- 1Department of Microbiology, Cornell University, Ithaca, NY, United States
- 2Division of Commercial Fisheries, Alaska Department of Fish and Game, Juneau, AK, United States
- 3Department of Biology, Simon Fraser University, Vancouver, BC, Canada
- 4Department of Applied Fisheries, University of Alaska Southeast, Sitka, AK, United States
- 5Sciences and Mathematics, Division of School of Interdisciplinary Arts and Sciences, University of Washington, Tacoma, WA, United States
Flaviviruses cause some of the most detrimental vertebrate diseases, yet little is known of their impacts on invertebrates. Microbial activities at the animal-water interface are hypothesized to influence viral replication and possibly contribute to pathology of echinoderm wasting diseases due to hypoxic stress. We assessed the impacts of enhanced microbial production and suboxic stress on Apostichopus californicus associated flavivirus (PcaFV) load in a mesocosm experiment. Organic matter amendment and suboxic stress resulted in lower PcaFV load, which also correlated negatively with animal mass loss and microbial activity at the animal-water interface. These data suggest that PcaFV replication and persistence was best supported in healthier specimens. Our results do not support the hypothesis that suboxic stress or microbial activity promote PcaFV replication, but rather that PcaFV appears to be a neutral or beneficial symbiont of Apostichopus californicus.
Introduction
Over two decades of research have highlighted the crucial importance of viruses to aquatic ecosystem function, where they cause significant microbial mortality (Bergh et al., 1989; Suttle et al., 1990; Fuhrman, 1999). High throughput sequencing technologies have enabled discovery of a vast diversity of viruses associated with marine invertebrates through metagenomic and transcriptomic approaches (Breitbart et al., 2002; Ng et al., 2009; Thurber et al., 2009; Shi et al., 2016). These studies have widened viral family host ranges and provided insight into viral evolutionary history, circumventing cell culture biases which affect the majority of invertebrate taxa (Holmes, 2016). The viral metagenome approach and mining host transcriptomes (e.g., RNASeq) has also identified putative agents of invertebrate holobiont abnormalities (Hewson et al., 2014; Soffer et al., 2014; Work et al., 2021; Grupstra et al., 2022), and the detection of metazoan viruses that may be shed into overlying waters or surrounding benthic habitats (Allen et al., 2017; Wolf et al., 2020; Dominguez-Huerta et al., 2022; Solomon and Hewson, 2022; Zayed et al., 2022). Despite the increasing application of metagenomic approaches to metazoan-associated viruses, few studies have investigated the autecology of novel viruses in concert with host biology (e.g., growth, death, and physiology).
Over a decade of scientific investigation failed to identify a single microbial agent associated with Asteroid Idiopathic Wasting (AIW; also known as Sea Star Wasting Disease) (Mccracken et al., 2023). A wide diversity of DNA and RNA viruses were identified in metagenomic survey of grossly normal and AIW-affected specimens (Hewson et al., 2014; Hewson et al., 2018; Hewson et al., 2020a; Jackson et al., 2020). Aquino et al. (2021) associated AIW with microbial activities on exposed tissues at the animal-water interface. They found signs consistent with AIW could be elicited by suboxic conditions in aquaria, and also by organic substrate amendment that increased bacterial abundance (and presumably respiration) on their exposed surfaces. Importantly, viral richness increased in tissues over time as animals transitioned from grossly normal to abnormal (Hewson et al., 2020a). Another study of the asteroid Pycnopodia helianthoides during challenge experiment with tissue homogenates filtered through 0.2 µm filters found that the sea star associated densovirus (SSaDV) increased in abundance when the host was incubated with tissue homogenates of the sea star (Hewson et al., 2014). Yet, SSaDV was not associated with AIW symptoms in a wide suite of specimens (Hewson et al., 2018). This observation supports that enhanced microbial activity stimulated by tissue homogenates may lead to greater replication of SSaDV and other viruses. Because enhanced microbial activity may reduce O2 concentrations at the animal-water interface, it is possible that suboxic conditions may lead to viral proliferation. Indeed, several viruses replicate faster under hypoxic host cell conditions in culture by hijacking hypoxia inducible factors (Vassilaki and Frakolaki, 2017; Frakolaki et al., 2018). Whether enhanced viral load enhances the effects of hypoxic stress on tissues or through buildup of toxic compounds under suboxic conditions (Alessandra et al., 2003) remains unclear.
A metagenomic study of grossly abnormal Apostichopus californicus (Giant Pacific Sea Cucumber; formerly Parastichopus californicus) from Ketchikan, Alaska, United States in 2016 revealed the presence of a flavivirus, which was among the first enveloped RNA virus ever recovered in surveys of aquatic invertebrates (Hewson et al., 2020b). Flaviviridae are pathogens of humans and other vertebrates, but also include related families that infect invertebrates and plants. The majority of invertebrate flaviviruses do not have defined pathogenesis, and indeed there are few studies of entry, replication, and egress of invertebrate flaviviruses in living hosts. The Apostichopus californicus associated flavivirus (PcaFV) was most closely related to several genomes recovered during RNA viral surveys of a tunicate (NCBI Genbank accession CAI5758347), a shark (the Wenzhou Shark flavivirus) and a crab (Portunus trituberculatus) in China, discovered through whole organism transcriptomes (Shi et al., 2016). Together with these close relatives, this virus forms a clade of viruses together with the Tamana Bat virus, which was recovered from bat guano in a cave in Trinidad (De Lamballerie et al., 2002; Hewson et al., 2020b). This group of “Aquatic Invertebrate Flaviviruses” (aiFVs) includes members that have been recovered from fish and mollusks (Parry and Asgari, 2019), suggesting that it has evolved in marine habitats separately from insect or other terrestrial invertebrate flaviviruses. The virology of aiFVs is not yet known since their recovery comes from single timepoint genomic/transcriptomic surveys. Despite this, aiFVs may be useful markers to examine how extracellular hypoxia may affect viral load in invertebrate tissues since they are easily distinguishable from other RNA viruses which occur widely in echinoderm tissues (Jackson et al., 2018; Hewson et al., 2020b).
In this study, we tested the hypothesis that water column and animal surface hypoxia positively affect PcaFV load in tissues by reducing oxygen concentrations and enriching organic matter amendment in experimental mesocosms. Enrichment of organic matter stimulates microbial activity, which we hypothesized would lead to microbiome variation favoring facultative and strict anaerobes, and that depleted O2 would likewise favor bacteria with anaerobic metabolisms. We found that suboxic conditions and organic matter amendment had no impact on PcaFV load within tissues, but rather that PcaFV load correlates strongly and positively with various indicators of animal health.
Materials and methods
Specimen collection to assess PcaFV tissue distribution
Sea cucumbers were collected at 2 locations (Juneau, Alaska and Friday Harbor, Washington, USA) to assess the presence and load of PcaFV and other aquatic invertebrate flaviviruses (Supplemental Table 1). Twenty individual Apostichopus californicus specimens were collected by SCUBA divers during routine fisheries surveys by Alaska Department of Fish and Game from near southeastern Alaska in June/July 2021. Specimens were immediately placed into a refrigerated hold on the ship in zip-lock bags before transport to the lab in Juneau later in the day. There, specimens were frozen at -20°C and couriered overnight to Cornell University (Ithaca, NY, USA) where they were frozen at -80°C on arrival. Specimens (n = 9) from the Friday Harbor Laboratory were collected by dredge in waters adjacent to the lab and frozen immediately at -80°C before transport to the laboratory at Cornell University. Additionally, samples of A. californicus with grossly abnormal lesions (n = 5) were collected from Nanoose Bay, British Columbia, Canada on 1 September 2021 (Supplemental Table 1; Lim et al., 2023). Samples were refrigerated upon collection and frozen at -20°C before being couriered to Cornell University. In addition to A. californicus, 20 specimens of Cucumaria miniata from the Juneau sampling in June 2021 were also collected and processed as described above.
Prior to dissection, sea cucumber carcasses were thawed at room temperature until they were pliable to touch. Small subsamples (~ 200 mg) were removed from the dorsal and ventral body wall, longitudinal muscle, oral tentacle, respiratory tree, and digestive tract, and immediately placed into 2 mL Bead Basher tubes (Zymo Research) and frozen at -80°C. RNA was extracted from all specimens in this study using the Zymo Tissue and Insect RNA kit following manufacturer’s protocols. Extracted RNA was quantified on an ND-1000 Nanodrop spectrophotometer.
Reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) of PcaFV and comparison to qRT-PCR
RT-LAMP was performed in duplicate in 25 µL reactions containing 1X WarmStart Colorimetric RT-LAMP Master Mix (New England Biolabs), 0.5 µL of fluorescent dye, 2.5 µL primers designed using the New England BioLabs primer tool around a portion of the PcaFV NS5 amplicon (GenBank accession MT949664), and 5 µL of extracted RNA. The primer master mix comprised 16 μL of the 100uM PcaFV_NS5_FIP and PcaFV-NS5_BIP primers, 2 μL of 100uM PcaFV_NS5_F3 and PcaFV_NS5_B3 primers, 4 μL of the 100uM PcaFV_NS5_LoopF and PcaFV_NS5_Loop B primers (Supplemental Table 2), as well as 56 μL of nuclease-free water (diH2O). Reactions were performed in optically clear quantitative PCR tubes (Applied Biosystems). Negative controls comprised 5 μL of diH2O, while positive controls used 5 μL of PcaFV synthetic gene fragment. The tube strips were incubated in a water bath at 65°C for 30 min. After incubation, tubes were removed from the bath and scored based on color change. Positive detections were scored when the reaction appeared yellow and negative detections were scored when the reaction appeared red. Positive controls were consistently red, and negative controls were consistently yellow. When positive controls had not turned yellow, reactions were further incubated for 15 min in the bath, and then reassessed. Possible detections were scored when the reactions turned orange. Fifty-two RNA extracts which represented positive detection (RT-LAMP reactions appeared yellow), possible detection (RT-LAMP reactions appeared orange) and no detection (RT-LAMP reactions appeared red) were further subject to qRT-PCR using primers PcaFV_NS5_F3, PcaFV_NS5_R3, and PcaFV_NS5_Pr probe (see below).
Organic matter amendment and depleted oxygen experimental design
Forty-two specimens of Apostichopus californicus were collected by SCUBA divers in Thimbleberry Bay (57.0297N, 135.2283W), near Sitka, Alaska on 10 November 2021 and transported together in plastic tubs to the lab at the University of Alaska Southeast (Japonski Island, Sitka). Specimens were immediately weighed, photographed, and placed into individual mesh containers within 7 x 1200 L outdoor mesocosms (6 specimens per mesocosm) filled with seawater from the nearby Sitka Channel. Two mesocosms served as controls (no amendment), 4 mesocosms were subject to daily organic matter (20 µM) amendment, and 1 mesocosm was continuously sparged with N2 (Airgas, medical grade; the other 6 mesocosms were bubbled continuously with air). Seawater was subject to 50% volume water change daily and specimens were not fed during captivity. Mesocosms were covered while not sampled (i.e., they were light limited). We selected two organic matter substrates (glucose and peptone) based on their ability to stimulate microbial activity in prior work (Bianchi et al., 1998) in addition to two common constituents of dissolved organic matter in coastal environments (N-acetylglucosamine and fucose + rhamnose). We monitored dissolved oxygen levels in each mesocosm using continuous submersible HOBO loggers.
Tissue sampling
Surface swabs of each individual were collected daily by rubbing a Puritan polyester sterile swab over a ~ 1 cm2 area of epidermis. Swabs were cut using clean scissors to remove the polyester tip and placed into 2 mL cryovials containing RNALater. Tube feet samples were collected from each specimen daily using disposable plastic forceps, and feet were placed immediately into cryovials containing RNALater. A 5mm biopsy punch of the dorsal body wall was collected from half of the individuals in each mesocosm at 0, 1, 3, and 6 d, which were then preserved in RNALater. All specimens were photographed and weighed daily. All RNALater preserved samples were frozen at -80°C and transported in liquid N2 to the laboratory at Cornell University. Animal carcasses were frozen at -20°C and transported on blue ice to Cornell.
Quantitative reverse transcriptase PCR of PcaFV
Because metagenomic detection of PcaFV in Apostichopus californicus was based on body wall homogenates (Hewson et al., 2020a), we first sought to understand tissue tropism of this virus. We assessed two molecular biology approaches to detect and quantify PcaFV in sea cucumber tissues based on reverse transcriptase loop mediated isothermal amplification (RT-LAMP) and quantitative reverse transcriptase PCR (qRT-PCR) and applied these to several tissue samples from field-collected specimens (Supplemental Materials). Based on these results, we used qRT-PCR to detect PcaFV in specimens obtained during the suboxia and organic matter amendment experiment.
Quantitative reverse transcriptase PCR (qRT-PCR) using primers PcaFV_NS5_F3 (5’- CCA GCC ATG GAT GAG TAA TG-3’) and PcaFV_NS5_R3 (5’- GCT GAA CTG CTC CTG AAA CC-3’), and probe PcaFV_NS5_Pr3 (5’-[FAM]CAC GAA TGT ACG GCA ACG GAC G[TAMRA]-3’) was used to determine PcaFV load within individual tissue RNA extracts. qRT-PCR reactions were performed in duplicate for each specimen and compared against duplicate reactions of oligonucleotide standards spanning the amplicon region (from 102 to 107 copies µl-1) and using 4 negative controls (nuclease free H2O only). PCR reactions (20 µL) contained 1 X Luna Universal Probe One-Step Reaction Mix (New England Biolabs), 1X Luna WarmStart RT Enzyme Mix (New England Biolabs), 8 µmol each of primers and probes, and 1 µL of template RNA. Reactions were thermal cycled in an ABI StepOne qPCR machine. Reactions were initially held at 55°C for 10 min, followed by an initial denaturation step at 95°C for 1 min. Reactions were then subject to 45 cycles of denature at 95°C and anneal at 56°C, with fluorescence data collected after the annealing step. Linearity of standards was checked during downstream analysis of qPCR data.
Relative microbial activity via bromodeoxyuridine incorporation
Relative microbial activity was measured in each mesocosm and from the surface of 2 specimens per mesocosm by bromodeoxyuridine (BrDU) incorporation as described previously (Nelson and Carlson, 2005). Five 2 mL water samples were retrieved from each mesocosm by pipette and placed into 2 ml cryovials. Water samples (5 replicates) were collected from the surface of each of two haphazardly chosen specimens by pressing the pipette tip against the dorsal epidermis and slowly retrieving sample, which was then placed into 2 mL cryovials. Water samples were inoculated with 0.04 nmol BrDU and incubated in the mesocosm for 8 h. Incubations were then retrieved, and flash frozen in liquid N2.
A standard for comparison between BrDU blots was made by amending 50 mL of water collected from an aquarium with 0.04 nmol BrDU. A single sheet of chromatography paper was placed in a Tupperware container soaked with 6X SSC Buffer with a nylon transfer membrane on top and allowed to soak for 10 minutes. The nylon transfer filter was then mounted in a slot blot manifold. Samples (0.5 mL) were pipetted into slot wells, where each membrane included 6 standards, then vacuum filtered through the membrane. The membrane was placed sample side down onto a stack of 3 sheets of chromatography paper with the lysis solution, then immediately turned sample side up and incubated for 10 min. This was repeated in the same fashion for neutralization solution. The membrane was then placed sample-side down on a piece chromatography paper soaked in FixDenat for 15 s, followed by 30 min incubation on the FixDenat stack. The membrane was placed between two sheets of chromatography paper and baked at 80°C for ~1 h.
The dried membrane was placed in a plastic tray, 25 mL Blocking Solution was added, and the apparatus incubated on a rotating incubator at 30 rev min-1 for 1 h. After this time, the blocking solution was poured off and 6 mL of Antibody Incubation Solution was added to the tray, which was placed on a rotating incubator at 30 rev min-1 for 3 h. The antibody incubation solution was poured off and 25 mL wash solution was added, which was incubated on rotating incubator for 5 min. The wash step was repeated. After washing, the wash solution was poured off, and 25 mL of maleic acid buffer was added and incubated on rotating incubator for 5 min. This maleic acid buffer treatment then was repeated. The nylon transfer membrane was placed in a plastic tray, sample side up, to which 4 mL of the Supersignal West Femto was added and incubated for 2 minutes. The membrane was visualized in a BioRad ChemiDoc system at high sensitivity and 4 x 4 binning with an exposure time of 600 s.
Tissue protein content
Protein content was measured in body wall tissues using RIPA buffer extraction and Bradford reagent spectrophotometry. Two biopsy punch (5 mm) tissue samples were harvested from frozen animal carcasses and placed into 2 mL microcentrifuge tubes. RIPA buffer (2 mL; 150mM NaCl, 10 mM Tris, 1 mM EDTA, 1% Triton X-100, 0.01% sodium doecyl sulfate, and 0.01% sodium deoxycholate) was added to each sample, and samples were homogenized in a mortar and pestle for 2 min or until the tissue was completely homogenized. Samples were incubated at room temperature in a shaking incubator for 2 h. Following this, tubes were centrifuged at 16,000 x g at 4°C for 20 min, and supernatant removed into a new microcentrifuge tube. Protein quantity was determined against a bovine serum albumin standard following the approach of (Gotham et al., 1988). Bradford reagent (1500 µL) was added to 30 µL sample, allowed to incubate at room temperature for 15 min, then absorbance at 595 nm measured on a spectrophotometer.
Tissue lipid content
Lipid content was assessed following the approach of Lee et al. (1996). Two biopsy punch (5 mm) tissue samples were harvested from animal carcass dorsal body wall and placed into 15 mL centrifuge tubes. Tissue samples were weighed, amended with 12 mL chloroform:methanol, then homogenized in a mortar and pestle until the tissue had completely disintegrated and resembled a paste. The homogenates were centrifuged at 3,000 x g for 10 min in a benchtop centrifuge and the solvent fraction removed into fresh centrifuge tubes. Three microcentrifuge tubes per sample were weighed, and then 2 mL of the solvent fraction from each tissue sample homogenate was placed into the tubes. Tubes were then placed into a rotating vacuum desiccator until they were dry. Sample tubes were then re-weighed, and mass difference calculated on aggregate for all replicate tubes per sample as a percentage of wet weight.
Statistical analyses
All statistical analyses of significance on tissue biochemistry, microbial activity, animal mass loss, and PcaFV load were performed in XLStat (Addinsoft SARL). Normality of data distribution was tested for each parameter by the Shapiro-Wilk test. With the exception of mass loss and PcaFV load, all other parameters did not follow a normal distribution. Hence in all downstream comparisons we used nonparametric tests of association. A correlation matrix was calculated across all variables by Kendall statistic. The effects of treatment on individual parameters were compared by Kruskal-Wallis tests.
16S rRNA amplicon sequencing and bioinformatic analysis
Surface swabs were collected from 42 animals over the course of 7 d (sampled on t = 0 d, 1 d, 3 d, 5 d, and 7 d) and frozen at -20°C until further processing. DNA was extracted from frozen swabs using Zymo Quick-DNA Fungal/Bacterial kits (Zymo Research) as per the manufacturer’s protocol. Bacterial communities in sample extracts were identified using dual-barcoded PCR (polymerase chain reaction) amplification and sequencing of the V4 region of the 16S rRNA gene (Kozich et al., 2013; Apprill et al., 2015; Parada et al., 2016). Each 40 µL PCR reaction comprised 1X PCR master mix (One-Taq Quick-Load 2x Master Mix with Standard; New England Biolabs), 0.125 µM of each barcoded primer (515f; 5’-GTG YCA GCM GCC GCG GTA A-3’ and 806r; 5’-GGA CTA CNV GGG TWT CTA AT-3’), and 2 uL of template (swab extract). 16S rRNA amplicons were pooled at equimolar concentrations using SequalPrep Normalization Plate kit (Invitrogen) and sequenced on one lane of Illumina MiSeq (2 x 250 paired end) at the Cornell University Biotechnology Research Center. 16S rRNA amplicon sequences were submitted to NCBI (BioProject accession number PRJNA947521).
The raw sequencing data were processed in QIIME2 (v. 2022.8) (Bolyen et al., 2019) where the following files were generated - an amplicon sequence variant (ASV) table, phylogenetic tree, and taxonomy table. After importing the raw sequences, the forward and reverse reads were joined using the vsearch join-pairs command (Rognes et al., 2016). Joined reads were then quality filtered using quality-filter q-score and denoised using the Deblur plugin to create an ASV table (Amir et al., 2017). A Deblur trim length of 250 nt was chosen based on Phred quality. A phylogenetic tree was then constructed using the SEPP q2-fragment-insertion plugin with the Greengenes 13-8 reference database (Janssen et al., 2018), and taxonomic assignments were made using the SILVA database 138 release and a naive bayes classifier plugin trained on the V4 hypervariable region of the 16s gene (Bokulich et al., 2018).
All 16S rRNA analyses were performed using the R statistical language (v. 4.2.1) (R Core Team, 2022). The ASV table, phylogenetic tree, and taxonomy table were imported using qiime2r (v. 0.99.6) (Bisanz, 2018) and combined into a single object using phyloseq (v. 1.40.0) (Mcmurdie and Holmes, 2013). Alpha diversity analysis was calculated using Shannon diversity and chao1 richness indexes phyloseq (v. 1.40.0) and statistical comparison was done using a Wilcox using ggpubr (0.6.0). Beta diversity was calculated using weighted UniFrac distances and ordinated using a principal coordinate analysis (Lozupone and Knight, 2005). Statistical testing on weighted UniFrac distances was performed with a permutational analysis of variance (PERMANOVA) using the adnois2 function in the Vegan package (v. 2.6-4) (Dixon, 2003). Differential abundance was determined using the Phylofactor package (v. 0.0.1) (Washburne et al., 2017).
Results
Detection method validation and tissue distribution
We developed a reverse-transcriptase isothermal amplification (RT-LAMP; Notomi et al., 2000) approach to rapidly screen RNA extracts for PcaFV across tissue types. The RT-LAMP approach was validated by PCR amplification and sequencing of a region within RT-LAMP amplicons which matched 100% nucleotide identity with the target sequence, and by comparing RT-LAMP detections with PcaFV copy number, determined by quantitative reverse transcriptase PCR (qRT-PCR; Supplemental Figure 1). While PcaFV load (via qRT-PCR) of possible detection, combined possible, and negative detections were not significantly different from negative detections, positive PcaFV detections via RT-LAMP had significantly higher PcaFV copies than both negative detections (p = 0.040, ANOVA) and possible detections (p = 0.04, ANOVA) (Supplemental Figure 1).
RT-LAMP detected PcaFV in 50% of Juneau specimens and 78% of Friday Harbor specimens, but not in any of the 15 tissue samples collected from 5 grossly abnormal specimens from Nanoose Bay, nor in any Cucumaria miniata tissues from Juneau in June 2021 (Supplemental Table 1). PcaFV was detected across all tissue types sampled but in varying prevalence. In both Juneau and Friday Harbor, PcaFV was most consistently in ventral body wall samples (Supplemental Figure 2; ns between tissue types; Fisher’s Exact Test). Quantitative PCR across tissue types revealed the greatest load in body wall specimens, followed by respiratory tree, but lowest abundance in gonad and digestive tract samples (p<0.001 for all comparisons, Kruskal-Wallis test).
Impact of organic matter amendment and suboxic conditions on A. californicus
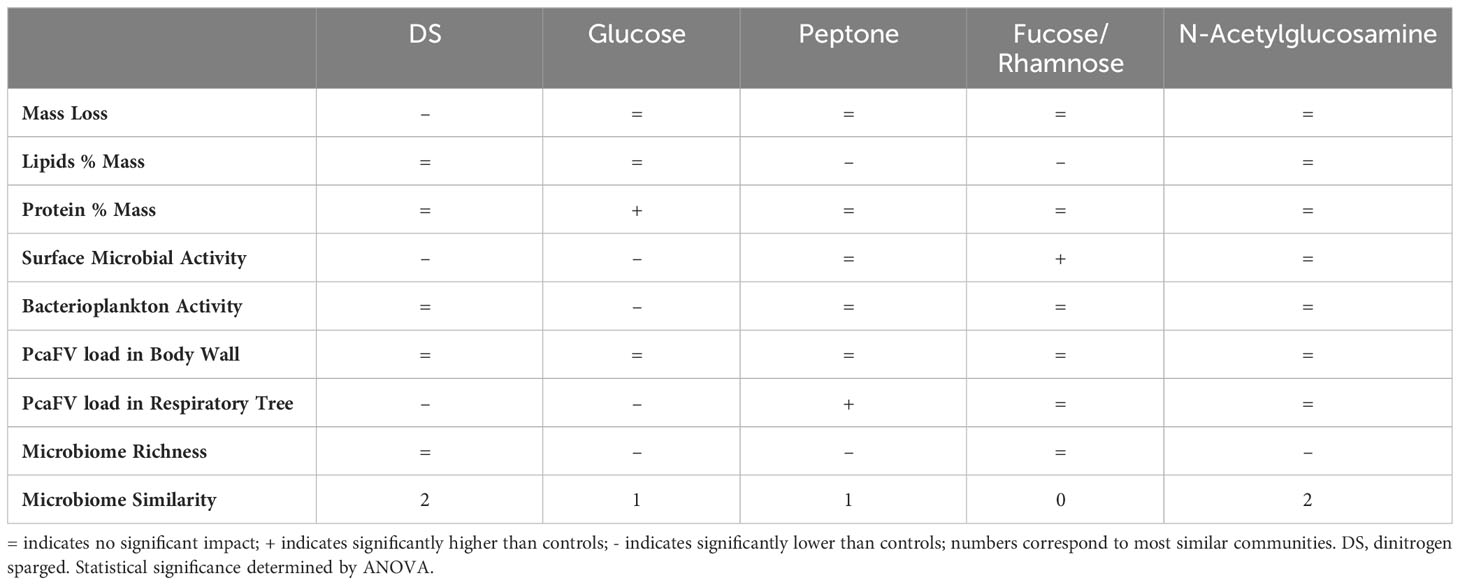
Suboxic conditions induced by dinitrogen sparging (DS), and organic matter (OM) amendment had variable impact on A. californicus (Table 1). Water column oxygen concentrations in glucose treatments were no different from controls and were higher (11.8-11.9 g L-1 or saturated) than DS treatment (3.5g L-1), while peptone amendment resulted in lower oxygen conditions compared to controls (10.8 g L-1). DS treated specimens had significantly greater mass loss over the 7-d experiment (p<0.001, Kruskal-Wallis test), but mass loss by specimens in all other treatments were not different to controls (Figure 1). Body wall lipid content was not significantly different between control and treatments (ns, Kruskal-Wallis test). Tissue protein contents were significantly higher in glucose and fucose+rhamnose amendments than controls (p<0.01 for both, Kruskal-Wallis test). Microbial activity on animal surfaces was significantly less in DS and glucose treated cucumbers than controls (p<0.01 for both, Kruskal-Wallis test; Figure 1). However, there was no difference in microbial productivity between treatments.
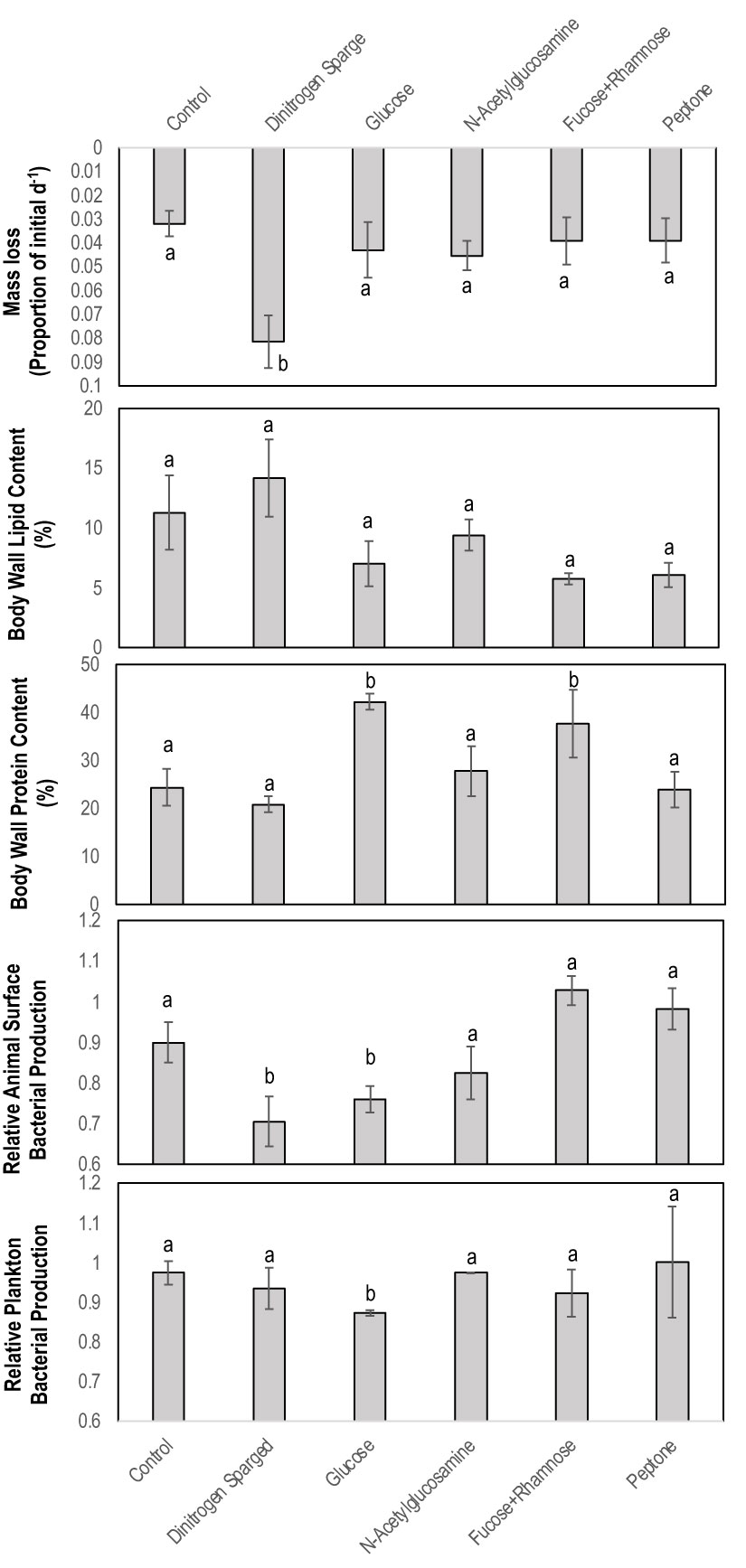
Figure 1 Physical and chemical characteristics of Apositchopus californicus body wall tissues during organic matter amendment and suboxic stress experiment. Top to bottom: Mass loss as a proportion of initial mass over the 7 d of the experiment; Body wall lipid concentration determined by chloroform extraction; Body wall protein concentration determined by Bradford assay; Microbial activity on animal surfaces as determined by BrDU incorporation; Microbial activity in plankton samples of incubation tanks. Microbial activity values are expressed relative to a single standard prepared from artificial seawater from an aquarium. a,b denotes significant difference to control (Kruskal Wallis Test, df=5).
Impact of OM amendment and DS on PcaFV load
OM amendment and DS had no impact on tissue PcaFV loads in the body wall (Figure 2). PcaFV loads in the respiratory tree of DS, N-acetylglucosamine and glucose treated specimens were significantly lower (p<0.05, ANOVA) than controls but were not different for Fucose+Rhamnose. PcaFV loads generally decreased in tube feet samples over time (Figure 3); however, treatments had no impact on PcaFV load variation over time. There was a significant positive correlation between microbial activity and body wall PcaFV load (R2 = 0.83, Pearson, p = 0.041), and a significant negative correlation between body wall protein content and body wall PcaFV load (R2 = -0.84, Pearson, p = 0.035) when specimens were aggregated per treatment (Table 2). When individual specimens were considered separately, there were significant and positive correlations between microbial activity on animal surfaces and overall (R2 = 0.23, Kendall, p = 0.040) and last -3-day mass loss (R2 = 0.22, Kendall, p = 0.050), and between microbial activity and respiratory tree viral load (R2 = 0.26, Kendall, p = 0.023). There were also significant and positive correlations between body wall PcaFV load and body wall lipid content (R2 = 0.41, Kendall, p = 0.002) and microbial activity (R2 = 0.44, Kendall, p = 0.010). Body wall lipid content also significantly and positively correlated with microbial activity (R2 = 0.31, Kendall, p = 0.010), but protein content significantly and negatively correlated with microbial activity (R2 = -0.28, Kendall, p = 0.007). Respiratory tree PcaFV load positively and nonlinearly correlated with mass load in tissues (Figure 4).
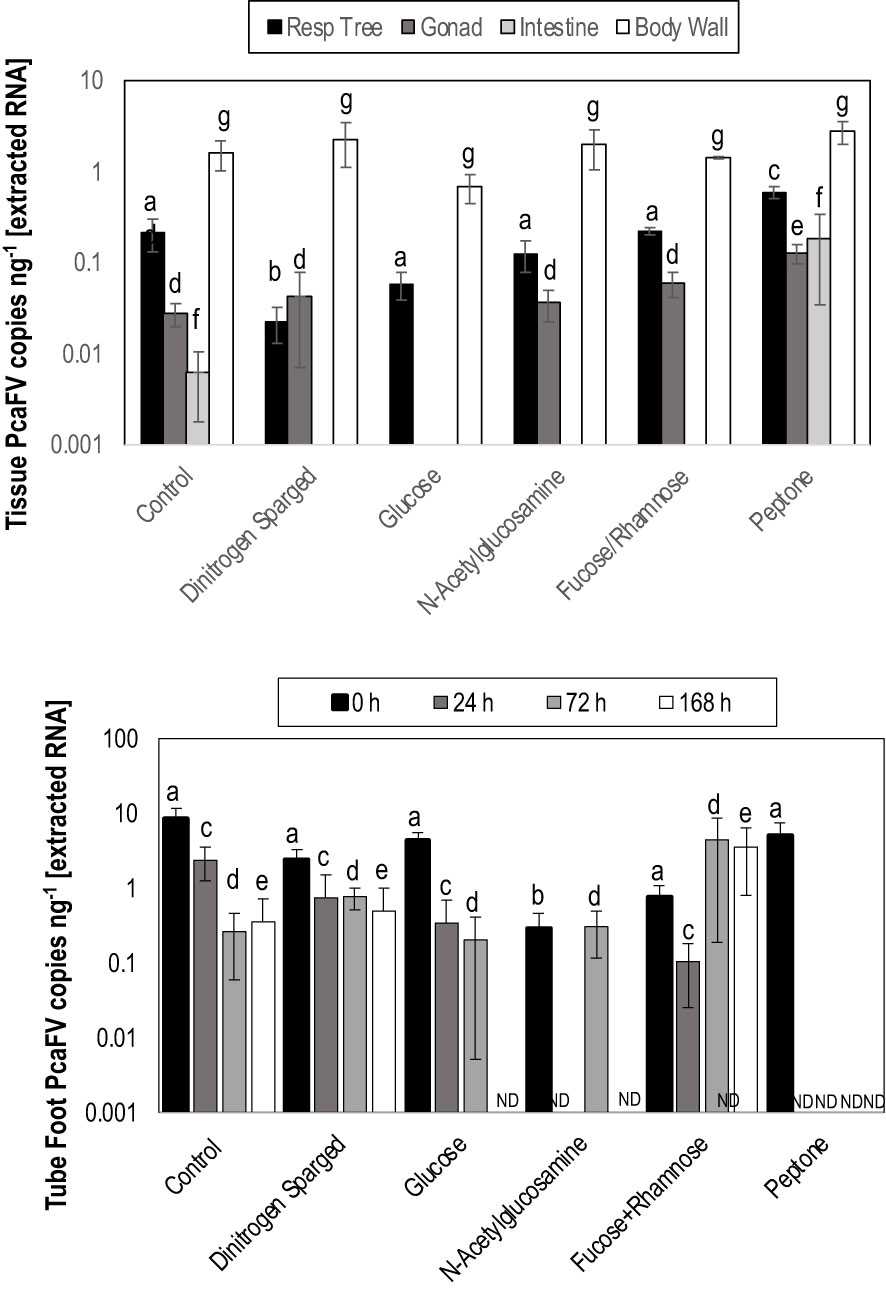
Figure 2 Tissue (top) level response to treatment with organic matter amendments and Tube foot (bottom) response over time to organic matter amendment and dinitrogen sparging. The only significant explanatory variable for treatment was tube foot PcaFV load at 0 h (ANOVA, df=429, p<0.0001). a – f denotes significant difference at p< 0.05 (Kruskal-Wallis Test).
Figure 3 Mean PcaFV load in Apostichopus californicus tube feet in specimens that had >10 copies ng-1 PcaFV at the start of the experiment.
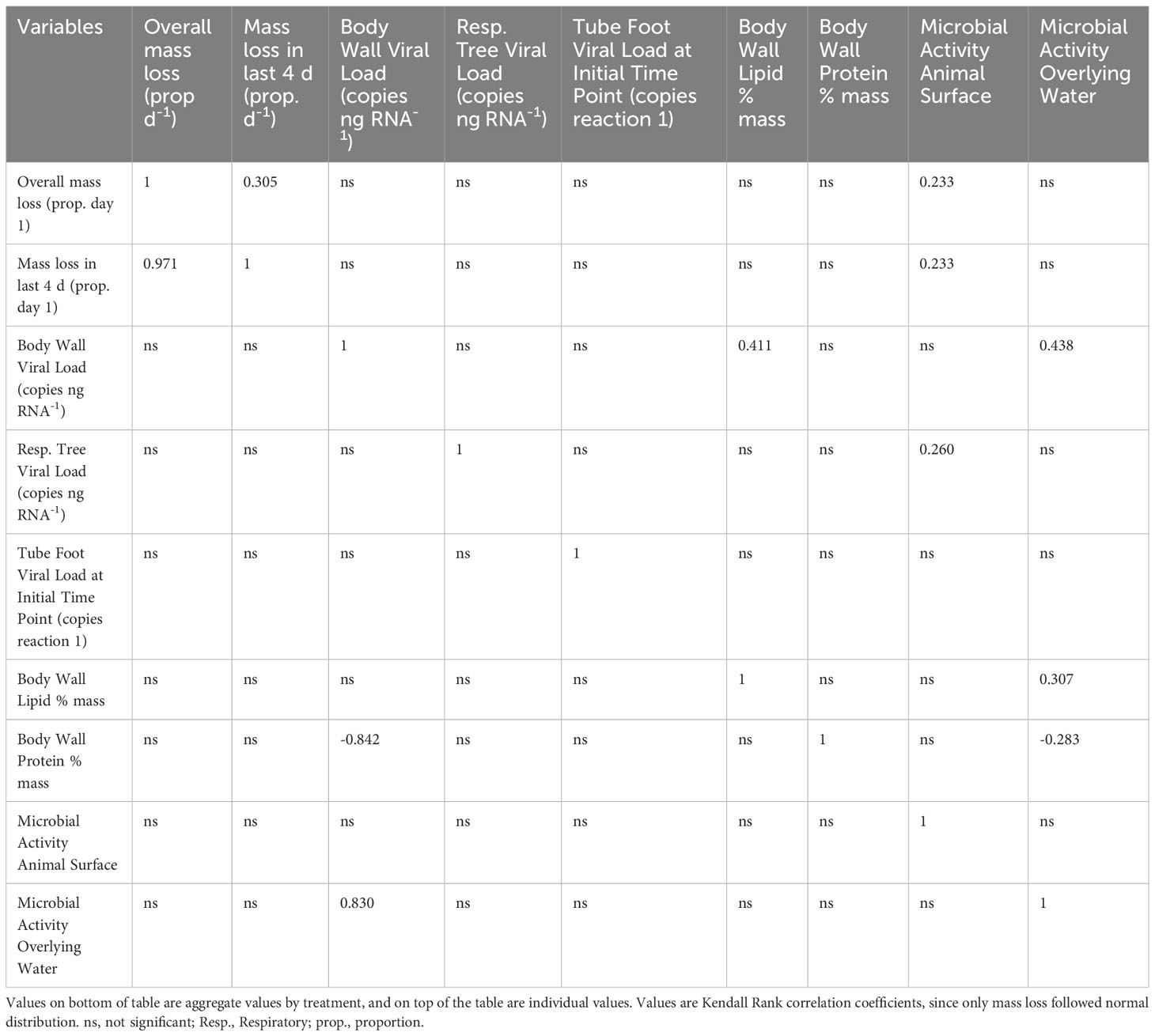
Table 2 Correlation matrix of biochemical, microbial productivity, and viral load parameters measured in the OM amendment and DS experiment.
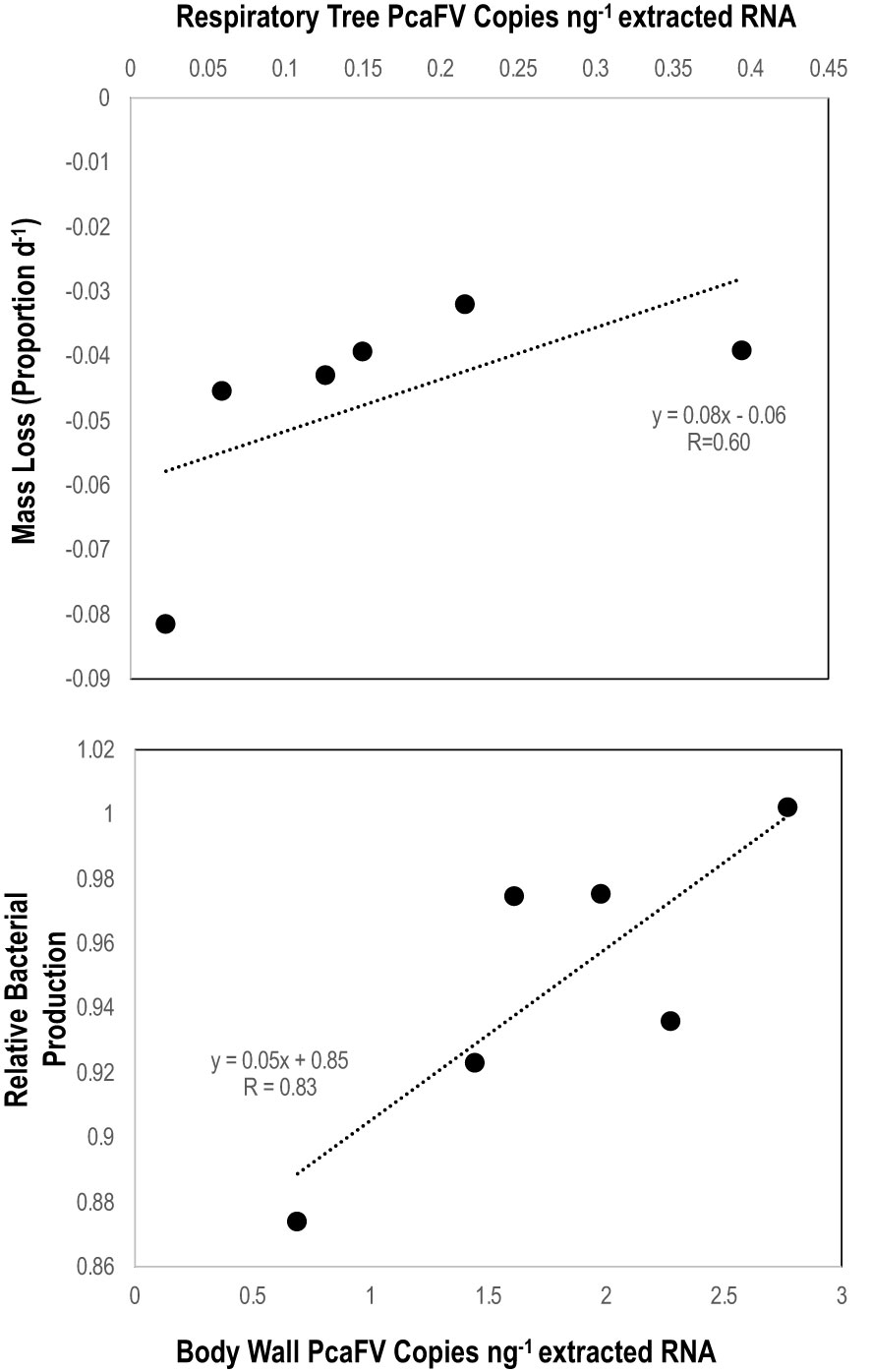
Figure 4 Linear regression between respiratory tree PcaFV load with 7-day mass loss (top) and between body wall (bottom) PcaFV load and relative microbial activity in mesocosm water. PcaFV loads, mass loss, and microbial activities represent the mean values by treatment (i.e., each data point indicates the mean values across all replicates in those treatments).
Impact of organic matter amendment on microbiome composition
Microbiome composition varied strongly at the start of the experiment in both alpha and beta diversity (Figures 5 and 6). After 7d incubation, both Chao1 and Shannon Diversity were significantly higher in controls than in amendments of Peptone and N-Acetylglucosamine, and in Chao1 alone in Glucose treated specimens (Figure 6). Beta diversity also varied with treatment. The beta diversity between treatments was not statistically different at time zero (Weighted Unifrac; Permanova: p = 0.302) (Figure 5). By day 7, except for fructose + rhamnose, the microbiomes of sea cucumbers treated with organic matter and DS shifted away from the controls, which had similar microbial communities. Glucose, peptone and N-acetylglucosamine, DS treatments both formed respective clusters, signaling similar compositional shifts. We also found a negative correlation between ASVs from the family Arcobacteraceae and animal productivity (Figure 7). However, we did not find statistically significant correlations between the microbiome composition and any other parameters.
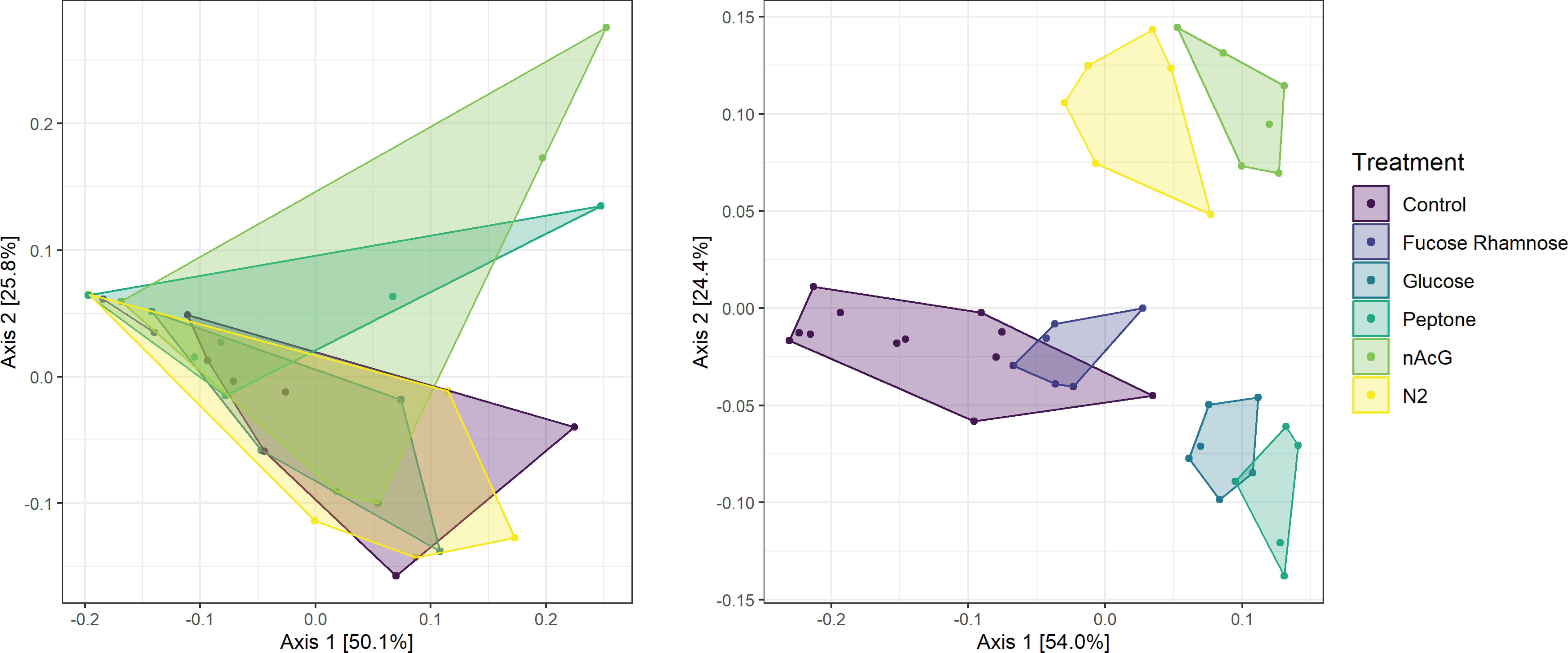
Figure 5 Principal coordinate analysis of Weighted UniFrac distances of bacterial communities (i.e., 16S rRNA amplicon libraries) of Apostichopus californicus surfaces in response to organic matter amendment and dinitrogen sparging (N2) at experiment initiation (t = 0 d; left; PERMANOVA, p = 0.301) and after 7 d incubation (right; PERMANOVA, p = 0.001).
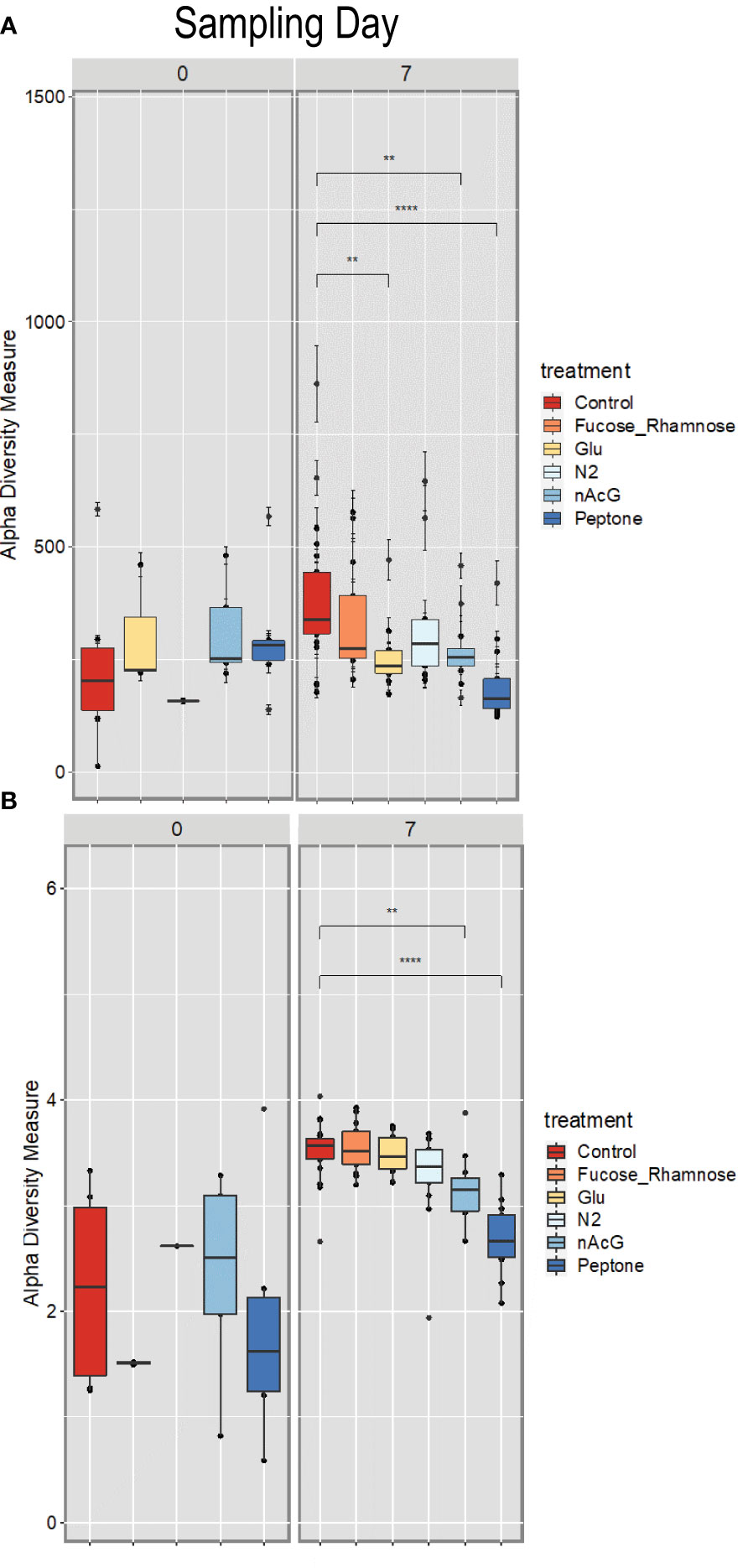
Figure 6 Alpha diversity of Apostichopus californicus surface-associated microbiomes as measured by 16S rRNA amplicon sequencing, (A) Shannon Index and (B) Chao1 Estimator in response to organic matter amendment and dinitrogen sparging at 0 and 7 d incubation. **p< 0.01, ***p< 0.0001. Fucose_Rhamnose, Fucose + Rhamnose addition; Glu, Glucose amendment; N2, dinitrogen sparged; NAcG, N-acetylglucosamine.
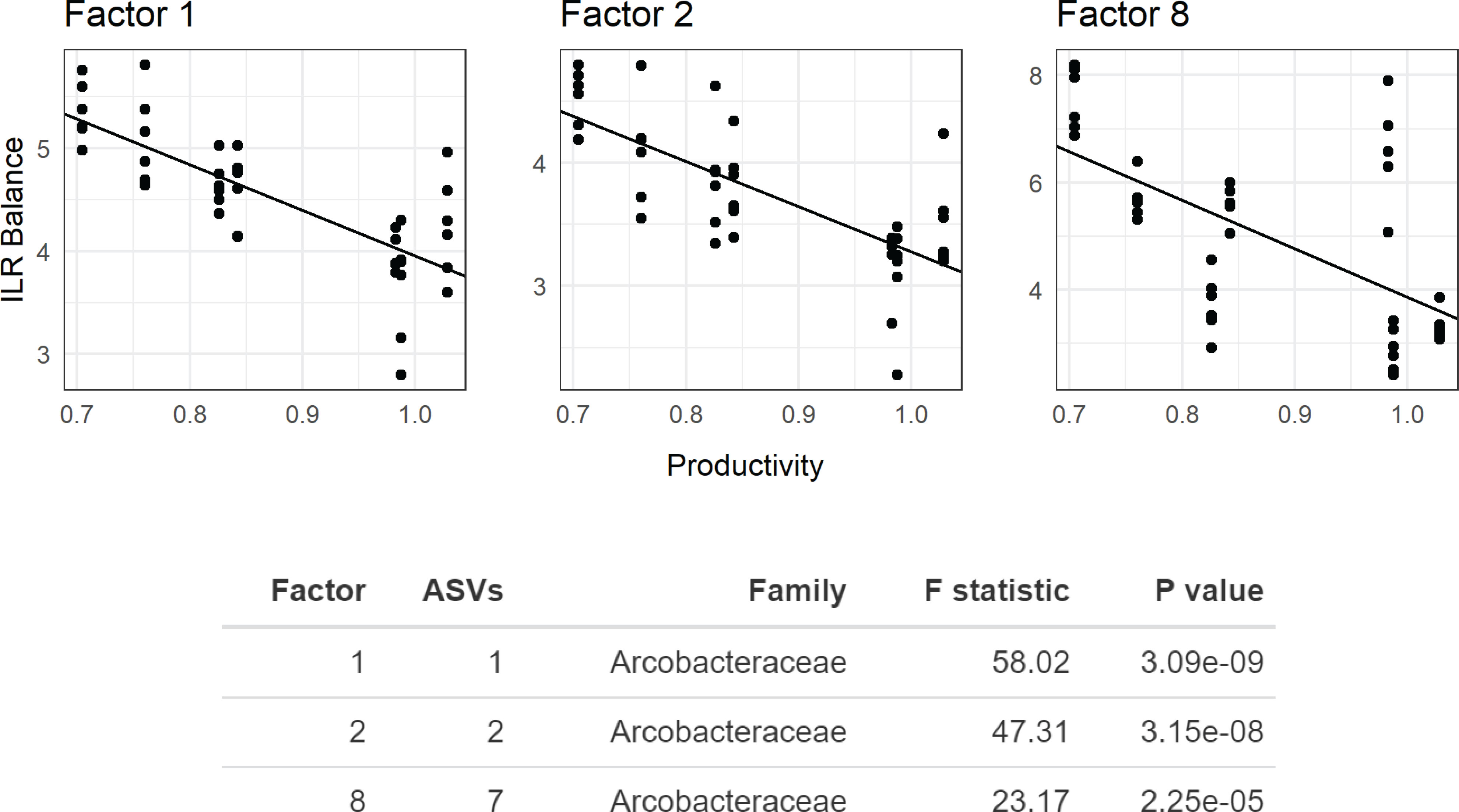
Figure 7 Phylofactor analysis of Apostichopus californicus-associated microbial assemblages after 7 d incubation during the organic matter amendemend and dinitrogen sparging experiment with respect to microbial productivity. Phylofactor uses a generalized linear model to regress the isometric log ratio between opposing clades (clades separated by an edge) in a phylogenetic tree. This is performed iteratively, where the first factor, Factor 1, represents the edge that maximizes a user defined metric. Here, we maximized the F statistic, and highlight three factors (Factors 1, 2, and 8) that show the relationship between members in the family Arcobacteraceae and productivity near the animal surface. The y-axis shows the isometric long ratio (ILR), and the x-axis shows the degree of productivity. The number of amplicon sequence variants (ASVs) indicates how many bacterial strains belong to each factor, where a value of 1 indicates a phylogenetic branch tip (1 species).
Discussion
Our results suggest that PcaFV likely does not cause acute pathology in A. californicus. Rather, our results suggest that PcaFV may behave like an endogenized virus that is supported by grossly normal tissues (Lequime and Lambrechts, 2017) or perhaps a symbiont similar to other microorganisms in echinoderm tissues (de Ridder and Foret, 2001). The variable PcaFV load and prevalence within populations (Supplemental Materials) indicates that PcaFV is not itself an endogenized virus. However, our results are consistent with the idea that most metagenomically discovered viruses recovered from grossly normal tissues do not cause gross pathology (Gudenkauf et al., 2014; Gudenkauf and Hewson, 2016).
Tissue distribution of PcaFV
The pattern of PcaFV occurrence and abundance within different tissue types suggest that surfaces with greatest contact with surrounding waters harbor the highest PcaFV infection. PcaFV was most frequently detected and had the highest load in the body wall, followed by the respiratory tree. RT-LAMP frequently detected PcaFV in the longitudinal muscle tissues of Friday harbor specimens, and in the respiratory trees of Juneau specimens. Variation in detection between Friday Harbor and Juneau specimens may suggest unmeasured environmental variables may affect the distribution of PcaFV between tissue types. Our results show PcaFV has a different occurrence pattern to the Apostichopus japonicus nodavirus, which was found primarily in digestive tract specimens (Wang et al., 2021), and the un-named virus discovered by microscopy in both digestive tract and respiratory tree tissues (Deng et al., 2008). To date there are no reports of any virus recovered from body wall tissues. Our data support that tissues exposed to surrounding seawater or water exchange internally may bear the greatest burden of PcaFV infection.
Impact of suboxic stress and organic matter load on PcaFV and Apositchopus californicus
The overall profile of A. californicus responses (Table 1) to suboxic conditions induced by DS suggests that this species is sensitive to low oxygen stress, which results in decreased microbial activity on their surfaces. While body wall PcaFV load was not reduced in the DS treatment, it was significantly lower in the respiratory tree, suggesting that in internal, seawater-filled spaces the impacts of hypoxia may be more pronounced than in external surfaces, consistent with studies of host gene expression in this organ (Huo et al., 2017). Holothurians may compensate for hypoxic conditions through accelerated respiratory rate (Ru et al., 2020), but long-term hypoxia may result in respiratory tree degradation in the absence of viruses or other pathogens (Huo et al., 2018). Our results therefore do not support our hypothesis that hypoxic conditions lead to proliferation of viruses in animal tissues.
The results of organic substrate amendment were variable and suggest that organic substrates that are atypical of natural dissolved organic matter generated the largest response in both animal biochemistry and PcaFV load. Glucose, which is usable by around half of bacterioplankton taxa (Giovannoni and Rappe, 2000), led to significantly elevated protein content of tissues, lower surface microbial and plankton activity, and lower PcaFV loads in both body wall and respiratory tree tissues. Peptone, which is commonly used in marine bacterial culture media (Moebus, 1980), significantly decreased body wall lipid content, but significantly increased PcaFV load in respiratory tree tissues, and had no impact on other measured parameters. The response pattern to glucose was most like the DS treatment, although measured oxygen levels were not different from controls. In contrast, oxygen levels were reduced in the peptone treatment and produced an opposite effect to DS treatment. Hence, these data do not support that lower oxygen conditions alone resulted in the lower PcaFV load observed in glucose treatments. The elevated PcaFV load in peptone treatments, however, may indicate that lower oxygen conditions leading to animal mortality may progress past an intermediate stage in which marginally lower oxygen conditions result in enhanced PcaFV load, but as the animals progress to death PcaFV loads may decline.
Of the natural organic matter mimics there were no effects on PcaFV load, and only marginal effects on host and surrounding microbiota. N-acetylglucosamine, which is a subcomponent of bacterial and eukaryotic cell walls and highly abundant in coastal DOM (Riemann and Azam, 2002), led to no significant changes in productivity, tissue biochemistry, nor PcaFV loads in any tissue. Fucose and rhamnose, which are subcomponents of macroalgal detritus (Bayu et al., 2021), led to greater protein content in tissues, but had no impact on other biochemical parameters, mass loss, microbial activity or PcaFV load in any tissues. These results suggest that the overall load of natural organic matter mimics was insufficient to generate changes in microbial activity that would lead to oxygen depletion.
In contrast to animal biochemical parameters and PcaFV load, the response of A. californicus microbiomes provide evidence that OM amendment of several substrates mimics the impacts of decreased oxygen. Despite high variation in microbiome composition at the onset of experimental treatment between treatments, microbial communities converged based on treatment by 7d incubation. The microbiome composition in the glucose amendment was most like the peptone amendment, both of which had significant but juxtaposed impacts on PcaFV load in respiratory trees. The most similar community to the DS treatment was N-acetylglucosamine, which did not exhibit effects on animal biochemistry, nor PcaFV load in tissues. Microbiomes had significantly lower amplicon sequence variant richness in glucose, peptone, and N-acetylglucosamine, suggesting that these substrates led to dominance of few taxa or the loss of taxa through competitive exclusion or toxicity. Of microbiome constituents, only ASV annotations of Arcobacter, a microaerophilic genus (Roalkvam et al., 2015), correlated negatively with surface microbial activity, and there were no other correlations with any parameter measured. The negative correlation with microbial activity is consistent with our observation that low oxygen concentrations in the DS treatment and glucose amendment led to lower microbial activity, presumably due to less efficient metabolism under anaerobic conditions. Some strains of Arcobacter use sulfur as an electron acceptor and can grow organ heterotrophically on complex OM (Roalkvam et al., 2015), and Arcobacter spp. oxidize sulfides in microaerophilic conditions. Overall, these data suggest that the treatment impacts on the microbiome are different from the impacts on animal biochemistry and PcaFV load in tissues. However, the similar response of glucose to DS in terms of both microbiome composition and animal biochemistry and PcaFV load may point to glucose causing low oxygen conditions at the animal-water interface.
Potential role in epidermal disease process
The presence of PcaFV, which as yet has no known pathology in its host, primarily in the body wall of A. californicus (Supplemental Materials) is intriguing since this is the most frequently abnormal tissue type in described diseases. Ulcerative or deteriorating (wasting) body wall, commonly referred to as skin ulcerative diseases (SKUDs), are believed to be caused by a wide variety of pathogens and environmental stressors including viruses. However, current studies have been unable to replicate infection of suspected pathogens in vivo, or to separate these conditions from environmental stressors such as hypoxia (Delroisse et al., 2020). SKUDs have been reported across a wide geographic region. One study reported a cross-species infection event of covert mortality nodavirus (CMNV), a virus believed to be found in shrimp, infecting Apostichopus japonicus which led to wasting and ulceration of the intestinal epithelium (Wang et al., 2021). A study of an A. japonicus mass mortality event in an indoor farming facility near the Dalian coast (China) proposed a potential synergistic role of viral and bacterial co-infection contributing to a weakening of the respiratory tree, alimentary canal, and water vascular system endoplasmic reticulum and mitochondria (Deng et al., 2008). Additional studies show bacterial pathogens may contribute to disease and wasting states in sea cucumbers. Eight bacterial cultures isolated from an indoor sea cucumber farm challenged A. japonicus resulted in skin ulceration and viscera ejection syndrome (Deng et al., 2009). These studies indicate that a wide variety of pathogens and environmental conditions may result in similar ulcerative lesions, and that these gross observations may not be indicative of any unique or individual pathogen or the impact of environmental stressors.
While it is tempting to ascribe PcaFV as a potential cause for SKUDs, our results suggest that PcaFV is not a pathogenic agent in holothurian body wall lesions. Specimens of A. californicus with body wall lesions that were collected during a mass mortality event in Nanoose Bay, British Columbia (Lim et al., 2023), did not bear PcaFV. In the OM amendment and DS study presented in this report, loads of PcaFV in tube feet samples collected from almost all animals over the 7d incubation declined over time, and PcaFV load over time was only explained by initial timepoint tube foot PcaFV load. Importantly, respiratory tree PcaFV load correlated positively with animal mass loss over time (i.e., animals that lost less weight and were presumably more normal than individuals with greater weight loss, had the greatest PcaFV load). If PcaFV generates host pathology, it was expected that its detection and load would correspond inversely with abnormal (greater) weight loss. Body wall PcaFV load positively correlated with relative microbial activity. Since microbial activity variation is presumably driven by animal excretion (Uthicke, 2001), this trend may be explained by normal specimens excreting greater amounts of material than abnormal specimens. Furthermore, body wall PcaFV load correlated significantly and positively with body wall lipid content, which is a marker of tissue health. Taken together, our data supports the idea that PcaFV does not correlate with poor animal health, but instead it appears that PcaFV may proliferate in healthy tissues.
Conclusions
The results of this study provide new information about flavivirus distribution and dynamics within a marine host. We found no evidence of any negative impact of PcaFV on A. californicus. Rather, PcaFV appears related positively with animal health, indicating that it may have limited pathology under the conditions it was observed or may represent an attenuated viral strain that may lead to pathologies under stressful environmental conditions, when new virulent strains emerge, or host immunity is compromised. While some OM amendment and DS had significant impacts on tissue biochemistry and microbial productivity on and within A. californicus, and impacts on microbiome composition and richness, the response pattern of measured variables suggests that PcaFV load does not respond positively to DS. Overall, these data do not support the hypothesis that PcaFV replication and persistence is positively associated with suboxic conditions, but rather that conditions influencing the health state of its host play a role in PcaFV dynamics within tissues. The mechanisms by which PcaFV replicates and environmental and biological factors which may influence its dynamics in tissues remain unclear, however our study suggests that OM load in surrounding waters may change the A. californicus microbiome composition and potentially O2 conditions on their epidermis. The overall condition of A. californicus may be influenced by environmental conditions, and this in turn may influence both productivity of surrounding water (and surface microbial activity), and perhaps PcaFV load. However, these effects appear to be very small in magnitude. Key questions remain as to whether PcaFV elicits pathology under other, untested environmental variations, such as elevated temperature. However, our work does not support that PcaFV causes pathology or deleterious health consequences under low oxygen conditions, and most impacts on PcaFV load are a downstream consequence of factors influencing the overall health of the animals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA947521.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because invertebrates are not subject to ethical approval.
Author contributions
JC: Formal Analysis, Investigation, Visualization, Writing – original draft. AA: Formal analysis, Investigation, Writing – review & editing. CD: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Writing – review & editing. KH: Formal analysis, Investigation, Methodology, Resources, Writing – review & editing. JM: Conceptualization, Formal analysis, Investigation, Resources, Writing – review & editing. EL: Formal analysis, Investigation, Resources, Writing – review & editing. KP: Formal analysis, Investigation, Methodology, Writing – review & editing. JR: Formal analysis, Investigation, Software, Writing – review & editing. MS: Formal analysis, Investigation, Resources, Writing – review & editing. BV-C: Formal analysis, Investigation, Software, Writing – review & editing. EW: Formal analysis, Investigation, Writing – review & editing. IH: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NSF grant OCE- 2049225 awarded to IH.
Acknowledgments
Sampling of Apostichopus californicus specimens in Sitka was performed under Alaska Department of Fish and Game permit CF-21-105. Specimens from Friday Harbor Laboratory were collected under the blanket approval granted to the lab manager in the Marine Biological Preserve of San Juan County and Cypress Island. MBP Specimens collected from Nanoose Bay were collected under Fisheries and Oceans Canada scientific license number XR 153 2021. The authors thank Anthony Walloch and Ethan Christiansen for collection of specimens and the faculty and students at the University of Alaska Southeast for assistance during the organic matter enrichment experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1295276/full#supplementary-material
References
Alessandra A. N., Marco P., Chiara L., Annamaria V. G. (2003). Ammonia as confounding factor in toxicity tests with the sea urchin Paracentrotus lividus (Lmk). Toxicol. Environ. Chem. 85, 183–191. doi: 10.1080/02772240410001665418
Allen L. Z., Mccrow J. P., Ininbergs K., Dupont C. L., Badger J. H., Hoffman J. M., et al. (2017). The baltic sea virome: Diversity and transcriptional activity of DNA and RNA viruses. mSystems 2, e00125–e00116. doi: 10.1128/mSystems.00125-16
Amir A., Mcdonald D., Navas-Molina J. A., Kopylova E., Morton J. T., Xu Z. Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191–e00116. doi: 10.1128/mSystems.00191-16
Apprill A., Mcnally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Aquino C. A., Besemer R. M., Derito C. M., Kocian J., Porter I. R., Raimondi P. T., et al. (2021). Evidence that microorganisms at the animal-water interface drive sea star wasting disease. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.610009
Bayu A., Warsito M. F., Putra M. Y., Karnjanakom S., Guan G. (2021). Macroalgae-derived rare sugars: Applications and catalytic synthesis. Carbon Res. Convers 4, 150–163. doi: 10.1016/j.crcon.2021.04.002
Bergh O., Borsheim K. Y., Bratbak G., Heldal M. (1989). High abundance of viruses found in aquatic environments. Nature 340, 467–468. doi: 10.1038/340467a0
Bianchi A., Van Wambeke F., Garcin J. (1998). Bacterial utilization of glucose in the water column from eutrophic to oligotrophic pelagic areas in the eastern North Atlantic Ocean. J. Mar. System 14, 45–55. doi: 10.1016/S0924-7963(98)00012-8
Bisanz J. E. (2018). qiime2R: Importing QIIME2 artifacts and associated data into R sessions. 13. Available at: https://github.com/jbisanz/qiime2R.
Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. doi: 10.1186/s40168-018-0470-z
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Breitbart M., Salamon P., Andresen B., Mahaffy J., Segall A., Mead D., et al. (2002). Genomic analysis of uncultured marine viral communities. Proc. Nat. Acad. Sci. U.S.A. 99, 14250–14255. doi: 10.1073/pnas.202488399
De Lamballerie X., Crochu S., Billoir F., Neyts J., De Micco P., Holmes E. C., et al. (2002). Genome sequence analysis of Tamana bat virus and its relationship with the genus Flavivirus. J. Gen. Virol. 83, 2443–2454. doi: 10.1099/0022-1317-83-10-2443
Delroisse J., Van Wayneberghe K., Flammang P., Gillan D., Gerbaux P., Opina N., et al. (2020). Epidemiology of a SKin Ulceration Disease (SKUD) in the sea cucumber Holothuria scabra with a review on the SKUDs in Holothuroidea (Echinodermata). Sci. Rep. 10, 22150. doi: 10.1038/s41598-020-78876-0
Deng H., He C., Zhou Z., Liu C., Tan K., Wang N., et al. (2009). Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture 287, 18–27. doi: 10.1016/j.aquaculture.2008.10.015
Deng H., Zhou Z.-C., Wang N.-B., Liu C. (2008). The syndrome of sea cucumber (Apostichopus japonicus) infected by virus and bacteria. Virol. Sin. 23, 63–67. doi: 10.1007/s12250-008-2863-9
de Ridder C., Foret T. W. (2001). Non-parasitic symbioses between echinoderms and bacteria. Echinoderm Stud. 6, 111–169.
Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Veget. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dominguez-Huerta G., Zayed A. A., Wainaina J. M., Guo J., Tian F., Pratama A. A., et al. (2022). Diversity and ecological footprint of global ocean RNA viruses. Science 376, 1202–1208. doi: 10.1126/science.abn6358
Frakolaki E., Kaimou P., Moraiti M., Kalliampakou K. I., Karampetsou K., Dotsika E., et al. (2018). The role of tissue oxygen tension in Dengue Virus replication. Cells 7, 241. doi: 10.3390/cells7120241
Fuhrman J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Giovannoni S., Rappe M. (2000). “Evolution, diversity and molecular ecology of marine prokaryotes,” in Microbial Ecology of the Oceans. Ed. Kirchman. D. (New York: Wiley-Liss), 47–84.
Gotham S. M., Fryer P. J., Paterson W. R. (1988). The measurement of insoluble proteins using a modified Bradford assay. Analyt. Biochem. 173, 353–358. doi: 10.1016/0003-2697(88)90199-6
Grupstra C. G. B., Howe-Kerr L. I., Veglia A. J., Bryant R. L., Coy S. R., Blackwelder P. L., et al. (2022). Thermal stress triggers productive viral infection of a key coral reef symbiont. ISME J. 16, 1430–1441. doi: 10.1038/s41396-022-01194-y
Gudenkauf B. M., Eaglesham J. B., Aragundi W. M., Hewson I. (2014). Discovery of urchin-associated densoviruses (Parvoviridae) in coastal waters of the Big Island, Hawaii. J. Gen. Virol. 95, 652–658. doi: 10.1099/vir.0.060780-0
Gudenkauf B. M., Hewson I. (2016). Comparative metagenomics of viral assemblages inhabiting four phyla of marine invertebrates. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00023
Hewson I., Aquino C. A., Derito C. M. (2020a). Virome variation during sea star wasting disease progression in Pisaster ochraceus (Asteroidea, Echinodermata). Viruses 12, 1332. doi: 10.3390/v12111332
Hewson I., Bistolas K. S. I., Quijano Cardé E. M., Button J. B., Foster P. J., Flanzenbaum J. M., et al. (2018). Investigating the complex association between viral ecology, environment, and Northeast Pacific sea star wasting. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00077
Hewson I., Button J. B., Gudenkauf B. M., Miner B., Newton A. L., Gaydos J. K., et al. (2014). Densovirus associated with sea-star wasting disease and mass mortality. Proc. Nat. Acad. Sci. U.S.A. 111, 17276–17283. doi: 10.1073/pnas.1416625111
Hewson I., Johnson M. R., Tibbetts I. R. (2020b). An unconventional flavivirus and other RNA viruses in the sea cucumber (Holothuroidea; Echinodermata) virome. Viruses 12, 1057. doi: 10.3390/v12091057
Holmes E. C. (2016). The expanding virosphere. Cell Host Microbe 20, 279–280. doi: 10.1016/j.chom.2016.08.007
Huo D., Sun L., Li X., Ru X., Liu S., Zhang L., et al. (2017). Differential expression of miRNAs in the respiratory tree of the sea cucumber Apostichopus japonicus under hypoxia stress. G3 (Bethesda) 7, 3681–3692. doi: 10.1534/g3.117.1129
Huo D., Sun L., Ru X., Zhang L., Lin C., Liu S., et al. (2018). Impact of hypoxia stress on the physiological responses of sea cucumber Apostichopus japonicus: Respiration, digestion, immunity and oxidative damage. PeerJ 6, e4651. doi: 10.7717/peerj.4651
Jackson E. W., Pepe-Ranney C., Debenport S. J., Buckley D. H., Hewson I. (2018). The Microbial landscape of sea stars and the anatomical and interspecies variability of their microbiome. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01829
Jackson E. W., Pepe-Ranney C., Johnson M. R., Dietel D. L., Hewson I. (2020). A highly prevalent and pervasive densovirus discovered among sea stars from the North American Atlantic Coast. Appl. Environ. Microbiol. 86, e02723–e02719. doi: 10.1128/AEM.02723-19
Janssen S., Mcdonald D., Gonzalez A., Navas-Molina J. A., Jiang L., Xu Z. Z., et al. (2018). Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems 3, e00021–e00018. doi: 10.1128/mSystems.00021-18
Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lee C. M., Trevino B., Chaiyawat M. (1996). A simple and rapid solvent extraction method for determining total lipids in fish tissue. J. AOAC Int. 79, 487–492. doi: 10.1093/jaoac/79.2.487
Lequime S., Lambrechts L. (2017). Discovery of flavivirus-derived endogenous viral elements in Anopheles mosquito genomes supports the existence of Anopheles-associated insect-specific flaviviruses. Virus Evol. 3, vew035. doi: 10.1093/ve/vew035
Lim E., Reed K. J., Campbell J. A., Côté I. M. (2023). Localised mass mortality of giant California sea cucumbers in Western Canada. Mar. Biol. 170, 86. doi: 10.1007/s00227-023-04230-3
Lozupone C., Knight R. (2005). UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Mccracken A. R., Christensen B. M., Munteanu D., Case B. K. M., Lloyd M., Herbert K. P., et al. (2023). Microbial dysbiosis precedes signs of sea star wasting disease in wild populations of Pycnopodia helianthoides. Mar. Sci. 10. doi: 10.3389/fmars.2023.1130912
Mcmurdie P. J., Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Moebus K. (1980). A method for the detection of bacteriophages from ocean water. Helgoland Meeres 34, 1–14. doi: 10.1007/BF01983537
Nelson C. E., Carlson C. A. (2005). A nonradioactive assay of bacterial productivity optimized for oligotrophic pelagic environments. Limnol. Oceanogr. Meth. 3, 211–220. doi: 10.4319/lom.2005.3.211
Ng T. F. F., Manire C., Borrowman K., Langer T., Ehrhart L., Breitbart M. (2009). Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J. Virol. 83, 2500–2509. doi: 10.1128/JVI.01946-08
Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucl. Acid Res. 28, e63–e63. doi: 10.1093/nar/28.12.e63
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Parry R., Asgari S. (2019). Discovery of novel crustacean and cephalopod flaviviruses: Insights into the evolution and circulation of flaviviruses between marine invertebrate and vertebrate hosts. J. Virol. 93, e00432–e00419. doi: 10.1128/JVI.00432-19
R Core Team. (2022). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Riemann L., Azam F. (2002). Widespread N-acetyl-D-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol. 68, 5554–5562. doi: 10.1128/AEM.68.11.5554-5562.2002
Roalkvam I., Drønen K., Stokke R., Daae F. L., Dahle H., Steen I. H. (2015). Physiological and genomic characterization of Arcobacter anaerophilus IR-1 reveals new metabolic features in Epsilonproteobacteria. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00987
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Ru X., Zhang L., Liu S., Yang H. (2020). Plasticity of respiratory function accommodates high oxygen demand in breeding sea cucumbers. Front. Physiol. 11. doi: 10.3389/fphys.2020.00283
Shi M., Lin X.-D., Tian J.-H., Chen L.-J., Chen X., Li C.-X., et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. doi: 10.1038/nature20167
Soffer N., Brandt M. E., Correa A. M. S., Smith T. B., Thurber R. V. (2014). Potential role of viruses in white plague coral disease. ISME J. 8, 271–283. doi: 10.1038/ismej.2013.137
Solomon C., Hewson I. (2022). Putative invertebrate, plant, and wastewater derived ssRNA viruses in plankton of the anthropogenically impacted Anacostia River, District of Columbia, USA. Microbes Environ. 37, ME21070. doi: 10.1264/jsme2.ME21070
Suttle C. A., Chan A. M., Cottrell M. T. (1990). Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347, 467–469. doi: 10.1038/347467a0
Thurber R. V., Haynes M., Breitbart M., Wegley L., Rohwer F. (2009). Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4, 470–483. doi: 10.1038/nprot.2009.10
Uthicke S. (2001). Nutrient regeneration by abundant coral reef holothurians. J. Exper. Mar. Biol. Ecol. 265, 153–170. doi: 10.1016/S0022-0981(01)00329-X
Vassilaki N., Frakolaki E. (2017). Virus-host interactions under hypoxia. Microbes Infect. 19, 193–203. doi: 10.1016/j.micinf.2016.10.004
Wang C., Yao L., Wang W., Sang S., Hao J., Li C., et al. (2021). First report on natural infection of nodavirus in an Echinodermata, sea cucumber (Apostichopus japonicas). Viruses 13, 636. doi: 10.3390/v13040636
Washburne A. D., Silverman J. D., Leff J. W., Bennett D. J., Darcy J. L., Mukherjee S., et al. (2017). Phylogenetic factorization of compositional data yields lineage-level associations in microbiome datasets. PeerJ 5, e2969. doi: 10.7717/peerj.2969
Wolf Y. I., Silas S., Wang Y., Wu S., Bocek M., Kazlauskas D., et al. (2020). Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol. 5, 1262–1270. doi: 10.1038/s41564-020-0755-4
Work T. M., Weatherby T. M., Landsberg J. H., Kiryu Y., Cook S. M., Peters E. C. (2021). Viral-like particles are associated with endosymbiont pathology in Florida corals affected by stony coral tissue loss disease. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.750658
Keywords: Apostichopus, Echinodermata, oxygen, microbiome, flavivirus
Citation: Crandell JG, Altera AK, DeRito CM, Hebert KP, Lim EG, Markis J, Philipp KH, Rede JE, Schwartz M, Vilanova-Cuevas B, Wang E and Hewson I (2023) Dynamics of the Apostichopus californicus-associated flavivirus under suboxic conditions and organic matter amendment. Front. Mar. Sci. 10:1295276. doi: 10.3389/fmars.2023.1295276
Received: 15 September 2023; Accepted: 30 October 2023;
Published: 17 November 2023.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Jing-zhe Jiang, Chinese Academy of Fishery Sciences (CAFS), ChinaAmy Long, Fisheries and Oceans Canada (DFO), Canada
Copyright © 2023 Crandell, Altera, DeRito, Hebert, Lim, Markis, Philipp, Rede, Schwartz, Vilanova-Cuevas, Wang and Hewson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian Hewson, aGV3c29uQGNvcm5lbGwuZWR1
†Present address: Jameson G. Crandell, Department of Immunobiology, Yale University, New Haven, CT, United States
 Jameson G. Crandell1†
Jameson G. Crandell1† Christopher M. DeRito
Christopher M. DeRito Katherine H. Philipp
Katherine H. Philipp Jordan E. Rede
Jordan E. Rede Brayan Vilanova-Cuevas
Brayan Vilanova-Cuevas Ian Hewson
Ian Hewson