
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 03 January 2024
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1283893
 YuJin Jeong1
YuJin Jeong1 JunMo Lee1,2*
JunMo Lee1,2*Diatoms (Bacillariophyta) are abundant phytoplankton groups in marine environments, which contribute approximately 20% of global carbon fixation through photosynthesis. Moreover, diatoms exhibit the highest species diversity (approximately 18,000 diatom species) among marine photosynthetic eukaryotes, which were identified by morphological characteristics. Molecular phylogenetic analyses could shed new insights into the evolutionary relationships of diverse diatom species. Nevertheless, a comprehensive understanding of the phylogenetic relationships of diatom species still remains unclear because the available molecular data are insufficient compared with their high species diversity. Furthermore, several novel diatom species were reported from field samples with no molecular evidence. In particular, the phylogenies of diatom species constructed using organelle genomes revealed that several diatom genera are paraphyletic with high supporting values. We constructed high-resolution phylogenetic trees of diatom species using organelle genomes (plastids and mitochondria) and compared the morphologies in several paraphyletic diatom genera. Especially, the clades Nitzschia and Thalassiosira include several different diatom genera with high phylogenetic supports. Our study demonstrated that some morphological characteristics (e.g., genus characters) of several diatom genera could not represent current genus boundaries. Based on the results, we highlight the necessity for taxonomic reinvestigation. To reestablish this in diatoms, it will be essential to incorporate more genome data from a broader range of taxon samples, along with a comparison of morphological characteristics.
Diatoms (Bacillariophyta) are unicellular photosynthetic eukaryotes widely distributed in marine environments (Mann et al., 2017). A total of 18,374 diatom species have been reported based on morphological characteristics (Hasle and Syvertsen, 1997; Finkel and Kotrc, 2010), which indicates the highest species diversity among photosynthetic algae (e.g., red algae: 7,538, green algae: 7,766, and brown algae: 2,120; Algaebase; Guiry and Guiry, 2023). Nonetheless, a comprehensive understanding for molecular phylogenies of diatom species still remains unaddressed because available molecular data (e.g., molecular markers and organelle genomes) is insufficient to construct a high-resolute phylogenetic tree of diatom species despite their high species diversity (Visco et al., 2015; Wang et al., 2022a). Furthermore, several novel diatom species were reported with no molecular evidence because they were observed from field samples (Kryk et al., 2021; Kulikovskiy et al., 2021). Mann et al. (2021) mentioned about controversial molecular data from morphologically unverified diatom species, which are available in the public databases (e.g., NCBI). Therefore, to establish their phylogenetic relationships, sufficient molecular data are required from morphologically verified diatom species.
Genomic studies are increasingly in diverse taxa due to the significantly reduced costs of genome sequencing approaches (e.g., next-generation sequencing technologies). As a result, huge amounts of genomic information are currently available. The phylogenetic analysis using single or several nuclear marker genes generally show insufficient phylogenetic resolution with low support values (Winchell et al., 2004; Bruder and Medlin, 2007; Dong et al., 2010). In addition, nuclear genetic variation could result from hybridization and introgression through sexual reproduction (Baack and Rieseberg, 2007; Colbeck et al., 2011; Harrison and Larson, 2016). In contrast, the organelle genomes are maternally inherited, and as a result, they generally have conserved gene contents, which are useful for phylogenetic studies (Timmis et al., 2004; Lee et al., 2016; Liu et al., 2021). In addition, the organelle genomes are relatively easier to handle in the generation and analysis of sequencing data compared to complete nuclear genome data (Cunha et al., 2009; Song et al., 2016). Consequently, the organelle genomes have much more available data than complete nuclear genomes. In diatoms, for example, 141 plastid and 81 mitochondrial genomes were reported in the NCBI database, while complete nuclear genomes were reported from only seven species (November 2023; NCBI database). Phylogenetic analyses using concatenated genes from organelle genomes could provide high-resolution phylogenetic relationship in diverse taxa (Lemieux et al., 2007; Lee et al., 2016; Jeong and Lee, 2021; Liu et al., 2021). Through these phylogenetic approaches, traditionally established taxonomic relationships were revised in several taxonomic groups (Wang et al., 2022b; Park et al., 2023). To study phylogenetic relationships in diatom species, therefore, current studies have been frequently conducted using organelle genome (Jeong and Lee, 2021; Wang et al., 2022a). Interestingly, the phylogeny analysis using organelle genomes shows that several diatom genera exhibit paraphyletic relationships with high phylogenetic support values (Liu et al., 2021; Wang et al., 2022a). For instance, the genus Nitzschia are paraphyletic with respect to the genera Cylindrotheca, Pseudo-nitzschia, and Fragilariopsis (Wang et al., 2022a). In particular, the genus Nitzschia exhibits a high species diversity among diatom genera (878 Nitzschia species are reported; Algaebase; Guiry and Guiry, 2023). To better understand the phylogenetic relationships in diatoms, more organelle genome data are required in diverse diatom taxa. However, the number of available organelle genomes is still insufficient to cover their high species diversity, estimated at about 100,000 species (Mann et al., 2017). In addition, further discussion is required to address the conflict between phylogenetic relationships and morphology-based genus boundaries.
In this study, we newly report the complete organelle genomes (plastids and mitochondria) from Entomoneis umbratica, Navicula avium, Pleurosigma inscriptura, Nitzschia dissipatoides, Nit. anomalus sp. nov., and Nit. reversa var. latus var. nov. We constructed multigene phylogenies using available organelle genomes of diatom species, and discussed about the morphological similarity in several diatom genera, which exhibit paraphyletic with high phylogenetic supports. We suggest a potential for taxonomic revisions of the diatom taxa (or genera).
Target diatoms were collected from field samples in Korean coastal waters. Entomoneis umbratica (MEG002), Navicula avium (MEG004), Pleurosigma inscriptura (MEG005), and Nitzschia reversa var. latus (MEG012) were isolated from Gijang (Busan, Korea; 35°21′82.93″N, 129°22′97.99″E) on June 12, 2020. Nit. anomalus (MEG011) and Nit. dissipatoides (MEG028) were isolated from Homigot (Pohang, Korea; 36°02′19.1′′N 129°34′49.4′′E; May 17, 2020) and Shinan-gun (Jeollanam-do, Korea; 34°83′20.36″N, 126°36′64.67″E; December 5, 2020), respectively (Figure 1). Single-cell isolation method was performed using customized Pasteur pipettes (glass) under light microscopic observation. The isolated strains were cultured in L1 medium at 20°C (Marine Ecological Genomics Lab. at Kyungpook National University).
The morphological characteristics of the target diatoms were observed using a light microscope (LM; Nikon ECLIPSE Ni-U, Nikon, Tokyo, Japan), scanning electron microscope (SEM; Hitachi SU8220; Hitachi Ltd., Tokyo, Japan), and transmission electron microscope (TEM; Hitachi HT7700, Hitachi Ltd., Tokyo, Japan). The diatom cultures were fixed with 2.5% glutaraldehyde, and the organic matters of diatom cells were removed using sodium hypochlorite (NaClO; Vilhena et al., 2021). The type specimens (permanent slides) of diatom cells (Nit. anomalus sp. nov. MABIK DI00043462 and Nit. reversa var. latus var. nov. MABIK DI00043463) prepared in this study were deposited to the National Marine Biodiversity Institute of Korea (MABIK; http://www.mabik.re.kr).
Genomic DNA was extracted using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Genome sequencing was performed using Illumina Novaseq 6000 (Illumina, San Diego, CA; 150 bp paired-end library), and the sequencing raw reads were assembled using SPAdes assembler (v3.14.2; Bankevich et al., 2012). To predict 18S rDNA regions from the assembled genomes, local BLASTn searches (e-value cutoff = 1.e-05) were performed with 18S rDNA sequences of Phaeodactylum tricornutum (JF489968.1).
Organelle (plastid and mitochondrion) contigs were selected by local BLASTn search (e-value cutoff = 1.e-05) using their comparable organelle genomes (MF997419.1, NC056793.1, MT383639.1, NC038001.1, NC056794.1, and MW971520.1), and reassembled as circular organelle genomes using Geneious Prime (v2023.0.4). Protein-coding regions in the organelle genomes were manually predicted by BLASTx search (NCBI nr database; e-value cutoff = 1.e-05) with codon table 11 (plastid) and 4 (mitochondria), and annotated using Geneious Prime (v2023.0.4.). Transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) were analyzed using tRNA scan-SE 2.0 (Lowe and Chan, 2016; Chan and Lowe, 2019) and barrnap 0.9 (https://github.com/tseemann/barrnap), respectively.
The 18S rDNA phylogenies were constructed using the BLASTn top 1,000 hits (e-value cutoff = 1.e-05; NT database in NCBI) of 18s rDNA sequences from the target diatoms, excluding unclassified strains (Supplementary Figures S1–S4).
A total of 147 plastid and 86 mitochondrial genomes were used in this study (Supplementary Tables S1, S2). Homologous groups of organelle genes were identified by local BLASTp search (e-value cutoff = 1.e-05). Each homologous gene set was aligned using MAFFT (v7.313; Katoh and Toh, 2008) with the default options. Concatenated alignments were constructed using 123 plastid and 34 mitochondrial genes, respectively. Maximum-likelihood (ML) trees constructed using each concatenated alignment were analyzed using the IQ-tree program (v1.6.12; Nguyen et al., 2015) with the following options: gene partition information (-q), model test (-m TEST), and ultrafast bootstrapping with 1,000 replications (-bb 1,000).
To clearly distinguish morphological features, we compared six target diatoms with their morphologically similar counterparts as follows: E. umbratica MEG002 was compared with E. umbratica, E. infula, E. adriatica, E. punctulate, E. tenera, E. gracilis, E. pusilla, and E. vilicicii (Osada and Kobayasi, 1990; Mejdandžić et al., 2017; Mejdandžić et al., 2018). Navicula avium MEG004 was compared with N. avium, N. babeiensis, N. pseudokuseliana, N. coraliana, N. nielsfogedii, N. bergstromiana, N. kuseliana, N. serotina, and N. zanonii (Hustedt, 1949; Metzeltin and Lange-Bertalot, 1998; Rumrich et al., 2000; Kulikovskiy et al., 2012; Taylor et al., 2016; Li et al., 2017a; Sabbe et al., 2019; Chudaev et al., 2020; Kulikovskiy et al., 2021). P. inscriptura MEG005 was compared with P. inscriptura, P. obtusum, P. strigosum, P. rigidum, and P. williamsii (Sterrenburg, 2001; Reid, 2002; Stidolph, 2002; Sterrenburg, 2003; Harper et al., 2009). Nit. reversa var. latus MEG012 was compared with Nit. reversa, Nit. droebakensis, Nit. gobbii, Nit. acicularis, Nit. decipiens, Nit. draveillensis, Nit. kavirondoensis, Nit. longissima, and Nit. ventricosa (Cleve and Grunow, 1880; Smith, 1853; Hasle, 1964; Coste and Ricard, 1980; Hasle and Syvertsen, 1997; Sitoki et al., 2013; Giulietti et al., 2021). Nit. dissipatoides MEG028 was compared with Nit. dissipatoides, Nit. erosa, Nit. thienemanni, Nit. fragilariiformis, Nit. hybridaeformis, Nit. pseudoamphioxys, Nit. serrata, Nit. yunchengensis, and Nit. albicostalis (Hustedt, 1938; Hustedt, 1942; Bourrelly and Manguin, 1952; Hustedt, 1955; Giffen, 1966; Archibald, 1982; Compère, 1986; Li and Volcani, 1987; Shu-qi and Ting, 1994). Nit. anomalus MEG011 was compared with Nit. nanodissipata, Nit. volvendirostrata, Nit. aestatis (Giffen, 1973; Witkowski et al., 2016).
To visualize and compare the morphological characteristics of paraphyletic diatom genera, we redrew the illustrations of several representative diatom species based on their previously reported morphological descriptions and images as follows: Pseudo-nitzschia simulans (Li et al., 2017b; Ajani et al., 2020), Pseudo-nitzschia micropora (Priisholm et al., 2002; Rivera-Vilarelle et al., 2013), Pseudo-nitzschia multiseries (Hasle, 1995; Evans et al., 2004), Pseudo-nitzschia pungens (Lim et al., 2012; Kim et al., 2015), Pseudo-nitzschia americana (Lundholm et al., 2002a; Rivera-Vilarelle et al., 2013), Fragilariopsis cylindrus (Von Quillfeldt, 2001; Cefarelli et al., 2010), Nitzschia inconspicua (Sonneman et al., 2000; Trobajo et al., 2013), Nitzschia alba (Lewin and Lewin, 1967), Nitzschia palea (Crowell et al., 2019; Wang et al., 2020), Cylindrotheca closterium (Reimann and Lewin, 1964; Ryabushko et al., 2019), Tryblionella apiculata (Gregory, 1857; Yamamoto et al., 2017), Psammodictyon constrictum (Yamamoto et al., 2017), Nitzschia traheaformis (Witkowski et al., 2016), Nitzschia dissipatoides (Archibald, 1982; this study), Bacillaria paxillifer (B. paxillifera; Jahn and Schmid, 2007), Skeletonema marinoi (Jung et al., 2009), Thalassiosira nordenskioeldii (Cleve, 1873; Shevchenko et al., 2020), Discostella pseudostelligera (Houk and Klee, 2004; Guerrero and Echenique, 2006), Minidiscus spinulatus (Li et al., 2020), Cyclotella tecta (Prasad and Nienow, 2006), Thalassiosira pseudonana (Horvát et al., 2021), and Conticribra weissflogii (Cavalcante et al., 2013).
We classified and described the morphological characteristics of six diatom species (E. umbratica, N. avium, P. inscriptura, Nit. reversa var. latus, Nit. dissipatoides, and Nit. anomalus), which are observed using LM, SEM, and TEM (See Methods). We also conducted phylogenetic analyses of 18S rDNA sequences from the target diatoms to discuss the molecular taxonomy of the target species.
One multilobed plate plastid was observed in the central region of living cells (Figure 2A). Frustules are panduriform, with a markedly constricted bilobate keel in the middle part of the valve (Figure 2B). Cells are torsional around the apical axis and have numerous girdle bands (Figure 2C). The valve shape is linear-lanceolate, with a length (apical axis) of 18–32 μm (avg. 24 μm, n = 100) and a width (transapical axis) of 8–15 μm (avg. 11 μm, n = 100; Figure 2C). Striae density in the valve and keel was observed 45 in 10 μm (Figure 2D). Striae are closed by hymens with parallel-located roundish-to-elliptical perforations (Figure 2E). The copula and valve copula have similar ultrastructure, showing 47 striae in 10 μm (Figure 2F; Mejdandžić et al., 2018).
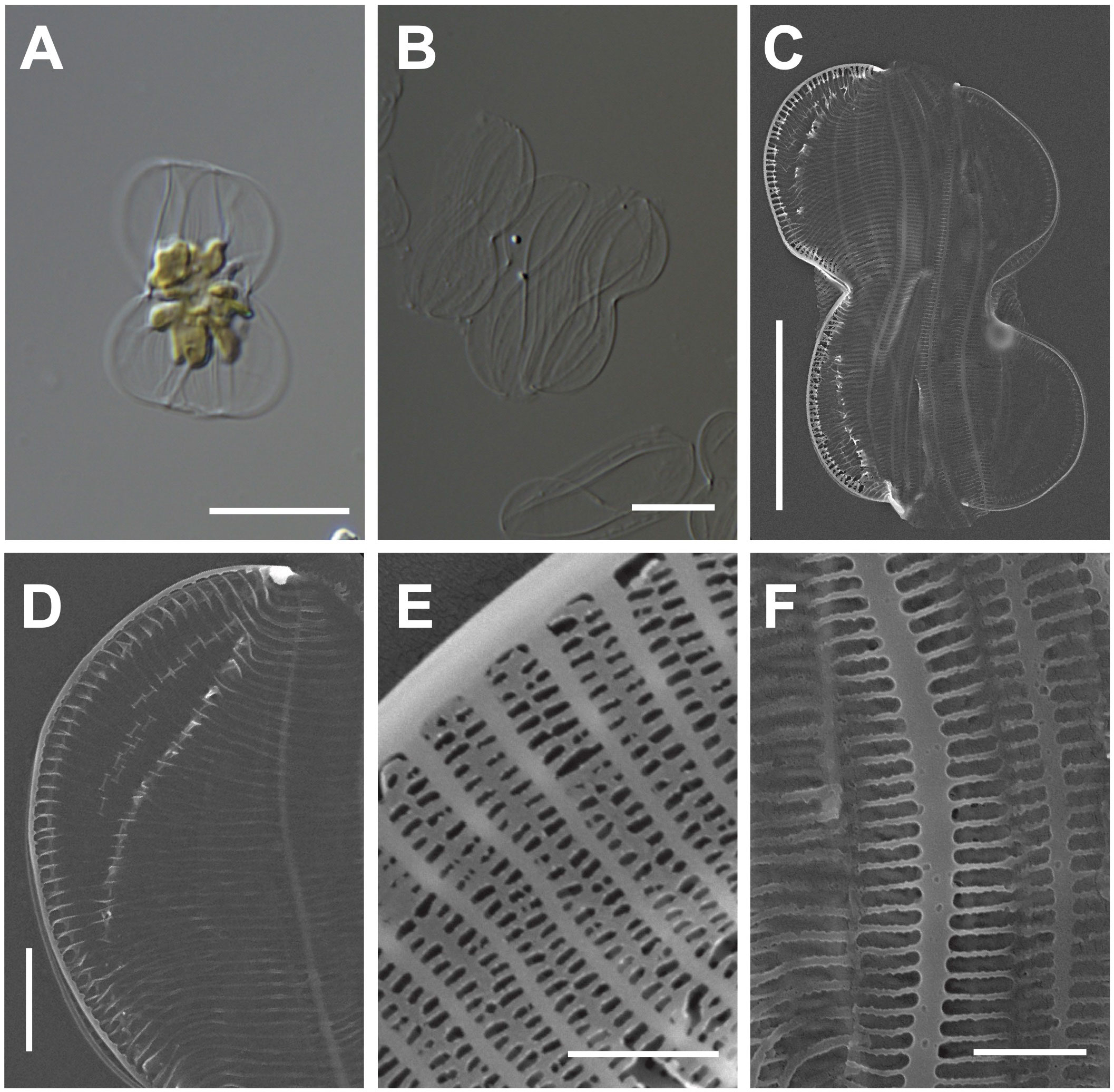
Figure 2 Light and electron microscopic images of Entomoneis umbratica (MEG002). (A) Panduriform frustule containing one multilobed plate plastid. (B) Cleaned frustules. (C) Numerous girdle bands in the external valve view. (D) Keel structures. (E) Striae with parallel-located roundish-to-elliptical perforations (F) Valvocopula structures. Scale bars: (A, B) = 5 μm, (C) = 10 μm, (D) = 2 μm, and (E) = 0.5 μm.
Entomoneis umbratica (MEG002) demonstrated similar valve outlines with those of E. infula and E. adriatica, but its cell length (apical axis) and valve width (transapical axis) were larger than those of others (Supplementary Table S3). Moreover, the striae in E. infula and E. adriatica were denser than those E. umbratica (Supplementary Table S3). The BLASTn search of E. umbratica (MEG002; OR398973) revealed 99.88% identity with the 18S rDNA sequences of E. umbratica (MF000604.1), and their phylogenetic analysis with homologous sequences (top 500 matches of BLASTn search, e-value cutoff = 1.e-05) revealed a monophyletic clade of Entomoneis species (Supplementary Figure S1).
The valve shape is narrowly lanceolate with acute apices, and the valve has a straight raphe (Figures 3A–C). Cells are 20–27 μm (avg. 24 μm, n = 100) in length and 7–11 μm (avg. 11 μm, n = 100) in width (Figures 3C, D). The external valve face exhibits continuous longitudinal strips and two pore-like extensions at the external central raphe endings (Figures 3D, E). In the internal valve view, the accessory rib is parallel to the raphe slit (Figure 3F). Small helictoglossae are found at the tip of the terminal raphe slits, and the region around helictoglossa is thickened with three pores (Figure 3G). The transverse striae are arranged parallel or radiate, and their density is 12 in 10 μm with apically elongated linear areola (Sterrenburg et al., 2015; Li et al., 2017a).
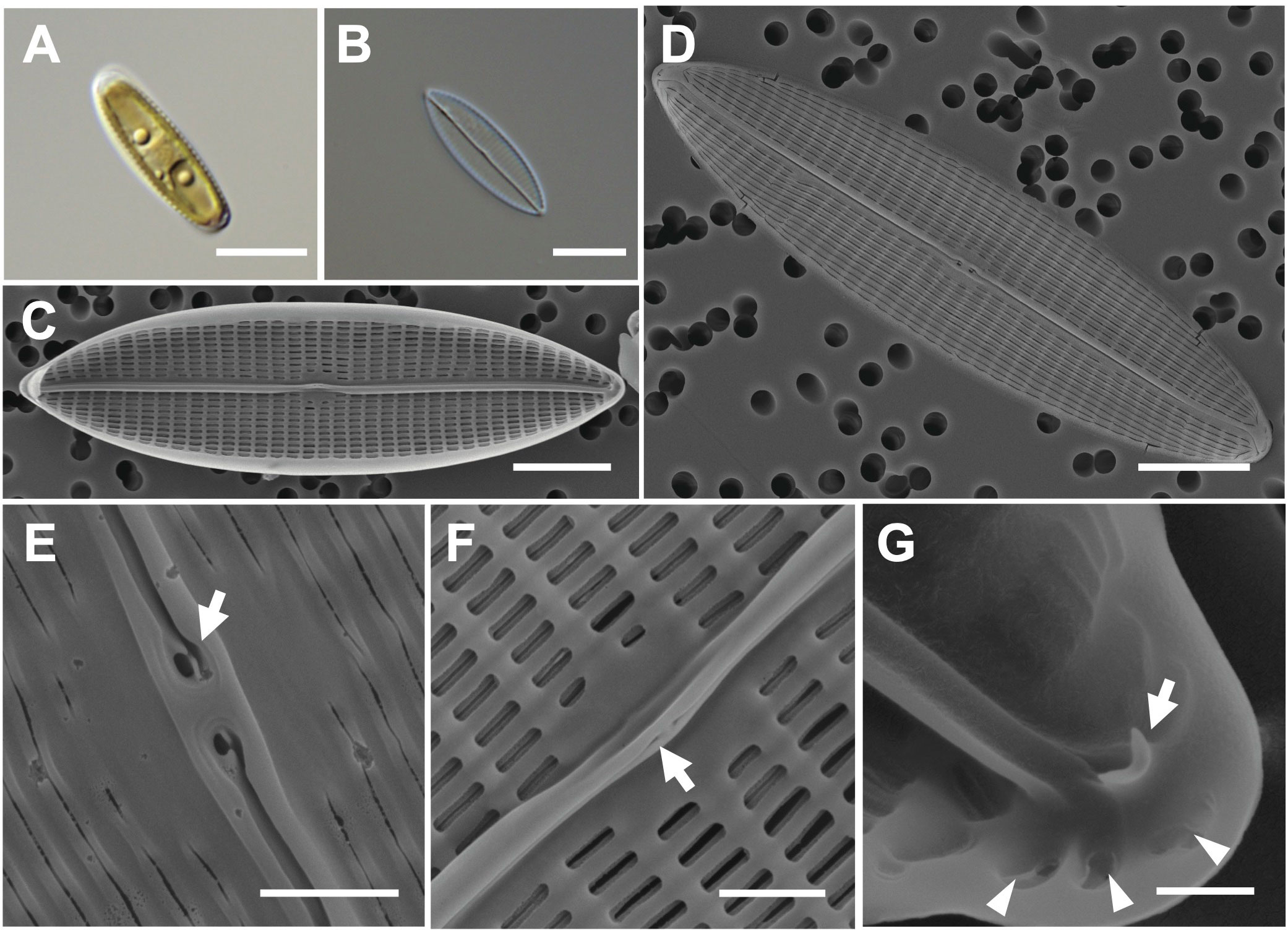
Figure 3 Light and electron microscopic images of Navicula avium (MEG004). (A) Narrowly lanceolate cell containing two plastids. (B) Cleaned frustule. (C) Straight raphe in the internal valve view. (D) Acute apex in the external valve view. (E) External central raphe endings with two pore-like extensions (arrow). (F) Accessory rib (arrow) in the internal valve view. (G) Internal terminal raphe slits with helictoglossa (arrow) around three pores (arrowhead). Scale bars: (A–D) = 5 μm, (E, F) = 1 μm, and (G) = 0.5 μm.
Navicula avium (MEG004) displays a smaller cell size (20–27 μm) than previously reported N. avium (Li et al., 2017a; 32.4–62.5 μm), but their 18S rDNA sequences are identical (Supplementary Table S4); hence, the difference is considered cell size variation in N. avium. The valve outlines of N. avium and N. babeiensis are similar (narrowly lanceolate with broadly rounded), but the shape of the valve apices in N. babeiensis is slightly subrostrate (Supplementary Table S4). Moreover, the striae density in 10 μm of N. avium (7–11) was lower than that of N. babeiensis (17–19; Supplementary Table S4). The phylogenetic analysis conducted using the 18S rDNA sequnces of N. avium (MEG004; OR398987) with homologous sequences (top 500 matches of BLASTn search, e-value cutoff = 1.e-05) revealed a monophyletic clade of N. avium (Supplementary Figure S2).
Cells have lanceolate valves with rounded apices and sigmoid raphe with a 3.6° raphe angle (Figures 4A–C). Cells measure 104–115 μm (avg. 111 μm, n = 100) in length and 15–25 μm (avg. 4 μm, n = 100) in width. Each cell has a small oval central nodule with no hyaline (Figure 4D). Transverse striae are 20 in 10 μm. The internal polar raphe ends appear as small triangles (Figure 4E; Harper et al., 2009; Jeong and Lee, 2021).
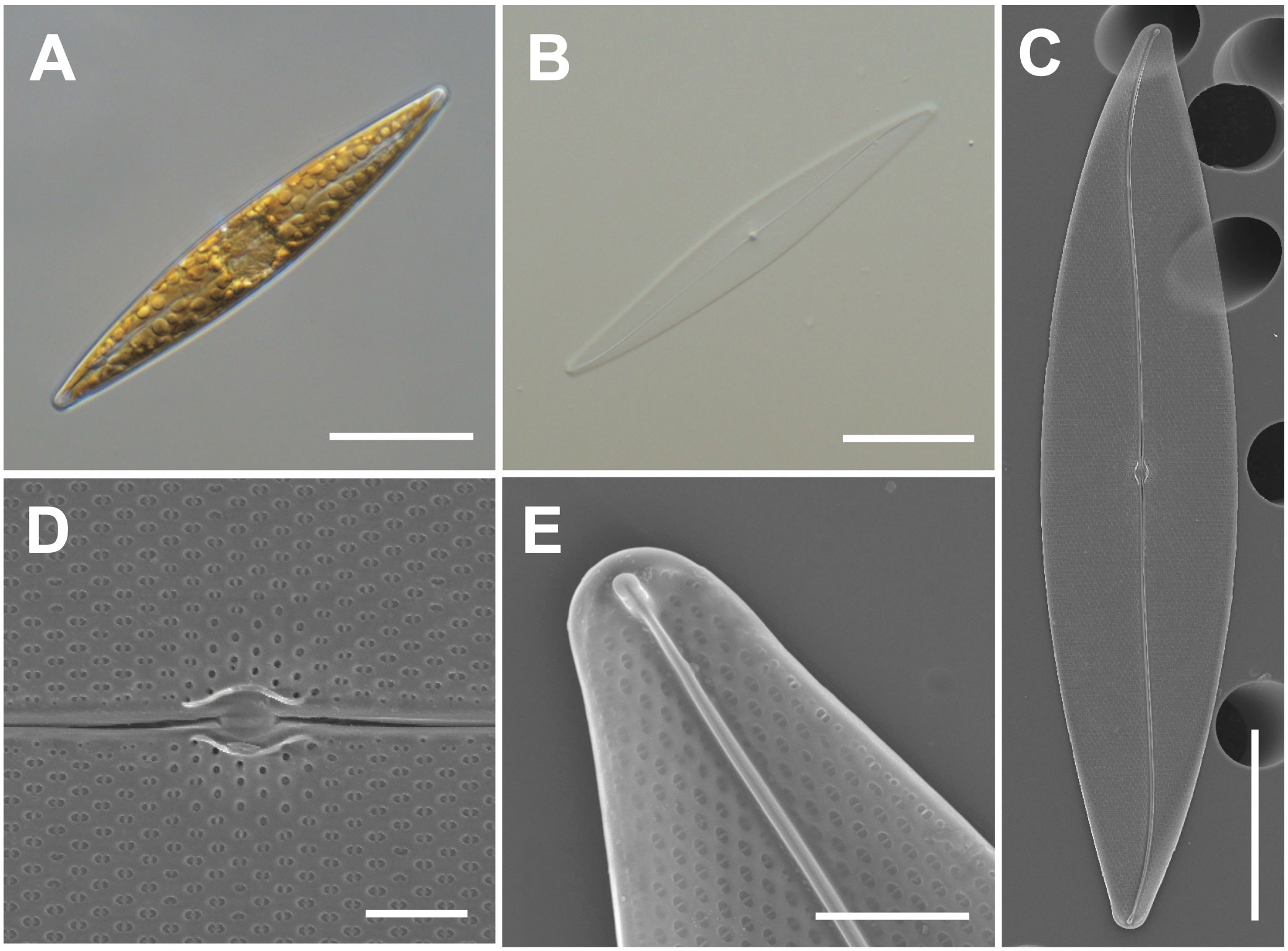
Figure 4 Light and electron microscopic images of Pleurosigma inscriptura (MEG005). (A) Lanceolate cell. (B) Cleaned frustule. (C) Sigmoid raphe in the external valve view. (D) Small oval central nodule in the external valve view. (E) Internal polar raphe ends with small triangles. Scale bars: (A–C) = 20 μm, (E) = 3 μm, and (D) = 2 μm.
Pleurosigma inscriptura (MEG005) shows a similar size as that of P. obtusum, but P. obtusum exhibits a rhombic value shape (Supplementary Table S5). The BLASTn search of 18S rDNA sequences of P. inscriptura (MEG005; OR398989) revealed 97.90% identity with the 18S rDNA sequences of Pleurosigma sp. (KX981840.1), and their phylogenetic analysis with homologous sequences (top 500 matches of BLASTn search, e-value cutoff = 1.e-05) revealed a monophyletic clade of P. intermedium (Supplementary Figure S3).
Cells are 38–50 μm (avg. 46 μm, n = 100) in length and 2–5 μm (avg. 4 μm, n = 100) in width (Figures 5A, B). The valve outline is lanceolate-fusiform with a slightly sigmoid and subrostrate shape (Figures 5C, D). The raphe ends are slightly curved at the poles, and the raphe canal is eccentric (Figures 5C, D). The striae of cells are 54 in 10 μm, and square poroids with rounded corners are 7 in 1 μm with finely perforated areolae (Figures 5D–F). Fibulae are randomly arranged as 9–15 in 10 μm, and the interval in the middle part is particularly wide (Figure 5G). Helictoglossae were observed at the tip of the terminal raphe slits (Figure 5H). The central nodule is visualized in the internal view (Figure 5I).
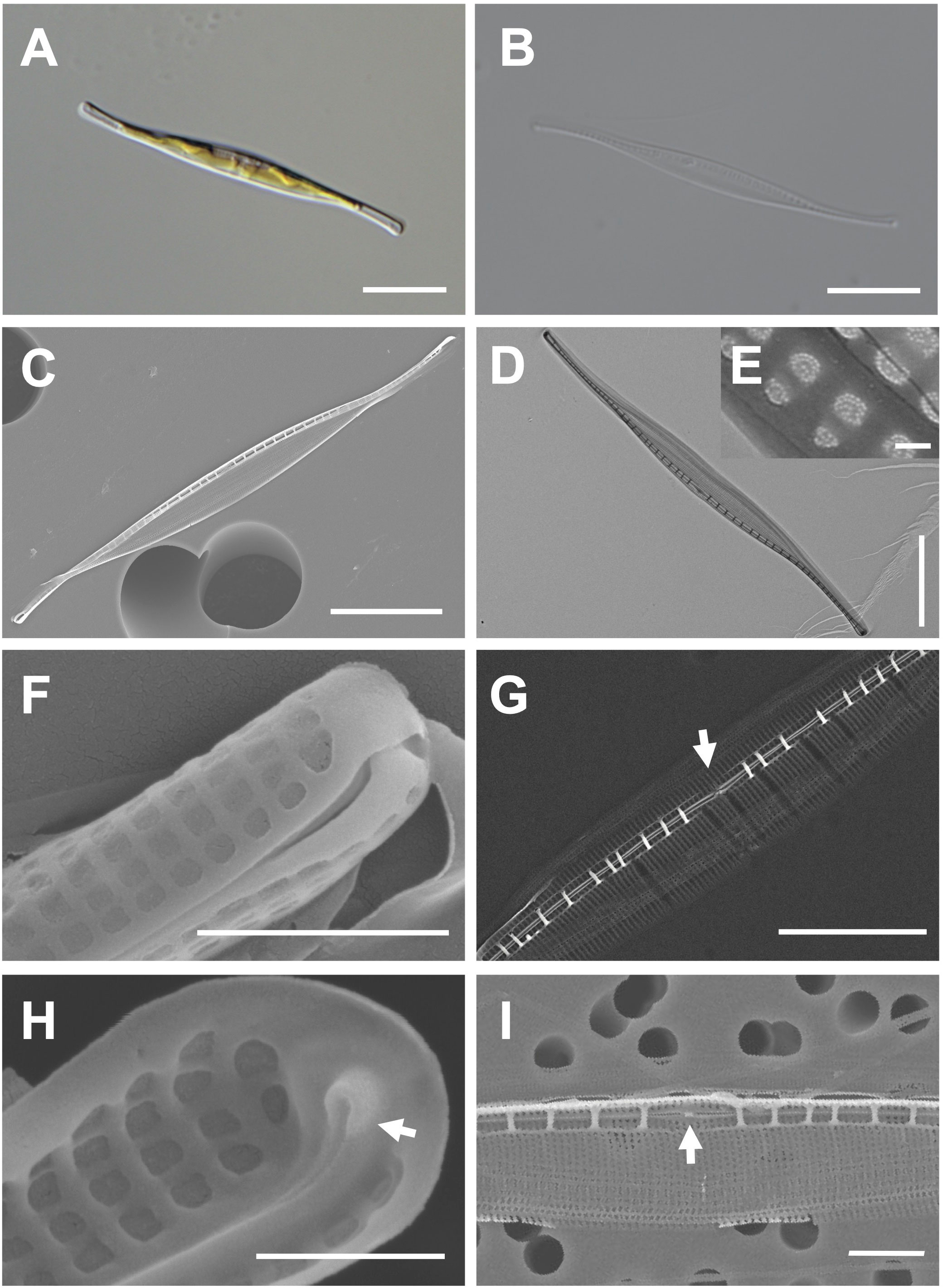
Figure 5 Light and electron microscopic images of Nitzschia reversa var. latus Jeong & Lee var. nov. (MEG012). (A) Lanceolate-fusiform cell. (B) Cleaned frustule. (C) Eccentric raphe canal in the external valve view. (D) Frustule (TEM). (E) Perforate hymenate areolae (TEM). (F) Round to square poroids in the external valve view. (G) Randomly arranged fibulae in the internal valve view, and particularly wide interval of fibulae (arrow) in the central area. (H) Internal terminal raphe slits with helictoglossae (arrow). (I) Central nodule (arrow) in the internal valve view. Scale bars: (A–D) = 10 μm, (G) = 5 μm, (I) = 2 μm, (F) = 1 μm, (H) = 500 nm, and (E) = 100 nm.
Etymology: The name refers to a distinctly wider interval between fibulae than that in Nitzschia reversa.
Comparison with similar species: The morphology with a lanceolate central portion and fine protrusions is commonly found in the genus Nitzschia (Hasle, 1964; Giulietti et al., 2021). Nitzschia reversa var. latus (MEG012) displays similar morphological characteristics as those of Nit. reversa such as the cell size, raphe canal, and central nodule (Supplementary Table S6; Smith, 1853). However, the fibulae density of Nitzschia reversa var. latus (MEG012) is larger than that of Nit. reversa; thus, this species (MEG012) is considered a variety of Nit. reversa (Supplementary Table S6). Most morphological characteristics of Nit. reversa var. latus were also similar to those of Nit. droebakensis without fibulae density (Supplementary Table S6; Hasle, 1964; Giulietti et al., 2021). Nonetheless, all the described morphological characteristics (cell size, valve outline, raphe canal, central nodule, and fibulae density) of Nit. droebakensis were almost identical to those of Nit. reversa (Supplementary Table S6). Therefore, Nit. droebakensis Hasle (1964) is considered a synonym of Nit. reversa Smith (1853) because of priority in nomenclature. Nit. reversa var. latus displays a sigmoid valve outline, whereas other comparable Nitzschia species exhibit a linear valve outline (e.g., Nit. gobbii; Supplementary Table S6). Furthermore, the valve length of Nit. reversa var. latus (38–50 μm) is smaller than that of Nit. draveillensis (55–110 μm), Nit. longissima (ca. 200 μm), and Nit. ventricose (100–650 μm; Supplementary Table S6).
Cells are 20–30 μm (avg. 25 μm, n = 100) in length and 3–6 μm (avg. 4 μm, n = 100) in width (Figures 6A, B) and have numerous girdle bands (Figure 6C). The valve shape is lanceolate constricted in the middle part of the keel (Figure 6D). The rostrate apices are slightly curved (Figure 6D). The raphe canal is eccentric (Figure 6D). Fibulae are 10 in 10 μm but are irregularly arranged in interval (Figure 6E). A central nodule was observed in the internal valve view (Figure 6F). Striae density is 35 in 10 μm on the elevated virgae (Figure 6G).
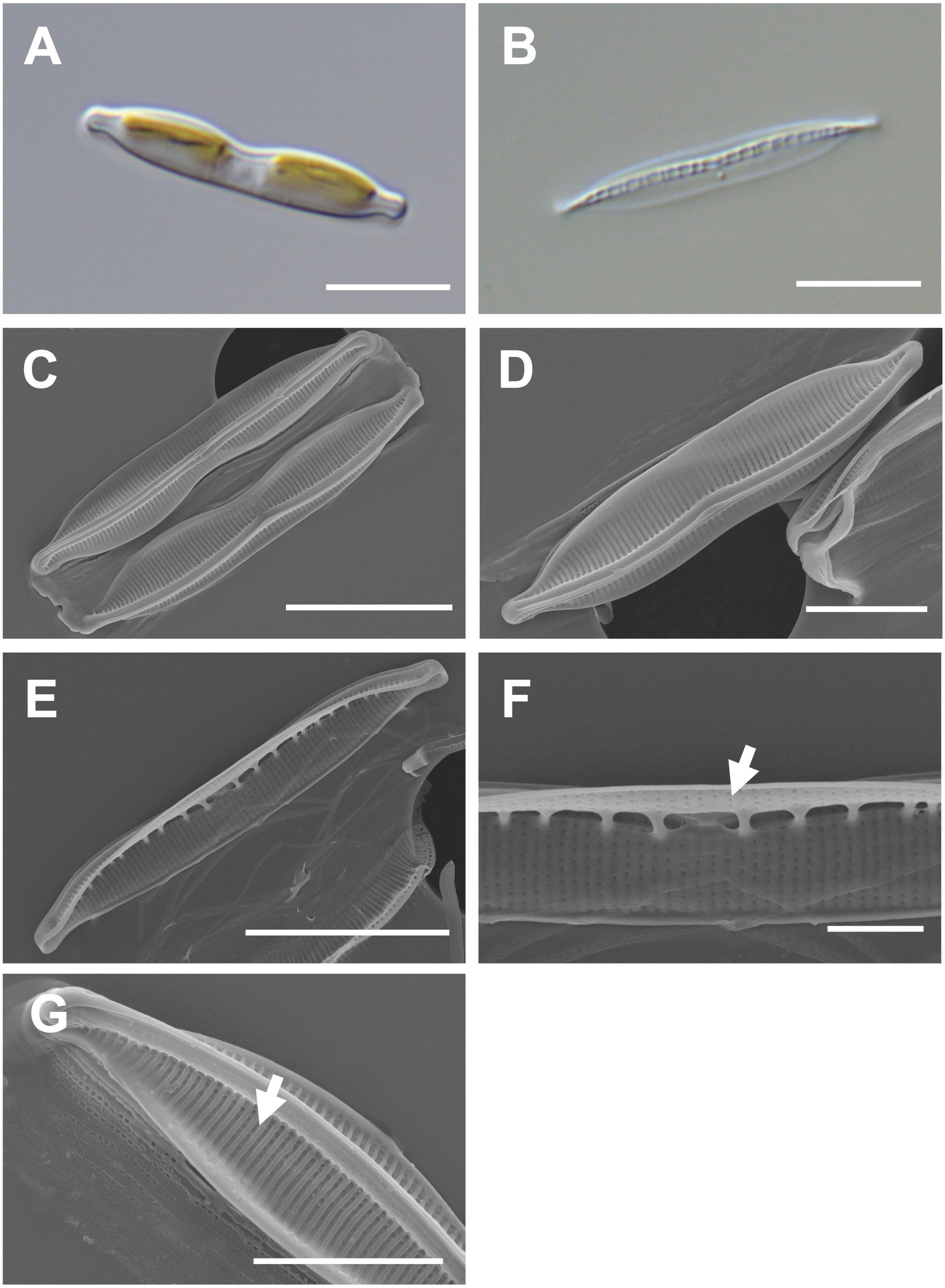
Figure 6 Light and electron microscopic images of Nitzschia dissipatoides (MEG028). (A) Lanceolate cell. (B) Cleaned frustule. (C) Lanceolate valve constricted in the middle part of the keel (external valve view). (D) Rostrate apices in the external valve view. (E) Fibulae structures in the internal valve view. (F) Central nodule (arrow) in the internal valve view. (G) Elevated virgae (arrow) in external valve view. Scale bars: (A, B, D), and (F) = 5 μm, (C, E) = 10 μm, and (G) = 2 μm.
Nitzschia dissipatoides (MEG028) exhibited similar morphological characteristics as those of the previously reported Nit. dissipatoides (Archibald, 1982; Supplementary Table S7). The valve outline of Nit. dissipatoides (lanceolate) is different from those (linear) of Nit. erosa, Nit. fragilariiformis, Nit. hybridaeformis, Nit. pseudoamphioxys, and Nit. yunchengensis (Supplementary Table S7). The valve length of Nit. dissipatoides (21.21–30.35 μm) is smaller than that of Nit. thienemanni (47–90 μm) and Nit. hybridaeformis (60–93 μm; Supplementary Table S7). The striae density of Nit. dissipatoides (35 in 10 μm) is larger than that of Nit. serrata (25 in 10 μm) and Nit. pseudoamphioxys (24 in 10 μm) but smaller than that of Nit. yunchengensis (60 in 10 μm; Supplementary Table S7).
The valve is lanceolate with a slightly rounded apices (Figures 7A, B). Numerous girdle bands are present (Figures 7C, D). Cells are 18–33 μm (avg. 26 μm, n = 100) in length and 4–7 μm (avg. 11 μm, n = 100) in width (Figure 7D). The external valve has a slightly curved central raphe canal (Figure 7E). The internal valve reveals a central raphe canal (slightly curved) with 8 fibulae in 10 μm (Figures 7F, G). The striae density is 37 in 10 µm, and they are arranged on elevated virgae parallel to the transapical axis (Figure 7H). Helictoglossae were detected in the internal terminal raphe slits (Figure 7I). The areolae with silicified rings are randomly arranged on the striae (Figure 7J). The central nodule is absent (Figure 7K).
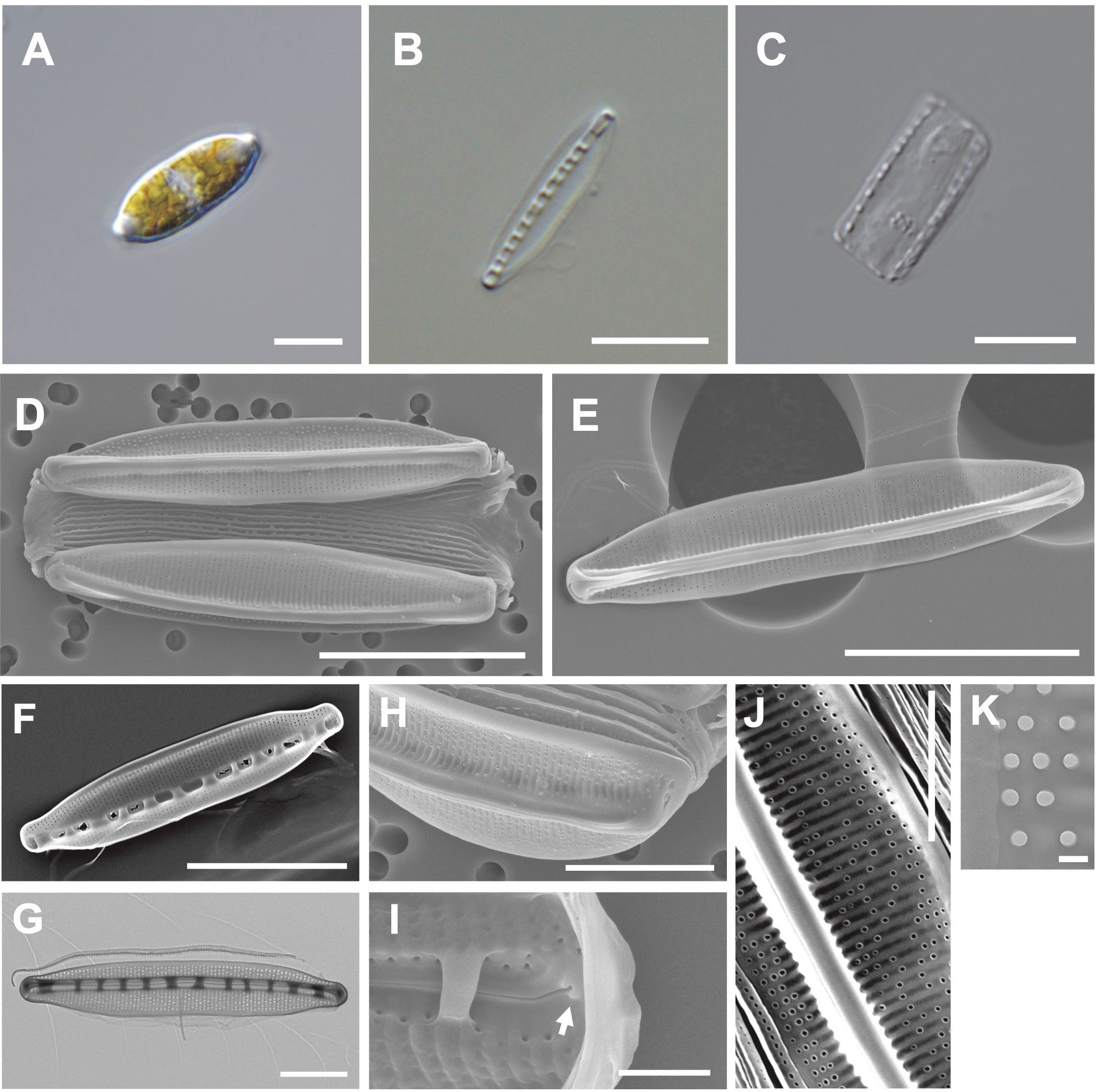
Figure 7 Light and electron microscopic images of Nitzschia anomalus Jeong & Lee sp. nov. (MEG011). (A) Lanceolate cell. (B) Cleaned frustule. (C) Numerous girdle bands in cleaned frustule. (D) Slightly rounded apices in the external valve view. (E) Slightly curved central raphe canal in the external valve view. (F) Fibulae structures in the internal valve view. (G) Frustule (TEM). (H) Randomly arranged areolae in the external valve view. (I) Internal terminal raphe slits with helictoglossae (arrow). (J) Elevated virgae in the external valve view. (K) Valve areolae (TEM). Scale bars: (A–C, G, H) = 5 μm, (D–F) = 10 μm, (J) = 3 μm, (I) = 1 μm, and (K) = 200 nm.
Etymology: The name refers to randomly distributed areolae on the striae.
Comparison with similar species: Nitzschia anomalus (MEG011) displays a similar lanceolate valve outline to that of Nit. nanodissipata (Witkowski et al., 2016), but their valve apices, cell size, and striae density are different (Supplementary Table S8). Nit. anomalus exhibits a larger valve in length (17.69–32.97 μm) than Nit. volvendirostrata (7–11.5 μm). Furthermore, the striae density of Nit. volvendirostrata (80–90 in 10 μm) is larger than that of Nit. anomalus (37 in 10 μm; Supplementary Table S8). Nit. anomalus exhibits a lanceolate valve outline and a central raphe canal, while Nit. aestatis displays a rectangular-linear valve outline and an eccentric raphe canal. Moreover, Nit. anomalus shows a smaller valve in length (18–33 μm) than Nit. aestatis (44–50 μm; Supplementary Table S8).
The phylogenetic analysis of 18S rDNA sequences from the Nitzschia species (OR418054, OR418055, and OR418056) with their homologous sequences (top 500 matches of BLASTn search, e-value cutoff = 1.e-05) revealed a monophyletic relationship with other Nitzschia species (Supplymentary Figure S4). However, the phylogenetic clade includes other diatom genera (e.g., Cylindrotheca, Tryblionella, and Bacillaria) with morphologically unverified diatom species; hence, further studies without controversial molecular data are required (Mann et al., 2021).
The 18S rDNA phylogeny reveals that Enotomoneis species exhibit a paraphyletic relationship with respect to the genera Amphiprora, Auricula, and Epithemia (Supplementary Figure 1). In the phylogenetic analysis using 18S rDNA sequences, the genus Navicula shows a paraphyletic relationship with respect to the genera Haslea, Minutocellus, and Seminavis (Supplementary Figure 2). The genus Pleurosigma shows a monophyletic clade based on the 18S rDNA phylogeny (Supplementary Figure 3). The genus Nitzschia shows paraphyletic relationship with respect to the genera Phaeodactylum, Pseudo-nitzschia, Fragilariopsis, Cymbella, Conticribra, Durinskia, Peridinium, Amphora, Hantzschia, Achnanthidium, Achnanthes, Bacillaria, Navicula, Tryblionella, and Cylindrotheca in the 18S rDNA phylogeny (Supplementary Figure 4).
We postulate that several types of nuclear genetic variation (Baack and Rieseberg, 2007; Colbeck et al., 2011; Harrison and Larson, 2016) could introduce confusion in phylogenetic signals. Consequently, the paraphyletic relationships in several diatom genera may be attributed to use of single or several nuclear marker genes (e.g., 18S rDNA). Another potential cause of the paraphyletic relationships could arise from misidentified or morphologically unverified species (Kooistra et al., 2008; Nguyen et al., 2011; Cimarelli et al., 2015; Mann et al., 2021). Although more taxon samples from morphologically verified species with their complete nuclear genome data can help resolve this issue, available nuclear genomes in diatoms are still insufficient. Therefore, we attempted to address this using diatom organelle genomes in this study.
We completely constructed the circular organelle (plastid and mitochondrial) genomes of the six diatom species (Supplementary Table S9). The plastid genomes of these diatoms range from 121 to 177 kbp in length and contain 127 to 140 protein-coding sequences (CDS), 30 to 31 tRNAs, and 6 ribosomal RNAs (rRNAs). The mitochondria genomes of six diatoms range from 36 to 61 kbp in length and contain 33 to 36 CDS, 19 to 25 tRNAs, and 2 rRNAs (Supplementary Table S9).
Several diatom genera showed paraphyletic relationships in the plastid phylogeny with high bootstrap support values (BS). For example, the genus Navicula showed a paraphyletic relationship with respect to Seminavis robusta (MH356727), and this clade showed a 100% BS (Figure 8). The genus Seminavis has two plate-like plastids that typically lie along each side of the girdle, with valves exhibiting a strong dorsiventral orientation, distinguishing it from the genus Navicula (Bruder and Medlin, 2008; Li et al., 2022). Seminavis robusta also possesses distinct morphological characteristics compared with those of the closely related Navicula species (e.g., N. veneta and N. arenaria) in the phylogenetic clade (Figure 8; Donkin, 1858; Kützing, 1844; Danielidis and Mann, 2002; Chudaev et al., 2022). However, to clarify the phylogenetic and taxonomic position of the genus Seminavis, more taxon samples from this genus are required. In the plastid phylogeny, Pleurosigma inscriptura (MEG005) and Entomoneis umbratica (MEG002) are clustered with their sister species, respectively (BS: 100%; Figure 8). Interestingly, the Nitzschia species (yellow blocks in Figure 8) exhibit a paraphyletic relationship with respect to the genera Pseudo-nitzschia, Fragilariopsis, Cylindrotheca, Tryblionella, Psammodictyon, and Bacillaria (BS: 95%; Figure 8), even though taxon samples to compare inter-genera relationship are adequate only from the genera Nitzschia and Pseudo-nitzschia. In particular, Nit. anomalus (MEG011) and Bacillaria paxillifer (NC_061049) present an early diverged subclade within the Nitzschia clade (Figure 8). Although the mitochondrial phylogeny constructed using diatom species is slightly different in the topologies of Nitzschia species compared with the plastid phylogeny, the Nitzschia clade (BS: 100%) exhibits paraphyletic relationship with respect to the genera Tryblionella, Cylindrotheca, and Pseudo-nitzschia (Figure 9). Diatom species in the Nitzschia clade have relatively conserved gene contents in plastid and mitochondrial genomes (Supplementary Tables S10, S11). However, several plastid genes such as psaE, psaI, serC, syfB, and ycf35 were not found in most Pseudo-nitzschia species (Supplementary Table S10). In addition, only Nitzschia palea (MH113811) and Tryblionella apiculata (NC_056791) possessed ycf91 and tyrC, respectively (Supplementary Table S10). The non-photosynthetic diatom Nitzschia alba has lost most plastid genes (Supplementary Table S10). Therefore, the molecular and evolutionary histories of several different diatom genera are closely related to each other despite the distinctly different morphological characteristics, such as genus boundaries. The development of morphological features could be influenced by various types of interactive nuclear-encoded genes. For example, in plants, the transcription factor MADS-box genes are involved in flower morphogenesis (Coen and Meyerowitz, 1991; Saedler et al., 2001). Therefore, morphological characteristics and their related genes in diatoms may not accurately represent their phylogenetic and evolutionary relationships.

Figure 8 Multigene phylogeny constructed using a concatenated alignment of 123 plastid genes from diatom species (only ≥ 50 bootstrap supporting values are shown).
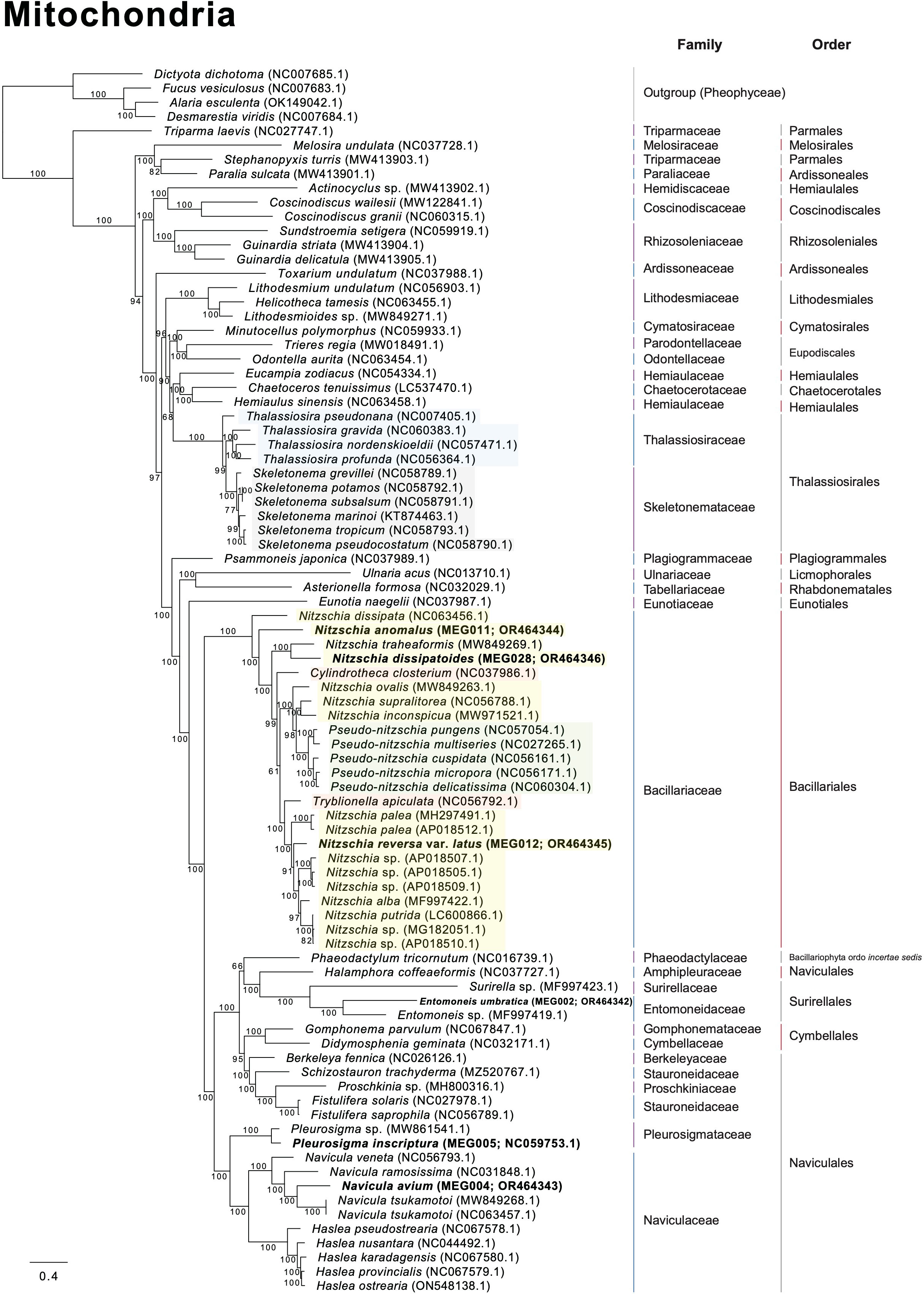
Figure 9 Multigene phylogeny constructed using a concatenated alignment of 34 mitochondrial genes from diatom species (only ≥ 50 bootstrap supporting values are shown).
In the plastid phylogeny, Thalassiosira species (blue blocks in Figure 8) are also clustered with other genera such as Discostella, Minidiscus, Skeletonema, and Cyclotella (BS: 100%; Figure 8). The mitochondrial phylogeny also revealed that Thalassiosira species shows paraphyletic relationship with respect to Skeletonema species (BS: 100%; Figure 9). These diatom species in the Thalassiosira clade exhibit conserved gene contents in plastid and mitochondrial genomes (Supplementary Tables S12, S13). Based on our results, we postulate that the morphological characteristics of several different diatom genera, which are currently established, were independently diverged within the genetically close diatom groups.
The family Thalassiosiraceae exhibits paraphyletic relationship with respect to the families Stephanodiscaceae and Skeletonemataceae in the plastid phylogeny (Supplementary Figure S5). The family Chaetoceroaceae exhibits paraphyletic relationship with respect to the family Hemiaulaceae in the plastid phylogeny (Supplementary Figure S5). In the mitochondrial phylogeny, these families (Thalassiosiraceae-Skeletonemataceae and Chaetoceroaceae-Hemiaulaceae) are also paraphyletic (Supplementary Figure S5). The order Naviculales exhibits paraphyletic relationship with respect to the orders Surirellales and Cymbellales in both plastid and mitochondrial phylogenies (Supplementary Figure S6). The order Thalassiosirales exhibits paraphyletic relationship with respect to the order Stephanodiscale in the plastid phylogeny (Supplementary Figure S6). The order Chaetocerotales displays a paraphyletic relationship with respect to the order Hemiaulales in both plastid and mitochondrial phylogenies (Supplementary Figure S6). Paraphyletic relationships at the family- and order-level in diatoms are completely supported by both plastid and mitochondrial phylogenies (BS 100% as red dots in Supplementary Figures S5, S6).
Approximately 800 Nitzschia species are currently reported (Guiry and Guiry, 2023) based on their morphological characteristics as follows: symmetric nitzschoid cell shape, position of the raphe, transverse extension of the fibula, and the number of striae and aleolae (Mann, 1981; Morales et al., 2020). Interestingly, several species from the genera Pseudo-nitzschia, Fragilariopsis, Cylindrotheca, Tryblionella, and Bacillaria had been reported as Nitzschia species, but these were reclassified by morphological characteristics such as colony formations and raphe structures (Reimann and Lewin, 1964; Hasle, 1994; Li et al., 2007; Schmid, 2007; Cefarelli et al., 2010; Cavalcante et al., 2013). These diatom genera were also clustered as the Nitzschia clade in this study (Figures 8, 9). Moreover, as one of the early diverged taxa in the Nitzschia clade of the plastid phylogeny, Nit. anomalus displays typical morphological characteristics of the genus Nitzschia, but this is clustered with Bacillaria paxillifer (Figure 10). Individual cells of B. paxillifer exhibit similar morphological features as those of Nitzschia species, but B. paxillifer colonies exhibit a gliding motion (Schmid, 2007; image no. 1 and 2 in Figure 10). Moreover, in the mitochondrial phylogeny, Nit. dissipata (NC_063456) is an earlier diverged taxon (this study) than Nit. anomalus in the Nitzschia clade (Figure 10). Therefore, we postulate that the typical genus characters of Nitzschia species could be considered phylogenetically early diverged features. Psammodictyon constrictum and Tryblionella apiculata are similar to the Nitzschia species in possessing symmetry of the raphe on a frustule, wide valves, and eccentric raphe (Round et al., 1990; Yang et al., 2020). However, the genera Psammodictyon and Tryblionella are distinguished from Nitzschia by the differences in valve shape and plastid position (Round et al., 1990; Yang et al., 2020). Moreover, these genera exhibit different characteristics in valve outline and arrangement and structure of areolae (e.g., Psammodictyon: panduriform valve with distinctively centrally constricted; Tryblionella: linear-lanceolate or elliptic in shape, with only slightly centrally constricted; Round et al., 1990; Yang et al., 2020). Although the morphological characteristics of Cylindrotheca closterium, such as valve shape, are similar to those of sigmoid Nitzschia species (e.g., Nitzschia longissima and Nitzschia reversa var. latus) under light microscopic observation, Cylindrotheca closterium has twisted frustules in the apical axis and slightly silicified raphe in the valves, which are different from those of Nitzschia species (Reimann and Lewin, 1964; Li et al., 2007). Pseudo-nitzschia and Fragilariopsis species exhibit a paraphyletic relationship with respect to the Nitzschia species (BS 100%; Figure 10). The genera Pseudo-nitzschia and Fragilariopsis demonstrate morphological differences in colony formation (Pseudo-nitzschia species: stepped colonies; Fragilariopsis species: ribbon-shape colonies; Hasle, 1994; Lundholm et al., 2002b) compared with Nitzschia. However, Pseudo-nitzschia americana exhibits a unicellular formation (Figure 10; Lundholm et al., 2002a).

Figure 10 Comparison of representative morphological characteristics based on the phylogenetic relationship of the Nitzschia clade (Figures 8, 9). The illustrations of diatom species were re-drawn based on previous studies including original descriptions (See Supplementary Table S14).
Thalassiosira species display paraphyletic relationship with respect to the genera Cyclotella, Minidiscus, Discostella, and Skeletonema (Figure 11). In the Thalassiosira clade, the early diverged taxa are T. pseudonana and Cyclotella species. Although T. pseudonana was previously reported as Cyclotella nana, these two genera present morphological differences in the central area of the valve surface (Figure 11; Lowe and Busch, 1975). T. nordenskioeldii is the type (i.e., representative) species of the genus Thalassiosira, which clustered with Thalassiosira sp., T. gravida, T. profunda, and T. tenera, but their sister clade includes Discostella pseudostelligera and Minidiscus spinulatus, which exhibit different morphological characteristics in marginal fultoportulae, location of rimoportula, and central area (Figure 11; Houk and Klee, 2004). Nevertheless, several morphological features (e.g., cell size and marginal fultoportulae) to distinguish genus boundaries are still controversial (Park et al., 2017). Furthermore, the Thalassiosira and Skeletonema species exhibit completely different morphological characteristics. For instances, Skeletonema has cylindrical cells with a ring of long processes emerging from the edge of the valve face, and the colony is linked by elongate tubular processes (Jung et al., 2009; Spaulding et al., 2021). However, Thalassiosira has disk-shaped valves, and the colony is linked by chitin fibrils (Johansen and Fryxell, 1985).
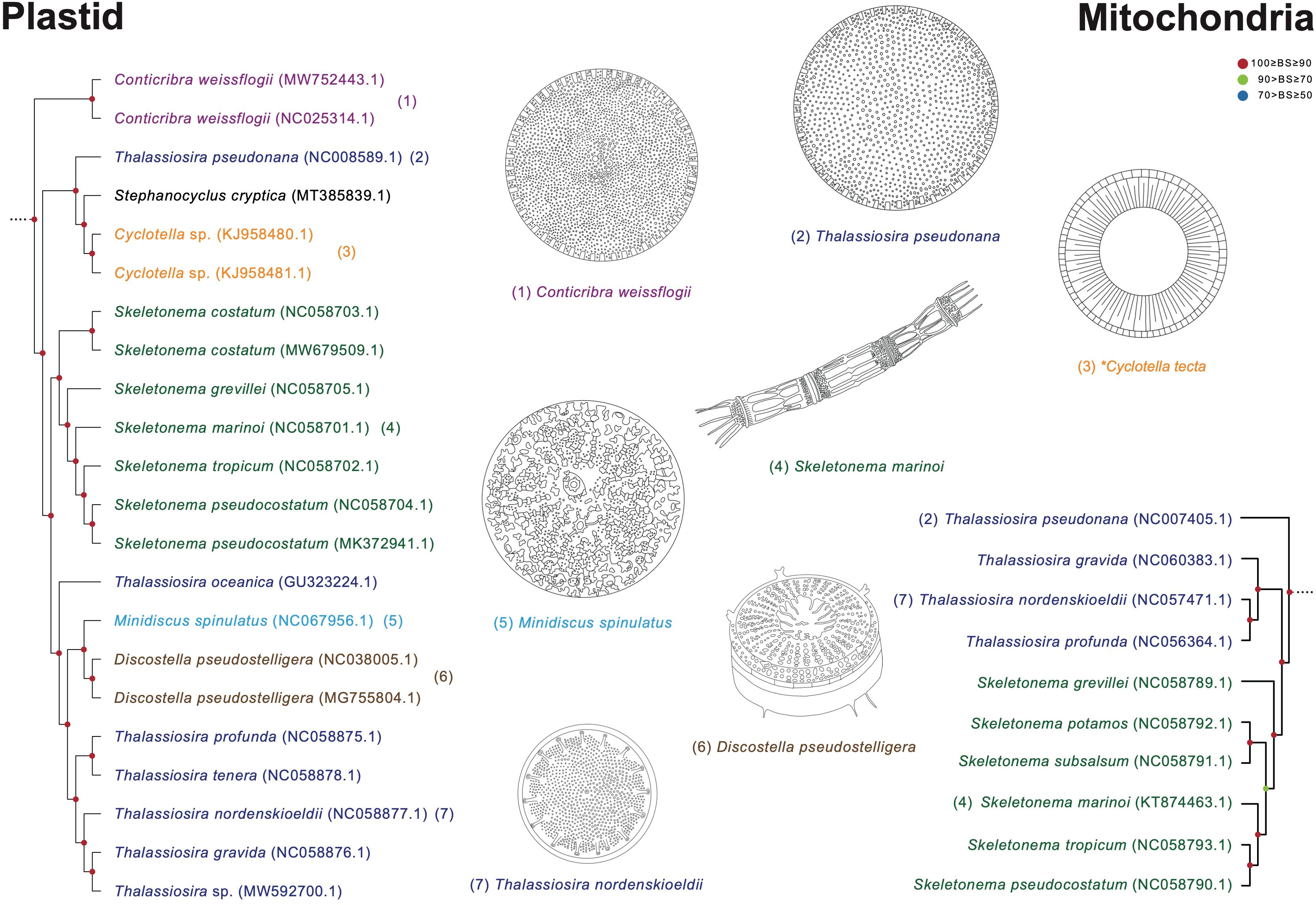
Figure 11 Comparison of representative morphological characteristics based on the phylogenetic relationship of the Thalassiosira clade (Figures 8, 9). The illustrations of diatom species were re-drawn based on previous studies including original descriptions (See Supplementary Table S14).
Paraphyletic relationships are recognized in diverse taxonomic levels of diatoms based on the highly supported organelle phylogenies (Figures 8, 9). As a result, we believe that taxonomic reinvestigation in diatoms is required. However, almost all genome data have no morphological descriptions and images of the target diatoms, which is a critical limitation for further discussion.
We isolated six diatom species from field samples, and each extracted DNA sample was sequenced and assembled to make organelle (plastid and mitochondrial) genomes. Based on the highly supported plastid and mitochondrial phylogenies, we demonstrate that several morphologically similar diatom genera show paraphyletic relationships, thus suggesting that the morphology-based genus boundaries and phylogeny relationships frequently conflict in diatoms. To reestablish the conflict relationship in diatoms, there is a need for more organelle and nuclear genomes from broader taxon samples of diatom species with a comparison of morphological characteristics. In particular, photographic materials (especially SEM images) of target diatoms are necessary, along with morphological descriptions, when new genomic references are generated. This ensures that many researchers can avoid the confusion caused by several pieces of genome information generated from misidentified or morphologically unverified species. Finally, highly supported phylogenetic trees (e.g., organelle phylogenies) and their accompanying photographic materials in diatoms can lead to active discussion for taxonomic reinvestigation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YJ: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Korea Environment Industry & Technology Institute (KEITI) through the Project to make multi-ministerial national biological research resources more advanced, funded by Korea Ministry of Environment (MOE) (2021003420004; JML), and the National Research Foundation of Korea (NRF) grant (RS-2023-00209930; JML) funded by the Korean government (MSIT).
We thank for technical supports to HyeonShik Cho (Marine Ecological Genomics Lab., Department of Oceanography, Kyungpook National University, Daegu, South Korea).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1283893/full#supplementary-material
Ajani P. A., Lim H. C., Verma A., Lassudrie M., McBean K., Doblin M. A., et al. (2020). First report of the potentially toxic marine diatom Pseudo-nitzschia simulans (Bacillariophyceae) from the East Australian Current. Phycol. Res. 68, 254–259. doi: 10.1111/pre.12421
Archibald R. E. M. (1982). Diatoms of South Africa 1. New species from the sundays river (Eastern cape province). Bacillaria. 5, 23–42.
Baack E. J., Rieseberg L. H. (2007). A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17 (6), 513–518. doi: 10.1016/j.gde.2007.09.001
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bourrelly P., Manguin E. (1952). Algues d'eau douce de la Guadeloupe et dépendances (Paris, France: Centre National de la Recherche Sientifique, Société d'Edition d Enseignement Supérieur).
Bruder K., Medlin L. (2007). Molecular assessment of phylogenetic relationships in selected species/genera in the naviculoid diatoms (Bacillariophyta). I. The genus Placoneis. Nova Hedwigia 85, 331. doi: 10.1127/0029-5035/2007/0085-0331
Bruder K., Medlin L. K. (2008). Morphological and molecular investigations of naviculoid diatoms. III. Hippodonta and Navicula ss. Diatom Res. 23, 331–347. doi: 10.1080/0269249X.2008.9705760
Cavalcante K. P., Tremarin P. I., Ludwig T. A. V. (2013). Taxonomic studies of centric diatoms (Diatomeae): unusual nanoplanktonic forms and new records for Brazil. Acta Bot. Bras. 27, 237–251. doi: 10.1590/S0102-33062013000200001
Cefarelli A. O., Ferrario M. E., Almandoz G. O., Atencio A. G., Akselman R., Vernet M. (2010). Diversity of the diatom genus Fragilariopsis in the Argentine Sea and Antarctic waters: morphology, distribution and abundance. Polar Biol. 33, 1463–1484. doi: 10.1007/s00300-010-0794-z
Chan P. P., Lowe T. M. (2019). tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. Clifton NJ 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chudaev D. A., Jüttner I., Glushchenko A. M., Kuli-kovskiy M. S., Gurung S., Williams D. (2020). On the geographical distribution of Navicula nielsfogedii J.C. Taylor & Cocquyt. Diatom Res. 35, 185–192. doi: 10.1080/0269249X.2020.1758794
Chudaev D. A., Juttner I., Taylor J. C. (2022). The genus Navicula (bacillariophyta) from the por'ya guba estuary (Murmansk region, Russia) with description of a new species and reinvestigation of the type materials of N. eidrigiana and N. pseudosalinarum. Новости систематики низшихрастений 56, 55–70. doi: 10.31111/nsnr/2022.56.1
Cimarelli L., Singh K. S., Mai N. T. N., Dhar B. C., Brandi A., Brandi L., et al. (2015). Molecular tools for the selective detection of nine diatom species biomarkers of various water quality levels. Int. J. Environ. Health Res. 12 (5), 5485–5504. doi: 10.3390/ijerph120505485
Cleve P. T., Grunow A. (1880). Beiträge zur Kenntniss der arctischen Diatomeen. K. Svenska Velensk. Akad. Handl. 17, 1–121.
Coen E. S., Meyerowitz E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353 (6339), 31–37. doi: 10.1038/353031a0
Colbeck G. J., Turgeon J., Sirois P., Dodson J. J. (2011). Historical introgression and the role of selective vs. neutral processes in structuring nuclear genetic variation (AFLP) in a circumpolar marine fish, the capelin (Mallotus villosus). Mol. Ecol. 20 (9), 1976–1987. doi: 10.1111/j.1365-294X.2011.05069.x
Compère P. (1986). Nitzschia fragilariiformis a new species from NW Sudan forming ribbon-like colonies. Proc. Eight Int. Diatom Symposium 1984, 253–258.
Coste M., Ricard M. (1980). Observations en microscopie photonique de quelques Nitzschia nouvelles ou intéressantes dont la striation est à la limite du pouvoir de résolution. Cryptogam. Algol. 1, 187–212.
Crowell R. M., Nienow J. A., Cahoon A. B. (2019). The complete chloroplast and mitochondrial genomes of the diatom Nitzschia palea (Bacillariophyceae) demonstrate high sequence similarity to the endosymbiont organelles of the dinotom Durinskia baltica. J. Phycol. 55, 352–364. doi: 10.1111/jpy.12824
Cunha R. L., Grande C., Zardoya R. (2009). Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol. Biol. 9, 1–16. doi: 10.1186/1471-2148-9-210
Danielidis D. B., Mann D. G. (2002). The systematics of Seminavis (Bacillariophyta): the lost identities of Amphora angusta, A. ventricosa and A. macilenta. Eur. J. Phycol. 37, 429–448. doi: 10.1017/S0967026202003724
Dong S., Shen Z., Xu L., Zhu F. (2010). Sequence and phylogenetic analysis of SSU rRNA gene of five microsporidia. Curr. Microbiol. 60, 30–37. doi: 10.1007/s00284-009-9495-7
Donkin A. S. (1858). On the marine Diatomaceae of Northumberland, with a description of eighteen new species. Trans. Microscopical Soc. 5, 12–34.
Evans K. M., Bates S. S., Medlin L. K., Hayes P. K. (2004). Microsatellite marker development and genetic variation in the toxic marine diatom Pseudo-nitzschia multiseries (Bacillariophyceae). J. Phycol. 40, 911–920. doi: 10.1111/j.1529-8817.2004.04027.x
Finkel Z. V., Kotrc B. (2010). Silica use through time: macroevolutionary change in the morphology of the diatom fustule. Geomicrobiol. J. 27, 596–608. doi: 10.1080/0149045100370294
Giffen M. H. (1966). Contributions to the diatom flora of South Africa 3. Nova Hedwigia 13, 245–292.
Giffen M. H. (1973). Diatoms of the marine littoral of Steenberg’s Cove in st. Helena bay, Cape Province, South Africa. Bot. Mar. 16, 32–48. doi: 10.1515/botm.1973.16.1.32
Giulietti S., Totti C., Romagnoli T., Siracusa M., Bacchiocchi S., Accoroni S. (2021). Nitzschia gobbii sp. nov. (Bacillariophyceae): a common but overlooked planktonic diatom species from the northwestern Adriatic Sea. Phycologia 60, 558–571. doi: 10.1080/00318884.2021.1952513
Gregory W. (1857). On the post-tertiary diatomaceous sands of Glenshira (Scotland). Trans. Micr. Soc London Ser. 2. 5, 67–88.
Guerrero J. M., Echenique R. O. (2006). Discostella taxa (Bacillariophyta) from the Río limay basin (northwestern patagonia, Argentina). Eur. J. Phycol. 41, 83–96. doi: 10.1080/09670260500512363
Guiry M. D., Guiry G. M. (2023)AlgaeBase. In: World-wide electronic publication (AlgaeBase). Available at: https://www.algaebase.org (Accessed Mar. 13, 2023).
Harper M. A., Patterson J. E., Harper J. F. (2009). New diatom taxa from the world’s first marine Bioblitz held in New Zealand: Skeletomastus a new genus, Skeletomastus coelatus nov. comb. and Pleurosigma inscriptura a new species. Acta Bot. Croat. 68, 339–349.
Harrison R. G., Larson E. L. (2016). Heterogeneous genome divergence, differential introgression, and the origin and structure of hybrid zones. Mol. Ecol. 25 (11), 2454–2466. doi: 10.1111/mec.13582
Hasle G. R. (1964). Nitzschia and Fragilariopsis species studied in the light and electron microscopes. I. Some marine species of the groups Nitzschiella and Lanceolatae. Skrift. Norske Vidensk. Akad. 16, 1–48.
Hasle G. R. (1994). Pseudo-nitzschia as a genus distinct from Nitzschia (Bacillariophyceae). J. Phycol 30, 1036–1039. doi: 10.1111/j.0022-3646.1994.01036.x
Hasle G. R. (1995). Pseudo-nitzschia pungens and P. multiseries (Bacillariophyceae): nomenclatural history, morphology, and distribution. J. Phycol. 31, 428–435. doi: 10.1111/j.0022-3646.1995.00428.x
Hasle G. R., Syvertsen E. E. (1997). Identifying Marine Phytoplankton. Ed. Tomas C. R. (San Diego: Academic Press).
Horvát S., Fathima A., Görlich S., Schlierf M., Modes C. D., Kröger N. (2021). Computational analysis of the effects of nitrogen source and sin1 knockout on biosilica morphology in the model diatom Thalassiosira pseudonana. Discover Materials 1, 1–10. doi: 10.1007/s43939-021-00008-w
Houk V., Klee R. (2004). The stelligeroid taxa of the genus Cyclotella (Kützing) Brébisson (Bacillariophyceae) and their transfer into the new genus Discostella gen. nov. Diatom Res. 19, 203–228. doi: 10.1080/0269249X.2004.9705871
Hustedt F. (1938). Systematische und ökologische Untersuchungen über die Diatomeen-Flora von Java, Bali und Sumatra nach dem Material der Deutschen Limnologischen Sunda-Expedition. Allgemeiner Teil. Arch. Hydrobiol. Suppl. 15, 393–506.
Hustedt F. (1942). Diatomeen aus der Umgebung von Abisko in Schwedisch-Lappland. Arch. Hydrobiol 39, 82–174.
Hustedt F. (1949). “Süsswasser-Diatomeen aus dem Albert-Nationalpark in Belgisch-Kongo,” in Exploration du Parc National Albert – Mission H. Dumans, (1935–1936) (Hayez, Bruxelles: Institut des parcs nationaux du Congo belge).
Hustedt F. (1955). Marine littoral diatoms of Beaufort, North Carolina (Cambridge: Cambridge University Press).
Jahn R., Schmid A. M. M. (2007). Revision of the brackish-freshwater diatom genus Bacillaria Gmelin (Bacillariophyta) with the description of a new variety and two new species. Eur. J. Phycol. 42, 295–312. doi: 10.1080/09670260701428864
Jeong Y., Lee J. (2021). The complete mitochondrial genome of the benthic diatom Pleurosigma inscriptura. Mitochondrial DNA B: Resour. 6, 2584–2586. doi: 10.1080/23802359.2021.1945970
Johansen J. R., Fryxell G. A. (1985). The genus Thalassiosira (Bacillariophyceae): studies on species occurring south of the Antarctic Convergence Zone. Phycologia 24, 155–179. doi: 10.2216/i0031-8884-24-2-155.1
Jung S. W., Yun S. M., Lee S. D., Kim Y. O., Lee J. H. (2009). Morphological characteristics of four species in the genus Skeletonema in coastal waters of South Korea. Algae 24, 195–203. doi: 10.4490/ALGAE.2009.24.4.195
Katoh K., Toh H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9, 286–298. doi: 10.1093/bib/bbn013
Kim J. H., Park B. S., Kim J. H., Wang P., Han M. S. (2015). Intraspecific diversity and distribution of the cosmopolitan species Pseudo-nitzschia pungens (Bacillariophyceae): morphology, genetics, and ecophysiology of the three clades. J. Phycol. 51, 159–172. doi: 10.1111/jpy.12263
Kooistra W. H., Sarno D., Balzano S., Gu H., Andersen R. A., Zingone A. (2008). Global diversity and biogeography of Skeletonema species (Bacillariophyta). Protist 159 (2), 177–193. doi: 10.1016/j.protis.2007.09.004
Kryk A., Witkowski W., Ribeiro L., Kociolek J. P., Mayama S., Wróbel R. J., et al. (2021). Novel Diatoms (Bacillariophyta) from tropical and temperate marine littoral habitats with the description of Catenulopsis gen. nov., and two Catenula species. Diatom Res. 36, 265–280. doi: 10.1080/0269249X.2021.1974572
Kulikovskiy M., Chudaev D., Glushchenko A., Kuznetsova I., Frolova L., Kociolek J. P. (2021). Two new species of the diatom genus Navicula Bory (Bacillariophyceae) from Vietnam (Southeast Asia). Diatom Res. 36, 61–73. doi: 10.1080/0269249X.2020.1853608
Kulikovskiy M. S., Lange-Bertalot H., Metzeltin D., Witkowski A. (2012). Lake Baikal: Hotspot of endemic diatoms I. Iconogr. Diatomol. 23, 7–608.
Kützing F. T. (1844). Die kieselschaligen Bacillarien oder Diatomeen. Ed. Förstemann F. (Nordhausen, Germany: zu finden bei W. Köhne).
Lee J. M., Cho C. H., Park S. I., Choi J. W., Song H. S., Yoon H. S. (2016). Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 14, 1–16. doi: 10.1186/s12915-016-0299-5
Lemieux C., Otis C., Turmel M. (2007). A clade uniting the green algae Mesostigma viride and Chlorokybus atmophyticus represents the deepest branch of the Streptophyta in chloroplast genome-based phylogenies. BMC Biol. 5, 1–17. doi: 10.1186/1741-7007-5-2
Lewin J., Lewin R. A. (1967). Culture and nutrition of some apochlorotic diatoms of the genus Nitzschia. Microbiology 46, 361–367. doi: 10.1099/00221287-46-3-361
Li Y., Chen X., Sun Z., Xu K. (2017a). Taxonomy and molecular phylogeny of three marine benthic species of Haslea (Bacillariophyceae), with transfer of two species to Navicula. Diatom Res. 32, 451–463. doi: 10.1080/0269249X.2017.1401008
Li Y., Guo X. H., Lundholm N. (2020). Molecular phylogeny and taxonomy of the genus Minidiscus (Bacillariophyceae), with description of Mediolabrus gen. nov. J. Phycol. 56, 1443–1456. doi: 10.1111/jpy.13038
Li Y., Huang C. X., Xu G. S., Lundholm N., Teng S. T., Wu H., et al. (2017b). Pseudo-nitzschia simulans sp. nov. (Bacillariophyceae), the first domoic acid producer from Chinese waters. Harmful Algae 67, 119–130. doi: 10.1016/j.hal.2017.06.00
Li C., Tan L., Gao Y., Li X., Sun L., Liang J., et al. (2022). Morphology of Amphora lunulata AH Wachnicka & EE Gaiser and its transfer to Seminavis DG Mann. Diatom Res. 37 (1), 63–69. doi: 10.1080/0269249X.2022.2041490
Li C. W., Volcani B. E. (1987). Four new apochlorotic diatoms. Br. Phycological J. 22, 375–382. doi: 10.1080/00071618700650441
Li H., Yang G., Sun Y., Wu S., Zhang X. (2007). Cylindrotheca closterium is a species complex as was evidenced by the variations of rbcL gene and SSU rDNA. J. Ocean Univ. China. 6, 167–174. doi: 10.1007/s11802-007-0167-6
Lim H. C., Lim P. T., Su S. N. P., Kotaki Y., Leaw C. P. (2012). Morphological observation of two species of Pseudo-nitzschia (Bacillariophyceae). Coast. Mar. Sci. 35, 52–57. doi: 10.15083/00040632
Liu K., Chen Y., Cui Z., Liu S., Xu Q., Chen N. (2021). Comparative analysis of chloroplast genomes of Thalassiosira species. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.788307
Lowe R. L., Busch D. E. (1975). Morphological observations on two species of the diatom genus Thalassiosira from fresh-water habitats in Ohio. Trans. Am. Micros. Soc. 94, 118–123. doi: 10.2307/3225537
Lowe T. M., Chan P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Lundholm N., Daugbjerg N., Moestrup Ø. (2002b). Phylogeny of the Bacillariaceae with emphasis on the genus Pseudo-nitzschia (Bacillariophyceae) based on partial LSU rDNA. Eur. J. Phycol. 37, 115–134. doi: 10.1017/S096702620100347X
Lundholm N., Hasle G. R., Fryxell G. A., Hargraves P. E. (2002a). Morphology, phylogeny and taxonomy of species within the Pseudo-nitzschia americana complex (Bacillariophyceae) with descriptions of two new species, Pseudo-nitzschia brasiliana and Pseudo-nitzschia linea. Phycologia 41, 480–497. doi: 10.2216/i0031-8884-41-5-480.1
Mann D. G. (1981). A new species of sigmoid Nitzschia (Bacillariophyta). Isr. J. Plant Sci. 30, 1–10. doi: 10.1080/0021213X.1981.10676899
Mann D., Crawford R., Round F. (2017). “Bacillariophyta”, in Handbook of the Protists, vol. 7 . Ed. Archibald J., Simpson AGB., Slamovits C. H., Margulis L., Melkonian M., Chapman D. J., et al (Cham: Springer), 205–266. doi: 10.1007/978-3-319-32669-6_29-1
Mann D. G., Trobajo R., Sato S., Li C., Witkowski A., Rimet F., et al. (2021). Ripe for reassessment: A synthesis of available molecular data for the speciose diatom family Bacillariaceae. Mol. Phylogenet. Evol. 158, 106985. doi: 10.1016/j.ympev.2020.106985
Mejdandžić M., Bosak S., Nakov T., Ruck E., Orlić S., Gligora Udovič M., et al. (2018). Morphological diversity and phylogeny of the diatom genus Entomoneis (Bacillariophyta) in marine plankton: six new species from the Adriatic Sea. J. Phycol 54, 275–298. doi: 10.1111/jpy.12622
Mejdandžić M., Bosak S., Orlić S., Gligora Udović M., Štefanić P. P., Špoljarić I., et al. (2017). Entomoneis tenera sp. nov., a new marine planktonic diatom (Entomoneidaceae, Bacillariophyta) from the Adriatic Sea. Phytotaxa 292, 1–18. doi: 10.11646/phytotaxa.292.1.1
Metzeltin D., Lange-Bertalot H. (1998). “Tropical diatoms of South America I: About 700 predominantly rarely known or new taxa representative of the neotropical flora,” in Iconographia Diatomologica. Annotated Diatom Micrographs, vol. 5 . Ed. Lange-Bertalot H. (Diversity–Taxonomy–Geobotany. Königstein, Germany: Koeltz Scientific Books).
Morales E. A., Wetzel C. E., Novais M. H., Morais M. M., Ector L. (2020). Nitzschia transtagensis sp. nov.(Bacillariophyceae) from a spring in Southern Portugal. Bot. Lett. 167, 32–41. doi: 10.1080/23818107.2019.1688676
Nguyen T. N. M., Berzano M., Gualerzi C. O., Spurio R. (2011). Development of molecular tools for the detection of freshwater diatoms. J. Microbiol. Methods 84 (1), 33–40. doi: 10.1016/j.mimet.2010.10.008
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Osada K., Kobayasi H. (1990). “Observations on the forms of the diatom Entomoneis paludosa and related taxa,” in Proceedings of the tenth international diatom symposium. Ed. Simola H. (Koenigstein: Koeltz Scientific Book).
Park S. I., Cho C. H., Ciniglia C., Huang T. Y., Liu S. L., Bustamante D. E., et al. (2023). Revised classification of the Cyanidiophyceae based on plastid genome data with descriptions of the Cavernulicolales ord. nov. and Galdieriales ord. nov. (Rhodophyta). J. Phycol. 59, 444–466. doi: 10.1111/jpy.13322
Park J. S., Jung S. W., Ki J. S., Guo R., Kim H. J., Lee K. W., et al. (2017). Transfer of the small diatoms Thalassiosira proschkinae and T. spinulata to the genus Minidiscus and their taxonomic re-description. PloS One 12, e0181980. doi: 10.1371/journal.pone.0181980
Prasad A. K., Nienow J. A. (2006). The centric diatom genus Cyclotella, (Stephanodiscaceae: Bacillariophyta) from Florida Bay, USA, with special reference to Cyclotella choctawhatcheeana and Cyclotella desikacharyi, a new marine species related to the Cyclotella striata complex. Phycologia 45, 127–140. doi: 10.2216/05-13.1
Priisholm K., Moestrup Ø., Lundholm N. (2002). Taxonomic notes on the marine diatom genus Pseudo-nitzschia in the Andaman Sea near the island of Phuket, Thailand, with a description of Pseudo-nitzschia micropora sp. nov. Diatom Res. 17, 153–175. doi: 10.1080/0269249X.2002.9705536
Reid G. (2002). Four new species of Pleurosigma (Bacillariophyta) from Alexandria, Egypt. Bot. J. Linn. Soc. 140, 77–92. doi: 10.1046/j.1095-8339.2002.00084.x
Reimann B. E., Lewin J. C. (1964). The diatom genus Cylindrotheca Rabenhorst. J. R. Microsc. Soc. 83, 283–296. doi: 10.1111/j.1365-2818.1964.tb00542.x
Rivera-Vilarelle M., Quijano-Scheggia S., Olivos-Ortiz A., Gaviño-Rodríguez J. H., Castro-Ochoa F., Reyes-Herrera A. (2013). The genus Pseudo-nitzschia (Bacillariophyceae) in manzanillo and santiago bays, Colima, Mexico. Botanica Marina 56, 357–373. doi: 10.1515/bot-2012-0176
Round F. E., Crawford R. M., Mann D. G. (1990). The diatoms: biology and morphology of the genera (Cambridge: Cambridge University Press).
Rumrich U., Lange-bertalot H., Rumrich M. (2000). Diatoms of the Andes. From Venezuela to Patagonia/Tierra del Fuego and two additional contributions. Iconographia Diatomologica 9, 1–673.
Ryabushko L. I., Balycheva D. S., Bondarenko A. V., Zheleznova S. N., Begun A. A., Stonik I. V. (2019). Different aspects of studying a diatom Cylindrotheca closterium (Ehrenberg) Reimann et Lewin 1964 in natural and laboratory conditions. Mar. Biol. 4, 52–62. doi: 10.21072/mbj.2019.04.2.06
Sabbe K., Vyverman W., Ector L., Wetzel C. E., John J., Hodgson D. A., et al. (2019). On the identity of Navicula gottlandica (Bacillariophyta), with the description of two new species Navicula eileencoxiana and Navicula bergstromiana from the Australo-Pacific region. Plant Ecol. Evol. 152, 313–326. doi: 10.5091/PLECEVO.2019.1607
Saedler H., Becker A., Winter K. U., Kirchner C., Theißen G. (2001). MADS-box genes are involved in floral development and evolution. Acta Biochim. Pol. 48 (2), 351–358. doi: 10.18388/abp.2001_3920
Schmid A. M. M. (2007). The “paradox” diatom Bacillaria paxillifer (Bacillariophyta) revisited 1. J. Phycol 43, 139–155. doi: 10.1111/j.1529-8817.2006.00299.x
Shevchenko O. G., Shulgina M. A., Shulkin V. M., Tevs K. O. (2020). The long-term dynamics and morphology of the diatom Thalassiosira nordenskioeldii Cleve 1873 (Bacillariophyta) from the coastal waters of peter the great bay, sea of Japan. Russ. J. Mar. Biol. 46, 284–291. doi: 10.1134/S1063074020040069
Shu-qi X., Ting L. (1994). A new diatom species of salt lake from Shanxi Province. J. Syst. Evol. 32, 273.
Sitoki L., Kofler W., Rott E. (2013). Planktonic needle-shaped Nitzschia species from Lake Victoria, Africa, revisited. Diatom Res. 28, 165–174. doi: 10.1080/0269249X.2013.765509
Song H. J., Lee J. M., Graf L., Rho M., Qiu H., Bhattacharya D., et al. (2016). A novice’s guide to analyzing NGS-derived organelle and metagenome data. Algae 31, 137–154. doi: 10.4490/algae.2016.31.6.5
Sonneman J. A., Sincock A., Fluin J., Reid M., Newall P., Tibby J., et al. (2000). An illustrated guide to common stream diatom species from temperate Australia (Albury, NSW: Cooperative Research Centre for Freshwater Ecology).
Spaulding S. A., Potapova M. G., Bishop I. W., Lee S. S., Gasperak T. S., Jovanoska E., et al. (2021). Diatoms.org: supporting taxonomists, connecting communities. Diatom Res. 36, 291–304. doi: 10.1080/0269249X.2021.2006790
Sterrenburg F. A. S. (2001). Studies on the genera Gyrosigma and Pleurosigma: the types of Shadbolt and related taxa. Proc. Acad. Natural Sci. Philadelphia 151, 121–127. doi: 10.1635/0097-3157(2001)151[0121:SOTGPA]2.0.CO;2
Sterrenburg F. A. S. (2003). Studies on the diatom genera Gyrosigma and Pleurosigma (Bacillariophyceae): Pleurosigma strigosum W. Smith and some presumptive relatives. Micropaleontology 49, 159–169. doi: 10.2113/49.2.159
Sterrenburg F. A., Tiffany M. A., Hinz F., Herwig W. E., Hargraves P. E. (2015). Seven new species expand the morphological spectrum of Haslea. A comparison with Gyrosigma and Pleurosigma (Bacillariophyta). Phytotaxa 207, 143–162. doi: 10.11646/phytotaxa.207.2.1
Stidolph S. R. (2002). Observations and remarks on the morphology and taxonomy of the diatom genera Gyrosigma Hassall and Pleurosigma W. Smith. 5. Pleurosigma types of A. Mann, (1925): a critical re-investigation. Micropaleontology 48, 273–284. doi: 10.1661/0026-2803(2002)048[0273:OAROTM]2.0.CO;2
Taylor J. C., Cocquyt C., Mayama S. (2016). Navicula nielsfogedii J.C. Taylor & Cocquyt sp. nov., a new diatom (Bacillariophyta) from tropical and sub-tropical Africa. Fottea 16, 201–208. doi: 10.5507/fot.2016.015
Timmis J. N., Ayliffe M. A., Huang C. Y., Martin W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5 (2), 123–135. doi: 10.1038/nrg1271
Trobajo R., Rovira L., Ector L., Wetzel C. E., Kelly M., Mann D. G. (2013). Morphology and identity of some ecologically important small Nitzschia species. Diatom Res. 28, 37–59. doi: 10.1080/0269249X.2012.734531
Vilhena J. C., Amorim A., Ribeiro L., Ferreira I. M., Pombo M. (2021). Assessing the efficiency of low-cost cleaning methods for diatoms from intertidal sediment samples. J. Microbiol. Methods 187, 106255. doi: 10.1016/j.mimet.2021.106255
Visco J. A., Apothéloz-Perret-Gentil L., Cordonier A., Esling P., Pillet L., Pawlowski J. (2015). Environmental monitoring: inferring the diatom index from next-generation sequencing data. Environ. Sci. Technol. 49, 7597–7605. doi: 10.1021/es506158m
Von Quillfeldt C. H. (2001). Identification of some easily confused common diatom species in Arctic spring blooms. Bot. Mar. 44, 375–389. doi: 10.1515/BOT.2001.048
Wang T., Chen L. L., Shu H. J., You F., Liang X. L., Li J., et al. (2022b). Fortunella venosa (Champ. ex Benth.) C. C. Huang and F. hindsii (Champ. ex Benth.) swingle as independent species: evidence from morphology and molecular systematics and taxonomic revision of Fortunella (Rutaceae). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.867659
Wang Y., Liu S., Wang J., Yao Y., Chen Y., Xu Q., et al. (2022a). Diatom biodiversity and speciation revealed by comparative analysis of mitochondrial genomes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.749982
Wang Y., Mu W., Sun X., Lu X., Fan Y., Liu Y. (2020). Physiological response and removal ability of freshwater diatom Nitzschia palea to two organophosphorus pesticides. Chem. Ecol. 36, 881–902. doi: 10.1080/02757540.2020.1784883
Winchell C. J., Martin A. P., Mallatt J. (2004). Phylogeny of elasmobranchs based on LSU and SSU ribosomal RNA genes. Mol. Phylogenet. Evol. 31 (1), 214–224. doi: 10.1016/j.ympev.2003.07.010
Witkowski A., Li C., Zgłobicka I., Yu S. X., Ashworth M., Dąbek P., et al. (2016). Multigene assessment of biodiversity of diatom (Bacillariophyceae) assemblages from the littoral zone of the Bohai and Yellow Seas in Yantai Region of Northeast China with some remarks on ubiquitous taxa. J. Coast. Res. 74, 166–195. doi: 10.2112/SI74-016.1
Yamamoto M., Chiba T., Tuji A. (2017). Salinity responses of benthic diatoms inhabiting tidal flats. Diatom Res. 32, 243–250. doi: 10.1080/0269249X.2017.1366951
Keywords: diatom, Nitzschia, organelle genome, multigene phylogeny, genus boundary
Citation: Jeong Y and Lee J (2024) Comparative analysis of organelle genomes provides conflicting evidence between morphological similarity and phylogenetic relationship in diatoms. Front. Mar. Sci. 10:1283893. doi: 10.3389/fmars.2023.1283893
Received: 25 September 2023; Accepted: 05 December 2023;
Published: 03 January 2024.
Edited by:
Jin Sun, Ocean University of China, ChinaReviewed by:
Daniele De Luca, Anton Dohrn Zoological Station Naples, ItalyCopyright © 2024 Jeong and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: JunMo Lee, anVubW9sZWVAa251LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.