- National Engineering Laboratory of Marine Germplasm Resources Exploration and Utilization, Marine Science and Technology College, Zhejiang Ocean University, Zhoushan, Zhejiang, China
In the neuroendocrine system, corticotropin-releasing hormone (CRH) plays an important role in the hypothalamic–pituitary–adrenal/interrenal (HPA/HPI) axis. It exerts its effects by activating CRHRs, which belong to the class B G protein-coupled receptor family. Two characteristic genes of CRHR1 subtypes in the Larimichthys crocea genome were identified: LcCRHR1-1 and LcCRHR1-2. Alignments indicated that they were highly homologous to known and validated teleost CRHR1s. The CDS sequences of the two receptors were cloned into the pEGFP-N1 plasmid, and membrane localization of the fusion expressing LcCRHR1-1-EGFP and LcCRHR1-2-EGFP was revealed in HEK293 cells. Treatment with LcCRH could lead to two receptors internalization and trigger a significant increase in the secondary messenger cAMP and Ca2+ and mitogen-activated protein kinase phosphorylation in an LcCRH dose-dependent manner. Based on quantitative real-time PCR, LcCRHR1s were expressed in all examined tissues and highly expressed in the brain and ovaries. Furthermore, immunohistochemical findings showed the specific localization of CRHR1s in ovarian follicle cells. Collectively, our study identified two CRH receptors in L. crocea and suggested that the CRH/CRHR1 system is potentially involved in the neuroendocrine regulation of reproduction in this marine fish.
1 Introduction
Hypothalamic corticotropin-releasing hormone (CRH), also known as corticotropin-releasing factor (CRF), is a neurohormone that primarily controls the hypothalamic–pituitary–adrenal/interrenal (HPA/HPI) axis and plays a significant role in the stress response by activating its specific receptors (CRHR) (Denver, 2009; Henckens et al., 2016). CRH was first isolated and characterized in the hypothalamus of ovine in which it can activate CRHR in the pituitary gland and trigger the synthesis and secretion of adrenocorticotropin (ACTH) (Vale et al., 1981). CRH and its structurally similar peptides with related functions are collectively referred to CRH-family peptides for review (Lovejoy et al., 2014), which include urotensin I (Lederis et al., 1982), urocortin I (Vaughan et al., 1995; Cardoso et al., 2016), urocortin II (Reyes et al., 2001), urocortin III (Lewis et al., 2001), and sauvagine (Montecucchi et al., 1980). In many mammalian species, the 41-amino acid mature CRH peptide is formed by removing the 24-amino acid signal peptide from the precursor peptide with an amidated C-terminus (Dautzenberg and Hauger, 2002; Pangalos and Davies, 2002). Mature CRH peptides have a high degree of conservation across teleosts, amphibians, and mammals and mediate various biological activities that affect reproductive behavior, cardiovascular regulation, appetite control, and immune function in addition to stress response (Grammatopoulos and Chrousos, 2002; Ma et al., 2020). Corresponding to its diverse biological functions, CRH is ubiquitously expressed in the central nervous system (CNS) and peripheral organs, such as the ovaries, heart, spleen, liver, and gastrointestinal tract (Wang et al., 2014; Popov et al., 2021).
With advances in bioinformatics and molecular biology, two types of CRHRs, CRHR1 and CRHR2 have been identified and found to be highly conserved in vertebrates (Dautzenberg and Hauger, 2002; Hemley et al., 2007). Based on phylogenetic and functional characteristics, these two receptors share ~ 70% amino acid sequence homology in mammalian species (Lovejoy et al., 2014). A third CRHR, named CRHR3, has only been discovered in the catfish Ameirus nebulosus, receptor which shares 85% and 80% sequence homology with catfish CRHR1 and CRHR2, respectively (Arai et al., 2001). These three types of CRHRs, particularly the CRHR1 and CRHR2 in mammals, have been determined to be encoded by genes containing multiple exons and introns with unique splicing patterns and distinct pharmacological properties (Hillhouse and Grammatopoulos, 2006; Grammatopoulos, 2012). Compared to CRHR2, which mediates stress recovery and the restoration of homeostasis, the activation of CRHR1 aggravates stress responses and anxiety-like behaviors (Smith et al., 1998). Rodents lacking CRHR1 can prevent CRH-induced anxiogenic phenotypes and display reduced anxiety-like behaviors in anxiety tests (Bale et al., 2002). In teleosts, CRHR1 was reported to be involved in a broad range of physiological functions (Lovejoy et al., 2014). Therefore, studies on the molecular characteristics and activation mechanisms of CRH/CRHR1 are of great significance for understanding the biological regulation of external stimulation (Füzesi et al., 2016; Zalachoras et al., 2022).
In addition to its functions in the environmental stress response, the CRH/CRHR1 signaling system also has reproductive regulatory functions in mammals. The gene expression of CRHR1 was detectable in reproductive organs, such as the uterus and ovary (Petraglia et al., 2010). CRH inhibits the gonadotropin-releasing hormone (GnRH) release, thereby reducing the synthesis and secretion of gonadal steroid hormones (Melón and Maguire, 2016; Raftogianni et al., 2018). Accumulating evidence suggests that CRH may be involved in the reproductive development and ovarian maturation (Kiapekou et al., 2010; Wypior et al., 2011). Therefore, a deep understanding of the signaling and functional mechanisms of the CRH/CRHR1 system needs to be developed in teleosts. Scientists have elucidated the molecular properties of CRHR1 in teleost fish, such as the gene structural characters of the receptor in Cyprinus carpio (Huising, 2004; Manuel et al., 2014), protein structure and functions of the receptor in Astatotilapia burtoni (Chen and Fernald, 2008), and distribution profiles of the receptor in the ovary of Paralichthys olivaceus (Zhou et al., 2019), Fugu (Cardoso et al., 2003), and chum salmon (Pohl et al., 2001). CRHR expressions changed during ovarian development in the olive flounder (Zhou et al., 2019) and in the zebrafish (Zhou et al., 2021). In addition, treatment with 17 α, 20 β-dihydroxy-4-pregnen-3-one (DHP) increases CRHR1 mRNA expression in ovarian follicles from olive flounder (Zhou et al., 2019), and the two CRH identified in the zebrafish, decrease estradiol release from primary cultures of follicular cells in this species (Zhou et al., 2021). These data suggest a potential involvement of CRH system in teleost reproduction. However, the cell signaling system and physiological functions mediated by fish CRHR1 have rarely been clarified, particularly focusing on reproductive control.
In mammals, as others Class B1 G Protein-Coupled Receptors (GPCR), CRHR1 contains a distal N-terminal extracellular domain (ECD) that can specifically select and bind ligands, and the C-terminal tail can activate specific kinases to mediate a series of signaling cascades (Grammatopoulos, 2012). Activated by CRH, CRHR1 primarily couples to Gαs proteins, triggering the accumulation of cyclic AMP (cAMP) and the activation of the extracellular signal-regulated kinase-mitogen-activated protein kinase (cAMP-MAPK) pathway. In addition, the receptor could also couple the Gαq protein and lead to calcium mobilization by phospholipase C activation (Graziani et al., 2002; Gutknecht et al., 2009). These signals induce the transcription of downstream target genes by activating protein kinase A (PKA), cAMP-response element binding protein (CREB) (Inda et al., 2017), protein kinase B (Akt), and protein kinase C (PKC), depending on coupling with different G proteins, ligands, and cell types (Brar, 2017). In teleosts, the functional characteristics and cell signal transduction mechanisms of CRHR1 need to be explored.
The large yellow croaker (Larimichthys crocea) is a widely cultured and economically important marine fish found in the coastal areas of East and Southeast China (Chen et al., 2018). With the development of artificial mariculture in this species, biological research on the regulatory mechanisms underlying its physiological processes is becoming increasingly important, particularly regarding reproduction and growth. In this study, to clarify the functional roles of the CRH/CRHR1 signaling system in the reproduction of L. crocea, a mature LcCRH peptide was synthesized, and two genes of LcCRHR1s (LcCRHR1-1 and LcCRHR1-2) were functionally identified and characterized. Our study provides new insights for the bioactivity and regulatory mechanisms of the CRH/CRHR1 system in a marine teleost.
2 Materials and methods
2.1 Animals, sample collection, RNA extraction and cDNA synthesis
Female large yellow croaker were collected from Zhoushan Fishery Research Institute and stabilized in an indoor mariculture system for half a month (weight 390.5 ± 35.5 g, temperature range 25.0 ± 1.0°C, salinity range 24.38-25.45‰, NH4+ < 0.2 mg/L, and NO2− < 0.1 mg/L). The ovary tissues were fixed with 4% PFA for further histological analysis, and total RNA from the brain, heart, liver, spleen, ovary, intestine, retina, and head kidney tissues was extracted using TRIzol reagent (TaKaRa, Kusatsu, Japan). The quality of total RNA was checked by electrophoresis, and the concentration was measured using a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The total RNA (1 μg) was co-incubated with M-MLV Reverse Transcriptase (TaKaRa Kusatsu, Japan) and OligdT-18 (Promega Inc., Shanghai, China) at 42°C for 1 h to synthesize single-stranded cDNA. The cDNA samples were stored at -80°C for further experiments.
2.2 Sequence analysis and plasmid construction
The cDNA sequences of LcCRHR1-1 and LcCRHR1-2 (NCBI accession numbers: XP-019133265.1 and XP-010752790, respectively) were obtained from the NCBI database using the online BLAST tool (http://blast.ncbi.nlm.nih.gov). The amino acid sequence was deduced using Bio Eidt 7.2. Phosphorylation sites and the signal peptide were located using the NetNGlyc1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/). Transmembrane proteins were analyzed using Tmpred (http://www.cbs.dtu.dk/services/TMHMM/), and secondary structures were analyzed using the transmembrane prediction Protter server (http://wlab.ethz.ch/protter/start/). The protein 3D structures of LcCRHR1-1 and LcCRHR1-2 were predicted using the SWISS-MODEL software (http://swissmodel.expasy.org/). The deduced amino acid sequences were aligned using ClustalW (http://www.bioinformatics.org/sms/). The color alignment was generated using the Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/color_align_prop.html). Species sequence identity was calculated using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). A phylogenetic tree was constructed using the maximum likelihood (ML) method of MEGA 5.0 with 1000 bootstrap trials. The GenBank accession numbers for the amino acid sequences used for phylogenetic tree analysis are listed in Supplementary Table 1.
Sequences of CRH and CRHR1 were searched and downloaded from zebrafish, mouse, and human in NCBI. Then, a local BLAST comparison of these sequences with the entire amino acid sequence of the L. crocea genome (GCF_000972845.2_L_crocea_2.0_genomic) was performed. The parameters were set as follows: e-value = 0.00001, maximum match number = 5. The most similar sequence was selected based on the BLAST results and then confirmed using NCBI Blastp.
The forward (LcCRHR1-1 or LcCRHR1-2-seq-F) and reverse primers (LcCRHR1-1 or LcCRHR1-2-seq-R) used for the coding sequence (CDS) cloning are listed in Supplementary Table 2, using cDNA from whole brain as a template for PCR. After verifying the sequence of the cloned full-length LcCRHR1-1 or LcCRHR1-2 CDS by TA cloning and pyrosequencing, the plasmids LcCRHR1-1-EGFP and LcCRHR1-2-EGFP were synthesized by LeQi-Biomedicine (Hangzhou) Co. Ltd. The reading frame, orientation, and sequence fidelity were verified by PCR product sequencing.
Mature CRH peptide sequences of L. crocea (XP_010732512.1) were obtained from genome (Supplementary Figure 1), synthesized by GL Biochem Shanghai (China), and purified using preparative reverse-phase high-performance liquid chromatography. Fractions containing the appropriate peptides were pooled and lyophilized, and the purity of the final product was 95.51%. The feasibility of solid-phase synthesis and the ability of chemically modified peptides to activate their receptors have been verified (Fredriksson et al., 2006; Yang et al., 2013).
2.3 Receptor localization and internalization assay in HEK293 cells
HEK293 cells were grown to 80-90% density in high-glucose medium (DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone, Waltham, MA, USA), 1% penicillin (Beyotime, Shanghai, China), and 1% streptomycin (Beyotime, Shanghai, China), and transfected with plasmids (LcCRHR1-1 or LcCRHR1-2-EGFP) using X-treme GENE HP DNA Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA). All cells were cultured in a humidified environment at 37 °C and 5% CO2.
Cells expressing LcCRHR1-1 or LcCRHR1-2-EGFP were seeded onto cell slides (14 mm diameter) and attached overnight. For receptor expression analysis, cells were stained with the membrane probe DiI (Beyotime, Shanghai, China) for 25-30 min at 37°C, then fixed with 4% paraformaldehyde for 10 min, and finally incubated with DAPI for 5-10 min. For receptor internalization assays, HEK293 cells were treated with LcCRH (100 nM) for 0, 5, 15, 30, and 60 min at 37°C, fixed with 4% paraformaldehyde for 15 min, and finally treated with DAPI for 5 min. DMEM was used as the control. Cells were visualized by the confocal microscope using an HCX PL APO λ blue 63×1.4 oil immersion lens.
2.4 cAMP measurement
HEK293 cells transiently co-transfected with the pCRE-luciferase system and LcCRHR1s-EGFP plasmids were reseeded in 96-well plates overnight (Wang et al., 2019). Cells were treated with serum-free medium containing different concentrations of LcCRH (10-13-10-5 M) for 4 h and lysed for 10 min after discarding the medium. Luciferase activity was measured using the firefly luciferase assay kit (Kenreal, Shanghai, China). Simultaneously, cAMP concentrations were detected using a competitive binding enzyme-linked immunosorbent assay (ELISA) kit (Parameter cAMP assay, R&D Systems, Minneapolis, MN, USA).
2.5 Intracellular calcium measurement
HEK293 cells transiently expressing LcCRHR1-1 or LcCRHR1-2 were reseeded overnight in 50-mm cell culture dishes. The cells were digested with PBS containing 0.02% EDTA for 5 min and suspended in Hanks balanced salt solution (HBSS). The cell suspension was incubated with 3.0 μM Fura-2/AM (the fluorescent Ca2+ indicator) in an CO2 incubator (37°C, 5% CO2) for 30 min and washed twice in Hanks’ solution. Cells were stimulated with the indicated concentrations of LcCRH (1 μM, 10 nM, and 100 pM). Finally, the ratio of the excitation wavelengths at 340 nm and 380 nm was measured to determine the intracellular calcium flux within 60 s using a fluorescence spectrometer (Infinite 200 PRO, Tecan, Mannedorf, Switzerland).
2.6 Extracellular signal‐regulated kinase phosphorylation assay
LcCRHR1-1 or LcCRHR1-2-EGFP expressing cells were seeded in 24-well plates for 18-24 h and starved in a high-glucose medium without FBS for 2 h to eliminate the effect of FBS. HEK293 cells expressing LcCRHR1-1 and LcCRHR1-2 were stimulated by 100 nM LcCRH for 0, 2, 5, 10, 15, 20, 30 and 45 min. Based on the time-course results, different concentrations of LcCRH (10-12-10-6 M) were tested during the strongest phosphorylated signal time (5 min). To elucidate the signaling pathways, DMEM containing 1‰ DMSO (control: solvent for antagonist), 10 μM H89 (PKA antagonist) or 10 μM ESI09 (EPAC antagonist) was pretreated for 1 h separately in expressing LcCRHR1-1 or LcCRHR1-2 cells. Then, different concentrations of LcCRH (control, 10 nM, 100 nM, and 1 μM) were used to stimulate the cells for 5 min, and the phosphorylated ERK1/2 levels were detected. Finally, the cells were treated with RIPA buffer (Beyotime, Shanghai, China) containing 1% protease inhibitor phenylmethylsulfonyl fluoride (PMSF) for half an hour to achieve a full release of intracellular protein. The protein samples were fractionated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% SDS-PAGE). The target protein on the gel was transferred onto a polyvinylidene fluoride membrane (PVDF; Millipore, Shanghai, China) using a transfer device. The PVDF membrane was blocked with a blocking solution (TBST containing 5% nonfat milk) at room temperature for 1 h. The PVDF membrane was incubated with anti-rabbit phospho-p44/42 ERK1/2 antibody (4370S; Cell‐Signaling Technology, USA) and anti-rabbit p44/42 ERK1/2 antibody (4695S; Cell Signaling Technology, USA) at 4°C overnight and then incubated with anti-rabbit-HRP secondary antibody (1:2000, Beyotime, Shanghai, China) at 25°C for 2 h. Finally, the membrane was detected with a luminescent substrate to detect protein signal intensity and analyzed using ImageJ software.
2.7 Real-time quantitative PCR
cDNA samples from the brain, heart, liver, spleen, ovary, intestine, retina, and head kidney of L. crocea were synthesized for qRT-PCR. Primers were first tested to ensure that a single discrete band was amplified without primer dimers. The qRT-PCR assays were performed using SYBR PrimeScript™ RT Reagent Kit (TaKaRa) and ABI 7500 Software v.2.0.6 (Applied Biosystems) (Supplementary Table 4). Relative levels of gene expression were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). Significance was evaluated using one-way analysis of variance (ANOVA), followed by Tukey’s post-hoc test using PASW Statistics 18.00 (SPSS Inc.).
2.8 Immunohistochemistry assay
The ovaries of adult female L. crocea were dissected and fixed in a 4% paraformaldehyde fixative solution (Solarbio, Beijing, China). The tissues were dehydrated and placed into paraffin at 4°C overnight. The tissue embedded in the paraffin block was sectioned (6 μm thick) with a microtome (Leica Biosystems, Wetzlar, Germany). The sections were dewaxed, rehydrated, subjected to antigen retrieval treatment, and then incubated with anti-rabbit CRHR1 antibody (PA5-27121; 1:500, human/mouse CRHR1, ThermoFisher, USA) at 4°C overnight. The control group, except for the incubation with PBS instead of CRHR1 antibody, follows the same experimental steps as the CRHR1 experimental group. The corresponding secondary antibodies (1:500, Cy3 labeled, Beyotime, Shanghai, China) were incubated with the slides at room temperature for 2 h. Images of the sections were captured by confocal microscope using a Leica TCS SP5II laser scanning confocal microscope.
2.9 Statistical analysis
All data are presented as mean ± standard error of the mean (SEM), and statistical significance was assessed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test using GraphPad Prism 6.0. All experimental data were obtained from at least three independent replicates (*, P < 0.05; ** P < 0.01; ***, P < 0.001).
3 Results
3.1 Molecular characterization and phylogenetic analysis of LcCRHR1-1 and LcCRHR1-2
The full-length CDS of LcCRHR1-1 and LcCRHR1-2 encoded two L. crocea CRH receptors containing 436 and 429 amino acids, respectively (Supplementary Figures 2, 3). Multiple sequence alignment analysis of these amino acid sequences was performed to evaluate the homologous relationships. The two predicted L. crocea CRHR1s amino acid sequences showed similarities to vertebrate CRHR1s (LcCRHR1-1 with 76.11-86.27% identity, LcCRHR1-2 with 73.54-84.98% identity) and revealed a high level of conservation in the transmembrane domains of (Figure 1; Supplementary Table 3).
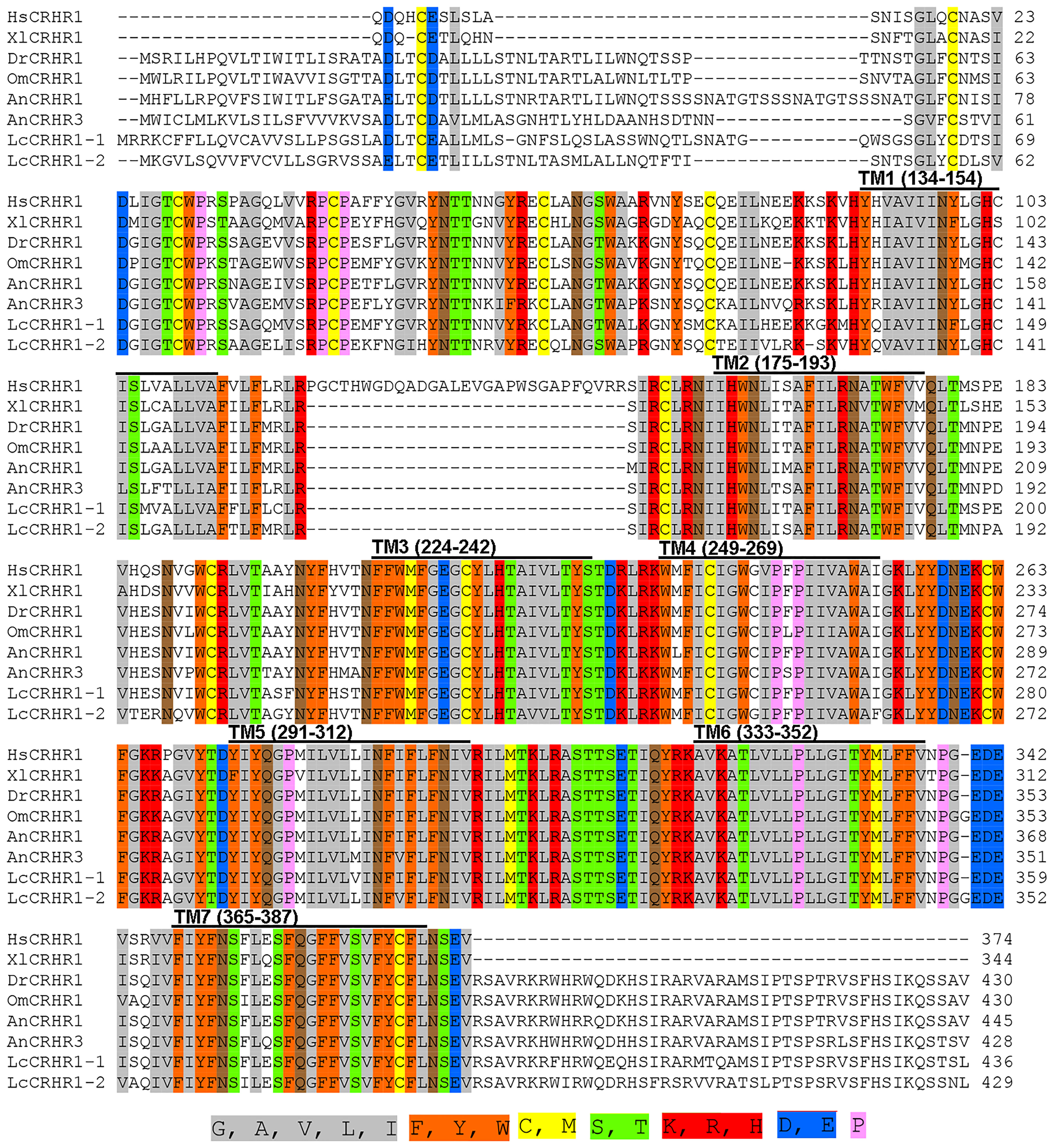
Figure 1 Multiple alignments of LcCRHR1s amino acid sequence with sequences from other species. CRHR1s sequences include Homo sapiens, Xenopus laevis, Danio rerio, Oncorhynchus mykiss, and A. nebulosus. The GenBank accession numbers are as follows: L. crocea1-1 (XP_019133265.1), L. crocea1-2 (XP_010752790.1), H. sapiens (NP_001138618.1), X. laevis (NP_001079049.1), D. rerio (XP_696346.3), O. mykiss (XP_021480247.1), A. nebulosus (AF229359, AF229361). ClustalW was used to assess the alignment, and the online Sequence Manipulation Suite was used to generate the color aligns attribute. The percentage of sequences that must agree for identity or similarity coloring was set as 80%. The seven transmembrane domains (TM1-TM7) are marked with a black horizontal line above the sequence alignment. The conserved amino acid residues with identical chemical properties are highlighted with the corresponding color.
Based on protein secondary structure prediction, both receptors had seven transmembrane domains and six N-linked glycosylation sites in the extracellular domain, which is a typical feature of membrane glycoproteins for ligand recognition. 1 The N-terminus of LcCRHR1-1 or LcCRHR1-2 has a signal peptide consisting of 24 or 21 amino acids, respectively (Figures 2A, B). These two amino acid sequences contained 37 potential phosphate sites: LcCRHR1-1 contained 22 serine, 10 threonine, and 5 tyrosine residues, and LcCRHR1-2 had 18 serine, 12 threonine, and 7 tyrosine residues. Homology modeling revealed that these proteins were like 6pb0.1. A and 6p9x.1. E from the Protein Data Bank.
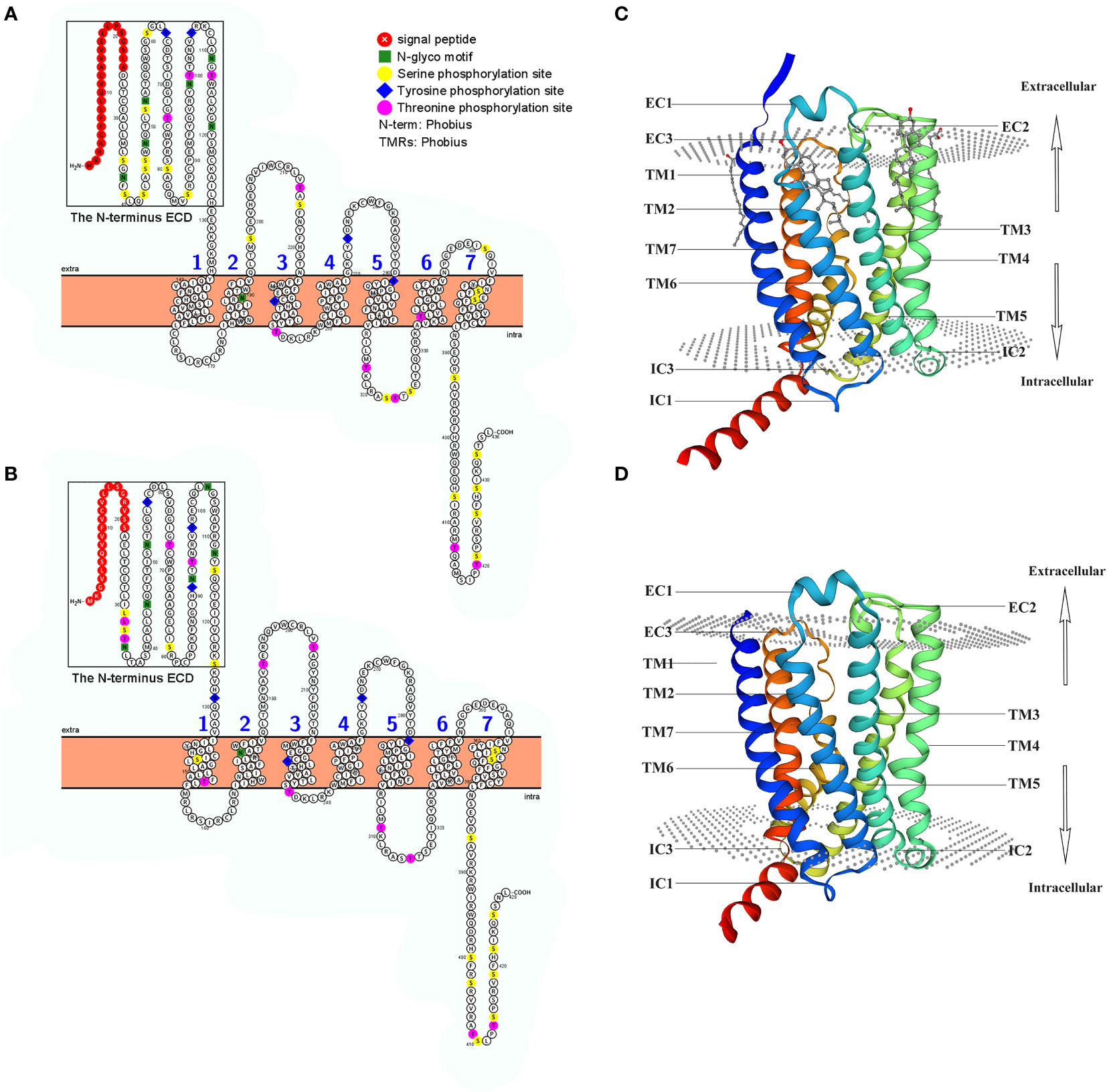
Figure 2 Predicted sequence characteristics of LcCRHR1s. Schematic diagram of the transmembrane structure of LcCRHR1-1 (A) and LcCRHR1-2 (B). The predicted 3D structure of the LcCRHR1-1 (C) and LcCRHR1-2 (D). The 2D structure was generated by using Protter, and the 3D structure of the protein was generated using the SWISS model. Seven transmembrane domains (TM1-TM7), three extracellular (EC), and three intracellular (IC) loops are marked. N-terminal ECD is marked with a black rectangle.
To investigate the relationship between LcCRHR1s and CRHRs from different species, phylogenetic tree analysis of CRHR1 protein sequences from different species was performed using the maximum likelihood (ML) method in Mega 5.0 (Figure 3). The results showed that CRHR could be divided into two groups (CRHR1 and CRHR2), whereas the neuropeptide Y (NPY) receptors were clustered as an out-group, apart from the CRHR sequences. The two predicted LcCRHR1s clustered alongside the reported CRHR1s from other species.
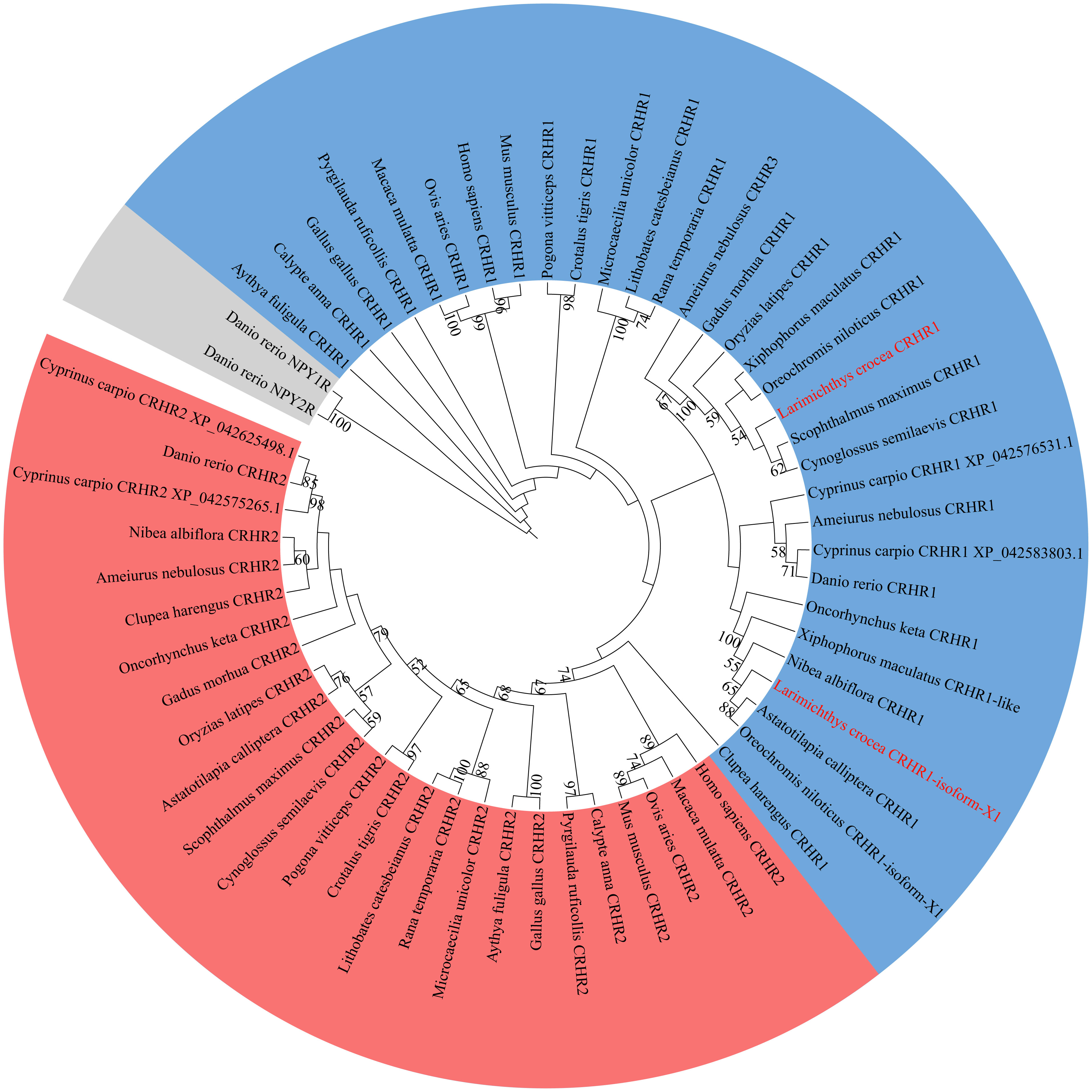
Figure 3 Phylogenetic analysis of LcCRHR1s. The phylogenetic tree was generated by the Maximum Likelihood (ML) algorithms and bootstrapped 1000 times using MEGA 5.0. Bootstrap values > 50% are indicated above branches. Colors indicate individual groups. The sequence of LcCRHR1-1 and LcCRHR1-2 are marked in red. LcCRHR1-1 is expressed as “Larimichthys crocea CRHR1” and LcCRHR1-2 is expressed as “Larimichthys crocea CRHR1-isoform-X1” in the phylogenetic tree. Sequence ID numbers for CRHRs used in phylogenetic analyses are detailed in Supplementary Table 1.
3.2 Cellular localization and internalization of LcCRHR1-1 and LcCRHR1-2
Results detected using confocal microscopy revealed significant colocalization of GFP and DiI on the cell membrane in the absence of ligand stimulation, indicating the location of these two receptors (Figure 4A). After treating cells expressing LcCRHR1-1 or LcCRHR1-2 with 100 nM LcCRH for 5 min, we found that the green fluorescent protein was originally transported into the cytoplasm in a time-dependent manner, largely aggregated in the perinuclear region, and reached its peak within 60 min (Figure 4B).
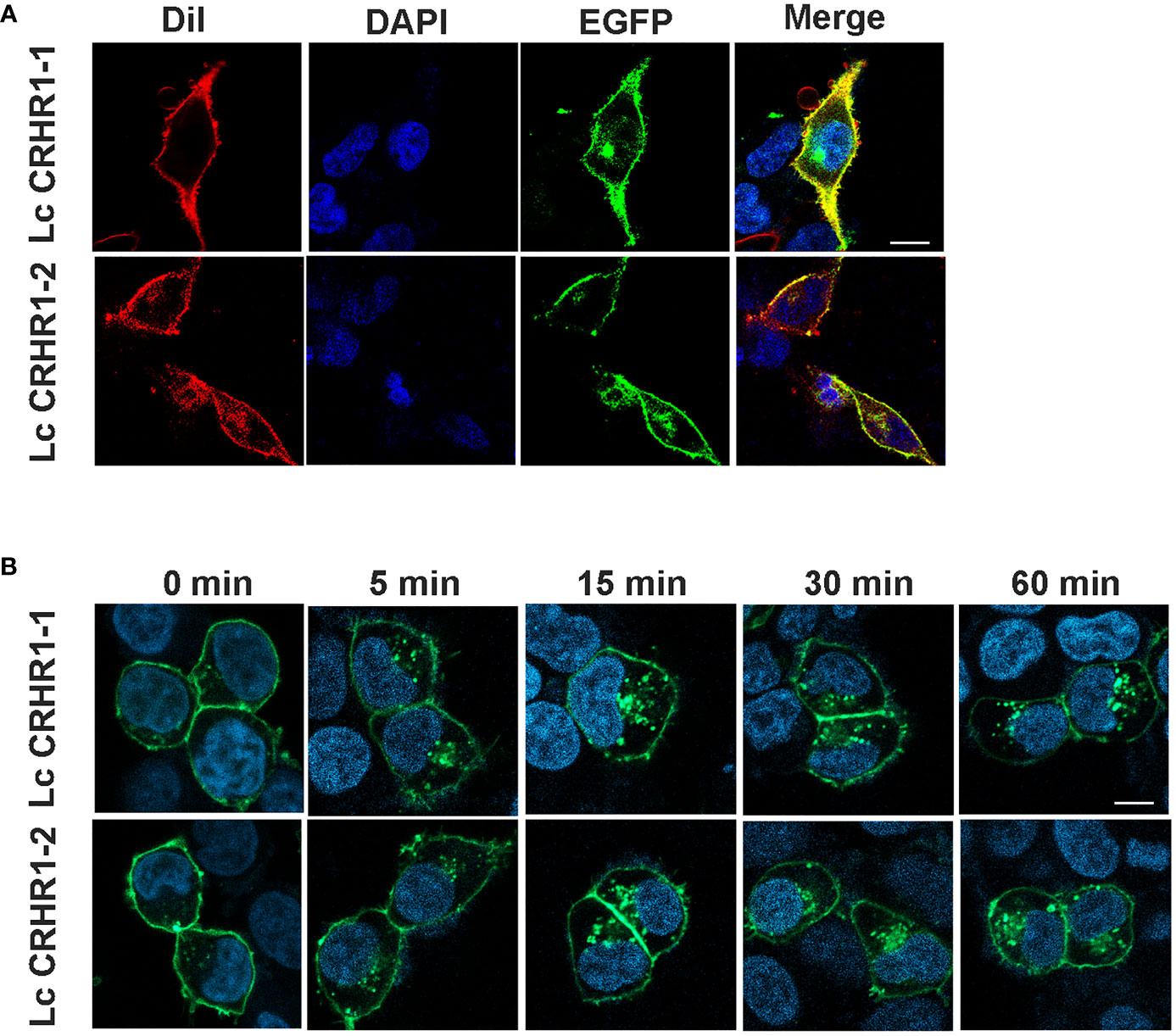
Figure 4 Confocal microscopy of HEK293 cells expressing the LcCRHR1s-EGFP fusion protein. (A) LcCRHR1-1 and LcCRHR1-2 distribution in HEK293 cells stained with membrane plasma probe (DiI) and nuclear stain (DAPI) (scale=10 μm). (B) Internalization of LcCRHR1-1 and LcCRHR1-2 in HEK293 cells. The cells transfected with LcCRHR1-1-EGFP or LcCRHR1-2-EGFP were stimulated by LcCRH (100 nM) for 0, 5, 15, 30, and 60 min (scale=15 μm). DMEM was used as a control group for 0 min. All images are representative of at least three independent experiments.
3.3 cAMP accumulation in LcCRHR1-1 or LcCRHR1-2-expressing cells stimulated by LcCRH
Previous studies have shown intracellular cAMP accumulation of the activated CRHRs in many species (Bayatti et al., 2005)). Results showed that the luciferase activity was enhanced after LcCRH stimulation in a dose-dependent manner. (Figures 5A, B). To further verify that cAMP production was induced by LcCRH through the two receptors, ELISA was performed, and the results were consistent with those obtained from the CRE/Luc system (Figures 5C, D). These results illustrated that the intracellular cAMP accumulation induced through LcCRHR1s by ligand activation, demonstrating Gαs protein-coupled cell signaling.
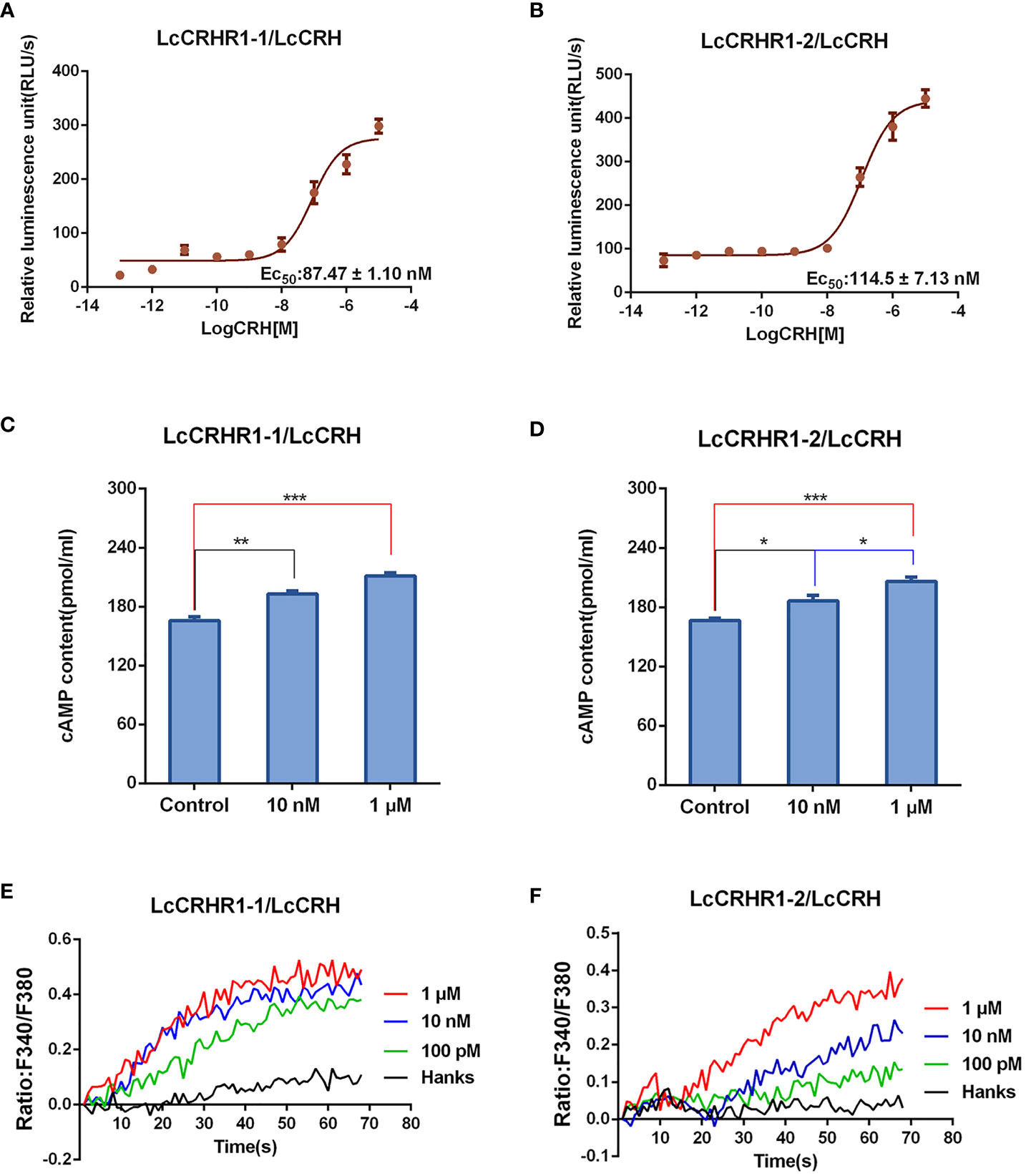
Figure 5 Functional characteristics of LcCRHR1s. (A, B) The CREB activation was induced by LcCRH in HEK293 cells expressing LcCRHR1-1 or LcCRHR1-2. CREB activation in the cells was assayed in response to different doses of LcCRH for 4 h, which indirectly reflected the level of intracellular cAMP. (C, D) cAMP accumulation in the cells expressing LcCRHR1-1 or LcCRHR1-2. Cells were stimulated with indicated concentrations of LcCRH for 15 min using the cAMP detection kits (R&D Systems). (E, F) Intracellular Ca2+ mobilization in the cells expressing LcCRHR1-1 or LcCRHR1-2 was measured in response to LcCRH after loading with Fura-2/AM. Hanks was used as the control. All data are shown as mean ± SEM from at least three independent experiments. Data were analyzed using the one‐way ANOVA, followed by Tukey’s multiple comparison tests (*, P <0.05; **, P < 0.01; ***, P < 0.001).
3.4 Ca2+ mobilization in LcCRHR1-1 or LcCRHR1-2-expressing cells activated by LcCRH
Studies have reported a secondary messenger Ca2+ cascade when CRHR1s are activated by CRH in many vertebrate species (Gutknecht et al., 2009). As illustrated in Figures 5E, F, LcCRH-induced Ca2+ mobilization occurred in a dose-dependent manner.
3.5 LcCRHR1-1 and LcCRHR1-2 mediate ERK1/2 phosphorylation upon LcCRH stimulation
After 10 min of stimulation with LcCRH (100 nM), significant ERK1/2 phosphorylation was detected in LcCRHR1-1 or LcCRHR1-2-expressing cells but not in blank or control (Figure 6A). Furthermore, treatment with LcCRH (100 nM) induced highly pronounced ERK1/2 phosphorylation in LcCRHR1-1 or LcCRHR1-2-expressing cells over time. Stimulation with LcCRH elicited transient activation kinetics, with maximal phosphorylation evident at 2-5 min in HEK293 cells expressing LcCRHR1s that returned to nearly basal levels by 15 min (Figure 6B). In addition, ligand exposure induced intracellular ERK1/2 phosphorylation within 5 min in a ligand-dose-dependent manner and an EC50 value of 16.17 ± 1.33 and 139.5 ± 18.0 nM (Figure 6C). The results demonstrated that the LcCRHR1-1 and LcCRHR1-2-mediated activation of ERK1/2 was significantly blocked by the EPAC inhibitor ESI09 but not influenced by the PKA inhibitor H89 (Figures 6D, E), confirming that MAPK cascade was initiated by LcCRH activation via the Gαs/cAMP/EPAC/MAPK signaling pathway (Figure 6F). LcCRH is a major agonist of these two receptors.
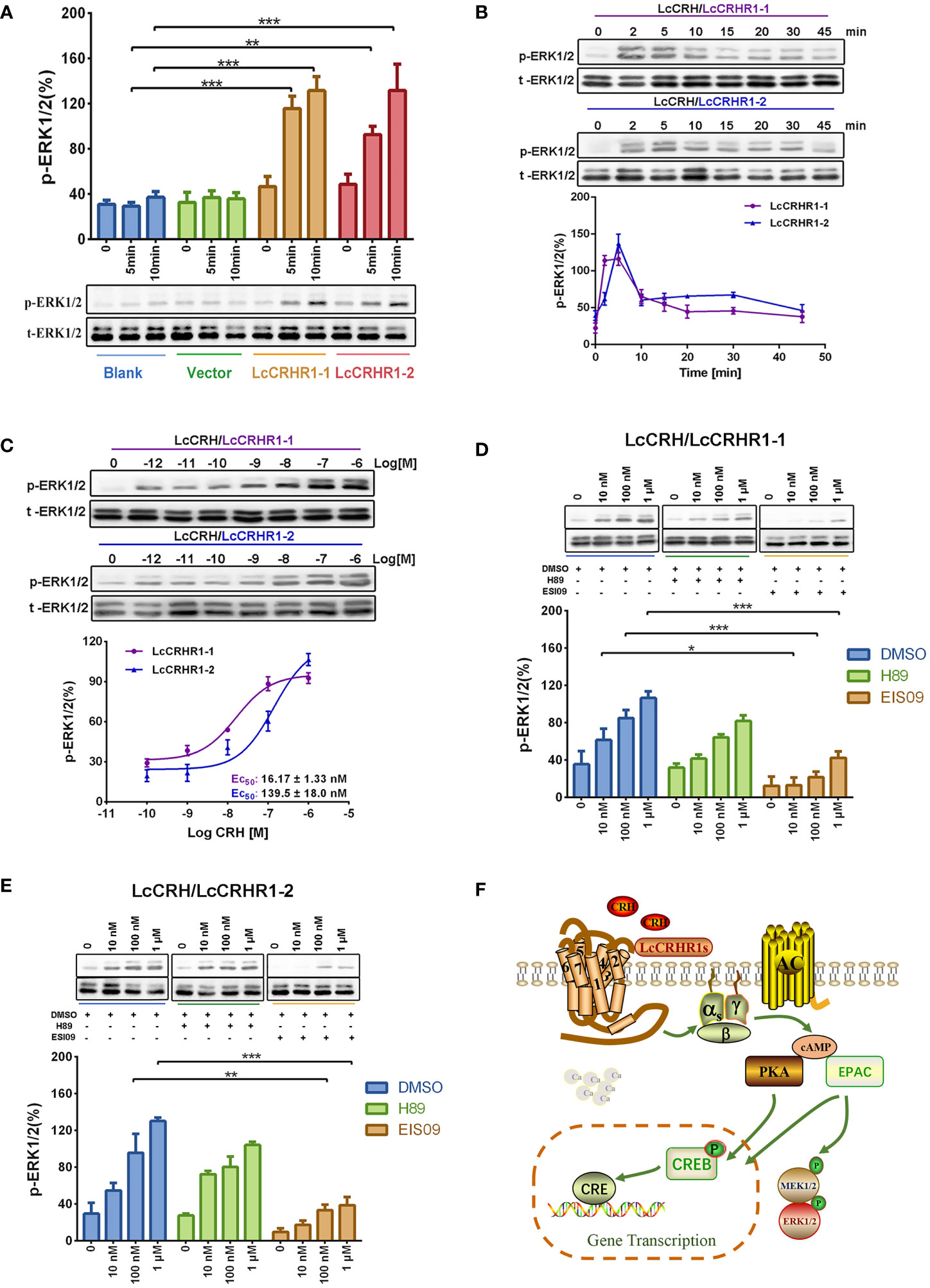
Figure 6 ERK1/2 activation mediated by LcCRHR1s. (A) Blank HEK293 cells, cells expressing EGFP-vector, and cells expressing LcCRHR1-1 or LcCRHR1-2-EGFP were stimulated with LcCRH (100 nM) for 0-10 min. (B) Time course of ERK1/2 induced by LcCRH (100 nM) stimulation in the cells expressing LcCRHR1-1 or LcCRHR1-2. The p‐ERK was normalized to loading control (total‐ERK1/2). (C) Concentration dependence of LcCRH stimulated phosphorylation of extracellular signal‐regulated kinase (ERK1/2) in the cells expressing LcCRHR1-1 or LcCRHR1-2 for 5 min. (D, E) ERK1/2 phosphorylation activated by LcCRH was blocked by PKA or EPAC inhibitors. The cells expressing LcCRHR1-1 or LcCRHR1-2-EGFP were pretreated with DMSO, PKA inhibitor (H89, 10 μM), or EPAC inhibitor (ESI09, 10 μM) for 1 h before LcCRH stimulation. (F) Schematic diagram of LcCRHR1-1 or LcCRHR1-2 activation. Ligand LcCRH binding to LcCRHR1-1 or LcCRHR1-2 activates Gαs family of heterotrimeric G proteins, which leads to Gα dissociation from the G protein subunits, activating adenylate cyclase activity and leading to intracellular cAMP accumulation and MAPK cascade. MEK1/2: MAP kinase kinase 1; MAPK: mitogen-activated protein kinase All data are shown as mean ± SEM from at least three independent experiments. Data were analyzed using the one‐way ANOVA, followed by Tukey’s multiple comparison tests (*, P < 0.05, **, P < 0.01, ***, P < 0.001).
3.6 Expression characteristics of LcCRHR1-1 and LcCRHR1-2 in L. crocea
qRT-PCR analysis revealed the expression patterns of LcCRHR1-1 and LcCRHR1-2 in various tissues of mature L. crocea. These two genes were highly expressed in the brain and ovary and ubiquitously expressed in the heart, liver, spleen, intestine, retina, and head kidney (Figure 7A). To further determine the distribution of LcCRHR1s in the ovaries of L. crocea, an anti-rabbit CRHR1 antibody was used to conduct immunohistochemical assays to reveal the detailed localization of these receptors. The results showed that LcCRHR1s were distributed in the follicular cells of the ovary of L. crocea (Figure 7B).
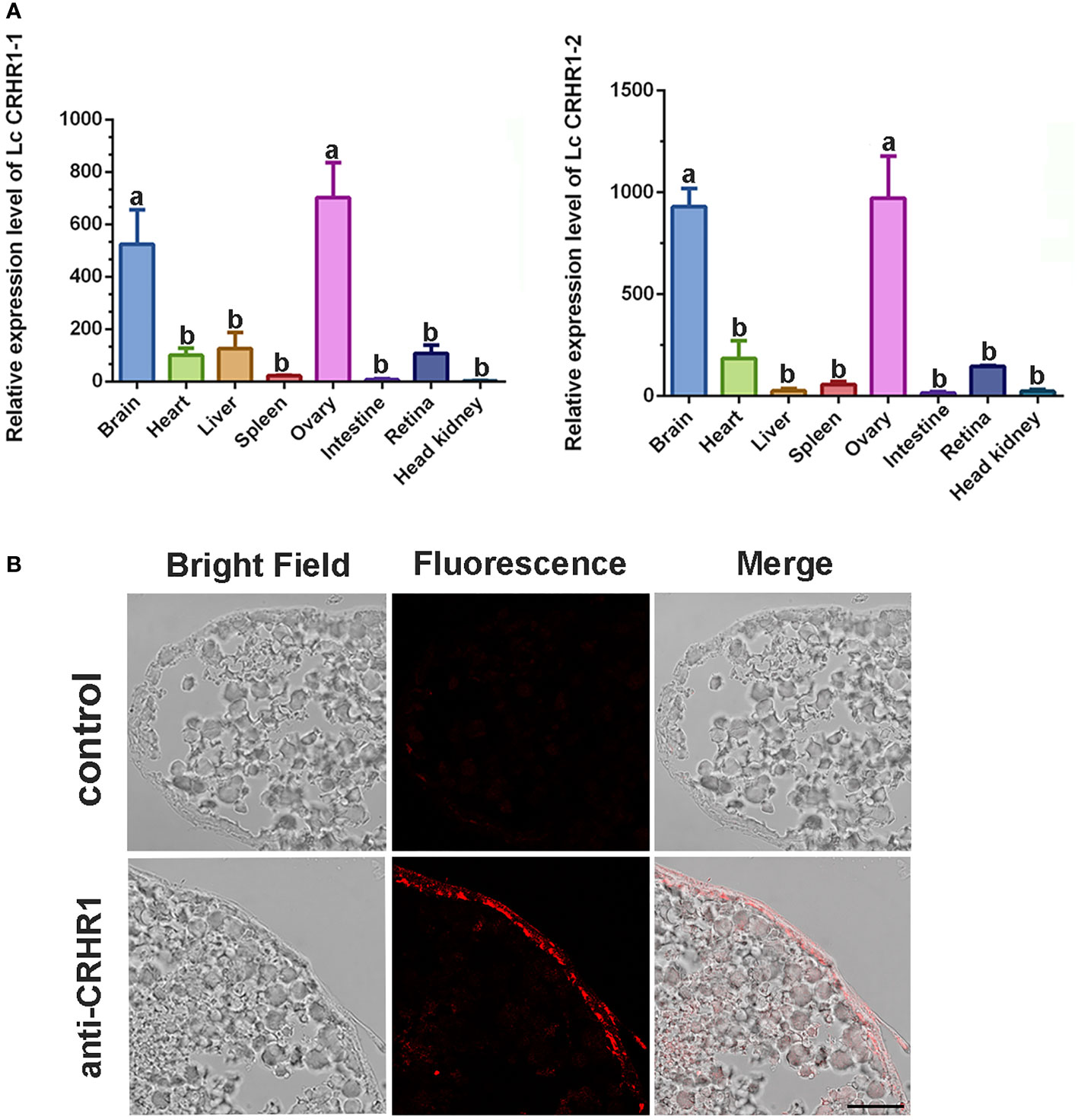
Figure 7 Distribution of LcCRHR1s in multiple tissues of L. crocea. (A) Relative expression of LcCRHR1-1 and LcCRHR1-2 in different tissues of L. crocea. Expression was normalized against the expression of the internal control gene (β-actin). Each value represents the mean ± SEM (n=3). Different lowercase letters above the bars indicate significant differences (P < 0.05) between different tissues. (B) Distribution of LcCRHR1s analyzed by immunohistochemistry in the ovary of L. crocea. Immuno-labeled CRHR1 (red) in ovary is shown by the representative photomicrographs. “anti-CRHR1” indicates that ovary sections were incubated with anti-CRHR1 antibody and bonded with the corresponding secondary antibody labeled Cy3 (fluorescence). “Control” indicates sections were incubated with PBS (scale=100 μm).
4 Discussion
CRH is the most crucial hypothalamic neuropeptide for the stress response. It induces the production of ACTH from the anterior pituitary, followed by cortisol release from the adrenal or interrenal cascade, known as the HPA/HPI axis endocrine cascade (Kim et al., 2019). The large yellow croaker (L. crocea), one of the most important mariculture species in China, exhibits seasonal reproduction; artificial breeding and the reproduction of this fish are critical for aquaculture. In this study, we systematically cloned, bioinformatically analyzed and functionally investigated the two CRHR1s in L. crocea to explore their physiological roles in teleost fish, especially in reproductive control.
This is the first study to report the cloning of two CRH receptors in L. crocea. A comparison of the exon-intron organization of LcCRHR1s indicated the distribution of 14 exons, which were structurally similar to H. sapiens, X. laevis, and D. rerio (Supplementary Figure 4). The deduced amino acid sequences indicated that they belonged to the secretin subfamily of GPCRs (Bockaert et al., 2002). Bioinformatics investigation of LcCRHR1-1 and LcCRHR1-2 revealed 24 and 21 amino acid signal peptides respectively, speculated to be associated with ligand recognition (Teichmann et al., 2012; Owji et al., 2018). Furthermore, both receptors contained seven N-linked glycosyl groups and 37 phosphorylation sites primarily involved in protein localization, function, signal transduction, and desensitization (Arnold et al., 2007; Miedlich and Abou-Samra, 2008). We also identified a short consensus repeat fold (ECD) in the N-terminal domain, which is a typical characteristic of class B1 GPCRs, in both receptors (Pioszak et al., 2008; Pal et al., 2012). The N-terminal ECD in LcCRHR1s demonstrates its ability to dock peptides, binding-related agonists, and antagonists (Liang et al., 2020). Our multiple sequence alignment showed that the two L. crocea CRH receptors had 76.1% homology and a high degree of conservation in seven transmembrane domains. And they also share the respectively 69.89% and 65.66% similarity with the genomic predicted L. crocea CRH receptor-2 (Gene ID XM_010729216.3, cloning is ongoing). These features are critical for ligand-affinity binding, membrane stability, G protein activation, and signal transduction. In addition, significant cell surface expression of these two LcCRHRs was revealed by confocal microscope, which is consistent with the above bioinformatics prediction.
To date, with the identification of CRH receptors in different species, the classification and evolutionary origin have been well elucidated (Nordström et al., 2009; On et al., 2019). There are two major CRH receptor subtypes, CRHR1 and CRHR2; only one CRHR3 from brown bullhead catfish (A. nebulosus) has been discovered thus far (Arai et al., 2001). Subsequent phylogenetic analysis of 57 homologous genes, including 14 species of fish and 13 other vertebrates, showed that they clustered into two groups (CRHR1 and CRHR2, or CRF1 and CRF2) and the two CRH receptors reported in L. crocea were grouped with CRHR1-type members, including duplicates of CRHR1 from two Percomorpha representatives, the Nile tilapia (Oreochromis niloticus) and the platyfish (Xiphophorus maculatus) (Cardoso et al., 2014). Therefore, they were referred to as LcCRHR1-1 and LcCRHR1-2 (LcCRHR1a or LcCRHR1b) respectively. Incidentally, the only catfish CRHR3 is also included in the CRHR1 subfamily, as revealed in a previous study (Cardoso et al., 2003). Because the two L. crocea CRH receptors are two different genes (located on different chromosomes based on genomic data), both structurally resemble each other, similar to the catfish cfCRHR1 and cfCRHR3. Moreover, these two L. crocea CRH receptors, cfCRHR1 and cfCRHR3 were phylogenetically clustered in the CRHR1 group, and we tended to nominate the two receptors as CRHR1 subtypes and propose cfCRHR3 in the same situation. If this hypothesis could be confirmed by future research on other teleost fish, it would further support whole genome duplication (WGD) in vertebrate evolution, particularly the second WGD in fish (Putnam et al., 2008; Cardoso et al., 2016). And synteny analysis on genomic regions needs to be conducted to further explore CRHR gene evolution in teleosts and determine the impact of the 3R (teleost-specific WGD) event.
Vertebrate CRH-like peptides can be divided into the CRH/Ucn I and Ucn II/Ucn III subgroups (Cardoso et al., 2016), and CRHR1 has a high affinity for both CRH and Ucn I, whereas Ucn II and Ucn III are unique CRHR2 ligands (Takefuji and Murohara, 2019). The results of LcCRHR1s activation by LcCRH using cAMP assay demonstrated that both receptors couple to the Gαs protein, stimulate the adenylate cyclase activity, and increase intracellular cAMP concentration in a CRH dose-dependent manner as in other teleost characterized CRH receptors (Arai et al., 2001; Pohl et al., 2001). In addition, the two receptors had nanomolar EC50 affinity with LcCRH and LcUcn I; however, LcUcn III could not achieve activation within a comparable range (unpublished data), which is a classic feature of CRHR1, supporting that the two identified L. crocea CRH receptors belong to the CRHR1 subfamily (Chen et al., 1993; Hsu and Hsueh, 2001; Lewis et al., 2001). Furthermore, calcium fluxes were also detected in a ligand dose-dependent manner after LcCRHR1s activation, which has been frequently reported in model organisms (Gutknecht et al., 2009; Brar, 2017) but rarely in fish CRHRs. This Ca2+ comes possibly from the Gαq/PLC signaling by LcCRHR1 activation as that in hamsters and humans (Fazal et al., 1998; Wiesner, 2003) or likely from the Gαs/EPAC pathway or the extracellular pool induced by membrane Ca2+ channel opening (Kang et al., 2003; Dautzenberg et al., 2004; Gutknecht et al., 2009), all of which require more evidence for clarification, particularly in the fish cells.
MAPK activation is a key effector of the biological actions of the CRH/CRHR system in numerous cell models (Grammatopoulos, 2012). In this study, we detected the increase in cAMP level by the CRE/luciferase reporter system as studied in humans (McEvoy et al., 2002) and the ERK1/2 phosphorylation of MAPK cascade in a ligand dose‐dependent manner following LcCRHR1-1 or LcCRHR1-2 activation. Previous study showed ERK1/2 activation could be significantly inhibited by the EPAC inhibitor ESI09 (Zhu et al., 2015), which is re-verified in our result. But our data indicate that ERK phosphorylation was not influenced by the PKA inhibitor H89. This result is inconsistent with the traditional Gαs/cAMP/PKA/ERK cascade, but they indicate that LcCRHR1s activate ERK1/2 through the Gαs/cAMP/EPAC/ERK cascade. As a relatively new signal pathway (Bos, 2003; Maymó et al., 2012; Robichaux and Cheng, 2018), it has been reported involved in CRF-induced ERK phosphorylation by CRF1 receptor activation in AtT20 cells (Van Kolen et al., 2010). This is a new finding for LcCRH receptor signal transduction and requires further verification in fish cellular systems.
In vertebrates, CRH and its related peptides play essential roles in reproductive, neuropsychiatric, gastrointestinal, and immunological functions, as well as in the development of tumors, and in the well-known adaptation response to external challenges and stress (Suda et al., 2004; Phumsatitpong et al., 2020). CRHR1 is distributed throughout the central nervous system and peripheral tissues and mediates diverse physiological functions (Chen, 2022). According to LcCRHR1-1 or LcCRHR1-2 gene expression data, both these receptors are abundantly expressed in the brain, which is comparable with data from other fish, including cichlid fish (A. burtoni) (Chen and Fernald, 2008), common carp (C. carpio) (Huising, 2004), takifugu rubripes (Fugu rubripes) (Cardoso et al., 2003), chum salmon (Oncorhynchus keta) (Pohl et al., 2001), and catfish (A. nebulosus) (Arai et al., 2001). They have also been proposed to regulate reproductive processes directly at the level of the gonads and secondary sex organs. For instance, Studies have reported that CRH systems are expressed in ovarian tissue in mammals (Asakura et al., 1997; Dinopoulou et al., 2013) and in teleosts (chum salmon: Pohl et al., 2001; fugu: Cardoso et al., 2003; Astatotilapia: Chen and Fernald, 2008; olive flounder: Zhou et al., 2019). In addition to the CRH-mediated estrogen production in human placental trophoblasts (You et al., 2006), a recent study also showed that the highest CRHα mRNA levels were detected in mature follicles of D. rerio, and the highest CRHR1 mRNA levels were detected in mid-vitellogenin (MV) and early vitellogenic (EV) stages, indicating their critical role in reproductive development (Zhou et al., 2021). In this study, our immunohistochemical data validated the expression of LcCRHR1-1 or LcCRHR1-2 in ovarian follicle cells, which may imply and support the view that the CRH/CRHR system has a considerably broader range of physiological roles in fish. Therefore, further research is required to investigate the role of the CRH/CRHR neuroendocrine system in regulating fish reproduction.
5 Conclusions
In summary, the full-length coding sequences of two candidate CRH receptors were screened using genomic data in L. crocea. Their protein structures were constructed, and a phylogenetic tree was established using bioinformatics, which proved both receptors, LcCRHR1-1, and LcCRHR1-2, belong to the CRHR1 subtypes. When activated by LcCRH, these two receptors are internalized and boost intracellular cAMP and Ca2+ levels while activating the MAPK signaling pathway in a CRH dose-dependent manner. These results confirm the biological activity of the two candidate receptors. Not only are these two receptors highly expressed in the brain, but expression was also detected in ovarian follicular cells implying that they are potentially involved in sexual maturation and reproduction. Further studies are still needed to confirm and clarify the biological roles and the evolution origin for these important CRH receptors.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
JY, TW, and XL contributed to experimental design, statistical analysis, and manuscript preparation. XL, JF, and GZ performed the experiments. ZJ contributed to the experimental preparation and data analysis. JY, TW, XX, and JW contributed to the revision of the manuscript. JY and TW contributed to funding acquisition. All the authors contributed to the manuscript and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 42076130 and 32072952), “Pioneer” and “Leading Goose” R&D Program of Zhejiang (2022C02040) and the Science and Technology Program of Zhoushan (2019C21020).
Conflict of interest
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1184792/full#supplementary-material
References
Arai M., Assil I. Q., Abou-Samra A. B. (2001). Characterization of three corticotropin-releasing factor receptors in catfish: a novel third receptor is predominantly expressed in pituitary and urophysis. Endocrinology 142 (1), 446–454. doi: 10.1210/endo.142.1.7879
Arnold J. N., Wormald M. R., Sim R. B., Rudd P. M., Dwek R. A. (2007). The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25 (1), 21–50. doi: 10.1146/annurev.immunol.25.022106.141702
Asakura H., Zwain I. H., Yen S. S. C. (1997). Expression of genes encoding corticotropin-releasing factor (CRF), type 1 CRF receptor, and CRF-binding protein and localization of the gene products in the human ovary. J. Clin. Endocrinol. Metab. 82 (8), 2720–2725. doi: 10.1210/jcem.82.8.4119
Bale T. L., Picetti R., Contarino A., Koob G. F., Vale W. W., Lee K.-F. (2002). Mice deficient for both corticotropin-releasing factor receptor 1 (CRFR1) and CRFR2b have an impaired stress response and display sexually dichotomous anxiety-like behavior. J. Neurosci. 22 (1), 193–199. doi: 10.1523/jneurosci.22-01-00193.2002
Bayatti N., Hermann H., Lutz B., Behl C. (2005). Corticotropin-releasing hormone-mediated induction of iintracellular signaling pathways and brain-derived neurotrophic factor expression is inhibited by the activation of the endocannabinoid system. Endocrinology 146 (3), 1205–1213. doi: 10.1210/en.2004-1154
Bockaert J., Claeysen S., Bécamel C., Sylvie P., Dumuis A. (2002). G Protein-coupled receptors: dominant players in cell–cell communication. Int. Rev. Cytol. 212, 63–136. doi: 10.1016/s0074-7696(01)12004-8
Bos J. L. (2003). Epac: a new cAMP target and new avenues in cAMP research. Nat. Rev. Mol. Cell Biol. 4 (9), 733–738. doi: 10.1038/nrm1197
Brar B. (2017). Corticotropin-releasing hormone receptor signaling. Ref. Modul. Neurosci. Biobehav. Psychol, 1–21. doi: 10.1016/B978-0-12-809324-5.03380-0
Cardoso J. C. R., Bergqvist C. A., Félix R. C., Larhammar D. (2016). Corticotropin-releasing hormone family evolution: five ancestral genes remain in some lineages. J. Mol. Endocrinol. 57 (1), 73–86. doi: 10.1530/jme-16-0051
Cardoso J. C., Felix R. C., Bergqvist C. A., Larhammar D. (2014). New insights into the evolution of vertebrate CRH (corticotropin-releasing hormone) and invertebrate DH44 (diuretic hormone 44) receptors in metazoans. Gen. Comp. Endocrinol. 209, 162–170. doi: 10.1016/j.ygcen.2014.09.004
Cardoso J. C. R., Power D. M., Elgar G., Clark M. S. (2003). Isolation and characterisation of the corticotropin releasing factor receptor 1 (CRFR1) gene in a teleost fish. Fugu rubripes. DNA Sequence. 14 (3), 215–218. doi: 10.1080/1042517031000112624
Chen X. (2022). Functional diversity of corticotropin-releasing hormone. Nat. Rev. Endocrinol. 18, 333–333. doi: 10.1038/s41574-022-00663-w
Chen C.-C., Fernald R. D. (2008). Sequences, expression patterns and regulation of the corticotropin-releasing factor system in a teleost. Gen. Comp. Endocrinol. 157 (2), 148–155. doi: 10.1016/j.ygcen.2008.04.003
Chen R., Lewis K. A., Perrin M. H., Vale W. W. (1993). Expression cloning of a human corticotropin-releasing-factor receptor. Proc. Natl. Acad. Sci. 90 (19), 8967–8971. doi: 10.1073/pnas.90.19.8967
Chen S., Su Y., Hong W. (2018). Aquaculture of the large yellow croaker. aquaculture in China. Success stories modern trends., 297–308. doi: 10.1002/9781119120759.ch3_10
Dautzenberg F. M., Gutknecht E., der Linden I. V., Olivares-Reyes J. A., Dürrenberger F., Hauger R. L. (2004). Cell-type specific calcium signaling by corticotropin-releasing factor type 1 (CRF1) and 2a (CRF2(a)) receptors: phospholipase c-mediated responses in human embryonic kidney 293 but not SK-N-MC neuroblastoma cells. Biochem. Pharmacol. 68 (9), 1833–1844. doi: 10.1016/j.bcp.2004.07.013
Dautzenberg F. M., Hauger R. L. (2002). The CRF peptide family and their receptors: yet more partners discovered. Trends Pharmacol. Sci. 23 (2), 71–77. doi: 10.1016/s0165-6147(02)01946-6
Denver R. J. (2009). Structural and functional evolution of vertebrate neuroendocrine stress systems. Ann. New York Acad. Sci. 1163 (1), 1–16. doi: 10.1111/j.1749-6632.2009.04433.x
Dinopoulou V., Partsinevelos G. A., Mavrogianni D., Anagnostou E., Drakakis P., Makrigiannakis A., et al. (2013). The effect of CRH and its inhibitor, antalarmin, on in vitro growth of preantral mouse follicles, early embryo development, and steroidogenesis. Endocrinology 154 (1), 222–231. doi: 10.1210/en.2012-1838
Fazal N., Slominski A., Choudhry M. A., Wei E. T., Sayeed M. M. (1998). Effect of CRF and related peptides on calcium signaling in human and rodent melanoma cells. FEBS Lett. 435 (2-3), 187–190. doi: 10.1016/s0014-5793(98)01067-9
Fredriksson R., Sjödin P., Larson E. T., Conlon J. M., Larhammar D. (2006). Cloning and characterization of a zebrafish Y2 receptor. Regul. Peptides. 133 (1-3), 32–40. doi: 10.1016/j.regpep.2005.09.013
Füzesi T., Daviu N., Wamsteeker Cusulin J. I., Bonin R. P., Bains J. S. (2016). Hypothalamic CRH neurons orchestrate complex behaviours after stress. Nat. Commun. 7, 11937. doi: 10.1038/ncomms11937
Grammatopoulos D. K. (2012). Insights into mechanisms of corticotropin-releasing hormone receptor signal transduction. Br. J. Pharmacol. 166 (1), 85–97. doi: 10.1111/j.1476-5381.2011.01631.x
Grammatopoulos D. K., Chrousos G. P. (2002). Functional characteristics of CRH receptors and potential clinical applications of CRH-receptor antagonists. Trends Endocrinol. Metab. 13 (10), 436–444. doi: 10.1016/s1043-2760(02)00670-7
Graziani G., Tentori L., Portarena I., Barbarino M., Tringali G., Pozzoli G., et al. (2002). CRH inhibits cell growth of human endometrial adenocarcinoma cells via CRH-receptor 1-mediated activation of cAMP-PKA pathway. Endocrinology 143 (3), 807–813. doi: 10.1210/endo.143.3.8694
Gutknecht E., van der Linden I., Van Kolen K., Verhoeven K. F. C., Vauquelin G., Dautzenberg F. M. (2009). Molecular mechanisms of corticotropin-releasing factor receptor-induced calcium signaling. Mol. Pharmacol. 75 (3), 648–657. doi: 10.1124/mol.108.050427
Hemley C., McCluskey A., Keller P. (2007). Corticotropin releasing hormone - a GPCR drug target. Curr. Drug Targets 8 (1), 105–115. doi: 10.2174/138945007779315542
Henckens M. J. A. G., Deussing J. M., Chen A. (2016). Region-specific roles of the corticotropin-releasing factor–urocortin system in stress. Nat. Rev. Neurosci. 17 (10), 636–651. doi: 10.1038/nrn.2016.94
Hillhouse E. W., Grammatopoulos D. K. (2006). The molecular mechanisms underlying the regulation of the biological activity of corticotropin-releasing hormone receptors:implications for physiology and pathophysiology. Endocr. Rev. 27 (3), 260–286. doi: 10.1210/er.2005-0034
Hsu S. Y., Hsueh A. J. W. (2001). Human stresscopin and stresscopin-related peptide are selective ligands for the type 2 corticotropin-releasing hormone receptor. Nat. Med. 7 (5), 605–611. doi: 10.1038/87936
Huising M. (2004). Structural characterisation of a cyprinid (Cyprinus carpio l.) CRH, CRH-BP and CRH-R1, and the role of these proteins in the acute stress response. J. Mol. Endocrinol. 32 (3), 627–648. doi: 10.1677/jme.0.0320627
Inda C., Armando N. G., dos Santos Claro P. A., Silberstein S. (2017). Endocrinology and the brain: corticotropin-releasing hormone signaling. Endocr. Connect. 6 (6), R99–R120. doi: 10.1530/ec-17-0111
Kang G., Joseph J. W., Chepurny O. G., Monaco M., Wheeler M. B., Bos J. L., et al. (2003). Epac-selective cAMP analog 8-pCPT-2′-O-Me-cAMP as a stimulus for Ca2+-induced Ca2+ release and exocytosis in pancreatic β-cells. J. Biol. Chem. 278 (10), 8279–8285. doi: 10.1074/jbc.m211682200
Kiapekou E., Zapanti E., Mastorakos G., Loutradis D. (2010). Update on the role of ovarian corticotropin-releasing hormone. Ann. New York Acad. Sci. 1205 (1), 225–229. doi: 10.1111/j.1749-6632.2010.05685.x
Kim J. S., Han S. Y., Iremonger K. J. (2019). Stress experience and hormone feedback tune distinct components of hypothalamic CRH neuron activity. Nat. Commun. 10 (1), 5696. doi: 10.1038/s41467-019-13639-8
Lederis K. A., Letter A., McMaster D., Moore G., Schlesinger D. (1982). Complete amino acid sequence of urotensin I, a hypotensive and corticotropin-releasing neuropeptide from catostomus. Science 218 (4568), 162–165. doi: 10.1126/science.6981844
Lewis K., Li C., Perrin M. H., Blount A., Kunitake K., Donaldson C., et al. (2001). Identification of urocortin III, an additional member of the corticotropin-releasing factor (CRF) family with high affinity for the CRF2 receptor. Proc. Natl. Acad. Sci. 98 (13), 7570–7575. doi: 10.1073/pnas.121165198
Liang Y.-L., Belousoff M. J., Zhao P., Koole C., Fletcher M. M., Truong T. T., et al. (2020). Toward a structural understanding of class b GPCR peptide binding and activation. Mol. Cell. 77 (3), 656–668. doi: 10.1016/j.molcel.2020.01.012
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lovejoy D. A., Chang B. S., Lovejoy N. R., del Castillo J. (2014). CRH/CRH receptors. J. Mol. Endocrinol. 52, T43–T60. doi: 10.1530/jme-13-0238
Ma S., Shen Q., Zhao L.-H., Mao C., Zhou X. E., Shen D.-D., Xu H. (2020). Molecular basis for hormone recognition and activation of corticotropin-releasing factor receptors. E.Mol. Cell. 77 (3), 669–680. doi: 10.1016/j.molcel.2020.01.013
Manuel R., Metz J. R., Flik G., Vale W. W., Huising M. O. (2014). Corticotropin-releasing factor-binding protein (CRF-BP) inhibits CRF-and urotensin-i-mediated activation of CRF receptor-1 and-2 in common carp. Gen. Comp. Endocrinol. 202, 69–75. doi: 10.1016/j.ygcen.2014.04.010
Maymó J. L., Pérez Pérez A., Maskin B., Dueñas J. L., Calvo J. C., Sánchez Margalet V., et al. (2012). The alternative Epac/cAMP pathway and the MAPK pathway mediate hCG induction of leptin in placental cells. PloS One 7 (10), e46216. doi: 10.1371/journal.pone.0046216
McEvoy A. N., Bresnihan B., Fitzgerald O., Murphy E. P. (2002). Corticotropin-releasing hormone signaling in synovial tissue vascular endothelium is mediated through the cAMP/CREB pathway. Ann. New York Acad. Sci. 966 (1), 119–130. doi: 10.1111/j.1749-6632.2002.tb04209.x
Melón L., Maguire J. (2016). GABAergic regulation of the HPA and HPG axes and the impact of stress on reproductive function. J. Steroid Biochem. Mol. Biol. 160, 196–203. doi: 10.1016/j.jsbmb.2015.11.019
Miedlich S. U., Abou-Samra A. B. (2008). Eliminating phosphorylation sites of the parathyroid hormone receptor type 1 differentially affects stimulation of phospholipase c and receptor internalization. Am. J. Physiology-Endocrinology Metab. 295 (3), E665–E671. doi: 10.1152/ajpendo.00036.2008
Montecucchi P. C., Anastasi A., de Castiglione R., Erspamer V. (1980). Isolation and amino acid composition of sauvagine: an active polypeptide from methanol extracts of the skinof the south American frog Phyllomedusa sauvagei. Int. J. Pept. Protein Res. 16 (3), 191–199. doi: 10.1111/j.1399-3011.1980.tb02952.x
Nordstrom K. J. V., Lagerstrom M. C., Waller L. M. J., Fredriksson R., Schioth H. B. (2009). The secretin GPCRs descended from the family of adhesion GPCRs. Mol. Biol. Evol. 26 (1), 71–84. doi: 10.1093/molbev/msn228
On J. S. W., Arokiaraj A. W. R., Chow B. K. C. (2019). Molecular evolution of CRH and CRHR subfamily before the evolutionary origin of vertebrate. Peptides 120, 170087. doi: 10.1016/j.peptides.2019.04.014
Owji H., Nezafat N., Negahdaripour M., Hajiebrahimi A., Ghasemi Y. (2018). A comprehensive review of signal peptides: structure, roles, and applications. Eur. J. Cell Biol. 97 (6), 422–441. doi: 10.1016/j.ejcb.2018.06.003
Pal K., Melcher K., Xu H. E. (2012). Structure and mechanism for recognition of peptide hormones by class b G-protein-coupled receptors. Acta Pharmacologica Sinica. 33 (3), 300–311. doi: 10.1038/aps.2011.170
Pangalos M. N., Davies C. H. (2002). Corticotropin releasing factor receptors. Understanding G protein-coupled receptors and their role in the CNS (Oxford University Press). 25, 505–526
Petraglia F., Imperatore A., Challis J. R. G. (2010). Neuroendocrine mechanisms in pregnancy and parturition. Endocr. Rev. 31 (6), 783–816. doi: 10.1210/er.2009-0019
Phumsatitpong C., De Guzman R. M., Zuloaga D. G., Moenter S. M. (2020). A CRH receptor type 1 agonist increases GABA transmission to GnRHneurons in a circulating-estradiol-dependent manner. Endocrinology 161 (11). doi: 10.1210/endocr/bqaa140
Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008). Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1. J. Biol. Chem. 283 (47), 32900–32912. doi: 10.1074/jbc.m805749200
Pohl S., Darlison M. G., Clarke W. C., Lederis K., Richter D. (2001). Cloning and functional pharmacology of two corticotropin-releasing factor receptors from a teleost fish. Eur. J. Pharmacol. 430 (2-3), 193–202. doi: 10.1016/s0014-2999(01)01391-7
Popov S. V., Prokudina E. S., Mukhomedzyanov A. V., Naryzhnaya N. V., Ma H., Zurmanova J. M., et al. (2021). Cardioprotective and vasoprotective effects of corticotropin-releasing hormone and urocortins: receptors and signaling. J. Cardiovasc. Pharmacol. Ther. 26 (6), 575–584. doi: 10.1177/1074248420985301
Putnam N. H., Butts T., Ferrier D. E. K., Furlong R. F., Hellsten U., Kawashima T., et al. (2008). The amphioxus genome and the evolution of the chordate karyotype. Nature 453 (7198), 1064–1071. doi: 10.1038/nature06967
Raftogianni A., Roth L. C., García-González D., Bus T., Kühne C., Monyer H., et al. (2018). Deciphering the contributions of CRH receptors in the brain and pituitary to stress-induced inhibition of the reproductive axis. Front. Mol. Neurosci. 11. doi: 10.3389/fnmol.2018.00305
Reyes T. M., Lewis K., Perrin M. H., Kunitake K. S., Vaughan J., Arias C. A., et al. (2001). Urocortin II: a member of the corticotropin-releasing factor (CRF) neuropeptide family that is selectively bound by type 2 CRF receptors. Proc. Natl. Acad. Sci. 98 (5), 2843–2848. doi: 10.1073/pnas.051626398
Robichaux W. G., Cheng X. (2018). Intracellular cAMP sensor EPAC: physiology, pathophysiology, and therapeutics development. Physiol. Rev. 98 (2), 919–1053. doi: 10.1152/physrev.00025.2017
Smith G. W., Aubry J.-M., Dellu F., Contarino A., Bilezikjian L. M., Gold L. H., et al. (1998). Corticotropin releasing factor receptor 1–deficient mice display decreased anxiety, impaired stress response, and aberrant neuroendocrine development. Neuron. 20 (6), 1093–1102. doi: 10.1016/s0896-6273(00)80491-2
Suda T., Kageyama K., Sakihara S., Nigawara T. (2004). Physiological roles of urocortins, human homologues of fish urotensin I, and their receptors. Peptides 25 (10), 1689–1701. doi: 10.1016/j.peptides.2004.03.027
Takefuji M., Murohara T. (2019). Corticotropin-releasing hormone family and their receptors in the cardiovascular system. Circ. J. 83 (2), 261–266. doi: 10.1253/circj.cj-18-0428
Teichmann A., Rutz C., Kreuchwig A., Krause G., Wiesner B., Schülein R. (2012). The pseudo signal peptide of the corticotropin-releasing factor receptor type 2A prevents receptor oligomerization. J. Biol. Chem. 287 (32), 27265–27274. doi: 10.1074/jbc.m112.360594
Vale W., Spiess J., Rivier C., Rivier J. (1981). Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science 213 (4514), 1394–1397. doi: 10.1126/science.6267699
Van Kolen K., Verstraeten K., Royaux I., De Hoogt R., Gutknecht E., Peeters P. J. (2010). Corticotropin releasing factor-induced ERK phosphorylation in AtT20 cells occurs via a cAMP-dependent mechanism requiring EPAC2. Neuropharmacology 58 (1), 135–144. doi: 10.1016/j.neuropharm.2009.06.022
Vaughan J., Donaldson C., Bittencourt J., Perrin M. H., Lewis K., Sutton S., et al. (1995). Urocortin, a mammalian neuropeptide related to fish urotensin I and to corticotropin-releasing factor. Nature 378 (6554), 287–292. doi: 10.1038/378287a0
Wang T., Liang J., Xiang X., Chen X., Zhang B., Zhou N., et al. (2019). Pharmacological characterization, cellular localization and expression profile of NPY receptor subtypes Y2 and Y7 in large yellow croaker, Larimichthys crocea. comparative biochemistry and physiology part b. Biochem. Mol. Biol. 238, 110347. doi: 10.1016/j.cbpb.2019.110347
Wang T., Zhou C., Yuan D., Lin F., Chen H., Wu H., et al. (2014). Schizothorax prenanti corticotropin-releasing hormone (CRH): molecular cloning, tissue expression, and the function of feeding regulation. Fish Physiol. Biochem. 40 (5), 1407–1415. doi: 10.1007/s10695-014-9935-6
Wiesner B. (2003). Intracellular calcium measurements of single human skin cells after stimulation with corticotropin-releasing factor and urocortin using confocal laser scanning microscopy. J. Cell Sci. 116 (7), 1261–1268. doi: 10.1242/jcs.00301
Wypior G., Jeschke U., Kurpisz M., Szekeres-Bartho J. (2011). Expression of CRH, CRH-related peptide and CRH receptor in the ovary and potential CRH signaling pathways. J. Reprod. Immunol. 90 (1), 67–73. doi: 10.1016/j.jri.2011.04.009
Yang J., Huang H., Yang H., He X., Jiang X., Shi Y., et al. (2013). Specific activation of the G protein-coupled receptor BNGR-A21 by the neuropeptide corazon from the silkworm, Bombyx mori, dually couples to the gq and gs signaling cascades. J. Biol. Chem. 288 (17), 11662–11675. doi: 10.1074/jbc.m112.441675
You X., Yang R., Tang X., Gao L., Ni X. (2006). Corticotropin-releasing hormone stimulates estrogen biosynthesis in cultured human placental trophoblasts1. Biol. Reprod. 74 (6), 1067–1072. doi: 10.1095/biolreprod.105.049361
Zalachoras I., Astori S., Meijer M., Grosse J., Zanoletti O., Suduiraut I. G., et al. (2022). Opposite effects of stress on effortful motivation in high and low anxiety are mediated by CRHR1 in the VTA. Sci. Adv. 8, eabj9019. doi: 10.1126/sciadv.abj9019
Zhou H., Chen A., Lu W. (2021). Corticotropin-releasing hormone reduces basal estradiol production in zebrafish follicular cells. Mol. Cell. Endocrinol. 527, 111222. doi: 10.1016/j.mce.2021.111222
Zhou H., Ge C., Chen A., Lu W. (2019). Dynamic expression and regulation of urotensin I and corticotropin-releasing hormone receptors in ovary of olive flounder paralichthys olivaceus. Front. Physiol. 10. doi: 10.3389/fphys.2019.01045
Keywords: stress, reproduction, neuroendocrine, Larimichthys crocea, ovary, corticotropin-releasing hormone receptor
Citation: Liu X, Feng J, Jiang Z, Zhang G, Xu X, Wang J, Yang J and Wang T (2023) Functional characterization of two corticotropin-releasing hormone receptors in Larimichthys crocea. Front. Mar. Sci. 10:1184792. doi: 10.3389/fmars.2023.1184792
Received: 12 March 2023; Accepted: 03 May 2023;
Published: 17 May 2023.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Geng Qin, Chinese Academy of Sciences (CAS), ChinaKarine Rousseau, Muséum National d’Histoire Naturelle, France
Copyright © 2023 Liu, Feng, Jiang, Zhang, Xu, Wang, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Yang, eWFuZ2ppbmd3ZW5AempvdS5lZHUuY24=; Tianming Wang, d2FuZ3RpYW5taW5nQHpqb3UuZWR1LmNu
 Xue Liu
Xue Liu Jiaqian Feng
Jiaqian Feng Jingwen Yang
Jingwen Yang Tianming Wang
Tianming Wang