
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 01 August 2024
Sec. Aquatic Physiology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1444929
This article is part of the Research Topic Endocrine Regulation and Physiological Adaptation of Stress Response in Aquatic Organisms, Volume II View all 13 articles
As one of the major limiting environment factors in aquaculture, ammonia nitrogen brings severe threat to the growth and survival of aquatic animals, especially mollusk in benthic zones. However, the molecular mechanism underlying the toxic response and tolerance of mollusks to ammonia nitrogen remain unclear. In this study, transcriptome, metabolome and physiological indicators were combined to investigate the metabolic mechanism of adult razor clam (Sinonvacula constricta), which was exposed to ~46mg/L ammonia nitrogen for 1 day (A1) and 10 days (A10). It was observed that compared with A1, the contents of free amino acids, including taurine (Tau), alanine (Ala) and arginine (Arg), the activities of immune-related enzymes acid phosphatase (ACP) and alkaline phosphatase (AKP), and antioxidation-related enzymes superoxide dismutase (SOD) and glutathione peroxidase (GPX) in hepatopancreas, were significantly increased in A10, while the content of malondialdehyde (MDA) was significantly decreased (P < 0.05). Furthermore, the contents of glucose and pyruvate in hepatopancreas, foot and hemolymph urea nitrogen (HUN) were significantly changed (P < 0.05). Meantime, the comparative transcriptome analysis between A1 and A10 groups revealed the effects of ammonia stress on immune defense, antioxidant system and metabolic pathway. Likewise, metabolomic analysis showed that ammonia exposure interfered with amino acid metabolism, lipid metabolism and carbohydrate metabolism, with metabolism related-genes changed according to RNA-seq analysis. By comparing the metabolites and transcripts profiles of A10 and A1, the expression of some genes involved in detoxification and ammonia excretion was significantly changed. Combined with the changes in metabolites, we speculated that the convert endogenous ammonia to alanine, alanine-glucose cycle and urea synthesis might be adaptive strategies of the razor clam after ammonia stress. Collectively, the combination of physiological, transcriptome and metabolome will greatly contribute to the progressively understand the toxicity of ammonia exposure and the defense mechanism of razor clam against ammonia toxicity, and provide new sights on the potential molecular mechanisms of ammonia adaptive strategies in benthic mollusk.
Over the past decades, ammonia nitrogen, a common pollutant in aquatic ecosystem, has been one of the major limiting factors affecting farmed animals in semi-intensive and intensive farming systems. It is worth noting that un-ionized form (NH3-N) and ion form (NH4+) are two existing forms of ammonia nitrogen in water. Among them, NH3-N exhibits stronger toxicity due to its ability to diffuse through cell membrane (in this study, the term “ammonia nitrogen” refers to the sum of NH3-N and NH4+) (Cong et al., 2019). Generally, ammonia nitrogen can be kept at a very low concentration in most aquatic ecosystems (< 2mg/L) (Romano and Zeng, 2013). However, ammonia nitrogen in marine sediments can exceed 39mg/L (Romano and Zeng, 2013) and reach as high as 46mg/L (Cui et al., 2022) during semi-intensive and intensive farming processes due to increased anthropogenic activities such as agricultural fertilizers. In aquaculture systems, with the input of a large amount of feed and the excretion of cultured animals, ammonia accumulated over time, seriously affecting the growth and survival of farmed animals (Zhao et al., 2020). So far, the level of ammonia nitrogen in aquaculture has become a common concern and an urgent problem to be solved (Bernasconi and Uglow, 2011).
Many studies have been carried out based on the toxicity of ammonia nitrogen to aquatic animals, and some detoxication metabolic mechanisms of ammonia tolerance and adaptation have been found in fish. Among them, the gulf toadfish (Opsanus beta) (Veauvy et al., 2005), Magadi tilapia (Alcolapia graham (Ip and Chew, 2010), Atlantic hagfish (Myxine glutinosa) (Edwards et al., 2015) and other fish have found that the synthesis of glutamine and urea, the role of Rhesus glycoproteins (Rh protein) play an important role in the detoxification metabolism of ammonia nitrogen. In addition, decapod crustaceans have also evolved related ammonia excretion or metabolic mechanisms to avoid ammonia accumulation in blood. For example, studies on green shore crab (Carcinus maenas) (Weihrauch et al., 2002), Chinese mitten-handed crab (Eriocheir sinensis) (Hong et al., 2007), Amazon river shrimp (Macrobrachium amazonicum) (Pinto et al., 2016) and swimming crab (Portunus trituberculatus) (Meng et al., 2021) have shown that ammonia can be actively excreted along an inwardly directed gradient through the coordinated action of several transporters, such as Na+/K+-ATPase (NKA), V-type H+-ATPase (VHA) and K+-channels. Not only that, the internal ammonia can be converted into free amino acids, especially glutamine, which can participate in a variety of metabolic and synthetic biochemical reactions in the body (Hong et al., 2007). However, the related study on invertebrates, especially bivalves, is still in a primary stage.
The razor clam (Sinonvacula constricta), is a marine bivalve with important economic and ecological significance, which is loved by consumers because of its delicious taste and rich nutrition. In 2022, the annual production of razor clam is 847,626 tons (from China Fishery Statical Yearbook, 2023). In the current mixed farming model of aquaculture system, the clams with fish, shrimp, crabs were widely used to improve economic benefits (Sun et al., 2021a). In this farming mode, with a large amount of deposition and decay of surplus food, as well as dead organisms in the sediment, the razor clam, as a benthic bivalve, often lives in mudflats or under 30–40cm of mud in ponds, faces a more severe ammonia nitrogen environment than other aquatic animals. In the previous study, we found that the median lethal concentration of the razor clam was 244.55mg/L for 96h (Lv et al., 2022), while the semi-lethal concentration of other bivalves was relatively low, such as Asiatic hard clam (Meretrix meretrix) was 92.37mg/L (Chen et al., 2010), and Venus clam (Cyclina sinensis) was 65.79mg/L (Ge et al., 2021). These findings indicated that the razor clam has a strong tolerance to ammonia nitrogen. Moreover, our study also found that synthesis of glutamine was an important ammonia detoxification strategy for the razor clam after ammonia exposure (Zhang et al., 2020a). However, why do razor clams have such a high tolerance for ammonia nitrogen? Is the detoxification mechanism of mollusk different from that of other aquatic animals? These questions have not been definitively answered. In this study, physiological index measurement, transcriptomic and metabolomic were analyzed to study the tolerance and detoxification mechanism of the razor clam to ammonia nitrogen stress.
The razor clam (shell length of 61.34 ± 3.21mm, wet weight 13 ± 1.05g), consisting of 1200 individuals at age of one year, were collected in June 2022 in the Genetic Breeding Research Center of Zhejiang Wanli University, China, and all experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University, China. Then, the clams were aerated and acclimated in a 500L recirculating seawater tank with temperature of 23 ± 0.5°C, salinity of 22 ± 0.5, and fed the microalgae (Chaetoceros muelleri) twice a day.
After seven days domestication, clams were divided into six tanks (100L, N = 200 for per tank), among which three tanks were treated as ammonia nitrogen stress group (AG) and the other three tanks were treated as control group (CG) without ammonia nitrogen. It is worth noting that ammonium chloride (NH4Cl, BBI, Sangon, Shanghai, China) was used as raw material to prepare total ammonia nitrogen. According to relevant reports, the concentration of ammonia nitrogen in intensive culture system can reach up to 46mg/L (Cui et al., 2022). For the present study, total concentrations of ammonia nitrogen of 0mg/L (CG) and 46mg/L (AG) were used to understand the adverse effects of high ammonia nitrogen on razor clam in culture environment. Notably, the actual concentrations of CG and AG were 0.13 ± 0.38 and 46 ± 3.17mg/L, respectively. The method for determination of ammonia nitrogen concentration in water was described by Zhang et al. (2020a). The ammonia concentration in the AG was regulated by adding NH4Cl solution to the seawater. Throughout the experiment, the pH of the water was checked by HACH HQ30d pH meter (Hach, Loveland, CO, United States) and kept at a relatively constant value, that is, when the pH was greater than or less than 8, it was regulated by sodium dihydrogen phosphate (NaH2PO4) and 2mg/L sodium bicarbonate (NaHCO3), respectively (Zhang et al., 2020a). All the clams were monitored and the dead individuals were removed in time. The stress lasted for 10 days (240h). Then, the hepatopancreas, foot and hemolymph of 12 clams from each group were collected at 0, 6, 12, 24, 48, 72, 144, and 240h for the determination of physiological indexes (including free amino acids, enzyme activities, malondialdehyde content, glucose and pyruvate levels and changes in hemolymph urea nitrogen). In addition, the hepatopancreas of another six clams in each group at 24 and 240h were frozen at -80°C liquid nitrogen for subsequent transcriptome and metabolome analysis.
Hepatopancreas from AG and CG at 0 (C0), 24 (A1) and 240h (A10) were selected for free amino acids (FAAs) content analysis. Before detection, the hepatopancreas tissue was freeze-dried for 24h, and then grinded into powder. Then, 5mL of 0.01mol/L HCl was added to every 0.1g sample for hydrolysis for 30min and centrifuged at 12000g for 2min. The supernatant after centrifugation was added with 8% sulfosalicylic acid and mixed overnight. After overnight, the sample were centrifuged again at 12000g for 2min. The supernatant was collected and diluted with 0.02mol/L HCl. Then the diluted supernatant was filtered by 0.22μm filter membrane and analyzed by L-8900 automatic amino acid analyzer (Hitachi High-Tech Corporation, Japan) (Zhu et al., 2024).
Hepatopancreas and foot from each group were collected at 0, 6, 12, 24, 48, 72, 144, and 240h, respectively, and the corresponding tissue extracts suggested by the kit were homogenized in the refrigerator at the ratio of 1:9 by weight (g) to volume (mL). Different mixtures were centrifuged according to the instructions of different kits. Then, the supernatant was taken for enzyme activity and malondialdehyde content in hepatopancreas and glucose and pyruvate content in hepatopancreas and foot. Next, the activity of enzymes (immune-related enzymes acid phosphatase (ACP) and alkaline phosphatase (AKP), and antioxidation-related enzymes superoxide dismutase (SOD) and glutathione peroxidase (GPX)) and the content of MDA, glucose and pyruvate were detected using corresponding kits (Note that in addition to pyruvate assay kits from Sangon Shanghai, China, the rest of the kits were from Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer’s instructions. Likewise, the hemolymph urea nitrogen (HUN) was also determined according to the kit instructions (Nanjing Jiancheng Bioengineering Institute, China).
Total RNA was extracted from hepatopancreas of each group by Trizol reagent (Omega, Norcross, GA, United States) according to the instructions of the manufacturer. Then, the concentration and purity of the total RNA was quantified by NanoDrop spectrophotometers (Thermo Scientific, Waltham, MA, United States) and Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, United States). Next, all RNA-seq procedures were performed on Illumina Hiseq 2000 platform as described by the Novogene Company (Beijing, China).
Clean data were filtered to remove joint sequence and low-quality reads. Meanwhile, Q20, Q30 and GC content of the clean data were all calculated. HISAT2 software was used to compare clean reads with the reference genome of the razor clam (Dong et al., 2020), and the location information of reads on the reference genome was obtained.
Gene expression levels were estimated using RSEM (Li and Dewey, 2011). The featureCounts software (Liao et al., 2014) was used to filter out genes with low read counts before performing gene expression analysis. Genes between AG and CG with an adjusted P value < 0.05 found by DESeq2 were assigned as differentially expressed. In addition, the fold change > 2 and false discovery rate (FDR) of 0.05 were used as key indicators for differentially expressed gene (DEGs) screening.
GO and KEGG enrichment analysis were performed by clusterProfiler R package (Yu et al., 2012). The enrichment analysis was based on hypergeometric distribution principle, in which the differential gene sets were the differential genes obtained from significant difference analysis and annotated into Go and KEGG database, with a cutoff q-value of 0.05.
To verify the results of RNA-seq, seven DEGs were randomly selected for quantitative real time PCR (qRT-PCR). The specific experimental methods of qRT-PCR, including total RNA extraction, cDNA synthesis, selection of internal reference genes (ribosomal protein S9, RS9) (Zhao et al., 2018) (Table 1) and data processing were consistent with those previous reported by Sun et al. (2021b).
To gain a more effective understanding of the mechanisms of ammonia nitrogen reactions and tolerance, differential metabolites in hepatopancreas tissues of clams (N = 6) from each group were evaluated through non-targeted GC-MS metabolomics techniques. Tissues (100mg) were ground with liquid nitrogen, and the homogenate was resuspended with pre-cooled 80% methanol through the well vortex. The sample were incubated on ice for 5min and centrifuged at 4°C for 15000g for 20min. The supernatant was diluted with LC-MS grade water to a final concentration of 53% methanol. The sample were then transferred to fresh Eppendorf tubes and centrifuged at 15000g, 4°C for 20min. Finally, the supernatant was injected into LC-MS/MS system for analysis (Want et al., 2012). More details were available in Supplementary Material 1.
The enzyme activity and content of MDA in hepatopancreas were evaluated by one-way ANOVA and all data were presented as means ± standard of the means (SE). All statistical analyses were performed in SPSS 22 (International Business Machines (IBM) Corporation, Chicago, United States) and the results were considered significantly different when P < 0.05.
The results of the determination of total free amino acids in hepatopancreas of the razor clam after ammonia nitrogen stress showed that among the 16 amino acids detected, except histidine (His, no significant changes), the other 15 amino acids had significant changes of varying degrees (Table 2). Compared with C0, aspartic acid (Asp), threonine (Thr), serine (Ser), glutamic acid (Glu), glycine (Gly), alanine (Ala), valine (Val), methionine (Met), isoleucine (Ile), leucine (Leu), tyrosine (Tyr), phenylalanine (Phe) and lysine (Lys) were significantly down-regulated in A1 (P < 0.05). In A10, Asp, Thr, Ser, Glu, Met, Ile, Leu, Tyr, Phe and Lys were significantly down-regulated (P < 0.05), while taurine (Tau), Ala and arginine (Arg) were significantly up-regulated (P < 0.05).
The activities of four enzymes including ACP, AKP, SOD, GPX and the content of MDA and HUN at different times after ammonia nitrogen stress were shown in Figure 1. The enzyme activity of two immune-related enzymes (AKP and ACP) showed no significant change at 0~24h, and both showed a significant increase at 48h (P < 0.05), and then continued until the end of the experiment (Figure 1A). The activity of SOD increased significantly at 0~48h (P < 0.05), and reached the highest level at 72h, and then increased at the later stage (Figure 1B). GPX activity increased significantly at 0~24h (P < 0.05), decrease significantly at 48h (P < 0.05), and then increased rapidly at 72h until the end of the experiment (P < 0.05) (Figure 1B). The content of MDA increased continuously from the beginning to 24h, and decreased significantly at 48h (P < 0.05) and continued until the end of the experiment (Figure 1C). The content of HUN increased from the outset to 12h (P < 0.05), decreased from 24~48h (P < 0.05), and then showed an upward trend at 72h and continued until the end of the experiment (Figure 1D).
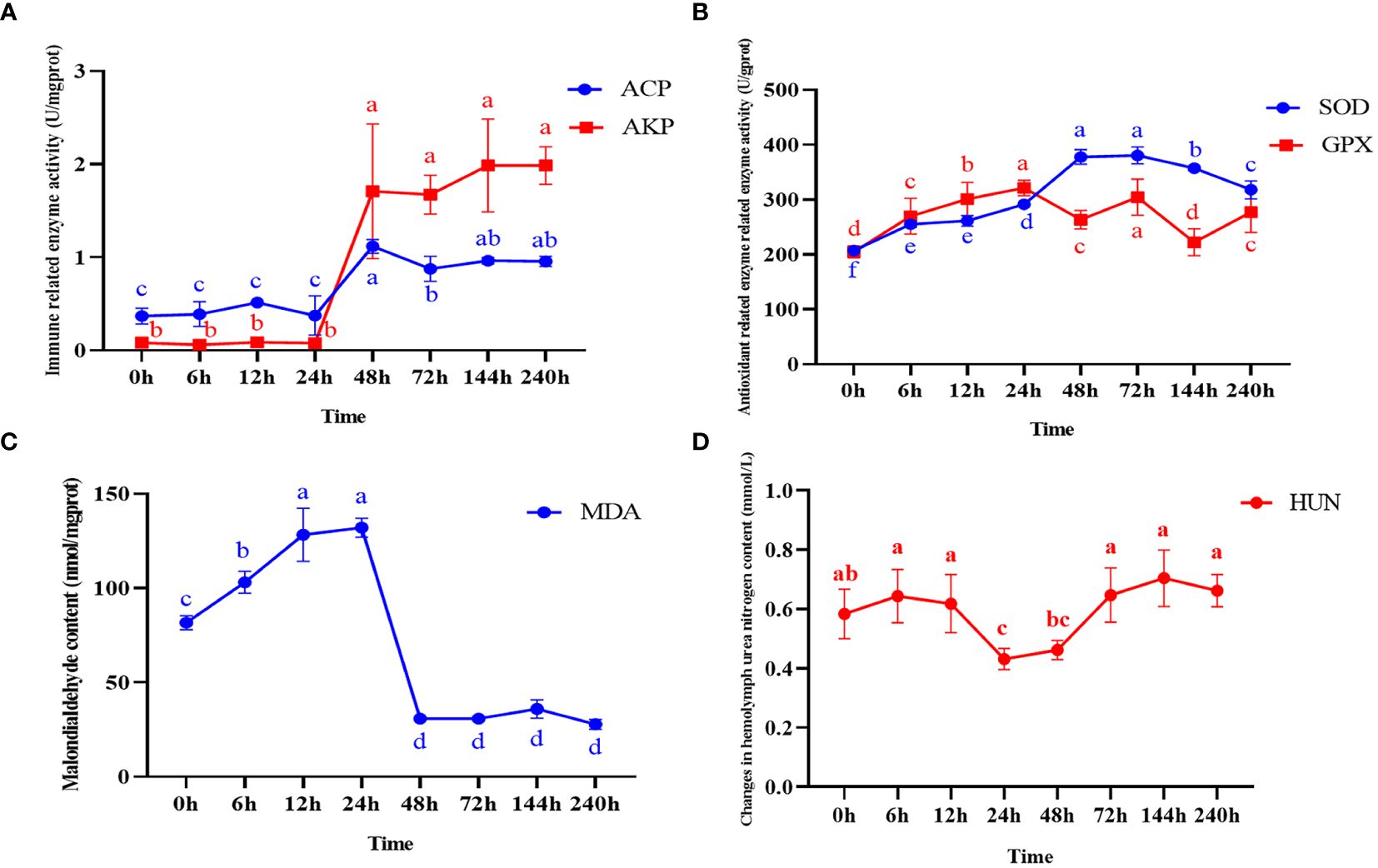
Figure 1 Enzyme activity and malondialdehyde content in hepatopancreas and changes in hemolymph urea nitrogen content after ammonia nitrogen stress. (A) Change in immune-related enzyme activity. (B) Antioxidant related enzyme activity changes. (C) Malondialdehyde content changes. (D) Changes in hemolymph urea nitrogen content. Values were expressed as mean ± S. (D) (N = 6). Data with different letters showed significant difference among tissues using one-way ANOVA (P < 0.05).
The changes of glucose and pyruvate in hepatopancreas and foot at different time after ammonia stress were shown in Figure 2. As the results shows, the glucose in the foot increased significantly at 0~48h (P < 0.05), and then decreased at 72h (P < 0.05) but was still significantly higher than the initial ammonia stress content (Figure 2A). Likewise, the content of pyruvate in the foot increased significantly at 0~72h (P < 0.05), reached the peak at 72h, and then showed a decreasing trend until the end of the experiment (Figure 2B). The glucose content in hepatopancreas increased gradually in the first 12h (P < 0.05), and then decreased briefly at 24h (P < 0.05) and then stabilized (Figure 2C). The content of pyruvate in hepatopancreas showed significantly high expression at 12h (P < 0.05) and 144h (P < 0.05), and the expression level in other time periods was generally the same, but still significantly higher than that at the initial stage of the experiment (Figure 2D).
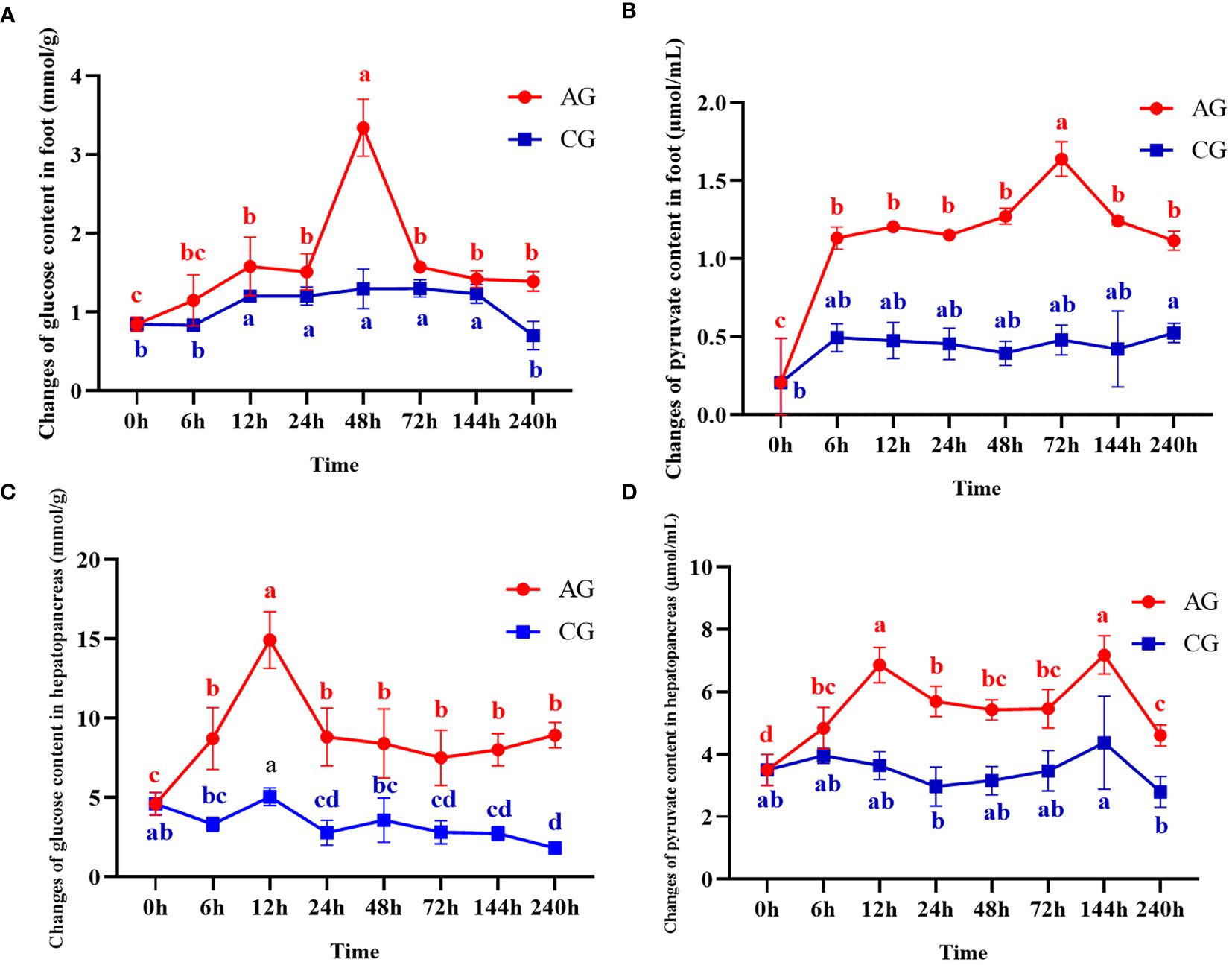
Figure 2 Changes of glucose and pyruvate contents after ammonia nitrogen stress. (A) Change of glucose content in foot. (B) Changes of pyruvate content in foot. (C) Change of glucose content in hepatopancreas. (D) Changes of pyruvate content in hepatopancreas. Values were expressed as mean ± S. (D) (N = 6). Data with different letters showed significant difference among tissues using one-way ANOVA (P < 0.05).
The data quality of each sample in this experiment was summarized in Supplementary Material 2 and the raw data were submitted to NCBI (accession number: PRJNA1083772, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1083772). A total of 44,339,646 clean reads (93.44% of raw data) were obtained from the reference transcriptome after low quality sequences filtering by pruning the sequencing adapters/poly-N. The G and C bases in clean data accounted for 40.25~42.84%. The percentage of Q30 bases was higher than 94.29% and the radio of mapped reads was all more than 79.15%. A total of 2,777 DEGs were identified between A1 and A10 (adjusted p-value ≤ 0.05 and absolute log2 fold change ≥ 1), including 1,283 upregulated genes and 1494 downregulated genes after ammonia nitrogen stress (Figure 3A).
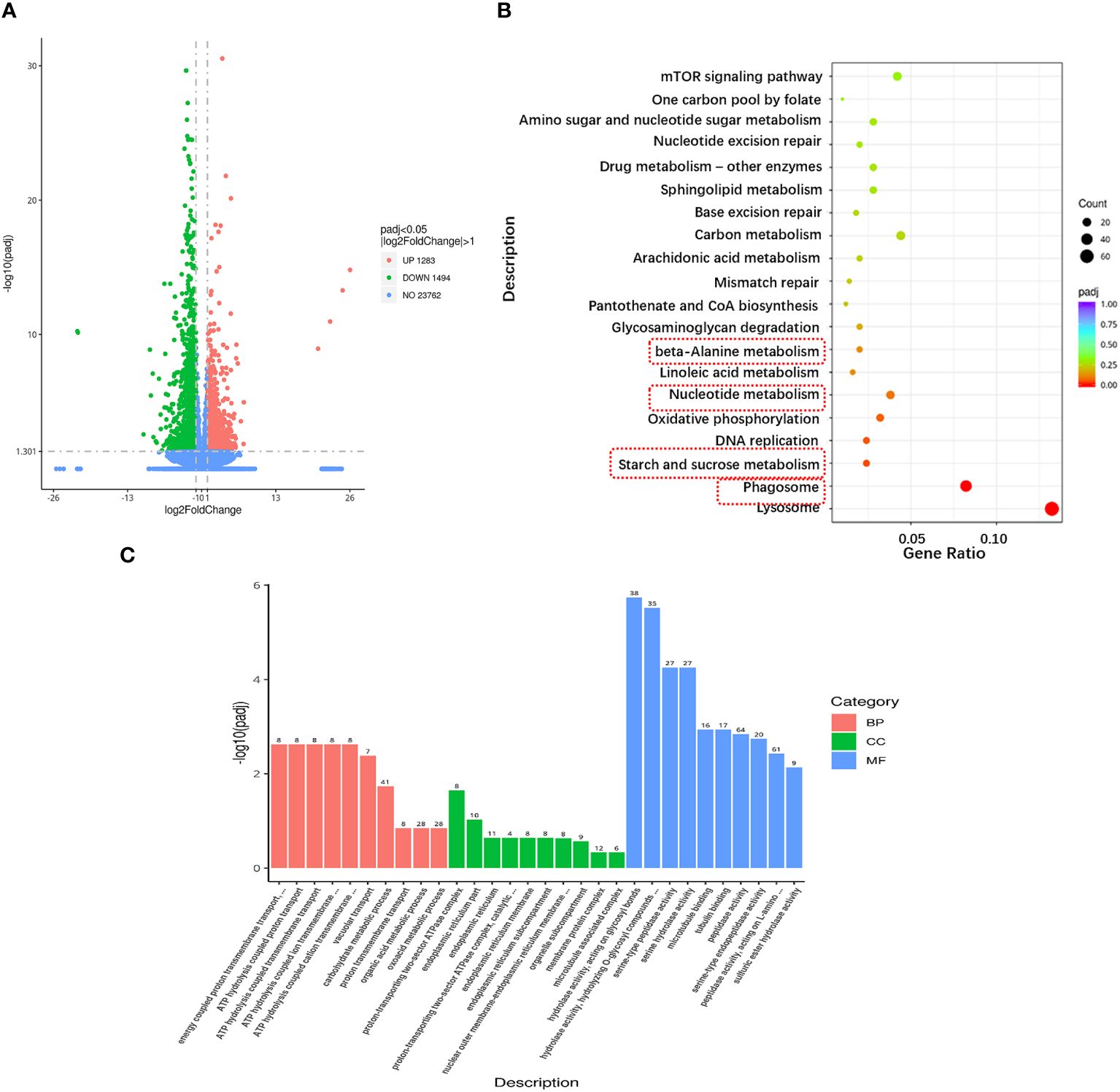
Figure 3 Transcriptional changes in hepatopancreas of the razor clam under ammonia nitrogen stress. (A) The volcano plot showing gene expression in hepatopancreas of the razor clam after ammonia nitrogen stress. Red and green dots represent significantly upregulated and downregulated genes, grey dots indicate no significant difference. (B) KEGG pathway enrichment analysis of the DEGs between A1 and A10. (C) GO enrichment analysis of the DEGs between A1 and A10.
To elucidate the biochemical pathways affected by ammonia nitrogen, DEGs were mapped with KEGG database (Figure 3B). Most DEGs were related to the “lysosome”, followed by “phagosome”, “starch and sucrose metabolism”, “DNA replication”, “oxidative phosphorylation”, “nucleotide metabolism” and “linoleic acid metabolism”. According to the NR annotation, most DEGs were involved in phagocytosis [hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) and C-type mannose receptor 2 (CMR2)], apoptosis (caspase recruitment domain (CARD) and inhibitor of apoptosis-promoting Bax1 (BI-1)), immune system (interleukin-17 (IL-17) and toll-like receptor 8 (TLR8)), oxidative damage (cytochrome P450 (CYP450) and glutathione peroxidase (GPX)) and molecular chaperones (heat shock protein beta-1 (HSPβ1), heat shock protein 70 (HSP70), heat shock protein 90 (HSP90)), and so on (Table 3). In addition, the genes involved in ammonia detoxification and ammonia excretion, including glutamate pyruvate transaminase2 (GPT2), xanthine dehydrogenase (XDH), xanthine oxidase (XO), carbonic anhydrase (CA), arginase 1 (Arg 1), ammonium transporter Rh type B (Rh-B), V-type proton ATPase (VHA) and Na+/H+ exchanger (NHE) were also changed significantly after ammonia exposure (Table 3). In order to further validate the transcriptome data, the expression levels of seven DEGs were detected by qRT-PCR, and the results showed similar expression patterns to those of RNA-seq (Figure 4).
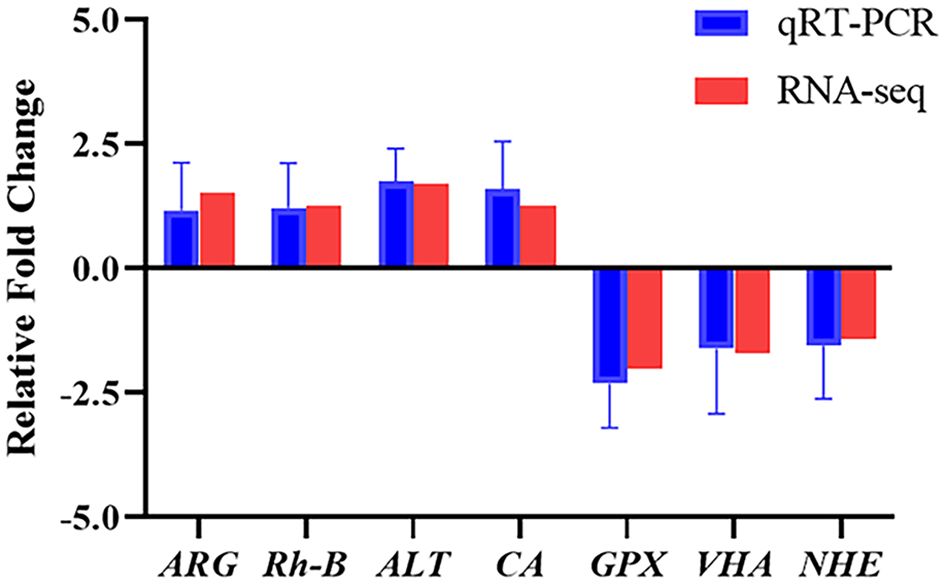
Figure 4 Expression of seven DEGs from the transcriptome analysis validated by qRT-PCR. Data were normalized to RS9 as the reference and presented as a fold to validate the transcriptomic analysis results. ARG, arginase 1; Rh-B, ammonium transporter Rh type B; ALT, alanine aminotransferase; CA, carbonic anhydrase; GPX, glutathione peroxidase; VHA, V-type proton ATPase; NHE, Na+/H+ exchanger.
To further understand the biological significance of DEGs, we classified the DEGs in hepatopancreas as GO and categorized them by cellular component, molecular function and biological process (Figure 3C). Of the three GO categories, the molecular functions were the largest group, followed by biology processes and cellular components. These terms in molecular functions mainly include “hydrolase activity, acting on glycosyl bonds”, “hydrolase activity, hydrolyzing O-glycosyl compounds”, “serine-type peptidase activity”, “serine hydrolase activity”, “microtubule binding”, “tubulin binding”, “peptidase activity”, “serine-type endopeptidase activity”, “peptidase activity, acting on L-amino acid peptides” and “sulfuric ester hydrolase activity”.
Untargeted metabolomics technology (LC-MS/MS) was used to investigate the metabolic changes in hepatopancreas of the razor clam, and 1,393 different metabolites were identified. The PCA result showed a significant separation (account for 29.52% of the variation) on PC1 between AG and CG (Figure 5A). The PLS-DA results of AG and CG, with R2Y=0.94 and Q2Y=-0.11, showed that the AG were clearly separated from the CG, also suggesting significant changes of metabolites between these two groups (Figure 5B). According to VIP > 1.0, FC > 1.2 or FC < 0.833 and P < 0.05, the differential metabolites were screened. In comparison group A10 and A1, 119 metabolites were significantly different, 61 metabolites were up-regulated and 58 metabolites were down-regulated. In addition, according to the classification coefficient, the model is stable and can be used to fitness and forecast (Figure 5C).
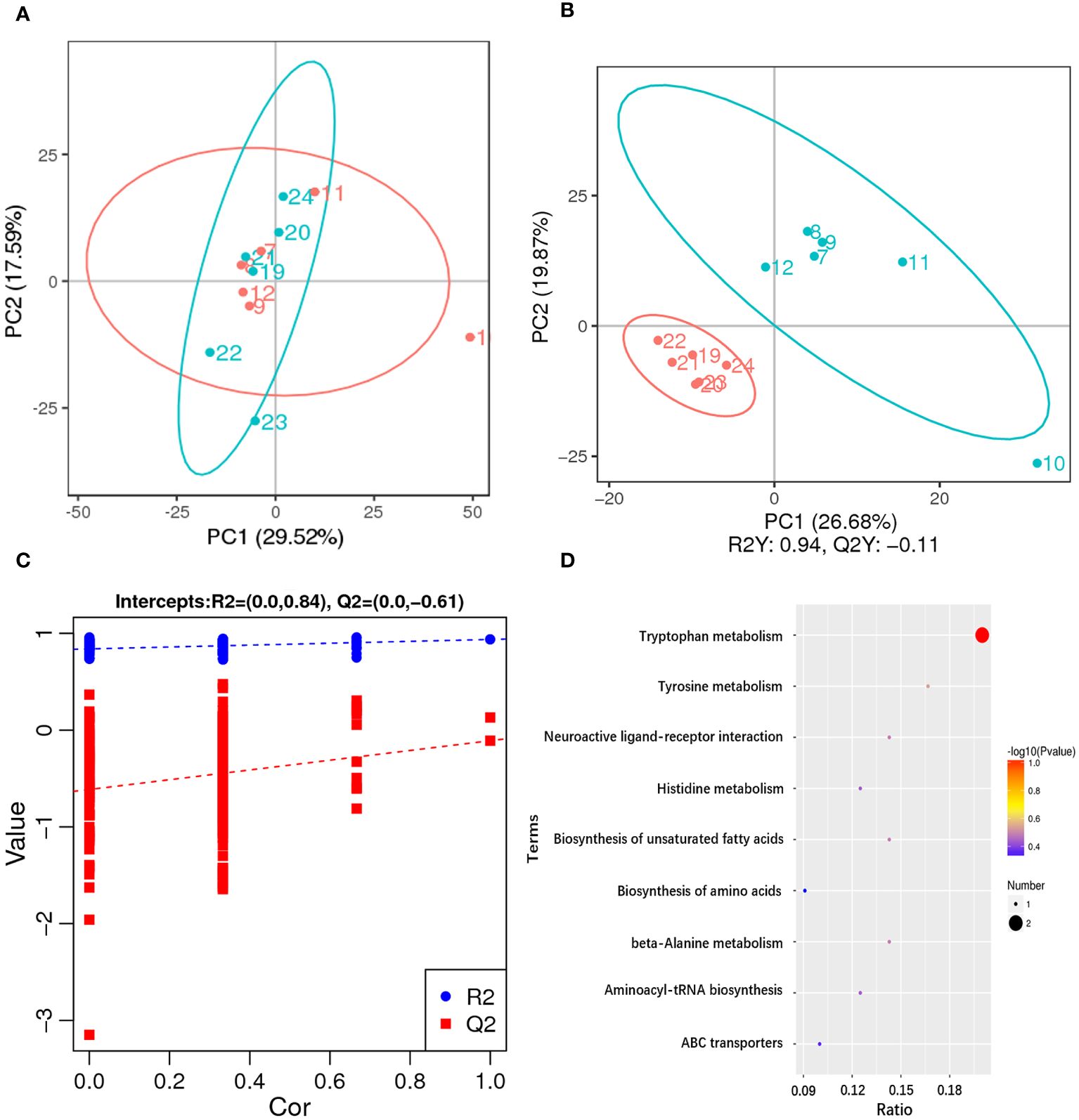
Figure 5 Metabolome changes in hepatopancreas of the razor clam after ammonia stress. (A) The PCA scores plot for the razor clam in CG (blue) and AG (red). (B) PLS-DA score plots for the razor clam in CG (blue) and AG (red). (C) PLS-DA permutation test. (D) KEGG enrichment analysis of differential metabolites.
Among these metabolites, the classes involved in “amino acid metabolism (tryptophan metabolism, histidine metabolism, tyrosine metabolism, cysteine and methionine metabolism, alanine, aspartate and glutamate metabolism, arginine biosynthesis and phenylalanine, tyrosine and tryptophan biosynthesis)”, “fatty acid metabolism (biosynthesis of unsaturated fatty acids, glycerophospholipid metabolism and alpha-linolenic acid metabolism)” and “carbohydrate metabolism (glyoxylate and dicarboxylate metabolism and citrate cycle)” were overrepresented (Figure 5D). Some amino acids including arginine, proline, histidine, lysine and so on, showed obvious increased, which was a response to ammonia stress. According to the results of transcriptome and metabolome, the related metabolites such as citrulline and ornithine increase significantly in alanine-glucose cycle and urea cycle, and the expression level of related gene (ARG, alanine aminotransferase (ALT), etc.), changes significantly, indicating that ammonia detoxification and metabolism occur in the body of razor clam after ammonia stress (Figure 6).
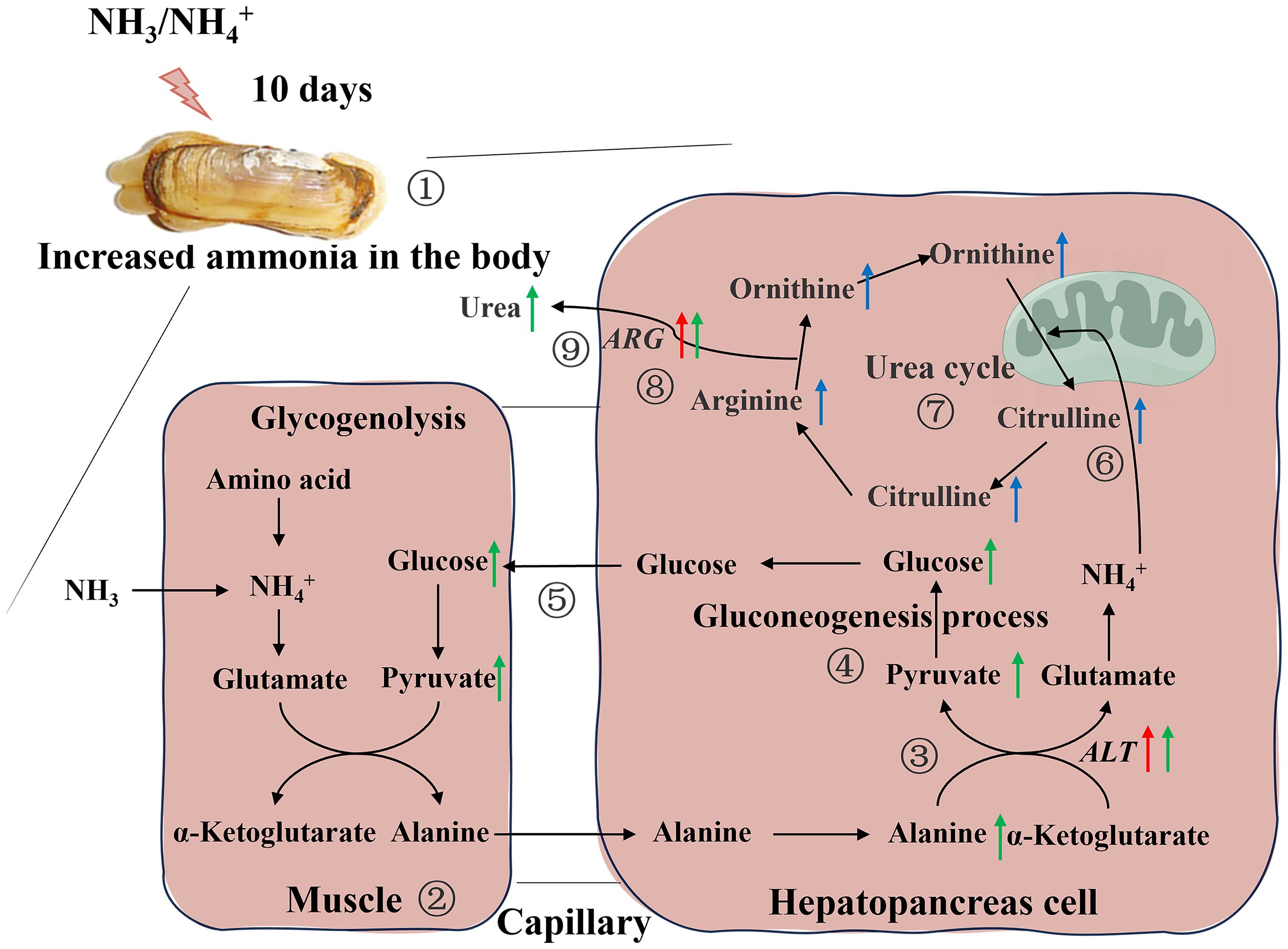
Figure 6 Schematic overview of the alteration in physiological parameters, transcriptome and metabolome in hepatopancreas of the razor clam after ammonia stress. Red, blue and green arrows represent changes in genes, metabolites, and physiological parameters, respectively. 1. The razor clam was exposed to ~46mg/L ammonia nitrogen environment for 10days, and the increase of stress time leads to an increase ammonia in the body; 2. In muscle cell, glucose was produced by glycolysis to pyruvate, and further to alanine; 3. Alanine enters the hepatopancreas through the hemolymph and was catalyzed by alanine aminotransferase (ALT) to produce pyruvate and glutamate; 4. Pyruvate was converted to glucose during gluconeogenesis process, which was reused by tissues and muscles; 5. The glucose from the hepatopancreas re-enters in the muscle cells and notably that four pathways, 2, 3, 4 and 5, constitute the alanine-glucose cycle; 6, 7, 8. The ammonia produced by glutamate metabolism in the alanine-glucose cycle enters the urea cycle and activates this cycle (the content of arginine and citrulline were significantly changed, and the mRNA expression of arginase (ARG) and hemolymph urea nitrogen were increased).
The high concentration of ammonia nitrogen in aquaculture water is a serious threat to the life and safety of aquatic organisms. Here, the studies investigated the toxicity of ammonia to marine aquatic invertebrates, such as Asian clam (Corbicula fluminea) (Zhang et al., 2019), Manila clam (Ruditapes philippinarum) (Cong et al., 2021), Chinese mitten crab (Eriocheir sinensis) (Wang et al., 2021) and Pacific white shrimp (Litopenaeus vannamei) (Cui et al., 2022). Among them, in our previous studies, the toxicity and tolerant mechanism to ammonia nitrogen in the razor clam were also discussed (Zhang et al., 2020a; Sun et al., 2021b; Hu et al., 2023), but there was no comprehensive and complete molecular mechanism to explain its high tolerance to ammonia nitrogen. In the current study, a combination of transcriptome, metabolome and physiological indicators were used to explore the molecular response mechanism of the razor clam to ammonia stress. Understanding the toxic mechanism of ammonia to the mollusks represented by razor clam is of great significance for marker-assisted breeding and breeding of ammonia-tolerant variety in the razor clam.
Lysosomes are organelles that receive and degrade macromolecules through endocytosis, phagocytosis and autophagy (Luzio et al., 2007). The phagocytosis of phagosomes plays an important role as an immune barrier against the entry of foreign harmful substances into the body. Exposure to 2.0 mg/L ammonia nitrogen for one week significantly reduced lysosomal integrity (up to 33%) in green-lipped mussel (Perna viridis) (Fang et al., 2008). Likewise, studies in R. philippinarum have shown that ammonia exposure could significantly reduce the integrity of lysosomes in a dose-dependent manner (Cong et al., 2017). In addition, studies on rockfish (Sebastes schlegelii) have also confirmed that ammonia exposure significantly reduces phagocytosis and lysosome activity (Kim et al., 2015). In this study, there were significantly changes in the lysosome and phagosome pathways (most of which were reduced in gene expression), which may further demonstrate the disruption of immune defenses following ammonia stress. Comparative transcriptomic analysis showed significant changes in several immune-related genes such as interleukin-17, toll-like receptor 8, toll-like receptor tollo and AKP. Similarly, in physiological experiments, the activity of AKP and ACP were also increased significantly in the comparison between A10 and A1. We speculated that the increased expression of immune-related genes is related to the destruction of the immune system of razor clam after prolonged exposure to ammonia, and similar studies have also confirmed this conjecture (Cui et al., 2017; Qi et al., 2017; Zhang et al., 2018).
Oxidative stress was one of the causes of cytotoxicity in aquatic animals caused by environmental stress such as temperature and ammonia (Zhang et al., 2020b). In this study, the activity of SOD increased due to ammonia exposure, while the activity of GPX increased first and then decreased. SOD breaks down superoxide (O2-) into oxygen (O2) and hydrogen peroxide (H2O2), and GPX breaks down H2O2 into O2 and H2O (Meng et al., 2021). With the extension of ammonia stress time, the increase of SOD activity can lead to the increase of H2O2, while the decrease of GPX activity can lead to accumulation of H2O2. Elevated levels of MDA after a period of ammonia exposure indicate that the accumulation of H2O2 levels can cause damage to lipids. Previous studies have shown that heat shock proteins 70 (HSP70) and HSP90 can play an important role in protecting the body from oxidative stress under environmental stress conditions (Liang et al., 2020). Our study found that ammonia exposure changed the transcription levels of HSP70 and HSP90 genes in the razor clam, similarly to the trend of HSP70 and HSP90 genes in mud eel (Takifugu obscurus) (Hangzo et al., 2017) and Chinese strip-necked turtle (Mauremys sinensis) (Liang et al., 2020) when exposed to ammonia nitrogen. In addition, oxidative stress may induce DNA damage (Meng et al., 2021). The expression of two important genes, proliferating cell nuclear antigen (PCNA) and growth arrest and DNA damage-inducible protein GADD45 alpha (GADD45α), were significantly increased between A1 and A10. PCNA is a homologous trimer protein that can participate in different pathways of DNA repair, including nucleotide and base excision repair, as well as mismatch repair (Scovassi and Prosperi, 2006). GADD45 family genes can regulate DNA damage in cell under various stress stimuli, and GADD45α is a key enzyme in the antioxidant defense system, and the direct interaction with other necessary replication factors can coordinate the promotion of nucleotide excision repair of damaged DNA (Siafakas and Richardson, 2009). The significant upregulation of these two genes suggests that ammonia exposure may cause DNA damage and trigger DNA repair mechanisms in the razor clam.
Some fish could excrete part of an ammonia load against high ambient levels. It appears that the giant mudskipper (Periophthalmodon schlosseri), is able to survive both high ammonia levels in the water and terrestrial exposure by actively excreting ammonium ions (Ip et al., 2001). And related studies have shown that ammonium ion clearance on the gills of giant mudskipper may be the result of a variety of ion transport genes (NKA, VHA, CA and NHE, etc.) (Randall and Tsui, 2002). While the mRNA expression levels of VHA and NHE were down-regulated in this study. This result seems unexpected, possibly because they mainly play an important role in the process of gill ammonia excretion, and similar results have been reported in crabs, other marine invertebrates and fish (Martin et al., 2011). Our previous results also confirmed that the mRNA expression levels of VHA and NHE were indeed increased in gills after ammonia nitrogen stress (Lv et al., 2022).
Ammonia in fish is mainly produced by amino acid metabolism, so they can reduce the production of ammonia by reducing amino acid metabolism to prevent the increase of ammonia concentration in the body. Some fish, such as pond loach (Misgurnus anguillicaudatus) (Chew et al., 2001; Tsui et al., 2002) and swamp eel (Monopterus albus) (Ip et al., 2004), reduce the ammonia content in the body by reducing the amino acid metabolism, which is one of the effective strategies to deal with ammonia nitrogen toxicity, and this strategy does not require the participation of external energy (Chew and Ip, 2014). However, this situation also inhibits the use of amino acids as an energy source by certain aquatic animals. Therefore, some fish (e.g., P. schlosseri) will metabolize some amino acids while inhibiting protein hydrolysis and amino acids metabolism to ensure the energy supply of the body (Ip et al., 2001). Certain amino acids (Glutamine, Arg, His, Pro, etc.) can be converted into Glu, and Glu under the action of transamination to produce α-ketoglutaric acid (α-KG), α-KG can be converted to malic acid through the Krebs cycle, malic acid is converted to pyruvate under the action of malic enzyme, pyruvate plus Glu under the action of ALT to produce Ala without the release of ammonia (Ip et al., 2001). If a continuous supply of pyruvate is available, transamination will continue to produce Ala to reduce internal contamination caused by endogenous ammonia produce in the body. In this study, amino acid metabolism in the transcriptome and metabolome of razor clam at A1 and A10 were significantly changed, and the contents of Arg, Pro, His and Lys in the differential metabolites were significantly up-regulated (P < 0.05), which was consistent with the content changes of Arg and Lys in the free amino acid content determination experiment. In addition, the content of Ala occupies the highest proportion in the determination of free amino acids, and it is also significantly up-regulated in A10 vs A1 (P < 0.05). What’s more, the content of pyruvate in hepatopancreas increased significantly after ammonia nitrogen stress. In conclusion, it can be speculated that the razor clam may reduce the ammonia concentration in the body by converting endogenous ammonia to Ala through partial amino acid metabolism.
In vertebrates, when adverse conditions (limited water availability, anaerobic metabolism, etc.) increase ammonia production or impair excretion/detoxification, alanine is produced in their extrahepatic tissues and is consumed within the liver as an NH4+ carrier as part of alanine-glucose cycle (also referred to as the Cahill cycle, Felig et al., 1970; Felig, 1973). In this cycle, glucose in the extrahepatic tissues is glycolyzed to produce pyruvate, which is then transaminated by ALT to form alanine. The newly synthesized alanine enters the liver with the blood for deamination, and the free ammonia is excreted through the urea cycle. The pyruvate in liver cells is converted to glucose by gluconeogenesis, and then transported to muscle by blood circulation, and then decomposed to produce pyruvate (Felig et al., 1970; Felig, 1973). In the present study, transcriptome results showed that the expression of genes related to glycogen metabolism (e.g., hexokinase, enolase 4, phosphoglucomutase and trehalase) was significantly upregulated in the comparison of A1 and A10 (P < 0.05). Interestingly, the expression levels of ALT and ARG genes were also significantly up-regulated in A1 and A10 (P < 0.05, this was confirmed by subsequent qRT-PCR results). Combined with the differential metabolites of A1 and A10, it was found that arginine, ornithine, citrulline and other metabolites related to urea cycle increased. Based on the above findings, it can be inferred that the body of razor clam may also reduce the accumulation of ammonia through the alanine-glucose cycle and urea cycle. To verify this hypothesis, the content of glucose, pyruvate in hepatopancreas and foot and HUN were measures after ammonia nitrogen stress. The results showed that the contents of glucose, pyruvate and HUN in different tissues increased significantly after ammonia nitrogen stress (P < 0.05). Similar results were found in mangrove crab (Helice formosensis), which may use a combination of urea cycle and ammonia detoxification to maintain nitrogen homeostasis in the presence of adverse factors (Allen et al., 2021). In addition, one study in Asian seabass (Lates calcariffer) also found a reciprocal relation between the muscle and liver of the fish to remove excess ammonia (Jahanbani et al., 2023). Based on our previous experimental results (including the functional verification of glutamate dehydrogenase, glutamine synthetase and glutaminase after ammonia nitrogen stress) (Zhang et al., 2020a; Sun et al., 2021a, Sun et al., 2021b; Lv et al., 2022), we hypothesized that the alanine-glucose cycle and synthesis of glutamine in the muscles and hepatopancreas of the razor clam manages and detoxify the increased ammonia by activating a number of enzymes and the urea cycle.
In conclusion, this study investigated the toxicity of hepatopancreas to ammonia and the tolerance mechanism of ammonia in the razor clam under ~46mg/L ammonia nitrogen stress for 1 and 10 days by means of physiological indicators, transcriptome and metabolome methods. The results showed that exposure to ammonia nitrogen could significantly reduce phagocytosis and lysosomal activity of razor clam, induce oxidative stress and activate innate immune response. Faced with the challenge of ammonia stress, the razor clam takes active measures to deal with and resist the toxic reaction caused by the increase of ammonia nitrogen stress time. According to our experimental results, the convert endogenous ammonia to alanine, the enhancement of alanine-glucose cycle and urea synthesis may be the possible mechanisms for the tolerance of razor clam to ammonia nitrogen. These results provide a new perspective for understanding the tolerance mechanism of ammonia nitrogen in benthic bivalves.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University, China. The study was conducted in accordance with the local legislation and institutional requirements.
GS: Data curation, Validation, Writing – original draft. LL: Funding acquisition, Writing – review & editing. HY: Funding acquisition, Writing – review & editing. ZL: Funding acquisition, Writing – review & editing. NX: Methodology, Writing – review & editing. YD: Methodology, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (32202918), Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069–7), Ningbo Major Project of Science and Technology (2021Z114) and National Marine Genetic Resource Center Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1444929/full#supplementary-material
Allen G. J. P., Wang M. C., Tseng Y. C., Weihrauch D. (2021). Effects of emersion on acid-base regulation, osmoregulation, and nitrogen physiology in the semi-terrestrial mangrove crab, Helice formosensis. J. Comp. Physiol. B. 191, 455–468. doi: 10.1007/s00360-021-01354-0
Bernasconi C., Uglow R. J. (2011). Purineolytic capacity response of Nephrops norvegicus to prolonged emersion: an ammonia detoxification process. Aquat. Biol. 11, 263–270. doi: 10.3354/ab00320
Chen J., Huang H., Xu H., He H., Fan W. (2010). Ammonia effects the survival and the energy budget of clam Meretrix meretrix. Mar. Sci. 34, 40–46. doi: 10.3724/SP.J.1077.2010.01263
Chew S. F., Ip Y. K. (2014). Excretory nitrogen metabolism and defence against ammonia toxicity in air-breathing fishes. J. Fish Biol. 84, 603–638. doi: 10.1111/jfb.12279
Chew S. F., Jin Y., Ip Y. K. (2001). The loach Misgurnus anguillicaudatus reduces amino acid catabolism and accumulates alanine and glutamine during aerial exposure. Physiol. Biochem. Zoology. 74, 226–237. doi: 10.1086/319663
Cong M., Li Y., Xu H., Lv J., Wu H., Zhao Y. (2021). Ammonia nitrogen exposure caused structural damages to gill mitochondria of clam Ruditapes philippinarum. Ecotoxicology Environ. Safety. 222, 112528. doi: 10.1016/j.ecoenv.2021.112528
Cong M., Wu H., Cao T., Ji C., Lv J. (2019). Effects of ammonia nitrogen on gill mitochondria in clam Ruditapes philippinarum. Environ. Toxicoloogy Pharmacol. 65, 46–52. doi: 10.1016/j.etap.2018.12.003
Cong M., Wu H., Yang H., Zhao J., Lv J. (2017). Gill damage and neurotoxicity of ammonia nitrogen on the clam Ruditapes philippinarum. Ecotoxicology. 26, 459–469. doi: 10.1007/s10646-017-1777-4
Cui Y., Ren X., Li J., Zhai Q., Feng Y., Xu Y., et al. (2017). Effects of ammonia-N stress on metabolic and immune function via the neuroendocrine system in Litopenaeus vannamei. Fish Shellfish Immunol. 64, 270–275. doi: 10.1016/j.fsi.2017.03.028
Cui Y., Zhao N., Wang C., Long J., Chen Y., Deng Z., et al. (2022). Acute ammonia stress-induced oxidative and heat shock responses modulated by transcription factors in Litopenaeus vannamei. Fish Shellfish Immunol. 128, 181–187. doi: 10.1016/j.fsi.2022.07.060
Dong Y., Zeng Q., Ren J., Yao H., Lv L., He L., et al. (2020). The chromosome-level genome assembly and comprehensive transcriptomes of the razor clam (Sinonovacula constricta). Front. Genet. 11. doi: 10.3389/fgene.2020.00664
Edwards S. L., Arnold J., Blair S. D., Pray M., Bradley R., Erikson O., et al. (2015). Ammonia excretion in the Atlantic hagfish (Myxine glutinosa) and responses of an Rhc glycoprotein. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 308, R769–R778. doi: 10.1152/ajpregu.00355.2014
Fang J. K., Wu R. S., Chan A. K., Yip C. K., Shin P. K. (2008). Influences of ammonia-nitrogen and dissolved oxygen on lysosomal integrity in green-lipped mussel Perna viridis: laboratory evaluation and field validation in Victoria harbour, Hongkong. Mar. pollut. Bulletin. 56, 2052–2058. doi: 10.1016/j.marpolbul.2008.08.003
Felig P. (1973). The glucose-alanine cycle. Metabolism 22, 179–207. doi: 10.1016/0026-0495(73)90269-2
Felig P., Pozefsky T., Marliss E., Cahill G. F. Jr. (1970). Alanine: key role in gluconeogenesis. Science. 167, 1003–1004. doi: 10.1126/science.167.3920.1003
Ge H., Liang X., Liu J., Cui Z., Guo L., Li L., et al. (2021). Effects of acute ammonia exposure on antioxidant and detoxification metabolism in clam Cyclina sinensis. Ecotoxicology Environ. Safety. 15, 211:111895. doi: 10.1016/j.ecoenv.2021.111895
Hangzo H., Banerjee B., Saha S., Saha N. (2017). Ammonia stress under high environmental ammonia induces Hsp70 and Hsp90 in the mud eel, Monopterus cuchia. Fish Physiol. Biochem. 43, 77–88. doi: 10.1007/s10695-016-0269-4
Hong M., Chen L., Sun X., Gu S., Zhang L., Chen Y. J. (2007). Metabolic and immune responses in Chinese mitten-handed crab (Eriocheir sinensis) juveniles exposed to elevated ambient ammonia. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 145, 363–369. doi: 10.1016/j.cbpc.2007.01.003
Hu C., Dai W., Zhu X., Yao H., Lin Z., Dong Y., et al. (2023). Expression and functional analysis of AMT1 gene responding to high ammonia stress in razor clam (Sinonovacula constricta). Anim. (Basel). 13, 1638. doi: 10.3390/ani13101638
Ip Y. K., Chew S. F. (2010). Ammonia production, excretion, toxicity, and defense in fish: a review. Front. Physiol. 1. doi: 10.3389/fphy.2010.00134
Ip Y. K., Lim C. B., Chew S. F., Wilson J. M., Randall D. J. (2001). Partial amino acid catabolism leading to the formation of alanine in Periophthalmodon schlosseri (mudskipper): a strategy that facilitates the use of amino acids as the energy source during locomotory activity on land. J. Exp. Biol. 204, 1615–1624. doi: 10.1242/jeb.204.9.1615
Ip Y. K., Tay A. S., Lee K. H., Chew S. F. (2004). Strategies for surviving high concentrations of environmental ammonia in the swamp eel Monopterus albus. Physiol. Biochem. Zoology. 77, 390–405. doi: 10.1086/383510
Jahanbani A., Shahriari A., Mohammadian T. (2023). Ureagenesis of Asian seabass (Lates calcarifer) under ammonia stress and overcrowding. Aquaculture. 576, 739810. doi: 10.1016/j.aquaculture.2023.739810
Kim S. H., Kim J. H., Park M. A., Hwang S. D., Kang J. C. (2015). The toxic effects of ammonia exposure on antioxidant and immune responses in rockfish, Sebastes schlegelii during thermal stress. Environ. Toxicol. Pharmacol. 40, 954–959. doi: 10.1016/j.etap.2015.10.006
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Liang L., Huang Z., Li N., Wang D., Ding L., Shi H., et al. (2020). Effects of ammonia exposure on antioxidant function, immune response and NF-κB pathway in Chinese strip-necked turtle (Mauremys sinensis). Aquat. Toxicology. 229, 105621. doi: 10.1016/j.aquatox.2020.105621
Liao Y., Smyth G. K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 230, 923–930. doi: 10.1093/bioinformatics/btt656
Luzio J. P., Pryor P. R., Bright N. A. (2007). Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632. doi: 10.1038/nrm2217.v
Lv L., Ren J., Zhang H., Sun C., Dong Y., Lin Z. (2022). Transcriptomic analysis of gill and hepatopancreas in razor clam (Sinonovacula constricta) exposed to acute ammonia. Front. Mar. Science. 9. doi: 10.3389/fmars.2022.832494
Martin M., Fehsenfeld S., Sourial M. M., Weihrauch D. (2011). Effects of high environmental ammonia on branchial ammonia excretion rates and tissue Rh-protein mRNA expression levels in seawater acclimated Dungeness crab Metacarcinus magister. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 160, 267–277. doi: 10.1016/j.cbpa.2011.06.012
Meng X., Jayasundara N., Zhang J., Ren X., Gao B., Li J., et al. (2021). Integrated physiological, transcriptome and metabolome analyses of the hepatopancreas of the female swimming crab Portunus trituberculatus under ammonia exposure. Ecotoxicology Environ. Safety. 228, 113026. doi: 10.1016/j.ecoenv.2021.113026
Pinto M. R., Lucena M. N., Faleiros R. O., Almeida E. A., McNamara J. C., Leone F. A. (2016). Effects of ammonia stress in the Amazon river shrimp Macrobrachium amazonicum (Decapoda, Palaemonidae). Aquat. Toxicology. 170, 13–23. doi: 10.1016/j.aquatox.2015.10.021
Qi X. Z., Xue M. Y., Yang S. B., Zha J. W., Wang G. X., Ling F. (2017). Ammonia exposure alters the expression of immune-related and antioxidant enzymes-related genes and the gut microbial community of crucian carp (Carassius auratus). Fish Shellfish Immunol. 70, 485–492. doi: 10.1016/j.fsi.2017.09.043
Randall D. J., Tsui T. K. (2002). Ammonia toxicity in fish. Mar. pollut. Bulletin. 45, 17–23. doi: 10.1016/S0025-326X(02)00227-8
Romano N., Zeng C. (2013). Toxic effects of ammonia, nitrite, and nitrate to decapod crustaceans: a review on factors influencing their toxicity, physiological consequences, and coping mechanisms. Rev. Fisheries Sci. Aquaculture. 21, 1–21. doi: 10.1080/10641262.2012.753404
Scovassi A. I., Prosperi E. (2006). Analysis of proliferating cell nuclear antigen (PCNA) associated with DNA. Methods Mol. Biol. 314, 457–475. doi: 10.1385/1-59259-973-7:457
Siafakas A. R., Richardson D. R. (2009). Growth arrest and DNA damage-45 alpha (GADD45α). Int. J. Biochem. Cell Biol. 41, 986–989. doi: 10.1016/j.biocel.2008.06.018
Sun G., Dong Y., Sun C., Yao H., Lin Z. (2021b). Vital role of glutamate dehydrogenase gene in ammonia detoxification and the association between its SNPs and ammonia tolerance in Sinonovacula constricta. Front. Physiol. 12. doi: 10.3389/fphys.2021.664804
Sun G., Sun C., He J., Yao H., Dai W., Lin Z., et al. (2021a). Characterizing the role of glutamine synthetase gene on ammonia nitrogen detoxification metabolism of the razor clam Sinonovacula constricta. Front. Mar. Science. 8. doi: 10.3389/fmars.2021.793118
The ministry of agriculture and fishery of the People’s Republic of China (2023). Chinese fishery statistical yearbook 2023 (Beijing: China Agriculture Press).
Tsui T. K., Randall D. J., Chew S. F., Jin Y., Wilson J. M., Ip Y. K. (2002). Accumulation of ammonia in the body and NH3 volatilization from alkaline regions of the body surface during ammonia loading and exposure to air in the weather loach. Misgurnus anguillicaudatus. J. Exp. Biol. 205, 651–659. doi: 10.1242/jeb.205.5.651
Veauvy C. M., McDonald M. D., Van Audekerke J., Vanhoutte G., Van Camp N., van der Linden A., et al. (2005). Ammonia affects brain nitrogen metabolism but not hydration status in the gulf toadfish (Opsanus beta). Aquat. Toxicol. 74, 32–46. doi: 10.1016/j.aquatox.2005.05.003
Wang T., Yang C., Zhang T., Liang H., Ma Y., Wu Z., et al. (2021). Immune defense, detoxification, and metabolic changes in juvenile Eriocheir sinensis exposed to acute ammonia. Aquat. Toxicology. 240, 105989. doi: 10.1016/j.aquatox.2021.105989
Want E. J., Masson P., Michopoulos F., Wilson I. D., Theodoridis G., Plumb R. S., et al. (2012). Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 8, 17–32. doi: 10.1038/nprot.2012.135
Weihrauch D., Ziegler A., Siebers D., Towle D. W. J. (2002). Active ammonia excretion across the gills of the green shore crab Carcinus maenas: participation of Na+/K+- ATPase, V-type H+-ATPase and functional microtubules. J. Exp. Biol. 205, 2765–2775. doi: 10.1242/jeb.205.18.2765
Yu G., Wang L. G., Han Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang M., Li M., Wang R., Qian Y. (2018). Effects of acute ammonia toxicity on oxidative stress, immune response and apoptosis of juvenile yellow catfish Pelteobagrus fulvidraco and the mitigation of exogenous taurine. Fish Shellfish Immunol. 79, 313–320. doi: 10.1016/j.fsi.2018.05.036
Zhang H., Sun G., Lin Z., Yao H., Dong Y. (2020a). The razor clam Sinonovacula constricta uses the strategy of conversion of toxic ammonia to glutamine in response to high environmental ammonia exposure. Mol. Biol. Rep. 47, 9579–9593. doi: 10.1007/s11033-02006018-w
Zhang T., Yan Z., Zheng X., Fan J., Wang S., Wei Y., et al. (2019). Transcriptome analysis of response mechanism to ammonia stress in Asian clam (Corbicula fluminea). Aquat. Toxicology. 214, 105235. doi: 10.1016/j.aquatox.2019.105235
Zhang T., Yan Z., Zheng X., Wang S., Fan J., Liu Z. (2020b). Effects of acute ammonia toxicity on oxidative stress, DNA damage and apoptosis in digestive gland and gill of Asian clam (Corbicula fluminea). Fish Shellfish Immunol. 99, 514–525. doi: 10.1016/j.fsi.2020.02.046
Zhao X., Fu J., Jiang L., Zhang W., Shao Y., Jin C., et al. (2018). Transcriptome-based identification of the optimal reference genes as internal controls for quantitative RT-PCR in razor clam (Sinonovacula constricta). Genes Genomics 40, 603–613. doi: 10.1007/s13258-018-0661-9
Zhao M., Yao D., Li S., Zhang Y., Aweya J. J. (2020). Effects of ammonia on shrimp physiology and immunity: a review. Rev. Aquaculture. 12, 2194–2211. doi: 10.1111/raq.12429
Keywords: Sinonovacula constricta, ammonia challenge, multi-omics, alanine-glucose cycle, adaptive strategies
Citation: Sun G, Lv L, Yao H, Lin Z, Xu N and Dong Y (2024) Integrated application of multi-omics and biochemical analysis revealed the physiological response mechanism of ammonia nitrogen tolerance in the razor clam (Sinonovacula constricta). Front. Mar. Sci. 11:1444929. doi: 10.3389/fmars.2024.1444929
Received: 06 June 2024; Accepted: 17 July 2024;
Published: 01 August 2024.
Edited by:
Yiming Li, Fishery Machinery and Instrument Research Institute, ChinaReviewed by:
Basanta Kumar Das, Central Inland Fisheries Research Institute (ICAR), IndiaCopyright © 2024 Sun, Lv, Yao, Lin, Xu and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinghui Dong, ZG9uZ3lpbmdodWkxMThAMTI2LmNvbQ==; Nianjun Xu, eHVuaWFuanVuQG5idS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.