- 1Institute of Marine Environment and Ecology, National Taiwan Ocean University, Keelung, Taiwan
- 2Center of Excellence for the Oceans, National Taiwan Ocean University, Keelung, Taiwan
Many anthozoans, e.g., sea anemones and corals, have remarkable capacities for asexual reproduction and regeneration, creating complete individual polyps from pieces of parental polyp tissue. Notably, polyps produced asexually (or regenerated) from somatic tissues that originally contain no gonads are capable of sexual reproduction. However, how somatically derived polyps can reproduce remains largely unexplored. Here, using a sea anemone, Exaiptasia diaphana, we show that potential germline progenitor cells are present in the extra-gonadal tissues, and that they are allocated to newly produced individuals during asexual reproduction/regeneration. Histological analysis of E. diaphana demonstrated that polyps asexually produced from pedal lacerates can create mature gametes. Amputation experiments showed that polyps regenerated from the head, which originally lacked gonadal tissue, formed gonads and initiated gametogenesis. Immunohistochemical analysis with a germline marker, vasa, demonstrated the presence of vasa-positive cells in various polyp tissues. These findings suggest that vasa-positive cells in extra-gonadal tissues may be a reservoir of germline progenitor cells to ensure sexual reproductive capacity in individuals produced asexually or regenerated from somatic tissues.
1 Introduction
Anthozoans (corals, sea anemones, sea pens, sea pansies, etc.) are marine organisms that belong to the phylum Cnidaria, a group of animals with specialized stinging cells called cnidocytes (Harrison and Wallace, 1990; Ruppert et al., 2003). The class Anthozoa comprises more than 5,000 species and forms the largest group in the phylum. They are widely distributed globally from shallow waters in the tropics and subtropics to deep waters in cool temperate regions (Harrison and Wallace, 1990; Ruppert et al., 2003; Shikina and Chang, 2018). Reef structures formed by corals serve as nurseries for many marine animals and have great ecological and economic importance (Odum and Odum, 1955).
Many anthozoans have the ability to produce genetically identical individuals (polyps) through asexual reproduction, allowing them to increase colony or population sizes relatively rapidly (Harrison and Wallace, 1990; Bocharova and Kozevich, 2011). When polyps reach a certain size (or age), they initiate sexual reproduction to produce genetically diverse offspring. Gametes are generally produced at specific sites in gastrodermal tissues, also called gonads (Harrison and Wallace, 1990; Bocharova and Kozevich, 2011). Fertilized eggs develop into free-swimming larvae, metamorphose into primary polyps after settling on appropriate substrates, and then initiate asexual reproduction to form colonies (Harrison and Wallace, 1990; Shikina and Chang, 2018). Anthozoans lack a medusa phase and live exclusively as polyps (Harrison and Wallace, 1990; Technau and Steele, 2011; Shikina and Chang, 2018).
Anthozoans have a remarkable capacity for asexual reproduction and regeneration. Various modes of asexual reproduction have been reported, with budding and fission (longitudinal or transverse) being the major modes (Harrison and Wallace, 1990; Bocharova and Kozevich, 2011; Bocharova, 2016). In addition, in many cases, complete polyps are generated from parts of parental somatic tissues such as tentacles, pedal disks, and coenosarc (Bocharova and Kozevich, 2011; Bocharova, 2016; Toh and Ng, 2016). Sexual reproductive capacity is also observed in polyps that were asexually produced or were regenerated from somatic tissues that originally contained no gonads. However, how polyps derived from such somatic tissues can reproduce sexually has not yet been explored in depth, and underlying cellular mechanisms remain largely unknown.
One of the prerequisites for sexual reproduction is the presence of germline cells or cells that are capable of producing germline cells, such as stem cells. In this study, we hypothesized that such cells are present in extra-gonadal tissues, and that they are transmitted to newly developing polyps during asexual reproduction or regeneration to ensure sexual reproductive capacity. To verify this hypothesis, this study focused on a gonochoric sea anemone, Exaiptasia diaphana, previously known as Exaiptasia pallida or Aiptasia pallida. Exaiptasia diaphana is distributed globally in temperate and tropical marine shallow-water environments (Grajales and Rodríguez, 2014). Exaiptasia diaphana is easy to culture in the laboratory, and it is an excellent model to study cnidarian-dinoflagellate symbioses (Weis et al., 2008). Importantly, E. diaphana actively reproduces asexually by pedal disc laceration (Hunter, 1984; Clayton, 1985). After growth, newly created polyps undergo sexual reproduction, even under artificial rearing conditions (Grawunder et al., 2015). Therefore, this animal has desirable characteristics as a model to study the sexual reproductive capacity of polyps derived from somatic tissues.
In this study, we first established clonal strains of E. diaphana collected from coastal areas of Northern Taiwan, and characterized their asexual and sexual reproductive capacity. We then established a gonad transcriptome to select a gene for use as a germline/stem cell marker, and examined its expression profiles. Finally, to verify our hypothesis, we investigated the presence of germline or stem cells in various polyp tissues using the identified marker.
2 Materials and methods
2.1 Sampling and cultivation of E. diaphana
Sampling of E. diaphana was performed in a ditch (25°09′04.5′′N, 121°46′41.1′′ E) near the Aquatic Animal Center at National Taiwan Ocean University (NTOU), northern Taiwan (Figures 1A-C). Four large individuals (5~7 cm in height) were collected with a spoon from the ditch wall, and were transported to our laboratory at NTOU. The four sea anemones were rinsed several times with seawater, and cultured separately in four 40-L aquaria under T5 lights (6500k; light intensity, 20-40 μmol/m2/s, 12-hr light/dark cycle). Each aquarium was equipped with an external filtration system, and the water temperature was set at 28°C. Artemia naupli were provided as food, five times a week, and one-third of the seawater in the tanks was replaced every week. We then established two clonal strains (one male and one female) by allowing them to reproduce asexually. Developing testes/ovaries were examined in dissected polyps under a stereo microscope (EZ4, Leica Microsystems, Mannheim, Germany). To observe development of lacerated pieces (pedal fragment) into small anemones, collected pieces were cultured in 6-well plates containing 5 mL of 0.22 μm-filtered seawater per well. Micrographs were taken with a digital camera (RX100II, Sony, Tokyo, Japan) under the stereo microscope. All experiments were carried out in accordance with principles and procedures approved by the Institutional Animal Care and Use Committee, NTOU.
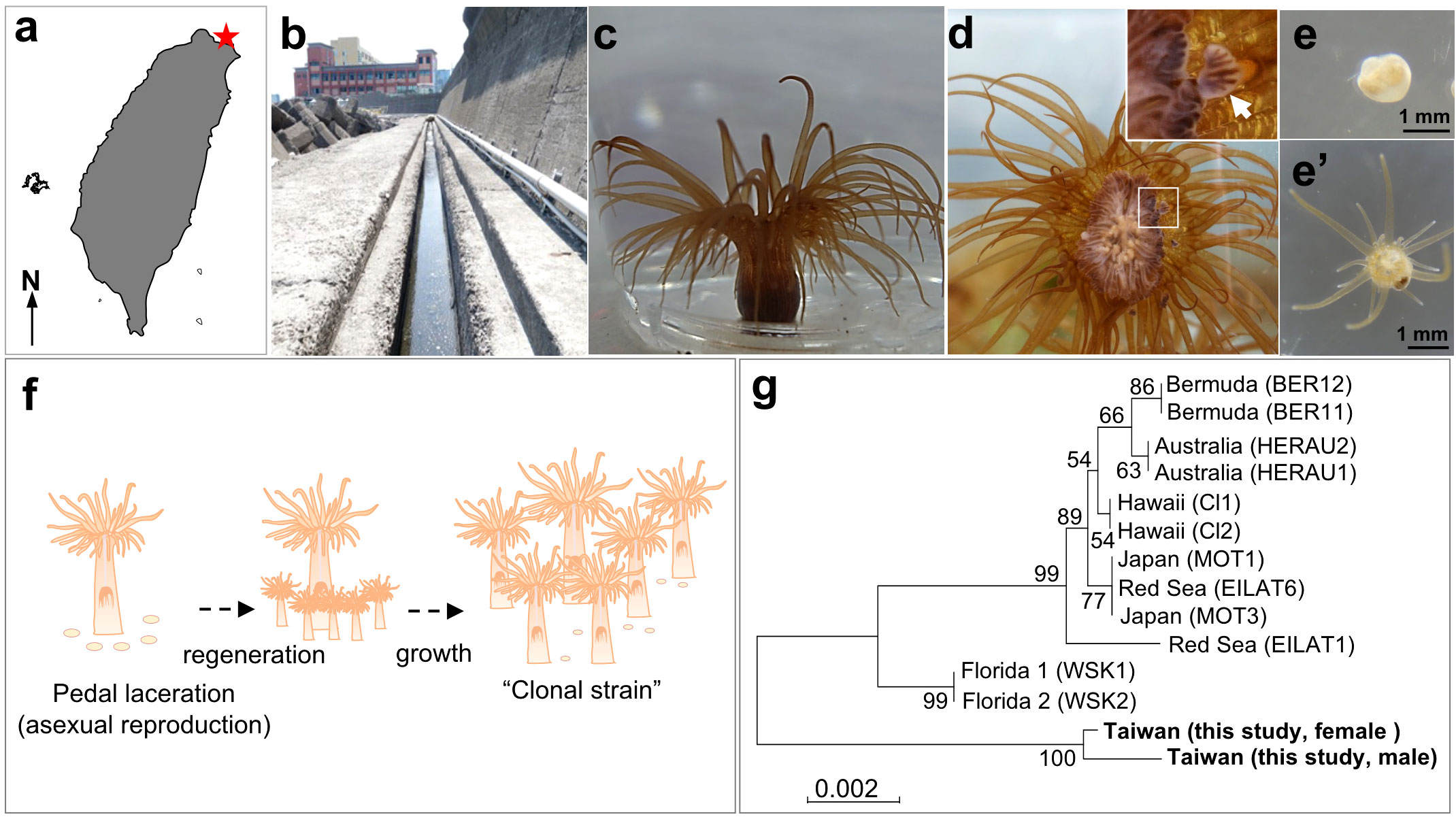
Figure 1 Establishment of E. diaphana clonal strains from Taiwan. (A) Map of Taiwan. A red star indicates the sampling location. (B) The ditch near the Aquatic Animal Center of National Taiwan Ocean University (NTOU), where parental E. diaphana were collected. (C) Appearance of E. diaphana maintained in an aquarium. (D) Initial stage of pedal laceration (asexual reproduction) of E. diaphana. The anemone attached to the glass surface of the aquarium wall was photographed from the bottom (pedal disc side). The box indicates the source of the inset (higher magnification view). An arrow indicates a lacerated piece observed at the peripheral margin of pedal disc. [(E) and e’] Polyp formation from a lacerated piece. The lacerated piece (E) was isolated from the aquarium wall and maintained in a 6-well plate. Regeneration into a juvenile polyp e’) was observed within 14 days of culture. (F) Schematic diagram of the process for establishing a clonal strain. (G) Phylogenetic analysis of E. diaphana strains using SCAR markers. Location names and names in parentheses indicate the location where the strains were established and the name of the strain, respectively.
2.2 Genomic DNA extraction and analysis of phylogenetic relationships
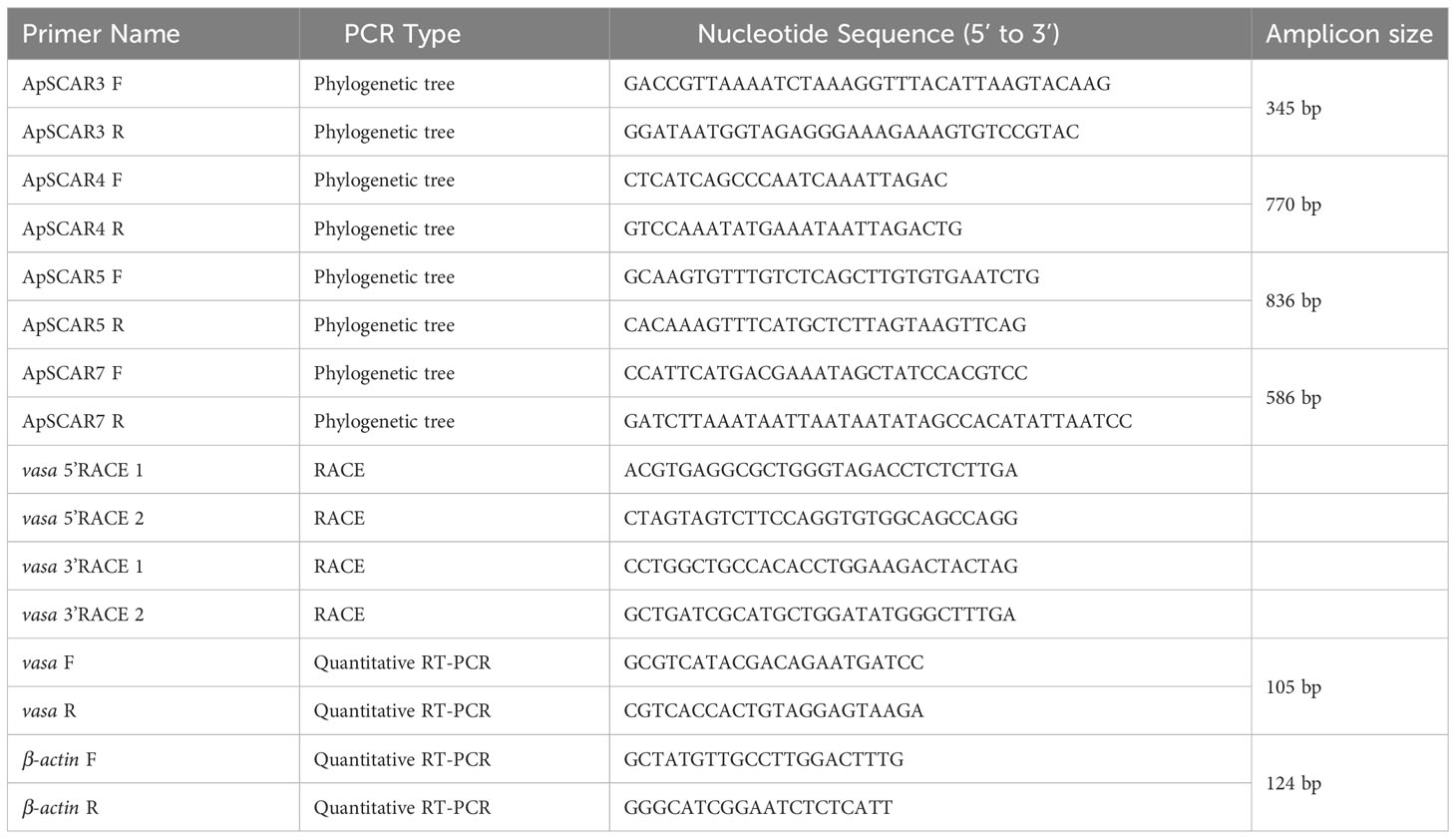
Three individuals were sampled from each male and female strain and homogenized with a homogenizer (IKA Ultra-Turrax, Sigma-Aldrich, St. Louis, USA) in lysis solution supplied with Wizard Genomic DNA Purification Kit (Promega, Madison, WI). Genomic DNA was extracted according to the manufacturer’s protocol. To compare phylogenetic relationships of the established E. diaphana strains and existing strains, a set of four E. diaphana-specific inter-simple sequence repeat (ISSR)-derived Sequence Characterized Amplified Region (SCAR) markers were used (Thornhill et al., 2013). PCR was performed with specific primers for each of the four SCAR markers (Table 1). PCR products were resolved by electrophoresis, subcloned, and sequenced as previously described (Shikina et al., 2012). Sequences of SCAR markers of existing E. diaphana strains were retrieved from GenBank, and phylogenetically compared with those of E. diaphana strains established in this study. Sequence alignment was performed using Muscle software, and results were used to create a 10,000 bootstrap replicate phylogenetic tree (neighbor-joining method) using MEGA7 (Kumar et al., 2016).
2.3 Histological analysis
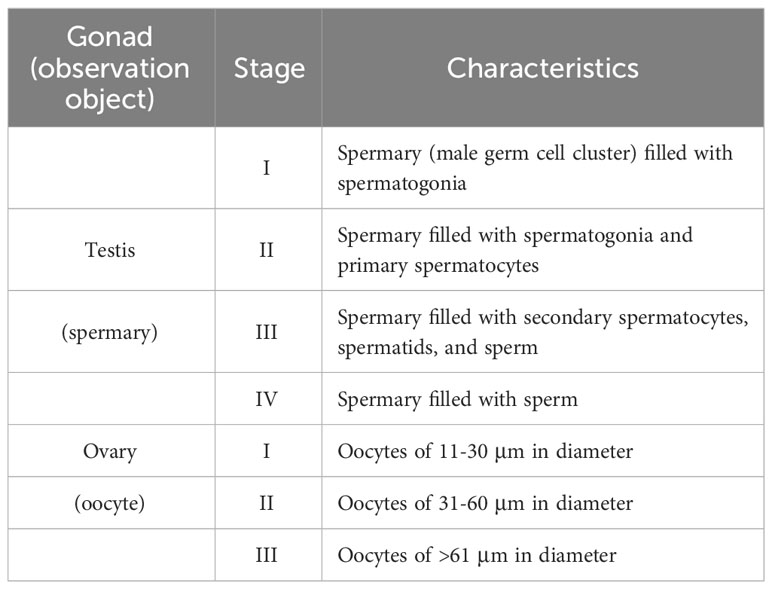
Specimens of E. diaphana having different pedal disc-sizes (1-2 mm, 3-4 mm, 5-6 mm, 7-8 mm, and 9-10 mm) were sampled from male and female strains, and fixed with Zinc Formal-Fixx (Thermo Shandon, Pittsburgh, PA) diluted 1:5 in 0.22-μm-filtered seawater for 16 h. Pedal diameters were measured with a scale when sea anemones were attached to the sides of the glass aquaria. For both male and female strains, 3 individuals were collected in each size range. Samples were embedded in Paraplast plus (Sherwood Medical, St. Louis, MO) and 4-µm serial sections were prepared with a microtome (Thermo Shandon). Hydrated sections were stained with Hematoxylin and Eosin Y (Thermo Shandon), and micrographs were taken under a microscope (IX71SF1, Olympus, Tokyo, Japan). To compare the progress of oogenesis in E. diaphana of different pedal-sizes, oocyte diameters were measured and recorded as the average of two measurements (the largest dimension of an oocyte and the second-largest dimension at a right angle to the first). Approximately 50-300 oocytes were analyzed from each individual, and average values were calculated. Then, based on diameters of mature oocytes (Chen et al., 2008), oocyte development was arbitrarily classified into 3 stages (Stages I-III) (Table 2). To compare progress of spermatogenesis among E. diaphana of different pedal disc-sizes, male germ cells comprising the spermary were observed. Their development was classified into 4 stages (Stages I-IV) (Table 2). Approximately 100 spermaries were analyzed from each individual. All image analyses were performed with Image J software (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA; https://imagej.nih.gov/ij).
2.4 Amputation experiment
Large E. diaphana (5~7 mm in pedal diameter) were cut horizontally into two pieces with scissors, and the resulting head and trunk were retained. Then, the heads and trunks were put separately in two 40-L aquaria (~20 heads or trunks per aquarium) and cultured for 3 months under conditions described above. Survivorship of heads and trunks was determined by dividing numbers at the end of culture by initial numbers. The experiment was conducted in triplicate, with a total of approximately 60 individuals (parts) cultured in each group. Presence or absence of gonads and gametogenesis was investigated in regenerated individuals (n=9 for each group). Histological sections were stained with Hematoxylin and Eosin Y (Thermo Shandon), and observed under a microscope (BX51, Olympus).
2.5 Gonad transcriptome
Testes/ovaries were isolated with forceps under a stereo microscope (EZ4, Leica Microsystem) from sea anemones with pedal disc diameters >7 mm and pooled in a Petri dish (9-cm diameter) containing 50 mL of 0.22-μm-filtered seawater. After several washes with 0.22 μm-filtered seawater, isolated gonads were snap-frozen with liquid nitrogen and stored at -80°C until use. Total RNA of the 2 samples (pooled testis and ovary samples) was extracted with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). Then, DNase I-treated RNA samples were sent to the Beijing Genomics Institute (BGI, Shenzhen, China) and qualified using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA) with an RNA 6,000 Labchip kit (Agilent Technologies). cDNA libraries were constructed from high-quality RNAs (RIN > 9) using TruSeq RNA Sample Prep Kits (Illumina, San Diego, USA), according to the manufacturer’s protocol. Libraries were subjected to 150-bp paired-end Illumina Hiseq sequencing. Illumina adaptors and low-quality bases or reads (QV< 20) were trimmed from raw sequences. Only high-quality data were used for the subsequent transcriptome assembly. Trimmed reads obtained above were mapped to the genomic reference sequence of E. diaphana (Baumgarten et al., 2015). Unmappable trimmed reads of each sample were assembled using Trinity v2.0.6 software (Grabherr et al., 2011). CD-HIT-EST was used to remove contig sequence redundancy (Li and Godzik, 2006). Obtained unigenes were annotated to the NCBI non-redundant protein (nr) database using the alignment algorithm, RAPSearch2, with a cut-off E-value <= -3.
2.6 Identification of germline markers
To identify genes that could be used as markers for germline cells or cells capable of producing germline cells, i.e., stem cell-like cells, genes that are reportedly expressed in cnidarian germline cells or multipotent stem cells were used as queries in the E. diaphana gonadal transcriptome, based on the literature (Mochizuki et al., 2001; Seipel et al., 2004; Extavour et al., 2005; Rebscher et al., 2008; Alié et al., 2011; Leclère et al., 2012; Nishimiya-Fujisawa and Kobayashi, 2012; Shikina et al., 2012; Shikina et al., 2015; Praher et al., 2017). We utilized the following two strategies: 1) Gene names or keyword searches were performed in annotation results; and 2) Full-length cDNA sequences of genes in other animals were retrieved from Genbank (NCBI), and local BLAST searches were conducted (BLASTP, cut-off -E value 1e-5) against translated sequences of E. diaphana gonadal transcriptome.
2.7 Phylogenetic analysis of vasa-related genes
Sequences of vasa and PL10 subfamily members from various animals were retrieved from GenBank, and phylogenetically compared with E. diaphana vasa and PL10 obtained in the present study. Sequence Alignment was performed using Muscle, and results were used to create a 10,000-bootstrap-replicate phylogenetic tree (neighbor-joining method) using MEGA7 (Kumar et al., 2016).
2.8 Quantitative RT-PCR
Different polyp tissues, i.e., tentacles, testes, ovaries, and mesenterial filaments, were microscopically isolated from E. diaphana. Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s protocol. First‐strand cDNA was synthesized from 2 μg of DNase‐treated RNA using SuperScript III reverse transcriptase (Thermo Fisher Scientific). Transcript levels were analyzed with quantitative real-time RT-PCR using a Bio-Rad CFX Connect™ Real-Time PCR detection system (Bio-Rad Laboratories, Hercules, CA). The reaction was performed with iQ™ SYBR Green Supermix (Bio-Rad Laboratories) under the following amplification conditions: 95°C for 5 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. Exaiptasia diaphana β-actin was used as a reference gene. Calculations were performed with the 2−▵▵Ct method (Livak and Schmittgen, 2001). Primers (Table 1) were designed using Primer Express 3.0 software (Applied Biosystems, USA). Primer efficiencies were determined with a 5-fold dilution series of template cDNA. Only primer pairs that worked at 90-100% efficiency were used for the analysis.
2.9 Western blotting
Proteins of tentacles, testes, ovaries, and mesenterial filaments were extracted with TRIzol reagent (Invitrogen) following the manufacturer’s protocol. Protein concentrations were determined using a bicinchoninic assay (BCA) protein assay kit (Pierce Biotechnology, Rockford, Il, USA). Protein samples (10 μg) were subjected to SDS-PAGE and Western blotting, as previously described (Shikina et al., 2012). For the primary antibody reaction, an antibody against Euphyllia ancora (a stony coral, recently renamed Fimbriaphyllia ancora Luzon et al., 2017) vasa was used (Shikina et al., 2012). The antibody was diluted 1:20,000 in Tris-buffered saline and 0.1% Tween 20 (TBST) with 1% skim milk and used for the reaction. For the secondary antibody reaction, alkaline phosphatase-conjugated goat anti-rabbit IgG antibody (AnaSpec, San Jose, California; 0.25 μg/mL in TBST with 1% skim milk) was used. Visualization of immunoreactive signals on membranes was performed using NBT/BCIP solution (Sigma-Aldrich).
2.10 Immunohistochemical staining
Immunohistochemical staining was performed according to methodology described elsewhere (Shikina et al., 2012). Briefly, hydrated sections were incubated for 30 min with HistoVT ONE (Nacalai Tesque, Inc, Kyoto, Japan) for antigen retrieval. After washing with phosphate-buffered saline containing 0.1% Tween 20 (PBT), sections were incubated for 10 min in 3% H2O2, and for 1 h in in 5% skim milk for blocking. Sections were then incubated for 16 h in anti-E. ancora vasa antibody (1:4,000 in PBT with 2% skim milk) (Shikina et al., 2012) at 4°C. For the secondary antibody reaction, sections were incubated with a biotinylated goat anti-rat IgG antibody (Vector Laboratories, Burlingame, USA; diluted 1: 2,000 in PBT with 2% skim milk) for 30 min. Immunoreactive signals were visualized with avidin–biotin–peroxidase complex (ABC) solution (Vector Laboratories), and 3,30-diaminobenzidine (DAB; Sigma-Aldrich). Sections were counterstained with haematoxylin. Observations were performed under a microscope (BX51, Olympus).
2.11 Statistics
All data are presented as means ± standard errors (SE). For comparisons among more than three groups, statistical significance was determined using one-way ANOVA followed by Tukey’s test, with statistical significance at P<0.05. All analyses were performed using SPSS software.
3 Results
3.1 Establishment of E. diaphana clonal strains and characterization of their asexual reproductive capacity
A number of E. diaphana were found in a ditch near the aquatic animal center of National Taiwan Ocean University in northern Taiwan (Figures 1A, B). We collected several specimens and reared them in aquaria in the laboratory (Figure 1C). Exaiptasia diaphana underwent active asexual reproduction following pedal laceration (Figure 1D). Pedal laceration began when the anemones formed small fragments (1-2 mm in diameter) by separating part of the peripheral margin of pedal disc tissue extended on the glass surface of aquaria (Figure 1D). The lacerated pieces (pedal fragments) transformed into small anemones within about 1-2 weeks (Figures 1E, e’). Exaiptasia diaphana that were cultured for more than 2 years in the laboratory, continued to grow and produced many individuals asexually. Four to eight lacerated pieces were produced per week per individual. Eventually, we were able to establish two clonal strains (one male and one female) (Figure 1F). Phylogenetic analysis with SCAR markers showed that established clones were genetically different from known E. diaphana strains (Figure 1G).
3.2 Characterization of sexual reproductive capacity
To investigate the sexual reproductive capacity of these established E. diaphana strains, some large individuals (> 5-6 mm in pedal disc diameter) were dissected and examined anatomically and histologically. Ovaries and testes were formed in mesenterial tissues of some large females and males, respectively (Figures 2A, B). Microscopically, ovaries were faint yellow, whereas testes were white. Developing germ cells were confirmed histologically (Figure 2C, D). Subsequent investigation of the relationship between polyp (pedal disc) size and gametogenesis showed that larger individuals of both sexes tend to have more mature gametes (Figures 2E, F). Females with a pedal disc diameter > 5-6 mm had larger mature oocytes (Figure 2E). Mean diameters of oocytes in females with pedal sizes of 5-6, 7-8, and 9-10 mm were 37.2 ± 1.7 μm (n=137 oocytes), 45.0 ± 7.9 μm (n= 686 oocytes), and 59.8 ± 7.8 μm (n=935 oocytes), respectively. Male E. diaphana with pedal disc diameters > 3-4 mm had mature sperm (Figure 2F). Gametogenesis was not observed in females < 3-4 mm or males < 1-2 mm in pedal disc diameter (Figures 2E, F).
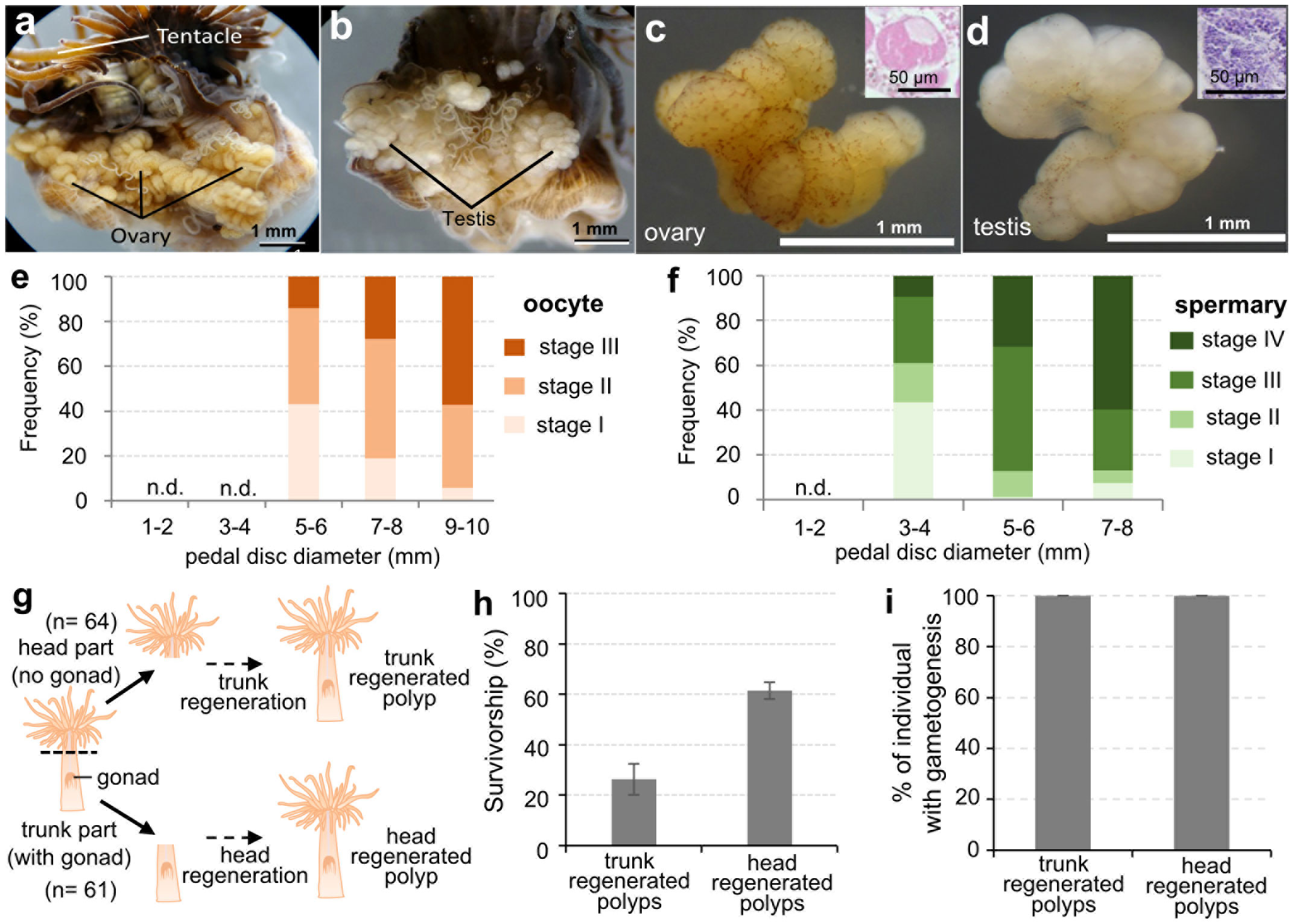
Figure 2 Characterization of the sexual reproductive capacity of established E. diaphana strains. (A) A dissected female E. diaphana with developed ovaries. (B) A dissected male E. diaphana with developed testes. (C) An isolated ovary. (D) An isolated testis. Insets in (C, D) are histological sections of the ovary and testis, respectively. (E) The frequency distribution of oocyte stages among female E. diaphana with different pedal diameters (n = 3 individuals for each group, in total 15 individuals). (F) The frequency distribution of spermatogenic stages among male E. diaphana with different pedal diameters (n = 3 individuals for each group, in total 12 individuals). Stage classification was performed according to criteria in Table 2. (G) Schematic illustration of the amputation experiment to further characterize the sexual reproductive capacity of E. diaphana. Polyps (64 individuals) were cut in two horizontally, and the resulting heads and trunks were cultured in separate tanks for 3 months. Three of the 64 trunk samples were lost for unknown reasons just prior to the experiment, so the final number of trunk samples in the experiment was 61. (H) Survivorship (%) of trunk-regenerated polyps and head-regenerated polyps. Data are expressed as means ± SEs (triplicate experiments. Sixty-four and sixty-one individuals were analyzed for trunk-regenerated and head-regenerated polyps, respectively). (I) Percentage of regenerated E. diaphana undergoing gametogenesis. Presence or absence of gonads and gametogenesis were histologically investigated in regenerated individuals (n=9 for each group).
3.3 Characterization of the sexual reproductive capacity of regenerated polyps
To further understand the sexual reproduction capacity of E. diaphana, we conducted an amputation experiment. Exaiptasia diaphana (5~7 mm in pedal diameter) were cut horizontally in two, and the resulting heads and trunks were cultured in separate tanks (Figure 2G). Heads (upper part of the polyp) were mainly composed of tentacles, the mouth, the upper part of the pharynx, and body wall, and contained no gonadal tissues. In contrast, trunks (lower part of the polyp) were composed of gonads, part of the pharynx, mesenterial filaments, body wall, and pedal disk (Figure 2G). After a few months of culture, all surviving individuals had regenerated their lost body parts. Survivorship of trunk-regenerated and head-regenerated polyps was 26.3 ± 6.1% and 61.5 ± 3.4%, respectively (Figure 2H). Particularly, in trunk-regenerated polyps, which originally had no gonads, formation of gonads and gametes was histologically confirmed in all individuals, as in head- regenerated polyps (Figure 2I).
3.4 Gonad transcriptome and identification of a germline marker, vasa, in E. diaphana
To efficiently identify marker genes for germline cells of E. diaphana, we performed gonad transcriptome sequencing. Testes and ovaries were isolated from E. diaphana with pedal disc diameters 7-8 mm (one individual for each sex), and were subjected to RNA-seq (Table S1). Approximately 234 million raw reads comprising 28 Gb of clean, transcriptomic, sequencing data were obtained by Illumina paired-end sequencing. De novo assembly of all clean reads produced 26,932 unigenes with an average size of 1,226 bp and an N50 of 1,660 bp (Table S1). Sequences were deposited in NCBI BioProject under accession numbers PRJNA1004228.
After annotation, genes reportedly expressed in germline cells of sea anemones were searched and identified in the established gonadal transcriptomes (Table S2). We then focused on vasa, and performed molecular identification and characterization. Full-length E. diaphana vasa cDNA was successfully determined by PCR, followed by RACE-PCR (GenBank No. ON375355). The deduced amino acid sequence was confirmed to contain conserved motifs characteristic of Vasa proteins, including the Q-motif, ATPase motifs, motifs involved in ATP binding and cleavage, RNA unwinding motifs, and the helicase C domain (Figure 3A). A phylogenetic analysis of DEAD-box protein family members showed that E. diaphana Vasa belonged to the Vasa family, and is not associated with PL10 proteins (Figure 3B). Tissue distribution analysis of E. diaphana vasa transcripts by quantitative RT-PCR showed that the transcripts were expressed significantly more abundantly in ovary than in other tissues (Figure 3C). Transcripts were also present in testis, although the statistical significance was not determined (Figure 3C). Subsequent Western blotting detected expression of E. diaphana Vasa protein primarily in testes and ovaries. The immunoreactive band was also faintly detected in tentacles and mesenterial filaments of both sexes (Figure 3D). The size of the immunoreactive band was approximately 82 kDa, close to the predicted molecular mass of 81 kDa of E. diaphana Vasa (Figure 3D).
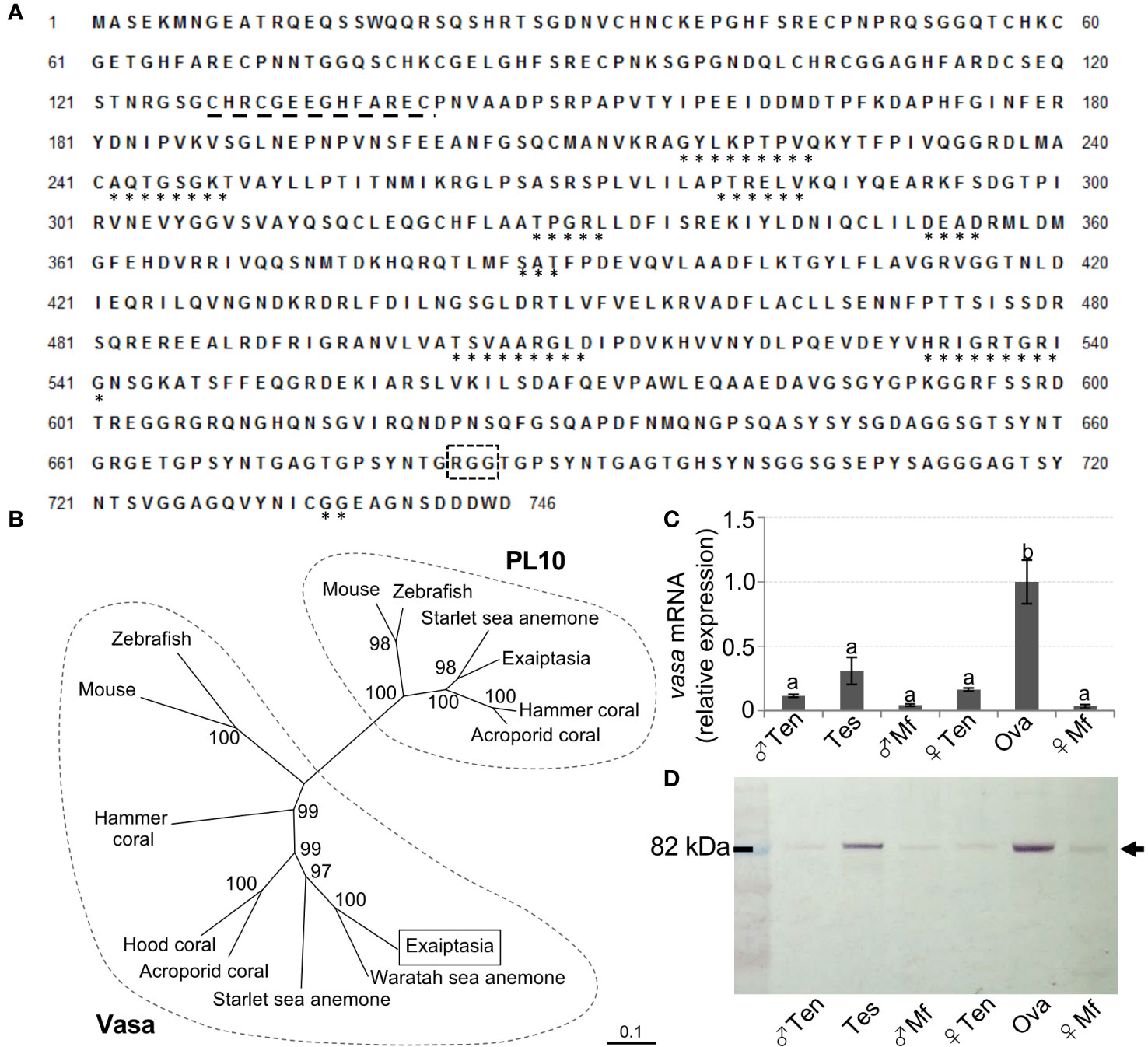
Figure 3 Molecular identification and characterization of E. diaphana vasa. (A) The deduced amino acid sequence of a cloned cDNA encoding a E. diaphana vasa (GenBank accession no. ON375355). *, motifs conserved among DEAD-box protein family members; black dashed line, CCHC zinc fingers; black box with dashed line, RGG motif. (B) A phylogenetic tree of amino acid sequences of Vasa- and PL10-related proteins constructed using the neighbor-joining method. Numbers adjacent to nodes represent bootstrap probability (%). Branches correspond to values of 50% and higher. Lengths of lines indicate genetic distances. Names and corresponding GenBank accession numbers of proteins analyzed are as follows: (left) Vasa, Mouse (Mus musculus, EDL18409), Zebrafish (Danio rerio, AAI29276), E. diaphana (This study, WCJ13147), Waratah sea anemone (Actinia tenebrosa XP_031556804), Starlet sea anemone (Nematostella vectensis, AFP87471), Acroporid coral, (Acropora digitifera, XP_015780252), Hood coral (Stylophora pistillata, XP_022779027), and Hammer coral (Euphyllia ancora, AFP52950); (right) PL10, Mouse (Mus musculus, AAA39942), Zebrarish (Danio rerio, NP_571016), Starlet sea anemone (Nematostella vectensis, XP_001627306), Exaiptasia (Exaiptasia pallida, XP_020899200), Hammer coral (Euphyllia ancora, AFP52949), and Acroporid corals (Acropora millepora, XP_029207734). (C) Distribution of E. diaphana vasa transcripts in various male and female polyp tissues, as analyzed by quantitative RT-PCR. Data are shown as means ± SEs (5 males and 5 females) of relative values with that of the ova group. Groups with different letters are significantly different (p < 0.05). (D) Tissue distribution analysis of E. diaphana vasa protein in male and female samples, as assessed by Western blotting. Molecular weight markers are shown on the left. Ten, tentacles; Tes, testis; Mf, mesenterial filaments; Ova, ovary.
Immunohistochemical analysis of gonads demonstrated that E. diaphana Vasa is expressed in oogonia and developing oocytes in the ovary (Figures 4A, B). Vasa was also expressed in spermatogonia (Figures 4C, D), spermatocytes, and spermatids in the testes, (Figures 4E, F). Immunoreactivity was faint or almost undetectable in mature sperm (Figure 4F). These results demonstrated that vasa can be used as a marker of germline cells.
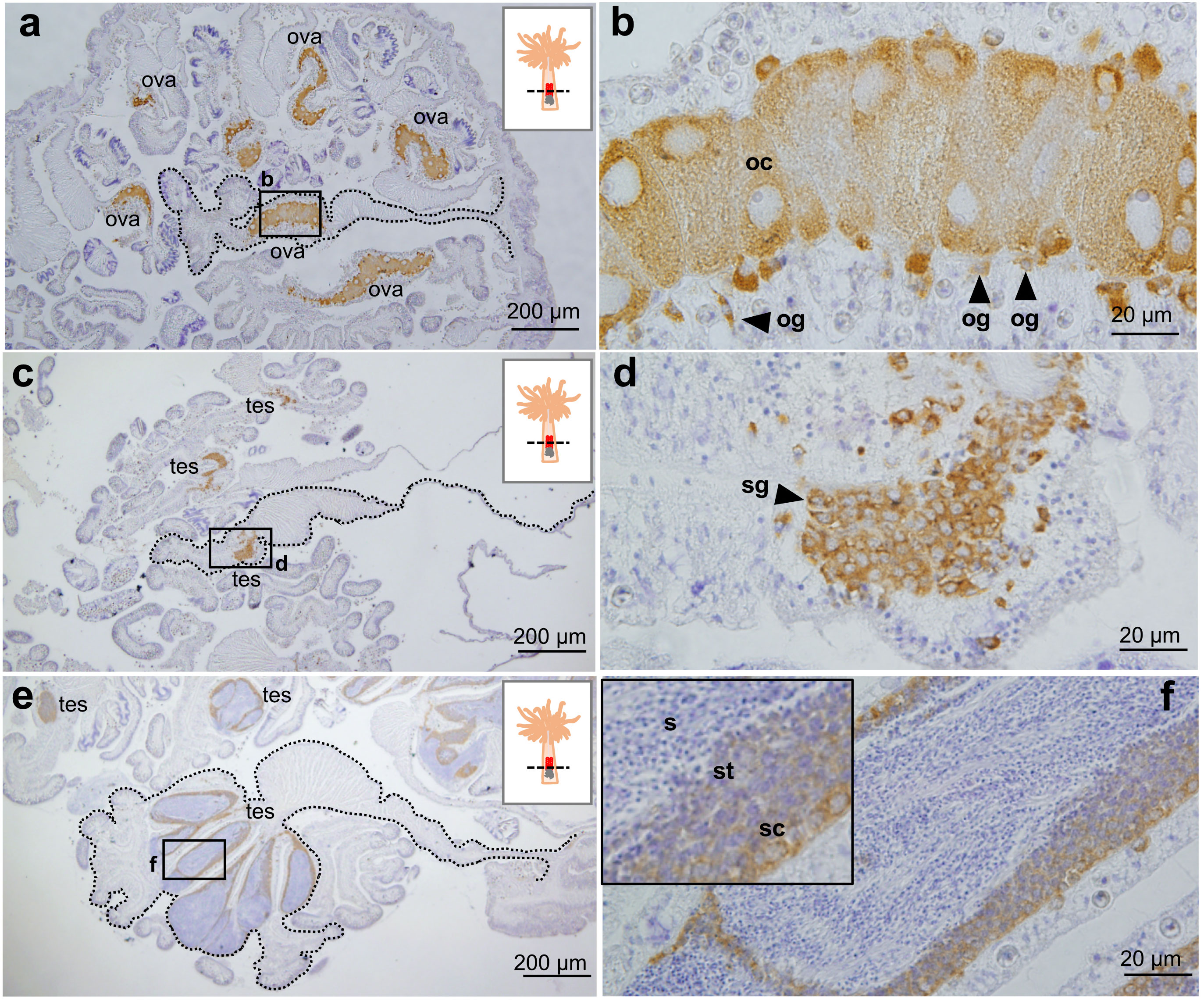
Figure 4 Immunohistochemical characterization of Vasa protein expression in E. diaphana female and male germ cells. (A, B). Anti- E. diaphana Vasa immunoreactivity in oogonia (og) and developing oocytes (oc) in ovaries (ova). The section was prepared from an E. diaphana with a pedal size of 7-8 mm. (C, D). Anti- E. diaphana Vasa immunoreactivity in spermatogonia (sg) in testis (tes). The section was prepared from a male E. diaphana with a pedal size of 3-4 mm. (E, F) Anti-E. diaphana Vasa immunoreactivity in spermatocytes (sc) and spermatids (st) in testis (tes). The section was prepared from male E. diaphana with a pedal size of 7-8 mm. Note that immunoreactivity was detected in spermatocytes and spermatids, but was faint or almost undetectable in sperm (inset). Boxed regions in (A, C, E) indicate the source of higher magnification views [(B, D, F) respectively]. In (A, C, E), broken lines indicate the shape of the mesentery tissues.
3.5 Detection of Vasa-positive cells in extra-gonadal tissues
Finally, we investigated germline cells or cells capable of producing germline cells in various somatic tissues in polyps using vasa expression as a marker. In tentacles, vasa-positive cells were detected in patchy clusters in the gastrodermis (Figures 5A, a’). The morphology of vasa-positive cells in tentacles was different from that in gonads. Vasa-positive cells in tentacles exhibited irregular shapes and had small nuclei that stain well with hematoxylin (Figure 5a’). In contrast, there are few vasa-positive cells in pharynx and mesenteries (Figures 5B, b’), mesenterial filaments (Figures 5C, c’), pedal disc (Figures 5D, d’), or lacerated pieces (Figures 5E, e’). Vasa-positive cells in those tissues were present singly, and were morphologically similar to early-stage germ cells that were observed in gonads of juvenile anemones (Figures 5F, f’).
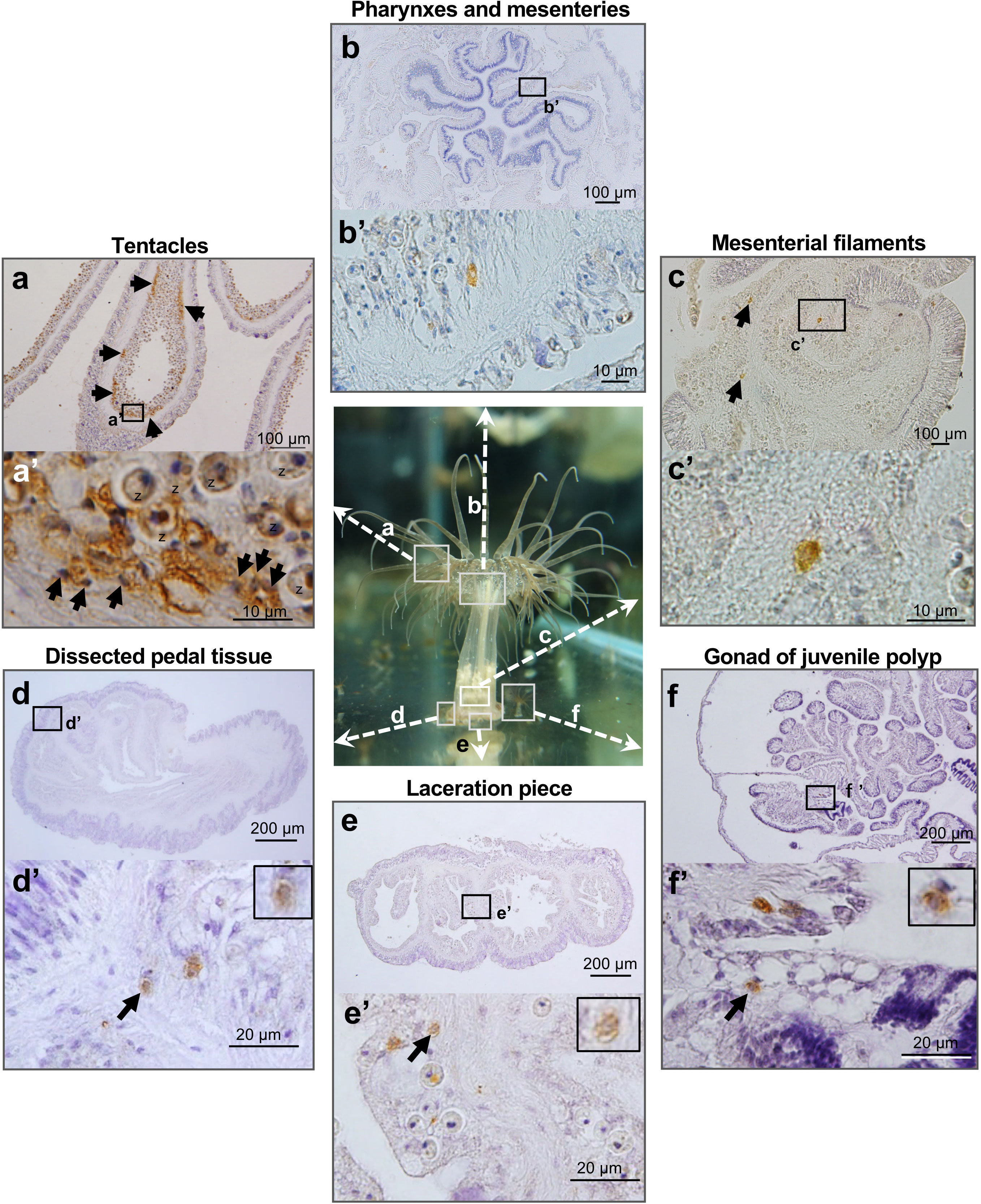
Figure 5 Marker (Vasa protein)-assisted detection of potential germline cells or cells capable of producing germline cells in various somatic tissues of E. diaphana. Vasa-positive cells in tentacles (A), pharynxes and mesenteries (B), mesenterial filaments (C), dissected pedal tissue (D), lacerated pieces (E), and gonads of juvenile polyps (F). In tentacles, vasa-positive cells are mosaically distributed, forming clusters in the gastrodermal region (arrows). They exhibited irregular shapes and had small nuclei, well stained by hematoxylin. z, zooxanthellae. In contrast, in other polyp tissues, there are only a few vasa-positive cells, which exhibited morphological similarities to early-stage germ cells present in gonads of juvenile polyps. Boxed regions in (A–F) indicate the source of higher magnification views (a’-f’, respectively).
4 Discussion
Anthozoans employ both asexual and sexual reproduction to ensure both population growth and genetic diversity. However, mechanisms that ensure sexual reproductive capacity in asexually reproducing or regenerating individuals have been little studied, and remain largely unknown. In this study, we verified that germline cells or cells capable of producing germline cells exist in extra-gonadal tissues, and that they are allocated to newly produced/regenerating individuals during asexual reproduction/regeneration.
First, we established new E. diaphana strains and demonstrated their flexible sexual reproductive capacity after asexual reproduction/tissue regeneration. Subsequently, gonadal transcriptomes were established, and the vasa gene was isolated as a germline marker. Finally, we demonstrated the presence of vasa-positive cells in various polyp tissues, as well as lacerated pieces using immunohistochemical analysis. Data obtained supported our hypothesis. Vasa-positive cells found in extra-gonadal tissues may function as a reservoir of progenitors of germline cells to ensure sexual reproductive capacity in polyps that are asexually produced or regenerated from somatic tissues. This study provides new insight into sea anemone reproductive biology.
Individuals that were asexually produced from pedal discs initiated gametogenesis at pedal sizes >3-4 mm for males and >5-6 mm for females. In addition, our amputation experiment showed that E. diaphana has high regenerative and flexible sexual reproductive capacity. These results strongly suggest germline cells or cells capable of producing germline cells in tissues other than gonads.
We then employed marker-assisted cell identification to identify such cells. In hydras and hydractinians, vasa protein is expressed not only in germline cells, but also in multipotent stem cells, e.g., interstitial cells (i-cells), which can self-renew and generate multiple cell lineages, such as nematocytes, nerve cells, gland cells, and germline cells (Nishimiya-Fujisawa and Kobayashi, 2012). With reference to these studies, E. diaphana vasa was then cloned and the distribution of vasa-positive cells was investigated by immunohistochemical staining. As a result, a small number of vasa-positive cells were found in the pedal region and gastrodermal tissues of the lacerated pieces. These vasa-positive cells were morphologically similar to germline cells found in gonads of juvenile polyps. Given their distribution, location, and morphological characteristics, it appears that vasa-positive cells in the pedal and lacerated pieces are germline cells that may have migrated to the gonadal region during the dynamic morphogenetic process that transformed lacerated pieces into juvenile polyps (Figure 6). However, we cannot rule out the possibility that the somatic population dedifferentiates and generates germline cells. In the future, our hypothesis will be verified by establishing transgenic sea anemones, e.g., vasa-GFP sea anemones with the germ cell lineage specifically labeled, and by observing germ cell distribution and migration in detail using live imaging and germ cell transplantation techniques.
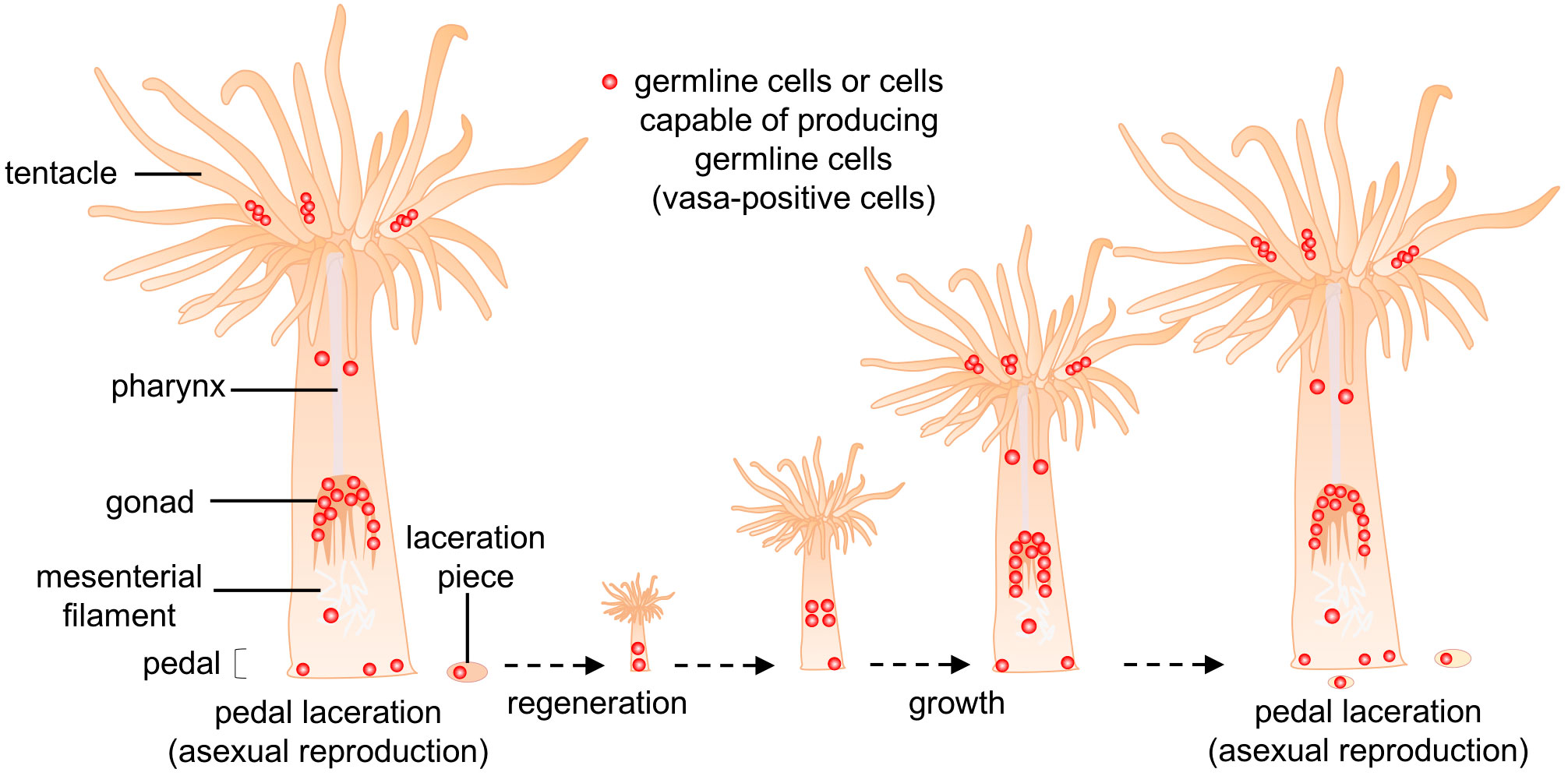
Figure 6 Schematic diagram of the proposed cellular mechanism for germline-cell allocation during asexual reproduction (pedal laceration). Small numbers of germline cells or cells capable of producing germline cells (vasa-positive cells, red dot) exist in pedal tissue, and are allocated to lacerated pieces during asexual reproduction. These allocated cells may eventually produce gametes, and contribute to gametogenesis in new polyps.
In many organisms, gametes generally originate from germline cells, e.g., primordial germ cells or germ line stem cells in embryonic/adult gonads (Wylie, 1999). In contrast, in sea anemone germ cells have been thought to originate from gastrodermal (gonadal) epithelial cells, and spermatogonia or oogonia have been assumed to differentiate de novo from those somatic cells (Bocharova and Kozevich, 2011). Although many studies describing sea anemone gametogenesis have been published, to the best of our knowledge, there is no experimental evidence for reprograming of gastrodermal (gonadal) epithelial cells into germ cells. In the present study, immunohistochemical analysis demonstrated that vasa-positive cells are constantly present in the gastrodermis of lacerated pieces, and in the gonadal region (between mesenterial filaments and retractor muscles) from juvenile to adult polyps. These findings suggest instead that gastrodermal (gonadal) somatic cells most likely do not differentiate into germline cells, but rather that germline cells are always present in gonads at least from juvenile to adult polyps for gametogenesis in sea anemones.
Germline cells in cnidarians are assumed to originate from interstitial cells (i-cells), although interstitial cells have so far been demonstrated experimentally only in hydrozoans (Technau and Steele, 2011; Gold and Jacobs, 2013). In Hydras, various studies have demonstrated that i-cell populations are primarily composed of multipotent stem cells (MPSCs) and germline stem cells (GSCs) (Nishimiya-Fujisawa and Kobayashi, 2012). MPSCs can self-renew and generate multiple cell lineages, such as nerve cells, gland cells, and GSCs. In contrast, GSCs can self-renew and generate cells of the germline lineage, but not the somatic lineage (Nishimiya-Fujisawa and Kobayashi, 2012). In the colonial marine hydroid, H. echinata, i-cells give rise to all cell types, including ectodermal and endodermal epithelial cells and germline cells (Müller et al., 2004; Künzel et al., 2010; Plickert et al., 2012). Interestingly, vasa genes are expressed in i-cell populations and germline cells in Hydra and Hydractinia. Most recently, the existence of an adult Vasa2+/Piwi1+ multipotent stem-like cell population, which can produce both germline and somatic lineages, was demonstrated in the sea anemone, Nematostella vectensis (Miramón-Puértolas and Steinmetz, 2023). Taking these reports into account, vasa-positive cells found in extra-gonadal tissues in E. diaphana may be cells having potencies similar to those of i-cells or multipotent stem-like cell population. Gene silencing by RNA interference (RNAi) (Agrawal et al., 2003; Wilson and Doudna, 2013) will allow us to explore functions of the vasa gene and presence of i-cells in E. diaphana.
In this study, we collected E. diaphana from a ditch near the aquatic animal culture center at National Taiwan Ocean University, located on the northern coast of Taiwan. Because the center directly uses seawater from the ocean to cultivate marine organisms, a variety of planktonic organisms are brought from the ocean into the facility. Exaiptasia diaphana may also be brought into the center with seawater, and was eventually found growing naturally in the ditch where we sampled. Our phylogenetic analysis using SCAR markers (Thornhill et al., 2013) showed that clonal strains established in this study were genetically different from known E. diaphana strains. Currently, E. diaphana is being used as a model organism to investigate the symbiotic relationship between corals (cnidaria) and dinoflagellates at cellular and molecular levels (Weis et al., 2008; Davy et al., 2012; Sproles et al., 2020; Cui et al., 2023). PCR analysis of strains established in this study revealed that they possess mainly Clade B symbionts (data not shown). Our strains could also be used as material to study cnidarian endosymbiosis.
Exaiptasia diaphana offers various advantages as an experimental animal. For example, they are easy and inexpensive to maintain in the laboratory (Weis et al., 2008; Davy et al., 2012). Monthly spawning can be induced under certain conditions (Grawunder et al., 2015). The genome of E. diaphana has been sequenced (Baumgarten et al., 2015), and fundamental techniques for gene functional analyses are being developed (Jones et al., 2018). Moreover, adult E. diaphana are large enough to allow us to isolate specific types of polyp tissue, e.g., testis and ovary, from living polyps. This enables us to conduct tissue-specific transcriptome/proteome analysis. Since E. diaphana belongs to the same class as scleractinian corals (Class Anthozoa), they share many morphological and physiological characteristics with scleractinian corals, e.g., symbiosis. Exaiptasia diaphana may be useful as a model to study molecular and cellular mechanisms underlying gametogenesis and spawning in anthozoans. Comparisons of a recently established coral gonad transcriptome (Chiu et al., 2020) with the gonadal transcriptome established in this study will likely reveal similarities and differences that underlie gametogenesis in sea anemones and corals. This approach may give us valuable molecular insights into reproductive biology of anthozoans.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal studies were approved by National Taiwan Ocean University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
SS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. P-JS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. Y-LC: Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – review & editing. P-HT: Investigation, Methodology, Project administration, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by a grant from the Ministry of Science and Technology, Taiwan (103-2621-B-019-006-MY3 to SS).
Acknowledgments
The authors gratefully acknowledge colleagues who helped us to collect samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1278022/full#supplementary-material
References
Agrawal N., Dasaradhi P. V., Mohmmed A., Malhotra P., Bhatnagar R. K., Mukherjee S. K. (2003). RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67 (4), 657–685. doi: 10.1128/MMBR.67.4.657-685.2003
Alié A., Leclère L., Jager M., Dayraud C., Chang P., Le Guyader H., et al. (2011). Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of "germline genes" with stemness. Dev. Biol. 350 (1), 183–197. doi: 10.1016/j.ydbio.2010.10.019
Baumgarten S., Simakov O., Esherick L., Liew Y., Lehnert E., Michell C., et al. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112 (38), 11893–11898. doi: 10.1073/pnas.1513318112
Bocharova E. S. (2016). “Reproduction of sea anemones and other hexacorals,” in The Cnidaria, Past, Present and Future. Eds. Goffredo S., Dubinsky Z. (Cham: Springer), 239–248. doi: 10.1007/978-3-319-31305-4_15
Bocharova E. S., Kozevich I. A. (2011). Modes of reproduction in sea anemone (Cnidaria, Anthozoa). Biol. Bull. 38 (9), 849–860. doi: 10.1134/S1062359011090020
Chen C., Soong K., Chen C. A. (2008). The smallest oocytes among broadcast-spawning actiniarians and a unique lunar reproductive cycle in a unisexual population of the sea anemone, Aiptasia pulchella (Anthozoa: Actiniaria). Zool. Stud. 47 (1), 37–45.
Chiu Y. L., Shikina S., Yoshioka Y., Shinzato C., Chang C. F. (2020). De novo transcriptome assembly from the gonads of a scleractinian coral, Euphyllia ancora: molecular mechanisms underlying scleractinian gametogenesis. BMC Genom. 21 (1), 732. doi: 10.1186/s12864-020-07113-9
Clayton W. S. (1985). Pedal laceration by the anemone Aiptasia pallida. Mar. Ecol. Prog. Ser. 21, 75–80. doi: 10.3354/meps021075
Cui G., Konciute M. K., Ling L., Esau L., Raina J. B., Han B., et al. (2023). Molecular insights into the Darwin paradox of coral reefs from the sea anemone Aiptasia. Sci. Adv. 9 (11), p.eadf7108. doi: 10.1126/sciadv.adf7108
Davy S. K., Allemand D., Weis V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76 (2), 229–261. doi: 10.1128/mmbr.05014-11
Extavour C. G., Pang K., Matus D. Q., Martindale M. Q. (2005). vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol. Dev. 7 (3), 201–215. doi: 10.1111/j.1525-142X.2005.05023.x
Gold D. A., Jacobs D. K. (2013). Stem cell dynamics in Cnidaria: are there unifying principles? Dev. Genes. Evol. 223 (1-2), 53–66. doi: 10.1007/s00427-012-0429-1
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi: 10.1038/nbt.1883
Grajales A., Rodríguez E. (2014). Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea). Zootaxa 3826 (1), 55–100. doi: 10.11646/zootaxa.3826.1.2
Grawunder D., Hambleton E. A., Bucher M., Wolfowicz I., Bechtoldt N., Guse A. (2015). Induction of gametogenesis in the cnidarian endosymbiosis model Aiptasia sp. Sci. Rep. 5 (1), 15677. doi: 10.1038/srep15677
Harrison P. L., Wallace C. C. (1990). “Reproduction, dispersal and recruitment of scleractinian corals,” in Ecosystems of the World 25 Coral Reefs. Eds. Dubinsky Z., Stambler N. (New York: Elsevier), 133–207.
Hunter T. (1984). The Energetics of asexual reproduction: pedal laceration in the symbiotic sea anemone Aiptasia pulchella (Carlgren 1943). J. Exp. Mar. Biol. Ecol. 83 (2), 127–147. doi: 10.1016/0022-0981(84)90041-8
Jones V., Bucher M., Hambleton E. A., Guse A. (2018). Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Sci. Rep. 8 (1), 16437. doi: 10.1038/s41598-018-34773-1
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Künzel T., Heiermann R., Frank U., Müller W., Tilmann W., Bause M., et al. (2010). Migration and differentiation potential of stem cells in the cnidarian Hydractinia analyzed in eGFP-transgenic animals and chimeras. Dev. Biol. 348 (1), 120–129. doi: 10.1016/j.ydbio.2010.08.017
Leclère L., Jager M., Barreau C., Chang P., Le Guyader H., Manuel M., et al. (2012). Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev. Biol. 364 (2), 236–248. doi: 10.1016/j.ydbio.2012.01.018
Li W., Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 (13), 658–1659. doi: 10.1093/bioinformatics/btl158
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Luzon K. S., Lin M. F., Ablan, Lagman M. C. A., Licuanan W. R. Y., Chen C. A. (2017). Resurrecting a subgenus to genus: molecular phylogeny of Euphyllia and Fimbriaphyllia (order Scleractinia; family Euphyllidae; clade V). PeerJ 5, e4074. doi: 10.7717/peerj.4074
Miramón-Puértolas P., Steinmetz P. R. H. (2023). An adult stem-like cell population generates germline and neurons in the sea anemone Nematostella vectensis. bioRxiv. doi: 10.1101/2023.01.27.525880
Mochizuki K., Nishimiya-Fujisawa C., Fujisawa T. (2001). Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev. Genes. Evol. 211 (6), 299–308. doi: 10.1007/s004270100156
Müller W. A., Teo R., Frank U. (2004). Totipotent migratory stem cells in a hydroid. Dev. Biol. 275 (1), 215–224. doi: 10.1016/j.ydbio.2004.08.006
Nishimiya-Fujisawa C., Kobayashi S. (2012). Germline stem cells and sex determination in Hydra. Int. J. Dev. Biol. 56 (6-8), 499–508. doi: 10.1387/ijdb.123509cf
Odum H. T., Odum E. P. (1955). Trophic structure and productivity of a windward coral reef community on Eniwetok Atoll. Ecol. Monog. 25 (3), 291–320. doi: 10.2307/1943285
Plickert G., Frank U., Müller W. A. (2012). Hydractinia, a pioneering model for stem cell biology and reprogramming somatic cells to pluripotency. Int. J. Dev. Biol. 56 (6-8), 519–534. doi: 10.1387/ijdb.123502gp
Praher D., Zimmermann B., Genikhovich G., Columbus-Shenkar Y., Modepalli V., Aharoni R., et al. (2017). Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol. 14 (12), 1727–1741. doi: 10.1080/15476286.2017.1349048
Rebscher N., Volk C., Teo R., Plickert G. (2008). The germ plasm component Vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia eChinata. Dev. Dyn. 237 (6), 1736–1745. doi: 10.1002/dvdy.21562
Ruppert E. E., Fox R. S., Barnes R. D. (2003). “A functional evolutionary approach,” in Invertebrate zoology, 7th ed (California: Brooks/Cole Thompson Learning), 111–180.
Seipel K., Yanze N., Schmid V. (2004). The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int. J. Dev. Biol. 48 (1), 1–7. doi: 10.1387/ijdb.15005568
Shikina S., Chang C. F. (2018). “Cnidaria,” in Encyclopedia of Reproduction, 2nd edition, vol. 6 . Ed. Skinner M. K. (Netherlands: Elsevier Academic Press), 491–497. doi: 10.1016/B978-0-12-809633-8.20597-9
Shikina S., Chen C. J., Liou J. Y., Shao Z. F., Chung Y. J., Lee Y. H., et al. (2012). Germ cell development in the scleractinian coral Euphyllia ancora (Cnidaria, Anthozoa). PloS One 7 (7), e41569. doi: 10.1371/journal.pone.0041569
Shikina S., Chung Y. J., Wang H. M., Chiu Y. L., Zhao Z. F., Lee Y. H., et al. (2015). Localization of early germ cells in a stony coral, Euphyllia ancora: potential implications for a germline stem cell system in coral gametogenesis. Coral Reefs. 34 (2), 639–653. doi: 10.1007/s00338-015-1270-6
Sproles A. E., Oakley C. A., Krueger T., Grossman A. R., Weis V. M., Meibom A., et al. (2020). Sub-cellular imaging shows reduced photosynthetic carbon and increased nitrogen assimilation by the non-native endosymbiont Durusdinium trenchii in the model cnidarian Aiptasia. Environ. Microbiol. 22 (9), 3741–3753. doi: 10.1111/1462-2920.15142
Technau U., Steele R. E. (2011). Evolutionary crossroads in developmental biology: Cnidaria. Development 138 (8), 1447–1458. doi: 10.1242/dev.066712
Thornhill D. J., Xiang Y., Pettay D. T., Zhong M., Santos S. R. (2013). Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 22 (17), 4499–4515. doi: 10.1111/mec.12416
Toh T. C., Ng C. S. L. (2016). Tentacular autotomy and polyp regeneration in the scleractinian coral Euphyllia glabrescens. Coral Reefs. 35 (3), 819. doi: 10.1007/s00338-016-1433-0
Weis V. M., Davy S. K., Hoegh-Guldberg O., Rodriguez-Lanetty M., Pringle J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23 (7), 369–376. doi: 10.1016/j.tree.2008.03.004
Wilson R. C., Doudna J. A. (2013). Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239. doi: 10.1146/annurev-biophys-083012-130404
Keywords: sea anemone, Exaiptasia diaphana, Vasa, pedal laceration, reproduction
Citation: Shao P-J, Chiu Y-L, Tsai P-H and Shikina S (2023) Possible germline progenitor cells in extra-gonadal tissues of the sea anemone, Exaiptasia diaphana. Front. Mar. Sci. 10:1278022. doi: 10.3389/fmars.2023.1278022
Received: 15 August 2023; Accepted: 31 October 2023;
Published: 14 November 2023.
Edited by:
Rui Rosa, University of Lisbon, PortugalReviewed by:
Pedro Martinez, University of Barcelona, SpainEldon Ball, Australian National University. Australia
Copyright © 2023 Shao, Chiu, Tsai and Shikina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shinya Shikina, c2hpa2luYUBtYWlsLm50b3UuZWR1LnR3
†These authors have contributed equally to this work
 Pei-Jung Shao1†
Pei-Jung Shao1† Yi-Ling Chiu
Yi-Ling Chiu Shinya Shikina
Shinya Shikina