
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 06 December 2023
Sec. Marine Biotechnology and Bioproducts
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1321196
This article is part of the Research Topic Omics Applied to the Body Colour Formation and Pigmentation of Aquatic Animals View all 3 articles
 Xi Liu1†
Xi Liu1† Mouyan Jiang1,2,3†
Mouyan Jiang1,2,3† Simin Wen1
Simin Wen1 Kaixiang Zhang1
Kaixiang Zhang1 Yucong Hong4
Yucong Hong4 Kaihui Sun4
Kaihui Sun4 Xianwu Deng1
Xianwu Deng1 Yang Huang1,2,3
Yang Huang1,2,3 Gang Shi1,2,3
Gang Shi1,2,3 Changxu Tian1,2,3
Changxu Tian1,2,3 Huapu Chen1,2,3
Huapu Chen1,2,3 Guangli Li1,2,3
Guangli Li1,2,3 Chunhua Zhu1,2,3,5*
Chunhua Zhu1,2,3,5*MicroRNAs (miRNAs) are endogenous small non-coding RNAs that play important roles in several biological processes, including the regulation of body color. Leopard coral grouper (Plectropomus leopardus) is a valuable marine aquaculture fish; however, at present, there are no published reports on its early body color development. The skin color of P. leopardus undergoes a transition from transparent to red from 26 days post-hatching (dph) to 30 dph. In this study, we performed miRNA sequencing on 26 dph (Transparent, PT) and 30 dph (Red, PR) skin samples of P. leopardus to explore the molecular mechanism underlying red color formation. A total of 44.31 M and 37.55 M clean tags were obtained from PT and PR group, respectively. Among these tags, 981 miRNAs were identified, including 493 known and 488 novel miRNAs. A total of 106 differentially expressed miRNAs (DEMs) were identified in PT vs. PR, with 43 up-regulated and 63 down-regulated miRNAs in the PR group, compared to the PT group (|fold change| > 2 and p-value < 0.05). A miRNA-mRNA network based on 18 candidate miRNAs and 53 target genes related to pigmentation, and KEGG enrichment analysis of the target genes of all DEMs, revealed that miRNAs involved in the formation of red skin color were mainly related to: 1) the inhibition of melanin synthesis (miR-141-z, miR-206-z, miR-206-y, miR-27-z, miR-137-y, miR-204-x, miR-204-y, miR-211-x, miR-211-z); 2) chromatophore development (miR-206-z, miR-206-y, miR-499-y, miR-1-z, miR-2188-x, miR-423-x); and 3) carotenoid metabolism (miR-204-x, miR-204-y, miR-499-y). This study demonstrates the potential role of miRNAs in red color formation and lays the foundation for the molecular mechanism of body color polymorphism in P. leopardus.
The body color of animals plays an important role in predator, camouflage, interspecific communication, courting, growth and development (Hill, 1991; Stuart-Fox and Moussalli, 2009; Stevens and Ruxton, 2011; Li-Yan, 2013; Figon and Casas, 2018). Fish have the most diverse color patterns among vertebrates. To adapt to environmental changes, they can undergo reversible changes in body color through long-term or short-term changes in morphology or physiological color (Aspengren et al., 2008; Leclercq et al., 2009). The regulation of body color is a complex process impacted by genetics, environment, physiology and diet etc. (Ninwichian et al., 2022; Toomey et al., 2022; Zhao et al., 2022). Genetics is a major factor, while changes in other factors are accompanied by changes in the genetically related factors. For example, the skin color of P. leopardus (Song et al., 2022a) and Malaysian red tilapia (Wang et al., 2020) varies in different backgrounds, as do the expression levels of genes involved in pigmentation (scarb1, tyr, pomc, mch). However, the molecular mechanisms underlying color changes in teleost fishes have remained largely elusive.
Color patterns in fish are determined by the types, position, and interactions between chromatophores (Yamanaka and Kondo, 2014; McGowan and Barsh, 2016). Chromatophores are derived from neural crest cells and migrate to specific locations after delamination and differentiation to form the final color pattern (Kelsh et al., 2021). To data, at least six pigment cells have been identified, including melanophores, erythrophores, xanthophores, iridophores, leucophores and cyanophores (Kelsh, 2004). These chromatophores play a vital role in the formation of body color. Unlike mammals and birds, only eumelanin has been identified in teleost fishes. The existence of pheomelanin is highly controversial in fishes, although slc7a11/xCT, which is required for pheomelanin formation in mammals (Chintala et al., 2005), has been identified in most fishes (Wang et al., 2018; Wang et al., 2019; Zhi Weng Josiah et al., 2022). Therefore, the red color of fish is thought to be produced by erythrophores with carotenoids and pteridine. Carotenoids cannot be synthesized de novo in fish; they are obtained from the diet, absorbed in the intestine, and transported through the bloodstream by grouping into lipoproteins, which are then deposited in target tissues such as the skin, fins, and scales to produce the final body color (Parker, 1996; Matthews et al., 2006). Based on color, carotenoids can be divided into two groups: yellow carotenoids (zeaxanthin, β-carotene, β-cryptoxanthin, lutein) and red keto-carotenoids (astaxanthin, canthaxanthin). The red color is produced either by the direct deposition of keto-carotenoids in the skin or by the deposition of yellow carotenoids converted to keto-carotenoids (Inouye, 2007; Lafountain et al., 2015). It is generally believed that the skin pigmentation can be significantly improved by dietary supplementation with astaxanthin, such as in P. leopardus (Zhang et al., 2023), Malabar snappers (Lutjanus malabaricus) (Zhi Weng Josiah et al., 2022) and Atlantic Salmon (Sufmo sulur, L.) (Schmeisser et al., 2021). In relation to keto-carotenoids conversion, Toomey et al. (Toomey et al., 2022) reported that zeaxanthin and β-carotene can be converted to astaxanthin and canthaxanthin, respectively, within cyp2j19 and bdh1l1 co-expressed in birds. cyp2ae2 and bdh1a have similar functions to cyp2j19 and bdh1l1 in Danio albolineatus (Huang et al., 2021). This conversion is also observed in crustaceans (Wade et al., 2015); however, the molecular mechanisms are not clear. Pteridine biosynthesis involves gch1, pts and spr, which in turn catalyze the formation of BH4 from GTP. Interestingly, mutations in melanin biosynthesis genes are also associated with the production of red body color. For example, disruption of oca2, which plays a role in melanophore differentiation in zebrafish (Beirl et al., 2013), can cause abnormal red skin in Yellow River carp (Cyprinus carpio haematopterus) (Jiang et al., 2022). mitfa-/- and mitfb-/- double mutations reportedly resulted in red and yellow skin in adult Nile tilapia (Wang et al., 2022). Recently, Kelsh et al. (Kelsh et al., 2021) proposed a cyclical fate restriction hypothesis, to explain the origin of red coloration in Yellow River carp and Nile tilapia (Jiang et al., 2022; Wang et al., 2022).
MicroRNAs(miRNAs) are 18-25nt small non-coding RNAs that affect transcription and translation by targeting mRNA (Nilsen, 2007; Bizuayehu and Babiak, 2014). miRNAs play a role in multiple biological processes, including osmotic regulation, cell proliferation and the innate immune response by influencing gene expression (Hwang and Mendell, 2007; Martín-Gómez et al., 2014; Lingyu et al., 2023). Studies have shown that miRNAs are involved in the regulation of body color. For example, miR-21a-5p targeting sox5 (Wang et al., 2016), miR-27a-3p targeting wnt3a (Zhao et al., 2015), and miR-137 targeting c-Kit and tyrp2 (Jiang et al., 2016) have been reported to affect melanogenesis in mammals. In teleost fishes, overexpression of miR-206 and miR-430b was reported to inhibit melanogenesis and carotenoid levels by targeting mc1r and scarb1, respectively, in koi carp (Cyprinus carpio L.) (Dong et al., 2020; Tian et al., 2022). In rainbow trout (Oncorhynchus mykiss), miR-382 decreased melanogenesis by targeting dct, and miR-330 increased carotenoid accumulation by targeting bco2 (Wu et al., 2022; Wu et al., 2023b). These studies demonstrated that miRNAs are involved in the regulation of body color. However, the process of miRNA regulation is complicated because one miRNA can target multiple mRNAs, and one mRNA can be regulated by multiple miRNAs.
Plectropomus leopardus is a valuable coral reef fish that comes in a variety of colors including black, red and pink. As the red fish has high economic and ornamental value, its market price is significantly higher than that of black fish. However, blackening is becoming more and more common in intensive aquaculture. To explore the molecular mechanisms underlying body color variation in P. leopardus, comparative analyses between black and red skin adults were performed using multi-omics (Wang et al., 2015; Yang et al., 2020; Ruijuan et al., 2021; Ruijuan et al., 2022; Song et al., 2022b). In the process of cultivating P. leopardus seedlings, we found that the skin was transparent at 26 days post-hatching (dph). Then, the skin color changed to red with the production of a large number of erythrophores at 30 dph (Figure 1), which can be used as an ideal model for body color studies. To explore the molecular mechanisms of the transparent to red color transition in P. leopardus, transparent and red-colored skin was submitted to comparative miRNA-seq analysis. A miRNA-mRNA network was constructed based on candidate miRNAs and their putative target genes related to pigmentation, and KEGG enrichment analysis of the target genes of all DEMs was performed. The results revealed that miRNAs play a potential role in the red coloration of P. leopardus. This finding lay the foundation for an understanding of the molecular mechanisms of body color variation in P. leopardus.
A total of 150 P. leopardus were collected from the wild ponds of Hainan Dongfang Star Technology Co.,Ltd. at 26 dph (transparent, 71.27 ± 2.31 mg, 18.80 ± 0.13 mm) and 30 dph (red, 112.9 ± 3.11 mg, 21.71 ± 0.19 mm), respectively. Transparent skin (PT) and red skin (PR) were carefully collected with sharp tweezers and immediately transferred to liquid nitrogen. They were then stored at -80°C for RNA extraction. Each sample was prepared by mixing the skins of 50 larval P. leopardus in a bowl. Before sampling, the P. leopardus larvae were anesthetized with MS-222. All experiments were conducted in accordance with the guidelines and approval of the respective Animal Care and Use Committee of Guangdong Ocean University, China.
Total RNA from six skin samples was extracted using a TRIzol reagent kit (Invitrogen, USA) according to the manufacturer’s instructions. RNA purity and integrity were tested using the NanoDrop spectrophotometer (Thermo Scientific, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, USA), respectively.
For miRNA library construction, 18-30nt small RNAs (sRNAs) were enriched by 6% polyacrylamide gel electrophoresis (PAGE) from high-quality total RNA; 3’ and 5’ adaptors were added at the 3’ and 5’ ends, respectively. 140-160 bp fragments were enriched by PAGE after reverse transcription and PCR amplification. Subsequently, six miRNA libraries were prepared for sequencing by Gene Denovo Biotechnology Co. (Guangzhou, China) using an Illumina HiSeq X ten.
The clean tags were obtained by filtering the raw reads using the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) according to the following rules: 1) filter out reads without the 3’ adaptor and extract the sequence before the 3’ adaptor; 2) filter out reads with the 5’ adapter; 3) filter out low-quality reads with no insert fragments and insert fragment length < 18 nt; 4) filter out reads containing poly A. Then, the clean tags were aligned to the Genebank (http://www.ncbi.nlm.nih.gov) and Rfam (http://rfam.xfam.org/) databases to remove rRNA, scRNA, sonRNA, snRNA and tRNA. The clean tags were also blasted to the P. leopardus reference genome (PRJDB9154) (Yang et al., 2020) to remove the reads mapped in exons and introns. The final clean tags were then submitted to miRbase (Griffiths-Jones, 2006) to identify the existing miRNAs, and the unannotated tags were aligned to the reference genome. The novel miRNAs were identified based on the localization of the tags in the genome and the hairpin structure predicted by the miRDeep software (Friedländer et al., 2011).
Due to a lack of P. leopardus information in the miRbase database, the total miRNAs consisted of known and novel miRNAs. According to the miRNA expression in each sample, the miRNAs expression levels were normalized by tags per million (TPM=actual miRNA tags*106/total counts of cleans tags). edgeR software (http://www.r-project.org/) was used to identify DEMs according to the following threshold: |fold change| > 2 and a p-value < 0.05.
Based on the known and novel miRNA sequences, three software programs, i.e., TargetScan (http://www.targetscan.org), miRanda (http://www.microrna.org) and RNAhybird (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) were utilized to predict the target genes of the DEMs. The overlaps of these three software were programs served as the final result. Subsequently, all target genes of the DEMs were annotated by checking their Gene Ontology (GO) functions and Kyto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment. GO terms and KEGG pathways with a p-value < 0.05 were defined as significantly enriched.
The relationships between the DEMs and target genes were assessed by the Pearson correlation coefficient (PCC). Based on previous research reports, 53 target genes related to pigmentation were selected, and miRNAs closely related to these genes were selected as candidate miRNAs. Then, a miRNA-mRNA network between the candidate miRNAs and target genes related to pigmentation was constructed and visualized with Cytoscape (v3.10.0) software.
10 DEMs were used to validate the quality of the sequencing data. The high-quality RNA obtained above was used to synthesize first-stand miRNA, according to the instruction of the TransScript® miRNA First-Strand cDNA Synthesis SuperMix kit (Trans, AT351-01). The qPCR reaction was then performed on a 384-well plate using the Roche LightCycler 480 SystemII (Roche, Manheim, Germany). The 10 ul reaction volume consisted of 5ul Green qPCR SuperMix, 1ul miRNA, 0.5ul forward/reverse primer, and 3ul ddH2O. All of miRNA primers were designed using Primer Premier 6 software. The primer information is provided in Table S1. miR-23a was used as an internal control (Zhu et al., 2015). Three biological replicates were used for the PT and PR groups, respectively, and each biological replicate contained three technical replicates. The relative expression levels of the miRNAs were calculated using the 2-△△ct method (Livak and Schmittgen, 2001).
SPSS 27.0 and Graphpad Prism 9 software were used for data analysis and visualization, respectively. All parameters are presented as the mean ± SD. p-value < 0.05 was considered as a significant difference.
A total of 45.02 M and 38.18 M clean reads were obtained in the PT and PR groups, respectively, by filtering the preliminary raw reads. After filtering out low-quality reads from all samples, the proportion of high-quality reads exceeded 98.87%, and 44.31 M and 37.55 M clean tags in the PT and PR groups, respectively, were obtained through further quality control (Table 1). The length of the clean tags were mainly distributed in 22 nt (Figure 2). These results indicate that the quality of the sequencing results meets the required standards for next step of analysis.
After removing the tags of rRNA, scRNA, snoRNA, snRNA and tRNA by aligning the tags to Genebank and Rfam databases, the remaining tags were mapped to the P. leopardus reference genome. A total of 73,308,113 tags were mapped, with a mapping rate of around 89.55% (Table 2). Then, the tags positioned in the exons and introns were removed. In total, 981 miRNAs were identified, including 493 known and 488 novel miRNAs. The miRNA expression profiles are shown in Table S2.
Principal component analysis (PCA) was performed prior to the identification of the DEMs. The results showed good within-group repeatability and good discrimination between the groups (Figure 3A). After normalization of the miRNAs expression levels by TPM, a total of 106 miRNAs were identified as DEMs on the basis of a |fold change| > 2 and p-value < 0.05. Compared to the PT group, there were 43 up-regulated and 63 down-regulated miRNAs in the PR group, including 52 known and 54 novel miRNAs (Figure 3B). Analysis using TargetScan, miRanda, and RNAhybird software resulted in the prediction of a total of 22,433 target genes for the above 106 DEMs, giving rise to 255,643 miRNA-target pairs (Table S3).
18 candidate miRNAs were selected from the 106 DEMs based on the 53 target genes related to pigmentation. These 53 target genes include genes involved in carotenoid metabolism (scarb1, apod, fabp7, bco2 etc), pteridine biosynthesis (gch1, pts, spr), melanogenesis (tyr, tyrp1, dct etc) and chromatophore development (pax3, pax7, sox10 ect.). Based on the above 18 miRNAs and 53 target genes, 216 miRNA-mRNA pairs were constructed and selected for further analysis. The constructed miRNA-mRNA network is shown in Figure 4A.
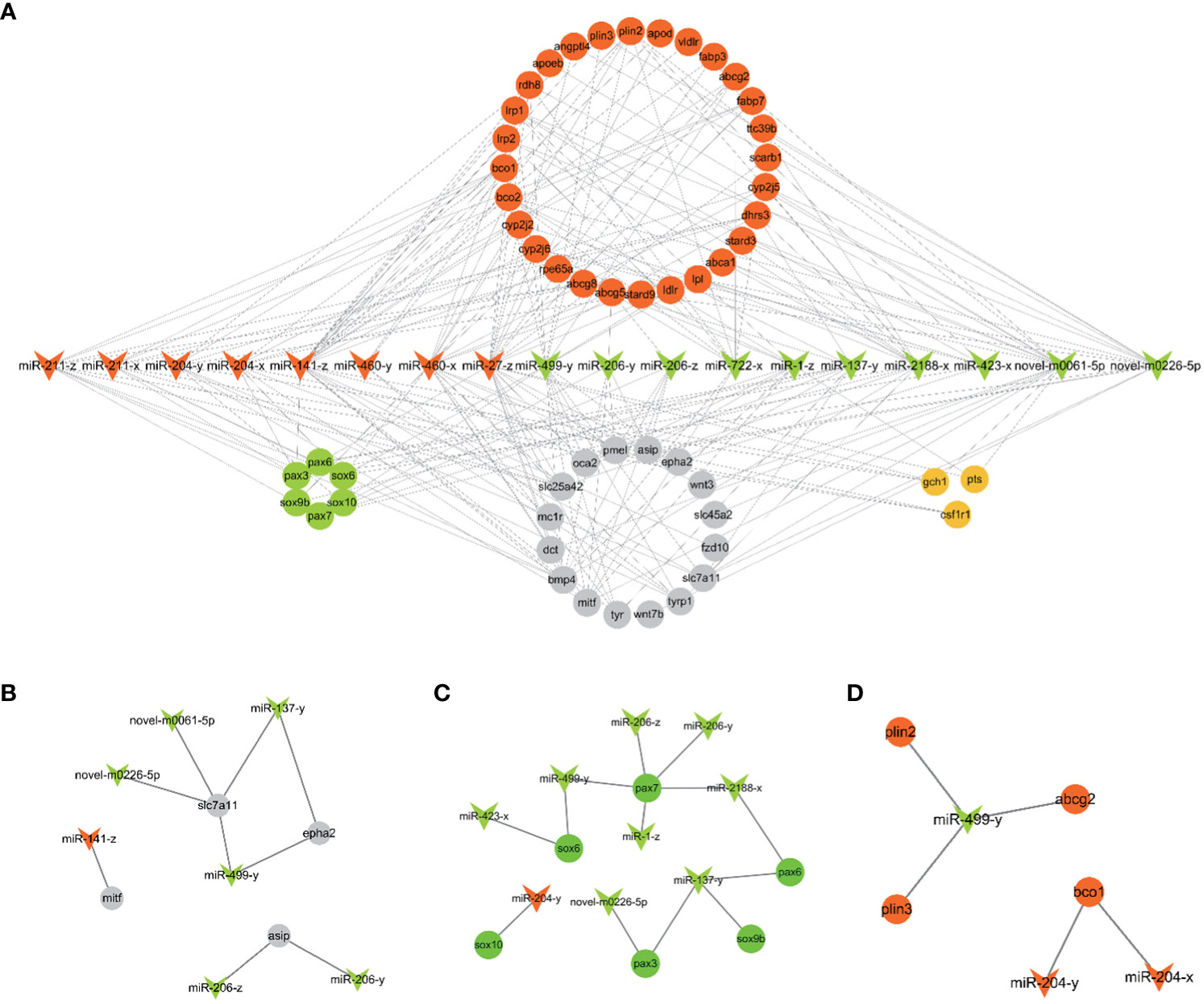
Figure 4 (A) The putative miRNA-mRNA network related to pigmentation in PT vs. PR. (B) The pairs of melanogenesis. (C) The network of chromatophore development. (D) The pairs of carotenoids metabolism. The arrowhead represents candidate miRNAs (red: up-regulated and green: down-regulated in PR vs. PT). The circle represents target genes of candidate miRNAs associated with pigmentation (red: carotenoids metabolism; orange: pteridine biosynthesis; grey: melanogenesis; green: chromatophores development).
To better understand the molecular functions of the target genes, all targets of the DEMs were evaluated for further GO and KEGG enrichment analysis. GO enrichment analysis showed that 193 GO terms were significantly enriched (p < 0.05), including 168, 15 and 10 in biological process, cellular component and molecular function, respectively. The most abundant GO terms in the three GO categories were cellular process (GO:0009987) and single-organism process (GO:0044699) in biological process; binding (GO:0005488) and catalytic activity (GO:0003824) in molecular function, and cell (GO:0005623), cell part (GO:0044464) and organelle (GO:0043226) in cellular component (Figure 5).
For the KEGG pathway analysis, a total of 352 pathways were enriched. 20 pathways related to pigmentation and immunity are presented in Figure 6, including pathways related to melanogenesis (MAPK, cAMP, and Wnt signaling pathway), pathways related to lipid metabolism (Bile secretion, Retinol metabolism and Cholesterol metabolism), and pathways related to the immune response (microRNAs in cancer, Pathways in cancer and Insulin resistance).
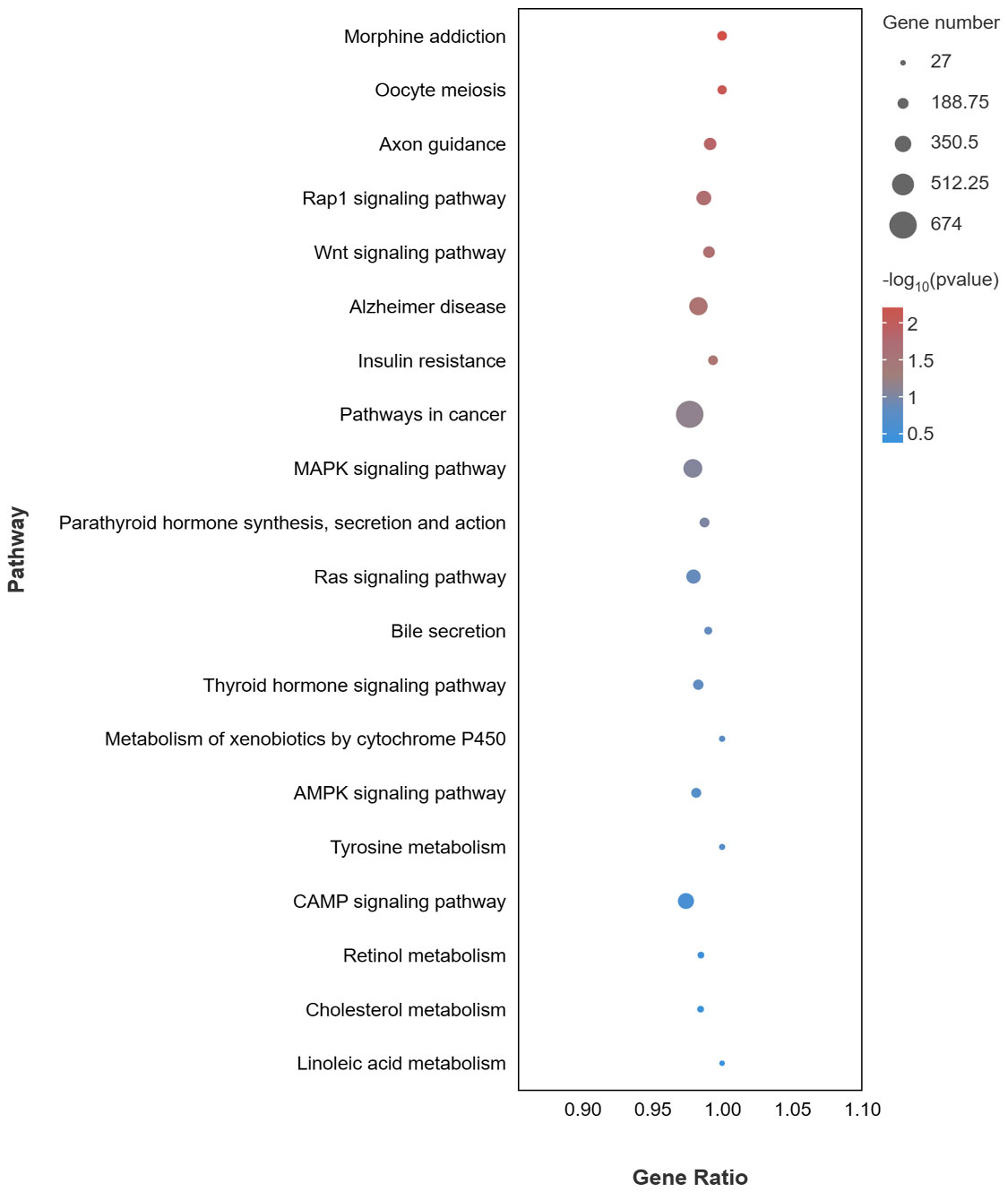
Figure 6 KEGG enrichment pathways of target genes of DEMs associated with pigmentation and immune of DEMs in PT vs. PR.
To validate the miRNA-seq data quality, 10 DEMs (miR-137-y, miR-206-y, miR-458-y, miR-551-x, miR-2188-x, miR-8159-x, miR-27-z, miR-204-x, miR-455-x, miR-460-x) were randomly selected for qPCR validation. The qPCR results were consistent with the miRNA-seq results, indicating the reliability of the sequencing data (Figure 7).
Fish possess high economic and ornamental value due to the bright red color of their body surface. An increasing number of studies have explored the molecular mechanisms of red color formation (Zhu et al., 2016; Ruijuan et al., 2022). At present, it is known that carotenoids play a crucial role in the production of red skin color in most fish, including Atlantic salmon, Malabar snapper, and Oujiang color common carp (Zorić, 2017; Du et al., 2021; Zhi Weng Josiah et al., 2022). In goldfish and guppies, red coloring is determined by pteridine pigment (Masada et al., 1990; Grether et al., 2001). Before that, largely increasing erythrophores is basis for the formation of red, which involves specification, migration and development of chromatophores (Kelsh et al., 2021). In addition, inhibition of melanophores and melanin production contributes to redness (Wu et al., 2023a). The main phenotype of P. leopardus comprises red and black coloration, and astaxanthin is the major carotenoids of red skin in P. leopardus (Maoka et al., 2017). In previous study, a series of genes (ldlr, bco2, angptl2, scd) related to carotenoid metabolism were identified by transcriptome and genome analysis based on red and black color skin in adults fish (Yang et al., 2020; Zhu et al., 2021; Xin et al., 2022). In this study, miRNA-seq of 26 dph (PT) and 30 dph (PR) skin was used to characterize the molecular mechanisms of early red color formation in P. leopardus. The miRNA-mRNA network based on candidate miRNAs and their putative target genes related to pigmentation, and the enrichment analysis of target genes of all DEMs, further revealed that the miRNAs involved in red formation mainly involves melanin synthesis (Figure 4B), chromatophore development (Figure 4C) and carotenoid metabolism (Figure 4D).
Body color is closely related to melanin synthesis. A large of studies have demonstrated that the inhibition of melanin synthesis results in lightening of black coloring, albinism and even redness (Dong et al., 2012; Jiang et al., 2022; Wang et al., 2022). Melanin synthesis is a complex process; upstream signaling pathways or transcription factors acting on mitf (a master regulator of melanin synthesis and melanophore development), promote or inhibit melanin synthesis by regulating the expression levels of downstream members of the tyr gene family (tyr, tyrp1 and dct) (Levy et al., 2006). The MAPK, Wnt and cAMP signaling pathways have been shown to be involved in melanin synthesis in teleost fish (Ruijuan et al., 2021; Song et al., 2022b). In this study, the MAPK, Wnt and cAPM signaling pathways were enriched, suggesting that miRNAs may be involved in melanin synthesis.
Several miRNAs involved in melanin synthesis were identified in this study, including miR-141-z, miR-206-z, miR-206-y, miR-27-z, miR-137-y, miR-204-x, miR-204-y, miR-211-x and miR-211-z. Among these, miR-141-3p expression was found to be significantly reduced in α-msh-stimulated B16-4A5 cells (mouse melanophore), and miR-141-3p overexpression significantly reduces MITF protein and tyrosinase activity by targeting Mitf, thus inhibiting melanin synthesis (Itoh et al., 2020). Furthermore, previous studies have demonstrated that miR-141 is more abundant in white alpaca skin than black alpaca skin (Tian et al., 2012), and higher expression levels have been observed in common carp with red skin as compared to those with white skin (Yan et al., 2013). In another study, P. leopardus skin cultured on different backgrounds showed significant differences (Feibiao et al., 2023). Moreover, miR-141-5p was significantly up-regulated in whole pink (WP) skin compared with pink with scattered black spots (PB) and pink with scattered red spots (PR) in red tilapia (Wang et al., 2018). In the present study, miR-141-z was significantly up-regulated in PR compared with PT, and this may result in the inhibition of melanin synthesis by targeting mitf.
Inhibition of miR-206 expression was found to significantly increased mc1r (which promotes the expression of mitf and downstream genes such as tyr and tyrp1 by increasing cAMP levels) mRNA and protein levels in koi carp (Dong et al., 2020). Although miR-206-z and miR-206-y were not predicted to have a target relationship with mc1r in this study, asip, as a target gene for miR-206-z and miR-206-y, may be associated with α-MSH, which competitively binds to mc1r, thereby reducing the downstream expression of mitf (Suzuki et al., 1997). The down-regulated miR-206-z and miR-206-y may inhibit melanin synthesis by promoting the expression of asip and inhibiting the expression of tyr and dct. miR-27a-3p functions as a melanin inhibitor in mammals. MiR-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting wnt3a (Zhao et al., 2015). Meanwhile, the expression level of miR-27-3p was found to be six times higher in white alpaca skin than in brown alpaca skin (Tian et al., 2012), and these differences have also been observed in the skin of P. leopardus cultured in different backgrounds (Feibiao et al., 2023). In this study, miR-27-z was up-regulated in the PR group compared to the PT group, suggesting that miR-27-z may play a role in inhibiting melanin synthesis. Previous studies have demonstrated that the overexpression of miR-137 in mice results in a lighter grey phenotype, and that miR-137 affects the expression of mitf and its downstream tyr gene family members (Dong et al., 2012). In addition, overexpression of miR-137 can inhibit tyrp2 (dct) expression and melanin synthesis by targeting c-KIT in mouse melanocytes (Jiang et al., 2016). However, in the present study, miR-137-y was down-regulated, suggesting it may play other roles. The specific regulatory mechanisms require further investigation.
miR-204 and miR-211 have been shown to inhibit the growth of melanoma cell lines (Galasso et al., 2018), and miR-211 inversely regulates tumor growth factor (TGF-β) and decreases the expression of tyr and tyrp1 (Dai et al., 2014). In this study, miR-204-x, miR-204-y, miR-211-x and miR-211-z were up-regulated in the PR group, suggesting that they may be involved in the inhibition of melanin synthesis. In addition, slc7a11 and epha2 were predicted to be the target genes of miR-499-y, miR-137-y, novel-m0061-5p and novel-m0226-5p. Slc7a11, which is required for pheomelanin formation in mammals, has been shown to be expressed at significantly higher levels in PR red tilapia than in PB and WP tilapia (Wang et al., 2019). epha2 is significantly up-regulated in the red skin of P. leopardus (Wu et al., 2023a), and may inhibit melanogenesis by blocking the MAPK and AKT signaling pathways in human lens epithelial (HLE) cells (Ma et al., 2017). In this study, epha2 was also enriched in several pathways, including Axon guidance, Rap1 signaling pathway and MAPK signaling pathway. These results suggest that miR-499-y, miR-137-y, novel-m0061-5p and novel-m0226-5p may inhibit eumelanin synthesis and contribute to the formation of red coloration by targeting epha2 and slc7a11.
Chromatophores originate from the neural crest. The early development of chromophores (xanthophores, melanophores, iridophores) during embryonic development has been well studied in model fish (Tsunogai et al., 2021); however, there are few research reports on erythrophores. It has been reported that xanthophores and erythrophores share a common progenitor (Huang et al., 2021), suggesting that erythrophore development, similar to xanthophores, may depend on pax and sox gene family members.
In our study, miR-206-z, miR-206-y, miR-499-y, miR-1-z, and miR-2188-x were predicted to target pax7; miR-137-y, novel-m0226-5p were predicted to target pax3; miR-137-y was predicted to target sox9b, and miR-204-y was predicted to target sox10. In zebrafish, pax7 and pax3 are required for xanthophore development, and pax7a/pax7b double mutants and pax3 knockdown resulted in complete depletion and reduction of xanthophores, respectively (Minchin and Hughes, 2008; Nord et al., 2016). In addition, pax7 play a role in inhibiting melanophores by regulating mitf to promote the differentiation of xanthophores lineage rather than melanophores. Mutations in pax7 resulted in an increase of melanophores in model fish (Motohiro et al., 2023). Sox10 is a key regulator of xanthophore specification. sox10a mutants and sox10a/sox10b double mutants exhibit a reduction and severe defect in the number of xanthophore, respectively (Tsunogai et al., 2021; Subkhankulova et al., 2023). In addition, sox9b has also been shown to contribute to the formation of pigment cells in madaka (Tsunogai et al., 2021). Although the above studies focused on embryogenesis, the expression levels of sox10 and pax7 have been found to be significantly higher in most red and yellow skin adult fish than in black skin in adult fish (Santos et al., 2016; Dick et al., 2018; Sušnik Bajec et al., 2022). Thus, these miRNAs may be involved in erythrophore development by targeting pax and sox gene family members.
Red, orange and yellow body colors are produced by carotenoids due to their unique conjugated double-bond system (Böhm, 2019). Fish are not able to synthesize carotenoids de novo, they have to be obtained from the diet. The process of carotenoid metabolism involves absorption and transport, binding and deposition and degradation (Toews et al., 2017; Toomey et al., 2022). Only a few studies have reported that miRNAs are involved in the regulation of carotenoid metabolism. For example, miR-430b was found to reduce the red skin pigmentation in koi carp by targeting scarb1 (Tian et al., 2022), and miR-330 overexpression was found to significantly reduces bco2 expression and promotes carotenoid accumulation in rainbow trout skin (Wu et al., 2023b).
The present study identified several miRNAs that may be involved in carotenoid metabolism, such as miR-204-x and miR-204-y targeting bco1 and miR-499-y targeting plin2, plin3 and abcg2. Two oxygenases, β, β-carotene 15,15’ dioxygenase (bco1) and β, β-carotene 9,10’ dioxygenase (bco2), are known to mediate the degradation of carotenoids. bco1l and bco1 have β-carotene catalytic activity in Atlantic salmon and zebrafish, respectively (Lampert et al., 2003; Helgeland et al., 2019). Loss of bco2 results in a deep carotenoid color in mammals (Våge and Boman, 2010; Thomas et al., 2023). The plin gene family may be related to carotenoid deposition. Mutation in piln6 resulted in a decrease in xanthophores carotenoids and carotenoid droplet dispersion in zebrafish (Granneman et al., 2017). Moreover, abcg2 has been reported to be involved in the proliferation of erythrophores in the cherry shrimp (Neocaridina denticulata sinensis) (Lu et al., 2022). Thus, miR-204-x, miR-204-y and miR-499-y may play a potential role in carotenoid metabolism and may promote redness.
In addition, target genes (such as ldlr, apoe, abca1, bco1, stard3 and abcg2/5/8) of DEMs related to carotenoid metabolism were enriched in bile secretion, retinol metabolism and cholesterol metabolism signaling pathways. Furthermore, members of the cytochrome P450 enzyme family, cyp2j19 and cyp2ae2, are closely related to the site of keto-carotenoid formation in birds and zebrafish, respectively (Toomey et al., 2022). In this study, cyp2j were enriched in pathways. This indicates that miRNAs may functions in keto-carotenoids formation in P. leopardus. However, the exact molecular mechanism of carotenoid metabolism remains unclear and requires further investigation.
In this study, comparative miRNA-seq analysis was performed on 26dph (PT) and 30dph (PR) skin in P. leopardus. A total of 981 miRNAs were identified, including 493 known miRNAs and 488 novel miRNAs. 18 out of 106 DEMs were selected as candidate miRNAs related to red color formation. The results suggested that miRNAs involved in red color formation are mainly involve the inhibition of melanin synthesis, chromatophore development and carotenoid metabolism. The finding of this study offer insight into the potential molecular mechanism underlying the role of miRNAs in the early formation of red skin color in P. leopardus.
The data presented in this study are deposited in the SRA repository, the accession number is PRJNA839546.
The animal studies were approved by Animal Care and Use Committee of Guangdong Ocean University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
XL: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft. MJ: Formal Analysis, Conceptualization, Funding acquisition, Visualization, Writing – review & editing, Validation. SW: Formal Analysis, Investigation, Writing – original draft, Data curation. KZ: Data curation, Formal Analysis, Investigation, Writing – original draft. YCH: Funding acquisition, Resources, Writing – original draft. KS: Funding acquisition, Resources, Writing – original draft. XD: Data curation, Formal Analysis, Investigation, Writing – original draft. YH: Resources, Writing – original draft. GS: Resources, Writing – original draft. CT: Formal Analysis, Visualization, Writing – original draft. HC: Formal Analysis, Visualization, Writing – original draft. GL: Project administration, Resources, Writing – original draft. CZ: Writing – review & editing, Funding acquisition, Project administration, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the National Natural Science Foundation of China (Nos. 32002368); the Zhan jiang city science and technology projects (2022A01015); Guangdong Basic and Applied Basic Research Foundation (2021A1515010430); the science and technology plan of Guangdong Province (2023B0202010016); the talent team tender grant of Zhanjiang marine equipment and biology (2021E05035); the program for scientific research start-up funds of Guangdong Ocean University (060302022101), and the cooperative project from Guangdong Provincial Key Laboratory of Aquatic Larvae Feed.
Authors YH and KS were employed by company Guangdong Yuequn Biotechnology Co., Ltd.,.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1321196/full#supplementary-material
Aspengren S., Sköld H. N., Wallin M. (2008). Different strategies for color change. Cell. Mol. Life Sci. 22 (9), 464–468. doi: 10.1016/j.tig.2006.06.010
Beirl A. J., Linbo T. H., Cobb M. J., Cooper C. D. (2013). oca2 Regulation of chromatophore differentiation and number is cell type specific in zebrafish. Pigment Cell Melanoma Res. 27 (2), 178–189. doi: 10.1111/pcmr.12205
Bizuayehu T. T., Babiak I. (2014). MicroRNA in teleost fish. Genome Biol. Evolution. 6 (8), 1911–1937. doi: 10.1093/gbe/evu151
Chintala S., Li W., Lamoreux M. L., Ito S., Wakamatsu K., Sviderskaya E. V., et al. (2005). Slc7a11 gene controls production of pheomelanin pigment and proliferation of cultured cells. Proc. Natl. Acad. Sci. United States America. 102 (31), 10964–10969. doi: 10.1073/pnas.0502856102
Dai X., Rao C., Li H., Chen Y., Fan L., Geng H., et al. (2014). Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-β receptor 2. Pigment Cell Melanoma Res. 28 (2), 217–222. doi: 10.1111/pcmr.12334
Dick C., Reznick D. N., Hayashi C. Y. (2018). Sex-biased expression between guppies varying in the presence of ornamental coloration. PeerJ 6, e5782. doi: 10.7717/peerj.5782
Dong Z., Luo M., Wang L., Yin H., Zhu W., Fu J. (2020). MicroRNA-206 regulation of skin pigmentation in koi carp (Cyprinus carpio L.). Front. Genet. 11. doi: 10.3389/fgene.2020.00047
Dong C., Wang H., Xue L., Dong Y., Yang L., Fan R. (2012). Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA 18 (9), 1679–1686. doi: 10.1261/rna.033977.112
Du J., Chen H., Mandal B. K., Wang J., Shi Z., Lu G., et al. (2021). HDL receptor/Scavenger receptor B1-Scarb1 and Scarb1-like mediate the carotenoid-based red coloration in fish. Aquaculture 545, 737208. doi: 10.1016/j.aquaculture.2021.737208
Feibiao S., Zihang Y., Liping S., Da Z., Huan L., Lei W., et al. (2023). Transcriptome analysis reveals candidate miRNAs involved in skin color differentiation of juvenile Plectropomus leopardus in response to different background colors. Comp. Biochem. Physiol. D: Genomics Proteomics. 48, 101141. doi: 10.1016/j.cbd.2023.101141
Figon F., Casas J. (2018). Morphological and physiological colour changes in the animal kingdom. eLS 1–11. doi: 10.1002/9780470015902.a0028065
Friedländer M. R., Mackowiak S. D., Li N., Chen W., Rajewsky N. (2011). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40 (1), 37–52. doi: 10.1093/nar/gkr688
Galasso M., Morrison C., Minotti L., Corrà F., Zerbinati C., Agnoletto C., et al. (2018). Loss of miR-204 expression is a key event in melanoma. Mol. Cancer. 17 (1), 71. doi: 10.1186/s12943-018-0819-8
Granneman J. G., Kimler V. A., Zhang H., Ye X., Luo X., Postlethwait J. H., et al. (2017). Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. eLife 6, e21771. doi: 10.7554/eLife.21771
Grether G. F., Hudon J., Endler J. A. (2001). Carotenoid scarcity, synthetic pteridine pigments and the evolution of sexual coloration in guppies (Poecilia reticulata). Proc. R. Soc. B: Biol. Sci. 268 (1473), 1245–1253. doi: 10.1098/rspb.2001.1624
Griffiths-Jones S. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34 (Database issue), D140–D144. doi: 10.1093/nar/gkj112
Helgeland H., Sodeland M., Zoric N., Torgersen J. S., Grammes F., Von Lintig J., et al. (2019). Genomic and functional gene studies suggest a key role of beta-carotene oxygenase 1 like (bco1l) gene in salmon flesh color. Sci. Rep. 9 (1), 20061. doi: 10.1038/s41598-019-56438-3
Hill G. E. (1991). Plumage coloration is a sexually selected indicator of male quality. Nature 350, 337–339. doi: 10.1038/350337a0
Huang D., Lewis V. M., Foster T. N., Toomey M. B., Corbo J. C., Parichy D. M. (2021). Development and genetics of red coloration in the zebrafish relative Danio albolineatus. eLife 10, e70253. doi: 10.7554/eLife.70253
Hwang H. W., Mendell J. T. (2007). MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer. 94 (6), 776–780. doi: 10.1038/sj.bjc.6603023
Inouye C. Y. (2007). Bird coloration. Volume I: mechanisms and measurements. Condor: Ornithol. Appl. 109 (2), 482–483. doi: 10.1093/condor/109.2.482
Itoh T., Fukatani K., Nakashima A., Suzuki K. (2020). MicroRNA-141-3p and microRNA-200a-3p regulate α-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting microphthalmia-associated transcription factor. Sci. Rep. 10 (1), 2149. doi: 10.1038/s41598-020-58911-w
Jiang Y., Li B., Yu M., Chang S., Li S., Xu J. (2022). Genome-wide association study and gene editing reveals the causal gene responsible for abnormal red skin color in Yellow River carp. Aquaculture 560, 738530. doi: 10.1016/j.aquaculture.2022.738530
Jiang S., Yu X., Dong C. (2016). MiR-137 affects melanin synthesis in mouse melanocyte by repressing the expression of c-Kit and Tyrp2 in SCF/c-Kit signaling pathway. Biosci. Biotechnol. Biochem. 80 (11), 2115–2121. doi: 10.1080/09168451.2016.1200455
Kelsh R. N. (2004). Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 17 (4), 326–336. doi: 10.1111/j.1600-0749.2004.00174.x
Kelsh R. N., Camargo Sosa K., Farjami S., Makeev V., Dawes J. H. P., Rocco A. (2021). Cyclical fate restriction: a new view of neural crest cell fate specification. Development 148 (22), dev176057. doi: 10.1242/dev.176057
Lafountain A. M., Prum R. O., Frank H. A. (2015). Diversity, physiology, and evolution of avian plumage carotenoids and the role of carotenoid-protein interactions in plumage color appearance. Arch. Biochem. Biophys. 572, 201–212. doi: 10.1016/j.abb.2015.01.016
Lampert J. M., Holzschuh J., Hessel S., Driever W., Vogt K., Von Lintig J. (2003). Provitamin A conversion to retinal via the beta,beta-carotene-15,15'-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development 130 (10), 2173–2186. doi: 10.1242/dev.00437
Leclercq E., Taylor J. F., Migaud H. (2009). Morphological skin colour changes in teleosts. Fish Fish. 11, 159–193. doi: 10.1111/j.1467-2979.2009.00346.x
Levy C., Khaled M., Fisher D. E. (2006). MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12 (9), 406–414. doi: 10.1016/j.molmed.2006.07.008
Lingyu W., Xiaoyan Z., Haishen W., Xin Q., Donglei S., Xueqi L., et al. (2023). Integrated miRNA and mRNA analysis in gills of spotted sea bass reveals novel insights into the molecular regulatory mechanism of salinity acclimation. Aquaculture 575, 739778. doi: 10.1016/j.aquaculture.2023.739778
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li-Yan Z. (2013). Observation and analysis of growth dynamics in Oujiang color common carp with different pigmentation types. J Shanghai Fish Univ. 22:341–348.
Lu X., Zhang L., Wang G., Huang S. (2022). Functional analysis of ABCG2 gene in pigment transport of Neocaridina denticulata sinensis. Gene 844, 146810. doi: 10.1016/j.gene.2022.146810
Ma X., Ma Z., Jiao X., Hejtmancik J. F. (2017). Functional non-coding polymorphism in an EPHA2 promoter PAX2 binding site modifies expression and alters the MAPK and AKT pathways. Sci. Rep. 7 (1), 9992. doi: 10.1038/s41598-017-10117-3
Maoka T., Sato W., Nagai H., Takahashi T. (2017). Carotenoids of red, brown, and black specimens of plectropomus leopardus, the coral trout (Suziara in Japanese). J. Oleo Sci. 66 (6), 579–584. doi: 10.5650/jos.ess16179
Martín-Gómez L., Villalba A., Kerkhoven R. H., Abollo E. (2014). Role of microRNAs in the immunity process of the flat oyster Ostrea edulis against bonamiosis. Infect. Genet. Evolution. 27, 40–50. doi: 10.1016/j.meegid.2014.06.026
Masada M., Matsumoto J., Akino M. (1990). Biosynthetic pathways of pteridines and their association with phenotypic expression in vitro in normal and neoplastic pigment cells from goldfish. Pigment Cell Res. 3 (2), 61–70. doi: 10.1111/j.1600-0749.1990.tb00324.x
Matthews S. J., Ross N. W., Lall S. P., Gill T. A. (2006). Astaxanthin binding protein in Atlantic salmon. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 144 (2), 206–214. doi: 10.1016/j.cbpb.2006.02.007
McGowan K. A., Barsh G. S. (2016). How the zebrafish got its stripes. eLife 5, e14239. doi: 10.7554/eLife.14239
Minchin J. E. N., Hughes S. M. (2008). Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 317 (2), 508–522. doi: 10.1016/j.ydbio.2008.02.058
Motohiro M., Hiroyuki T., Akiko S., Tetsuaki K., Ikuko W., Hikaru K., et al. (2023). Pax3 and Pax7 function in combination with Mitf to generate melanophores and xanthophores in medaka and zebrafish. bioRxiv - Dev. Biol. 150 (19), dev202114. doi: 10.1242/dev.202114
Nilsen T. W. (2007). Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 23 (5), 243–249. doi: 10.1016/j.tig.2007.02.011
Ninwichian P., Phuwan N., Limlek P. (2022). Effects of tank color on the growth, survival rate, stress response, and skin color of juvenile hybrid catfish (Clarias macrocephalus × Clarias gariepinus). Aquaculture 554, 738129. doi: 10.1016/j.aquaculture.2022.738129
Nord H., Dennhag N., Muck J., Von Hofsten J. (2016). Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol. Biol. Cell. 27 (11), 1853–1862. doi: 10.1091/mbc.E15-12-0821
Parker R. S. (1996). Absorption, metabolism, and transport of carotenoids. FASEB J. 10 (5), 542–551. doi: 10.1096/fasebj.10.5.8621054
Ruijuan H., Xiaowen Z., Changxu T., Chunhua Z., Guangli L. (2022). Analysis of body color formation of leopard coral grouper Plectropomus leopardus. Front. Mar. Sci. 48, 101138. doi: 10.1016/j.cbd.2023.101138
Ruijuan H., Xiaowen Z., Changxu T., Mouyan J., Yang H., Chunhua Z. (2021). Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus). Aquaculture 548, 737575. doi: 10.1016/j.aquaculture.2021.737575
Santos M. E., Baldo L., Gu L., Boileau N., Musilova Z., Salzburger W. (2016). Comparative transcriptomics of anal fin pigmentation patterns in cichlid fishes. BMC Genomics 17 (1), 712. doi: 10.1186/s12864-016-3046-y
Schmeisser J., Verlhac-Trichet V., Madaro A., Lall S. P., Torrissen O., Olsen R. E. (2021). Molecular mechanism involved in carotenoid metabolism in post-smolt atlantic salmon: astaxanthin metabolism during flesh pigmentation and its antioxidant properties. Mar. Biotechnol. 23 (4), 653–670. doi: 10.1007/s10126-021-10055-2
Song F., Shi L., Yao F., Gu Y., Zheng D., Zhang W., et al. (2022a). The effect of background color on skin color variation of juvenile plectropomus leopardus. Animals 12 (23), 3349. doi: 10.3390/ani12233349
Song F., Wang L., Yang Z., Shi L., Zheng D., Zhang K., et al. (2022b). Transcriptome analysis reveals the complex regulatory pathway of background color in juvenile plectropomus leopardus skin color variation. Int. J. Mol. Sci. 23 (19), 11186. doi: 10.3390/ijms231911186
Stevens M., Ruxton G. D. (2011). Linking the evolution and form of warning coloration in nature. Proc. R. Soc. B: Biol. Sci. 279 (1728), 417–426. doi: 10.1098/rspb.2011.1932
Stuart-Fox D., Moussalli A. (2009). Camouflage, communication and thermoregulation: lessons from colour changing organisms. Philos. Trans. R. Soc. B: Biol. Sci. 364 (1516), 463–470. doi: 10.1098/rstb.2008.0254
Subkhankulova T., Camargo Sosa K., Uroshlev L. A., Nikaido M., Shriever N., Kasianov A. S., et al. (2023). Zebrafish pigment cells develop directly from persistent highly multipotent progenitors. Nat. Commun. 14 (1), 1258. doi: 10.1038/s41467-023-36876-4
Sušnik Bajec S., Djurdjevič I., Linares Andújar C., Kreft M. E. (2022). Genetic and correlative light and electron microscopy evidence for the unique differentiation pathway of erythrophores in brown trout skin. Sci. Rep. 12 (1), 1015. doi: 10.1038/s41598-022-04799-7
Suzuki I., Tada A., Ollmann M. M., Barsh G. S., Im S., Lamoreux M. L., et al. (1997). Agouti signaling protein inhibits melanogenesis and the response of human melanocytes to alpha-melanotropin. J. Invest. Dermatol. 108 (6), 838–842. doi: 10.1111/1523-1747.ep12292572
Thomas L. D., Ramkumar S., Golczak M., Von Lintig J. (2023). Genetic deletion of Bco2 and Isx establishes a golden mouse model for carotenoid research. Mol. Metab. 73, 101742. doi: 10.1016/j.molmet.2023.101742
Tian X., Jiang J., Fan R., Wang H., Meng X., He X., et al. (2012). Identification and characterization of microRNAs in white and brown alpaca skin. BMC Genomics 13, 555. doi: 10.1186/1471-2164-13-555
Tian X., Peng N.-N., Ma X., Wu L., Shi X., Liu H.-F., et al. (2022). microRNA-430b targets scavenger receptor class B member 1 (scarb1) and inhibits coloration and carotenoid synthesis in koi carp (Cyprinus carpio L.). Aquaculture 546, 737334. doi: 10.1016/j.aquaculture.2021.737334
Toews D. P. L., Hofmeister N. R., Taylor S. A. (2017). The evolution and genetics of carotenoid processing in animals. Trends Genet. 33 (3), 171–182. doi: 10.1016/j.tig.2017.01.002
Toomey M. B., Marques C. I., Araújo P. M., Huang D., Zhong S., Liu Y., et al. (2022). A mechanism for red coloration in vertebrates. Curr. Biol. 32 (19), 4201–4214.e12. doi: 10.1016/j.cub.2022.08.013
Tsunogai Y., Miyadai M., Nagao Y., Sugiwaka K., Kelsh R. N., Hibi M., et al. (2021). Contribution of sox9b to pigment cell formation in medaka fish. Develop. Growth Different. 63 (9), 516–522. doi: 10.1111/dgd.12760
Våge D. I., Boman I. A. (2010). A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 11, 10. doi: 10.1186/1471-2156-11-10
Wade N. M., Gabaudan J., Glencross B. D. (2015). A review of carotenoid utilisation and function in crustacean aquaculture. Rev. Aquacult. 9, 141–156. doi: 10.1111/raq.12109
Wang L.-M., Bu H.-Y., Song F.-B., Zhu W.-B., Fu J.-J., Dong Z.-J. (2019). Characterization and functional analysis of slc7a11 gene, involved in skin color differentiation in the red tilapia. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 236, 110529. doi: 10.1016/j.cbpa.2019.110529
Wang C., Kocher T. D., Wu J., Li P., Liang G., Lu B., et al. (2022). Knockout of microphthalmia-associated transcription factor (mitf) confers a red and yellow tilapia with few pigmented melanophores. Aquaculture 565, 739151. doi: 10.1016/j.aquaculture.2022.739151
Wang L.-M., Luo M.-K., Yin H.-R., Zhu W.-B., Fu J.-J., Dong Z.-J. (2020). Effects of background adaptation on the skin color of Malaysian red tilapia. Aquaculture 521, 735061. doi: 10.1016/j.aquaculture.2020.735061
Wang L., Yu C., Guo L., Lin H., Meng Z. (2015). In silico comparative transcriptome analysis of two color morphs of the common coral trout (Plectropomus leopardus). PloS One 10 (12), e0145868. doi: 10.1371/journal.pone.0145868
Wang P., Zhao Y., Fan R., Chen T., Dong C. (2016). MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting sox5 in mouse skin melanocytes. Int. J. Mol. Sci. 17 (7), 959. doi: 10.3390/ijms17070959
Wang L., Zhu W., Dong Z., Song F., Dong J., Fu J. (2018). Comparative microRNA-seq Analysis Depicts Candidate miRNAs Involved in Skin Color Differentiation in Red Tilapia. Int. J. Mol. Sci. 19 (4), 1209. doi: 10.3390/ijms19041209
Wu H.-Y., Chen K.-S., Huang Y.-S., Hsieh H.-Y., Tsai H. (2023a). Comparative transcriptome analysis of skin color-associated genes in leopard coral grouper (Plectropomus leopardus). BMC Genomics 24 (1), 5. doi: 10.1186/s12864-022-09091-6
Wu S., Huang J., Li Y., Liu Z., Zhao L. (2022). MiR-382 Functions on the Regulation of Melanogenesis via Targeting dct in Rainbow Trout (Oncorhynchus mykiss). Mar. Biotechnol. 24 (4), 776–787. doi: 10.1007/s10126-022-10143-x
Wu S., Zhao L., Huang J., Li Y., Liu Z., Zhang D. (2023b). miR-330 targeting BCO2 is involved in carotenoid metabolism to regulate skin pigmentation in rainbow trout (Oncorhynchus mykiss). BMC Genomics 24 (1), 124. doi: 10.1186/s12864-023-09173-z
Xin W., Haizhan T., Mengling Z., Min Y., Jie H., Junchi L., et al. (2022). Genome-wide association study of red skin color in leopard coral grouper (Plectropomus leopardus) based on genome resequencing. Aquaculture 563, 739014. doi: 10.1016/j.aquaculture.2022.739014
Yamanaka H., Kondo S. (2014). In vitro analysis suggests that difference in cell movement during direct interaction can generate various pigment patterns in vivo. Proc. Natl. Acad. Sci. United States America. 111 (5), 1867–1872. doi: 10.1073/pnas.1315416111
Yan B., Liu B., Zhu C.-D., Li K.-L., Yue L.-J., Zhao J.-L., et al. (2013). microRNA regulation of skin pigmentation in fish. J. Cell Sci. 126 (Pt 15), 3401–3408. doi: 10.1242/jcs.125831
Yang Y., Null N., Wu L.-N., Chen J.-F., Wu X., Xia J.-H., et al. (2020). Whole-genome sequencing of leopard coral grouper (Plectropomus leopardus) and exploration of regulation mechanism of skin color and adaptive evolution. Zool. Res. 41 (3), 328–340. doi: 10.24272/j.issn.2095-8137.2020.038
Zhang J., Tian C., Zhu K., Liu Y., Zhao C., Jiang M., et al. (2023). Effects of natural and synthetic astaxanthin on growth, body color, and transcriptome and metabolome profiles in the leopard coral grouper (Plectropomus leopardus). Animals 13 (7), 1252. doi: 10.3390/ani13071252
Zhao N., Ge X., Jiang K., Huang J., Wei K., Sun C., et al. (2022). Ultrastructure and regulation of color change in blue spots of leopard coral trout Plectropomus leopardus. Front. Endocrinol. 13. doi: 10.3389/fendo.2022.984081
Zhao Y., Wang P., Meng J., Ji Y., Xu D., Chen T., et al. (2015). MicroRNA-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting wnt3a. Int. J. Mol. Sci. 16 (5), 10921–10933. doi: 10.3390/ijms160510921
Zhi Weng Josiah P., Xueyan S., Joseph Angelo U., Celestine T., Shi Wei Gavin C., Jose A. D. (2022). Comparative transcriptome analysis reveals factors involved in the influence of dietary astaxanthin on body colouration of Malabar Snapper (Lutjanus malabaricus). Aquaculture 562, 738874. doi: 10.1016/j.aquaculture.2022.738874
Zhu X., Hao R., Tian C., Zhang J., Zhu C., Li G. (2021). Integrative transcriptomics and metabolomics analysis of body color formation in the leopard coral grouper (Plectropomus leopardus). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.726102
Zhu X., Li Y.-L., Chen D.-X., Wu P., Yi T., Chen T., et al. (2015). Selection of reference genes for microRNA quantitative expression analysis in Chinese perch, Siniperca chuatsi. Int. J. Mol. Sci. 16 (4), 8310–8323. doi: 10.3390/ijms16048310
Zhu W., Wang L., Dong Z., Chen X., Song F., Liu N. (2016). Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in red tilapia. Sci. Rep. 6, 31347. doi: 10.1038/srep31347
Keywords: miRNAs, chromatophore development, carotenoid metabolism, red color, Plectropomus leopardus
Citation: Liu X, Jiang M, Wen S, Zhang K, Hong Y, Sun K, Deng X, Huang Y, Shi G, Tian C, Chen H, Li G and Zhu C (2023) Comparative miRNA-seq analysis revealed molecular mechanisms of red color formation in the early developmental stages of Plectropomus leopardus. Front. Mar. Sci. 10:1321196. doi: 10.3389/fmars.2023.1321196
Received: 13 October 2023; Accepted: 21 November 2023;
Published: 06 December 2023.
Edited by:
Bruno Cavalheiro Araújo, University of Mogi das Cruzes, BrazilReviewed by:
Liang Guo, Hunan Normal University, ChinaCopyright © 2023 Liu, Jiang, Wen, Zhang, Hong, Sun, Deng, Huang, Shi, Tian, Chen, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunhua Zhu, Y2h6NDE2QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.