- 1School of Life and Health Sciences, Hainan University, Haikou, China
- 2Department of Biology, Hong Kong Baptist University, Hong Kong, Hong Kong SAR, China
Mitochondrial genomes serve as valuable markers for phylogenetic and evolutionary studies across diverse invertebrate taxa, but their application within Annelida remains limited. In this study, we report the mitochondrial genomes of seven species from four genera of Chaetopteridae (Annelida), obtained by high-throughput sequencing. Phylogenetic analysis was performed using cox1, 18S, 28S and all mitochondrial genes. Our results reveal Chaetopterus and Mesochaetopterus as well-supported monophyletic sister clades, while Phyllochaetopterus and Spiochaetopterus appear paraphyletic, with species from both genera in a mixed clade sister to Chaetopterus + Mesochaetopterus. While mitochondrial gene orders remain conserved within Chaetopteridae, they appear substantially different from those of the ancestral patterns in Annelida. All 13 protein-coding genes found in Chaetopteridae evolved under strong purification selection, although Phyllochaetopterus exhibited the highest base-substitution rate for most of them, suggesting a more relaxed purified selection. Overall, our study provides molecular resources for phylogenetic studies of Chaetopteridae, highlighting the necessity for a comprehensive revision of the family, particularly dealing with the paraphyletic Phyllochaetopterus and Spiochaetopterus.
1 Introduction
Chaetopteridae is a small family of the phylum Annelida, whose species live in self-secreted membranous tubes and are commonly found in different habitats from the intertidal to the deep sea (Moore et al., 2017; Britayev and Martin, 2019; Rouse et al., 2022). While most of them live buried in soft sediment, some are attached to rocks, either living alone or in groups, and one species is holoplanktonic (Blake, 1996; Osborn et al., 2007; Nishi and Hsieh, 2009; Nishi et al., 2009). Chaetopterids measure from less than 1 cm to more than 40 cm and usually have less than 60 segments (Moore et al., 2017). Members of the family are easy to be identified by having bodies usually divided into three highly distinct regions. To date, Chaetopteridae contains 79 species belonging to four genera: Chaetopterus Cuvier, 1830, Mesochaetopterus Potts, 1914, Phyllochaetopterus Grube, 1863, and Spiochaetopterus Sars, 1856 (Read and Fauchald, 2023).
The phylogenetic position of Chaetopteridae in the tree of life of Annelida has not been stable over the last three decades. Based on morphology, they were considered a family within Spionida, together with the Spionidae, Poecilichaetidae, and Trochochaetidae, among others, with Trochochaetidae as its sister family (Rouse and Fauchald, 1997). In molecular phylogenetic analyses based on 18S rRNA, Chaetopterus variopedatus (Chaetopteridae) was recovered as sister to a clade containing two species of Brachiopoda and two of Phoronida, which altogether, were sister to Dodecaceria concharum (Spionidae) (McHugh, 2000). Later studies with more genes (Colgan et al., 2006; Rosset et al., 2007), including transcriptomes (Struck, 2011; Weigert et al., 2014) or mitochondrial genomes (mtgenome) (Weigert et al., 2016; Struck et al., 2023), usually recovered Chaetopteridae as one of the basal groups of Annelida. However, the relationships among the basal annelid groups (i.e., Amphinomidae, Magelonidae, Myzostomida, Oweniidae, Sipuncula) and between annelids and other lophotrochozoans (i.e., Nemertea, Brachiopoda, Mollusca) remained unstable.
Within Chaetopteridae, there have also been inconsistencies in the monophyly of the four traditionally recognized genera, as well as in the phylogenetic relationships among them. Chaetopterus and Mesochaetopterus were paraphyletic based on cox1 only (Morineaux et al., 2010; Zhang et al., 2015), but monophyletic based on the combined dataset of cox1 and 18S (Martin et al., 2022) and cox1, 18S and 28S genes (Zhang et al., 2015; Moore et al., 2017), while the same authors found Phyllochaetopterus and Spiochaetopterus as paraphyletic, forming a sister clade to Mesochaetopterus + Chaetopterus (Osborn et al., 2007; Zhang et al., 2015). Therefore, more taxon sampling within each chaetopterid genus and more genetic data for each species are required to provide a well-resolved phylogenetic tree of Chaetopteridae and to properly place this family in the annelid tree of life.
Several studies have utilized transcriptomes and mtgenomes to reconstruct the phylogeny of annelids. However, within Chaetopteridae, only one species each in Chaetopterus, Phyllochaetopterus, and Mesochaetopterus have a sequenced mtgenome (Weigert et al., 2016; Yang et al., 2022), which limits assessing its utility both in annelid phylogeny or mtgenome evolution, such as gene order rearrangement, gene duplication and loss, codon usage and mutation rate (Bernt et al., 2013b; Halanych, 2016; Li et al., 2019). The gene order of mitogenomes was initially considered conserved in Annelida, including in the deep-sea tube worm Riftia pachyptila Jones, 1981, which shows remarkable body structure changes, such as loss of the digestive tract (Jennings and Halanych, 2005). However, more and more annelid mtgenomes have been found to deviate from the ancestral annelid gene order arrangements, such as in Owenia fusiformis (Weigert et al., 2016), Serpulidae (Seixas et al., 2017; Sun et al., 2021), Syllidae (Aguado et al., 2015, 2016), and deep-sea polynoids (Zhang et al., 2018).
In this study, we conducted high-throughput low-coverage sequencing using the Illumina platform to obtain the mtgenomes of seven species of Chaetopteridae, attempting to contribute to a better understanding of their evolution within the family, as well as of phylogenetic relationships among the four genera.
2 Materials and methods
2.1 Sample collection and genome sequencing
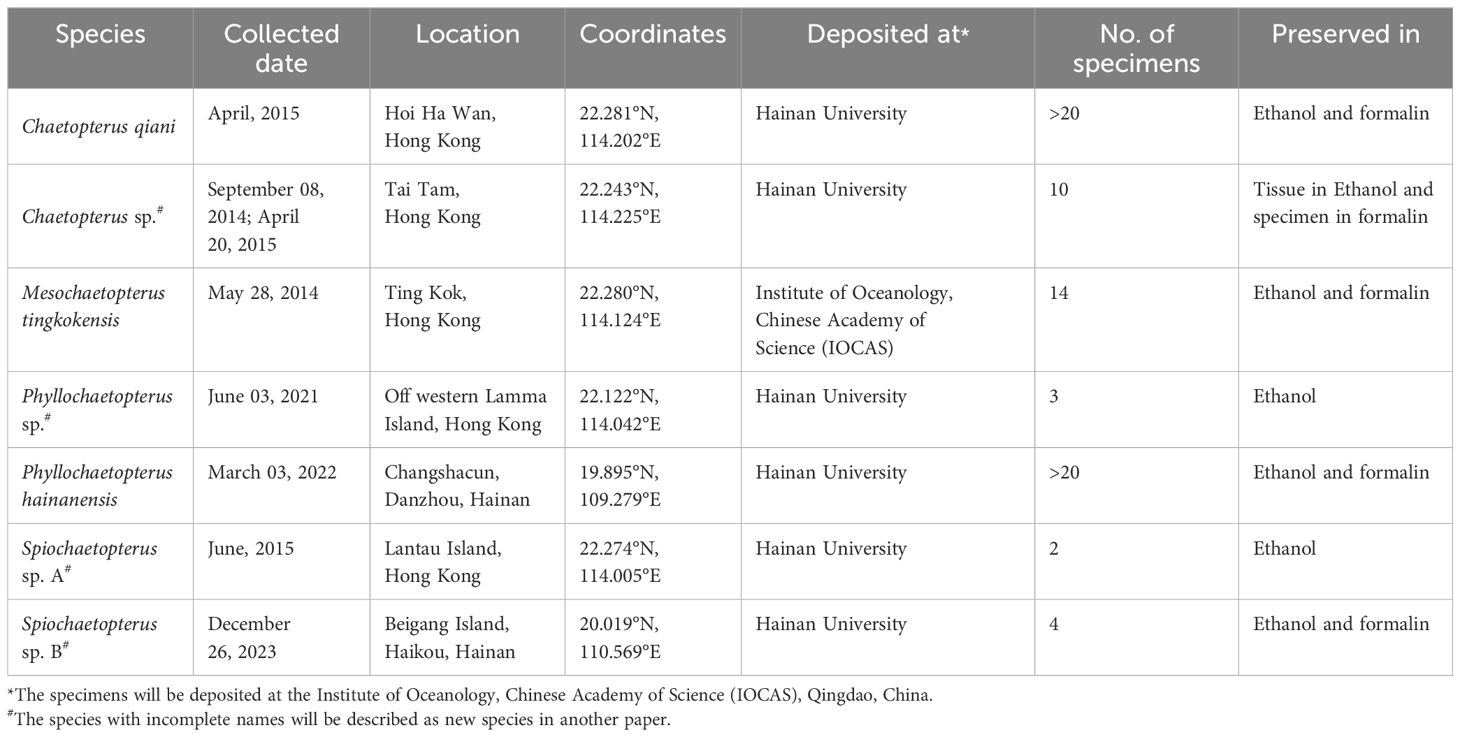
Specimens of Chaetopterus qiani Sun and Qiu, 2014, Chaetopterus sp., Phyllochaetopterus sp., Phyllochaetopterus hainanensis Wang and Li, 2017, Mesochaetopterus tingkokensis Zhang et al., 2015, Spiochaetopterus sp. A and Spiochaetopterus sp. B were collected from the intertidal and subtidal of Hong Kong and Hainan, China (Table 1, Figure 1) and preserved in 95% ethanol for DNA extraction or 4% formalin for morphological observations. Genomic DNA for each species was extracted using the CTAB method. Paired-end sequencing on the Illumina platform was performed at Novegene or Sangon to obtain 3.67 to 8.86 Gb 150 bp data (Supplementary Table 1).

Figure 1 Species sequenced in this study. (A), Spiochaetopterus sp. A; (B), Spiochaetopterus sp. B; (C), Mesochaetopterus tingkokensis; (D), Phyllochaetopterus sp.; (E), Phyllochaetopterus hainanensis; (F), Chaetopterus qiani; (G), Chaetopterus sp.
2.2 Mtgenome assembly
Assembly for all species except P. hainanensis and Spiochaetopterus sp. B was performed using CLC Genomics Workbench v7.03 (CLCbio, Arhus, Denmark). For P. hainanensis and Spiochaetopterus sp. B, SPAdes-3.15.4 (Prjibelski et al., 2020) was applied for contig assembly using multiple kmer settings (kmer = 21, 33, 55, 77, 99, 127), and NOVOPlasty v2.7.0 (Dierckxsens et al., 2017) and GetOrganelle (Jin et al., 2020) were run under default settings to build scaffolds, one of which was found to be the full mtgenome.
2.3 Mtgenome annotation
The chaetopterid mitochondrial genes or mtgenome or 18S/28S rRNA were detected using BLAST V2.13.0 (Camacho and Madden, 2013). The mitogenomes were annotated on the MITOS web server using genetic code 05 for invertebrates (Bernt et al., 2013c). The boundaries of protein-coding genes (PCGs) and rRNA genes were manually examined and adjusted based on alignment with published mtgenomes of this family. The GC-skew and AT-skew were defined according to Perna and Kocher (1995): AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C).
2.4 Phylogenetic analyses
A three-gene dataset (mitochondrial cox1, nuclear 18S and 28S rRNA genes) contained 50 species, including 41 published in Moore et al. (2017) and Britayev et al. (2017), seven sequenced in this study, and two as outgroups, Eurythoe complanata (Pallas, 1766) and Owenia fusiformis Delle Chiaje, 1844, were used in our phylogenetic analyses (Supplementary Table 2). The sequences of each gene were aligned using the MAFFT version 7 web server (Katoh et al., 2017), and concatenated using PhyloSuite (Zhang et al., 2020). Poorly arranged locations and very dispersive regions were removed using less stringent selection settings of Gblocks Server which include smaller final blocks, gap positions within the final blocks, and less strict flanking positions (Talavera and Castresana, 2007). The best nucleotide evolutionary model for each partition was selected based on the Akaike information criterion (AIC) (Darriba et al., 2012) of the PartitionFinder2 module in PhyloSuite. Phylogenetic analysis was performed using Maximum Likelihood (ML) and Bayesian inference (BI) were respectively executed using the IQ-TREE module in PhyloSuite with 10,000 Ultrafast Bootstrap (UFBoot) replicates with the SH-aLRT test (Minh et al., 2013) and the MrBayes module in PhyloSuite with 1000 Sampling Freq replicates.
Phylogenetic analyses were also conducted using all mitochondrial genes (13 PCGs, 2 rRNAs, and 22 tRNAs) for all mtgenomes, except in Spiochaetopterus sp. A which contained only 3 PCGs and 11 tRNAs. Eurythoe complanata (Pallas, 1766) (accession number KT726962.1) and Owenia fusiformis (Delle Chiaje, 1844) (accession number NC_028712.1) were used as the outgroups. The saturation of genetic sequences was assessed using DAMBE (Xia, 2018). The methods for sequence alignment, concatenations, removal of poorly aligned locations, model selection and phylogenetic tree construction were the same as those mentioned above.
To assess the phylogenetic placement of Chaetopteridae in Annelida, we used all protein-coding amino acid datasets of 240 species in Struck et al. (2023) and the seven here sequenced, with Terebratulina retusa (Linnaeus, 1758) as outgroup (Supplementary Table 2). The methods for model selection and ML phylogenetic tree construction were the same as the above, while BI was executed with two chains for 5,000,000 generations using Mpi-MrBayes v3.2 (Ronquist et al., 2012). In ML, a bootstrap >90 was considered strong clade support, 70–90 as moderate, and < 70 as weak support (Krenz et al., 2005). For BI, posterior probabilities > 0.95 were considered strong support (Jacobsen et al., 2010).
2.5 Mitochondrial gene order rearrangement
CREx2 was used to assess the mitogenomic rearrangement (Bernt et al., 2007), and the TreeRex (Bernt and Middendorf, 2011) allowing deducing the ancestral gene order of the inner nodes for Chaetopteridae and predicting the evolution process of present species based on the given phylogenetic tree. The parameters were set following the software recommendations: -s (strong consistency method), -w (weak consistency method), and -W (parsimonious weak consistency method).
2.6 Genetic distance and base substitution rates of mitochondrial genes
The pairwise genetic distance analyses of amino acid sequences of each mitochondrial PCG were conducted using MEGA X (Kumar et al., 2018). Base-substitution rates for the 13 PCGs of all chaetopterid mtgenomes were calculated followed by Sun et al. (2021). In short, the sequences for each gene were aligned using default parameters of the Muscle module within MEGA X. The non-synonymous to synonymous rate ratio (Ka/Ks) was then calculated using the YN method implemented in KaKs_Calculator 2.0, with Ka/Ks indicating the strength of the selective pressure as > 1 positive selection, = 1 neutral evolution and < 1, purified selection (Yang and Nielsen, 2000). Spiochaetopterus sp. A was excluded from this analysis due to its incomplete mtgenome.
3 Results
3.1 Chaetopterid mtgenomes
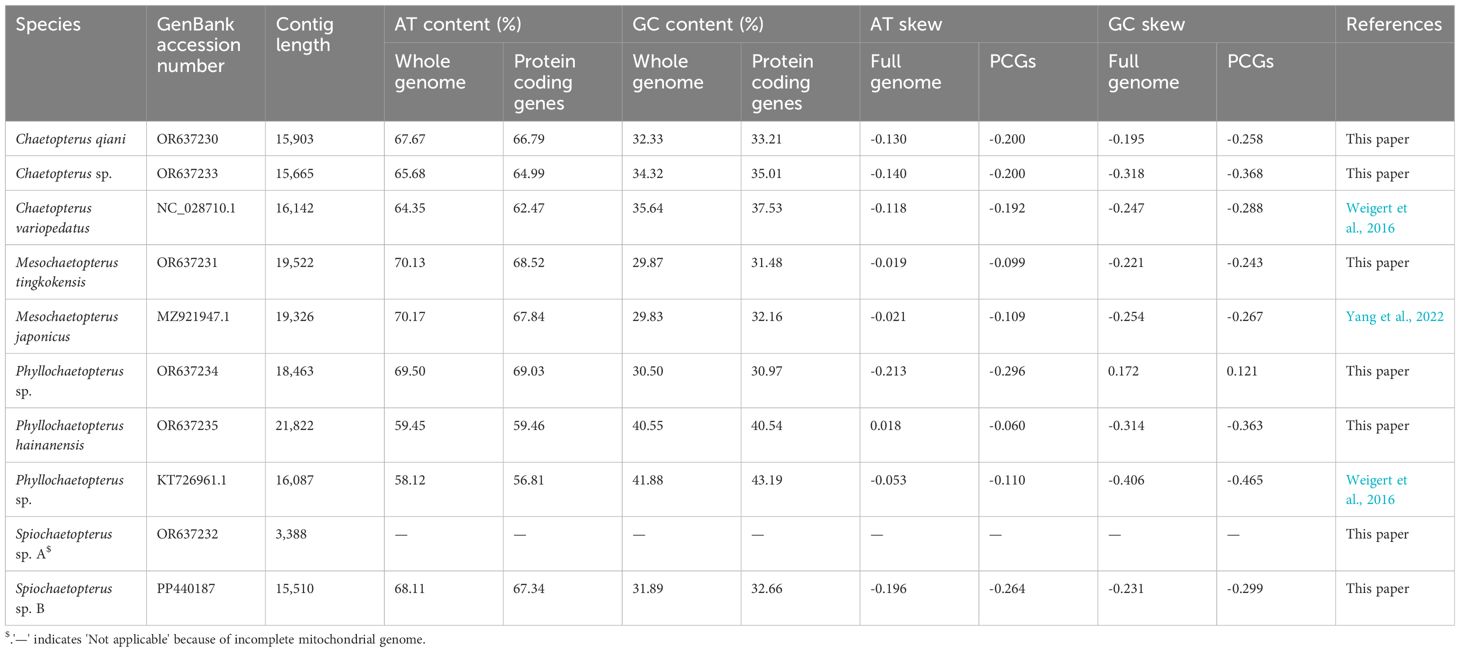
Sequence assembly resulted in six complete or nearly complete mtgenomes ranging from 15,665 bp to 21,822 bp (Table 1) except for Spiochaetopterus sp. A which resulted in a mitochondrial contig with 3,388 bp with only three potential PCGs (cox3, nad2, nad3) and 11 tRNAs (Figure 2; Supplementary Table 3). The start gene protein-coding codon for the species of Chaetopteridae was ATG, ATA and ATT, while the stop codon was either TAA, TAG, or defective TA or T (Supplementary Table 4).

Figure 2 Comparison of gene orders of chaetopterid mtgenomes. Conserved gene clusters of annelids are marked by different color bars. White circles represent non-coding regions with >100 bp sequences between genes. White blocks represent the location of control region in mtgenome. Triangles stand for a long non-coding region within a gene. Missing genes are indicated by black blocks.
All chaetopterid mtgenomes exhibit a high base bias and are AT-rich (58.2–70.17%), with only P. hainanensis (59.45%) and Phyllochaetopterus sp. (58.12%) having lower values but also close to 60% A+T (Table 2; Supplementary Table 5). The 13 PCGs (63.65%) and the third codon (63.45%) were also AT-rich. The full mtgenome and the PCGs showed negative AT-skew (-0.213–0.018, -0.296–0.06) and negative GC-skew (-0.318–0.172, -0.465–0.121) in all chaetopterid species except for the full mtgenome of P. hainanensis whose AT-skew (0.018) was greater than GC-skew (-0.314) (Supplementary Table 5). The tRNAs and rRNAs GC-skew are also negative, while their AT-skew were positive in all species except Phyllochaetopterus sp. (Supplementary Table 5).
3.2 Mtgenomes gene order
Three conserved gene blocks are present in all chaetopterids except Spiochaetopterus sp. B: rrnS-trnV-rrnL-trnL1-nad3-trnS1-nad2, trnI-nad6-trnG-atp6-trnQ-trnW, and cob-trnS2-nad4L-nad4-trnH-nad5, with those in Chaetopterus and Mesochaetopterus being identical except that the location of trnY and trnF (Figure 2). >100 bp non-coding regions between different genes occur in different mtgenome locations but show differences between genera (Figure 2; Supplementary Table 6): only 0–1 in Chaetopterus, 2–4 in Mesochaetopterus and Phyllochaetopterus, two in Spiochaetopterus sp. B and five in the incomplete mtgenome of Spiochaetopterus sp. A. In addition, the non-coding regions interrupted several PCGs: cox1 in most Mesochaetopterus and Chaetopterus spp., Phyllochaetopterus sp.; nad6 and nad2 in M. tingkokensis and M. japonicus.
3.3 Phylogenetic analysis based on cox1, 18S and 28S
Phylogenetic analysis with 48 ingroups was conducted based on concatenated fragments of cox1, 18S, and 28S with a total length of 3,179 bp, including 649 bp for cox1, 1,653 by for 18S, and 877 bp for 28S (Supplementary Table 7). Chaetopteridae appears as a well-supported clade with ML/BI = 100/1 (Figure 3). Chaetopterus and Mesochaetopterus are both monophyletic sister groups, while Phyllochaetopterus and Spiochaetopterus form a well-supported clade (ML/BI: 100/1) with two moderately-supported sub-clades (ML/BI/83/0.95; ML/BI: 85/#) including species from two genera so that both are paraphyletic.
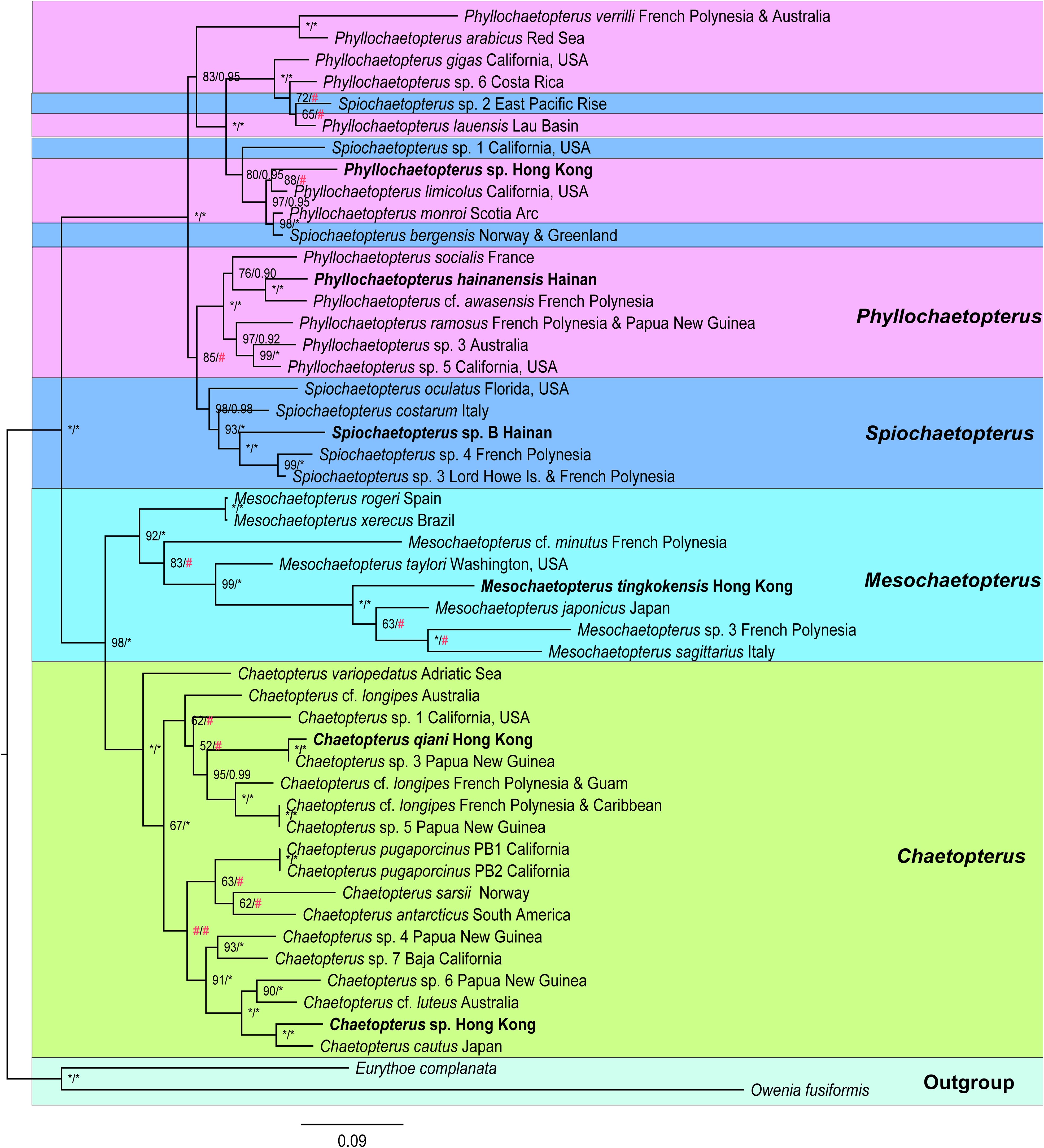
Figure 3 Phylogenetic tree of Chaetopteridae reconstructed based on cox1, 18S and 28S rRNA genes by ML/BI methods. The ML tree was displayed. The species newly sequenced for this study are in bold. Bootstrap support values of ML (left) and posterior probability of BI (right) are indicated above the nodes. Asterisks stand for maximum supports and pound keys stand for < 50 or unsupported values.
3.4 Phylogeny tree based on all mtgenome genes
The phylogenetic analyses were based on a data matrix containing 16,939 characters, including 9,900 bp from 13 PCGs, 2,687 bp from two rRNA genes and 1,630 bp from 22 tRNA genes (Figure 4; Supplementary Table 7). Five out of 13 PCGs showed significant saturation but were included because they contained phylogenetic information (Supplementary Table 8). The best models selected for the phylogenetic analyses were shown in Supplementary Table 9 and they produced similar tree topologies to those based on the three-gene dataset (Figure 4). All species of Chaetopteridae form a well-supported clade (ML/BI: 100/1) with two sub-clades, one with three species of Chaetopterus and two of Mesochaetopterus, both well supported (ML/BI: 100/1) consistently with the three-gene dataset results, in which Chaetopterus and Mesochaetopterus are also monophyletic. Another well-supported sub-clade consisted of species from Phyllochaetopterus and Spiochaetopterus, but this showed the three species of Phyllochaetopterus as paraphyletic with species of Spiochaetopterus and Phyllochaetopterus from the same location clustered together.
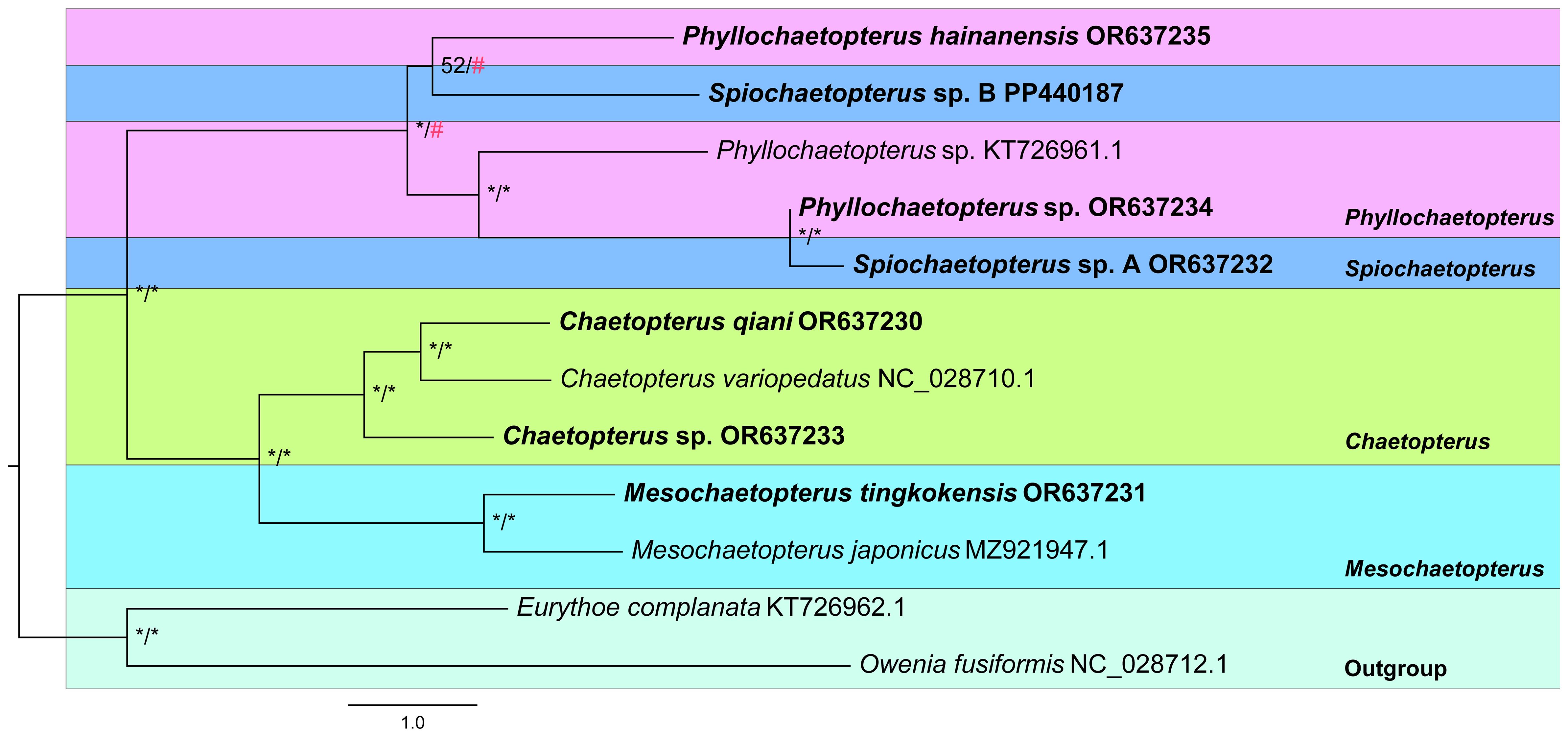
Figure 4 Phylogenetic tree of Chaetopteridae constructed based on all mitochondrial genes by ML/BI methods. The ML tree was displayed. The species newly sequenced for this study are in bold. Bootstrap support values of ML (left) and posterior probability of BI (right) are indicated above nodes. Asterisks stand for maximum supports and pound keys stand for unsupported values.
3.5 Mitochondrial gene order rearrangements
There were seven gene order patterns in the chaetopterid mtgenomes, including three in Chaetopterus and Mesochaetopterus, three in Phyllochaetopterus and one in Spiochaetopterus (Figure 5). The gene order distance ranges from 1 to 7 (Supplementary Table 10). Those between Chaetopterus and Mesochaetopterus are similar (1 to 2) but they are substantially larger than with Phyllochaetopterus (4 to 7).

Figure 5 Hypothetical ancestral gene order of mtgenomes for Chaetopteridae and gene order rearrangement scenarios. R= Reversion and T= transposition, with the related genes being marked by lines and triangles, respectively.
The gene order evolutionary patterns were predicted by TreeREx based on the phylogenetic tree (Figure 5; Supplementary Table 11). Reversions were detected in the most recent common ancestor of the M. tingkokensis-M. japonicus clade (trnE and trnF), in C. qiani (trnY and trnM), when compared with its most recent common ancestor with Chaetopterus. Two transpositions were detected in the most recent common ancestor of two Phyllochaetopterus sp. (OR637234 and KT726961.1) (trnN and trnK), and P. hainanensis and Spiochaetopterus sp. B (trnD and trnN), compared with its most recent hypothetical common ancestral gene order. One reversion (trnA-trnP and trnR-trnD)was detected in Phyllochaetopterus sp (KT726961.1) and two transpositions (trnC and trnM-rrnS) and one reversion (trnP and trnA) in Spiochaetopterus sp. B, compared with that of its most recent common ancestor.
3.6 Genetic divergence of chaetopterid mtgenome PCGs
Pairwise genetic distance analysis was performed using PCG amino acids. Cob, cox1, cox2 and cox3 exhibit low genetic divergence, with the lowest value in cox1 (0.014–0.283) and the highest in atp8 (0.255–0.804), followed by nad2 (0.363–0.712) (Figure 6). Compared to Chaetopterus and Mesochaetopterus, Phyllochaetopterus has a higher genetic divergence in several PCGs, including atp8, cob, cox1 and nad2.
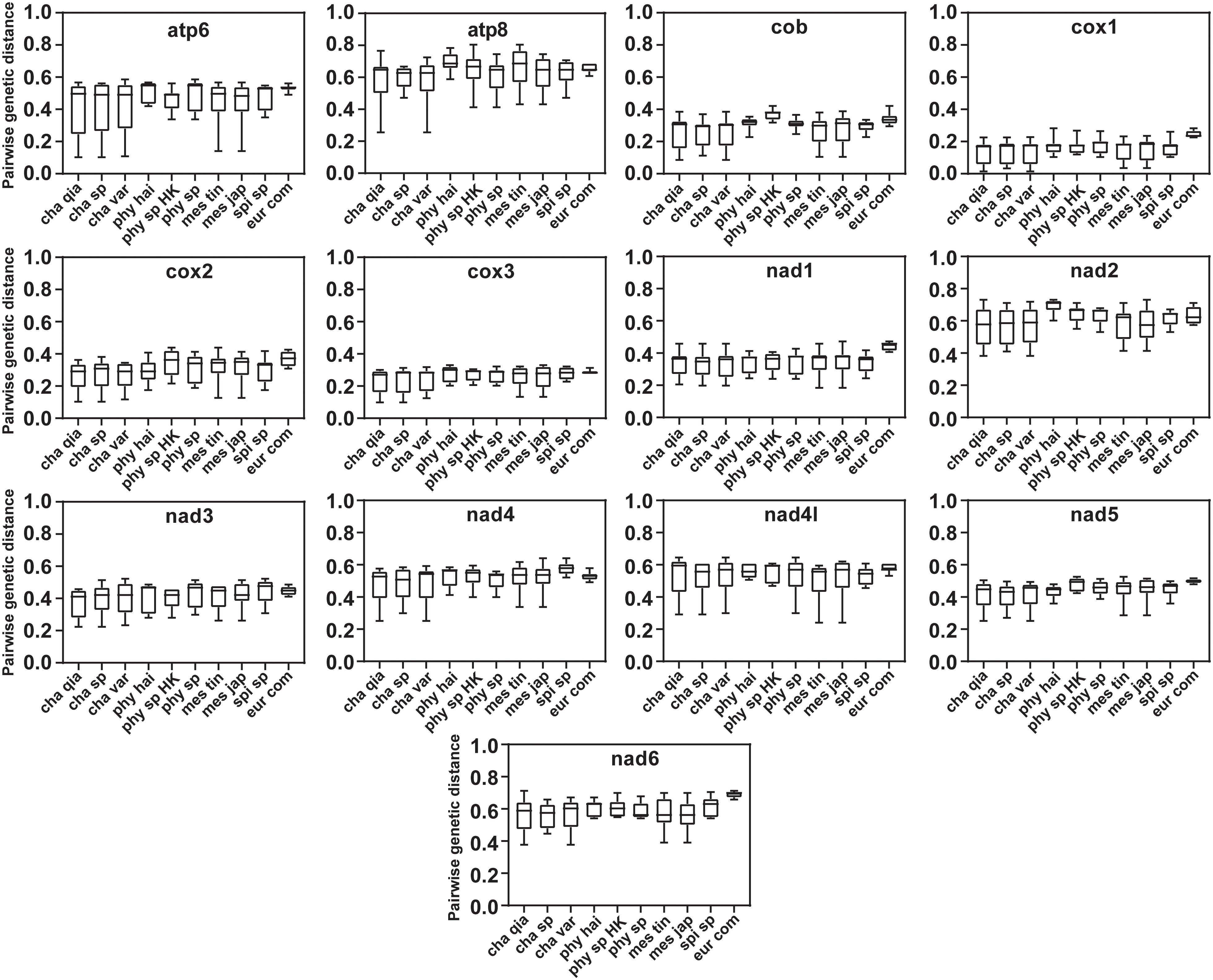
Figure 6 Pairwise genetic distances of mitochondrial protein-coding genes (amino acids) among species of Chaetopteridae. cha qia, Chaetopterus qiani; cha sp HK, Chaetopterus sp.; cha var, Chaetopterus variopedatus; phy hai, Phyllochaetopterus hainanensis; phy sp HK, Phyllochaetopterus sp.; phy sp, Phyllochaetopterus sp.; mes tin, Mesochaetopterus tingkokensis; mes jap, Mesochaetopterus japonicus; spi sp, Spiochaetopterus sp. B; eur com, Eurythoe complanata.
3.7 Purifying selection of 13 mitochondrial PCGs
The Ka/Ks of the 13 chaetopterid mitochondrial PCGs are all lower than 0.5, suggesting that they have undergone strong purification selection. Nevertheless, the Ka/Ks of atp8 (0.378) and nad4 (0.423) are higher than those of the other PCGs (Figure 7A), with the smallest in the complex IV (i.e. cox1 = 0.018, cox2 = 0.051, cox3 = 0.035) and nad5 (0.070) (Figure 7A). Compared with Chaetopterus and Mesochaetopterus, Ka/Ks was higher in Phyllochaetopterus for all mitochondrial PCGs except nad4 (Figure 7B), indicating a general relaxation of purifying selection in the mtgenomes of this genus.
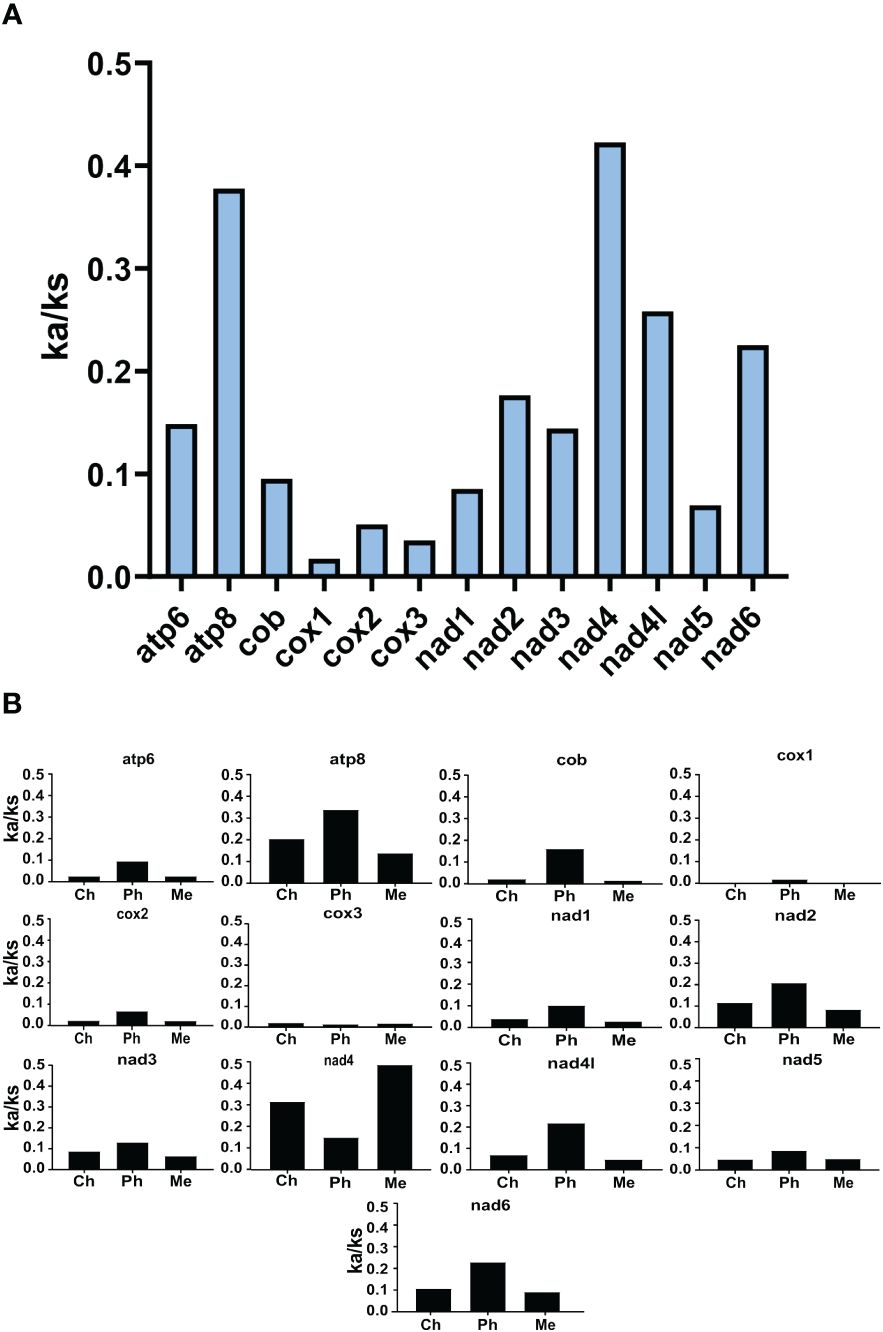
Figure 7 Base substitution rate of the 13 mitochondrial PCGs found in Chaetopteridae. (A) Ka/Ks for each mitochondrial gene. (B) Ka/Ks of each mitochondrial gene compared among different genera. Ka/Ks <1 indicates purification selection. Ch, Chaetopterus; Ph, Phyllochaetopterus; Me, Mesochaetopterus.
3.8 Phylogenetic position of Chaetopteridae in the annelid tree of life
The ML/BI phylogenetic tree was constructed using the concatenated datasets of the amino acid sequences of 13 PCGs of mitogenome, with 247 annelid terminals (Supplementary Table 2B, Supplementary Figure 1). Our results show the Oweniidae and Magelonidae as sister groups, forming a sister clade to Chaetopteridae + all other annelid families (ML/BI: 95/#).
4 Discussion
4.1 Structural features of the chaetopterid mtgenomes
The mitochondrial intergenic gaps of metazoans are usually composed of very short non-coding regions or very few overlapping bases (Zhong et al., 2008). Therefore, the length of metazoan mtgenome is generally stable, especially in vertebrates. Conversely, annelid mtgenomes vary substantially from 14kb (Aguado et al., 2016) to 25kb (Seixas et al., 2017; Sun et al., 2021), although chaetopterid mtgenomes show a smaller range from 15kb (Chaetopterus) to 21kb (Phyllochaetopterus). Longer annelid mtgenomes obey either an enormous control region (D-loop) (as in Spirobranchus and Siboglinum) or non-coding region within the cox1 gene (the group II intron) and many huge intergenic gaps (as in Glycera, Hydroides and Polynoidae). Longer mitogenomes generally contain more long non-coding regions and a non-conserved gene order, as in Hydroides spp. (Sun et al., 2021), Siboglinidae (Siboglinum fiordicum Webb, 1963, 19,502 bp; Li et al., 2015). However, there are some exceptions, such as among Glyceridae (Glycera unicornis Lamarck, 1818, 20,366 bp; Richter et al., 2015). In Phyllochaetopterus and Mesochaetopterus, large mitogenomes were either due to the group II introns among the cox1, to the non-coding region within the nad2, or numerous huge intergenic non-coding regions. Long non-coding regions were produced probably during changes in gene structure in Chaetopteridae, since intergenic non-coding spaces have been suggested to facilitate the inception of replication (Pons et al., 2014), or as remnants of gene order change (such as TDRL and transpositions) (Seixas et al., 2017), strands of the template mispairing, or imprecise termination during replication (Bernt et al., 2013b; Aguado et al., 2016).
The mtgenome gene orders of Chaetopteridae greatly differ from those of the ancestral annelid and most other annelids, although it is relatively conserved and shows three conserved gene blocks in most species (Figure 2). Among the ten mtgenomes, those of Chaetopterus and Mesochaetopterus show very similar gene orders, with only one reversion in the position of trnY and trnM or trnE and trnF. The Phyllochaetopterus and Spiochaetopterus mtgenomes are also very similar, with several unstable positions of tRNA (i.e. trnR, trnD, trnP, trnN, trnK, trnC) and a fragment of trnM-rrnS. These results are consistent with the close phylogenetic relationships between Chaetopterus and Mesochaetopterus, and between Spiochaetopterus and Phyllochaetopterus, respectively.
4.2 Phylogenetic relationships and pairwise genetic distance analysis
Our study shows the Chaetopteridae as a sister family to most annelid families, being recovered as sisters to the Oweniidae/Magelonidae clade. Transcriptome and mtgenome studies have all recovered Chaetopteridae as one of the basal annelid families, although its relationships with other basal families are unstable (Rousset et al., 2007; Weigert et al., 2016; Struck et al., 2023). Our ML result (i.e. ((Oweniidae + Magelonidae) + (Chaetopteridae + other annelid families)) agrees with the result in Struck et al. (2023), while the BI analyses did not favor these relationships (Struck et al., 2023). Our Chaetopteridae clade was well supported, in agreement with the previous results based on universal biomarker genes (cox1, 18S, 28S) (Martin et al., 2008; Osborn et al., 2007; Morineaux et al., 2010; Weigert et al., 2014; Andrade et al., 2015; Zhang et al., 2015; Britayev et al., 2017; Moore et al., 2017; Martin et al., 2022; Yang et al., 2022), while differing from morphological studies based on larvae and adults, which classified Chaetopteridae as a family within Sabellida (Rouse and Fauchald, 1997; Brown et al., 1999; Rouse, 1999). (Apistobranchidae + (Psammodrilidae + Chaetopteridae)) formed the clade of Chaetopteriformia based on phylogenetic analysis using transcriptomes (Helm et al., 2018), however, only Chaetopteridae was included in the later report on mtgenome evolution in Annelida (Struck et al., 2023). The mtgenomes of Apistobranchidae and Psammodrilidae are needed to further explore the ancestral mtgene order of Chaetopteridae considering their close evolutional relationship with Chaetotperidae.
The monophyly of Chaetopterus and Mesochaetopterus was not supported by morphology and BI based on 28S (Osborn et al., 2007; Morineaux et al., 2010). In this study, Chaetopterus appears as a well-supported sister taxon to Mesochaetopterus, in agreement with Moore et al. (2017); Martin et al. (2022) and Zhang et al. (2015). The paraphyletic relationship of Phyllochaetopterus + Spiochaetopterus is consistent with the results of the three scholars above. Major revisions are needed to define more distinguishing characteristics of these two genera instead of the present/absent of notopodial cirrus of A1.
Among the PCGs, atp8 has the highest pairwise genetic distance (Figure 6), followed by nad2 and nad6, suggesting faster substitution rates compared to other PCGs, as previously shown in deep-sea Polynoidae (Zhang et al., 2018), in which atp8 is the fastest-evolving mitochondrial gene (Sun et al., 2021). Additionally, the PCG genetic distance in Phyllochaetopterus was generally greater than that of the other genera, suggesting different selective pressures and faster mutation rates in its mitogenomes. In addition, the branch length of the Phyllochaetopterus + Spiochaetopterus clade was longer than that of the Chaetopterus + Mesochaetopterus clade (Figures 3, 4), indicating a faster substitution rate in the former.
4.3 Mtgenome rearrangement
Chaetopteridae exhibits distinct mtgenome gene order arrangement patterns compared to other annelids. Gene orders were initially considered as conserved among annelid mtgenomes (Jennings and Halanych, 2005), which is still true for many families (Weigert et al., 2016). However, substantial gene order arrangements have been found among Syllidae (Aguado et al., 2015, 2016), deep-sea Polynoidae (Zhang et al., 2018) and Serpulidae (Sun et al., 2021). Chaetopteridae shows different mtgenomes than the ancestral annelids (excluding Oweniidae and Magelonidae), mainly in the atp6, nad1, nad6, cox3, cox1, and several tRNAs positions (Supplementary Figure 2). However, its gene order is relatively conserved, with three conserved regions in all species except Spiochaetopterus (Figure 2). The uncertain evolutional relationships between Chaetopteridae and all other polychaetes, the gene order of its most recent ancestor cannot be determined. Therefore, it is not clear how the mitochondrial gene order of the common ancestor of Chaetopterus and Mesochaetopterus or Phyllochaetopterus and Spiochaetopterus evolved from that of the chaetopterid common ancestor.
4.4 Base-substitution rates of mtgenomes
The diversity of gene orders is positively related to the substitution rate (Bernt et al., 2013a, b; Luo et al., 2015). The conserved functions in the respiration of mitochondrial genes explain why they are undergoing purifying selection (Stewart et al., 2008). Our study shows the Ka/Ks for all chaetopterid protein-coding genes as being lower than 1, indicating purifying selection (Figure 7), while being quite fast evolving compared to other polychaete (Zhang et al., 2018) or invertebrates (Ren et al., 2010). Phyllochaetopterus shows higher base-substitute rates for most mitochondrial PCGs than Chaetopterus or Mesochaetopterus, indicating faster mutation rate that is consistent with their longer branch length in phylogenetic trees. Different mutation rates have also been shown among Polynoidae, with the deep-sea genera Branchipolynoe and Branchinotogluma, having high mutation rates that suggest adaptation values (Zhang et al., 2018).
Overall, our results show different mitochondrial gene orders in Chaetopteridae compared to the conserved Annelida pattern, despite most species in this family sharing three singular conserved regions. Chaetopterus and Mesochaetopterus are sister groups forming well-supported monophyletic clades, which, together, are sister to a paraphyletic Phyllochaetopterus clade with Spiochaetopterus as subclade. The mitochondrial PCG genes in all examined chaetopterids have undergone purifying selection, but in most cases, those of Phyllochaetopterus show a higher base-substitution rate than those of the other genera.
Data availability statement
Sequence data of the mitochondrial genomes first reported in this study are available in the NCBI GenBank OR637230 - OR63235, PP440187.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XW: Writing – review & editing, Writing – original draft, Visualization, Formal analysis, Data curation. XS: Writing – review & editing, Visualization, Methodology. YW: Writing – review & editing, Visualization, Methodology. CH: Writing – review & editing, Visualization, Methodology. J-WQ: Writing – review & editing, Writing – original draft, Conceptualization. YZ: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (42106118), Hainan Province Science and Technology Special Fund (ZDYF2024SHFZ101) and the Hainan University Research Start-up Fund Project (KYQD(ZR)-21144).
Acknowledgments
The authors would like to thank Prof. Jin Sun from Ocean University of China, and Deyuan Yang from Xiamen University for their guidance on sequence processing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1382212/full#supplementary-material
References
Aguado M. T., Glasby C. J., Schroeder P. C., Weigert A., Bleidorn C. (2015). The making of a branching annelid: an analysis of complete mitochondrial genome and ribosomal data of Ramisyllis multicaudata. Sci. Rep. 17, 12072. doi: 10.1038/srep12072
Aguado M. T., Richter S., Sontowski R., Golombek A., Struck T. H., Bleidorn C. (2016). Syllidae mitochondrial gene order is unusually variable for Annelida. Gene 594 1, 89–96. doi: 10.1016/j.gene.2016.08.050
Andrade S. C. S., Novo M., Kawauchi G. Y., Worsaae K., Pleijel F., Giribet G., et al. (2015). Articulating “archiannelids”: Phylogenomics and annelid relationships, with emphasis on meiofaunal taxa. Mol. Biol. Evol. 32, 2860–2875. doi: 10.1093/molbev/msv157
Bernt M., Bleidorn C., Braband A., Dambach J., Donath A., Fritzsch G., et al. (2013a). A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol. Phylogenet. Evol. 69, 352–364. doi: 10.1016/j.ympev.2013.05.002
Bernt M., Braband A., Schierwater B., Stadler P. F. (2013b). Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 69, 328–338. doi: 10.1016/j.ympev.2012.10.020
Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., et al. (2013c). MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Bernt M., Merkle D., Ramsch K., Fritzsch G., Perseke M., Bernhard D., et al. (2007). CREx: inferring genomic rearrangements based on common intervals. Bioinformatics 23, 2957–2958. doi: 10.1093/bioinformatics/btm468
Bernt M., Middendorf M. (2011). A method for computing an inventory of metazoan mitochondrial gene order rearrangements. BMC Bioinform. 12, S6. doi: 10.1186/1471-2105-12-S9-S6
Blake J. A. (1996). “Family Chaetopteridae Malmgren 1867,” in The Annelida. Part3 Po/chaeta: Orbiniidae to Cossurdae, vol. 6 . Eds. Blake J. A., Hilbig B., Scott P. H. (Santa Barbara Museum of Natural History, Santa Barbara), 233–252.
Britayev T. A., Martin D. (2019). “Chapter 5.3. Chaetopteridae Audouin & Milne Edwards 1833,” in Volume 1: Annelida basal groups and Pleistoannelida, Sedentaria; in: Purschke, G., Böggermann, M., Westheide, W. (Eds.), Annelida; in: Schmidt-Rhaesa, A. (Ed.), Handbook of Zoology. Eds. Purschke G., Böggermann M., Westheide W. (De Gruyter, Berlin, Boston), 156–174.
Britayev T. A., Mekhova E., Deart Y., Martin D. (2017). Do syntopic host species harbour similar symbiotic communities? The case of Chaetopterus spp. (Annelida: Chaetopteridae). PeerJ 5, e2930. doi: 10.7717/peerj.2930
Brown S., Rouse G. W., Hutchings P. A., Colgan D. J. (1999). Assessing the usefulness of histone H3, U2 snRNA and 28S rDNA in analyses of polychaete relationships. Aust. J. Zool. 47, 499–516. doi: 10.1071/ZO99026
Camacho C., Madden T. (2013). “BLAST+ Release Notes. Mar 12 [Updated 2023 Apr 24],” in BLAST® Help (National Center for Biotechnology Information (US). 2008-, Bethesda (MD). Available at: https://www.ncbi.nlm.nih.gov/books/NBK131777/.
Colgan D. J., Hutchings P. A., Braune M. (2006). A multigene framework for polychaete phylogenetic studies. Organisms. Diversity Evol. 6, 220–235. doi: 10.1016/j.ode.2005.11.002
Cuvier G. (1830). Le règne animal distribué d’apres son organisation, pour servir de base à l’histoire naturelle des animaux et d’introduction a l’anatomie comparée, Volume 3 [avec Polychaeta]. 2nd edition (Paris: Déterville et Crochard), 504 pp.
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9, 772–772. doi: 10.1038/nmeth.2109
Delle Chiaje S. (1844). Descrizione e notomia degli animali invertebrati della Sicilia citeriore osservati vivi negli anni 1822–1830 (Naples: Batteli & Co.), 1–8.
Dierckxsens N., Mardulyn P., Smits G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Grube E. (1863). Beschreibung neuer oder wenig bekannter Anneliden. Archiv. für. Naturgeschichte. 29, 37–69. doi: 10.5962/bhl.part.9306
Halanych K. M. (2016). How our view of animal phylogeny was reshaped by molecular approaches. Org. Divers. Evol. 16, 319–218. doi: 10.1007/s13127-016-0264-8
Helm C., Beckers P., Bartolomaeus T., Drukewitz S., Kourtesis I., Weigert A., et al. (2018). Convergent evolution of the ladder-like ventral nerve cord in Annelida. Front. Zool. 15, 36. doi: 10.1186/s12983-018-0280-y
Jacobsen F., Friedman N. R., Omland K. E. (2010). Congruence between nuclear and mitochondrial DNA: combination of multiple nuclear introns resolves a well-supported phylogeny of New World orioles (Icterus). Mol. Phylogenet. Evol. 56, 419–427. doi: 10.1016/j.ympev.2010.03.035
Jennings R. M., Halanych K. (2005). Mitochondrial genomes of Clymenella torquata (Maldanidae) and Riftia pachyptila (Siboglinidae): Evidence for conserved gene order in Annelida. Mol. Biol. Evol. 2, 210–222. doi: 10.1093/molbev/msi008
Jin J. J., Yu W. B., Yang J. B., Song Y., DePamphilis C. W., Yi T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 1-31. doi: 10.1186/s13059-020-02154-5
Jones M. L. (1981). Riftia pachyptila, new genus, new species, the vestimentiferan worm from the Galápagos Rift geothermal vents. Proc. Biol. Soc Wash. 93, 1295–1313.
Katoh K., Rozewicki J., Yamada K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Krenz J. G., Naylor G. J., Shaffffer H. B., Janzen F. J. (2005). Molecular phylogenetics and evolution of turtles. Mol. Phylogenet. Evol. 37, 178–191. doi: 10.1016/j.ympev.2005.04.027
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lamarck J. B. (1818). [volume 5 of] Histoire naturelle des Animaux sans Vertèbres, préséntant les caractères généraux et particuliers de ces animaux, leur distribution, leurs classes, leurs familles, leurs genres, et la citation des principales espèces qui s’y rapportent; precedes d’une Introduction offrant la determination des caracteres essentiels de l’Animal, sa distinction du vegetal et desautres corps naturels, enfin, l’Exposition des Principes fondamentaux de la Zoologie. Paris. Deterville. 5, 612.
Li Y., Kocot K. M., Schander C., Santos S. R., Thornhill D. J., Halanych K. M. (2015). Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol. Phylogenet. Evol. 85, 221–229. doi: 10.1016/j.ympev.2015.02.008
Li Y., Kocot K. M., Tassia M. G., Cannon J. T., Bernt M., Halanych K. M. (2019). Mitogenomics reveals a novel genetic code in Hemichordata. Genome Biol. Evol. 11, 29–41. doi: 10.1093/gbe/evy254
Linnaeus C. (1758). Systema Naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio. Decima. Reformata. 1, 824.
Luo Y. J., Satoh N., Endo K. (2015). Mitochondrial gene order variation in the brachiopod Lingula anatina and its implications for mitochondrial evolution in lophotrochozoans. Mar. Genomic. 24, 31–40. doi: 10.1016/j.margen.2015.08.005
Martin D., Mecca M., Meca M. A., van Moorsel G., Romano C. (2022). Citizen science and integrative taxonomy reveal a great diversity within Caribbean Chaetopteridae (Annelida), with the description of one new species. Invertebr. Syst. 36, 581–607. doi: 10.1071/IS21081
Martin J., Gil J., Carreras-Carbonell J. (2008). Description of a new species of Mesochaetopterus (Annelida, Polychaeta, Chaetopteridae), with re-description of M. xerecus and an approach to the phylogeny of the family. Zool. J. Linn. Soc 152, 201–225. doi: 10.1111/j.1096-3642.2007.00342.x
McHugh D. (2000). Molecular phylogeny of the annelida. Can. J. Zool. 78, 1873–1884. doi: 10.1139/z00-141
Minh B. Q., Nguyen M. A. T., von Haeseler A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Moore J. M., Nishi E., Rouse G. W. (2017). Phylogenetic analyses of chaetopteridae (Annelida). Zool. Scr. 46, 596–610. doi: 10.1111/zsc.12238
Morineaux M., Nishi E., Ormos A., Mouchel O. (2010). A new species of Phyllochaetopterus (Annelida: Chaetopteridae) from deep-sea hydrothermal Ashadze-1 vent Fifield, Mid-Atlantic Ridge: taxonomical description and partial COI DNA sequence. Cah. Biol. Mar. 51, 239–248.
Nishi E., Hickman Jr. C. P., Bailey-Brock J. H. (2009). Chaetopterus and Mesochaetopterus (Polychaeta: Chaetopteridae) from the Galapagos Islands, with descriptions of four new species. Proc. Acad. Nat. Sci. 158, 239–259.
Nishi E., Hsieh H. L. (2009). Chaetopterid polychaetes from Taiwan and Okinawa Island (Japan), with descriptions of two new species. Zool. Stud. 48, 370–379.
Osborn K. J., Rouse G. W., Goffredi S. K., Robison B. H. (2007). Description and relationships of Chaetopterus pugaporcinus, an unusual pelagic polychaete (Annelida, Chaetopteridae). Biol. Bull. 212, 40–54. doi: 10.2307/25066579
Pallas P. S. (1766). Miscellanea zoologica. Quibus novae imprimis atque obscurae animalium species describuntur et observationibus iconibusque illustrantur. Petrum van Cleef. Hagí Comitum. xii 224, 14.
Perna N.T., Kocher T.D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41, 353–358.
Pons J., Bauzà-Ribot M. M., Jaume D., Juan C. (2014). Next-generation sequencing, phylogenetic signal and comparative mitogenomic analyses in Metacrangonyctidae (Amphipoda: Crustacea). BMC Genom. 15, 566. doi: 10.1186/1471-2164-15-566
Potts F. A. (1914). Polychaeta from the N. E. Pacific: the Chaetopteridae. With an account of the phenomenon of asexual reproduction in Phyllochaetopterus and the description of two new species of Chaetopteridae from the Atlantic. Proc. Zool. Soc Lond. 67, 955–994.
Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 70, e102. doi: 10.1002/cpbi.102
Read G., Fauchald K. (2023). World polychaeta Database. Chaetopteridae Audouin & Milne Edwards 1833 (World Register of Marine Species). Available at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=918.
Ren J., Liu X., Jiang F., Guo X., Liu B. (2010). Unusual conservation of mitochondrial gene order in Crassostrea oysters: evidence for recent speciation in Asia. BMC Evol. Biol. 10, 1–14. doi: 10.1186/1471-2148-10-394
Richter S., Schwarz F., Hering L., B¨oggemann M., Bleidorn C. (2015). The utility of genome skimming for phylogenomic analyses as demonstrated for glycerid relationships (Annelida, Glyceridae). Genome Biol. Evol. 7, 3443–3462. doi: 10.1093/gbe/evv224
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rouse G. W. (1999). Trochophore concepts: ciliary bands and the evolution of larvae in spiralian Metazoa. Biol. J. Linn. Soc 66, 411–464. doi: 10.1111/bij.1999.66.issue-4
Rouse G. W., Fauchald K. (1997). Cladistics and polychaetes. Zool. Scr. 26, 139–204. doi: 10.1111/j.1463-6409.1997.tb00412.x
Rouse G. W., Pleijel F., Tilic E. (2022). Annelida (Oxford: Oxford University Press). doi: 10.1093/oso/9780199692309.001.0001
Rousset V., Pleijel F., Rouse G. W., Erséus C., Siddall M. E. (2007). A molecular phylogeny of annelids. Cladistics 23, 41–63. doi: 10.1111/j.1096-0031.2006.00128.x
Seixas V. C., Russo C. A. M., Paiva P. C. (2017). Mitochondrial genome of the Christmas tree worm Spirobranchus giganteus (Annelida: Serpulidae) reveals a high substitution rate among annelids. Gene 605, 43–53. doi: 10.1016/j.gene.2016.12.024
Stewart J. B., Freyer C., Elson J. L., Wredenberg A., Cansu Z., Trifunovic A., et al. (2008). Strong purifying selection in transmission of mammalian mitochondrial DNA. PloS Biol. 6, e10. doi: 10.1371/journal.pbio.0060010
Struck T. H. (2011). Direction of evolution within Annelida and the definition of Pleistoannelida. J. Zool. 49, 340–345. doi: 10.1111/j.1439-0469.2011.00640.x
Struck T. H., Golombek A., Hoesel C., Dimitrov D., Elgetany A. H. (2023). Mitochondrial genome evolution in Annelida-A systematic study on conservative and variable gene orders and the factors influencing its evolution. Syst. Biol. 72, 925–945. doi: 10.1093/sysbio/syad023
Sun Y., Daffe G., Zhang Y., Pons J., Qiu J. W., Kupriyanova E. K. (2021). Another blow to the conserved gene order in Annelida: Evidence from mitochondrial genomes of the calcareous tubeworm genus Hydroides. Mol. Phylogenet. Evol. 160, 107–124. doi: 10.1016/j.ympev.2021.107124
Sun Y., Qiu J. W. (2014). A new species of Chaetopterus (Annelida, Chaetopteridae) from Hong Kong. Mem. Mus. Vic. 71, 303–309. doi: 10.24199/j.mmv.2014.71.23
Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Wang Y., Li X. (2017). A new species of Phyllochaetopterus Grube 1863 (Polychaeta: chaetopteridae) from hainan island, South China sea. Chin. J. Oceanol. Limnol. 35, 360–366. doi: 10.1007/s00343-016-5256-1
Webb M. (1963). Siboglinum fiordicum sp. nov. (Pogonophora) from the Raunefjord, western Norway. Sarsia 13, 33–43. doi: 10.1080/00364827.1963.10409515
Weigert A., Golombek A., Gerth M., Schwarz F., Struck T. H., Bleidorn C. (2016). Evolution of mitochondrial gene order in Annelida. Mol. Phylogenet. Evol. 94, 196–206. doi: 10.1016/j.ympev.2015.08.008
Weigert A., Helm C., Meyer M., Nickel B., Arendt D., Hausdorf B., et al. (2014). Illuminating the base of the annelid tree using transcriptomics. Mol. Biol. Evol. 31, 1391–1401. doi: 10.1093/molbev/msu080
Xia X. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Yang Z., Nielsen R. (2000). Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17, 32–43. doi: 10.1093/oxfordjournals.molbev.a026236
Yang M., Wang W., Li X., Sui J. (2022). The complete mitochondrial genome of Mesochaetopterus japonicus (Sedentaria: Chaetopteridae). Mitochondrial. DNA B. Resour. 7, 838–840. doi: 10.1080/23802359.2022.2073840
Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W. X., et al. (2020). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang Y., Rouse G. W., Qiu J. W. (2015). A new species of Mesochaetopterus (Annelida, Chaetopteridae) from Hong Kong with comments on the phylogeny of the family. Zootaxa 3974, 495–506. doi: 10.11646/zootaxa.3974.4
Zhang Y., Sun J., Rouse G. W., Wiklund H., Pleijel F., Watanabe H. K., et al. (2018). Phylogeny, evolution and mitochondrial gene order rearrangement in scaleworms (Aphroditiformia, Annelida). Mol. Phylogenet. Evol. 125, 220–231. doi: 10.1016/j.ympev.2018.04.002
Keywords: base substitution, phylogenetics, gene order rearrangements, genetic distance, polychaetes
Citation: Wu X, Su X, Wang Y, He C, Qiu J-W and Zhang Y (2024) Unraveling the phylogeny of Chaetopteridae (Annelida) through mitochondrial genome analysis. Front. Mar. Sci. 11:1382212. doi: 10.3389/fmars.2024.1382212
Received: 05 February 2024; Accepted: 20 May 2024;
Published: 05 June 2024.
Edited by:
Junlong Zhang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Daniel Martin, Spanish National Research Council (CSIC), SpainJenna Moore, Leibniz-Institut zur Analyse des Biodiversitätswandels (LIB), Germany
Yueyun Wang, Ministry of Natural Resources, China
Copyright © 2024 Wu, Su, Wang, He, Qiu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjie Zhang, eWFuamllemhhbmdAaGFpbmFudS5lZHUuY24=
 Xiu Wu
Xiu Wu Xuechun Su
Xuechun Su Yu Wang1
Yu Wang1 Caifang He
Caifang He Jian-Wen Qiu
Jian-Wen Qiu Yanjie Zhang
Yanjie Zhang