
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 12 May 2023
Sec. Marine Pollution
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1172942
This article is part of the Research TopicMarine Biofilms on Natural and Artificial Surfaces : Impacts and Novel SolutionsView all 4 articles
 Jeff Shimeta1*†
Jeff Shimeta1*† Gemma Wilding-McBride1†
Gemma Wilding-McBride1† Nathan J. Bott1
Nathan J. Bott1 Richard Piola2
Richard Piola2 Rene Santander3
Rene Santander3 Martin Leary3
Martin Leary3 Andrew J. Scardino2
Andrew J. Scardino23D printing has become widely used to rapidly prototype and manufacture novel or bespoke objects or replacement components in a wide range of marine industries, engineering, and research. 3D-printed objects are subject to marine biofouling, impacting their operation and longevity. Application of antifouling paints or coatings adds costly and time-consuming steps and may interfere with the function of fine surface features, counteracting some of the benefits of 3D-printing technology. We measured the antifouling performance of two 3D-printing thermoplastics embedded with antifouling biocides to create 3D-printed materials with inherent antifouling properties: 1) polycaprolactone (PCL) mixed with the organic biocide dichlorooctylisothiazolinone (DCOIT) and extruded as 3D-printing filament, and 2) a commercial polylactic acid (PLA) 3D-printing filament with embedded copper powder. Settlement plates printed from these thermoplastics (“PCL-DCOIT” and “PLA-Cu”, respectively) and deployed in temperate, coastal marine water for 17 weeks during summer remained free of macrofouling. A biofilm developed, and 16S and 18S rRNA metabarcoding analyses revealed that early stage biofilms (at 5 and 12 weeks) had dramatically altered assemblage structures of both prokaryotes and eukaryotes compared to natural biofilms. The assemblage on PCL-DCOIT had reduced microbial diversity, strong dominance of Proteobacteria and chlorophytes, and almost complete absence of Flavobacteriia, Cyanobacteria, and diatoms. In contrast, the biofilm on PLA-Cu had a dominance of Flavobacteriia over Proteobacteria, and resistance to chlorophytes, yet similar to PCL-DCOIT it resisted Cyanobacteria and diatoms. Such alterations to biofilm microbial assemblages could influence microbial dynamics, biofilm growth, and settlement cues to which biofouler propagules respond. At 17 weeks, the two biocide-embedded thermoplastics completely resisted macrofouling, equally well as three commercial antifouling coatings (Intercept 8500, Hempaguard X7, Hempasil X3); however, PCL-DCOIT was more extensively covered by a microalgal film (79%, evidently chlorophytes) than were the commercial coatings, and PLA-Cu had the most settled detritus (100% cover). Biofilm assemblages on the commercial coatings were investigated for comparison, with PCL-DCOIT standing out due to its almost complete resistance to Flavobacteriia. Thermoplastic 3D-printing filaments with embedded biocides show promise for producing 3D-printed objects with inherent antifouling properties, avoiding or lessening the need to apply antifouling coatings, and possibly extending their service lifetime.
3D printing has rapidly become a widely used method to produce materials and equipment parts for numerous applications in the marine environment. For various maritime operations and industries, 3D printing allows rapid production of prototype and replacement parts for machinery, infrastructure, and oceanographic instrumentation (Mohammed, 2016). Some examples in industries, engineering, and research are components of autonomous underwater and surface vehicles (Staiano et al., 2016; Odetti et al., 2019), pressure housings (Breddermann et al., 2016), custom inserts for vessel sea chests (Leary et al., 2016; Piola et al., 2022), artificial reef structures and bioshelters (Zavoleas et al., 2020; Matus et al., 2021), coral reef restoration material (Albalawi et al., 2021), experimental substrates to study reef biodiversity and biofouling (Wolfe and Mumby, 2020; Ly et al., 2021), splints for marine veterinary uses (Christiansen et al., 2014), and marine mammal tags (Frankfurter et al., 2019).
Like all artificial materials in the marine environment, 3D-printed objects are subject to biofouling, which can interfere with the function of infrastructure and equipment (Delgado et al., 2021), facilitate the transport of invasive species (Piola et al., 2009), and necessitate costly cleaning or replacement (Schultz et al., 2011; Fitridge et al., 2012). The most common fouling-preventative measure for surfaces in the marine environment is the application of antifouling (AF) coatings that either contain biocides or have material properties that facilitate fouling release (FR) (Lejars et al., 2012; Gizer et al., 2023). Common biocides in AF paints and other coatings include metals such as Cu, Zn, Ni, and Pb, as well as organic compounds such as pyrithione, DCOIT (dichlorooctylisothiazolinone), diuron, and many others (Yebra et al., 2004; Bao et al., 2011). Biocides are gradually revealed at the surface of the coating by self-polishing from water motion or by controlled release. Common FR coatings are superhydrophobic, superhydrophilic, or amphiphilic (Gittens et al., 2013), and/or they may have modified surface topography (Carve et al., 2019; Védie et al., 2021), which are properties that prevent organisms from attaching firmly enough to resist shear forces from water flow. However, the need to apply antifouling coatings to 3D-printed objects before deploying them in water offsets the advantages of 3D printing for making parts quickly and cheaply, particularly parts with fine and complex geometries.
Many different materials such as thermoplastics and metals, and various additives such as colorants, plasticizers, flame retardants, etc. can be used in 3D printing. This allows creation of new composite materials that may have inherent antifouling properties, reducing the need for post-manufacture application of AF coatings. Research on 3D-printed materials with inherent antibacterial properties for medical applications is advanced (González-Henríquez et al., 2019), but little work has been done to develop materials for the marine environment. Notable examples are Bouranta et al. (2022), who made 3D-printed acrylonitrile butadiene styrene (ABS) infused with micro- or nano-ZnO that was effective in inhibiting microalgal fouling for aquaculture applications; and Dong et al. (2021), who used a hydrophobic ink to 3D-print superhydrophobic objects that may have marine antifouling applications.
Here we evaluated the marine antifouling performance of 3D-printed thermoplastic polymers that were infused prior to printing with powdered DCOIT (4,5-dichloro-2-n-octyl-4-isothiazolin-3-one) or copper, two of the most common biocides in AF paints and coatings. DCOIT was developed to replace organotin paints such as TBT, as it is highly toxic and nonspecific to marine organisms yet has been purported (controversially) to degrade rapidly in the environment (Chen and Lam, 2017). We developed a method, described here, of embedding powdered DCOIT into polycaprolactone (PCL) and extruding it as filament for fused filament fabrication (FFF) 3D printing. DCOIT is an appropriate biocide for this application because, when applied at sufficiently high concentrations (~10%), it is able to migrate through polymers and be exposed and released at the surface (Nichols, 2004). PCL was chosen as the thermoplastic because it is a hydrophobic, biodegradable polyester, and for FFF fabrication its recommended extrusion temperature (100-130 C) and printing temperature (120-140 C) are well below the temperature at which DCOIT volatilizes and/or becomes inactive (170 C). The other biocide used here, copper, is the most common toxic metal in AF paints (Yebra et al., 2004). Recent research has improved the effectiveness of both these biocides in AF coatings such as epoxies, polyurethanes, and polydimethylsiloxane (PDMS) (Ma et al., 2016; Fu et al., 2019; Ali et al., 2021; Padmavathi et al., 2021), but they have not been used previously in 3D printing to produce composite materials with inherent antifouling properties for marine applications. The copper-infused thermoplastic (polylactic acid, PLA) used here is available commercially as a printer filament for FFF 3D printing to make hard objects with a cast metal appearance. We have previously shown that the marine antifouling performance of test coupons printed from this filament is directly related to the amount of copper content, with the most concentrated form (80% by weight) remaining free of macrofouling after 24 months of exposure in the ocean (Piola et al., 2021). Another previous biofouling study of copper-infused plastic using a different technology was Vucko et al. (2012), who used cold spray techniques to embed copper into high-density polyethylene (HDPE), which remained free of hard fouling (shell-bearing animals) after 8 months in the field.
We evaluated the inherent antifouling nature of these 3D-printed materials by measuring early-stage biofouling (up to 17 weeks), including biofilms (microfouling) and macrofouling, focusing largely on the taxonomic composition of biofilms using 16S and 18S rRNA metabarcoding. The recent application of metabarcoding to marine biofouling analysis has revealed an unprecedented level of detail concerning taxonomic assemblage structure, especially of microbes within biofilms. This approach has led to rapid advances in understanding the effects of environmental factors, substrates, and even AF coatings on assemblage structure and succession (Chen et al., 2013; Salta et al., 2013; Muthukrishnan et al., 2014; Lawes et al., 2016; Zaiko et al., 2016; Briand et al., 2017; Briand et al., 2018; Dobretsov et al., 2018; Kirstein et al., 2018; Pollet et al., 2018; von Ammon et al., 2018; Winfield et al., 2018; Abed et al., 2019; Catão et al., 2019; Pinto et al., 2019; Antunes et al., 2020; Catão et al., 2021a; Catao et al., 2021b; Briand et al., 2022; Lemonnier et al., 2022; Portas et al., 2022). Although commonly used AF coatings resist macrofouling well, a biofilm still grows on both biocidal and FR coatings (Cassé and Swain, 2006; Molino and Wetherbee, 2008; Chen et al., 2013; Muthukrishnan et al., 2014; Briand et al., 2017; Winfield et al., 2018; Briand et al., 2022). Knowledge of biofilm assemblage structure is important because these microbes influence the subsequent colonization by macrofoulers and may be an important mechanism by which some AF technologies operate indirectly on invertebrates and algae (Qian et al., 2007; Hadfield, 2011; Lema et al., 2019). We therefore assessed macrofouling assemblages at the end of the study to attempt to relate biofilm dynamics to the subsequent macrofouling development.
We determined the influences of the biocide-infused, 3D-printed thermoplastics on fouling assemblages and fouling extent by comparing them to control thermoplastics. We also made comparisons to three different types of commonly used, commercial AF coatings: a self-polishing copper and zinc biocidal paint, a silicone hydrogel FR coating, and a combined copper-biocide and silicone hydrogel FR coating.
Two types of control settlement plates were printed from PCL (polycaprolactone, purchased from DIYKits Australia) and PLA (polylactic acid, purchased from ColorFABB), respectively (Table 1). Experimental treatment plates were: 1) PCL infused with the biocide DCOIT (KATHON 287T, Dow, hereafter “PCL-DCOIT”); 2) commercial PLA containing 80% copper powder by weight (CopperFill, colorFabb B.V., hereafter “PLA-Cu”); and PLA coated with a commercial antifoulant, either 3) Intercept 8500 LPP Red (Akzo Nobel, hereafter “INTER”), 4) Hempaguard X7 89909 (Hempel, hereafter “X7”), or 5) Hempasil X3 + 87500 (Hempel, hereafter “X3”).
Powdered DCOIT and raw PCL pellets were mixed 1:10 by weight. 1.1 kg of this blend was placed in a Banbury mixer (DS1-5, Moriyama Mfg. Works) with 5 g of black PCL pigment pellets for colouration and mixed for 10 min at 32 C. At this temperature, DCOIT is sufficiently tacky to coat the polymer but not fuse the PCL pellets into clumps. When mixing was complete, the PCL-DCOIT blend was removed and stored in an airtight-sealed bag until the time of extrusion. PCL-DCOIT filament was produced using a Noztek Xcalibur extruder (Noztek) at a diameter of 1.75 mm. Filament was cooled by a combination of a water bath and air-cooling (Filabot Airpath, Filabot). Extruded filament was spooled using a Filabot Spooler. If multi-pass extrusion was required, filament was re-pelletised using either a 3devo SHR3D IT plastic recycler or a Filabot Pelletizer.
Settlement plates were 3D printed by fused filament fabrication with Prusa Research MK3S and BCN3D Sigmax R19 printers. Plates were printed in two sizes, which were intended for either sampling biofilms (75 mm x 25 mm) or documenting macrofouling (110 mm x 110 mm). Plates were soaked in reverse-osmosis purified water for 7 days to leach excess chemicals.
Settlement plates were attached to panels and hung vertically at 1 m depth beneath a raft at Williamstown, Port Phillip Bay, Australia (36.8710°S, 144.8850°E). Replicate plates of each experimental treatment and control were interspersed on the panels and spaced 2 cm apart such that adjacent plates differed. On 13 November 2020, 12 replicates of the smaller plates and 4 replicates of the larger plates, for each treatment and control, were deployed. At weeks 2, 3, and 5 following deployment, 4 replicate small plates of each treatment and control were removed and sampled for biofilms. The biofilm growth obtained at weeks 2 and 3 was insufficient for DNA extraction, so the 4 replicate large plates of each treatment and control were sacrificed at 12 weeks for an additional date of biofilm sampling. A new set of large plates (4 replicates of each treatment and control) was deployed on 18 December 2020 and collected after 17 weeks for analysis of macrofouling.
Biofilms were sampled by wiping each settlement plate with a Speci-Sponge (Whirl-Pak) dampened with UltraPure distilled water (Invitrogen), placed in a Whirl Pack bag. Samples were stored at -20°C until processing. Biofilm material was removed from each sponge by adding 40 mL of UltraPure water, processing in a stomacher for 2 min, and separating by centrifugation. DNA was extracted from the supernatant using a DNA PowerSoil Pro kit (Qiagen) as per the manufacturer’s protocol. A NanoDrop One (ThermoFisher) was used to determine the concentration and confirm the quality of extracted DNA.
For 16S rRNA sequencing, primers specific for the 16S V4 region (Caporaso et al., 2011) with Illumina overhang adapters were used for DNA amplification: 515F 5’-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GTG CCA GCM GCC GCG GTA A-3’, 806R 5’-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGG ACT ACH VGG GTW TCT AAT-3’. DNA amplification was based on Afshari et al. (2020), using Amplitaq Gold, with PCR conditions of 95 C (10 min), 30 cycles of 95 C (30 sec), 55 C (30 sec), 72 C (1 min), then 72 C (7 min) and 4 C (infinite hold). For 18S rRNA sequencing, primer sequences specific for the 18S V4 region (Zhan et al., 2013) with Illumina overhang adapters were used for DNA amplification: Uni18SF 5’-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG AGG GCA AKY CTG GTG CCA GC-3’, Uni18SR 5’-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGR CGG TAT CTR ATC GYC TT-3’. DNA was amplified in the presence of Amplitaq Gold 360 (ThermoFisher) under the following PCR conditions: 95 C (10 min), 30 cycles of 95 C (30 sec), 56 C (30 sec), 72 C (1 min), then 72 C (7 min) and 4 C (infinite hold). PCR products were visualized on SYBR-Safe agarose gels and concentration determined using a Qubit fluorometer (ThermoFisher). 16S and 18S rRNA sequencing was conducted on an Illumina MiSeq platform by the Ramaciotti Centre for Genomics, Sydney, Australia.
The amplicon sequence data were processed using the Greenfield Hybrid Amplicon Pipeline (GHAP) v2.1 (Greenfield, 2017), built around tools from USEARCH (Edgar, 2013) and RDP (Cole et al., 2014). Merged reads were trimmed and clustered into operational taxonomic units (OTUs) at 97% similarity. Post-merge, 18S amplicons smaller than 435 bp were excluded, while 16S amplicons smaller than 285 bp were excluded and longer amplicons trimmed to 295 bp. 18S OTUs were then taxonomically classified through a curated SILVA reference set (Quast et al., 2013), while 16S OTUs were classified based on the RDP classifier and RefSeq 16S reference set. Data were further processed prior to subsequent analysis: OTUs were excluded if not taxonomically assigned (and could not be identified through subsequent manual BLAST analysis), if BLAST identity scores were less than 97%, or if total read count was less than 10.
Percentages of phyla and of classes were fourth-root transformed and converted to Bray-Curtis dissimilarity for non-metric Multidimensional Scaling (nMDS), Analysis of Similarity (ANOSIM), and Similarity Percentage (SIMPER) analysis in PRIMER 7.
Macrofouling plates collected after 17 weeks of deployment were photographed while submerged in seawater with a Lumix G digital camera (model DMC-G7). Images were analyzed for macrofouling taxa and coverage using Coral Point Count (CPCe v4.1) software. A 0.7-cm wide border at the edge of each plate was excluded from analysis to avoid edge effects. One hundred analysis points, each randomly positioned within a cell of a 10 x 10 grid, were overlain within the analysis frame, and the object under each point was identified to the following categories: sponge, solitary ascidian, colonial ascidian, arborescent bryozoan, encrusting bryozoan, barnacle, tube worm, skeleton shrimp, gammarid amphipod tube, microalgae, macroalgae, and unknown fouling, as well as detritus, bare substrate, and indistinguishable.
After 5 weeks of deployment, the 16S rRNA phylum-level assemblage of natural biofilms on the two controls (untreated PCL and PLA) were very similar to each other, being dominated by Proteobacteria (48-63%, almost entirely class Alphaproteobacteria with a small amount (5%) of Gammaproteobacteria), followed by Bacteroidetes (32-40%, almost entirely class Flavobacteriia) (Figures 1A, B).

Figure 1 Mean percentages of prokaryotic taxa (including plastids of eukaryotes) in biofilm samples from 16S rRNA sequencing after 5 weeks (top: A, B) and 12 weeks (bottom: C, D) of deployment; phyla (left: A, C) and classes (right: B, D). Taxa contributing <1% are not shown in the legends. N = 4, except in (A, B) where n = 0 for X7 and X3, and in (C, D) where n = 3 for X7 and n = 1 for X3, due to insufficient biofilm growth on some replicates.
The assemblage on PCL-DCOIT was dramatically different from the PCL control in being strongly dominated by 98.5% Proteobacteria [with much more in class Gammaproteobacteria (54%) and less in class Alphaproteobacteria (44%)], and only 1% Bacteroidetes (Figures 1A, B). In contrast, the assemblage on PLA-Cu differed less from its PLA control and in the opposite direction: it had much less Proteobacteria (29%, due to reduction of class Alphaproteobacteria) and was dominated by Bacteroidetes (64%, entirely class Flavobacteriia).
Comparing the biocide-infused polymers to the commercial coatings, the assemblage on PLA-Cu was very similar to that on INTER (dominated to 58-64% by Bacteroidetes, class Flavobacteriia), whereas the assemblage on PLC-DCOIT stood out with its 98.5% dominance by Proteobacteria (Figures 1A, B). Note that no sequences were retrieved from the X7 or X3 treatments at week 5 due to insufficient biofilm growth.
On the phylum-level nMDS plot for week 5 (Figure 2A), the assemblage structure on PCL-DCOIT was strongly separated from all other treatments and from the controls, although all pairwise comparisons were significantly different (Table 2). SIMPER confirmed that the greatest average assemblage dissimilarities at the phylum level were between PCL-DCOIT and all other treatments and controls (42 - 52% dissimilarity), and that most of this dissimilarity was due to differences in the relative amount of Bacteriodetes (Table S1).
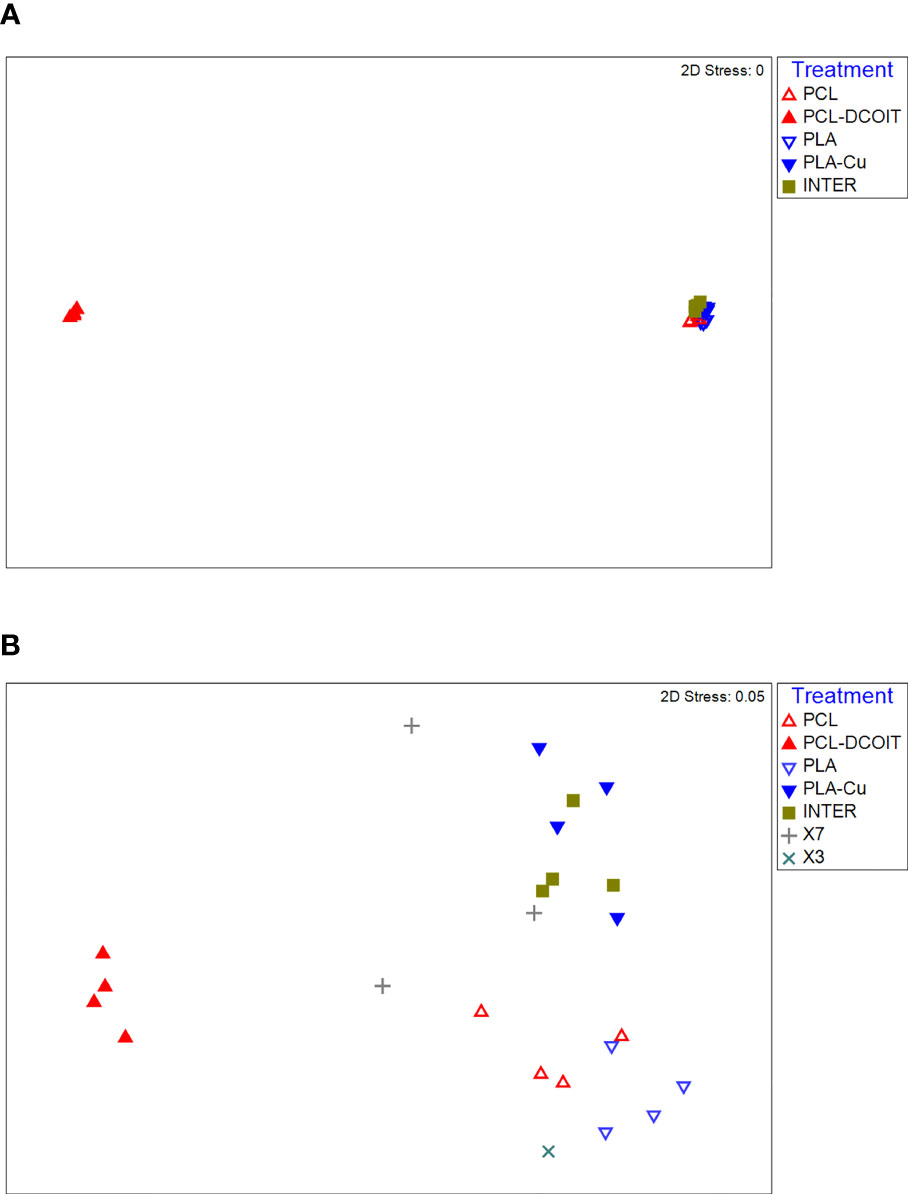
Figure 2 nMDS plots of biofilm assemblages at the phylum level from 16S rRNA sequencing after (A) 5 weeks of deployment, (B) 12 weeks of deployment. Sample sizes are as in Figure 1.
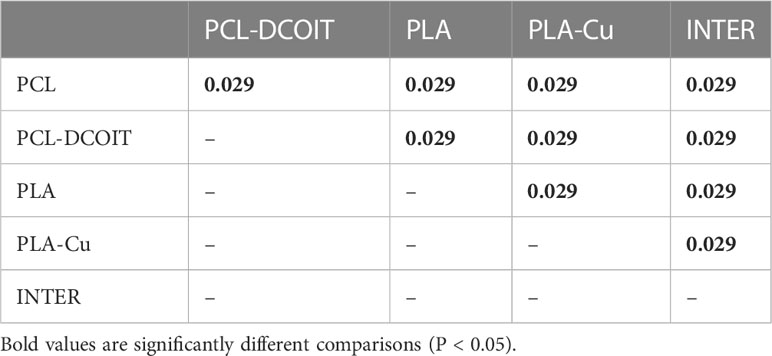
Table 2 Pairwise ANOSIM results (global R = 0.922, P = 0.001, 999 permutations) from 16S rRNA phylum data collected from samples after 5 weeks of deployment.
After 12 weeks, the assemblages across all treatments and controls were generally more diverse, with the category Cyanobacteria/Chloroplasts having increased considerably, yet they remained mostly dominated by the phyla Proteobacteria and Bacteroidetes (Figure 1C). The natural biofilms on the two controls (untreated PCL and PLA) were still very similar to each other, with Proteobacteria being most abundant (45-50%, largely class Alphabacteria), followed by Bacteroidetes (26-30%, entirely class Flavobacteriia), and now Cyanobacteria/Chloroplast at 11-13% (with roughly equal components of Cyanobacteria and eukaryotic algae) (Figures 1C, D). The two controls were the only plates in the entire experiment that had notable amounts of Cyanobacteria, whereas all antifouling treatments inhibited the cyanobacteria. The PLA control also had a notable amount of Thaumarchaeota (11%).
The assemblage on PCL-DCOIT remained the least diverse of all treatments and controls, being dominated to 99% by two phyla: Proteobacteria (62%, now with a switch to more in class Alphaproteobacteria than in class Gammaproteobacteria) and Cyanobacteria/Chloroplast (37%, entirely eukaryotic algae, which was unique among all treatments and controls) (Figures 1C, D). PCL-DCOIT was the most effective of all treatments in resisting Cyanobacteria. In contrast to PCL-DCOIT, the assemblage on PLA-Cu still showed the opposite divergence from its control (PLA), in that it was dominated by Bacteroidetes (54%, entirely class Flavobacteriia), and it also had less Cyanobacteria/Chloroplasts and less Thaumarchaeota than the PLA control.
Comparing the biocide-infused polymers to the commercial coatings, the assemblage on PLA-Cu remained very similar to that on INTER at both the phylum and class levels, except that PLA-Cu had less of class Gammaproteobacteria (2%) than did INTER (10%) (Figures 1C, D). PLA-Cu differed from X7 and X3 in having more Bacteroidetes (54% vs. 24-31%, all class Flavobacteriia), less Proteobacteria (38% vs. 57-58%), and less Cyanobacteria/Chloroplast (3% vs. 10%, almost entirely eukaryotic algae). Furthermore, PLA-Cu had a class makeup within Proteobacteria that differed from that on X7 and X3: PLA-Cu had less Gammaproteobacteria (2% vs. 10-13%) and less Betaproteobacteria (2% vs. 11-18%). In fact, X7 and X3 stood out in having far more Betaproteobacteria than any other treatment or control. Finally, PCL-DCOIT stood out from all the commercial coatings in its dominance by Proteobacteria and eukaryotic algae, and its absence of Bacteroidetes.
On the phylum-level nMDS plot for week 12 (Figure 2B), the PCL-DCOIT assemblage was still strongly separate, but it was less dissimilar to the other treatments and controls than it had been at week 5. Most pairwise comparisons were significantly different, with notable exceptions being similarities between some of the copper-containing treatments (PLA-Cu vs. INTER, and INTER vs. X7), and that the assemblage on X3 was not significantly different from any other treatments or controls (Table 3). SIMPER confirmed that the greatest average assemblage dissimilarities at the phylum level were still between PCL-DCOIT and all other treatments and controls (30 - 41% dissimilarity) (Table S2). However, reflecting the increased diversity compared to week 5, the leading contributors to dissimilarity in pairwise comparisons were more variable (Table S2).
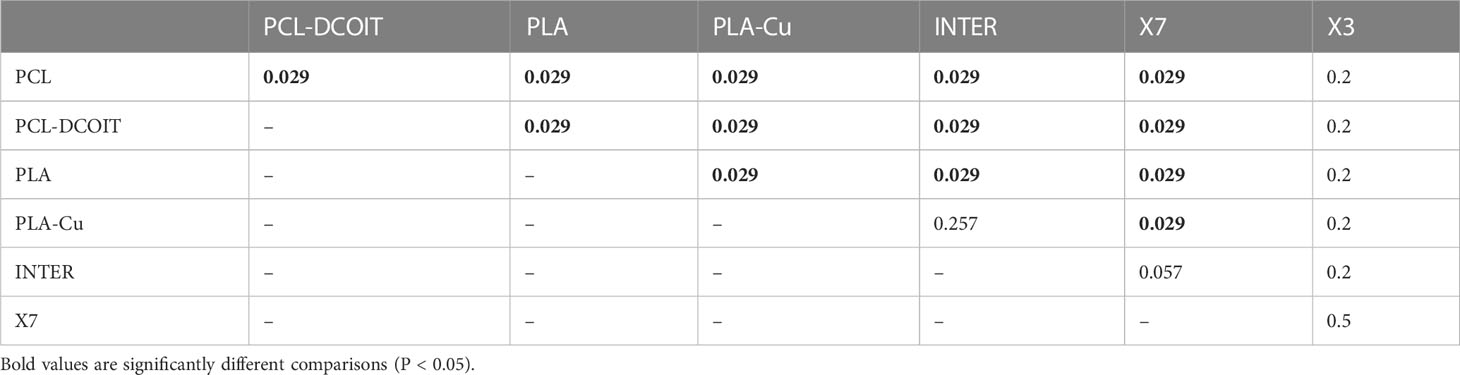
Table 3 Pairwise ANOSIM results (global R = 0.812, P = 0.001, 999 permutations) from 16S rRNA phylum data collected from samples after 12 weeks of deployment.
After 5 weeks of deployment, the 18S rRNA phylum-level assemblage of natural biofilms on the two controls (untreated PCL and PLA) were very similar to each other, being dominated by 81-88% Arthropoda (mostly class Malacostraca, the largest class of crustaceans including decapods, followed by class Maxillopoda, which includes barnacles) (Figures 3A, B).
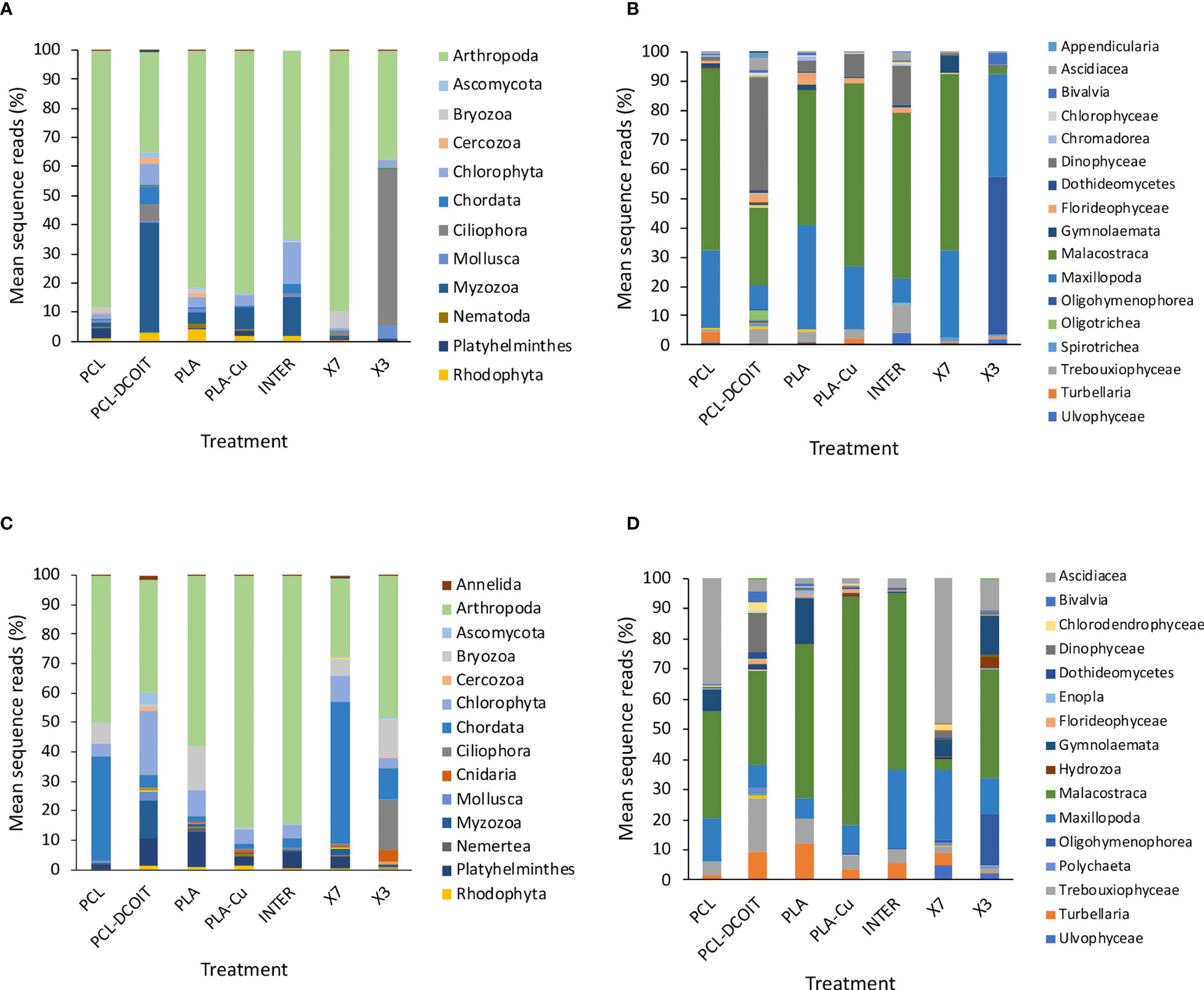
Figure 3 Mean percentages of eukaryotic taxa in biofilm samples from 18S rRNA sequencing after 5 weeks (top: A, B) and 12 weeks (bottom: C, D) of deployment; phyla (left: A, C) and classes (right: B, D). Taxa contributing <1% are not shown in the legends. N = 4, except in A and B where n = 2 for X7 and n = 3 for X3, and in (C, D) where n = 3 for X7, due to insufficient biofilm growth on some replicates.
The assemblage on PCL-DCOIT was quite different from the PCL control in being dominated by 38% Myzozoa (entirely the class Dinophyceae), with Arthropoda reduced to 34% (and more dominated by class Malacostraca over class Maxillopoda) (Figures 3A, B). PCL-DCOIT had the smallest amount of Maxillopoda (8%) among all the treatments and controls, being similar only to INTER. The assemblage on PCL-DCOIT was also more diverse than that on the PCL control, including Chlorophyta, Chordata (mostly class Ascidiacea), and Ciliophora (mostly class Oligotrichea), each at 5-7%.
In contrast to PCL-DCOIT, the assemblage on PLA-Cu was very similar to that on its control (PLA), with both dominated by 81-84% Arthropoda (classes Malacostraca and Maxillopoda) (Figures 3A, B). The only notable difference between PLA-Cu and the control was more Myzozoa (class Dinophyceae) on the former (8% vs. 4%).
The assemblages on the two biocide-infused polymers thus differed greatly from each other, and they each differed considerably from the three commercial coatings, which were quite variable among themselves (Figures 3A, B). The assemblage on INTER had an intermediate amount of Arthropoda (65%), with the lowest amount of class Maxillopoda (8%) among all treatments and controls except for the same amount being on PCL-DCOIT. INTER also had the largest amount of Chlorophyta (14%) and the second largest amount of Myzozoa (14%, class Dinophyceae). The assemblage on X7 was similar to the controls in being heavily dominated by Arthropoda (90%, mostly class Malacostraca), but it was notable as the only treatment or control with considerable Byrozoa at this point (6%, all class Gymnolaemata). The assemblage on X3 was quite different from all others, with the largest member being Ciliophora (54%, entirely class Oligohymenophorea), and with the reduced Arthropoda (37%) being almost entirely class Maxillopoda.
On the phylum-level nMDS plot for week 5 (Figure 4A), all treatments and controls showed some overlap in their assemblage distribution except for X3 which clustered separately. ANOSIM showed that the assemblages on PCL-DCOIT and PLA-Cu each differed significantly from their respective control; the three copper-containing treatments (PLA-Cu, INTER, and X7) were all statistically similar to each other; and the assemblage on X3 differed significantly from all other treatments and controls except X7 (Table 4). SIMPER confirmed that the greatest average assemblage dissimilarities at the phylum level were between X3 and all other treatments and controls (41 - 44% dissimilarity) except X7, and between PCL-DCOIT and PCL (42% dissimilarity), much of which was attributed to variations in Myzozoa or Ciliophora (Table S3).
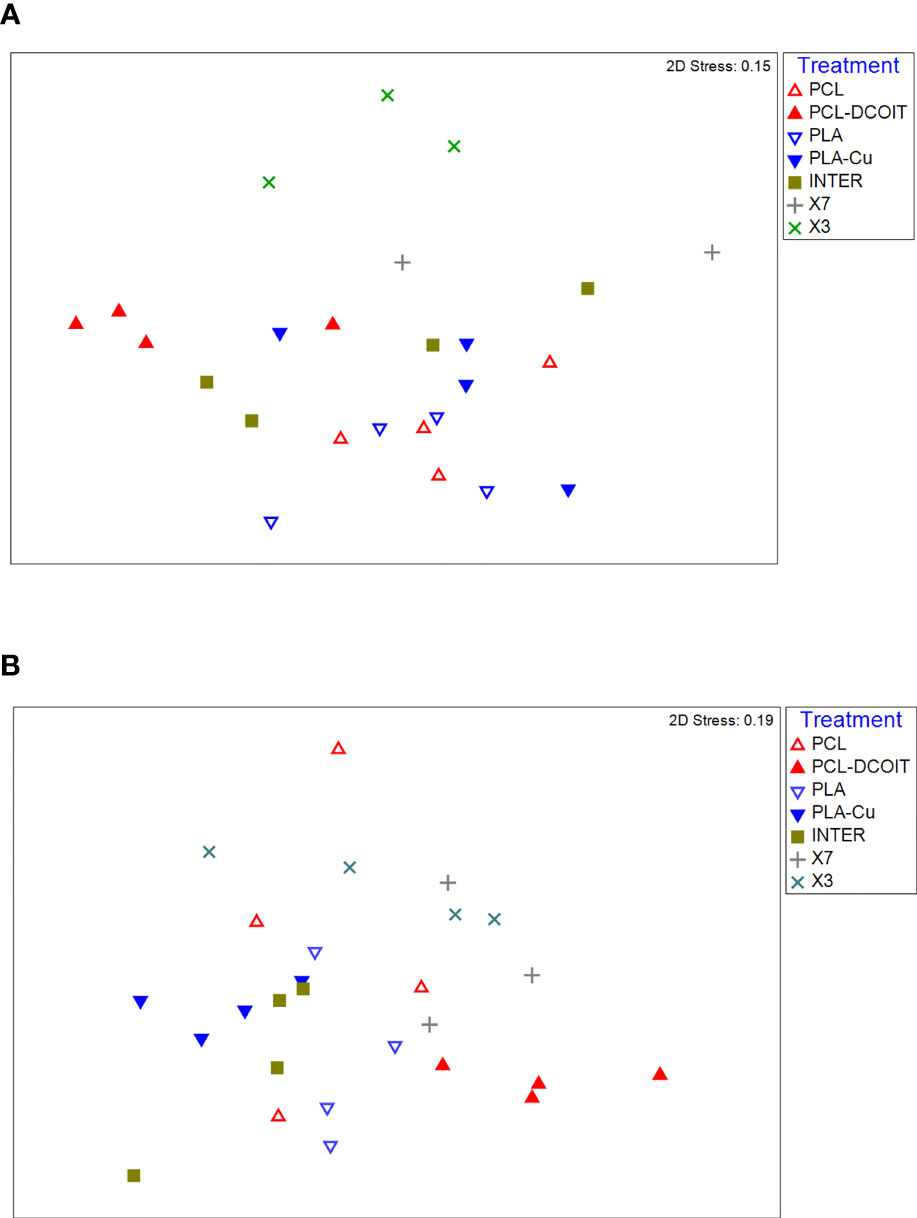
Figure 4 nMDS plots of biofilm assemblages at the phylum level from 18S rRNA sequencing after (A) 5 weeks of deployment, (B) 12 weeks of deployment. Sample sizes are as in Figure 3.
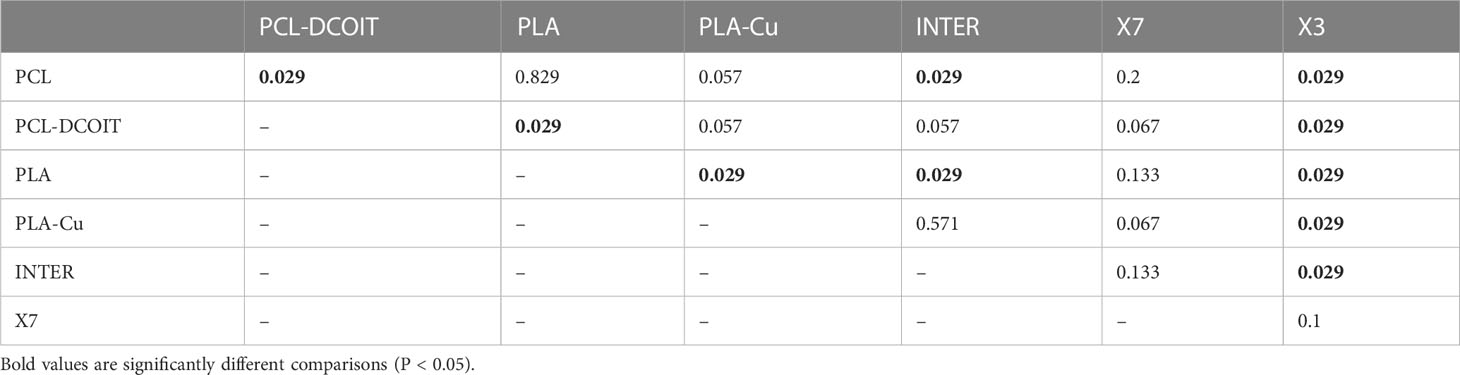
Table 4 Pairwise ANOSIM results (global R = 0.56, P = 0.001, 999 permutations) from 18S rRNA phylum data collected from samples after 5 weeks of deployment.
After 12 weeks, the eukaryotic assemblages changed in variable ways, with most becoming more diverse, and differences between treatments and controls changing (Figures 3C, D). Some notable, common aspects of succession were as follows. There was an increase of Chordata (entirely class Ascidiacea) on all treatments and controls, with the greatest contributions on X7 (48%) and on the PCL control (35%). There was also at least 1% Platyhelminthes (entirely turbellarians) on all treatments and controls except X3, with a maximum of 12% on the PLA control. Bryozoa (entirely class Gymnolaemata) was present in at least 1% on everything except PLA-Cu and INTER, with the largest presence (7-15%) on the two controls (PCL and PLA) and X3. Notably, all of the treatments and controls had <1% of phylum Bacillariophyta (diatoms).
The natural biofilm assemblages on the two controls (untreated PCL and PLA) remained quite similar to each other, except that PCL had more Chordata (35%, all class Ascidiacea) and less Platyhelminthes (2%) than PLA, which had only 2% Chordata but 12% Platyhelminthes (Figures 3C, D). Both controls had considerable contributions from Bryozoa (7% and 15%, respectively).
PCL-DCOIT had the most diverse assemblage of all. Compared to the PCL control, it most notably had more Chlorophyta (22%) and Myzozoa (12%, all class Dinophyceae), less Arthropoda (38%, mostly Malacostraca with some Maxillopoda), and much less Chordata (4%) (Figures 3C, D). PCL-DCOIT still had (almost) the lowest amount of Maxillopoda (7%) among the treatments and controls, except for the PLA control which had 6%. It was also unique in having the most Annelida, Ascomycota, Cercozoa, Chlorophyta, Mollusca, and Myzozoa.
The assemblage on PLA-Cu diverged considerably from that on its PLA control, and again in quite different ways than PCL-DCOIT differed from its control (Figures 3C, D). PLA-Cu differed from the control in being much more dominated by Arthropoda (85% vs. 58%), having negligible Bryozoa (<1% vs. 15%), and less Platyhelminthes (3% vs. 12%). PLA-Cu differed from PCL-DCOIT in that the former had far more Arthropoda and less Chlorophyta, Chordata, Myzozoa, and Platyhelminthes; these differences reflected the greater diversity of the assemblage on PCL-DCOIT.
The assemblages on the two biocide-infused polymers thus still differed greatly from each other, and the only strong similarity between either of them and the commercial coatings was between PLA-Cu and INTER (Figures 3C, D). These two treatments had similar dominance by Arthropoda (85% in each case, mostly class Malacostraca), and only differed notably in that INTER had more Platyhelminthes (6% vs. 3%). The assemblage on PCL-DCOIT bore little resemblance to any of the commercial coatings, except for having at least a moderate amount of Arthropoda. The other copper-containing treatment, X7, was quite unique in being dominated by Chordata (48%, entirely class Ascidiacea) and having the lowest amount of Arthropoda (27%, which were unusual in being mostly class Maxillopoda). The assemblage on X3 had more Arthropoda (48%, but mostly class Malacostraca) and was unique among all treatments and controls in still having a considerable amount of Ciliophora (17%, all class Oligohymenophorea) and now Cnidaria (4%, all class Hydrozoa), while being the only antifouling treatment with Bryozoa at a similar level to the controls (13% vs. 7-15%).
The phylum-level nMDS for week 12 (Figure 4B) showed more overlapping clusters of assemblages than a week 5. ANOSIM confirmed that PCL-DCOIT was still significantly different from its PCL control; PLA-Cu and INTER were still statistically similar but no longer significantly different from the PLA control; X7 had become significantly different from several other treatments, including the other two copper-containing treatments (PLA-Cu and INTER); and X3 remained significantly different from most other assemblages (Table 5). SIMPER showed that the greatest total assemblage dissimilarity at the phylum level was between PCL-DCOIT and the PCL control (Table S4). Reflecting the increased diversity compared to week 5, the leading contributors to dissimilarity in pairwise comparisons were highly variable.
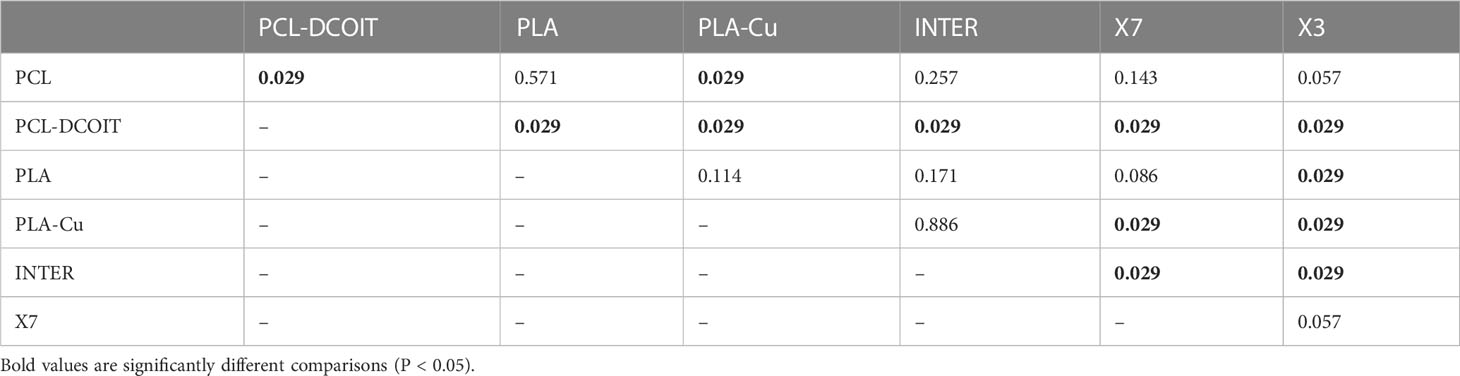
Table 5 Pairwise ANOSIM results (global R = 0.531, P = 0.001, 999 permutations) from 18S rRNA phylum data collected from samples after 12 weeks of deployment.
After 17 weeks of deployment, the PCL and PLA controls had 39% and 61% coverage, respectively, of diverse and common macrofouling assemblages including solitary and colonial ascidians, encrusting and arborescent bryozoans, barnacles, tube worms, skeleton shrimp, amphipod tubes, and macroalgae (Figures 5, 6). None of the 3D-printed or commercial antifouling treatments had any macrofouling. A microalgal film was visible on the PCL-DCOIT and X3 treatments, and all treatments and controls had at least some settled detritus. PCL-DCOIT had the most extensive microalgal film (79% coverage), and PLA-Cu was completely covered (100%) by detritus.
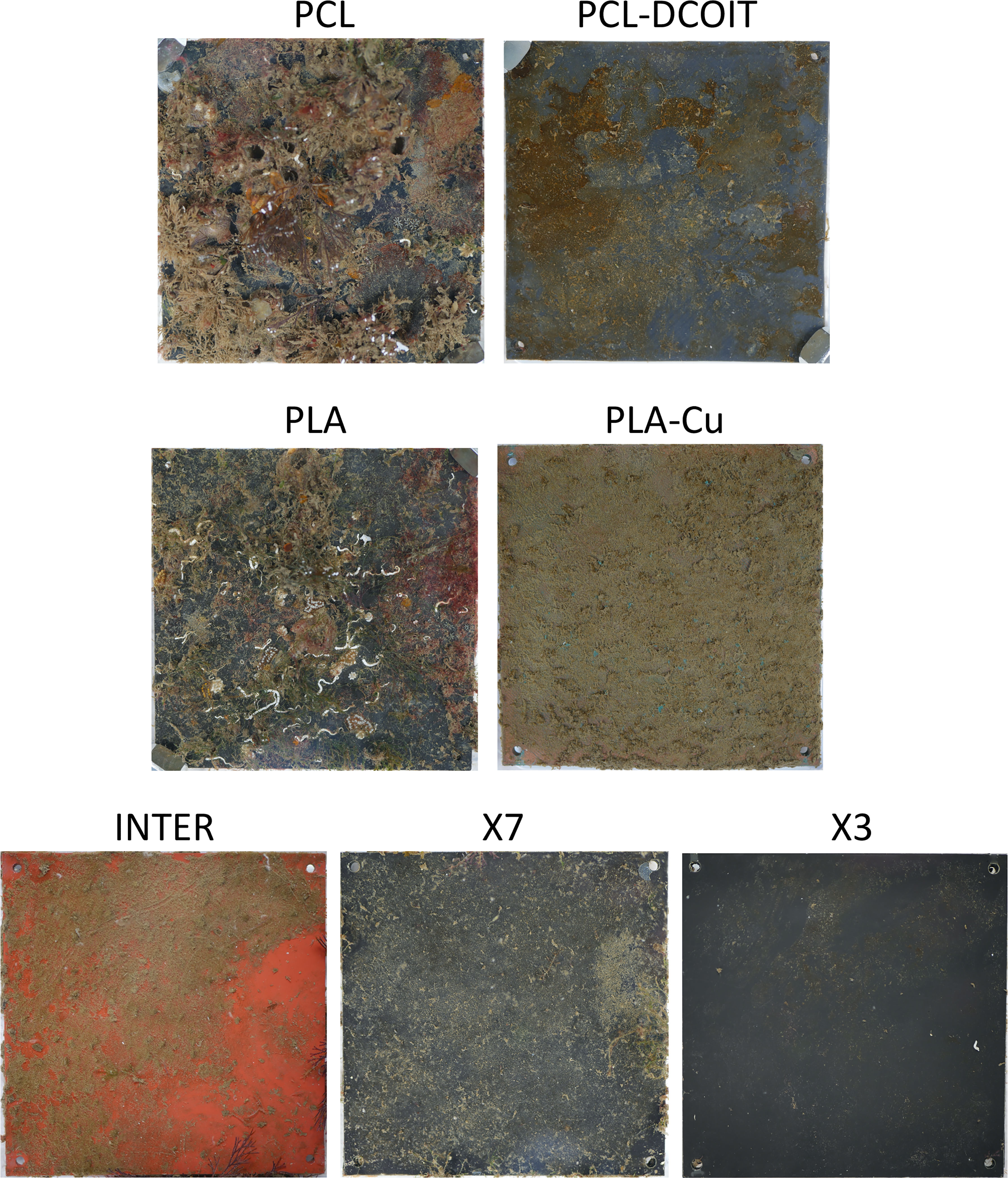
Figure 5 Settlement plates after 17 weeks of deployment; one typical replicate shown for each control and treatment.
The PCL-DCOIT thermoplastic we produced with embedded DCOIT completely resisted macrofouling as of 17 weeks in the ocean. In several other trials at various marine locations, we have found this material to be free of macrofouling for at least 18 months—often 24 months (unpublished data). Measurements of DCOIT leaching rates from this material submerged in water yielded 5 µg cm-2 d-1 after a 15-day stabilization period, and >1 µg cm-2 d-1 after 3 years (R. Piola, unpublished data). These release rates are similar to the 2-5 µg cm-2 d-1 for DCOIT-based antifouling paints reviewed by Takahashi (2009).
The PCL-DCOIT material nonetheless developed a biofilm of microfouling, which was visible at 17 weeks as a film of microalgae covering 79% of the plate, and was evident in the biofilm sequencing results as being predominantly chlorophytes. This biofilm was strongly influenced by DCOIT compared to the control, in that it had reduced microbial diversity and a shift to being heavily dominated by Proteobacteria and chlorophytes. The PCL-DCOIT proved especially resistant to Bacteroidetes (including Flavobacteriia), Cyanobacteria, and diatoms. Despite the absence of macrofouling at the end of the study, the 18S rRNA sequencing revealed a great diversity of eukaryotes in the early biofilm, including several animal phyla. These sequences could have been from propagules of macrofoulers that failed to survive or to grow, or they could have been from extracellular DNA that merely became associated with the biofilm. It is notable that the biofilm on PCL-DCOIT had the least amount of barnacle DNA of all the AF treatments in this study.
DCOIT is well known as a broad-spectrum biocide that, as a component of AF paint, is highly effective against bacteria, diatoms and other microalgae, macroalgae, and macrofouling animals, although a biofilm does typically develop over it (Ali et al., 2021; Bressy et al., 2022; Roepke et al., 2022). Ours is the first study to our knowledge that characterises the influence of DCOIT on assemblage structure in biofilms. The extreme inhibition of Bacteroidetes (namely the class Flavobacteriia) compared to the controls and to all other AF treatments is notable because Flavobacteriia have recently been recognised as abundant members of natural, coastal marine biofilms (Dang and Lovell, 2016; Pollet et al., 2018). Furthermore, several species of Bacteroidetes, or of Flavobacteriia in particular, have been shown to induce settlement of the tubeworm Hydroides elegans (Huang and Hadfield, 2003), the bryozoan Bugula neritina (Dobretsov and Qian, 2006), and the green alga Ulva (Tait et al., 2009). Therefore, the resistance to Bacteroides colonization afforded by DCOIT could have a follow-on indirect effect of reducing the settlement-inducing nature of the biofilm with regards to a variety of macrofouling species. Certainly, DCOIT can reduce macrofouling by its direct toxicity to organisms (Chen and Lam, 2017), but mechanisms of fouling resistance by biocidal additives in paints and coatings are not fully understood, and it is possible that indirect effects play an important role. For example, McElroy et al. (2017) showed that a copper-based paint reduced macrofouling more via alterations to the microbial biofilm than via direct toxicity to macrofoulers. Some macrofouling can develop on DCOIT-containing paints (Bressy et al., 2022), and the biofilm alterations seen here may potentially influence the nature and extent of that macrofouling growth.
Numerous species of Gammaproteobacteria are commonly observed as early biofilm colonizers (Pollet et al., 2018; Briand et al., 2022) and are known to either induce or inhibit settlement of various macrofoulers (Holmström et al., 2002; Hadfield, 2011; Salta et al., 2013). Several types of biocidal AF coatings have been shown to affect the relative abundance of Gammaproteobacteria in marine biofilms (Briand et al., 2017; Dobretsov et al., 2018; von Ammon et al., 2018; Briand et al., 2022), which therefore has the potential to influence subsequent settlement of macrofouling propagules. Although we found a strong enhancement of Gammaproteobacteria on the PCL-DCOIT treatment at week 5, this effect was gone by week 12. At that point, Gammaproteobacteria showed little variation among the AF treatments and controls, so they are unlikely to have influenced the macrofouling or 18S sequencing results.
Cyanobacteria were almost completely excluded from the biofilm by PCL-DCOIT, but the implications of this for subsequent settlement of macrofoulers are unknown because little work has been done on Cyanobacteria as settlement cues (Qian et al., 2007).
The technique we developed here for infusing DCOIT into PCL and extruding an effective antifouling, 3D-printing filament could be used with other biocides in attempts to maximize the AF effectiveness. Copper pyrithione or Cu2O, alone or in combination with DCOIT, are promising candidates. In fact, Bressy et al. (2022) found that paint containing DCOIT and Cu2O extended the AF effectiveness from 7 to 16 months compared to paint containing DCOIT alone. A similar additive effect might be achieved by combining multiple biocides in a thermoplastic to create inherently antifouling material.
The PLA-Cu material, made from commercially available copper-infused 3D-printing filament, was equally effective as the PCL-DCOIT material in completely resisting macrofouling as of 17 weeks in the ocean, and we showed previously that this PLA-Cu product remained free of macrofouling as of 24 months (Piola et al., 2021). The product is available with several different copper concentrations (30%, 50%, and 80%, the latter being what was tested here), and we showed that the AF effectiveness was directly related to the amount of copper content (Piola et al., 2021). Mechanisms of exposure and replenishment of copper at the surface-water interface of this material were discussed in Piola et al. (2021).
Other methods of embedding copper particles into materials or AF coatings have also been effective. For example, Vucko et al. (2012) showed that embedding copper powder into HDPE thermoplastic by cold spraying prevented macrofouling growth for over 8 months. Padmavathi et al. (2021) embedded CuO nanoparticles into a PDMS FR coating, which enhanced the coating’s effectiveness by reducing microalgal growth and total biofouling mass as of 90 days in the field. An advantage of the copper-embedded 3D-printing filament, however, is that objects can be printed with an inherent AF property, avoiding the need for any coating.
The influences of PLA-Cu on biofilm assemblage structure were very different from those of PCL-DCOIT. Whereas PCL-DCOIT caused almost a complete elimination of Bacteroidetes (class Flavobacteriia) from the biofilm and a dominance of Protobacteria and chlorophytes, PLA-Cu had the opposite effect of facilitating Flavobacteriia to the point that they dominated the biofilm, and it largely resisted the growth of chlorophytes or any other microalgae. Similar to PCL-DCOIT, the PLA-Cu was resistant to Cyanobacteria and diatoms. The dominance of Flavobacteriia on the commercial PLA-Cu product is evidently temporary, however, because we previously found that after 16 months in the ocean, the biofilm on samples of this material was dominated by Proteobacteria at roughly 85-90%, being mostly the class Alphaproteobacteria with a smaller amount of class Gammaproteobacteria (Piola et al., 2021). Nonetheless, the facilitation of Flavobacteriia during at least the period of 5-12 weeks could favor the settlement of propagules from macrofoulers such as tubeworms, bryozoans, and green algae that have been shown to be stimulated by these bacteria (Huang and Hadfield, 2003; Dobretsov and Qian, 2006; Tait et al., 2009). On the other hand, Gammaproteobacteria, many of which stimulate macrofouler settlement (Hadfield, 2011; Salta et al., 2013), were reduced compared to the control at 12 weeks, although even in the control they were only a very small proportion of the assemblage (8%). Finally, as mentioned above, any consequences of the strong resistance to cyanobacterial growth, which was similar on both PLA-Cu and PCL-DCOIT, for the subsequent settlement of macrofoulers is unknown. Although we detected a large diversity of eukaryotic 18S rRNA sequences in the early biofilms on PLA-Cu, including macrofouling species, none of them were visible on the plates at 17 weeks, either because they failed to survive or grow, or because their detection had been due to extracellular DNA associated with the biofilms.
The effects of PLA-Cu on the microbial assemblage of the biofilm were consistent with some other recent studies of copper-containing coatings. Muthukrishnan et al. (2014) found that several coatings containing Cu2O and either zinc or copper pyrithione had an enhancement and in many cases a dominance of Flavobacteriia in the biofilms sampled after one year. Winfield et al. (2018) found that three AF coatings containing Cu2O and copper pyrithione had biofilms dominated by Bacteroidetes (mostly Flavobacteriia), sampled within 13-27 weeks. Briand et al. (2022) found that by 3 weeks in the field, Flavobacteria began increasing in the biofilm on an AF coating containing Cu2O and copper pyrithione, and they ultimately dominated the assemblage by 11 weeks. They also found, similar to our PLA-Cu, an absence of Cyanobacteria on the copper-based coatings compared to controls. In contrast, other studies have found different results, such as Dobretsov et al. (2018), who found that biofilms on paints containing Cu2O and fungicides were strongly dominated (≥90%) by Proteobacteria, with ≤10% Flavobacteria. Also, von Ammon et al. (2018) found that biofilms on a paint containing Cu2O and ZnO were dominated by Proteobacteria, sampled at 17 weeks; and Briand et al. (2017) found that a coating containing only copper pyrithione as the biocide had a reduced component of Bacteroidetes in the biofilm after 4 weeks. Biofilm assemblages may be highly sensitive in complex ways to the differing biocidal components of these coatings (cf. Muthukrishnan et al., 2014), and the mechanisms influencing assemblage structure are still not well understood.
The two biocide-embedded 3D-printing thermoplastics performed equally well as the three commercial AF coatings in completely resisting macrofouling, although they were significantly fouled by chlorophyte microalgae (79% coverage on PCL-DCOIT) or covered by detritus (100% on PLA-Cu). This fouling is undesirable but perhaps could be eliminated with future formulations of the thermoplastics containing multiple biocides, such as a combination of DCOIT and copper powder, possibly with copper pyrithione. While the microalgal film on PCL-DCOIT could perhaps be eliminated directly by biocidal action, the heavy coverage of detritus on PLA-Cu may be more challenging to address. The mechanism behind this effect is unknown, but it may be that an aspect of the biofilm assemblage structure, and especially the extracellular matrix of the biofilm, may have caused it to become more adhesive (chemically or electrostatically) to detritus. The adhesive effect could possibly be reversed by further additives to the thermoplastic, although it would need to be accomplished by trial and error unless the mechanism can be revealed by further research. However, the adhesion might not be problematic for applications where materials are exposed to strong flow that prevents detrital accumulation, or in areas of clear water.
The 16S rRNA sequencing showed that assemblages in biofilms were quite similar among the three commercial AF coatings, although more Betaproteobacteria were detected on the two FR coatings (Hempaguard X7 and Hempasil X3) than on the purely biocidal coating, Intercept 8500. The assemblage on 3D-printed PLA-Cu was statistically similar to both Intercept 8500 and Hempasil X3, and it only differed from Hempaguard X7 mainly in the amount of Proteobacteria. Therefore, while PLA-Cu strongly altered the biofilm assemblage compared to a natural biofilm on an untreated control, its effects were generally similar to those of the commercial AF coatings. The 3D-printed PCL-DCOIT, in contrast, had a much different effect on the biofilm from that of the commercial coatings, in that PCL-DCOIT almost eliminated the Flavobacteriia and greatly enhanced the relative number of chlorophytes. Thus, PCL-DCOIT has the greatest potential of all antifoulants tested here to influence microbial biofilm composition, succession, and interactions with macrofouling propagules. Whether its effects are similar to those of DCOIT in paints and other coatings (as opposed to the 3D-printed material here) is unknown, as our study is believed to be the first to characterize prokaryotic assemblage structure on any DCOIT-treated material in the marine environment.
Although the 18S rRNA sequencing showed numerous differences in eukaryotic assemblage structure in the biofilms among all the AF treatments, the significance of these differences is unknown because no macrofoulers were visible at 17 weeks. One potentially important finding, however, was the great abundance of ciliate sequences on Hempasil X3 at weeks 5 and 12. Ciliates could have remained abundant and possibly important members of the biofilm at later times, but they would not have been detected by our imaging of visible fouling. The presence of ciliates and their grazing activities can strongly influence the physical structure and microbial dynamics in biofilms (Huws et al., 2005), and they can influence settlement (positively or negatively) of various macrofouling invertebrates (Shimeta et al., 2012; Watson et al., 2016). Ciliates rapidly colonize marine biofilms (Gong et al., 2005; Watson et al., 2015a), and Watson et al. (2015b) found that biocidal and FR coatings can dramatically alter ciliate abundances and assemblage structure. Similar to our findings here, Watson et al. (2015b) showed that Hempasil X3 (as well as another non-biocidal FR coating, Intersleek 970) had much more abundant ciliates in the biofilms than did two biocidal coatings, as well as having significantly different species assemblages of ciliates. The indirect effects of antifouling coatings on microbial biofilm dynamics and macrofouler settlement, via the coatings’ influence on ciliates, warrants further research.
The choice of polymer to use in the technique reported here—embedding biocide for 3D printing—is important not only to ensure the melting points of the thermoplastic and the biocide are compatible for mixing, extruding, and printing, but also because thermoplastics themselves vary in their inherent tendency to foul. This tendency could contribute to the ultimate antifouling effectiveness. Many studies have shown differences in microbial biofilm assemblages among various artificial materials, including plastics, in the marine environment (Kirstein et al., 2018; Pinto et al., 2019; Lemonnier et al., 2022), although few studies have included thermoplastics commonly used in 3D printing. Here we found the PCL and PLA controls to have significantly different assemblages of 16S rRNA sequences at the phylum level at both sampling times. Ryley et al. (2021) compared marine biofouling on PCL and two other 3D-printing thermoplastics (VisiJet® SL Clear and VeroClear™), finding not only differences in microbial and macrofouling assemblages among these materials and compared to PDMS (a common FR material), but also that PCL had the least fouling coverage of all the tested materials after 12 weeks in the ocean. This finding suggests that PCL is a promising candidate for further research into producing inherently antifouling, 3D-printing materials.
Objects that were 3D printed from thermoplastic filament with embedded biocide (DCOIT in PCL or copper powder in PLA) proved equally resistant to macrofouling as three commercial antifouling coatings (biocidal, fouling release, and hybrid) after 17 weeks of immersion in the ocean. PCL-DCOIT, however, had more microalgal growth, and PLA-Cu had more settled detritus, than did the coatings. The embedded biocides significantly altered the taxonomic assemblage structure of both prokaryotes and eukaryotes in the early biofilms compared to natural biofilms, with the two biocides having differing effects. These alterations to the biofilm assemblages may influence the microbial dynamics, biofilm growth, and settlement cues to which biofouler propagules respond. Thermoplastic 3D-printing filaments with embedded biocides show promise for producing 3D-printed objects with inherent antifouling properties for marine applications, avoiding or at least lessening the need to apply antifouling coatings, and possibly extending their service lifetime.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SAMN33295129-SAMN33295224.
Conceptualization, JS, NB, RP, ML, AS. Methodology, JS, GW-M, NB, RP, ML, RS. Formal analysis, GW-M. Investigation, GW-M, RP, RS. Resources, JS, NB, RP, ML. Data curation, JS, GW-M. Supervision, JS, RP, ML. Project administration, JS, RP, ML. Funding acquisition, JS, AS. Writing - original draft, JS, GW-M. Visualization, JS, GW-M. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was funded by Australian Defence Science & Technology (MyIP 7273) and RMIT University.
The authors thank Mark Ciacic and Jim Dimas (Defence Science & Technology) assistance in the field; Roya Ahsari (RMIT University) for providing primers for 16S rRNA amplification; Luke Norbury, Cecilia Power, and Tahnee Manning (RMIT University) for assistance in the lab and with bioinformatics and statistical analysis; and BAE Systems for use of facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1172942/full#supplementary-material
Abed R. M. M., Fahdi D. A., Muthukrishnan T. (2019). Short-term succession of marine microbial fouling communities and the identification of primary and secondary colonizers. Biofouling 35, 526–540. doi: 10.1080/08927014.2019.1622004
Afshari R., Pillidge C. J., Read E., Rochfort S., Dias D. A., Osborn M., et al. (2020). New insights into cheddar cheese microbiota-metabolome relationships revealed by integrative analysis of multi-omics data. Sci. Rep. 10, 3164. doi: 10.1038/s41598-020-59617-9
Albalawi H. I., Khan Z. N., Valle-Pérez A. U., Kahin K. M., Hountondji M., Alwazani H., et al. (2021). Sustainable and eco-friendly coral restoration through 3D printing and fabrication. Sustain. Chem. Eng. 9, 12634–12645. doi: 10.1021/acssuschemeng.1c04148
Ali A., Song L., Hu J., Jiang J., Rao Q., Shoaib M., et al. (2021). Synthesis and characterization of caprolactone based polyurethane with degradable and antifouling performance. Chin. J. Chem. Eng. 34, 299–306. doi: 10.1016/j.cjche.2020.11.007
Antunes J., Sousa A., Azevedo J., Rego A., Leão P., Vasconcelos V. (2020). Distinct temporal succession of bacterial communities in early marine biofilms in a Portuguese Atlantic port. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01938
Bao V. W. W., Leung K. M. Y., Qiu J.-W., Lam M. H. W. (2011). Acute toxicities of five commonly used antifouling booster biocides to selected subtropical and cosmopolitan marine species. Mar. Poll. Bull. 62, 1147–1151. doi: 10.1016/j.marpolbul.2011.02.041
Bouranta A., Tudose I. V., Georgescu L., Karaiskou A., Vrithias N. R., Viskadourakis Z., et al. (2022). 3D printed metal oxide-polymer composite materials for antifouling applications. Nanomaterials 12, 917. doi: 10.3390/nano12060917
Breddermann K., Drescher P., Polzin C., Seitz H., Paschen M. (2016). Printed pressure housings for underwater applications. Ocean Eng. 113, 57–63. doi: 10.1016/j.oceaneng.2015.12.033
Bressy C., Briand J.-F., Lafond S., Davy R., Mazeas F., Tanguy B., et al. (2022). What governs marine fouling assemblages on chemically-active antifouling coatings? Prog. Org. Coatings 164, 106701. doi: 10.1016/j.porgcoat.2021.106701
Briand J.-F., Barani A., Garnier C., Réhel K., Urvois F., Lepoupon C., et al. (2017). Spatio-temporal variations of marine biofilm communities colonizing artificial substrata including antifouling coatings in contrasted French coastal environments. Microb. Ecol. 74, 585–598. doi: 10.1007/s00248-017-0966-2
Briand J.-F., Pochon X., Wood S. A., Bressy C., Garnier C., Réhel K., et al. (2018). Metabarcoding and metabolomics offer complementarity in deciphering marine eukaryotic biofouling community shifts. Biofouling 34, 657–672. doi: 10.1080/08927014.2018.1480757
Briand J.-F., Pollet T., Misson B., Garnier C., Lejars M., Maintenay M., et al. (2022). Surface characteristics together with environmental conditions shape marine biofilm dynamics in coastal NW Mediterranean locations. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.746383
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carve M., Scardino A., Shimeta J. (2019). Effects of surface texture and interrelated properties on marine biofouling: a systematic review. Biofouling 35, 597–617. doi: 10.1080/08927014.2019.1636036
Cassé F., Swain G. W. (2006). The development of microfouling on four commercial antifouling coatings under static and dynamic immersion. Int. Biodeter. Biodegrad. 57, 179–185. doi: 10.1016/j.ibiod.2006.02.008
Catao E. C. P., Gallois N., Fay F., Misson B., Briand J.-F. (2021b). Metal resistance genes enrichment in marine biofilm communities selected by biocide-containing surfaces in temperate and tropical coastal environments. Environ. Poll. 268, 115835. doi: 10.1016/j.envpol.2020.115835
Catão E. C. P., Pollet T., Garnier C., Barry-Martinet R., Rehel K., Linossier I., et al. (2021a). Temperate and tropical coastal waters share relatively similar microbial biofilm communities while free-living or particle-attached communities are distinct. Mol. Ecol. 30, 2891–2904. doi: 10.1111/mec.15929
Catão E. C. P., Pollet T., Misson B., Garnier C., Ghiglione J.-F., Barry-Martinet R., et al. (2019). Shear stress as a major driver of marine biofilm communities in the NW Mediterranean Sea. Front. Microbiol. 10. doi: 10.3389/fmicb.2019
Chen L., Lam J. C. W. (2017). SeaNine 211 as antifouling biocide: a coastal pollutant of emerging concern. J. Environ. Sci. 61, 68–79. doi: 10.1016/j.jes.2017.03.040
Chen C.-L., Maki J. S., Rittschof D., Teo S. L.-M. (2013). Early marine bacterial biofilm on a copper-based antifouling paint. Int. Biodeter. Biodegrad. 83, 71–76. doi: 10.1016/j.ibiod.2013.04.012
Christiansen E. F., Marcellin-Little D. J., Issacs A., Horn T. J., Aman R. L., Legner C., et al. (2014). “A customized 3D-printed splint for stabilization of an open front flipper fracture in a green sea turtle (Chelonia mydas),” in International Association for Aquatic Animal Medicine, Gold Coast, Australia. 21–25. Available at: https://www.vin.com/doc/?id=6251888
Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucl. Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Delgado A., Briciu-Burghina C., Regan F. (2021). Antifouling strategies for sensors used in water monitoring: review and future perspectives. Sensors 21, 389. doi: 10.3390/s21020389
Dobretsov S., Abed R. M. M., Muthukrishnan T., Sathe P., Al-Naamani L., Queste B. Y., et al. (2018). Living on the edge: biofilms developing in oscillating environmental conditions. Biofouling 34, 1064–1077. doi: 10.1080/08927014
Dobretsov S., Qian P.-Y. (2006). Facilitation and inhibition of larval attachment of the bryozoan Bugula neritina in association with mono-species and multi-species biofilms. J. Exp. Mar. Biol. Ecol. 333, 263–274. doi: 10.1016/j.jembe.2006.01.019
Dong Z., Vuckovac M., Cui W., Zhou Q., Ras R. H. A., Levkin P. A. (2021). 3D printing of superhydrophobic objects with bulk nanostructure. Adv. Mater. 33, 2106068. doi: 10.1002/adma.202106068
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fitridge I., Dempster T., Guenther J., de Nys R. (2012). The impact and control of biofouling in marine aquaculture: a review. Biofouling 28, 649–669. doi: 10.1080/08927014.2012.700478
Frankfurter G., Beltran R. S., Hoard M., Burns J. M. (2019). Rapid prototyping and 3D printing of antarctic seal flipper tags. Wildl Soc. Bull. 43, 313–316. doi: 10.1002/wsb.964
Fu Y., Wang W., Zhang L., Vinokurov V., Stavitskaya A., Lvov Y. (2019). Development of marine antifouling epoxy coating enhanced with clay nanotubes. Materials 12, 4195. doi: 10.3390/ma12244195
Gittens J. E., Smith T. J., Suleiman R., Akid R. (2013). Current and emerging environmentally-friendly systems for fouling control in the marine environment. Biotechnol. Adv. 31, 1738–1753. doi: 10.1016/j.biotechadv.2013.09.002
Gizer G., Önal U., Ram M., Sahiner N. (2023). Biofouling and mitigation methods: a review. Biointerface Res. Appl. Chem. 13, 185. doi: 10.33263/BRIAC132.185
Gong J., Song W. B., Warren A. (2005). Periphytic ciliates colonization: annual cycle and responses to environmental conditions. Aquat. Microb. Ecol. 39, 159–170. doi: 10.3354/ame039159
González-Henríquez C. M., Sarabia-Vallejos M. A., Hernandez J. R. (2019). Antimicrobial polymers for additive manufacturing. Int. J. Mol. Sci. 20, 1210. doi: 10.3390/ijms20051210
Greenfield P. (2017). Greenfield Hybrid Analysis Pipeline (GHAP). CSIRO Software Collection. doi: 10.4225/08/59f98560eba25
Hadfield M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Holmström C., Egan S., Franks A., McCloy S., Kjelleberg S. (2002). Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41, 47–58. doi: 10.1111/j.1574-6941.2002.tb00965.x
Huang S. Y., Hadfield M. G. (2003). Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 260, 161–172. doi: 10.3354/meps260161
Huws S. A., McBain A. J., Gilbert P. (2005). Protozoan grazing and its impact upon population dynamics in biofilm communities. J. Appl. Microbiol. 98, 238–244. doi: 10.1111/j.1365-2672.2004.02449.x
Kirstein I. V., Wichels A., Krohne G., Gerdts G. (2018). Mature biofilm communities on synthetic polymers in seawater - specific or general? Mar. Environ. Res. 142, 147–154. doi: 10.1016/j.marenvres.2018.09.028
Lawes J. C., Neilan B. A., Brown M. V., Clark G. F., Johnston E. L. (2016). Elevated nutrients change bacterial community composition and connectivity: high throughput sequencing of young marine biofilms. Biofouling 32, 57–69. doi: 10.1080/08927014.2015.1126581
Leary M., Piola R., Shimeta J., Toppi S., Mayson S., McMillan M., et al. (2016). Additive manufacture of anti-biofouling inserts for marine applications. Rapid Prototyping J. 22, 416–434. doi: 10.1108/RPJ-02-2014-0022
Lejars M., Margaillan A., Bressy C. (2012). Fouling release coatings: a nontoxic alternative to biocidal antifouling coatings. Chem. Rev. 112, 4347–4390. doi: 10.1021/cr200350v
Lema K. A., Constancias F., Rice S. A., Hadfield M. G. (2019). High bacterial diversity in nearshore and oceanic biofilms and their influence on larval settlement by Hydroides elegans (Polychaeta). Environ. Microbiol. 21, 3472–3488. doi: 10.1111/1462-2920.14697
Lemonnier C., Chalopin M., Huvet A., Le Roux F., Labreuche Y., Petton B., et al. (2022). Time-series incubations in a coastal environment illuminates the importance of early colonizers and the complexity of bacterial biofilm dynamics on marine plastics. Environ. Poll. 312, 119994. doi: 10.1016/j.envpol.2022.119994
Ly O., Yoris-Nobile A. I., Sebaibi N., Blanco-Fernandez E., Boutouil M., Castro-Fresno D., et al. (2021). Optimisation of 3D printed concrete for artificial reefs: biofouling and mechanical analysis. Construct. Build. Mater. 272, 121649. doi: 10.1016/j.conbuildmat.2020.121649
Ma C., Xu W., Pan J., Xie Q., Zhang G. (2016). Degradable polymers for marine antibiofouling: optimizing structure to improve performance. Ind. Eng. Chem. Res. 55, 11495–11501. doi: 10.1021/acs.iecr.6b02917
Matus I. V., Alves J. L., Gois J., da Rocha A. B., Neto R., Mota C. D. S. (2021). Effect of 3D printer enabled surface morphology and composition on coral growth in artificial reefs. Rapid Prototyp. J. 27, 692–706. doi: 10.1108/RPJ-07-2020-0165
McElroy D. J., Hochuli D. F., Doblin M. A., Murphy R. J., Blackburn R. J., Coleman R. A. (2017). Effect of copper on multiple successional stages of a marine fouling assemblage. Biofouling 33, 904–916. doi: 10.1080/08927014.2017.1384468
Mohammed J. S. (2016). Applications of 3D printing technologies in oceanography. Methods Oceanogr. 17, 97–117. doi: 10.1016/j.mio.2016.08.001
Molino P. J., Wetherbee R. (2008). The biology of biofouling diatoms and their role in the development of microbial slimes. Biofouling 24, 365–379. doi: 10.1080/08927010802254583
Muthukrishnan T., Abed R. M. M., Dobretsov S., Kidd B., Finnie A.A (2014). Long-term microfouling on commercial biocidal fouling control coatings. Biofouling 30, 1155–1164. doi: 10.1080/08927014.2014.972951
Nichols D. (2004). “Biocides in plastics,” in Rapra review reports, vol. 15. (UK: Thor Overseas Limited).
Odetti A., Altosole M., Bruzzone G., Caccia M., Viviani M. (2019). Design and construction of a modular pump-jet thruster for autonomous surface vehicle operations in extremely shallow water. J. Mar. Sci. Eng. 7, 222. doi: 10.3390/jmse7070222
Padmavathi A. R., Murthy P. S., Das A., Rao T. S. (2021). Enhanced antifouling property of polydimethylsiloxane-CuO nanocomposite in marine environment. Materials Lett. 301, 130342. doi: 10.1016/j.matlet.2021.130342
Pinto M., Langer T. M., Huffer T., Hofmann T., Herndl G. J. (2019). The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PloS One 14, e0217165. doi: 10.1371/journal.pone.0217165
Piola R. F., Dafforn K. A., Johnston E. L. (2009). The influence of antifouling practices on marine invasions. Biofouling 25, 633–644. doi: 10.1080/08927010903063065
Piola R., Grandison C., Shimeta J., Del Frate A., Leary M. (2022). Can vessel sea chest design improve fouling-control coating performance? Ocean Eng. 256, 111426. doi: 10.1016/j.oceaneng.2022.111426
Piola R., Leary M., Santander R., Shimeta J. (2021). Antifouling performance of copper-containing fused filament fabrication (FFF) 3D printing polymer filaments for marine applications. Biofouling 37, 206–221. doi: 10.1080/08927014.2021.1892085
Pollet T., Berdjeb L., Garnier C., Durrieu G., Le Poupon C., Misson B., et al. (2018). Prokaryotic community successions and interactions in marine biofilms: the key role of flavobacteriia. FEMS Microbiol. Ecol. 94, fiy083. doi: 10.1093/femsec/fiy083
Portas A., Quillien N., Culioli G., Briand J.-F. (2022). Eukaryotic diversity of marine biofouling from coastal to offshore areas. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.971939
Qian P.-Y., Lau S. C. K., Dahms H.-U., Dobretsov S., Harder T. (2007). Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotechnol. 9, 399–410. doi: 10.1007/s10126-007-9001-9
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Roepke L. K., Brefeld D., Soltmann U., Randall C. J., Negri A. P., Kunzmann A. (2022). Antifouling coatings can reduce algal growth while preserving coral settlement. Sci. Rep. 12, 15935. doi: 10.1038/s41598-022-19997-6
Ryley M., Carve M., Piola R., Scardino A. J., Shimeta J. (2021). Comparison of biofouling on 3D-printing materials in the marine environment. Int. Biodeter. Biodegrad. 164, 105293. doi: 10.1016/j.ibiod.2021.105293
Salta M., Wharton J. A., Blache Y., Stokes K. R., Briand J.-F. (2013). Marine biofilms on artificial surfaces: structure and dynamics. Environ. Microbiol. 15, 2879–2893. doi: 10.1111/1462-2920.12186
Schultz M. P., Bendick J. A., Holm E. R., Hertel W. M. (2011). Economic impact of biofouling on a naval surface ship. Biofouling 27, 87–98. doi: 10.1080/08927014.2010.542809
Shimeta J., Cutajar J., Watson M. G., Vlamis T. (2012). Influences of biofilm-associated ciliates on the settlement of marine invertebrate larvae. Mar. Ecol. Prog. Ser. 449, 1–12. doi: 10.3354/meps09638
Staiano G., Gloria A., Ausanio G., Lanzotti A., Pensa C., Martorelli M. (2016). Experimental study on hydrodynamic performances of naval propellers to adopt new additive manufacturing processes. Int. J. Interact. Design Manuf. 12, 1–14. doi: 10.1007/s12008-016-0344-1
Tait K., Williamson H., Atkinson S., Williams P., Cámara M., Joint I. (2009). Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ. Microbiol. 11, 1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x
Takahashi K. (2009). “Release rates of biocides from antifouling paints,” in Ecotoxicology of antifouling biocides. Eds. Arai T., Harino H., Ohji M., Langston W. J. (Tokyo: Springer), 3–22. doi: 10.1007/978-4-431-85709-9_1
Védie E., Brisset H., Briand J.-F., Bressy C. (2021). Bioinspiration and microtopography as nontoxic strategies for marine bioadhesion control. Adv. Mater. Interfaces 8, 2100994. doi: 10.1002/admi.202100994
von Ammon U., Wood S. A., Laroche O., Zaiko A., Tait L., Lavery S., et al. (2018). The impact of artificial surfaces on marine bacterial and eukaryotic biofouling assemblages: a high-throughput sequencing analysis. Mar. Environ. Res. 133, 57–66. doi: 10.1016/j.marenvres.2017.12.003
Vucko M. J., King P. C., Poole A. J., Carl C., Jahedi M. Z., de Nys R. (2012). Cold spray metal embedment: an innovative antifouling technology. Biofouling 28, 239–248. doi: 10.1080/08927014.2012.670849
Watson M. G., Scardino A. J., Zalizniak L., Shimeta J. (2015a). Colonisation and succession of marine biofilm-dwelling ciliates in response to environmental variation. Aquat. Microb. Ecol. 74, 95–105. doi: 10.3354/ame01731
Watson M. G., Scardino A. J., Zalizniak L., Shimeta J. (2015b). Colonisation and succession of microbial biofilm assemblages on antifouling and foul-release coatings in temperate Australia. Biofouling 31, 709–720. doi: 10.1080/08927014.2015.1105221
Watson M. G., Scardino A. J., Zalizniak L., Shimeta J. (2016). Inhibition of invertebrate larval settlement by biofilm ciliates. Mar. Ecol. Prog. Ser. 557, 77–90. doi: 10.3354/meps11825
Winfield M. O., Downer A., Longyear J., Dale M., Barker G. L. A. (2018). Comparative study of biofilm formation on biocidal antifouling and fouling-release coatings using next-generation DNA sequencing. Biofouling 34, 464–477. doi: 10.1080/08927014.2018.1464152
Wolfe K., Mumby P. J. (2020). RUbble biodiversity samplers: 3D-printed coral models to standardize biodiversity censuses. Methods Ecol. Evol. 11, 1395–1400. doi: 10.1111/2041-210X.13462
Yebra D. M., Kiil S., Dam-Johansen K. (2004). Antifouling technology-past, present and future steps towards efficient and environmentally friendly antifouling coatings. Prog. Organ. Coatings 50, 75–104. doi: 10.1016/j.porgcoat.2003.06.001
Zaiko A., Schimanski K., Pochon X., Hopkins G. A., Goldstien S., Floerl O., et al. (2016). Metabarcoding improves detection of eukaryotes from early biofouling communities: implications for pest monitoring and pathway management. Biofouling 32, 671–684. doi: 10.1080/08927014.2016.1186165
Zavoleas Y., Haeusler M., Dunn K., Bishop M., Dafforn K., Schaefer N., et al. (2020). Designing bio-shelters: improving water quality and biodiversity in the bays precinct through dynamic data-driven approaches. J. Digital Land. Architect. 5521–532. doi: 10.14627/537690053
Keywords: marine biofouling, marine biofilms, microbial assemblage, metabarcoding, antifouling, 3D printing, DCOIT
Citation: Shimeta J, Wilding-McBride G, Bott NJ, Piola R, Santander R, Leary M and Scardino AJ (2023) Growth of marine biofilms and macrofouling organisms on biocide-infused, 3D-printed thermoplastics. Front. Mar. Sci. 10:1172942. doi: 10.3389/fmars.2023.1172942
Received: 24 February 2023; Accepted: 27 April 2023;
Published: 12 May 2023.
Edited by:
Maria Salta, University of Portsmouth, United KingdomReviewed by:
Sriyutha Murthy, Bhabha Atomic Research Centre, IndiaCopyright © 2023 Shimeta, Wilding-McBride, Bott, Piola, Santander, Leary and Scardino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeff Shimeta, amVmZi5zaGltZXRhQHJtaXQuZWR1LmF1
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.