to host a diverse array of microorganisms within their tissues, including bacteria and fungi (Beniddir et al., 2022). Interestingly, during the last decade, Haplosclerida produced multiple compounds either from the hosts or their associated microbes. Previous research found Dictyoceratida and Haplosclerida were reported with more species when compared with other sponge orders (Turon et al., 2009) (Table 1).
It is noted that there was a fluctuation in the number of genera discovered to produce new compounds annually within the Dictyoceratida order, ranging from 5 to 14 genera (Figure 6). This variability highlights a significant diversity of genera within this specific order, with the peak number of 14 recorded in both 2015 and 2017. Between 2011 and 2020, there were fluctuations in the number of genera within the orders Haplosclerida, Poecilosclerida, Tetractinellida, and Verongiida that were found to produce new compounds. The range of genera varied each year as follows: Haplosclerida (6-9), Poecilosclerida (4-12), Tetractinellida (4-9), and Verongiida (1-6) (Hong et al., 2022).
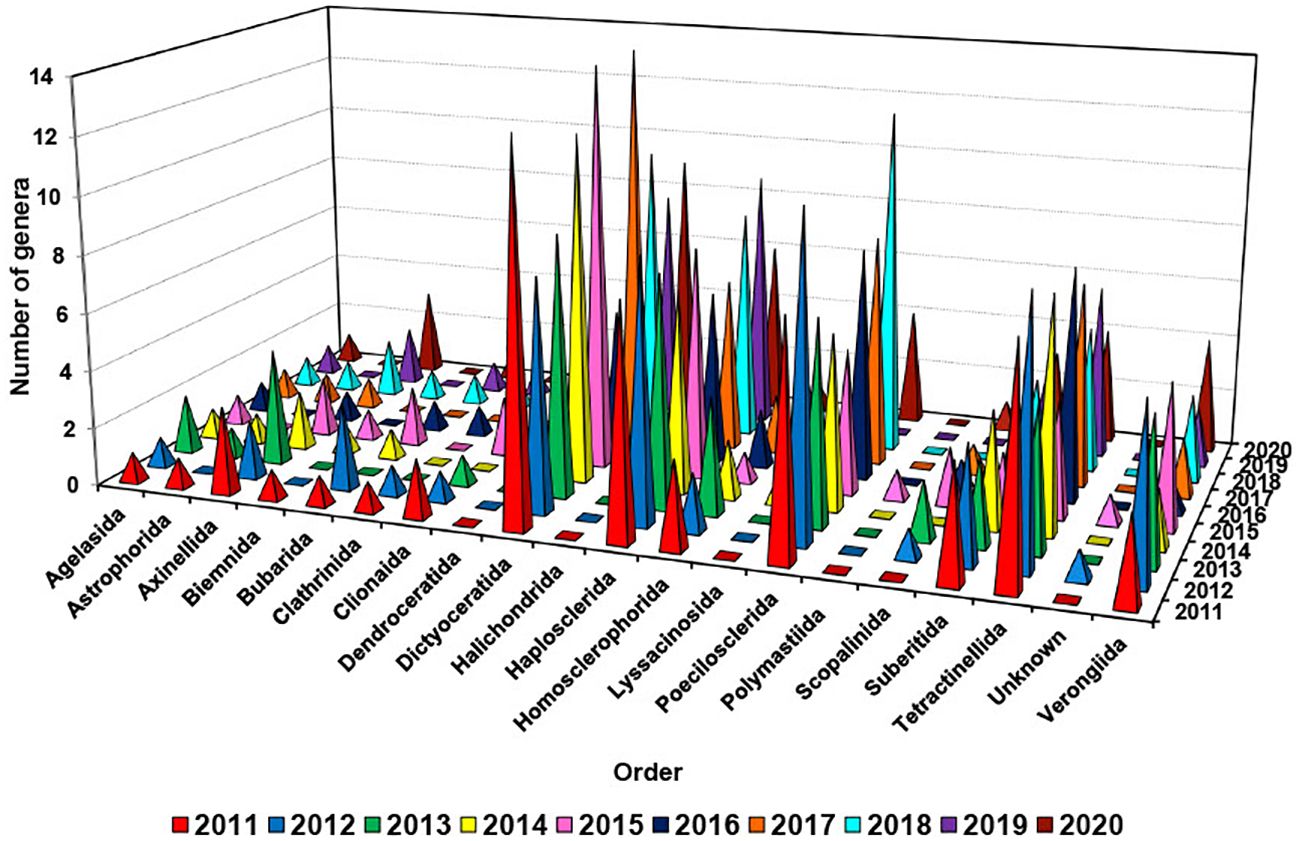
Figure 6. Number of different genera used for isolation of new compounds from different orders of marine sponges from 2011 to 2020.
Between 2011 and 2020, individual species from various sponge orders typically contributed an average of two to four new compounds, except Lyssacinosida, which contributed an average of 11 compounds per species (Figure 7). On average, species from the orders Bubarida, Dictyoceratida, Homosclerophorida, Poecilosclerida, and Verongiida each contributed approximately four compounds per species. This observation suggests that regardless of the order, individual species have the potential to contribute a similar number of new compounds. However, Lyssacinosida stands out as warranting further study due to its notably higher average number of new compounds contributed per species.
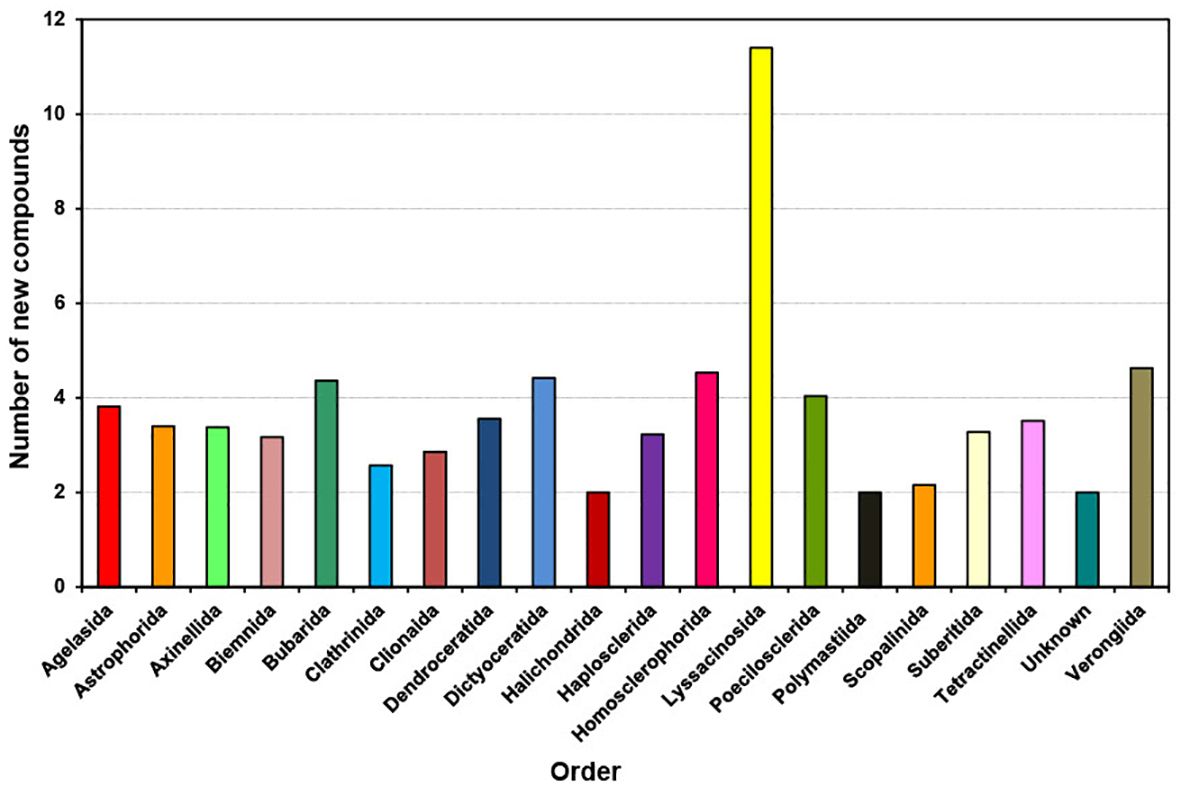
Figure 7. Average number of new compounds per species isolated from different orders of marine sponges from 2011 to 2020.
The chemical classes considered for this manuscript followed Blunt et al. and Carroll et al. from Natural Product Reports of 2012 to 2022 (Blunt et al., 2012; Blunt et al., 2013; Blunt et al., 2014; Blunt et al., 2015; Blunt et al., 2016; Blunt et al., 2017; Blunt et al., 2018; Carroll et al., 2019; Carroll et al., 2020; Carroll et al., 2022) as mentioned earlier. However, there are some overlaps in macrolide and polyketide, sterol and steroid, and fatty acid and lipid.
Sponges are remarkable organisms that have evolved to thrive in various aquatic environments. They are known to produce a wide array of secondary metabolites, including alkaloids, terpenes, terpenoids, peptides, and lipids.
The trend in chemical classes during the last decade (2011 to 2020) has not changed from the previous decade (2001 to 2010). However, the number of compounds has increased particularly in the top three chemical classes compared to the last decade. The dominant chemical classes (Figure 8) were (total number of new compounds in brackets) Alkaloids (609), Terpenes (392), Terpenoids (264), Peptides (140), Quinones (123), and Lipids (116). Alkaloids contributed 23.4% of the new compounds during the last decade (Supplementary Figure S4) whereas Terpenes and Terpenoids together contributed 25.2% of the new compounds. Therefore, almost 50% of the new compounds belong to these three chemical classes.
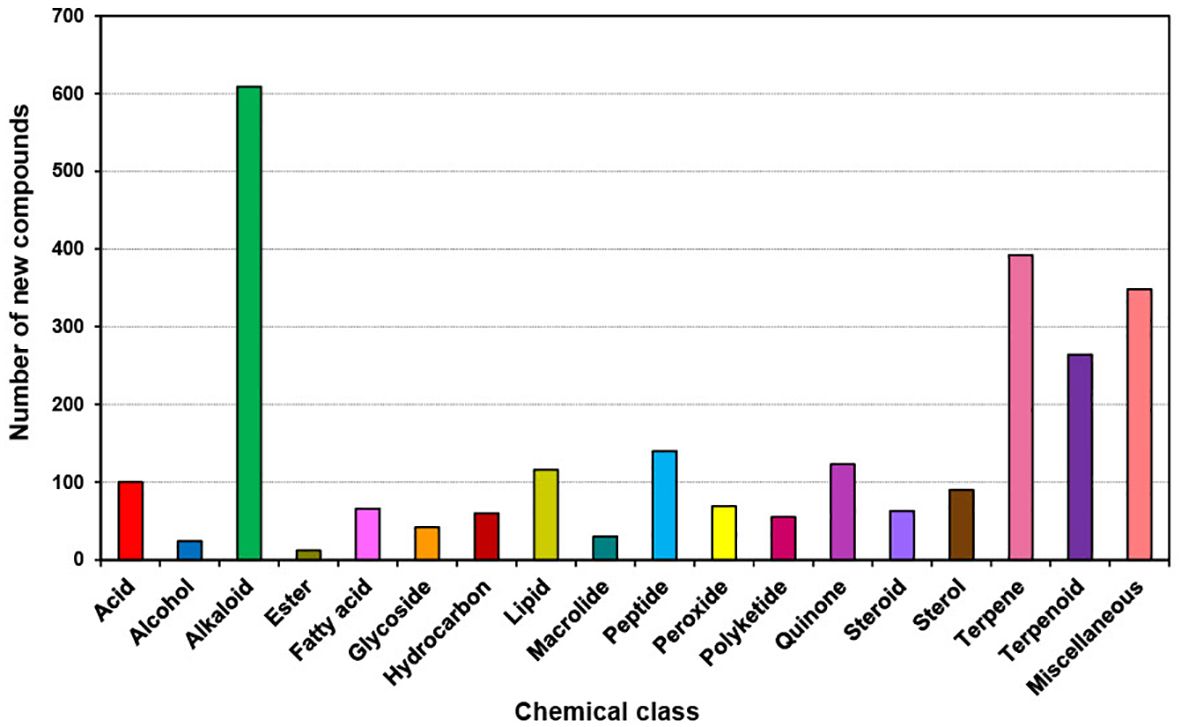
Figure 8. Chemical Class of new compounds isolated from different marine sponge orders from 2011 to 2020.
Alkaloids, ubiquitous in sponge species, serve multifaceted roles in chemical defense against predators and pathogens. Recent research highlights the pharmacological importance of sponge-derived alkaloids, showing promise for cancer treatment, pain management, and neurological disorders (Sobarzo-Sánchez, 2015; Cragg and Pezzuto, 2016; Bian et al., 2020). The production of alkaloids by sponges is thought to be an evolutionary adaptation to the competitive and predatory pressures of their environment, enhancing their survival and reproductive success.
A recent study reviewed 149 marine alkaloids extracted from sponges, examining their structures and reported bioactivities between 2015 and 2020. The study also analyzed the marine sponge habitats rich in alkaloids, which include indole alkaloids, carboline alkaloids, guanidine alkaloids, bromotyrosine alkaloids, pyrrole alkaloids, diterpene alkaloids, and others. Additionally, the research investigated the habitats of marine sponges containing potential alkaloids and explored the biosynthetic pathways of pyrrole and guanidine alkaloids from the marine sponges Agelas and Monanchora (Bian et al., 2020).
Manzamines represent a significant alkaloid family, with the first member, Manzamine A, isolated from Haliclona sp (Sakai et al., 1986). This group of compounds displays a range of bioactivities, including cytotoxicity and antibacterial effects (Kim et al., 2017). Bromopyrrole alkaloids are exclusive to the Porifera phylum. Oroidin, featuring pyrrole-imidazole and bromopyrrole carboxamide components, stands as the pioneering compound in this class. These alkaloids are predominantly found in the families Agelasidae, Axinellidae, and Halichondridae, where they likely serve as a deterrent against predators (Lindel et al., 2000). Pyrrole−imidazole alkaloids are a large and chemically complex class of sponge natural products with over 150 congeners (Mohanty et al., 2020). Natural product researchers are particularly intrigued by this alkaloid class due to its diverse range of biological activities, including insecticidal, antibacterial, and cytotoxic properties (Edrada et al., 1996).
Similarly, terpenes and terpenoids, extensively produced by sponges, play crucial roles in chemical defense mechanisms and ecological interactions. Previous studies emphasized the antimicrobial, antifungal, and anticancer properties of sponge-derived terpenes and terpenoids, as well as their potential as leads for drug development (Minale, 2012; Ruiz-Ruiz et al., 2017; Martignago et al., 2023). Sponges synthesize these compounds as a response to environmental stressors, including predation and competition, highlighting their adaptive significance in marine ecosystems.
Peptides are another abundant class of compounds in sponges, known for their diverse structures and biological activities. The antimicrobial, antiviral, and anticancer properties of sponge-derived peptides, along with their potential applications in biomedicine has been discussed previously by several researchers (Tincu and Taylor, 2004; Vitali, 2018). Sponges produce peptides as a defense mechanism against microbial colonization and predation, underscoring their importance in maintaining symbiotic relationships and ecological balance in marine communities (Wulff, 2006; Flórez et al., 2015).
Moreover, sponges are rich sources of various lipids, including fatty acids, sterols, and lipid-soluble compounds. Lipids are important for the structural and functional properties of sponges which contribute to membrane integrity, energy storage, and intercellular signaling (Koutsouveli et al., 2022). The production of lipids by sponges are essential for their physiological processes and adaptation to fluctuating environmental conditions, illustrating their ecological and evolutionary importance (Blunt et al., 2012; Bennett et al., 2018).
Sponge-derived quinones represent a significant chemical class with diverse ecological and pharmacological importance. These compounds, abundant in marine sponges, play pivotal roles in the chemical defense mechanisms of these organisms, acting as deterrents against predators and competitors. Additionally, sponge-derived quinones exhibit antimicrobial properties, contributing to the defense against microbial colonization and infection. Past study highlighted the ecological significance of sponge-derived quinones, emphasizing their role in maintaining the ecological balance of marine ecosystems (Hong et al., 2022). Furthermore, quinones from sponges have drawn attention from the pharmaceutical industry due to their potential as lead compounds for drug discovery and development (Motti et al., 2007). Their diverse biological activities, including antimicrobial, antifungal, and anticancer properties, make them promising candidates for further pharmacological research, offering potential applications in various therapeutic areas.
In this review, the bioactivates of new compounds from different sponge orders are classified into 10 distinct types. Among these, nine have definitive activities, which are presented in Table 2, while all other activities fall under the miscellaneous group. Compounds with no activity reported were excluded from this table. For compounds that exhibit more than one activity, each is considered individually.
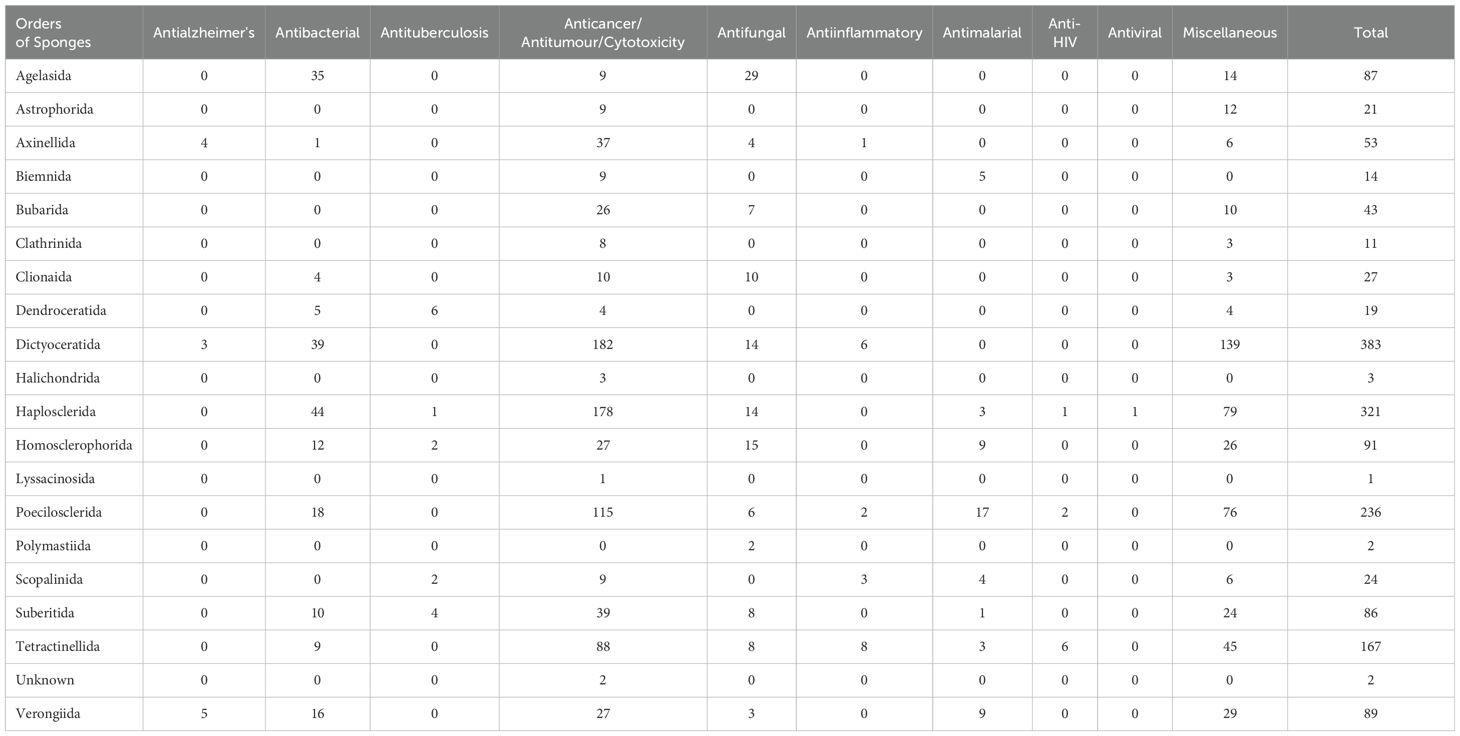
Table 2. Total number of new compounds isolated from different orders of sponges with different bioactivities from 2011 to 2020.
The highest number of compounds with activities (383) was found in Dictyoceratida sponges during the last decade 2011 to 2020. Among these, anticancer/antitumor/cytotoxicity activity was dominant (182). Additionally, this order also exhibited activities against bacteria (39), fungi (14), inflammation (6), Alzheimer’s, and miscellaneous (139). Mehbub et al. (2014) mentioned that during the decade from 2001 to 2010, compounds reported from the order Dictyoceratida exhibited a diverse range of bioactivities, including antibacterial, anticancer, antifungal, anti-HIV, anti-inflammatory, cytotoxicity, Gsk-3β inhibitor, inhibitor of phosphatase (Cdc25B), inhibitor of PLA2, inhibitor of protein kinase, and other miscellaneous activities (Mehbub et al., 2014). However, in the decade from 2011 to 2020, compounds were extracted from fewer genera compared to the previous decade, with similar genera of sponges, except Petrosaspongia, which was not evident in the previous decade.
The order Haplosclerida contains the greatest biodiversity of sponges, with representatives inhabiting all oceans and habitat types, including freshwater bodies, on every continent in the world (Goudie et al., 2013). The abundance of bioactive compounds in Haplosclerida sponges can be attributed to several factors. Firstly, these sponges often host diverse microbial communities, including bacteria, archaea, fungi, and microalgae, which can produce bioactive compounds either independently or in symbiosis with the sponge host (Taylor et al., 2007) which also revealed in the present study. Secondly, sponges, including those in the Haplosclerida order, rely on bioactive compounds as chemical defenses against predators, competitors, and pathogens, driving the production of diverse secondary metabolites with biological activities (Pawlik et al., 1995). Sponge falling under the order Haplosclerida are renowned as some of the most abundant reservoirs of bioactive natural products. These include alkaloids, polyacetylenes, and terpene derivatives. Thirdly, Haplosclerida sponges inhabit various marine environments, such as coral reefs and shallow coastal waters, where the production of bioactive compounds may aid in adaptation to specific environmental conditions or ecological niches (Goudie et al., 2013). Notably, this group stands out for its exceptional diversity both in terms of species numbers and habitats. Moreover, these sponges possess complex metabolic pathways capable of synthesizing a wide range of secondary metabolites, which often serve ecological roles, such as allelopathy or antifouling, in addition to their bioactivity against microbes. The relationship between Haplosclerida sponges and microbes is intricate, with symbiotic interactions, microbial diversity, chemical communication, and bioprospecting potential all playing significant roles in shaping the production and diversity of bioactive compounds within these sponge-microbe ecosystems (Thomas et al., 2010). Interestingly, members of the Haplosclerida also possess relatively few distinguishing morphological characteristics (Tribalat et al., 2016).
The second-largest group showcasing diverse bioactivities is affiliated with the Haplosclerida order of sponges, which contributed 321 bioactivities between 2011 and 2020 (Figure 9) (Table 2). While Haplosclerida sponges showed no activity against inflammation, they did demonstrate activity against malaria, HIV, and viruses. Otherwise, their activities were largely comparable to those of the Dictyoceratida order and mostly exhibited cytotoxicity similar to Dictyoceratida. The Poecilosclerida order encompasses 25 families, with species recorded in all marine habitats characterized by the presence of both organic (fiber) and inorganic (specular) skeletal components. An interesting observation from these sponges is that they exhibited distinguished antimalarial activity in 17 instances (see Table 2), comprising a total of 236 bioactivities. Tetractinellida sponges are distributed across a variety of marine habitats worldwide (Fromont et al., 2016), frequently inhabiting intertidal zones, coral reefs, and deep-sea environments. They display a diverse array of structural complexity in their compounds, offer refuge and breeding grounds for various species, thus promoting biodiversity and supporting the balance of the marine ecosystems. This sponge order produced compounds that exhibited the highest activity against HIV, and hold great promise from a pharmaceutical perspective (Al-Khayri et al., 2022). The other two orders that demonstrated these activities are Poecilosclerida and Haplosclerida.
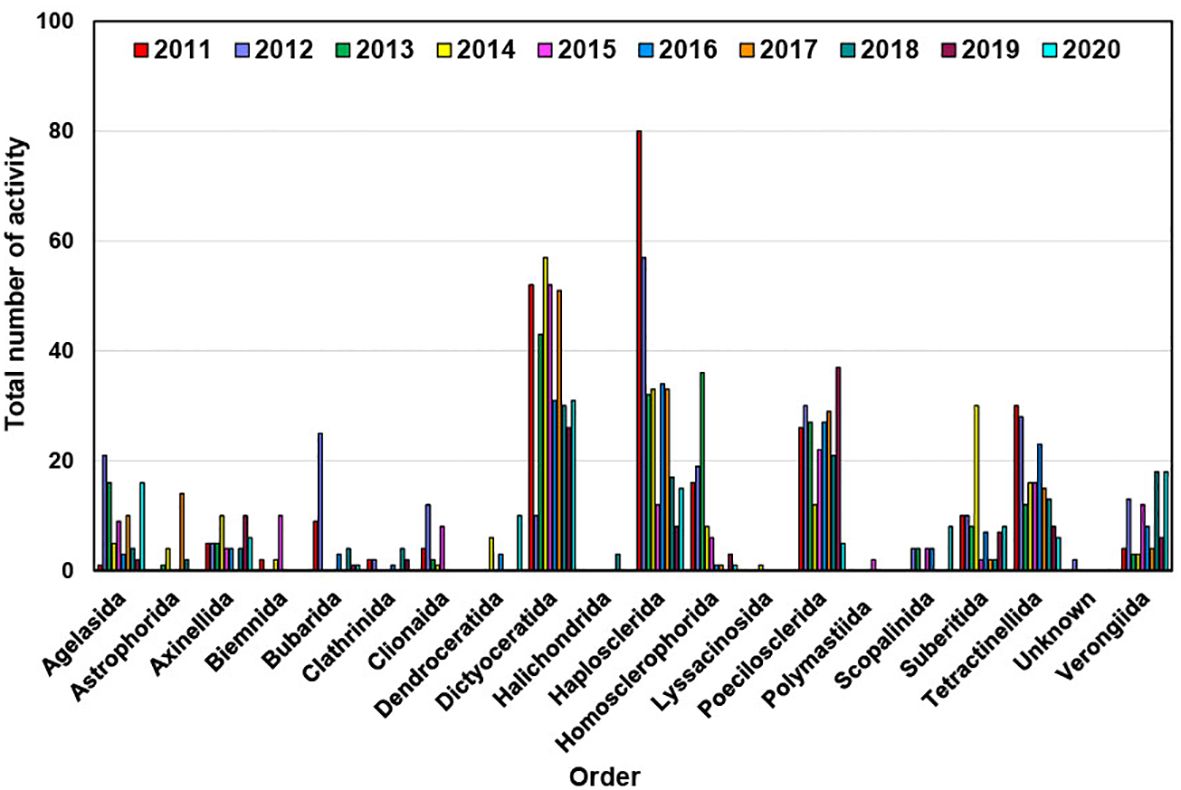
Figure 9. Annual distribution of bioactivity of new compounds isolated from different marine sponge orders from 2011 to 2020.
During the last decade it has been found that most of the bioactivities shown by new compounds from different orders of marine sponges are anticancer/antitumor/cytotoxicity (Figure 10). Astrophorida, Axinellida, Biemnida, Clathrinida, Dictyoceratida, Halichondrida, Haplosclerida, Lyssacinosida, Poecilosclerida, Suberitida and Tetractinellida order of sponges when tested for bioactivity showed more than 50% bioactivity as anticancer/antitumor/cytotoxicity (Figure 11). The expanding understanding of the bioactivities of compounds isolated from marine sponges to target different stages of the carcinogenic process implies their potential utility as a source of anticancer agents because of their potential to inhibit tumor growth through various mechanisms, including cell-cycle regulation, anti-inflammatory effects, apoptosis induction, initiation of ER stress, and interaction with multiple targets implicated in cancer progression, such as the mitochondrial membrane, PARP, cytochrome c, Akt, and caspases (Calcabrini et al., 2017). It has been reported that Petrosia, Haliclona (Haplosclerida), Rhabdastrella (Tetractinellida), Coscinoderma, and Hippospongia (Dictyoceratida) have emerged as the most promising genera due to their ability to generate novel bioactive compounds. Many of these newly discovered compounds and their synthetic counterparts have exhibited cytotoxic and pro-apoptotic effects in vitro against a range of tumor and cancer cell lines (Mioso et al., 2017).
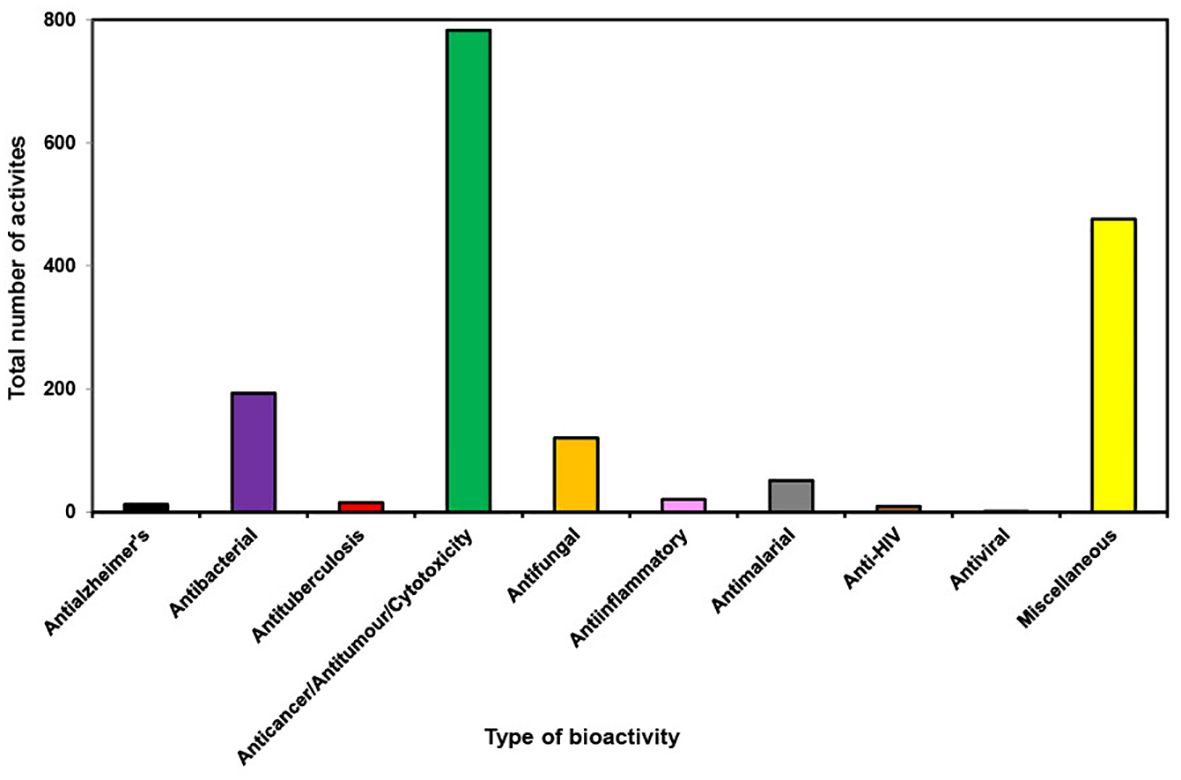
Figure 10. Total number of bioactivities shown by new compounds from different orders of marine sponges from 2011 to 2020.
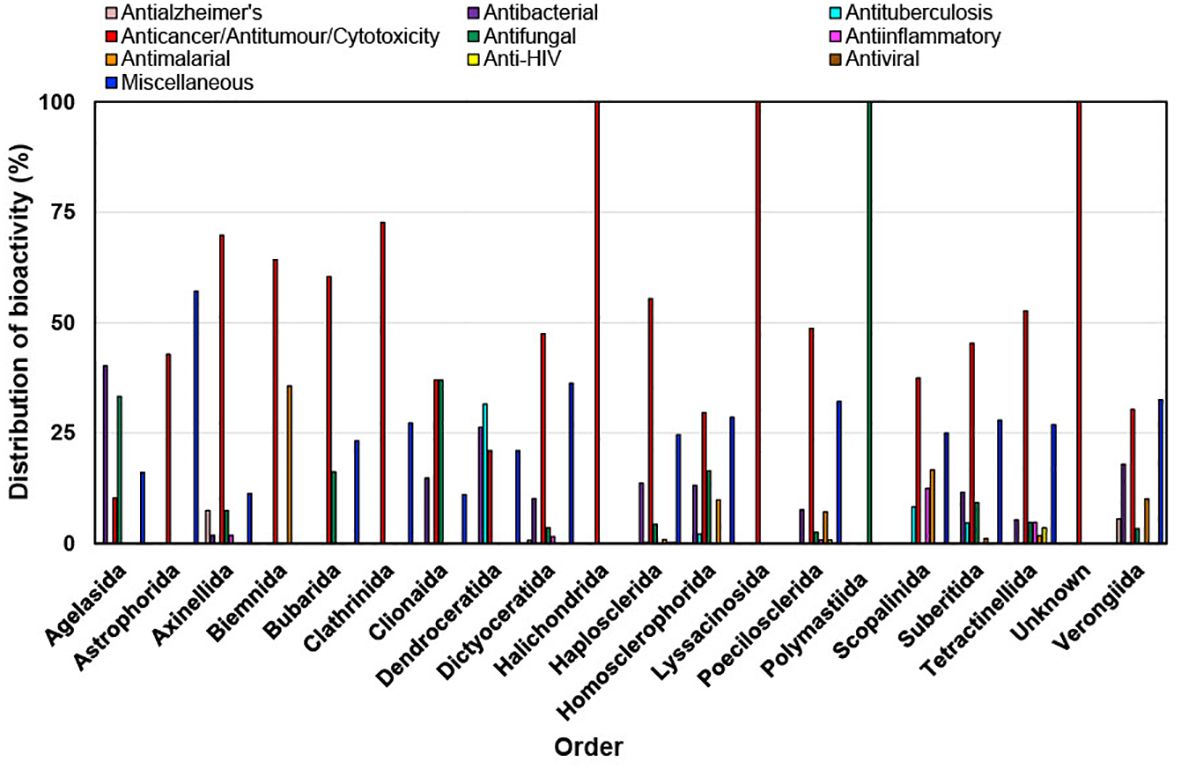
Figure 11. Distribution of bioactivities of new compounds isolated from different orders of marine sponges from 2011 to 2020.
Marine sponges have been increasingly recognized for their potential to provide natural sources of antibacterial agents. It was hypothesized that sponge-associated microbes particularly bacteria are responsible for producing those bioactive compounds (Anand et al., 2006; Bibi et al., 2020; Nasser Alahmari et al., 2022). Studies have revealed that extracts and compounds derived from marine sponges exhibit significant antibacterial activity against various pathogens, including both Gram-positive and Gram-negative bacteria (Cita et al., 2017). These antibacterial properties are attributed to the diverse chemical compounds present in the sponges, such as peptides, alkaloids, terpenoids, and polyketides (Varijakzhan et al., 2021).
Furthermore, the unique marine environment in which sponges thrive offers a rich biodiversity that may contribute to the discovery of novel antibacterial agents (Bayona et al., 2020). The exploration of marine sponge microbiomes has also revealed the presence of symbiotic microorganisms that produce bioactive compounds with antibacterial properties (Brinkmann et al., 2017).
There have been many new sponge-derived compounds with unusual structures and potent bioactivity which were discovered during the last decade of 2011-2020. Some of these representative compounds are illustrated in Table 3.
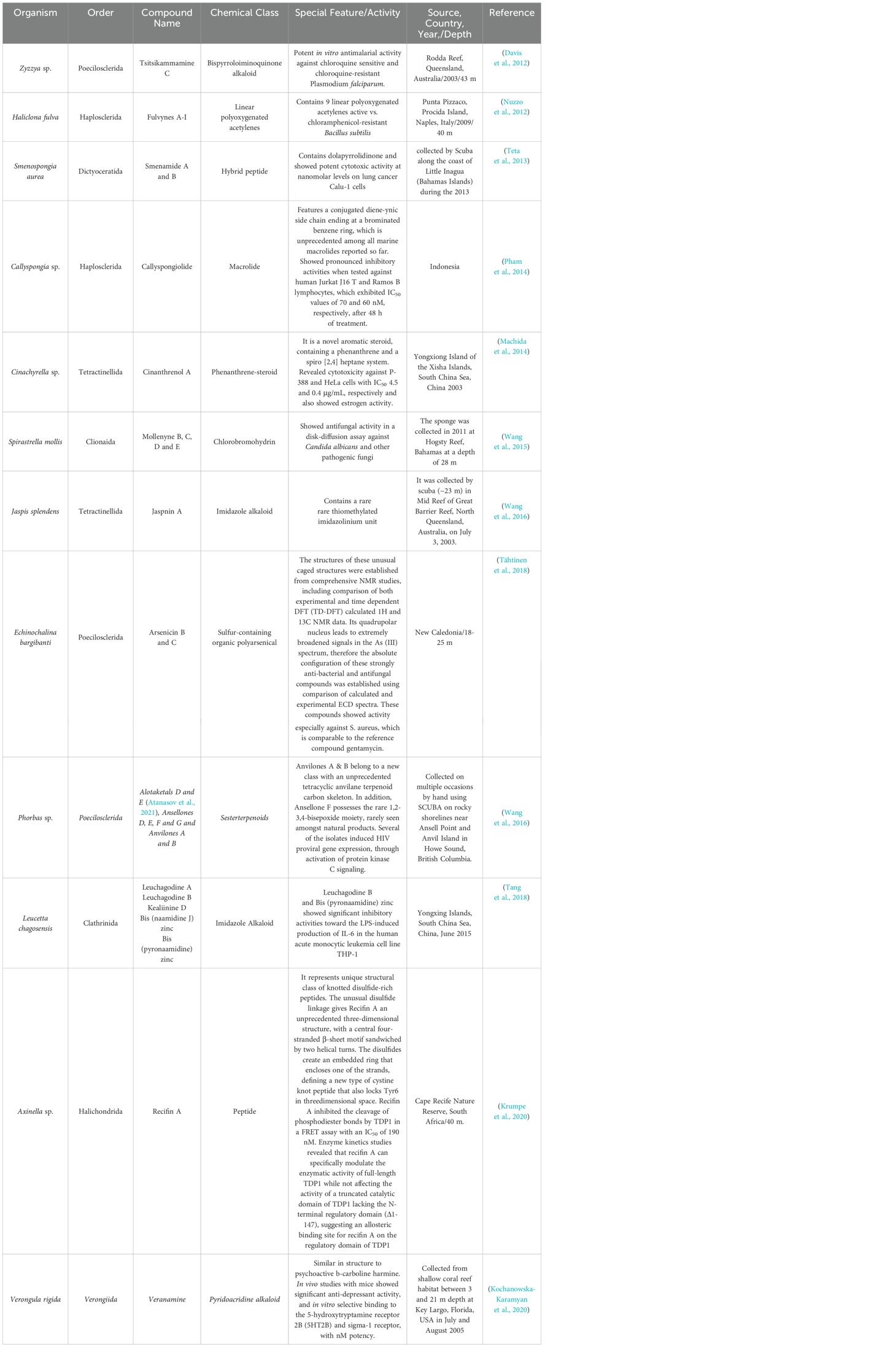
Table 3. Selected representative new compounds with unusual structure and significant bioactivity from sponges discovered between 2011-2020.
The geographic location and its local water environment play a significant role in shaping the metabolite profiles present in marine sponges around the world (Lee et al., 2021) with diverse biological activities (Folkers and Rombouts, 2020). Specific location can directly influence the variety and abundance of metabolites present (Zhang et al., 2022). Environmental factors, such as temperature, salinity, nutrient availability, light penetration, and water currents, vary across different geographic locations leading to variations in metabolite production (Pita et al., 2018). Geographic location can also influence the ecological interactions of marine sponges with other organisms, such as predation and competition (Wulff, 2012) and thus impacts on the production of metabolites by marine sponges (Tan, 2023).
The extent of scientific sampling efforts varies across locations, with less explored regions potentially harboring undiscovered species with unique metabolite profiles. International collaboration and sharing of research findings support sponge research globally, while conservation efforts aimed at protecting marine habitats preserve ecosystems crucial for sponge diversity and the discovery of new compounds.
Over the last decade (2011 to 2020), new compounds were isolated from sponges collected from 59 identified locations, including countries, continents, provinces, seas, and unspecified locations (Supplementary Figure S5). Among these, sponges from China emerged as the primary source, yielding 561 new compounds (accounting for 21.5% of all new compounds discovered). Additionally, significant contributions came from other leading locations such as Japan, Indonesia, and Australia, each providing over 200 new compounds individually. Korea and Vietnam also contributed substantially, with each yielding more than 100 new compounds.
An intriguing shift occurred during the decade from 2011 to 2020, with China emerging as the new hotspot for the discovery of sponge-derived compounds, surpassing Australia, Indonesia, and Japan, which held the positions of fourth, third and second hotspot locations (Figure 12).
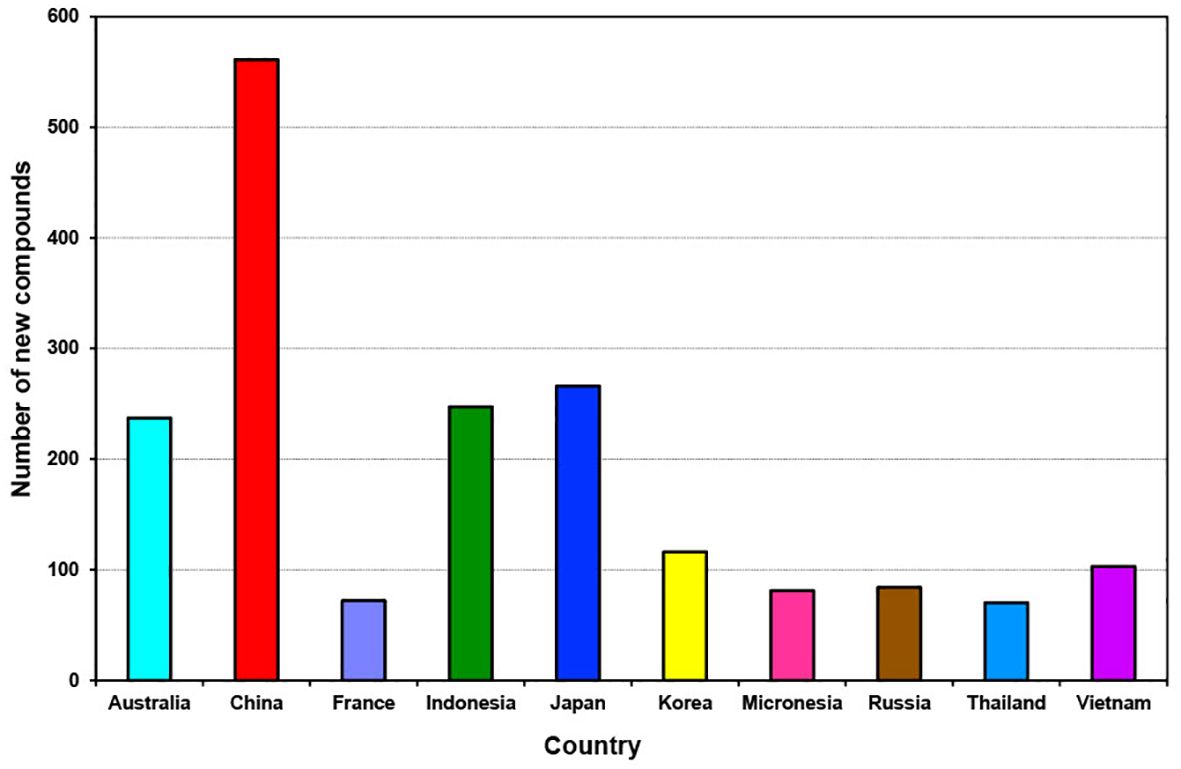
Figure 12. Total number of new compounds isolated from marine sponges from the top 10 source countries from 2011 to 2020.
At least 40 new compounds each year from 2011 to 2020 were isolated from Chinese sponge samples, with the highest output occurring in 2014 and 2011 with 83 and 79 new compounds (Supplementary Table S1). In contrast, the top three ranking locations are Japan, Indonesia and China contributed at least 9% of new compounds discovered during the previous decade 2001 to 2010 (Mehbub et al., 2014).
China has been recognized as one of the top “megadiverse” nations, boasting an abundance of marine genetic resources that rank among the richest and most varied worldwide (Huang et al., 2021). The significant discovery of numerous new compounds from sponges in China can be attributed to several factors. Firstly, China’s extensive coastline and diverse marine ecosystems provide a rich habitat for sponge biodiversity, offering ample opportunities for researchers to explore and uncover new species. Secondly, advancements in marine biodiscovery research infrastructure in China have facilitated more extensive and systematic studies of marine organisms, including sponges. Furthermore, increased collaboration and investment in marine science and bioprospecting projects have enabled scientists in China to conduct comprehensive surveys and expeditions, leading to the discovery of novel compounds.
As we unlock the chemical diversity of sponge and sponge-associated microbial communities, it is essential to consider the conservation implications of our discoveries and the need to protect these delicate ecosystems in the face of environmental change. Sponges are highly susceptible to anthropogenic stressors, including habitat destruction, pollution, overfishing, and climate change, which threaten the stability of sponge-microbe symbioses and the ecological services they provide. Moreover, the extraction of sponge-derived natural products for commercial pharmaceutical and biotechnological purposes raises ethical concerns regarding biodiversity conservation and sustainable use of marine resources.
It has been widely recognised that marine invertebrates, particularly sponges, host a wide-ranging and varied array of associated microorganisms, many of which are genuine symbionts (Gomes et al., 2016). Efforts to conserve sponge-microbe symbioses and their associated biodiversity require a multifaceted approach that integrates scientific research, policy development, and stakeholder engagement. Marine protected areas, sustainable fishing practices, and ecosystem-based management strategies can help mitigate the impacts of human activities on sponge habitats and promote the resilience of sponge-associated microbial communities. Furthermore, initiatives aimed at raising public awareness and fostering community stewardship of marine ecosystems are essential for fostering a culture of conservation and promoting sustainable use of natural resources for many generations to come.
The production of natural compounds produced by microbes living in sponges present a promising, and likely sustainable avenue for commercial development including medical and other marine bioproducts for industrial applications. Prior research has shown that these microbes, including bacteria, fungi, and algae, produce metabolites that can be utilized in medicine, cosmetics, environmental protection, and manufacturing. Both temporary and permanent bacteria make up the microbial community of sponges, with the latter significantly contributing to the production of secondary metabolites. The diverse and rich microbial communities within sponges have various symbiotic relationships, which attract attention due to their potential to overcome the supply issues of marine-sponge-derived compounds for medicinal and other uses.
A recent study as well as present study identified several sponge orders as sources of sponge-associated microbes including Agelasida, Axinellida, Bubarida, Chondrillida, Chondrosiida, Choristida, Clionaida, Dictyoceratida, Hadromerida, Haplosclerida, Poecilosclerida, Scopalinida, Sphaerocladina, Suberitida, Tethyida, Tetractinellida, and Verongiida (Samirana et al., 2022). The discovery of new compounds from these sponge orders is crucial for advancing drug development and understanding microbial symbiosis. Techniques such as genome mining and synthetic bioengineering provide avenues for sustainable supply and application development of these new compounds (Rastegari et al., 2020; Rotter et al., 2021). Among all the sponge orders explored from 2011 to 2020, Haplosclerida, Dictyoceratida, Axinellida, Suberitida, and Tetractinellida appear to be particularly promising.
More efforts are needed to explore the resources of marine sponge-associated microbes. Collaboration at national and international levels is vital for establishing dedicated facilities for marine-based drug discovery and preserving microbial cultures. Despite challenges such as accessing harsh marine environments and maintaining laboratory conditions, the benefits of marine microbial resources continue to drive new discovery. This underscores the importance of ongoing research and innovation in this field.
In conclusion, sponges remain an important, second largest source of new marine natural product discovery during the decade 2011-2020, but only accounting for around one-third of the largest source, marine microbes and phytoplankton. The pivot towards marine microbes, similarly sponge-associated microbes as a prolific source of bioactive compounds marks a paradigmatic change in natural product discovery, propelled by advances in metagenomics, metabolomics, and bioinformatics. By unraveling the chemical complexity of sponge-microbe interaction, researchers are uncovering a treasure trove of bioactive molecules with pharmaceutical potential and biotechnological applications. However, as we harness the therapeutic and industrial promise of sponge-derived natural products, it is imperative that we prioritize the conservation of sponge habitats and protect the delicate balance of symbiotic relationships that underpin these valuable ecosystems. Only through collaborative efforts and holistic approaches we can fully unlock the potential of sponge-associated microbes and their biosynthetic capacity for the sustainable benefit of human health, industry and the environment.
MM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. QY: Data curation, Writing – review & editing. YC: Data curation, Writing – review & editing. CF: Conceptualization, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. WZ: Conceptualization, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors acknowledge the support from Flinders University and all the staff supporting our work at the Centre for Marine Bioproducts Development.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1462825/full#supplementary-material
Supplementary Figure 1 | Total number of new compounds discovered from different marine sources from 2001 to 2020.
Supplementary Figure 2 | Distribution of new compounds isolated from different groups of marine organisms/resources as a percentage found within the year, 2011–2020.
Supplementary Figure 3 | Distribution of new compounds isolated from different orders of marine sponges as a percentage found within the year, 2011–2020.
Supplementary Figure 4 | The distribution of chemical classes of new compounds isolated from different marine sponges from 2011 to 2020.
Supplementary Figure 5 | Total number of new compounds isolated from different marine sponges and their source locations from 2011 to 2020.
Aguiar-Pulido V., Huang W., Suarez-Ulloa V., Cickovski T., Mathee K., Narasimhan G. (2016). Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol. Bioinf. Online 12, 5–16. doi: 10.4137/ebo.S36436
Alencar A., Bourgeois B., Butscher J., Debitus C., Ekins M., Fleurisson D., et al. (2017). Sponges of polynesia. Papeete (PYF) IRD, 1–827.
Al-Khayri J. M., Asghar W., Khan S., Akhtar A., Ayub H., Khalid N., et al. (2022). Therapeutic potential of marine bioactive peptides against human immunodeficiency virus: recent evidence, challenges, and future trends. Mar. Drugs 20, 477. doi: 10.3390/md20080477
Anand T. P., Bhat A. W., Shouche Y. S., Roy U., Siddharth J., Sarma S. P. (2006). Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of south east India. Microbiological Res. 161, 252–262. doi: 10.1016/j.micres.2005.09.002
Atanasov A. G., Zotchev S. B., Dirsch V. M., Orhan I. E., Banach M., Rollinger J. M., et al. (2021). Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discovery 20, 200–216. doi: 10.1038/s41573-020-00114-z
Bakus G. J., Abbott D. P. (1980). Porifera: The Sponges. In: Morris R. H., Abbott D. P., Haderlie E. C. Eds. Intertidal inverte-brates of California. (Stanford University Press: Stanford, California), 21–39 p.
Bayona L. M., van Leeuwen G., Erol Ö., Swierts T., van der Ent E., de Voogd N. J., et al. (2020). Influence of geographical location on the metabolic production of Giant Barrel sponges (Xestospongia spp.) revealed by metabolomics tools. ACS Omega 5, 12398–12408. doi: 10.1021/acsomega.0c01151
Beniddir M. A., Le Moyec L., Triba M. N., Longeon A., Deville A., Blond A., et al. (2022). Metabolomics with multi-block modelling of mass spectrometry and nuclear magnetic resonance in order to discriminate Haplosclerida marine sponges. Analytical Bioanalytical Chem. 414, 5929–5942. doi: 10.1007/s00216-022-04158-5
Bennett H., Bell J. J., Davy S. K., Webster N. S., Francis D. S. (2018). Elucidating the sponge stress response; lipids and fatty acids can facilitate survival under future climate scenarios. Global Change Biol. 24, 3130–3144. doi: 10.1111/gcb.14116
Bergmann W., Feeney R. J. (1950). The isolation of a new thymine pentoside from sponges. J. Am. Chem. Soc. 72, 2809–2810. doi: 10.1021/ja01162a543
Bergmann W., Feeney R. J. (1951). Contributions to the study of marine products. Xxxii. The nucleosides of sponges. I. J. Organic Chem. 16, 981–987. doi: 10.1021/jo01146a023
Bergquist P. R. (1978). Sponges (Berkely & Los Angeles: Hutchinson: London, University of California Press).
Bernot J., Ahyong S., Boyko C. B., Bailly N., Bieler R., Brandão S. N., et al. (2024). World Register of Marine Species (Worms) (WoRMS Editorial Board). Available at: https://www.marinespecies.org. [cited 2024-03-10].
Bian C., Wang J., Zhou X., Wu W., Guo R. (2020). Recent advances on marine alkaloids from sponges. Chem. Biodiversity 17, e2000186. doi: 10.1002/cbdv.202000186
Bibi F., Faheem M I., Azhar E., Yasir M., Alvi S A., Kamal M A., et al. (2017). Bacteria from marine sponges: a source of new drugs. Curr. Drug Metab. 18, 11–15. doi: 10.2174/1389200217666161013090610
Bibi F., Yasir M., Al-Sofyani A., Naseer M. I., Azhar E. I. (2020). Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. Ea348. Saudi J. Biol. Sci. 27, 1139–1147. doi: 10.1016/j.sjbs.2020.02.002
Blunt J. W., Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2018). Marine natural products. Natural Product Rep. 35, 8–53. doi: 10.1039/c7np00052a
Blunt J. W., Copp B. R., Hu W. P., Munro M. H., Northcote P. T., Prinsep M. R. (2009). Marine natural products. Natural Product Rep. 26, 170–244. doi: 10.1039/b805113p
Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2012). Marine natural products. Natural Product Rep. 29, 144–222. doi: 10.1039/C2NP00090C
Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2013). Marine natural products. Natural Product Rep. 30, 237–323. doi: 10.1039/C2NP20112G
Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2014). Marine natural products. Natural Product Rep. 31, 160–258. doi: 10.1039/C3NP70117D
Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2015). Marine natural products. Natural Product Rep. 32, 116–211. doi: 10.1039/C4NP00144C
Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2016). Marine natural products. Natural Product Rep. 33, 382–431. doi: 10.1039/C5NP00156K
Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. (2017). Marine natural products. Natural Product Rep. 34, 235–294. doi: 10.1039/C6NP00124F
Brinkmann C. M., Marker A., Kurtböke D. I. (2017). An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity 9, 40. doi: 10.3390/d9040040
Brusca R. C., Brusca G. J. (2004). Invertebrates: Systematic Biology. Sunderland Massachussetts: Sinauer Associates Inc 53, 664–666.
Buttachon S., Ramos A. A., Inácio Â., Dethoup T., Gales L., Lee M., et al. (2018). Bis-Indolyl Benzenoids, Hydroxypyrrolidine derivatives and other constituents from cultures of the marine sponge-associated fungus Aspergillus candidus Kufa0062. Mar. Drugs 16, 119. doi: 10.3390/md16040119
Calcabrini C., Catanzaro E., Bishayee A., Turrini E., Fimognari C. (2017). Marine sponge natural products with anticancer potential: a updated review. Mar. Drugs 15, 310. doi: 10.3390/md15100310
Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2019). Marine natural products. Natural Product Rep. 36, 122–173. doi: 10.1039/C8NP00092A
Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2020). Marine natural products. Natural Product Rep. 37, 175–223. doi: 10.1039/C9NP00069K
Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2021). Marine natural products. Natural Product Rep. 38, 362–413. doi: 10.1039/D0NP00089B
Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2022). Marine natural products. Natural Product Rep. 39, 1122–1171. doi: 10.1039/D1NP00076D
Carroll A. R., Copp B. R., Davis R. A., Keyzers R. A., Prinsep M. R. (2023). Marine natural products. Natural Product Rep. 40, 275–325. doi: 10.1039/D2NP00083K
Cita Y. P., Suhermanto A., Radjasa O. K., Sudharmono P. (2017). Antibacterial activity of marine bacteria isolated from sponge Xestospongia testudinaria from Sorong, Papua. Asian Pacific J. Trop. Biomedicine 7, 450–454. doi: 10.1016/j.apjtb.2017.01.024
Cragg G. M., Pezzuto J. M. (2016). Natural products as a vital source for the discovery of cancer chemotherapeutic and chemopreventive agents. Med. Principles Pract. 25, 41–59. doi: 10.1159/000443404
Curdt F., Schupp P. J., Rohde S. (2022). Light availability affects the symbiosis of sponge specific cyanobacteria and the common blue aquarium sponge (Lendenfeldia chondrodes). Anim. (Basel) 12, 1283. doi: 10.3390/ani12101283
Davis R. A., Buchanan M. S., Duffy S., Avery V. M., Charman S. A., Charman W. N., et al. (2012). Antimalarial activity of Pyrroloiminoquinones from the Australian marine sponge Zyzzya sp. J. Medicinal Chem. 55, 5851–5858. doi: 10.1021/jm3002795
Debbab A., Aly A. H., Lin W. H., Proksch P. (2010). Bioactive compounds from marine bacteria and fungi. Microbial Biotechnol. 3, 544–563. doi: 10.1111/j.1751-7915.2010.00179.x
Debitus C., Guella G., Mancini I., Waikedre J., Guemas J.-P., Nicolas J. L., et al. (1998). Quinolones from a bacterium and Tyrosine metabolites from its host sponge, Suberea creba from the Coral Sea. J. Mar. Biotechnol. 6, 136–141.
Devi P., Wahidullah S., Rodrigues C., Souza L. D. (2010). The sponge-associated bacterium Bacillus licheniformis Sab1: a source of antimicrobial compounds. Mar. Drugs 8, 1203–1212. doi: 10.3390/md8041203
Edrada R. A., Proksch P., Wray V., Witte L., Müller W., Van Soest R. W. (1996). Four new bioactive manzamine-type alkaloids from the Philippine marine sponge Xestospongia ashmorica. J. Natural Products 59, 1056–1060. doi: 10.1021/np9604083
El-Samak M., Zakeer S., Hanora A., Solyman S. (2023). Metagenomic and metatranscriptomic exploration of the Egyptian Red Sea sponge Theonella sp. associated microbial community. Mar. Genomics 70, 101032. doi: 10.1016/j.margen.2023.101032
Erwin P. M., Thacker R. W. (2008). Phototrophic nutrition and symbiont diversity of two Caribbean sponge–cyanobacteria symbioses. Mar. Ecol. Prog. Ser. 362, 139–147. doi: 10.3354/meps07464
Flórez L. V., Biedermann P. H., Engl T., Kaltenpoth M. (2015). Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Natural Product Rep. 32, 904–936. doi: 10.1039/C5NP00010F
Folkers M., Rombouts T. (2020). “Sponges revealed: A synthesis of their overlooked ecological functions within aquatic ecosystems,” in Youmares 9 - the Oceans: Our Research, Our Future: Proceedings of the 2018 Conference for Young Marine Researcher in Oldenburg, Germany. Eds. Jungblut S., Liebich V., Bode-Dalby M. (Springer International Publishing, Cham), 181–193.
Fromont J., Abdul Wahab M. A., Gomez O., Ekins M., Grol M., Hooper J. N. A. (2016). Patterns of sponge biodiversity in the Pilbara, Northwestern Australia. Diversity 8, 21. doi: 10.3390/d8040021
Gao Z.-M., Zhou G.-W., Huang H., Wang Y. (2017). The Cyanobacteria-dominated sponge Dactylospongia elegans in the South China Sea: prokaryotic community and metagenomic insights. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01387
Gomes N. G. M., Dasari R., Chandra S., Kiss R., Kornienko A. (2016). Marine invertebrate metabolites with anticancer activities: solutions to the “supply problem. Mar. Drugs 14, 98. doi: 10.3390/md14050098
Goudie L., Norman M. D., Finn J. (2013). Sponges: A Museum Victoria Field Guide: Museums Victoria. Australia: Museum Victoria.
Gunasekera S. P., Gunasekera M., Longley R. E., Schulte G. K. (1990). Discodermolide: A new bioactive polyhydroxylated lactone from the marine sponge Discodermia dissoluta. J. Organic Chem. 55, 4912–4915. doi: 10.1021/jo00303a029
Han B.-N., Hong L.-L., Gu B.-B., Sun Y.-T., Wang J., Liu J.-T., et al. (2019). “Natural products from sponges,” in Symbiotic Microbiomes of Coral Reefs Sponges and Corals. Ed. Li Z. (Springer Netherlands, Dordrecht), 329–463.
Harvey A. L., Edrada-Ebel R., Quinn R. J. (2015). The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discovery 14, 111–129. doi: 10.1038/nrd4510
Helber S. B., Hoeijmakers D. J. J., Muhando C. A., Rohde S., Schupp P. J. (2018). Sponge chemical defenses are a possible mechanism for increasing sponge abundance on reefs in Zanzibar. PLoS One 13, e0197617. doi: 10.1371/journal.pone.0197617
Hentschel U., Piel J., Degnan S. M., Taylor M. W. (2012). Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641–654. doi: 10.1038/nrmicro2839
Hill R. T., Hamann M. T., Peraud O., Kasanah N. (2004). Manzamine-producing actinomycetes. Google Patents.
Hong L.-L., Ding Y.-F., Zhang W., Lin H.-W. (2022). Chemical and biological diversity of new natural products from marine sponges: a review (2009–2018). Mar. Life Sci. Technol. 4, 356–372. doi: 10.1007/s42995-022-00132-3
Huang G., Ping X., Xu W., Hu Y., Chang J., Swaisgood R. R., et al. (2021). Wildlife conservation and management in China: achievements, challenges and perspectives. Natl. Sci. Rev. 8, 1–5. doi: 10.1093/nsr/nwab042
Imhoff J. F., Stöhr R. (2003). Sponge-associated bacteria: general overview and special aspects of bacteria associated with Halichondria panicea. Sponges (Porifera) 37, 35–57. doi: 10.1007/978-3-642-55519-0_2
Jin L., Quan C., Hou X., Fan S. (2016). Potential pharmacological resources: natural bioactive compounds from marine-derived fungi. Mar. Drugs 14, 76. doi: 10.3390/md14040076
Karimi E., Keller-Costa T., Slaby B. M., Cox C. J., da Rocha U. N., Hentschel U., et al. (2019). Genomic blueprints of sponge-prokaryote symbiosis are shared by low abundant and cultivatable Alphaproteobacteria. Sci. Rep. 9, 1999. doi: 10.1038/s41598-019-38737-x
Kim S. K., Dewapriya P. (2012). “Chapter 8 - bioactive compounds from marine sponges and their symbiotic microbes: A potential source of nutraceuticals,” in Advances in Food and Nutrition Research, vol. 65 . Ed. Kim S.-K. (Academic Press), 137–151.
Kim C.-K., Riswanto R., Won T. H., Kim H., Elya B., Sim C. J., et al. (2017). Manzamine alkaloids from an Acanthostrongylophora sp. sponge. J. Natural Products 80, 1575–1583. doi: 10.1021/acs.jnatprod.7b00121
Kiran G. S., Sekar S., Ramasamy P., Thinesh T., Hassan S., Lipton A. N., et al. (2018). Marine sponge microbial association: towards disclosing unique symbiotic interactions. Mar. Environ. Res. 140, 169–179. doi: 10.1016/j.marenvres.2018.04.017
Kochanowska-Karamyan A. J., Araujo H. C., Zhang X., El-Alfy A., Carvalho P., Avery M. A., et al. (2020). Isolation and synthesis of Veranamine, an antidepressant lead from the marine sponge Verongula Rigida. J. Natural Products 83, 1092–1098. doi: 10.1021/acs.jnatprod.9b01107
Koutsouveli V., Balgoma D., Checa A., Hedeland M., Riesgo A., Cárdenas P. (2022). Oogenesis and Lipid metabolism in the deep-sea sponge Phakellia ventilabrum (Linnaeus, 1767). Sci. Rep. 12, 6317. doi: 10.1038/s41598-022-10058-6
Krumpe L. R. H., Wilson B. A. P., Marchand C., Sunassee S. N., Bermingham A., Wang W., et al. (2020). Recifin a, initial example of the tyr-lock peptide structural family, is a selective allostericinhibitor of Tyrosyl-DNA phosphodiesterase I. J. Am. Chem. Soc. 142, 21178–21188. doi: 10.1021/jacs.0c10418
Lee Y.-J., Cho Y., Tran H. N. K. (2021). Secondary metabolites from the marine sponges of the genus Petrosia: a literature review of 43 years of research. Mar. Drugs 19, 122. doi: 10.3390/md19030122
Lee Y.-K., Lee J.-H., Lee H.-K. (2001). Microbial symbiosis in marine sponges. J. Microbiol. 39, 254–264.
Li P., Lu H., Zhang Y., Zhang X., Liu L., Wang M., et al. (2023). The natural products discovered in marine sponge-associated microorganisms: structures, activities, and mining strategy. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1191858
Lindel T., Hoffmann H., Hochgürtel M., Pawlik J. R. (2000). Structure–activity relationship of inhibition of fish feeding by sponge-derived and synthetic pyrrole–imidazole alkaloids. J. Chem. Ecol. 26, 1477–1496. doi: 10.1023/A:1005591826613
Machida K., Abe T., Arai D., Okamoto M., Shimizu I., de Voogd N. J., et al. (2014). Cinanthrenol a, an estrogenic steroid containing phenanthrene nucleus, from a marine sponge Cinachyrella sp. Organic Lett. 16, 1539–1541. doi: 10.1021/ol5000023
Maldonado M., Ribes M., van Duyl F. C. (2012). “Chapter three - nutrient fluxes through sponges: biology, budgets, and ecological implications,” in Advances in Marine Biology, vol. 62 . Eds. Becerro M. A., Uriz M. J., Maldonado M., Turon X. (Academic Press), 113–182.
Martignago C. C. S., Soares-Silva B., Parisi J. R., Silva L., Granito R. N., Ribeiro A. M., et al. (2023). Terpenes extracted from marine sponges with antioxidant activity: A Systematic Review. Natural Products Bioprospecting 13, 23. doi: 10.1007/s13659-023-00387-y
Maslin M., Gaertner-Mazouni N., Debitus C., Joy N., Ho R. (2021). Marine sponge aquaculture towards drug development: an ongoing history of technical, ecological, chemical considerations and challenges. Aquaculture Rep. 21, 100813. doi: 10.1016/j.aqrep.2021.100813
Mehbub M. (2015). Development and validation of a novel approach for delineating the role of sponge-bacteria interactions: Aplysilla rosea as a model sponge. PhD thesis (Adelaide (SA: Flinders University).
Mehbub M. F., Franco C. M., Zhang W. (2015). “Secondary metabolites from microorganisms isolated from marine sponges from 2000 to 2012,” in Antimicrobials: Synthetic and Natural Compounds (Taylor & Francis Group), 279–316.
Mehbub M. F., Lei J., Franco C., Zhang W. (2014). Marine sponge derived natural products between 2001 and 2010: trends and opportunities for discovery of bioactives. Mar. Drugs 12, 4539–4577. doi: 10.3390/md12084539
Mehbub M. F., Tanner J. E., Barnett S. J., Bekker J., Franco C. M. M., Zhang W. (2018). A controlled aquarium system and approach to study the role of sponge-bacteria interactions using Aplysilla rosea and Vibrio natriegens. Sci. Rep. 8, 11801. doi: 10.1038/s41598-018-30295-y
Mehbub M. F., Tanner J. E., Barnett S. J., Franco C. M. M., Zhang W. (2016). The role of sponge-bacteria interactions: the sponge Aplysilla rosea challenged by its associated bacterium Streptomyces Act-52A in a controlled aquarium system. Appl. Microbiol. Biotechnol. 100, 10609–10626. doi: 10.1007/s00253-016-7878-9
Minale L. (2012). Terpenoids from marine sponges. Marine natural products. Chemical and biological perspectives 1. Paul J. Scheuer 1, 175. doi: 10.1007/978-90-481-3834-0_4
Mioso R., Marante F. J. T., Bezerra R. D. S., Borges F. V. P., Santos B., Laguna I. (2017). Cytotoxic compounds derived from marine sponges. a review (2010–2012). Molecules 22, 208. doi: 10.3390/molecules22020208
Mohanty I., Moore S. G., Yi D., Biggs J. S., Gaul D. A., Garg N., et al. (2020). Precursor-guided mining of marine sponge metabolomes lends insight into biosynthesis of pyrrole–imidazole alkaloids. ACS Chem. Biol. 15, 2185–2194. doi: 10.1021/acschembio.0c00375
Mohapatra B., Bapuji M. (1998). Characterization of acetylcholinesterase from Arthrobacter ilicis associated with the marine sponge (Spirastrella sp.). J. Appl. Microbiol. 84, 393–398. doi: 10.1046/j.1365-2672.1998.00360.x
Mohapatra B., Bapuji M., Sree A. (2003). Production of industrial enzymes (Amylase, Carboxymethylcellulase and Protease) by bacteria isolated from marine sedentary organisms. Acta Biotechnologica 23, 75–84. doi: 10.1002/abio.200390011
Morrow C., Cárdenas P. (2015). Proposal for a revised classification of the Demospongiae (Porifera). Front. Zoology 12, 7. doi: 10.1186/s12983-015-0099-8
Motti C. A., Bourguet-Kondracki M.-L., Longeon A., Doyle J. R., Llewellyn L. E., Tapiolas D. M., et al. (2007). Comparison of the biological properties of several marine sponge-derived sesquiterpenoid quinones. Molecules 12, 1376–1388. doi: 10.3390/12071376
Muñoz-Miranda L. A., Iñiguez-Moreno M. (2023). An extensive review of marine pigments: sources, biotechnological applications, and sustainability. Aquat. Sci. 85, 68. doi: 10.1007/s00027-023-00966-8
Nagai K., Kamigiri K., Arao N., Suzumura K., Kawano Y., Yamaoka M., et al. (2003). Ym-266183 and Ym-266184, Novel Thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge. I. taxonomy, fermentation, isolation, physico-chemical properties and biological properties. J. Antibiotics 56, 123–128. doi: 10.7164/antibiotics.56.123
Nasser Alahmari A., Hassoubah S A., Ali Alaidaroos B. (2022). Sponges-associated marine bacteria as sources of antimicrobial compounds. Novel Res. Microbiol. J. 6, 1742–1767. doi: 10.21608/nrmj.2022.267424
Naughton L. M., Romano S., O’Gara F., Dobson A. D. W. (2017). Identification of secondary metabolite gene clusters in the Pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01494
Nuzzo G., Ciavatta M. L., Villani G., Manzo E., Zanfardino A., Varcamonti M., et al. (2012). Fulvynes, Antimicrobial polyoxygenated acetylenes from the Mediterranean sponge Haliclona fulva. Tetrahedron 68, 754–760. doi: 10.1016/j.tet.2011.10.068
Pabel C. T., Vater J., Wilde C., Franke P., Hofemeister J., Adler B., et al. (2003). Antimicrobial activities and matrix-assisted laser desorption/ionization mass spectrometry of Bacillus isolates from the marine sponge Aplysina aerophoba. Mar. Biotechnol. 5, 424–434. doi: 10.1007/s10126-002-0088-8
Palomo S., González I., de la Cruz M., Martín J., Tormo J. R., Anderson M., et al. (2013). Sponge-derived Kocuria and Micrococcus spp. as sources of the new Thiazolyl peptide antibiotic Kocurin. Mar. Drugs 11, 1071–1086. doi: 10.3390/md11041071
Parte S., Sirisha V. L., D’Souza J. S. (2017). “Chapter four - biotechnological applications of marine enzymes from algae, bacteria, fungi, and sponges,” in Advances in Food and Nutrition Research, vol. 80 . Eds. Kim S.-K., Toldrá F. (Academic Press), 75–106.
Pawlik J. R., Chanas B., Toonen R. J., Fenical W. (1995). Defenses of Caribbean sponges against predatory reef fish. I. chemical deterrency. Mar. Ecol. Prog. Ser. 127, 183–194. doi: 10.3354/meps127183
Pham C.-D., Hartmann R., Böhler P., Stork B., Wesselborg S., Lin W., et al. (2014). Callyspongiolide, a cytotoxic macrolide from the marine sponge Callyspongia sp. Organic Lett. 16, 266–269. doi: 10.1021/ol403241v
Pineda M.-C., Strehlow B., Duckworth A., Doyle J., Jones R., Webster N. S. (2016). Effects of light attenuation on the sponge holobiont- implications for dredging management. Sci. Rep. 6, 39038. doi: 10.1038/srep39038
Pita L., Rix L., Slaby B. M., Franke A., Hentschel U. (2018). The sponge holobiont in a Changing ocean: from microbes to ecosystems. Microbiome 6, 46. doi: 10.1186/s40168-018-0428-1
Rastegari A. A., Yadav A. N., Yadav N. (2020). New and future developments in microbial biotechnology and bioengineering: Trends of Microbial Biotechnology for Sustainable Agriculture and Biomedicine Systems: Diversity and Functional Perspectives (Elsevier).
Reveillaud J., Maignien L., Eren A. M., Huber J. A., Apprill A., Sogin M. L., et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8, 1198–1209. doi: 10.1038/ismej.2013.227
Rotter A., Barbier M., Bertoni F., Bones A. M., Cancela M. L., Carlsson J., et al. (2021). The essentials of marine biotechnology. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.629629
Ruiz-Ruiz F., Mancera-Andrade E I., MN Iqbal H. (2017). Marine-derived bioactive peptides for biomedical sectors: a review. Protein Pept. Lett. 24, 109–117. doi: 10.2174/0929866523666160802155347
S P. D. (2003). Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14, 303–310. doi: 10.1016/S0958-1669(03)00067-3
Sakai R., Higa T., Jefford C. W., Bernardinelli G. (1986). Manzamine a, a Novel antitumor alkaloid from a sponge. J. Am. Chem. Soc. 108, 6404–6405. doi: 10.1021/ja00280a055
Samirana P. O., Jenie R. I., Murti Y. B., Setyowati E. P. (2022). Application of metabolomics on marine sponges and sponge-associated microorganisms: a review. J. Appl. Pharm. Sci. 12, 018–033. doi: 10.7324/JAPS.2022.120702
Sathiyanarayanan G., Saibaba G., Kiran G. S., Yang Y.-H., Selvin J. (2017). Marine sponge-associated bacteria as a potential source for polyhydroxyalkanoates. Crit. Rev. Microbiol. 43, 294–312. doi: 10.1080/1040841X.2016.1206060
Schiefenhövel K., Kunzmann A. (2012). Sponge farming trials: survival, attachment, and growth of two Indo-Pacific sponges, Neopetrosia sp. and Stylissa massa. J. Mar. Biol. 2012, 417360. doi: 10.1155/2012/417360
Schneemann I., Kajahn I., Ohlendorf B., Zinecker H., Erhard A., Nagel K., et al. (2010a). Mayamycin, a cytotoxic polyketide from a Streptomyces strain isolated from the marine sponge Halichondria panicea. J. Natural Products 73, 1309–1312. doi: 10.1021/np100135b
Schneemann I., Ohlendorf B., Zinecker H., Nagel K., Wiese J., Imhoff J. F. (2010b). Nocapyrones a– D, Γ-Pyrones from a Nocardiopsis strain isolated from the marine sponge Halichondria panicea. J. Natural Products 73, 1444–1447. doi: 10.1021/np100312f
Selvin J., Ninawe A., Seghal Kiran G., Lipton A. (2010). Sponge-microbial interactions: ecological implications and bioprospecting avenues. Crit. Rev. Microbiol. 36, 82–90. doi: 10.3109/10408410903397340
Selvin J., Shanmughapriya S., Gandhimathi R., Seghal Kiran G., Rajeetha Ravji T., Natarajaseenivasan K., et al. (2009). Optimization and production of novel antimicrobial agents from sponge associated marine Actinomycetes Nocardiopsis dassonvillei Mad08. Appl. Microbiol. Biotechnol. 83, 435–445. doi: 10.1007/s00253-009-1878-y
Sipkema D., Franssen M. C. R., Osinga R., Tramper J., Wijffels R. H. (2005). Marine sponges as pharmacy. Mar. Biotechnol. 7, 142–162. doi: 10.1007/s10126-004-0405-5
Skropeta D., Wei L. (2014). Recent advances in deep-sea natural products. Natural product Rep. 31, 999–1025. doi: 10.1039/C3NP70118B
Sobarzo-Sánchez E. (2015). Alkaloids: Biosynthesis, Biological Roles and Health Benefits (Nova Science Publishers, Incorporated).
Steffen K., Indraningrat A. A. G., Erngren I., Haglöf J., Becking L. E., Smidt H., et al. (2022). Oceanographic setting influences the prokaryotic community and metabolome in deep-sea sponges. Sci. Rep. 12, 3356. doi: 10.1038/s41598-022-07292-3
Tähtinen P., Guella G., Saielli G., Debitus C., Hnawia E., Mancini I. (2018). New sulfur-containing polyarsenicals from the New Caledonian sponge Echinochalina Bargibanti. Mar. Drugs 16, 1–14. doi: 10.3390/md16100382
Tan L. T. (2023). Impact of marine chemical ecology research on the discovery and development of new pharmaceuticals. Mar. Drugs 21, 174. doi: 10.3390/md21030174
Tang W.-Z., Yang Z.-Z., Wu W., Tang J., Sun F., Wang S.-P., et al. (2018). Imidazole alkaloids and their Zinc complexes from the calcareous marine sponge Leucetta Chagosensis. J. Natural Products 81, 894–900. doi: 10.1021/acs.jnatprod.7b01006
Taylor M. W., Radax R., Steger D., Wagner M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/mmbr.00040-06
Taylor M. W., Tsai P., Simister R. L., Deines P., Botte E., Ericson G., et al. (2012). ‘Sponge-specific’ bacteria are widespread (but rare) in diverse marine environments. ISME J. 7, 438–443. doi: 10.1038/ismej.2012.111
Teta R., Irollo E., Della Sala G., Pirozzi G., Mangoni A., Costantino V. (2013). Smenamides a and B, Chlorinated peptide/polyketide hybrids containing a Dolapyrrolidinone unit from the Caribbean sponge Smenospongia aurea. evaluation of their role as leads in antitumor drug research. Mar. Drugs 11, 4451–4463. doi: 10.3390/md11114451
Thakur A. N., Thakur N. L., Indap M. M., Pandit R. A., Datar V. V., Müller W. E. (2005). Antiangiogenic, antimicrobial, and cytotoxic potential of sponge-associated bacteria. Mar. Biotechnol. 7, 245–252. doi: 10.1007/s10126-004-4085-y
Thomas T. R., Kavlekar D. P., LokaBharathi P. A. (2010). Marine drugs from sponge-microbe association–a review. Mar. Drugs 8, 1417–1468. doi: 10.3390/md8041417
Tincu J. A., Taylor S. W. (2004). Antimicrobial peptides from marine invertebrates. Antimicrobial Agents Chemotherapy 48, 3645–3654. doi: 10.1128/aac.48.10.3645-3654.2004
Tribalat M.-A., Marra M., McCormack G., Thomas O. (2016). Does the chemical diversity of the order Haplosclerida (phylum Porifera: class Demospongia) fit with current taxonomic classification? Planta Med. 82, 843–856. doi: 10.1055/s-0042-105879
Turon X., Martí R., Uriz M. J. (2009). Chemical bioactivity of sponges along an environmental gradient in a Mediterranean cave. Scientia Marina 73, 387–397. doi: 10.3989/scimar.2009.73n2387
Varijakzhan D., Loh J.-Y., Yap W.-S., Yusoff K., Seboussi R., Lim S.-H. E., et al. (2021). Bioactive compounds from marine sponges: fundamentals and applications. Mar. Drugs 19, 246. doi: 10.3390/md19050246
Versluis D., Nijsse B., Naim M. A., Koehorst J. J., Wiese J., Imhoff J. F., et al. (2018). Comparative genomics highlights symbiotic capacities and high metabolic flexibility of the marine genus Pseudovibrio. Genome Biol. Evol. 10, 125–142. doi: 10.1093/gbe/evx271
Vitali A. (2018). Antimicrobial peptides derived from marine sponges. Am. J. Clin. Microbiol. Antimicrobials 1, 1006.
Wang X., Duggan B. M., Molinski T. F. (2015). Mollenynes B–E from the Marine sponge Spirastrella mollis. Band-selective heteronuclear single quantum coherence for discrimination of bromo–chloro regioisomerism in natural products. J. Am. Chem. Soc. 137, 12343–12351. doi: 10.1021/jacs.5b07858
Wang D., Feng Y., Murtaza M., Wood S., Mellick G., Hooper J. N. A., et al. (2016). A grand challenge: unbiased phenotypic function of metabolites from Jaspis splendens against Parkinson’s disease. J. Natural Products 79, 353–361. doi: 10.1021/acs.jnatprod.5b00987
Wang M., Tietjen I., Chen M., Williams D. E., Daoust J., Brockman M. A., et al. (2016). Sesterterpenoids isolated from the sponge Phorbas sp. activate latent Hiv-1 provirus expression. J. Organic Chem. 81, 11324–11334. doi: 10.1021/acs.joc.6b02312
Wulff J. L. (2006). Ecological interactions of marine sponges. Can. J. Zoology 84, 146–166. doi: 10.1139/z06-019
Wulff J. (2012). Ecological interactions and the distribution, abundance, and diversity of sponges. Adv. Mar. Biol. 61, 273–344. doi: 10.1016/B978-0-12-387787-1.00003-9
Zhang J., Liu F., Wang Q., Gong Q., Gao X. (2023). Effect of light wavelength on biomass, growth, photosynthesis and pigment content of Emiliania huxleyi (Isochrysidales, Cocco-Lithophyceae). J. Mar. Sci. Eng. 11, 456. doi: 10.3390/jmse11020456
Zhang S., Song W., Nothias L.-F., Couvillion S. P., Webster N., Thomas T. (2022). Comparative metabolomic analysis reveals shared and unique chemical interactions in sponge holobionts. Microbiome 10, 22. doi: 10.1186/s40168-021-01220-9
Zhang H., Zhao Z., Wang H. (2017). Cytotoxic natural products from marine sponge-derived microorganisms. Mar. Drugs 15, 68. doi: 10.3390/md15030068
Keywords: Porifera, sponge, sponge-associated microbes, marine natural products (MNPs), secondary metabolites, bioactivity
Citation: Mehbub MF, Yang Q, Cheng Y, Franco CMM and Zhang W (2024) Marine sponge-derived natural products: trends and opportunities for the decade of 2011-2020. Front. Mar. Sci. 11:1462825. doi: 10.3389/fmars.2024.1462825
Received: 10 July 2024; Accepted: 06 August 2024;
Published: 26 August 2024.
Edited by:
Bin Wu, Zhejiang University, ChinaReviewed by:
Yueying Li, University of Oklahoma, United StatesCopyright © 2024 Mehbub, Yang, Cheng, Franco and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Ferdous Mehbub, bW9oYW1tYWQubWVoYnViQHJtaXQuZWR1LmF1; Christopher Milton Mathew Franco, Y2hyaXMuZnJhbmNvQGZsaW5kZXJzLmVkdS5hdQ==; Wei Zhang, d2VpLnpoYW5nQGZsaW5kZXJzLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.