
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 17 August 2023
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1251251
This article is part of the Research Topic The Response, Adaptation, and Evolution of Marine Molluscs View all 6 articles
 Xiaoya Shen1,2,3†
Xiaoya Shen1,2,3† Yige Chen1,2,3†
Yige Chen1,2,3† Liang Jia1,2,3
Liang Jia1,2,3 Wang He1
Wang He1 Xiaofeng Chen1
Xiaofeng Chen1 Yiwen Chen1
Yiwen Chen1 Xingrong Xuan1
Xingrong Xuan1 Jiale Li1,2,3
Jiale Li1,2,3 Zhiyi Bai1,2,3*
Zhiyi Bai1,2,3* Wenjuan Li1,2,3*
Wenjuan Li1,2,3*Biomineralization is a widespread phenomenon in marine mollusks and is responsible for the production of shells and pearls. However, the regulatory mechanisms governing the adaptive immune responses in the mollusk mantle and visceral mass during mineralization remain unclear. In this work, we examined the mantle and visceral mass immune responses of Hyriopsis cumingii during pearl culture using high-throughput sequencing techniques. A mantle transcriptome database was established using transcriptome sequencing technology and reference to the major databases. Digital gene expression profiling was used to identify the differentially expressed genes of mantle and visceral mass at different insertion periods. Moreover, quantitative real-time PCR was used to verify the expression of five immune-related genes. Transcriptome sequencing results showed 257,457 unigenes were identified. Digital gene expression profiles showed 1389, 3572, 1888, and 2613 differentially expressed genes (DEGs) in the mantle and visceral mass at 5, 20, 50, and 90 d after insertion, respectively, with the highest number at 20 d and the lowest at 5 d after insertion (q < 0.05). A cluster analysis of the DEGs showed similar clustering and expression features in the mantle to the control group, and at 5, 50 and 90 d, after mantle insertion. The DEGs in the visceral mass showed similar clustering and expression features to the control group and at 5, 20 and 50 d after insertion. We also screened 22 immune-related DEGs in the mantle and visceral mass during the same pearl culture period, including serine/threonine-protein kinase NLK, C-type lectin, and galectin. The greatest number of DEGs was found 90 d after insertion. Compared with the mantle, more immune-related DEGs were down-regulated than up-regulated in the visceral mass during pearl culture, indicating that the immune regulatory mechanisms in the visceral mass and the mantle differ during pearl culture, and that the visceral mass is liable to higher infection and mortality rates. Quantitative real-time PCR results showed that the expression of five immune-related genes was consistent with DGE results. Our findings will further knowledge of the immune systems that are present in the mantle and visceral mass during pearl culture.
Biomineralization is a widespread phenomenon in marine mollusks. Their shells and pearls are important products of this process and their formation is regulated at the genetic level (Jin et al., 2019; Liu et al., 2019). During the pearl culture process, pieces of the mantle are inserted into the surrounding shell causing the cells at the nucleus insertion site to proliferate and divide to form pearl sacs, which secrete nacre to produce pearls (Hua and Neves, 2011). Research into practical ways to produce the same effect in the viscera, imitating the mantle process, has made gradual progress in mass pearl culture, but the technology is still far from perfect.
Studies have shown that both the mantle and visceral mass nucleus insertion sites cause extensive immune responses in pearl culture conditions (Huang et al., 2019). At the organizational level, pearl oyster Pinctada fucata mortality occurred after nucleation of the mantle during the resting period (Weng et al., 2012). In the triangular sail mussel Hyriopsis cumingii the hemocyte density increased significantly and there was a relative increase in the number of granulocytes and a relative decrease in the number of hyaline cells after nucleation of the visceral mass, as immune defense responses to external stimulation (He et al., 2010). Shi et al. found that the regulation of immune defenses was enhanced in Hyriopsis cumingii after nucleation of the visceral mass, with significant changes in the function of the immune-related enzymes acyl carrier protein (ACP) and sodium dismutase (SOD) in the blood (Li et al., 2010). Huang et al. found that blood uric acid levels were significantly higher after implantation of the visceral mass compared to the control group (Hang et al., 2013). Additionally, after nucleation during pearl culture, there were comparable variations in the levels of immune-related gene expression. After implantation of the visceral mass in Hyriopsis cumingii the expression of the immune-related factor gene alpha-2 macroglobulin (α2M) increased significantly (Li et al., 2010), the expression of tumor necrosis factor receptor-associated factor 6 (TRAF6) fluctuated significantly in the pearl sac, gills, and hemocytes after mantle nucleation (Huang et al., 2018), and the expression levels of galectin, allograft inflammatory factor-1 (AIF-1) were similarly changed in the pearl sacs (Bai et al., 2016; Li et al., 2016). In summary, further studies on the immunomodulatory mechanisms during nucleus insertion into the mantle and visceral masses during pearl culture are still required.
In this study, we analyzed the immune regulation process in the mantle and visceral mass of Hyriopsis cumingii at the mRNA level during pearl culture by constructing a transcriptome platform and digital gene expression profiles, and by analyzing the DEGs related to immune responses in the mantle and visceral mass during the different stages of pearl culture. The study of the adaptive immunomodulatory processes of the mantle and visceral masses of mollusks during pearl culture will have a theoretical foundation thanks to the work presented here.
Healthy Hyriopsis cumingii (2 years old and of consistent individual size) were purchased from the Weimin Pearl Farm in Jinhua City, Zhejiang Province, China. The mantles of five of the mussels, where the pearls were cultivated, were removed, put in liquid nitrogen and kept at −80°C. In addition, 200 mussels were temporarily kept in the laboratory ready for the next nucleation experiment.
According to the manufacturer’s instructions, individual mantles were lysed in 1 ml of Trizol Reagent (TaKaRa, Tokyo, Japan) for total RNA extraction. On 1% agarose gels, RNA deterioration and contamination were observed. Using a NanoDropND-2000C (Thermo, Waltham, MA, USA), RNA purity was examined. A Qubit1RNA Assay Kit (Life Technologies, Carlsbad, CA, USA) Fluorometer was used to measure the amount of RNA. An RNA Nano 6000 Assay Kit was used in a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA) to assess the RNA Integrity Number (RIN). Transcriptome analysis was then performed on samples prepared using an Illumina kit (Illumina Inc., San Diego, CA, USA), following the manufacturer’s instructions. Finally, the library was sequenced using an Illumina HiSeq TM2000 (Illumina, San Diego, CA, USA).
Trinity (https://github.com/trinityrnaseq/trinityrnaseq/wiki) was used to connect distinct contigs into scaffolds, where ambiguities (Ns) were used to indicate unknown bases between adjacent contigs. To obtain sequences with the fewest Ns that could not be expanded on either end and were referred to as unigenes, gaps in the scaffolds were filled with paired-end reads. Finally, we used BLASTX (with an E-value of 10-5) to search our unigenes against various protein databases, including Nr, Nt, Pfam, KOG/COG, Swiss-Prot, KEGG, and GO. The sequence direction of the unigenes was determined using the best hits. When a unigene did not align with any of the entries in these databases, ESTScan v.3.0.3 was used to predict its coding regions and to determine its sequence direction.
Two hundred mussels of uniform size were randomly selected from those temporarily reared in the laboratory. Twenty were assigned to a control group without insertion and 180 were assigned to the experimental group with insertion, and both were cultured in the same conditions in natural water in a nearby river. The insertion surgery was performed using the ‘bead nucleus with small pieces’ method. In the experimental group, the mussel was secured to the stand so that the back was facing to the right and the sail was down; the shell was pried up slightly with a shell opener, 3 cm diameter bead nuclei were autoclaved and dried, and then inserted into the visceral mass and the posterior part of the mantle, and then small pieces of the outer surface of the mantle were attached over the surface of the nucleus. The mussels were stood up in buckets for temporary rearing, and two days later were suspended in the pond. At 3-5 d after the insertion of the nucleus, we can judge whether the mussel spits out the nucleus, dies or heals the wound; at 20-30 d after the insertion of the nucleus, the pearl sac is basically established; at 50-60 d after the insertion of the nucleus, the pearl sac stabilizes and produces the nacre; at 90 d after the surgery, the comprehensive coverage of the surface of the nucleus with the initial completion of the nacre (Wang et al., 2022). Therefore, we chose these periods of insertion for our study. Six mussels at 5, 20, 50 and 90 d post-insertion and six in the control group were selected, and 80 –100 mg of samples were collected from the insertion sites in the mantle and visceral mass, treated with Trizol, and stored in a refrigerator at −80°C. Total RNA was extracted from the samples and stored in refrigerator at −80°C. The cDNA library produced after total RNA extraction was sequenced using an Illumina HiSeqTM2000. Base calling was used to convert the raw image data into sequence data.
Total RNA was collected from 10 samples from the mantle and visceral mass of each mussel at five time points. All of the samples were homogenized separately for RNA isolation. Total RNA was extracted, analyzed qualitatively and quantitatively as described in 2.2.
Tag library construction for the 10 Hyriopsis cumingii samples (M_Con, M_5 d, M_20 d, M_50 d, M_90 d, V_Con, V_5 d, V_20 d, V_50 d and V90 d, where M and V denote the mantle and viscera) was performed in parallel, and sequencing was performed as described in 2.2.
The data obtained after high-throughput sequencing were termed the raw data. Clean reads were obtained after removing reads with adapters, poly-N, and low-quality reads, and subsequent analysis was based on these. The guanine-cytosine (GC) content, Q20, and Q30 values of the clean reads were also calculated. The mussel transcriptome data measured in our laboratory were selected as the reference sequence. The clean reads of each sample were mapped to reference sequences using Bowtie v.0.12.9. and the results of the comparison are then tallied using RSEM.
The access data for the DEGs are the read-count data obtained from the gene expression level analysis. DEG analysis was performed using DESeq v.1.12.0 and genes with corrected p values < 0.05 were identified as DEGs. The relative expression level values log2 (ratio) of DEGs were clustered using H-cluster, K-means, and SOM.
Gene Ontology-enrichment analysis of DEGs was performed using GOSeq Release v.2.12. GO terms with corrected p values < 0.05 were identified as enrichment terms for DEGs. KOBAS v.2.0 was used to complete the KEGG enrichment analysis of DEGs, and identify the most important biochemical metabolic pathways and signal transduction pathways associated with the DEGs. Similarly, pathways with corrected p values < 0.05 were identified as enriched for DEGs.
To investigate the mechanisms governing immune regulation at different insertion sites during pearl formation, we analyzed the immune-related DEGs in the culture sites in the mantle and visceral mass at the different time points, and in the mantle and visceral mass during the same time period.
To validate the DEGs results, we randomly selected five immune-related genes for quantitative real-time reverse transcription PCR (qRT-PCR) analysis. cDNA was synthesized using a Superscript III first-strand synthesis system (Invitrogen, ThermoFisher, Waltham, MA, USA) according to the manufacturer’s protocol. Specific primers for qRT-PCR were designed using Premier Primer 5.0 (Table 1). The amplification was performed in triplicate on a Bio-Rad CFX96 (Bio-Rad, Hercules, CA, USA) using TB Green ® Premix Ex Taq ™ (TaKaRa, Tokyo, Japan). Cycling parameters were 95°C for 5 min, then 40 cycles of 95°C for 5 s and 57°C for 20 s. Melting curve analyses were performed following the amplifications to verify specific amplification. Relative gene expression data was analyzed using the 2−ΔΔCT method to obtain the expression of each sample relative to the internal reference gene EF1α (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). All reactions were performed in three replicates using six biological samples. The data were analyzed using the t-test software in SPSS v.26.0 with p < 0.05.
Illumina sequencing yielded 65.3 million raw reads, and 63.6 million reads remained after filtering out those with adapters, low quality, and N ratios greater than 10%. The error rate was 0.03%, and the Q20 and Q30 percentages were 96.95% and 90.68%, respectively. For the clean reads, 40.38% of the content was GC.
A total of 257,457 unigenes with a length of 193 million nucleotides were created from the filtered reads (Table 2). The mean length of a unigene was 751 nucleotides and lengths ranged from 18–20,429 bp. Among these unigenes, 165,546 (64.30%) of the clean unigenes were smaller than 500 bp in length, 41,398 (16.08%) were between 500–1,000 bp, 25,838 (10.03%) were between 1,000–2,000 bp, and 24,675 (9.58%) were longer than 2,000 bp. The length distribution of the assembled unigenes is shown in Figure 1.
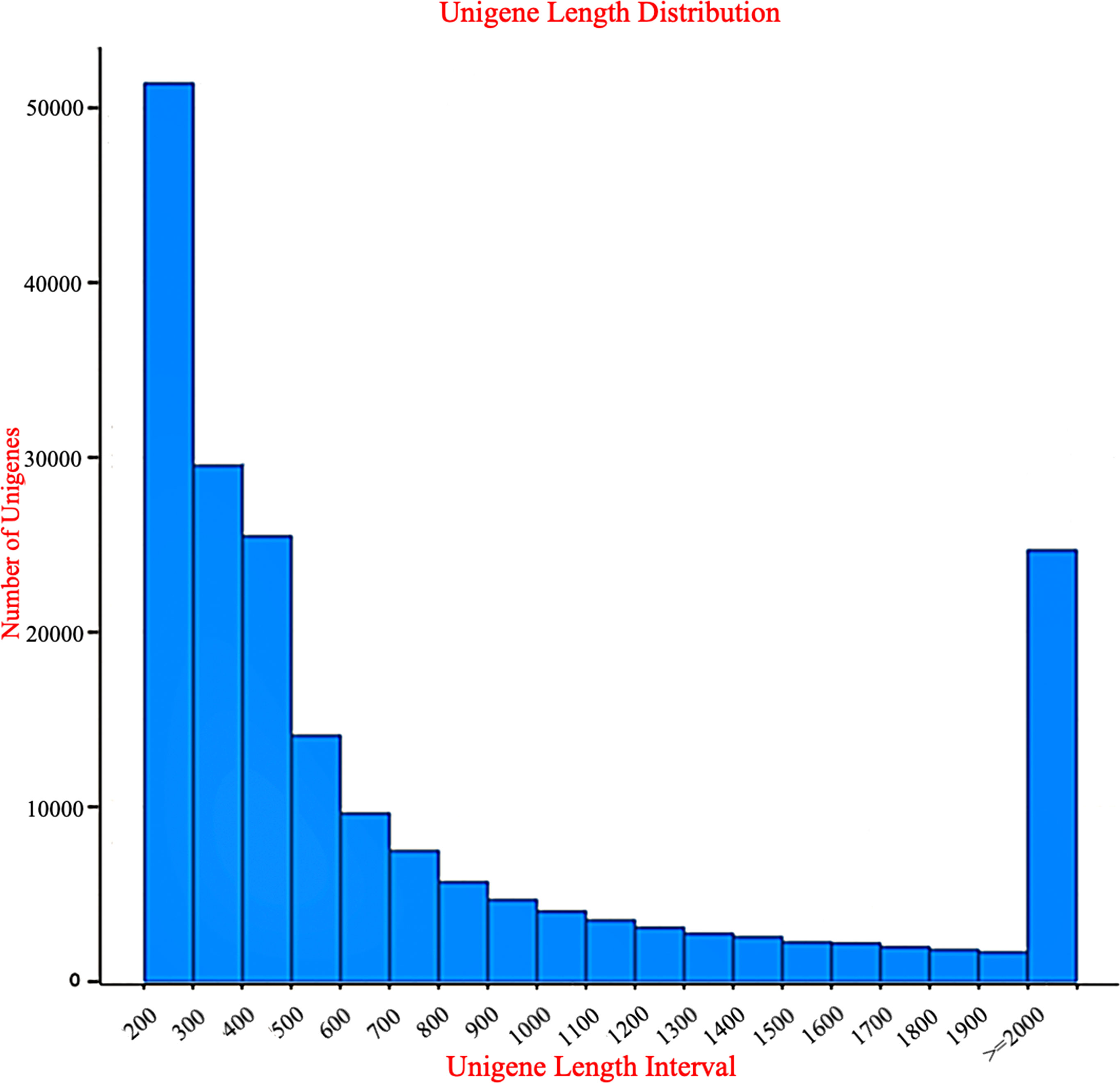
Figure 1 Data for the length distributions of unigenes The X-axis indicates the sequence length (bp), and the Y-axis indicates the number of assembled unigenes.
The unigene sequences were aligned using BlastN and BlastX searches with an E-value of less than 10-5 against the Nt, Nr, Swiss-Prot, KO, GO, KOG, and PFAM databases. Of the 257,457 unigenes, 67,821 (26.34%) were annotated to at least one database using BLAST matches and 2114 (0.82%) were annotated across all of the databases consulted. The GO database, which has annotations for 19.49% of all unigenes, was followed by annotations for 19.13% of unigenes in the PFAM database, 17.40% in the Nr database, and just 4.29% of unigenes in the Nt database.
The DEG sequencing was performed on the mantle and visceral masses of the mussels at the 5, 20, 50, and 90 d points after surgery and on the control group, and 10 DEG databases were obtained. More than 7 million raw reads were included in each database, and after filtering out splice sequences, duplicate sequences, and low quality reads, more than 7 million clean reads remained, with Q20 and Q30 both above 95% (Table 3). The results showed that the sequencing results were reliable and could be safely analyzed in the next step.
We evaluated the two DEG libraries from the various tissues at the same time point in order to determine the differentially expressed and non-differentially expressed genes in the mantle and visceral mass at various stages of pearl culture. As shown in Figure 2, a total of 2172 DEGs (1048 upregulated and 1124 downregulated, q < 0.05) were identified in the control group, compared with the visceral mass and mantle membrane. Five days after insertion, a total of 1389 DEGs (583 up-regulated and 806 down-regulated, q < 0.05) were identified, the lowest numbers recorded. The greatest number of DEGS were identified after 20 d, with a total of 3572 (1367 up-regulated and 2205 down-regulated, q < 0.05). After 50 and 90 d, 1888 (1086 up-regulated and 802 down-regulated, q < 0.05) and 2613 DEGs (1614 up-regulated and 999 down-regulated, q < 0.05) were identified, respectively. As shown in Figure 3, the clustering and expression features of DEGs obtained from the control group were similar to the mantle at 5, 50, and 90 d, while for the visceral masses, they were similar to the control group at 5, 20, and 50 d. These results indicate that the gene expression patterns of the mantle and visceral mass during pearl culture were different during the same time period.
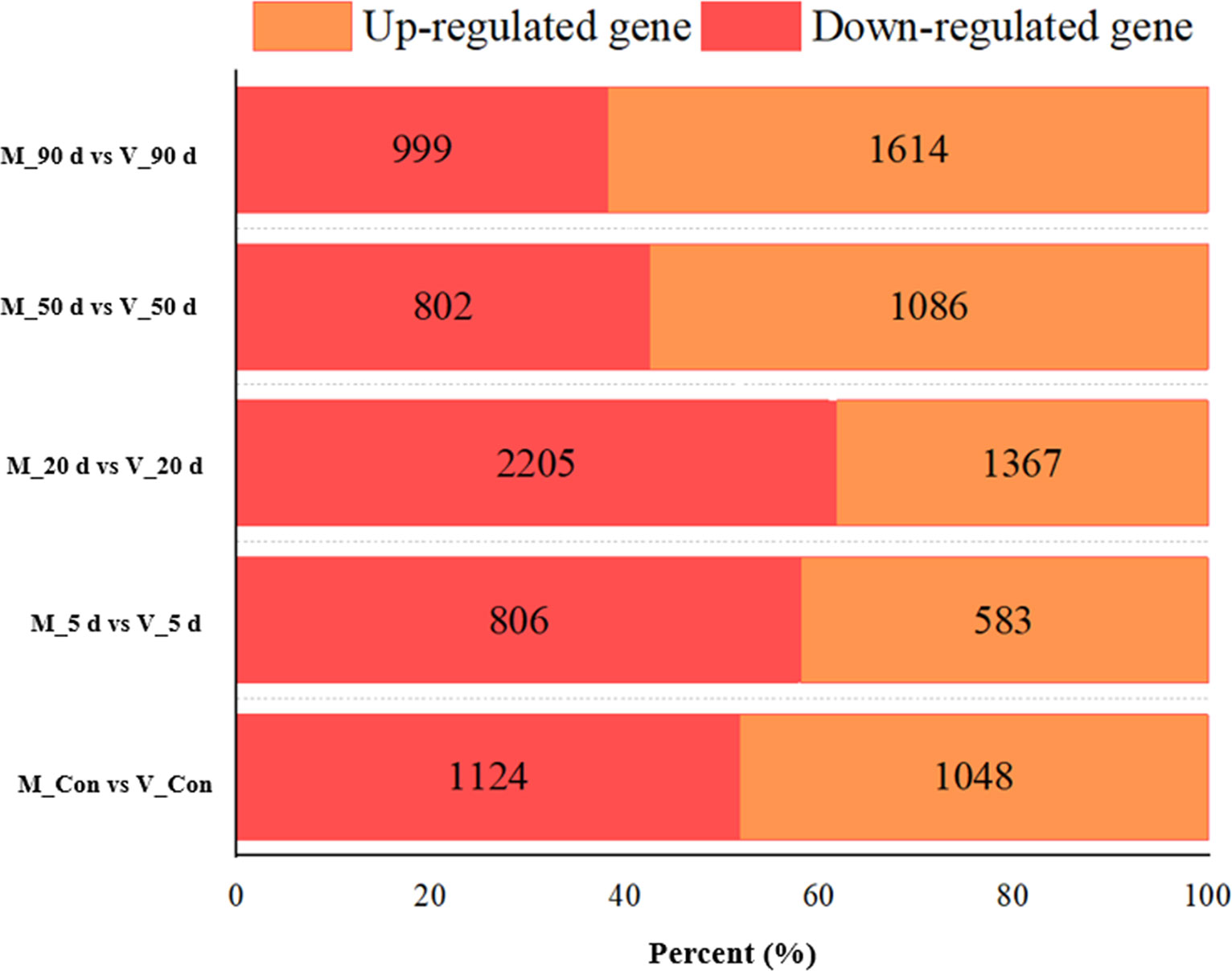
Figure 2 Statistics of differentially expressed genes between mantle and visceral mass at different days of nucleation.
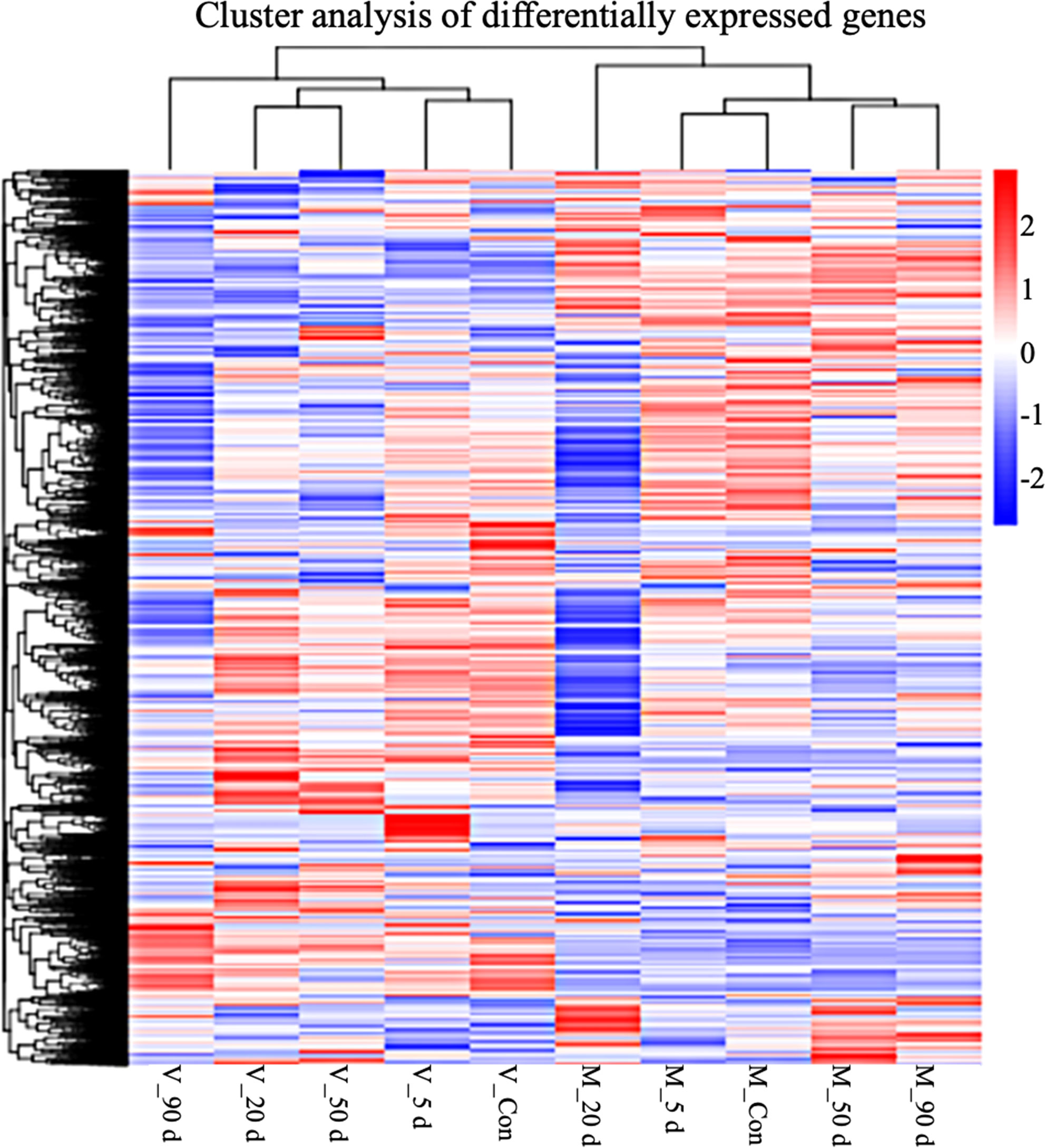
Figure 3 Overall reads per kilobase of transcript per million mapped reads (rpkm) hierarchical clustering map. Clustering was performed using log10RPKM values. Red indicates high expression genes and blue indicates low expression genes. Color gradations from blue to red indicate log10 (RPKM) transitions from small to large.
A differential significance enrichment analysis was performed by comparing the DEGs found in the data from two mantle and visceral mass samples taken at the same time point. Figure 4 shows the top 10 significant clusters in the three GO categories; biological processes (BP), cellular components (CC), and molecular functions (MF). Two clusters were highly significant: carbohydrate derivative metabolic process (CP), and extracellular region (CC). The three clusters calcium ion binding, carbohydrate binding, and chitin binding (MF) were also significant. In addition, there were more DEGs at 20 and 90 d after nucleation in most of the enrichment clusters. Moreover, in the first ten important clusters in each GO category, many were related to immunity, such as the response to oxidative stress (BP), extracellular matrix (CC), and actin cytoskeleton, and peroxidase activity (MF).
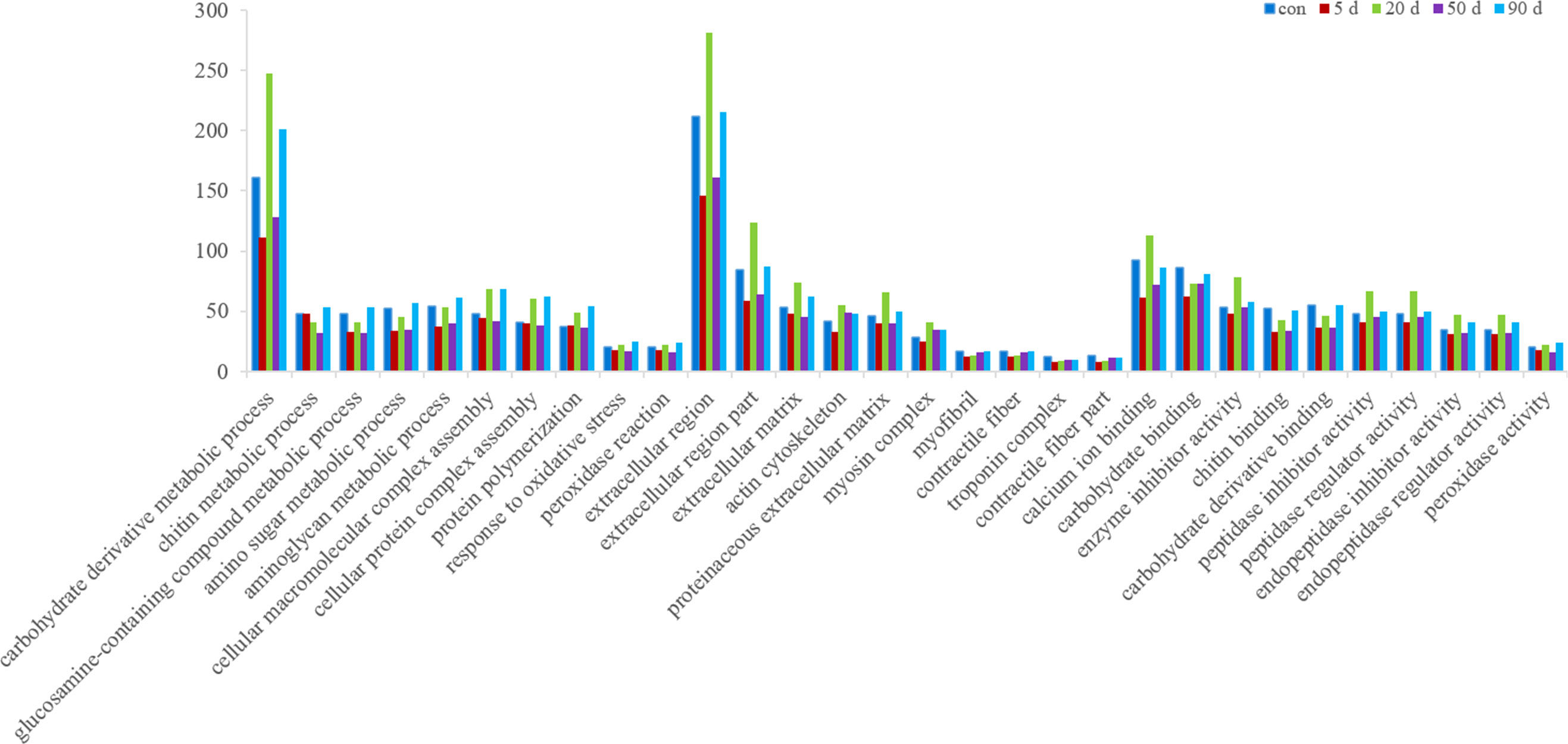
Figure 4 GO functional annotation of the DEGs between the mantle and visceral mass of Hyriopsis cumingii at different days after insertion.
Different genes perform their biological functions in organisms in coordination with each other. And the most crucial biochemical metabolic and signal transduction pathways in which DEGs participate can be found using significant pathway enrichment analysis. Scatter plots were used to present the results of the KEGG enrichment analysis. The 20 most significantly enriched pathways that we choose to display, but if there were less than 20, all were shown (Figure 5). Five days after nucleation, DEGs were mainly involved in viral myocarditis, tight junctions, proteoglycans in cancer, phagosomes, pathogenic E. coli infection, and focal adhesion. At 20 d after nucleation, DEGs were involved in these same pathways, except for focal adhesion and those associated with ribosomes. At 50 d after nucleation, DEGs were enriched in the same major pathways as the control group, while at 90 d the DEGs included those associated with viral myocarditis, tight junctions, ribosomes, phagosomes, and pathogenic E. coli infection, as well as bacterial invasion of epithelial cells, Vibrio cholerae infection, shigellosis, Hippo signaling pathway, phototransduction, leukocyte transendothelial migration, adhesion junctions, gap junctions, Parkinson’s disease, influenza A, oxidative phosphorylation, arrhythmogenic right ventricular cardiomyopathy (ARVC), complement and coagulation cascades, and myocardial contraction. All these pathways are related to immunity except for photoconduction.
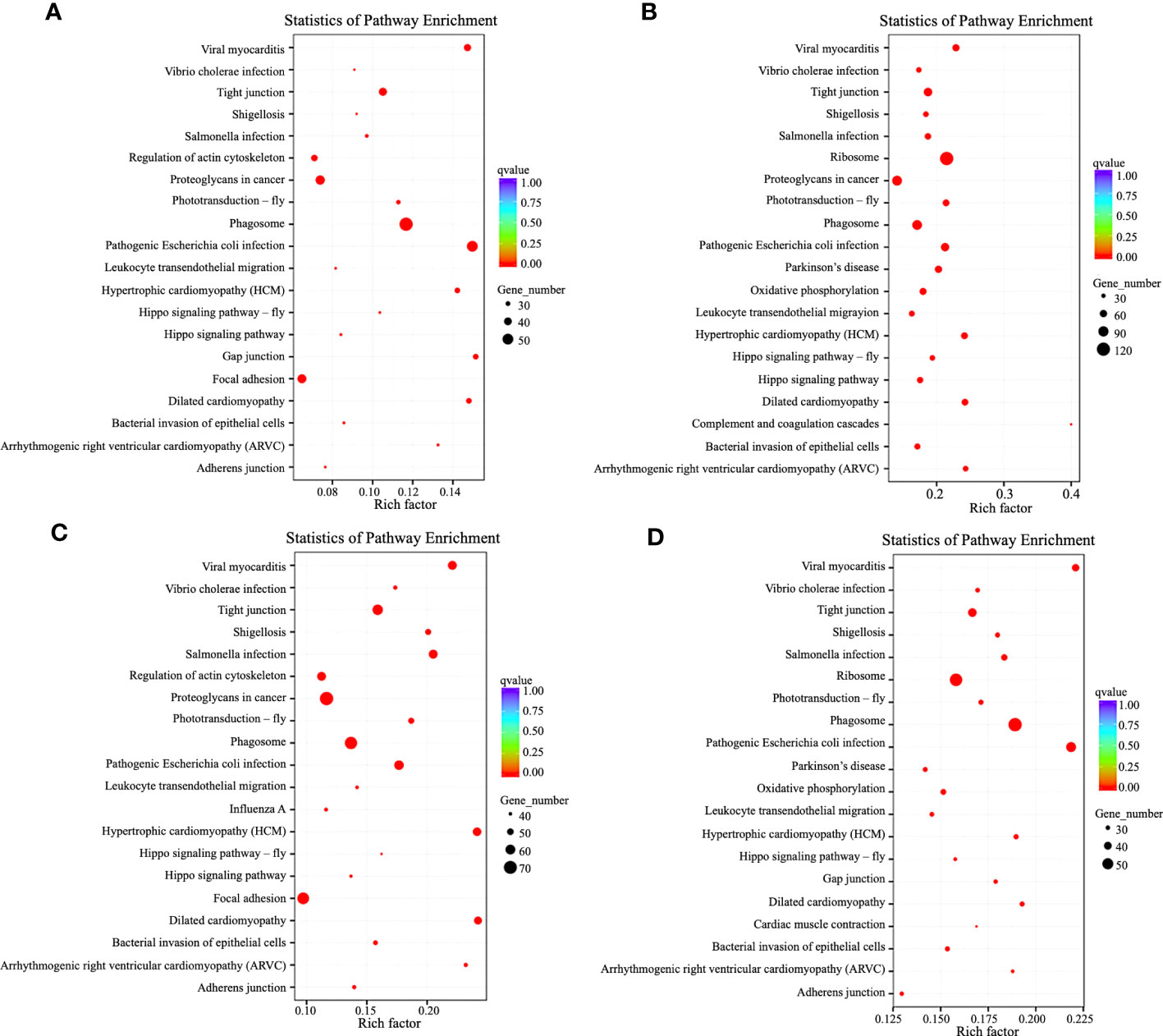
Figure 5 KEGG pathway enrichment analysis of DEGs in the mantle and visceral mass of Hyriopsis cumingii at different times after insertion. The vertical axis shows the pathway name and the horizontal axis shows the Rich factor. The size of the dots indicates the number of DEGs in that pathway, and the color of the dots corresponds to different q-value ranges. The parts of the diagram show KEGG pathway enrichment analysis of DEGs between the mantle and visceral mass: (A) 5 d after insertion; (B) 20 d after insertion; (C) 50 d after insertion; and (D) 90 d after insertion.
In order to better understand the immune system regulation in the mantle and visceral mass during pearl culture, we analyzed the differential expression of immune-related genes at different times at the same insertion site. As shown in Figure 6, the immune-related genes were up-regulated to a greater degree in the mantle during pearl culture compared with the control group, and, in contrast, more down-regulated in the visceral mass. These results indicate that immune system regulation differs between the mantle and the visceral mass during pearl culture.
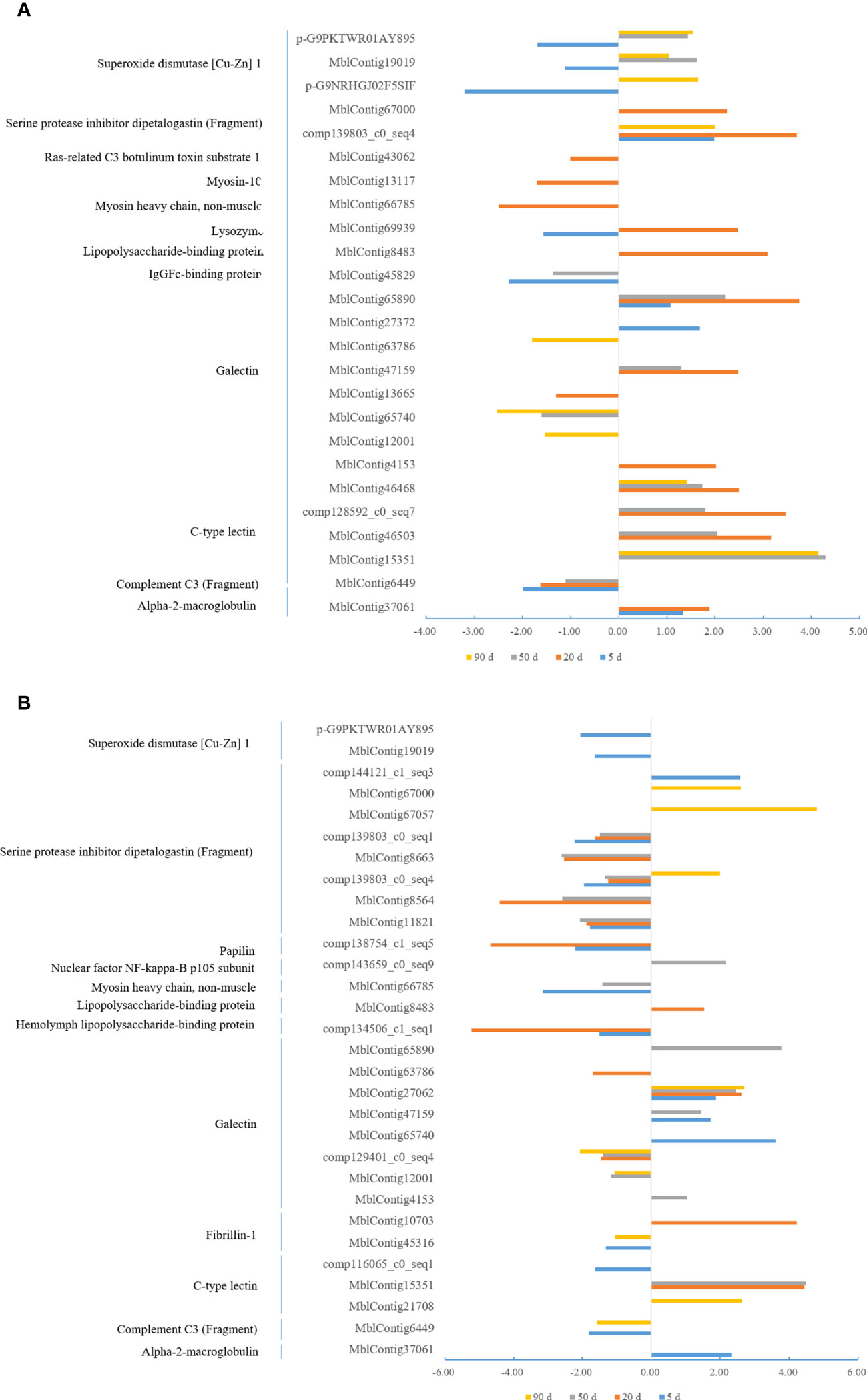
Figure 6 Analysis of DEGs at different time points in the same tissue in Hyriopsis cumingii. The horizontal axes represent Log2 (r_V/M/r_Con), where r_V and r_M are the read count values of a gene in the mantle and visceral mass tissues after insertion, r_Con are control group. (q < 0.05) (A) Mantle. (B) Visceral mass.
We screened for immune-related DEGs among those identified between the mantle and visceral mass, as shown in Figure 7 (q < 0.05), including ras-related C3 botulinum toxin substrate 1, alpha-2-macroglobulin, prothrombin, plasma kallikrein, IgGFc-binding protein, hemolymph lipopolysaccharide-binding protein, myosin-10, complement component C3, galectin, C-type lectin, mannan-binding lectin serine protease 2, lysozyme, lipopolysaccharide-binding protein, actin, serine/threonine-protein kinase NLK, superoxide dismutase [Cu-Zn] 1, serine protease inhibitor dipetalogastin (Fragment), paramyosin myosin heavy chain, non-muscle, antileukoproteinase (Fragment), myosin-10, nuclear factor NF-kappa-B p105 subunit, receptor tyrosine-protein kinase erbB-2.The highest expression of immune-related DEGs was at 90 d after insertion, with 35 unigenes (Figure 7E), followed by 32, 32, and 31 unigenes at 50 d (Figure 7D), control group (Figure 7A), and 20 d (Figure 7C), respectively, and the lowest at 5 d after insertion, with 20 unigenes (Figure 7B). Moreover, the expressions of immune-associated DEGs were more down-regulated than up-regulated in the visceral mass relative to the mantle, at different times in both the control and experimental groups.

Figure 7 Analysis of immune-related DEGs after nucleation of the mantle and visceral mass insertions in Hyriopsis cumingii. The horizontal axes represent Log2 (r_V/r_M), where r_V and r_M are the read count values of a gene in the mantle and visceral mass tissues after insertion, respectively. (q < 0.05). (A) Control group. (B) 5 days after insertion. (C) 20 days after insertion. (D) 50 days after insertion. (E) 90 days after insertion.
Based on the DEG data, we used qRT-PCR to validate five immune-related genes: C-type lectin (CLEC), complement component C3 (C3), lysozyme (Lyso), mannan-binding lectin serine protease 2 (MASP2), and superoxide dismutase [Cu-Zn] (SOD1). The results showed that the expression of 5 genes in different nucleation periods of mantle and visceral mass was consistent with DGE analysis. Compared with the mantle, the expression of C3 was up-regulated at 20 days after nucleation in visceral mass (Figure 8A), MASP2 was down-regulated at 50 days after nucleation in visceral mass (Figure 8C), Lyso (Figure 8B), CLEC (Figure 8D) and SOD1 (Figure 8E) were down-regulated at all time periods after nucleation in visceral mass.
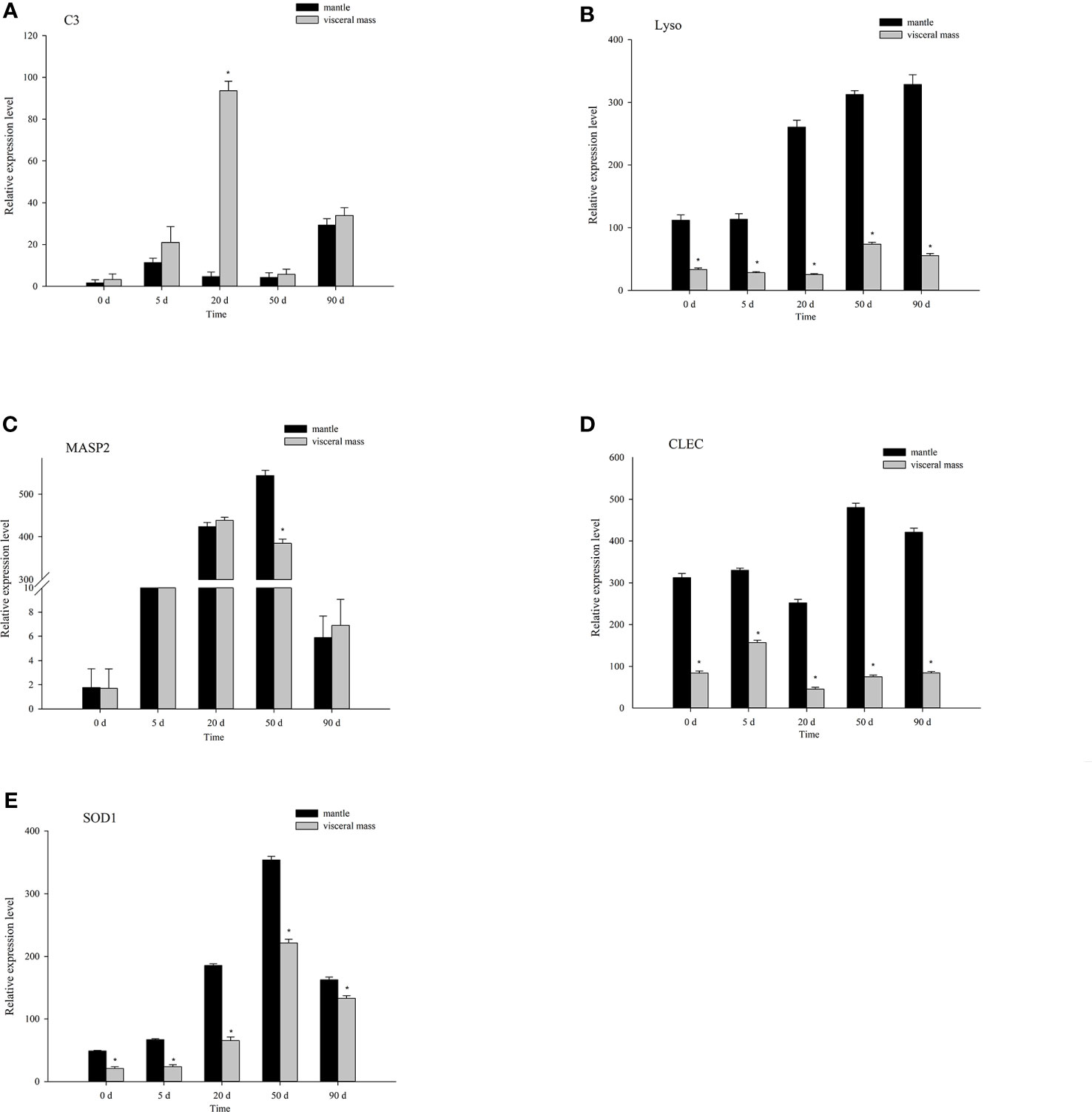
Figure 8 Quantitative real-time PCR was used to analyze the relative expression levels of five immune-related genes in the mantle and visceral mass of Hyriopsis cumingii on different days after nucleation. * indicates p < 0.05. (A) Complement component C3 (C3). (B) Lysozyme (Lyso). (C) Mannan-binding lectin serine protease 2 (Masp2). (D) C-type lectin (CLEC). (E) Superoxide dismutase [Cu-Zn] (SOD1).
In this study, we used DGE techniques to identify DEGs in the mantle and visceral mass of Hyriopsis cumingii at different times after nucleus insertion. The formation of pearls is a biomineralization process with immune regulation. When the nucleus and the small pieces of the mantle of the donor mussel enter the mantle or the visceral mass of the host mussel, the host cells are stimulated to secrete nacre to form a pearl sac, and the deposited nacre wraps the foreign body and gradually forms a pearl. The nucleus insertion process promotes a transplantation tolerance reaction, prompting an immune rejection of the small piece of tissue and nucleus in the donor shell (Wang et al., 2016). The mantle is commonly used as a pearl production site, but most of the pearls it produces are small and of poor quality. The visceral mass facilitates the cultivation of large, nucleated pearls due to its larger size. However, insertion into the visceral mass in large scale pearl production can increase the infection rate (Li et al., 2010) and reduce the survival rate of the mussels, as well as lowering the rate of production of nucleated pearls (Liang et al., 2015).
The cluster analysis of DEGs showed different gene expression patterns in the mantle and visceral mass, indicating that the two tissues were differently regulated during pearl culture. In addition, the GO cluster analysis results showed that there were more DEGs at 20 d and 90 d after insertion, which may be due to the fact that these two periods occurred relatively early and late in the process of pearl formation, respectively (Bai et al., 2017), indicating that the regulatory mechanisms of mantle and visceral mass pearl formation were different during these two important periods. The KEGG cluster analysis results showed that DEGs were clustered into immune-related pathways. Therefore, we screened the immune-related DEGs of the mantle and visceral mass at different periods after insertion, in order to verify the cluster analysis results and explore the immune regulatory mechanism of the visceral mass after insertion, laying a groundwork for greater understanding of the formation of visceral pearl clusters.
The immune system of invertebrates differs from that of vertebrates in that they only show innate immunity and no acquired immunity. As invertebrates, mollusks have no specific immune cells and corresponding antibodies and rely on humoral factors and hemocytes in the hemolymph to defend against foreign invasion, so-called ‘humoral and cellular immunity’ (Li et al., 2022a; Roch, 1999). One of the earliest immune system components, the complement system is crucial to both innate and acquired immunity (Hajishengallis et al., 2017). One of the more well researched complement activation pathways is the lectin pathway, which is crucial for pathogen conditioning, chemotaxis, leukocyte activation, direct pathogen killing, and inflammation regulation (Kinoshita, 1991). C3, mannose-binding lectin (MBL), and MASP are essential components in the lectin pathway (Matsushita, 2018). A novel MBL has been identified in Pacific oysters Crassostrea gigas and has been reported to bind to MASP-1, promoting C3 cleavage and thus activating the complement system, which is involved in the perforation and growth inhibition of bacteria (Sun et al., 2021). Complement C3 and Masp2 are both enriched in the complement and coagulation cascades pathway, along with two other enriched DEGs, Prothrombin and plasma kallikrein. Prothrombin not only plays a role in hemostasis and coagulation, but also induces a series of inflammatory responses (Luyendyk et al., 2019). Prothrombin has also been demonstrated to degrade fibrinogen to create fibrin clots, which inhibit the spread of pathogens in vertebrates (Sun, 2005). Plasma kallikrein plays an important role in the inflammatory response after tissue injury and is considered to be an important new drug target in anti-inflammatory immunopharmacology (Wang et al., 2019). In this study, both C3 and Masp2 were downregulated after visceral mass nucleation relative to the mantle. In contrast, Prothrombin and plasma kallikrein were upregulated.
Cultured pearl formation has been linked to wound healing and increased immunity to invading pathogens (Huang et al., 2018). An accurate immune recognition mechanism and a perfect immune regulation system are necessary for effective defense against invading invaders. Both cellular and humoral immunity rely heavily on lectins. Lectins on the surface of mollusks help cells to recognize pathogens and activate cell chemotaxis so that they can move towards pathogens, and adsorp and eventually engulf them (Kuchel et al., 2010). It has been found that the galectin HcGal1 of Hyriopsis cumingii and the C-type lectin HdClec of the Halitosis discus hannai both play a role in promoting hemocyte phagocytosis of pathogenic bacteria (Bai et al., 2016; Lv et al., 2022). In this study, we found that, C-type lectins and galectins in the mantle were differentially expressed at each stage after insertion and were more down-regulated than up-regulated. IgGFc-binding protein was identified as the Fc part of the binding site of the Ig G molecule, which may play a role in cell protection and anti-inflammatory processes in tissues (Xiong et al., 2014). We found that expression of the gene for the Fc gamma binding protein (FcGBP) was lower in the visceral mass than in the mantle after nucleus insertion. FcGBP may play a role in cell protection and anti-inflammatory responses during formation of the pearl sac.
In addition to the above, the humoral immunity of mollusks includes oxidative enzymes, hydrolytic enzymes, and other immune functional proteins, which play an important role in resisting foreign invasion and inflammatory reactions (Teng et al., 2003). One of the most crucial lysosome enzymes is lyso. By destroying and dissolving microscopic bacteria, it plays a part in filtering saltwater microorganisms and avoiding diseases. Lyso cannot only dissolve pathogen cell walls, but also partially or completely inhibit their survival or normal development. It has been discovered that lyso plays a crucial part in how mollusks respond to inflammation. (Cajaraville and Pal, 1995). Liu et al. reported that Lyso can dissolve the cell walls of pathogens and partially or completely inhibit their growth and reproduction (Li et al., 2021; Gonzalez et al., 2022). SOD1 is an important antioxidant enzyme that can remove oxygen free radicals and protect organisms from free radical attacks. Studies have shown that its activity is strongly tied to the immune ability of aquatic organisms, and it is crucial for enhancing the defensive abilities of phagocytic cells and, by extension, the body’s overall immune system. (Xie et al., 2018; Liu et al., 2022). Li et al. found that SOD1 activity in the hemolymphatic system of Hyriopsis cumingii increased significantly compared with a control group after visceral mass insertion (Li et al., 2022b). In this study, we found that these immune-related genes were mostly down-regulated in the visceral mass after nucleation, relative to the mantle.
The regulation of the release of inflammatory mediators and apoptosis may be beneficial to the healing of epithelial cells and the formation of the pearl sac after insertion (Li et al., 2022a). In this study, the lipopolysaccharide-binding protein, serine/threonine-protein kinase NLK, antileukoproteinase (fragment), nuclear factor NF-kappa-B p105 subunit, and fibrillin-1 were related to inflammatory responses and cell apoptosis and were differentially expressed. Through the heterodimer p65-p50, the typical NF-κB signaling pathway controls hundreds of distinct genes, including those that produce pro-inflammatory cytokines, chemokines, adhesion molecules, and enzymes. The NFKB1 gene produces the protein p105, which the proteasome can cleave into p50. The p50 protein most often binds to p65, and together they form the heterodimer that drives pro-inflammatory gene expression. Increased activation of NF-κB signaling leads to increased deposition of extracellular matrix at the site of injury and contributes to injury repair (Best et al., 2019). Antileukoproteinase regulates inflammation and immune responses after bacterial and intracellular parasitic infections. It plays a role in regulating NF-κB activation and inflammatory responses, preventing tissue damage and promoting wound healing by limiting protease activity (Klimenkova et al., 2014). The toll-like receptor signaling pathway, which can control NF-B-related molecules and take part in inflammatory reactions, contains a protein called lipopolysaccharide-binding protein (Won et al., 2021). Fibrillin-1 regulates inflammatory responses by participating in the TGF-β signaling pathway (Lien et al., 2019). The serine/threonine protein kinases NLK play a critical role in determining cell fate and control cell death. They control several transcription factors, take part in a number of signaling pathways, including Notch, MAPK, and Wnt, etc. (Zhang et al., 2015; Shi et al., 2019). In this study, the expression levels of these genes in the visceral mass tissue after nucleation were mostly down-regulated compared with the mantle.
In summary, we identified key immune-related differential genes during insertion into the mantle and the visceral mass using transcriptome sequencing and digital gene expression analysis, and found that the expression of immune-related genes was mostly down-regulated after insertion into the visceral mass, compared with the mantle. This may be related to the fact that the mantle, as the first line of autoimmune defense in Hyriopsis cumingii, is the main immune system organ. Moreover, we also confirmed that Hyriopsis cumingii was more susceptible to infection and even death after visceral mass nucleation. We hypothesize that the immunomodulatory mechanism after insertion into the viscera is different from that of the mantle. Nonetheless, these efforts are insufficient and the hypotheses that have been derived require further research and validation. Molecular biology techniques, such as overexpression or knockdown, are needed to verify the functions of relevant genes after nucleation and to gain a deeper understanding of the immune molecular mechanisms of the pearl formation process. In the case of visceral mass cultured pearls, the expression of immune-related genes can be regulated to reduce the rate of pearl spitting and infection, and to improve the yield of visceral mass cultured pearls.
The data involved in this study have been uploaded to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). The project number is PRJNA992764, and the transcriptome sequencing data are numbered SRR25206151 through SRR25206163. This information has been provided in the Supplementary Material.
The animal study was reviewed and approved by Shanghai Ocean university. The study was conducted in accordance with the local legislation and institutional requirements.
XS: Investigation, Writing – original draft, writing – review & editing. YGC: Conceptualization, writing – review & editing. LJ: Review & editing. HW: Formal analysis. XC: Investigation. YWC: Software. XX: Data curation. JL: Methodology. ZB: Methodology, supervision, project administration. WL: Methodology, writing – review & editing, supervision, project administration. All authors contributed to the article and approved the submitted version.
Our work was supported by the National Key Research and Development Program of China (2022YFD2400105), the earmarked fund for China Agriculture Research System of MOF and MARA (CARS-49), the National Natural Science Foundation of China (31872565).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1251251/full#supplementary-material
Bai Z., Zhao L., Chen X., Li Q., Li J. (2016). A galectin from Hyriopsis cumingii involved in the innate immune response against to pathogenic microorganism and its expression profiling during pearl sac formation. Fish Shellfish Immunol. 56, 127–135. doi: 10.1016/j.fsi.2016.07.006
Bai Z., Zhao L., Chen X., Li Q., Li J. (2017). A galectin contributes to the innate immune recognition and elimination of pathogens in the freshwater mussel Hyriopsis cumingii. Dev. Comp. Immunol. 73, 36–45. doi: 10.1016/j.dci.2017.03.008
Best K. T., Lee F. K., Knapp E., Awad H. A., Loiselle A. E. (2019). Deletion of NFKB1 enhances canonical NF-kappaB signaling and increases macrophage and myofibroblast content during tendon healing. Sci. Rep. 9, 10926. doi: 10.1038/s41598-019-47461-5
Cajaraville M. P., Pal S. G. (1995). Morphofunctional study of the haemocytes of the bivalve mollusc Mytilus galloprovincialis with emphasis on the endolysosomal compartment. Cell Structure Funct. 20, 355–367. doi: 10.1247/csf.20.355
Gonzalez R., Gonzalez D., Stambuk F., Ramirez F., Guzman F., Mercado L., et al. (2022). A g-type lysozyme from the scallop Argopecten purpuratus participates in the immune response and in the stability of the hemolymph microbiota. Fish Shellfish Immunol. 123, 324–334. doi: 10.1016/j.fsi.2022.03.015
Hajishengallis G., Reis E. S., Mastellos D. C., Ricklin D., Lambris J. D. (2017). Novel mechanisms and functions of complement. Nat. Immunol. 18, 1288–1298. doi: 10.1038/ni.3858
Hang K., Shi Z., Li W., Li q. (2013). Effects of different inserting-nucleus locations on body physiological metabolism in visceral mass of Hyriopsis cumingii, China. Acta Hydrobiologica Sin. 37, 1085–1093. doi: 10.7541/2013.147
He X., Shi Z., Chen X., Cheng Q. (2010). Effect pearl-nucleus-inserting operation in visceral mass on hemocytes of Hyriopsis cumingii, China. Acta Hydrobiologica Sinica. 34, 410–417. doi: 10.3724/SP.J.1035.2010.00410
Hua D., Neves R. J. (2011). Captive survival and pearl culture potential of the pink heelsplitter potamilus alatus. North Am. J. Aquaculture. 69, 147–158. doi: 10.1577/a05-108.1
Huang D., Bai Z., Shen J., Zhao L., Li J. (2018). Identification of tumor necrosis factor receptor-associated factor 6 in the pearl mussel Hyriopsis cumingii and its involvement in innate immunity and pearl sac formation. Fish Shellfish Immunol. 80, 335–347. doi: 10.1016/j.fsi.2018.06.035
Huang D., Shen J., Li J., Bai Z. (2019). Integrated transcriptome analysis of immunological responses in the pearl sac of the triangle sail mussel (Hyriopsis cumingii) after mantle implantation. Fish Shellfish Immunol. 90, 385–394. doi: 10.1016/j.fsi.2019.05.012
Jin C., Zhao J., Pu J., Liu X., Li J. (2019). Hichin, a chitin binding protein is essential for the self-assembly of organic frameworks and calcium carbonate during shell formation. Int. J. Biol. Macromol. 135, 745–751. doi: 10.1016/j.ijbiomac.2019.05.205
Kinoshita T. (1991). Biology of complement: The overture. Immunol. Today 12, 291–295. doi: 10.1016/0167-5699(91)90001-A
Klimenkova O., Ellerbeck W., Klimiankou M., Unalan M., Kandabarau S., Gigina A., et al. (2014). A lack of secretory leukocyte protease inhibitor (SLPI) causes defects in granulocytic differentiation. Blood. 123, 1239–1249. doi: 10.1182/blood-2013-06-508887
Kuchel R. P., Raftos D. A., Birch D., Vella N. (2010). Haemocyte morphology and function in the Akoya pearl oyster, Pinctada imbricata. J. Invertebr Pathol. 105, 36–48. doi: 10.1016/j.jip.2010.04.011
Li Q., Bai Z., Zhao L., Li J. (2016). Characterization of allograft inflammatory factor-1 in Hyriopsis cumingii and its expression in response to immune challenge and pearl sac formation. Fish Shellfish Immunol. 59, 241–249. doi: 10.1016/j.fsi.2016.10.037
Li L., Cardoso J. C. R., Felix R. C., Mateus A. P., Canario A. V. M., Power D. M. (2021). Fish lysozyme gene family evolution and divergent function in early development. Dev. Comp. Immunol. 114, 103772. doi: 10.1016/j.dci.2020.103772
Li X., Feng S., Xuan X., Wang H., Shen X., Chen Y., et al. (2022a). A proteomic approach reveals biomineralization and immune response for mantle to pearl sac in the freshwater pearl mussel (Hyriopsis cumingii). Fish Shellfish Immunol. 127, 788–796. doi: 10.1016/j.fsi.2022.06.057
Li W., Shi Z., He X. (2010). Study on immune regulation in Hyriopsis cumingii Lea: effect of pearl-nucleus insertion in the visceral mass on immune factors present in the hemolymph. Fish Shellfish Immunol. 28, 789–794. doi: 10.1016/j.fsi.2010.01.019
Li W., Yang J., Feng S., LI X., Chen Y., Shen X., et al. (2022b). Analysis of proteomic differences and immune factors between Hyriopsis cumingii mantle and pearl sac, China. J. Of Shanghai Ocean University. 31, 379–390. doi: 10.12024/jsou.2021060349
Liang F., Xie S., Lin W. (2015). Progress on pearl production from Hyriopsis Cumingii, China. Trans. Oceanology LIimnology. 2, 113–118. doi: 10.13984/j.cnki.cn37-1141.2015.02.016
Lien H. C., Lee Y. H., Juang Y. L., Lu Y. T. (2019). Fibrillin-1, a novel TGF-beta-induced factor, is preferentially expressed in metaplastic carcinoma with spindle sarcomatous metaplasia. Pathology. 51, 375–383. doi: 10.1016/j.pathol.2019.02.001
Liu Y., Bao Z., Lin Z., Xue Q. (2022). Genome-wide identification and characterization of superoxide dismutases in four oyster species reveals functional differentiation in response to biotic and abiotic stress. BMC Genomics 23, 378. doi: 10.1186/s12864-022-08610-9
Liu X., Liu Z., Jin C., Li H., Li J. L. (2019). A novel nacre matrix protein hic24 in Hyriopsis cumingii is essential for calcium carbonate nucleation and involved in pearl formation. Biotechnol. Appl. Biochem. 66, 14–20. doi: 10.1002/bab.1690
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Luyendyk J. P., Schoenecker J. G., Flick M. J. (2019). The multifaceted role of fibrinogen in tissue injury and inflammation. Blood. 133, 511–520. doi: 10.1182/blood-2018-07-818211
Lv X., Chen Y., Cai Y., Lv C., Bi X., Wang M., et al. (2022). A single-CRD C-type lectin from Haliotis discus hannai acts as pattern recognition receptor enhancing hemocytes opsonization. Fish Shellfish Immunol. 125, 17–25. doi: 10.1016/j.fsi.2022.04.049
Matsushita M. (2018). The complement system of agnathans. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01405
Roch P. (1999). Defense mechanisms and disease prevention in farmed marine invertebrates. Aquaculture. 172, 125–145. doi: 10.1016/S0044-8486(98)00439-6
Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shi C., Xu L., Tang Z., Zhang W., Wei Y., Ni J., et al. (2019). Knockdown of Nemo−like kinase promotes metastasis in non−small−cell lung cancer. Oncol. Rep. 42, 1090–1100. doi: 10.3892/or.2019.7226
Sun H. (2005). The interaction between pathogens and the host coagulation system. Physiology. 21, 281–288. doi: 10.1152/physiol.00059.2005
Sun J., Wang L., Yang W., Li Y., Jin Y., Wang L., et al. (2021). A novel C-type lectin activates the complement cascade in the primitive oyster Crassostrea gigas. J. Biol. Chem. 297, 101352. doi: 10.1016/j.jbc.2021.101352
Teng L., Wang Y., Hong S., Chen J., Meng Q., Liu L. (2003). Purification and properties of superoxide dismutase( SOD); in allium sativum. Chem.res.chinese 19, 442–445. doi: 1005-9040(2003)-04-442-04
Wang H., Huang K., Feng S., Li X., Wang L., Xuan X., et al. (2022). Effect on physiological responses and cultured pearl biology of different nuclei insertion positions in the visceral mass of freshwater pearl mussel (Hyriopsis cumingii). Aquaculture Res. 53, 6782–6796. doi: 10.1111/are.16145
Wang Z., Wang B., Chen G., Jian J., Lu Y., Xu Y., et al. (2016). Transcriptome analysis of the pearl oyster (Pinctada fucata) hemocytes in response to Vibrio alginolyticus infection. Gene. 575, 421–428. doi: 10.1016/j.gene.2015.09.014
Wang B., Yan X., Chen F., Yang A., Lu Y., Wu Y. (2019). Plasma kallikrein contributes to ambient particulate matter-induced lung injury. Biochem. Biophys. Res. Commun. 518, 409–415. doi: 10.1161/ATVBAHA.119.313503
Weng X., Li Y., Fan H. C., Jiang S., You H., Zhang D., et al. (2012). Effect of different shapes of mantle piece on death rate and nucleus spitting rate during the recuperation period of pearl oyster Pinctada fucata nucleus insertion Vol. 23 (China: Guangdong Agricultural Sciences). doi: 10.16768/j.issn.1004-874x.2012.23.069
Won Y., Yang J. I., Park S., Chun J. S. (2021). Lipopolysaccharide binding protein and CD14, cofactors of toll-like receptors, are essential for low-grade inflammation-induced exacerbation of cartilage damage in mouse models of posttraumatic osteoarthritis. Arthritis Rheumatol. 73, 1451–1460. doi: 10.1002/art.41679
Xie Y., Chen H., Zheng S., Zhang X., Mu S. (2018). Molecular characterization of cu/Zn SOD gene in Asian clam Corbicula fluminea and mRNA expression and enzymatic activity modulation induced by metals. Gene. 663, 189–195. doi: 10.1016/j.gene.2018.04.044
Xiong L., Wen Y., Miao X., Yang Z. (2014). NT5E and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal transition (EMT) are associated with tumor progression and survival of patients with gallbladder cancer. Cell Tissue Res. 355, 365–374. doi: 10.1007/s00441-013-1752-1
Keywords: Hyriopsis cumingii, pearl culture, mantle, visceral mass, immune-related differentially expressed genes
Citation: Shen X, Chen Y, Jia L, He W, Chen X, Chen Y, Xuan X, Li J, Bai Z and Li W (2023) Transcriptome and digital gene expression analysis reveal immune responses of mantle and visceral mass pearl culturing in Hyriopsis cumingii. Front. Mar. Sci. 10:1251251. doi: 10.3389/fmars.2023.1251251
Received: 04 July 2023; Accepted: 02 August 2023;
Published: 17 August 2023.
Edited by:
Yingxiang Lim, University of Michigan, United StatesReviewed by:
Zheng Yuan, China Academy of Chinese Medical Sciences, ChinaCopyright © 2023 Shen, Chen, Jia, He, Chen, Chen, Xuan, Li, Bai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Li, d2psaUBzaG91LmVkdS5jbg==; Zhiyi Bai, enliYWlAc2hvdS5lZHUuY24=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.