- 1Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 2Master of Science Program in Applied Microbiology (International Program), Department of Biology, Faculty of Science, Chaing Mai University, Chiang Mai, Thailand
- 3Department of Chemical Engineering and Materials Science, Yuan Ze University, Taoyuan, Taiwan
- 4Multidisciplinary Research Institute, Chiang Mai University, Chiang Mai, Thailand
- 5Center of Excellence in Microbial Diversity and Sustainable Utilization, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 6Environmental Science Research Center, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
Microalgae, especially spirulina, have been globally used as a food supplement due to their rich protein content, safety for human consumption, and provision of enhanced immunomodulatory capabilities. There are, however, few reports that have investigated the immunomodulatory properties of spirulina protein hydrolysate. Consequently, this study aims to optimize the best extraction techniques for spirulina protein hydrolysate and characterize its antioxidant activities and immunomodulation properties in vitro. The results indicated that protein hydrolysate with Flavourzyme and alkaline extraction after ultrasonication and pre-enzymatic assistant with cellulase exhibited superior antioxidant properties compared to other methods. Additionally, all the protein extracts demonstrated a dose-dependent inhibition of nitric oxide production without significantly impacting cell viability. Furthermore, in vitro immunomodulatory properties were evaluated using Candida albicans (DMST 5815) as the test pathogen, with phagocytic activity and index measurements conducted. Notably, the results correlated with the previous assessments, wherein the protein hydrolysate with Flavourzyme displayed the highest phagocytic percentage, measuring 52.3% at a concentration of 10 mg/mL. These findings suggest that enzymatically derived protein hydrolysates from spirulina could serve as a potential source for enhancing immunostimulant activity. Thus, they hold promise as natural bioactive ingredients for therapeutic purposes and the development of functional foods.
1 Introduction
Proteins play a crucial role in every stage of life, from infancy to old age. Insufficient protein consumption can lead to various consequences, including growth failure and a higher rate of infection (Jayawardena et al., 2020). According to the Centers for Disease Control and Prevention, individuals with compromised immune systems are more susceptible to illnesses and infections. Moreover, protein deficiency has become evident in many parts of the world due to food insecurity. Consequently, numerous researchers have sought alternative protein sources to reduce dependency on existing agricultural and food production systems, which may struggle to meet the growing food demand from the increasing global population (Rösch et al., 2019; Béné et al., 2015).
Microalgae might be a good source due to less competition for space and resources during production. Moreover, they contain all essential amino acids and are rich in other compounds such as lipids and carbohydrates, making them suitable as food supplements (Mathur, 2018). In the past, microalgae powder was encapsulated into edible tablets for therapeutic purposes; however, the inclusion of all compounds in these tablets could compromise the properties of some individual components. Consequently, many researchers have focused on extracting specific components, with protein extract garnering considerable attention due to the aforementioned reasons. While several microalgae species boast high protein content in their dry weight, such as Chlorella vulgaris, with 51%–58% protein content (Liu and Hu, 2013), Haematococcus pluvialis with 26% protein content, and Dunaliella salina with 26%–29% protein content (Ba et al., 2016), spirulina stands out as one of the most promising potential protein sources with 70% protein content (El-Kassas et al., 2015).
Spirulina, also known as the biomass of cyanobacterium Arthrospira platensis, has been globally used as a food supplement for many years, owing to its high nutritional values and safety for human consumption. Notably, its protein content in dry weight surpasses that of soybean by 20 times, corn by 40 times, and beef by a staggering 400 times. Additionally, the proteins derived from spirulina have been intensively documented for their numerous health benefits, including enhancement of antioxidant properties, anti-inflammatory effects, and immunomodulatory capabilities (Sedighi et al., 2019). Furthermore, it is important to consider that various protein extraction techniques may influence the bioactive metabolite content of spirulina. For instance, hydrolyzation yields and bioactive peptides from spirulina have been reported to exhibit anticancer properties and effectiveness against immunodeficiency diseases caused by HIV and other viral infections (Sathasivam et al., 2019).
Basically, proteins contain two to 20 amino acids breaking down during gastrointestinal digestion, food processing, or fermentation into peptides (Cian et al., 2012b; Sathya et al., 2021; Qian et al., 2008). Since microalgae have complex structures of carbohydrate-rich cell walls, pretreatment, either mechanically or nonmechanically, is required to support the disruption of the microalgae cell wall (Geada et al., 2021). Depending on the algal species, the specific pretreatments have to be optimized to achieve high protein yields (O’Connor et al., 2022). Once the protein is extracted, enzymatic hydrolysis synthesizes bioactive peptides from microalgal proteins, which are used for clinical applications since it depletes the harmful substances to give a good yield (Sathya et al., 2021). Moreover, different protease enzymes have different preferred cleavage sites, resulting in different bioactive properties. For example, immune peptides are responsible for modulating both lymphocyte proliferation and increasing the phagocytic activity of macrophages in humans (Ahn et al., 2008), and previous studies of macroalgae called Porphyra columbina exhibited that enzymatic hydrolysis using Alcalase and trypsin had immunosuppressive effects on rat splenocytes as they enhanced IL-10 production while the production of TNF-α and IFN-γ was inhibited (Cian et al., 2012b).
Despite previous studies that have reported on anti-inflammatory activities of aqueous spirulina extracts under full-range solar spectrum or controlled light conditions (Tzachor et al., 2021), to our knowledge, no investigation into the immune activities of both crude extracts and protein hydrolysate from A. platensis has been documented previously. This highlights the novelty and significance of exploring the immunomodulatory properties of spirulina-derived extracts, providing a new avenue for potential health applications. Therefore, in this study, we meticulously optimized the methods of cell disruption to enhance the production of protein hydrolysate from spirulina (Arthrospira platensis). Subsequently, we conducted a comprehensive characterization of the spirulina protein to assess its immunomodulatory properties in an in vitro study.
2 Materials and methods
2.1 Spirulina biomass
Dried spirulina (Arthrospira platensis) powder provided by the Algal and Cyanobacterial Research Laboratory, Faculty of Science, Chiang Mai University, Thailand, was used for the extraction of proteins.
2.2 Protein extraction from spirulina
2.2.1 Cell-free lysate by ultrasonic-assisted cell disruption
This method is achieved by means of ultrasonication (Vibra-cell VC505, Sonics & Materials Inc., USA), which disrupts the cell wall completely or partially, and then a mixture of cellular components is released from the spirulina cells into the surrounding medium phosphate buffer (1:20 (w/v) ratio, pH 7). This mechanism could be denoted as cell-free lysate (CFL). After that, centrifugation at 2,000×g for 45 min by Hettich (REF 1206, SN 0027600-03) was done to separate the biomass from the supernatant. The supernatant was taken and lyophilized until it was dried, and this method was regarded as a cell-free lysate by ultrasonic-assisted cell disruption (CFL).
2.2.2 Cell-free lysate by ultrasonic and cellulase-pretreated cells
This method is achieved by means of ultrasonication (Vibra-cell VC505, Sonics & Materials Inc., USA), which disrupts the cell wall completely or partially, and then a mixture of cellular components is released from the spirulina cells into the surrounding medium phosphate buffer (1:20 (w/v) ratio, pH 7). After that, the cellulase purchased from iKnowZyme was added at a concentration of 50 U/mL and incubated at 50°C for 90 min (Le Nguyen Doan et al., 2022). Subsequently, centrifugation at 2,000×g for 45 min by Hettich (REF 1206, SN 0027600-03) was done to separate the biomass from the supernatant. The supernatant was then subjected to lyophilization until it reached a completely dried state, and this method was named as a cell-free lysate by ultrasonic and cellulase-pretreated cells (CFL+C).
2.2.3 Cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells
The last pretreatment technique was referred to as cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells (AAE). Following the incubation of the cell suspension with ultrasonication and cellulase, centrifugation was performed at 2,000×g for 45 min by Hettich (REF 1206, SN 0027600-03). After that, the obtained supernatant was solubilized at alkaline pH using 1 M of NaOH (pH 10). Subsequently, another centrifugation was conducted at 2,000×g for 45 min with the intention of isolating the proteins from the supernatant. It was achieved by treating with 1 M of HCL (pH 3), leading to protein precipitation, and only the pellet was collected for further analysis (Parimi et al., 2015). The entire process is identified as alkaline/acid extraction of ultrasonicated and cellulase-pretreated cells (AAE).
All of the protein samples extracted from various methods were stored at −20°C until use. The cell disruption flow chart is illustrated in Figure 1.
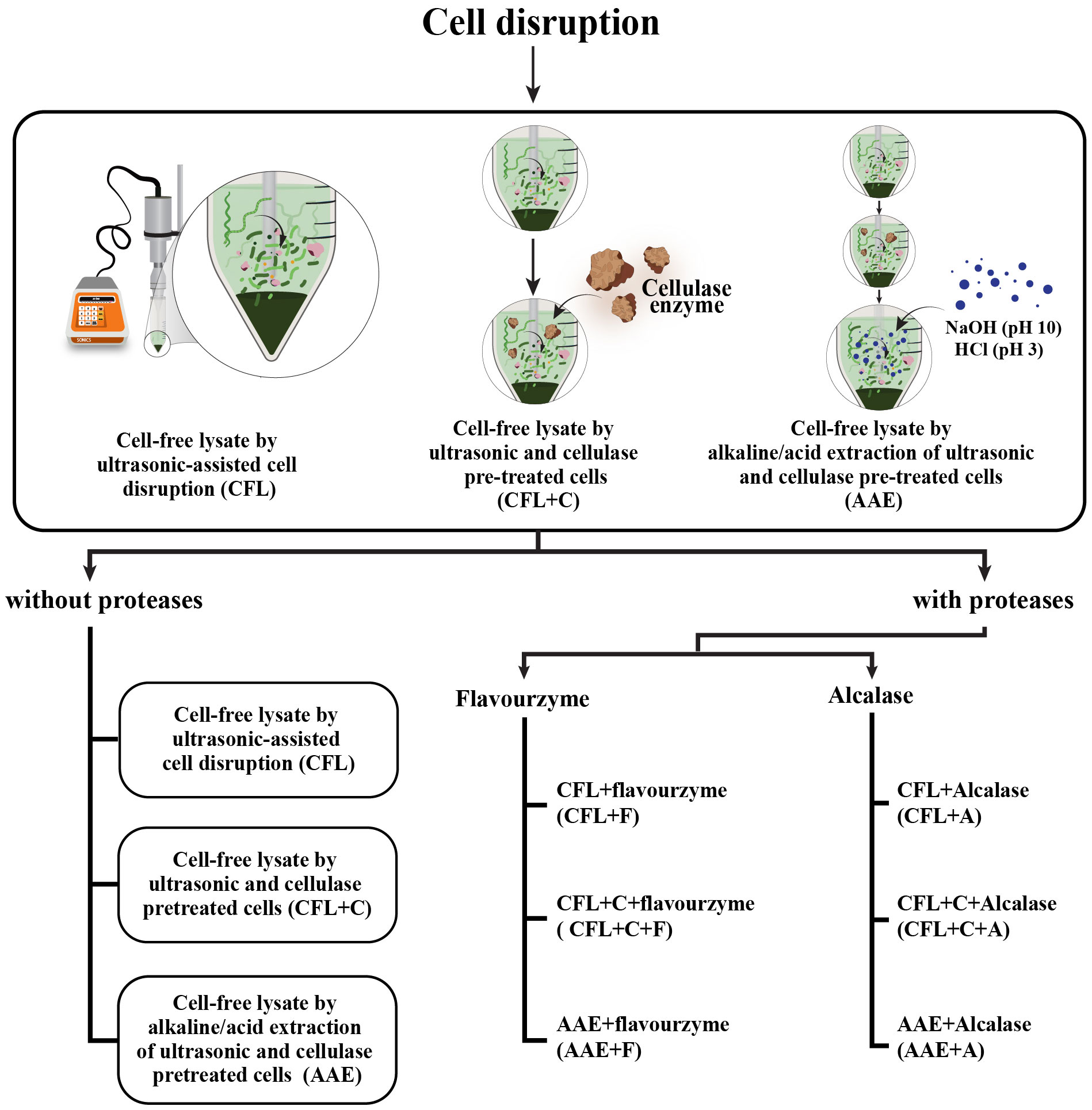
Figure 1 The flowchart depicts the procedural details of the methodology applied in obtaining nine treatments. As regards the first, three treatments called CFL, CFL+C, and AAE were obtained through cell disruption. Subsequently, these three treatments were hydrolyzed using two proteases—Flavourzyme and Alcalase. As a consequence, six additional treatments named CFL+F, CFL+C+F, and AAE+F were obtained using Flavourzyme, whereas CFL+A, CFL+C+A, and AAE+A were derived using Alcalase.
2.3 Preparation of protein hydrolysates
The lyophilized protein obtained from the methods (CFL, CFL+C, and AAE) was dissolved in phosphate buffer (pH 7) to achieve a final concentration of 3.89 mg/mL. Enzymatic hydrolysis employed two different proteases, Alcalase (70 U/mL) and Flavourzyme (≥ 20,000 U/mg), purchased from Novo Nordisk (Bagsverd, Denmark). Hydrolysis was conducted at pH 6.5, a temperature of 63°C, and for a duration of 1 h and 12 min, with an enzyme-to-substrate ratio of 1% (v/v). To ensure enzyme inactivation, the hydrolyzed samples were exposed to a temperature of 90°C for 20 min. Subsequently, centrifugation at 6,000×g at 4°C for 20 min separated the supernatant from other components. The resulting supernatant was stored at −20°C until further experimentation (Pekkoh et al., 2021).
2.4 Analytical methods
2.4.1 Total protein content analysis
The hydrosoluble protein content was determined using the Lowry protein assay with minor modifications. The principle of the Lowry protein assay depends on the complexity of copper with tryptophan and tyrosine residues. Bovine serum albumin (BSA) was set as a standard curve, and nine protein extracts were dissolved in distilled water (1 mg/mL), and the absorbance was measured at 610 nm. The protein concentration was then calculated using the standard curve obtained from the BSA calibration (Lowry et al., 1951).
2.4.2 Total nonprotein component analysis
2.4.2.1 Total sugar content assay
The amount of sugar in each extract was determined by using phenol-sulfuric acid, with two main chemicals—96% sulfuric acid and 5% phenol (DuBois et al., 1956). In the presence of phenol and concentrated sulfuric acid, the sugars, their methyl derivatives, oligosaccharides, and polysaccharides could be determined. The glucose solution was set on a standard curve with various concentrations (0.01–0.25 mg/mL). Briefly, 0.5 mL glucose or protein extracts from spirulina, 0.5 mL of 5% phenol, and 2.5 mL of 96% sulfuric acid were added and incubated at room temperature for 10 min. Blank is performed by using distilled water, and the absorbance was measured at 490 nm.
2.4.2.2 Reducing sugar assay
The presence of reducing sugars in the extracts can be determined by using 3,5-dinitrosalicylic acid (DNS), which detects the presence of a carboxyl group. In brief, the aldehyde group is oxidized into a carboxyl group, and simultaneously, 3,5-dinitrosalicylic acid (DNS) is reduced to 3-amino,5-nitrosalicylic acid under alkaline conditions. The glucose solution was set as a standard curve with various concentrations (0.02–1 mg/mL). Briefly, 0.5 mL of glucose or protein extracts from spirulina and 0.5 mL of DNS were added and incubated at 98°C for 15 min. After that, they were diluted with 5 mL of distilled water. Blank is performed by using distilled water, and the absorbance was measured at 540 nm (DuBois et al., 1956).
2.4.2.3 Total phenolic content assay
The total phenolic content was measured using Folin–Ciocalteu. The Folin–Ciocalteu is an electron transfer-based assay that has a reduced capacity and is expressed as phenolic content. By using a 96-well plate, the phenolic compounds present in different extracts from spirulina were detected. The calibration curve was performed with gallic acid from 0.02mg/mL to 0.2 mg/mL. In brief, 20 µL of nine extracts from spirulina were mixed with 100 µL of a 10% Folin–Ciocalteu solution and incubated at room temperature for 5 min. Thereafter, 80 µL of a 5% sodium carbonate solution was added and incubated for another 1 h (Khan Yusufzai et al., 2018; Aryal et al., 2019). The absorbance was then measured at 765 nm, and the total phenolic content was calculated using Equation 1.
2.5 Analysis of the degree of hydrolysis
The degree of hydrolysis (DH) is highly dependent on the number of peptide bonds after hydrolysis and is related to substrate availability and the specific enzymatic sites of protease that are determined by a specific amino acid composition and sequence. In this study, DH was assessed based on the ratio of amino nitrogen before and after the hydrolysis of protein based on the reaction of primary amino nitrogen with o-phthaldialdehyde (OPA). Briefly, the OPA solution was freshly prepared by dissolving 0.160 g of o-phthaldialdehyde (C8H6O2) in 4 mL of ethanol. Another solution was prepared by dissolving 7.62 g of sodium tetraborate decahydrate (NaC12H25SO4) and 0.2 g of sodium dodecyl sulfate (NaCl2H25SO4) into 150 mL of DI water. Ultimately, 0.176 g of dithiothreitol (C4H10O2S2) was dissolved in 5 mL of DI as the third solution. These solutions were mixed in a volumetric flask and adjusted to the final volume by adding 200 mL of DI, 150 µL of an OPA reagent, and 50 µL of the sample, control blank, and sample blank to the 96-well plates and incubated exactly for 2 min (Pekkoh et al., 2021; Bahari et al., 2020). The absorbance was measured at 340 nm, and the DH was calculated based on Equation 2.
Where, A is the absorbance of the sample after hydrolysis, B is the absorbance of the sample blank after hydrolysis, C is the absorbance of control blank, D is the absorbance of the sample before hydrolysis, and E is the absorbance of a sample blank before hydrolysis.
2.6 Analyzing the biological activities
2.6.1 Potassium ferricyanide-reducing antioxidant power
Reducing the ferricyanide complex (Fe3+) to its ferrous form plays a crucial role in food antioxidants.
Phosphate buffer at 0.2 M (pH 6.6) was prepared, and 30 mL of buffer was added in 1% (w/v) potassium ferricyanide solution. Subsequently, 10% (w/v) trichloroacetic acid (TCA) solution and 0.1% (w/v) ferric chloride FeCl3 were dissolved in 30 mL of DI, 3 g and 0.03 g, respectively. The gallic acid was set as a standard curve (Benzie and Strain, 1999). The experimental procedure involved using 96-well plates, where 145 µL of 0.2 M phosphate buffer and 1% K3Fe(CN)6 were mixed with 60 µL of samples, and the mixture was incubated at 50°C for 20 min. After that, 145 µL of 10% TCA solution was added. The resulting mixture was centrifuged at 3,000×g for 10 min. Following this, 500 µL of DI and 100 µL of 0.1% FeCl3 were added to the supernatant. However, sample control was prepared in the same volume with only 290 µL of buffer, 745 µL of DI, and 60 µL of samples. The absorbance was measured at 700 nm, and increased adsorption of the reaction mixture indicates an increase in reducing power. The activity was calculated using Equation 3.
2.6.2 Antioxidant activity determined by radical cations
The ABTS assay was conducted according to the reference method (Lomakool et al., 2023). In brief, 0.0192 ABTS was dissolved in 5 mL of distilled water. Subsequently, 0.3784 g of potassium persulfate was also dissolved in 10 mL of distilled water. The two solutions were mixed into a 1:1 ratio and kept in the dark for 16 h before use. Next, the mixture was diluted with distilled water, and the absorbance was measured at 734 nm to a value of 0.70 ± 0.02. Trolox was used to establish a standard curve. Using 96-well plates, the ABTS assay was conducted as follows: 10 µL of the sample was mixed with 190 µL of radical cation (ABTS+·) solution and left at room temperature for exactly 6 min. After the incubation, the absorbance was measured at 734 nm. The ABTS+· radical inhibition capacity of the extract was then compared with the Trolox standard to determine its Trolox Equivalent Antioxidant Capacity (TEAC) using Equation 4.
2.6.3 Metal chelating activity
The metal chelating activity was determined following the procedure outlined in reference (Gulcin and Alwasel, 2022). In brief, 2 mM of iron (II) chloride tetrahydrate was dissolved in distilled water to detect this activity. After that 5 mM of ferrozine was also dissolved in distilled water. The EDTA was employed as a standard curve. Using 96-well plates, 400 µL of sample was mixed with 50 µL of FeCl2 and 200 µL of ferrozine. As soon as this step was done, they were incubated at room temperature for 10 min, and the absorbance was measured at 562 nm. The activity was calculated using Equation 5.
2.7 Anti-inflammation activity
2.7.1 Cell culture
RAW264.7 cells obtained from 2711, the Division of Microbiology, Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand, were cultured in Dulbecco’s modified Eagle’s medium (DMEM) Gibco (Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 µg/mL). The cell culture was incubated at 37°C in a humidified atmosphere in a 5% CO2 incubator for 48 h. After this, the cells were seeded in a 96-well plate (2 × 105 cells/well) and kept at 37°C in a humidified atmosphere with a 5% CO2 incubator for 24 h to ensure cell adherence and readiness for further experiments (Flores Hernandez et al., 2017). All reagents and chemicals used were of analytical grade.
2.7.2 Inhibitory effects on nitric oxide production
After being incubated for 24 h, the cells were stimulated with 1 µg/mL LPS (50 µL) for 10 min. The cells were then treated with various concentrations of five different extracts derived from spirulina that showed the highest antioxidant activities during the screening process (0.03–5 mg/mL). The extracts were diluted with DMEM without phenol red (Grand Island, NY, USA) and supplemented with 10% FBS. The untreated cells were set as a control. The cells were then incubated for 24 h at 37°C in a humidified atmosphere with 5% CO2. After incubation, the supernatant from the cells was taken, and nitric oxide production was estimated using the Griess reaction. In brief, the supernatant was mixed with Griess reagent and incubated at room temperature for 15 min. After that, the absorbance of each well was measured at 540 nm by a microplate reader, and the inhibitory effects of the various extracts on nitric oxide (NO) production were measured using Equation 6 (Maruyama et al., 2010).
Where, A0 is the absorbance of the control and A1 is the absorbance of the extract/standard.
2.7.3 Cytotoxicity assay on RAW264.7 cells
Cell viability was measured by 93-(4,5-dimethylthiazol-1-yl)-2,5-diphenyl tetrazolium bromide) MTT assay from Bio Basic (Amherst, NY, USA). The metabolically active cells possess a mitochondrial enzyme called succinate dehydrogenase, which reduces MTT into insoluble purple formazan crystals. The cell viability was measured by using a spectrophotometer. In brief, 30 µL of 2 mg/mL MTT solution was added to each well after the supernatant was removed and incubated for 3 h. Subsequently, 200 µL of dimethyl sulfoxide (DMSO) was added to each well and incubated for another 10 min. The absorbance was measured at 540 nm with a reference wavelength of 630 nm by a microplate reader (Maruyama et al., 2010), and cell viability was measured using Equation 7.
Where, A0 is the absorbance of the control and A1, the absorbance of the extract/standard.
2.8 Characterization of immunomodulatory properties in vitro
2.8.1 Preparation of test pathogen
Candida albicans (DMST 5815), obtained from SCB 2711, the Department of Biology, Faculty of Science, Chiang Mai University, Thailand, was used as a test pathogen for the phagocytosis assay. Sabouraud agar was used as a growth medium for this strain,which was incubated at 37°C for 24–48 h. They are transferred to the same medium without agar to prepare them for the in vitro assay. After that, it was mixed with 1x phosphate buffer (pH 7.4), and the cells were counted and adjusted to 1 × 106 cells/mL by hematocytometer, ensuring a standardized number of cells for the phagocytosis assessment.
2.8.2 Phagocytosis assay in vitro
The phagocytosis assay, as described by the reference, was performed as an in vitro assay (Gabhe et al., 2006). First of all, the blood was collected by the puncture of the caudal vein. As soon as the blood was obtained, it was mixed with ethylenediaminetetraacetic acid (EDTA), an anticoagulant. Next, the blood is diluted with 1× phosphate buffer in a 1:1 ratio (pH 7.4). The white blood cell layer was then collected and separated polymorphonuclear leucocyte (PMN) by Ficoll–Hypaque media with the same ratio (PMN:Ficoll–Hypaque media) and centrifuged at 800×g for 20 min by SANTRIFUJ (Bench-Top Centrifuge) NF800. After that, only PMN was selected and mixed with 1× phosphate buffer in a 1:1 ratio (pH 7.4) three times and centrifuged at 600×g for 7 min in each round.
After that, only PMN was mixed with Candida albicans (DMST 5815). After mixing, various concentrations of four different extracts from spirulina that exhibited the highest antioxidant activities during screening (1–10 mg/mL) were added and incubated at room temperature for 90 min. The control was set without the extract from spirulina. After incubation, cytosmears were prepared, fixed, and stained with Wright–Giemsa stain. The phagocytic cells were visualized under an optical microscope under ×100 using immersion oil, and the phagocytic number and phagocytic index were calculated using Equations 8, 9.
2.9 Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis
The molecular mass distributions of all treatments were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using the TGX Stain-Free™ FastCast™ Acrylamide Kit from Bio-Rad (Hercules, CA, USA). To begin, the samples were suspended in phosphate buffer (pH 7) and mixed with 6% SDS dye. After that, the mixture was incubated at 95°C for 3 min to cleave noncovalent bonds and centrifuged at 6,000×g for 4 min. The Precision Plus Protein™ Dual Color Standards markers ranging from 2 kDa to 250 kDa (Bio-Rad, Hercules, CA, USA) were applied as a reference for the molecular weight of protein bands in the samples. Electrophoresis conditions were set at a constant current setting of 120 V for 120 min (Pekkoh et al., 2021; Meinlschmidt et al., 2016a).
2.10 Statistical analysis
Means of three replicates of determination ± standard deviation (SD) was used. One-way ANOVA using Duncan’s multiple range test (p< 0.05) was used to assess differences in significance values between treatments. Statistical analyses were conducted using SPSS for Windows software (Version 10, Chicago, IL, USA) to determine the significant differences. For the phagocytosis assay, an Analysis of Variance (ANOVA) test was performed in R studio to assess the statistical significance of differences among the means of multiple groups.
3 Results
3.1 Total protein concentration, phenolic content, and sugar assay
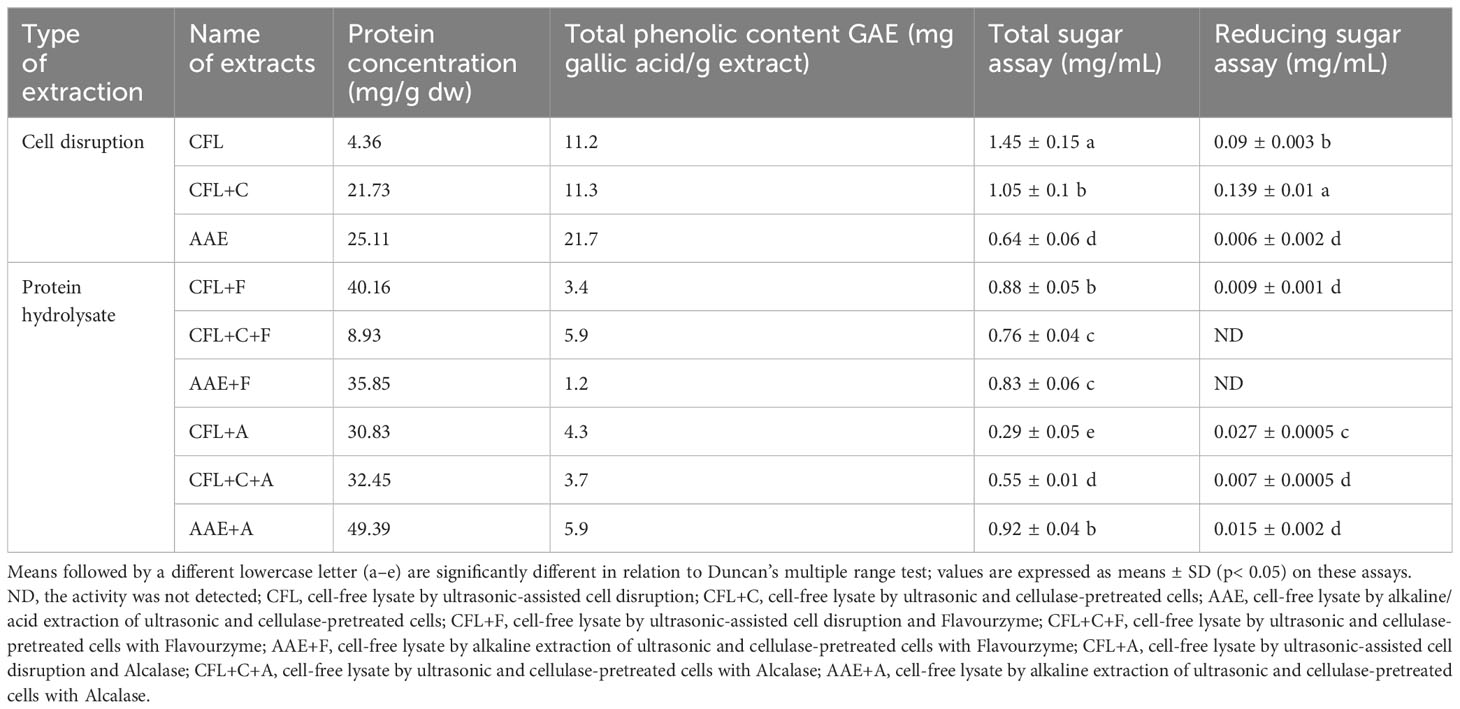
In this section, an assessment is undertaken to quantify the presence of potential interferents that could impact biological activities. This is motivated by the findings of a preceding study indicating that proteins are commonly co-extracted alongside other interferents, such as sugar or phenolic compounds (Peter et al., 2021). Consequently, the detection of phenolic and sugar assays is deemed essential to ascertaining whether the observed activities can be attributed to proteins or interfering substances. Table 1 presents data on total protein concentration, phenolic content, total sugar assay, and reducing sugar assay. The results reveal elevated protein concentrations in the protein hydrolysate compared to other interferents. Consequently, it can be inferred that the bioactive properties, to be discussed subsequently, are primarily derived from proteins, despite potential disruptions from phenolic and sugar content.
3.2 Effect of degree of hydrolysis
In this study, DH was assessed based on the ratio of amino nitrogen before and after the hydrolysis of protein based on the reaction of primary amino nitrogen with o-phthaldialdehyde (OPA). According to the experimental results exhibited in Figure 2, after hydrolysis, the DH increases significantly from 0.018 to 10 with Flavourzyme and Alcalase in the case of CFL. Similarly, CFL+C also demonstrated a sharp increase from 0.025 before and 2.3 with Flavourzyme and Alcalase. There was also a substantial increase from 0.023 to around 7 with Flavourzyme and almost 12 with Alcalase.
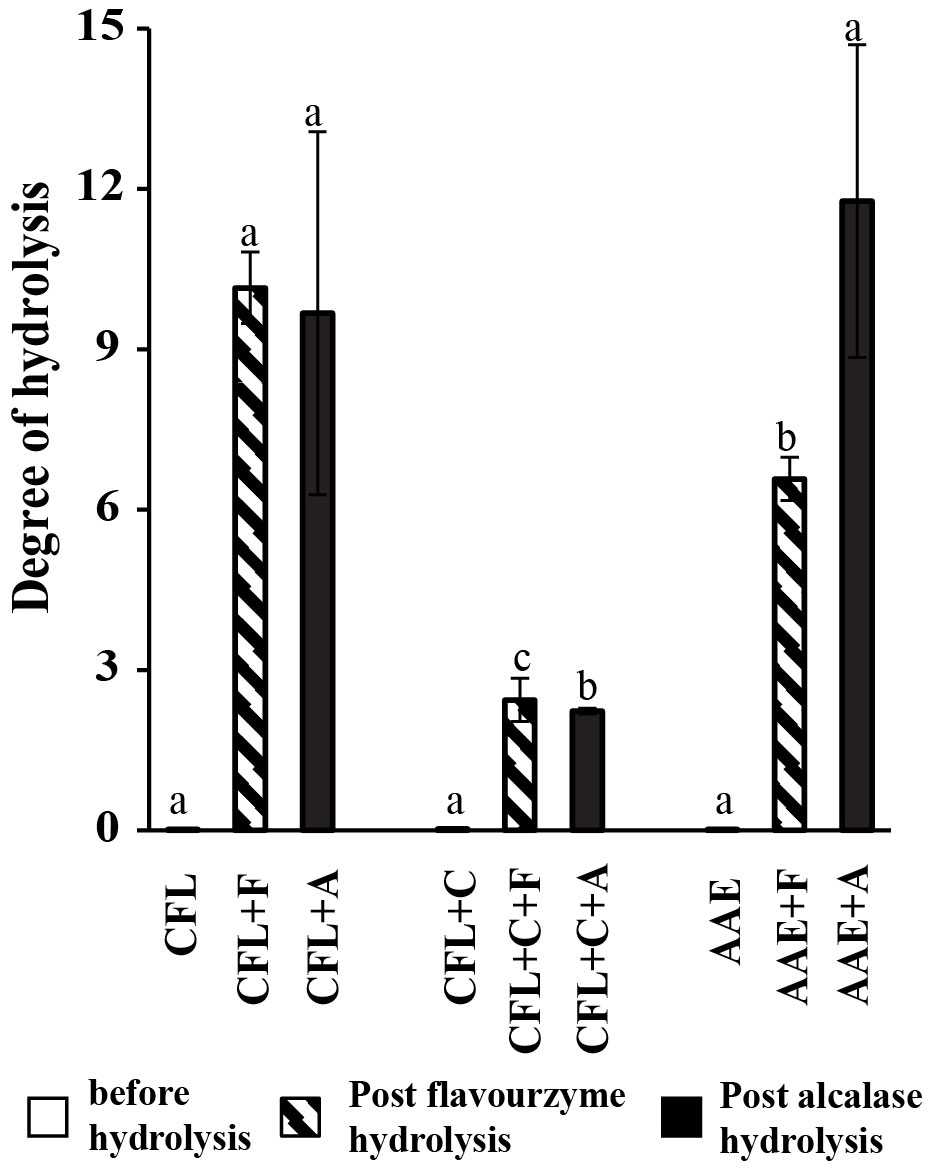
Figure 2 Degree of hydrolysis. Means followed by a different letter (a–c) are significantly different in relation to Duncan’s multiple range test; values are expressed mean ± SD (p > 0.05) before hydrolysis, (p< 0.05) on post-Flavourzyme hydrolysis and post-Alcalase hydrolysis. CFL, cell-free lysate by ultrasonic-assisted cell disruption; CFL+C, cell-free lysate by ultrasonic and cellulase-pretreated cells; AAE, cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells; CFL+F, cell-free lysate by ultrasonic-assisted cell disruption and Flavourzyme, CFL+C+F, cell-free lysate by ultrasonic and cellulase-pretreated cells with Flavourzyme; AAE+F, cell-free lysate by alkaline extraction of ultrasonic and cellulase-pretreated cells with Flavourzyme; CFL+A, cell-free lysate by ultrasonic-assisted cell disruption and Alcalase; CFL+C+A, cell-free lysate by ultrasonic and cellulase-pretreated cells with Alcalase; AAE+A, cell-free lysate by alkaline extraction of ultrasonic and cellulase-pretreated cells with Alcalase.
3.3 Effects on antioxidant activities
3.3.1 Potassium ferricyanide-reducing antioxidant power
The result of the antioxidant activity that was determined by means of potassium ferricyanide-reducing antioxidant power (PFRAP) in relation to a standard curve revealed that the extracts that encountered hydrolysis showed higher antioxidant activities than just a protein extraction. An equal gallic acid equivalent antioxidant capacity was found in AAE and CFL+C+F, with 1.2 in each, followed by CFL+F, which contributed to 0.8. The rest of the six treatments were not more than 0.5, as illustrated in Figure 3A.
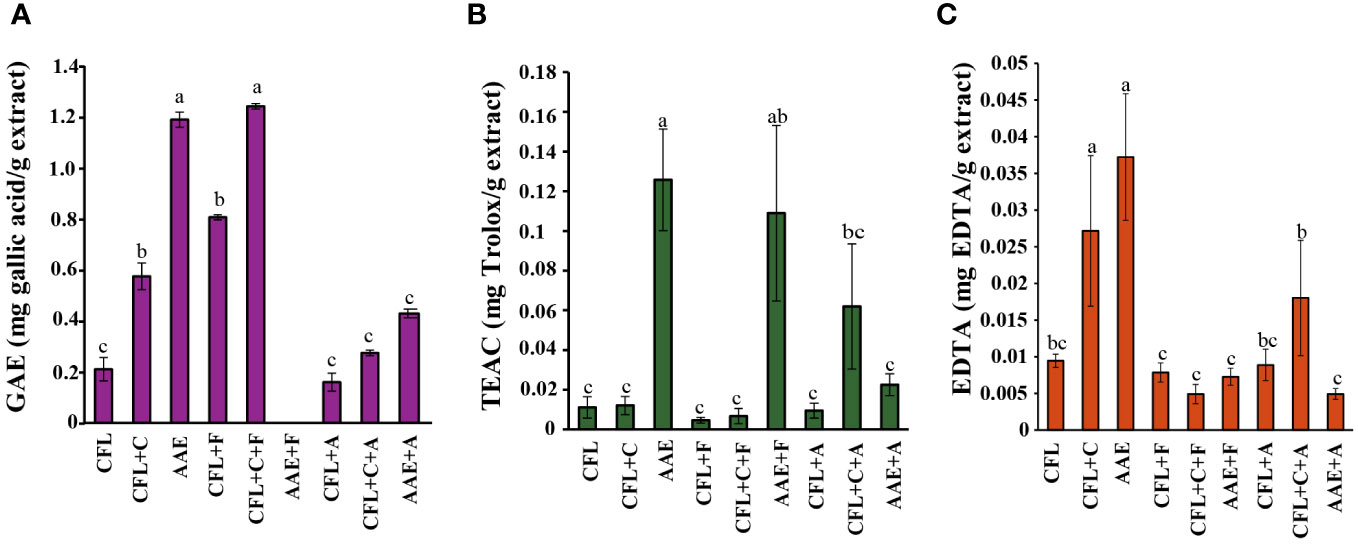
Figure 3 Effects of antioxidant properties. (A) Potassium ferricyanide-reducing antioxidant power (PFRAP), p< 0.05 of GAE (mg gallic acid/g extract), (B) ABTS radical scavenging activity, p< 0.05 of TEAC (mg Trolox/g extract), (C) metal chelating activity, p< 0.05 of EDTA (mg EDTA/g of extract). Means followed by a different letter (a–c) are significantly different in relation to Duncan’s multiple range test. Value are expressed mean ± SD. CFL, cell-free lysate by ultrasonic-assisted cell disruption; CFL+C, cell-free lysate by ultrasonic and cellulase-pretreated cells; AAE, cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells; CFL+F, cell-free lysate by ultrasonic-assisted cell disruption and Flavourzyme; CFL+C+F, cell-free lysate by ultrasonic and cellulase-pretreated cells with Flavourzyme; AAE+F, cell-free lysate by alkaline extraction of ultrasonic and cellulase-pretreated cells with flavozyme; CFL+A, cell-free lysate by ultrasonic-assisted cell disruption and Alcalase; CFL+C+A, cell-free lysate by ultrasonic and cellulase-pretreated cells with Alcalase; AAE+A, cell-free lysate by alkaline extraction of ultrasonic and cellulase-pretreated cells with Alcalase.
3.3.2 Antioxidant activity determined by radical cations
The ABTS radical scavenging activity (Figure 3B) is used to evaluate the ABTS+ stable radical cation, whose intensity decreases in the presence of antioxidants. Similar to the PFRAP method, the Trolox equivalent antioxidant capacity revealed that the maximum antioxidant activity was obtained from AAE, AAE+F, and CFL+C+A, with 0.13, 0.19, and 0.06, respectively.
3.3.3 Metal chelating activity
Metal chelating activity can be considered the most putative and common way to detect antioxidant activity. In the biological system, an excess of free ions could perform induction and form free radicals. The result of the antioxidant activity that was determined by means of metal chelating activity in relation to a standard curve was in accordance with the previous methods where AAE, CFL+C+A, and C had strong chelating activity, amounting to 0.04, 0.02, and 0.03 as depicted in Figure 3C.
3.4 Effects of anti-inflammation
3.4.1 Inhibitory effects of spirulina extracts on nitric oxide production
The five extracts on NO production were estimated using the Griess reagent on RAW246.7 cells at various concentrations from 0.039 mg/mL to 0.16 mg/mL. After being induced by LPS for 24 h, the nitrite content of the supernatants was calculated. The result illustrated that CFL+F contributed the highest percentage of inhibition. It was apparent that the lowest percentage of inhibition came from CFL, not more than 15%.
3.4.2 Cytotoxicity of spirulina extracts on RAW264.7 cells
The cytotoxicity effects of five spirulina extracts on Raw264.7 cell macrophages were evaluated by MTT assay. Various concentrations of samples (0.039–0.156 mg/mL) were tested, and results indicated that all the extracts did not significantly affect cell viability. Consequently, all of the extracts were used for the subsequent experiments since they are considered to be highly safe and noncytotoxic to RAW264.7 cells.
3.5 Effects of spirulina extracts on phagocytic activity
Four extracts (CFL+C+F, CFL+F, AAE, and CFL+A) that exhibited the highest antioxidant and anti-inflammatory properties were further evaluated for phagocytic activity at concentrations of 1 mg/mL, 2 mg/mL, 4 mg/mL, 6 mg/mL, and 10 mg/mL and compared to the control group. When there is an invading pathogen, neutrophils serve as part of the innate immune response and respond at a fast rate. Therefore, the stimulation of neutrophils could result in an increase in the immediate cellular immune response. The result showed that there was a significant increase in the phagocytic activity in CFL+F and CFL+C+F. The former showed the highest activity at concentrations of 6 mg/mL (36.61%) and 10 mg/mL (52.33%), while the latter exhibited the highest activity at the same concentrations, with 39.98% and 42.12%, respectively. However, the other two extracts, AAE and CFL+A, did not show any significant increase in the percentage of phagocytosis versus the control group (Figures 4, 5).
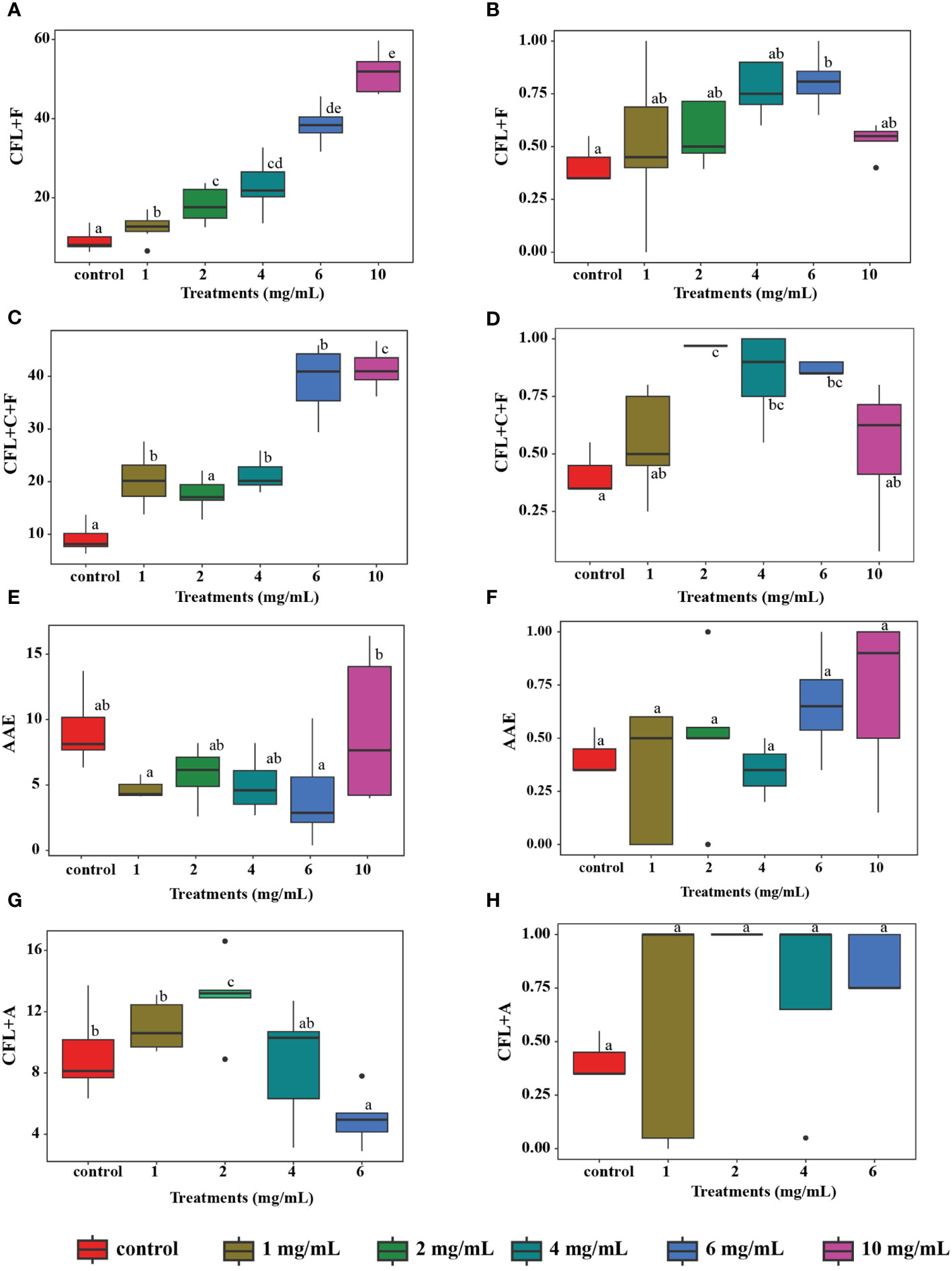
Figure 4 In vitro phagocytosis test: (A, C, E, G) Phagocytic number of different extracts (p< 0.05); (B, D, F, H) The percentage of the phagocytic index of different extracts (p< 0.05). Means followed by a different letter (a–c) are significantly different in relation to Tukey’s HSD, n = 100 cells in each sample. AAE, cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells; CFL+F, cell-free lysate by ultrasonic-assisted cell disruption and Flavourzyme; CFL+C+F, cell-free lysate by ultrasonic and cellulase-pretreated cells with Flavourzyme; CFL+A, cell-free lysate by ultrasonic-assisted cell disruption and Alcalase.
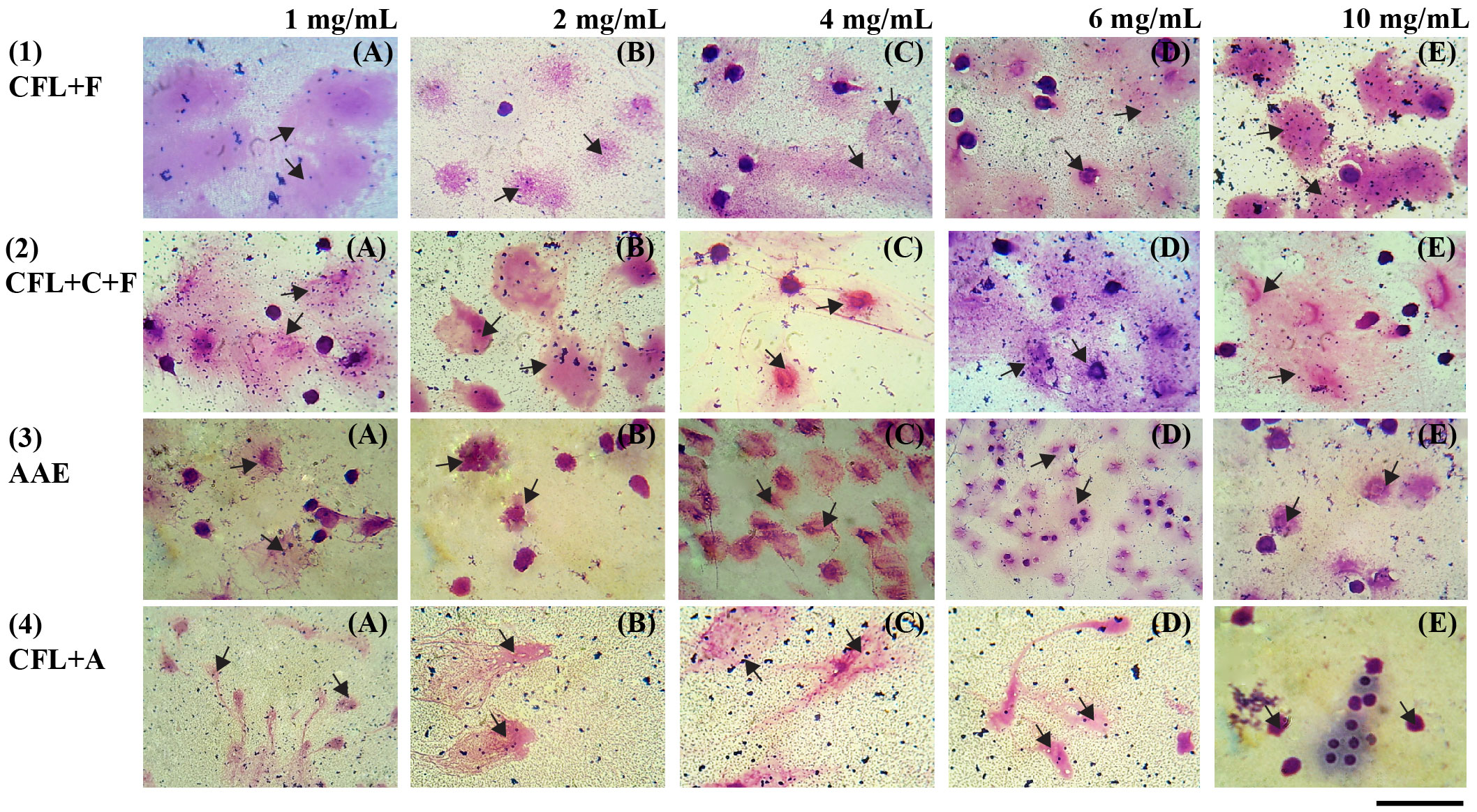
Figure 5 Wright–Giemsa-stained white blood cells (×1,000) show phagocytosis activity at different concentrations: (A) 1 mg/mL, (B) 2 mg/mL, (C) 4 mg/mL, (D) 6 mg/mL, and (E) 10 mg/mL; scale bar (-) = 3µm; arrowhead = phagocytic cells engulfed the yeasts. AAE, cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells; CFL+F, cell-free lysate by ultrasonic-assisted cell disruption and Flavourzyme; CFL+C+F, cell-free lysate by ultrasonic and cellulase-pretreated cells with Flavourzyme; CFL+A, cell-free lysate by ultrasonic-assisted cell disruption and Alcalase.
3.6 SDS-PAGE analysis
The molecular weight distribution of all nine samples was estimated through gel electrophoresis. As shown in Figure 6, the electrophoretic pattern of the CFL, C, and AAE samples exhibited prominent bands with high intensity at molecular weights (Mw) of 75 kDa, 50 kDa, 37 kDa, 25 kDa, 20 kDa, and 15 kDa. Notably, the most intense bands were detected within the 15–25-kDa range. However, the electrophoretic profiles of proteins hydrolyzed using Flavourzyme and Alcalase enzymes displayed distinct characteristics that exhibited around 15 kDa in Flavourzyme-treated samples and a single weak band at Mw< 10 kDa in Alcalase-treated samples.
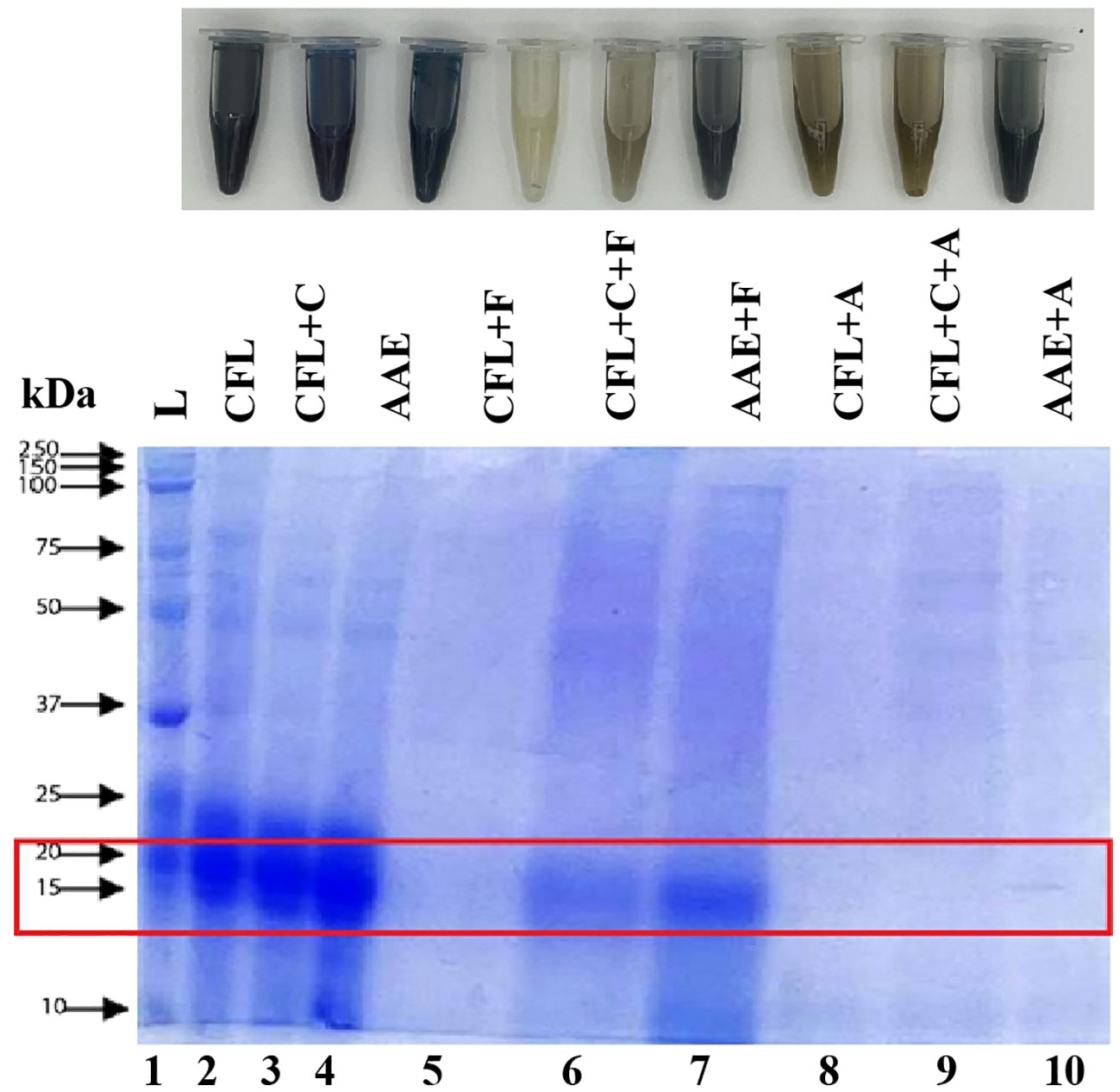
Figure 6 SDS-PAGE analysis. Lane 1 displays Precision Plus Protein™ Dual Color Standards markers ranging from 2 kDa to 250 kDa. Lanes 2–10 are ordered as follows based on their labels: cell-free lysate by ultrasonic-assisted cell disruption (CFL), cell-free lysate by ultrasonic and cellulase-pretreated cells (CFL+C), cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells (AAE), cell-free lysate by ultrasonic-assisted cell disruption and Flavourzyme (CFL+F), cell-free lysate by ultrasonic and cellulase-pretreated cells with Flavourzyme (CFL+C+F), cell-free lysate by alkaline extraction of ultrasonic and cellulase-pretreated cells with Flavourzyme (AAE+F), cell-free lysate by ultrasonic-assisted cell disruption and Alcalase (CFL+A), cell-free lysate by ultrasonic and cellulase-pretreated cells with Alcalase (CFL+C+A), and cell-free lysate by alkaline extraction of ultrasonic and cellulase-pretreated cells with Alcalase (AAE+A).
3.7 Comparison with previous studies on the anti-inflammatory and immunomodulatory assays (in vitro)
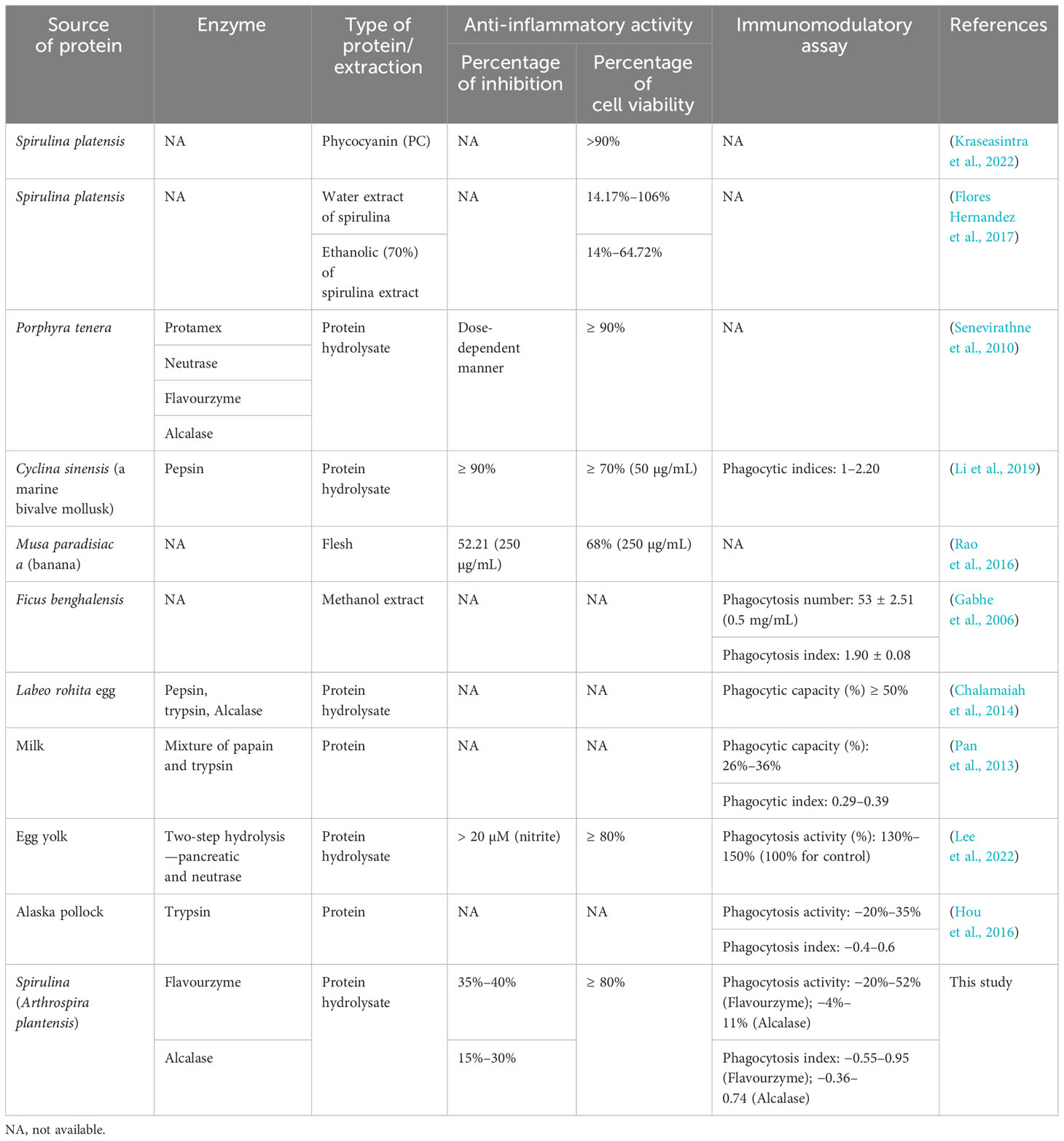
Table 2 shows a comparative analysis of our study’s findings on two different bioactive properties, namely anti-inflammatory activity and immunomodulatory activity, with previous findings. Similar to the bioactive properties obtained from protein hydrolysate of plant- and animal-based sources regarding anti-inflammatory and immunomodulatory properties, protein hydrolysate from spirulina could also provide those properties, indicating that algae-based products hold the potential to serve as a valuable protein source with beneficial properties.
4 Discussion
The percentage of protein content in spirulina is relatively higher than that of lipids, carbohydrates, and ash, standing notably at 70% (Parimi et al., 2015). Additionally, the proteins derived from spirulina have been intensively documented for their numerous health benefits, including enhanced antioxidant properties, anti-inflammatory effects, and immunomodulatory capabilities (Sedighi et al., 2019). Consequently, there has been a remarkable increase in the investigation of protein properties. As depicted in Figure 1, protein extraction was performed by means of ultrasonication as the first treatment, with the aim of disrupting cell wall polysaccharides. The resulting suspension underwent a secondary treatment with cellulase, which has been characterized as a food-degrade approach to breaking down the cell wall. Alkaline extraction was followed as the third treatment. After the cell suspension was subjected to ultrasonication and cellulase, the solution was subjected to NaOH, which is usually used to extract proteins and is used in various food matrices (Naseri et al., 2020). Both the values of the second and third treatments exceeded the protein content achieved through ultrasonication, which was 4.4 mg/g. These findings could deduce that cell disruption by ultrasonication assisted in protein extraction only partially. More protein content was obtained from ultrasonic-assisted cellulase-pretreated cells (21.73 mg/g), which could break down into short-chain cellulose, promoting the release of more intracellular biomolecules after ultrasonication since the cell wall of spirulina is mainly composed of cellulose (Le Nguyen Doan et al., 2022). The protein content showed the best when all the parameters were simultaneously involved as the third treatment, or AAE, with 25 mg/g, where some of the protein might have alkaline-soluble properties when treated with NaOH. It is logical that a higher protein content is obtained with AAE because they can only be extracted in an alkaline environment, unlike the previous two treatments at pH 7, where the solubility of proteins is not high, and therefore the extraction is weaker. Additionally, other compounds are present (simple sugars and phenols). As depicted in Table 1, all of these three parameters did not encounter enzymatic hydrolysis and were denoted as without proteases.
After protein extractions were performed by the three different methods mentioned above, all of the treatments mentioned above encountered enzymatic hydrolysis by using two proteases named Flavourzyme and Alcalase and obtained six further treatments. Enzymatic hydrolysis is more specific to the desired peptides, results in higher peptide yield, and becomes biologically active. Since the DH is highly dependent on the number of peptide bonds after hydrolysis and is related to substrate availability and the specific enzymatic sites of protease that are determined by a specific amino acid composition and sequence, this method was applied in our study. As depicted in Figure 2, the degree of hydrolysis of the spirulina was increased when compared to unhydrolyzed samples at enzyme/substrate ratio (1%) and hydrolysis time (1 h and 12 min). Previous reports by Pekkoh et al. (2021) mentioned that a lower enzyme/substrate ratio with increasing hydrolyzing time will support an increase in the number of peptides cleaved by protease enzymes (Pekkoh et al., 2021). This result was in alignment with the previous report using two different proteases: pepsin, which exhibited a higher degree of hydrolysis (16%), whereas protein hydrolysate with pancreatin showed an 11% degree of hydrolysis in spirulina (Arthrospira platensis) (Mohammadi et al., 2022a). Furthermore, Cian et al. (2012a) showed that protein hydrolysate with Flavourzyme increased the degree of hydrolysis in 7 h to 14% in Porphyra columbina (red seaweed). Similarly, values of the index in the hydrolysates of spirulina (Arthrospira platensis) with Alcalase were revealed (42.4%) (Akbarbaglu et al., 2022).
To elucidate this mechanism, a hypothesis may be proposed that, in the case of the sample of CFL+C, two types of cell disruption were encountered. Ultrasonication partially disrupts the cell wall of the spirulina; subsequently, cellulase enzyme is added in order to hydrolyze the cellulose into short-chain cellulose, promoting the release of intracellular biomolecules, leading to an increased amount of protein in the extract (Le Nguyen Doan et al., 2022). Proteins obtained from this stage are considered intact proteins; as a consequence, the amount of proteins in this sample is 21.73. mg/g dw when detecting with Lowry assay.
On the other hand, when this sample of CFL+C was hydrolyzed with Flavourzyme to obtain CFL+C+F, the Flavourzyme hydrolyzed the intact proteins, which could bind to the active site of the protease Flavourzyme to obtain peptides (Sun et al., 2019). Moreover, Flavourzyme contains a mixture of aminopeptidase, carboxypeptidase, and endoprotease, exhibiting complex cleavage specificity (Xu et al., 2022). The aminopeptidase and carboxypeptidase cleavage peptide bonds in the N-terminus or C-terminus, while the endoprotease breaks the protein molecules, providing more –NH2 and –COOH sites for the action of aminopeptidase and carboxypeptidase (Zhou et al., 2021; Zhang et al., 2023).
Additionally, the electrophoretic pattern of the CFL+C was detected within the 25–25-kDa range, corresponding to the subunits of C-phycocyanin (C-PC), whereas electrophoretic profiles of proteins hydrolyzed using Flavourzyme displayed distinct characteristics where low molecular weight was less visible in Flavourzyme (Figure 6). This might be due to the ability of Flavourzyme, which favors the formation of single amino acids rather than longer polypeptides, which may not be visible on the gel or may be obscured by the dye front (Aiello et al., 2019). Thus, it seems that a complex type of mechanism was established where detecting at wavelength 660 nm by means of a Lowry assay was not accomplished, and further work is required to elucidate it and to understand the possible contributions of protein content in these samples.
Proteins are generally co-extracted with other interferents, such as sugar or phenolic compounds (Peter et al., 2021), so it is often suggested to purify by means of ultrafiltration, ionic-exchange chromatography, and dialysis. However, our study did not conduct purification, and as a consequence, it is important to ensure the target bioactive properties mainly come from the proteins. Table 1 shows the analysis of protein yields together with sugar assay and phenolic compounds, and the result confirmed that the amount of proteins is relatively higher than that of other interferents (sugars and phenolic compounds).
All of the treatments were lyophilized and screened for antioxidant properties as superior activity. Antioxidants play a crucial role in maintaining human health and preventing and treating diseases because of their ability to reduce oxidative stress, which occurs when there is an imbalance between pro-oxidants and antioxidants (Munteanu and Apetrei, 2021). Among the treatments without proteases, AAE had the highest antioxidant activities, which could conclude that protein extraction by conventional methods with slight modifications had the ability against the harmful ABTS+ free radicals through the oxidation reaction (Pekkoh et al., 2022; Conde et al., 2021), PFRAP, which occurs via the action of electron-donating antioxidants as reductants by breaking the free radical chains by donating a hydrogen atom (Giannoglou et al., 2022; Harnedy and FitzGerald, 2013), and metal chelating activity, where molecules in extracts chelate iron ions with biologically active –OH and –OCH3 groups (Ak and Gülçin, 2008a). Spirulina contains various antioxidant compounds, where the subunits of C-phycocyanin (C-PC) the predominant protein in spirulina (20% of the dry biomass), are more soluble in alkaline solutions. Notably, the SDS-polyacrylamide gel electrophoretogram of AAE showed intense bands within the 15–25-kDa range, corresponding to the subunits of C-PC (Aiello et al., 2019), suggesting these compounds could relate with higher antioxidant properties.
The result of antioxidant properties in enzymatic hydrolysis with two proteases—Flavourzyme and Alcalase—exhibited different variations in different methods. For example, CFL+F and CFL+C+F showed the highest antioxidant activities by means of PFRAP whereas AAE+F and CFL+C+A showed the best in ABTS+· hazardous molecules, and all of the treatments did not show significant antioxidant properties in metal chelating activity.
The outcomes are illustrated in Figure 6. The electrophoretic pattern of hydrolyzed proteins is 10–15 kDa, where proteins are cleaved into specific peptide bonds with desired functional properties, and the protein content is relatively higher than that without proteases. However, the processing conditions employed during hydrolysis, such as enzymatic or chemical treatment, pH, and reaction time, influence the availability and reactivity of active fragments required for antioxidant activities. In addition, it is very crucial to note that the complexity of protein hydrolysates, including various peptide fragments and amino acids, could result in elaborate interactions between different functional groups. Consequently, the specific arrangement of these groups within the peptide sequences results in lower antioxidant properties compared to those without proteases (Rao et al., 2016).
Antioxidants play a crucial role in maintaining human health and preventing and treating diseases because of their ability to reduce oxidative stress, which occurs when there is an imbalance between pro-oxidants and antioxidants (Munteanu and Apetrei, 2021). In this study, three different antioxidant properties were screened as superior properties: PFRAP, ABTS radical scavenging activity, and metal chelating activity. Firstly, PFRAP detects the ability of an antioxidant to transfer an electron to reduce metallic ions, carbonyl groups, and free radicals, so this mechanism is known as a single electron transfer reaction, or SET.
Additionally, it is based primarily on the deprotonation and ionization potential of the reactive functional group. Therefore, SET reactions are pH-dependent, and PFRAP was done under acidic conditions. However, it is suggested to test with other methods to distinguish the dominant mechanisms for different antioxidants since this method has a low relation with the process of radical extinction (HAT mechanism). Unlike PFRAP, the ABTS radical scavenging activity can be screened over a wide pH range, and this test offers the determination of a large variety of antioxidant substances. For example, ABTS+· radical reacts rapidly with both synthetic and natural antioxidant substances (i.e., phenols, amino acids, peptides, vitamin E, and vitamin C) in food components. Moreover, it is interesting to screen metal chelating properties that link existing ions or molecules of a ligand to a central metal atom or ion through an acyclic or ring-like coordination bond. For instance, in this study, molecules in different treatments chelate iron ions with biological compounds containing two or more –OH, –COOH, –SH, –OCH3, –C=O, –PO3H2, –NR2, –O, and –S functional groups in a suitable function structure. Consequently, it can be concluded that the extracts that revealed the best antioxidant properties in all methods are potentially more effective in protecting biological systems from oxidative stress and related damage. By neutralizing free radicals and reducing oxidative stress, other undesirable consequences, such as the activation of proinflammatory transcription factors and reducing the production of inflammatory mediators such as cytokines, might be prohibited.
After the superior antioxidant properties screening, the top five extracts that exhibited the highest antioxidant properties were further screened for the production of nitric oxide on Raw264.7 cells. In murine macrophage RAW 264.7 cells, stimulation with LPS only has been indicated to induce iNOS transcription and its protein synthesis, which could raise NO production (Flores Hernandez et al., 2017). Macrophages, as important components of the immune system, play an essential role in controlling various immunopathological phenomena, for example, the overproduction of pro-inflammatory cytokines and inflammatory cytokines like IL-1ß, IL-6, NO, iNOS, COX-2, and TNF during inflammation in response to LPS.
Moreover, the bioactive compounds from macroalgae and microalgae have been potential candidates for exhibiting iNOS inhibitory activity (Figure 7), which can prevent inflammation-related diseases due to the overproduction of NO. Our result also indicated that all the extracts were able to inhibit NO production in a dose-dependent manner (0.039–0.16 mg/mL), where CFL+F and CFL+C+F indicated the highest percentage from 25% to 40%.
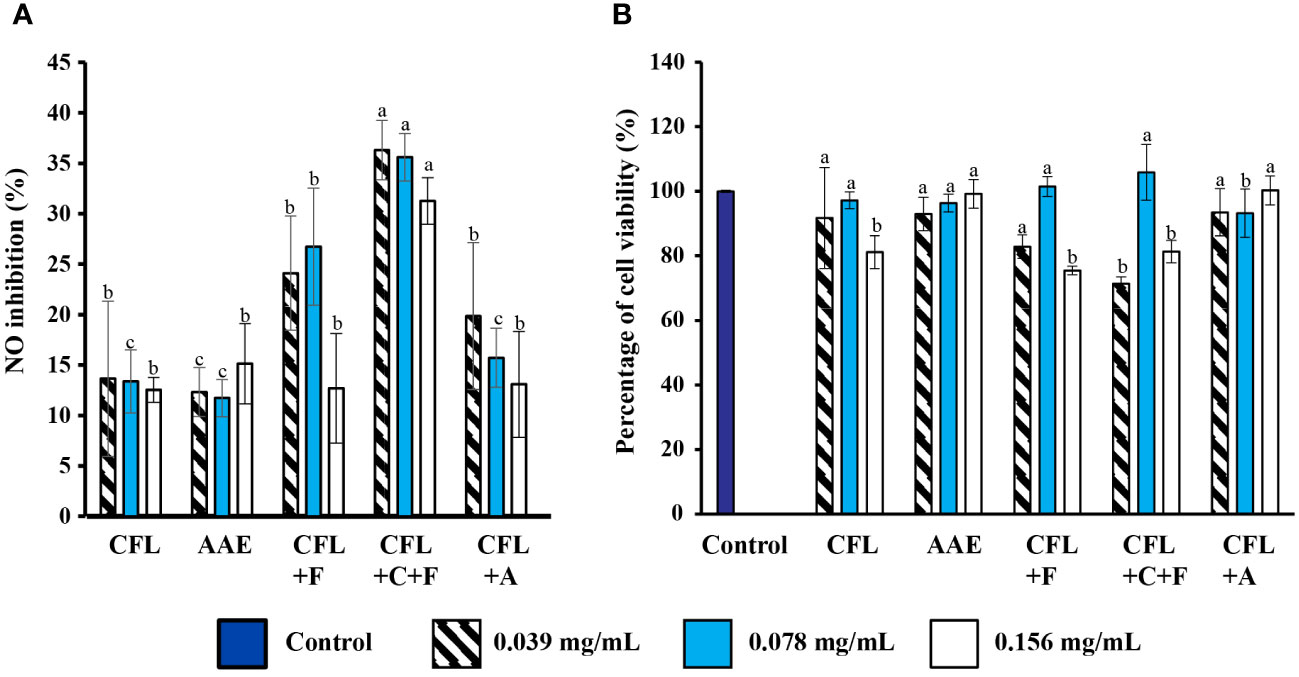
Figure 7 Effect of anti-inflammation properties. (A) Inhibitory effects of extracts on NO production in various concentrations (p< 0.05) on NO production. (B) Percentage of cell viability (p > 0.05) on cell viability. Values are expressed mean ± SD. CFL, cell-free lysate by ultrasonic-assisted cell disruption; AAE, cell-free lysate by alkaline/acid extraction of ultrasonic and cellulase-pretreated cells; CFL+F, cell-free lysate by ultrasonic-assisted cell disruption and Flavourzyme; CFL+C+F, cell-free lysate by ultrasonic and cellulase-pretreated cells with Flavourzyme; CFL+A, cell-free lysate by ultrasonic-assisted cell disruption and Alcalase.
Similar to current results, two previous reports studied a marine bivalve mollusk (Cyclina Sinensis), and egg yolk also demonstrated the production of NO. The former case exhibited the production of NO (µM/L) in a dose-dependent manner when hydrolyzed with pepsin. Additionally, the latter produced a NO of around 25 µM when hydrolyzed with pancreatin and neutrase (Li et al., 2019; Lee et al., 2022). Furthermore, Flores Hernandez et al. (2017) reported that the NO inhibition from banana Musa paradisiaca ranged from 19% to 52% based on the extracts (Rao et al., 2016) (Table 2). Moreover, the cytotoxic effect of the five extracts on RAW264.7 cells was determined by an MTT assay, as shown in Figure 7B. The result demonstrated that our five extracts did not significantly affect the cell viability. Our result was in accordance with previous reports extracting phycocyanin (PC) from Spirulina platensis, where the percentage of cell viability is around 90%, which did not significantly affect the cell viability (Kraseasintra et al., 2022). Similarly, the water and ethanolic (70%) extracts from Spirulina platensis also showed similar results on the cytotoxicity assay (Flores Hernandez et al., 2017).
In order to understand the role of macrophages, which are important components of the immune system and play an essential role in controlling various immunopathological phenomena, for example, the overproduction of proinflammatory cytokines and inflammatory cytokines like IL-1ß, IL-6, NO, iNOS, COX-2, and TNF (Chen et al., 2016), a phagocytosis assay was detected in vitro using the peripheral blood. The function of macrophages involved in the innate immune system is to respond at a fast rate whenever there is a pathogen. After mixing white blood cells with Candida albicans (DMST 5815), the top four extracts that exhibited the highest anti-inflammatory properties were used, and the percentage of phagocytic activity and phagocytic index were evaluated according to Gabhe et al. (2006) (Gabhe et al., 2006). As shown in Figure 4, compared to negative control containing only pathogens and white blood cells, all four extracts showed phagocytic activity in a dose-dependent manner, with CFL+F and CFL+C+F showing the best phagocytosis activity. Although AAE exhibited the highest percentage of antioxidant properties, this was surpassed by the two extracts called CFL+F and CFL+C+F in terms of anti-inflammatory and immunomodulatory activity in vitro. Further identification of active fragments, for example, through purification or molecular weight cut-off filtration, is needed for a better understanding of the ability of generated bioactive peptides.
Although our study did not conduct purification, electrophoretic mobility and the measurement of the degree of hydrolysis for monitoring the progress of before and after hydrolysis were measured. The electrophoretic pattern of the CFL, C, and AAE samples exhibited prominent bands with high intensity at molecular weights (Mw) of 75 kDa, 50 kDa, 37 kDa, 25 kDa, 20 kDa, and 15 kDa. This observation parallels previous research on isolated spirulina proteins Mohammadi et al. (2022), which indicated that unhydrolyzed proteins displayed multiple bands ranging from 85 kDa to 25 kDa (Mohammadi et al., 2022a). Notably, the most intense bands were detected within the 25–25-kDa range, corresponding to the subunits of C-PC, the predominant protein in spirulina, accounting for 20% of the dry biomass (Aiello et al., 2019). However, the electrophoretic profiles of proteins hydrolyzed using Flavourzyme and Alcalase enzymes displayed distinct characteristics where low molecular weight was less visible in Flavourzyme compared to Alcalase (Figure 6). This might be due to the ability of Flavourzyme, which favors the formation of single amino acids rather than longer polypeptides, which may not be visible on the gel or may be obscured by the dye front. Additionally, the endo-peptidase enzyme, Alcalase, could dominate the quaternary and tertiary conformations of proteins by cleaving peptide bonds within individual or aggregated proteins to produce smaller peptides (Meinlschmidt et al., 2016a).
However, to the best of our knowledge, no previous studies have reported the hydrolysis of spirulina protein with Flavourzyme. Consequently, the DH values were compared from different proteases and macroalgae. The previous report used two different proteases: pepsin, which exhibited a higher degree of hydrolysis (16%), whereas protein hydrolysate with pancreatin showed an 11% degree of hydrolysis in spirulina (Arthrospira platensis) (Mohammadi et al., 2022a). Furthermore, Cian et al. (2012) showed that protein hydrolysate with Flavourzyme increased the degree of hydrolysis in 7 h to 14% in Porphyra columbina (red seaweed) (Cian et al., 2012b). Moreover, values of the index in the hydrolysates of spirulina (Arthrospira platensis) with Alcalase were revealed (42.4%) (Akbarbaglu et al., 2022). In vivo study is necessary to investigate the efficiency, safety, and potential toxicity of these two extracts, CFL+F and CFL+C+F, by investigating the different blood parameters and validating the immune-enhancing effects in animal experiments and clinical trials. Furthermore, the active fragments are also required to be confirmed by amino acid profiling to elucidate the bioactive properties of each peptide involved in enzymatic hydrolysis.
5 Conclusion
In this study, protein extraction was performed prior to protein hydrolysate, with the aim of achieving a higher concentration of active bioactive compounds. The antioxidant property was first investigated in all extracts in response to oxidative stress. Anti-inflammation was then followed by selecting the optimal extracts that displayed superior antioxidant properties. As the last stage, an in vitro assay of immunomodulatory properties was carried out, and the phagocytic activity and index were quantified. As expected, the protein hydrolysate generated using proteases revealed the highest phagocytic activity compared to other extracts. Although there could have been some interferents, such as sugar and phenolic compounds, during protein extraction, the analysis showed lower levels of sugar content and phenolic compounds, suggesting that most of the bioactive properties come from proteins. In addition, the two extracts, CFL with Flavourzyme and CFL with cellulase and Flavourzyme, showed the highest phagocytic percentage; further study through in vivo by animal model is necessary to investigate the efficiency, safety, and potential toxicity of these extracts with more precision and accuracy.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by approved by the Institutional Animal Care and Use Committee of the Biology Department, Faculty of Science, Chiang Mai University (Re: 001/23). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
NH: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Writing – original draft. OK: Methodology, Writing – review & editing. WB: Methodology, Resources, Writing – review & editing. TK: Methodology, Resources, Writing – review & editing. JP: Funding acquisition, Methodology, Resources, Writing – review & editing. YT: Methodology, Resources, Writing – review & editing. KK: Writing – review & editing. SC: Resources, Writing – review & editing. SS: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. CP: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Research Council of Thailand (NRCT), the CMU Presidential Scholarship Academic Year 2022, and the Graduate School of Chiang Mai University. This research work was partially supported by Chiang Mai University. Additionally, the first and seventh authors were supported by the NSTC International Internship Pilot Program (IIPP).
Acknowledgments
I would like to express my sincere gratitude for having a chance to study the Master’s Degree Program in Applied Microbiology, Faculty of Science, Chiang Mai University, under the CMU Presidential Scholarship. My special thanks go to Mr. Nitiphong Kaewman, Mrs. Kritsana Duangjan, Ms. Kittiya Phinyo, and Ms. Srithip Sensupa for their assistance with technical support. Additionally, thank you very much to Ms. Panumat Piosinsak and Ms. Phornphan Phrompanya for their assistance with the phagocytosis assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahn G., Hwang I., Park E., Kim J., Jeon Y. J., Lee J., et al. (2008). Immunomodulatory effects of an enzymatic extract from ecklonia cava on murine splenocytes. Mar. Biotechnol. 10, 278–289. doi: 10.1007/s10126-007-9062-9
Aiello G., Li Y., Boschin G., Bollati C., Arnoldi A., Lammi C. (2019). Chemical and biological characterization of spirulina protein hydrolysates: Focus on ACE and DPP-IV activities modulation. J. Funct. Foods. 63, 103592. doi: 10.1016/j.jff.2019.103592
Ak T., Gülçin İ. (2008a). Antioxidant and radical scavenging properties of curcumin. Chem. Biol. Interact. 174, 27–37. doi: 10.1016/j.cbi.2008.05.003
Akbarbaglu Z., Ayaseh A., Ghanbarzadeh B., Sarabandi K. (2022). Techno-functional, biological and structural properties of Spirulina platensis peptides from different proteases. Algal Res. 66, 102755. doi: 10.1016/j.algal.2022.102755
Aryal S., Baniya M. K., Danekhu K., Kunwar P., Gurung R., Koirala N. (2019). Total phenolic content, flavonoid content and antioxidant potential of wild vegetables from western Nepal. Plants 8, 96. doi: 10.3390/plants8040096
Ba F., Ursu A. V., Laroche C., Djelveh G. (2016). Haematococcus pluvialis soluble proteins: Extraction, characterization, concentration/fractionation and emulsifying properties. Bioresour Technol. 200, 147–152. doi: 10.1016/j.biortech.2015.10.012
Bahari A. N., Bahari A. N., Saari N., Salim N., Salim N., Salim N., et al. (2020). Response factorial design analysis on papain-generated hydrolysates from Actinopyga lecanora for determination of antioxidant and antityrosinase activities. Molecules 25 (11), 2663. doi: 10.3390/molecules25112663
Béné C., Barange M., Subasinghe R., Pinstrup-Andersen P., Merino G., Hemre G. I., et al. (2015). Feeding 9 billion by 2050 – Putting fish back on the menu. Food Secur. 7, 261–274. doi: 10.1007/s12571-015-0427-z
Benzie I. F. F., Strain J. J. (1999). Ferric reducing/antioxidant power assay: Direct measure of total antioxidant activity of biological fluids and modified version for simultaneous measurement of total antioxidant power and ascorbic acid concentration. Meth. Enzymol. 299, 15–27. doi: 10.1016/S0076-6879(99)99005-5
Chalamaiah M., Hemalatha R., Jyothirmayi T., Diwan P. V., Uday Kumar P., Nimgulkar C., et al. (2014). Immunomodulatory effects of protein hydrolysates from rohu (Labeo rohita) egg (roe) in BALB/c mice. Food Res. Int. 62, 1054–1061. doi: 10.1016/j.foodres.2014.05.050
Chen Y. F., Wang K., Zhang Y. Z., Zheng Y. F., Hu F. L. (2016). In vitro anti-inflammatory effects of three fatty acids from royal jelly. Mediators Inflamm. 2016, 3583684. doi: 10.1155/2016/3583684
Cian R. E., López-Posadas R., Drago S. R., Sánchez de Medina F., Martínez-Augustin O. (2012a). A Porphyra columbina hydrolysate upregulates IL-10 production in rat macrophages and lymphocytes through an NF-κB, and p38 and JNK dependent mechanism. Food Chem. 134, 1982–1990. doi: 10.1016/j.foodchem.2012.03.134
Cian R. E., Martínez-Augustin O., Drago S. R. (2012b). Bioactive properties of peptides obtained by enzymatic hydrolysis from protein byproducts of Porphyra columbina. Food Res. Int. 49, 364–372. doi: 10.1016/j.foodres.2012.07.003
Conde T. A., Neves B. F., Couto D., Melo T., Neves B., Costa M., et al. (2021). Microalgae as sustainable bio-factories of healthy lipids: evaluating fatty acid content and antioxidant activity. Mar. Drugs 19, 357. doi: 10.3390/md19070357
DuBois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
El-Kassas H. Y., Heneash A. M. M., Hussein N. R. (2015). Cultivation of Arthrospira (Spirulina) platensis using confectionary wastes for aquaculture feeding. J. Genet. Eng. Biotechnol. 13, 145–155. doi: 10.1016/j.jgeb.2015.08.003
Flores Hernandez F. Y., Khandual S., Ramírez López I. G. (2017). Cytotoxic effect of Spirulina platensis extracts on human acute leukemia Kasumi-1 and chronic myelogenous leukemia K-562 cell lines. Asian Pac J. Trop. Biomed. 7, 14–19. doi: 10.1016/j.apjtb.2016.10.011
Gabhe S., Tatke P., Khan T. (2006). Evaluation of the immunomodulatory activity of the methanol extract of Ficus benghalensis roots in rats. Indian J. Pharmacol. 38, 271–275. doi: 10.4103/0253-7613.27024
Geada P., Moreira C., Silva M., Nunes R., Madureira L., Rocha C. M. R., et al. (2021). Algal proteins: Production strategies and nutritional and functional properties. Bioresour Technol. 332, 125125. doi: 10.1016/j.biortech.2021.125125
Giannoglou M., Andreou V., Thanou I., Markou G., Katsaros G. (2022). High pressure assisted extraction of proteins from wet biomass of Arthrospira platensis (spirulina) – A kinetic approach. Innovative Food Sci. Emerg Technol. 81, 103138. doi: 10.1016/j.ifset.2022.103138
Gulcin İ., Alwasel S. H. (2022). Metal ions, metal chelators and metal chelating assay as antioxidant method. Processes 10, 132. doi: 10.3390/pr10010132
Harnedy P. A., FitzGerald R. J. (2013). Extraction of protein from the macroalga Palmaria palmata. LWT Food Sci. Technol. 51, 375–382. doi: 10.1016/j.lwt.2012.09.023
Hou H., Fan Y., Wang S., Si L., Li B. (2016). Immunomodulatory activity of Alaska pollock hydrolysates obtained by glutamic acid biosensor – Artificial neural network and the identification of its active central fragment. J. Funct. Foods. 24, 37–47. doi: 10.1016/j.jff.2016.03.033
Jayawardena R., Sooriyaarachchi P., Chourdakis M., Jeewandara C., Ranasinghe P. (2020). Enhancing immunity in viral infections, with special emphasis on COVID-19: A review. Diabetes Metab. Syndrome: Clin. Res. Rev. 14, 367–382. doi: 10.1016/j.dsx.2020.04.015
Khan Yusufzai S., Rafatullah M S. A. N. I., Sarjadi M., Shaheen Khan M., Sani Sarjadi M., Razlan M. (2018). Determination of total phenolic content, total flavonoid content and antioxidant activity of various organic crude extracts of licuala spinosa leaves from Sabah, Malaysia. ASM Sci. J. 11, 449–454. doi: doi: 10.1016/j.jtusci.2014.11.001
Kraseasintra O., Tragoolpua Y., Pandith H., Khonkarn R., Pathom-aree W., Pekkoh J., et al. (2022). Application of phycocyanin from Arthrospira (Spirulina) platensis as a hair dye. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.1024988
Lee J. H., Lee J. E., Paik H. D. (2022). Immunomodulatory activity of egg yolk protein hydrolysates prepared by novel two-step hydrolysis: A study of mechanism and stability after in vitro digestion model. Poult Sci. 101, 101802. doi: 10.1016/j.psj.2022.101802
Le Nguyen Doan D., Phan The D., Le T. T. H., Nguyen T. X. Q., Nguyen D. N. (2022). Protein extraction from spirulina platensis with the cellulase enzyme assistance. J. Tech. Educ. Sci. 70B), 25–32. doi: 10.54644/jte.70B.2022.1213
Li W., Ye S., Zhang Z., Tang J., Jin H., Huang F., et al. (2019). Purification and characterization of a novel pentadecapeptide from protein hydrolysates of cyclina sinensis and its immunomodulatory effects on RAW264.7 cells. Mar. Drugs 17, 30.
Liu J., Hu Q. (2013). “Chlorella: industrial production of cell mass and chemicals,” in Handbook of Microalgal Culture (John Wiley & Sons, Ltd, Oxford, UK), 327–338.
Lomakool S., Ruangrit K., Jeerapan I., Tragoolpua Y., Pumas C., Srinuanpan S., et al. (2023). Biological activities and phytochemicals profiling of different cyanobacterial and microalgal biomass. Biomass Convers Biorefin. 13, 4195–4211. doi: 10.1007/s13399-021-01974-0
Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275. doi: 10.1016/S0021-9258(19)52451-6
Maruyama H., Sakamoto T., Araki Y., Hara H. (2010). Anti-inflammatory effect of bee pollen ethanol extract from Cistus sp. of Spanish on carrageenan-induced rat hind paw edema. BMC Complementary Altern. Med. 10, 30. doi: 10.1186/1472-6882-10-30
Mathur M. (2018). Bioactive molecules of spirulina: A food supplement. In reference series in phytochemistry. 1–22. doi: 10.1007/978-3-319-54528-8_97-1
Meinlschmidt P., Sussmann D., Schweiggert-Weisz U., Eisner P. (2016a). Enzymatic treatment of soy protein isolates: effects on the potential allergenicity, technofunctionality, and sensory properties. Food Sci. Nutr. 4, 11–23. doi: 10.1002/fsn3.253
Mohammadi M., Soltanzadeh M., Ebrahimi A. R., Hamishehkar H. (2022a). Spirulina platensis protein hydrolysates: Techno-functional, nutritional and antioxidant properties. Algal Res. 65, 102739. doi: 10.1016/j.algal.2022.102739
Munteanu I. G., Apetrei C. (2021). Analytical methods used in determining antioxidant activity: A review. Int. J. Mol. Sci. 22 (7), 3380. doi: 10.3390/ijms22073380
Naseri A., Marinho G. S., Holdt S. L., Bartela J. M., Jacobsen C. (2020). Enzyme-assisted extraction and characterization of protein from red seaweed Palmaria palmata. Algal Res. 47, 101849. doi: 10.1016/j.algal.2020.101849
O’Connor J., Garcia-Vaquero M., Meaney S., Tiwari B. K. (2022). Bioactive peptides from algae: traditional and novel generation strategies, structure-function relationships, and bioinformatics as predictive tools for bioactivity. Mar. Drugs 20, 317. doi: 10.3390/md20050317
Pan D. D., Wu Z., Liu J., Cao X. Y., Zeng X. Q. (2013). Immunomodulatory and hypoallergenic properties of milk protein hydrolysates in ICR mice. J. Dairy Sci. 96, 4958–4964. doi: 10.3168/jds.2013-6758
Parimi N. S., Singh M., Kastner J. R., Das K. C., Forsberg L. S., Azadi P. (2015). Optimization of protein extraction from spirulina platensis to generate a potential co-product and a biofuel feedstock with reduced nitrogen content. Front. Energy Res. 3. doi: 10.3389/fenrg.2015.00030
Pekkoh J., Phinyo K., Thurakit T., Lomakool S., Duangjan K., Ruangrit K., et al. (2022). Lipid profile, antioxidant and antihypertensive activity, and computational molecular docking of diatom fatty acids as ACE inhibitors. Antioxidants 11, 186. doi: 10.3390/antiox11020186
Pekkoh J., Ruangrit K., Pumas C., Duangjan K., Chaipoot S., Phongphisutthinant R., et al. (2021). Transforming microalgal Chlorella biomass into cosmetically and nutraceutically protein hydrolysates using high-efficiency enzymatic hydrolysis approach. Biomass Convers Biorefin. 6299–6315. doi: 10.1007/s13399-021-01622-7
Peter A. P., Chew K. W., Koyande A. K., Yuk-Heng S., Ting H. Y., Rajendran S., et al. (2021). Cultivation of Chlorella vulgaris on dairy waste using vision imaging for biomass growth monitoring. Bioresour Technol. 341, 125892. doi: 10.1016/j.biortech.2021.125892
Qian Z. J., Jung W. K., Kim S. K. (2008). Free radical scavenging activity of a novel antioxidative peptide purified from hydrolysate of bullfrog skin, Rana catesbeiana Shaw. Bioresour Technol. 99, 1690–1698. doi: 10.1016/j.biortech.2007.04.005
Rao U. S. M., Ahmad B. A., Mohd K. S. (2016). Aktiviti pemerangkapan nitrik oksida dan anti-radang secara in vitro oleh ekstrak pelarut berbeza dari pelbagai bahagian Musa paradisiaca. Malaysian J. Analytical Sci. 20, 1191–1202. doi: 10.17576/mjas
Rösch C., Roßmann M., Weickert S. (2019). Microalgae for integrated food and fuel production. GCB Bioenergy 11, 326–334. doi: 10.1111/gcbb.12579
Sathasivam R., Radhakrishnan R., Hashem A., Abd Allah E. F. (2019). Microalgae metabolites: A rich source for food and medicine. Saudi J. Biol. Sci. 26, 709–722. doi: 10.1016/j.sjbs.2017.11.003
Sathya R., MubarakAli D., MohamedSaalis J., Kim J. W. (2021). A systemic review on microalgal peptides: bioprocess and sustainable applications. Sustainability 13, 3262. doi: 10.3390/su13063262
Sedighi M., Jalili H., Darvish M., Sadeghi S., Ranaei-Siadat S. O. (2019). Enzymatic hydrolysis of microalgae proteins using serine proteases: A study to characterize kinetic parameters. Food Chem. 284, 334–339. doi: 10.1016/j.foodchem.2019.01.111
Senevirathne M., Ahn C. B., Je J. Y. (2010). Enzymatic extracts from edible red algae, Porphyra tenera, and their antioxidant, anti-acetylcholinesterase, and anti-inflammatory activities. Food Sci. Biotechnol. 19, 1551–1557. doi: 10.1007/s10068-010-0220-x
Sun S., Xu X., Sun X., Zhang X., Chen X., Xu N. (2019). Preparation and identification of ACE inhibitory peptides from the marine macroalga ulva intestinalis. Mar. Drugs 17 (3), 179. doi: 10.3390/md17030179
Tzachor A., Rozen O., Khatib S., Jensen S., Avni D. (2021). Photosynthetically controlled spirulina, but not solar spirulina, inhibits TNF-α Secretion: potential implications for COVID-19-related cytokine storm therapy. Mar. Biotechnol. 23, 149–155. doi: 10.1007/s10126-021-10020-z
Xu Q., Zheng L., Huang M., Zhao M. (2022). Exploring structural features of potent dipeptidyl peptidase IV (DPP-IV) inhibitory peptides derived from tilapia (Oreochromis niloticus) skin gelatin by an integrated approach of multivariate analysis and Gly-Pro-based peptide library. Food Chem. 397, 133821. doi: 10.1016/j.foodchem.2022.133821
Zhang X., Yang J., Suo H., Tan J., Zhang Y., Song J. (2023). Identification and molecular mechanism of action of antibacterial peptides from Flavourzyme-hydrolyzed yak casein against Staphylococcus aureus. J. Dairy Sci. 106, 3779–3790. doi: 10.3168/jds.2022-22823
Keywords: anti-inflammation, hydrolysate, immunomodulation, protein, microalgae, spirulina
Citation: Htoo NYM, Kraseasintra O, Buncharoen W, Kaewkod T, Pekkoh J, Tragoolpua Y, Khoo KS, Chaipoot S, Srinuanpan S and Pumas C (2024) In vitro immunomodulation activity of protein hydrolysate from spirulina (Arthrospira platensis): the ingredient of future foods. Front. Mar. Sci. 11:1303025. doi: 10.3389/fmars.2024.1303025
Received: 27 September 2023; Accepted: 01 February 2024;
Published: 22 February 2024.
Edited by:
Jelena Vladic, NOVA University of Lisbon, PortugalReviewed by:
Ljiljana Popovic, University of Novi Sad, SerbiaFiaz Ahmad, Northwestern Polytechnical University, China
Copyright © 2024 Htoo, Kraseasintra, Buncharoen, Kaewkod, Pekkoh, Tragoolpua, Khoo, Chaipoot, Srinuanpan and Pumas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sirasit Srinuanpan, c2lyYXNpdC5zQGNtdS5hYy50aA==; Chayakorn Pumas, Y2hheWFrb3JuLnB1bWFzQGdtYWlsLmNvbQ==
 Nang Yee Mon Htoo
Nang Yee Mon Htoo Oranit Kraseasintra
Oranit Kraseasintra Wararut Buncharoen1
Wararut Buncharoen1 Thida Kaewkod
Thida Kaewkod Jeeraporn Pekkoh
Jeeraporn Pekkoh Kuan Shiong Khoo
Kuan Shiong Khoo Sirasit Srinuanpan
Sirasit Srinuanpan Chayakorn Pumas
Chayakorn Pumas