
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 17 May 2024
Sec. Marine Ecosystem Ecology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1408958
This article is part of the Research Topic The Cycling of Biogenic Elements and Their Microbial Transformations in Marine Ecosystems View all 12 articles
 Pengbing Pei1
Pengbing Pei1 Muhammad Aslam1,2
Muhammad Aslam1,2 Chunyou Yang1
Chunyou Yang1 Peilin Ye1
Peilin Ye1 Xiao Ke1
Xiao Ke1 Zhanhua Liang1
Zhanhua Liang1 Tangcheng Li3
Tangcheng Li3 Weizhou Chen1
Weizhou Chen1 Hong Du1,4*
Hong Du1,4*Diazotrophic communities contribute inorganic nitrogen for the primary productivity of the marine environment by biological nitrogen fixation (BNF). They play a vital role in the biogeochemical cycle of nitrogen in the marine ecological environment. However, there is still an incomplete understanding of BNF and diazotrophs in artificial seaweed farms. Therefore, this study comprehensively investigated the temporal variations of BNF associated with Gracilariopsis lemaneiformis, as well as the diazotrophic communities associated with macroalgae and its surrounding seawater. Our results revealed that a total of 13 strains belonging to Proteobacteria and Bacteroidetes were identified as N2-fixing bacteria using azotobacter selective solid medium and nifH gene cloning. Subsequently, BNF and diazotrophic communities were characterized using the acetylene reduction method and high-throughput sequencing of the nifH gene, respectively. The results showed that nitrogenase activity and nifH gene abundance of epiphytic bacteria on G. lemaneiformis varied significantly among four different cultivation periods, i.e., Cultivation Jan. (CJ), Cultivation Feb. (CF), Cultivation Mar. (CM), Cultivation Apr. (CA). Among them, the nitrogenase activity and nifH gene abundance of epiphytic bacteria on G. lemaneiformis in CM were significantly higher than those in CJ, CF, and CA, indicating that the BNF of eiphytic bacteria on G. lemaneiformis was markedly enhanced. Combined with the data on environmental factors, it was found that the low concentration of nitrogen and phosphorus in CM might considerably boost the BNF of epiphytic bacteria in G. lemaneiformis. The sequencing results of the nifH gene showed that the α-diversity of diazotrophic communities associated with G. lemaneiformis and seawater in CM was higher than that in other cultivation periods. In addition, the diazotrophic communities on G. lemaneiformis were significantly different in CJ, CF, CM, and CA, and they were significantly diverse from diazotrophic communities in seawater. LEfSe analysis indicated that Rhodobacterales, Hyphomonadaceae, Robiginitomaculum, and Robiginitomaculum antarcticum within α-proteobacteria played a remarkable role in BNF in response to nitrogen nutrient deficiency. Taken together, these results provide a unique insight into the interaction between macroalgae and its epiphytic bacteria and lay a foundation for further research on the mechanism of action of nitrogen-cycling microorganisms associated with macroalgae.
Seaweed farms are important ecosystems along subtropical coastlines, playing a vital role in improving the quality of coastal waters and maintaining the ecological balance of subtropical regions (Kim et al., 2017; Zhang et al., 2017a). The carbon fixed by cultivated seaweed may either be buried in sediments or exported to the deep sea, thus establishing seaweed farms ecosystem as a major contributor of marine carbon sink (Duarte et al., 2017; Zhang et al., 2017b; Hasselström et al., 2018). To a certain extent, seaweed cultivation can mitigate wave energy, protect shorelines, enhance pH, and supply oxygen to the waters. This contribution aids in climate change adaptation by locally reducing the effects of ocean acidification and de-oxygenation (Duarte et al., 2017). In subtropical marine environments, seaweeds are considered crucial primary producers of organic matter (Neori, 2008). However, despite the richness of organic matter in seaweed farm ecosystems, they tend to become oligotrophic environments due to the significant consumption of inorganic nitrogen nutrients in the water caused by large-scale seaweed cultivation (Yang et al., 2015).
Microorganisms are a crucial component of seaweed farm ecosystems, and there is mounting evidence highlighting their importance in biogeochemical cycling and the overall health of these ecosystems (Azam and Malfatti, 2007; Rousk and Bengtson, 2014; Egan and Gardiner, 2016). These microorganisms primarily include epiphytic microbes in seaweed and seawater, which significantly regulate the circulation of marine nutrient elements (Dang and Lovell, 2016). Functional microbes have garnered substantial attention among these microorganisms due to their pivotal roles. Nitrogen-fixing microorganisms, also known as diazotrophs, are important functional microorganisms in marine environments (Cardini et al., 2017). Although diazotrophs may differ significantly in taxonomy, they harbor similar functional genes that enable them to perform the same ecological functions. BNF performed by diazotrophs, involves the reduction of atmospheric N2 to bioavailable ammonium (Caffin et al., 2018). This process constitutes a significant source of bioavailable nitrogen in numerous marine and freshwater environments (Bonnet et al., 2016; Geisler et al., 2019). In addition, BNF by marine diazotrophs compensates for the loss of nitrogen through sinking and denitrification, thereby sustaining the primary productivity of marine ecosystems (Yang et al., 2019). Diazotrophic communities are affected by many key factors, such as energy to sustain nitrogenase activity, availability of vitamins and micronutrients, oxidized environments, and concentrations of dissolved inorganic nitrogen (Geisler et al., 2019). For instance, adequate inorganic nitrogen to meet the nitrogen requirements of diazaotrophs means that consequential rates of marine BNF process do not occur (Knapp, 2012).
Many coral reefs still hold relatively high biological productivity despite low dissolved inorganic nitrogen (DIN) concentrations in marine environments (Messer et al., 2017). This seemingly paradoxical phenomenon can be attributed to the crucial role played by associated BNF in providing bioavailable nitrogen to coral reef habitats (Zhang et al., 2016; Lesser et al., 2018). Additionally, diazotrophs have been identified as a significant component of coral symbionts (Olson and Lesser, 2013; Zhang et al., 2016). In our previous observations, we find that the growth status of G. lemaneiformis maintains normal in the marine environment with low concentration of inorganic nitrogen (data not shown). This discrepancy between the availability of inorganic nutrients and the growth of G. lemaneiformis can be understood to mean that there are new bioavailable nitrogen sources besides the inorganic nutrients to sustain the growth of macroalgae. New nitrogen sources may be derived from the BNF drove by associated diazotrophs in many marine and freshwater environments (Sohm et al., 2011; Geisler et al., 2019). Although this inconsistent phenomenon has been demonstrated in coral reef habitats, the role of associated diazotrophs and BNF in the cultivation environment of G. lemaneiformis farms remains unclear.
Diazotrophic communities form specific species associations with their hosts (Lema et al., 2012), and differences in diazotrophic communities are observed under different environmental conditions (Lema et al., 2014). The significant effects of environmental conditions on diazotrophs’ diversity and community structure may alter their ability to perform BNF (Feng et al., 2018). It is worth noting that the environmental conditions for the growth of G. lemaneiformis may vary during the entire cultivation period. Therefore, it is imperative to study the nitrogenase activity, nifH gene abundance of epiphytic bacteria on G. lemaneiformis, and the variations of diazotrophic community associated with G. lemaneiformis and seawater.
In recent years, high-throughput sequencing has provided a more comprehensive view of microbial communities in different environments and has been employed to investigate the diversity and composition of bacterial communities associated with seaweed farms (Xie et al., 2017; Liang et al., 2019; Selvarajan et al., 2019; Wang et al., 2020, 2021; Pei et al., 2021). The functional nifH gene encodes a conserved subunit of nitrogenase ferritin and is present in all known diazotrophs (Zehr et al., 2003). The nifH gene is consistent with 16S rRNA gene phylogeny and serves as an ideal molecular target (Gaby and Buckley, 2012). It is widely used to study the diversity and community composition of diazotrophs (Yang et al., 2019). However, the nifH gene has seldom been studied in high-throughput sequencing of diazotrophic communities associated with G. lemaneiformis and seawater, and comprehensive knowledge about the role of environmental variables in shaping the diazotrophic communities of G. lemaneiformis and seawater is limited. Thus, it is necessary to investigate the nitrogenase activity, nifH gene abundance of epiphytic bacteria on G. lemaneiformis, and the diversity and composition of diazotrophic communities associated with G. lemaneiformis and seawater to enhance our understanding of seaweed farms ecosystem functioning.
In this study, we aim to achieve the following objectives: (i) isolate and identify culturable N2-fixing bacteria by plate streaking and PCR identification; (ii) measure nitrogenase activity and nifH gene abundance in epiphytic bacteria on G. lemaneiformis using acetylene reduction and qPCR; and (iii) explore the diversity and composition of diazotrophic communities associated with G. lemaneiformis and seawater through high-throughput sequencing of the nifH gene. This study provides a scientific foundation for further understanding of the BNF function of diazotrophs associated with the macroalgae G. lemaneiformis.
Samples of G. lemaneiformis and seawater were collected seasonally (Cultivation in January, CJ; Cultivation in February, CF; Cultivation in March, CM; Cultivation in April, CA) from artificial seaweed farms located in Zoumapu Village, Nan’ao Island of Guangdong Province (117°2′39″E, 23°28′32″N). At each month, three replicate G. lemaneiformis were collected for composition of diazotrophic communities and BNF analyses, respectively. In addition, three replicate seawater at each month were collected for composition of diazotrophic communities analyses. Three replicate surface (0.5 m depth) seawater were collected at each month (0.5 L) for physicochemical analyses. Full details of the sampling information can be found in Supplementary Tables S1 and S2. The cultivation of G. lemaneiformis has been shown to affect various dissolved and particulate forms of nitrogen and phosphorus in seaweed farms (Huang et al., 2017; Zhang et al., 2018). Therefore, differences in nutrient loads during various cultivation periods may influence the diversity of diazotrophic communities associated with seaweed and seawater. The latitude and longitude were measured by GPS (Garmin GPS72H) from the sampling site. The algal samples were stored in sterile polyethylene bags with surrounding seawater, while the seawater samples were stored in sterile polyethylene bottles. Algal and seawater samples were stored at 4°C and transported to the laboratory within two hours.
In the laboratory, G. lemaneiformis samples were washed three times with autoclaved seawater to eliminate loosely attached epiphytes, sand particles and other attached settlements (Stabili et al., 2017; Karthick and Mohanraju, 2018). After rinsing, sterile cotton buds were used to swab repeatedly firmly attached epiphytic bacteria from algae. They were spread on azotobacter selective solid medium (ASSM), or algae was spread directly on the ASSM. The ASSM consists of 0.1 g K2HPO4, 0.005 g Na2MoO4·2H2O, 0.05 g CaSO4·2H2O, 0.2 g MgSO4·7H2O, 0.03 g FeCl3·6H2O, 5.0 g sucrose, 0.5 g yeast extract, 15.0 g agar, and 1.0 L filtered seawater (Head and Carpenter, 1975). Additionally, 0.1 mL seawater sample was dropped onto the ASSM, and the bacteria solution was evenly spread using a sterile spreader. The plates were then incubated at 25°C for 4 days. Morphological i.e. (size, shape, color etc) different bacterial colonies were picked with inoculating loop to make streak on plates for getting purified colonies. This step was repeated twice in order to obtain pure individual colonies (22 colonies from G. lemaneiformis, 6 colonies from seawater), which were then preserved at -80°C in marine broth supplemented with 25% sterile glycerol.
The DNA of pure bacterial colonies was extracted following the procedures described in the DNA extraction kit (TIANGEN Biotech, Beijing). The 16S rRNA gene was amplified using the universal primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’) (Pei et al., 2024). PCR was carried out in 30 μL reaction mixture containing 1.0 μL genomic DNA, 15.0 μL Super Mix (Sigma), 1.0 μL of each primer, and 12.0 μL H2O under thermal cycle of 96°C for 5 min, 35 cycles of 20 s at 96°C, 30 s at 62°C and 10 min at 72°C, followed by 72°C for 10 min in a Bio-rad T100™ Thermal Cycler (Bio-Rad, USA). The quality of PCR products was verified by 1% agarose electrophoresis gel. The 3730XL DNA Analyzer (ABI, USA) was used for sequencing of pure isolates. The sequencing primers used for the 16S rRNA gene were V4-515F (5’-GTGCCAGCAGCCGCGGTAA-3’) and V4-806R (5’-GGACTACCAGGGTATCTAA-3’). The genetic relationship between different samples and known bacterial species were determined according to blast results. The most closely related bacterial species was selected as the species identification information of the sample. Different strains with identical description of matched species and accession number were considered to be the same species. The sequence data reported in this study has been deposited in the NCBI GenBank database under the accession number SUB11205054. All data are available at: https://submit.ncbi.nlm.nih.gov/subs/?search=SUB11205054.
The nifH gene of potential N2-fixing bacteria was amplified using the universal primers PolF (5’-TGCGAYCCSAARGCBGACTC-3’) and PolR (5’-ATSGCCATCATYTCRCCGGA-3’) (Poly et al., 2001). PCR was carried out in 20 μL reaction mixture containing 2.0 μL genomic DNA, 1.0 μL of each primer, 10.0 μL 2×Master Mix, and 6.0 μL H2O under thermal cycle of 98°C for 30 s, 35 cycles of 10 s at 98°C, 30 s at 68°C and 30 s at 72°C, followed by 72°C for 2 min in a Bio-rad T100™ Thermal Cycler (Bio-Rad, USA). PCR products were verified by running them on a 2% agarose electrophoresis gel and subsequently subjected to sequencing by using a 3730XL DNA Analyzer (ABI, USA). The primers used for sequencing of the nifH gene were the same as those used for amplification of the nifH gene.
The nifH gene sequence was translated to protein sequence (https://web.expasy.org/translate/). The nifH gene sequences of various N2-fixing bacteria were blasted to check their sequence homology against other sequences from NCBI GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The protein sequences of nifH gene of 23 known N2-fixing bacteria with the highest homology were selected. The aligned protein sequences were used to construct the phylogenetic trees with the neighbor joining method using the MEGA 11.0.13 software. The sequences were compiled and aligned using ClustalW embedded in MEGA 11.0.13. For reliability, the bootstrap test was performed with 1000 replications in the phylogenetic trees (Aslam et al., 2023).
We employed the standard method, the reduction of C2H2 to C2H4 (Cardini et al., 2017), to assess the nitrogenase activity of epiphytic bacteria on G. lemaneiformis during a full dark-light cycle (D: L=12 h: 12 h) incubation. Nitrogenase catalyzes not only the reduction of N2 to NH3, but also the reduction of C2H2 to C2H4. C2H2 and N2 have similar affinity for nitrogenase, and their reduction processes are parallel related. Since C2H2 and C2H4 but not N2 and NH3 can be separated using gas chromatography, the reduction of C2H2 to C2H4 can be used to indirectly estimate N2 fixation by organisms. The reaction formulas of N2 + 6H++6e-→2NH3 and 3C2H2 + 6H++6e-→3C2H4 show that the reduction of 3 grams C2H2 is equivalent to the reduction of 1 gram N2, so the amount of N2 fixation can be calculated. First, 1.0 g (fresh weight, FW) of the G. lemaneiformis sample was accurately weighed and placed in a 20 mL headspace vial. Then, 7 mL artificial seawater filtered by 0.22 μm filter was injected into the vial. The artificial seawater composition included 26.518 g NaCl, 2.447 g MgCl2, 3.305 g MgSO4, 1.141 g CaCl2, 0.725 g KCl, 0.202 g NaHCO3, 0.083 g NaBr, 1.0 L dH2O, pH=7.0-7.2. Following the addition of 1.5 mL C2H2 and sealing the vial with pliers, the vials were incubated in a constant temperature illumination incubator for 48 h (temperature: 20 ± 0.5°C, light intensity: 100 ± 5 μmol·m-2·s-1, light cycle: L: D=12 h: 12h). The content of ethylene (C2H4) in gas sample was detected by gas chromatograph (Agilent 7890B GC-FID, Agilent USA). Nitrogenase activity was expressed as nmol C2H4 produced per sample per hour.
The total DNA from G. lemaneiformis and its epiphytic bacteria was extracted using the procedures described by Pei et al. (2021). The primers PolF and PolR were used to quantify the number of nifH gene copies. Appropriate nifH gene sequences were obtained from the NCBI database and synthesized by BGI. Bacteria containing the nifH gene were inoculated in the liquid medium with ampicillin resistance, and the nifH gene plasmid was extracted using TIANprep Mini Plasmid Kit (DP103-03, TIANGEN). Standard curves of nifH gene copy numbers were established by serially diluting plasmids containing the nifH gene, resulting in final concentrations ranging from 102 to 108 copies/μL. The qPCR was carried out in 20 μL reaction mixture containing 2.0 μL plasmid DNA, 0.6 μL of each primer, 10.0 μL 2×Talent qPCR Premix, and 6.8 μL RNase-free ddH2O under thermal cycle of 95°C for 3 min, 40 cycles of 5 s at 95°C, 15 s at 60°C and 15 s at 60°C in a Real-time Fluorescence Quantitative PCR System (qTOWER3G, Jena). The specificity of amplification products was verified by melting-curve analysis, and the amplified fragments were verified by electrophoresis on a 2% agarose gel to confirm the expected sizes of the amplicon. The qPCR efficiency (E) was 86.17%, and the R2 of standards was 0.9996. The total DNA from G. lemaneiformis and its epiphytic bacteria was substituted for plasmid DNA for the qPCR experiment. All samples and standard reactions were performed in triplicate, and average values were calculated. The number of nifH copies was ultimately expressed as per gram of algal fresh weight.
The seawater temperature (Temp.), salinity (Sal.), pH, and dissolved oxygen (DO), were measured using an In-Situ SMARTPOLL MP (U.S.A). Briefly, seawater samples were filtered with a 0.45 μm mixed cellulose ester microporous filter membrane (MF-Millipore HAWP04700, USA) within 2 hours. The filtered seawater samples were added with chloroform (2‰, v/v=chloroform: seawater) and stored at low temperatures for subsequent detection. Ammonia (NH4-N), nitrate (NO3-N), nitrite (NO2-N), total nitrogen (TN), reactive phosphorus (PO4-P), and total phosphorus (TP) levels in the seawater samples were analyzed according to standard methods described by AQSIQ (2007). The content of chlorophyll a (Chl-a) was determined by acetone extraction and spectrophotometry as described in the national standard method (GB17378.7-2007).
Total DNA from G. lemaneiformis and its epiphytic bacteria was extracted using a DNA extraction kit (JL-50, Guangzhou Jianlun Biological Technology Co., LTD.). Total DNA from seawater was extracted using CTAB method (Pei et al., 2021). The concentration and purity were measured using the NanoDrop One (Thermo Fisher Scientific, MA, USA).
The diazotrophic communities were characterized by the nifH gene, which was amplified using the universal primers containing 12 bp barcode PolF and PolR (Poly et al., 2001). The PCR was carried out in 50 μL reaction mixture, containing 25.0 μL 2×Premix Taq (Takara Biotechnology, Dalian Co. Ltd., China), 1.0 μL each primer, 3.0 μL DNA template, and 20 μL RNase-free ddH2O under thermal cycle of 94°C for 5 min, 30 cycles of 30 s at 94°C, 30 s at 52°C and 30 s at 72°C, followed by 72°C for 10 min in a BioRad S1000 Thermal Cycler (Bio-Rad Laboratory, CA, USA). The length and concentration of the PCR products were detected by 1% agarose gel electrophoresis. PCR products were mixed in equidensity ratios according to the GeneTools Analysis Software (Version4.03.05.0, SynGene). The mixture of PCR products was purified with E.Z.N.A. Gel Extraction Kit (Omega, USA). Sequencing libraries were generated using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA). The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific, MA, USA). At last, the library was sequenced on an Illumina Nova6000 platform and 250 bp paired-end reads were generated (Guangdong Magigene Biotechnology Co. Ltd., Guangzhou, China). The sequence data reported in this study have been deposited in the NCBI GenBank database under the accession number SRP368355. All data are available at: https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA824550.
The primers were removed by using cutadapt software (https://github.com.marcelm/cutadapt/) according to the primer information at the beginning and end of sequence to obtain the paired-end clean reads. Paired-end clean reads were merged using usearch -fastq_mergepairs (V10, http://www.drive5.com/usearch/) according to the relationship of the overlap between the paired-end reads. Fastp (version 0.14.1, https://github.com/OpenGene/fastp) was used to control the quality of the raw data by sliding window (-W 4 -M 20) to obtain the paired-end clean tags.
OTUs were clustered based on the UPARSE method by removing the chimera sequence, singleton OTU, and the OTU annotated as chloroplasts or mitochondria (16S amplicons). For each representative sequence, the silva (https://www.arb-silva.de/) database was used to annotate taxonomic information by usearch -sintax (set the confidence threshold to default to ≥ 0.8). The taxonomy of the species annotation was divided into seven levels: kingdom, phylum, class, order, family, genus, and species. R software was used to analyze the common and endemic species, community composition, and richness of species. In order to study phylogenetic relationship of different OTUs, the KRONA software (http://sourceforge.net/projects/krona/) was used to visualize the results of individual sample annotations.
The Richness and Shannon indices were calculated using usearch-alpha_div (V10, http://www.drive5.com/usearch/) and plotted using OriginPro 9.0 software. The differences in the alpha diversity index between groups (GL and SW) were analyzed using R software.
Linear Discriminant Analysis Effect Size (LEfSe) was used to find the biomarker of each group based on homogeneous OTU_table. Initially, a non-parametric factorial Kruskal Wallis (kw) sum-rank test was used to detect species with significant difference in abundance among different groups. Secondly, Wilcoxon rank sum test was used to judge the difference between the two groups. Finally, a linear discriminant analysis (LDA) was used to evaluate the impact of significant species (LDA score) by setting LDA score ≥ 2 and obtained the biomarkers in different groups.
The nitrogenase activity and nifH gene abundance of epiphytic bacteria on G. lemaneiformis, the physicochemical factors of seawater, and the alpha diversity of diazotrophic communities associated with G. lemaneiformis and seawater were analyzed using one-way ANOVA (Pei et al., 2021). The analysis of significant difference of data was carried out using SPSS 19.0 software, with the significant threshold set to 0.05. The differences among the diazotrophic communities associated with G. lemaneiformis and seawater in different cultivation periods were performed by the Permutational Multivariate Analysis of Variance (PERMANOVA) with Adonis function from the vegan package in R software (Xie et al., 2017).
A total of 28 single colonies were screened from G. lemaneiformis and seawater, out of which 16 bacterial species were identified based on 16S rRNA gene. As shown in Table 1, at the phylum level, these isolates were classified into Proteobacteria (75%) and Bacteroidetes (25%).
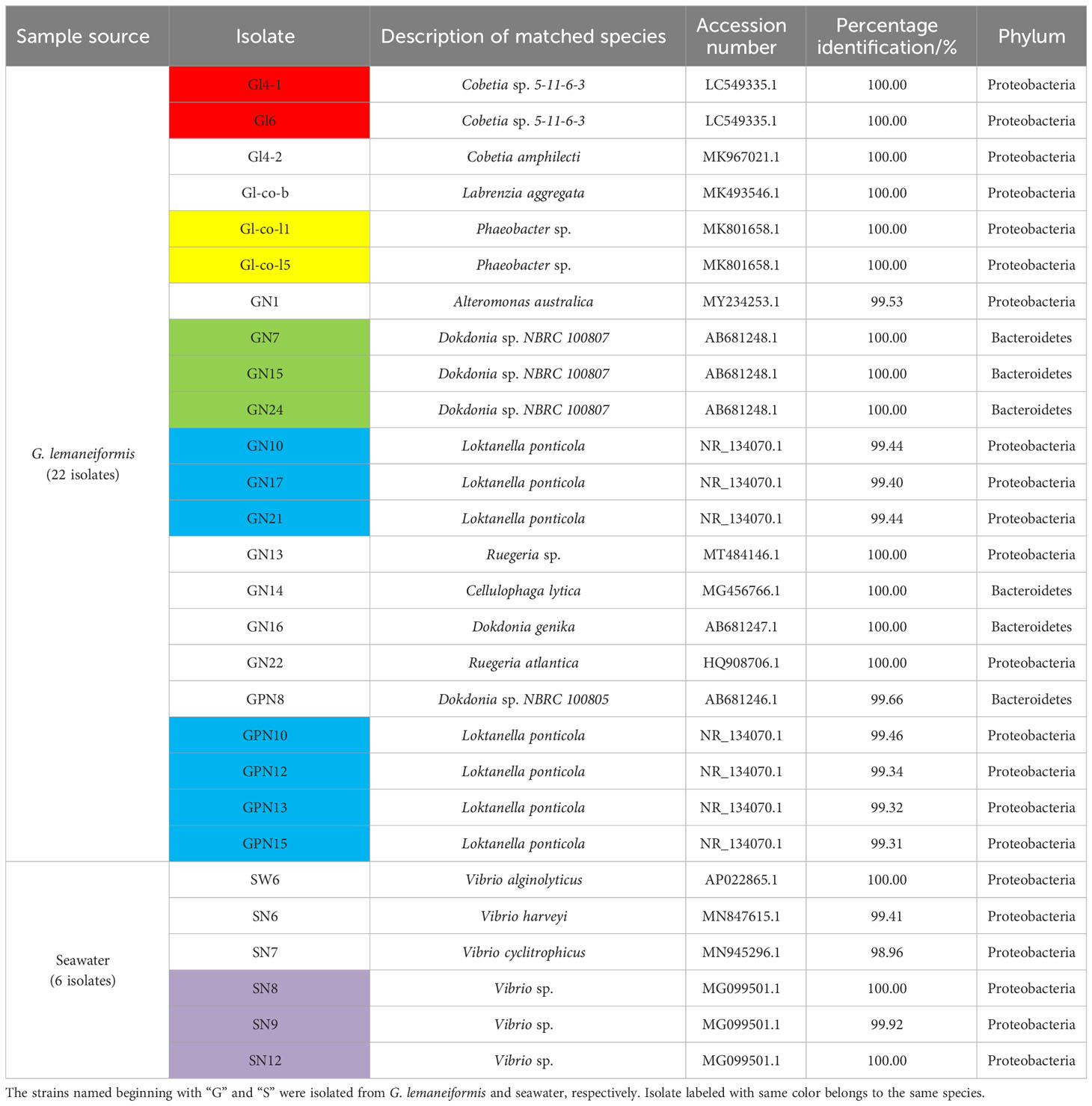
Table 1 16S rRNA gene sequence identity of twenty-eight bacterial isolates obtained from G. lemaneiformis and seawater.
The target gene nifH was analyzed to identify N2-fixing bacteria in various environment. The nifH gene target band of 23 strains was amplified by PCR and was about 360 bp in size (Supplementary Figure S1), consistent with the expected result. These strains with targeted bands were preliminarily believed to contain the nifH gene. To further verify the presence of nifH gene in these strains, 20 μL PCR product was sequenced using the PolF forward primer. Sequencing results showed that 23 strains were successfully sequenced.
Combined with the results of 16S species identification, 13 N2-fixing bacteria were formally obtained from G. lemaneiformis and its seawater and classified into Cobetia (2 species, Gl4-1, Gl6, Gl4-2), Labrenzia (1 species, Gl-co-b), Phaeobacter (1 species, Gl-co-l1, Gl-co-l5), Loktanella (1 species, GN10, GN17, GN21, GPN10, GPN12, GPN13, GPN15), Dokdonia (3 species, GN16, GN24, GPN8), Ruegeria (1 species, GN22), Vibrio (4 species, SW6, SN6, SN7, SN8, SN9, SN12) at the genus level. The nitrogenase gene (nifH) sequence details of all N2-fixing bacteria are shown in Supplementary Table S3. The amino acid sequence details of the nifH gene of 13 N2-fixing bacteria are shown in Supplementary Table S4.
The phylogenetic tree constructed for the N2-fixing bacteria represents their closest relatives as obtained from the GeneBank (Figure 1). The nifH gene sequences of most N2-fixing bacteria were closely related to those of mangrove root bacterium HS001 species, but were estranged from GPN8.
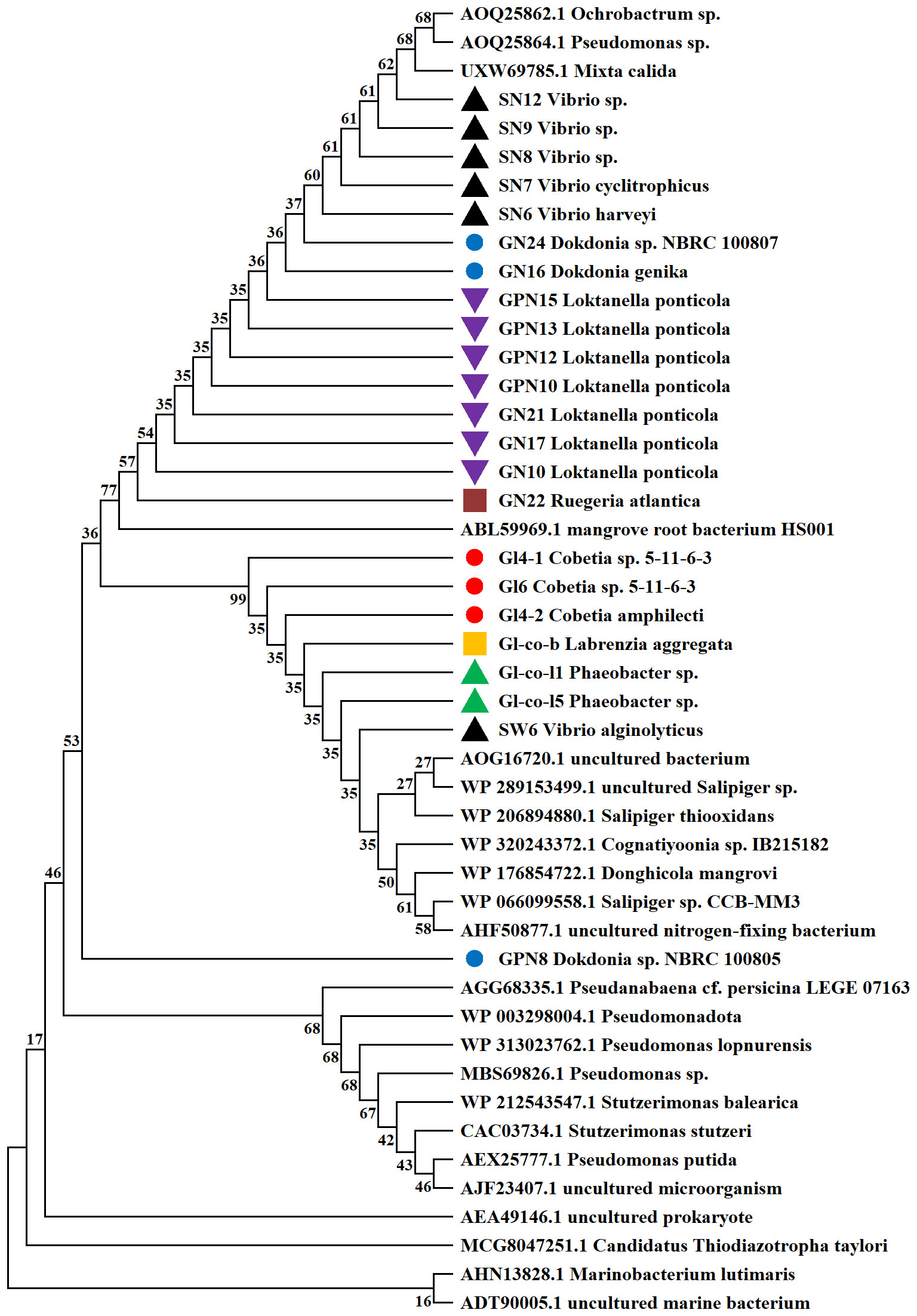
Figure 1 Phylogenetic tree of 13 various N2-fixing bacteria created with MEGA 11.0.13 using the Neighbor-Joining method. Strains harboring the same color belong to the same genus.
The physicochemical factors of seawater from different cultivation periods of study areas including Temp., pH, Sal., DO, NH4-N, NO3-N, NO2-N, dissolved inorganic nitrogen (DIN), PO4-P, Chl-a, TN, and TP levels are shown in Figure 2. The Temp. and pH of CJ were significantly (p < 0.05) lower than those of CF, CM, and CA (Figure 2A), whereas the Sal. and DO of CJ were significantly (p < 0.05) higher than those of CF, CM, and CA (Figure 2B). The concentrations of NH4-N, NO3-N, NO2-N, DIN, PO4-P, and TP in CJ were significantly (p < 0.05) higher than those in CF, CM, and CA (Figures 2C–F). There were no significant differences (p > 0.05) in TN between different cultivation periods (Figure 2F).
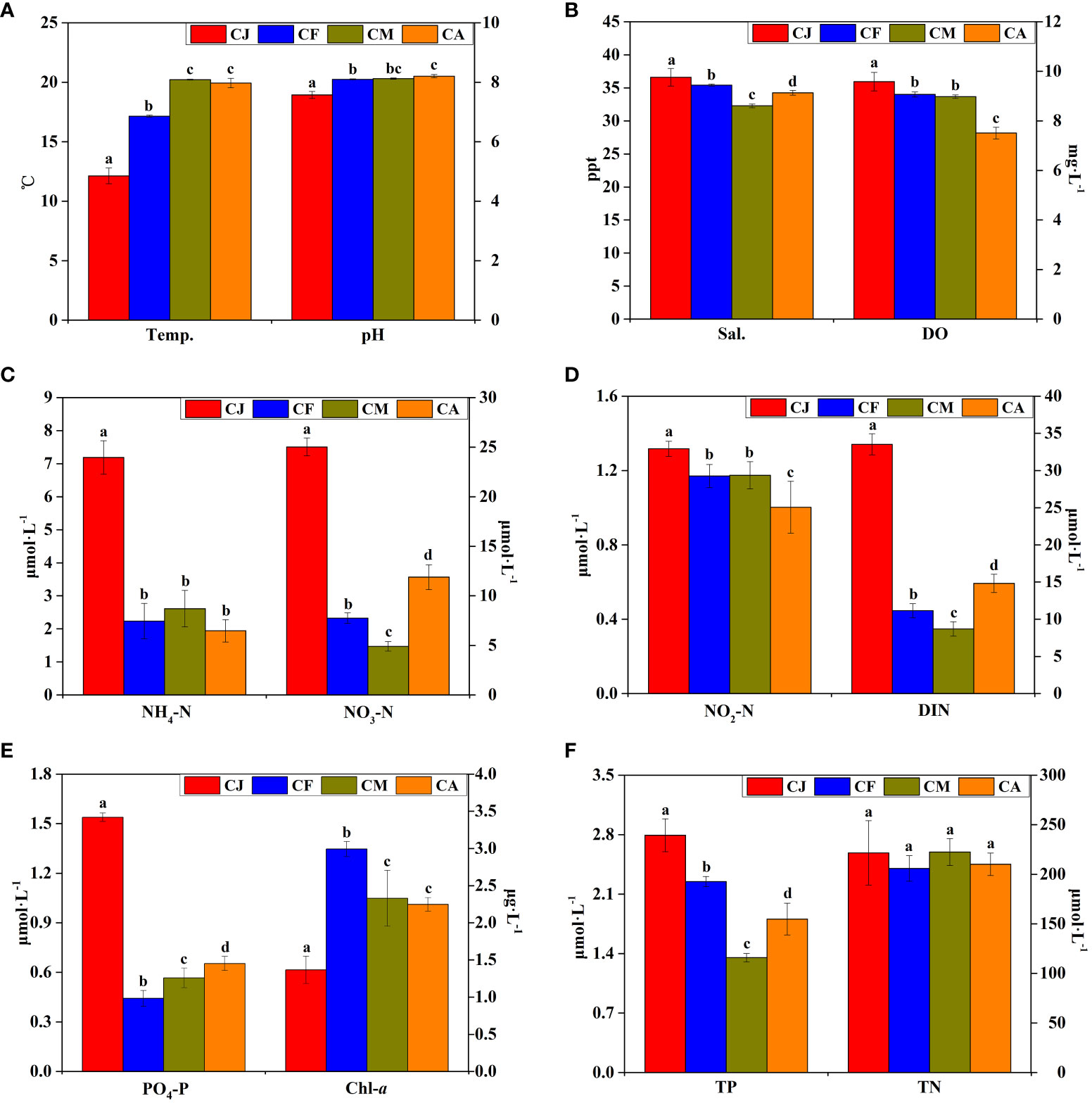
Figure 2 Main physicochemical factors of seawater during different cultivation periods. CJ, Cultivation Jan.; CF, Cultivation Feb.; CM, Cultivation Mar.; CA, Cultivation Apr.; Temp., temperature; Sal., salinity; DO, dissolved oxygen; NH4-N, ammonia; NO3-N, nitrate; NO2-N, nitrite; DIN, dissolved inorganic nitrogen; PO4-P, reactive phosphorous; Chl-a, chlorophyll a; TN, total nitrogen; TP, total phosphorous. Different letters (a, b, c, d) denote significant (p < 0.05) differences in physicochemical factors of seawater between different cultivation periods. Conventional physicochemical factors (A, B), inorganic nitrogen concentration (C, D), phosphate and chlorophyll (E), total nitrogen and phosphorus concentration (F) of seawater.
Considerable variations in nitrogenase activity and nifH gene abundance of epiphytic bacteria on G. lemaneiformis were observed during different cultivation periods, as shown in Figure 3. The nitrogenase activity of epiphytic bacteria in CM was the highest, reaching 1.96 ± 0.20 nmol C2H4 g-1 frond h-1, which was significantly (p < 0.05) higher than that in CJ, CF, and CA (Figure 3A). Similarly, the nifH gene abundance of epiphytic bacteria in CM was the highest, reaching 2.22×105 copies·(g·frond)-1, which was significantly (p < 0.05) higher than that in CJ, CF, and CA (Figure 3B). Seawater physicochemical factors affected nitrogenase activity and nifH gene abundance differently. Notably, Temp., pH, and Chl-a positively influenced nitrogenase activity, while Sal., NH4-N, NO3-N, DIN, PO4-P, and TP had a negative effect (Table 2). In addition, Sal. and TP had a negative effect on nifH gene abundance (Table 2).
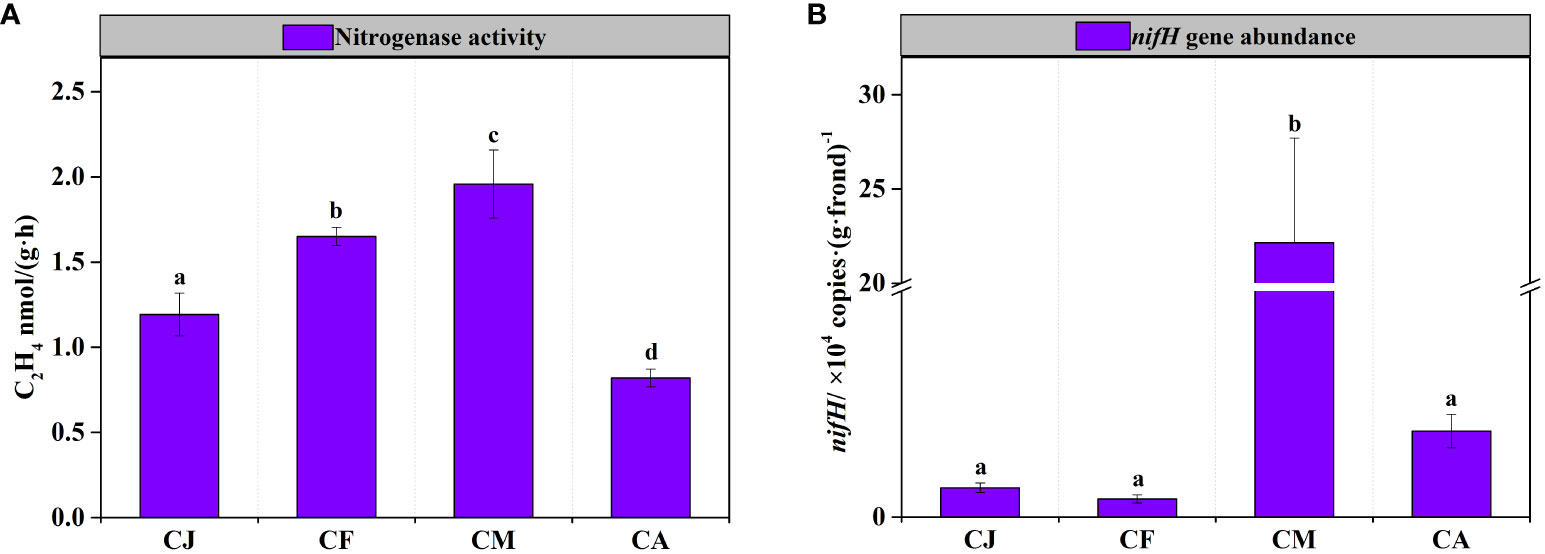
Figure 3 BNF of epiphytic bacteria on G. lemaneiformis during different cultivation periods. CJ, Cultivation Jan.; CF, Cultivation Feb.; CM, Cultivation Mar.; CA, Cultivation Apr. Different letters (a, b, c, d) denote significant (p < 0.05) differences in nitrogenase activity (A) and nifH gene abundance (B) of epiphytic bacteria on G. lemaneiformis between different cultivation periods.
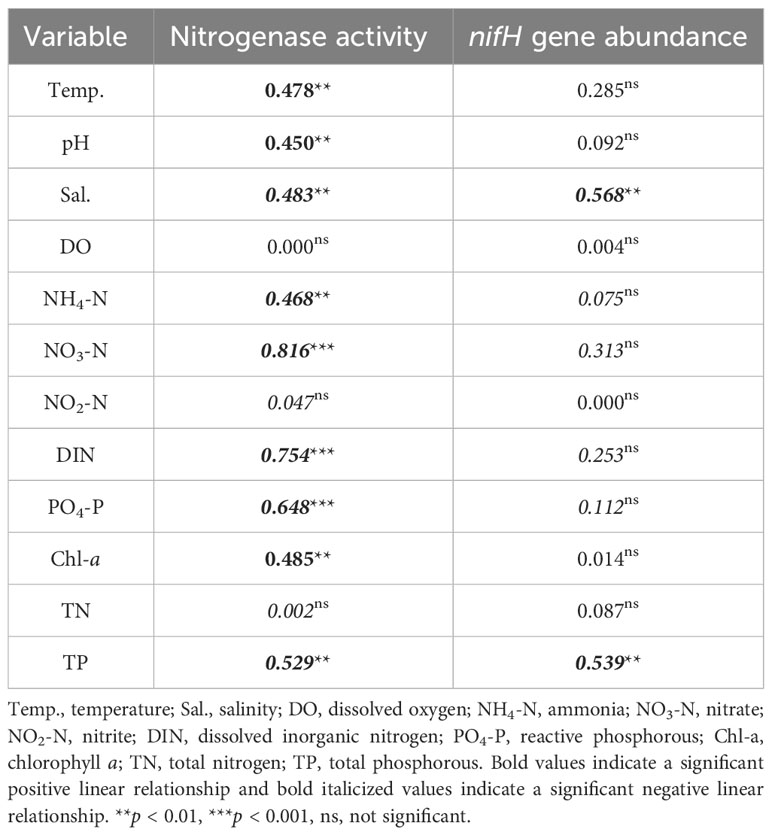
Table 2 Linear regression analysis (presented as R2 values) of the influence of seawater physicochemical factors on nitrogenase activity and nifH gene abundance.
Twelve G. lemaneiformis samples (i.e., 3 GL.CJ, 3 GL.CF, 3 GL.CM, 3 GL.CA) and twelve seawater samples (i.e., 3 SW.CJ, 3 SW.CF, 3 SW.CM, 3 SW.CA) were analyzed using Novaseq sequencing of nifH gene. A total 2,143,939 effective sequences were obtained with an average of 89,331 tags per sample (n=24). Sequences were clustered into operational taxonomic units (OTUs) at the 97% similarity level to generate 10,021 OTUs from the 32 samples. The OTUs were classified into 25 phyla, 46 classes, 82 orders, 128 families, and 150 genera. The Richness of GL.CM was 141.50 ± 2.12 per sample, which was significantly (p < 0.05) higher than that of GL.CJ (45.00 ± 12.73), GL.CF (59.00 ± 9.54), and GL.CA (75.00 ± 5.66) (Figure 4). Similarly, the Richness of SW.CM was significantly (p < 0.05) higher than that of SW.CJ, SW.CF, and GL.CA (Figure 4). Besides, the Shannon was the highest in GL.CM compared with SW.CJ, SW.CF, and GL.CA (Figure 4). Likewise, SW.CM had the highest Shannon compared with SW.CJ, SW.CF, and GL.CA (Figure 4). Notably, the diversity of diazotrophic communities associated with seawater was much higher than that of G. lemaneiformis.
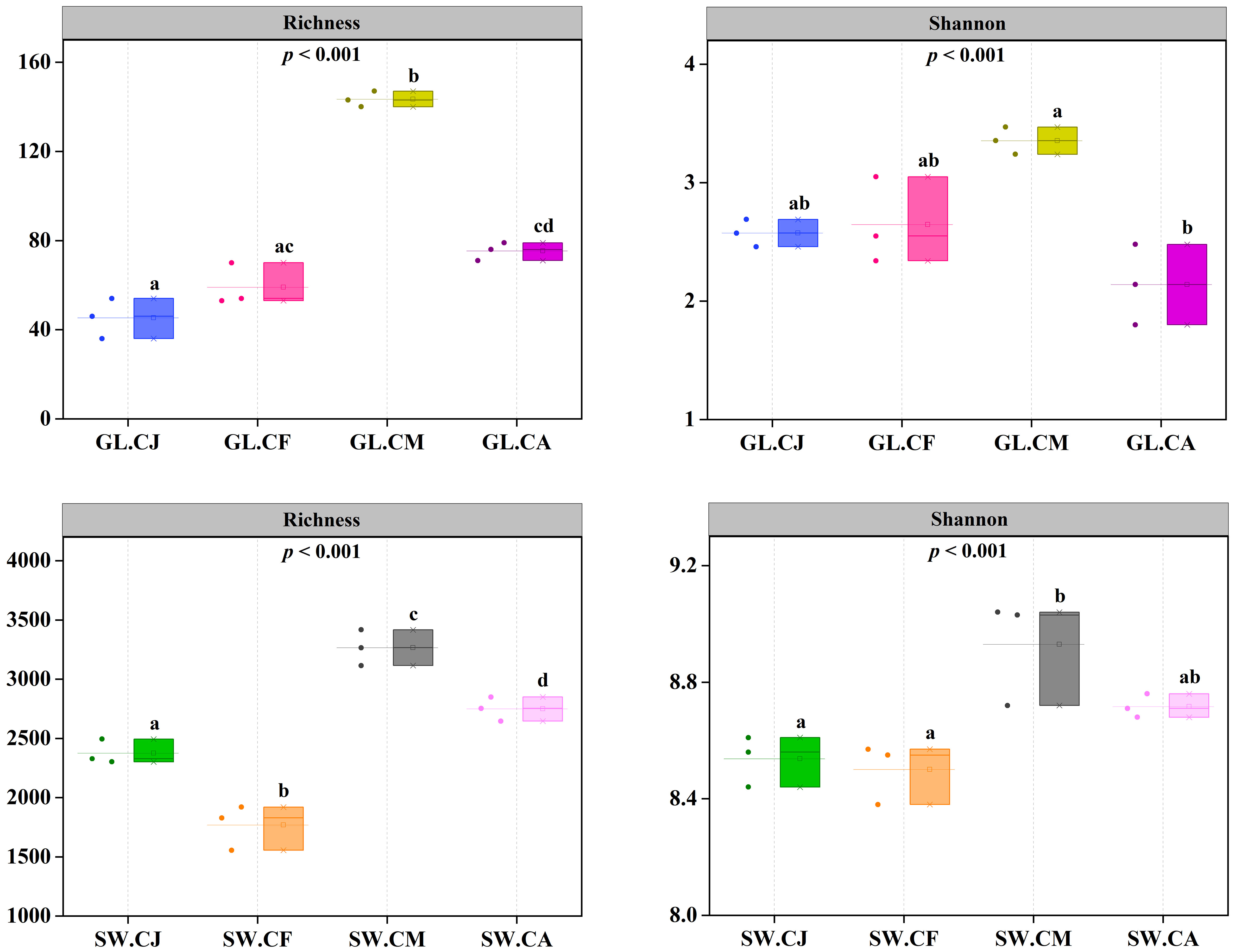
Figure 4 The diversity of diazotrophic communities in different cultivation periods. CJ, Cultivation Jan.; CF, Cultivation Feb.; CM, Cultivation Mar.; CA, Cultivation Apr.; GL, G. lemaneiformis; SW, seawater. Different letters (a, b, c, d) denote significant (p < 0.05) differences in diversity of diazotrophic communities associated with G. lemaneiformis and seawater between different cultivation periods.
Based on the analysis of OTUs, an nMDS (non-metric multidimensional scaling) ordination biplot indicated clear clustering of diazotrophic communities (Figure 5A). The stress value was 0.102. The diazotrophic communities of G. lemaneiformis samples generally clustered together, while with a strong clustering was observed at seawater samples. Notably, the diazotrophic communities of G. lemaneiformis were obviously distinct with that of seawater.
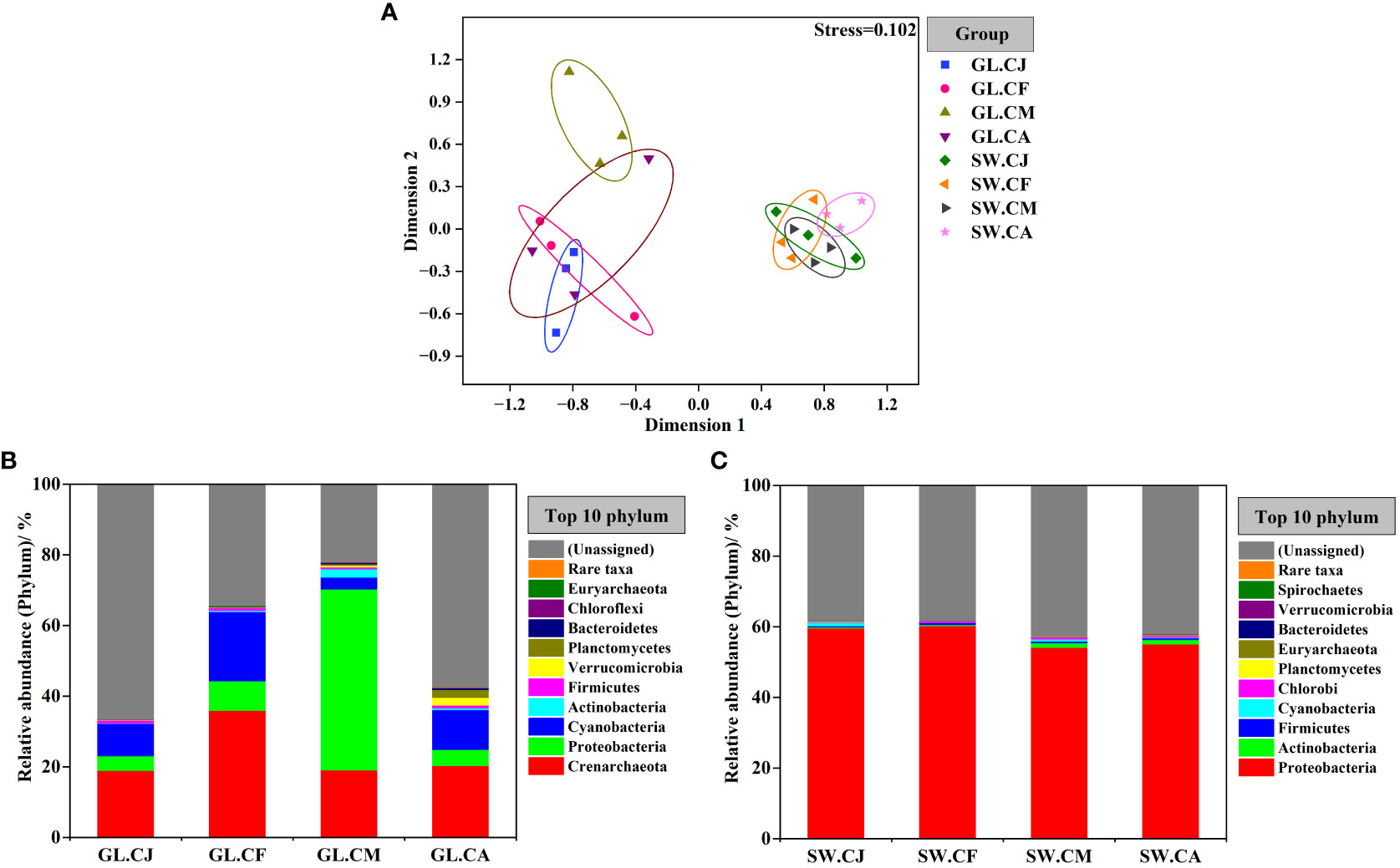
Figure 5 Difference comparison and temporal variations of the diazotrophs communities composition. Non-metric multidimensional scaling (nMDS) based on Bray-Curtis measure (A). The communities composition of diazotrophs associated with G. lemaneiformis (B) and seawater (C) at phylum level in different cultivation periods. CJ, Cultivation Jan.; CF, Cultivation Feb.; CM, Cultivation Mar.; CA, Cultivation Apr.; GL, G. lemaneiformis; SW, seawater.
The community composition of diazotrophs associated with G. lemaneiformis differed considerably in various cultivation periods (Figure 5B and Supplementary Figure S2). In contrast, the community composition of diazotrophs associated with seawater was similar in diverse cultivation periods (Figure 5C and Supplementary Figure S2). Proteobacteria was the most predominant phylum in GL.CM, with a relative abundance of 51.259%, followed by Crenarchaeota (19.049%) and Cyanobacteria (3.349%). Proteobacteria was also the most predominant phylum in SW.CJ, SW.CF, SW.CM, and SW.CA, but there were no significant differences among them. G. lemaneiformis collected from GL.CM exhibited significantly (p < 0.05) disparate diazotrophic communities compared to GL.CJ, GL.CF, and GL.CA. For instance, Crenarchaeota was the most predominant phylum in GL.CF, with a relative abundance of 35.959%, was significantly (p < 0.05) higher than that in GL.CM. The second dominant phylum was Cyanobacteria, with relative abundances of 9.188% in GL.CJ, 19.597% in GL.CF, and 11.239% in GL.CA, were significantly (p < 0.05) higher than that in GL.CM. On the other hand, there is a relative abundance of Proteobacteria and Actinobacteria in GL.CM was significantly (p < 0.05) higher than that in GL.CJ, GL.CF, and GL.CA.
LEfSe was used to find biomarkers with marked differences in abundance between two or more groups. In different cultivation periods, there were significant changes in the biomarkers associated with G. lemaneiformis and seawater (Figures 6 and 7). In SW.CJ, SW.CF, and SW.CA, the top 3 biomarkers significantly influencing the difference between groups (GL versus SW) were identical (Supplementary Figure S3). Likewise, the top 3 biomarkers significantly influencing the difference between groups (GL versus SW) were similar in GL.CJ, GL.CF, and GL.CA (Supplementary Figure S3). In CM, 31 biomarkers were significantly enriched in diazotrophic communities associated with seawater, among which γ-proteobacteria, δ-proteobacteria and Nitrosomonadales had a more significant influence on the difference between groups (GL.CM vs SW.CM) (Figure 6). However, there were 11 significantly enriched biomarkers in diazotrophic communities associated with G. lemaneiformis, and 5 (α-proteobacteria, Robiginitomaculum antarcticum, Robiginitomaculum, Hyphomonadaceae, and Rhodobacterales) of 11 had greater influence on difference between groups (GL.CM vs SW.CM) (Figure 6). Interestingly, 4 biomarkers that significantly enriched in diazotrophic communities associated with G. lemaneiformis in four cultivation periods (CJ, CF, CM, CA) were Rhodobacterales, Hyphomonadaceae, Robiginitomaculum, Robiginitomaculum antarcticum and had a more significant influence on difference between groups (GL) (Figure 7).
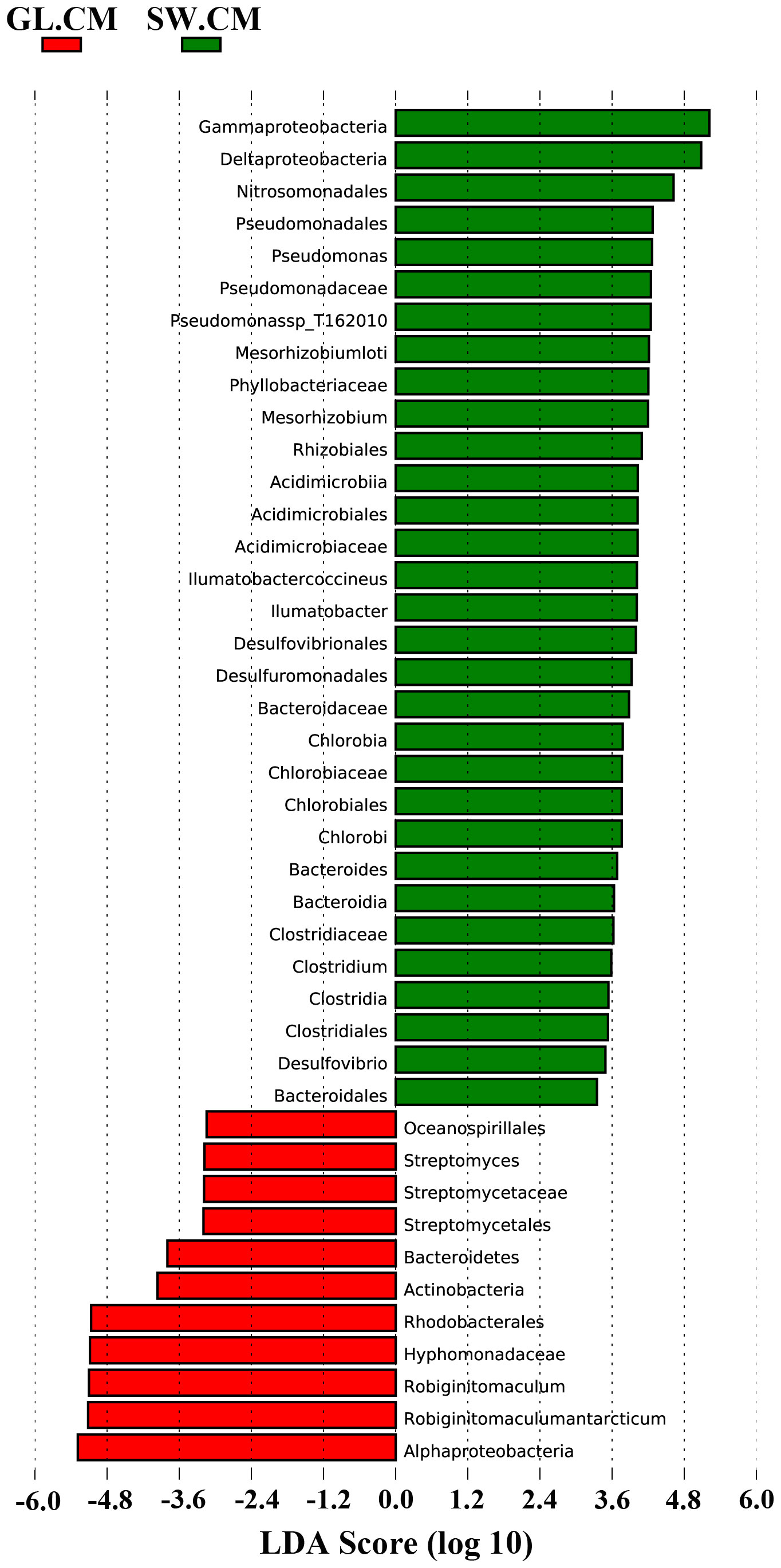
Figure 6 The LDA score histogram of significantly different biomarkers associated with G. lemaneiformis and seawater in CM. CM, Cultivation Mar.; GL, G. lemaneiformis; SW, seawater.
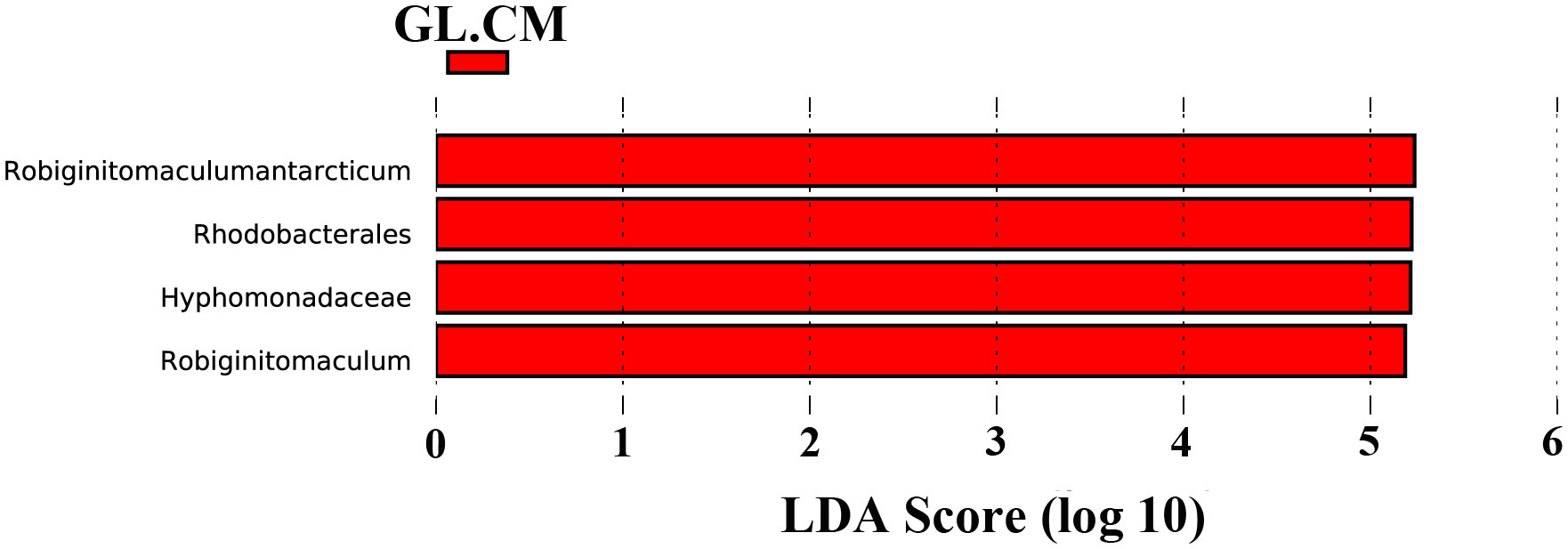
Figure 7 The LDA score histogram of significantly different biomarkers associated with G. lemaneiformis in different cultivation periods. CM, Cultivation Mar.; GL, G. lemaneiformis.
With the large-scale cultivation of seaweed, the coastal marine environment rich in N resources will become a N-limited environment. The concentration of N2 in the atmosphere is much higher than that of other inorganic N in the ocean (Bernhard, 2010). Thus this N source becomes potential nutrient source for marine primary productivity. The conversion of N2 into ammonium relies heavily on the diazotrophic communities (Bernhard, 2010). In recent times, an increasing number of studies have emphasized diazotrophs in seawater (Gradoville et al., 2017; Delmont et al., 2018; Chen et al., 2019), but there are few reports on the epiphytic diazotrophs in marine plants. Furthermore, the relationships between diazotrophs in marine plants and those in seawater remain largely unexplored. This study employed isolation and identification techniques, acetylene reduction, qPCR, and high-throughput nifH gene sequencing technology to analyze nitrogenase activity, nifH gene abundance in epiphytic bacteria on G. lemaneiformis, as well as variations in the diversity and composition of diazotrophic communities associated with G. lemaneiformis and seawater during different cultivation periods.
The current study observed a higher diversity of N2-fixing bacteria derived from the surface of macroalgae (G. lemaneiformis) compared to the surrounding seawater. Our previous findings have reported that the number of functional bacteria isolated from seaweed capable of growth on marine agar was higher than those isolated from the surrounding seawater (Pei et al., 2024). We assume that bacteria attached to the surface of algae are more likely to grow on marine agar media than planktonic bacteria. This hypothesis is supported by Jensen and Fenical (1994), who believed that the adaptations evolved by bacteria on algae may enhance their ability to form colonies on artificial media. Moreover, the harvested seaweed can be used as raw material for agar production (Yang et al., 2015), which makes it easier for these bacteria on algae to grow on agar media. Previous studies point out that the survival strategies evolved by seawater bacteria, including responses to starvation, may dramatically reduce their ability to form colonies on nutrient-rich agar media (Roszak and Colwell, 1987). Large portions of bacterial populations are dormant due to the reduced size and activity of bacteria led by starvation.
Most of the N2-fixing bacteria genera identified in our study have not been reported in previous studies, which may be closely related to the type of sample and the environment (Shieh et al., 1989; Bentzon-Tilia et al., 2014; Castellano-Hinojosa et al., 2016). Bentzon-Tilia et al. (2014) isolated 16 strains of N2-fixing bacteria from low-oxygen waters in the Baltic Sea, identified as Pseudomonas stutzeri 2A38. Castellano-Hinojosa and co-workers isolated 21 strains of N2-fixing bacteria from the rhizosphere of Lolium perenne, belonging to Bacillus, Paenibacillus, Pseudoxanthomonas, Burkholderia, Staphylococcus (Castellano-Hinojosa et al., 2016). In another study by Shieh, 7 distinct groups of N2-fixing bacteria were isolated from Zostera marina, primarily falling into the Vibrio and Photobacterium genera (Shieh et al., 1989). Genus Vibrio, belonging to the γ-proteobacteria class, has consistently been identified as a core member of diazotrophic communities associated with various coral species, as evidenced by nifH gene sequencing in previous studies (Olson et al., 2009; Zhang et al., 2016). Our current study isolated 4 Vibrio (Vibiro alginolyticus, Vibrio harveyi, Vibrio cyclitrophicus, Vibrio sp.) from the seawater surrounding G. lemaneiformis macroalgae. These findings suggest that these Vibrio bacteria likely represent diazotrophic communities associated with the seawater, as supported by molecular identification of the nifH gene. As heterotrophs, Vibrio can readily use labile sugars derived from macroalgae as a carbon source to sustain their growth (Cárdenas et al., 2018), which may partly explain high abundance of culturable Vibrio in the seawater microbiome. In addition, ecology of Vibrio has also been reported in other coastal ecosystems (Haas et al., 2016; Kelly et al., 2022). For instance, in the ecosystem of coral reefs, Vibrionaceae with support of DOC released by algae, which ultimately dominate in bacterial communities (Haas et al., 2016). The molecular structure of these compounds released by algae is one of the main factors affecting the microbial community that metabolize them (Kelly et al., 2022).
The findings mentioned above unequivocally establish the presence of diazotrophic microorganisms in G. lemaneiformis. To gain insights into how these diazotrophic communities associated with G. lemaneiformis react to environmental variations across distinct cultivation periods, we employed acetylene reduction and qPCR techniques to assess both nitrogenase activity and nifH gene abundance in association with G. lemaneiformis. The results indicate that the highest nitrogenase activity of epiphytic bacteria associated with G. lemaneiformis was observed in the CM cultivation period (Figure 8), reaching 1.958 nmol C2H2 g-1·algae·h-1. Based on the established correlation, where the reduction of 3 grams of C2H2 is equivalent to the reduction of 1 gram of N2 (Cardini et al., 2017), it can be estimated that epiphytic bacteria carried by 1 ton of G. lemaneiformis can fix approximately 0.531×1010 nmol of N2 annually. Notably, it has been reported that the annual yield of Gracilaria reached 610,824 tons in 2022, as per data from the 2023 China Fishery Statistical Yearbook. Since G. lemaneiformis significantly contributes to this yield, it is estimated to fix around 0.185×107 moles of N2 annually. This observation suggests that the diazotrophs associated with G. lemaneiformis convert atmospheric N2 into a new nitrogen source for the marine environment through BNF. This newly introduced N source serves a dual purpose. On the one hand, it can be used as an additional N source for marine organisms. On the other hand, it serves as a supplementary N source for marine organisms, particularly in conditions of N deficiency.
Our study found that the N and P levels in the CM seawater were significantly lower compared to CJ, CF, and CA (Figure 8). According to the preliminary data of G. lemaneiformis harvested by local farmers every month, it can be seen that the yield of G. lemaneiformis reaches the maximum in the CM period. Yang et al. (2006) have pointed out that the cultivated G. lemaneiformis harbors a high productivity and can absorb a large amount of N and P. Therefore, this significant reduction regarding the N and P levels in CM seawater can be attributed to the substantial consumption of N and P by G. lemaneiformis. Furthermore, the growth environment of G. lemaneiformis is characterized as oligotrophic (DIN < 9 μmol·L-1, PO4-P < 0.6 μmol·L-1), which potentially prompts diazotrophs associated with G. lemaneiformis to fix more N2 to meet the N demands of the entire marine environment. Cardini et al. (2017) demonstrated that epiphytic N2 fixation, such as in seagrass like Halophila stipulacea, is crucial in supplying N to marine plants in oligotrophic environments. Similarly, N2 fixation associated with Posidonia oceanica significantly contributes to N requirements for primary productivity (Agawin et al., 2017). These findings highlight that nutrient deficiency in the marine environment may enhance the N2-fixing capabilities of diazotrophs in preparation for nutrient stress. Moreover, during the CM period, the seawater temperature reached its maximum of 20.23°C, which is also optimal for indoor G. lemaneiformis cultivation (Liu et al., 2019b, 2019a). Indeed, previous research by Wahbeh and Mahasneh (1984) revealed that heterotrophic bacteria, which also include heterotrophic diazotrophs, predominantly dominated the epiphytic bacteria on the roots, stems, and leaves of halophile plants at higher temperatures. Building on this, subsequent work by Cardini et al. (2017) demonstrated that under elevated temperature, the heterotrophic bacteria residing on the roots, stems, and seagrass leaves exhibited higher rates of N2 fixation. Furthermore, it’s worth noting that long-term warming has been shown to significantly increase the abundance of diazotrophs by 86.3%, as reported by Feng et al. (2019). This information leads us to speculate that the elevated nitrogenase activity observed with G. lemaneiformis in the CM period may be attributed to accumulating a higher number of heterotrophic diazotrophs on the surface of G. lemaneiformis.
We measured quantitative nifH gene abundance associated with G. lemaneiformis in various cultivation periods to test this hypothesis. Our findings demonstrated that during the CM period, the nifH gene abundance in epiphytic bacteria on G. lemaneiformis reached 22.15×104 copies/g, a significantly higher level than CJ, CF, and CA. These results strongly suggest that microorganisms containing the nifH gene became notably enriched on the surface of G. lemaneiformis during the CM period (Figure 8), and they played a significant role in BNF under conditions of nutrient deficiency and elevated temperature, as depicted in Figure 3. Prior studies have indicated that temperature, light, and nutrient availability can affect nitrogenase activity, nifH gene transcription, and N2 fixation rate in N2-fixing cyanobacteria (Church et al., 2005). Nitrogenase activity and nifH gene abundance associated with G. lemaneiformis were low in CJ. This could be attributed to a few factors. Firstly, it’s possible that planktonic microorganisms, including diazotrophs, had not firmly adhered to the surface of G. lemaneiformis during CJ, and the catalytic activity of nitrogenase decreased significantly due to the lower temperature (12.13°C). Secondly, the high concentration of inorganic nutrients in the seawater (DIN: 33.53 μmol·L-1; NH4-N: 7.19 μmol·L-1; PO4-P: 1.54 μmol·L-1) could have contributed to the weakening of BNF associated with G. lemaneiformis. Hence, we propose that the nutrients needed by G. lemaneiformis and its epiphytic bacteria to complete their life activities can be provided by sufficient inorganic nutrients in seawater, and little depends on BNF associated with G. lemaneiformis. This hypothesis is supported by numerous previous studies which supposed that BNF of Cyanobacteria and Proteobacteria decreases significantly with the increase of DIN concentration (Knapp, 2012; Bentzon-tilia et al., 2015), indicating that DIN can maintain their growth (Masuda et al., 2013; Bentzon-tilia et al., 2015). Furthermore, DIN was significantly negatively correlated with N2 fixation in tropical estuarine eutrophication regions and was independent of season (Jabir et al., 2020). NH4-N, the main component of DIN in some estuary regions (Jabir et al., 2020), controls the N2 fixation rate by inhibiting the synthesis of new nitrogenase (Rees et al., 2006). In the present study, NH4-N concentration in seawater reached 7.19 μmol·L-1 in CJ, significantly higher than in CF, CM, and CA. Hence, it can be seen that the nitrogenase activity and nifH gene abundance associated with G. lemaneiformis in CJ may also be inhibited by higher ammonium concentration. Although the relationship between DIN availability and N2 fixation in marine environments is very complicated, DIN plays an essential role in the construction of diazotrophic communities (Messer et al., 2017), which in turn may influence BNF associated with G. lemaneiformis.
Although high-throughput sequencing technology is widely used to study the microbial community associated with macroalgae such as G. lemaneiformis (Xie et al., 2017; Pei et al., 2021), it has not been broadly applied to the nifH gene to explore the diazotrophic communities associated with G. lemaneiformis. Our previous research only obtained a tiny amount of information about diazotrophs (Pei et al., 2021), and we could not fundamentally understand the diazotrophs associated with G. lemaneiformis and its surrounding seawater, and the role of functional microorganisms in the cultivation of ecological environment remained to be studied. The current study showed that the diazotrophic communities associated with G. lemaneiformis significantly differed in various cultivation periods, especially in CM. In contrast, the diazotrophic communities associated with seawater have little change in distinct cultivation periods. To the best of our knowledge, only some reports on diazotrophs are associated with macroalgae. However, more attention has been paid to diazotrophs associated with coral (Zhang et al., 2016), mangrove rhizosphere (Zhang et al., 2017a), and seawater (Messer et al., 2017). Therefore, this is the first study that used nifH gene high-throughput sequencing to explore the diazotrophs associated with red macroalgae G. lemaneiformis. Studies have shown that α-Proteobacteria, δ-Proteobacteria and γ-Proteobacteria are the main diazotroph groups associated with most corals (Lema et al., 2012; Zhang et al., 2016). Moreover, δ-proteobacteria and γ-proteobacteria are the most predominant diazotrophic communities associated with mangrove rhizosphere (Alzubaidy et al., 2016; Zhang et al., 2017a). In the present study, α-proteobacteria and Thermoprotei, the main diazotrophic groups associated with G. lemaneiformis differed significantly in various cultivation periods (Supplementary Figure S1A). However, α-proteobacteria, δ-proteobacteria and γ-proteobacteria were the main diazotrophic groups associated with seawater, similar in different cultivation periods (Supplementary Figure S1B). The above results indicate that these diazotrophic groups may play a vital role of BNF in the cultivation environment of G. lemaneiformis. Findings show that diazotrophic communities associated with marine organisms (e.g. corals) are species-specific and environment-specific due to the differences of species and environment conditions (Lema et al., 2012). Notably, the relative abundance of α-proteobacteria associated with G. lemaneiformis in CM reached 47.972%, much higher than that in CJ, CF, and CA. We speculate that the higher abundance of α-proteobacteria could contribute to the significant increase in nitrogenase activity and nifH gene abundance associated with G. lemaneiformis. LEfSe analysis results showed that the biomarkers with more significant influence on difference between groups (GL vs SW) remained the same in CJ, CF, and CA (Supplementary Figures S2A, B, D). However, the biomarkers with more significant influence on difference between groups (GL vs SW) had noticeable changes in CM, reflected primarily in α-proteobacteria, Robiginitomaculum, and Robiginitomaculum antarcticum (Figure 6). Consequently, α-proteobacteria plays a significant role of BNF in response to oligotrophic conditions, reflected in the remarkable increase of nitrogenase activity and nifH gene abundance in CM.
To further explore the diazotrophic communities composition under the class α-proteobacteria, a comprehensive analysis was conducted at the order, family, and genus level. The current study observed significant differences in the dominant diazotrophic communities associated with G. lemaneiformis at the order, family, and genus levels in different cultivation periods (Supplementary Figures S1C, E, G), while the dominant diazotrophic communities associated with seawater were similar (Supplementary Figures S1D, F, H). Microbial groups with higher relative abundance found in α-proteobacteria, including Rhodobacterales (36.915%), Hyphomonadaceae (36.743%), and Robiginitomaculum (36.722%), were dominant diazotrophic groups associated with G. lemaneiformis in CM. Predictive functional results from Lesser and coworkers uncovered that genera within the Rhodobacterales with putative N2-fixing capabilities (Lesser et al., 2018). These findings are confirmed by a previous study indicating that the nifHDK genes were detected in the Rhodobacterales genome (Tsoy et al., 2016). The existence of significant BNF associated with G. lemaneiformis in CM was further supported by the above studies. These bacteria exhibit extreme metabolic flexibility under environmental conditions related to variability in pO2, light availability, and carbon source (Lesser et al., 2018). The family Hyphomonadaceae encompasses several members (Lee et al., 2005, 2007) that are widely distributed in diverse environments, including surface water (Deng et al., 2013), freshwater (Driscoll et al., 2017), deep-sea (Zhang et al., 2013), and marine sediment (Zhao et al., 2019). The family Hyphomonadaceae is currently composed of eleven well-defined genera: Hyphomonas, Maricaulis, Hirschia, Oceanicaulis, Robiginitomaculum, Woodsholea, Hellea, Henriciella, Ponticaulis, Litorimonas, and Algimonas (Lee et al., 2007; Deng et al., 2013). LefSe analysis results displayed that the biomarkers with more significant influence on difference between groups (GL.CJ vs GL.CF vs GL.CM vs GL.CA) were Rhodobacterales, Hyphomonadaceae, Robiginitomaculum, Robiginitomaculum antarcticum (Figure 7). As a result, Rhodobacterales, Hyphomonadaceae, Robiginitomaculum, and Robiginitomaculum antarcticum within the α-proteobacteria play a significant role in BNF in response to oligotrophic conditions (Figure 8).
A study on symbiotic diazotrophic bacterial communities has indicated that Rhizobiales belonging to the class α-proteobacteria are stable and dominant diazotrophic groups associated with Acropora coral species (Lema et al., 2012). In our study, there were no variations in the relative abundance of Rhizobiales (SW.CJ: 2.382%; SW.CF: 2.931%; SW.CM: 1.821%: SW.CA: 1.953%) within different cultivation periods, indicating that Rhizobiales are also stable and dominant diazotrophic groups associated with seawater. Besides, we also found two dominant diazotrophic groups from δ-proteobacteria associated with seawater, such as Desulfuromonadale and Desulfovibrionales, belonging to the sulfate-reducing bacteria (Zhang et al., 2017a). It has been reported that the order Desulfovibrionales has strong adaptability to marine environmental stresses, such as anthropogenic heavy metal contamination and oil pollution (Varon-Lopez et al., 2014), which suggests that Desulfovibrionales may contribute substantially to BNF in coastal seawater. Pseudomonas within the γ-proteobacteria is not only a diazotrophic genus (Zhang et al., 2016), but also the dominant bacterial genus associated with seawater in this study.
Thirteen N2-fixing bacteria carrying the nifH gene were isolated, screened, and identified from G. lemaneiformis and its surrounding seawater. Notably, G. lemaneiformis exhibited the highest nitrogenase activity and nifH gene abundance in the CM period, underlining the substantial contribution of epiphytic bacteria to BNF. Linear regression assessments revealed strong negative correlations between nitrogenase activity and several variables, including Sal., NH4-N, NO3-N, DIN, PO4-P, and TP, while positive correlations were evident with Temp., pH, and Chl-a. Furthermore, nifH gene abundance exhibited negative associations with Sal. and TP. These results emphasize the intricate interplay between diazotrophic activity and environmental conditions. Our results also showed the highest alpha-diversity of diazotrophic communities associated with G. lemaneiformis and seawater during the CM period. The diazotrophic communities linked to G. lemaneiformis displayed pronounced differences among cultivation periods, distinguishing them from seawater-associated ones. Conversely, the diazotrophic communities in seawater appeared relatively stable across different cultivation phases. Of particular significance, our findings highlighted specific taxa, such as the order Rhodobacterales, the family Hyphomonadaceae, and the genus Robiginitomaculum and the species Robiginitomaculum antarcticum within the α-proteobacteria class, as significant contributors to BNF, particularly in response to oligotrophic environmental conditions. These results significantly advance our understanding of diazotrophs associated with G. lemaneiformis, providing a foundation for future investigations into the underlying mechanisms and ecological implications of their activities in the marine environment.
The datasets presented in this study can be found online repositories, the names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
PP: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MA: Methodology, Software, Validation, Writing – review & editing. CY: Investigation, Methodology, Validation, Writing – review & editing. PY: Investigation, Methodology, Validation, Writing – review & editing. XK: Investigation, Methodology, Validation, Writing – review & editing. ZL: Investigation, Methodology, Validation, Writing – review & editing. TL: Validation, Writing – review & editing. WC: Methodology, Validation, Writing – review & editing. HD: Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by several sources, including Project supported by Southern Marine Science and Engineering Guangdong Laboratory(Zhuhai) (SML2023SP204), the Program for University Innovation Team of Guangdong Province (2022KCXTD008), the Science and Technology Plan Projects of Guangdong Province (2021B1212050025), and the China Agriculture Research System of MOF and MARA (CARS-50).
We thank Guangdong Magigene Biotechnology Co. Ltd. for providing sequencing services, and the Shaowu Zhu villager of artificial seaweed farms located in Zoumapu Village, Nan’ao Island of Guangdong Province for his help with sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1408958/full#supplementary-material
Agawin N. S. R., Ferriol P., Sintes E., Moyà G. (2017). Temporal and spatial variability of in situ nitrogen fixation activities associated with the Mediterranean seagrass Posidonia oceanica meadows. Limnol. Oceanogr. 62, 2575–2592. doi: 10.1002/lno.10591
Alzubaidy H., Essack M., Malas T. B., Bokhari A., Motwalli O., Kamanu F. K., et al. (2016). Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene 576, 626–636. doi: 10.1016/j.gene.2015.10.032
AQSIQ (2007). Specifications for Oceanographic Survey. Part 4: Survey of Chemical Parameters in Sea Water (Beijing: Stand. Press China), 16–26.
Aslam M., Pei P., Ye P., Li T., Liang H., Zhang Z., et al. (2023). Unraveling the Diverse Profile of N-Acyl Homoserine Lactone Signals and Their Role in the Regulation of Biofilm Formation in Porphyra haitanensis-Associated Pseudoalteromonas galatheae. Microorganisms 11, 2228. doi: 10.3390/microorganisms11092228
Azam F., Malfatti F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Bentzon-Tilia M., Farnelid H., Jürgens K., Riemann L. (2014). Cultivation and isolation of N2-fixing bacteria from suboxic waters in the Baltic Sea. FEMS Microbiol. Ecol. 88, 358–371. doi: 10.1111/fem.2014.88.issue-2
Bentzon-tilia M., Severin I., Hansen L. H., Riemann L. (2015). Genomics and ecophysiology of heterotrophic nitrogen-fixing bacteria isolated from estuarine surface water. MBio 6, 1–11. doi: 10.1128/mBio.00929-15
Bernhard A. (2010). The nitrogen cycle: processes, players, and human impact. Nat. Educ. Knowl. 2, 1–8.
Bonnet S., Berthelot H., Turk-Kubo K., Cornet-Barthaux V., Fawcett S., Berman-Frank I., et al. (2016). Diazotroph derived nitrogen supports diatom growth in the South West Pacific: A quantitative study using nanoSIMS. Limnol. Oceanogr. 61, 1549–1562. doi: 10.1002/lno.10300
Caffin M., Moutin T., Ann Foster R., Bouruet-Aubertot P., Michelangelo Doglioli A., Berthelot H., et al. (2018). N2 fixation as a dominant new N source in the western tropical South Pacific Ocean (OUTPACE cruise). Biogeosciences 15, 2565–2585. doi: 10.5194/bg-15-2565-2018
Cárdenas A., Neave M. J., Haroon M. F., Pogoreutz C., Rädecker N., Wild C., et al. (2018). Excess labile carbon promotes the expression of virulence factors in coral reef bacterioplankton. ISME J. 12, 59–76. doi: 10.1038/ismej.2017.142
Cardini U., van Hoytema N., Bednarz V. N., Al-Rshaidat M. M. D., Wild C. (2017). N2 fixation and primary productivity in a red sea Halophila stipulacea meadow exposed to seasonality. Limnol. Oceanogr. 63, 786–798. doi: 10.1002/lno.10669
Castellano-Hinojosa A., Correa-Galeote D., Palau J., Bedmar E. J. (2016). Isolation of N2-fixing rhizobacteria from Lolium perenne and evaluating their plant growth promoting traits. J. Basic Microbiol. 56, 85–91. doi: 10.1002/jobm.201500247
Chen T., Chen Y. L., Sheu D., Chen H., Lin Y. (2019). Community and abundance of heterotrophic diazotrophs in the northern South China Sea : Revealing the potential importance of a new alphaproteobacterium in N2 fixation. Deep. Res. Part I 143, 104–114. doi: 10.1016/j.dsr.2018.11.006
Church M. J., Short C. M., Jenkins B. D., Karl D. M., Zehr J. P. (2005). Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71, 5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Delmont T. O., Quince C., Shaiber A., Esen Ö.C., Lee S. T., Rappé M. S., et al. (2018). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813. doi: 10.1038/s41564-018-0176-9
Deng W., Zhang Y., Xie X., Zhao Z., Fu Y. (2013). Euryhalocaulis caribicus gen. nov., sp. nov., a new members of the family Hyphomonadaceae isolated from the Caribbean Sea. Curr. Microbiol. 66, 606–612. doi: 10.1007/s00284-013-0314-9
Driscoll C. B., Otten T. G., Brown N. M., Dreher T. W. (2017). Towards long-read metagenomics: Complete assembly of three novel genomes from bacteria dependent on a diazotrophic cyanobacterium in a freshwater lake co-culture. Stand. Genomic Sci. 12, 1–16. doi: 10.1186/s40793-017-0224-8
Duarte C. M., Wu J., Xiao X., Bruhn A., Krause-Jensen D. (2017). Can seaweed farming play a role in climate change mitigation and adaptation? Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00100
Egan S., Gardiner M. (2016). Microbial dysbiosis: Rethinking disease in marine ecosystems. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00991
Feng J., Penton C. R., He Z., Nostrand J. D., Yuan M. M., Wu L. (2019). Long-term warming in Alaska enlarges the diazotrophic community in deep soils. MBio 10, 1–12. doi: 10.1128/mBio.02521-18
Feng G., Sun W., Zhang F., Orlić S., Li Z. (2018). Functional transcripts indicate phylogenetically diverse active ammonia-scavenging microbiota in sympatric sponges. Mar. Biotechnol. 20, 131–143. doi: 10.1007/s10126-018-9797-5
Gaby J. C., Buckley D. H. (2012). A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7, e42149. doi: 10.1371/journal.pone.0042149
Geisler E., Bogler A., Rahav E., Bar-Zeev E. (2019). Direct detection of heterotrophic diazotrophs associated with planktonic aggregates. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-019-45505-4
Gradoville M. R., Bombar D., Crump B. C., Letelier R. M., Zehr J. P., White A. E. (2017). Diversity and activity of nitrogen-fixing communities across ocean basins. Limnol. Oceanogr. 62, 1895–1909. doi: 10.1002/lno.10542
Haas A. F., Fairoz M. F. M., Kelly L. W., Nelson C. E., Dinsdale E. A., Edwards R. A., et al. (2016). Global microbialization of coral reefs. Nat. Microbiol. 1, 1–7. doi: 10.1038/nmicrobiol.2016.42
Hasselström L., Visch W., Gröndahl F., Nylund G. M., Pavia H. (2018). The impact of seaweed cultivation on ecosystem services-a case study from the west coast of Sweden. Mar. pollut. Bull. 133, 53–64. doi: 10.1016/j.marpolbul.2018.05.005
Head W. D., Carpenter E. J. (1975). Nitrogen fixation associated with the marine macroalga Codium fragile. Limnol. Oceanogr. 20, 815–823. doi: 10.4319/lo.1975.20.5.0815
Huang Y. S., Ou L. J., Yang Y. F. (2017). Nutrient competition between macroalgae Gracilaria lemaneiformis and phytoplankton in coastal waters of Nan’ao island, Guangdong. Oceanol. Limnol. Sin. 48, 806–813. doi: 10.11693/hyhz20170200031
Jabir T., Vipindas P. V., Jesmi Y., Valliyodan S., Parambath P. M., Singh A., et al. (2020). Nutrient stoichiometry (N:P) controls nitrogen fixation and distribution of diazotrophs in a tropical eutrophic estuary. Mar. pollut. Bull. 151, 110799. doi: 10.1016/j.marpolbul.2019.110799
Jensen P. R., Fenical W. (1994). Strategies for the discovery of secondary metabolites from marine bacteria: Ecological perspectives. Annu. Rev. Microbiol. 48, 559–584. doi: 10.1146/annurev.mi.48.100194.003015
Karthick P., Mohanraju R. (2018). Antimicrobial potential of epiphytic bacteria associated with seaweeds of little Andaman, India. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00611
Kelly L. W., Nelson C. E., Petras D., Koester I., Quinlan Z. A., Arts M. G. I., et al. (2022). Distinguishing the molecular diversity, nutrient content, and energetic potential of exometabolomes produced by macroalgae and reef-building corals. Proc. Natl. Acad. Sci. U. S. A. 119, e2110283119. doi: 10.1073/PNAS.2110283119
Kim J. K., Yarish C., Hwang E. K., Park M., Kim Y. (2017). Seaweed aquaculture: Cultivation technologies, challenges and its ecosystem services. Algae 32, 1–13. doi: 10.4490/algae.2017.32.3.3
Knapp A. N. (2012). The sensitivity of marine N2 fixation to dissolved inorganic nitrogen. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00374
Lee K., Lee H. K., Choi T. H., Cho J. C. (2007). Robiginitomaculum antarcticum gen. nov., sp. nov., a member of the family Hyphomonadaceae, from Antarctic seawater. Int. J. Syst. Evol. Microbiol. 57, 2595–2599. doi: 10.1099/ijs.0.65274-0
Lee K. B., Liu C., Anzai Y., Kim H., Aono T., Oyaizu H. (2005). The hierarchical system of the “Alphaproteobacteria”: Description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55, 1907–1919. doi: 10.1099/ijs.0.63663-0
Lema K. A., Willis B. L., Bourne D. G. (2014). Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the Acropora millepora microbiome. Environ. Microbiol. 16, 3345–3359. doi: 10.1111/1462-2920.12366
Lema K. A., Willis B. L., Bourneb D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Lesser M. P., Morrow K. M., Pankey S. M., Noonan S. H. C. (2018). Diazotroph diversity and nitrogen fixation in the coral Stylophora pistillata from the Great Barrier Reef. ISME J. 12, 813–824. doi: 10.1038/s41396-017-0008-6
Liang Z., Liu F., Wang W., Zhang P., Sun X., Wang F., et al. (2019). High-throughput sequencing revealed differences of microbial community structure and diversity between healthy and diseased Caulerpa lentillifera. BMC Microbiol. 19, 1–15. doi: 10.1186/s12866-019-1605-5
Liu X., Wen J., Chen W., Du H. (2019a). Physiological effects of nitrogen deficiency and recovery on the macroalga Gracilariopsis lemaneiformis (Rhodophyta). J. Phycol. 55, 830–839. doi: 10.1111/jpy.12862
Liu X., Wen J., Zheng C., Jia H., Chen W., Du H. (2019b). The impact of nitrogen deficiency and subsequent recovery on the photosynthetic performance of the red macroalga Gracilariopsis lemaneiformis. J. Appl. Phycol. 31, 2699–2707. doi: 10.1007/s10811-019-1745-x
Masuda T., Furuya K., Kodama T., Takeda S., Harrison P. J. (2013). Ammonium uptake and dinitrogen fixation by the unicellular nanocyanobacterium Crocosphaera watsonii in nitrogen-limited continuous cultures. Limnol. Oceanogr. 58, 2029–2036. doi: 10.4319/lo.2013.58.6.2029
Messer L. F., Brown M. V., Furnas M. J., Carney R. L., McKinnon A. D., Seymour J. R. (2017). Diversity and activity of diazotrophs in great barrier reef surface waters. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00967
Neori A. (2008). Essential role of seaweed cultivation in integrated multi-trophic aquaculture farms for global expansion of mariculture: An analysis. J. Appl. Phycol. 20, 567–570. doi: 10.1007/s10811-007-9206-3
Olson N. D., Ainsworth T. D., Gates R. D., Takabayashi M. (2009). Diazotrophic bacteria associated with Hawaiian Montipora corals : Diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Bio. Ecol. 371, 140–146. doi: 10.1016/j.jembe.2009.01.012
Olson N. D., Lesser M. P. (2013). Diazotrophic diversity in the Caribbean coral, Montastraea cavernosa. Arch. Microbiol. 195, 853–859. doi: 10.1007/s00203-013-0937-z
Pei P., Aslam M., Du H., Liang H., Wang H., Liu X., et al. (2021). Environmental factors shape the epiphytic bacterial communities of Gracilariopsis lemaneiformis. Sci. Rep. 11, 1–15. doi: 10.1038/s41598-021-87977-3
Pei P., Aslam M., Wang H., Ye P., Li T., Liang H., et al. (2024). Diversity and ecological function of urease-producing bacteria in the cultivation environment of Gracilariopsis lemaneiformis. Microb. Ecol. 87, 35. doi: 10.1007/s00248-023-02339-y
Poly F., Monrozier L. J., Bally R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. doi: 10.1016/S0923-2508(00)01172-4
Rees A. P., Law C. S., Woodward E. M. S. (2006). High rates of nitrogen fixation during an in-situ phosphate release experiment in the Eastern Mediterranean Sea. Geophys. Res. Lett. 33, 2–5. doi: 10.1029/2006GL025791
Roszak D. B., Colwell R. R. (1987). Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51, 365–379. doi: 10.1128/mr.51.3.365-379.1987
Rousk J., Bengtson P. (2014). Microbial regulation of global biogeochemical cycles. Front. Microbiol. 5, 1–3. doi: 10.3389/fmicb.2014.00103
Selvarajan R., Sibanda T., Venkatachalam S., Ogola H. J. O., Christopher Obieze C., Msagati T. A. (2019). Distribution, interaction and functional profiles of epiphytic bacterial communities from the rocky intertidal seaweeds, South Africa. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-56269-2
Shieh W. Y., Simidu U., Maruyama Y. (1989). Enumeration and characterization of nitrogen-fixing bacteria in an eelgrass (Zostera marina) bed. Microb. Ecol. 18, 249–259. doi: 10.1007/BF02075812
Sohm J. A., Webb E. A., Capone D. G. (2011). Emerging patterns of marine nitrogen fixation. Nat. Rev. Microbiol. 9, 499–508. doi: 10.1038/nrmicro2594
Stabili L., Rizzo L., Pizzolante G., Alifano P. (2017). Spatial distribution of the culturable bacterial community associated with the invasive alga Caulerpa cylindracea in the Mediterranean Sea. Mar. Environ. Res. 125, 90–98. doi: 10.1016/j.marenvres.2017.02.001
Tsoy O. V., Ravcheev D. A., Čuklina J., Gelfand M. S. (2016). Nitrogen fixation and molecular oxygen: Comparative genomic reconstruction of transcription regulation in Alphaproteobacteria. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01343
Varon-Lopez M., Dias A. C. F., Fasanella C. C., Durrer A., Melo I. S., Kuramae E. E., et al. (2014). Sulphur-oxidizing and sulphate-reducing communities in Brazilian mangrove sediments. Environ. Microbiol. 16, 845–855. doi: 10.1111/1462-2920.12237
Wahbeh M. I., Mahasneh A. M. (1984). Heterotrophic bacteria attached to leaves, rhizomes and roots of three seagrass species from Aqaba (Jordan). Aquat. Bot. 20, 87–96. doi: 10.1016/0304-3770(84)90029-9
Wang J., Mao Y., Du G., Li X., Tang X. (2021). On microbial community of Pyropia haitanensis by metagenomic analysis. J. Oceanol. Limnol. 39, 1091–1102. doi: 10.1007/s00343-020-0189-0
Wang W., Wu L., Xu K., Xu Y., Ji D., Chen C., et al. (2020). The cultivation of Pyropia haitanensis has important impacts on the seawater microbial community. J. Appl. Phycol. 32, 2561–2573. doi: 10.1007/s10811-020-02068-6
Xie X., He Z., Hu X., Yin H., Liu X., Yang Y. (2017). Large-scale seaweed cultivation diverges water and sediment microbial communities in the coast of Nan’ao Island, South China Sea. Sci. Total Environ. 598, 97–108. doi: 10.1016/j.scitotenv.2017.03.233
Yang Y., Chai Z., Wang Q., Chen W., He Z., Jiang S. (2015). Cultivation of seaweed Gracilaria in Chinese coastal waters and its contribution to environmental improvements. Algal Res. 9, 236–244. doi: 10.1016/j.algal.2015.03.017
Yang Q. S., Dong J., Ahmad M., Ling J., Zhou W. G., Tan Y. H., et al. (2019). Analysis of nifH DNA and RNA reveals a disproportionate contribution to nitrogenase activities by rare plankton-associated diazotrophs. BMC Microbiol. 19, 1–12. doi: 10.1186/s12866-019-1565-9
Yang Y. F., Fei X. G., Song J. M., Hu H. Y., Wang G. C., Chung I. K. (2006). Growth of Gracilaria lemaneiformis under different cultivation conditions and its effects on nutrient removal in Chinese coastal waters. Aquaculture 254, 248–255. doi: 10.1016/j.aquaculture.2005.08.029
Zehr J. P., Jenkins B. D., Short S. M., Steward G. F. (2003). Nitrogenase gene diversity and microbial community structure: A cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Zhang X. Y., Li G. W., Wang C. S., Zhang Y. J., Xu X. W., Li H., et al. (2013). Marinicauda pacifica gen. nov., sp. nov., a prosthecate alphaproteobacterium of the family Hyphomonadaceae isolated from deep seawater. Int. J. Syst. Evol. Microbiol. 63, 2248–2253. doi: 10.1099/ijs.0.046656-0
Zhang A., Wen X., Yan H., He X., Su H., Tang H., et al. (2018). Response of microalgae to large-seaweed cultivation as revealed by particulate organic matter from an integrated aquaculture off Nan’ao Island, South China. Mar. pollut. Bull. 133, 137–143. doi: 10.1016/j.marpolbul.2018.05.026
Zhang Y., Yang Q., Ling J., Van Nostrand J. D., Shi Z., Zhou J., et al. (2016). The shifts of diazotrophic communities in spring and summer associated with coral Galaxea astreata, Pavona decussata, and Porites lutea. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01870
Zhang Y., Yang Q., Ling J., Van Nostrand J. D., Shi Z., Zhou J., et al. (2017a). Diversity and structure of diazotrophic communities in mangrove rhizosphere, revealed by high-throughput sequencing. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02032
Zhang Y., Zhang J., Liang Y., Li H., Li G., Chen X., et al. (2017b). Carbon sequestration processes and mechanisms in coastal mariculture environments in China. Sci. China Earth Sci. 60, 2097–2107. doi: 10.1007/s11430-017-9148-7
Keywords: nifH gene, biological nitrogen fixation (BNF), diazotrophic communities, cultivation periods, Gracilariopsis lemaneiformis
Citation: Pei P, Aslam M, Yang C, Ye P, Ke X, Liang Z, Li T, Chen W and Du H (2024) Temporal variations of biological nitrogen fixation and diazotrophic communities associated with artificial seaweed farms. Front. Mar. Sci. 11:1408958. doi: 10.3389/fmars.2024.1408958
Received: 29 March 2024; Accepted: 30 April 2024;
Published: 17 May 2024.
Edited by:
Junfu Dong, Shandong University, ChinaCopyright © 2024 Pei, Aslam, Yang, Ye, Ke, Liang, Li, Chen and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, aGR1QHN0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.