
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 30 November 2023
Sec. Microbial Symbioses
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1278837
This article is part of the Research Topic Degradation, Conservation and Ecological Restoration of Seagrass Beds under Intensifying Global Changes View all 8 articles
 Anthony S. Randell1
Anthony S. Randell1 Jason E. Tanner2
Jason E. Tanner2 Melissa L. Wos-Oxley3
Melissa L. Wos-Oxley3 Sarah R. Catalano2,4,5
Sarah R. Catalano2,4,5 Gunnar Keppel1,6
Gunnar Keppel1,6 Andrew P. A. Oxley7*
Andrew P. A. Oxley7*Introduction: Seagrass roots harbour diverse assemblages of microorganisms that likely benefit the growth and survival of meadows. Yet, restoration efforts rarely consider their effect on developing seagrass seedlings. Sediment origin should determine the types of rhizosphere and root-colonising (rhizoplane) microorganisms and thus the performance of seedlings during restoration, particularly for slow growing climax species like Posidonia. Recent Posidonia restoration attempts in South Australia used commercially sourced 'play pit sand' for seedling propagation and planting, but have been impacted by high mortality. More natural substrates like seagrass meadow sediment have not been previously investigated for suitability over conventional substrates with regard to seedling growth and survival.
Methods: To assess the relevance of seagrass associated microorganisms in the growth of Posidonia angustifolia seedlings, we investigated the bacterial communities from tank-raised seedlings propagated in autoclave treated and untreated 'play pit sand' and meadow mix (comprising a 1:3 ratio of natural meadow sediment and beach sand) over a 12-week period. Autoclave treatment was adopted in order to diminish the bacterial load prior to planting and thus inform their contribution (if any) on early seedling growth. Samples for bacterial community analysis and seedling growth measurements (numbers and total length of roots/leaves, root diameter, seedling weight, starch reserves) were taken at 4 and 12 weeks. Bacterial assemblages were surveyed from DNA extracts from bulk and rhizosphere sediments and root tissues, as well as from swabs of P. angustifolia fruit, established meadow sediment and water samples prior to seedling propagation, by constructing Illumina 16S rRNA gene libraries.
Results: While most growth measurements did not vary significantly between sediment type or treatment, proportional growth of roots versus leaves (as expressed as a pseudo root:shoot ratio) was significantly related to treatment, sediment type and seed length. Seedlings from meadow mix invested more in leaves, regardless of treatment, when compared to play sand. Autoclave treatment increased investment in roots for play sand but increased the investment in leaves for meadow mix. Bacterial communities differed significantly between sediments and between sample types (bulk, rhizosphere and roots), with the roots from meadow mix seedlings containing an increased abundance of various potentially beneficial bacterial taxa.
Discussion: While such changes appear to affect the early development of seedlings, bacterial community dynamics are also likely coupled to changes in nutrient availability. Further research is thus required to disentangle host seedling growth-nutrient-bacterial community dynamics with the view to identifying microbes that may support the growth and vigour of seedlings under different nutrient conditions as part of future restoration efforts.
Seagrass meadows are one of the most productive systems on earth (Nordlund et al., 2016), and are fundamental to the broader dynamics of marine ecosystems. They provide important nursery grounds for a multitude of fish species (Blandon and zu Ermgassen, 2014), are highly efficient carbon sequesters (Fourqurean et al., 2012; Duarte and Krause-Jensen, 2017), and protect coastlines from storm surges and erosion (Paul, 2018). Seagrass meadows also harbour diverse, complex assemblages of sedimentary microorganisms (Jankowska et al., 2015; Sun et al., 2015; Cúcio et al., 2016) that, like in terrestrial soils, underpin the health and critical services of these ecosystems by supporting plant growth and vigour. Central to this supporting function are microbes that occur in close proximity to seagrass roots (rhizosphere), or are more intimately associated with root surfaces (rhizoplane) or underlying tissues (endosphere), where they form important components of the 'seagrass holobiont' (Ugarelli et al., 2017). Bacteria are thought to be dominant constituents of the seagrass microbiome where, among other roles, they act to fix nitrogen, solubilise essential nutrients like phosphorus (Jose et al., 2014), and detoxify the surrounding sediment by oxidising sulfide (Garcias-Bonet et al., 2012; Sun et al., 2015; Martin et al., 2019). In certain seagrasses, like species of Posidonia that form extensive meadows and have a long evolutionary history with coastal waters (Aires et al., 2011), root-associated bacteria have been shown to be highly diverse and more efficient at nitrogen fixation (by as much as 10x) than bacterial communities associated with the leaves (phyllosphere) (Garcias-Bonet et al., 2012; Lehnen et al., 2016). The seagrass holobiont may include endosymbionts thought to improve the capacity of seagrasses to thrive in nutrient poor environments (Mohr et al., 2021). Furthermore, regressing P. oceanica seagrass meadows have been linked to declining bacterial communities in sediments (García-Martínez et al., 2009), suggesting an important role for microorganisms in seagrass growth and survival.
Seagrass-bacterial community dynamics are thought to be shaped by a range of different plant-host and environmental factors. Alongside changes in the surrounding environmental conditions (e.g. light availability, meadow health, geographical location) (García-Martínez et al., 2009; Bourque et al., 2015; Martin et al., 2017), this may include the emission of oxygen and exudates from the root tips which create microhabitats for a plethora of both aerobic and anaerobic bacteria (Jensen et al., 2007; Kilminster and Garland, 2009; Martin et al., 2019). In terrestrial plants, root exudates produced throughout the plant life cycle have been suggested to influence the occurrence of different subsets of microbes with specific functions (Chaparro et al., 2014), leading to the formation of juvenile and adult plant-specific assemblages (Edwards et al., 2018). Modulation of these communities through the inoculation of roots with bacteria that possess health- and growth-promoting traits (Hayat et al., 2010), has been reported to increase seedling growth, survival and nutrient availability with potential benefits for agriculture and revegetation projects (Reed et al., 2005; Thrall et al., 2005; Wang et al., 2017). Though an intriguing prospect yet to be realised for seagrasses, stimulation of sediment bacterial activity through organic matter enrichment and remineralisation has been reported to increase belowground biomass of P. australis seedlings, enhancing their root branching and stability in sediments (Fraser et al., 2015). Furthermore, the inoculation of experimental mesocosms with meadow sediment (presumably containing beneficial microbes) has been anecdotally noted to increase stem density compared to controls in Eelgrass (Zostera marina) seedlings (Ort et al., 2014). Observations such as this imply that improving our understanding of the seagrass microbiome and its role in seedling establishment may provide useful insights into their role in supporting restoration attempts.
Indeed, with the mounting loss of seagrass meadows occurring throughout the world due to anthropogenic disturbance (Waycott et al., 2009), there has been a significant investment in restoration efforts in recent years. For this, seeds, seedlings or cuttings are either placed directly into the sediment (Van Katwijk et al., 2016; Valdez et al., 2020) or, in turbulent areas, anchored to the sediment using various means, e.g. sediment filled hessian sacks (Irving et al., 2010; Zhang et al., 2015; Unsworth et al., 2019). Thus far, restoration attempts have had varied success, with notable challenges arising from, among others, propagule supply and survival (Tan et al., 2020; Vanderklift et al., 2020; Boudouresque et al., 2021). For environmentally and economically important seagrass meadow systems in Australia, those occurring off the Adelaide metropolitan coast in South Australia, (which have seen losses of ~6,200 ha since 1949) have been the focus of recent efforts (Tanner et al., 2014). Compared to most local seagrasses, the genus Posidonia has suffered a greater reduction in its Adelaide distribution, with only Amphibolis displaying greater susceptibility (Bryars et al., 2006a). This is concerning because Posidonia meadows are climax communities that can take many decades to recover naturally and require an absence of ocean swell for seedlings to establish (Bryars and Neverauskas, 2004). Currently, for restoration attempts employed in this region, fruit are collected and stored in onshore tanks until the seedling (germinated seed) is released. The seedlings are then transplanted offshore into sand-filled hessian sacks by divers (Tanner, 2015). While initial findings have been promising, there is high plant mortality (~86%) within 3 to 4 years (Tanner and Theil, 2016). Improving our understanding of the early development of Posidonia seedlings is critical for enhanced restoration outcomes. Recent efforts have primarily focused on investigating sediment composition and nutrient addition effects on growth and survival (Statton et al., 2013; Statton et al., 2014; Fraser et al., 2015; Tanner and Theil, 2016; Tanner and Theil, 2019; Tanner, 2023). Further advances, however, may be gained by improving our understanding of the microbiome and the identification of microbes that may support early vigour. However, such knowledge is currently limited for species like Posidonia in Australia (Ugarelli et al., 2017; York et al., 2017).
Here, we examine the rhizosphere and root-associated bacterial community assemblages of Posidonia angustifolia seedlings, a dominant and ecologically significant meadow species endemic to Australian temperate waters that typically inhabits sandy sediments, and which has suffered widespread loss (Cambridge and Kuo, 1979; Carruthers et al., 2007; Tanner et al., 2014). Specifically, we sought to compare the bacterial community diversity of seedlings grown in a commercial 'play pit sand' currently used in restoration efforts, with a mixture that included locally sourced seagrass meadow sediment. Bacterial communities and potential effects on seedling growth (as assessed using an Illumina 16S rRNA gene deep-sequencing approach, and changes in root and leaf length metrics respectively) were studied through comparisons between autoclave treated and untreated sediments. This study is one of the first to investigate the potential influence of the microbiome on seagrass seedlings, where such knowledge could be used to identify potentially beneficial bacterial species that may support future restoration programs by improving seedling growth.
Meadow sediment required for seedling propagation (~20 kg) was freshly collected prior to the experiment on the 23rd of December 2018 from the edge of the intertidal and subtidal zone of Lady Bay, Normanville, South Australia (35°28'10.07"S, 138°17'27.41"E). The meadow here is protected by a shore platform of limestone conglomerate and consists largely of P. angustifolia with scattered P. sinuosa and a patch of Amphibolis sp. (Figure 1A). The sediment was collected above the Posidonia rhizomes, sieved to remove molluscs (> 4 mm), and then submerged in a flow-through ~50 cm deep seawater tank until required. Any remaining molluscs that may predate on the seagrass seedlings were opportunistically removed during a two-week holding period prior to the commencement of the experiment. To characterise the bacterial assemblages associated with the native meadow sediment, bulk sediment samples (n = 3) were also collected for DNA extraction from just above the P. angustifolia rhizomes (to a depth of ~5 cm) at Lady Bay using sterile 6 ml syringes with the ends cut off. In addition, to identify the variation (if any) in the root-associated bacterial community composition between mature plants from the meadow and tank-raised seedlings grown in different substrates, root samples were also obtained from six adult P. angustifolia by carefully removing the plants from the sediment and trimming the tips of the primary and secondary roots (~1 cm) using scissors cleaned with 70% ethanol solution. Ten randomly selected root cuttings were rinsed thoroughly in 0.22 µm filter sterilised seawater to remove any sediment particles and placed into sterile centrifuge tubes. A seawater sample (1 L) was also collected from between the leaves of the P. angustifolia to control for the surrounding environmental bacterial communities. All samples were placed on ice for transport (<1 hr), and then the sediment and root samples were stored at -20°C and the water at 4°C until DNA extraction.

Figure 1 Study site and sampling method. A particularly low tide at (A) Lady Bay, South Australia, has exposed Posidonia seagrass in the background near the rocky platform, in the foreground a transition from Amphibolis to the Posidonia, where some samples were taken, is shown. The circulating sea water (B) tank with potted seedlings. (C) Seedling for growth measurements divided into leaves, seed and roots. (D) Microscope image of seed starch stained with iodine. Scale bar = 5 mm. (E) Example seedling sampled for bacterial community analysis, line indicates portion of a root used for rhizosphere and root sampling, rectangle shows approximate location for bulk sediment sample. (F) A colourised Environmental Scanning Electron Microscope (ESEM) image of microorganisms found adhering to the root (~0.2mm from the tip) of a seedling grown in untreated meadow mix.
In the absence of mature P. angustifolia fruits at Lady Bay, seedlings required for the experiment were obtained from beach-cast fruits collected from West Beach (34°57'2.01"S, 138°30'10.62"E) near Adelaide, South Australia on the 28th of December 2018, where large P. angustifolia meadows are dominant offshore (Cambridge and Kuo, 1979; Bryars et al., 2006b). To identify the role (if any) of fruit-associated bacteria in seedling root colonisation, 10 randomly collected fruits were rinsed thoroughly with 0.22 µm filter sterilised seawater and the entire surface swabbed using FLOQSwabs® (Copan Diagnostics, Murrieta, CA, United States). Swabs were placed into sterile centrifuge tubes on ice, and stored at -20°C until DNA extraction. Fruits collected for seedling propagation were placed in a semi-submersible mesh tray in the same seawater tank used to hold the meadow sediment to dehisce (open) before seedlings could be separated and planted.
The effects of two propagating substrates and their associated bacterial assemblages on P. angustifolia seedling growth were investigated over a 12-week period during summer (January-April 2019). One substrate was a commercial 'play pit sand' (PS) ('Play Sand' 20 kg bags, Richgro Garden Products, Jandakot, WA, Australia) of a coastal, yet terrestrial origin (Richgro pers. comms.) which was similar to sand currently used in local restoration programs in the region. The other substrate, a 'meadow mix' (MM), consisted of a 1:3 ratio of seagrass meadow sediment and beach sand obtained from a local beach (West Beach, SA, Australia). Beach sand was incorporated into this mix in order to avoid minimise the logistical difficulties and impacts on the collection of large quantities (~80 kg) of sediment from the meadow. To elucidate the contribution of sediment bacterial communities in early seedling growth, half of each of the propagating substrates were autoclaved at 121°C for 52 min to diminish the bacterial load prior to planting. Propagating substrates (representing four treatments: MM autoclave treated, MM untreated, PS autoclave treated and PS untreated) were evenly distributed into fine mesh-lined 575 ml super tube plant pots (DanBar Plastics, Horsham, VIC, Australia) as used elsewhere (Statton et al., 2013; Statton et al., 2014). Undamaged dehisced seedlings (n = 230), regardless of size, were randomly allocated and planted into the individual experimental pots, which were then randomly placed in trays within a flow-through ~50 cm deep seawater tank (Figure 1B), with unfiltered water sourced directly from the adjacent Gulf St Vincent (the same water body from which the seagrass fruits were obtained). A flow rate of ~10 L min-1, giving a turnover time of ~ 3 h, was maintained throughout the experiment. A single tank was used to avoid tank effects which can occur when samples are separated into multiple tanks (Statton et al., 2014). The tank was covered with 75% shade cloth. This shading mimics the light levels at ~8 m depth, which is within the depths P. angustifolia is known to occur naturally and reduces water temperature in the shallow tanks, helping to slow the rate of algal growth. The trays were moved a quarter clockwise around the tank weekly to reduce any further bias that may arise from variable light and water circulation. Seedlings were regularly cleaned carefully by hand to reduce epiphyte growth. At the time of planting, three untreated meadow mix samples were collected (pre-potting) for bacterial comparison with the native meadow sediment. A pre-experiment seawater sample (1 L) from the middle of the tank was also collected to control for the surrounding environmental bacterial communities. Sediment samples were stored at -20°C and the water at 4°C until DNA extraction.
Seedling growth measurements (total leaf and root length/count, lateral root count, root diameter, and seedling weight) were taken destructively in weeks 4 and 12. A total of 37 seedlings were evaluated for each substrate/treatment (17 in week 4, and 20 in week 12), except for the autoclaved PS, where n = 32 due to plant loss (17 in week 4, and 15 in week 12). Seed length was also measured as a covariate. For measurements, seedlings were removed in small batches (to reduce dehydration shrinkage), cleaned of loosely adherent sediment by rinsing in water, and gently dried with absorbent tissue. Seedlings were then weighed and separated into component parts (i.e. seed, leaves and roots), which were measured lengthwise (Figure 1C). The primary root diameter was determined ~1 cm from the base, where the root meets the stem, using callipers. To explore the treatment effects on above and belowground resource allocation, the ratio of total root length to total leaf length (herein termed the pseudo root:shoot ratio, PRSR) was estimated. Furthermore, at the end of the experiment, microscopic observations of surplus seedling roots (as directly removed from the pots without rinsing in water) were undertaken using a Nikon SMZ745T dissecting microscope to examine the impacts of treatment on gross morphology, with representative images captured using a Nikon DS-Fi2 digital camera (Supplementary Figure S1: Datasheet 2).
To investigate the usage of seed starch reserves by the seedlings, the seeds were halved longitudinally at the end of week 12 and one half was stained (1:5 ratio Lugol's iodine to water) to highlight the remaining endosperm, which was photographed using a Nikon SMZ745T dissecting microscope and Nikon DS-Fi2 digital camera (Figure 1D). ImageJ software (version 1.52i) (Schindelin et al., 2012) was used to measure total area (mm2) of the seed and remaining starch area (mm2), as indicated by a dark brown-black colouration.
Seedling growth was investigated using multiple regressions for: 1) total biomass (seedling weight); 2) resource allocation into above- and below-ground compartments (PRSR); and 3) degree of establishment (number of lateral roots). Each regression was implemented with the "lm" command in R, version 4.0.2 (R Core Team, 2020) and included substrate type (PS or MM), treatment (autoclaved or untreated), seed length (mm, log10 transformed), growth stage (week 4 or 12), and their interactions as explanatory variables; i.e., Response Variable ~ Sediment Type * Treatment * log(Seed Length) * Growth Stage. Growth stage was excluded as a variable for lateral roots, as lateral roots had not yet developed in week 4. Models including all variables and interactions were simplified by removing components that were not significant (p > 0.05). The assumptions of normally distributed and homogeneous residuals were verified by visual inspection of Q-Q plots and residuals plotted against fitted values for the final models (including only significant variables) (Supplementary Figures S2–S4: Datasheet 2).
To test whether parameters measuring plant growth differed between weeks 4 and 12, we used mixed linear models with week as the fixed effect and treatment and sediment as random effects, implemented using the lme4 (Bates et al., 2015) and lmerTest (Kuznetsova et al., 2017) packages. If the response variable consisted of count data, we implemented generalised mixed models with a Poisson error distribution using the 'glmer' command.
Autoclave treatment of sediment has been previously reported to influence nutrient levels and grain size (Lotrario et al., 1995; Otte et al., 2018). Therefore, at the end of the seedling growth experiment, five replicates from each substrate treatment group were analysed for calcium carbonate, organic carbon, nitrogen and phosphorus. For this, sediment was collected from the centre of the pot (excluding the top and bottom 2 cm) after removal of seedling, to best represent nutrients around the roots. The nitrogen, total carbon and organic carbon were assessed using Dumas high temperature combustion, rapid titration was used for inorganic carbon and equivalent calcium carbonate, and reverse aqua regia microwave assisted digestion for phosphorus (Rayment and Lyons, 2011). Tests were conducted by the Australian Precision Ag Laboratory (APAL), Hindmarsh, South Australia. Individual nutrient types (calcium carbonate, organic carbon, nitrogen and phosphorus) were compared between sediment and treatment by two-way analysis of variance (ANOVA) tests.
Particle size was determined using a Malvern Mastersizer 3000 (Malvern Panalytical Ltd., Malvern, UK) with water as the dispersant at the University of South Australia, Mawson Lakes, South Australia. Three samples from each sediment treatment were used to confirm particle size. The output was classified using the Wentworth (1922) scale in order to describe sediment characteristics. Grain particle size was compared with a permutational multivariate analysis of variance (PERMANOVA) with 9999 permutations under a reduced model, allowing for type III (partial) sums of squares and a fixed effects sum to zero for mixed terms.
To assess changes in the microbiome (bacterial communities) of the seedlings, five seedlings from each substrate treatment group were randomly selected at each growth stage (in week 4 and 12; n = 10 in total per treatment group). Roots were sampled by carefully removing the plants from the sediment and trimming the root tip (~2 cm) using scissors cleaned with 70% ethanol (Figure 1E). For some seedlings where the roots extended through the bottom of the pots at week 12, the tips were removed and a portion of the roots still in contact with the sediment was collected. To distinguish the rhizosphere from the more intimately associated rhizoplane (or endosphere) constituents, the root tips (n=10) were first placed in individual Lysing Matrix E tubes (MP Biomedicals) containing 978 µl sodium phosphate buffer and manually agitated with sterilised forceps to collect the loosely adhering sediment (rhizosphere) for downstream DNA extraction. The washed root tips (comprising the rhizoplane/endosphere constituents) were then cut into smaller pieces using a sterile scalpel, and the fragments from each placed into individual Lysing Matrix E tubes (MP Biomedicals) containing 978 µl sodium phosphate buffer for DNA extraction. The occurrence of adherent (rhizoplane-associated) communities was verified by the direct visualisation of fresh roots using the FEI Quanta 450 FEG Environmental Scanning Electron Microscope (ESEM) through Adelaide Microscopy (University of Adelaide, South Australia) (Figure 1F; Supplementary Materials: Datasheet 2 and Methods).
To distinguish the root-associated (rhizosphere and rhizoplane/endosphere) communities from the surrounding sediment, bulk sediment was also collected from three seedlings for each treatment, which was taken ~1.5 cm away from, but horizontal to, the root tips (Figure 1E). Aliquots of ~250 mg of bulk sediment were placed into individual Lysing Matrix E tubes (MP Biomedicals) for downstream DNA extraction. In addition, a seawater sample (1 L) was collected from the middle of the tank in weeks 4 and 12 to assess the contribution of the environmental bacterial assemblages. Rhizosphere and seawater samples were stored at 4°C and the bulk sediment and root samples at -20°C prior to DNA extraction.
To analyse the global bacterial community structure, DNA was extracted from propagating substrates and root samples using the FastDNA™ Spin Kit for Soil (MP Biomedicals) according to the manufacturer's instructions. To ensure efficient lysis, a further bead-beating step was performed for all root samples using the same default Fast Prep-24™ 5G Instrument settings (MP Biomedicals). In addition, to evaluate the surrounding environmental bacterial assemblages, DNA was extracted from the seawater samples following filtration onto 0.22 µM Nalgene™ Rapid-Flow™ filters (ThermoFisher Scientific) using the same kit according to the manufacturer's instructions. Sample DNA was re-eluted in 100 μl of DES (MP Biomedicals), quantified using the NanoDrop™ 2000 spectrophotometer (ThermoFisher Scientific) and stored at -20°C prior to down-stream library preparation.
The V1-V2 hypervariable region of the 16S rRNA gene was amplified from DNA extracts using a multi-step PCR approach, with pre-enrichment using universal eubacterial primers 27F and 338R as described previously (Camarinha-Silva et al., (2014). More specifically, for Illumina library generation, ~25 ng of each DNA sample was first subjected to 20 cycles of PCR, whereby 1 µl of this reaction was used as the template in a second 15 cycle PCR containing individual sample 6 nt barcodes and Illumina specific adaptors. One microlitre of this reaction was subsequently used in a final 10 cycle PCR for incorporating the Illumina multiplexing sequencing and index primers. The resultant PCRs were visualised via agarose gel electrophoresis and products of the expected size (~438 bp) were purified using Agencourt AMPure XP beads (Beckman Coulter). Samples were quantified using the Quant-iT™ Picogreen® dsDNA kit (Life Technologies) following the manufacturer's instructions. All samples (n = 133) were then pooled in equimolar ratios and sequenced by the Australian Genome Research Facility (AGRF, North Melbourne, VIC, Australia) on the Illumina MiSeq platform using 250 nt paired-end sequencing chemistry. Amplicons generated from a single bacterial species (Lactobacillus reuteri) were also sequenced alongside the samples as controls.
Approximately 16 million raw sequence reads were obtained from a total of 133 samples comprising: 23 seagrass meadow 'environment' samples (1x seawater, 3x sediment, 9x fruit swabs, and 10x seagrass roots); and 110 experimental samples (3x seawater, 27x bulk sediment, 40x rhizosphere and 40x roots) (Supplementary Table S1). Reads were assembled using PEAR (version 0.9.5; Zhang et al., 2014), and the primers identified and removed. Trimmed sequences were processed using Quantitative Insights into Microbial Ecology (QIIME version 1.8; Caporaso et al., 2010), USEARCH (version 8.0.1623; Edgar, 2010), and UPARSE software (Edgar, 2013). Using USEARCH tools, sequences were quality filtered to remove low-quality reads, full-length duplicate sequences and singletons. Sequences were clustered into operational taxonomic units (OTUs) at a minimum identity of 97%, with putative chimeras removed using the RDP-gold database as a reference (Cole et al., 2014).
A total of 6,475,702 high quality, paired-end reads were clustered into 37,674 OTUs (mean = 48,689 ± 15,968 reads/sample; min = 20,261; max = 105,995). These OTUs were further filtered as conducted previously (Legrand et al., 2018) where only those that contributed to > 0.01% dataset were retained. The resultant OTUs were interrogated against the RDP and SILVA databases (Wang et al., 2007; Quast et al., 2013), whereby taxonomic lineages based on the SILVA taxonomy and the best hit from RDP were assigned for each OTU (Supplementary Datasheet 1). A further 55 chloroplast and mitochondrial OTUs were removed, leaving a total of 1,532 OTUs for downstream analysis. Rarefaction curves were used to assess (retrospectively) sampling depth (Supplementary Figure S5: Datasheet 2).
In order to explore for patterns across the global bacterial communities, a data matrix comprising the percent standardised abundances of 1,532 OTUs across all 133 samples was constructed, where samples were then ordinated using non-metric multidimensional scaling (nMDS) with 50 random restarts (Clarke and Warwick, 2001) and principal co-ordinates analysis (PCoA) with 2 axes using the Bray-Curtis algorithm (Bray and Curtis, 1957). Multivariate dispersion indices (MVDISP) were calculated in Primer-E to gauge the degree of variation among replicate samples within sample groups (i.e. MM and PS bulk sediment, rhizosphere and roots). Groups of samples were evaluated for significant differences using both one-way and two-way permutational multivariate analysis of variance (PERMANOVA) with 9999 permutations, allowing for type III (partial) sums of squares, fixed effects sum to zero for mixed terms, and exact p-values generated using unrestricted permutation of raw data (Anderson, 2001). Groups of samples were considered significantly different if the p-value falls < 0.05. Pairwise tests in PERMANOVA were used to determine which predefined categories were significantly different (using unadjusted p-values). The multivariate analyses, bacterial class plots and rarefaction curves were generated using PRIMER (v.7.0.11), PRIMER-E, Plymouth Marine Laboratory, UK (Clarke and Warwick, 2001). Venn diagrams were used to visualise shared and unique OTUs between sample groups (i.e. bulk vs rhizosphere vs roots) for untreated MM and PS, and between the two substrates for each sample group (i.e. bulk MM vs bulk PS; rhizosphere MM vs rhizosphere PS; root MM vs root PS). The entire sampling period (i.e. weeks 4 and 12) was considered in determining the numbers of unique and shared OTUs among sample groups, with those unique to untreated MM and PS roots reported in tables.
Measures of species diversity were calculated using algorithms for OTU richness (S), Pielou's evenness (J'), Shannon diversity (H') and Simpson diversity (1-λ), along with algorithms of taxonomic distinctness for diversity (delta+) and evenness (lambda+), using PRIMER (v.7.0.11) (Clarke and Warwick, 2001). These indicators of diversity (S, J', H', 1-λ, delta+, lambda+) were compared between groups of samples using one- and two-way ANOVA (i.e. fruit swabs vs bulk meadow sediment vs bulk meadow mix vs roots; and sample type [bulk/rhizosphere/root] vs sediment type [MM/PS] respectively) (Prism v. 7.01, Graphpad Software Inc.). To determine the impact of autoclave treatment on the bacterial communities, comparisons of OTU richness and average taxonomic distinctness (delta+) between autoclave treated/untreated substrates (MM/PS) and sampling time (growth stages: weeks 4 and 12) were also assessed using two-way ANOVA. Tukey's post-hoc multiple comparisons was also performed on significantly different (p < 0.05) variables in Prism. Diversity indicators were plotted in PRIMER.
Differential abundance analysis based on Linear Discriminant Analysis (LDA) Effect Size (LEfSe) was also conducted in MicrobiomeAnalyst (Dhariwal et al., 2017) to discern the top 20 significant families and OTUs contributing to the observed differences among treatments; as determined using the Kruskal-Wallis rank test (adjusted p-value cut off = 0.001), with the Log LDA Score value adjusted to 2.0 and significant taxa/OTUs given in descending order from the highest to lowest LDA score.
The OTU table and seedling growth data used for the associated analyses is presented in Supplementary Datasheet S1 and Supplementary Table S2, respectively. Sequences from individual samples were deposited within the NCBI SRA repository under accession numbers SAMN28854748 – SAMN28854880.
PS was calcareous and unsorted, consisting primarily of fine (grain size = > 125 – 250 μm; 37%), medium (> 250 – 500 μm; 44%) and coarse (> 0.5 – 1 mm; 15%) sand. MM was siliceous and moderately well sorted with fine (60%) and medium (35%) sand representing the majority of the sediment grains. Grain size was not changed by autoclave treatment (one-way PERMANOVA: pseudo-F = 1.38, p = 0.298 for PS; pseudo-F = 0.88, p = 0.607 for MM; Supplementary Figure S6: Datasheet 2). PS had higher concentrations of phosphorus (two-way ANOVA: P - F1,16 = 855.98, p < 0.001), nitrogen (N - F1,16 = 64.29, p < 0.001), inorganic carbon (inorgC - F1,16 = 3978.12, p < 0.001), calcium carbonate (CaCO3 - F1,16 = 14457.58, p < 0.001) and total carbon (C - F1,16 = 855.98, p < 0.001) compared to MM (Table 1). Autoclave treatment did not significantly alter nutrient levels (P - F1,16 = 1.34, p = 0.264; N - F1,16 = 0.29, p = 0.600; inorgC - F1,16 = 0.47, p = 0.502; CaCO3 - F1,16 = 2.89, p = 0.108; C - F1,16 = 1.34, p = 0.265).
A total of 30 seedlings, six in autoclaved treated MM and eight in each of the other treatments, failed to thrive (rotten or damaged seed) and were excluded from analyses. Summary statistics for key seedling growth parameters at the end of the experiment (week 12) are given in Supplementary Table S2. Overall, leaf count declined (mean 4.63 ± 0.66 to 3.34 ± 0.53 SD; z = -3.84) and combined length of leaves increased significantly (mean 237.41 ± 44.17 to 335.22 mm ± 77.22 SD; t = 9.34, p < 0.001) from week 4 to 12. Limited change in root count was observed (3.07 ± 0.71 to 2.80 ± 0.82; z = -0.95, p = 0.340) but combined length of roots increased (186.13 ± 68.38 to 279.86 mm ± 92.67 SD; t = -1.39, p = 0.170), with some roots restricted by the size of the pot by week 12. Root diameter showed negligible change between the sample periods (0.78 ± 0.14 to 0.75 mm ± 0.11 SD). Root hairs in PS seemed to be longer, more abundant, and entwined with sediment particles (Supplementary Figures S1A, B: Datasheet 2), while the number of lateral roots was greater in MM. The elongation zones (area free of root hairs where young cells increase in size before becoming part of the maturation zone) of MM roots were often notably longer than PS. Rust coloured root surfaces were observed for the untreated MM, though were absent from the autoclaved treatment groups (Supplementary Figures S1C, D: Datasheet 2). Mean seedling weight increased by 62% from week 4 (0.29 g ± 0.08 SD) to 12 (0.47 g ± 0.14 SD; t = 8.94, p < 0.001).
Multiple regressions suggested that total biomass (seedling weight) had a linear and positive relationship with age (weeks 4 and 12; t = 11.27, p < 0.001) and the size of the seed (t = 7.33, p < 0.001). No other factors (treatment and sediment type) or interactions were significant. A three-way interaction between treatment, sediment and seed length was a significant predictor of the pseudo root:shoot ratio (PRSR; ratio of total root to total shoot length; t = 2.16, p = 0.032), with PRSR being larger in PS than in MM, especially for autoclaved sediment (Figure 2A). In addition, seed length had a significant positive linear relationship with PRSR in PS (r = 0.35, p = 0.003) but not in MM (r = 0.01, p = 0.910) (Figure 2B), and in autoclaved (r = 0.24, p = 0.045) but not untreated (r = 0.13, p = 0.270) sediment (Figure 2C). Growth stage had no influence on PRSR, either individually or interacting with other terms. The interaction between sediment type and seed length was a significant (t = 2.90, p = 0.004) predictor of the number of lateral roots, with the number of lateral roots increasing significantly with seed size in PS (r = 0.50, p = 0.002) but not in MM (r = 0.14, p = 0.370; Supplementary Figure S7A: Datasheet 2). Treatment did not influence the number of lateral roots. The remaining starch area was positively and significantly related to the total seed area and was not influenced by sediment type or treatment (multiple regression: p < 0.001) (Supplementary Figure S7B: Datasheet 2).
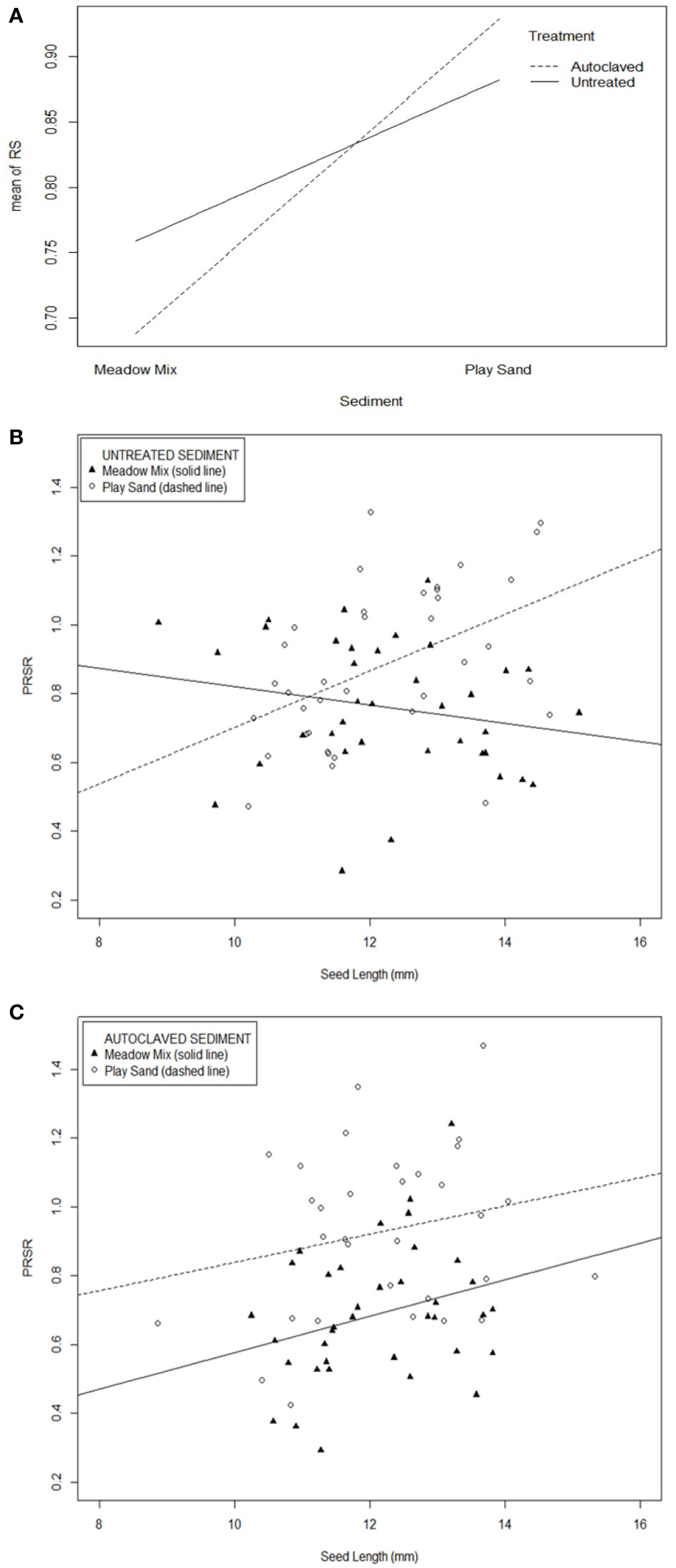
Figure 2 Relationship of the root:shoot ratio (RS) to sediment type, autoclave treatment and seed length (the three-way interaction of which was a significant predictor of RS). (A) Interaction plot between sediment type and treatment, Relationship of the pseudo root:shoot ratio (PRSR) with seed length for the two sediment types (meadow mix and play sand) in (B) untreated and (C) autoclave treated sediment.
To establish the bacterial communities associated with the seagrass 'environment', from which inferences could then be drawn from the seedling growth experiment, the diversity of the Lady Bay Posidonia meadow sediment, seawater, adult P. angustifolia roots and beach cast fruit, and the MM prepared for the experiment were first evaluated. Bacterial assemblages of the Lady Bay Posidonia meadow sediment, MM (pre-potted), seawater, mature seagrass roots and beach-cast fruit clustered according to their sources (Figure 3A). This was confirmed by a one-way PERMANOVA (pseudo-F = 13.02, p < 0.001), with pairwise comparisons revealing significant differences among all sample types (t = 2.58 – 4.52; p < 0.001) except for bulk meadow sediment and MM (t = 3.30; p = 0.096). Overall, samples largely comprised taxa belonging to seven bacterial classes (representing ~89% of the total sequence reads across all samples), with a further 47 ('Other') classes accounting for the remaining ~11%. Proteobacteria appeared to be a dominant component of all samples, with δ-proteobacteria notably abundant in roots and (albeit to a lesser extent) in sediment samples (Figure 3B). In contrast, α-proteobacteria dominated the bacterial communities in seawater and on fruit. Roots also appeared to have a lower abundance of γ-proteobacteria and Acidimicrobiia (Actinobacteria), and a greater abundance of Anaerolineae (Chloroflexi) compared to all other sample types.
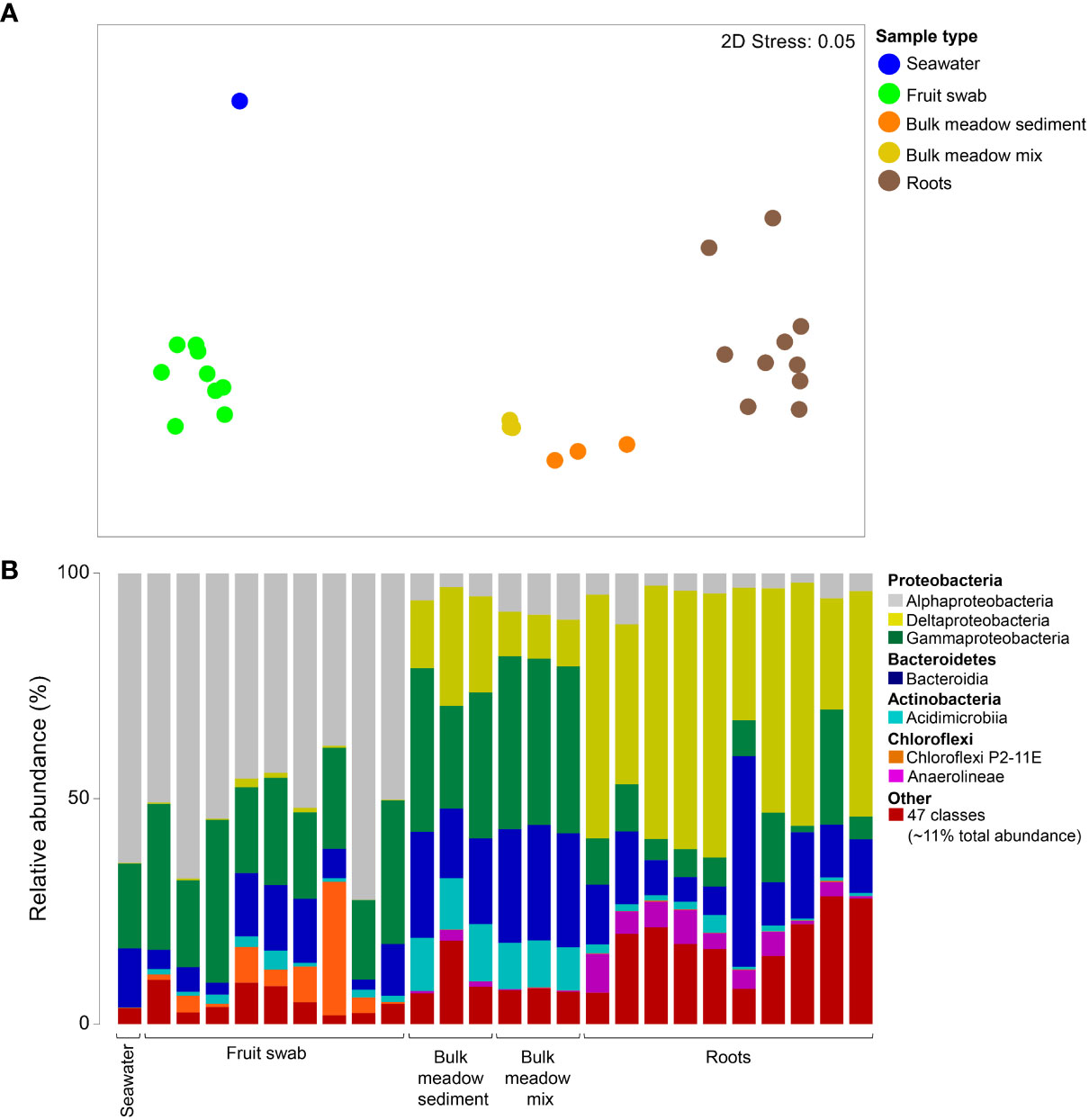
Figure 3 Differences in the bacterial assemblages of the seagrass environment. (A) Ordination plot representing the global bacterial community structure from field-collected seawater, seagrass fruit swabs, bulk meadow sediment and mature seagrass root samples; and bulk (pre-potted) meadow mix used in the growth experiments, as assessed by non-metric multidimensional scaling (nMDS) using Bray–Curtis dissimilarity. (B) Mean relative abundances of the predominant 7/54 bacterial classes (accounting for 89% of the total sequence reads) detected from the samples, where "Other" represents the remaining 47 classes (accounting for 11% of the total sequence reads).
Measures of diversity (OTU richness, Shannon's and Simpson's diversity, Pielou's evenness) all followed a similar trend, with sediment samples (bulk meadow sediment and pre-potted MM) having higher values and fruit swabs the lowest (Supplementary Figure S8A: Datasheet 2). For each diversity metric, significant differences were observed between all sample groups (one-way ANOVA: df = 3, F = 11110 – 40.82, p < 0.001) except for between MM and meadow sediment (all diversity measures), and between MM/sediment and roots (Simpson's index of diversity only). In plotting average taxonomic distinctness (delta+) as a function of variation in taxonomic distinctness (lambda+), greater taxonomic breadth (based on higher values of delta+) were observed for meadow sediment, pre-potted MM, and roots. Furthermore, while both meadow sediment and pre-potted MM also had a more even distribution of OTUs representing each of the bacterial lineages (based on lower values of lambda+) roots were more varied and comprised samples that were dominated by a few distinct bacterial lineages while still being taxonomically diverse (based on higher values for lambda+). In contrast, fruit samples were less taxonomically diverse with a more taxonomically uneven distribution of OTUs among lineages, indicating the presence of select bacterial taxa (Supplementary Figure S8B: Datasheet 2). While this was confirmed by one-way ANOVA (df = 3; F = 10.24 – 42.64; p < 0.001), pairwise differences using Tukey's post-hoc test were observed only between fruit swabs and all other samples (p < 0.001). Differential abundance analysis revealed several significantly different bacterial families and OTUs associated with the differences among sample groups (p < 0.001) (Supplementary Figures S9A, B: Datasheet 2). The most notable included Desulfobacteraceae (OTUs 82, 653, 131 and 2488: Desulfatitalea sp.), Sedimenticolaceae, and Spirochaetaceae in roots; Bacteroidetes BD2-2, Thiotrichaceae (OTU_98: unclassified sp.), uncultured Actinomarinales, and Desulfobulbaceae (OTU_118: unclassified sp.) in meadow sediment; and Rhodobacteraceae (OTU_2: Thalassococcus sp; OTU_73 unclassified sp.), unclassified Chloroflexi P2-11E (OTU_15), Methylophilaceae (OTU_50: Methylotenera sp.), and Burkholderiaceae (OTU_81: Burkholderia sp.) in fruit. The addition of beach sand to meadow sediment (which created the MM used in the growth experiment) did have some effect on particular families/OTUs, with those from the meadow sediment apparent but less pronounced in the MM. This was also accompanied by a greater abundance of Flavobacteriaceae (OTU_29: Muriicola sp.), Woeseiaceae (OTU_45: Woeseia sp.), Idiomarinaceae (OTU_311: Idiomarina sp.) and γ-proteobacteria BD7 8 (OTU_13: unclassified sp.) in the MM compared to the meadow sediment. For seawater, particular SAR clades (namely α-proteobacteria SAR11 1a and Puniceispirillales SAR116) appeared to be dominant features, and were more evident from fruit compared to the other sample groups. Interestingly, a number of taxa that were predominant from fruit (namely Methylophilaceae and unclassified Chloroflexi P2-11E), were also more abundant in roots compared to the other samples.
In order to determine the differences in the contribution of the surrounding environmental microbial consortia on the establishment of seagrass specific associations during early growth, we evaluated the bacterial communities from the bulk sediment, rhizosphere and roots of seedlings grown in two different propagating substrates (PS and MM). Seedlings grown in untreated MM separated from those grown in PS, with bulk sediment, rhizosphere and roots clustering independently of one another (Figure 4A). Highly significant differences were observed between sample types (bulk sediment, rhizosphere and roots) and substrates (MM and PS) (pseudo-F = 7.18, p < 0.001, pseudo-F = 30.09, p < 0.001, respectively). Despite a similar trend across sample types (where pairwise differences were observed between all groups, p < 0.001), there was a significant interaction effect (pseudo-F = 6.87, p < 0.001), indicating that the changes were substrate specific. The greatest variation among groups of samples (as indicated by higher MVDISP values) was observed for MM compared to PS, whereby variation among samples increased from bulk sediment to rhizosphere to roots (Figure 4A, inset key table). Overall, OTUs were comprised of at least 32 phyla, 53 classes, 162 orders, 265 families, and 513 genera (Supplementary Datasheet 1). However, samples were largely represented by taxa belonging to five phyla and nine bacterial classes (representing ~94% of the total sequence reads across all samples), with a further 44 ('Other') classes accounting for the remaining ~6% (Figure 4B). In general, like the meadow environment, samples from seedlings grown in different propagating substrates were largely dominated by Proteobacteria. Compared to MM, PS had a higher abundance of γ- and α-proteobacteria and Actinobacteria and a lower abundance of Epsilonbacteraeota (Campylobacteria) and Bacteroidetes, while MM had higher abundances of δ-proteobacteria, Bacteroidetes (Bacteriodia) and Acidimicrobiia. Rhizosphere and root samples were more similar than either was to bulk sediment for both substrates, with those from MM having notably higher abundances of Campylobacteria and, to a lesser extent, Ignavibacteria (Bacteroidetes) and Spirochaetia (Spirochaetes) compared to PS.
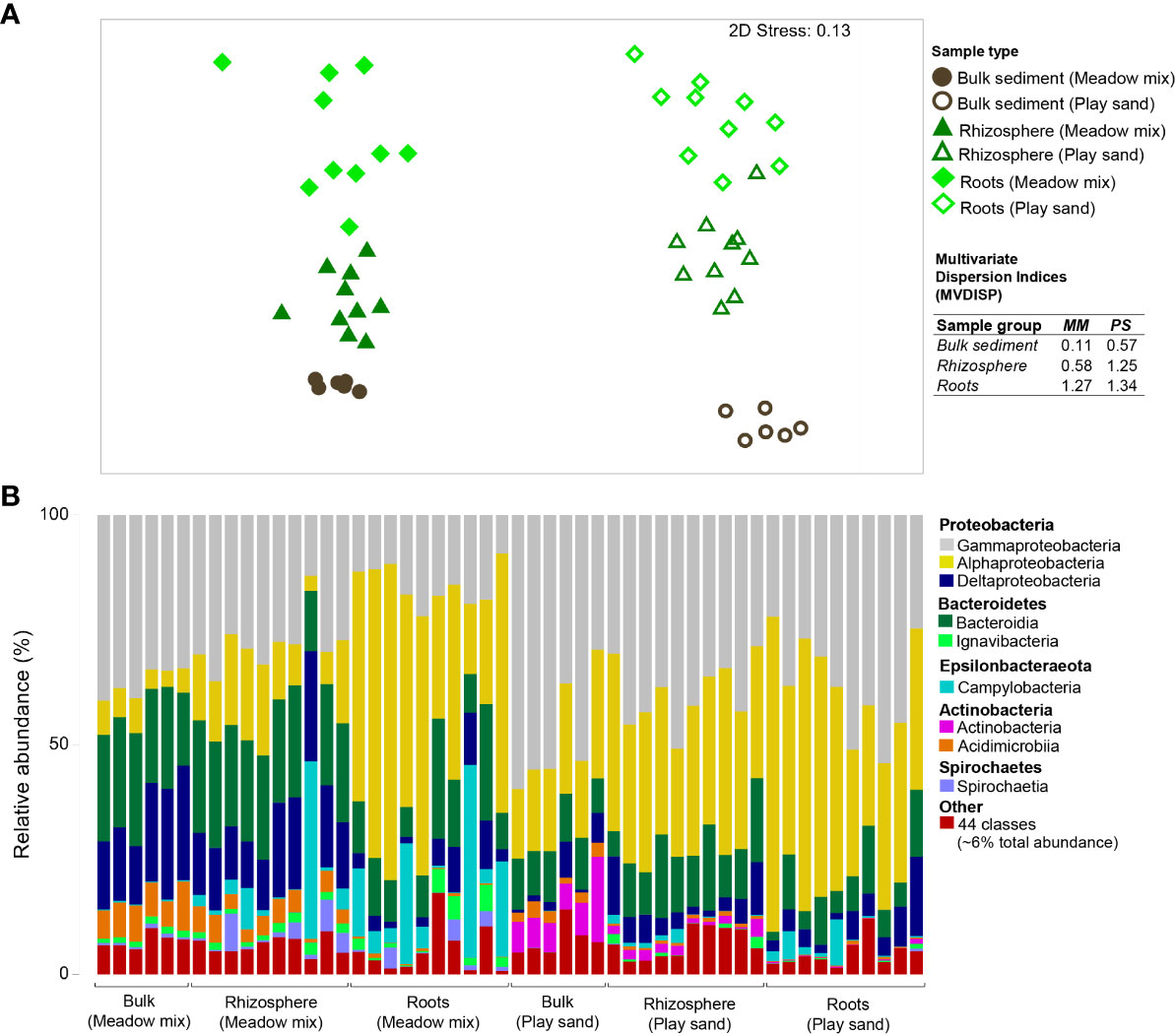
Figure 4 Differences in the bacterial communities of seagrass seedlings grown in different propagating substrates. (A) Ordination plot representing the global bacterial community structure from bulk sediment, rhizosphere and seagrass root samples collected from the untreated experimental meadow sediment mix and play sand used in the growth experiments, as assessed by non-metric multidimensional scaling (nMDS) using Bray–Curtis dissimilarity. Corresponding multivariate dispersion indices (MVDISP) representing the global variation in the bacterial community composition among replicate samples are given for each sample group (inset key table), where higher values represent greater variation and lower values less variation among samples. (B) Mean relative abundances of the predominant 9/53 bacterial classes (accounting for 94% of the total sequence reads) detected from the samples, where "Other" represents the remaining 44 classes (accounting for 6% of the total sequence reads).
Both substrates comprised a large number of common (core) OTUs, with > 50% of the total OTUs shared between the different sample types for both MM (659/1182) and PS (564/1065) (Figure 5A). Similar numbers of unique OTUs were also apparent among the different sample types for both substrates, with the largest numbers occurring within the rhizosphere (170 in MM and 125 in PS) and the least in the roots (46 in MM and 42 in PS). For both substrates, rhizosphere and root samples shared a greater number of OTUs compared to bulk sediment. Of the OTUs found to be specifically associated with each sample type (i.e. either in bulk sediment, rhizosphere or roots), only a small number were unique to either MM or PS (Figure 5B). For the rhizosphere, this included 35 from MM and 41 from PS (Supplementary Tables S3 and S4), and for the roots 14 from MM and 15 from PS (Supplementary Tables S5 and S6). As many as ~60% of these unique OTUs were also detected in the seagrass meadow environmental samples, with several being abundant or exclusively associated with mature roots or fruit. Most notably, across both substrates this included several OTUs belonging to the α-proteobacteria (namely Rhodobacteraceae and Hyphomonadaceae) that were largely abundant in fruit. Alongside this were other fruit associated OTUs that were detected solely in the roots of either PS or MM. Within MM this included OTU_215 Marinobacterium (Nitrincolaceae; γ-proteobacteria) and OTU_570 Spirochaeta 2 (Spirochaetaceae; Spirochaetes), and in PS OTU_910 uncultured Micavibrionaceae (α-proteobacteria), OTU_8302 Methylotenera (Methylophilaceae; γ-proteobacteria) and OTU_8835 unclassified Babeliales UBA12409 (Dependentiae) (Supplementary Tables S5 and S6).
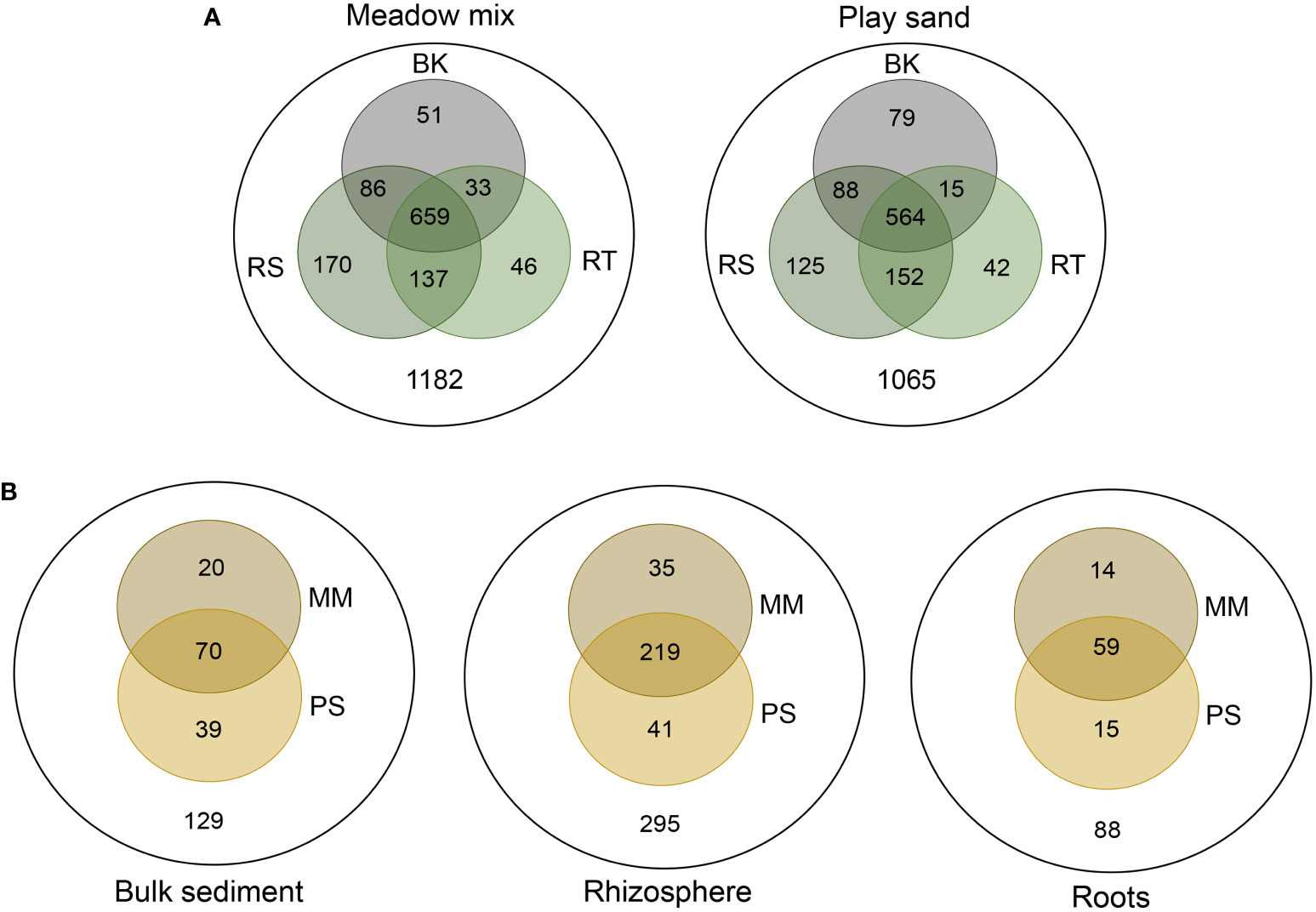
Figure 5 Venn diagrams depicting the distribution of shared and unique OTUs in (A) bulk (BK), rhizosphere (RS) and root (RT) samples from meadow mix and play sand; and (B) meadow mix (MM) and play sand (PS) from each sample group. Total OTUs are denoted in the outer circles.
Overall, the highest species (OTU) richness was observed for MM, particularly within the rhizosphere (Figure 6A). For roots, however, richness and other measures of diversity (Shannon's and Simpson's diversity, Pielou's evenness) were substantially reduced. For each diversity metric, significant differences were observed between all three sample types (i.e. bulk sediment, rhizosphere and roots) (two-way ANOVA: df = 2, F > 13, p < 0.001). There were also significant differences in species richness, Shannon's diversity and taxonomic distinctness metrics between substrates (i.e. MM and PS) (two-way ANOVA: df = 1, F > 8, p < 0.005). There was an interaction effect in Pielou's evenness, Shannon's diversity and variation in taxonomic distinctness (two-way ANOVA: df = 2, F > 7, p < 0.004), indicating that the changes across sample types (from bulk to rhizosphere to roots) were substrate specific.
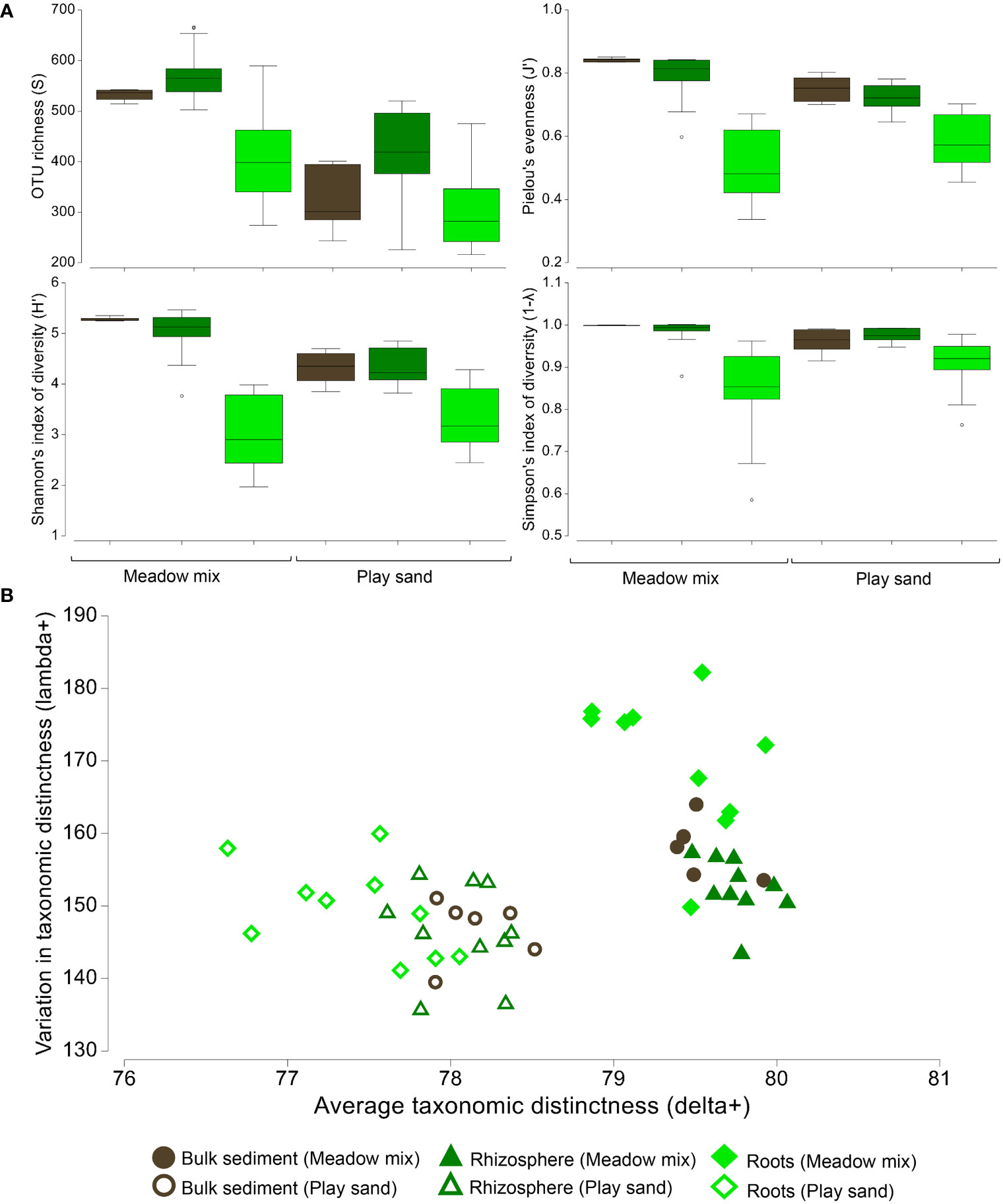
Figure 6 Measures of diversity from seagrass seedlings grown in different substrates. (A) Box plots representing the median, interquartile ranges (IQR) and distribution of measures of OTU richness, Shannon's and Simpson's indices of diversity and Pielou's evenness from bulk sediment, rhizosphere and seagrass root samples collected from the untreated meadow mix and play sand used in the growth experiments. (B) Scatter plot charting the average taxonomic distinctness (delta+) as a function of variation in taxonomic distinctness (lambda+) for each of the sample groups.
In plotting average taxonomic distinctness (delta+) as a function of variation in taxonomic distinctness (lambda+), all MM samples comprised greater taxonomic breadth (based on higher values of delta+) compared to PS (Figure 6B). Furthermore, bulk sediment and rhizosphere samples of the MM had a more even distribution of OTUs representing each of the different bacterial lineages (based on lower values of lambda+), while the roots were dominated by a few distinct bacterial lineages while still being taxonomically diverse. In contrast, PS samples were less taxonomically diverse with a more taxonomically even distribution of OTUs among lineages. The roots of PS established the lowest level of bacterial taxonomic diversity. Differential abundance analysis revealed several significantly different bacterial families and OTUs associated with the differences among sample groups (p < 0.001) (Supplementary Figures S10A, B: Datasheet 2). MM samples were enriched with Flavobacteriaceae, Desulfobacteraceae, Woeseiaceae, uncultured Actinomarinales, Desulfobulbaceae, Thiovulaceae, Marinilabiliaceae, and Chromatiaceae, while PS samples were enriched with Solimonadaceae, Burkholderiaceae, Micrococcaceae, Xanthomonadaceae, Marinomonadaceae, and Tenderiaceae. Most notable in the rhizosphere and/or roots of seedlings grown in MM were Rhizobiaceae (OTU_5: Lentibacter sp.), Arcobacteriaceae (OTU_32: Arcobacter sp.), Marinilabiliaceae (OTU_9: Labilibacter sp.), Magnetospiraceae (OTU_59: Magnetovibrio sp.), and Methylophagaceae (OTUs 163 and 2764: Methylophaga sp., unclassified sp.); and in PS Rhodobacteraceae (OTUs 2 and 4: Thalassococcus sp., unclassified sp.), Methylophagaceae (OTU_3: Methylophaga sp.); Rhizobiaceae (OTU_21: Lentibacter sp.), and Devosiaceae (OTU_40: Devosia sp.).
Given that root samples likely represent the closest relationships with seagrass seedlings and the surrounding microbial consortia, we investigated these bacterial communities at the different growth stages (weeks 4 and 12) in untreated samples only (Figure 7A). Significant differences between groups were evaluated using a two-way PERMANOVA which crossed substrate type (MM and PS) with sampling period (week 4 and week 12), indicating that there were highly significant differences between substrates and between sampling periods (pseudo-F = 11.10, p < 0.001, pseudo-F = 3.50, p = 0.001, respectively). However, there was a significant interaction effect (pseudo-F = 2.67, p = 0.006), indicating that the changes across growth stages were substrate specific, with pair-wise differences revealing only a significant change in the PS roots across growth stages (t = 2.26; p = 0.007). While ordination of the seedling root samples with those obtained from mature (adult) meadow roots from Lady Bay revealed a distinct separation between groups, MM roots were closer to the Lady Bay samples than those from PS (Figure 7A). In evaluating the diversity metrics among sample groups, no significant differences were apparent for the MM roots across growth stages or with the adult meadow roots (Figure 7B). However, PS roots differed significantly for Pielou's evenness and Shannon's diversity, with both measures increasing substantially between weeks 4 and 12 (one-way ANOVA: df = 4; F = 3.43 – 49.12; p < 0.023) (Supplementary Table S4D). Furthermore, unlike MM and adult roots, PS roots also had a significantly reduced, though slightly more even, distribution of OTUs across bacterial lineages (based on lower values for delta+ and lambda+, respectively).
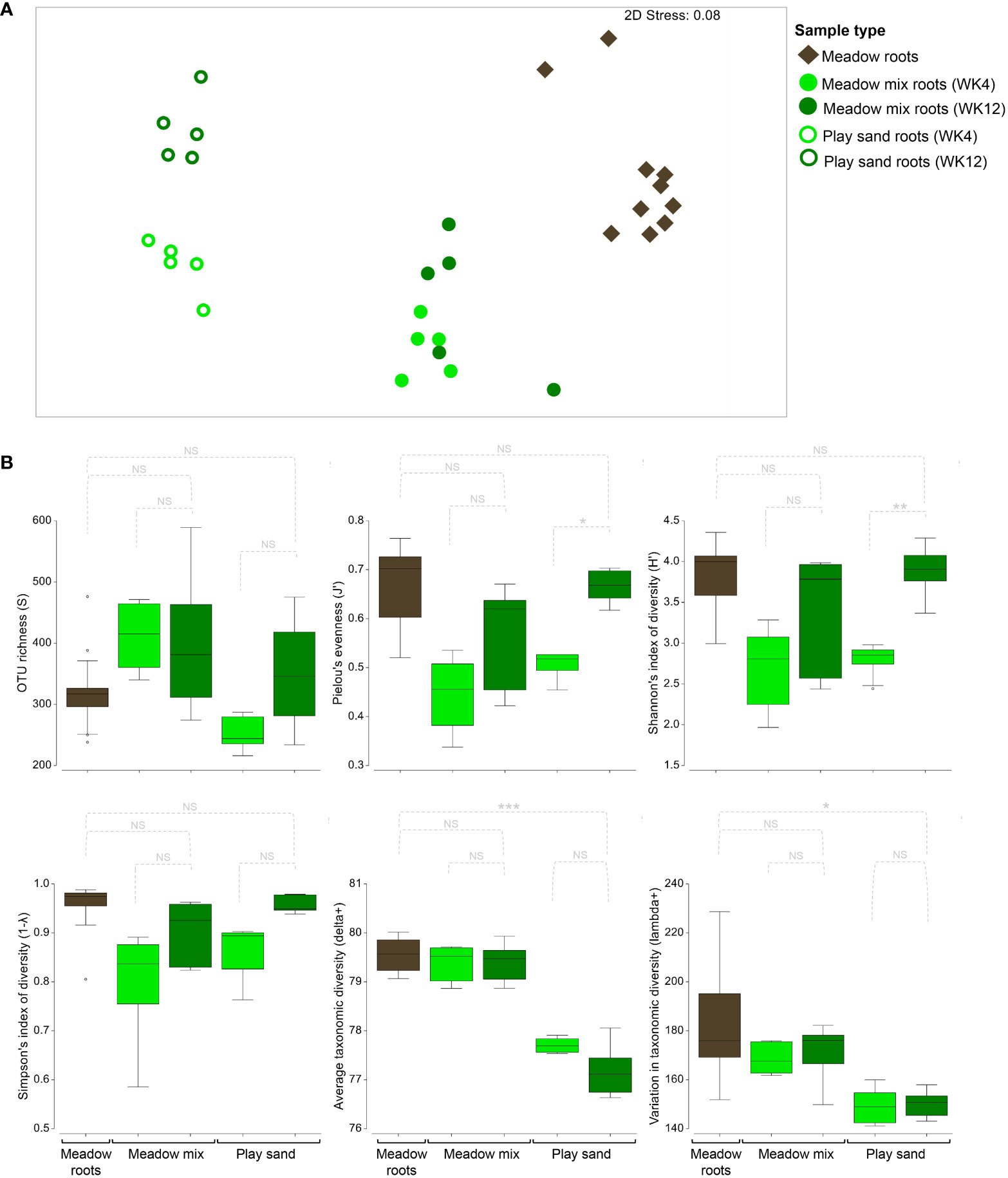
Figure 7 Differences in the root bacterial communities of seagrass seedlings across growth stages. (A) Ordination plot representing the global bacterial community structure from mature seagrass roots obtained from Lady Bay (South Australia) and seagrass seedling roots grown in untreated meadow mix and play sand at weeks 4 and 12 (WK 4, WK12), as assessed by non-metric multidimensional scaling (nMDS) using Bray–Curtis dissimilarity. (B) Box plots representing the median, interquartile ranges (IQR) and distribution of measures of OTU richness, Shannon's and Simpson's indices of diversity, Pielou's evenness, and average and variation in taxonomic distinctness (delta+, lambda+) from each of the sample groups.
Differential abundance analysis revealed several significantly different bacterial families and OTUs associated with the differences among sample groups (p < 0.001) (Supplementary Figures S11A, B). Unlike PS, bacterial root communities of seedlings grown in MM were somewhat more similar to adult root communities, with several that were predominant in MM also having the correspondingly highest abundance in adult roots. Most notably, this included Desulfobacteraceae (OTUs 82, 653 and 131: Desulfatitalea sp.), Bacteroidetes BD2-2, Sedimenticolaceae, Spirochaetaceae, and uncultured Ardenticatenales. However, a number of other families/OTUs that were also predominant in MM roots had instead the lowest abundance in adult roots, including Rhizobiaceae, Arcobacteraceae (OTUs 16 and 32: Arcobacter sp.), Flavobacteriaceae, Magnetospiraceae, and Marinilabiliaceae (OTU_9: Labilibacter sp.). PS roots were enriched by taxa that generally occurred in the least abundance in adult roots (e.g. Rhodobacteraceae and Methylophagaceae) (Supplementary Figure S11A: Datasheet 2).
To better gauge the contribution of the surrounding microbial consortia on the early growth of seagrass seedlings and the establishment of the more intimate root-associated (rhizoplane) communities, we also investigated the bacterial assemblages from the roots of seedlings grown in autoclave treated and untreated MM and PS. Significant differences between groups were evaluated using two-way PERMANOVA which crossed treatment (autoclave treated and untreated sediment) with sampling period (week 4 and week 12). While autoclave treatment had a significant impact on the root bacterial communities of both MM (pseudo-F = 6.65, p < 0.001) and PS (pseudo-F = 2.03, p = 0.046), the shift in the bacterial communities was much more pronounced in the MM compared to the PS (Figures 8A, B). More specifically, in the MM, greater separation occurred between autoclave treated and untreated samples, followed by sampling periods, while for PS there was a greater separation between sampling periods, followed by treatment. The pronounced change in beta diversity of MM samples with treatment was also reflected in significant changes in alpha diversity, such as species (OTU) richness (two-way ANOVA: F = 31.03, p < 0.001) and average taxonomic distinctness (delta+) (F = 14.40, p = 0.002). Most notably, as much as ~50% of the OTUs were lost from the MM root samples following autoclave treatment at week 4, with a notable reduction in the distribution of OTUs across bacterial lineages (based on lower values for delta+) at week 12. For PS, significant differences in these measures were only observed between sampling periods rather than treatment (richness: F = 9.59, p = 0.007; delta+: F = 6.66, p = 0.020), with an increase in richness and a reduction in the distribution of OTUs across bacterial lineages (based on lower values for delta+) from week 4 to week 12.
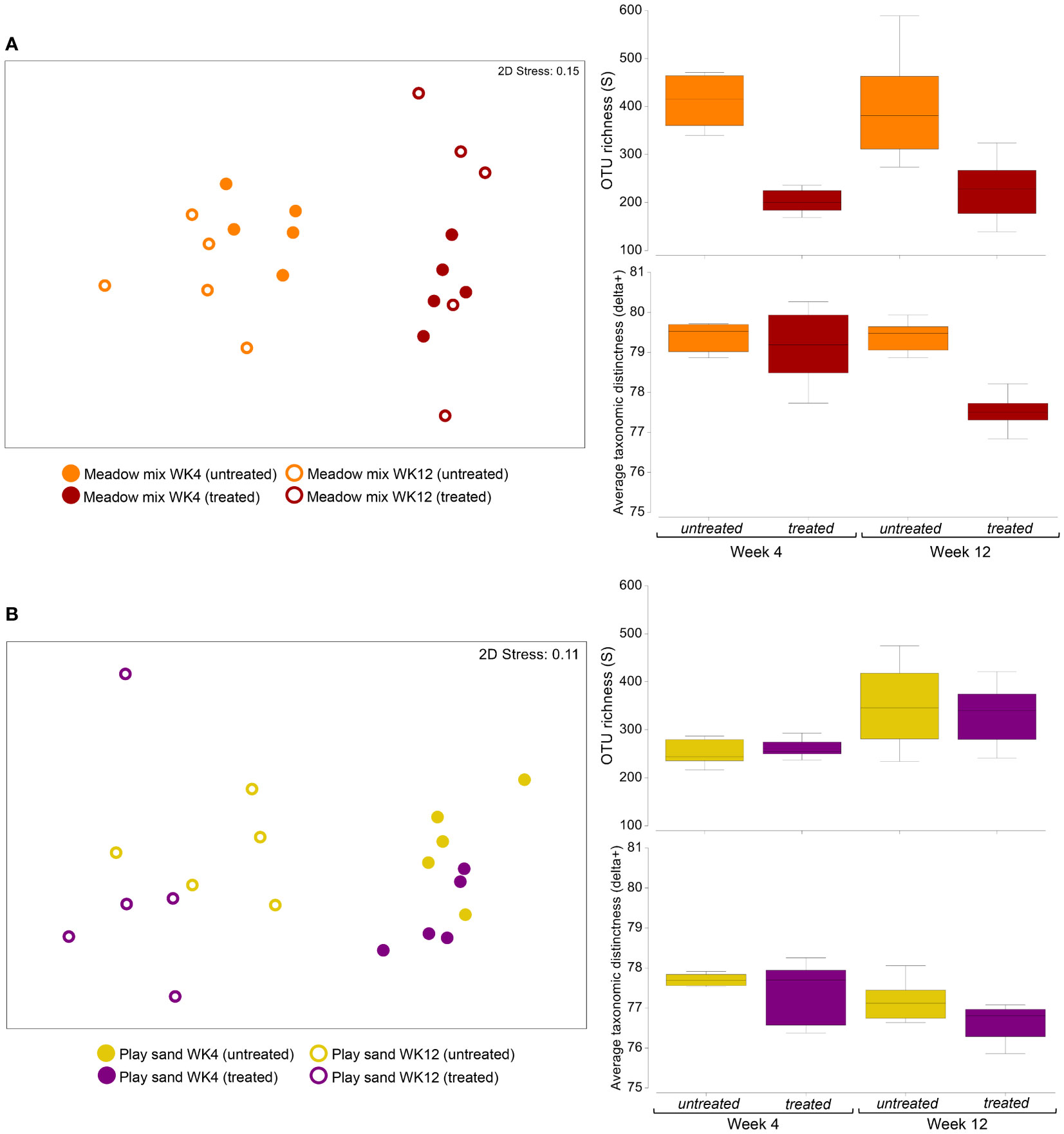
Figure 8 Differences in the root bacterial communities of seagrass seedlings following autoclave treatment of propagating substrates. Ordination (nMDS) plots based on Bray-Curtis dissimilarity representing the global bacterial community structure between autoclave treated and untreated root samples at weeks 4 and 12, and corresponding box plots representing the median, interquartile ranges (IQR) and distribution of measures of OTU richness and average taxonomic distinctness from each of the sample groups for (A) meadow mix and (B) play sand.
Differential abundance analysis revealed several significantly different bacterial families and OTUs associated with the differences among sample groups (p < 0.001) (Supplementary Figures S12A, B: Datasheet 2). Most notable in MM was a decrease in abundance of Rhizobiaceae (OTU_23517: Lentilitoribacter sp.), Thiovulvaceae, Bacteroidetes BD2-2 (OTU_97: unclassified sp.), Magnetospiraceae (OTU_59: Magnetovibrio sp.), Melioribacteraceae, Sedimenticolaceae (OTU_138: Candidatus Thiodiazotropha), and Calditrichaceae, and an increase in Rhodobacteraceae (OTU_4: unclassified sp.), Cellvibrionaceae, Arcobacteraceae (OTUs 23 and 32: Arcobacter sp.), Desulfobacteraceae, and Desulfovibrionaceae. While changes in PS roots were generally not as pronounced, certain taxa predominant in untreated MM became enriched or decreased in abundance following treatment (namely Magnetospiraceae [OTU_59 Magnetovibrio sp.] and Thiovulvaceae, respectively).
Our findings illustrate the complexity of the microbiome (bacterial assemblages) of seagrass seedlings and that manipulation of the microbiome could potentially lead to better outcomes for Posidonia restoration. The bacterial communities of seedlings differed significantly between meadow mix – MM (a more 'natural' propagating substrate) and the predominantly used play sand - PS, with the composition of taxa in the rhizosphere and roots being unique for each propagating substrate. The meadow mix contained a larger diversity of bacteria compared to play sand, suggesting that seagrass seedlings in this substrate likely have a greater 'pool' of potentially beneficial microbes that could be selected to support growth (Ugarelli et al., 2017; Conte et al., 2021). Growth measurements offered little support for the hypothesis that bacterial communities influence the growth rate of seagrass seedling. However, when considering the pseudo root:shoot ratio (PRSR), seed length had an effect as well as substrate type and autoclave treatment, suggesting that the seedlings may be responding to differences in the microbiome from as early as four weeks after planting.
We observed marked differences in the bacterial communities of samples collected from the natural Posidonia meadow environment and the untreated propagating substrates used in the growth experiment. Meadow sediment and root communities were distinct to those observed from the seawater and fruit, and when meadow sediment was incorporated with beach sand (in preparing the meadow mix used in the growth experiment), it yielded a substrate that was considerably more diverse and comprised a greater proportion of δ-proteobacteria, Bacteriodia (Bacteroidetes) and Acidimicrobiia (Actinobacteria) compared to play sand. Most notable in the meadow mix was the enrichment of taxa previously reported from other seagrass meadows that are important constituents in biogeochemical cycling of sulfur (Jørgensen et al., 2019). This included the sulfate reducing δ-proteobacteria families Desulfobacteraceae and Desulfobulbaceae (Kuever, 2014a; Kuever, 2014b), as well as the sulfide oxidising families Thiovulaceae (Campylobacteria) (Waite et al., 2017), Thiotrichaceae (γ-proteobacteria) (Garrity et al., 2005) and Chromatiaceae (purple sulfur bacteria) (Imhoff, 2014). In addition, alongside other ubiquitous marine sediment associated taxa like Woesiaceae (which are purported denitrifiers) (Mußman et al., 2017), the Bacteroidetes (namely Flavobacteriaceae and Bacteroidetes BD2-2), a group assumed to contribute to the cycling of carbon through the breakdown of complex polysaccharides of algae and accumulated plant deposits (Fernández-Gómez et al., 2013; Gavriilidou et al., 2020), occurred only in substrates that included meadow sediments. In contrast, play sand (despite sharing a large proportion of OTUs with meadow mix) was less diverse and had notably higher abundances of γ- and α-proteobacteria and Actinobacteria (Actinobacteria) and less Bacteroidetes. This included the occurrence of certain families such as Solimonadaceae that are typically associated with soils and other aquatic ecosystems (Zhou et al., 2014), and likely reflects the coastal, yet terrestrial, source of the play sand used here. The types of bacterial assemblages observed thus appear to be primarily a function of the origins of the propagating substrates, where the incorporation of meadow sediment into beach sand is more likely to support assemblages represented in the natural environment, which may promote greater resilience and better growth. For example, communities with a high ratio of Proteobacteria and/or Actinobacteria compared to Bacteroidetes may reduce a plant's resistance to environmental stresses (Pérez-Jaramillo et al., 2018), with a reduction of sediment associated Bacteroidetes thought to be a precursor to a decline in Posidonia oceanica meadows (García-Martínez et al., 2009). Furthermore, at least in terrestrial systems, Bacteroidetes have also been attributed to pathogen suppression and the mobilisation of other essential and limited nutrients like phosphorous which are important for metabolism in seagrasses (Touchette and Burkholder, 2000; Lidbury et al., 2021). Given the relatively low concentrations of total phosphorous in the meadow mix compared to the play sand here (by at least an order of magnitude), higher proportions of Bacteroidetes in the meadow mix may be of particular benefit.
Despite the presence of a large number of shared OTUs, partitioning of the communities between the bulk sediment, rhizosphere and roots was evident for both untreated substrates, with the rhizosphere and roots being more similar to one another than the bulk sediment, a trend that is widely reported among angiosperms (Fitzpatrick et al., 2018). This suggests that the rhizosphere and roots constitute distinct niches that are strongly influenced by the surrounding sediment. In seagrasses, these microenvironments are suggested to be driven by root exudates (Martin et al., 2017; Martin et al., 2019). For Posidonia oceanica, such exudates occur primarily as sucrose, which is secreted in large (μM) concentrations into the rhizosphere together with inhibitory phenolic compounds which, alongside its role in balancing local osmolarity, is thought to regulate the surrounding bacterial communities and attract species that may support the health of the plant (Sogin et al., 2022). Interestingly, of the select few rhizosphere constituents reported by Sogin et al., (2022) to represent putative sucrose specialists with the capacity to breakdown otherwise inhibitory phenolic compounds (based on metagenome-assembled genomes and sediment metatranscriptomes), OTUs associated with two orders (namely Desulfobacterales and Xanthomonadales) were observed here. Specifically, in the meadow mix the Desulfobacterales families Desulfobacteraceae and Desulfobulbaceae were enriched in the bulk sediment and rhizosphere, while in the play sand we observed the enrichment of the Xanthomonadales family Xanthomonadaceae. The Desulfobacteraceae and Desulfobulbaceae are typically anaerobic sulfate reducers, though some species are also capable of nitrogen fixation (Kuever, 2014a; Kuever, 2014b), and are frequently reported from meadow sediments and the rhizosphere of seagrasses (Jensen et al., 2007; García-Martínez et al., 2009; Cúcio et al., 2016; Hurtado-McCormick et al., 2019; Markovski et al., 2022). Supported by the highly reduced, sulfate rich conditions typical of these systems, particularly in sediments that are naturally composed of finer particles (Martinez-Garcia et al., 2015), such taxa not only play an important role in the decomposition of organic matter, but in supporting seagrass productivity as potential diazotrophs (Herbert, 1999; Welsh, 2000; Sun et al., 2015). In contrast, the Xanthomonadaceae are obligately aerobic and are one of the largest groups of phytopathogens and endophytes (Saddler and Bradbury, 2015; Assis et al., 2017). Given their reliance on oxygen for respiration, their enrichment in play sand, rather than meadow mix, suggests that the surrounding conditions of the play sand are likely to have been at least partially oxic (despite the rhizosphere of seagrasses usually being anoxic) (Borum et al., 2006). Though oxygen concentrations were not measured here, grain size strongly influences the permeability and movement of oxygen into the sediment, with more coarse sediments associated with increased pore water advection (Ahmerkamp et al., 2017). Of course, while other factors cannot be excluded (e.g. radial diffusion of oxygen from the roots) (Borum et al., 2006; Brodersen et al., 2018), the majority (~60%) of the play sand was made up of medium-coarse grains (unlike the meadow mix which was largely fine grained), and thus it seems plausible that varied conditions arising from differences in grain size may have led to the enrichment of specific assemblages in these substrates. Indeed, in other seagrasses like Enhalus acoroides it has also been speculated that differences in grain size may have an impact on the rhizosphere communities, with fine sediments supporting a stronger "rhizosphere effect" due to the limited diffusion of exudates away from the root (Zhang et al., 2022). However, given the clear partitioning of the communities among the microenvironments of both substrates, grain size does not appear to be the sole contributing factor.
Interestingly, alongside CO2 fixation, the Xanthomonadaceae also have the capacity to metabolise H2 (Sogin et al., 2022), a product generated from nitrogen fixation (Teng et al., 2019) and fermentation by ubiquitous sedimentary bacterial taxa like Woeseiaceae, which is reported to accumulate in high concentrations in the pore water of permeable (sandy) seagrass sediments (Kessler et al., 2019). With other nitrogen fixers (e.g. Rhizobiaceae) (Carareto Alves et al., 2014) and fermentative bacteria (Woeseiaceae) also occurring in the play sand (albeit it in lower abundances compared to meadow mix), the generation of H2 required for supporting such populations is likely, and may be further supported here by the enrichment of other aerobic H2 utilising organisms in the play sand.
Various Burkholderiaceae (β-proteobacteria) genera, such as Acidovorax, Hydrogenophaga, and Polaromonas, all of which comprise species capable of the oxidation of hydrogen (Willems et al., 1989; Willems et al., 1990; Sizova and Panikov, 2007), were detected somewhat unexpected as the Burkholderiaceae are typically reported from the leaves rather than the rhizosphere of seagrass (Hurtado-McCormick et al., 2019). Generally, such taxa have been observed in association with sediments contaminated with polycyclic aromatic hydrocarbons (PAHs). For example, Hydrogenophaga was found to be enriched in PAH-treated sediments of Halophilis ovalis (Ahmad et al., 2021). Intriguingly, alongside other β-proteobacteria like Acidovorax and Polaromonas, the Xanthomonads are also well known degraders of petroleum hydrocarbons (Chang and Zylstra, 2010; Tan and Parales, 2019). In Australia, the occurrence of PAHs in nearshore marine sediments is well documented and likely arises from industrial discharge, bushfires, and urban, recreational and agricultural activities (Maher and Aislabie, 1992). Their impact on seagrasses is varied, but in sub-lethal concentrations they can be incorporated into the tissues and reduce their tolerance to other stressors (Zieman et al., 1984; Runcie et al., 2005). For Posidonia australis, a decrease in photosynthetic oxygen production has been reported in the presence of PAHs together with other chemical dispersants (Hatcher and Larkum, 1982). While the occurrence of such species (particularly in the play sand) thus raises some concerns regarding the possibility of PAH contamination, evidence from terrestrial and other marine plants indicates a possible role for such constituents in the broader plant functioning to include the rhizoremediation of petroleum contaminants (Kotoky et al., 2018; Chen et al., 2019; Sampaio et al., 2019).
Differences in the bacterial communities of the root (rhizoplane) samples were also observed and included the varied enrichment of specific taxa and small numbers of unique OTUs in both of the untreated substrates. As reported for other plant species and in seagrasses, the rhizoplane is considered an exclusive niche that comprises a smaller subset of taxa that are selected for in a stepwise manner from the surrounding sediment and rhizosphere, leading to the formation of more intimate associations likely fostered by the host plant (Edwards et al., 2015; Zhang et al., 2022). In our study, greater intimacy was reflected by microbial cells adhering tightly to the root surfaces of P. angustifolia seedlings (as observed by ESEM) and by considerably reduced species (OTU) richness and diversity in the roots compared to the rhizosphere and bulk sediment. Though the rapid dehydration of seedling Martinez-Garciaroot specimens prior to ESEM visualisation impeded Martinez-Garciaour ability to obtain a detailed assessment of microbial cell densities, cell coverage was sparse and is consistent with the lower rates of colonisation observed previously on the roots of the closely related Australian species P. australis and P. sinuosa, compared to the Mediterranean species P. oceanica (García-Martínez et al., 2005). This may be further exemplified by the notable variation observed among root samples compared to the rhizosphere or bulk sediment, particularly in the meadow mix. Unlike play sand, bacterial root communities of seedlings grown in meadow mix were somewhat more similar to adult roots and were represented by OTUs that, despite covering a considerable breadth of taxa (based on high values for delta+), were dominated by a few distinct bacterial lineages (based on high values for lambda+). This higher taxonomic diversity (delta+) of the meadow mix seedlings may afford greater resilience to disturbances that affect their bacterial communities, as increased diversity improves the resilience and recovery time of a natural system after disturbance (Hector and Bagchi, 2007; Cardinale et al., 2012). However, phylogenetic conservatism in bacterial lineages indicates that many related OTUs may have similar functional traits (nitrogen fixation, sulfate-reduction etc) (Martiny et al., 2013), implying that functional diversity should also be considered using complementary omics-based approaches (e.g. metatranscritptomics) or assays that directly measure specific activities (e.g. nitrogen fixation).
The high productivity of seagrass meadows is linked to their inability to resorb nutrients before discarding aging foliage (Hocking et al., 1981; Hemminga et al., 1999). Consequently, seagrasses have high nutrient (nitrogen, phosphorus etc) demands and a strong reliance on microbes, associated with both the leaves and roots, to sequester nutrients in the often nutrient poor marine environment (Ugarelli et al., 2017; Tarquinio et al., 2019). For example, P. oceanica root bacteria can be up to 10 times more efficient at nitrogen fixation than those found on the leaves (Lehnen et al., 2016). Several bacterial taxa known to be important for nutrient acquisition were particularly common in meadow mix samples. Alongside the occurrence of saccharolytic bacteria like Flavobacteriaceae and Spirochaeta (Leschine et al., 2006; Breznack and Warnecke, 2008; Gavriilidou et al., 2020), which likely play a role in the remineralisation and processing of organic carbon (e.g. from decaying plant matter or exudates), we also observed taxa associated with nitrogen cycling. These taxa included Rhizobiaceae (Lentilitoribacter spp.) and Arcobacteraceae (Arcobacter spp.), which are commonly associated with nitrogen fixation (Crump et al., 2018; Tedersoo et al., 2018), and anaerobic sulfate-reducing bacteria, which provide seagrasses with a significant portion of their required nitrogen (Welsh, 2000; Tarquinio et al., 2019). Most notably, like in the rhizosphere (as stated above), high abundances of Desulfobacteraceae (Desulfatitalea spp.) and Desulfobulbaceae were observed and these taxa can, in addition to nitrogen fixation, also remove toxic methanol (Dekas et al., 2014; Crump et al., 2018), a potential exudate of the growing seedling (Abanda-Nkpwatt et al., 2006). Interestingly, other γ-proteobacteria capable of utilising toxic methanol were also observed in the roots of seedlings (though in both substrates) and included the methylotrophic lineages Methylophagaceae (Methylophaga spp.) and Methylophilaceae (Methylotenera spp). Alongside their role in mitigating abiotic stresses in plants, they also contribute to nitrogen fixation (albeit indirectly through ammonia production) and secrete phytohormones, like auxins, that are required for seed germination and plant root growth (Trotsenko et al., 2001; Kumar et al., 2019).
Intriguingly, Methylophilaceae were also notably abundant in the fruit swab samples as well as the adult roots in Lady Bay, and together with Methylophaga species have been previously reported from the fruits of Halophila ovalis (Tarquinio et al., 2021). Given the distinct sources of the substrates used for the propagation of seedlings, the occurrence of such taxa in the roots of both play sand and meadow mix samples may reflect their common origins, and may be indicative of a prospective role for the reproductive tissues in supplying microbes that may support early seedling establishment as potential phytosymbionts. Nevertheless, other pertinent fruit associated taxa observed here and reported from the fruits and leaves of H. ovalis (namely Rhodobacteraceae) (Tarquinio et al., 2021), were predominantly associated with the roots of seedlings grown in play sand rather than meadow mix, suggesting that other factors likely contribute to their selection. The Rhodobacteraceae are a group of largely aerobic bacteria that exhibit diverse nutritional strategies (as photo- or chemoheterophic, or purple non-sulfur bacteria), colonise surfaces (as epiphytes) and may form symbioses with various hosts (Pujalte et al., 2014). Of the Rhodobacteraceae associated OTUs that were detected, Thalassococcus sp. (Roseobacter clade) was previously isolated from corals and can reduce nitrate to nitrite (Lee et al., 2007). Because sandy (permeable) sediments provide improved penetration of oxygen and nutrients that favour aerobic denitrification (Gao et al., 2009; Marchant et al., 2014), the occurrence of such organisms in the play sand (given its larger particle sizes compared to meadow mix) was not surprising. Nitrate reduction is an important process in terrestrial and marine ecosystems, and though often associated with N loss, is also important in N-recycling and assimilation by plants (through the conversion of nitrate to ammonium) (Herbert, 1999; Liu et al., 2022). Seagrasses obtain their nitrogen through leaf absorption and root uptake from the sediment pore water, whereby nitrate reductases catalyse the important first step in its assimilation (Touchette and Burkholder, 2000; Alexandre et al., 2004). Interestingly, in semiaquatic plants, like rice, the inoculation of seeds with denitrifying bacteria has been associated with improved growth and nitrogen use efficiency, and was attributed to the nitrate reductase activity of the bacteria (Wang et al., 2017). Given the low nutrient concentrations in Posidonia meadows (Cambridge and Hocking, 1997; Gobert et al., 2002), the relevance of such organisms in supporting the nitrogen demands of seagrass is thus intriguing but requires further investigation.
The occurrence of other putative denitrifying bacteria was also evident in the roots from seedlings grown in meadow mix. This included OTUs associated with the Ignavibacteria (Bacteroidetes), a metabolically versatile group that are facultatively anaerobic and, as revealed from the analysis of environmental MAGs, are capable of dissimilatory nitrate reduction to ammonium (DNRA) and the reduction of N2O to N2 (Bei et al., 2021). Widely distributed in paddy soils and reported from marine sediments surrounding H. ovalis (Tarquinio et al., 2021), the Ignavibacteria have been suggested to thrive in N-cycling consortia, contributing to their denitrifying capacity (Liu et al., 2012) and potentially supporting the growth of N2 fixing bacteria around the root zone. In addition, the Ignavibacteria are also reported to decompose complex polysaccharides (e.g. cellulose and hemicellulose), degrade phenylacetate and synthesise trehalose (Bei et al., 2021). Interestingly, phenylacetic acid (PAA - the conjugate acid of phenylacetate) is a growth promoting plant hormone and antimicrobial (Cook, 2019), which naturally occurs in the shoots and on the fruits and seeds of higher plants (Wightman and Lighty, 1982). When added exogenously, it induces root hair growth and upregulates the expression of pathogen defence and growth-related genes (Sumayo et al., 2018).
We observed marked differences in the root architecture of seedlings grown in different substrates. Roots from seedlings grown in play sand had strikingly longer root hairs that adhered tightly to the sediment particles, while those from meadow mix had less root hairs but had greater numbers of lateral roots and elongation zones that were noticeably larger. The formation of extensive root hairs has been previously reported for P. oceanica and suggested to represent an early anchoring mechanism on rocky surfaces, despite being also observed in sandy substrates (Badalamenti et al., 2015). Like root hairs, lateral root formation is also evident in Posidonia, with P. australis forming extensive root systems with long laterals that make up the bulk of the root length and are suggested to act as important anchoring mechanisms, particularly in response to hydrodynamic exposure (Hovey et al., 2012). The processes governing lateral root development in plants is complex, and is tightly coupled to local nitrate concentrations and hormonal (e.g. auxin and abscisic acid) signalling pathways, whereby the presence of nitrate stimulates lateral root elongation, while its accumulation at the root (shoot) interface reduces their formation (Zhang H. et al., 1999; Mantelin and Touraine, 2004). Mantelin and Touraine (2004) proposed that the occurrence of prospective plant growth promoting (denitrifying) bacteria may interfere in these mechanisms by reducing the local nitrate pool sufficiently enough to increase the numbers of lateral roots while limiting the effects on their elongation.
In rice field soils, the phenylacetate degrading capacity of Ignavibacteria is induced together with the synthesis of trehalose under micro-oxic conditions, which is thought to act to protect the cells against osmotic stress (Paul, 2018; Bei et al., 2021). This may be particularly important in the root zone of Posidonia where high amounts of plant exudates like sucrose may accumulate (as stated above) (Sogin et al., 2022), and are less likely to diffuse away from the roots in finer sediments (Gupta and Mukerj, 2002). Hypothetically, in the case of Ignavibacteria, its putative denitrifying capacity would thus also help to support lateral root branching (as needed for improved seagrass anchorage) in exchange for an increased supply of phenylacetate and other carbon sources needed to support its populations. Furthermore, given that PAA may also be synthesised by other bacteria and fungi as a defence mechanism (for a review see Cook, 2019), its relevance as a phenylacetate degrader in the control of other microbial populations in the root microenvironment may also be relevant.
Enriched in the roots of seedlings were also OTUs representing taxa associated with other biogeochemical processes that are important for supporting conditions that promote seagrass growth and productivity, including the cycling of sulfur and iron. Those connected to the sulfur cycle included representatives of the sulfate-reducing bacteria (as detailed above), as well as known or putative sulfur-oxidising bacteria, such as Arcobacteraceae, Thiovulaceae and Sedimenticolaceae (Waite et al., 2017; Crump et al., 2018; Martin et al., 2019). Reported from the roots of Zostera marina (Wang et al., 2021), sulfur-oxidising bacteria are thought to support seagrasses by reducing the toxic sulfide that accumulates from sulfate-reducing bacteria activity and is common in vegetated marine sediments (Borum et al., 2005; Lamers et al., 2013). While play sand roots also contained Arcobacteraceae and Thiovulaceae (albeit to a lesser extent), the Sedimenticolaceae were largely observed in roots from seedlings grown in meadow mix. Moreover, unlike the other sulfur-oxidising bacteria, this family was abundant in the mature roots from Lady Bay. Represented by an OTU most closely associated with Ca. Thiodiazotropha, these bacteria are symbionts of lucinid bivalves and have been reported from other seagrasses, including P. australis (Martin et al., 2020). Its association with roots is attributed to the close affiliation lucinids have with seagrasses (Van Der Geest et al., 2020) and its capacity to survive outside the host (Martin et al., 2020). Lucinid bivalves occur in P. australis meadows around Australia (Barnes and Hickman, 1999) and, although any obvious molluscs were removed from the collected sediment, their occurrence suggests that they were introduced directly from the meadow sediment when preparing the meadow mix, which is further supported by their absence from the play sand.
The colonisation of the roots by sulfur-oxidising bacteria is thought to be influenced by radial oxygen loss (ROL) from primarily young, actively growing root tips, which create oxic-anoxic microzones that also favour the growth of filamentous cable bacteria (Martin et al., 2019) – as likely represented here by the detection of Ca. Electrothrix, an organism originally detected from marine sediments (Trojan et al., 2016). Cable bacteria perform electrogenic sulfur oxidation (e-SOx) which not only mediates the impacts of toxic sulfide, but also increases the availability and mobilisation of essential nutrients like iron and phosphorous through dissolution of minerals like FeS from sediment acidification (Brodersen et al., 2017; Martin et al., 2019). Interestingly, Ca. Electrothrix was also recently reported from the root surfaces of P. australis seedlings and other aquatic plants (potentially as endophytes) where, in oxygen-releasing seedling shoots, it was accompanied by the formation of red iron oxide precipitates on the root surfaces (Scholz et al., 2021). This is consistent with our findings, where similar red deposits were observed on the seedling root surfaces, though only on those from untreated meadow mix. These iron 'plaques' have also been reported from rice and other aquatic plants, and act as a buffer against the uptake of toxic sulfide and heavy metals like cadmium and arsenic, and as a sink for phosphorous (Zhang X. et al., 1999; Liu et al., 2004; Seitaj et al., 2015; Fu et al., 2018; Scholz et al., 2021). Iron redox changes in the rhizosphere environments of rice and wetland plants are also suggested to create niches for iron bacteria (Emerson et al., 1999; Maisch et al., 2019), and may be reflected here by the detection of OTUs associated with particular magnetotactic bacteria belonging to the family Magnetospiraceae (namely Magnetovibrio sp.). As organisms that use soluble forms of iron (i.e., Fe2+ and Fe3+) for the synthesis of their magnetosomes (Lefèvre and Bazylinski, 2013), their activity is likely coupled to the acidic dissolution activity of FeS by e-SOx bacteria (Sulu-Gambari et al., 2016). While iron is important for growth of plants, it may also be toxic at high concentrations (Mehraben et al., 2008). Although there is no evidence to support this, the uptake of excess dissolved iron by magnetotactic bacteria may ameliorate potential toxicity. Furthermore, given that some Magnetovibrio species may support nitrogenase activity (Bazylinski et al., 2013), they may support seedling growth by fixing nitrogen. Given the benefits of iron additions on seagrass survival and growth rates of shoots (Marbà et al., 2007), these and other bacteria contributing to the cycling of iron and other nutrients are potentially important for seagrass health.
Despite the limited duration of the growth experiment (12 weeks), changes in the root bacterial assemblages of seedlings were evident between the sampling periods (weeks 4 and 12), indicating that these communities are dynamic during early growth. The most pronounced changes occurred in those grown in play sand, where a marked increase in species (OTU) diversity and evenness was observed between weeks 4 and 12. In contrast, the meadow mix root communities did not change significantly over the sampling period, but instead remained more similar to those belonging to the adult meadow plants, where OTUs represented a greater breadth of bacterial lineages (based on measures of delta+) compared to play sand. As mentioned above, given the differences in the origins of the substrates, such a finding was not unexpected, and likely reflects the differences in the richness and diversity of their respective bacterial 'pools' from which taxa may be sourced for supporting seedling growth. For play sand, where community diversity was already low compared to meadow mix, such marked changes in diversity were likely caused by the introduction and/or enrichment of various taxa following propagation and extended submersion of the potted medium in seawater. This may include organisms like Rhodobacteraceae and Methylophagaceae that, as discussed above, are known to be surface colonisers of the fruits and/or leaves of other seagrasses like H. ovalis, and could have established on the seedlings either before or shortly after release from the fruit (Tarquinio et al., 2021). Their marked enrichment in the play sand samples shortly following propagation (week 4), particularly when compared to the meadow mix samples, may be reflective of this and may be linked to reduced niche competition due to lower initial bacterial species richness (Chu et al., 2021).
Interestingly, while there was a lack of significant changes in measures of diversity in the meadow mix samples, prominent shifts in the abundance of various taxa were observed over the sample period. Alongside the enrichment of certain taxa in week 12 that were also predominant features in the adult meadow roots (namely Desulfobacteraceae, Bacteriodetes BD-2, Sedimenticolaceae, Spirochaetacea and uncultured Ardenticatenales), this included a number of taxa that were conversely less abundant in the adult roots (namely Flavobacteriaceae, Magnetospiraceae, Thiovulaceae and Rhizobiaceae). Together, these observations support the notion of seagrasses having transitional bacterial communities dependent on life stage and plant needs, as has already been shown in terrestrial plants (Chaparro et al., 2014; Edwards et al., 2018). For example, during early growth, the enrichment of important nitrogen fixing taxa like Rhizobiaceae may be important for the generation of above ground (leaf) tissues, which then generate a sustained supply of energy from photosynthesis, supporting both these early microbial colonisers and its further growth and development. As the plant grows, increases in organic carbon deposition from e.g. root exudation, may lead to changes in the surrounding sediments that stimulate sulfate reduction and the colonisation of the root tissues by sulfate-reducing bacteria, like Desulfobacteraceae, which, as stated above, may also serve as potential diazotrophs to further support seagrass productivity.
Indeed, while both sediment types favoured roots over shoots and displayed ratios like those reported for other Posidonia seedlings using traditional root:shoot biomass ratios (Statton et al., 2014), seedlings grown in meadow mix showed greater investments in leaf growth (based on lower pseudo root:shoot ratios) compared to those in play sand, which instead allocated more energy to growing roots. Despite both strategies being advantageous, where greater resource allocation to the roots may increase seedling anchorage and access to sedimentary nutrients, more leaves provide greater photosynthetic potential and increased access to water soluble nutrients. Nonetheless, the occurrence of such varied growth strategies was somewhat unexpected given that seed size was the most important predictor of growth in our study; a feature that likely reflects the reliance of Posidonia seedlings on initial seed reserves, as reported elsewhere (Hocking et al., 1981; Statton et al., 2013; Statton et al., 2014). Accordingly, it is tempting to postulate that the mode of growth during early establishment is thus likely driven by other substrate type and the associated microbial consortia. This may include the enrichment of nitrogen fixing bacteria together with other taxa that may support metabolic activity (e.g. the iron bacteria Magnetospiraceae, as stated above). Indeed, given the comparably reduced abundance of these and other putatively beneficial taxa (e.g. Bacteroidetes) from the roots of seedlings grown in the autoclave treated meadow mix (and a concomitant shift towards greater investment in leaves rather than roots in this treatment group), such a prospect is intriguing and requires further elucidation. This follows the ability of autoclaving to alter community composition by reducing the number of viable bacterial cells and their activity in marine sediments (Otte et al., 2018). However, given that autoclaving can also affect nutrient bioavailability (despite the lack of obvious impacts on macronutrients here), and is intrinsically coupled with the surrounding bacterial consortia (Hayat et al., 2010), further efforts are required to disentangle the mechanisms that underpin the adoption of a specific growth strategy. This may include assessing whether Posidonia seedlings potentially 'nurture' the selection and enrichment of certain microbes that may support growth under altered nutrient conditions.
Our two sample periods showed a gradual, yet significant, change to the bacterial communities that was possibly driven by the individual seedling's exudates. There is growing evidence that the microbiome is highly dynamic during a plant's seedling phase and relatively stable when the plant reaches maturity (Chaparro et al., 2014; Edwards et al., 2018). This indicates the importance of early exposure to diverse or known beneficial marine bacteria in seagrass revegetation sediment and these bacteria could stay with the plant for life and vary in response to the plant's needs. Indeed, our results highlight important effects of adding sediment from natural seagrass stands on the microbiome of P. australis seedlings, increasing diversity and adding many potentially beneficial taxa. They also document clear impact on the growth patterns of the seagrass. However, these potentially positive effects should be carefully interpreted. Local in situ restoration trials in the Greater Adelaide metropolitan area over the last four years that added organic matter show that, while sediment type may influence growth, it did not affect growth or survival (Irving et al., 2010; Tanner and Theil, 2016). While it is possible that the sudden addition of carbon in these trials disrupted the microbes and forced a rapid change in the community structure that was detrimental to the developing seedlings, this highlights that further experiments are needed to confirm the relevance of adding sediment from natural seagrass stands for conservation success. Such experiments should move beyond the 12-week stage of seedlings. Furthermore, Posidonia seedlings naturally recruit to restored Amphibolis patches (Tanner and Theil, 2016) and can grow well in the presence of macroalgae (Pereda-Briones et al., 2018). Hence, Posidonia spp. are climax species (Bryars and Neverauskas, 2004), i.e., arrive at the later stages of ecological succession, and setting root among other marine plants may allow species of this genus to gain parts of the bacterial community that has developed during the earlier successional stages. Therefore, it may be necessary to utilise the natural succession process during restoration. Nevertheless, with the findings from our limited growth experiment also pointing to the involvement of specific bacterial constituents in early growth, it is tempting to speculate on a future role for the direct application of such microbes (as potential plant probiotics) for supporting seedling vigour and improved resilience. However, given the limited cultivability of environmental microbes, renewed efforts will likely be required to overcome the inherent challenges associated with their isolation and maintenance (Kapinusova et al., 2023).
Modification of root bacterial assemblages, whether through changes in the propagation substrate or through alteration by autoclaving, appears to affect the growth of P. angustifolia seedlings. Bacterial communities therefore appear to be an important consideration for the restoration of this late-successional species. Our findings suggest that early exposure of seedlings to the bacteria found in a seagrass community may provide benefits for future Posidonia restoration. Seedlings grown in a more natural substrate that was amended with seagrass meadow sediment (a meadow mix) possessed communities that were highly diverse and more similar to the natural environment, with many taxa likely involved in essential nutrient cycling processes. In contrast, the less natural play sand sediment had lower diversity and comprised taxa that were not typical of marine systems, and which may have varied functions. Future restoration efforts therefore may benefit from using propagating substrates that incorporate natural microbial populations associated with healthy seagrass meadows to promote the formation of rhizobiomes that comprise beneficial bacteria that will support early seedling growth. Given the limited duration of our trial, the relevance of the early establishment of bacterial assemblages on seedling growth and vigour requires further elucidation (beyond 3 months) and should be supported by their performance under different experimental conditions (e.g. varied temperature, salinity) as well as in the natural environment following their transplantation as part of restoration trials. This includes the functional assessment of particular bacterial taxa in key nutrient cycling processes, with the view to identify those that may enhance restoration, perhaps in the form of an inoculum that could be added to the seedling or hessian sacks before deployment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AR: Conceptualization, Formal Analysis, Methodology, Writing – original draft, Writing – review & editing, Investigation. JT: Conceptualization, Methodology, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision. MW: Methodology, Writing – review & editing, Formal Analysis. SC: Formal Analysis, Writing – review & editing, Investigation. GK: Formal Analysis, Writing – review & editing, Methodology, Supervision. AO: Formal Analysis, Methodology, Supervision, Writing – review & editing, Conceptualization, Data curation, Project administration, Resources, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding was provided by the Adelaide & Mount Lofty Ranges Natural Resources Management Board through the South Australian Research and Development Institute as part of the seagrass rehabilitation project. Additional funding was provided by honours scholarships, awarded to AR, from the Playford Memorial Trust and the Natural Resources Management Research and Innovation Network (NRM RaIN). Supplementary funding was provided from the School of Natural and Built Environments (now UniSA STEM), University of South Australia.
We acknowledge the traditional owners of the land that our research was conducted on, the Kaurna people. A special thanks to Rob Aebi for his assistance with logistics and scouting for a suitable seagrass meadow, and Victoria Zawko for assisting with fieldwork. We also thank Ken Neubauer from Adelaide Microscopy for ESEM use and guidance. We acknowledge the contribution of AGRF, APAL, University of South Australia, SARDI, and Richgro staff for their help. AR was supported by scholarships from the Playford Memorial Trust and NRM RaIN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1278837/full#supplementary-material
Abanda-Nkpwatt D., Musch M., Tschiersch J., Boettner M., Schwab W. (2006). Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J. Exp. Bot. 57 (15), 4025–4032. doi: 10.1093/jxb/erl173
Ahmad M., Ling J., Yang Q., Sajjad W., Zhou W., Yin J., et al. (2021). Insight into bacterial community responses to polycyclic aromatic hydrocarbons and the degradation potentials of three bacterial isolates in seagrass Halophila ovalis sediments. Curr. Microbiol. 78, 4084–4097. doi: 10.1007/s00284-021-02670-y
Ahmerkamp S., Winter C., Krämer K., de Beer D., Janssen F., Friedrich J., et al. (2017). Regulation of benthic oxygen fluxes in permeable sediments of the coastal ocean. Limnol. Oceanogr. 62 (5), 1935–1954. doi: 10.1002/lno.10544
Aires T., Marbà N., Cunha R. L., Kendrick G. A., Walker D. I., Serrão E. A., et al. (2011). Evolutionary history of the seagrass genus Posidonia. Mar. Ecol. Prog. Ser. 421, 117–130. doi: 10.3354/meps08879
Alexandre A., Silva J., Santos R. (2004). The maximum nitrate reductase activity of the seagrass Zostera noltii (Hornem.) varies along its vertical distribution. J. Exp. Mar. Biol. Ecol. 302 (1), 127–135. doi: 10.1016/j.jembe.2004.02.002
Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26 (1), 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Assis R., de A. B., Polloni L. C., Patané J. S. L., Thakur S., Felestrino E. B., et al. (2017). Identification and analysis of seven effector protein families with different adaptive and evolutionary histories in plant-associated members of the Xanthomonadaceae. Sci. Rep. 7, 16133. doi: 10.1038/s41598-017-16325-1
Badalamenti F., Alagna A., Fici S. (2015). Evidence of adaptive traits to rocky substrates undermine paradigm of habitat preference of the Mediterranean seagrass Posidonia oceanica. Sci. Rep. 5, 8804. doi: 10.1038/srep08804
Barnes P. A. G., Hickman C. S. (1999). “Lucinid bivalves and marine angiosperms: a search for causal relationships,” in The Seagrass Flora and Fauna of Rottnest Island, Western Australia. Eds. Walker D. I., Wells F. E. (Perth: Western Australian Museum), 215–238.
Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67 (1), 1–48. doi: 10.18637/jss.v067.i01
Bazylinski D. A., Williams T. J., Lefevre C. T., Trubitsyn D., Fang J., Beveridge T. J., et al. (2013). Magnetovibrio blakemorei gen. nov., sp. nov., a magnetotactic bacterium (Alphaproteobacteria: Rhodospirillaceae) isolated from a salt marsh. Int. J. System. Evol. Microbiol. 63 (5), 1824–1833. doi: 10.1099/ijs.0.044453-0
Bei Q., Peng J., Liesack W. (2021). Shedding light on the functional role of the Ignavibacteria in Italian rice field soil: A meta-genomic/transcriptomic analysis. Soil Biol. Biochem. 163, 108444. doi: 10.1016/j.ecss.2014.01.009
Blandon A., and zu Ermgassen P. S. E. (2014). Quantitative estimate of commercial fish enhancement by seagrass habitat in southern Australia. Estuarine. Coast. Shelf Sci. 141, 1–8. doi: 10.1016/j.ecss.2014.01.009
Borum J., Pedersen O., Greve T. M., Frankovich T. A., Zieman J. C., Fourqurean J. W., et al. (2005). The potential role of plant oxygen and sulphide dynamics in die-off events of the tropical seagrass, Thalassia testudinum. J. Ecol. 93 (1), 148–158. doi: 10.1111/j.1365-2745.2004.00943.x
Borum J., Sand-Jensen K., Binzer T., Pedersen O., Greve T. M. (2006). “Oxygen movement in seagrasses”, in Seagrasses: Biology, Ecology and Conservation. Ed. Larkum A. W. D. (Netherlands: Springer), 255–270. doi: 10.1007/1-4020-2983-7_10
Boudouresque C.-F., Blanfuné A., Pergent G., Thibaut T. (2021). Restoration of seagrass meadows in the Mediterranean Sea: a critical review of effectiveness and ethical issues. Water 13 (8), 1034. doi: 10.3390/w13081034
Bourque A. S., Vega-Thurber R., Fourqurean J. W. (2015). Microbial community structure and dynamics in restored subtropical seagrass sediments. Aquat. Microb. Ecol. 74 (1), 43–57. doi: 10.3354/ame01725
Bray J. R., Curtis J. T. (1957). An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27 (4), 325–349. doi: 10.2307/1942268
Breznack J. A., Warnecke F. (2008). Spirochaeta cellobiosiphila sp. nov., a facultatively anaerobic, marine spirochaete. Int. J. System. Evol. Microbiol. 58 (12), 2762–2768. doi: 10.1099/ijs.0.2008/001263-0
Brodersen K. E., Koren K., Moβhammer M., Ralph P. J., Kühl M., Santner J. (2017). Seagrass-mediated phosphorous and iron solubilization in tropical sediments. Environ. Sci. Technol. 51, 14155–14163. doi: 10.1021/acs.est.7b03878
Brodersen K. E., Siboni N., Nielsen D. A., Pernice M., Ralph P. J., Seymour J., et al. (2018). Seagrass rhizosphere microenvironment alters plant-associated microbial community composition. Environ. Microbiol. 20 (8), 2854–2864. doi: 10.1111/1462-2920.14245
Bryars S., Collings G., Nayar S., Westphalen G., Miller D., O’Loughlin E., et al. (2006a). "Assessment of the effects of inputs to the Adelaide coastal waters on the meadow forming seagrasses, Amphibolis and Posidonia. Task EP 1 Final Technical Report" (Adelaide, South Australia: South Australian Research and Development Institute).
Bryars S., Miller D., Collings G., Fernandes M., Mount G., Wear R. (2006b). “Field surveys 2003-2005: Assessment of the quality of Adelaide’s coastal waters, sediments and seagrasses,” in Adelaide Coastal Waters Study Technical Report No. 14 (Adelaide: South Australian Research and Development Institute: Aquatic Sciences).
Bryars S., Neverauskas V. (2004). Natural recolonisation of seagrasses at a disused sewage sludge outfall. Aquat. Bot. 80 (4), 283–289. doi: 10.1016/j.aquabot.2004.09.001
Camarinha-Silva A., Jauregui R., Chaves-Moreno D., Oxley A. P., Schaumburg F., Becker K., et al. (2014). Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ. Microbiol. 16 (9), 2939–2952. doi: 10.1111/1462-2920.12362
Cambridge M. L., Hocking P. J. (1997). Annual primary production and nutrient dynamics of the seagrasses Posidonia sinuosa and P. australis in south-western Australia. Aquat. Bot. 59, 277–295. doi: 10.1016/S0304-3770(97)00062-4
Cambridge M. L., Kuo J. (1979). Two new species of seagrasses from Australia, Posidonia sinuosa and P. angustifolia (Posidoniaceae). Aquat. Bot. 6 (C), 307–328. doi: 10.1016/0304-3770(79)90071-8
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335. doi: 10.1038/nmeth.f.303
Carareto Alves L. M., Marcondes de Souza J. A., de Mello Varani A., de Macedo Lemos E. G. (2014). “"The family rhizobiaceae,",” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 419–437. doi: 10.1007/978-3-642-30197-1_297
Cardinale B. J., Duffy J. E., Gonzalez A., Hooper D. U., Perrings C., Venail P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486 (7401), 59–67. doi: 10.1038/nature11148
Carruthers T. J. B., Dennison W. C., Kendrick G. A., Waycott M., Walker D. I., Cambridge M. L. (2007). Seagrasses of south-west Australia: A conceptual synthesis of the world’s most diverse and extensive seagrass meadows. J. Exp. Mar. Biol. Ecol. 350, 21–45. doi: 10.1016/j.jembe.2007.05.036
Chang H.-K., Zylstra G. J. (2010). “Xanthomonads”, in Handbook of Hydrocarbon and Lipid Microbiology. Ed. Timmis K. N. (Berlin Heidelberg: Springer-Verlag), 1806–1811. doi: 10.1007/978-3-540-77587-4_131
Chaparro J. M., Badri D. V., Vivanco J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. Int. Soc. Microb. Ecol. 8 (4), 790–803. doi: 10.1038/ismej.2013.196
Chen S., Ma Z., Li S., Waigi M. G., Jiang J., Liu J., et al. (2019). Colonization of polycyclic aromatic hydrocarbon-degrading bacteria on roots reduces the risk of PAH contamination in vegetables. Environ. Int. 132, 105081. doi: 10.1016/j.envint.2019.105081
Chu X. L., Zhang Q. G., Buckling A., Castledine M. (2021). Interspecific niche competition increases morphological diversity in multi-species microbial communities. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.699190
Clarke K. R., Warwick R. M. (2001). Change in marine communities: an approach to statistical analysis and interpretation (Plymouth: Plymouth marine laboratory).
Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Conte C., Rotini A., Manfra L., D'Andrea M. M., Winters G., Migliore L. (2021). The seagrass holobiont: What we know and what we still need to disclose for its possible use as an ecological indicator. Water 13, 406. doi: 10.3390/w13040406
Cook S. D. (2019). An historical review of phenylacetic acid. Plant Cell Physiol. 60 (2), 243–254. doi: 10.1093/pcp/pcz004
Crump B. C., Wojahn J. M., Tomas F., Mueller R. S. (2018). Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00388
Cúcio C., Engelen A. H., Costa R., Muyzer G. (2016). Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00440
Dekas A. E., Chadwick G. L., Bowles M. W., Joye S. B., Orphan V. J. (2014). Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environ. Microbiol. 16 (10), 3012–3029. doi: 10.1111/1462-2920.12247
Dhariwal A., Chong J., Habib S., King I. L., Agellon L. B., Xia J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45 (W1), W180–W188. doi: 10.1093/nar/gkx295
Duarte C. M., Krause-Jensen D. (2017). Export from seagrass meadows contributes to marine carbon sequestration. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00013
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 (19), 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 (10), 996. doi: 10.1038/nmeth.2604
Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N. K., Bhatnagar S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Edwards J. A., Santos-Medellin C. M., Liechty Z. S., Nguyen B., Lurie E., Eason S., et al. (2018). Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PloS Biol. 16 (2), e2003862. doi: 10.1371/journal.pbio.2003862
Emerson D., Weiss J. V., Megonigal J. P. (1999). Iron-oxidizing bacteria are associated with ferric hydroxide precipitates (Fe-plaque) on the roots of wetland plants. Appl. Environ. Microbiol. 65 (6), 2758–2761. doi: 10.1128/aem.65.6.2758-2761.1999
Fernández-Gómez B., Richter M., Schüler M., Pinhassi J., Acinas S. G., González J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1073/pnas.1717617115
Fitzpatrick C. R., Copeland J., Wang P. W., Guttman D. S., Kotanen P. M., Johnson M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115 (6), E1157–E1165. doi: 10.1073/pnas.1717617115
Fourqurean J. W., Duarte C. M., Kennedy H., Marbà N., Holmer M., Mateo M. A., et al. (2012). Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 5 (7), 505–509. doi: 10.1038/ngeo1477
Fraser M. W., Statton J., Hovey R. K., Laverock B., Kendrick G. A. (2015). Seagrass derived organic matter influences biogeochemistry, microbial communities, and seedling biomass partitioning in seagrass sediments. Plant Soil 400 (1-2), 133–146. doi: 10.1007/s11104-015-2721-0
Fu Y., Yang X., Shen H. (2018). Root iron plaque alleviates cadmium toxicity to rice (Oryza sativa) seedlings. Ecotoxicol. Environ. Saf. 161, 534–541. doi: 10.1016/j.ecoenv.2018.06.015
Gao H., Schreiber F., Collins G., Jensen M. M., Kostka J. E., Lavik G., et al. (2009). Aerobic denitrification in permeable Wadden Sea sediments. ISME J. 4, 417–426. doi: 10.1038/ismej.2009.127
García-Martínez M., Kuo J., Kilminster K., Walker D., Rosselló-Mora R., Duarte C. M. (2005). Microbial colonization in the seagrass Posidonia spp. roots. Mar. Biol. Res. 1, 388–395. doi: 10.1080/17451000500443419
García-Martínez M., López-López A., Calleja M. L., Marbà N., Duarte C. M. (2009). Bacterial community dynamics in a seagrass (Posidonia oceanica) meadow sediment. Estuaries Coasts 32 (2), 276–286. doi: 10.1007/s12237-008-9115-y
Garcias-Bonet N., Arrieta J. M., de Santana C. N., Duarte C. M., Marba N. (2012). Endophytic bacterial community of a Mediterranean marine angiosperm (Posidonia oceanica). Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00342
Garrity G. M., Bell J. A., Lilburn T. (2005). “Order V. Thiotrichales ord. nov,” in Bergey’s Manual of Systematic Bacteriology: (The Proteobacteria), part B (The Gammaproteobacteria), 2nd Edn, vol. 131 . Eds. Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (New York, NY: Springer). doi: 10.1007/0-387-28022-7_5
Gavriilidou A., Gutleben J., Versluis D., Forgiarini F., van Passel M. W. J., Ingham C. J., et al. (2020). Comparative genomic analysis of Flavobacteriaceae: insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genomics 21, 569. doi: 10.1186/s12864-020-06971-7
Gobert S., Laumont N., Bouquegneau J.-M. (2002). Posidonia oceanica meadow: a low nutrient high chlorophyll (LNHC) system? BMC Ecol. 2, 9. doi: 10.1186/1472-6785-2-9
Gupta R., Mukerj K. G. (2002). “Root exudate—Biology”, in Techniques in Mycorrhizal Studies. Eds. Mukerji K. G., Manoharachary C., Chamola B. P. (Netherlands: Kluwer Academic Publishers), 103–31.
Hatcher A. I., Larkum A. W. D. (1982). The effects of short term exposure to Bass Strait Crude Oil and Corexit 8667 on benthic community metabolism in Posidonia australis Hook.f. dominated microcosms. Aquat. Bot. 12, 219–227. doi: 10.1016/0304-3770(82)90018-3
Hayat R., Ali S., Amara U., Khalid R., Ahmed I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60 (4), 579–598. doi: 10.1007/s13213-010-0117-1
Hector A., Bagchi R. (2007). Biodiversity and ecosystem multifunctionality. Nature 448 (7150), 188–190. doi: 10.1038/nature05947
Hemminga M. A., Marba N., Stapel J. (1999). Leaf nutrient resorption, leaf lifespan and the retention of nutrients in seagrass systems. Aquat. Bot. 65 (1-4), 141–158. doi: 10.1016/S0304-3770(99)00037-6
Herbert R. A. (1999). Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 23, 563–590. doi: 10.1111/j.1574-6976.1999.tb00414.x
Hocking P. J., Cambridge M. L., McComb A. J. (1981). The nitrogen and phosphorus nutrition of developing plants of two seagrasses, Posidonia australis and Posidonia sinuosa. Aquat. Bot. 11 (C), 245–261. doi: 10.1016/0304-3770(81)90064-4
Hovey R. K., Cambridge M. L., Kendrick G. A. (2012). Season and sediment nutrient additions affect root architecture in the temperate seagrasses Posidonia australis and P. sinuosa. Mar. Ecol. Prog. Ser. 446, 23–30. doi: 10.3354/meps09483
Hurtado-McCormick V., Kahlke T., Petrou K., Jeffries T., Ralph P. J., Seymour J. R. (2019). Regional and microenvironmental scale characterization of the Zostrea muelleri seagrass microbiome. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01011
Imhoff J. F. (2014). “The family chromatiaceae”, in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 75–86. doi: 10.1007/978-3-642-38922-1_295
Irving A., Tanner J., Seddon S., Miller D., Collings G., Wear R., et al. (2010). Testing alternate ecological approaches to seagrass rehabilitation: links to life-history traits. J. Appl. Ecol. 47 (5), 1119–1127. doi: 10.HH/j.1365-2664.2010.01852.x
Jankowska E., Jankowska K., Wlodarska-Kowalczuk M. (2015). Seagrass vegetation and meiofauna enhance the bacterial abundance in the Baltic Sea sediments (Puck Bay). Environ. Sci. pollut. Res. 22, 14372–14378. doi: 10.1007/s11356-015-5049-7
Jensen S. I., Kuhl M., Prieme A. (2007). Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 62 (1), 108–117. doi: 10.1111/j.1574-6941.2007.00373.x
Jørgensen B. B., Findlay A., Pellerin A. (2019). The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00849
Jose P., Sundari I., Sivakala K., Jebakumar S. (2014). Molecular phylogeny and plant growth promoting traits of endophytic bacteria isolated from roots of seagrass Cymodocea serrulata. Indian J. Of Geo Marine Sci. 43 (4), 571–579.
Kapinusova G., Lopez Marine M. A., Uhlik O. (2023). Reaching unreachables: Obstacles and successes of microbial cultivation and their reasons. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1089630
Kessler A. J., Chen Y. J., Waite D. W., Hutchinson T., Koh S., Popa M. E., et al. (2019). Bacterial fermentation and respiration processes are uncoupled in anoxic permeable sediments. Nat. Microbiol. 4, 1014–1023. doi: 10.1038/s41564-019-0391-z
Kilminster K., Garland J. (2009). Aerobic heterotrophic microbial activity associated with seagrass roots: effects of plant type and nutrient amendment. Aquat. Microb. Ecol. 57, 57–68. doi: 10.3354/ame01332
Kotoky R., Rajkumari J., Pandey P. (2018). The rhizosphere microbiome: Significance in rhizoremediation of polyaromatic hydrocarbon contaminated soil. J. Environ. Manage. 217, 858–870. doi: 10.1016/j.jenvman.2018.04.022
Kuever J. (2014a). “The family desulfobacteraceae”, in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 45–74. doi: 10.1007/978-3-642-39044-9_266
Kuever J. (2014b). “The family desulfobulbaceae,” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 75–86. doi: 10.1007/978-3-642-39044-9_267
Kumar M., Kour D., Yadav A. N., Saxena R., Rai P. K., Jyoti A., et al. (2019). Biodiversity of methylotrophic microbial communities and their potential role in mitigation of abiotic stresses in plants. Biologia 74, 287–308. doi: 10.2478/s11756-019-00190-6
Kuznetsova A., Brockhoff P. B., Christensen R. H. B. (2017). lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 82 (13), 1–26. doi: 10.18637/jss.v082.i13
Lamers L. P., Govers L. L., Janssen I. C., Geurts J. J., van der Welle M. E., Van Katwijk M. M., et al. (2013). Sulfide as a soil phytotoxin-a review. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00268
Lee O. O., Tsoi M. Y. M., Li X., Wong P.-K., Qian P.-Y. (2007). Thalassococcus halodurans gen. nov., sp. nov., a novel halotolerant member of the Roseobacter clade isolated from the marine sponge Halichondria panicea at Friday Harbor, USA. Int. J. System. Evol. Microbiol. 57 (8), 1919–1924. doi: 10.1099/ijs.0.64801-0
Lefèvre C. T., Bazylinski D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77 (3), 497–526. doi: 10.1128/MMBR.00021-13
Legrand T. P. R. A., Catalano S. R., Wos-Oxley M. L., Stephens F., Landos M., Bansemer M. S., et al. (2018). The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02664
Lehnen N., Marchant H. K., Schwedt A., Milucka J., Lott C., Weber M., et al. (2016). High rates of microbial dinitrogen fixation and sulfate reduction associated with the Mediterranean seagrass Posidonia oceanica. System. Appl. Microbiol. 39 (7), 476–483. doi: 10.1016/j.syapm.2016.08.004
Leschine S., Paaster B. J., Canale-Parola E. (2006). “"Free-living saccharolytic spirochetes: the genus spirochaeta",” in The Prokaryotes. Eds. Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (New York: Springer), 195–210. doi: 10.1007/0-387-30747-8_7
Lidbury I. D. E. A., Borsetto C., Murphy A. R. J., Bottril A., Jones A. M. E., Bending G. D., et al. (2021). Niche-adaptation in plant associated Bacteroidetes favours specialisation in organic phosphorous mineralisation. ISME J. 15, 1040–1055. doi: 10.1038/s41396-020-00829-2
Liu Z., Frigaard N.-U., Vogl K., Iino T., Ohkuma M., Overmann J., et al. (2012). Complete genome of Ignavibacterium album, a metabolically versatile, flagellated, facultative anaerobe from the phylum Chlorobi. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00185
Liu X., Hu B., Chu C. (2022). Nitrogen assimilation in plants: current status and future prospects. J. Genet. Genomics 49 (5), 394–404. doi: 10.1016/j.jgg.2021.12.006
Liu W. J., Zhu Y. G., Smith F., Smith S. (2004). Do phosphorous nutrition and iron plaque alter arsenate (As) uptake by rice seedlings in hydroponic culture? New Phytol. 162, 481–488. doi: 10.1111/j.1469-8137.2004.01035.x
Lotrario J. B., Stuart B. J., Lam T., Arands R. R., O'Connor O. A., Kosson D. S. (1995). Effects of sterilization methods on the physical characteristics of soil: Implications for sorption isotherm analyses. Bull. Environ. Contamination Toxicol. 54 (5), 668–675. doi: 10.1007/bf00206097
Maher W. A., Aislabie J. (1992). Polycyclic aromatic hydrocarbons in nearshore marine sediments of Australia. Sci. Total Environ. 112, 143–164. doi: 10.1016/0048-9697(92)90184-T
Maisch M., Lueder U., Kappler A., Schmidt C. (2019). Iron lung: how rice roots induce iron redox changes in the rhizosphere and create niches for microaerophilic Fe(II)-oxidizing bacteria. Environ. Sci. Technol. Lett. 6 (10), 600–605. doi: 10.1021/acs.estlett.9b00403
Mantelin S., Touraine B. (2004). Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J. Exp. Bot. 55 (394), 27–34. doi: 10.1093/jxb/erh010
Marbà N., Calleja M. L., Duarte C. M., Álvarez E., Díaz-Almela E., Holmer M. (2007). Iron additions reduce sulfide intrusion and reverse seagrass (Posidonia oceanica) decline in carbonate sediments. Ecosystems 10 (5), 745–756. doi: 10.1007/s10021-007-9053-8
Marchant H. K., Lavik G., Holtappels M., Kuypers M. M. M. (2014). The fate of nitrate in intertidal permeable sediments. PloS One 9 (8), e104517. doi: 10.1371/journal.pone.0104517
Markovski M., Najdek M., Herndl G. J., Korlević M. (2022). Compositional stability of sediment microbial communities during a seagrass meadow decline. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.966070
Martin B. C., Bougoure J., Ryan M. H., Bennett W. W., Colmer T. D., Joyce N. K., et al. (2019). Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J. 13 (3), 707–719. doi: 10.1038/s41396-018-0308-5
Martin B. C., Gleeson D., Statton J., Siebers A. R., Grierson P., Ryan M. H., et al. (2017). Low light availability alters root exudation and reduces putative beneficial microorganisms in seagrass roots. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02667
Martin B. C., Middleton J. A., Fraser M. W., Marshall I. P. G., Scholz V. V., Hausl B., et al. (2020). Cutting out the middle clam: lucinid endosymbiotic bacteria are also associated with seagrass roots worldwide. ISME J. 14 (11), 2901–2905. doi: 10.1038/s41396-020-00771-3
Martinez-Garcia E., Carlsson M. S., Sanchez-Jerez P., Sánchez-Lizaso J. L., Sanz-Lazaro C., Holmer M. (2015). Effect of sediment grain size and bioturbation on decomposition of organic matter from aquaculture. Biogeochemistry 125, 133–148. doi: 10.1007/s10533-015-0119-y
Martiny A. C., Treseder K., Pusch G. (2013). Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7 (4), 830–838. doi: 10.1038/ismej.2012.160
Mehraben P., Zadeh A. A., Sadeghipour H. R. (2008). Iron toxicity in rice (Oryza sativa L.), under different potassium nutrition. Asian J. Plant Sci. 7, 251–259. doi: 10.3923/ajps.2008.251.259
Mohr W., Lehnen N., Ahmerkamp S., Marchant H. K., Graf J. S., Tschitschko B., et al. (2021). Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 600, 105–109. doi: 10.1038/s41586-021-04063-4
Mußman M., Pjevac P., Kruger K., Dyksma S. (2017). Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 11 (5), 1276–1281. doi: 10.1038/ismej.2016.185
Nordlund L. M., Koch E. W., Barbier E. B., Creed J. C. (2016). Seagrass ecosystem services and their variability across genera and geographical regions. PloS One 11 (10), 1–23. doi: 10.1371/journal.pone.0163091
Ort B. S., Cohen C. S., Boyer K. E., Reynolds L. K., Tam S. M., Wyllie-Echeverria S. (2014). Conservation of eelgrass (Zostera marina) genetic diversity in a mesocosm-based restoration experiment. PloS One 9 (2), e89316. doi: 10.1371/journal.pone.0089316
Otte J. M., Blackwell N., Soos V., Rughoft S., Maisch M., Kappler A., et al. (2018). Sterilization impacts on marine sediment—Are we able to inactivate microorganisms in environmental samples? FEMS Microbiol. Ecol. 94 (12), fiy189. doi: 10.1093/femsec/fiy189
Paul M. (2018). The protection of sandy shores - Can we afford to ignore the contribution of seagrass? Mar. pollut. Bull. 134, 152–159. doi: 10.1016/j.marpolbul.2017.08.012
Pereda-Briones L., Tomas F., Terrados J. (2018). Field transplantation of seagrass (Posidonia oceanica) seedlings: Effects of invasive algae and nutrients. Mar. pollut. Bull. 134, 160–165. doi: 10.1016/j.marpolbul.2017.09.034
Pérez-Jaramillo J. E., Carrion V. J., de Hollander M., Raaijmakers J. M. (2018). The wild side of plant microbiomes. Microbiome 6 (1), 143. doi: 10.1186/s40168-018-0519-z
Pujalte M. J., Lucena T., Ruvira M. A., Arahal D. R., Macian M. C. (2014). “The family rhodobacteraceae,” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 439–512. doi: 10.1007/978-3-642-30197-1_377
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (Database issue), D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2020). R: A language and environment for statistical computing. R Foundation Stat. Computing (Vienna, Austria). Available at: https://www.R-project.org/.
Rayment G. E., Lyons D. J. (2011). Soil chemical methods Australasia (Collingwood, Vic: CSIRO Publishing).
Reed M. L., Warner B. G., Glick B. R. (2005). Plant growth-promoting bacteria facilitate the growth of the common reed Phragmites australisin the presence of copper or polycyclic aromatic hydrocarbons. Curr. Microbiol. 51 (6), 425–429. doi: 10.1007/s00284-005-4584-8
Runcie J., Macinnis-Ng C., Ralph P. (2005). “The toxic effects of petrochemicals on seagrasses: Literature Review,” in Report prepared by Institute for Water and Environmental Resource Management (Canberra: University of Technology, Sydney, for Australian Maritime Safety Authority). Available at: https://www.amsa.gov.au/file/334/download?token=zaGzDLLn.
Saddler G. S., Bradbury J. F. (2015). “"Xanthomonadaceae fam. nov.",” in Bergey's Manual of Systematics of Archaea and Bacteria. eds. Trujillo M. E., Dedysh S., DeVos P., Hedlund B., Kämpfer P., Rainey F. A., et al (John Wiley & Sons). doi: 10.1002/9781118960608.fbm00237
Sampaio C. J. S., de Souza J. R. B., Damião A. O., Bahiense T. C., Roque M. R. A. (2019). Biodegradation of polycyclic aromatic hydrocarbons (PAHs) in a diesel oil-contaminated mangrove by plant growth-promoting rhizobacteria. 3 Biotech. 9 (4), 155. doi: 10.1007/s13205-019-1686-8
Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676. doi: 10.1038/nmeth.2019
Scholz V. V., Martin B. C., Meyer R., Schramm A., Fraser M. W., Nielsen L. P., et al. (2021). Cable bacteria at oxygen-releasing roots of aquatic plants: a widespread and diverse plant–microbe association. New Phytol. 232, 2138–2151. doi: 10.1111/nph.17415
Seitaj D., Schauer R., Sulu-Gambari F., Hidalgo-Martinez S., Malkin S. Y., Burdorf L. D. W., et al. (2015). Cable bacteria generate a firewall against euxinia in seasonally hypoxic basins. Proc. Natl. Acad. Sci. U.S.A. 112, 13278–13283. doi: 10.1073/pnas.1510152112
Sizova M., Panikov N. (2007). Polaromonas hydrogenivorans sp. nov., a psychrotolerant hydrogen-oxidizing bacterium from Alaskan soil. Int. J. System. Evol. Microbiol. 57 (3), 616–619. doi: 10.1099/ijs.0.64350-0
Sogin E. M., Michellod D., Gruber-Vodicka H. R., Bourceau P., Geier B., Meier D. V., et al. (2022). Sugars dominate the seagrass rhizosphere. Nat. Ecol. Evol. 6, 866–877. doi: 10.1038/s41559-022-01740-z
Statton J., Cambridge M. L., Dixon K. W., Kendrick G. A. (2013). Aquaculture of Posidonia australis seedlings for seagrass restoration programs: effect of sediment type and organic enrichment on growth. Restor. Ecol. 21 (2), 250–259. doi: 10.1111/j.1526-100X.2012.00873.x
Statton J., Kendrick G. A., Dixon K. W., Cambridge M. L. (2014). Inorganic nutrient supplements constrain restoration potential of seedlings of the seagrass, Posidonia australis. Restor. Ecol. 22 (2), 196–203. doi: 10.1111/rec.12072
Sulu-Gambari F., Seitaj D., Behrends T., Banerjee D., Meysman F. J. R., Slomp C. P. (2016). Impact of cable bacteria on sedimentary iron and manganese dynamics in a seasonally-hypoxic marine basin. Geochemica Cosmochimica Acta 192, 49–69. doi: 10.1016/j.gca.2016.07.028
Sumayo M. S., Son J.-S., Ghim S.-Y. (2018). Exogenous application of phenylacetic acid promotes root hair growth and induces the systemic resistance of tobacco against bacterial soft-rot pathogen Pectobacterium carotovorum subsp. carotovorum. Funct. Plant Biol. 45 (11), 1119–1127. doi: 10.1071/FP17332
Sun F., Zhang X., Zhang Q., Liu F., Zhang J., Gong J. (2015). Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and culfur cycling. Appl. Environ. Microbiol. 81 (19), 6901–6914. doi: 10.1128/AEM.01382-15
Tan Y. M., Dalby O., Kendrick G. A., Statton J., Sinclair E. A., Fraser M. W., et al. (2020). Seagrass restoration is possible: Insights and lessons from Australia and New Zealand. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00617
Tan W. A., Parales R. E. (2019). “"Hydrocarbon degradation by betaproteobacteria",” in Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes, Handbook of Hydrocarbon and Lipid Microbiology. Ed. McGenity T. J. (Cham: Springer), 125–141. doi: 10.1007/978-3-030-14796-9_18
Tanner J. E. (2015). Restoration of the seagrass Amphibolis Antarctica - temporal variability and long-term success. Estuaries Coasts 38, 668–678. doi: 10.1007/s12237-014-9823-4
Tanner J. E. (2023). “"New life for our coastal environment seagrass rehabilitation project: 2019-2022",” in Final Report to the Department for Environment and Water (Adelaide: South Australian Research and Development Institute (Aquatic Sciences). SARDI Publication No. F2022/000383-1. SARDI Research Report Series No. 1167.
Tanner J. E., Irving A. D., Fernandes M., Fotheringham D., McArdle A., Murray-Jones S. (2014). Seagrass rehabilitation off metropolitan Adelaide: a case study of loss, action, failure and success. Ecol. Manage. Restor. 15 (3), 168–179. doi: 10.1111/emr.12133
Tanner J. E., Theil M. (2016). "Adelaide Seagrass Rehabilitation Project: 2014-2016" (Adelaide, South Australia: South Australian Research and Development Institute).
Tanner J. E., Theil M. (2019). “"Adelaide seagrass rehabilitation project: 2017-2019",” in Final report prepared for the Adelaide and Mount Lofty Ranges Natural Resources Management Board (Adelaide: South Australian Research and Development Institute (Aquatic Sciences). SARDI Publication No. F2009/000210-3. SARDI Research Report Series No. 1025.
Tarquinio F., Attlan O., Vanderklift M. A., Berry O and Bissett A. (2021). Distinct endophytic bacterial communities inhabiting seagrass seeds. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.703014
Tarquinio F., Hyndes G. A., Laverock B., Koenders A., Sawstrom C. (2019). The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 366 (6), fnz057. doi: 10.1093/femsle/fnz057
Tedersoo L., Laanisto L., Rahimlou S., Toussaint A., Hallikma T., Pärtel M. (2018). Global database of plants with root-symbiotic nitrogen fixation: Nod DB. J. Veget. Sci. 29 (3), 560–568. doi: 10.1111/jvs.12627
Teng Y., Xu Y., Wang X., Christie P. (2019). Function of biohydrogen metabolism and related microbial communities in environmental bioremediation. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00106
Thrall P. H., Millsom D. A., Jeavons A. C., Waayers M., Harvey G. R., Bagnall D. J., et al. (2005). Seed inoculation with effective root-nodule bacteria enhances revegetation success. J. Appl. Ecol. 42 (4), 740–751. doi: 10.1111/j.1365-2664.2005.01058.x
Touchette B. W., Burkholder J. M. (2000). Review of nitrogen and phosphorous metabolism in seagrasses. J. Exp. Mar. Biol. Ecol. 250 (1-2), 133–167. doi: 10.1016/S0022-0981(00)00195-7
Trojan D., Schreiber L., Bjerg J. T., Bøggild A., Yang T., Kjeldsen K. U., et al. (2016). A taxonomic framework for cable bacteria and proposal of the candidate genera Electrothrix and Electronema. System. Appl. Microbiol. 39, 297–306. doi: 10.1016/j.syapm.2016.05.006
Trotsenko Y. A., Ivanova E. G., Doronina N. V. (2001). Aerobic methylotrophic bacteria as phytosymbionts. Microbiology 70, 623–632. doi: 10.1023/A:1013167612105
Ugarelli K., Chakrabarti S., Laas P., Stingl U. (2017). The seagrass holobiont and its microbiome. Microorganisms 5 (4), 81. doi: 10.3390/microorganisms5040081
Unsworth R. K. F., Bertelli C. M., Cullen-Unsworth L. C., Esteban N., Jones B. L., Lilley R., et al. (2019). Sowing the seeds of seagrass recovery using hessian bags. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00311
Valdez S. R., Zhang Y. S., van der Heide T., Vanderklift M. A., Tarquinio F., Orth R. J., et al. (2020). Positive ecological interactions and the success of seagrass restoration. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00091
Van Der Geest M., van der Heide T., Holmer M., De Wit R. (2020). First field-based evidence that the seagrass-lucinid mutualism can mitigate sulfide stress in seagrasses. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00011
Vanderklift M. A., Doropoulos C., Gorman D., Leal I., Minne A. J. P., Statton J., et al. (2020). Using propagules to restore coastal marine ecosystems. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00724
Van Katwijk M. M., Thorhaug A., Marbà N., Orth R. J., Duarte C. M., Kendrick G. A., et al. (2016). Global analysis of seagrass restoration: the importance of large-scale planting. J. Appl. Ecol. 53 (2), 567–578. doi: 10.1111/1365-2664.12562
Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., et al. (2017). Comparative genomic analysis of the Class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00682
Wang L., English M. K., Tomas F., Mueller R. S. (2021). Recovery and community succession of the Zostera marina rhizobiome after transplantation. Appl. Environ. Microbiol. 87, e02326–e02320. doi: 10.1128/AEM.02326-20
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 (16), 5261–5267. doi: 10.1128/AEM.00062-07
Wang W., Zhai Y., Cao L., Tan H., Zhang R. (2017). Improvement of rice seedling growth and nitrogen use efficiency by seed inoculation with endophytic denitrifiers. Environ. Sci. pollut. Res. 24 (16), 14477–14483. doi: 10.1007/s11356-017-9064-8
Waycott M., Duarte C., Carruthers J., Orth R., Dennison W., Olyarnik S., et al. (2009). Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. U. States America 106 (30), 12377–12381. doi: 10.1073/pnas.0905620106
Welsh D. T. (2000). Nitrogen fixation in seagrass meadows: Regulation, plant–bacteria interactions and significance to primary productivity. Ecol. Lett. 3 (1), 58–71. doi: 10.1046/j.1461-0248.2000.00111.x
Wentworth C. K. (1922). A scale of grade and class terms for classic sediments. J. Geol. 30 (5), 377–392.
Wightman F., Lighty D. L. (1982). Identification of phenylacetic acid as a natural auxin in the shoots of higher plants. Physiologia Plantarum 55 (1), 17–24. doi: 10.1111/j.1399-3054.1982.tb00278.x
Willems A., Busse J., Goor M., Pot B., Falsen E., Jantzen E., et al. (1989). Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (formerly Pseudomonas pseudoflava and “Pseudomonas carboxy do flava”), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis). Int. J. System. Evol. Microbiol. 39 (3), 319–333. doi: 10.1099/00207713-39-3-319
Willems A., Falsen E., Pot B., Jantzen E., Hoste B., Vandamme P., et al. (1990). Acidovorax, a new genus for Pseudomonas facilis, Pseudomonas delafieldii, E. Falsen (EF) group 13, EF group 16, and several clinical isolates, with the species Acidovorax facilis comb. nov., Acidovorax delafieldii comb. nov., and Acidovorax temperans sp. nov. Int. J. System. Bacteriology 40 (4), 384–398. doi: 10.1099/00207713-40-4-384
York P. H., Smith T. M., Coles R. G., McKenna S. A., Connolly R. M., Irving A. D., et al. (2017). Identifying knowledge gaps in seagrass research and management: An Australian perspective. Mar. Environ. Res. 127, 163–172. doi: 10.1016/j.marenvres.2016.06.006
Zhang P. D., Fang C., Liu J., Xu Q., Li W. T., Liu Y. S. (2015). An effective seed protection method for planting Zostera marina (eelgrass) seeds: Implications for their large-scale restoration. Mar. pollut. Bull. 95, 89–99. doi: 10.1016/j.marpolbul.2015.04.036
Zhang H., Jennings A., Barlow P. W., Forde B. G. (1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. U.S.A. 96, 6529–6534. doi: 10.1073/pnas.96.11.6529
Zhang J., Kobert K., Flouri T., Stamatakis A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30 (5), 614–620. doi: 10.1093/bioinformatics/btt593
Zhang X., Liu S., Jiang Z., Wu Y., Huang X. (2022). Gradient of microbial communities around seagrass roots was mediated by sediment grain size. Ecosphere 13 (2), e3942. doi: 10.1002/ecs2.3942
Zhang X., Zhang F., Mao D. (1999). Effect of iron plaque outside roots on nutrient uptake by rice (Oryza sativa L.): Phosphorus uptake. Int. J. Plant-Soil Relat. 209 (2), 187–192. doi: 10.1023/A:1004505431879
Zhou Y., Lai R., Li W. J. (2014). “"The family solimonadaceae",” in The Prokaryotes. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 627–638. doi: 10.1007/978-3-642-38922-1_373
Keywords: seagrass restoration, Posidonia, bacterial communities, meadow sediment, rhizosphere, rhizoplane, seed starch, seedling growth
Citation: Randell AS, Tanner JE, Wos-Oxley ML, Catalano SR, Keppel G and Oxley APA (2023) The root of influence: root-associated bacterial communities alter resource allocation in seagrass seedlings. Front. Mar. Sci. 10:1278837. doi: 10.3389/fmars.2023.1278837
Received: 22 August 2023; Accepted: 15 November 2023;
Published: 30 November 2023.
Edited by:
Juan D. Gaitan-Espitia, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Bovern Suchart Arromrak, The University of Hong Kong, Hong Kong SAR, ChinaCopyright © 2023 Randell, Tanner, Wos-Oxley, Catalano, Keppel and Oxley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew P. A. Oxley, YW5kcmV3Lm94bGV5QGRlYWtpbi5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.