
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 23 September 2024
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1450142
 Lei Xu1,2,3
Lei Xu1,2,3 Jinxia Zhang1,4
Jinxia Zhang1,4 Xuehui Wang1,3,5
Xuehui Wang1,3,5 Qinghua Chen6
Qinghua Chen6 Quehui Tang1,2
Quehui Tang1,2 Lianggen Wang1,2
Lianggen Wang1,2 Jiajia Ning1,2
Jiajia Ning1,2 Yafang Li1,2,3
Yafang Li1,2,3 Shuangshuang Liu1,2
Shuangshuang Liu1,2 Delian Huang1,2
Delian Huang1,2 Feiyan Du1,2*
Feiyan Du1,2*Grasping the genetic structure of marine fish populations is vital for comprehending species connectivity patterns and determining the appropriate spatiotemporal scales for conservation management strategies. Here, we analyzed the population genetics of the Orangefin Ponyfish (Photopectoralis bindus Valenciennes, 1835) by examining a portion of the gene coding for the mitochondrial cytochrome c oxidase subunit I. The aim was to evaluate the haplotype pattern, genetic structure, demographic history, as well as the influence of ecological connectivity through the Qiongzhou Strait on the distribution patterns of this species in the northern South China Sea and the Beibu Gulf. In total, 257 specimens yielded only 13 haplotypes, with the predominant haplotype present at all sampling locations. The analysis revealed a “star-like” haplotype pattern, indicating low levels of both haplotype and nucleotide diversity. Additionally, a small but significant genetic structure was observed between the coastal regions flanking the Leizhou Peninsula. These patterns in the haplotype network and genetic structure may be significantly influenced by contemporary currents, particularly through the connectivity of the Qiongzhou Strait. Tajima’s D and Fu’s Fs demonstrated pronouncedly negative values, along with a unimodal mismatch distribution, suggested a recent demographic expansion of Photopectoralis bindus during the late Pleistocene, likely influenced by fluctuations in sea levels.
Genetic diversity, both within and among populations, plays a significant role in a species’ ability to adapt to environmental changes, influencing their long-term capacity to withstand ecological disruptions (Camurugi et al., 2021). High level of population genetic divergence can arise from various mechanisms, including geographic distance, reproductive isolation, biogeographic barriers (such as hydrological processes or ecological bridges), and specific behavioral traits (Cowman and Bellwood, 2013). Conversely, a lack of genetic structure has long been assumed to result from unrestricted gene flow between populations, particularly in species with higher dispersal capacity (Waples, 1998). However, quantifying dispersal is notoriously challenging. The traits that define dispersal, such as the frequency of organism movement and the distances they cover, are anticipated to affect the dispersal of species, their population genetic structure across landscapes, as well as how they respond to selective pressures in varying spatial gradients (Manel et al., 2003; Richardson et al., 2014). Marine organisms are often thought to lack distinct geographic differentiation compared to terrestrial organisms, mainly because physical barriers are often perceived to be absent in marine ecosystems that impede gene flow between populations in “open” oceanic areas. Furthermore, several biological traits weaken population differentiation in marine organisms. For instance, their large population sizes and strong reproductive capacities reduce genetic drift in small populations. Additionally, wide population distributions and the robust dispersal capabilities of adults or larvae facilitate gene flow between populations (Palumbi, 1992; Kenchington et al., 2003; Luttikhuizen et al., 2003; Selkoe et al., 2008). Furthermore, marine organisms typically have relatively recent population histories following postglacial recolonizations, which has allowed little time for the establishment of migration-drift equilibria (Bradbury et al., 2008). However, research conducted over the past two decades has revealed that despite the absence of distinct physical barriers, marine ecosystems exhibit high connectivity. Marine organisms also demonstrate strong abilities for both active migration and passive dispersal. Despite these characteristics, significant genetic differentiation between populations persists. Overall, the genetic structure of marine populations reflects a combination of moderate structuring and strong gene flow (Ward et al., 1994; Baus et al., 2005; Ellis et al., 2017; Diopere et al., 2018). Marine fish, known for their high mobility in both juvenile and adult stages, are considered to have greater dispersal potential compared to other marine organisms (Hauser and Carvalho, 2008; Hemmer-Hansen et al., 2019). Nevertheless, the extensive dispersal capabilities of marine fish do not consistently lead to substantial levels of effective gene flow. Despite this, pronounced genetic differentiation in marine fish has been observed across various geographical scales, ranging from large oceanic differences to more regional patterns, and even to small geographical scales showing genetic differentiation over distances of just a few tens of kilometers (Beheregaray and Sunnucks, 2001; Rolland et al., 2007; Gaither et al., 2010). Numerous studies have documented strong genetic differentiation among marine fish populations, which could be attributed to a combination of genetic drift, migration, demographic history, and locally divergent selection (Beheregaray and Sunnucks, 2001; Planes et al., 2001; Diopere et al., 2018).
Located within the South China Sea (SCS), the Qiongzhou Strait acts as a divider between Hainan Island and the Leizhou Peninsula in southern China. This strait runs in an east-west direction, forming a crucial connection between the northern South China Sea (NSCS) and the Beibu Gulf. It spans approximately 80 kilometers in length from west to east and 20 to 40 kilometers in width from south to north, covering an area of roughly 2400 square kilometers. It serves as a tidal channel linking the SCS with the Beibu Gulf on the NSCS shelf. Around 2.5 million years ago, before the Quaternary period, the Leizhou Peninsula and Hainan Island were joined (Yao et al., 2009). According to the latest research, starting from the late Tertiary period (25 to 2.5 million years ago), due to crustal fractures and differential movements of tectonic plates, the land between the Leizhou Peninsula and Hainan Island fractured and subsided, forming a graben-like depression. During the post-glacial period, the sea level rose, and the depression was inundated by seawater. Through repeated coast erosion and marine transgression, the Qiongzhou Strait as we know it today was eventually formed (Zhao et al., 2007; Zhu, 2016).The east-west flow field environment in the Qiongzhou Strait provides an ecological corridor for the northward expansion and migration of fish populations from the waters east of Hainan Island to the Beibu Gulf. This has significant impacts on the dynamics, community structure, diversity, and genetic structure of fish populations in the Beibu Gulf and the NSCS.
Photopectoralis bindus (Valenciennes, 1835), commonly known as orangefin ponyfish, is a fascinating marine species with a wide distribution range and unique ecological characteristics. This species is a tropical and brackish water species and can be found in the Indian Ocean as well as the western Pacific Ocean, inhabiting demersal environments such as shallow coastal waters near coral reefs and sandy bottoms. The distribution range of P. bindus spans from the eastern coast of Africa to the western Pacific Ocean, encompassing areas like the Arabian Sea, the Persian Gulf, the Red Sea, and waters around Indonesia and northern Australia. This wide distribution range indicates the species’ ability to adapt to diverse marine environments. It is typically found at depths ranging from a few meters to around 100 meters, although it can occasionally be found at greater depths. It prefers areas with sandy or muddy bottoms near coral reefs, where it can find shelter and food. The species is benthic, meaning it spends most of its time near the ocean floor, feeding on small fish and crustaceans. As in other Southeast Asian countries like Malaysia or Thailand, these small-sized species can serve purposes beyond human consumption, such as for the production of manure, fish meal, and as a protein source (Rao et al., 2015; Klangnurak and True, 2022).
In China, P. bindus is primarily distributed along the southern coastline, from the East China Sea to the Beibu Gulf. Its mainly distribution extends from Hainan Island’s southern coastline to the coastal areas of southern China, including Guangdong, Fujian, Zhejiang, and Jiangsu provinces. In the Beibu Gulf, overfishing has been identified as the primary cause of significant declines in demersal fish stocks. This has led to a shift from high-value, larger-sized, and high trophic level species to low-value, smaller-sized, and low trophic level ones. As a result, the small-sized demersal species P. bindus has become one of the dominant species in the area (Geng et al., 2018; Su et al., 2022). However, previous studies on P. bindus have mainly focused on aspects such as reproductive biology, ecological distribution, and fish stock assessments, primarily in the coastal regions of India (Rao et al., 2015; Rawat et al., 2019; Klangnurak and True, 2022). Here, this work aimed to perform a population genetics analysis utilizing a segment of the COI gene, which codes for cytochrome c oxidase subunit I. This analysis was intended to evaluate the haplotype pattern, genetic structure, demographic history, as well as to estimate the expansion time of P. bindus in the NSCS and the Beibu Gulf. It also sought to enhance our understanding of the ecological connectivity of the Qiongzhou Strait and its influence on the distribution patterns of P. bindus in the NSCS and the Beibu Gulf. Additionally, it also aimed to provide a reliable basis for stock assessments and the formulation of protection policies.
The SCS stands as most extensive semi-enclosed sea within the tropical and subtropical regions of the northwestern Pacific Ocean, spanning an expanse of around 3.3 million square kilometers. The northern half of the SCS, often referred to as the northern South China Sea (NSCS), is primarily located in subtropical zone. It is primarily the first level of three-step ladder of the SCS continental shelf. Mainland China, Indo-china Peninsula, Luzon Island, and Taiwan Island form the boundaries of the NSCS. Westward, the Beibu Gulf extends, a naturally formed semi-enclosed gulf spanning approximately 128,000 square kilometers. The NSCS is linked to the East China Sea (ECS) and the western Pacific Ocean to its east via the Luzon Strait and the Taiwan Strait, respectively (Morton and Blackmore, 2001; Nan et al., 2011). The Qiongzhou Strait, a narrow waterway just 20 kilometers wide, located north of Hainan Island, separating it from mainland China.
Samples of fish were gathered from 16 locations along the coastline of the northern SCS, encompassing the distribution range of P. bindus (Figure 1). All the samples were obtained using bottom trawl nets aboard the commercial fishing vessel “Guibeiyu20168” during survey of fishery resources in the NSCS. These surveys took place from February to April and September to October 2021. Following trawling surveys, 20 samples were collected from each sampling site for morphological identification and subsequent DNA analysis. Either muscle tissue or dorsal fins were extracted from each specimen, followed by preservation in 100% ethanol at -20°C.
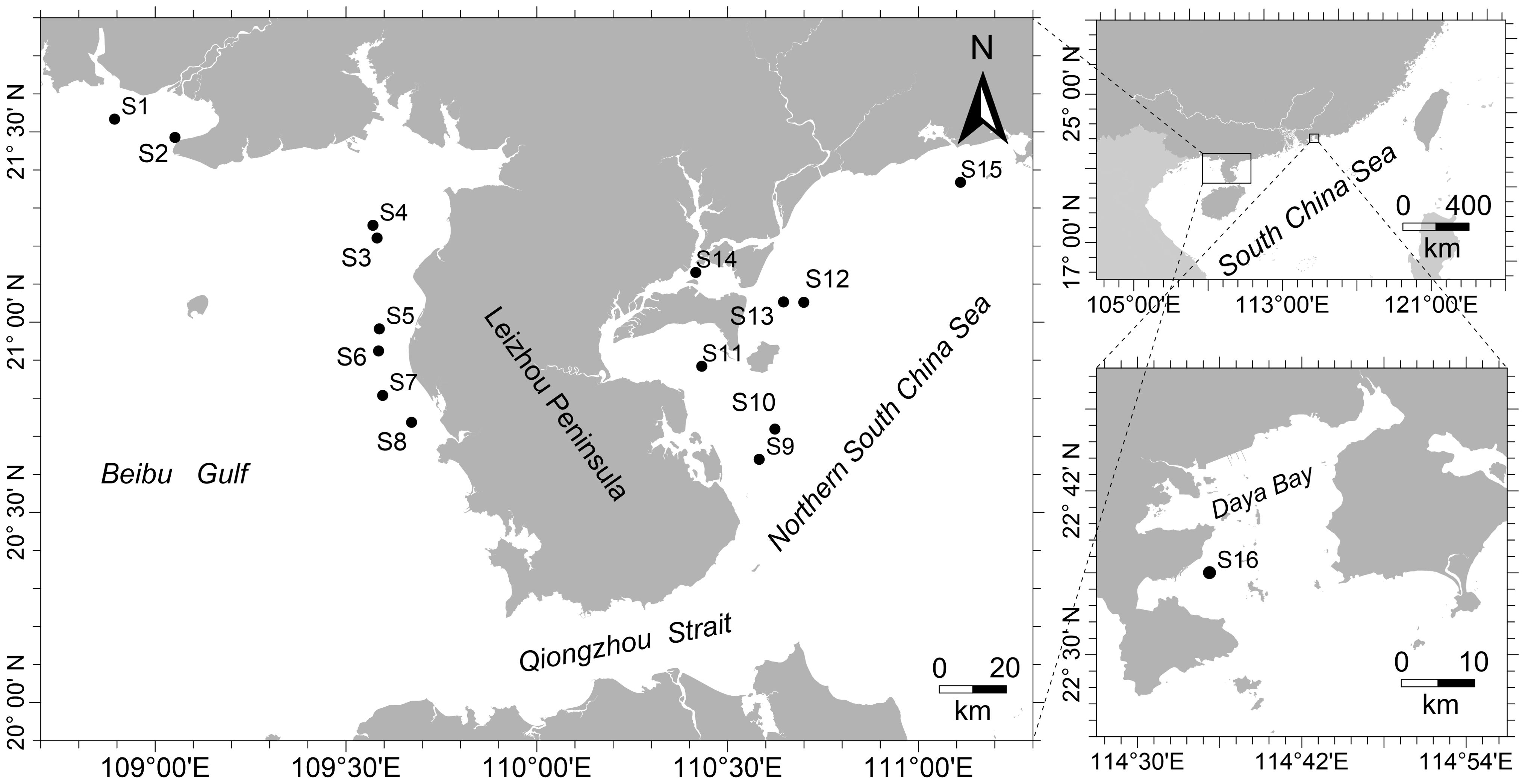
Figure 1. A map illustrating the study region with 16 sampling locations in the northern South China Sea and the Beibu Gulf.
Genomic DNA extraction was performed on dorsal fin tissue samples of the specimens stored in 100% ethanol, following the manufacturer’s instructions for the TIANamp Marine Animals DNA Kit (TIANGEN, China). We employed a UV-Vis Spectrophotometer NanoDrop ND-1000 from NanoDrop Technologies in the United States, to measure the concentration of DNA in each sample. We chose DNA samples that had A260/A280 ratios falling within the 1.8 to 2.0 range to serve as templates for subsequent PCR amplification. The COI gene of the mitochondrial genome was amplified employing the universal fish primer sets FishR1 and FishF1 (Ward et al., 2005).
The PCR reaction volume was 20 µL, the components included 2 µL of 10 ×PCR buffer, 0.2 µL of 25 µM of each primer, 0.8 µL of 10 µM dNTP, 2 µL DNA template, 0.2 µL Taq DNA polymerase (Takara Bio. Inc), and 14.6 µL ddH2O. The amplification protocol proceeded: an initial denaturation at 95°C for 3 minutes, followed by 35 cycles at 95°C for 45 seconds, 55°C for 60 seconds, and 72°C for 2 minutes, with a final extension at 72°C for 3 minutes. This was carried out using a 2720 Thermal Cycler (Applied Biosystems, USA). Subsequently, bidirectional sequencing of all selected PCR products was performed on an ABI 3730XL sequencer.
The sequences were initially visualized and assembled in Bioedit, and subsequently, an automated alignment was carried out using the default settings (Hall, 1999). To ensure consistency, the initial 20 and final 10 base pairs, which were absent in some sequences, were excluded. Unclear sequences were subsequently trimmed post-alignment. DnaSP 5.0 was employed to assess the genetic diversity indices of COI sequences, which included calculating the number of haplotypes (H) and polymorphic sites (S), haplotype (Hd) and nucleotide (π) diversity (Librado and Rozas, 2009).
For the graphical representation of connections among the mtDNA haplotypes in each P. bindus individual, we analyzed the network using Haploviewer (Salzburger et al., 2011). This tool converts phylogenetic trees created using traditional methods into haplotype genealogies. The maximum likelihood method was used in PhyML 3.0 to build the phylogenetic tree. The GTR model was employed as the substitution model with four gamma-distributed rate categories (Guindon et al., 2010). To test to what extent seascape genetic structure in this species was structured by Leizhou Peninsula and Qiongzhou Strait, we estimated molecular differentiation among P. bindus sampling sites using mtDNA (ΦST, using an infinite allele model) markers and conducted an Analysis of Molecular Variance (AMOVA) to explore the distribution of genetic variability, both analyses were carried out utilizing Arlequin version 3.5 (Excoffier and Lischer, 2010). The AMOVA quantified the genetic differentiation at three levels: regional level (eastern and western regions of the Leizhou Peninsula), among populations within each region, and within individual populations. In this analysis the sampling site S1 to S8 are considered to be western group, while sampling site S9 to S16 are eastern group.
Spatial variation in the genetic structure was evaluated by principal coordination analysis (PCoA) based on linearized pairwise ΦST (as ΦST/(1- ΦST)) (Slatkin, 1995). This method was used to assess the genetic structure difference in population between eastern and western waters of Leizhou Peninsula. PCoA was performed in R statistical platform (https://www.r-project.org) by employing the ‘cmdscale’ function from the ‘vegan’ package (Oksanen et al., 2019). To estimate migration rates between the eastern and western regions of the Leizhou Peninsula, we used Migrate 4.4.3 to conduct a maximum likelihood analysis with the coalescence approach. The calculated parameters were expressed as 𝒫ij = θiMij, where 𝒫ij represents the effective quantity of migrants from i population to j population, Mij is calculated as mij/μ (mij, representing the rate of immigration from i population to j population, and μ, the rate of mutation per generation), θ denotes the population size scaled by mutation rate (Beerli and Felsenstein, 1999).
Tajima’s D and Fu’s Fs statistics in Arlequin 3.5 were employed to assess population demographic changes, such as expansions or bottlenecks, in P. bindus (Tajima, 1989; Fu, 1997; Excoffier and Lischer, 2010). This analysis, with 10,000 permutations, aimed to determine if the data for P. bindus in the NSCS and the Beibu Gulf matched the expectations of the neutral theory model, indicating a potential population bottleneck or expansion. Negative Tajima’s D and Fu’s Fs values may suggest that the population experienced expansion for neutral markers. We proceeded to analyze the mismatch distribution of haplotypes following a sudden expansion model to assess its smoothness. We employed parametric bootstrapping with 1000 replicates in Arlequin 3.5 to assess how well the observed mismatch distribution fit the anticipated distribution assumed by the sudden expansion model. We used the degree of raggedness and the cumulative squared deviations for this assessment (Excoffier, 2004).
We also calculated the timing of population expansion for P. bindus in the NSCS and the Beibu Gulf. The estimation of the population expansion time involved analyzing the mismatch distribution through the τ (tau) statistic, which was subsequently transformed into actual time in years (t) utilizing the τ = 2ut equation (Rogers and Harpending, 1992). Here, u represents the sequence’s mutation rate, determined by u = 2µk, with µ being the rate of mutation for the mtDNA gene per generation, and k being the nucleotide count in the sequence. Mutation rates ranging from 1% to 3% per million years, as reported by Cantatore et al. (1994), were chose for our mitochondrial analysis. Additionally, the demographic history of P. bindus was assessed using Bayesian skyline plot (BSP) within BEAST 1.8.0 (Drummond and Rambaut, 2007).
After alignment, we obtained a total of 257 high-quality COI gene sequences from P. bindus, with a length of 624 bp. The average composition of bases was 24.36% A, 28.85% C, 16.67% G, and 30.13% T. These analyses revealed a limited genetic diversity in P. bindus, 257 sequences only yield 13 haplotypes. The mean nucleotide diversity was 0.00071 (range 0-0.00109), and haplotype diversity was 0.399 (range 0-0.542). The population’s genetic diversity in terms of nucleotide diversity and haplotype diversity in the western area of the Leizhou Peninsula are significantly higher than those of the population in the eastern area of the Leizhou Peninsula (Table 1).
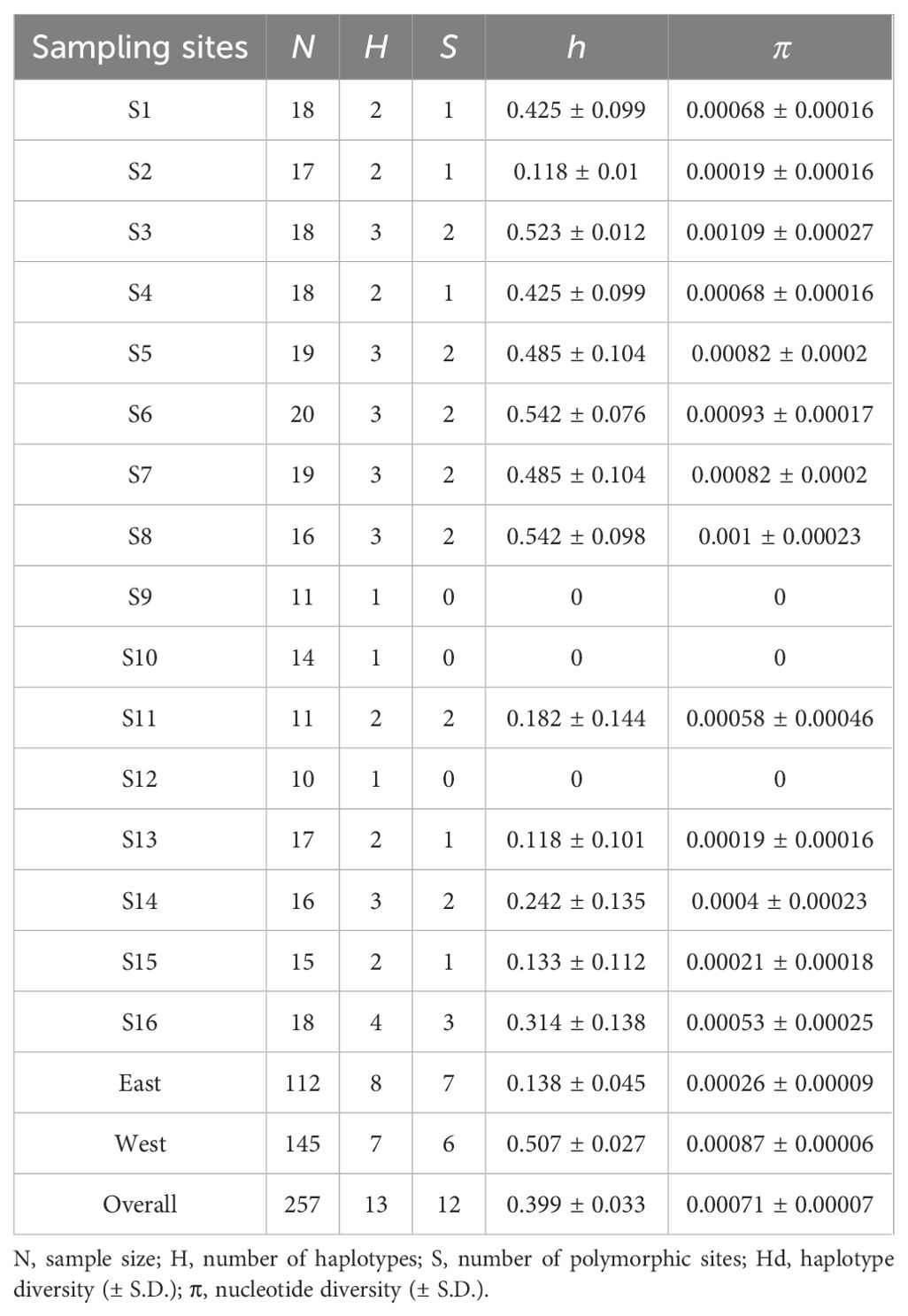
Table 1. Analyzing the genetic diversity of Photopectoralis bindus from the northern South China Sea and the Beibu Gulf through descriptive statistics using COI sequence data.
The haplotype network of P. bindus from the NSCS and the Beibu Gulf revealed a star-shaped pattern, with one central haplotype exhibiting high frequency (75.1%). This central haplotype was shared among all the sampling sites. The other haplotype, which also had a relatively high frequency (19.5%), was only distributed in the western waters of the Leizhou Peninsula. All other haplotypes were observed at low frequencies, surrounding the two high-frequency haplotypes, with only one mutation separating them from these high-frequency haplotypes (Figure 2). The 13 haplotype sequences from this study have been submitted to GenBank and can be accessed using the accession numbers PP789680-PP789692. Among the thirteen haplotypes, nine were limited to specific sampling sites, while four were found at multiple sites. Notably, two of the more common haplotypes were identified across numerous sites, suggesting a wider geographical spread. Haplotype H1, the prevailing genetic variant, was extensively present in all 16 sampling sites, establishing dominance within each population. This haplotype was found at sampling sites that were over 700 kilometers apart from each other. Haplotype H2 was more regionally distributed, primarily found in the western waters of the Leizhou Peninsula. The haplotypes H1 and H2 already account for 94.6% of the total. Site 9, site 10, and site 12 each harbor a single haplotype, while the other sites exhibit 2-3 haplotypes, with site 16 containing 4 haplotypes (Figure 3).
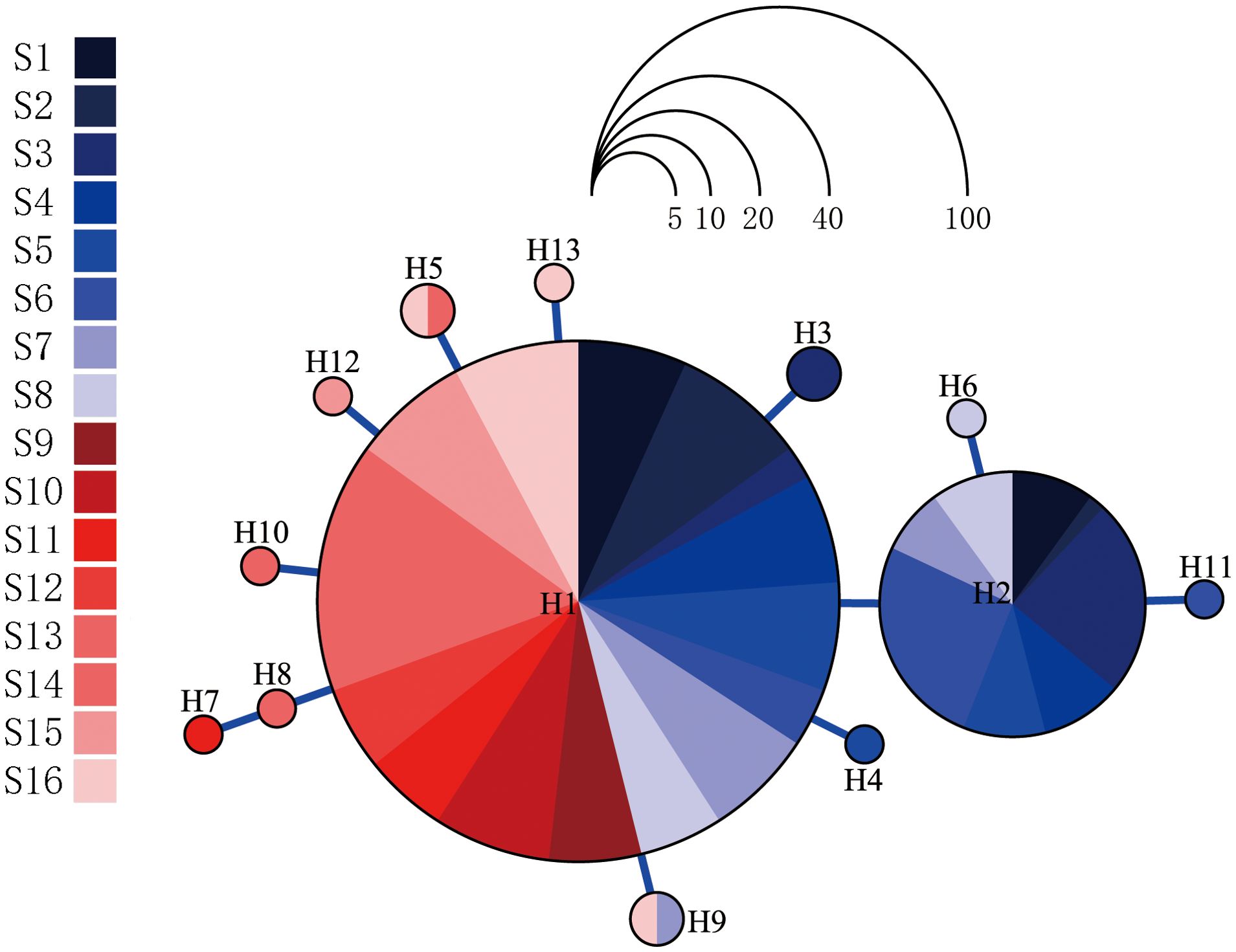
Figure 2. A haplotype network was constructed for Photopectoralis bindus based on genetic differences found in the nucleotides of the cytochrome c oxidase subunit I (COI) gene. The size of each circle denotes the frequency of individuals sharing a specific haplotype, with every circle representing a unique genetic variation. Branches that involve multiple mutational steps are labeled accordingly. The blue series represents samples from the western part of the Leizhou Peninsula (Beibu Gulf), while the red series represents samples from the eastern part of the Leizhou Peninsula (northern South China Sea).
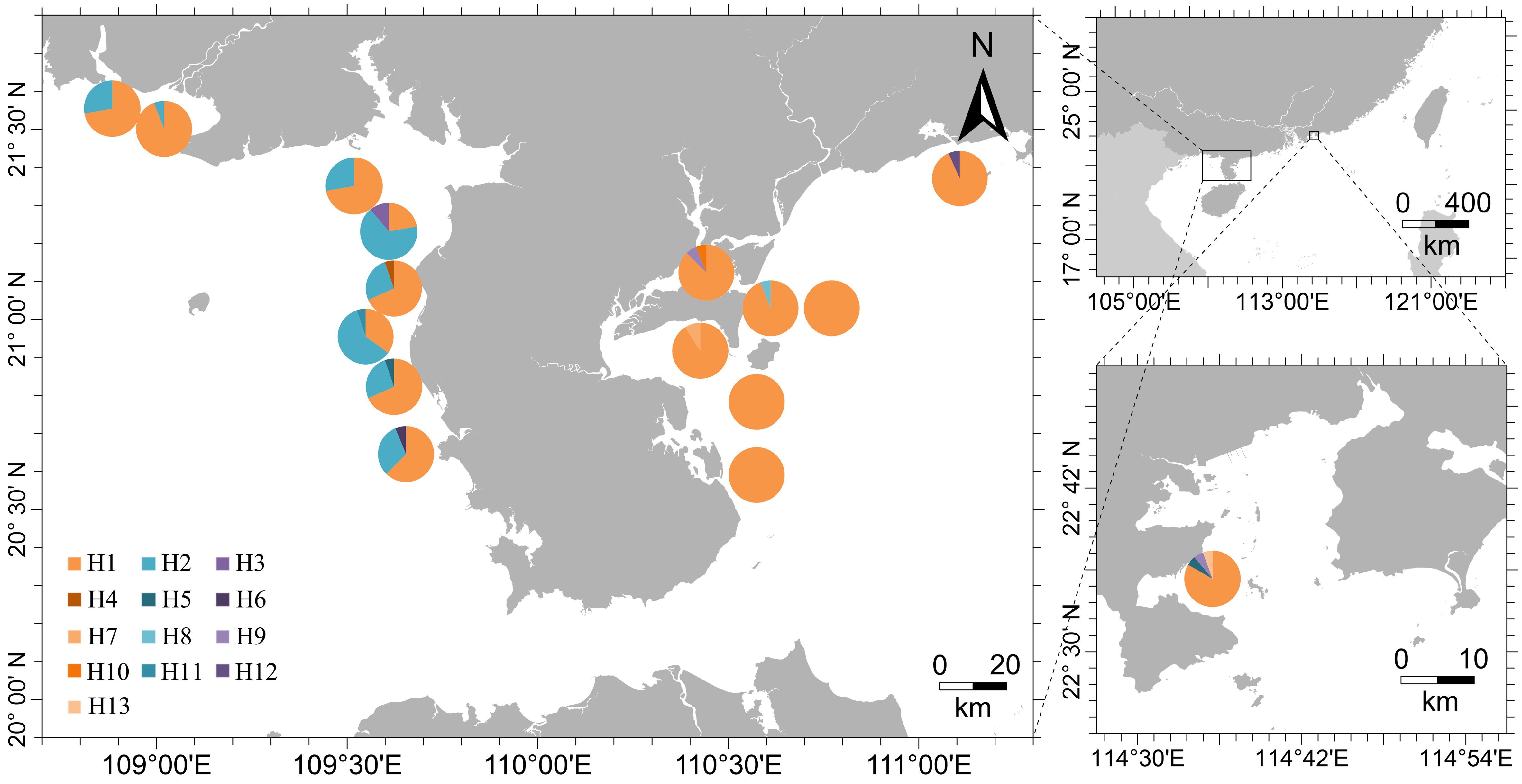
Figure 3. Pie charts illustrating the proportional distribution of different haplotypes among the sixteen sampling sites (total distinct haplotypes = 13). Each segment’s size in the pie chart reflects the percentage of individuals carrying a specific haplotype.
Analysis of molecular variance (AMOVA) revealed a relatively distinct genetic structure between habitats within the NSCS and the Beibu Gulf. Approximately 22.6% of the total genetic variation in mitochondrial DNA came from the western and the eastern groups of Leizhou Peninsula. In other words, a significant genetic structure was detected over the western and eastern regions of the Leizhou Peninsula, albeit small in scale. The majority (69.2%) of the total genetic variance stemmed from variance within populations (Table 2). The average genetic differentiation ΦST of P. bindus within the NSCS and the Beibu Gulf was 0.121 (range, −0.059 to 0.528). P. bindus also exhibited significant pairwise ΦST values between eastern and western populations of the Leizhou Peninsula (Table 3). PCoA suggested that the population’s genetic structure in eastern waters was significantly different from that in western waters of Leizhou Peninsula (Figure 4). Furthermore, a larger spatial variation in the genetic structure was observed in the eastern waters compared to the western waters. The first dimension explained 65.12%, while the second dimension explains 13.44% of the total variance. The effective number of migrants per generation from eastern populations to western populations of the Leizhou Peninsula was 14.21, whereas the flow in the opposite direction was only 1.69 (see Supplementary Tables).
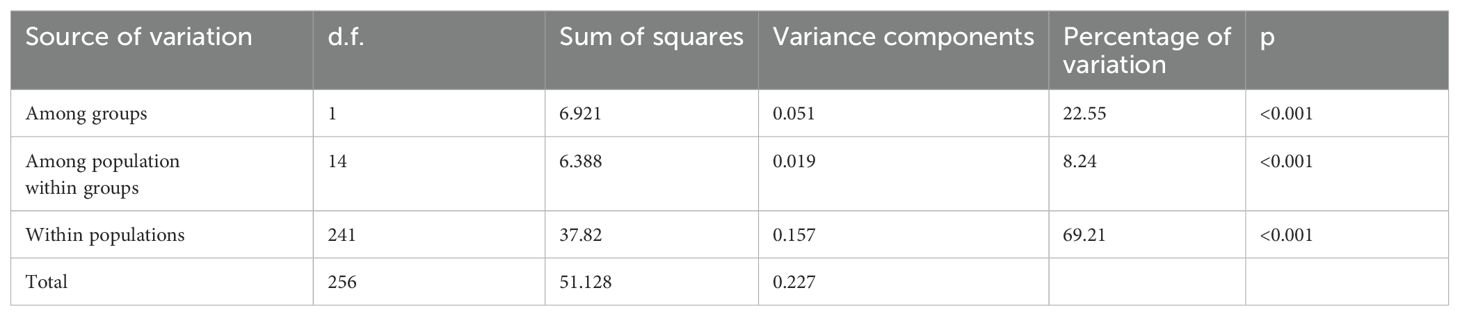
Table 2. Results of the analysis of molecular variance (AMOVA), partitioning overall mitochondrial DNA (mt-FST, using an infinite allele model) within and among groups.
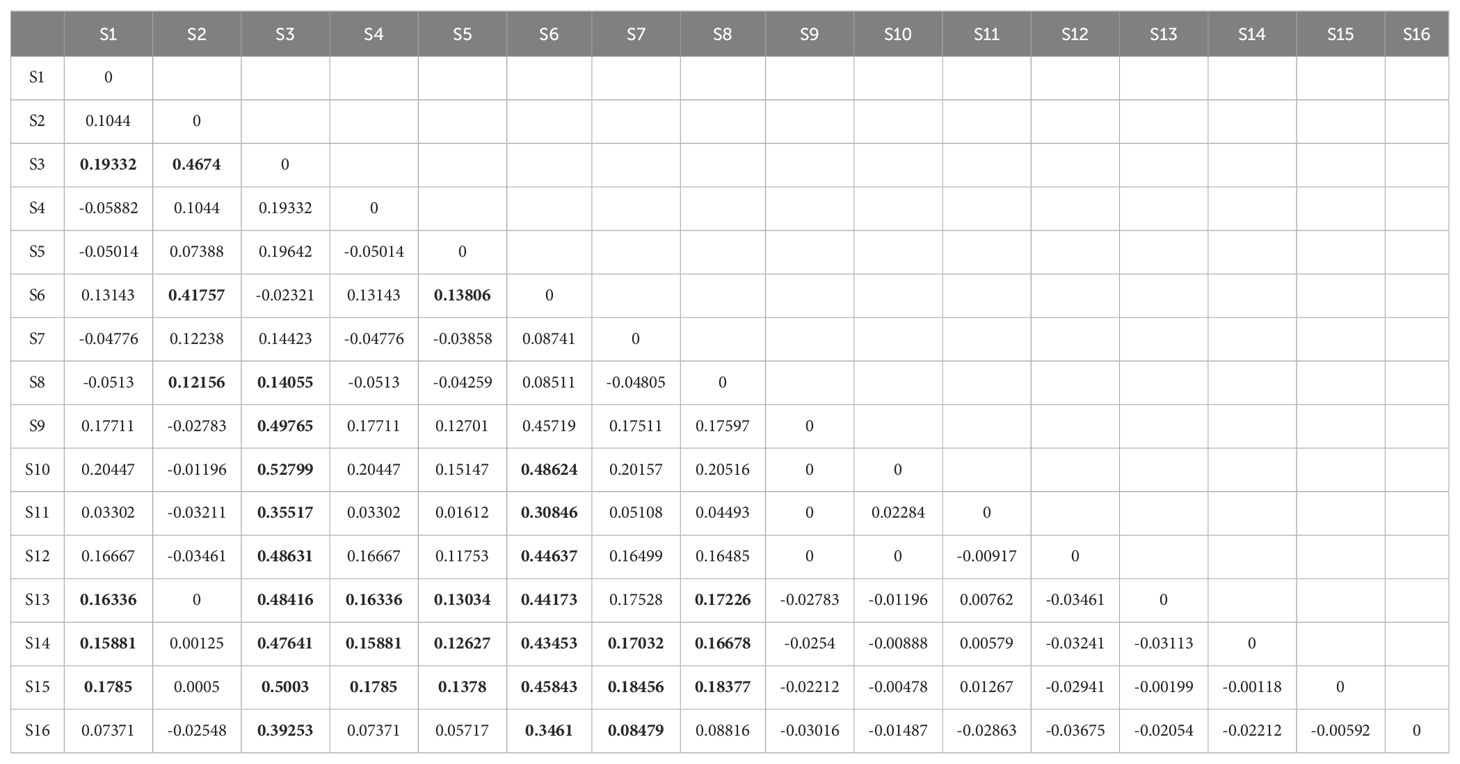
Table 3. Pairwise differentiation ΦST of Photopectoralis bindus sampled from northern South China Sea and the Beibu Gulf based on COI sequence data (Bold indicates significant Pairwise ΦST).
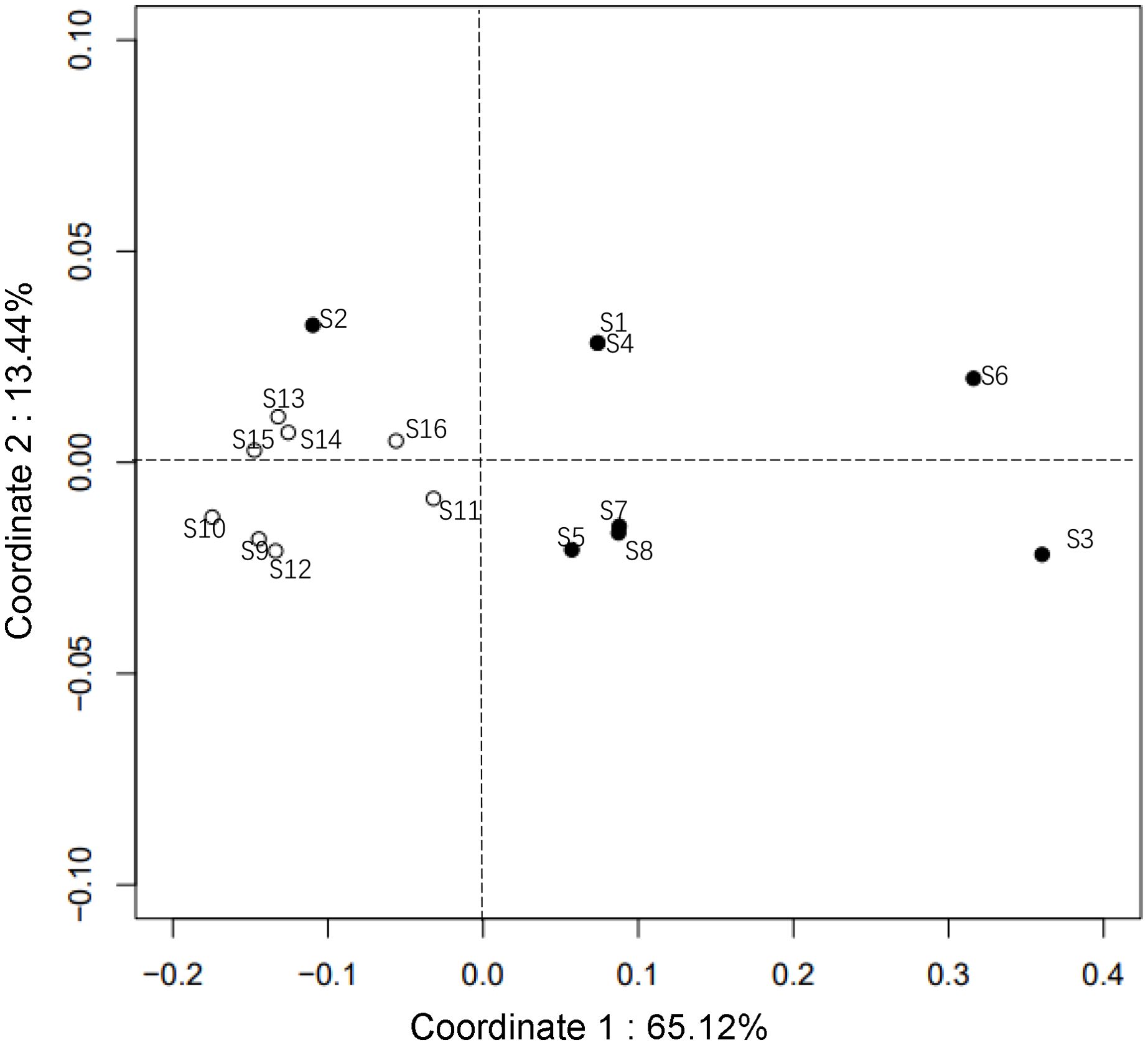
Figure 4. Biplot of principle coordination analysis showing the linearized pairwise ΦST. Solid circles represent the sampling locations in the eastern waters and open circle represents the sampling locations in the western waters of Leizhou Peninsula.
The Tajima’s D and Fu’s Fs values for P. bindus in the NSCS and the Beibu Gulf both showed statistically significant negativity (Tajima’s D = -1.828, p < 0.005; Fu’s Fs =-12.564, p < 0.005) (Table 4). These notable negative values, signifying an abundance of rare variants at nucleotide sites compared to the anticipated under a neutral evolutionary model, can stem from both natural selection and demographic expansion. The mismatch distribution exhibited a unimodal pattern (Figure 5), in line with the anticipated dispersal pattern predicted by a rapid expansion model (Table 4) (R index = 0.161, p = 0.29). We combined all the samples for the historical demographic analysis, resulting in a τ value of 0.516 (95% confidence interval: 0.357 - 0.695). Given that the mitochondrial genes mutate at a rate of 1% to 3% per million years (Cantatore et al., 1994), we estimated that the expansion time for P. bindus in the NSCS and the Beibu Gulf occurred approximately 7-14 thousand years ago. The Bayesian Skyline Plot (BSP) analysis revealed that the haplotypes of P. bindus in the NSCS and the Beibu Gulf coalesced approximately 13 thousand years ago, also assuming mutation rate of mitochondrial genes is 1-3% per million years. Furthermore, a continuous population expansion occurred between 3 and 10 thousand years ago (Figure 6).
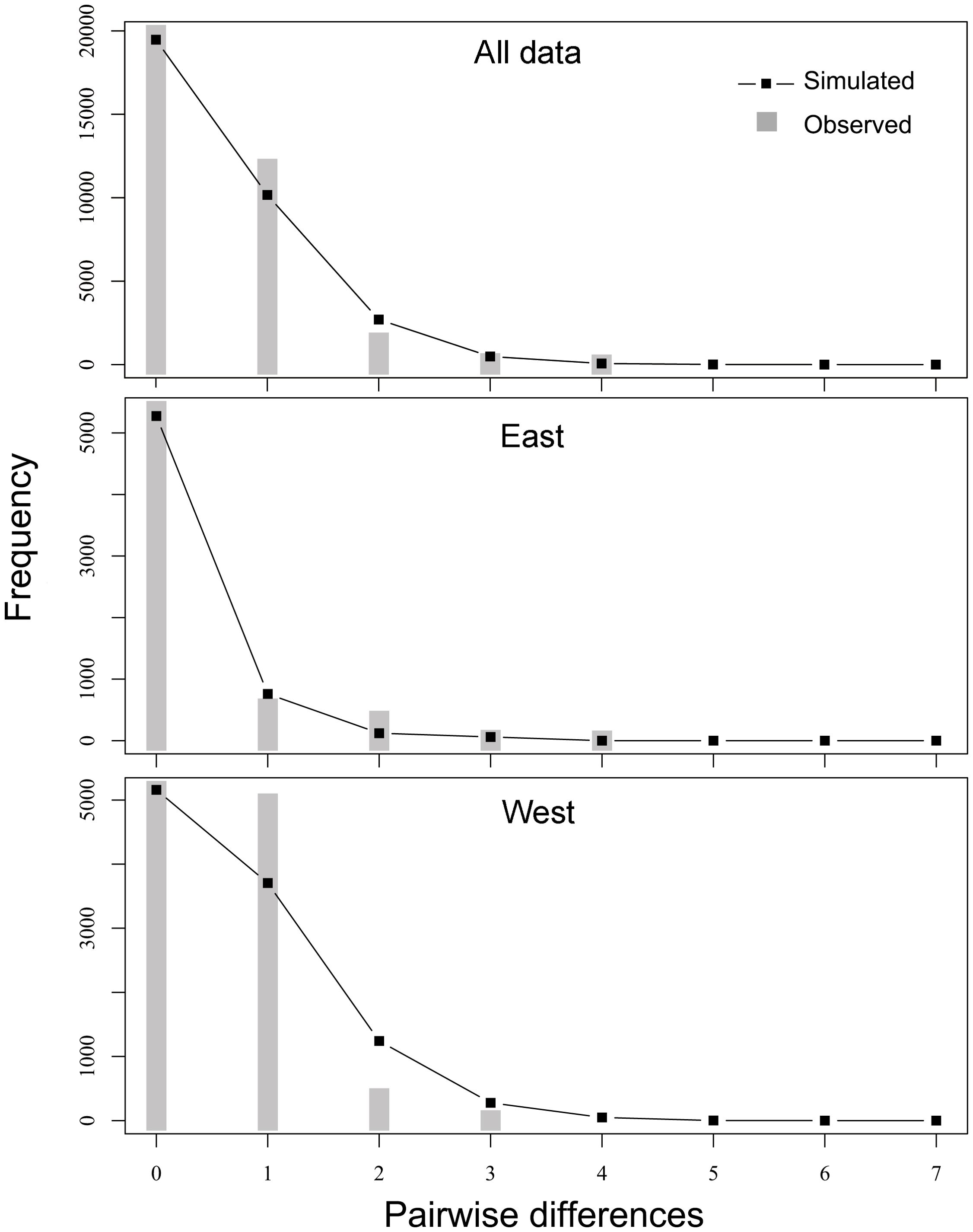
Figure 5. Expected mismatch distributions under sudden expansion model (line) and observed mismatch distributions (bars) of COI for Photopectoralis bindus from the northern South China Sea and the Beibu Gulf.
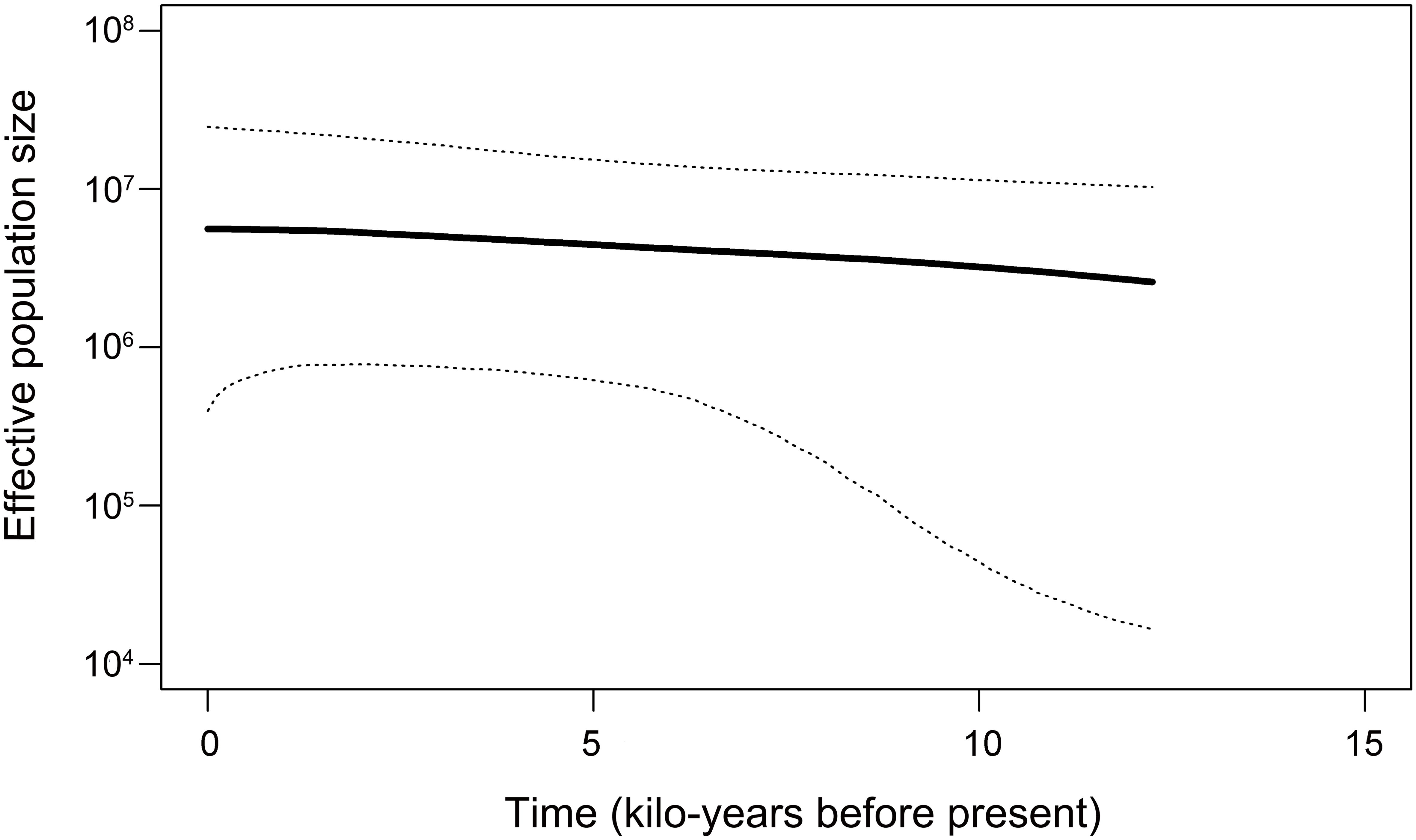
Figure 6. The demographic trends of Photopectoralis bindus in the northern South China Sea and the Beibu Gulf are depicted through a Bayesian skyline plot. The dark solid line represents the median estimate based on the assumption of a per site mutate at a rate of 2% per million years, with the dashed lines indicating the 95% highest posterior density (HPD) limits.
Population connectivity refers to the degree to which populations are connected or isolated from each other, influencing gene flow and genetic diversity. Numerous previous studies have demonstrated that mitochondrial markers are very efficient in uncovering genetic diversity and population connectivity in marine fish, especially for coastal species or populations distributed in bays (Beheregaray and Sunnucks, 2001; Alvarado-Bremer et al., 2005; Craig et al., 2007; Malcher et al., 2023). The present study provides the first insights into the population genetics and historical demography of P. bindus in the NSCS and the Beibu Gulf base on mitochondrial DNA variability.
In our study, P. bindus exhibited a “star-like” haplotype pattern along with low levels of both nucleotide and haplotype diversity. This pattern likely reflects a recent swift population increase following a period of reduced effective population size. Identifying the factors contributing to the low levels of genetic diversity in P. bindus is challenging. Overexploitation could be one contributing factor. Multiple studies have definitively connected overexploitation to the diminished genetic diversity observed in fish populations. Examples include the deep-water darkblotched rockfish (Sebastes crameri) from the coast of North America (Gomez-Uchida and Banks, 2006), snapper (Pagrus auratus) from the Southern Pacific Ocean (Hauser et al., 2002), and cod (Gadus morhua) from the North Sea (Hutchinson et al., 2003). Pinsky and Palumbi (2014) conducted a more comprehensive study by compiling information from 11,049 loci spanning 140 marine fish species. Their findings revealed that overexploited populations exhibited reduced allelic richness in 9 out of 12 genera and families, indicating that the decrease in genetic diversity across a broad spectrum of marine fish is driven by overharvesting. Overexploitation exacerbates genetic drift in various species, potentially leading to the loss of crucial alleles from multiple populations. While marine fishes often have large population sizes, sometimes in the millions, the intensity of genetic drift is influenced by the effective population size. Therefore, a large population size does not always correspond to a large effective population size (Ryman et al., 1995; Hare et al., 2011). Based on the FAO report, Teh et al. (2017) found that most of the assessed populations were either overexploited or fully fished. The most severe overfishing occurred in the western, shallower fishing grounds on the continental shelf of China and Vietnam. Notably, the fisheries resources in the NSCS were in the worst condition, characterized by the highest proportion of overexploited species, particularly demersal species. Over the past forty years, the rapid increase in fishing vessel power and fishing intensity has made overfishing in the NSCS and the Beibu Gulf highly evident. Major commercial fish species have exhibited signs of becoming smaller body size, earlier sexual maturity, and a shift from species at higher trophic levels to those at lower trophic levels (Chen and Qiu, 2005; Wang et al., 2019; Su et al., 2022). In addition to overexploitation, various other human activities, such as the destruction of coastal mangroves, pollution from marine aquaculture, illegal fishing, and habitat degradation, also pose significant threats to fish populations and the marine ecosystem in the NSCS and the Beibu Gulf. The impact is particularly evident on coastal and offshore species, significantly reducing their effective population size (Qiu et al., 2010). Despite improvements in fishing techniques and the implementation of fishing moratoriums, the catch in the NSCS and the Beibu Gulf has continued to decline gradually over the past few years.
An intriguing discovery from this study is the different distribution pattern of two dominant haplotypes. The central haplotype of the “star-like” haplotype pattern in P. bindus was distributed across all sampling sites from east of the Pearl River Estuary to the Beibu Gulf, spanning a range of over 700 kilometers. Another prevalent haplotype was exclusively found in the Beibu Gulf, particularly in the western region of the Leizhou Peninsula. Approximately 22.6% of the total genetic variation (AMOVA) in mitochondrial DNA was associated with distinctions between the eastern and western groups of the Leizhou Peninsula. In other words, a small but significant genetic structure was observed between the eastern and western regions of the Leizhou Peninsula. These patterns of genetic structure are strongly influenced by migration and, consequently, by the changing connectivity between suitable habitat areas over time. Currents are the most significant factor affecting the connectivity of fish populations in both larval and adult phases. During winter, the coastal waters of the NSCS are marked by currents flowing southwestward. Affected by the northeasterly monsoon, these currents stretch from the southwestern part of the Taiwan Strait to the eastern part of the Qiongzhou Strait. Upon reaching the eastern part of the Qiongzhou Strait, the southwestward coastal currents divide into two branches: one flows in a westerly direction into the Beibu Gulf via the Qiongzhou Strait, while the other veers southward along Hainan Island’s southern coast. In summer, the coastal currents west of the Zhujiang River mouth primarily flow westward. Additionally, a portion of this water flows in a westerly direction into the Beibu Gulf via the Qiongzhou Strait. This pattern of coastal currents in the Qiongzhou Strait differs from the traditional current system in the SCS, which typically flows eastward during the summer but westward during the winter. The coastal current in the Qiongzhou Strait is consistently westward, possibly turning northeast only temporarily when the southwest wind is strong (Chen et al., 2009; Ding et al., 2020). Shi et al. (2002) discovered a persistent westward mean flow through the Qiongzhou Strait throughout the year. The outflow from the Qiongzhou Strait has the potential to refresh up to 44% of the Beibu Gulf’s water annually, significantly impacting the seasonal circulation within the gulf. The Qiongzhou Strait serves as a conduit for water transport to the Gulf of Beibu. Our results on migration rates also indicated high connectivity between habitats within the NSCS and the Beibu Gulf. Populations in the eastern sea area of the Leizhou Peninsula receive few (1.69) and export many (14.21) individuals, whereas populations in the west of the Leizhou Peninsula show the opposite pattern. This was also somewhat consistent with the observation that the Qiongzhou Strait current flows westward year-round, influenced by the coastal current along the western coast of Guangdong. Larvae and adults of P. bindus with the dominant haplotype H1 were capable of long-distance movement with currents from the east to the west of the Leizhou Peninsula through the Qiongzhou Strait, but not vice versa.
Furthermore, mitochondrial DNA markers are particularly well-suited for deducing historical demographic events that might explain the present-day geographical distribution of marine fish. This is due to mitochondrial markers are more susceptible to genetic drift compared to nuclear markers and possess a comparatively smaller effective population size. In this study on P. bindus, the analysis of mtDNA genes from the NSCS and the Beibu Gulf revealed a “star-like” haplotype network, with the most dominant haplotype being shared across all localities. Similar results have been observed in other marine taxa from various coastal waters, such as Centropomus irae and Centropomus unidecimalis in the tropical Western Atlantic (Malcher et al., 2023), Terapon jarbua in the Indian Ocean (Lavergne et al., 2014), Larimichthys polyactis in the Western Pacific Ocean (Wang et al., 2013), and Evynnis cardinalis in SCS (Xu et al., 2021). In all cases, the observed population expansion follows a distinct growth pattern. The authors concluded that this expansion is linked to notable changes in sea level throughout the Pleistocenee. The hypothesis of a recent demographic expansion of P. bindus in the NSCS and the Beibu Gulf is further supported by the results of Fu’s Fs and Tajima’s D tests (significantly negative values), as well as mismatch distribution analyses (unimodal mismatch distribution). Based on a mitochondrial gene mutation rate ranging from 1% to 3% per million years, we estimated that the expansion time for P. bindus in the NSCS and the Beibu Gulf occurred approximately 7-14 thousand years before present, during the late Pleistocene epoch. Based on the Bayesian Skyline Plot (BSP) analysis, the P. bindus haplotypes in the NSCS and the Beibu Gulf coalesced approximately 13 thousand years ago. This event was followed by a consistent population growth between 3 and 10 thousand years before present. The congruence in population expansion estimates from both methods is primarily attributed to sea level fluctuations in the NSCS region throughout the Pleistocene. P. bindus is a typical species of tropical and brackish waters, commonly inhabiting shallow coastal waters near coral reefs and sandy bottoms. The fluctuation of sea level directly influences the changes in its distribution range. During the Pleistocene, successive glaciations alternated with interglacial periods, leading to significant fluctuations in global sea levels (Ludt and Rocha, 2015). Indeed, in the Pleistocene epoch, specifically during the Last Glacial Maximum (LGM), the SCS experienced isolation from the Indian Ocean due to substantially reduced global sea levels, leading to the formation of a semi-closed basin. Absolutely, the NSCS and the Beibu Gulf were integrated parts of the South China continent, with Hainan Island linked to mainland China before geographical changes occurred. After the last glacial period of the Late Pleistocene, sea level began to rise but were still 30 meters lower than they are today. The opening of the Qiongzhou Strait allowed seawater to inundate the Beibu Gulf (Voris, 2000; Lu et al., 2003). The rising sea level had altered shorelines, oceanic circulations, and upwelling patterns, leading to a rapid expansion of available coastal habitat. Therefore, it is likely that the surviving P. bindus rapidly expanded from potential refugia, for instance, the semi-enclosed SCS, into the Beibu Gulf. This study, however, focused solely on the mitochondrial COI gene marker, thus limiting the analysis to the maternal mode of inheritance. Future research should therefore include codominant markers, such as microsatellites and single nucleotide polymorphisms, with larger sample sizes from various marine regions to provide a more comprehensive genetic perspective on biparental inheritance.
The first insights into the population genetics and historical demography of P. bindus in the NSCS and the Beibu Gulf based on mitochondrial DNA variability revealed: (1) a “star-like” haplotype pattern along with low levels of both haplotype and nucleotide diversity. (2) a small but significant genetic structure was observed between the eastern and western regions of the Leizhou Peninsula. These patterns are primarily due to a rapid increase in population size following a period of small effective population size and the varying genetic exchange of P. bindus during reproduction due to changing connectivity between suitable habitat areas over time. Our work also suggests that P. bindus populations in the NSCS and the Beibu Gulf likely expanded and colonized new areas during the late Pleistocene, possibly in response to fluctuations in sea levels. The connectivity of the Qiongzhou Strait offers novel insights into the migration processes and population structure of P. bindus. This information is crucial for defining stock boundaries and evolutionary units, thereby aiding in the management and conservation of P. bindus. Our study enhances the understanding of the mechanisms behind oceanic environmental variation and the evolutionary patterns of migratory marine fish species. This knowledge is crucial for predicting future biodiversity changes and guiding adaptive management practices.
The data presented in the study are deposited in the NCBI GenBank repository, accession number PP789680-PP789692.
The animal study was approved by South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
LX: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. JZ: Data curation, Formal analysis, Writing – original draft. XW: Funding acquisition, Project administration, Resources, Writing – original draft. QC: Data curation, Formal analysis, Writing – original draft. QT: Data curation, Formal analysis, Writing – original draft. LW: Data curation, Formal analysis, Writing – original draft. JN: Data curation, Formal analysis, Writing – original draft. YL: Data curation, Formal analysis, Writing – original draft. SL: Data curation, Formal analysis, Writing – original draft. DH: Data curation, Formal analysis, Writing – original draft. FD: Funding acquisition, Project administration, Resources, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by Hainan Provincial Natural Science Foundation of China (324MS131, 323QN333), and Guangdong Basic and Applied Basic Research Foundation (2022B1515250005).
We thank Mr. Xin Liang, Dr. Yuezhong Wang for helping in fish identification. We thank all colleagues and students for their help with sampling. We thank Dr. Diego Fontaneto from National Research Council of Italy, Institute of Ecosystem Study, Dr. Kay Van Damme from Ghent University for providing useful comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1450142/full#supplementary-material
Alvarado-Bremer J. R., Mejuto J., Gómez-Márquez J., Boán F., Carpintero P., Rodríguez J. M., et al. (2005). Hierarchical analyses of genetic variation of samples from breeding and feeding grounds confirm the genetic partitioning of northwest Atlantic and South Atlantic populations of swordfish (Xiphias gladius L.). J. Exp. Mar. Biol. Ecol. 327, 167–182. doi: 10.1016/j.jembe.2005.06.022
Baus E., Darrock D. J., Bruford M. W. (2005). Gene-flow patterns in Atlantic and Mediterranean populations of the Lusitanian sea star Asterina gibbosa. Mol. Ecol. 14, 3373–3382. doi: 10.1111/j.1365-294X.2005.02681.x
Beerli P., Felsenstein J. (1999). Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152, 763–773. doi: 10.1093/genetics/152.2.763
Beheregaray L. B., Sunnucks P. (2001). Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Mol. Ecol. 10, 2849–2866. doi: 10.1046/j.1365-294X.2001.t01-1-01406.x
Bradbury I. R., Laurel B., Snelgrove P. V. R., Bentzen P., Campana S. E. (2008). Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proc. R. Soc B. 275, 1803–1809. doi: 10.1098/rspb.2008.0216
Camurugi F., Gehara M., Fonseca E. M., Zamudio K. R., Haddad C. F. B., Colli G. R., et al. (2021). Isolation by environment and recurrent gene flow shaped the evolutionary history of a continentally distributed Neotropical treefrog. J. Biogeogr. 48, 760–772. doi: 10.1111/jbi.14035
Cantatore P., Roberti M., Pesole G., Ludovico A., Milella F., Gadaletal M. N., et al. (1994). Evolutionary analysis of cytochrome b sequences in some perciformes: Evidence for a slower rate of evolution than in mammals. J. Mol. Evol. 39, 589–597. doi: 10.1007/BF00160404
Chen C., Li P., Shi M., Zuo J., Chen M., Sun H. (2009). Numerical study of the tides and residual currents in the Qiongzhou Strait. Chin. J. Oceanol. Limnol. 27, 931–942. doi: 10.1007/s00343-009-9193-0
Chen Z., Qiu Y. (2005). Stock variation of Parargyrops edita Tanaka in Beibu Gulf. South China Fisheries Sci. 1, 21–31. doi: 10.3969/j.issn.2095-0780.2005.03.004
Cowman, P.F., and Bellwood D.R. (2013). Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proc. R. Soc. B. 280 (1768), 20131541. doi: 10.1098/rspb.2013.1541
Craig M. T., Eble J. A., Bowen B. W., Robertson D. R. (2007). High genetic connectivity across the Indian and Pacific oceans in the reef fish Myripristis berndti (Holocentridae). Mar. Ecol. Prog. Ser. 334, 245–254. doi: 10.3354/meps334245
Ding Y., Yao Z., Zhou L., Bao M., Zang Z. (2020). Numerical modeling of the seasonal circulation in the coastal ocean of the Northern South China Sea. Front. Earth Sci. 14, 90–109. doi: 10.1007/s11707-018-0741-9
Diopere E., Vandamme S. G., Hablützel P. I., Cariani A., Van Houdt J., Rijnsdorp A., et al. (2018). Seascape genetics of a flatfish reveals local selection under high levels of gene flow. ICES J. Mar. Sci. 75, 675–689. doi: 10.1093/icesjms/fsx160
Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Ellis C. D., Hodgson D. J., Daniels C. L., Collins M., Griffiths A. G. F. (2017). Population genetic structure in European lobsters: implications for connectivity, diversity and hatchery stocking. Mar. Ecol. Prog. Ser. 563, 123–137. doi: 10.3354/meps11957
Excoffier L. (2004). Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol. Ecol. 13, 853–864. doi: 10.1046/j.1365-294X.2003.02004.x
Excoffier L., Lischer H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fu Y.-X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Gaither M. R., Toonen R. J., Robertson D. R., Planes S., Bowen B. W. (2010). Genetic evaluation of marine biogeographical barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus). J. Biogeogr. 37, 133–147. doi: 10.1111/j.1365-2699.2009.02188.x
Geng P., Zhang K., Chen Z., Xu Y., Sun M. (2018). Interannual change in biological traits and exploitation rate of Decapterus maruadsi in Beibu Gulf. South China Fisheries Sci. 14, 1–9. doi: 10.12131/20180106
Gomez-Uchida D., Banks M. A. (2006). Estimation of effective population size for the long-lived darkblotched rockfish sebastes crameri. J. Hered. 97, 603–606. doi: 10.1093/jhered/esl042
Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hall T. A. (1999). BioEdit: a user-friendly biological sequence algnment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Hare M. P., Nunney L., Schwartz M. K., Ruzzante D. E., Burford M., Waples R. S., et al. (2011). Understanding and estimating effective population size for practical application in marine species management. Conserv. Biol. 25, 438–449. doi: 10.1111/j.1523-1739.2010.01637.x
Hauser L., Adcock G. J., Smith P. J., Bernal Ramírez J. H., Carvalho G. R. (2002). Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Natl. Acad. Sci. 99, 11742–11747. doi: 10.1073/pnas.172242899
Hauser L., Carvalho G. R. (2008). Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish 9, 333–362. doi: 10.1111/j.1467-2979.2008.00299.x
Hemmer-Hansen J., Hüssy K., Baktoft H., Huwer B., Bekkevold D., Haslob H., et al. (2019). Genetic analyses reveal complex dynamics within a marine fish management area. Evol. Appl. 12, 830–844. doi: 10.1111/eva.12760
Hutchinson W. F., Oosterhout C.v., Rogers S. I., Carvalho G. R. (2003). Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua). Proc. R. Soc Lond. B. Biol. Sci. 270, 2125–2132. doi: 10.1098/rspb.2003.2493
Kenchington E., Heino M., Nielsen E. E. (2003). Managing marine genetic diversity: time for action? ICES J. Mar. Sci. 60, 1172–1176. doi: 10.1016/S1054-3139(03)00136-X
Klangnurak W., True J. D. (2022). Spatial–temporal shifts of ponyfish (Perciformes: Leiognathidae) diversity in by-catch of trawl fishery, including morph variation of Photopectoralis bindus. J. Appl. Ichthyol. 38, 34–43. doi: 10.1111/jai.14290
Lavergne E., Calvès I., Meistertzheim A. L., Charrier G., Zajonz U., Laroche J. (2014). Complex genetic structure of a euryhaline marine fish in temporarily open/closed estuaries from the wider Gulf of Aden. Mar. Biol. 161, 1113–1126. doi: 10.1007/s00227-014-2404-z
Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lu B., Huang S., Li G., Zhang F. (2003). Vertical variations of core sound velocity: evidence of paleooceanographic history since the Pleistocene epoch. Mar. Georesour. Geotec. 21, 63–71. doi: 10.1080/716100485
Ludt W. B., Rocha L. A. (2015). Shifting seas: the impacts of Pleistocene sea-level fluctuations on the evolution of tropical marine taxa. J. Biogeogr. 42, 25–38. doi: 10.1111/jbi.12416
Luttikhuizen P. C., Drent J., Baker A. J. (2003). Disjunct distribution of highly diverged mitochondrial lineage clade and population subdivision in a marine bivalve with pelagic larval dispersal. Mol. Ecol. 12, 2215–2229. doi: 10.1046/j.1365-294X.2003.01872.x
Malcher G., Amorim A. L., Ferreira P., Oliveira T., Melo L., Rêgo P. S., et al. (2023). First evaluation of the population genetics and aspects of the evolutionary history of the Amazonian snook, Centropomus irae, and its association with the Amazon plume. Hydrobiologia 850, 2115–2125. doi: 10.1007/s10750-023-05223-5
Manel S., Schwartz M. K., Luikart G., Taberlet P. (2003). Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197. doi: 10.1016/S0169-5347(03)00008-9
Morton B., Blackmore G. (2001). South China sea. Mar. pollut. Bull. 42, 1236–1263. doi: 10.1016/s0025-326x(01)00240-5
Nan F., He Z., Zhou H., Wang D. (2011). Three long-lived anticyclonic eddies in the northern South China Sea. J. Geophys. Res. 116 (C5), 1–15. doi: 10.1029/2010JC006790
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019). vegan: Community Ecology Package.
Palumbi S. R. (1992). Marine speciation on a small planet. Trends Ecol. Evol. 7, 114–118. doi: 10.1016/0169-5347(92)90144-Z
Pinsky M. L., Palumbi S. R. (2014). Meta-analysis reveals lower genetic diversity in overfished populations. Mol. Ecol. 23, 29–39. doi: 10.1111/mec.12509
Planes S., Doherty P. J., Bernardi G. (2001). Strong genetic divergence among populations of a marine fish with limited dispersal, Acanthochromis polyacanthus, within the Great Barrier Reef and the Coral Sea. Evolution 55, 2263–2273. doi: 10.1111/j.0014-3820.2001.tb00741.x
Qiu Y., Lin Z., Wang Y. (2010). Responses of fish production to fishing and climate variability in the northern South China Sea. Prog. Oceanogr. 85, 197–212. doi: 10.1016/j.pocean.2010.02.011
Rao Y. P., Veni N. K. D., Rukmini Sirisha I. (2015). Biology of orange fin pony fish, Photopectoralis bindus (Valenciennes 1835), off Visakhapatnam, East coast of India. Int. J. Environ. Sci. 5, 1159–1171. doi: 10.6088/ijes.2014050100110
Rawat S., Kumar J., Benakappa S., Sonwal M. C., Naik K. S. T. (2019). Reproductive biology of the orangefin ponyfish Photopectoralis bindus (Valenciennes 1835) off Mangaluru coast, Karnataka. Indian J. Fish. 66, 120–124. doi: 10.21077/IJF.2019.66.2.81002-17
Richardson J. L., Urban M. C., Bolnick D. I., Skelly D. K. (2014). Microgeographic adaptation and the spatial scale of evolution. Trends Ecol. Evol. 29, 165–176. doi: 10.1016/j.tree.2014.01.002
Rogers A. R., Harpending H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569. doi: 10.1093/oxfordjournals.molbev.a040727
Rolland J. L., Bonhomme F., Lagardère F., Hassan M., Guinand B. (2007). Population structure of the common sole (Solea solea) in the Northeastern Atlantic and the Mediterranean Sea: revisiting the divide with EPIC markers. Mar. Biol. 151, 327–341. doi: 10.1007/s00227-006-0484-0
Ryman N., Utter F., Laikre L. (1995). Protection of intraspecific biodiversity of exploited fishes. Rev. Fish Biol. Fish. 5, 417–446. doi: 10.1007/BF01103814
Salzburger W., Ewing G. B., Von Haeseler A. (2011). The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol. Ecol. 20, 1952–1963. doi: 10.1111/j.1365-294X.2011.05066.x
Selkoe K. A., Henzler C. M., Gaines S. D. (2008). Seascape genetics and the spatial ecology of marine populations. Fish Fish. 9, 363–377. doi: 10.1111/j.1467-2979.2008.00300.x
Shi M., Chen C., Xu Q., Lin H., Liu G., Wang H., et al. (2002). The role of Qiongzhou strait in the seasonal variation of the south China sea circulation. J. Phys. Oceanogr. 32, 103–121. doi: 10.1175/1520-0485(2002)032<0103:TROQSI>2.0.CO;2
Slatkin M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139, 457–462. doi: 10.1093/genetics/139.1.457
Su L., Xu Y., Qiu Y., Sun M., Zhang K., Chen Z. (2022). Long-term change of a fish-based index of biotic integrity for a semi-enclosed bay in the Beibu gulf. Fishes 7 (3), 1–10. doi: 10.3390/fishes7030124
Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Teh L. S. L., Witter A., Cheung W. W. L., Sumaila U. R., Yin X. (2017). What is at stake? Status and threats to South China Sea marine fisheries. Ambio 46, 57–72. doi: 10.1007/s13280-016-0819-0
Voris H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27, 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x
Wang L., Liu S., Zhuang Z., Guo L., Meng Z., Lin H. (2013). Population genetic studies revealed local adaptation in a high gene-flow marine fish, the small yellow croaker (Larimichthys polyactis). PloS One 8, e83493. doi: 10.1371/journal.pone.0083493
Wang X., Qiu Y., Du F., Liu W., Sun D., Chen X., et al. (2019). Roles of fishing and climate change in long-term fish species succession and population dynamics in the outer Beibu Gulf, South China Sea. Acta Oceanologica Sin. 38, 1–8. doi: 10.1007/s13131-019-1484-5
Waples R. S. (1998). Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J. Hered. 89, 438–450. doi: 10.1093/jhered/89.5.438
Ward R. D., Woodwark M., Skibinski D. O. F. (1994). A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. J. Fish Biol. 44, 213–232. doi: 10.1111/j.1095-8649.1994.tb01200.x
Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. (2005). DNA barcoding Australia's fish species. Phil. Trans. R. Soc B. 360, 1847–1857. doi: 10.1098/rstb.2005.1716
Xu L., Wang X., Wang L., Ning J., Li Y., Huang D., et al. (2021). Threadfin porgy (Evynnis cardinalis) haplotype pattern and genetic structure in Beibu gulf, South China Sea. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.726533
Yao Y., Harff J., Meyer M., Zhan W. (2009). Reconstruction of paleocoastlines for the northwestern South China Sea since the Last Glacial Maximum. Sci. China Ser. D: Earth Sci. 52, 1127–1136. doi: 10.1007/s11430-009-0098-8
Zhao H., Wang L., Yuan J. (2007). Origin and time of Qiongzhou stait. Mar. Geol. Quat. Geol. 27, 33–40.
Keywords: haplotype pattern, genetic structure, Leizhou Peninsula, The South China Sea, historical demography
Citation: Xu L, Zhang J, Wang X, Chen Q, Tang Q, Wang L, Ning J, Li Y, Liu S, Huang D and Du F (2024) Ecological connectivity of the Qiongzhou Strait: a case form Orangefin Ponyfish (Photopectoralis bindus) haplotype diversity and genetic structure. Front. Mar. Sci. 11:1450142. doi: 10.3389/fmars.2024.1450142
Received: 17 June 2024; Accepted: 26 August 2024;
Published: 23 September 2024.
Edited by:
Jin Sun, Ocean University of China, ChinaReviewed by:
Longshan Lin, State Oceanic Administration, ChinaCopyright © 2024 Xu, Zhang, Wang, Chen, Tang, Wang, Ning, Li, Liu, Huang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feiyan Du, ZmVpeWFuZWdnQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.