
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 26 April 2024
Sec. Marine Conservation and Sustainability
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1366971
 Paige Strudwick1*†
Paige Strudwick1*† David J. Suggett2†
David J. Suggett2† Justin R. Seymour1†
Justin R. Seymour1† Matthew Z. DeMaere3
Matthew Z. DeMaere3 Amanda Grima1†
Amanda Grima1† John Edmondson4†
John Edmondson4† Alicia McArdle5
Alicia McArdle5 Freda Nicholson5
Freda Nicholson5 Emma F. Camp1†
Emma F. Camp1†Significant threats to the long-term persistence of coral reefs have accelerated the adoption of coral propagation and out-planting approaches. However, how materials commonly used for propagation structures could potentially affect coral-associated bacterial communities remains untested. Here, we examined the impact of metal propagation structures on coral-associated bacterial communities. Fragments of the coral species Acropora millepora were grown on aluminium, sand/epoxy-coated steel (Reef Stars), and uncoated steel (rebar) structures. After 6 months, the functional and taxonomic profiles of coral-associated bacterial communities of propagated corals and reef colonies were characterised using amplicon (16S rRNA gene) and shotgun metagenomic sequencing. No differences in the phylogenetic structure or functional profile of coral-associated bacterial communities were observed between propagated corals and reef colonies. However, specific genes and pathways (e.g., lipid, nucleotide, and carbohydrate metabolism) were overrepresented in corals grown on different materials, and different taxa were indicative of the materials. These findings indicate that coral propagation on different materials may lead to differences in the individual bacterial taxa and functional potential of coral-associated bacterial communities, but how these contribute to changed holobiont fitness presents a key question to be addressed.
Reef restoration approaches that involve propagation, out-planting, and/or substrate stabilisation are commonly being applied to assist natural recovery and retain reef resilience in the face of mounting local and global stressors (Boström-Einarsson et al., 2018; GBRMPA, 2019; Hein et al., 2021). Coral propagation and out-planting approaches facilitate increases in coral biomass and cover at target sites (Hein et al., 2020; Suggett et al., 2020; Roper et al., 2022; Howlett et al., 2023), whereas reef stabilisation techniques aid substrate consolidation but similarly enhance coral biomass when corals are attached to the structures (Fox et al., 2019; Williams et al., 2019; Ceccarelli et al., 2020; Kenyon et al., 2023). There is a strong motivation to boost the abundance of resilient corals during propagation to improve long-term survival under future climate scenarios (Caruso et al., 2021; Camp, 2022). However, this is often conducted empirically by selecting coral stock that has survived previous stress events (Caruso et al., 2021). Arguably, the efficacy of such a proactive approach would be increased if bolstered by knowledge of the underlying coral biological dynamics during restoration (Voolstra et al., 2021; Shaver et al., 2022). However, whilst such restoration approaches continue to be adopted with accelerating enthusiasm (Boström-Einarsson et al., 2020; McAfee et al., 2021; Suggett and Van Oppen, 2022), how the processes and materials used impact coral biology (and related resilience) has only recently been considered (Morikawa and Palumbi, 2019; Strudwick et al., 2022, 2023, 2024; Nuñez Lendo et al., 2023).
Multifaceted communities of microorganisms—the coral “microbiome”—are central to the health of the coral host, and in totality, the host and associated microorganisms are referred to as the coral “holobiont” (Rohwer et al., 2002; Reshef et al., 2006; Rosenberg et al., 2007). Recently, the focus of restoration research has expanded to consider approaches to “conserve the holobiont” to improve restoration success (Carthey et al., 2020; Voolstra et al., 2021; Peixoto et al., 2022). Such an approach is important for coral reefs where associated microorganisms can impact reef resilience (Putnam and Gates, 2015; Rosado et al., 2018; Peixoto et al., 2022) and thus present an opportunity (e.g., engineering of an optimal microbial consortium; Peixoto et al., 2017) or risk (e.g., disease outbreaks; Rosenberg et al., 2007; Moriarty et al., 2020) in restoration activities. Coral-associated bacterial communities are known to be impacted by transfer between distinct environments during nursery propagation and out-planting (Strudwick et al., 2022, 2023). However, the extent to which materials used during propagation influence the coral environment and subsequently contribute—negatively or positively—to bacterial community changes is relatively unexplored.
A range of materials are used in reef restoration, from concrete to chemical adhesives, metals, plastics, ropes, and natural fibres (Nedimyer et al., 2011; Meesters et al., 2015; Williams et al., 2019; Boström-Einarsson et al., 2020; Ceccarelli et al., 2020; Suggett et al., 2020; Dehnert et al., 2022). Materials are often selected due to their low cost, structural integrity, scalability, and ease of deployment (Ceccarelli et al., 2020), rather than considering how they may detrimentally impact or benefit propagated corals and the other members of their holobiont. The use of artificial materials may create a unique biogeochemical interface for corals that could act as a potential source of essential resources, such as trace elements (Ray et al., 2010; Reich et al., 2023), or harbour distinct bacterial communities and consequently impact coral-associated bacterial communities (Ceccarelli et al., 2020). Recent work has revealed that the type of plastic material used to secure corals to substrates does not differentially impact coral-associated bacterial communities, yet some zip-tie materials have greater coral retention rates (Strudwick et al., 2024). However, whether metal structures influence environmental conditions for propagated corals and subsequently influence coral-associated bacterial communities remains untested.
Across multiple studies, we have recorded changes in the bacterial communities of Acropora millepora when transferred from native reef to aluminium nursery structures during restoration (Strudwick et al., 2022, 2024). Aluminium is generally considered to be inert (although alloys can also corrode, Ezuber et al., 2008), whereas steel corrodes and releases iron oxides (Ray et al., 2010) in marine environments. Iron (Fe) plays essential roles in the physiological processes of both the coral host and associated microorganisms (Duckworth et al., 2009; Patel et al., 2020; Rubio-Portillo et al., 2020) and has the potential to leach into the environment at the coral–material interface as steel structures interact with the marine environment (Procópio, 2019). Thus, compared to aluminium, it is plausible to expect that steel structures will differentially impact the coral holobiont during propagation. We therefore hypothesise that coral propagated on steel structures will (i) host distinct bacterial communities, (ii) have different functional potential compared to those propagated on aluminium structures, and (iii) specifically involve overrepresentation of genes related to Fe cycling or with Fe requirements (e.g., as cofactors) and/or higher abundance of putatively pathogenic bacterial taxa. To test these hypotheses, we compared the functional and taxonomic characteristics of associated bacterial communities of the coral species A. millepora propagated on aluminium frames, sand/epoxy-coated steel modular structures (Reef Stars; Williams et al., 2019), and uncoated steel stakes.
The study period spanned 6 months from February to August 2022 and was conducted on the northern Great Barrier Reef (GBR), at the site “Long Bommie” on Opal Reef (16°22′17.2″S 145°87′60.6″E, Supplementary Figures S1A, B). Long Bommie was impacted by Cyclone Ita in 2014, which caused high structural degradation over most of the site and substantial areas of unconsolidated rubble. Despite widespread coral bleaching on the GBR in 2016/2017, 2020, and 2022, good recovery has been observed over the last 2 years on the reef flat and crest (Edmondson personal obs, Supplementary Figures S1C, D). Coral cover has particularly increased in the shallow (< 3 m depth) areas of the site (Edmondson personal obs, Supplementary Figures S1C, D). However, recovery has been relatively slow within the rubble, and subsequently, areas of unconsolidated rubble remain on the southern side of the bommie (Edmondson personal obs, Supplementary Figure S1E). Two 2.0 × 1.2-m aluminium diamond-mesh frames were installed approximately 30–40 cm above the sand, secured to and held in place with 4 × 9 kg Besser blocks (Figure 1A). Six hexagonal Reef Stars (coated steel) 54 cm in diameter × 33 cm in height, constructed from rebar coated with fibreglass and sand (Figure 1B), were installed directly over coral rubble and secured with steel rods (as per Williams et al., 2019; Figure 1B). Six uncoated 60-cm rebar stakes were hammered ~ 15 cm into rubble (Figure 1C). All structures were installed at the start of the experiment without preconditioning and were located 20–40 m away from source colonies on the reef.
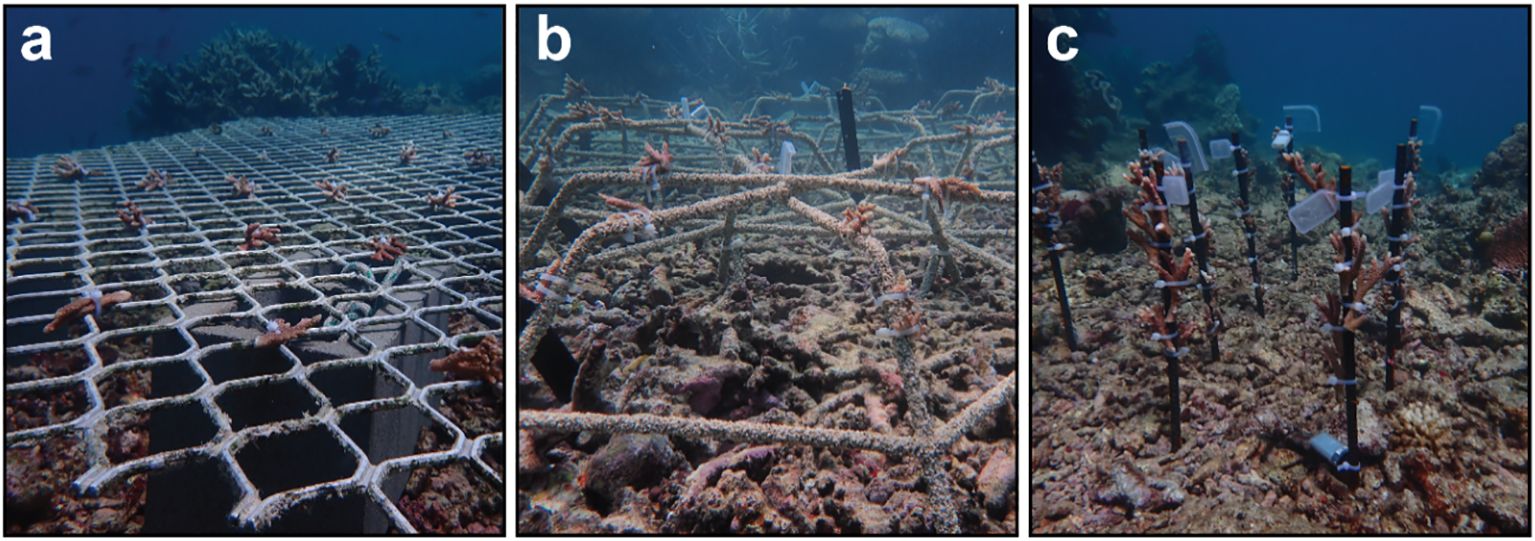
Figure 1 Coral fragments at Long Bommie are secured to (A) an aluminium frame, (B) Reef Stars, and (C) rebar stakes.
Coral sampling methods were designed to minimise any trace metal contamination (as per Grima et al., 2022). In brief, clear polypropylene plastics were prepared via a series of wash steps (as per Rodriguez et al., 2016) and used to mark and transfer the corals, whilst wooden chisels (or new bone cutters wrapped in parafilm) were used to sample coral. Acropora millepora was the chosen study species because the Acropora genus is commonly used in reef restoration (Boström-Einarsson et al., 2020) and has previously demonstrated variable coral-associated bacterial communities during propagation and out-planting on the Great Barrier Reef (Strudwick et al., 2022, 2023, 2024). The location of six A. millepora source colonies (≥ 55 cm diameter)—representing biological replicates—on the native reef was marked with transparent polypropylene plastic tags. Sixteen fragments (≤ 5 cm) were taken from each source colony. One fragment was retained in a sterile Whirl-Pak® bag for T0 microbial community characterisation, whilst the remaining 15 fragments were then divided into three plastic baskets. Fragments were then transported to the three different structures by SCUBA: five fragments from each of the six source colonies were taken to each of the aluminium frames, rebar stakes, or the Reef Stars (Supplementary Figure S2). Coral fragments were evenly divided and attached to the two aluminium frames (to account for potential frame effect), six Reef Stars, and six rebar stakes with clear cable ties (Supplementary Figure S2). On the aluminium frames, each row of corals corresponded to one source colony; fragments from only one source colony were attached to each structure for rebar stakes and Reef Stars. Clear tags were attached to the individual structures (or rows on the aluminium frames) to identify the respective source colonies (Figures 1A–C; Supplementary Figure S2).
At the start of the experiment, one (5–10 cm) fragment from each source colony was placed in an individual sterile Whirl-Pak® bag and immediately taken to the operation vessel to be snap-frozen in liquid nitrogen to characterise the bacterial community of source colonies at “time zero” (T0) (Supplementary Figure S2B). Following T0, corals on the propagation structures (aluminium frames, Reef Stars, rebar stakes) and source colonies were resampled at 6 months (T6m) (Supplementary Figure S2C). After 6 months, coral fragments (5–10 cm) were either removed from the propagation structures by detaching the cable tie (or cut using bone cutters wrapped in parafilm in cases where fragments had begun to overgrow the structure) or from the source colony in the natural reef environment using parafilm-wrapped bone cutters. Samples were placed into sterile Whirl-Pak® bags and snap-frozen in liquid nitrogen as per sampling at T0.
In total, 30 coral samples were collected, and up to two DNA extractions were conducted from each sample to provide DNA for shotgun metagenomics and amplicon (16S rRNA gene) sequencing. To increase the yield of coral-associated bacterial DNA, a phenol-chloroform extraction with an endonuclease step to remove the coral host and Symbiodiniaceae DNA was used for shotgun metagenomic applications (detailed in Appendix 1). This approach is not required for amplicon sequencing, where the target region is amplified via PCR prior to sequencing; hence, we proceeded with a previously successful DNA extraction kit for amplicon sequencing.
In some cases, there was not adequate material for two DNA extractions, and replication of n = 3 for each treatment across consistent source colonies (e.g., colonies 1, 2, and 3 sampled at both T0 and T6m for source reef colonies) for metagenomic sequencing was prioritised (Supplementary Figure S7). DNA was extracted from 15 samples for shotgun metagenomic analyses: three replicates × one time point for each of the propagation structures (n = 9) and three replicates × two time points for the source reef colonies (n = 6). DNA was extracted from 25 samples for amplicon (16S rRNA gene) sequencing: five to six replicates × two time points for source reef colonies (T0 n = 5 and T6m n = 5), four to six replicates × one time point for propagated corals (T6m: aluminium frames n = 4, Reef Stars n = 4, and rebar stakes n = 6) (Supplementary Figure S7), and one blank DNA extraction to identify laboratory contaminants.
All samples were transported from the field site to the laboratory in a dry shipper and then stored at −80°C for 3–9 months so that all DNA extractions could be conducted simultaneously. To minimise sample bias based on fragment size, preliminary analysis of the coral elementome (specifically Fe content) was performed (as per Grima et al., 2022) to identify a location on the coral branch (tip, middle, or base) for standardised metagenomics sampling. Results from this testing did not indicate any areas of host tissue or symbiotic algae with significant Fe enrichment. Thus, a 3–4-cm fragment was subsampled 1 cm from the base (structural contact point) of the propagated coral (Supplementary Figures S8A, B), and a 3–4-cm fragment of the source colony sample was placed in sterile zip-lock bags, and coral tissue was removed from the coral skeleton via airbrushing with sterile pipette tips into 2 mL of autoclaved PBS (3×, pH 7.4). The tissue slurry was transferred to a 2-mL microcentrifuge tube and centrifuged at 8,000×g for 5 min. The supernatant was removed, and the pellet was stored at −80°C for 2 days until DNA extraction. Coral tissue pellets were resuspended in 3× PBS prior to a series of homogenisation steps (as per Voolstra et al., 2022, detailed in Appendix 1). Following all homogenisation steps, a 250-µL aliquot of tissue slurry was used for phenol-chloroform DNA extraction that included an incubation with benzonase to remove free host and Symbiodiniaceae DNA (as per Voolstra et al., 2022, detailed in Appendix 1).
Prior to DNA extraction for amplicon (16S rRNA gene) sequencing, coral tissue was removed from the coral skeleton, using an airbrushing technique. For each sample, a 3–4-cm fragment was subsampled from the same area as before and placed in a sterile zip-lock bag to be airbrushed with sterile pipette tips into 4 mL of autoclaved PBS (3×, pH 7.4). The tissue slurry was divided across two 2-mL microcentrifuge tubes and centrifuged at room temperature for 5 min at 8,000×g. The supernatant was removed, and DNA was extracted from approximately 100 µL of the coral tissue pellet using a DNeasy Blood and Tissue kit (Qiagen—July 2020 version) following the manufacturer’s protocols with a total elution volume of 40 µL. A kit negative (no sample material added) was included in the DNA extraction to identify any kit contaminants. For both the shotgun metagenomic and 16S amplicon DNA extraction methods, extracted DNA was quality checked, and the concentration was quantified using a NanoDrop spectrophotometer prior to sequencing.
DNA samples were stored at −80°C for 2–4 days until transportation on dry ice to the Ramaciotti Centre for Genomics (Sydney, NSW, Australia) for 16S rRNA gene amplicon and shotgun metagenomic sequencing to characterise taxonomic and functional profiles of the coral-associated bacterial communities. To taxonomically characterise coral-associated bacterial communities, the hypervariable V3 to V4 region of the bacterial 16S rRNA gene was amplified using the primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) (Klindworth et al., 2013), and the amplicons were sequenced using the Illumina MiSeq v3 2 × 300 bp platform. To characterise the functional profile of the coral-associated bacterial communities, shotgun metagenomic sequencing was performed using the Illumina NovaSeq 6000 SP 2 × 150 bp Flowcell platform (Ramaciotti Centre for Genomics). Raw reads from 16S rRNA amplicon sequencing and shotgun metagenomics were deposited in the NCBI Sequence Read Archive (SRA) in FASTQ format under Bioproject number PRJNA988823 and will be released upon publication or by request.
Raw demultiplexed sequencing data were analysed with the Quantitative Insights into Microbial Ecology (QIIME 2, version 2020.6) platform (Callahan et al., 2016). The data were denoised (with the DADA2 plugin) prior to taxonomic assignment against the SILVA v138 database using the classify-sklearn classifier (Pedregosa et al., 2011). After denoising, 3,666,205 reads were generated from 25 samples. Mitochondrial or chloroplast amplicon sequence variants (ASVs) were filtered from the dataset. Nine ASVs that comprised 100% of the sequences in the kit negative DNA extraction were removed for subsequent analyses using the filter command in R (version 4.2.2). Two samples were removed before diversity analyses due to poor sequencing outputs, resulting in low read numbers after quality filtering and contaminant removal (< 55 ASVs) (Supplementary Figure S7). For beta diversity analyses, the raw read ASV table was converted to relative abundances, scaled to 20,000 (McKnight et al., 2019), and square root transformed.
Filtering of low-quality reads, trimming of adapter and low-quality sequences, and deduplication was performed on raw reads using fastp (v0.23.2) (Chen et al., 2018). Removal of contaminating host A. millepora (acc:GCF_013753865.1) and human (acc:GCF_013753865.1) DNA was performed using HoCoRT (v1.0.0) (Rumbavicius et al., 2023, in press). Read-based taxonomic profiles were generated from cleaned read sets using Sourmash (v4.8.2) (Brown and Irber, 2016) against the Genome Taxonomy Database (GTDB) (release 207) (Parks et al., 2022). Overlapping pairs were first merged for each read-set using fastp, and a (pooled) coassembly was then constructed using MEGAHIT (v1.2.9), where merged reads were passed as single-ended (Li et al., 2015). Coassembled contigs were passed as a user-supplied assembly to the SqueezeMeta (v1.6.1) pipeline (analysis mode: coassembly), and read-sets for each sample were aligned to the coassembly to predict genes, annotate gene functions and estimate the abundance of individual genes per sample (Tamames and Puente-Sánchez, 2019).
Differences in bacterial community structure and dispersion (beta diversity patterns) of source reef corals over time (T0 to T6m) and between source colonies and propagated corals after 6 months (T6m) were analysed using the Bray–Curtis dissimilarity distance metric. Patterns in bacterial community structure were visualised using nonmetric multidimensional scaling (nMDS) plots. Differences in community structure were tested for significance with permutational multivariate analysis of variance (PERMANOVA; perm = 999) of Bray–Curtis dissimilarities using the adonis function of the “vegan” R package and subsequent pairwise comparisons (if significant) with pairwise.adonis, p-values were adjusted by applying a Benjamini and Hochberg correction, and all padj values < 0.05 were considered significant. Permutation tests for homogeneity of multivariate dispersion (PERMDISP) of the coral-associated bacterial community were calculated using the betadisper function of the “vegan” R package (perm = 999), p-values were adjusted by applying a Benjamini and Hochberg correction, and all padj values < 0.05 were considered significant. The core_members function of the “microbiome” R package was used to identify core bacterial community members (present in > 75% samples with relative abundance (RA) > 0.1%) for source reef corals and propagated fragments at T0 and T6m. To identify bacterial taxa significantly associated with different propagation structures, we applied an indicator species analysis with the multipatt function of the R “indicspecies” package. Results were cross-referenced against the relative abundance (RA) of each ASV and retained if present in > 75% of replicates.
Metagenome reads were assigned gene functions and pathways based on the database structure of the Kyoto Encyclopedia of Genes and Genomes (KEGG). To quantify the abundance of KEGG Orthologs (KOs), transcripts per million (tpm) values were calculated. Functional analysis at the KO level was conducted using a Bray–Curtis dissimilarity distance metric on the KO tpm matrix. Patterns in the diversity of functions at the community level were visualised using nMDS plots and differences between treatments were tested for significance with PERMANOVA of Bray–Curtis dissimilarities using the adonis function of vegan.
To identify functions unique to each treatment, a presence/absence analysis was conducted. KOs were isolated based on their presence in source colonies at T6m (in all replicates) and simultaneous absence (0% RA in) in a given propagated coral (e.g., in all replicates and for each structure separately) or their absence in source colonies at T6m (in all replicates) and simultaneous presence in a given propagated coral (in all replicates) at T6m. Functional analysis was also conducted at the path level for pathways that were related to KOs identified in the absence/presence analysis. All KOs were grouped by KEGG-classified pathway, and differences between treatments were visualised with bar plots. Differences between treatments for each pathway were tested for significance using a Kruskal–Wallis test, where a significant pairwise Wilcox post-hoc test was applied with Benjamini and Hochberg p-value adjustment, all padj values < 0.05 were considered significant.
Taxonomic profiles of coral-associated bacterial communities generated from shotgun metagenomic sequencing (Supplementary Figure S8) did not match those acquired from the (more directed) amplicon sequencing of the 16S rRNA gene (Supplementary Figure S7A). Such contrasting outcomes are potentially from the two different databases used for the taxonomic classification of reads and/or two different extraction methods used to prepare the DNA for sequencing. Considering the taxonomic profiles acquired from the amplicon sequencing were more comprehensive (2,899 ASVs vs. 96 unique taxa)—and to capture any differences in rare taxa that were not quantified in the shotgun sequencing output—all subsequent analysis of the phylogenetic characteristics of coral-associated bacterial communities was conducted using the amplicon (16S rRNA gene) dataset.
No significant changes were observed for the taxonomic structure of bacterial communities associated with A. millepora source colonies over the course of the experiment (T0 to T6m: PERMANOVABray–Curtis, F = 1.156, padj = 0.182, Supplementary Figures S7A, B). Consequently, all comparisons between the source colony and propagated corals were conducted only at the 6-month sampling point (T6m). Two ASVs (Synechococcus genus and Endozoicomonas acroporae, mean relative abundance (RA) = 5.13% and 14.92%, respectively) were identified as “core” members of the bacterial communities of source colonies at the start of the experiment (T0) and were retained throughout the 6-month study period. After 6 months, an additional ASV (Psychrobacter pacificensis, mean RA = 6.61%) was identified as the core of source reef colonies (Supplementary Table S2).
After 6 months, no consistent differences were observed between the taxonomic structure of the coral-associated bacterial community of propagated corals across all propagation materials or versus source reef colonies (T6m) (PERMANOVABray–Curtis, padj > 0.05, Figures 2A, B; Supplementary Table S1). No differences were found for the core bacterial taxa of source reef colonies (T6m) versus all propagated coral (regardless of propagation structure material). The core included one P. pacificensis species ASV, one Synechococcus genus ASV, and one E. acroporae species ASV (Supplementary Table S2). The mean RA of the E. acroporae ASV was higher in aluminium and rebar corals (mean RA = 33.33% and 34.79%, respectively) than source reef colonies (T6m mean RA = 24.88%) but lower in Reef Star corals (mean RA = 2.38%). Highlighting minor differences in core bacterial community members across propagation structures. Reef Star corals formed a discrete cluster on the nMDS plot (Figure 2B) and exhibited significantly less multivariate dispersion compared to source reef colonies (at T6m) and rebar corals (pairwise PERMUTEST, padj = 0.041 and padj = 0.048, respectively), but no significant differences in taxonomic structure between Reef Star coral and source reef colonies or other propagated corals were observed (PERMANOVABray–Curtis, padj > 0.05). Thus, differences in coral-associated bacterial communities between propagation structures may have been too small to detect. Consequently, we further assessed bacterial communities to identify any discriminating factors at the ASV level using indicator species analysis.
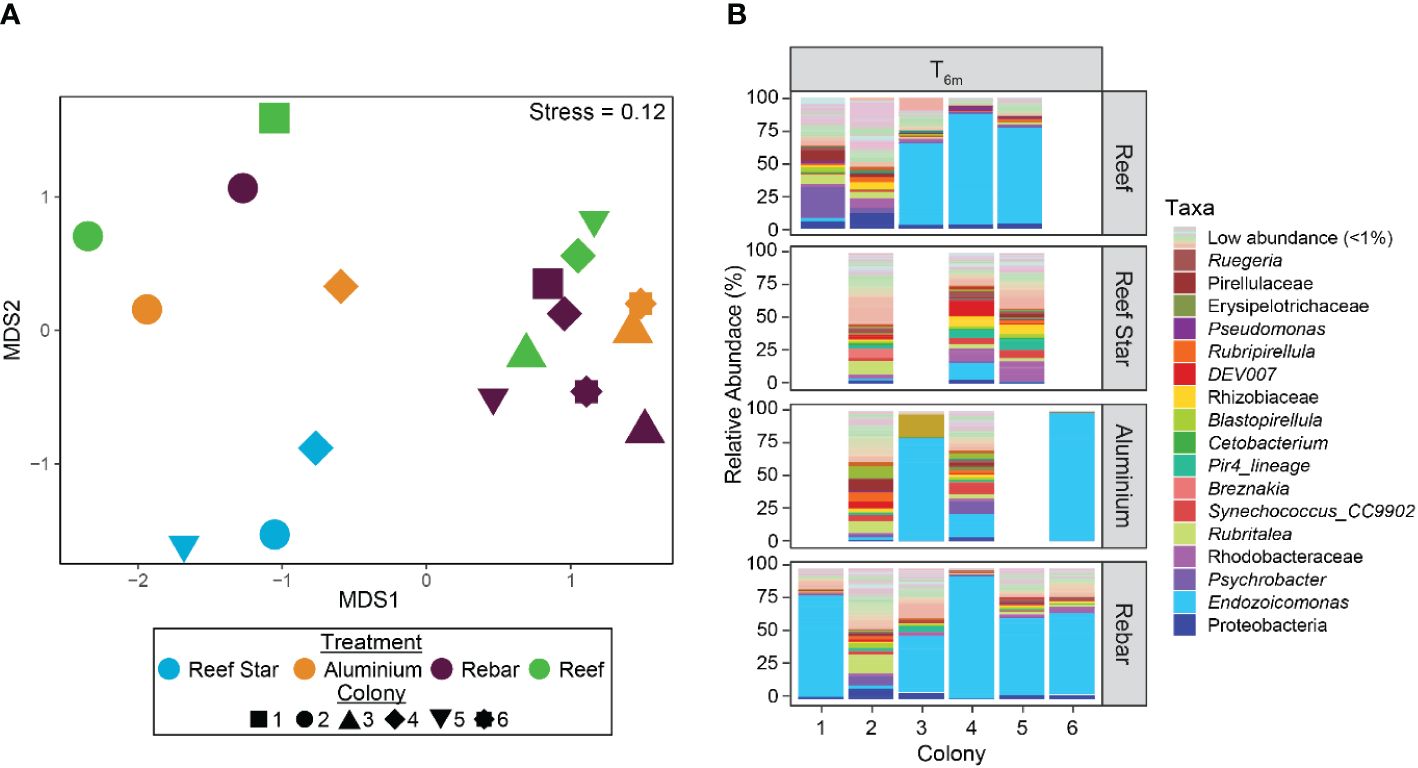
Figure 2 (A) Bacterial community structure and relative dispersion of the microbial communities of the source reef colonies and corals propagated on different metal structures (Reef Stars, rebar stakes, and aluminium frames) after 6 months (T6m). The plot is based on nonmetric multidimensional scaling (nMDS) of Bray–Curtis distances of bacterial community structure from 16S rRNA gene taxonomy profiles. (B) Bacterial community composition (relative abundances) by genus* of source reef colonies and of corals grown on three different propagation structures after 6 months (T6m). Pastel colours represent genera with an average relative abundance of < 0.1% in all samples; a full legend is provided as supplemental data (Supplementary Data S1). *Family/class classifications were used when the genus was unknown.
Although no community-level (beta diversity) differences were observed between propagated corals, seven different ASVs were significant “indicators” for bacterial communities associated with propagated corals across the different metal structures and source reef colonies at T6m. Three ASVs from the Ruegeria and Trichodesmium_IMS101 genera (mean RA = 1.91% and 1.10%, respectively) and from the family Rhizobiaceae (mean RA = 1.45%) were indicative of Reef Star coral-associated bacterial communities. Three ASVs of the E. acroporae species were indicators of rebar coral-associated bacterial communities (mean RA = 1.19%, 2.41%, and 0.33%). One ASV of the Prosthecochloris vibrioformis species was indicative of aluminium frame coral-associated bacterial communities (mean RA = 4.47%). One ASV of the Flavobacteriaceae family was indicative of source reef coral bacterial communities (mean RA = 0.91%).
No differences in the functional profiles of coral-associated bacterial communities were observed in source colonies over time (from T0 to T6m) (PERMANOVABray–Curtis, F = 0.915, padj = 0.5, Supplementary Figure S7A). Consequently (as with the taxonomic data), all analysis of bacterial community function between the source colony and propagated corals was conducted at the 6-month sampling point (T6m). Overall, there were no differences in the functional profiles of coral-associated bacterial communities between corals grown on different metal propagation structures and source reef colonies at T6m, nor were there any differences between propagation structures (PERMANOVABray–Curtis, padj > 0.05, Supplementary Figure S7B).
To resolve any high-resolution differences in coral-associated bacterial community functioning potential, e.g., from a more nuanced response to the propagation materials (that might not be detected when comparing the whole profiles) and could be linked to differences in indicator taxa, we analysed the abundance of individual KOs. At the 6-month time point (T6m), a presence/absence analysis highlighted 22 KOs (groups of genes performing the same functions) of the 5,125 assigned KOs were present in propagated corals and completely absent in source reef colonies. One KO was present in the source reef colonies but was absent from the aluminium frame coral, and no KOs were present in the source reef colonies and absent in rebar and Reef Star coral. Hence, we focussed on the KOs present in coral-associated bacterial communities across different metal propagation structures (Figure 3A). Four KOs were present in the metagenomes of corals grown on aluminium propagation structures (and absent in source reef colonies) assigned to genetic information processing (KEGG classification 2) and environmental information processing pathways (KEGG classification 3.2, Supplementary Table S3; Appendix 2). Three KOs were present in Reef Star coral metagenomes (and absent in source reef colonies) relating to environmental information processing (KEGG classification 3.1), metabolic pathways (KEGG classification 1.2), and one unknown pathway (Supplementary Table S3; Appendix 2). Fifteen KOs were present in rebar coral-associated bacterial communities (and absent in source reef colonies), four were related to genetic information processing pathways (KEGG classification 2.1, 2.3, and 2.4), two KOs were related to environmental information processing pathways (KEGG classification 3.1), four KOs were related to metabolic pathways (KEGG classification 1.1, 1.4), one KO was related to a signalling and cellular processing pathway (KEGG classification 4.1), and three KOs were from poorly described pathways (Supplementary Table S3; Appendix 2).
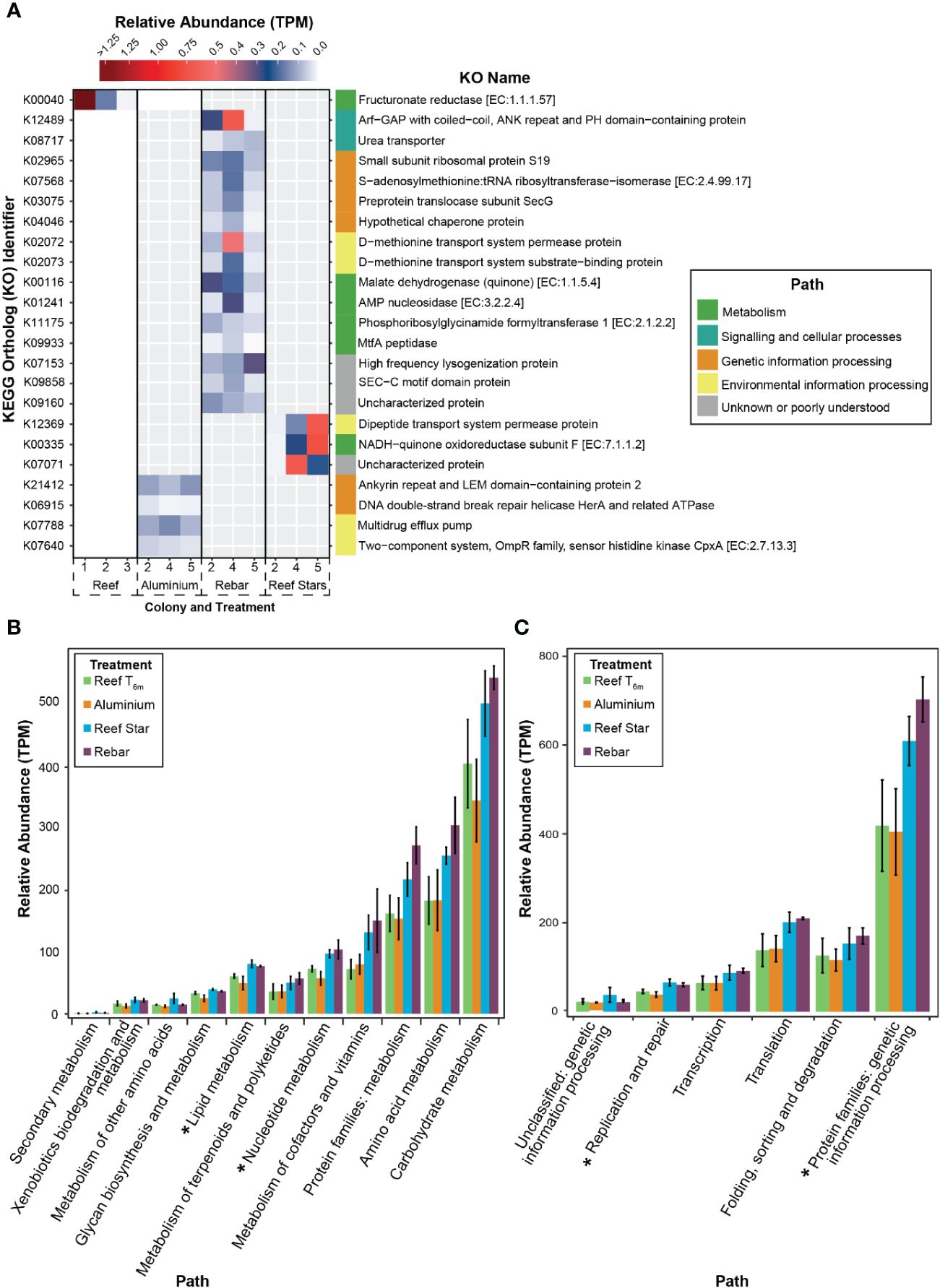
Figure 3 (A) Presence/absence analysis used to identify propagation material-related functions in coral-associated bacterial communities of propagated corals. The heatmap shows KEGG orthologs (KO) in the metagenome of corals propagated on different materials (coated steel: Reef Stars, uncoated steel: Rebar and aluminium frames) and source reef colonies after 6 months (T6m). The colour gradient corresponds to the transcripts per million (tpm) (higher tpm = higher abundance). The KO identifier is provided on the left of the heatmap, and the respective name is on the right of the heat plot for each KO. KO names are coloured by their related functional paths, and entries are organised by these functions. KOs are only shown if they are present in 100% of replicates for each treatment. (B) KEGG metabolism and (C) genetic information processing pathways in the metagenomes of source reef colonies or corals propagated on different material propagation structures after 6 months. KOs were grouped by path, and a mean of the total tpm values is plotted with standard error bars. The asterisk indicates where the Kruskal–Wallis test on each pathway by treatment showed p < 0.05.
Across restoration structures, the KOs present in propagated corals but absent in source reef colonies were components of carbohydrate and nucleotide metabolic pathways, translation, membrane transport and folding, sorting and degradation pathways, and signalling pathways (Figure 3A). Therefore, to determine whether entire functional pathways were elevated in propagated corals, we analysed the functional profiles at the pathway level. All metabolic and genetic information processing pathways (KEGG classifications 1 and 2) were overrepresented in the metagenomes of corals propagated on Reef Stars and rebar stakes compared to source reef colonies and aluminium corals (Figures 3B, C), apart from energy metabolism (KEGG classification 1.2), which was underrepresented in rebar coral compared to coral propagated on other metal structures and source reef colonies (Supplementary Figure S8). Differences across propagated corals and source reef colonies were significant for only two categories of metabolic pathways (KEGG classification 1.3—Lipid metabolism, Kruskal–Wallis, χ2 = 8.5385, df = 3, p = 0.0361 and KEGG classification 1.4—Nucleotide metabolism, Kruskal–Wallis, χ2 = 8.641, df = 3, p = 0.0345, Figure 3B) and two genetic information processing pathways (KEGG classification 2.4—Replication and repair, Kruskal–Wallis, χ2 = 8.641, df = 3, p = 0.03446; Genetic information processing protein families, Kruskal–Wallis, χ2 = 9.0513, df = 3, p = 0.02862, Figure 3C). No significant differences were observed in the post-hoc tests (pairwise Wilcox post-hoc, padj > 0.05).
Various materials are being used in ever-scaling reef restoration approaches (Nedimyer et al., 2011; Meesters et al., 2015; Williams et al., 2019; Boström-Einarsson et al., 2020; Ceccarelli et al., 2020; Suggett et al., 2020; Dehnert et al., 2022) that have the potential to alter coral holobiont fitness by influencing the composition and functioning of coral-associated bacterial communities (Ceccarelli et al., 2020; Reich et al., 2023). Yet, it remains unclear if the use of different metals positively or negatively affects coral-associated bacterial communities during propagation and, in turn, how this regulates reef restoration success (van Oppen and Blackall, 2019; Voolstra et al., 2021; Peixoto et al., 2022; Strudwick et al., 2022, 2023). Here, we show that across different propagation structures after 6 months, minor differences in individual bacterial taxa and the functional potential of coral-associated bacterial communities can emerge. Specifically, propagated corals exhibited several KOs that were absent in the bacterial communities of source reef colonies, and coral propagated on steel structures exhibited an overrepresentation of various metabolic and genetic information processing pathways. Furthermore, distinct bacterial taxa were indicative of coral-associated bacterial communities across the different metal propagation structures. Together, the minor differences in functional potential and individual members of coral-associated bacterial communities demonstrate that propagated coral may have been under distinct environmental conditions (Ziegler et al., 2017; Maher et al., 2019; Camp et al., 2020, 2019). Consequently, further testing is required to resolve the confounding findings of differences in functional potential and lack of differences in community composition of coral-associated bacterial communities, to determine whether Fe from steel structures plays a role in these changes, and to identify the implications of such differences in coral-associated bacterial communities on the fitness and resilience of the holobiont.
We compared three propagation structures to identify whether the type of metal used differentially impacts coral-associated bacterial communities. No differences in the structure of coral-associated bacterial communities were recorded between A. millepora source reef colonies and propagated corals across structures. Restructuring of coral-associated bacterial communities of Acropora spp. is typically seen during transplantation between distinct environments (Ziegler et al., 2019; Haydon et al., 2021), and for A. millepora, particularly during reef restoration on the GBR (Strudwick et al., 2022, 2024). Previously, coral-associated bacterial community differences have been recorded after 4 to 6 months of propagation on aluminium structures at other sites at Opal Reef (Strudwick et al., 2022, 2024). Therefore, it is perhaps surprising that no differences in coral-associated bacterial communities were observed between source reef colonies and propagated corals on aluminium structures (or any other structure) after 6 months in our current study. The observed lack of changes in coral-associated bacterial communities between source reef colonies and propagated coral may be linked to environmental site differences (Haydon et al., 2021). High flow, for example, has previously been linked to more stable coral-associated bacterial communities (Lee et al., 2017), and the location of our current study (Long Bommie, Opal Reef) experiences greater flow and wave action than Rayban and Mojo of Opal Reef in previous studies (Edmondson personal obs.; Strudwick et al., 2022, 2024). At Long Bommie, corals were also grown on structures positioned 30–40 cm above the substrate, whereas mid-water aluminium frames suspended 2–3 m above the benthos were used at Rayban and Mojo (Strudwick et al., 2022, 2024). The different positioning of propagation structures in relation to the substrate likely resulted in exposure to lower water flow (Perkol-Finkel et al., 2008) for propagated corals, or more importantly, similar environmental conditions (such as temperature and water quality) to source reef colonies at Long Bommie. The unique positioning of the structures may have also altered the interaction of the structure material with the environment, which in turn could influence the coral-associated bacterial community dynamics [e.g., varied flow rates can alter the rate of steel corrosion in a marine environment (Peng et al., 2024) and Fe has important roles in coral and microorganism health (Duckworth et al., 2009; Patel et al., 2020; Rubio-Portillo et al., 2020)]. Whilst this notion remains to be verified, it is consistent with the growing evidence of site-specific environmental conditions as drivers of coral-associated bacterial communities (Maher et al., 2019; Ziegler et al., 2019; Camp et al., 2020; Osman et al., 2020). It also highlights the importance of further characterising coral-associated bacterial communities during restoration across diverse sites and potentially on longer time frames to combine knowledge of species-specific and site-specific bacterial community trends necessary to tailor local protocols as approaches scale geographically.
Whilst propagation on different materials did not yield changes in the structure of the coral-associated bacterial communities, distinct taxa were identified at the individual ASV level as representative of the different coral environments. Three ASVs of the putative coral symbiont Endozoicomonas acroporae (Tandon et al., 2022) were identified as indicator taxa for rebar coral. Three bacterial taxa that were putative symbionts and/or had nitrogen-fixing and iron-binding abilities were identified as indicator taxa for Reef Star coral (Ruegeria, Kitamura et al., 2021; Trichodesmium_IMS101, Capone et al., 1997; Held et al., 2022; Rhizobiaceae, Rincon-Rosales et al., 2010). Finally, one photosynthetic green sulphur bacterium, Prosthecochloris Vibrioformis (Nie et al., 2023) ASV, was indicative of coral grown on aluminium frames. We hypothesised that pathogenic bacterial taxa may proliferate on steel structures, due to the role of Fe in pathogenesis and virulence (Kelly et al., 2012; Rubio-Portillo et al., 2020; Gnanagobal and Santander, 2022), but importantly, no putative pathogens were identified as indicator species across any of the propagation structures. However, differences in indicator taxa suggest propagated coral may be under distinct environmental conditions. Thus, our results indicate that the unique biogeochemical niche surrounding each propagation structure had a subtle influence on propagated coral and affected individuals within coral-associated bacterial communities for the period of growth in this study.
No differences were observed in the overall functional profile of coral-associated bacterial communities across propagation structures and source reef colonies. However, several KOs were elevated in propagated corals that were absent from source reef colonies, and various pathways were differentially represented between all corals (propagated and source reef colonies). Given our hypothesis that coral propagated on steel structures would display enrichment of genes associated with Fe within associated bacterial communities, we specifically investigated whether overrepresented KOs were related to Fe. Coral propagated on rebar exhibited the most overrepresented Kos, and these were components of nucleotide (purine and adenosine monophosphate) and carbohydrate (pyruvate and glucose) metabolism pathways and genetic information processing (translation, membrane transport and folding, sorting, and degradation) pathways. Though only components of these pathways were enriched, the overrepresentation of carbohydrate metabolism KOs (malate dehydrogenase and MtfA peptidase) and nucleotide metabolism KOs (AMP nucleosidase and phosphoribosylglycinamide formyltransferase 1) in rebar coral compared to source reef colonies suggests that rebar coral-associated bacterial communities could have altered or increased metabolic potential (Zhang et al., 2015). In fact, both rebar and (to a lesser extent) Reef Star coral-associated bacterial communities exhibited an overrepresentation of several KEGG metabolic pathways (carbohydrate, lipid, amino acid, and nucleotide metabolism) compared to both aluminium coral and source reef colonies. The metabolic potential of coral-associated bacterial communities has been shown to reflect local oceanographic conditions (Kelly et al., 2014; Zhang et al., 2015), and metabolic demands can increase in response to Fe enrichment (Rädecker et al., 2017). Consequently, the possible increase in metabolic potential may be linked to different environmental conditions (e.g., nutrient availability; Kelly et al., 2014; Rädecker et al., 2017) between propagation structures and source reef colonies. Furthermore, Fe leaching rates may differ between steel structures due to the rust preventative on Reef Stars (Williams et al., 2019) and the lack thereof on rebar stakes, which could explain the different extent of metabolic pathway overrepresentation between Reef Star and rebar corals. However, whether steel structures (coated or not) alter Fe availability in the coral microenvironment and whether Fe availability is responsible for the altered functional potential is yet to be resolved.
Only three KOs were present in Reef Star coral and absent in source reef colonies, yet two of these had putative links to Fe transport or utilisation (a periplasmic transport protein that binds Fe-containing molecules (hemes), Hogle et al., 2014; and an oxidative phosphorylation protein that shuttles electrons from NADH via iron–sulphur centres in the respiratory chain, Leif et al., 1995). The presence of these KOs in Reef Star coral indicates the potential contribution of Fe to the coral microenvironment from the steel propagation structures. Our results suggest that depending on propagation structure material, there are implications for the functional potential of coral-associated bacterial communities, but whether this directly relates to Fe and the extent to which this impacts holobiont fitness requires further testing.
Successful preservation of beneficial coral–microorganism associations will likely be essential for maintaining coral health and resilience during reef restoration activities (Peixoto et al., 2022) and, more importantly, optimal ecosystem functioning (Lima et al., 2023). The materials used to manufacture propagation structures and to secure corals during propagation are diverse (Boström-Einarsson et al., 2018), yet their impacts on coral-associated bacterial communities have until now been untested. In this study, we demonstrate that propagation on different materials can lead to subtle differences in individual bacteria and the functional potential of coral-associated bacterial communities; however, the implications of these differences on holobiont fitness and the implications for ecosystem functions of a restored reef using these materials are unknown. Restoration practitioners may operate with the understanding that structure material does not significantly impact coral-associated bacterial community composition for A. millepora in the short term. However, KOs that were present in Reef Star coral and pathways overrepresented in both Reef Star and rebar corals point to differing conditions in the coral local environment between steel structures and source reef colonies or aluminium structures; therefore, further testing is required to determine the role of Fe and its impacts on the holobiont health. Considering low-level nutrient (e.g., Fe) enrichment can enhance coral thermal performance (Becker et al., 2021), we suggest future research should assess Fe accumulation within the coral elementome during propagation on steel structures and any related changes in stress tolerance (Becker et al., 2021; Reich et al., 2023). In conclusion, our findings highlight that propagation structure material has minimal impacts on the taxonomic composition of coral-associated bacterial communities but may have implications for the functional potential of these communities that deserves further attention.
The datasets presented in this study can be found in online repositories: https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA988823.
The manuscript presents research on animals that do not require ethical approval for their study.
PS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft. DS: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. JS: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing. AG: Methodology, Project administration, Writing – review & editing. MD: Data curation, Project administration, Writing – review & editing. JE: Methodology, Project administration, Writing – review & editing. AM: Methodology, Project administration, Writing – review & editing. FN: Methodology, Project administration, Writing – review & editing. EC: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by an Australian Government Research Training Programme (RTP) Fee Offset Scholarship and Stipend to PS; Australian Coral Reef Society Terry Walker Award to PS; the University of Technology Sydney Chancellor’s Postdoctoral Research Fellowship and ARC Discovery Early Career Research Award (DE190100142); a donation from David and Susan Rockefeller to EC; Mars Sustainable Solutions Funding to DS and EC; and an Australian Research Council Grant (DP210101610) to JS. Field operations at Opal Reef were supported by funding to the Coral Nurture Program from the Australian and Queensland Governments (“Solving the bottleneck of reef rehabilitation through boosting coral abundance: Miniaturising and mechanising coral out-planting”) to DS, EC, JE.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1366971/full#supplementary-material
Becker D. M., Putnam H. M., Burkepile D. E., Adam T. C., Vega Thurber R., Silbiger N. J. (2021). Chronic low-level nutrient enrichment benefits coral thermal performance in a fore reef habitat. Coral Reefs 40, 1637–1655. doi: 10.1007/s00338-021-02155-1
Boström-Einarsson L., Babcock R. C., Bayraktarov E., Ceccarelli D., Cook N., Ferse S. C. A., et al. (2020). Coral restoration – A systematic review of current methods, successes, failures and future directions. PloS One 15, e0226631. doi: 10.1371/journal.pone.0226631
Boström-Einarsson L., Ceccarelli D., Russel C., Bayraktarov E., Cook N., Harrison P., et al. (2018). Coral Restoration in a changing world A global synthesis of methods and techniques (Cairns: Reef and Rainforest Research Centre Ltd). Available at: https://researchonline.jcu.edu.au/59790/1/59790_2019%20nesp%20project%204.3%20technical%20report.pdf, ISBN: ISBN: 9781925514315. Sponsored by NESP Tropical Water Quality Hub.
Brown C. T., Irber L. (2016). sourmash: a library for MinHash sketching of DNA. J. Open-source Softw. 1, 27. doi: 10.21105/joss.00027
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camp E. F. (2022). Contingency planning for coral reefs in the Anthropocene; The potential of reef safe havens. Emerging Topics Life Sci. 6, 107–124. doi: 10.1042/etls20210232
Camp E. F., Suggett D. J., Pogoreutz C., Nitschke M. R., Houlbreque F., Hume B. C. C., et al. (2020). Corals exhibit distinct patterns of microbial reorganisation to thrive in an extreme inshore environment. Coral Reefs 39, 701–716. doi: 10.1007/s00338-019-01889-3
Capone D. G., Zehr J. P., Paerl H. W., Bergman B., Carpenter E. J. (1997). Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229. doi: 10.1126/science.276.5316.1221
Carthey A. J., Blumstein D. T., Gallagher R. V., Tetu S. G., Gillings M. R. (2020). Conserving the holobiont. Funct. Ecol. 34, 764–776. doi: 10.1111/1365-2435.13504
Caruso C., Hughes K., Drury C. (2021). Selecting heat-tolerant corals for proactive reef restoration. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.632027
Ceccarelli D. M., McLeod I. M., Boström-Einarsson L., Bryan S. E., Chartrand K. M., Emslie M. J., et al. (2020). Substrate stabilisation and small structures in coral restoration: State of knowledge, and considerations for management and implementation. PloS One 15, e0240846. doi: 10.1371/journal.pone.0240846
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Dehnert I., Saponari L., Isa V., Seveso D., Galli P., Montano S. (2022). Exploring the performance of mid-water lagoon nurseries for coral restoration in the Maldives. Restor. Ecol. 30, e13600. doi: 10.1111/rec.13600
Duckworth O. W., Bargar J. R., Jarzecki A. A., Oyerinde O., Spiro T. G., Sposito G. (2009). The exceptionally stable cobalt(III)–desferrioxamine B complex. Mar. Chem. 113, 114–122. doi: 10.1016/j.marchem.2009.01.003
Ezuber H., El-Houd A., El-Shawesh F. (2008). A study on the corrosion behavior of aluminum alloys in seawater. Mater. Design 29, 801–805. doi: 10.1016/j.matdes.2007.01.021
Fox H. E., Harris J. L., Darling E. S., Ahmadia G. N., Razak T. B. (2019). Rebuilding coral reefs: success (and failure) 16 years after low-cost, low-tech restoration. Restor. Ecol. 27, 862–869. doi: 10.1111/rec.12935
GBRMPA (2019) Great Barrier Reef Outlook Report 2019. Available online at: http://hdl.handle.net/11017/3474.
Gnanagobal H., Santander J. (2022). Host–pathogen interactions of marine gram-positive bacteria. Biology 11. doi: 10.3390/biology11091316
Grima A. J., Clases D., Gonzalez de Vega R., Nitschke M. R., Goyen S., Suggett D. J., et al. (2022). Species-specific elementomes for scleractinian coral hosts and their associated Symbiodiniaceae. Coral Reefs 41, 1115–1130. doi: 10.1007/s00338-022-02259-2
Haydon T. D., Seymour J. R., Raina J. B., Edmondson J., Siboni N., Matthews J. L., et al. (2021). Rapid shifts in bacterial communities and homogeneity of Symbiodiniaceae in colonies of Pocillopora acuta transplanted between reef and mangrove environments. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.756091
Hein M. Y., Beeden R., Birtles A., Gardiner N. M., Le Berre T., Levy J., et al. (2020). Coral restoration effectiveness: Multiregional snapshots of the long-term responses of coral assemblages to restoration. Diversity 12, 153. doi: 10.3390/D12040153
Hein M. Y., Staub F., Banaszack A., Dallison T., Deri W., Grimsditch G., et al. (2021). ICRI-Mapping the global funding landscape for coral reef restoration Vol. 23 (International Coral Reef Initiative Forum). Available at: www.icriforum.org.
Held N. A., Waterbury J. B., Webb E. A., Kellogg R. M., McIlvin M. R., Jakuba M., et al. (2022). Dynamic diel proteome and daytime nitrogenase activity supports buoyancy in the cyanobacterium Trichodesmium. Nat. Microbiol. 7, 300–311. doi: 10.1038/s41564-021-01028-1
Hogle S. L., Barbeau K. A., Gledhill M. (2014). Heme in the marine environment: from cells to the iron cycle. Metallomics 6, 1107–1120. doi: 10.1039/c4mt00031e
Howlett L., Camp E. F., Edmondson J., Hosp R., Taylor B., Coulthard P., et al. (2023). Active coral propagation outcomes on coral communities at high-value Great Barrier Reef tourism sites. Biol. Conserv. 279, 109930. doi: 10.1016/j.biocon.2023.109930
Kelly L. W., Barott K. L., Dinsdale E., Friedlander A. M., Nosrat B., Obura D., et al. (2012). Black reefs: iron-induced phase shifts on coral reefs. ISME 6, 638–649. doi: 10.1038/ismej.2011.114
Kelly L. W., Williams G. J., Barott K. L., Carlson C. A., Dinsdale E. A., Edwards R. A., et al. (2014). Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc. Natl. Acad. Sci. United States America 111, 10227–10232. doi: 10.1073/pnas.1403319111
Kenyon T. M., Doropoulos C., Wolfe K., Webb G. E., Dove S., Harris D., et al. (2023). Coral rubble dynamics in the Anthropocene and implications for reef recovery. Limnol. Oceanogr. 68, 110–147. doi: 10.1002/lno.12254
Kitamura R., Miura N., Ito M., Takagi T., Yamashiro H., Nishikawa Y., et al. (2021). Specific detection of coral-associated ruegeria, a potential probiotic bacterium, in corals and subtropical seawater. Mar. Biotechnol. 23, 576–589. doi: 10.1007/s10126-021-10047-2
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, 1–11. doi: 10.1093/nar/gks808
Lee S. T., Davy S. K., Tang S. L., Kench P. S. (2017). Water flow buffers shifts in bacterial community structure in heat-stressed Acropora muricata. Sci. Rep. 7, 43600. doi: 10.1038/srep43600
Leif H., Sled V. D., Ohnishi T., Weiss H., Friedrich T. (1995). Isolation and characterization of the proton-translocating NADH: ubiquinone oxidoreductase from Escherichia coli. Eur. J. Biochem. 230, 538–548. doi: 10.1111/j.1432-1033.1995.tb20594.x
Lima L. F. O., Alker A. T., Papudeshi B., Morris M. M., Edwards R. A., de Putron S. J., et al. (2023). Coral and seawater metagenomes reveal key microbial functions to coral health and ecosystem functioning shaped at reef scale. Microbial Ecol. 86, 392–407. doi: 10.1007/s00248-022-02094-6
Maher R. L., Rice M. M., McMinds R., Burkepile D. E., Vega Thurber R. (2019). Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci. Rep. 9, 6834. doi: 10.1038/s41598-019-43274-8
McAfee D., Costanza R., Connell S. D. (2021). Valuing marine restoration beyond the ‘too small and too expensive. Trends Ecol. Evol. 36, 968–971. doi: 10.1016/j.tree.2021.08.002
McKnight D. T., Huerlimann R., Bower D. S., Schwarzkopf L., Alford R. A., Zenger K. R. (2019). Methods for normalizing microbiome data: An ecological perspective. Methods Ecol. Evol. 10, 389–400. doi: 10.1111/2041-210x.13115
Meesters H. W. G., Boomstra B., Hurtado-Lopez N., Montbrun A., Virdis F. (2015) Coral restoration Bonaire: an evaluation of growth, regeneration and survival. IMARES Report, C152/15, 35. Available online at: https://library.wur.nl/webquery/wurpubs/fulltext/364844.
Moriarty T., Leggat W., Huggett M. J., Ainsworth T. D. (2020). Coral disease causes, consequences, and risk within coral restoration. Trends Microbiol. 28, 793–807. doi: 10.1016/j.tim.2020.06.002
Morikawa M. K., Palumbi S. R. (2019). Using naturally occurring climate resilient corals to construct bleaching-resistant nurseries. Proc. Natl. Acad. Sci. 116, 10586–10591. doi: 10.1073/pnas.1721415116
Nedimyer K., Gaines K., Roach S. (2011). Coral Tree Nursery©: An innovative approach to growing corals in an ocean-based field nursery. Aquac. Aquarium Conserv. Legislation 4, 442–446.
Nie Z., Tang K., Wang W., Wang P., Guo Y., Wang Y., et al. (2023). Comparative genomic insights into habitat adaptation of coral-associated Prosthecochloris. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1138751
Nuñez Lendo C. I., Suggett D. J., Boote C., McArdle A., Nicholson F., Fisher E., et al. (2023). Carbonate budgets induced by coral restoration of a great barrier reef site following cyclone damage. doi: 10.3389/fmars.2023.1298411
Osman E. O., Suggett D. J., Voolstra C. R., Pettay D. T., Clark D. R., Pogoreutz C., et al. (2020). Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome 8, 1–16. doi: 10.1186/s40168-019-0776-5
Parks D. H., ChuvoChina M., Rinke C., Mussig A. J., Chaumeil P. A., Hugenholtz P. (2022). GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, 785–794. doi: 10.1093/nar/gkab776
Patel N. P., Shimpi G. G., Haldar S. (2020). Evaluation of heterotrophic bacteria associated with healthy and bleached corals of Gulf of Kutch, Gujarat, India for siderophore production and their response to climate change factors. Ecol. Indic. 113, 106219. doi: 10.1016/j.ecolind.2020.106219
Pedregosa F., Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O. (2011). sklearn. model_selection. GridSearchCV. Journal of Machine Learning Research.
Peixoto R. S., Rosado P. M., Leite D. C. D. A., Rosado A. S., Bourne D. G. (2017). Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00341
Peixoto R. S., Voolstra C. R., Sweet M., Duarte C. M., Carvalho S., Villela H., et al. (2022). Harnessing the microbiome to prevent global biodiversity loss. Nat. Microbiol. 7, 1726–1735. doi: 10.1038/s41564-022-01173-1
Peng J., Xiao J., Yang Y., Dong Y., Zhang J. (2024). Long-term experimental study and prediction of the mechanical performance on corroded prestressing steel strands subjected to marine salt spray environment. Constr. Build. Mater. 425, 136069. doi: 10.1016/j.conbuildmat.2024.136069
Perkol-Finkel S., Zilman G., Sella I., Miloh T., Benayahu Y. (2008). Floating and fixed artificial habitats: Spatial and temporal patterns of benthic communities in a coral reef environment. Estuar. Coast. Shelf Sci. 77, 491–500. doi: 10.1016/j.ecss.2007.10.005
Procópio L. (2019). The role of biofilms in the corrosion of steel in marine environments. World J. Microbiol. Biotechnol. 35, 73. doi: 10.1007/s11274-019-2647-4
Putnam H. M., Gates R. D. (2015). Preconditioning in the reef-building coral Pocillopora damicornis and the potential for trans-generational acclimatization in coral larvae under future climate change conditions. J. Exp. Biol. 218, 2365–2372. doi: 10.1242/jeb.123018
Rädecker N., Pogoreutz C., Ziegler M., Ashok A., Barreto M. M., Chaidez V., et al. (2017). Assessing the effects of iron enrichment across holobiont compartments reveals reduced microbial nitrogen fixation in the Red Sea coral Pocillopora verrucosa. Ecol. Evol. 7, 6614–6621. doi: 10.1002/ece3.3293
Ray R. I., Lee J. S., Little B. J. (2010). Iron-oxidizing bacteria: a review of corrosion mechanisms in fresh water and marine environments. Corrosion.
Reich H. G., Camp E. F., Roger L. M., Putnam H. M. (2023). The trace metal economy of the coral holobiont: supplies, demands and exchanges. Biol. Rev. 98, 623–642. doi: 10.1111/brv.12922
Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x
Rincon-Rosales R., Lloret L., Ponce E., Martínez-Romero E. (2009). Rhizobia with different symbiotic efficiencies nodulate Acaciella angustissima in Mexico, including Sinorhizobium chiapanecum sp. nov. which has common symbiotic genes with Sinorhizobium mexicanum. FEMS Microbiol. Ecol. 67, 103–117. doi: 10.1111/j.1574-6941.2008.00590.x
Rodriguez I. B., Lin S., Ho J., Ho T. Y. (2016). Effects of trace metal concentrations on the growth of the coral endosymbiont Symbiodinium kawagutii. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00082
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Roper C. D., Camp E. F., Edmondson J., Suggett D. J. (2022). Combined impacts of natural recruitment and active propagation for coral population recovery on the Great Barrier Reef. Mar. Ecol. Prog. Ser. 700, 95–109. doi: 10.3354/meps14184
Rosado P. M., Leite D. C. A., Duarte G. A. S., Chaloub R. M., Jospin G., Nunes da Rocha U., et al. (2018). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME 13, 921–936. doi: 10.1038/s41396-018-0323-6
Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rubio-Portillo E., Martin-Cuadrado A. B., Caraballo-Rodríguez A. M., Rohwer F., Dorrestein P. C., Antón J. (2020). Virulence as a side effect of interspecies interaction in Vibrio coral pathogens. mBio 11, 1–16. doi: 10.1128/mbio.00201-20
Rumbavicius I., Rounge T. B., Rognes T. (2023). HoCoRT: Host contamination removal tool. bioRxiv. doi: 10.1101/2022.11.18.517030
Shaver E. C., McLeod E., Hein M. Y., Palumbi S. R., Quigley K., Vardi T., et al. (2022). A roadmap to integrating resilience into the practice of coral reef restoration. Global Change Biol. 28, 4751–4764. doi: 10.1111/gcb.16212
Strudwick P., Camp E. F., Seymour J., Roper C., Edmondson J., Howlett L., et al. (2024). Assessing efficacy of plastic-free alternative ties for coral propagation in reef restoration. Environ. Microbiol. Rep. 16, e13229. doi: 10.1111/1758-2229.13229
Strudwick P., Seymour J., Camp E. F., Edmondson J., Haydon T., Howlett L., et al. (2022). Impacts of nursery-based propagation and out-planting on coral-associated bacterial communities. Coral Reefs 41, 95–112. doi: 10.1007/S00338-021-02207-6
Strudwick P., Seymour J., Camp E. F., Roper C., Edmondson J., Howlett L., et al. (2023). Bacterial communities associated with corals out-planted on the Great Barrier Reef are inherently dynamic over space and time. Mar. Biol. 170, 1–17. doi: 10.1007/s00227-023-04235-y
Suggett D. J., Edmondson J., Howlett L., Camp E. F. (2020). Coralclip®: a low-cost solution for rapid and targeted out-planting of coral at scale. Restor. Ecol. 28, 289–296. doi: 10.1111/rec.13070
Suggett D. J., Van Oppen M. J. H. (2022). Horizon scan of rapidly advancing coral restoration approaches for 21st century reef management. Emerging Topics Life Sci. 6, 125–136. doi: 10.1042/etls20210240
Tamames J., Puente-Sánchez F. (2019). SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03349
Tandon K., Chiou Y. J., Yu S. P., Hsieh H. J., Lu C. Y., Hsu M. T., et al. (2022). Microbiome restructuring: dominant coral bacterium Endozoicomonas species respond differentially to environmental changes. MSystems 7, e00359-22. doi: 10.1128/msystems.00359-22
van Oppen M. J. H., Blackall L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Voolstra C. R., Perna G., Cárdenas A., Colin L., Dörr M. S., Fiesinger A. (2022). DNA preservation & DNA extraction protocol for field collection of coral samples suitable for host-, marker gene-, and metagenomics-based sequencing approaches (1.2). Zenodo. doi: 10.5281/zenodo.6962735
Voolstra C. R., Suggett D. J., Peixoto R. S., Parkinson J. E., Quigley K. M., Silveira C. B., et al. (2021). Extending the natural adaptive capacity of coral holobionts. Nat. Rev. Earth Environ. 2, 747–762. doi: 10.1038/s43017-021-00214-3
Williams S. L., Sur C., Janetski N., Hollarsmith J. A., Rapi S., Barron L., et al. (2019). Large-scale coral reef rehabilitation after blast fishing in Indonesia. Restor. Ecol. 27, 447–456. doi: 10.1111/rec.12866
Zhang Y., Ling J., Yang Q., Wen C., Yan Q., Sun H., et al. (2015). The functional gene composition and metabolic potential of coral-associated microbial communities. Sci. Rep. 5, 16191. doi: 10.1038/srep16191
Ziegler M., Grupstra C. G. B., Barreto M. M., Eaton M., BaOmar J., Zubier K., et al. (2019). Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-10969-5
Keywords: coral, microbiome, propagation, restoration, bacterial communities
Citation: Strudwick P, Suggett DJ, Seymour JR, DeMaere MZ, Grima A, Edmondson J, McArdle A, Nicholson F and Camp EF (2024) Assessing how metal reef restoration structures shape the functional and taxonomic profile of coral-associated bacterial communities. Front. Mar. Sci. 11:1366971. doi: 10.3389/fmars.2024.1366971
Received: 08 January 2024; Accepted: 08 April 2024;
Published: 26 April 2024.
Edited by:
Elisabeth Marijke Anne Strain, University of Tasmania, AustraliaReviewed by:
Cory J. Krediet, Eckerd College, United StatesCopyright © 2024 Strudwick, Suggett, Seymour, DeMaere, Grima, Edmondson, McArdle, Nicholson and Camp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paige Strudwick, cGFpZ2Uuc3RydWR3aWNrQHV0cy5lZHUuYXU=
†ORCID: Paige Strudwick, orcid.org/0000-0003-3053-5041
David J. Suggett, orcid.org/0000-0001-5326-2520
Justin R. Seymour, orcid.org/0000-0002-3745-6541
Amanda Grima, orcid.org/0000-0003-0230-9266
John Edmondson, orcid.org/0009-0007-0200-4485
Emma F. Camp, orcid.org/0000-0003m-1962-1336
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.