
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 31 May 2024
Sec. Microbial Symbioses
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1408979
The objective of this study was to identify effective strategies for disease prevention and control that circumvent antimicrobial resistance (AMR) issues, thereby facilitating sustainable aquaculture practices. This research focused on isolating probiotic Bacillus strains from the intestinal tract of healthy large yellow croaker (Larimichthys crocea). Among the 11 strains tested, four demonstrated protease and lipase activities. Further testing identified one strain, Lysinibacillus sp. (LYD11), with strong inhibitory activity against Vibrio harveyi and Vibrio alginolyticus, two major pathogens of the large yellow croaker. The screened strains showed more than 45% co-aggregation with five pathogens and were capable of inhibiting pathogen colonization through competition, repulsion, and substitution adhesion of 20.56 to 48.45%, 40.37 to 75.62% and 55.51 to 85.30%, respectively. The hydrophobicity and self-aggregation rates of LYD11 were 82.73% and 49.47%, respectively, indicating a potent ability for adhesion and colonization within the fish intestine. LYD11 demonstrated robust growth under the NaCl 0.5–3.5% and pH range of 6–8 and was unaffected by the gastrointestinal environment. Safety evaluations at high concentrations in grouper, in both injected and fed groups, indicated no adverse effects, as evidenced by the absence of morbidity or mortality. Consequently, Lysinibacillus sp. LYD11 emerges as a potential probiotic candidate for the prevention and control of bacterial diseases in the large yellow croaker.
China stands as the leading nation in both aquaculture production and consumption, with its aquaculture industry experiencing ongoing expansion. However, this growth has been marred by frequent disease outbreaks, leading to substantial economic losses (Wu et al., 2022). At present, disease management poses a significant challenge and an important factor restricting the healthy and sustainable development of aquaculture in China (Yuan et al., 2022). Furthermore, the adverse effects associated with the application of antibiotics and chemical agents (Kumar et al., 2016; Kavitha et al., 2018) have intensified the pursuit of innovative, antimicrobial-free strategies for disease prevention and control within aquaculture, marking a critical area of research in aquatic disease management.
Probiotics, originating from the Greek word meaning “for life” (El-Saadony et al., 2021), are characterized by the World Health Organization (WHO) as “live microorganisms that, when accumulated in certain amounts, are beneficial to the health of the host” (El-Saadony et al., 2021; Kasozi et al., 2023). Their application in human nutrition and animal husbandry has expanded significantly, demonstrating efficacy in the prevention and treatment of a variety of diseases (Chauhan and Singh, 2019; Wang et al., 2021; Iorizzo et al., 2022). However, despite their proven efficacy in humans and other mammals, the integration of probiotics into aquaculture remains in its infancy, with limited exploration of related mechanisms (Martínez Cruz et al., 2012). In aquaculture, probiotics are employed as safe dietary supplements, boosting host health by promoting growth, providing nutrients, regulating intestinal bacterial flora, maintaining intestinal epithelial barrier integrity, improving immune responses, increasing feed utilization, enhancing digestive and absorptive enzyme activities, improving water quality, and increasing disease resistance in fish (He et al., 2013; El-Saadony et al., 2021; Jahan et al., 2021). Bacillus species are notable among probiotics for their superior characteristics, stemming from their ability to synthesize antimicrobial compounds that modulate microbial activity without toxicity or pathogenicity, and their spore-forming capabilities, which confer enhanced heat resistance and extended viability, offering a considerable survival advantage in different environments (Geng et al., 2012; Kavitha et al., 2018; Kuebutornye et al., 2019; Van Doan et al., 2020). Research has documented the improvement of disease resistance in various aquatic species, such as rainbow trout (Neissi et al., 2024), tilapia (Liao et al., 2023), and white shrimp (Proespraiwong et al., 2023), following dietary supplementation with Bacillus, suggesting potential viability as an alternative to antibiotics.
Bacillus species applied in aquaculture originate from various sources, including soil, water, decaying organic matter, commercial products, and the gastrointestinal tracts of fish and other vertebrates (Sankar et al., 2017; Sumon et al., 2018; Adorian et al., 2019). Previous studies have demonstrated that host-associated probiotics can confer beneficial effects, including enhanced growth performance and disease resistance (Peixoto et al., 2021). However, most probiotics utilized in aquaculture are commercially sourced and often fail to colonize the fish gut or deliver probiotic benefits due to their non-aquatic origins (Kavitha et al., 2018; Mortezaei et al., 2020). As such, the aquaculture industry needs to adopt probiotics isolated from aquatic organisms (Kuebutornye, 2020).
The large yellow croaker (Larimichthys crocea), which is primarily distributed in the southern Yellow Sea, East China Sea, northern South China Sea, and Taiwan Strait, inhabits the lower and middle layers of coastal and offshore waters at depths up to 80 m. This species is highly valued by consumers for its delicate and flavorful flesh, golden scales and lips, and aesthetically pleasing appearance (Huang et al., 2023). Despite being the most significant marine fish in China in terms of aquaculture production, the large yellow croaker is plagued by a variety of bacterial diseases. In the current study, we investigated a novel approach for preventing and controlling bacterial diseases in the large yellow croaker, focusing on methods that do not lead to pathogen resistance. We isolated and evaluated probiotic Bacillus strains from the intestinal tract of healthy large yellow croakers, examining their ability to produce enzymes, inhibit bacterial growth, and prevent the adhesion of pathogenic bacteria.
All animal experiments were approved by the Animal Ethics Committee of Jimei University (permit number JMULAC2011–58) and were carried out in compliance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Healthy fish (300–400 g body weight) were euthanized by immersion in a tank containing MS-222 (100 ppm, West Gene, China) for 15 min. Subsequently, the fish were rinsed several times in sterile water and their body surface was disinfected with 75% alcohol (XiLong, China). The abdomen was opened using sterile scissors and the gastrointestinal tract was removed and rinsed with sterile saline to remove intestinal contents. The weight of the extracted intestines was recorded before the addition of saline (Concentration of 0.9%) at a rate of 0.5 g/ml and homogenization on ice. The resulting homogenate was then subjected to a 30 min water bath at 60°C, followed by serial dilution and plating on LB agar. After incubation for 24 h at 37°C, single colonies on the LB agar plates were isolated and purified by plate streaking to obtain pure cultures. The purified isolates were stored in Luria-Bertani (LB) broth supplemented with 20% glycerol (Macklin, China) at −80°C.
The extracellular activities of protease, amylase, and lipase in the selected bacterial strains were quantitatively estimated using protease, amylase, and lipase test agar plates following previous published methods (Leyva-Madrigal et al., 2011). The formula of the medium used for the enzyme-producing capacity assay was: Protease agar plate: beef paste (Macklin, China) 3g, peptone (Macklin, China) 10g, NaCl (Macklin, China) 5g, agar powder (Macklin, China) 16g, skimmed milk powder (Biosharp, China) 2g, distilled water 1000ml. Amylase agar plate: beef paste 3g, peptone 10g, NaCl 5g, soluble starch (Macklin, China) 10g, agar powder 16g, distilled water 1000ml. Lipase agar plate: peptone 10g, calcium chloride monohydrate 0.1g, Tween-80 (Macklin, China) 10ml, agar powder 16g, distilled water 1000ml. Following incubation for 24 h at 37°C in a thermostat, the presence of hydrolysis and precipitation zones was determined, and their diameters were calculated.
The antimicrobial activity of isolated strained was tested following previous study (Wang et al., 2022). Indicator strains, including Vibrio harveyi, Vibrio alginolyticus, Pseudomonas plecoglossicida, Aeromonas hydrophila, and Edwardsiella tarda The strains used for the antimicrobial test were taken from growth stationary phase cells and cultured three times in passages, with their optical density at 600 nm (OD600) adjusted to 0.3. The bacterial culture samples of V. harveyi, V. alginolyticus, P. plecoglossicida, A. hydrophila, and E. tarda were mixed with LB agar medium at a volume ratio of 1:9. Three small holes (approximately 8 mm in diameter) were created in the medium, and added 100 μL of isolated bacterial culture samples. After incubation at 37°C for 24 h in a constant temperature incubator, the diameter of the inhibition zone was measured.
Bacterial agglutination ability was determined following previously published methods (Wang et al., 2022). In brief, bacterial cultures were incubated at 37°C for 16–18 h, then centrifuged at 4 500 rpm for 10 min and washed twice with sterile phosphate-buffered saline (PBS, Biosharp, China). The bacteria were suspended in Heps-Hank’s buffer with Hepes (HHBS: 10 mM Hepes; pH 7.4. ShangHai MaoKang, China) (1 × 108 CFU/mL) and stored at 4°C. Mucins were prepared following previously described protocols (Xin et al., 2022). The gastrointestinal tract of healthy large yellow croakers was harvested and washed with PBS containing 0.01% gelatin (Macklin, China), with the inner surface of the intestine then scraped to obtain the mucinous protein mixture, which was transferred to HHBS. The supernatant was collected after centrifugation and its concentration was adjusted to 0.5 mg/mL, followed by storage at 20°C. Hoechst 33258 staining solution (Beyotime, China) was added to the bacterial suspension to a final concentration of 2 μg/mL. After 30 min of dark staining at 37°C, the collected bacteria were twice washed with PBS and suspended in HHBS by centrifugation. After adjustment to a concentration of 1 × 108 CFU/mL, the bacterial suspension was stored at 4°C.
Mucin was added to 96-well black polystyrene culture plates and fixed at 4°C for 24 h. After washing the culture plate with 200 μL of HHBS, 100 μL of stained bacteria (1.0 × 108 CFU/mL) was added to each well and incubated at 37°C for 1 h. After rewashing the plate, 200 μL of 1% sodium dodecyl-sulfate (SDS, Macklin, China)-0.1 mol/L NaOH (XiLong, China) lysate was added to each well and incubated at 60°C for 1 h. For the negative control, 100 μL of unlabeled bacteria (1.0 × 108 CFU/mL) was added to each well. For the positive control, 100 μL of labeled bacteria (1.0 × 108 CFU/mL) was added to each well. Fluorescence intensities of the negative control (t0), positive control (t1), and experimental group (t2) were measured using an automatic fluorescence microplate reader, with an excitation wavelength of 340 nm and fluorescence emission wavelength of 460 nm. The adhesion rate was determined using the equation adhesion rate (%) = [(t2 − t0)/(t1− t0)] × 100.
The effect of bacterial adhesion was observed according to previous research (Yang et al., 2023). Mucus (20 μL) was evenly spread onto a glass slide (22 × 22 mm, Biosharp, China) and fixed with 200 μL of 4% methanol (Beyotime, China) for 30 min, after which 200 μL of bacterial suspension (1.0 × 108 CFU/mL) was evenly spread over the mucus. After incubation at 28°C for 2 h in a humidified chamber, the slides were washed three times with PBS to remove non-adhered cells. The adhered bacterial cells were fixed using 200 μL of 4% methanol for 30 min, stained with 0.1% crystal violet for 3 min, rinsed with PBS, and finally observed under a Leica microscope. (×1 000).
Morphological observations and Gram staining were used for preliminary identification of bacteria, which were cultured in LB liquid medium at 37°C for 24 h. DNA extraction was performed using a FastPure Cell DNA Isolation Mini Kit (Vazyme, China), in accordance with the manufacturer’s instructions. Using polymerase chain reaction (PCR) with the molecular identification targeting 16S rRNA genes primers sequences 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGCTACCTTGTTACGACTT-3’). The PCR products were detected by electrophoresis on a 1% agarose gel containing ethidium bromide. The sequencing of the PCR products was carried out by Shanghai Shengong Bioengineering Co., Ltd. (China). Sequence analysis was performed using BLAST (National Institutes of Health, USA; http://blast.ncbi.nlm.nih.gov/blast.cgi). Multisequence alignments were obtained using CLUSTALW implemented in MEGA 7.0. Phylogenetic reconstructions of sequencing data were performed using the neighbor-joining (NJ) method, with branching reliability tested using bootstrap resampling with 1 000 pseudo-replicates.
Bacterial agglutination ability was determined following existing methods (Jlidi et al., 2022), with minor modifications. In brief, V. harveyi, V. alginolyticus, P. plecoglossicida, A. hydrophila, and E. tarda were selected as indicator bacteria. Bacterial cells were collected by centrifugation at 5 000 rpm for 10 min and washed twice with sterile saline (0.9%). The cells were then resuspended to a final concentration of 1.0 × 108 CFU/mL (A0). Equal volumes of the LYD11 and indicator bacterial suspensions were combined, vortexed for 1 min, and allowed to stand at room temperature. The upper layer of the solution was collected, and OD600 was measured for 2 h (At). Sterile saline was used as a blank control.
Where A0 represents absorbance at 0 h and At represents absorbance at 2 h.
The bacterial adhesion assay was performed in reference to existing methods (Wang et al., 2022), with some modifications.
For the competitive inhibition adhesion assay, 100 μL of mucin was placed in a 96-well black polystyrene culture plate and fixed at 4°C for 24 h. The culture plate was washed twice with 200 μL of HHBS to remove non-adherent mucin, followed by the addition of 100 μL of stained (1.0 × 108 CFU/mL) and unstained bacteria (1.0 × 108 CFU/mL). For the positive control, wells were inoculated with 100 μL of fluorescently labeled bacterial pathogens (1.0 × 108 CFU/mL) and incubated at 37°C for 1 h, followed by two washes with HHBS buffer. Afterwards, 200 μL of 1% SDS-0.1 mol/L NaOH lysate was introduced to each well and incubated at 60°C for 1 h to detach and elute the adherent bacteria. In the negative control, 100 μL of non-fluorescently labeled bacteria (1.0 × 108 CFU/mL) and pathogenic bacteria (1.0 × 108 CFU/mL) were added, with subsequent fluorescence measured using an automated fluorescence microplate reader.
In the rejection inhibition adhesion assay, 100 μL of mucin was placed in a 96-well black polystyrene culture plate and incubated at 4°C for 24 h. The plate was then washed twice with 200 μL of HHBS, to remove non-adherent mucin, followed by the addition of 100 μL of unstained bacteria (1.0 × 108 CFU/mL). After incubation at 37°C for 1 h, the plates were washed twice with HHBS, followed by the addition of 100 μL of stained bacterial strains (1.0 × 108 CFU/mL). For the positive control, wells were inoculated with 100 μL of fluorescently labeled bacterial pathogens (1.0 × 108 CFU/mL), incubated at 37°C for 1 h, and subsequently washed twice with HHBS. Then, 200 μL of 1% SDS-0.1 mol/L NaOH lysate was introduced to each well and incubated at 60°C for 1 h to detach and collect adherent bacteria. In the negative control, 100 μL of non-fluorescent bacteria (1.0 × 108 CFU/mL) and non-fluorescent pathogenic bacteria (1.0 × 108 CFU/mL) were added, with fluorescence subsequently measured using an automated fluorescence microplate reader.
In the substitution inhibition adhesion assay, 100 μL of mucin was placed in a 96-well black polystyrene culture plate and incubated at 4°C for 24 h. The plate was then washed twice with 200 μL of HHBS, to remove the non-adherent mucin, followed by the addition of 100 μL of stained pathogenic bacteria (1.0 × 108 CFU/mL) and incubation at 37°C for 1 h. The plate was then washed twice with HHBS, followed by the addition of 100 μL of unstained bacterial solution (1.0 × 108 CFU/mL). For the positive control, wells were inoculated with 100 μL of fluorescently labeled bacterial pathogens (1.0 × 108 CFU/mL), incubated at 37°C for 1 h, and washed twice with HHBS buffer. Subsequently, 200 μL of 1% SDS-0.1 mol/L NaOH lysate was added to each well and incubated at 60°C for 1 h to elute the adherent bacteria. In the negative control, 100 μL of non-fluorescent bacteria (1.0 × 108 CFU/mL) and non-fluorescent pathogenic bacteria (1.0 × 108 CFU/mL) were introduced to the wells, with fluorescence then measured using an automated fluorescence microplate reader.
Bacterial cell surface hydrophobicity (CSH%) was assessed in accordance with previously established methods (Kuebutornye, 2020), with some modifications. In brief, bacterial cells were harvested by centrifugation at 5 000 rpm for 10 min, washed twice with sterile saline, and resuspended to achieve a concentration of 1.0 × 108 CFU/mL. The bacterial suspension (3 mL) was mixed with 1 mL of xylene (Macklin, China), pre-incubated at room temperature for 10 min, then vortexed for 2 min. After allowing the mixture to stand for 40 min at room temperature, the aqueous phase was separated and OD600 was measured, using sterile saline as the blank control. The hydrophobicity was determined based on set criteria (Santos et al., 1990), where a CSH% greater than 50% indicates high hydrophobicity, a CSH% ranging from 20% to 50% suggests moderate hydrophobicity, and a CSH% less than 20% denotes low hydrophobicity.
Where A0 represents absorbance at 0 h and At represents absorbance at 40 min.
The self-aggregation ability of selected Bacillus strains was analyzed based on previously published methods (Kuebutornye, 2020), with slight modifications. In brief, bacterial cells were collected by centrifugation at 5 000 rpm for 10 min and subsequently washed twice with sterile saline. The cells were then resuspended to achieve a concentration of 1.0 × 108 CFU/mL. The bacterial suspension (1.8 mL) was then placed in a 2-mL EP tube, vortexed for 30 s, and allowed to stand at room temperature for 2 h. The OD600 of the upper layer of the solution was measured, using sterile saline as a control.
Where A0 represents absorbance at 0 h and At represents absorbance at 4 h.
Healthy groupers (average weight of 19 ± 1.0 g) were randomly divided into two groups containing 30 fish each. The aquarium was continuously aerated during the experimental period and the water temperature was maintained at 27°C. To maintain water quality, one-third of the aquarium water was replaced twice daily. The fish were fed a basic diet, and fecal matter was regularly removed. Following a one-week acclimatization period, the fish were prepared for the experiment.
Use Jianma brand grouper compound feed, purchased from Fujian Tianma Technology Group Co., Ltd. The LYD11 bacterial cells were harvested by centrifugation at 5 000 rpm for 10 min, washed twice with sterile saline, and resuspended to achieve a concentration 1.0 × 1010 CFU/ml. Then mixed with the basal diet and dried at low temperature to obtain the LYD11 diet (bacterial content 1.0 × 109 CFU/g). The basic feed containing equal amount of sterile saline (0.9%) was prepared as a control. Each group of fish was fed once a day (8.5 g per group) for 7 days. Intestinal tissue samples were collected from the intestinal tract on days 7 and 10 following the commencement of the feeding regimen. These tissues were homogenized in sterile saline (0.9%) to achieve a mass-to-volume ratio of 1 g/mL, then incubated in a water bath at 60°C for 30 min. Subsequently, the homogenized samples were coated on LB agar medium to assess the presence of bacterial colonies.
The salinity of the LB liquid medium was modified to varying NaCl concentrations (0.5%, 1.5%, 2.5%, 3.5%, 4.5%, 5.5%, and 6.5%). Bacterial cultures were then incubated overnight under constant temperature (37°C) with shaking. Subsequently, the cultures were diluted 1:50 into LB liquid medium at the designated salinity levels. After a 5 min stabilization period, incubation proceeded for 24 h with continuous shaking and then the OD600 of the bacterial culture samples was measured.
The pH levels of the LB liquid medium were adjusted to 2, 4, 6, 8, and 10 with the addition of dilute hydrochloric acid (HCl, Luqiao, China) solution and 1 mol/L NaOH solution. The bacterial cultures to be tested were incubated overnight under a constant temperature (37°C) with shaking. Inoculate 1:50 into different concentrations of LB liquid medium, start timing after 5 min and incubate for 24 h with shaking and then the OD600 of the bacterial culture samples was measured.
The resilience of strains to simulated gastric and intestinal fluids was evaluated using a modified method (Zhang et al., 2022). To prepare the simulated gastric fluid, 16.4 mL of 10% HCl and 1 mg/mL pepsin (Solarbio, China) were mixed into LB liquid medium, with the pH adjusted to 3, followed by sterilization through a 0.22 μm membrane filter. For the simulated intestinal fluid, 1.36 g of KH2PO4 was dissolved in 100 mL of LB liquid medium, with trypsin added to achieve a concentration of 10 mg/mL (Solarbio, China) and the pH adjusted to 6.8, following by sterilization using a 0.22 μm membrane filter. The bacteria to be tested underwent overnight incubation at a constant temperature (37°C) with shaking. The bacteria were then diluted at a ratio of 1:50 into LB medium adjusted for the respective assays, with timing starting 5 min later and incubation continuing for 4 h with shaking, and then the OD600 of the bacterial culture samples was measured.
LB liquid medium was prepared with concentrations of porcine bile salts (Biosharp, China) at 0.15, 0.3, and 0.45 mg/mL. A control medium without any additions was also prepared. Both media were then filtered using a 0.22 μm filter. The bacterial cultures designated for evaluation were inoculated into the prepared LB medium at a 1:50 dilution and subjected to incubation with shaking. The OD600 of the bacterial culture samples was recorded after a 4 h incubation period.
Hemolytic activity was assessed according to earlier research (Chang et al., 2000). In brief, bacterial colonies were streaked onto blood agar plates supplemented with 5% defibrinated sheep blood (Vazyme, China) and incubated at 37°C for 24–48 h. The presence of transparent zones around the bacterial colonies on the agar plate suggests β-hemolysis, while green zones signify α-hemolysis, and the absence of any change indicates γ-hemolysis.
The antibiotic susceptibility of the selected isolates was evaluated based on previous research (Thankappan et al., 2015). The screened strains were inoculated in LB liquid medium and incubated at 37°C for 18–24 h. Subsequently, 100 μL of bacterial suspension was evenly spread on an LB solid medium plate, and a drug-sensitive paper sheet (HangWei, China) was placed on the surface of the petri dish medium. The plates were incubated at a constant temperature (37°C) for 24 h. The sensitivity of the strains to antibiotics was assessed based on the diameter size of the final inhibition ring. The test results were evaluated using the WS/T125–1999 Standard for Antimicrobial Drug Sensitivity Test by Paper Slice Method.
Healthy groupers (average weight of 19 ± 1.0 g) were randomly divided into two groups containing 30 fish each. The aquarium was kept continuously aerated during the experimental period and the water temperature was maintained at 27°C. To maintain water quality, one-third of the aquarium water was replaced twice daily. The fish were fed a basic diet, and fecal matter was regularly removed. Following a one-week acclimatization period, the fish were prepared for the experiment.
The bacterial strains were incubated at 37°C until reaching the exponential growth phase, followed by centrifugation at 6 000 rpm for 10 min. The concentration of the bacterial suspension was then adjusted to 1.0 × 109 CFU/mL for safety evaluation using both injection and feeding approaches. Each fish received an injection of 0.1 mL of the bacterial solution. Post-injection, the groupers were returned to their original aquatic environment for further cultivation. Feed infused with bacteria (1.0 × 109 CFU/g) was prepared, while the control group received a similar quantity of blank feed (sterile saline) daily (8.5 g/group) for a duration of 7 days. Mortality rates were documented throughout a 15 day observation period.
After 15 days of observation, three fish were randomly selected from each group and anesthetized with MS-222 (100 ppm). The liver, spleen, and blood were then removed under aseptic conditions. All samples, apart from the blood, were weighed and homogenized in 0.9% sterile saline at a mass-volume ratio of 1 g/mL. The homogenized samples were then coated on LB agar medium, with the absence of bacterial growth indicative of probiotic safety. Other liver and spleen tissues were fixed in Dietrich’s fixative (70% paraformaldehyde, 5% glacial acetic acid, and 25% anhydrous ethanol). After dehydration and paraffin embedding, 10 μm serial sections were created using a microtome. Pathological changes were assessed via hematoxylin-eosin (H&E) staining.
After 7 days of feeding, the entire intestine was removed. Midgut tissues were collected and fixed in Dietrich’s fixative. After dehydration and paraffin embedding, 10 μm serial sections were prepared using a microtome and examined using H&E staining. The presence of intestinal tissue damage was assessed.
Statistical analyses were conducted using one-way analysis of variance (ANOVA) with SPSS v22.0. Duncan’s multiple comparison test was used to evaluate differences among groups. A P-value of less than 0.05 was considered significant. All experiments were performed in triplicate.
A total of 11 strains were isolated from the intestinal tract of the yellow croaker, among which four strains were screened to produce protease and lipase (Table 1). These four strains formed substantial discoloration zones on more than one type of medium after 24 h of incubation. Thus, these four strains, demonstrating high enzyme production capacities, were selected for further analysis.
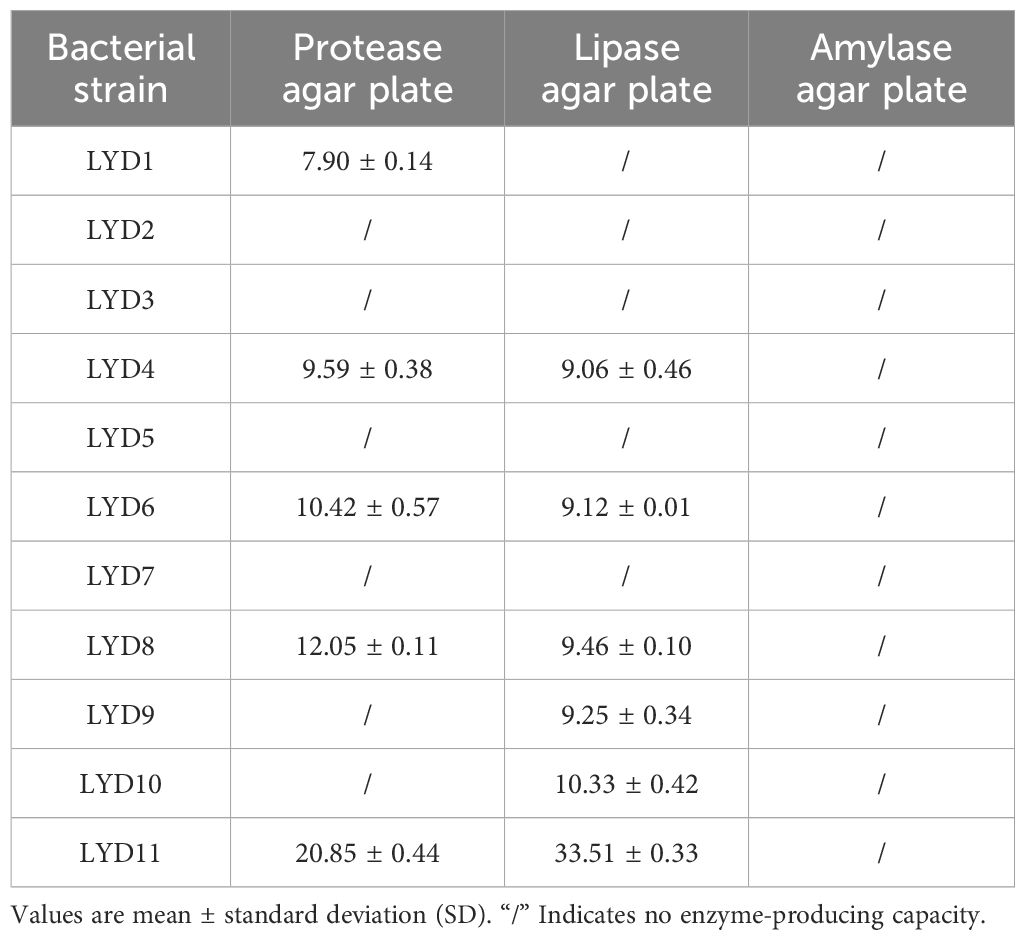
Table 1 Diameters (mm) of enzymatic circles of each isolated strain on protease, lipid, and amylase agar plates.
The results from in vitro assays evaluating the antagonistic potential against common pathogens are shown in Table 2 and Figure 1. These results indicate that the LYD11 strain exhibited inhibitory capabilities against two significant pathogens after 24 h of incubation, characterized by inhibition zones extending beyond 22 mm in diameter. Conversely, the LYD6 strain exhibited antagonistic properties against only one common pathogen.
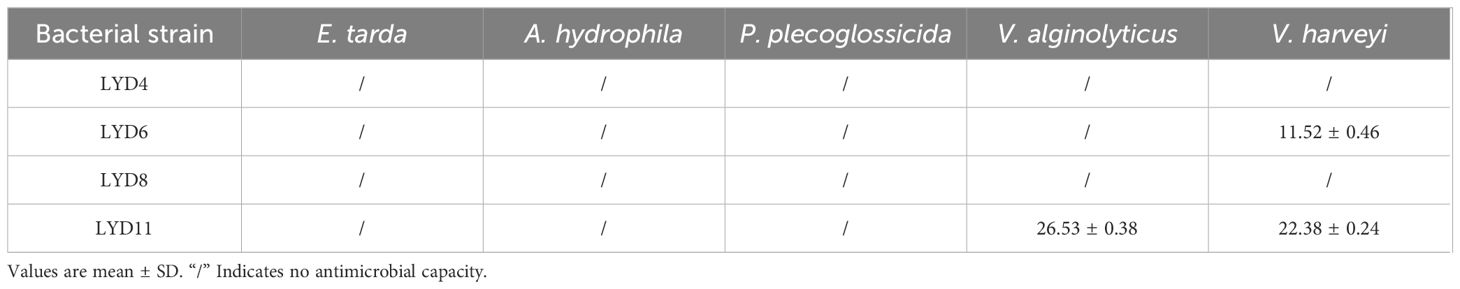
Table 2 Diameter (mm) of inhibition zones for each isolate against common pathogens of large yellow croaker.
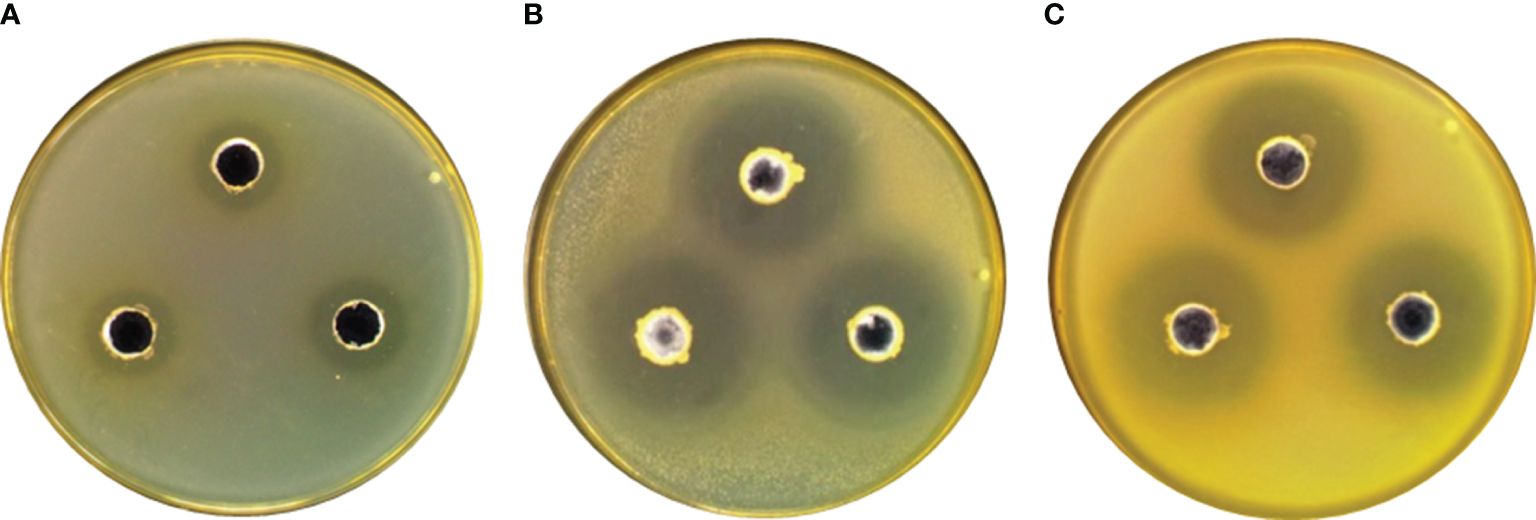
Figure 1 Inhibitory effects of (A) LYD6 strain against V. harveyi, (B) LYD11 strain against V. harveyi, and (C) LYD11 strain against V. alginolyticus..
Figure 2 presents the adhesion rates of the LYD6 and LYD11 strains to the mucus protein of the large yellow croaker. Results indicated that the mucus protein adhesion rate of LYD11 (13.57%) was higher than that of LYD6 (P< 0.05). Based on comparative analysis of adhesion capabilities, the LYD11 strain was chosen for further experimentation.
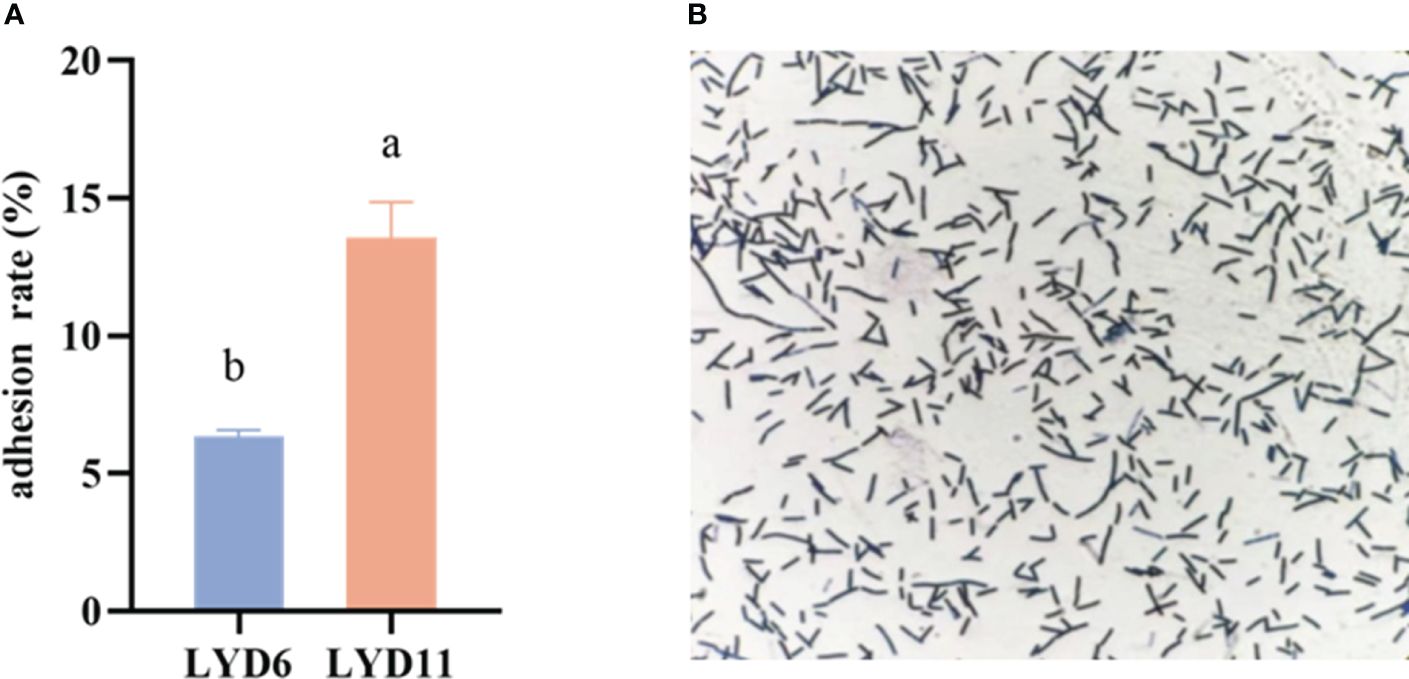
Figure 2 (A) In vitro adhesion results of LYD6 and LYD11 strains. (B) Bacterial LYD11 adhesion images.
Strain morphology was characterized by observational analysis. The LYD11 colonies formed on the LB solid medium were white, with a raised center and a rough and irregular morphology. Gram staining and microscopic examination identified LYD11 as a gram-positive bacterium with a short rod-shaped morphology (Figure 3A, B). The 16S rDNA sequences of the strains were compared with BLAST sequences in GenBank, revealing a homology of more than 99.8%. Phylogenetic analysis based on the 16S rDNA sequences was conducted using the proximity method. Results showed that the LYD11 strain was clustered with Lysinibacillus species, forming a distinct branch. Thus, comparison of the 16S rDNA gene sequences and phenotypic characteristics confirmed the identification of LYD11 as Lysinibacillus sp. (Figure 3C).
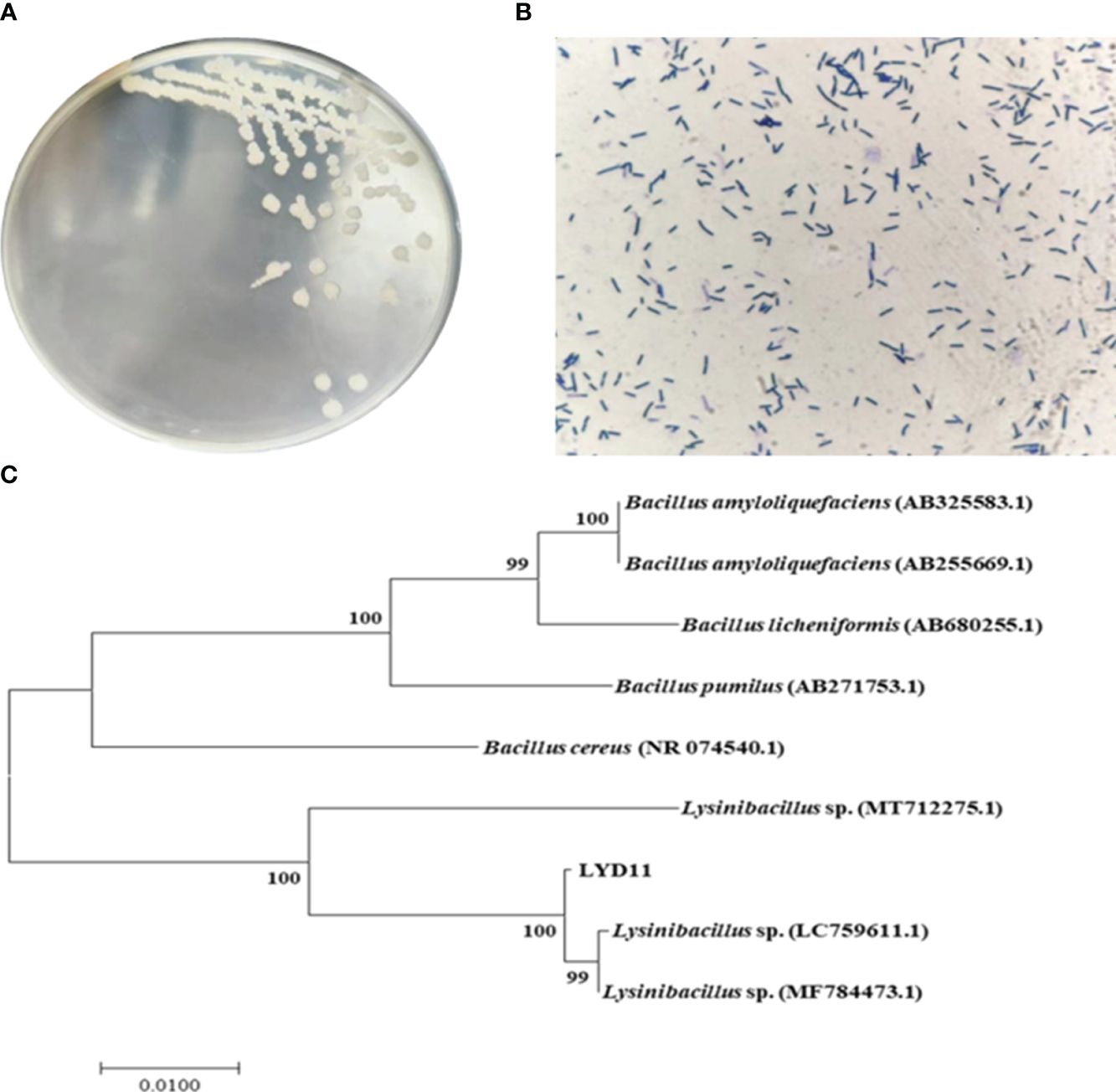
Figure 3 (A) Colony morphology and (B) Gram staining results of LYD11 strain. (C) Phylogenetic tree of LYD11 strain isolated from intestinal tract of large yellow croaker.
The LYD11 strain demonstrated significant co-aggregation capabilities with various pathogens, with co-aggregation percentages surpassing 40% (Figure 4A). Specifically, LYD11 exhibited the highest co-aggregation percentages against E. tarda (57.21%), P. plecoglossicida (54.08%), and A. hydrophila (47.15%) following 2 h of incubation. Additionally, LYD11 showed notable co-aggregation ability against V. alginolyticus (45.16%) and V. harveyi (43.77%).
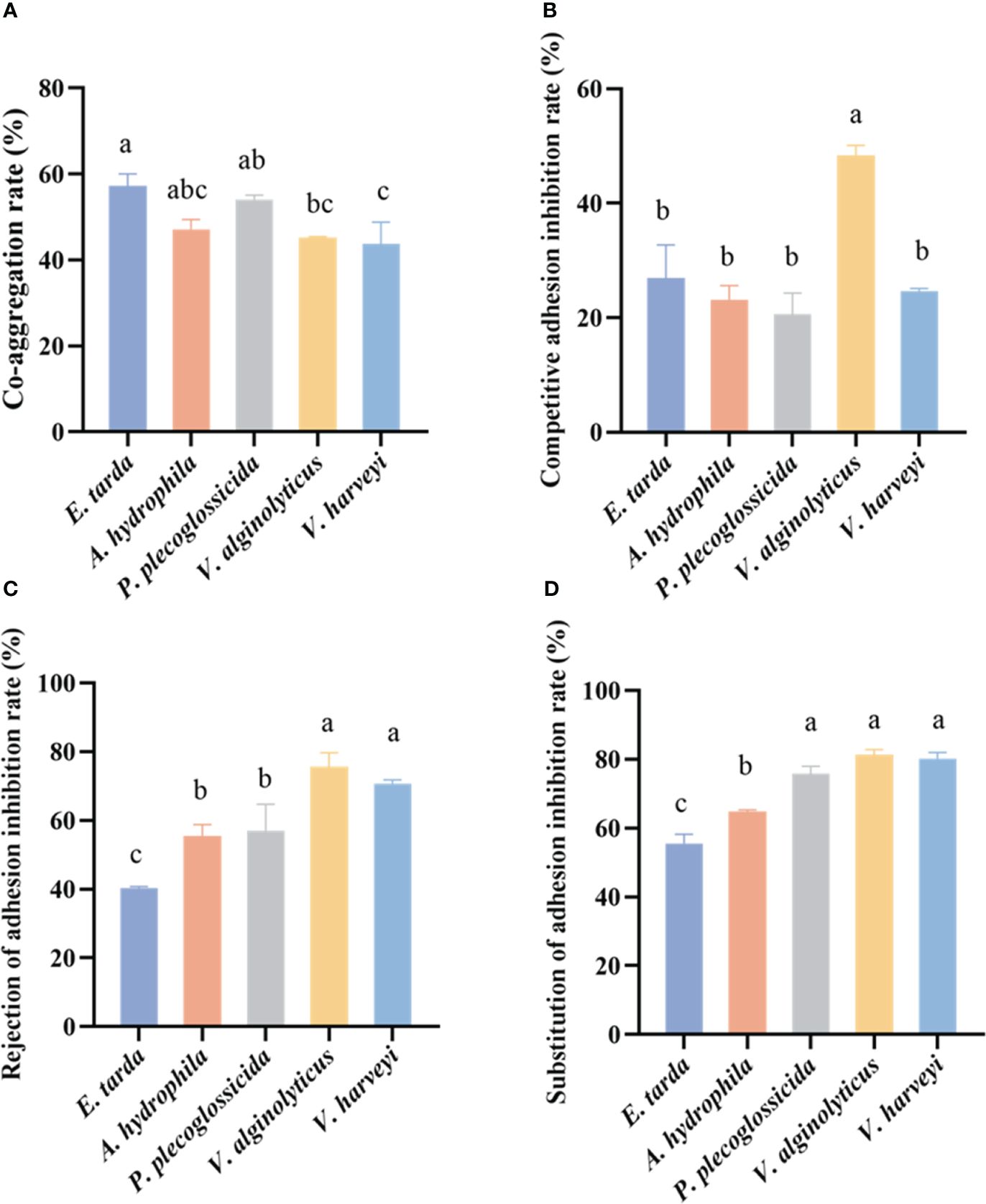
Figure 4 (A) Co-aggregation results of LYD11 strain with five pathogenic bacteria. (B) Rate of adhesion inhibition of LYD11 strain against five pathogenic bacteria by competition. (C) Rate of adhesion inhibition of LYD11 strain against five pathogenic bacteria by rejection. (D) Rate of adhesion inhibition of LYD11 strain against five pathogenic bacteria by substitution. Different letters indicate significant differences between groups (P < 0.05, ANOVA and Duncan’s multiple comparison test).
The capacity of the isolated LYD11 strain to engage in competition, rejection, and substitution with five pathogenic bacteria, E. tarda, A. hydrophila, P. plecoglossicida, V. alginolyticus, and V. harveyi, was further explored (Figures 4B–D, 5). Notably, LYD11 exhibited strong competition inhibition against all five pathogens, with a significantly greater inhibitory effect on V. alginolyticus than on the other four pathogens (P< 0.05), reaching approximately 50%. The LYD11 strain also exhibited strong rejection inhibition against all five pathogenic bacteria, with a significantly higher inhibitory effect on V. alginolyticus and V. harveyi than on the other pathogenic bacteria, exceeding 60% (P< 0.05). LYD11 also exhibited significant and varying substitution inhibition against all five pathogens, with the highest inhibitory effect on V. harveyi, V. alginolyticus, and P. plecoglossicida, reaching approximately 80%, and the weakest inhibitory effect on E. tarda, still achieving over 50% (P< 0.05).
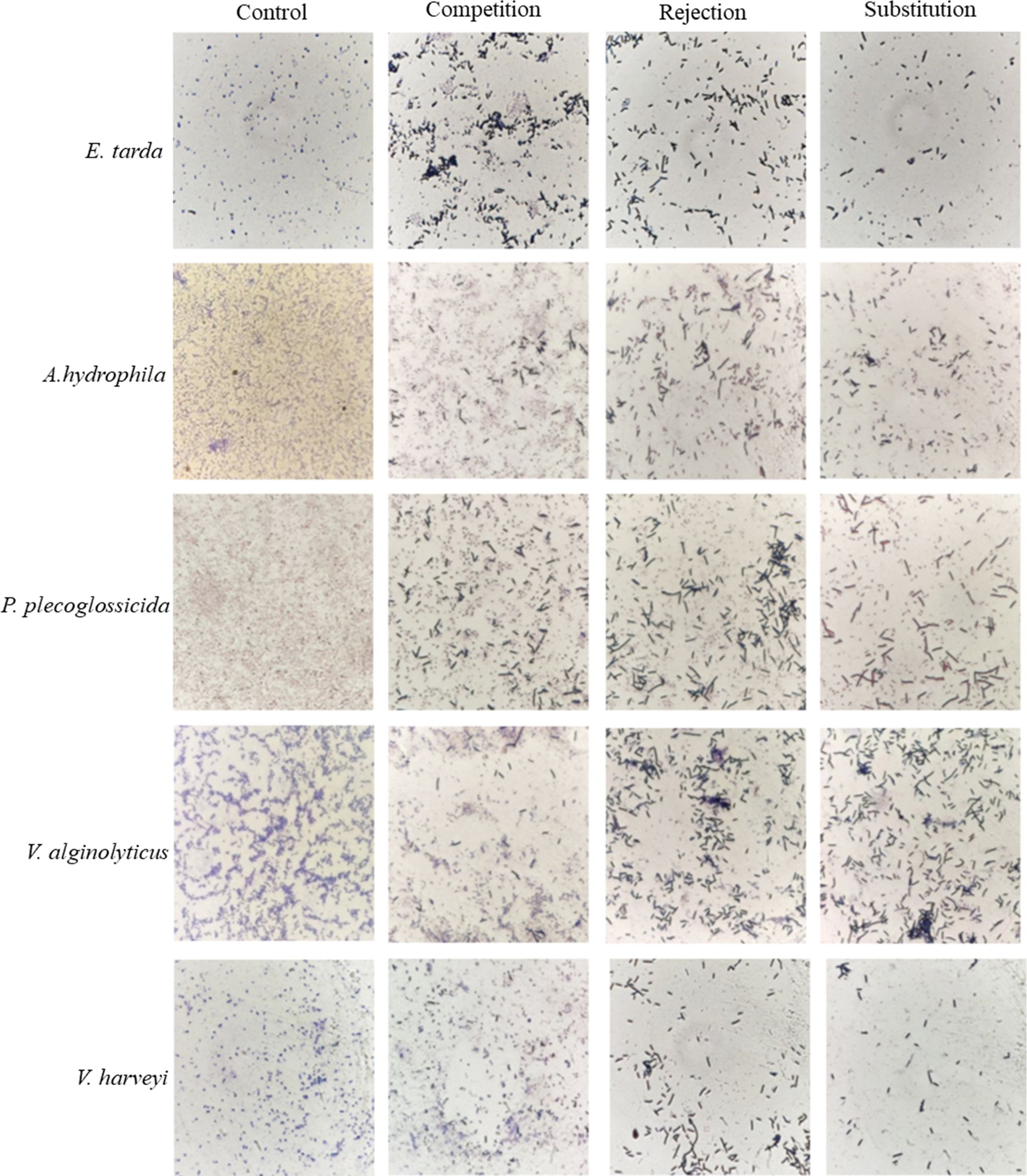
Figure 5 Competition, rejection, and substitution adhesion inhibitory effects of LYD11 strain against five pathogenic bacteria. LYD11 strain is the larger and darker colored bacterium. (100×).
The LYD11 strain demonstrated high survival across various tolerance tests during the study (Figure 6), including a wide salt tolerance range. Notably, LYD11 growth remained robust at NaCl concentrations ranging from 0.5% to 3.5% but decreased significantly (P< 0.05) at levels exceeding 3.5%.
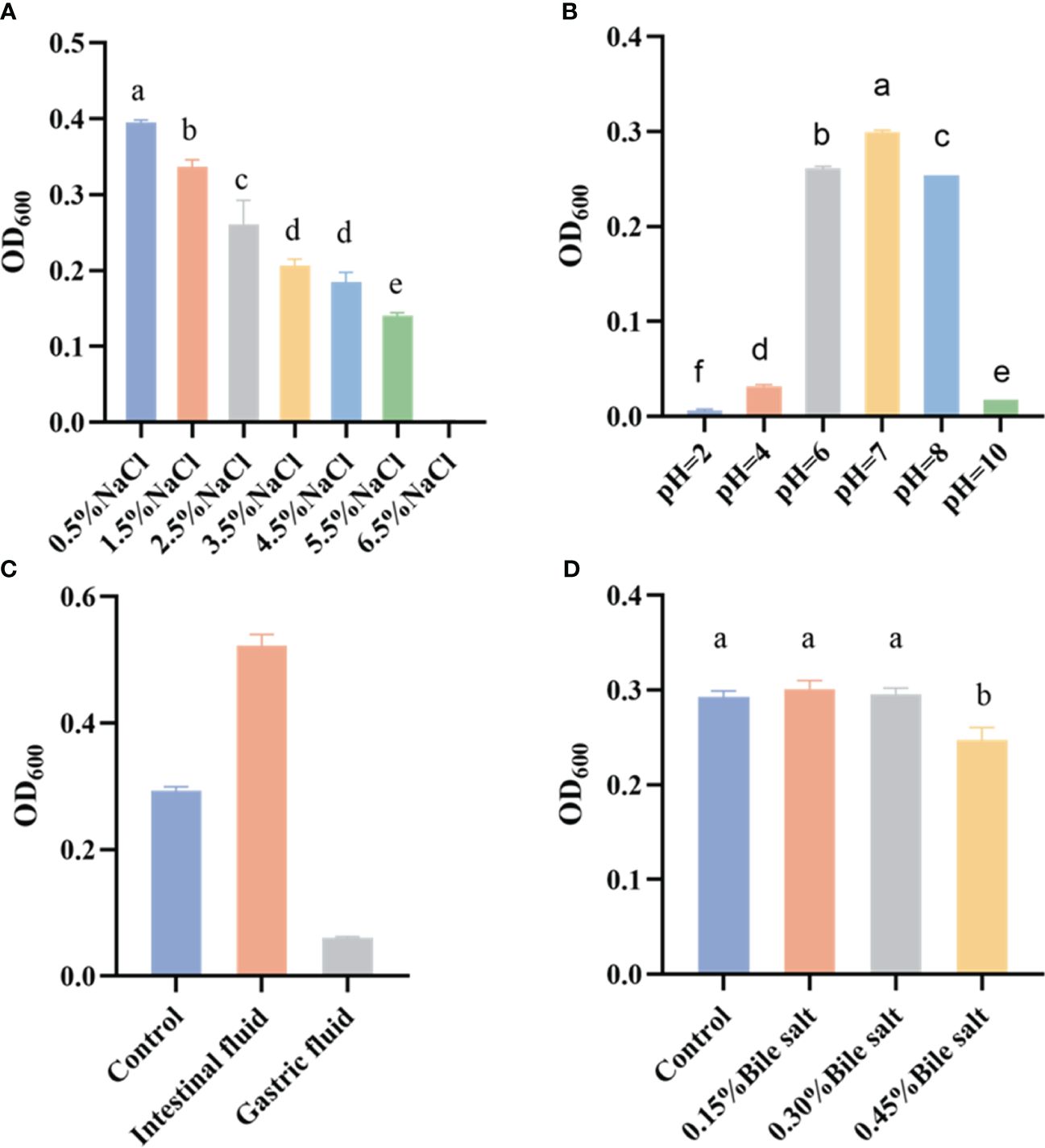
Figure 6 Growth of LYD11 strain under different (A) NaCl concentrations, (B) pH conditions, (C) simulated gastric and intestinal fluids, and (D) bile salt concentrations. Different letters indicate significant differences between groups (P < 0.05, ANOVA and Duncan’s multiple comparison test).
The screened strains were also subjected to pH tolerance tests under different culture conditions. Results showed that LYD11 exhibited broad adaptability to both acidic and alkaline environments, thriving in a pH range of 2–10, with optimal growth observed between 6 and 8. While the growth capabilities of LYD11 were significantly diminished at the extremes of pH 2, 4, and 10, it still managed weak growth.
The resilience of the strains in simulated gastric and intestinal fluids was tested to gauge their survival potential within the gastrointestinal tract of the host. Results showed that LYD11 grew slower in the simulated gastric fluid after 4 h, but grew significantly faster in the simulated intestinal fluid, indicating potential preferential adaptation to this environment.
LYD11 also showed good tolerance to bile salt exposure at concentrations of 0.15%, 0.30%, and 0.45% over 4 h. At a bile salt concentration of 0.15%, LYD11 growth did not differ significantly from the control. While bacterial growth was influenced at a bile salt concentration of 0.45%, growth activity remained high.
As detailed in the surface hydrophobicity and agglutination tests (Figure 7A), the LYD11 strain exhibited high hydrophobicity (82.73%) after 40 min, as well as strong self-agglutination capability (49.47%) after 2 h.
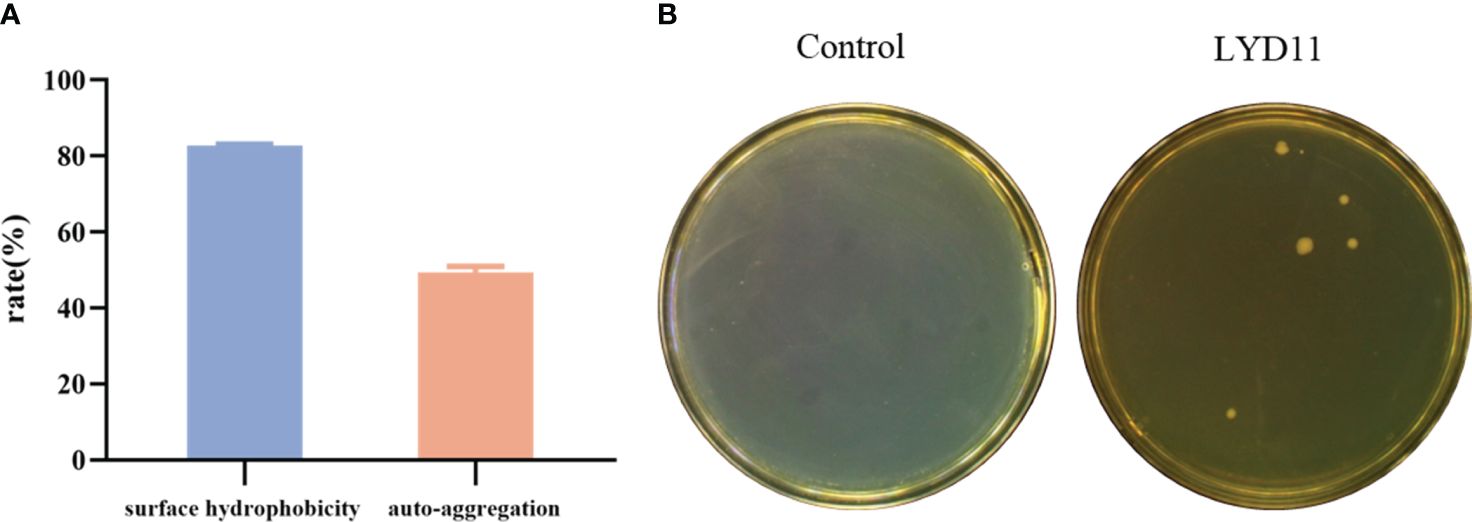
Figure 7 (A) Hydrophobicity and self-agglutination rate of LYD11 strain. (B) Intestinal homogenate smear results of grouper after 10 days of LYD11-strain feeding.
Intestinal tissues were collected on days 7 and 10 following one-week administration of a bacterial-containing diet. Analysis of homogenized samples revealed the absence of Bacillus colonies in the control group, while the group that received the LYD11 strain diet showed the presence of colonies. Colonies exhibiting similar morphological characteristics were selected and confirmed as Lysinibacillus strains by DNA sequencing (100% similarity). While the number of colonies decreased with increasing days, a small amount of LYD11 was still detectable in the intestines on day 10 post-feeding (Figure 7B). These findings suggest a robust colonization ability of the strain, providing a solid foundation for its beneficial effects.
Figure 8A illustrates the hemolytic behavior of the LYD11 strain, which did not produce zones around the colonies, classifying it as γ-hemolytic. In vitro antibacterial tests indicated that LYD11 was highly sensitive to 11 antibiotics, moderately sensitive to 15, and resistant to four (Table 3). In vivo biosafety testing showed no mortality in either the experimental or control groups, maintaining a survival rate of 100%. There were no noticeable changes in the physical or anatomical findings between the control and experimental groups (Figure 9). Cultures from blood, liver, or spleen homogenates did not yield any bacterial colonies. Further, histological analysis of liver and spleen tissue sections revealed the absence of lesions, with cells appearing closely packed and intact, suggesting a high safety level of LYD11 for grouper application (Figure 10). The survival rate for grouper fed with feed containing LYD11 also stood at 100%. Histological examination of grouper midgut sections (Figure 8B) showed no pathological changes. Notably, there was an increase in the thickness of the muscularis layer in the midgut for the group treated with LYD11, suggesting improved nutrient absorption, unlike in the control group.
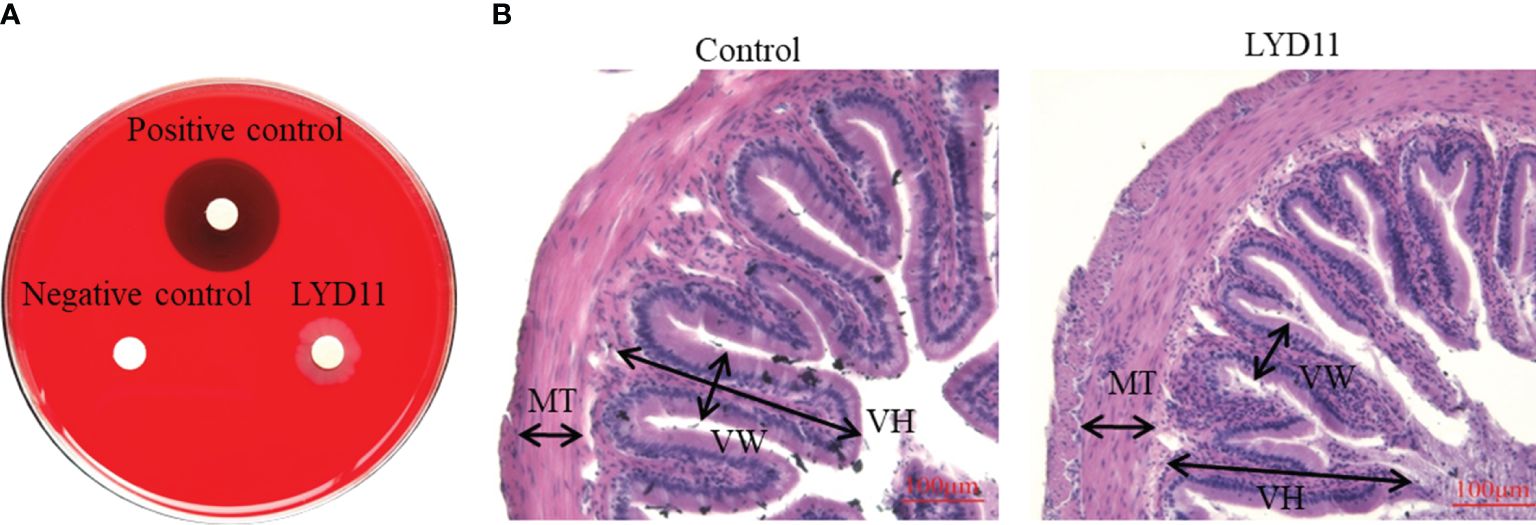
Figure 8 (A) Hemolytic activity of LYD11 strain. (B) H&E staining of midgut tissues after feeding LYD11 strain to grouper.
The origin of probiotics plays a critical role in their screening and application, directly affecting their efficacy, with inappropriate strains potentially damaging host intestines upon introduction. The diminished effectiveness of many commercial aquatic micro-ecological products may stem from their non-aquatic origins, leading to inefficiencies within the intestinal tracts and aquatic environments of their intended hosts (Cerezuela et al., 2012). Aquatic and terrestrial animals differ significantly in their living environments, growth, development, and intestinal flora composition (Vine et al., 2004). The aquatic animal gut constitutes a dynamic ecosystem, hosting a multitude of microorganisms vital for animal health, yet largely unexplored (Kuebutornye, 2020; Medina et al., 2020; Jlidi et al., 2022b). The gastrointestinal microbiota is considered a prime candidate for the isolation and screening of potential probiotics, with those derived from the host itself being safer and more effective than those screened from other animals and environments (Zorriehzahra et al., 2016; Yamashita et al., 2020; Santos et al., 2021). In this study, we isolated Bacillus species from the intestinal tract of large yellow croaker and evaluated their probiotic potential and safety, aiming to identify the most suitable strain for disease prevention and control in large yellow croaker culture.
Bacterial diseases continue to constrain the development of large yellow croaker aquaculture, with probiotics emerging as a viable alternative to manage these diseases (Khan et al., 2021). In the present study, key pathogenic bacteria in large yellow croaker aquaculture were selected as indicator bacteria for inhibition experiments, revealing V. alginolyticus and V. harveyi as the most sensitive to the Lysinibacillus LYD11 strain. Vibrio species, known as opportunistic pathogens in aquatic environments, pose a threat to marine fish, with most species being vulnerable to at least one Vibrio species (Khan et al., 2023). As a result, Vibrio infections have become economically significant in marine aquaculture, adversely affecting many fish and farmed species (Uzun and Ogut, 2015). Previous research has highlighted the efficacy and beneficial activity of Lysinibacillus as a feed additive in tilapia (Reda et al., 2018), as well as its antimicrobial activity against Gram-positive and Gram-negative bacteria and fungi. Reda et al. (Reda et al., 2018) isolated Lysinibacillus strains from the intestinal tract of Nile tilapia, demonstrating potential probiotic properties and antagonistic activity against specific pathogens, including A. mildewii, A. hydrophila, P. aeruginosa, P. malodorata, and Staphylococcus aureus. Furthermore, Rosland et al. (Rosland et al., 2021) identified Lysinibacillus strains A-1 and C1–3, which exhibited inhibitory effects on V. harveyi and V. parahaemolyticus, consistent with our findings. However, this investigation marks the first isolation of a Lysinibacillus species with probiotic potential from the intestinal tract of the large yellow croaker. In the previous literature, no isolation of Lysinibacillus with potential probiotic properties has been reported from the intestinal tract of large yellow croaker, and our study may be the first time that a Lysinibacillus species with potential probiotic properties has been isolated from the intestinal tract of large yellow croaker.
An important prerequisite for the efficacy of probiotics is their ability to colonize the host gut, with adhesion representing the initial phase of colonization, immune activation, and antagonistic activity against pathogens. Many pathogenic bacteria establish infections by attaching to the mucosal layer of the gastrointestinal tract, whereas probiotics benefit the host by colonizing the gut and competing with pathogenic bacteria for adhesion sites. Consequently, we selected strains that exhibit both potent bacterial inhibition and substantial adhesion ability to maximize their potential in blocking the adhesion and colonization of pathogens. Many Bacillus species, including Bacillus KUAQ1, Bacillus KUAQ2, Bacillus C-3102, Bacillus subtilis, and B. velezensis LF01, have demonstrated the capacity to compete for adhesion sites on the gastrointestinal epithelium of the Nile tilapia (He et al., 2013; Zhang et al., 2019; Sookchaiyaporn et al., 2020; Tabassum et al., 2021), with B. pumilus, B. firmus, B. subtilis, and B. cereus found to effectively inhibit the growth of V. harveyi, V. parahaemolyticus, V. alginolyticus, and Mycobacterium species in Nile tilapia when used as probiotic bacteria (Das et al., 2010). Consistent with these findings, our studied strains exhibited inhibitory activity against all five pathogenic bacteria through competitive, substitutive, and rejection mechanisms, with substitutive adhesion inhibition proving particularly effective.
The capability for co-aggregation is regarded as a mechanism for pathogen exclusion from the host, enhancing competition for epithelial cell receptors and potentially safeguarding the intestines against pathogenic microbial invasions (Balakrishna, 2013; Dutta et al., 2018; Kaktcham et al., 2018; Meidong et al., 2018; Rungsirivanich et al., 2020). In the present study, all evaluated strains demonstrated the ability to co-aggregate with pathogenic bacteria, achieving inhibition rates exceeding 45%. Notably, the LYD11 strain exhibited the highest co-aggregation rates with E. tarda (57.21%) and P. plecoglossicida (54.08%) after 2 h of incubation. These results align with earlier studies documenting co-aggregation rates of 43.4% for B. HLJ1 and 36.7% for B. C1 with V. parahaemolyticus following a 2-h incubation period (Jiang et al., 2023). Overall, these findings highlight the considerable potential of LYD11 in managing bacterial infections among aquatic animals.
Assessing hydrophobicity and auto-aggregation of bacteria serves as an indirect measure of the adhesion potential of a probiotic strain. Significant correlations between hydrophobicity, auto-aggregation, and cellular adhesion have been noted, underpinning the role these properties play in enabling probiotic bacteria to colonize aquatic animals. Moreover, probiotic aggregation is key not only to intestinal adhesion and colonization but also to immune system modulation (Balakrishna, 2013; Dutta et al., 2018; Kaktcham et al., 2018; Khan et al., 2021). In the present study, the evaluated strain showed 82.73% hydrophobicity after 40 min of incubation and 82.73% self-aggregation capacity after 2 h of incubation, mirroring previously documented findings. The capacity for probiotic strains to bind to mucus on the intestinal wall is deemed essential for their activity, serving as a critical mechanism for probiotic function (Adams, 2010). The fish colonization results were consistent with those of the indirect assessment, suggesting that prolonged intestinal retention post-feeding facilitates the probiotic efficacy of the strains.
The ability of microorganisms to survive in a simulated gastrointestinal environment is an effective indicator for evaluating probiotics. To navigate the complex digestive tract, probiotics need to possess resilience against the low pH of the gastrointestinal environment, withstand destruction by gastric and intestinal fluids, and adapt to elevated bile concentrations for successful survival and colonization (Zhang et al., 2022). Guo et al. (2016) reported that except for B. subtilis, which was isolated from the intestinal tract of grass carp and capable of tolerating strong acidic environments (pH 2.0), the growth of all other Bacillus species was inhibited to varying degrees at the same pH level. Similarly, our study showed that the growth of the LYD11 strain was very slow but persisted after 4 h under both pH 2.0 and artificial gastric fluid conditions. The presence of trypsin in the intestinal fluid, which hydrolyzes bacterial proteins, did not appear to hinder the cultivation of LYD11, with no inhibition of growth observed. Bile salts, antimicrobial compounds present in the digestive system, can pose a significant challenge to bacterial survival and function (Nguyen et al., 2023; Saravanakumar et al., 2023). In this study, the addition of bile salts did not significantly affect the growth of LYD11, with growth inhibition only occurring at a concentration of 0.45%. Therefore, the strains investigated in this study demonstrate considerably adaptability to the extreme environments of the aquatic animal digestive tract, providing strong evidence for their potential as probiotics in aquaculture.
While the effectiveness of probiotics is often the primary focus of research, assessing their safety and suitability for use in animal feeding remains essential (Fu et al., 2023). The antibiotic resistance profile of probiotics is important from a safety perspective (Emam and Dunlap, 2020). In this study, we observed no formation of hemolytic rings on blood agar plates with Bacillus, and antibiotic sensitivity tests revealed that LYD11 was sensitive to 26 antibiotics. Moreover, no instances of fish morbidity or mortality were recorded in the subsequent fish injection and feeding trials, indicating that LYD11 is safe for fish, warranting further investigation. Interestingly, after 7 days of feeding with LYD11, a significant increase in the thickness of the intestinal muscular layer was observed, promoting intestinal digestion and demonstrating beneficial probiotic effects.
This study identified a strong adhesion-capable Lysinibacillus strain from the intestinal tract of the large yellow croaker with high enzyme-producing capacity and strong inhibitory activity against prevalent Vibrio species. This strain demonstrated effective co-aggregation and adhesion exclusion capabilities, crucial for preventing the adhesion and colonization of pathogenic bacteria. Such characteristics make it a viable candidate for combating diseases caused by common aquatic pathogens in cultured fish. Capable of thriving across various NaCl concentrations, pH levels, bile salts, and digestive enzymes, the strain also possesses notable hydrophobicity and self-aggregation capacity and does not harm the host, enabling it to safely colonize the gastrointestinal tract of the host. Furthermore, the strain also promotes the thickening of the intestinal muscular layer, thereby enhancing the digestion and absorption of nutrients. Collectively, these findings underscore the promising probiotic properties and potential practical application of the identified strain as a beneficial agent in the large yellow croaker culture.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author/s.
The animal study was approved by Animal Ethics Committee of Jimei University (permit number JMULAC2011-58). The study was conducted in accordance with the local legislation and institutional requirements.
YY: Writing – original draft, Investigation. XW: Writing – original draft, Investigation. XL: Writing – review & editing, Resources, Conceptualization. JW: Writing – original draft, Resources. PW: Writing – original draft, Investigation. CZ: Writing – review & editing, Resources. QY: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China under contract No. 32373181, Science and Technology Plan Project of Fujian Province under contract No. 2022L3059, Natural Science Foundation of Fujian Province under contract No. 2022J02044, Science and Technology Plan Project of Fuzhou City under contract No. 2022-P-014, Open fund of Fujian Province Key Laboratory of Special Aquatic Formula Feed under contract No. TMKJZ2302.
Authors Chuanzhong Zhu was/were employed by Fujian Dabeinong Aquatic Sci. & Tech. Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams C. A. (2010). The probiotic paradox: live and dead cells are biological response modifiers. Nutr. Res. Rev. 23, 37–46. doi: 10.1017/S0954422410000090
Adorian T. J., Jamali H., Farsani H. G., Darvishi P., Hasanpour S., Bagheri T., et al. (2019). Effects of probiotic bacteria bacillus on growth performance, digestive enzyme activity, and hematological parameters of Asian sea bass, lates calcarifer (Bloch). Probio. Antimicro. Prot. 11, 248–255. doi: 10.1007/s12602-018-9393-z
Balakrishna A. (2013). In vitro evaluation of adhesion and aggregation abilities of four potential probiotic strains isolated from guppy (Poecilia reticulata). Braz. Arch. Biol. Technol. 56, 793–800. doi: 10.1590/S1516-89132013000500010
Cerezuela R., Guardiola F. A., González P., Meseguer J., Esteban M.Á. (2012). Effects of dietary Bacillus subtilis, Tetraselmis chuii, and Phaeodactylum tricornutum, singularly or in combination, on the immune response and disease resistance of sea bream (Sparus aurata L.). Fish Shellf. Immunol. 33, 342–349. doi: 10.1016/j.fsi.2012.05.004
Chang C., Liu W., Shyu C. (2000). Use of prawn blood agar hemolysis to screen for bacteria pathogenic to cultured tiger prawns Penaeus monodon. Dis. Aquat. Org. 43, 153–157. doi: 10.3354/dao043153
Chauhan A., Singh R. (2019). Probiotics in aquaculture: a promising emerging alternative approach. Symbiosis 77, 99–113. doi: 10.1007/s13199-018-0580-1
Das S., Ward L. R., Burke C. (2010). Screening of marine Streptomyces spp. for potential use as probiotics in aquaculture. Aquaculture 305, 32–41. doi: 10.1016/j.aquaculture.2010.04.001
Dutta D., Banerjee S., Mukherjee A., Ghosh K. (2018). Potential gut adherent probiotic bacteria isolated from rohu, Labeo rohita (Actinopterygii: Cypriniformes: Cyprinidae): Characterisation, exo-enzyme production, pathogen inhibition, cell surface hydrophobicity, and bio-film formation. Acta Ichthyol. Piscat. 48, 221–233. doi: 10.3750/AIEP/02251
El-Saadony M. T., Alagawany M., Patra A. K., Kar I., Tiwari R., Dawood M. A. O., et al. (2021). The functionality of probiotics in aquaculture: An overview. Fish Shellf. Immunol. 117, 36–52. doi: 10.1016/j.fsi.2021.07.007
Emam A. M., Dunlap C. A. (2020). Genomic and phenotypic characterization of Bacillus velezensis AMB-y1; a potential probiotic to control pathogens in aquaculture. Antonie van Leeuwenhoek 113, 2041–2052. doi: 10.1007/s10482-020-01476-5
Fu C., Shah A. A., Khan R. U., Khan M. S., Wanapat M. (2023). Emerging trends and applications in health-boosting microorganisms-specific strains for enhancing animal health. Microbial. Pathogen. 183, 106290. doi: 10.1016/j.micpath.2023.106290
Geng X., Dong X.-H., Tan B.-P., Yang Q.-H., Chi S.-Y., Liu H.-Y., et al. (2012). Effects of dietary probiotic on the growth performance, non-specific immunity and disease resistance of cobia, Rachycentron canadum: Effect of dietary probiotic on growth and immunity of cobia. Aquacult. Nutr. 18, 46–55. doi: 10.1111/anu.2011.18.issue-1
Guo X., Chen D.-D., Peng K.-S., Cui Z.-W., Zhang X.-J., Li S., et al. (2016). Identification and characterization of Bacillus subtilis from grass carp (Ctenopharynodon idellus) for use as probiotic additives in aquatic feed. Fish Shellf. Immunol. 52, 74–84. doi: 10.1016/j.fsi.2016.03.017
He S., Zhang Y., Xu L., Yang Y., MArubashi T., Zhou Z., et al. (2013). Effects of dietary Bacillus subtilis C-3102 on the production, intestinal cytokine expression and autochthonous bacteria of hybrid tilapia Oreochromis niloticus ♀×Oreochromis aureus ♂. Aquaculture 412–413, 125–130. doi: 10.1016/j.aquaculture.2013.06.028
Huang L., Shui X., Wang H., Qiu H., Tao C., Yin H., et al. (2023). Effects of Bacillus halophilus on growth, intestinal flora and metabolism of Larimichthys crocea. Biochem. Biophys. Rep. 35, 101546. doi: 10.1016/j.bbrep.2023.101546
Iorizzo M., Albanese G., Letizia F., Testa B., Tremonte P., Vergalito F., et al. (2022). Probiotic potentiality from versatile lactiplantibacillus plantarum strains as resource to enhance freshwater fish health. Microorganisms 10, 463. doi: 10.3390/microorganisms10020463
Jahan N., Islam S. M. M., Rohani M. F., Hossain M. T., Shahjahan M. (2021). Probiotic yeast enhances growth performance of rohu (Labeo rohita) through upgrading hematology, and intestinal microbiota and morphology. Aquaculture 545, 737243. doi: 10.1016/j.aquaculture.2021.737243
Jiang N., Hong B., Luo K., Li Y., Fu H., Wang J. (2023). Isolation of Bacillus subtilis and Bacillus pumilus with Anti-Vibrio parahaemolyticus Activity and Identification of the Anti-Vibrio parahaemolyticus Substance. Microorganisms 11, 1667. doi: 10.3390/microorganisms11071667
Jlidi M., Akremi I., Ibrahim A. H., Brabra W., Ali M. B., Ali M. B. (2022). Probiotic properties of Bacillus strains isolated from the gastrointestinal tract against pathogenic Vibriosis. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.884244
Kaktcham P. M., Temgoua J.-B., Zambou F. N., Diaz-Ruiz G., Wacher C., Pérez-Chabela M. D. L. (2018). In vitro evaluation of the probiotic and safety properties of bacteriocinogenic and non-bacteriocinogenic lactic acid bacteria from the intestines of Nile tilapia and common carp for their use as probiotics in aquaculture. Probio. Antimicro. Prot. 10, 98–109. doi: 10.1007/s12602-017-9312-8
Kasozi N., Iwe G. D., Walakira J., Langi S. (2023). Integration of probiotics in aquaponic systems: an emerging alternative approach. Aquacult. Int. 32, 2131–2150. doi: 10.1007/s10499–023-01261-x
Kavitha M., Raja M., Perumal P. (2018). Evaluation of probiotic potential of Bacillus spp. isolated from the digestive tract of freshwater fish Labeo calbasu (Hamilton 1822). Aquacult. Rep. 11, 59–69. doi: 10.1016/j.aqrep.2018.07.001
Khan M. N., Bashir S., Imran M. (2023). Probiotic characterization of Bacillus species strains isolated from an artisanal fermented milk product Dahi. Folia Microbiol. 68, 757–769. doi: 10.1007/s12223-023-01048-w
Khan Md. I.R., Kamilya D., Choudhury T. G., Tripathy P. S., Rathore G. (2021). Deciphering the Probiotic Potential of Bacillus amyloliquefaciens COFCAU_P1 Isolated from the Intestine of Labeo rohita Through In Vitro and Genetic Assessment. Probio. Antimicro. Prot. 13, 1572–1584. doi: 10.1007/s12602-021-09788-2
Kuebutornye F. K. A. (2020). In vitro Assessment of the Probiotic Characteristics of Three Bacillus Species from the Gut of Nile Tilapia, Oreochromis niloticus. Probiotics Antimicrob. Proteins. 12, 412–424. doi: 10.1007/s12602-019-09562-5
Kuebutornye F. K. A., Abarike E. D., Lu Y. (2019). A review on the application of Bacillus as probiotics in aquaculture. Fish Shellf. Immunol. 87, 820–828. doi: 10.1016/j.fsi.2019.02.010
Kumar V., Roy S., Meena D. K., Sarkar U. K. (2016). Application of probiotics in shrimp aquaculture: importance, mechanisms of action, and methods of administration. Rev. Fish. Sci. Aquacult. 24, 342–368. doi: 10.1080/23308249.2016.1193841
Leyva-Madrigal K. Y., Luna-González A., Escobedo-Bonilla C. M., Fierro-Coronado J. A., Maldonado-Mendoza I. E. (2011). Screening for potential probiotic bacteria to reduce prevalence of WSSV and IHHNV in whiteleg shrimp (Litopenaeus vannamei) under experimental conditions. Aquaculture 322–323, 16–22. doi: 10.1016/j.aquaculture.2011.09.033
Liao Z., Liu Y., Wei H., He X., Wang Z., Zhuang Z., et al. (2023). Effects of dietary supplementation of Bacillus subtilis DSM 32315 on growth, immune response and acute ammonia stress tolerance of Nile tilapia (Oreochromis niloticus) fed with high or low protein diets. Anim. Nutr. 15, 375–385. doi: 10.1016/j.aninu.2023.05.016
Martínez Cruz P., Ibáñez A. L., Monroy Hermosillo O. A., Ramírez Saad H. C. (2012). Use of probiotics in aquaculture. ISRN Microbiol. 2012, 1–13. doi: 10.5402/2012/916845
Medina M., Sotil G., Flores V., Fernández C., Sandoval N. (2020). In vitro assessment of some probiotic properties and inhibitory activity against Yersinia ruckeri of bacteria isolated from rainbow trout Oncorhynchus mykiss (Walbaum). Aquacult. Rep. 18, 100447. doi: 10.1016/j.aqrep.2020.100447
Meidong R., Khotchanalekha K., Doolgindachbaporn S., Nagasawa T., Nakao M., Sakai K., et al. (2018). Evaluation of probiotic Bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of Pla-mong Pangasius bocourti. Fish Shellf. Immunol. 73, 1–10. doi: 10.1016/j.fsi.2017.11.032
Mortezaei F., Royan M., Allaf Noveirian H., Babakhani A., Alaie Kordghashlaghi H., Balcázar J. L. (2020). In vitro assessment of potential probiotic characteristics of indigenous Lactococcus lactis and Weissella oryzae isolates from rainbow trout ( Oncorhynchus mykiss Walbaum). J. Appl. Microbiol. 129, 1004–1019. doi: 10.1111/jam.14652
Neissi A., Majidi Zahed H., Roshan R. (2024). Probiotic performance of B. subtilis MS. 45 improves aquaculture of rainbow trout Oncorhynchus mykiss during acute hypoxia stress. Sci. Rep. 14, 3720. doi: 10.1038/s41598–024-54380–7
Nguyen N. H. K., Giang B. L., Truc T. T. (2023). Isolation and Evaluation of the Probiotic Activity of Lactic Acid Bacteria Isolated from Pickled Brassica juncea (L.) Czern. et Coss. Foods 12, 3810. doi: 10.3390/foods12203810
Peixoto R. S., Harkins D. M., Nelson K. E. (2021). Advances in microbiome research for animal health. Annu. Rev. Anim. Biosci. 9, 289–311. doi: 10.1146/annurev-animal-091020-075907
Proespraiwong P., Mavichak R., Imaizumi K., Hirono I., Unajak S. (2023). Evaluation of Bacillus spp. as Potent Probiotics with Reduction in AHPND-Related Mortality and Facilitating Growth Performance of Pacific White Shrimp (Litopenaeus vannamei) Farms. Microorganisms 11, 2176. doi: 10.3390/microorganisms11092176
Reda R. M., Selim K. M., El-Sayed H. M., El-Hady M. A. (2018). In vitro selection and identification of potential probiotics isolated from the gastrointestinal tract of Nile tilapia, oreochromis niloticus. Probio. Antimicro. Prot. 10, 692–703. doi: 10.1007/s12602-017-9314-6
Rosland N.-A., Ikhsan N., Min C. C., Yusoff F. M., Karim M. (2021). Influence of symbiotic probiont strains on the growth of amphora and chlorella and its potential protections against vibrio spp. in artemia. Curr. Microbiol. 78, 3901–3912. doi: 10.1007/s00284-021-02642-2
Rungsirivanich P., Supandee W., Futui W., Chumsai-Na-Ayudhya V., Yodsombat C., Thongwai N. (2020). Culturable Bacterial Community on Leaves of Assam Tea (Camellia sinensis var. assamica) in Thailand and Human Probiotic Potential of Isolated Bacillus spp. Microorganisms 8, 1585. doi: 10.3390/microorganisms8101585
Sankar H., Philip B., Philip R., Singh I. S. B. (2017). Effect of probiotics on digestive enzyme activities and growth of cichlids, Etroplus suratensis (Pearl spot) and Oreochromis mossambicus (Tilapia). Aquacult. Nutr. 23, 852–864. doi: 10.1111/anu.2017.23.issue-4
Santos Y., Bandin I., Nieto T. P., Bruno D. W., Ellis A. E., Toranzo A. E. (1990). “Comparison of the cell surface hydrophobicity of bacterial fish pathogens by different procedures,” in Pathology in Marine Science (San Diego, California: Elsevier), 101–115. doi: 10.1016/B978–0-12–550755–4.50016–1
Santos R. A., Oliva-Teles A., Pousão-Ferreira P., Jerusik R., Saavedra M. J., Enes P., et al. (2021). Isolation and characterization of fish-gut bacillus spp. as source of natural antimicrobial compounds to fight aquaculture bacterial diseases. Mar. Biotechnol. 23, 276–293. doi: 10.1007/s10126-021-10022-x
Saravanakumar S., Prabakaran N. N., Ashokkumar R., Jamuna S. (2023). Unlocking the Gut’s Treasure: Lipase-Producing Bacillus subtilis Probiotic from the Intestine of Microstomus kitt (Lemon sole). Appl. Biochem. Biotechnol. doi: 10.1007/s12010-023-04749-7
Sookchaiyaporn N., Srisapoome P., Unajak S., Areechon N. (2020). Efficacy of Bacillus spp. isolated from Nile tilapia Oreochromis niloticus Linn. on its growth and immunity, and control of pathogenic bacteria. Fish Sci. 86, 353–365. doi: 10.1007/s12562-019-01394-0
Sumon M. S., Ahmmed F., Khushi S. S., Ahmmed M. K., Rouf M. A., Chisty Md. A.H., et al. (2018). Growth performance, digestive enzyme activity and immune response of Macrobrachium rosenbergii fed with probiotic Clostridium butyricum incorporated diets. J. King Saud Univ. - Sci. 30, 21–28. doi: 10.1016/j.jksus.2016.11.003
Tabassum T., Sofi Uddin Mahamud A. G. M., Acharjee T. K., Hassan R., Akter Snigdha T., Islam T., et al. (2021). Probiotic supplementations improve growth, water quality, hematology, gut microbiota and intestinal morphology of Nile tilapia. Aquacult. Rep. 21, 100972. doi: 10.1016/j.aqrep.2021.100972
Thankappan B., Ramesh D., Ramkumar S., Natarajaseenivasan K., Anbarasu K. (2015). Characterization of Bacillus spp. From the Gastrointestinal Tract of Labeo rohita—Towards to Identify Novel Probiotics Against Fish Pathogens. Appl. Biochem. Biotechnol. 175, 340–353. doi: 10.1007/s12010-014-1270-y
Uzun E., Ogut H. (2015). The isolation frequency of bacterial pathogens from sea bass (Dicentrarchus labrax) in the Southeastern Black Sea. Aquaculture 437, 30–37. doi: 10.1016/j.aquaculture.2014.11.017
Van Doan H., Hoseinifar S. H., Ringø E., Ángeles Esteban M., Dadar M., Dawood M. A. O., et al. (2020). Host-associated probiotics: A key factor in sustainable aquaculture. Rev. Fish. Sci. Aquacult. 28, 16–42. doi: 10.1080/23308249.2019.1643288
Vine N. G., Leukes W. D., Kaiser H. (2004). In vitro growth characteristics of five candidate aquaculture probiotics and two fish pathogens grown in fish intestinal mucus. FEMS Microbiol. Lett. 231, 145–152. doi: 10.1016/S0378-1097(03)00954-6
Wang P., Chen S., Wei C., Yan Q., Sun Y.-Z., Yi G., et al. (2021). Monascus purpureus M-32 improves growth performance, immune response, intestinal morphology, microbiota and disease resistance in Litopenaeus vannamei. Aquaculture 530, 735947. doi: 10.1016/j.aquaculture.2020.735947
Wang J., Wu Z., Wang S., Wang X., Zhang D., Wang Q., et al. (2022). Inhibitory effect of probiotic Bacillus spp. isolated from the digestive tract of Rhynchocypris Lagowskii on the adhesion of common pathogenic bacteria in the intestinal model. Microbial. Pathogen. 169, 105623. doi: 10.1016/j.micpath.2022.105623
Wu X.-Y., Xiong J.-B., Fei C.-J., Dai T., Zhu T.-F., Zhao Z.-Y., et al. (2022). Prior exposure to ciprofloxacin disrupts intestinal homeostasis and predisposes ayu (Plecoglossus altivelis) to subsequent Pseudomonas plecoglossicida-induced infection. Zool. Res. 43, 648–665. doi: 10.24272/j.issn.2095–8137.2022.159
Xin G., Zhao L., Zhuang Z., Wang X., Fu Q., Huang H., et al. (2022). Function of the rpoD gene in Pseudomonas plecoglossicida pathogenicity and Epinephelus coioides immune response. Fish Shellf. Immunol. 127, 427–436. doi: 10.1016/j.fsi.2022.06.045
Yamashita M. M., Ferrarezi J. V., Pereira G. D. V., Bandeira G., Côrrea Da Silva B., Pereira S. A., et al. (2020). Autochthonous vs allochthonous probiotic strains to Rhamdia quelen. Microbial. Pathogen. 139, 103897. doi: 10.1016/j.micpath.2019.103897
Yang D., Zhao L., Li Q., Huang L., Qin Y., Wang P., et al. (2023). flgC gene is involved in the virulence regulation of Pseudomonas plecoglossicida and affects the immune response of Epinephelus coioides. Fish Shellf. Immunol. 132, 108512. doi: 10.1016/j.fsi.2022.108512
Yuan B., Zhao L.-M., Zhuang Z.-X., Wang X.-R., Fu Q., Huang H.-B., et al. (2022). Transcriptomic and metabolomic insights into the role of the flgK gene in the pathogenicity of Pseudomonas plecoglossicida to orange-spotted grouper (Epinephelus coioides). Zool. Res. 43, 952–965. doi: 10.24272/j.issn.2095-8137.2022.216
Zhang D., Gao Y., Ke X., Yi M., Liu Z., Han X., et al. (2019). Bacillus velezensis LF01: in vitro antimicrobial activity against fish pathogens, growth performance enhancement, and disease resistance against streptococcosis in Nile tilapia (Oreochromis niloticus). Appl. Microbiol. Biotechnol. 103, 9023–9035. doi: 10.1007/s00253-019-10176-8
Zhang Y., Wu Z., Yu M., Zhang D., Wang Q., Lin L., et al. (2022). Evaluating the probiotic potential and adhesion characteristics of Bacillus spp. isolated from the intestine of Rhynchocypris lagowskii Dybowski. Aquacult. Int. 30, 747–772. doi: 10.1007/s10499-022-00840-8
Keywords: bacterial diseases, probiotics, Lysinibacillus sp., adhesion, colonization, safety assessment
Citation: Yao Y, Wang X, Lin X, Wu J, Wang P, Zhu C and Yan Q (2024) Isolation and characterization of probiotic Lysinibacillus species from the gastrointestinal tract of large yellow croaker (Larimichthys crocea). Front. Mar. Sci. 11:1408979. doi: 10.3389/fmars.2024.1408979
Received: 15 April 2024; Accepted: 16 May 2024;
Published: 31 May 2024.
Edited by:
Francesca Turroni, University of Parma, ItalyReviewed by:
Maria Touraki, Aristotle University of Thessaloniki, GreeceCopyright © 2024 Yao, Wang, Lin, Wu, Wang, Zhu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingpi Yan, eWFucXBAam11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.