
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 08 January 2024
Sec. Microbial Symbioses
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1279849
This article is part of the Research TopicMarine Microbial Symbioses: Host-Microbe Interaction, Holobiont’s Adaptation to Niches and Global Climate ChangeView all 12 articles
Bacteria associated with marine invertebrate play a fundamental role in the biology, ecology, development and evolution of their hosts. Although many studies have been focused on the microbial populations of benthic and pelagic habitats, little is known about bacteria colonizing tube-dwelling polychaete. In this context, the current study provided the first characterization of the Melinna palmata Grube, 1870 microbiome based on Illumina sequencing of 16S rRNA gene of the polychaete tissue and proximate sediments collected from the Black Sea, Romania, along a 24.2 m – 45.4 m depth-gradient. The diversity, taxonomic composition and deduced functional profile of the tissue and sediments associated bacterial communities were compared and analyzed in relation with the environmental parameters. This polychaete harbored a distinct bacterial assemblage as compared to their sediments and independent on the depth of their habitat, including 8 phyla in tissues dominated by Proteobacteria, and 12 phyla in sediments majorly represented by Actinobacteriota, respectively. At order level, Synechococcales, Rhodobacterales and Actinomarinales were highly represented in the M. palmata microbiome, while Microtrichales, Anaerolineales and Caldilineales were mostly found in sediments. A significant correlation was observed between Cyanobacteria taxa and the dissolved oxygen concentrations in shallow waters impacted by the Danube inputs. Meanwhile, this phylum showed a positive correlation with Planctomycetota colonizing the invertebrate tissues, and a negative one with Actinobacteriota and Chloroflexi found in sediments. The deduced functional profile of these bacterial assemblages suggested the prevalence of the amino acid and carbohydrate metabolism for both analyzed matrices. This pioneering report on the M. palmata microbiome highlighted the environment contribution to bacterial species enrichment of the polychaete, and provided a glimpse on the putative role of microbial communities associated with this marine organism.
Marine benthic organisms are naturally colonized by microorganisms, which play a fundamental role in the biology of their hosts (Kelman et al., 2009; Ketchum et al., 2018). The diversity of bacterial communities associated with marine organisms plays an important role in immunity, metabolism, physiology (Mcfall-Ngai et al., 2013; Bordenstein and Theis, 2015; Theis et al., 2016), as well as in host development, adaptation and evolution (Rosenberg et al., 2007; Zilber-Rosenberg and Rosenberg, 2008) of these invertebrates. The bacterial-host relationship has aroused considerable interest in the field of marine biology (Piel, 2004); some of these microorganisms may be symbionts, pathogens or can create mutualism relations (Kelman et al, 2009). Symbiotic bacteria are found in many biological systems, influencing the host’s evolution and suggesting that microorganisms are involved in limiting pathogen colonization through antimicrobial compounds and competition for food resources (Felbeck and Distel, 2004; Desriac et al., 2014). As a result of co-evolving with their specific hosts, microbial symbionts exhibit a diverse array of custom-tailored biochemical traits. This renders them a reservoir of secondary metabolites characterized by distinct bioactivities that hold significant medical and commercial appeal (Zhang et al., 2015). Filtering and deposit feeding invertebrates that can concentrate bacteria from water and sediments also play an important role in aquatic microbiome contribution to the ecosystem, representing biomarkers for the microbial diversity in aquatic environments (Burkhardt et al., 1992; Graczyk et al., 2003; Marino et al., 2005).
Marine ecosystems, among the most complex environments, harbour a significant part of the global microbial population. Certain bacterial species within these ecosystems are essential contributors to many biogeochemical cycles (Muriel-Millán et al., 2021). Many microorganisms, such as taxa belonging to Proteobacteria, engage in symbiotic relationships with various marine invertebrates, influencing processes such as nutrient cycling, digestion, and defence mechanisms (Kunihiro et al., 2011; Summers et al., 2013; Von Borzyskowski et al., 2019). Cyanobacteria are crucial contributors to primary production and often create symbiotic relationships with sponges, corals or molluscs, providing their hosts with fixed carbon through photosynthesis (Erwin and Thacker, 2008; Zhukova et al., 2022). In the broader marine ecosystem, Actinobacteriota’s metabolic activities, including the breakdown of complex organic compounds, have a significant impact on the availability of nutrients and the carbon cycling (Goodfellow and William, 1983; Stevens et al., 2007).
Various environmental factors, including salinity, temperature, pH, organic matter content, and oxygen levels, play an important role in shaping the distribution and abundance of microbial communities Dang and Lovell, 2016; Guo et al., 2022a). These microorganisms’ interactions with environmental parameters serve as indicators that reflect the function and structure of marine ecosystems, as highlighted in previous studies (Boucher et al., 2006). For instance, the community structure and dynamics of bacteria belonging to Proteobacteria, Firmicutes, and Actinobacteriota phyla appeared to be affected by various factors such as salinity, dissolved oxygen, ammonia, phosphate and silicate concentrations (Guo et al., 2022b). Although variations in microbial community can be substantial across different habitat types, primarily due to the impact of environmental gradients (Martiny et al., 2006), some studies suggested consistency in community composition across similar habitats, regardless of geographical distance (Lauber et al., 2009).
Deep screening of free-living bacterial assemblages in the marine environments (Pommier et al., 2010; Costas-Selas et al., 2022; Ruginescu et al., 2022) and of bacteria associated with different organisms (Musella et al., 2020; Li et al., 2023) was successfully carried out based on next-generation sequencing of 16S rRNA genes (Webster and Taylor, 2012), overcoming some of the challenges previously associated with determining microbial diversity (Sogin et al., 2006; Winand et al., 2019).
For the last decades, investigations of bacteria-invertebrate associations from various marine environments became of high interest for the scientific community (Goffredi et al., 2007; Gilbertson et al., 2012; Lo Giudice and Rizzo, 2022). Many studies demonstrated that interactions between these organisms create symbioses leading to production of secondary metabolites with antimicrobial activities by the invertebrates-associated microbiome (Thomas et al., 2010; Graca et al., 2013; Abdelmohsen et al., 2014; Kuo et al., 2019). Also, symbiotic bacteria from bivalves were involved in the host defense mechanisms (Defer et al., 2013), and could contribute to adaptation to environmental stress conditions (Leite et al., 2017). Various methods, such as biochemical, enzymatic, molecular, and transmission electron microscopy techniques confirmed the presence of chemoautotrophic symbiotic bacteria in these organisms, supporting the idea that these bacteria play a crucial role in nutrition (Conway et al., 1992; Eisen et al., 1992). Microbes inhabiting tunicates were able to produce metabolites with anti-tumor effect (Schmidt et al., 2005), and for various biotechnologies and pharmaceutical applications (Paul and Ritson-Williams, 2008; Erwin et al., 2010). Moreover, recent studies have emphasized the potential of symbiotic bacteria found in polychaetes as a novel reservoir for biosurfactant-producing microbes (Rizzo et al., 2013; Markande et al., 2014).
Bacterial communities of tube-building worms could have an important contribution to the biogeochemical processes that occur at the interface between benthic organisms and their habitat. Thus, burrowing organisms appeared to be essential contributors to the biogeochemistry of the benthic environment through excavation, grazing and excretion (Konhauser et al., 2020). Furthermore, polychaetes host symbiotic microorganisms involved in transformation, degradation and detoxification processes, such as Ophelina species that are characterized by a high content of metal-resistant bacteria (Neave et al., 2012). Also, Capitella teleta was reported to be associated with highly abundant microbes able to degrade polyaromatic hydrocarbons which potentially allows the species to survive in polluted environments or serve as indicators of pollution (Hochstein et al., 2019).
Melinna palmata Grube, 1870 is a deposit-feeder tube-dwelling ampharetid polychaeta, with a length of 3- 4 cm (Hunt, 1925; Mare, 1942; Fauchald and Jumars, 1979). It builds a mucus-lined tube from which it can emerge to spread its tentacular palate on the sediment surface inhabiting the Black Sea generally at depths ranging from 30 to 40 m (Băcescu et al., 1971). Although this species did not occupy a well-established habitat in the Romanian Back Sea shelf until 1970, its populations started to reach significant densities and biomass a decade later (Gomoiu, 1982). Currently, M. palmata and/or Spisula subtruncata constitute the engineering species of the Circalittoral mud habitat, reaching an average density of 3,450 specimen m−2 in the Danube influenced area (Teacă et al., 2020).
Several studies in the Black Sea have been focused on investigating the microbial populations of sediments (Schulz et al, 1999; Thamdrup et al, 2000; Leloup et al, 2007; Schäfer et al, 2007; Coolen and Shtereva, 2009; Schippers et. al, 2012) and pelagic habitats (Jørgensen et al, 1991; Sorokin et al, 1995; Glaubitz et al, 2010; Bryukhanov et al, 2015; Ruginescu et al., 2022). In contrast, only one study characterized bacteria associated with the bivalve Mytilaster lineatus (Onishchenko and Kiprianova, 2006), while no research has been carried out so far on bacteria colonizing polychaete species in order to untangle the role of these symbionts in the invertebrate marine adaptation and possible microbiome exchanges with the seabed sediments.In this context, the current investigation focused on identifying the bacterial communities associated with the tube-building polychaete M. palmata and its surrounding sediments from the Romanian Black Sea shelf based on 16S rRNA gene Illumina sequencing. Comparative analyses of the microbial diversity, community composition and putative functional profile of the polychaete tissue and sediments, and correlation of bacterial taxa with the physicochemical parameters of the sea water were carried out in order to determine if the M. palmata bacterial assembly reflects the partial recruitment of surrounding representatives depending on environmental variables. To our knowledge, this study provides the first characterization of the microbiome of the marine polychaete M. palmata.
The North-Western part of the Black Sea receives 80% of the basin’s total freshwater input originating from the Dnieper, Dniester and Danube, the latter being the major contributor of freshwater and sediment inputs (Mikhailov and Mikhailova, 2008). The Danube River is the second European largest river basin, covering territories of 18 states including EU-Member States, Accession Countries and other countries (ICPDR, 2004). Romanian coastal waters are subject to Danube inputs, which provide significant organic and inorganic matter, the mean annual river flow recorded form all three main branches being of 6500 m3 s−1. The direction, range and depth of dispersion of the Danube waters are dependent on the intensity of water flow and wind direction. North and northeast winds are dominant during spring, reaching average speeds of 5.5 m s-1, while during autumn, north winds predominate, with an average speed of 8.0 m s-1 (Tescari et al., 2006). According to Panin and Jipa (2002), the average sediment discharge from the Danube into the Black Sea is estimated to range between 25 and 35 million tones/year, with sandy material accounting 4-6 million tonnes/year. The study area is located within the marine protected area ROSCI0066 (Danube Delta - marine zone), included in Natura 2000 European ecological network (Figure 1A).
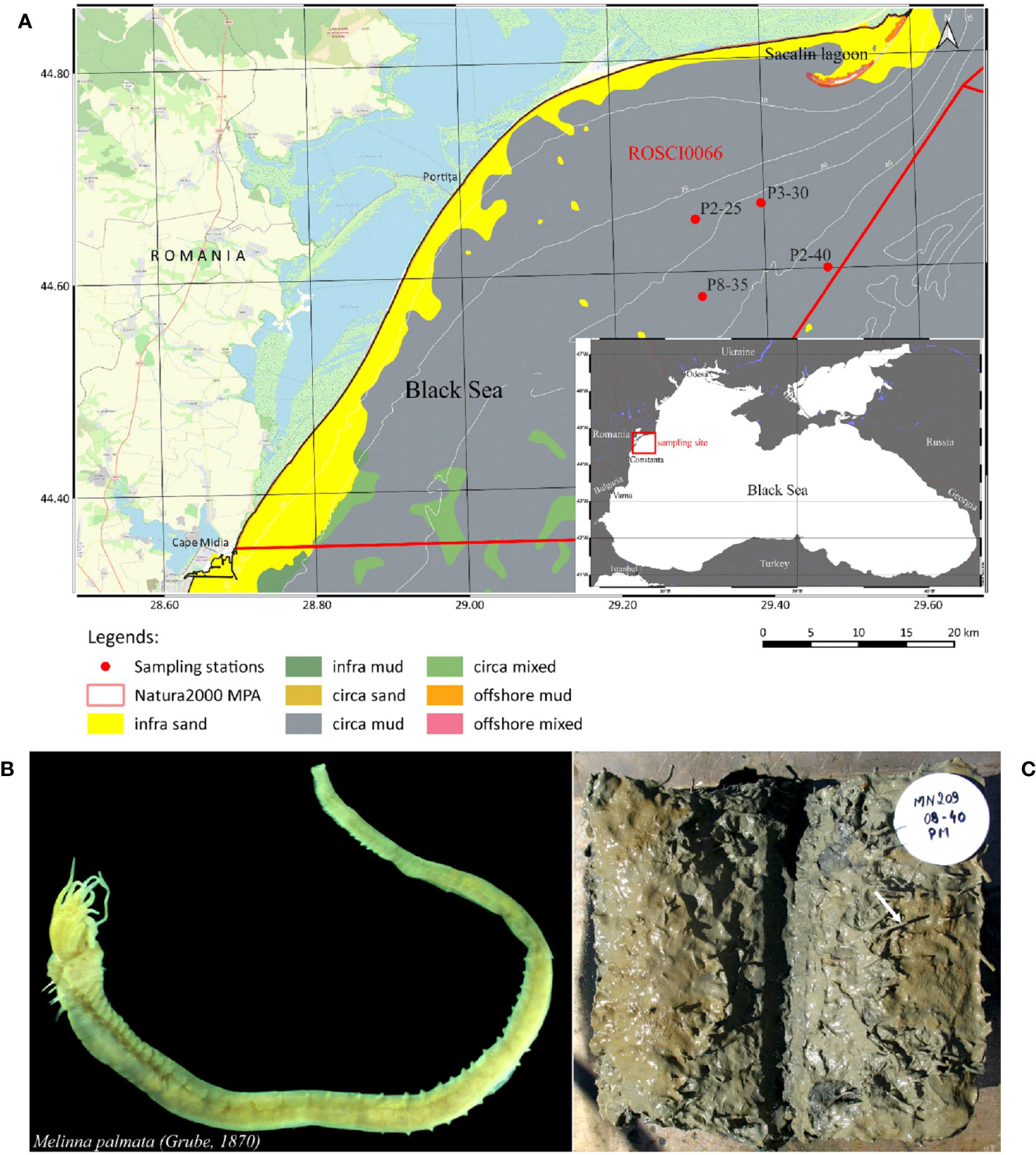
Figure 1 Study area and investigated organism. (A) Map of Black Sea sampling locations and detailed sampling sites area (B) Melinna palmata specimens collected from the Black Sea (photo by photo by A. Teacă); (C) Sediments collected from the Black Sea studied area containing M. palmata (photo by photo by A. Teacă).
The major habitat of the investigated area is represented by Circalittoral muds with Melinna palmata. The total benthic communities of this area comprise a diverse assemblage of deposit feeders and suspension filters, such as oligochaetes, polychaetes (Mellina palmata, Heteromastus filiformis), molluscs (Mytilus galloprovincialis, Spisula subtruncata, Abra nitida, Pitar rudis, Acanthocardia paucicostata), and bacteriouvorus nematodes (Terschellingia longicaudata, Sabatieria pulchra, S. abyssalis, Neochromadora izhorica, Desmolaimus sp.) (Mureşan and Teacă, 2019; Teacă et al., 2020). Among these, M. palmata represented more than 42% of the total average macrozoobenthic density (Mureşan et al., 2019; Mureşan and Teacă, 2019; Teacă et al., 2019; Teacă et al., 2020).
Sediment samples were collected using a Box Corer with a surface of 0.1 m2 (Todorova and Konsulova, 2005) from four different sites located in the Romanian northwestern continental shelf of the Black Sea, during June 2020 (Figure 1A). In order to assess the microbial communities from M. palmata individuals (Figure 1B) and sediments (Figure 1C), two subsamples were collected from each site. The sedimentary material was collected in sterile containers after discarding the fraction in contact with the sampling equipment, and stored at -20°C. For polychaete isolation, sediment particles were removed using 250 μm and 125 μm mesh sieve. Each specimen selected was washed with sterile water and stored in 200 μl Tris-EDTA pH 8 buffer at −20°C for genetic analyses (Ross et al., 1990).
In situ measurements of temperature, salinity, dissolved oxygen (DO) and pH of water above the seabed (bottom layer) were performed for each location with an EXO2 multi-parameter probe (YSI Incorporated, Yellow Springs, USA). The salinity was expressed in practical salinity units (PSU) corresponding to parts per thousand (PPT).
Total DNA was extracted from two sediment samples and two Melinna palmata individuals from each site using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following an optimized protocol that includes an initial cell disruption step (Iancu et al., 2015). Tissue samples were resuspended into Tris-EDTA buffer pH 8 and homogenized at 20°C for 12 min in a SpeedMill PLUS cell homogenizer at 50 Hz (Analytik Jena, Jena, Germany) in the presence of 5 ZR BashingBead 0.2 mm lysis matrix (Zymo Research, Irvine, CA, USA), and further processed according to the manufacturer’s protocol. For sediment samples, DNA was isolated using DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany) following the manufacturer’s instruction.
Sequencing of the amplified V3-V4 region of the 16S rRNA genes was performed using 341F/805R primer pair (Takahashi et al., 2014) and an Illumina MiSeq 300PE platform (Macrogen, Seoul, South Korea).
The resulted DNA sequences were processed using the DADA2 package v1.8) implemented in R (v4.0.2) (Callahan et al., 2016). After removing the forward and reverse primers sequences using cutadapt (v4.2.2) (Martin, 2011), the sequences were trimmed and filtered. Amplicon Sequence Variants (ASVs) were inferred from de-replicated sequences, and chimeras were removed using the “consensus” method. Taxonomic assignment of the ASVs was performed using the Silva v138 16S rRNA database (silva.nr.v138). Analyses were carried out using MicrobiomeAnalyst 2.0 (Lu et al., 2023).
Pattern Search was used to for sequence assignment to phylum Cyanobacteria. The putative functional profile was obtained by mapping the gene abundance as predicted from Tax4Fun2 according to KEGG metabolism (Lu et al., 2023).
The 16S rRNA gene sequences of the bacteria from M. palmata tissues and adjacent sediments of the 16 samples (MP1-MP16) (Table 1) were deposited in the NCBI SRA Sequence Read Archive under the BioProject PRJNA923146.
Alpha and Beta diversities were calculated using the pyloseq package (McMurdie and Holmes, 2013). Alpha diversity of ASVs was evaluated based on Chao1, Shannon, Evenness, and Fisher indices. Bray-Curtis dissimilarity was used to assess the Beta diversity, in order to compare the diversity between samples and microbial communities. Non-metric multidimensional scaling (NMDS) was used to visualize the 2-D matrix where each point represents the entire microbiome of a single sample. The statistical significance (p < 0.05) of the clustering pattern in ordination plots was evaluated using Analysis of group Similarities (ANOSIM) and Permutational multivariate analysis of variance (PERMANOVA). Multiple Linear Regression with Covariate Adjustment was used to find associations between the microbial community structure and the physicochemical water parameters using MaAsLin2. All statistical analyses were conducted using MicrobiomeAnalyst 2.0 (Lu et al., 2023).
A correlation analysis based on SECOM (Pearson 1) between abundances in the two matrices was performed (Lin et al., 2022). The principal Component Analysis (PCA) was performed using tidyverse and ggplot2 packages in R v4.2.3. (Wickham, 2016; Wickham et al., 2019; R Core Team, 2023). Student’s t-test was used to compare the means of gene abundance between the two types of samples (Wickham, 2016; Mishra et al., 2019; R Core Team, 2023).
Sediment samples containing M. palmata Grube, 1870 specimens were collected from 4 sites which formed a perimeter with an area of about 25 km2, with the minimum distance of 6.8 km (between P2-25 and P3-30) and a maximum distance of 14 km (between P8-35 and P2-40) located along the Romanian shore of the Black Sea, where the water depth ranged from 24.2 m to 45.5 m (Figure 1A, Table 1).
The seabed from all sampling locations consisted of mud sediments originating from alluvial deposits made of silt and clay particles.
The physicochemical properties of the sea water showed little variations between the sites (Table 2). Water temperature varied between 9.05°C (P2-40) and 14.80°C (P2-25), with an average of 11.63 (± 2.40) °C. The average salinity was of 17.97 (± 0.38) PSU, with a minimum of 17.60 PSU at station P2-25 and a maximum of 18.49 PSU at station P2-40 (Table 2). The dissolved oxygen concentration had a slight increasing trend with the water depth, between 4.40 mg L-1 and 6.48 mg L-1 at P3-30 and P2-40, respectively, with an average of 5.32 (± 0.94) mg L-1. A slightly alkaline pH was measured all 4 stations with an average value of 8.05(± 0.12), varying between 7.90 (at P3-30 station) and 8.20 (at P8-35 station) (Table 2).
The microbial diversity and community structure of M. palmata tissues and surrounding sediments were determined from the duplicate samples (Table 1) collected from the Romanian Black Sea coast based on Illumina sequencing of the 16S rRNA gene. The total number of DNA sequences (134975) corresponded to 1444 unique bacterial amplicon sequence variants (ASVs). Rarefaction curves of both analyzed matrices showed a complete identified bacterial community, thus capturing a representative portion of the microbial diversity present in samples (Supplementary Figure S1).
Alpha diversity analysis of the tissue and sediment associated bacteria based on Chao1 index showed significant differences between the analyzed group (p-value 0.0249; Table 3, Supplementary Figure S2). The microbiome of all sediment samples exhibited higher microbial diversity, with Chao1 index values ranging from 401 to 647.1 and a mean of 554.8 ± 27. Meanwhile, this diversity index varied within the 295.6 - 628.1 interval, with a mean value of 425.9 ± 43.6 in the case of tissue-associated bacterial communities (Table 3). No major variation was observed between bacterial diversity at different depths (Supplementary Figure S2B). The corresponding calculated Fisher indices (Table 3) reflected a similar trend (p-value 0.0229), showing a slightly lower diversity for M. palmata associated bacteria (average of 97.37 ± 11,19) as compared to that of the adjacent sediments (average 130.3 ± 6.38). Slight variation of these diversity indices was observed between samples and within the replicate groups (Table 3). The calculated Shannon indices showed higher values for both tissue and sediment associated bacteria with no significant differences. Furthermore, no statistically significant variations between these communities along the depth gradient was observed (Chao1 p-value: 0.61761; [ANOVA] F-value: 0.61619; Shannon p-value: 0.47727; [ANOVA] F-value: 0.88328).
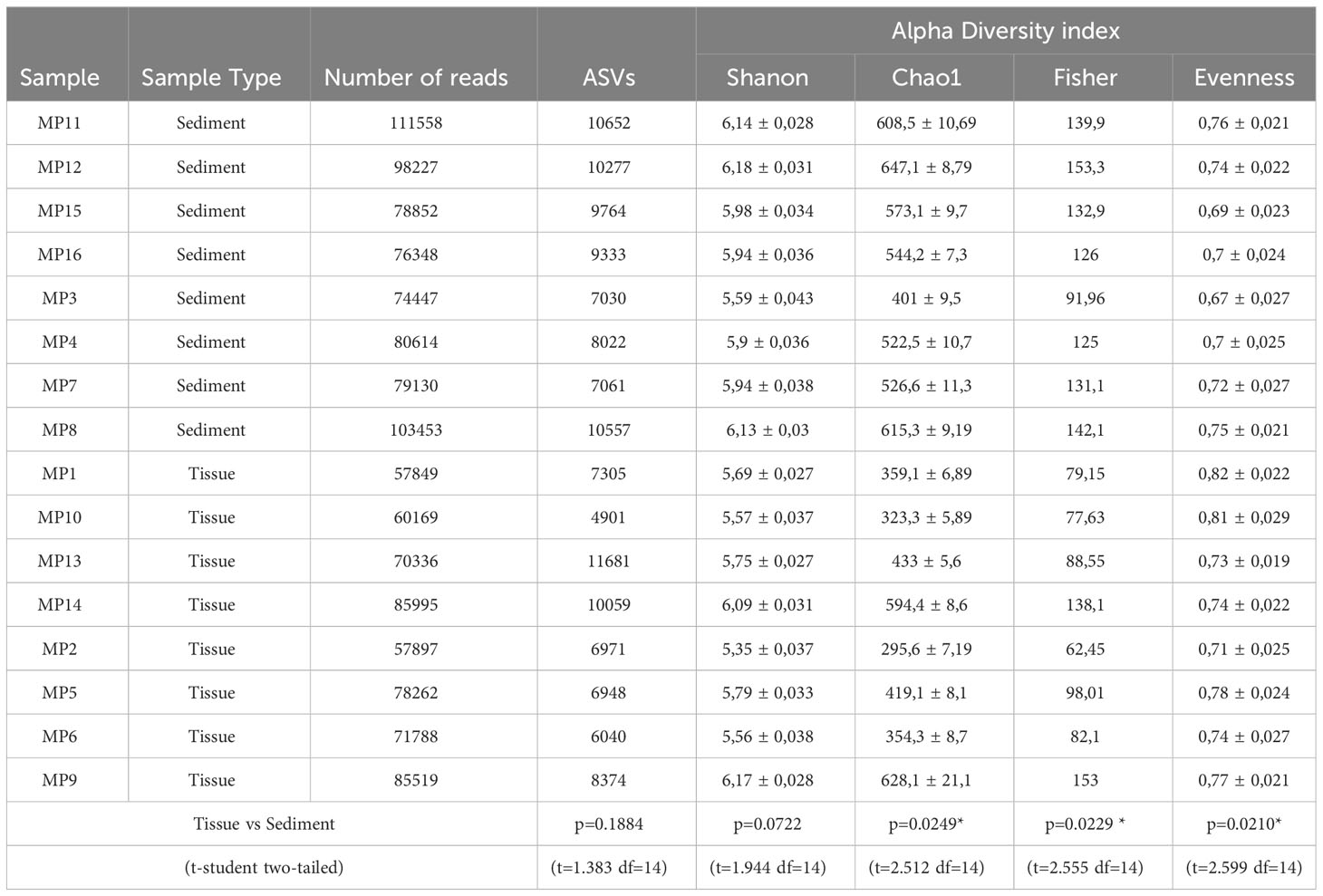
Table 3 Number of reads, ASVs and alpha diversity indices of bacterial communities from M. palmata tissue and associated sediments with statistical comparison (p-values).
The multidimensional beta diversity analysis of the microbial communities in relation with the type of matrices showed significantly different diversity and relative abundance between the polychaete tissue and the sediment samples ([ANOSIM] R: 0.77623; p-value < 0.001 [NMDS] Stress = 0.061336) (Figure 2), and [PERMANOVA] F-value: 4.6741; R-squared: 0.2503; p-value: 0.001.
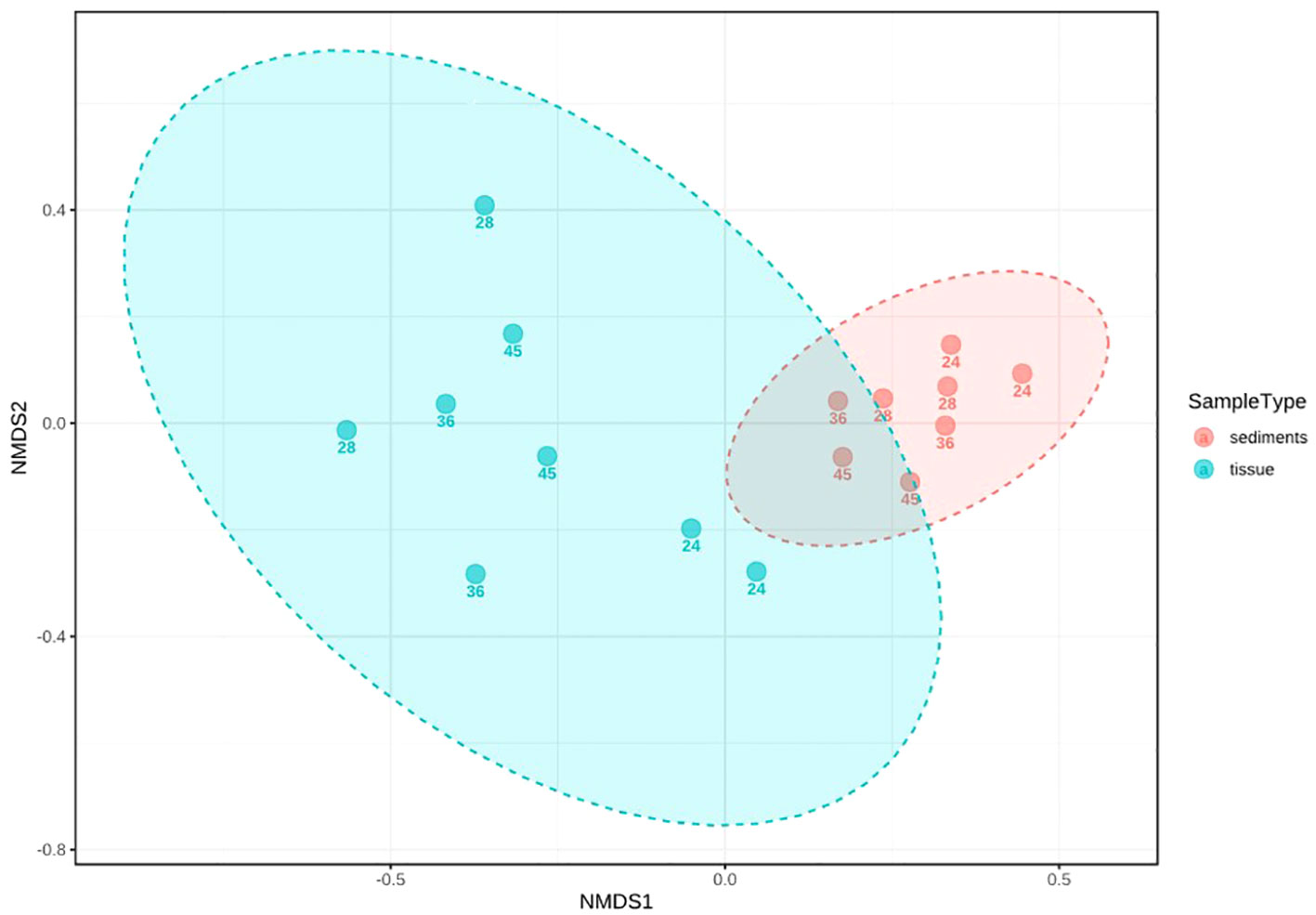
Figure 2 Multidimensional analysis (ANOSIM) of the beta diversity of bacterial communities from tissues and sediments, based on ASVs. The sample depth (number) is indicated for each sample.
[NMDS] Stress = 0.061336 (Supplementary Figure S3), with higher differences among tissue samples.
Taxonomic assignment of bacterial community from M. palmata tissue led to identification of 12 phyla, 14 classes, 26 orders, 26 families and 30 genera, while the adjacent sediments microbiota contained 12 phyla, 19 classes, 34 orders, 29 families and 36 genera. Among these, 14 classes were common to the invertebrate and its habitat, while Fusobacteriia, ABY1, Thermodesulfovibrionia, Parcubacteria and Phycisphaerae were identified only in sediments.
At phylum level, the tissue-colonizing bacteria were dominated by Proteobacteria (16541 ASVs), followed by Actinobacteriota (14171 ASVs), Cyanobacteria (11419 ASVs) and Chloroflexi (6330 ASVs), accounting for a total of 62276 ASVs (Figure 3A). Meanwhile, the sediment communities totaling 72696 ASVs was mostly represented by Actinobacteriota (28581 ASVs) and Chloroflexi (18968 ASVs), with a lower abundance of Proteobacteria (7682 ASVs) and Campylobacterota (6924 ASVs). The relative content of phylum Fimicutes was similar in both tissues and sediments, accounting for 2576 and 2572 ASVs of total reads, respectively, while Planctomycetota taxa were more prominently represented in tissues (5951 ASVs vs. 2887 ASVs, respectively). Also, low presence of phyla Fusobacteria, Nitrospirota and Patescibacteria (less than 250 ASVs for each group) have been observed in sediments (Figure 3A).
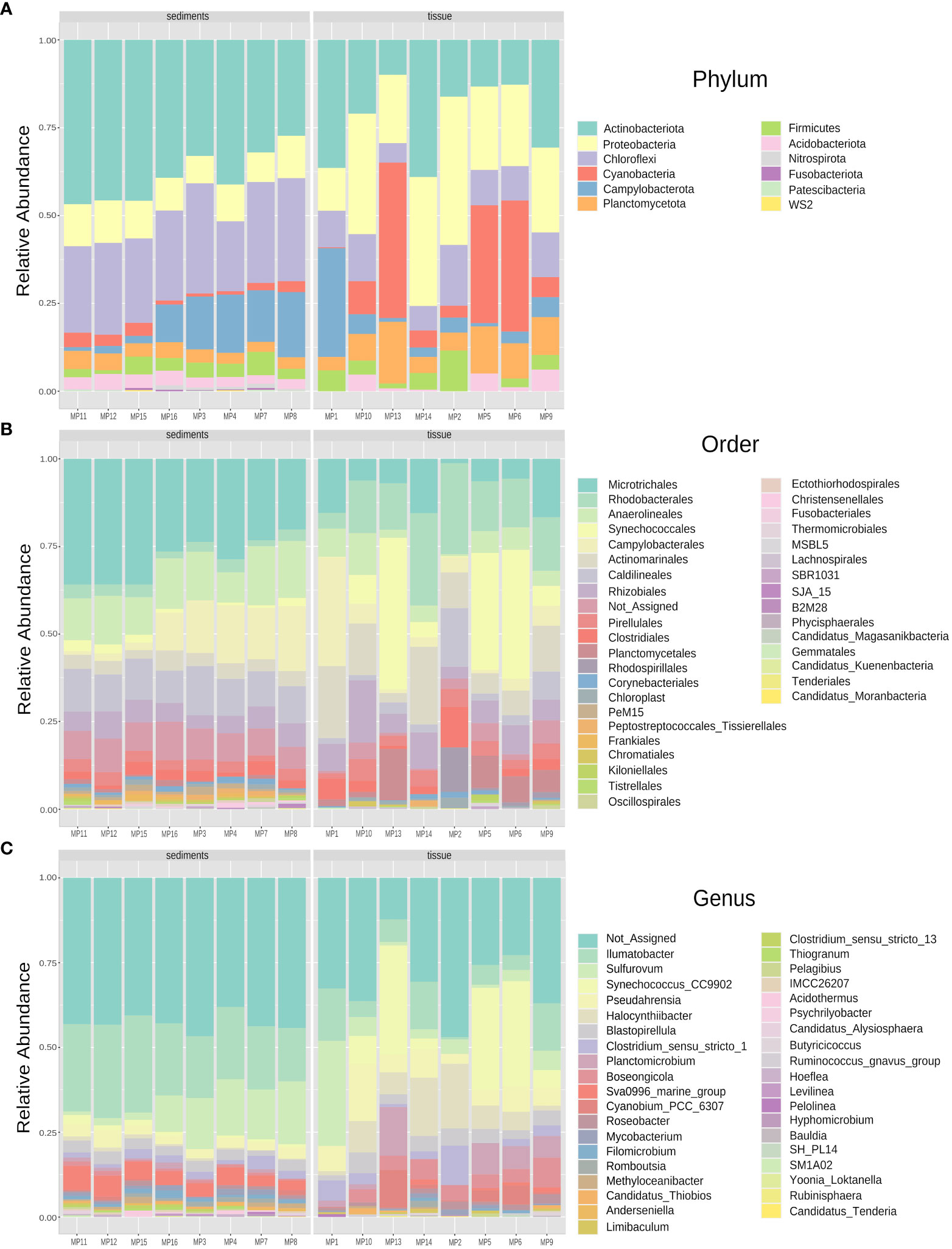
Figure 3 Relative abundance profile of bacterial communities colonizing sediments and tissues of M. palmata at Phylum (A), Order (B), and Genus (C) levels.
At order level, 24 taxa were found in both polychaete and sediments, with Rhodospirillales and Lachnospirales exclusively identified in tissues, and Christensenellales, SJA_15, B2M28, Phycisphaerales, Ectothiorhodospirales, Frankiales, Candidatus Kuenenbacteria, Candidatus Moranbacteria, Candidatus_Magasanikbacteria and Fusobacteriales only in sediments. At family level, 23 taxa were shared, while Butyricicoccaceae, Lachnospiraceae and Magnetospiraceae representative were specific for tissues and Ectothiorhodospiraceae, Acidothermaceae, Phycisphaeraceae, Hungateiclostridiaceae, Fusobacteriaceae and Christensenellaceae found only in sediments. Among these, Synechococcales (10948 ASVs), Rhodobacterales (10173 ASVs), Actinomarinales (7414 ASVs) were the most prominent representatives in M. palmata tissues, while Micotrichales (21239 ASVs), Anaerolineas (9660 ASVs), Caldilineales (8042 ASVs) and Campylobacterales (6924 ASVs) recorded higher relative abundances in sediments (Figure 3B).
From the 36 identified bacterial genera, 28 were common to both types of samples. The tissue-colonizing bacteria were assigned to 30 genera Butyricicoccus and Ruminococcus species were present only in the polychaete tissues, while those associated with sediments belonged to 36 different genera with Hoeflea, Clostridium_sensu_stricto_13S, M1A02, Candidatus Alysiosphaera, Psychrilyobacter, Levilinea, Thiogranum and Acidothermus species exclusively found in these samples (Figure 3C). Among these, species belonging to genera Ilumatobacter (163210ASVs) and Sulfurovum (6924 ASVs) were dominant in the sediments, while notable abundance of Synechocossus-CC9902 (8866 ASVs) was recorded in tissues (Figure 3C).
To assess the effect of the habitat depth on the M. palmata-associated bacterial communities and related sediments, the core microbiome of phyla with a higher prevalence than 10% from both tissue and sediments was analyzed for each sampling site (Supplementary Figure S4). No notable differences between the relative abundance of major taxa were observed between communities collected from variable water depth, with the highest presence in all cases of Actinobacteriota taxa, followed by Chloroflexi, Proteobacteria, and Planctomycetota, and the lowest representation by phylum Nitrospirota (Supplementary Figure S4).
Pearson analysis of phyla abundances from the two types of samples showed the correlation between the top features (bacterial taxa) ranked by their significance according to the cutoff p-value (0.05) and correlation threshold (0.3) (Figure 4). Specifically, Verrucomicrobiota taxa positively correlated with Planctomycetota, both much more abundant in tissues, while bacteria belonging to phylum Desulfobacterota positively correlated with Nitrospinota and Sumerlaeota taxa highly represented in sediments. Meanwhile, a negative correlation was observed between phyla Nitrospirota and Planctomycetota, as well as between Verrucomicrobiota and Actinobacteriota (Figure 4).
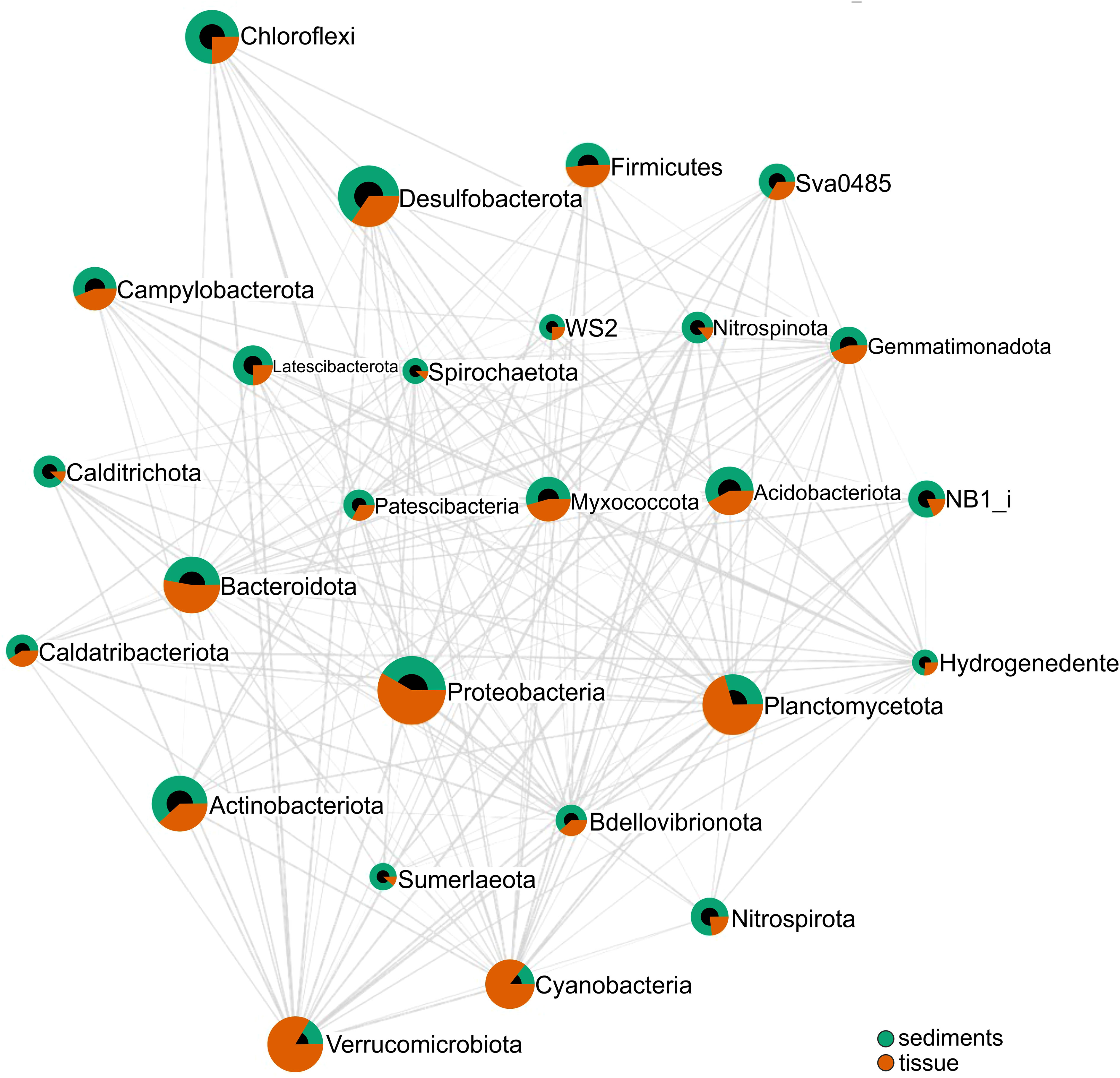
Figure 4 Pearson correlation analysis of relative abundance of bacterial phyla associated with sediments and tissues. Phyla connected by an edge (line) correspond to a correlation between them for p-value cutoff (0.05) and correlation threshold (0.3). The connection length reflects the magnitude of the significant correlation (shorter lengths corresponding to stronger correlations), and the diameter corresponds to the relative abundance of each taxon.
The impact of sea water parameters on the community structure was evaluated by a Multiple Linear Regression analysis showing a significant correlation between Cyanobacteria representatives and DO concentration (p-value 0.0188), with considerable relative abundance of these communities registered at 4.75 mg L-1 dissolved oxygen in shallow waters (Figure 5). Meanwhile, no significant correlation between other taxa and physicochemical parameters was observed (Supplementary Table S1).

Figure 5 Correlation between Cyanobacteria phylum distribution and dissolved oxygen (DO) concentrations of sea water (abundance is expressed in ASVs).
Moreover, a positive pattern correlation between Cyanobacteria phylum and representatives of phylum Planctomycetota colonizing tissues has been identified (Pearson r = 0.89083; p-value: 3.6972e-06), while a negative correlation has been observed between the former group and Actinobacteriota (Pearson r = 0.76177; p-value: 0.00060532) and with Chroloflexi (Pearson r = -0.62317; p-value: 0.0099101) from sediments, respectively (Supplementary Figure S5).
Based on the taxonomic profile and sequence analysis of the ASVs from 16S rRNA metagenomics, an estimated metabolic profile of both M. palmata tissue and sediments bacterial communities was generated, in order to evaluate putative contribution of the sediment substrate to the polychaete microbiota (Figure 6).
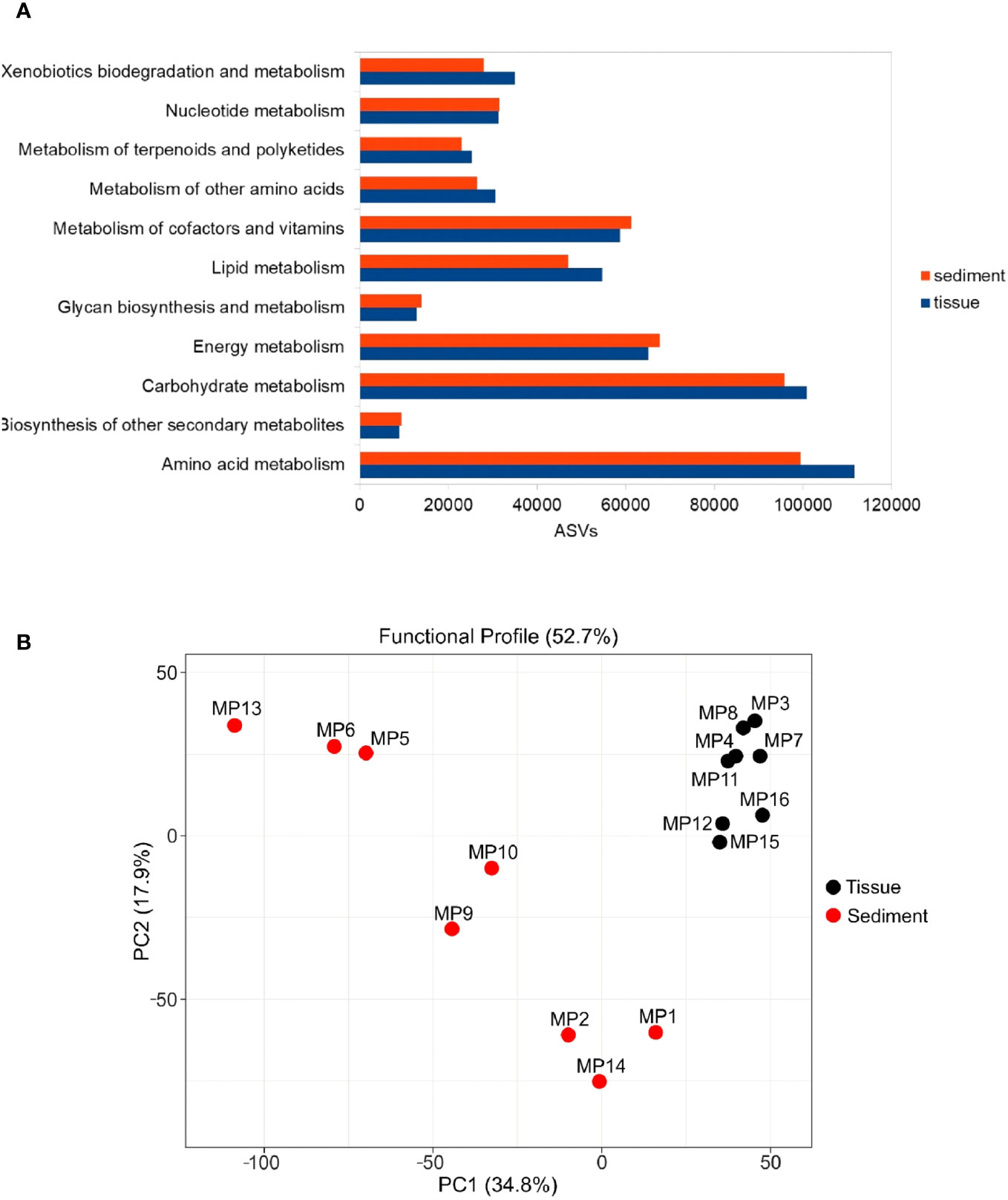
Figure 6 Predicted functional profile of bacterial communities from tissues and sediments based on the gene abundance from KEGG metabolic pathways. (A) Number of predicted functional genes based on amplicon sequence variants (ASVs) from the analyzed matrices (B) diversity of functional genes in the analyzed matrices based on PCA analysis.
Analysis of the predicted functional profile of these microbiomes showed no major differences between the distribution of relative abundance of various genes in accordance with the KEGG (Kyoto Encyclopedia of Genes and Genomes) metabolic pathways, with the prevalence of the amino acid and carbohydrate metabolism across all examined samples (Figure 6A).
Nevertheless, the statistical analysis (Student’s t-test) indicated a higher content of genes involved in carbohydrates, amino acids, lipid, terpenoids and polyketides metabolism along with the those associated with biodegradation and metabolism of xenobiotics (p-value < 0.05) in the polychaete symbiotic bacteria relative to the prokaryotic community from adjacent sediments. Meanwhile, genes related to biosynthesis and metabolism of lipids, terpenoids, polyketides, nucleotides, cofactors, vitamins, energy, glycans, and other secondary metabolites were evidenced in both the tissue-associated and sediment microbiomes (Figure 6A).
A Principal Component Analysis (PCA) of predicted functional genes indicated differences between the gene functionality diversity of the analyzed matrices (Figure 6B). These findings are in accordance with the outcomes of the NMDS analysis (Figure 2), providing further support for the observed differentiation in genes functionality resulting from the comparison between distinct communities, namely tissues and sediments.
Despite the significant diversity differences between M. palmata and sediments-associated bacteria, their predicted metabolic profile based on 16S rRNA taxonomic data could suggest only limited variances between the two communities. Further investigations of these communities by metagenomics and metatranscriptomics are required to highlight the metabolic variability of the polychaete microbiome in relation with that of its substrate, and untangle their putative ecological role.
The current investigation reported original data on the tissue microbiome from the marine polychaete Melinna palmata from the Black Sea, Romania, in relation with the bacterial community of the adjacent sediments. For this invertebrate, a recent study conducted in Arcachon Bay (French Atlantic coast) (Massé et al., 2019) described the aerobic bacterial community composition colonizing the surrounding sediments of this ampharetid polychaete, while so far no investigations have been carried out on the tissue-associated microbiome. In this survey, we applied a culture-independent method based on 16S rRNA gene Illumina sequencing of the bacterial community inhabiting the tissues of M. palmata collected from various depths in the Romanian Black Sea sediments in comparison with that of the sediments of collected specimens. The community composition of both analyzed matrices did not show notable changes along the sea depth gradient between 24.2 and 45.8 mm, most likely as a result of similar environmental characteristics in the studied areas (Teacă et al., 2020).
Our findings demonstrated that M. palmata harbors a distinct microbiome as compared to that of its habitat, which is consistent with other similar surveys on three sipunculan worms species (Sipunculus nudus, Siphonosoma australe, Phascolosoma arcuatum) from Beibu Gulf, Changhua River and Guangcun that showed a significantly lower microbial diversity as compared to that of their surrounding sediments (Li et al., 2023; Liu et al., 2023). The reported diversity of microorganisms associated with benthic invertebrates, such as worms, was generally lower as compared to its surrounding sediments (Liu et al., 2023). Thus, the tube-dwelling organisms often create microenvironments with particular conditions which might favor the colonization with specific bacterial communities (Fuirst et al., 2021). Moreover, sediments belong to a more heterogeneous and diverse set of microhabitats, creating a range of niches that can support a wider diversity of bacterial communities (Liu et al., 2023). It has been previously demonstrated that nematodes and macroinvertebrates contributed to the dynamics of bacterial populations (De Mesel et al., 2004; Plante et al., 2022). Free-living nematodes, mainly those inhabiting the rich organic sediments, are non-selective or selective deposit bacterivorous feeders that excrete mineral or readily mineralizable forms that otherwise would have been locked up in the microbial biomass after nutrients ingestion, which exceeds in general the requirements of nematodes (Bonaglia et al., 2014). Moreover, their mucus was shown to stimulate the nitrogen production which, in turn, provided substrate for microorganisms’ activity and growth (Moens et al., 2005). In contrast, macrozoobenthic deposit feeders processed large volumes of sediment. Recent studies showed that the selective and non-selective deposit feeder nematodes and macrozoobenthic species were the dominant species, both as diversity and abundance, of habitats directly impacted by the Danube’s inputs (Mureşan and Teacă, 2019; Teacă et al., 2020). Bioturbating organisms, such as Melinna sp., were reported to play a substantial role in the nitrogen cycling of sediments and exchanges between sediment and seawater (Laverock et al., 2011). This could be due to their burrow ventilation and particle reworking activities which lead to the redistribution of organic material and increase of sediment–water interface (Kristensen et al., 2012). Recent data indicated that the aerobic bacterial community of M. palmata habitats was more diverse as compared to that of the undisturbed sediments (Massé et al., 2019). Moreover, the mucus secreted by invertebrates appeared to stimulate the growth of bacterial and archaeal ammonia oxidizers, fostering a more abundant and distinct microbial community (Dale et al., 2019).
In M. palmata tissues, the orders Synechococcales, Rhodobacterales, Actinomarinales, Rhizobiales, Planctomycetales, Pirellulales, and Clostridiales showed a higher relative abundance, while sediments were dominated by Microtrichales, Anaerolineales, Caldilineales and Campylobacterales species. Synechococcales (Cyanobacteria) species that inhabit various ecological habitats are able to perform nitrogen fixation (Foster and O’Mullan, 2008) and create symbiotic relationships with other benthic invertebrates (Kumar and Kumar, 2020) such as sponges (Erwin and Thacker, 2008), ascidians (Tianero et al., 2015), and mollusks (Zhukova et al., 2022). In the current study, high content of Cyanobacteria taxa was found in shallow water sediments strongly influenced by the Danube inputs, at low dissolved oxygen levels. As photoautotrophic primary producers, they contribute to sediment enrichment with organic matter, while reducing the levels of atmospheric and hydrospheric carbon dioxide and bicarbonate (Golubic et al., 2000). In addition, Siynechococcus species were found associated with sipunculan worms, which might be an important food source for these species (Li et al., 2023; Liu et al., 2023). Therefore, the prevalence of these microorganisms in the M. palmata tissues might be also linked to the dietary requirements of this invertebrate. As deposit-feeding organisms, they acquire substantial amounts of sediment, potentially incorporating organic debris and microbes along with it (Lopez and Levinton, 1987). Furthermore, frequent algal blooms in the Danube influence area could affect the sea water microbiome during summer (Raport privind starea mediului in Romania in anul 2019, 2020). Due to this phenomenon, the occurrence of phylum Planctomycetota, represented in tissues by phylotypes belonging to orders Planctomycetales and Pirellulales, was also expected. These bacteria able to use polysaccharides as carbon and energy sources, were also detected in other marine organisms such as sponges (Kallscheuer et al., 2020), fish (Kormas et al., 2022), and other polychaete (Liu et al., 2023).
The presence of Rhodobacterales and Rhizobiales taxa belonging to phylum Proteobacteria could be associated with their involvement in the carbon cycling, being able to assimilate glyocolate, one of the most abundant organic carbon sources in the ocean (Von Borzyskowski et al., 2019). Rhodobacterales representatives were identified in different invertebrate species such as Capitella sp. and its surrounding organic-enriched sediments (Kunihiro et al., 2011). In the case of M. palmata, the prevalence of this group could be associated with its specific habitat represented by rich organic sediments (Bucşe et al., 2020; Teacă et al., 2020), suggesting the possible involvement in decomposing and incorporating the organic matter within the organically enriched sediments. Rhodobacterales were positively correlated with some of the most abundant organic pollutants from sediments such as polycyclic aromatic hydrocarbons (PAHs) (Rodríguez et al., 2021). These compounds were detected in high concentrations in benthic fish populating the Romanian Black Sea coast (Damir et al., 2022) that feed also with Melinna, which could indicate the functional role of Rhodobacterales within tissues. Moreover, Anaerolineales order (Chloroflexi) that was higher represented in sediments has been reported as highly abundant in polluted sediments (Rodríguez et al., 2021). Therefore, these bacteria may be used as bioindicators of high levels of particular pollutants. Moreover, some invertebrates (i.e. the marine polychaete Meganerilla bactericol) were reported to obtain bioavailable nitrogen from symbioses with diazotrophs such as Rhizobiales (Summers et al., 2013) by temporary or permanent intracellular interactions with symbionts passed between generations (Fiore et al., 2010). Also, the higher presence of Clostridiales in Melinna tissues was in accordance with the previously reported dominance of Firmicutes taxa in the gut microbiome of several marine invertebrates (Priscilla et al., 2022).
A high representation of Actionomarinales in Melinna tissue could be due to the adaptation of some of Actinobacteria taxa to colonize marine organisms such as sponges, molluscs, and polychaete tubes, constituting important sources for natural products with pharmaceutical applications (Konig et al., 2006; Fuirst et al., 2021). Meanwhile, the order Microtrichales of Actinobacteria showed a higher relative abundance within sediments. A recent study (Miksch et al., 2021) revealed that members of these heterotroph bacteria, along with Actinomarinales, inhabit muddy bottoms, suggesting their putative involvement in the carbon mineralization processes. The occurrence of Campylobacterales might be related to the low dissolved oxygen concentrations of the study area (Table 2), which is in line with other reports of their presence in marine sediments, usually in habitats characterized by low oxygen/sulfide ratios (Wirsen et al., 2002).
Based on the 16S rRNA gene sequence data obtained, the TAX4FUN tool was used for predicting the functional profiles of bacterial communities associated with the polychaete and sediments, while many questions are raised in the ecology of this ecosystem requiring information on the community function in addition to their taxonomic composition. However, the accuracy of this approach depends on the available genomic information from public databases, which, in many cases, do not represent the microorganisms of the investigated ecosystem (Wemheuer et al., 2020). In addition, the approach used in our survey cannot differentiate the DNA sequences of live and dead organisms, which may impact the accurate inference of the metabolic profile of the analyzed microbial communities, with consequential ecological implications (Wang et al., 2023).
Although the predicted functional profile did not reflect the taxonomic differences found between the communities associated with the polychaete and sediments, the deduced metabolic profile of the M. palmata bacterial community was in accordance with previous data demonstrating that several marine bacteria play a significant role in amino acid cycling and utilization (Mudryk et al., 2005; Wünsch et al., 2019). In this context, antimicrobial peptides were isolated from various marine invertebrates such as the worms Arenicola marina (Ovchinnikova et al., 2004) and Nereis diversicolor (Tasiemski et al., 2007), and from the oyster Magallana gigas (Zhang et al., 2018). The current investigation also evidenced the amino acid metabolism as the dominant function among bacteria colonizing both matrices. The dominant species identified in M. palmata tissue belonging to Rhodobacterales and Synechoccocales are known to be involved in the cofactors metabolism, being capable to synthesize thiamine (vitamin B1), riboflavin (vitamin B2) and cobalamin (vitamin B12) (Cooper et al., 2018; Mills et al., 2020). Moreover, representatives of Verrucomicrobiota, mainly present in Melinna tissues, could play an important role in nutrient cycling, with putative applicative potential in biotechnologies (Cardman et al., 2014), and represent good candidates for production of secondary metabolites and exopolysaccharides (EPS) (Feng et al., 2021).
The higher prevalence of genes involved in carbohydrate, amino acid, and lipid metabolism within tissues could suggest an enhanced capacity for nutrient processing, thereby contributing to the energy metabolism of the polychaete. This metabolic capacity likely plays a crucial role in supporting the organism’s growth, reproduction, and other physiological processes. Previous studies indicated that genes associated with terpenoid and polyketide metabolism are frequently linked to the production of secondary metabolites, which serve various ecological functions such as defense against predators or competition with other organisms (Osbourn, 2010). Also, microorganisms such as bacteria and fungi have distinct properties that allow them to metabolize xenobiotic substances either partially or entirely in various ecosystems (Miglani et al., 2022). Therefore, the presence of genes involved in xenobiotics biodegradation and metabolism suggests that the polychaete may have evolved a strategy for detoxification and tolerance to potentially harmful substances and expression of these genes could be a response to exposure to pollutants present in sediments.
The current data revealed that M. palmata harbors a distinct but significantly lower bacterial diversity as compared with its surrounding sediments, with no prominent differences between the communities along the depth gradient of their habitat. Phylum Proteobacteria dominated the tissue-colonizing bacteria, while Actinobacteriota prevailed in the sediment communities. This investigation showed that bacterial communities colonizing the invertebrate and its surrounding sediments are involved in various metabolic processes, from biosynthesis to degradation of different xenobiotic substances. Overall, this first characterization of the M. palmata microbiome can serve as a starting point for revealing the role of associated bacterial communities in the polychaete metabolism and their interchange dynamics within marine habitats.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA923146/.
The manuscript presents research on animals that do not require ethical approval for their study.
SM: Conceptualization, Formal Analysis, Investigation, Methodology, Writing – original draft. PL: Data curation, Formal Analysis, Software, Writing – original draft. TB: Funding acquisition, Investigation, Methodology, Resources, Writing – review & editing. MM: Investigation, Resources, Writing – original draft. AT: Investigation, Software, Writing – review & editing. CP: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was financially supported by the UEFISCDI ERANET-MARTERA-MOBILTOX-2-224/2021 grant and PNCDI III AMBI AQUA no. 23PFE grant, the Romanian Academy project RO1567-IBB05/2022, the Romanian Ministry of Research CORE Programme project PN 23300202; the European Union Horizon Europe project MARCO-BOLO, grant agreement no. 101082021, the European Union H2020 project DOORS, grant agreement no. 101000518, and the European Commission POIM project SMIS Code 123322, contract no. 253/18.06.201.
The authors thank Lavinia Iancu for technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1279849/full#supplementary-material
Abdelmohsen U. R., Bayer K., Hentschel U. (2014). Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 31, 381–399. doi: 10.1039/C3NP70111E
Băcescu M., Müller G., Gomoiu M. T. (1971). Cercetări de ecologie bentală în Marea Neagră. Analiza cantitativă calitativă şi comparată faunei bentale pontice. Ecologie marina. 4, 1–357.
Bonaglia S., Nascimento F. J. A., Bartoli M., Kiawonn I., Brüchert V. (2014). Meiofauna increases bacterial denitrification in marine sediments. Nat. Commun. 5, 5133. doi: 10.1038/ncomms6133
Bordenstein S. R., Theis K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS. Biol. 13, e1002226. doi: 10.1371/journal.pbio.1002226
Boucher D., Jardillier L., Debroas D. (2006). Succession of bacterial community composition over two consecutive years in two aquatic systems: a natural lake and a lake-reservoir. FEMS Microbiol. Ecology. 55 (1), 79–97. doi: 10.1111/j.1574-6941.2005.00011.x
Bryukhanov A. L., Korneeva V. A., Pimenov N. V. (2015). Detection of anaerobic sulfate reducing bacteria in oxygen containing upper water layers of the black and baltic seas. Moscow Univ. Biol.Sci. Bull. 70 (4), 184–188. doi: 10.3103/S0096392515040057
Bucşe A., Vasiliu D., Balan S., Parvulescu O. C., Dobre T. (2020). Heavy metal spatial distribution and pollution assessment in the surface sediments of the north – western black sea shelf. Rev. Chim. 71 (4), 155–170. doi: 10.37358/Rev.Chim.1949
Burkhardt W., Watkins W. D., Rippey S. R. (1992). Seasonal effects on accumulation of microbial indicator organisms by Mercenaria mercenaria. Appl. Environ. Microbiol. 58, 826–831. doi: 10.1128/aem.58.3.826-831.1992
Callahan B. J., Mcmurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cardman Z., Arnosti C., Durbin A., Ziervogel K., Cox C., Steen A. D., et al. (2014). Verrucomicrobia are candidates for polysaccharide-degrading bacterioplankton in an arctic fjord of Svalbard. Appl. Environ. Microbiol. 80 (12), 3749–3756. doi: 10.1128/AEM.00899-14
Conway N. M., Howes B. L., McDowell Capuzzo J. E., Turner R. D., Cavanaugh C. M. (1992). Characterization and site description of Solemya borealis (Bivalvia; Solemydae), antoher bivalve-bacteria symbiosis. Mar. Biol. 112, 601–613. doi: 10.1007/BF00346178
Coolen M. J. L., Shtereva G. (2009). Vertical distribution of metabolicaly active eukaryotes in the water column and sediments of the Black Sea. FEMS Microbiology Ecology 70(3):525–39. doi: 10.1111/j.1574-6941.2009.00756.x
Cooper M. B., Kazamia E., Helliwell K. E., Kudahl U. J., Sayer A., Wheeler G. L., et al. (2018). Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 13 (2), 334–345. doi: 10.1038/s41396-018-0274-y
Costas-Selas C., Martínez-García S., Logares R., Hernández-Ruiz M., Teira E. (2022). Role of bacterial community composition as a driver of the small-sized phytoplankton community structure in a productive coastal system. Microb. Ecol. 86 (2), 777–794. doi: 10.1007/s00248-022-02125-2
Dale H., Taylor J. D., Solan M., Lam P., Cunliffe M. (2019). Polychaete mucopolysaccharide alters sediment microbial diversity and stimulates ammonia-oxidising functional groups. FEMS Microbiol. Ecol. 95 (2), 1. doi: 10.1093/femsec/fiy234
Damir N. A., Coatu V., Pantea E. D., GalaŢchi M., Botez E., Birghilă S. (2022). Assessment of polycyclic aromatic hydrocarbons content in marine organisms of commercial interest from the Romanian black sea coast. Polycyclic Aromatic Compounds. 42 (10), 7595–7606. doi: 10.1080/10406638.2021.2006243
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Defer D., Desriac F., Henry J. (2013). Antimicrobial peptides in oyster hemolymph: the bacterial connection. Fish Shellfish Immunol. 34 (6), 1439–1447. doi: 10.1016/j.fsi.2013.03.357
De Mesel I., Derycke S., Moens T., van der Gucht K., Vincx M., Swings J. (2004). Top-down impact of bacterivorous nematodes on the bacterial community structure: a microcosm study. Environ. Microbiol. 6 (7), 733–744. doi: 10.1111/j.1462-2920.2004.00610.x
Desriac F., Le Chevalier P., Brillet B. (2014). Exploring the hologenome concept in marine bivalvia: haemolymph microbiota as a pertinent source of probiotics for aquaculture. FEMS Microbiol. Lett. 350, 107–116. doi: 10.1111/1574-6968.12308
Eisen J. A., Smith S. W., Cavanaugh C. M. (1992). Phylogenetic Relationships of Chemoautotrophic Bacterial Symbionts of Solemya velum Say (Mollusca: Bivalvia) determined by 16S rRNA Gene Sequence Analysis. J. Bacteriol. 174 (10), 3416–3421. doi: 10.1128/jb.174.10.3416-3421.1992
Erwin P. M., López-Legentil S., Schuhmann P. W. (2010). The pharmaceutical value of marine biodiversity for anti-cancer drug discovery. Ecol. Econmics 70 (2), 445–451. doi: 10.1016/j.ecolecon.2010.09.030
Erwin P. M., Thacker R. W. (2008). Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Mol. Ecol. 17 (12), 2937–2947. doi: 10.1111/j.1365-294X.2008.03808.x
Fauchald K., Jumars P. A. (1979). : The diet of worms: a study of polychaete feeding guilds. Oceanogr. Mar. Biol. Ann. Rev. 17, 193–284.
Felbeck H., Distel L. D. (2004). “Prokaryotic symbionts of marine invertebrates,” in The prokaryotes: An evolving electronic resource for the microbiological community. Ed. Dwrorkin M. (New York: Springer-Verlag).
Feng X., Gong Y., Ye M. Q., Du Z. J. (2021). Antibiotic Modulation of Capsular Exopolysaccharide in Pelagicoccus enzymogenes sp. nov. Isolated From Marine Sediment. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.655735
Fiore C. L., Jarett J. K., Olson N. D., Lesser M. P. (2010). Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18 (10), 455–463. doi: 10.1016/j.tim.2010.07.001
Foster R. A., O’Mullan G. D. (2008). “Nitrogen-fixing and nitrifying symbioses in the marine environment,” in Nitrogen in the Marine Environment. Eds. Capone D. G., Bronk D. A., Mulholland M. R., Carpenter E. J. (London: Academic Press Inc.), 1197–1218.
Fuirst M., Ward C. S., Schwaner C., Diana Z., Schultz T., Rittschof D. (2021). Compositional and functional microbiome variation between tubes of an intertidal polychaete and surrounding marine sediments. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.656506
Gilbertson W. W., Gilbertson Solan M., Prosser J. I. (2012). Differential effects of microorganism–invertebrate interactions on benthic nitrogen cycling. FEMS Microbiol. Ecol. 82, 11–22. doi: 10.1111/j.1574-6941.2012.01400.x
Glaubitz S., Labrenz M., Jost G., Jurgens K. (2010). Diversity of active chemolithoautotrophic prokaryotes in the sulfidic zone of a Black Sea pelagic redox cline as determined by rRNA-based stable isotope probing. FEMS Microbiol. Ecol. 74 (1), 32–41. doi: 10.1111/j.1574-6941.2010.00944.x
Goffredi S. K., Johnson S. B., Vrijenhoek R. C. (2007). Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl. Environ. Microbiol. 73 (7), 2314–2323. doi: 10.1128/AEM.01986-06
Golubic S., Seong-Joo L., Browne K. M. (2000). “Cyanobacteria: Architects of Sedimentary Structures,” in Microbial Sediments. Eds. Riding R. E., Awramik S. M. (Berlin: Springer-Verlag).
Gomoiu M. T. (1982). On the populations of Melinna palmata Grube at the Romanian littoral of the Black Sea. Cercetari Marine IRCM. 15, 115–131.
Goodfellow M., Williams S. T. (1983). Ecology of Actinomycetes. Annual Review of Microbiology. 37, 189–216. doi: 10.1146/annurev.mi.37.100183.001201
Graca A. P., Bondoso J., Gaspar H., Xavier J. R., Monteiro M. C., de la Cruz M., et al. (2013). Antimicrobial activity of heterotrophicbacterial communities from the Marine Sponge Erylus discophorus (Astrophorida, Geodiidae). PloS One 8 (11), e78992. doi: 10.1007/s00436-002-0729-x
Graczyk T. K., Conn D. B., Marcogliese D. J., Graczyk H., DeLafontaine Y. (2003). Accumulation of human waterborne parasites by zebra mussels (Dreissena polymorpha) and Asian freshwater clams (Corbicula fluminea). Parasitol Res. 89 (2), 107–112. doi: 10.1007/s00436-002-0729-x
Guo R., Ma X., Zhang J., Liu C., Thu C. A., Win T. N., et al. (2022a). Microbial community structures and important taxa across oxygen gradients in the Andaman Sea and eastern Bay of Bengal epipelagic waters. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1041521
Guo Y., Wu C., Sun J. (2022b). Pathogenic bacteria significantly increased under oxygen depletion in coastal waters: A continuous observation in the central Bohai Sea. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1035904.’rft
Hochstein R., Zhang Q., Sadowsky M. J., Forbes V. E. (2019). The deposit feeder Capitella teleta has a unique and relatively complex microbiomes likely supporting its ability to degrade pollutants. Sci. Total Environ. 670, 547–554. doi: 10.1016/j.scitotenv.2019.03.255
Hunt O. (1925). The food of the bottom fauna of the plymouth fishing grounds. J. Mar. Biol. Assoc. United Kingdom. 13 (3), 560–599. doi: 10.1017/S0025315400008079
Iancu L., Carter D. O., Junkins E. N., Purcarea C. (2015). Using bacterial and necrophagous insect dynamics for postmortem interval estimation during cold season: Novel case study in Romania. Forensic Sci. Int. 254, 106–117. doi: 10.1016/j.forsciint.2015.07.024
ICPDR (2004). The Danube River Basin District. River basin characteristics, impact of human activities and economic analysis required under Article 5, Annex II and Annex III, and inventory of protected areas required under Article 6, Annex IV of the EU Water Framework Directive, (2000/60/EC) Part A – Basin-wide overview Short: “Danube Basin Analysis (WFD Roof Report 2004). (International Commission for the Protection of the Danube River. Vienna International Centre D0412.)
Jørgensen B. B., Fossing H., Wirsen C. O., Jannasch H. W. (1991). Sulfide oxidation in the anoxic Black Sea chemocline. International Commission for the Protection of the Danube River. Vienna International Centre D0412. Deep-Sea Res. 38, S1083–S1103. doi: 10.1016/S0198-0149(10)80025-1
Kallscheuer N., Wiegand S., Heuer A., Rensink S., Boersma A. S., Jogler M., et al. (2020). Blastopirellula retiformator sp. nov. isolated from the shallow-sea hydrothermal vent system close to Panarea Island. Antonie Van Leeuwenhoek. 113 (12), 1811–1822. doi: 10.1007/s10482-019-01377-2
Kelman D., Kashman Y., Hill R., Rosenberg E., Loya Y. (2009). Chemical warfare in the sea: the search for antibiotics from red sea corals and sponges. Pure. Appl. Chem. 81 (6), 1113–1121. doi: 10.1351/PAC-CON-08-10-07
Ketchum R. N., Smith E. G., Vaughan G. O., Phippen B. L., McParland D., Al-Mansoori N., et al. (2018). DNA extraction method plays a significant role when defining bacterial community composition in the marine invertebrate echinometra mathaei. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00255
Konhauser K. O., Hao W., Li U., Gunten K., Bishop B. A., Alessi D. S., et al. (2020). Diopatra cuprea worm burrow parchment: a cautionary tale of infaunal surface reactivity. Lethaia. 53, 47–61. doi: 10.1111/let.12335
Konig G. M., Kehraus S., Seibert S. F., Abdel-Lateff A., Muller D. (2006). Natural products from marine organisms and their associated microbes. ChembioChem. 7 (2), 229–238. doi: 10.1002/cbic.200500087
Kormas K., Nikouli E., Kousteni V., Damalas D. (2022). Midgut bacterial microbiota of 12 fish species from a marine protected area in the aegean sea (Greece). Microb. Ecol. 86, 1405–1415. doi: 10.1007/s00248-022-02154-x
Kristensen E., Penha-Lopes G., Delefosse M., Valdemarsen T., Quintana C. O., Banta G. T. (2012). What is bioturbation? The need for a precise definition for fauna in aquatic sciences. Mar. Ecol. - Prog. Series. 446, 285–302. doi: 10.3354/meps09506
Kumar A., Kumar P. A. (2020). “Database resources for cyanobacterial research,” in Advances in Cyanobacterial Biology. Eds. Kumar P. S., Kumar A., Kumar V. S., Kumar A. S. (London: Academic Press), 47–54. doi: 10.1016/B978-0-12-819311-2.00004-8
Kunihiro T., Takasu H., Miyazaki T., Uramoto Y., Kinoshita K., Yodnarasri S., et al. (2011). Increase in Alphaproteobacteria in association with a polychaete, Capitella sp., in the organically enriched sediment. ISME J. 5 (11), 1818–1831. doi: 10.1038/ismej.2011.57
Kuo J., Yang Y.-T., Lu M.-C., Wong T.-Y., Sung P.-J., Huang Y.-S. (2019). Antimicrobial activity and diversity of bacteria associated with Taiwanese marine sponge Theonella swinhoei. Ann. Microbiol. 69, 253–265. doi: 10.1007/s13213-018-1414-3
Lauber C., Hamady M., Knight R., Fierer N. (2009). Pyrosequencing based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. ApplEnvironMicrobiol. 75 (15), 5111–5120. doi: 10.1128/AEM.00335-09
Laverock B., Gilbert J. A., Tait K., Osborn A. M., Widdicombe S. (2011). Bioturbation: Impact on the marine nitrogen cycle. Biochem. Soc. Trans. 39 (1), 315–320. doi: 10.1042/BST0390315
Leite L., Jude-Lemeilleur F., Raymond N., Henriques I., Garabetian F., Alves A. (2017). Phylogenetic diversity and functional characterization of the Manila clam microbiota: a culture-based approach. Environ. Sci. pollut. Res. 24, 21721–21732. doi: 10.1007/s11356-017-9838-z
Leloup J., Loy A., Knab N. J., Borowski C., Wagner M., Jørgensen B. B. (2007). Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 9 (1), 131–142. doi: 10.1111/j.1462-2920.2006.01122.x
Li J., Chen S., Wu P., Zhu C., Hu R., Li T., et al. (2023). Insights into the relationship between intestinal microbiota of the aquaculture worm Sipunculus nudus and surrounding sediments. Fishes. 8 (1), 32. doi: 10.3390/fishes8010032
Lin H., Eggesbo M., Peddada S. D. (2022). Linear and nonlinear correlation estimators unveil undescribed taxa interactions in microbiome data. Nat. Commun. 13 (1), 4946. doi: 10.1038/s41467-022-32243-x
Liu C., Liu C., Gao F., Wang A., Wang H., Yang Y., et al. (2023). Composition of Particulate Matter and Bacterial Community in Gut Contents and Surrounding Sediments of Three Sipunculan Species (Siphonosoma australe, Phascolosoma arcuatum, and Sipunculus nudus). Int. J. Mol. Sci. 24 (6), 6001. doi: 10.3390/ijms24066001
Lo Giudice A., Rizzo C. (2022). Bacteria associated with benthic invertebrates from extreme marine environments: promising but underexplored sources of biotechnologically relevant molecules. Mar. Drugs 20 (10), 617. doi: 10.3390/md20100617
Lopez G. R., Levinton J. S. (1987). Ecology of deposit-feeding animals in marine sediments. Q. Rev. Biol. 62 (3), 235–260. doi: 10.1086/415511
Lu Y., Zhou G., Ewald J., Pang Z., Shiri T., Xia J. (2023). MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 51 (W1), W310–W318. doi: 10.1093/nar/gkad407
Mare M. F. (1942). A study of a marine benthic community with special reference to the micro-organisms. J. Mar. Biol. Assoc. UK. 25 (3), 517–554. doi: 10.1017/S0025315400055132
Marino A., Lombardo L., Fiorentino C., Orlandella B., Monticelli L., Nostro A., et al. (2005). Uptake of Escherichia coli, Vibrio cholerae non-O1 and Enterococcus durans by, and depuration of mussels (Mytilus galloprovincialis). Int. J. Food Microbiol. 99 (3), 281–286. doi: 10.1016/j.ijfoodmicro.2004.09.003
Markande A. R., Mikaelyan A., Bhusan Nayak B., Patel K. D., Vachharajani N. B., Vennila A., et al. (2014). Analysis of midgut bacterial community structure of neanthes chilkaensis from polluted mudflats of Gorai, Mumbai, India. Adv. Microbiol. 4, 906–918. doi: 10.4236/aim.2014.413101
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martiny J. B. H., Bohannan B. J. M., Brown J. H., Colwell R. K., Fuhrman J. A., Green J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4 (2), 102–112. doi: 10.1038/nrmicro1341
Massé C., Garabetian F., Deflandre B., Maire O., Costes L., Mes-mer-Dudons N., et al. (2019). Feeding ethology and surface sediment reworking bythe ampharetid polychaete Melinna palmata Grube 1870: Effects onsediment characteristics and aerobic bacterial community composition. J. Exp. Mar. Biol. Ecology. 512, 63–77. doi: 10.1016/j.jembe.2018.12.009
Mcfall-Ngai M., Hadfield M. G., Bosch T. C., Carey H. V., Domazet-Lošo T., Douglas A. E. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Nat. Acad. Sci. U.S.A. 110 (9), 3229–3236. doi: 10.1073/pnas.1218525110
McMurdie P. J., Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Miglani R., Parveen N., Kumar A., Ansari M. A., Khanna S., Rawat G., et al. (2022). Degradation of xenobiotic pollutants: an environmentally sustainable approach. Metabolites. 12 (9), 818. doi: 10.3390/metabo12090818
Mikhailov V. N., Mikhailova M. V. (2008). “River mouths,” in The Black Sea Environment: The Handbook of Environmental Chemistry, vol. 5 . Eds. Kostianoy A. G., Kosarev A. N. (Berlin: Springer), 91–133.
Miksch S., Meiners M., Meyerdierks A., Probandt D., Wegener G., Titschack J., et al. (2021). Bacterial communities in temperate and polar coastal sands are seasonally stable. ISME Commun. 1, 29. doi: 10.1038/s43705-021-00028-w
Mills L. A., McCormick A. J., Lea-Smith D. J. (2020). Current knowledge and recent advances in understanding metabolism of the model cyanobacterium Synechocystis sp. PCC 6803. Biosci. Rep. 40 (4), BSR20193325. doi: 10.1042/BSR20193325
Mishra P., Singh U., Pandey C. M., Mishra P., Pandey G. (2019). Application of student's t-test, analysis of variance, and covariance. Ann. Card Anaesth. 22 (4), 407–411. doi: 10.4103/aca.ACA_94_19
Moens T., Santos G. A. P., Thompson F., Swings J., Fonsêca-Genevois V., Vincx M., et al. (2005). Do nematode mucus secretions affect bacterial growth? Aquat. Microb. Ecol. 40, 77–83. doi: 10.3354/ame040077
Mudryk Z. J., Podgórska B., Dwulit M. (2005). Bacterial utilization of amino acids and carbohydrates in a marine beach. Baltic Coast. Zone. 9, 29–41.
Mureşan M., Teacă A., Popa A., Begun T. (2019). Free-living marine nematodes community structural changes within a post-dredging site at the Romanian shelf. J. Environ. Prot. Ecology. 20 (2), 753–760.
Mureşan M., Teacă A. (2019). The free-living nematode community structure within the Romanian circalittoral habitats. Geo-Eco-Marina. 25, 7–13. doi: 10.5281/zenodo.3607208
Muriel-Millán L. F., Millán-López S., Pardo-López L. (2021). Biotechnological applications of marine bacteria in bioremediation of environments polluted with hydrocarbons and plastics. Appl. Microbiol. Biotechnol. 105 (19), 7171–7185. doi: 10.1007/s00253-021-11569-4
Musella M., Wathsala R., Tavella T., Rampelli S., Barone M., Palladino G., et al. (2020). Tissue-scale microbiota of the Mediterranean mussel (Mytilus galloprovincialis) and its relationship with the environment. Sci. Total Environment. 717, 137209. doi: 10.1016/j.scitotenv.2020.137209
Neave M. J., Streten-Joyce C., Glasby C. J., McGuinness K. A., Parry D. L., Gibb K. S. (2012). The bacterial community associated with the marine polychaete Ophelina sp. (Annelida: Opheliidae) is altered by copper and zinc contamination in sediments. Microb. Ecol. 63, 639–650. doi: 10.1007/s00248-011-9966-9
Onishchenko O. M., Kiprianova E. A. (2006). Shewanella genus bacteria isolated from the Black Sea water and molluscs. Mikrobiol Z. 68 (2), 12–21.
Osbourn A. (2010). Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet. 26 (10), 449–457. doi: 10.1016/j.tig.2010.07.001
Ovchinnikova T. V., Aleshina G. M., Balandin S. V., Krasnosdembskaya A. D., Markelov M. L., Frolova E. I., et al. (2004). Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett. 577 (1-2), 209–214. doi: 10.1016/j.febslet.2004.10.012
Panin N., Jipa D. (2002). Danube river Sediment Input and its Interaction with the North-western Black Sea. Estuar. Coast. Shelf Sci. 54 (3), 551–562. doi: 10.1006/ecss.2000.0664
Paul V. J., Ritson-Williams R. (2008). Marine chemical ecology. Nat. Prod. Rep. 25 (4), 662–695. doi: 10.1039/b702742g
Piel J. (2004). Metabolites from symbiotic bacteria. Nat. Prod. Rep. 21, 519–538. doi: 10.1039/B703499G
Plante C. J., Hill-Spanik K. M., Emerson R. (2022). Inputs don't equal outputs: bacterial microbiomes of the ingesta, gut, and feces of the keystone deposit feeder Ilyanassa obsolete. FEMS Microbiol. Ecol. 99 (1), fiac152. doi: 10.1093/femsec/fiad010
Pommier T., Neal P. R., Gasol J. M., Coll M., Acinas S. G., Pedrós-Alió C. (2010). Spatial patterns of bacterial richness and evenness in the NW Mediterranean Sea explored by pyrosequencing of the 16SrRNA. Aquat. Microb. Ecol. 61 (3), 221–233. doi: 10.3354/ame01484
Priscilla L., Rajeev M., Pandian S. K., Malathi E. (2022). Gut associated culturable bacterial community in intertidal polychaete worms (Annelida: Polychaeta), their characterization and implications in captive shrimp aquaculture. Regional Stud. Mar. Science. 52, 102274. doi: 10.1016/j.rsma.2022.102274
Raport privind starea mediului in Romania in anul 2019 (2020). AgenŢia NaŢională pentru ProtecŢia Mediului (Bucuresti: Ministerul Mediului).
R Core Team (2023). R: A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org.
Rizzo C., Michaud L., Hörmann B., Gerçe B., Syldatk C., Hausmann R., et al. (2013). Bacteria associated with sabellids (Polychaeta: Annelida) as a novel source of surface active compounds. Mar. pollut. Bulletin. 70 (1-2), 125–133. doi: 10.1016/j.marpolbul.2013.02.020
Rodríguez J., Gallampois C. M. J., Haglund P., Timonen S., Rowe O. (2021). Bacterial communities as indicators of environmental pollution by POPs in marine sediments. Environ. pollut. 268 (Pt A), 115690. doi: 10.1016/j.envpol.2020.115690
Rosenberg E., Koren O., Reshef L. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5 (5), 355–362. doi: 10.1038/nrmicro1635
Ross K. S., Haites N. E., Kelly K. F. (1990). Repeated freezing and thawing of peripheral blood and DNA in suspension: effects on DNA yield and integrity. J. Med. Genet. 27 (9), 569–570. doi: 10.1136/jmg.27.9.569
Ruginescu R., Lavin P., Iancu L., Menabit S., Purcarea C. (2022). Bioprospecting for novel bacterial sources of hydrolytic enzymes and antimicrobials in the Romanian littoral zone of the black sea. Microorganisms. 10, 2468. doi: 10.3390/microorganisms10122468
Schäfer H., Ferdelman T. G., Fossing H., Muyzer G. (2007). Microbial diversity in deep sediments of the Benguela Upwelling System. Aquat. Microb. Ecol. 50 (1), 1–9. doi: 10.3354/ame01164
Schippers A., Kock D., Höft C., Köweker G., Siegert M. (2012). Quantification of microbial communities in subsurface marine sediments of the Black Sea and off Namibia. Frontiers in Microbiology. 3. doi: 10.3389/fmicb.2012.00016
Schmidt E. W., Nelson J. T., Rasko D. A., Sudek S., Eisen J. A., Haygood, et al. (2005). Patellamide A and C biosynthesis by amicrocin-like pathway in Prochloron didemnid, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. .Sci. U.S.A. 102, 7315–7320. doi: 10.1073/pnas.0501424102
Schulz H. N., Brinkhoff T., Ferdelman T. G., Mariné M. H., Teske A., Jørgensen B. B. (1999). Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science. 284 (5413), 493–495. doi: 10.1126/science.284.5413.493
Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., et al. (2006). Microbial diversity in the deepsea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103 (32), 12115–12120. doi: 10.1073/pnas.0605127103
Sorokin Y. I., Sorokin P. Y., Avdeev V. A., Sorokin D. Y., Ilchenko S. V. (1995). Biomass, production and activity of bacteria in the Black Sea, with special reference to chemosynthesis and the sulfur cycle. Hydrobiologia. 308, 61–76. doi: 10.1007/BF00037788
Stevens H., Brinkhoff T., Rink B., Vollmers. J., Simon M. (2007). Diversity and abundance of Gram positive bacteria in a tidal flat ecosystem. Environ. Microbiol. 9 (7), 1810–1822. doi: 10.1111/j.1462-2920.2007.01302.x
Summers M. M., Katz S., Allen E. E., Rouse G. W. (2013). Association of rhizobia with a marine polychaete. Environ. Microbiol. Rep. 5 (4), 492–498. doi: 10.1111/1758-2229.12043
Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M. (2014). Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PloS One 9 (8), e105592. doi: 10.1371/journal.pone.0105592
Tasiemski A., Schikorski D., Le Marrec-Croq F., Pontoire-Van Camp C., Boidin-Wichlacz C., Sautière P. E. (2007). Hedistin: a novel antimicrobial peptide containing bromotryptophan constitutively expresses in NK cells-like of the marine annelid, Nereis diversicolor. Dev. Comp. Immunol. 31, 749–762. doi: 10.1016/j.dci.2006.11.003
Teacă A., Mureşan M., Begun T., Popa A., Ion G. (2019). Marine benthic habitats within a physical disturbed site from the Romanian Coast of the Black Sea. J. Environ. Prot. Ecology. 20 (2), 723–732.
Teacă A., Mureşan M., Menabit S., Bucse A., Begun T. (2020). Assessment of Romanian circalittoral soft bottom benthic habitats under Danube River influence. Regional Stud. Mar. Sci. 40, 101523. doi: 10.1016/j.rsma.2020.101523
Tescari S., Umgiesser G., Ferrarin C., Stanica A. (2006). Current circulation and sediment transport in the coastal zone in front of the Danube Delta. Geo-Eco-Mar. (Coast. Zones Deltas). 12, 5–16.
Thamdrup B., Rosselló-Mora R., Amann R. (2000). Microbial manganese and sulfate reduction in Black Sea shelf sediments. Appl. Environ. Microbiol. 66 (7), 2888–2897. doi: 10.1128/AEM.66.7.2888-2897.2000
Theis K. R., Dheilly N. M., Klassen J. L., Brucker R. M., Baines J. F., Bosch T. C. G. (2016). Getting the hologenome concept right: An ecoevolutionary framework for hosts and their microbiomes. mSystems. 1 (2), e00028–e00016. doi: 10.1128/mSystems.00028-16
Thomas T. R. A., Kavlekar D. P., Loka Bharathi P. A. (2010). Marine drugs from sponge-microbe association—a review. Mar. Drugs 8 (4), 1417–1468. doi: 10.3390/md8041417
Tianero M. D. B., Kwan J. C., Wyche T. P., Presson A. P., Koch M., Barrows L. R., et al. (2015). Species specificity of symbiosis and secondary metabolism in ascidians. ISME J. 9 (3), 615–628. doi: 10.1038/ismej.2014.152
Todorova V., Konsulova T. (2005). Manual for Quantitative Sampling and Sample Treatment of Marine Soft-Bottom Macrozoobenthos (PO Box 152, 9000 Varna, Bulgaria: Institute of Oceanology, Bulgarian Academy of Sciences). Available at: www.blacksea-commission.org. 38 pp.
Von Borzyskowski L. S., Severi F., Kruger K., Hermann L., Gilardet A., Sippel F., et al. (2019). Marine proteobacteria metabolize glycolate via the B-hydrozyaspartate cycle. Nature 575, 500–504. doi: 10.1038/s41586-019-1748-4
Wang Y., Thompson K. N., Yan Y., Short M. I., Zhang Y., Franzosa E. A., et al. (2023). RNA-based amplicon sequencing is ineffective in measuring metabolic activity in environmental microbial communities. Microbiome. 11 (1), 1–15. doi: 10.1186/s40168-022-01449-y
Webster N. S., Taylor M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14 (2), 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Wemheuer F., Taylor J. A., Daniel R., Johnston E., Meinicke P., Thomas T., et al. (2020). Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome. 15 (1), 1–12. doi: 10.1186/s40793-020-00358-7
Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis (New York, USA: Springer-Verlag), 260.
Wickham H., Averick M., Bryan J., Chang W., McGowan L. D., François R., et al. (2019). Welcome to the tidyverse. J. Open Source Software 4 (43), 1686. doi: 10.21105/joss.01686
Winand R., Bogaerts B., Hoffman S., Lefevre L., Delvoye M., Braekel J. V., et al. (2019). Targeting the second (Illumina) and third (Oxford Nanopore Technologies) Generation Sequencing Technologies. Int. J. Mol. Sci. 21 (1), 298. doi: 10.3390/ijms21010298
Wirsen C. O., Sievert S. M., Cavanaugh C. M., Molyneaux S. J., Ahmad A., Taylor L. T., et al. (2002). Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68 (1), 316–325. doi: 10.1128/AEM.68.1.316-325.2002
Wünsch D., Trautwein K., Scheve S., Hinrichs C., Feenders C., Blasius B., et al. (2019). Amino acid and sugar catabolism in the marine bacterium phaeobacter inhibens DSM 17395 from an energetic viewpoint. Appl. Environ. Microbiol. 27 (85(24)), e02095–e02019. doi: 10.1128/AEM.02095-19
Zhang Y., Cui P., Wang Y., Zhang S. (2018). Identification and bioactivity analysis of a newly identified defensin from the oyster Magallana gigas. Dev. Comp. Immunol. 85, 177–187. doi: 10.1016/j.dci.2018.04.014
Zhang X., Wei W., Tan R. (2015). Symbionts, a promising source of bioactive natural products. Sci. China Chem. 58, 1097–1109. doi: 10.1007/s11426-015-5398-6
Zhukova N. V., Eliseikina M. G., Balakirev E. S., Ayala F. J. (2022). Multiple bacterial partners in symbiosis with the nudibranch mollusk Rostanga alisae. Sci. Rep. 12, 169. doi: 10.1038/s41598-021-03973-7
Keywords: Melinna palmata microbiome, invertebrates bacteria, Black Sea sediments, marine polychaete, illumina sequencing, 16S rRNA gene
Citation: Menabit S, Lavin P, Begun T, Mureşan M, Teacă A and Purcarea C (2024) First screening of bacteria assemblages associated with the marine polychaete Melinna palmata Grube, 1870 and adjacent sediments. Front. Mar. Sci. 10:1279849. doi: 10.3389/fmars.2023.1279849
Received: 21 August 2023; Accepted: 18 December 2023;
Published: 08 January 2024.
Edited by:
Zhiyong Li, Shanghai Jiao Tong University, ChinaReviewed by:
Xiaoshou Liu, Ocean University of China, ChinaCopyright © 2024 Menabit, Lavin, Begun, Mureşan, Teacă and Purcarea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Purcarea, Y3Jpc3RpbmEucHVyY2FyZWFAaWJpb2wucm8=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.