
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 May 2023
Sec. Marine Biotechnology and Bioproducts
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1183744
This article is part of the Research Topic Biological Macromolecules from Marine Organisms: Isolation, Characterization and Pharmacological Activities View all 6 articles
 Jianqing Wen1,2
Jianqing Wen1,2 Jinyu Gong2,3
Jinyu Gong2,3 Pengwei Li1
Pengwei Li1 Penghui Deng1,2
Penghui Deng1,2 Mengsi Sun4
Mengsi Sun4 Yujie Wu4
Yujie Wu4 Chenxi Tian2,3
Chenxi Tian2,3 Hao Wang1*
Hao Wang1* Yunchen Bi1,2,5*
Yunchen Bi1,2,5*Elasmobranchs are crucial for comparative studies of evolution, as they belong to the most ancient vertebrate lineages that survived numerous extinction events and persist until today. The immunoglobulin new antigen receptor (IgNAR) found in sharks and heavy-chain-only antibody (HCAb) found in camelidae are products of convergent evolution. Although it was previously believed that IgNAR emerged 220 million years ago, before the divergence of sharks and skates, there is limited evidence to support this. In this study, we provide data supporting the existence of IgNAR in the ocellate spot skate (Okamejei kenojei) mononuclear cell transcriptome and peripheral blood serum. Additionally, we characterize the germline gene configuration of the ocellate spot skate IgNAR V domain. The ocellate spot skate IgNAR structure prediction and VNAR crystal structure exhibit high similarity to their shark counterparts. These data strongly suggest that IgNAR in both sharks and skates share a common ancestor. Sequencing of the ocellate spot skate VNAR repertoire provided crucial data for further understanding of the IgNAR generation. Notably, we discovered that approximately 99% of the ocellate spot skate VNARs belonged to type IV. This represents an exceptionally high proportion of type IV within the VNAR repertoire, which has not been documented in previously studied elasmobranchs. This unique characteristic of the ocellate spot skate VNAR adds essential structural diversity to the naïve VNAR library from elasmobranchs and could potentially benefit the development of pharmaceutical drugs.
Cartilaginous fish are evolutionarily significant as they are among the oldest vertebrate species. Cartilaginous fish are believed to have evolved 450 million years ago and represent one of the earliest vertebrate groups with an adaptive immune system (Rumfelt et al., 2004b). Elasmobranchs, a major lineage in the cartilaginous fish tree, consists of two subclasses: Batoidea (skates, rays, and sawfish) and Selachii (sharks). The ancient adaptive immune system of elasmobranchs comprises a variety of antigen receptors, such as IgM, IgW, and immunoglobulin new antigen receptor (IgNAR). This composition of receptors differs significantly from mammals and birds (Flajnik, 2018).
IgNARs are naturally occurring heavy-chain-only antibodies (HCAb) (Greenberg et al., 1995). In addition to the absence of light chains, the study of shark IgNAR furthered our understanding of the heavy-chain-only immunoglobulin gene germline configuration paradigm. Unlike HCAb in Camelidae, IgNAR has the clustered organization of its gene, with four to 200 IgNAR gene clusters or miniloci scattered across different shark genomes (Shamblott and Litman, 1989; Greenberg et al., 1995; Malecek et al., 2008; Wei et al., 2021). A complete IgNAR minilocus comprises consecutive exons for V, D, and J segments, as well as the five constant domains, secretory and transmembrane tails (Flajnik, 2018). Compared to studies on shark immunoglobulins, research on skate immunoglobulins is relatively limited. Nonetheless, the findings from these investigations have provided valuable insights. The study of skate immunoglobulin light chain gene clusters led to the discovery of the first germline-joined immunoglobulin gene. In little skate (Raja erinacea), the absence of combinatorial and junctional diversity of its type I light chain gene clusters have been reported. This absence was considered to have special functions in adult life (Anderson et al., 1995; Lee et al., 2002).
IgNAR was first reported in the nurse shark in 1995 (Greenberg et al., 1995). Over the past two decades, shark IgNAR has been extensively studied. However, skate IgNAR has remained somewhat elusive; the evidences of its presence are an early cDNA deposition (Rumfelt et al., 2004a) and Southern blot verifications in cownose ray as well as little skate (Criscitiello et al., 2006). The existence of IgNAR in skates is significant because it suggests that IgNAR emerged prior to the evolutionary separation of Batoidea and Selachii (Rumfelt et al., 2004a; Flajnik et al., 2011; Barelle and Porter, 2015). Therefore, clear identification and proper characterization of skate IgNAR are crucial for understanding the evolution of adaptive immune systems.
IgNARs have variable domains capable of functioning independently in antigen recognition and binding. These variable domains can be used as single-domain antibodies (sdAb) called VNAR (Stanfield et al., 2004). The diversity of single-domain antibody repertoire in elasmobranchs also results from variations in VNAR structures, in addition to variations in CDR sequences. There are four types of VNARs, which differ based on the position and number of cysteine residues (Zielonka et al., 2015). Type III is typically expressed in newborn nurse sharks within their first year (Diaz et al., 2002; Streltsov et al., 2004). Type I VNAR has one pair of canonical cysteines in FR1 and FR3, as well as four noncanonical cysteines in FR2, CDR3, and FR4. This type of VNAR has only been identified in nurse sharks (Ginglymostoma cirratum) thus far (Streltsov et al., 2004; Feng et al., 2019). Type II VNAR has one pair of both canonical and noncanonical cysteines and is the most prevalent type found in most shark species with IgNAR identification (Greenberg et al., 1995; Nuttall et al., 2001; Diaz et al., 2002; Liu et al., 2007a; Liu et al., 2007b; Camacho-Villegas et al., 2013; Crouch et al., 2013; Ohtani et al., 2013). Type IV VNAR has only one pair of cysteines in FR1 and FR3, forming the canonical disulfide bond that stabilizes the main frame of the VNAR structure. It has been identified in a few shark species, such as spiny dogfish sharks, spotted wobbegong sharks, and bamboo sharks, but has never been found to be the dominant type (Nuttall et al., 2001; Liu et al., 2007b; Crouch et al., 2013; Zhang et al., 2020; Wei et al., 2021). Despite the insignificant contribution of type IV VNAR to the natural antibody repertoire of sharks, several type IV VNAR have been found to have exceptional functional properties (Kovalenko et al., 2013; Fernandez-Quintero et al., 2022; Li et al., 2022; Burciaga-Flores et al., 2023). Type IV VNAR was even preferred during nanobody screening due to its similarity to human immunoglobulin counterparts, which also have one pair of canonical cysteines (Li et al., 2022).
In this study, we present clear evidence of the existence of IgNAR in the peripheral blood serum and mononuclear cell transcriptome of the ocellate spot skate. We also cloned and annotated the germline configuration of the ocellate spot skate IgNAR V domain. Additionally, we characterized the VNAR repertoire of the ocellate spot skate and identified that type IV VNAR was the absolute dominant isotype. This represents a new VNAR isotype composition paradigm that has not been reported in other elasmobranchs.
An adult male ocellate spot skate (Okamejei kenojei) was obtained from the Yellow Sea, China. The skate was anesthetized with MS-222 (TCI). Peripheral blood, spleen, and epigonal organs were collected. Lymphocytes from the peripheral blood and spleen were isolated using mononuclear cell preparation kits (Solarbio, Beijing, China, P8610) following the manufacturer’s protocols. A dilute solution consisting of PBS and urea was prepared to match the osmolarity of skate cells. The epigonal organ tissues were homogenized in TRIzol Reagent (Invitrogen, CA, USA) and stored at −80°C until use. All animal experiments in the study were approved by the Ethics Committee of the Institute of Oceanology, Chinese Academy of Sciences (No. MB2208-3).
The mRNA extraction, library construction, sequencing, and basic analysis were performed by a contracted NGS service-providing company, Novogene Co. Ltd., Beijing, China Briefly, total RNA was isolated from peripheral blood with TRIzol reagent (Invitrogen, CA, USA) following the manufacturer’s procedure. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. The library was prepared using the NEBNext® Ultra RNA Library Prep Kit for Illumina (NEB) and sequenced by the Illumina NovaSeq 6000. Paired-end reads of 150 bp were generated. After obtaining clean reads, de novo assembly of the transcriptome was performed with Trinity 2.6.6 (Grabherr et al., 2011), as the ocellate spot skate does not have genome data and annotations. To obtain the ocellate spot skate IgNAR sequence, shovelnose guitarfish IgNAR (GenBank: AY524298.1) was used as a reference sequence to identify the ocellate spot skate IgNAR from the transcriptome library by local BLAST, and subsequent research was based on this result.
Total RNA was extracted from mononuclear cells of the spleen and peripheral blood, as well as whole tissue of the epigonal organ, using TRIzol reagent (Invitrogen, CA, USA) following the manufacturer’s procedure. For the first-strand cDNA synthesis, RT-PCR was performed using the HiScript III 1st strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The RT-PCR products were used as templates for PCR amplification. The transmembrane and secretory IgNARs were amplified with PCR primers (Supplementary Table S1). The PCR reactions were performed using the Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) according to the manufacturer’s protocols.
The program consisted of initial denaturation at 95°C for 5 min, 35 cycles of 15 s at 95°C, 15 s at 58°C, and 1 min of 15 s at 72°C, followed by a final extension at 72°C for 5 min. The fragments were recovered using the FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China) from agarose gel after electrophoresis. The recovered fragments were ligated into the pCE2 TA/Blunt-Zero vector using the 5 min TA/Blunt-Zero Cloning Kit (Vazyme, Nanjing, China) and sequenced.
Protein sequences of different IgNAR clones were aligned with ClustalW (Thompson et al., 1994). Phylogenetic analysis was performed by the maximum likelihood method with 1,000 bootstrap replicates in MEGA v.11 software (Thompson et al., 1994; Tamura et al., 2021).
Whole-genome DNA was extracted from skate muscle tissue using the FastPure Cell/Tissue DNA Isolation Mini Kit (Vazyme, Nanjing, China), following the manufacturer’s procedure. The VNAR germline sequence was amplified using specific primers (Supplementary Table S1).
The PCR reactions were performed using the Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) according to the manufacturer’s protocols. The program consisted of initial denaturation at 95°C for 5 min, 35 cycles of 15 s at 95°C, 15 s at 63.8°C, and 50 s at 72°C, followed by a final extension at 72°C for 5 min. The fragments were recovered using the FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China) from agarose gel after electrophoresis. The recovered fragments were ligated into the pCE2 TA/Blunt-Zero vector using the 5 min TA/Blunt-Zero Cloning Kit (Vazyme, Nanjing, China) and sequenced.
Multiple sequence alignments were performed by Jalview 2.11.2.6 and ESPript 3 (Waterhouse et al., 2009; Robert and Gouet, 2014).
Blood samples were centrifuged at 300×g for 10 min to separate the serum at 4°C. One microliter of serum was analyzed on 10% SDS-PAGE (NCM Biotech) under reducing conditions and stained with Coomassie blue. Bands around 70 and 100 kDa were excised from the gel and analyzed with mass spectrometry on an LTQ Orbitrap Elite System (ThermoFisher, MA, USA). In brief, the excised bands were digested with trypsin and subsequently analyzed using mass spectrometry. The detected peptide masses were matched against a reference protein database to identify IgNAR. This reference database included deduced AA sequences of IgNAR Sec1, IgNAR Sec2 (Figure 1A), and O. kenojei (Taxonomy ID: 432798) proteins archived in UniProt.
The coding sequence of the VNARs for further expression was amplified from previously cloned IgNAR sequences in the pCE2 TA/Blunt-Zero vector. PCR products were inserted into the pET-22b plasmid with a C-terminal His-tag and transformed into BL21 (DE3)-competent E. coli (Tsingke, Beijing, China). The strains containing recombinant VNARs were cultured for protein expression in 2YT media with 100 µg/ml ampicillin at 37°C until the OD600 reached approximately 0.8. Protein expression was induced with 0.4 mM IPTG, and the cultures were shaken for 20 h at 28°C. The collected bacteria were centrifuged and lysed by osmotic shock, and proteins were initially purified on a Ni-NTA Sepharose column (GE Healthcare, IL, USA) followed by size-exclusion chromatography (SEC) using a SuperdexTM 75 increase 10/300 GL column (Cytiva, MA, USA) for further purification. The eluted fractions were evaluated by SDS-PAGE for purity and then concentrated using ultrafiltration spin columns with a molecular mass cutoff of 10 kDa (Merck, NJ, USA). The protein concentration was quantified using the NanoPhotometer NP80 (Implen, Munich, Germany).
The urea-induced and temperature-induced unfolding of VNARs were evaluated using the UNcle system (Unchained Labs, CA, USA). The intrinsic fluorescence signal excited at 266 nm was recorded from 250 to 500 nm to monitor the evolution of protein structures. For the urea-induced unfolding, 1 mg/ml VNARs were treated with various concentrations of urea overnight at 25°C and then tested at 25°C. The temperature-induced unfolding was measured from 25°C to 95°C at a rate of 1°C/min. Data analysis of both chemical and thermal unfolding experiments was performed using UNcle Analysis V6.01.
VNAR1 (9.8 mg/ml) was set up in 0.5 µl sitting drops using Crystal Screen (Hampton Research). Successful conditions were scaled up to 1 µl sitting drops. Plates were incubated at 4°C. The final crystallization conditions were 0.2 M lithium sulfate monohydrate, 0.1 M Tris, pH 8.0, and 26% w/v polyethylene glycol 4000. Diffraction-quality crystals were obtained after 24 h. Crystals were harvested and flash-frozen in liquid nitrogen with 15% glycerol as a cryoprotectant.
Complete X-ray diffraction data sets were collected using XtaLAB Synergy (Rigaku, Tokyo, Japan). Diffraction images were processed with CrysAlisPro. The structure of VNAR1 was determined by molecular replacement using the shark IgNAR variable domain (PDB: 4HGK) as a model. Model building and crystallographic refinement were carried out in Coot and PHENIX. Detailed data collection and refinement statistics are listed in Supplementary Table S2.
Figures were generated with PyMOL. The coordinates have been deposited in the Protein Data Bank under the accession code 8HT3.
cDNAs of mononuclear cells from the spleen and peripheral blood, and tissue from epigonal organs were used as templates. VNARs were PCR amplified using the primer pairs: OkIgNAR-F (5′-ATGAATATTTTTTGTGTTTCAATACTTTTGGCC-3′) and OkVNAR-J-R (5′-AGGTTTCACAGTCAGCTTTG-3′), respectively. The PCR reactions were performed with Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) according to the manufacturer’s protocols. The program consisted of initial denaturation at 95°C for 5 min, 30 cycles of 15 s at 95°C, 15 s at 58.8°C, and 20 s at 72°C, and a final extension at 72°C for 5 min. The VNAR DNA fragments at 300–400 bp were recovered using a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) from agarose gel after electrophoresis. The VNAR DNA fragments were then A-tailed and ligated with the full-length adapter for Illumina sequencing, followed by further PCR amplification for library preparation. The PCR products were purified using the AMPure XP system (Beckman Coulter, CA, USA) and sequenced with the Illumina NovaSeq 6000 using a PE250 strategy. A brief summary of the datasets is provided in Supplementary Table S4.
After sequencing, all reads were trimmed using Trimmomatic (version 0.39) (Bolger et al., 2014) to remove adapter sequences and low-quality reads. The clean paired-end reads were merged using FLASH (version 1.2.11) (Magoc and Salzberg, 2011), based on approximately 150 bp of overlap between the paired-end sequences. A laboratory-developed script was used to split the fasta files into spike-in sequences and sample sequences according to specific site information.
To remove sequence errors introduced by library preparation and sequencing strategy, a sequence filtering approach was established. First, merged sequence reads were aligned to the V and J gene region primers of Okamejei kenojei IgNARs, and the matching degree of each sequence was calculated. Sequences that didn’t cover the V and J primer regions were discarded based on specific parameters. Then, the count of duplicate reads for each sequence was calculated, and only unique sequences with at least two representative reads were retained. The above steps were completed using pRESTO (Vander Heiden et al., 2014) (version 0.7.1). The reads meeting the filter criteria were translated into protein sequences, and sequences with early stop codons were removed. Amino acid sequences were required to start with “VHV” at the N-terminus and end with “VKP” at the C-terminus to be considered full-length VNARs (Feng et al., 2019). Finally, these protein sequences were clustered by cd-hit (version 4.8.1) (Li and Godzik, 2006) according to their similarity to remove redundant sequences. The remaining amino acid sequences and corresponding original DNA sequences were considered unique sequences.
To analyze the library’s diversity, unique sequences (defined as having one differing amino acid in sequence) and diversity (defined as having one differing amino acid in the CDR/HV region) were counted. The VNAR sequences were further classified into different subtypes based on residue numbers and positions. Type IV and all VNAR sequences were also separately analyzed for CDR3 lengths, total cysteine numbers, and CDR3 cysteine numbers. All VNAR sequences were aligned using muscle (version 5.1) (Edgar, 2021), and the conserved amino acids of these sequences were displayed through TBtools (Chen et al., 2020) software.
To find recombination signal sequences (RSS) of Okamejei kenojei V-domain germline sequences, several conserved heptamer and nonamer sequences were collected from previously published research. A custom Python script was built to identify potential RSS in the IgNAR V-domain germline sequences. Two and three nucleotide mismatches were allowed to align heptamer and nonamer sequences to the genome, respectively. Based on the results of the integrated analysis and detailed manual calibration, the RSS sequences of IgNAR V-domain sequences were determined (Wei et al., 2021). Finally, the four IgNAR V-domain germline configurations of Okamejei kenojei were depicted with detailed localizations of V-D-J and the structural features of RSS.
As the genome and gene annotations of the ocellate spot skate were unavailable, we conducted transcriptome sequencing of mononuclear cells from the ocellate spot skate’s (Okamejei kenojei (O. kenojei)) blood. Through de novo assembly of the sequencing data, we obtained several clusters that were highly similar to other elasmobranch IgNARs. We selected the longest sequence with a leader sequence and secretory tail homologous to the other IgNARs as a reference for primer design and further cloning. We then amplified potential IgNAR transcripts from O. kenojei blood mononuclear cell cDNA, cloned them, and sequenced them using Sanger sequencing. The deduced amino acid (AA) sequences are shown in Figure 1A. By performing multiple sequence alignment among the amino acid sequences from our cDNA and other cartilaginous IgW, IgM, and IgNAR sequences, we identified the cloned cDNA transcripts as O. kenojei IgNARs, with a 68.1% similarity to amino acids of P. products IgNAR (Supplementary Figure S1).
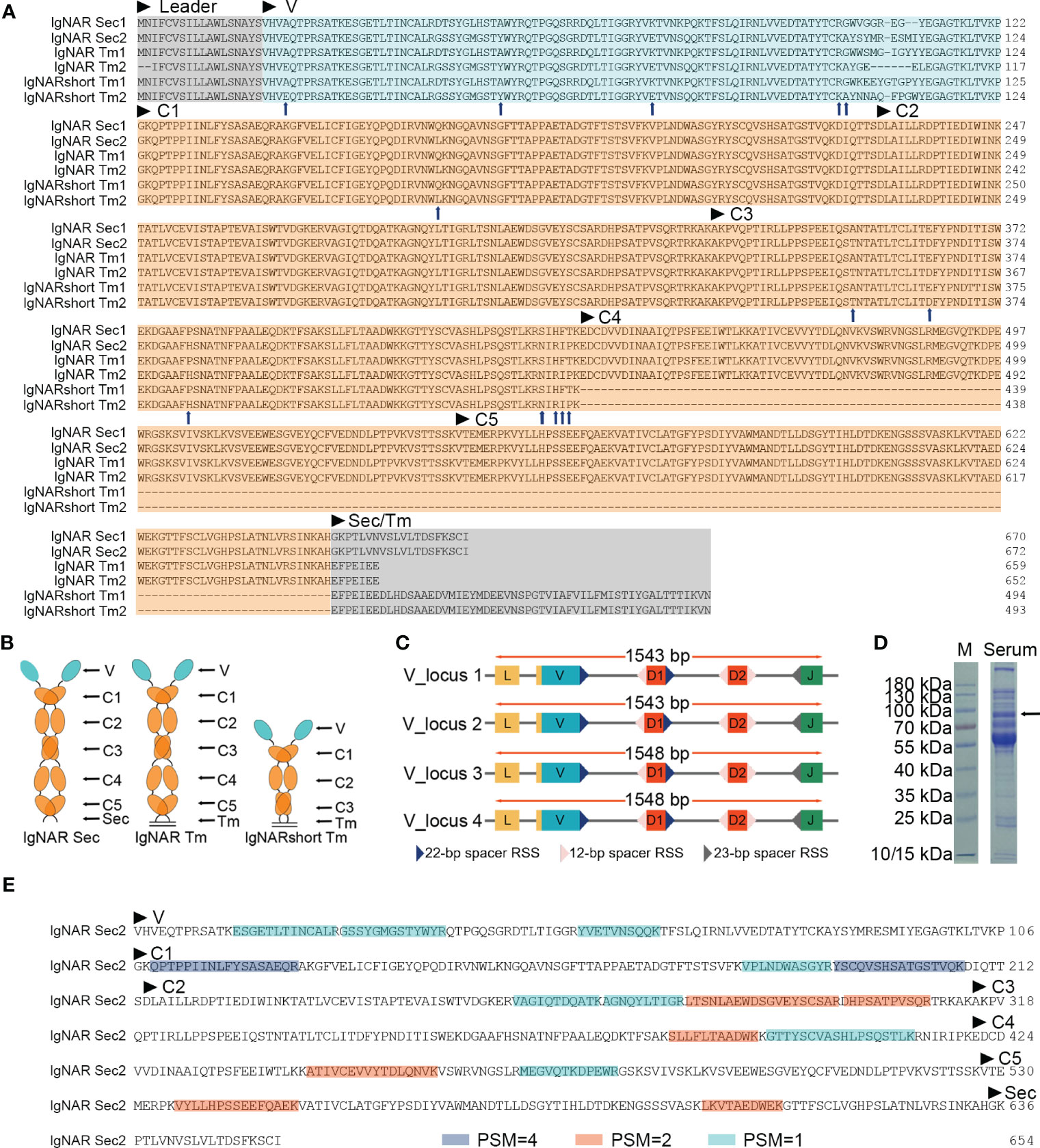
Figure 1 Identification of the ocellate spot skate IgNAR. (A) Multiple sequence alignment (MSA) by Jalview, with arrowheads, indicates differential amino acids of sequences from the two V-domain loci. Sec, secretory tail; Tm, transmembrane tail; short, without C4 and C5; “IgNARshort” equals “IgNARshort(C3Tm)” in the main text; 1 or 2 following “Sec” or “Tm,” means this sequence is theoretically transcripted from V-domain locus 1 or 2, based on nucleotide sequence alignment. (B) Scheme of different forms of IgNAR. (C) Germline configuration of the four V-domain loci, with RSS. (D) SDS-PAGE of the ocellate spot skate serum; the arrow indicates the band positive for IgNAR verified by mass spectrometry. (E) Summary of mass spectrometry result; peptides captured in MS are highlighted. Blue, PSM = 4; orange, PSM = 2; cyan, PSM = 1. PSM, peptide-spectrum match. The accession numbers of all the sequences included in this figure are as follows: IgNAR Sec1, OQ793031; IgNAR Sec2, OQ793032; IgNAR Tm1, OQ793033; IgNAR Tm2, OQ793034; IgNARshort Tm1, OQ793035; and IgNARshort Tm2, OQ793036. The raw data generated from transcriptome sequencing are archived in the NCBI SRA (https://www.ncbi.nlm.nih.gov/sra) with the BioProject accession number PRJNA953513.
Germline sequences of the O. kenojei IgNAR variable domain were cloned by high-fidelity PCR on muscle genomic DNA using one set of primers (Supplementary Table S1), designed based on the above IgNAR transcripts. We obtained 15 clones, and Sanger sequencing results showed there were four different variable domain germline sequences with a minimum of nine bases of difference from each other. These four V-domain germline sequences likely came from four different IgNAR miniloci, and we named them V_locus 1, 2, 3, and 4. To locate the recombination signal sequence (RSS) in cloned germline DNA, we used previously published RSS data and employed sequence alignment and manual calibration to identify the signal. We then annotated RSS, V, D, and J segments on V-domain germline sequences based on our analysis of the O. kenojei IgNAR cDNA, germline sequences, and previous reports of several IgNARs from different shark species (Figure 1C; Supplementary Figure S2). An intron is present downstream of the V segment, separating the V from the D segments. Similar to spiny dogfish shark IgNAR loci, two D segments were downstream of the V segment, flanked by 12 or 22/23 bp spacer RSSs. Prejoined D segments were not observed in our germline V-domain clones.
The full-length O. kenojei IgNAR cDNAs encode a leader peptide, variable domains (V segment, D segment, and J segment), constant domains C1 to C5, and a transmembrane tail or a secretory tail, consistent with its other cartilaginous homologs, and with an estimated molecular weight of 72 kDa. Electrophoresis results of O. kenojei blood serum in SDS-PAGE under reducing conditions displayed a prominent band around 70–100 kDa, similar to the molecular weight of the monomeric full-length O. kenojei IgNAR (Figure 1D). The band between 70 and 100 kDa was carefully excised from the gel and analyzed by mass spectrometry. The results revealed 16 unique peptides that could be assigned to our cloned O. kenojei IgNAR, encompassing all key components from the variable domain to the C5 domain, with at least one peptide detected for each domain (Figure 1E). These findings confirmed the presence of IgNAR in the ocellate spot skate blood mononuclear cell cDNA library and its existence in blood serum.
Phylogenetic trees were constructed to evaluate the position of O. kenojei IgNAR within the evolutionary history of the immunoglobulin superfamily. The analysis demonstrated that O. kenojei IgNAR clusters with P. products IgNAR and is closely related to Triakis scyllium (Figure 2A). In comparison to other IgNARs, sequence conservation between O. kenojei IgNAR and cartilaginous IgM and IgW is considerably lower (Supplementary Figure S1). Furthermore, the O. kenojei IgNAR V domain demonstrates the highest homology with other cartilaginous IgNAR V domains and exhibits potential homology to the nurse shark NAR-TCR V domain (Figure 2B). This finding implicated that the NAR-TCR V domain may have been a possible precursor to the IgNAR V domain. Similarly, the previously reported nurse shark IgNAR V-domain clustered with human TCR V domains (Greenberg et al., 1995).
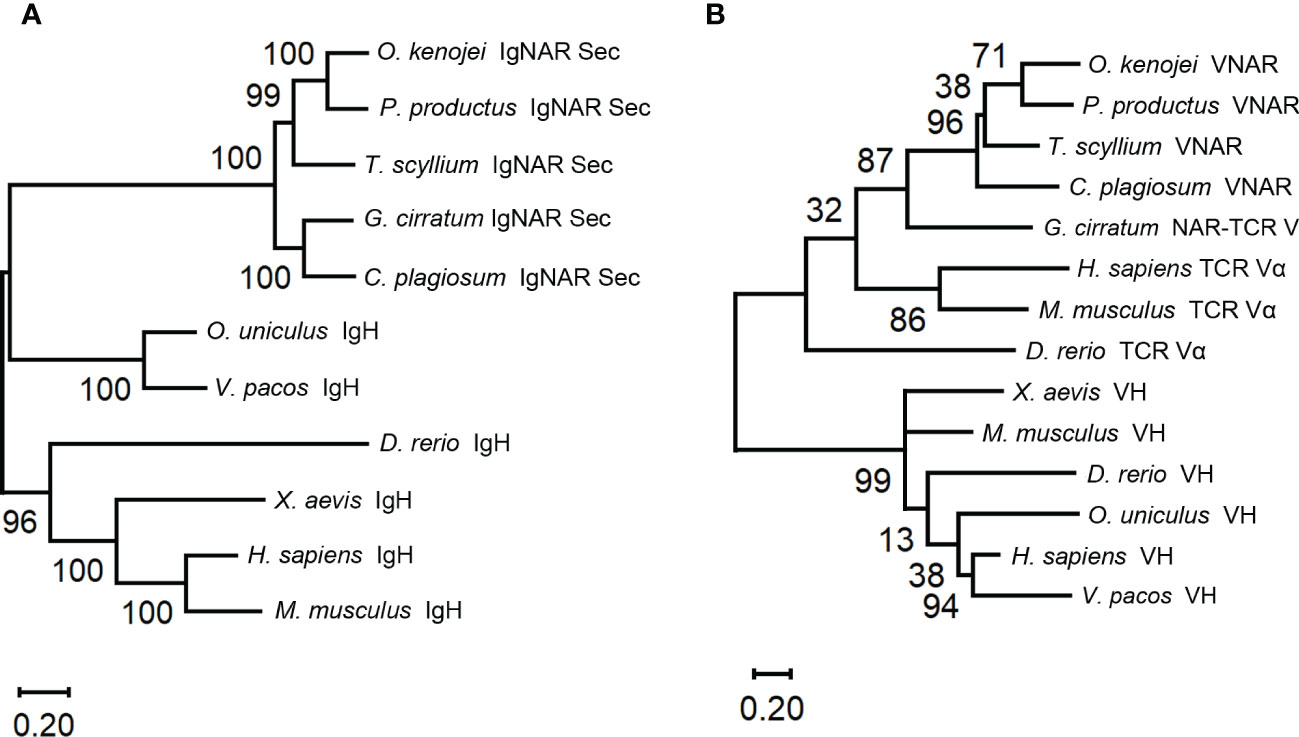
Figure 2 Phylogenetic trees comparing the ocellate spot skate VNAR/IgNAR with other immunoglobulins. Trees were constructed by MEGA V11 using the maximum likelihood method with the WAG+G+I model and the WAG+G model for (A), (B), respectively. The numbers at the branch points indicate the confidence levels of the paired sequences, given by bootstrap statistical analysis. Branch lengths matched with the number of substitutions per site (Scales are as indicated). (A) Comparing the full-length secretory form IgNAR with immunoglobulin heavy chains. (B) Comparing the V domain of the ocellate spot skate IgNAR (VNAR) with other VNARs, TCR V domains, and heavy-chain V domains. The accession numbers of all the sequences included in this figure are summarized in Supplementary Table S3.
During the analysis of the full-length (Tm) O. kenojei IgNAR transcript amplification, smaller DNA bands were detected on the electrophoresis gel that differed from the expected five-constant domain IgNAR (data not shown). Cloning and Sanger sequencing revealed several clones with a shorter composition that lacked the C4 to C5 domains, similar to other cartilaginous IgNARs such as Chiloscyllium plagiosum, as reported previously (Figures 1A, B) (Rumfelt et al., 2004a; Zhang et al., 2020). The deduced amino acid sequence of the two short compositions is identical to the two full-length IgNAR transmembrane form sequences, respectively (Figure 1A), except for the CDR and HV regions. Therefore, we designated these short compositions as alternative splicing forms of O. kenojei IgNAR, which spliced C3 to Tm, hereafter referred to as IgNARshort(C3Tm) (Figure 1A).
Structural predictions of the O. kenojei IgNAR sec1 homodimer and IgNARshort(C3Tm) Tm1 homodimer (monomer sequences as shown in Figure 1A) were performed by Alphafold2 Multimer v2 (Figure 3A; Supplementary Figure S3A) (Jumper et al., 2021; Evans et al., 2022). The overall configuration of the IgNAR sec1 homodimer was consistent with previously reported characteristics of shark IgNARs (Feige et al., 2014). The cysteines between C3 and C4 were predicted to form interchain disulfide bonds, enabling IgNAR sec1 to form dimers (Figure 3A). The IgNARshort(C3Tm) homodimer structure predicted by Alphafold2 Multimer v2 showed a similar configuration to the full-length structures but lacked interchain disulfide bonds (Supplementary Figure S3A).
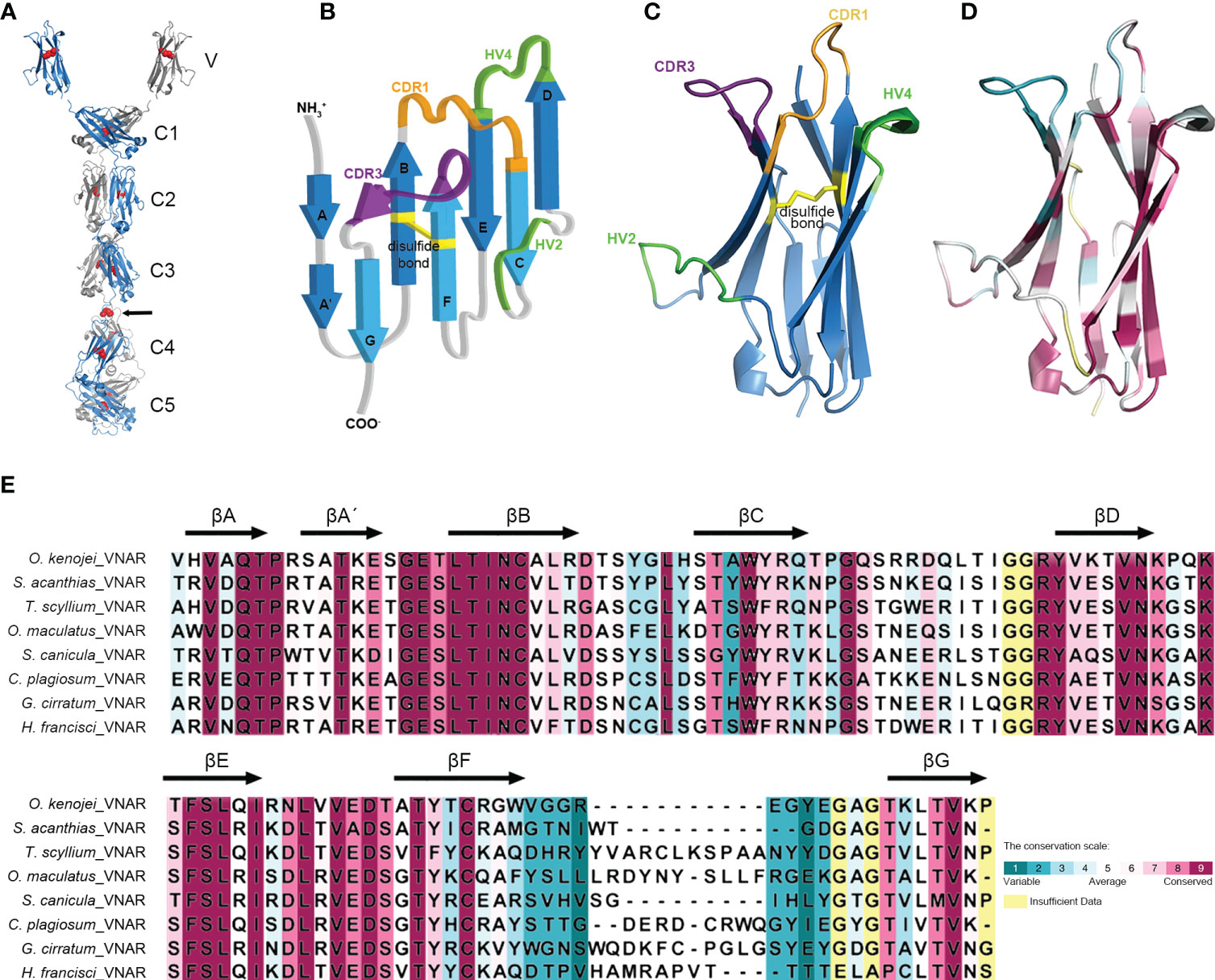
Figure 3 Structure of the ocellate spot skates IgNAR and VNAR. (A) Alphafold2 predicted the structure of full-length IgNAR sec1, with the crystal structure of VNAR stitched to the loop linking the V domain and C1. Disulfide bonds are shown in red spheres. The arrow indicates the interchain disulfide bond. (B) Topology diagram of the ocellate spot skate VNAR. (C) Crystal structure of VNAR (PDB ID: 8HT3). (D) Consurf analysis showing in 8HT3 of MSA between the ocellate spot skate VNAR and other VNARs. (E) Consurf analysis showing VNAR amino acid sequence conservation colored to the conservation scale.
In the structural prediction of the transmembrane form IgNAR, the transmembrane domain did not appear as transmembrane helices but rather as a loop. However, DeepTMHMM predicted the existence of transmembrane helices at the end of the Tm (Supplementary Figure S3C). Recent reports have shown that B-cell receptors usually have a transmembrane α-helix domain that participates in their assembly (Dong et al., 2022). Given that the hydrophobic core of a typical cell membrane is 3–4 nm in thickness (Frallicciardi et al., 2022), the predicted 21-amino acid transmembrane domain in Tm, forming a roughly 3-nm-long α-helix, should be able to attach the IgNARshort(C3Tm) to the B-cell membrane (Supplementary Figures S3A, C). To compensate for the insufficient part of the dimer prediction, we manually replaced the original part in the homodimer and stitched the transmembrane helices structure to it with dashed lines. The stitched transmembrane helices were also generated by Alphafold2 solely on the Tm amino acid sequences (Supplementary Figure S3A).
Multiple sequence alignment results revealed that the typical hydrophobic core present in the constant domains of most known antibodies is also conserved in O. kenojei IgNAR. Using O. kenojei IgNAR C1 (from IgNAR sec1) as an example, W43 and W80 are highly conserved and surrounded by several hydrophobic amino acids, such as P36, I39, V41, V75, and V90. The nearby C29 and C88 are also evolutionarily conserved features in known antibody C domains. In O. kenojei IgNAR, they are predicted to form an intradomain disulfide bond, stabilizing the C-domain structure (Supplementary Figure S4B).
To elucidate the structure of the variable domain (VNAR) of O. kenojei IgNAR, we derived VNAR1 from IgNAR sec1 (Figure 1), which was obtained from a wild-caught ocellate spot skate. VNAR1 was expressed in Escherichia coli and purified using affinity chromatography and size-exclusion chromatography (Supplementary Figure S4A). The structure of VNAR1 was determined at 2.5 Å resolution by X-ray crystallography. The overall crystal structure exhibited considerable similarity to other VNARs identified in sharks (Figures 3B, C). The CDR1, HV2, HV4, and CDR3 amino acids formed loops stringed by consecutive β-strands in the conserved framework regions. The eight β-strands roughly adopted a two-layer sandwich structure, covalently connected by a buried disulfide bond. Consurf (Yariv et al., 2023) alignment of O. kenojei VNAR1 with several shark VNARs from different species demonstrated that the most variable sequences in the alignment were narrowed in the CDR and HV regions, while the remaining framework regions were conserved. The amino acids in frameworks FR1 and FR3b exhibited the highest conservation, including the canonical cysteines (Figures 3D, E). The highly conserved framework and highly variable CDR illustrated by Consurf analysis of different elasmobranch VNARs suggest that shark and skate VNARs share a common ancestor.
The CDR3 length of VNAR1 was nine amino acids, which was significantly shorter than the CDR3 length in previously published VNAR structures, except for the VNAR against human serum albumin (HSA) (PDB ID: 4HGK) from spiny dogfish sharks (Squalus acanthias) (Supplementary Figure S3B). The VNAR against HSA also had a nine AA-long CDR3. It utilized an atypical mode involving HV2, CDR3, and part of the framework to bind HSA with nanomolar affinity (Kovalenko et al., 2013). Therefore, the short CDR3 length should not hinder O. kenojei IgNAR and VNAR from binding to antigens.
These results suggest that O. kenojei IgNAR and IgNARshort(C3Tm) adopt the standard homodimer structure of shark IgNARs. This classical and conserved immunoglobulin structural configuration could facilitate their antigen-binding capabilities.
To gain a comprehensive understanding of the O. kenojei VNAR repertoire and assess its potential for producing functional single-domain antibodies, we performed next-generation sequencing (NGS) on VNAR libraries generated from O. kenojei peripheral blood, spleen, and epigonal organ, respectively. We employed a PE250 strategy in sequencing, producing approximately 2 million paired-end reads in each library. After merging the forward and reverse sequences, we filtered the reads based on the following criteria: (1) discarding reads without the forward primer and reverse primer used in VNAR amplification; (2) discarding reads with a copy number of one; (3) discarding reads with a stop codon; (4) requiring the deduced amino acid sequence to begin with “VHV” and end with “VKP”; (5) clustering sequences with no more than one AA mismatch. These criteria are stringent to minimize false diversity introduced by amplification and sequencing errors. Following these procedures, we obtained 24,827 VNAR unique sequences (AA) from peripheral blood, 27,137 VNAR unique sequences (AA) from the spleen, and 9,574 VNAR unique sequences (AA) from the epigonal organ.
The primer setting of VNAR library amplification constrained our results to only include VNAR transcripts generated from the four V-domain loci mentioned earlier or other IgNAR miniloci compatible with the primers. Given that sharks exhibit immunoglobulin heavy-chain exclusion (Eason and Litman, 2002; Malecek et al., 2008), we focused on the cloned four V-domain loci to examine if there was a locus preference in O. kenojei IgNAR allelic exclusion. We extracted and analyzed unique nucleotide sequences with an exact match to the four V-segment loci. The results indicated that unique VNAR transcripts were predominantly generated from V_locus 1 to V_locus 2 in all three tissues, accounting for 94% to 95%. The strongest preference was found in the epigonal organ, where 59% of VNAR transcripts originated from V_locus 2 (Figure 4A). Despite the limited coverage of IgNAR miniloci, these findings suggest a strong locus preference in O. kenojei IgNAR.
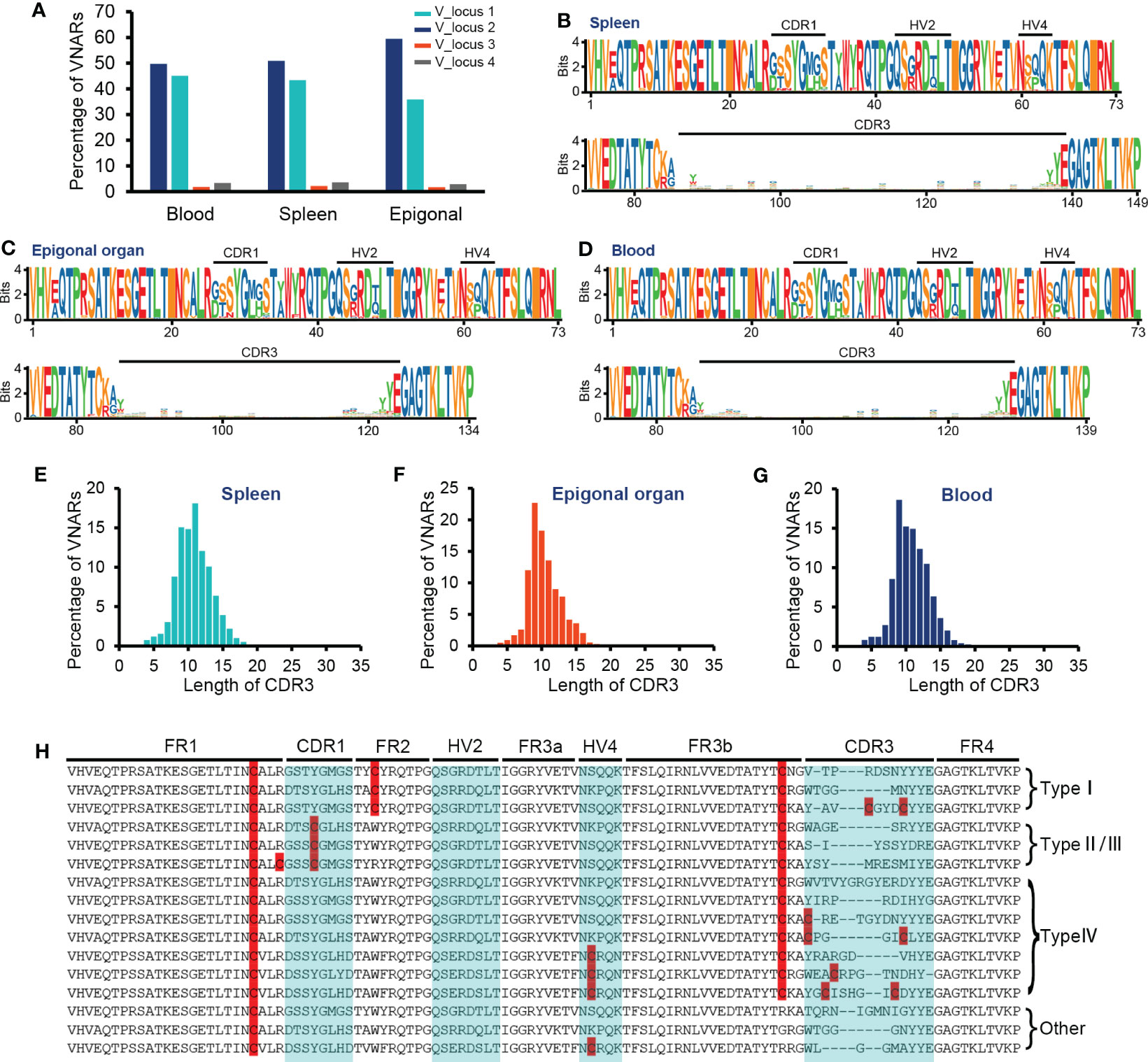
Figure 4 Characteristics of VNAR libraries from the spleen, peripheral blood, and epigonal organ. (A) Locus usage of the four V-domain loci. (B–D) Amino acid variation analysis. The height of the letter positively correlates to the AA frequency in the library. (E–G) Statistics on CDR3 length in each library. (H) Several examples of VNARs from the ocellate spot skate VNAR repertoire. Cysteines were highlighted. In terms of the four types of VNAR classification, in our cases, type I VNAR must have C22, C36, and C83; type II/III VNAR must have C22, C29, and C83; and type IV VNAR must have C22 and C83. The raw sequencing data generated from high-throughput sequencing were archived in the NCBI SRA (https://www.ncbi.nlm.nih.gov/sra) with the BioProject accession number PRJNA953503.
The CDR3 lengths of most VNARs in nurse sharks range from 10 to 30 AA and from 12 to 21 AA in white-spotted bamboo sharks (Feng et al., 2019; Wei et al., 2021). We observed the relatively short CDR3 length in the crystal structure of O. kenojei VNAR. Therefore, we conducted an analysis of CDR3 lengths in the three libraries from different tissues (Figures 4E–G). The majority of CDR3 lengths ranged from eight to 15 AA in the blood, spleen, and epigonal organs. Compared to the other two tissues, the epigonal organ had the shortest average CDR3 length of 10.42 AA, with a median value of 10 AA. The average and median values of CDR3 length in the spleen were both the highest among the three libraries, although still shorter than those of shark VNARs. In cartilaginous fish, B cells are generated in the epigonal organ, and the spleen is the major secondary lymphoid organ (Rumfelt et al., 2002; Dooley, 2018). Our results suggest that, in O. kenojei B-cell development, CDR3 length may slightly increase due to B-cell clonal selection.
Amino acid variability analysis of all VNAR unique sequences revealed that, in O. kenojei primary and secondary lymphoid organs, VNAR diversity was mainly constituted by CDR3 diversity. The CDR1, HV2, and HV4 regions showed much lower variability compared to the CDR3 region. Within the CDR3 region, there was no amino acid preference except for the RSS-related junction regions (Figures 4B–D, H; Supplementary Figure S5). Gaps caused by multiple sequence alignments were removed to prevent false-positive amino acid conservation.
VNARs are classified into four types based on the number and location of cysteines: types I, II, III, and IV (Zielonka et al., 2015). We analyzed the VNAR unique sequences (AA) in the three libraries to determine cysteine number and distribution. The data showed that approximately 99% of the VNARs in all three libraries belonged to type IV, which had only one pair of canonical disulfide bonds connecting β-strand B and β-strand F (Figures 5A–C). Types I, II, and III were also discovered in the VNAR libraries and were classified by the presence of Cys29 or Cys36. However, no classical type I VNAR with six cysteines was found. The structural diversity of the O. kenojei VNAR library was therefore limited by the almost uniform framework structure. As a result, the diversity of O. kenojei IgNAR should be primarily determined by variable domain amino acid sequence diversity. In the context of VNARs from elasmobranchs, with the prevalent type II VNARs from sharks, O. kenojei VNAR would greatly compensate for the rare naïve type IV compositions.
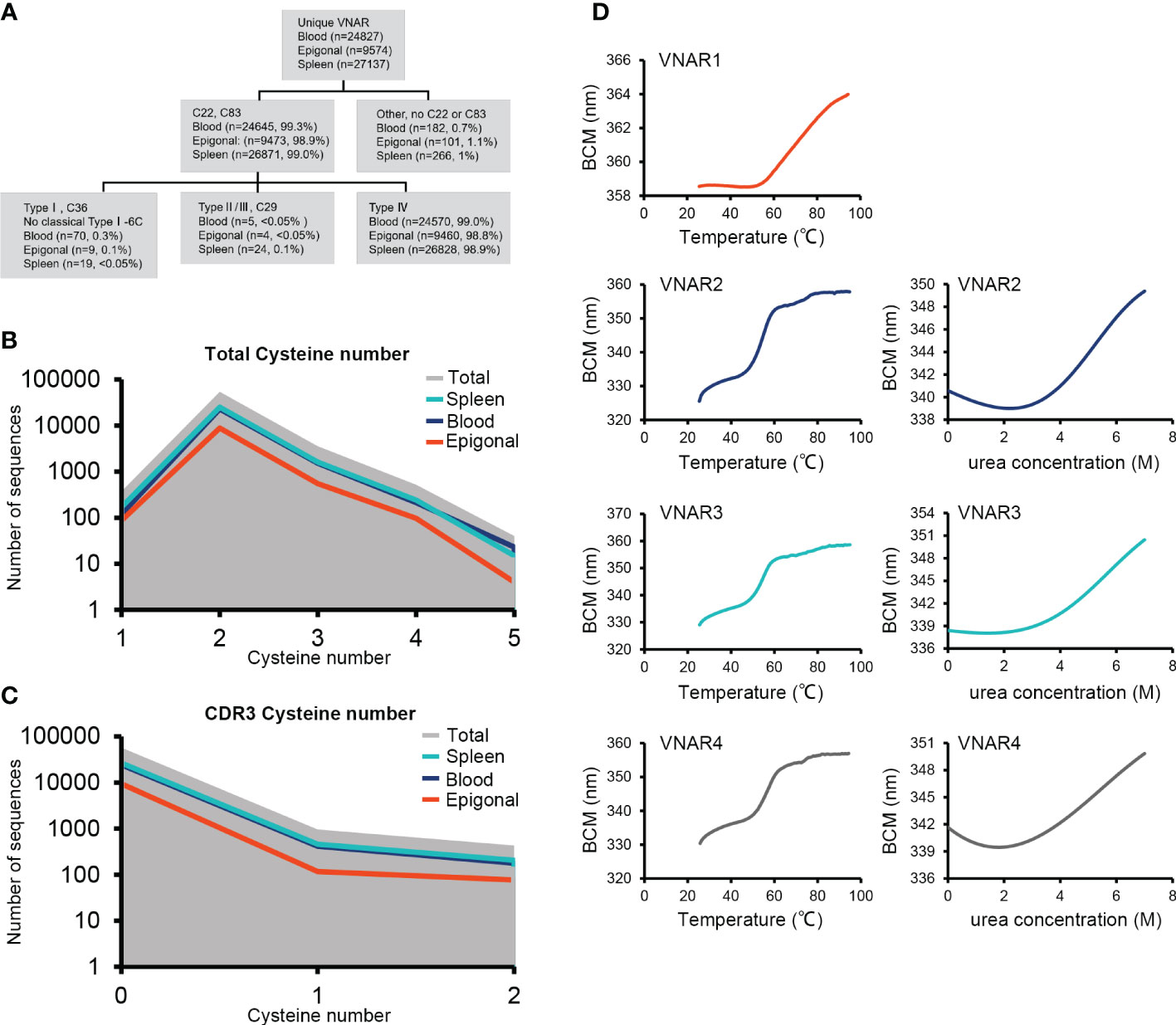
Figure 5 VNAR classification and thermal and chemical stability measurements. (A) Detailed analysis of the ocellate spot skate VNAR classification and cysteine distribution. (B) Full-length VNAR cysteine number distribution plots of all VNARs and in three tissues, respectively. (C) CDR3 cysteine number distribution plots of all VNARs and in three tissues, respectively. (D) Thermal stability (melting temperature) and chemical stability (urea, Cm) measurement plots. VNAR1 did not show obvious denaturation under 0 to 8 M urea treatment during the test (data not shown). The missed signal could be due to the special tryptophan distribution of VNAR1. The Trp86, located at the VNAR1 hydrophilic area, generated the strong fluorescence that caused the high baseline emission. During VNAR1 unfolding, the Trp36 that is covered in the hydrophobic core will be exposed, while Trp86 could be buried by conformational change. The resulting Trp36 fluorescence is offset by the vanished Trp86 signal and eventually causes the loss of an obvious denaturation point.
Type IV VNAR contains only the canonical disulfide bond, which stabilizes the two β-sheet sandwich conformation. To determine the thermal and chemical stability of O. kenojei VNAR, we cloned, expressed, and purified three additional type IV VNAR proteins from our library, named VNAR2, VNAR3, and VNAR4 (Supplementary Figure S6). Melting temperature and chemical denaturation midpoint (Cm) were measured by gradient heating and urea treatment, respectively (Figure 5D; Table 1). O. kenojei VNAR showed melting temperature values ranging from 54°C to 63°C, a moderate range compared to all four types of shark VNARs (Liu et al., 2014). In terms of chemical stability, O. kenojei VNAR had a Cm value of around 5 M urea. The thermal stability of O. kenojei VNAR was within the normal range of shark single-domain antibodies. The absence of extra disulfide bonds between CDR1 and CDR3 did not compromise the stability of O. kenojei VNAR but instead made O. kenojei VNAR a potentially good repertoire of sdAb with more flexible paratopes (Zielonka et al., 2015).
In summary, we have presented evidence of cDNA transcripts, variable domain germline sequence, and MS analysis for the identification of O. kenojei IgNAR. The crystal structure of O. kenojei VNAR showed high similarity to shark VNARs. The cloning of full-length and short-form O. kenojei IgNAR, as well as the high-throughput sequencing of the VNAR library, has provided valuable insights into the sequence diversity and structural diversity of O. kenojei IgNAR.
The comparative study of elasmobranchs’ adaptive immune systems will further our understanding of the origin and evolution of adaptive immunity. One of the most intriguing immunological macromolecules found in elasmobranchs is IgNAR, which is naturally devoid of the light chain and has a single-domain variable region that can function properly independently of the constant domains. Phylogenetic analysis of shark IgNAR and camelid HCAb showed that this immunoglobulin innovation evolved through convergent evolution in two distant vertebrate groups (Flajnik et al., 2011). IgNAR is believed to have appeared before the divergence of sharks and skates, with only one IgNAR cDNA transcript identified in skates (Rumfelt et al., 2004a; Flajnik et al., 2011; Barelle and Porter, 2015). In the current study, we provided evidence and analysis of O. kenojei IgNAR from blood serum, mononuclear cell transcriptome, and somatic genome, demonstrating that O. kenojei IgNAR was highly homologous to shark IgNAR. These findings support the notion that IgNAR is one of the most unique and ancient representatives of the adaptive immune system.
The discovery of IgNAR not only provides fresh insight into immunoglobulin composition but also presents a great opportunity for the modern pharmaceutical industry in developing biologics based on immunoglobulin variable domains. The O. kenojei VNARs are mostly constituted by type IV VNARs, which are rarely seen in other reported elasmobranchs. This type IV O. kenojei VNAR repertoire could greatly enhance the structural and clonal diversity of the elasmobranch single-domain antibody naïve library, which means previously difficult antigens or epitopes could have a better chance to generate specific and functional antibodies.
From a functional perspective, the O. kenojei VNAR crystal structure showed high similarity to shark VNARs, especially in the framework structures, suggesting that O. kenojei VNAR may function as a single-domain antibody to bind with antigens. A previous report has shown that a type IV VNAR against human serum albumin binds to the antigen in an atypical mode that involves the CDR3, HV2, and a portion of the framework (Kovalenko et al., 2013). We look forward to determining if O. kenojei, with its unique VNAR isotype composition, could provide new insights into antibody–antigen interactions.
It has been reported that both the framework and CDRs contribute to sdAb stability (Goldman et al., 2017). To investigate the stability of O. kenojei VNAR, we tested the melting temperature and urea resistance of several Type IV VNARs and compared them to Type II VNARs with two pairs of disulfide bonds (Liu et al., 2014). Surprisingly, these type IV O. kenojei VNARs, which have only one pair of canonical disulfide bonds, exhibited high melting temperature and Cm values and did not appear to compromise stability, despite the common practice of introducing additional disulfide bonds to enhance antibody stability (Goldman et al., 2017). The O. kenojei VNAR’s framework and CDR amino acid composition may therefore serve as important references for antibody stability improvement.
Certain aspects should be addressed in further studies on O. kenojei IgNAR. The complete and accurate identification of O. kenojei IgNAR miniloci could not be accomplished solely through standard molecular cloning approaches. The publication of genome sequencing results will enable us to determine the exact chromosomal location and copy number of IgNAR miniloci. Previous studies have demonstrated that IgNAR homodimers can aggregate and multimerize in blood serum (Smith et al., 2012). O. kenojei IgNAR-specific monoclonal antibodies, accompanied by mass spectrometry, would be suitable for further elucidating the natural multimeric state of O. kenojei IgNAR.
The NGS data of VNAR from different organs presented in the study are deposited in NCBI repository, accession numbers are: BioProject: PRJNA953503, SRA: SRR24109324 (epigonal organ), SRR24109325 (spleen), and SRR24109326 (blood). The NGS data of blood PBMC transcriptome presented in the study are deposited in NCBI repository, accession number are: BioProject: PRJNA953513 and SRA: SRR24110829. The cloned cDNA sequences of IgNAR presented in the study are deposited in GeneBank NCBI, accession number are: IgNAR Sec1: OQ793031, IgNAR Sec2: OQ793032, IgNAR Tm1: OQ793033, IgNAR Tm2: OQ793034, IgNARshort Tm1: OQ793035, IgNARshort Tm2: OQ793036. The cloned genomic sequence of VNAR miniloci presented in the study are deposited in GeneBank NCBI, accession number are: V_locus 1: OQ793037, V_locus 2: OQ793038, V_locus 3: OQ793039, V_locus 4: OQ793040.
All animal experiments in this study were approved by the Ethics Committee of the Institute of Oceanology, Chinese Academy of Sciences (No. MB2208-3).
JW identified the IgNAR and VNAR and generated all the samples for structure determination and stability assays. JW and PD generated the sample for sequencing. JG and CT performed the sequencing data processing and analyzed the VNAR diversity. PL determined the VNAR structure. MS and YW performed and analyzed the stability assays. JW, JG, HW and YB contributed to preparing the manuscript. All authors contributed to the article and approved the submitted version.
This work is financed by the National Natural Science Foundation of China (Grant No. 41876168), the Taishan Young Scholar Program of Shandong Province (tsqn201812108), and the Users with Excellence Program of Hefei Science Center, CAS (Grant No. 2020HSC-UE018).
We thank the staff at the Intelligent Simulator of Marine Ecosystems (ISME) and the staff at the UNcle System at the Shenzhen Bay Laboratory for instrument support and technical assistance. We thank Haitao Xiang and Jiahai Shi for kindly providing a collection of RSS query sequences. This work is also supported by the Oceanographic Data Center, IOCAS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1183744/full#supplementary-material
Anderson M. K., Shamblott M. J., Litman R. T., Litman G. W. (1995). Generation of immunoglobulin light chain gene diversity in raja erinacea is not associated with somatic rearrangement, an exception to a central paradigm of b cell immunity. J. Exp. Med. 182, 109–119. doi: 10.1084/jem.182.1.109
Barelle C., Porter A. (2015). VNARs: an ancient and unique repertoire of molecules that deliver small, soluble, stable and high affinity binders of proteins. Antibodies 4, 240–258. doi: 10.3390/antib4030240
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burciaga-Flores M., Marquez-Aguirre A. L., Duenas S., Gasperin-Bulbarela J., Licea-Navarro A. F., Camacho-Villegas T. A. (2023). First pan-specific vNAR against human TGF-beta as a potential therapeutic application: in silico modeling assessment. Sci. Rep. 13, 3596. doi: 10.1038/s41598-023-30623-x
Camacho-Villegas T., Mata-Gonzalez T., Paniagua-Solis J., Sanchez E., Licea A. (2013). Human TNF cytokine neutralization with a vNAR from heterodontus francisci shark: a potential therapeutic use. MAbs 5, 80–85. doi: 10.4161/mabs.22593
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Criscitiello M. F., Saltis M., Flajnik M. F. (2006). An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc. Natl. Acad. Sci. 103, 5036–5041. doi: 10.1073/pnas.0507074103
Crouch K., Smith L. E., Williams R., Cao W., Lee M., Jensen A., et al. (2013). Humoral immune response of the small-spotted catshark, scyliorhinus canicula. Fish Shellfish Immunol. 34, 1158–1169. doi: 10.1016/j.fsi.2013.01.025
Diaz M., Stanfield R. L., Greenberg A. S., Flajnik M. F. (2002). Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): identification of a new locus preferentially expressed in early development. Immunogenetics 54, 501–512. doi: 10.1007/s00251-002-0479-z
Dong Y., Pi X., Bartels-Burgahn F., Saltukoglu D., Liang Z., Yang J., et al. (2022). Structural principles of b cell antigen receptor assembly. Nature 612, 156–161. doi: 10.1038/s41586-022-05412-7
Dooley H. (2018). “Chondrichthyes: The immune system of cartilaginous fishes,” in Advances in comparative immunology (Springer International Publishing), 659–685. doi: 10.1007/978-3-319-76768-0_18
Eason D. D., Litman G. W. (2002). Haplotype exclusion: the unique case presented by multiple immunoglobulin gene loci in cartilaginous fish. Semin. Immunol. 14, 145–152. doi: 10.1016/S1044-5323(02)00038-6
Edgar R. C. (2021). MUSCLE v5 enables improved estimates of phylogenetic tree confidence by ensemble bootstrapping. bioRxiv 2021.06.20.449169. doi: 10.1101/2021.06.20.449169
Evans R., O’neill M., Pritzel A., Antropova N., Senior A., Green T., et al. (2022). Protein complex prediction with AlphaFold-multimer. bioRxiv 2021. 10.04.463034. doi: 10.1101/2021.10.04.463034
Feige M. J., Grawert M. A., Marcinowski M., Hennig J., Behnke J., Auslander D., et al. (2014). The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl. Acad. Sci. U.S.A. 111, 8155–8160. doi: 10.1073/pnas.1321502111
Feng M., Bian H., Wu X., Fu T., Fu Y., Hong J., et al. (2019). Construction and next-generation sequencing analysis of a large phage-displayed V(NAR) single-domain antibody library from six naive nurse sharks. Antib Ther. 2, 1–11. doi: 10.1093/abt/tby011
Fernandez-Quintero M. L., Fischer A. M., Kokot J., Waibl F., Seidler C. A., Liedl K. R. (2022). The influence of antibody humanization on shark variable domain (VNAR) binding site ensembles. Front. Immunol. 13, 953917. doi: 10.3389/fimmu.2022.953917
Flajnik M. F. (2018). A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 18, 438–453. doi: 10.1038/s41577-018-0003-9
Flajnik M. F., Deschacht N., Muyldermans S. (2011). A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PloS Biol. 9, e1001120. doi: 10.1371/journal.pbio.1001120
Frallicciardi J., Melcr J., Siginou P., Marrink S. J., Poolman B. (2022). Membrane thickness, lipid phase and sterol type are determining factors in the permeability of membranes to small solutes. Nat. Commun. 13, 1605. doi: 10.1038/s41467-022-29272-x
Goldman E. R., Liu J. L., Zabetakis D., Anderson G. P. (2017). Enhancing stability of camelid and shark single domain antibodies: an overview. Front. Immunol. 8, 865. doi: 10.3389/fimmu.2017.00865
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Greenberg A. S., Avila D., Hughes M., Hughes A., Mckinney E. C., Flajnik M. F. (1995). A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374, 168–173. doi: 10.1038/374168a0
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kovalenko O. V., Olland A., Piche-Nicholas N., Godbole A., King D., Svenson K., et al. (2013). Atypical antigen recognition mode of a shark immunoglobulin new antigen receptor (IgNAR) variable domain characterized by humanization and structural analysis. J. Biol. Chem. 288, 17408–17419. doi: 10.1074/jbc.M112.435289
Lee S. S., Tranchina D., Ohta Y., Flajnik M. F., Hsu E. (2002). Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 16, 571–582. doi: 10.1016/S1074-7613(02)00300-X
Li D., English H., Hong J., Liang T., Merlino G., Day C. P., et al. (2022). A novel PD-L1-targeted shark V(NAR) single-domain-based CAR-T cell strategy for treating breast cancer and liver cancer. Mol. Ther. Oncolytics 24, 849–863. doi: 10.1016/j.omto.2022.02.015
Li W., Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Liu J. L., Anderson G. P., Delehanty J. B., Baumann R., Hayhurst A., Goldman E. R. (2007a). Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol. Immunol. 44, 1775–1783. doi: 10.1016/j.molimm.2006.07.299
Liu J. L., Anderson G. P., Goldman E. R. (2007b). Isolation of anti-toxin single domain antibodies from a semi-synthetic spiny dogfish shark display library. BMC Biotechnol. 7, 78. doi: 10.1186/1472-6750-7-78
Liu J. L., Zabetakis D., Brown J. C., Anderson G. P., Goldman E. R. (2014). Thermal stability and refolding capability of shark derived single domain antibodies. Mol. Immunol. 59, 194–199. doi: 10.1016/j.molimm.2014.02.014
Magoc T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Malecek K., Lee V., Feng W., Huang J. L., Flajnik M. F., Ohta Y., et al. (2008). Immunoglobulin heavy chain exclusion in the shark. PloS Biol. 6, e157. doi: 10.1371/journal.pbio.0060157
Nuttall S. D., Krishnan U. V., Hattarki M., De Gori R., Irving R. A., Hudson P. J. (2001). Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol. 38, 313–326. doi: 10.1016/S0161-5890(01)00057-8
Ohtani M., Hikima J., Jung T. S., Kondo H., Hirono I., Takeyama H., et al. (2013). Variable domain antibodies specific for viral hemorrhagic septicemia virus (VHSV) selected from a randomized IgNAR phage display library. Fish Shellfish Immunol. 34, 724–728. doi: 10.1016/j.fsi.2012.11.041
Robert X., Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rumfelt L. L., Diaz M., Lohr R. L., Mochon E., Flajnik M. F. (2004a). Unprecedented multiplicity of ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol. 173, 1129–1139. doi: 10.4049/jimmunol.173.2.1129
Rumfelt L. L., Lohr R. L., Dooley H., Flajnik M. F. (2004b). Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 5, 8. doi: 10.1186/1471-2172-5-8
Rumfelt L. L., Mckinney E. C., Taylor E., Flajnik M. F. (2002). The development of primary and secondary lymphoid tissues in the nurse shark ginglymostoma cirratum: b-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol. 56, 130–148. doi: 10.1046/j.1365-3083.2002.01116.x
Shamblott M. J., Litman G. W. (1989). Genomic organization and sequences of immunoglobulin light chain genes in a primitive vertebrate suggest coevolution of immunoglobulin gene organization. EMBO J. 8, 3733–3739. doi: 10.1002/j.1460-2075.1989.tb08549.x
Smith L. E., Crouch K., Cao W., Muller M. R., Wu L., Steven J., et al. (2012). Characterization of the immunoglobulin repertoire of the spiny dogfish (Squalus acanthias). Dev. Comp. Immunol. 36, 665–679. doi: 10.1016/j.dci.2011.10.007
Stanfield R. L., Dooley H., Flajnik M. F., Wilson I. A. (2004). Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 305, 1770–1773. doi: 10.1126/science.1101148
Streltsov V. A., Varghese J. N., Carmichael J. A., Irving R. A., Hudson P. J., Nuttall S. D. (2004). Structural evidence for evolution of shark ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc. Natl. Acad. Sci. U.S.A. 101, 12444–12449. doi: 10.1073/pnas.0403509101
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Vander Heiden J. A., Yaari G., Uduman M., Stern J. N., O'connor K. C., Hafler D. A., et al. (2014). pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30, 1930–1932. doi: 10.1093/bioinformatics/btu138
Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009). Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wei L., Wang M., Xiang H., Jiang Y., Gong J., Su D., et al. (2021). Bamboo shark as a small animal model for single domain antibody production. Front. Bioeng Biotechnol. 9, 792111. doi: 10.3389/fbioe.2021.792111
Yariv B., Yariv E., Kessel A., Masrati G., Chorin A. B., Martz E., et al. (2023). Using evolutionary data to make sense of macromolecules with a "face-lifted" ConSurf. Protein Sci. 32, e4582. doi: 10.1002/pro.4582
Zhang W., Qin L., Cai X., Juma S. N., Xu R., Wei L., et al. (2020). Sequence structure character of IgNAR sec in whitespotted bamboo shark (Chiloscyllium plagiosum). Fish Shellfish Immunol. 102, 140–144. doi: 10.1016/j.fsi.2020.04.037
Keywords: IgNAR, VNAR, skate, VNAR structure, antibody diversity
Citation: Wen J, Gong J, Li P, Deng P, Sun M, Wu Y, Tian C, Wang H and Bi Y (2023) Identification and characterization of IgNAR and VNAR repertoire from the ocellate spot skate (Okamejei kenojei). Front. Mar. Sci. 10:1183744. doi: 10.3389/fmars.2023.1183744
Received: 10 March 2023; Accepted: 04 May 2023;
Published: 24 May 2023.
Edited by:
Zedong Jiang, Jimei University, ChinaReviewed by:
Andor J. Kiss, Miami University, United StatesCopyright © 2023 Wen, Gong, Li, Deng, Sun, Wu, Tian, Wang and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Wang, d2FuZ2hhb0BxZGlvLmFjLmNu; Yunchen Bi, eXVuY2hlbmJpQHFkaW8uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.