
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 05 April 2024
Sec. Marine Biology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1368315
 Jing Yang1
Jing Yang1 Zhimeng Xu1*
Zhimeng Xu1* Yi Chen1
Yi Chen1 Huo Xu1
Huo Xu1 Zuyuan Gao1
Zuyuan Gao1 Xiaodong Zhang1
Xiaodong Zhang1 Mengwen Pang1
Mengwen Pang1 Shuwen Zhang2
Shuwen Zhang2 Hongbin Liu1,3*
Hongbin Liu1,3*Zooplankton grazers, like copepods, can feed on toxic microalgae and live normally. We hypothesize that gut microbial communities (GMCs) may contribute to the detoxification of the host by changing their compositions and recruiting more beneficial bacteria. Here, we measured the physiological responses of two copepod species (Acartia sp. and Paracalanus sp.) fed with toxic (Alexandrium tamarense) and non-toxic (Alexandrium andersonii) dinoflagellates, respectively. Both copepods maintained consistently high survival rates but slightly reduced ingestion rates when feeding upon the toxic dinoflagellate (when compared to the non-toxic one), suggesting a compensatory mechanism. The compositional variation of copepod GMCs, at the amplicon sequence variant (ASV) level, was mostly significantly different among copepod host species (R = 0.83, by ANOSIM test), while diet type played minor but significant roles. Under the toxic diet, Acartia sp. enriched only five ASVs while Paracalanus sp. recruited a wide range of taxa (38 ASVs) mostly belonging to Alphaproteobacteria (e.g., Rhodobacteraceae) and Gammaproteobacteria (e.g., Alteromonadaceae). In contrast, when clustering GMCs by predicted functions, diet type was the key regulating factor, suggesting the functional convergence of copepod GMCs in response to algal toxins. This can be explained by the fact that most of the enriched bacteria under the toxic diet have similar functions on detoxification and maintaining the host homeostasis. This study deepens our understanding of the roles of GMC in the detoxification and adaptation mechanisms of copepods during harmful algal blooms.
Harmful algal blooms (HABs) caused by dinoflagellates are a significant global issue that can have serious ecological and economic impacts. Ecologically, dinoflagellate HABs can lead to the depletion of oxygen levels in the water, the creation of dead zones, and the alteration of food webs (Turner, 2014). Economically, HABs can lead to the closure of fisheries, the loss of tourism revenue, and the costs associated with monitoring and mitigating HAB outbreaks (Anderson et al., 2000). For example, the high PSP toxins levels produced by toxic Alexandrium tamarense caused high mortality of caged salmon in Nova Scotia (Cembella et al., 2002). Additionally, the transfer of PSP toxins through the food chain has put the western North Atlantic right whale Eubalaena glacialis at risk of reduced reproduction rates (Doucette et al., 2006). Humans have also been impacted by this issue, with reports of poisonings from consuming crabs contaminated with PSP toxins (Noguchi et al., 2011). Several marine dinoflagellate species such as Alexandrium spp., Pyrodinium spp., and Gymnodinium spp., can produce paralytic shellfish poisoning (PSP) toxins (Anderson et al., 1990; Band-Schmidt et al., 2019). PSP is a foodborne illness that results from consuming seafood products contaminated with neurotoxins. These toxins include several derivatives, with gonyautoxins (GTX) 1–4, saxitoxin (STX), and neosaxitoxin (NEO) being the most potent (Etheridge, 2010). These neurotoxins bind to voltage-gated sodium channels, preventing the passage of sodium ions and attenuating action potentials, progressively inhibit neurotransmission, relax smooth muscle, and lead to paralysis and respiratory failure in humans, as well as mussels, fishes, and seabirds (Tester et al., 2000; Hallegraeff, 2003; Turner, 2014), though several studies showed no evidence of these effects on some marine zooplankton (Van Dolah, 2000; Etheridge, 2010; Finiguerra et al., 2014; Roncalli et al., 2017). Shellfish, zooplankton, and herbivorous fish consume these toxic microalgae and act as vectors to humans directly or transfer through the food web to higher trophic levels (Van Dolah, 2000).
Copepods are the dominant components of mesozooplankton in the ocean, playing a vital role in marine food webs by transferring energy from primary producers to higher trophic levels (Sommer and Stibor, 2002; Schminke, 2007; De Troch et al., 2012). Interestingly, studies have shown that copepods can graze on toxic microalgae without experiencing any apparent negative effects (Turner and Anderson, 1983; Uye, 1986; Turner and Tester, 1989; Turner, 2014). For instance, studies have shown that Acartia bifilosa can consume toxic Dinophysis spp. which causes diarrhetic shellfish poisoning (DSP) without any food selectivity, regardless of the concentration of the toxic microalgae (Kozlowsky-Suzuki et al., 2006; Reguera et al., 2014). Both Acartia sp. and Temora sp. do not suffer any negative effects on survival, egg production, or hatching success when consuming toxic Pseudo-nitzschia spp., which is responsible for amnesic shellfish poisoning, and Acartia sp. even had a higher ingestion rate when consuming toxic P. multiseries (Lincoln et al., 2001). Meanwhile, the okadaic acid, a diarrhetic toxin, excreted by copepods was lower than what copepods ingested, suggesting that copepods are efficient in detoxification and do not bring toxins to upper consumers (Kozlowsky-Suzuki et al., 2006; Reguera et al., 2014). However, mechanisms for the adaptation of marine copepods to toxic microalgae, especially toxic dinoflagellates, are still unclear.
Gut microbes play a crucial role in the detoxification of toxic diets for a wide range of animals including mammals and arthropods. For instance, thanks to their gut flora, the freshwater crustacean Daphnia magna can tolerate toxic cyanobacteria and the black soldier fly (Hermetia illucens) larvae can consume toxic seaweed Rugulopteryx okamurae without experiencing any adverse effects (Macke et al., 2017; Patón et al., 2023). These benefits can be attributed to the ability of gut microbes to regulate the host’s expression of detoxifying enzymes, such as cytochrome P450 and glutathione S-transferase (GST), or directly digest toxic compounds before they diffuse from the gut into other tissues of the host (De Wit et al., 2018; Sun et al., 2022). However, little is known about the roles of gut microbes in helping copepods adapt to toxic algal diet and maintain homeostasis (i.e., stable physiology performance) during HAB events. One study reported that the abundance of the mlrA gene, a microcystin-degrading gene, was positively correlated with the density of toxic cyanobacteria, suggesting that gut bacteria carrying mlrA gene can contribute to the biodegradation of toxins ingested by the host and facilitate nutrition (Gorokhova et al., 2021). Still, the detailed changes of GMCs, both in terms of composition and function, were much less studied and require comprehensive analysis to understand the detoxification mechanisms.
The copepod gut creates a distinct acidic and suboxic-anoxic microhabitat that promotes the colonization of microbiota, supporting unique carbon and nutrient sources (Tang et al., 2011). Bacteria are introduced into the gut when copepods feed on detritus or plankton prey that have bacteria attached to them (Moisander et al., 2015; Scavotto et al., 2015). Shoemaker & Moisander (2017) found that large amounts of active bacteria were isolated from fecal pellets and released into the surrounding seawater, potentially playing a significant role in marine biogeochemical cycles, sedimentation, and enhancing nutrient regeneration if these bacteria have specialized functions. Many kinds of bacteria, such as Bacteroidetes, Gammaproteobacteria, and Alphaproteobacteria, which are commonly associated with phytoplankton, can enter the copepod GMC through ingested food (Teeling et al., 2012). Due to the wide distribution of copepods, such as through vertical migration and passive dispersal by ocean currents, the microbial communities within their guts contribute greatly to the distribution of bacterioplankton in all oceans.
The impact of different copepod host species and ambient environmental conditions on copepod GMC are the main focus of recent studies. Researchers have shown that differences in copepod host species played the greatest role in the composition of copepod GMC (Skovgaard et al., 2015; Datta et al., 2018; Wäge et al., 2019), mainly regulated by the different physiology and life history of the hosts (Tang, 2005; Skovgaard et al., 2015; Almada and Tarrant, 2016). However, other reports have suggested that environmental conditions, especially food supply, may be the dominant factor regulating zooplankton GMC (Tang et al., 2011; Bickel et al., 2014; De Corte et al., 2014). For instance, seasonal variations may bring different food sources, leading to dramatic alterations in copepod GMC (Shoemaker and Moisander, 2017). The significant difference in the gut content of Calanus finmarchicus among the North Atlantic regions has been attributed to diverse environmental conditions (Yeh et al., 2020). Field diets can contain various toxins and these toxic diets can lead to significant changes of GMC as a detoxification strategy of hosts, which has been reported in terrestrial arthropods (Shukla and Beran, 2020). However, the changes of copepod GMC in response to toxic dinoflagellate diet, which is vital to the understanding of their detoxifying mechanisms, has received little attention and less is known about the regulating factors, such as species difference of copepods (as host) and dinoflagellates (as food).
In this study, we hypothesize that GMCs may help the host with the detoxification of toxic dinoflagellate as a diet by changing their compositions and recruiting more beneficial bacteria. To test this hypothesis, we ask the following questions: (1) Do copepod GMC change temporally and significantly in response to food supply? Further, can GMC be different or similar between toxic and non-toxic microalgae diets? (2) Do diet-induced GMC changes differ between copepod species? (3) How do GMC functionally help copepods detoxify? To answer these questions, we used two copepod species, Acartia sp. and Paracalanus sp., in our study and fed them two dinoflagellates species from the same genus, the toxic Alexandrium tamarense (as toxic microalgae) and the non-toxic Alexandrium andersonii (as control). The two copepods and dinoflagellates are typically abundant and frequently coexist in subtropical coastal waters (Liu et al., 2021; Xiang et al., 2021). In particular, the two copepod species are from different taxonomic families and have different feeding modes (Acartia sp.: ambush and filter feeding, Paracalanus sp.: filter feeding) (Turner, 1984; Hansen et al., 1994; Behrends and Schneider, 1995; Saiz and Kiørboe, 1995), providing insights into the host differences in feeding strategies may align with differences in host GMCs. The two dinoflagellate species are from the same genus, which minimizes the influence of other factors such as nutritional values. In addition to the measurement of the physiologic performance of copepods, we revealed the copepod GMCs using the high throughput sequencing of 16S rRNA gene at the individual level and performed comprehensive analyses on the compositional and functional changes of GMC in response to the toxic diet. We showed the significant changes in GMCs of copepods under different diets which confirmed our hypothesis and identified a number of enriched bacterial taxa which may contribute to the detoxification and homeostasis of the host.
Copepods were collected from the Port Shelter (Hong Kong SAR, China) using a 200-µm mesh size zooplankton net, and immediately taken back to the laboratory. Two copepod species, Acartia sp. and Paracalanus sp. were identified under a dissection microscope (Olympus IX51, Japan), and adults (n = 500 for each species) with similar body lengths were picked and used for experiments immediately. Experimental copepods were divided into field, toxic, and non-toxic algal diet groups. The field groups were performed with gut dissection immediately after collection, while the toxic and non-toxic algal diet groups were fed with the corresponding dinoflagellates, and cultured in triplicated 5-L beakers filled with 0.2-μm filtered local coastal seawater with continuously pumped air to provide sufficient oxygen. Algal debris and copepod carcasses were removed by droppers to maintain good water quality in culturing flasks.
Alexandrium tamarense (strain TIO855, isolated from Sishili Bay in Yantai, China) and Alexandrium andersonii (strain TIO940, isolated from the coastal water of Koh Mak, Thailand) were selected as the toxic and non-toxic algal diet for copepods, respectively. They were cultured in 800-mL culture flasks containing 0.2-μm filtered sterilized seawater supplemented with f/2 medium (Guillard, 1975). Cultures were maintained in a temperature-controlled walk-in chamber at 23.5°C with a 14h:10h light: dark cycle, 60 μmol m-2 s-1 light intensity and transferred to fresh medium every two weeks (Tillmann et al., 2009). To ensure consistent results and minimize the influence of external factors, such as toxin accumulation during algae cultivation (particularly in the post-exponential phase), we provided copepods with the algae consistently at the exponential growth stage (i.e., days 10 and 7 following transfer, for A. tamarense and A. andersonii, respectively) daily throughout the experiment period.
Grazing rate depends greatly on prey concentration and can be largely biased when the prey concentration is too high. Thus, preliminary experiments were conducted to determine the minimal dinoflagellate cell concentration leading to maximal copepod grazing rates to be used in the grazing experiment. Four algal concentrations (~ 500, 1000, 1500, 2000 cells mL-1) of A. tamarense and A. andersonii were added separately to experimental bottles (500 mL) with 10 copepods and control bottles (500 mL) without copepods, both in triplicates. After incubating at room temperature in the dark and on a rotating plankton wheel (0.5 rpm) for 8h, the number of surviving copepods was recorded and 500 μL algal samples from each bottle were collected and fixed with 2% (final concentration) acid Lugol’s solution. Cell counts were obtained in 24-well cell culture plates under an inverted microscope (Olympus IX51, Japan). Dinoflagellate cells were counted from top to bottom and from left to right, counting 5 fields of view for each direction, obtaining 10 fields of view. The ingestion rate of copepods under different algal concentrations was calculated following the formula (Frost, 1972):
where I (cells cop-1 d-1) is the ingestion rate of the copepod, Cc1 (cell mL-1) and Ce1 (cell mL-1) are the algae concentrations of the control group and the experimental group at the end of the experiment separately, Ce0 (cell mL-1) is the algae concentration of the experimental group at the beginning of the experiment, V (mL) is the volume of the algae solution for foraging experiments, N is the number of surviving copepods, and t (Day) is the duration of the grazing experiment.
Grazing experiments were performed separately by feeding copepods toxic or non-toxic dinoflagellates on day 0, 1, 2, and 3 with the minimal cell concentrations achieving maximal ingestion rate (determined in the preliminary experiment) (Supplementary Table 1). The experimental settings such as the number of copepods, grouping, temperature, and time are the same as those in the preliminary experiment. The experiments were conducted in triplicates and grazing rates were estimated as explained above.
The copepod survival rate under different algal diets was measured daily by randomly picking out 10 individuals from the culture beaker of each treatment and culturing them in a 500 mL polycarbonate (PC) bottle with the same prey concentration for 24 h. The number of alive copepods was counted and then removed. Survival rate was calculated by:
where S (%) is the survival rate of copepods, and n is the number of surviving copepods. If there was no movement within 60 s after the mechanical stimulus, the copepod was defined as dead and excluded from the calculation of survival rate (da Costa et al., 2008).
Concurrently with the grazing experiments, 200 mL of exponentially grown algae were collected and filtered using a GF/F glass fiber membrane (Whatman). Filters were added with 0.5 mL of 0.1 mol L-1 acetic acid solution, extracted by ultrasonic breaking, and centrifuged at 12,000 rpm for 5 min. The supernatant was taken to another reagent bottle, by adding 0.5 mL of extractant to the residue, and repeated the above steps to combine the supernatant liquid. Then a C-18 solid-phase extraction cartridge was used for purification. The supernatant was eluted with 0.5 mL of 0.1 mol L-1 acetic acid solution after passing through the column. The liquid and eluent passing through the column were obtained and adjusted to 2 mL with 0.1 mol L-1 acetic acid solution and then passed through a 0.22-μm filter membrane to run on the LC-MS/MS machine (LCMS-8040, Japan) for toxin detection. Here, paralytic shellfish poisoning (PSP) toxins were measured, including gonyautoxins (GTX 1-4 and GTX 5-b: formerly B1), saxitoxin (STX), neosaxitoxin (NEO), N-sulfocarbamoyl gonyautoxins (C1 and C2).
For each combination of copepod species and toxic/non-toxic dinoflagellates, 15 copepod individuals were collected from the culturing beakers on day 0 and day 2. Gut dissection was performed on each individual copepod, with sterilized (i.e., autoclaved and 70% ethanol steeped) dissection tweezers (Regine 5, Switzerland) in a sterile Petri dish under the stereomicroscope (Stemi 305, Germany) (Supplementary Figure 1) and stored in a 1.5 mL tube with small amount of 70% ethanol (~ 10 μL). All samples were kept at − 80°C for DNA extraction.
DNA was extracted from the tubes containing copepod gut using the DNeasy Blood & Tissue Kit (QIAGEN, Germany) following manufacturer protocol. Briefly, 180 μL proteinase K was added to each 1.5-mL microcentrifuge tube containing one copepod gut. Tubes were then vortexed and incubated at 56°C until complete lysis (around 1 h, vortexed every 20 mins), followed by the addition of 200 μL Buffer AL and vortexing. Copepod gut samples were incubated at 56°C for 10 min, then 200 μL absolute ethanol was added and vortexed. Afterward, the mixture was transferred to a DNeasy Mini spin column and placed into a 2 mL collection tube. These mixtures were centrifuged at 8,000 rpm for 2 min, then the flow-through and collection tube were discarded, the spin column was placed in a new 2 mL collection tube and added 500 μL Buffer AW1. The centrifugation procedure was repeated once but added 500 μL Buffer AW2. At last, the mixture was centrifuged at 14,000 rpm for 5 min and the spin column was transferred to a new 1.5 mL microcentrifuge tube. The extracted DNA was then eluted into 60 μL AE buffer for PCR amplification.
PCR was conducted to amplify the 16S rRNA gene of bacteria in the copepod gut with the forward primer 341F (5′-CCTACGGGRSGCAGCAG-3′) (Muyzer et al., 1993) and reverse primer 787R (5′-CTACNRGGGTATCTAA-3′) (Buchholz-Cleven et al., 1997). The PCR cycling program was performed as follows: pre-denaturing at 95 °C for 5 min; 30 cycles of denaturing at 95 °C for 45 s, annealing at 55 °C for 45 s, extension at 72 °C for 60 s; and a final extension at 72 °C for 10 min. The PCR reactions were conducted in triplicates to ensure the reliability of the results. The PCR products of 16S rDNA were pooled together and sequenced by a Hiseq 2500 System (Illumina) with 2 × 250 bp paired end read configurations.
R raw reads were processed using QIIME2 (https://docs.qiime2.org/) with tutorial pipelines (Bolyen et al., 2019). After removing the barcode and primer, the sequences (with paired ends for each sample) were imported into QIIME2 (v 2019.10), visually inspected for sequence quality after demultiplexing, and processed with DADA2 to remove contamination, trim reads, correct errors, merge read pairs and remove chimeras. Representative ASV (amplicon sequence variants) sequences and their abundances were extracted by feature table (file “table.qza”). Then, the representative ASV sequences were classified with detailed taxonomy information using the trained classifier. ASVs assigned to chloroplast and mitochondria (75 of 6773 ASVs) were removed. ASV abundance of each sample was obtained, annotated to different taxonomic levels, and rarefied to the same depth using the “rrarefy” function in the “vegan” package of R software (Oksanen et al., 2007) (https://www.r-project.org/). GMCs of copepods were constructed by the ASV abundance (number of sequences) shared by samples.
After rarefied to the same depth, the α-diversity (Richness and Shannon) of GMC was calculated using the “estimate” function in the “vegan” package of R. The values of α-diversity in different treatments were compared by the Wilcoxon test after the Shapiro–Wilk normality test. Community similarity was calculated by Bray-Curtis’s distance using the “vegdist” function. To show the effects of copepod species and diet type on the GMC, grouping the GMC based on community similarity was shown by Non-metric Multidimensional Scaling (nMDS) maps using the “monoMDS” function in the “vegan” package of R, with the difference and significance tested by analysis of similarity (ANOSIM) (Clarke, 1993).
A similarity percentage (SIMPER) analysis was conducted to identify the distinct ASVs between two experimental groups (i.e., toxic vs. non-toxic) of GMC with significance (p-value) by using the “vegdist” and “anosim” functions in the “vegan” package of R. The mean relative abundance of each ASV was calculated and compared between two groups, with largely and significantly changed ones (i.e., both |fold-change| > 2 and SIMPER p-value< 0.05) regarded as distinct ASVs (between two groups).
PICRUSt2 (phylogenetic investigation of communities by reconstruction of unobserved states) (Douglas et al., 2020) was used for the functional prediction of GMCs. In brief, sequencing data of the 16S rRNA marker gene were annotated using the phylogenetic tree from the software and matched with the microbial metagenomic database to obtain the functional information. The abundance of predicted functions was normalized by the sequence abundance of each ASV. The functional annotation results were shown in both KEGG Orthology (KO) (Kanehisa and Goto, 2000) and Enzyme Code (EC). ANOSIM tests were performed to analyze the effects of both copepod species and algal diet types on the functions of GMC.
Saxitoxin, neosaxitoxin, and N-sulfocarbamoyl gonyautoxins were not detected in the two dinoflagellates, while gonyautoxins were detected only in A. tamarense including GTX-1, GTX-2, GTX-3, and GTX-4 with concentrations of 1.10 ± 0.54, 0.03 ± 0.01, 0.01 ± 0.01, and 0.34 ± 0.17 ng per cell, respectively.
Positive correlations between dinoflagellate cell concentration and copepod ingestion rate were observed in our preliminary experiment of grazing (Supplementary Figure 2). All the ingestion rates were saturated at the highest dinoflagellate cell concentration (2000 cells mL-1). For instance, when the two copepod species fed on the toxic A.tamarense, their feeding rates reached stable at the cell concentration of 1000 cells mL-1 (Supplementary Figures 2A, C) while the saturating point occurred at ~ 2000 cells mL-1 for the non-toxic A.andersonii (Supplementary Figures 2B, D). These cell concentrations were used for the later daily measurement of the grazing rate of copepods fed on different dinoflagellate diets (Supplementary Table 1).
In the daily-measured grazing experiment, both copepod species showed a stable ingestion rate on the non-toxic diet throughout the period (7.05 ± 1.06 × 104 cells copepod-1 day-1 for Acartia sp., 7.35 ± 0.64 ×104 cells copepod-1 day-1 for Paracalanus sp.) (Figures 1B, D). However, when fed the toxic diet, the ingestion rate of both copepod species showed a slight but significant decline after the second day (p< 0.05, by Kruskal-Wallis test) (Figures 1A, C).
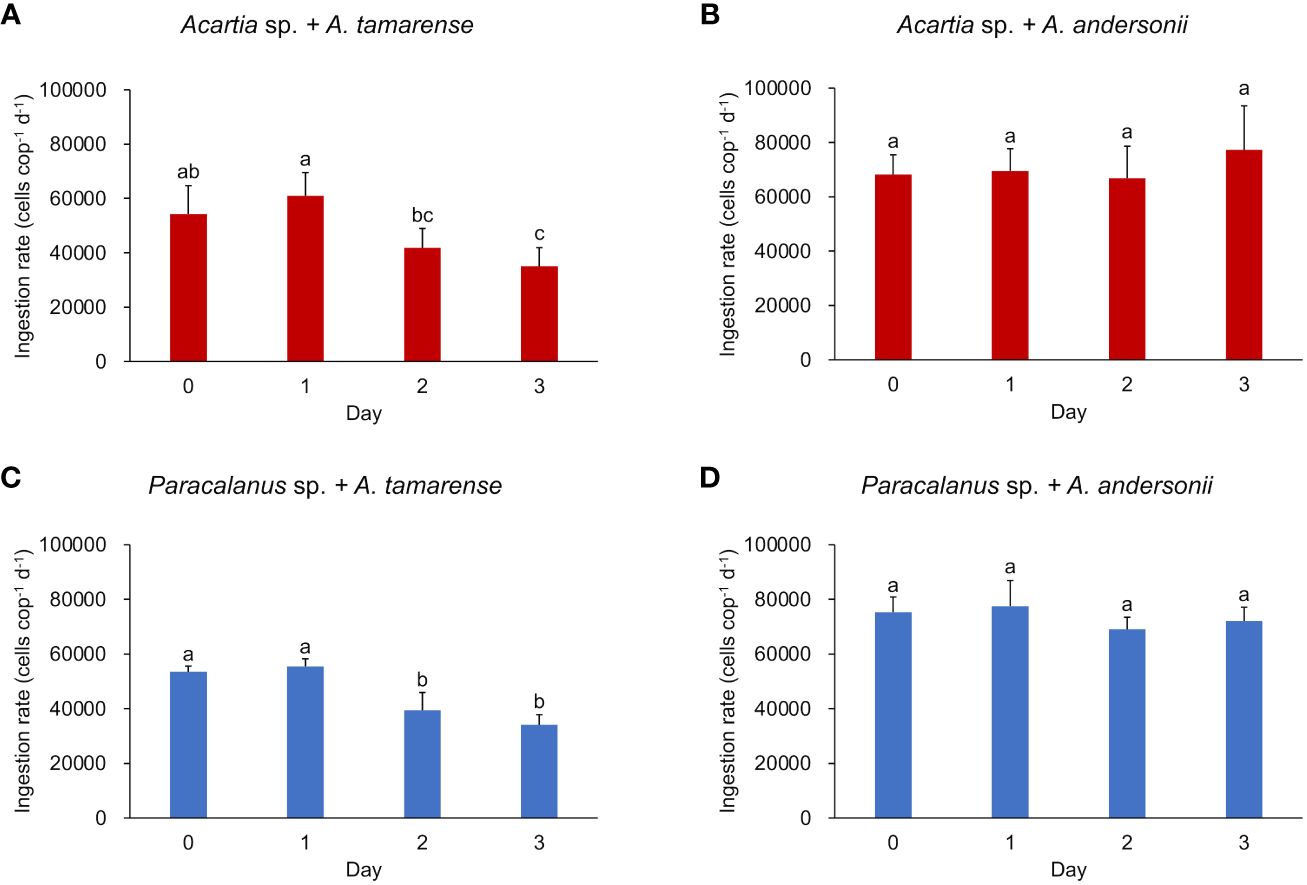
Figure 1 Ingestion rate of two copepod species under different algal diets. (A, B) show the ingestion rate of Acartia sp. fed with toxic algae Alexandrium tamarense and non-toxic algae Alexandrium andersonii, respectively; (C, D) show the ingestion rate of Paracalanus sp. fed with Alexandrium tamarense and Alexandrium andersonii, respectively. Ingestion rate was measured in triplicates and shown by average value with error bar (SD). Kruskal-Wallis tests were used to test the significant differences between groups marked with different letters. Histograms marked with the same letter have no significant change, while histograms marked with different letters are significantly different from each other, p< 0.05.
Survival rate of the two copepod species was consistently high (> 80%) during the incubation period (Figure 2). In general, survival rates of Acartia sp. were relatively lower with larger variations (88% ± 10% fed on toxic diet, 86% ± 7% fed on non-toxic diet) when compared to Paracalanus sp. which showed consistently higher values (93% ± 4% fed on toxic A. tamarense, 98% ± 1% fed on non-toxic A. andersonii). Except for Acartia sp., which had a significant increase in survival rate when eating toxic dinoflagellate in day 2, the survival rates of other groups had no significant changes.
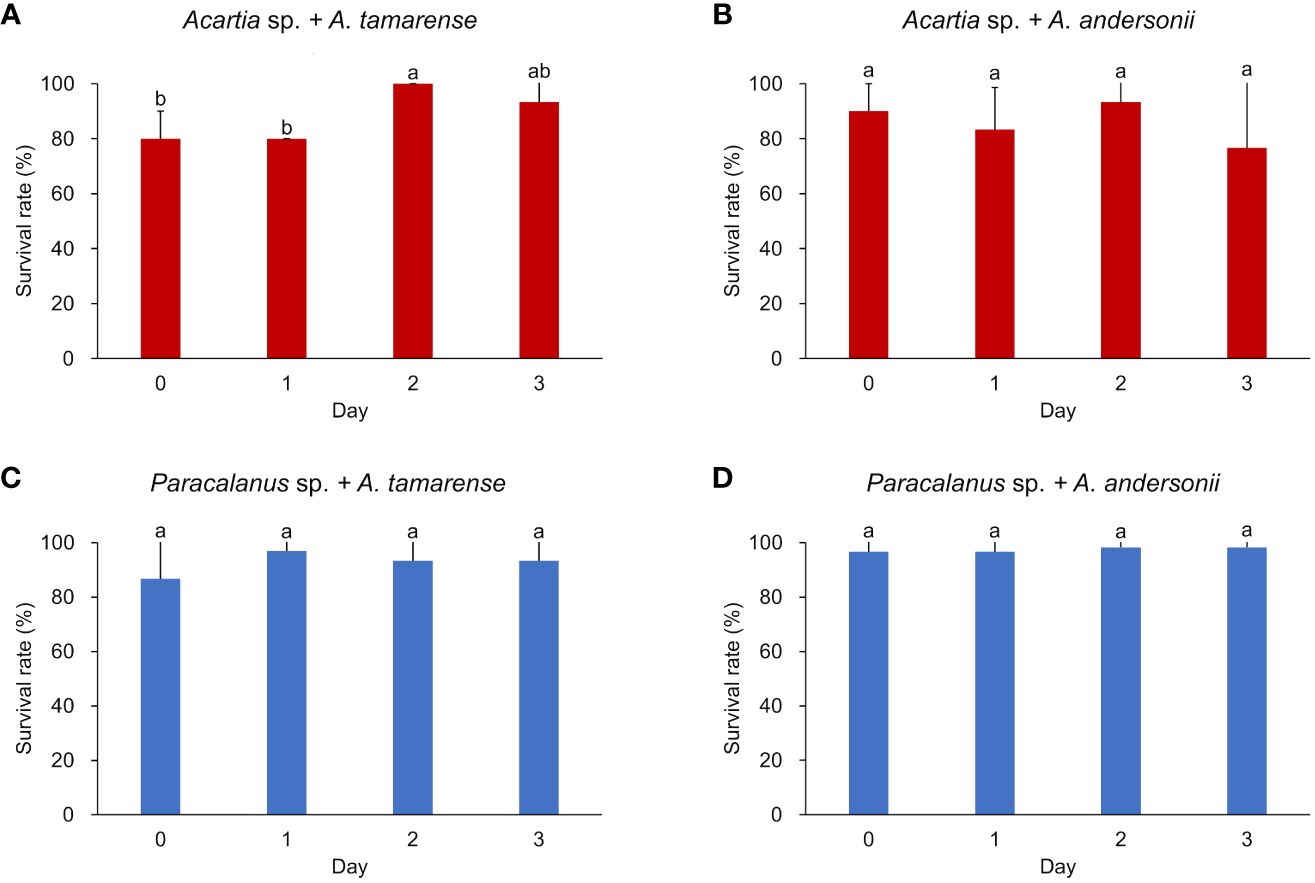
Figure 2 Survival rate of two copepod species under different algal diets. (A, B) show the survival rate of Acartia sp. fed with toxic algae Alexandrium tamarense and non-toxic algae Alexandrium andersonii, respectively; (C, D) show the survival rate of Paracalanus sp. fed with Alexandrium tamarense and Alexandrium andersonii, respectively. Survival rate was measured in triplicated (each had 10 individuals of copepods) and shown by average value with error bar (standard deviation, SD). Significant differences between groups were analyzed by Kruskal-Wallis tests and marked with different letters. Histograms marked with the same letter have no significant change, while histograms marked with different letters are significantly different from each other, p< 0.05.
A total of 66 GMCs were constructed by 16S rDNA sequencing data (Figure 3). In general, at the class level, Alphaproteobacteria were the most dominant component across all communities (53.80%, on average). For Acartia sp. GMC, apart from Alphaproteobacteria, Actinobacteria (17.77%, on average) was the most abundant group in the field samples, while remarkable proportions of Campylobacteria (8.32%, on average) and Saccharimonadia (6.27%, on average) were found in the non-toxic and toxic diets, respectively. For Paracalanus sp. GMC, however, no apparently consistent change of bacterial class was observed between treatments, which was mainly due to the large heterogeneity of community composition among individuals, suggesting that the difference of GMC should be compared at finer taxonomic levels (i.e., lower than class level).
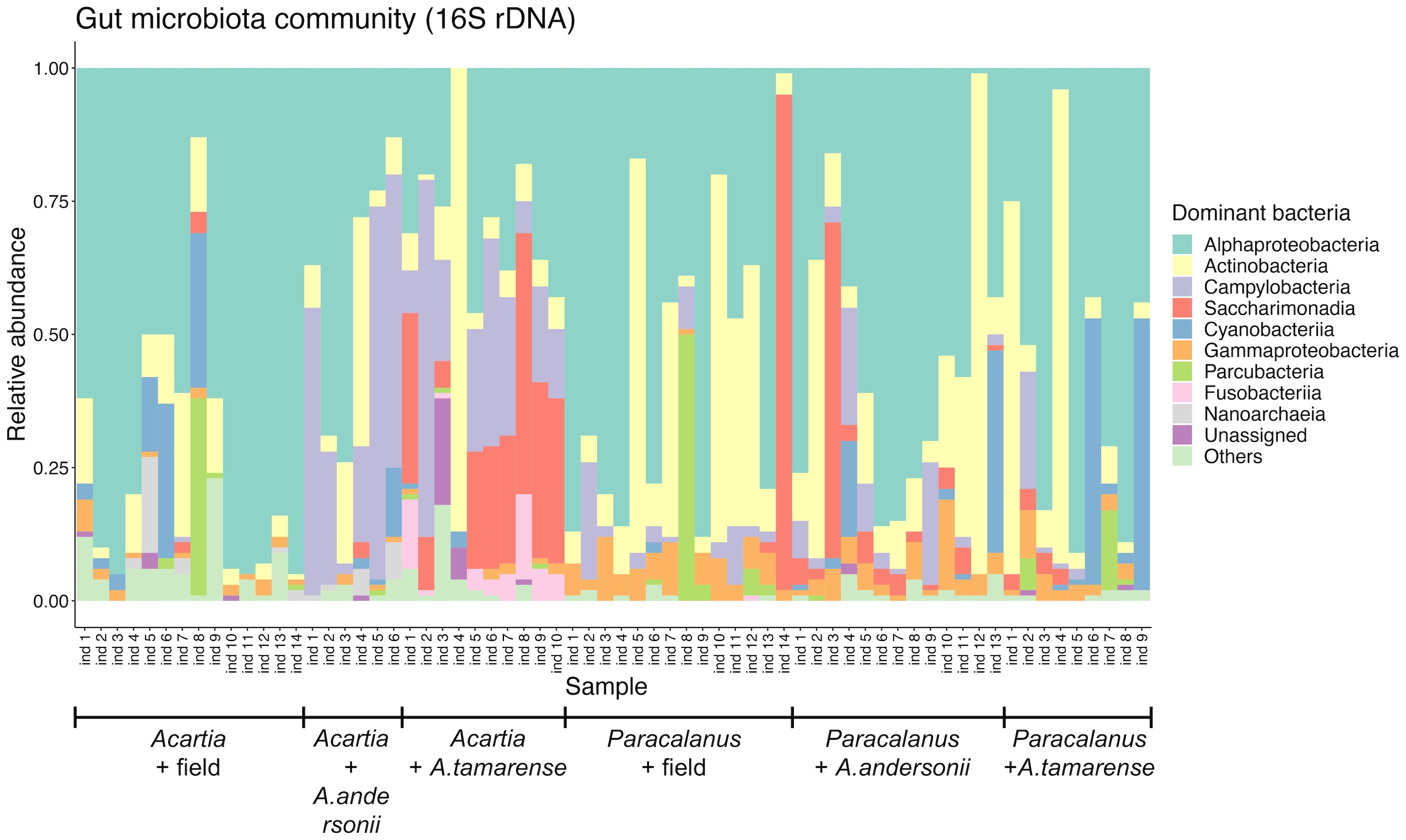
Figure 3 Relative abundance of major bacterial groups in the copepod GMC. Community compositions of copepod gut microbes were shown at the class level, with a relative abundance of sequences of the most abundant ten classes. Each community was constructed from the gut of one individual copepod (Acartia sp. or Paracalanus sp.) that was treated differently (i.e., field sampled, toxic or non-toxic diet treatment). Alexandrium tamarense: toxic diet; Alexandrium andersonii: non-toxic diet.
Alpha diversity of copepod GMC was compared between different groups using the indices of species richness (i.e., observed ASV number in a sample) and Shannon diversity (Supplementary Figure 3). In general, no significant difference was found between groups by two-sample Wilcoxon tests, except the significantly higher richness observed in the toxic diet treatment for Acartia sp. compared with the non-toxic diet (Supplementary Figure 3C).
Beta diversity of copepod GMC was calculated by the Bray-Curtis distance between any two samples. While there were no significant differences between any two groups by Wilcoxon test, the average value of community dissimilarity for each group is high (> 0.8), suggesting that the GMC varied greatly among individuals and should be revealed at the individual level for copepods (Supplementary Figure 4).
Effects of copepod species and algal diet type on GMC were shown and compared on the nMDS maps which were constructed based on community similarity (Bray Curtis distance) (Figure 4). A clear separation of GMC between Acartia sp. and Paracalanus sp. was observed (ANOSIM-R = 0.83, p< 0.001) (Figure 4A), suggesting the strong effects of copepod species on the structure of GMC. Significant effects of three treatments on GMC were shown for each copepod species separately (Figures 4B, C).
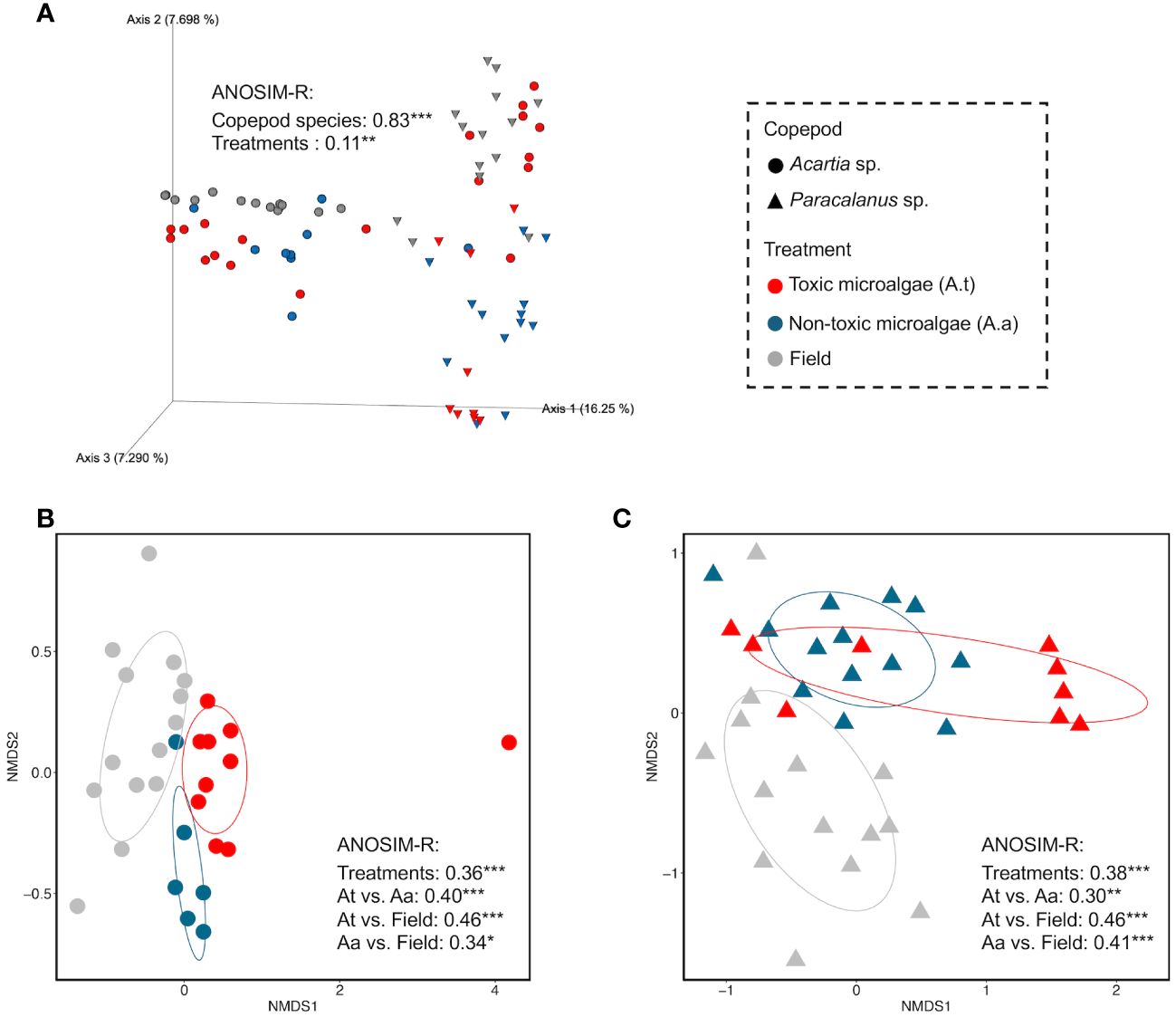
Figure 4 Effects of copepod species and algal diet on GMC compositions based on community similarity analysis. Community similarity between any two samples was calculated by Bray-Curtis distance at the ASV level and visualized in non-metric multidimensional scaling (nMDS). (A) showed the effects of copepod species and diet on GMC compositions, using a 3D map. (B, C) showed the diet effects (field, toxic, and non-toxic algal diet) on the GMC of Acartia sp. and Paracalanus sp., respectively. Analysis of similarities (ANOSIM) was used to test the significant difference between different groups of GMCs (i.e., significant influence of factors on GMCs). A higher ANOSIM-R value indicates a larger difference between the two groups.
Compared to non-toxic dinoflagellate diet, GMC of Acartia sp. fed on toxic dinoflagellate diet had a total of 5 ASVs significantly increased (Log2FoldChange > 1, p< 0.05) and 2 ASVs significantly decreased (Log2FoldChange< -1, p< 0.05) (Figure 5A). Notably, all the increased ASVs had a large fold-change value over 4, which were from different classes including Actinobacteria, Alphaproteobacteria, Campylobacteria and Parcubacteria (Supplementary file 2).
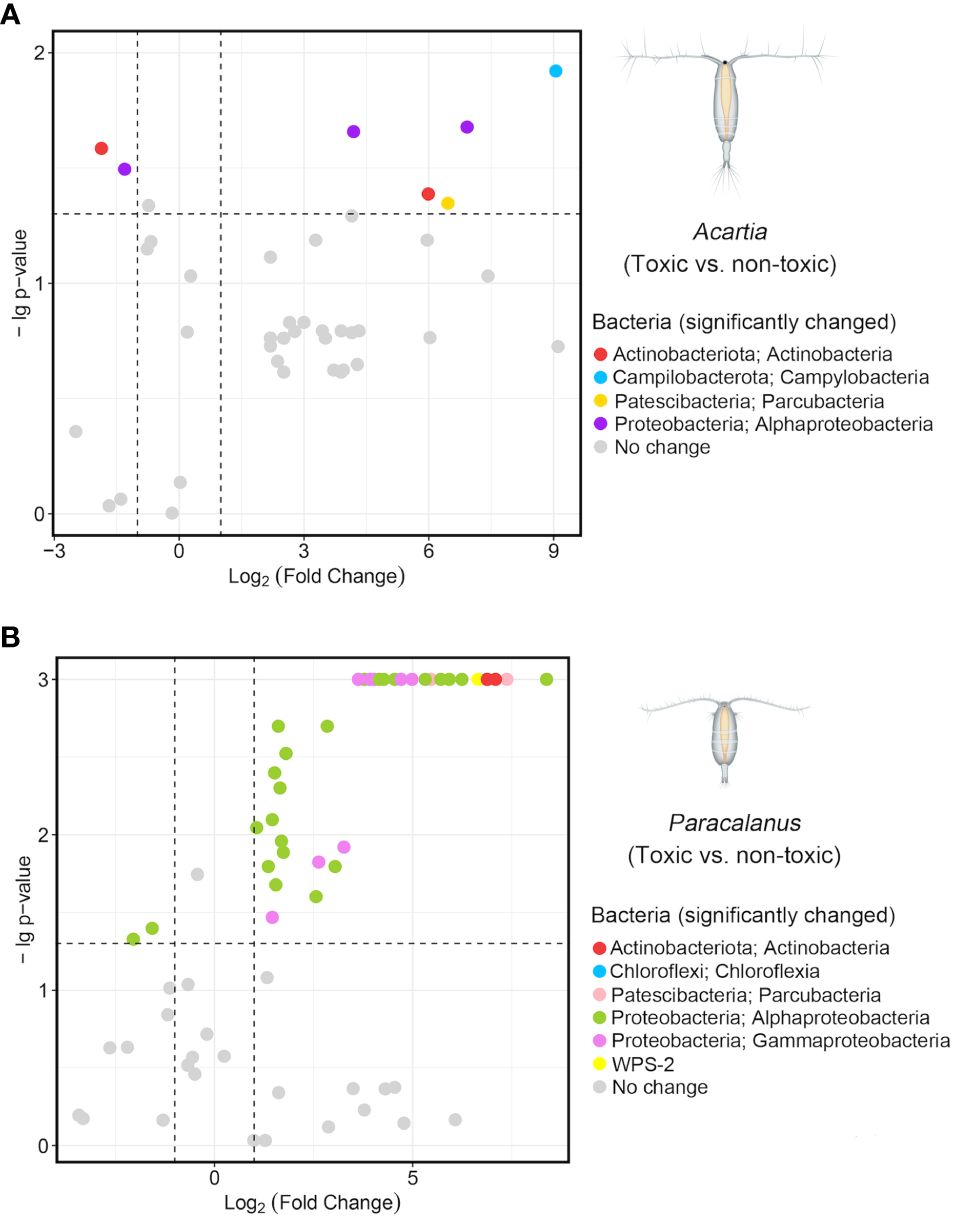
Figure 5 Gut bacteria with significant changes under a toxic diet. A similarity analysis (SIMPER) was conducted to identify all the distinct ASVs between two groups (i.e., toxic and non-toxic) of GMC with significance (p-value) for Acartia sp. (A) and Paracalanus sp. (B). Fold Changes of these distinct ASVs were calculated based on their relative abundance in each group (i.e., toxic and non-toxic diet) and log2 transformed. The two dashed lines on the x-axis were x = 1 and x = -1, representing the increasing (x > 1) and decreasing (x< -1) trend of gut microbes after feeding toxic algae (vs. non-toxic), respectively. The dashed line on the y-axis was y = 1.3, and ASV with a p-value< 0.05 (i.e., -lg(p-value) > 1.3) was considered as significantly changed and colored according to its taxonomy (Phylum, Class).
For Paracalanus sp., GMC under toxic diet, compared to non-toxic, had a large number of significantly increased ASVs (n = 38) which were mainly from Alphaproteobacteria (n = 23, mainly from the order Rhodobacterales), followed by Gammaproteobacteria (n = 8) (Figure 5B). Three ASVs belonged to Actinobacteria increased significantly with a high fold-change value over 6 (Supplementary file 2). Similar to Acartia sp., only 2 ASVs belonging to Alphaproteobacteria (order Rhodobacterales) were detected as significantly decreased in the toxic dinoflagellate diet treatment for the GMC of Paracalanus sp.
Functions of GMC were predicted in terms of KEGG Orthology (KO) and Enzyme Code (EC). Functional similarity of GMC was calculated by Bray-Curtis distance of the two indices between two communities and visualized by nMDS maps (Figure 6). Algal diet type had a more significant effect on copepod GMC (ANOSIM-R = 0.15 for both KO and EC, p< 0.001) than copepod host species (ANOSIM-R = 0.068 for KO, ANOSIM-R = 0.048 for EC, both p< 0.05) for both functional indices.
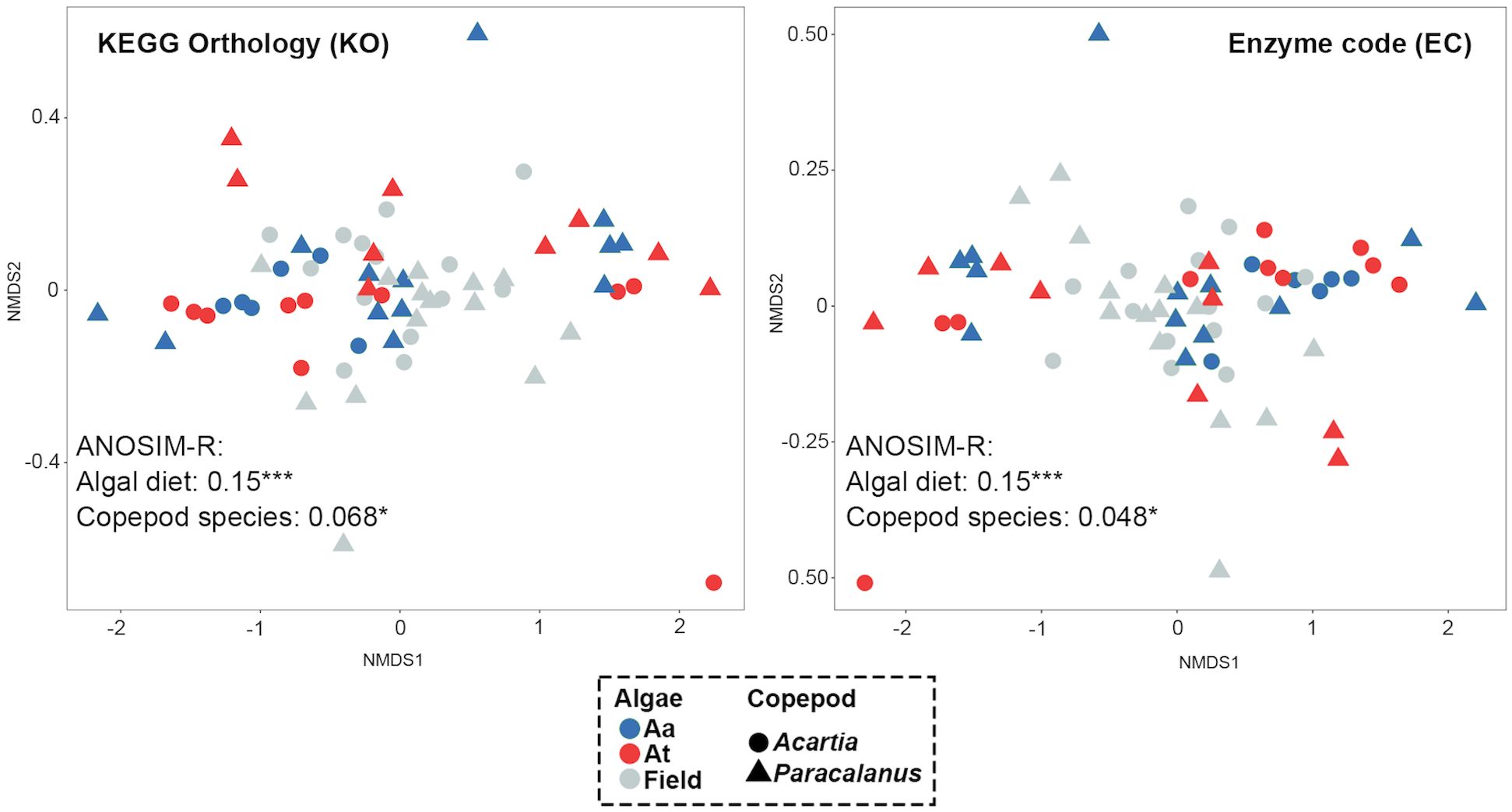
Figure 6 Effects of copepod species and algal diet on the GMC functions. Copepod GMC functions were predicted by the QIIME2-Picrust2 plugin and represented by KEGG Orthology (KO) and enzyme code (EC). Similarity of the two GMCs was calculated by the Bray-Curtis distance between them and visualized by nMDS maps. Each dot represents one GMC, with color indicating algal diet type and shape representing copepod species. A significant difference between groups of GMCs was found, with a higher R-value indicating the greater difference between groups (i.e., higher impacts of the grouping factor on GMC functions).
We observed that the ingestion rate of both copepods significantly decreased after two days of feeding on a toxic dinoflagellate diet. This finding is in accord with the reports of Acartia hudsonica feeding upon toxic Alexandrium fundyense, Euterpina acutifrons and Acartia grani feeding upon toxic Gymnodinium catenatum (Colin and Dam, 2003; da Costa et al., 2012), but different from the study of Calanus finarchicus feeding on toxic Alexandrium fundyense which showed no significant change of ingestion rate (Roncalli et al., 2016). Despite the reduced ingestion rate, both copepod species had consistently high survival rates (> 80% in all treatments), suggesting a compensatory mechanism in response to the algal toxins. It is supported by a previous study that showed copepod Acartia tonsa only consumed a few algae during an A. tamarense bloom, to maintain their basic metabolism and survive (Xu et al., 2018). The decrease in their grazing rate may be due to the reduced activity, as well as increased prey rejection and regurgitation of ingested toxic cells such as mouthpart twitching in Calanus pacificus, a passive filter-feeder (Landry, 1980; Sykes and Huntley, 1987; Guisande et al., 2002; Xu et al., 2018). This compensatory mechanism, by reducing the feeding rate of toxic algae to ensure survival, has also been reported in many other animals including arthropods (e.g., daphnids and insects) and mammals (e.g., rats) (Allen et al., 1995; Wheeler and Isman, 2001; Kohl et al., 2014).
In addition to the compensatory mechanism, detoxification by copepod GMCs may also play an important role in maintaining the high survival rate of copepods under the toxic diet. This is supported by the significant increase in the relative abundance of bacterial taxa at the ASV level in the toxic diet treatment when compared to the non-toxic group. In general, there was a relatively higher number of ASVs with an increased trend compared to those showing a decreased trend, suggesting the positive recruitment of beneficial bacterial taxa rather than the removal of harmful or functionally redundant ones. This finding is further supported by our results showing no significant change in bacterial richness among treatments, except for a significant increase of GMC richness in the toxic diet treatment of Acartia sp. (compared to the non-toxic diet, Supplementary Figure 3C).
Our results showed that host difference had a much higher impact than diet on the composition (ASV relative abundance based) of copepod GMCs (Figure 4A). Previous studies reported that copepod GMCs may be affected by various host factors, such as differences in feeding behavior, feeding status, or sex (Wäge et al., 2019). Similarly, the two copepod species used in our experiment have different feeding modes, which could contribute to the variations in their GMCs. Acartia sp. acts as a feeding-current feeder or coarse-filter feeder when feeding on small phytoplankton cells and as an active ambush-feeder when feeding on motile prey, using its mechanosensory setae on the antennules to detect the prey (Behrends and Schneider, 1995; Saiz and Kiørboe, 1995). In contrast, Paracalanus sp. is a fine-filter feeder that uses its antennules as open rakes to filter whole parcels of water containing food particles from the water, not just single food particles on the basis of size (Turner, 1984; Hansen et al., 1994; Behrends and Schneider, 1995). Therefore, Paracalanus sp. may need to filter more ambient water than Acartia sp. to obtain adequate food, which may lead to variations in the GMC. Moreover, the two copepod species have different body sizes (Acartia sp.: ~ 1.2 mm, Paracalanus sp.: ~ 0.9 mm) and biomass, which may reflect their different metabolic rates, such as excretion rate of ammonium and phosphate, leading to variations in their gut GMC (Ikeda et al., 2001). Additionally, studies have shown that many of the bacterial sequences were found only in one of the two copepod species, suggesting host-specific affiliation of bacteria (Dziallas et al., 2013).
For each copepod species individually, we showed that diet type had significant effects on the composition of GMC, which is consistent with previous studies on Daphnia (Freese and Schink, 2011; Callens et al., 2016; Akbar et al., 2020). Shoemaker & Moisander (2017) also demonstrated that food sources varied due to seasonal change which brought copepod GMC differences. In our experiment, to eliminate the potential effects from other factors influencing GMC, such as different nutritional values between algal taxa, we utilized two species of dinoflagellates from the same genus and at the same growth stage. We measured their toxin productions during the experiment and showed that A. tamarense had a remarkable production of gonyautoxins which was not detected in A. andersonii. Therefore, we argue that the significant change in copepod GMC under the toxic diet compared to the non-toxic diet or field samples could be attributed to the response to the algal toxin. Similarly, Macke et al. (2017) showed that when exposed to toxic cyanobacteria, the gut bacteria of both tolerant and susceptible genotypes of Daphnia tended to be more diverse, with some bacteria having the ability to degrade microcystin (Dziga et al., 2013; Mou et al., 2013). These findings suggest that changes in GMC in response to toxic diets may reflect copepods’ adaptive strategies to cope with the toxic effects of HAB species.
For Acartia sp., when fed with a toxic diet, remarkably lower numbers of ASVs (n = 7) were found to show significant changes in the GMC, compared to that of Paracalanus sp. (n = 40), suggesting the host differences in the recruitment of specific bacterial taxa for detoxification. These enriched bacterial taxa were previously reported to have special or strong abilities in detoxification, especially for algal toxins. For instance, Brevibacterium (Micrococcales, Actinobacteria) species have highly active and multiple proteolytic enzymes, with the ability to completely degrade neurotoxins (e.g., neurotoxic ochratoxin A, a mycotoxin produced by fungi species) (Sørhaug, 1981; Fernández et al., 2000; Rodriguez et al., 2011; Hathout and Aly, 2014).
Notably, for Paracalanus sp. fed by a toxic diet, 20 ASVs belonging to the family Rhodobacteraceae (Rhodobacterales; Alphaproteobacteria) displayed significantly increased patterns, suggesting their crucial roles in copepods’ detoxification. While most of the Rhodobacteraceae ASVs detected in our study were previously reported to have detoxifying abilities on synthetic chemicals or heavy metals (i.e., methylmercury) (Pujalte et al., 2014; Lin et al., 2023), their roles in detoxifying algal toxins remain poorly understood and require further investigation.
In addition to Rhodobacteraceae, other bacteria taxa within Actinobacteria, Alphaproteobacteria, and Gammaproteobacteria also displayed increased patterns in the GMC of Paracalanus sp. under a toxic dinoflagellate diet. Specially, three ASVs belonging to Actinobacteria were strongly enriched (with more than 6-fold change values), together with Brevibacterium sp. that is mentioned above for Acartia sp., further highlighting the key roles of Actinobacteria in copepod detoxification of HAB algae. Within Gammaproteobacteria, the genus Pseudomonas can produce an enzyme responsible for the transformation into less toxic products (Škrinjar et al., 1996). A similar function was also reported in the genus Stenotrophomonas (Gammaproteobacteria) which can strongly degrade Aflatoxin B1 and completely degrade two microcystin variants simultaneously (Guan et al., 2008; Yang et al., 2014).
Our study also revealed that the unchanged survival rate of copepods under the toxic diet could be attributed to the increases of some bacterial groups, such as Rhodobacteraceae, Rhizobiaceae, and Parcubacteria, which have functions in maintaining host homeostasis through various mechanisms (Strompl et al., 2003; Dogs et al., 2017; Barak-Gavish et al., 2018). For instance, intestine bacteria from Rhodobacteraceae play an important role in the growth and health of arthropod hosts, such as shrimps (Wang et al., 2022). Species from Rhizobiaceae can supply auxiliary as well as essential metabolites to their host (Erlacher et al., 2015). Parcubacteria species have also been shown to play an important role in celiac disease-related pathology (Primec et al., 2019). Furthermore, researchers have demonstrated that the bacteria-free Daphnia were smaller, less fecund, and had higher mortality than those with microbiota (Sison-Mangus et al., 2015), and copepods with bacteria consumed more algae than those without bacteria (Gyllenberg, 1980). Similarly, microbiota has been shown to increase the host resistance to toxic environments, as evidenced by the insecticide resistance in the bean bug Riptortus pedestris and facilitated intake of plant toxins in herbivores (Kikuchi et al., 2012; Kohl et al., 2014). All these studies support our finding that gut bacteria play necessary roles in maintaining the homeostasis of copepods.
We acknowledge that functional prediction with 16S rDNA data using software, such as PICRUSt2 or Tax4Fun2, was widely used in the studies of GMCs (Sadaiappan et al., 2021; Chen et al., 2022; Guo et al., 2022; Luo et al., 2022; Xu et al., 2023), however, the results may suffer from the incomplete database of bacterial genomes, especially for field samples with diverse bacteria taxa (Douglas et al., 2020). We argue that this bias would be minor for the comparison of effects between host and diet on copepod GMC functions in our results, as the same method and database were used. We showed that GMCs, constructed by predicted functional data (i.e., Enzyme codes and KEGG Orthologs), were highly regulated by diet type compared to host effects, which is opposite to the pattern observed in the communities with ASV relative abundance data. We regard this as a functional convergence of GMC when exposed to toxic algae. This could be explained by the fact that different bacteria taxa can perform the same or similar functions. This was supported by the above-mentioned species enriched in the toxic diet treatment, most of which have the similar function of detoxification or maintaining the homeostasis of hosts. Further study combined with metatranscriptomics data will be conducted to provide a more comprehensive understanding of the mechanisms, such as gene regulation, underlying the detoxification of algal toxins by copepods, and the roles of specific bacterial groups in this process.
GMC plays an important role in the physiology performance (e.g., fitness, growth, survival, and reproduction) of zooplankton host and greatly influences the biogeochemical cycles in the ocean (Roura et al., 2017; Callens et al., 2018; Li et al., 2021). Therefore, disentangling the major factors regulating the spatiotemporal dynamics of GMC in zooplankton remains a key issue in recent studies.
Our results found high variations in GMC among copepod individuals in the same treatment, as evident from the differences in community composition (at the class level) and beta diversity (Bray-Curtis dissimilarity, at the ASV level). This finding is in line with a previous study that reported significant variation in the GMCs of single samples of identified bacterial taxa at the order level in Temora sp. and Acartia sp., which indicated that copepods should be considered as individuals rather than a group when studying their GMC (Wäge et al., 2019).
Datta et al. (2018) also reported that the most abundant operational taxonomic units (OTUs), such as Flavobacteriaceae, Oceanospirillaceae, and Pseudoalteromonadaceae, were not consistently present in the gut of Calanus finmarchicus. These OTUs showed patchy distributions with relative abundances varying greatly among individual copepods. Similar observations were made in studies of other arthropod species, including Daphnia and black tiger shrimp (Penaeus monodon) (Rungrassamee et al., 2014; Hegg et al., 2021; Maszczyk et al., 2022; Rajarajan et al., 2023). Therefore, we suggest that using individual gut to do PCR amplification and analyze GMC composition is more accurate than using the bulk community of copepods.
Despite their small body size and even smaller gut compared to other animals, we successfully dissected the entire gut of each copepod and performed PCR amplification. This demonstrates the feasibility of examining copepod GMCs at the individual level. Moreover, with a high number of replicates (maximum of 15) used in the statistical analyses (e.g., ANOSIM tests), we showed that variations in GMC were relatively higher between treatments than within a certain treatment, reflecting the significant effects from either host or diet.
Overall, we provided the first study of copepod GMC at the individual level to disentangle the roles and mechanisms of gut microbes on the detoxification of copepods in response to toxic dinoflagellates. We showed that copepod species difference had a much greater impact than algal diet type on shaping copepod GMC compositions. However, from the functional aspect, the pattern was the opposite (i.e., diet type predominated). Under a toxic diet, the number and taxonomy of enriched bacteria were largely different between two copepod species, and most of these taxa were reported to have the function of degrading algal toxins (e.g., species from Brevibacterium, Pseudomonas, Stenotrophomonas and Alteromonadaceae) and maintaining the homeostasis of hosts (e.g., species from Rhodobacteraceae, Rhizobiaceae, and Parcubacteria). This indicates the potentially functional convergence of GMC under algal toxins for different copepod species. Our study highlights the key role of gut microbes in the detoxification of copepods, offering new insights into the food webs, biogeochemical cycles, and management of HABs. Further studies, such as transcriptomic analysis, will be performed to investigate the detailed gene and pathway regulation of copepod GMCs.
Raw sequencing reads for analyses in this study were deposited in the online open database of the National Center for Biotechnology Information Search database (NCBI) with the projection accession number PRJNA1032266. R scripts used for analysis can be found on the website: https://github.com/JingYang00/Copepod-gut-micorbes.git.
The manuscript presents research on animals that do not require ethical approval for their study.
JY: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. ZX: Conceptualization, Formal analysis, Methodology, Project administration, Visualization, Writing – review & editing. YC: Data curation, Investigation, Validation, Writing – review & editing. HX: Methodology, Resources, Validation, Writing – review & editing. ZG: Investigation, Methodology, Writing – review & editing. XZ: Methodology, Resources, Writing – review & editing. MP: Methodology, Validation, Writing – review & editing. SZ: Conceptualization, Methodology, Writing – review & editing. HL: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2022YFC3105301) and the Hong Kong Research Grants Council (16101622).
The authors sincerely thank Dr. Qian Wang and Dr. Meng Chen from Xiamen University for measuring algal toxins. We would like to acknowledge Prof. Haifeng Gu for providing the dinoflagellate cultures. We are grateful to Ms. Oi Ting KWOK, Ms. Yin Ki TAM, and Dr. Zhiyuan Shi from the Department of Ocean Science, HKUST, for sharing all kinds of experimental experience, equipment, and platforms.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1368315/full#supplementary-material
Akbar S., Gu L., Sun Y., Zhou Q., Zhang L., Lyu K., et al. (2020). Changes in the life history traits of Daphnia magna are associated with the gut microbiota composition shaped by diet and antibiotics. Sci. Total Environ. 705, 135827. doi: 10.1016/j.scitotenv.2019.135827
Allen Y., Calow P., Baird D. J. (1995). A mechanistic model of contaminant-induced feeding inhibition in Daphnia magna. Environ. Toxicol. Chemistry: Int. J. 14, 1625–1630. doi: 10.1002/etc.5620140923
Almada A. A., Tarrant A. M. (2016). Vibrio elicits targeted transcriptional responses from copepod hosts. FEMS Microbiol. Ecol. 92, fiw072. doi: 10.1093/femsec/fiw072
Anderson D. M., Hoagland P., Kaoru Y., White A. W. (2000). Estimated annual economic impacts from harmful algal blooms (HABs) in the United States. Massachusetts, USA: Woods Hole Oceanographic Institution. doi: 10.1575/1912/96
Anderson D. M., Kulis D. M., Sullivan J. J., Hall S. (1990). Toxin composition variations in one isolate of the dinoflagellate Alexandrium fundyense. Toxicon 28, 885–893. doi: 10.1016/0041-0101(90)90018-3
Band-Schmidt C. J., Duran-Riveroll L. M., Bustillos-Guzman J. J., Leyva-Valencia I., Lopez-Cortes D. J., Nunez-Vazquez E. J., et al. (2019). Paralytic toxin producing dinoflagellates in Latin America: Ecology and physiology. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00042
Barak-Gavish N., Frada M. J., Ku C., Lee P. A., DiTullio G. R., Malitsky S., et al. (2018). Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci. Adv. 4, eaau5716. doi: 10.1126/sciadv.aau5716
Behrends G., Schneider G. (1995). Impact of Aurelia aurita medusae (Cnidaria, Scyphozoa) on the standing stock and community composition of mesozooplankton in the Kiel Bight (western Baltic Sea). Mar. Ecol. Prog. Ser. 127, 39–45. doi: 10.3354/meps127039
Bickel S. L., Tang K. W., Grossart H.-P. (2014). Structure and function of zooplankton-associated bacterial communities in a temperate estuary change more with time than with zooplankton species. Aquat. Microbial Ecol. 72, 1–15. doi: 10.3354/ame01676
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Buchholz-Cleven B. E. E., Rattunde B., Straub K. L. (1997). Screening for genetic diversity of isolates of anaerobic Fe (II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst. Appl. Microbiol. 20, 301–309. doi: 10.1016/S0723-2020(97)80077-X
Callens M., Macke E., Muylaert K., Bossier P., Lievens B., Waud M., et al. (2016). Food availability affects the strength of mutualistic host–microbiota interactions in Daphnia magna. ISME J. 10, 911–920. doi: 10.1038/ismej.2015.166
Callens M., Watanabe H., Kato Y., Miura J., Decaestecker E. (2018). Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0444-1
Cembella A. D., Quilliam M. A., Lewis N. I., Bauder A. G., Dell’Aversano C., Thomas K., et al. (2002). The toxigenic marine dinoflagellate Alexandrium tamarense as the probable cause of mortality of caged salmon in Nova Scotia. Harmful Algae 1, 313–325. doi: 10.1016/S1568-9883(02)00048-3
Chen M., Yue Y., Bao X., Feng X., Ou Z., Qiu Y., et al. (2022). Effects of polystyrene nanoplastics on oxidative stress, histopathology and intestinal microbiota in largemouth bass (Micropterus salmoides). Aquac Rep. 27, 101423. doi: 10.1016/j.aqrep.2022.101423
Clarke K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Colin S. P., Dam H. G. (2003). Effects of the toxic dinoflagellate Alexandrium fundyense on the copepod Acartia hudsonica: a test of the mechanisms that reduce ingestion rates. Mar. Ecol. Prog. Ser. 248, 55–65. doi: 10.3354/meps248055
da Costa R. M., Pereira L. C. C., Fernández F. (2008). Effects of toxic Alexandrium minutum strains on the feeding and survival rates of pelagic marine copepods Acartia grani and Euterpina acutifrons. Hydrobiologia 614, 55–63. doi: 10.1007/s10750-008-9536-4
da Costa R. M., Pereira L. C. C., Ferrnández F. (2012). Deterrent effect of Gymnodinium catenatum Graham PSP-toxins on grazing performance of marine copepods. Harmful Algae 17, 75–82. doi: 10.1016/j.hal.2012.03.002
Datta M. S., Almada A. A., Baumgartner M. F., Mincer T. J., Tarrant A. M., Polz M. F. (2018). Inter-individual variability in copepod microbiomes reveals bacterial networks linked to host physiology. ISME J. 12, 2103–2113. doi: 10.1038/s41396-018-0182-1
De Corte D., Lekunberri I., Sintes E., Garcia J. A. L., Gonzales S., Herndl G. J. (2014). Linkage between copepods and bacteria in the North Atlantic Ocean. Aquat. Microbial Ecol. 72, 215–225. doi: 10.3354/ame01696
De Troch M., Boeckx P., Cnudde C., Van Gansbeke D., Vanreusel A., Vincx M., et al. (2012). Bioconversion of fatty acids at the basis of marine food webs: insights from a compound-specific stable isotope analysis. Mar. Ecol. Prog. Ser. 465, 53–67. doi: 10.3354/meps09920
De Wit P., Yamada K., Panova M., André C., Johannesson K. (2018). Diet-dependent gene expression highlights the importance of Cytochrome P450 in detoxification of algal secondary metabolites in a marine isopod. Sci. Rep. 8, 16824. doi: 10.1038/s41598-018-34937-z
Dogs M., Wemheuer B., Wolter L., Bergen N., Daniel R., Simon M., et al. (2017). Rhodobacteraceae on the marine brown alga Fucus spiralis are abundant and show physiological adaptation to an epiphytic lifestyle. Syst. Appl. Microbiol. 40, 370–382. doi: 10.1016/j.syapm.2017.05.006
Doucette G. J., Cembella A. D., Martin J. L., Michaud J., Cole T. V. N., Rolland R. M. (2006). Paralytic shellfish poisoning (PSP) toxins in North Atlantic right whales Eubalaena glacialis and their zooplankton prey in the Bay of Fundy, Canada. Mar. Ecol. Prog. Ser. 306, 303–313. doi: 10.3354/meps306303
Douglas G. M., Maffei V. J., Zaneveld J. R., Yurgel S. N., Brown J. R., Taylor C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Dziallas C., Grossart H.-P., Tang K. W., Nielsen T. G. (2013). Distinct communities of free-living and copepod-associated microorganisms along a salinity gradient in Godthåbsfjord, West Greenland. Arct Antarct Alp Res. 45, 471–480. doi: 10.1657/1938-4246.45.4.471
Dziga D., Wasylewski M., Wladyka B., Nybom S., Meriluoto J. (2013). Microbial degradation of microcystins. Chem. Res. Toxicol. 26, 841–852. doi: 10.1021/tx4000045
Erlacher A., Cernava T., Cardinale M., Soh J., Sensen C. W., Grube M., et al. (2015). Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00053
Etheridge S. M. (2010). Paralytic shellfish poisoning: seafood safety and human health perspectives. Toxicon 56, 108–122. doi: 10.1016/j.toxicon.2009.12.013
Fernández J., Mohedano A. F., Gaya P., Medina M., Nuñez M. (2000). Purification and properties of two intracellular aminopeptidases produced by Brevibacterium linens SR3. Int. Dairy J. 10, 241–248. doi: 10.1016/S0958-6946(00)00046-7
Finiguerra M., Avery D. E., Dam H. G. (2014). No evidence for induction or selection of mutant sodium channel expression in the copepod Acartia husdsonica challenged with the toxic dinoflagellate Alexandrium fundyense. Ecol. Evol. 4, 3470–3481. doi: 10.1002/ece3.1197
Freese H. M., Schink B. (2011). Composition and stability of the microbial community inside the digestive tract of the aquatic crustacean Daphnia magna. Microb. Ecol. 62, 882–894. doi: 10.1007/s00248-011-9886-8
Frost B. W. (1972). Effects of size and concentration of food particles on the feeding behavior of the marine planktonic copepod Calanus pacificus 1. Limnol Oceanogr 17, 805–815. doi: 10.4319/lo.1972.17.6.0805
Gorokhova E., El-Shehawy R., Lehtiniemi M., Garbaras A. (2021). How copepods can eat toxins without getting sick: Gut bacteria help zooplankton to feed in cyanobacteria blooms. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.589816
Guan S., Ji C., Zhou T., Li J., Ma Q., Niu T. (2008). Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 9, 1489–1503. doi: 10.3390/ijms9081489
Guillard R. R. L. (1975). Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of marine invertebrate animals: proceedings—1st conference on culture of marine invertebrate animals greenport (Boston: Springer US), 29–60. doi: 10.1007/978-1-4615-8714-9_3
Guisande C., Frangópulos M., Carotenuto Y., Maneiro I., Riveiro I., Vergara A. R. (2002). Fate of paralytic shellfish poisoning toxins ingested by the copepod Acartia clausi. Mar. Ecol. Prog. Ser. 240, 105–115. doi: 10.3354/meps240105
Guo X., Lv M., Li J., Ding J., Wang Y., Fu L., et al. (2022). The distinct toxicity effects between commercial and realistic polystyrene microplastics on microbiome and histopathology of gut in zebrafish. J. Hazard Mater 434, 128874. doi: 10.1016/j.jhazmat.2022.128874
Gyllenberg G. (1980). Feeding of the copepod Eurytemora hirundoides (Crustacea) on different algal cultures. Annales Zoologici Fennici (JSTOR) 17, 181–184. Available at: https://www.jstor.org/stable/23734049.
Hallegraeff G. M. (2003). Harmful algal blooms: a global overview. Manual harmful Mar. microalgae 33, 1–22.
Hansen B., Bjornsen P. K., Hansen P. J. (1994). The size ratio between planktonic predators and their prey. Limnol Oceanogr 39, 395–403. doi: 10.4319/lo.1994.39.2.0395
Hathout A. S., Aly S. E. (2014). Biological detoxification of mycotoxins: a review. Ann. Microbiol. 64, 905–919. doi: 10.1007/s13213-014-0899-7
Hegg A., Radersma R., Uller T. (2021). A field experiment reveals seasonal variation in the Daphnia gut microbiome. Oikos 130, 2191–2201. doi: 10.1111/oik.08530
Ikeda T., Kanno Y., Ozaki K., Shinada A. (2001). Metabolic rates of epipelagic marine copepods as a function of body mass and temperature. Mar. Biol. 139, 587–596. doi: 10.1007/s002270100608
Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kikuchi Y., Hayatsu M., Hosokawa T., Nagayama A., Tago K., Fukatsu T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. 109, 8618–8622. doi: 10.1073/pnas.1200231109
Kohl K. D., Weiss R. B., Cox J., Dale C., Denise Dearing M. (2014). Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246. doi: 10.1111/ele.12329
Kozlowsky-Suzuki B., Carlsson P., Rühl A., Granéli E. (2006). Food selectivity and grazing impact on toxic Dinophysis spp. by copepods feeding on natural plankton assemblages. Harmful Algae 5, 57–68. doi: 10.1016/j.hal.2005.05.002
Landry M. R. (1980). Detection of prey by Calanus pacificus: Implications of the first antennae 1. Limnol Oceanogr 25, 545–549. doi: 10.4319/lo.1980.25.3.0545
Li Y., Xu Z., Liu H. (2021). Nutrient-imbalanced conditions shift the interplay between zooplankton and gut microbiota. BMC Genomics 22, 1–18. doi: 10.1186/s12864-020-07333-z
Lin W., He Y., Li R., Mu C., Wang C., Shi C., et al. (2023). Adaptive changes of swimming crab (Portunus trituberculatus) associated bacteria helping host against dibutyl phthalate toxification. Environ. pollut. 324, 121328. doi: 10.1016/j.envpol.2023.121328
Lincoln J. A., Turner J. T., Bates S. S., Léger C., Gauthier D. A. (2001). Feeding, egg production, and egg hatching success of the copepods Acartia tonsa and Temora longicornis on diets of the toxic diatom Pseudo-nitzschia multiseries and the non-toxic diatom Pseudo-nitzschia pungens. Hydrobiologia 453, 107–120. doi: 10.1023/A:1013163816771
Liu Y., Chen Z., Gao Y., Zou J., Lu S., Zhang L. (2021). Identifying the source organisms producing paralytic shellfish toxins in a subtropical bay in the South China Sea. Environ. Sci. Technol. 55, 3124–3135. doi: 10.1021/acs.est.0c06991
Luo M., An R., Fu J., Wan S., Zhu W., Wang L., et al. (2022). Comparative analysis of the gut microbiota in bighead carp under different culture patterns. J. Appl. Microbiol. 132, 1357–1369. doi: 10.1111/jam.15248
Macke E., Callens M., De Meester L., Decaestecker E. (2017). Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 8, 1608. doi: 10.1038/s41467-017-01714-x
Maszczyk P., Kiersztyn B., Gozzo S., Kowalczyk G., Jimenez-Lamana J., Szpunar J., et al. (2022). Combined effects of polystyrene nanoplastics and enrofloxacin on the life histories and gut microbiota of Daphnia magna. Water (Basel) 14, 3403. doi: 10.3390/w14213403
Moisander P. H., Sexton A. D., Daley M. C. (2015). Stable associations masked by temporal variability in the marine copepod microbiome. PloS One 10, e0138967. doi: 10.1371/journal.pone.0138967
Mou X., Lu X., Jacob J., Sun S., Heath R. (2013). Metagenomic identification of bacterioplankton taxa and pathways involved in microcystin degradation in Lake Erie. PloS One 8, e61890. doi: 10.1371/journal.pone.0061890
Muyzer G., De Waal E. C., Uitterlinden A. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Noguchi T., Miyazawa K., Daigo K., Arakawa O. (2011). Paralytic shellfish poisoning (PSP) toxin-and/or tetrodotoxin-contaminated crabs and food poisoning by them. Toxin Rev. 30, 91–102. doi: 10.3109/15569543.2011.640743
Oksanen J., Kindt R., Legendre P., O’Hara B., Stevens M. H. H., Oksanen M. J., et al. (2007). The vegan package. Community Ecol. Package 10, 719.
Patón D., García-Gómez J. C., Loring J., Torres A. (2023). Composting the invasive toxic seaweed Rugulopteryx okamurae using five invertebrate species, and a mini-review on composting macroalgae. Waste Biomass Valorization 14, 167–184. doi: 10.1007/s12649-022-01849-z
Primec M., Klemenak M., Di Gioia D., Aloisio I., Cionci N. B., Quagliariello A., et al. (2019). Clinical intervention using Bifidobacterium strains in celiac disease children reveals novel microbial modulators of TNF-α and short-chain fatty acids. Clin. Nutr. 38, 1373–1381. doi: 10.1016/j.clnu.2018.06.931
Pujalte M. J., Lucena T., Ruvira M. A., Arahal D. R., Macián M. C. (2014). The family rhodobacteraceae. In Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F.. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria (Berlin, Heidelberg: Springer Berlin Heidelberg), 439–512. doi: 10.1007/978-3-642-30197-1_377
Rajarajan A., Wolinska J., Walser J.-C., Dennis S. R., Spaak P. (2023). Host-associated bacterial communities vary between Daphnia galeata genotypes but not by host genetic distance. Microb. Ecol. 85, 1578–1589. doi: 10.1007/s00248-022-02011-x
Reguera B., Riobó P., Rodríguez F., Díaz P. A., Pizarro G., Paz B., et al. (2014). Dinophysis toxins: causative organisms, distribution and fate in shellfish. Mar. Drugs 12, 394–461. doi: 10.3390/md12010394
Rodriguez H., Reveron I., Doria F., Costantini A., De Las Rivas B., Munoz R., et al. (2011). Degradation of ochratoxin A by Brevibacterium species. J. Agric. Food Chem. 59, 10755–10760. doi: 10.1021/jf203061p
Roncalli V., Lenz P. H., Cieslak M. C., Hartline D. K. (2017). Complementary mechanisms for neurotoxin resistance in a copepod. Sci. Rep. 7, 14201. doi: 10.1038/s41598-017-14545-z
Roncalli V., Turner J. T., Kulis D., Anderson D. M., Lenz P. H. (2016). The effect of the toxic dinoflagellate Alexandrium fundyense on the fitness of the calanoid copepod Calanus finmarchicus. Harmful Algae 51, 56–66. doi: 10.1016/j.hal.2015.11.003
Roura Á., Doyle S. R., Nande M., Strugnell J. M. (2017). You are what you eat: a genomic analysis of the gut microbiome of captive and wild Octopus vulgaris paralarvae and their zooplankton prey. Front. Physiol. 8. doi: 10.3389/fphys.2017.00362
Rungrassamee W., Klanchui A., Maibunkaew S., Chaiyapechara S., Jiravanichpaisal P., Karoonuthaisiri N. (2014). Characterization of intestinal bacteria in wild and domesticated adult black tiger shrimp (Penaeus monodon). PloS One 9, e91853. doi: 10.1371/journal.pone.0091853
Sadaiappan B., PrasannaKumar C., Nambiar V. U., Subramanian M., Gauns M. U. (2021). Meta-analysis cum machine learning approaches address the structure and biogeochemical potential of marine copepod associated bacteriobiomes. Sci. Rep. 11, 3312. doi: 10.1038/s41598-021-82482-z
Saiz E., Kiørboe T. (1995). Predatory and suspension feeding of the copepod Acartia tonsa in turbulent environments. Mar. Ecol. Prog. Ser. 122, 147–158. doi: 10.3354/meps122147
Scavotto R. E., Dziallas C., Bentzon-Tilia M., Riemann L., Moisander P. H. (2015). Nitrogen-fixing bacteria associated with copepods in coastal waters of the North Atlantic Ocean. Environ. Microbiol. 17, 3754–3765. doi: 10.1111/1462-2920.12777
Schminke H. K. (2007). Entomology for the copepodologist. J. Plankton Res. 29, i149–i162. doi: 10.1093/plankt/fbl073
Shoemaker K. M., Moisander P. H. (2017). Seasonal variation in the copepod gut microbiome in the subtropical North Atlantic Ocean. Environ. Microbiol. 19, 3087–3097. doi: 10.1111/1462-2920.13780
Shukla S. P., Beran F. (2020). Gut microbiota degrades toxic isothiocyanates in a flea beetle pest. Mol. Ecol. 29, 4692–4705. doi: 10.1111/mec.15657
Sison-Mangus M. P., Mushegian A. A., Ebert D. (2015). Water fleas require microbiota for survival, growth and reproduction. ISME J. 9, 59–67. doi: 10.1038/ismej.2014.116
Skovgaard A., Castro-Mejia J. L., Hansen L. H., Nielsen D. S. (2015). Host-specific and pH-dependent microbiomes of copepods in an extensive rearing system. PloS One 10, e0132516. doi: 10.1371/journal.pone.0132516
Škrinjar M., Rašić J. L., Stojičić V. (1996). Lowering of ochratoxin A level in milk by yoghurt bacteria and bifidobacteria. Folia Microbiol. (Praha) 41, 26–28. doi: 10.1007/BF02816335
Sommer U., Stibor H. (2002). Copepoda–Cladocera–Tunicata: the role of three major mesozooplankton groups in pelagic food webs. Ecol. Res. 17, 161–174. doi: 10.1046/j.1440-1703.2002.00476.x
Sørhaug T. (1981). Comparison of peptide hydrolases from six strains of Brevibacterium linens. Milchwissenschaft 36, 137–139. Available at: https://www.cabidigitallibrary.org/doi/full/10.5555/19810469449.
Strompl C., Hold G. L., Lunsdorf H., Graham J., Gallacher S., Abraham W.-R., et al. (2003). Oceanicaulis alexandrii gen. nov., sp. nov., a novel stalked bacterium isolated from a culture of the dinoflagellate Alexandrium tamarense (Lebour) Balech. Int. J. Syst. Evol. Microbiol. 53, 1901–1906. doi: 10.1099/ijs.0.02635-0
Sun Z., Liu Y., Xu H., Yan C. (2022). Genome-wide identification of P450 genes in Chironomid Propsilocerus akamusi reveals candidate genes involved in gut microbiota-mediated detoxification of Chlorpyrifos. Insects 13, 765. doi: 10.3390/insects13090765
Sykes P. F., Huntley M. E. (1987). Acute physiological reactions of Calanus pacificus to selected dinoflagellates: direct observations. Mar. Biol. 94, 19–24. doi: 10.1007/BF00392895
Tang K. W. (2005). Copepods as microbial hotspots in the ocean: effects of host feeding activities on attached bacteria. Aquat. Microbial Ecol. 38, 31–40. doi: 10.3354/ame038031
Tang K. W., Glud R. N., Glud A., Rysgaard S., Nielsen T. G. (2011). Copepod guts as biogeochemical hotspots in the sea: Evidence from microelectrode profiling of Calanus spp. Limnol Oceanogr 56, 666–672. doi: 10.4319/lo.2011.56.2.0666
Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Sci. (1979) 336, 608–611. doi: 10.1126/science.1218344
Tester P. A., Turner J. T., Shea D. (2000). Vectorial transport of toxins from the dinoflagellate Gymnodinium breve through copepods to fish. J. Plankton Res. 22, 47–62. doi: 10.1093/plankt/22.1.47
Tillmann U., Alpermann T. L., da Purificação R. C., Krock B., Cembella A. (2009). Intra-population clonal variability in allelochemical potency of the toxigenic dinoflagellate Alexandrium tamarense. Harmful Algae 8, 759–769. doi: 10.1016/j.hal.2009.03.005
Turner J. T. (1984). Zooplankton feeding ecology: Contents of fecal pellets of the copepods Eucalanus pileatus and Paracalanus quasimodo from continental shelf waters of the Gulf of Mexico. Marine ecology progress series. Oldendorf 15, 27–46. doi: 10.3354/meps015027
Turner J. T. (2014). Planktonic marine copepods and harmful algae. Harmful Algae 32, 81–93. doi: 10.1016/j.hal.2013.12.001
Turner J. T., Anderson D. M. (1983). Zooplankton grazing during dinoflagellate blooms in a cape cod embayment, with observations of predation upon tin-tinnids by copepods. Mar. Ecol. 4, 359–374. doi: 10.1111/j.1439-0485.1983.tb00119.x
Turner J. T., Tester P. A. (1989). “Zooplankton feeding ecology: copepod grazing during an expatriate red tide,” in Phytoplankton Blooms, eds. Cosper E. M., Bricelj V. M., Carpenter E. J.. (Berlin: Springer-Verlag), 359–374. doi: 10.1007/978-3-642-75280-3
Uye S. (1986). Impact of copepod grazing on the red-tide flagellate Chattonella antiqua. Mar. Biol. 92, 35–43. doi: 10.1007/BF00392743
Van Dolah F. M. (2000). Marine algal toxins: origins, health effects, and their increased occurrence. Environ. Health Perspect. 108, 133–141. doi: 10.1289/ehp.00108s1133
Wäge J., Strassert J. F. H., Landsberger A., Loick-Wilde N., Schmale O., Stawiarski B., et al. (2019). Microcapillary sampling of Baltic Sea copepod gut microbiomes indicates high variability among individuals and the potential for methane production. FEMS Microbiol. Ecol. 95, fiz024. doi: 10.1093/femsec/fiz024
Wang R., Chen H., Zhu Y., Al-Masqari Z. A., Yan M., Wang G., et al. (2022). Survival status of Penaeus vannamei is associated with the homeostasis and assembly process of the intestinal bacterial community. Aquaculture 558, 738398. doi: 10.1016/j.aquaculture.2022.738398
Wheeler D. A., Isman M. B. (2001). Antifeedant and toxic activity of Trichilia americana extract against the larvae of Spodoptera litura. Entomol Exp. Appl. 98, 9–16. doi: 10.1046/j.1570-7458.2001.00751.x
Xiang C., Ke Z., Li K., Liu J., Zhou L., Lian X., et al. (2021). Effects of terrestrial inputs and seawater intrusion on zooplankton community structure in Daya Bay, South China Sea. Mar. pollut. Bull. 167, 112331. doi: 10.1016/j.marpolbul.2021.112331
Xu Y., Cheng H., Meng J., Xu B., Li X., Shen P., et al. (2023). Study on the quality and symbiotic microbial composition of Artemia nauplii in three main producing areas. J. Freshw. Ecol. 38, 1–12. doi: 10.1080/02705060.2022.2158140
Xu J., Nielsen L. T., Kiørboe T. (2018). Foraging response and acclimation of ambush feeding and feeding-current feeding copepods to toxic dinoflagellates. Limnol Oceanogr 63, 1449–1461. doi: 10.1002/lno.10782
Yang F., Zhou Y., Yin L., Zhu G., Liang G., Pu Y. (2014). Microcystin-degrading activity of an indigenous bacterial strain Stenotrophomonas acidaminiphila MC-LTH2 isolated from Lake Taihu. PloS One 9, e86216. doi: 10.1371/journal.pone.0086216
Keywords: harmful algal blooms, dinoflagellates, copepods, gut microbes, detoxification, toxic microalgae
Citation: Yang J, Xu Z, Chen Y, Xu H, Gao Z, Zhang X, Pang M, Zhang S and Liu H (2024) Community changes of gut microbes highlight their importance in the adaptation of copepods to toxic dinoflagellates. Front. Mar. Sci. 11:1368315. doi: 10.3389/fmars.2024.1368315
Received: 10 January 2024; Accepted: 22 March 2024;
Published: 05 April 2024.
Edited by:
Paulo Sergio Salomon, Federal University of Rio de Janeiro, BrazilReviewed by:
Catharina Alves-de-Souza, University of Concepcion, ChileCopyright © 2024 Yang, Xu, Chen, Xu, Gao, Zhang, Pang, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhimeng Xu, enh1YmVAY29ubmVjdC51c3QuaGs=; Hongbin Liu, bGl1aGJAdXN0Lmhr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.