
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 11 April 2023
Sec. Marine Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1174176
This article is part of the Research Topic Interactions between the Adaptability of Marine Organisms and Environmental Factors View all 5 articles
Introduction: The rapid development of the Litopenaeus vannamei industry largely depends on the acquisition of high-quality larvae. Shrimp larval development is a unique metamorphic process that results in mortality due to disease vulnerability. Currently, studies on innate immunity in shrimp are mainly focused on the adult stage, but little has been done on innate immunity in larvae.
Methods: In this study, utilizing a high-throughput Illumina sequencing approach, we compare the transcriptome of L. vannamei Zoea larvae and adults infected by Vibrio parahaemolyticus.
Results: A total of 487,887,650 high quality reads were obtained, assembled, and aggregated into 23,802 genes, among which 3315 were identified as differentially expressed genes. Then GO term enrichment analysis, KEGG pathway enrichment analysis and functional gene analysis were performed. We found that pathways that are involved in the innate immune response, such as mTOR signaling pathway, MAPK signaling pathway, and Notch signaling pathway, were more enriched in the Zoea stage compared to the adult stage, suggesting that innate immunity against Vibrio vulnificus may exist during the Zoea larvae stage.
Discussion: were more enriched in the Zoea stage compared to the adult stage, suggesting that innate immunity against Vibrio vulnificus may exist during the Zoea larvae stage. These findings will provide useful information for the study of innate immunity in shrimp larvae.
The Pacific white shrimp, Litopeneaus vannamei, is one of the most common aquaculture species worldwide, reaching a production of 4,966,200 MT (FAO, 2020). The cultivation of Pacific white shrimp is hampered mostly by poor larvae quality and frequent disease outbreaks (Wang Y. et al., 2020). Larval nursery refers to the phases of shrimp larval development, including nauplius, Zoea, mysis, and early postlarvae (Hertzler and Freas, 2009). It is a critical process that largely determines the success of L. vannamei culture. In successive shrimp culture phases, the growth, development, and stress/disease resistance of the shrimp are directly proportional to the quality of the larvae (Angthong et al., 2021).
During the early stages of shrimp development, their morphology, physiology and ecology undergo profound changes. At larvae stages, especially from the Zoea to postlarvae, shrimps have low immunity and are susceptible to many pathogens, which could result in mass mortalities in the shrimp hatchery (Zheng et al., 2016). Vibrio Parahemolyticus is gram-negative bacteria that has strong pathogenicity and high transmissibility for aquatic animals (Velázquez-Lizárraga et al., 2019). Infection with Vibrio parahaemolyticus in the early stages of shrimp, particularly the Zoea stage, often results in huge death of larvae (Angthong et al., 2021). In China, an unique illness known as “translucent post-larvae disease” (TPD) or “glass post-larvae disease” (GPD) caused by highly pathogenic vibrios with virulence has lately caused significant economic losses (Zou et al., 2020).
Same with other invertebrates, shrimps depend on innate immunity consisting of humoral and cellular responses to combat infections. Upregulation of the prophenoloxidase system (ProPO), clotting proteins, melanization, and antimicrobial peptides are all components of the humoral immune response (Mycology et al., 1998). Cellular immune responses eliminate pathogens by hemocyte processes such as phagocytosis, apoptosis, nodule formation, and encapsulation (Jiravanichpaisal et al., 2006). However, the majority of these immune responses were detected in adult stages of L. vannamei, and little is known about the innate immune system in the early developmental stages of L. vannamei. In this study, we analyzed the transcriptome of Zoea larvae and adults infected with Vibrio parahaemolyticus in L. vannamei using Illumina high-throughput sequencing technique. In order to uncover immune-related genes and biological processes involved with pathogen resistance responses in Zoea larvae stages, adult stage transcriptome data were compared to larval stage transcriptome data. This study will provide insights into the innate immune response in Zoea larvae of L. vannamei to pathogenic microbial infections.
The L. vannamei in nauplii (stages 4–5) and adult shrimp (mean weight 10.55 ± 1.97g) were collected from Hengxing shrimp farm in Zhanjiang city, Guangdong, China. No specific permissions were required for the sampling locations and activities, and the studies did not involve endangered or protected species and locations.
The nauplii were acclimated, thoroughly rinsed with seawater, disinfected with iodine (100 μl L-1) and maintained in a 200 L glass aquarium at 30–31°C with 0.45 μm filtered seawater. From Zoea 1, they were fed with a mixture of Isochrisis galbana (30%) and Chaetoceros muelleri (70%) at a minimum density of 75,000 cells ml−1. The water was exchanged 30% daily, and the larvae were kept in a 12-h light dark photoperiod. Before each experiment, the nauplii were observed with a microscope to evaluate activity, deformities, yolk sac conditions, parasites, and debris adherence.
Shrimps were cultured for acclimation for one week in the laboratory. The water in the culture tank was maintained at a temperature of 26 ± 0.5 °C, salinity at 3‰, and pH at 7.2 ± 0.2 and one-third was replaced every 48h. Aeration was provided to maintain dissolved oxygen at a concentration of approximately 8 mg/L. The shrimp were fed with a formula feed at 1% of their body weight at 8:00 AM and 5:00 PM every day.
Vibrio parahaemolyticus was obtained from the Guangdong Microbiological Collection Center, GDMCC No. 1.306. Overnight, Vibrio parahaemolyticus was inoculated in Luria–Bertani broth at 37 °C with agitation at 150 rpm. The colonies were picked up and suspended in terile normal saline (0.9% NaCl) and centrifuged at 6000×g for 5 min at 20°C. Adjusting the bacterial suspension to an optical density of 1.0 at 610 nm and serially diluted to achieve a density 105 CFU ml−1.
The Vibrio parahaemolyticus-infected Zoea larvae experiment was divided into a control group (LC) and a Vibrio parahaemolyticus-infected group (LE). One hundred Zoea larvae were subjected for 24h to 1 ml of 104 cfu/ml concentration of Vibrio parahaemolyticus and 1 ml of PBS, respectively. Without any water changes throughout the experiment, each treatment was repeated five times. Five milliliters of microalgae were added every six hours. After feeding the larvae microalgae, a single inoculation of a bacterial solution was performed. Using a stereomicroscope, surviving larvae were counted after 24 hours. At 0 and 24 hours, samples of surviving larvae from the control and experimental groups were immediately stored in liquid nitrogen and then stored at -80°C.
Shrimp (mean weight 10.55 ± 1.97 g, n=50) were divided into two groups, control (SC) and Vibrio parahaemolyticus challenge (SE). Each group had four tanks with 50 shrimp in 200 L of UV-sterilized, aerated water. In the same way, Vibrio parahaemolyticus was cultivated with a final bacterial concentration of 2 x 103 CFU/mL. Injecting 50 ul of bacterial solution and PBS intramuscularly into the shrimp. The hepatopancreas from surviving shrimp in the control and experimental groups was taken for 24 hours and samples were immediately submerged in liquid nitrogen and then stored at -80°C.
Total RNA was extracted from each sample (hundred of larvae; six of shrimp) using Trizol reagent (Invitrogen, CA, USA) following the manufacturer’s procedure. Bioanalyzer 2100 and RNA 1000 Nano LabChip Kit (Agilent, CA, USA) were used to analyze the quantity and purity of total RNA having a RIN number >7.0. Poly(A) RNA is purified from total RNA(5ug) using poly-T oligo-attached magnetic beads using two rounds of purification. Following purification, the mRNA is fragmented into small pieces using divalent cations under elevated temperature. Then the cleaved RNA fragments were reverse-transcribed to create the final cDNA library in accordance with the protocol for the mRNASeq sample preparation kit (Illumina, San Diego, USA), the average insert size for the paired-end libraries was 300 bp ( ± 50 bp). At last, we performed the 2×150bp paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) following the vendor’s recommended protocol.
Firstly, Cutadapt (Kechin et al., 2017) and perl scripts in house were used to remove the reads that contained adaptor contamination, low quality bases and undetermined bases. Then sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), including the Q20, Q30 and GC-content of the clean data. All downstream analyses were based on clean data of high quality. De novo assembly of the transcriptome was performed with Trinity 2.4.0 (Grabherr et al., 2011). Trinity groups transcripts into clusters based on shared sequence content. Such a transcript cluster is very loosely referred to as a ‘gene’. The longest transcript in the cluster was chosen as the ‘gene’ sequence (Unigene).
All assembled Unigenes were aligned against the non-redundant (Nr) protein database (http://www.ncbi.nlm.nih.gov/), Gene ontology (GO) (http://www.geneontology.org), SwissProt(http://www.expasy.ch/sprot/), Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) and eggnog (http://eggnogdb.embl.de/) databases using DIAMOND (Buchfink et al., 2014) with a threshold of evalue<0.00001.
Salmon (Patro et al., 2017) was used to perform expression level for Unigenes by calculating transcript per millions (TPM) (Mortazavi et al., 2008). The differentially expressed Unigenes were selected with statistical significance (p value < 0.05) by R package edgeR (Robinson et al., 2009). Next, GO and KEGG enrichment analysis were again performed on the differentially expressed Unigenes by perl scripts in house.
QPCR was performed to validate gene expression patterns obtained from RNA-seq. Q-PCR was conducted with the SYBR® PrimeScript™ RT-PCR Kit (TaKaRa, China) according to the manufacturer’s protocol. Each reaction contained 1× SYBR Premix Ex Taq, 10 μM each primer, and 2 μL of cDNA (50 ng/mL) in a final volume of 25 μL. Purity and concentration of cDNA samples were determined by using NanoDrop (ND-8000) spectrophotometer. Eleven immune-related genes were selected for qPCR validation. Specific primer of each gene was designed using Primer Premier Program (Table S2). Each qPCR reaction contained 100 ng of cDNA template, 0.2 µM of each primer and 1X SYBR Green SsoAdvanced (BioRad). The cycle parameters were as follows; initial denaturation at 95°C for 30 s to activate the DNA polymerase, followed by 40 cycles of 5 s at 95°C, 30 s at 55°C, and 30 s at 72°C. The expression profile of each gene was calculated using 2−ΔΔCT method (Livak and Schmittgen, 2001). Relative gene expression analysis was normalized to that the housekeeping gene (β-actin) as an internal control. All qPCRs were performed in three biological replicates (n = 3).
After the metamorphosis of the nauplius larvae to zoea larvae, the body appears to be segmented and mouth open for feeding. Normal zoea larvae have transparent cephalothoracic armor and movable appendages that do not attach to objects (Figure 1A). After 24 h of Vibrio parahaemolyticus infection, a large amount of black substances were observed in the cephalothoracic armor area of Zoea larvae as compared to healthy larvae. The larvae had declined activity, weakened feeding ability, and the larvae body became sticky (Figure 1B). Compared to normal shrimp, adult shrimp infected with Vibrio parahaemolyticus exhibited atrophied hepatopancreas and unclear intestines (Figures 1C, D).
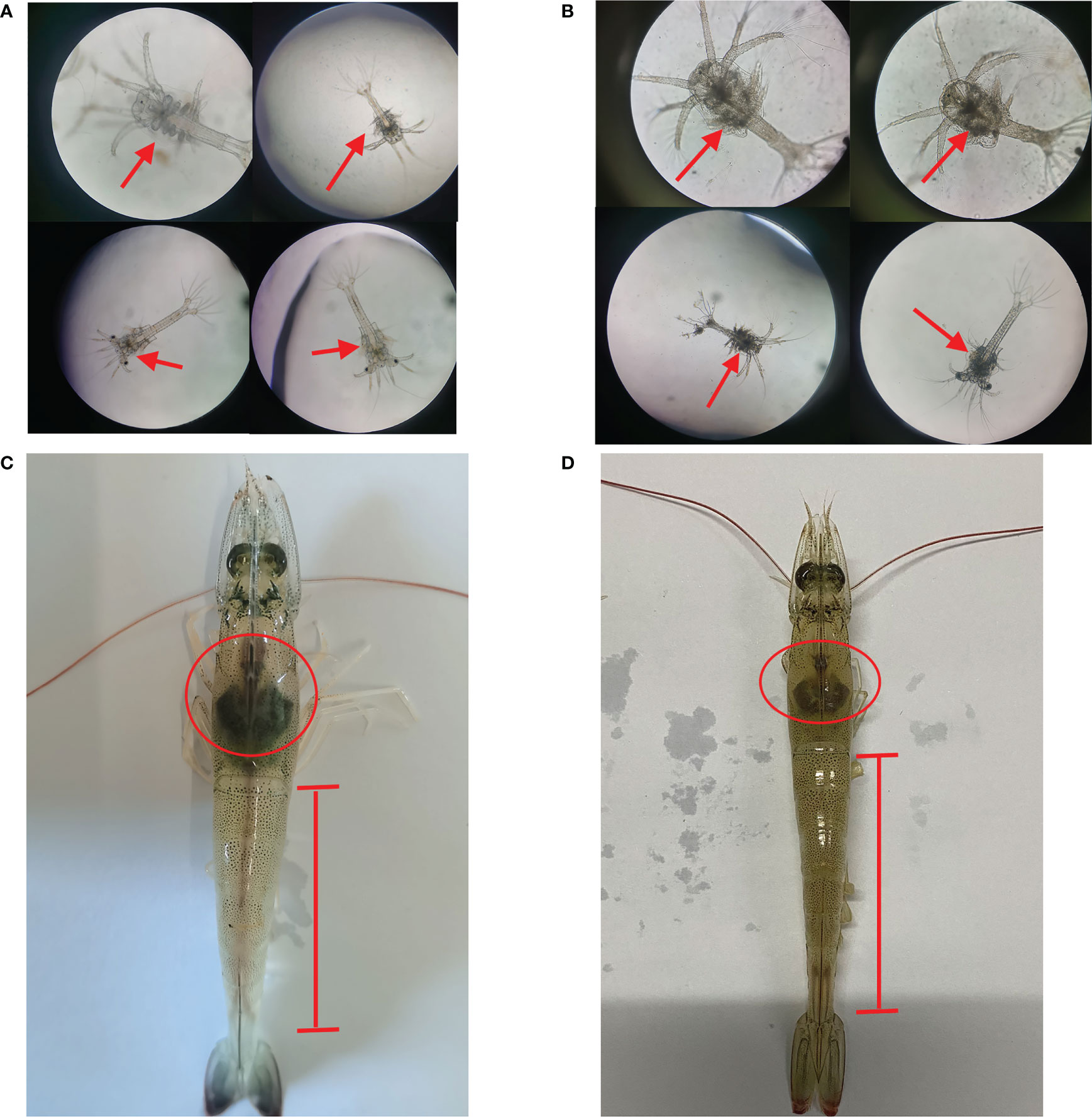
Figure 1 External signs of Zoea larvae and adults shrimp infected with Vibrio parahaemolyticus. (A) Normal Zoea larvae. (B) Larvae infected with Vibrio parahaemolyticus. (C) Normal adult shrimp. (D) Adult shrimp infected with Vibrio parahaemolyticus.
Using the Illumina Novaseq™ 6000 sequencer, larvae and adult shrimp samples from both the control group and the infection group infected with Vibrio parahaemolyticus were subjected to high throughput transcriptome sequencing. In each library, between 39,359,790 and 66,776,776 raw readings were generated. After removing reads containing adapters or poly-N and low-quality reads from raw data, 33,005,612 to 48,153,928 valid reads were filtered out (Table 1). Quality of the data was ensured with Q20 (base sequencing error probability < 1%) > 99% and Q30 (base sequencing error probability < 0.1%) > 96% in each library (Table 1).
To determine the gene expression profile of the Zoea larvae and adult shrimp after pathogen infection, four differentially expressed gene (DEG) libraries were constructed from larvae control (LC), adult control (SC), larvae experiment (LE) and adult experiment (SE). DEGs were identified using a Bayesian approach after gene expression levels were standardized to reads per kilobase of transcript sequence per million mapped reads (RPKM). By comparing the transcriptome libraries of pathogen-infected and control groups, a total of 3,315 differentially expressed genes (DEGs) were discovered. There were 1156 up-regulated genes and 578 down-regulated genes within the LC-LE group. The SC-SE group had a total of 1245 up-regulated genes and 336 down-regulated genes (Figure 2A). The study of the Venn diagrams of the DEGs for the various groups revealed that 81 DEGs overlapped between the LC-LE and SC-SE groups, but 1653 DEGs in the LC-LE group were distinct from 1500 DEGs in the SC-SE group (Figure 2B). 36 upregulated DEGs were overlapping between the LC-LE and SC-SE groups, whereas 1209 upregulated DEGs in SC-SE were distinct from 1120 upregulated DEGs in LC-LE (Figure 2C). 3 downregulated DEGs were overlapping between the LC-LE and SC-SE groups, while the 575 downregulated DEGs in LC-LE differed from the 333 downregulated DEGs in SC-SE (Figure 2D).
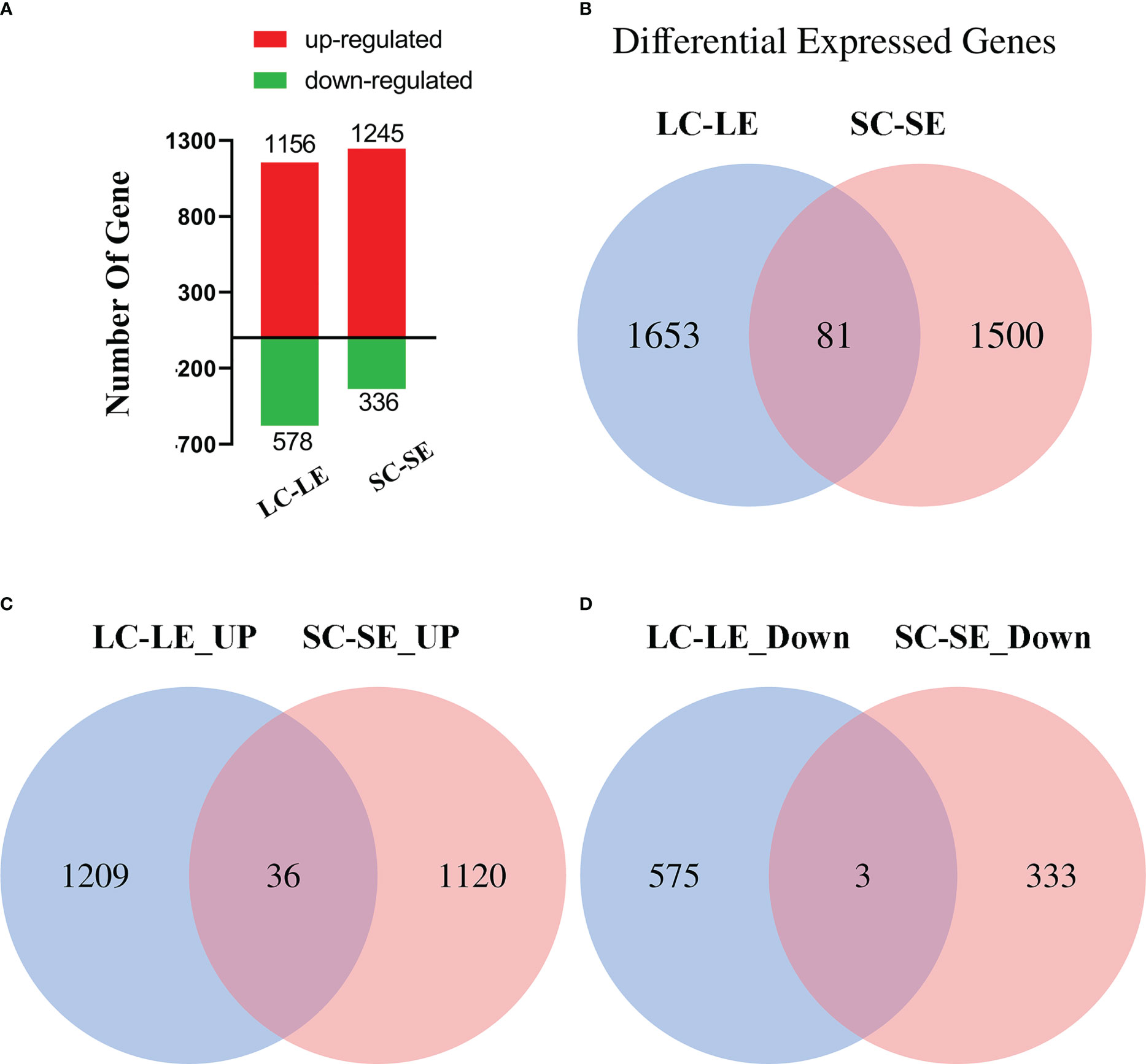
Figure 2 Statistical results of different expressed genes (DEGs). (A) The number of the differentially expressed genes in workers, LC-LE and SC-SE. Red bar for up-regulated genes, green bar for down-regulated genes. (B) Venn diagram shows 2-fold expression of transcripts in the LC-LE and SC-SE transcriptome. (C) Venn diagram shows the up-regulated expression of transcripts in the LC-LE and SC-SE transcriptome. (D) Venn diagram shows the down-regulated expression of transcripts in the LC-LE and SC-SE transcriptome. Statistical analysis results were all Padj < 0.05.
To further evaluate the functions of DEGs and the signaling pathways in which they engage, 3315 DEGs were classified into biological process (BP), molecular function (MF), and cellular component (CC) categories (Figures 3A, B). Among the transcripts of the LC-LE group, transcripts show the following enriched terms for biological process: proteolysis (46 transcripts), chitin metabolic process (30 transcripts) and protein phosphorylation (27 transcripts). For molecular function the enriched terms were: protein binding (47 transcripts), ATP binding (45 transcripts) and metal ion binding (39 transcripts). Integral component of membrane (122 transcripts), cytoplasm (114 transcripts) and plasma membrane (93 transcripts) were the GO keywords for cellular components (Figure 3A). Among the SC-SE group, terms for the transcripts showed the following terms, for biological process: proteolysis (61 transcripts), oxidation-reduction process (37 transcripts) and cell adhesion (25 transcripts); for molecular function: protein binding (50 transcripts), metal ion binding (48 transcripts) and calcium ion binding (48 transcripts); for cellular component: cytoplasm (139 transcripts), integral component of membrane (124 transcripts) and plasma membrane (105 transcripts) (Figure 3B).

Figure 3 Gene ontology assignments for differentially expressed genes (DEGs) in Zoea larvae and adult after infection with V. parahaemolyticus. The GO classification results summarized in three main GO categories (cellular component, molecular function, and biological process). (A) GO analysis for the genes upon V. parahaemolyticus infected Zoea larvae. (B) GO analysis for the genes upon V. parahaemolyticus infected adult.
KEGG pathway database is a database to analyze gene product during metabolism process and related gene functions in the cellular processes. Pathway-based analysis helps us to further understand the biological functions of genes (Yi et al., 2020). 551 DEGs found in LC-LE groups were linked to 236 known KEGG pathways in this investigation. 402 DEGs were linked to 178 known KEGG pathways for SC-SE groups. On the basis of an earlier study of the Venn diagram of DEGs, KEGG enrichment analyses for both up- and down-regulated DEGs in each group were conducted (Figures 4A–D). DEGs specifically upregulated in the LC-LE group were mainly enriched in immune pathways such as MAPK signaling pathway, Complement and coagulation cascades and mTOR signaling pathway (Figure 4A). Up-regulation of DEGs specific to the SC-SE group, mainly enriched in immune pathways including ECM-receptor interaction, Lysosome, Toll and Imd signaling pathway, VEGF signaling pathway and MAPK signaling pathway (Figure 4B). DEGs that were particularly downregulated in the LC-LE group were concentrated in the Phototransduction – fly, Purine metabolism and Fructose and mannose metabolism pathway (Figure 4C). The specific down-regulated DEGs in the SC-SE group were mainly clustered in the Lysosome, Arginine and proline metabolism and Pentose and glucuronate interconversions pathways (Figure 4D). A total of 36 DEGs were upregulated in the LC-LE and SC-SE groups (Table 2), among which the immune-related genes were: serine protease inhibitor dipetalogastin-like, C-type lectin, Phenoloxidase-activating factor 3-like, Phenoloxidase-activating factor 1-like, prophenoloxidase-activating enzyme 2a and putative antimicrobial peptide. The immune pathways enriched in both upregulated genes are: ECM-receptor interaction, Endocytosis, NF-kappa B signaling pathway, Toll and Imd signaling pathways and phagosome. Three genes were common down-regulated in the LC-LE and SC-SE groups (Table 3), tropomyosin-1, uncharacterized protein LOC113817585 and uncharacterized protein LOC113812737.
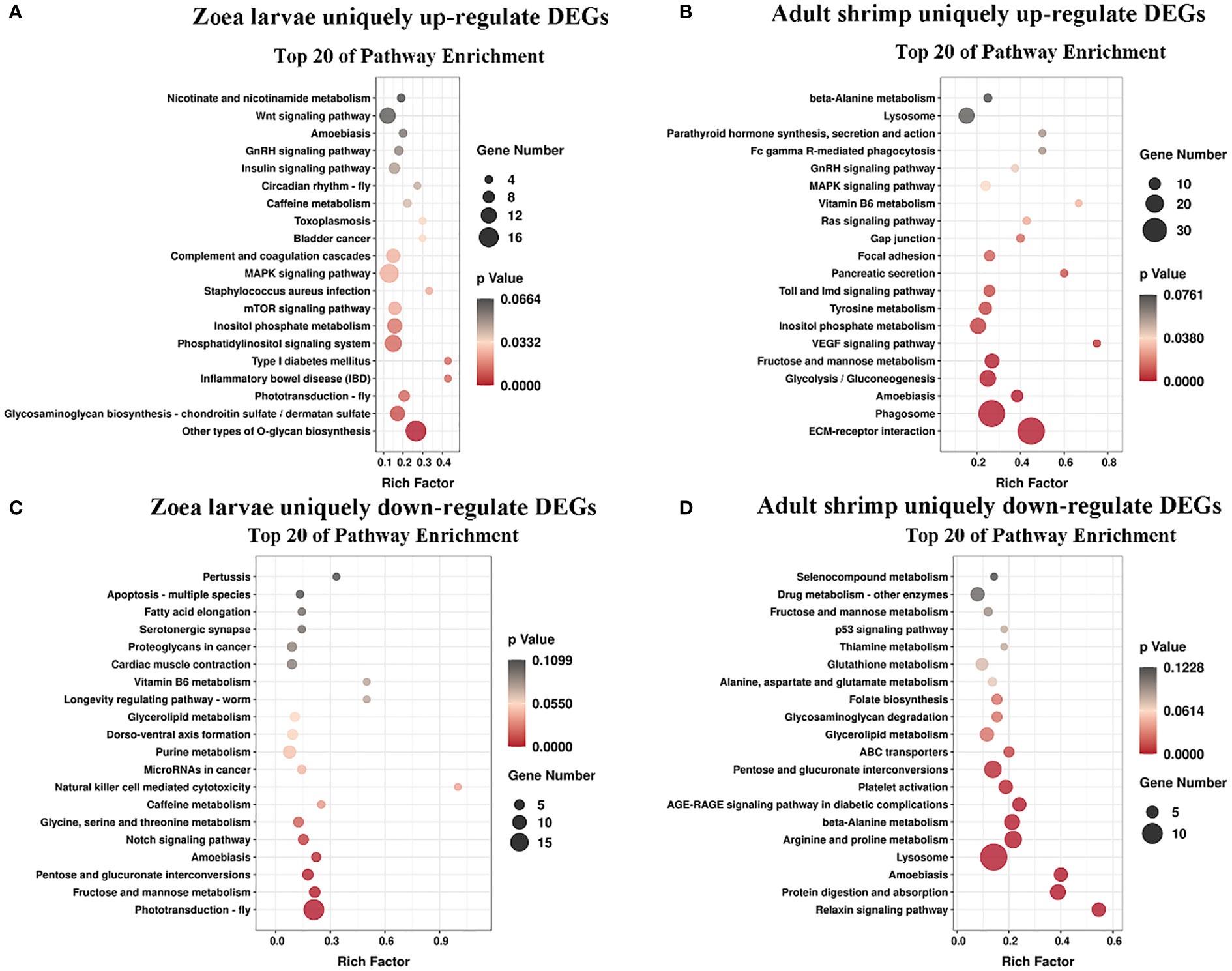
Figure 4 KEGG pathway enriched scatterplot. Axis represents the name of the pathway, the horizontal axis represents the corresponding rich factor of the pathway, the size of the p value is represented by the color of the point. The number of differential genes is indicated by the size of the point. (A) Enrichment of KEGG terms with unique up-regulated genes after Vibrio parahaemolyticus infection of Zoea larvae. (B) KEGG terminology enrichment of unique up-regulated genes after Vibrio parahaemolyticus infection of adults. (C) Enrichment of KEGG terms with unique down-regulated genes after Vibrio parahaemolyticus infection of Zoea larvae. (D) Enrichment of KEGG terms unique to down-regulated genes after Vibrio parahaemolyticus infection of adults.
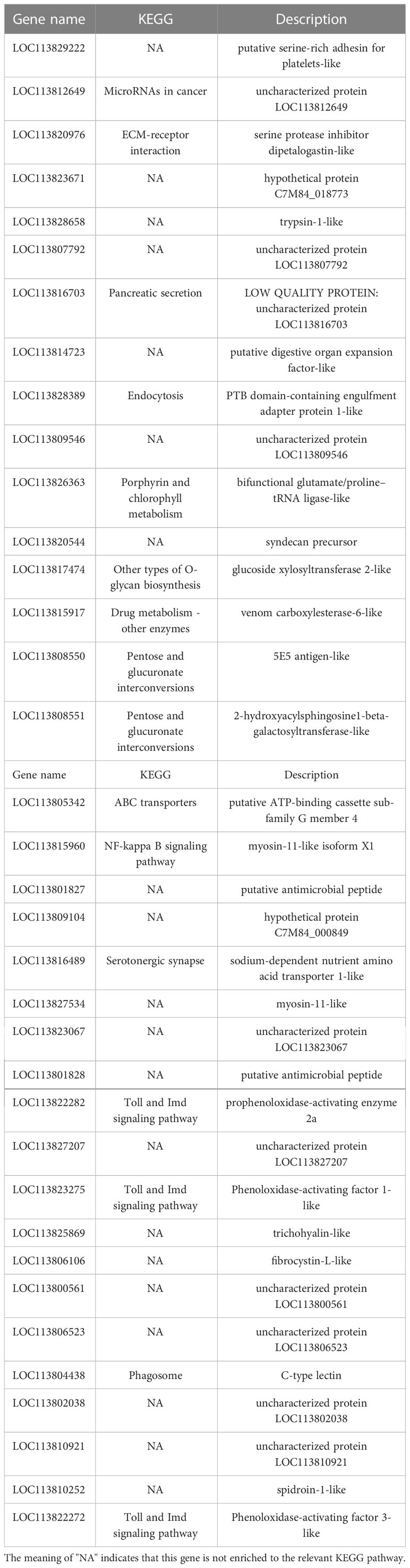
Table 2 The infection of Zoea larvae and adults with Vibrio parahaemolyticus co-up-regulates 36 genes.
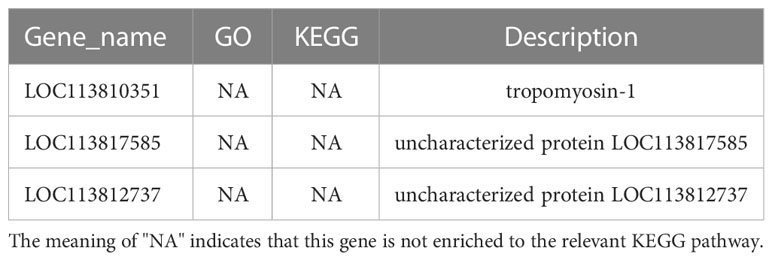
Table 3 The infection of Zoea larvae and adults with Vibrio parahaemolyticus co-down-regulates 3 genes.
To validate the expression patterns of DEGs after infected with Vibrio parahaemolyticus in Zoea larvae and adult shrimp, we selected 8 DEGs for qRT-PCR analysis (Figure 5) including 5 up-regulated and 3 down-regulated genes. The expression profiles of all selected IRDs from qPCR were compared with the expression profiles from RNA-seq. Dynamic changes in the levels of gene expression of these 8 DEGs after stimulation for 24 hours: β-Arrestin1, Protein yippee-like 1, beta-1,3-glucan-binding protein precursor, anti-lipopolysaccharide factor-like and phenoloxidase-activating factor 3-like were significantly up-regulated, while the expression level of adenosylhomocysteinase-like isoform X1, glutathione peroxidase-like and trypsin-1-like were significantly down-regulated.
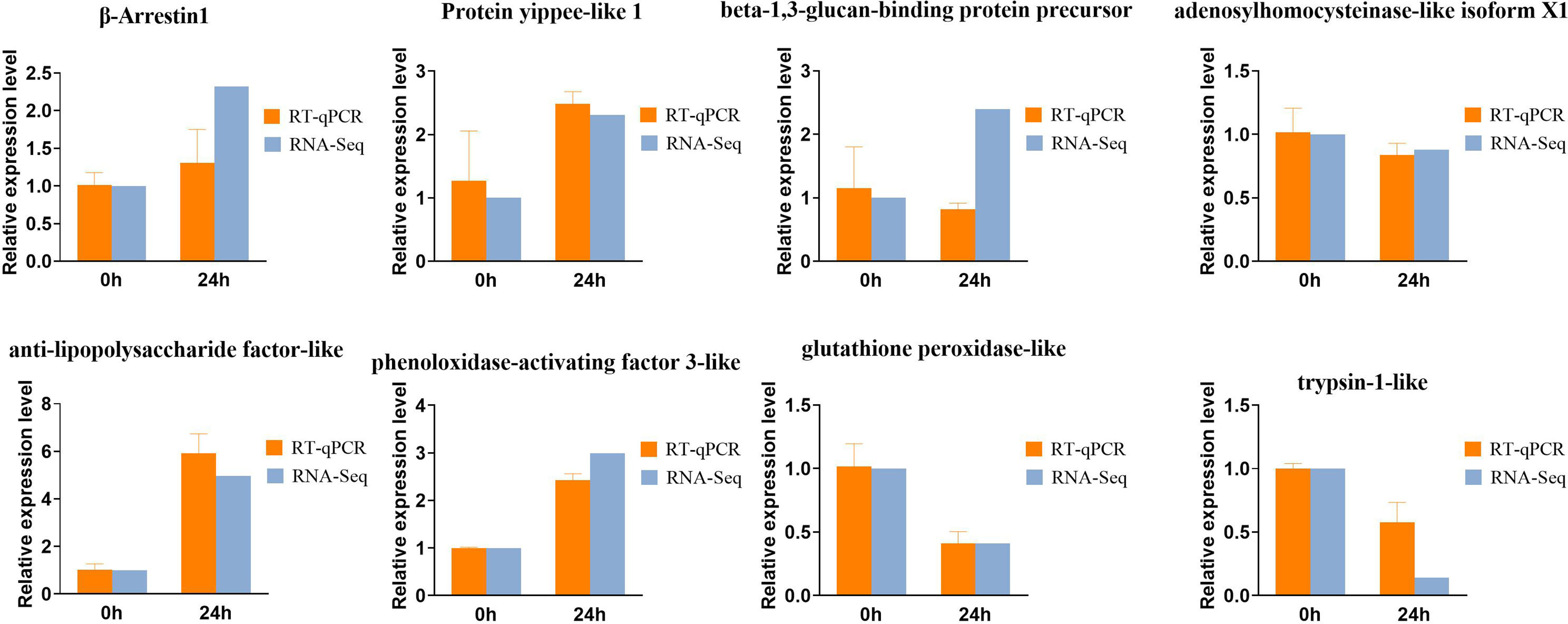
Figure 5 Transcripts from RNAseq data were verified by quantitative real-time PCR (qPCR), where the genes associated on Zoea larvae were beta-Arrestin1, Protein yippee-like 1, beta-1, 3-glucan-binding protein precursor, adenosine homocysteinase-like isoform X1, and on adults were anti-lipopolysaccharide factor-like, phenol oxidase activator 3-like, glutathione peroxidase-like and trypsin-like. The error bars indicate standard error of the mean from biological triplicates. Different letters show significant different by ANOVA (p value < 0.05).
As a member of Crustacea, Pacific white shrimp (Litopenaeus vannamei) undergoes several metamorphic developments in its early stages (Wei et al., 2014). This unique early developmental mode leads to dramatic changes in both nutritional patterns and morphological structure through metamorphosis and they are more susceptible to disease due to their underdeveloped immune systems. Particularly, the nutritional mode changes dramatically from yolk reserve to predation nutrition among nauplius to Zoea (Aguirre-Guzmán et al., 2001). During the period of Zoea larvae, the larvae rely on external food sources and are more sensitive to external environmental impacts, particularly infections by pathogenic Vibrio, which hinder larval growth. It has been reported that transcriptomic sequencing analysis has been performed throughout the early developmental stages of shrimp with the aim of comparatively analyzing the physiological changes during shrimp metamorphosis (Wei et al., 2014; Angthong et al., 2021). At present, majority of the research have focused on the adult stage, with few studies investigating the anti-vibrio immunity in the early stages of shrimp.
In this study, we performed transcriptomic analysis of Litopenaeus vannamei infected with Vibrio parahaemolyticus at both the larva and adult stage. In the sequencing study, whole larvae were utilized, whereas hepatopancreas tissue was taken from adults. The hepatopancreas is the primary organ for immunoprophylaxis in shrimp (Ji et al., 2009), and since the larvae were too tiny to sample the larval hepatopancreas, the larvae had to be collected in their whole. GO annotations of unigenes were classified into three functional categories: biological process, cellular component and molecular function (Figure 3B). The overrepresented GO terms for biological process were proteolysis and oxidation-reduction process. In the category of cellular component, three GO terms were considerably enriched: cytoplasm, integral component of membrane and plasma membrane. Protein binding and metal ion binding were the most prevalent molecular functions. Activation of the shrimp immune response depends on a variety of mechanisms, including the antioxidant system, hemolymph, wound repair and apoptosis-related proteins (Tassanakajon et al., 2013). During normal aerobic metabolism, all organisms produce reactive oxygen species (ROS). Microbial infection with stressful conditions can lead to oxygen deficiency and increase ROS, resulting in intracellular oxidative stress (Livingstone, 2001). Increased levels of ROS causes macromolecules damage which in turn affect membranes and enzymes structure and nucleic acids integrity (Indo et al., 2007). After adult infection with Vibrio parahaemolyticus, these differentially up- and down-regulated genes were examined independently for enrichment in the KEGG pathway (Figure 4B). KEGG-enriched pathways such as Toll (Huang et al., 2010; Yang et al., 2007), immunodeficiency (Imd) (Lan et al., 2013; Wang et al., 2019), MAPK signaling pathway (Yan et al., 2013) and phagosome pathway (Liu et al., 2020) have been shown to be involved in the immune response of shrimp to invading pathogens. This is consistent with the findings of the pathway enriched with immune genes in Vibrio parahaemolyticus-infected adult shrimp in this investigation. The pathway and genes that were upregulated the most in shrimp infected with Vibrio is the ECM-receptor interaction pathway. In crustaceans, some reports suggest that the ECM-receptor interaction pathway may be involved in the immune response against bacterial infection (Yang et al., 2018), white spot syndrome virus (WSSV) infection (Zhong et al., 2017) and heavy metal exposure (Meng et al., 2015). The vascular endothelial growth factor (VEGF) signaling pathway is known to play a crucial role in endothelial cell proliferation, migration, angiogenesis, vascular permeability, inhibition of apoptosis, and viral infection (Li et al., 2017; Wang et al., 2019). In this study, upregulated genes were enriched to this pathway, and it is possible that this pathway is associated with anti-vibrio immunity.
Vibrios infection is one of the main causes of mortality in early stages of shrimp. In this study, Vibrio parahaemolyticus was selected to infect Zoea larvae and adult shrimp. We aimed to identify any identical or distinct genes and pathways in response to Vibrios infections by larva or adult shrimp. The most significant GO biological process phrase for the LC-LE group was “proteolysis”, followed by “chitin metabolic process” and “protein phosphorylation”. In the category of cellular component, three GO terms integral component of membrane, cytoplasm and plasma membrane were significantly enriched. Protein binding, ATP binding, and metal ion binding were the most prevalent molecular functions (Figure 3A). Infection of Zoea larvae with Vibrio parahaemolyticus generates ATP through the body’s own oxidative reactions, which drive protein synthesis and degradation and activate protein phosphorylation and other activities to resist pathogenic invasion (Tang et al., 2022). In mammals, chitin has been found to activate innate immune cells and trigger airway inflammation in mice (Koller et al., 2011). It is also a major component of the exoskeleton of crabs and shrimp, and its metabolic activity is particularly high during the early development of shrimp, which require multiple molts. Chitin may be utilized as a feed supplement to improve the lifespan of shrimp infected with Vibrio lysis (Cheng et al., 2021), and chitin-binding protein works as a modulator or pattern recognition receptor to generate an antimicrobial immune response in shrimp (Hakimi et al., 2017).
The defense of shrimp against infectious agents begins with a humoral response, which relies on pattern recognition receptors (PRR) such as C-type lectins (Wang X. W. et al., 2020) that induce the synthesis of immune effectors such as antimicrobial peptides (AMP) by recognizing signals that trigger Toll-like receptors (TLR) and immunodeficiency (IMD) pathways (Tassanakajon et al., 2013). In contrast, cellular immunity is initiated by a series of phenoloxidase (proPO) activation systems, leading to phagocytosis, encapsulation, coagulation and melanization of invading pathogens (Cerenius et al., 2008). In this work, Vibrio parahaemolyticus-infected Zoea larvae and adults co-expressed 36 up-regulated genes (Table 2). Among them, those related to cellular immunity included C-type lectins, putative antimicrobial peptides. The C-type lectins are a large class of pattern recognition proteins (PRR) and have been reported to be involved in invertebrate innate immunity, such as cell adhesion, bacterial clearance, phagocytosis, activation of prophenoloxidase and encapsulation (Bi et al., 2020). Antimicrobial peptides (AMP) are direct effector molecules for the elimination and clearance of pathogenic infections, which are generally generated via the Toll and IMD pathways, and they play a crucial part in crustacean humoral immunity (Tassanakajon et al., 2018). In cellular immunity, pathogen invasion triggers the prophenoloxidase activation system, which is composed of endogenous trypsin-like serine proteases also known as phenoloxidase activators (PPAF) such as: phenoloxidase activator 3-like, phenoloxidase activator 1-like, prophenoloxidase activator 2a. These PPAFs then convert phenol oxidase (pro-PO) to the active form of phenol oxidase (PO),which blackens the pathogen (Söderhäll et al., 1994). Serine protease inhibitors (Serpins) are a large family of protease inhibitors that are involved in many critical bio-logical processes, including blood clotting, fibrinolysis, programmed cell death, development and innate immunity (Kong et al., 2013). MjSerp1 has been shown to play a direct effector role in the bacterial clearance of M. japonicus (Zhao et al., 2014). The immune-related pathways are ECM-receptor interactions, endocytosis, NF-kappa B signaling pathway, Toll and Imd signaling pathways and phagosome, which were consistently seen in both Zoea larvae and adults, implying that innate immunity against bacterial invasion may exist in zoea larvae, which is consistent with the findings of (Angthong et al., 2021).
For the LC-LE group, MAPK signaling pathway, Complement and coagulation cascades, and mTOR signaling pathway were among the 1209 immune-relevant pathways enriched in elevated DEGs (P<0.05). Several studies have demonstrated that activation of the mTOR signaling pathway by adding substances to the feed can regulate the intestinal health of shrimp (Duan et al., 2017). Additionally, the (mTORC1) signaling pathway can affect the replication of shrimp white spot virus (Hong et al., 2022). However, little research has been conducted on the anti-vibrio aspects of shrimp, and additional research is required to determine whether it is involved in innate immunity in shrimp larvae. In mammals, however, it is essential for intestinal inflammation and epithelial cell morphogenesis, as well as for the development and function of innate and adaptive immune cells (Mafi et al., 2022).
The pathways enriched for the respective unique down-regulated genes in LC-LE and SC-SE are significantly different. Uniquely down-regulated genes in the LC-LE group were mostly enriched in metabolic pathways, and immune-related pathways such as the Notch signaling system. In mammals, the Notch signaling pathway is a highly evolutionarily conserved signaling pathway that regulates number of biological processes such as cell differentiation, tissue development and immune responses (Lai, 2004). Notch inhibition in mature T cells hampers their antifungal action (Neal et al., 2017). Notch’s significance in immunity is not restricted to vertebrates; it plays a significant part in the immune system of shrimp. L. vannamei Notch (LvNotch) regulates the formation of reactive oxygen species (ROS) in blood cells and is implicated in the immunological response. In the study, Vibrio parahaemolyticus-infected Zoea larvae exhibited downregulation of genes involved in the Notch signaling system, which is consistent with the discovery of (Zhao et al., 2021) that in vivo silencing of LvNotch increased the expression of genes involved in the NF- b pathway. Lysosomal pathways are among the immunological pathways enriched for differentially downregulated genes in the SC-SE group. Lysosomes maintain cellular homeostasis, act as signaling centers, integrate metabolism with cellular clearance, and are involved in many fundamental cellular processes such as plasma membrane repair, immune response, and cell death (Yang and Wang, 2017). The above results indicate that the immune genes and pathways that may be down-regulated after Vibrio infection during larvae period are not consistent with those of adults.
Our study described the transcriptomic changes due to Vibrio parahaemolyticus infections during either the Zoea larvae or adult stages. We compared the anti-vibrio immunity between the Zoea larvae and adult stages to investigate the innate resistance to Vibrio at the Zoea larvae stage. By RNA-seq data analysis, we detected the presence of a few immune-related signaling pathways such as mTOR signaling pathway, MAPK signaling pathway and Notch signaling pathway at the Zoea larvae stage, suggesting that Litopenaeus vannamei might be capable of producing innate immune responses against the infections of Vibrio during the Zoea larvae. This study provides useful information for further understanding mechanisms of antimicrobial immunity in Zoea larvae.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA950912.
NC: writing-original draft. YC and YH: formal analysis. JJ: formal analysis and editing. YS and SL: writing-review & editing. All authors contributed to the article and approved the submitted version.
This research was supported by Supported by China Agriculture Research System of MOF and MARA(CARS-48). We are especially grateful for the critical comments from the anonymous reviewers.
Authors YC, YH, and SL were employed by the company Guangdong Evergreen Feed Industry.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1174176/full#supplementary-material
Aguirre-Guzmán G., Vázquez-Juárez R., Ascencio F. (2001). Differences in the susceptibility of American white shrimp larval substages (Litopenaeus vannamei) to four vibrio species. J. Invertebr. Pathol. 78, 215–219. doi: 10.1006/jipa.2001.5073
Angthong P., Uengwetwanit T., Arayamethakorn S., Rungrassamee W. (2021). Transcriptomic analysis of the black tiger shrimp (Penaeus monodon) reveals insights into immune development in their early life stages. Sci. Rep. 11, 1–14. doi: 10.1038/s41598-021-93364-9
Bi J., Ning M., Xie X., Fan W., Huang Y., Gu W., et al. (2020). A typical c-type lectin, perlucin-like protein, is involved in the innate immune defense of whiteleg shrimp litopenaeus vannamei. Fish Shellfish Immunol. 103, 293–301. doi: 10.1016/j.fsi.2020.05.046
Buchfink B., Xie C., Huson D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Cerenius L., Lee B. L., Söderhäll K. (2008). The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. doi: 10.1016/j.it.2008.02.009
Cheng A. C., Shiu Y. L., Chiu S. T., Ballantyne R., Liu C. H. (2021). Effects of chitin from daphnia similis and its derivative, chitosan on the immune response and disease resistance of white shrimp, litopenaeus vannamei. Fish Shellfish Immunol. 119, 329–338. doi: 10.1016/j.fsi.2021.10.017
Duan Y., Zhang Y., Dong H., Wang Y., Zhang J. (2017). Effects of dietary poly-β-hydroxybutyrate (PHB) on microbiota composition and the mTOR signaling pathway in the intestines of litopenaeus vannamei. J. Microbiol. 55, 946–954. doi: 10.1007/s12275-017-7273-y
FAO. (2020). The state of world fisheries and aquaculture (SOFIA) 2020 REPORT[J]. Integr. Environ. Assess. Manage. 2020 (5), 16.
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hakimi M., Kisaka T., Sietsema K., Kihara Y., Wasserman K., Budoff M. (2017). The prognostic value of cpet: Association with coronary artery calcium score. J. Am. Coll. Cardiol. 69, 1830. doi: 10.1016/s0735-1097(17)35219-1
Hertzler P. L., Freas W. R. (2009). Pleonal muscle development in the shrimp penaeus (Litopenaeus) vannamei (Crustacea: Malacostraca: Decapoda: Dendrobranchiata). Arthropod Struct. Dev. 38, 235–246. doi: 10.1016/j.asd.2008.12.003
Hong P. P., Li C., Niu G. J., Zhao X. F., Wang J. X. (2022). White spot syndrome virus directly activates mTORC1 signaling to facilitate its replication via polymeric immunoglobulin receptormediated infection in shrimp. PloS Pathog. 18, 1–26. doi: 10.1371/journal.ppat.1010808
Huang X. D., Yin Z. X., Jia X. T., Liang J. P., Ai H. S., Yang L. S., et al. (2010). Identification and functional study of a shrimp dorsal homologue. Dev. Comp. Immunol. 34, 107–113. doi: 10.1016/j.dci.2009.08.009
Indo H. P., Davidson M., Yen H. C., Suenaga S., Tomita K., Nishii T., et al. (2007). Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion 7, 106–118. doi: 10.1016/j.mito.2006.11.026
Ji P. F., Yao C. L., Wang Z. Y. (2009). Immune response and gene expression in shrimp (Litopenaeus vannamei) hemocytes and hepatopancreas against some pathogen-associated molecular patterns. Fish Shellfish Immunol. 27, 563–570. doi: 10.1016/j.fsi.2009.08.001
Jiravanichpaisal P., Lee B. L., Söderhäll K. (2006). Cell-mediated immunity in arthropods: Hematopoiesis, coagulation, melanization and opsonization. Immunobiology 211, 213–236. doi: 10.1016/j.imbio.2005.10.015
Kechin A., Boyarskikh U., Kel A., Filipenko M. (2017). CutPrimers: A new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143. doi: 10.1089/cmb.2017.0096
Koller B., Müller-Wiefel A. S., Rupec R., Korting H. C., Ruzicka T. (2011). Chitin modulates innate immune responses of keratinocytes. PloS One 6, 1–7. doi: 10.1371/journal.pone.0016594
Kong H. J., Lee Y. J., Park I. S., Lee W. W., Kim Y. O., Nam B. H., et al. (2013). Molecular and functional characterizations of a kunitz-type serine protease inhibitor FcKuSPI of the shrimp fenneropenaeus chinensis. Fish Shellfish Immunol. 35, 1025–1029. doi: 10.1016/j.fsi.2013.06.023
Lai E. C. (2004). Notch signaling: Control of cell communication and cell fate. Development 131, 965–973. doi: 10.1242/dev.01074
Lan J. F., Zhou J., Zhang X. W., Wang Z. H., Zhao X. F., Ren Q., et al. (2013). Characterization of an immune deficiency homolog (IMD) in shrimp (Fenneropenaeus chinensis) and crayfish (Procambarus clarkii). Dev. Comp. Immunol. 41, 608–617. doi: 10.1016/j.dci.2013.07.004
Li S., Wang Z., Li F., Yu K., Xiang J. (2017). A novel vascular endothelial growth factor receptor participates in white spot syndrome virus infection in litopenaeus vannamei. Front. Immunol. 8. doi: 10.3389/fimmu.2017.01457
Liu J., Zhou T., Wang C., Wang W., Chan S. (2020). Comparative transcriptomics reveals eyestalk ablation induced responses of the neuroendocrine-immune system in the pacific white shrimp litopenaeus vannamei. Fish Shellfish Immunol. 106, 823–832. doi: 10.1016/j.fsi.2020.08.029
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Livingstone D. R. (2001). Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar. pollut. Bull. 42, 656–666. doi: 10.1016/S0025-326X(01)00060-1
Mafi S., Mansoori B., Taeb S., Sadeghi H., Abbasi R., Cho W. C., et al. (2022). mTOR-mediated regulation of immune responses in cancer and tumor microenvironment. Front. Immunol. 12. doi: 10.3389/fimmu.2021.774103
Meng X., Tian X., Nie G., Wang J., Liu M., Jiang K., et al. (2015). The transcriptomic response to copper exposure in the digestive gland of Japanese scallops (Mizuhopecten yessoensis). Fish Shellfish Immunol. 46, 161–167. doi: 10.1016/j.fsi.2015.05.022
Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mycology P., Biology C., Pld L. P. S. (1998). Role of the prophenoloxidase-activating system in invertebrate immunity Kenneth S6derh ill and lage cerenius. Curr. Opin. Immunol. 10, 23–28. doi: 10.1016/S0952-7915(98)80026-5
Neal L. M., Qiu Y., Chung J., Xing E., Cho W., Malachowski A. N., et al. (2017). T Cell–restricted notch signaling contributes to pulmonary Th1 and Th2 immunity during cryptococcus neoformans infection. J. Immunol. 199, 643–655. doi: 10.4049/jimmunol.1601715
Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Robinson M. D., McCarthy D. J., Smyth G. K. (2009). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Söderhäll K., Cerenius L., Johansson M. W. (1994). The prophenoloxidase activating system and its role in invertebrate defence. Ann. N. Y. Acad. Sci. 712, 155–161. doi: 10.1111/j.1749-6632.1994.tb33570.x
Tang X., Liu T., Li X., Sheng X., Xing J., Chi H., et al. (2022). Protein phosphorylation in hemocytes of fenneropenaeus chinensis in response to white spot syndrome virus infection. Fish Shellfish Immunol. 122, 106–114. doi: 10.1016/j.fsi.2022.01.038
Tassanakajon A., Rimphanitchayakit V., Visetnan S., Amparyup P., Somboonwiwat K., Charoensapsri W., et al. (2018). Shrimp humoral responses against pathogens: antimicrobial peptides and melanization. Dev. Comp. Immunol. 80, 81–93. doi: 10.1016/j.dci.2017.05.009
Tassanakajon A., Somboonwiwat K., Supungul P., Tang S. (2013). Discovery of immune molecules and their crucial functions in shrimp immunity. Fish Shellfish Immunol. 34, 954–967. doi: 10.1016/j.fsi.2012.09.021
Velázquez-Lizárraga A. E., Juárez-Morales J. L., Racotta I. S., Villarreal-Colmenares H., Valdes-Lopez O., Luna-González A., et al. (2019). Transcriptomic analysis of pacific white shrimp (Litopenaeus vannamei, Boone 1931) in response to acute hepatopancreatic necrosis disease caused by vibrio parahaemolyticus. PloS One 14, 1–28. doi: 10.1371/journal.pone.0220993
Wang Z., Li S., Yu Y., Yu K., Zhang X., Xiang J., et al. (2019). Identification and characterization of two novel vascular endothelial growth factor genes in litopenaeus vannamei. Fish Shellfish Immunol. 84, 259–268. doi: 10.1016/j.fsi.2018.10.019
Wang X. W., Vasta G. R., Wang J. X. (2020). The functional relevance of shrimp c-type lectins in host-pathogen interactions. Dev. Comp. Immunol. 109, 103708. doi: 10.1016/j.dci.2020.103708
Wang Y., Wang K., Huang L., Dong P., Wang S., Chen H., et al. (2020). Fine-scale succession patterns and assembly mechanisms of bacterial community of litopenaeus vannamei larvae across the developmental cycle. Microbiome 8, 1–16. doi: 10.1186/s40168-020-00879-w
Wei J., Zhang X., Yu Y., Huang H., Li F., Xiang J. (2014). Comparative transcriptomic characterization of the early development in pacific white shrimp litopenaeus vannamei. PloS One 9, 1–13. doi: 10.1371/journal.pone.0106201
Yan H., Zhang S., Li C. Z., Chen Y. H., Chen Y. G., Weng S. P., et al. (2013). Molecular characterization and function of a p38 MAPK gene from litopenaeus vannamei. Fish Shellfish Immunol. 34, 1421–1431. doi: 10.1016/j.fsi.2013.02.030
Yang H., Gao X., Li X., Zhang H., Chen N., Zhang Y., et al. (2018). Comparative transcriptome analysis of red swamp crayfish (Procambarus clarkia) hepatopancreas in response to WSSV and aeromonas hydrophila infection. Fish Shellfish Immunol. 83, 397–405. doi: 10.1016/j.fsi.2018.09.051
Yang C., Wang X. (2017). Cell biology in China: Focusing on the lysosome. Traffic 18, 348–357. doi: 10.1111/tra.12483
Yang L.-S., Yin Z.-X., Liao J.-X., Huang X.-D., Guo C.-J., Weng S.-P., et al. (2007). A toll receptor in shrimp. Mol. Immunol. 44, 1999–2008. doi: 10.1016/j.molimm.2006.09.021
Yi Y., Fang Y., Wu K., Liu Y., Zhang W. (2020). Comprehensive gene and pathway analysis of cervical cancer progression. Oncol. Lett. 19, 3316–3332. doi: 10.3892/ol.2020.11439
Zhao Y. R., Xu Y. H., Jiang H. S., Xu S., Zhao X. F., Wang J. X. (2014). Antibacterial activity of serine protease inhibitor 1 from kuruma shrimp marsupenaeus japonicus. Dev. Comp. Immunol. 44, 261–269. doi: 10.1016/j.dci.2014.01.002
Zhao W., Zheng Z., Aweya J. J., Wang F., Li S., Tuan T. N., et al. (2021). Litopenaeus vannamei notch interacts with COP9 signalosome complex subunit 1 (CNS1) to negatively regulate the NF-κB pathway. J. Proteomics 232, 104074. doi: 10.1016/j.jprot.2020.104074
Zheng Y., Yu M., Liu Y., Su Y., Xu T., Yu M., et al. (2016). Comparison of cultivable bacterial communities associated with pacific white shrimp (Litopenaeus vannamei) larvae at different health statuses and growth stages. Aquaculture 451, 163–169. doi: 10.1016/j.aquaculture.2015.09.020
Zhong S., Mao Y., Wang J., Liu M., Zhang M., Su Y. (2017). Transcriptome analysis of kuruma shrimp (Marsupenaeus japonicus) hepatopancreas in response to white spot syndrome virus (WSSV) under experimental infection. Fish Shellfish Immunol. 70, 710–719. doi: 10.1016/j.fsi.2017.09.054
Keywords: Zoea larvae, Vibrio parahaemolyticus, shrimp, RNA-seq, immune
Citation: Chen N, Jin J, Chen Y, Hu Y, Shen Y and Li S (2023) The transcriptome of Litopenaeus vannamei in zoea larvae and adults infected by Vibrio parahaemolyticus. Front. Mar. Sci. 10:1174176. doi: 10.3389/fmars.2023.1174176
Received: 26 February 2023; Accepted: 27 March 2023;
Published: 11 April 2023.
Edited by:
Pengzhi Qi, Zhejiang Ocean University, ChinaReviewed by:
Chaozheng Li, Sun Yat-sen University, ChinaCopyright © 2023 Chen, Jin, Chen, Hu, Shen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuchun Shen, c2hlbnl1Y2h1bkAxNjMuY29t; Sedong Li, bGlzZWRvbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.