- 1Department of Biology, College of Science, United Arab Emirates University, Al-Ain, Abu-Dhabi, United Arab Emirates
- 2Khalifa Center for Genetic Engineering and Biotechnology (KCGEB), United Arab Emirates University, Al Ain, United Arab Emirates
- 3Department of Life and Environmental Sciences, College of Natural and Health Sciences, Zayed University, Dubai, United Arab Emirates
- 4International Center of Biosaline Agriculture, Dubai, United Arab Emirates
- 5National Water and Energy Center, United Arab Emirates University, Al Ain, United Arab Emirates
Global food production intensification presents a major hurdle to ensuring food security amidst a growing world population. Widespread use of chemical fertilizers in recent decades has risked soil fertility, compounded by the challenges posed by climate change, particularly in arid regions. To address these issues, adopting plant growth-promoting (PGP) bacteria stands out as a promising solution, offering multifaceted benefits to arid agroecosystems. We isolated a bacterial strain, SW7, from mangrove sediment, characterised the entire genome followed by phylogenetic analyses, and evaluated its in-vitro PGP activity. Subsequently, we examined its impact on tomato seed germination and plant growth. The strain SW7 exhibited growth on 11% NaCl, survival at 50°C, and possessed multiple PGP traits such as significant increase in seed germination rate (60.60 ± 38.85%), phosphate (83.3 g L−1) and potassium (39.6 g L−1) solubilization and produced indole acetic acid (3.60 ppm). Additionally, strain SW7 tested positive for ammonia, catalase, and oxidase enzyme production. The strain SW7 genome consists of 5.1 MB with 35.18% G+C content. Through genome-based phylogenetic and orthoANI analyses, the strain was identified as a novel Bacillus species, designated herein as Bacillus sp. SW7. In an eight-week shade-house experiment, inoculation of strain SW7 improved, leaf number, leaf density, leaf area index and mass water of tomatoes. Additional parameters, like chlorophyll a, chlorophyll b and carotenoids were not affected in SW7-inoculated tomatoes. In conclusion, Bacillus sp. SW7 exhibits multiple PGP traits and an adaptive capacity to high temperature and salinity, positioning it as a potential candidate for elevating the productivity of arid agroecosystems.
1 Introduction
Global food production intensification is a major challenge in achieving food security (Devaux et al., 2020) amid a growing world population (Arndt et al., 2023). The increase in human population creates a hurdle in achieving food safety, this has led to use of large quantities of chemical fertilizer for the past decades, despite its harmful effects on the environment and humans (Yadav et al., 2018). Also, the climate changes escalate the situation, due to rise in temperature the percentage of arid land is increasing. The semi- to hyper-arid region of the world faces several issues that will possibly continue to endanger food security in the upcoming years (Ammar et al., 2023). To meet the growing demand for sustainable agriculture in arid regions, enhancing food security becomes attainable through the efficient utilization of limited water resources to boost domestic food production. However, intensive cropping needs sufficient water supply which is not adequate in arid regions (Goddek et al., 2023). At the moment, most of the countries in the arid regions are facing water shortages because their water supply is below the water poverty line of 1000 m3/capita/year, compared with the world average of 7240 m3/capita/year (Hejazi et al., 2023). Furthermore, cultivating crops in arid regions requires extensive usage of synthetic fertilizers to enhance productivity, which ultimately deteriorates the ecosystem (Liu et al., 2020). As a result, innovative alternate approaches are required in such regions to concurrently increase food production, water sustainability and reduce ecosystem deterioration (Abdullah et al., 2022).
Promoting sustainable agriculture involves gradually reducing reliance on chemical fertilizers and embracing bio-waste-derived substances, alongside leveraging the potential of microorganisms (Basu et al., 2021). Biofertilizers, microbial-based products, and alternatives like organic extracts and vermicompost teas have emerged to counteract the negative impacts of toxic agrochemicals (Yatoo et al., 2021). Among these microbial solutions, known for enhancing plant growth, disease control, mitigating plant stress and environmental friendliness, offer a viable path to improve soil fertility and mitigate agrochemical harm (Nosheen et al., 2021). Utilizing microbial formulations as fertilizers in agriculture holds promise for enhancing soil fertility while reducing the detrimental effects of agrochemicals (Sumbul et al., 2020). Employing effective PGP bacteria as biofertilizers and biological control agents presents a suitable alternative to lessen reliance on chemical fertilizers in crop production (Zainab et al., 2021).
In the Middle-Easter and North African (MENA) region, PGP bacteria has a vital role in enhancing plant growth and crop yields by reducing the direct and indirect effects of biotic and abiotic stresses like high temperature, and salinity (Abdelaal et al., 2021; Reang et al., 2022). PGP bacteria mitigate the salinity damage in several crops through various mechanisms, including improved water uptake and osmolyte accumulation (glutamate, glycine betaine, proline and soluble sugars). Also, PGP bacteria increases antioxidant levels, synthesis of ACC deaminase, exopolysaccharides production and ionic homeostasis, which in turn enhances agricultural productivity even in saline-alkaline soil (Mishra et al., 2023). Currently, studies in various parts of the world are reporting improvements in the growth and production of crops such as tomatoes (Fan et al., 2016; Astorga-Eló et al., 2021), maize (Ojuederie and Babalola, 2023), and wheat (Shankar and Prasad, 2023), attribute to the use of PGP bacteria. The utilization of PGP bacteria as inoculants has substantial potential for application in farming, and lowering the rate of chemical fertilizers (Bittencourt et al., 2023). PGP bacteria have drawn more interest recently, due to their potential usage as a biofertilizer to improve crop development and production. However, there is an increasing need to find novel PGP bacteria that adapt and grow in extreme conditions with new phyto-beneficial traits (Lutfullin et al., 2022).
The mangroves are true extremophiles, which evolved to grow in extreme ecological conditions such as arid and hot intertidal environments (Friis et al., 2023). The mangrove tree is known to tolerate high salinity and plays an important role in the carbon sequestration, nutrient cycle, conservation and production of coastal ecosystems (Osland et al., 2018; Costa et al., 2022). Microbes associated with the mangrove are known to tolerate high salinity. Several studies have demonstrated that bacterial isolates from mangrove sediments and rhizosphere have PGP potentials, make them potential sources for isolating PGP strains (Mishra et al., 2023).
This study aimed to: (i) isolate bacteria from mangrove sediment collected from Khor Al Yeefrah, Umm Al Quwain, UAE; (ii) assess their PGP traits in in-vitro conditions; (iii) characterize the whole genome of strain SW7 to identify taxonomy, phylogeny, and gene clusters responsible for PGP traits in extreme conditions; and (iv) evaluate the effect of strain SW7 on seed germination, as well as the growth and productivity of tomatoes (Solanum Lycopersicum) in a shade-house field trial condition.
2 Materials and methods
2.1 Study site and sampling collection
The mangrove forest in Umm Al Quwain extends approximately 12 km along the coast with a homogeneous mangrove species Avicennia marina. The sediment samples were collected from Khor Al Yeefrah (25°32’07.8” N, 55°38’04.2” E) in the Umm Al Quwain mangrove forest (east coast, UAE). The sediment samples were promptly transported to the lab and processed for bacterial screening within 24 hr under cooling conditions. Various physicochemical properties of mangrove sediment samples were analysed and reported in Tounsi-Hammami et al., 2024.
2.2 Bacterial isolation, identification, NaCl and heat stress analyses
Out of total 41 bacterial isolates, obtained from mangrove sediments, which exhibited multiple PGP traits (Tounsi-Hammami et al., 2024), we selected one promising bacterial strain SW7 for further testing characterisation and in-vitro assays. Additionally, we subjected the strain to salt tolerance assays (NaCl 2-11% w/v) and heat stress tests following the method described by Mahdi et al. (2020).
2.3 In vitro assessment of multiple PGP traits
2.3.1 Phosphate and potassium solubilization
Phosphate solubilization activity was assessed by spot-inoculating pure colonies on PVK agar medium and incubating them for 3 days at 30°C. The Phosphate Solubilization Index (PSI) was calculated as described by Teymouri et al. (2016). For potassium solubilization, the Aleksandrov medium (5.0 g/L glucose, 0.5 g/L MgSO4. 7H2O, 0.1 g/L CaCO3, 0.006 g/L FeCl3, 2.0 g/L Na2 HPO4, and feldspath 2.0 g/L) was used as the sole K source (Rana et al., 2011). After 24 hr, the colony was taken with a sterile plastic pipette, inoculated on solid Aleksandrov medium, and incubated at room temperature for 5 days. The formation of a clear zone around the growing colony indicated potassium solubilizing ability, and the solubilization index was calculated using the formula by Sadiq et al. (2013).
2.3.2 Ammonia and indole acetic acid production
Ammonia production was determined by growing strain in peptone water at 30°C for 4 days. Nessler’s reagent was used to assess ammonia production, with a faint yellow colour indicating a small quantity and a deep yellow to brownish colour signifying maximum ammonia production (Sharma et al., 2021). Indole acetic acid (IAA) production was assessed by inoculating strain into Jensen’s broth containing 2 mg mL−1 tryptophan. The culture was incubated at 28°C for 48 hr with continuous shaking at 125 rpm. After centrifugation, a 1 mL aliquot of the supernatant was mixed with Salkowski’s reagent and incubated at room temperature for 20 minutes in darkness, as described by Islam et al. (2016). The concentration of IAA was determined by measuring the absorbance at 530 nm and using a standard curve from pure IAA solutions.
2.3.3 Hydrogen cyanide production
Hydrogen cyanide (HCN) production was performed using a nutrient agar medium with 0.44% glycine. Filter paper strips were dipped in 2% sodium carbonate solution and 0.5% picric acid and stored at 30°C for 72 hr. Colour change from orange to red to brown on the filter paper indicated the level of HCN production by the isolate, and quantitative analysis was done by examining by dipping the altered filter paper strips in double distilled water and observing at 625 nm, following Lotfi et al. (2022).
2.3.4 Siderophore assay
Siderophore assay was conducted using Chrome azurol S agar medium as described by Petrillo et al. (2021). The presence of a yellow-orange halo surrounding bacterial growth indicated siderophore production. The siderophore production index (SPI) was calculated using the formula.
2.3.5 Oxidase and catalase enzyme
Oxidase and catalase enzyme activities were assessed using Kovacks’s reagent and a 6% H2O2 solution, respectively. Positive results for oxidase were indicated by a lavender colour, while catalase activity was confirmed by the formation of air bubbles during the test (Prajakta et al., 2019).
2.4 Seed preparation, surface coating, in-vitro/in-vivo germination assay under shade-house conditions
We collected seeds of tomato (Solanum Lycopersicum, F1 hybrid; 96 seeds) and coated them following the method described earlier (Mahdi et al., 2020). In brief, we surface sterilized seeds with 5% sodium hypochlorite solution for 1 min, shaken in 70% ethanol for 1 min and washed 5 times in sterilized distilled water followed by air-drying under a laminar hood. The culture pellets (OD600 nm = 0.8) were taken from fresh cultures via centrifugation at 10,000 rpm for 5 min. The bacterial pellets were resuspended in 10 mL of sterile distilled water, vortexed for 10 s, and used for seed treatment. The culture suspensions were used as seed dry-out with a ratio of 10 mL per (120, 96, 56) seeds for 1 hr. Next, seeds were air-dried and placed on petri plates filled with sterile filter paper. Thirty seeds were kept on each plate. The triplicates of each treatment were sustained, and seeds treated with sterile distilled water were used as a negative control. The petri plates were kept at 25°C in the dark and germination rates were observed after the 14 days of post-incubation. The total length and fresh and dry weight were measured. The germination rate and vigor index were calculated using Equations 1, 2 (Islam et al., 2016):
2.5 Preparation of coated tomato seeds for greenhouse assay
The tomato seeds were first put through the float test to determine their viability (Degwale et al., 2022). Seeds were placed in sterilized water and observed for their floating characteristics. Floating seeds were discarded as it may indicate their inability to germinate, however, seeds that sunk were considered viable and continued to the next stage of the sowing. Next, 5 centrifuge tubes (5 ml) were prepared for strain SW7, they were then labelled accordingly and filled with 1ml of the SW7 strain inoculum (108 cfu/ml) using a micropipette, followed by adding 10 seeds to each tube using sterilized forceps. Afterwards, the centrifuge tubes containing the inoculum and seeds were placed in the shaker at 37°C to incubate for 24 hours. In the final step, coated seeds were removed from the incubation and the inoculum was removed using the pipette which left the 50 coated seeds ready for sowing.
Once the seeds were coated, the sowing preparations began. The seedling potting trays with 50 pits each were prepared with peat moss substrate. Each tray was labelled according to treatment and placed inside the greenhouse at the International Centre of Biosaline Agriculture (ICBA, Dubai, UAE) to start the sowing process. The conditions of the greenhouse were similar to those described by (Rath and Ghosal, (2020). Small dents were made in the soil and sterilized forceps were then used to pick up each seed and plant one in each pit to cover it with soil. The trays were then irrigated and left to start the germination process in greenhouse conditions. The trays were watered 3 times daily, just enough to always keep the soil moist for seeds to germinate under ideal conditions. Once the seedlings started growing the amount of irrigation was slightly increased to accommodate the growth.
After 4 weeks of monitoring the seedling growth and collecting data under the greenhouse conditions, the well-grown seedlings were selected and transplanted from the potting trays straight to the ground inside the shade house to receive near-constant irrigation to allow continuous growth for another 4 weeks. Shade house conditions were like described by (Quamruzzaman et al., 2021). Shade house conditions were not strictly controlled, it was based on the weather, temperature, and humidity of the day. A saganizer garden hand was used to make small holes in the soil then filled with a mixture of NPK fertilizer and natural humic substance (organic soil conditioner) with a ratio of 1:3 to further help seedling growth and help transplanted seedlings grow well under the shade house field trial conditions. The seedlings were irrigated twice a day until eight weeks as described earlier (Savva and Frenken, 2002). The shoot height was measured once in a week upto eight weeks to compare the influence of strain on the growth of tomato plants (Supplementary Figure S1).
2.6 In vitro plant growth promotion attributes
The soil plant analysis development (SPAD) analyses were conducted to assess the nutritional status of the fully matured leaves (three replicates per plant and 5 plants per group) using a 502P chlorophyllometer (Konica Minolta, Inc., Tokyo, Japan). We also calculated the leaf area index (LAI) by measuring the leaf length (L) and width (W) using the following formula (L × W × 0.75; where 0.75 represents the leaf compensating factor. We collected three leaves per plant and measured the water content by calculating fresh (Fw) and dry (Dw) weight recorded after incubating at 104°C for 2 and 80°C for 72 hr and following this formula: leaf water content (%) = (Fw−Dw)/Fw∗100. Leaf density was measured using the gravimetric method, where leaf mass is divided by leaf area, typically expressed in grams per square meter (g/m²). For chlorophyll contents we collected three leaves per plant. Briefly, we mixed 0.2 g of frozen leaf samples with 4 mL dimethyl sulphoxide for 24 hr followed by the absorbance measurement at wavelengths of 663 nm, 645 nm, and 470 nm. Chl a, Chl b, Chl a + b and carotenoid contents were determined by using the following equations described by Batool et al. (2020). The statistical analyses of control vs inoculated samples were carried out using student’s t-test and the results were considered significant at P < 0.05.
2.7 DNA extraction and whole genome characterization
Genomic DNA was extracted from the overnight grown culture of strain SW7 using E.Z.N.A soil DNA isolation kit, with minor modifications. Further to perform whole genome sequencing we constructed the library using Illumina Nextera XT sample prep and index kits, as well as the MiSeq Reagent Micro Kit v2 (Illumina, San Diego, CA, USA). We performed 150 bp x 2 paired-end sequencing using MiSeq Illumina sequencer. The quality of raw reads was assessed with the FastQC V 0.12.1 quality control tool, and then using sickle the sequences were “Filter by quality” tool to remove low-quality reads (with a Phred score below 20) as well as reads with a 90% high-quality score distribution. We used SPAdes in two different ways for assembling the contigs 1. The trimmed reads were assembled using the default parameter for isolates (–isolates) and 2. We also used SPAdes from the Partic server (http://www.bv-brc.org) (Davis et al., 2019) to join forward and reverse reads and then assemble them into larger contigs using a 60-bp overlap and a 300-bp length threshold. Then the assembled contigs were polished using RACON (Vaser et al., 2017) and Pilon (Walker et al., 2014) with 2 and 4 iterations respectively. Based on the Quast 5.2.0 (Gurevich et al., 2013) assembly statistics, the assembly using Patric SPAdes had better quality with less number of contigs and mismatches. Future ragtag program (Alonge et al., 2019) was used to reorder the assembled contigs into a complete genome by filling the gaps in the sequences using a closed genome as a reference. Annotation was performed with Prokka by using the assembled genome (Seemann, 2014) and Rapid Annotation using Subsystem Technology (RAST) (Overbeek et al., 2014).
The functional annotation of predicted genes was carried out based on COGclassifier (https://pypi.org/project/cogclassifier), the amino acid sequences obtained from rast annotations was used to predict the functional genes using Kyoto Encyclopaedia of Genes and Genomes database (KEGG; http://www.genome.jp/kegg/; https://www.kegg.jp/blastkoala/; https://www.genome.jp/eg-bin/find_pathway_object), and the Gene Ontology database (GO; http://www.geneontology.org/). As a final step, DNA sequences, genes, and predicted non-coding RNA sequences were integrated into the GenBank file format and the circle map of the genome was created using the Circos version 0.69-6 (Krzywinski et al., 2009). The raw sequencing reads and assembly were deposited in the National Centre of Biotechnology Information (NCBI) database under the accession number PRJNA1068844 and the assembled genome was deposited under the accession number CP150440.
2.8 Multilocus sequence typing and phylogenomic analysis
We performed MLST analysis to identify the strain SW7 using seven house-keeping genes such as glpF (glycerol uptake facilitator protein), gmk (guanylate kinase, putative), ilvD (dihydroxy-acid dehydratase), pta (phosphate acetyltransferase), pur (phosphoribosylaminoimidazolecarboxamide), pycA (pyruvate carboxylase) and tpi (triosephosphate isomerase) (Priest et al., 2004). To confirm the taxonomic identification, we conducted a phylogenetic analysis using the Type (Strain) Genome server (TYGS) (https://tygs.dsmz.de). The genome sequences were pairwise compared with closely related genomes using Genome BLAST Distance Phylogeny (GBDP) (Camacho et al., 2009) along with B. anthracis Ames (three other strains), B. cereus ATCC 4342 (11 other Bacillus strains), 11 Bacillus sp. isolated from the international space station (ISS) (Venkateswaran et al., 2017b) and with two PGP reported Bacillus species (Bacillus sp. RZAMS9 (Bonatelli et al., 2020), and Bacillus sp. D5 (PGP) (Magotra et al., 2021) and one plasmidless B. megaterium QM B1515 (Eppinger et al., 2011). The distance from the genome blast was employed to determine the closest genome to the strain SW7. Subsequently, GGDC 3.0 was utilized to calculate the digital DNA-DNA hybridization (DDH) value and confidence intervals. These intergenomic distances were then used to infer a balanced minimum evolution tree with branch support via FASTME (V2.1.6.1), including SPR post-processing (Athmika et al., 2021), with 100 pseudo-bootstrap replicates. The rooted tree was constructed and visualized using PhyD3 (Kim et al., 2017). The OrthoANI value with the closest strain was calculated using the FastANI software (Lee et al., 2016). Also, we pairwise compare the Bacillus sp. SW7 using mauve aligner (Darling et al., 2010) with other PGP Bacillus sp. RZ2MS9 (PGPB), non-PGP Bacillus cereus (Q1 strain and S1-R2T1-FB) and Bacillus anthracis str Ames.
3 Results
3.1 Isolation, biochemical characterization and identification of bacterial strain
The strain SW7 is a gram-positive bacterium with a circular shape, cream colour, with raised elevation (7 nm diameter) on an agar plate. Biochemical tests showed that the strain was positive for oxidase, and catalase activity whereas negative for urease, decarboxylase, and ornithine decarboxylase (Supplementary Table S1). Similarly, the strain tested negative for lysine, citrate utilization, H2S production, sorbitol, rhamnose, sucrose melibiose and amygdalin. The strain utilized various carbon sources such as ortho-Nitrophenyl-β-galactoside (ONPG), arginine, tryptophan, gelatinase, glucose, mannitol, inositol and arabinose. It displayed both phosphate (clear zone 8.00 ± 0.00 mm; 83.3 g/L-1) and potassium (23.00 ± 0.57 mm; 39.6 g/L-1) solubilization activity. The strain tested positive for ammonia (0.5 ppm) and IAA (3.60 ppm) production and no HCN production and siderophore activity were recorded. The salt and heat stress assay showed the strain was able to tolerate 11% NaCl (Figure 1A) and can grow up to 50°C (Figure 1B).
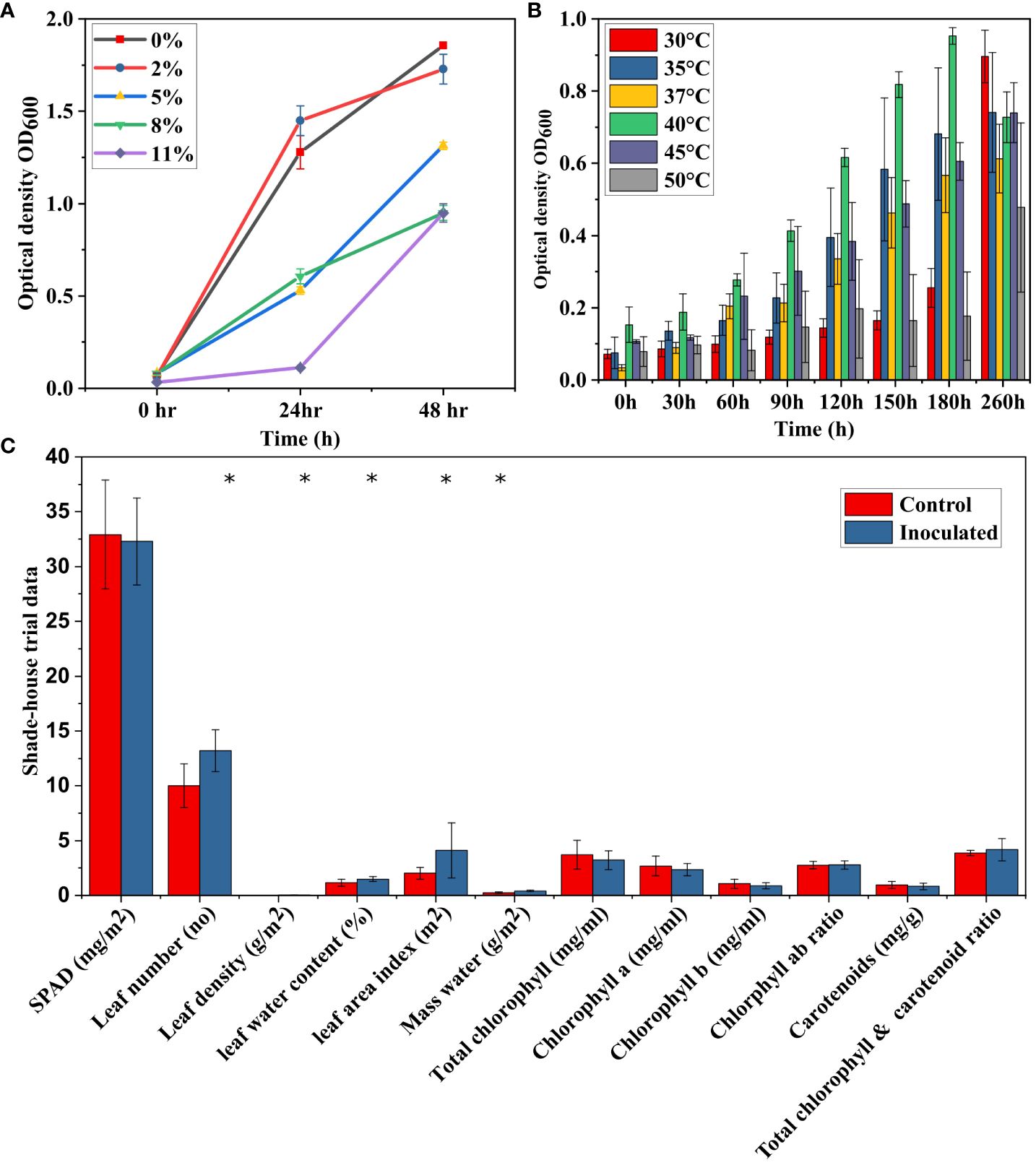
Figure 1 The ability of Bacillus sp. SW7 strains to tolerate salinity ranging from 0-11% NaCl (A) and their heat stress tolerance, showing growth up to 50°C (B). (C) Effect of Bacillus sp. SW7 inoculation on tomato plant growth parameters. The experiment consisted of 5 trials, each performed in triplicate, and data were analyzed using Student’s t-test. Shade-house evaluation of Bacillus sp. SW7 inoculation on various growth parameters in tomato plants, with * indicating significant differences.
3.2 Seed germination and shade house assay
The strain SW7 inoculated seeds (60.60 ± 38.85%) had a significantly higher germination rate compared to the control (50.6 ± 35.04%; p-value = 0.02). However, the seedling length (4.86 ± 1.38 cm vs. 3.99 ± 0.14 cm, p-value = 0.38) and vigor index (297.1 ± 10.4 vs. 378.8 ± 107.6, p-value = 0.31) didn’t differ significantly between control and inoculated seeds.
The SW7 exhibits similar values for most plant growth parameters, including Chlorophyll a and b, total chlorophyll, carotenoids, and chlorophyll-carotenoid ratio. For example, the SPAD value varied only 0.6% between the SW7-inoculated and control plant groups. However, SW7-inoculated tomato plants exhibited significantly higher (p > 0.05) leaf numbers, density, and mass water (Figure 1C). Also, increased leaf area index and leaf water content were observed in the SW7-inoculated plants, suggesting broader leaves. These findings strongly suggest a positive impact of the inoculation treatment on the overall growth and physiological aspects of the tomato plants.
3.3 Whole-genome characterization of strain SW7
The assembled genome of strain SW7 comprises a circular genome of 5,155,809 bp with G+C content of 35.17% (Figure 2). In total, the genome consists of 5,424 predicted protein-coding sequences (CDSs), 02 rRNA, and 69 tRNA genes. The assembled genome has a completeness of 98.56% with zero percent contamination. Approximately, 4,104 protein-coding genes were predicted to be functional, and 1,320 hypothetical proteins. Among the predicted CDSs, around 3,736 CDSs (68.88%) were assigned to the Clusters of Orthologous Groups (COG) database. For more details on COG categories, KEGG pathways, Enzymatic (Reactions) Commission numbers (EC) and GO terms assigned to the strain SW7 (refer Supplementary Results S1, Supplementary Figure S2).
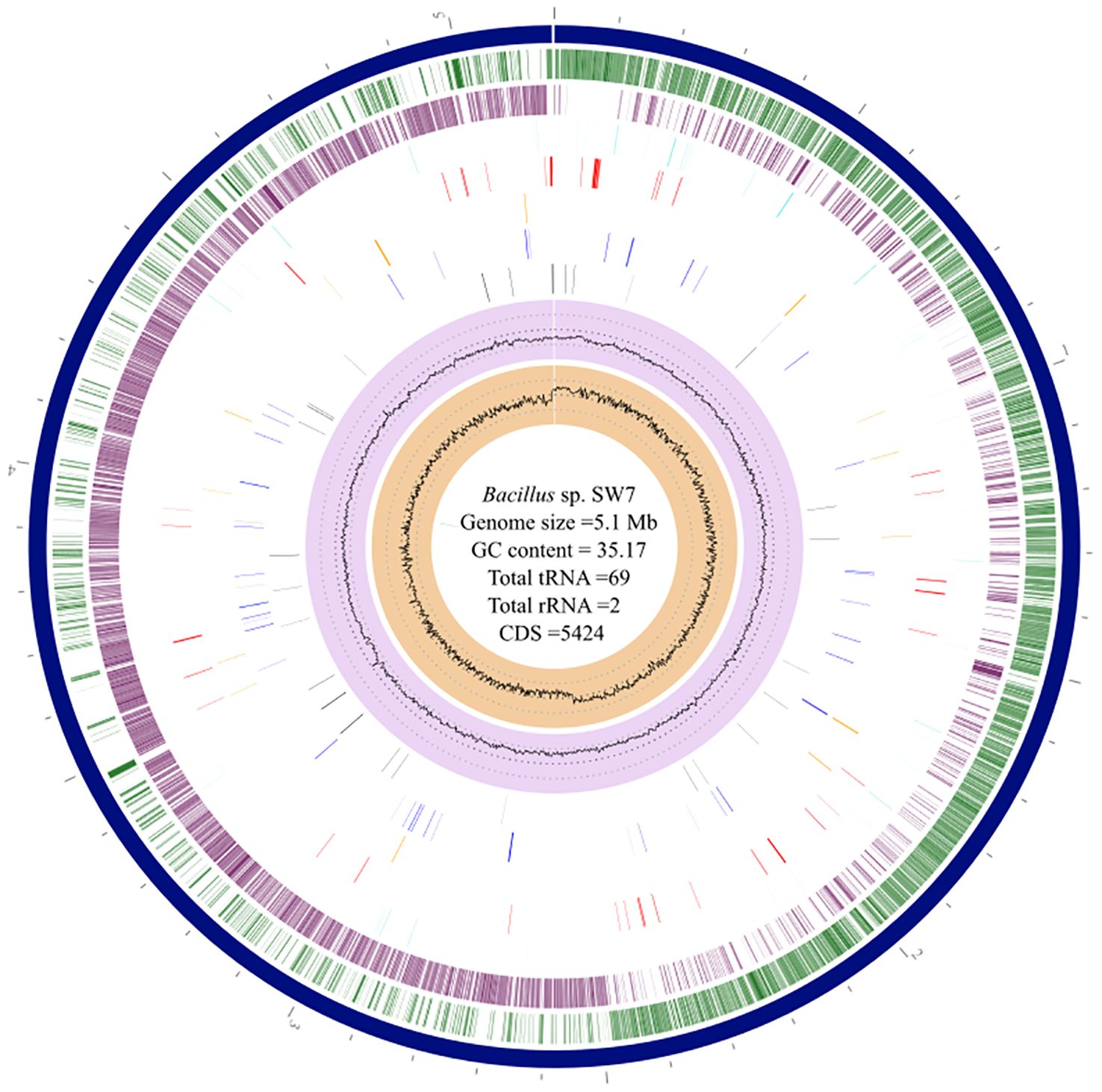
Figure 2 Circos plot representing the genome of Bacillus sp. SW7. The innermost circle is shown in sandy brown for GC skew, while the second innermost circle, in light violet represents for GC content. Drug target and transporter regions are indicated by black and blue lines respectively. Orange and red lines highlight virulence factor and AMR genes. The second (green) and third (purple) circles from the outside represent the CDS in the forward and reverse direction, followed by non-coding sequences represented in light cyan.
3.4 Molecular evidence of PGP potential of strain Bacillus sp. SW7
The functional genes responsible for plant growth-promoting traits were identified in the genome sequence of Bacillus sp. SW7. Bacillus sp. SW7 is identified as a non-pathogenic strain since it lacks plasmids pXO1 and pXO2, which carry the capBCAD operon for capsule protein and the gene lef, cya, and pagA responsible for the lethal factor (LF) or edema factor (EF), respectively. The genome of strain Bacillus sp. SW7 includes genes responsible for phosphate solubilization (pstA, pstB, pstC), potassium solubilization (ackA and mdh), siderophore production (dhbACBEF), indole acetic acid (ipdC), nitrogen fixation (nifS), hydrogen cyanide production (hcnA, hcnB, hcnC), catalase (katE), oxidase (qoxC), arginine decarboxylase (speA_1), beta-galactosidase (lacA), inositol (iolA & C) and sorbitol (sdhA, sdhB).
3.5 Genes conferring for PGP fitness
Bacillus sp. SW7 genome comprised of genes responsible for heat shock proteins (dnaJ, K, GrpE and groS, L), cold shock proteins (cspA, and D), and osmoregulated proline transporter gene (OpuE, opeU). Stress response protein SCP2 (yceC), general stress protein (yceD_1 and 2) are part of the genome. The genome also has some antioxidants genes (sodA1, sodA2, katE and qoxC), reflecting the need for survival under the extreme environments from where they have been isolated. We also found the xerC gene that is responsible for recombinase, which is critical for the PGP bacteria to be an effective rhizosphere colonizer.
3.6 Phylogenetic analysis
The whole genome-based taxonomic analysis using Type Strain Genome Server (TYGS) revealed that strain SW7 is closely related to the Bacillus anthracis str. Ames (Figure 3), with dDDH (d0) of 83.4%, and relatively close to other Bacillus cereus and Bacillus species isolated from the international space station with high (≥ 82%) dDDH value (Supplementary Table S2).
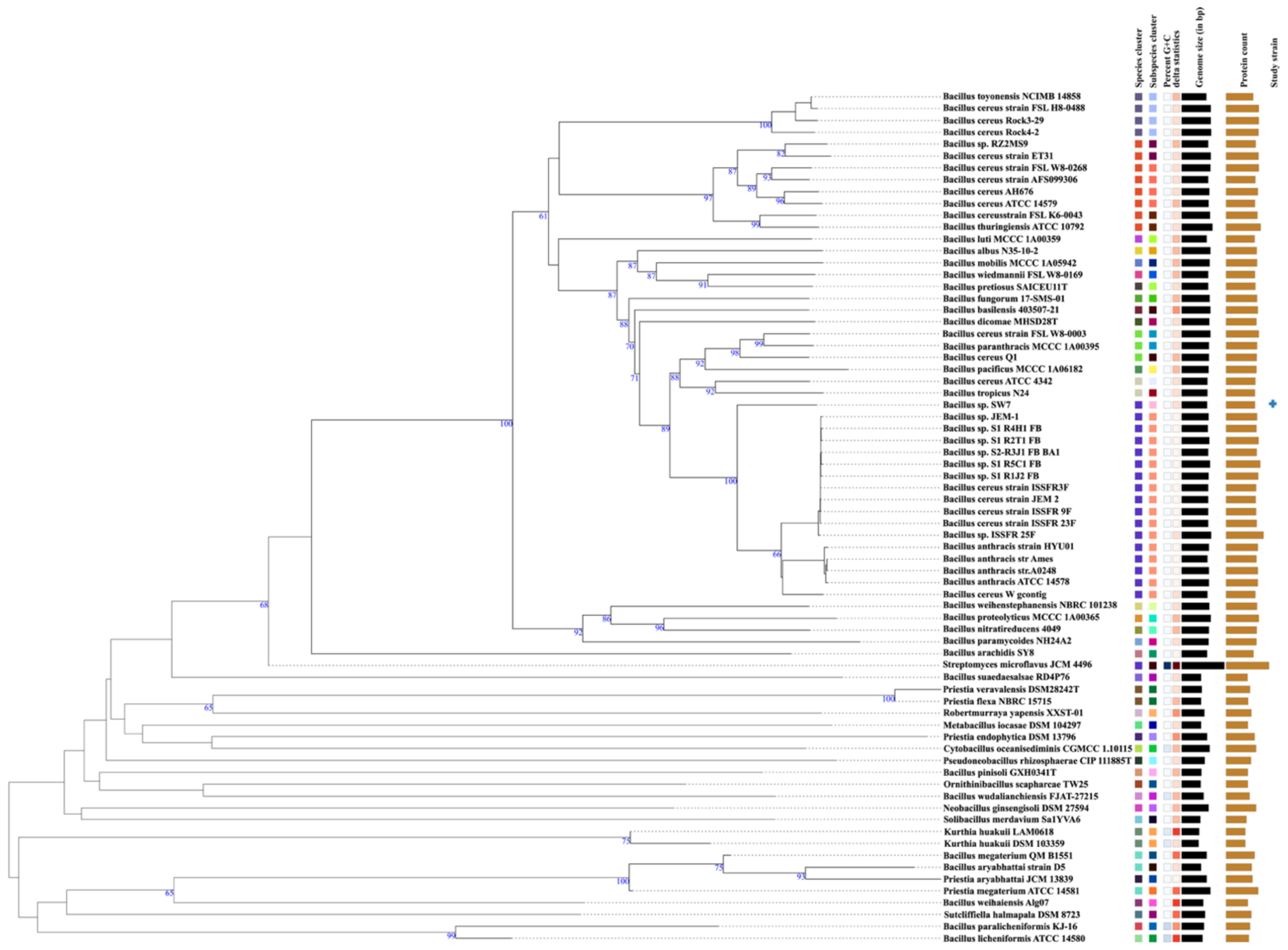
Figure 3 Phylogenetic identification of strain Bacillus sp. SW7. The tree was plotted using GBDP distance and inferred with FastME 2.1.6.1. GBDP distance formula d5 is used as a scale for branch length with tree rooted at the midpoint. A whole genome sequence-based tree with an average branch support of 54.4% (with δ statistical value of 0.226).
3.7 MLST analysis and ANI
Although the phylogenetic analyses suggested that the strain is closely related to Bacillus anthracis str Ames, due to the absence of pXO1 and pXO2 biomarkers, we would like to perform MLST analysis using seven housekeeping genes. This analysis reveals that the strain SW7 is identified as Bacillus cereus, with the following housekeeping genes: glp(3), gmk (23), ilv (132), pta (152), pur (38), pyc (43), and tpi (99). To assess the consistency of evolutionary distance, the SW7 genome was compared using average nucleotide identity (ANI), which shows an ANI value of 97.32% against Bacillus-sp.S1-R2T1-FB (Bacillus isolated from international space station) (Supplementary Table S3). Moreover, other Bacillus species isolated from the ISS and non-virulent Bacillus anthracis exhibit greater than 97% average nucleotide identity (ANI) to Bacillus sp. SW7 (Supplementary Table S3). Utilizing the mauve aligner, we identified that Bacillus sp. SW7 shares more homologous regions with Bacillus-sp.S1-R2T1-FB (ISS), followed by Bacillus anthracis str. Ames. Additionally, significant homology was observed with the PGP Bacillus sp. RZ2MS9 (Supplementary Figure S3).
4 Discussion
The study shows a positive effect of strain SW7 inoculation on tomato seed germination, plant leaf number, density and mass water. Moreover, the strain displays multiple PGP traits such as the ability to produce ammonia, and IAA, solubilize potassium and phosphate. It can withstand 11% NaCl salinity and 50°C of temperature. Whole-genome analysis confirms the presence of multiple PGP and stress-alleviating genes, making the strain a potential candidate as a PGP bacterium to mitigate stress in marginal environments. Furthermore, the phylogenetic analysis reveals that the strain belongs to a novel Bacillus sp.
4.1 Bacillus sp. SW7 inoculation enhanced the tomato seed germination and plant growth
We observed a significantly higher seed germination rate and higher vigor index in tomatoes after the inoculation of Bacillus sp. SW7. This is in agreement with a previous study showing that inoculation of Bacillus species such as Bacillus pumilus and Bacillus firmus enhances seed germination of tomatoes under both saline and non-saline conditions (Patani et al., 2023). Fan et al. (2016) demonstrated that the inoculation of B. megaterium increased the seed germination and vigor index in salt-affected tomato plants. Notably, a consortium involving other PGP bacteria (Paenibacillus polymyxa and Brevibacillus), B. subtilis, and B. altitudinis also increases the seed germination rate in tomatoes (Astorga-Eló et al., 2021).
Furthermore, the inoculation of Bacillus sp. SW7 proved to enhance various growth parameters of tomato plants after 8 weeks in a shade-house experiment. Leaf number, density, and mass water in the Bacillus sp. SW7 inoculated tomatoes significantly increased compared to the control group. These findings align with previous studies that demonstrated the inoculation of Bacillus species enhancing various physiological parameters in tomatoes grown in high salt conditions (Patani et al., 2023). Similarly, Bacillus cereus was found to improve plant growth parameters in soybean plants (Khan et al., 2020) and increase leaf number, length, and area index in peanuts (Arachis hypogaea) (Gohil et al., 2022), which is consistent with the findings of this study, suggesting that Bacillus sp. SW7 is a promising candidate for PGP, capable of mitigating arid conditions and enhancing plant growth.
4.2 Genomic analysis for PGP traits in Bacillus sp. SW7 and in-vitro confirmation through shade-house experiment
The genome analysis of strain SW7 revealed a size of 5.15 Mb with a GC content of 35.17%, encompassing 5,424 CDSs, consistent with previously reported genomes of related isolates in the NCBI database (Leontidou et al., 2020). A comparable genome size, GC content, and number of Coding DNA Sequences (CDS) were observed in Bacillus sp. strain RZ2MS9, known for its multiple PGP traits (Bonatelli et al., 2020).
PGP bacterial strains, in general, play a crucial role in enhancing plant development, contributing to activities like phosphate and potassium solubilization, nitrogen fixation, HCN production, and the secretion of essential plant hormones such as IAA, abscisic acid, and cytokinin etc (Hakim et al., 2021). Bacillus sp. SW7 was found to produce plant growth promoting factor NH3, demonstrated phosphate and potassium solubilization activity and produce the hormone IAA. Genome analysis uncovered the presence of genes associated with multiple PGP traits, encompassing phosphate (operon pstABC), and potassium solubilization (ackA and mdh), nitrogen fixation (only nifS), as well as the production of IAA (ipdC). Various studies have highlighted the diverse PGP potentials of Bacillus species. For example, Bacillus sp. PG-8, isolated from Panchagavya, significantly improved plant growth by solubilizing insoluble phosphate and producing IAA (Gohil et al., 2022). Furthermore, Bacillus sp. SW7 possesses a two-component signal transduction system for phosphate uptake (phoBPR genes, and the phnXW) genes crucial for the solubilization of organophosphorus phosphonate compounds, making them bioavailable for the plant. Similarly, complete traits of phosphonate solubilization gene clusters were reported in PGP Bacillus firmus I-1582, which promotes tomato growth and decreases nematode counts in the rhizosphere (Susič et al., 2020). Notably, various Bacillus species are known to have phosphate solubilization genes that promote plant growth, such as Bacillus sp. strain D5 (Bonatelli et al., 2020), Bacillus sp. Strain RZ2MS9 (Magotra et al., 2021) and B. subtilis EA-CB0575 (Franco-Sierra et al., 2020).
Phytohormones, particularly IAA, are vital for regulating the growth, division, differentiation, and response of plant cells to light and gravity (Asim et al., 2023) further promoting root expansion and nutrient uptake. Earlier studies have demonstrated the positive effects of Bacillus species on plant growth. For instance the inoculation of Bacillus sp. PG-8 (Gohil et al., 2022), Bacillus thuringiensis (Batista et al., 2021), and Bacillus siamensis (Saleem et al., 2021), has been demonstrated to enhance the growth of peanuts, tomatoes and cotton respectively. These Bacillus strains were found to alleviate stress by producing IAA. The genome of B. megaterium RmBm31 (Dahmani et al., 2020), encodes genes for multiples PGP traits including IAA, HCN, phosphate/potassium solubilization, aligning with the findings of the present study.
The Bacillus sp. SW7 harbours other PGP genes, including HCN (hcnA, hcnB, hcnC) and siderophore genes (dhbACBEF). However, both HCN production and siderophore activity was not observed in our in-vitro assessment, possibly due to the specific condition provided during the experiment. Typically, siderophores are produced under unfavourable or low-iron conditions to capture iron from the surrounding environment (Sultana et al., 2021). Studies have shown that the addition of siderophores can more effectively improve plant growth than the addition of Fe(III) (De Serrano, 2017). Genome analysis of various Bacillus strains with PGP potential, including B. subtilis EA-CB0575 (Franco-Sierra et al., 2020), B. megaterium RmBm31 (Dahmani et al., 2020), has revealed the presence of siderophore genes. Additionally, the application of Bacillus amyloliquefaciens Ba13 increased beneficial bacteria in the rhizosphere and rhizoplane of tomatoes by producing siderophores, which compete for soluble Fe(III). This competition leads to a decrease in soil fungi, and genome analysis revealed that B. amyloliquefaciens has two genes encoding siderophores (Bacillibactin) (Ji et al., 2021).
4.3 Strain SW7 phylogenetically different from B. anthracis and B. cereus and represent novel species
The whole-genome blast analysis revealed that the strain SW7 is closely related to Bacillus anthracis str. Ames. However, the strain SW7 lacks the pOX1 and pOX2 plasmids. To confirm the taxonomy, we performed MLST, which indicated that SW7 belongs to the Bacillus cereus group based on the housekeeping genes. Further, the phylogenetic analysis revealed that the strain SW7 is placed in a different clade from Bacillus anthracis str. Ames and other Bacillus cereus species, including isolates from the International Space Station (Venkateswaran et al., 2017b). About the recent phylogenetic investigation of the family Bacillaceae which are polyphyletic, it is difficult to trace the evolutionary lineages (Patel and Gupta, 2020). Herein we place the strain SW7 as a novel Bacillus species based on phylogenetics and MLST, which is phylogenetically different from B. anthracis and B. cereus. Although B. anthracis is a common soil bacterium, which are obligate and opportunistic animal pathogen belonging to Bacillus cereus sensu lato, and B. thuringiensis, some Bacillus anthracis strains lack the virulent plasmids pOX1 as well as pOX2 contributed positively to plant growth (Azeem et al., 2023).
5 Conclusion
Our study reveals that the newly identified PGP bacteria Bacillus sp. SW7 from mangrove sediments significantly improves tomato seed germination rates and increases the number of leaves and density, enhancing leaf area index compared to the untreated control group. The in-vitro and genome analysis confirm the presence of multiple PGP functions in Bacillus sp. SW7, including IAA, and NH3 production, phosphate and potassium solubilization, which contribute to tomato plant growth promotion. Furthermore, the genome encompasses stress-tolerant genes related to heat and salinity, suggesting the suitability of the strain to arid environments. These experimental evidences and genomic traits data positioned the Bacillus sp. SW7, as a promising candidate for arid agroecosystems, offers potential benefits for crop growth in marginal environments using a sustainable and environmentally friendly alternative. This study recommends the utilization of the Bacillus sp. SW7 to enhance crop growth and productivity, particularly in challenging arid agricultural settings. However, before considering the SW7 strain for larger-scale field trials, it is crucial to conduct further preliminary studies under greenhouse conditions. This should involve selecting different tomato seed varieties, assessing seed viability, and optimizing SW7 inoculation rates. Furthermore, it is recommended to evaluate the effectiveness of the SW7 strain on tomato seedling growth under varying arid agroecosystem marginal environmental conditions, such as salinity, water availability, and reduced synthetic fertilization rates. Addressing these aspects will provide a more comprehensive understanding of the SW7 strain’s potential and inform its practical implementation in agriculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MA: Writing – original draft, Data curation, Methodology, Visualization. BS: Methodology, Visualization, Writing – original draft, Formal analysis, Software, Validation, Writing – review & editing. AN: Formal analysis, Methodology, Writing – review & editing, Validation. HM: Validation, Writing – review & editing, Visualization, Data curation. MK: Methodology, Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Project administration. SM: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – review & editing, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors than UAE University for providing UAEU-ZU Joint Research project (R22024; 12R139) and the Zayed University for Research Incentive Fund (RIF) project-R21074.
Acknowledgments
We acknowledge the UAE University for providing a UAEU-ZU (12R139/R22024) research grant to SM and MK and Zayed University Research Incentive Fund (RIF) project-R21074 awarded to MK and HM. We thank Amna Almarri, Meera Ashkanani, Aroosa Zeb, and Aneesa Rasheed Anwar for helping with SW7 strain seed coating, salt and heat stress assay; AlSayeda Zahra Salman for preliminary characterization of PGP traits isolates and Qurban Ali for his help in preparing new figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1379439/full#supplementary-material.
References
Abdelaal K., AlKahtani M., Attia K., Hafez Y., Király L., Künstler A. (2021). The role of plant growth-promoting bacteria in alleviating the adverse effects of drought on plants. Biology 10, 520. doi: 10.3390/biology10060520
Abdullah M. M., Assi A., Zubari W. K., Mohtar R., Eidan H., Al Ali Z., et al. (2022). Revegetation of native desert plants enhances food security and water sustainability in arid regions: Integrated modeling assessment. Sci. Total Environ. 806, 151295. doi: 10.1016/j.scitotenv.2021.151295
Alonge M., Soyk S., Ramakrishnan S., Wang X., Goodwin S., Sedlazeck F. J., et al. (2019). RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20, 224. doi: 10.1186/s13059-019-1829-6
Ammar K. A., Kheir A. M. S., Ali B. M., Sundarakani B., Manikas I. (2023). Developing an analytical framework for estimating food security indicators in the United Arab Emirates: A review. Environ. Dev. Sustain. 26, 5689–5708. doi: 10.1007/s10668-023-03032-3
Arndt C., Diao X., Dorosh P., Pauw K., Thurlow J. (2023). The Ukraine war and rising commodity prices: Implications for developing countries. Global Food Secur. 36, 100680. doi: 10.1016/j.gfs.2023.100680
Asim M., Zhang Y., Liu W. (2023). Cooperation of gene regulatory networks and phytohormones in cell development and morphogenesis. Front. Plant Sci. 14, 1290538. doi: 10.3389/fpls.2023.1290538
Astorga-Eló M., Gonzalez S., Acuña J. J., Sadowsky M. J., Jorquera M. A. (2021). Rhizobacteria from ‘flowering desert’events contribute to the mitigation of water scarcity stress during tomato seedling germination and growth. Sci. Rep. 11, 13745. doi: 10.1038/s41598-021-93303-8
Athmika, Ghate S. D., Arun A. B., Rao S. S., Kumar S. A., Kandiyil M. K., et al. (2021). Genome analysis of a halophilic bacterium Halomonas malpeensis YU-PRIM-29T reveals its exopolysaccharide and pigment producing capabilities. Sci. Rep. 11, 1749. doi: 10.1038/s41598-021-81395-1
Azeem M. A., Ali F., Ullah A., Iqbal M., Ali K., Al Farraj D. A., et al. (2023). Exploration of plant growth promoting traits and regulatory mechanisms of Bacillus anthracis PM21 in enhancing salt stress tolerance in maize. Environ. Sci. pollut. Res. 30, 77499–77516. doi: 10.1007/s11356-023-27820-6
Basu A., Prasad P., Das S. N., Kalam S., Sayyed R. Z., Reddy M. S., et al. (2021). Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: recent developments, constraints, and prospects. Sustainability 13, 1140. doi: 10.3390/su13031140
Batista B. D., Dourado M. N., Figueredo E. F., Hortencio R. O., Marques J. P. R., Piotto F. A., et al. (2021). The auxin-producing Bacillus thuringiensis RZ2MS9 promotes the growth and modifies the root architecture of tomato (Solanum lycopersicum cv. Micro-Tom). Arch. Microbiol. 203, 3869–3882. doi: 10.1007/s00203-021-02361-z
Batool T., Ali S., Seleiman M. F., Naveed N. H., Ali A., Ahmed K., et al. (2020). Plant growth promoting rhizobacteria alleviates drought stress in potato in response to suppressive oxidative stress and antioxidant enzymes activities. Sci. Rep. 10, 16975. doi: 10.1038/s41598-020-73489-z
Bittencourt P. P., Alves A. F., Ferreira M. B., da Silva Irineu L. E. S., Pinto V. B., Olivares F. L. (2023). Mechanisms and applications of bacterial inoculants in plant drought stress tolerance. Microorganisms 11, 502. doi: 10.3390/microorganisms11020502
Bonatelli M. L., Li J., Dumaresq A., Quecine M. C., Settles M. L. (2020). Complete genome sequence of bacillus sp. Strain RZ2MS9, a multitrait plant growth promoter. Microbiol. Resour Announc 9, e00623–e00620. doi: 10.1128/MRA.00623-20
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Costa M. T., Ezcurra E., Ezcurra P., Salinas-de-León P., Turner B., Kumagai J., et al. (2022). Sediment depth and accretion shape belowground mangrove carbon stocks across a range of climatic and geologic settings. Limnology Oceanography. 67(S2):S104–S117. doi: 10.1002/lno.12241
Dahmani M. A., Desrut A., Moumen B., Verdon J., Mermouri L., Kacem M., et al. (2020). Unearthing the Plant Growth-Promoting Traits of Bacillus megaterium RmBm31, an Endophytic Bacterium Isolated From Root Nodules of Retama monosperma. Front. Plant Science. 11. doi: 10.3389/fpls.2020.00124
Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147. doi: 10.1371/journal.pone.0011147
Davis J. J., Wattam A. R., Aziz R. K., Brettin T., Butler R., Butler R. M., et al. (2019). The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–D612. doi: 10.1093/nar/gkz943
Degwale A., Tesfa T., Meseret B., Fantaw S. (2022). Seed extraction methods affect the physiological quality of tomato seed and developing seedlings. Int. J. Vegetable Sci. 29, 16–24. doi: 10.1080/19315260.2022.2083042
De Serrano L. O. (2017). Biotechnology of siderophores in high-impact scientific fields. Biomolecular Concepts 8, 169–178. doi: 10.1515/bmc-2017-0016
Devaux A., Goffart J.-P., Petsakos A., Kromann P., Gatto M., Okello J., et al. (2020). Global food security, contributions from sustainable potato agri-food systems. potato crop: Its agricultural Nutr. Soc. contribution to humankind, 3–35. doi: 10.1007/978-3-030-28683-5_1
Eppinger M., Bunk B., Johns M. A., Edirisinghe J. N., Kutumbaka K. K., Koenig S. S. K., et al. (2011). Genome sequences of the biotechnologically important bacillus megaterium strains QM B1551 and DSM319. J. Bacteriol 193, 4199–4213. doi: 10.1128/JB.00449-11
Fan P., Chen D., He Y., Zhou Q., Tian Y., Gao L. (2016). Alleviating salt stress in tomato seedlings using Arthrobacter and Bacillus megaterium isolated from the rhizosphere of wild plants grown on saline–alkaline lands. Int. J. Phytoremediation 18, 1113–1121. doi: 10.1080/15226514.2016.1183583
Franco-Sierra N. D., Posada L. F., Santa-María G., Romero-Tabarez M., Villegas-Escobar V., Álvarez J. C. (2020). Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct. Integr. Genomics 20, 575–589. doi: 10.1007/s10142-020-00736-x
Friis G., Smith E. G., Lovelock C. E., Ortega A., Marshell A., Duarte C. M., et al. (2023). Rapid diversification of gray mangroves (Avicennia marina) driven by geographic isolation and extreme environmental conditions in the Arabian Peninsula. Authorea. (preprint). doi: 10.22541/au.165451878.80994363/v4
Goddek S., Körner O., Keesman K. J., Tester M. A., Lefers R., Fleskens L., et al. (2023). How greenhouse horticulture in arid regions can contribute to climate-resilient and sustainable food security. Global Food Secur. 38, 100701. doi: 10.1016/j.gfs.2023.100701
Gohil R. B., Raval V. H., Panchal R. R., Rajput K. N. (2022). Plant growth-promoting activity of Bacillus sp. PG-8 isolated from fermented panchagavya and its effect on the growth of arachis hypogea. Front. Agron. 4, 805454. doi: 0.3389/fagro.2022.805454
Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hakim S., Naqqash T., Nawaz M. S., Laraib I., Siddique M. J., Zia R., et al. (2021). Rhizosphere engineering with plant growth-promoting microorganisms for agriculture and ecological sustainability. Front. Sustain. Food Syst. 5, 617157. doi: 10.3389/fsufs.2021.617157
Hejazi M., Santos Da Silva S. R., Miralles-Wilhelm F., Kim S., Kyle P., Liu Y., et al. (2023). Impacts of water scarcity on agricultural production and electricity generation in the Middle East and North Africa. Front. Environ. Sci. 11, 157. doi: 10.3389/fenvs.2023.1082930
Islam S., Akanda A. M., Prova A., Islam M. T., Hossain M. M. (2016). Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front. Microbiol. 6, 1360. doi: 10.3389/fmicb.2015.01360
Ji C., Zhang M., Kong Z., Chen X., Wang X., Ding W., et al. (2021). Genomic analysis reveals potential mechanisms underlying promotion of tomato plant growth and antagonism of soilborne pathogens by bacillus amyloliquefaciens ba13. Microbiol. Spectr. 9, e01615–e01621. doi: 10.1128/Spectrum.01615-21
Khan M. A., Asaf S., Khan A. L., Jan R., Kang S.-M., Kim K.-M., et al. (2020). Thermotolerance effect of plant growth-promoting Bacillus cereus SA1 on soybean during heat stress. BMC Microbiol. 20, 175. doi: 10.1186/s12866-020-01822-7
Kim A.-Y., Shahzad R., Kang S.-M., Seo C.-W., Park Y.-G., Park H.-J., et al. (2017). IAA-producing Klebsiella variicola AY13 reprograms soybean growth during flooding stress. J. Crop Sci. Biotechnol. 20, 235–242. doi: 10.1007/s12892-017-0041-0
Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lee I., Ouk Kim Y., Park S.-C., Chun J. (2016). OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Systematic Evolutionary Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Leontidou K., Genitsaris S., Papadopoulou A., Kamou N., Bosmali I., Matsi T., et al. (2020). Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 10, 14857. doi: 10.1038/s41598-020-71652-0
Liu X., Shi L., Qian H., Sun S., Wu P., Zhao X., et al. (2020). New problems of food security in Northwest China: A sustainability perspective. Land Degrad Dev. 31, 975–989. doi: 10.1002/ldr.3498
Lotfi N., Soleimani A., Çakmakçı R., Vahdati K., Mohammadi P. (2022). Characterization of plant growth-promoting rhizobacteria (PGPR) in Persian walnut associated with drought stress tolerance. Sci. Rep. 12, 12725. doi: 10.1038/s41598-022-16852-6
Lutfullin M. T., Lutfullina G. F., Pudova D. S., Akosah Y. A., Shagimardanova E. I., Vologin S. G., et al. (2022). Identification, characterization, and genome sequencing of Brevibacterium sediminis MG-1 isolate with growth-promoting properties. 3 Biotech. 12, 326. doi: 10.1007/s13205-022-03392-z
Magotra S., Bhagat N., Ambardar S., Ali T., Hurek B. R., Hurek T., et al. (2021). Field evaluation of PGP Bacillus sp. strain D5 native to Crocus sativus, in traditional and non traditional areas, and mining of PGP genes from its genome. Sci. Rep. 11, 5454. doi: 1038/s41598-021-84585-z
Mahdi I., Fahsi N., Hafidi M., Allaoui A., Biskri L. (2020). Plant growth enhancement using rhizospheric halotolerant phosphate solubilizing bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 isolated from Chenopodium quinoa willd. Microorganisms 8, 948. doi: 10.3390/microorganisms8060948
Mishra R. K., Sahu P. K., Mishra V., Jamal H., Varma A., Tripathi S. (2023). Isolation and characterization of halotolerant plant growth promoting rhizobacteria from mangrove region of Sundarbans, India for enhanced crop productivity. Front. Plant Sci. 14, 1122347. doi: 10.3389/fpls.2023.1122347
Nosheen S., Ajmal I., Song Y. (2021). Microbes as biofertilizers, a potential approach for sustainable crop production. Sustainability 13, 1868. doi: 10.3390/su13041868
Ojuederie O. B., Babalola O. O. (2023). Growth enhancement and extenuation of drought stress in maize inoculated with multifaceted ACC deaminase producing rhizobacteria. Front. Sustain. Food Systems. 6. doi: 10.3389/fsufs.2022.1076844
Osland M. J., Feher L. C., López-Portillo J., Day R. H., Suman D. O., Menéndez J. M. G., et al. (2018). Mangrove forests in a rapidly changing world: Global change impacts and conservation opportunities along the Gulf of Mexico coast. Estuarine Coast. Shelf Sci. 214, 120–140. doi: 10.1016/j.ecss.2018.09.006
Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Patani A., Prajapati D., Ali D., Kalasariya H., Yadav V. K., Tank J., et al. (2023). Evaluation of the growth-inducing efficacy of various Bacillus species on the salt-stressed tomato (Lycopersicon esculentum Mill.). Front. Plant Sci. 14, 1168155. doi: 10.3389/fpls.2023.1168155
Patel S., Gupta R. S. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Systematic Evolutionary Microbiol. 70, 406–438. doi: 10.1099/ijsem.0.003775
Petrillo C., Castaldi S., Lanzilli M., Selci M., Cordone A., Giovannelli D., et al. (2021). Genomic and physiological characterization of Bacilli isolated from salt-pans with plant growth promoting features. Front. Microbiol. 12, 715678. doi: 10.3389/fmicb.2021.715678
Prajakta B. M., Suvarna P. P., Raghvendra S. P., Alok R. R. (2019). Potential biocontrol and superlative plant growth promoting activity of indigenous Bacillus mojavensis PB-35 (R11) of soybean (Glycine max) rhizosphere. SN Appl. Sci. 1, 1143. doi: 10.1007/s42452-019-1149-1
Priest F. G., Barker M., Baillie L. W. J., Holmes E. C., Maiden M. C. J. (2004). Population structure and evolution of the bacillus cereus group. J. Bacteriol 186, 7959–7970. doi: 10.1128/JB.186.23.7959-7970.2004
Quamruzzaman A. K. M., Islam F., Mallick S. R. (2021). Effect of different shade house on quality seedling raising of high value vegetables. Am. J. Plant Sci. 12, 7. doi: 10.4236/ajps.2021.121002
Rana A., Saharan B., Joshi M., Prasanna R., Kumar K., Nain L. (2011). Identification of multi-trait PGPR isolates and evaluating their potential as inoculants for wheat. Ann. Microbiol. 61, 893–900. doi: 10.1007/s13213-011-0211-z
Rath J. R., Ghosal M. K. (2020). Studies on environmental control of greenhouse for crop production. Int. J. Chem. Stud. 8, 674–676. doi: 10.22271/chemi.2020.v8.i5j.10377
Reang L., Bhatt S., Tomar R. S., Joshi K., Padhiyar S., Vyas U. M., et al. (2022). Plant growth promoting characteristics of halophilic and halotolerant bacteria isolated from coastal regions of Saurashtra Gujarat. Sci. Rep. 12, 4699. doi: 10.1038/s41598-022-08151-x
Sadiq H. M., Jahangir G. Z., Nasir I. A., Iqtidar M., Iqbal M. (2013). Isolation and characterization of phosphate-solubilizing bacteria from rhizosphere soil. Biotechnol. Biotechnol. Equip. 27:4248–4255. doi: 10.5504/BBEQ.2013.0091
Saleem S., Iqbal A., Ahmed F., Ahmad M. (2021). Phytobeneficial and salt stress mitigating efficacy of IAA producing salt tolerant strains in Gossypium hirsutum. Saudi J. Biol. Sci. 28, 5317–5324. doi: 10.1016/j.sjbs.2021.05.056
Savva A. P., Frenken K. (2002). Crop water requirements and irrigation scheduling (FAO Sub-Regional Office for East and Southern Africa Harare). Available at: https://www.fao.org/3/ai593e/ai593e.pdf (Accessed 20, 2024).
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shankar A., Prasad V. (2023). Potential of desiccation-tolerant plant growth-promoting rhizobacteria in growth augmentation of wheat (Triticum aestivum L.) under drought stress. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1017167
Sharma A., Dev K., Sourirajan A., Choudhary M. (2021). Isolation and characterization of salt-tolerant bacteria with plant growth-promoting activities from saline agricultural fields of Haryana, India. J. Genet. Eng. Biotechnol. 19, 99. doi: 10.1186/s43141-021-00186-3
Sultana S., Alam S., Karim M. M. (2021). Screening of siderophore-producing salt-tolerant rhizobacteria suitable for supporting plant growth in saline soils with iron limitation. J. Agric. Food Res. 4, 100150. doi: 10.1016/j.jafr.2021.100150
Sumbul A., Ansari R. A., Rizvi R., Mahmood I. (2020). Azotobacter: A potential bio-fertilizer for soil and plant health management. Saudi J. Biol. Sci. 27, 3634–3640. doi: 10.1016/j.sjbs.2020.08.004
Susič N., Žibrat U., Sinkovič L., Vončina A., Razinger J., Knapič M., et al. (2020). From genome to field—observation of the multimodal nematicidal and plant growth-promoting effects of Bacillus firmus I-1582 on tomatoes using hyperspectral remote sensing. Plants. 9, 592. doi: 10.3390/plants9050592
Teymouri M., Akhtari J., Karkhane M., Marzban A. (2016). Assessment of phosphate solubilization activity of Rhizobacteria in mangrove forest. Biocatalysis Agric. Biotechnol. 5, 168–172. doi: 10.1016/j.bcab.2016.01.012
Tounsi-Hammami S., Ali Khan M., Aroosa Z., Rasheed Anwar A., Arora N., Naseem M., et al. (2024). Harnessing bacterial strains from mangrove sediments and roots to develop native inoculant for enhancing tomato (Solanum lycopersicum) seedling growth at a reduced dose of NPK synthetic fertilization. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1356545
Vaser R., Sović I., Nagarajan N., Šikić M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Venkateswaran K., Singh N. K., Checinska Sielaff A., Pope R. K., Bergman N. H., Van Tongeren S. P., et al. (2017b). Non-Toxin-Producing Bacillus cereus Strains Belonging to the B. anthracis Clade Isolated from the International Space Station. mSystems 2, e00021–e00017. doi: 10.1128/mSystems.00021-17
Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Yadav R. S., Singh V., Pal S., Meena S. K., Meena V. S., Sarma B. K., et al. (2018). Seed bio-priming of baby corn emerged as a viable strategy for reducing mineral fertilizer use and increasing productivity. Scientia Hortic. 241, 93–99. doi: 10.1016/j.scienta.2018.06.096
Yatoo A. M., Ali M., Baba Z. A., Hassan B. (2021). Sustainable management of diseases and pests in crops by vermicompost and vermicompost tea. A review. Agron. Sustain. Dev. 41, 7. doi: 10.1007/s13593-020-00657-w
Zainab R., Shah G. M., Khan W., Mehmood A., Azad R., Shahzad K., et al. (2021). Efficiency of plant growth promoting bacteria for growth and yield enhancement of maize (Zea mays) isolated from rock phosphate reserve area Hazara Khyber Pakhtunkhwa, Pakistan. Saudi J. Biol. Sci. 28, 2316–2322. doi: 10.1016/j.sjbs.2021.01.025
Keywords: arid agroecosystem, Bacillus species, mangrove, plant growth-promoting bacteria, whole genome sequencing
Citation: Afridi M, Sadaiappan B, Nassar AS, Mahmoudi H, Khan MA and Mundra S (2024) Mangrove sediments-associated bacterium (Bacillus sp. SW7) with multiple plant growth-promoting traits promotes the growth of tomato (Solanum Lycopersicum). Front. Mar. Sci. 11:1379439. doi: 10.3389/fmars.2024.1379439
Received: 31 January 2024; Accepted: 08 May 2024;
Published: 28 May 2024.
Edited by:
Luiz Drude Lacerda, Federal University of Ceara, BrazilReviewed by:
Devendra Singh, Dr. Rajendra Prasad Central Agricultural University, IndiaArti Mishra, University of Delhi, India
Copyright © 2024 Afridi, Sadaiappan, Nassar, Mahmoudi, Khan and Mundra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunil Mundra, c3VuaWxtdW5kcmFAdWFldS5hYy5hZQ==; Munawwar Ali Khan, TXVuYXd3YXIuS2hhbkB6dS5hYy5hZQ==
†These authors have contributed equally to this work
 Mahideen Afridi
Mahideen Afridi Balamurugan Sadaiappan
Balamurugan Sadaiappan Amna Saood Nassar3
Amna Saood Nassar3 Henda Mahmoudi
Henda Mahmoudi Munawwar Ali Khan
Munawwar Ali Khan Sunil Mundra
Sunil Mundra