
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 08 February 2024
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1254806
In order to explore the antagonistic mechanism of Lysinibacillus sphaericus AEB18 against Nocardia seriolae, the whole genome of strain AEB18 was sequenced, and the effect of strain AEB18 in controlling nocardiosis of hybrid snakehead was verified in vitro and in vivo. Strain AEB18 genome was a circular DNA of 4,653,716 base pairs (bp) and GC content of 37.4%, predicting 4,529 coding genes, 108 tRNAs, 37 rRNAs, and 95 non-coding (nc)RNAs. The eggNOG (Non-supervised Orthologous Groups), GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes) and the CAZy (Carbohydrate-Associated Enzyme) database annotated the strain AEB18 genome to 3,965, 3,325, 2,061, and 112 genes, respectively. Furthermore, nine gene clusters, including polyketones, non-ribosomal synthetic peptides, β-lactones, terpenes, ribosomal synthetic peptides, and other secondary metabolites with bacteriostatic effect were obtained through the prediction of secondary metabolites of strain AEB18. After the addition of L. sphaericus AEB18 (1 × 109 CFU/g) with a mass fraction of 1% to the basal diet for 21 days, the weight gain rate and the activity of amylase in intestinal chyme of hybrid snakehead were significantly increased (P< 0.05). In addition, hybrid snakehead fed with L. sphaericus AEB18 supplemented diets showed significantly reduced mortality rate (P< 0.05) after N. seriolae infection, compared with the control group (mortality rate was 100%). Overall, L. sphaericus AEB18 positively affected hybrid snakeheads, promoted growth and reduced mortality rate of hybrid snakeheads with nocardiosis. Taken together, the L. sphaericus AEB18 possesses great potential as a biocontrol agent, which can be commercially developed to improve disease control in freshwater aquaculture.
The rapid development of aquaculture has propagated aquaculture in China, and the output has reached 75% of the world (Pauly and Zeller, 2017). As fish production increases, fish diseases (such as Nocardia, Aeromonas, and Edwardsiella) have become a serious problem in aquaculture, often causing major economic losses (Bondad-Reantaso et al., 2005; Lee et al., 2014; Watts et al., 2017). Previously, chemical drugs such as antibiotics have been the common choice for preventing fish diseases, but the high use of antibiotics has caused problems of drug residues, multi-drug resistance, and environmental pollution (Watts et al., 2017), as well as damage to the immune system of fish and adversely affecting the digestive tract flora of the fish (Lee et al., 2014; Capkin et al., 2017). The intestinal environment of fish is a critical protective barrier against the occurrence of disease, and the dynamic interaction between the intestinal mucosa and pathogenic microorganisms induces inflammatory responses and alters the barrier function of the intestine (Honda and Littman, 2012). Some studies have shown that probiotics prevent the colonization of pathogenic bacteria by regulating the intestinal mucosal barrier function of fish, competitively inhibiting the growth of fish intestinal pathogenic bacteria, stimulating the specific or non-specific immune function, promoting animal growth, improving the feed conversion rate, enhancing the animal immune function, and improving ecological environment in vivo (Zhang et al., 2019a). Therefore, probiotics, as one of the safe alternatives to antibiotics, have been widely used in aquaculture in the form of feed additives in recent years (Selim and Reda, 2015; Mukherjee and Ghosh, 2016; Yi et al., 2018). Compared to Lactobacillus, Bacillus can form endospores that protect it from environmental influence, which is more advantageous in the prevention of fish diseases.
Lysinibacillus sphaericus, discovered in 1965, was first proposed as a new genus associated with Bacillus (formerly known as Bacillus sphaericus) and is gaining importance due to the commercial use of some members for the biological control of vector-borne diseases (Baumann et al., 1991). Subsequently, L. sphaericus was widely used as a reference for the study of larvicidal activity of Lactobacillus coccidiosis (Lozano and Dussán, 2013), metal tolerance (Davidson et al., 1977; Lozano and Dussán, 2013; Peña-Montenegro et al., 2015), surface protein structures such as the s layer (Brown et al., 2004), and biosurfactant production comparison points (Lacey, 2007). However, at present, no studies have reported that L. sphaericus microorganisms promote growth and prevent bacterial diseases in fish. In addition, only a few studies have yet reported the whole genome analysis related to L. sphaericus.
Whole genome sequencing can detect the entire genome sequence for a species and can decipher the encompassed genetic mechanism information by analyzing the base sequence of the species comprehensively and accurately; it has the advantages of high sequencing coverage and strong accuracy (Bartram et al., 2011). Third-generation sequencing technology refers to Oxford Nanopore Technologies, nanopore technologies nanopore, single-molecule sequencing, and SMRT technology developed by PacBio, which does not require polymerase chain reaction (PCR) amplification for the sequencing process and has the characteristics of sequencing read length and high sequencing efficiency (Clarke et al., 2009). With the rapid development of technology, third-generation whole genome sequencing has become an efficient detection method, widely used in medicine, biology, and other fields, and plays a critical role in the study of the biological characteristics of environmental microorganisms and intestinal microflora (Peña-Montenegro et al., 2015; Finton et al., 2020; Jose et al., 2020).
This study, for the first time, a potential probiotic strain AEB18 with significant inhibition of N. seriolae was isolated from healthy fish intestines, and the whole genome of this strain was sequenced on the Nanopore platform. Subsequently, the antibacterial gene cluster of strain AEB18 was analyzed by gene function annotation and antiSMASH online tool to further explore its antibacterial mechanism. In addition, after feeding with AEB18 supplemented diets, the effects of promoting growth and preventing nocardiosis of hybrid snakehead were determined.
All experimental protocols were approved by the Animal Ethics Committee of South China Agricultural University, Guangzhou, Guangdong, China (approval ID: SYXK-2019-0136). This study does not involve protected or endangered species.
Hybrid snakeheads (13.0 ± 2.3 g) used in the trial were obtained from Rongda Farm in Guangzhou, Guangdong, China, which were healthy and had not been fed antibiotics or other drugs. Fish were placed in an aquarium for 7 days before the test. The experimental glass water tank was 100 L, and the water temperature was 19.6–28.8°C during the breeding period. The water temperature and the dissolved oxygen were measured (the dissolved oxygen should be maintained at > 6 mg/L) daily at 9:00 and 14:00. The flow system was used to suck up the pollution once in the morning and evening, respectively.
The intestines of healthy hybrid snakehead were cut aseptically, and the contents were mixed with 1 mL of normal saline. The mixture was diluted 100-fold with sterile water. Then, 100 μL of diluted test solution was spread on Luria-Bertani (LB) agar (Haibo, Qindao, Shandong, China), and the plate was incubated at 37°C overnight. Subsequently, the morphology of individual colonies on the medium, including the color, size, colony edges, and surface roughness, was observed. The suspected Bacillus colonies were selected for Gram staining. Gram-positive bacteria were selected by microscopic examination and stored as suspected Bacillus strains. The purified cultures were scribed and incubated at 37°C overnight. Finally, single colonies were picked and enriched in LB liquid medium (Haibo, Qindao, Shandong, China) for 12 h, followed by glycerol storage at −80°C.
The Kirby-Bauer method (K-B method) was used in the experiment. Initially, the pathogenic bacterium Nocardia seriolae was grown in Trypticase Soy Broth (TSB, Haibo, China) with 2% fetal bovine serum (FBS) for 72 hours and then rinsed with 0.85% physiological saline. The concentration of the bacterial broth was adjusted to 1 × 108 CFU/mL using Maxwell’s turbidimetric tube, and 0.1 mL of the bacterial suspension was evenly spread on Trypticase Soy Agar (TSA, Haibo, China). A sterile filter paper was placed on the surface of the TSA medium, and the suspected Bacillus strains (test strains) were prepared into a bacterial suspension at a specific concentration of 1 × 109 CFU/mL. Next, 10 μL of this suspension was spotted onto the paper, and the cultures were incubated at 28°C for 48-72 hours. Finally, the diameter of the zone of inhibition was measured.
The suspected strain was used as the template, and primers 27F/1492R (27F, 5’-agagtttgatcctgtgctcag-3’; 1492R, 5’-ggttaccttggactt-3’) were amplified by PCR (Polymerase chain reaction) to obtain the target sequence. The strain information was confirmed by BLAST (Basic Local Alignment Search Tool) analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Then, the 16S rRNA gene sequences of L. sphaericus strains and other strains were selected, and the phylogenetic tree was constructed by using MEGA software (version 7.2).
DNA from L. sphaericus AEB18 was obtained by using the Rapid Bacterial Genomic DNA Extraction Kit (Sigma-Aldrich, ShangHai, China), according to the manufacturer’s instructions. After extracting the total DNA, the purity and concentration of the genomic DNA of L. sphaericus AEB18 were detected by 0.35% agarose gel electrophoresis (Liuyi, Beijing, China) and on a Nanodrop micro-spectrophotometer (Thermo, Shanghai, China), respectively.
Whole genome sequencing of strain AEB18 was carried out on the nanopore sequencing platform of Biotech Co., Ltd (Beijing, China). After dismounting data were quality controlled, the sequence splicing assembly was performed using Canu v1.5 (Koren et al., 2017) assembly software, and then optimized to obtain a genome with high accuracy for subsequent analysis. The whole genome sequence of L. sphaericus AEB18 was compared and analyzed with databases of eggNOG (Non-supervised Orthologous Groups) (Powell et al., 2014), GO (Gene Ontology) (Conesa et al., 2005), KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al., 2004) and CAZy (carbohydrate-associated enzymes) (Cantarel et al., 2009), respectively, and the annotation information of genome function was obtained. Then, the antiSMASH (https://dl.secondarymetabolites.org/releases/4.0.2/) (Blin et al., 2016) software was used to predict the secondary metabolite synthesis gene cluster of the sample and obtain the metabolite prediction results.
A total of 120 hybrid snakeheads were randomly and equally divided into two groups with three replicates (20 replicates each, 60 fish in each group). The two groups were fed different feeds: the hybrid snakehead in the control groups was fed with basal diet and the experimental group was fed an additional L. sphaericus AEB18 (1 × 109 CFU/g) with a mass fraction of 1% based on the same amount of basal diet. After 21 days, the effects of L. sphaericus on the growth performance (weight) and intestinal digestive enzyme (intestinal α-amylase) activity of hybrid snakehead were analyzed.
Enzyme assay: Eighteen fish were randomly taken from each group and dissected on an ice tray. The intestinal (containing contents) tissue was rinsed with deionized water at 4°C and stored at −20°C for later use. Subsequently, 0.5 g of midgut chyme was mixed with 4 mL of pre-chilled (4°C) ionized water, and the supernate was collected by centrifugation at 4000 rpm at 4°C for 30 min. The α-amylase activity was determined using the amylase kit (Nanjing Institute of Bioengineering, Nanjing, China), and the absorbance (660 nm) was measured on a microplate reader (Merck, Darmstadt, Germany).
According to the above grouping method, 180 hybrid snakeheads were divided into three groups randomly, and each group consisted of triplicates (20 each, a total of 60 fish in each group). The experimental group was fed a basal diet with L. sphaericus AEB18 for 21 days before challenge test (1 × 107 CFU/mL of N. seriolae suspension 100 μL injected intraperitoneally in each fish). The blank group was treated without poisoning after 21 days of the basal diet (intraperitoneal injection of 0.85% normal saline per fish). The control group was given a basal diet for 21 days, followed by challenge treatment (intraperitoneal injection of 1 × 107 CFU/mL of N. seriolae suspension 100 μL per tail).
The specific operation of the challenge treatment was to prepare N. seriolae (isolated from the liver tissue of hybrid snakehead in the Rongda breeding base in Guangzhou, China) into a bacterial suspension with a concentration of 1×107 CFU/mL and infect hybrid snakehead by abdominal fin injection. The hybrid snakeheads were observed daily for 2 weeks, and the deaths were recorded.
The original data were sorted out by Microsoft Excel 2003, and the results were expressed as mean ± standard error (SD) of the mean. GraphPad Prism 8.0 software was used for the data analysis. The significance one-way analysis of variance (ANOVA), P< 0.05 indicates a significant difference.
Multiple strains were isolated from healthy fish intestines and suspected Bacillus colonies were selected for Gram staining and microscopic examination. Then, Gram-positive bacteria were selected and stored, the antagonistic strains against N. seriolae were screened by K-B method, and finally, a suspected Bacillus strain with the best inhibitory effect against N. seriolae in vitro was obtained, as shown in Figure 1.
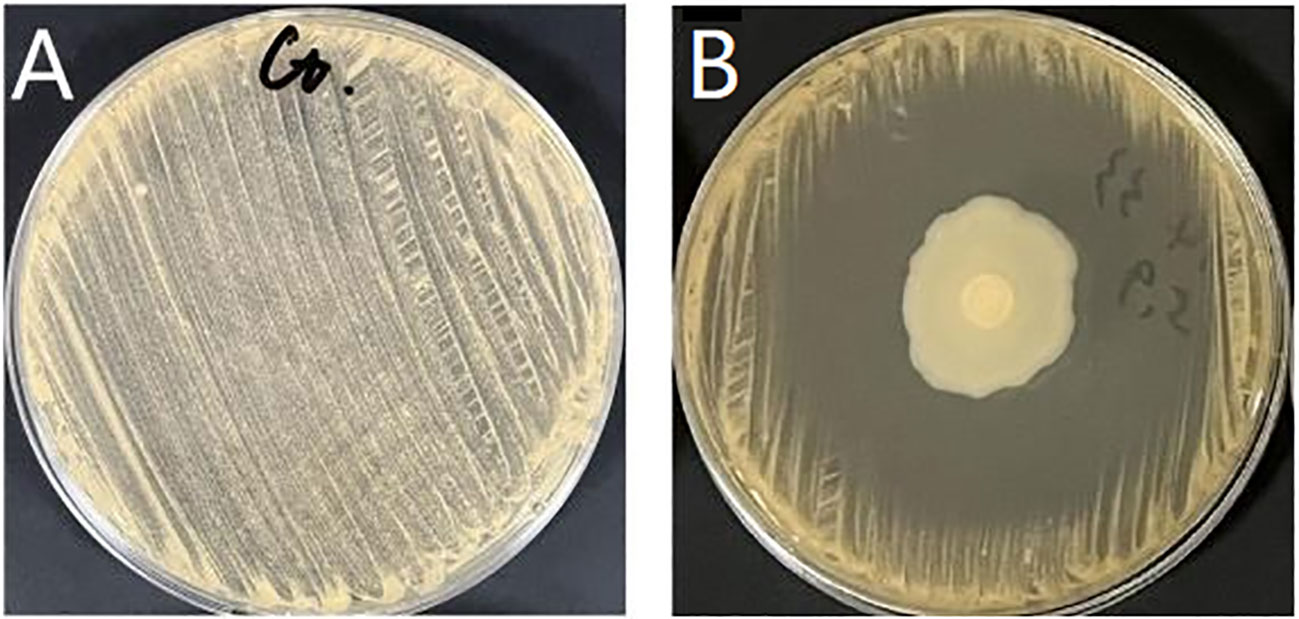
Figure 1 Determination of inhibitory activity of antagonistic bacteria against N. seriolae. (A) N. seriolae (control); (B) Diameter of inhibition zone of AEB18 strain (35 ± 0.87 mm).
PCR amplification of antagonistic AEB18 using universal primers 27F and 1,492R (16S rRNA) resulted in specific bands around 1,500 bp, and the fragments of 1,552 bp were sequenced. The sequencing results were compared by BLAST, and 22 representative strains were selected with the AEB18 strain for phylogenetic tree construction, using MEGA 7.2 software and neighbor-joining model, as shown in Figure 2 AEB18 and L. sphaericus DSM28 (NR042073.1) are associated genetically with about 99.36% homology; therefore, it was termed L. sphaericus AEB18.
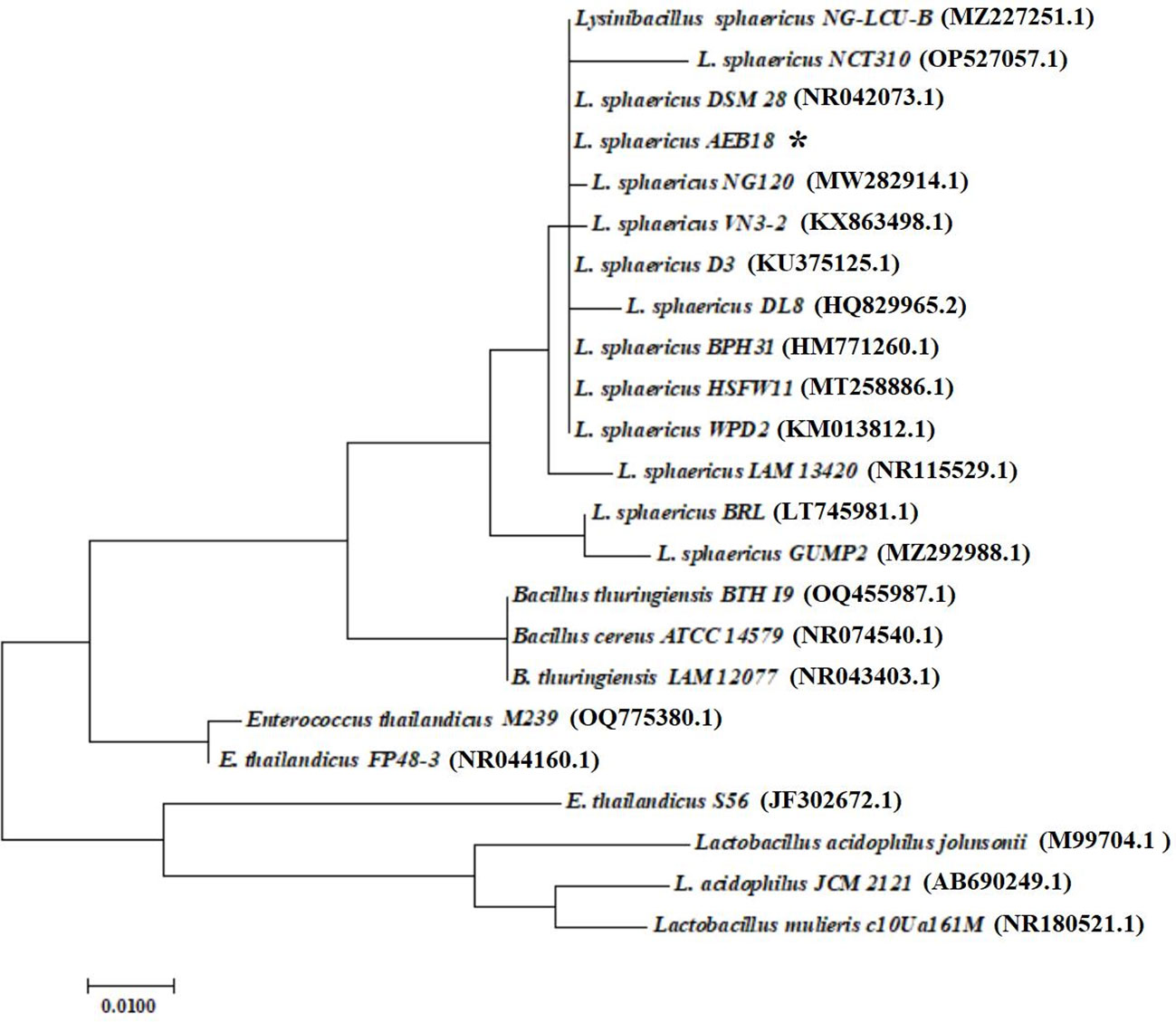
Figure 2 An antagonistic L. sphaericus AEB18 phylogenetic tree constructed based on 16S rRNA gene sequences. Bootstrap times are set to 1,000; Ruler represents 0.01% of sequence evolutionary differences, and the parentheses represent the GenBank accession number.
In our study, the L. sphaericus AEB18 was subjected to whole genome sequencing on the Nanopore platform. The results showed that the total length of L. sphaericus AEB18 gene sequence was 4,653,716 bp, the length of N50 (bp) was 23,336, the length of N90 (bp) was 6,536, and the GC content was 37.4%, including 108 tRNAs, 37 rRNAs, 95 ncRNAs, and 4,529 coding genes; these accounted for 83.44% of the total genome, and the genome circle diagram is shown in Figure 3.
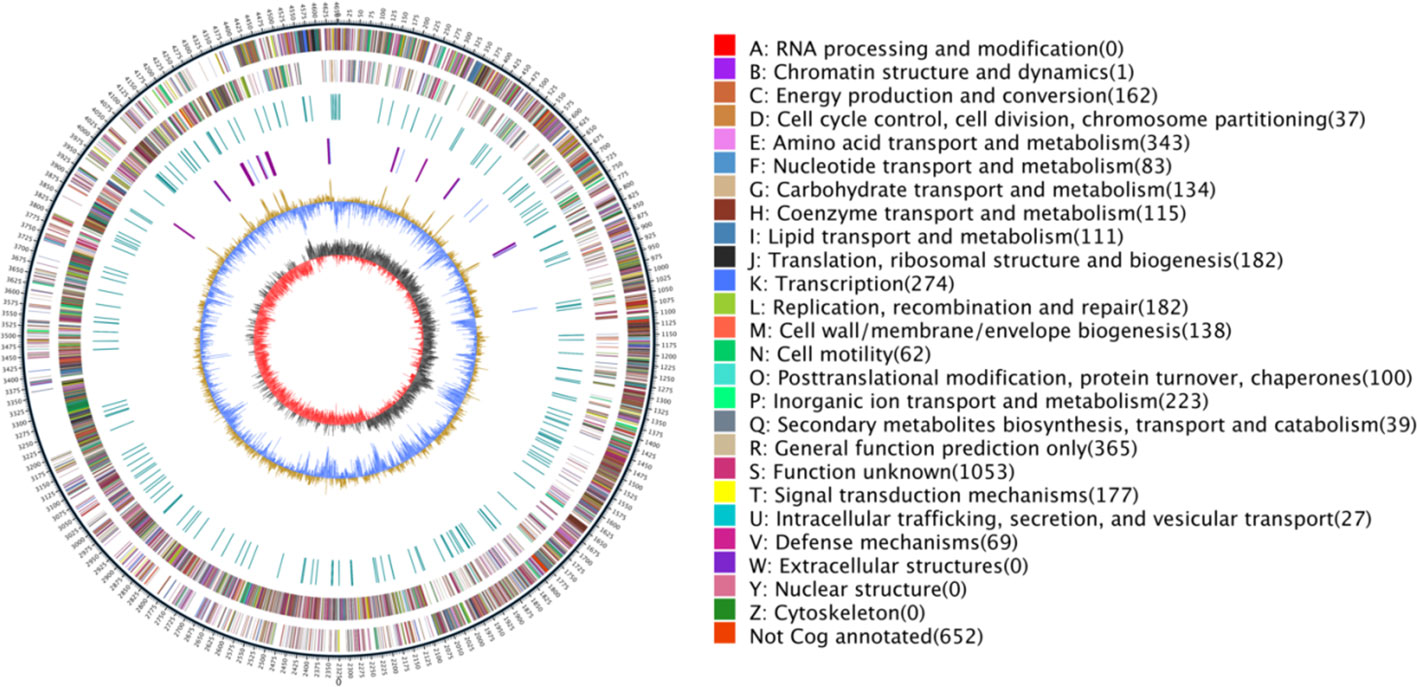
Figure 3 Genome circle. External to internal: the outermost circle is the marker of genome size (5 kb). The second and third circles are genes on the positive and negative strands of the genome (color by COG category), respectively. The fourth circle is a repetitive sequence. The fifth circle is RNA gene (tRNA blue and rRNA purple). The sixth circle is GC content. The innermost ring is GC-skew.
The comparison of eggNOG, NR, SWISS-PROT, PFAM, KEGG, TrEMBL_Annotation, and GO databases revealed the corresponding annotation information, and the detailed annotation statistical results are shown in Table 1. Among them, eggNOG database can be divided into 21 categories according to function, and 3,965 proteins were successfully annotated by comparison with eggNOG database, and the statistical results of eggNOG of L. sphaericus AEB18 are shown in Figure 4 Class B (chromatin structure and dynamics) and class U (intracellular trafficking, secretion, and vesicular transport) have the lowest amount of proteins, displaying that the structure and dynamics of chromatin and intracellular transport and that the secretion and vesicle transport of the AEB18 has less protein content and low cell motility. Furthermore, class Q with 55 proteins (secondary metabolites biosynthesis, transport, and catabolism) indicated that AEB18 strain can produce secondary metabolites. Finally, the most abundant class S with 1,053 proteins (function unknown) indicated that the functions of many proteins in AEB18 are unknown but could produce new compounds.
The analysis of the protein function of L. sphaericus AEB18 genome facilitated the annotation of a total of 3,325 genes in GO database, and the results are shown in Figure 5. The GO-annotated classification of L. sphaericus AEB18 mainly focuses on the functions of cells, membrane components, cellular processes, metabolic processes, ligation, and catalytic activity. Among these, 1,186 and 1,135 genes were involved in membrane and membrane components in cell components, 1,802 and 1,324 genes were related to catalytic activity and binding function in molecular functions, and 1,483 and 1,213 genes were involved in metabolism and cellular processes in biological pathways, respectively. The KEGG annotation results revealed 2,061 genes, accounting for about 46.0% of the total gene numbers of bacteria and 48 metabolic pathways; the specific statistical results are shown in Figure 6.
CAZy is a special database of carbohydrate enzymes, which consists of five categories, including carbohydrate esterases (CEs), glycosidases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs) and auxiliary activity (AAs). In addition, it also includes carbohydrate-binding modules (CBMs) related to carbohydrates. In this study, AEB18 predicted a total of 112 carbohydrate-related enzymes (CREs), among which CEs were most abundant 42 (37.5%); the specific results are shown in Figure 7.
antiSMASH (https://antismash.secondarymetabolites.org) software was used for the analysis and prediction of biosynthetic gene clusters (BGCs) in bacteria, fungi, or plants (Blin et al., 2016). The platform was also utilized to annotate and analyze the genomic data of L. sphaericus AEB18 to predict the associated synthetic gene clusters of active secondary metabolites of this strain. Importantly, 9 gene clusters, involving polyketones, non-ribosomal synthetic peptides, β-lactones, terpenes, and ribosomal synthetic peptides and other secondary metabolites were obtained in the prediction of secondary metabolites of strain AEB18 (Table 2). These peptidoglycan and polyketoglycan resistance compounds are crucial for the prevention and control of hybrid snakehead nocardiosis; the similarity of the kijanimicin gene cluster in L. sphaericus AEB18 was only 4%, and it might produce new bacteriostatic substances that need to be investigated in depth. Also, three unknown secondary metabolite synthesis gene clusters (cluster 3, 6, 9) were identified in the genome, indicating that the L. sphaericus AEB18 may have new gene clusters related to antagonists that could have a significant application potential in aquaculture.
Hybrid snakeheads were fed basal diet in the control group, while the experimental groups were given the same amount of basal diet with an additional mass fraction of 1% L. sphaericus AEB18 (1 × 109 CFU/g). After 21 days of feeding, the results showed that the growth performance and intestinal chyme digestive enzyme activity of hybrid snakehead in the AEB18 experimental group were significantly different from the control group (P< 0.05) (Tables 3, 4). The above results showed that the addition of L. sphaericus AEB18 promoted the growth and development of hybrid snakeheads.
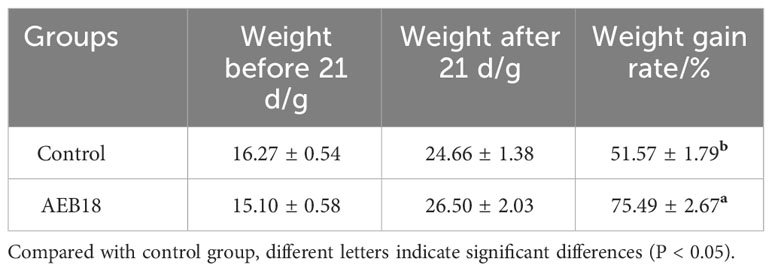
Table 3 Effect of the L. sphaericus AEB18 antagonism on weight gain rate of hybrid snakehead ( ± SD, n=60).
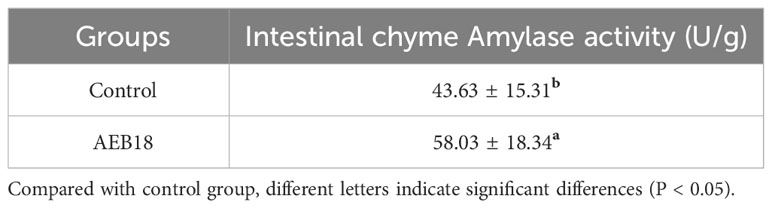
Table 4 Effects of L. sphaericus AEB18 on amylase activity in the intestine of hybrid snakehead ( ± SD; n = 18).
The experimental group was fed a basal diet with L. sphaericus AEB18 for 21 days before challenge test (1 × 107 CFU/mL of N. seriolae suspension 100 μL injected intraperitoneally in each fish); the negative control group was treated without poisoning after 21 days of the basal diet (intraperitoneal injection of 0.85% normal saline per fish); the positive control group was given a basal diet for 21 days, followed by challenge treatment (intraperitoneal injection of 1 × 107 CFU/mL of N. seriolae suspension 100 μL per tail). The results showed that the mortality rate of fish in the negative control group was 0%. Compared to the positive control group, the time of fish death in the experimental group fed L. sphaericus AEB18 was delayed by 4 days, which further deferred the occurrence of death and allowed time for control. On the 14th day after challenge, the mortality rate of the positive control group was 100% (all the fish died on the 8th day). However, the mortality rate of the L. sphaericus AEB18 + N. seriolae group was 75%, which significantly reduced the death caused by N. seriolae infection (P< 0.05) (Figure 8). The above results showed that the L. sphaericus AEB18 has a positive effect on preventing and treating diseases caused by N. seriolae.
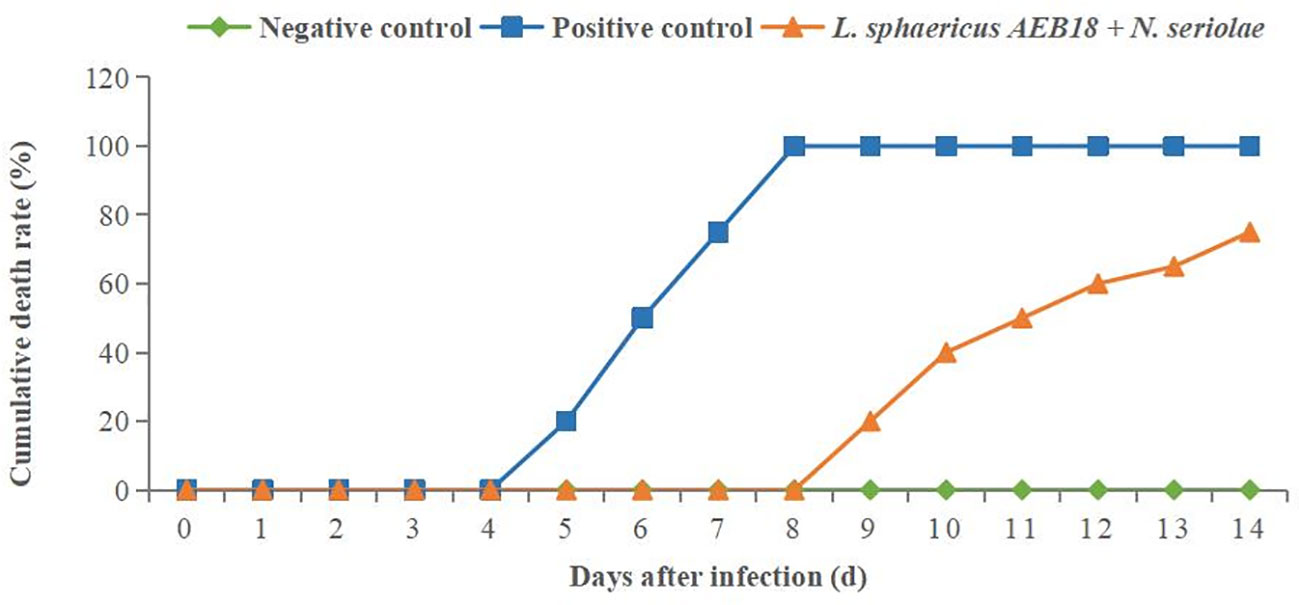
Figure 8 Protection rate of L. sphaericus AEB18 against hybrid snakeheads infected with N. seriolae.
The isolation of strains from the breeding environment including that of the host is the most effective way to screen probiotics. The majority of aquatic probiotics are screened according to the effect of inhibiting specific pathogenic bacteria. This antagonistic screening method has become a specific mode to identify probiotic strains. Several studies are focused on wild fish from an Indian maior carp, Catla catla (Hamilton) (Mukherjee and Ghosh, 2016), Atlantic cod (Gadus morhua) (Gildberg and Mikkelsen, 1998), turbot (Scophthalmus maximus) (Chen et al., 2016), goldfish (Carassius auratus) (Ghosh, 2008), gray mullet (Mugil cephalus L.) (Lin et al., 2013), and carp (Cyprinus carpio L.) (Feng et al., 2019) isolated from the intestines of probiotics that have antagonistic effects on common aquatic pathogens. In this study, we isolated microorganisms from healthy hybrid snakehead intestines and screened strains that have antagonistic effects on N. seriolae, of which Bacillus exerted a significant inhibitory effect, seeming to be a promising probiotic. Meanwhile, the 16S rRNA identification method was used to identify the strain of Bacillus AEB18 in depth. The results showed that the genetic correlation between AEB18 and L. sphaericus DSM 28 exhibited a 99.36% homology, indicating that AEB18 belongs to L. sphaericus. Studies have shown that L. sphaericus is widely used in larvicide activity and metal tolerance (Lozano and Dussán, 2013), however, its role in the prevention and control of aquaculture diseases has not been reported. Therefore, we screened and identified L. sphaericus AEB18 with potential probiotic effects for follow-up research.
Currently, the nanopore sequencing technology (third-generation sequencing technology) has been widely used for whole-genome sequencing on bacteria isolated from varied environmental media. Sher et al. (2020) performed whole-genome sequencing of one strain of bacteria carrying polymetallic genes isolated from industrial wastewater on the Illumina and Nanopore sequencing platforms, then, a complete sequence was assembled and identified as Micrococcus luteus, which was named AS2 based on 16S rRNA sequence, with a genome size of 2.86 Mbp and GC content of 72.8%. Jose et al. (2020) used nanopore sequencing technology to sequence the whole genome of one water-based strain of Vibrio cholerae, MinION sequencer produced 17,901 reads and 151,833,237 base pairs, with an average length of 8,551 bp and the longest sequence length of 86,745 bp, and the assembled sequence GC content was 47.72%, the N50 value was 2,984 kbp. The full-length genome of V. cholerae was successfully obtained by nanopore sequencing. Finton et al. (2020) performed whole-genome sequencing of antibiotic-resistant bacteria isolated from campus pond water samples through the second-generation sequencing platform Illumina MiSeq and the third-generation sequencing platform Oxford Nanopore MinION and identified Escherichia coli, Klebsiella pneumoniae, and Holderia parapot. Whole-genome sequencing data analysis showed that Enterobacter carried β-lactamase antibiotic-resistance genes, including blaCTX-M, blaTEM, blaCMY, blaSHV, blaKPC-like, and other genes (Finton et al., 2020). Sorokin et al. isolated a new halophilic strain AB-CW3 from the high-saline lake of Kulunda Steppe, assembled the complete genome of AB-CW3 by combining Nanopore and Illumina sequencing technologies, and found that it expressed an antimicrobial peptide antibiotic that had not yet been produced by Gram-positive bacteria (Sorokin et al., 2020). In our study, the whole genome of L. sphaericus AEB18 was first sequenced on the Nanopore sequencing platform, and the total length of the L. sphaericus AEB18 gene sequence was 4,653,716 bp, the length of N50 (bp) was 23,336, the length of N90 (bp) was 6,536, the GC content was 37.4%, and it contained 108 tRNAs, 37 rRNAs, and 95 ncRNAs, assembled into a complete genome of AEB18.
Secondary metabolites are a series of macromolecular non-essential substances synthesized by microorganisms in a specific growth period with primary metabolites as precursors, such as antibiotics, toxins, hormones, and pigments (Harwood et al., 2018). In this study, the antiSMASH online tool was used to predict the secondary metabolite gene clusters of strain AEB18, and a total of 9 lipopeptide and polyketone antibiotic gene clusters were obtained, involving polyketones, non-ribosomal synthetic peptides, β-lactones, terpenes, and ribosomal synthetic peptides. Among them, 6 antibiotic synthetic gene clusters (bacillibactin, kijanimicin, molybdenum cofactor, petrobactin, bacillopaline, and fengycin) were less similar to the currently known antibiotic gene clusters, of which one antibiotic gene cluster (butirosin) was only 7% similar to the currently known antibiotic gene cluster; it also included three gene clusters with unknown functions. The study has shown that bacillibactin is considered virulent against animal pathogens (Miethke et al., 2008), but its role in microbial competition remains to be elucidated. In addition to antimicrobial activity, kijanimicin also has antitumor activity (Bradner et al., 1983). Petrobactin exerts a protective effect against oxidative stress and improves spore-producing efficiency in Bacillus anthracis (Hagan et al., 2018). Fengycin is a bacteriostatic substance synthesized by Bacillus subtilis that targets the immune response to fungal infections and disrupts the cell membranes of the target cells (Vanittanakom et al., 1986). Based on the above information, it is speculated that the AEB18 can achieve biological control by producing a variety of bacteriostatic substances and CEs, and has great potential for aquatic applications.
Several studies have shown improved growth performance and survival rate of fish, which is a direct embodiment of the effect of probiotics. For example, Bjornsdottir et al. (2010) found that artemia was fortified with fish-derived probiotics (Vibrio sp., V. splendidus, and Pseudoalteromonas elyakovii) (107 CFU/g) that was then fed to open-stage Atlantic halibut (Hippoglossus hippoglossus L.), after 57 days, the survival rate and body weight of juvenile fish were significantly higher than the control group. Avella et al. (2010) reported that feeding three strains of Bacillus (B. subtilis, B. licheniformis, and B. pumilus) strengthened rotifer (Brachionus plicatilis) in the breeding of Sparus aurata, and the length and weight of larvae in the probiotic group increased significantly. Mohapatra et al. compared the probiotic compound feeding methods and found that feeding three active probiotics B. subtilis, Saccharomyces cerevisiae and Lactococcus lactis significantly improved the immunity of Labeo rohita species and the survival rate after Aeromonas hydrophila infection (Mohapatra et al., 2014). In this study, the results showed that compared to the control group, the L. sphaericus AEB18 (1 × 109 CFU/g) significantly (P< 0.05) promoted the growth of hybrid snakehead and increased the activity of intestinal digestive enzymes after 21 days applications, indicating an improved food utilization efficiency of hybrid snakeheads. This application effect was similar to that reported previously (Avella et al., 2010; Bjornsdottir et al., 2010; Mohapatra et al., 2014).
Accumulating evidences suggested that adding probiotics (e.g. Bacillus and Lactobacillus) to aquatic feed can inhibit the occurrence of bacterial diseases in fish (Bahi et al., 2017; Srisapoome and Areechon, 2017; Zhang et al., 2019b). The bacteriostatic activity of probiotics is manifested in the active substances, which exert antibacterial effects on a variety of fish pathogenic microorganisms. Additionally, probiotics improve the host’s disease resistance through direct antagonism to pathogenic bacteria. Antagonism is indispensable in maintaining the balance between potential probiotics and pathogenic bacteria in fish (Pandiyan et al., 2013). The growth inhibition effect of probiotics on pathogenic bacteria may be achieved by producing bacteriocins, lysozyme, proteases, siderophores, and other antimicrobial metabolites alone or in combination with other probiotics or prebiotics or by altering the pH in the intestine or competing with pathogenic bacteria for essential nutrients (Verschuere et al., 2000). On the other hand, probiotics regulate the balance of fish intestinal microflora, elevate the dominant flora and bacteria content in the intestine, compete with pathogenic bacteria for nutrient space, increase the number of beneficial bacteria, reduce the number of harmful bacteria, and enhance the function of the fish immune system, thereby inhibiting the pathogenicity of pathogenic microorganisms to fish (Kuebutornye et al., 2020). Similarly, in the present study, L. sphaericus AEB18 had a specific effect on the prevention and control of hybrid snakehead nocardiosis. The results showed that compared to the positive control group, the L. sphaericus AEB18 + N. seriolae group significantly reduced the mortality rate of hybrid snakeheads (P< 0.05). Thus, we speculated that the inhibitory effect of L. sphaericus AEB18 on pathogenic bacteria may be achieved by producing antibacterial metabolites, such as bacillibactin, kijanimicin, petrobactin, fengycin, and esterase, but its specific inhibitory mechanism needs to be investigated in depth.
In summary, the strain L. sphaericus AEB18, derived from the healthy intestine of hybrid snakeheads, demonstrates a notable inhibitory effect on N. seriolae in laboratory conditions. The genome of L. sphaericus AEB18 consists of a circular DNA with 4,653,716 base pairs (bp) and a GC content of 37.4%. Through the prediction of secondary metabolites, nine gene clusters associated with various bacteriostatic secondary metabolites, such as polyketones, non-ribosomal synthetic peptides, β-lactones, terpenes, and ribosomal synthetic peptides, were identified in L. sphaericus AEB18. Moreover, in vivo studies have shown that L. sphaericus AEB18 significantly enhances the growth of hybrid snakeheads and increases the activity of intestinal digestive enzymes. Additionally, it has a positive impact on the survival rate of fish infected with N. seriolae. These findings suggest that L. sphaericus AEB18 holds significant potential as a biocontrol agent and could be utilized to enhance disease management in freshwater aquaculture.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The animal study was approved by [please insert the full name and affiliation of the ethics committee/institutional review board: the Animal Ethics Committee of South China Agricultural University, Guangzhou, Guangdong, China (approval ID: SYXK-2019-0136). The study was conducted in accordance with the local legislation and institutional requirements.
TZ: Funding acquisition, Writing – original draft, Writing – review & editing, Project administration. PC: Data curation, Writing – original draft. JL: Investigation, Writing – original draft. ZL: Writing – review & editing. XD: Resources, Validation, Writing – review & editing. XH: Resources, Validation, Writing – review & editing. XZ: Resources, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors declare that this study received funding from China Postdoctoral Science Foundation (2022M720844). The funder had the following involvement in the study: Funding acquisition, Writing - original draft, Writing - review & editing, Project administration.
The authors would like to thank PC and JL for taking care of the snakehead and extracting test samples. Thanks to ZL and XD for helping with the experimental guidance. Special thanks to XH and XZ for helping with the manuscript revise.
Authors TZ, PC, JL and ZL were employed by the company Guangdong Haid Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Avella M. A., Gioacchini G., Decamp O., Makridis P., Bracciatelli C., Carnevali O. (2010). Application of multi-species of Bacillus in sea bream larviculture. Aquaculture 305 (1-4), 12–19. doi: 10.1016/j.aquaculture.2010.03.029
Bahi A., Guardiol F. A., Messina C., Mahdhi A., Cerezuela R., Santulli A., et al. (2017). Effects of dietary adminstration of fenugreek seeds, alone or in combination with probiotics, on growth performance parameters, humoral immune response and gene expression of gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 60, 50–58. doi: 10.1016/j.fsi.2016.11.039
Bartram A. K., Lynch M. D., Stearns J. C., Moreno-Hagelsie G., Neufeld J. D. (2011). Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 77 (11), 3846–3852. doi: 10.1128/AEM.02772-10
Baumann P., Clark M. A., Baumann L., Broadwell A. H. (1991). Bacillus sphaericus as a mosquito pathogen: properties of the organism and its toxins. Microbiol. Rev. 55 (3), 425–436. doi: 10.1128/mr.55.3.425-436.1991
Bjornsdottir R., Karadottir E. G., Johannsdottir J., Thorarinsdottir E. E., Smaradottir H., Sigurgisladottir S., et al. (2010). Selection of bacteria and the effects of bacterial treatment of Atlantic halibut (Hippoglossus hippoglossus L.) eggs and larvae. Aquaculture 302 (3-4), 219–227. doi: 10.1016/j.aquaculture.2010.02.026
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y. (2016). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research(W1). 45, 555–559. doi: 10.1093/nar/gkw960
Bondad-Reantaso M. G., Subasinghe R. P., Arthur J. R., Ogawa K., Chinabut S., Adlard R., et al. (2005). Disease and health management in Asian aquaculture. Veterinary Parasitol. 132 (3/4), 249–272. doi: 10.1016/j.vetpar.2005.07.005
Bradner W. T., Claridge C. A., Huftalen J. B. (1983). Antitumor activity of kijanimicin. J. Antibiot. 36, 1078–1079. doi: 10.7164/antibiotics.36.1078
Brown M. D., Watson T. M., Carter J., Purdie D. M., Kay B. H., Watson T. M., et al. (2004). Toxicity of VectoLex (Bacillus sphaericus) products to selected Australian mosquito and nontarget species. Econ. Entomol. 97 (1), 51–58. doi: 10.1603/0022-0493-97.1.51
Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37 (1), D233–D238. doi: 10.1093/nar/gkn663
Capkin E., Ozcelep T., Kayis S., Altinok I. (2017). Antimicrobial agents, triclosan, chloroxylenol, methylisothiazolinone and borax, used in cleaning had genotoxic and histopathologic effects on rainbow trout. Chenosphere 182, 720–729. doi: 10.1016/j.chemosphere.2017.05.093
Chen Y., Li J., Xiao P., Li G. Y., Mo Z. L. (2016). Isolation and characterization of Bacillus spp. M001 for potential application in turbot (Scophthalmus maximus L.) against Vibrio Anguillarum. Aquaculture Nutr. 22 (2), 374–381. doi: 10.1007/s11274-015-1833-2
Clarke J., Wu H. C., Jayasinghe L., Patel A., Reid S., Bayley H. (2009). Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4 (4), 265–270. doi: 10.1038/nnano.2009.12
Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 (18), 3674–3676. doi: 10.1093/bioinformatics/bti610
Davidson E. W., Morton H. L., Moffett J. O., Singer S. (1977). Effect of Bacillus sphaericus strain SSII-1 on honey bees, Apis mellifera. J. Invertebr. Pathol. 29 (3), 344–346. doi: 10.1016/S0022-2011(77)80041-4
Feng J. C., Chang X. L., Zhang R. Y., Yan X., Zhang J. X., Nie G. X. (2019). Effects of Lactococcus lactis from Cyprinus carpio L. as probiotics on growth performance, innate immune response and disease resistance against Aeromonas hydrophila. Fish Shellfish Immunol. 93, 73–81. doi: 10.1016/j.fsi.2019.07.028
Finton M. D., Meisal R., Porcellato D., Brandal L. T., Lindstedt B. A. (2020). Whole genome sequencing and characterization of multidrug-resistant (MDR) bacterial strains isolated from a Norwegian university campus pond. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01273
Ghosh S. (2008). Bioaugmentation in the growth and water quality of livebearing omamental fishes. Aquaculture Int. 16 (5), 393–403. doi: 10.1007/s10499-007-9152-8
Gildberg A., Mikkelsen H. (1998). Effects of supplementing the feed to Atlantic cod (Gadus morhua) fry with lactic acid bacteria and innumo-stimulating peptides during a challenge trial with Vibrio Anguillarum. Aquaculture 167, 103–113. doi: 10.1016/S0044-8486(98)00296-8
Hagan A. K., Plotnick Y. M., Dingle R. E., Mendel Z. I., Hanna P. C. (2018). Petrobactin protects against oxidative stress and enhances sporulation efficiency in Bacillus anthracis sterne. mBio 9 (6), e02079-18. doi: 10.1128/mBio.02079-18
Harwood C. R., Mouillon J-M., Pohl S., Arnau J. (2018). Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 42 (6), 721–738. doi: 10.1093/femsre/fuy028
Honda K., Littman D. R. (2012). The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30, 759–795. doi: 10.1146/annurev-immunol-020711-074937
Jose V. L., Pileggi M. T., Alam M. T., Ali A., Wong A. C. N. (2020). Draft genome sequence of an environmental Vibrio cholerae strain 2012Env-25, obtained using nanopore sequencing technology. Microbiol. Resour Announc. 9 (32), e00625–e00620. doi: 10.1128/MRA.00625-20
Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The kegg resource for deciphering the genome. Nucleic Acids Res. 32 (1), D277–D280. doi: 10.1093/nar/gkh063
Koren S., Walenz B. P., Berlin K., Miller J. R., Phillippy A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27 (5), 722–736. doi: 10.1101/gr.215087.116
Kuebutornye F. K. A., Wang Z. W., Lu Y. S., Abarike E. D., Sakyi M. E., Li Y., et al. (2020). Effects of three host-associated Bacillus species on mucosal immunity and gut health of Nile tilapia, Oreochromis niloticus and its resistance against Aeromonas hydrophila infection. Fish Shellfish Immunol. 97), 83–95. doi: 10.1016/j.fsi.2019.12.046
Lacey L. A. (2007). Bacillus thuringiensis serovariety Israelensis and Bacillus sphaericus for mosquito control. J. Am. Mosq. Control Assoc. 23 (2), 133–163. doi: 10.2987/8756-971x(2007)23
Lee D. H., Lim S. R., Han J. J., Lee S.-W., Ra C.-S., Kim J.-D. (2014). Effects of dietary garlic powder on growth, feed utilization and whole body composition changes in fingerling sterlet sturgeon, Acipenser ruthenus. Asian-Australasian J. Anim. Sci. 27 (9), 1303–1310. doi: 10.5713/ajas.2014.14087
Lin Y. H., Chen Y. S., Wu H. C., Pan S. F., Yu B., Chiang C. M., et al. (2013). Screening and characterization of LAB-produced bacteriocin-like substances from the intestine of grey mullet ( Mugil cephalus L.) as potential biocontrol agentsin aquaculture. J. Appl. Microbiol. 114 (2), 299–307. doi: 10.1111/jam.12041
Lozano L. C., Dussán J. (2013). Metal tolerance and larvicidal activity of Lysinibacillus sphaericus. World J. Microbiol. Biotechnol. 29 (8), 1383–1389. doi: 10.1007/s11274-013-1301-9
Miethke M., Schmidt S., Marahiel M. A. (2008). The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J. Bacteriol. 190, 5143–5152. doi: 10.1128/JB.00464-08
Mohapatra S., Chakraborty T., Prusty A. K., Prasad K. P., Mohanta K. N. (2014). Dietary multispecies probiotic supplementation enhances the immunohematological responses and reduces mortality by Aeromonas hydrophila in Labeo rohita fingerlings. J. World Aquaculture Society. 45 (5), 532–544. doi: 10.1111/jwas.12144
Mukherjee A., Ghosh K. (2016). Antagonism against fish pathogens by cellular components and verification of probiotic properties in autochthonous bacteria isolated from the gut of anIndian maior carp, Catla catla (Hamilton). Aquaculture Res. 47 (7), 2243–2255. doi: 10.1111/are.12676
Pandiyan P., Balaraman D., Thirunavukkarasu R., George E. G.J., Subaramaniyan K., Manikkam S., et al. (2013). Probiotics in aquaculture. Drug Invention Today 5 (1), 55–59. doi: 10.1016/j.dit.2013.03.003
Pauly D., Zeller D. (2017). Comments on FAOs state of word fisheries and aquaculture (SOFIA 2016). Mar. Policy 77, 176–181. doi: 10.1016/j.marpol.2017.01.006
Peña-Montenegro T. D., Lozano L., Dussán J. (2015). Genome sequence and description of the mosquitocidal and heavy metal tolerant strain Lysinibacillus sphaericus CBAM5. Stand Genomic Sci. 10, 2. doi: 10.1186/1944-3277-10-2
Powell S., Forslund K., Szklarczyk D., Trachana K., Roth A., Huerta-Cepas J., et al. (2014). eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 42, 231–239. doi: 10.1093/nar/gkt1253
Selim K. M., Reda R. M. (2015). Improvement of immunity and disease resistance in the Niletilapia, Oreochromis niloticus, by dietary supplementation with Bacillus amyloliquefaciens. Fish Shellfish Immunol. 44 (2), 496–503. doi: 10.1016/j.fsi.2015.03.004
Sher S., Hussain S. Z., Rehman A. (2020). Phenotypic and genomic analysis of multiple heavy metal-resistant Micrococcus luteus strain AS2 isolated from industrial waste water and its potential use in arsenic bioremediation. Appl. Microbiol. Biotechnol. 104 (5), 2243–2254. doi: 10.1007/s00253-020-10351-2
Sorokin D. Y., Mosier D., Zorz J. K., Dong X., Strous M. (2020). Wenzhouxiangella strain AB-CW3, a proteolytic bacterium from hypersaline soda lakes that preys on cells of gram-positive bacteria. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.597686
Srisapoome P., Areechon N. (2017). Efficacy of viable Bacillus pumilus isolated from farmed fish on immune responses and increased disease resistance in Nile tilapia (Oreochromis niloticus): laboratory and on farm trials. Fish Shellfish Immunol. 67, 199–210. doi: 10.1016/j.fsi.2017.06.018
Vanittanakom N., Loeffler W., Koch U., Jung G. (1986). Fengycin—a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot (Tokyo) 39 (7), 888–901. doi: 10.7164/antibiotics.39.888
Verschuere L., Rombaut G., Sorgeloos P., Verstraete W. (2000). Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Review. 64 (4), 655–671. doi: 10.1128/MMBR.64.4.655-671.2000
Watts J. E. M., Scjreier H. J., Lanska L., Hale M. S. (2017). The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar. Drugs 15 (6), 158. doi: 10.3390/md15060158
Yi Y. L., Zhang Z. H., Zhao F., Liu H., Yu L. J., Zha J. W., et al. (2018). Probiotic potential of Bacillus svelezensis JW: antimicrobial activity against fish pathogenic bacteria and immue enhancement effects on Carassiu sauratus. Fish Shellfish Immuology. 78, 322–330. doi: 10.1016/j.fsi.2018.04.055
Zhang D. F., Gao Y. X., Ke X. L., Yi M. M., Liu Z. G., Han X. Q., et al. (2019a). Bacillus velezensis LF01: in vitro antimicrobial activity against fish pathogens, growth performance enhancement, and disease resistance against streptococcosis in Nile tilapia (Oreochromis niloticus). Appl. Microbiol. Biotechnol. 103 (21/22), 9023–9035. doi: 10.1007/s00253-019-10176-8
Keywords: Lysinibacillus sphaericus AEB18, genome sequence, hybrid snakehead, growth, probiotic
Citation: Zhou T, Cai P, Li J, Li Z, Dan X, Huang X and Zhang X (2024) Whole genome analysis of intestinal source Bacillus and its effect on the prevention and control of hybrid snakehead (Channa maculata ♀ × Channa argus ♂) nocardiosis. Front. Mar. Sci. 11:1254806. doi: 10.3389/fmars.2024.1254806
Received: 06 October 2023; Accepted: 05 January 2024;
Published: 08 February 2024.
Edited by:
Benjamin Costas, University of Porto, PortugalReviewed by:
Arabinda Mahanty, National Rice Research Institute (ICAR), IndiaCopyright © 2024 Zhou, Cai, Li, Li, Dan, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyong Zhang, emhhbmd4aWFveW9uZ0BzY2F1LmVkdS5jbg==; Xiande Huang, aHVhbmd4ZEBzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.