- 1Shenzhen Key Laboratory of Marine Archaea Geo-Omics, Department of Ocean Science and Engineering, Southern University of Science and Technology, Shenzhen, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 3Department of Biology, HADAL, Nordcee & DIAS, University of Southern Denmark, Odense M, Denmark
- 4Shanghai Sheshan National Geophysical Observatory, Shanghai, China
- 5Key Laboratory of Physical Oceanography, Ministry of Education, Ocean University of China, Qingdao, China
Branched glycerol dialkyl glycerol tetraethers (brGDGTs) are bacterial membrane lipids that are widely used in terrestrial paleoclimatic reconstructions. Recent studies have reported that brGDGTs can also be produced by marine bacteria. However, the environmental factors influencing marine-derived brGDGTs and their source organisms remain largely unknown. Here, we investigated the distribution and composition of brGDGTs and a suite of their putative derivatives called overly branched GDGTs (obGDGTs) in the Mariana Trench core sediments (water depth 8300 m, core length 320 cm), as well as the composition of bacterial communities. The ratio of the branched over isoprenoid tetraethers (BIT) was 0.03-0.21 (average 0.07; SD = 0.04; n = 21) and the ratio ΣIIIa/ΣIIa of brGDGTs was 0.93-7.47 (average 3.39; SD = 1.73; n = 21), which support the in situ production of brGDGTs. Co-occurrence network analysis revealed that a total of 33 types of bacteria at the order level (e.g., Armatimonadota DG-56, Proteobacteria Rhodospirillales, Chloroflexi SAR202_clade) were closely related to the distribution of brGDGTs and obGDGTs, which could be potential sources for these compounds. The abrupt increase in brGDGT and obGDGT concentrations in deeper oxygen-depleted sediments and their good correlations with anaerobic bacterial abundances suggest that these brGDGTs and obGDGTs may be produced by anaerobic bacteria residing in the anoxic sediments. Considerable variation in the degrees of methylation and cyclization of brGDGTs (obGDGTs) under different redox conditions indicate that sediment oxygen levels may have a profound impact on the presence and abundance of brGDGTs and obGDGTs, which should be considered when applying them for paleo-temperature or pH reconstructions. This study shows that brGDGTs and obGDGTs obtained from the Mariana Trench were probably produced by a variety of bacterial phyla indigenous in the hadal ocean, which are different from Acidobacteria commonly considered to be major terrestrial sources of brGDGTs.
1 Introduction
Molecular biomarkers preserved in sedimentary archives provide valuable information on changes in paleoenvironments and climates (Summons et al., 2022). A suite of bacterial cell membrane lipids called branched glycerol dialkyl glycerol tetraethers (brGDGTs) are among these biomarkers that are ubiquitously distributed in virtually all ecosystems, including soils, peats, rivers, lakes, and marine environments (Tierney and Russell, 2009; Schouten et al., 2013; Günther et al., 2014; Zheng et al., 2016). BrGDGTs consist of two alkyl chains bound by ether bonds to two glycerol moieties, which are distinguished by the number and position of methyl branches and cyclopentane rings on the alkyl chains (Sinninghe Damsté et al., 2000; De Jonge et al., 2013). Several proxies based on brGDGT compositions, such as the Branched over Isoprenoid Tetraether (BIT) index (Hopmans et al., 2004; De Jonge et al., 2015) and the Methylation and Cyclization of Branched Tetraether (MBT and CBT) indexes (Weijers et al., 2007a), have been developed and widely employed to estimate organic carbon source, temperature and soil pH in modern and ancient geological settings (Weijers et al., 2007b; Wang et al., 2013; Wang et al., 2014; Yang et al., 2014; Sun et al., 2016; Wang et al., 2016; Zheng et al., 2016).
Despite widespread applications of brGDGTs in paleoclimatic research, their source organisms remain elusive. The stereochemistry of the glycerol units in brGDGTs points to a bacterial rather than archaeal origin (Sinninghe Damsté et al., 2000). Based on environmental studies in soils and peats, the phylum Acidobacteria was assumed to be the most likely biological sources of brGDGTs (Weijers et al., 2006; Weijers et al., 2009). This hypothesis was subsequently supported by the detection of abundant iso-diabolic acids, as brGDGT precursor lipids in strains of acidobacterial subgroups 1 and 3, and the identification of trace amount of brGDGT-Ia in two subgroup 1 cultures (Sinninghe Damsté et al., 2018).
Halamka et al. (2021) reported that oxygen limitation triggers brGDGT production in at least one subgroup 1 strain and confirmed the biosynthesis of three brGDGT compounds. More recently, Chen et al. (2022) and Halamka et al. (2023) identified another brGDGT-producing Acidobacterium (Candidatus Solibacter usitatus), and found that it can make a large portion of its cellular membrane out of structurally diverse brGDGTs. These studies are of great significance for understanding the mechanism of brGDGT-based proxies under laboratory conditions. Chen et al. (2022) specifically identified C5 methylated brGDGTs-producers by screening the homologs of archaeal tetraether synthase (Tes), which was initially proposed by Zeng et al. (2022) as a key protein responsible for synthesizing archaeal isoprenoid GDGTs. This approach offers a viable means of identifying potential brGDGTs producers in environmental samples. However, the structural variation of brGDGTs in diverse environments is much greater than that observed in the acidobacterial cultures, indicating that other phyla of bacteria could be possible brGDGT-producers (Liu et al., 2012b; Zhang et al., 2012; Zhang et al., 2013; Liu et al., 2014; Xie et al., 2014; Chen et al., 2018a; Weber et al., 2018; De Jonge et al., 2019; Xiao et al., 2022).
A challenging issue for the interpretation of brGDGT-based proxies arises from the mixing of brGDGTs from various sources into sedimentary archives (Peterse et al., 2009; Zell et al., 2014; De Jonge et al., 2015; Sinninghe Damsté, 2016). BrGDGTs were traditionally thought to be specific to terrestrial bacteria. However, mounting evidence suggests that brGDGTs can also be produced by marine bacteria (Zhang et al., 2012; Liu et al., 2014; Weijers et al., 2014; Zell et al., 2014; Sinninghe Damsté, 2016; Xiao et al., 2016; Xu et al., 2020). By compiling globally distributed soils and marine sediments, Xiao et al. (2016) developed a proxy, as expressed by the abundance ratio of hexamethylated to pentamethylated brGDGT (ΣIIIa/ΣIIa), to constrain the marine-derived brGDGTs with significantly higher ΣIIIa/ΣIIa values. Sinninghe Damsté (2016) proposed a threshold of #ringstetra > 0.7 as an indicative criterion for predominantly marine-sourced brGDGTs, primarily based on shelf sediments. The composition and environmental implication of marine-derived brGDGTs remain elusive, preventing their applications in marine environments. Additionally, microbial community producing brGDGTs can be expected to vary remarkably between terrestrial and marine settings (Bahram et al., 2018; Hoshino et al., 2020). The reported pure cultures that can biosynthesize brGDGTs, however, almost exclusively belong to terrestrial bacteria (Sinninghe Damsté et al., 2018; Halamka et al., 2021; Chen et al., 2022; Halamka et al., 2023) and little is known about the biological sources of marine-derived brGDGTs.
In addition to the commonly measured brGDGTs, the so-called overly-branched GDGTs (obGDGTs) have been detected in a semi-global range of marine surface sediments (Liu et al., 2012a; Liu et al., 2014; Xie et al., 2014; Becker, 2015). ObGDGTs differ from brGDGTs by having a higher degree of methylation (Liu et al., 2012a). ObGDGTs in marine sediments are thought to be primarily derived from anaerobic planktonic microbes (Liu et al., 2014; Xie et al., 2014). To date, very limited studies have reported the environmental distribution pattern and laboratory cultivation of obGDGTs, making it difficult to determine their biological sources and functional roles. Recently, Connock et al. (2022) presented a wide array of biomarkers in core samples from the North Atlantic Ocean, and showed the fractional abundance of obGDGTs abruptly increased during Oceanic Anoxic Event 2, implying that obGDGTs may be suitable to infer low oxygen conditions. The proxy potential of obGDGTs would benefit from detailed investigations of obGDGT distributions in marine sediments.
While it is commonly viewed that temperature and pH play a significant role in regulating the distribution of brGDGTs in natural environments (Weijers et al., 2007a; Peterse et al., 2012; Xiao et al., 2015; De Jonge et al., 2021), oxygen levels were also increasingly assumed to have an impact on brGDGTs (Weber et al., 2018; Halamka et al., 2021; Wu et al., 2021). Marine sediments are generally characterized by a redox gradient from oxic at surface to anoxic at deeper depths caused by gradual consumption of oxygen (Glud, 2008; Jørgensen et al., 2022). This gradient enables us to investigate the impact of oxygen levels on these brGDGTs and obGDGTs produced in situ. Additionally, bacterial communities vary significantly with sediment depth in conjunction with the redox zonation (Hiraoka et al., 2019; Hoshino et al., 2020). By comparing bacterial community compositions and brGDGT and obGDGT distributions, and relating their variations to sediment depth, we can better understand the potential candidates contributing to these bacterial lipids and constrain their ecophysiology in marine sediments.
Here, we investigated the vertical distribution of brGDGTs and obGDGTs and of microbial 16S rRNA genes retrieved from a Mariana Trench sediment core MT03. As one of the most remote places on Earth, the Mariana Trench offers an ideal opportunity to distinguish GDGTs produced in situ from a terrestrial origin that usually muddles the interpretation of offshore sediments. We aimed to 1) determine the composition and distribution of brGDGTs and obGDGTs from the Mariana Trench core sediments, 2) constrain their possible sources, and 3) assess their environmental and biological implications.
2 Materials and methods
2.1 Study area and samples
The Mariana Trench is formed as the subduction of the Pacific plate beneath the eastern edge of the Philippine Sea plate. It has a total length of ca. 2500 km and a mean width of 70 km (Jamieson, 2015). Our study area is located in the southern section of the Mariana Trench covering the Challenger Deep (~11,000 m depth). The Mariana Trench is overlain by extremely oligotrophic waters. However, sediments in the Challenger Deep were found to support elevated microbial activity compared to adjacent abyssal plains (Glud et al., 2013).
During an expedition aboard RV DongFang-Hong 2 (March 2019), a sediment core (MT03, 142.32°E, 11.12°N, water depth 8300 m, core length 320 cm) was retrieved from the Mariana Trench (Figure 1A), which was subsequently sliced into 2 cm segments at approximately 10 cm intervals. A total of 21 samples were collected at 9-287 cm depth intervals between the top and the bottom of the sediment core (320 cm depth). All sediment samples were freeze-dried at −40°C and homogenized by steel spatulas.
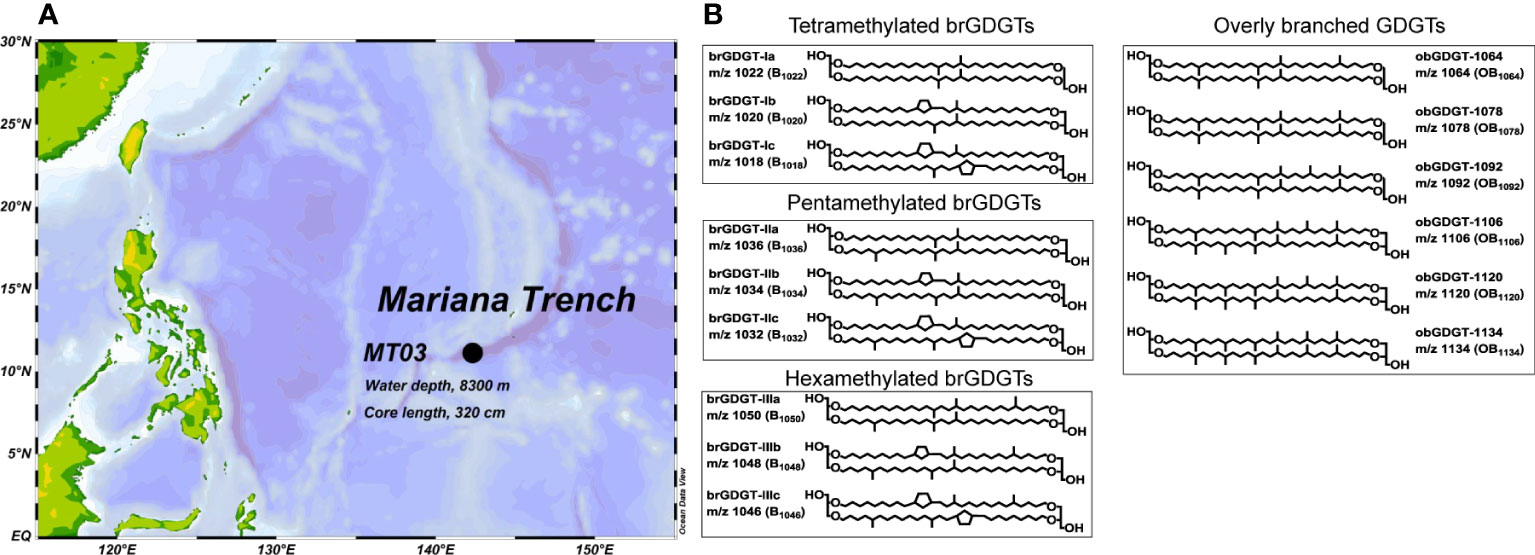
Figure 1 (A) Location of study site in the Mariana Trench. (B) Molecular structures of studied biomarkers in this work.
2.2 Lipid extraction and analysis
A portion of the freeze-dried sediment samples was ground into powder manually. Powdered sediments (3-5 g) were dispensed for lipids extraction by a modified Bligh and Dyer method (Sturt et al., 2004). Samples were extracted twice ultrasonically using a mixture of methanol (MeOH)/dichloromethane (DCM)/phosphate buffer (pH 7.4; 2:1:0.8, v/v/v) for 15 min each, and centrifuged at 3000 rpm for 5 min. Samples were then extracted with MeOH/DCM/trichloroacetic acid buffer (pH 2; 2:1:0.8, v/v/v) for two more times. All supernatants were collected; DCM and water were then added to achieve a final ratio of 1:1:0.8 for MeOH/DCM/water. The DCM phase was collected as total lipid extracts (TLEs) and dried under a N2 stream. 50 μl synthesized C46-GTGT (1.189 ng/μl) was added as an internal standard before lipid analysis.
The TLEs were re-dissolved in 200 μl methanol and centrifuged at 14,000 g for 5 min to remove large particles. GDGTs were analyzed by a Waters ACQUITY I-Class ultra-performance ion mobility liquid chromatography (UPLC) coupled with a SYNAPT G2-Si quadrupole time-of-flight high-resolution mass spectrometry (qTOF-HRMS) equipped with an electrospray ionization (ESI) source operated at positive mode. Separation of compounds was achieved using an ACE SuperC18 column (2 μm, 2.1 × 150 mm; Advanced Chromatography technologies LTD) kept at 45°C modified from Zhu et al. (2013). The injection volume was 10 μl. The solvent A was methanol (100%) and the solvent B was isopropanol (100%). Each flow phase was added with 0.04% formic acid (98%, Sigma Aldrich, Germany) and 0.1% ammonia (>25% NH3, Sigma Aldrich, Germany). During the initial 5 min, the lipids were eluted with 100% A. Subsequently, the proportion of B was increased to 24% within the next 5 min, further increased to 60% during the 10-36 min. The proportion of solvent B was ultimately raised to 90% during the 36-45 min. Subsequently, the column was re-equilibrated with 100% A. The flow rate was maintained at 0.35 ml min-1 throughout the experiment.
Detection of GDGTs was achieved in Fast-DDA mode with mass ranges of m/z 100−2000 for MS1 and 50−2000 for MS2 and scan time for 0.2 s. The five most abundant ions at the scan were fragmented through collision-induced dissolution (CID) to perform data-dependent MS2 analysis. MS setting was as follows: Capillary 2.5 kV, source temperature 120 °C, sampling cone 45, source offset 80, desolvation gas flow 800 l/hr at 350 °C, cone gas flow 50 l/hr, nebulizer gas flow 6.5 bar.
The regular brGDGTs and obGDGTs were identified by accurate molecular mass and isotope pattern as well as MS2 mass spectra according to Liu et al. (2012b), with representative MS2 spectra shown in Supplementary Figure S3. The peaks were integrated by extracting ion chromatograms with ± 0.05 Da mass width including adducts of [M+H]+, [M+NH4]+ and [M+Na]+. Concentrations of brGDGTs and obGDGTs were calculated by comparing the peak areas of target compounds and peak areas of the known amount of C46-GTGT internal standard (Huguet et al., 2006). The chemical structure diagram of brGDGTs identified in this study is shown in Figure 1B.
2.3 GDGT-derived parameters
The BIT and ΣIIIa/ΣIIa indexes were adopted from Hopmans et al. (2004) and Xiao et al. (2016), respectively. The CBT and MBT indexes were calculated according to the definitions of Weijers et al. (2007a). The MIob and MIob/br indexes were determined to calculate the degree of methylation of obGDGTs and obGDGTs/brGDGTs with the modifications of Liu et al. (2014).
2.4 DNA extraction and 16S rRNA gene sequencing
DNA was extracted from 21 samples using the FastDNA™ SPIN Kit for Soil (MP Bio) according to manufacturer’s protocols. The quantitative PCR (qPCR) was performed using a thermocycler PCR system (Quantstudio 5, ABI, United States). Bacterial 16S rRNA gene was amplified with primers Bac349F (5’–AGGCAGCAGTDRGGAAT–3’) and Bac806R (5’–GGACTACYVGGGTATCTAAT–3’) (Takai and Horikoshi, 2000). The real-time polymerase chain reaction was performed with the following program (PCR stage): 5 s for denaturation at 95°C, 30 s at 60°C for annealing, and 1 min at 72°C for extension. The whole process went through 35 cycles. The standard product was diluted into six gradients, with the highest concentration being 6.5×109 and the lowest concentration being 6.5×104. Triplicates analyses were performed for each sample and the standard. The R2 values for the standard curve were > 0.98 and the efficiency was between 98–100%.
The universal primers used for archaeal and bacterial 16S rRNA gene sequencing were 515FmodF (5’-GTGYCAGCMGCCGCGGTAA-3’) and 806RmodR (5’- GGACTACNVGGGTWTCTAAT-3’) (Caporaso et al., 2011; Parada et al., 2016). Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform/NovaSeq PE250 platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
After demultiplexing, the resulting sequences were merged by using FLASH (v1.2.11) (Magoc and Salzberg, 2011) and quality filtered by using fastp (0.19.6) (Chen et al., 2018b). Then the high-quality sequences were de-noised using DADA2 plugin in the Qiime2 (version 2020.2) pipeline with recommended parameters, which obtained single nucleotide resolution based on error profiles within samples. DADA2-denoised sequences are usually called amplicon sequence variants (ASVs). Taxonomic assignment of ASVs was performed using the Naive bayes consensus taxonomy classifier implemented in Qiime2 and the SILVA 16S rRNA database (v138). The bacterial sequences were retained to perform downstream analysis. To minimize the effects of sequencing depth, every sample was sequenced with a minimum of 100,000 clean reads. Analyses of the 16S rRNA microbiome sequencing data were performed using the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Pearson correlation coefficients between bacterial orders and relative abundances of brGDGTs and obGDGTs were calculated using the R software (version 4.0.1, http://www.r-project.org). The significant positive correlations indicated that the shift of bacterial community might exert a significant impact on GDGT distribution by potentially producing brGDGTs and obGDGTs. The Positive correlations (r > 0.65, p < 0.05) were visualized as networks in Gephi (version9.0, https://gephi.org/). We further examined the presence of Tes homolog protein (e-value< 1e−50, identity >20%) in bacterial orders that showed strong positive relationship with the relative abundance of brGDGTs and obGDGTs. The bacterial genomes were not determined in our samples; instead, the Tes homolog (MA_1486) protein sequence (Zeng et al., 2022) was searched from the deposited genomes in NCBI database, which belonged to the same order of bacteria from our samples. The presence of Tes homolog indicates bacteria that are potentially capable of synthesizing brGDGTs, which may hold significant implications for identifying the biological sources of brGDGTs in the Mariana Trench in the future.
2.5 Bulk organic carbon analysis
After being freeze-dried and ground, ca. 1 g of homogenized sediment powder was digested in 5% HCl for 24 h to remove inorganic carbon. Total organic carbon (TOC) contents were obtained using Thermo Scientific FLASH 2000 CHNS/O Elemental Analyzer. The standard error of three times determination of standard element sample was less than 0.3%.
3 Results
3.1 Bulk organic carbon content
The TOC content (expressed as weight percentage of TOC to dry weight sediment) varies in a narrow range from 0.04% to 0.18%, with an average of 0.11 ± 0.03% (avg. ± std). The highest TOC content is at 106 cm depth, and the lowest is at 136 cm depth (Figure 2A).
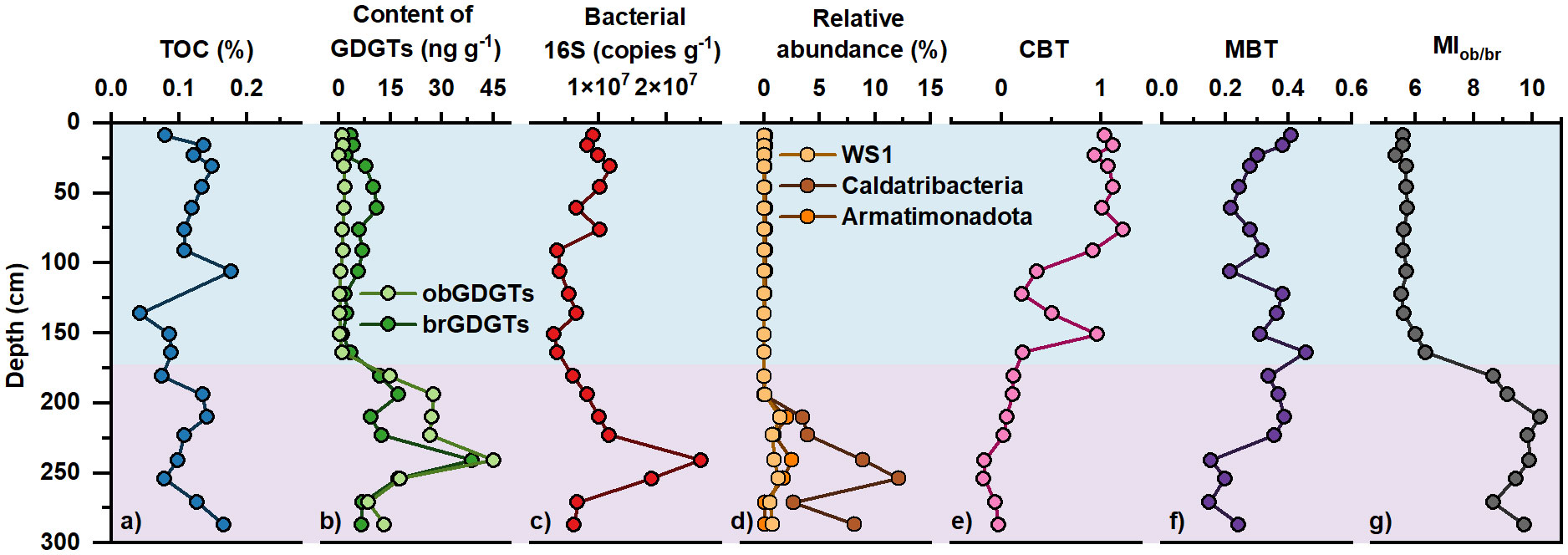
Figure 2 Vertical distribution of (A) TOC content, (B) absolute concentration of brGDGTs and obGDGTs, (C) abundance of bacterial 16S rRNA genes, (D) relative abundance of anaerobic bacteria, (E) CBT index, (F) MBT index, and (G) MIob/br index in the Mariana Trench core sediments.
3.2 Composition and distribution of brGDGTs
A suite of brGDGTs and obGDGTs compounds were detected in the MT03 core. The overall variation of concentration and composition of both brGDGTs and obGDGTs along sediment depth were shown in Figure 3. The concentration and composition of brGDGTs varied significantly with sediment depth. The concentration of brGDGTs varied between 0.92 and 38.95 ng g-1 dry sediment (the unit “ ng g-1” was used in subsequent text.) in MT03 core, with an average value of 8.86 ± 8.16 ng g-1 (Figures 2B, 3A). The concentration of brGDGTs increased from 5.03 ± 3.10 ng g-1 at 9 – 164 cm depth to 15.08 ± 9.83 ng g-1 at 164 – 287 cm depth (Figures 2B, 3A). BrGDGT-IIIa was the dominant compound (40.06 ± 12.26% of total brGDGTs), followed by brGDGT-Ia (20.87 ± 6.90%) and brGDGT-IIa (14.46 ± 6.89%) (Table S2). The proportions of brGDGTs with one (Ib, IIb and IIIb) and two (Ic, IIc and IIIc) cyclopentyl rings were 19.64 ± 14.16% and 4.97 ± 4.79%, respectively (Figure 3C). The relative abundance of acyclic brGDGTs showed a gradual decreasing trend down core, whereas brGDGTs with one or two cyclopentyl rings exhibited a gradual increasing trend down core (Figure 3C). The classification based on the number of methyl groups showed the dominance of hexamethylated brGDGTs (IIIa, IIIb and IIIc; 47.31 ± 10.95%) over tetramethylated (Ia, Ib and Ic; 30.24 ± 8.32%) and pentamethylated (IIa, IIb and IIc; 22.45 ± 9.01%) brGDGTs.
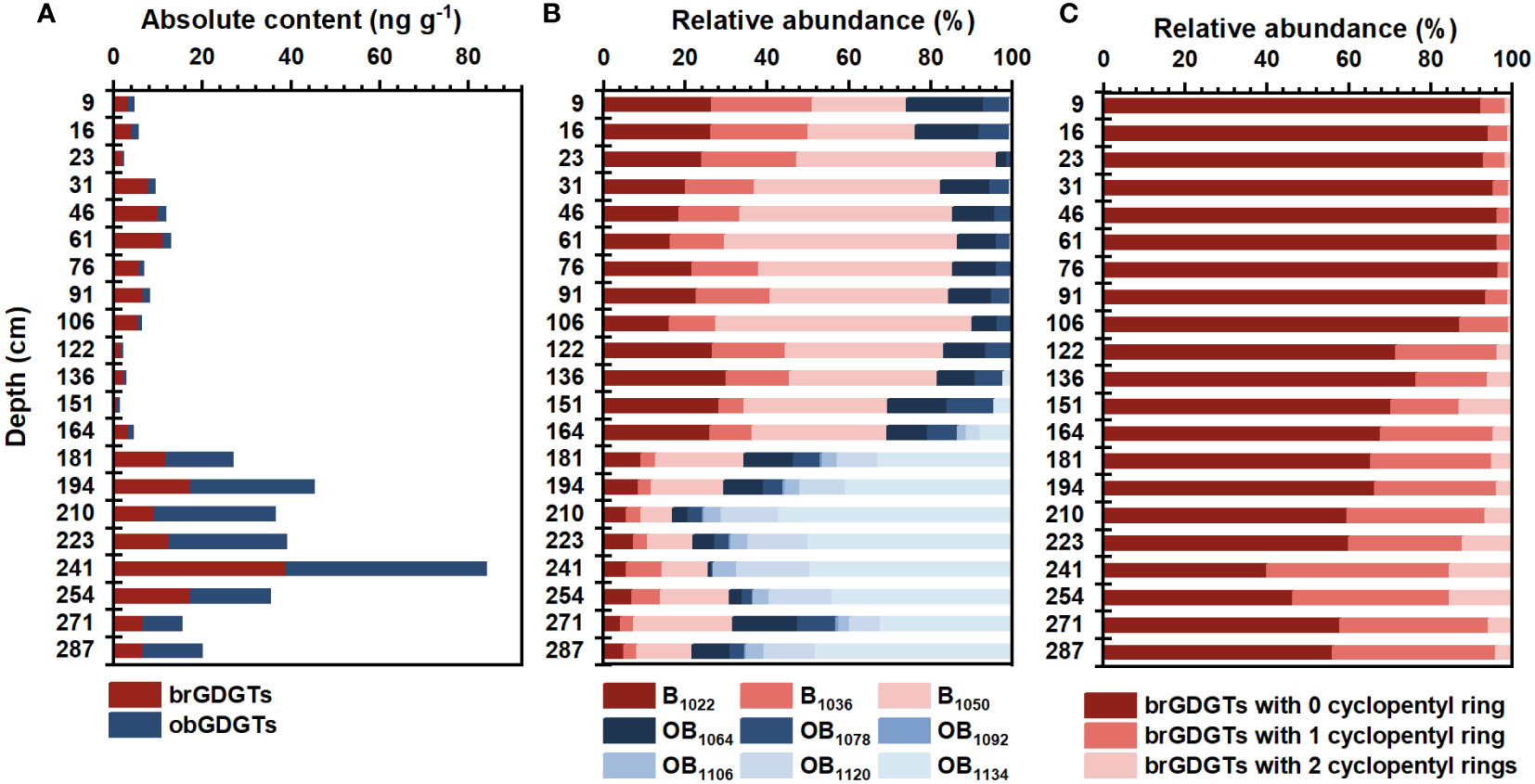
Figure 3 (A) Absolute concentration of brGDGTs and obGDGTs, and (B) relative abundance of acyclic brGDGTs and obGDGTs in the Mariana Trench core sediments; (C) relative abundance of brGDGTs with 0-, 1- and 2 cyclopentyl rings in the Mariana Trench core sediments. Note: The term “GDGTs” in the figure refers to the GDGTs included in the legend of the figure.
The BIT index varied between 0.03 and 0.21 (0.07 ± 0.04), showing an overall decreasing trend with sediment depth. Whereas the ΣIIIa/ΣIIa index varied between 0.93 and 7.47 (3.39 ± 1.73), showing an overall increasing trend with large variations from the core top to the maximum sediment depth. The CBT index ranged from -0.18 to 1.22 (0.50 ± 0.49). It was high (0.87 ± 0.32) in the upper 151 cm sediment layer with fluctuations at 106 – 136 cm depth, and then gradually decreased from 0.96 to -0.03 towards the core bottom (Figure 2E). The MBT index varied between 0.15 and 0.46 (0.30 ± 0.08), and displayed dramatic variations along depth (Figure 2F).
3.3 Composition and distribution of obGDGTs
All six known obGDGTs (obGDGT1064, 1078, 1092, 1106, 1120 and 1134) were detected in the core sediments. The concentration of obGDGTs varied between 0.08 and 45.08 ng g-1 (9.19 ± 12.48 ng g-1) (Figure 3A). The composition of obGDGTs was dominated by the obGDGT1064 (42.77 ± 27.29% of total obGDGTs), followed by the obGDGT1134 (26.27 ± 27.30%), obGDGT1078 (21.05 ± 12.61%), obGDGT1120 (7.29 ± 8.72%), obGDGT1106 (2.44 ± 2.89%) and obGDGT1092 (0.19 ± 0.33%) (Table S2).
The concentration and composition of obGDGTs were highly variable with sediment depth. The concentration of obGDGTs increased from less than 2 ng g-1 at 9 – 164 cm depth to above 8 ng g-1 at 164 – 287 cm depth, with the highest value occurring at 241 cm depth (45.08 ng g-1) (Figure 3A). ObGDGT1064 and 1078 were the most abundant compounds above 151 cm (65.17 ± 7.92% and 30.75 ± 4.93%, respectively), but decreased sharply to 4.57% and 4.48%, respectively, at 210 cm depth, and then remained at a low level with large variability (9.47 ± 7.68% and 5.43 ± 4.34%, respectively) towards the core bottom. In contrast, obGDGT1092, 1106, 1120 and 1134 exhibited opposite downcore profiles. Specifically, they had much lower abundance above 151 cm (<0.01%, <0.01%, 0.14 ± 0.45% and 3.94 ± 4.41%, respectively), but increases strongly to 0.42%, 5.08%, 16.87 and 68.58%, respectively, at 210 cm depth, and then remained at a high level (0.43 ± 0.43%, 5.67 ± 1.14%, 18.56 ± 4.69% and 60.45 ± 6.89%, respectively) towards the core bottom (Table S2).
The MIob/br index in the core MT03 varied between 5.34 and 10.27 (7.13 ± 1.87%). It was the lowest at 9 – 151 cm depth (5.34 – 6.02), and showed a rapidly increasing trend until 210 cm depth (10.27), and then remained at a high level (8.67 – 9.91) toward the bottom of the core (Figure 2G).
3.4 Composition and distribution of microbial community
The Illumina sequencing of the bacterial and archaeal 16S rRNA genes resulted in a total of 4,052,760 high-quality sequences. These sequences were clustered into 7539 ASVs with 6426 ASVs being classified to bacteria. The bacterial sequences were assigned to 382 orders, 158 classes and 57 phyla at different phylogenetic levels. Bacterial abundance varied obviously across the profile (3.4×106 – 2.5×107 copies/g) (Figure 2C). Proteobacteria (69% – 87%) was the overwhelmingly dominant bacterial phylum in the MT03 sediment core, while other major groups included Chloroflexi (4% – 13%), Actinobacteriota (2% – 4%) and Planctomycetota (1% – 4%) (Figure 4).
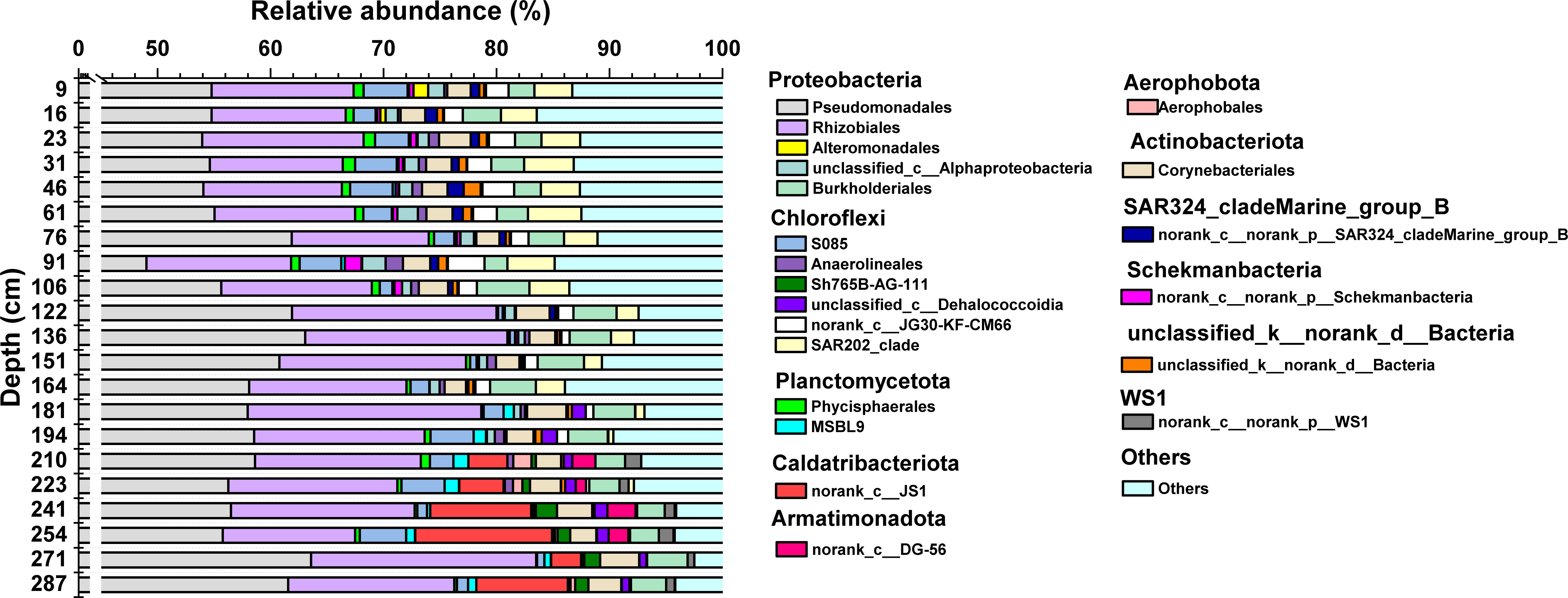
Figure 4 Relative abundance of bacterial communities at the order level. The legend on the right shows the phylum category belonging to different orders (e.g., Proteobacteria and Aerophobota). Groups with relative abundance < 1% were summarized as “others”.
A depth-dependent variation of bacterial community composition was observed. In the top section of the sediment core (9 – 194 cm depth), proteobacterial orders of Pseudomonadales, Rhizobiales, and Burkholderiales and the Chloroflexi order of SAR202_clade were most abundant. The proportions of WS1 (order WS1) (Ferrer et al., 2011; Lee et al., 2013), Caldatribacteriota (order JS1) (Carr et al., 2015; Nobu et al., 2016; Liu et al., 2019; Katayama et al., 2020) and Armatimonadota (order DG-56) (Zhao et al., 2018; Ismail et al., 2021), which are reported as anaerobic bacteria in previous studies, remarkably increased at 194 –287 cm depth (Figure 2D).
Co-occurrence network analysis was performed to correlate relative abundance of bacterial communities to brGDGTs and obGDGTs. The results based on positive Pearson correlation (r > 0.65, p < 0.05) showed that 22 orders with the Tes gene were significantly related to brGDGTs and 11 orders with Tes were significantly related to obGDGTs (Figure 5). These orders contained the Tes homolog protein with relatively high alignment scores (e-value < 1e−50, identity > 20%), including Armatimonadota DG-56, Proteobacteria Rhodospirillales, and the Chloroflexi SAR202_clade (Figure 5).
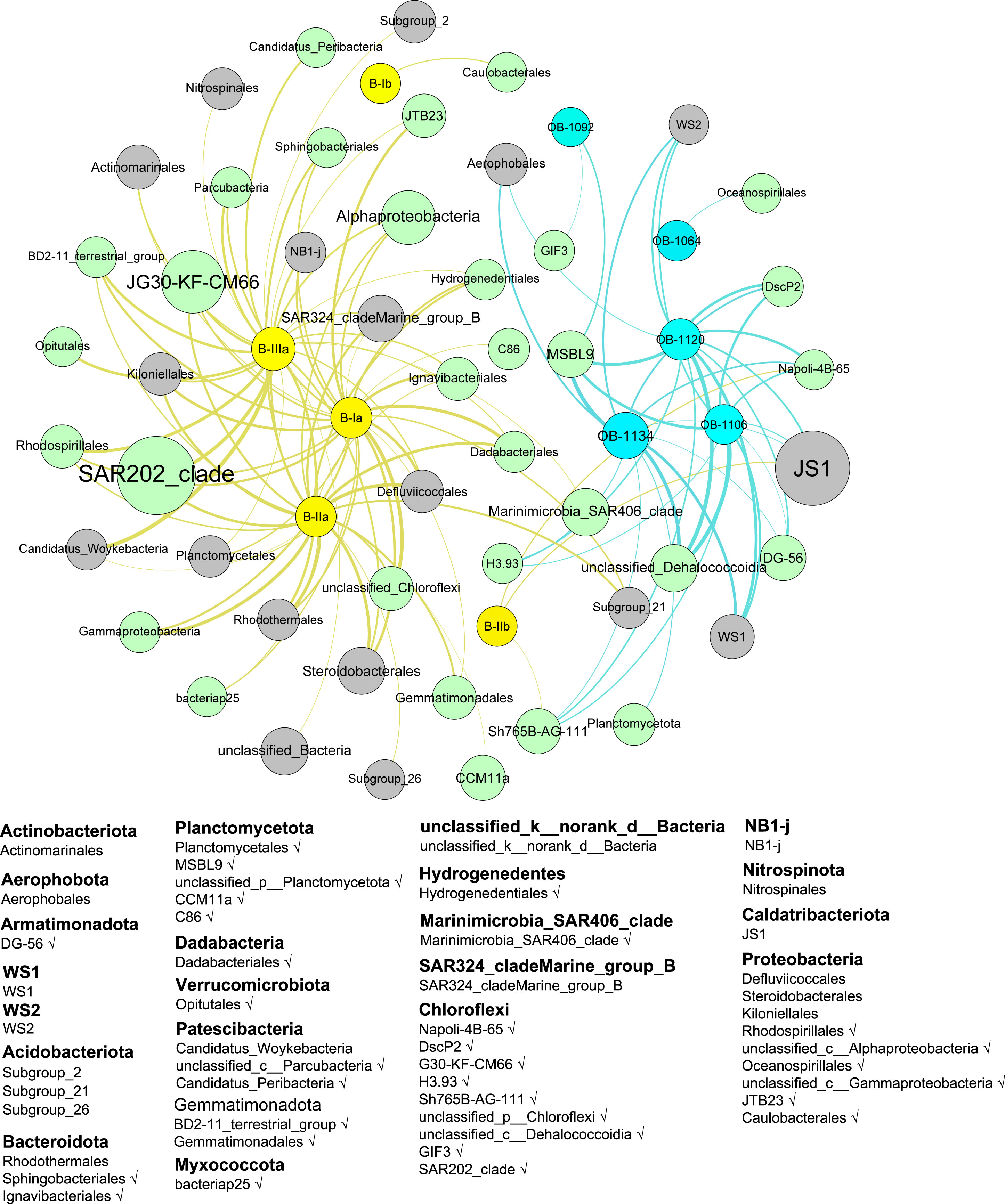
Figure 5 Co-occurrence network between composition of bacterial communities and relative abundance of brGDGTs and obGDGTs. The network is established based on positive Pearson correlation (R > 0.65, p< 0.05). The yellow, blue, green and gray nodes represent brGDGTs, obGDGTs, bacteria groups with Tes and bacteria groups without Tes, respectively. The larger the node, the higher the fractional abundance; the thicker the connection line between two nodes, the stronger the correlation. The bottom legend shows the phylum category belonging to different orders (e.g., Actinobacteria, Aerophobota). Groups marked with ‘√‘ have the Tes gene for the synthesis of GDGTs.
4 Discussion
4.1 In situ production of brGDGTs and obGDGTs in the Mariana Trench sediments
The BIT index is widely used for tracking changes in the relative contribution of terrestrial and marine organic matter. Its value is typically < 0.15 for sediments from open marine settings (Schouten et al., 2013). Based on compilation of globally distributed soils and marine sediments, Xiao et al. (2016) proposed that ΣIIIa/ΣIIa index can serve as an indicator for the source of brGDGTs; it is < 0.59 in 90% of soils but increased to > 0.92 in marine sediments without significant terrestrial inputs. For the MT03 sediments, the low BIT (0.07 ± 0.04) and high ΣIIIa/ΣIIa (3.39 ± 1.73) values strongly point to a predominately marine source for brGDGTs. These results are in line with previous studies conducted in the Mariana Trench (Ta et al., 2019; Xiao et al., 2020), and are consistent with reported sedimentological and geochemical data (Luo et al., 2017; Lai et al., 2023).
It is noteworthy that the #ringstetra index, proposed by Sinninghe Damsté (2016), has been utilized as an indicator for marine-derived brGDGTs and has found widespread application in continental shelf systems. However, emerging evidence from deep-ocean investigations, such as those conducted in the Mariana Trench, South China Sea, and Brazilian continental margin, highlights the necessity for caution when employing the #ringstetra index in open-ocean deep water environments. Therefore, we only employed the BIT and ΣIIIa/ΣIIa indices to assess the in situ production of brGDGTs in the Mariana Trench sediments.
Marine-derived brGDGTs can come from both planktonic and benthic bacteria (Liu et al., 2014; Xiao et al., 2022). While the extent to which these sources contribute to the sedimentary brGDGT pool remains debated, growing evidence supports that marine-derived brGDGTs preserved in sediments are primarily biosynthesized by benthic bacteria, especially in deep-ocean sediments (Weijers et al., 2014; Xiao et al., 2022). The strong correlation observed between brGDGT concentrations and the abundance of bacterial 16S rRNA genes (r2 = 0.67, p < 0.01) supports the notion that production of brGDGTs in the sediments of the Mariana Trench may be predominately endemic.
Comparing to our knowledge on brGDGTs, studies on obGDGTs are much limited. The investigation of obGDGTs was performed on suspended particulate matter from the oxygen minimum zone of the North Pacific Ocean and from the anoxic marine water columns of Black Sea and Cariaco Basin, which led to the hypothesis that their producers are marine sources, most likely anaerobic planktonic microbes (Liu et al., 2014; Xie et al., 2014). Becker (2015) investigated a variety of lipids in the Eastern Mediterranean Sea, and showed that obGDGTs were not present in the water column samples but were found in the sediments. In addition, obGDGTs in intact polar lipids were abundant in deep core sediments of the Discovery Basin (Becker, 2015). These studies show that organisms living in oxygen-depleted water column and sediments can both produce obGDGTs, while the latter possibly are the main contributors to sedimentary obGDGTs because respiration of organic matter in sediments is more likely to make the environment anaerobic. In the MT03 core, the co-occurrence of obGDGT concentrations with the abundance of bacterial 16S rRNA genes (r2 = 0.47, p < 0.01) indicates that obGDGTs might be primarily biosynthesized within sediments, similar to the origin of brGDGTs.
4.2 Oxygen-induced changes of brGDGTs and obGDGTs
At the water-sediment interface, dissolved oxygen is gradually lost through aerobic microbial respiration and chemical species oxidation, resulting in the transition from oxic to anoxic conditions with sediment depth (Glud, 2008; Jørgensen et al., 2022). Unfortunately, due to the extreme water depth, the in situ oxygen profile of the MT03 core was not directly measured. Nevertheless, Liu et al. (2019) reported the pore water chemistry of the MT sediments with maximum O2 penetration depth of 80 to 108 cm, and suggested an anaerobic condition in the deeper part of the sediments. Further, Lai et al. (2023) conducted a study on the mineralogy of the same core and reported redox changes that align with the evidence presented in this study (see below).
First, the core is characterized by prominent color variation with depth: The red-brown sediments occur in the upper layer (0 – 200 cm) and the olive-dark gray sediments in the deep layer (200 – 300 cm). According to Deaton and Balsam (1991) and Schwertmann (1993), this color change may be related to redox. Second, the strictly anaerobic bacteria Caldatribacteriota (order JS1) and Armatimonadota (order DG-56) are only prevalent in sediments deeper than 200 cm. Third, the pyrite content in the core exhibits an increase from the upper to the lower sections, accompanied by a decrease in the content of goethite, as reported by Lai et al. (2023). This observation suggests a shift towards a reducing sedimentary environment in the lower regions of the core. Taken together, these findings provide evidence supporting the presence of anoxic conditions in deeper depth of the MT03 core (Figure 4).
BrGDGT and obGDGT concentrations generally rise with increasing sediment depth, with their peak concentrations occurring at 241 cm depth, suggesting that they were likely to be produced in anoxic sediments. This is consistent with earlier studies that brGDGTs and obGDGTs are preferentially biosynthesized by anaerobic bacteria in soils, peats, lakes and marine environments (Weijers et al., 2006; Weijers et al., 2009; Wu et al., 2021; Xiao et al., 2022). The abrupt shift in brGDGT and obGDGT concentrations starting at 164 cm depth suggests that brGDGTs and obGDGTs could also be produced under low oxygen conditions, which is consistent with Halamka et al. (2021).
In addition to concentration, the composition of brGDGTs and obGDGTs is also greatly influenced by oxygen conditions. The MBT values of the upper layer sediments (0 – 164 cm depth, 0.32 ± 0.07) are slightly higher than those of the deep layer sediments (181 – 287 cm depth, 0.27 ± 0.09). However, the methylation degree of obGDGTs varies dramatically with much lower methylated obGDGT1064 and 1078 at 0 – 164 cm depth (92.89 ± 11.34% of total obGDGTs), and much higher methylated obGDGT1092, 1106, 1120 and 1134 at 181 – 287 cm depth (83.42 ± 10.85%). The deep layer sediments had significantly higher MIob/br values (9.46 ± 0.55 at 181 – 287 cm depth vs. 5.69 ± 0.24 at 0 – 164 cm depth) due to the elevated methylation degree of obGDGTs and their higher increase in abundance compared to brGDGTs (Figure 2G). A similar phenomenon was observed in marine water columns in the Black Sea and Cariaco Basin (Liu et al., 2014). Previous studies have demonstrated that changing oxygen concentrations alter the brGDGT methylation number in dependent of temperature. For instance, Wu et al. (2021) discovered that the methylation degree of brGDGTs in lake sediments decreased with decreasing dissolved oxygen content of the bottom water. Our study indicates that a higher degree of methylation of brGDGTs may tend to occur toward increasing anoxic conditions.
There may be a concern regarding the potential influence of environmental changes during geological history on the distribution and composition of brGDGTs and obGDGTs in the core sediments of the Mariana Trench. The lack of suitable dating materials hinders the establishment of an accurate age framework for this study. Nevertheless, utilizing the excess Pb210 data from Glud et al. (2013) and Luo et al. (2017), the sedimentation rates in the southern slope (6,037 m) and the axis (10,810 m) of the Challenger Deep were estimated to be 0.02 and 0.04 cm yr-1, respectively, which allowed for a rough calculation of the age of the bottom sediments in core MT03 in the range of approximately 5 – 20 kyr. Within this time span, significant alterations in the temperature and redox conditions of the bottom water in the Mariana Trench are unlikely, rendering it arduous to account for the noteworthy variations in brGDGT-based proxies and the strong correlation between brGDGT/obGDGT concentrations and bacterial 16S rRNA genes. Consequently, we propose that the observed changes in brGDGTs and obGDGTs primarily reflect the influence of oxygen levels within the sediment porewater.
4.3 Implications for paleoenvironmental reconstructions
The reliability of the empirical relationships between brGDGT-based proxies and environmental parameters is crucial for the use of brGDGTs in paleoclimatic research. It is widely recognized that the methylation and cyclization degree of terrestrial-derived brGDGTs are mainly controlled by temperature and pH, though other elements like oxygen levels, seasonality, nutrient availability, and soil chemistry may also have an impact (Weijers et al., 2007a; Xiao et al., 2015; Wu et al., 2021). However, knowledge on the primary environmental factors influencing marine-derived brGDGTs is very limited. In the MT03 core, brGDGTs are suggested to be produced within sediments by marine bacteria, hence the methylation degree of brGDGTs may be unaffected by changes in deep water temperature (2 – 4°C). The large variations, which range from -4.6°C to 14.7°C (4.3 ± 5.7°C), in the MBT/CBT-inferred estimates of temperature using the empirical correlation of Weijers et al. (2007b) do not match the bottom water temperature. The temperature may not have been the primary factor in the vertical variation of the methylation degree of brGDGTs in the MT03 core. We suggest that brGDGTs’ ability to function as a temperature indicator in marine environments may be severely hampered by the impact of oxygen levels. Similarly, the CBT-inferred estimates of pH in the MT03 core using the empirical correlation of Weijers et al. (2007b) ranged from 5.5 to 9.2 (7.4 ± 1.3), also suggesting that the relationship between brGDGT cyclization and pH may be masked by oxygen levels.
Therefore, brGDGTs and obGDGTs that are primarily produced by bacteria in situ may serve as important tools for tracing oxygen levels in marine sediments. In the MT03 core, a significant rise in the methylation degree of brGDGTs and obGDGTs was observed under oxygen limitation. The highly methylated obGDGTs (like obGDGT1120 and 1134) are relatively more sensitive to the changes in oxygen levels. Based on the vertical variation in MIob/br values, we propose 6.5 as a threshold value to identify oxic and anoxic conditions. This notion is supported by the vertical profiles of brGDGTs and obGDGTs of the suspended particulate matter in the Black Sea and Cariaco Basin (Liu et al., 2014; Xie et al., 2014). In these two regions, the MIob/br values are, respectively, 6.1 ± 0.4 and 5.8 ± 0.6 for oxic water columns, and 8.0 ± 0.6 and 7.0 ± 0.6 for anoxic water columns. Further research is necessary to confirm the viability of the MIob/br index in indicating oxygen levels in marine sediments.
4.4 Implications for source organisms of brGDGTs and obGDGTs
Our knowledge on biological sources of brGDGTs has been largely improved with the detection of C5 methylation brGDGTs in some strains belonging to acidobacterial subgroups 1 and 3 (Sinninghe Damsté et al., 2018; Halamka et al., 2021; Chen et al., 2022; Halamka et al., 2023). Acidobacteria are currently considered as the only confirmed sources of brGDGTs in environments where they were abundant, e.g., acidic soils and peat bogs. However, increasing evidence revealed that the potential brGDGT-producers may extend to a suite of bacteria other than Acidobacteria alone in various environmental settings (Chen et al., 2018a; De Jonge et al., 2019). On the basis that both brGDGTs and obGDGTs are probably produced in situ by bacteria thriving in the sediments as we discussed above, we first explored the relationship between distribution of brGDGTs (obGDGTs) and relative abundance of bacterial community in the sediment core. The results showed that the 49 bacterial groups at the order level positively related to the distribution of brGDGTs and obGDGTs (Figure 5). These bacteria included but are not limited to Caldatribacteriota (JS1), Proteobacteria (e.g., Alphaproteobacteria and Rhodospirillales), and Chloroflexi (e.g., G30-KF-CM66 and Napoli-4B-65). Our results show that the biological sources of these lipids could be related to diverse benthic bacterial groups in intact forms (alive or dormant) in the cold deep water marine environments, which can be significantly different in community structure from water column plankton (Jing et al., 2022; Tian et al., 2018). The biological sources of brGDGTs and obGDGTs may also be different from those observed in soils (Chen et al., 2018a; De Jonge et al., 2019) or lacustrine water column and sediments (Weber et al., 2018; Wu et al., 2021).
To date, the brGDGTs producers that have been experimentally confirmed were all classified to phylum Acidobacteria. However, Acidobacteria only accounted for 0.60% of the total bacterial community in our samples, and they were mainly enriched in the upper layer sediments. A total of 7 subgroups of Acidobacteria have been reported, of which only 3 subgroups (2, 21, 26) are associated with brGDGTs, and all three subgroups do not contain Tes homologs. Therefore, we suggest that Acidobacteria may not be the main producers of brGDGTs in the Mariana Trench sediments. Based on co-occurrence with brGDGTs and potential indication of Tes homologs in our study, we suggest that bacteria Chloroflexi, Proteobacteria and Dadabacteria could be the potential producers of brGDGTs and Armatimonadota, Planctomycetota and Chloroflexi may probably be the producers of obGDGTs in the Mariana Trench sediments. However, it requires cultivation experiments to confirm the biosynthesis of brGDGTs and obGDGTs in these microorganisms.
5 Summary and conclusion
We examined brGDGTs and obGDGTs as well as bacterial communities in a sediment core from the Mariana Trench. The rapid increase in concentrations of brGDGTs and obGDGTs with sediment depth and their good correlation with the abundance of anaerobic bacteria suggest that brGDGTs and obGDGTs in the Mariana Trench sediments may be biosynthesized by anaerobic bacteria (either alive or dormant) in the sediments. The anaerobic sediments were characterized by significantly higher degrees of methylation and cyclization, suggesting that brGDGTs and obGDGTs may be indicative of oxygen availability. Network analysis suggests that brGDGTs in the Mariana Trench sediments may be more likely to be produced by bacterial phyla such as Chloroflexi, Proteobacteria and Dadabacteria; whereas, obGDGTs may be more likely to be produced by Armatimonadota, Planctomycetota and Chloroflexi. Our study indicates that the use of brGDGT-based proxies in paleotemperature and pH reconstructions using marine sedimentary archives may be significantly influenced by the impact of changes in bacterial community compositions and oxygen levels in the sediment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below BioProject, PRJNA980304.
Author contributions
ZZ, WX, FZ, YZ, and CZ conceived the study and designed the experiments. FZ and JT collected the samples. ZZ, WX, FZ, and YC performed the analyses and created the figures. ZZ and YC analyzed sequence data and provided interpretation of organic geochemical data. FZ, WX and CZ acquired funding. ZZ, WX, FZ, and CZ drafted the manuscript, and all authors contributed to writing and approved the submitted version.
Acknowledgments
We would like to thank Wei He for help drawing the figures, Yanwei Chen for help in qPCR analysis, Wenxiu Wang for analysis of dissolved organic matter (DOM) present in the pore water, and the captain and crew of R/V “Dong Fang Hong 2” for their sampling efforts during the cruise of the Mariana Trench. Financial supports for this research were provided by the National Natural Science Foundation of China (42141003, 42003063, 42206040), the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. K19313901), the Stable Support Plan Program of Shenzhen Natural Science Fund (20200925173954005), the Guangdong-Shenzhen Joint Fund (2021B1515120080), the Shenzhen Key Laboratory of Marine Archaea Geo-Omics, Southern University of Science and Technology (ZDSYS201802081843490), and the Shanghai Sheshan National Geophysical Observatory (2020Z01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1233560/full#supplementary-material
References
Bahram M., Hildebrand F., Forslund S. K., Anderson J. L., Soudzilovskaia N. A., Bodegom P. M. V., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Becker K. W. (2015). Biogeochemical significance and biomarker potential of novel glycerolipids and respiratory quinones in the marine environment. Doctoral Dissertation (University of Bremen: Bremen).
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108 (supplement_1), 4516–4522. doi: 10.1073/pnas.1000080107
Carr S. A., Orcutt B. N., Mandernack K. W., Spear J. R. (2015). Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00872
Chen Y. F., Zheng F., Chen S. Z., Liu H. D., Phelps T. J., Zhang C. L. (2018a). Branched GDGT production at elevated temperatures in anaerobic soil microcosm inCubations. Org. Geochem. 117, 12–21. doi: 10.1016/j.orggeochem.2017.11.015
Chen Y. F., Zheng F., Yang H., Yang W., Wu R. J., Liu X. Y., et al. (2022). The production of diverse brGDGTs by an Acidobacterium providing a physiological basis for paleoclimate proxies. Geochim. Cosmochim. Acta 337, 155–165. doi: 10.1016/j.gca.2022.08.033
Chen S. F., Zhou Y. Q., Chen Y. R., Gu J. (2018b). Fastp.: Ultra–fast All–in–one FASTQ Preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Connock G. T., Owens J. D., Liu X. L. (2022). Biotic induction and microbial ecological dynamics of Oceanic Anoxic Event 2. Commun. Earth Environ. 3, 136. doi: 10.1038/s43247-022-00466-x
Deaton B. C., Balsam W. L. (1991). Visible spectroscopy; a rapid method for determining hematite and goethite concentration in geological materials. J. Sedimentary Res. 61, 628–632. doi: 10.1306/D4267794-2B26-11D7-8648000102C1865D
De Jonge C., Hopmans E. C., Stadnitskaia A., Rijpstra W. I. C., Hofland R., Tegelaar E. W., et al. (2013). Identification of novel penta- and hexamethylated branched glycerol dialkyl glycerol tetraethers in peat using HPLC-MS2, GC-MS and GC-SMB-MS. Org. Geochem. 54, 78–82. doi: 10.1016/j.orggeochem.2012.10.004
De Jonge C., Kuramae E., Radujković D., Weedon J. T., Janssens I. A., Peterse F. (2021). The influence of soil chemistry on branched tetraether lipids in mid– and high latitude soils: Implications for brGDGT– based paleothermometry. Geochim. Cosmochim. Acta 310, 95–112. doi: 10.1016/j.gca.2021.06.037
De Jonge C., Radujković D., Sigurdsson B. D., Sigurdsson B. D., Weedon J. T., Weedon J. T., et al. (2019). Lipid biomarker temperature proxy responds to abrupt shift in the bacterial community composition in geothermally heated soils. Org. Geochem. 137, 103897. doi: 10.1016/j.orggeochem.2019.07.006
De Jonge C., Stadnitskaia A., Hopmans E. C., Cherkashov G., Fedotov A., Streletskaya I. D., et al. (2015). Drastic changes in the distribution of branched tetraether lipids in suspended matter and sediments from the Yenisei River and Kara Sea (Siberia): Implications for the use of brGDGT–based proxies in coastal marine sediments. Geochim. Cosmochim. Acta 165, 200–225. doi: 10.1016/j.gca.2015.05.044
Ferrer M., Guazzaroni M–E, Richter M., García-Salamanca A., Yarza P., Suárez–Suárez A., et al. (2011). Taxonomic and functional metagenomic profiling of the microbial community in the anoxic sediment of a sub–saline shallow lake (Laguna de Carrizo, Central SPAIN). Microbial. Ecol. 62, 824–837. doi: 10.1007/s00248-011-9903-y
Glud R. N. (2008). Oxygen dynamics of marine sediments. Mar. Biol. Res. 4, 243–289. doi: 10.1080/17451000801888726
Glud R. N., Wenzhöfer F., Middelboe M., Oguri K., Turnewitsch R., Canfield D. E., et al. (2013). High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nat. Geosci. 6, 284–288. doi: 10.1038/ngeo1773
Günther F., Thiele A., Gleixner G., Xu B. Q., Yao T. D., Schouten S. (2014). Distribution of bacterial and archaeal ether lipids in soils and surface sediments of Tibetan lakes: Implications for GDGT-based proxies in saline high mountain lakes. Org. Geochem. 67, 19–30. doi: 10.1016/j.orggeochem.2013.11.014
Halamka T. A., McFarlin J., Younkin A. D., Depoy J., Dildar N., Kopf S. H. (2021). Oxygen limitation can trigger the production of branched GDGTs in culture. Geochem. Perspect. Lett. 19, 36–39. doi: 10.7185/geochemlet.2132
Halamka T. A., Raberg J. H., McFarlin J. M., Younkin A. D., Mulligan C., Liu X–L, et al. (2023). Production of diverse brGDGTs by Acidobacterium Solibacter usitatus in response to temperature, pH, and O2 provides a culturing perspective on brGDGT proxies and biosynthesis. Geobiology 21, 102–118. doi: 10.1111/gbi.12525
Hiraoka S., Hirai M., Matsui Y., Makabe A., Minegishi H., Tsuda M., et al. (2019). Microbial community and geochemical analyses of trans–trench sediments for understanding the roles of hadal environments. ISME J. 14, 740–756. doi: 10.1038/s41396-019-0564-z
Hopmans E. C., Weijers J. W. H., Schefuss E., Herfort L., Sinninghe Damsté J. S., Schouten S. (2004). A novel proxy for terrestrial organic matter in sediments based on branched and isoprenoid tetraether lipids. Earth Planetary Sci. Lett. 224, 107–116. doi: 10.1016/j.epsl.2004.05.012
Hoshino T., Doi H., Uramoto G–I, Wörmer L., Adhikari R., Xiao N., et al. (2020). Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U.S.A. 117, 27587–27597. doi: 10.1073/pnas.1919139117
Huguet C., Hopmans E. C., Febo–Ayala W., Thompson D. H., Sinninghe Damsté J. S., Schouten S. (2006). An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org. Geochem. 37, 1036–1041. doi: 10.1016/j.orggeochem.2006.05.008
Ismail S., Elreedy A., Fujii M., Ni S–Q, Tawfik A., Elsamadony M. (2021). Fatigue of anammox consortia under long–term 1,4–dioxane exposure and recovery potential: N–kinetics and microbial dynamics. J. Hazardous Mater. 414, 125533. doi: 10.1016/j.jhazmat.2021.125533
Jamieson A. J. (2015). The Hadal Zone: Life in the Deepest Oceans (Cambridge: Cambridge University Press).
Jing H. M., Xiao X., Zhang Y., Li Z. Y., Jian H. H., Luo Y. F., et al. (2022). Composition and ecological roles of the core microbiome along the abyssal-hadal transition zone sediments of the mariana trench. Microbiol. Spectr. 10, 3. doi: 10.1128/spectrum.01988-21
Jørgensen B., Wenzhöfer F., Egger M., Glud R. N. (2022). Sediment oxygen consumption: Role in the global marine carbon cycle. Earth-Sci. Rev. 228, 103987. doi: 10.1016/j.earscirev.2022.103987
Katayama T., Nobu M. K., Kusada H., Meng X–Y, Hosogi N., Uematsu K., et al. (2020). Isolation of a member of the candidate phylum ‘Atribacteria’ reveals a unique cell membrane structure. Nat. Commun. 11. doi: 10.1038/s41467-020-20149-5
Lai W., Liu X., Tian J., Wang H., Zhang J., Huang J., et al. (2023). Mineralogy of sediments in the Mariana Trench controlled by environmental conditions of the West Pacific since the Last Glacial Maximum. J. Asian Earth Sci. 245, 105553. doi: 10.1016/j.jseaes.2023.105553
Lee J. W., Kwon K., Azizi A., Oh H–M, Kim W., Bahk J–J, et al. (2013). Microbial community structures of methane hydrate–bearing sediments in the Ulleung Basin, East Sea of Korea. Mar. Petroleum Geol. 47, 136–146. doi: 10.1016/j.marpetgeo.2013.06.002
Liu X. L., Lipp J. S., Simpson J. H., Lin Y. S., Summons R. E., Hinrichs K–U. (2012a). Mono– and dihydroxyl glycerol dibiphytanyl glycerol tetraethers in marine sediments: Identification of both core and intact polar lipid forms. Geochim. Et Cosmochim. Acta 89, 102–115. doi: 10.1016/j.gca.2012.04.053
Liu Y. F., Qi Z., Shou L. B., Liu J. F., Yang S. Z., Gu J. D., et al. (2019). Anaerobic hydrocarbon degradation in candidate phylum ‘Atribacteria’ (JS1) inferred from genomics. ISME J. 13, 2377–2390. doi: 10.1038/s41396-019-0448-2
Liu X. L., Summons R. E., Hinrichs K–U. (2012b). Extending the known range of glycerol ether lipids in the environment: structural assignments based on tandem mass spectral fragmentation patterns. Rapid Commun. Mass Spectrometry: RCM 26 (19), 2295–2302.doi: 10.1002/rcm.6355
Liu X. L., Zhu C., Wakeham S. G., Hinrichs K–U. (2014). In situ production of branched glycerol dialkyl glycerol tetraethers in anoxic marine water columns. Mar. Chem. 166, 1–8. doi: 10.1016/j.marchem.2014.08.008
Luo M., Gieskes J., Chen L. Y., Shi X. F., Chen D. F. (2017). Provenances, distribution, and accumulation of organic matter in the southern Mariana Trench rim and slope: Implication for carbon cycle and burial in hadal trenches. Mar. Geol. 386, 486–498. doi: 10.1016/j.margeo.2017.02.012
Magoc T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi: 10.1093/bioinformatics/btr507
Nobu M. K., Dodsworth J. A., MurugapIran S. K., Rinke C., Gies E. A., Webster G., et al. (2016). Phylogeny and physiology of candidate phylum ‘Atribacteria’ (OP9/JS1) inferred from cultivation–independent genomics. ISME J. 10, 273–286. doi: 10.1038/ismej.2015.97
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18 (5), 1403–1414. doi: 10.1111/1462-2920.13023
Peterse F., Kim J–H, Schouten S., Kristensen D. K., Koç N., Sinninghe Damsté J. S. (2009). Constraints on the application of the MBT/CBT palaeothermometer at high latitude environments (Svalbard, Norway). Org. Geochem. 40, 692–699. doi: 10.1016/j.orggeochem.2009.03.004
Peterse F., van der Meer J., Schouten S., Weijers J. W. H., Fierer N., Jackson R. B., et al. (2012). Revised calibration of the MBT–CBT paleotemperature proxy based on branched tetraether membrane lipids in surface soils. Geochim. Et Cosmochim. Acta 96, 215–229. doi: 10.1016/j.gca.2012.08.011
Schouten S., Hopmans E. C., Sinninghe Damsté J. S. (2013). The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: A review. Org. Geochem. 54, 19–61. doi: 10.1016/j.orggeochem.2012.09.006
Schwertmann U. (1993). Relations between iron oxides, soil color, and soil formation. Soil Color 31, 51–69. doi: 10.2136/sssaspecpub31.c4
Sinninghe Damsté J. S. (2016). Spatial heterogeneity of sources of branched tetraethers in shelf systems: The geochemistry of tetraethers in the Berau River delta (Kalimantan, Indonesia). Geochim. Cosmochim. Acta 186, 13–31. doi: 10.1016/j.gca.2016.04.033
Sinninghe Damsté J. S., Hopmans E. C., Pancost R. D., Schouten S., Geenevasen J. A. J. (2000). Newly discovered non–isoprenoid glycerol dialkyl glycerol tetraether lipids in sediments. Chem. Commun. 2000, 1683–1684. doi: 10.1039/b004517i
Sinninghe Damsté J. S., Rijpstra W. I. C., Foesel B. U., Huber K. J., Overmann J., Nakagawa S., et al. (2018). An overview of the occurrence of ether– and ester–linked iso–diabolic acid membrane lipids in microbial cultures of the Acidobacteria: Implications for brGDGT paleoproxies for temperature and pH. Org. Geochem. 124, 63–76. doi: 10.1016/j.orggeochem.2018.07.006
Sturt H. F., Summons R. E., Smith K. E., Elvert M., Hinrichs K–U. (2004). Intact polar membrane lipids in prokaryotes and sediments deciphered by high–performance liquid chromatography/electrospray ionization multistage mass spectrometry—-new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrometry: RCM 18 6, 617–628. doi: 10.1002/rcm.1378
Summons R. E., Welander P. V., Gold D. A. (2022). Lipid biomarkers: molecular tools for illuminating the history of microbial life. Nat. Rev. Microbiol. 20, 174–185. doi: 10.1038/s41579-021-00636-2
Sun C. R., Zhang C. L., Li F. Y., Wang H. Y., Liu W. G. (2016). Distribution of branched glycerol dialkyl glycerol tetraethers in soils on the Northeastern Qinghai-Tibetan Plateau and possible production by nitrite-reducing bacteria. Sci. China Earth Sci. 59, 1834–1846. doi: 10.1007/s11430-015-0230-2
Ta K. W., Peng X. T., Xu H. C., Du M. R., Chen S., Li J. W., et al. (2019). Distributions and sources of glycerol Dialkyl Glycerol Tetraethers in sediment cores from the mariana subduction zone. J. Geophysical Res.: Biogeosci. 124, 857–869. doi: 10.1029/2018JG004748
Takai K., Horikoshi K. (2000). Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072. doi: 10.1128/AEM.66.11.5066-5072.2000
Tian J., Fan L., Liu H., Liu J., Li Y., Qin Q., et al (2018). A nearly uniform distributional pattern of heterotrophic bacteria in the Mariana Trench interior. Deep Sea Res. Part I Oceanogr. Res. Papers 142, 116–126. doi: 10.1016/j.dsr.2018.10.002
Tierney J. E., Russell J. M. (2009). Distributions of branched GDGTs in a tropical lake system: Implications for lacustrine application of the MBT/CBT paleoproxy. Org. Geochem. 40 (9), 1032–1036. doi: 10.1016/j.orggeochem.2009.04.014
Wang H. Y., Dong H. L., Zhang C. L., Jiang H. C., Liu W. G. (2016). A 12-kyr record of microbial branched and isoprenoid tetraether index in Lake Qinghai, northeastern Qinghai-Tibet Plateau : Implications for paleoclimate reconstruction. Sci. China Earth Sci. 59 (5), 951–960. doi: 10.1007/s11430-015-5213-4
Wang H. Y., Liu W. G., Zhang C. L. (2014). Dependence of the cyclization of branched tetraethers on soil moisture in alkaline soils from arid–subhumid China: implications for palaeorainfall reconstructions on the Chinese Loess Plateau. Biogeosciences 11, 6755–6768. doi: 10.5194/bg-11-6755-2014
Wang H. Y., Liu W. G., Zhang C. L., Liu Z. H., He Y. X. (2013). Branched and isoprenoid tetraether (BIT) index traces water content along two marsh-soil transects surrounding Lake Qinghai: Implications for paleo-humidity variation. Org. Geochem. 59, 75–81. doi: 10.1016/j.orggeochem.2013.03.011
Weber Y., Sinninghe Damsté J. S., Zopfi J., De Jonge C., Gilli A., Schubert C. J., et al. (2018). Redox–dependent niche differentiation provides evidence for multiple bacterial sources of glycerol tetraether lipids in lakes. Proc. Natl. Acad. Sci. 115, 10926–10931. doi: 10.1073/pnas.1805186115
Weijers J. W. H., Panoto E., van Bleijswijk J. D. L., Schouten S., Rijpstra W. I. C., Balk M., et al. (2009). Constraints on the biological source(s) of the orphan branched tetraether membrane lipids. Geomicrobiol. J. 26, 402–414. doi: 10.1080/01490450902937293
Weijers J. W. H., Schefuß E., Kim J–H, Sinninghe Damsté J. S., Schouten S. (2014). Constraints on the sources of branched tetraether membrane lipids in distal marine sediments. Org. Geochem. 72, 14–22. doi: 10.1016/j.orggeochem.2014.04.011
Weijers J. W. H., Schefuß E., Schouten S., Sinninghe Damsté J. S. (2007b). Coupled thermal and hydrological evolution of tropical Africa over the last deglaciation. Science 315, 1701–1704. doi: 10.1126/science.1138131
Weijers J. W. H., Schouten S., Donker J. C., Hopmans E. C., Sinninghe Damsté J. S. (2007a). Environmental controls on bacterial tetraether membrane lipid distribution in soils. Geochim. Cosmochim. Acta 71, 703–713. doi: 10.1016/j.gca.2006.10.003
Weijers J. W. H., Schouten S., Hopmans E. C., Geenevasen J. A. J., David O. R. P., Coleman J. M., et al. (2006). Membrane lipids of mesophilic anaerobic bacteria thriving in peats have typical archaeal traits. Environ. Microbiol. 8 (4), 648–657. doi: 10.1111/j.1462-2920.2005.00941.x
Wu J., Yang H., Pancost R. D., Naafs B. D. A., Qian S., Dang X. Y., et al. (2021). Variations in dissolved O2 in a Chinese lake drive changes in microbial communities and impact sedimentary GDGT distributions. Chem. Geol. 579, 120348. doi: 10.1016/j.chemgeo.2021.120348
Xiao W. J., Wang Y. S., Liu Y. S., Zhang X., Shi L., Xu Y. P. (2020). Predominance of hexamethylated 6–methyl branched glycerol dialkyl glycerol tetraethers in the Mariana Trench: source and environmental implication. Biogeosciences 17, 2135–2148. doi: 10.5194/bg-17-2135-2020
Xiao W. J., Wang Y. H., Zhou S. Z., Hu L. M., Yang H., Xu Y. P. (2016). Ubiquitous production of branched glycerol dialkyl glycerol tetraethers(brGDGTs) in global marine environments: a new source indicator for brGDGTs. Biogeosciences 13, 5883–5894. doi: 10.5194/bg-13-5883-2016
Xiao W. J., Xu Y. P., Ding S., Wang Y. H., Zhang X. Y., Yang H., et al. (2015). Global calibration of a novel, branched GDGT–based soil pH proxy. Org. Geochem. 89, 56–60. doi: 10.1016/j.orggeochem.2015.10.005
Xiao W. J., Xu Y. P., Lin J., Zeng Z. Y., Liu Y. S., Zhang H. R., et al. (2022). Global scale production of brGDGTs by benthic marine bacteria: Implication for developing ocean bottom environmental proxies. Global Planetary Change 211, 103783. doi: 10.1016/j.gloplacha.2022.103783
Xie S. T., Liu X. L., Schubotz F., Wakeham S. G., Hinrichs K–U. (2014). Distribution of glycerol ether lipids in the oxygen minimum zone of the Eastern Tropical North Pacific Ocean. Org. Geochem. 71, 60–71. doi: 10.1016/j.orggeochem.2014.04.006
Xu Y. P., Jia Z. H., Xiao W. J., Fang J. S., Wang Y. S., Luo M., et al. (2020). Glycerol dialkyl glycerol tetraethers in surface sediments from three Pacific trenches: Distribution, source and environmental implications. Org. Geochem. 147, 104079. doi: 10.1016/j.orggeochem.2020.104079
Yang H., Pancost R. D., Dang X. Y., Zhou X. Y., Evershed R. P., Xiao G. Q., et al. (2014). Correlations between microbial tetraether lipids and environmental variables in Chinese soils: Optimizing the paleo-reconstructions in semi-arid and arid regions. Geochim. Cosmochim. Acta 126, 49–69. doi: 10.1016/j.gca.2013.10.041
Zell C., Kim J–H, Hollander D. J., Lorenzoni L., Baker P. A., Silva C. G., et al. (2014). Sources and distributions of branched and isoprenoid tetraether lipids on the Amazon shelf and fan: Implications for the use of GDGT–based proxies in marine sediments. Geochim. Cosmochim. Acta 139, 293–312. doi: 10.1016/j.gca.2014.04.038
Zeng Z. R., Chen H., Yang H., Chen Y. F., Yang W., Feng X., et al. (2022). Identification of a protein responsible for the synthesis of archaeal membrane–spanning GDGT lipids. Nat. Commun. 13. doi: 10.1038/s41467-022-29264-x
Zhang C. L., Wang J. X., Dodsworth J. A., Willianms A. J., Zhu C., Hinrichs K–U, et al. (2013). In situ production of branched glycerol dialkyl glycerol tetraethers in a great basin hot spring (USA). Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00181
Zhang C. L., Wang J. X., Wei Y. L., Zhu C., Huang L. Q., Dong H. L. (2012). Production of branched tetraether lipids in the lower pearl river and estuary: effects of extraction methods and impact on bGDGT proxies. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00274
Zhao Y. P., Liu S. F., Jiang B., Feng Y., Zhu T., Tao H. C., et al. (2018). Genome–centered metagenomics analysis reveals the symbiotic organisms possessing ability to cross–feed with anammox bacteria in anammox consortia. Environ. Sci. Technol. 52 (19), 11285–11296. doi: 10.1021/acs.est.8b02599
Zheng F., Zhang C. L., Chen Y. F., Li F. Y., Ma C. L., Pu Y., et al. (2016). Branched tetraether lipids in Chinese soils: Evaluating the fidelity of MBT/CBT proxies as paleoenvironmental proxies. Sci. China Earth Sci. 59 (7), 1353–1367. doi: 10.1007/s11430-016-5268-x
Keywords: branched GDGTs, overly branched GDGTs, in situ production, anaerobic bacteria, Mariana Trench
Citation: Zeng Z, Xiao W, Zheng F, Chen Y, Zhu Y, Tian J and Zhang C (2023) Enhanced production of highly methylated brGDGTs linked to anaerobic bacteria from sediments of the Mariana Trench. Front. Mar. Sci. 10:1233560. doi: 10.3389/fmars.2023.1233560
Received: 02 June 2023; Accepted: 21 July 2023;
Published: 18 August 2023.
Edited by:
Junfu Dong, Shandong University, ChinaReviewed by:
Mengyuan Wang, Sun Yat-sen University, ChinaRonnakrit Rattanasriampaipong, Texas A and M University, United States
Copyright © 2023 Zeng, Xiao, Zheng, Chen, Zhu, Tian and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengfeng Zheng, emhlbmdmZkBzdXN0ZWNoLmVkdS5jbg==; Chuanlun Zhang, emhhbmdjbEBzdXN0ZWNoLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Zhiyu Zeng
Zhiyu Zeng Wenjie Xiao
Wenjie Xiao Fengfeng Zheng
Fengfeng Zheng Yufei Chen1,2
Yufei Chen1,2 Jiwei Tian
Jiwei Tian Chuanlun Zhang
Chuanlun Zhang