Abstract
Nitrate is mainly converted via aerobic nitrite oxidation during the second step of nitrification, which is catalyzed by nitrite-oxidizing bacteria (NOB) and the recently discovery complete ammonia oxidizers (comammox). Members of the genus Nitrospira are the most diverse and widespread known NOB and comammox. However, the community assembly of Nitrospira in estuary and coastal wetland and the major environmental shaping factors remain understudied. Here in this study, we investigated the geographical distribution pattern of Nitrospira along the large-scale coastal wetlands of China. The results showed that the abundance of Nitrospira ranged from 4.96×106 - 3.88×107 copies/g dry sediment, significantly (more than one order of magnitude) higher than amoA gene of ammonia-oxidizers. The identified Nitrospira belong to Nitrospira lineage IV (50%), lineage I and II. The adaptability of the three lineages to environmental factors (such as temperature, pH, salinity and particle size) are different, which leads to the diversity of its distribution composition in different estuaries. Network analysis showed that the cooperation takes greater portion than competition in the relationship of Nitrospira population. This study revealed the abundance and community composition of Nitrospira bacteria, as well as the major environmental driving factors in coastal wetland ecosystems, which deepens our understanding of the niche separation of Nitrospira with the nitrogen cycling.
Introduction
Nitrogen is an essential component of all living organism and considered as a growth limiting factor in many ecosystems. As a major source of bioavailable nitrogen, nitrate is mainly converted by aerobic nitrite oxidation during the second step of nitrification, which is catalyzed by chemolithoautotrophic nitrite-oxidizing bacteria (NOB) (Kuypers et al., 2018).
To date, the known NOB were found in seven genera (Nitrobacter, Nitrococcus, Nitrospina, Nitrospira, Nitrotoga, Nitrolancea, and Candidatus Nitromaritima) (Daims et al., 2016). Among these NOB, Nitrospira is the most diverse genus and consists of six phylogenetically diverged sub-lineages (Daims et al., 2016). Nitrospira is globally distributed in natural and engineered ecosystems, including soil (Pester et al., 2014), ocean (Watson et al., 2004; Pachiadaki et al., 2017), freshwater habitats (Altmann et al., 2003), geothermal springs (Lebedeva et al., 2005; Edwards et al., 2013), wastewater treatment plants (WWTPs) (Daims et al., 2001; Kruse et al., 2013; Ushiki et al., 2013; Daims et al., 2015; Gruber-Dorninger et al., 2015; Annavajhala et al., 2018), saline-alkaline lakes (Daebeler et al., 2020), and aquaculture biofilters (Keuter et al., 2011; van Kessel et al., 2015). The Nitrospira lineage I and II exhibited dominance in WWTPs (Okabe and Watanabe, 1999; Daims et al., 2001; Kruse et al., 2013), with ventilation intensity emerging as a significant factor influencing their relative abundance (Park and Noguera, 2008). Furthermore, within marine ecosystems, it is the linage IV rather than linage II that assumes dominance among NOB (Pester et al., 2014).
Nevertheless, Nitrospira have a reputation for being difficult to isolate and grow under laboratory conditions so that progress in Nitrospira-related research is lagging (Daims et al., 2016). Remarkably, the hitherto known lineage I Nitrospira were mainly detected in the activated sludge of WWTPs (Lucker et al., 2010; Fujitani et al., 2014).
Nitrospira had been regarded as obligate chemolithoautotrophic organisms that gain growth energy solely from nitrite oxidation for a long time. However, the discovery of complete ammonia oxidizers (comammox) belonging to Nitrospira lineage II challenged our perception (Daims et al., 2015; van Kessel et al., 2015). Meanwhile, some Nitrospira were physiologically confirmed of being able to grow with various organic substrates. For example, nitrite oxidizers of the genus Nitrospira can hydrolyze urea or cyanate to NH3 and CO2, then provides ammonia-oxidizing bacteria (AOB) with ammonia. Subsequently, they can acquire nitrite from the AOB. This interaction between NOB and AOB called “reciprocal feeding” (Koch et al., 2015; Palatinszky et al., 2015). Moreover, genomic analysis and physiological experiment prove that several Nitrospira can grow aerobically with hydrogen or formate as sole energy source. Meanwhile, under anoxic conditions, they are capable of reducing nitrate to nitrite with hydrogen or formate as electron donor (Koch et al., 2014; Koch et al., 2015). All of these studies revealed previously unrevealed metabolic potentials of Nitrospira beyond the autotrophic nitrite-oxidizing lifestyle.
Due to the sea-land interaction, estuaries are highly productive environments that harbor active and diverse microbial communities so that have always been a hotspot for the study of biochemical cycles. Meanwhile, estuaries also continuously enrich reactive nitrogen from the upstream of the rivers (Hou et al., 2015; Osburn et al., 2016; Hou et al., 2018b). Therefore, the process and mechanism of nitrogen biogeochemical cycling are the focus of estuarine research. China has a wide latitude range of eastern coastline, the researching to Nitrospira of the coastal wetlands may expand our knowledge of ecological niche differentiation among Nitrospira genus. In this study, we aimed to examine the diversity and distribution of Nitrospira in the eastern estuaries of China, and determine the key environmental factors shaping the community structure of Nitrospira.
Experimental procedures
Sample collection
Sediment samples were collected from 16 estuarine intertidal zones of China in May 2019 (Sun et al., 2020), including LH, BDH, HH, YR, SYH, BCYH, CJ, JJ, OJ, MJ, MLX, JLJ, YFX, ZJ, YJ and NLJ (Figure 1). The collected sediment samples were sealed in sterile plastic bags, subsequently refrigerated and transported back to the laboratory. Then aliquots of the sediment samples were used for enrichment cultures and physicochemical properties analyses, the others were stored at -20°C for molecular analyses.
Figure 1

Sampling site and Nitrospira nxrB Gene abundance in the coastal wetlands of China. This figure illustrates the geographical location of our sampling site within the coastal wetlands of China, as referenced in Sun et al. (2020). The pie chart inset depicts the relative abundance of the nxrB gene across three lineages of Nitrospira: Lineage I, Lineage II, and Lineage IV. Each segment of the pie chart is proportionally representative of the respective lineage's abundance within the sampled population.
DNA extraction and quantitative PCR
DNA was extracted in duplicate from the sediment samples according to the standard protocol of the FastDNA Spin Kit for Soil (QBIOgene, Carlsbad, CA, United States). The nxrB gene of Nitrospira was amplified using nxrB169F (TACATGTGGTGGAACA and nxrB638R (CGGTTCTGGTCRATCA) primers. Reactors for quantitative PCR were prepared with Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The reaction comprised 10 µL Hieff ® qPCR SYBR Green Master Mix with Low Rox Plus (Yeasan, China), 1 µL template DNA (approximately 5 ng), 0.4 µL of forward and reverse mixed primers (10 µM) and 9.2 µL of ddH2O. Samples were run in triplicates on an ABI 7500 sequence detection system (Applied Biosystems, Canada). Standard curve for quantitative PCR (qPCR) was created by purifying PCR products containing the target gene fragments with the QIAquick PCR Purification Kit (Qiagen, Germany) according to manufacturer’s instructions, the concentration was measured with the Nanodrop-2000 Spectrophotometer (Thermo, United States) and serially 10-fold diluted before qPCR was conducted. The correlation coefficient (r2) value for standard curve was greater than 0.99, and the amplification efficiency was between 96% and 105%. The amplicon specificity was assessed by the melting curve. The qPCR results were showed as the copy numbers calculated as: Copy number = (C × 10-9/MW) × NA (C represents template concentration (ng/µL); MW indicates template molecular weight in Daltons; NA indicates Avogadro’s constant, 6.022 × 1023). For each qPCR assay, negative controls were performed by the no-template DNA for ruling out any possible contamination.
Phylogenetic analysis
For amplicon sequencing, DNA was extracted from sedimental samples and used as template for PCR with primers targeting the nxrB gene of Nitrospira. Then PCR products for Illumina MiSeq sequencing were performed at Shanghai Majorbio Biomedical Technology Company (Shanghai, China). The processing of raw data was performed by Quantitative Insight into Microbial Ecology (QIIME) (Caporaso et al., 2010). Paired-end reads were stitched with FLASH plugin based on matched overlapping regions. Then the sequences were grouped into operational taxonomic units (OTU) with a similarity threshold of 95% (Schloss, 2013). The closest reference strain to each OTU was searched using the NCBI Web BLASTn program. All of the sequences we obtained were aligned using ClusalX (Thompson et al., 1997). Neighbor-joining phylogenetic trees from one representative OTU sequence and its closest reference sequence retrieved from GenBank (Kumar et al., 2004) were created using MEGA 7.0 with 1000 bootstrap replicates to confirm the tree topologies (Tamura et al., 2007).
Statistical analysis
The alpha diversity was calculated for the numbers of OTU, richness estimator (Chao1) and diversity indices (Shannon-Weiner and Simpson) using the Vegan packages V2.5-4 in R V3.4 (Okasen et al., 2007; R Core Team, 2020). Non-metric multidimensional scaling (NMDS) ordination was performed in R (R Core Team, 2020), and principal coordinate analysis (PCoA) was conducted in QIIME, both of them were employed to explore the community classification of Nitrospira. The correlation between community structure and environmental parameters was determined by canonical correspondence analysis (CCA) using Canoco 4.5 (Braak and Šmilauer, 2002). Pearson correlation in SPSS 22.0 was used to uncover the relationship between the abundance, diversity and sediment physiochemical parameters. For network analysis, the highly representative OTUs were selected. The network analysis was conducted using the “Hmisc” package in R and was visualized in Gephi V0.9.2.
Results
Abundance and diversity of Nitrospira in coastal wetlands of China
Based on the qPCR results, Nitrospira was discovered in all of the sediment samples from 16 sites, and the nxrB gene of Nitrospira with an abundance between 4.96×106 - 3.88×107 copies/g dry sediment (Figure 1), was significantly (more than one magnitude) higher than amoA gene of ammonia-oxidizers (Sun et al., 2020). Among these studied areas, MLX has the most copy number of nxrB gene, similar as comammox Nitrospira amoA gene (Sun et al., 2020). Meanwhile, the abundance of the Nitrospira nxrB gene was lower in the southern than in the northern and middle estuaries.
Overall, 141,688 high-quality were generated from the 16 sediment samples after trimming and chimera moving. There Nitrospira nxrB gene sequences were further clustered into 451 OTUs based on 95% nucleotide similarity cutoff threshold, and the number of OTUs for each sample ranges from 53 to 201 (Table S1). The richness of Chao1 ranged from 64.14 (for the NLJ site) to 234.39 (for the JLJ site). For diversity indices, the Shannon indices ranged from 1.01 (for the BCYH site) to 3.39 (for the JLJ site) and Simpson indices ranged from 0.32 (for the BCYH site) to 0.93 (for the JLJ site).
Phylogeny of Nitrospira nxrB gene
The neighbor-joining tree based amino acid was constructed with 25 representative OTUs which are more than 0.5% abundance together constituting 85.26% of the total sequences (Figure 2). In sediment samples from 16 estuaries, we detected diverse Nitrospira phylotypes affiliated within lineage I, II and IV. Nitrospira lineage II and IV were more prevalent and abundant than lineage I, with the lineage I mainly present in 5 samples (YR, CJ, MJ, MLX and JLJ).
Figure 2
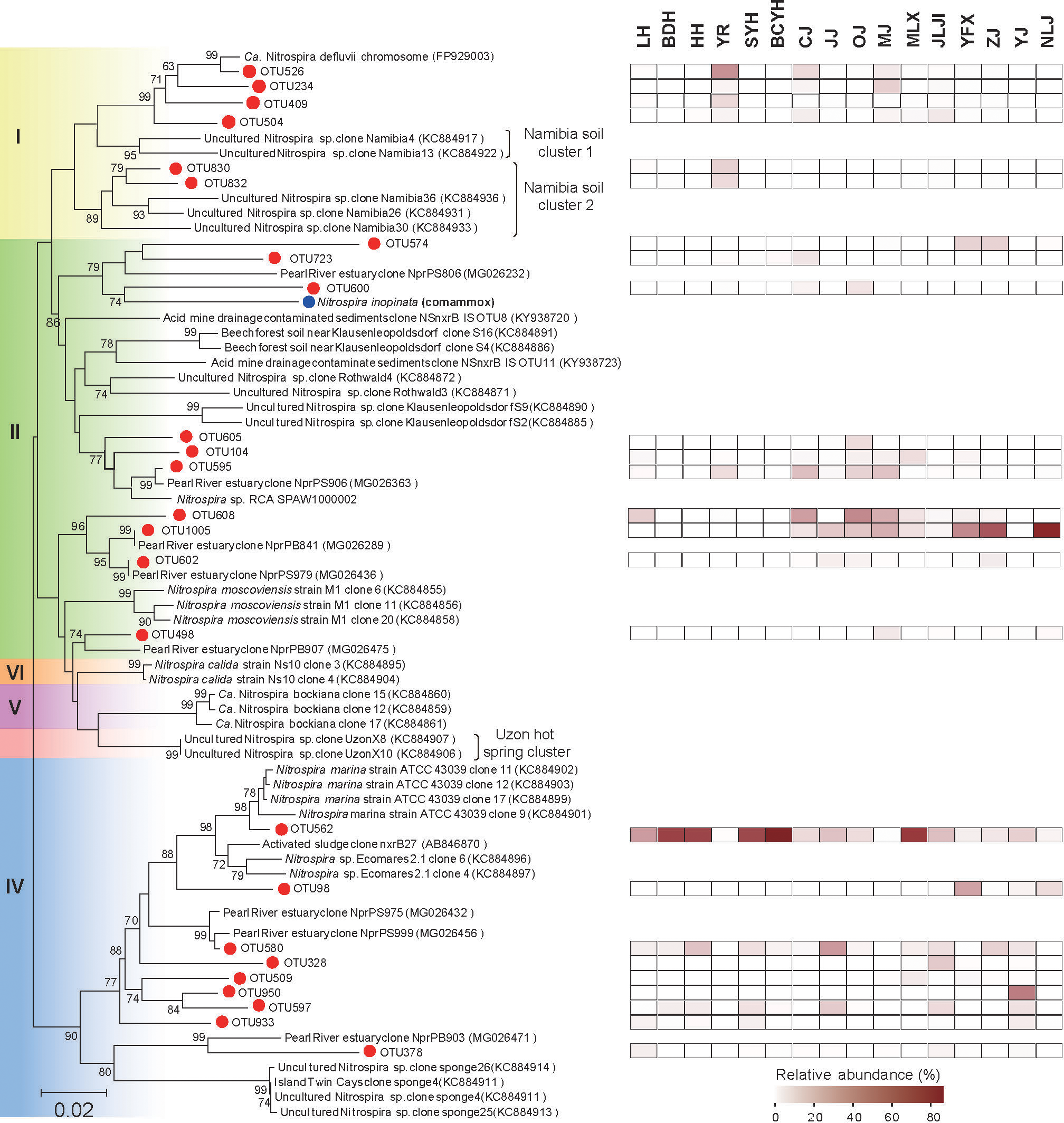
Neighbor-joining phylogenetic tree of Nitrospira nxrB genes. Sequences were collected from this study and the GenBank database. Bootstrap values were given on the branches of the tree. The scale bar represents 2% nucleic acid sequence divergence.
OTU 526 was mainly presented in YR and was the major OTU in lineage I, closely related to Ca. Nitrospira defluvii. Besides, the remaining four representative OTUs of lineage I clustered without any reported Nitrospira species, among them, OTU 830 and OTU 832 possessed a relatively low abundance and were only detected at YR. OTU 1005 was the most abundant OTU in Nitrospira lineage II and was related to Pearl River estuary clone NprPB841, similarly, OTU 1005 reached high relative abundance in Nanliu River (NLJ) and Pearl River (ZJ) estuaries. However, OTU 602, which is also closely related to Pearl River Estuary Clone NprPS979, was nearly absent in all sample sites. OTU 562 was dominant in Nitrospira lineage IV and was detected in most sample sites but YR. Interestingly, lineage I was not detected in the samples with higher relative abundance of lineage IV, such as BDH, HH, SYH, BCYH and MLX. Conversely, lineage IV was absent at YR and MJ, where lineage I was more abundant.
Community structure of Nitrospira
PCoA and NMDS analyses based on the Bray-Curtis distances of Nitrospira OTUs were performed to evaluate the differences of Nitrospira community structure in acidic areas (pH: 6-7) versus alkaline areas (pH: 7-9), and both of them exhibited a significant separation of Nitrospira community (Figure 3). In order to illustrate the occurrence pattern among Nitrospira, network was constructed according to the significance (Spearman, p<0.05) and correlation coefficients (Spearman, |x|>0.6) of highly representative Nitrospira OTUs. The network showed a complicated structure containing 42 nodes and 100 edges with the average degree of 4.762 and the average clustering coefficient of 0.573. In general, majority of the nodes belonged to Nitrospira lineage IV (50%), and the ratio of positive edges was higher (66%). Therein, the correlation between lineage I and lineage II was mainly positive, while both of them competed with lineage IV. Especially, there were no positive edges in OTU 622 belonging to Nitrospira lineage IV, suggesting the different physiological characteristics from most Nitrospira.
Figure 3
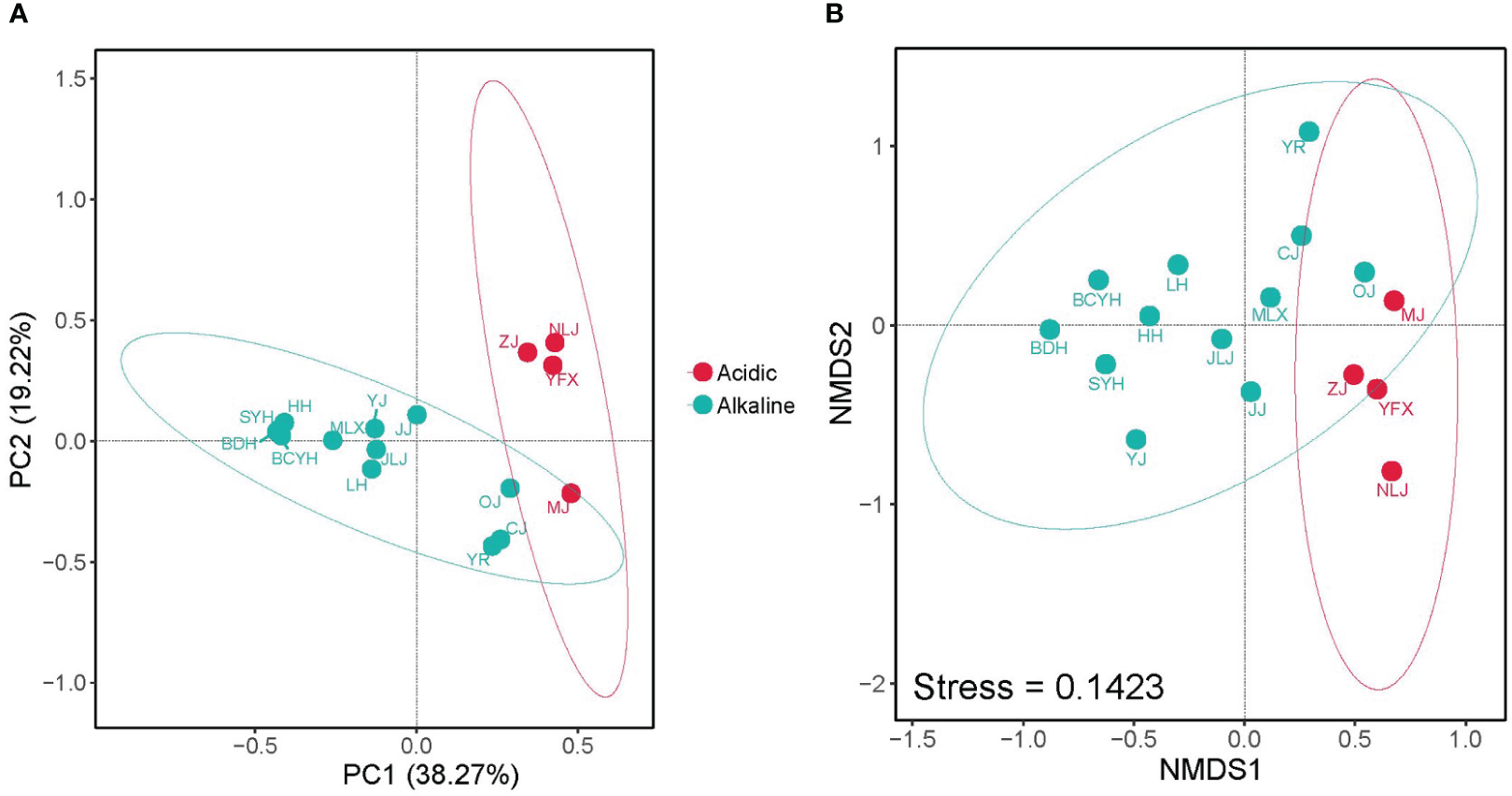
(A) PCoA plots of Nitrospira community dissimilarities among sampling sites based on Bray-Curtis distance; (B) NMDS plots showing the Nitrospira community composition separation. Red solid circle, acidic (pH: 6-7); Green solid circle, alkaline (pH: 7-9).
The effects of physiochemical parameters
The environmental factors of the sample sites refer to previous studies (Table S2). The correlations of Nitrospira community structure with physiochemical parameters were tested by CCA (Figure 4). The first two CCA dimensions explained 44.06% of the cumulative variance of the Nitrospira community-environment correlation. The results confirmed that Nitrospira community structures had significant correlation with pH (P=0.004, F=2.9), salinity (P=0.028, F=1.6) and particle size (P=0.046, F=2.5), theseparameters provided 38.3% of total CCA explanatory power. By contrast, other parameters had little effect on community structure (P>0.064), and they accounted for 50.9% of CCA explanatory power.
Figure 4
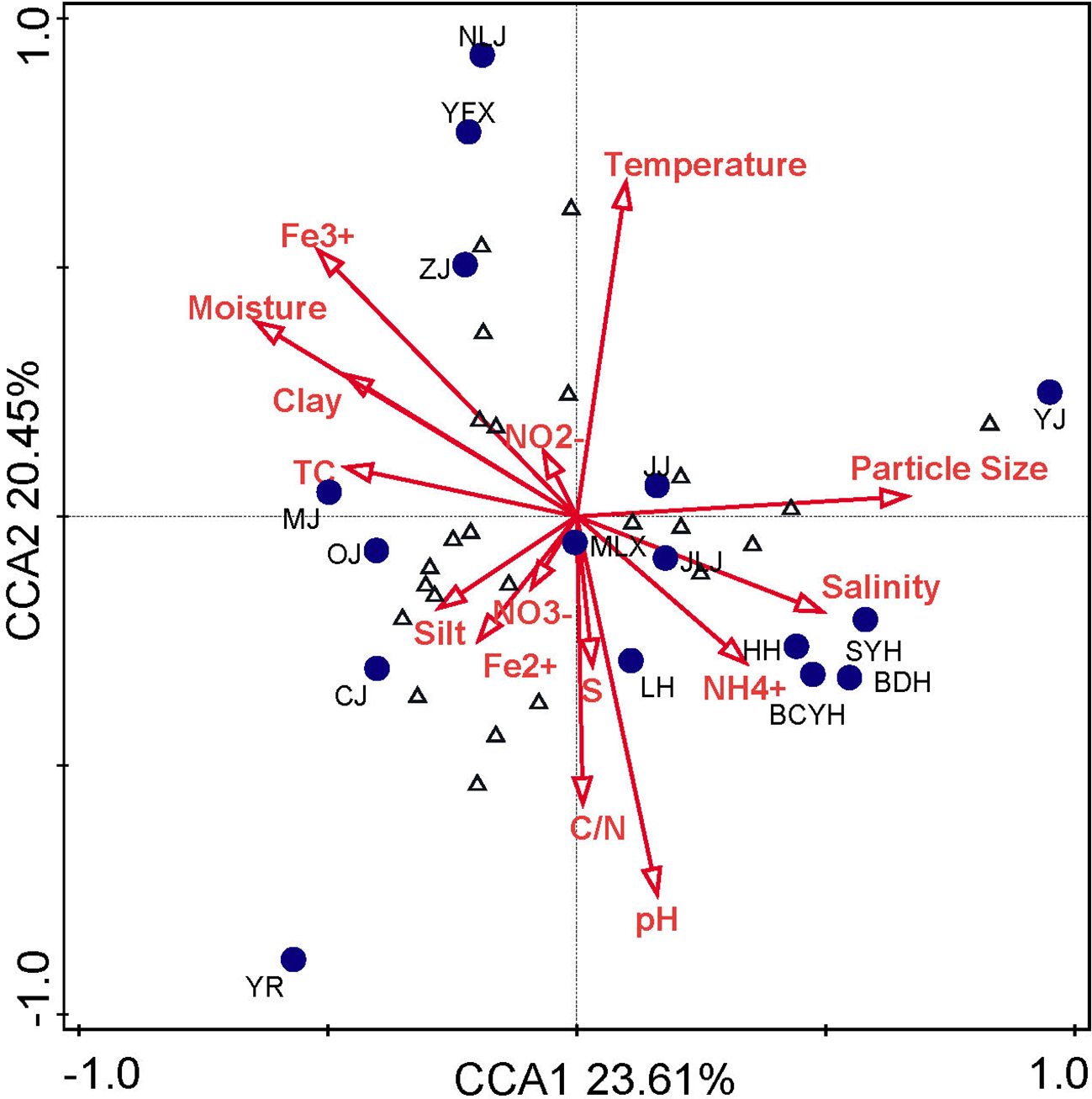
CCA plots for the first two principal dimensions of the relationship between the Nitrospira community pattern and the environmental parameters. Blue circles, samples; Blue triangles, Nitrospira OTUs; Red arrows, environmental factors.
Environmental impacts on Nitrospira diversity indices including the Chao1, Shannon, Simpson and OTU numbers were evaluated by Pearson correlation analysis (Table S3). The results demonstrated that Chao1 richness varied with salinity, but other indices seemed stable among different salinity. In addition, Shannon index, Simpson index and OTU numbers were negatively correlated to temperature, the lower diversity at higher temperature indicating that only a few Nitrospira species could tolerate high temperature. Shannon index and Simpson index also showed negative correlation with nitrate concentration, but no significant correlation with nitrite concentration. Meanwhile, OTU numbers were significantly correlated to ammonia and ferric ion content, additionally, the correlation between other parameters we studied and Nitrospira diversity indices was not significant.
Correlation between abundance of 12 dominant Nitrospira OTUs and environment parameters was performed by Spearman correlation analysis (Figure 5), which showed a strong association between the representative OTUs and selective physiochemical properties such as temperature, pH value, particle size, moisture and Fe3+. In general, the correlation of abundance with environment parameters was inverse between Nitrospira lineage I and lineage II. For example, OTUs affiliating with lineage II were negatively correlated with pH value, particle size and the content of sulfur, while the correlation with lineage I was positive. The content of Fe3+, total nitrogen, total organic carbon showed positive correlation with the relative abundance of lineage II, but negative correlation with lineage I. Besides, temperature was positively correlated to the OTUs belonging to lineage IV but negatively correlated to lineage I, suggesting the different adaptations to temperature among them.
Figure 5
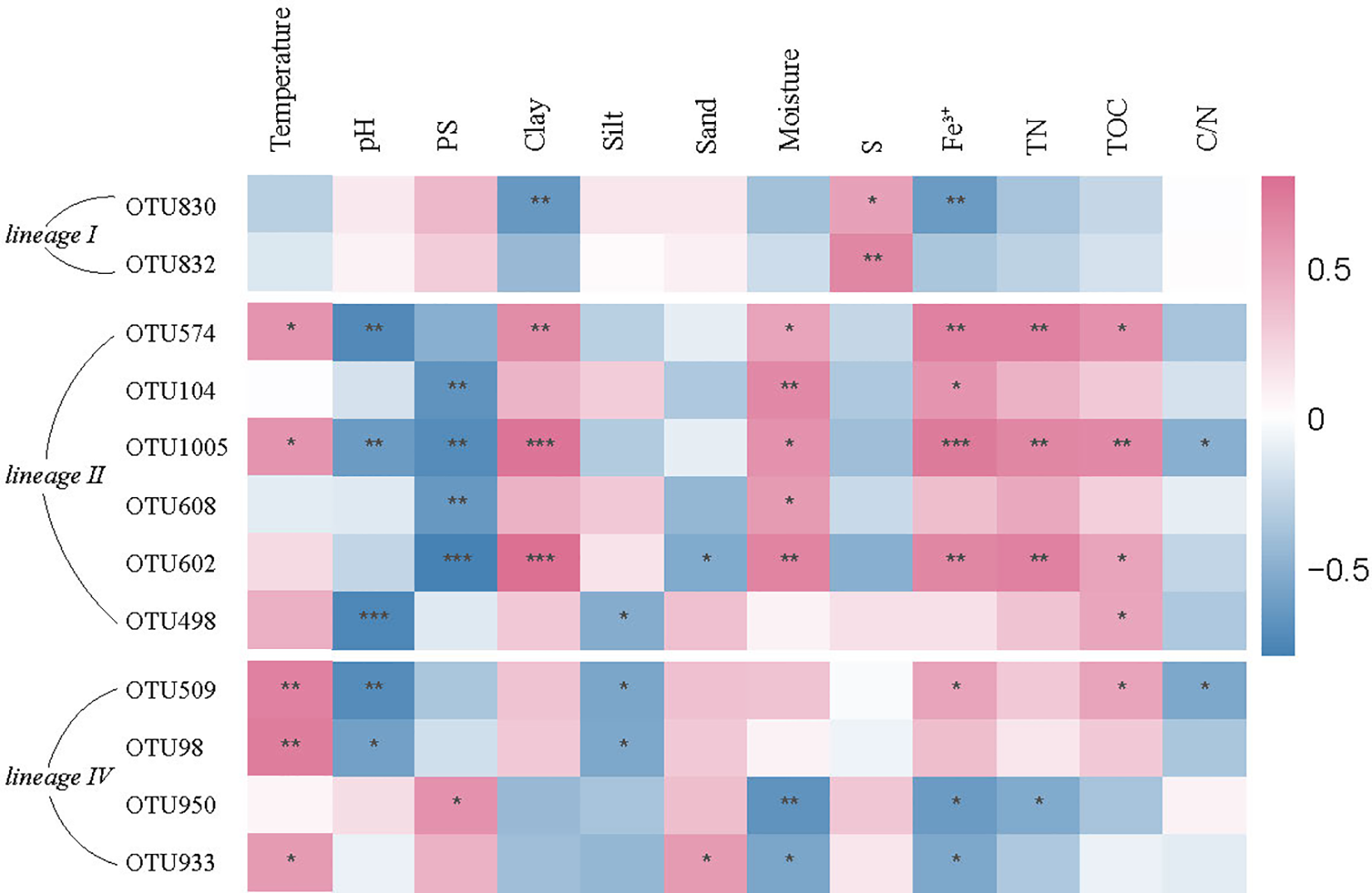
Correlation of relative abundance of 12 dominant Nitrospira OTUs with environment parameters. *P<0.05, **P<0.01, ***P<0.001. PS, particle size; S, sulfur; TN, total nitrogen; TOC, total organic carbon; C/N, ratio of carbon to nitrogen.
Discussion
Community pattern of Nitrospira bacteria in coastal wetlands of China
Members of the Nitrospira bacteria were considered as the most diverse and widespread known NOB. However, distribution pattern of Nitrospira in large-scale coastal wetland are missing, and a systematic assessment of multi-factor effects their different niche adaptation strategies is scarce (Daims et al., 2016). In this study, we investigated the abundance, diversity and community pattern of Nitrospira in sediments along the eastern estuary and coastal line of China and identified the major driving environmental factors of their niche partitions.
Quantification results showed that the nxrB gene of Nitrospira was detected in all of the sediment samples, and was more abundant than ammonia oxidizers (Sun et al., 2020). This result was in line with one previous study showing the high proportion of NOB in the detected environment (Winkler et al., 2012), and the growth on alternative energy source from hydrogen or formate utilization may sometimes explain higher in situ abundances of NOB than their ammonia-oxidizing counterparts (Koch et al., 2014; Koch et al., 2015). According to the phylogenetic analysis, Nitrospira lineage IV was present at all sites except YR at which lineage I was dominant. Meanwhile, the relative abundance of lineage IV was higher than other two lineage at most sites, suggesting a dominant but fluctuating role of lineage IV in the nitrite oxidizing of coastal wetlands. Hitherto, strains belonging to Nitrospira lineage I and lineage II were mainly obtained from engineering systems, but rarely founded in marine (Ushiki et al., 2013; Fujitani et al., 2014; Gruber-Dorninger et al., 2015). In contrast, Nitrospira lineage IV was usually present in marine or hypersaline systems (Watson et al., 2004; Daebeler et al., 2020; Bayer et al., 2021). Due to the influence of the ocean, most of the sample sites we studied showed significant marine characteristics, therefore lineage IV was more abundant. As a large amount of sediment from the upstream deposited in the estuary, the riverbed at the YR site uplifted and turned into the elevated stream, thus was relatively less affected by seawater, this may be the reason why lineage IV was not found here. In addition to the large amount of nutrients carried by sediment, the excessive application of fertilizers in the downstream of the Yellow River caused inorganic nitrogen was enriched in the estuary, therefore lineage I was dominant here.
The network analysis showed that the positive correlations were more dominant among Nitrospira (Figure 6), suggesting the cooperation is more than competition in the relationship of Nitrospira population. Interestingly, OTU 622 belonging to Nitrospira lineage IV was negatively corelated with the OTUs of lineage I, II and IV, suggesting its unique characteristic in Nitrospira.
Figure 6
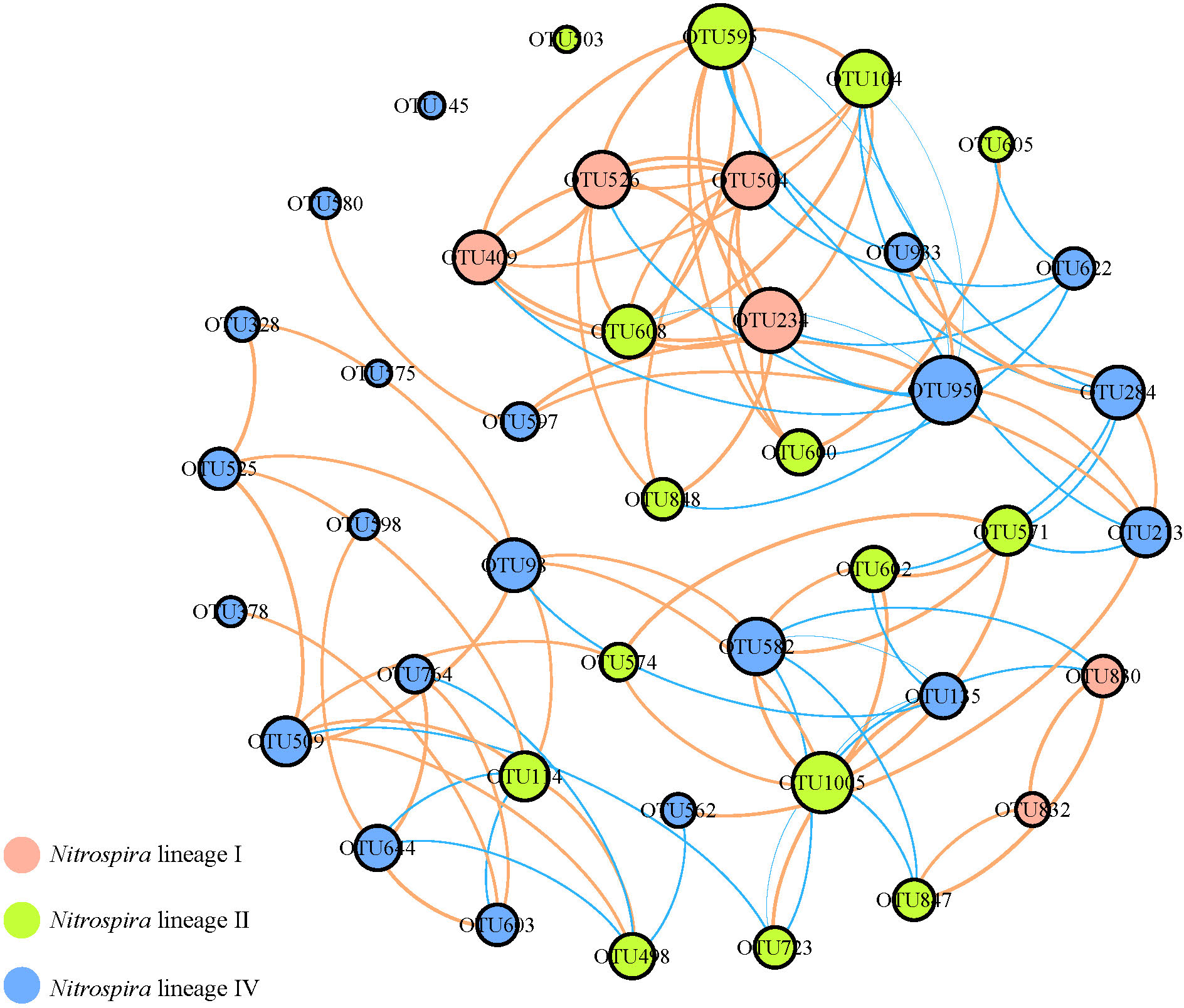
Co-occurrence network analysis of representative Nitrospira OTUs. The size of circles means relative abundance; orange lines represent negative interactions; blue lines represent positive interactions.
Influence of environmental factors on the distribution of Nitrospira bacteria in coastal wetlands of China
Previous study proved that the concentration of nitrite was a key factor affecting the population and niche separation, the growth of some Nitrospira would suspend with the increasing nitrite concentration (Wagner et al., 2002). However, the correlation between nitrite concentration with abundance or diversity was not significant in this study, which may be explained by the limited accumulation of nitrite in natural environment. Besides, we investigated more other physicochemical parameters contributing to the community construction, which broaden our understanding of niche differentiation of Nitrospira.
It is generally accepted that Nitrospira strains grew in the optimum temperature between 25 and 31 (Spieck and Lipski, 2011), while nitrite oxidation by these organisms was still detected in a broad temperature range (10 to 65 °C) (Alawi et al., 2009). Comparatively, Nitrospira lineage I has weak tolerance to higher temperature, and Nitrospira found in geothermal springs was mainly affiliated with lineage II and lineage IV (Lebedeva et al., 2011; Marks et al., 2012; Edwards et al., 2013). Additionally, the studies performed in the Yangtze River showed that, with the increase of ambient temperature, the relative abundance of NOB-Nitrospira lineage I or lineage II decreased, while that of comammox-Nitrospira increased (Liu et al., 2020). Nitrospira showed different adaptation characteristics to temperature, only a few strains were able to tolerate the higher temperature (Alawi et al., 2009). Similarly, Nitrospira diversity tended to decrease with the elevating temperature in this study. Meanwhile, the positive correlation between the relative abundance of lineage IV OTUs and temperature suggesting lineage IV was more adaptable to higher temperature conditions.
The recent study reported that Nitrospira population was decreased with the salinity gradient (Cébron and Garnier, 2005), whereas another study showed the richness of Nitrospira in the high saline regions of Pearl River estuary (Hou et al., 2018a). Members of Nitrospira lineage IV were mostly detected in marine and hypersaline environments so far (Watson et al., 2004; Foesel et al., 2008; Keuter et al., 2011; Haaijer et al., 2013; Daebeler et al., 2020), but there were no members of lineage I and II found in these habitats (Santos et al., 2017; Sun et al., 2020), suggesting the different adaption of Nitrospira lineage to salinity condition. Here, we discovered a higher abundance of Nitrospira lineage I and II in the estuaries that salinity was low, while lower than lineage IV when the salinity was above 1 ppt. The result might guarantee the tolerance of Nitrospira lineage IV to hypersaline habitats and the role of salinity in niche separation between Nitrospira lineage IV and lineage I, II.
In addition to salinity, pH was another environmental factor that affects the abundance and diversity of Nitrospira. Previous study reported that Nitrospira defluvii belonging to lineage I grew best at pH 7.3 (Hupeden et al., 2016). Besides, study of Nitrospira community in the saline-alkaline lakes showed the decrease of Nitrospira diversity with the increasing pH and the Nitrospira lineage IV enrichment cultures, “Candidatus Nitrospira alkalitolerans”, could oxidize nitrite at the condition of neutral to pH 10.5 (Daebeler et al., 2020). In this study, both PCoA and NMDS analysis showed a clear separation of the Nitrospira communities with the pH conditions (Figure 3). According to the results of CCA analysis, the relative abundance of Nitrospira OTUs was significantly correlated with pH. The result of Pearson correlation showed that OTUs number was significantly negative corelated with pH value, indicating that only specific Nitrospira species tolerate the highly alkaline conditions. The negative correlation between relative abundance of Nitrospira lineage II and pH might explain the vacancies of Nitrospira lineage II detected in alkaline environments. These results suggested a strong influence of pH on the community pattern of Nitrospira bacteria.
As a chemical catalyst, iron could enhance the cell membrane permeability and hence accelerate the nutrients transmission rate. Meanwhile, iron presents in the center of various active enzymes which function in the molecular oxygen transportation and the nitrogen species’ transformation in nitrifying microbes. Fe3+ in NOB also was hypothesized to act as alternative terminal electron acceptors for nitrite oxidation in the oxygen minimum zone (Lam and Kuypers, 2011). Here, our study showed that Fe3+ content was positively correlated with the relative abundance of Nitrospira lineage II (Figure 5), whereas this correlation tended to be negative with lineage I. Besides, the significantly negative correlation between Fe3+ content and Nitrospira OTUs was showed by the Pearson correlation. All of the results suggested Fe3+ might play different role in Nitrospira lineages, and further physiological verification is needed.
Particle size directly influences the physicochemical property of the sediments and thereby affects the distribution of microbial community (Dang et al., 2010). We found that Nitrospira lineage II abundance was negatively correlated with particle size of sediments. In addition, growth of Nitrospira could be inhibited under high concentration of nitrate. For instance, growth of N.moscoviensis ceased to grow in the presence of 75mM nitrate (Ehrich et al., 1995), and the marine lineage IV Nitrospira ecomares 2.1 80mM nitrate (Keuter et al., 2011). Consistently, our study showed that Nitrospira diversity decreased with the increasing nitrate concentration. Moreover, previous study showed the high concentration of TOC would mutually strengthen the effect on nitrification in low oxygen conditions (Ling and Chen, 2005). Here, with the low oxygen concentration of estuarine tidal flats, there were a positive correlation between TOC and the relative abundance of Nitrospira lineage II, suggesting the community composition of Nitrospira was affected by TOC, whereas the interaction mechanism requires further research. As for any other parameters, clay and moisture had a remarkable influence on the abundance of Nitrospira OTUs, whereas sulfur content seemed to only significantly effect Nitrospira lineage I. These factors might exhibit a role in niche differentiation of Nitrospira lineage I and II, therefore the experimental tests are needed.
Concluding remarks
Nitrospira was prevalent and abundant in the estuarine tidal flat wetlands of China and was higher than ammonia oxidizers in abundance. Meanwhile, Nitrospira phylotypes were affiliated with Nitrospira lineage I, II and IV, in which lineage IV was dominant but fluctuating at different sites.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
QZ: Conceptualization, Data curation, Formal analysis, Investigation, Software, Visualization, Writing - original draft, Writing - review & editing. DS: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing - review & editing. XT: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing - review & editing. LH: Conceptualization, Data curation, Funding acquisition, Investigation, Resources, Writing - review & editing. ML: Conceptualization, Funding acquisition, Investigation, Resources, Writing - review & editing. PH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (Nos. 42371064, 41807465 and 42030411) and the Starting Research Program of Suzhou University of Science and Technology (No. 332214803).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1288142/full#supplementary-material
References
1
Alawi M. Off S. Kaya M. Spieck E. (2009). Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Env. Microbiol. Rep.1, 184–190. doi: 10.1111/j.1758-2229.2009.00029.x
2
Altmann D. Stief P. Amann R. De Beer D. Schramm A. (2003). In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol.5, 798–803. doi: 10.1046/j.1469-2920.2003.00469.x
3
Annavajhala M. K. Kapoor V. Santo-Domingo J. Chandran K. (2018). Comammox functionality identified in diverse engineered biological wastewater treatment systems. Environ. Sci. Technol. Lett.5, 110–116. doi: 10.1021/acs.estlett.7b00577
4
Bayer B. Saito M. A. Mcilvin M. R. Lucker S. Moran D. M. Lankiewicz T. S. et al . (2021). Metabolic versatility of the nitrite-oxidizing bacterium Nitrospira marina and its proteomic response to oxygen-limited conditions. ISME J.15, 1025–1039. doi: 10.1038/s41396-020-00828-3
5
Braak C. J. F. T. Šmilauer P. (2002). CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5). Ithaca: Microcomputer Power, p500.
6
Caporaso J. G. Kuczynski J. Stombaugh J. Bittinger K. Bushman F. D. Costello E. K. et al . (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods7, 335–336. doi: 10.1038/nmeth.f.303
7
Cébron A. Garnier J. (2005). Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: detection, quantification and growth along the lower Seine River (France). Water Res.39 20, 4979–4992. doi: 10.1016/j.watres.2005.10.006
8
Daebeler A. Kitzinger K. Koch H. Herbold C. W. Steinfeder M. Schwarz J. et al . (2020). Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant Nitrospira. ISME J.14, 2967–2979. doi: 10.1038/s41396-020-0724-1
9
Daims H. Lebedeva E. V. Pjevac P. Han P. Herbold C. Albertsen M. et al . (2015). Complete nitrification by Nitrospira bacteria. Nature528, 504–509. doi: 10.1038/nature16461
10
Daims H. Lucker S. Wagner M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol.24, 699–712. doi: 10.1016/j.tim.2016.05.004
11
Daims H. Nielsen J. L. Nielsen P. H. Schleifer K. H. Wagner M. (2001). In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol.67, 5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001
12
Dang H. Li J. Chen R. Wang L. Guo L. Zhang Z. et al . (2010). Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl. Environ. Microbiol.76, 4691–4702. doi: 10.1128/AEM.02563-09
13
Edwards T. A. Calica N. A. Huang D. A. Manoharan N. Hou W. Huang L. et al . (2013). Cultivation and characterization of thermophilic Nitrospira species from geothermal springs in the US Great Basin, China, and Armenia. FEMS Microbiol. Ecol.85, 283–292. doi: 10.1111/1574-6941.12117
14
Ehrich S. Behrens D. Lebedeva E. Ludwig W. Bock E. (1995). A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol.164, 16–23. doi: 10.1007/BF02568729
15
Foesel B. U. Gieseke A. Schwermer C. Stief P. Koch L. Cytryn E. et al . (2008). Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol. Ecol.63, 192–204. doi: 10.1111/j.1574-6941.2007.00418.x
16
Fujitani H. Ushiki N. Tsuneda S. Aoi Y. (2014). Isolation of sublineage I Nitrospira by a novel cultivation strategy. Environ. Microbiol.16, 3030–3040. doi: 10.1111/1462-2920.12248
17
Gruber-Dorninger C. Pester M. Kitzinger K. Savio D. F. Loy A. Rattei T. et al . (2015). Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J.9, 643–655. doi: 10.1038/ismej.2014.156
18
Haaijer S. C. Ji K. Van Niftrik L. Hoischen A. Speth D. Jetten M. S. et al . (2013). A novel marine nitrite-oxidizing Nitrospira species from Dutch coastal North Sea water. Front. Microbiol.4, 60. doi: 10.3389/fmicb.2013.00060
19
Hou L. J. Wang R. Yin G. Y. Liu M. Zheng Y. L. (2018b). Nitrogen fixation in the intertidal sediments of the Yangtze Estuary: occurrence and environmental implications. J. Geophys. Res.-Biogeo.123, 936–944. doi: 10.1002/2018JG004418
20
Hou L. Xie X. B. Wan X. H. Kao S. J. Jiao N. Z. Zhang Y. (2018a). Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the Pearl River estuary to the South China Sea. Biogeosciences15, 5169–5187. doi: 10.5194/bg-2018-189
21
Hou L. Yin G. Liu M. Zhou J. Zheng Y. Gao J. et al . (2015). Effects of sulfamethazine on denitrification and the associated N2O release in estuarine and coastal sediments. Environ. Sci. Technol.49, 326–333. doi: 10.1021/es504433r
22
Hupeden J. Wegen S. Off S. Lucker S. Bedarf Y. Daims H. et al . (2016). Relative abundance of Nitrotoga spp. in a biofilter of a cold-freshwater aquaculture plant appears to be stimulated by slightly acidic pH. Appl. Environ. Microbiol.82, 1838–1845. doi: 10.1128/AEM.03163-15
23
Keuter S. Kruse M. Lipski A. Spieck E. (2011). Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ. Microbiol.13, 2536–2547. doi: 10.1111/j.1462-2920.2011.02525.x
24
Koch H. Galushko A. Albertsen M. Schintlmeister A. Gruber-Dorninger C. Lucker S. et al . (2014). Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science345, 1052–1054. doi: 10.1126/science.1256985
25
Koch H. Lucker S. Albertsen M. Kitzinger K. Herbold C. Spieck E. et al . (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. U.S.A.112, 11371–11376. doi: 10.1073/pnas.1506533112
26
Kruse M. Zumbragel S. Bakker E. Spieck E. Eggers T. Lipski A. (2013). The nitrite-oxidizing community in activated sludge from a municipal wastewater treatment plant determined by fatty acid methyl ester-stable isotope probing. Syst. Appl. Microbiol.36, 517–524. doi: 10.1016/j.syapm.2013.06.007
27
Kumar S. Tamura K. Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform.5, 150–163. doi: 10.1093/bib/5.2.150
28
Kuypers M. M. M. Marchant H. K. Kartal B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol.16, 263–276. doi: 10.1038/nrmicro.2018.9
29
Lam P. Kuypers M. M. (2011). Microbial nitrogen cycling processes in oxygen minimum zones. Ann. Rev. Mar. Sci.3, 317–345. doi: 10.1146/annurev-marine-120709-142814
30
Lebedeva E. V. Alawi M. Fiencke C. Namsaraev B. Bock E. Spieck E. (2005). Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol. Ecol.54, 297–306. doi: 10.1016/j.femsec.2005.04.010
31
Lebedeva E. V. Off S. Zumbragel S. Kruse M. Shagzhina A. Lucker S. et al . (2011). Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol. Ecol.75, 195–204. doi: 10.1111/j.1574-6941.2010.01006.x
32
Ling J. Chen S. L. (2005). Impact of organic carbon on nitrification performance of different biofilters. Aquacult. Eng.33, 150–162. doi: 10.1016/j.aquaeng.2004.12.002
33
Liu S. F. Wang H. Y. Chen L. M. Wang J. W. Zheng M. S. Liu S. T. et al . (2020). Comammox Nitrospira within the Yangtze River continuum: community, biogeography, and ecological drivers. ISME J.14, 2488–2504. doi: 10.1038/s41396-020-0701-8
34
Lucker S. Wagner M. Maixner F. Pelletier E. Koch H. Vacherie B. et al . (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A.107, 13479–13484. doi: 10.1073/pnas.1003860107
35
Marks C. R. Stevenson B. S. Rudd S. Lawson P. A. (2012). Nitrospira-dominated biofilm within a thermal artesian spring: a case for nitrification-driven primary production in a geothermal setting. Geobiology10, 457–466. doi: 10.1111/j.1472-4669.2012.00335.x
36
Okabe S. Watanabe H. S. Y. (1998). In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of Microelectrodes. Appl. Environ. Microbiol.65, 3182–3191. doi: 10.1128/AEM.65.7.3182-3191.1999
37
Okasen J. Kindt R. Legendre P. O’Hara R. B. (2007). Vegan: community ecology package version. 1.8–6. Available at: https://cran.rproject.org/web/packages/vegan.
38
Osburn C. L. Handsel L. T. Peierls B. L. Paerl H. W. (2016). Predicting sources of dissolved organic nitrogen to an estuary from an agro-urban coastal watershed. Environ. Sci. Technol.50, 8473–8484. doi: 10.1021/acs.est.6b00053
39
Pachiadaki M. G. Sintes E. Bergauer K. Brown J. M. Record N. R. Swan B. K. et al . (2017). Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science358, 1046–1051. doi: 10.1126/science.aan8260
40
Palatinszky M. Herbold C. Jehmlich N. Pogoda M. Han P. Von Bergen M. et al . (2015). Cyanate as an energy source for nitrifiers. Nature524, 105–108. doi: 10.1038/nature14856
41
Park H.-D. Noguera D. R . (2008). Nitrospira community composition in nitrifying reactors operated with two different dissolved oxygen levels. J. Microbiol. Biotechnol.18, 1470–1474. doi: 10.1007/s10295-008-0366-1
42
Pester M. Maixner F. Berry D. Rattei T. Koch H. Lucker S. et al . (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol.16, 3055–3071. doi: 10.1111/1462-2920.12300
43
Santos J. P. Mendes D. Monteiro M. Ribeiro H. Baptista M. S. Borges M. et al . (2017). Salinity impact on ammonia oxidizers activity and amoA expression in estuarine sediments. Estuar. Coast. Shelf S.211, 177–187. doi: 10.1016/j.ecss.2017.09.001
44
Schloss P. D. (2013). Secondary structure improves OTU assignments of 16S rRNA gene sequences. ISME J.7, 457–460. doi: 10.1038/ismej.2012.102
45
Spieck E. Lipski A. (2011). Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria. Methods Enzymology: Res. Nitrification Related Processes486, 109–130. doi: 10.1016/B978-0-12-381294-0.00005-5
46
Sun D. Tang X. Zhao M. Zhang Z. Hou L. Liu M. et al . (2020). Distribution and diversity of comammox Nitrospira in coastal wetlands of China. Front. Microbiol.11, 589268. doi: 10.3389/fmicb.2020.589268
47
Tamura K. Dudley J. Nei M. Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol.24, 1596–1599. doi: 10.1093/molbev/msm092
48
R. Core Team . (2020). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
49
Thompson J. D. Gibson T. J. Plewniak F. Jeanmougin F. Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res.25, 4876–4882. doi: 10.1093/nar/25.24.4876
50
Ushiki N. Fujitani H. Aoi Y. Tsuneda S. (2013). Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ.28, 346–353. doi: 10.1264/jsme2.ME13042
51
van Kessel M. A. Speth D. R. Albertsen M. Nielsen P. H. Op Den Camp H. J. Kartal B. et al . (2015). Complete nitrification by a single microorganism. Nature528, 555–559. doi: 10.1038/nature16459
52
Wagner M. Loy A. Nogueira R. Purkhold U. Lee N. Daims H. (2002). Microbial community composition and function in wastewater treatment plants. Anton. Leeuw. Int. J. G.81, 665–680. doi: 10.1023/A:1020586312170
53
Watson S. W. Bock E. Valois F. Waterbury J. Schlosser U. (2004). Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol.144, 1–7. doi: 10.1007/BF00454947
54
Winkler M. K. Bassin J. P. Kleerebezem R. Sorokin D. Y. Van Loosdrecht M. C. (2012). Unravelling the reasons for disproportion in the ratio of AOB and NOB in aerobic granular sludge. Appl. Microbiol. Biotechnol.94, 1657–1666. doi: 10.1007/s00253-012-4126-9
Summary
Keywords
coastal wetlands, Nitrospira , nitrification, distribution pattern, environmental driving factors
Citation
Zhao Q, Sun D, Tang X, Hou L, Liu M and Han P (2023) An investigation of Nitrospira bacteria in coastal wetlands of China: distribution pattern and related environmental driving factors. Front. Mar. Sci. 10:1288142. doi: 10.3389/fmars.2023.1288142
Received
03 September 2023
Accepted
03 November 2023
Published
17 November 2023
Volume
10 - 2023
Edited by
Xue-Wei Xu, Ministry of Natural Resources, China
Reviewed by
Jiapeng Wu, Guangzhou University, China; Heng-Lin Cui, Jiangsu University, China; Hongchen Jiang, China University of Geosciences Wuhan, China
Updates
Copyright
© 2023 Zhao, Sun, Tang, Hou, Liu and Han.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Han, phan@geo.ecnu.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.