Abstract
Citizen science involves non-professionals assisting with scientific research, contributing data, and conducting experiments under professional guidance. In this paper, we describe the citizen science project DNA & life, based at the Natural History Museum of Denmark, which actively engages high school students in both fieldwork and advanced laboratory analyses by collecting and analyzing eDNA samples from marine environments across Denmark. The analysis is performed via species-specific real-time PCR and in a case study we present data gathered in the project from 2017-2023 with focus on the potential of eDNA detecting in use for species monitoring. The results include seasonal occurrence of Mnemiopsis leidyi and national distributions of selected species of particular ecological interest Perca fluviatilis, Anguilla anguilla, Neogobius melanostomus and Alexandrium ostenfeldii. In addition to the eDNA case study, we present a new way of conducting citizen science and eDNA analysis, as a development of the established DNA & life project. Local DNA laboratories are created to shorten the path from sampling site to laboratory facility, creating the possibility for Danish high schools to participate in a new version of the project. In close collaboration with the academic staff of the DNA & life project both teachers and students help develop facilities, protocols and laboratory analyses in an extreme citizen science and co-creation approach, where the participants are involved in a higher level of laboratory work and data analysis. Allowing high school students to perform intricate molecular lab analyzes through an extreme citizen science approach has demonstrated encouraging outcomes and potential for data of high quality in terms of sampling and analyzing eDNA with the purpose of species monitoring and conservation.
1 Introduction
1.1 Environmental DNA for species monitoring
Engaging and training citizen scientists in advanced laboratory analysis, such as the monitoring of environmental DNA (eDNA) in water samples, has promising applications for large-scale national eDNA surveillance of aquatic species that can be used in governmental mapping and conservation efforts (Tøttrup et al., 2021; Knudsen et al., 2023). In this paper we will showcase the development of an existing citizen science project with focus on eDNA monitoring towards a more comprehensive method for engaging volunteers in field sampling and laboratory analysis.
The use of eDNA extracted from various sample types is a well-established tool for early and non-invasive detection of species in different environments. It serves as a screening tool for species monitoring and involves detecting genetic material released by living organisms into their surroundings (Thomsen and Willerslev, 2015; Beng and Corlett, 2020). eDNA monitoring can be performed in several ways, by species-specific detection using real-time PCR (or quantitative PCR) or by a metabarcoding or metagenomic approach to target a broader range of species within a specific taxonomic group. Initially, the species-specific approach has been focusing on freshwater species (Thomsen et al., 2012; Biggs et al., 2015; Sigsgaard et al., 2015), but in later years there is a growing use of marine eDNA (Garlapati et al., 2019; Knudsen et al., 2019; Reinholdt Jensen et al., 2021).
The advantage of using eDNA for monitoring of aquatic environments, relative to traditional fisheries surveillance tools, is its reduced reliance on taxonomic expertise (Jerde et al., 2011). Also collecting samples is relatively straightforward and accessible for individuals with minimal training (Bohmann et al., 2014; Thomsen and Willerslev, 2015; Beng and Corlett, 2020). Several research teams have developed eDNA analysis protocols, which have yielded a range of methods for detecting the genetic material of aquatic organisms of a variety of different taxonomic groups and in different environments (Goldberg et al., 2016; Tsuji et al., 2019). The easy accessibility of eDNA methods have facilitated their use in citizen science programs where samples of water have been collected and analyzed (Tøttrup et al., 2021; Agersnap et al., 2022; Knudsen et al., 2023).
Whether eDNA monitoring is conducted by trained scientists or untrained citizen scientists, the risk for error is inherent. In eDNA monitoring, this risk involves either falsely indicating the presence of a species when it is absent (false positive) or incorrectly suggesting the absence of a species when it is actually present (false negative). These false detections typically arise from sample contamination with unrelated DNA or sample mix-ups, occurring possibly both in the field and laboratory settings. To address challenges in eDNA monitoring within a citizen science framework, it is vital to systematically address the potential for false positive and false negative detections (Goldberg et al., 2016; Agersnap et al., 2022; Knudsen et al., 2023).
1.2 Using citizen science and high school students to investigate marine environments
The concept of citizen science holds many definitions, interpretations and ways of understanding, and we will refer to the principles described by the European Citizen Science Association (ECSA, 2015). Here we use ‘citizen science’ as scientific activities where individuals who are not professional scientists willingly engage in the processes of gathering, analyzing, and sharing data for a scientific purpose (Cohn, 2008; Silvertown, 2009). Traditionally, citizen science has primarily found its application in biodiversity monitoring due to its capacity for facilitating extensive data gathering (Pocock et al., 2018), and in terms of combining citizen science with eDNA studies, several projects have succeeded in engaging participants in fieldwork and sample collection, such as eDNA monitoring of the great crested newt in the UK (Biggs et al., 2015), collection of pond water for eDNA analysis (Buxton et al., 2018) and metabarcoding of marine eDNA collected in coastal areas (Agersnap et al., 2022). Nonetheless, engaging citizens in activities beyond data collection is still in its blooming stages since a higher level of volunteer engagement most likely implies a higher level of challenges regarding the scientific process (Senabre et al., 2021).
Several research groups have found great success in inviting school students to participate in citizen science projects with examples of primary and secondary schools collecting insects via Malaise traps (Steinke et al., 2017), testing the ability of schoolchildren to accurately estimate the strength of biotic interactions (Castagneyrol et al., 2020), letting high school students identify archaeological leather fragments with protein analysis (Brandt et al., 2022) or when high school students conduct eDNA analysis in order to detect distribution of amphibians (Knudsen et al., 2023). The schools represent a vast number of participants, students feel engaged in the work and have a positive scientific learning outcome when the teachers are able to combine the citizen science labor with school curriculum. The scientific aspect of citizen science can also give teachers a possibility for professional development (Harlin et al., 2018). The Danish citizen science project DNA & life has since 2014 invited more than 10,000 high school students into the laboratory to conduct scientific analyses of eDNA samples. Here, the students not only contribute by collecting water samples across the country, but they also take a great part in preparing the laboratory analyses for real-time PCR to investigate aquatic eDNA from selected species (Tøttrup et al., 2021). The project is anchored at the Natural History Museum of Denmark (NHMD) and the University of Copenhagen, which has shown to be a great asset in building a bridge between established researchers and the future generation of high school students (Tøttrup et al., 2021). From 2017 and onwards the project has worked with a well-tested and standardized protocol of collecting eDNA samples in the field for later real-time PCR analysis in the laboratory. High school students participate in the project either by collecting water samples of eDNA in the field, or by participating in a full-day program at the NHMD where they conduct real-time PCR analysis of eDNA samples. This laboratory work is facilitated and instructed by academic staff from the project group to make sure that the students correctly handle the equipment and conduct the laboratory methods according to scientific standards.
Over the years, DNA & life has pioneered a novel approach to citizen science and eDNA research, fostering a stable model for letting high school students work with eDNA sampling and laboratory analyses for generating data on aquatic species distribution to give a snapshot of the marine environment and insight into an otherwise inaccessible part of nature. The project has a great focus on data quality and has developed procedures of data control to obtain data and results of high standards, comparable in quality to those conducted by trained researchers within the field of eDNA (Tøttrup et al., 2021; Agersnap et al., 2022; Knudsen et al., 2023). Furthermore, DNA & life has established collaborative partnerships with numerous research projects and national government-led species monitoring programs, creating a sustainable method for involving citizen scientists in scientific activities.
1.3 Case study of species monitoring in DNA & life
Throughout the years of 2017-2023 the DNA & life project has worked with detecting different species in eDNA samples from marine environments. In this paper we will showcase eDNA results from real-time PCR analysis of marine eDNA samples with focus on results from five different species, selected for this case study, and the scientific importance of knowledge about their distribution in Danish waters:
Sea walnut [Mnemiopsis leidyi (Agassiz, 1865)]. Danish scientists, researchers, and authorities rigorously track seasonal fluctuations in Mnemiopsis leidyi populations. Understanding its interactions with marine species and ecosystem impacts is vital for investigating its potential influence on aquatic biodiversity. This research can help to develop effective strategies for managing invasive species, ensuring the health and balance of marine environments in Denmark and beyond (Carstensen and Jakobsen, 2023).
European perch [Perca fluviatilis (Linnaeus, 1758)]. monitoring in brackish/saltwater environments is important for understanding the status of key species. This will provide new insights to coastal ecosystem dynamics and the recreational and commercial value of the species (Overton et al., 2008).
European eel [Anguilla anguilla (Linnaeus, 1758)] is critically endangered in Denmark and the decline requires monitoring migration patterns and population trends. Identification of factors that affect the decline contributes to aid of conservation efforts for freshwater and coastal ecosystems (Sonne et al., 2021).
Round goby [Neogobius melanostomus (Pallas, 1814)], an invasive species for Danish waters, can disrupt ecosystems, alter food web dynamics, and spread to new areas. Monitoring is crucial for early detection and intervention to limit its impact and further distribution (van Deurs et al., 2021).
Sea fire [Alexandrium ostenfeldii ((Paulsen) Balech & Tangen, 1985)], linked to harmful algal blooms, poses threats to marine life and human health through toxin-contaminated shellfish. Regular monitoring in Danish waters is vital for public health and fisheries management (Kremp et al., 2009; Carstensen and Jakobsen, 2023).
The results from these species in the case study will outline the potential of using eDNA combined with a citizen science approach in terms of detecting the presence of selected species, yielding new information of species distribution and habitat or knowledge of the seasonal change in occurrence.
1.4 New method: taking DNA & life to extreme citizen science
In 2022 the original DNA & life project took a new direction for engaging citizens to a higher level of participation, We will refer to the topological levels of citizen science as described and defined by Haklay (2013), and use Haklays’ term “extreme citizen science” (also referred as co-creation) when the participants are able to choose their level of engagement and can be potentially involved in the analysis and publication or utilization of results. This extreme citizen science version of DNA & life will for the rest of the paper be referred to as Extreme DNA & life, whereas the original concept and project of DNA & life will be referred to as Original DNA & life. These two DNA & life projects have been co-existing since 2022 and run simultaneously. The project aims for an extreme citizen science approach, and includes collaborative partnerships with the high schools, the teachers and most certainly the high school students. The scientific staff in the project act as facilitators, in addition to their role as experts. Defining the framework for conducting the scientific work carried out by the participants is an essential part of working with the project in an extreme citizen science approach – both for the participants to be aware of the scientific impact of their actions, but also for creating convincing results that are built on reliable data sets (Brandt et al., 2022).
In this method, development of the DNA & life project from a participatory level to an extreme level of participation with a greater focus on standardization and quality control is undoubtedly essential to conduct consistent laboratory work and data (Haklay, 2013). While citizen science facilitates the buildup of extensive datasets, concerns about data quality have been raised (Burgess et al., 2017). Typically, ensuring data quality has been linked to a robust experimental design and volunteer training (Bonney et al., 2014). When volunteers undertake tasks similar to those performed by professionals, data quality assessments become essential for scientific rigor (Castagneyrol et al., 2020). Even though conducting school-based citizen science has its disadvantages in terms of data quality, conflicts of interest regarding the project outcome, and simple logistics, citizen science can empower students to take ownership of their own learning and science education if the citizen science project uses an extreme citizen science approach (Roche et al., 2020).
Extreme citizen science is the most extensive type of public involvement in research where citizen science-based projects strive to involve citizens in every stage and decision throughout the entire research journey (Heinisch, 2021).To conduct such a high level or participant engagement it is essential to establish training sessions targeting facilitators and participants in citizen science. This approach ensures successful achievement of the projects’ objectives. Offering such training not only facilitates the necessary scientific outcomes but also enhances participants’ scientific competences and raises awareness of the actual research question (Lorke et al., 2019). Therefore, we will also describe the steps of training high school students and high school teachers to be able to solely perform the practical, scientific work related to the research project. We will explore the potential of eDNA analysis in an extreme citizen science approach and discuss the future prospect and potential for inviting citizen scientists to engage in more than the sole data collection.
2 Materials and equipment
2.1 Collection of eDNA samples
High school classes received field kits to collect eDNA from water samples at local marine environments chosen by the class in agreement with the academic staff at the NHMD. Each field kit consisted of essential equipment and comprehensive instructions of the sampling process.
A sample of water from the specific environment was collected in a larger container. The sample volume varied from 0.5 L to 1.5 L. The water was filtered through a Sterivex™-GP (Merck Millipore) filter unit cartridges with a pore size of 0.22 µm using a 50 mL syringe. After water filtration the filter unit was emptied for water and filled with 96% ethanol to conserve the collected eDNA within the filter, using a 5 mL syringe for ethanol application. Finally, the cartridge was sealed with matching plugs. Each class conducted a minimum of two eDNA sampling via. Sterivex filtration at each location site to produce a set of two replicates. Use of ethanol conservation and selling the filter units with plugs allows easy and convenient storage and transportation of the filters. To reduce field contamination, the kit also included latex gloves that students were instructed to make use of while filtering water. Our setup is targeting specific species which means that human and other eukaryotic DNA in the sampled habitat is less prone to influence the subsequent analysis. Students were also guided to record supplementary information about the sampling site. This included the date, latitude and longitude coordinates, volume of water filtered (Supplementary File 2), and any other relevant observations made during the sampling process. The Sterivex filters, fixed with ethanol, were shipped to the NHMD and stored at -18°C until DNA extraction.
2.2 Extraction of DNA
Before any extraction work was initiated all table surfaces in the laboratory, Sterivex filter unit cartridges and equipment were cleaned with a 5% chlorine solution followed by 70% ethanol. These measures were implemented to reduce the risk of getting foreign DNA contaminating the samples. The plugs at both ends of the filter unit were removed, allowing ethanol to drain. The filter unit cartridge was opened, revealing the filter within the plastic cylinder to ease the extraction process (Figure 1). The filter unit was removed from the cylinder and divided into minor pieces. This is to ensure that as much filter paper as possible is exposed to extraction liquids, thus increasing the possibility of a higher DNA yield. Filter pieces were transferred to a microcentrifuge tube, and the extraction process began. DNA was extracted using the NucleoSpin® DNA RapidLyse kit (AH Diagnostics) and manufactures’ protocol. DNA concentration was measured with Qubit High Sensitivity protocol before the eDNA sample was used for later real-time PCR.
Figure 1

Process of opening Sterivex® filter unit cartridges in order to access the filter paper itself. (A) Essential tools for opening the cartridge, which all have been sterilized. (B) Cutting cartridge with pipe cutter. (C) Removing filter unit. (D) Exposing the white filter paper. (E) The filter paper is cut vertically and (F) horizontally at both ends. (G) The paper is removed from the cylinder with a tweezer and (H) placed in a petri dish. (I) With a scissor the paper is cut into minor fragments, and (J) ready for DNA extraction.
2.3 Real-time PCR analysis of eDNA sample
The DNA extracted from the eDNA sample was amplified by species-specific real-time PCR. One real-time PCR reaction of 25 μL consisted of a mastermix of 10 μL TaqMan® Environmental Master Mix 2.0 (Life Technologies), 9 μL double-distilled H2O, 1 μL of each primer and probe and 3 μL of extracted DNA template in addition to the mastermix. The list of sequences for primers and probes for each species-specific system included in the case study is listed in Supplementary File 1. In the real-time PCR setup, each species-specific real-time PCR analysis consisted of four real-time PCR reactions in four separate tubes, where the DNA template differs from each tube consisting of a positive control, a negative control and two replicates of the extracted eDNA sample from a single water sample (Figure 2). A complete real-time PCR set up consisted of a maximum of 24 sets of four-tube real-time PCR species-specific assays, allowing for a screening of eDNA from up to 12 different species, yielding two repetitions of each species-specific assay per PCR set up. With this setup, similar to previously described setups for the Original DNA & Life (Knudsen et al., 2023), every single one of the individual 24 assays comprised a positive and negative control, which later allows for evaluating if the reagents and templates mixed by students had been added correctly.
Figure 2
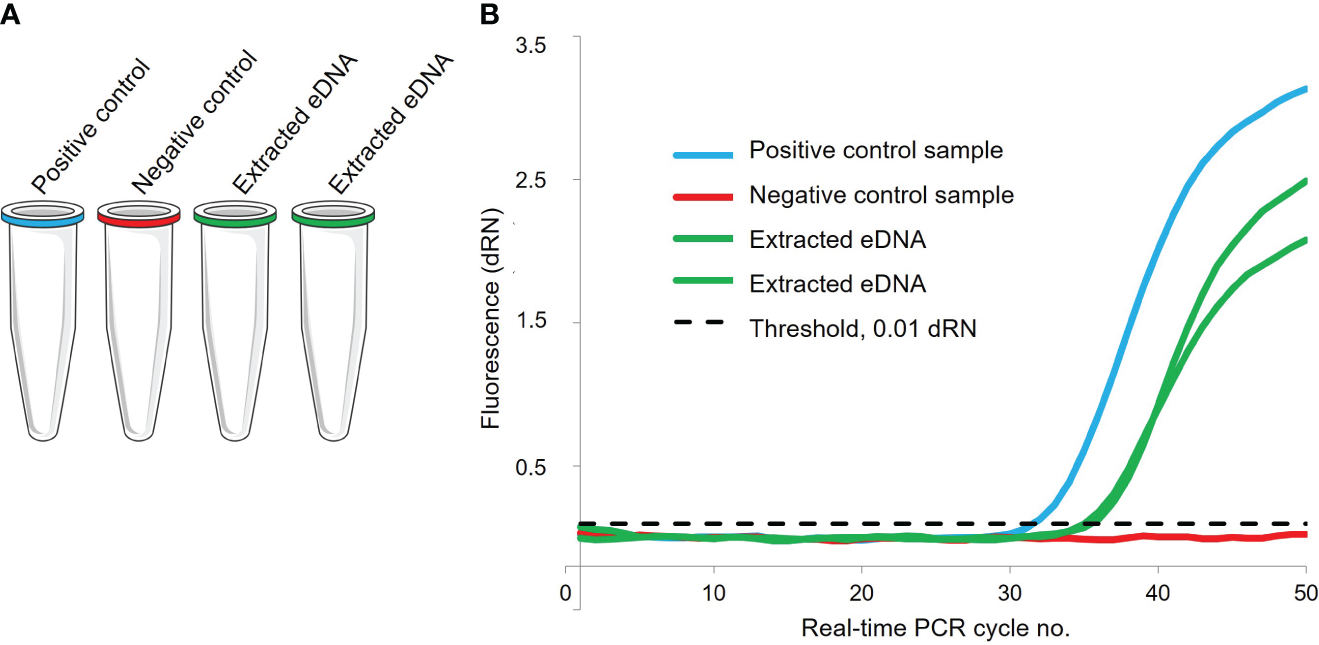
(A) Each species-specific real-time PCR analysis consists of four tubes with varying DNA template: The positive control sample (blue) is a purified and diluted target DNA molecule obtained from the target species. The negative control sample (red) is double-distilled sterile water. The eDNA sample (green) is extracted DNA from the Sterivex filter unit. (B) The sigmoid amplification plot obtained from the PCR reaction is a typical diagram and result of a real-time PCR assay. The increase in relative fluorescence levels reflects a higher number of DNA molecules present in the reaction tube for each cycle of amplification in the real-time PCR. Figure is adapted from Tøttrup et al. (2021).
The control samples were included to eliminate false positive and false negative detections. Real-time PCR analysis was performed with AriaMx real-time PCR System (AH Diagnostic). Thermal settings for the real-time PCR included 5 minutes initial denaturation at 50°C, then 10 minutes at 95°C, 50 cycles alternating between 95°C for 30 seconds and 60°C for 1 minute. When the real-time PCR program was finished, the results were visualized by amplification plots (Figure 2).
Since our objective was to do numerous parallel individual assays within a single real-time PCR setup, using assays that all originally were designed in silico with an annealing and extension temperature of 60°C (Spens et al., 2017; Ruvindy et al., 2018; Knudsen et al., 2019; 2022; Supplementary File 1), we did not perform an optimization test for the annealing temperature. With multiple specific assays operating in parallel in the same PCR plate the annealing temperature was consistently kept at 60°C.
We designed the real-time PCR setup to deviate from the conventional approach, which typically targets a single species and incorporates a standard dilution series to determine the limit of detection (LOD) and the limit of quantification (LOQ). The specialized setup applied here is identical to setups in our previous studies (Tøttrup et al., 2021; Knudsen et al., 2023), which allows for using several species-specific real-time PCR assays in parallel in the same plate and enables each class of students to analyze multiple assays using their own extracted water sample as template. We chose this plate setup to allow each class to run all species-specific assays concurrently in a single real-time PCR experiment, rather than limiting them to one species assay per setup.
3 Method
3.1 The DNA & life in an extreme citizen science approach
In 2022 the Original DNA & life as described from (Tøttrup et al., 2021) expands the participation of high school teachers and students to engage from a participatory level of citizen science to an extreme level of citizen science, as defined by (Haklay, 2013), in the project Extreme DNA & life. A project group was formed including academic staff at the NHMD and two volunteer board members from the Society of Danish Biologists (SDB), in Danish referred to as ‘Foreningen af Danske Biologer’. The members of SDB mainly consist of high school teachers within the subjects of biology and biotechnology. The SDB was an invaluable collaboration partner for the new development of the Extreme DNA & life project to engage with as many biology teachers as possible. In addition, the project group’s volunteer board members of SDB are also co-authoring in this paper.
The project group established two local DNA laboratories for conducting the development of the project. The two DNA laboratories for extreme citizen science are located geographically distant from the university and the original citizen science DNA laboratory at the NHMD, but in closer distance to rural areas of Denmark, shortening the distance from sample site to laboratory facilities. These laboratories were situated at two selected high schools, Hjørring Gymnasium and Herning Gymnasium respectively (Figure 3).
Figure 3

Location of DNA laboratories in the DNA & life project. Blue square represents the Original DNA & life laboratory at the Natural History Museum of Denmark and red triangles represent Extreme DNA & life DNA laboratories at Hjørring Gymnasium and Herning Gymnasium respectively.
The aim for the extreme citizen science laboratories were:
-
To bring laboratory facilities closer to all high schools of Denmark.
-
To involve high school students and teachers in a higher level of participation.
-
To conduct eDNA sampling and analysis with a higher yield in terms of sample volume and data quality.
To ensure that the work in the laboratory would be conducted correctly, and be able to compare data from different laboratory, following actions were made:
-
Uniform laboratory protocols and structure of real-time PCR assays.
-
Thorough training program for the participating high school teachers.
-
Administration of samples, data and logistics of material centered at the NHMD.
The two local DNA laboratories in the Extreme DNA & life project were equipped with all necessary inventory and material for conduction DNA extractions and real-time PCR analyses. The academic staff of the project at the NHMD made sure to provide the DNA laboratories with the correct operating material and reagents. Each of the two DNA laboratories has a local teacher and employee of the school as a lab manager, who is responsible for introducing each visiting high school class to the laboratory, facilities, and machinery use. This lab manager is also the direct contact to the academic staff at the NHMD and will provide the project group with data created at each DNA laboratory.
All work prior to data analysis and interpretation is solely performed by high school students, and in combination with their teacher, the students have a high level of influence regarding several elements in the scientific process such as fieldwork, DNA extraction and real time-PCR analysis which, helped develop the Original DNA & life to an extreme citizen science approach (Figure 4).
Figure 4
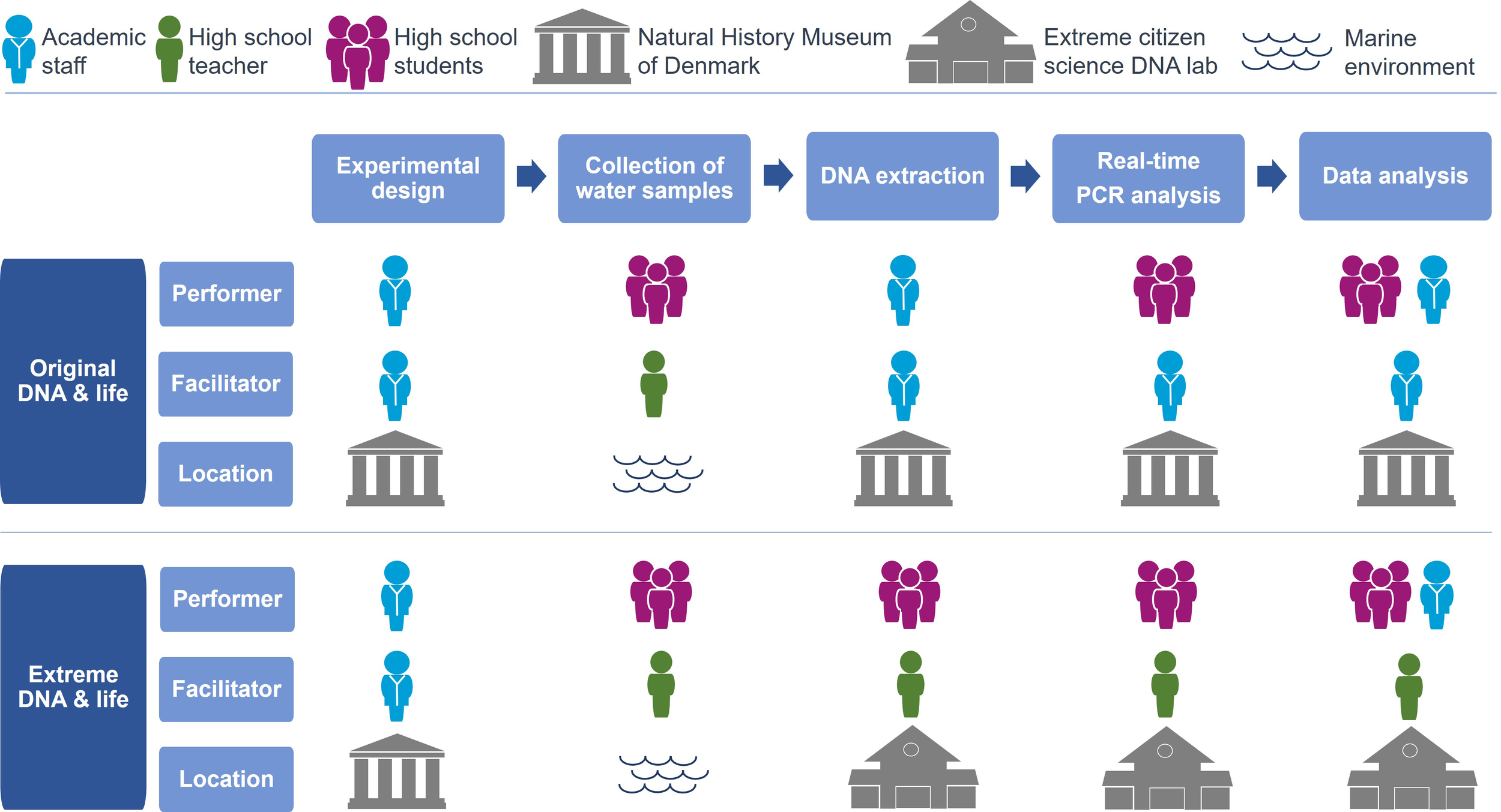
Development and distinction between the Original DNA & life project and the Extreme DNA & life project. The person/group performing or facilitating different phases of the scientific process, at different locations, is emphasized for both projects.
The first two phases of the scientific process in the citizen science project Original DNA & life and the development into Extreme DNA & life are identical in terms of the performing and facilitating persons/groups. Scientific staff at the NHMD ensure that the experimental design follows the guidelines and recommendations of working with marine eDNA samples, and that all material and protocols are distributed to the high schools. The students collect water samples of eDNA in a marine environment under guidance from their teacher. Here they have the decision of which location to choose, which part of the coastline they focus on and how distant from shore and to the seabed they collect the water sample. To create synergy between the two projects there is no difference regarding fieldwork and collection of water samples and later laboratory protocols used for DNA extraction and real-time PCR analysis.
The phase of DNA extraction shows the clear development of the Original DNA & life from a citizen science project to an extreme citizen science project in terms of the performer and facilitator of the DNA extractions (Figure 4). In the Original DNA & life DNA extraction is performed by academic staff at the NHMD, and in the Extreme DNA & life this step is performed by high school students, facilitated and under guidance of their teacher, located at one of two extreme citizen science DNA laboratories.
In the Original DNA & life project the school program at the NHMD invites high school students to perform the next phase of real-time PCR analysis of the extracted eDNA samples, followed by the final phase of data analysis of real-time PCR results. This is facilitated by academic staff but in the Extreme DNA & life this laboratory work and data analysis of real-time PCR results also takes place at the two DNA laboratories for the project’s extreme citizen science version where the class teacher is the facilitator while the high school students perform the all laboratory work. The students and the teachers are in charge of ensuring that the eDNA samples are handled correctly, choosing the DNA extract aliquot with highest amount of DNA concentration, making sure that the real-time PCR set up with all 24 species-specific PCR assays is carried through the whole process of preparation and analyzing in the real-time PCR machine and data collection. This comprehensive approach ensures that students actively participate in critical elements of the scientific process, from DNA collection and extraction to the precise analysis of eDNA samples.
The extreme citizen science DNA laboratories are open to all high schools in Denmark, and every teacher has the possibility to bring their class to the laboratory for a day of working with eDNA analysis without any entry cost. To ensure the success of these labs, teachers, who act as gatekeepers, complete a training course for the local laboratory procedures. Subsequently, these teachers take students to the field to collect water samples, which are then brought back to the lab for DNA extraction.
3.2 Training of high school teachers and students
For preparing the teachers to use the facilities and perform the methods correctly, a one-day training course is mandatory prior to participating in the Extreme DNA & life’s laboratory analyses. During the course teachers are instructed in how to use the equipment and how to work with the methods. The teachers undergo the same laboratory procedure and analysis as when they later bring their own class, so that they have a complete understanding of where equipment is placed, how to handle the machinery and how to conduct the DNA extraction and real-time PCR analysis.
During the course day a team of facilitators from the project group is present. This group includes the local lab manager and the two volunteer board members of the SDB. After the training course the teachers have the ability to contact the project group at the NHMD for sparring and guidance prior to their own day in one of the extreme citizen science DNA laboratories. The academic staff at NHMD also facilitates an online webinar for the teachers who have completed the training course. The purpose of the webinar is to brush up on the teachers’ knowledge of how to work in the lab, give any insight into the research focus and to be available for any questions the teachers might have. All this combined creates a thorough and thoughtful training of the high school teachers that act as crucial gatekeepers when high school students work within the Extreme DNA & life project.
When trained teachers visit one of the extreme citizen science DNA laboratories for a full day of laboratory work, including DNA extraction and real-time PCR, they are responsible for how the students work in the laboratory. The project group at the NHMD provides online protocols and guides for the students and teachers to read and prepare the work in advance. In addition, the project group have created guide videos for some of the most essential laboratory procedures including:
-
How to clean a lab bench.
-
How to handle a micropipette.
-
How to cut and open the Sterivex filter unit.
-
How to measure the DNA concentration via Qubit Fluorometer.
-
How to handle the real-time PCR machine and export data.
These videos were visual and auditive guides for both students and teachers to prepare the technical work in advance but also to use directly while performing the laboratory work.
During the day in the laboratory the students receive both theoretical and practical training, instructed by the class teacher. Initially, they go through an introduction covering aspects of working with eDNA, real-time PCR and molecular methods, including species-specific primers and probes, and they are provided with instructions on preparing and interpreting a real-time PCR analysis. Students complete training in sterile techniques to prevent sample cross-contamination and learn how to work in a molecular laboratory. Upon completion of the training, the students initiated extraction of eDNA from the Sterivex filter following measurement of DNA concentration from the eDNA extract and finally preparation and conduction of real-time PCR analysis. Notably, all these activities are conducted without direct involvement from the project group at the NHMD, but only via protocols and video guides created by the academic staff in the project group.
3.3 Minimization of contamination
To minimize the risk of contamination between samples and reagents the following precautions have been made:
-
Students never work with stock reagents. The reagents have been aliquoted in portion size corresponding to one filter extraction or one species-specific real-time PCR analysis.
-
Before mixing of reagents took place all work surfaces were washed with 5% chlorine solution followed by ethanol wash.
-
In the DNA laboratory at the NHMD the extraction of eDNA samples is in a separate room from the where the students perform real-time PCR analysis.
-
At the extreme citizen science DNA laboratories, the DNA extraction and real-time PCR analysis are separated by distinct workspaces since both laboratories are located in single room facilities. The students perform a thorough cleaning procedure with chlorine and ethanol between working with extraction and working with real-time PCR reagents. Though separate rooms for each laboratory procedure would be most preferred.
-
In all DNA laboratories each day is allocated to perform real-time PCR analysis of only one eDNA sample, in order to minimize potential cross-contamination between samples.
-
Post-PCR tubes were never opened in the DNA laboratories but were discarded straight after data collection to further minimize the risk of getting cross contamination from previous PCR setups.
-
A discussion with students, instructors, and teachers explored the significance of contamination. The conversation provided diverse perspectives, enhancing awareness of the implications of various contaminations.
3.4 Quality control of data
Both the Original DNA & life and the Extreme DNA & life follow identical protocols for collecting water samples, conducting DNA extractions and performing real-time PCR analysis as described in the Material and equipment section. Therefore, the produced data also follows the same guideline for quality control. To ensure high data quality the students make replicates of each eDNA sample, and all students include a positive and a negative control in the PCR set up (Figure 2). Interpretation of real-time PCR plots is a collaborative effort between academic staff in the project group and students where each analysis was validated to confirm the detection of eDNA from specific species in the eDNA samples. All real-time PCR data is collected by the academic staff of the project group at the NHMD. Here the data is handled via a uniform procedure to be able to work with a large dataset.
In order to validate the real-time PCR results, data was handled by a conservative cut off in order to reduce the likelihood of misinterpreting false positives. Any amplification signals observed after PCR cycle 41 and falling below a relative fluorescence threshold of 0.01 dRN were disregarded. This criterion was applied in all PCR analyses to create uniform data validation. The combination of data from the positive control and the negative control for each species-specific assay (Figure 2A) established the results for each single assay. A real-time PCR result was determined as ‘valid’ if the PCR signal for the positive control was above the relative fluorescence threshold of 0.01 dRN and occurred before PCR cycle 41 and no signal was detected for the negative control (Figure 2B). Other signal combinations for the positive and negative control were categorized as ‘invalid’. If one or both extracted eDNA replicates resulted in a relative fluorescence threshold of 0.01 dRN and occurred before PCR cycle 41 the eDNA signal was determined as positive and the occurrence of species-DNA was categorized as ‘present’. If the eDNA replicates show no PCR signal of amplification the species-DNA was categorized as ‘absent’. An overview of samples collected and processed by real-time PCR analysis in the Original DNA & life and the Extreme DNA & life, with indication of distribution of valid and invalid results, is presented in Supplementary File 3.
4 Results
4.1 Potential of eDNA data from case study
Results represent data from eDNA samples collected from 2017-2023 (Supplementary File 2), by 3,300 high school students and 140 teachers, from Danish marine environments in the Original DNA & life project. Samples cover most of the Danish coastline and fjord systems (Figure 5) which showcase the potential of citizen science to yield a high coverage of samples for an entire country. Several marine species were included in the project’s real-time PCR analysis of eDNA samples where 2,900 high school students and 170 teachers participated in the Original DNA & life teaching program to conduct laboratory analysis of the extracted DNA from the eDNA samples. This paper and presentation of results will outline data from five selected species as described in the introduction case study, thereof seasonal change in occurrence of Mnemiopsis leidyi and distribution of Perca fluviatilis, Anguilla anguilla, Alexandrium ostenfeldii and Neogobius melanostomus. An overview of the success of real-time PCR analysis from these species assays is illustrated in Supplementary File 4.
Figure 5
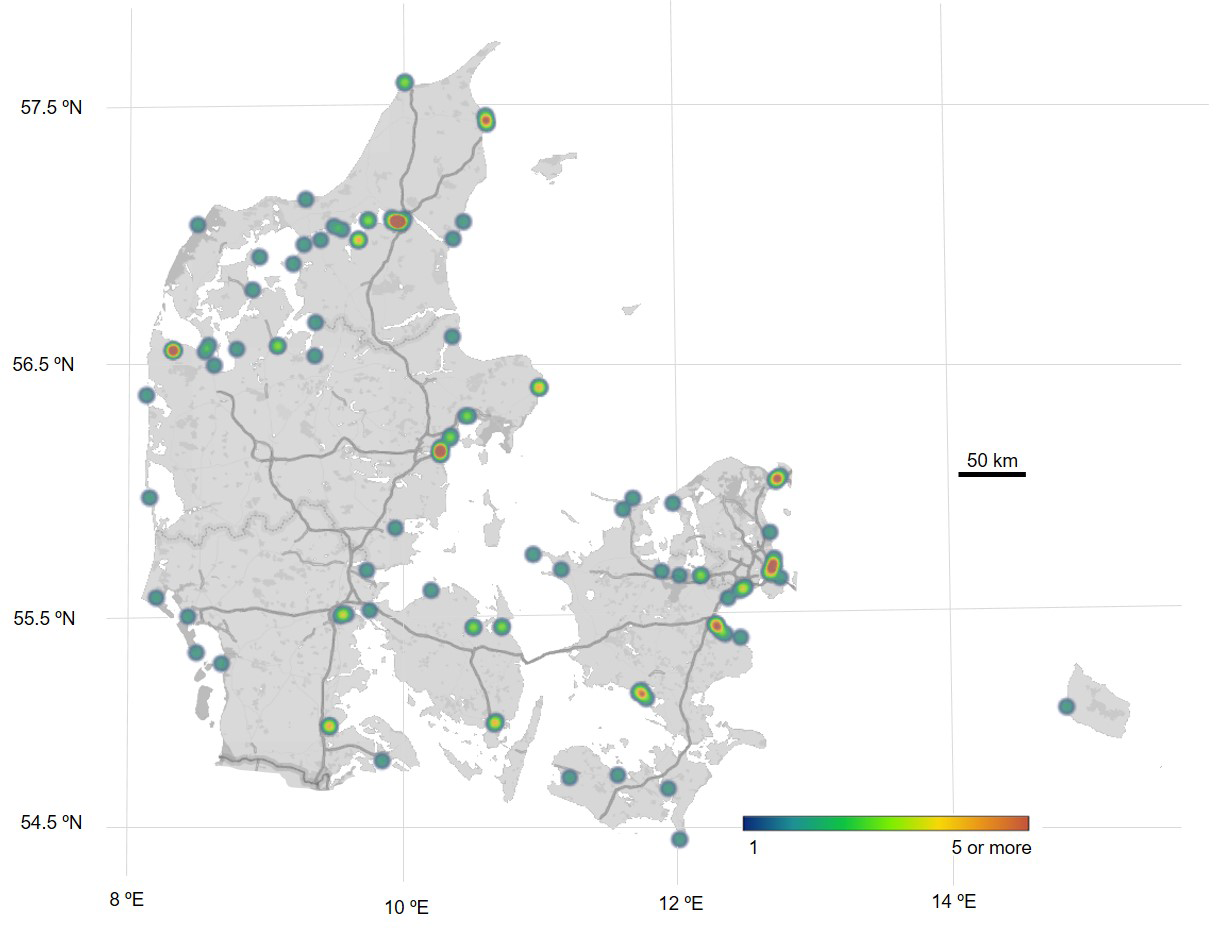
Distribution of Danish, marine eDNA samples collected within the Original DNA & life project during the years of 2017-2023. Each dot represents a collection site and color variations indicate the volume of samples at each site. Cold color indicates a lower sample volume and warm color indicates high sample volume. Map generated from data in Supplementary File 2.
The eDNA from Mnemiopsis leidyi showed seasonal fluctuations spanning the years 2017 to 2023 (Figure 6). The percentages reflect positive observations relative to the total samples collected within this period. The results reveal the highest occurrence of Mnemiopsis leidyi eDNA presence during the months of September to November. During months characterized by lower temperatures in Danish water (December to May), we observed a lower level of presence of Mnemiopsis leidyi eDNA.
Figure 6
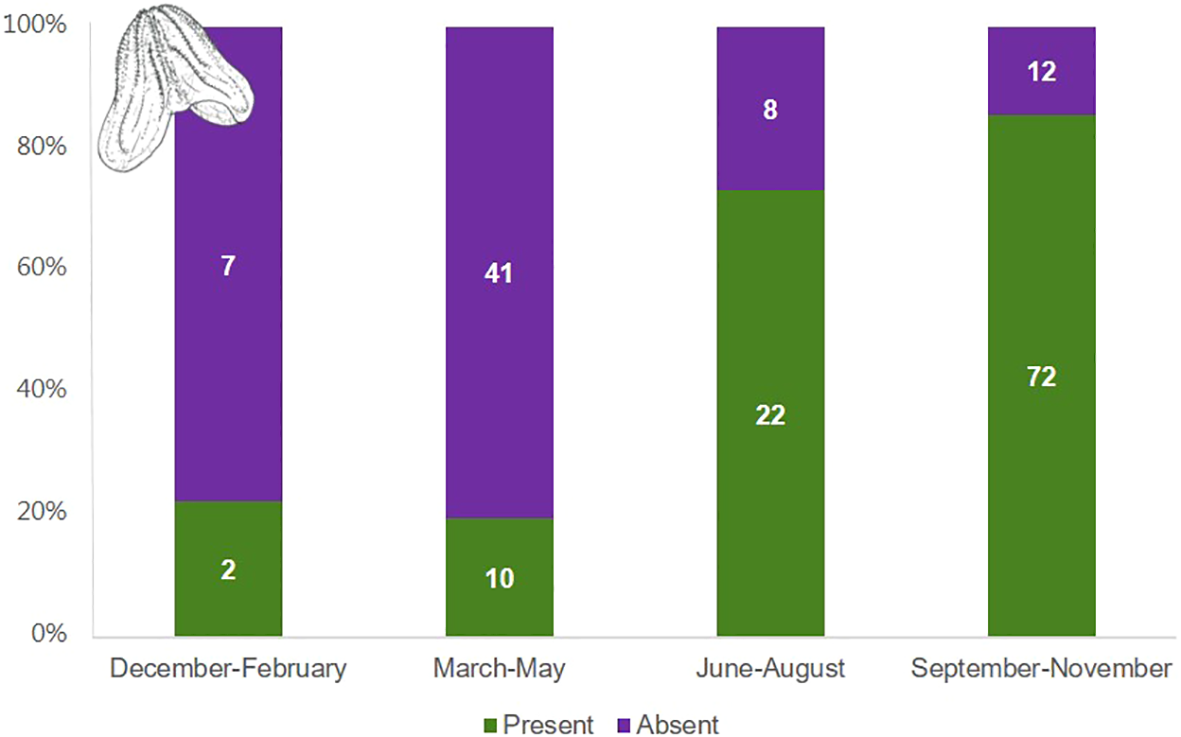
Mnemiopsis leidyi seasonal presence and absence in species-specific real-time PCR analysis of eDNA samples collected in the Original DNA & life project from 2017-2023. The columns represent the distribution between ‘present’ and ‘absent’ results for only assays that have passed the quality control and therefore are categorized as ‘valid’. Invalid PCR results have been discarded. Numbers indicate the volume of eDNA samples from the specific period. Drawings of Mnemiopsis leidyi by Maria Rytter.
The eDNA detected from the common Perca fluviatilis (Figure 7A) is mainly from the eastern part of Denmark where samples originate from open, brackish waters. The rare and endangered Anguilla anguilla return positive eDNA detections in more than half of the sampling sites (Figure 7B). The results illustrate a widespread distribution in Danish waters, both in marine environments but also in the many fjord systems of Denmark. Observations of both presence and absence at certain sites occur when eDNA samples are collected over a yearly distribution covering seasonal variations. The continuous monitoring of eDNA from Neogobius melanostomus provides a comprehensive view of this nonindigenous species in Danish waters (Figure 7C), revealing a distribution across open waters and in a fjord in the northern part of Denmark. The distribution of positive eDNA detections from the toxic algae Alexandrium ostenfeldii in samples collected between 2022 and 2023 (Figure 7D), show that this alga has a broad distribution across Danish waters.
Figure 7
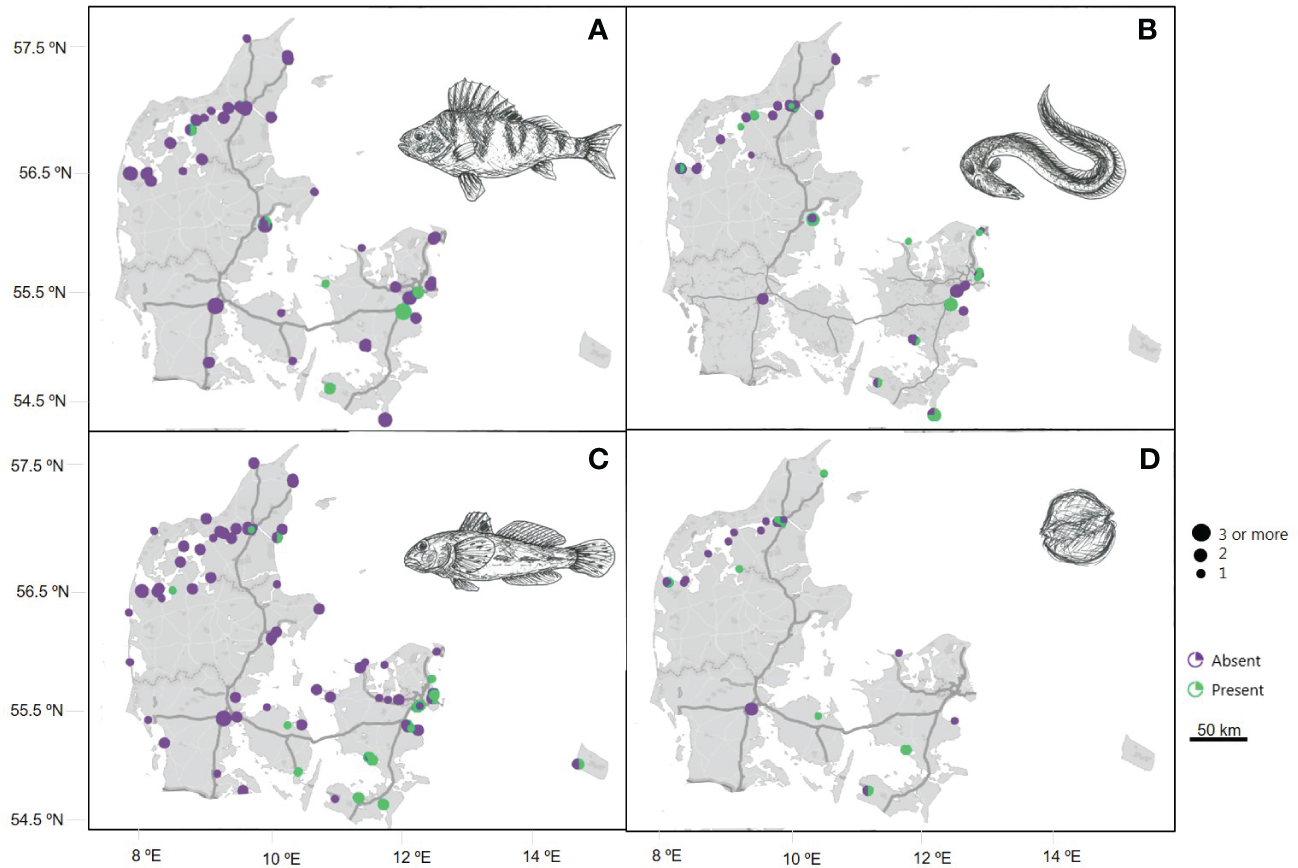
Maps of species distribution in Denmark based on species-specific real-time PCR analysis of marine eDNA samples collected in the Original DNA & life project from 2017-2023, except for figure D where samples were collected from 2022-2023. Linear scale bar indicates distance over land. Dot size corresponds to the number of samples analyzed from a specific location, with the scale bar of dots representing sample volume from ‘one’ to ‘three or more’ samples. Circle color indicates absence (purple) or presence (green) of eDNA from four of the species included in the DNA & life project: (A)Perca fluviatilis, (B)Anguilla anguilla, (C)Neogobius melanostomus and (D)Alexandrium ostenfeldii. Drawings by Maria Rytter.
4.2 Potential of eDNA data from extreme citizen science
In the Extreme DNA & life project the sampling of marine eDNA was performed identical as for the Original DNA & life project, but in this version the whole subsequent process of laboratory work and data analysis was performed at the two DNA laboratories for extreme citizen science. Since the two DNA laboratories were established in 2022 in total 170 high school students and ten teachers have participated in the Extreme DNA & life laboratory analysis. From this we presented a selection of results from real-time PCR analysis created in the Extreme DNA & life project (Figure 8), where we have chosen to focus om the same species as for the case study of species distribution in Figure 7 where results illustrate the potential of eDNA data. The selected results are based on eDNA samples collected in the year 2022 and 2023, and later handled and analyzed in an extreme citizen science DNA laboratory (Figure 8), show both presence and absence of eDNA detections.
Figure 8
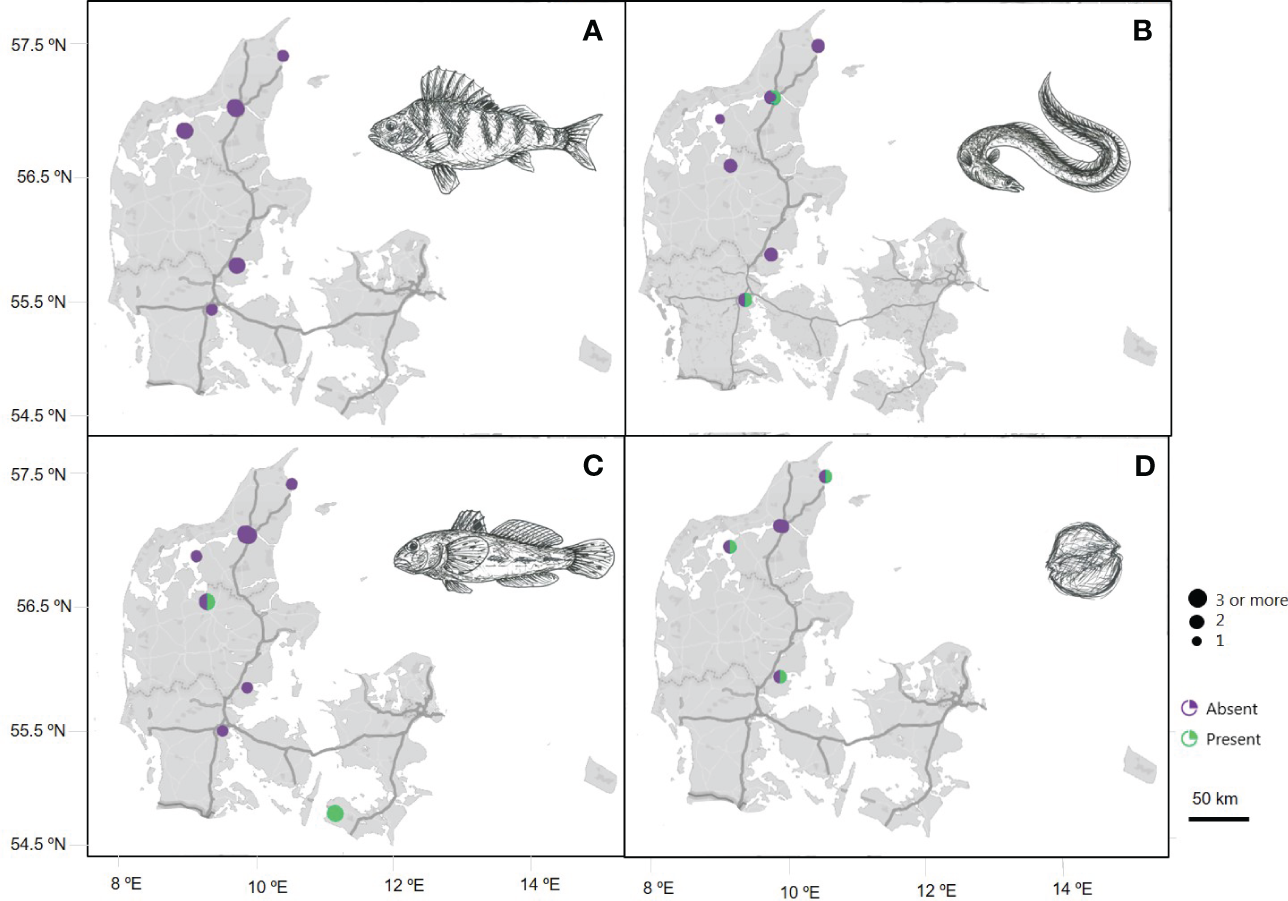
Maps of species distribution in Denmark based on species-specific real-time PCR analysis of marine eDNA samples collected and analyzed in the Extreme DNA & life project from 2022-2023. Linear scale bare indicates distance over land. Dot size corresponds to the number of samples analyzed from a specific location, with the scale bar of dots representing sample volume from ‘one’ to ‘three or more’ samples. Circle color indicates absence (purple) or presence (green) of eDNA from four of the species included in the DNA & life project: (A)Perca fluviatilis, (B)Anguilla anguilla, (C)Neogobius melanostomus and (D)Alexandrium ostenfeldii. Drawings by Maria Rytter.
The species distribution shows the potential of the data created in the new development of the project, as the data has been subjected to the same level of quality control as the data generated in the Original DNA & life project. Though, the data volume in the Extreme DNA & life is not nearly compatible with data gathered in the Original DNA & life project since the project still is in its early phase of development. The results are a preliminary window to the potential of monitoring species distribution with eDNA via an extreme citizen science approach, and we expect to generate more data in the following years. The data will be in resemblance to results generated in the Original DNA & life, and one of the project goals is to gather enough data to compare the distribution results and data quality between the Original DNA & life and the Extreme DNA & life.
5 Discussion
This study outlines the citizen science project Original DNA & life that until 2022 has been performed under control of academic staff at the NHMD. We highlight the successful engagement of students in the scientific process, by showcasing examples of some of the results from eDNA monitoring by species-specific real-time PCR analysis. One of the primary scientific goals of the project is to analyze the distribution of these species and mapping out their prevalence in specific geographic areas. This geographical insight is invaluable for understanding the ecological landscape and planning conservation strategies, especially when focusing on species categorized as problematic or red-listed. The volume of eDNA samples, geographical coverage, level of quality control and knowledge about species distribution has the potential of supporting scientific research and informing environmental management in Denmark for sustainable development. In the realm of policy and scientific development, these findings contribute valuable information, enhancing understanding, supporting evidence-based decision-making, and guiding future research initiatives.
The presented case study provides insights into the distribution and abundance of normal, rare, toxic, and invasive species in Danish waters. The eDNA method emerges as a non-invasive and effective screening tool, particularly valuable for detecting species that are challenging to monitor. Continuous surveillance of Neogobius melanostomus, Alexandrium ostenfeldii, Anguilla anguilla, and Perca fluviatilis reveals their distribution across a wide geographic range, and for Mnemiopsis leidyi the eDNA monitoring here also indicates patterns of abundance over time. Information on distribution and shifts in abundance is crucial for effective conservation management. Notably, the eDNA of Neogobius melanostomus presence in northern regions of Denmark challenges existing assumptions about species habitats, contributing to a more nuanced understanding of the species’ ecological role in Danish waters (Puntila et al., 2018). The occurrence of Perca fluviatilis in open waters in eastern part of Denmark explores the traditional perceptions of the species’ habitat preferences for freshwater environment or brackish water, as for previous study has highlighted the potential for recruitment of the species in coastal waters in the western Baltic Sea (Christensen et al., 2016), indicating a broad ecological adaptability for Perca fluviatilis. The widespread distribution of Alexandrium ostenfeldii emphasizes potential environmental risks linked to toxic algae, necessitating water quality assessments to mitigate the impact of harmful algal blooms on public health. The conservation status of Anguilla anguilla is marked as potentially critically endangered, underscoring the need for conservation measures and policies to protect its population in Danish waters. Furthermore, the presence and abundance of the species in the case study act as indicators of overall ecosystem health, enabling early detection of ecological imbalances and facilitating proactive measures to maintain the health and resilience of Danish aquatic ecosystem as presented in previous study by Knudsen et al. (2022). This targeted approach aids in the early detection of issues related to these species, contributing to their conservation and management. It challenges assumptions about species habitats, aids in invasive species management, highlights potential environmental risks, and prompts reconsideration of conservation strategies.
There is a significant potential for broadening the scope of our analyses by including several other species. This expansion allows for a more comprehensive understanding of the interactions and dynamics among different species within the ecosystem. In summary, the data’s potential lies in its ability to inform a thorough and adaptive approach to environmental management, conservation, and policy development in Danish marine waters.
Our study is a preliminary method study where the eDNA method is used as a screening tool with potential as an early-warning system. In this study we excluded standard dilution series to follow a PCR set up that was more suitable in an educational aspect, where the students were analyzing their own eDNA sample by multiple species-specific real-time PCR assays. This set up made it possible to test multiple species, yielding results that are used as an early detection of eDNA from a selected variety of species rather than investigating the quantitative aspect of the eDNA sample. Though the inclusion of a series of standard dilutions with known concentrations for each species assay would enable the determination of LOD and LOQ and ultimate make the results more comparable with similar eDNA studies of same species or same origin of eDNA sample.
It is evident that a systematic application is crucial if the method is to stand alone as the sole indicator for rare species. While the method effectively identifies DNA from organisms, this necessarily does not mean the detection of the actual individual. Therefore, supplementary methods are necessary to form a comprehensive picture of biodiversity and actual species occurrence. It is important to clarify that this study focuses on investigating the potential of the method as a screening tool in combination with a citizen science approach and not on presenting new scientific findings alone. The intention is to evaluate the utility of the eDNA method as a preliminary approach. Concerning our case study, it is crucial to outline any reservations pertaining to the results.
The new method of exploring an extreme citizen science approach in the Extreme DNA & life has yielded insight into the potential of letting high school students and teachers conduct all laboratory work in the scientific process, which have created successful results in terms of eDNA monitoring of selected species based on real-time PCR analysis. The great focus on training teachers and students, along with a thorough and uniform protocol and guidelines for minimization of contaminations, have shown that high school classes are able to take responsibility for advanced molecular methods to produce results of species distribution based on eDNA sampling and analysis. The quality control of the real-time PCR set up and data validation made it possible to distinguish between successful datasets and invalid results. The focus for the coming project period will be to investigate the similarities between results created at the controlled environment at the NHMD DNA laboratory in the Original DNA & life versus the results obtained from the two extreme citizen science DNA laboratories in the Extreme DNA & life project. It is crucial to have comparability and consistency between samples, whether they have been analyzed under facilitation of academic staff at the NHMD or by high school teachers alone.
Though the process of establishing and anchoring the Extreme DNA & life has not been entirely without challenges. The extensive administrative work and logistics around the laboratories and experimental design is not without importance, and the fact that the project is anchored at the NHMD and with SDB as the main collaborative partner has been an absolute necessity to create continuity, uniform working procedure and to reach the target group of high school teachers and students. Just because several Danish high schools now are within closer proximity of highly advanced laboratory facilities, that even is free of charge, doesn’t necessarily lead to greater interest for participation in the project. Several teachers, who have completed the training course, still find the laboratory methods too advanced and complicated for them to be comfortable taking responsibility for conducting a full-day program in one of the extreme citizen science DNA laboratories. The project group is therefore in close contact with the teachers and is in an ongoing process of developing the scope of the laboratory procedure to be suitable, not only for the purpose of the research strategy but also for the convenience for participating teachers and students. In this way the high school teachers together with the students have a great impact and influence on the evolution of the project and future developments.
The new method of conducting eDNA analysis via extreme citizen science opens the door to continuous and widespread citizen science-based biodiversity monitoring in both marine environments and other aquatic environments. The involvement of citizen scientists in laboratory activities not only carries educational and training benefits but also serves as a motivational factor for the participants to follow the research process more closely and comprehensively. The prospects and potential in letting high school students conduct complex molecular laboratory analyses via a citizen science approach have shown promising results. The Extreme DNA & life project continues until the end of 2024 where all data, results and the exploration of the citizen science aspect will be evaluated. In conclusion, exploration of the potential of both the Original DNA & life and the Extreme DNA & life have proven effective and demonstrated promising signs, though it is important to account for several scientific issues and risks when working both with citizen science and eDNA. Altogether, this study implies further development of engaging students in extreme citizen science and our preliminary results could serve as foundation for further specialized monitoring efforts.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The requirement of ethical approval was waived by Natural History Museum of Denmark for the studies involving humans. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
FL: Writing – original draft, Writing – review and editing. MR: Writing – original draft, Writing – review and editing. ML: Writing – review and editing. BR: Writing – review and editing. CR: Writing – review and editing. JO: Writing – review and editing. MO: Writing – review and editing. PM: Writing – review and editing. NL: Writing – review and editing. SK: Writing – review and editing. AT: Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors declare that this the extreme citizen science development of the project DNA & life received funding from the Novo Nordisk Foundation (Grant J.nr.NNF21OC0072554). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We would like to thank all the high school students and teachers for their willingness to take part in the project and contribute with labor in the field and in the laboratories. The authors would like to thank Anne Hjortlund Hansen and Emil Lau Holmstrup for help with operational tasks in the laboratory, and Daniel Geneser and Birgit Kristine Hougaard for engagement as laboratory managers at Hjørring Gymnasium and Herning Gymnasium respectively. The project was not able to succeed without the major support from the Society of Danish Biologists (Foreningen af Danske Biologer).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1347298/full#supplementary-material
References
1
Agersnap S. Sigsgaard E. E. Jensen M. R. Avila M. D. P. Carl H. Møller P. R. et al . (2022). A national scale ‘BioBlitz’ Using citizen science and eDNA metabarcoding for monitoring coastal marine fish. Front. Mar. Sci.9, 824100. doi: 10.3389/fmars.2022.824100
2
Beng K. C. Corlett R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodivers Conserv.29, 2089–2121. doi: 10.1007/s10531-020-01980-0
3
Biggs J. Ewald N. Valentini A. Gaboriaud C. Dejean T. Griffiths R. A. et al . (2015). Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv.183, 19–28. doi: 10.1016/j.biocon.2014.11.029
4
Bohmann K. Evans A. Gilbert M. T. P. Carvalho G. R. Creer S. Knapp M. et al . (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol.29, 358–367. doi: 10.1016/j.tree.2014.04.003
5
Bonney R. Shirk J. L. Phillips T. B. Wiggins A. Ballard H. L. Miller-Rushing A. J. et al . (2014). Next steps for citizen science. Science343, 1436–1437. doi: 10.1126/science.1251554
6
Brandt L.Ø. Lillemark M. R. Toftdal M. Andersen V. L. Tøttrup A. P. (2022). Are we betting on the wrong horse? Insignificant archaeological leather fragments provide the first evidence for the exploitation of horsehide in renaissance Denmark. Herit5, 972–990. doi: 10.3390/heritage5020053
7
Burgess H. K. DeBey L. B. Froehlich H. E. Schmidt N. Theobald E. J. Ettinger A. K. et al . (2017). The science of citizen science: exploring barriers to use as a primary research tool. Biol. Conserv.208, 113–120. doi: 10.1016/j.biocon.2016.05.014
8
Buxton A. Groombridge J. Griffiths R. (2018). Comparison of two citizen scientist methods for collecting pond water samples for environmental DNA studies. Citiz Sci.3, 2. doi: 10.5334/cstp.151
9
Carstensen J. Jakobsen H. H. (2023). Harmful algae in limfjorden: A desktop study. Advisory Memorandum DCA, 59.
10
Castagneyrol B. Valdés-Correcher E. Bourdin A. Barbaro L. Bouriaud O. Branco M. et al . (2020). Can school children support ecological research? Lessons from the oak bodyguard citizen science project. Citiz Sci.5, 10. doi: 10.5334/cstp.267
11
Christensen E. Skovrind M. Olsen M. Carl H. Gravlund P. Møller P. (2016). Hatching success in brackish water of Perca fluviatilis eggs obtained from the western Baltic Sea. Cybium: J. Ichthyol.40, 133–135.
12
Cohn J. P. (2008). Citizen science: can volunteers do real research? BioScience58 (3), 192–197. doi: 10.1641/B580303
13
Deurs M. Moran N. P. Plet-Hansen K. S. Dinesen G. E. Azour F. Carl H. et al . (2021). Impacts of the invasive round goby (Neogobius melanostomus) on benthic invertebrate fauna: A case study from the baltic sea. NeoBiota68, 19–30. doi: 10.3897/neobiota.68.67340
14
ECSA (2015). ECSA 10 principles of citizen science. European citizen science association. Center For Open Sci. doi: 10.17605/OSF.IO/XPR2N
15
Garlapati D. Charankumar B. Ramu K. Madeswaren P. Ramana Murthy M. V. (2019). A review on the applications and recent advances in environmental DNA (eDNA) metagenomics. Rev. Environ. Sci. Biotechnol.18, 389–411. doi: 10.1007/s11157-019-09501-4
16
Goldberg C. S. Turner C. R. Deiner K. Klymus K. E. Thomsen P. F. Murphy M. A. et al . (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol.7, 1299–1307. doi: 10.1111/2041-210X.12595
17
Haklay M. (2013). Citizen science and volunteered geographic information: overview and typology of participation. Crowdsourcing Geographic Knowledge, 105–122. doi: 10.1007/978-94-007-4587-2_7
18
Harlin J. Kloetzer L. Patton D. Leonhard C. Leysin American School high school students (2018). “Turning students into citizen scientists,” in Citizen Science: Innovation in Open Science, Society and Policy. Eds. HeckerS.HaklayM.BowserA.MakuchZ.VogelJ.BonnA. (London: UCL Press), 410–428.
19
Heinisch B. (2021). Reaching the limits of co-creation in citizen science — Exemplified by the linguistic citizen humanities project ‘On everyone’s mind and lips — German in Austria’. J. Sci. Commun.20, A05. doi: 10.22323/2.20060205
20
Jerde C. L. Mahon A. R. Chadderton W. L. Lodge D. M. (2011). “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett.4, 150–157. doi: 10.1111/conl.2011.4.issue-2
21
Knudsen S. W. Ebert R. B. Mortensen P. B. Kuntze F. Hesselsøe M. Hassingboe J. et al . (2019). Species-specific detection and quantification of environmental DNA from marine fishes in the baltic sea. J. Exp. Mar. Bio Ecol.510, 31–45. doi: 10.1016/j.jembe.2018.09.004
22
Knudsen S. W. Hesselsøe M. Rytter M. Lillemark M. R. Tøttrup A. P. Rahbek C. et al . (2023). Detection of environmental DNA from amphibians in Northern Europe applied in citizen science. Environ. DNA5, 1–20. doi: 10.1002/edn3.462
23
Knudsen S. W. Hesselsøe M. Thaulow J. Agersnap S. Hansen B. K. Jacobsen M. W. et al . (2022). Monitoring of environmental DNA from introduced species of algae, dinoflagellates and animals in the North Eastern Atlantic. Sci. Total Environ.821, 153093. doi: 10.1016/j.scitotenv.2022.153093
24
Kremp A. Lindholm T. Dreßler N. Erler K. Gerdts G. Eirtovaara S. et al . (2009). Bloom forming alexandrium ostenfeldii (Dinophyceae) in shallow waters of the åland archipelago, Northern Baltic Sea. Harmful Algae8, 318–328. doi: 10.1016/j.hal.2008.07.004
25
Lorke J. Golumbic Y. N. Ramjan C. Atias O. (2019). Training needs and recommendations for citizen science participants, facilitators and designers. COST Action., 1–28.
26
Overton J. L. Overton M. B. Paulsen H. Wang T. (2008). Salinity tolerance of cultured eurasian perch, perca fluviatilis L.: effects on growth and on survival as a function of temperature. Aquaculture277, 282–286. doi: 10.1016/j.aquaculture.2008.02.029
27
Pocock M. J. O. Chandler M. Bonney R. Thornhill I. Albin A. August T. et al . (2018). A vision for global biodiversity monitoring with citizen science. Adv. Ecol. Res.59, 169–223. Academic Press. doi: 10.1016/bs.aecr.2018.06.003
28
Puntila R. Strake S. Florin A.-B. Naddafi R. Lehtiniemi M. Behrens J. W. et al . (2018). Abundance and distribution of round goby (Neogobius melanostomus): HELCOM Baltic Sea Environment Fact Sheet 2018. HELCOM., 1–10.
29
Reinholdt Jensen M. Egelyng Sigsgaard E. Agersnap S. Jessen Rasmussen J. Baattrup-Pedersen A. Wiberg-Larsen P. et al . (2021). Seasonal turnover in community composition of stream-associated macroinvertebrates inferred from freshwater environmental DNA metabarcoding. Environ. DNA3, 861–876. doi: 10.1002/edn3.193
30
Roche J. Bell L. Galvão C. Golumbic Y. N. Kloetzer L. Knoben N. et al . (2020). Citizen science, education, and learning: challenges and opportunities. Front. Sociol.5. doi: 10.3389/fsoc.2020.613814
31
Ruvindy R. Bolch C. J. MacKenzie L. Smith K. F. Murray S. A. (2018). qPCR assays for the detection and quantification of multiple paralytic shellfish toxin-producing species of alexandrium. Front. Microbiol.9. doi: 10.3389/fmicb.2018.03153
32
Senabre E. Perelló J. Becker F. Bonhoure I. Legris M. Cigarini A. (2021). “Participation and co-creation in citizen science,” in The Science of Citizen Science. Ed. VohlandK. (Springer, Cham).
33
Sigsgaard E. E. Carl H. Møller P. R. Thomsen P. F. (2015). Monitoring the near-extinct european weather loach in Denmark based on environmental DNA from water samples. Biol. Conserv.183, 46–52. doi: 10.1016/j.biocon.2014.11.023
34
Silvertown J. (2009). A new dawn for citizen science. Trends Eco.l Evol.24, 467–471. doi: 10.1016/j.tree.2009.03.017
35
Sonne C. Peng X. Alstrup A. K. O. Lam S. S. (2021). European eel population at risk of collapse. Science. 372, 1271. doi: 10.1126/science.abj3359
36
Spens J. Evans A. R. Halfmaerten D. Knudsen S. W. Sengupta M. E. Mak S. S. T. et al . (2017). Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods Ecol. Evol.8, 635–645. doi: 10.1111/2041-210X.12683
37
Steinke D. Breton V. Berzitis E. Hebert P. D. N. (2017). The school malaise trap program: coupling educational outreach with scientific discovery. PloS Biol.15, e2001829. doi: 10.1371/journal.pbio.2001829
38
Thomsen P. F. Kielgast J. Iversen L. L. Møller P. R. Rasmussen M. Willerslev E. et al . (2012). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PloS One7, e41732. doi: 10.1371/journal.pone.0041732
39
Thomsen P. F. Willerslev E. (2015). Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv.183, 4–18. doi: 10.1016/j.biocon.2014.11.019
40
Tøttrup A. P. Svenningsen L. Rytter M. Lillemark M. R. Møller P. Knudsen S. W. (2021). Citizens in the lab: performance and validation of eDNA results. Citiz Sci.6, 35. doi: 10.5334/cstp.382
41
Tsuji S. Takahara T. Doi H. Shibata N. Yamanaka H. (2019). The detection of aquatic macroorganisms using environmental DNA analysis—A review of methods for collection, extraction, and detection. Environ. DNA.1, 99–108. doi: 10.1002/edn3.21
Summary
Keywords
extreme citizen science, co-creation, environmental DNA, marine species, high school
Citation
Leerhøi F, Rytter M, Lillemark MR, Randeris B, Rix C, Olesen J, Olsen MT, Møller PR, Lundholm N, Knudsen SW and Tøttrup AP (2024) Exploring the potential of extreme citizen science with Danish high school students using environmental DNA for marine monitoring. Front. Mar. Sci. 11:1347298. doi: 10.3389/fmars.2024.1347298
Received
30 November 2023
Accepted
13 February 2024
Published
05 March 2024
Volume
11 - 2024
Edited by
Serena Lucrezi, North-West University, South Africa
Reviewed by
David Stanković, National Institute of Biology, Slovenia
Ben McAteer, Queen’s University Belfast, United Kingdom
Updates
Copyright
© 2024 Leerhøi, Rytter, Lillemark, Randeris, Rix, Olesen, Olsen, Møller, Lundholm, Knudsen and Tøttrup.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frederik Leerhøi, frederik.leerhoei@snm.ku.dk
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.