
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
POLICY AND PRACTICE REVIEWS article
Front. Mar. Sci., 27 September 2023
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1224471
 Alex David Rogers1,2*
Alex David Rogers1,2* Hannah Appiah-Madson3
Hannah Appiah-Madson3 Jeff A. Ardron4,5
Jeff A. Ardron4,5 Nicholas J. Bax6
Nicholas J. Bax6 Punyasloke Bhadury7
Punyasloke Bhadury7 Angelika Brandt8
Angelika Brandt8 Pier-Luigi Buttigieg9,10
Pier-Luigi Buttigieg9,10 Olivier De Clerck11
Olivier De Clerck11 Claudia Delgado12
Claudia Delgado12 Daniel L. Distel3
Daniel L. Distel3 Adrian Glover13
Adrian Glover13 Judith Gobin14
Judith Gobin14 Maila Guilhon15
Maila Guilhon15 Shannon Hampton16
Shannon Hampton16 Harriet Harden-Davies17
Harriet Harden-Davies17 Paul Hebert18
Paul Hebert18 Lisa Hynes2
Lisa Hynes2 Miranda Lowe13
Miranda Lowe13 Sandy MacIntyre2
Sandy MacIntyre2 Hawis Madduppa19
Hawis Madduppa19 Ana Carolina de Azevedo Mazzuco20
Ana Carolina de Azevedo Mazzuco20 Anna McCallum21
Anna McCallum21 Chris McOwen22
Chris McOwen22 Tim Wilhelm Nattkemper23
Tim Wilhelm Nattkemper23 Mika Odido24
Mika Odido24 Tim O’Hara21
Tim O’Hara21 Karen Osborn25,26
Karen Osborn25,26 Angelique Pouponneau27
Angelique Pouponneau27 Pieter Provoost28
Pieter Provoost28 Muriel Rabone13
Muriel Rabone13 Eva Ramirez-Llodra1,2
Eva Ramirez-Llodra1,2 Lucy Scott29
Lucy Scott29 Kerry Jennifer Sink30
Kerry Jennifer Sink30 Daniela Turk22
Daniela Turk22 Hiromi Kayama Watanabe31
Hiromi Kayama Watanabe31 Lauren V. Weatherdon22
Lauren V. Weatherdon22 Thomas Wernberg32
Thomas Wernberg32 Suzanne Williams13
Suzanne Williams13 Lucy Woodall2,33,34
Lucy Woodall2,33,34 Dawn J. Wright35
Dawn J. Wright35 Daniela Zeppilli36
Daniela Zeppilli36 Oliver Steeds2
Oliver Steeds2Ocean Census is a new Large-Scale Strategic Science Mission aimed at accelerating the discovery and description of marine species. This mission addresses the knowledge gap of the diversity and distribution of marine life whereby of an estimated 1 million to 2 million species of marine life between 75% to 90% remain undescribed to date. Without improved knowledge of marine biodiversity, tackling the decline and eventual extinction of many marine species will not be possible. The marine biota has evolved over 4 billion years and includes many branches of the tree of life that do not exist on land or in freshwater. Understanding what is in the ocean and where it lives is fundamental science, which is required to understand how the ocean works, the direct and indirect benefits it provides to society and how human impacts can be reduced and managed to ensure marine ecosystems remain healthy. We describe a strategy to accelerate the rate of ocean species discovery by: 1) employing consistent standards for digitisation of species data to broaden access to biodiversity knowledge and enabling cybertaxonomy; 2) establishing new working practices and adopting advanced technologies to accelerate taxonomy; 3) building the capacity of stakeholders to undertake taxonomic and biodiversity research and capacity development, especially targeted at low- and middle-income countries (LMICs) so they can better assess and manage life in their waters and contribute to global biodiversity knowledge; and 4) increasing observational coverage on dedicated expeditions. Ocean Census, is conceived as a global open network of scientists anchored by Biodiversity Centres in developed countries and LMICs. Through a collaborative approach, including co-production of science with LMICs, and by working with funding partners, Ocean Census will focus and grow current efforts to discover ocean life globally, and permanently transform our ability to document, describe and safeguard marine species.
The ocean comprises the greatest area of wilderness on Earth (Jones et al., 2018) and is its largest ecosystem, comprising 1.3 billion km3 of water covering 71% of the surface of our planet. It harbours an astonishing diversity of species especially at a deep phylogenetic level. For Metazoa, at the phylum level, 15 of 33 accepted animal phyla (WoRMS, 2023) are comprised exclusively of marine species, an additional 8 are dominated by marine species while only one has currently no known marine members (May, 1994; Rogers et al., 2022a; see Table 1). The ocean is home to some of the largest and smallest organisms ever to have existed on earth. Smaller organisms constitute the greatest potential for taxonomic discovery, accounting for the largest proportion of marine diversity and biomass (Bar-On et al., 2018). Life is found everywhere in the ocean and has adapted to the most extreme marine environments including the deepest trenches (e.g. Challenger Deep in the Marianas Trench; Jamieson, 2015; Jamieson et al., 2023), anoxic hypersaline basins (Danovaro et al., 2010), and high temperature deep-sea hydrothermal vents (Van Dover, 2000). Ocean life has evolved over nearly 4 billion years (e.g. Tashiro et al., 2017) and provides numerous benefits to humankind (e.g. Worm et al., 2006; Barbier, 2017; Lotze, 2021; see Supplementary Material 1.0), many of which we are only just uncovering (e.g. marine genetic resources; Blasiak et al., 2020; see Supplementary Material 1.3). Yet, compared to terrestrial systems, we still only have a basic understanding of how life is distributed in the ocean (e.g. Gagné et al., 2020), how it contributes to the functioning of marine ecosystems (Thurber et al., 2014; Gamfeldt et al., 2015) as part of the Earth’s life-support systems, and how it underpins human society and the well-being of its citizens. Of an estimated 1 to 2 million species of marine life between 75% to 90% remain undescribed, and many geographic regions and ecosystems remain poorly explored (Webb et al., 2010; Mora et al., 2011; Ramirez-Llodra et al., 2011; Appeltans et al., 2012; Bouchet et al., 2023). As a result, human society cannot fully comprehend the vital importance of a healthy ocean and the indispensability of Earth’s marine life in supporting human well-being.
Global economic prosperity and long-lasting economic growth has been a recent achievement, but it has come at the expense of society (Moranta et al., 2022), climate stability and biodiversity which has declined by an estimated 40% (Dasgupta, 2021). In the ocean, defaunation may have started later than on land and there are fewer recorded extinctions, but evidence suggests that there has been a significant loss of biodiversity with cascading ecological changes across food webs, altering the function of marine ecosystems (McCauley et al., 2015; Cowie et al., 2022; Edgar et al., 2023) and their capacity to provide goods and services to millions of people in the world (Smale et al., 2019; Isbell et al., 2023; Supplementary Material 1.0). Assessment of the IUCN Red List of species, a metric that quantitatively estimates extinction risk in organisms (IUCN, 2017), suggests that the proportion of marine species threatened with extinction is 11-46% and spans the range for species from terrestrial groups (20-25%; Webb and Mindel, 2015; Rogers et al., 2020). It is likely that the poor level of observation and monitoring of ocean species and, more fundamentally, less taxonomic knowledge compared to terrestrial ecosystems means that many extinctions and declines have not been recorded (e.g. Webb and Mindel, 2015; Rogers et al., 2020; Cowie et al., 2022). The ocean biodiversity crisis is driven both by the decline of species (e.g. more than three quarters of oceanic sharks and rays are threatened with extinction; Pacoureau et al., 2021) and the degradation and collapse of ecosystems (e.g. tropical reefs have lost half of their coral cover since the 1870s; Wilkinson et al., 2016a; IPCC, 2019).
An important key to halting the biodiversity crisis, sustainably managing human activities in the ocean and protecting nature’s contributions to society is transforming our understanding of the diversity and distribution of life in the ocean. This transformation is not currently proceeding at sufficient speed because taxonomy has largely been reliant on methods first developed in the 18th and 19th Centuries (Rogers et al., 2022a). Even though this process is now assisted by DNA sequencing, it is still very slow and often requires examination of the original samples on which previous species descriptions were based (type specimens). Limited capacity in terms of numbers of taxonomists across all groups of marine organisms is also a key challenge (Engel et al., 2021; Mammola et al., 2023). As a result of this, the rate of description of new species has barely changed since the 1840s (see Figure 1; Rogers et al., 2022a; Bouchet et al., 2023). On average, the length of time to describe a species from when it is first collected from the ocean has been estimated at 13.5 years (Bouchet et al., 2023). Because taxonomic studies are intrinsically time consuming and processed slowly, the lack of robust systematic frameworks that build upon an accurate taxonomy is negatively affecting our ability to address many big questions in marine and broader biological sciences including an understanding of global patterns and processes, as well as in ocean management for which a knowledge of species diversity is crucial (e.g. Godfray, 2002; Godfray and Knapp, 2004; Godfray, 2007; Rogers et al., 2022a). As a result, marine taxonomy has fallen out of favour with scientific journals and funding agencies (e.g. Hamilton et al., 2021; Pinto et al., 2021), leading to a decline in taxonomic science and the current taxonomic crisis (Godfray, 2002). This decline is particularly visible in some regions such as Africa, South and South-east Asia, where the flow of funding support for taxonomy-related research has fallen dramatically over the years. Unfortunately, this has coincided with the global biodiversity crisis, when knowledge on the diversity of marine species and where they live is urgently required to inform efforts to conserve, restore or sustainably manage them (Knapp and Boxshall, 2010; Britz et al., 2020) or, in the event they become extinct in the near future, preserve the knowledge of them, including their genomic data (Dubois et al., 2021; Engel et al., 2021; Cowie et al., 2022). This lends a new urgency to accelerate collection, documentation and description of ocean life (e.g. Maddison et al., 2012; Dubois et al., 2021).
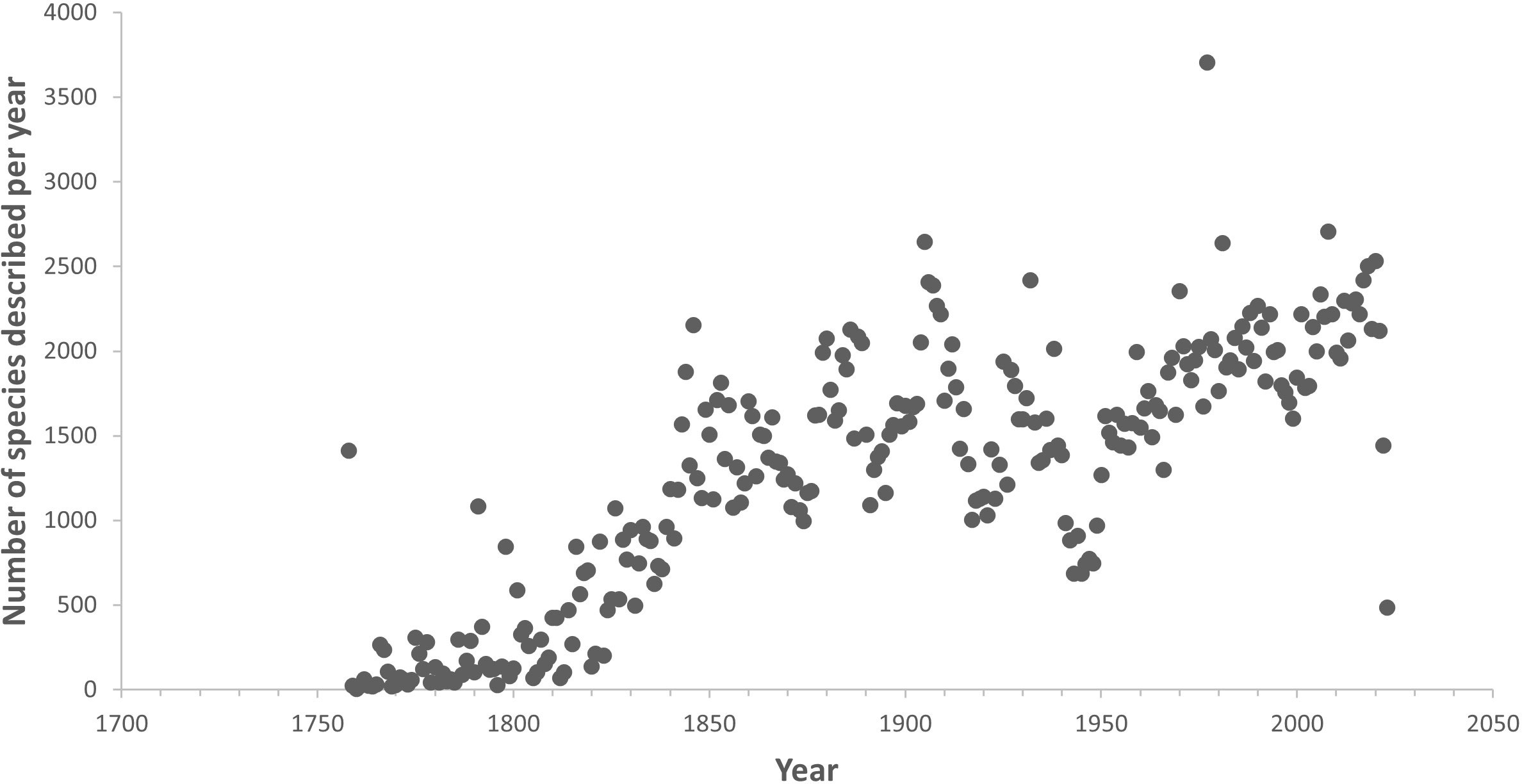
Figure 1 Rate of species discovery in the ocean. Data from World Register of Marine Species (https://www.marinespecies.org/), downloaded 14th April, 2023.
To resolve the taxonomic crisis as it manifests for ocean species, The Nippon Foundation (https://www.nippon-foundation.or.jp/en) and the Nekton Foundation (https://nektonmission.org/) assembled four international Expert Working Groups on: Marine Taxonomy and Species Discovery; Digitisation; Public Engagement; and Capacity Development & Legacy. These Working Groups collectively developed a strategy to accelerate the rate of ocean species discovery and description, the goal being to assist in addressing the biodiversity crisis in an inclusive, diverse and equitable manner, and by managing, storing, sharing and curating this new knowledge so that it is accessible to all stakeholders. This strategy involves solving both technical and social challenges as well as adopting already proven approaches to accelerate species discovery, the accessibility of taxonomic data and its application to current societal needs. It will form the basis of a new programme to accelerate the discovery and description of marine species: Ocean Census.
A literature review was undertaken to understand the current state of marine taxonomy and biodiversity research, its historical development, and potential new approaches to accelerating species discovery and description (Rogers et al., 2022a). This review formed an initial document for subsequent deliberations by the four working groups. Expert Working Groups were drawn globally to ensure representation from both high income and low- and middle-income countries (LMICs), as well as equal representation in terms of gender and including both early career and more senior researchers. Two virtual workshops were undertaken, resulting in the development of two Interim Reports (Rogers et al., 2021b; c) and a Feasibility Report (Rogers et al., 2022b). These reports were reviewed and commented upon by a team of Senior Experts (see Acknowledgements) and finalised following revision. The present paper summarises the findings of the Interim Reports and Feasibility Report prepared by the Expert Working Groups and provides a holistic view of how to accelerate species discovery in the ocean, how to translate biodiversity data to knowledge that can be applied in decision making with respect to human activities in the ocean, and how to expedite future biodiversity monitoring in the ocean.
Solving the current taxonomy crisis has been subject to wide discussion and controversy in the literature (e.g. Godfray, 2002; Godfray and Knapp, 2004; Godfray, 2007; Ebach et al., 2011; Dupérré, 2020; Dubois et al., 2021; Engel et al., 2021). In particular, there has been polarisation between those wanting to maintain traditional approaches to species description versus those advocating methods based solely or largely on DNA sequencing and/or the use of high-resolution videos or photographs, the latter in cases where preservation destroys the appearance and characteristics of the organism (e.g. Ebach and Holdrege, 2005; Gutiérrez and Pine, 2017; Dupérré, 2020; Engel et al., 2021). We suspect that part of this dispute has resulted from a polarisation in taxonomy between research focused on understanding evolution in species at a range of scales from deep phylogeny in the tree of life to mechanisms of speciation and radiation (evolutionary taxonomists) versus that focused on trying to understand recent diversity and distribution of life and its drivers, both natural and anthropogenic (eco-taxonomists; see Figure 2 for explanation). Evolutionary taxonomists tend to require highly detailed integrative phylogenetic descriptions with a large number of characters and as much genomic information as feasible whereas eco-taxonomists tend to require the minimum number of morphological characters and genetic information possible in order to define and identify species. The semantics and volcabulary used in evolutionary versus ecological studies are also different (Pavoine and Bonsall, 2011).
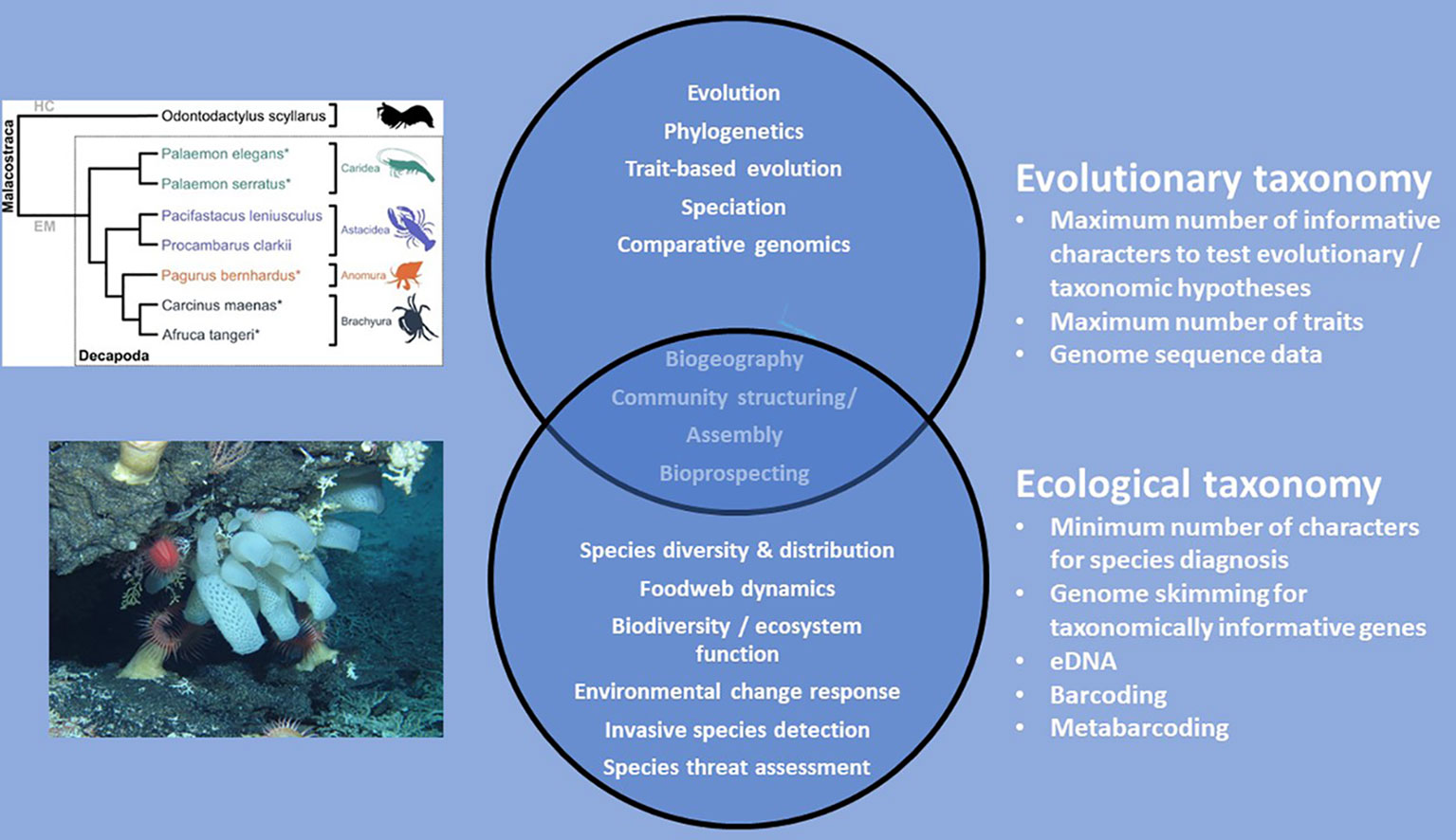
Figure 2 Polarisation in the thematic areas of scientific research deployed by evolutionary taxonomists and eco-taxonomists. Some science themes are common to both types of taxonomy and therefore these overlap. Phylogeny is taken from Drerup and How, 2021 under Creative Commons License BY 4.0. Photograph is from NERC Cruise JC66 part of the NERC/IUCN Seamounts project. Shows a mixed community of deep-sea invertebrates on Atlantis Bank seamount, South West Indian Ridge, southern Indian Ocean.
Here we favour an integrated approach to accelerating taxonomy whereby conventional species description is improved, guided and accelerated using modern technologies (e.g. high-resolution digital imaging and DNA sequencing; e.g. Wheeler, 2018; Rogers et al., 2022a), albeit with an additional step of rigorous standardised approaches to digitisation and by minimising the number of characters needed for species descriptions. We also recognise that, whilst specialist taxonomists may be needed for final species descriptions (e.g. Britz et al., 2020), many aspects of morphological description comprise time-consuming but non-specialist genetic sequencing, imaging and specimen vouchering, tasks which can be adequately carried out by skilled scientists, technicians, curators or research students (Vences, 2020). Likewise, field samples can be sorted into recognisable groups by parataxonomists, i.e. non-scientists or technicians trained to sort biodiversity samples into putative taxa, also known as informal names or morphospecies (e.g. Krell, 2004). Sequencing barcode genes also provides in-depth data that allows for sorting specimens into evolutionary significant units that can then be studied more formally. This frees up taxonomists to devote their time to the hypothesis-driven science of taxonomy, as well as associated phylogenetic and other analyses to address wider questions regarding evolution, biogeography and biodiversity (approaches are summarised in Figure 3). Importantly, this integrative approach may accelerate ocean biodiversity-related research and renewed funding support in the LMICs where many habitats, including shallow coastal biotopes are yet to be fully explored.
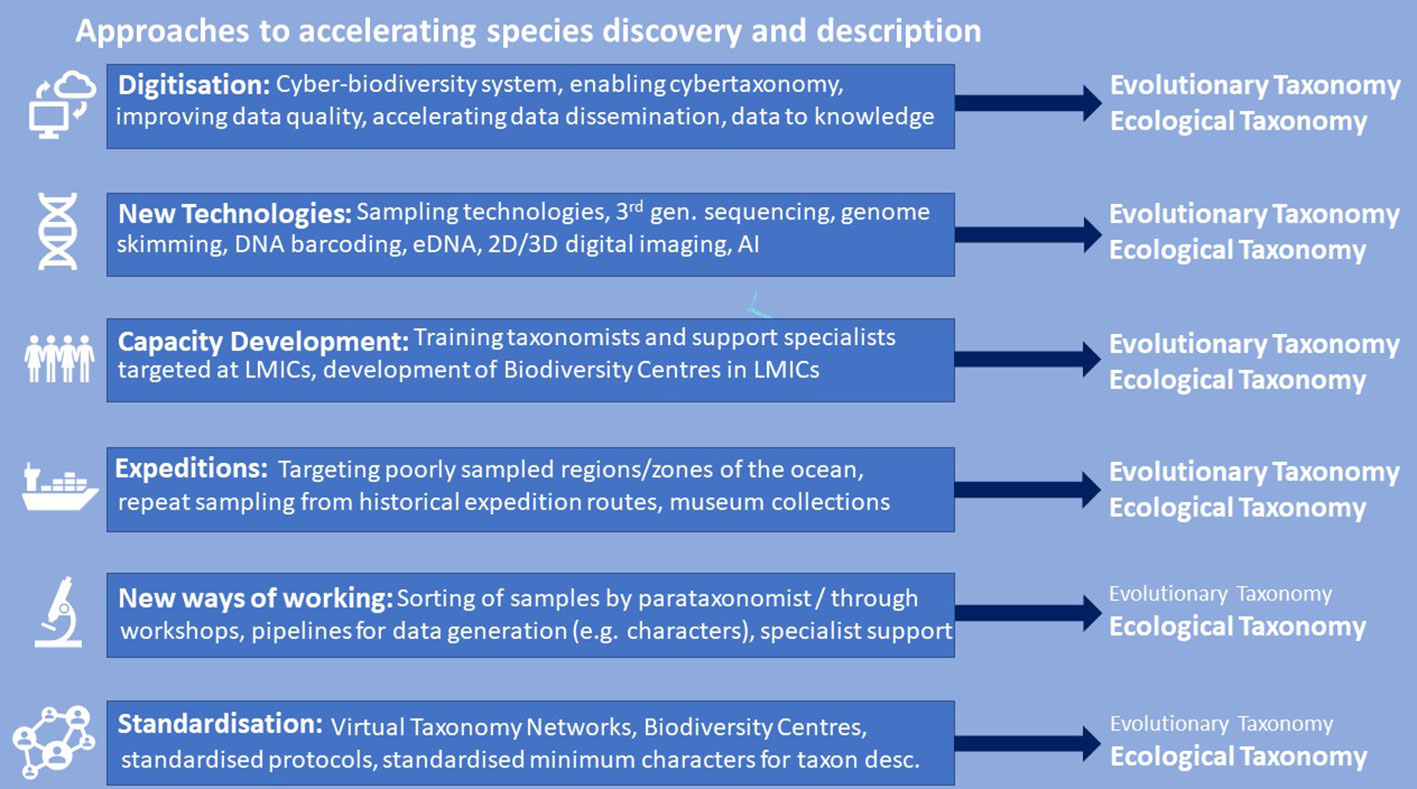
Figure 3 A summary of Ocean Census approaches to accelerating species discovery and description. We note that many of these approaches benefit studies of both evolutionary and ecological taxonomy although some are more relevant to the latter (e.g. New Ways of Working and Standardisation). Together these represent the Ocean Census integrated taxonomic strategy.
We identify four key areas which, together, will assist in accelerating the discovery of species. These are:
● Digitisation
∘ Digitisation of all data associated with specimen collection
∘ Generation of a Digital Life Form (DLF)
∘ Development of an integrative cyber-biodiversity system for dissemination of data that uses and builds on existing infrastructures (e.g. WoRMS, OBIS, GBIF)
● Adoption of new technologies
∘ Technologies that accelerate the identification and description of new species
∘ Technologies that enable cybertaxonomy
∘ Technologies that enable subsequent biodiversity monitoring
● Capacity development
∘ Training and employment of more taxonomists and support specialists, especially from LMICs, to increase the rates of species discovery and description
∘ Creation of a global open network of taxonomists working towards species discovery and description in the ocean
∘ Funding and support to develop taxonomic institutions in LMICs
∘ Promote and subsidise access to specialist equipment/facilities/knowledge at established institutions to support knowledge transfer
● Expeditions targeting poorly studied parts of the ocean with respect to biodiversity
∘ Global ocean expeditions from the intertidal zone to deep-ocean trenches
∘ Continued effort to describe species in existing collections from past expeditions
Core to the concept that has been developed is the initial creation of a Digital Life Form (DLF) in the field or in the laboratory which is associated with a collected specimen (Figures 4, 5). We use the term DLF as it has no taxonomic connotations, although the specimen to which it pertains is identified as far as possible using a standardised Open Nomenclature (ON) method (Sigovini et al., 2016; Horton et al., 2021). Additionally, a specimen is assigned a Globally Unique Identifier (GUID) which in the case of Ocean Census will usually refer to an expedition, sampling event and identification number (Rabone et al., 2019; Horton et al., 2021) and, where appropriate, a parent identification (many species are found living in or on other species, in which case the latter is the parent). As well as the specimen information, a rigorous standardised approach to the digitisation of all information related to the collection of a specimen in the field is adopted (see below). This will include use of the Taxonomic Databases Working Group (TDWG) standards and vocabularies embodied in the Access to Biological Collection Data (ABCD) and Darwin Core (DwC) aimed to support the interoperability of collection and observation data from physical to digital (Berendsohn et al., 2011; Wieczorek et al., 2012). Usage of these community-led, globally accepted data standards is critical to making data Findable, Accessible, Interoperable and Reusable (FAIR; Wilkinson et al., 2016b), and therefore to incorporation into environmental policy and widespread public use downstream (see Rabone et al., 2023 for discussion). Further digital information will be acquired whilst a specimen is processed from a collection in a museum or other institution. Data should include (see Figure 5):
● A Globally Unique Identifier (GUID) for the specimen or sample (e.g. Rabone et al., 2019; Horton et al., 2021)
● Date and location of collection including precise day, time, latitude, longitude, and depth
● Maps or bathymetric charts of sampling location and habitat description (e.g., Mayer et al., 2018; Gerovasileiou et al., 2019)
● Measurements of essential biodiversity and ocean variables (e.g. physics, biogeochemistry, biology and ecosystems; Pereira et al., 2013; Constable et al., 2016; Muller-Karger et al., 2018; Sloyan et al., 2018; Woodall et al., 2018; Centurioni et al., 2019; Jetz et al., 2019; Levin et al., 2019; Estes et al., 2021; Global Ocean Observing System, 2022)
● High-resolution stills and video imagery of the specimen (preferably living and in an anaesthetised, ‘relaxed’ state where good photos cannot be obtained from living animals), the sampling locality and, where appropriate, the sampling event itself (e.g., Katija et al., 2022).
● DNA sequence data for universal barcoding genes (as minimum)
● Specimen characterization such as biomass, dimensions, colour, state of health, maturity
● Details of parent sample and sub-sampling of specimen (e.g. tissue subsamples for DNA sequencing, see Rabone et al., 2019)
● Any other relevant information which may support better understanding of species biology and ecology
● Details of sample processing, preservation, and final repository (institution, registration number)
● Policy-relevant information (e.g. number of the permit for collection, number of CITES export/import permits, jurisdiction where sampling took place)
● Culturally relevant information (indigenous or local name for organism; indigenous and local knowledge and uses for organism; e.g. Sigwart et al., 2021)
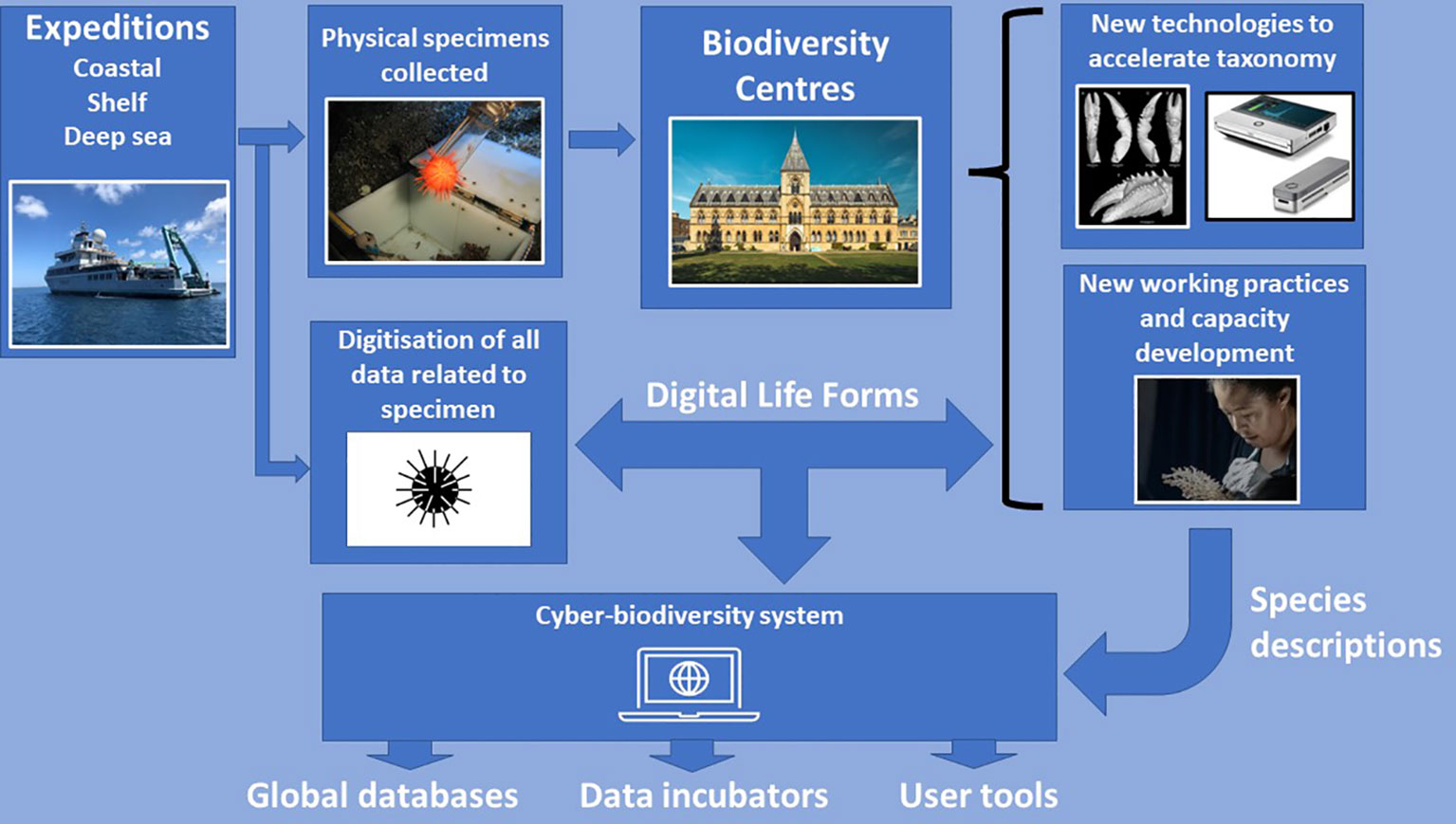
Figure 4 Diagram depicting the workflow of Ocean Census to accelerate species discovery and description (Digitisation, Adoption of New Technologies, Capacity Development, Expeditions). Image of MinION sequencer is from Oxford Nanopore Technologies (https://nanoporetech.com/products/minion). Micro-CT scan image is from Landscoff et al. (2018). Urchin icon is available under a Creative Commons License 3.0 from Iconfinder (https://www.iconfinder.com/).
The DLF created is maintained indefinitely and enables the addition of a full species description in the future and subsequent updating of information on distribution and other aspects of ecology and taxonomy (e.g. morphological variability). In this respect, this concept bears some similarities to the idea of quantum taxonomy (Maddison et al., 2012) and to the Digital Extended Specimen concept described by Hardisty et al. (2022). Data collected with specimens are uploaded to appropriate global databases as soon as possible following collection (e.g. species occurrence records to the Ocean Biodiversity Information System – OBIS https://obis.org/; genetic data to the International Nucleotide Sequence Database Collaboration – INSDC https://www.insdc.org/; taxonomic data to the World Register of Marine Species – WoRMS https://www.marinespecies.org/; genomic sample availability to the Global Genome Biodiversity Network – GGBN https://www.ggbn.org/ggbn_portal/). Where such databases are not available (e.g. for image data), data incubators will be developed to hold and allow access to data while the community of scientists who specialise in the data type can develop standards for data acquisition, metadata, storage and access. This should build towards obtaining the funds for a new global database.
Our suggested approach to marine species discovery is an example of integrated taxonomy which introduces an intermediate step between the sampling process and final species description. The introduction of the DLF enables ecological and other studies to be undertaken on consistently identified organisms prior to formal description. A proof of concept of this approach has been demonstrated through the Moorea Biocode Project (see Supplementary Material 2.0), whereby an effort was made to produce both genetic (DNA) and morphological barcodes (digital keys), dynamically linked to images and other data to create a model system for biodiversity science (Meyer et al., 2006). Using these data, ecological studies, such as on food webs, have been possible in the absence of described species but by using consistent identification based on DNA barcodes (e.g. Leray et al., 2012; Leray et al., 2015). A similar initiative for wholly image-based workflows is the SMarTaR-ID marine taxon reference image database (Howell et al., 2019).
DLFs will be useful for short-term and finer-scale ecological studies, but crucially we see the digitisation and accessibility of species data as a tool to assist taxonomists in the ultimate aim of naming and describing species. Naming species with reference to related species will reconcile provisional species discoveries with the knowledge built over the last 250 years using morphological characters. This final step is important to provide knowledge of biodiversity over large spatial and temporal scales. Since Linnaeus first devised a system and language to describe biodiversity, taxonomists have named, defined and refined an inventory of life on Earth, including more than 240,000 marine species (WoRMS, 2023) and counting. Most of these species do not have DNA sequences (Aylagas et al., 2016; Hestetun et al., 2020), and thus an integrated approach using both DNA and morphology is essential to build on our existing knowledge of biodiversity. Both DNA and morphological data on species are useful, DNA sequences allow for a statistically rigorous method for species delimitation and evolutionary relationships, while morphological data can provide information on ecosystem function, ecology and anatomy. Functional diversity studies, which have come to the fore in trying to understand how biological communities are assembled, how they respond to global and local human pressures, and consequences in terms of ecosystem function, require input of genetic and morphological information (Pavoine and Bonsall, 2011; Lauretoa et al., 2015). Both are important as they provide insight into community change and their drivers across different spatial and temporal scales (Pavoine and Bonsall, 2011).
Fundamental to our digitisation concept is that data are, where feasible, interoperable across institutional, national, regional and global databases, and that they are quality assured and traceable. This is essential to ensure that they are viewed by all stakeholders as trustworthy for application to science, ocean conservation and management. Accessibility of taxonomic and biodiversity data to end users is key to resolving the current taxonomy crisis and to ensuring long-term security for taxonomists and their institutions (Ebach et al., 2011; Saunders, 2020). It is also important that digitisation is aligned with international principles such as the FAIR (Findability, Accessibility, Interoperability, and Reusability; Wilkinson et al., 2016b), CARE (Collective Benefit, Authority to Control, Responsibility, and Ethics; Carroll et al., 2022) and FPIC (Free, Prior and Informed Consent; Perrault, 2004) principles to guide data collection and sharing practices in a way that is ethical, equitable and respectful of communities’ and data providers’ rights. This approach therefore requires the infrastructure, tools and capacity for near-real-time recording of the discovery of ocean life worldwide, with the ‘digital fingerprint’ necessary to improve data quality, interoperability, accessibility and more equitable and fairer use by communities including indigenous and local communities, scientists, and policy- and decision-makers.
Ocean Census will develop a Cyber-Biodiversity System providing the technological infrastructure, including the workflows and tools, required to ensure that all Ocean Census data are transparently and efficiently managed. The system will utilise existing data workflows and technologies (e.g WoRMS, OBIS, GBIF and DarwinCore vocabulary) and integrate them. The Ocean Census Cyber-Biodiversity System will be cloud-based, providing on-demand access to data and digital services for digitising, processing, storing and sharing marine biodiversity data, information and knowledge using standard operating procedures. This system will be built in close consultation with Ocean Census partners and will support delivery of the discovery and capacity development objectives of the programme by co-designing the workflows and tools necessary to enable more effective collection, curation, and translation of data on ocean life for scientists and other users from all countries. A modular approach will be used to develop the Cyber-Biodiversity System (Figure 5).
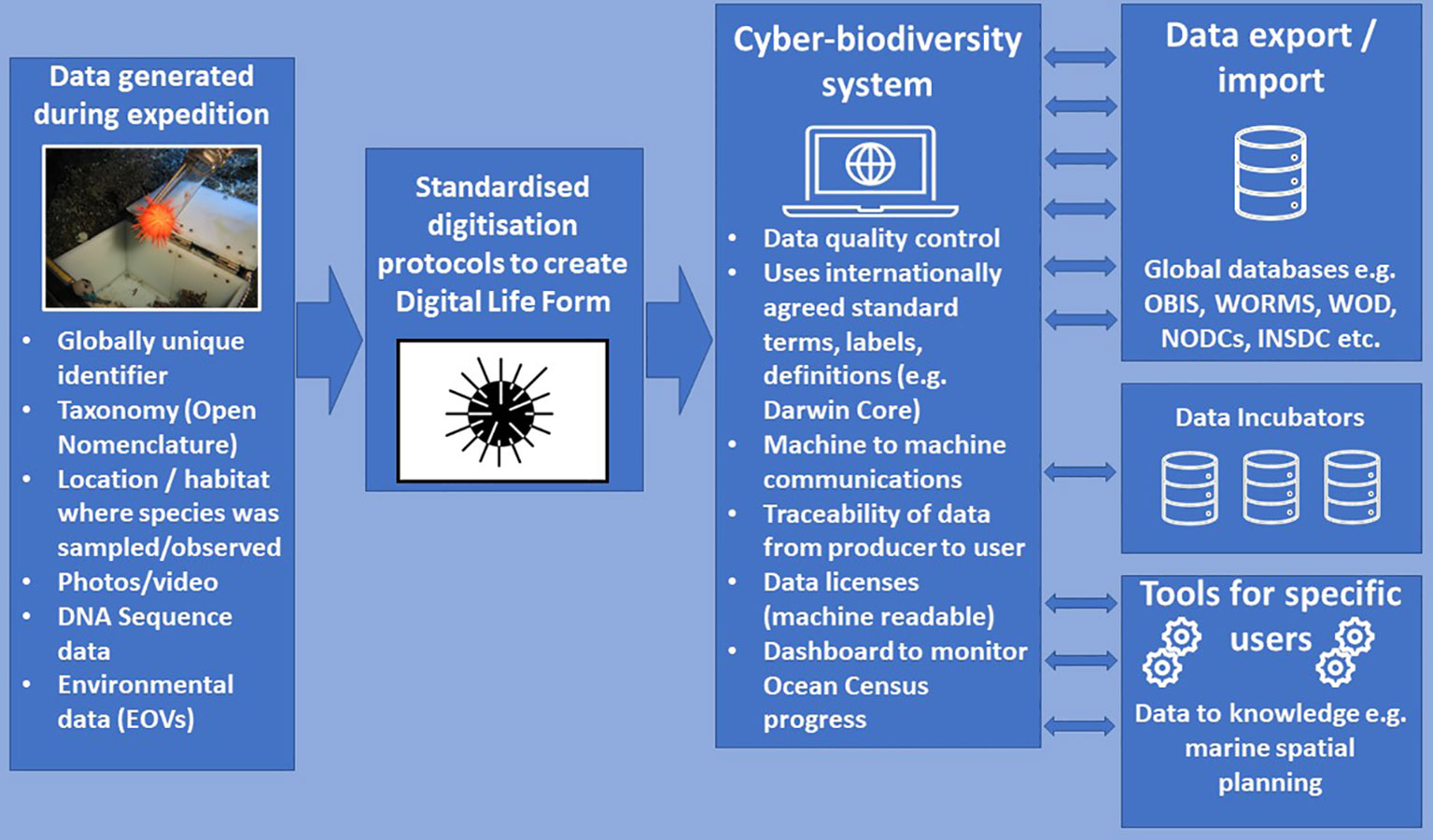
Figure 5 Diagram illustrating the digitisation of data related to the collection of a marine organism and the generation of Digital Life Forms, the flow of data into the Ocean Census Cyber-Biodiversity System and flows of data to global databases and data incubators (and return of data to the Cyber-Biodiversity System). Tools will be generated to translate data to actionable knowledge for different user groups (e.g. policy makers, scientists, industry, civil society). OBIS, Ocean Biodiversity Information System; WORMS, World Register of Marine Species; WOD, World Ocean Database; NODCs, National Oceanographic Data Centres; INSDC, International Nucleotide Sequence Database Collaboration. Urchin icon is available under a Creative Commons License 3.0 from Iconfinder (https://www.iconfinder.com/).
A Digitisation and Data Staging Module will provide a secure and high-performance space to gather, clean, quality control, and standardise all Ocean Census data and information before it is distributed for processing or archiving. The digital output of each Ocean Census expedition will be formidable: images, acoustics, video recordings, gene and genome sequences, sensor readings, and written observations are just a handful of the (meta)data types that Ocean Census will generate and synthesise. Rapidly ‘staging’ or hosting, cleaning, and organising data so they can be efficiently processed, combined with other data, and delivered to users or long-term archives will be essential to the efficient discovery of new biodiversity as well as the digital legacy of the Ocean Census programme. The Digitisation and Data Staging Module will use cloud-based services to securely host the data generated and needed by the “scientific” community and other users in a ‘Data as a Service’ (DaaS) model (see below).
All partners, including those who do not have capacity to host and mobilise large data collections locally, will have the capacity to analyse data as it streams in from global expeditions and integration activities. Importantly, this module does not seek to be a long-term archive itself, but a working space to promote standardised data handling to speed discovery and transparency. For data types with matching global long-term archives/databases (e.g. INSDC or OBIS), this module provides a local, shared storage solution to prepare Ocean Census data products for submission. For data types without a long-term, multinational archive, this module will provide mid-term storage and preservation of data in the Ocean Census Digital Incubator until viable long-term options are available (see below).
The Data Processing Module will host the software and services to analyse data from Ocean Census, its partners and/or external sources before these data are distributed to partners, decision-makers and the public. The Data Processing Module will contain collections of software and tools for Ocean Census personnel and collaborators to transform, integrate, analyse, visualise, and communicate the results of the programme’s digitisation efforts and it will be combined with data imported through the Cyber-Biodiversity System. It will rely on standardised data in the Digitisation and Data Staging Module and the standard operating procedures developed in the early stages of the programme.
This module will offer ‘software as a service’ (SaaS) via the cloud and will ensure that consistent, traceable, and reproducible methods are used on data and information in the Ocean Census Cyber-Biodiversity System. For partners without local computing capacity, the Data Processing Module will provide a shared processing facility, while also providing tools that can be used in others’ systems to achieve the same Ocean Census-grade standardisation, quality assurance, and interoperability with external systems. We view this as essential in the democratisation of species discovery for the global marine taxonomy community.
Building on the two prior modules, the Digital Exchange Module will facilitate access to and the delivery of Ocean Census data between all partners to online repositories and platforms. The Digital Exchange Module will be designed to facilitate delivery of ‘gold standard’ marine biodiversity data, as well as to ensure that the programme’s digital reach and impact are tracked.
The Ocean Census Digital Incubator will support communities with developing and adopting new technologies to transform how they digitally capture and communicate data on ocean life. While some biodiversity data are well-managed globally (e.g. sequence information), other data types have no internationally agreed archives, governance, or standards (e.g. image data). Acquisition of ocean life data often uses time-consuming manual methods that depend on individual’s activities and expertise, slowing down data sharing. Thus, data are often underutilised or lost after projects conclude. Moreover, limited human, technological or financial capacity can limit adoption of promising technological solutions in local, national or regional contexts.
The aim of this activity is primarily to help ‘level the playing field’ in the application of data management and science in marine biodiversity research, and to address known gaps in knowledge transfer—both in terms of data types (e.g. large-scale storage, automation and application of machine learning to imagery data; see below) and country- or region-specific capacity development to adopt or integrate data management and science into workflows, supported by the Ocean Census network of institutions and researchers.
We recognise that other cyber-taxonomy platforms exist (e.g. the European Distributed Institute of Taxonomy [EDIT] Platform for Cybertaxonomy; Berendsohn et al., 2011) and, following the philosophy to make use of what has already been developed rather than repeat work, we will integrate the Ocean Census Cyber-Biodiversity System with these or at the very least ensure it is interoperable with them. We also note that the Ocean Census Cyber-Biodiversity System will be developed through extensive consultation on all aspects of the platform to ensure the system meets the needs of scientists, ocean managers, industry and policymakers. Once the system is established, applications can be further developed to meet the needs of these user communities. Options may include enhancing use of Ocean Census data in leading decision-support tools and platforms, tailored to public or private sector decision-making needs, or harnessing citizen science initiatives to generate data on ocean life in collaboration with indigenous and local communities. These pathways could be established by strengthening the ocean knowledge base of existing policy- and decision-support platforms, such as the Exploring Natural Capital Opportunities, Risks and Exposure (ENCORE) or the UN Biodiversity Lab, among many others.
This will require deeper curation and processing of decision-grade derivative products from Ocean Census data (e.g., species occurrence records, abundance, biomass, etc.) to provide applicable indicators for the diverse range of users (e.g., ecosystem service indicators, biodiversity seascape maps for marine spatial planning, species, populations or stocks). For example, based on the knowledge generated through Ocean Census, platforms such as Ocean+ Habitats could be enhanced through near-real-time connections with OBIS for taxonomic coverage, transparently visualising the ‘known unknowns’—i.e. locations that have not been surveyed for species or habitats —by country or region. This application could help to guide targeting of Ocean Census expeditions to focus sampling activities, support governments in identifying which areas of marine biodiversity importance require protection or restoration or will be able to direct overseas development aid to the locations that need it most. Solutions will also be co-developed in community-based or low-income contexts. For example, by coordinating Ocean Census with community science initiatives and other discovery activities with indigenous peoples and local communities, this activity could support integration of community insights and local or traditional knowledge into policy processes and indicators for biodiversity and ecosystem conservation and sustainable management. This approach could also pave the way towards developing blended blue financing schemes in order to support blue economy initiatives, in particular for coastal countries representing LMICs.
A variety of technologies have been developed or are in development which increase the quality of species descriptions and can potentially accelerate taxonomy over the longer term through their applications to cybertaxonomy (defined in Rajmohana and Bijoy, 2012). Some of these technologies are reviewed in Rogers et al. (2022a) and include advanced, high-resolution 2D and 3D imaging, DNA sequencing including the use of 3rd generation sequencing technologies in the field and artificial intelligence/machine learning.
High resolution digital 3D imaging techniques such as photogrammetry, confocal laser scanning microscopy, optical projection tomography, magnetic resonance imaging and micro-computed tomography (micro-CT), can rapidly create high-resolution morphological and anatomical data in 3D (e.g. Boistel et al., 2011; Faulwetter et al., 2013a; b; Nguyen et al., 2014; Medina et al., 2020). Multispectral imaging has also been used to produce high resolution images of the colours and shapes of organisms (e.g. Chan et al., 2022). 3D and multispectral images can be used to produce virtual type specimens from original type material. This is a major enabler of cybertaxonomy, allowing taxonomists to examine a specimen via the web for comparative purposes without access to the actual specimen, thus saving time and cost and avoiding risk in sending specimens from museums or negating the need for researchers to travel to where a sample is stored. Whilst some argue that such technologies do not yet have the resolution to produce virtual types (Dupérré, 2020) or that some information is not stored by these methods (e.g. CT or SEM scans do not record colour), technology is improving rapidly.
Molecular data can assist in the identification and classification of new taxa (Rogers et al., 2022a). The simplest approaches are based on a single or a few barcoding genes (e.g. Hebert et al., 2003a; b). However, 3rd generation high-throughput sequencing technologies enable a larger portion of the genome to be analysed for taxonomic purposes (Rogers et al., 2022a). Low coverage genome sequencing followed by analysis of a number of taxonomically informative nuclear, mitochondrial and chloroplast genes can provide a large amount of data for phylogenetic analysis and diagnosis of new taxa (genome skimming; Krehenwinkel et al., 2019). DNA metabarcoding approaches have been applied by terrestrial taxonomists on large numbers of specimens with clustering of sequence data being used to focus the work on undescribed species (e.g. Riedel and Narakusumo, 2019; Srivathsan et al., 2019). These technologies are now highly portable, making them amenable to undertaking sequencing studies in the field allowing the of training researchers in metabarcoding or metagenomic approaches (Krehenwinkel et al., 2019; Watsa et al., 2020; Ames et al., 2021). This can be especially useful in LMICs where fully equipped molecular biology laboratories may not be available to local scientists.
New technologies can also enable sampling of organisms that are highly fragile, and which are destroyed by more traditional sampling methods such as trawls. A good example of this are gelatinous zooplankton, especially groups such as physonect siphonophores, lobate ctenophores and larvaceans (Robison et al., 2017; Hetherington et al., 2022). Remotely operated vehicles (ROVs) or submersibles have been equipped with detritus samplers (D-samplers) which consist of an open tube which can be manoeuvred to enclose an organism then closed at the top and bottom (Robison et al., 2017; Teoh et al., 2018). More recently, a rotary actuated dodecahedron made of polymer has been developed which can also be employed by an ROV or submersible to enclose small to medium-sized gelatinous organisms (Teoh et al., 2018). Even when these organisms are sampled intact, the process of preservation can destroy them through fragmentation/disintegration or damage delicate structures and modify colours which are important characters for taxonomy (e.g. the external feeding filters of larvaceans; Robison et al., 2017). For such organisms, in-situ photographs/video of the animal of sufficient resolution to image taxonomically important morphological characteristics are important (Matsumoto et al., 2003; Robison et al., 2017). Large gelatinous organisms remain a significant problem for sampling and in the future a combination of a tissue sample, DNA sequence data and high resolution in-situ 3D imaging may be a useful approach for taxonomy (Vences, 2020). Because of the morphological plasticity in some gelatinous taxa, cases have been uncovered where species diagnoses are only possible through including DNA sequence data (Lawley et al., 2022; Montenegro et al., 2023). Ecological studies of zooplankton are now often carried out using imaging combined with machine learning to identify taxa (Irisson et al., 2022; Rogers et al., 2022a).
Artificial intelligence (AI) approaches, inclusive of both machine learning and deep-learning methods, such as artificial neural networks, are being widely applied to the identification of taxa from images as well as from active (biological echosounder) and passive acoustic data (reviewed in Rogers et al., 2022a). Machine learning has been applied to specific identification projects or tasks (e.g. identification of fish or plankton from image data or whales from acoustic monitoring data; Goodwin et al., 2022; Katija et al., 2022). There are developing initiatives to generate global databases for some organisms, including plankton and benthic organisms (Goodwin et al., 2022; Irisson et al., 2022). However, the development of ever more powerful computing capacity, new approaches to pattern recognition and larger collections of images/videos could enable AI to play a greater role in species identification (e.g. MacLeod et al., 2010) if taxonomist-curated images were available in the large quantities necessary for producing robust training data. To address this bottleneck problem of labelled high quality training data, the machine learning community has proposed new learning algorithms such as self-supervised learning that can make use of non-annotated data for improving performance (for instance in plankton image analysis; Midtvedt et al., 2022). Access to data on the web via web scraping technologies have made large quantities of identified and unidentified images of marine organisms available for deep-learning approaches to species identification (e.g. Langenkämper et al., 2017; Jarić et al., 2020; Stringham et al., 2020), especially where unsupervised clustering techniques are employed (Schroeder et al., 2020). Coupled with large-scale citizen science projects dedicated to collecting such imagery, specifically for biodiversity documentation such as iNaturalist, (https://www.inaturalist.org/) these technologies could offer a powerful approach to identification of species, especially megafauna and macroalgae. This could include documentation of unidentified or undescribed species which could help to target future sample collecting efforts.
More than two-thirds of countries surveyed by the International Oceanographic Commission of UNESCO have a national strategy in place to achieve the UN Agenda 2030 for Sustainable Development, but only a fifth have a specific strategy for Sustainable Development Goal 14 (Life Below Water; IOC-UNESCO, 2020). Ocean research capacity, researchers and infrastructure, are disproportionately concentrated in high-income countries and within the hands of men (IOC-UNESCO, 2020), illustrating inequalities in the capacity to understand, manage and sustainably use our connected ocean (e.g. Rogers et al., 2021a). On the other hand, many threats to the ocean and its biodiversity including warming, acidification, pollution, fishing and invasive species are global issues affecting all marine ecosystems. Addressing such imbalances in scientific capacity and access to the global ocean will be a fundamental component in improving its governance and ultimately its health (Hornidge et al., 2023).
Increasing capacity for marine species discovery and taxonomy is required to meet the current deficit in our knowledge of ocean biodiversity (e.g. Ebach et al., 2011; Fontaine et al., 2012; Baldini et al., 2021; Engel et al., 2021). This is not just human capacity but also institutional capacity to process samples and to store, maintain and curate specimen collections, including, for example, cryogenic storage of tissues for genomic DNA extraction (e.g. Paknia et al., 2015; Baldini et al., 2021) and associated images. For example, there are limited efforts underway to develop DNA tissue banks regionally in the Bay of Bengal of the Northern Indian Ocean and the Ocean Census can help accelerate such initiatives over the long term. Capacity development activities must be fully inclusive and equitable, with a particular focus on biodiversity rich LMICs (e.g. Barber et al., 2014; Paknia et al., 2015; Titley et al., 2017; Britz et al., 2020; Prathapan and Rajan, 2020). Such countries should be able to advance the development of science and knowledge through the employment, training and development of local experienced taxonomists and support staff through professional valorisation and stability within their respective countries. This will enable species discovery and biodiversity monitoring to be led locally to meet specific economic needs as well as contributing to international commitments under treaties such as the Convention for Biological Diversity and the new treaty on Biodiversity Beyond National Jurisdiction (Prathapan and Rajan, 2020; Saunders, 2020). Our approach in Ocean Census is to foster co-production of taxonomy and biodiversity research between scientists and to assist in developing the research infrastructure for species discovery, the housing of collections of biological material (Barber et al., 2014; Paknia et al., 2015; Tolochko and Vadrot, 2021; Woodall et al., 2021) and the management of a correlated curated image repository accessible to all stakeholders. Ocean Census also recognises the need to strengthen international networks of taxonomists for purposes of knowledge exchange, making decisions on standardisation of methods and for pooling of resources and effort to achieve ambitious goals for species discovery targeting regions and ecosystems that have been poorly sampled (Bax et al., 2018; Tolochko and Vadrot, 2021; Woodall et al., 2021). Such networks are also important in maintaining dialogue and engagement with scientists from LMICs to ensure that capacity development activities have a long-lasting legacy (Bax et al., 2018). Building expertise across LMICs may also help to reduce the need for transboundary movement of specimens and therefore any impact of the permitting processes under the Nagoya Protocol, aimed at combating biopiracy, but which have seriously impacted international biodiversity science (e.g. Ebach et al., 2011; Prathapan and Rajan, 2020).
An overriding goal in developing global capacity in marine taxonomy and biodiversity science will be to maximise the opportunities for scientists trained in this program to remain employed in marine science and biodiversity discovery and its conservation beyond the duration of the program. It can be challenging for expert taxonomists without additional areas of expertise to remain employed. Therefore, we propose to build capacity across the biodiversity value chain from foundational research to knowledge products and policy advice. This approach will improve uptake of research and the retention and employability of those whose capacity has been developed and self-sufficiency for countries to lead, from the collection, identification and processing of samples to the translation of science into decision-making processes. Explicit requirements and opportunities in all elements of this program for training and development are essential to ensure appropriate capacity in marine biodiversity discovery, data science and communication is achieved and is done so within the specific context of the partner institutions and countries.
Through building capacity across the biodiversity value chain, we will also provide individual countries with the capability and opportunity to lead surveys in their own waters (providing tailored support as needed) including, for example, for biodiversity discovery and environmental assessment and enhance their capability to engage in management and policy activities. Skills development will be tailored to meet the needs and aspirations of the individuals. Such skills development will likely consist of a mix of hands-on training held during expeditions, general training at national and regional centres, and more specific skills training at the Ocean Census Biodiversity Centres. Technical skills training (e.g. sample processing, database management, communication) will be extended to participants at national and regional centres, while specific specialist training at the Biodiversity Centres will develop specific skills and knowledge (potentially as part of a higher degree).
Monitoring, evaluating and learning processes will be an essential component of capacity development under Ocean Census and should help identify, in conjunction with the registries of practices in the literature, needs and opportunities to be further explored by Ocean Census and its funders. As a starting point, there will be an annual review of progress against the Ocean Census Equity Principles (see Supplementary Material 3.0) as well as against KPIs to be developed. At the end of Year 3 there will be a more extensive review of the progress of Ocean Census and further development of a capacity development component more strategically aligned with needs and expectations captured.
The urgency that the decline of biodiversity places on species discovery means that a significant effort in field collection of organisms is required (e.g. Dubois et al., 2021; Cowie et al., 2022). Many parts of the ocean remain poorly sampled, either because they lie in or adjacent to the waters of LMICs, they are inaccessible (e.g. the high seas) or lie in extreme environments (e.g. the deep sea; Glover et al., 2018; Hughes et al., 2021; or polar marine ecosystems; e.g. Brandt et al., 2007; Verhaegen et al., 2021; Ramirez-Llodra et al., 2022). Only small fractions of many deep-sea ecosystems have been explored by biologists (Ramirez-Llodra et al., 2011; Rogers, 2015); with the deep pelagic being a particular challenge (Webb et al., 2010) because of its size, remoteness and the fragile nature of the organisms that inhabit it making them difficult to sample (e.g. Raskoff et al., 2003). As well as geographic biases in collections of taxa, there are also taxonomic biases in the effort on their taxonomy (e.g. Troudet et al., 2017; Hughes et al., 2021; reviewed in Rogers et al., 2021a). Groups such as vertebrates are well sampled (Appeltans et al., 2012; Scheffers et al., 2012), whereas for some groups of marine invertebrates it is estimated that less than 20% have been described (generally small organisms; Appeltans et al., 2012). In these cases, tens of thousands or even more than a hundred thousand species may remain unsampled and/or undescribed (e.g. Nematoda, Gastropoda, Peracarida; Appeltans et al., 2012; Bouchet et al., 2016). More complete sampling of taxa is also important in testing taxonomic hypotheses and identifying global patterns and processes and lack of adequate coverage of taxa could potentially lead to erroneous conclusions which impact on our understanding of the evolution and biogeography of the marine realm (see Dubois et al., 2021 for examples from recent amphibians; Sands et al., 2021 for an example on Antarctic ophiuroids). Description of species from single individual specimens (singletons) is also problematic, as the range of morphological and genetic variation cannot be documented (e.g. Fontaine et al., 2012). This can lead to mis-informed and unstable species descriptions (e.g. Hippocampus guttulatus; Vasil Eva, 2007), although such specimens are key targets for further investigation and study. More field collections can help to address many of these issues.
We also identified that repeat sampling of stations from some of the global and regional expeditions of European and US vessels in the 19th and 20th centuries (e.g. the Challenger Expedition) would be worthwhile to obtain fresh specimens of species that they described, especially where those descriptions were incomplete. A particular priority will be to collect genetic data from type-localities from original descriptions. This is because inadequate descriptions from these expeditions acts as a significant constraint on efforts to describe new species, often requiring the study of museum types with all that implies in terms of travel, costs and time (Riedel et al., 2013). Even where such material still exists in museums, particularly for deep-sea species, it is often a single, damaged specimen that was not live collected (e.g. Williams et al., 2020) and is unlikely to be amenable to modern integrated taxonomic approaches, limiting its use. It may also provide samples to study change in some of these sampling localities following severe impacts on the ocean from industrialisation, including pollution, overfishing and climate disruption such as changing carbonate chemistry (ocean acidification). Such repeat sampling will enable LMICs to “reclaim” their biodiversity through co-produced expeditions. There is a growing fleet of Ocean-Class research vessels with mechanisms to enable participation or leadership of cruises by scientists from LMICs, including privately owned platforms (Rogers et al., 2021a).
Expeditions will be undertaken to discover marine species from the intertidal zone, coastal subtidal environments, and the deep sea, including the hadal zone, from polar regions to the tropics, and will incorporate both benthic and pelagic ecosystems. These expeditions will deploy appropriate sampling technologies and infrastructure to these different ecosystems with implications for expedition organisations and logistics as well as costs (the further from shore and the deeper the ecosystem the larger the cost).
Finally, we also recognise that large collections of undescribed and in many cases uninvestigated samples from past expeditions exist in museums and other institutions. Ocean Census will also include these specimens in efforts to describe marine species. Priority will have to be based on condition of samples, documentation of their collection and subsequent curation as well as on geographic regions which are poorly sampled.
Ocean Census has been initially funded by The Nippon Foundation, an organisation that funds global projects in the areas of social innovation including ocean sciences (e.g. The Nippon Foundation-GEBCO Seabed 2030; Mayer et al., 2018; https://seabed2030.org/). The project will be administered by the Nekton Foundation who were initially tasked with developing the scientific approach to the programme. A wide network of partners are being engaged with the programme with collaborations including: the provision of seagoing infrastructure (e.g. research vessels, deep-submergence platforms, other equipment), co-production of science (e.g. Museums, other biodiversity institutions, sequencing centres), development of cyber-infrastructure (e.g. UNEP World Conservation Monitoring Centre), provision of other forms of technical assistance and training (e.g. industry partners) and other forms of direct and indirect funding or support. Partnerships will also be established with governments, governmental institutions, academic institutions, non-governmental institutions and civil society, including indigenous communities within countries in whose waters Ocean Census will operate.
Ocean Census will be organised as an open network with the aim to promote inclusivity and equity across the global community of marine taxonomists, taxonomy support specialists and trainee or early career researchers (Figure 6). The programme will initially be established at Biodiversity Centres located at national natural history museums or other institutions undertaking marine species discovery and description activities and able to arrange national and international loans of material. These institutions have established collections of ocean species and significant infrastructure for taxonomic studies including imaging, sequencing and other advanced technologies discussed above. Their primary activities will include:
● Sorting and digitisation of samples collected during Ocean Census Expeditions including identification of known species.
● DNA sequencing of species collected during Ocean Census Expeditions.
● High resolution 2D and 3D imaging of specimens collected during Ocean Census Expeditions including through application of advanced imaging technologies such as microcomputed tomography (micro-CT).
● Description of new species collected during Ocean Census Expeditions.
● Undertake training and capacity development activities.
● To act as resource centres for use of advanced facilities for Ocean Census where they may not be generally available for the global marine taxonomy community (e.g. high-throughput DNA sequencing, micro-CT).
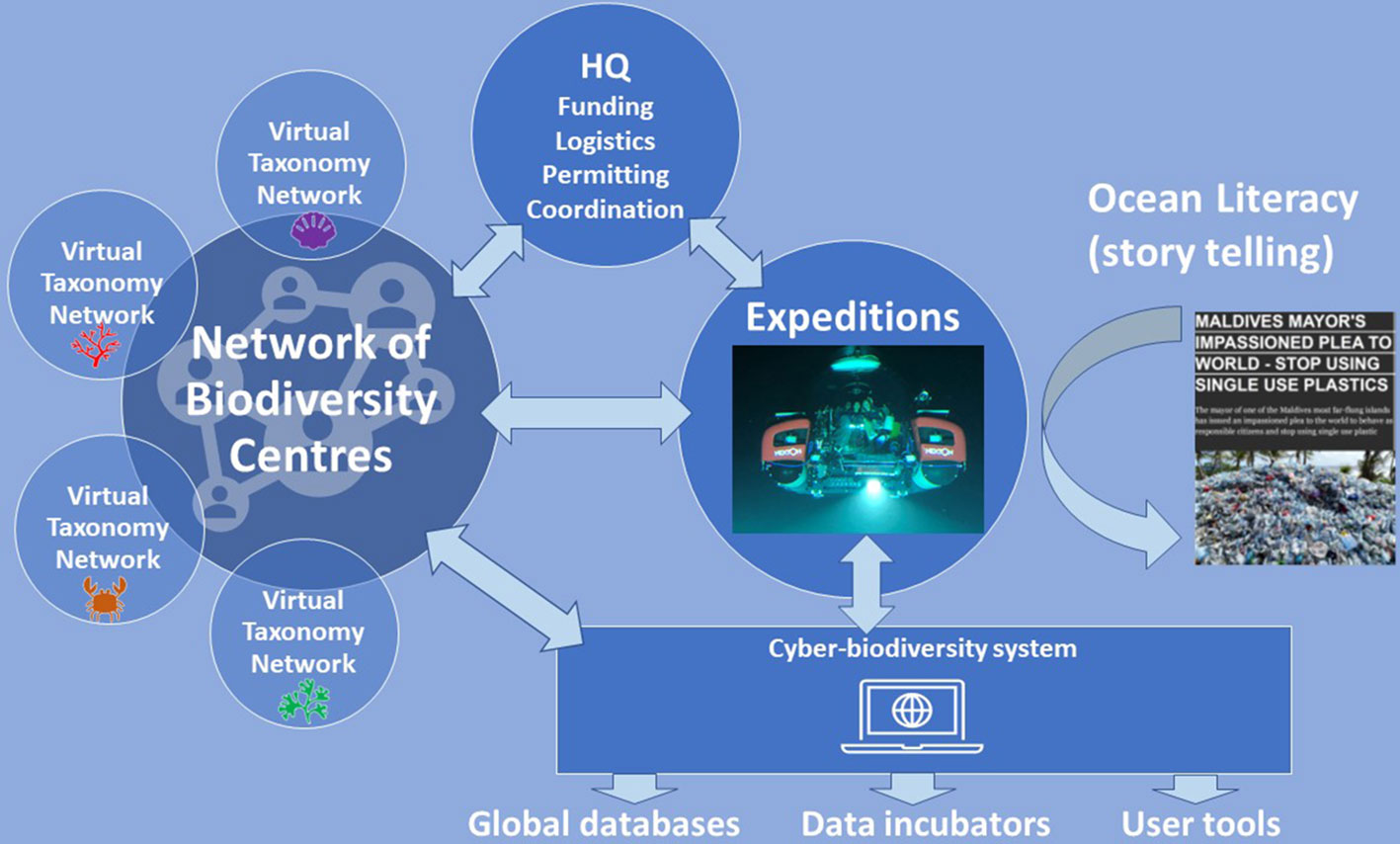
Figure 6 The open network structure of the Ocean Census programme. Core to developing a lasting legacy in acceleration of discovery and description of marine species is the development of a network of Biodiversity Centres and Virtual Taxonomy Networks enabling taxonomists to cooperate and coordinate their activities globally. Expeditions generate samples for species discovery and also stories for ocean literacy. Data generated by expeditions and taxonomists are communicated to the Ocean Census Cyber-Biodiversity System. Ocean Census Headquarters (HQ) will coordinate the programme including expeditions, expedition logistics, permitting, some funding bids as well as take on reporting functions to funders.
As the programme develops, Biodiversity Centres will be identified in regions where such institutions have historically been under-supported, usually in LMICs. Assistance will be provided to these institutions to develop a business plan for each potential centre, to contribute to species discovery within the Ocean Census programme as well as providing facilities for capacity development as laid out above. All Biodiversity Centres will be encouraged to develop complementary skills across the biodiversity value chain to ensure longevity of these institutions and their personnel once the funded Ocean Census programme has been completed. The Biodiversity Centres will together form the Ocean Census Global Marine Biodiversity Network.
The marine species discovery pipeline will be considerably enhanced by the coordination of taxon-specific expertise at a global scale. An accelerated and scaled species discovery process will need to draw upon all available expertise, including expertise that resides outside the Ocean Census Biodiversity Centres, in universities, regional museums, government research organisations, non-governmental research organisations, and the general community (citizen scientists). The Ocean Census solution is to create a series of taxon-specific virtual networks that will act as a bastion of knowledge for species identification, species descriptions, and support the Biodiversity Centres with taxonomic training and mentorship. We envisage ten or more such networks that could grow from informal taxonomy guilds that already exist for organism groups such as isopods, bivalves, sea urchins, worms, red algae etc. This has proven the most effective way of harnessing taxonomic expertise for projects such as the WoRMS database.
An experienced taxonomist within each network will be resourced to organise discussion groups, identification/training workshops (based on specimens from Ocean Census Expeditions), mentorship of emerging experts such as early career researchers, establishment of minimum standards of species description and publication, validation of new species discoveries and descriptions generated by Ocean Census, validation of sequence and distribution records in global databases, preparation of biogeographic datasets, and to organise the sequencing or imaging of pivotal specimens (e.g. types/topotypes) in museum collections.
These networks will be funded either directly from the program or via the Ocean Census Biodiversity Centres. Annual re-funding will be contingent on meeting key performance indicators set for the previous year.
The global ocean observing community is providing guidance and incentives for researchers to standardise and share their measurements. The Framework for Ocean Observing published in 2012 (Lindstrom et al., 2012) outlines a series of standards, and a process through which Essential Ocean Variables (EOVs) can be identified and progressed from conceptual through prototype to a mature level where they support global observation and reporting (Constable et al., 2016). Alongside collection of specimens, Ocean Census will collect as many EOVs as feasible within the limitations of specific expeditions. This will range from measurements of temperature using instrumentation carried by SCUBA divers (e.g. from dive computer data) to a full range of physical measurements achievable on an Ocean Class research vessel capable of deploying a CTD and water sampling rosette, mapping the seafloor using multibeam bathymetry and undertaking in-situ measurements using submersibles, remotely operated vehicles or autonomous platforms. Such data will add to global data archives on EOVs but will also enable the modelling of the distribution of species and whole communities to better understand current distribution and the future effects of climate change.
The usefulness of the Ocean Biodiversity Information System as a general repository for marine biodiversity data is being substantially upgraded (e.g. De Pooter et al., 2017). Collaboration with the Marine Biodiversity Observation Network and the Smithsonian’s MarineGEO observatory (Duffy et al., 2013; Muller-Karger et al., 2014) is helping to develop an integrated and practical pathway toward a Global Ocean Observing System (GOOS) that includes biological measurements supported by existing physical and biogeochemical EOVs. Environmental DNA (eDNA) is already forming an important tool to assess the relative levels of biodiversity in different localities (reviewed in Rogers et al., 2022a). However, this approach is limited by the lack of sequences that are attributable to known species (Rogers et al., 2022a). Because of the integrated approach used by Ocean Census in terms of collection of sequence data alongside species, the programme will greatly assist in populating the reference sequence libraries within the Barcode of Life Data System (BOLD; https://www.boldsystems.org/) and global DNA databases (INSDC). This will enhance the use of eDNA approaches for assessment of species diversity and how it changes over time as a result of global or local anthropogenic stressors or as a result of management interventions. eDNA is increasingly being used as a biomonitoring tool, enabling the identification and quantification of species within different marine ecosystems (e.g. Thomsen et al., 2016; Hunter et al., 2018; Shelton et al., 2019; Sanchez et al., 2022) and the assessment of water quality (Apothéloz-Perret-Gentil et al., 2017; Yang and Zhang, 2020). Deployment of well-designed eDNA surveys will be more cost effective than large-scale field sampling surveys using morphological identification (Apothéloz-Perret-Gentil et al., 2017; Gold et al., 2021; Jo et al., 2022) potentially enabling LMICs to undertake biodiversity assessment and monitoring where at present they are unable because of limited human capacity and other resources. Ocean Census will include such molecular tools in its capacity development activities with a particular focus on low-cost portable sequencing technologies (Rogers et al., 2022a).
Ocean Census will enhance data collection of EOVs and molecular data through the deployment of a “Tool Kit” deployable through a containerised laboratory that can be transported to coastal localities or onto large research vessels. This will include equipment for sample sorting and preservation as well as measurement of environmental parameters and for DNA sequencing.
There is a major deficit in people’s awareness of ocean life and its importance in supporting human society. Ocean Census therefore aims to raise and inspire public and partner awareness of ocean life and the programme to accelerate its discovery. Highlighting the critical nature of the biodiversity crisis and why it is important to all eight billion people needs a carefully crafted communications strategy which does not hide from the reality of how hard it is to gain media coverage, or the attention of policymakers, existing regional frameworks and the public. The thesis is simple - widespread attention leads to knowledgeable support at scale and that, in turn, triggers global action and positive change. Such a communications campaign can take a long time to bear fruit as has been illustrated by the length of time it has taken to create public awareness and increasingly gain broad, if largely passive, public support over the climate crisis.
The recent June 2022 survey by The Nippon Foundation of 10,000 Japanese people’s relationship with the ocean provides an excellent barometer of the need to pull every media lever possible to maintain an emotional connection with the ocean (Nippon Foundation, 2022). In the wake of the COVID pandemic, the survey showed 45% of respondents had not physically been to the ocean in the past year - and as a result their ‘connection’ and ‘affection’ for the ocean declined and fewer of those people felt ‘that the ocean is important’. The good news in the survey was that overall awareness of ocean issues and participation in activities to protect it had risen since 2019. It is reasonable to conclude that if the ocean issue and the importance of biodiversity within it can be kept in the public eye we can, in the words of The Nippon Foundation Chairman Mr Yohei Sasakawa, help young people ‘feel affection toward the ocean and understand the role that the bounty of the ocean plays in their daily lives’.
To gain an audience, to inform, educate and entertain that audience, and to build its trust and loyalty can only be done through establishing a constant emotional connection. Maintaining audience attention in such a ferociously fought battle for our precious time, when fewer people are experiencing the ocean first hand, means serving a constant diet of stories that appeals to their taste for facts and quenches their thirst for knowledge. To succeed in this strategy Ocean Census aims to simultaneously address policymakers and vast numbers of people right from the start by:
● Creating a constant flow of entertaining and informative content.
● Harnessing a cost-effective and proven global distribution partnership with paid distribution through trusted news sources and a paid social media amplification partner.
Content will be created for Ocean Census outreach activities in the following ways:
▪ Nautilus Expedition content: providing the main pillars for content creation, public and partner engagement throughout the year.
▪ People at the centre: put the voices of real people who others can relate to at the centre of the storytelling - creating an emotional connection with the audience and thereby gain attention in a “shared” media economy that will accelerate our ability to build awareness and ultimately garner active support to encourage and enact change.
▪ Different content for difference audiences: content will be created/repackaged for different audiences on different platforms – i.e. content for news media distribution is very different to social media
▪ Innovative ocean storytelling: the greatest area of opportunity for innovation – which will be a key aspect of the strategy – will be achieved during Ocean Census expeditions, utilising new technologies and live broadcast links.
▪ Community generated content: harvest and repackage content about ocean life from the scientific, civil society, public, indigenous and local communities as well as citizen science communities for distribution through the news and digital media.
▪ Partnership: harness the investment and storytelling skills of the storytelling industry to tell the story of ocean life and Ocean Census (e.g. broadcaster commissions a series/documentary about Ocean Census rather than Ocean Census trying to produce and distribute its own content).
Ocean Census will focus these activities through an Ocean Life Media Centre that will be the core of engagement activities with the following objectives and activities:
▪ To create content: anchored from Ocean Census Expeditions and from community generated content.
▪ To create awareness of Ocean Census and raise public awareness of the startling diversity of ocean life through:
∘ Distribution of news content via partnership and paid distribution.
∘ To build an online community: through distribution of social media content amplified by paid promotions.
▪ To engage key partners to collaborate with Ocean Census: through successful launch and direct marketing and engagement of key stakeholder groups (scientific, expedition, technology, civil society etc).
▪ To participate on the global policy stage: through Ocean Census leadership (Director, Science Director, Digitisation Lead, Communications Director and Expedition Director) participating at high profile international ocean events as keynote speakers (e.g. UN Oceans, Economist World Ocean Summit).
▪ To build partnerships with the storytelling industry: to harness their skills, resources and investment to tell the story of Ocean Census and ocean life (e.g. broadcast series, immersive installations).
▪ To innovate ocean storytelling during Ocean Census Expeditions.
▪ To inspire and engage young people: initially with live educational events from Ocean Census Expeditions and, as the programme develops, with the production of new ocean life educational resources, freely available for schools, teachers and students with partnerships with professional education companies.
▪ To operationalise and coordinate communications activities across all of Ocean Census – by maximising media and public engagement from visuals and data harvested on Expeditions and developing frictionless, cost-effective workflows, both on expeditions and onshore, with science and data teams.
The content generation strategy builds on the experience of the Nekton Foundation with science expeditions in the western Indian Ocean co-produced with in-country scientists and policymakers. An important element of the outreach for these expeditions has been the participation and storytelling of local scientists, policymakers and members of civil society (see for the Maldives Expedition, 2022: https://nektonmission.org/missions/maldives/the-voice-of-the-maldives).
In 2010, Wheeler asked the intriguing question of what NASA would do if it encountered a planet where only 2 million out of 10 million species were known, and a large number of these species were facing extinction. He concluded that NASA would design a mission to learn as much as possible about life on this hypothetical planet including prioritization of scientific questions, clarifying the goals of the mission and detailing what infrastructure was needed to achieve it, using this as a basis to seek funding (Wheeler, 2010). Proportionately we probably know less about the species in the ocean but it is widely accepted that they are under threat from both local and global anthropogenic stressors (Rogers et al., 2022a). What we have proposed here is the equivalent of a NASA Large Strategic Science Mission (Cangi et al., 2019). These missions typically cost between $1.3 billion (Earth Science Aqua) to $9.2 billion (Hubble Space Telescope; Cangi et al., 2019). Smaller projects in the Discovery and New Frontiers programmes are capped at $450 million and $850 million respectively but these costs exclude launch and post-launch costs (Cangi et al., 2019).
The largest programme to date aiming to document marine biodiversity was the Census of Marine Life (CoML) which ran from 2000 to 2010. CoML was initiated by the Alfred P Sloan Foundation with a grant that amounted to $78 million by the end of the project (https://sloan.org/programs/completed-programs/census-of-marine-life). However, with in kind and other cash contributions from governments, international organisations and maritime industries, the total funding for the programme amounted to approximately $650 million (http://www.coml.org/about-census/). Fourteen Field Projects catalogued about 30 million species distribution records and by the end of the programme 1,200 new species were discovered with a further estimated 5,000 still to be described (http://www.coml.org/about-census/). Whilst this number seems small given the size of the programme it should be considered that CoML was focused on documenting ecosystems and communities as well as their ecology and led to more than 2,600 scientific papers by its completion and also founded OBIS (http://www.coml.org/about-census/).
As with the Census of Marine Life, Ocean Census has been initiated with a multi-year core grant, in this case from The Nippon Foundation. We expect that partner contributions will significantly boost this funding through the provision of infrastructure, equipment and consumables for the programme. Sea time is especially expensive with research vessels costing between $25,000 to $87,500 a day or more excluding fuel costs (Rogers et al., 2021a). Contributions of places on existing research vessels or donation of dedicated vessel time for Ocean Census is therefore a particularly valuable source of funding for the programme. As Ocean Census continues, we anticipate contributions from many partners including governments, foundations, industry, academic institutions and from civil society. However, we regard Ocean Census as a Large-Scale Strategic Science Mission, the largest ever to be undertaken on ocean biodiversity, and as such it is anticipated funding from all sources, including cash and in-kind support will exceed $1 billion. Furthermore, Ocean Census has deliberately been designed to continue beyond the 10 years of planned programme to deliver a permanent legacy in terms of approaches, infrastructure and human capacity to discover, describe and monitor the biodiversity of our largest ecosystem, the ocean.
Ocean Census will become the single largest global programme in history to discover and document ocean life and fulfil a major role in the great scientific challenge to describe and sequence all life on Earth, playing a key role in the protection of the ocean, and thus the Earth and human society that relies on it. Ocean Census will transform our understanding of the diversity and distribution of life in the ocean by accelerating species discovery through four main strategies:
● Digitisation of all data associated with the collection of marine species to promote the use of cybertaxonomy including the development of an intermediate step between specimen collection and description of species termed here the Digital Life Form. The Ocean Census Cyber-Biodiversity System will be an integral part of this digitisation strategy collecting, storing and disseminating species data to appropriate existing global databases, archiving data for which such databases do not exist and converting data to knowledge useful to specific user communities (e.g. scientists, ocean managers, policymakers, industry and civil society).
● The adoption of new technologies and working practices that accelerate the process of species description. This will involve the development of large teams of taxonomists and support specialists (e.g. molecular biologists, imaging specialists) at Biodiversity Centres. Also, the development of networks of taxonomists including both academic institutions and citizen scientists globally working towards the same goal of species discovery and description.
● Increase the global human capacity and infrastructure to discover and protect marine life including through Ocean Census Expeditions and specific training activities.
● Conduct Ocean Census Expeditions targeted at collecting biological specimens (and when feasible related physical and biochemical measurements from poorly investigated regions e.g. areas beyond national jurisdiction, the deep sea) thereby contributing to increased coverage of Essential Ocean Variables, and global biodiversity monitoring efforts.
● Raise and inspire public and partner awareness of ocean life and the programme to accelerate its discovery and Ocean Literacy.
Ocean Census will be organised as an open network of scientists and other specialists focused on the discovery and description of species in the ocean. This will be anchored by Biodiversity Centres and will be accompanied by Virtual Taxonomic Networks which focus on specific taxonomic groups to fulfil the Ocean Census mission. This open network approach will be central to democratisation of marine taxonomy/biodiversity research globally and is supported by the digitisation and data policies and capacity-building activities through Expeditions and Biodiversity Centres. Together these initiatives are essential to maximise human effort on species discovery.
Ocean Census also comprises a major public outreach and education element with storytelling on the ocean focused on scientists, policymakers and citizens living in coastal countries hosting Expeditions. Outreach will be managed by professional communicators and will be distributed through partner and paid organisations to ensure maximum reach and impact amongst audiences at local to global levels.
Initial funding for Ocean Census has been granted by The Nippon Foundation and will be supplemented directly and in-kind through a network of partners. It is a Large-Scale Strategic Science Mission that has been planned for delivery over 10 years, but which is also designed to live beyond this time providing a permanent legacy in acceleration of marine species discovery and biodiversity monitoring. Over the decade of Ocean Census, it is our intention to accelerate species discovery 10-fold and deliver fundamental scientific understanding of the distribution of life in the ocean. This will provide the science required to understand how the ocean is changing and how it can be better managed as a place of importance to the planet and wonder to humans.
This paper was developed through four working groups (Science, Digitisation, Communications and Capacity Development) in two successive workshops with contributions to the discussions, Interim and Feasibility Reports from all authors. Working Groups were led by TO and KO (Science), P-LB and LVW (Digitisation), SM and LH (Communication) and NB and AP (Capacity Development). Workshops were organised and coordinated by ER-L, AR and OS. Writing of this manuscript was led by AR with direct contributions to the text from other authors and content drawn from Interim and Feasibility (Internal) Reports for Ocean Census. OS secured funding to support this work from The Nippon Foundation. All authors contributed to the article and approved the submitted version.
Funding for this study was provided by the Nippon Foundation and administered by the Nekton Foundation.
AR and OS would like to thank The Nippon Foundation for funding the Ocean Census Feasibility Study and for providing constant and stimulating intellectual input and support to the programme development from 2021–2023. Special thanks is also given to the Working Groups on Science, Digitisation and Capacity Development who graciously devoted their time and effort to generation of ideas and concepts for Ocean Census. These are included as authors on the present paper but also include Ward Appeltans from the Ocean Biodiversity Information System and Peter Ng from the Department of Biological Sciences, National University of Singapore. AR would like to give special thanks to Sir Charles Godfray, Dr Patricia Miloslavich, Dr Paul Snelgrove and Dr Jyotika Virmani for providing expert advice through the development of Interim Reports 1 and 2 of Ocean Census. SM and OS would like to thank the following for assistance on development of the education and outreach plans for Ocean Census: Paula Carlson, Emily Caroe, Bill Gardner, Alan Gerschenfeld, James Honeybourne, Dr Alan Jamieson, Lotte Jones, Richard Nockles, Dr Helen Scales, Richard Vevers and Dr Carlie Wiener. AR and ER-L would also like to thank REV Ocean for extended secondment to the Nekton Foundation during the development of Ocean Census.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor JI declared a past co-authorship with the author DZ.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1224471/full#supplementary-material
Ames C. L., Ohdera A. H., Colston S. M., Collins A. G., Fitt W. K., Morandini A. C., et al. (2021). Fieldable environmental DNA sequencing to assess jellyfish biodiversity in nearshore waters of the Florida Keys, United States. Front. Mar. Sci. 8, 640527. doi: 10.3389/fmars.2021.640527
Apothéloz-Perret-Gentil L., Cordonier A., Straub F., Iseli J., Esling P., Pawlowski J. (2017). Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol. Ecol. Resour. 17, 1231–1242. doi: 10.1111/1755-0998.12668
Appeltans W., Ahjong S. T., Anderson G., Angel M. V., Artoiset T., Bailly N., et al. (2012). The magnitude of global marine species diversity. Curr. Biol. 22, 2189–2202. doi: 10.1016/j.cub.2012.09.036
Aylagas E., Borja A., Irigoien X., Rodriguez-Ezpeleta N. (2016). Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment. Front. Mar. Sci. 3, 96. doi: 10.3389/fmars.2016.00096
Baker C. M., Buckman-Young R. S., Costa C. S., Giribet G. (2021). Phylogenomic analysis of velvet worms (Onychophora) uncovers an evolutionary radiation in the neotropics. Mol. Biol. Evol. 38, 5391–5404. doi: 10.1093/molbev/msab251
Baldini R. M., Cota-Sánchez J. H., Aedo C. (2021). Is the demise of plant taxonomy in sight? Maybe yes, maybe no… Webbia. J. Plant Taxon. Geogr. 76, 3–10. doi: 10.36253/jopt-10802
Barber P. H., Ablan-Lagman M. C. A., Ambariyanto, Berlinck R. G. S., Cahyani D., Crandall E. D., et al. (2014). Advancing biodiversity research in developing countries: the need for changing paradigms. Bull. Mar. Sci. 90, 187–210. doi: 10.5343/bms.2012.1108
Barbier E. B. (2017). Marine ecosystem services. Curr. Biol. 27, R507–R510. doi: 10.1016/j.cub.2017.03.020
Bar-On Y. M., Phillips R., Milo R. (2018). The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511. doi: 10.1073/pnas.1711842115
Bax N. J., Appeltans W., Brainard R., Duffy J. E., Dunstan P., Hanich Q., et al. (2018). Linking capacity development to GOOS monitoring networks to achieve sustained ocean observation. Front. Mar. Sci. 5, 346. doi: 10.3389/fmars.2018.00346
Berendsohn W. G., Güntsch A., Hoffmann N., Kohlbecker A., Luther K., Müller A. (2011). Biodiversity information platforms: From standards to interoperability. ZooKeys 150, 71–87. doi: 10.3897/zookeys.150.2166
Blasiak R., Wynberg R., Grorud-Colvert K., Thambisetty S., Bandarra N., Canário A. V. M., et al. (2020). The ocean genome and future prospects for conservation and equity. Nat. Sustain. 3, 588–596. doi: 10.1038/s41893-020-0522-9
Boistel R., Swoger J., Fernandez V., Gillet B., Reynaud E. G. (2011). The future of three-dimensional microscopic imaging in marine biology. Mar. Ecol. 32, 438–452. doi: 10.1111/j.1439-0485.2011.00442.x
Bouchet P., Bary S., Heros V., Marani G. (2016). “How many species of molluscs are there in the world’s oceans, and who is going to describe them?,” in Tropical deep-Sea Benthos 29. Eds. Heros V., Strong E., Bouchet P. (Paris: Museum National d’Histoire Naturelle), 9–24.
Bouchet P., Decock W., Lonneville B., Vanhoorne B., Vandepitte L. (2023). Marine biodiversity discovery: The metrics of new species descriptions. Front. Mar. Sci. 10, 929989. doi: 10.3389/fmars.2023.929989
Brandt A., Gooday A. J., Brandão S. N., Brix S. B., Brökeland W., Cedhagen T., et al. (2007). The Southern Ocean deep sea: first insights into biodiversity and biogeography. Nature 447, 307–311. doi: 10.1038/nature05827
Britz R., Hundsdörfer A., Fritz U. (2020). Funding, training, permits—the three big challenges of taxonomy. Megataxa 1, 49–52. doi: 10.11646/megataxa.1.1.10
Cangi E., Gibson J., Luebbers M. (2019). Mission costs: Past, Present, Future. (Presentation available at Laboratory for Atmospheric and Space Physics, University of Colorado at Boulder). Available at: https://lasp.colorado.edu/mop/files/2019/11/Mission-costs.pdf Last accessed 18/9/2023.
Carroll S. R., Plevel R., Jennings L. L., Garba I., Sterling R., Cordova-Marks F. M., et al. (2022). Extending the CARE Principles from tribal research policies to benefit sharing in genomic research. Front. Genet. 13, 1052620. doi: 10.3389/fgene.2022.1052620
Centurioni L. R., Turton J., Lumpkin R., Braasch L., Brassington G., Chao Y., et al. (2019). Global in situ observations of essential climate and ocean variables at the air–sea interface. Front. Mar. Sci. 6, 419. doi: 10.3389/fmars.2019.00419
Chan W.-P., Childers R. R., Ashe S., Tsai C.-C., Elson C., Keleher K. J., et al. (2022). A high-throughput multispectral imaging system for museum specimens. Commun. Biol. 5, 1318. doi: 10.1038/s42003-022-04282-z
Constable A. J., Costa D. P., Schofield O., Newman L., Urban E. R. Jr., Fulton E. A., et al. (2016). Developing priority variables (“ecosystem Essential Ocean Variables” — eEOVs) for observing dynamics and change in Southern Ocean ecosystems. J. Mar. Syst. 161, 26–41. doi: 10.1016/j.jmarsys.2016.05.003
Cowie R. H., Bouchet P., Fontaine B. (2022). The Sixth Mass Extinction: fact, fiction or speculation? Biol. Rev. 97, 640–663. doi: 10.1111/brv.12816
Danovaro R., Dell'Anno A., Pusceddu A., Gambi C., Heiner I., Kristensen R. M. (2010). The first metazoa living in permanently anoxic conditions. BMC Biol. 8, 30. doi: 10.1186/1741-7007-8-30
Dasgupta P. (2021). The Economics of Biodiversity: The Dasgupta Review (London, U.K.: HM Treasury), 602.
De Pooter D., Appeltans W., Bailly N., Bristol S., Deneudt K., Eliezer M., et al. (2017). Toward a new data standard for combined marine biological and environmental datasets - expanding OBIS beyond species occurrences. Biodivers. Data J. 5, e10989. doi: 10.3897/BDJ.5.e10989
Drerup C., How M. (2021). Polarization contrasts and their effect on the gaze stabilisation of crustaceans. J. Exp. Biol. 224, jeb229898. doi: 10.1242/jeb.229898
Dubois A., Ohler A., Pyron R. A. (2021). New concepts and methods for phylogenetic taxonomy and nomenclature in zoology, exemplified by a new ranked cladonomy of recent amphibians (Lissamphibia). Megataxa 5, 1–738. doi: 10.11646/megataxa.5.1.1
Duffy J. E., Amaral-Zettler L. A., Fautin D. G., Paulay G., Rynearson T. A., Sosik H. M., et al. (2013). Envisioning a marine biodiversity observation network. BioScience 63, 350–361. doi: 10.1525/bio.2013.63.5.8
Dupérré N. (2020). Old and new challenges in taxonomy: what are taxonomists up against? Megataxa 1, 59–62. doi: 10.11646/megataxa.1.1.12
Ebach M. C., Holdrege C. (2005). More taxonomy, not DNA barcoding. Bioscience 55, 822–823. doi: 10.1641/0006-3568(2005)055[0823:MTNDB]2.0.CO;2
Ebach M. C., Valdecasas A. G., Wheeler Q. D. (2011). Impediments to taxonomy and users of taxonomy: accessibility and impact evaluation. Cladistics 27, 550–557. doi: 10.1111/j.1096-0031.2011.00348.x
Edgar G. J., Stuart-Smith R. D., Heather F. J., Barrett N. S., Turak E., Sweatman H., et al. (2023). Continent-wide declines in shallow reef life over a decade of ocean warming. Nature 615, 858–865. doi: 10.1038/s41586-023-05833-y
Engel M. S., Ceríaco L. M. P., Daniel G. M., Dellapé P. M., Löbl I., Marinov M., et al. (2021). The taxonomic impediment: a shortage of taxonomists, not the lack of technical approaches. Zool. J. Linn. Soc 193, 381–387. doi: 10.1093/zoolinnean/zlab072
Estes M. Jr., Anderson C., Appeltans W., Bax N., Bednaršek N., Canonico G., et al. (2021). Enhanced monitoring of life in the sea is a critical component of conservation management and sustainable economic growth. Mar. Policy 132, 104699. doi: 10.1016/j.marpol.2021.104699
Faulwetter S., Dailianis T., Vasileiadou A., Arvanitidis C. (2013a). Contrast enhancing techniques for the application of micro-CT in marine biodiversity studies. Microsc. Anal. Tomography Suppl. 2013, S4–S7. doi: 10.1007/978-3-319-17001-5_53-1
Faulwetter S., Vasileiadou A., Kouratoras M., Dailianis T., Arvanitidis C. (2013b). Micro-computed tomography: introducing new dimensions to taxonomy. Zookeys 263, 1–45. doi: 10.3897/zookeys.263.4261
Fontaine B., Perrard A., Bouchet P. (2012). 21 years of shelf life between discovery and description of new species. Curr. Biol. 22, R943–R944. doi: 10.1016/j.cub.2012.10.029
Gagné T. O., Reygondeau G., Jenkins C. N., Sexton J. O., Bograd S. J., Hazen E. L., et al. (2020). towards a global understanding of the drivers of marine and terrestrial biodiversity. PloS One 15, e0228065. doi: 10.1371/journal.pone.0228065
Gamfeldt L., Lefcheck J. S., Byrnes J. E. K., Cardinale B. J., Duffy J. E. (2015). Marine biodiversity and ecosystem functioning: what's known and what's next? Oikos 124, 252–265. doi: 10.1111/oik.01549
Gerovasileiou V., Smith C. J., Sevastou K., Papadopoulou N., Dailianis T., Bekkby T., et al. (2019). Habitat mapping in the European Seas - is it fit for purpose in the marine restoration agenda? Mar. Policy 106, 103521. doi: 10.1016/j.marpol.2019.103521
Global Ocean Observing System. (2022). Essential Ocean Variables. Available at: https://www.goosocean.org/index.php?option=com_content&view=article&layout=edit&id=283&Itemid=441.
Glover A. G., Wiklund H., Chen C., Dahlgren T. G. (2018). Managing a sustainable deep-sea ‘blue economy’ requires knowledge of what actually lives there. eLife 7, e41319. doi: 10.7554/eLife.41319.005
Godfray H. C. J., Knapp S. (2004). Introduction. Phil. Trans. R. Soc B 359, 559–569. doi: 10.1098/rstb.2003.1457
Gold Z., Sprague J., Kushner D. J., Marin E. Z., Barber P. H. (2021). eDNA metabarcoding as a biomonitoring tool for marine protected areas. PloS One 16, e0238557. doi: 10.1371/journal.pone.0238557
Goodwin M., Halvorsen K. T., Jiao L., Knausgard K. M., Martin A. H., Moyano N., et al. (2022). Unlocking the potential of deep learning for marine ecology: overview, applications, and outlook. ICES J. Mar. Sci. 79, 319–336. doi: 10.1093/icesjms/fsab255
Gutiérrez E. E., Pine R. H. (2017). Specimen collection crucial to taxonomy. Science 355, 1275. doi: 10.1126/science.aan0926
Hamilton C. A., Shockley F. W., Simmons R., Smith A., Ware J., Zaspel J. M. (2021). The future for a prominent taxonomy. Insect Syst. Divers. 5, 1–2. doi: 10.1093/isd/ixaa020
Hardisty A. R., Ellwood E. R., Nelson G., Zimkus B., Buschbom J., Addink W., et al. (2022). Digital Extended Specimens: enabling an extensible network of biodiversity data records as integrated digital objects on the internet. BioScience 72, 978–987. doi: 10.1093/biosci/biac060
Hebert P. D. N., Cywinska A., Ball S. L., deWaard J. R. (2003a). Biological identifications through DNA barcodes. Proc. R. Soc Lond. B 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert P. D. N., Ratnasingham S., deWaard J. R. (2003b). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc Lond B (Suppl), S96–S99. doi: 10.1098/rsbl.2003.0025
Hestetun J. T., Bye-Ingebrigtsen E., Nilsson H., Glover A. G., Johansen P.-O., Thomas G. Dahlgren T. G. (2020). Significant taxon sampling gaps in DNA databases limit the operational use of marine macrofauna metabarcoding. Mar. Biodivers. 50, 70. doi: 10.1007/s12526-020-01093-5
Hetherington E. D., Choy C. A., Thuesen E. V., Haddock S. H. D. (2022). Three distinct views of deep pelagic community composition based on complementary sampling approaches. Front. Mar. Sci. 9, 864004. doi: 10.3389/fmars.2022.864004
Hornidge A.-K., Partelow S., Knopf K. (2023). “Knowing the ocean: epistemic inequalities in patterns of science collaboration,” in Ocean Governance: Knowledge Systems, Policy Foundations and Thematic Analyses. Eds. Partelow S., Hadjimichael M., Hornidge A.-K. (Cham: Springer International Publishing), 25–45.
Horton T., Marsh L., Bett B. J., Gates A. R., Jones D. O. B., Benoist N. M. A., et al. (2021). Recommendations for the standardisation of open taxonomic nomenclature for image-based identifications. Front. Mar. Sci. 8, 620702. doi: 10.3389/fmars.2021.620702
Howell K. L., Davies J. S., Allcock A. L., Braga Henriques A., Buhl-Mortensen P., Carreiro-Silva M., et al. (2019). A framework for the development of a global standardised marine taxon reference image database (SMarTaR-ID) to support image-based analyses. PloS One 14, e0218904. doi: 10.1371/journal.pone.0218904
Hughes A. C., Orr M. C., Ma K., Costello M. J., Waller J., Provoost P., et al. (2021). Sampling biases shape our view of the natural world. Ecography 44, 1–11. doi: 10.1111/ecog.05926
Hunter M. E., Meigs-Friend G., Ferrante J. A., Kamla A. T., Dorazio R. M., Diagne L. K., et al. (2018). Surveys of environmental DNA (eDNA): a new approach to estimate occurrence in Vulnerable manatee populations. Endang. Species Res. 35, 101–111. doi: 10.3354/esr00880
IOC-UNESCO. (2020). Global Ocean Science Report 2020–Charting Capacity for Ocean Sustainability. Ed. Isensee K. (Paris, France: UNESCO Publishing), 245.
IPCC. (2019). “IPCC Special Report on the Ocean and Cryosphere in a Changing Climate,” in International Panel on Climate Change, Geneva, SWITZERLAND. Eds. Pörtner H. O., Roberts D. C., Masson-Delmotte V., Zhai P., Tignor M., Poloczanska E., et al. (Cambridge, U.K.: Cambridge University Press), 755.
Irisson J.-O., Ayata S.-D., Lindsay D. J., Karp-Boss L., Stemmann L. (2022). Machine learning for the study of plankton and marine snow from images. Annu. Rev. Mar. Sci. 14, 277–301. doi: 10.1146/annurev-marine-041921-013023
Isbell F., Balvanera P., Mori A. S., He J.-S., Bullock J. M., Regmi G. R., et al. (2023). Expert perspectives on global biodiversity loss and its drivers and impacts on people. Front. Ecol. Environ. 21, 94–103. doi: 10.1002/fee.2536
IUCN. (2017). Guidelines for Using the IUCN Red List Categories and Criteria. Version 13 (Gland, Switzerland: IUCN, Standards and Petitions Subcommittee), 107.
Jamieson A. (2015). The Hadal Zone: Life in the Deepest Oceans (Cambridge, U.K.: Cambridge University Press), 372.
Jamieson A. J., Lindsay D. J., Kitazato H. (2023). Maximum depth extensions for Hydrozoa, Tunicata and Ctenophora. Mar. Biol. 170, 1–6. doi: 10.1007/s00227-023-04177-5
Jarić I., Correia R. A., Brook B. W., Buettel J. C., Courchamp F., Di Minin E., et al. (2020). iEcology: harnessing large online resources to generate ecological insights. Trends Ecol. Evol. 35, 630–639. doi: 10.1016/j.tree.2020.03.003
Jetz W., McGeoch M. A., Guralnick R., Ferrier S., Beck J., Costello M. J., et al. (2019). Essential biodiversity variables for mapping and monitoring species populations. Nat. Ecol. Evol. 3, 539–551. doi: 10.1038/s41559-019-0826-1
Jo T., Takao K., Minamoto T. (2022). Linking the state of environmental DNA to its application for biomonitoring and stock assessment: Targeting mitochondrial/nuclear genes, and different DNA fragment lengths and particle sizes. Environ. DNA. 4, 271–283. doi: 10.1002/edn3.253
Jones K. R., Klein C. J., Halpern B. S., Venter O., Grantham H., Kuempel C. D., et al. (2018). The location and protection status of Earth’s diminishing marine wilderness. Curr. Biol. 28, 2506–2512. doi: 10.1016/j.cub.2018.06.010
Katija K., Orenstein E., Schlining B., Lundsten L., Barnard K., Sainz G., et al. (2022). FathomNet: A global image database for enabling artificial intelligence in the ocean. Sci. Rep. 12, 15914. doi: 10.1038/s41598-022-19939-2
Knapp S., Boxshall G. A. (2010). Biodiversity and Systematics: how have we fared in the International Year of Biodiversity? Syst. Biodivers. 8, 419–422. doi: 10.1080/14772000.2010.543008
Krehenwinkel H., Pomerantz A., Prost S. (2019). Genetic biomonitoring and biodiversity assessment using portable sequencing technologies: current uses and future directions. Gene 10, 858. doi: 10.3390/genes10110858
Krell F.-T. (2004). Parataxonomy vs. taxonomy in biodiversity studies – pitfalls and applicability of ‘morphospecies’ sorting. Biodivers. Conserv. 13, 795–812. doi: 10.1023/B:BIOC.0000011727.53780.63
Langenkämper D., Zurowietz M., Schoening T., Nattkemper T. W. (2017). BIIGLE 2.0 - Browsing and annotating large marine image collections. Front. Mar. Sci. 4, 83. doi: 10.3389/fmars.2017.00083
Lauretoa L. M. O., Cianciaruso M. V., Samiac D. S. M. (2015). Functional diversity: an overview of its history and applicability. Nat. Conserv. 13, 112–116. doi: 10.1016/j.ncon.2015.11.001
Lawley J. W., Gamero-Mora E., Maronna M. M., Chiaverano L. M., Stampar S. N., Hopcroft R. R., et al. (2022). Morphology is not always useful for diagnosis, and that’s ok: Species hypotheses should not be bound to a class of data. Reply to Brown and Gibbons. S. Afr. J. Sci. 118, 12590. doi: 10.17159/sajs.2022/14495
Leray M., Boehm J. T., Mills S. C., Meyer C. P. (2012). Moorea BIOCODE barcode library as a tool for understanding predator–prey interactions: insights into the diet of common predatory coral reef fishes. Coral Reefs 31, 383–388. doi: 10.1007/s00338-011-0845-0
Leray M., Meyer C. P., Mills S. C. (2015). Metabarcoding dietary analysis of coral dwelling predatory fish demonstrates the minor contribution of coral mutualists to their highly partitioned, generalist diet. Peer J. 3, e1047. doi: 10.7717/peerj.1047
Levin L. A., Bett B. J., Gates A. R., Heimbach P., Howe B. M., Janssen F., et al. (2019). Global observing needs in the deep ocean. Front. Mar. Sci. 6, 241. doi: 10.3389/fmars.2019.00241
Lindstrom E., Gunn J., Fischer A., McCurdy A., Glover L. K., Alverson K., et al. (2012). A Framework for Ocean Observing. Task Team for an Integrated Framework for Sustained Ocean Observing, UNESCO, IOC/INF-1284 rev., United Nations Educational (Paris, France: Scientific and Cultural Organization-Intergovernmental Oceanographic Commission), 25. doi: 10.5270/OceanObs09-FOO
Lotze H. K. (2021). Marine biodiversity conservation. Curr. Biol. 31, R1190–R1195. doi: 10.1016/j.cub.2021.06.084
MacLeod N., Benfield M., Culverhouse P. (2010). Time to automate identification. Nature 467, 154–155. doi: 10.1038/467154a
Maddison D. R., Guralnick R., Hill A., Reysenbach A.-L., McDade L. A. (2012). Ramping up biodiversity discovery via online quantum contributions. Trends Ecol. Evol. 27, 72–77. doi: 10.1016/j.tree.2011.10.010
Mammola S., Fukushima C. S., Biondo G., Bongiorni L., Cianferoni F., Domenici P., et al. (2023). How much biodiversity is concealed in the word ‘biodiversity’? Curr. Biol. 33, R59–R60. doi: 10.1016/j.cub.2022.12.003
Matsumoto G., Raskoff K., Lindsay D. J. (2003). Tiburonia granrojo, a new mesopelagic scyphomedusa from the Pacific Ocean representing the type of a new subfamily (Class Scyphozoa, Order Semaeostomae, Family Ulmaridae, Subfamily Tiburoniinae subfam nov.). Mar. Biol. 143, 73–77. doi: 10.1007/s00227-003-1047-2
May R. M. (1994). Biological diversity: differences between land and sea. Phil. Trans. R. Soc Lond. B 343, 105–111. doi: 10.1098/rstb.1994.0014
Mayer L., Jakobsson M., Allen G., Dorschel B., Falconer R. V., Ferrini V., et al. (2018). The Nippon Foundation—GEBCO Seabed 2030 Project: The quest to see the world’s oceans completely mapped by 2030. Geosciences 8, 63. doi: 10.3390/geosciences8020063
McCauley D. J., Pinsky M. L., Palumbi S. R., Estes J. A., Joyce F. H., Warner R. R. (2015). Marine defaunation: animal loss in the global ocean. Science 347, 1255641. doi: 10.1126/science.1255641
Medina J. J., Maley J. M., Sannapareddy S., Medina N. N., Gilman C. M., McCormack J. E. (2020). A rapid and cost-effective pipeline for digitization of museum specimens with 3D photogrammetry. PloS One 15, e0236417. doi: 10.1371/journal.pone.0236417
Meyer C., Davies N., Meyer J.-Y., Moritz C., Planes S., Roderick G. (2006). “The moorea biocode project,” in Saillan M., Charlat S., Murphy H., Davies N. (Eds.) International Joint Symposium: Tropical Island Ecosystems and Sustainable Development (Moorea. French Polynesia) December 2-7, 2006, Moorea, French Polynesia, Centre de Recherches Insulaires et Observatoire de l’Environnement (CRIOBE) and Richard B. Gump South Pacific Research Station (University of California Berkeley, California, USA, 21st COE Program of University of the Ryukyus: Nishihara, Okinawa, Japan) S1-B, 1.
Midtvedt B., Pineda J., Skärberg F., Olsén E., Bachimanchi H., Wesén E., et al. (2022). Single-shot self-supervised object detection in microscopy. Nat. Commun. 13, 7492. doi: 10.1038/s41467-022-35004-y
Montenegro J., Collins A. G., Hopcroft R. R., Questel J. M., Thuesen E. V., Bachtel T. S., et al. (2023). Heterogeneity in diagnostic characters across ecoregions: A case study with Botrynema (Hydrozoa: Trachylina: Halicreatidae). Front. Mar. Sci. 9, 1101699. doi: 10.3389/fmars.2022.1101699
Mora C., Tittensor D. P., Adl S., Simpson A. G. B., Worm B. (2011). How many species are there on Earth and in the ocean? PloS Biol. 9, e1001127. doi: 10.1371/journal.pbio.1001127
Moranta J., Torres C., Murray I., Hidalgo M., Hinz H., Gouraguine A. (2022). Transcending capitalism growth strategies for biodiversity conservation. Conserv. Biol. 36, e13821. doi: 10.1111/cobi.13821
Muller-Karger F. E., Kavanaugh M. T., Montes E., Balch W. M., Breitbart M., Chavez F. P., et al. (2014). A framework for a marine biodiversity observing network within changing continental shelf seascapes. Oceanography 27, 18–23. doi: 10.5670/oceanog.2014.56
Muller-Karger F. E., Miloslavich P., Bax N. J., Simmons S., Costello M. J., Pinto I. S., et al. (2018). Advancing marine biological observations and data requirements of the complementary Essential Ocean Variables (EOVs) and Essential Biodiversity Variables (EBVs) frameworks. Front. Mar. Sci. 5, 211. doi: 10.3389/fmars.2018.00211
Nguyen C. V., Lovell D. R., Adcock M., La Salle J. (2014). Capturing natural-colour 3D models of insects for species discovery and diagnostics. PloS One 9, e94346. doi: 10.1371/journal.pone.0094346
Nippon Foundation (2022). Available at: https://www.nippon-foundation.or.jp/en/news/articles/2022/20220715-75332.html (Accessed 19/4/2023).
Pacoureau N., Rigby C. L., Kyne P. M., Sherley R. B., Winker H., Carlson J. K., et al. (2021). Half a century of global decline in oceanic sharks and rays. Nature 589, 567–571. doi: 10.1038/s41586-020-03173-9
Paknia O., Rajaei H., Koch A. (2015). Lack of well-maintained natural history collections and taxonomists in megadiverse developing countries hampers global biodiversity exploration. Org. Divers. Evol. 15, 619–629. doi: 10.1007/s13127-015-0202-1
Pavoine S., Bonsall M. B. (2011). Measuring biodiversity to explain community assembly: a unified approach. Biol. Rev. 86, 792–812. doi: 10.1111/j.1469-185X.2010.00171.x
Pereira H. M., Ferrier S., Walters M., Geller G. N., Jongman R. H., Scholes R. J., et al. (2013). Essential biodiversity variables. Science 339, 277–278. doi: 10.1126/science.1229931
Perrault A. (2004). Facilitating prior informed consent context of genetic resources and traditional knowledge. Sustain. Dev. Law Pol. 4, 21–26. Available at: https://digitalcommons.wcl.american.edu/sdlp/vol4/iss2/7/.
Pinto Â.P., Mejdalani G., Mounce R., Silveira L. F., Marinoni L., Rafael J. A. (2021). Are publications on zoological taxonomy under attack? R. Soc Open Sci. 8, 201617. doi: 10.1098/rsos.201617
Prathapan K. D., Rajan P. D. (2020). Advancing taxonomy in the global south and completing the grand Linnaean enterprise. Megataxa 1, 73–77. doi: 10.11646/megataxa.1.1.15
Rabone M., Harden-Davies H., Collins J. E., Zajderman S., Appeltans W., Droege G., et al. (2019). Access to marine genetic resources (MGR): raising awareness of best-practice through a new agreement for biodiversity beyond national jurisdiction (BBNJ). Front. Mar. Sci. 6, 520. doi: 10.3389/fmars.2019.00520
Rabone M., Horton T., Jones D. O. B., Simon-Lledó E., Glover A. G. (2023). A review of the International Seabed Authority database DeepData from a biological perspective: challenges and opportunities in the UN Ocean Decade. Database 2023, 1–17. doi: 10.1093/database/baad013
Rajmohana K., Bijoy C. (2012). “Cybertaxonomy: a novel tool in biodiversity science,” in Biodiversity: Utilization, Threats and Cultural Linkages. Eds. Kumar A. B., Nayar M. P., Varma R. V., Peethambaran C. K. (Delhi, India: Narendra Publishing House), 55–64.
Ramirez-Llodra E., Argentino C., Baker M., Boetius A., Costa C., Dahle H., et al. (2022). Hot vents beneath an icy ocean: The Aurora Vent Field, Gakkel Ridge, revealed. Oceanography 36, 6–17. doi: 10.5670/oceanog.2023.103
Ramirez-Llodra E., Tyler P. A., Baker M. C., Bergstad O. A., Clark M. R., Escobar E., et al. (2011). Man and the last great wilderness: human impact on the deep sea. PloS One 6, e22588. doi: 10.1371/journal.pone.0022588
Raskoff K. A., Sommer F. A., Hamner W. M., Cross K. M. (2003). Collection and culture techniques for gelatinous zooplankton. Biol. Bull. 204, 68–80. doi: 10.2307/1543497
Riedel A., Narakusumo R. P. (2019). One hundred and three new species of Trigonopterus weevils from Sulawesi. ZooKeys 828, 1–153. doi: 10.3897/zookeys.828.32200
Riedel A., Sagata K., Suhardjono Y. R., Tänzler R., Balke M. (2013). Integrative taxonomy on the fast track—towards more sustainability in biodiversity research. Front. Zool. 10, 15. doi: 10.1186/1742-9994-10-15
Robison B. H., Reisenbichler K. R., Sherlock R. E. (2017). The coevolution of midwater research and ROV technology at MBARI. Oceanography 30, 26–37. doi: 10.5670/oceanog.2017.421
Rogers A. D. (2015). Environmental change in the deep ocean. Annu. Rev. Environ. Res. 40, 1–38. doi: 10.1146/annurev-environ-102014-021415
Rogers A., Aburto-Oropeza O., Appeltans W., Assis J., Balance L. T., Cury P., et al. (2020). “Blue Paper 10: Critical Habitats and Biodiversity: Inventory, Thresholds, and Governance,” in Report prepared for the (Norwegian) Prime Minister’s High Level Panel on a Sustainable Ocean Economy (Washington DC, USA: World Resources Institute), 85. Available at: https://www.oceanpanel.org/blue-papers/critical-habitats-and-biodiversity-inventory-thresholds-and-governance.
Rogers A. D., Appeltans W., Assis J., Balance L. T., Cury P., Duarte C., et al. (2022a). Discovering marine biodiversity in the 21st Century. Adv. Mar. Biol. 93, 23–115. doi: 10.1016/bs.amb.2022.09.002
Rogers A. D., Baco-Taylor A., Currie D., Escobar-Briones E., Gjerde K., Gobin J., et al. (2021a). Marine genetic resources in areas beyond national jurisdiction: promoting marine scientific research and enabling equitable benefit sharing. Front. Mar. Sci. 8, 667274. doi: 10.3389/fmars.2021.667274
Rogers A. D., Bax N., Buttigieg P. L., Guilhon M., Hynes L., MacIntyre S., et al. (2021c). Project Nautilus Interim Report 2 (Begbroke, Oxfordshire, U.K: Nekton Foundation), 168.
Rogers A. D., Bax N., Buttigieg P. L., Guilhon M., Hynes L., MacIntyre S., et al. (2022b). Project Nautilus Feasibility Report (Begbroke, Oxfordshire, U.K: Nekton Foundation), 478.
Rogers A. D., Bax N., Buttigieg P. L., Hynes L., MacIntyre S., O'Hara T., et al. (2021b). Project Nautilus Interim Report 1 (Begbroke, Oxfordshire, U.K: Nekton Foundation), 42. 63pp Appendices.
Sanchez L., Boulanger E., Arnal V., Boissery P., Dalongeville A., Dejean T., et al. (2022). Ecological indicators based on quantitative eDNA metabarcoding: the case of marine reserves. Ecol. Indic. 140, 108966. doi: 10.1016/j.ecolind.2022.108966
Sands C. J., O’Hara T. D., Martín-Ledo R. (2021). Pragmatic assignment of species groups based on primary species hypotheses: the case of a dominant component of the Southern Ocean benthic fauna. Front. Mar. Sci. 8, 723328. doi: 10.3389/fmars.2021.723328
Saunders T. E. (2020). Taxonomy at a crossroads: communicating value, building capability, and seizing opportunities for the future. Megataxa 1, 63–66. doi: 10.11646/megataxa.1.1.13
Scheffers B. R., Joppa L. N., Pimm S. L., Laurance W. F. (2012). What we know and don’t know about Earth’s missing biodiversity. Trends Ecol. Evol. 27, 501–510. doi: 10.1016/j.tree.2012.05.008
Schroeder S.-M., Kiko R., Koch R. (2020). MorphoCluster: efficient annotation of plankton images by clustering. Sensors 20, 3060. doi: 10.3390/s20113060
Shelton A. O., Kelly R. P., O'Donnell J. L., Parka L., Schwenk P., Greene C., et al. (2019). Environmental DNA provides quantitative estimates of a threatened salmon species. Biol. Conserv. 237, 383–391. doi: 10.1016/j.biocon.2019.07.003
Sigovini M., Keppel E., Tagliapietra D. (2016). Open Nomenclature in the biodiversity era. Methods Ecol. Evol. 7, 1217–1225. doi: 10.1111/2041-210X.12594
Sigwart J. D., Wong N. L. W. S., Esa Y. (2021). Global controversy in oyster systematics and a newly described species from SE Asia (Bivalvia: Ostreidae: Crassostreinae). Mar. Biodivers. 51, 83. doi: 10.1007/s12526-021-01203-x
Sloyan B. M., Roughan M., Hill K. (2018). “The global ocean observing system,” in New Frontiers in Operational Oceanography. Eds. Chassignet E., Pascual A., Tintoré J., Verron J. (Tallahassee, Florida, USA: GODAE OceanView), 75–90. doi: 10.17125/gov2018.ch03
Smale D. A., Wernberg T., Oliver E. C. J., Thomsen M., Harvey B. P., Straub S. C., et al. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Clim. Change 9, 306–312. doi: 10.1038/s41558-019-0412-1
Srivathsan A., Hartop E., Puniamoorthy J., Lee W. T., Kutty S. N., Kurina O., et al. (2019). Rapid, large-scale species discovery in hyperdiverse taxa using 1D MinION sequencing. BMC Biol. 17, 96. doi: 10.1186/s12915-019-0706-9
Stringham O. C., Toomes A., Kanishka A. M., Mitchell L., Heinrich S., Ross J. V., et al. (2020). A guide to using the internet to monitor and quantify the wildlife trade. Conserv. Biol. 35, 1130–1139. doi: 10.1111/cobi.13675
Tashiro T., Ishida A., Hori M., Igisu M., Koike M., Méjean P., et al. (2017). Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 549, 516–518. doi: 10.1038/nature24019
Teoh Z. E., Phillips B. T., Becker K. P., Whittredge G., Weaver J. C., Hoberman C., et al. (2018). Rotary-actuated folding polyhedrons for midwater investigation of delicate marine organisms. Sci. Robot 3, eaat5276. doi: 10.1126/scirobotics.aat5276
Thomsen P. F., Møller P. R., Sigsgaard E. E., Knudsen S. W., Jørgensen O. A., Willerslev E. (2016). Environmental DNA from seawater samples correlate with trawl catches of Subarctic, deep-water fishes. PloS One 11, e0165252. doi: 10.1371/journal.pone.0165252
Thurber A. R., Sweetman A. K., Narayanaswamy B. E., Jones D. O. B., Ingels J., Hansman R. L. (2014). Ecosystem function and services provided by the deep sea. Biogeosciences 11, 3941–3963. doi: 10.5194/bg-11-3941-2014
Titley M. A., Snaddon J. L., Turner E. C. (2017). Scientific research on animal biodiversity is systematically biased towards vertebrates and temperate regions. PloS One 12, e0189577. doi: 10.1371/journal.pone.0189577
Tolochko P., Vadrot A. B. M. (2021). The usual suspects? Distribution of collaboration capital in marine biodiversity research. Mar. Policy 124, 104318. doi: 10.1016/j.marpol.2020.104318
Troudet J., Grandcolas P., Blin A., Vignes-Lebbe R., Legendre F. (2017). Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 7, 9132. doi: 10.1038/s41598-017-09084-6
Van Dover C. L. (2000). The Ecology of Deep-Sea Hydrothermal Vents (Princeton, New Jersey, U.S.A.: Princeton University Press), 448.
Vasil Eva E. D. (2007). Seahorse species (genus Hippocampus, Pisces) described by C. Linné. Folia Zool. 56, 319–327.
Vences M. (2020). The promise of next-generation taxonomy. Megataxa 1, 35–38. doi: 10.11646/megataxa.1.1.6
Verhaegen G., Cimoli E., Lindsay D. J. (2021). Life beneath the ice: jellyfish and ctenophores from the Ross Sea, Antarctica, with an image-based training set for machine learning. Biodivers. Data J. 9, e69374. doi: 10.3897/BDJ.9.e69374
Watsa M., Erkenswick G. A., Pomerantz A., Prost S. (2020). Portable sequencing as a teaching tool in conservation and biodiversity research. PloS Biol. 18, e3000667. doi: 10.1371/journal.pbio.3000667
Webb T. J., Mindel B. L. (2015). Global patterns of extinction risk in marine and non-marine systems. Curr. Biol. 25, 506–511. doi: 10.1016/j.cub.2014.12.023
Webb T. J., Vanden Berghe E., O’Dor R. (2010). biodiversity’s big wet secret: the global distribution of marine biological records reveals chronic under-exploration of the deep pelagic ocean. PloS One 5, e10223. doi: 10.1371/journal.pone.0010223
Wheeler Q. (2010). What would NASA do? Mission-critical infrastructure for species exploration. Syst. Biodivers. 8, 11–15. doi: 10.1080/14772001003628075
Wheeler Q. (2018). Blank canvas: The case for descriptive taxonomy. Integr. Comp. Biol. 58, 1118–1121. doi: 10.1093/icb/icy067
Wieczorek J., Bloom D., Guralnick R., Blum S., Döring M., Giovanni R., et al. (2012). Darwin core: an evolving community-developed biodiversity data standard. PloS One 7, e29715. doi: 10.1371/journal.pone.0029715
Wilkinson M. D., Dumontier M., Aalbersberg I. J., Appleton G., Axton M., Baak A., et al. (2016b). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 1–9. doi: 10.1038/sdata.2016.18
Wilkinson C., Salvat B., Eakin C. M., Brathwaite A., Francini-Filho R., Webster N., et al. (2016a). “Tropical and sub-tropical coral reefs,” in The First Global Integrated Marine Assessment. World Ocean Assessment I. Eds. Iniss L., Simcock A., Ajawin A. Y., et al. (New York, U.S.A: United Nations Division for Ocean Affairs and Law of the Sea), 42.
Williams S. T., Kano Y., Warén A., Herbert D. G. (2020). Marrying molecules and morphology: first steps towards a re-evaluation of solariellid (Gastropoda, Trochoidea) genera in the light of molecular phylogenetic studies. J. Molluscan Stud. 86, 1–26. doi: 10.1093/mollus/eyz038
Woodall L. C., Andradi-Brown D. A., Brierley A. S., Clark M. R., Connelly D., Hall R. A., et al. (2018). A multidisciplinary approach for generating globally comparable data on mesophotic, deep-pelagic and bathyal biological communities. Oceanography. 31, 76–89. doi: 10.5670/oceanog.2018.301
Woodall L. C., Talma S., Steeds O., Stefanoudis P., Jeremie-Muzungaile M.-M., de Comarmond A. (2021). Co-development, coproduction and co-dissemination of scientific research: a case study to demonstrate mutual benefits. Biol. Lett. 17, 20200699. doi: 10.1098/rsbl.2020.0699
Worm B., Barbier E. B., Beaumont N., Duffy J. E., Folke C., Halpern B. S., et al. (2006). Impacts of biodiversity loss on ocean ecosystem services. Science 314, 787–790. doi: 10.1126/science.1132294
WoRMS. (2023). World Register of Marine Species: Statistics. Available at: https://www.marinespecies.org/aphia.php?p=stats (Accessed 19/4/2023).
Keywords: biodiversity, integrated taxonomy, biodiversity crisis, capacity development, ocean literacy, species discovery, DNA barcoding, cybertaxonomy
Citation: Rogers AD, Appiah-Madson H, Ardron JA, Bax NJ, Bhadury P, Brandt A, Buttigieg P-L, De Clerck O, Delgado C, Distel DL, Glover A, Gobin J, Guilhon M, Hampton S, Harden-Davies H, Hebert P, Hynes L, Lowe M, MacIntyre S, Madduppa H, Mazzuco ACdA, McCallum A, McOwen C, Nattkemper TW, Odido M, O’Hara T, Osborn K, Pouponneau A, Provoost P, Rabone M, Ramirez-Llodra E, Scott L, Sink KJ, Turk D, Watanabe HK, Weatherdon LV, Wernberg T, Williams S, Woodall L, Wright DJ, Zeppilli D and Steeds O (2023) Accelerating ocean species discovery and laying the foundations for the future of marine biodiversity research and monitoring. Front. Mar. Sci. 10:1224471. doi: 10.3389/fmars.2023.1224471
Received: 17 May 2023; Accepted: 07 September 2023;
Published: 27 September 2023.
Edited by:
Jeroen Ingels, Florida State University, United StatesReviewed by:
Maikon Di Domenico, Federal University of Paraná, BrazilCopyright © 2023 Rogers, Appiah-Madson, Ardron, Bax, Bhadury, Brandt, Buttigieg, De Clerck, Delgado, Distel, Glover, Gobin, Guilhon, Hampton, Harden-Davies, Hebert, Hynes, Lowe, MacIntyre, Madduppa, Mazzuco, McCallum, McOwen, Nattkemper, Odido, O’Hara, Osborn, Pouponneau, Provoost, Rabone, Ramirez-Llodra, Scott, Sink, Turk, Watanabe, Weatherdon, Wernberg, Williams, Woodall, Wright, Zeppilli and Steeds. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alex David Rogers, YWxleC5yb2dlcnNAcmV2b2NlYW4ub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.