- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 3College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 4Laboratory of Marine Protozoan Biodiversity and Evolution, Marine College, Shandong University, Weihai, China
- 5College of Fisheries, Ocean University of China, Qingdao, China
Ciliates of the genus Pleuronema are a speciose and ubiquitous group. Recent studies suggest that there may be a sizable amount of undiscovered species diversity. In the present study, two new Pleuronema species have been isolated from the subtropical coastal waters of China and characterized using morphological and taxonomical methods: Pleuronema pulchra n. sp. is characterized by a body size of 75–90 × 25–40 µm in vivo, 32–48 somatic kineties, four to seven preoral kineties, somatic kinety 1 composed of about 80 kinetids, and membranelle 2a with a single-rowed mid-portion and hook-like posterior portion. Pleuronema warreni n. sp. is defined by a body size of 55–80 × 25–45 µm in vivo, 35–42 somatic kineties, three to six preoral kineties, somatic kinety 1 with approximately 75 kinetids, and a mid-portion of membranelle 2a that is single-rowed and extremely long (occupying > 75% length of M2a). The phylogenetic analysis of the small subunit ribosomal RNA genes of Pleuronema members, including those of the two novel species, shows that the genus Pleuronema is a polyphyletic group. Both new species form a cluster with Pleuronema binucleatum KT033424, P. elegans KF840518, and “P. coronatum” JX310014 (identification to be verified). Additionally, we provide an illustrated key for 20 “coronatum-type” Pleuronema species, including the two new ones.
1 Introduction
Scuticociliates are a speciose group that can live freely in aquatic and terrestrial environments or as parasites or endocommensal forms in animals. Usually, scuticociliates are characterized by a relatively small body size, one or more caudal cilia, and a complex three-membraned oral apparatus. Their great similarity and overlapping of features in vivo often challenge the identification of species (Song, 2000; Lynn, 2008; Fan et al., 2011; Gao et al., 2013; Whang et al., 2013; Hao et al., 2022). The genus Pleuronema is characterized by an expansive oral apparatus and a sail-like paroral membrane; it is a common and cosmopolitan genus of scuticociliates classified within the order Pleuronematida, subclass Scuticociliatia (Stock et al., 2012; Kalinowska, 2013; Azovsky and Mazei, 2018). Since Dujardin (1841) first established the genus Pleuronema, 37 nominal species have been described. However, only in the last 15 years 13 new species have been reported, which indicates that the species diversity of Pleuronema is underestimated, and further studies are thus still required (Wang et al., 2008a; Wang et al., 2008b; Wang et al., 2009; Pan et al., 2015; Pan X. et al., 2015; Pan et al., 2016; Liu et al., 2022; Zhang et al., 2023). Moreover, the results of phylogenetic analyses based on the nuclear and mitochondrial data have demonstrated that Pleuronema is polyphyletic (Yi et al., 2010; Gao et al., 2013; Antipa et al., 2016; Gao et al., 2017; Antipa et al., 2020). However, Schizocalyptra is nested in the Pleuronema with relatively low support in the studies mentioned above, which implies that more morphological and molecular data are needed for further analysis.
The taxonomy of Pleuronema currently involves some unresolved issues, i.e.: (i) only approximately two-thirds of Pleuronema species have detailed descriptions or reliable illustrations depicting their ciliary patterns after silver staining; other species, especially those reported in the last century, lack distinct ciliary pattern information (Dujardin, 1841; Roux, 1901; Kahl, 1926; Kahl, 1931; Dragesco, 1960; Small, 1964; Dragesco, 1968; Pätsch, 1974; Foissner et al., 1994); (ii) approximately half of the nominal species with a detailed ciliary pattern description lack information about their small subunit ribosomal RNA (SSU rRNA) gene. Moreover, many sequences in GenBank have no related morphological description, and thus their identification needs to be confirmed (Yi et al., 2009; Gao et al., 2013). Consequently, further studies to increase the taxon sampling within the genus Pleuronema, using both morphological and molecular methods, are needed to expand our knowledge of this group.
In this work, we present two new Pleuronema species discovered in brackish waters in Ningbo, eastern China. Both species, named Pleuronema pulchra n. sp. and Pleuronema warreni n. sp., were investigated using a combination of morphological and molecular methods. Additionally, we provide a key to the identification of 20 Pleuronema species with the posterior end of membranelle 2a hook-like (i.e., “coronatum-type”).
2 Materials and methods
2.1 Sample collection and identification
Both species were collected from the subtropical brackish waters of the East China Sea (Ningbo, China) (Figures 1A, B). Pleuronema pulchra n. sp. was isolated from a sandy beach (29°46′7.95″N, 121°55′21.81″E) on 6 April 2022, with a water temperature of approximately 15°C and a salinity of 14.3 PSU (practical salinity units) (Figure 1C). Holes with a depth of roughly 10 cm were dug in the sand during the ebb tide (Figure 1C). Once the water permeated into the holes, it was mixed with the sand at the bottom of the holes, and 300 ml of water samples mixed with sand were collected and then transferred into a 400 ml sampling bottle. Pleuronema warreni n. sp. was collected on 27 May 2022 from a brackish lake (29°44′1.22″N, 121°53′47.34″E), with a water temperature of approximately 26.5°C and a salinity of 15.1 PSU (Figure 1D). Approximately 300 ml of water sample, including wilted leaves and sediment, was placed into a 400 ml sampling bottle using a 30 ml plastic dropper. In both cases, samples were transported to the laboratory and kept at room temperature (ca. 25°C). For each sample, raw cultures of the targeted species were established in 90-mm Petri dishes, and rice grains were added to promote the growth of bacteria as a food source for ciliates. All cultured ciliates were kept in the laboratory for approximately one week, and sufficient individuals were isolated for the detailed morphological description of the two species (Zhao et al., 2022).
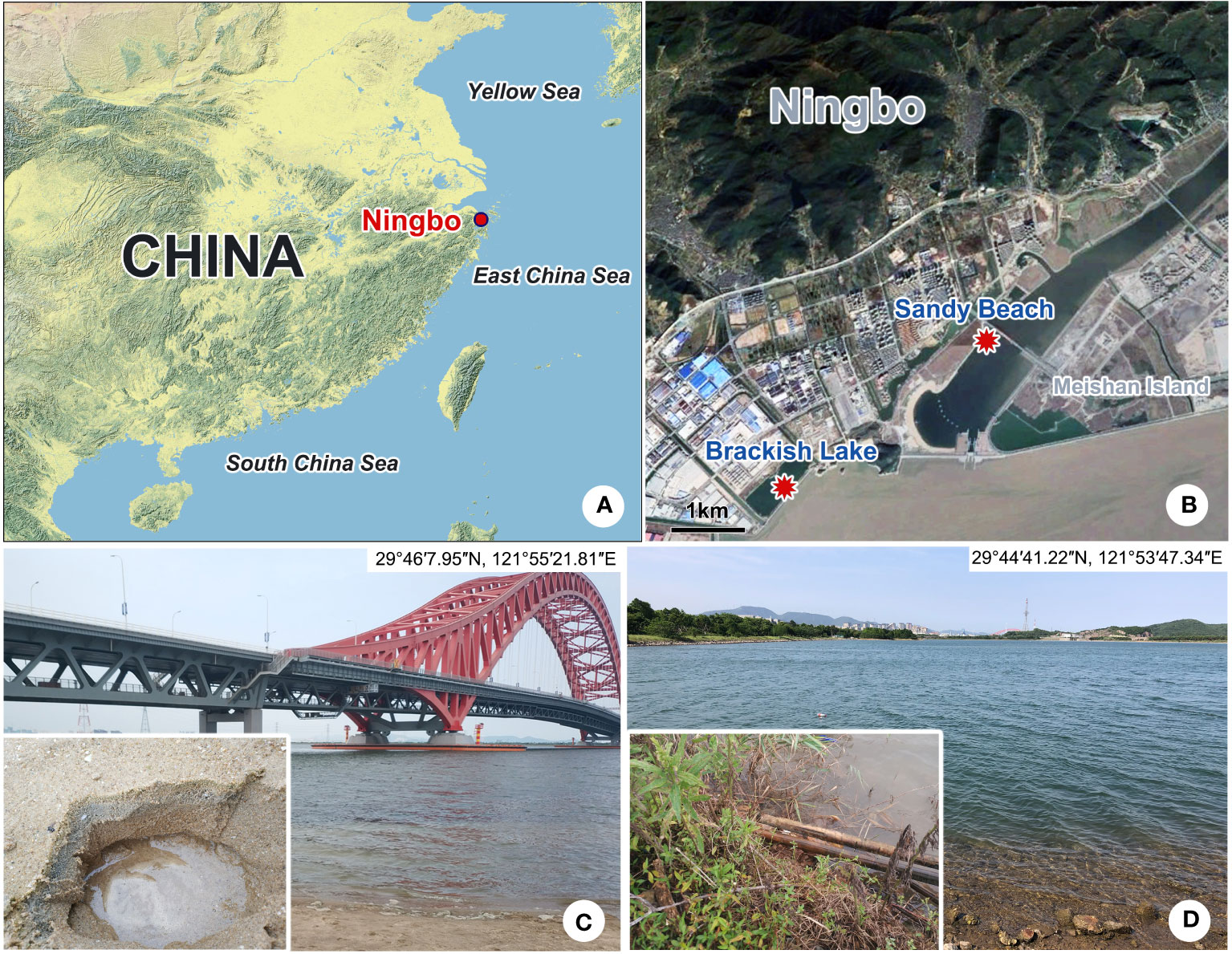
Figure 1 Sampling locations and habitats (A–D). (A) Map of China; the red dot shows the location of Ningbo. (B) Geographic location of Meishan Island and the sampling sites (red dots). (C) Sandy beach near the Meishan Bay Wetland, where Pleuronema pulchra n. sp. was collected. (D) Brackish lake, where Pleuronema warreni n. sp. was collected.
Living cells were observed using bright-field and differential interference contrast (DIC) microscopy (Leica DM2500, Germany) at 400–1000× magnification. The protargol impregnation method by Wilbert (1975) was used to display the ciliature and nuclear pattern. Counts, measurements, and drawings of living specimens were performed at 1000× magnification. Drawings of silver-stained cells of each species were made using Paint Tool SAI Ver. 2 and Adobe Photoshop. Terminology follows Zhang et al. (2023), and systematics follows Lynn (2008) and Gao et al. (2016).
2.2 DNA extraction, PCR amplification, and sequencing
Eight to ten cells for each species were isolated from the raw cultures using a micropipette under the stereomicroscope. The cells were washed three to five times using filtered in situ water (0.22 µm pore size membrane, Millipore, United States) and then divided into five Eppendorf tubes (Axygen, USA), sequentially containing one, two, or multiple individuals, and the first three tubes were used for PCR. The genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The SSU rRNA gene was amplified using the primers 18S-F (5’-AAC CTG GTT GAT CCT GCC AGT-3’), 18S-R (5’-TGA TCC TTC TGC AGG TTC ACC TAC-3’), and 82F (5’-GAA ACT GCG AAT GGC TC-3’) (Medlin et al., 1988; Jerome et al., 1996). To minimize the possibility of substitution errors originating from PCR, Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs, USA) was used as DNA polymerase.
The PCR conditions for the amplification of SSU rRNA gene sequences were as follows: initial denaturation at 98°C for 30 s, followed by 18 cycles of amplification (98°C, 10 s; 69–51°C touch-down, 30 s; 72°C, 60 s), another 18 cycles of amplification (98°C, 10 s; 51°C, 30 s; 72°C, 60 s), and a final elongation step at 72°C for 5 min (Jiang et al., 2021; Chi et al., 2022). PCR products were sequenced by Tsingke Biological Technology Company (Hangzhou, China), and the sequences from three tubes for each species were consistent.
2.3 Phylogenetic analyses
The SSU rRNA gene dataset for phylogenetic analyses consisted of 74 sequences, including P. pulchra n. sp., P. warreni n. sp., and 72 sequences downloaded from the National Center for Biotechnology Information (NCBI) database. Four sequences from the order Colpodida, namely Colpoda magna EU039896, Colpoda minima EU039897, Bresslaua vorax MN160328, and Bresslauides discoideus OM278536, were set as the outgroup taxa. All sequences were aligned using the GUIDANCE2 Server (http://guidance.tau.ac.il) with default parameters (Penn et al., 2010), and both ends were manually trimmed in BioEdit 7.0.5.3 (Hall, 1999), following Ye et al. (2022). Maximum likelihood (ML) analysis with 1000 bootstrap replicates was performed on the CIPRES Science Gateway server using RAxML-HPC2 on XSEDE v8.2.12 (Stamatakis, 2014) with the GTRGAMMA model. Bayesian inference (BI) analysis was performed using MrBayes on XSEDE v3.2.6 (Ronquist and Huelsenbeck, 2003). The GTR + I + G model selected by MrModeltest v2.2 was applied in the BI analysis (Nylander, 2004). The chain was run for 106 generations and sampled every 100th generation, and the first 2500 trees were discarded as burn-in. Trees were visualized using MEGA v.7.0 and TreeView v.1.6.6 (Page, 1996; Kumar et al., 2016).
3 Results
Subclass: Scuticociliatia Small, 1967.
Order: Pleuronematida Fauré-Fremiet in Corliss, 1956.
Family: Pleuronematidae Kent, 1881.
Genus: Pleuronema Dujardin, 1841.
3.1 Pleuronema pulchra n. sp.
3.1.1 Diagnosis
Cell size approximately 75–90 µm × 25–40 µm in vivo. Right ventrolateral side basically straight, with four to seven preoral kineties, 32–48 somatic kineties. Usually one large, spherical macronucleus, one or two micronuclei. Mid-portion of membranelle 2a (M2a) single-rowed, with posterior end hook-like. Anterior end of membranelle 2b (M2b) approximately at same level as posterior end of M2a.
3.1.2 Etymology
The species-group name pulchra (Latin adjective, beautiful) refers to the regular and attractive appearance of the cell.
3.1.3 Type locality and habitat
Brackish water from a sandy beach in Meishan Bay Wetland, Ningbo (29°46′7.95″N, 121°55′21.81″E), China. The water temperature was 15°C, and the salinity was 14.3 PSU at the time of sampling.
3.1.4 Type materials
The slide (registration number: WZH-20220406-01) with the protargol-impregnated holotype specimen circled in black ink, plus seven slides (registration numbers: WZH-20220406-02, 03… 08) with protargol-impregnated paratype specimens, has been deposited at the Laboratory of Protozoology, Ocean University of China.
3.1.5 SSU rRNA gene sequence
The SSU rRNA gene sequence of Pleuronema pulchra n. sp. was deposited in GenBank under the accession number OR658910 (1635 base pairs long and 43.36% G + C content).
3.1.6 ZooBank registration
urn:lsid:zoobank.org:act:7FF0E2FD-56D9-4766-AA8B-7770ABFBB217.
3.1.7 Description
Cells in vivo approximately 75–90 µm × 25–40 µm, oval or elliptical in outline (Figure 2A). Slightly reniform with anterior and posterior ends broadly rounded (Figures 3B, D). Right ventrolateral side basically straight, with dorsal side convex (Figure 3D). Buccal field broad, occupying 65–80% of body length (Figures 2A, 3B; Table 1). Oral cilia approximately 25 µm long. Pellicle slightly notched with shallow longitudinal grooves (Figure 3F, arrow). Bar-shaped extrusomes 2.5–3.0 µm in length and densely distributed beneath pellicle (Figure 3E, arrowheads). Cytoplasm colorless to slightly grayish, containing refractile globules (approximately 1.2 µm across) and irregularly shaped crystals (approximately 2 µm long) (Figures 3A, D). Numerous food vacuoles in middle and posterior portions of cell (Figures 2A, 3D, arrowhead, 3E). Single contractile vacuole subterminally located ca. 12 µm across, pulsating at intervals of approximately 80–95 s (Figures 2A, arrow; 3C, arrow). Somatic cilia usually 9–10 µm long (Figures 2A, 3A, arrowhead, 3B). Thirteen or 14 long and rather stiff caudal cilia approximately 15–25 µm in length (Figures 2A, 3A, arrows). Locomotion moderately fast, rotating around the main body axis, then remaining immobile for some time.
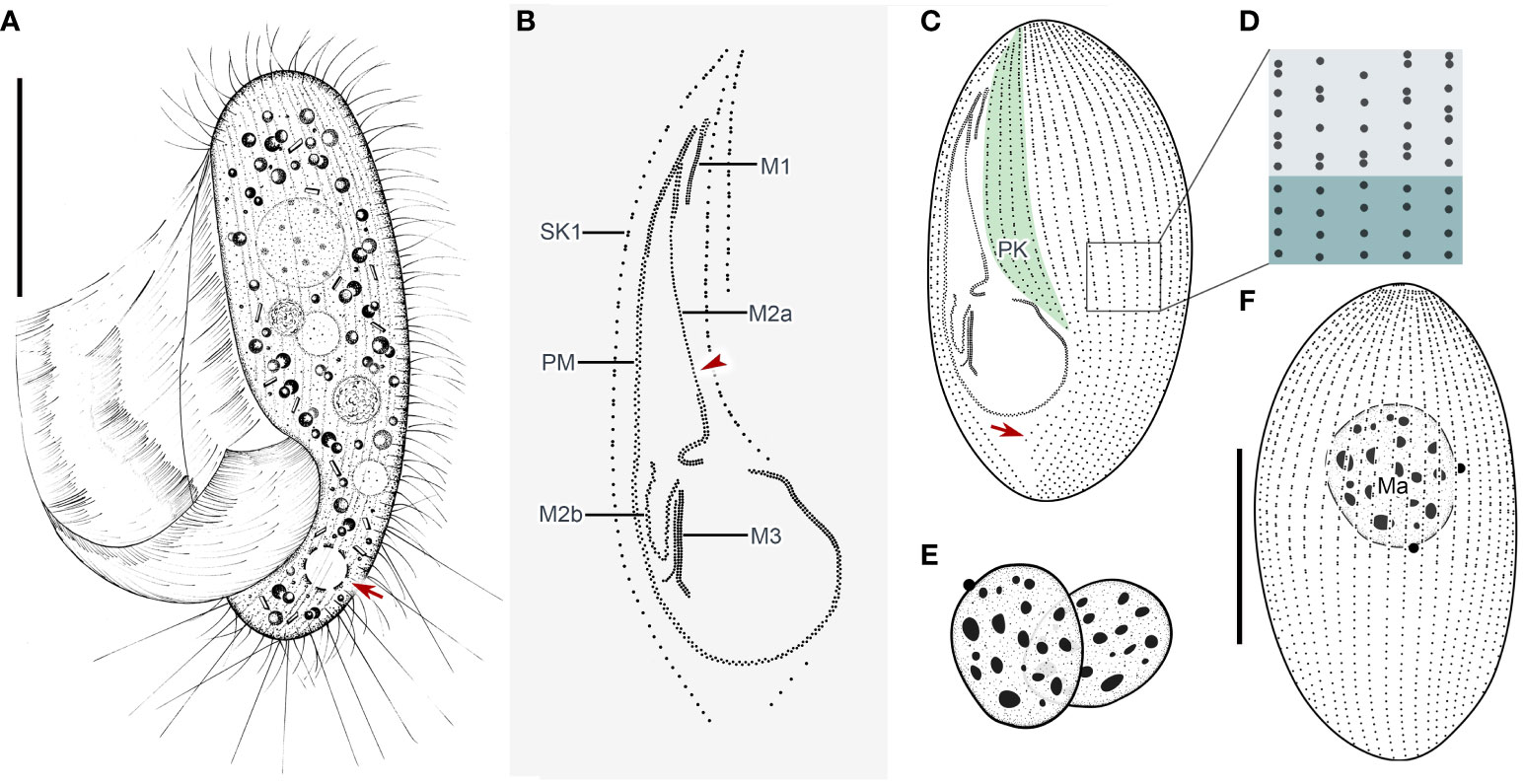
Figure 2 Pleuronema pulchra n. sp. in vivo (A) and after protargol impregnation (B–F). (A) Ventrolateral view of a representative cell; the arrow shows the contractile vacuole. (B) Detail of the oral cilia. The arrowhead marks the single row in the mid-portion of membranelle 2a. (C, F) Ciliature of ventral and dorsal sides of the holotype specimen; the arrow in (C) shows the postoral kinety. (D) Detail of the somatic kineties. The anterior half of the cell consists of dikinetids mixed with monokinetids, and the remaining part of the cell consists of monokinetids. (E) Detail of two macronuclei as seen in some individuals. M1, membranelle 1; M2a, membranelle 2a; M2b, membranelle 2b; M3, membranelle 3; Ma, macronucleus; PK, preoral kineties; PM, paroral membrane; SK1, somatic kinety 1. Scale bars: 30 µm.
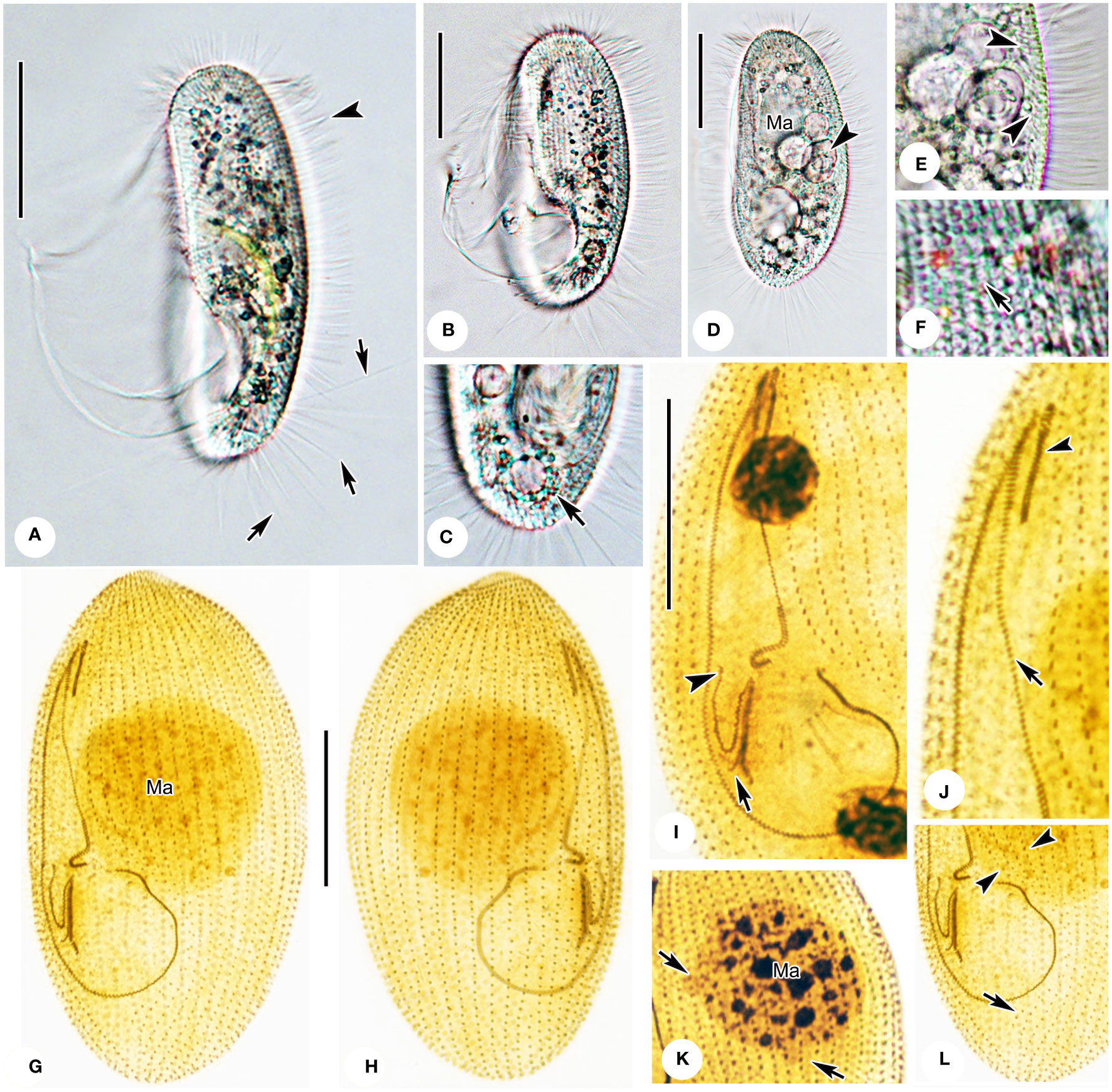
Figure 3 Photomicrographs of Pleuronema pulchra n. sp. in vivo (A–F) and after protargol impregnation (G–L). (A) Ventrolateral view of an individual; arrows point to the caudal cilia, the arrowhead shows the somatic cilia. (B, D) Different shapes of individuals; the arrowhead in D shows the food vacuoles. (C) Dorsolateral view of the posterior portion; the arrow denotes the contractile vacuole. (E) Detail of the cortex; arrowheads mark extrusomes below the pellicle. (F) Detail showing the cortical grooves on the cell surface (arrow) (G, H) Ventrolateral (G), and dorsolateral (H) views of the holotype specimen show the ciliature and macronucleus. (I) Detail of the oral structure. The arrow depicts the rightmost row of membranelle 3. Arrowhead marks the V-shaped membranelle 2b. (J) View of the anterior oral portion. The arrowhead shows membranelle 1. The arrow indicates the single-rowed portion of membranelle 2a. (K) Nuclear apparatus; the micronuclei adjacent to the macronucleus (arrows) should be noted. (L) Posterior oral portion, with arrowheads indicating preoral kineties and the arrow showing the postoral kinety. Ma, macronucleus. Scale bars: 30 µm.
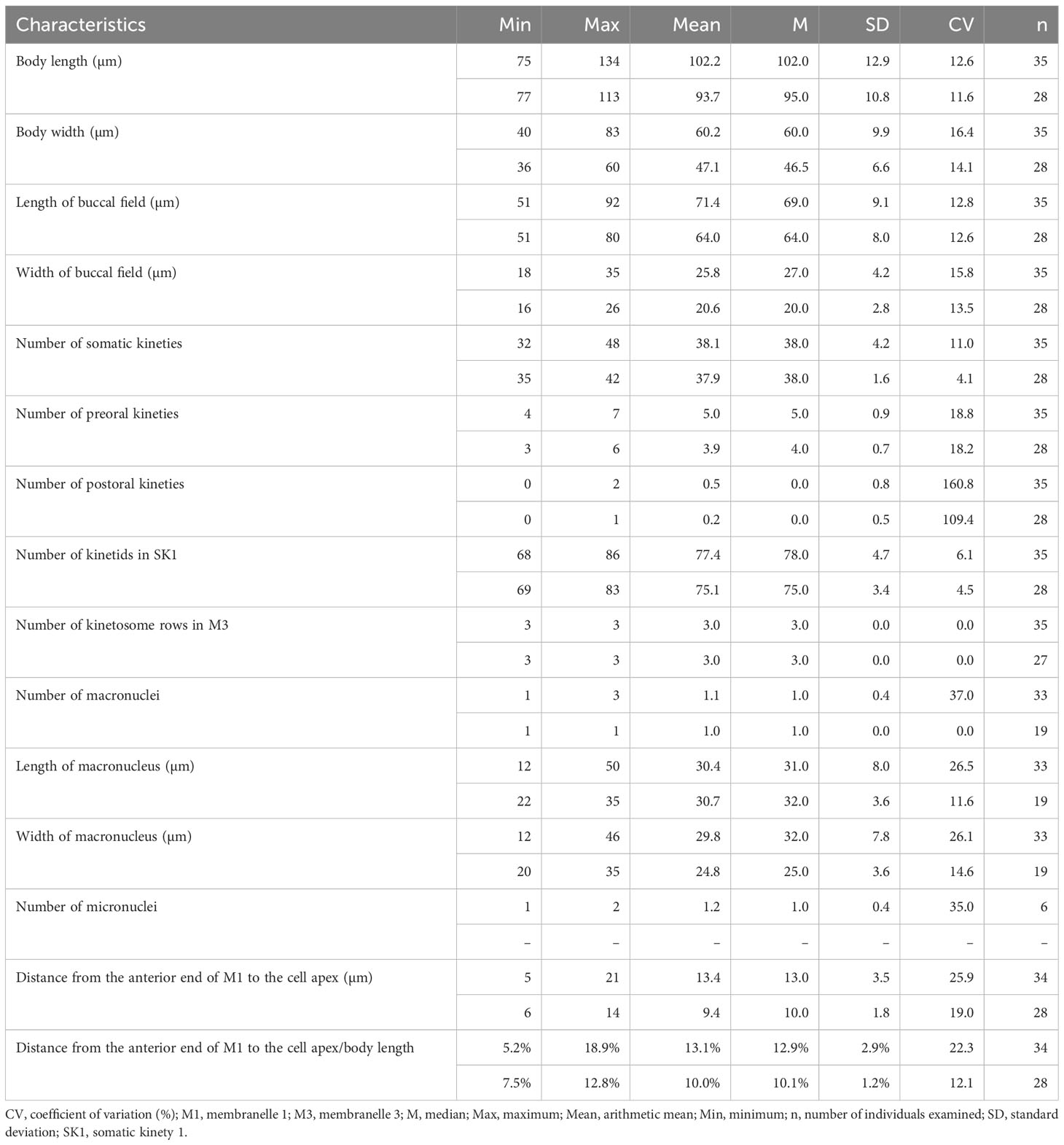
Table 1 Morphometric data of Pleuronema pulchra n. sp. (upper line) and P. warreni n. sp. (lower line) based on randomly selected protargol-stained specimens.
Thirty-two to 48 somatic kineties extending along whole body length (Figures 2C, F, 3G, H; Table 1). Anterior half of somatic kineties consisting of mixed monokinetids and dikinetids, posterior half composed only of monokinetids (Figures 2C, D, F, 3G, H). Somatic kinety 1 (SK1, first somatic kinety located on right side of oral apparatus) consists of 68–86 kinetids. Four to seven preoral kineties (Figures 2C, 3G, I; Table 1). Postoral kinety absent in most specimens, i.e., out of 35 cells examined, 24 lacked any postoral kineties, six presented two postoral kineties, and five showed one postoral kinety. One, rarely two or three, spherical macronuclei, 25–30 µm in diameter after protargol staining, slightly smaller (15–20 µm in diameter) when more than one macronucleus presenting (Figures 2E, F, 3G, H, K; Table 1). One or two (usually one) spherical micronuclei (3–4 µm across) adjacent to the macronucleus (Figure 3K, arrows; Table 1).
Oral apparatus: membranelle 1 (M1) three-rowed in anterior fifth, two-rowed in remaining portion (Figures 2B, C; 3H, J, arrowhead); membranelle 2a (M2a) two-rowed in anterior and posterior portions and single-rowed in mid-posterior portion, occupying three-fifths of length of M2a, and having a hook-like posterior end (Figures 2B, arrowhead, 2D; 3G, I, J, arrow); membranelle 2b (M2b) V-shaped, with kinetosomes arranged in zig-zag pattern and anterior end of M2b approximately at same level as posterior end of M2a (Figures 2B, C; 3G, I, arrowhead, 3L); membranelle 3 three-rowed, with posterior end of rightmost row notably distinct from other rows; basal bodies of paroral membrane (PM) arranged in zig-zag pattern, occupying 65–75% of cell length (Figures 2B, C; 3G, I, arrow, 3J, L).
3.2 Pleuronema warreni n. sp.
3.2.1 Diagnosis
Cells in vivo approximately 55–80 µm × 25–45 µm with elliptical body shape. Right ventrolateral side basically straight, with three to six preoral kineties and 35–42 somatic kineties. One spherical macronucleus usually located in anterior half of cell. Mid-portion of membranelle 2a (M2a) single-rowed, and posterior end hook-like. Anterior end of membranelle 2b commencing significantly above level of posterior end of M2a.
3.2.2 Etymology
The species-group name warreni is dedicated to Dr. Alan Warren of the Natural History Museum, United Kingdom, who has contributed enormously to research in the field of ciliate biodiversity.
3.2.3 Type locality and habitat
A brackish artificial lake in Ningbo (29°44′1.22″N, 121°53′47.34″E), China. The water temperature was 26.5 °C and the salinity was 15.1 PSU.
3.2.4 Type materials
The slide (registration number: WZH-20220527-05) with the protargol-impregnated holotype specimen circled in black ink and four slides (registration numbers: WZH-20220527-01, 02… 04) with protargol-impregnated paratype specimens has been deposited at the Laboratory of Protozoology, Ocean University of China.
3.2.5 SSU rRNA gene sequence
The SSU rRNA gene sequence of Pleuronema warreni n. sp. was deposited in GenBank under the accession number OR658911 (1635 base pairs long and 43.43% G + C content).
3.2.6 ZooBank registration
urn:lsid:zoobank.org:act:835371D1-3B03-4FFB-BC8D-4E0A95D79B0B.
3.2.7 Description
Cells in vivo approximately 55–80 µm × 25–45 µm. Body shape elliptical with broadly rounded anterior end and slightly tapered posterior end (Figures 4A, 5A, B, D). Laterally flattened approximately 3:2, featuring slightly straight ventral side and convex dorsal side. Buccal field occupying 65–75% of body length. Oral cilia approximately 20 µm long. Pellicle slightly notched, with shallow longitudinal grooves (Figures 5A, B). Bar-shaped extrusomes ca. 3 µm long, densely distributed beneath pellicle (Figure 5E, arrowheads). Cytoplasm transparent to grayish, containing many greasy, shining globules approximately 1.5 µm in diameter. Irregularly shaped crystals (approximately 2.5 µm long) scattered throughout cell, and several food vacuoles usually in middle of cell (Figures 5A, B, D). One contractile vacuole subterminal approximately 9 µm in diameter, when fully expanded, pulsating at intervals of approximately 100–120 s (Figures 4A, arrow; 5B, arrowhead, 5D; Table 1). Somatic cilia approximately 9–10 µm in length (Figures 4A, 5A–D). Twelve to 17 caudal cilia approximately 20–25 µm in length (Figures 4A; 5A, D, arrows). Locomotion by rotation around main body axis and sometimes remaining motionless for short periods of time.
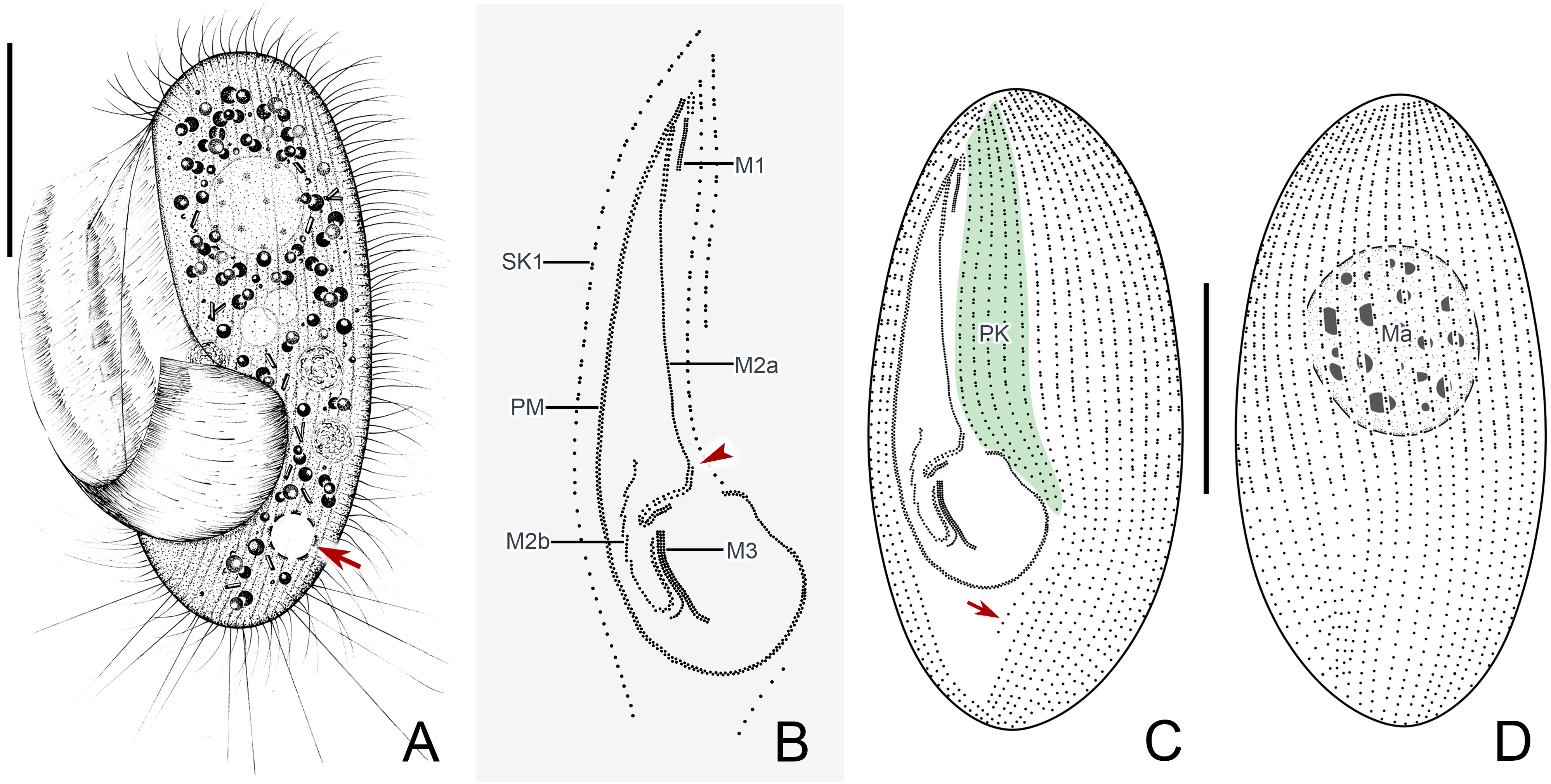
Figure 4 Pleuronema warreni n. sp. in vivo (A) and after protargol impregnation (B–D). (A) Ventrolateral view of a representative cell; the arrow points to the contractile vacuole. (B) Detail of the oral apparatus. The arrowhead marks the two-rowed posterior part of membranelle 2a. (C, D) Ciliature of the ventral (C) and dorsal (D) sides of the holotype specimen; the arrow shows the postoral kinety. M1, membranelle 1; M2a, membranelle 2a; M2b, membranelle 2b; M3, membranelle 3; Ma, macronucleus; PK, preoral kineties; PM, paroral membrane; SK1, somatic kinety 1. Scale bars: 30 µm.
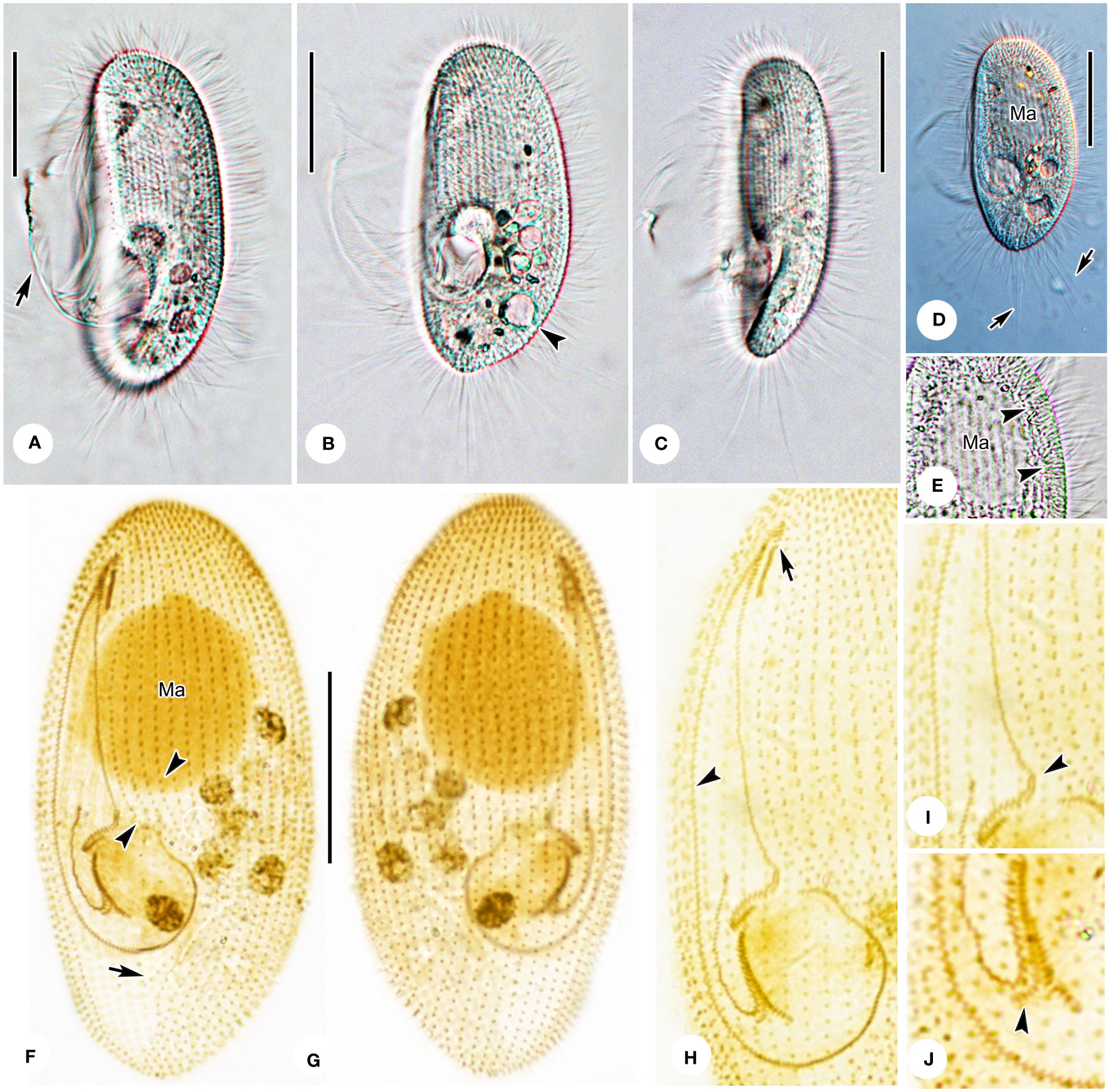
Figure 5 Photomicrographs of Pleuronema warreni n. sp. in vivo (A–E) and after protargol impregnation (F–J). (A, B) Ventrolateral views of representative cells; the arrow in A denotes oral cilia, and the arrowhead in B points to the contractile vacuole. (C) Lateral view, showing the tapered caudal portion. (D) Ventrolateral view of a representative specimen; note the long caudal cilia (arrows). (E) Detail of the cortex; arrowheads point to the extrusomes beneath the pellicle. (F, G) Ventrolateral (F) and dorsolateral (G) views of the holotype specimen, showing the ciliature and macronucleus. The arrowheads point to the preoral kineties, and the arrow points to the postoral kinety. (H) Detailed view of the oral apparatus. Arrow shows membranelle 1. The arrowhead marks the paroral membrane. (I, J) Posterior oral portions of two representative specimens. The arrowhead in (I) points to the two-rowed part of membranelle 2a. The arrowhead in J shows the detached rightmost row of membranelle 3. Ma, macronucleus. Scale bars: 30 µm.
Thirty-five to 42 somatic kineties (SK) extend along nearly entire length of body, except for bald apical plate at anterior end (Figures 4C, D, 5F, G). Anterior half of each SK composed of mixed monokinetids and dikinetids, and posterior half composed of monokinetids only (Figures 4C, D, 5F–H). Somatic kinety 1 (SK1) consisting of 69–83 kinetids. Three to six preoral kineties at left of buccal field; one postoral kinety (PoK) in 13 out of 28 cells examined, while remaining cells locked PoK (arrow in Figures 4C, 5F). One spherical macronucleus, 25–35 µm × 20–35 µm after protargol staining, located one-third of length of body in all 19 individuals observed (Figures 4A, D, 5F, G; Table 1). Micronucleus not detected.
Oral apparatus: M1 three-rowed, composed of one short and two long rows of kinetosomes, approximately one-third of length of M2a, and mid-row of M1 clearly separated into two parts (Figures 4B, C, 5F, H, arrow). Anterior and posterior portions of M2a two-rowed, with mid-portion single-rowed and occupying approximately 75% of length of M2a, and posterior portion hook-like (Figures 4B, arrowhead, 5F, H, I, arrowhead). Membranelle 2b V-shaped, with kinetosomes of right anterior half arranged in consecutive groups of single rows, each consisting of three to six kinetosomes and connected to each other (Figures 4B, C, 5F, H). Anterior end of M2b in a more apical position than posterior end of M2a (Figures 4B, C, 5F, H, I). Membranelle 3 consisted of three closely packed rows, with posterior end of rightmost row conspicuously separated at a right angle from other rows (Figures 4B, C, 5F, H, J, arrowhead). Paroral membrane (PM) consisted of two-rowed kinetosomes and arranged in zig-zag pattern, occupying approximately 60–70% of cell length (Figures 4B, C, 5F, H).
3.3 Molecular data and phylogenetic analyses
The topologies of the maximum likelihood (ML) and Bayesian Inference (BI) trees are nearly concordant; therefore, only the ML tree is presented here with support values from both algorithms (Figure 6). According to the SSU rRNA gene tree, the order Pleuronematida is a fully supported (ML/BI: 100%/1.00) monophyletic group. In the present study, eight families within the order Pleuronematida are divided into three major clades. Three families, namely Pleuronematidae, Peniculistomatidae, and Histiobalantiidae, form one clade, and the families Ctedoctematidae and Eurystomatidae form another fully supported clade sister to the previous clade. The families Ancistridae, Hemispeiridae, and Cyclidiidae form the third clade with strong support by both methods (ML/BI: 96%/1.00). Four of the eight families, i.e., Peniculistomatidae, Histiobalantiidae, Ctedoctematidae, and Eurystomatidae, are monophyletic. The family Peniculistomatidae is nested within the family Pleuronematidae, with some sequences of the genera Peniculistoma, Mytilophilus, and Schizocalyptra scattered within Pleuronema, making both the family Pleuronematidae and the genus Pleuronema polyphyletic.
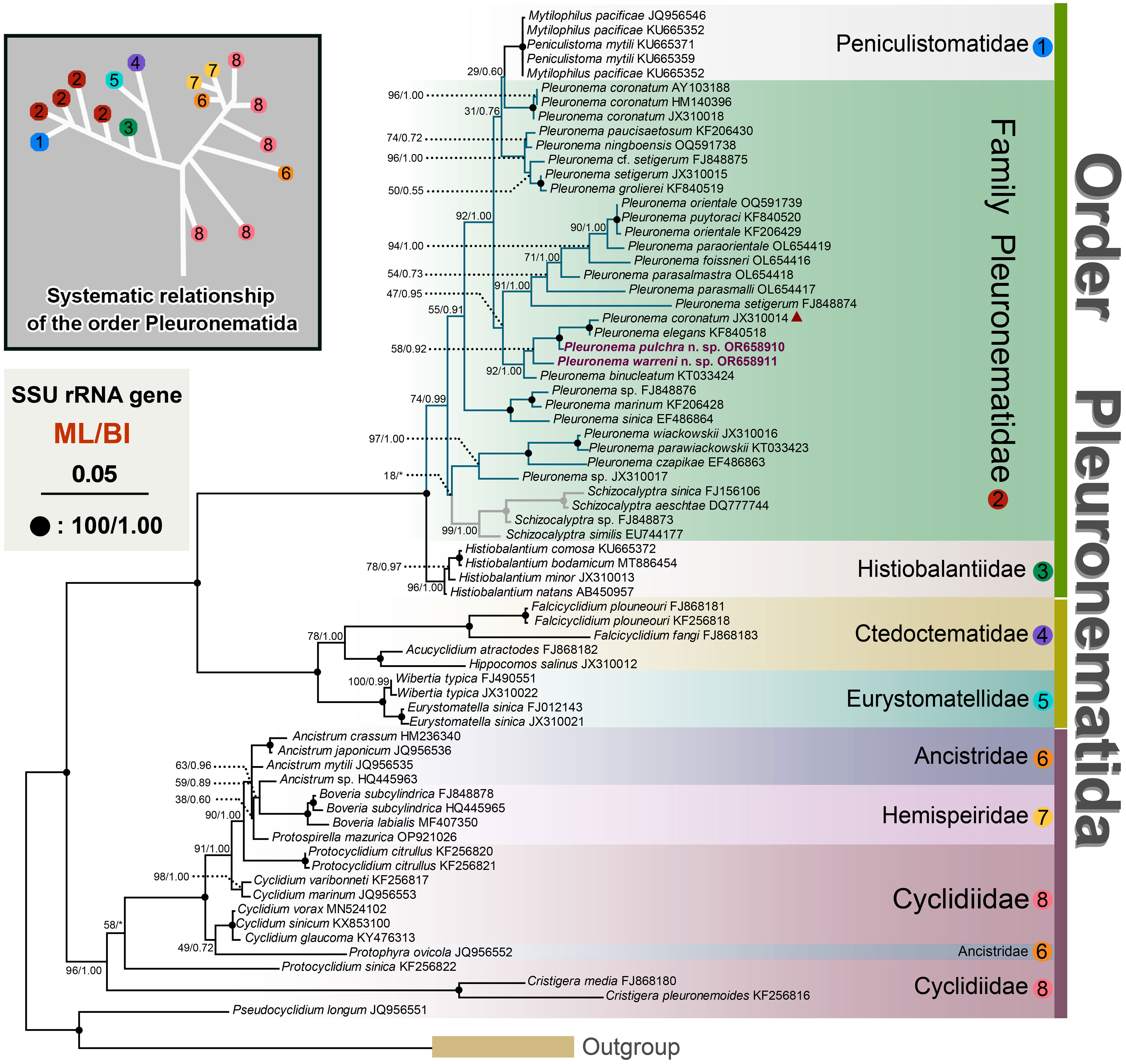
Figure 6 Maximum likelihood (ML) tree inferred from SSU rRNA gene sequences, showing the phylogenetic positions of Pleuronema pulchra n. sp. OR658910 and Pleuronema warreni n. sp. OR658911 (sequences in bold). The inset in the top left corner shows the major clades of Pleuronematida. Numbers at nodes denote ML bootstrap values/Bayesian inference (BI) posterior probabilities. Fully supported (100%/1.00) branches are marked with solid circles. The scale bar corresponds to five substitutions per 100 nucleotide positions. The red triangle marks the deviation of Pleuronema coronatum JX310014 from the other three P. coronatum sequences.
Both sequences of the two newly described species, P. pulchra n. sp. and P. warreni n. sp., nested within other Pleuronema species in the well-supported clade (ML/BI: 92%/1.00) of the family Pleuronematidae, clustering with P. coronatum JX310014, P. elegans KF840518, and P. binucleatum KT033424 (Figure 6). In the SSU rRNA gene tree, P. pulchra n. sp. OR658910 is sister to a fully supported clade that includes P. coronatum JX310014 and P. elegans KF840518, followed by P. warreni n. sp. OR65891 and P. binucleatum KT033424.
A comparison of the SSU rRNA gene sequences between the two new species and their closest relatives in the genus Pleuronema shows that P. pulchra n. sp. OR658910 is largely similar to P. warreni n. sp. OR658911, P. binucleatum KT033424, P. elegans KF840518, and P. coronatum JX310014 with 36–37 unmatched nucleotides (corresponding to 97.7–97.8% similarity) (Figures 7A, B). Additionally, Pleuronema warreni n. sp. OR658911 is most similar to P. binucleatum KT033424 and P. pulchra n. sp. OR658910, with 30 and 36 differing nucleotides (98.2% and 97.8% sequence identity), respectively (Figures 7A, B). Although P. warreni n. sp. OR658911 clusters in a clade with P. coronatum JX310014, it appears to be more divergent from this isolate (63 unmatched nucleotides) than from P. coronatum JX310018 and P. coronatum AY103188 (56 and 61 unmatched nucleotides, respectively) (Figure 7).
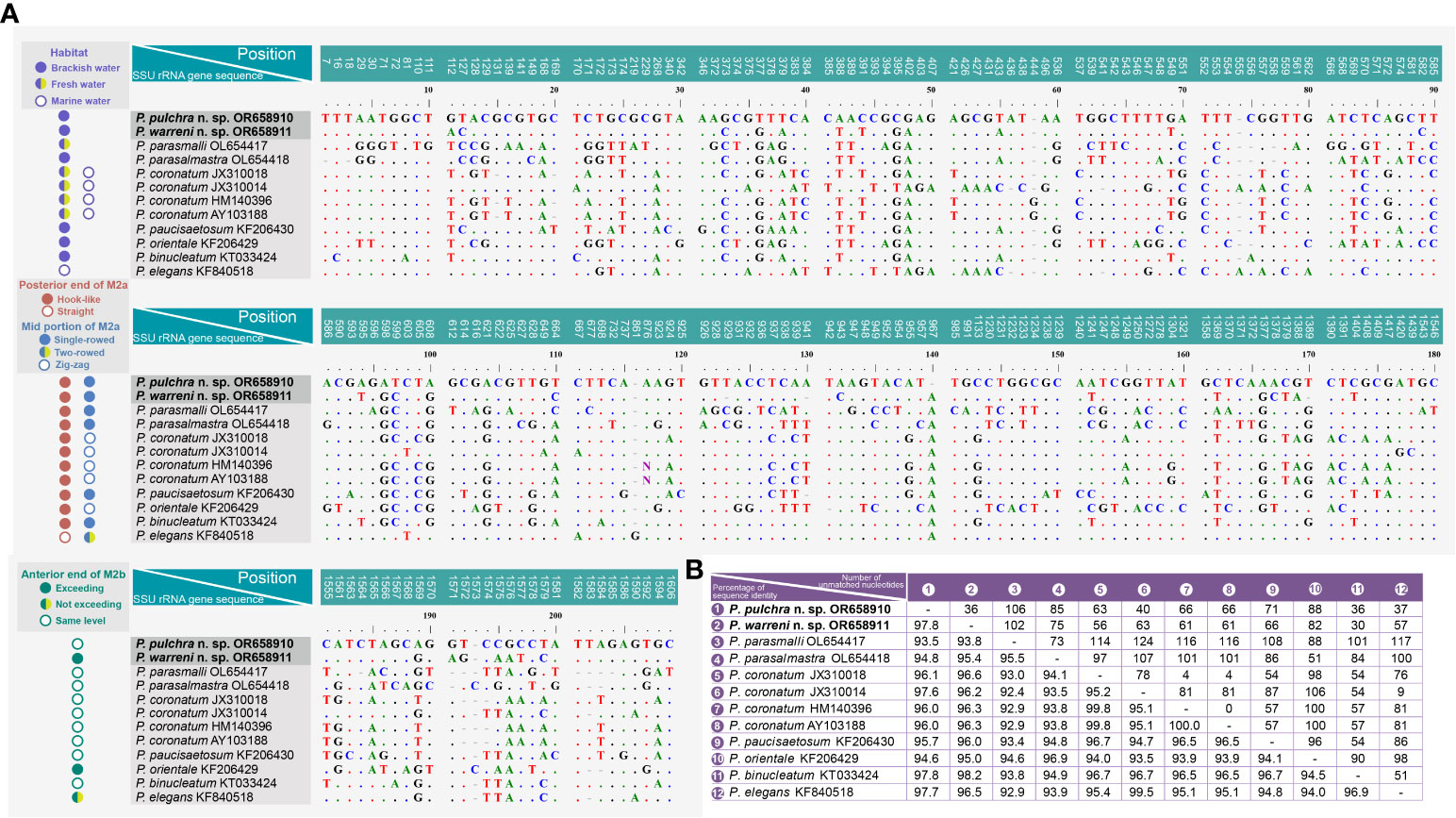
Figure 7 Sequence comparison of the SSU rRNA gene of Pleuronema pulchra n. sp. and P. warreni n. sp. with related taxa. (A) Comparison of the SSU rRNA genes and their main characteristics. The numbers at the top of each column indicate the positions of unmatched columns in the alignment. Alignment gaps (-) are introduced to account for insertions and deletions. (B) Comparison matrix of the SSU rRNA gene sequences. The percentage of sequence identity (%) is displayed in the bottom left of the matrix, while the number of unmatched nucleotides is shown in the upper right. “Exceeding” in the anterior end of M2b means that M2b commences above the level of the posterior end of M2a, while “not exceeding” means it commences below the level of the posterior end of M2a. M2a, membranelle 2a; M2b, membranelle 2b.
4 Discussion
4.1 Comparison of the new species with congeners
4.1.1 Morphological comparison of the two new species with congeners lacking detailed information on the oral apparatus
Pleuronema is a diverse and widely distributed genus, consisting of approximately 40 recognized species, which has been widely reported in different aquatic environments since the establishment of the genus by Dujardin (Dujardin, 1841; Dragesco, 1960; Borror, 1963; Dragesco, 1968; Dragesco and Dragesco-Kernéis, 1986; Foissner et al., 1994; Song, 2000; Lynn, 2008; Pan et al., 2015; Pan et al., 2016; Liu et al., 2022; Zhang et al., 2023). The oral structure serves as the crucial criterion for differentiating species within this genus. Therefore, Wang et al. (2008a) proposed that the species of Pleuronema can be separated into two types: (i) the “coronatum-type” with a hook-shaped posterior end of M2a, and (ii) the “marinum-type” with a straight posterior end of M2a. This separation into two types received widespread recognition (Wang et al., 2008a; Wang et al., 2009; Pan et al., 2015; Zhang et al., 2023). According to this classification, both P. pulchra n. sp. and P. warreni n. sp. belong to the “coronatum-type”.
Ten species, namely Pleuronema anodontae, P. arenicola, P. balli, P. borrori, P. crassum, P. grassei, P. prunulum, P. roscoffensis, P. simplex, and P. smalli, lack detailed descriptions or photomicrographs of the oral apparatus, which is the vital characteristic for identification. However, our two new species can be distinguished from all of them by at least one characteristic. Pleuronema anodontae and P. prunulum were only recorded by Kahl; P. anodontae has only one caudal cilium, while two new species have at least twelve caudal cilia. P. prunulum differs from the two new species by the length of its caudal cilia (which are equal to the body length in P. prunulum vs. one-third of the body length) (Kahl, 1926; Kahl, 1931). Pleuronema simplex is characterized by having only one-rowed M1, M2b, and M3 (as shown in the schematic diagram by Dragesco), which distinguishes it from all other Pleuronema species (Dragesco, 1960). While P. crassum has been reported several times, it mainly differs from the two new species by the lack of caudal cilia (vs. present). Unfortunately, photomicrographs in vivo and the detailed ciliature patterns of P. crassum were not available (Dujardin, 1841; Roux, 1901; Kahl, 1931; Gajewskaja, 1933; Vuxanovici, 1960; Chorik, 1968; Pätsch, 1974; Foissner et al., 1994; Dinçer, 2016). Pleuronema grassei has been established as a new species based on its extremely large body length in vivo (140–210 µm), which differs from P. pulchra n. sp. and P. warreni n. sp. (75–90 µm and 55–80 µm, respectively) (Dragesco, 1960). Pleuronema arenicole and P. roscoffensis were both only described by Dragesco. According to the description and schematic diagrams, Pleuronema arenicole differs from P. pulchra n. sp. and P. warreni n. sp. by the arrangement of M2b (U-shaped in the former vs. V-shaped in the two new species) and the rows of M3 (2 in the former vs. 3) (Dragesco, 1960). Pleuronema roscoffensis can be distinguished from our two species by the number of PK (2 vs. 4–7 and 3–6, in P. pulchra n. sp. and P. warreni n. sp., respectively) and rows of M1 (two in P. roscoffensis vs. three) (Dragesco, 1968).
Wang et al. (2008a) regarded Pleuronema balli, P. borrori, and P. smalli to be synonyms of P. coronatum, while Liu et al. (2022) concluded that only P. balli is a synonym of P. coronatum, and P. borrori and P. smalli are valid species due to their morphological divergences with P. coronatum. Unfortunately, no morphological information in vivo or SSU rRNA gene sequences of these populations are available. Pleuronema warreni n. sp. differs from these species due to the position of the anterior end of M2b (significantly above the posterior end of M2a in P. warreni n. sp. vs. the same level as the posterior end of M2a). Pleuronema borrori is distinguished from P. pulchra n. sp. by the length ratio of oral/body (0.55 vs. 0.59–0.85 in P. pulchra n. sp.) and having more somatic kineties (41–46, 43 on average vs. 32–48, 38 on average in P. pulchra n. sp.) (Dragesco, 1968), while Pleuronema smalli can be easily distinguished from P. pulchra n. sp. by body size after protargol-staining (40–70 × 25–40 µm vs. 75–135 × 40–85 µm in P. pulchra n. sp.).
4.1.2 Morphological comparison of Pleuronema pulchra n. sp. with related congeners
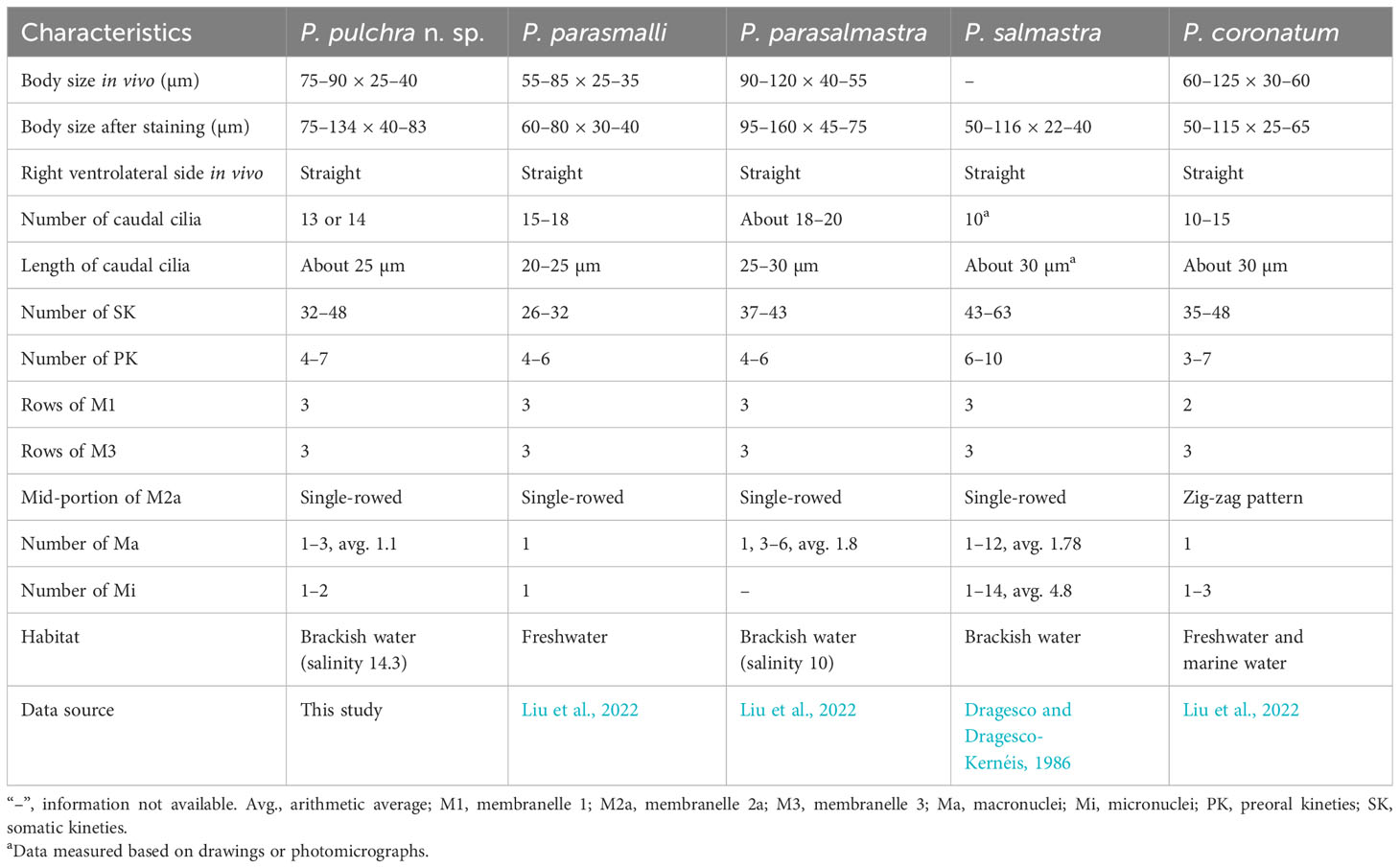
Based on the body size and shape (75–90 μm, right ventrolateral side straight), number of somatic kineties (32-48), posterior end of M2a hook-like, M2b V-shaped (slightly above or at the same level of the posterior end of M2a) and three-rowed M3, four species should be compared with P. pulchra n. sp., namely P. coronatum Kent, 1881, P. parasalmastra Liu et al., 2022, P. parasmalli Liu et al., 2022, and P. salmastra Dragesco and Dragesco-Kernéis, 1986 (Figure 8; Table 2).
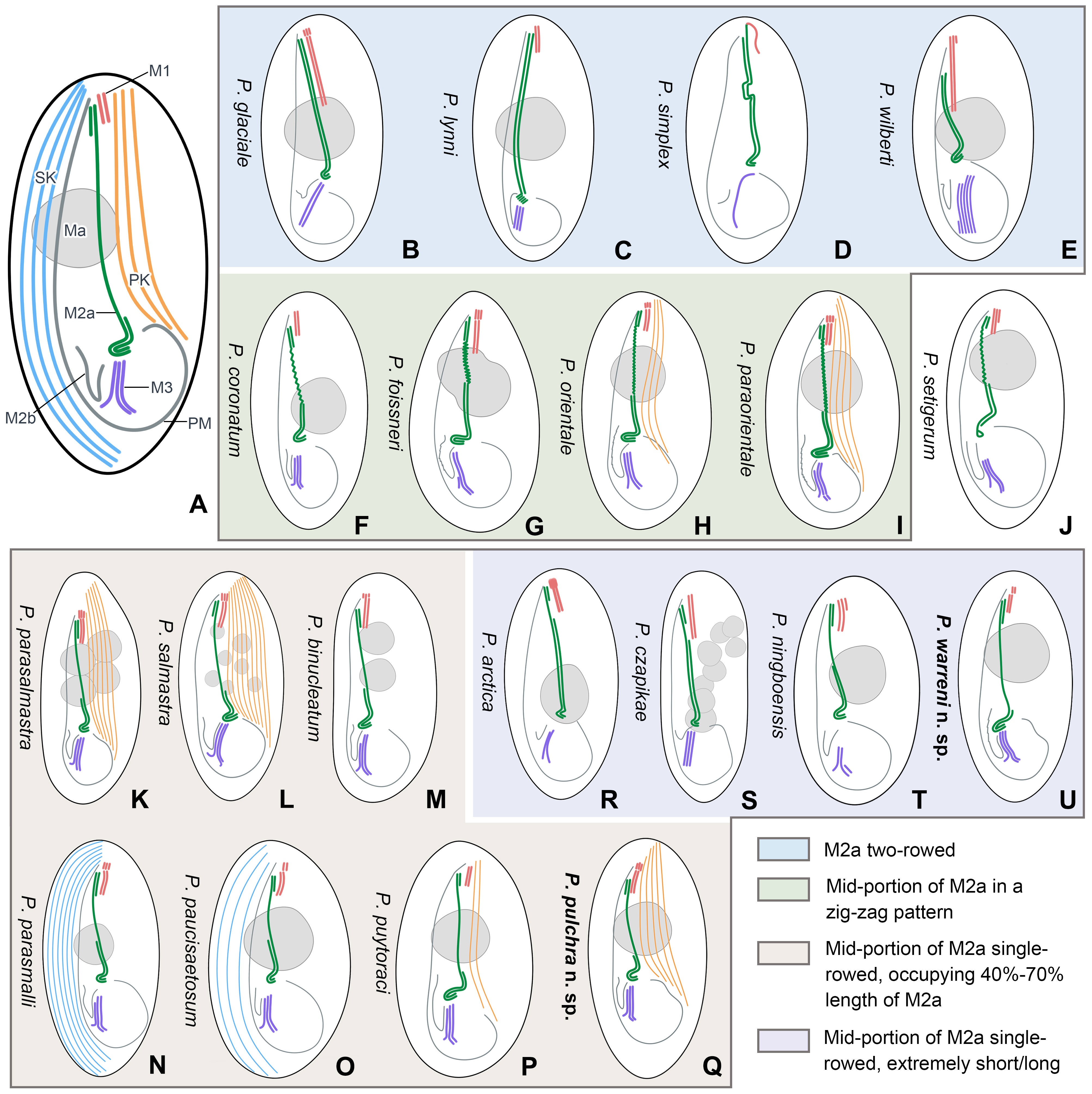
Figure 8 Schematic illustrations of the ciliature of 20 Pleuronema species with the posterior end of membranelle 2a hook-like. (A) Diagrammatic drawing of the genus Pleuronema, showing the main characteristic structures. (B–E) Four species with fully two-rowed M2a. (F–I) Four species with a zig-zag pattern in the mid-portion of M2a. (J) P. setigerum is the only species with a ring-like structure in the posterior end of M2a. (K–Q) Seven species with M2a single-rowed in the mid-portion. (R–U) Four species show extremely short or long single-rowed parts in the mid-portion of M2a. M1, membranelle 1; M2a, membranelle 2a; M2b, membranelle 2b; M3, membranelle 3; Ma, macronucleus; PK, preoral kineties; PM, paroral membrane; SK, somatic kineties.
Pleuronema coronatum is a well-known species and has been collected from Europe, East Asia, and America since Kent first discovered it in 1881 (Kent, 1881; Kahl, 1931; Dragesco, 1960; Dragesco, 1968; Foissner et al., 1994; Song, 2000; Wang et al., 2008a). The detailed descriptions of its living cells and the whole ciliary pattern have been widely studied. However, the gradual expansion of ranges of different morphological traits (e.g., body size, preoral and somatic kinety numbers, habitats), and the lack of a clear diagnosis have made “P. coronatum” a complex of species prone to misidentifications and the potential presence of cryptic species. Efforts should be made not only to define and improve the diagnosis of P. coronatum but also to detect possible misidentifications. An example of this is the work of Liu et al. (2022), who concluded that P. balli is a synonym of P. coronatum after reinvestigating the morphological information of several related populations of Pleuronema. A major problem stemming from this situation is the fact that the molecular data of several alleged Pleuronema coronatum isolates currently available from GenBank are not linked to morphological information, and thus their validity cannot be proven. In particular, P. coronatum JX310014 differs greatly from the other three P. coronatum sequences (JX310018, HM140396, and AY103188) with 78–81 unmatched nucleotides (Figure 7A). Consequently, the identification of these sequences remains uncertain.
Pleuronema pulchra n. sp. resembles P. coronatum in characteristics such as body size, the number of somatic kineties, and preoral kineties. However, Pleuronema pulchra n. sp. can be clearly distinguished by a basically straight mid-portion of M2a (vs. a mid-portion in a zig-zag pattern in P. coronatum) and more kinetids in somatic kinety 1 (68–86 vs. 21–30) (Foissner et al., 1994; Song, 2000; Liu et al., 2022).
Pleuronema pulchra n. sp. most closely resembles P. parasmalli in body size, outline in vivo, and structure of the oral apparatus. However, the former species can be easily distinguished from the latter species in terms of more somatic kineties (32–48 in P. pulchra n. sp. vs. 26–32) and the different habitat (brackish water in P. pulchra n. sp. vs. freshwater) (Liu et al., 2022). Moreover, the comparison of SSU rRNA gene sequences shows that the new species is clearly separated from P. parasmalli with 106 unmatched nucleotides (93.5% in sequence identity) (Figures 7A, B).
Pleuronema pulchra n. sp. is similar to P. parasalmastra in the number of SK, preoral kineties, and the structure of the oral apparatus. Nevertheless, the new species can be separated from P. parasalmastra by the body size in vivo (75–90 × 25–40 µm in P. pulchra n. sp. vs. 90–120 µm × 40–55 µm) and the lower number of caudal cilia (13–14 vs. 18–20). Moreover, the distance between the cell apex and the anterior end of M1 occupies a smaller portion of the body length in P. pulchra n. sp. than in P. parasalmastra (8–13% vs. 16–25% of body length, respectively) (Liu et al., 2022). In addition, the SSU rRNA gene sequence of P. pulchra n. sp. is largely different from the gene sequence of P. parasalmastra, with 85 unmatched nucleotides (94.8% in sequence identity) (Figures 7A, B).
When compared with Pleuronema salmastra, P. pulchra n. sp. has fewer somatic kineties (32–48, 38 on average vs. 43–63, 53 on average) and fewer preoral kineties (4–7, 5 on average vs. 6–10, 7.6 on average) (Dragesco and Dragesco-Kernéis, 1986).
4.1.3 Morphological comparison of Pleuronema warreni n. sp. with related congeners
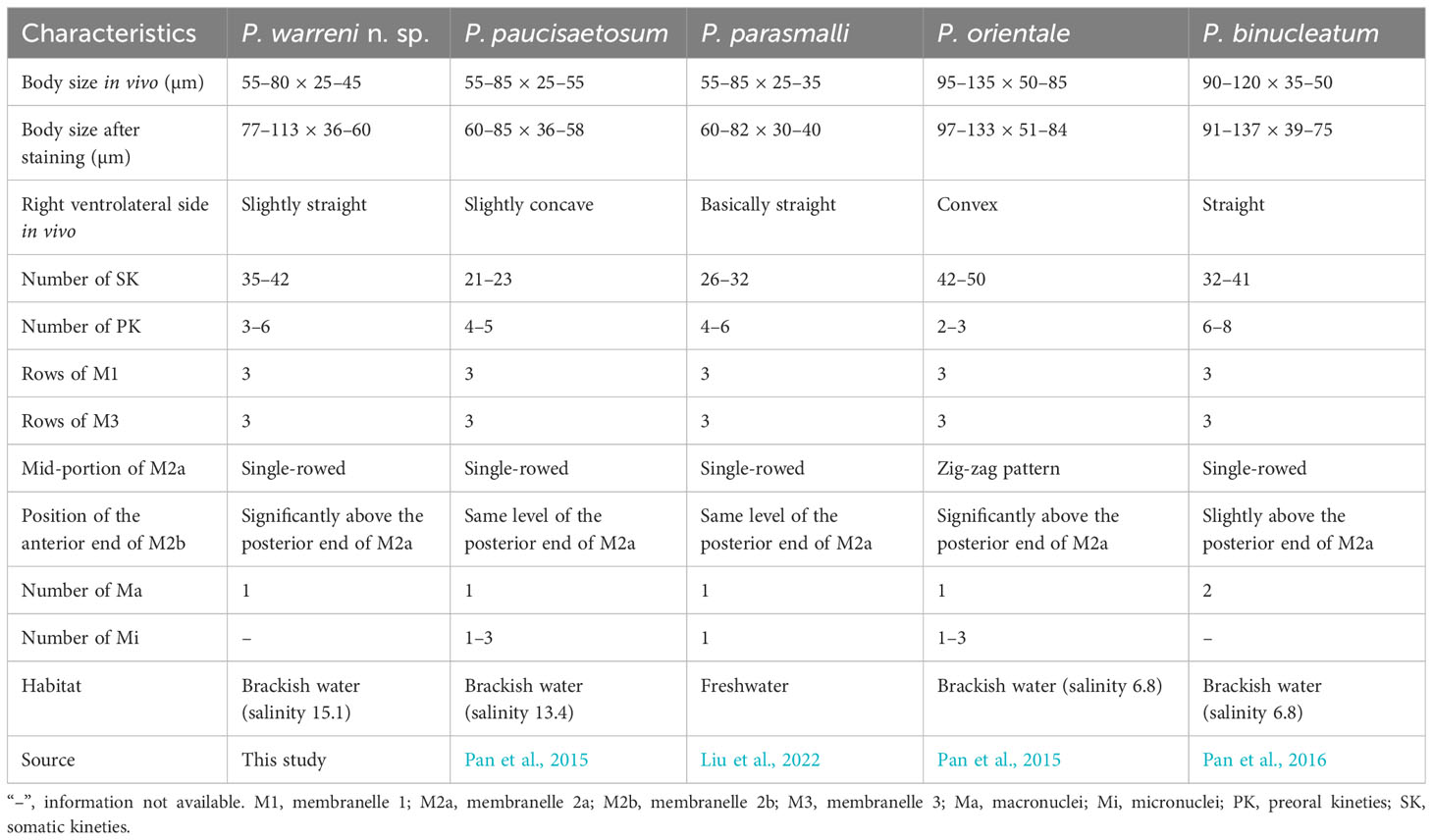
Considering the number of somatic kineties (35-42), spherical macronucleus, posterior end of M2a hook-like, and M3 three-rowed, four species can be compared with Pleuronema warreni n. sp., namely P. binucleatum Pan et al., 2016, P. orientale Pan et al., 2015, P. parasmalli Liu et al., 2022, and P. paucisaetosum Pan et al., 2015 (Figure 8; Table 3).
Although Pleuronema warreni n. sp. is similar to P. paucisaetosum in body size in vivo, in the number and shape of the macronucleus (i.e., one spherical macronucleus), and in the number of preoral kineties, they differ in the shape of the right ventrolateral side of the cell in vivo (slightly straight in P. warreni n. sp. vs. slightly concave) and in the number of somatic kineties (33–42 in P. warreni n. sp. vs. 21–23). Moreover, M2b of P. warreni n. sp. commences in a more anterior position than the posterior end of M2a (vs. at the same level as the posterior end of M2a in P. paucisaetosum) (Pan et al., 2015). The SSU rRNA gene sequences of these two species exhibit a difference of 66 nucleotides, supporting their molecular divergence as two different species (Figure 7A).
Pleuronema warreni n. sp. resembles P. parasmalli in body size and shape in vivo. However, the former species differs from P. parasmalli in the number of somatic kineties (35–42 vs. 26–32), the position of the anterior end of M2b (significantly above the posterior end of M2a vs. the same level of the posterior end of M2a), and the habitat (brackish water vs. freshwater) (Liu et al., 2022). Additionally, the differences in SSU rRNA gene sequences between these two Pleuronema species (102 unmatched nucleotides) support their separation as different species (Figure 7A).
When compared with Pleuronema orientale, Pleuronema warreni n. sp. has an extremely similar V-shaped M2b, which is divided into several single-rowed groups. Pleuronema warreni n. sp. can be clearly separated from P. orientale based on body length in vivo (55–80 µm vs. 95–135 µm), the number of SK (35–42 vs. 42–50), and the mid-pattern of M2a (single-rowed vs. zig-zag pattern) (Pan et al., 2015). The nucleotide differences (82 unmatched nucleotides) between the sequences of P. warreni n. sp. and P. orientale also support the molecular separation between these species (Figure 7).
Pleuronema warreni n. sp. can be distinguished from P. binucleatum by having a smaller body length in vivo (55–80 µm vs. 90–120 µm), a single macronucleus (as opposed to two macronuclei), and fewer preoral kineties (3–6 vs. 6–8) (Pan et al., 2016). Moreover, Pleuronema warreni n. sp. has more kinetids in SK1 (69–83 vs. about 60) (Pan et al., 2016). Although the SSU rRNA gene sequence of Pleuronema warreni n. sp. OR658911 is rather similar to that of P. binucleatum KT033424, they differ by 30 unmatched nucleotides, indicating that they can be classified as two different species (Figure 7).
4.1.4 Comments on Pleuronema species with hook-like type of membranelle 2a
The crucial importance of oral structures in the identification of Pleuronema species has been widely emphasized in previous studies (Dragesco, 1968; Foissner et al., 1994; Song, 2000; Wang et al., 2008a; Wang et al., 2008b; Wang et al., 2009; Pan et al., 2015; Pan X. et al., 2015; Pan et al., 2016; Liu et al., 2022; Zhang et al., 2023). Based on the structure of M2a, the members of the genus Pleuronema can be categorized into two distinct groups: the “coronatum-type”, with the posterior portion of the M2a hook-like, and the “marinum-type”, with the posterior portion of the M2a straight or lightly curved (Wang et al., 2008a; Wang et al., 2009; Pan et al., 2015; Zhang et al., 2023).
The genus Pleuronema contains 25 nominal species (including the two new species in this work), which have a hook-like posterior end of M2a and thus belong to the “coronatum-type”. Pleuronema balli, P. borrori, and P. smalli are widely discussed species, as they are similar to P. coronatum (Small, 1964; Dragesco, 1968; Wang et al., 2008a; Liu et al., 2022). After a detailed comparison, Liu et al. (2022) concluded that P. balli should be a synonym of P. coronatum, but treated both P. smalli and P. borrori as valid species due to the body width after protargol, the ratio of buccal field to body length, or the number of somatic kineties. Despite this, it should be noted that the oral apparatus of these three species remains uncertain.
Additionally, P. crassum and P. grassei are two more Pleuronema species without the complete ciliature pattern; P. crassum can be clearly distinguished by the lack of caudal cilia (Dujardin, 1841; Kahl, 1931; Vuxanovici, 1960; Chorik, 1968; Pätsch, 1974; Foissner et al., 1994; Dinçer, 2016), while the most remarkable feature of P. grassei is its largest body length in vivo (approximately 140–210 µm). However, since Dragesco (1960) first described P. grassei, it has never been reported again, and its ciliature pattern remains unknown.
In the present work, we provide a comprehensive illustrated key, based on a detailed study of the oral structures, for the identification of the two new species along with 18 nominal Pleuronema species that have a hook-like posterior end of M2a (Illustrations of selected key characteristics are in Figure 8). These 20 species can be divided into four main types of M2a: (I) M2a is totally two-rowed, including Pleuronema glaciale, P. lynni, P. simplex, and P. wilberti (which, however, can be differentiated from each other by the number of rows in M3) (Figures 8B–E) (Dragesco, 1960; Corliss and Snyder, 1986; Fernandez-Leborans and Novillo, 1994; Wang et al., 2009); (II) the mid-portion of M2a in a zig-zag pattern, including five species, namely P. coronatum, P. foissneri, P. orientale, P. paraorientale, and P. setigerum. Pleuronema setigerum can be easily distinguished by the ring-like posterior end of M2a (Figures 8F–J) (Song, 2000; Pan et al., 2010; Pan et al., 2015; Liu et al., 2022). P. foissneri differs from the other four species by its body shape (anterior end slightly narrowed in P. foissneri vs. anterior end rounded) and the shape of the macronucleus (usually ellipsoidal to spherical in shape, notched in the mid-portion in P. foissneri vs. generally spherical, without any notch) (Figure 8G) (Liu et al., 2022). When compared with P. orientale and P. paraorientale (the anterior end of M2b is conspicuously more anteriorly located than the posterior end of M2a), P. coronatum has an M2b with its anterior end slightly below the level of the posterior end of M2a. Moreover, the primary distinction between P. orientale and P. paraorientale is the number of preoral kineties (2–3 vs. 3–5, respectively) (Figures 8H, I) (Song, 2000; Pan et al., 2015; Liu et al., 2022);
(III) M2a two-rowed with a single-rowed mid-portion that is remarkably short (< 40% of the length of M2a) or long (> 70% of the length of M2a), includes four species, namely P. arctica, P. cazpikae, P. ningboensis, and P. warreni n. sp. Pleuronema arctica and P. cazpikae have an extremely short single-rowed portion (< 20% of the length of M2a) and can be distinguished from each other by the number of rows in M3 (two-rowed vs. three-rowed) (Figures 8R, S) (Agatha et al., 1993; Wang et al., 2008b); P. ningboensis has a single-rowed portion of M2a that is less than 40% of the length of M2a (on average 35%), and a particular M3 (three-rowed, where the leftmost row is shortened) (Figure 8T) (Zhang et al., 2023); and P. warreni n. sp. has a very long single-rowed portion of M2a, starting from the anterior hook-like portion (single-rowed portion > 70% of the length of M2a) (Figure 8U);
(IV) posterior and anterior portions of M2a two-rowed with a single-rowed mid-portion occupying 40–70% of the length of M2a. This group includes seven species, namely P. binucleatum, P. parasalmastra, P. parasmalli, P. paucisaetosum, P. pulchra n. sp., P. puytoraci, and P. salmastra, with a similar M2a structure (the posterior and anterior portions of M2a two-rowed and single-rowed in the mid portion, which occupies 40%–70% of the length of M2a) (Figures 8K–Q) (Dragesco and Dragesco-Kernéis, 1986; Pan et al., 2011; Pan et al., 2015; Pan et al., 2016; Liu et al., 2022). Pleuronema binucleatum, P. parasalmastra, and P. salmastra show obvious differences from the other four species in the number of macronuclei (multiple macronuclei vs. a single macronucleus). Among them, P. binucleatum has two spherical macronuclei, while P. parasalmastra and P. salmastra have 1–12 macronuclei (both species usually have one, with an average of 1.8 in P. parasalmastra and 1.68 on average in P. salmastra). However, Pleuronema parasalmastra is distinguished from P. salmastra by having fewer preoral kineties (4–7 vs. 6–10) (Figures 8K–M) (Dragesco and Dragesco-Kernéis, 1986; Pan et al., 2016; Liu et al., 2022). Pleuronema puytoraci can be distinguished by M2b commencing conspicuously anteriorly than the posterior end of M2a (vs. at the same level as the posterior end of M2a in P. parasmalli, P. paucisaetosum, and P. pulchra n. sp.). Moreover, Pleuronema puytoraci differs from P. pulchra n. sp. by having fewer preoral kineties (1–3 vs. 4–7). Pleuronema parasmalli mainly differs from P. paucisaetosum by having more somatic kineties (26–32 vs. 21–23). When compared with Pleuronema pulchra n. sp., P. parasmalli has fewer somatic kineties (26–32 vs. 32–48). Meanwhile, Pleuronema paucisaetosum also has fewer somatic kineties than P. pulchra n. sp. (21–23 vs. 32–48) (Figures 8N–Q) (Pan et al., 2011; Pan et al., 2015; Liu et al., 2022).
4.2 Phylogenetic analyses
The molecular phylogeny of Pleuronema was first recognized by Baroin-Tourancheau et al. (1992) through the sequencing of the 28S rRNA gene of P. marinum. As phylogenetic studies have progressed, the relationships among members of the Pleuronematidae are becoming clearer (Gao et al., 2013). In recent years, several novel Pleuronema members have undergone comprehensive taxonomy investigations, accompanied by corresponding molecular phylogenetic analyses (Pan et al., 2015; Pan X. et al., 2015; Pan et al., 2016; Liu et al., 2022; Zhang et al., 2023). All the results indicate that the genus Pleuronema is polyphyletic; however, the Schizocalyptra sequences nest within Pleuronema at low support values, so the relationship between Pleuronema and Schizocalyptra is still uncertain.
In the SSU rRNA gene trees, the “marinum-type” SSU rRNA gene sequences of Pleuronema cazpikae EF486863, Pleuronema parawiasckowskii KT033423, and Pleuronema wiackowskii JX310016 fell into the same clade, followed by another clade including Pleuronema marinum KF206428 + Pleuronema sinica EF486864, also of the “marinum-type”. However, in the remaining sequenced members of the genus Pleuronema, the evolutionary relationships among species are inconsistent with this morphological distinction, as evidenced by the fact that two sequences of “marinum-type” species, i.e., Pleuronema elegans KF840518 and Pleuronema grolierei KF840519, cluster within sequences of species of the “coronatum-type”. Therefore, the posterior portion of M2a does not correspond to the phylogenetic analyses of the genus Pleuronema. Unfortunately, almost half of the Pleuronema sequences lack a related morphological description, and their identification needs to be confirmed. Greater taxon sampling and the incorporation of molecular data are essential to improving the phylogenetic resolution within the genus Pleuronema.
The previous phylogenetic analyses indicated a close relationship between Pleuronema and Schizocalyptra, and members of the genus Pleuronema formed one clade including the genus Schizocalyptra (Miao et al., 2008; Fan et al., 2009; Yi et al., 2009; Gao et al., 2013; Gao et al., 2014; Zhang et al., 2019; Zhang and Vďačný, 2021). However, the SSU rRNA gene analyses consistently reveal that the sequences of Pleuronema species fall into two clades with the genus Schizocalyptra nested among them (Pan et al., 2015; Pan X. et al., 2015; Pan et al., 2016; Liu et al., 2022; Zhang et al., 2023). Our phylogenetic results are in agreement with previous studies based on the SSU rRNA gene, with Schizocalyptra nesting with Pleuronema although with low support from ML (18%) (Figure 6). Additional morphological data are needed to shed light on the relationship between Schizocalyptra and Pleuronema and to provide a better resolution of their phylogenetic positions.
In previous phylogenetic analyses of Pleuronematida, the family Pleuronematidae had a close relationship with the family Peniculistomatidae, according to Antipa et al. (2016). Subsequent studies obtained the same results, and Zhang et al. (2019) hypothesized that members of the Peniculistomatidae are supposed to have evolved from Pleuronema-like ancestors (Zhang et al., 2019; Antipa et al., 2020; Liu et al., 2022). The phylogenetic analyses in the present study also show the close relationship between the families Peniculistomatidae and Pleuronematidae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
ZW: Investigation, Visualization, Writing – original draft. ML: Methodology, Resources, Writing – review & editing. TY: Investigation, Resources, Writing – review & editing. XZ: Methodology, Resources, Writing – review & editing. FW: Investigation, Writing – review & editing. YJ: Methodology, Supervision, Writing – review & editing. XC: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundation of China (Project numbers: 32270473, 31970398, and 32100404), the Natural Science Foundation of Shandong Province of China (Project No. ZR2021QC045), and the Postdoctoral Innovation Program of Shandong Province.
Acknowledgments
We greatly appreciate the editor and reviewers for constructive comments and feedback.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agatha S., Spindler M., Wilbert N. (1993). Ciliated protozoa (Ciliophora) from Arctic Sea ice. Acta Protozool. 32, 261–268.
Antipa G. A., Dolan J. R., Lynn D. H., Obolkina L. A., Strüder-Kypke M. C. (2016). Molecular phylogeny and evolutionary relationships between the ciliate genera Peniculistoma and Mytilophilus (Peniculistomatidae, Pleuronematida). J. Eukaryot. Microbiol. 63, 642–650. doi: 10.1111/jeu.12316
Antipa G. A., Strüder-Kypke M. C., Lynn D. H. (2020). Molecular phylogeny, taxonomic relationships and north American distribution of Conchophthirus (Conchophthiridae, Scuticociliatia). Aquat. Ecosyst. Health Manage. 23, 58–68. doi: 10.1080/14634988.2020.1735919
Azovsky A. I., Mazei Y. A. (2018). Diversity and distribution of free-living ciliates from high-Arctic Kara Sea sediments. Protist 169, 141–157. doi: 10.1016/j.protis.2018.01.001
Baroin-Tourancheau A., Delgado P., Perasso R., Adoutte A. (1992). A broad molecular phylogeny of ciliates: identification of major evolutionary trends and radiations within the phylum. Proc. Natl. Acad. Sci. U. S. A. 89, 9764–9768. doi: 10.1073/pnas.89.20.9764
Borror A. C. (1963). Morphology and ecology of the benthic ciliated protozoa of Alligator Harbor, Florida. Arch. Protistenkd. 106, 465–534.
Chi Y., Wang Z., Ye T., Wang Y., Zhao J., Song W., et al. (2022). A new contribution to the taxonomy and phylogeny of the ciliate genus Spirostomum (Alveolata, Ciliophora, Heterotrichea), with comprehensive descriptions of two species from wetlands in China. Water Biol. Secur. 1, 100031. doi: 10.1016/j.watbs.2022.100031
Chorik F. P. (1968). Planktonwimpertiere kleiner Gewässer in Moldavien (Kischinjow: Akademie der Wissenschaften).
Corliss J. O. (1956). On the evolution and systematics of ciliated protozoa. Syst. Zool. 5, 68–91. doi: 10.2307/2411926
Corliss J. O., Snyder R. A. (1986). A preliminary description of several new ciliates from the Antarctica, including Cohnilembus grassei n. sp. Protistologica 22, 39–46.
Dinçer S.Ç. (2016). Freshwater ciliates from Beytepe Pond in Ankara with new records for Turkey. Turk. J. Zool. 40, 663–674. doi: 10.3906/zoo-1508-57
Dragesco J. (1960). Les Ciliés mésopsammiques littoraux (Systématique, morphologie, écologie). Trav. Stat. Biol. Roscoff. 12, 1–356.
Dragesco J. (1968). Les genres Pleuronema Dujardin, Schizocalyptra nov. gen. et Histiobalantium Stokes (ciliés holotriches hyménostomes). Protistologica 4, 85–106.
Dragesco J., Dragesco-Kernéis A. (1986). Ciliés libres de l’Afrique intertropicale: Introduction à la Connaissance et à l’étude des Ciliés. Faune Tropicale, Vol. 26. 1–559. (Paris: Institut de Recherche pour le Développement).
Dujardin F. (1841). Histoire naturelle des Zoophytes: Infusoires, comprenant la physiologie et la classification de ces animaux, et la Manière de les étudier à l’aide du microscope (Paris: Ouvrage Accompagné de Planches).
Fan X., Hu X., Al-Farraj S. A., Clamp J. C., Song W. (2011). Morphological description of three marine ciliates (Ciliophora, Scuticociliatia), with establishment of a new genus and two new species. Eur. J. Protistol. 47, 186–196. doi: 10.1016/j.ejop.2011.04.001
Fan X., Miao M., Al-Rasheid K. A., Song W. (2009). A new genus of marine scuticociliate (Protozoa, Ciliophora) from northern China, with a brief note on its phylogenetic position inferred from small subunit ribosomal DNA sequence data. J. Eukaryot. Microbiol. 56, 577–582. doi: 10.1111/j.1550-7408.2009.00436.x
Fernandez-Leborans G., Novillo A. (1994). Morphology and taxonomic position of two marine pleuronematine species: Pleuronema lynni and Schizocalyptra marina (Protozoa, Ciliophora). J. Zoo. Lond. 233, 259–275. doi: 10.1111/j.1469-7998.1994.tb08587.x
Foissner W., Berger H., Kohmann F. (1994). Taxonomische und ökologische revision der Ciliaten des Saprobiensystems – Band III: Hymenostomata, Prostomatida, Nassulida Vol. 1 (Informationsber. Bayer. Landesamtes Wasserwirtschaft), 1–548.
Gajewskaja N. (1933). Zur Ökologie, morphologie und systematik der infusorien des baikalsees. Zoologica 32, 1–298.
Gao F., Gao S., Wang P., Katz L. A., Song W. (2014). Phylogenetic analyses of cyclidiids (Protista, Ciliophora, Scuticociliatia) based on multiple genes suggest their close relationship with thigmotrichids. Mol. Phylogenet. Evol. 75, 219–226. doi: 10.1016/j.ympev.2014.01.032
Gao F., Huang J., Zhao Y., Li L., Liu W., Miao M., et al. (2017). Systematic studies on ciliates (Alveolata, Ciliophora) in China: progress and achievements based on molecular information. Eur. J. Protistol. 61, 409–423. doi: 10.1016/j.ejop.2017.04.009
Gao F., Katz L. A., Song W. (2013). Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol. 68, 55–63. doi: 10.1016/j.ympev.2013.03.018
Gao F., Warren A., Zhang Q., Gong J., Miao M., Sun P., et al. (2016). The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 6, 24874. doi: 10.1038/srep24874
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.12691/ajmr-3-2-1
Hao T., Song Y., Li B., Pan X. (2022). Morphology and molecular phylogeny of three freshwater scuticociliates, with establishments of one new genus and three new species (Ciliophora, Oligohymenophorea). Eur. J. Protistol. 86, 125918. doi: 10.1016/j.ejop.2022.125918
Jerome C. A., Simon E. M., Lynn D. H. (1996). Description of Tetrahymena empidokyrea n. sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can. J. Zool. 74, 1898–1906. doi: 10.1139/z96-214
Jiang L., Wang C., Zhuang W., Li S., Hu X. (2021). Taxonomy, phylogeny, and geographical distribution of the little-known Helicoprorodon multinucleatum Dragesco 1960 (Ciliophora, Haptorida) and key to species within the genus. Eur. J. Protistol. 78, 125769. doi: 10.1016/j.ejop.2021.125769
Kahl A. (1926). Neue und wenig bekannte Formen der holotrichen und heterotrichen Ciliaten. Arch. Protistenkd. 55, 197–438.
Kahl A. (1931). Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) 2. Holotricha. Tierwelt Dtl. 21, 181–398.
Kalinowska K. (2013). Community structure of psammon ciliates in sandy beaches of lakes. Oceanol. Hydrobiol. Stud. 42, 14–21. doi: 10.2478/s13545-013-0051-5
Kent W. S. (1881). A manual of the Infusoria: Including a description of all known flagellate, ciliate, and tentaculiferous Protozoa, British and foreign, and an account of the organization and affinities of the sponges (London, UK: David Bogue).
Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu M., Liu Y., Zhang T., Lu B., Gao F., Gu J., et al. (2022). Integrative studies on the taxonomy and molecular phylogeny of four new Pleuronema species (Protozoa, Ciliophora, Scuticociliatia). Mar. Life Sci. Technol. 4, 179–200. doi: 10.1007/s42995-022-00130-5
Lynn D. H. (2008). The ciliated Protozoa: Characterization, classification, and guide to the literature, 3rd Edn (Dordrecht: Springer). doi: 10.1007/978-1-4020-8239-9
Medlin L., Elwood H. J., Stickel S., Sogin M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Miao M., Warren A., Song W., Wang S., Shang H., Chen Z. (2008). Analysis of the internal transcribed spacer 2 (ITS2) region of scuticociliates and related taxa (Ciliophora, Oligohymenophorea) to infer their evolution and phylogeny. Protist 159, 519–533. doi: 10.1016/j.protis.2008.05.002
Nylander J. A. A. (2004). MrModeltest (Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University).
Page R. D. M. (1996). Treeview: an application to display phylogenetic trees on personal computers. Bioinformatics 12, 357–358. doi: 10.1093/bioinformatics/12.4.357
Pan H., Hu J., Jiang J., Wang L., Hu X. (2016). Morphology and phylogeny of three Pleuronema species (Ciliophora, Scuticociliatia) from Hangzhou Bay, China, with description of two new species, P. binucleatum n. sp. and P. parawiackowskii n. sp. J. Eukaryot. Microbiol. 63, 287–298. doi: 10.1111/jeu.12277
Pan H., Hu J., Warren A., Wang L., Jiang J., Hao R. (2015a). Morphology and molecular phylogeny of Pleuronema orientale spec. nov. and Pleuronema paucisaetosum spec. nov. (Ciliophora, Scuticociliata) from Hangzhou Bay, China. Int. J. Syst. Evol. Microbiol. 65, 4800–4808. doi: 10.1099/ijsem.0.000651
Pan H., Huang J., Hu X., Fan X., Al-Rasheid K. A., Song W. (2010). Morphology and SSU rRNA gene sequences of three marine ciliates from Yellow Sea, China, including one new species, Uronema heteromarinum nov. spec. (Ciliophora, Scuticociliatida). Acta Protozool. 49, 45–59.
Pan X., Huang J., Fan X., Ma H., Al-Rasheid K. A. S., Miao M., et al. (2015b). Morphology and phylogeny of four marine scuticociliates (Protista, Ciliophora), with descriptions of two new species: Pleuronema elegans spec. nov. and Uronema orientalis spec. nov. Acta Protozool. 54, 31–43. doi: 10.4467/16890027AP.15.003.2190
Pan X., Shao C., Ma H., Fan X., Al-Rasheid K. A., Al-Farraj S. A., et al. (2011). Redescriptions of two marine scuticociliates from China, with notes on stomatogenesis in Parauronema longum (Ciliophora, Scuticociliatida). Acta Protozool. 50, 301–310. doi: 10.4467/16890027AP.11.027.0064
Pätsch B. (1974). Die Aufwuchsciliaten des Naturlehrparks Haus Wildenrath. Monographische Bearbeitung der Morphologie und Ökologie. Arb. Inst. Landw. Zool. Bienenkd. Universität Bonn 1, 1–82.
Penn O., Privman E., Ashkenazy H., Landan G., Graur D., Pupko T. (2010). GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38, W23–W28. doi: 10.1093/nar/gkq443
Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Roux J. (1901). Faune infusorienne des eaux stagnates des environs de Genève. Mem. Inst. natn. génev. 19, 1–148.
Small E. B. (1964). An analysis of morphology and ontogenesis in the genus Pleuronema Dujardin 1841, a description of two new species and a new genus, and a comparative analysis of stomatogenesis (Los Angeles: University of California).
Small E. B. (1967). The Scuticociliatida, a new order of the class Ciliatea (phylum Protozoa, subphylum Ciliophora). Trans. Am. Microsc. Soc. 86, 345–370. doi: 10.2307/3224258
Song W. (2000). Morphological and taxonomical studies on some marine scuticociliates from China Sea, with description of two new species, Philasterides armatalis sp. n. and Cyclidium varibonneti sp. n. (Protozoa: Ciliophora: Scuticociliatida). Acta Protozool. 39, 295–322.
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stock A., Breiner H. W., Pachiadaki M., Edgcomb V., Filker S., Cono V. L., et al. (2012). Microbial eukaryote life in the new hypersaline deep-sea basin Thetis. Extremophiles 16, 21–34. doi: 10.1007/s00792-011-0401-4
Vuxanovici A. (1960). Noi contributii la studiul ciliatelor dulcicole din Republica Popularä Rominä (Nota I). Studii Cerc. Biol. 12, 355–381.
Wang Y., Hu X., Long H., Al-Rasheid K. A. S., Al-Farraj S. A., Song W. (2008a). Morphological studies indicate that Pleuronema grolierei nov. spec. and P. coronatum Kent 1881 represent different sections of the genus Pleuronema (Ciliophora: Scuticociliatida). Eur. J. Protistol. 44, 131–140. doi: 10.1016/j.ejop.2007.08.008
Wang Y., Song W., Hu X., Warren A., Chen X., Al-Rasheid K. A. S. (2008b). Descriptions of two new marine species of Pleuronema, P. czapikae sp. n. and P. wiackowskii sp. n. (Ciliophora: Scuticociliatida), from the Yellow Sea, North China. Acta Protozool. 47, 35–45. doi: 10.1016/j.ejop.2008.06.001
Wang Y., Song W., Warren A., Al-Rasheid K. A. S., Al-Quraishy S. A., Al-Farraj S. A., et al. (2009). Descriptions of two new marine scuticociliates, Pleuronema sinica n. sp. and P. wilberti n. sp. (Ciliophora: Scuticociliatida), from the Yellow Sea, China. Eur. J. Protistol. 45, 29–37. doi: 10.1016/j.ejop.2008.06.001
Whang I., Kang H. S., Lee J. (2013). Identification of scuticociliates (Pseudocohnilembus persalinus, P. longisetus, Uronema marinum and Miamiensis avidus) based on the cox1 sequence. Parasitol. Int. 62, 7–13. doi: 10.1016/j.parint.2012.08.002
Wilbert N. (1975). Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos 64, 171–179.
Ye T., Jiang Y., Chen S., Xu Y., Li L., Shin M. K., et al. (2022). The widely reported but poorly studied ciliate family Folliculinidae (Protozoa, Ciliophora, Heterotrichea): a revision with notes on its taxonomy, morphology and phylogenetic relationships. Mar. Life Sci. Technol. 4, 471–492. doi: 10.1007/s42995-022-00152-z
Yi Z., Song W., Gong J., Warren A., Al-Rasheid K. A. S., Al-Arifi S., et al. (2009). Phylogeny of six oligohymenophoreans (Protozoa, Ciliophora) inferred from small subunit rRNA gene sequences. Zool. Scr. 38, 323–331. doi: 10.1111/j.1463-6409.2008.00371.x
Yi Z., Wang Y., Lin X., Al-Rasheid K. A., Song W. (2010). Phylogeny of subclass Scuticociliatia (Protozoa, Ciliophora) using combined data inferred from genetic, morphological, and morphogenetic evidence. Chin. J. Oceanol. Limnol. 28, 778–784. doi: 10.1007/s00343-010-9100-8
Zhang T., Fan X., Gao F., Al-Farraj S. A., El-Serehy H. A., Song W. (2019). Further analyses on the phylogeny of the subclass Scuticociliatia (Protozoa, Ciliophora) based on both nuclear and mitochondrial data. Mol. Phylogenet. Evol. 139, 106565. doi: 10.1016/j.ympev.2019.106565
Zhang T., Vďačný P. (2021). Re-discovery and novel contributions to morphology and multigene phylogeny of Myxophyllum steenstrupi (Ciliophora: Pleuronematida), an obligate symbiont of terrestrial pulmonates. Zool. J. Linn. Soc 192, 1–23. doi: 10.1093/zoolinnean/zlaa095
Zhang H., Zhao X., Ye T., Wu Z., Wu F., Chen X., et al. (2023). Taxonomic and phylogenetic studies of two brackish Pleuronema species (Protista, Ciliophora, Scuticociliatia) from subtropical coastal waters of China, with report of a new species. Microorganisms 11, 1422. doi: 10.3390/microorganisms11061422
Zhao X., Zhang H., Zhang Q., Qu Z., Warren A., Wu D., et al. (2022). A case study of the morphological and molecular variation within a ciliate genus: taxonomic descriptions of three Dysteria species (Ciliophora, Cyrtophoria), with the establishment of a new species. Int. J. Mol. Sci. 23, 1764. doi: 10.3390/ijms23031764
Keywords: ciliated protozoa, morphology, new taxa, pleuronematids, scuticociliates, SSU rRNA gene sequence
Citation: Wu Z, Liu M, Ye T, Zhao X, Wu F, Jiang Y and Chen X (2023) New contributions to the taxonomy and phylogeny of the ciliate genus Pleuronema (Ciliophora, Scuticociliatia), with descriptions of two new species collected from the subtropical coastal wetlands in China. Front. Mar. Sci. 10:1320684. doi: 10.3389/fmars.2023.1320684
Received: 12 October 2023; Accepted: 04 December 2023;
Published: 22 December 2023.
Edited by:
Xiaofeng Lin, Xiamen University, ChinaReviewed by:
Xinpeng Fan, East China Normal University, ChinaHongbo Pan, Shanghai Ocean University, China
Copyright © 2023 Wu, Liu, Ye, Zhao, Wu, Jiang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangrui Chen, Y2hlbnhpYW5ncnVpQG5idS5lZHUuY24=
†These authors have contributed equally to this work
 Zehao Wu
Zehao Wu Mingjian Liu
Mingjian Liu Tingting Ye4
Tingting Ye4 Xiangrui Chen
Xiangrui Chen