- 1Ministry of Education Key Laboratory of Protein Sciences, School of Life Sciences, Tsinghua University, Beijing, China
- 2Department of Biotechnology and Biomedicine, Yangtze Delta Region Institute of Tsinghua University, Jiaxing, Zhejiang, China
- 3Zhejiang Provincial Key Laboratory of Applied Enzymology, Yangtze Delta Region Institute of Tsinghua University, Jiaxing, China
- 4Taizhou Innovation Center, Yangtze Delta Region Institute of Tsinghua University, Taizhou, Zhejiang, China
Mollusk shells contain biominerals with remarkable mechanical properties enabled by a small fraction of embedded organic matrix proteins. However, the specific molecular functions of most shell proteins have remained elusive. Traditional genomics and functional studies are extremely laborious to identify key components. To address this, we developed an in-silico pipeline integrating protein structure modeling, molecular dynamics simulations, and machine learning to elucidate the critical ion protein interactions governing shell formation. Using the pearl oyster Pinctada fucata as a test case, our framework successfully recapitulated known protein functions and predicted roles of uncharacterized proteins to guide future experiments. Moreover, the pipeline’s modular design enables versatile applications for rapidly elucidating structure-function relationships in diverse biomineralization systems, complementing conventional wet-lab methods. Overall, this computational approach leverages automatic simulations and analytics to unlock molecular insights into shell protein ion dynamics, accelerating the discovery of key crystallization regulators for bioinspired materials design.
Introduction
Biominerals are biogenic composites of inorganic constituents and embedded organic matters. The latter endows biominerals extraordinary mechanical properties despite their low content and therefore is of great interests for scientists to design new materials following similar strategy (Huang et al., 2019; Zhao et al., 2020; Zhou et al., 2022). The organic components in biominerals, also refer to organic matrix, play a pivotal role in mineral deposition such as crystal nucleation, orientation, polymorph selection and morphology modification (Addadi and Weiner, 2014) Understanding how organic matrix precisely affect the crystallization is crucial to bioinspired material synthesis.
Mollusks are masters in producing plentiful biominerals with various microstructures, textures, and shapes. Their functions include protection, feeding, buoyancy, mating and vision, making them a great reservoir for bioinspired materials (Lowenstam and Weiner, 1989). For example, the shiny nacreous layer in some bivalves, gastropods and cephalopods, namely the mother of pearl, have been extensively studied both on the fundamental basis of its fracture-resistance and architectural assembly, and on the biomimetic applications of nacre-like materials (Finnemore et al., 2012; Cartwright, 2016). Despite the large progress in making nacre-like structure of improved mechanical properties, the natural nacre is far beyond completely replicated, especially regarding the stiffness and toughness, which may due to the unique crystallization process in nacre.
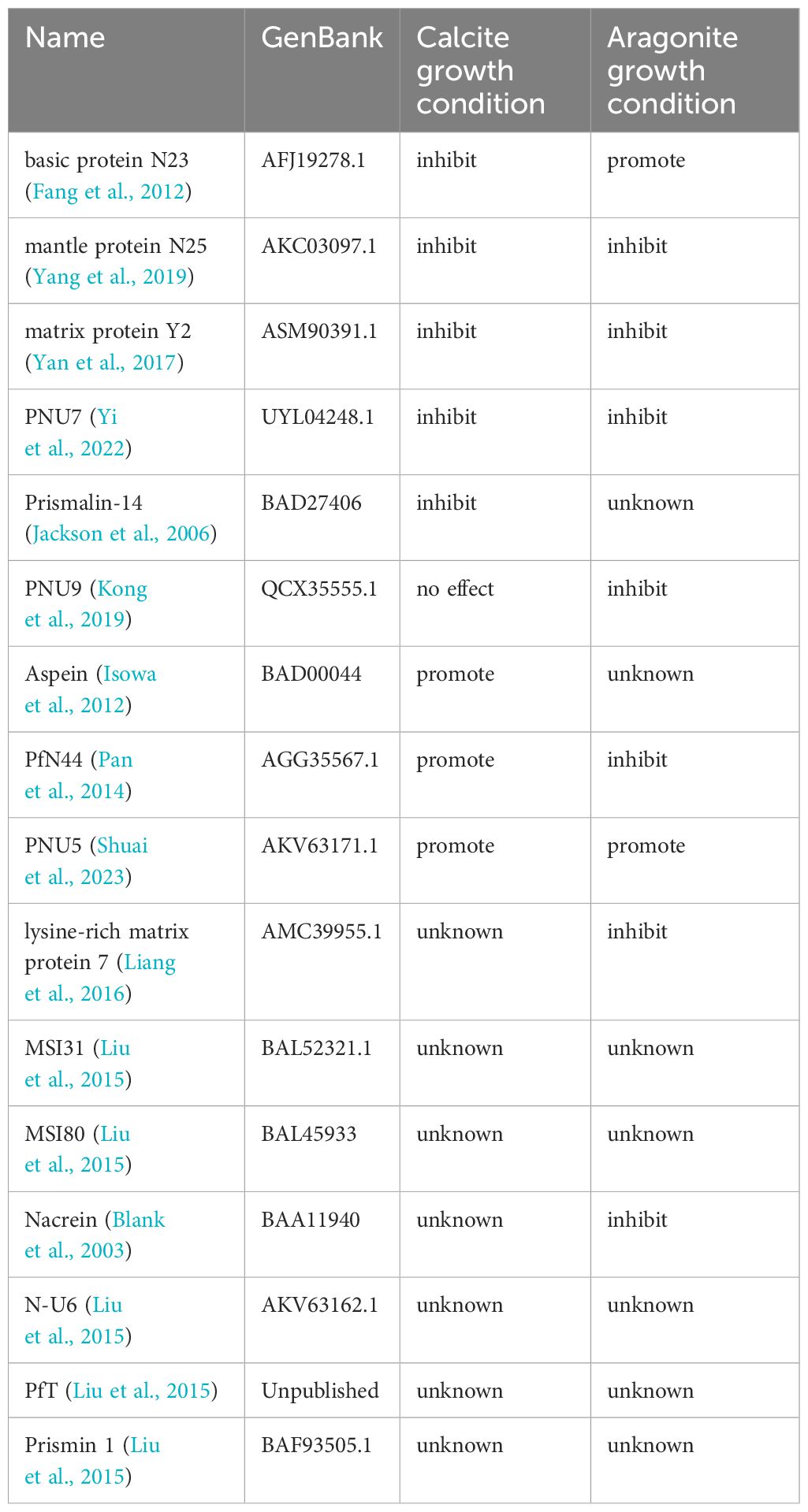
The formation of shell structure in mollusks is regulated by the shell matrix, which contains polysaccharides, proteins, and lipids (Marin et al., 2012). Shell proteins are the key components controlling shell formation, as revealed by intensive studies in the Pinctada genus (Liu et al., 2015; Kintsu et al., 2017; Mariom et al., 2019). In the past three decades, hundreds of shell proteins have been identified by biochemical methods or by omics tools (Liu and Zhang, 2021). Yet their exact roles in controlling CaCO3 crystallization are not clear, except for a few members. This may be ascribed to the time and effort consuming procedures of functional characterization of shell proteins, as well as the relatively limited methods to explore the protein’s function. Basically, the function of a shell protein is revealed by its amino acid sequence and their performance in in vitro CaCO3 crystallization, which may further be supported by the expression of the related gene either under normal situation or under treatments of shell damage or RNAi knock-down. Such methods have been successful in characterizing the potential roles of shell proteins in crystallization or in some particular cases in immunity (Bahn et al., 2017; Yang et al., 2020).
However, the rapid expansion of numbers of shell proteins identified by omics tools, especially proteomics, in the recent years has challenged the traditional characterizing methods, making it a really tough and nearly inaccessible task to reveal the roles of all the identified shell proteins (Liu and Zhang, 2021). Moreover, the traditional characterization methods cannot fully elucidate the molecular mechanism of the crystallization controlling by the shell proteins at molecular or even atomic level. Molecular dynamics is a time-saving method for studying the movement patterns and interactions of proteins at the atomic level, and can be used to explore the intricate structure of protein molecules, the relationships between atoms, and how these relationships affect the physical properties and biological functions of proteins (Eastman et al., 2017). For example, by understanding the dynamic behavior of proteins, scientists can predict and design new drug molecules that can effectively interact with specific proteins, thereby achieving the goal of treating diseases (Sinha et al., 2022). Moreover, molecular dynamics simulations revealed that cement protein MrCP20 from barnacle Megabalanus rosa can sequester free Ca2+ and CO32− ions on its highly charged surface through disorder−order interplay of the protein and ions, and thus regulating calcite deposition in the barnacle base plate (Harini et al., 2019). To fill the gap between the biomineralization genome and the protein functions, here we presented a systematic framework integrating the state-of-the-art protein folding algorithm, molecular dynamics (MD) simulations and machine learning analysis, in attempt to describe the interaction among proteins and the inorganic ions (calcium, bicarbonate, and magnesium).
Methods
Sequence analysis
We firstly retrieved all the protein sequences of Pinctada fucata from Uniprot, with a total of 1116 entries (up to August of 2023). Out of all proteins, we did a literature search and gathered all the cases with experimental results for influence on calcite or aragonite crystallization (see Table 1 and Supplement Material for details). Also, we included some potentially functional proteins from previous in-depth proteomic analysis of shell matrix proteins (Liu et al., 2015) to examine their functions with our protocol. Then we processed the sequences with two approaches: MAFFT (Katoh et al., 2002) method to perform the multiple sequence alignment and obtain the distance matrix; and large protein sequence language model ESM2b (Lin et al., 2023) to process the sequences and obtained the embedding vectors. Then, the distance matrix and the embedding vectors were subject to t-SNE (Van der Maaten and Hinton, 2008) for visualization using Scikit-learn package (Pedregosa et al., 2011).
Structure modeling and molecular dynamics simulations
All protein structures used for MD simulations were predicted using Uni-Fold (Li et al., 2022). To perform large scale MD simulations with various settings, we developed an automated pipeline named ProtIon based on OpenMM (Eastman et al., 2017), and the source code can be found here: (https://github.com/Dongwentao96/ProtIon). The standard protocol is shown in Figure 1: the input structure was firstly examined to repair the missing residues or atoms with PDBfixer (https://github.com/openmm/pdbfixer), while determining the protonation states depending on the pre-defined pH value. This procedure is particularly useful if experimental structures are used, which often contain missing residues or atoms. Then, the protein was put into a box with 1.0 nm buffer padding and the ions of choices were added at the pre-defined concentration [in our case 8mM CaCl2 and 16mM NaHCO3 for calcite and an additional 50mM MgCl2 for aragonite based on our previous study (Yi et al., 2022)], along with counter-ions to maintain the electro-neutrality of the systems. For simulations, the energy minimization was firstly performed, followed by equilibration under NVT ensemble for 100ps and NPT (300K, 1bar) ensemble for 500ps, with heavy atoms fixed using Langevin dynamics (Zhang et al., 2019) and Monte-Carlo barostat (Chow and Ferguson, 1995), Amber FF14SB (Maier et al., 2015) forcefield and TIP3P (Jorgensen et al., 1983) water model. Finally, production simulations were performed under NPT conditions (300K, 1 bar). In our case, 100ns simulations were performed for each case to capture the local distribution of ions. Upon simulation completion, the integrated script can automatically generate the density map of ion distribution throughout the simulation, as well as perform routine trajectory analysis. The package support various solvent or small organic molecules, with GAFF2 (Wang et al., 2004) forcefield by default and parameters from Li et. al. (Li et al., 2015) for ions to automatically do the parameterization. The trajectory was processed with MDTraj (McGibbon et al., 2015) and MDanalysis (Michaud-Agrawal et al., 2011), and all visualization was performed using VMD (Humphrey et al., 1996).
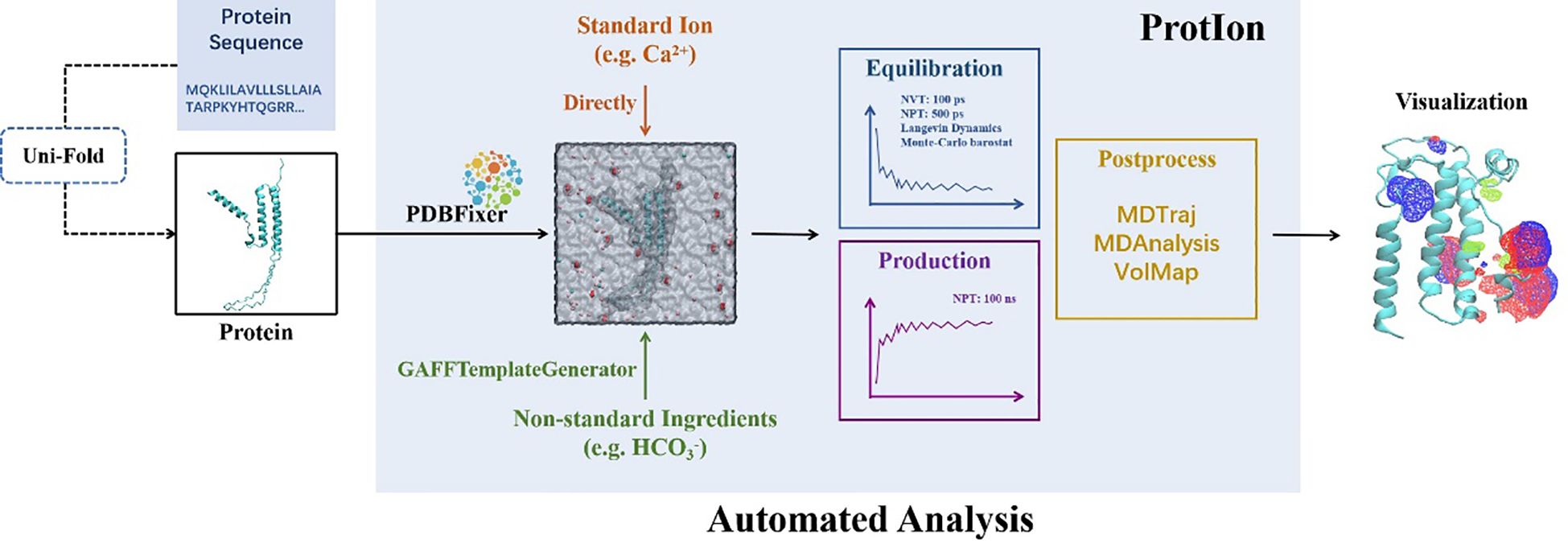
Figure 1 ProtIon overall workflow. The input sequences can be processed with protein folding algorithm like Uni-Fold. Then, the structures were processed by PDBFixer, and the MD systems were built with ions and other molecules added and the whole system solvated. After equilibration and production runs, the trajectories were processed to display the ion density maps and subject to further analysis.
Ion distribution calculation
To analyze the ion distribution over the simulations, we applied a 4 Å cut-off around each amino acid and counted the ion numbers for each snapshot as upper bound considering the van der waals radius of the studying ions. Then the count numbers were divided by the total number of ions then the average counts over all snapshots were calculated for further analysis and visualization. The Volmap module in VMD was used to perform the density calculation.
Machine learning analysis
We used the statistical domain features from the Python package TSFEL (Barandas et al., 2020) to systematically embed the residue-level ion enrichment capability for each system from the molecular dynamics simulation trajectories into feature vectors of the same length, with the domain set to statistical, fs set to 1, window_spliter set to False, overlap set to 0, and the rest as default values. The features include histogram, interquartile range, mean absolute deviation, median absolute deviation, root mean square, standard deviation, variance, ECDF percentile count and ECDF slope. Similar to sequence analysis, we used the t-SNE method with the Python package Scikit-learn for visualization using Matplotlib (Hunter, 2007) and Seaborn (Waskom, 2021) packages.
Results
Proteins usually exert physiological functions through some specific side chains of the amino acid residues. Therefore, elucidating the ion interaction maps of shell proteins is essential to deciphering the molecular mechanisms of shell proteins on biomineralization. Our computational simulations aimed to capture this key biophysical process and predict protein functions from the perspectives of structure, dynamics and ion distributions. However, most shell proteins identified so far in mollusk lack known homologous proteins, because only a few mollusk species have relatively complete draft genomes. At the same time, most of them do not have the three-dimensional structure analyzed by experiment. Recent advancement in protein folding algorithm paved the way for structural and dynamic studies towards our goal, since most matrix proteins are poorly characterized, and with no resolved experimental structures. Among several state-of-the-art folding algorithms (Baek et al., 2021; Li et al., 2022; Lin et al., 2023), we used Uni-Fold (Li et al., 2022) to process our sequences on the online platform Hermite (https://hermite.dp.tech). With the predicted structures we employed our ProtIon pipeline to perform MD simulations in calcite and aragonite crystallization solutions with corresponding ion concentrations (see Methods). Usually, the folded structures contain uncertainties, especially in the loop regions, but such deviation can be restored within the simulation process governed by the physical forcefield (Wang et al., 2022). Indeed, we observed the RMSD may change significantly but reached plateau within 100ns simulation time in almost all systems (Supplementary Figure 1 and 2). Subsequent analysis of the trajectories revealed distinct ion binding capabilities correlated with promotion or inhibition of crystal growth.
The results show that proteins known to promote calcite or aragonite growth exhibited localized enrichments of both Ca2+ and HCO3- ions in certain regions (Figure 2). In contrast, proteins inhibiting mineralization showed accumulation of single ion types or spatially distant enrichment regions. Although other factors may also affect the specific functions such as the interactions between the proteins and the substrate or the geometric shape or size of the proteins. While most proteins of interests contain rich charged, especially acidic amino acids in the sequences, it is only through the 3D structures, combined with the MD simulations that can we examine whether the protein can gather both ions (Ca2+ and HCO3-) to the same regions.
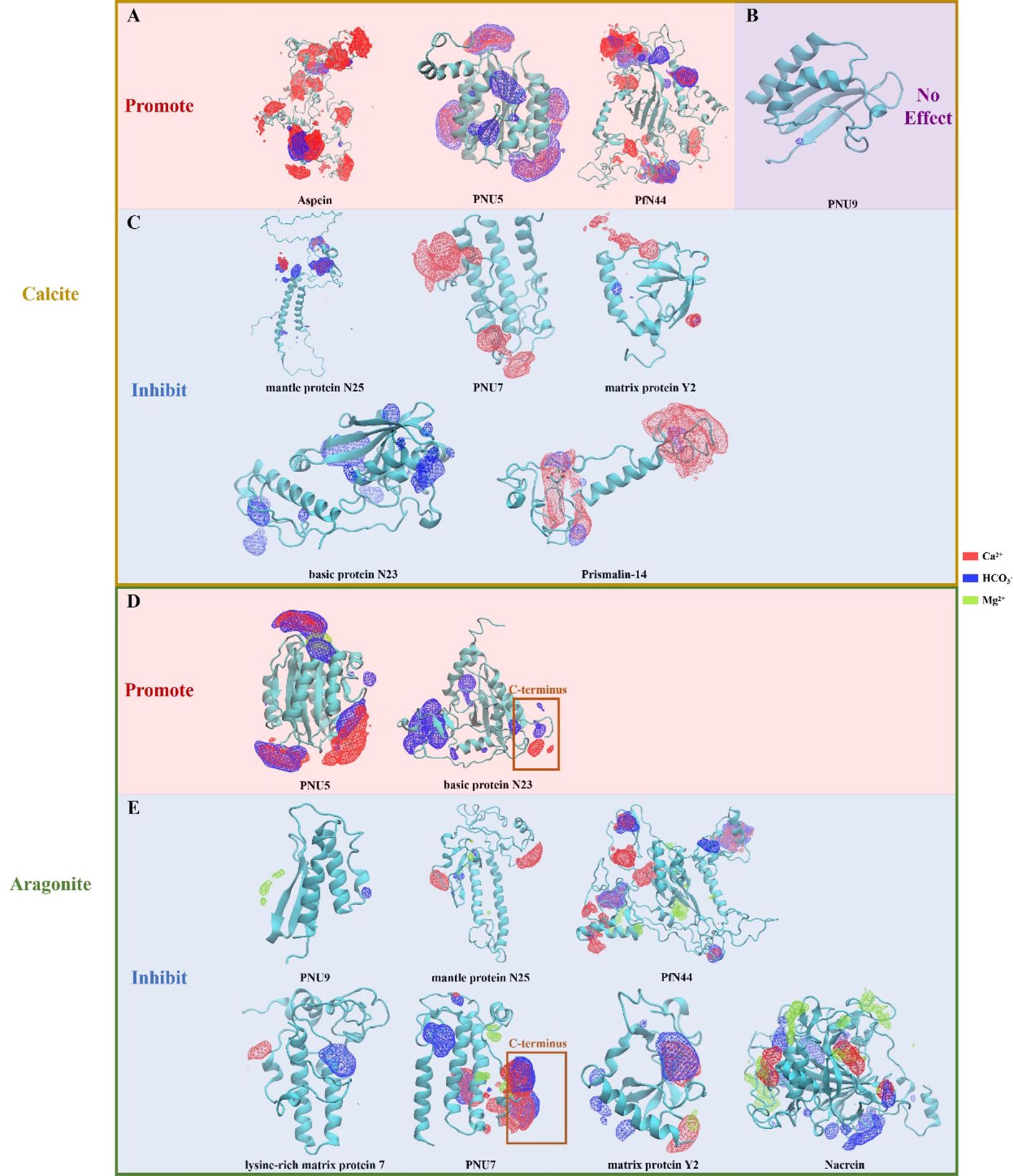
Figure 2 Ionic density volmaps captured by molecular dynamics simulation. (A) Proteins promote growth of calcite. (B) Protein PNU 9, which has no effects on growth of calcite. (C) Proteins inhibit growth of calcite. (D) Proteins promote growth of aragonite. (E) Proteins inhibit growth of aragonite. For N23 and PNU7, the C-terminal domains were highlighted due to their distinct and important role in aragonite formation as mentioned in the main text.
MD simulations explains past functional studies
In the past few decades, the impact of some matrix proteins on CaCO3 precipitation have been characterized (see Table 1). Then, we examined these matrix proteins in the table by molecular dynamics model. It should be noted that, Aspein, PNU5, and PfN44 all displayed concurrent accumulations of large amounts of Ca2+ and HCO3- locally (Figure 2A), agreeing with their reported promoting role in the calcite growth (Isowa et al., 2012; Pan et al., 2014; Shuai et al., 2023). Moreover, PNU5 showed co-enrichment of Ca2+, Mg2+, and HCO3- (Figure 2D), consistent with its reported aragonite crystallization promotion (Shuai et al., 2023). It is particularly interesting to see that when Mg2+ ion was present, the binding affinity of PfN44 protein in the system altered significantly, due to the Mg2+ ions binding to the central beta sheet and thus disrupting the overall scaffold (Figure 2E). This phenomenon is in alignment with previous study reporting that PfN44 can stabilize magnesium-calcite to inhibit the crystallization of aragonite, explaining its opposite functions in calcite and aragonite formation (Pan et al., 2014).
PfN23 is a basic SMP identified from the P. fucata shell, which can specifically induce the crystallization of aragonite, and its positively charged C-terminal is supposed to be the key functional region (Fang et al., 2012). Coincide with this, we found that the C-terminus of basic protein N23 selectively accumulated Ca2+ and HCO3- only in the aragonite solution (Figure 2C). Compared to its aragonite conformation, N23 in the calcite solution without Mg2+ exhibited larger structural changes and lost the C-terminal Ca2+ enrichment, and instead accumulate HCO3- in more regions. This change in anion binding might be involved in the previous reported promotion of calcite dissolution (Fang et al., 2012).
Similarly, mantle protein N25 also displayed localized Ca2+ and HCO3- enrichments, but with lower overall levels and more directional focus compared to the abovementioned proteins when subjected to calcite crystallization solution (Figure 2C). Previous evidence indicates that N25 can block growing sites for forming crystal layer, thus increasing the energy cost for deposition and decreasing growth rates of some crystal faces of CaCO3 (Yang et al., 2019). Therefore, the observed ion binding regions of N25 may be involved in interaction with the crystal layer. Additionally, N25 and lysine-rich matrix protein 7 showed localized single ion enrichments in the aragonite solution, consistent with their reported inhibition of aragonite crystal growth (Liang et al., 2016).
PNU9 showed no significant ion accumulation in calcite crystallization solution (Figure 2B), agreeing with the functional study reported previously (Kong et al., 2019). In the Mg2+-containing aragonite solution, PNU9 exhibited only minor, dispersed enrichments of Mg2+ and HCO3-. Similarly, matrix protein PfY2 displayed local accumulation of single ion types in calcite. Such discrete single ion binding likely underlies their inhibiting effect of crystal growth (Yan et al., 2017).
Likewise, nacrein in the aragonite forming condition showed the ability to locally enrich single ions in multiple separated regions (Figure 2E), consistent with its reported inhibition of aragonite crystal growth (Blank et al., 2003). Prismalin-14 was expressed only at the mantle edge and primarily present in the prismatic layer composed of columnar calcite surrounded by organic matrices (Suzuki et al., 2004). Our results show that Prismalin-14 has two minor HCO3- enriching regions sharing a planar Ca2+ site (Figure 2C). Another distinct region with high single Ca2+ accumulation may modulate the inhibition of calcium carbonate crystallization.
In the calcite solution, PNU7 only showed Ca2+ enrichment (Figure 2C), and is predicted to play an inhibition role. However, in the aragonite simulation, PNU7’s C-terminus exhibited two sites with high Ca2+/HCO3- accumulation and minor surrounding Mg2+ enrichment (Figure 2E). It also displayed a region near the N-terminus with high HCO3- accumulation. In vitro crystallization experiment showed that PNU7 can modify calcite morphology and stabilize large vaterite particles when Mg2+ is absent, while at lower Mg2+ concentration, large amounts of tiny crystals were formed (Yi et al., 2022). When the C-terminus of PNU7 was deleted, the growth of calcite and vaterite were inhibited. Our observed binding patterns of this protein agree with these reported functions.
In the aragonite solution, matrix protein Y2 exhibited localized co-enrichment of Ca2+ and HCO3-, along with minor co-enrichment of Ca2+ and Mg2+ (Figure 2E). Purified recombinant rPfY2 protein has been found to significantly suppressed CaCO3 precipitation rate and participated in the crystal nucleation process (Yan et al., 2017). Additionally, the morphology of crystals was modified, and the transformation of amorphous calcium carbonate (ACC) to calcite or aragonite was inhibited. We hypothesize the functional domains enriching Ca2+ and HCO3- can regulate CaCO3 precipitation rate, representing a more specialized form of growth inhibition.
PCA analysis on MD simulation data well separated proteins of different functions
Comparing with previous studies on the shell matrix proteins, we found that proteins with different functions exhibit highly correlated results in molecular dynamics simulations, suggesting that molecular dynamics models can be used to deduce the potential functions of some uncharacterized shell proteins and to predict their possible effects on the growth of calcite or aragonite. Thus, we recorded the frequency of ion occurrences within proximity to each residue during the calcite and aragonite crystallization simulations as a characterization of the ion enrichment capability for each residue (Supplementary Figures 3 and 4). We then used TSFEL to extract features as protein representations (See Methods). Additionally, we selected traditional distance matrices based on sequence alignment and embeddings from pre-trained protein language models as alternative protein representations.
Subsequently, we utilized t-SNE for unsupervised dimensionality reduction of the protein representations from the three methods. As seen in Figures 3A, D, the matrix proteins were grouped into two groups of promoting crystallization/inhibiting crystallization by using the multi-sequence comparison distance matrix as a parameter, and there was a certain degree of differentiation between the two types of proteins under aragonite crystallization condition, but it was not ideal in the calcite environment. When embedding vectors are used as parameters, the resulting groupings have a large overlap, both in the calcite and aragonite growth conditions (Figures 3B, E). When molecular dynamics simulations of locus ion enrichment were used to describe the function of proteins, the two protein groups were well separated, with very little overlap between proteins with promoting or inhibiting crystallization properties (Figures 3C, F). Therefore, the ion enrichment capability representations have a natural advantage for depicting proteins affecting the growth of calcite versus aragonite.
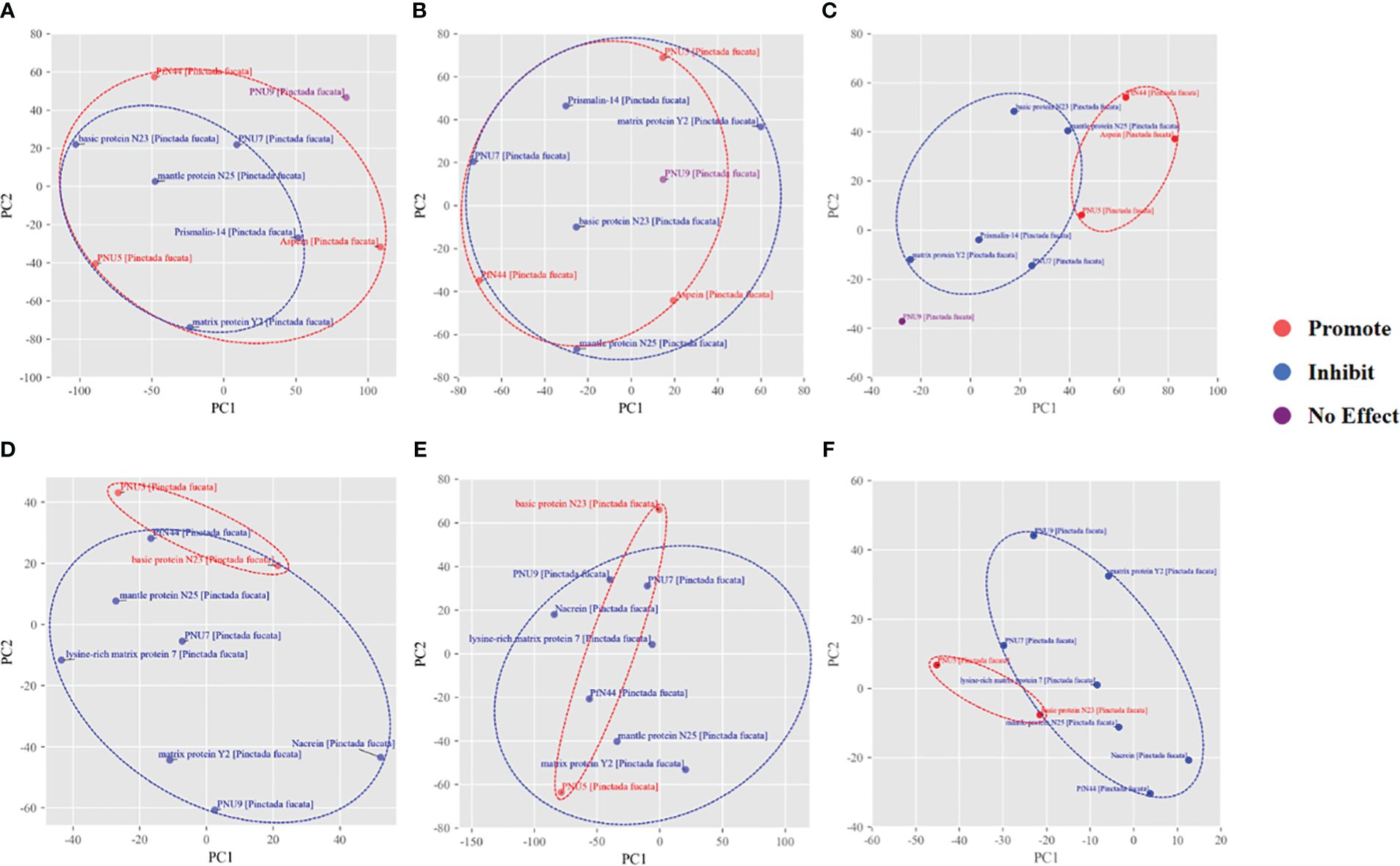
Figure 3 t-SNE results of mineralization related proteins. The t-SNE dimensional reduction applied to (A, D) multisequence comparison distance matrix (B, E) esm2_t30_150M_UR50D embedding vectors (C, F) the molecular dynamics simulation trajectory extracted feature vectors, for calcite/aragonite respectively.
ProtIon combined with machine learning to explore biomineralization proteins
Based on the molecular dynamics simulation features, we employed SVM (Figure 4) to build a model to classify the known proteins and applied to other proteins potentially related to biomineralization that were previously reported (Liu et al., 2015). Interestingly, PNU7, N23 and PfN44 are very close to the hyperplane, and these three have subtle and contradictory roles in calcite and aragonite, with both Ca2+ and HCO3- bind to a specific domain and/or greatly affected by the presence of Mg2+.
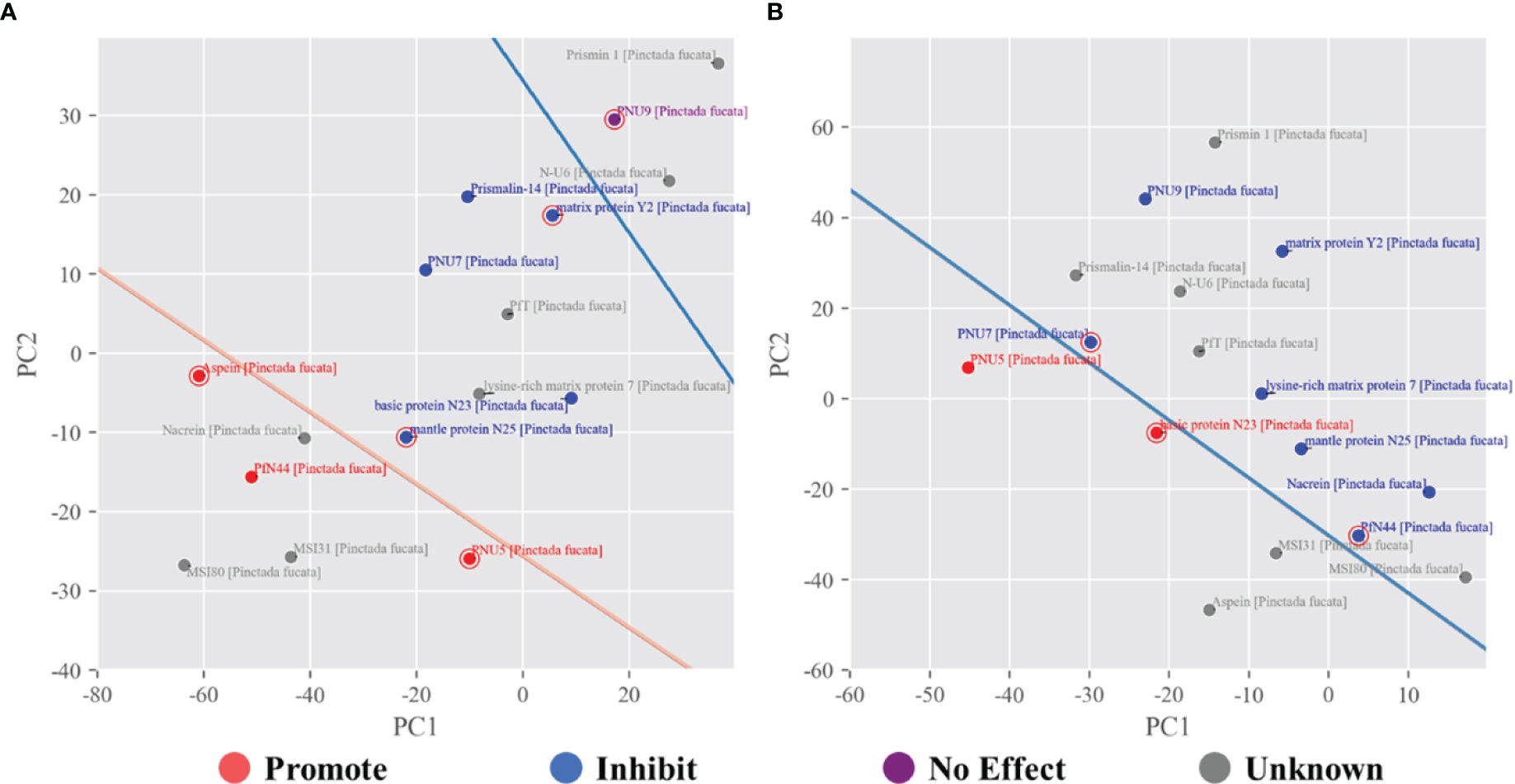
Figure 4 SVM classification of shell proteins under calcite (A) and aragonite (B) growth conditions. For calcite the promote, inhibit and no effect proteins were separated by two hyperplanes shown as two lines. For aragonite, only the promote and inhibit proteins were separated by the hyperplanes shown as a line, since no ‘no effect’ proteins were reported. In both cases, other unknown proteins were projected to predict their potential role.
According to the SVM predictions (Figure 4) and ionic density maps, we speculated that MSI31, MSI80 and Nacrein (Figure 5A and Supplementary Table 1) may promote calcite growth. They exhibited multiple regions of co-enrichment of Ca2+ and HCO3- ions and relatively large size, which are consistent with the aforementioned cases of Aspein, PNU5 and PfN44 under calcite growth condition (Figure 2). As to Lysine-rich matrix protein 7 (KRMP7), it only showed enrichment of HCO3- ions and is likely to inhibit the growth of calcite. PfT has three small regions of co-enrichment of Ca2+ and HCO3- (Figure 5B), as well as a significant Ca2+ ion enrichment region, which is relatively similar to the simulation result of Prismalin-14, indicating potential functions of changing crystal morphology and inhibiting calcite growth.

Figure 5 Ion density maps of unknown potential proteins (A) the potentially promoting proteins MSI31, Nacrein and MSI80 for calcite formation; (B) to possibly inhibiting proteins lysine-rich matrix protein 7 and PfT for calcite formation; (C) the potentially promoting proteins Aspein and MSI31 for aragonite formation and (D) the potentially inhibiting proteins MSI80 and PfT for aragonite formation.
In terms of the impact on aragonite crystals, Aspein and MSI31 may promote aragonite growth, as predicted by the SVM model. As shown in Figure 5D, they both have regions of co-enrichment of Ca2+, HCO3- and Mg2+, similar to the simulation results of PNU5 and N23. Meanwhile, MSI80 and PfT do not have significant large co-enrichment regions, also consistent with the prediction.
However, it should be noted that the SVM model may have limited predictive potential for some proteins. For example, there are weak cases like N-U6 and Prismin-1 (see Supplementary Figure 5 and Supplementary Table 1) under calcite growth condition, both having some ion enrichment regions but were predicted to have no effect on calcite crystal growth, probably due to the relatively small protein size that prevent the stable binding to the substrate. Also, when under aragonite growth condition, N-U6, Prismalin-14 and Prismin-1 all have some regions of co-enrichment of Ca2+ and HCO3-, but no Mg2+ was accumulated around these regions. Their predicted inhibition on aragonite growth may be ascribed to the relatively small size and ion enrichment region, or due to the attachment to the newly formed crystal layer, or reduce the precipitation rate of CaCO3 as reported in previous studies (Yang et al., 2019; Yi et al., 2022).
Discussion
Mollusk shell formation is a complex biomineralization process precisely controlled by the organism to generate intricate composite materials with remarkable mechanical properties. The shell matrix comprises proteins, polysaccharides, and other macromolecules that regulate mineral deposition and crystal growth at multiple levels (Marin et al., 2007a, 2007, 2012). At the heart of this process is the interaction between the organic matrix and the inorganic ions that make up the mineral phase. The availability and transport of ions, particularly calcium, carbonate, and magnesium, are critical determinants of shell construction (Cartwright and Checa, 2007). Calcium and bicarbonate ions are taken up from seawater by the mantle epithelium and transported to the calcification site via the extrapallial fluid (Marin et al., 2012). The organic matrix creates localized ion-rich environments to stimulate crystal nucleation. Matrix proteins selectively bind ions via acidic residues, acting as scaffolds for oriented crystal growth (Falini et al., 1996). Specific macromolecular conformations and ion binding motifs direct the polymorph selection between calcite and aragonite.
The precise spatial and temporal regulation of ion concentrations by the organic matrix is key to controlling crystallization kinetics and directing the assembly of composite shell microstructures (Nudelman and Sommerdijk, 2012). Tracking the dynamics of ion accumulation reveals mechanistic insights into how proteins influence crystal nucleation, orientation, phase, morphology and material properties. As Marin et al. discussed, the protein-ion interactions that direct crystallization occur at multiple length scales, from the nanoscale ion binding sites within individual proteins to the larger-scale accumulation patterns shaped by the 3D matrix scaffold (Marin et al., 2007b).
Although the full mechanism of biomineral formation is complicated (Yang et al., 2020), involving enzymatic processes, spatial patterning, and controlled ion accumulation, the general picture is that matrix proteins attach to a chitin-silk fibroin substrate and then concentrate Ca2+ and HCO3- from the environment to form shell layers. This requires the proteins to stably bind both ions while presenting a large enough surface for crystal nucleation. If the protein can only gather anion or cation, it will drive the crystallization kinetics towards dissociation. The results in the present study indicate that the interactions between the side chain of the shell proteins and the inorganic ions are related to the regulatory role of the proteins. Magnesium can directly affect the precipitation of calcite and aragonite (Morse et al., 1997). Our study showed that some shell proteins, especially those from aragonitic nacre layers, interacted with Mg2+ differently, which may explain their control of crystal polymorph selection during shell formation. Previous studies have shown that proteins rich in aspartic acid are thought to bind calcium ions, and proteins containing the EF-hand domain can also bind calcium ions (Hattan et al., 2001; Gotliv et al., 2005; Takeuchi et al., 2008). The binding of calcium or magnesium ions in turn changes the structure of the matrix protein. In addition, Lia Addadi and Steve Weiner’s group have shown that the regular arrangement of amino acid residues in the tandem repeats containing asparagine is well matched to the crystal lattice of calcium carbonate with a specific crystal polymorph, thus determining the CaCO3 polymorph (Addadi and Weiner, 1985). Consistently, tandem repeats containing DDRK can significantly affect the deposition of calcium carbonate (Tah et al., 2024).
The automated simulation and analysis tool ProtIon based on Python package OpenMM to study protein-ion interactions and performed consistency validation on some important proteins in the field of biomineralization, proving a high consistency between simulation results and experimental results. Based on the simulation analysis results, we can identify residues in proteins that have a greater impact on mineralization. The rich structural information provided by our method can be further used for understanding the detailed mechanism for shell formation protein design and domain fusion to incubate better varieties, as well as protein design and domain fusion to incubate better varieties.
The machine learning model we here developed to predict the function of matrix proteins is only theoretically tested, which warrant more experimental evidence in the future to support it. For example, the ion enrichment of specific amino acid sequence can be analyzed by truncating the protein to obtain mutant. Alternatively, we can also directly express the domain with ion binding affinity to verify its role. If the validity of our model can be verified experimentally, its application can be expanded. In the present study, we only use one shell protein in a single stimulation, but under natural biomineralization conditions, many proteins interact and regulate CaCO3 precipitation synergistically. Nevertheless, many shell proteins have disordered regions which usually exhibit as loops and coiled coils. Previous studied have shown that these disordered regions are vital in shell mineralization (Ndao et al., 2010; Brown et al., 2014), which is further supported by the results present here revealing the interaction of the disordered regions and the inorganic ions. However, we excluded some shell proteins with large proportion of disordered regions and have no 3D structures in this study (e.g. MSI60 and shematrin family) due to technical difficulties in the subsequent MD simulation. Because our machine learning model depends on the 3D structure of the target proteins, this shortcoming of our method limits its application to all shell proteins. Moreover, the training dataset of our model was relatively limited due to the fact that only a small part of the identified molluscan shell proteins has been characterized via in vitro crystallization experiment. Excluding shell protein other than P. fucata shell proteins also led to the limited training data. Therefore, more efforts are warranted to establish a routine method that is applicable to most shell protein from various molluscan genera.
Conclusion
On the top of the proteomic studies (Connors et al., 2012; Liu et al., 2015; Du et al., 2017), our framework can perform simulations under various conditions and reveal the ensemble average ion distributions over the protein surfaces, which in turn can be used to determine the particular role of the protein in shell formation. In particular, we tested some well-studied shell proteins and a few potentially functional proteins, for the cases of calcite and aragonite formation. Our simulations successfully captured the ion accumulation and correlate well with the experiments. In addition, we build up a machine learning model to further identify potential proteins with certain promoting or inhibiting mineral formation functions. Moreover, our molecular dynamics simulation module was based on OpenMM (Eastman et al., 2017), an open source and versatile simulation framework, allowing for convenient adjustment according to specific case.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
WD: Writing – original draft, Writing – review & editing. LX: Funding acquisition, Supervision, Writing – review & editing. RZ: Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China Grants 32072951.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1362131/full#supplementary-material
References
Addadi L., Weiner S. (1985). Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc. Natl. Acad. Sci. U.S.A. 82, 4110–4114.
Addadi L., Weiner S. (2014). Biomineralization: mineral formation by organisms. Physica. Scripta. 89, 098003. doi: 10.1088/0031-8949/89/9/098003
Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. doi: 10.1126/science.abj8754
Bahn S. Y., Jo B. H., Choi Y. S., Cha H. J. (2017). Control of nacre biomineralization by Pif80 in pearl oyster. Sci. Adv. 3. doi: 10.1126/sciadv.1700765
Barandas M., Folgado D., Fernandes L., Santos S., Abreu M., Bota P., et al. (2020). TSFEL: time series feature extraction library. SoftwareX 11, 100456. doi: 10.1016/j.softx.2020.100456
Blank S., Arnoldi M., Khoshnavaz S., Treccani L., Kuntz M., Mann K., et al. (2003). The nacre protein perlucin nucleates growth of calcium carbonate crystals. J. Microsc. 212, 280–291. doi: 10.1111/j.1365-2818.2003.01263.x
Brown A. H., Rodger P. M., Evans J. S., Walsh T. R. (2014). Equilibrium conformational ensemble of the intrinsically disordered peptide n16n: linking subdomain structures and function in nacre. Biomacromolecules 15, 4467–4479.
Cartwright J. H. E. (2016). Directed self-assembly, genomic assembly complexity and the formation of biological structure, or, what are the genes for nacre? Philos. Trans. R. Soc. A.: Mathematical. Phys. Eng. Sci. 374, 20150449.
Cartwright J. H. E., Checa A. G. (2007). The dynamics of nacre self-assembly. J. R. Soc. Interface 4, 491–504. doi: 10.1098/rsif.2006.0188
Chow K.-H., Ferguson D. M. (1995). Isothermal-isobaric molecular dynamics simulations with Monte Carlo volume sampling. Comput. Phys. Commun. 91, 283–289. doi: 10.1016/0010-4655(95)00059-O
Connors M. J., Ehrlich H., Hog M., Godeffroy C., Araya S., Kallai I., et al. (2012). Three-dimensional structure of the shell plate assembly of the chiton Tonicella marmorea and its biomechanical consequences. J. Struc. Biol. 177, 314–328.
Du X., Fan G., Jiao Y., Zhang H., Guo X., Huang R., et al. (2017). The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. GigaScience 6.
Eastman P., Swails J., Chodera J. D., McGibbon R. T., Zhao Y., Beauchamp K. A., et al. (2017). OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PloS Comput. Biol. 13, e1005659. doi: 10.1371/journal.pcbi.1005659
Falini G., Albeck S., Weiner S., Addadi L. (1996). Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271, 67–69. doi: 10.1126/science.271.5245.67
Fang D., Pan C., Lin H., Lin Y., Zhang G., Wang H., et al. (2012). Novel basic protein, PfN23, functions as key macromolecule during nacre formation. J. Biol. Chem. 287, 15776–15785. doi: 10.1074/jbc.M112.341594
Finnemore A., Cunha P., Shean T., Vignolini S., Guldin S., Oyen M., et al. (2012). Biomimetic layer-by-layer assembly of artificial nacre. Nat. Commun. 3, 966. doi: 10.1038/ncomms1970
Gotliv B., Kessler N., Sumerel J. L., Morse D. E., Tuross N., Addadi L., et al. (2005). Asprich: A Novel Aspartic Acid-Rich Protein Family from the Prismatic Shell Matrix of the Bivalve Atrina rigida. ChemBioChem. 6, 304–314.
Harini M., Akshita K., Chandra S. V., Konstantin P., Ali M. (2019). Three-dimensional structure of Megabalanus rosa Cement Protein 20 revealed by multi-dimensional NMR and molecular dynamics simulations. Philosoph. Transact. R. Soc B: Biol. Sci., 374.
Hattan S. J., Laue T. M., Chasteen N. D. (2001). Purification and Characterization of a Novel Calcium-binding Protein from the Extrapallial Fluid of the Mollusc, Mytilus edulis. J. Biol. Chem. 276, 4461–4468.
Huang W., Restrepo D., Jung J. Y., Su F. Y., Liu Z. Q., Ritchie R. O., et al. (2019). Multiscale toughening mechanisms in biological materials and bioinspired designs. Adv. Mater. 31. doi: 10.1002/adma.201901561
Humphrey W., Dalke A., Schulten K. (1996). VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Hunter J. D. (2007). Matplotlib: A 2D graphics environment. Computing. Sci. Eng. 9, 90–95. doi: 10.1109/MCSE.2007.55
Isowa Y., Sarashina I., Setiamarga D. H., Endo K. (2012). A comparative study of the shell matrix protein aspein in pterioid bivalves. J. Mol. Evol. 75, 11–18. doi: 10.1007/s00239-012-9514-3
Jackson D. J., McDougall C., Green K., Simpson F., Wörheide G., Degnan B. M. (2006). A rapidly evolving secretome builds and patterns a sea shell. BMC Biol. 4, 40. doi: 10.1186/1741-7007-4-40
Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. doi: 10.1063/1.445869
Katoh K., Misawa K., K.i. and Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kintsu H., Okumura T., Negishi L., Ifuku S., Kogure T., Sakuda S., et al. (2017). Crystal defects induced by chitin and chitinolytic enzymes in the prismatic layer of Pinctada fucata. Biochem. Biophys. Res. Commun. 489, 89–95. doi: 10.1016/j.bbrc.2017.05.088
Kong J., Liu C., Yang D., Yan Y., Chen Y., Liu Y., et al. (2019). A novel basic matrix protein of Pinctada fucata, PNU9, functions as inhibitor during crystallization of aragonite. CrystEngComm 21, 1250–1261. doi: 10.1039/C8CE02194E
Li P., Song L. F., Merz K. M. Jr. (2015). Systematic parameterization of monovalent ions employing the nonbonded model. J. Chem. Theory Comput. 11, 1645–1657. doi: 10.1021/ct500918t
Li Z., Liu X., Chen W., Shen F., Bi H., Ke G., et al. (2022). Uni-Fold: an open-source platform for developing protein folding models beyond AlphaFold. bioRxiv. 2022.2008. 2004.502811. doi: 10.1101/2022.08.04.502811
Liang J., Xie J., Gao J., Xu C. Q., Yan Y., Jia G. C., et al. (2016). Identification and characterization of the lysine-rich matrix protein family in pinctada fucata: indicative of roles in shell formation. Mar. Biotechnol. (NY). 18, 645–658. doi: 10.1007/s10126-016-9724-6
Lin Z., Akin H., Rao R., Hie B., Zhu Z., Lu W., et al. (2023). Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130. doi: 10.1126/science.ade2574
Liu C., Li S. G., Kong J. J., Liu Y. J., Wang T. P., Xie L. P., et al. (2015). In-depth proteomic analysis of shell matrix proteins of Pinctada fucata. Sci. Rep-Uk. 5. doi: 10.1038/srep17269
Liu C., Zhang R. (2021). Biomineral proteomics: A tool for multiple disciplinary studies. J. Proteomics 238, 104171. doi: 10.1016/j.jprot.2021.104171
Lowenstam H. A., Weiner S. (1989). On biomineralization (New York and London: Oxford University Press). doi: 10.1093/oso/9780195049770.001.0001
Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C. (2015). ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. doi: 10.1021/acs.jctc.5b00255
Marin F., Luquet G., Marie B., Medakovic D. (2007a). Molluscan shell proteins: primary structure, origin, and evolution, current topics in developmental biology. Acad. Press. pp, 209–276. doi: 10.1016/S0070-2153(07)80006-8
Marin F., Pokroy B., Luquet G., Layrolle P., De Groot K. (2007b). Protein mapping of calcium carbonate biominerals by immunogold. Biomaterials 28, 2368–2377. doi: 10.1016/j.biomaterials.2007.01.029
Marin F., Roy N. L., Marie B. (2012). The formation and mineralization of mollusk shell. Front. Biosci. (Schol. Ed). 4, 1099–1125. doi: 10.2741/s321
Mariom, Take S., Igarashi Y., Yoshitake K., Asakawa S., Maeyama K., et al. (2019). Gene expression profiles at different stages for formation of pearl sac and pearl in the pearl oyster Pinctada fucata. BMC Genomics 20, 240. doi: 10.1186/s12864-019-5579-3
McGibbon R. T., Beauchamp K. A., Harrigan M. P., Klein C., Swails J. M., Hernández C. X., et al. (2015). MDTraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 109, 1528–1532. doi: 10.1016/j.bpj.2015.08.015
Michaud-Agrawal N., Denning E. J., Woolf T. B., Beckstein O. (2011). MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327. doi: 10.1002/jcc.21787
Morse J. W., Wang Q., Tsio M. Y. (1997). Influences of temperature and Mg: Ca ratio on CaCO3 precipitates from seawater. Geology 25, 85–87. doi: 10.1130/0091-7613(1997)025<0085:IOTAMC>2.3.CO;2
Ndao M., Keene E., Amos F. F., Rewari G., Ponce C. B., Estroff L., et al. (2010). Intrinsically disordered mollusk shell prismatic protein that modulates calcium carbonate crystal growth. Biomacromolecules 11, 2539–2544.
Nudelman F., Sommerdijk N. A. (2012). Biomineralization as an inspiration for materials chemistry. Angew. Chem. Int. Ed. Engl. 51, 6582–6596. doi: 10.1002/anie.201106715
Pan C., Fang D., Xu G., Liang J., Zhang G., Wang H., et al. (2014). A novel acidic matrix protein, PfN44, stabilizes magnesium calcite to inhibit the crystallization of aragonite. J. Biol. Chem. 289, 2776–2787. doi: 10.1074/jbc.M113.504027
Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. (2011). Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830. doi: 10.5555/1953048.2078195
Shuai B., Deng T., Xie L., Zhang R. (2023). A novel matrix protein PNU5 facilitates the transformation from amorphous calcium carbonate to calcite and aragonite. Int. J. Biol. Macromol. 224, 754–765. doi: 10.1016/j.ijbiomac.2022.10.163
Sinha S., Tam B., Wang S. M. (2022). Applications of molecular dynamics simulation in protein study. Membranes, 12.
Suzuki M., Murayama E., Inoue H., Ozaki N., Tohse H., Kogure T., et al. (2004). Characterization of Prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster ( Pinctada fucata ). Biochem. J. 382, 205–213.
Tah B., Upcher A., Berman A., Goldstein-Goren S. (2024). A simple periodic peptide derived from Pinctada fucata Pif80 protein induces aragonite nucleation in magnesium absence. ChemRxiv.
Takeuchi T., Sarashina I., Iijima M., Endo K. (2008). In vitro regulation of CaCO 3 crystal polymorphism by the highly acidic molluscan shell protein Aspein. FEBS Lett. 582, 591–596.
Wang D., Wang Y., Chang J., Zhang L., Wang H., Weinan E. (2022). Efficient sampling of high-dimensional free energy landscapes using adaptive reinforced dynamics. Nat. Comput. Sci. 2, 20–29. doi: 10.1038/s43588-021-00173-1
Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A. (2004). Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174. doi: 10.1002/jcc.20035
Waskom M. L. (2021). Seaborn: statistical data visualization. J. Open Source Software. 6, 3021. doi: 10.21105/joss.03021
Yan Y., Yang D., Yang X., Liu C., Xie J., Zheng G., et al. (2017). A novel matrix protein, pfY2, functions as a crucial macromolecule during shell formation. Sci. Rep. 7, 6021. doi: 10.1038/s41598-017-06375-w
Yang D., Yan Y., Yang X., Liu J., Zheng G., Xie L., et al. (2019). A basic protein, N25, from a mollusk modifies calcium carbonate morphology and shell biomineralization. J. Biol. Chem. 294, 8371–8383. doi: 10.1074/jbc.RA118.007338
Yang X., Yang D., Yan Y., Li S. G., Han Z. M., Ji Y. H., et al. (2020). A novel matrix protein PfX regulates shell ultrastructure by binding to specific calcium carbonate crystal faces. Int. J. Biol. Macromol. 156, 302–313. doi: 10.1016/j.ijbiomac.2020.04.016
Yi L., Zou B., Xie L., Zhang R. (2022). A novel bifunctional protein PNU7 in CaCO(3) polymorph formation: Vaterite stabilization and surface energy minimization. Int. J. Biol. Macromol. 222, 2796–2807. doi: 10.1016/j.ijbiomac.2022.10.059
Zhang Z., Liu X., Yan K., Tuckerman M. E., Liu J. (2019). Unified efficient thermostat scheme for the canonical ensemble with holonomic or isokinetic constraints via molecular dynamics. J. Phys. Chem. A. 123, 6056–6079. doi: 10.1021/acs.jpca.9b02771
Zhao C. Q., Zhang P. C., Zhou J. J., Qi S. H., Yamauchi Y., Shi R. R., et al. (2020). Layered nanocomposites by shear-flow-induced alignment of nanosheets. Nature 580, 210–21+. doi: 10.1038/s41586-020-2161-8
Keywords: biomineralization, molecular dynamics simulation, machine learning, shell proteins, Pinctada fucata
Citation: Dong W, Xie L and Zhang R (2024) Integrative computational framework to decipher the functions of shell proteins in biomineralization. Front. Mar. Sci. 11:1362131. doi: 10.3389/fmars.2024.1362131
Received: 27 December 2023; Accepted: 26 June 2024;
Published: 19 July 2024.
Edited by:
Chuang Liu, Hohai University, ChinaReviewed by:
Zhi Liao, Zhejiang Ocean University, ChinaFelipe Aguilera, University of Concepcion, Chile
Copyright © 2024 Dong, Xie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongqing Zhang, cnF6aGFuZ0BtYWlsLnRzaW5naHVhLmVkdS5jbg==; Liping Xie, bHB4aWVAbWFpbC50c2luZ2h1YS5lZHUuY24=
 Wentao Dong
Wentao Dong Liping Xie1*
Liping Xie1* Rongqing Zhang
Rongqing Zhang