
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 21 May 2024
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1403909
Sediment bacterial communities are decisive drivers of nutrient cycling processes in aquaculture ecosystems and are readily affected by surrounding environmental factors. However, the knowledge of sediment nutrient accumulations and bacterial community structure is limited in the emerging polyculture systems. Herein, we investigated the profiles of sediment properties and bacterial communities in six typical polyculture ponds and primarily explored the influence of total nitrogen and phosphorus on the bacterial species and diversity. In almost all sediment samples, Proteobacteria, Chloroflexi, and Bacteroides were the dominant species at the phylum level, and the five most abundant bacterial genera were Sulfurovum, Woeseia, Ilumatobacter, Robiginitalea, and Cyanobium_PCC-6307. A clear different bacterial community was observed with the most dominant bacterial phylum Firmicutes and the lowest bacterial diversity in TZ1 pond sediment; meanwhile, the TZ1 pond also showed the highest TN and TP concentrations. Notably, sediments from WZ1 and WZ2 ponds in low-latitude regions exhibited higher bacterial richness and diversity. Based on Pearson’s correlation analysis, bacterial α-diversity indices showed significant negative correlation with sediment TP content, and TN content contributed the most to the abundance of sediment dominant bacterial genus, indicating that the bacterial community is highly associated with sediment nutrient concentrations. Moreover, co-occurrence network analysis further revealed some keystone taxa that exhibited high correlations with other bacterial species, especially the high-abundance genus Robiginitalea bridging a large number of connections. Compared to traditional mono-mariculture pattern, our study provided direct evidence of lower nutrient loadings and different bacterial communities in the polyculture ponds. This could assist polyculture practitioners in developing effective strategies for detailed nutritional management.
The global aquaculture industry has expanded rapidly in the past two decades and contributes dramatically to the socioeconomic development (Kim et al., 2022; Naylor et al., 2021). China as the world’s largest aquaculture producer reached approximately 62.8 million tons of aquatic products in 2020, accounting for more than 57% of global yield. Because of the ever-increasing demand for high-quality seafood, intensive and high-density aquaculture has shown a rapid upward trend in China (Chang Z. Q. et al., 2020). Correspondingly, it has also led to some inevitable environmental issues, such as water quality deterioration and eutrophication, greenhouse gas emission, hazardous chemical residues, and bacterial antibiotic resistance (Mavraganis et al., 2020; Lin and Lin, 2022). In particular, excessive emission of aquaculture tailwater rich in nitrogen (N) and phosphorus (P) severely threatens the offshore marine ecosystem health (Jing et al., 2023). Meanwhile, increased breeding density and unscientific strategies could lead to the deterioration of fishery water, further poisoning aquaculture animals (Li et al., 2018). Water pollution has been recognized as the most important factor hindering the sustainable development of fisheries (Ahmad et al., 2022). Thus, in recent years, an integrated multi-trophic aquaculture (IMTA) mode named “polyculture” has been quietly emerging (Fernandez-Gonzalez et al., 2018; Omont et al., 2020). Aquaculture practice also confirmed that polyculture can produce higher-quality aquatic products and gain more considerable economic benefits than conventional mode (Biswas et al., 2020). Aquaculture organisms for polyculture in coastal areas of China are usually composed of shrimp (Penaeus vannamei or Exopalaemon carinicauda), crab (Portunid or Scylla Serrata), and shellfish (Sinonovacula constricta, clam, or Arca granosa) (Guan et al., 2020; Zhang et al., 2020), and in most circumstances, shrimp are the major economic species. The large quantity of high protein feed input for the diet of shrimp was directly related to pond water quality. Conversely, crabs and shellfish as benthic organisms typically play vital roles in ingesting the excessive nutrients and metabolic waste, thereby improving pond water environment. Undeniably, polyculture as an eco-friendly aquaculture model has been widely popularized around the world (Khanjani et al., 2022).
Large amounts of commercial feeds introduced to ponds end up depositing into the pond sediments (Boyd et al., 2010; Gál et al., 2016), and thus, pond sediments are regarded as a net reservoir of nutrients originated from feed remnants, excrement, and dead animal bodies (Liu et al., 2021; Zhang et al., 2021b). It is well known that pond sediments are the ideal setting for species-driven nutrient cycling. In polyculture ponds, benthic organisms (e.g., crab and shellfish) are the most affected by excessive nutrient accumulation sediment via directly exposing to sediment and ingesting contaminated particles (Adámek and Maršálek, 2013). Moreover, both biological and abiotic matter are frequently exchangeable between sediment and aquatic habitat and may lead to pollutants in the sediment migrating and transforming into an aquatic environment (Murphy et al., 2016; Wu et al., 2019), which could increase potential disease risks to fish, shrimp, and other aquatic organisms. Therefore, sediment eco-environmental quality could greatly represent the health status of the aquaculture pond ecosystem.
Sediment N and P as essential biogenic elements play important roles in pond system functions by affecting sediment properties, organism growth, and microbial activities (Li et al., 2015; Xu et al., 2022). However, the undesirable and excess N and P stored in pond sediments can enter the cultural water body through nitrification, ammonification, dissolution, and mineralization; consequently lead to water quality deterioration; and could even cause eutrophication, endangering pond aquaculture organisms (Bouwman et al., 2013; Kroupová et al., 2018). Microbes are an important component for driving the biogeochemical cycling of carbon (C), N, and P in aquaculture ecosystems (Nho et al., 2018; Shen et al., 2020). Simultaneously, it has been reported that sediment bacterial communities contribute significantly to regulate sediment quality and maintain pond ecological functions (Zhang et al., 2021b). Notably, some pathogenic bacterial species, such as Vibrio parahaemolyticus, Vibrio alginolyticus, Vibrio splendidus, Aeromonas hydrophila, and Aeromonas caviae, are frequently associated with disease outbreak of shrimp, crab, and shellfish (Hong To et al., 2020; Liao et al., 2022; Zhou et al., 2019). Bacterial community composition and diversity in pond sediment are thereby recognized as potential indicators for pond ecosystem health (Su et al., 2018; Wu et al., 2023).
In the long-running polyculture pond, sediment physicochemical characteristics and bacterial community structure are the important factors that assess the impact on the health and sustainability of pond ecosystems, especially during the later culture stages (i.e., maturation and harvesting periods) due to the large amount of nutrient input (Shen et al., 2020). In this study, we focus on the sediment ecological environment of polyculture ponds after shrimp growth at a maturation period, and the objectives are as follows: (1) to analyze the several critical sediment parameters for six typical polyculture ponds of the Zhejiang Province, southern China; (2) to investigate their dissimilarities in bacterial community composition and diversity from a geographical perspective; and (3) to explore the relationships between selected nutrient contents and sediment bacterial communities. The results of this study can provide fundamental data and a valuable reference for sediment nutrient accumulations and bacterial response in polyculture patterns, which could help managers to optimize the stocking densities and improve the aquaculture yield maximally.
In August 2022, a total of 18 sediment samples were obtained from six shrimp–crab–shellfish polyculture ponds situated in eastern coastal cities of Zhoushan, Taizhou, and Wenzhou in Zhejiang province, China (Figure 1). All surface sediments during the shrimp maturation and harvesting stages were collected with a metallic core sampler. For each pond, sediment taken from the nine locations was thoroughly mixed into a composite sample. Each pond sampling was performed in triplicate. All collected sediments were placed in sterile plastic bags and transported to the laboratory in a cooler within 24 h. Each sample was divided into two parts. One part for C, N, and P analysis was dried with a vacuum freeze dryer (FreeZone®, Labconco, USA), passed through a stainless-steel sieve after impurity removal and ground in a porcelain mortar. The other part for bacterial 16S rRNA analysis was placed in a 50-mL sterile centrifuge tube and stored at −80°C after liquid nitrogen freezing.
Total nitrogen (TN) was measured by the potassium persulfate (K2S2O8) oxidation analysis method (Feng et al., 2023). A digestion module was constructed to handle approximately 0.05 g of sediment sample, 10 mL of ammonia-free water, and 5 mL of 50 g·L−1 K2S2O8 solution in a 25-mL colorimetric tube. The solution was digested at 121°C for 30 min and then flushed into 1.0 mL of 3.0 mol·L−1 HCl solution after cooling down the sample. The mixture was diluted to 25 mL with ammonia-free water and was subsequently shaken. After natural settlement for at least 2 h, the suspension was extracted to determine TN using an ultraviolet spectrophotometer (Cary 50, VARIAN, USA).
Likewise, the total phosphorus (TP) content was analyzed by using the K2S2O8 oxidation method (Feng et al., 2023). A total of 0.05 g of sediment was added to 25 mL of distilled water and 4 mL of 50 g·L−1 K2S2O8 solution in a 50-mL colorimetric tube. After high-temperature digestion and natural cooling, the extracting solution was adjusted to a final volume of 50 mL, mixed well, and settled. The final concentration of TP in the supernatant was detected by applying a V-1600PC spectrophotometer (MAPADA, China).
Total organic carbon (TOC) of the sediments was determined using the potassium dichromate oxidation–reduction method (Tian et al., 2020). Briefly, 0.4 g of freeze-dried unground sediment was reacted with 0.1 g of Ag2SO4 and 10 mL of 0.4 mol·L−1 K2Cr2O7-H2SO4 standard solution for 5 min in a 175°C oil bath to convert TOC into CO2. The cooled reaction mixture was transferred to a 250-mL Erlenmeyer flask and was then diluted to 60–70 mL volume with distilled water. Phosphoric acid solution (5 mL; H3PO4:H2O = 1:1) was added and 0.2 mol·L−1 FeSO4 solution was used to remove the remaining K2Cr2O7. When the yellow has faded almost completely, two to three drops of the N-phenylanthranilic acid indicator was supplemented and the solution continued to be titrated with FeSO4 solution until a color change from purple to green is reached. Finally, the TOC content was calculated according to the K2Cr2O7 consumption.
The pH of the sediment samples was measured in situ using a TP310 pH meter (TIMEPOWER, Beijing, China). The determination of heavy metals (Pb, Cd, Cr, Cu, and Zn) in the sediments was carried out according to the specification of the marine monitoring part 5 (GB 17378.5–2007, China). Cu, Zn, and Cr were detected using the flame atomic absorption spectrometry (FAAS) after acid digestion, while Pb and Cd were determined by a graphite furnace atomic absorption spectrometer at their respective wavelengths. Analysis of the metal Ni in the sediment was conducted using inductively coupled plasma mass spectrometry (ICP-MS) according to the standard method described by Zhang et al. (2021a).
Genomic DNA was extracted from the sediment samples using HiPure Soil DNA Isolation Kits (Magen, Guangzhou, China) according to the manufacturer’s guidelines. The specific primer pairs of 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′- GGACTACHVGGGTATCTAAT-3′) were designed to amplify the V3–V4 hypervariable regions of bacterial 16S rRNA genes. Polymerase chain reaction (PCR) was conducted as follows: 5 min at 95°C (initial denaturation); 30 cycles of 1 min at 95°C (denaturation), 1 min at 60°C (annealing), 1 min at 72°C (elongation); and 7 min at 72°C (final extension). The PCR reaction mixture (50 μL) contained template DNA (50 ng), 5× reaction buffer (10 μL), 5× high GC enhancer (10 μL), 2.5 mM dNTPs (1.5 μL), 10 μM oligonucleotide primers (each 1.5 μL), and High-Fidelity DNA polymerase (0.2 μL). PCR products were separated by 2% agarose gel electrophoresis, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) following the manufacturer’s protocol, and further quantified in an ABI StepOnePlus Real-Time PCR System (Life Technologies, Foster City, USA).
The paired-end sequencing (PE250) for purified amplicons was conducted by using the Novaseq 6000 platform (Illumina, San Diego, CA, USA) by Genedenovo Biotechnology Co. Ltd. (Guangzhou, China). A strict quality control process was performed as follows: (i) raw reads with >10% of unknown nucleotides and ≥50% of bases (Phred quality score ≤ 20) were quality-filtered; (ii) filtered reads with >10 bp of overlap and <2% of nucleotide mismatch ratio were merged as raw tags; (iii) low-quality raw tags were further filtered to obtain clean tags under specific conditions as described by Bokulich et al. (2013); and (iv) pre-processed clean reads were clustered into operational taxonomic units (OTUs) with 97% similarity cutoff, chimeric tags were removed, and finally effective tags were obtained for further bioinformatics analysis. An accession number of PRJNA1064545 was acquired after submitting raw data to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database.
FASTP software (v.0.18.0) was used to filter the raw sequence reads (Chen et al., 2018), and paired-end clean reads were merged by FLASH software (v.1.2.11) (Magoč and Salzberg, 2011). The UPARSE (v.9.2.64) pipeline was used to define OTUs (Edgar, 2013), and then chimeric tags were identified and removed using UCHIME algorithm (Edgar et al., 2011). The OTUs’ representative sequences were screened and annotated for taxonomic information with the SILVA (v.132) and UNITE (v.8.0) databases using the Ribosomal Database Project (RDP) tool based on a confidence threshold value of 80% (Wang et al., 2007). The taxonomic α-diversity (Sobs, Chao1, ACE, Shannon, Simpson, and PD whole tree) was calculated with the QIIME (v.1.9.1) (Caporaso et al., 2010) and picante (v.1.8.2) software (Kembel et al., 2010). Non-metric multidimensional scaling (NMDS) analysis of the Bray–Curtis distance was used to identify the differences in the bacterial community structure among all sediment samples. The ANOSIM test was performed for a better understanding of statistically different characteristics among biological samples. The correlations among physicochemical properties, bacterial communities, and α-diversity were analyzed via Pearson’s analysis (SPSS Institute, Chicago, USA). Origin Pro 2018 was used to draw the correlation heatmap and abundance circle graph. The other figures (bubble plots, NMDS, RDA, and network diagrams) were conducted with the igraph and ggplot2 packages in R software. For all statistical analyses, p-values lower than 0.05 were considered statistically significant.
The characteristics of sediments from six polyculture ponds are summarized in Supplementary Table S1. The concentrations of TN and TP varied from 513.37 to 676.56 mg N·kg−1 and from 404.64 to 686.45 mg P·kg−1, respectively. Sediment TOC ranged from 3.73 to 6.88 g C·kg−1, wherein the highest TN and TP contents were found at the TZ1 pond, and WZ2 had the maximum TOC content and the minimum TP content. The minimum TN and TOC values were obtained at ZS2. Pearson correlation analysis of all sediment measured factors was performed, and the results are shown in Figure 2. TOC exhibited a significant negative correlation with TP (coefficient = −0.59, p < 0.05) and yet was significantly positively correlated with TN (coefficient = 0.62, p < 0.05). The pH displayed significant positive correlations with TN and TOC. Moreover, TP and TOC significantly negatively correlated with many heavy metals (Zn, Cr, Cu, and Ni).
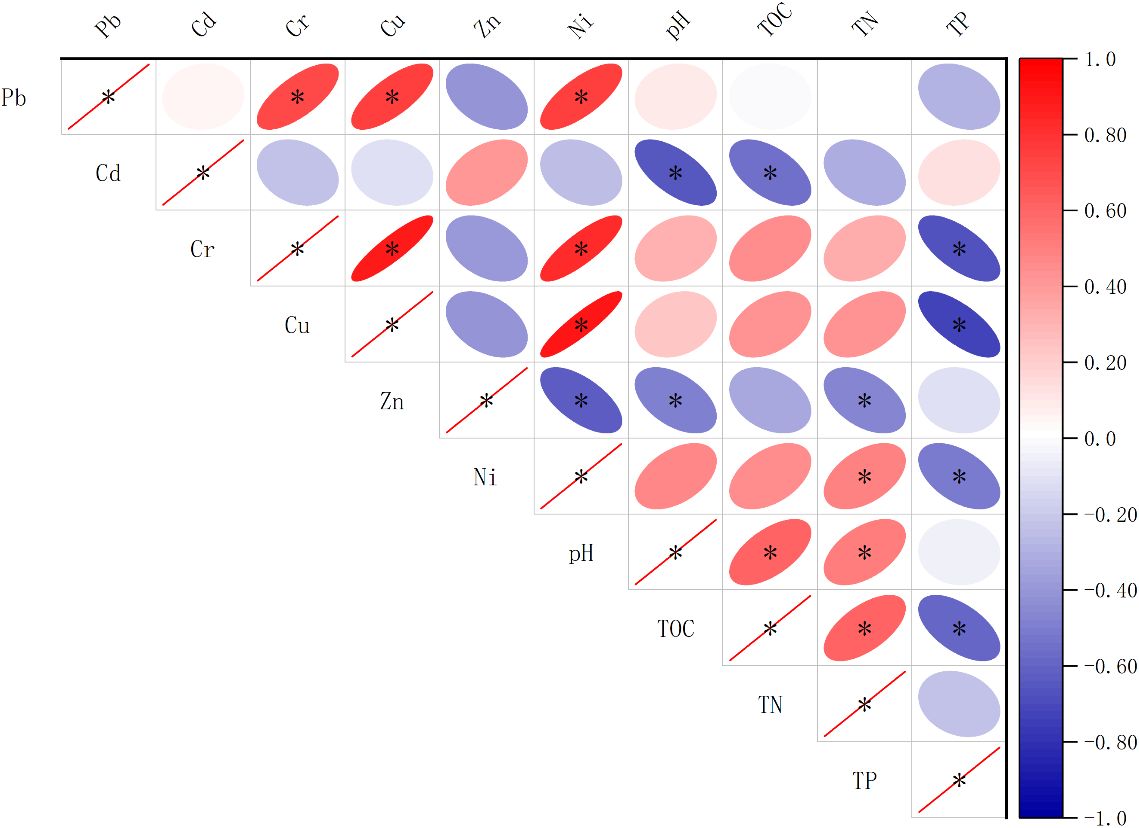
Figure 2 Associations among sediment physicochemical properties based on Pearson correlation analysis. The color intensity and elliptic size represent the correlation coefficients, *p < 0.05.
A total of 84,170 OTUs were classified into 67 phyla, 192 classes, 399 orders, 484 families, and 667 different genera in 18 sediment samples. At the phylum level (Figure 3A), Proteobacteria, Chloroflexi, and Bacteroidota were the three most dominant phyla in all samples, accounting for 12.52%–23.79%, 5.68%–18.93%, and 4.70%–15.72%, respectively. Chloroflexi was the most abundant phylum in the three polyculture ponds, with proportions of 18.93% in ZS1, 17.33% in ZS2, and 15.33% in TZ2. However, Proteobacteria had the largest relative abundance in WZ1 (23.48%) and WZ2 (23.79%). Notably, the phylum Firmicutes with the highest relative abundance of 21.38% was identified in TZ1. These results indicated that much differences of dominant phyla were found among sediments obtained in different regional polyculture ponds.
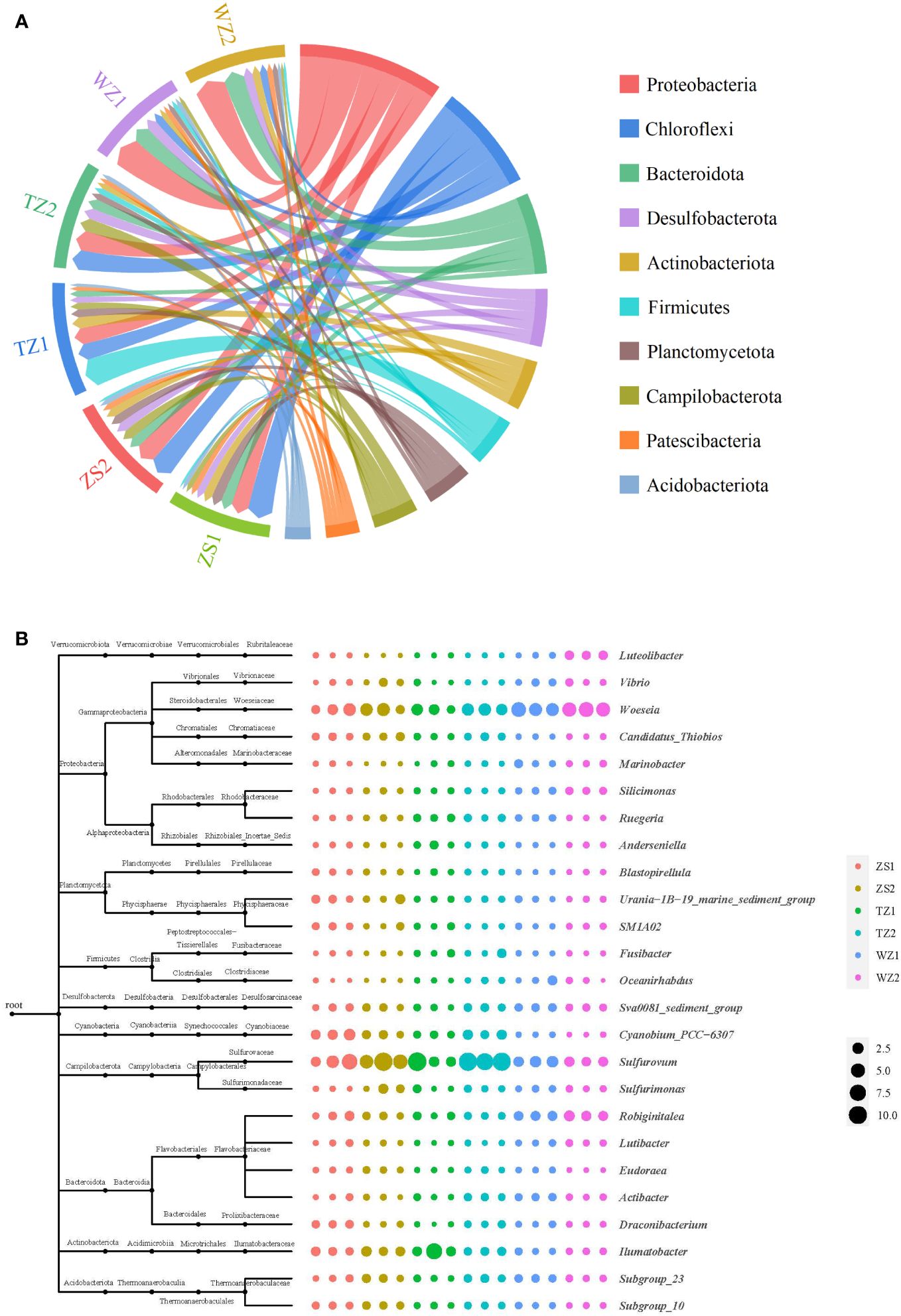
Figure 3 Relative abundance of sediment bacterial communities in six polyculture ponds: (A) circle diagram at the phylum level; (B) bubble plots at the genus level.
At the genus level (Figure 3B), the relative abundances of the top five genera were 1.56%–9.48% for Sulfurovum, 1.83%–5.16% for Woeseia, 0.46%–3.43% for Ilumatobacter, 0.61%–1.79% for Robiginitalea, and 0.11%–2.01% for Cyanobium_PCC-6307. Woeseia belonging to phylum Proteobacteria was the most abundant and largest genus species in ponds WZ1 and WZ2, displaying consistent community distributions at the phylum level. In contrast, Sulfurovum affiliated with the phylum Campilobacterota was the most dominant genus in other sediments, which showed a slight inconsistency with the composition of the dominant bacterial phyla. Additionally, some composition variation details of bacterial genera among different sediment samples were also obtained; for example, Robiginitalea was more abundant in ponds WZ1 and WZ2, while Cyanobium_PCC-6307 was more prevalent in sites ZS1 and TZ2.
To explore the bacterial diversity in the studied polyculture ponds, six α-diversity metrics (Sobs, Chao1, ACE, Shannon, Simpson, and PD whole tree) were calculated, and the profile is shown in Supplementary Table S2. Good’s coverage for all samples exceeded 98%, suggesting that the 16S sequencing results with sufficient depth had covered the majority of the bacterial community. The richness indices, Sobs, Chao1, and ACE, were 3,992–5,237, 4,142–5,360, and 4,244–5,475, respectively. Richness and evenness indices Shannon and Simpson were 8.755–10.440 and 0.9795–0.9967, respectively. The PD whole tree index representing for phylogenetic diversity ranged from 657.1 to 864.2. The highest values of all indices were observed in pond WZ1, followed in site WZ2, and in contrast, the least diverse bacterial communities were consistently detected in site TZ1. The results well reflected that pond sediments from ponds WZ1 and WZ2 located in low-latitude regions had higher bacterial diversity and richness. In addition, Pearson correlation analysis (Figure 4A) between physicochemical factors and bacterial α-diversity metrics demonstrated that diversity indices significantly negatively correlated with the TP and showed strong positive relationships with some heavy metal (Cr and Cu) contents. Meanwhile, several physicochemical properties (i.e., TOC, Pb, Ni, and Zn) displayed the weak and positive correlations with α-diversity indices.
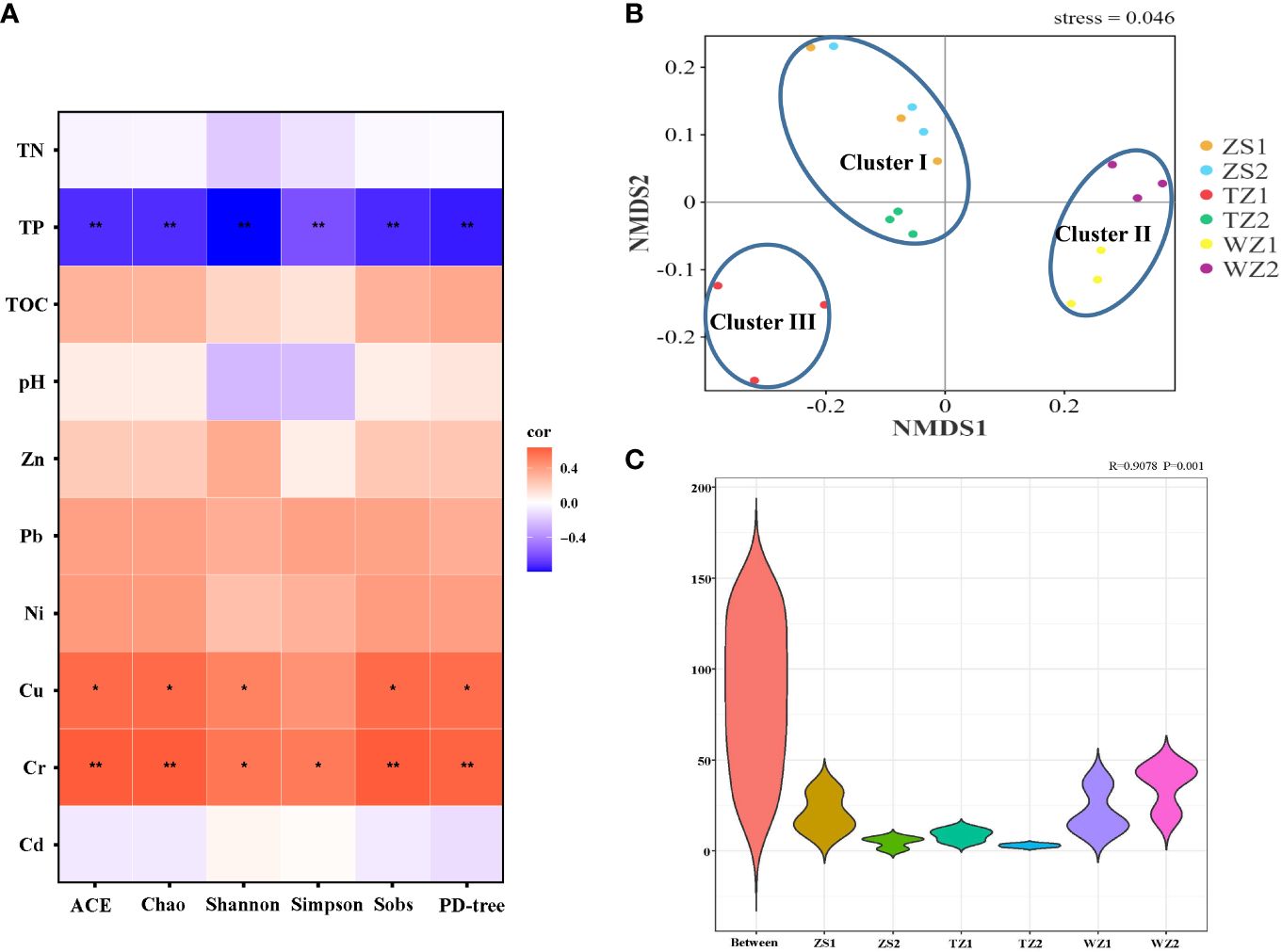
Figure 4 Analyses of bacterial α- and β-diversity in the different sediment samples. (A) The correlation heatmap illustrates the relationship between physicochemical variables and bacterial α-diversity metrics. *p < 0.05, **p < 0.01. (B) NMDS analyses of bacterial communities based on a Bray–Curtis distance algorithm at OTU levels. (C) ANOSIM test reveals clear differences between the different groups.
The variation in the bacterial community among the different pond sediments was visualized by NMDS based on a Bray–Curtis distance with the stress of 0.046 (Figure 4B). Bacterial community compositions were distinctly separated into three clusters, including cluster I (ZS1, ZS2, and TZ2), cluster II (WZ1 and WZ2) and cluster III (TZ1). Generally, samples with high similarity among bacterial communities were clustered together, and apparently, sediment bacterial community structure in pond TZ1 was significantly distinct with other sites, which was consistent with the dominant bacteria distributions. The ANOSIM test further demonstrated the obvious variance between the sediment groups with r = 0.9078, p = 0.001 (Figure 4C).
To reveal the environmental parameters that shape microbial community structures, four representative factors (pH, TN, TP, and TOC) and bacterial communities of different pond sediments were used for the redundancy analysis (RDA) (Figure 5A). The first two axes (RDA1 and RDA2) explained 61.59% of the total variation, indicating that the bacterial community structure of different sampling sites can be clearly separated from each other. Sediment TP (envfit analysis, r = 0.74, p < 0.005) and TN (envfit analysis, r = 0.53, p < 0.005) were considered to be the principal environmental factors influencing bacterial communities. TOC was positively correlated with the bacterial communities in WZ1 and WZ2. TP had a weak positive correlation with the bacterial community structure in ZS1, ZS2, and TZ2; however, bacterial communities from ZS1, ZS2, and TZ2 were strongly negatively correlated with TN. Regarding bacterial communities in TZ1, they were marginally influenced by sediment pH.
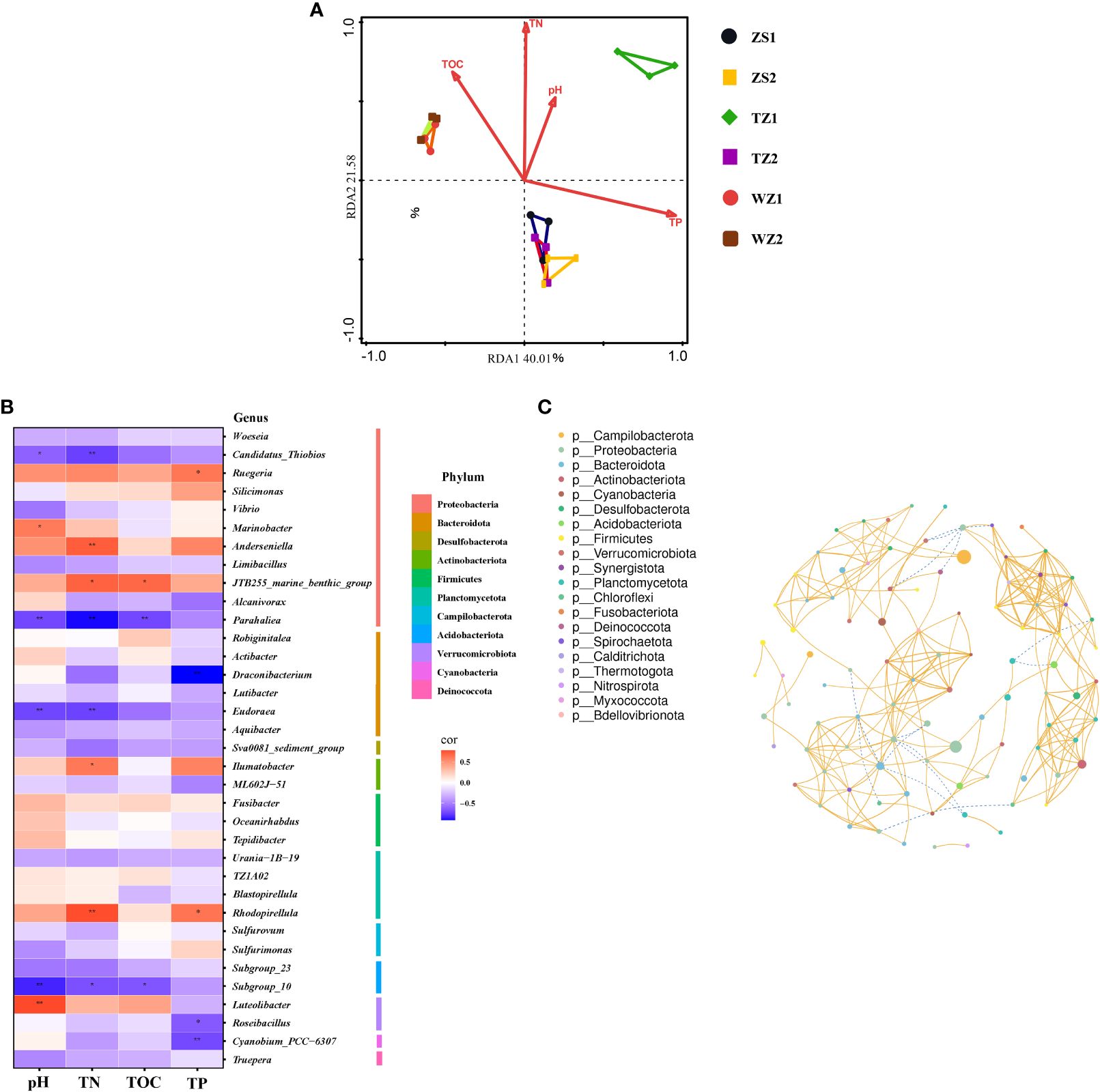
Figure 5 Correlation analysis and co-occurrence network tests. (A) Redundancy analysis (RDA) showing the contribution of sediment environmental factors to bacterial community structure. (B) Pearson correlation heatmap illustrating the relationship between the top 35 bacterial genera and environmental factors. *p < 0.05, **p < 0.01. (C) Co-occurrence networks identifying the bacterial interactions. A connection was based on the criteria with a strong (Pearson’s coefficient r > 0.7) and significant (p < 0.05) correlation; the size of the node represents the abundances of each bacterial genus, and nodes of the same color indicate the members from the same bacterial phylum; yellow and green lines represent positive and negative correlations, respectively.
Heatmap plot was further used to visualize the correlation between four chemical indices and the top 35 bacterial genera, which were classified into 11 phyla (Figure 5B). The results indicated that bacterial communities might be most sensitive to the TN content. Notably, TN strongly positively correlated with the dominant genus Ilumatobacter, and significantly negatively correlated with the dominant genera Subgroup_10 and Candidatus_Thiobios. TP and pH were considered as the other two important parameters that influence bacterial communities, because the highly abundant species Cyanobium_PCC-6307, Ruegeria, and Draconibacterium showed significant correlations with TP, and pH had a significantly negative correlation with the dominant genera Subgroup_10 and Candidatus_Thiobios. Moreover, the significantly negative correlation with the TOC and the dominant genera Subgroup_10 was also observed in this study.
Co-occurrence network was constructed to describe sediment bacterial interactions and ecological assembly rules. A total of 95 nodes and 265 edges were registered in the network (Figure 5C). The nodes were divided into 20 phyla, of which 3 dominant phyla (Proteobacteria, Bacteroidota, and Firmicutes) at all sediment samples accounted for 21.05%, 14.74%, and 11.58% of all the nodes, respectively. Keystone taxa were defined as critical bacterial species with a high degree (>2% of total interactions). Results of network analysis showed some keystone taxa, including Silicimonas, RBG-16–49-21, Robiginitalea, Altererythrobacter, SC103, Vallitalea, Blvii28_wastewater-sludge_group, and Dethiosulfovibrio. There were high correlations between these keystone bacteria, for example, the correlation between SC103 and RBG-16–49-21 (r = 0.91, p < 0.001), SC103 and Dethiosulfovibrio (r = 0.94, p < 0.001), and Blvii28_wastewater-sludge_group and Vallitalea (r = 0.97, p < 0.001). Notably, the genus Silicimonas (with relative abundances ranging from 0.15% to 0.95%) was the core keystone taxon, bridging the maximum number of nodes (n = 16) in the network. Robiginitalea (n = 12) with the ability to reduce N2O to N2 (El Hamouti et al., 2023) had the highest relative abundance among these keystone taxa identified, and thus might be considered as a potential indicator bacterium. However, there were low correlations between keystone and the top 10 most abundant bacteria, except for Robiginitalea and Woeseia (r = 0.77, p < 0.001), Silicimonas and Woeseia (r = 0.74, p < 0.001), and Altererythrobacter and Woeseia (r = 0.72, p < 0.001). In particular, the bacterial genus Sulfurovum, which is the most abundant for all sediment samples. was only directly linked to the other two genera, Desulfobacter and Desulfosarcina.
The polyculture pond is a complex and interactive ecosystem in which aquaculture animals, phytoplankton, benthic organisms, and microorganisms interact with each other to regulate the overall ecological balance (Dong et al., 2023). The intensive cultivation with high protein feed input threatens the ecosystem stability, since the majority of nutrients cannot be assimilated by aquaculture animals (Avnimelech and Ritvo, 2003; Pouil et al., 2019). As a result, the aquaculture pattern leads to heavy nutrient loading and large amounts of uneaten feeds rich in nitrogen and phosphorus are deposited on the pond sediment (Chatvijitkul et al., 2017; Sugiura, 2018), which eventually may cause contamination and eutrophication of aquaculture environments (Shen et al., 2020). In the present study, sediment nutrient contents showed significant differences in six polyculture ponds. The highest concentrations of TN and TP in TZ1 pond sediment may reflect more nutrient accumulations, which were largely attributed to aquaculture practices with the highest rate of commercial feed input (more than 200 kg per pond per day). Compared with the traditional shrimp ponds (Hou et al., 2021; Wu et al., 2023), the polyculture pond has a relatively lower nutrient loading because of a lower determination content in pond sediments. Not surprisingly, low nutrient accumulations in sediments from polyculture ponds are associated with filter-feeding benthic shellfish that can consume excess nutrients to optimize environmental quality (Waite, et al., 2014). Our results provided direct evidence that polyculture is an eco-friendly aquaculture model with a high efficiency of nutrient utilization.
Microorganisms as primary participants in biogeochemical cycling of essential nutrients in the aquaculture pond are closely associated with the stability of the pond ecosystem (Deng et al., 2020; Dai et al., 2021). As an indispensable part of the pond ecosystem, sediment bacterial communities have proved to be important indicators of the health status of the aquaculture ecosystem (Nho et al., 2018; Zhang et al., 2021b). Many previous studies have indicated that bacterial phyla Proteobacteria, Chloroflexi, and Bacteroidota were the most common and most abundant species in the aquaculture sediments (Shen et al., 2020; Wu et al., 2023; Xu et al., 2022), and the dominant phyla for polyculture pond sediments are also the case in this study. Although the majority of abundant bacterial phyla were highly similar in six pond sediments, small differences were also observed. For instance, Firmicutes, which often plays an important role for organic decomposition and fermentation (Chen et al., 2016), was determined as the most predominant phylum in TZ1 pond sediment. It could be attributed to the higher nutrient content that was significantly distinct from other ponds. Sulfurovum and Woeseia were the two most abundant bacterial genera in six polyculture sediments, which displayed great differences in bacterial community structure with the traditional mono-mariculture pattern (Hou et al., 2021). Most of two genera are chemoautotrophic bacteria (Wang et al., 2023). Members of Sulfurovum can oxidize–reduce sulfur compounds to gain energy (Inagaki et al., 2003; Sun et al., 2020), and the genus Woeseia is widely involved in dissimilatory sulfur oxidation and denitrification (Du et al., 2016; Mußmann et al., 2017). Therefore, the results implied that the polyculture ponds possessed a high degree of S and N metabolic functional potential.
The bacterial diversity and community composition in aquaculture sediments are an essential basis for sediment nutrient decomposition and utilization, which are extremely susceptible to variations of sediment nutrient levels (Moncada et al., 2019; Santander-de Leon et al., 2017; Wang et al., 2021). Several factors such as feed, antibiotics, cultivated species, and density in aquaculture practices have be confirmed to affect sediment microbial communities by altering sediment properties (Liu et al., 2020). The results of the correlation analysis revealed that bacterial α-diversity was negatively correlated with sediment TP content, in accordance with the observations of Ling et al. (2017) that N and P input could result in the decline in richness and diversity. Evidently, sediments in TZ1 showed the maximum nutrient content and minimum bacterial diversity, which was also supported by the previous report (Lin and Lin, 2022) that microbial community richness and diversity in coastal wetland sediment were significantly lost with the increasing bait input via enhancing a series of sediment properties (including sediment TN, sulfide, and TOC). Hence, TZ1 aquaculture practitioners should pay more attention to nutrition supply management because the loss of bacterial diversity poses a great threat to the health of aquaculture and adjacent ecosystems. Different sediment chemical properties also cause remarkable differences in the bacterial community assembly. The present study indicated that the top 35 genera in pond sediments were mainly affected by sediment TN content, followed by TP. TN and TP in other sediment environments had frequently exhibited close relationships with the dominant bacterial taxa. For example, Chang W. et al. (2020) found that the most abundant bacterial genus in the lake sediments was positively associated with TN and TP. Xie et al. (2016) reported that the “core” bacterial communities in the river sediments showed a negative correlation with the amount of TN and TP. Herein, increasing concerns regarding nitrogen and phosphorus input and output in this polyculture ecosystem need to be addressed, which will be conducive to guide aquaculture practices through scientific management including nutrient load regulation, antibiotic drug usage, and water management.
Nitrogen metabolism is a key process in intensive polyculture ecosystems, and sediment functional microbes involved in N fixation, assimilation, and mineralization are directly related to N cycling functional potential (Deng et al., 2020). A previous study (Zhang et al., 2023) has shown that intensive mariculture activities can lead to seasonal deoxygenation, which exerts a profound impact on sediment functional microbial communities involved in nitrate reduction processes. Our study also revealed that the TN content was largely associated with sediment bacterial diversity and many predominant bacterial taxa, including Ilumatobacter and Subgroup_10, which are part of the top 10 genera. Here, we further investigated the common nitrification and denitrification functional microbes that drive ultimate nitrogen removal. The results indicated that 12 main bacteria and archaea were found to be active in nitrification and denitrification processes (Figure 6). Nitrospina and Nitrospira as nitrite-oxidizing bacteria (NO2−→NO3−) were present in high abundance. The low-abundance bacteria Nitrosomonas and Nitrosospire that can oxidize ammonia (NH4+) to nitrite (NO2−) are evenly distributed across all the polyculture ponds. Notably, the archaea Candidatus_Nitrosopumilus was the statistically predominant ammonia-oxidizing microorganisms in pond WZ1, implying a higher ammonia-oxidizing potential for AOB than AOA in this pond system, and the similar findings were also supported by other researchers (Lu et al., 2015; Deng et al., 2020). Additionally, the majority of sediment-denitrification bacteria were affiliated with the phylum Proteobacteria, which is consistent with previous results (Ji et al., 2015). Pseudomonas and Bacillus, both highly abundant, were considered as the dominant bacterial genera driving denitrification in polyculture pond sediments. However, there are potential limitations in our present research to elucidate the feedback mechanisms of bacteria to detailed nutrient changes in pond sediments. Further studies will attempt to conduct comprehensive investigations of microorganisms and functional genes associated with the cycling of C, N, and P for the polyculture pond.
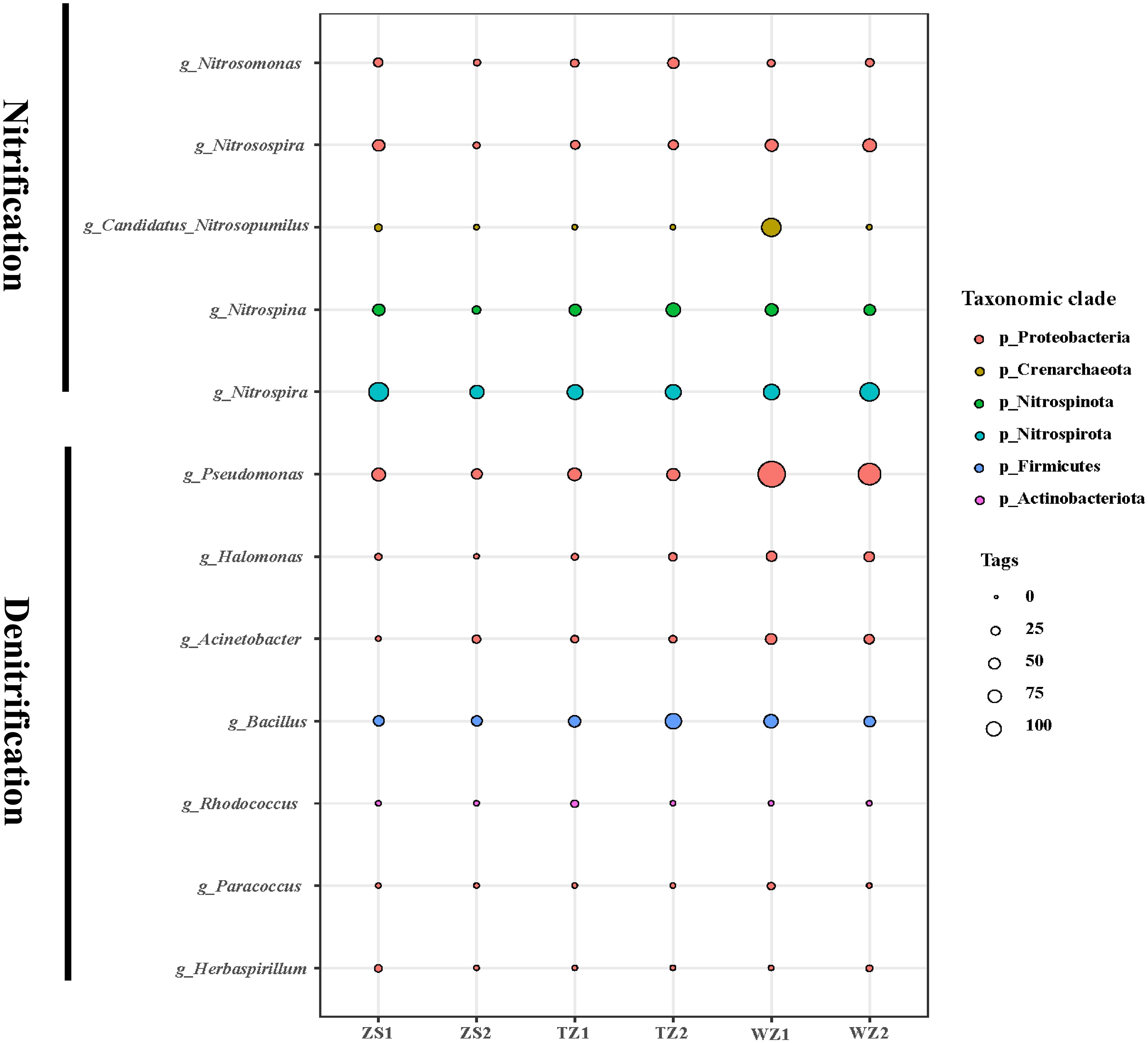
Figure 6 Relative abundances of nitrification and denitrification bacteria and archaea in six polyculture ponds. Circles of the same color indicate the members from the same bacterial phylum.
The present study proposed a new framework that describes sediment bacterial community responses to the different nutrient concentrations in shrimp, crabs, and the shellfish polyculture system. Our results indicate that the concentrations of C, N, and P nutrients in the polyculture pond sediments are significantly lower than those in traditional ponds, which may be largely attributed to the sensible nutrient utilization by multi-trophic cultured species. Immoderate nutrient availability from aquaculture practices might result in excessive nutrient accumulation in pond sediments and decreased bacterial richness and diversity, just as this study revealed in the TZ1 pond sediment. The principal contribution to differences in sediment bacterial diversity may derive from TP content, which exhibited a significant negative relationship between them. However, TN is the decisive factor affecting the abundance of sediment bacterial communities. Many nitrification and denitrification functional microbes with high abundance in sediment, such as Nitrospina, Nitrospira, Pseudomonas, and Bacillus, play major roles in N removal processes in the polyculture ecosystem. Overall, our findings provide a deep understanding on sediment microbial ecology and facilitate nutrient management for a healthy polyculture.
The data presented in the study are deposited in the the National Center for Biotechnology Information (NCBI) repository, accession number PRJNA1064545.
YanZ: Visualization, Validation, Software, Formal analysis, Data curation, Writing – original draft. TL: Conceptualization, Project administration, Resources, Supervision, Writing – original draft. GL: Project administration, Resources, Writing – original draft. TY: Writing – original draft, Resources, Project administration. YaoZ: Writing – original draft, Validation, Software, Formal analysis, Data curation. LJ: Writing – original draft, Visualization, Supervision, Methodology, Investigation, Funding acquisition, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the Basic Commonweal Research Program of Zhejiang Province, China (No. LTGS24C030003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1403909/full#supplementary-material
Adámek Z., Maršálek B. (2013). Bioturbation of sediments by benthic macroinvertebrates and fish and its implication for pond ecosystems: a review. Aquac. Int. 21, 1–17. doi: 10.1007/s10499-012-9527-3
Ahmad A. L., Chin J. Y., Harun M. H. Z. M., Low S. C. (2022). Environmental impacts and imperative technologies towards sustainable treatment of aquaculture wastewater: a review. J. Water Process. Eng. 46, 102553. doi: 10.1016/j.jwpe.2021.102553
Avnimelech Y., Ritvo G. (2003). Shrimp and fish pond soils: processes and management. Aquaculture 220, 549–567. doi: 10.1016/S0044-8486(02)00641-5
Biswas G., Kumar P., Ghoshal T. K., Kailasam M., De D., Bera A., et al. (2020). Integrated multi-trophic aquaculture (IMTA) outperforms conventional polyculture with respect to environmental remediation, productivity and economic return in brackishwater ponds. Aquaculture 516, 734626. doi: 10.1016/j.aquaculture.2019.734626
Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bouwman L., Beusen A., Glibert P. M., Overbeek C., Pawlowski M., Herrera J., et al. (2013). Mariculture: significant and expanding cause of coastal nutrient enrichment. Environ. Res. Lett. 8, 44026. doi: 10.1088/1748-9326/8/4/044026
Boyd C. E., Wood C. W., Chaney P. L., Queiroz J. F. (2010). Role of aquaculture pond sediments in sequestration of annual global carbon emissions. Environ. Pollut. 158, 2537–2540. doi: 10.1016/j.envpol.2010.04.025
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chang W., Sun J., Pang Y., Zhang S., Gong L., Lu J., et al. (2020). Effects of different habitats on the bacterial community composition in the water and sediments of lake taihu, China. Environ. Sci. Pollut. R. 27, 1–12. doi: 10.1007/s11356-020-10376-0
Chang Z. Q., Neori A., He Y. Y., Li J. T., Qiao L., Preston S. I., et al. (2020). Development and current state of seawater shrimp farming, with an emphasis on integrated multi-trophic pond aquaculture farms, in China-a review. Rev. Aquac. 12, 2544–2558. doi: 10.1111/raq.12457
Chatvijitkul S., Boyd C. E., Davis D. A., McNevin A. A. (2017). Pollution potential indicators for feed-based fish and shrimp culture. Aquaculture 477, 43–49. doi: 10.1016/j.aquaculture.2017.04.034
Chen S., Cheng H., Wyckoff K. N., He Q. (2016). Linkages of firmicutes and bacteroidetes populations to methanogenic process performance. J. Ind. Microbiol. Biotechnol. 43, 771–781. doi: 10.1007/s10295-016-1760-8
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Dai L., Liu C., Peng L., Song C., Li X., Tao L., et al. (2021). Different distribution patterns of microorganisms between aquaculture pond sediment and water. J. Microbiol. 59, 1–13. doi: 10.1007/s12275-021-0635-5
Deng M., Hou J., Song K., Chen J., Gou J., Li D., et al. (2020). Community metagenomic assembly reveals microbes that contribute to the vertical stratification of nitrogen cycling in an aquaculture pond. Aquaculture 520, 734911. doi: 10.1016/j.aquaculture.2019.734911
Dong S. L., Gao Q. F., Li L. (2023). “Aquaculture ecosystem,” in Aquaculture ecology. (Singapore: Springer Nature Singapore), 33–91.
Du Z. J., Wang Z. J., Zhao J. X., Chen G. J. (2016). Woeseia oceani gen. Nov., sp. Nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 66, 107–112. doi: 10.1099/ijsem.0.000683
Edgar R. C. (2013). Uparse: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). Uchime improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194. doi: 10.1093/bioinformatics/btr381
El Hamouti C., Castellano-Hinojosa A., Mabrouki Y., Chaouni B., Ghazal H., Boukhatem N., et al. (2023). Anthropogenic nitrate contamination impacts nitrous oxide emissions and microbial communities in the marchica lagoon (Morocco). Sustainability 15, 4077. doi: 10.3390/SU15054077
Feng L., Zhang Z., Yang G., Wu G., Yang Q., Chen Q. (2023). Microbial communities and sediment nitrogen cycle in a coastal eutrophic lake with salinity and nutrients shifted by seawater intrusion. Environ. Res. 225, 115590. doi: 10.1016/j.envres.2023.115590
Fernandez-Gonzalez V., Toledo-Guedes K., Valero-Rodriguez J. M., Agraso M. M., Sanchez-Jerez P. (2018). Harvesting amphipods applying the integrated multitrophic aquaculture (IMTA) concept in off-shore areas. Aquaculture 489, 62–69. doi: 10.1016/j.aquaculture.2018.02.008
Gál D., Pekár F., Kerepeczki É. (2016). A survey on the environmental impact of pond aquaculture in Hungary. Aquac. Int. 24, 1543–1554. doi: 10.1007/s10499-016-0034-9
Guan X., Wang B., Duan P., Tian J., Dong Y., Jiang J., et al. (2020). The dynamics of bacterial community in a polyculture aquaculture system of Penaeus chinensis, Rhopilema esculenta and Sinonovacula constricta. Aquac. Res. 51, 1789–1800. doi: 10.1111/are.14528
Hong To T. T., Yanagawa H., Khanh Thuan N., Hiep D. M., Cuong D. V., Khai L. T.L., et al. (2020). Prevalence of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease of shrimp in shrimp, molluscan shellfish and water samples in the mekong delta, Vietnam. Biology 9, 312. doi: 10.3390/biology9100312
Hou D., Zhou R., Zeng S., Wei D., Deng X., Xing C., et al. (2021). Stochastic processes shape the bacterial community assembly in shrimp cultural pond sediments. Appl. Microbiol. Biotechnol. 105, 1–10. doi: 10.1007/S00253-021-11378-9
Inagaki F., Takai K., Kobayashi H., Nealson K. H., Horikoshi K. (2003). Sulfurimonas autotrophica gen. Nov., Sp. Nov., A novel sulfur-oxidizing ϵ-Proteobacterium isolated from hydrothermal sediments in the mid-okinawa trough. Int. J. Syst. Evol. Microbiol. 53, 1801–1805. doi: 10.1099/ijs.0.02682-0
Ji B., Yang K., Zhu L., Jiang Y., Wang H., Zhou J., et al. (2015). Aerobic denitrification: a review of important advances of the last 30 years. Biotechnol. Bioprocess Eng. 20, 643–651. doi: 10.1007/s12257-015-0009-0
Jing X., Zhang J., Li Y., Li Q., Shen Y., Liu J., et al. (2023). Cyanometallate framework templated synthesis of hierarchically porous la(oh)3 for high-efficient and stable phosphorus removal from tailwater. Chem. Eng. J. 465, 142789. doi: 10.1016/j.cej.2023.142789
Kembel S. W., Cowan P. D., Helmus M. R., Cornwell W. K., Morlon H., Ackerly D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464.
Khanjani M. H., Zahedi S., Mohammadi A. (2022). Integrated multitrophic aquaculture (IMTA) as an environmentally friendly system for sustainable aquaculture: functionality, species, and application of biofloc technology (BFT). Environ. Sci. pollut. Res. 29, 67513–67531. doi: 10.1007/s11356-022-22371-8
Kim D. Y., Shinde S. K., Kadam A. A., Saratale R. G., Saratale G. D., Kumar M., et al. (2022). Advantage of species diversification to facilitate sustainable development of aquaculture sector. Biology 11, 368. doi: 10.3390/biology11030368
Kroupová H. K., Valentová O., Svobodová Z., Šauer P., Máchová J. (2018). Toxic effects of nitrite on freshwater organisms: a review. Rev. Aquac. 10, 525–542. doi: 10.1111/raq.12184
Li H., Zhang C., Han X., Shi X. (2015). Changes in concentrations of oxygen, dissolved nitrogen, phosphate, and silicate in the southern yellow sea 1980–2012: sources and seaward gradients. Estuar. Coast. Shelf S. 163, 44–55. doi: 10.1016/j.ecss.2014.12.013
Li Z., Peng F., Niu B., Li G., Wu J., Miao Z. (2018). Water quality prediction model combining sparse auto-encoder and lstm network. IFAC PapersOnLine. 51, 831–836. doi: 10.1016/j.ifacol.2018.08.091
Liao G., Wu Q., Mo B., Zhou J., Li J., Zou J., et al. (2022). Intestinal morphology and microflora to Vibrio alginolyticus in pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 121, 437–445. doi: 10.1016/j.fsi.2022.01.026
Lin G., Lin X. (2022). Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Res. 218, 118520. doi: 10.1016/j.watres.2022.118520
Ling N., Chen D., Guo H., Wei J., Bai Y., Shen Q., et al. (2017). Differential responses of soil bacterial communities to long-term n and p inputs in a semi-arid steppe. Geoderma 292, 25–33. doi: 10.1016/j.geoderma.2017.01.013
Liu X., Shao Z., Cheng G., Lu S., Gu Z., Zhu H., et al. (2021). Ecological engineering in pond aquaculture: a review from the whole-process perspective in China. Rev. Aquac. 13, 1060–1076. doi: 10.1111/RAQ.12512
Liu Z., Iqbal M., Zeng Z., Lian Y., Zheng A., Zhao M., et al. (2020). Comparative analysis of microbial community structure in the ponds with different aquaculture model and fish by high-throughput sequencing. Microb. Pathog. 142, 104101. doi: 10.1016/j.micpath.2020.104101
Lu S., Liao M., Xie C., He X., Li D., He L., et al. (2015). Seasonal dynamics of ammonia-oxidizing microorganisms in freshwater aquaculture ponds. Ann. Microbiol. 65, 651–657. doi: 10.1007/s13213-014-0903-2
Magoč T., Salzberg S. L. (2011). Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mavraganis T., Constantina C., Kolygas M., Vidalis K., Nathanailides C. (2020). Environmental issues of aquaculture development. Egypt. J. Aquat. Biol. Fisher. 24, 441–450. doi: 10.21608/ejabf.2020.85857
Moncada C., Hassenrück C., Gärdes A., Conaco C. (2019). Microbial community composition of sediments influenced by intensive mariculture activity. Fems. Microbiol. Ecol. 95, fiz006. doi: 10.1093/femsec/fiz006
Mußmann M., Pjevac P., Krüger K., Dyksma S. (2017). Genomic repertoire of the Woeseiaceae/jtb255, cosmopolitan and abundant core members of microbial communities in marine sediments. Isme J. 11, 1276–1281. doi: 10.1038/ismej.2016.185
Murphy A. E., Emery K. A., Anderson I. C., Pace M. L., Brush M. J., Rheuban J. E. (2016). Quantifying the effects of commercial clam aquaculture on c and n cycling: an integrated ecosystem approach. Estuar. Coast. 39, 1746–1761. doi: 10.1007/s12237-016-0106-0
Naylor R., Troell M., Little D., Hardy R., Buschmann A. H. (2021). Naylor etal.2021.nature- a 20-year retrospective review of global aquaculture. Nature 591, 551. doi: 10.1038/S41586-021-03308-6
Nho S. W., Abdelhamed H., Paul D., Park S., Mauel M. J., Karsi A., et al. (2018). Taxonomic and functional metagenomic profile of sediment from a commercial catfish pond in mississippi. Front. Microbiol. 9, 2855. doi: 10.3389/fmicb.2018.02855
Omont A., Elizondo-González R., Quiroz-Guzmán E., Escobedo-Fregoso C., Hernández-Herrera R., Peña-Rodríguez A. (2020). Digestive microbiota of shrimp Penaeus vannamei and Oyster crassostrea gigas co-cultured in integrated multi-trophic aquaculture system. Aquaculture 521, 735059. doi: 10.1016/j.aquaculture.2020.735059
Pouil S., Samsudin R., Slembrouck J., Sihabuddin A., Sundari G., Khazaidan K., et al. (2019). Nutrient budgets in a small-scale freshwater fish pond system in Indonesia. Aquaculture 504, 267–274. doi: 10.1016/j.aquaculture.2019.01.067
Santander-de Leon S. M. S., Nu Al S. N., Wei H., Yoshikawa T., Maeda H. (2017). Bacterial community composition shifts in sediments influenced by fish feeds. Aquac. Res. 48, 4380–4389. doi: 10.1111/are.2017.48.issue-8
Shen X., Xu M., Li M., Zhao Y., Shao X. (2020). Response of sediment bacterial communities to the drainage of wastewater from aquaculture ponds in different seasons. Sci. Total Environ. 717, 137180. doi: 10.1016/j.scitotenv.2020.137180
Su Z., Dai T., Tang Y., Tao Y., Huang B., Mu Q., et al. (2018). Sediment bacterial community structures and their predicted functions implied the impacts from natural processes and anthropogenic activities in coastal area. Mar. pollut. Bull. 131, 481–495. doi: 10.1016/j.marpolbul.2018.04.052
Sugiura S. H. (2018). Phosphorus, aquaculture, and the environment. Rev. Fish. Sci. Aquac. 26, 515–521. doi: 10.1080/23308249.2018.1471040
Sun Q. L., Zhang J., Wang M. X., Cao L., Du Z. F., Sun Y. Y., et al. (2020). High-throughput sequencing reveals a potentially novel Sulfurovum species dominating the microbial communities of the seawater-sediment interface of a deep-sea cold seep in South China sea. Microorganisms 8, 687. doi: 10.3390/microorganisms8050687
Tian Y., An J., Tao S. (2020). Determination of organic carbon content in soil by potassium dichromate oxidation-spectrophotometry. Mod. Chem. Ind. 40, 231–235. doi: 10.16606/j.cnki.issn0253-4320.2020.04.049
Waite R., Beveridge M., Brummett R., Castine S., Chaiyawannakarn N., Kaushik S., et al. (2014). Improving productivity and environmental performance of aquaculture. WorldFish.
Wang C., Wang Y., Liu P., Sun Y., Song Z., Hu X. (2021). Characteristics of bacterial community structure and function associated with nutrients and heavy metals in coastal aquaculture area. Environ. Pollut. 275, 116639. doi: 10.1016/j.envpol.2021.116639
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive bayesian classifier for rapid assignment of Rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang Y., Zhang Y., Jia C., Xu Q., Rong Y., Xu Z., et al. (2023). Comparative analysis of gut microbial community structure of three tropical sea cucumber species. Diversity 15, 855. doi: 10.3390/d15070855
Wu J., Su Y., Deng Y., Guo Z., Mao C., Liu G., et al. (2019). Prevalence and distribution of antibiotic resistance in marine fish farming areas in Hainan, China. Sci. Total Environ. 653, 605–611. doi: 10.1016/j.scitotenv.2018.10.251
Wu L., Yang P., Zhang L., Luo L., Hong Y., Zhu W., et al. (2023). Sediment sulfate content determines assembly processes and network stability of bacteria communities of coastal land-based shrimp aquaculture ponds. Aquaculture 563, 738953. doi: 10.1016/J.aquaculture.2022.738953
Xie Y., Wang J., Wu Y., Ren C., Song C., Yang J., et al. (2016). Using in situ bacterial communities to monitor contaminants in river sediments. Environ. pollut. 212, 348–357. doi: 10.1016/j.envpol.2016.01.031
Xu M., Xu R. Z., Shen X. X., Gao P., Xue Z. X., Huang D. C., et al. (2022). The response of sediment microbial communities to temporal and site-specific variations of pollution in interconnected aquaculture pond and ditch systems. Sci. Total Environ. 806, 150498. doi: 10.1016/j.scitotenv.2021.150498
Zhang D., Tian X., Dong S., Chen Y., Feng J., He R., et al. (2020). Carbon dioxide fluxes from two typical mariculture polyculture systems in coastal China. Aquaculture 521, 735041. doi: 10.1016/j.aquaculture.2020.735041
Zhang X., Yao C., Zhang B., Tan W., Gong J., Wang G. Y., et al. (2023). Dynamics of benthic nitrate reduction pathways and associated microbial communities responding to the development of seasonal deoxygenation in a coastal mariculture zone. Environ. Sci. Technol. 57, 15014–15025. doi: 10.1021/acs.est.3c03994
Zhang Y., Li H., Yin J., Zhu L. (2021a). Risk assessment for sediment associated heavy metals using sediment quality guidelines modified by sediment properties. Environ. pollut. 275, 115844. doi: 10.1016/j.envpol.2020.115844
Zhang Z., Deng Q., Wan L., Cao X., Zhou Y., Song C. (2021b). Bacterial communities and enzymatic activities in sediments of long-term fish and crab aquaculture ponds. Microorganisms 9, 501. doi: 10.3390/MICROORGANISMS9030501
Keywords: polyculture, pond sediment, bacterial assembly, TN and TP, co-occurrence network
Citation: Zhang Y, Li T, Li G, Yuan T, Zhang Y and Jin L (2024) Profiling sediment bacterial communities and the response to pattern-driven variations of total nitrogen and phosphorus in long-term polyculture ponds. Front. Mar. Sci. 11:1403909. doi: 10.3389/fmars.2024.1403909
Received: 20 March 2024; Accepted: 24 April 2024;
Published: 21 May 2024.
Edited by:
Xianbiao Lin, Ocean University of China, ChinaReviewed by:
Genmei Lin, Sun Yat-sen University, ChinaCopyright © 2024 Zhang, Li, Li, Yuan, Zhang and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Jin, amlubGVpMjM4OEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.