
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 11 August 2023
Sec. Marine Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1195253
 Zhidong Zhang1
Zhidong Zhang1 Xuefeng Sun1,2
Xuefeng Sun1,2 Aihua Chen1
Aihua Chen1 Jiaxin Yang2
Jiaxin Yang2 Suhua Chen1
Suhua Chen1 Yu Zhang1
Yu Zhang1 Yi Cao1
Yi Cao1 Yuheng Peng1
Yuheng Peng1 Yanqing Zhu1,2
Yanqing Zhu1,2 Yangping Wu1*
Yangping Wu1*For the razor clam Solen grandis, siphonal autotomy and regeneration is a necessary means of facing external stress, which is accompanied by a large amount of energy consumption. To explore the function of the ENO4 gene in the siphonal autotomy and regeneration of S. grandis, the full-length cDNA of the ENO4 in S. grandis was cloned and characterized. The full-length cDNA of SgENO4 was 2,773 base pairs (bp), with an 1,851 bp coding sequence (CDS) encoding 616 amino acids. Bioinformatic analysis revealed a slightly lower codon usage bias and a relatively stable nucleotide composition in the SgENO4 sequence. Three conserved motifs were found from the SgENO4 sequence. These conserved motifs vary in length from 14 to 27 amino acids. Moreover, in the siphonal pre-autotomy of razor clams, expression level of the SgENO4 gene was significantly higher in the hepatopancreas and siphonal base than other tissues (P < 0.05). However, the expression of the SgENO4 gene in the siphonal base decreased significantly in the siphonal post-autotomy 7 hours and gradually recovered as the regeneration process continued. Linear correlation analysis showed that there was a significant linear correlation between specific weight of siphon and relative expression levels of SgENO4 (R2 = 0.826). RNA interference (RNAi) result showed that the specific weight (the proportion of siphonal weight to body weight) is significantly lower than other groups on the seventh day (P < 0.05), suggesting that interfering with the expression of the SgENO4 gene could inhibit the siphonal regeneration of S. grandis. These results further demonstrate that SgENO4 is a conserved sequence and that it plays a crucial role in the siphonal autotomy and regeneration of S. grandis, advancing further understanding in exploring molecular mechanisms in the siphonal autotomy and regeneration.
Solen grandis, commonly known as the razor clam, inhabits the intertidal zone at depths of 20 - 40 cm and is naturally distributed along the west coast of the Pacific Ocean (Yuan et al., 2010). In the Chinese seafood market, S. grandis is one of the most expensive (100 CNY/kg) shellfish because of its rich and flavorful meat (Yuan et al., 2012). However, during aquaculture, transport and sales, S. grandis is highly sensitive to external stimuli such as tides and the agitation of symbiotic organisms, collisions, and shaking, causing the razor clam siphon to be prone to autotomy (Wu et al., 2022b). Similar to spiders, geckos, and crabs, S. grandis is able to perform autotomy of its appendage (siphon) and hence, it avoids being preyed (Mykles, 2001; Wrinn and Uetz, 2007; Kaczmarski et al., 2020; Wu et al., 2022b). After siphonal autotomy, razor clams are weak and may even die, unable to dive deep into the sand, which has serious effects on survival, feeding, and digestion (Wu et al., 2022b). Fortunately, autotomized appendages may be replaced by regeneration in S. grandis.
Regeneration is a process in which the whole or organ of an organism is damaged by external forces and part of it is lost, and on the basis of the remaining part, a structure identical in form and function to the lost part is grown (Barr et al., 2020). The ability of siphonal regeneration in clams may be due to the successful wound healing (Wang et al., 2021). Wound healing is necessarily accompanied by stress, immunity and cell proliferation, which need consume a large amount of energy (Wang et al., 2021). Although some progresses have been made in these areas, the mechanism of regeneration in mollusks remains largely unclear (Bely et al., 2014).
As the rate-limiting enzyme in the glycolysis process, enolase can catalyze the conversion of glycerol phosphate (PGA) to phosphoenolpyruvate (PEP), and can also catalyze its reverse reaction process (Feo et al., 2000; Sun et al., 2022). As a result, enolases play an important role in the energy metabolism of cells. Once, enolase has long been regarded as a single functional and evolutionarily conserved ancient enzyme (Straeten et al., 1991). However, A growing body of research has demonstrated that enolase belongs to a new family of proteins called part-time functional proteins, which have a variety of functions related to cell pathophysiology and other activities, such as involvement in cell apoptosis, inflammatory responses and bacterial infection (Yu et al., 2012; Luo et al., 2021). For instance, enolase gene is a key gene related to the growth of Litopenaeus vannamei (Li et al., 2011). The expression and distribution of enolase in Helicoverpa armigera is related to the pathological environment of the cell and the degree of vigorous metabolism. Enolase may play a role in the body’s energy sources in life activities such as worm growth and development and nutrient metabolism (Hu, 2017). Regeneration, therefore, as a particular form of growth, and we doubt whether enolase plays any role in the siphonal regeneration of S. grandis. But to the best of our knowledge, no studies have investigated the molecular properties and function of enolase in S. grandis.
In this study, we investigated the ENO4 gene in S. grandis by cloning its full-length cDNAs and examining its expression levels during siphonal autotomy and regeneration in adult tissue. Then, we compared siphonal growth rate (weight) and gene expression levels of the clam under developmental stage of siphon. The results of this study will improve our understanding of the function of SgENO4 and the mechanism of siphonal regeneration, and can better guide aquaculture efforts for S. grandis.
The razor clams S. grandis were collected from the Jiangsu fine breeding farm of Solen grandis (Nantong, China). Adult clams (2-year age) were selected in July with shell length of 105.62 ± 2.60 mm for following experiments. Before the experiment, all the razor clams were temporarily kept in the tank for two days at a salinity of 22~23‰, a pH of 8.1-8.3, and a temperature of 26~29°C.
Analytical methods for total RNA isolation, cDNA synthesis, and full-length cloning referred to the methods published in previous studies (Chen et al., 2017; Wu et al., 2022a). The 5’ RACE and 3 ‘RACE specific primers were designed on the known intermediate fragments (Table 1). Real-time fluorescent quantitative PCR was used to analyze the expression levels of SgENO4 gene (Applied Biosystems, USA). The amplification system was configured according to the instruction of Tiangen fluorescence quantitative premix kit eferred to the methods published in previous studies. The β-actin gene was used as an internal control gene (Table 1). Dissociation curves of amplified products were analyzed at the end of each PCR analysis (Chen et al., 2017; Wu et al., 2022a).
After the sequence splicing, the sequence analysis was performed with slight modifications as published in previous studies (Chen S. et al., 2017; Wu et al., 2022a; Yan et al., 2022; Zhang et al., 2022). Multiple expectation maximization for motif elicitation (MEME) program in TBtools software (version 1.09) was used to analyze the conserved motifs of the ENO4 genes (Chen et al., 2020; Yu et al., 2022). Nucleotide composition and synonymous codons were analyzed using Codon W software (version 1.4.2) (Chen Y. et al., 2017; Hou, 2020).
Additionally, a total of 80 razor clams were kept in four polyethylene tanks (diameter = 100 cm, height 80 cm, each), and then induced to remove siphons. Prior to culture trials, all prepared aquariums were cleaned, filled with sand-filtered seawater, and laid with 20 cm thick soil. During the rearing period, clam diet was established utilizing a mixture of three microalgae (50 Isochrysis galbana cell per μL, 25 Pavlova lutheri cell per μL and 25 Chaetoceros calcitrans cell per μL) twice a day (Trigos et al., 2018). The rearing water was changed regularly and removed dead individuals in a timely manner. Five razor clams were randomly selected to measure live body weight and siphonal weight in pre-autotomy, 7h, 7d, 14d and 21d after autotomy, respectively, and the liver, gill, foot, gonad and siphonal base tissues were dissected into sterile tubes, respectively(1.5mL). Each sample was washed with 0.01 M phosphate buffered saline, immediately frozen in liquid nitrogen, and stored at -80°C until and qRT-PCR.
Razor clams were removed siphon and reared in the polyethylene tanks (blank control group). 0.9 mL depc-water was injected into the siphonal roots of S. grandis by micro syringe (depc-water control group). Similarly, 0.9 mL depc-water with 1 μg dsSgENO4 (experimental group) and nonsense dsSgENO4 (negative control group) were injected into the siphonal base of S. grandis, respectively. After successful injection, three randomly selected razor clams were taken on days 1, 3, 7, and 10 to record and assay siphonal weight and relative expression levels of SgENO4.
The relative copy number of fluorescence quantitative data was calculated according to 2–ΔΔCt (Livak and Schmittgen, 2002). In order to detect statistically significant differences, all quantitative data were subjected to ANOVA after homogeneity and normality tests using SPSS software (version 20.0). It was used to determine significant differences among means (P < 0.05) by Duncan’ s multiple range test.
The full-length cDNA sequence of SgENO4 gene was cloned using RACE. SgENO4 was 2,773 bp long (Genbank accession number: ON526756.1), which consist of a 244 bp 5’- untranslated region (UTR), a 678 bp 3’- untranslated region (UTR) and an 1,851 bp coding sequence (CDS). The CDS encoded 616 amino acids with a predicted molecular weight and theoretical pI of 117.47 kDa and 5.03, respectively. The SMART program results showed that SgENO4 gene possessed N-terminal and C- terminal TIM bucket domain of enolase (Figure 1). The predicted secondary and tertiary structures of the SgENO4 protein is shown in Figures 2A, B. SgENO4 protein consisted of 22 α-helixes; 13 β-strands; and 34 coils, respectively.
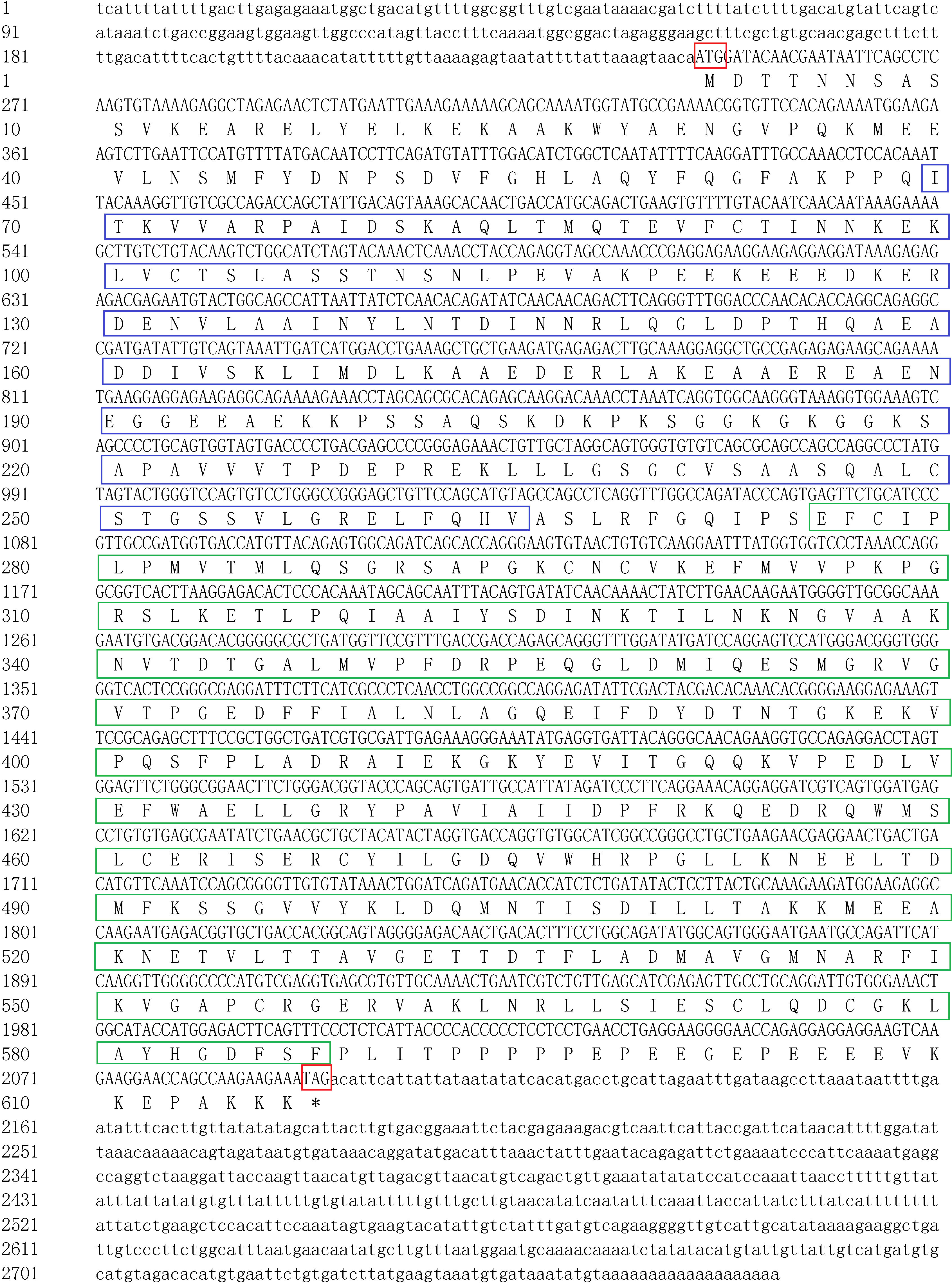
Figure 1 Nucleotide and predicted protein sequence of SgENO4. The uppercase letters in cDNA represent the coding sequence (CDS), and the lowercase letters represent the 5′ and 3′ noncoding regions. The start (ATG) and stop (TGA) codons are presented in the red box. The blue box represents the N-terminal domain of enolase, and the green box represents the C-terminal TIM bucket domain.
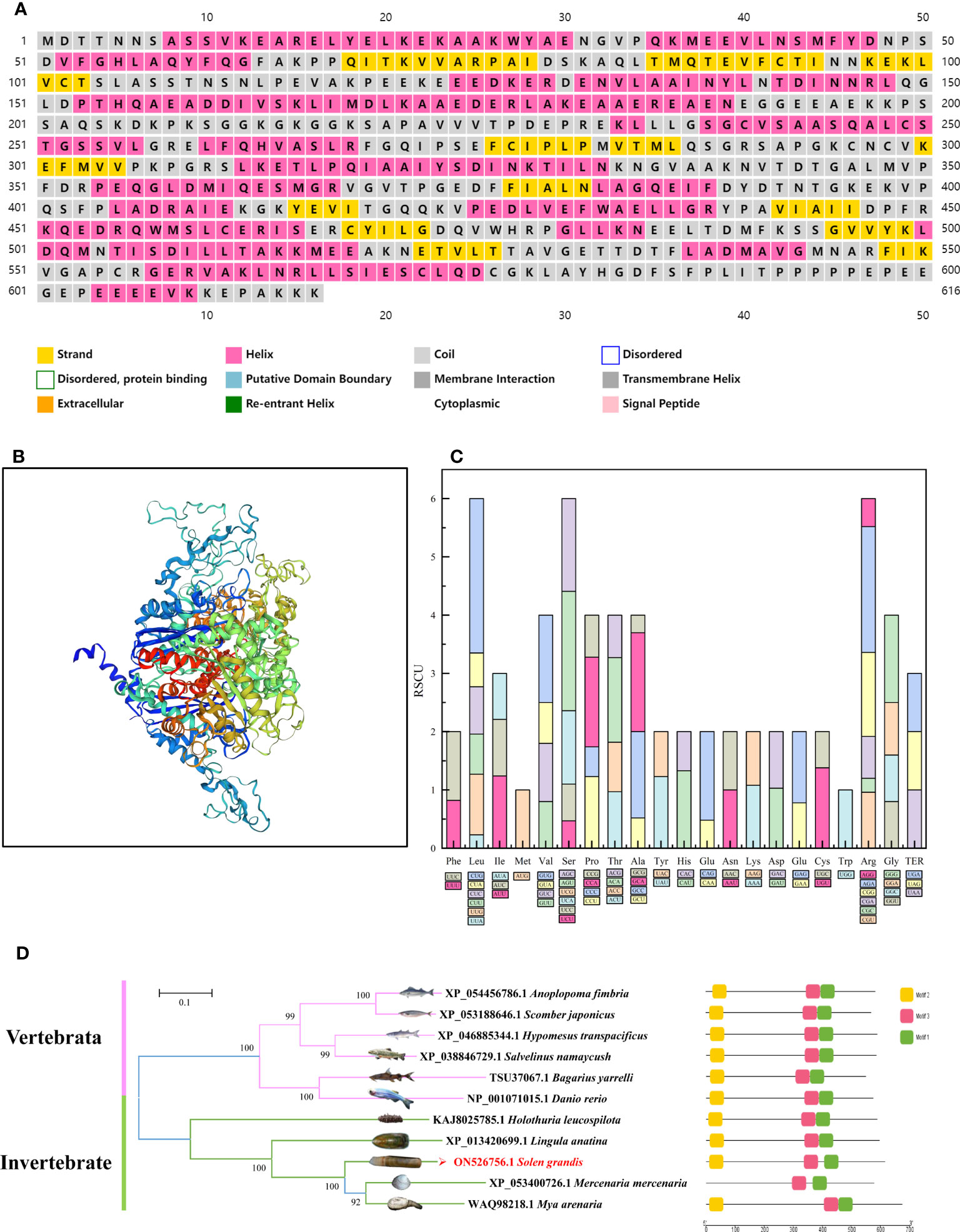
Figure 2 Bioinformatic analysis based on the ENO4 sequences. Predicted secondary structure of SgENO4 protein (A) Predicted tertiary structure of SgENO4 protein (B) Codon distribution in SgENO4 (C) motif analysis of ENO4 (D). All motifs were identified by the MEME database with complete amino acid sequence of ENO4. Lengths of motifs for each ENO4 protein are displayed proportionally.
Nucleotide composition analysis revealed that SgENO4 had the highest compositional value of U% (31.66), which was followed by A% (25.50) and C% (24.16), the lowest compositional value of G% (18.68). It appears that there may be more usage bias of U nucleotides among SgENO4 codons. At the third codon position (A3, U3, G3, C3, GC3), C3% (36.58) was the highest, followed by U3% (32.46), and similar A3% (30.64) and G3% (29.15). Interestingly, mostly over-represented and under-represented codons are C-ended and G-ended, respectively, which also reveals that the preferred codons are influenced by compositional constraints. However, GC% and GC3% composition is 49.0% and 50.1%, respectively, suggesting that there is slightly lower codons usage bias in the SgENO4 sequence.
To explore the extent of the codons usage bias, the relative synonymous codon usage (RSCU) values of the overall 64 codons, including 59 sense codons, 3 termination codons (TER) and 2 no-bias codons (AUG and UGG), were calculated (Figure 2C). The results showed that the most frequently used codon is GAG (Glu, 39 times), and the least frequently used codon is UCG (Ser, 0 times). Among the 23 most abundantly used codons (RSCU >1), 10 codons are G/C-ended and 13 codons are A/U-ended, which evidently confirmed that there exists low codon usage bias in the SgENO4 sequence. Furthermore, the effective number of codons (ENC) was also used to quantify the absolute codon usage bias of SgENO4 sequence. The value of ENC is 54.36, which reveals a relatively stable nucleotide composition and slightly lower (ENC>40) codon usage bias.
The predicted SgENO4 protein shares a sequence identity with Mya arenaria (54.66% similarity), Mercenaria mercenaria (52.47% similarity), Lingula anatina (50.51% similarity) (Figure 3). A phylogenetic tree was formed using ENO4 protein sequences and other species (Figure 2D). The phylogenetic tree presents that SgENO4 have a close evolutionary relationship with Mya arenaria, Mercenaria mercenaria and Lingula anatina, while are far away from vertebrate ENO4 members, providing firm phylogenetic evidence for the identities of the Solen grandis ENO4 gene. Then, all motifs were identified by the MEME database with complete amino acid sequence of ENO4. The differentiation of the ENO4 protein across species is not obvious, and three conserved motifs were found (Figure 2D). The length of these conserved motifs varied from 14 to 27 amino acids (Table 2).

Figure 3 The multiple sequence alignment of SgENO4 with other species. The different colors indicate the conserved properties of the amino acid sequence, with black being the highest, followed by pink and blue being the lowest.
Our results indicate that SgENO4 is ubiquitously expressed in all tissues and stages of siphonal autotomy and regeneration (Figure 4). The relative expression level of SgENO4 in the hepatopancreas is higher than other tissues in the stage of siphonal pre-autotomy, followed by siphonal base (P<0.05). However, the expression levels of SgENO4 in the gill, gonad, and foot are low. Moreover, with tissue regeneration in the siphonal post-autotomy, the relative expression level of SgENO4 in the siphonal base gradually increases to the maximum (on the 21st day) and is significantly higher than that in other tissues (P<0.05).
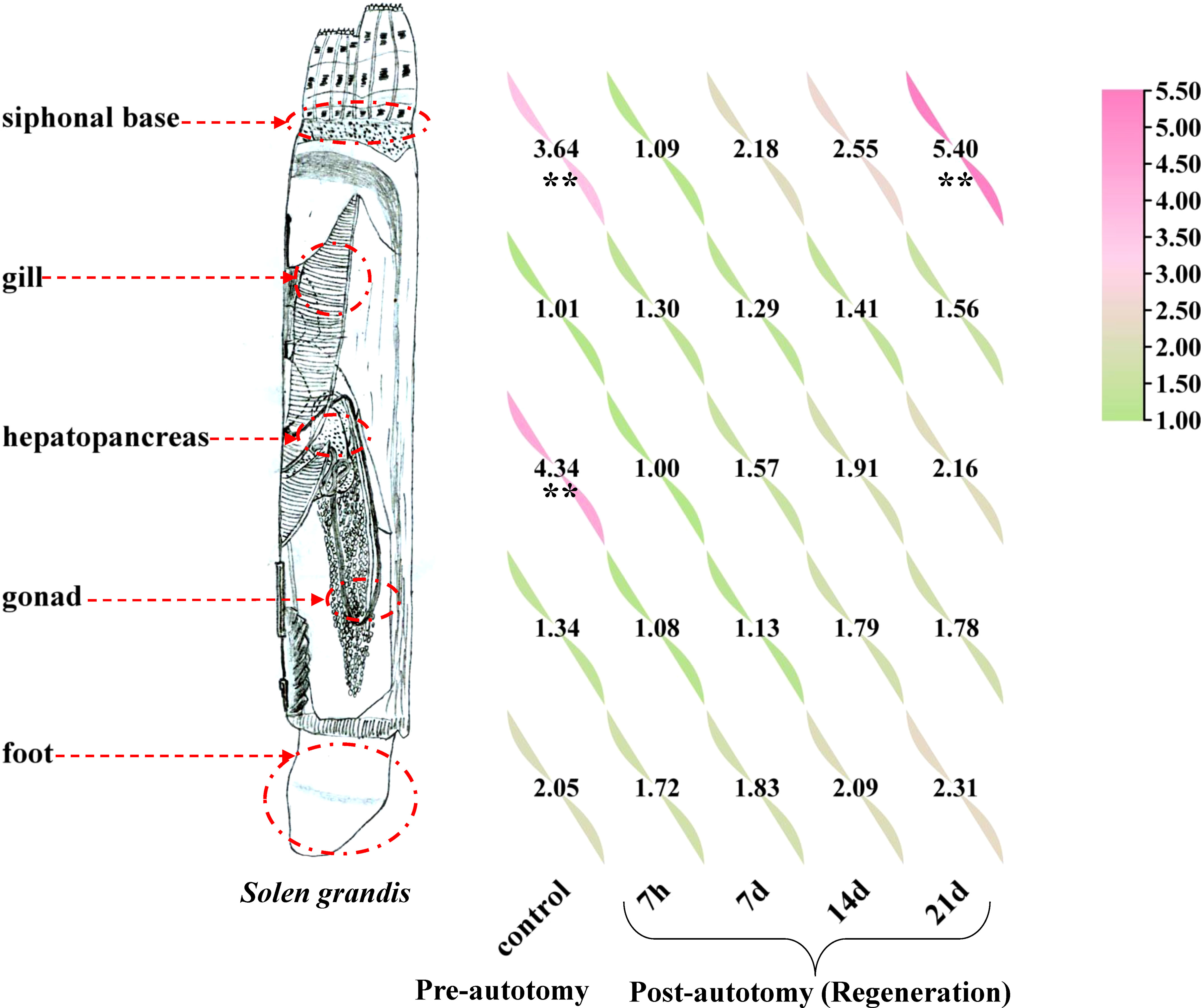
Figure 4 Relative expression levels of SgENO4 gene in different tissues and siphonal autotomy and regeneration stages. The asterisk (**) indicate a significant difference at P < 0.05.
As presented in Table 3, the siphon can regenerate to the pre-autotomy status about 21 days after siphonal autotomy of razor clam. On the 21st day, the siphonal weight of S. grandis accounted for 9.25% of the live body weight. It is noted that the relative expression levels of SgENO4 was consistent with the trend of specific weight of siphon (Figure 5A). Therefore, Linear regression models were simultaneously constructed with specific weight as the x-axis and relative expression levels of SgENO4 as the y-axis. The result showed that there was a significant linear correlation between specific weight of siphon and relative expression levels of SgENO4 (R2 = 0.826, Figure 5B).
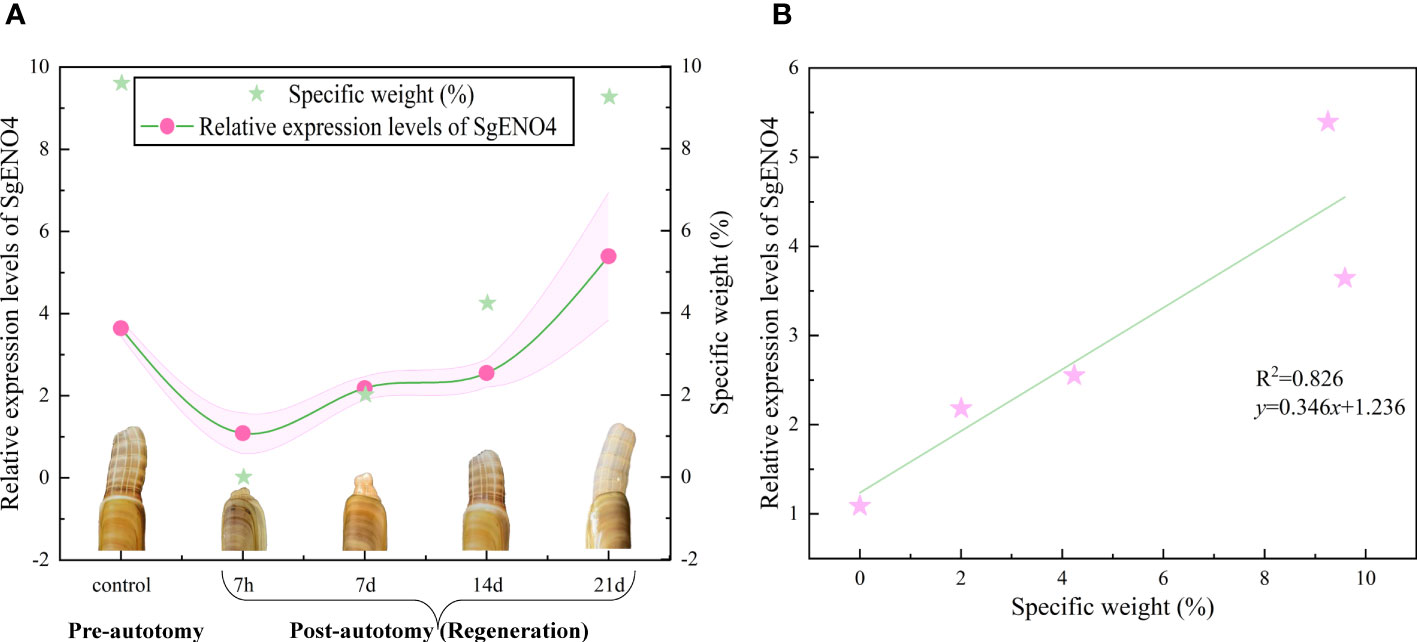
Figure 5 (A) Effect of siphonal autotomy and regeneration on the relative expression levels of SgENO4. (B) Relationship between siphonal regeneration and relative expression levels of SgENO4.
In order to verify the role of SgENO4 gene in the siphonal regeneration of Solen grandis, the RNAi experiment was performed. The results showed that the relative expression level of SgENO4 first decreased and then increased and reached the lowest value on the seventh day, indicating that the expression of SgENO4 was inhibited successfully (Figure 6A). The specific weight (the proportion of siphonal weight to body weight) is significantly lower than other groups on the seventh day (P < 0.05), suggesting that interfering with the expression of the SgENO4 gene could inhibit the siphonal regeneration of S. grandis (Figure 6B).
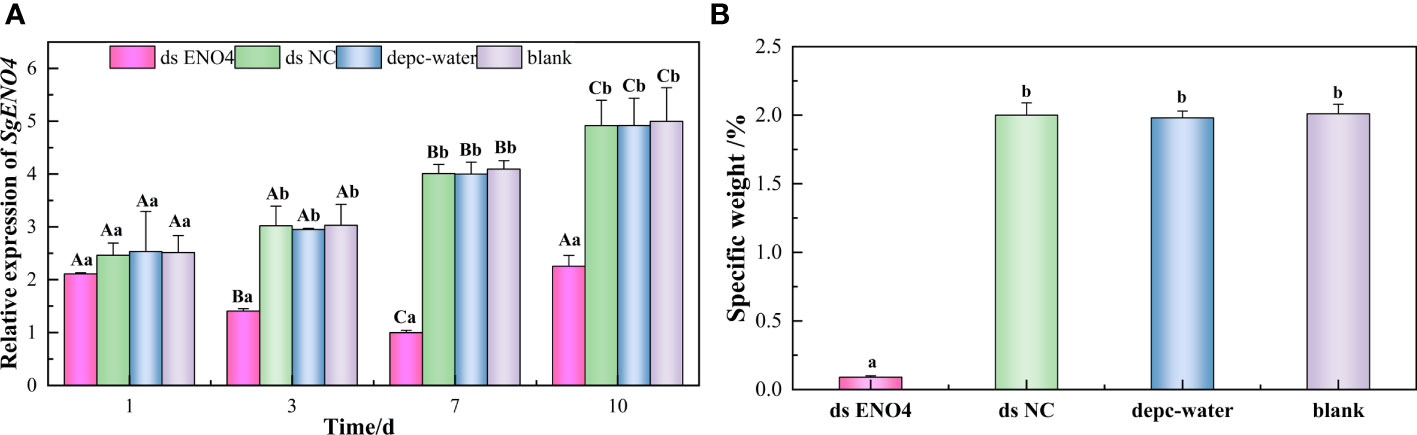
Figure 6 Effects of RNAi on (A) the relative expression of SgENO4 gene and (B) specific weight of Solen grandis after 7 days. Different lower-case letters indicate significant differences between different groups at the same time, and different upper-case letters indicate significant differences at different times within the same group.
The unique ENO4 transcript sequences were obtained by comparing the internal S. grandis transcriptome data (unpublished data) with NCBI database. Our study cloned enolase4 of S. grandis, namely SgENO4 gene. The molecular characteristics and the potential function in the siphonal autotomy and regeneration of S. grandis. The predicted amino acid sequence and tertiary structure of S. grandis are less similar to those of other species, which should not be considered as weakly conserved types. The main reason is that there is less information about the ENO4 gene in the existing database. In addition, SgENO4 gene possessed N-terminal and C-terminal TIM bucket domain of enolase. All motifs were identified by the MEME database with complete amino acid sequence of ENO4. The differentiation of the ENO4 protein across species is not obvious, and three conserved motifs were found. To gain further insight into the influence of nucleotide content in shaping the SgENO4 codon usage pattern, the nucleotide compositions were calculated. The results show that there is slightly lower codons usage bias in the SgENO4 sequence and the value of ENC is 54.36, which reveals a relatively stable nucleotide composition and slightly lower (ENC>40) codon usage bias. These results further demonstrate that ENO4 is a conserved ancient enzyme (Feo et al., 2000; Chen Y. et al., 2017; Hou, 2020).
As a key enzyme in glycolysis regulation, enolase indirectly drives mitochondria to produce large amounts of energy to sustain the organism’s development and metabolism. The enolase gene has been confirmed to be highly expressed in a variety of cancer tissues and has become a potential biomarker of early-stage cancer, which is closely related to the proliferation and spread of cancer cells (Yu et al., 2012; Pan et al., 2020). Previous studies showed that the expression and distribution of enolase in Helicoverpa armigera is related to the pathological environment of the cell and the degree of vigorous metabolism. Enolase may play a role in the body’s energy sources in life activities such as worm growth and development and nutrient metabolism (Hu, 2017). In the present study, the SgENO4 gene is expressed in all tissues, with significantly higher levels of expression in the hepatopancreas and siphonal base than in other tissues. This may be related to the fact that the hepatopancreas is a site of energy metabolism, and the siphonal recovery and regeneration requires a lot of energy. In addition, the high energy requirements of the siphon, the filter feeding organ of razor clams, are also responsible for the high expression of the SgENO4 gene (Luo et al., 2021; Zhang et al., 2022).
Enolase functions as a part-time protein with regional expression specificity and complex versatility. In addition to its role in energy metabolism during glycolysis, enolase plays an important role in tumor, immunity, infection, regeneration and other processes. many studies have shown that acute stress significantly affects energy metabolism processes in organisms and razor clam siphons are prone to autotomy because of external stimuli, such as tides and the agitation of symbiotic organisms (Wu et al., 2022b). Based on the expression patterns of oxidative phosphorylation related genes during liver regeneration in rats, previous study found that liver regeneration has stages, with gene expression levels decreasing in the early and middle stages of liver regeneration and gradually increasing in the middle and late stages (Lin et al., 2009). During tissue regeneration, enolase may participate in the recovery and regeneration of damaged tissue through the interaction of tubulin with microtubules (Keller et al., 2007; Sun et al., 2022). In the present study, the expression of the SgENO4 gene in the siphonal base decreased significantly within 7 hours of post-autopsy siphon base removal and gradually recovered as the regeneration process continued. This could also explain why razor clams are prone to die after low immunity. It has been speculated that the energy metabolism level of S. grandis drops sharply during the early stages of siphonal regeneration in response to wound. As the energy demand increases during regeneration, the energy metabolism level gradually increases to meet the wound healing and regeneration demand (Hu, 2017; Wang et al., 2021).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
All applicable International, National, and/or Institutional Guidelines for the Care and Use of Animals (invertebrates) were followed.
ZZ: Wrote manuscript and analyzed the data. XS: Performed the qRT-PCR experiment. AC: Conceived the idea. JY: Guided the experiment. YZ and YP: Performed the rearing experiment. YC and YZ: Collected and prepared samples. SC: Performed the RACE experiment. YW: Provided subsidization. All authors contributed to the article and approved the submitted version.
This work was supported by the Nantong Science and Technology Innovation Fund (Grant No. JC2021012).
The constructive comments of the reviewers on an earlier version of this manuscript are greatly appreciated.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Barr J. I., Somaweera R., Godfrey S. S., Gardner M. G., Bateman P. W. (2020). When one tail isn't enough: abnormal caudal regeneration in lipid osaurs and its potential ecological impacts. Biol. Rev. Camb Philos. Soc 95, 1479–1496. doi: 10.1111/brv.12625
Bely A. E., Zattara E. E., Sikes J. M. (2014). Regeneration in spiralians: evolutionary patterns and developmental processes. Int. J. Dev. Biol. 58, 623–634. doi: 10.1387/ijdb.140142ab
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen S., Qiao H., Fu H., Sun S., Zhang W., Jin S., et al. (2017). Molecular cloning, characterization, and temporal expression of the clock genes period and timeless in the oriental river prawn Macrobrachium nipponense during female reproductive development. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 207, 43–51. doi: 10.1016/j.cbpa.2017.02.011
Chen Y., Xu Q., Tan C., Li X., Chi X., Cai B., et al. (2017). Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Senecavirus A evolution. Microb. Pathog. 112, 313–319. doi: 10.1016/j.micpath.2017.09.040
Feo S., Arcuria D., Piddinia E., Passantino R., Giallongo A. (2000). ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Letters 473, 47–52. doi: 10.1016/S0014-5793(00)01494-0
Hou W. (2020). Characterization of codon usage pattern in SARS-CoV-2. Virol. J. 17, 138. doi: 10.1186/s12985-020-01395-x
Hu R. (2017). Cloning, Expression and Functional Analysis of enolase gene from Helicoverpa armigera (Lepidoptera: Noctuidae). Cent. China Normal Univ. 1–5. doi: 10.27159/d.cnki.ghzsu.2017.000139
Kaczmarski M., Ziemblinska K., Tryjanowski P. (2020). Sand lizards Lacerta agilis with higher digit ratios are more likely to autotomy. J. Anat 237, 1103–1113. doi: 10.1111/joa.13277
Keller A., Peltzer J., Carpentier G., Horváth I., Oláh b J., Duchesnay A., et al. (2007). Interactions of enolase isoforms with tubulin and microtubules during myogenesis. Biochim. Biophys. Acta 1770, 919–926. doi: 10.1016/j.bbagen.2007.01.015
Li W., Gao Y., Sheng X., Chen X., Xiong J. (2011). Clone and expression analysis of Enolase in Litopenaeus vannamei. Guangdong Agric. Sci. 5, 168–174. doi: 10.16768/j.issn.1004-874x.2011.05.075
Lin F., Mu R., Zhao H., Zhu H., Xu C. (2009). Expression patterns of the oxidative phosphorylation-associated genes during rat liver regeneration. Acta Agric. universitatis jiangxi en sis 31, 1107–1112. doi: 10.3969/j.issn.1000-2286.2009.06.026
Livak K., Schmittgen T. (2002). Analysis of relative gene expression data using real-time quantitative PCR. Methods. 254, 402–408. doi: 10.1006/meth.2001.1262
Luo X., Ma L., Liu S., Hai L., Ding X. (2021). Study on the expression and correlation of alpha-enolase (eno1) in the serum of patients with hepatocellular carcinoma. Res. square. doi: 10.21203/rs.3.rs-279041/v1
Mykles D. L. ;. (2001). Interactions between limb regeneration and molting in decapod crustaceans. Am. Zoologist 41, 399–406. doi: 10.1093/icb/41.3.399
Pan X., Wu H., Chen G., Li W. (2020). Prognostic value of enolase gene family in colon cancer. Med. Sci. Monitor: Int. Med. J. Exp. Clin. Res. 26, e922980. doi: 10.12659/MSM.922980
Straeten D., Rodrigues R., Goodman H., Montagu M. (1991). Plant enolase: gene structure, expression, and evolution. Am. Soc. Plant Physiologists 3, 719–735. doi: 10.2307/3869253
Sun H., Mo J., Cheng R., Li F., Li Y., Guo Y., et al. (2022). ENO1 expression and Erk phosphorylation in PDAC and their effects on tumor cell apoptosis in a hypoxic microenvironment. Cancer Biol. Med. 19, 1598–1616. doi: 10.20892/j.issn.2095-3941.2022.0451
Trigos S., Vicente N., Prado P., Espinós F. J. (2018). Adult spawning and early larval development of the endangered bivalve Pinna nobilis. Aquaculture. 483, 102–110. doi: 10.1016/j.aquaculture.2017.10.015
Wang Z., Bai Y., Nie H., Xu Q., Yin Z., Zhang Y., et al. (2021). Molecular mechanisms of wound healing and regeneration of siphon in the Manila clam Ruditapes philippinarum revealed by transcriptomic analysis. Genomics. 113, 1011–1025. doi: 10.1016/j.ygeno.2021.02.010
Wrinn K., Uetz G. (2007). Impacts of leg loss and regeneration on body condition, growth, and development time in the wolf spider Schizocosa ocreata. Can. J. Zoology 85, 823–831. doi: 10.1139/Z07-063
Wu Y., Chen S., Chen A., Li Q., Zhang Y., Cao Y., et al. (2022a). Molecular cloning and expression analysis of CDK genes during selective and wild populations of juvenile Meretrix meretrix. Aquaculture Res. 53, 5445–5454. doi: 10.1111/are.16025
Wu Y., Chen A., Zhang Y., Zhang Z., Cao Y., Chen S., et al. (2022b). Metabolomics approach to assess the effect of siphonal autotomy on metabolic characteristics of razor clam Solen grandis. Sci. Rep. 12, 5528. doi: 10.1038/s41598-022-09562-6
Yan F., Xiao X., Tang L., Song Y., Han C., Wang C., et al. (2022). Molecular cloning, expression patterns and functional characterization of Gpr3 in the orange-spotted grouper (Epinephelus coioides). Aquaculture Rep. 23, 1–7. doi: 10.1016/j.aqrep.2022.101050
Yu Z. L., Feng J., Song H., Yang M. J., Hu Z., Zhou C., et al. (2022). Identification and characterization of olfactory receptor genes and olfactory perception in rapa whelk Rapana venosa (Valenciennes, 1846) during larval settlement and metamorphosis. Gene. 825, 146403. doi: 10.1016/j.gene.2022.146403
Yu L., Shi J., Cheng S., Zhu Y., Zhao X., Yang K., et al. (2012). Estrogen promotes prostate cancer cell migration via paracrine release of ENO1 from stromal cells. Mol. Endocrinol. 26, 1521–1530. doi: 10.1210/me.2012-1006
Yuan Y., Li Q., Kong L., Yu H. (2012). The complete mitochondrial genome of the grand jackknife clam, Solen grandis (Bivalvia: Solenidae): a novel gene order and unusual non-coding region. Mol. Biol. Rep. 39, 1287–1292. doi: 10.1007/s11033-011-0861-8
Yuan Y., Li Q., Kong L., Zheng X. (2010). Isolation and characterization of polymorphic microsatellite loci in the grand jackknife clam Solen grandis (Bivalvia: Veneroida). Genes Genomics 32, 191–197. doi: 10.1007/s13258-009-0780-4
Keywords: Solen grandis, SgENO4 gene, regeneration, siphon, autotomy
Citation: Zhang Z, Sun X, Chen A, Yang J, Chen S, Zhang Y, Cao Y, Peng Y, Zhu Y and Wu Y (2023) Molecular bioinformatic and functional analysis of Enolase4 gene during siphonal autotomy and regeneration of razor clam Solen grandis. Front. Mar. Sci. 10:1195253. doi: 10.3389/fmars.2023.1195253
Received: 28 March 2023; Accepted: 25 July 2023;
Published: 11 August 2023.
Edited by:
Yuehuan Zhang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Hui Qiao, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2023 Zhang, Sun, Chen, Yang, Chen, Zhang, Cao, Peng, Zhu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangping Wu, Mjc5MDE5MzU1OEBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.