- 1Department of Aquaculture, National Taiwan Ocean University, Keelung, Taiwan
- 2Study Program of Aquaculture, Mataram University, Mataram, Indonesia
- 3Department of Aquaculture, National Pingtung University of Science and Technology, Pingtung, Taiwan
The health of the host is significantly influenced by the gut microbiota. Penaeus vannamei (white shrimp) is one of the most profitable aquaculture species globally. Synbiotics are typically used as a beneficial diet supplement for raising aquaculture species’ growth capacities and enhancing immunity against pathogenicity. However, the effects of synbiotics on the white shrimp intestinal microbiota remain poorly understood. In the present study, we targeted the V3–V4 region of 16S rRNA genes to analyze the effects of synbiotics on white shrimp gut microbiota. Dietary synbiotics, having Lactobacillus acidophilus, and Moringa oleifera leaf extract were added to the white shrimps’ feed in various proportions in the present study. In total, 490 operational taxonomic units yielding 23 phyla, 41 classes, 94 orders, 151 families, and 250 genera of microorganisms were obtained. The diet containing L. acidophilus at 1 × 107 CFU/g and M. oleifera at 2.5 g/kg led to an increase in the relative abundance of beneficial microorganisms through a significant decrease in the α diversity. Moreover, it upregulated several physiological pathways such as carbohydrate metabolism, signal transduction, lipid metabolism, nucleotide metabolism, amino acid metabolism, and environmental adaptation, which led to the upregulation of the AMPK, MAPK, P13K-Akt, lysosome, peroxisome, and ferroptosis signaling pathways; this enhanced growth and immunity in white shrimp. Whether a single species or a combination of different microorganisms improves growth and immunity remains unclear till now. Nevertheless, our results will facilitate further in-depth investigation into beneficial microbial communities for upliftment of white shrimp aquaculture.
1 Introduction
White shrimp, one of the most significant aquaculture crustaceans accounts for approximately 52.9% of the global aquaculture production (Babinska et al., 2005). As an euryhaline species, white shrimp may be found in a variety of aquatic environments, making it the most prevalent and successful aquaculture species worldwide (FAO, 2016). Nevertheless, diseases due to several pathogens, such as bacteria, viruses, fungi, and parasites, threaten the shrimp farming sector. However, the frequent rise in shrimp bacterial infections, such as early mortality syndrome (EMS), acute hepatopancreatic necrosis disease (AHPND), hepatopancreas necrosis syndrome (HPNS), and white feces syndrome (WFS), has led to considerable economic losses annually across the globe (Sriurairatana et al., 2014; Lee et al., 2015; Choi et al., 2017; Huang et al., 2017; Xiong et al., 2017). Bacterial diseases are mostly seen in shrimp, and vibriosis is one of those diseases that continues to hinder shrimp farming development and cause large financial losses (Amatul-Samahah et al., 2020; FAO, 2020). In general, shrimp farmers use various antibiotics to reduce losses caused by Vibrio spp. However, excessive antibiotic use has resulted in the occurrence of antibiotic residues in food; it has also led to the emergence of antibiotic-resistant bacteria. Both these factors have decreased the quality of aquaculture products (Shinn et al., 2018). The use of probiotics (Anderson and Nelson, 2003; Wang A. et al., 2019; Prabawati et al., 2022) and natural extracts (Wu et al., 2021; Huang et al., 2022) can address the growing concern about eco-friendly aquaculture farm management. Several studies (Zhu et al., 2016; Hou et al., 2018; Huang et al., 2020; Zeng et al., 2020) have indicated that changes in the host gut microbiota are directly related to bacterial infections prevalence in white shrimp.
The host health state is fundamentally maintained by the intestinal microbiota (Hooper and Gordon, 2001; Boulangé et al., 2016). The identification of the host’s gut microbial profile and the related contributing factors is crucial. In aquaculture, many diseases are connected to the dysbiosis of their intestinal microbiota (Li et al., 2017; Dai et al., 2020; Huang et al., 2020). Moreover, their trophic level, developmental stage, and feed composition considerably affect their intestinal microbiota (Rungrassamee et al., 2013; Yan et al., 2016; Xiong et al., 2017; Liu et al., 2018; Wilkes Walburn et al., 2019).
In general, the use of specific probiotics in aquaculture can improve aquatic animals’ growth abilities to and help them respond to immunological stimuli, resist sickness, and maintain a healthy gut microbial population (Chien et al., 2020; Ringø et al., 2020). The probiotic bacterial species Lactobacillus spp. can boost the disease resistance of Penaeus monodon, a tiger shrimp, against Vibrio harveyi infection (Gobi et al., 2018). Tiger shrimp’s survival, immunity, and resistance to V. harveyi were also increased when they were given an oral dose of Bacillus subtilis BP11 (Rengpipat et al., 2003; Utiswannakul et al., 2011)). Lee et al. (2021) reported that dietary Limosilactobacillus fermentus SWP-AFFS02 reduced possible infections caused by Vibrionaceae and Enterobacteriaceae bacteria and increased survival, growth performance, and immunity in white shrimp. Similarly, white shrimp fed synbiotics with Lactobacillus plantarum and galacto-oligosaccharide exhibited a low prevalence of V. harveyi and Photobacterium damselae infections (Huynh et al., 2019).
Currently, next-generation sequencing (NGS) technology has the ability to sequence millions of DNA fragments at the same time. This technique allows for the prompt identification of microbial communities directly from materials, eliminating the requirement for culturing (Parlapani et al., 2018). NGS has numerous benefits, such as exceptional precision in identifying certain bacteria, the capacity to uncover previously unidentified species, extensive examination of the genome under study, and the utilization of the 16S rRNA gene (Gu et al., 2019). The application of high-throughput NGS technique using metagenomics, with a specific focus on the 16S rRNA gene, enables the thorough detection and characterization of the whole bacterial community found in the intestines of both healthy and ill shrimp (Ye et al., 2014). In the current study, we examined the structure and composition of the gut microbiota of white shrimp fed with six combinations of experimental synbiotic diets but grown under identical growth conditions in terms of temperature and oxygen supply. All experiments were performed in triplicates. Our results may also enhance the current knowledge on relationships between gut microbiota and white shrimp growth and immunity against pathogen. In particular, by Illumina-based NGS, we examined the variety and composition of the gut bacteria and the association of the gut bacterial structure with the growth of the shrimp fed with different diets. A thorough understanding of the bacterial ecology in white shrimp gut may aid in increasing the sustainability and productivity of white shrimp aquaculture.
2 Materials and methods
2.1 Experimental design
Here, we obtained six formulated diets with various pre-and probiotic concentrations, as indicated by Abidin et al. (2022), all of which led to differential growth performances (Supplementary Table 1). The following criteria were maintained while culturing the above samples: 18 75-L aquariums were randomly filled with 180 healthy juvenile white shrimp (2.04 ± .05 g), and the shrimps were divided into the six diet groups, with three replications per group. The shrimps were raised in a recirculating system with 50% water exchange daily and were fed with the experimental diets three times a day at 5% of their body weight for the course of the growth trial, which lasted 60 days. Every 2 weeks, the diet amount was changed based on the shrimp’s weight. The following water quality parameter were used: temperature = 28°C–29°C, oxygen content > 6 mg/L, salinity = 30–33, and pH = 6.8–7.6 (to preserve the water’s purity).
2.2 Sample collection and DNA extraction
After disinfecting the shrimp’s surface with 70% ethanol, three shrimps were randomly selected from each group. Next, by using sterile tools, we aseptically dissect the intestines and collected gut content. Finally, by using the Quick-DNA Fecal/Soil Microbe Miniprep kit (Zymo Research, USA), microbial DNA was extracted from gut content.
2.3 16S rRNA sequencing and data analysis
By using universal primers 341F and 805R (which have been used for investigating the microbiota of white shrimp), we performed amplicon sequencing of the V3–V4 region of the bacterial 16S rRNA gene. Illumina NovaSeq 6000 (Illumina, USA) was used for sequencing, and the Illumina TruSeq DNA Library kit was used to create a barcoded library. The sequencing data were uploaded to the NCBI database; the SRA accession numbers are SRR22923665–SRR22923684.
Analysis was performed in accordance with QIIME2’s (Quantitative Insight into Microbial Ecology) guidelines (v2019.10) (Bolyen et al., 2019). By using in-house scripts, the paired-end reads were overlapped to build the sequences. The sequence pre-processing was done using FastQC. The data were then demultiplexed by an internal script and trimmed by Trimmomatics (v0.39) for quality assurance. Cutadapt (v3.3) was then used with these guidelines: read length ≥ 150 bp and default error rate of 0.1. Preprocessing was then carried out using DADA2 (v1.12), which included eliminating chimera to filter amplicon sequence variants (ASVs), combining paired-end reads, and filtering out noisy sequences (denoising). By using the Usearch tool, the candidate sequences were grouped into operational taxonomic units (OTUs) based on a 97% sequence similarity and analysis using principal coordinates. By using the vegan function in R (version 3.4.4; https://www.r-project.org/), principal coordinate analysis (PCoA) plots based on unweighted UniFrac metrics were used to assess beta diversity. Through dimension reduction based on Euclidean and other distances, the probable principal components that could influence the variation in sample community composition were identified. A number of statistical analysis indicators are included in the alpha diversity package to quantify species diversity and abundance in the surrounding ecosystem. The indices, Chao and Ace (http://www.mothur.org/wiki/Chao and http://www.mothur.org/wiki/Ace), were used to determine community richness. Shannon (http://www.mothur.org/wiki/Shannon) and Simpson (http://www.mothur.org/wiki/Simpson) indices were employed to calculate community diversity. Predictions of the gut microbiota’s functional potentials were made using the PICRUSt2 (phylogenetic investigation of communities by reconstruction of observed states, v2.1.0-b) pipeline. Using the picrust2_pipeline.py software, functional profiles were predicted, creating a table of KEGG orthologs (KOs). By using the KEGG Mapper, we reconstructed KEGG reference categories (KEGG level 1) and modules (KEGG level 2) according to the KO annotations.
3 Results
In the current study, we assessed the effects of five dietary synbiotics treatments and a control diet on the structure, diversity, and uniqueness of the bacterial communities in the white shrimps. The diets were as follows (group A= Control; group B = L. acidophilus 1 × 107 CFU/g; group C = M. oleifera 2.5 g/kg; group D = M. oleifera 2.5gm/kg and L. acidophilus 1 × 107 CFU/g; group E = M. oleifera 5 g/kg; and group F consists of M. oleifera 5 g/kg and L. acidophilus 1 × 107 CFU/g). Despite being raised in identical environments and having the same initial weights, considerable growth differences were found among the six diet groups. Supplementary Table 4 represents each group’s average final weights, weight gain, specific growth rate, average daily growth, and feed efficiency (Abidin et al., 2022). After pooling three samples from each group, we performed bacterial community analyses.
3.1 16SrRNA sequence analysis
We prepared 20 libraries from the 6 groups (n = 3–4 group) after sequencing on Illumina NovaSeq 6000 platform and obtained 2,355,035 raw sequences with an average of 117,751 sequences per sample (Table 1). Next, 2,136,094 high-quality sequences were selected after quality filtering and were used to create OTUs. Table 1 provides a summary of the raw and filtered sequences acquired at each data processing stage.
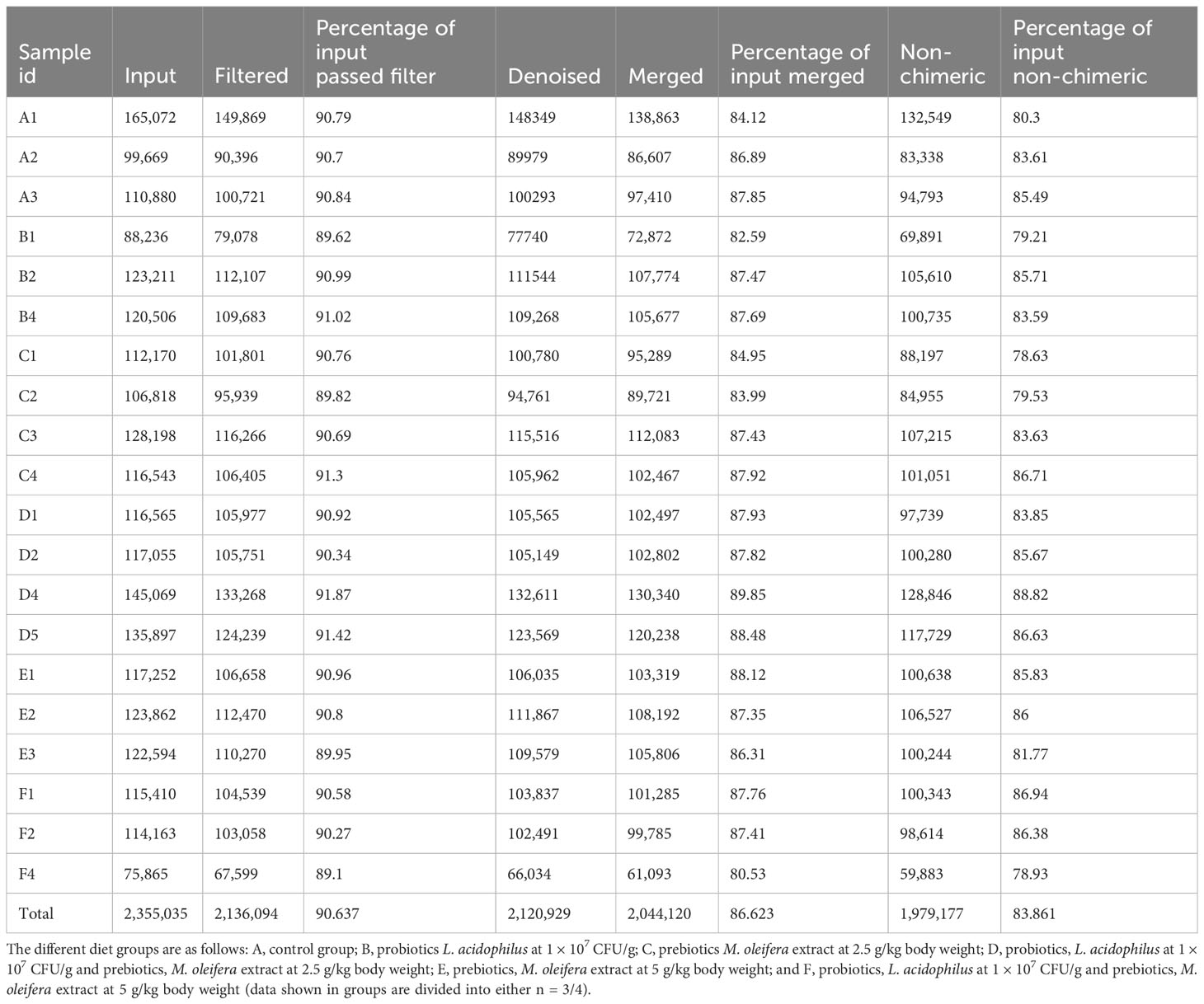
Table 1. 16S rRNA sequencing reads obtained from P. vannamei gut content fed with different experimental diets.
The highest and lowest numbers of sequences were generated from group A and F, respectively. Good’s coverage revealed that >90% of all OTUs were obtained for various treatments, indicating that the sequencing effort accurately reflected most bacterial communities in the group F. Consequently, at the obtained sequencing depth, the rarefaction curves also indicated outstanding bacterial community resolution (Supplementary Figure 1). After applying frequency filters, we obtained 490 OTUs, of which 97.51% were shared by all six groups, and all OTUs shared 97% similarity. Of these 490 OTUs, only 374 could be assigned to putative bacteria under various taxonomical categories.
3.2 Core shrimp gut bacteria
Taxonomic annotation corresponding to the OTUs was created to illustrate bacterial community structures and diversities. Groups A, B, C, D, E, and F demonstrated 135, 121, 191, 186, 171, and 318 OTUs, respectively; group F demonstrated the highest OTU number and diversity. The 374 OTUs were classified into the domain bacteria under 23 phyla (Acidobacteria, Actinobacteria, Armatimonadetes, Bacteroidetes, Chlamydiae, Chloroflexi, Cloacimonetes, Cyanobacteria, Dadabacteria, Deferribacteres, Deinococcus-Thermus, Dependentiae, Epsilonbacteraeota, Firmicutes, Fusobacteria, Nitrospirae, Patescibacteria, Planctomycetes, Proteobacteria, Rokubacteria, Spirochaetes, Tenericutes, and Verrucomicrobia), 41 classes, 94 orders, 151 families, and 250 genera (Supplementary Table 2).
Of the aforementioned phyla, five were the prevalent core phyla because they represented >1% of all sequences. Proteobacteria represented 38% of the total sequences in white shrimp, followed by Bacteroidetes (30%), Tenericutes (17%), Firmicutes (13%), Patescibacteria (1%), and others (1%) (Figure 1A). The most frequent bacterial genus was also revealed by the association between the 30 most abundant genera (Figure 1B).
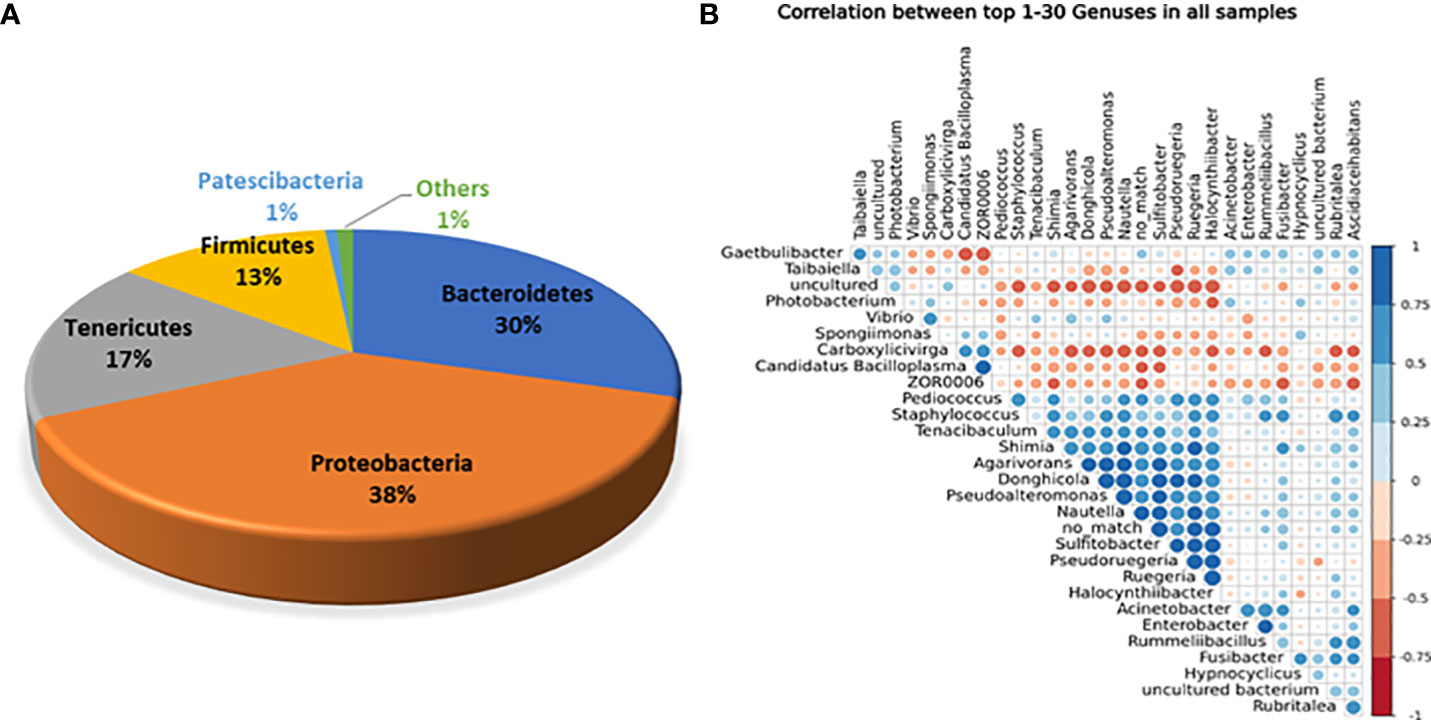
Figure 1. (A) Abundance of overall core bacterial phyla present in the gut of white shrimp, P. vanammei. (B) Correlation among core bacterial genera present in gut of white shrimp, P. vannamei.
3.3 Diversity and structure of bacteria within and between groups
Through treatment-wise sampling, which indicates the microbial community richness and variety (Figure 2), we determined the diversity of the bacterial community and specific taxa within the groups. The results indicated that group F had much higher OTU richness and more species than the other groups. Moreover, group F demonstrated much higher species richness and evenness than did the other groups, as measured using the Shannon index. A strong correlation was also noted between the gut bacteria and shrimp’s body weight. The beta diversity of the bacterial communities in the white shrimp gut from the six groups was examined through PCoA (Figure 3). A total of 39% of the variation was explained by the first two components (PC1, 21%; PC2, 18%). Moreover, groups D and F had a broader dispersion than the other groups, which indicates the significant variations among the shrimp groups. Our analysis of similarities (ANOSIM) revealed no significant difference among the six groups (Table 2). The results of the Multiple Response Permutation Procedure (MRPP) results were comparable to those obtained using ANOSIM (Table 2).
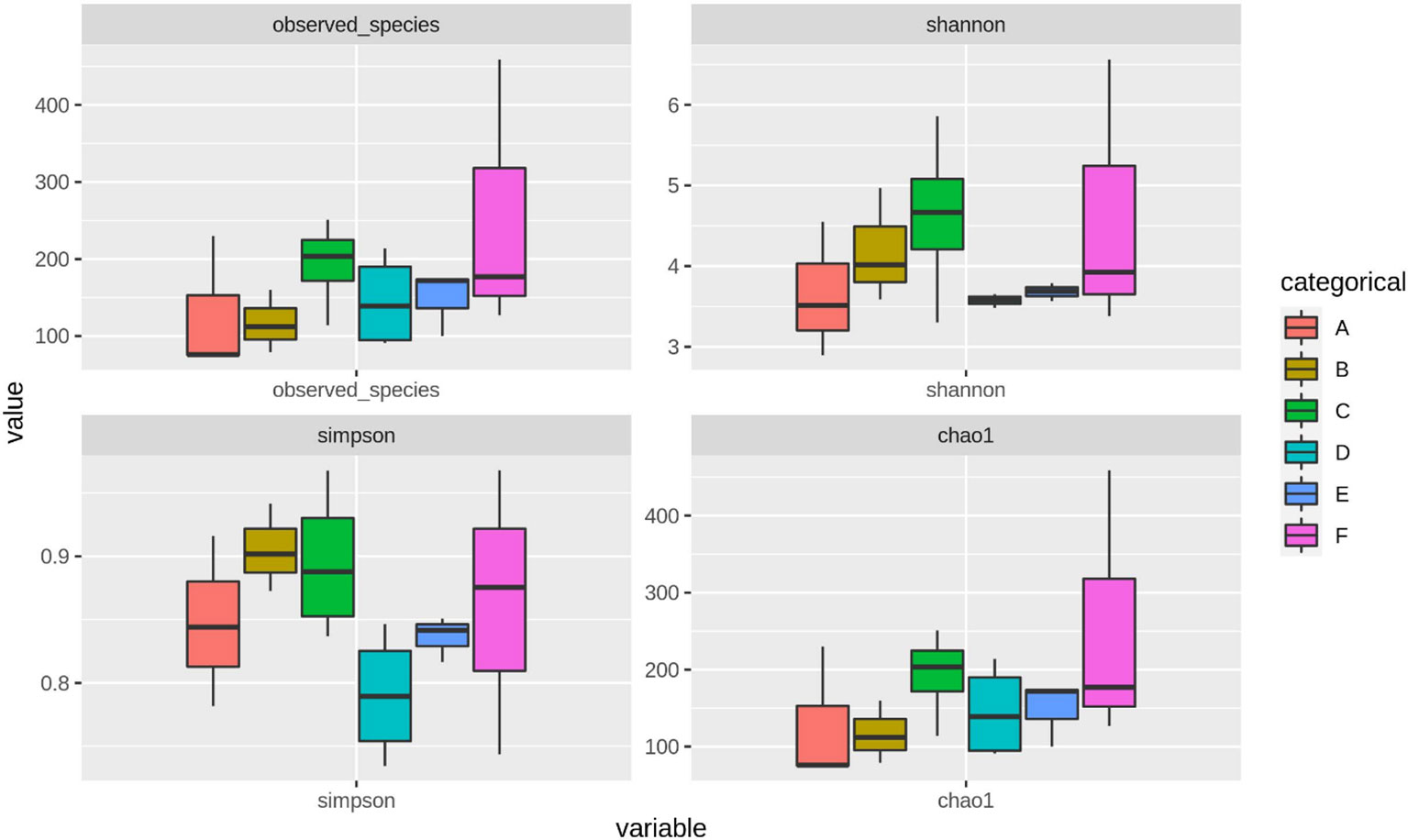
Figure 2. The α-diversity indices throughout different experimental diets of P. vannamei. The experimental diets are as follows: A: Control group, B: probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at 5g/kg body weight.
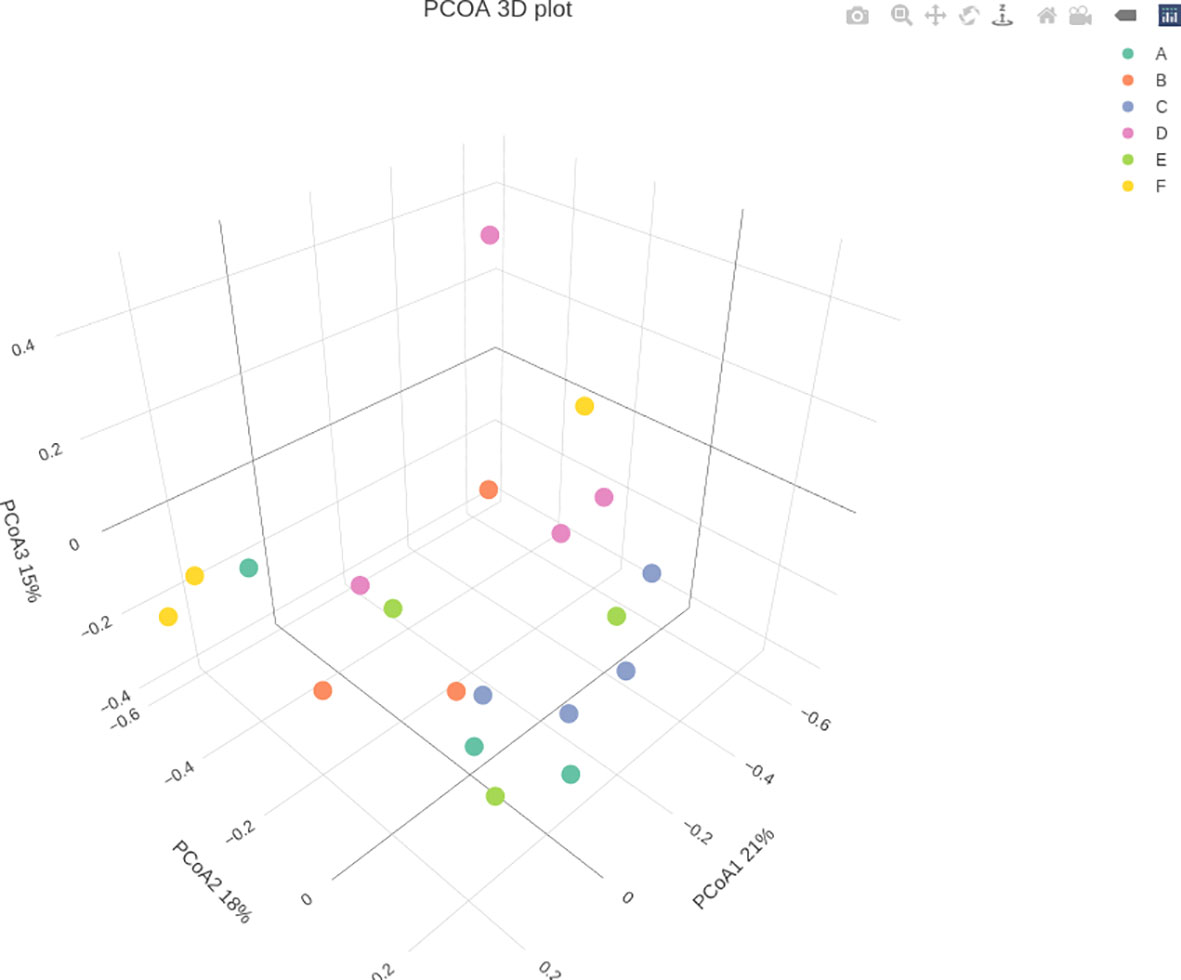
Figure 3. Principal Coordinates Analysis (PCoA) of P. vannamei at different growth rates of the bacterial community associated with the guts of different experimental diets. The experimental diets are as follows: A: Control group, B: probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at5g/kg body weight.
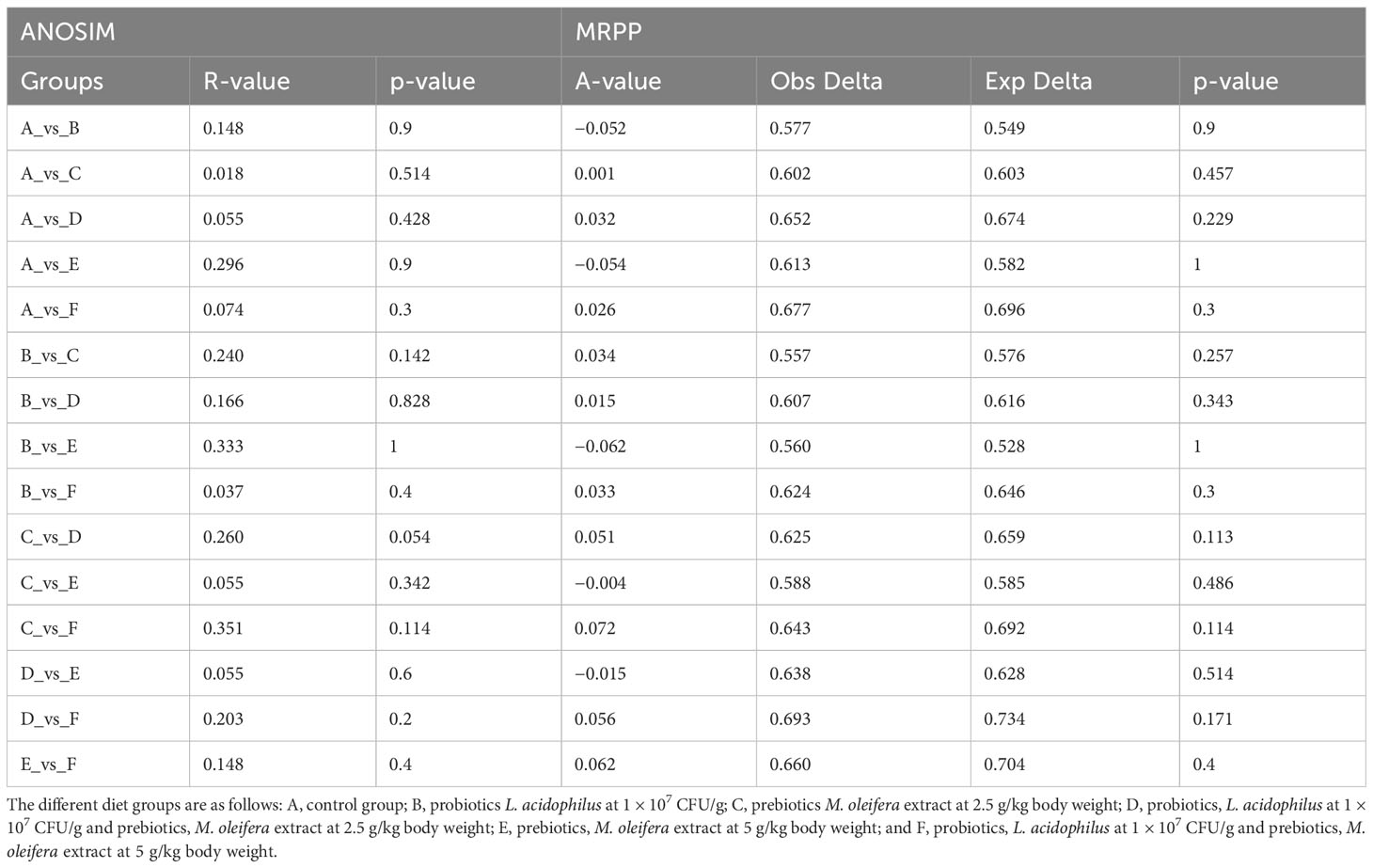
Table 2. Comparative analysis of ANOSIM and MRPP of gut microbiota among different experimental diets.
3.4 Bacterial signature is diet and growth dependent in shrimps
Bacterial phylum compositions in the white shrimp gut in all six groups are shown in the Figure 4A. The dominant phyla in group A were Bacteroidetes (34.79%) and Proteobacteria (29.52%) representing approximately 65% of the total gut microbiota. Moreover, groups B, C, and E demonstrated an abundance of Proteobacteria (37.15%, 63.32%, and 43.83%, respectively) and Bacteroidetes (29.62%, 21.83%, and 28.32%, respectively). Compared with the higher growth groups, groups D and F shows an abundance of slightly different bacterial phyla. Group F shows an abundance of Bacteroidetes (44.17%) followed by that of Proteobacteria (38.05%). Moreover, group D shows an abundance of Tenericutes (29.22%), followed by that of Firmicutes (26.24%; Figure 4A).
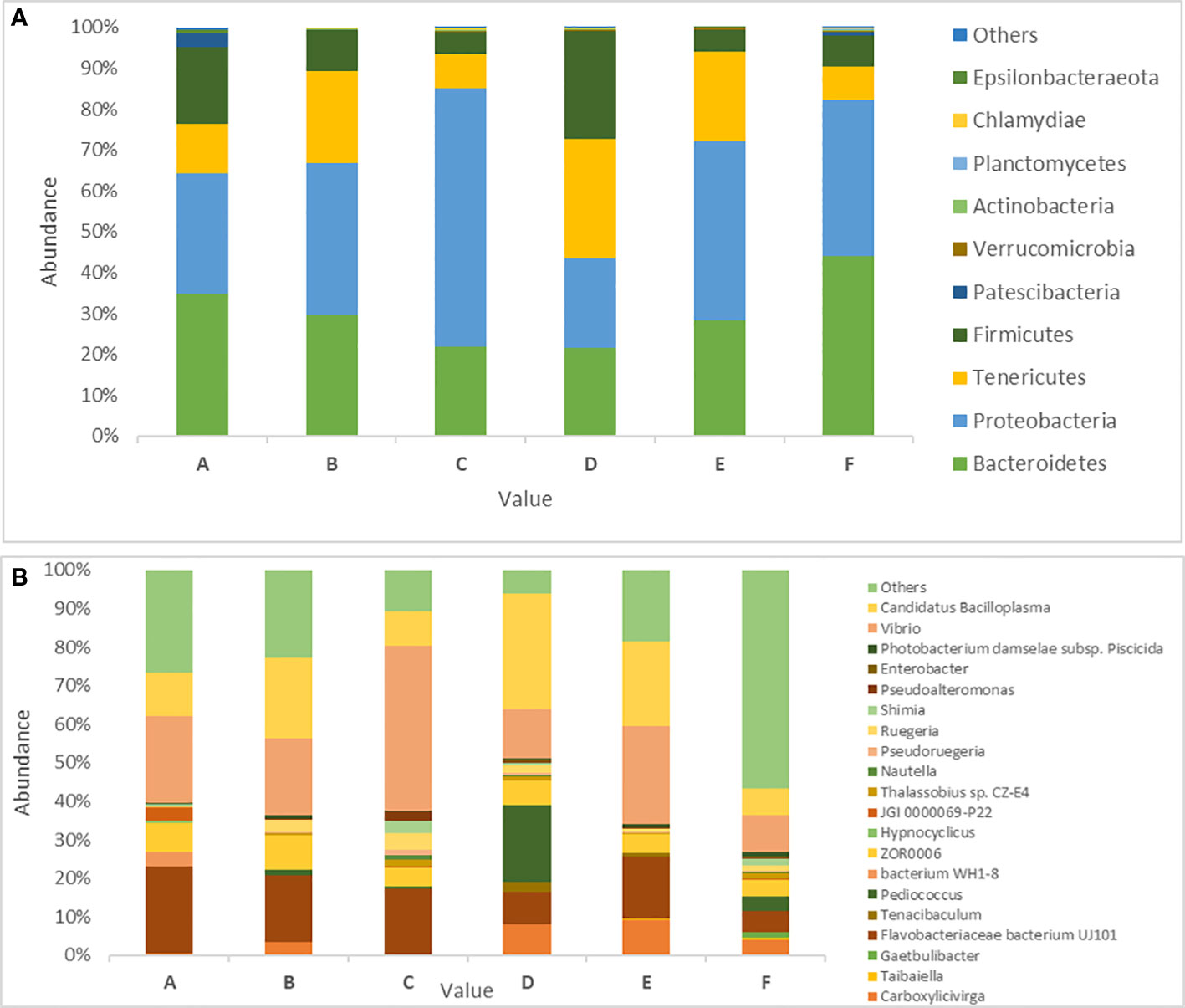
Figure 4. Prevalence of gut microbiota in P. vannamei in six different experimental diets. (A) Abundance of 10 dominant Bacterial phyla across the six different treatments in P. vanammei. (B) Showing abundance of Bacterial genus across the five different treatment and control. The experimental diets are as follows: A: Control group, B: probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at 5g/kg body weight.
In total, 250 genera were identified from all six groups. Group A demonstrated the highest abundance of Spongiimonas (Flavobacteriaceae bacterium UJ101), followed by Vibrio, Candidatus bacilloplasma, Firmicutes bacterium ZOR0006, and Fusibacter. Group B demonstrated the highest abundance of C. bacilloplasma, followed by Vibrio, Spongiimonas, Photobacterium, and Firmicutes bacterium ZOR0006. Group C had an abundance of Vibrio followed by Spongiimonas, C. bacilloplasma, Firmicutes bacterium ZOR006, and Ruegeria. Group E had the highest abundance of Vibrio followed by C. bacilloplasma, Spongiimonas, Photobacterium, and Carboxylicivirga. Group F exhibited the highest abundance of uncultured Flavobacteriaceae bacterium, followed by Photobacterium, Vibrio, C. bacilloplasma, and Spongiimonas. Finally, group D demonstrated the highest abundance of C. bacilloplasma, followed by Peddiococcus, Vibrio, Spongiimonas, and Carboxylicivirga (Figure 4B). Significant between-group differences were found in the abundance of the pathogenic Vibrionaceae bacteria; the occurrence was higher in groups A, B, C, and E (slow-growth groups) but lower in groups D and F (the high growth groups). Three novel Vibrionaceae genera Vibrio fortis, Enterovibrio, and uncultured bacterium were found only in the slow-growth group and group A (Figure 5A). In addition, a decline was recorded in the abundance of other pathogenic bacteria in groups D and F (the high-growth groups; Figure 5B).
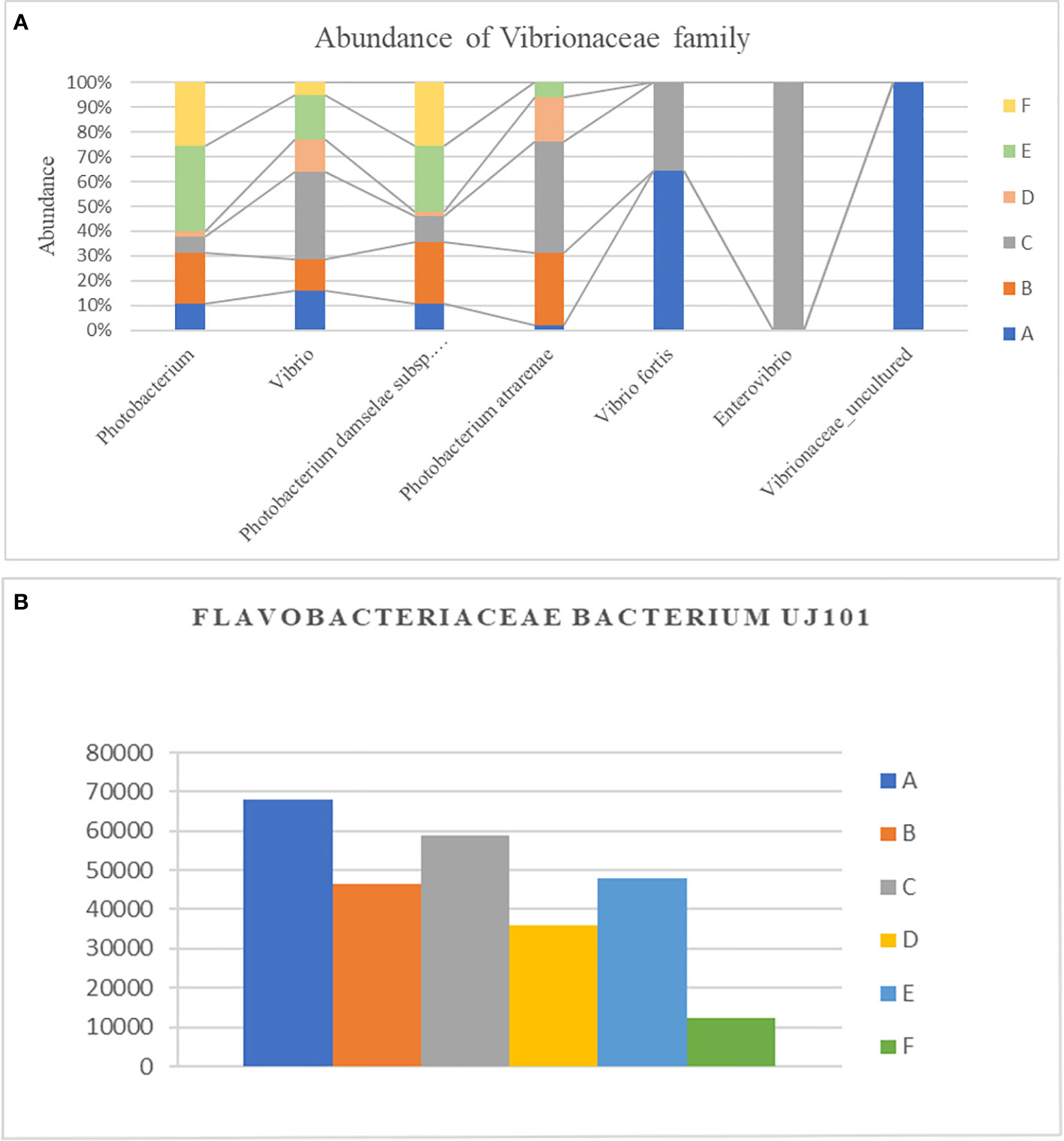
Figure 5. Effect of different experimental diets on opportunistic pathogens. (A) Difference in relative abundance of Vibrionaceae family along different genera in different experimental diets. (B) Difference in relative abundance of Flavobacteriaceae bacterium UJ101 with different experimental diets. The groups are as follows: A: Control group, B: probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at5g/kg body weight.
3.5 Diet-specific shared and unique gut microbiota in white shrimp
To identify the unique gut microbiota among five groups and one control diet group, species abundance of the individual treatment group was first characterized at the taxonomic level of the genus so as to reflect the white shrimp’s adaptation to the treatments (Figure 2). In general, the larger the treatment plot, the higher is the shrimp gut capacity to accommodate different types of microbes. Venn diagrams demonstrating the unique phyla and genera and those shared between the groups are shown in Supplementary Figures 1A, B). Venn diagram of OTUs for phylum shows at least 8 out of 23 phyla shared between all the groups. Moreover, Fusobacteria was unique to group A; Dadabacteria and Dependentiae were unique to group C; and Armatimonadetes, Acidobacteria, Deferribacteres, Nitrospirae, Chloroflexi, Cloacimonetes, and Deinococcus-Thermus were unique to group F (Supplementary Figure 2A and Supplementary Table 3A).
Similarly, as shown in the Venn diagram, 51 genera were shared by all groups but at different abundance levels (Supplementary Figure 2B and Supplementary Table 3B). Moreover, groups A, B, C, D, E, and F had 26, 10, 33, 35, 18, and 133 unique genera, respectively.
3.6 KEGG functional annotation analysis
Our KEGG analysis give rise to six functional categories at level 1 across all the groups: cellular processes, metabolism, organismal system, human diseases, genetic information processing, and environmental information processing. These six categories could be subdivided into 44 functional categories at level 2. The top 10 level 2 functional categories were the biosynthesis of other secondary metabolites, signal transduction, endocrine system, metabolism of terpenoids and polyketides, lipid metabolism, carbohydrate metabolism, amino acid metabolism, glycan biosynthesis and metabolism, and metabolism of cofactors and vitamins. Then, by using online SRPlots (http://www.bioinformatics.com.cn/en?p=1), statistically distinct aspects of functional categories based on KEGG (level 2) were analyzed between the six diet groups. Group A had a higher abundance of infectious diseases, namely, bacterial and parasitic; endocrine system; and folding sorting and degradation. These groups were demonstrated to have a significantly lower abundance in groups F and D. In particular, group D showed an increase in abundance of carbohydrate metabolism, lipid metabolism, transcription, nucleotide metabolism, membrane transport, etc. (Figure 6A).
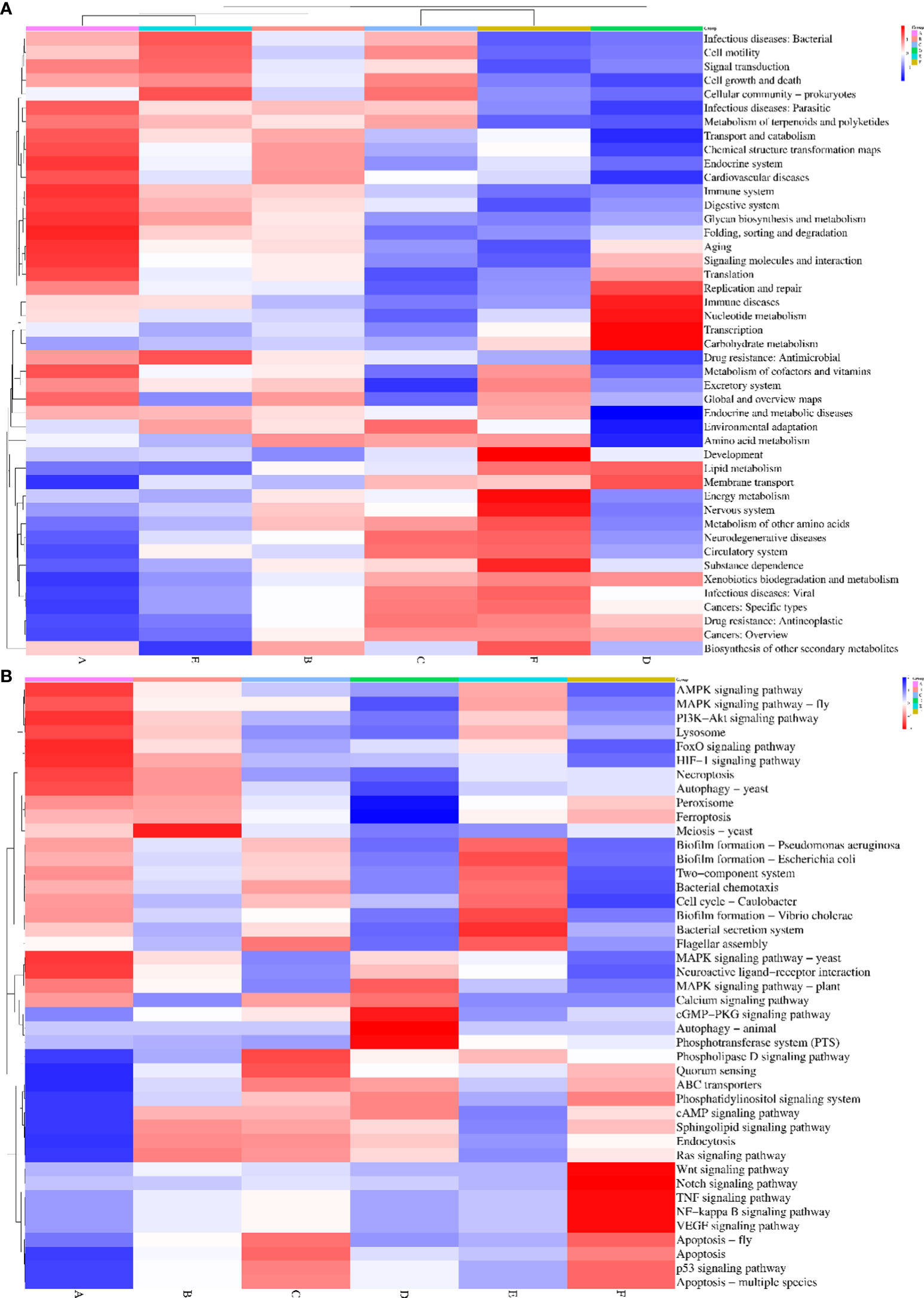
Figure 6. Heatmap showing the alteration of KEGG metabolic pathways after administration of six different experimental diet (A) Heatmap showing functional pathways of different groups in Level 2. (B) Heatmap showing functional pathways of different groups in Level 3. The different diet groups are as follows: A: Control group, B: probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at5g/kg body weight.
The 44 level 2 functional categories could be subdivided into 346 different level 3 functional groups by using the KO terms. The most commonly occurring functional category, namely, biosynthesis of other secondary metabolites, could be divided into 22 pathways under 22 KO terms: KO00525, KO009,65, KO00232, KO00332, KO00331, KO00944, KO00941, KO00966, KO00901, KO00943, KO00950, KO00261, KO00524, KO00401, KO00311, KO00405, KO00940, KO00333, KO00404, KO00945, KO00521, and KO00960. The functional category of signal transduction could be divided into 20 pathways showing considerable differences among groups A, D, and F (Figure 6B). Level-3 KEGG pathway assessment with reference to signal transduction revealed the upregulation of the AMPK, MAPK, and P13k-Akt signaling pathways; peroxisome; lysozyme; and ferroptosis in diet group D, whereas, these pathways were downregulated in group A (control).
4 Discussion
The rapid increase in the global population has led to an increase in the demand for seafood. Aquaculture seafood production has increased significantly. However, extensive aquaculture might foster the growth of potential pathogens, which could result in frequent disease outbreaks and high rates of mortality. Traditional methods of disease prevention, such as the use of antibiotics and chemotherapy, were once widely employed in many countries, but their usage has since been outlawed due to public knowledge and worries about food safety. This necessitated the discovery of novel, ecologically acceptable methods to reduce the vulnerability of the animals to diseases. In the aquaculture sector, probiotic use is strongly promoted as a risk-free, ecologically friendly approach for preventing diseases, reducing the harmful effects of diseases and enhancing the growth of aquaculture animals. Therefore, in this study, we fed white shrimp with the probiotics L. acidophilus combined with the prebiotics M. oleifera extract and assessed their effects on the gut microbiota of the white shrimp. Understanding the effects of microbiota on shrimp health and sickness is crucial for managing its composition under challenging situations that can have an adverse impact on farm production. In hosts, various factors, including the host’s developmental cycle, food preferences, intestinal pH, and geographic location, influence the richness of the gut microbiome (Kim and Jazwinski, 2018). Aquatic microbiologists were more interested in studying the gut microbiota because it plays a significant role in the growth of its hosts through a variety of mechanisms, such as producing digestive enzymes for food digestion (Amin, 2018), excreting vitamins (Chen J. et al., 2017), and producing short-chain fatty acids (SCFAs) (Hoseinifar et al., 2017). Dietary management, such as probiotic and prebiotics supplementation, affects the gut microbiota composition (Gbadebo et al., 2019; Amoah et al., 2020; Munaeni et al., 2020). The growth performance of shrimp fed with diets containing a combination of a probiotic and a prebiotic is higher than that of shrimp fed with diets containing a probiotic or prebiotic alone (Abidin et al., 2022) Moreover, many researchers also found an increase in growth performance by using multiple probiotics (Lee et al., 2022). In the current study, the highest growth performance was demonstrated by group D (2.5 g/kg body weight M. oleifera and 1 × 107 CFU/g body weight L. acidophilus). However, as stated by Abidin et al. (2022) the feed containing 5g/kg body weight M. oleifera prebiotics with or without probiotics did not lead to significant growth.
4.1 Core microbiota of the gut
We observed that the structure and composition of gut bacteria were related to white shrimp growth, emphasizing the essential role of gut bacteria in shrimp growth. Across all gut content samples, we identified at least 23 phyla, out of which Proteobacteria dominates the bacterial phyla with occurrence of 38%, followed by Bacteriodetes 30%, Tenericutes 17%, Firmicutes 13%, Patescibacteria 1%, and others. Several authors have noted that Proteobacteria predominated in the shrimp gut. On the other hand, He et al. (2020); Zhang et al. (2019), and Zoqratt et al. (2018) have reported that Proteobacteria represent 44.12%, 38.94%, and 50%–80% of all shrimp gut bacteria. Amin et al. (2022) reported that in a white shrimp culture in Indonesia, 53.99% of the gut bacteria were Proteobacteria. Similarly, Gao S. et al. (2019) and Hasyimi et al. (2020) reported the predominance of Proteobacteria in the gut of white shrimp raised in Indonesia with Bacteroides and Firmicutes being the second and third most prevalent.
Fan J. et al. (2019) reported a relatively low prevalence of Proteobacteria (40.83%) in the gut of white shrimp raised in China; nevertheless, the bacterial species remained the predominant gut microbe, followed by Bacteroidetes (19.96%), Verrucomicrobia (8.26%), Firmicutes (6.17%), and Actinobacteria (1.59%). Finally, Huynh et al. (2019) also noted that Proteobacteria were predominant in the white shrimp gut in both the indoor (86.6s%) and outdoor (51.8%) pond culture systems.
The shrimp gut bacterial population appears to vary genus-wise and be influenced by a variety of factors, such as shrimp age, life stage, feeds, culture system, and geographical location (Fan J. et al., 2019; Fan L. et al., 2019; Gao S. et al., 2019; Huynh et al., 2019; Hasyimi et al., 2020). The current findings showed 250 bacterial genera in the total shrimp gut content. Of these genera, only 30 were considered to be included in the core white shrimp gut microbiome because of their frequency in all samples, out of which Vibrio, Candidatus bacilloplasma, Pseudoalteromonas, Donghicola, Agarivorans, Sulfitobacter, Rugeria, Enterobacter, Shimia, etc. show a higher abundance throughout all the white shrimp gut content. These results are remarkably comparable to those of Gao S. et al. (2019) [Vibrio (31%), Photobacterium (23.9%) and, C. Bacillophora (7.6%)]. Hasyimi et al. (2020) earlier reported several dominating genera from shrimp grown in Indonesia, with the five most prevalent genera being Nocardiodes, Neptunomonas, Spongiimonas, Desulfopila, and Bryobacter. These data strongly imply the presence of a wide range for the proportions of different genera of gut bacteria.
In particular, some genera including Photobacterium, Spongiimonas, and Neptunomonas were also frequently reported as the predominant genera in white shrimp gut.
4.2 Bacterial signature is diet and growth dependent in shrimps
Here, we noted that groups D and F demonstrated a high bacterial diversity based on the Shannon index and species richness index. In comparing with the high growth group, group D shows a fewer diversity and yet shows a high growth rate. These findings might imply that particular community of microbes with high diversity and species richness are preferable, particularly in terms of growth rate. This is in contrast with the results of Lee et al. (2022): less diversity was associated with high growth performance. Daniels et al. (2013) indicated that the gut of shrimp with a high growth rate demonstrated a larger variety of bacterial species and a higher Shannon index. The current findings are in contrast with those of Fan J et al. (2019): white shrimp with higher growth performance demonstrated a higher Shannon index. Huynh et al. (2019), on the other hand, came to the conclusion that no significant relationship between the Shannon index and the growth of white shrimps was observed, since there was no discernible difference in the average values of the Shannon index between shrimp with rapid and sluggish growth. These findings could suggest that other variables, such as the dominating genus or species level, influence the growth of white shrimps in addition to species richness and variety of gut bacteria. In the present study, the most dominant bacterial phylum in the high growth group, i.e., group D, was Tenericutes, followed by Firmicutes, Proteobacteria, and Bacteroidetes. In group F, the Bacteroidetes and Proteobacteria were reported to have high percentage in occurrence. In the present study, we also found a significantly high occurrence of firmicutes in group D in comparison to the other five groups. Candidatus bacilloplasma dominates the intestines of healthy shrimp, but changes in its abundance may cause a change in the disease-causing bacterial population in the intestinal microbial communities in shrimp infecting with many diseases (Chen W. Y. et al., 2017; Chen Y et al., 2017; Huang et al., 2020; Wang et al., 2020b). In this study, the increase in abundance of C. bacilloplasma might influence the growth status of shrimp. However, to date, the functions of C. bacilloplasma in the shrimp intestine are still unknown. In addition, in group D, another genus commonly found was Pedicoccus and was found in significantly higher occurrence when compared to other groups. Given that Firmicutes make up a large portion of aquaculture probiotics (Wang Y. C. et al., 2019), it is possible that these organisms may significantly influence the growth of white shrimps, albeit more research is still needed. Another bacterial genus with high occurrence is Vibrio, being an opportunistic bacterium; it has been found in almost every group irrespective of the diets. According to reports (Huang et al., 2018; Fan L. et al., 2019; Shao et al., 2019), Vibrio was one of the most common genera found in shrimp intestines, and its abundance in shrimp ponds was far higher than that of the surrounding water or soil. Our result shows that the percentage of Vibrio has been decreased in groups D and F where probiotics and M. oleifera have been used significantly. Additionally, it has been discovered that an excess of Vibrio in the gut could alter the shrimp’s health status by increasing the possibility of disease outbreaks (Xiong et al., 2017; Hou et al., 2018). To lessen the risk of disease outbreaks, it is crucial to regularly monitor the prevalence of Vibrio in certain areas because shrimps are susceptible to infections from the water. High abundances of Vibrio in the water can reach the shrimp’s intestinal tract.
4.3 Functional annotation
To explore the mechanisms underlying the biochemical and physiological changes in shrimp, we performed functional annotation. The KEGG level 2 annotation results demonstrated five dominant functional category groups in group D: carbohydrate metabolism, nucleotide metabolism, lipid metabolism, transcription, and membrane transport (Schweinitzer and Josenhans, 2010; LeBlanc et al., 2017). This is in line with numerous earlier studies on environmental and intestinal microbiota. Moreover, our KEGG level 2 analysis showed that gut microbiota were enriched in carbohydrate metabolism. Studies in shrimp, panda, and turbot (Zhu et al., 2011; Xing et al., 2013; Gao M. et al., 2019) have indicated that carbohydrates may be a primary nutrient supplied to microbiota. Our KEGG level 3 annotation with respect to signal transduction revealed the upregulation of MAPK signaling pathway in group D. Members of the MAPK family’s cascade are crucial in controlling immune response and cell viability. In animals ranging from yeast to humans, MAPKs are evolutionary-conserved signaling pathways that act in response to stimulating signal transduction (Zhang and Dong, 2007; Huang et al., 2017). A variety of phosphorylation processes work together to activate MAPKs. When phosphorylated, MAPK affects a variety of transcription factors, enzymes, and other proteins that regulate cellular activity by phosphorylating a variety of substrates in the cytosol and nucleus (Fujiwara and Denlinger, 2007; Morrison, 2012; Huang et al., 2017). Another metabolic pathway that shows an upregulation is the AMP-activated protein kinase (AMPK) signaling pathway in group D. AMPK controls energy metabolism and serves as a sensor for intracellular energy in terrestrial vertebrates (Hardie, 2003). The role of AMPK in the energy metabolism of aquatic species, particularly shrimp, is, on the other hand, less well understood. Magnoni et al. (2012) indicated that AMPK is critical for promoting glucose uptake in fish skeletal muscles; thus, it might have a role in controlling energy responses to hypoxia in fish.
Ferroptosis is a unique reactive oxygen species (ROS)–dependent cell death process characterized by an iron overload and lipid peroxide formation. The physical signs of ferroptosis include shrinking of the mitochondria, a decrease in or absence of the mitochondrial cristae, and an increase in mitochondrial membrane density. Similar to other cell death processes, glutathione (GSH) synthesis, lipid peroxidation, cysteine transport, iron homeostasis, and NADPH all play vital roles in the regulation of ferroptosis (Zheng and Conrad, 2020). In the present investigation, we have seen comparatively high upregulation of ferroptosis in group D than in group A (control group).
In addition, noticeable upregulation of ferroptosis in group D could be related to the high number of beneficial bacteria present in this groups. In contrast to apoptosis, necrosis, and autophagy, ferroptosis is a type of intracellular iron-dependent cell death. Numerous studies have indicated that ferroptosis is crucial for the inhibition of tumors, opening up new therapeutic possibilities for cancer (Zhang et al., 2022). The molecular mechanisms that distinguish between these two distinct types of cell death remain obscure; mitochondrial ROS are crucial for both the production of apoptosis and ferroptosis (Wang et al., 2020a; Lee et al., 2020; Li et al., 2021). Depending on its substrate, AMP-activated protein kinase (AMPK), a key regulator of ATP homeostasis, has two distinct roles in ferroptosis. While AMPK-mediated ACACA phosphorylation inhibits ferroptosis by inhibiting fatty acid biosynthesis, AMPK-mediated BECN1 phosphorylation promotes ferroptosis by inhibiting SLC7A11 activity or inducing autophagy. This suggests that energy status may affect lipid biosynthesis and peroxidation during ferroptosis (Song et al., 2018; Zhang et al., 2018). Additionally, group D contained more peroxisomes, which are capable of performing various metabolic tasks, such as ROS production and catabolism, the biosynthesis of ether lipids and bile acids (Lodhi and Semenkovich, 2014), and the catabolism of very long chain fatty acids, branched chain fatty acids, D-amino acids, and polyamines. Lysozymes were also identified at larger concentrations; this may be related to peroxisome interactions with lysosomes for free cholesterol transfer (Chu et al., 2015). In group D, Vibrio was less prevalent than that in other groups. Moreover, Vibrio possessed large amounts of “bacterial chemotaxis,” “bacterial secretion system,” and “quorum sensing,” all of which were associated with bacterial virulence (Krukonis and DiRita, 2003; Green and Mecsas, 2016; Kovacikova and Skorupski, 2022). A crucial mechanism that might control the physiological and metabolic activities of the microbial community was quorum sensing. Furthermore, quorum sensing is frequently used to virulence factor secretion (Suntharalingam and Cvitkovitch, 2005; Li and Tian, 2012).
Finally, we noted different results for the beneficiary pathways when the prebiotics concentration was >5 g/kg body weight. Therefore, the ideal feed for balanced gut microbiota that may facilitate growth and enhance the immune system was group D feed, comprising 1 × 107 CFU/g body weight L. acidophillus as the probiotics and 2.5 g/kg of body weight M. oleifera as the prebiotics.
5 Conclusion
In the present study, we examined the underlying mechanisms of prebiotic (M. oleifera) and probiotic (L. acidophilus)-induced growth and immunity enhancement in white shrimp by identifying 250 microbial genera in the white shrimp gut. Moreover, the feed containing 1 × 107 CFU/g body weight L. acidophilus (probiotics) and 2.5 g/kg of body weight M. oleifera prebiotics lead to the highest gut microbiota abundance, composition, and performance, along with the highest growth and upregulation of many immune signaling pathways, in white shrimp. In conclusion, alterations in probiotic and prebiotic concentrations in the feed can lead to the upregulation of many metabolic pathways for growth enhancement and upregulation of immune signaling pathways, thus providing considerable health benefit to white shrimp in terms of immunity against pathogens. These findings expand our understanding of the utilization of synbiotics, including probiotics (e.g., L. acidophilus) and prebiotics (e.g., M. oleifera), for white shrimp aquaculture and disease resistance and in scientific basis for usage as feed additives in aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article as follows SRR22923665–SRR22923684. https://www.ncbi.nlm.nih.gov/bioproject/916636.
Ethics statement
The animal study was approved by National Taiwan Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SD: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. ZA: Data curation, Resources, Writing – review & editing, Methodology. HH: Formal analysis, Methodology, Writing – review & editing. YL: Data curation, Investigation, Writing – review & editing. CH: Data curation, Investigation, Writing – review & editing. YW: Formal analysis, Writing – review & editing. YH: Conceptualization, Supervision, Writing – review & editing, Writing – original draft. FN: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported financially by the Council of Agriculture, Taiwan (112-18.3.1-FA-F5).
Acknowledgments
We thank the Animal Aquatic Center, National Taiwan Ocean University, for preparing the shrimp that was used in this experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1342708/full#supplementary-material
Supplementary Figure 1 | Rarefaction measurements of alpha diversity indices. The groups are as follows: A: Control group, B: Probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at 5g/kg body weight.
Supplementary Figure 2 | Venn diagram showing unique and shared bacteria along different experimental diet groups. a) genus level unique and shared bacteria b) phylum level shared and unique bacteria in P. vannamei. The groups are as follows: A: Control group, B: probiotics L. acidophilus at 1X107 CFU/g, C: prebiotics M. oleifera extract at 2.5g/kg body weight, D: Probiotics, L. acidophilus at 1X107 CFU/g and Prebiotics, M. oleifera extract at 2.5g/kg body weight, E: prebiotics, M. oleifera extract at 5g/kg body weight and F: probiotics, L. acidophilus at 1X107 CFU/g and prebiotics, M. oleifera extract at5g/kg body weight.
References
Abidin Z., Huang H. T., Hu Y. F., Chang J. J., Huang C. Y., Wu Y. S., et al. (2022). Effect of dietary supplementation with Moringa oleifera leaf extract and Lactobacillus acidophilus on growth performance, intestinal microbiota, immune response, and disease resistance in whiteleg shrimp (Penaeus vannamei). Fish Shellfish Immunol. 127, 876–890. doi: 10.1016/j.fsi.2022.07.007
Amatul-Samahah M. A., Omar W. H. H. W., Ikhsan N. F. M., Azmai M. N. A., Zamri-Saad M., Ina-Salwany M. Y. (2020). Vaccination trials against vibriosis in shrimp: A review. Aquac. Rep. 18, 100471. doi: 10.1016/j.aqrep.2020.100471
Amin M. (2018). Marine protease-producing bacterium and its potential use as an abalone probiont. Aquac. Rep. 12, 30–35. doi: 10.1016/j.aqrep.2018.09.004
Amin M., Kumala R. R. C., Mukti A. T., Lamid M., Nindarwi D. D. (2022). Metagenomic profiles of core and signature bacteria in the guts of white shrimp, Litopenaeus vannamei, with different growth rates. Aquaculture 550, 737849. doi: 10.1016/j.aquaculture.2021.737849
Amoah K., Dong X. H., Tan B. P., Zhang S., Chi S. Y., Yang Q. H., et al. (2020). Administration of probiotic Bacillus licheniformis induces growth, immune and antioxidant enzyme activities, gut microbiota assembly and resistance to Vibrio parahaemolyticus in Litopenaeus vannamei. Aquac. Nutr. 26 (5), 1604–1622. doi: 10.1111/anu.13106
Anderson A. D., Nelson J. M., Rossiter S. H., Angulo F. J. (2003). Public health consequences of use of antimicrobial agents in food animals in the United States. Microb. Drug Resist. 9 (4), 373–379. doi: 10.1089/107662903322762815
Babinska I., Rotkiewicz T., Otrocka-Domagala I. (2005). The effect of Lactobacillus acidophilus and Bifidobacterium spp. administration on the morphology of the gastrointestinal tract, liver and pancreas in piglets. Polish J. Vet. Sci. 8 (1), 29–35.
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Boulangé C. L., Neves A. L., Chilloux J., Nicholson J. K., Dumas M. E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8, 1–12. doi: 10.1186/s13073-016-0303-2
Chen Y., Huang X., Wang J., Li C. (2017). Effect of pure microcystin-LR on activity and transcript level of immune-related enzymes in the white shrimp (Litopenaeus vannamei). Ecotoxicology 26, 702–710. doi: 10.1007/s10646-017-1802-7
Chen W. Y., Ng T. H., Wu J. H., Chen J. W., Wang H. C. (2017). Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis disease. Sci. Rep. 7 (1), 9395. doi: 10.1038/s41598-017-09923-6
Chen J., Vestergaard M., Jensen T. G., Shen J., Dufva M., Solem C., et al. (2017). Finding the needle in the haystack—the use of microfluidic droplet technology to identify vitamin-secreting lactic acid bacteria. MBio 8 (3), e00526–e00517. doi: 10.1128/mbio.00526-17
Chien C. C., Lin T. Y., Chi C. C., Liu C. H. (2020). Probiotic, Bacillus subtilis E20 alters the immunity of white shrimp, Litopenaeus vannamei via glutamine metabolism and hexosamine biosynthetic pathway. Fish Shellfish Immunol. 98, 176–185. doi: 10.1016/j.fsi.2020.01.014
Choi M., Stevens A. M., Smith S. A., Taylor D. P., Kuhn D. D. (2017). Strain and dose infectivity of Vibrio parahaemolyticus: the causative agent of early mortality syndrome in shrimp. Aquac. Res. 48 (7), 3719–3727. doi: 10.1111/are.13197
Chu B. B., Liao Y. C., Qi W., Xie C., Du X., Wang J., et al. (2015). Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 161 (2), 291–306. doi: 10.1016/j.cell.2015.02.019
Dai W., Sheng Z., Chen J., Xiong J. (2020). Shrimp disease progression increases the gut bacterial network complexity and abundances of keystone taxa. Aquaculture 517, 734802. doi: 10.1016/j.aquaculture.2019.734802
Daniels C. L., Merrifield D. L., Ringø E., Davies S. J. (2013). Probiotic, prebiotic and synbiotic applications for the improvement of larval European lobster (Homarus gammarus) culture. Aquaculture 416, 396–406. doi: 10.1016/j.aquaculture.2013.08.001
Fan J., Chen L., Mai G., Zhang H., Yang J., Deng D., et al. (2019). Dynamics of the gut microbiota in developmental stages of Litopenaeus vannamei reveal its association with body weight. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-018-37042-3
Fan L., Wang Z., Chen M., Qu Y., Li J., Zhou A., et al. (2019). Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci. Total Environ. 657, 1194–1204. doi: 10.1016/j.scitotenv.2018.12.069
FAO (2020). The state of world fisheries and aquaculture 2020, Sustainability in Action (Rome, Italy: Food and Agriculture Organization (FAO)).
Fujiwara Y., Denlinger D. L. (2007). p38 MAPK is a likely component of the signal transduction pathway triggering rapid cold hardening in the flesh fly Sarcophaga crassipalpis. J. Exp. Biol. 210 (18), 3295–3300. doi: 10.1242/jeb.006536
Gao S., Pan L., Huang F., Song M., Tian C., Zhang M. (2019). Metagenomic insights into the structure and function of intestinal microbiota of the farmed Pacific white shrimp (Litopenaeus vannamei). Aquaculture 499, 109–118. doi: 10.1016/j.aquaculture.2018.09.026
Gao M., Yi J., Zhu J., Minikes A. M., Monian P., Thompson C. B., et al. (2019). Role of mitochondria in ferroptosis. Mol. Cell 73 (2), 354–363. doi: 10.1016/j.molcel.2018.10.042
Gbadebo A. O., Okareh O. T., Ogunjobi A. A., Dada A. O. (2019). Effects of Moringa oleifera lam. leaf powder on bifidobacteria and Escherichia coli in the gut of albino rats. J. Adv. Microbiol. 18, 1–11. doi: 10.9734/jamb/2019/v18i230168
Gobi N., Vaseeharan B., Chen J. C., Rekha R., Vijayakumar S., Anjugam M., et al. (2018). Dietary supplementation of probiotic Bacillus licheniformis Dahb1 improves growth performance, mucus and serum immune parameters, antioxidant enzyme activity as well as resistance against Aeromonas hydrophila in tilapia Oreochromis mossambicus. Fish Shellfish Immunol. 74, 501–508. doi: 10.1016/j.fsi.2017.12.066
Green E. R., Mecsas J. (2016). Bacterial secretion systems-An overview. Microbiol. Spectr. 4:VMBF-0012-2015. doi: 10.1128/microbiolspec
Gu W., Miller S., Chiu C. Y. (2019). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol.: Mech. Dis. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Hardie D. G. (2003). Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology 144 (12), 5179–5183. doi: 10.1210/en.2003-0982
Hasyimi W., Widanarni W., Yuhana M. (2020). Growth performance and intestinal microbiota diversity in Pacific white shrimp Litopenaeus vannamei fed with a probiotic bacterium, honey prebiotic, and synbiotic. Curr. Microbiol. 77, 2982–2990. doi: 10.1007/s00284-020-02117-w
He Z., Pan L., Zhang M., Zhang M., Huang F., Gao S. (2020). Metagenomic comparison of structure and function of microbial community between water, effluent and shrimp intestine of higher place Litopenaeus vannamei ponds. J. Appl. Microbiol. 129 (2), 243–255. doi: 10.1111/jam.14610
Hooper L. V., Gordon J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292 (5519), 1115–1118. doi: 10.1126/science.1058709
Hoseinifar S. H., Sun Y. Z., Caipang C. M. (2017). Short-chain fatty acids as feed supplements for sustainable aquaculture: an updated view. Aquac. Res. 48 (4), 1380–1391. doi: 10.1111/are.13239
Hou D., Huang Z., Zeng S., Liu J., Wei D., Deng X., et al. (2018). Intestinal bacterial signatures of white feces syndrome in shrimp. Appl. Microbiol. Biotechnol. 102, 3701–3709. doi: 10.1007/s00253-018-8855-2
Huang K. C., Lee J. W., Shiu Y. L., Ballantyne R., Liu C. H. (2022). Micro-Aid Liquid 10 promotes growth performance and health status of white shrimp, Litopenaeus vannamei. J. Mar. Sci. Eng. 10 (1), 49. doi: 10.3390/jmse10010049
Huang F., Pan L., Song M., Tian C., Gao S. (2018). Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl. Microbiol. Biotechnol. 102, 8585–8598. doi: 10.1007/s00253-018-9229-5
Huang L., Xu Y. B., Xu J. X., Ling J. Y., Chen J. L., Zhou J. L., et al. (2017). Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere 168, 1107–1114. doi: 10.1016/j.chemosphere.2016.10.096
Huang Z., Zeng S., Xiong J., Hou D., Zhou R., Xing C., et al. (2020). Microecological Koch’s postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8, 1–13. doi: 10.1186/s40168-020-00802-3
Huynh T. G., Hu S. Y., Chiu C. S., Truong Q. P., Liu C. H. (2019). Bacterial population in intestines of white shrimp, Litopenaeus vannamei fed a synbiotic containing Lactobacillus plantarum and galacto oligosaccharide. Aquac. Res. 50 (3), 807–817. doi: 10.1111/are.13951
Kim S., Jazwinski S. M. (2018). The gut microbiota and healthy aging: a mini-review. Gerontology 64 (6), 513–520. doi: 10.1159/000490615
Kovacikova G., Skorupski K. (2002). Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46 (4), 1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x
Krukonis E. S., DiRita V. J. (2003). From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6 (2), 186–190. doi: 10.1016/S1369-5274(03)00032-8
LeBlanc J. G., Chain F., Martín R., Bermúdez-Humarán L. G., Courau S., Langella P. (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Factories 16 (1), 1–10. doi: 10.1186/s12934-017-0691-z
Lee C. T., Chen I. T., Yang Y. T., Ko T. P., Huang Y. T., Huang J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. 112 (34), 10798–10803. doi: 10.1073/pnas.1503129112
Lee J. W., Chiu S. T., Wang S. T., Liao Y. C., Chang H. T., Ballantyne R., et al. (2022). Dietary SYNSEA probiotic improves the growth of white shrimp, Litopenaeus vannamei and reduces the risk of Vibrio infection via improving immunity and intestinal microbiota of shrimp. Fish Shellfish Immunol. 127, 482–491. doi: 10.1016/j.fsi.2022.06.071
Lee B. H., Hsu W. H., Chen Y. Z., Hsu K. T., Pan T. M. (2021). Limosilactobacillus fermentum SWP-AFFS02 improves the growth and survival rate of white shrimp via regulating immunity and intestinal microbiota. Fermentation 7 (3), 179. doi: 10.3390/fermentation7030179
Lee H., Zandkarimi F., Zhang Y., Meena J. K., Kim J., Zhuang L., et al. (2020). Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat. Cell Biol. 22 (2), 225–234. doi: 10.1038/s41556-020-0461-8
Li T., Li H., Gatesoupe F. J., She R., Lin Q., Yan X., et al. (2017). Bacterial signatures of “Red-Operculum” disease in the gut of crucian carp (Carassius auratus). Microb. Ecol. 74, 510–521. doi: 10.1007/s00248-017-0967-1
Li Y. H., Tian X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors 12 (3), 2519–2538. doi: 10.3390/s120302519
Li C., Zhang Y., Liu J., Kang R., Klionsky D. J., Tang D. (2021). Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy 17 (4), 948–960. doi: 10.1080/15548627.2020.1739447
Liu H., Li J., Guo X., Liang Y., Wang W. (2018). Yeast culture dietary supplementation modulates gut microbiota, growth and biochemical parameters of grass carp. Microb. Biotechnol. 11 (3), 551–565. doi: 10.1111/1751-7915.13261
Lodhi I. J., Semenkovich C. F. (2014). Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19 (3), 380–392. doi: 10.1016/j.cmet.2014.01.002
Magnoni L. J., Vraskou Y., Palstra A. P., Planas J. V. (2012). AMP-activated protein kinase plays an important evolutionary conserved role in the regulation of glucose metabolism in fish skeletal muscle cells. PloS One 7 (2), e31219. doi: 10.1371/journal.pone.0031219
Morrison D. K. (2012). MAP kinase pathways. Cold Spring Harbor Perspect. Biol. 4 (11), a011254. doi: 10.1101/cshperspect.a011254
Munaeni W., Yuhana M., Setiawati M., Wahyudi A. T. (2020). Impact of dietary supplementation with Eleutherine bulbosa (Mill.) Urb. on intestinal microbiota diversity and growth of white shrimp, Litopenaeus vannamei. Aquaculture 528, 735466. doi: 10.1016/j.aquaculture.2020.735466
Parlapani F. F., Michailidou S., Anagnostopoulos D. A., Sakellariou A. K., Pasentsis K., Psomopoulos F., et al. (2018). Microbial spoilage investigation of thawed common cuttlefish (Sepia officinalis) stored at 2 C using next generation sequencing and volatilome analysis. Food Microbiol. 76, 518–525. doi: 10.1016/j.fm.2018.08.004
Prabawati E., Hu S. Y., Chiu S. T., Balantyne R., Risjani Y., Liu C. H. (2022). A synbiotic containing prebiotic prepared from a by-product of king oyster mushroom, Pleurotus eryngii and probiotic, Lactobacillus plantarum incorporated in diet to improve the growth performance and health status of white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 120, 155–165. doi: 10.1016/j.fsi.2021.11.031
Rengpipat S., Tunyanun A., Fast A. W., Piyatiratitivorakul S., Menasveta P. (2003). Enhanced growth and resistance to Vibrio challenge in pond-reared black tiger shrimp Penaeus monodon fed a Bacillus probiotic. Dis. Aquat. Organisms 55 (2), 169–173. doi: 10.3354/dao055169
Ringø E., Van Doan H., Lee S. H., Soltani M., Hoseinifar S. H., Harikrishnan R., et al. (2020). Probiotics, lactic acid bacteria and bacilli: interesting supplementation for aquaculture. J. Appl. Microbiol. 129 (1), 116–136. doi: 10.1111/jam.14628
Rungrassamee W., Klanchui A., Chaiyapechara S., Maibunkaew S., Tangphatsornruang S., Jiravanichpaisal P., et al. (2013). Bacterial population in intestines of the black tiger shrimp (Penaeus monodon) under different growth stages. PloS One 8 (4), e60802. doi: 10.1371/journal.pone.0060802
Schweinitzer T., Josenhans C. (2010). Bacterial energy taxis: a global strategy? Arch. Microbiol. 192, 507–520. doi: 10.1007/s00203-010-0575-7
Shao J., Jiang K., Wang L. (2019). Litopenaeus vannamei fed diets with different replacement levels of fish meal by fish silage: a molecular approach on intestinal microbiota. Aquac. Nutr. 25 (3), 721–728. doi: 10.1111/anu.12900
Shinn A. P., Pratoomyot J., Griffiths D., Trong T. Q., Vu N. T., Jiravanichpaisal P., et al. (2018). Asian shrimp production and the economic costs of disease. Asian Fishery Sci. S 31, 29–58. doi: 10.33997/j.afs.2018.31.S1.003
Song X., Zhu S., Chen P., Hou W., Wen Q., Liu J., et al. (2018). AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system Xc–activity. Curr. Biol. 28 (15), 2388–2399. doi: 10.1016/j.cub.2018.05.094
Sriurairatana S., Boonyawiwat V., Gangnonngiw W., Laosutthipong C., Hiranchan J., Flegel T. W. (2014). White feces syndrome of shrimp arises from transformation, sloughing and aggregation of hepatopancreatic microvilli into vermiform bodies superficially resembling gregarines. PloS One 9 (6), e99170. doi: 10.1371/journal.pone.0099170
Suntharalingam P., Cvitkovitch D. G. (2005). Quorum sensing in streptococcal biofilm formation. Trends Microbiol. 13 (1), 3–6. doi: 10.1016/j.tim.2004.11.009
Utiswannakul P., Sangchai S., Rengpipat S. (2011). Enhanced growth of black tiger shrimp Penaeus monodon by dietary supplementation with Bacillus (BP11) as a probiotic. J. Aquac. Res. Dev. 2 (Special issue). doi: 10.4172/2155-9546.S1-006
Wang Y. C., Hu S. Y., Chiu C. S., Liu C. H. (2019). Multiple-strain probiotics appear to be more effective in improving the growth performance and health status of white shrimp, Litopenaeus vannamei, than single probiotic strains. Fish Shellfish Immunol. 84, 1050–1058. doi: 10.1016/j.fsi.2018.11.017
Wang H., Liu C., Zhao Y., Gao G. (2020a). Mitochondria regulation in ferroptosis. Eur. J. Cell Biol. 99 (1), 151058. doi: 10.1016/j.ejcb.2019.151058
Wang A., Ran C., Wang Y., Zhang Z., Ding Q., Yang Y., et al. (2019). Use of probiotics in aquaculture of China—a review of the past decade. Fish Shellfish Immunol. 86, 734–755. doi: 10.1016/j.fsi.2018.12.026
Wang H., Wan X., Xie G., Dong X., Wang X., Huang J. (2020b). Insights into the histopathology and microbiome of Pacific white shrimp, Penaeus vannamei, suffering from white feces syndrome. Aquaculture 527, 735447. doi: 10.1016/j.aquaculture.2020.735447
Wilkes Walburn J., Wemheuer B., Thomas T., Copeland E., O’Connor W., Booth M., et al. (2019). Diet and diet-associated bacteria shape early microbiome development in Yellowtail Kingfish (Seriola lalandi). Microb. Biotechnol. 12 (2), 275–288. doi: 10.1111/1751-7915.13323
Wu P. S., Liu C. H., Hu S. Y. (2021). Probiotic Bacillus safensis NPUST1 administration improves growth performance, gut microbiota, and innate immunity against Streptococcus iniae in Nile tilapia (Oreochromis niloticus). Microorganisms 9 (12), 2494. doi: 10.3390/microorganisms9122494
Xing M., Hou Z., Yuan J., Liu Y., Qu Y., Liu B. (2013). Taxonomic and functional metagenomic profiling of gastrointestinal tract microbiome of the farmed adult turbot (Scophthalmus maximus). FEMS Microbiol. Ecol. 86 (3), 432–443. doi: 10.1111/1574-6941.12174
Xiong J., Zhu J., Dai W., Dong C., Qiu Q., Li C. (2017). Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ. Microbiol. 19 (4), 1490–1501. doi: 10.1111/1462-2920.13701
Yan Q., Li J., Yu Y., Wang J., He Z., Van Nostrand J. D., et al. (2016). Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ. Microbiol. 18 (12), 4739–4754. doi: 10.1111/1462-2920.13365
Ye L., Amberg J., Chapman D., Gaikowski M., Liu W. T. (2014). Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J. 8 (3), 541–551. doi: 10.1038/ismej.2013.181
Zeng S., Zhou R., Bao S., Li X., Deng Z., Hou D., et al. (2020). Identification of multigene biomarker for shrimp white feces syndrome by full-length transcriptome sequencing. Front. Genet. 11, 71. doi: 10.3389/fgene.2020.00071
Zhang Y., Dong C. (2007). Regulatory mechanisms of mitogen-activated kinase signaling. Cell. Mol. Life Sci. 64, 2771–2789. doi: 10.1007/s00018-007-7012-3
Zhang C., Liu X., Jin S., Chen Y., Guo R. (2022). Ferroptosis in cancer therapy: a novel approach to reversing drug resistance. Mol. Cancer 21 (1), 47. doi: 10.1186/s12943-022-01530-y
Zhang M., Pan L., Huang F., Gao S., Su C., Zhang M., et al. (2019). Metagenomic analysis of composition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments under different culture modes. Aquaculture 506, 280–293. doi: 10.1016/j.aquaculture.2019.03.038
Zhang Z., Yao Z., Wang L., Ding H., Shao J., Chen A., et al. (2018). Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy 14 (12), 2083–2103. doi: 10.1080/15548627.2018.1503146
Zheng J., Conrad M. (2020). The metabolic underpinnings of ferroptosis. Cell Metab. 32 (6), 920–937. doi: 10.1016/j.cmet.2020.10.011
Zhu J., Dai W., Qiu Q., Dong C., Zhang J., Xiong J. (2016). Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microb. Ecol. 72, 975–985. doi: 10.1007/s00248-016-0831-8
Zhu L., Wu Q., Dai J., Zhang S., Wei F. (2011). Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. 108 (43), 17714–17719. doi: 10.1073/pnas.1017956108
Keywords: synbiotics, P. vannamei, gut microbiota, 16S rRNA, immune pathways
Citation: Das SP, Abidin Z, Huang H-T, Lin Y-R, Huang C-Y, Wu Y-S, Hu Y-F and Nan F-H (2024) Deciphering the influence of dietary synbiotics in white shrimp gut and its effects in regulating immune signaling pathways. Front. Mar. Sci. 10:1342708. doi: 10.3389/fmars.2023.1342708
Received: 22 November 2023; Accepted: 22 December 2023;
Published: 23 January 2024.
Edited by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaReviewed by:
Basanta Sahu, The University of Hong Kong, Hong Kong SAR, ChinaSoumya Prasad Panda, Central Inland Fisheries Research Institute (ICAR), India
Copyright © 2024 Das, Abidin, Huang, Lin, Huang, Wu, Hu and Nan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yeh-Fang Hu, eWVoZmFuZ2h1QGVtYWlsLm50b3UuZWR1LnR3; Fan-Hua Nan, ZmhuYW5AbWFpbC5udG91LmVkdS50dw==
 Sofia Priyadarsani Das
Sofia Priyadarsani Das Zaenal Abidin
Zaenal Abidin Huai-Ting Huang1
Huai-Ting Huang1 Chih-Yang Huang
Chih-Yang Huang Yu-Sheng Wu
Yu-Sheng Wu Yeh-Fang Hu
Yeh-Fang Hu Fan-Hua Nan
Fan-Hua Nan