
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 29 February 2024
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1346535
Globally, groupers (Serranidae) are commercially important fish species. Hybridization within this family has been reported in captivity and natural environments, usually among congeneric species. However, intergeneric hybridization has been reported only occasionally. Given the commercial interest in these fish, artisanal or recreational fishermen easily notice “weird-looking” individuals. This is the case of an unidentified grouper noticed by a spearfisher in Lebanese Mediterranean waters. Visual comparisons raised the possibility that the observed specimen was a hybrid. Molecular analysis based on two mitochondrial and three nuclear markers was performed to investigate this potential hybrid’s affinity. The discordance of the phylogenetic gene trees of the mitochondrial and nuclear markers indicates that E. marginatus and M. rubra are potential parent species of a hybrid individual in our samples. The subsequent haploweb analysis based on the S7 nuclear marker reveals the affinity of the hybrid individual to both E. marginatus and M. rubra. In this specific hybridization case, the nuclear marker S7 emerges as a valuable diagnostic tool for species delineation and hybrid detection.
Hybridizations between grouper (family: Serranidae) species have been reported in both natural and artificial environments. Some of the hybridization cases include species from different genera (Chen et al., 2017). The genetic similarities (p-distance, based on GenBank COI sequences) between each natural or artificial hybridizing parent grouper species pair range from 0.00 to 0.17 (He et al., 2022), and they include many different genera such as Epinephelus, Cephalopholis, and Cromileptes (Glamuzina et al., 1999; James et al., 1999; Liu et al., 2007; Addin and Senoo, 2011; Kiriyakit et al., 2011; Wang et al., 2014; Payet et al., 2016; Sun et al., 2016; Chen et al., 2017; Ching et al., 2018; Qu et al., 2018; He et al., 2022).
Unlike other hybridizing marine fishes in Pomacentridae (Hobbs et al., 2009; Tea et al., 2020), Pomacanthidae (Kemp, 2000), Acanthuridae (DiBattista et al., 2016), and Chaetodontidae (Montanari et al., 2012, 2014; DiBattista et al., 2015), some species from Serranidae have relatively higher market values (Craig et al., 2011). Hence, the Serranidae hybrids have a good chance of being noticed by artisanal or recreational fishers. A similar case has been documented on another group of high-economic value fishes. Fishermen from the Baltic Sea, who extensively trade flatfishes (family: Pleuronectidae) in Scandinavian fish markets, first noticed the hybridization between Pleuronectes platessa and Platichthys flesus (He and Mork, 2015). They even gave the name “Leps” (Danish) to the hybrid to separate it from their purebred parent species before scientists studied the hybridization case and developed identification tools (Sick et al., 1963; Kijewska et al., 2009; He et al., 2020).
Several approaches have been utilized for hybrid detection. According to previous studies, intermediate coloration (Kemp, 2000; Marie et al., 2007; Hobbs et al., 2013; Hobbs and Allen, 2014; DiBattista et al., 2015), morphology comparison (Wang et al., 2014; He et al., 2019a; He et al., 2020), isozyme electrophoretic patterns (Sick et al., 1963; He and Mork, 2015), diagnostic nuclear marker (van der Meer et al., 2012; Bernardi et al., 2013; Walter et al., 2014; Qu et al., 2018; He et al., 2018a; He et al., 2019b), microsatellites (Harrison et al., 2014; He et al., 2019a; He et al., 2019b; He et al., 2019c), and even genome-wide sequence comparison (Montanari et al., 2014) are all good candidates for identifying and confirming hybridization cases. Diagnostic nuclear markers are one of the most cost-efficient and robust methods with relatively high accuracy for hybridization detection purposes (He et al., 2018a).
A hybrid grouper was noticed by one of the authors (MB) in a photograph posted on social media in February 2017. It showed a spear fisher from Beirut holding three groupers, one of which seemed unusual (Figure 1A). Unfortunately, the fish was consumed but visual identification indicated a possible hybrid between Epinephelus marginatus and Mycteroperca rubra (Figure 1A). Approximately 15 months later, a similar hybrid grouper specimen was speared from the same area (Figure 1B). It was immediately purchased and processed.

Figure 1 (A) Specimen of Epinephelus marginatus (left), hybrid (middle), and Mycteroperca rubra (right) speared near Beirut, Lebanon in February 2017. (B) Hybrid grouper was sampled from off the Beirut port and used in the current study (AUBM OS3979).
In the current study, several genetic markers from mitochondrial and nuclear regions were sequenced on the hybrid and parent species. The parent species candidates were chosen according to phylogenetic relatedness (Ma and Craig, 2018), previous hybridization records (Glamuzina et al., 1999), and visual identification results. The hybridization case could be further supported by subsequent molecular analysis based on the sequenced regions. The diagnostic markers could also be found and used as species delineation and hybridization detection tools for the grouper hybridization case.
The grouper hybrid specimen was collected from off the Beirut port (33.907723°N, 35.504040°E) on May 2, 2018 (Figure 1B). The whole specimen was preserved and deposited in the marine collection of the American University of Beirut (accession number: AUBM-OS3979). Potential parent species (E. costae n=3, E. marginatus n=3, E. aeneus n=3, and M. rubra n=4) were collected from the same region on June 11 and 19, 2018. Fin clips were taken from fresh specimens at the time of collection, preserved in 96% ethanol, and stored at -20°C. A subsample (approximately 2 mm2) of each tissue was retrieved and used for DNA extraction, and samples were extracted using Qiagen DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany; Production Cat. No./ID: 69504).
To investigate potential diagnostic markers for clear discrimination between purebred parent species, nuclear single-copy marker TMO-4c4, nuclear ribosomal protein gene S7, and bone morphogenetic protein gene BMP4 were amplified using TMO-4C4F/TMO-4C4R (Lyons et al., 1997), S7RPEX1F/S7RPEX2R (Chow and Hazama, 1998), and BMP4Cos2FA/BMP4Cos2R primers (Cooper et al., 2009), respectively. To investigate maternal contributions of the hybrid, mitochondrial COI fragments and Cytb fragments were amplified with FishF2/FishR2 (Ward et al., 2005) and Cytb9/Cytb7 (Song et al., 1998). Additional nuclear marker primers, including RAG1 (Lovejoy and Collette, 2001), RAG2 (Cooper et al., 2009), ETS2 (Lyons et al., 1997), and RYR3 (Qu et al., 2018), could not be amplified across all the species. Therefore, they are not discussed further (Appendix file 1).
The QIAGEN Multiplex PCR Kit (Qiagen) was used for PCR. The PCR cycling parameters were as follows: initial denaturation at 95°C for 15 min; followed by 35 cycles of 94°C for 45 sec, annealing for 60 sec (optimal annealing temperatures in Appendix file 1), and 72°C for 60 sec; with a final elongation step of 72°C for 10 min. The PCR products were checked under UV light after running in 1% agarose gel at 90 V for 45 min. All PCR products were cleaned by incubating with exonuclease I and FastAP™ Thermosensitive Alkaline Phosphatase (ExoStar; USB, Cleveland, OH, USA) at 37°C for 60 min followed by incubation at 85°C for 15 min. The final products were sequenced in forward and reverse directions with fluorescently labeled dye terminators following the manufacturer’s protocols (BigDye, Applied Biosystems Inc., Foster City, CA, USA), and they were analyzed using an ABI 3130XL Genetic Analyzer (Applied Biosystems).
The sequences were aligned using the software Geneious Prime (Biomatters Ltd., Auckland, New Zealand), and they were uploaded to GenBank (COI: ON303310-23; Cytb: PP234656-68; TMO-4c4: ON329845-58; S7: ON329831-44; BMP4: ON329859-72). COI fragments from each species were blasted on GenBank, and they were found to be 98%-100% identical to voucher sequences from their respective species (Accessions: MH707768, MH707833, JX456389, MN729687). The mitochondrial DNA sequences of Plectropomus leopardus (COI : MG253533 and Cytb: MG253561, same individual) were downloaded and used as outgroup for subsequent phylogenetic analysis.
When hybridization, gene flow, or incomplete lineage sorting occurs, MrBayes’s Bayesian inference phylogenetic analysis (Ronquist et al., 2012) would show gene tree discordance between different gene markers. Observing the discordance between mitochondrial and nuclear gene trees, introduced by potential hybrid samples, helps indicate the parent species involved in hybridization among the collected samples.
In that regard, separate phylogenetic trees were generated using the MrBayes phylogenetic tree-building plug-in of Geneious prime (Biomatters Ltd., Auckland, New Zealand) based on the concatenated mitochondrial and nuclear gene fragments. The substitution model for each marker (COI: HKY+I; Cytb: HKY+G; BMP4: T92; S7: T92; TMO-4c4: JC69) was obtained using MEGA 6 (Tamura et al., 2013). The MCMC chain length was set up for 1,000,000 generations with a burn-in of 100,000. The average standard deviation of split frequencies (ASDSF) were monitored using the MrBayes plug-in (Ronquist et al., 2012), and they were found to be less than 0.009 when the chains stopped. The genetic difference (p-distance) between purebred species was calculated using MEGA 6 (Tamura et al., 2013).
Haploweb method was used to delineate the affinity of the hybrid individual. Haplotypes of each nuclear marker were inferred using the Bayesian analysis PHASE 2.1 (Stephens and Donnelly, 2003), and they were implemented in DnaSP 5.0 (Librado and Rozas, 2009). Each run was performed with a burn-in of 10,000 generations followed by 1,000,000 generations. Samples with double peaks in sequence chromatograms were found to be heterozygotes (Flot and Tillier, 2006). Two haplotypes of each sequence were separated using SeqPHASE (Flot, 2010). After delineation, heterozygote specimens were represented by two haplotype sequences in the alignments used for network construction. Median-joining networks showing relationships among the haplotypes were generated using NETWORK v4.6.1.3 (Bandelt et al., 1999). Haplowebs were derived from median-joining networks by drawing curved lines to connect haplotypes that co-occurred in heterozygous hybrids (Flot et al., 2010).
The major clades in the Bayesian phylogenetic tree based on mitochondrial fragments were well supported by high posterior probabilities (1), and the tree topology was consistent with a previous study (Ma and Craig, 2018). The hybrid was well clustered with E. marginatus samples (Figure 2), indicating that the maternal material of this hybrid came from a female E. marginatus. The major clades in the nuclear phylogenetic tree were well supported by high posterior probabilities (Figure 2). The clustering result of hybrid specimens showed inconsistency with the mitochondrial marker based tree. The hybrid specimen clustered with M. rubra individuals, indicating that the paternal material came from this species of grouper (Figure 2). Although E. aeneus hybridized with E. marginatus in artificial environments (Glamuzina et al., 1999), this was not the case in the present study. No evidence indicated that E. costae was involved in the hybridization process; however, due to its close phylogenetic relations to hybrid parent species, it was included in the haplotype network/haploweb analysis as an outgroup.
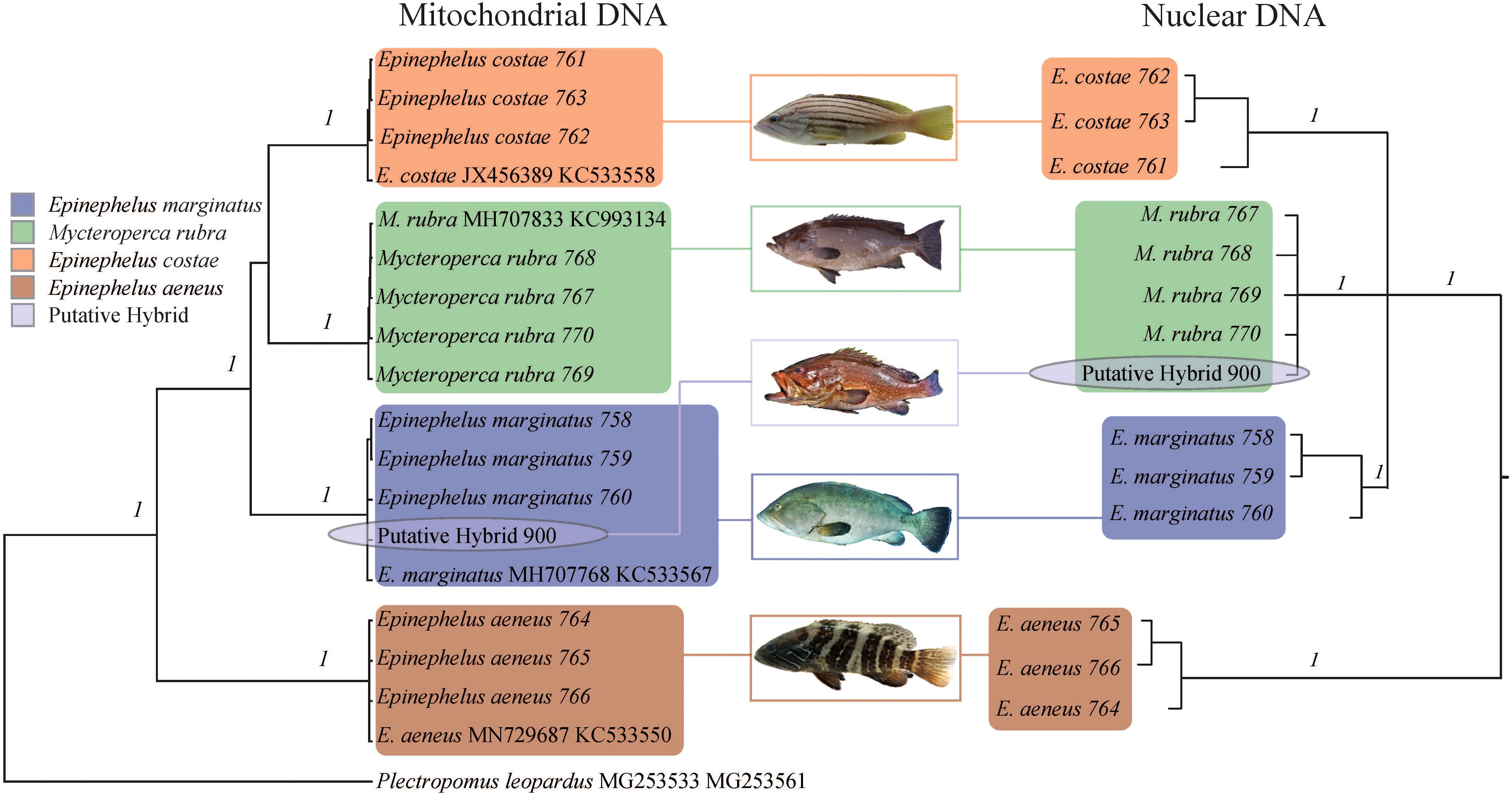
Figure 2 Phylogenetic relationships of Epinephelus marginatus, Mycteroperca rubra, E. costae, E. aeneus, and the hybrid represented in the Bayesian phylogenetic tree based on mitochondrial DNA (COI and Cytb) and three concatenated nuclear DNA markers (S7, BMP4, and TMO-4c4). Each branch represents an individual; the species name and sample ID (for the samples collected during the current study)/GenBank accession number (for downloaded reference data) are indicated in tip labels. Statistical support on each species clade (flagged with a colored rectangle) of the phylogenetic tree was given by Bayesian inference posterior probabilities. Colors denote species as indicated by the legend.
Grouper samples were well discriminated by approximately 60 and 80 substitutions in the 635 bp COI fragments (93 mutations in total, haplotype diversity 0.78) and 703 bp Cytb fragments (126 mutations in total, haplotype diversity 0.98) haplotype networks, respectively. These samples formed reciprocally monophyletic clades in the haplowebs. The hybrid was well blended into the E. marginatus gene pool on mitochondrial markers (Figure 3). The p-distance between purebred E. marginatus and M. rubra (based on COI sequences) was 0.10, which falls in the range of other purebred grouper species involved in previously confirmed hybridization cases (p-distance: 0.00 - 0.17).
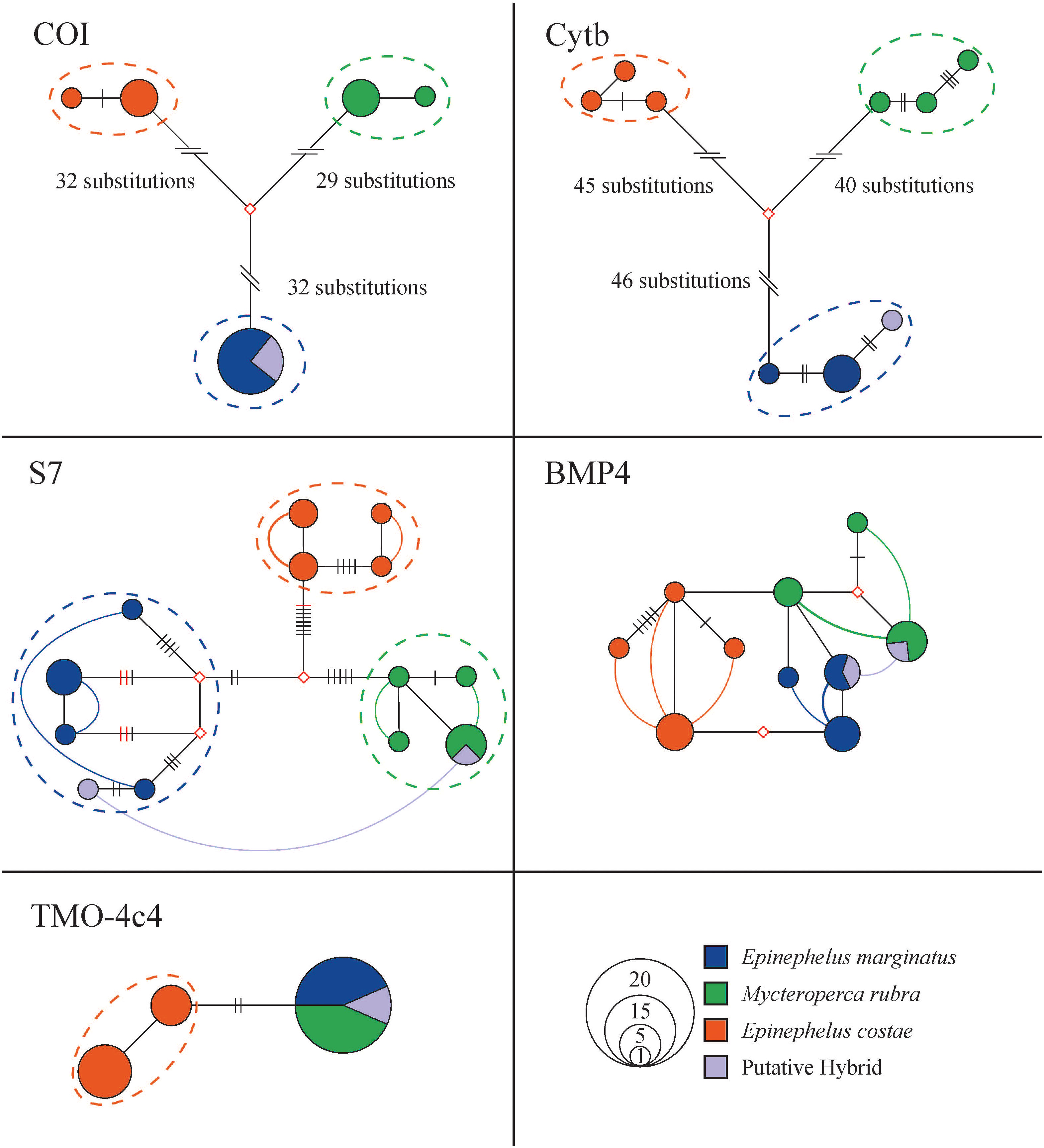
Figure 3 Phylogenetic relationships of Epinephelus marginatus, Mycteroperca rubra, E. costae, E. aeneus, and the hybrid haplotypes represented in network-based haploweb of COI, Cytb, S7, BMP4, and TMO-4c4. Each circle represents a unique haplotype, and the size of the circle is proportional to its total frequency. The red rhombus represents the missing haplotype. Each branch connecting different circles represents a single nucleotide change, and black cross-bars represent an additional nucleotide change. Red cross-bars in the haploweb represent deletion/insertion. The dashed line represents mutually exclusive species gene pools. Curves connecting haplotypes indicate these haplotypes occur in heterozygous individuals. If the curves connect haplotypes within a color, it indicates a heterozygous purebred individuals. The curve connect haplotypes of different colors indicate an individual with potential hybrid lineage. Colors denote sample species as indicated by the legend.
More than 10 mutations well discriminated among E. costae, E. marginatus, and M. rubra individuals in the haploweb based on 512 bp S7 fragments (35 mutations in total, haplotype diversity 0.94. Figure 3). The purebred samples formed reciprocally monophyletic species clades in the haploweb, with purebred heterozygous individuals linking diverse haplotypes within their respective mutually exclusive gene pools. The hybrid specimen, displaying heterozygosity with haplotypes from both E. marginatus and M. rubra clades, provided robust genetic evidence supporting its hybrid identity. Although the hybrid exhibited S7 haplotypes from different origins, it does not necessarily imply this individual is a first-generation hybrid; a recent hybridization event may be the alternative explanation, e.g., if alleles have not yet drifted (if S7 is a neutral marker) or have not yet been selected away (if S7 is a non-neutral marker). The hybrid’s connection of S7 gene pools from both species validates the occurrence of natural hybridization between E. marginatus and M. rubra. The nuclear marker S7 emerges as a valuable diagnostic tool for species delineation and hybrid detection in this specific hybridization context.
In the haploweb based on 483 bp BMP4 nuclear marker fragments (15 mutations in total, haplotype diversity 0.92), clear species boundaries among E. costae, E. marginatus, and M. rubra were not evident. Reciprocal monophyletic species clades were absent. The reticulate haplotype relationships suggest the possible existence of incomplete lineage sorting or hybridization. Even though purebred E. costae, E. marginatus, and M. rubra individuals exhibited distinct haplotypes, the presence of a putative hybrid with BMP4 haplotypes from E. marginatus and M. rubra does not definitively confirm hybridization on this marker. Therefore, the BMP4 fragment could not serve as diagnostic marker for this particular hybridization case (Figure 3).
The 453bp TMO-4c4 fragments from E. costae formed reciprocally monophyletic clade in the haploweb (4 mutations in total, haplotype diversity 0.48). No mutually exclusive gene pools observed between E. marginatus and M. rubra on the TMO-4c4 marker in the haplotype network. This nuclear marker was not suitable for species delineation for these two species (Figure 3).
The study confirmed natural hybridization between E. marginatus and M. rubra. Studying natural hybridization cases would increase the understanding of evolution and biodiversity. For economically valuable species involved in hybridization cases, misidentification of hybrid to parent species might have a negative influence on the effectiveness of catch quota setting. If hybrid offspring have reproduction deficiencies, the significant gamete waste for natural stocks could result due to groupers’ alpha male-dominated reproduction behavior. If the hybrid offsprings are fertile, subsequent repeat backcross to both or one side might cause reverse speciation or so-called hybrid extinction. Through the same route, the escape of artificial grouper hybrids from aquaculture facilities may introduce genetic pollution into the local stock, potentially leading to the extinction of native populations.
For the hybrid under study, maternal material came from the E. marginatus, and paternal material came from the M. rubra. More hybrids need to be sampled and identified using diagnostic markers for further investigation into the origin of reproduction material in this hybridization case to determine if the process is unidirectional or bidirectional.
The nuclear ribosomal protein gene S7 marker could be used as a diagnostic marker for species delineation and hybrid detection. This marker could reliably detect hybrids, including at least first-generation hybrids, between E. marginatus and M. rubra. However, the identification of backcrossing hybrids needs to be further tested. For example, the ability to detect backcrossing would require additional diagnostic markers and ensuring that they are in linkage equilibrium. Previous studies on Plectropomus leopardus and P. maculatus hybridization cases indicated that with two diagnostic markers, the identification accuracy of backcrossed hybrids was approximately 74% as these hybrids had the potential to regain homozygosity in diagnostic nuclear markers (He et al., 2018a).
The difference in genetic similarity (COI p-distance) between the purebred parents E. marginatus and M. rubra, compared with the p-distance of purebred parents in other hybridization cases within Serranidae, was not obvious. Artificial hybridization between E. polyphekadion and E. fuscoguttatus was documented with a similar COI p-distance (~0.11, According to James et al. (1999). And the largest known p-distance between purebred parents for a serranid hybrid is 0.17 (Chen et al., 2017). For predicting reproduction barriers, the current confirmed grouper hybridization cases offer a good reference range of purebred parents species genetic similarity. Future hybridization investigations could utilize this range to estimate reproduction possibilities.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON303310-23, https://www.ncbi.nlm.nih.gov/genbank/, ON329845-58, https://www.ncbi.nlm.nih.gov/genbank/, ON329831-44, https://www.ncbi.nlm.nih.gov/genbank/, ON329859-72, https://www.ncbi.nlm.nih.gov/genbank/, PP234656-68.
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because no live animal testing was performed during the study. The sampling process was only performed on samples from the fish market which were intended for consumption.
SH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. MB: Resources, Validation, Writing – review & editing. LP-A: Data curation, Writing – review & editing. MLB: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Financial support was provided by KAUST to MLB (baseline research funds) and Weida Sci. & Tech. Development Fund to SH.
We thank spearfisher, Mr. Mohamad A. Ibrahim, for providing the sample and sharing his observation on the hybrid grouper. We would also like to thank Mr. Elie Hashem for genetic sample transportation. We thank KAUST Bioscience Core Lab for lab assistance and Foshan Weefine Technology Co.,Ltd for technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Addin A. M., Senoo S. (2011). “Production of hybrid groupers: spotted grouper, Epinephelus polyphekadion X tiger grouper, E. fuscoguttatus and coral grouper, E. Corallicola X tiger grouper,” in International symposium on grouper culture - technological innovation and industrial development, Taiwan, Vol. 1. 1–65.
Bandelt H. J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bernardi G., Noguchi R., Anderson A. B., Floeter S. R., Ferreira C. E. L. (2013). Sargo amarelo, a traditionally recognized hybrid between two species of Brazilian reef fishes. Mar. Biodiversity 43, 255–256. doi: 10.1007/s12526-013-0169-0
Chen J., Ye Z., Yu Z., Li P., Chen X., Liu Y., et al. (2017). The complete mitochondrial genome of the hybrid grouper (Cromileptes altivelis♀× Epinephelus lanceolatus♂) with phylogenetic consideration. Mitochondrial DNA Part B 2, 171–172. doi: 10.1080/23802359.2017.1303346
Ching F. F., Othman N., Anuar A., Shapawi R., Senoo S. (2018). Natural spawning, embryonic and larval development of F2 hybrid grouper, tiger grouper Epinephelus fuscoguttatus × giant grouper E. lanceolatus. Int. Aquat. Res. 10, 391–402. doi: 10.1007/s40071-018-0214-5
Chow S., Hazama K. (1998). Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 7, 1255–1256.
Cooper W. J., Smith L. L., Westneat M. W. (2009). Exploring the radiation of a diverse reef fish family: phylogenetics of the damselfishes (Pomacentridae), with new classifications based on molecular analyses of all genera. Mol. Phylogen Evol. 52, 1–16. doi: 10.1016/j.ympev.2008.12.010
Craig M. T., Sadovy de Mitcheson Y., Heemstra P. C. (2011). Groupers of the World: A Field and Market Guide (Grahamstown, South Africa: NISC (Pty.) Ltd).
DiBattista J. D., Rocha L. A., Hobbs J. A., He S., Priest M. A., Sinclair-Taylor T. H., et al. (2015). When biogeographical provinces collide: hybridization of reef fishes at the crossroads of marine biogeographical provinces in the Arabian Sea. J. Biogeography 42, 1601–1614. doi: 10.1111/jbi.12526
DiBattista J. D., Whitney J., Craig M. T., Hobbs J. A., Rocha L. A., Feldheim K. A., et al. (2016). Surgeons and suture zones: hybridization among four surgeonfish species in the Indo-Pacific with variable evolutionary outcomes. Mol. Phylogenet. Evol. 101, 203–215. doi: 10.1016/j.ympev.2016.04.036
Flot J. F. (2010). SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol. Ecol. Resour. 10, 162–166. doi: 10.1111/j.1755-0998.2009.02732.x
Flot J. F., Couloux A., Tillier S. (2010). Haplowebs as a graphical tool for delimiting species: a revival of Doyle’s “field for recombination” approach and its application to the coral genus Pocillopora in Clipperton. BMC Evolutionary Biol. 10, 372. doi: 10.1186/1471-2148-10-372
Flot J.-F., Tillier S. (2006). “Molecular phylogeny and systematics of the scleractinian coral genus Pocillopora in Hawaii,” in Proceedings of the 10th International Coral Reef Symposium, Okinawa, Japan, Vol. 1. 24–29.
Glamuzina B., Koz˘ul V., Tutman P., Skaramuca B. (1999). Hybridization of Mediterranean groupers: Epinephelus marginatus ♀ X E. aeneus ♂ and early development. Aquaculture Res. 30, 625–628. doi: 10.1046/j.1365-2109.1999.00365.x
Harrison H. B., Feldheim K. A., Jones G. P., Ma K., Mansour H., Perumal S., et al. (2014). Validation of microsatellite multiplexes for parentage analysis and species discrimination in two hybridizing species of coral reef fish (Plectropomus spp., Serranidae). Ecol. Evol. 4, 2046–2057. doi: 10.1002/ece3.1002
He S., Green A. L., Samoilys M. A., Moussa Omar Y., Berumen M. L. (2022). Waif or hybrid? Observation records of rare coloration grouper in Djibouti. Environ. Biol. Fishes 105, 531–536. doi: 10.1007/s10641-022-01253-5
He S., Harrison H. B., Berumen M. L. (2018a). Species delineation and hybrid identification using diagnostic nuclear markers for Plectropomus leopardus and Plectropomus maculatus. Fisheries Res. 206, 287–291. doi: 10.1016/j.fishres.2018.05.022
He S., Johansen J. L., Hoey A. S., Pappas M. K., Berumen M. L. (2019a). Molecular confirmation of hybridization between Dascyllus reticulatus × Dascyllus aruanus from the Great Barrier Reef. Mar. Biodiversity 49, 395–404. doi: 10.1007/s12526-017-0819-8
He S., Mork J. (2015). Characteristic LDH isozyme electrophoretic patterns in six flatfish species in the Trondheimsfjord, Norway and their utility for the detection of natural species hybrids. Mar. Biol. Res. 11, 330–334. doi: 10.1080/17451000.2014.940976
He S., Mork J., Larsen W. B., Møller P. R., Berumen M. L. (2020). Morphology and genetic investigation of flatfish interspecies hybrids (Pleuronectes platessa X Platichthys flesus) from the Baltic Sea. Fisheries Res. 225, 105498. doi: 10.1016/j.fishres.2020.105498
He S., Planes S., Sinclair-Taylor T. H., Berumen M. L. (2019b). Diagnostic nuclear markers for hybrid Nemos in Kimbe Bay, PNG-Amphiprion chrysopterus x Amphiprion sandaracinos hybrids. Mar. Biodiversity 49, 1261–1269. doi: 10.1007/s12526-018-0907-4
He S., Robitzch V., Hobbs J. A., Travers M. J., Lozano-Cortés D., Berumen M. L., et al. (2019c). An examination of introgression and incomplete lineage sorting among three closely related species of chocolate-dipped damselfish (genus: Chromis). Ecol. Evol. 9, 5468–5478. doi: 10.1002/ece3.5142
Hobbs J.-P., Allen G. (2014). Hybridization among coral reef fishes at Christmas Island and the Cocos (Keeling) Islands. Raffles Bull. Zoology 30, 220–226.
Hobbs J. P., Frisch A. J., Allen G. R., Van Herwerden L. (2009). Marine hybrid hotspot at Indo-Pacific biogeographic border. Biol. Lett. 5, 258–261. doi: 10.1098/rsbl.2008.0561
Hobbs J.-P. A., van Herwerden L., Pratchett M. S., Allen G. R. (2013). Hybridization Among Butterflyfishes Biology of Butterfly Fish (FL: CRC Press Press), 48–69.
James C., Al-Thobaiti S., Rasem B., Carlos M. (1999). Potential of grouper hybrid (Epinephelus fuscoguttatus X E. polyphekadion) for aquaculture. Naga 22, 19–23.
Kemp J. M. (2000). Hybridization of the marine angelfishes Pomacanthus maculosus and P. semicirculatus in the Gulf of Aden. Fauna Arabia 18, 357–368.
Kijewska A., Burzyński A., Wenne R. (2009). Molecular identification of European flounder (Platichthys flesus) and its hybrids with European plaice (Pleuronectes platessa). ICES J. Mar. Sci. 66, 902–906. doi: 10.1093/icesjms/fsp110
Kiriyakit A., Gallardo W. G., Bart A. N. (2011). Successful hybridization of groupers (Epinephelus coioides x Epinephelus lanceolatus) using cryopreserved sperm. Aquaculture 320, 106–112. doi: 10.1016/j.aquaculture.2011.05.012
Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu X., Lin H., Huang G. (2007). Preliminary study on the hybrid redspotted grouper (Epinephelus akaara) ♂ X orange-spotted grouper (Epinephelus coioides) ♀. Acta Scientiarum Naturalium Universitatis Sunyatseni 46, 72–75.
Lovejoy N. R., Collette B. B. (2001). Phylogenetic relationships of new world needlefishes (Teleostei: Belonidae) and the biogeography of transitions between marine and freshwater habitats. Copeia 2001, 324–338. doi: 10.1643/0045-8511(2001)001[0324:PRONWN]2.0.CO;2
Lyons L. A., Laughlin T. F., Copeland N. G., Jenkins N. A., Womack J. E., O’Brien S. J. (1997). Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat. Genet. 15, 47–56. doi: 10.1038/ng0197-47
Ma K. Y., Craig M. T. (2018). An inconvenient monophyly: an update on the taxonomy of the groupers (Epinephelidae). Copeia 106, 443–456. doi: 10.1643/CI-18-055
Marie A. D., Van Herwerden L., Choat J. H., Hobbs J.-P. A. (2007). Hybridization of reef fishes at the Indo-Pacific biogeographic barrier: a case study. Coral Reefs 26, 841–850. doi: 10.1007/s00338-007-0273-3
Montanari S. R., Hobbs J. P. A., Pratchett M. S., Bay L. K., Van Herwerden L. (2014). Does genetic distance between parental species influence outcomes of hybridization among coral reef butterflyfishes? Mol. Ecol. 23, 2757–2770. doi: 10.1111/mec.12762
Montanari S. R., Van Herwerden L., Pratchett M. S., Hobbs J. P. A., Fugedi A. (2012). Reeffish hybridization: lessons learnt from butterflyfishes (genus Chaetodon). Ecol. Evol. 2, 310–328. doi: 10.1002/ece3.83
Payet S. D., Hobbs J. A., DiBattista J. D., Newman S. J., Sinclair-Taylor T., Berumen M. L., et al. (2016). Hybridization among groupers (genus Cephalopholis) at the eastern Indian Ocean suture zone: taxonomic and evolutionary implications. Coral Reefs 35, 1157–1169. doi: 10.1007/s00338-016-1482-4
Qu M., Tang W., Liu Q., Wang D., Ding S. (2018). Genetic diversity within grouper species and a method for interspecific hybrid identification using DNA barcoding and RYR3 marker. Mol. Phylogenet. Evol. 121, 46–51. doi: 10.1016/j.ympev.2017.12.031
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sick K., Frydenberg O., Nielsen J. T. (1963). Hemoglobin patterns of plaice, flounder, and their natural and artificial hybrids. Nature 198, 411–412. doi: 10.1038/198411b0
Song C. B., Near T. J., Page L. M. (1998). Phylogenetic relations among percid fishes as inferred from mitochondrial cytochromebDNA sequence data. Mol. Phylogenet. Evol. 10, 343–353. doi: 10.1006/mpev.1998.0542
Stephens M., Donnelly P. (2003). A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169. doi: 10.1086/379378
Sun Y., Guo C., Wang D., Li X. F., Xiao L., Zhang X., et al. (2016). Transcriptome analysis reveals the molecular mechanisms underlying growth superiority in a novel grouper hybrid (Epinephelus fuscogutatus♀× E. lanceolatus♂). BMC Genet. 17, 24–34. doi: 10.1186/s12863-016-0328-y
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tea Y. K., Hobbs J.-P. A., Vitelli F., DiBattista J. D., Ho S. Y. W., Lo N. (2020). Angels in disguise: sympatric hybridization in the marine angelfishes is widespread and occurs between deeply divergent lineages. Proc. R. Soc. London Ser. B 287, 20201459. doi: 10.1098/rspb.2020.1459
van der Meer M. H., Jones G. P., Hobbs J. P., van Herwerden L. (2012). Historic hybridization and introgression between two iconic Australian anemonefish and contemporary patterns of population connectivity. Ecol. Evol. 2, 1592–1604. doi: 10.1002/ece3.251
Walter R. P., Kessel S. T., Alhasan N., Fisk A. T., Heath D. D., Chekchak T., et al. (2014). First record of living Manta alfredi× Manta birostris hybrid. Mar. Biodiversity 44, 1–2. doi: 10.1007/s12526-013-0183-2
Wang Y., Zhang Y., Zhang H. F., Huang W., Xiao L., Li S. S., et al. (2014). Comparison of morphology between two kinds of hybrid groupers and their parents. J. Fisheries China 38, 778–785.
Keywords: grouper, hybridization, Mediterranean Sea, marine biodiversity, biogeography
Citation: He S, Bariche M, Pombo-Ayora L and Berumen ML (2024) Species delineation and hybrid identification using diagnostic nuclear markers for Mediterranean groupers Epinephelus marginatus and Mycteroperca rubra. Front. Mar. Sci. 11:1346535. doi: 10.3389/fmars.2024.1346535
Received: 30 November 2023; Accepted: 09 February 2024;
Published: 29 February 2024.
Edited by:
Anna Rita Rossi, Sapienza University of Rome, ItalyCopyright © 2024 He, Bariche, Pombo-Ayora and Berumen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song He, c29uZy5oZUBrYXVzdC5lZHUuc2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.