- 1Department of Chemistry and State Key Laboratory of Marine Pollution, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 2Golden Honu Services of Oceania, Honolulu, HI, United States
- 3Sea Turtle Association of Japan, Hirakata, Osaka, Japan
- 4AQUARIUM x ART átoa, Kobe, Hyogo, Japan
- 5Graduate School of Information Sciences, Tohoku University, Sendai, Miyagi, Japan
- 6Graduate School of Informatics, Kyoto University, Kyoto, Japan
- 7State Key Laboratory of Marine Environmental Science and College of Ocean & Earth Sciences, Xiamen University, Xiamen, Fujian, China
- 8National Museum of Marine Biology & Aquarium, Pingtung, Taiwan
- 9Department of Marine Biotechnology and Resources, National Sun Yat-Sen University, Kaohsiung, Taiwan
- 10Institute of Marine Ecology and Conservation, National Sun Yat-Sen University, Kaohsiung, Taiwan
- 11Biodiversity Research Centre, Academia Sinica, Taipei, Taiwan
- 12Biodiversity Program, Taiwan International Graduate Program, Academia Sinica and National Taiwan Normal University, Taipei, Taiwan
- 13TurtleSpot Taiwan, Pingtung, Taiwan
- 14Ocean&Fish Research, Busan, Republic of Korea
- 15Department of Ecology and Conservation, National Marine Biodiversity Institute of Korea, Seocheon, Republic of Korea
Understanding the current status and recent development of the population genetics and connectivity of sea turtles is crucial for effective conservation management of the species. Five sea turtle species, green turtle (Chelonia mydas), loggerhead turtle (Caretta caretta), hawksbill turtle (Eretmochelys imbricata), olive ridley turtle (Lepidochelys olivacea) and leatherback turtle (Dermochelys coriacea), are recorded in the East Asia Region situated in the western side of the North Pacific Ocean. We compiled information from 35 published genetic studies on the five sea turtle species, with a focus on green turtle and loggerhead turtle, which are the most studied species (in 30 studies) in view of their commonness and occurrence of nesting populations. We provided an overview of the key methods and findings of these previous studies, addressing two main objectives on genetic structure of the rookeries and their differences compared to other populations, and connectivity of the rookeries and foraging aggregations. By identifying information gaps and conservation needs, we discussed future developments for sea turtle genetic studies and conservation implications in the region.
1 Introduction
Five sea turtle species, green turtle (Chelonia mydas), loggerhead turtle (Caretta caretta), hawksbill turtle (Eretmochelys imbricata), olive ridley turtle (Lepidochelys olivacea) and leatherback turtle (Dermochelys coriacea), are recorded in the East Asia Region situated on the western side of the North Pacific Ocean. To date, a total of 35 genetic studies on the five sea turtle species have been published. Green turtle and loggerhead turtle are the most common species with occurrence of nesting populations and foraging aggregations in the region and hence are the primary focus in 30 of these studies.
Green turtles are widely distributed throughout the East Asia Region, including Japan, the Republic of Korea, Hong Kong, Taiwan, and Mainland China. Nesting sites of green turtles were recorded in numerous locations, such as the Ryukyu Islands and Ogasawara Islands of Japan (Kameda, 2013; Kondo et al., 2017; Okuyama et al., 2020), several islands of Taiwan (Chen and Cheng, 1995; Cheng et al., 2009, Cheng et al., 2015, Cheng et al., 2018), Hong Kong, Huidong of Guangdong Province in Mainland China (Ng et al., 2018) and the South China Sea, including Xisha (Paracel) Islands (Jia et al., 2019), Dongsha (Pratas) Island (Cheng, 1995) and Taiping (Itu Aba) Island (Cheng, 1996). No nesting of green turtle was documented in the Republic of Korea. Important in-water habitats such as migratory corridors of post-nesting green turtles and foraging grounds were identified in Japan, the Republic of Korea, and the South China Region (Cheng, 2000; Hatase et al., 2006; Cheng, 2007; Kuo et al., 2017; Cheng et al., 2018; Ng et al., 2018; Li et al., 2020; Hoh et al., 2022; Kim et al., 2022). These areas were utilized not only by green turtles originating from the East Asia Region but also by nesting green turtles from distant rookeries in the Pacific, as reported in a satellite tracking study by Kolinski et al. (2014). Green turtles in the East Asia Region fall under the Regional Management Units (RMUs) of the “Northwest Pacific” and the “West Pacific/Southeast Asia” identified by Wallace et al. (2010). A recent review initiated by Wallace et al. (2023) has restructured the global RMUs of all sea turtle species, categorizing green turtles in the East Asia Region into a geographically broader RMU of the “East Indian and Southeast Asia”.
Loggerhead turtles in the East Asia Region are found from Hokkaido of Japan in the north to the South China Sea in the south and are documented nesting almost only on Japanese sandy beaches, especially in southern Japan (Kamezaki et al., 2003). Nesting was recorded on Jeju Island in the Republic of Korea (Jung et al., 2012), but no nests were known from Mainland China (Kobayashi et al., 2011; Matsuzawa, 2012). Loggerhead turtles in the East Asia Region fall into the North Pacific RMU reported in both reviews conducted by Wallace et al. (2010) and Wallace et al. (2023). Post-hatchlings of loggerhead turtles disperse into the central and eastern Pacific on the Kuroshio Current and the North Pacific Current (Bowen et al., 1995; Kobayashi et al., 2008) and return to the western Pacific at sizes of approximately 50-70 cm carapace length (Ishihara et al., 2011; Kobayashi et al., 2011; Narazaki et al., 2015). After returning to the waters of East Asia, they undergo foraging migrations and eventually reach sexual maturity (Ishihara and Kamezaki, 2011).
To implement a more effective conservation approach, we aimed to synthesize our current understanding of the population genetics and connectivity of green turtles and loggerhead turtles by providing a literature review of the genetic studies conducted in the East Asia Region. Figure 1 displays the locations of the key places mentioned in this review. We summarized the key methods and findings of the 30 selected studies, identifying the information gaps that exist. Furthermore, we recommended future developments of sea turtle genetic studies and the implications for conservation efforts in the East Asia Region.
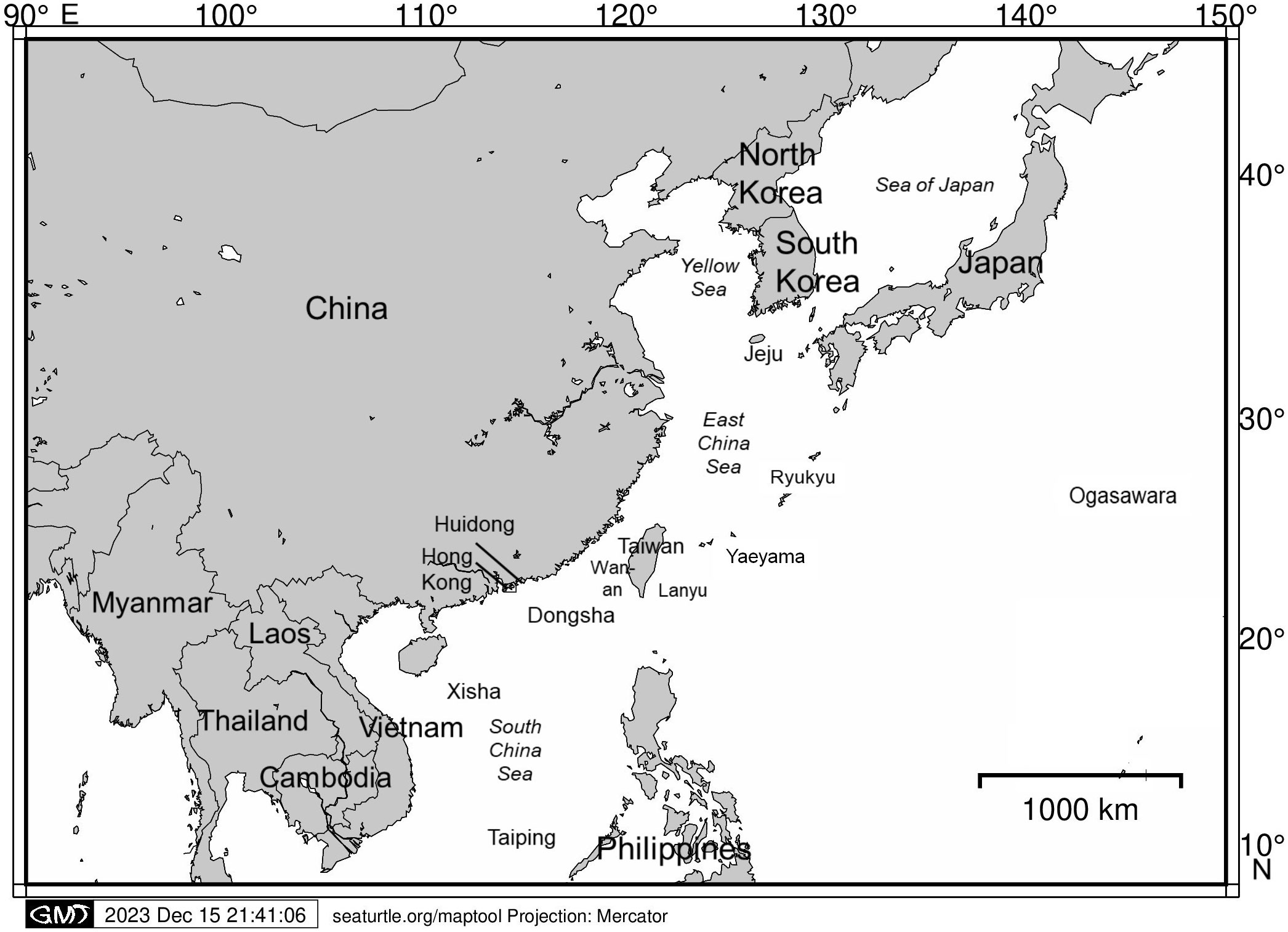
Figure 1 Locations of the key places in the East Asia Region and the vicinity mentioned in this review (basemap prepared by Maptool on http://seaturtle.org/). Please note that Ryukyu, Ogasawara, Jeju, Lanyu, Wan-an, Dongsha, Xisha, Taiping and Yaeyama are islands. Islands are omitted only for simplicity in the map presentation.
2 Literature review
Essential information and findings of the original research articles or master’s and doctoral dissertations on genetic studies conducted in the East Asia Region were analyzed in this literature review. Table 1 summarizes the information and findings from a total of 35 genetic studies on the five sea turtle species recorded in the region (detailed information available in the table in Supplementary Information). The studies were categorized with respect to species and study objectives that were divided into the following three categories: (i) Rookeries connectivity: to elucidate the genetic relationships among rookeries and natal philopatry, (ii) Foraging and rookery connectivity: to elucidate the genetic relationships between foraging grounds and rookeries, (iii) Species ID: to identify species from the genetic perspective. Green and loggerhead turtles were the target species in 30 of these studies with two main study objectives on rookeries connectivity and foraging and rookery connectivity. We, therefore, placed emphasis on the review of the current status and future developments of genetic research on green and loggerhead turtles in this review. On the other hand, the study objective on species ID concerned only olive ridley and hawksbill turtles (See Table 1 and Supplementary Information).
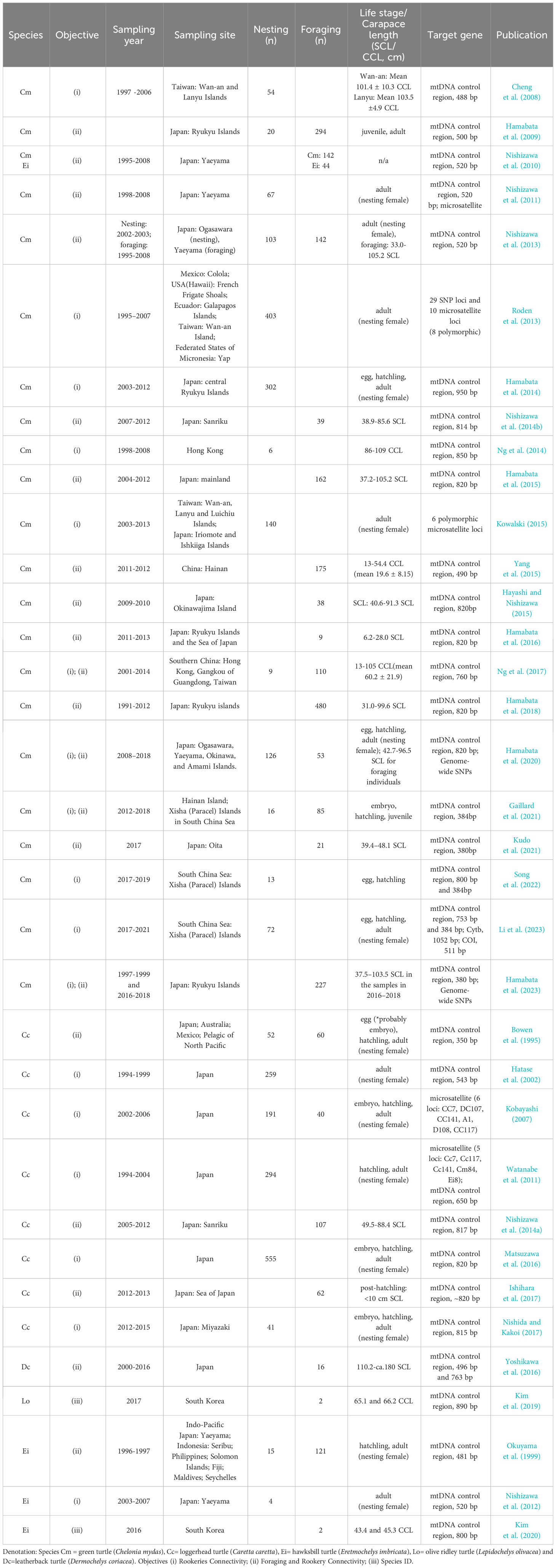
Table 1 Summary of the 35 genetic studies on five sea turtle species in the East Asia Region cited in this review.
3 Population genetics and connectivity of green turtles
A total of 22 studies covering the genetics of green turtles in Japan, Hong Kong, Taiwan, and Mainland China in the East Asia Region were published between 2008 and 2023. These studies are generally categorized into two distinct objectives: (i) Rookeries connectivity; (ii) Foraging and rookery connectivity. The target locus analyzed in 20 of these studies associated with both objectives was the mitochondrial DNA (mtDNA) control region, which allows comparison to a rich database of haplotype sequences based on mtDNA control region from most studies across time.
In the 11 studies focused on rookeries connectivity, skin biopsies, muscle tissue, and blood samples were collected from nesting females, hatchlings, and embryos between 1995 and 2023 in the nesting sites of Japan (such as Ryukyu Islands and Ogasawara Islands) and the South China Region (such as Wan-an Island and Lanyu Island of Taiwan, Gangkou of Guangdong, Sham Wan of Hong Kong and Xisha Islands in South China Sea). Several rookeries, including central Ryukyu Islands of Japan (Hamabata et al., 2014), Lanyu Island (Cheng et al., 2008), Sham Wan (Ng et al., 2014), Gangkou (Ng et al., 2017) and Xisha Islands (Gaillard et al., 2021; Song et al., 2022) in the South China Region, shared some haplotypes of mtDNA control region; for example CmP20 from Taiwan, Xisha Islands and Yaeyama Islands, CmP49 from Taiwan, Xisha Islands and central Ryukyu Islands, CmP18 from Hong Kong, Xisha Islands and Taiwan. CmP49 was also widely observed in Southeast Asia and the Eastern Indian Ocean. However, significantly different haplotype frequencies observed among rookeries in the East Asia Region indicated natal philopatry of female turtles within the groups. There are at least six management units (MUs), genetically distinct breeding populations of sea turtles (Komoroske et al., 2017) in the East Asia Region. MUs contain Yaeyama Islands, central Ryukyu Islands, Ogasawara Islands (Japan), Wan-an Island, Lanyu Island (Taiwan), and Xisha Islands. However, the number of MUs in Japan may increase after more comprehensive surveys using more samples and/or more sites. Importantly, the presence of endemic haplotypes not found elsewhere characterized certain rookeries in Ogasawara Island (Clade VII with CmP127, CmP128, CmP209 in Nishizawa et al., 2013; Hamabata et al., 2020), and Xisha Islands (Clade III with CmP244.1 in Song et al., 2022; Clade VIII with CmP250.1, CmP251.1, CmP252.1, CmP253.1 and CmP254.1 in Li et al., 2023). Habitat protection for these genetically unique rookeries should be accorded with high priority to sustain the essential populations.
In the 15 studies that investigated the source rookeries of the foraging green turtles in Japan and the South China Region, skin biopsies, muscle tissue, and blood samples were collected from bycatch, stranding, or direct take individuals (seen as foraging green turtles from the respective sampling locations) between 1991 and 2023. Specimens were also taken from nesting green turtles in four of the above studies in Japan to expand the baseline data for genetic analysis. Foraging green turtles in the South China Region and Ryukyu Islands of Japan shared similar natal rookeries in Southeast Asia, Yaeyama Islands of Japan, Micronesia and the Marshall Islands in the West Pacific (Nishizawa et al., 2013; Ng et al., 2017; Hamabata et al., 2018). Several studies confirmed that northern foraging green turtles in mainland Japan were primarily contributed by Japanese rookeries in the Ogasawara and Ryukyu Islands (Nishizawa et al., 2013; Nishizawa et al., 2014b; Hamabata et al., 2015, Hamabata et al., 2016, Hamabata et al., 2018). Whereas the southern foraging aggregations around the Yaeyama and Ryukyu Islands were sourced from the Yaeyama and Ogasawara Islands, and various Pacific rookeries in the West Pacific and Indian Oceans and Southeast Asia (Nishizawa et al., 2013; Hamabata et al., 2018). The flow of Kuroshio Current was likely a major factor that affected the composition of the northern and southern foraging aggregations (Nishizawa et al., 2013).
4 Population genetics and connectivity of loggerhead turtles
Eight genetic studies on loggerhead turtles in the East Asia Region were published from 1995 to 2017, and all samples were collected in Japan. Five articles focused on rookeries connectivity, and three articles characterized foraging and rookery connectivity. The genetic analysis targeted the mtDNA control region and/or the nuclear DNA (nDNA) microsatellite loci: where six articles analyzed with mtDNA, one article with nDNA, and one article with both. The length of the analyzed mtDNA control region was about 350 bp in earlier studies until Watanabe et al. (2011). It was extended to about 820 bp, including 350 bp of previous studies after Nishizawa et al. (2014a). Many of the samples for genetic analysis were skin tissues of the fore or hind limbs of nesting and bycatch turtles. Samples from hatchlings and embryos were predominantly tissues from dead specimens, although some studies collected tiny amounts of blood from living hatchlings.
Genetic studies on loggerhead turtles began with Bowen et al. (1995). This landmark study demonstrated that loggerhead turtles migrate across the Pacific Ocean by comparing mtDNA haplotypes of rookeries and foraging grounds on the Pacific scale, implicating Japan as the primary source of juvenile loggerhead turtles in the North Pacific Current and around Baja California. It was the first study to show trans-oceanic migration of sea turtles and brought significant impact on subsequent sea turtle studies and conservation efforts. The next article was published 17 years later in 2002, and subsequent seven publications were sporadic. Loggerhead turtles in the earlier life stages are distributed in the central to eastern Pacific, therefore genetic studies in the East Asia Region focused on rookeries and later life stages in foraging grounds. The five studies focused on rookeries connectivity indicated that nesting populations in Japan were genetically distinctive from rookeries in other RMUs (Hatase et al., 2002; Kobayashi, 2007; Watanabe et al., 2011; Matsuzawa et al., 2016; Nishida and Kakoi, 2017). Matsuzawa et al. (2016) identified three MUs, i.e., Ryukyu, Yakushima, and Mainland MUs, among the samples from 12 broadly selected Japanese loggerhead rookeries.
As to foraging and rookery connectivity, Nishizawa et al. (2014a) is the only published literature that analyzed the genetic structure of the habitual foraging ground of loggerhead turtles in the East Asia Region. The foraging aggregation at the Sanriku coast in Japan, the northern part of the foraging range, contained more than 82.1% of individuals originating from southern nesting sites in Japan. This suggested that loggerhead turtles in the North Pacific generally would not settle in the direct vicinity of their natal sites (Nishizawa et al., 2014a). The main natal nesting sites of post-hatchlings that irregularly mass-stranded in potential foraging grounds along the coast of the Sea of Japan were determined to be areas in southern Japan, including Okinawa Islands, Okinoerabu Island, and Yakushima Island (Ishihara et al., 2017).
5 Discussion
5.1 Knowledge gaps to be filled
For both green and loggerhead turtles in the East Asia Region, we identified several knowledge gaps that need to be addressed regarding sampling location and efforts, collection, and interpretation of ecological baseline information that complements genetic analysis. The specific recommendations are discussed below:
(1) The genetic structures of green and loggerhead turtle populations at major nesting sites have been revealed in the East Asia Region. However, based on the best available information from publications, genetic information is lacking for the foraging aggregations of green turtles in Jeju Island of the Republic of Korea and loggerhead turtles in Japan, other than Sanriku. A satellite telemetry study by Jang et al. (2018) on bycatch green turtles from pound nets revealed that green turtles from different regions, including China and Japan, may use the areas around Jeju Island for foraging, overwintering, and/or as a migratory corridor. Genetic analysis of these bycatch green turtles in Jeju should be conducted to investigate the connectivity of foraging aggregation and natal rookeries. Samples obtained from the foraging ground of loggerhead turtles are limited in location, as they are only from Sanriku in Japan. In addition to the coastal area of the Japanese archipelago, the East China Sea is an important foraging ground for loggerhead turtles nesting in the Ryukyu Islands (Oki et al., 2019; Okuyama et al., 2022). However, it is unclear whether this area is also used by individuals from other nesting sites or at other life stages. It is necessary to collect and analyze samples from a wider and more diverse range of regions and life stages in order to gain a holistic understanding of the relationship between rookeries and foraging grounds. Environmental DNA (eDNA) may also be considered as an alternative to conventional means of monitoring sea turtle populations when sea turtles are not visually observed in potential nesting sites and foraging grounds. This includes detecting and quantifying sea turtle eDNA using species-specific genetic assays in the environmental samples, such as water and nesting beach sand (Farrell et al., 2022). Results from satellite telemetry, mark and recapture, stable isotope ratio, and other approaches should be considered in conjunction with the results of genetic studies.
(2) Most studies in the East Asia Region investigated mtDNA control region sequences, which have been widely used worldwide. Earlier studies targeted mtDNA of about 350bp long in loggerhead turtles and 500bp long in green turtles. Since Nishizawa et al. (2014a) and Hamabata et al. (2014), longer sequences of about 820bp for both species have been sampled, which cover the 350bp and 500bp regions reported in previous studies. However, considering shared haplotypes, especially in loggerhead turtle nesting sites (Hatase et al., 2002; Matsuzawa et al., 2016), this region may not provide sufficient resolution to elucidate population differentiation and connectivity between foraging and nesting sites. The use of nDNA, including microsatellite loci or more recent genome-wide single nucleotide polymorphisms (SNPs), should be further explored. Analysis using microsatellites and SNPs was only reported in 7 studies in this review, but it has gained popularity recently as it offers higher resolution in genetic composition to identify the natal origin and define population structure at a finer scale. For example, Hamabata et al. (2020) and Hamabata et al. (2023) revealed that SNPs were useful in estimating the natal origins of green turtles from the Ogasawara Islands, whose origin was indistinguishable from the Ryukyu Islands according to mtDNA. The use of mitogenomic sequencing should also be considered to improve the resolution of population structure, particularly important in areas where nuclear gene flow along migratory corridors but fine-scale female natal homing might occur. This has been demonstrated for Caribbean green turtles (Shamblin et al., 2012) and Mediterranean loggerhead turtles (Tolve et al., 2023).
(3) Genetic studies with continuous sampling and collection of complementary information on demography are essential for monitoring the genetic diversity of populations and changes in the composition of foraging aggregations. For example, Hamabata et al. (2023) genetically examined the demographic change over twenty years in the foraging aggregation of green turtles around the Yaeyama Islands. They found an increase in the proportion of green turtles originating from Japanese rookeries, especially the local Yaeyama Islands. The relationship between the demographic change in the foraging aggregation and the increase in the number of nesting females in Japan remains unclear because the nesting trends in areas other than Japan are unknown. This emphasizes the importance of understanding demographic changes in other rookeries and foraging aggregations.
5.2 Conservation measures in need
Considering the broad geographic coverage and ecological connectivity of habitats used by sea turtles, as identified by genetic and movement information, it is imperative to establish and reinforce networks among stakeholders, including policymakers, managers, scientists, conservationists, and fisheries industries in the countries and regions concerned. The network of stakeholders should develop cooperation on goal-driven conservation strategies, namely the protection of a network of critical habitats, threat mitigation in activity hotspots and migratory corridors.
Besides the global impact of marine pollution, particularly plastic debris ingestion and entanglement (Savoca et al., 2022), bycatch and fisheries interaction are other imminent threats documented in the East Asia Region, where important in-water habitats, such as migratory corridors of post-nesting green turtles and foraging grounds of green and loggerhead turtles, are located (Cheng and Chen, 1997; Ishihara et al., 2014; Ng et al., 2018). In light of the connectivity and migratory corridors established from genetic and tracking studies, observer programs in collaboration with governments, scientists, and fisheries industries should be pursued. These programs can help identify areas of high bycatch risk in activity hotspots, examine potential interactions of bycatch with oceanography features and fisheries, and explore necessary mitigation measures. One potential approach can be an adaptive management tool similar to TurtleWatch developed by Howell et al. (2008); Howell et al. (2015) and the Turtle Escape Device.
To draw and connect people to conservation efforts, innovative and diverse means of communication among stakeholders, educators, and the public should be facilitated. Social media and communication tools can serve as interactive and effective platforms to call for conservation action and engage the public in citizen scientist programs. For example, TurtleSpot Taiwan is an interactive platform that collects sea turtle sighting data contributed by citizen scientists, where each turtle is identified at the individual level through unique facial-scute patterns and other physical characteristics (Hoh et al., 2022). In Japan, monitoring of nesting counts is conducted by citizen scientists, NGOs, voluntary organizations, students, academia, and local governments throughout the country. These groups mutually share information. Samples from nesting grounds for genetic studies and demographic information have been collected with the continuous support of this community-based monitoring program. Collaboration across sectors and engagement with local communities are essential to build a sustainable thrust in conservation.
Author contributions
CN: Conceptualization, Writing – original draft, Writing – review & editing. TI: Conceptualization, Writing – original draft, Writing – review & editing. TH: Writing – review & editing. HN: Writing – review & editing. ML: Writing – review & editing. J-HS: Writing – review & editing. TL: Writing – review & editing. CF: Writing – review & editing. DM: Writing – review & editing. IK: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Society of Entrepreneurs & Ecology Foundation (grant number: SEE-B-5619 and SEE-B-6158) to ML.
Acknowledgments
We would like to thank the many pioneers who have been involved in previous studies, including the authors and the people who collected the samples for the articles reviewed in this study, for their hard work and enormous efforts. We also thank the authorities that issued the permits for sample collections. The papers authored by CN cited in this review were based on the published work of her PhD study in the City University of Hong Kong and the review work was done in her current capacity as an independent researcher of Golden Honu Services of Oceania.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1325849/full#supplementary-material
References
Bowen B. W., Abreu-Grobois F. A., Balazs G. H., Kamezaki N., Limpus C. J., Ferl R. J. (1995). Trans-Pacific migrations of the loggerhead turtle (Caretta caretta) demonstrated with mitochondrial DNA markers. Proc. Natl. Acad. Sci. U.S.A. 92, 3731–3734. doi: 10.1073/pnas.92.9.3731
Chen T. H., Cheng I. J. (1995). Breeding biology of the green turtle, Chelonia mydas, (Reptilia: Cheloniidae) on Wan-an Island, Peng-Hu Archipelago, Taiwan. I. Nesting ecology. Mar. Biol. 124, 9–15. doi: 10.1007/BF00349141
Cheng I. J. (2000). Post-nesting migrations of green turtles (Chelonia mydas) at Wan-An Island, Penghu Archipelago, Taiwan. Mar. Biol. 137, 747–754. doi: 10.1007/s002270000375
Cheng I. J. (2007). Nesting ecology and postnesting migration of sea turtle on Taipin Tao, Nansha Archipelago, South China Sea. Chelonian Conserv. Biol. 6, 277–282. doi: 10.2744/1071-8443(2007)6[277:NEAPMO]2.0.CO;2
Cheng I. J., Chen T. H. (1997). The incidental capture of give species of sea turtles by coastal setnet fisheries in the eastern waters of Taiwan. Biol. Conserv. 82, 235–239. doi: 10.1016/S0006-3207(97)00027-X
Cheng I. J., Cheng W. H., Chan Y. T. (2018). Geographically closed, yet so different: Contrasting long-term trends at two adjacent sea turtle nesting populations in Taiwan due to different anthropogenic effects. PLoS One 13, e0200063. doi: 10.1371/journal.pone.0200063
Cheng I. J., Dutton P. H., Chen C. I., Chen H. C., Chen Y. H., Shea J. W. (2008). Comparison of the genetics and nesting ecology of two green turtle rookeries in Taiwan. J. Zool. 276, 375–384. doi: 10.1111/j.1469-7998.2008.00501.x
Cheng I. J., Huang C. T., Hung P. Y., Ke B. Z., Kuo C. W., Fong C. L. (2009). Ten years of monitoring the nesting ecology of the green turtle, Chelonia mydas, on Lanyu (Orchid Island), Taiwan. Zool. Stud. 48, 83–94.
Cheng I. J., Lin C. H., Tseng C. T. (2015). Factors influencing variations of oxygen content in nests of green sea turtles during egg incubation with a comparison of two nesting environments. J. Exp. Mar. 471, 104–111. doi: 10.1016/j.jembe.2015.05.013
Farrell J. A., Whitmore L., Mashkour N., Rollinson Ramia D. R., Thomas R. S., Eastman C. B., et al. (2022). Detection and population genomics of sea turtle species via noninvasive environmental DNA analysis of nesting beach sand tracks and oceanic water. Mol. Ecol. Resour. 22, 2471–2493. doi: 10.1111/1755-0998.13617
Gaillard D., Yeh F. C., Lin L., Chen H. Q., Zhang T., Luo S. J., et al. (2021). Lost at sea: determining geographic origins of illegally traded green sea turtles (Chelonia mydas) rescued on Hainan Island, China. Wildl. Res. 48, 55–63. doi: 10.1071/WR19127
Hamabata T., Hikida T., Ishihara T., Kawazu I., Nashiki Y., Oki K., et al. (2016). MtDNA analysis suggests local origin of pelagic-stage juvenile green turtles collected in Japanese coastal waters 1. Pac. Sci. 70, 45–54. doi: 10.2984/70.1.4
Hamabata T., Hikida T., Okamoto K., Watanabe S., Kamezaki N. (2015). Ontogenetic habitat shifts of green turtles (Chelonia mydas) suggested by the size modality in foraging aggregations along the coasts of the western Japanese main islands. J. Exp. Mar. 463, 181–188. doi: 10.1016/j.jembe.2014.12.007
Hamabata T., Kamezaki N., Hikida T. (2014). Genetic structure of green turtle (Chelonia mydas) peripheral populations nesting in the northwestern Pacific rookeries: evidence for northern refugia and postglacial colonization. Mar. Biol. 161, 495–507. doi: 10.1007/s00227-013-2352-z
Hamabata T., Kawata M., Kondo S., Matsuo A., Suyama Y., Suzuki K., et al. (2023). Twenty-year changes in the composition of a mixed stock of foraging green turtles in the Yaeyama Islands of Japan. Mar. Ecol. Prog. Ser. 716, 93–105. doi: 10.3354/meps14367
Hamabata T., Matsuo A., Sato M. P., Kondo S., Kameda K., Kawazu I., et al. (2020). Natal origin identification of green turtles in the North Pacific by genome-wide population analysis with limited DNA samples. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00658
Hamabata T., Nishida S., Kamezaki N., Koike H. (2009). Genetic structure of populations of the green turtle (Chelonia mydas) in Japan using mtDNA control region sequences. Bull. Grad. Sch. Soc Cult. Stud. Kyushu Univ. 15, 35–50. doi: 10.15017/13994
Hamabata T., Nishizawa H., Kawazu I., Kameda K., Kamezaki N., Hikida T. (2018). Stock composition of green turtles Chelonia mydas foraging in the Ryukyu Archipelago differs with size class. Mar. Ecol. Prog. Ser. 600, 151–163. doi: 10.3354/meps12657
Hatase H., Kinoshita M., Bando T., Kamezaki N., Sato K., Matsuzawa Y., et al. (2002). Population structure of loggerhead turtles, Caretta caretta, nesting in Japan: bottlenecks on the Pacific population. Mar. Biol. 141, 299–305. doi: 10.1007/s00227-002-0819-4
Hatase H., Sato K., Yamaguchi M., Takahashi K., Tsukamoto K. (2006). Individual variation in feeding habitat use by adult female green sea turtles (Chelonia mydas): are they obligately neritic herbivores? Oecologia 149, 52–64. doi: 10.1007/s00442-006-0431-2
Hayashi R., Nishizawa H. (2015). Body size distribution demonstrates flexible habitat shift of green turtle (Chelonia mydas). Glob. Ecol. Conserv. 3, 115–120. doi: 10.1016/j.gecco.2014.11.008
Hoh D. Z., Fong C. L., Su H., Chen P., Tsai C. C., Tseng K. W. H., et al. (2022). A dataset of sea turtle occurrences around the Taiwan coast. Biodivers. Data J. 10:e90196. doi: 10.3897/BDJ.10.e90196
Howell E. A., Hoover A., Benson S. R., Bailey H., Polovina J. J., Seminoff J. A., et al. (2015). Enhancing the TurtleWatch product for leatherback sea turtles, a dynamic habitat model for ecosystem-based management. Fish. Oceanogr. 24, 57–68. doi: 10.1111/fog.12092
Howell E. A., Kobayashi D. R., Parker D. M., Balazs G. H., Polovina J. J. (2008). TurtleWatch: a tool to aid in the bycatch reduction of loggerhead turtles Caretta caretta in the Hawaii-based pelagic longline fishery. Endanger. Species Res. 1033, 1–12. doi: 10.3354/esr00096
Ishihara T., Kamezaki N. (2011). Size at maturity and tail elongation of loggerhead turtles (Caretta caretta) in the North Pacific. Chelonian Conserv. Biol. 10, 281–287. doi: 10.2744/CCB-0893.1
Ishihara T., Kamezaki N., Matsuzawa Y., Ishizaki A. (2014). Assessing the status of Japanese coastal fisheries and sea turtle bycatch (in Japanese with English abstract). Wildlife Hum. Soc. 2, 23–35. doi: 10.20798/awhswhs.2.1_23
Ishihara T., Kamezaki N., Matsuzawa Y., Iwamoto F., Oshika T., Miyagata Y., et al. (2011). Reentery of juvenile and subadult loggerhead turtles into natal waters of Japan. Curr. Herpetol. 30, 63–68. doi: 10.5358/hsj.30.63
Ishihara T., Matsuzawa Y., Kamezaki N., Okamoto K., Hamabata T., Aoyagi A., et al. (2017). Mass-stranding suggests natal area and migration of loggerhead turtle hatchlings in the Sea of Japan. Jpn. J. Ecol. 67, 3–12. doi: 10.18960/seitai.67.1_3
Jang S., Balazs G. H., Parker D. M., Kim B., Kim M. Y., Ng C. K. Y., et al. (2018). Movements of green turtles (Chelonia mydas) rescued from pound nets near Jeju Island, Republic of Korea. Chelonian Conserv. Biol. 17, 236–244. doi: 10.2744/CCB-1279.1
Jia Y., Wang J., Balazs G. H., Liu M. (2019). Nest productivity for green turtles (Chelonia mydas) at Qilianyu of Xuande Islands, South China Sea, P.R. China: preliminary findings. Chelonian Conserv. Biol. 18, 116–120. doi: 10.2744/CCB-1307.1
Jung M. M., Moon D. Y., Kim S. H., Kim H. S., Kim J. W. (2012). Environmental conditions as accidental nesting place of sea turtle located in Jeju island of Korea. J. Fish. Mar. Sci. Edu. 24, 507–515. doi: 10.13000/JFMSE.2012.24.4.507
Kameda K. (2013). Green Turtle of Japan. (English translation: Mai Takase) (Hirakata, Japan: Sea Turtle Association of Japan).
Kamezaki N., Matsuzawa Y., Abe O., Asakawa H., Fujii T., Goto K., et al. (2003). ““Loggerhead turtles nesting in Japan,”,” in Loggerhead Sea Turtles. Eds. Bolten A. B., Witherington B. E. (Smithsonian Books, Washington, D.C), 210–217.
Kim I., Park I., Han D., Kim M., Park D., Moon D., et al. (2022). Movement patterns of juvenile loggerhead turtle (Caretta caretta L. 1758) and green turtles (Chelonia mydas L. 1758) hatched in captivity and released in the Korean waters. Animals 12, 2157. doi: 10.3390/ani12162157
Kim I., Yi C., Han D., Park D., Park J., Cho I., et al. (2020). First record of the hawksbill turtle (Eretmochelys imbricata, Reptilia: Testudines: Cheloniidae) from South Korea. J. Asia-Pac. Biodivers. 13, 151–155. doi: 10.1016/j.japb.2020.02.006
Kim I., Yi C., Lee J., Park D., Ho I., Han D., et al. (2019). First record of the olive ridley sea turtle Lepidochelys olivacea (Reptilia: Testudines: Cheloniidae) from South Korea. Curr. Herpetol. 38, 153–159. doi: 10.5358/hsj.38.153
Kobayashi D. R., Cheng I. J., Parker D. M., Polovina J. J., Kamezaki N., Balazs G. H. (2011). Loggerhead turtle (Caretta caretta) movement off the coast of Taiwan: characterization of a hotspot in the East China Sea and investigation of mesoscale eddies. ICES J. Mar. Sci. 68, 707–718. doi: 10.1093/icesjms/fsq185
Kobayashi D. R., Polovina J. J., Parker D. M., Kamezaki N., Cheng I., Uchida I., et al. (2008). Pelagic habitat characterization of loggerhead sea turtles, Caretta caretta, in the North Pacific Ocean, (1997-2006): Insights from satellite tag tracking and remotely sensed data. J. Exp. Mar. Bio. Ecol. 356, 96–114. doi: 10.1016/j.jembe.2007.12.019
Kobayashi E. (2007). Genetic diversity of loggerhead turtle (Caretta caretta) in Japan inferred from microsatellite markers and its application (Tokyo, Japan: Master of Science dissertation, University of Tokyo).
Kolinski S. P., Cruce J. A., Parker D. M., Balazs G. H., Clarke R. (2014). Migrations and conservation implications of post-nesting green turtles from Gielop Island, Ulithi Atoll, Federated States of Micronesia. Micronesica 2014-04, 1–9.
Komoroske L. M., Jensen M. P., Stewart K., Shamblin B. M., Dutton P. H. (2017). Advances in the application of genetics in marine turtle biology and conservation. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00156
Kondo S., Morimoto Y., Sato T., Suganuma H. (2017). Factors affecting the long-term population dynamics of green turtles (Chelonia mydas) in Ogasawara, Japan: Influence of natural and artificial production of hatchlings and harvest pressure. Chelonian Conserv. Biol. 16, 83–92. doi: 10.2744/CCB-1222.1
Kowalski P. (2015). Investigating population genetic structure among green turtle (Chelonia mydas) rookeries of the northwest pacific region (Keelung, Taiwan: Master of Science dissertation, National Taiwan Ocean University).
Kudo H., Nishizawa H., Uchida K., Sato K. (2021). Boldness-exploration behavioral syndrome in wild sub-adult green sea turtles caught at Oita, Japan. Appl. Anim. Behav. Sci. 236, 105216. doi: 10.1016/j.applanim.2021.105216
Kuo F. W., Fan T. Y., Ng C. K. Y., Cai Y., Balazs G. H., Li T. H. (2017). Tale of the unlucky tags: the story of a rescued, rehabilitated, and released green sea turtle (Chelonia mydas) in southern Taiwan. Bull. Mar. Sci. 96, 723–734. doi: 10.5343/bms.2016.1108
Li M., Zhang T., Liu Y., Li Y., Fong J. J., Yu Y., et al. (2023). Revisiting the genetic diversity and population structure of the endangered green sea turtle (Chelonia mydas) breeding populations in the Xisha (Paracel) Islands, South China Sea. PeerJ 11, e15115. doi: 10.7717/peerj.15115
Li T. H., Cai Y. R., Wu P. Y., Ng C. K. Y., Balazs G. H. (2020). Lesson to learn from an endangered green turtle (Chelonia mydas): marine debris ingestion, rehabilitation and satellite tracking. Indian J. Anim. Res. doi: 10.18805/ijar.B-1246
Matsuzawa Y. (2012). ““Breeding ecology - mating and nesting,”,” in Natural History of Sea Turtles in Japan. Ed. Kamezaki N. (Tokyo University Press, Tokyo), 115–140.
Matsuzawa Y., Kamezaki N., Ishihara T., Omuta K., Takeshita H., Goto K., et al. (2016). Fine scale genetic population structure of loggerhead turtles in the Northwest Pacific. Endanger. Species Res. 30, 83–93. doi: 10.3354/esr00724
Narazaki T., Sato K., Miyazaki N. (2015). Summer migration to temperate foraging habitats and active winter diving of juvenile loggerhead turtles Caretta caretta in the western North Pacific. Mar. Biol. 162, 1251–1263. doi: 10.1007/s00227-015-2666-0
Ng C. K. Y., Dutton P. H., Chan S. K. F., Cheung K. S., Qiu J. W., Sun Y. N. (2014). Characterization and conservation concern of green turtles (Chelonia mydas) nesting in Hong Kong, China. Pac. Sci. 68, 231–243. doi: 10.2984/68.2.5
Ng C. K. Y., Dutton P. H., Gu H. X., Li T. H., Ye M. B., Xia Z. R., et al. (2017). Regional conservation implications of green turtle (Chelonia mydas) genetic stock composition in China. Chelonian Conserv. Biol. 16, 139–150. doi: 10.2744/CCB-1253.1
Ng C. K. Y., Gu H. X., Li T. H., Ye M. B., Xia Z. R., Zhang F. Y., et al. (2018). Insights into identifying habitat hotspots and migratory corridors of green turtles in the South China Region. Aquat. Conserv.: Mar. Freshw. 28, 1181–1191. doi: 10.1002/aqc.2923
Nishida S., Kakoi Y. (2017). Mitochondrial DNA polymorphism analysis of loggerhead turtle nesting aggregation in Matsuzaki area, Miyazaki, Japan. Nat. Environ. Miyazaki 2, 36–40.
Nishizawa H., Abe O., Okuyama J., Kobayashi M., Arai N. (2011). Population genetic structure and implications for natal philopatry of nesting green turtles Chelonia mydas in the Yaeyama Islands, Japan. Endanger. Species Res. 14, 141–148. doi: 10.3354/esr00355
Nishizawa H., Naito Y., Suganuma H., Abe O., Okuyama J., Hirate K., et al. (2013). Composition of green turtle feeding aggregations along the Japanese archipelago: Implications for changes in composition with current flow. Mar. Biol. 160, 2671–2685. doi: 10.1007/s00227-013-2261-1
Nishizawa H., Narazaki T., Fukuoka T., Sato K., Hamabata T., Kinoshita M., et al. (2014a). Genetic composition of loggerhead turtle feeding aggregations: migration patterns in the North Pacific. Endanger. Species Res. 24, 85–93. doi: 10.3354/esr00588
Nishizawa H., Narazaki T., Fukuoka T., Sato K., Hamabata T., Kinoshita M., et al. (2014b). Juvenile green turtles on the northern edge of their range: mtDNA evidence of long-distance westward dispersals in the northern Pacific Ocean. Endanger. Species Res. 24, 171–179. doi: 10.3354/esr00592
Nishizawa H., Okuyama J., Abe O., Kobayashi M., Arai N. (2012). Mitochondrial DNA variation in hawksbill turtles (Eretmochelys imbricata) nesting on Ishigaki Island, Japan. Mar. Turtle Newslett. 132, 1–2.
Nishizawa H., Okuyama J., Kobayashi M., Abe O., Arai N. (2010). Comparative phylogeny and historical perspectives on population genetics of the Pacific hawksbill (Eretmochelys imbricata) and green turtles (Chelonia mydas), inferred from feeding populations in the Yaeyama Islands, Japan. Zool. Sci. 27, 14–18. doi: 10.2108/zsj.27.14
Oki K., Hamabata T., Arata T., Parker D. M., Ng C. K. Y., Balazs G. (2019). Inferred adult foraging grounds of two marine turtle species nesting at Amami-Oshima, Japan. Chelonian. Conserv. Biol. 18, 91–97. doi: 10.2744/CCB-1337.1
Okuyama T., Diaz-Fernandez R., Baba Y., Halim M., Abe O., Azeno N., et al. (1999). Genetic diversity of the hawksbill turtle in the Indo-Pacific and Caribbean regions. Chelonian Conserv. Biol. 3, 362–367.
Okuyama J., Ishii H., Tanizaki S., Suzuki T., Abe O., Nishizawa H., et al. (2020). Quarter-century, (1993–2018) nesting trends in the peripheral populations of three sea turtle species at Ishigakijima Island, Japan. Chelonian Conserv. Biol. 19, 101–110. doi: 10.2744/CCB-1428.1
Okuyama J., Watabe A., Takuma S., Tanaka K., Shirai K., Murakami-Sugihara N., et al. (2022). Latitudinal cline in the foraging dichotomy of loggerhead sea turtles reveals the importance of East China Sea for priority conservation. Divers. Distrib. 28, 1568–1581. doi: 10.1111/ddi.13531
Roden S. E., Morin P. A., Frey A., Balazs G. H., Zarate P., Cheng I. J., et al. (2013). Green turtle population structure in the Pacific: new insights from single nucleotide polymorphisms and microsatellites. Endanger. Species Res. 20, 227–234. doi: 10.3354/esr00500
Savoca M. S., Kühn S., Sun C., Avery-Gomm S., Choy C. A., Dudas S., et al. (2022). Towards a North Pacific Ocean long-term monitoring program for plastic pollution: A review and recommendations for plastic ingestion bioindicators. Environ. pollut. 310, 119861. doi: 10.1016/j.envpol.2022.119861
Shamblin B. A., Bjorndal K. A., Bolten A. B., Hillis-Star Z. M., Lundgren I., Naro-Maciels E., et al. (2012). Mitogenomic sequences better resolve stock structure of southern Greater Caribbean green turtle rookeries. Mol. Ecol. 21, 2330–2340. doi: 10.1111/j.1365-294X.2012.05530.x
Song J. H., Lin B. A., Jia Y. Y., Dutton P. H., Kang B., Balazs G. H., et al. (2022). New management unit for conservation of the endangered green turtle Chelonia mydas at the Xisha (Paracel) Islands, South China Sea. Endanger. Species Res. 47, 145–154. doi: 10.3354/esr01172
Tolve L., Iannucci A., Garofalo L., Ninni A., Dondona A. C., Ceciarini I., et al. (2023). Whole mitochondrial genome sequencing provides new insights into the phylogeography of loggerhead turtles (Caretta caretta) in the Mediterranean Sea. Mar. Biol. 171, 19. doi: 10.1007/s00227-023-04325-x
Wallace B. P., DiMatteo A. D., Hurley B. J., Finkbeiner E. M., Bolten A. B., Chaloupka M. Y., et al. (2010). Regional management units for marine turtles: A novel framework for prioritizing conservation and research across multiple scales. PLoS One 5, e15465. doi: 10.1371/journal.pone.0015465
Wallace B. P., Mast R., Posnik Z., Hurley B., Meyer. L., Brenner H., et al. (2023). A new coat of paint for sea turtle RMUs. SWOT Rep. XVIII. 12–15.
Watanabe K. K., Hatase H., Kinoshita M., Omuta K., Bando T., Kamezaki N., et al. (2011). Population structure of the loggerhead turtle, Caretta caretta, a large marine carnivore that exhibits alternative foraging behaviors. Mar. Ecol. Prog. Ser. 424, 273–283. doi: 10.3354/meps08989
Yang W., Wang Y., Chen M. (2015). Genetic structure and diversity of green sea turtle (Chelonia mydas) from South China Sea inferred by mtDNA control region sequence. Biochem. Syst. Ecol. 60, 95–98. doi: 10.1016/j.bse.2015.04.007
Keywords: North Pacific, East Asia, population genetics, connectivity, green turtle, loggerhead turtle
Citation: Ng CKY, Ishihara T, Hamabata T, Nishizawa H, Liu M, Song J-h, Li TH, Fong C-L, Moon DY and Kim IH (2024) Overview of the population genetics and connectivity of sea turtles in the East Asia Region and their conservation implications. Front. Mar. Sci. 11:1325849. doi: 10.3389/fmars.2024.1325849
Received: 22 October 2023; Accepted: 22 February 2024;
Published: 08 March 2024.
Edited by:
Andrea D. Phillott, Flame University, IndiaReviewed by:
Brian Michael Shamblin, University of Georgia, United StatesCopyright © 2024 Ng, Ishihara, Hamabata, Nishizawa, Liu, Song, Li, Fong, Moon and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Connie Ka Yan Ng, a2F5YW4ubmcuY29ubmllQGdtYWlsLmNvbQ==; Takashi Ishihara, aXNoaWhhcmFAdW1pZ2FtZS5vcmc=
 Connie Ka Yan Ng
Connie Ka Yan Ng Takashi Ishihara
Takashi Ishihara Tomoko Hamabata
Tomoko Hamabata Hideaki Nishizawa
Hideaki Nishizawa Min Liu
Min Liu Jia-hao Song
Jia-hao Song Tsung Hsien Li
Tsung Hsien Li Chia-Ling Fong11,12,13
Chia-Ling Fong11,12,13 Il Hun Kim
Il Hun Kim