
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 09 January 2024
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1338197
 Yinghui Qin1,2,3,4
Yinghui Qin1,2,3,4 Haixiang Liu1,2,3,4
Haixiang Liu1,2,3,4 Shuangshuang Mao1,2,3,4
Shuangshuang Mao1,2,3,4 Riying Deng1
Riying Deng1 Yuhang Wang1
Yuhang Wang1 Si Deng1,2,3,4
Si Deng1,2,3,4 Peipei Zhang1
Peipei Zhang1 Lunguang Yao1,2,3,4*
Lunguang Yao1,2,3,4*Largemouth bass virus (LMBV) poses a significant threat to largemouth bass farming, leading to substantial economic losses. In December 2022, massive largemouth bass juveniles died at a fish farm in the city of Xinxiang, China. Through a series of experiments, we conclusively identified LMBV as the causative pathogen. The affected fish displayed anorexia, lethargy, and hemorrhage at the pectoral and caudal fin base. No parasites or pathogenic bacteria were detected on the body surface or gills, or isolated from the diseased fish. Severe hemorrhage, lymphocyte infiltration, and extensive necrosis were observed in the liver, spleen, intestine, and stomach of the moribund fish. The tissue homogenate from the diseased fish induced epithelioma papulosum cyprini cells (EPC) cell death, while no such effects were observed in grouper spleen (GS) cells. Sequence similarity analysis of the major capsid protein (MCP) indicated the virus shared 100% similarity with the LMBV-FS2021 strain, placing it within the Ranavirus genus. Transmission electron microscopy (TEM) observations revealed plenty of hexagonal virions accumulated in the cytoplasm of infected EPC cells. Artificial infection demonstrated that LMBV-XX01 was highly fatal to Micropterus salmoides juveniles, with an LD50 of 103.081 TCID50/fish. RT-qPCR detection confirmed that LMBV appeared in all sampled tissues of the challenged largemouth bass, with significantly higher viral loads detected in the liver and heart compared to other tissues. Additionally, we successfully obtained a highly purified recombinant MCP of LMBV and developed two strains of monoclonal antibodies targeting MCP of LMBV-XX01. Overall, our findings provide valuable materials and insights for the design of prevention strategies and the development of detection methods for LMBV.
Largemouth bass (Micropterus salmoides) is a globally significant fish species that originated in the Mississippi River Basin, United States of America (Junjie and Shengjie, 2019). Largemouth bass has been widely farmed in various countries due to its advantages, such as rapid growth, delicious taste, lack of inter-muscular bones, and strong environmental adaptability (Wang et al., 2019). Since 1983, when largemouth bass was introduced in Guangdong province, its aquaculture scale has been continuously expanding in China, and its annual production in China reached 802,486 tons in 2022, representing a 14.3% increase compared to 2021 (China Fishery Statistical Yearbook, 2023). However, the continuous expansion of the scale and intensity of largemouth bass culture has caused frequent occurrence of viral diseases, resulting in significant economic losses and hindering industry development (Ma et al., 2013; Liu et al., 2023; Qin et al., 2023). To date, at least 10 kinds of viruses have been reported in largemouth bass, with largemouth bass virus (LMBV) and Micropterus salmoides rhabdovirus (MSRV) posing the greatest threats and causing mortalities of approximately 80% and 100%, respectively (Sibley et al., 2016; Godahewa et al., 2018; Waltzek et al., 2019; Zhang et al., 2019; Jia et al., 2020; Cai et al., 2022; Fu et al., 2022; Jin et al., 2022; Liu et al., 2023; Qin et al., 2023; Xu et al., 2023).
The first case of LMBV was reported in 1995 in largemouth bass at Santee-Cooper Reservoir, South Carolina, USA. Since then, LMBV has been detected in various countries and regions, such as Florida, Kansas, Texas, India, and China (Grizzle et al., 2002; Southard et al., 2009; George et al., 2015; Salazar et al., 2022). In addition, studies showed that LMBV can infect not only largemouth bass but also other fish species such as koi (Cyprinus carpio) and mandarin fish (Siniperca chuatsi) (George et al., 2015; Dong et al., 2017; Sivasankar et al., 2017). LMBV is an enveloped virus with an icosahedral structure, approximately 150 nm in diameter, belonging to the Ranavirus genus of the Iridoviridae family (Plumb et al., 1996). The genome of LMBV is composed of a 100 kb double-stranded linear DNA molecule, containing approximately 90 open read frames (ORFs) (GenBank accession nos. MW630113.1, ON936874.1, MK681855, and MK681856).
Vaccination is considered an ideal approach for preventing LMBV infection. Although some candidate vaccines and herbal extracts have been proven to be potent for defending against LMBV infection under laboratory conditions, there is no licensed vaccine or antiviral drug available for preventing or treating LMBV infection (Jia et al., 2022; Yao et al., 2022; Cheng et al., 2023; Wang et al., 2023; Zhang et al., 2023). Therefore, rapid detection and early diagnosis of LMBV remain crucial strategies to prevent the disease’s spread and mitigate economic loss. However, diagnosing LMBV solely based on clinical signs is challenging as it is influenced by the LMBV strains and the size of the infected fish (Liu et al., 2023). For instance, the LMBV-LS1809 strain causes severe ulceration and muscle necrosis, while the strain isolated by Liu et al. causes surface bleeding and spleen swelling in infected fish (Zhao et al., 2020; Liu et al., 2023). To date, some traditional and classical detection methods, such as RT-PCR, enzyme-linked apta-sorbent assay (ELASA), and quantitative polymerase chain reaction (qPCR), have been developed and utilized for LMBV diagnosis, but several limitations hinder their practical application in the field, such as being technically demanding, expensive instruments and reagents, and time-consuming procedures (Montelongo-Alfaro et al., 2019; Zhu et al., 2020; Guo et al., 2022; Zhang et al., 2022). Therefore, the development of new detection methods for LMBV is urgently needed. Immunoassay strips have increasingly garnered attention due to their rapidity, affordability, ease of use, and suitability for on-site detection (Li et al., 2021; Shen et al., 2021). The selection of targets and the production of antibodies are two crucial aspects in the development of immunoassay strips as they directly influence their specificity and sensitivity (Li et al., 2021; Shen et al., 2021). The MCP, which is the most abundant structural protein carrying virus-specific antigens, has emerged as an ideal target for genetic relationship identification, vaccine research, antibody production, development of detection methods, and systematic survey (Montelongo-Alfaro et al., 2019; Guo et al., 2022; Jia et al., 2022; Yao et al., 2022; Qin et al., 2023; Zhang et al., 2023). Thus, we developed the monoclonal antibodies against MCP and assessed their specificity, providing valuable material for the development of immunoassay strips for LMBV detection.
In this study, a strain of LMBV named LMBV-XX01 was isolated and identified from diseased juvenile largemouth bass collected in December 2022 at a fish farm in Xinxiang, Henan, China. We investigated the replication kinetics, tissue lesions, dynamic distribution, and pathogenicity of the LMBV-XX01 strain. Additionally, we successfully generated and characterized two strains of a monoclonal antibody targeting the MCP of LMBV. Our findings will contribute new information and resources for further exploring the pathogenesis of LMBV and for the development of detection methods for LMBV.
All animal experiments were strictly conducted following the guidelines of the Ethics Committee of Nanyang Normal University (Permit Number: NYSY007). To guarantee the best animal welfare and minimize pain, experimental fish and mice were anesthetized using tricaine methanesulfonate (MS-222) or ether before the experiment.
In December 2022, a fish farm in the city of Xinxiang, Henan province, China experienced a mass death of juvenile largemouth bass (~7 cm in length, ~5 g in weight). Moribund fish exhibited unnatural behaviors, including anorexia, insensitivity, abdominal distension, spiral swimming, and fin base hemorrhage. The diseased fish started dying on the second day, with the peak mortality occurring 5-7 days after the onset of abnormal behavior, and the cumulative mortality rate was approximately 70%. Diseased fish were immediately transported to the laboratory under low-temperature conditions for further examination. First, the gills and mucus were collected for parasite detection under a microscope. Subsequently, livers and spleens were sampled and homogenized using sterility tools. After centrifugation, the supernatant was used for bacteria detection and virus isolation. Additionally, tissues from diseased fish were sampled for section preparation and RT-PCR detection using previously described methods (Qin et al., 2023), and the tissues from healthy fish were utilized as negative controls.
The tissues from diseased or healthy fish were used to extract total RNA using a total RNA kit (NCM Biotech, China) according to the manufacturer’s directions, and the resulting RNA was used to synthesize cDNA using SweScript RT I First Strand cDNA Synthesis Kit (With gDNA Remover) (Servicebio, China). Then, the PCR assays were performed using the obtained cDNA with the specific primers of reported viruses in largemouth bass (Table 1). The resulting PCR products were electrophoretically separated with 2% agarose gel and visualized using an ultraviolet imaging system. Furthermore, the coding sequence (CDS) of the MCP of LMBV was obtained by PCR amplification using the primers designed according to the public genome of LMBV. The PCR product was purified using Gel DNA Extraction Kit (Servicebio, China), and digested with restriction endonucleases Hind III and Xho I. The resulting DNA was then fused into the pET28a plasmid to construct the pET28-MCP plasmid, which was transformed into E. coli DH5α competent cells (Sangon Biotech, China). Positive clones from colony PCR were further confirmed with DNA sequencing. The resulting sequence was analyzed through BLAST, and a phylogenetic analysis was performed with MEGA 11.
The RT-qPCR assay was carried out as previously described (Qin et al., 2023). The primers for RT-qPCR are listed in Table 1, and the 40s ribosomal (40s) protein served as an internal reference gene. The fold induction of the target gene was calculated and normalized to the mRNA level of the 40s gene using the 2- ΔΔCt algorithm. All experiments were carried out in triplicate and replicated independently three times.
Virus isolation was conducted as per a previous method with slight modifications (Qin et al., 2020). Briefly, tissues from moribund fish were homogenized in filtered phosphate buffer solution (PBS, pH=7.4) supplemented with Penicillin-Streptomycin of a final concentration of 100 U/mL (Biosharp, China) at a volume ratio of 1:9. After centrifugation at 12000×g for 15 min under 4°C, the supernatant was collected and filtered to remove bacteria. The filtered liquid was then diluted 1:10 and 1:100 in M199 or L15 medium and inoculated on confluent epithelioma papillosum cyprini cells (EPC) kindly bestowed by professor Xiuzhen Sheng (College of Fisheries, Ocean University of China) (Zhong et al., 2019) and grouper spleen (GS) cells kindly provided by Professor Qiwei Qin (College of Marine Sciences, South China Agricultural University) (Zheng et al., 2022), respectively. In the control EPC or GS cells, an equal volume of M199 or L15 medium was added, respectively. After a 2-hour attachment at 28°C, the supernatant was discarded, and 1 mL of maintenance medium was added. Then, the cells were maintained at 28°C and examined daily for cell status under an inverted microscope. When >80% of the cells showed cytopathic effect (CPE), the cells and supernatants were harvested and freeze-thawed for three cycles. After centrifugation (12000g, 10min), the supernatant was harvested and subsequently stored at -80°C.
Infected EPC cells showing clear CPE were harvested and fixed in a 2.5% glutaraldehyde buffer (pH=7.4) for 12 hours. Subsequently, they were immersed in a 1% osmium tetroxide buffer for 2 hours and subjected to gradient dehydration using ethanol. Then, the samples were entrapped into epoxy resin, cut, and dyed with lead citrate and uranyl acetate. Ultimately, virus particles were examined using a transmission electron microscope (Hitachi HT-7800, Japan).
To isolate and obtain a single LMBV clone, the plaque purification assay was performed as per a previously described method with minor modifications (Qin et al., 2023). Briefly, the LMBV stock suspension was 10-fold serially diluted using M199 medium and then added to an EPC cell monolayer. After a 2-hour attachment at 28°C, the virus was aspirated, and the cells were washed one time using serum-free M199 prior to being overlaid with M199 maintenance medium added with 1.5% sodium carboxymethyl cellulose (Biosharp, China). Once the plaques appeared, the overlaying media over the tagged plaques were transferred and mixed into 1 mL serum-free M199 medium. This process was repeated twice for further plaque purification. Finally, the resulting single clones were identified with RT-PCR assays.
The viral titers were determined in EPC cells using the 50% tissue culture infectious dose (TCID50) assay with slight modifications to a previous method (Qin et al., 2020). In brief, eight independent confluent EPC cell wells in 96-well plates (Labselect, China) were inoculated with 100 μL of 10-fold gradient virus dilutions (10-1-10-8). The CPE was counted daily under an inverted microscope for 7 days after infection and the titer was represented as TCID50 according to the Reed-Muench calculation method.
To facilitate studies, such as antiviral drug screening and investigation of antiviral mechanisms, it is crucial to understand the detailed growth patterns of the virus in permissive cells. Therefore, we conducted a systematic evaluation of LMBV-XX01 growth kinetics in EPC cells by examining intracellular viral RNA synthesis and extracellular infectious progeny virus. This experiment was undertaken as per a previous method (Qin et al., 2023). Briefly, 106 cells EPC cells were inoculated in 12-well plates for 12 hours before LMBV-XX01 infection at an MOI of 0.01. After adsorption for 2 hours, the viral inoculum was discarded, and unadsorbed viruses were removed by washing three times with an M199 medium. M199 maintenance medium containing 2% FBS was added to the infected EPC cells prior to being cultured at 28°C in an incubator containing 5% CO2. The cells and supernatants were collected at designated time points post-infection. Then, the total RNA extracted from the cell monolayers was used to determine intracellular LMBV RNA level through RT-qPCR, while the infectious virus in the supernatants was quantified using a TCID50 assay.
Healthy largemouth bass juveniles without obvious surface damage were purchased from a fish farm without a history of LMBV infection in the city of Nanyang, Henan province, China. Three fish were collected randomly and confirmed to be negative for LMBV, MSRV, NNV, ISKNV, VHSV, and SVCV by RT-PCR and temporarily reared in fresh water at 25°C for two weeks prior to the infection experiment. A total of 120 fish were divided randomly into six groups with 20 fish each. Fish in groups 1-5 were intraperitoneally injected with equal volume M199 medium containing 5×104, 104, 2×103, 4×102, or 8×101 TCID50 of the LMBV-XX01 strain, respectively. Group 6 served as uninfected control and was mock injected with an equal dose of M199 medium. After the challenge, these fish were kept at 25°C for 14 days, and their behaviors and mortality were recorded daily.
A parallel challenge experiment was conducted to investigate the tissue tropism and target organs of LMBV. Fish showing typical clinical signs were washed three times with sterile PBS and euthanized using MS-222. Tissues from the liver, spleen, kidney, intestine, gill, muscle, brain, heart, eye, and stomach were collected and stored in a Magzol medium at -80°C until used. Total RNA was extracted from the tissues, and the MCP mRNA levels of LMBV in the sampled tissues were quantified and compared using the RT-qPCR assay.
The CDS of MCP of LMBV was obtained by PCR amplification and purified using the Gel Extraction Kit (Sangon Biotech, China). The PCR production was then digested with restriction endonucleases Hind III and Xho I, and subsequently linked into pET28a plasmid to construct pET28-MCP fusion plasmid, which was then transformed into E. coli strain Trans5α (TransGen, China). After PCR verification and DNA sequencing, the positive plasmid was extracted from the positive clone and transformed into Trans BL21 (DE3) (TransGen, China). Then, the positive clone of BL21 containing the pET28-MCP expression vector was cultured to the logarithmic phase and 0.6 mM isopropyl β-D-1-thiogalactosidase (IPTG) was added to induce MCP expression. The MCP was subsequently purified using a Ni-NTA pre-packed gravity column, analyzed using an SDS-PAGE assay, and visualized with Coomassie Brilliant Blue R-250.
Three BALB/c mice were intraperitoneally injected with 100 μg MCP emulsified in Freund’s complete adjuvant (Biosharp, China). On the 14th day after the first immunization, a similar injection was performed using Freund’s incomplete adjuvant (Biosharp, China). Subsequently, two booster immunizations were administered with 100 μg of MCP without Freund’s adjuvant via the tail vein at an interval of 7 days. On the third day after the last immunization, a mouse was euthanized, and spleen cells were extracted and fused with SP2/0 myeloma cells using polyethylene glycol 1450 (Proteintech, USA). The fused cells were then resuspended in 1640 medium (Biosharp, China) containing 1% HAT (Sigma, USA) and seeded into 96-well culture plates (100 μL/well). After 14 days, the supernatant from those wells containing hybridomas was collected and screened using indirect enzyme-linked immunosorbent assay (ELISA), western blot, and indirect immunofluorescence assay (IFA) as per a previous method (Qin et al., 2022). The positive hybridomas were subcloned using the limited dilution method until all monoclonal cells’ supernatant reacted with MCP.
All data were represented as mean ± standard error (SEM) and analyzed using GraphPad Prism 9.5 software. Statistical differences between groups were calculated using one-way analysis of variance (ANOVA). Differences were considered significant if p < 0.05.
Diseased largemouth bass exhibited abnormal behaviors, including anorexia, insensitivity, abdominal distension, spiral swimming, and fin base hemorrhage (Figure 1A). No parasites or pathogenic bacteria were detected in moribund largemouth bass. Internally, the diseased fish displayed enlarged and white liver, splenomegaly, and mass ascites (Figure 1B). Furthermore, the hepatic sinusoids showed extensive congestion (Figure 1D). The spleen exhibited widespread congestion and hemorrhage (black arrow) as well as pronounced inflammatory infiltration (red arrow) (Figure 1F). The intestine revealed numerous villous atrophy erosion and detachment (Figure 1H). The stomach exhibited submucosal edema, along with significant lymphocytic infiltration (red arrow), slight congestion (black arrow), and severe necrosis and detachment of epithelia (blue arrow) (Figure 1J). No apparent lesions were observed in these tissues of healthy largemouth bass (Figures 1C, E, G, I).
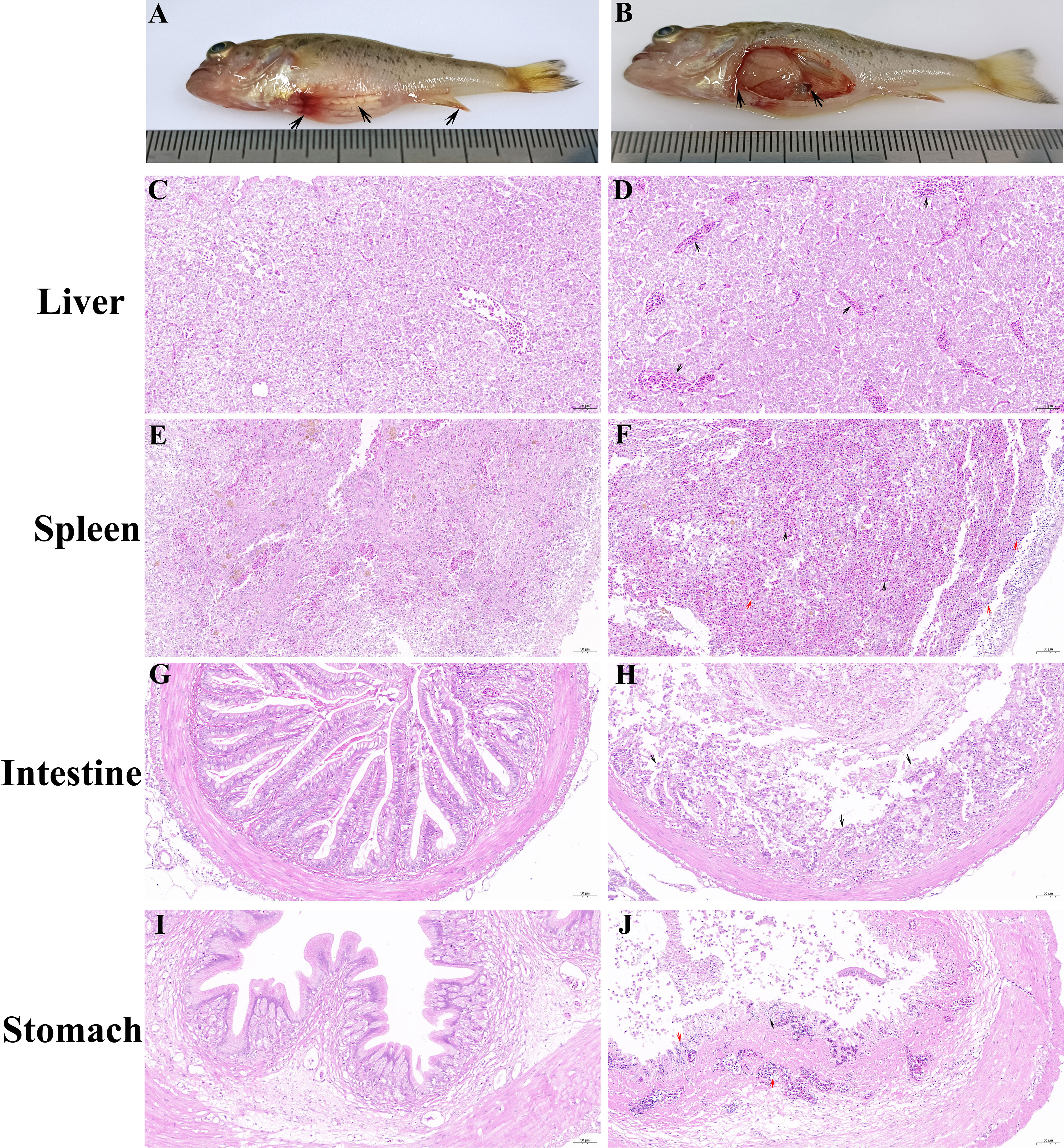
Figure 1 Clinical symptoms and histopathological observations of diseased largemouth bass. (A) Diseased fish exhibited abdominal distension and fin base hemorrhage. (B) Enlarged and white liver, splenomegaly, and mass ascites were observed in diseased fish. (D) Mass congestion occurred in the hepatic sinusoids of diseased fish. (F) Widespread congestion and hemorrhage (black arrow), along with pronounced inflammatory infiltration (red arrow), appeared in the spleen of diseased fish. (H) The intestine of diseased fish displayed numerous villous atrophy erosion and detachment. (J) Submucosal edema along with significant lymphocytic infiltration (red arrow), slight congestion (black arrow), and severe necrosis and detachment of epithelia (blue arrow) were observed in the stomach of diseased fish. No significant lesions were observed in the liver (C), spleen (E), intestines (G), and stomach (I) of healthy fish.
PCR assays were conducted using cDNA from moribund or healthy largemouth bass as templates to detect five common viruses of largemouth bass, with cDNA from healthy fish serving as a negative control. Agarose gel electrophoresis results showed the presence of an approximately 200 bp specific band in diseased fish using LMBV-specific primers (Figure 2A), while no specific bands were observed using primers for MSRV, NNV, ISKNV, VHSV, and SVCV, compared to those in healthy fish (Figure 2B).
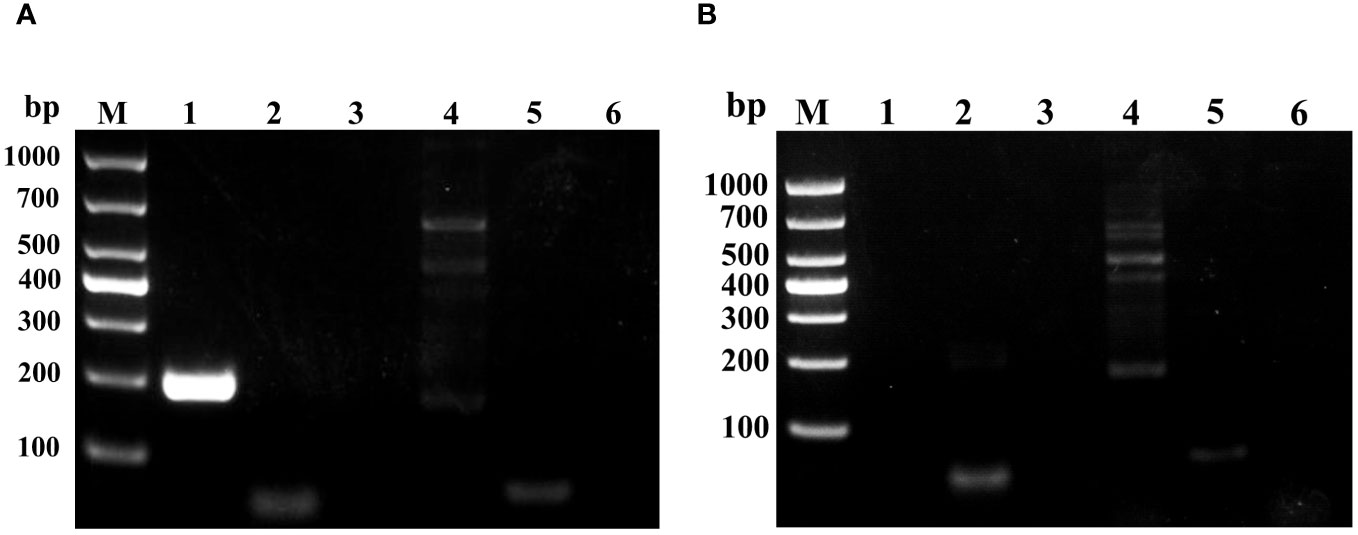
Figure 2 Agarose gel electrophoresis analysis of PCR products obtained from diseased or healthy fish using specific primers for LMBV, MSRV, NNV, ISKNV, and SVCV. (A) PCR products from diseased fish. (B) PCR products from healthy largemouth bass were used as a negative control. Lanes 1-6 are the PCR products using specific primers of LMBV, MSRV, NNV, ISKNV, and SVCV, respectively.
EPC and GS cells were used to isolate LMBV from mixed homogenates of the liver and spleen from diseased fish. EPC cells became round and shiny as early as 6 hours post-inoculation and displayed a typical cytopathic effect (CPE) characterized by grape-like clusters and cell detachment from 24 to 72 hours after incubation with the homogenate of diseased fish (Figure 3). No CPE was observed in GS cells or uninfected EPC cells.
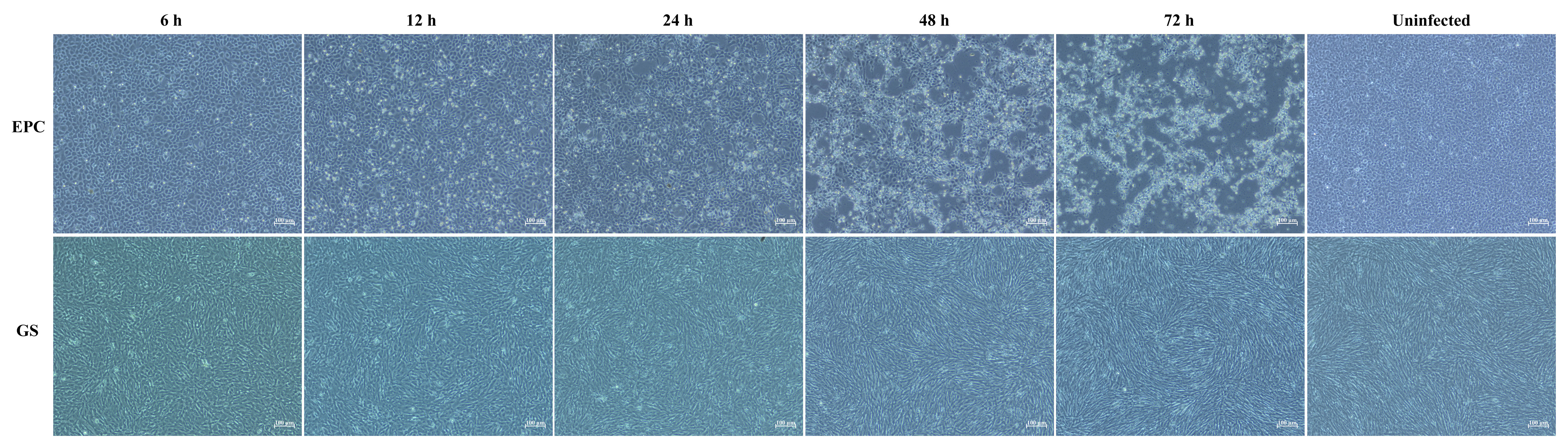
Figure 3 Isolation of virus on EPC cells and GS cells. Typical CPE characterized by cell rounding and shiny, grape-like clusters and detachment occurred in EPC cells after inoculation with tissue homogenate from diseased fish. No obvious CPE was observed in GS cells or uninfected EPC cells. The scale bars represent a length of 100 μm.
A single clone of LMBV was obtained through three successive rounds of plaque purification on EPC cells, and the clone was identified using RT-PCR. At 72 hours post LMBV infection, EPC cells and supernatants were harvested, and repeated freeze-thaw was performed for three cycles. After centrifugation (12000g, 10min), the supernatant was collected and the virus titer in the supernatant was determined to be 106.75 TCID50/mL.
TEM observation indicated that numerous hexagonal virus particles measuring 145-160 nm from vertex to vertex were scattered in the cytoplasm of infected EPC cells (Figure 4). Viruses at different assembly stages were observed in infected EPC cells, ranging from immature virus particles without or with incomplete electron-dense cores (Figure 4B, red arrows) to mature virus particles containing complete electron-dense cores (Figure 4B, black arrows).
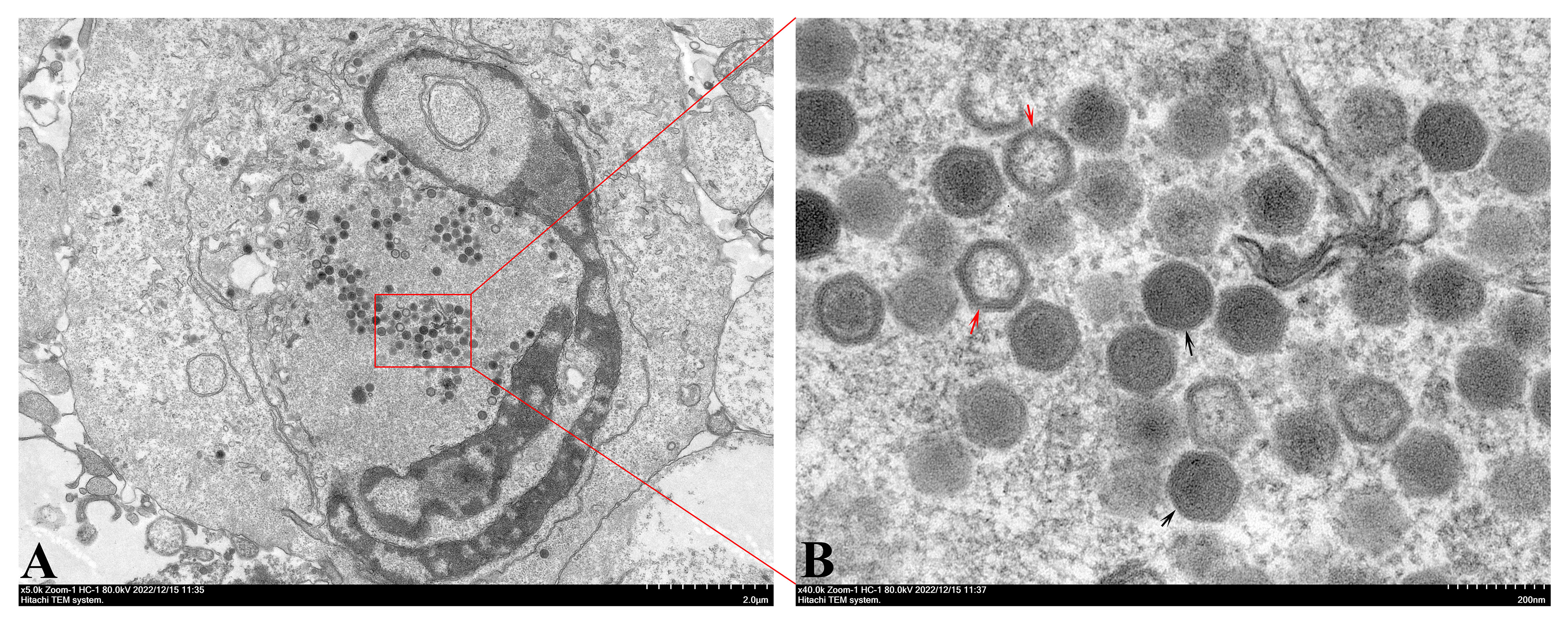
Figure 4 TEM observation of infected EPC cells. (A) Numerous virions accumulated in the cytoplasm of infected EPC cells. Bar=2.0 μM. (B) Different stages of virus assembly, including immature virions (red arrows) and mature virions (black arrows) were present in infected EPC cells. Bar=200 nm.
A 1389 bp DNA fragment corresponding to the CDS of the MCP gene was amplified by RT-PCR using LMBV-specific primers (Figure 5A). The resulting products were extracted and fused into a pET-28a vector (Figure 5B), followed by sequencing. BLAST analysis indicated that the CDS of the MCP gene of LMBV-XX01 shared 100% similarity with two LMBV strains: Santee-Cooper ranavirus isolate BG/TH/CU3 (GenBank accession no. NC_038508.1) and LMBV-FS2021 strain (GenBank accession no. ON418985.1). Subsequently, a phylogenetic tree was generated using the CDS of the MCP gene of LMBV-XX01 and 12 other MCP gene sequences of an iridovirus from different genera employing the NJ method. As shown in Figure 5C, LMBV-XX01 was clustered together with SCRV and LMBV-FS2021, both of which belong to the ranavirus genus. Based on these characteristics, the isolated virus was determined to be a strain of LMBV and provisionally named LMBV-XX01 strain.
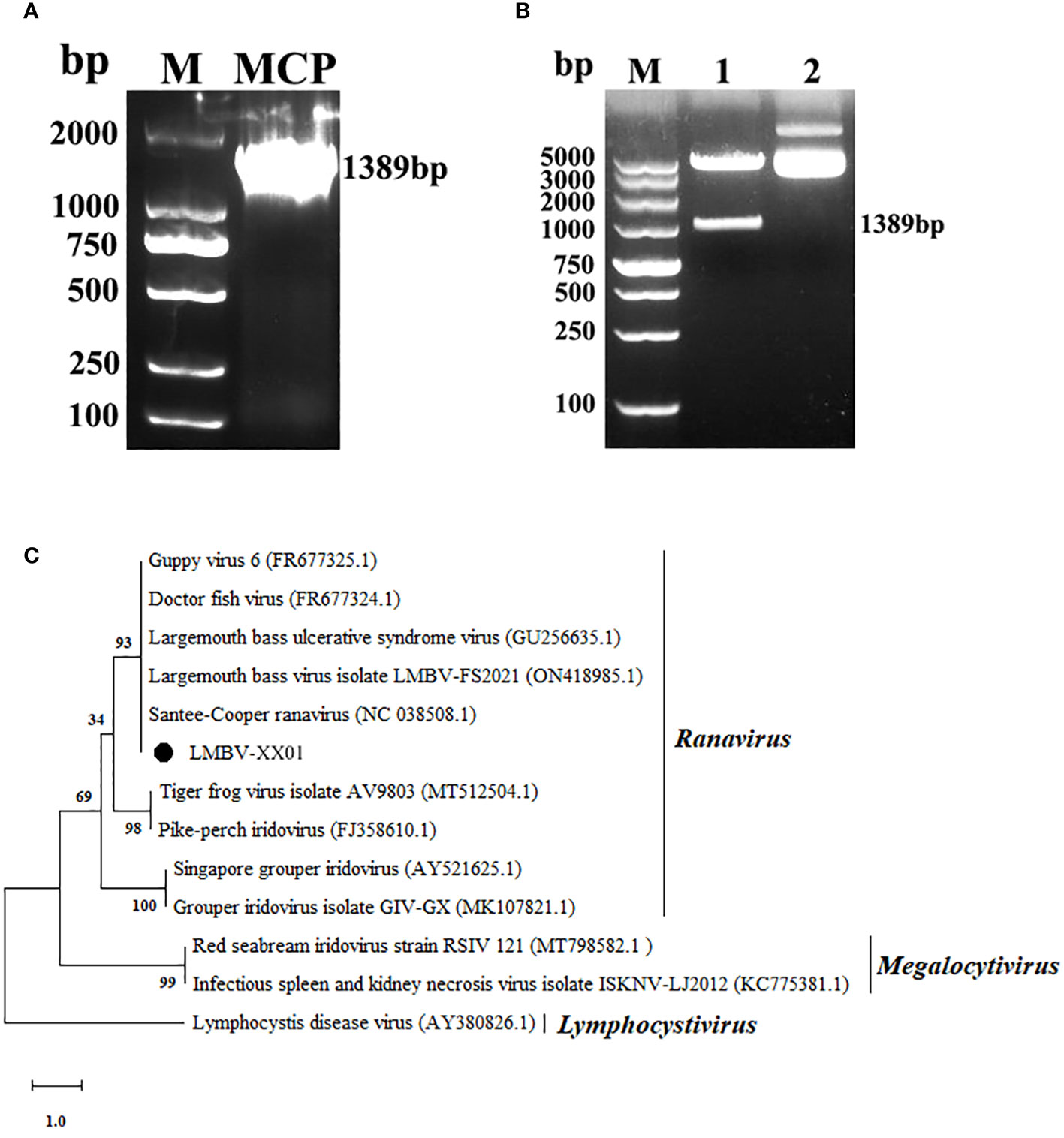
Figure 5 Gene amplification, sequencing, and phylogenetic analysis of LMBV MCP. (A) PCR amplification of LMBV MCP gene. Lane M: DL2000 DNA marker; Lane MCP: PCR product of MCP gene. (B) Recombinant pET-28a-MCP plasmid identified by double-enzyme digestion. Lane 1: The resulting plasmid containing the MCP gene was digested by Xho I and Hind III; Lane 2: The recombinant plasmid without double digestion; (C) Phylogenetic tree constructed using the CDS of the MCP gene of LMBV and other members of Iridoviridae family. The numbers on the tree nodes indicate bootstrap values derived from 1,000 replicates. The branch length on the tree is proportional to the number of nucleotide substitutions, as denoted by the scale bar beneath the tree, with each unit representing 0.1 nucleotide substitutions per site.
As depicted in Figure 6, virus titers in the supernatants and intracellular viral RNA levels rapidly increased from 12 hours to 60 hours post-infection, reaching their peak at 60 hours, and then remained stable until 96 hours, which is consistent with the development of CPE in EPC cells (Figure 3). Interestingly, the intracellular viral RNA levels rapidly increased during the 6-12 hour period post-infection, whereas the virus titers in the supernatants remained relatively unchanged (Figure 6).
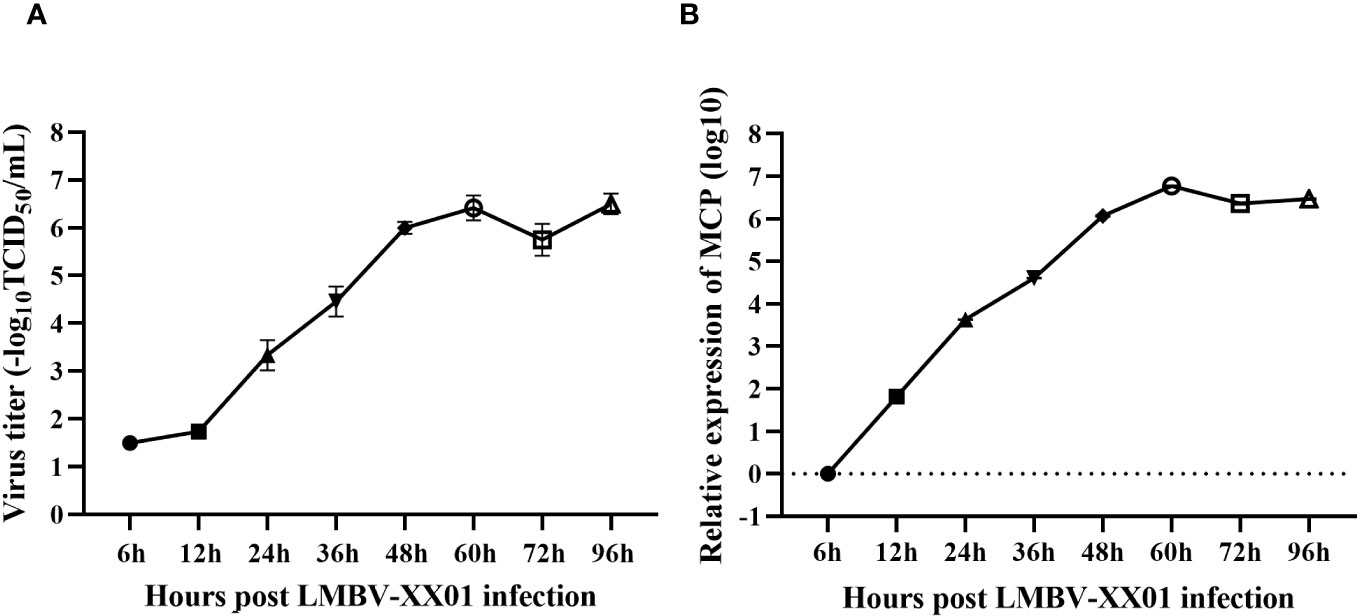
Figure 6 Replication kinetics of LMBV-XX01 on EPC cells. (A) The release process of infectious LMBV progeny from the cytoplasm into the culture medium. (B) Kinetics of MCP gene synthesis in LMBV-XX01 infected EPC cells.
The dying fish in the LMBV-XX01-injected group displayed clinical symptoms similar to those observed in naturally diseased largemouth bass from fish farms. These symptoms included anorexia, insensitivity, fin base hemorrhage, spiral swimming, and abdominal distension. Fish injected with a high dose of LMBV-XX01 exhibited abnormal behaviors such as anorexia, spiral swimming, lethargy, and redness of fin base starting on day 3 after the challenge. Massive mortality occurred 4 and 5 days after the challenge. In the low-dose injected groups, typical clinical symptoms began to appear on day 5 post-injection, and death occurred on day 7 post-injection. By the 14th day post-injection, the mortalities for fish challenged with different doses of LMBV-XX01 were as follows: 5×104 (100% mortality), 104 (80% mortality), 2×103 (45% mortality), 4×102 (40% mortality), and 8×101 TCID50 (20% mortality) (Figure 7A). No obvious symptoms or deaths were found in the mock-injected group throughout the entire experimental period (Figures 7A, B). The cumulative mortality curves were constructed for the challenged experiment (Figure 7A), and the lethal dose of 50% (LD50) in largemouth bass juveniles was calculated to be 1.0 × 103.081 TCID50/fish. Additionally, the RT-PCR detection results of livers and hearts collected from three individual LMBV-XX01 injected fish were all positive (Figure 7C). Typical CPE was induced in EPC cells after inoculation with pooled liver homogenate obtained from the LMBV-XX01 injected largemouth bass (Figure 7D). No CPE or specific PCR production was observed in the control group (Figures 7C, D).
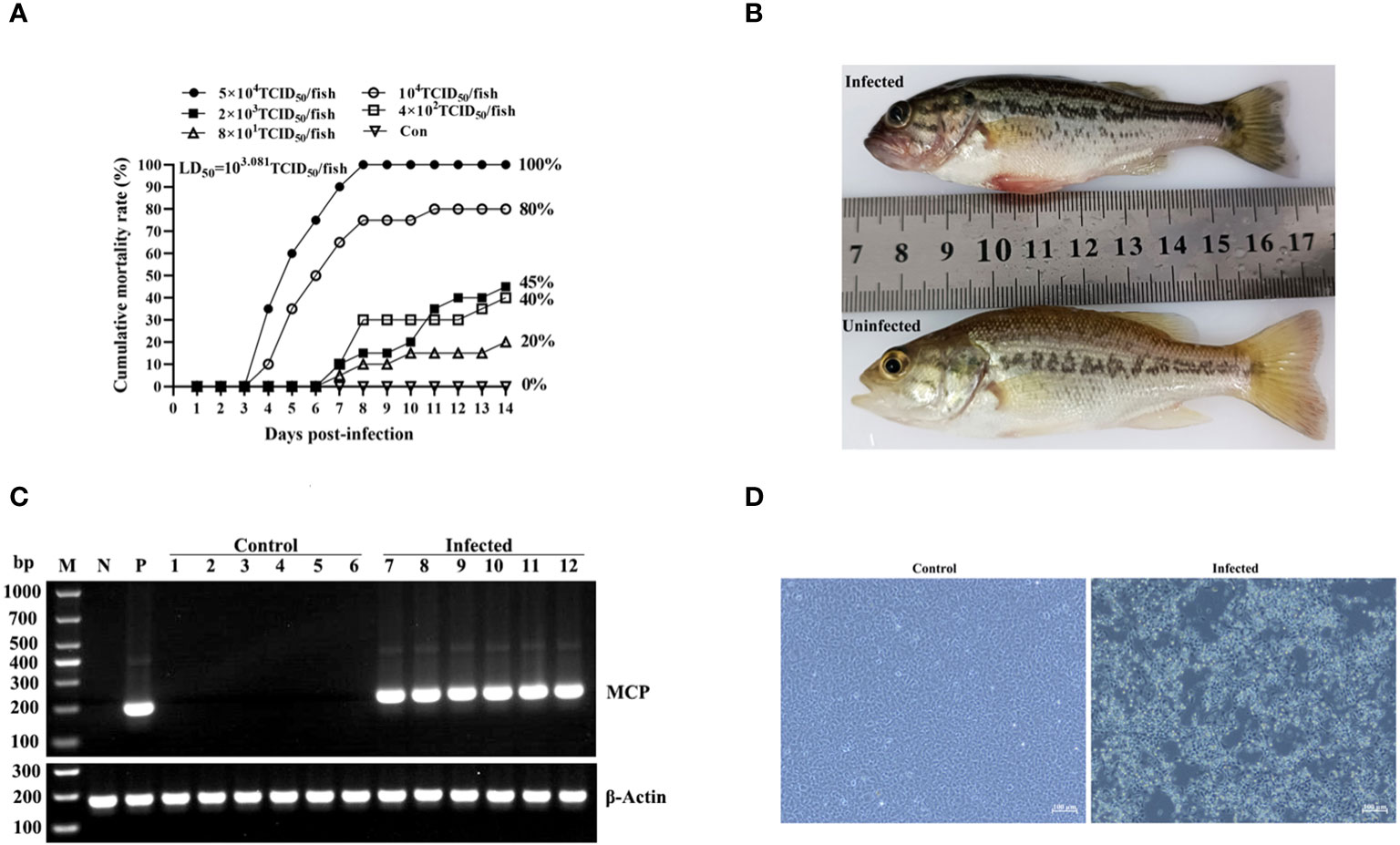
Figure 7 Regression infection of LMBV-XX01. (A) Mortality curves of largemouth bass juveniles injected with different doses of LMBV-XX01 over a period of 14 days. Each group consisted of 20 fish. (B) Clinical symptom of LMBV-XX01-injected or mock-injected largemouth bass. (C) RT-PCR examination of LMBV-XX01 in the livers and hearts of LMBV-injected or mock-injected largemouth bass juveniles. The lanes in the figure are labeled as follows: Lane M: DL1000 DNA marker; Lane N: negative control; Lane P: positive control; Lanes 1-3: liver tissues from mock-injected fish; Lanes 4-6: heart tissues from mock-injected fish; Lanes 7-9: liver tissues from LMBV-XX01-injected fish; Lanes 10-12: heart tissues from LMBV-XX01-injected fish. The largemouth bass β-actin serves as an internal control. (D) Virus isolation from LMBV-XX01-injected largemouth bass juveniles using EPC cells. Obvious CPE was observed in EPC cells incubated with liver homogenate from injected largemouth basses, while no CPE occurred in EPC cells incubated with homogenate from mock-injected fish. Scale bars = 100 μm.
RT-qPCR analysis revealed the presence of LMBV-XX01 in all 10 tested tissues, with the highest viral content observed in the liver, followed by the heart, intestine, and stomach. The virus content was lower in the kidney, muscle, spleen, brain, eye, and gill (Figure 8).
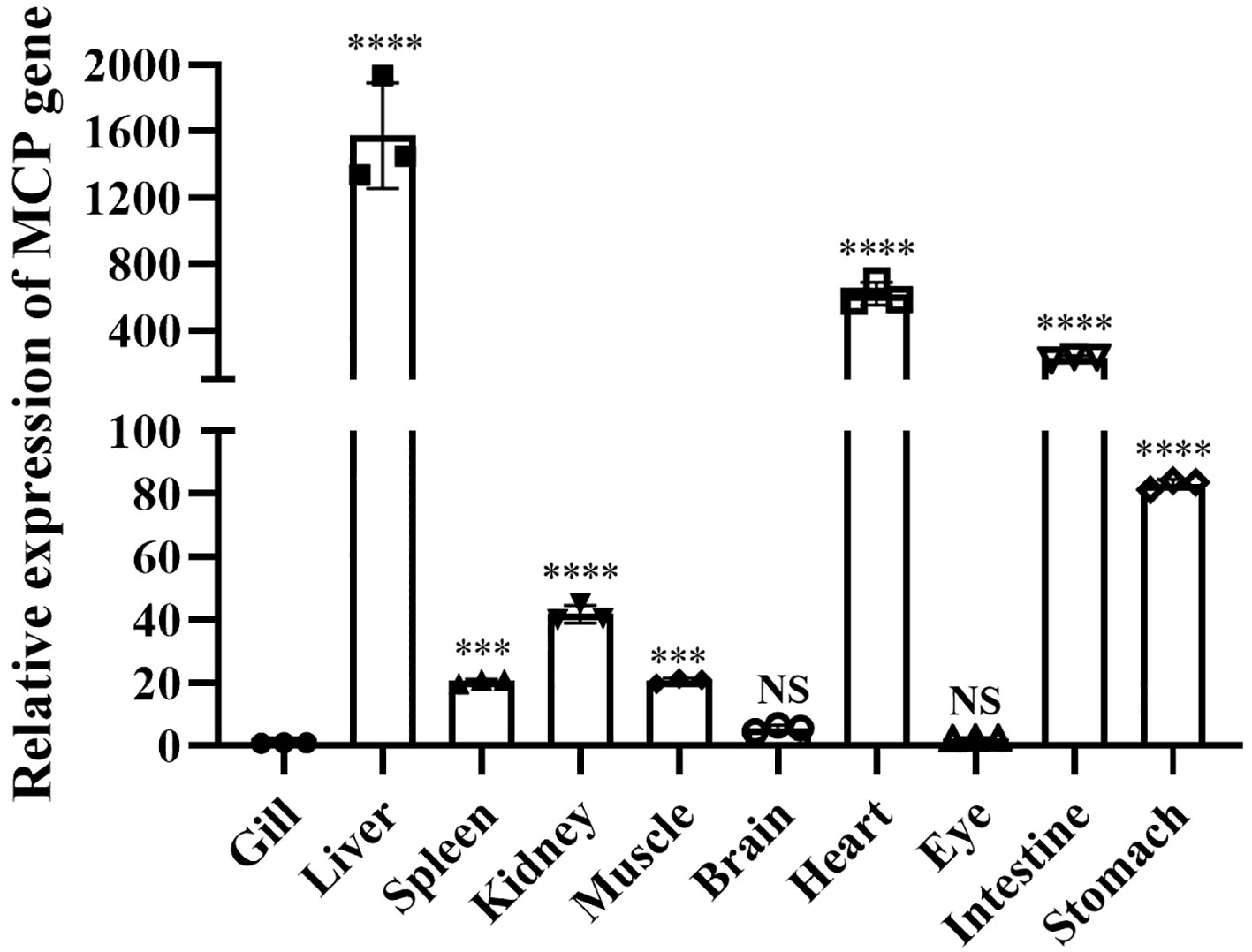
Figure 8 Tissue distribution of LMBV-XX01 in diseased largemouth bass. The relative LMBV-XX01 viral loads among infected fish tissues were quantified by measuring the mRNA level of the MCP gene. The MCP mRNA levels were calculated as fold induction normalized to the level in the gill tissue, which was set to 1. The experiments were repeated three times, and the resulting data were statistically analyzed, and the error bars mean the standard error of the mean (SEM). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. "NS" means "No significant difference".
SDS-PAGE analysis showed successful expression of a 50 kDa specific protein in E. coli BL21 (DE3) containing the pET-28a-MCP recombinant upon induction with IPTG, which coincided with the predicted molecular weight of the His-fusion expression MCP. After purification using His trap Ni2+ affinity chromatography, high-purity rMCP was obtained (Figure 9A, lane 5). Western blot confirmed the successful fusion expression of MCP with an N-terminal His tag, as the purified protein was recognized by anti-His mAb (Figure 9B).
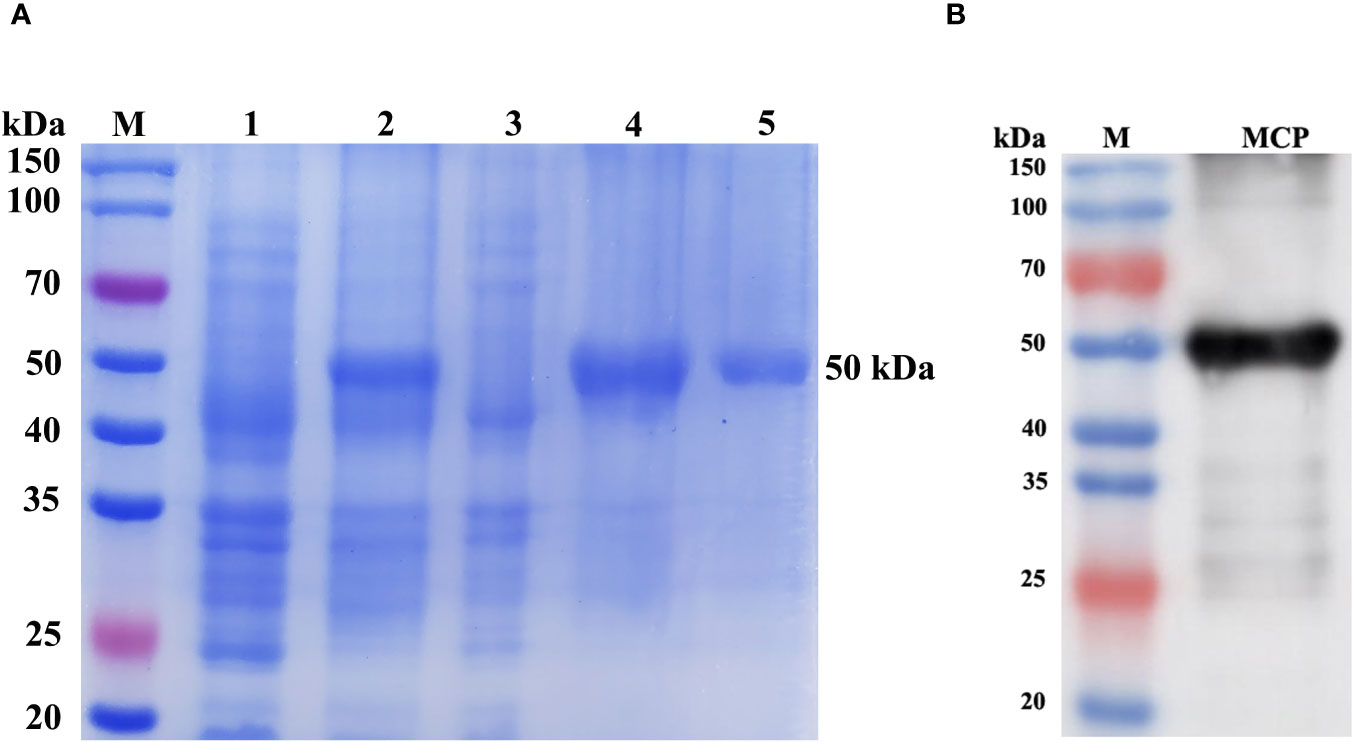
Figure 9 SDS-PAGE (A) and western blot analysis (B) of rMCP expressed by E. coli. Lane M: protein marker; Lane 1: E. coli transformed with the pET-28a-MCP recombinant plasmid without IPTG induction; Lane 2: E. coli containing pET-28a-MCP recombinant plasmid was induced with IPTG; The supernatant (Lane 3) and precipitate (Lane 4) of E. coli induced with IPTG after ultrasonication; Lane 5: purified rMCP. Lane MCP: western blot analysis of rMCP using anti-His tag monoclonal antibody.
A total of 60 monoclonal hybridomas cell wells were obtained on the 14th-day post-fusion, 40 of which were identified to have positive reactions with rMCP through indirect ELISA assays. Among the positive cells, two hybridomas named 2G11 and 4E8 exhibited positive reactions with rMCP and LMBV in western blot assay and IFA. These two hybridomas were subsequently subcloned by limited dilution until all the monoclonal cells showed positive results in indirect ELISA determination. The obtained positive clones were extended cultured and stored in liquid nitrogen.
Western blot assay revealed that the two mAbs specifically reacted with a 50 kDa protein in LMBV-infected EPC cells (Figure 10), which corresponded to the molecular mass of the MCP of LMBV. No positive band was observed in uninfected EPC cells. To determine whether the two mAbs could specifically react with LMBV, an indirect immunofluorescence assay (IFA) was performed using LMBV-infected EPC cells as the tested sample and the developed mAbs as the first antibody. As shown in Figure 11, strong green fluorescence appeared in LMBV-infected EPC cells when using 2G11 and 4E8 rather than myeloma culture supernatant as the first antibody, indicating these two mAbs specifically recognized LMBV and could be further utilized for the development of LMBV detection methods.
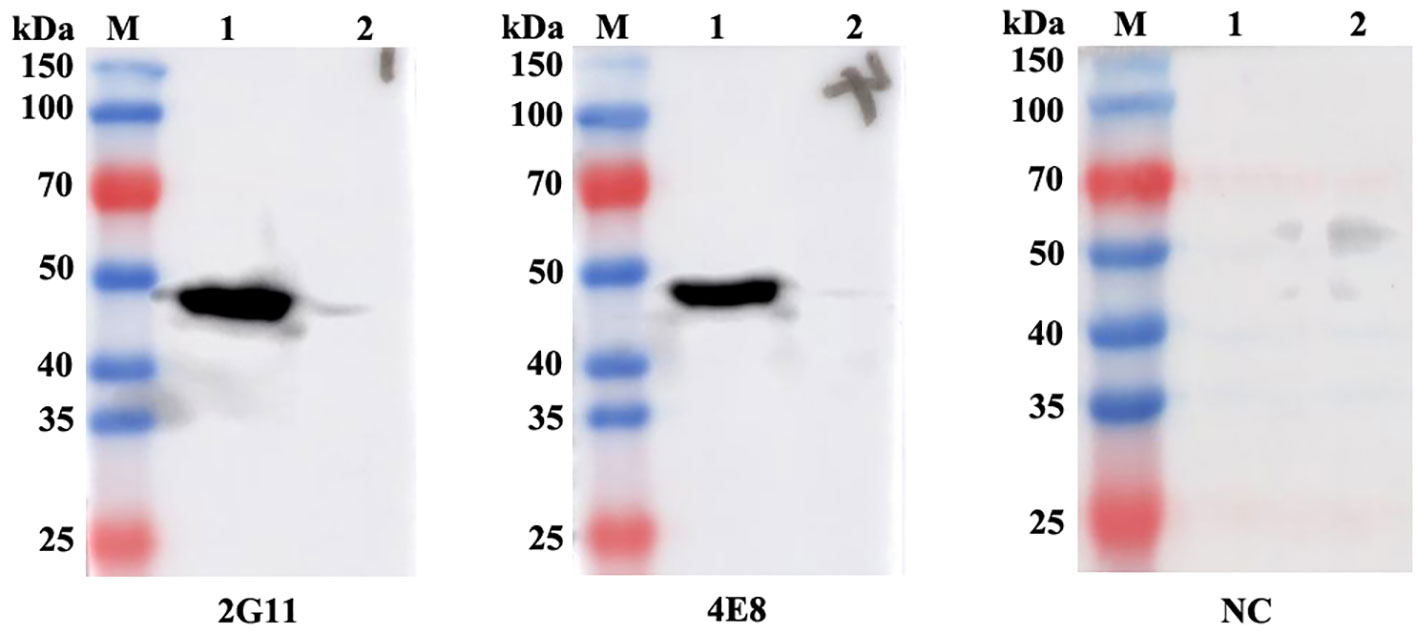
Figure 10 Western blot analysis of mAbs against the MCP of LMBV. Lane M: protein marker; Lane 1: LMBV-infected EPC cells; Lane 2: uninfected EPC cells. The labels 2G11 and 4E8 indicate the respective mAbs used. NC: Negative control means using the myeloma culture supernatant as the first antibody.
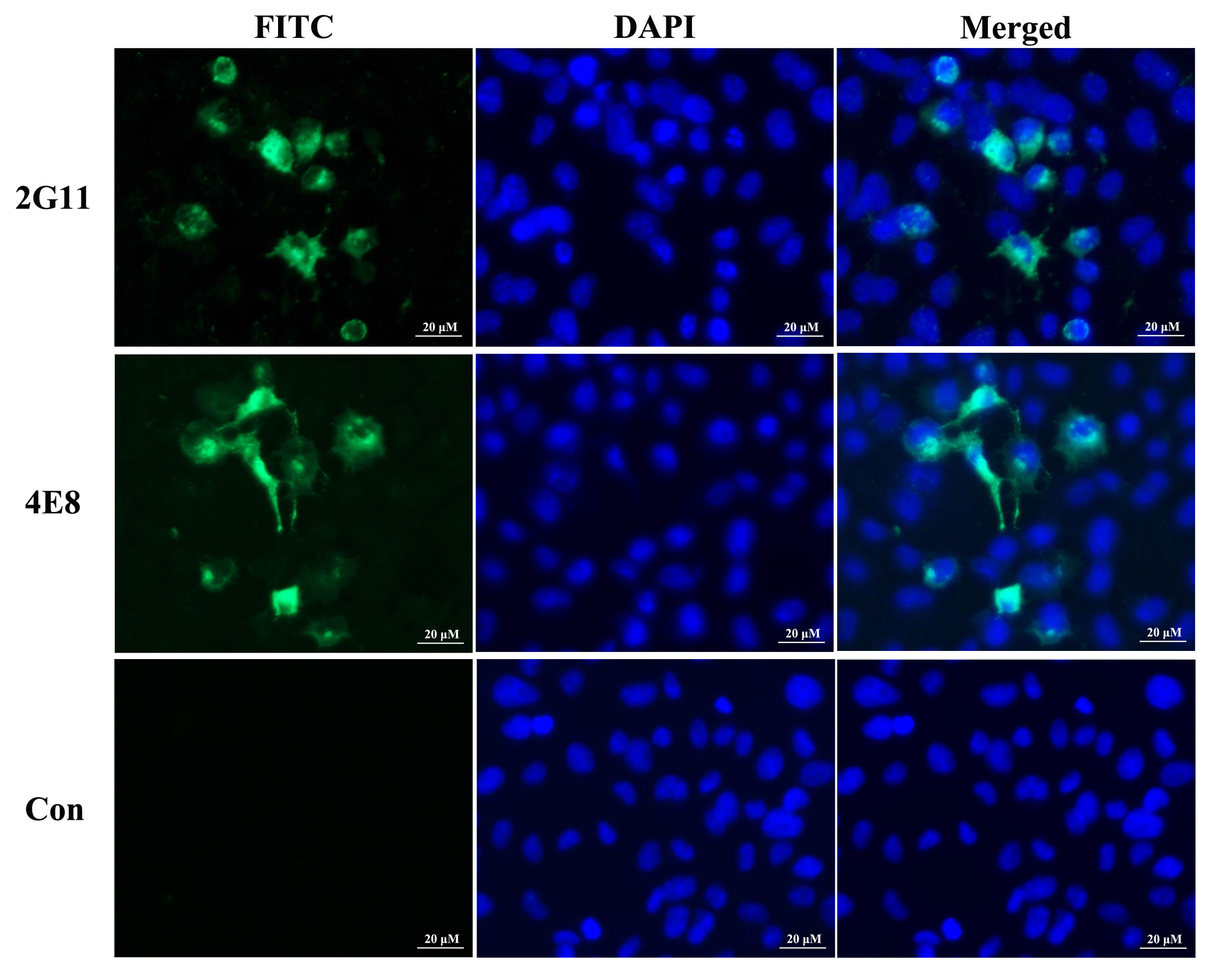
Figure 11 Indirect immunofluorescence assay of mAbs against MCP of LMBV in LMBV-XX01-infected EPC cells. Strong green fluorescence primarily occurred in the cytoplasm or membrane of infected cells incubated with the developed mAbs, while no positive signals appeared when using the myeloma culture supernatant as the first antibody. The labels 2G11 and 4E8 indicate the respective mAbs used. The nuclei of cells were stained with DAPI, showing blue fluorescence.
Largemouth bass (Micropterus salmoides) has gained significant importance in global aquaculture (Wang et al., 2019). The production of largemouth bass in 2022 in China reached 802,486 tons, representing a 14.3% increase compared to 2021, and largemouth bass has become the seventh largest farmed fish species in China (China Fishery Statistical Yearbook, 2023). However, the industry is facing challenges due to frequent outbreaks of lethal diseases caused by viruses and bacteria, leading to some serious issues such as antibiotic abuse, food quality deterioration, and substantial financial losses (Deng et al., 2011; Sibley et al., 2016; Cai et al., 2022; Liu et al., 2022; Qin et al., 2023).
Among these diseases, LMBV is a major threat to the largemouth bass industry, causing extensive mortality in largemouth bass of different sizes and ages (Plumb et al., 1996; Maceina and Grizzle, 2006; Liu et al., 2023). Although the first reported case of fish death caused by LMBV occurred in the Santee-Cooper Reservoir in 1995, it is controversial when and where LMBV was first isolated (Plumb et al., 1996; Grizzle et al., 2002). In 1999, Mao et al. confirmed that LMBV belonged to the Ranavirus genus within the Iridovindae family according to the molecular characterization of LMBV (Mao et al., 1999). Since then, multiple LMBV strains have been isolated from adult or juvenile largemouth bass in various countries and regions (Southard et al., 2009; Deng et al., 2011; Zhao et al., 2020; Jin et al., 2022). Notably, the clinical symptoms and mortality rates caused by LMBV vary among virus strains, fish species, and the sizes of infected fish (Grizzle et al., 2002; Maceina and Grizzle, 2006; Liu et al., 2023). For example, naturally diseased adult largemouth bass have appeared normal with no external lesions, except for losing balance and floating on the water surface (Plumb et al., 1996). In contrast, LMBV-infected juvenile largemouth bass documented by Liu et al. exhibited surface bleeding, ulceration, muscle necrosis, and splenomegaly (Liu et al., 2023). Except for the ulcer lesions, muscle necrosis, and bleeding on the body surface, LMBV-FS2021-infected fish also displayed white liver nodules and adherent intestines, which resemble the clinical signs of Nocardia seriolae infection (Jin et al., 2022; Liu et al., 2022). Additionally, many pathogens could also cause surface bleeding and ulceration and LMBV-infected fish sometimes show no obvious clinical symptoms, making clinical diagnosis difficult and necessitating further molecular identification (Sosa et al., 2007; Jin et al., 2022; Liu et al., 2022). The main symptoms of the naturally diseased or artificially infected fish in our study included anorexia, insensitivity, abdominal distension, spiral swimming, and body surface hemorrhage, which was not entirely consistent with the classical symptoms of iridovirus infection, such as ulcer lesions and muscle necrosis. Initially, we speculated that the pathogen may be MSRV rather than LMBV. To confirm the pathogen, we conducted a series of time-consuming experiments including parasite observation, bacterial isolation, PCR detection, virus isolation, TEM analysis, and regression infection, ultimately determining that the pathogen responsible for the diseased fish was LMBV.
While several vaccines and herbal extracts have shown efficacy in preventing LMBV infection, there are currently no approved vaccines or drugs available for its prevention or treatment (Yao et al., 2022; Cheng et al., 2023; Wang et al., 2023; Zhang et al., 2023). Moreover, infected fish with LMBV experience rapid mortality once symptoms appear (Zhao et al., 2020; Jin et al., 2022). Therefore, rapid diagnosis and early isolation are crucial in preventing the spread of LMBV and reducing economic loss. While numerous LMBV detection methods have been developed and utilized, they are time-consuming and not suitable for on-site diagnosis (Guo et al., 2022; Zhang et al., 2022). Immunostrips, based on the specific reaction between antibodies and antigens, are considered an ideal alternative for LMBV rapid detection, whether in the laboratory or field (Li et al., 2021). MCP is a predominant structural protein of iridovirus, accounting for 45% of all viral proteins (Black et al., 1981). Furthermore, the amino acid sequences of MCP are significantly conserved within the same genera and show lower homology between different genera within the family Iridoviridae (Tidona et al., 1998; Hyatt et al., 2000). MCP has been widely used in vaccine preparation, virus detection, taxonomy, and evolution analysis of iridoviruses (Maceina and Grizzle, 2006; Ohlemeyer et al., 2011; Yao et al., 2022). Therefore, we prepared high-purity MCP as an immunogen to immunize Balb/c mice and finally obtained two strains of anti-MCP mAbs. As shown in Figures 10 and 11, these two mAbs exhibited specific recognition of MCP and LMBV in infected EPC cells, indicating their potential for establishing an immunological detection method for LMBV.
Although we successfully obtained two strains of anti-MCP mAbs, certain challenges remained. For instance, while we obtained 40 positive monoclonal hybridomas recognizing rMCP through ELISA screening, only two of them were able to recognize native MCP in western blot and IFA using LMBV-infected EPC cells. The 5% positive rate was lower than that in our previous study (Qin et al., 2022). The low positive rate could be attributed to the recombinant MCP expressed in E.coil lacking similar posttranslational modifications as in fish or eukaryotes, which could lead to changes in epitopes related to the native MCP of LMBV (Zhao et al., 2007; Macek et al., 2019). This phenomenon suggests that the level of specific antibodies can better reflect the effectiveness of vaccine immunization compared to the level of total antibodies, making specific antibodies a better target for vaccine evaluation.
Electron microscopy observation revealed abundant hexagonal virus particles, approximately 150 nm in diameter, including empty capsids and mature virions, scattered throughout the cytoplasm of infected EPC cells, which is a typical characteristic of ranavirus and is consistent with previous reports (Deng et al., 2011; Zhao et al., 2020). The green fluorescence signals using anti-MCP mAbs as the first antibody predominantly appeared in the cytoplasm and cell membrane of LMBV-infected EPC cells (Figure 10), which corresponds to the distribution of LMBV virions observed under TEM (Figure 4).
Histopathology examination revealed extensive congestion and cell necrosis occurred in the liver, spleen, intestine, and stomach, which is supported by a previous study (Liu et al., 2023). RT-qPCR detection demonstrated the presence of LMBV in all 10 sampled tissues, indicating systemic infection in diseased fish. Among these tissues, higher virus loads were observed in the liver, heart, intestine, and stomach, which are considered the main target tissues for LMBV (Zhang et al., 2023). The higher virus loads and severe epithelial necrosis in the intestine and stomach may explain the observed anorexia and swollen intestines in LMBV-infected fish.
MSRV, another main pathogen of largemouth bass, induces obvious CPE and supports plaque formation in GS cells. Besides, Singapore grouper iridovirus (SGIV), also belonging to the genus Ranavirus, could propagate in GS cells (Li et al., 2020). Accordingly, we tried to isolate LMBV using GS cells. However, our results showed that LMBV fails to replicate in GS cells (Figure 4). The possible reasons behind the non-replication of LMBV in GS cells are as follows. First, although LMBV and SGIV both belong to the genus Ranavirus, the sequence similarity of MCP is only 71%, which carries virus-specific antigens and determines viral virulence. Besides, susceptible hosts of LMBV and SGIV are significantly different. There may be no receptors of LMBV on the surface of GS cells, which leads to LMBV failing to enter into GS cells.
In conclusion, we reported a natural mortality case in juvenile largemouth bass and determined the pathogen responsible was LMBV through a series of experiments, including TEM observation, RT-PCR, phylogenetic analysis, and regression infection. Additionally, we generated and characterized two strains of anti-MCP monoclonal antibodies. Our findings provide new insights and materials contributing to the design of prevention strategies and the development of detection methods for LMBV.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The animal study was approved by the Ethics Committee of Nanyang Normal University. The study was conducted in accordance with the local legislation and institutional requirements.
YQ: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. HL: Data curation, Investigation, Supervision, Writing – original draft. RD: Investigation, Validation, Writing – original draft. YW: Investigation, Validation, Writing – original draft. SM: Investigation, Validation, Writing – original draft. SD: Investigation, Writing – original draft. PZ: Investigation, Validation, Writing – original draft. LY: Conceptualization, Funding acquisition, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work received support from the Henan Provincial Science and Technology Research Project (222102110079), the National Natural Science Foundation of China (31870917; 31972834), the Nanyang Science and Technology Research Project (KJGG089), and the Foundation of Nanyang Normal University (2022ZX030; 2023PY018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Black P. N., Blair C. D., Butcher A., Capinera J. L., Happ G. M. (1981). Biochemistry and ultrastructure of iridescent virus type 29. J. Invertebrate Pathol. 38, 12–21. doi: 10.1016/0022-2011(81)90028-8
Cai J., Yu D., Xia H., Xia L., Lu Y. (2022). Identification and characterization of a nervous necrosis virus isolated from largemouth bass (Micropterus salmoides). J. Fish Dis. 45 (4), 607–611. doi: 10.1111/jfd.13576
Cheng Y., Liu M., Yu Q., Huang S., Han S., Shi J., et al. (2023). Effect of EGCG extracted from green tea against largemouth bass virus infection. Viruses 15 (1), 151. doi: 10.3390/v15010151
China Fishery Statistical Yearbook (2023). China fishery statistical yearbook Vol. 2023 (Beijing, China: China Agriculture Press), 22–25.
Deng G., Li S., Xie J., Bai J., Chen K., Ma D., et al. (2011). Characterization of a ranavirus isolated from cultured largemouth bass (Micropterus salmoides) in China. Aquaculture 312 (1-4), 198–204. doi: 10.1016/j.aquaculture.2010.12.032
Dong C., Wang Z., Weng S., He J. (2017). Occurrence of a lethal ranavirus in hybrid mandarin (Siniperca scherzerixSiniperca chuatsi) in Guangdong, South China. Vet. Microbiol. 203, 28–33. doi: 10.1016/j.vetmic.2017.02.006
Fu X., Luo M., Zheng G., Liang H., Liu L., Lin Q., et al. (2022). Determination and characterization of a novel birnavirus associated with massive mortality in largemouth bass. Microbiol. Spectr. 10 (2), e0171621. doi: 10.1128/spectrum.01716-21
George M. R., John K. R., Mansoor M. M., Saravanakumar R., Sundar P., Pradeep V. (2015). Isolation and characterization of a ranavirus from koi, Cyprinus carpio L., experiencing mass mortalities in India. J. Fish Dis. 38 (4), 389–403. doi: 10.1111/jfd.12246
Godahewa G. I., Lee S., Kim J., Perera N. C. N., Kim M. J., Kwon M. G., et al. (2018). Analysis of complete genome and pathogenicity studies of the spring viremia of carp virus isolated from common carp (Cyprinus carpio carpio) and largemouth bass (Micropterus salmoides): An indication of SVC disease threat in Korea. Virus Res. 255, 105–116. doi: 10.1016/j.virusres.2018.06.015
Grizzle J. M., Altinok I., Fraser W. A., Francis-Floyd R. (2002). First isolation of largemouth bass virus. Dis. Aquat Organ 50 (3), 233–235. doi: 10.3354/dao050233
Guo Y., Wang Y., Fan Z., Zhao X., Bergmann S. M., Dong H., et al. (2022). Establishment and evaluation of qPCR and real-time recombinase-aided amplification assays for detection of largemouth bass ranavirus. J. Fish Dis. 45 (7), 1033–1043. doi: 10.1111/jfd.13627
Hyatt A. D., Gould A. R., Zupanovic Z., Cunningham A. A., Hengstberger S., Whittington R. J., et al. (2000). Comparative studies of piscine and amphibian iridoviruses. Arch. Virol. 145 (2), 301–331. doi: 10.1007/s007050050025
Jia Y. J., Guo Z. R., Ma R., Qiu D. K., Wang G. X., Zhu B. (2020). Protective immunity of largemouth bass immunized with immersed DNA vaccine against largemouth bass ulcerative syndrome virus. Fish Shellfish Immunol. 107 (Pt A), 269–276. doi: 10.1016/j.fsi.2020.10.018
Jia Y. J., Xia J. Y., Jiang F. Y., Li Y., Chen G., Zhu B. (2022). Antigenic epitope screening and functional modification of mannose enhance the efficacy of largemouth bass virus subunit vaccines. J. Fish Dis. 45 (11), 1635–1643. doi: 10.1111/jfd.13686
Jin Y., Bergmann S. M., Mai Q., Yang Y., Liu W., Sun D., et al. (2022). Simultaneous isolation and identification of largemouth bass virus and rhabdovirus from moribund largemouth bass (Micropterus salmoides). Viruses 14 (8), 1643. doi: 10.3390/v14081643
Junjie B., Shengjie L. (2019). Genetic Breeding and Molecular Marker-Assisted Selective Breeding of Largemouth Bass (London: Academic Press).
Li C., Wang L., Liu J., Yu Y., Huang Y., Huang X., et al. (2020). Singapore grouper iridovirus (SGIV) inhibited autophagy for efficient viral replication. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01446
Li G., Wang A., Chen Y., Sun Y., Du Y., Wang X., et al. (2021). Development of a colloidal gold-based immunochromatographic strip for rapid detection of severe acute respiratory syndrome coronavirus 2 spike protein. Front. Immunol. 12. doi: 10.3389/fimmu.2021.635677
Lin Q., Fu X., Liu L., Liang H., Guo H., Yin S., et al. (2017). Application and development of a TaqMan real-time PCR for detecting infectious spleen and kidney necrosis virus in Siniperca chuatsi. Microb. Pathog. 107, 98–105. doi: 10.1016/j.micpath.2017.02.046
Liu X., Zhang Y., Zhang Z., An Z., Zhang X., Vakharia V. N., et al. (2023). Isolation, identification and the pathogenicity characterization of a Santee-Cooper ranavirus and its activation on immune responses in juvenile largemouth bass (Micropterus salmoides). Fish Shellfish Immunol. 135, 108641. doi: 10.1016/j.fsi.2023.108641
Liu Z. G., Dong J. J., Ke X. L., Yi M. M., Cao J. M., Gao F. Y., et al. (2022). Isolation, identification, and pathogenic characteristics of Nocardia seriolae in largemouth bass Micropterus salmoides. Dis. Aquat Organ 149, 33–45. doi: 10.3354/dao03659
Ma D., Deng G., Bai J., Li S., Yu L., Quan Y., et al. (2013). A strain of Siniperca chuatsi rhabdovirus causes high mortality among cultured Largemouth Bass in South China. J. Aquat Anim. Health 25 (3), 197–204. doi: 10.1080/08997659.2013.799613
Maceina M. J., Grizzle J. M. (2006). The relation of largemouth bass virus to largemouth bass population metrics in five alabama reservoirs. Trans. Am. Fisheries Soc. 135 (2), 545–555. doi: 10.1577/t05-100.1
Macek B., Forchhammer K., Hardouin J., Weber-Ban E., Grangeasse C., Mijakovic I. (2019). Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 17 (11), 651–664. doi: 10.1038/s41579-019-0243-0
Mao J., Wang J., Chinchar G. D., Chinchar V. G. (1999). Molecular characterization of a ranavirus isolated from largemouth bass Micropterus salmoides. Dis. Aquat Organ 37 (2), 107–114. doi: 10.3354/dao037107
Montelongo-Alfaro I. O., Amador-Cano G., Rabago-Castro J. L., Sanchez-Martinez J. G., Benavides-Gonzalez F., Gojon-Baez H. H. (2019). A preliminary study of largemouth bass virus in Mexico. J. Wildl Dis. 55 (2), 516–517. doi: 10.7589/2018-03-082
Ohlemeyer S., Holopainen R., Tapiovaara H., Bergmann S. M., Schutze H. (2011). Major capsid protein gene sequence analysis of the Santee-Cooper ranaviruses DFV, GV6, and LMBV. Dis. Aquat Organ 96 (3), 195–207. doi: 10.3354/dao02370
Plumb J. A., Grizzle J. M., Young H. E., Noyes A. D., Lamprecht S. (1996). An iridovirus isolated from wild largemouth bass. J. Aquat. Anim. Health 8 (4), 265–270. doi: 10.1577/1548-8667(1996)008<0265:AIIFWL>2.3.CO;2
Qin Y., Liu J., Liu W., Shi H., Jia A., Lu Y., et al. (2020). First isolation and identification of red-grouper nervous necrosis virus (RGNNV) from adult hybrid Hulong grouper (Epinephelus fuscoguttatus× Epinephelus lanceolatus) in China. Aquaculture 529, 735662. doi: 10.1016/j.aquaculture.2020.735662
Qin Y., Liu J., Lu Y., Liu X. (2022). Generation and characterization of monoclonal antibodies against red-spotted grouper nervous necrosis virus (RGNNV). J. Ocean Univ. China 21, 1061–1067. doi: 10.1007/s11802-022-4895-4
Qin Y., Zhang P., Zhang M., Guo W., Deng S., Liu H., et al. (2023). Isolation and identification of a new strain Micropterus salmoides rhabdovirus (MSRV) from largemouth bass Micropterus salmoides in China. Aquaculture 572, 739538. doi: 10.1016/j.aquaculture.2023.739538
Salazar V., Koch J. D., Neely B. C., Steffen C. J., Flores E., Steffen S. F. (2022). The effect of largemouth bass virus on bass populations in kansas impoundments. J. Aquat Anim. Health 34 (1), 38–44. doi: 10.1002/aah.10147
Shen L., Zhang Q., Luo X., Xiao H., Gu M., Cao L., et al. (2021). A rapid lateral flow immunoassay strip for detection of SARS-CoV-2 antigen using latex microspheres. J. Clin. Lab. Anal. 35 (12), e24091. doi: 10.1002/jcla.24091
Sibley S. D., Finley M. A., Baker B. B., Puzach C., Armien A. G., Giehtbrock D., et al. (2016). Novel reovirus associated with epidemic mortality in wild largemouth bass (Micropterus salmoides). J. Gen. Virol. 97 (10), 2482–2487. doi: 10.1099/jgv.0.000568
Sivasankar P., John K. R., George M. R., Mageshkumar P., Manzoor M. M., Jeyaseelan M. J. P. (2017). Characterization of a virulent ranavirus isolated from marine ornamental fish in India. Virusdisease 28 (4), 373–382. doi: 10.1007/s13337-017-0408-2
Sosa E. R., Landsberg J. H., Kiryu Y., Stephenson C. M., Cody T. T., Dukeman A. K., et al. (2007). Pathogenicity studies with the fungi Aphanomyces invadans, Achlya bisexualis, and Phialemonium dimorphosporum: induction of skin ulcers in striped mullet. J. Aquat Anim. Health 19 (1), 41–48. doi: 10.1577/H06-013.1
Southard G. M., Fries L. T., Terre D. R. (2009). Largemouth bass virus in Texas: distribution and management issues. J. Aquat Anim. Health 21 (1), 36–42. doi: 10.1577/H08-027.1
Tidona C. A., Schnitzler P., Kehm R., Darai G. (1998). Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 16 (1), 59–66. doi: 10.1023/a:1007949710031
Waltzek T. B., Subramaniam K., Leis E., Katona R., Fan Ng T. F., Delwart E., et al. (2019). Characterization of a peribunyavirus isolated from largemouth bass (Micropterus salmoides). Virus Res. 273, 197761. doi: 10.1016/j.virusres.2019.197761
Wang D., Yao H., Li Y. H., Xu Y. J., Ma X. F., Wang H. P. (2019). Global diversity and genetic landscape of natural populations and hatchery stocks of largemouth bass micropterus salmoides across American and Asian regions. Sci. Rep. 9 (1), 16697. doi: 10.1038/s41598-019-53026-3
Wang M., Yang B., Ren Z., Liu J., Lu C., Jiang H., et al. (2023). Inhibition of the largemouth bass virus replication by piperine demonstrates potential application in aquaculture. J. Fish Dis. 46 (3), 261–271. doi: 10.1111/jfd.13740
Xu F. F., Jiang F. Y., Zhou G. Q., Xia J. Y., Yang F., Zhu B. (2022). The recombinant subunit vaccine encapsulated by alginate-chitosan microsphere enhances the immune effect against Micropterus salmoides rhabdovirus. J. Fish Dis. 45 (11), 1757–1765. doi: 10.1111/jfd.13697
Xu Z., Liao J., Zhang D., Liu S., Zhang L., Kang S., et al. (2023). Isolation, characterization, and transcriptome analysis of an ISKNV-like virus from largemouth bass. Viruses 15 (2), 398. doi: 10.3390/v15020398
Yao J., Li C., Shi L., Lu Y., Liu X. (2020). Zebrafish ubiquitin-specific peptidase 5 (USP5) activates interferon resistance to the virus by increase the expression of RIG-I. Gene 751, 144761. doi: 10.1016/j.gene.2020.144761
Yao J. Y., Zhang C. S., Yuan X. M., Huang L., Hu D. Y., Yu Z., et al. (2022). Oral Vaccination With Recombinant Pichia pastoris Expressing Iridovirus Major Capsid Protein Elicits Protective Immunity in Largemouth Bass (Micropterus salmoides). Front. Immunol. 13. doi: 10.3389/fimmu.2022.852300
Zhang M., Chen X., Xue M., Jiang N., Li Y., Fan Y., et al. (2023). Oral Vaccination of Largemouth Bass (Micropterus salmoides) against Largemouth Bass Ranavirus (LMBV) Using Yeast Surface Display Technology. Anim. (Basel) 13 (7), 1183. doi: 10.3390/ani13071183
Zhang W., Deng H., Fu Y., Fu W., Weng S., He J., et al. (2023). Production and characterization of monoclonal antibodies against mandarinfish ranavirus and first identification of pyloric caecum as the major target tissue. J. Fish Dis. 46 (3), 189–199. doi: 10.1111/jfd.13733
Zhang W., Li Z., Xiang Y., Jia P., Liu W., Yi M., et al. (2019). Isolation and identification of a viral haemorrhagic septicaemia virus (VHSV) isolate from wild largemouth bass Micropterus salmoides in China. J. Fish Dis. 42 (11), 1563–1572. doi: 10.1111/jfd.13078
Zhang X., Zhang Z., Li J., Huang X., Wei J., Yang J., et al. (2022). A novel sandwich ELASA based on aptamer for detection of largemouth bass virus (LMBV). Viruses 14 (5), 945. doi: 10.3390/v14050945
Zhao R., Geng Y., Qin Z., Wang K., Ouyang P., Chen D., et al. (2020). A new ranavirus of the Santee-Cooper group invades largemouth bass (Micropterus salmoides) culture in southwest China. Aquaculture 526, 735763. doi: 10.1016/j.aquaculture.2020.735363
Zhao Z., Teng Y., Liu H., Lin X., Wang K., Jiang Y., et al. (2007). Characterization of a late gene encoding for MCP in soft-shelled turtle iridovirus (STIV). Virus Res. 129 (2), 135–144. doi: 10.1016/j.virusres.2007.07.002
Zheng J., Zhi L., Wang W., Ni N., Huang Y., Qin Q., et al. (2022). Fish TRIM21 exhibits antiviral activity against grouper iridovirus and nodavirus infection. Fish Shellfish Immunol. 127, 956–964. doi: 10.1016/j.fsi.2022.06.053
Zhong Y., Tang X., Sheng X., Xing J., Zhan W. (2019). Voltage-dependent anion channel protein 2 (VDAC2) and receptor of activated protein C kinase 1 (RACK1) act as functional receptors for lymphocystis disease virus infection. J. Virol. 93 (12), e00122-19. doi: 10.1128/JVI.00122-19
Keywords: largemouth bass, LMBV, isolation, molecular characterization, monoclonal antibody
Citation: Qin Y, Liu H, Mao S, Deng R, Wang Y, Deng S, Zhang P and Yao L (2024) Isolation, identification, and monoclonal antibody development of largemouth bass virus. Front. Mar. Sci. 10:1338197. doi: 10.3389/fmars.2023.1338197
Received: 16 November 2023; Accepted: 15 December 2023;
Published: 09 January 2024.
Edited by:
Ji Hyung Kim, Gachon University, Republic of KoreaReviewed by:
Sajal Kole, Chonnam National University, Republic of KoreaCopyright © 2024 Qin, Liu, Mao, Deng, Wang, Deng, Zhang and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lunguang Yao, bHVuZ3Vhbmd5YW9AMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.