- 1National Engineering Research Center for Marine Aquaculture, Zhejiang Ocean University, Zhoushan, Zhejiang, China
- 2Marine Science and Technical College, Zhejiang Ocean University, Zhoushan, Zhejiang, China
- 3Zhoushan Fisheries Research Institute, Zhoushan, Zhejiang, China
Hydroxychloroquine (HCQ) is an important public health therapeutic agent widely used in the prevention and treatment of malaria and autoimmune diseases, with some antiviral effects, as well as a common autophagy inhibitor. Its autophagy-inhibiting effect attracts great research interest in mammals but is still little studied in fish. We even have no idea about the effects of HCQ on different tissues of fish and what concentrations should be used for safety studies. This study investigated the effect of different concentration of HCQ treatments on the survival, tissue structure, and expressions of genes related to autophagy, apoptosis, and inflammation in the gill, spleen, testis, and ovary of Sebastiscus marmoratus. The results showed that the higher HCQ concentration (77.40 and 154.80 mg/mL) led to mass mortality within an hour. The half-lethal concentration (LC50, 24 h) of HCQ for S. marmoratus was approximately 48.95 mg/mL. When exposed to 30.96 mg/mL HCQ for 24 hours, autophagy was blocked as revealed by electron microscopy from gill, spleen and testis of fish. The expressions of autophagy-related genes (LC3/Cx43), apoptosis-related genes (Cas3/p53), and inflammation-related genes (TNF-α/IL8) exhibited tissue-specific and dose-dependent responses. The gonads showed preferential expression of all these genes and were found to be sensitive and regular after HCQ treatment. For example, at a concentration of 30.96 mg/mL, the testis demonstrated a regularity that suggests it is an ideal candidate tissue for studying the role of HCQ or autophagy. This study systematically revealed the response of S. marmoratus to different concentrations of HCQ and provided optional assay concentrations for key tissues, serving as an important reference for the future studies on HCQ and autophagy in S. marmoratus. Furthermore, the potential crosstalk between autophagy, apoptosis, and inflammatory pathways initially identified in this study could be helpful for the future research on autophagy regulation in marine fish.
1 Introduction
Chloroquine (CQ), for the first time synthesized in 1939, is a kind of anti-malarial drug (Tanenbaum and Tuffanelli, 1980). It interferes with the acid vesicle functions in the parasite by affecting endocytosis and proteolysis of hemoglobin and intracellular targeting of lysosomal enzymes (Krogstad and Schlesinger, 1987). However, due to its biological toxicity, CQ was soon replaced by its derivative, hydroxychloroquine (HCQ) for the treatment and prevention of malaria, which was first synthesized in 1944 (Patil et al., 2020; Purwati et al., 2021). HCQ is an antimalarial drug derived from 4-aminoquinoline. It has a hydroxyl group at the end of its side chain and a β-hydroxylated N-ethyl substituent. As a result, CQ is fat soluble, while HCQ is water soluble. Meanwhile, the effect and mechanism of HCQ remain similar to that of CQ, its toxicity is reduced by half (McChesney and Fitch, 1984). This makes HCQ easier to absorb in the gastrointestinal tract, and reduces its toxicity in the liver, eye, and gastrointestinal tissue (Ren et al., 2020), which also makes it an essential drug in the basic public health system in the standard list of Essential Medicines of the World Health Organization. However, data on the fate and behavior of HCQ in the environment are scarce. A monitoring study detected of HCQ in the surface sediment from tidal sections of a river in southeast China (Chen et al., 2013). Due to its good solubility and low biodegradation, derivatives of quinoline have become one of the common contaminants in groundwater and soil (Gosu et al., 2016; Mendonça-Gomes et al., 2021). Similar to other medicine consumed by humans, HCQ eventually reaches marine coastal environments through the sewage system and is subject to sudden increases (Dabić et al., 2019). This sudden increase raises serious questions, inquiries or concerns related to short- and long-term ecotoxicological effects on aquatic organisms and the environment.
In fact, the functions of CQ and its derivatives go beyond the treatment of malaria. They also have anti-inflammatory, immunity-regulating, anti-infective, light-filtering, and anti-coagulation properties. In 1989, researchers found a decreased incidence of Burkitt lymphoma in patients treated with CQ prophylactically for malaria (Geser et al., 1989; Belhassan et al., 2020). CQ and HCQ have also been proven to be effective in treating other inflammatory diseases such as rheumatoid arthritis (Montalvo-Casimiro et al., 2020; Nirk et al., 2020). In the past few decades, CQ and HCQ have been commonly used as autophagy inhibitors both in the lab and clinical settings. They block lysosomal acidification, autophagosome degradation, and inactivate autophagy, which further reduces the survival of tumor cells (Rubinsztein et al., 2007; Zhou and Wang, 2013; Li et al., 2020a; Martinez et al., 2020). In breast cancer cells, HCQ regulates the expression of apoptosis suppressor genes (Xlap, Livin, and Survivin), pro-apoptosis genes (Pten, Tceal17, Rassf2a, Arhi, Bax, and Caspase-3) and autophagy marker genes (Beclin1, LC3-II, Atg2b, Atg4d, and Atg9b), leading to apoptosis and autophagy (Dong et al., 2021). In human bladder cancer cells, it was found that both CQ and HCQ inhibit the proliferation of several bladder cell lines, increase caspase 3/7 activity, and cause apoptosis of the bladder cancer cells (Lin et al., 2017). In addition, researchers also found that CQ induced apoptosis in pancreatic neuroendocrine neoplasms via endoplasmic reticulum stress (Nakano et al., 2020). Not surprisingly, these CQ- or HCQ-induced changes in autophagy levels can cause a certain response in the apoptotic pathway (Kapuy and Korcsmáros, 2022).
As a lysosomal-dependent degradation pathway widely present in eukaryotic cells, autophagy is a highly conserved intracellular mechanism that plays a crucial role in self-protection, cell survival, regeneration, substance reuse, and homeostasis (Yorimitsu and Klionsky, 2005; Levine and Kroemer, 2008; Chiarelli et al., 2014). Autophagy was first discovered in the early 1960s (Klionsky, 2008), and since then, extensive research has been conducted on this process in various organisms, ranging from yeast to higher animals and plants (Moore et al., 2006; Ohsumi, 2014). The identification of autophagy genes in yeast and homologs in other organisms has revealed the conservation of the eukaryotic autophagy machinery, enabling researchers to use molecular genetics and biology to study the process in different model systems (Yorimitsu and Klionsky, 2005).
Up to now, most of the autophagy study were carried out in mammals. Studies inprove that autophagy plays an essential role in embryonic development (Mizushima and Komatsu, 2011), tissue renewal, and cell differentiation (Wu et al., 2020; Wada et al., 2014). Studies on mice have demonstrated that autophagy helps to remove inefficient organelles in cells during heart and liver development, thereby promoting cell differentiation (Gan et al., 2006; Dai et al., 2022). During times of stress or damage to the nervous system, autophagy activity is often boosted to help nerve cells cope with these unfavorable factors (Fimia et al., 2007). Under conditions of external stimuli threaten life, the level of autophagy also changes correspondingly (Ge et al., 2019; Yao et al., 2019). Starvation will cause autophagy and then cause body damage in mice (Komatsu et al., 2005). Moreover, under low oxygen, low energy, or other stress, autophagy is automatically activated to help the cell survive (Klionsky, 2005). Autophagy is crucial for the formation of the heart structure during zebrafish development, and inactivation of autophagy, specifically ATG5 and ATG7, can lead to increased cell death, structural heart defects, and decreased embryonic survival (Lee et al., 2014). In a study of zebrafish liver found that, hepatic glycogen was first mobilizing to supply energy, and then autophagy, especially mitophagy, played vital roles during nutrient deprivation (Chen et al., 2022; Dai et al., 2022). It is evident that autophagy plays an integral role throughout the process from embryonic development to the maintenance of homeostasis in the organism until the death of the individual (Tettamanti et al., 2019).
However, studies on autophagy in aquatic organisms are much fewer, which was still in its infancy (Seiliez et al., 2010; Xia et al., 2019). In a study of ovarian follicle atresia in Prochilodus argenteus and Leporinus taeniatus, researchers found that the intense heterophagy to engulf the yolk, and autophagy were detected in the follicular cells during advanced and late atresia (Santos et al., 2008). Autophagy and phagocytosis of European hake (Merluccius merluccius) follicular cells were similarly found to contribute to the digestion of oocyte yolk material during atresia (Nzioka et al., 2023). In Oncorhynchus mykiss, in vivo and in vitro studies showed that fasting fish for 14 days or serum depletion of trout myocytes strongly induces the expression of autophagy-related genes (LC3B, gabarapl1, atg12l, and atg4b) in the muscle (Seiliez et al., 2010). In vitro study on trout myocytes indicated that IGF1 induces FoxO3 phosphorylation but has a low or no effect on autophagy-related gene expression, suggesting a moderate role for this transcription factor on the autophagic/lysosomal pathway (Seiliez et al., 2010). In Seriola quinqueradiata cells, HSC70 and HSP70 were found to localize in lysosomes after high-temperature treatment, suggesting that molecular chaperon-mediated autophagy occurred in cells under heat shock conditions, and both heat shock proteins contribute to autophagy (Yabu et al., 2011). In Micropterus salmoides epithelioma papulosum cyprini cells, autophagy induced by the largemouth bass virus was found to inhibit virus replication and apoptosis (Deng et al., 2022). A study of Pelteobagrus fulvidraco found that autophagy could enable adaptive catabolism of cells under waterborne Zn exposure to reduce the stress response of organelles (such as endoplasmic reticulum) and maintain cell homeostasis (Wei et al., 2017). Another study of P. fulvidraco sperm found that CQ could improve sperm motility and fertilization efficiency by activating many signaling pathways including PI3K-AKT (Zhang et al., 2017). In the Epithelioma Papulosum Cyprinid cells, the researchers found that autophagy delayed apoptotic cell death caused by Siniperca chuatsi Rhabdo virus ( (Zhou et al., 2021). In Penaeus vannamei, TBC domain family 7-like has been found to enhance the tolerance to ammonia nitrogen by the up-regulation of autophagy (Wang et al., 2022). A study of Larimichthys crocea also found that the activation of autophagy relieved linoleic acid-Induced inflammation (Shi et al., 2007; Dai et al., 2022). In vitro and in vivo of Paralichthys olivaceus studies showed that PoPTEN affected autophagy activation via the AKT/mTOR pathway and also modulated the process of apoptosis (Johnstone and Chaves-Pozo, 2022). These studies have provided important insights into the role of autophagy in fish. However, there is still a lack of systematic knowledge of the potential effects and regulatory mechanisms of autophagy and its inhibitors in fish. Therefore, it is essential to investigate the scope of the autophagy inhibitor HCQ, its possible effects on different fish tissues, and its subsequent effects on other regulatory pathways (Xia et al., 2019).
Sebastiscus marmoratus is a bottom-carnivorous oviparous fish belonging to Scorpaeniformes, Scorpaenoidei, Scorpaenidae, and Sebastiscus. Its body is long elliptic, with big eyes on the upper side and a high eyeball positioned towards the back of the head (He et al., 2023). This species is indigenous to the nearshore seawaters of the West Pacific Ocean, including the Bohai Sea, Yellow Sea, East China Sea, and South China Sea, as well as the waters from southern Japan to eastern Korea (Shi et al., 2007; Bo et al., 2017), which is a typical reef sea fish. Owning to its small body size (approximately 25 g body weight for an adult), wide geographical distribution, restricted home range, high disease resistance, and uniform growth, S. marmoratus became a potential marker marine species for environmental studies (Bo et al., 2017). Recent studies have assessed the effects of various oceanic pollutants such as Lead (Pb), Petroleum hydrocarbon (PHC), Nonylphenol (NP), Cadmium (Cd), and their mixture on the reproductive endocrinology of male S. marmoratus (Zheng et al., 2016; Bo et al., 2017; Zheng et al., 2019). However, the effect of CQ and HCQ on S. marmoratus remains unclear.
The purpose of this study is to investigate the effect of different concentrations of HCQ on various tissues of S. marmoratus. We chose the gill, spleen, testis, and ovary to represent the respiratory, immune, and reproductive system. We determined the survival rate, LC50 (24 h), sub-microstructural changes, and the expression patterns of autophagy-related genes LC3 and Connexin 43 (Cx43), apoptosis-related genes Caspase 3 (Cas3) and Tumor protein 53 (p53), and inflammation-related genes Tumor necrosis factor-α (TNF-α) and Interlukin 8 (IL8) in the gill, spleen, testis, and ovary. By comparing their expression changes, peaking time, and elevation multiplier, the responses of autophagy, apoptosis, and inflammation pathways could be initially elucidated. The results would provide information on the optimal target tissues and the corresponding optimal detection windows for HCQ treatment in S. marmoratus. Furthermore, it would give evidence for the potential environmental impact of autophagy inhibitor HCQ, and serve as a basis for further research on autophagy in marine fish.
2 Materials and methods
2.1 Ethics committee permit and experimental procedure compliance
All procedures in this study were approved by the Animal Ethics Committee of Zhejiang Ocean University (Zhoushan, China), and carried out following the Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People’s Republic of China, 14 November 1988). All the experimental operations and procedures are legal and compliant. All experiments were carried out following the reference experimental guidelines and regulations and the US National Research Council’s guidelines for the Use and Care of Laboratory. Animals were followed, from animal transportation to the experiment itself and its completion, and there were no incidents of abuse or other infractions. Ethics Committee approval was obtained from the Institutional Ethics Committee of Zhejiang Ocean University to the commencement of the study. Approval Code: 2022095 and Approval Date: 16 June 2022. Thus, the experiment met ethical requirements.
2.2 Fish stocks
S. marmoratus were obtained from Zhoushan Fisheries Research Institute of Zhejiang Province, China. The weight and length of the fish were 25.13 ± 5.26 g and 10.44 ± 3.06 cm, respectively. The rearing conditions were as follows: salinity 31 ± 0.5, dissolved oxygen > 7.0 mg/L, pH 7.6 - 8.1, and photoperiod 12 h light: 12 h dark. The seawater was renewed every other day. The fish were fed twice a day (8:00 and 18:00) at about 5% of body weight (Ahmed et al., 2004). Residual bait was sucked out in time to prevent negative impact on water quality.
2.3 Experimental design and HCQ treatments
Used glass tanks (40 cm × 20 cm × 28 cm) to keep the fish in the aquarium at 20°C. Each tank contained 16 L of sand-filtrated seawater. The treatments were set according to the pretest and reference documents 1991-GB/T 13267–1991. There were total 72 individuals in 8 treatments, including Blank (blank control: nothing is injected), Ctrl (solvent control: an equal volume of 1×PBS was injected), 3.10 mg/mL, 7.74 mg/mL, 15.48 mg/mL, 30.96 mg/mL, 77.40 mg/mL, and 154.80 mg/mL HCQ groups. For each treatment, there set 3 tanks. Every timepoints, 3 individuals were collected randomly from each tank.
After intraperitoneally injection, deaths were recorded every 0.5 h until 24 h sampling. All of the fish were anesthetized using MS-222 (tricaine methanesulfonate; Sigma, USA), and then measured for total weight, body weight, length, liver weight, and gonad weight to make sure the growth situation and gonad stages were close to each other. The gill, spleen, testis, and ovary were quickly collected at the time point of 24 h or the time of death (<24 h), and frozen in liquid nitrogen rapidly to prevent RNA from degradation. All the samples underwent the same analysis.
2.4 Transmission electron microscope (TEM) assay
According to the mortality results, 30.96 mg/mL was the highest concentration that all S. marmoratus alive throughout the experiment. This concentration was also most closed to the LC50 (48.95 mg/mL). Therefore, samples in 30.96 mg/mL HCQ group were also chose for TEM assay to check the histological effects of HCQ to S. marmoratus. Total 9 individuals were collected for the gill, spleen, testis, and ovary. Then, 3 samples of each tissue we randomly sent for TEM analysis. The geometric center of gill, spleen, testis, and ovary were dissected and minced into fragments (< 1 mm) to represent for the developmental situation, and thereafter immersed by 2.5% glutaraldehyde in 1 × phosphate buffered saline (1×PBS) (pH 7.3). After fixation over 4 h at 4°C, the tissues were washed with 1×PBS for 3 times (10 min each) and post-fixed by 2% osmium tetroxide (OsO4) in 1×PBS over 4 h at 4°C. Then the tissues were washed repeatedly in distilled water and dehydrated by using a graded ethanol series (30%, 50%, 75%, 95%, 100% and 100%) for 10–20 min each.
For transmission electron microscopy (TEM), the samples were embedded in Epon-Araldite and kept in dark at 4°C. Resin and the sample sections of the testis were processed as previously described. Semi-thin sections (1.5 μm) were obtained by using a Reichert Ultracut E ultramicrotome and stained with methylene blue before the visualization and examination by a Nikon Ni-E light microscope with a Nikon DS-Ri2 imaging system. Ultrathin sections (75–80 nm) were obtained by using the the Ultracut E ultramicrotome and stained with urany-less EM stain, followed by counterstaining with lead citrate. The sections were then photographed by using a transmission electron microscope Hitachi H-7000 5 (Hitachi, Tokyo, Japan). Only histologic changes with a probability of greater than 75% are considered as typical changes.
2.5 Quantitative real-time PCR (qRT-PCR) analysis
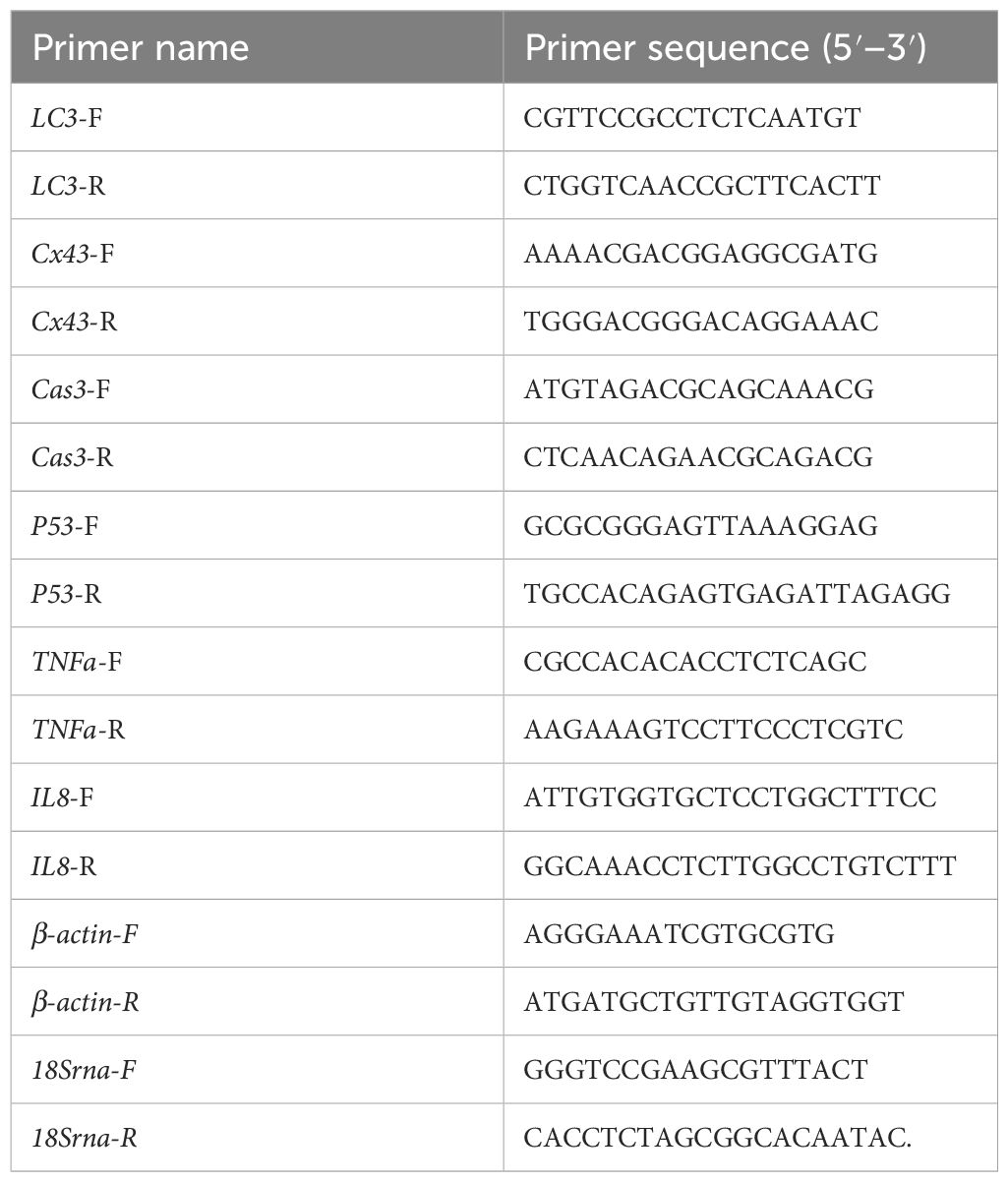
Total RNA was extracted from the gill, spleen, testis and ovary by using the Fastagene RNAfast 200 kit (Fastagen, China) and checked for both quantity and quality thereafter. Then, the cDNA was synthesized by using the PrimeScripe RT reagent kit (Takara, China) following the manufacturer’s instructions. Fluorescent real-time PCR was analyzed by using SYBR Premix real-time PCR kit (Takara, China) with an Bio-RAD@CFX connect system (Biorad, USA). The PCR reactions were performed in triplicate with a 20 μL reaction volume containing 10 μL Top Green qPCR Super Mix, 0.4 μL Passive Reference Dye I, 1 μL forward and reverse primer mix, 1 μL template cDNA, and 7.6 μL H2O.The reaction conditions were 94°C for 5 min, followed by 45 cycles at 94°C for 15 s, 55°C for 15 s, and 72°C for 20 s. The primers were designed using the Primer 5.0 program (Table 1). The reference genes β-actin and 18sRNA were used as the internal control (Yin and Qian, 2017; Zhang et al., 2009).
2.6 Statistical analysis
All data from this experiment are expressed as Mean ± SD (n = 9). Data from qRT-PCR were analyzed using the ΔΔCt method, and the 2 -ΔΔCt value was used as the relative expression of the target gene (Livak and Schmittgen, 2001). Statistical analysis was completed using SPSS 21.0 software (SPSS Inc., USA) using one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test for significance (p < 0.05) when comparing data between groups. Gene expression data in heat maps were analyzed after natural logarithmic transformation.
3 Results
3.1 Cumulative survival and mortality rates
The cumulative survival rate of S. marmoratus is shown in Figure 1. There were no deaths in the blank group, control group, 3.10 mg/mL group, 7.74 mg/mL group, 15.48 mg/mL group, and 30.96 mg/mL group within 24 h. When the HCQ concentration increased to 77.40 mg/mL, deaths occurred at 0.5 h (mortality 22.22 ± 11.11%), with the cumulative survival rate decreasing to 77.78%, then all the fish died at 1.0 h. In the 154.80 mg/mL group, a large number of deaths occurred at 0.5 h (88.89 ± 7.40%), and the cumulative survival rate decreased to 11.11%. Using the Karber method (Hamilton et al., 1977), the half-lethal concentration (LC50) of HCQ to S. marmoratus for 24 h was approximately 48.95 mg/mL, which was consistent with the probit method (56.76 mg/mL) and linear regression method (44.86 mg/mL) (Matsubara, 1962; Nath et al., 1996).
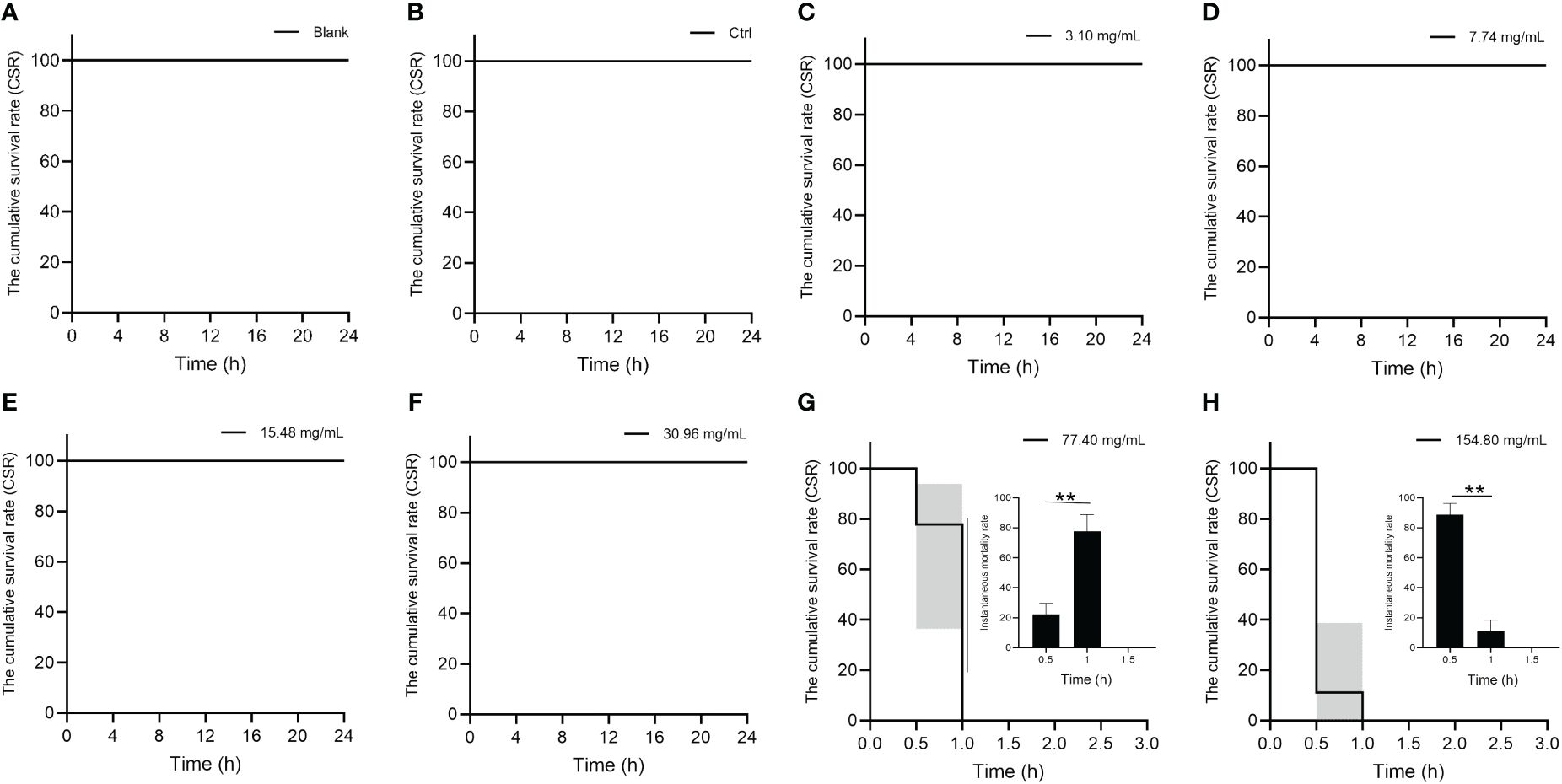
Figure 1 Cumulative survival rate (CSR) and instantaneous mortality of S. marmoratus under different concentration HCQ treatments. (A) Blank, no injection. (B) Control, injected 100 μL of 1 × PBS solution into the abdominal cavity per fish. (C) The 3.10 μg/mL HCQ group, injected 100 μL of 3.10 μg/mL HCQ solution prepared with 1 ×PBS solution into the abdominal cavity per fish. (D) The 7.74 μg/mL HCQ group. (E) The 15.48 μg/mL HCQ group. (F) The 30.96 μg/mL HCQ group. (G) The 77.40 μg/mL HCQ group. (H) The 150.80 μg/mL HCQ group. The values are means ± SD (n = 9). “**” denote significant differences (p < 0.05).
3.2 Electron microscope analysis
To examine the effects of HCQ on tissue structure, we checked the structural changes of gill, spleen, testis and ovary at 24 h in the 30.96 mg/mL group by transmission electron microscopy (TEM). The concentration of this group was the highest at which all the fish survived for 24 h. As shown in Figure 2, in the control group, the gill filament structure was intact. However, in the 30.96 mg/mL group, a mass of vacuolation, autophagy and apoptotic bodies could be found. In the spleen, the control group showed no special changes. In the 30.96 mg/mL group, mitochondrial autophagy occurred, with partial vacuolation and deeply stained autophagolysosomes appeared. In addition, the number of apoptotic bodies increased. For the testis in the control group, the normal state of spermatocytes and spermatocytes were concentrated, with the Sertoli cells and other somatic cells located mainly at the margins of the spermatogenic lobule, not far from the germ cells. However, in the 30.96 mg/mL group, vesicles in spermatocytes increased significantly, and the number of autophagosomes and apoptotic bodies also increased in Sertoli cells. The number of autophagosomes and apoptotic bodies around follicular cells and granulosa cells was increased in ovaries treated with 30.96 mg/mL HCQ compared to the control group.
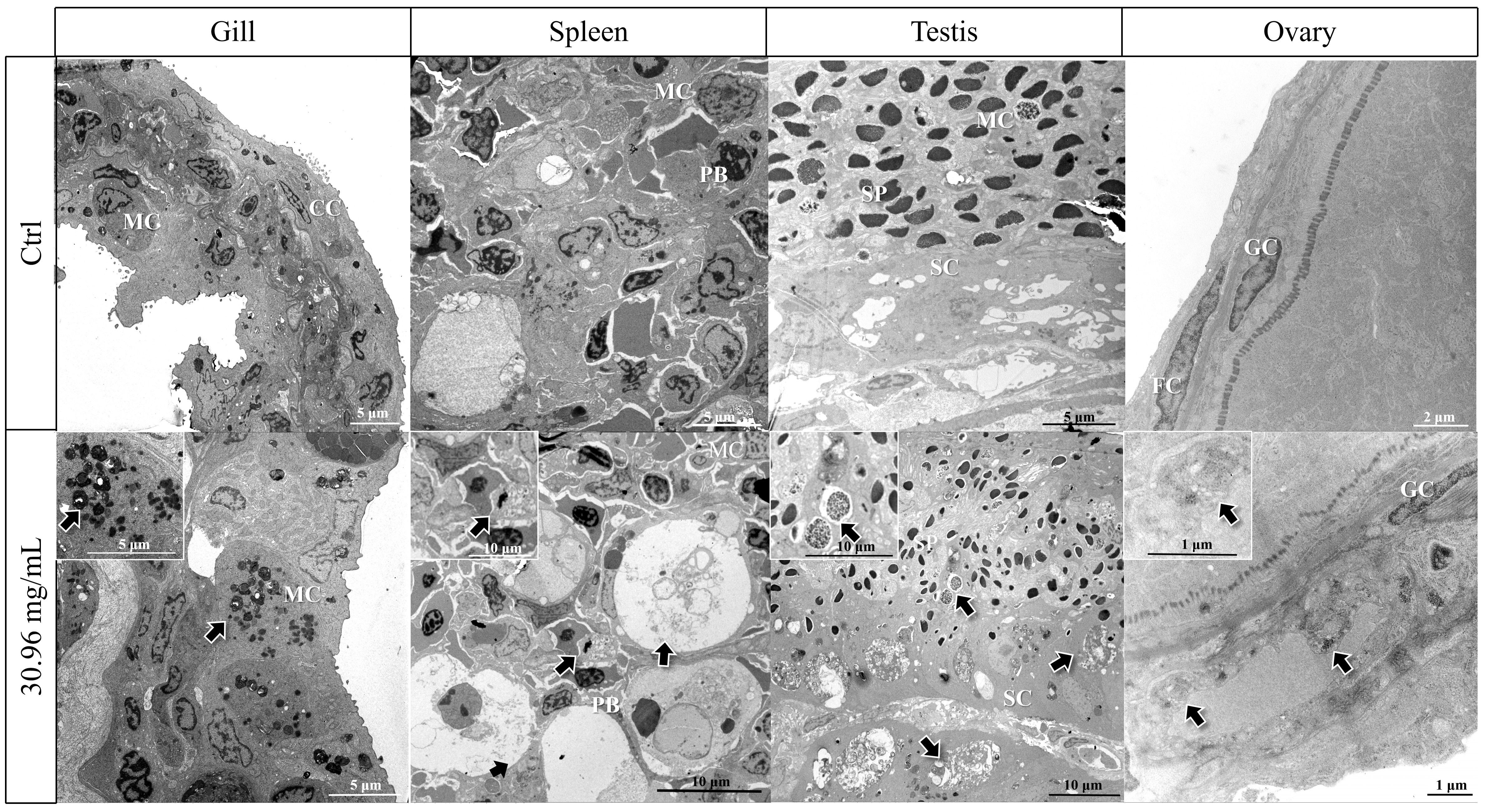
Figure 2 Transmission electron microscope (TEM) results of gill, spleen, testis and ovary of S. marmoratus after 30.96 mg/mL HCQ treatments, (SP, Sperm cell; SC, Sertoli cell; MC, Macrophage cel, CC, Central capillary, FC, Follicle cell; GC, Granulosa cells; PB, Plasmablast; Arrows, Accumulation of autophagosomes or autophagolysosomes).
3.3 Real-time fluorescence quantitative results
To determine whether HCQ treatment induces the autophagic response in S. marmoratus tissues, we analyzed the expression changes of autophagy-related genes LC3/Cx43, as shown in Figure 3. The expressions of LC3 and Cx43 showed similar trends in the gill, testis, and ovary. In the gills, the LC3 levels significantly increased in the 15.48 mg/mL and 77.40 mg/mL groups, but decreased in the 30.96 mg/mL group (p < 0.05). Cx43 also showed relatively high levels in the 15.48 mg/mL and 77.40 mg/mL groups, and low levels in the 30.96 mg/mL group, with the highest expression in the 3.10 mg/mL group. In the spleen, however, LC3 was most highly expressed in the group with the highest HCQ concentration (154.80 mg/mL), whereas Cx43 was more highly expressed in the low concentration group (3.10 mg/mL), and in the medium concentration group (15.48 mg/mL) than in the high concentration group (77.40 mg/mL) (p < 0.05). In testis, LC3 increased with increasing HCQ concentration, reaching maximum expression in the 30.96 mg/mL group and then declining in the higher concentration groups (p < 0.05). The 30.96 mg/mL group showed the greatest increase in LC3 gene expression compared to the control group, with an 8-fold increase. Cx43 likewise rose significantly in the 30.96 mg/mL group, while no significant change in expression was seen in the other groups. In the ovary, the expression of LC3 and Cx43 were much higher in the 3.10 mg/mL and 15.48 mg/mL groups, with Cx43 showing the highest expression in the 77.40 mg/mL group, about 14 times that of the control group (p < 0.05).
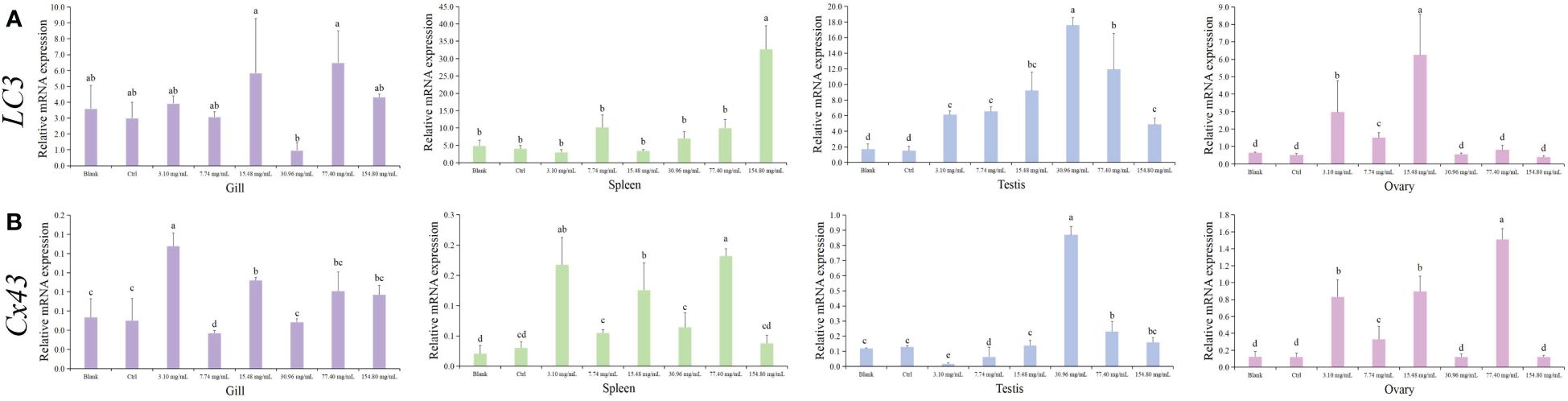
Figure 3 Expression of autophagy genes (LC3/Cx43) in different tissues of S. marmoratus under different concentration HCQ treatments. (A) LC3. (B) Cx43. Tissues are the gill (purple), spleen (green), testis (blue), and ovary (pink). The values are means ± SD (n = 9). Different letters denote significant differences between different groups (p < 0.05).
Next, we assessed the apoptosis response of S. marmoratus after HCQ treatment by measuring the expression changes of apoptosis-related genes Cas3 and p53, as shown in Figure 4. Both genes showed similar patterns. In the gills, as the concentration of HCQ increased, Cas3 and p53 first increased and the then decreased, with the highest levels in the 7.74 mg/mL group. In the spleen tissue, Cas3 and p53 showed higher expressions in the high concentration groups, with Cas3 peaking in the 77.40 mg/mL group, and p53 peaking in the 154.80 mg/mL group (p < 0.05). In the testis, both Cas3 and p53 first increased and then decreased, with their top expressions appearing in the 30.96 mg/mL group, and the maximum increase in Cas3 gene was about 40 times that of the control. In the ovary, the highest expression levels of Cas3 and p53 were found in the 77.40 mg/mL group, with p53 increasing about 7 times compared to the control group (p < 0.05).
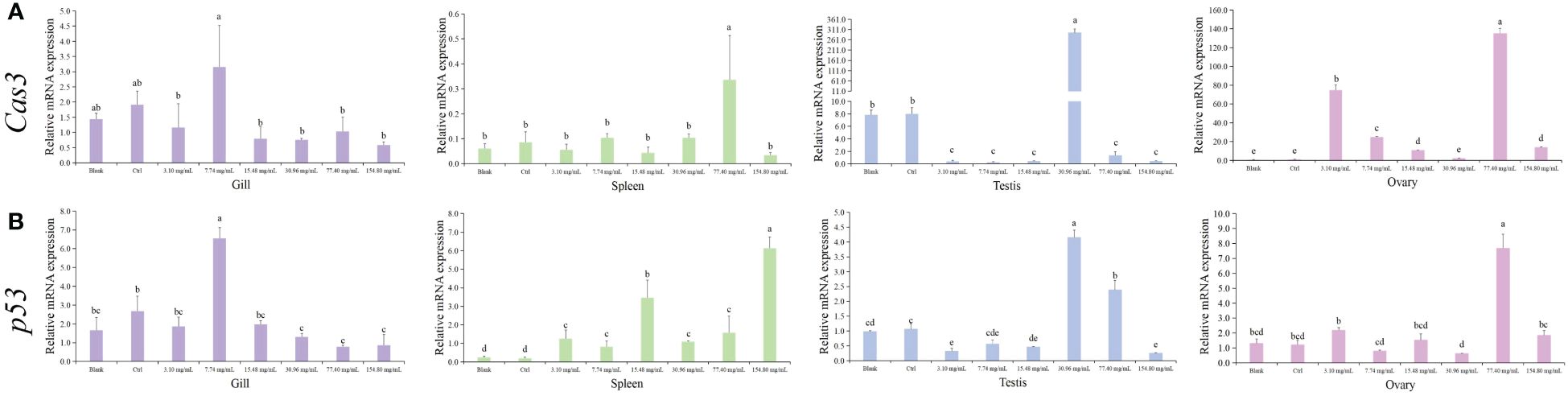
Figure 4 Expression of apoptosis genes (Cas3/p53) in different tissues of S. marmoratus under different concentration HCQ treatments. (A) Cas3. (B) p53. Tissues are the gill (purple), spleen (green), testis (blue), and ovary (pink). The values are means ± SD (n = 9). Different letters denote significant differences between different groups (p < 0.05).
The HCQ treatment also caused inflammatory reactions in S. marmoratus. As shown in Figure 5, the expressions of inflammatory-related genes TNFa and IL8 were measured. The trends of variation of these genes were similar in the spleen, testis, and ovary, respectively. In the spleen, TNFa and IL8 had higher expressions in the low HCQ concentration groups, with the highest expression of TNFa found in the 3.10 mg/mL group, and the top IL8 level appearing in the 7.74 mg/mL group (p < 0.05). In the ovary, both TNFa and IL8 were highly expressed in the 3.10 mg/mL group. In the testis, both TNFa and IL8 increased first and then decreased, with the peak expression in the mid-concentration group, the 30.96 mg/mL group (p < 0.05). However, in the gill, the changes in the two genes showed less significant changes between the different concentration groups, with no statistical difference in IL8, and only a slight decrease in TNFa in the 3.10 mg/mL group and a slight increase in the 154.80 mg/mL group (p < 0.05). Moreover, compared to the control group, the expression levels of TNFa and IL8 genes in ovary tissues elevated most dramatically in the 3.10 mg/mL group, which increased about 80 times and 8 times, respectively (p < 0.05).
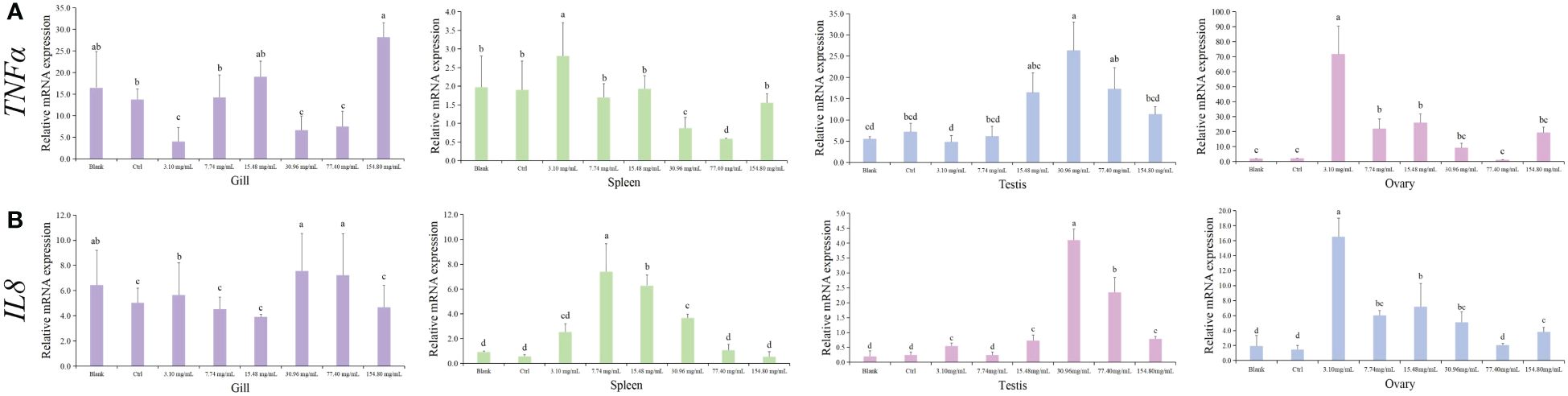
Figure 5 Expression of inflammatory genes (TNFα/IL8) in different tissues of S. marmoratus under different concentration HCQ treatments. (A) TNFα. (B) IL8. Tissues are the gill (purple), spleen (green), testis (blue), and ovary (pink). The values are means ± SD (n = 9). Different letters denote significant differences between different groups (p < 0.05).
3.4 Heat map of folds changes
To visualize the strength of gene response after different HCQ concentration treatments, we created heat maps of the change folds of gene expressions compared to their respective controls (Figure 6). The gonad showed the most significant elevations of gene expression. In the testis, the most significant elevation of gene expressions all appeared in the 30.96 mg/mL group. The maximum gene expressions were 9.39, 8.33, 36.62, 3.24, 3.77, and 4.22 times higher than their respective control. In the ovary, relatively higher elevations of the genes mainly appeared in the lower (3.10, 7.74, and 15.48 mg/mL) and higher (77.40 mg/mL) HCQ concentrations. In the spleen, the genes increased more significantly in the 7.74 and 77.40 mg/mL HCQ groups. However, the genes tested in the gill did not show a clear preference for a particular HCQ concentration.
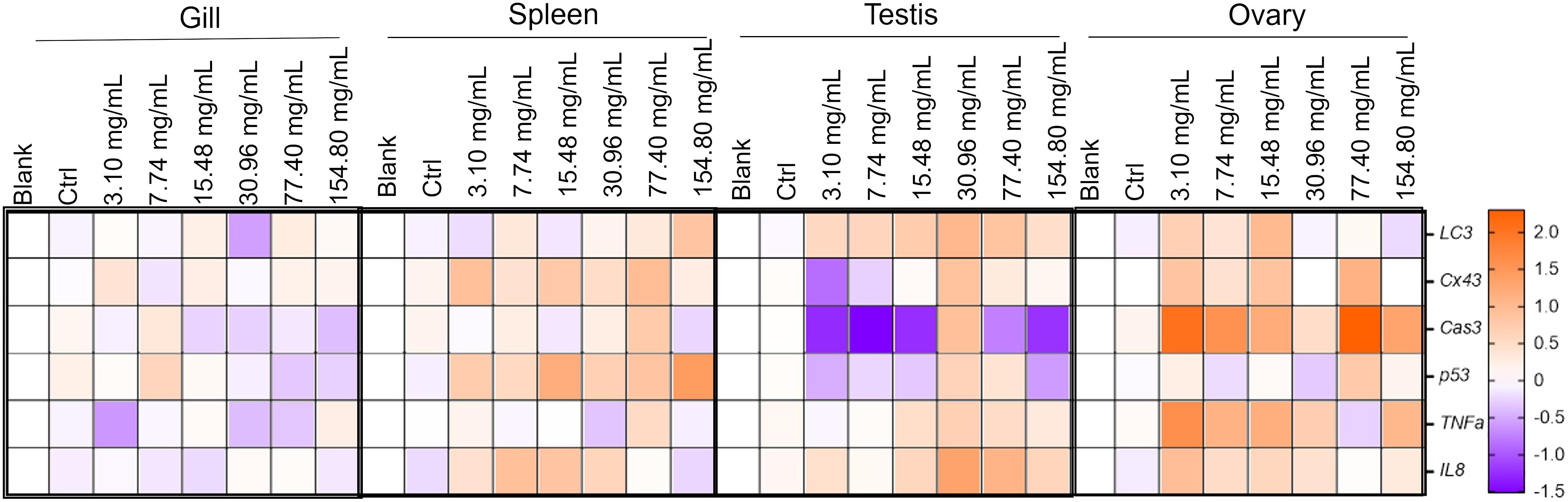
Figure 6 Heatmap of the expression elevated folds of LC3, Cx43, p53, Cas3, TNFα, and IL8 in different tissues of S. marmoratus under different concentration HCQ treatments. Brown color indicates that the expression of the gene is elevated at approximately higher multiplicity, while purple color indicates the opposite.
4 Discussion
Current research methods for HCQ administration in aquatic organisms include water baths, intraperitoneal injections, and altered rations. The intraperitoneal route of exposure increases the ability of the organisms tested to absorb toxic substances, affecting the gonads and other organs (Turner et al., 2011). Substances enter the organism directly and environmental and physiological variables have less influence on the results (Karami et al., 2011). Studies have used intraperitoneal injections of Pelteobagrus fulvidraco with different concentrations of CQ to study autophagy inhibition of its survival and physiological functions (Li et al., 2024). In a previous study, it was shown that intraperitoneal injection of different concentrations of CQ inhibited the mRNA levels of IL-8, NTF-α and apoptosis-related genes in Larimichthys crocea (Zhao et al., 2023). In the present study, we chose the intraperitoneal injections method to reduce the huge consumption of HCQ and environmental pressure caused by water baths. The changes in the cumulative survival rate of S. marmoratus were analyzed in response to increasing concentrations of HCQ. The results showed that the cumulative survival rate remained unchanged until the concentration reached 77.40 and 154.80 mg/mL. Obviously, high concentration of HCQ caused mass death within the first 1 hour. Similar phenomena have been observed in zebrafish (Huang, 2020) and Larimichthys crocea (Zhao et al., 2023), where higher concentration of HCQ (2.50 mg/mL) also caused mass death of zebrafish in the first 1 hour (Huang, 2020). In Larimichthys crocea, mass death occurred in the first 1–2 hours in higher HCQ concentration groups (50.32 and 251.60 μg/mL). It could be seen that in fish, the effect of HCQ was dose-dependent and the negative effect response caused by the high concentration acted rapidly, usually within 1–2 hours (Zhao et al., 2023). In fact, in mammals, HCQ also acted quickly, as observed in a Cynomolgus Macaques population pharmacokinetics model for HCQ, where physiological and biochemical indicators such as plasma concentration respond rapidly within 1 hour after treatment (Liu et al., 2021). Furthermore, HCQ itself has certain biological toxicity (Warhurst et al., 2003; Motarjemizadeh et al., 2015; Li et al., 2020b), which might contribute to the high mortality rate in the high concentration groups.
To investigate the effects of HCQ on tissue structure, we selected the highest concentration group (30.96mg/mL) from the survival groups and examined the gill, spleen, testis, and ovary after 24 h of treatment. Gills, as metabolically active tissues involved in gas exchange, are a major target of aquatic pollutants and are widely used to study the effects of environmental factors on fish. Numerous studies have shown that influencing the level of autophagy in gill tissues can have profound effects on their structure and function (Luzio et al., 2021). The spleen is physiologically important not only as the main site of specific immunity in the organism, but also for the synthesis and secretion of some important active substances, such as complement components and cytokines. Studies have shown that HCQ reduces the number and size of splenic white pulp follicles, the amount of apoptotic activity, and the number of T cell-rich cells (Liu et al., 2021; Alruwaili et al., 2023). HCQ-treated mice showed degeneration of spermatogonia, shedding of spermatogenic cells, testicular interstitial edema, and occasional spermatogenic arrest in testicular tissues (Alruwaili et al., 2023). And it has been noted that HCQ also inhibits autophagy in mouse ovaries (Furlong et al., 2016). Zhao et al. found that different concentrations of CQ had important effects on tissues such as the gonads of the large yellow croaker (Larimichthys crocea) (Zhao et al., 2023).
Our results showed a significant increase in the number of autophagosomes in the test tissues. Similar results have been reported in melanoma cell lines, Ara-C-resistant cell lines and cervical cancer SiHa cells (Mehnert et al., 2013). Specifically, in melanoma cell lines and animal models, HCQ administration inhibited the autophagy pathway as the increasing numbers of autophagic vesicles (AVs) and processing of the autophagosome marker LC3 from LC3-I to LC3-II (Mehnert et al., 2013). In Ara-C-resistant cell lines, HCQ triggers the activation of autophagy by increasing the number of autophagosomes (Kim et al., 2014). In cervical cancer SiHa cells, HCQ inhibited the autophagy by lysosomal acidification and subsequently blocking of the autophagosomes fusion, leading to the tumor cell death (Liu et al., 2014). Moreover, in studies of CQ and HCQ as antitumor agents, they have also been found to cause the accumulation of acidic vesicular organelles and interrupt autophagosome-lysosome fusion (Ferreira et al., 2021). In the present study, the amount of autophagosomes (or autophagolysosomes) and apoptotic bodies was significantly increased after 30.96 mg/mL HCQ treatment, indicating that HCQ strongly interfered with autophagy system in typical organizations of S. marmoratus, and might triggered subsequent apoptotic response. This destruction of organizations also led to impaired normal function. This is the first time that HCQ-induced autophagy blockade has been captured by TEM in fish, providing an intuitive sense of the effect of this autophagy inhibitor.
To further investigate the effects of HCQ on the tissues, we analyzed the gene expressions related to autophagy (LC3, Cx43), apoptosis (Cas3, p53), and inflammatory response (TNFa, IL8). Previous studies have shown that CQ can effectively inhibit protein degradation in autophagic lysozyme and induce the accumulation of LC3II, leading to the inhibition of autophagy in ovarian cancer cells A2780 and SKOV3 (Chen et al., 2022). Similarly, in breast cancer cells, CQ upregulated the expression of Caspase-3, Beclin1, and LC3-II significantly, thereby inducing autophagy (Dong et al., 2021). In bladder cancer cells treated with CQ and HCQ, autophagy was effectively interrupted by blocking the autophagic lysosomal fusion, leading to the inhibition of P62 and LC3-II degradation (Lin et al., 2017). In the present study, the expression patterns of LC3 and Cx43 genes were found to be synchronized in the gill, ovary, and testis of S. marmoratus. We found that HCQ treatment enhanced the expressions of autophagy marker genes (15.48 and 77.40 mg/mL in gills, 30.96 mg/mL in testis, and 3.10 and 15.45 mg/mL in ovary) and induced autophagy, which was also consistent with previous researches. Additionally, Cx43 was found to be directly involved in the initial stage of autophagosome formation in mouse liver cell lines (Bejarano et al., 2014). Our findings provided the first confirmation of a relatively synchronized expression pattern of Cx43 and LC3 in fish. However, further detailed work is required to elucidate the exact relationship between Cx43 and the autophagy and apoptosis pathways.
Numerous studies have confirmed the involvement of Cas3 in various cellular processes such as cell differentiation (Larsen et al., 2010), chemotactic substances production (Hou et al., 2019), wound healing and tissue regeneration (Moeller et al., 2004), neural regeneration and modulation (Contag et al., 2000; Ahn and Brown, 2008). The Cas3/p53 genes are closely linked to apoptosis (Yu et al., 2021). In a study on ventilator-related lung injury, ulinastatin was found to inhibit the Cas-1/3 and p53 pathway and ameliorate the injury caused by apoptosis (Wang and Ye, 2015). In Chinese perch brain (CPB) cells, p53 was shown to play a critical role in regulating autophagy (Zhang et al., 2022). Moreover, in studies on breast cancer cells, CQ was found to regulate the expression levels of apoptosis suppressor genes and pro-apoptotic genes, and induce apoptosis (Dong et al., 2021). Our study found that the expression of Cas3/p53 genes displayed good regularity. With the increase of HCQ concentration, the expression of Cas3/p53 genes first increased and then showed a downward trend. The top expression of Cas3/p53 genes occurred in the 7.74 mg/mL (lower) HCQ group for the gill, 30.96 mg/mL (medium) HCQ group for the testis, and 77.40 mg/mL (medium to high) HCQ group for the ovary. In the spleen, Cas3 peaked in the 77.40 mg/mL (medium to high) HCQ group and p53 peaked in the 154.80 mg/mL (highest) group. Thus, the apoptosis caused by HCQ also showed a strong concentration dependence. Our study supported previous findings in mammalian cell lines, indicating that the crosstalk between autophagy and apoptosis pathways is involved in the effect of HCQ (Toton et al., 2016; Ge et al., 2019; Saciloto et al., 2021). However, further work is needed to clarify the precise relationship between HCQ and apoptosis pathways.
It is well accepted that TNFa and IL8 are crucial pro-inflammatory cytokines that exert inflammatory and immunomodulatory activities essential in host defense (Cheong et al., 2006; Cormier et al., 2009). The defect of autophagy is a critical factor in the induction of inflammatory response and inflammatory disease (He et al., 2013). Therefore, the changes in the expression of TNFa and IL8 can reflect the intensity of the inflammatory response induced by HCQ. Our study found that the levels of TNFa/IL8 genes in the testis and ovary showed good regularities, with the highest expression observed in the 30.96 mg/mL group in the testis and 3.10 mg/mL in the ovary. Similar results were observed in chondrocytes and microglia, where curcumin was found to ameliorate the H2O2-induced chondrocyte inflammatory response by inducing autophagy activation (Chen et al., 2022), and the level of inflammation in microglia was regulated by autophagy (Su et al., 2016). Although the precise relationship between the inflammatory response and the autophagic system in mammals remains unclear, similar results in mammals support the validity of our novel findings in fish.
In fact, previous studies conducted in mammals have also reported that gonads were optimal organs for studying autophagy due to their inherent autophagic activity (Ghosh et al., 2018; Pang et al., 2018; Pajokh et al., 2019). Due to the close relationships between autophagy, apoptosis, and inflammation, it is natural for the gonads to have higher expression levels of marker genes associated with the latter two processes. Additionally, the gonad also showed higher fold-change multiples (refer to Figure 6), with all the tested genes in the testes displaying the highest expressions in the 30.96 mg/mL (medium) HCQ group. This suggestes that the testes are a sensitive and reliable candidate for HCQ detection. Moreover, our study has also provided novel basic data on the ideal HCQ concentration for testing in different tissues of S. marmoratus.
5 Conclusion
In the present study, we examined the effects of HCQ treatment on S. marmoratus by administering varying concentrations of the drug. Our findings revealed significant responses of different levels, including mortality, submicrostructure, and gene expressions. Higher concentrations of HCQ resulted in systemic changes in S. marmoratus, such as high mortality rates (all dead within 1 hour in the 77.40 and 154.80 mg/mL groups), accumulation of autophagosomes (or autophagolysosomes) in typical tissues (represented the blocked autophagy), and significant elevated levels of autophagy, apoptosis, and inflammation marker genes. These responses were rapid and remarkable, and the changes observed at different levels were consistent with each other, e.g. higher mortality corresponded to significant changes in histology and gene expression. Notably, the gonads, especially the testis, showed good sensitivity and consistency in both histology and gene expressions, making it a reliable candidate for HCQ detection. Meanwhile, we also found differences in the sensitivity of various tissues to different HCQ concentrations, which could be a valuable reference for future assays in both S. marmoratus and other marine fish. Additionally, our study identified the crosstalk between autophagy, apoptosis, and inflammatory pathways, with opens up avenues for future in-depth studies on the function of autophagy in marine fish.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Animal Ethics Committee of Zhejiang Ocean University (Zhoushan, China). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YL: Conceptualization, Methodology, Writing – review & editing. JL: Conceptualization, Data curation, Validation, Writing – original draft. CR: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. LC: Conceptualization, Investigation, Methodology, Software, Validation, Writing – review & editing. YZ: Data curation, Formal analysis, Investigation, Supervision, Visualization, Writing – review & editing. XY: Funding acquisition, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review & editing. BL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Zhejiang Provincial Natural Science Foundation of China (No. LY22D060001, LY20C190008), Zhoushan Public Welfare Science and Technology Project (No. 2021C31023).
Acknowledgments
We thank all members of the laboratory for their technical advice and helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed I., Khan M. A., Jafri A. K. (2004). Dietary threonine requirement of fingerling Indian major carp, Cirrhinus mrigala (Hamilton). Aquacult. Res. 35, 162–170. doi: 10.1111/j.1365-2109.2004.00997.x
Ahn G. O., Brown J. M. (2008). Matrix metalloproteinase-9 is required for tumor vasculogenesis but not for angiogenesis: role of bone marrow-derived myelomonocytic cells. Cancer Cell. 13, 193–205. doi: 10.1016/j.ccr.2007.11.032
Alruwaili M., Jarrar B., Jarrar Q., Alruwaili M., Goh K. W., Moshawih S., et al. (2023). Hydroxychloroquine toxicity in the vital organs of the body: In vivo study. Front. bioscience (Landmark edition) 28, 137. doi: 10.31083/j.fbl2807137
Bejarano E., Yuste A., Patel B., Stout R. F. Jr, Spray D. C., Cuervo A. M. (2014). Connexins modulate autophagosome biogenesis. Nat. Cell Biol. 16, 401–414. doi: 10.1038/ncb2934
Belhassan A., En-Nahli F., Zaki H., Lakhlifi T., Bouachrine M. (2020). Assessment of effective imidazole derivatives against SARS-CoV-2 main protease through computational approach. Life Sci. 262, 118469. doi: 10.1016/j.lfs.2020.118469
Bo J., Zheng R., Kuang W., Hong F., Xie Q., Zhang Y. (2017). The use of rockfish Sebastiscus marmoratus as a sentinel species to assess petroleum hydrocarbons pollution: A case study in Quanzhou Bay, China. Mar. pollut. Bull. 124, 984–992. doi: 10.1016/j.marpolbul.2017.01.025
Chen Y. S., Yu S., Hong Y. W., Lin Q. Y., Li H. B. (2013). Pharmaceutical residues in tidal surface sediments of three rivers in southeastern China at detectable and measurable levels. Environ. Sci. pollut. Res. Int. 20, 8391–8403. doi: 10.1007/s11356-013-1871-y
Chen H., Zhao F., Chen K., Guo Y., Liang Y., Zhao H., et al. (2022). Exposure of zebrafish to a cold environment triggered cellular autophagy in zebrafish liver. J. Fish Dis. 45, 991–1000. doi: 10.1111/jfd.13620
Cheong H. S., Shin H. D., Lee S. O., Park B. L., Choi Y. H., Lim G. I., et al. (2006). Polymorphisms in interleukin 8 and its receptors (IL8, IL8RA and IL8RB) and association of common IL8 receptor variants with peripheral blood eosinophil counts. J. Hum. Genet. 51, 781–787. doi: 10.1007/s10038-006-0021-5
Chiarelli R., Agnello M., Bosco L., Roccheri M. C. (2014). Sea urchin embryos exposed to cadmium as an experimental model for studying the relationship between autophagy and apoptosis. Mar. Environ. Res. 93, 47–55. doi: 10.1016/j.marenvres.2013.06.001
Contag C. H., Jenkins D., Contag P. R., Negrin R. S. (2000). Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2, 41–52. doi: 10.1038/sj.neo.7900079
Cormier C., Bossé Y., Mfuna L., Hudson T. J., Desrosiers M. (2009). Polymorphisms in the tumour necrosis factor alpha-induced protein 3 (TNFAIP3) gene are associated with chronic rhinosinusitis. J. Otolaryngol Head Neck Surg. 38, 133–141. doi: 10.2310/7070.2008.OA0220
Dabić D., Babić S., Škorić I. (2019). The role of photodegradation in the environmental fate of hydroxychloroquine. Chemosphere 230, 268–277. doi: 10.1016/j.chemosphere.2019.05.032
Dai T., Lu J., Tao X., Zhang X., Li M., Jin M., et al. (2022). Vitamin D3 alleviates high-fat induced hepatopancreas lipid accumulation and inflammation by activating AMPKkα/PINK1/Parkin-mediated mitophagy in Litopenaeus vannamei. Aquacult. Rep. 25, 101272. doi: 10.1016/j.aqrep.2022.101272
Deng L., Feng Y., OuYang P., Chen D., Huang X., Guo H., et al. (2022). Autophagy induced by largemouth bass virus inhibits virus replication and apoptosis in epithelioma papulosum cyprini cells. Fish Shellfish Immunol. 123, 489–495. doi: 10.1016/j.fsi.2022.03.026
Dong J. L., He Y., Sun Y. J. (2021). Effect of hydroxychloroquine combined with paclitaxel treatment on apoptosis and autophagy in breast cancer cells. Matern. Child. Hlth. Care China. 36, 3821–3824. doi: 10.19829/j.zgfybj.issn.1001-4411.2021.16.052
Ferreira P. M. P., Sousa R. W. R., Ferreira J. R. O., Militão G. C. G., Bezerra D. P. (2021). Chloroquine and hydroxychloroquine in antitumor therapies based on autophagy-related mechanisms. Pharmacol. Res. 168, 105582. doi: 10.1016/j.phrs.2021.105582
Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., et al. (2007). Ambra1 regulates autophagy and development of the nervous system. Nature. 447, 1121–1125. doi: 10.1038/nature05925
Furlong H. C., Wessels J. M., Guerra M. T., Stämpfli M. R., Foster W. G. (2016). Hydroxychloroquine attenuates cigarette smoke induced autophagic signaling in the mouse ovary. Reprod. Toxicol. (Elmsford N.Y.) 61, 105–113. doi: 10.1016/j.reprotox.2016.03.044
Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J. L. (2006). Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 175, 121–133. doi: 10.1083/jcb.200604129
Ge D., Gao J., Han L., Li Y., Liu H. H., Yang W. C., et al. (2019). Novel effects of sphingosylphosphorylcholine on the apoptosis of breast cancer via autophagy/AKT/p38 and JNK signaling. J. Cell Physiol. 234, 11451–11462. doi: 10.1002/jcp.27802
Geser A., Brubaker G., Draper C. C. (1989). Effect of a malaria suppression program on the incidence of African Burkitt's lymphoma. Am. J. Epidemiol. 129, 740–752. doi: 10.1093/oxfordjournals.aje.a115189
Ghosh A. K., Mau T., O'Brien M., Yung R. (2018). Novel role of autophagy-associated Pik3c3 gene in gonadal white adipose tissue browning in aged C57/Bl6 male mice. Aging. 10, 764–774. doi: 10.18632/aging.v10i4
Gosu V., Gurjar B. R., Zhang T. C., Surampalli R. Y. (2016). Oxidative degradation of quinoline using nanoscale zero-valent iron supported by granular activated carbon. J. Environ. 142, 04015047. doi: 10.1061/(ASCE)EE.1943-7870.0000981
Hamilton M. A., Russo R. C., Thurston R. V. (1977). Trimmed spearman-karber method for estimating median lethal concentrations in toxicity bioassays. Environ. Sci. Technol. 11, 714–719. doi: 10.1021/es60130a004
He X. H., He J., OuYang D. Y. (2013). Advances in the study of the interaction between cellular autophagy and inflammatory response. J. Jinan Univ. (Natural Sci. Med. Edition). 02, 125–128. doi: 10.3969/j.issn.1000-9965.2013.02.001
He Z., Shou C., Han Z. (2023). Transcriptome Analysis of Marbled Rockfish Sebastiscus marmoratus under Salinity Stress. Anim. (Basel). 13, 400. doi: 10.3390/ani13030400
Hou X., Yang S., Yin J. (2019). Blocking the REDD1/TXNIP axis ameliorates LPS-induced vascular endothelial cell injury through repressing oxidative stress and apoptosis. Am. J. Physiol. Cell Physiol. 316, C104–C110. doi: 10.1152/ajpcell.00313.2018
Huang Y. K. (2020). Autophagy Response of Zebrafish (Danio rerio) to Different Concentrations of Chloroquine. [master’s thesis]. ([Zhoushan (ZJ)]: Zhejiang Ocean University). doi: 10.27747/d.cnki.gzjhy.2020.000243
Johnstone C., Chaves-Pozo E. (2022). Antigen presentation and autophagy in teleost adaptive immunity. Int. J. Mol. Sci. 23, 4899. doi: 10.3390/ijms23094899
Kapuy O., Korcsmáros T. (2022). Chloroquine and COVID-19-A systems biology model uncovers the drug's detrimental effect on autophagy and explains its failure. PLoS One 17, e0266337. doi: 10.1371/journal.pone.0266337
Karami A., Christianus A., Ishak Z., Syed M. A., Courtenay S. C. (2011). The effects of intramuscular and intraperitoneal injections of benzo[a]pyrene on selected biomarkers in. Clarias gariepinus. Ecotoxicology Environ. Saf. 74, 1558–1566. doi: 10.1016/j.ecoenv.2011.05.012
Kim Y., Eom J. I., Jeung H., Jang J. E., Kim J. S., Cheong J., et al. (2014). Autophagy inhibition overcomes resistance to cytosine arabinoside in acute myeloid leukemia cells through inducing autophagic cell death and intrinsic pathway apoptosis. Blood 124, 909–909. doi: 10.1182/blood.V124.21.909.909
Klionsky D. J. (2005). The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18. doi: 10.1242/jcs.01620
Klionsky D. J. (2008). Autophagy revisited: a conversation with Christian de Duve. Autophagy. 4, 740–743. doi: 10.4161/auto.6398
Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434. doi: 10.1083/jcb.200412022
Krogstad D. J., Schlesinger P. H. (1987). The basis of antimalarial action: non-weak base effects of chloroquine on acid vesicle pH. Am. J. Trop. Med. Hyg. 36, 213–220. doi: 10.4269/ajtmh.1987.36.213
Larsen B. D., Rampalli S., Burns L. E., Brunette S., Dilworth F. J., Megeney L. A. (2010). Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc. Natl. Acad. Sci. U S A. 107, 4230–4235. doi: 10.1073/pnas.0913089107
Lee E., Koo Y., Ng A., Wei Y., Luby-Phelps K., Juraszek A., et al. (2014). Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. 10, 572–587. doi: 10.4161/auto.27649
Levine B., Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell. 132, 27–42. doi: 10.1016/j.cell.2007.12.018
Li Y., Ma Z., Zhang D. H., Ramachandran M., Lin T. (2020b). Abstract 1221: Novel strategies to target autophagy to improve cancer therapy. Cancer Res. 80, 1221. doi: 10.1158/1538-7445.AM2020-1221
Li X., Wang S., Zhang M., Li M. (2024). Enhancement of autophagy can alleviate oxidative stress, inflammation, and apoptosis induced by ammonia stress in yellow catfish Pelteobagrus fulvidraco. Fish shellfish Immunol. 149, 109582. doi: 10.1016/j.fsi.2024.109582
Li J. H., Xu Z. Y., Li M. J., Zheng W. L., Huang X. M., Xiao F., et al. (2020a). LC-MS based metabolomics reveals metabolic pathway disturbance in retinal pigment epithelial cells exposed to hydroxychloroquine. Chem. Biol. Interact. 328, 109212. doi: 10.1016/j.cbi.2020.109212
Lin Y. C., Lin J. F., Wen S. I., Yang S. C., Tsai T. F., Chen H. E., et al. (2017). Chloroquine and hydroxychloroquine inhibit bladder cancer cell growth by targeting basal autophagy and enhancing apoptosis. Kaohsiung J. Med. Sci. 33, 215–223. doi: 10.1016/j.kjms.2017.01.004
Liu Q., Bi G., Chen G., Guo X., Tu S., Tong X., et al. (2021). Time-dependent distribution of hydroxychloroquine in cynomolgus macaques using population pharmacokinetic modeling method. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.602880
Liu Q., Luo X. Y., Jiang H., Yang M. H., Yuan G. H., Tang Z., et al. (2014). Hydroxychloroquine facilitates autophagosome formation but not degradation to suppress the proliferation of cervical cancer SiHa cells. Oncol. Lett. 7, 1057–1062. doi: 10.3892/ol.2014.1879
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Luzio A., Parra S., Costa B., Santos D., Álvaro A. R., Monteiro S. M. (2021). Copper impair autophagy on zebrafish (Danio rerio) gill epithelium. Environ. Toxicol. Pharmacol. 86, 103674. doi: 10.1016/j.etap.2021.103674
Martinez G. P., Zabaleta M. E., Di Giulio C., Charris J. E., Mijares M. R. (2020). The role of chloroquine and hydroxychloroquine in immune regulation and diseases. Curr. Pharm. Des. 26, 4467–4485. doi: 10.2174/1381612826666200707132920
Matsubara H. (1962). On the synergisnic effect of various synergists on dimethrin. Sci. Insect Control. 27, 17–22.
McChesney E. W., Fitch C. D. (1984). “4-Aminoquinolines,” Antimalarial Drug II. ed Peters W., Richards W. H. G. (Berlin, Heidelberg: Springer Press), 68, 3–60. doi: 10.1007/978-3-642-69254-3_1
Mehnert J. M., Xie X., Rajpal M., Dudek L., Mehta M., Karantza V., et al. (2013). Abstract 3513: Hydroxychoroquine (HCQ) modulates autophagy in melanoma: preliminary results of a phase 0 trial in patients with resectable melanoma. Cancer Res. 73, 3513. doi: 10.1158/1538-7445.AM2013-3513
Mendonça-Gomes J. M., da Costa Araújo A. P., da Luz T. M., Charlie-Silva I., Braz H. L. B., Jorge R. J. B., et al. (2021). Environmental impacts of COVID-19 treatment: Toxicological evaluation of azithromycin and hydroxychloroquine in adult zebrafish. Sci. Total Environ. 790, 148129. doi: 10.1016/j.scitotenv.2021.148129
Mizushima N., Komatsu M. (2011). Autophagy: renovation of cells and tissues. Cell. 147, 728–741. doi: 10.1016/j.cell.2011.10.026
Moeller B. J., Cao Y., Li C. Y., Dewhirst M. W. (2004). Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 5, 429–441. doi: 10.1016/S1535-6108(04)00115-1
Montalvo-Casimiro M., González-Barrios R., Meraz-Rodriguez M. A., Juárez-González V. T., Arriaga-Canon C., Herrera L. A. (2020). Epidrug repurposing: discovering new faces of old acquaintances in cancer therapy. Front. Oncol. 10. doi: 10.3389/fonc.2020.605386
Moore M. N., Allen J. I., Somerfield P. J. (2006). Autophagy: role in surviving environmental stress. Mar. Environ. Res. 62, S420–S425. doi: 10.1016/j.marenvres.2006.04.055
Motarjemizadeh Q., Aidenloo N. S., Abbaszadeh M. (2015). Detection of hydroxychloroquine retinal toxicity by automated perimetry in 60 rheumatoid arthritis patients with normal fundoscopic findings. Glob J. Health Sci. 8, 59–64. doi: 10.5539/gjhs.v8n3p59
Nakano K., Masui T., Yogo A., Uchida Y., Sato A., Kasai Y., et al. (2020). Chloroquine induces apoptosis in pancreatic neuroendocrine neoplasms via endoplasmic reticulum stress. Endocr. Relat. Cancer. 27, 431–439. doi: 10.1530/ERC-20-0028
Nath K. A., Croatt A. J., Likely S., Behrens T. W., Warden D. (1996). Renal oxidant injury and oxidant response induced by mercury. Kidney Int. 50, 1032–1043. doi: 10.1038/ki.1996.406
Nirk E. L., Reggiori F., Mauthe M. (2020). Hydroxychloroquine in rheumatic autoimmune disorders and beyond. EMBO Mol. Med. 12, e12476. doi: 10.15252/emmm.202012476
Nzioka A., Valencia A., Atxaerandio-Landa A., Diaz de Cerio O., Hossain M. A., Korta M., et al. (2023). Apoptosis and autophagy-related gene transcription during ovarian follicular atresia in European hake (Merluccius merluccius). Mar. Environ. Res. 183, 105846. doi: 10.1016/j.marenvres.2022.105846
Ohsumi Y. (2014). Historical landmarks of autophagy research. Cell Res. 24, 9–23. doi: 10.1038/cr.2013.169
Pajokh M., Talaei-Khozani T., Bordbar H., Mesbah F. (2019). Apoptosis, autophagy, and necrosis in murine embryonic gonadal ridges and neonatal ovaries: an animal model. Iran J. Med. Sci. 44, 35–43. doi: 10.30476/ijms.2019.40622
Pang J., Li F., Feng X., Yang H., Han L., Fan Y., et al. (2018). Influences of different dietary energy level on sheep testicular development associated with AMPK/ULK1/autophagy pathway. Theriogenology. 108, 362–370. doi: 10.1016/j.theriogenology.2017.12.017
Patil V. M., Singhal S., Masand N. (2020). A systematic review on use of aminoquinolines for the therapeutic management of COVID-19: Efficacy, safety and clinical trials. Life Sci. 254, 117775. doi: 10.1016/j.lfs.2020.117775
Purwati, Miatmoko A., Nasronudin, Hendrianto E., Karsari D., Dinaryanti A., et al. (2021). An in vitro study of dual drug combinations of anti-viral agents, antibiotics, and/or hydroxychloroquine against the SARS-CoV-2 virus isolated from hospitalized patients in Surabaya, Indonesia. PLoS One 16, e0252302. doi: 10.1371/journal.pone.0252302
Ren L., Xu W., Overton J. L., Yu S., Chiamvimonvat N., Thai P. N. (2020). Assessment of hydroxychloroquine and chloroquine safety profiles: a systematic review and meta-analysis. MedRxiv. May 8. doi: 10.1101/2020.05.02.20088872
Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007). Potential therapeutic applications of autophagy. Nat. Rev. Drug Discovery 6, 304–312. doi: 10.1038/nrd2272
Saciloto B., Nicoletti N. F., Peletti-Figueiró M., Falavigna A. (2021). crosstalk between autophagy and apoptosis in intervertebral disc degeneration. J. Biosci. Medicines. 09, 15–29. doi: 10.4236/jbm.2021.912003
Santos H. B., Thomé R. G., Arantes F. P., Sato Y., Bazzoli N., Rizzo E. (2008). Ovarian follicular atresia is mediated by heterophagy, autophagy, and apoptosis in Prochilodus argenteus and Leporinus taeniatus (Teleostei: Characiformes). Theriogenology. 70, 1449–1460. doi: 10.1016/j.theriogenology.2008.06.091
Seiliez I., Gutierrez J., Salmerón C., Skiba-Cassy S., Chauvin C., Dias K., et al. (2010). An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 157, 258–266. doi: 10.1016/j.cbpb.2010.06.011
Shi G., Wang J. X., Liu X. Z., Wang R. X. (2007). Study on histology and histochemistry of digestive tract in Sebastiscus marmoratus. J. Fisheries China. 31, 293–302. doi: 10.3724/SP.J.00001
Su P., Zhang J., Wang D., Zhao F., Cao Z., Aschner M., et al. (2016). The role of autophagy in modulation of neuroinflammation in microglia. Neuroscience. 319, 155–167. doi: 10.1016/j.neuroscience.2016.01.035
Tanenbaum L., Tuffanelli D. L. (1980). Antimalarial agents. Chloroquine, hydroxychloroquine, and quinacrine. Arch. Dermatol. 116, 587–591. doi: 10.1001/archderm.116.5.587
Tettamanti G., Carata E., Montali A., Dini L., Fimia G. M. (2019). Autophagy in development and regeneration: role in tissue remodelling and cell survival. Eur. Zool. J. 86, 113–131. doi: 10.1080/24750263.2019.1601271
Toton E., Romaniuk A., Budzianowski J., Hofmann J., Rybczynska M. (2016). Zapotin (5,6,2',6'-tetramethoxyflavone) modulates the crosstalk between autophagy and apoptosis pathways in cancer cells with overexpressed constitutively active PKCϵ. Nutr. Cancer. 68, 290–304. doi: 10.1080/01635581.2016.1134595
Turner P. V., Brabb T., Pekow C., Vasbinder M. A. (2011). Administration of substances to laboratory animals: routes of administration and factors to consider. J. Am. Assoc. Lab. Anim. Sci. 50, 600–613. doi: 10.1080/07924259.2011.555619
Wada Y., Sun-Wada G. H., Kawamura N., Aoyama M. (2014). Role of autophagy in embryogenesis. Curr. Opin. Genet. Dev. 27, 60–66. doi: 10.1016/j.gde.2014.03.010
Wang F., Huang L., Liang Q., Liao M., Liu C., Dong W., et al. (2022). TBC domain family 7-like enhances the tolerance of Penaeus vannamei to ammonia nitrogen by the up-regulation of autophagy. Fish Shellfish Immunol. 122, 48–56. doi: 10.1016/j.fsi.2022.01.025
Wang L., Ye S. (2015). Mechanism of caspase 1/3 mediated apoptosis and intervention effect of ulinastatin in ventilator-induced lung injury. Med. Rev. 21 (08), 1477–1480. doi: 10.3969/j.issn.1006-2084.2015.08.048
Warhurst D. C., Steele J. C., Adagu I. S., Craig J. C., Cullander C. (2003). Hydroxychloroquine is much less active than chloroquine against chloroquine-resistant Plasmodium falciparum, in agreement with its physicochemical properties. J. Antimicrob. Chemother. 52, 188–193. doi: 10.1093/jac/dkg319
Wei C. C., Luo Z., Song Y. F., Pan Y. X., Wu K., You W. J. (2017). Identification of autophagy related genes LC3 and ATG4 from yellow catfish Pelteobagrus fulvidraco and their transcriptional responses to waterborne and dietborne zinc exposure. Chemosphere. 175, 228–238. doi: 10.1016/j.chemosphere.2017.02.042
Wu H., Du C., Yang F., Zheng X., Qiu D., Zhang Q., et al. (2020). Generation of hepatocyte-like cells from human urinary epithelial cells and the role of autophagy during direct reprogramming. Biochem. Biophys. Res. Commun. 527, 723–729. doi: 10.1016/j.bbrc.2020.03.119
Xia X., Wang X., Qin W., Jiang J., Cheng L. (2019). Emerging regulatory mechanisms and functions of autophagy in fish. Aquaculture. 511, 734212. doi: 10.1016/j.aquaculture.2019.734212
Yabu T., Imamura S., Mohammed M. S., Touhata K., Minami T., Terayama M., et al. (2011). Differential gene expression of HSC70/HSP70 in yellowtail cells in response to chaperone-mediated autophagy. FEBS J. 278, 673–685. doi: 10.1111/j.1742-4658.2010.07989.x
Yao G., Li R., Yao Y., et al. (2019). Effect and related mechanism of high-glucose level on the autophagy of retinal vascular endothelial cells. J. Clin. Ophthalmology. 27, 6. doi: 10.3969/j.issn.1006-8422.2019.04.021
Yin F., Qian D. (2017). Transcriptomic analysis reveals the key immune-related signalling pathways of Sebastiscus marmoratus in response to infection with the parasitic ciliate Cryptocaryon irritans. Parasit Vectors. 10, 576. doi: 10.1186/s13071-017-2508-7
Yorimitsu T., Klionsky D. J. (2005). Autophagy: molecular machinery for self-eating. Cell Death Differ. 12 Suppl 2, 1542–1552. doi: 10.1038/sj.cdd.4401765
Yu Y., Tong B., Liu Y., Liu H., Yu H. (2021). Bioaccumulation, histopathological and apoptotic effects of waterborne cadmium in the intestine of crucian carp Carassius auratus gibelio. Aquacult. Rep. 20, 100669. doi: 10.1016/j.aqrep.2021.100669
Zhang J., Ma W., Xie B., Gui J., Mei J. (2017). Beneficial effect and potential molecular mechanism of chloroquine on sperm motility and fertilizing ability in yellow catfish. Aquaculture. 468, 307–313. doi: 10.1016/j.aquaculture.2016.10.028
Zhang X., Ming Y., Fu X., Niu Y., Lin Q., Liang H., et al. (2022). PI3K/AKT/p53 pathway inhibits infectious spleen and kidney necrosis virus infection by regulating autophagy and immune responses. Fish Shellfish Immunol. 120, 648–657. doi: 10.1016/j.fsi.2021.12.046
Zhang J., Zuo Z., He C., Cai J., Wang Y., Chen Y., et al. (2009). Effect of tributyltin on testicular development in Sebastiscus marmoratus and the mechanism involved. Environ. Toxicol. Chem. 28, 1528–1535. doi: 10.1897/08-347.1
Zhao X., Yin X., Ma T., Song W., Jiang L., Zhang X., et al. (2023). The effect of chloroquine on large yellow croaker (Larimichthys crocea): From autophagy, inflammation, to apoptosis. Aquacult Rep. 28, 101457. doi: 10.1016/j.aqrep.2022.101457
Zheng R., Chen H., Bo J., Xie Q., Hong F., Zhang Y. (2016). Joint effects of crude oil and heavy metals on the gill filament EROD activity of marbled rockfish Sebastiscus marmoratus. Ecotoxicol Environ. Saf. 132, 116–122. doi: 10.1016/j.ecoenv.2016.06.002
Zheng R., Zhang Y., Fang C., Chen M., Hong F., Bo J. (2019). Joint effects of chronic exposure to environmentally relevant levels of nonylphenol and cadmium on the reproductive functions in male rockfish Sebastiscus marmoratus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 215, 25–32. doi: 10.1016/j.cbpc.2018.09.006
Zhou G. Z., Li J., Sun Y. H., Zhang Q., Zhang L., Pei C. (2021). Autophagy delays apoptotic cell death induced by siniperca chuatsi rhabdovirus in epithelioma papulosum cyprinid cells. Viruses. 13, 1554. doi: 10.3390/v13081554
Keywords: hydroxychloroquine (HCQ), autophagy, apoptosis, inflammation, sebastiscus marmoratus
Citation: Liu Y, Luo J, Ren C, Chen L, Zhao Y, Yin X and Liu B (2024) Autophagy, apoptosis, and inflammation response of Sebastiscus marmoratus to different concentrations of hydroxychloroquine. Front. Mar. Sci. 11:1404295. doi: 10.3389/fmars.2024.1404295
Received: 20 March 2024; Accepted: 17 June 2024;
Published: 01 July 2024.
Edited by:
Ana Varela Coelho, Universidade Nova de Lisboa, PortugalReviewed by:
Yan Fang, Ludong University, ChinaDongdong Xu, Zhejiang Marine Fisheries Research Institute, China
Copyright © 2024 Liu, Luo, Ren, Chen, Zhao, Yin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingjian Liu, liubingjian@zjou.edu.cn; Xiaolong Yin, xlyhndx@163.com
 Yifan Liu
Yifan Liu