
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 11 July 2024
Sec. Aquatic Physiology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1407743
Studying the effect of heating rate on upper thermal limit has gained considerable attention in enhancing our mechanistic understanding of how organisms respond to changing temperatures in the context of climate change. The present study aimed to investigate the effects of heating rate on upper thermal limit and understand the physiological and molecular mechanisms used by organisms to cope with thermal stress at different heating rates. Batillaria attramentaria snails were exposed to slow (3°C/h) or fast (9°C/h) heating rates. The median lethal temperature (LT50) of snails exposed to these varying heating rates was determined. Additionally, we assessed heart rate under constant heating and investigated the transcriptomic response at the temperature where the heart rate reaches zero (FLT). The results revealed that snails exhibit a higher upper thermal limit (approximately 1.5°C) during fast heating as compared to slow heating. On average, the heart rate of slowly heated snails was 11 beats per minute lower than that of fast heated snails when the temperature was below 45°C. The findings indicate that the metabolic rate is lower during slow heating compared to fast heating when subjected to the same level of thermal stress. When exposed to a temperature of FLT, snails initiated a typical heat shock response to thermal stress, which included the increased expression of genes encoding heat shock proteins (HSPs) and protein disulfide isomerase (PDIA5) involved in protein folding. Remarkably, the genes glucose-regulated protein 94 (GRP94) and Calnexin, which are associated with the binding of unfolded proteins, showed distinct up-regulation in snails that were heated slowly, indicating the accumulation of misfolded proteins. The accumulation of misfolded proteins, coupled with additional energy consumption, may contribute to the lower upper thermal limit observed at a slow heating rate. Our research provides valuable insights for determining the realistic upper limits of temperature tolerance and improving predictions of how organisms will be affected by climate change in the future.
Climate change, characterized by global warming (Hoegh-Guldberg et al., 2018) and more frequent extreme heat events (Frölicher et al., 2018), represents a pressing concern that amplifies the energy requirements of ectothermic organisms (Alton and Kellermann, 2023) and jeopardizes the long-term survival of these species (Dahlke et al., 2020). To better understand how organisms will respond to climate change, scientists have developed mechanistic modeling approaches to predict population extinction risks and future geographic distributions (Kearney and Porter, 2009). One mechanistic approach to assess an organism’s tolerance to future warming is by comparing its upper thermal limit to the highest temperature it encounters in the field (Sunday et al., 2014; Pinsky et al., 2019). Traditionally, quantification of the upper thermal limit involves controlled conditions utilizing the dynamic method (Lutterschmidt and Hutchison, 1997), wherein organisms are exposed to a continuous heating rate until they exhibit symptoms of thermal stress or reach the point of death (Fernández-Loras et al., 2019). However, a limitation of upper thermal limit studies arises from the influence of the heating rate during tolerance measurement on the obtained results (Dong et al., 2022). Understanding the physiological and molecular mechanisms governing organismal sensitivity, and assessing whether such sensitivity varies with heating rate, is crucial for enhancing predictions of organism vulnerability to future climate change.
In the range of examined heating rates (0.5°C/h - 60°C/h), faster heating resulted in an elevated upper thermal limit in diverse organisms, including ants (Atta sexdens rubropiosa, Ribeiro et al., 2012), tropical marine fishes (Acanthochromis polyacanthus, Amphiprion melanopus, and Lates calcarifer, Illing et al., 2020), and flies (Glossina pallidipes, Terblanche et al., 2007). Conversely, moth (Cydia pomonella, Chidawanyika and Terblanche, 2011) and crabs (Eurypanopeus abbreviatus and Menippe nodifrons, Vinagre et al., 2015) exhibited reverse correlations, as individuals subjected to faster heating displayed significantly reduced tolerance. Additionally, Antarctic fish Harpagifer antarcticus demonstrated no significant difference in upper thermal limits across various heating rates (Saravia et al., 2021). These findings highlight the species-specific nature of the relationship between heating rate and upper thermal limit, necessitating individualized analysis for each species when assessing realistic upper thermal limits in laboratory studies.
Furthermore, research has shown that the effect of heating rate on upper thermal limits depends on the characteristics of the habitat. Research on a variety of tropical marine ectotherms suggested that organisms inhabiting rapidly changing thermal environments, such as the upper shore, exhibit steeper relationships between thermal limits and temperature change rates compared to those living in less variable environments on the lower or subtidal shore (Nguyen et al., 2011). Moyen and colleagues (2019) demonstrated that the upper thermal limit of high-zone mussels was significantly higher at the faster heating rate compared to the slower heating rate. Consequently, the rate at which heating occurs is a critical factor in thermal acclimatization, emphasizing the need for a thorough understanding of heating rates in natural environments to accurately determine the maximum thermal limits of species (Han et al., 2023).
Previous studies have shown how heating rates affect upper thermal limits and emphasized the importance of considering these effects in models predicting population dynamics (Liao et al., 2021). However, little research has been done to investigate the physiological or molecular mechanisms used by organisms to deal with thermal stress at different heating rates (Sørensen et al., 2013; Illing et al., 2020; Saravia et al., 2021). The cellular stress response (CSR) is essential for organisms to manage thermal damage to proteins, nucleic acids, and membranes. When proteins unfold due to heat, they trigger the prompt activation of gene transcription for HSPs (Somero, 2020). In ectotherms, physiological responses are temperature-dependent, with metabolic rates generally increasing with body temperatures (Pörtner, 2001; Somero et al., 2017). Heat stress augments energy metabolism in ectotherms, and the increased tissue oxygen demand is satisfied by an elevation in heart rate (Christen et al., 2018). Consequently, heart rate serves as a potential indicator of metabolic rate (Hui et al., 2020). Thermal stress induces a range of transcriptomic responses, encompassing ATP-generating pathways, DNA and protein damage repair, and the elimination of cellular and molecular debris generated by stress (Sokolova et al., 2012). Transcriptional studies thus offer valuable insights into the molecular mechanisms employed by organisms under different heating rates.
In the present study, we tested dark unbanded shell type Batillaria attramentaria exposed to gradual temperature increase of 3°C/h (slow-heated) or 9°C/h (fast-heated) starting at 27°C. The snail is a dominant species within temperate tidal flats, playing a crucial role in the ecosystem due to its influence on carbon flows within the ecosystem (Kawasaki et al., 2019). The selected heating rates represented the typical slow and fast heating rates observed in the field (see Supplementary Figure S1 for details). Using median lethal temperature (LT50) as an index, we compared the upper thermal limit of snails exposed to slow and fast heating rates. Additionally, we measured the heart rate (HR) under constant heating at both slow and fast heating rates. Studies on the response of gastropods to heat stress have primarily concentrated on cardiac physiology, assessing the impact of sudden temperature variations on HR. Acute thermal stress typically leads to heightened metabolic requirements, with increased tissue oxygen demand being met through enhanced cardiac function (Marshall et al., 2011). And we investigated the transcriptomic response in snails at the temperature where heart rate falls to zero (FLT). Our key objective was to contribute insights into the physiological and molecular underpinnings of the observed variations in upper thermal limit measurements resulting from different heating rates. The implications of our findings extend to facilitating a more accurate assessment of upper thermal limits in laboratory investigations.
Snails were collected from a muddy shore located at Yangmadao Island (37°27′N, 121°36′E) during low tides in the daytime in summer (July 2023). After collection, snails were transported back to the laboratory within 1 h and acclimated in a plastic box. The snails were immersed under fresh seawater for 4 h every day. Before performing measurements, a 3-day common garden acclimation was conducted with a temperature of ~27°C. Snails were not fed during acclimation.
Snails were placed in empty test tubes with a diameter of 25 mm. The test tubes were then immersed in a water bath (Grant TXF 200, Grant, Shepreth, UK). The snails were exposed to temperature increases at rates of either 3°C/h or 9°C/h. An additional subset of snails (n=3) was randomly chosen to monitor body temperature. A small hole (0.8 mm diameter) was drilled in the shell at a position above the heart. Subsequently, a K-type thermocouple was inserted, and the snails were positioned at the bottom of an empty test tube. Body temperature of snails was recorded every 1 min. Survival in response to exposures at temperatures of 46, 49, 50, 51, 52, and 53°C was assessed across three groups, each consisting of 10 snails (∑N = 2 heating rates × 6 temperatures × 3 groups × 10 snails = 360 snails in total). After heating stress, the test tubes containing the snails were submerged in flow-through seawater at ~27°C during a 3-day recovery period. Snails that did not exhibit an opercular reflex (rapid and complete withdrawal into their shell) upon stimulation by a sharp probe on the foot were scored as dead. The median lethal temperature (LT50) at three days after heat stress was calculated with logistic analysis in R. Differences in LT50 between individuals subjected to fast and slow heating rates were analyzed by a two-sample t-test.
Snails were randomly selected to measure heart rate using a method described by Dong and colleagues (2021). An infrared sensor was affixed to the shell at a position above the heart (the upper left of the aperture) using Blu-Tac (Bostik, Australia). Subsequently, snail was placed on the bottom of an empty test tube and warmed in air by heating the test tube in a water bath. Experimental temperature was increased from 27°C at rates of either 3°C/h or 9°C/h until a temperature was reached where heart rate fell to zero. The heartbeat was detected by means of an infrared sensor affixed to the shell. The light-dependent current produced by the heartbeat underwent amplification through an amplifier (AMP03, Newshift, Leiria, Portugal) and was further filtered and recorded using a Powerlab AD converter (8/30, ADIstruments, Bella Vista, NSW, Australia). Data were viewed and analyzed using LabChart v7 (ADInstruments). The number of heartbeats was counted manually every one minute (expressed as beats per minute, ∑N = 2 heating rates × 20 replicates = 40 snails in total).
To determine the temperature at which heart rate initially increases, we firstly used the dplyr package in R to calculate the mean heart rate in 1-°C intervals (HR1°C) for each heating rate when temperature below 45°C (the temperature maximum heart rate achieved). Secondly, we performed Games-Howell tests in R to compare the differences of HR1°C between temperatures for each heating rate. The Arrhenius Breakpoint Temperature (ABT) for cardiac performance, which indicates the temperature at which there is a significant decrease in heart rate with progressive heating (usually from 40°C to FLT), was calculated using segmented regression methods in R software (Muggeo, 2008). The method generates the best-fit line on either side of a putative break point for the relationship of ln-transformed heart rate against the reciprocal value of absolute temperature. The Flatline Temperature (FLT) designates the temperature at which the heart rate drops to zero. Variations in maximum heart rate (MHR), ABT and FLT between individuals subjected to fast and slow heating rates were examined using a two-sample t-test.
To assess the transcriptomic response to different heating rates, we subjected snails to two heating rates in air: 3°C/h and 9°C/h, starting from an initial temperature of 27°C. When temperature increased to 49.0°C (approximating mean FLT), four snails from each heating rate were dissected, and foot muscle samples were immediately flash-frozen in liquid nitrogen. Concurrently, a control group consisting of four unheated snails was randomly selected to provide a baseline for comparison (∑N = 3 treatments × 4 replicates = 12 snails in total).
We extracted total RNA using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. The integrity of the RNA was assessed using the Fragment Analyzer 5400 (Agilent Technologies, CA, USA). The extracted total RNA served as the input material for subsequent RNA sample preparations. Sequencing libraries were crafted using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA), following the manufacturer’s recommended procedures, with index codes incorporated to uniquely identify sequences for each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq 6000 platform at Novogene (Tianjin, China), yielding 150 bp paired-end reads.
Raw reads were processed with trimmomatic v0.39 to remove the adapter sequences, and low-quality bases (Bolger et al., 2014). The clean reads were aligned to B. attramentaria draft genome (Patra et al., 2023; GCA_018292915.1) using HISAT2 v2.2.1 (Pertea et al., 2016), and sorted using SAMtools v1.6 (Li et al., 2009). All BAM files were merged using SAMtools, and transcripts were genome-guided assembled by Trinity v2.1.1 using the merged BAM file. These assembled transcripts were further bundled into a non-redundant set by PASA v2.5.2. Additionally, TransDecoder v5.7.1 was executed to identify coding regions within the transcripts. A Gene Transfer Format (GTF) annotation file for the mussel draft genome was generated, incorporating transcripts coding for more than 100 amino acids. Transcript abundances of each sample were estimated using StringTie v2.1.7 with the “-eB” option, based on GTF file. Read counts, essential for subsequent differential expression analysis, were obtained using the python script ‘‘prepDE.py’’ (http://ccb.jhu.edu/software/stringtie/dl/prepDE.py).
Differential expression analysis was carried out between fast and slow-heated snails and control snails using DESeq2 v1.28.1 (Love et al., 2014). Thresholds for screening differentially expressed genes (DEGs) were set at an absolute value of log2fold-change ≥ 1 and an adjusted P-value < 0.05. To elucidate common and specific responses of snails to fast and slow heating rates, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using compareCluster v3.0.4 (Wu et al., 2021) in R.
The mean LT50 values of snails at fast and slow heating rates were 50.5 ± 0.2°C and 49.0 ± 0°C, respectively (Figure 1). The results of two-sample t-test revealed a significantly higher LT50 for snails exposed to fast heating rate compared to those subjected to the slow heating rate (t= 12.35, df = 4, P < 0.001, Cohen’s d = 10.084).
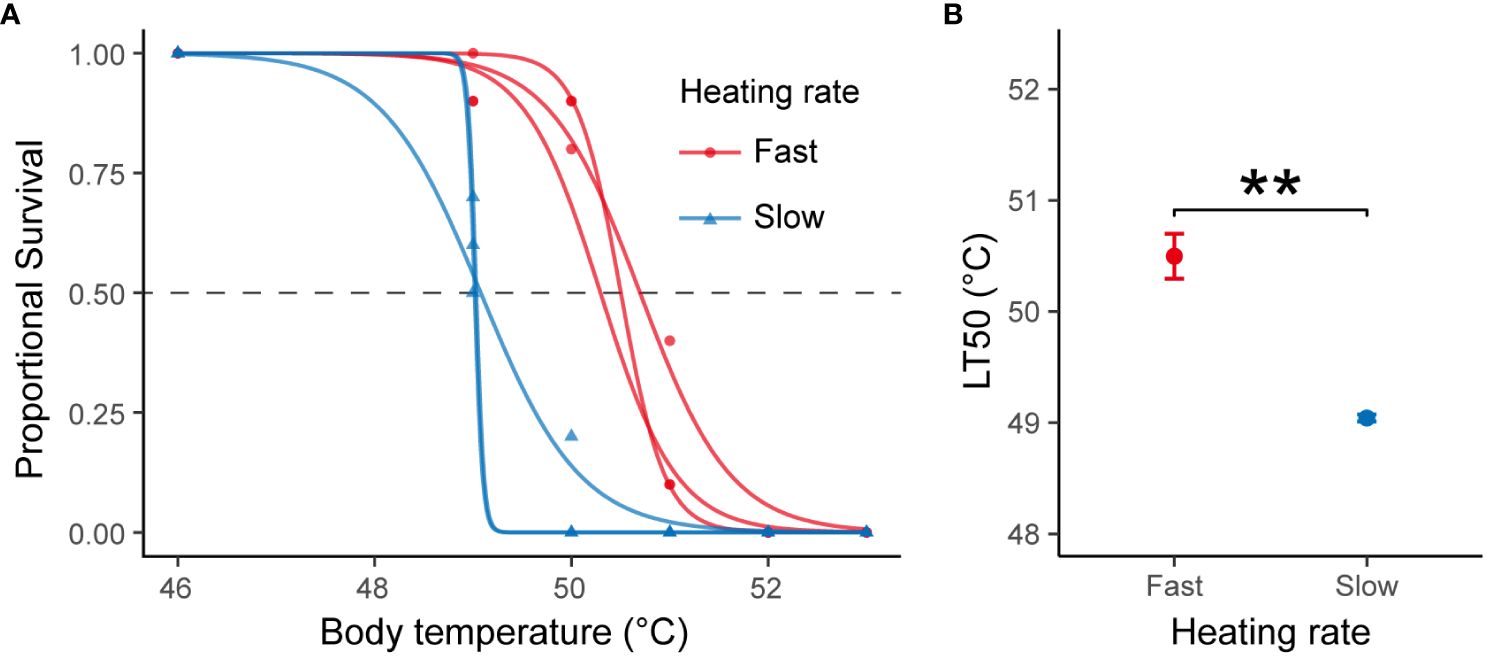
Figure 1 (A) Survival rate of B. attramentaria individuals after exposure to elevated body temperature. The red and blue curves were produced through logistic regression analysis for snails subjected to fast and slow heating rates, respectively. (B) Average LT50 for snails subjected to fast or slow heating rates (mean ± s.d., n=3). ** indicates significant differences (t-test, P < 0.001).
Under constant heating, the cardiac performance exhibited divergent patterns in snails subjected to fast and slow heating rates (Figure 2A). At a fast heating rate, there was a significant increase in HR1°C at 32°C (Figure 2B) accompanied by a plateau in the curve spanning temperatures from 34 to 40°C. Conversely, HR1°C remained constant between 28 and 40°C during slow heating. Upon reaching approximately 45°C, both fast- and slow-heated snails exhibited MHR. Below 45°C, the mean heart rate differed, with values of 68.6 ± 10.0 and 57.0 ± 9.2 beats per minute for fast- and slow-heated snails, respectively. The mean heart rate of individuals at fast heating rate was higher than that of individuals at slow heating rate (t = 3.845, df = 38, P < 0.001, Cohen’s d = 1.216).
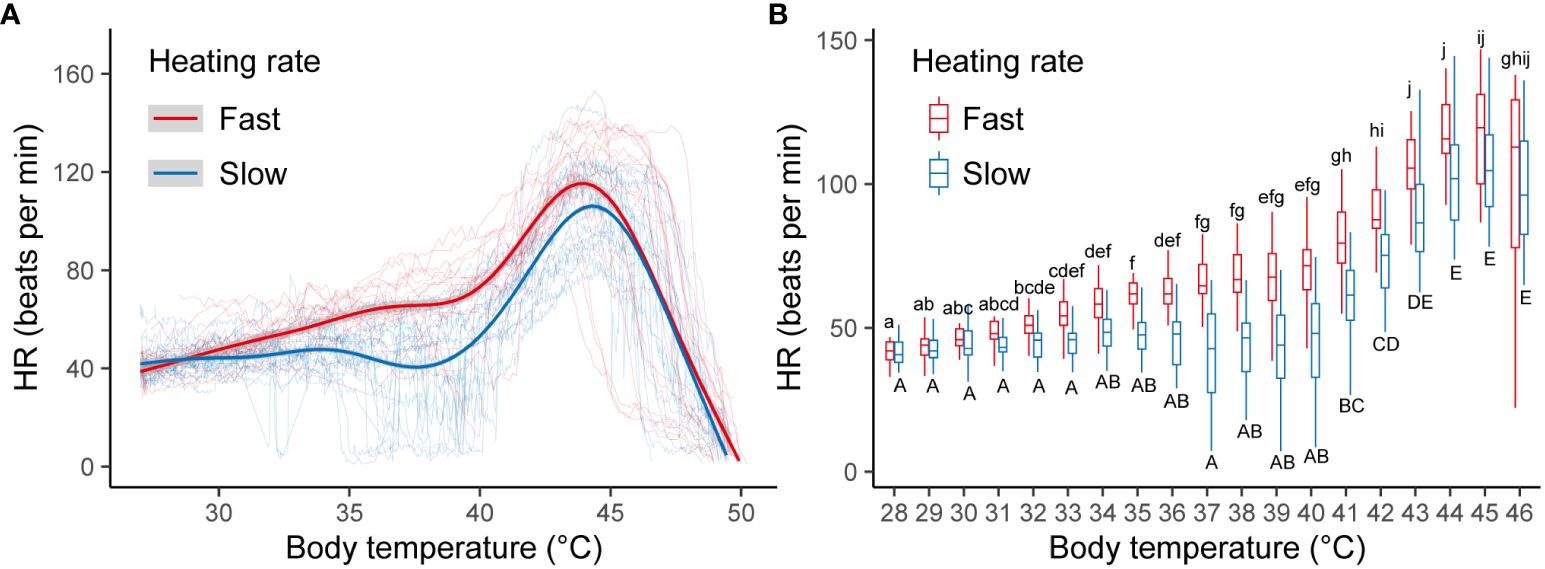
Figure 2 (A) Cardiac performance curves of B. attramentaria individuals over an acute warming ramp. Each thin transparent line represents the cardiac performance of a snail at fast or slow heating rate over different temperatures. The red and blue thick curves were fitted from GAMM for snails at fast (n=20) and slow (n=20) heating rates, respectively. The gray regions represent the 95% confidence intervals. (B) HR1°C in snails at fast versus slow heating rate when temperature was below 45°C. Significant differences between temperatures (n=20) for each heating rate were indicated by different letters (P<0.05).
The MHR exhibited a significant discrepancy between the two heating rates (t = 2.688, df = 38, P = 0.011, Cohen’s d = 0.850). Specifically, the average MHR was 125.4 ± 15.3 beats per minute during fast heating and 111.3 ± 17.6 beats per minute during slow heating (Figure 3A). In terms of the ABT, individuals displayed mean values of 45.5 ± 1.6 and 46.4 ± 1.1°C for fast and slow heating rates, respectively (Figure 3B). Notably, no statistically significant difference in ABT was observed between the two heating rates (t = -1.913, df = 38, P = 0.063, Cohen’s d = -0.605). The FLT among individuals subjected to fast heating surpassed that of individuals exposed to slow heating (t = 2.591, df = 50, P = 0.013, Cohen’s d = 0.724). The mean FLT for fast and slow heating rates was 49.4 ± 0.4 and 49.0 ± 0.6°C, respectively (Figure 3C).
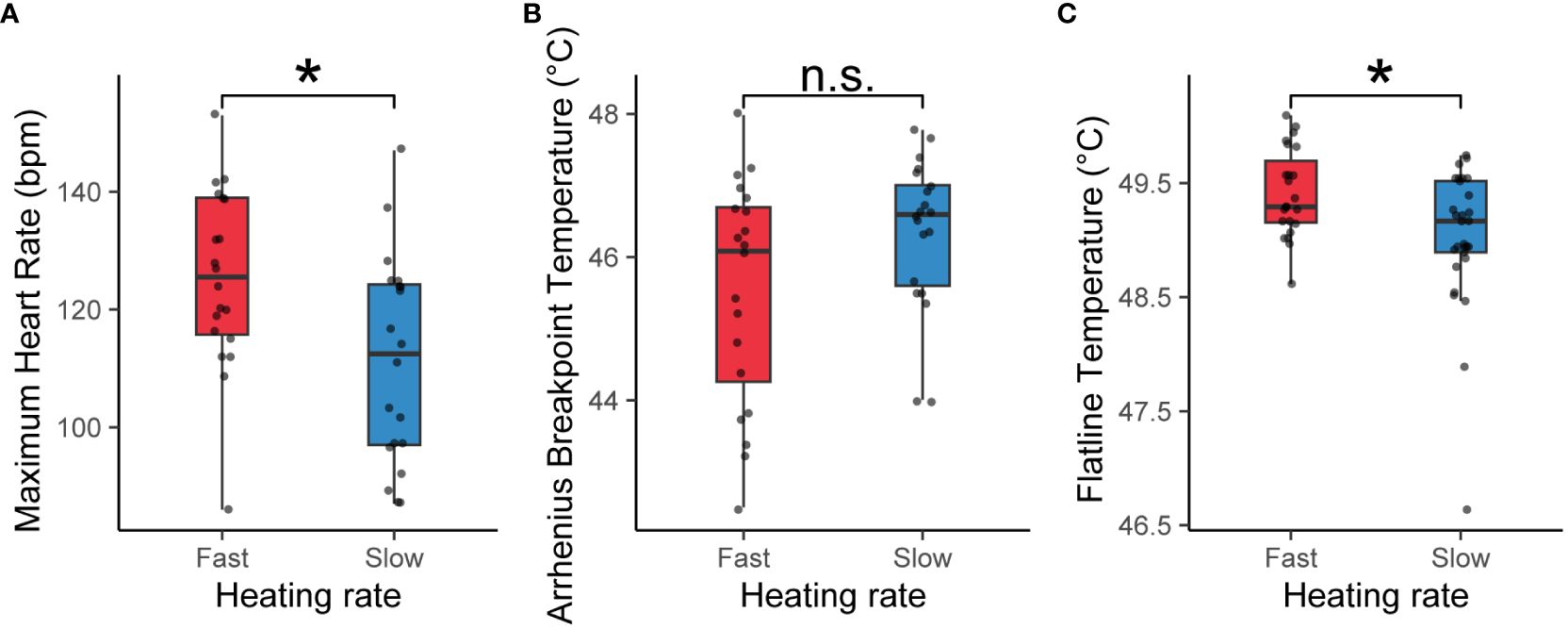
Figure 3 Maximum Heart Rate [MHR, (A)], Arrhenius Breakpoint Temperature [ABT, (B)] and Flatline Temperature [FLT, (C)] in snails at fast versus slow heating rate. Boxplots outline the 25th and 75th percentiles, and the midline indicates the median. Each data point represents an individual snail (n=20 at fast heating rate, n=20 at slow heating rate). * indicates significant differences (t-test, P<0.05).
A total of 75.128 Gb of clean bases were obtained from the transcriptomes of 12 individuals. The average mapping rate was 82.34%. Following assembly and identification of coding regions, a GTF file was generated, containing predictions for 35,328 genes. The expression levels of these 35,328 genes under each treatment condition (control, fast heating, or slow heating) were assessed by quantifying the reads mapped to the genome. Subsequent screening revealed 1,468 and 2,778 genes as DEGs in response to fast and slow heating, respectively. Specifically, during fast heating, 655 genes exhibited up-regulation, while 813 genes displayed down-regulation (Figure 4A). In contrast, 1,697 genes were up-regulated, and 1,081 genes were down-regulated during slow heating (Figure 4B). A total of 759 DEGs were commonly identified at both fast and slow heating rate. Additionally, 709 and 2019 DEGs were uniquely identified at fast and slow heating rate, respectively.
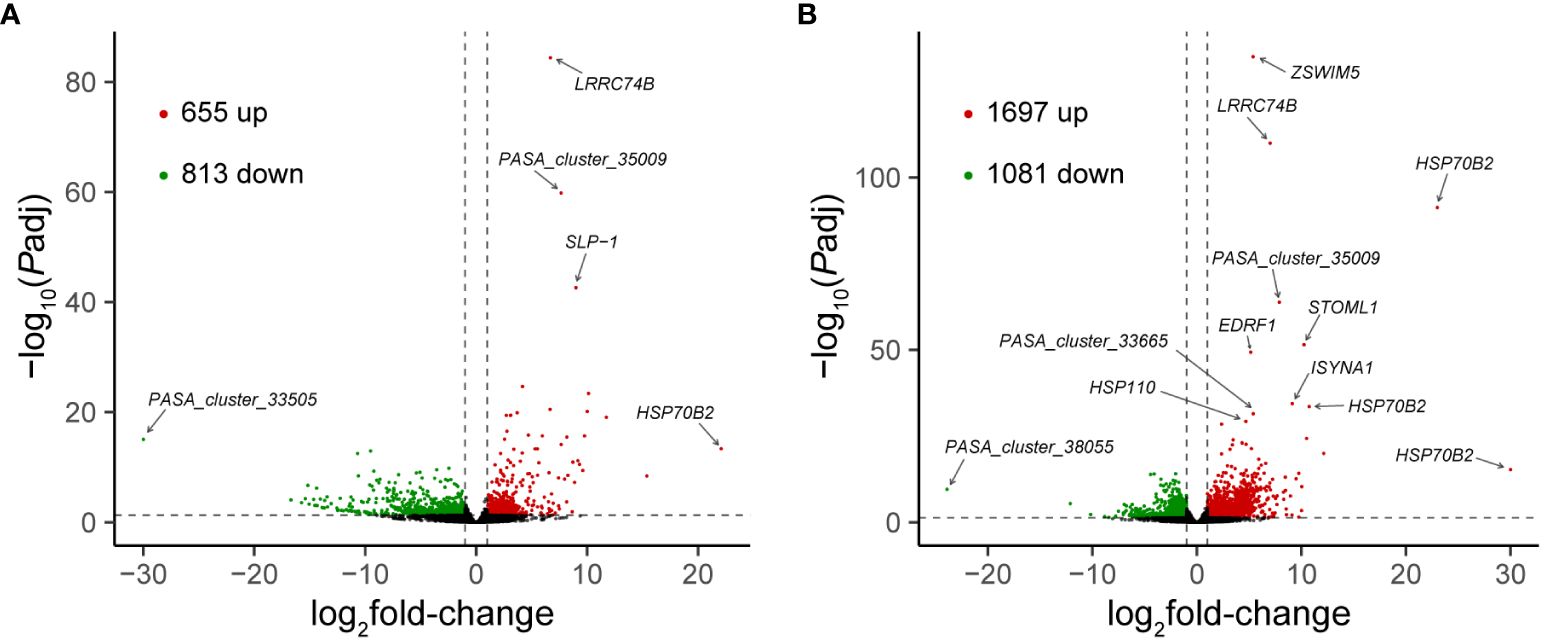
Figure 4 Volcano plots showing DEGs in fast heated (A) and slowly heated snails (B) versus control snails. The red and green dots represent up-regulated and down-regulated DEGs, respectively, and the black dots represent non-DEGs. Vertical dotted lines represent an absolute value of log2fold-change = 1, and horizontal dotted line represents the significance cut-off (Padj = 0.05).
Enrichment analyses conducted for GO terms unveiled common enrichment of DEGs in “ATP-dependent protein folding chaperone”, “serine-type endopeptidase inhibitor activity”, and “DNA-binding transcription factor activity” when comparing fast and slow heating rates (Figure 5A). And KEGG enrichment analyses indicated consistent enrichment of DEGs in “Antigen processing and presentation”, “Longevity regulation pathway”, “Estrogen signaling pathway”, and “Protein processing in the endoplasmic reticulum” (Figure 5B).

Figure 5 Functional analysis of differential gene expression. DEGs enriched in the GO terms (A) and KEGG pathways (B). The dot size represents the count of the number of DEGs enriched in each category. The adjusted P-value (Padj) are shown as different colours, with red being more significant than blue.
Further scrutiny of DEGs in fast-heated snails revealed unique enrichment in 13 GO terms, with the top 5 significantly enriched terms being “extracellular region”, “chitin binding”, “multicellular organism development”, “carbohydrate metabolic process”, and “cell-matrix adhesion”. Moreover, these DEGs exhibited unique enrichment in two KEGG pathways: “Amino sugar and nucleotide sugar metabolism” and the “MAPK signaling pathway”.
In contrast, DEGs in slow-heated snails displayed exclusive enrichment in six GO terms, encompassing “ribosome”, “structural constituent of ribosome”, “translation”, “endoplasmic reticulum”, “unfolded protein binding”, and “translation initiation factor activity”. Additionally, DEGs from slow heating rate demonstrated unique enrichment in three KEGG pathways: “Ribosome”, “Oxidative phosphorylation”, and “Thermogenesis”.
A total of 22 DEGs revealed enrichment in “ATP-dependent protein folding chaperone”, “protein folding”, and “unfolded protein binding” (Figure 6). Notably, all these genes exhibited up-regulation in response to a temperature increase to 49.0°C. Among these, three genes (Chaperonin Containing TCP1 Subunit 5, CCT5; Glucose-regulated protein 94, GRP94; and Calnexin) were uniquely up-regulated in slow-heated snails.
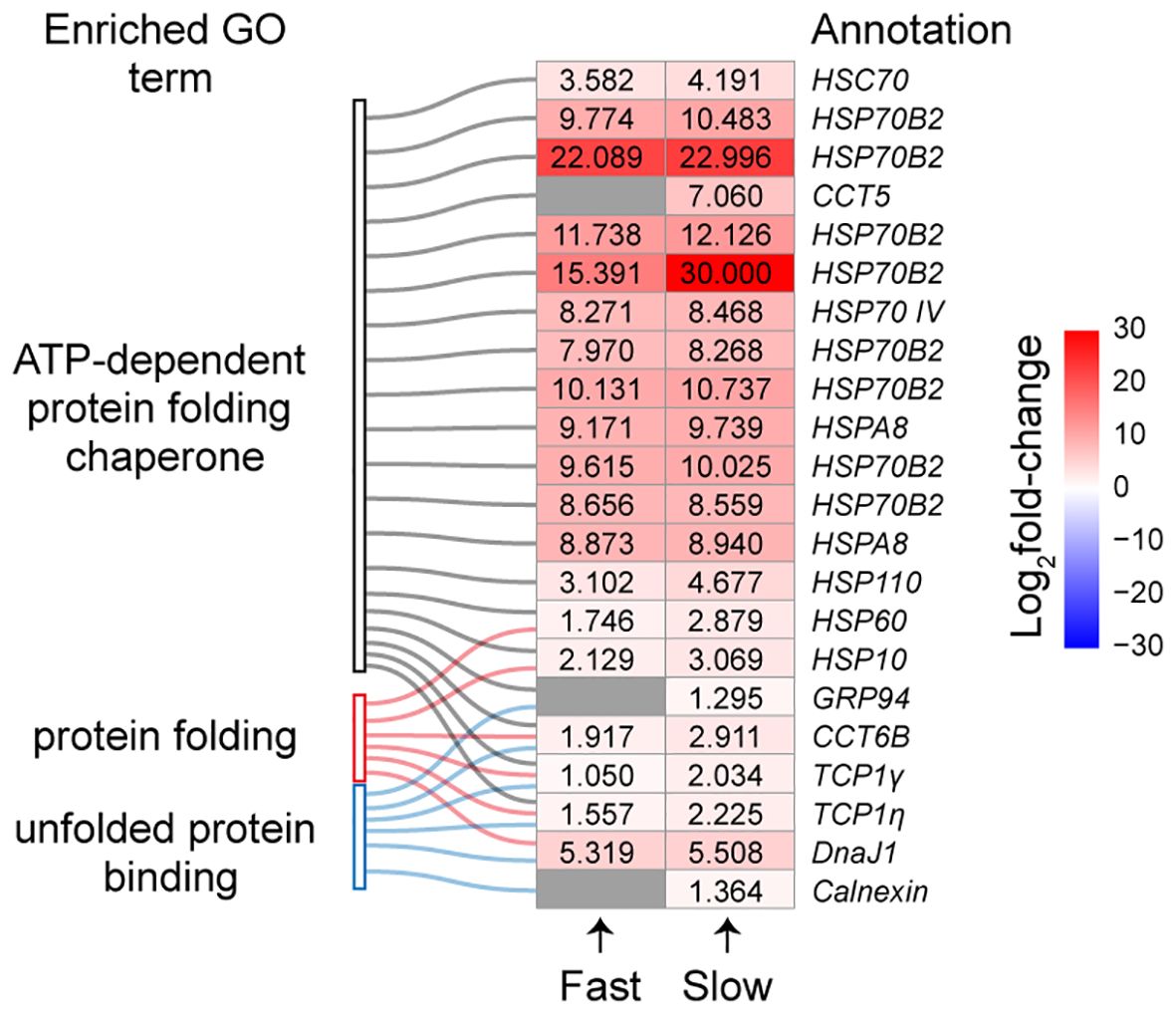
Figure 6 The expression levels of DEGs involved in protein folding and unfolded protein binding. Colour gradient represents the expression level of DEGs, and grey represents no significant difference. The numbers in heatmap are log2fold-change of DEGs.
Moreover, a total of 38 DEGs associated with the ribosome pathway were identified in slow-heated snails, whereas no DEGs were observed in the same pathway for fast-heated snails (Figure 7). The 38 DEGs from slow-heated snails comprised 23 large (RPL) and 15 small subunit ribosomal proteins (RPS), all of which were up-regulated except for RPL35e.

Figure 7 Heatmap of DEGs enriched in selected functional categories. Each DEG is represented by a single row, and the log2fold-change values are shown in different colours: red corresponds to an up-regulated gene, blue corresponds to a down-regulated gene and grey represents no significant difference.
The investigation of heating rate impacts on thermal limits has garnered significant attention in enhancing our mechanistic understanding of physiological responses to changing thermal conditions in the context of climate change. Nevertheless, there has been limited exploration of the physiological and molecular mechanisms employed by organisms to cope with thermal stress under varying heating rates. This study aims to fill this gap by demonstrating that the mudflat snail B. attramentaria reveals the physiological and molecular adaptations in response to distinct heating rates.
At a fast heating rate, heart rate of B. attramentaria exhibited a notable increase at 32°C, plateauing between 33 and 40°C. This observation implies that, during fast heating, B. attramentaria suppresses resting metabolism within the moderate thermal range, which is similar to other marine intertidal gastropods (Marshall et al., 2011; Chen et al., 2021). The metabolic rate depression serves as an adaptive strategy, enabling prolonged survival under thermal extremes by conserving energy stores while ensuring an adequate supply of ATP for essential physiological functions (Sokolova et al., 2012; Marshall and McQuaid, 2020). In contrast to the response in fast-heated snails, heart rate exhibited insensitivity to temperature variations during slow heating, with no significant increase between 27 and 40°C. The heart rate peaked at approximately 45°C. Thermal stress usually results in increased metabolic demands, and the increased requirement for tissue oxygen is met through improved cardiac performance (Marshall et al., 2011). On average, the heart rate of snails subjected to slow heating was 11 beats per minute lower than that of snails exposed to rapid heating when the temperature was below 45°C. A lower heart rate during slow heating signifies a decreased need for oxygen. Thus, our results indicate that the metabolic rate during slow heating is lower than that during rapid heating under comparable thermal stress conditions.
The MHR in ectotherms displays sensitivity to energy content, with individuals harboring greater energy stores exhibiting a heightened MHR under thermal stress (Hardison et al., 2023). When subjected to constant heating, snails experiencing slow heating manifested a lower MHR compared to fast-heated snails, indicative of diminished energy stores in slow-heated snails as the temperature reached 45°C. Despite the fact that snails exhibit a reduced metabolic rate at the equivalent temperature during slow heating, slow heating also prolongs exposure time to stressful conditions, resulting in higher energy consumption during slow heating as opposed to fast heating. In contrast to the ABT, slow-heated snails showed a significant decrease in their FLT compared to the fast-heated snails, similar to the difference observed in the LT50. These results suggest that FLT can be used as an effective measure to determine the maximum temperature tolerance in B. attramentaria snails.
When temperatures approach or surpass the FLT in intertidal mollusks, cellular damage becomes irreversible, as evidenced by the upregulation of genes responsible for encoding proteins involved in eliminating irreversibly damaged proteins from the cell (Gracey et al., 2008). Functional enrichment analyses revealed that DEGs were notably enriched in “ATP-dependent protein folding chaperones” during both fast and slow heating when the temperature increased to FLT. The translocation-associated proteins (TRAP), recognized as membrane channels facilitating protein transport during translation into the endoplasmic reticulum (ER) (Jaskolowski et al., 2023), and protein disulfide isomerase (PDIA5), another ER-resident protein (Wilkinson et al., 2005), were identified in the analyses. This suggests an augmentation in protein translocation into the ER and unfolding of ER proteins during both fast and slow heating.
ATP-dependent chaperones necessitate ATP hydrolysis to assist in protein folding, preventing the accumulation of aberrant protein conformations that may lead to proteotoxicity (Mitra et al., 2022). Our findings indicate that snails activate an energy-costly heat shock response during both fast and slow heating rates. The expression levels of most chaperones were slightly higher during slow heating than fast heating, except for one HSP70B2 gene, which exhibited a dramatic up-regulation during slow heating (Figure 6). These results suggest an increased production of chaperones aimed at removing irreversibly damaged proteins from the cell during slow heating when temperatures approach the FLT. Among these chaperones, we identified eight HSP70B2 genes with significantly increased expression under heat stress. It has been documented that HSP70B2 genes undergo expansion in intertidal invertebrates (Dong et al., 2023; Liang et al., 2023; Yuan et al., 2023). Furthermore, HSP70B2 expression is predominantly up-regulated during air exposure, indicating its pivotal role in intertidal adaptation (Yuan et al., 2023). Our results indicated the importance of HSP70B2 in response to heat stress in snail B. attramentaria.
The finding that a slower heating rate elicits a more pronounced CSR compared to a faster heating rate implies that the slower heating rate causes greater heat-induced damage at the cellular level. Specifically, two chaperones involved in binding unfolded proteins, GRP94 and Calnexin, were found to be up-regulated in slow-heated snails. GRP94 helps detect misfolded proteins in the endoplasmic reticulum and sends them for degradation (Buc Calderon et al., 2018). Calnexin is a molecular chaperone that assists in the folding and quality control of proteins associated with membranes and secretion (Paskevicius et al., 2023). These findings suggest that misfolded proteins accumulate at a slow heating rate, requiring additional energy for their degradation (Somero, 2002). Slow heating also leads to the enrichment of “oxidative phosphorylation” and “thermogenesis” pathways. Genes encoding proteins involved in redox regulation, such as cytochrome C oxidases (COX5A, COX5B), ATP synthases (ATP5MC2), and cytochrome reductase (Cytc), were up-regulated, indicating improved energy production when the temperature reaches the FLT (Figure 7). The accumulation of misfolded proteins and the increased energy consumption may contribute to the lower FLT and LT50 observed during slow heating.
The folding of newly synthesized proteins into their native conformation relies on the assistance of a network of heat shock protein chaperones. Initially, these proteins are produced as linear chains on ribosomes and must undergo folding to adopt their functional conformations (Hu et al., 2022). Our study found that the expression of several ribosomal protein genes was up-regulated in B. attramentaria when subjected to a slow heating rate. Ribosome activity has been identified as a potential biomarker for cellular stress. For instance, mudflat clams exposed to heat stress in mud-air conditions showed an increase in the expression of several ribosomal protein genes (Zhang et al., 2020). The up-regulation of ribosomal protein genes associated with both the large and small subunits can enhance the tolerance of fish to thermal stress (Quinn et al., 2011). Our findings suggest an increase in ribosome biogenesis in slowly heated snails, indicating that it plays a role in conferring tolerance to slow heating but not fast heating.
The GO terms “carbohydrate metabolic process,” “chitin binding,” and “chitin synthase activity,” as well as the KEGG pathway “amino sugar and nucleotide sugar metabolism,” were specifically enriched during fast heating. These categories are related to the metabolism of carbohydrates. Several DEGs were identified as being involved in chitin hydrolysis or synthesis, such as CHIT1, CDA8, and Hex. The expression of these genes decreased during fast heating, suggesting that chitin metabolism is temporally constrained when the temperature increases to FLT. Chitin is an insoluble linear polymer composed of β-(1,4) linked N-acetyl-D-glucosamine units and is commonly found in mollusk shells. For instance, the sea snail Conus inscriptus shell chitin has a yield of 21.65% on a dry weight basis (Mohan et al., 2019). The restricted chitin metabolism may conserve energy and allocate more resources to repair processes, which could contribute to the higher upper thermal limits of B. attramentaria during fast heating.
Four genes involved in carbohydrate metabolism, namely XYN1, BXL5, FUT1, and Fuc, were found to be upregulated in both treatment groups. XYN1 is a glycoside hydrolase that can cleave the β-1,4-glycoside linkages in the xylan backbone (Kim et al., 2023). BXL5 belongs to the exoglycosidase enzyme category and has a unique ability to break down xylo-oligosaccharides into xylose (Saleem et al., 2012). Fuc plays a role in catalyzing the hydrolysis of terminal fucose residues or trans-fucosylation (Wan et al., 2020). These findings suggest that xylan and fucose may serve as important energy sources for the snail B. attramentaria when exposed to high temperatures.
In conclusion, our study provides insights into the physiological and molecular foundations underlying the differences in upper thermal limit measurements obtained using different heating rates. We observed distinct patterns of cardiac performance in snails subjected to fast and slow heating rates. The heart rate during slow heating was lower compared to fast heating under equivalent thermal stress. However, slow heating also prolonged exposure to stressful temperatures, resulting in higher energy consumption. When the temperature reached the temperature of FLT, snails exhibited a common heat shock response characterized by the up-regulation of genes involved in protein folding, such as HSPs and PDIA5 (Figure 8). Interestingly, slow-heated snails uniquely up-regulated genes involved in unfolded protein binding, namely GRP94 and Calnexin, indicating the accumulation of misfolded proteins. This accumulation, along with the additional energy consumption, may contribute to the lower upper thermal limit observed in slowly heated snails. Our findings have implications for determining realistic upper thermal limits and improving predictions of organism vulnerability to future climate change.
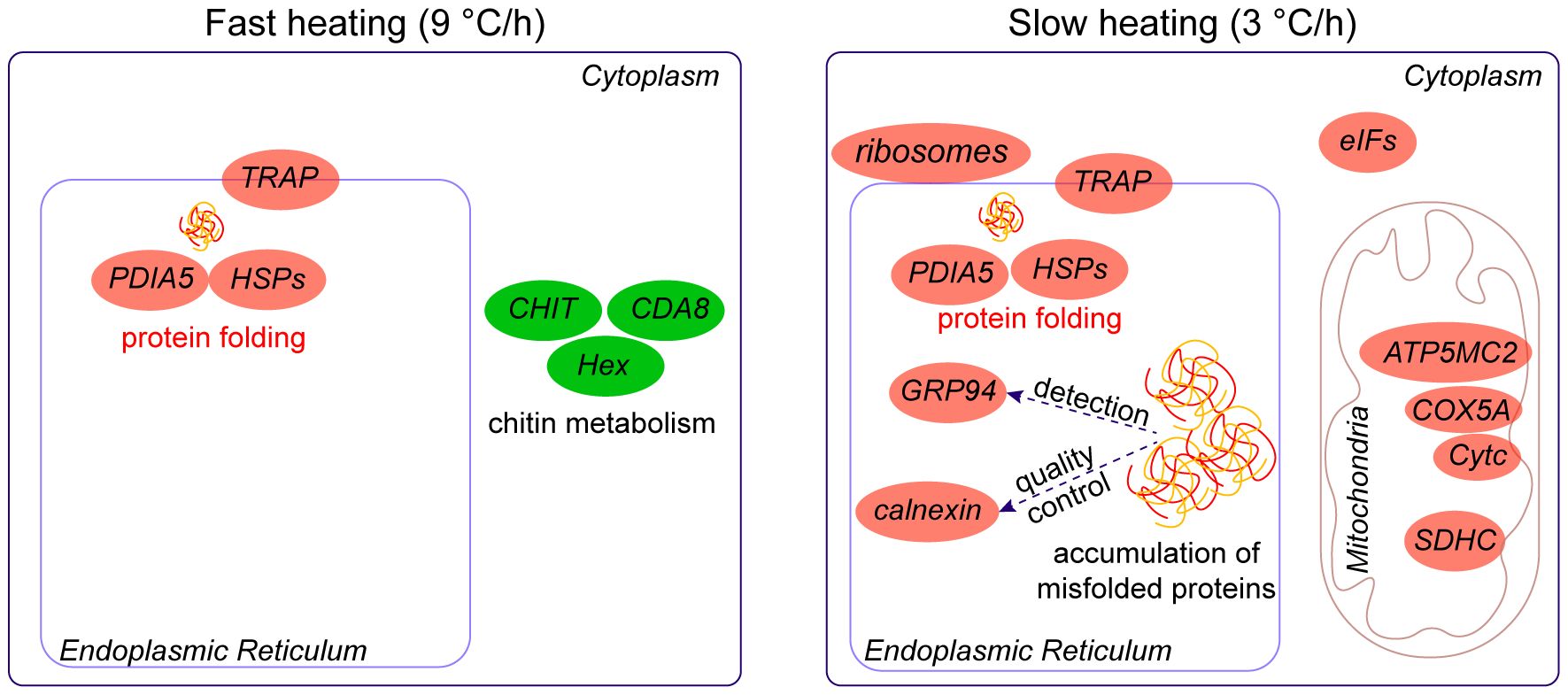
Figure 8 Summary of the molecular responses of B. attramentaria to thermal stress at different heating rates. Snails respond to thermal stress by activating a common heat shock response. This response involves the up-regulation of genes associated with protein folding, such as HSPs and PDIA5. During fast heating, genes involved in chitin hydrolysis or synthesis, such as CHIT1, CDA8, and Hex, showed a unique decrease, suggesting that chitin metabolism is temporally constrained. On the other hand, genes GRP94 and Calnexin, which are related to unfolded protein binding, were uniquely up-regulated in slowly heated snails, indicating the accumulation of misfolded proteins. Additionally, genes encoding proteins involved in redox regulation, such as ATP5MC2, COX5A, and Cytc, were up-regulated, suggesting improved energy production in slowly heated snails.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number can be found below: https://ngdc.cncb.ac.cn/gsa, CRA015574.
The animal study was approved by Ethics Committee for Experimental Animals of Yantai University. The study was conducted in accordance with the local legislation and institutional requirements.
L-ND: Formal analysis, Writing – original draft. Y-HD: Data curation, Formal analysis, Methodology, Writing – original draft. G-DH: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (42006107).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1407743/full#supplementary-material
Supplementary Figure 1 | The model temperature of Batillaria attramentaria. (A) Distribution of model temperature on all test days. (B) Changes in temperature in any given 10 min during temperature measurement. The horizontal axis shows the model temperature and the vertical axis shows the relative abundance (as normalized density). Data are from Han et al., 2023. Biomimetic models were positioned on the dry surface of a mud flat inhabited by snails at Yangmadao Island during sunny days in late July and early August 2022. The models were uniformly oriented and exposed to direct sunlight. The internal shell temperature was recorded every 10 seconds using a Modbus data logger (LCES_UDR_V62, Songyue, Guangdong, China). Changes in temperature within any given 10-minute interval for each model during temperature measurements were determined using a sliding window method (with a window size of 10 minutes and a jump size of 10 seconds).
Alton L. A., Kellermann V. (2023). Interspecific interactions alter the metabolic costs of climate warming. Nat. Climate Change 13, 382–388. doi: 10.1038/s41558-023-01607-6
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Buc Calderon P., Sennesael A.-L., Glorieux C. (2018). Glucose-regulated protein of 94 kDa contributes to the development of an aggressive phenotype in breast cancer cells. Biomed. Pharmacother. 105, 115–120. doi: 10.1016/j.biopha.2018.05.106
Chen Y., Wang J., Liao M., Li X., Dong Y. (2021). Temperature adaptations of the thermophilic snail Echinolittorina malaccana: Insights from metabolomic analysis. J. Exp. Biol. 224, jeb238659. doi: 10.1242/jeb.238659
Chidawanyika F., Terblanche J. S. (2011). Rapid thermal responses and thermal tolerance in adult codling moth Cydia pomonella (Lepidoptera: Tortricidae). J. Insect Physiol. 57, 108–117. doi: 10.1016/j.jinsphys.2010.09.013
Christen F., Desrosiers V., Dupont-Cyr B. A., Vandenberg G. W., Le François N. R., Tardif J.-C., et al. (2018). Thermal tolerance and thermal sensitivity of heart mitochondria: Mitochondrial integrity and ROS production. Free Radical Biol. Med. 116, 11–18. doi: 10.1016/j.freeradbiomed.2017.12.037
Dahlke F. T., Wohlrab S., Butzin M., Pörtner H.-O. (2020). Thermal bottlenecks in the life cycle define climate vulnerability of fish. Science 369, 65–70. doi: 10.1126/science.aaz3658
Dong Y., Han G., Li X. (2021). “Heart rate measurement in mollusks,” in Research Methods of Environmental Physiology in Aquatic Sciences. Eds. Gao K., Hutchins D. A., Beardall J. (Singapore: Science Press and Springer Nature Singapore Pte Ltd.), 327–334. doi: 10.1007/978-981-15-5354-7_38
Dong Y., Liao M., Han G., Somero G. N. (2022). An integrated, multi-level analysis of thermal effects on intertidal molluscs for understanding species distribution patterns. Biol. Rev. 97, 554–581. doi: 10.1111/brv.12811
Dong Z., Liu S., Yu H., Kong L., Li Q. (2023). Contraction of heat shock protein 70 genes uncovers heat adaptability of Ostrea denselamellosa. J. Ocean Univ. China 22, 1669–1676. doi: 10.1007/s11802-023-5641-2
Fernández-Loras A., Boyero L., Correa-Araneda F., Tejedo M., Hettyey A., Bosch J. (2019). Infection with Batrachochytrium dendrobatidis lowers heat tolerance of tadpole hosts and cannot be cleared by brief exposure to CTmax. PloS One 14, e0216090. doi: 10.1371/journal.pone.0216090
Frölicher T. L., Fischer E. M., Gruber N. (2018). Marine heatwaves under global warming. Nature 560, 7718. doi: 10.1038/s41586-018-0383-9
Gracey A. Y., Chaney M. L., Boomhower J. P., Tyburczy W. R., Connor K., Somero G. N. (2008). Rhythms of gene expression in a fluctuating intertidal environment. Curr. Biol. 18, 1501–1507. doi: 10.1016/j.cub.2008.08.049
Han G., Du Y., Du L. (2023). Effects of heating rate and shell colour on the cardiac thermal performance in a polymorphic gastropod Batillaria attramentaria. Mar. Environ. Res. 189, 106045. doi: 10.1016/j.marenvres.2023.106045
Hardison E. A., Schwieterman G. D., Eliason E. J. (2023). Diet changes thermal acclimation capacity, but not acclimation rate, in a marine ectotherm (Girella nigricans) during warming. Proc. R. Soc. B: Biol. Sci. 290, 20222505. doi: 10.1098/rspb.2022.2505
Hoegh-Guldberg O., Jacob D., Taylor M., Bindi M., Brown S., Camilloni I., et al. (2018). Impacts of 1.5°C of Global Warming on Natural and Human Systems (Cambridge, UK: Cambridge University Press).
Hu C., Yang J., Qi Z., Wu H., Wang B., Zou F., et al. (2022). Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 3, e161. doi: 10.1002/mco2.161
Hui T. Y., Dong Y., Han G., Lau S. L. Y., Cheng M. C. F., Meepoka C., et al. (2020). Timing metabolic depression: Predicting thermal stress in extreme intertidal environments. Am. Nat. 196, 501–511. doi: 10.1086/710339
Illing B., Downie A. T., Beghin M., Rummer J. L. (2020). Critical thermal maxima of early life stages of three tropical fishes: Effects of rearing temperature and experimental heating rate. J. Thermal Biol. 90, 102582. doi: 10.1016/j.jtherbio.2020.102582
Jaskolowski M., Jomaa A., Gamerdinger M., Shrestha S., Leibundgut M., Deuerling E., et al. (2023). Molecular basis of the TRAP complex function in ER protein biogenesis. Nat. Struct. Mol. Biol. 30, 6. doi: 10.1038/s41594-023-00990-0
Kawasaki K., Sasaki-Kinoshita A., Nakatsubo T. (2019). Annual respiration of Japanese mud snail Batillaria attramentaria in an intertidal flat: Its impact on ecosystem carbon flows. Landscape Ecol. Eng. 15, 113–120. doi: 10.1007/s11355-018-0365-y
Kearney M., Porter W. (2009). Mechanistic niche modelling: Combining physiological and spatial data to predict species’ ranges. Ecol. Lett. 12, 334–350. doi: 10.1111/j.1461-0248.2008.01277.x
Kim I. J., Kim S. R., Kim K. H., Bornscheuer U. T., Nam K. H. (2023). Characterization and structural analysis of the endo-1,4-β-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification. Sci. Rep. 13, 17332. doi: 10.1038/s41598-023-44495-8
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liang J., Cameron G., Faucher S. P. (2023). Development of heat-shock resistance in Legionella pneumophila modeled by experimental evolution. Appl. Environ. Microbiol. 89, e0066623. doi: 10.1128/aem.00666-23
Liao M., Li G., Wang J., Marshall D. J., Hui T. Y., Ma S., et al. (2021). Physiological determinants of biogeography: The importance of metabolic depression to heat tolerance. Global Change Biol. 27, 2561–2579. doi: 10.1111/gcb.15578
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lutterschmidt W. I., Hutchison V. H. (1997). The critical thermal maximum: History and critique. Can. J. Zool. 75, 1561–1574. doi: 10.1139/z97-783
Marshall D. J., Dong Y. W., McQuaid C. D., Williams G. A. (2011). Thermal adaptation in the intertidal snail Echinolittorina malaccana contradicts current theory by revealing the crucial roles of resting metabolism. J. Exp. Biol. 214, 3649–3657. doi: 10.1242/jeb.059899
Marshall D. J., McQuaid C. D. (2020). Metabolic regulation, oxygen limitation and heat tolerance in a subtidal marine gastropod reveal the complexity of predicting climate change vulnerability. Front. Physiol. 11. doi: 10.3389/fphys.2020.01106
Mitra R., Wu K., Lee C., Bardwell J. C. A. (2022). ATP-independent chaperones. Annu. Rev. Biophysics 51, 409–429. doi: 10.1146/annurev-biophys-090121-082906
Mohan K., Ravichandran S., Muralisankar T., Uthayakumar V., Chandirasekar R., Rajeevgandhi C., et al. (2019). Extraction and characterization of chitin from sea snail Conus inscriptus (Reeve 1843). Int. J. Biol. Macromol. 126, 555–560. doi: 10.1016/j.ijbiomac.2018.12.241
Moyen N. E., Somero G. N., Denny M. W. (2019). Impact of heating rate on cardiac thermal tolerance in the California mussel, Mytilus californianus. J. Exp. Biol. 222, jeb203166. doi: 10.1242/jeb.203166
Muggeo V. M. (2008). Segmented: An R package to fit regression models with broken-line relationships. R News 8, 20–25.
Nguyen K. D. T., Morley S. A., Lai C.-H., Clark M. S., Tan K. S., Bates A. E., et al. (2011). Upper temperature limits of tropical marine ectotherms: Global warming implications. PloS One 6, e29340. doi: 10.1371/journal.pone.0029340
Paskevicius T., Farraj R. A., Michalak M., Agellon L. B. (2023). Calnexin, more than just a molecular chaperone. Cells 12, 403. doi: 10.3390/cells12030403
Patra A. K., Ho P.-T., Jun S., Lee S. J., Kim Y., Won Y.-J. (2023). Genome assembly of the Korean intertidal mud-creeper Batillaria attramentaria. Sci. Data 10, 489. doi: 10.1038/s41597-023-02403-9
Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pinsky M. L., Eikeset A. M., McCauley D. J., Payne J. L., Sunday J. M. (2019). Greater vulnerability to warming of marine versus terrestrial ectotherms. Nature 569, 108–111. doi: 10.1038/s41586-019-1132-4
Pörtner H. (2001). Climate change and temperature-dependent biogeography: Oxygen limitation of thermal tolerance in animals. Naturwissenschaften 88, 137–146. doi: 10.1007/s001140100216
Quinn N. L., McGowan C. R., Cooper G. A., Koop B. F., Davidson W. S. (2011). Ribosomal genes and heat shock proteins as putative markers for chronic, sublethal heat stress in Arctic charr: Applications for aquaculture and wild fish. Physiol. Genomics 43, 1056–1064. doi: 10.1152/physiolgenomics.00090.2011
Ribeiro P. L., Camacho A., Navas C. A. (2012). Considerations for assessing maximum critical temperatures in small ectothermic animals: insights from leaf-cutting ants. PloS One 7, e32083. doi: 10.1371/journal.pone.0032083
Saleem M., Aslam F., Akhtar M. S., Tariq M., Rajoka M. I. (2012). Characterization of a thermostable and alkaline xylanase from Bacillus sp. And its bleaching impact on wheat straw pulp. World J. Microbiol. Biotechnol. 28, 513–522. doi: 10.1007/s11274-011-0842-z
Saravia J., Paschke K., Oyarzún-Salazar R., Cheng C.-H. C., Navarro J. M., Vargas-Chacoff L. (2021). Effects of warming rates on physiological and molecular components of response to CTMax heat stress in the Antarctic fish Harpagifer antarcticus. J. Thermal Biol. 99, 103021. doi: 10.1016/j.jtherbio.2021.103021
Sokolova I. M., Frederich M., Bagwe R., Lannig G., Sukhotin A. A. (2012). Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar. Environ. Res. 79, 1–15. doi: 10.1016/j.marenvres.2012.04.003
Somero G. N. (2002). Thermal physiology and vertical zonation of intertidal animals: Optima, limits, and costs of living. Integr. Comp. Biol. 42, 780–789. doi: 10.1093/icb/42.4.780
Somero G. N. (2020). The cellular stress response and temperature: Function, regulation, and evolution. J. Exp. Zool. Part A: Ecol. Integr. Physiol. 333, 379–397. doi: 10.1002/jez.2344
Somero G. N., Lockwood B. L., Tomanek L. (2017). Biochemical adaptation: Response to environmental challenges from life’s origins to the anthropocene. 1st ed (Massachusetts, USA: Sinauer Associates).
Sørensen J. G., Loeschcke V., Kristensen T. N. (2013). Cellular damage as induced by high temperature is dependent on rate of temperature change – investigating consequences of ramping rates on molecular and organismal phenotypes in Drosophila melanogaster. J. Exp. Biol. 216, 809–814. doi: 10.1242/jeb.076356
Sunday J. M., Bates A. E., Kearney M. R., Colwell R. K., Dulvy N. K., Longino J. T., et al. (2014). Thermal-safety margins and the necessity of thermoregulatory behavior across latitude and elevation. Proc. Natl. Acad. Sci. 111, 5610–5615. doi: 10.1073/pnas.1316145111
Terblanche J. S., Deere J. A., Clusella-Trullas S., Janion C., Chown S. L. (2007). Critical thermal limits depend on methodological context. Proc. R. Soc. B: Biol. Sci. 274, 2935–2943. doi: 10.1098/rspb.2007.0985
Vinagre C., Leal I., Mendonça V., Flores A. A. V. (2015). Effect of warming rate on the critical thermal maxima of crabs, shrimp and fish. J. Thermal Biol. 47, 19–25. doi: 10.1016/j.jtherbio.2014.10.012
Wan L., Zhu Y., Zhang W., Mu W. (2020). α-l-Fucosidases and their applications for the production of fucosylated human milk oligosaccharides. Appl. Microbiol. Biotechnol. 104, 5619–5631. doi: 10.1007/s00253-020-10635-7
Wilkinson B., Xiao R., Gilbert H. F. (2005). A structural disulfide of yeast protein-disulfide isomerase destabilizes the active site disulfide of the N-terminal thioredoxin domain. J. Biol. Chem. 280, 11483–11487. doi: 10.1074/jbc.M414203200
Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141. doi: 10.1016/j.xinn.2021.100141
Yuan J., Zhang X., Zhang X., Sun Y., Liu C., Li S., et al. (2023). An ancient whole-genome duplication in barnacles contributes to their diversification and intertidal sessile life adaptation. J. Advanced Res. doi: 10.1016/j.jare.2023.09.015
Keywords: heart rate, heat shock proteins, misfolded proteins, carbohydrate metabolism, heat stress
Citation: Du L-N, Du Y-H and Han G-D (2024) Effects of heating rate on upper thermal limit: insights from cardiac performance and transcriptomic response in mudflat snail Batillaria attramentaria. Front. Mar. Sci. 11:1407743. doi: 10.3389/fmars.2024.1407743
Received: 27 March 2024; Accepted: 24 June 2024;
Published: 11 July 2024.
Edited by:
Md Saydur Rahman, The University of Texas Rio Grande Valley, United StatesReviewed by:
Xianliang Meng, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2024 Du, Du and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-Dong Han, aGFuZ2RAeXR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.