
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 13 May 2024
Sec. Marine Molecular Biology and Ecology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1403559
This article is part of the Research Topic Endocrine Disruption in Marine Species: Unraveling Pollution and Climate Change Effects View all 5 articles
Salinity is a common abiotic stress in the culture of penaeid shrimp. Through post-transcriptional regulation of gene transcripts, microRNAs (miRNAs) play an important role in the adaptation to a stressful environment. However, the involvement of miRNAs in the salinity stress response of shrimp remains unclear. In the present study, the sequence and expression profile of miRNAs in the hepatopancreas of low-salinity-treated Penaeus monodon were obtained by the high-throughput sequencing technique. A total of 679 miRNAs were identified, including 167 miRNAs that were significantly differentially expressed after low-salinity exposure (p < 0.05). Remarkably, most of these miRNAs were downregulated, suggesting that a series of genes were activated to participate in stress response. In addition, 43 miRNAs differentially expressed at all treatment were selected as putative key modulators. Enrichment analysis of genes targeted by these miRNAs indicated that a network that consists of the nervous system, the immune system, and the endocrine system played a crucial role in maintaining the homeostasis of P. monodon under low-salinity stress. These findings may help contribute to a better understanding of the mechanism that regulates salinity tolerance in shrimp and provide valuable genetic information for subsequent studies.
The black tiger shrimp (Penaeus monodon) has been an economically important shrimp species cultured in China with a rapid growth in global demand. In general, P. monodon can survive in a wide range of salinities, and previous research showed that the optimal salinity for culture is 15–25 ppt (Ferraris et al., 1987). However, changes in abiotic and biotic factors may result in salinity fluctuation during the culture period, leading to osmotic pressure, which has a significant impact on the survival and growth of shrimps (Rahi et al., 2021). Drastic salinity changes could also affect the feed intake and induce higher energy utilization for osmoregulation, finally resulting in the poor growth of shrimps (Silva et al., 2010). In addition, changes in salinity might alter the physiological and biochemical parameters of shrimps in response to stress (Luo et al., 2022). Therefore, elucidating the adaptation mechanisms of P. monodon under different salinities is crucial for the development of shrimp culture.
Several vital genes responding to the low salinity of P. monodon have been identified in recent years, such as mitogen-activated protein kinase kinase (Fan et al., 2021), cold shock domain binding protein (Si et al., 2022a), and ETS-related transcriptional regulator (Si et al., 2022b). These genes played significant roles in stress resistance and immunity. Based on high-throughput sequencing technologies, Li et al. analyzed the transcriptome of P. monodon under chronic low-salinity stress. Results suggested that genes related to metabolism, immunity, and signal transduction were affected by stress (Li et al., 2023). The above findings might provide foundations for understanding the mechanism of low-salinity tolerance and resistance breeding research in P. monodon. Moreover, various studies have proved that microRNAs (miRNAs) participate in processes related to stress response in shrimps (Guo et al., 2018; He et al., 2019). However, few studies about miRNA in P. monodon could be detected, which are focused on development and disease (Zhao et al., 2018; Mondal et al., 2021).
miRNAs are single-stranded non-coding endogenous RNAs (approximately 22 nucleotides) that play essential regulatory roles by targeting mRNA for cleavage or translational repression (Farhadi et al., 2021). Generally, miRNAs function in a one-to-many transcripts relationship, which means that a single miRNA might regulate multiple target genes. The first miRNA (lin-4) was discovered in Caenorhabditis elegans in 1993, and the second (let-7) was identified 7 years later (Reinhart et al., 2000). With the development of high-throughput sequencing technologies, there has been a rapid growth of identified miRNAs in a diverse number of species (Saliminejad et al., 2019). Short read length and high coverage are especially suited for counting miRNA prevalence and calculating differential expression. High-throughput sequencing provides a genome-wide approach to explore miRNAs and explain the regulatory mechanism of living organisms. Recently, increasing studies related to aquatic animals’ miRNAs were reported, revealing the important function of miRNAs in several biological events such as cellular differentiation, proliferation, apoptosis, and immune system (Wang et al., 2018; Hao et al., 2022; Liu et al., 2023).
The hepatopancreas is an essential organ in crustaceans, which plays a vital role in metabolism, detoxification, and immunity (Vogt, 2019). To date, identification and characterization of P. monodon hepatopancreas miRNAs have been very limited. In this study, using high-throughput sequencing technologies, we identified and established the miRNA transcriptome library of the P. monodon hepatopancreas under low-salinity stress. The aim was to identify differentially expressed miRNAs (DEMs) associated with pathways that tolerate low salinity and provide new insights for understanding the molecular mechanism of salinity stress.
P. monodon for experiments was acquired from the Shenzhen Experimental Base of the South China Sea Fisheries Research Institute, Chinese Academy of Fisheries Sciences. Shrimps with a body mass of 10 ± 2 g were temporarily cultured in filtered aerated seawater at 28–30°C at a salinity of 30–32 psu and fed with commercial feed (Dongteng Feed, Guangdong, China) three times a day. Approximately one-third of the water in each tank was replaced every day.
Based on previous works, the salinity was determined as 3 psu, and the time was set to 96 h for acute low-salinity stress treatment (Fan et al., 2021; Si et al., 2022a). Experimental shrimps were randomly divided into two groups, which had salinity levels of 30 (control group) and 3 psu, respectively. Salinity was measured using a high-precision salinometer (ATAGO, Guangzhou). Each group was placed in triplicate plastic tanks, with 30 shrimps per replicate. Individuals were selected randomly and hepatopancreas tissues were collected at 0, 6, 24, and 96 h after stress. All samples were flash-frozen in liquid nitrogen overnight and then stored at −80°C for later use.
Total RNA was extracted by using the Trizol reagent kit (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Extracted total RNA were treated with RNase-free DNase I (Takara Bio, Shiga, Japan) for 30 min at 37°C to remove residual DNA. A NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, USA) was employed to estimate RNA concentration. The integrity of the RNA was determined by using agarose gel electrophoresis.
RNA molecules in a size range of 18–30 nt were enriched by polyacrylamide gel electrophoresis (PAGE). Then, adapters were added and ligated to the RNAs. After ligation, RNA products were reverse-transcribed by PCR amplification. PCR products with a length of 140–160 bp were enriched to generate a cDNA library and sequenced using Illumina Novaseq 6000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Raw reads obtained from Illumina sequencing were filtered according to the following rules: low-quality reads containing more than one low-quality (Q-value ≤ 20) base or containing unknown nucleotides (N), reads without 3′ adapters, reads containing 5′ adapters, reads without inserted fragments, reads containing polyA in small RNA fragment, and reads shorter than 18 nt (not including adapters). Filtered reads were aligned with small RNAs in the GeneBank database (Release 209.0) and Rfam database (Kalvari et al., 2018) (Release 11.0) to remove ribosomal RNA (rRNA), small conditional RNA (scRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), and transfer RNA (tRNA). Achieved reads were also BLAST searched against the reference genome to remove tags mapped to exons, introns, and repeat sequences. All clean reads were then searched against the miRBase database (Kozomara et al., 2019) (Release 22) to identify known miRNAs. In addition, novel miRNA candidates were identified by using miRDeep2 (Mackowiak, 2011) software with default parameters.
The miRNA expression level was calculated and normalized to transcripts per million (TPM) based on their counts in clean reads. We used the DESeq2 (Love et al., 2014) package in R to perform differential expression analysis between two groups. miRNAs with a fold change ≥ 2 and p-value < 0.05 in a comparison were identified as DEMs.
Target genes of miRNA were predicted by multiple tools, including miRanda (Betel et al., 2010) (version 3.3), TargetScan (Agarwal et al., 2015) (version 7.0), and RNAhybrid (Kruger and Rehmsmeier, 2006) (version 2.1.2). Parameters of miRanda were set to a score > 140 and free energy < −20 kcal/mol. The 2- to 8-nt sequences, which start from 5′ small RNA, were chosen as seed sequences to predict 3′-UTR of transcripts in the TargetScan program. For RNAhybrid, miRNAs with a maximum length of 24 nt and a free energy of less than −20 kcal/mol, having a helix from position 2 to 8 with respect to the query, were identified. The overlap of all three miRNA datasets were selected as predicted miRNA target genes.
Predicted target genes were subjected to Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway for enrichment analysis. Based on the hypergeometric test and FDR correction, terms or pathways with FDR less than 0.05 were defined as significantly enriched. The top 20 results were selected and visualized with the OmicShare tool, an online platform for data analysis (https://www.omicshare.com/tools/Home/Soft).
To estimate the accuracy of high-throughput sequencing, real-time quantitative PCR (qPCR) was performed for nine random miRNAs showing different expression patterns. As mentioned above, hepatopancreas from the control and treatment groups were used for stem-loop qPCR analysis. The miRNAs were first reverse-transcribed following the manufacturer’s protocol of the TaqMan miRNA Assay Kit (Applied Biosystems, Waltham, USA) as published in previous reports (Guo et al., 2018). Primers were designed according to the stem-loop miRNA primer design principle (Supplementary Table 1). To normalize the expression levels of miRNAs, U6 snRNA was selected as an internal control. The qPCR experiments were carried out in a LightCycler® 480 II Real-time PCR Instrument (Roche, Basel, Switzerland) using SYBR Green Master Mix (Qiagen, Hilden, Germany) following the manufacturer’s recommendations. Expression levels of miRNAs were calculated based on the 2−ΔΔCt method. Statistical analysis was performed using one-way ANOVA, followed by Tukey’s multiple range test, using SPSS statistics version 23.0 software (IBM, Armonk, USA).
To identify the miRNAs in response to low-salinity stress, small RNA libraries from shrimp hepatopancreas were conducted using high-throughput sequencing at 0, 6, 24, and 96 h after stress treatment, so-called CK, 6h, 24h, and 96h, respectively. A summary of all sequencing data is shown in Table 1. A total of 146,180,200 raw reads were generated. After filtering by multiple rules, 135,464,114 clean reads were obtained from all libraries, accounting for 92.67% of the total reads.
Analysis of length distribution showed that small RNAs of 21 nt size were the most common type, followed by those of 22 nt (Figure 1). All the clean reads were subjected to BLAST search against genome and various databases for annotation. Based on Figure 1, we can see that known miRNA is the major type identified in this research. The specific number and proportion of sequence tags aligned to different categories of small RNAs are shown in Table 2. Based on all libraries, a total of 679 miRNAs were identified and there was no significant difference in the number of miRNAs between all samples (Supplementary Figure 1).
Principal component analysis (PCA) is a common method used to reduce the dimensionality of large datasets, by transforming numerous variables into a smaller one that still contains most of the original information. To assess the correlations among the miRNA data, a PCA was performed for three replicates at each time point (Figure 2). The miRNA data were similar for the three replicates per treatment. Moreover, different treatments produced distinct results, suggesting that miRNA expression levels changed during an acute exposure to low-salinity stress.
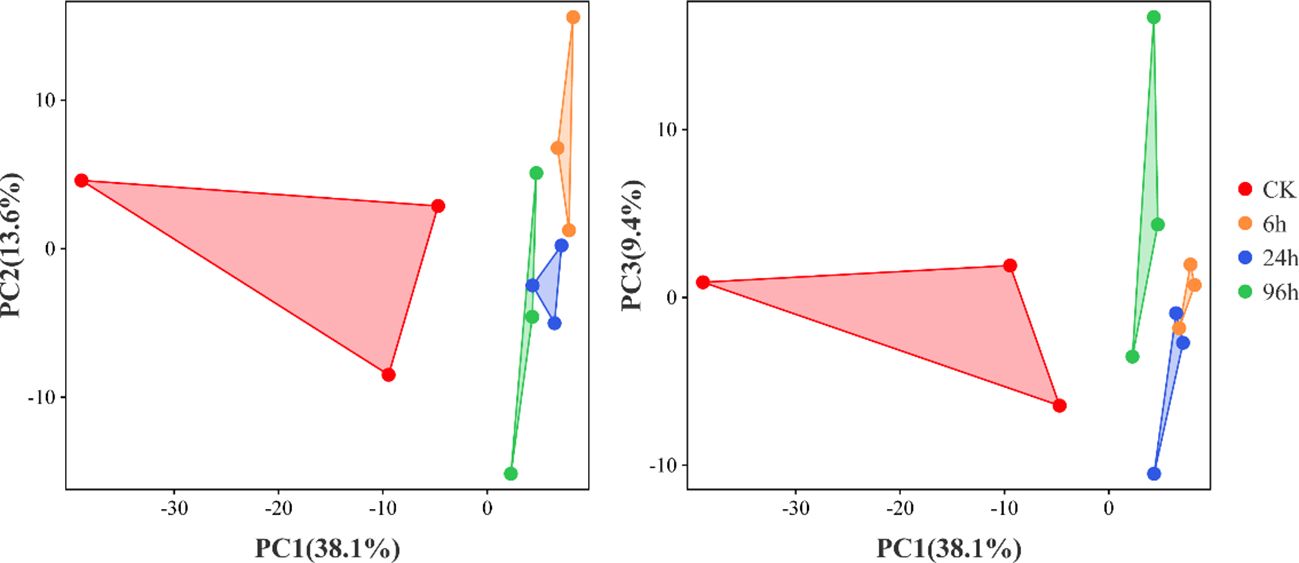
Figure 2 Principal component analysis of P. monodon miRNA data under low-salinity stress. Triangles with the same color represent the biological replicates for each treatment.
The miRNAs that were up- or downregulated under low-salinity treatments compared to the controls (CK) (log2|fold change| ≥ 1 and p-value <0.05) were determined as DEMs. In response to the 6h, 24h, and 96h low-salinity treatments, 9, 9, and 26 miRNAs exhibited upregulated expression, whereas 97, 66, and 97 miRNAs exhibited downregulated expression, respectively (Figure 3A). Venn diagram analysis of the DEMs revealed that different miRNAs were responsive to various low-salinity conditions, with 43 miRNAs being differentially expressed in response to all the treatments (Figure 3B).
Putative targets of the DEMs were predicted using the high-quality reference genome of P. monodon (GCF_015228065.2). For all 167 DEMs, a total of 5,967 transcripts were identified as targets (Supplementary Table 2), which means that miRNAs in P. monodon have more than one potential target gene. Further analysis was performed on 43 miRNAs differentially expressed at all treatments. Besides miR-124-3p, all the remaining miRNAs were downregulated (Supplementary Table 3). To gain an overview of the significantly enriched biological processes and pathways represented by the targets of these DEMs, GO and KEGG enrichment analysis were carried out. These putative targets were clustered into three GO terms, namely, biological processes, molecular functions, and cellular components. According to the FDR correction, anatomical structure morphogenesis (GO:0009653), multicellular organism development (GO:0007275), and nervous system development (GO:0007399) were recognized as the top GO biological process terms; protein binding (GO:0005515), phosphatidylinositol binding (GO:0035091) and phosphatidylinositol phosphate binding (GO:1901981) were the top terms in molecular functions; apicolateral plasma membrane (GO:0016327), main axon (GO:0044304), and spectrin-associated cytoskeleton (GO:0014731) were the top cellular component terms (Figure 4).
In addition, KEGG enrichment analysis showed that the target genes of DEMs were classified into several different categories, such as organismal systems, environmental information processing, cellular processes, and metabolism (Figure 5A). The top 20 significantly enriched pathways were also selected for drawing (Figure 5B). The most enriched pathways were mainly focused on immune response (chemokine signaling pathway, rap1 signaling pathway, hippo signaling pathway, and endocytosis), the nervous system (glutamatergic synapse, serotonergic synapse, GABAergic synapse, cholinergic synapse, axon guidance, axon regeneration, and synaptic vesicle cycle), environmental adaptation (oxytocin signaling pathway, relaxin signaling pathway, adrenergic signaling in cardiomyocytes, MAPK signaling pathway, and adherens junction), and the endocrine system (cortisol synthesis and secretion, parathyroid hormone synthesis, secretion and action, and retrograde endocannabinoid signaling).
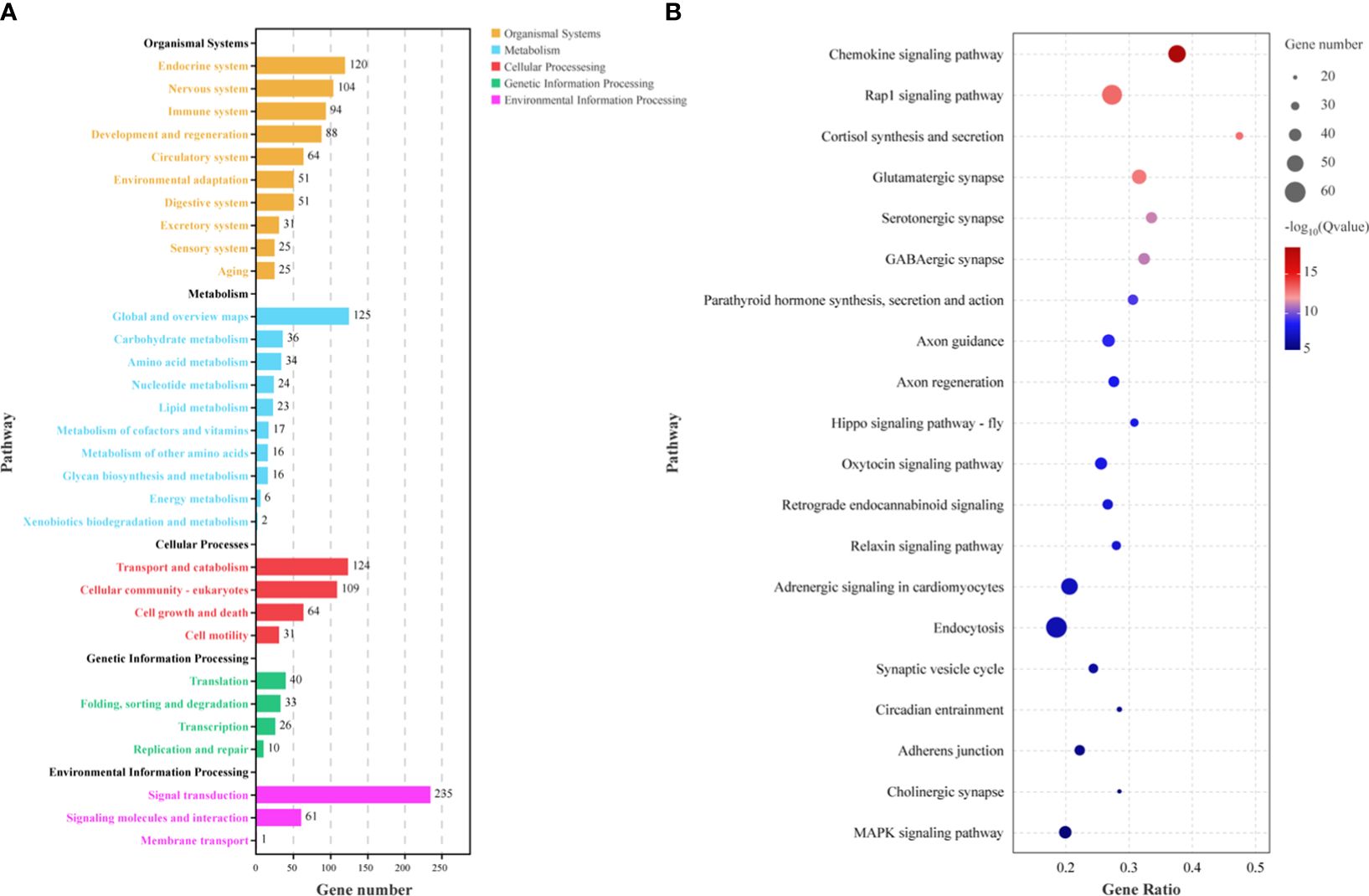
Figure 5 Pathway enrichment analysis of predicted target genes for differentially expressed miRNAs (A) and top 20 significantly enriched pathways (B).
To validate the accuracy of miRNA-seq data, expression levels of nine random miRNAs were analyzed by stem-loop qPCR. As shown in Figure 6, there was a good correlation between the transcript abundances based on the qPCR assay and the miRNA-seq data. The correlation coefficient of regression analysis between two approaches was calculated to be 0.8268 (p < 0.0001), indicating the reliability of high-throughput sequencing.
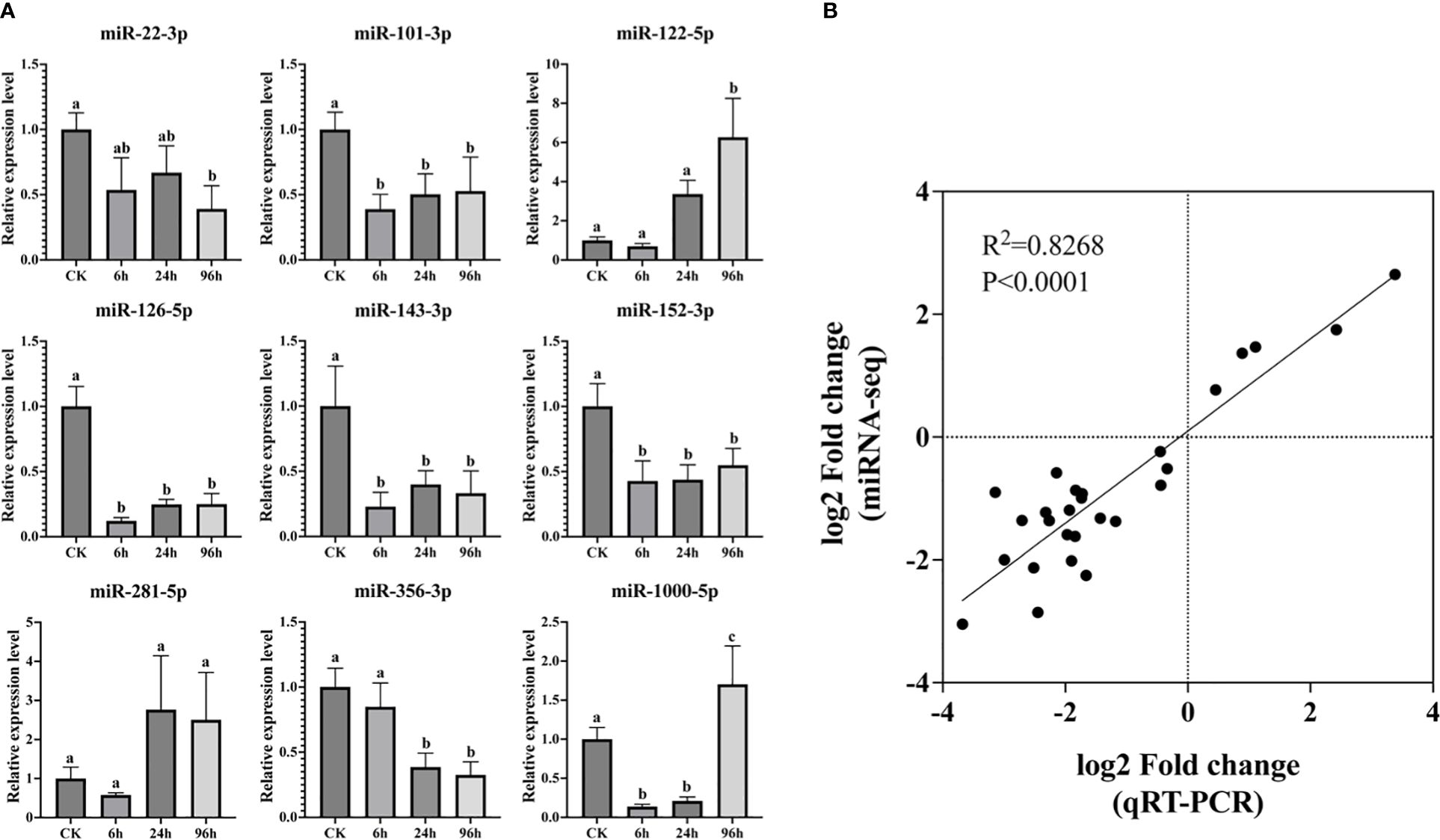
Figure 6 Relative expression level of nine miRNAs by qPCR assay. Bars with different letters indicate significant differences (A). Regression analysis between qPCR and miRNA-seq data (B).
Salinity is an important abiotic factor related to the growth, survival, and physiological function of shrimp (Rahi et al., 2021). Nevertheless, drought or heavy rain might alter the salinity of the aquatic water, restricting the productivity and sustainability of shrimp farming (Gao et al., 2012). Increasing scientific studies have been carried out to explain the molecular mechanism of low-salinity tolerance in shrimp (Fan et al., 2021; Li et al., 2023). Recently, considerable evidence has indicated that miRNAs play a vital role in response to abiotic stresses, such as hypoxia (Wang et al., 2019), heavy metal (Guo et al., 2018), pH (He et al., 2019), and virus (Kaewkascholkul et al., 2016). However, miRNAs participating in low-salinity tolerance are still limited in P. monodon. To gain global insights into the molecular mechanisms of P. monodon against low-salinity stress, we chose high-throughput sequencing technologies to identify miRNAs and target genes. A total of 167 DEMs were discovered following exposure to low-salinity conditions, the vast majority (~80%) of which were downregulated. In addition, 43 DEMs shared by all stress groups also showed downregulated expression except miR-124-3p. It has been proven that miRNAs could lead to the degradation or translational stagnation of target genes (Cai et al., 2009). The downregulation of DEMs might suggest that P. monodon could upregulate a variety of genes to survive when exposed to low-salinity stress. Similar results were also seen in Apostichopus japonicus, indicating that miRNAs are required to deal with environmental changes in salinity (Tian et al., 2019).
Enrichment analysis of putative target genes is an effective way to study the molecular mechanisms modulated by miRNAs under low-salinity stress. From KEGG results, pathways related to various synapses (glutamatergic synapse, serotonergic synapse, GABAergic synapse, and cholinergic synapse) and axon formation (axon guidance and axon regeneration) were found to be enriched. Combined with GO terms’ annotation, we can infer that the nervous system of P. monodon plays an important role in handling low-salinity stress. The crustacean nervous system consists basically of a brain, or supraesophageal ganglion, connected to a ventral nerve cord of ganglia, or nerve centers. A previous report implied that the nervous system could regulate many physiological processes to maintain the homeostasis of organism in both normal and stressful conditions (Ren et al., 2021). With the largest nervous system among invertebrates, penaeid shrimp showed that gene categories for neural development are significantly enriched for environment adaptation (Zhang et al., 2019). Similar results were also found in Litopenaeus vannamei, indicating that the neuroendocrine–immunoregulatory network plays a principal role in adapting to salinity changes (Zhao et al., 2016).
Chronic low salinity might weaken the immune system of shrimp, making it susceptible to pathogen infection. In this research, a series of pathways associated with immune response were significantly enriched by low-salinity treatment, such as chemokine signaling, rap1 signaling, hippo signaling, and endocytosis. The chemokine signal is transduced by chemokine receptors (G-protein-coupled receptors) expressed on the immune cells. As a component of innate immunity, the chemokine signaling pathway plays multiple roles in immune responses, which could help shrimps to survive under various stresses (Yu et al., 2019; Li et al., 2021). Rap1 and hippo signaling pathways are involved in the regulation of a range of immune functions. When Fenneropenaeus merguiensis are exposed to ammonia stress, differentially expressed genes were mainly related to immune response, with rap1 and hippo signaling pathway being over-represented (Wang et al., 2017). Endocytosis is the process by which cells take in substances engulfing a vesicle from the outside. These can include things like nutrients to support the cell or pathogens that immune cells engulf and destroy. Comparative proteome analysis of the hepatopancreas from L. vannamei showed that the protein related to endocytosis was significantly upregulated (Xu et al., 2017). These pathway changes indicate that various immune responses were activated to deal with acute low-salinity stress, which occurred in the hepatopancreas of P. monodon.
Facing environmental changes, the survival of each species depends on its ability to sense and respond to changes, which was called environmental adaptation. Five pathways that participated in environmental adaptation were found to be enriched, namely, oxytocin signaling, relaxin signaling, adrenergic signaling in cardiomyocytes, MAPK signaling, and adherens junction. The oxytocin signaling system could mediate a range of physiological functions that are important for metabolism, osmoregulation, memory, and learning (Liutkeviciute et al., 2016). From the genome of the kuruma shrimp, genes having a relation to the oxytocin signaling pathway were expanded, which might be related to environmental adaptation (Ren et al., 2022). As a polypeptide hormone, relaxin can reduce the accumulation of peroxide products and improve the antioxidant capacity of endothelial cells. In Eriocheir sinensis, the relaxin signaling pathway was activated under heat stress, which implied its essential role in reducing oxidative damage (Shen et al., 2023). For adrenergic signaling, the activation of this pathway could stimulate apoptosis. This activation was also detected in Oreochromis niloticus, showing a significant upregulation of the adrenergic signaling pathway under ammonia treatment (Mwaura et al., 2023). The MAPK signaling pathway can modulate several cellular physiological processes, leading to environmental adaptation. A previous study on P. monodon demonstrated that genes associated with the MAPK signaling pathway were upregulated to deal with low-salinity stress (Fan et al., 2021). Although there is no signal transduction function, adherens junction is also an important pathway for environmental adaptation. Together with tight junctions, adherens junctions form a regulated semipermeable diffusion barrier that allows the passive diffusion of certain ions and small hydrophilic molecules along concentration gradients and affected the osmotic equilibrium (Li et al., 2019b). From the above results, it can be inferred that P. monodon might activate a range of responses for environmental adaptation.
In crustaceans, the endocrine system forms a network along with the nervous system and the immune system, which plays a vital role in dealing with harsh environments. A typical stress response mediated by the endocrine system is the supply of energy. After perceiving the ammonia-N stress, L. vannamei can stimulate the release of cortisol (Si et al., 2019). As a stress hormone, the elevation of cortisol would promote gluconeogenesis to fit the energy demand for facing stress (Li et al., 2019a). In our study, the upregulation of cortisol synthesis and secretion also confirm this notion. Calcium is a necessary macro-element involved in many important physiological functions of crustaceans, including growth, exoskeleton mineralization, muscle contraction, osmoregulation and maintenance of cell integrity, and acid–base equilibrium (Xu et al., 2021). Secreted by the endocrine system, the parathyroid hormone is essential for the maintenance of calcium homeostasis. Therefore, synthesis of the parathyroid hormone might maintain the balance of calcium, contributing to osmoregulation and the growth of P. monodon. Endocannabinoids are key modulators of synaptic function. By activating cannabinoid receptors expressed in the central nervous system, these lipid messengers can regulate several neural functions and behaviors (Castillo et al., 2012). Emerging research proves that the endocannabinoid system is an integral regulator of the stress response (Lutz et al., 2015; Jia et al., 2021). Similar results were also found in this work, indicating the potential role of endocannabinoid signaling in shrimp under low salinity.
The miRNA sequencing files generated in this study have been deposited in the China Nucleic Acid Repository Database (CNGBdb, https://db.cngb.org/cnjb/) under accession CNP0005446.
The animal study was approved by Animal Care and Use Committee of the Chinese Academy of Fisheries Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
JS: Project administration, Visualization, Writing – original draft, Writing – review & editing. SJ: Investigation, Project administration, Writing – original draft. QY: Resources, Supervision, Writing – original draft. YL: Software, Validation, Writing – original draft. LY: Methodology, Software, Writing – original draft. JH: Data curation, Supervision, Writing – original draft. SGJ: Data curation, Supervision, Writing – original draft. FZ: Funding acquisition, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key R & D Program of China (2022YFD2400104), the Fund of Hainan Natural Science Foundation (322RC806), the Rural Revitalization Strategy Special Fund Seed Industry Revitalization Project of Guangdong Province (2022SPY00006 and 2022SPY00002), the China Agriculture Research System of MOF and MARA (CARS-48), the Central Public Interest Scientific Institution Basal Research Fund, CAFS (NO. 2023TD34), and the Central Public Interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (NO. 2023TS01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1403559/full#supplementary-material
Agarwal V., Bell G. W., Nam J.-W., Bartel D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005. doi: 10.7554/eLife.05005
Betel D., Koppal A., Agius P., Sander C., Leslie C. (2010). Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome. Biol. 11, R90. doi: 10.1186/gb-2010-11-8-r90
Cai Y., Yu X., Hu S., Yu J. (2009). A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinf. 7, 147–154. doi: 10.1016/S1672-0229(08)60044-3
Castillo P. E., Younts T. J., Chávez A. E., Hashimotodani Y. (2012). Endocannabinoid signaling and synaptic function. Neuron 76, 70–81. doi: 10.1016/j.neuron.2012.09.020
Fan H., Li Y., Yang Q., Jiang S., Yang L., Huang J., et al. (2021). Isolation and characterization of a MAPKK gene from Penaeus monodon in response to bacterial infection and low-salinity challenge. Aquacult. Rep. 20, 100671. doi: 10.1016/j.aqrep.2021.100671
Farhadi A., Cui W., Zheng H., Li S., Zhang Y., Ikhwanuddin M., et al. (2021). The regulatory mechanism of sexual development in decapod crustaceans. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.679687
Ferraris R. P., Parado-Estepa E. D., De Jesus E. G., Ladja J. M. (1987). Osmotic and chloride regulation in the hemolymph of the tiger prawn Penaeus monodon during molting in various salinities. Mar. Biol. 95, 377–385. doi: 10.1007/BF00409568
Gao W., Tan B., Mai K., Chi S., Liu H., Dong X., et al. (2012). Profiling of differentially expressed genes in hepatopancreas of white shrimp (Litopenaeus vannamei) exposed to long-term low salinity stress. Aquaculture 364–365, 186–191. doi: 10.1016/j.aquaculture.2012.08.024
Guo H., Lu Z., Zhu X., Zhu C., Wang C., Shen Y., et al. (2018). Differential expression of microRNAs in hemocytes from white shrimp Litopenaeus vannamei under copper stress. Fish Shellfish Immunol. 74, 152–161. doi: 10.1016/j.fsi.2017.12.053
Hao R., Zhu X., Tian C., Jiang M., Huang Y., Zhu C. (2022). Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus). Aquaculture 548, 737575. doi: 10.1016/j.aquaculture.2021.737575
He Y., Li Z., Zhang H., Hu S., Wang Q., Li J. (2019). Genome-wide identification of Chinese shrimp (Fenneropenaeus chinensis) microRNA responsive to low pH stress by deep sequencing. Cell Stress Chaperon. 24, 689–695. doi: 10.1007/s12192-019-00989-x
Jia R., Du J., Cao L., Feng W., He Q., Xu P., et al. (2021). Application of transcriptome analysis to understand the adverse effects of hydrogen peroxide exposure on brain function in common carp (Cyprinus carpio). Environ. pollut. 286, 117240. doi: 10.1016/j.envpol.2021.117240
Kaewkascholkul N., Somboonviwat K., Asakawa S., Hirono I., Tassanakajon A., Somboonwiwat K. (2016). Shrimp miRNAs regulate innate immune response against white spot syndrome virus infection. Dev. Comp. Immunol. 60, 191–201. doi: 10.1016/j.dci.2016.03.002
Kalvari I., Nawrocki E. P., Argasinska J., Quinones-Olvera N., Finn R. D., Bateman A., et al. (2018). Non-coding RNA analysis using the rfam database. Curr. Protoc. Bioinf. 62, e51. doi: 10.1002/cpbi.51
Kozomara A., Birgaoanu M., Griffiths-Jones S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Kruger J., Rehmsmeier M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454. doi: 10.1093/nar/gkl243
Li Z. S., Ma S., Shan H. W., Wang T., Xiao W. (2019a). Responses of hemocyanin and energy metabolism to acute nitrite stress in juveniles of the shrimp Litopenaeus vannamei. Ecotox. Environ. Safe. 186, 109753. doi: 10.1016/j.ecoenv.2019.109753
Li Y.-D., Si M.-R., Jiang S.-G., Yang Q.-B., Jiang S., Yang L.-S., et al. (2023). Transcriptome and molecular regulatory mechanisms analysis of gills in the black tiger shrimp Penaeus monodon under chronic low-salinity stress. Front. Physiol. 14. doi: 10.3389/fphys.2023.1118341
Li Z., Tang X., Li J., He Y. (2021). Comparative proteomic and transcriptomic analysis reveals high pH–induced expression signatures of Chinese shrimp Fenneropenaeus chinensis. Funct. Integr. Genomic. 21, 299–311. doi: 10.1007/s10142-021-00779-8
Li Z., Wang J., He Y., Hu S., Wang Q., Li J. (2019b). Comprehensive identification and profiling of Chinese shrimp (Fenneropenaeus chinensis) microRNAs in response to high pH stress using Hiseq2000 sequencing. Aquac. Res. 50, 3154–3162. doi: 10.1111/are.14269
Liu X., Jiang M., Wen S., Zhang K., Hong Y., Sun K., et al. (2023). Comparative miRNA-seq analysis revealed molecular mechanisms of red color formation in the early developmental stages of Plectropomus leopardus. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1321196
Liutkeviciute Z., Koehbach J., Eder T., Gil-Mansilla E., Gruber C. W. (2016). Global map of oxytocin/vasopressin-like neuropeptide signalling in insects. Sci. Rep. 6, 39177. doi: 10.1038/srep39177
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo Z., Yu Y., Zhang Q., Bao Z., Xiang J., Li F. (2022). Comparative transcriptome analysis reveals the adaptation mechanism to high salinity in litopenaeus vannamei. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.864338
Lutz B., Marsicano G., Maldonado R., Hillard C. J. (2015). The endocannabinoid system in guarding against fear, anxiety and stress. Nat. Rev. Neurosci. 16, 705–718. doi: 10.1038/nrn4036
Mackowiak S. D. (2011). Identification of Novel and Known miRNAs in Deep-Sequencing Data with miRDeep2. Curr. Protoc. Bioinf. 36, 12.10.1-12.10.15.. doi: 10.1002/0471250953.bi1210s36
Mondal D., Chakrabarty U., Dutta S., Mallik A., Mandal N. (2021). Identification and characterization of novel microRNAs in disease-resistant and disease-susceptible Penaeus monodon. Fish Shellfish Immunol. 119, 347–372. doi: 10.1016/j.fsi.2021.04.027
Mwaura J. G., Wekesa C., Ogutu P. A., Okoth P. (2023). Whole transcriptome analysis of differentially expressed genes in cultured nile tilapia (O. niloticus) subjected to chronic stress reveals signaling pathways associated with depressed growth. Genes 14, 795. doi: 10.3390/genes14040795
Rahi M., Azad K. N., Tabassum M., Irin H. H., Hossain K. S., Aziz D., et al. (2021). Effects of salinity on physiological, biochemical and gene expression parameters of black tiger shrimp (Penaeus monodon): potential for farming in low-salinity environments. Biology 10, 1220. doi: 10.3390/biology10121220
Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi: 10.1038/35002607
Ren X., Lv J., Liu M., Wang Q., Shao H., Liu P., et al. (2022). A chromosome-level genome of the kuruma shrimp (Marsupenaeus japonicus) provides insights into its evolution and cold-resistance mechanism. Genomics 114, 110373. doi: 10.1016/j.ygeno.2022.110373
Ren X., Wang Q., Shao H., Xu Y., Liu P., Li J. (2021). Effects of low temperature on shrimp and crab physiology, behavior, and growth: A review. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.746177
Saliminejad K., Khorram Khorshid H. R., Soleymani Fard S., Ghaffari S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 234, 5451–5465. doi: 10.1002/jcp.27486
Shen C., Feng G., Zhao F., Huang X., Wang M., Wang H. (2023). Integration of transcriptomics and proteomics analysis reveals the molecular mechanism of eriocheir sinensis gills exposed to heat stress. Antioxidants 12, 2020. doi: 10.3390/antiox12122020
Si M.-R., Li Y.-D., Jiang S.-G., Yang Q.-B., Jiang S., Yang L.-S., et al. (2022a). A CSDE1/Unr gene from Penaeus monodon: Molecular characterization, expression and association with tolerance to low salt stress. Aquaculture 561, 738660. doi: 10.1016/j.aquaculture.2022.738660
Si M.-R., Li Y.-D., Jiang S.-G., Yang Q.-B., Jiang S., Yang L.-S., et al. (2022b). Identification of multifunctionality of the PmE74 gene and development of SNPs associated with low salt tolerance in Penaeus monodon. Fish Shellfish Immunol. 128, 7–18. doi: 10.1016/j.fsi.2022.07.010
Si L., Pan L., Wang H., Zhang X. (2019). Ammonia-N exposure alters neurohormone levels in the hemolymph and mRNA abundance of neurohormone receptors and associated downstream factors in the gills of Litopenaeus vannamei. J. Exp. Biol. 222, jeb200204. doi: 10.1242/jeb.200204
Silva E., Calazans N., Soares M., Soares R., Peixoto S. (2010). Effect of salinity on survival, growth, food consumption and haemolymph osmolality of the pink shrimp Farfantepenaeus subtilis (Pérez-Farfante 1967). Aquaculture 306, 352–356. doi: 10.1016/j.aquaculture.2010.04.025
Tian Y., Shang Y., Guo R., Chang Y., Jiang Y. (2019). Salinity stress-induced differentially expressed miRNAs and target genes in sea cucumbers Apostichopus japonicus. Cell Stress Chaperon. 24, 719–733. doi: 10.1007/s12192-019-00996-y
Vogt G. (2019). Functional cytology of the hepatopancreas of decapod crustaceans. J. Morphol. 280, 1405–1444. doi: 10.1002/jmor.21040
Wang H., Wei H., Tang L., Lu J., Mu C., Wang C. (2018). Identification and characterization of miRNAs in the gills of the mud crab (Scylla paramamosain) in response to a sudden drop in salinity. BMC Genomics 19, 609. doi: 10.1186/s12864-018-4981-6
Wang W., Yang S., Wang C., Shi L., Guo H., Chan S. (2017). Gill transcriptomes reveal involvement of cytoskeleton remodeling and immune defense in ammonia stress response in the banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immunol. 71, 319–328. doi: 10.1016/j.fsi.2017.10.028
Wang W., Zhong P., Yi J.-Q., Xu A.-X., Lin W.-Y., Guo Z.-C., et al. (2019). Potential role for microRNA in facilitating physiological adaptation to hypoxia in the Pacific whiteleg shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 84, 361–369. doi: 10.1016/j.fsi.2018.09.079
Xu C., Li E., Liu Y., Wang X., Qin J. G., Chen L. (2017). Comparative proteome analysis of the hepatopancreas from the Pacific white shrimp Litopenaeus vannamei under long-term low salinity stress. J. Proteomics 162, 1–10. doi: 10.1016/j.jprot.2017.04.013
Xu L., Tian J., Wen H., Wu F., Zhang W., Gao W., et al. (2021). Dietary calcium requirement of red swamp crayfish (Procambarus clarkia). Aquacult. Nutr. 27, 153–162. doi: 10.1111/anu.13173
Yu J., Sun J., Zhao S., Wang H., Zeng Q. (2019). Transcriptome analysis of oriental river Prawn (Macrobrachium nipponense) Hepatopancreas in response to ammonia exposure. Fish Shellfish Immunol. 93, 223–231. doi: 10.1016/j.fsi.2019.07.036
Zhang X., Yuan J., Sun Y., Li S., Gao Y., Yu Y., et al. (2019). Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 10, 356. doi: 10.1038/s41467-018-08197-4
Zhao C., Fan S., Qiu L. (2018). Identification of microRNAs and their target genes associated with ovarian development in black tiger shrimp (Penaeus monodon) using high-throughput sequencing. Sci. Rep. 8, 11602. doi: 10.1038/s41598-018-29597-y
Keywords: low-salinity, high-throughput sequencing, microRNA, stress response, Penaeus monodon
Citation: Shi J, Jiang S, Yang Q, Li Y, Yang L, Huang J, Jiang S and Zhou F (2024) Identification of microRNAs in black tiger shrimp (Penaeus monodon) under acute low-salinity stress. Front. Mar. Sci. 11:1403559. doi: 10.3389/fmars.2024.1403559
Received: 19 March 2024; Accepted: 17 April 2024;
Published: 13 May 2024.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Ting Chen, Chinese Academy of Sciences (CAS), ChinaCopyright © 2024 Shi, Jiang, Yang, Li, Yang, Huang, Jiang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Falin Zhou, emhvdWZhbGluQGFsaXl1bi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.