- 1Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture and Rural Affairs, Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 2Department of Pediatric, Shanghai Sixth People’s Hospital, Shanghai, China
- 3Science and Technology Service Center, Ministry of Science and Technology, Shanghai Mugao Biotechnology Co., Ltd., Shanghai, China
Hyriopsis cumingii is a major freshwater pearl mussel in the world. In order to investigate the functional roles of the growth- and mineralization-related genes involved in the pearl formation process after the inserting nucleus into the mantle tissue of H. cumingii, we conducted a transcriptome analysis of data from different time periods after the pearl-nucleus insertion. We screened a total of 1,898 growth-related unigenes and 716 mineralization-related unigenes, including 12 growth-related differential genes and eight mineralization-related differential genes that showed sustained differential expression throughout the pearl formation. The results of the gene expression patterns among samples at different time points showed that the Mcon group and the M05d group clustered together, the M50d group and the M90d group clustered together, while the M20d group clustered separately. The overall density distribution of differential gene analysis at different periods after pearl-nucleus insertion of H. cumingii, cluster analysis, differential gene analysis, differential gene differential, and co-expression analysis together revealed that growth and mineralization-related genes have the same expression pattern and jointly regulate pearl formation. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment revealed that the same functional cluster and pathway were together present on the enrichment results of both growth- and mineralization-related genes, suggesting a synergistic effect between growth and mineralization. We found that genes such as bone morphogenetic proteins (BMPs) and calmodulin (CALM) are jointly involved in growth and mineralization processes. The study indicate that growth genes cooperate with mineralization genes to play biological roles in the early process of pearl formation in H. cumingii.
1 Introduction
The formation of pearls is a result of the intricate biomineralization process, and the artificial culture industry of saltwater and freshwater pearls is now widespread worldwide, with China accounting for more than 95% of the world’s freshwater pearl production (Siddique et al., 2024). The synergistic effect of the growth and mineralization of mantle is very important in the cultivation of pearls, by affecting the size, luster, and color of pearls (Blay et al., 2018; Le Luyer et al., 2019), but mechanisms are still unclear. Therefore, the research of the mechanisms involved in the formation of pearls are very necessary.
In artificial pearl cultivation, small pieces of the mantle tissue from donor mussels are inserted into the mantle of recipient mussels together with the nucleus of the pearl. After that, the mantle cells divide and proliferate quickly to form a pearl sac composed of single-layer columnar cells, and the pearl sac secretes bio-minerals continuously, thus forming a pearl (McDougall et al., 2013; Mariom et al., 2019). When a foreign matter enters the mantle cavity of the mussel, it will cause an immuno-inflammatory reactions of the mussel, and the mussel’s own immune rejection in the first week promotes the rapid division and proliferation of its own mantle cells in order to wrap the foreign matter and wound healing (Bai et al., 2016; Huang et al., 2019; Wu et al., 2022). The fifth day is the critical period of the pre-existing mantle cell division and formation. At 20 days, the complete structure of the pearl capsule has been formed (McDougall et al., 2013), cell division slows down, and the pearl capsule starts to secrete nacre with the help of its deposition (Zhang et al., 2018). At 50 days, the pearl layer starts to be deposited stably. At 90 days, the complete pearl layer has been formed on the surface of the pearl nucleus (Wang et al., 2022). Therefore, days 5, 20, 50, and 90 were selected for this study.
Some of the genes and signaling pathways related to growth and mineralization have been identified in marine pearl mussels in previous studies. The structural domain of epidermal growth factor (EGF), a shell matrix proteins (SMPs) key structural domain that affects the biomineralization of pearl mussels, has been identified in Crassostrea gigas (Shimizu et al., 2022). Meanwhile, it has been shown that both insulin-like growth factor IGF1 and its binding proteins, IGFBP1/5, are able to promote growth of the mantle and shell formation in C. gigas (Gricourt et al., 2003). Growth factor IGFs are involved in key regulatory mechanisms of the biomineralization in Pinctada martensii (Zheng et al., 2023). Bone-forming proteins (BMPs) were found in marine shellfish to regulate their biomineralization, as found in P. martensii and C. gigas (Du et al., 2017). Growth critical pathways such as NF-κB signaling pathway and Notch signaling pathway, which are found in vertebrates, have also been found to regulate biomineralization in marine shellfish (Auffret et al., 2020; Shuai et al., 2023). Shell matrix proteins (SMPs) play an important role in their biomineralization process (Jin et al., 2019). In the freshwater mussel Hyriopsis cumingii, 26 matrix proteins have been identified (Bai et al., 2021). It was found that many important matrix protein genes involved in pearl formation related to biomineralization, such as hic31 (Liu et al., 2015), which is involved in the formation of prismatic layer, and hic24 (Liu et al., 2019), which is involved in the formation of the nacreous layer, are expressed at high levels during the period of rapid division of epithelial cells prior to the formation of the pearl capsule. In addition, silkmaxin, a matrix protein gene involved in pearl layer formation, was also highly expressed in the mantle during the first 8–15 days after nuclear insertion (Jin et al., 2019). However, molecular studies related to growth and mineralization of marine and freshwater shellfish are both still very limited, and a large number of molecular mechanisms have yet to be discovered.
Hyriopsis cumingii is an important freshwater pearl mussel, which is widely cultured due to the excellent quality of the pearls that it produces. In addition, its pearls account for 80% of the world’s total freshwater pearl production value (Siddique et al., 2024), which is of great research value. In this study, H. cumingii was used as a research object to screen and analyze the role of genes related to growth and mineralization during the pearl formation through the transcriptome data constructed after the pearl-nucleus insertion of the mantle. It is expected that this study can enrich the biological theory of pearl formation and provide information for promoting the sustainable development and technological innovation of pearl culture.
2 Materials and methods
2.1 Experimental animals and sample collection
A total of 250 H. cumingii (2-year-old: length, 12±2 cm; width, 7±1 cm; height, 2.5±0.8 cm; weight, 225.23±30.77 g) of uniform size were selected (purchased from Weimin Pearl Farm, Jinhua City, Zhejiang Province, China), 50 mussels in the control group (without pearl-nucleus insertion) and 200 mussels in the experimental group. In the experimental group, pearl-nucleus insertion was carried out in the posterior part of the mantle of H. cumingii using a pearl-nucleus and a small piece of mantle tissue from the donor mussel (2 mm × 2 mm) (Jin et al., 2019). After the nucleation surgery, the mussels were temporarily housed in the 120-L circulating water aquarium in the laboratory at a temperature of 26°C, dissolved oxygen of 7–8 mg/L, pH 7.8, and normal light, and chlorella was fed twice a day. All experimental animals were cultured under the same conditions (Jin et al., 2019).
Tissue samples of control group and experimental group were obtained from the site of the pearl-nucleus insertion of the mantle in H. cumingii at days 5, 20, 50, and 90, and preserved in liquid nitrogen for subsequent experiments.
2.2 Transcriptome sequencing
The total RNA of the mantle tissue samples was extracted according to the optimized Trizol reagent RNA extraction process (Rio et al., 2010). The degradation and contamination of the extracted RNA was determined by gel electrophoresis. The concentration and purity of RNA were measured by a spectrophotometer (NanoDropND-2000); the samples with OD values (260/280) of 1.8–2.0 and OD (260/230) over 2.0 were considered to be qualified. The integrity of RNA was measured by Agilent Bioanalyzer 2100. After the samples were qualified, the RNA was mixed in equal amounts and used for transcriptome sequencing, which was carried out by Hangzhou Lianchuan Biotechnology Co., Ltd.
The total RNA was extracted from the mantle samples with different times mentioned above after pearl-nucleus insertion, the quality of the samples was checked by the above methods, and the transcriptome data were constructed by sequencing using Illumina HiSeq 2000.
2.3 Data mining and processing methods
The transcriptome data were evaluated for quality, and then data were assembled. The Unigene sequences obtained after filtering out the low-quality data splicing were compared with the databases such as Nr, Nt, Pfam, KOG/COG, Swiss-Prot, KEGG, and GO and annotated with the gene functions to get the target database that we wanted. Then, the bioinformatics correlation analysis was carried out.
In this paper, transcriptome sequenced data were used as reference library, unigenes associated with growth and mineralization were screened according to their annotation contents, and transcriptomic analysis was performed on the growth and mineralization related data.
We used the data analysis and visualization bioinformatics cloud platform (https://www.bioinformatics.com.cn) to plot the scatterplot of differential gene expression density distribution (Tang et al., 2023) and used the pheatmap package in R version 4.1.1 (https://www.r-project.org/) to perform heatmapping of growth, and mineralization-related gene expression data were heatmapped and the clustering of samples in the heatmap was observed. Differential gene analysis was performed after normalizing the data using the edgeR package (Robinson et al., 2010), setting the false discovery rate FDR < 0.05, log2FoldChange >1.5 or <−1.5 as differential genes. The obtained differential data were subjected to GO and KEGG enrichment analysis using the clusterProfiler package (Yu et al., 2012; Wu et al., 2021) and using the bioinformatics cloud platform for visualization. Differential and co-expression visualization of differential genes of different comparative groups was performed using TBtools (v2.085) (Chen et al., 2023) to analyze the shared and unique genes.
2.4 qRT-PCR of growth- and mineralization-related partial genes
To verify the reliability of the differential gene analysis results, we verified seven differential genes related to growth and mineralization. The genes are insulin-like growth factor 1 (IGF1), insulin-like growth factor 2 binding protein (IGF2BP), bone morphogenetic protein and activin membrane-bound inhibitor (BAMBI), bone morphogenetic protein 2/4 (BMP2_4), bone morphogenetic protein 5 (BMP5), bone morphogenetic protein 9/10 (BMP9_10), bone morphogenetic protein receptor type-2 (BMPR2), calmodulin (CALM), and calcium/calmodulin-dependent protein kinase II (CAMK2). We used Primer 5 to design specific primers for these genes (Table 1), in which EF1α was the internal reference (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). The amplification was performed in triplicate on a Bio-Rad CFX96 (Bio-Rad, Hercules, CA, USA) using TB Green ® Premix Ex Taq ™ (TaKaRa, Tokyo, Japan). Cycling parameters were 95°C for 5 min, then 36 cycles of 95°C for 5 s and 57°C for 20 s.
There were three biological replicates for each sample. Quantitative data were calculated by 2−ΔΔCT value, and data were shown mean ± se. IBM SPSS Statistics 27 was used for single-factor ANOVA analysis. The between-group comparisons were analyzed using Duncan’s multiple comparisons, and p <0.05 was considered statistically significant. GraphPad Prism 8 was used for histogram plotting.
3 Results and analysis
3.1 Screening and analysis of growth and mineralization transcriptome data after pearl-nucleus insertion
The transcriptome data of the mantle of H. cumingii was filtered and spliced to obtain 257,457 Unigenes, and a total of 223,345 Unigenes were annotated by comparing with the databases of Nr, Nt, Pfam, Swissprot, KO, GO, and KOG. We constructed a database of 1,898 growth-related Unigenes and 716 mineralization-related Unigenes based on the annotation information, pathway information, and keywords related to growth (including growth, cell division, and cell proliferation) and mineralization (including bone, bone morphogenetic protein, and mineralization).
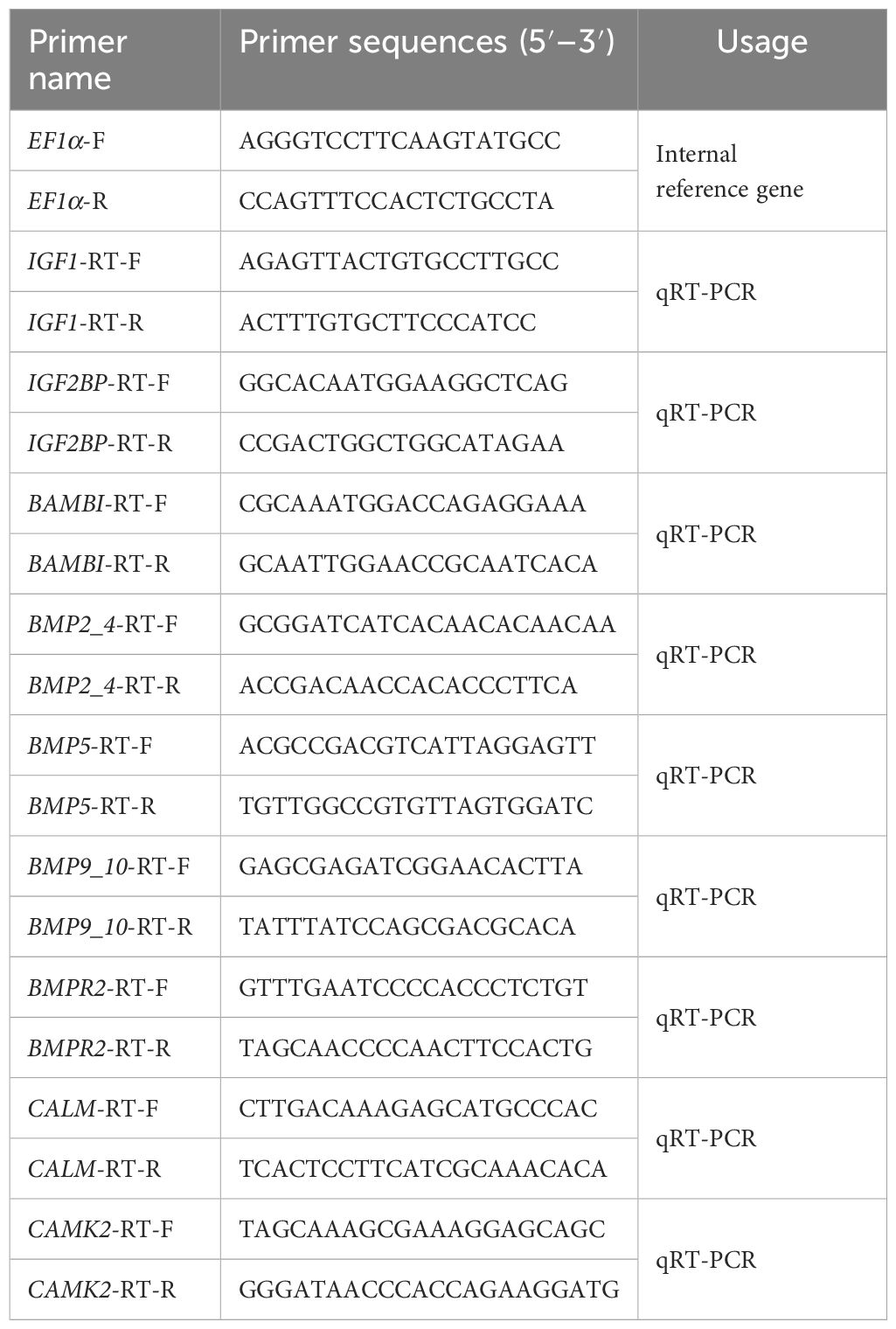
The results of Unigenes expression visualization of the above databases (Figure 1A) showed that the expression of growth- and mineralization-related genes was concentrated within 2,000, some genes had more than 2,000 expressions, and there were some differences in the expression of the samples in different periods, which showed dynamic changes. In addition, differential gene expression analysis of growth- and mineralization-related genes showed similar trends in overall density. After pearl-nucleus insertion, the highly expressed genes in the other groups increased significantly compared with the control group.
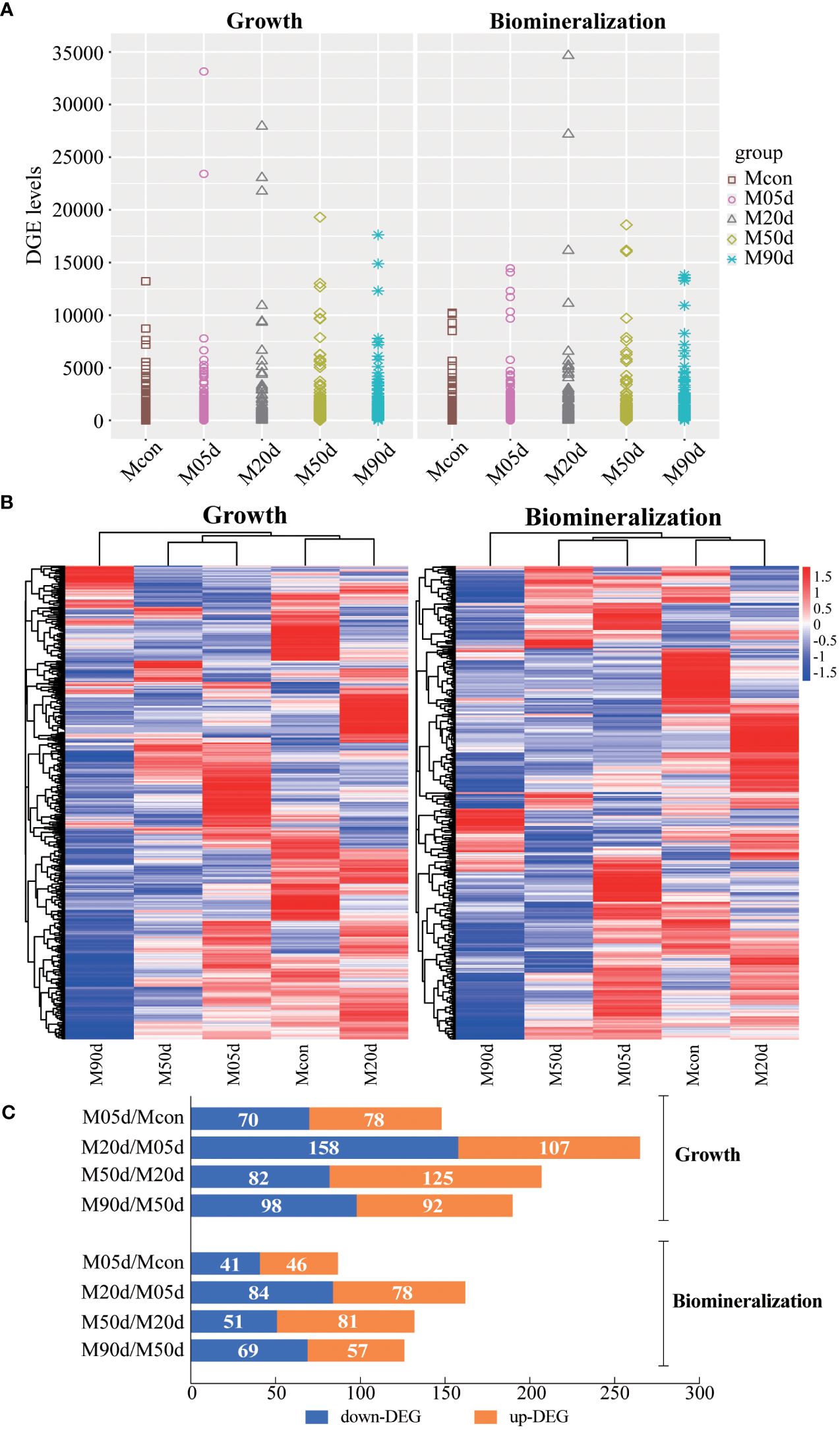
Figure 1 (A) Trend analysis of differential gene expression density distribution. (left) growth-related genes, (right) mineralization-related genes; horizontal coordinates are samples of different days, Mcon group (control group), M05d group (day 5 after pearl-nucleus insertion), M20 d group (day 20 after pearl-nucleus insertion), M50d group (day 50 after pearl-nucleus insertion), and M90d group (day 90 after pearl-nucleus insertion). (B) Clustering analysis of mantle growth and mineralization-related genes. (left) clustering heatmap of growth-related genes, (right) clustering heatmap of mineralization-related genes. Red color represents highly expressed genes, and blue color represents lowly expressed genes. (C) Differential gene expression analysis of mantle growth and mineralization during pearl formation. The horizontal coordinate is the number of differential genes; orange is upregulated genes, and blue is downregulated genes.
3.2 Clustering analysis of genes related to mantle growth and mineralization during pearl formation
As shown in Figure 1B, Mcon group and M5d group clustered together, M50d group and M90d group clustered together, and M20d group clustered separately for genes related to growth and mineralization of the mantle, indicating that the expression patterns of genes related to mantle growth and mineralization were the same during the same time period. Differential expression analysis of mantle growth and mineralization gene groups during pearl formation was performed.
Differential expression analysis of growth and mineralization gene groups of the mantle after pearl-nucleus insertion for different experimental days in H. cumingii is shown in Figure 1C. The number trend of differential genes screened in the mantle growth and mineralization gene groups was of similarity. The M05d/Mcon group was screened for the lowest number of differential genes, and the ratio of up- and downregulation of growth and mineralization genes was close to 1:1. The M20d/M05d group had the most differential genes and had more downregulated than upregulated genes. Among growth-related genes, 148 differential genes were found in the M05d/Mcon comparison group. Compared with the Mcon group, 78 genes were upregulated and 70 genes were downregulated in the M05d group. There were 265 differential genes in the M20d/M05d comparison group. Compared with M05d, 107 genes were upregulated and 158 genes were downregulated in the M20d group. There were 207 differential genes in the M50d/M20d comparison group. Compared with M20d, 125 genes were upregulated and 82 genes were downregulated in the M50d group. There were 190 differential genes in the M90d/M50d comparison group. Compared with M50d, 92 genes were upregulated and 98 genes were downregulated in M90d. Among mineralization-related genes, 87 differential genes were found in the M05d/Mcon comparison group. Compared with the Mcon group, 46 genes were upregulated and 41 genes were downregulated in the M05d group. A total of 162 differential genes were found in the M20d/M05d comparison group. Compared with M05d, 78 genes were upregulated and 84 genes were downregulated in M20d. There were 132 differential genes in the M50d/M20d comparison group. Compared with M20d, 81 genes were upregulated and 51 genes were downregulated in the M50d group. There were 126 differential genes in the M90d/M50d comparison group. Compared with M50d, 57 genes were upregulated and 69 genes were downregulated in M90d group.
3.3 Differential gene GO enrichment analysis of mantle growth and mineralization genes during pearl formation
The GO enrichment results of the comparison groups at different time periods after nucleus insertion (Figure 2) indicate that the biological processes (including cell division, serine family amino acid metabolic process, cell motility, growth cell proliferation, and chromosome segregation), the cellular component (including motile cilium), and the molecular function (including growth factor activity) remained highly enriched in growth-related genes throughout the pearl formation process. The biological processes (including calcium ion transport and ossification), the cellular component (including proteinaceous extracellular matrix), and the molecular function (including motor activity) remained highly enriched in mineralization-related genes throughout the pearl formation process. Interestingly, we found the presence of growth-related GO terms among the GO terms enriched in mineralization-related differential genes at different times after pearl-nucleus insertion, suggesting their involvement in the biomineralization and growth processes (the detailed enrichment GO term is shown in Supplementary Table S1).
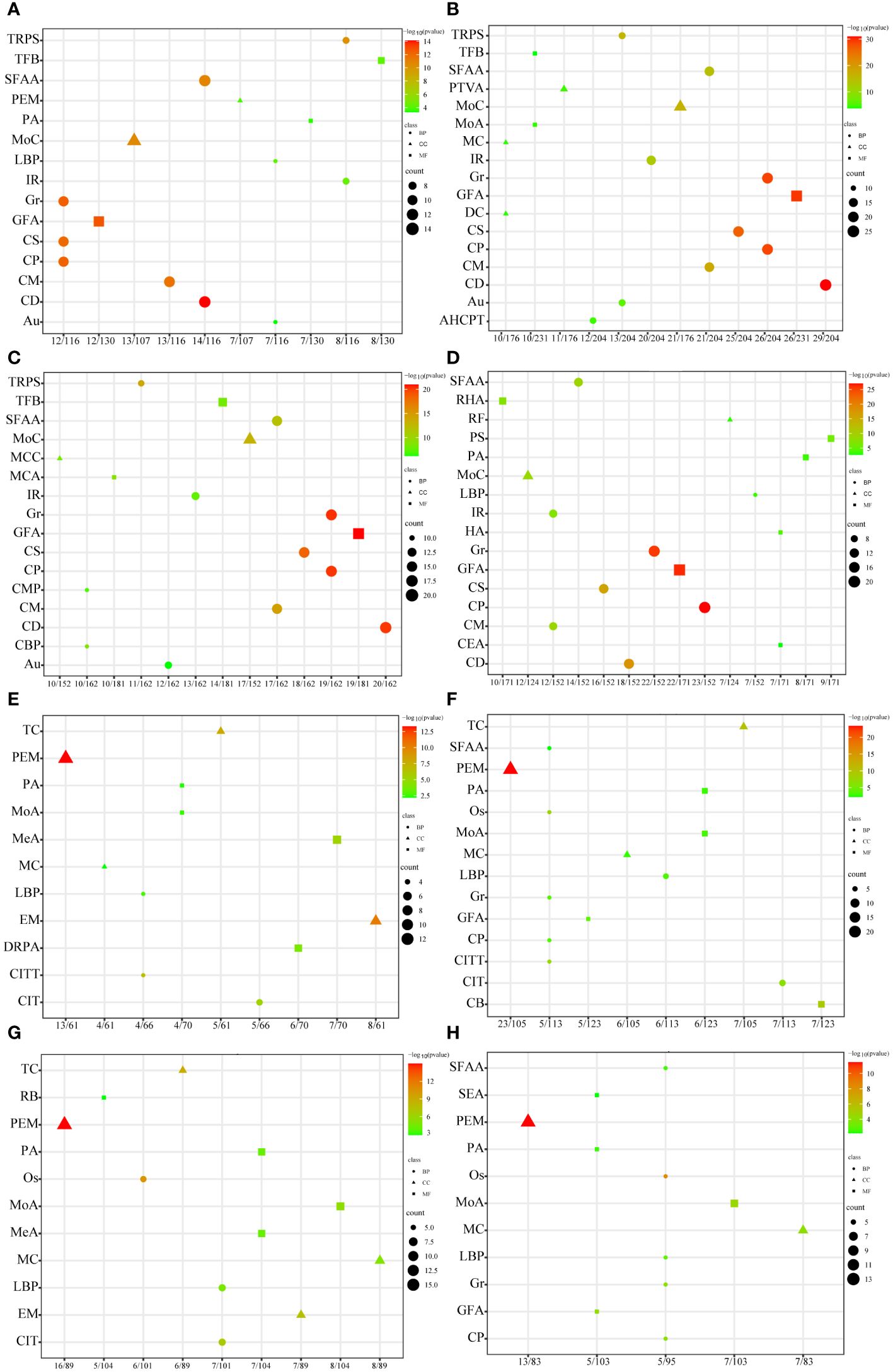
Figure 2 Differential gene GO enrichment analysis of growth and mineralization genes during pearl formation. Horizontal coordinate is the ratio of enriched genes; the vertical coordinate is the enriched GO term; from green to red represents the p-adjust value from large to small, i.e., −log10(p-adjust value) from small to large; the triangular, circular, and square dots in the figure represent BP, CC, and MF, respectively; and the size of dots represents the number of enriched genes. (A) Growth-related genes of the M05d group compared to the Mcon group, (B) growth-related genes of the M20d group compared to the M05d group, (C) growth-related genes of the M50d group compared to the M20d group, (D) growth-related genes of the M90d group compared to the M50d group; (E) mineralization-related genes of the M05d group compared to the Mcon group; (F) mineralization-related genes of the M20d group compared to the M05d group; (G) mineralization-related genes of the M50d group compared to the M20d group; (H) mineralization-related genes of the M90d group compared to the M50d group. Au, autophagy; CB, calmodulin binding; CD, cytokinesis; CIT, calcium transport; CITT, calcium transmembrane transport; CM, cell motility; CP, cell proliferation; CS, chromosome segregation; DR, DNA orientation RNA polymerase activity; DRPA, RNA polymerase activity; EM, extracellular matrix; GFA, growth factor activity; Gr, growth; IR, immune response; LBP, lipopolysaccharide biosynthesis process; MC, myosin complex; MoC, motile cilia; MeA, metalloendopeptidase activity; MoA, motility activity; Os, ossification; PA, phosphotransferase activity, alcohol group as receptor; PEM, protein extracellular matrix; RB, receptor binding; SEA, serine-type endopeptidase activity; SFAA, serine family amino acid metabolism; TC, troponin complex; TFB, transcription factor binding; TRPS, transmembrane receptor protein serine/threonine kinase signaling pathway; AHCPT, ATP hydrolysis-coupled proton translocation; PTVA, proton translocating V ATPase, V0 structural domain; DC, dynamin complex; RF, replication fork; RHA, RNA helicase activity; PS, protein serine/threonine kinase activity; HA, hydrolytic enzyme activity acting on acid anhydride; CEA, cysteine-type endopeptidase activity.
3.4 KEGG enrichment analysis of differential genes during pearl formation
As shown in Figure 3, growth- and mineralization-related genes showed a dynamic trend in pathway enrichment in the different days comparison groups, and there were a significant number of pathways that continued to be highly enriched throughout the pearl formation, including Ubiquitin-mediated proteolysis, NF-kappa B signaling pathway, PI3K-Akt signaling pathway, MAPK signaling pathway, apoptosis, oocyte meiosis, cell cycle, NOD-like receptor signaling pathway, neurotrophin signaling pathway, calcium signaling pathway, phosphatidylinositol signaling system, gastric acid secretion, and circadian entrainment. (The detailed pathway information is shown in the Supplementary Table S2). The information of these pathways was further categorized into five groups, namely, Genetic Information Processing, Environmental Information Processing, Cellular Processes, and Organismal Systems and Metabolism. The number of genes annotated in the apoptosis pathway was the highest among the growth-related genes, and the number of genes annotated in the calcium signaling pathway was the highest among the mineralization-related genes.
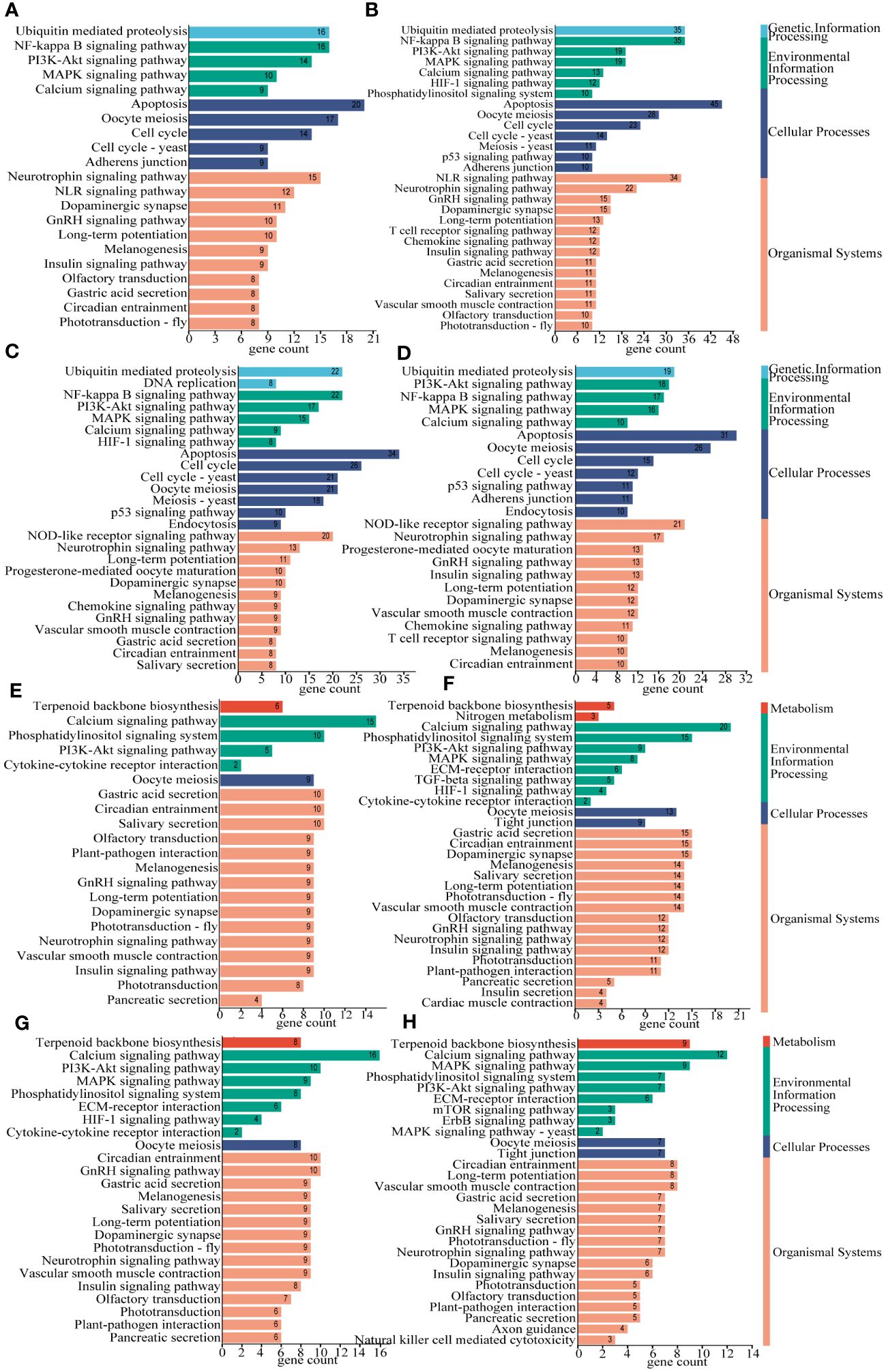
Figure 3 KEGG enrichment analysis of mantle growth and mineralization differential genes during pearl formation. The horizontal coordinate is the number of genes, the vertical coordinate is the pathway, and different colors represent the categories of pathways. (A) Growth-related genes of the M05d group compared with the Mcon group, (B) growth-related genes of the M20d group compared with the M05d group, (C) growth-related genes of the M50d group compared with the M20d group, (D) growth-related genes of the M90d group compared with the M50d group; (E) mineralization-related genes of the M05d group compared with the Mcon group, (F) mineralization-related genes of the M20d group compared with the M05d group, (G) mineralization-related genes of the M50d group compared with the M20d group, (H) mineralization-related genes of the M90d group compared with the M50d group.
3.5 Analysis of co-expression patterns of differential genes in different groups during pearl formation
The differential genes in four groups were analyzed for differential and co-expression (Figure 4A). Comparison with the process of pearl-nucleus insertion revealed that among the growth-related genes, there were 47 independent differential genes in the M05d/Mcon comparison group. There were 104 independent differential genes in the M20d/M05d comparison group, and it is the largest number of unique differential genes. There were 53 independent differential genes in the M50d/M20d comparison group and 80 independent differential genes in the M90d/M50d comparison group. Among the mineralization-related genes, there were 23 independent differential genes in the M05d/Mcon comparison group. There were 52 independent differential genes in the M20d/M05d comparison group, and it is still the largest number of unique differential genes. There were 27 independent differential genes in the M50d/M20d comparison group and 39 independent differential genes in the M90d/M50d comparison group. Among all the differential genes, the co-expressed differential genes were 12 growth-related genes and eight mineralization-related genes, and the specific pathways are shown in Supplementary Table S3.
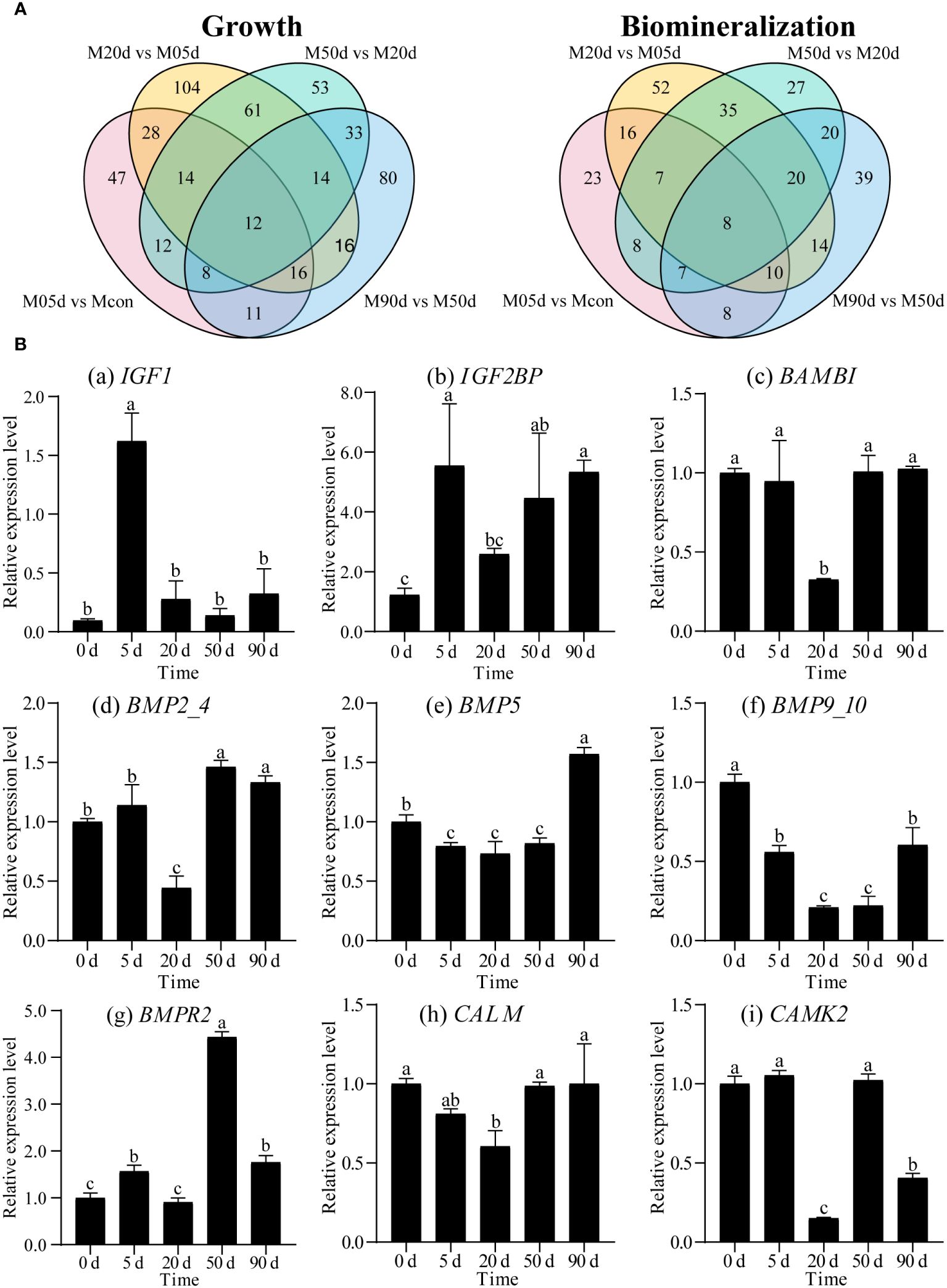
Figure 4 (A) Expression pattern analysis of differentially differentiated genes for mantle growth and mineralization during pearl formation. (left) Venn diagram of growth-related differential genes between samples with different days of mantle after insertion, (right) Venn diagram of mineralization-related differential genes between samples with different days of mantle after insertion. (B) Relative expression levels of genes related to growth and mineralization. Different letters indicate significant differences (p<0.05). (a)-(i) respectively represent the relative expression level of IGF1, IGF2BP, BAMBI, BMP2_4, BMP5, BMP9_10, BMPR2, CALM and CAMK2. The horizontal coordinate is the different time after nucleation.
Through co-expression analysis, we found that there were some genes in the mantle during pearl formation that were related to both growth and mineralization, and their names and pathway information involved in them are listed in Table 2.
3.6 Expression validation of mineralization- and growth-related partial genes
Based on differential genes expression data, we used qRT-PCR to verify nine differential genes associated with growth and mineralization (Figure 4B). The results showed that the expression of nine genes in different periods of nucleation in the mantle was consistent with differential gene data analysis. Among all the genes validated, the relative expression level of the 20d showed the most significant difference compared to the other groups. The relative expression levels of different genes showed dynamic changes at different intercalation periods.
4 Discussion
Pearl formation involves a series of complex biological processes, among which the functions played by growth- and mineralization-related genes are crucial. In this study, we investigated the roles of growth- and mineralization-related genes in different developmental stages of H. cumingii after pearl-nucleus insertion. Among them, Wang et al. have completed the observation experiments on HE sections of the pearl sac at different periods of nucleus insertion (Wang et al., 2022). It is consistent with the analysis results in this article, providing us with a more comprehensive and in-depth understanding.
The results of differential gene analysis density distribution showed the similarity of the overall distribution of growth- and mineralization-related differential genes. The results of clustering heat map analysis showed that the growth- and mineralization-related genes both clustered together in the M5d group and Mcon group after insertion of nuclei, the M50d group and the M90d group after insertion of nuclei, and the M20d group clustered alone after insertion of nuclei. During the first 2 weeks of pearl sac formation epithelial cells proliferate and differentiate rapidly, with elevated levels of expression of growth-related genes such as epidermal growth factor, and high expression of mineralization-associated matrix protein genes such as Nacrein and Pif during this time (Mariom et al., 2019). In this study, the results of differential gene analysis showed that the number of growth- and mineralization-related differential genes also had a consistent trend in different periods after pearl-nucleus insertion. In addition, the differential expression of growth- and mineralization-related genes at the 20th day after nucleation insertion was the largest among these five different time points of nuclei insertion, suggesting that the 20th day after nuclei insertion is a critical period for pearl formation. Studies have shown that in the pre-nucleation period, the mantle shows immune rejection and rapid cell proliferation, and after 20 days, the mantle cells form a pearl sac and begin to secrete nacre for biomineralization (Li et al., 2022; Wang et al., 2022; Shen et al., 2023). The number of differential genes in the growth and mineralization of the M50d group compared with the M20d group was less compared to the M20d group compared with the M5d group, but the numbers of their upregulated genes were all increasing and were the most numerous among the four comparative groups. Previous studies have shown that the inflammatory response in pearl sac formation is reduced in this period, cellular stress is weakened, and cellular proliferative capacity and biomineralization are stabilized (Li et al., 2022). The results of the above analysis revealed the similarity in the expression patterns of growth- and mineralization-related genes, providing evidence for their synergistic regulation of pearl formation.
The GO and KEGG enrichment analyses were further performed, and the results showed that numerous biological functions and biological pathways were significantly enriched at all stages after the pearl-nucleus insertion in the mantle, indicating the biological complexity of the pearl formation process. Among them, cell division, growth, cell proliferation, and chromosome segregation were highly enriched in growth-related differential genes; proteinaceous extracellular matrix and calcium ion transport were highly enriched in mineralization-related differential genes. In addition, we found that some of the growth-related GO terms were also present in the GO enrichment of mineralization-related genes. These results indicate that growth- and mineralization-related genes play important roles in the pearl formation process of H. cumingii and further confirm that growth- and mineralization-related genes synergistically regulate the pearl formation. MAPK signaling pathway, PI3K-Akt signaling pathway, NF-kappa B signaling pathway, cell cycle, oocyte meiosis, and apoptosis are widely enriched in growth-related genes. These pathways are involved in cell division, proliferation, migration, damage repair, and apoptosis (Chen et al., 2001; Cobrinik, 2005; Yang et al., 2003; Engelman et al., 2016; Haccard and Jessus, 2006; Oeckinghaus et al., 2011; Savitskaya and Onishchenko, 2015). Calcium signaling pathway, terpenoid backbone biosynthesis, ECM–receptor interaction, and other pathways are widely enriched in mineralization-related genes and enriched in mineralization-related genes, and these pathways are involved in biological functions such as storage of calcium ion, calcium ions stimulate contraction, and synthesis of different backbones and adhesion, and are involved in cell–cell interactions, formation of extracellular matrix, calcium-associated proteins, and skeleton (Berridge et al., 2003; Bosman and Stamenkovic, 2003; Martin, 2003). In higher organisms, MAPK and PI3K-Akt signaling pathways promote cell proliferation and apoptosis (Ornitz and Itoh, 2015; Zhao et al., 2021), in addition to bone formation, which is associated with biomineralization (Sun et al., 2020; Nagai et al., 2023). Previous studies have found that the NF-κB signaling pathway was found to be able to influence the formation and deposition of CaCO3 in Pinctada fucata through the regulation of matrix proteins such as Nacrein, which is involved in the regeneration of shells (Shuai et al., 2023) and in the formation of pearls (Sun et al., 2015). The MAPK pathway and PI3K/AKT pathway are activated by insulin-like peptides in P. fucata martensii to regulate the cell cycle and cell activity (Zhang and He, 2020). In H. cumingii, IGFs have been found to be involved in the PI3K/AKT pathway to regulate the downstream gene Cyclin D2, which is involved in the growth of the mantle cells (Feng et al., 2022; Li et al., 2022). Meanwhile, this study found that MAPK and PI3K-Akt signaling pathways were both enriched in growth and mineralization, suggesting that they play important roles in the post-nucleation growth and mineralization of the mussel. Differential and co-expression analyses of differential genes showed that both growth- and mineralization-related genes had the highest number of unique differential genes at 20 days after insertion of nuclei, which play a key role in co-regulating pearl formation. The above analyses indicated that growth- and mineralization-related genes acted synergistically in regulating pearl formation.
5 Conclusion
In conclusion, we investigated the expression patterns of growth and mineralization genes and their biological functions in different periods after the pearl-nucleus insertion in the mantle tissue of H. cumingii by transcriptome analysis, and the overall density distribution of differential gene expression analysis, clustering analysis, differential gene analysis, and differential gene differential and co-expression analysis.
Our study reveals the key role and importance of synergistic regulation of genes related to growth and mineralization in pearl formation and further provides more biological basis for the development of artificial pearl cultivation and breeding.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA992764, https://www.ncbi.nlm.nih.gov/, SRR25206151 to SRR25206163.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
YC: Investigation, Writing – original draft, Writing – review & editing. YY: Data curation, Software, Writing – review & editing. XS: Conceptualization, Writing – review & editing. YF: Writing – review & editing. XB: Methodology, Project administration, Supervision, Writing – review & editing. WL: Methodology, Project administration, Supervision, Writing – review & editing. SL: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2022YFD2400105), the earmarked fund for China Agriculture Research System of MOF and MARA (CARS-49), the National Natural Science Foundation of China (No. 31201991).
Acknowledgments
Thanks to Prof. Zhiyi Bai and Prof. Jiale Li for providing this experimental animal (Hyriopsis cumingii).
Conflict of interest
Author SL was employed by Shanghai Mugao Biotechnology Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1443863/full#supplementary-material
References
Auffret P., Le Luyer J., Sham Koua M., Quillien V., Ky C. L. (2020). Tracing key genes associated with the Pinctada margaritifera albino phenotype from juvenile to cultured pearl harvest stages using multiple whole transcriptome sequencing. BMC Genomics 21, 662. doi: 10.1186/s12864-020-07015-w
Bai Z., Zhao L., Chen X., Li Q., Li J. (2016). A galectin from Hyriopsis cumingii involved in the innate immune response against to pathogenic microorganism and its expression profiling during pearl sac formation. Fish Shellfish Immunol. 56, 127–135. doi: 10.1016/j.fsi.2016.07.006
Bai Z. Y., Yuan L., Liu X. J., Li J. L. (2021). Research progress of matrix proteins in Hyriopsis cumingii. J. Fisheries China 45, 982–991. doi: 10.11964/jfc.20200612302
Berridge M. J., Bootman M. D., Roderick H. L. (2003). Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529. doi: 10.1038/nrm1155
Blay C., Planes S., Ky C. L. (2018). Cultured pearl surface quality profiling by the shell matrix protein gene expression in the biomineralised pearl sac tissue of pinctada margaritifera. Mar. Biotechnol. (NY) 20, 490–501. doi: 10.1007/s10126-018-9811-y
Bosman F. T., Stamenkovic I. (2003). Functional structure and composition of the extracellular matrix. J. Pathol. 200, 423–428. doi: 10.1002/path.1437
Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., et al. (2001). MAP kinases. Chem. Rev. 101, 2449–2476. doi: 10.1021/cr000241p
Chen C., Wu Y., Li J., Wang X., Zeng Z., Xu J., et al. (2023). TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cobrinik D. (2005). Pocket proteins and cell cycle control. Oncogene 24, 2796–2809. doi: 10.1038/sj.onc.1208619
Du X., Fan G., Jiao Y., Zhang H., Guo X., Huang R., et al. (2017). The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 6, 1–12. doi: 10.1093/gigascience/gix059
Engelman J. A., Luo J., Cantley L. C. (2016). The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619. doi: 10.1038/nrg1879
Feng S., Wang H., Li X., Li W., Bai Z. (2022). Gene identification and functional analysis of a D-type cyclin (CCND2) in freshwater pearl mussel (Hyriopsis cumingii). Mol. Biol. Rep. 49, 6601–6611. doi: 10.1007/s11033-022-07501-2
Gricourt L., Bonnec G., Boujard D., Mathieu M., Kellner K. (2003). Insulin-like system and growth regulation in the Pacific oyster Crassostrea gigas: hrIGF-1 effect on protein synthesis of mantle edge cells and expression of an homologous insulin receptor-related receptor. Gen. Comp. Endocrinol. 134, 44–56. doi: 10.1016/S0016-6480(03)00217-X
Haccard O., Jessus C. (2006). Oocyte maturation, Mos and cyclins–a matter of synthesis: two functionally redundant ways to induce meiotic maturation. Cell Cycle 5, 1152–1159. doi: 10.4161/cc.5.11.2800
Huang D., Shen J., Li J., Bai Z. (2019). Integrated transcriptome analysis of immunological responses in the pearl sac of the triangle sail mussel (Hyriopsis cumingii) after mantle implantation. Fish Shellfish Immunol. 90, 385–394. doi: 10.1016/j.fsi.2019.05.012
Jin C., Zhao J. Y., Liu X. J., Li J. L. (2019). Expressions of shell matrix protein genes in the pearl sac and its correlation with pearl weight in the first 6 months of pearl formation in hyriopsis cumingii. Mar. Biotechnol. (NY) 21, 240–249. doi: 10.1007/s10126-019-09876-z
Le Luyer J., Auffret P., Quillien V., Leclerc N., Reisser C., Vidal-Dupiol J., et al. (2019). Whole transcriptome sequencing and biomineralization gene architecture associated with cultured pearl quality traits in the pearl oyster, Pinctada margaritifera. BMC Genomics 20, 111. doi: 10.1186/s12864-019-5443-5
Li X., Feng S., Xuan X., Wang H., Shen X., Chen Y., et al. (2022). A proteomic approach reveals biomineralization and immune response for mantle to pearl sac in the freshwater pearl mussel (Hyriopsis cumingii). Fish Shellfish Immunol. 127, 788–796. doi: 10.1016/j.fsi.2022.06.057
Liu X., Liu Z., Jin C., Li H., Li J. L. (2019). A novel nacre matrix protein hic24 in Hyriopsis cumingii is essential for calcium carbonate nucleation and involved in pearl formation. Biotechnol. Appl. Biochem. 66, 14–20. doi: 10.1002/bab.1690
Liu X., Zeng S., Dong S., Jin C., Li J. (2015). A novel matrix protein Hic31 from the prismatic layer of hyriopsis cumingii displays a collagen-like structure. PloS One 10, e0135123. doi: 10.1371/journal.pone.0135123
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mariom, Take S., Igarashi Y., Yoshitake K., Asakawa S., Maeyama K., et al. (2019). Gene expression profiles at different stages for formation of pearl sac and pearl in the pearl oyster Pinctada fucata. BMC Genomics 20, 240. doi: 10.1186/s12864-019-5579-3
Martin P. T. (2003). Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology 13, 55R–66R. doi: 10.1093/glycob/cwg076
McDougall C., Aguilera F., Moase P., Lucas J. S., Degnan B. M. (2013). Pearls. Curr. Biol. 23, R671–R673. doi: 10.1016/j.cub.2013.05.042
Nagai T., Sekimoto T., Kurogi S., Ohta T., Miyazaki S., Yamaguchi Y., et al. (2023). Tmem161a regulates bone formation and bone strength through the P38 MAPK pathway. Sci. Rep. 13, 14639. doi: 10.1038/s41598-023-41837-4
Oeckinghaus A., Hayden M. S., Ghosh S. (2011). Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 12, 695–708. doi: 10.1038/ni.2065
Ornitz D. M., Itoh N. (2015). The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev. Dev. Biol. 4, 215–266. doi: 10.1002/wdev.176
Rio D. C., Ares M. Jr., Hannon G. J., Nilsen T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, pdb prot5439. doi: 10.1101/pdb.prot5439
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Savitskaya M. A., Onishchenko G. E. (2015). Mechanisms of apoptosis. Biochem. (Mosc) 80, 1393–1405. doi: 10.1134/S0006297915110012
Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shen X., Chen Y., Jia L., He W., Chen X., Chen Y., et al. (2023). Transcriptome and digital gene expression analysis reveal immune responses of mantle and visceral mass pearl culturing in Hyriopsis cumingii. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1251251
Shimizu K., Takeuchi T., Negishi L., Kurumizaka H., Kuriyama I., Endo K., et al. (2022). Evolution of epidermal growth factor (EGF)-like and zona pellucida domains containing shell matrix proteins in mollusks. Mol. Biol. Evol. 39, msac148. doi: 10.1093/molbev/msac148
Shuai B., Deng T., Xie L., Zhang R. (2023). The regulatory effect of NF-kappaB signaling pathway on biomineralization and shell regeneration in pearl oyster, Pinctada fucata. Int. J. Biol. Macromol 253, 126956. doi: 10.1016/j.ijbiomac.2023.126956
Siddique M. F., Haque M. A., Barman A. C., Tanu M. B., Shahjahan M., Uddin M. J. (2024). Freshwater pearl culture in Bangladesh: Current status and prospects. Heliyon 10, e29023. doi: 10.1016/j.heliyon.2024.e29023
Sun K., Luo J., Guo J., Yao X., Jing X., Guo F. (2020). The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage 28, 400–409. doi: 10.1016/j.joca.2020.02.027
Sun J., Xu G., Wang Z., Li Q., Cui Y., Xie L., et al. (2015). The effect of NF-kappaB signalling pathway on expression and regulation of nacrein in pearl oyster, pinctada fucata. PloS One 10, e0131711. doi: 10.1371/journal.pone.0131711
Tang D., Chen M., Huang X., Zhang G., Zeng L., Zhang G., et al. (2023). SRplot: A free online platform for data visualization and graphing. PloS One 18, e0294236. doi: 10.1371/journal.pone.0294236
Wang H., Huang K., Feng S., Li X., Wang L., Xuan X., et al. (2022). Effect on physiological responses and cultured pearl biology of different nuclei insertion positions in the visceral mass of freshwater pearl mussel (Hyriopsis cumingii). Aquaculture Res. 53, 6782–6796. doi: 10.1111/are.16145
Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141. doi: 10.1016/j.xinn.2021.100141
Wu H., Yang C., Hao R., Liao Y., Wang Q., Deng Y. (2022). Lipidomic insights into the immune response and pearl formation in transplanted pearl oyster Pinctada fucata martensii. Front. Immunol. 13. doi: 10.3389/fimmu.2022.1018423
Yang S. H., Sharrocks A. D., Whitmarsh A. J. (2003). Transcriptional regulation by the MAP kinase signaling cascades. Gene 320, 3–21. doi: 10.1016/S0378-1119(03)00816-3
Yu G., Wang L. G., Han Y., He Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang H., He M. (2020). The role of a new insulin-like peptide in the pearl oyster Pinctada fucata martensii. Sci. Rep. 10, 433. doi: 10.1038/s41598-019-57329-3
Zhang R., Qin M., Shi J., Tan L., Xu J., Tian Z., et al. (2018). Molecular cloning and characterization of Pif gene from pearl mussel, Hyriopsis cumingii, and the gene expression analysis during pearl formation. 3 Biotech. 8, 214. doi: 10.1007/s13205-018-1233-z
Zhao A., Zhao K., Xia Y., Lyu J., Chen Y., Li S. (2021). Melatonin inhibits embryonic rat H9c2 cells growth through induction of apoptosis and cell cycle arrest via PI3K-AKT signaling pathway. Birth Defects Res. 113, 1171–1181. doi: 10.1002/bdr2.1938
Keywords: Hyriopsis cumingii, pearl-nucleus insertion, growth and mineralization, co-regulation, transcriptome
Citation: Chen Y, Yao Y, Shen X, Fu Y, Bian X, Li W and Liu S (2024) Transcription profiling reveals co-regulation mechanism of gene expression related to growth and mineralization induced by pearl cultivation in Hyriopsis cumingii. Front. Mar. Sci. 11:1443863. doi: 10.3389/fmars.2024.1443863
Received: 14 June 2024; Accepted: 22 July 2024;
Published: 08 August 2024.
Edited by:
Ming Li, Ningbo University, ChinaReviewed by:
Suvra Roy, Central Inland Fisheries Research Institute (ICAR), IndiaZhi Liao, Zhejiang Ocean University, China
Copyright © 2024 Chen, Yao, Shen, Fu, Bian, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Li, d2psaUBzaG91LmVkdS5jbg==; Xiangli Bian, Qmlhbl94aWFuZ19saUAxNjMuY29t
†These authors contributed equally to this work
 Yige Chen
Yige Chen Yuanbin Yao
Yuanbin Yao Xiaoya Shen
Xiaoya Shen Yuanshuai Fu
Yuanshuai Fu Xiangli Bian2*
Xiangli Bian2* Wenjuan Li
Wenjuan Li