
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 19 May 2023
Sec. Aquatic Microbiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1166287
This article is part of the Research TopicMicrobial Diversity and Resources in Tidal FlatsView all 17 articles
Marine microorganisms have long been acknowledged as a significant reservoir of enzymes required for industrial use. In this study, a novel extracellular protease HslHlyB derived from marine-originated haloarchaeon Halostella pelagica DL-M4T was identified. HslHlyB contained polycystic kidney disease (PKD) domain and pre-peptidase C-terminal (PPC) domain at the C-terminus. Truncation and replacement of the C-terminal extension (CTE) of HslHlyB demonstrated the importance of the CTE in maintaining the protease activity secreted by haloarchaeon. HslHlyB and HslHlyBΔCTE were expressed in Escherichia coli BL21(DE3), and purified by high-affinity column refolding and gel filtration chromatography. The molecular masses of HslHlyB and HslHlyBΔCTE were 42 kDa and 20 kDa, respectively. The optimum catalytic reaction conditions were 50°C, pH 8.5, NaCl 3.5 M and 50°C, pH 7.5, NaCl 3 M, respectively. They showed good stability and hydrolysis capabilities towards a wide range of protein substrates. HslHlyBΔCTE showed higher catalytic reaction rate and better thermal stability than the wild type against azocasein and tetrapeptide substrate. The hydrolysates of soybean protein hydrolyzed by HslHlyBΔCTE had smaller average molecular masses and shorter average peptide chain lengths than those by HslHlyB. These results indicated the diversity of halolysins from marine-originated haloarchaea to harness organic nitrogen in the marine environment and provided promising candidates for application in various industries.
It is widely recognized that marine is a reservoir of biological resources, harboring diverse microorganisms and functional enzymes. Intertidal zones with variable salinity, temperature and nutrient are kinds of important marine ecosystems including a variety of habitats, such as rocky shores, sandy beaches, soft sediments, salt marshes and mangroves (Kon et al., 2020). Haloarchaea are the most salt-requiring and salt-resistant microorganisms partly thriving in marine salt inhabitants, such as marine solar salterns, marine lagoons and costal sediment (Lee, 2013; Cui and Dyall Smith, 2021; Sun et al., 2023). The haloarchaea that inhabit this zone have adapted to extremely harsh environments and evolved the ability to produce enzymes that function optimally in high salt concentrations, even in extreme pH and temperatures (Amoozegar et al., 2017). It is increasingly interesting to look for specific enzymes with high thermal stability, high tolerance with wide range of salinities produced by haloarchaea to meet the needs of industrial applications.
Halolysin is a kind of proteases that are secreted extracellularly by haloarchaea and exert endo-protease activity. Halolysins reported so far all belong to serine proteases with molecular masses range of 21-130 kDa (Studdert et al., 1997; Elsztein et al., 2001; Studdert et al., 2001; Elbanna et al., 2015). Halolysin plays important ecological roles in organic nitrogen degradation in saline environments and provides its host with antagonistic and defensive activities against other haloarchaea for competitive advantage (Chen et al., 2021). Halolysins were considered as excellent candidates in industries such as food production (Chaveesuk et al., 1994; Yongsawatdigul et al., 2007; Akolkar et al., 2010), cleaning agent (Jaouadi et al., 2008), peptide synthesis (Ryu et al., 1994; Ruiz et al., 2010), detergent (Akolkar et al., 2008) and waste management (Lefebvre and Moletta, 2006).
A number of halolysins have been already reported with known coding genes, including halolysins 172P1 from Natrialba asiatica 172P1 (Kamekura et al., 1992), R4 from Haloferax mediterranei R4 (Kamekura et al., 1996), SptA and SptC from Natrinema sp. J7 (Zhang et al., 2014; Du et al., 2015), Nep from Natrialba magadii (De Castro et al., 2008), HlyA from Halococcus salifodinae (Hou et al., 2020), HlyHap from Haladaptatus sp. DYF46 (Hou et al., 2022). For protein domain organization, those halolysin precursors are usually made up of a Tat (twin-arginine translocation) signal peptide, an N-terminal propeptide, an S8/S53 family catalytic domain and a C-terminal extension (CTE). The CTE of halolysins are usually prepeptidase C-terminal (PPC) domain, except a combined polycystic kidney disease (PKD) domain and chitin-binding domain (ChBD) was reported in the CTE of SptC (Du et al., 2015). In recent years a marine halolysin HlyHap without CTE was discovered to prefer a low salinity in contrast to most halolysins (Hou et al., 2022). CTE domains have been shown to have a significant impact on the structure and function of a protease (Gao et al., 2010), especially those secreted by the marine bacteria (Huang et al., 2017). The halolysins thus far reported are much less diverse than those of proteases from bacteria, especially halolysins of marine origin that possess ecological significance and deserve to be explored more deeply.
Strain DL-M4T with ability of hydrolysis of gelatin and casein is the type strain of Halostella pelagica and was isolated from the salted seaweed produced in Dalian, Liaoning Province, China (Han and Cui, 2020). Strain DL-M4T may originate from marine solar salt or Laminaria farmed in tidal zone. In this study, the potential extracellular protease genes from Hsl. pelagica DL-M4T were identified, and the recombinant halolysins with different CTE domains were expressed in the heterologous host, Haloferax volcanii H1424, in order to explore the effect of different CTE on HslHlyB activity. Furthermore, HslHlyB and HslHlyBΔCTE were purified and characterized to explore the diversity of the catalytic properties of halolysins and the effect of CTE on enzymatic properties. At last, the potential application of HslHlyB and HslHlyBΔCTE in hydrolysis of soybean protein under high-salt concentration were evaluated. This study would enrich our knowledge of the diversity of marine halolysins and provide a promising halophilic protease.
Hsl. pelagica DL-M4T (=CGMCC 1.13603T = JCM 32954T, GenBank accession numbers: 16S rRNA gene MH062945, genome CP040678-CP040681) with the hydrolysis ability towards casein and gelatin was isolated from the salted brown alga Laminaria in the previous study (Han and Cui, 2020). Other halolysin producing strains of Hfx. mediterranei ATCC 33500T, Nab. magadii CGMCC 1.1966T and Hcc. salifodinae DSM 8989T were also used in this study. They were grown under aerobic conditions at 37°C in neutral haloarchaeal medium (NHM) (Cui et al., 2011). Hfx. volcanii H1424 used as the haloarchaeal host for expression was cultured in Hv-YPC medium (Stroud et al., 2012). Escherichia coli DH5α and BL21 (DE3) were used as host strains for cloning and protein expression, respectively. Luria-Bertani medium was used to cultivate the above E. coli strains at 37°C, if necessary, 100 μg mL−1 of ampicillin or 50 μg mL−1 of kanamycin was added.
The hmm model of Peptidase_S8 domain (Pfam accession: PF00082) was used to search for model-compatible proteins in Hsl. pelagica DL-M4T via the Hmmsearch of Hmmer software (Finn et al., 2011). The protein sequence of Nep from Nab. magadii (accession no. AAV66536) was also used to find halolysin genes in the genome of Hsl. pelagica DL-M4T by BlastP tool (Altschul et al., 1997). The resulting potential halolysins were also confirmed against Swiss-Prot databases. The theoretical molecular mass and isoelectric point were predicted using Prot Param (https://web.expasy.org/protparam/). Signal peptide prediction was performed using SignalP 5.0 (Armenteros et al., 2019). Protein domain prediction was performed using InterProScan (Quevillon et al., 2005). The C-terminal domain structure was modelled by SWISS-MODEL Workspace (https://swissmodel.expasy.org/). The structure of HslHlyB was predicted by AlphaFold2 (Jumper et al., 2021).
Three potential halolysin genes and HslHlyB without CTE or PKD domain encoding genes were amplified by PCR using Hsl. pelagica DL-M4T genomic DNA as the template. The encoding genes for chimeras of HslHlyB with different CTE domains were obtained using the overlapping extension PCR method as described previously (Hou et al., 2020) using Hsl. pelagica DL-M4T, Hfx. mediterranei ATCC 33500T, Nab. magadii CGMCC 1.1966 and Hcc. salifodinae DSM 8989T genomic DNA as the template. The PCR amplification was performed using the KOD-plus-neo DNA polymerase (Toyobo), and primer pairs shown in Table S1. The products were digested with a pUCm-T vector using EcoRI, BamHI/SphI or NcoI, XhoI as the restriction enzyme sites (the restriction endonucleases used for corresponding genes shown in Table S1). The purified amplicons of potential halolysin genes and encoding genes of HslHlyB variants were cloned into the haloarchaeal shuttle vector pTA under the strong promoter P.phaR (Liu et al., 2015) for expression in Hfx. volcanii. The purified amplicons of HslHlyB and HslHlyBΔCTE were cloned into the vector pET28a under Lac promoter for expression in E. coli BL21 (DE3). The colony PCR and Sanger sequencing were used to verify the inserts in the constructed plasmids.
There were two kinds of expressing hosts used in this study. For expressing HslHlyA, HslHlyB, HslHlyC and HslHlyB variants, the expression plasmid was transformed into Hfx. volcanii H1424. Cell cultures were grown in Hv-YPC medium at 37°C until the stationary phase. Culture supernatant (12,000 g, 10 min, 4°C) was used as the crude enzyme to assay the extracellular protease activity. To obtain purified enzymes, the HslHlyB and HslHlyBΔCTE precursors were expressed in E. coli BL21 (DE3) and in vitro refolded as previously described (Hou et al., 2021; Hou et al., 2022). For in vitro refolding, the recombinant cells were firstly resuspended in lysis buffer (8 M urea, 10 mM CaCl2, 50 mM Tris-HCl, 2 mM β-mercaptoethanol, pH 8.0). After sonicating and centrifuging, the supernatant containing denatured precursors was loaded onto the 2 mL Ni-column, then washed with lysis buffer containing 40 mM imidazole. The Ni-column bound precursors were incubated in refolding buffer (4 M NaCl, 10 mM CaCl2, 50 mM Tris-HCl, pH 8.0) for 24 h at 37°C to fully refold. After washing with the refolding buffer containing 20 mM imidazole, the target proteins were eluted using the refolding buffer containing 100 mM imidazole. The refolded halolysins were further purified by molecular sieves using the Superdex 200 10/300 GL column (GE healthcare).
The Hv-YPC agar plates added with 0.5% (w/v) skim milk or gelatin were used to evaluate the protein hydrolysis capacity of the recombinant haloarchaeal cells (inoculation amount: 5 μL). The plates were incubated at 37°C for 14 days for activity verification. Frazier’s reagent (g/L: HgCl2, 150 g; concentrated HCl, 200 mL) was used to detect the hydrolysis of gelatin.
Enzyme activity towards azocasein (Sigma-Aldrich, USA) and N-Succinyl-Ala-Ala-Pro-Phe p-nitroanilide (Suc-AAPF-pNA, Sigma-Aldrich, USA) was determined as previously described (Hou et al., 2021). For determination of azocaseinolytic activity, 250 μL of crude enzyme was reacted with an equal volume of 0.5% azocasein (buffer: 2 M NaCl, 50 mM Tris-HCl, pH 8.0) at 37°C for 1 h. The reaction was terminated by the addition of 500 μL of 10% TCA, left at room temperature for 15 min and then centrifuged at 10,000 g for 10 min. 300 μL of supernatant was added to an equal volume of 1 M NaOH, and then centrifuged to remove the precipitate. The absorbance of the supernatant at 440 nm (A440) was recorded. In the control experiments, solutions of the enzyme and substrate were incubated separately. Under specific reaction conditions the amount of enzyme required to increase OD440 by 0.01 was defined as 1 unit of enzyme activity. When purified enzyme was used for azocaseinolytic activity assay, 10 μg of enzyme was used in each reaction. For the enzyme activity towards Suc-AAPF-pNA, 300 μL of crude enzyme was mixed with 300 μL of Suc-AAPF-pNA (0.4 mM) in the buffer of 2 M NaCl, 50 mM Tris HCl (pH 8.0). The initial hydrolysis rate of the tetrapeptide substrate was measured at 410 nm using a DU800 Nucleic Acid Protein Analyzer with the temperature controlled at 37°C. One unit of protease activity (U) was defined as the amount of enzyme required to produce 1 µmol pNa (extinction coefficient of 8,480 M-1cm-1) per minute under certain conditions. Protein concentration was measured by Bradford assay (Beyotime Institute of Biotechnology, China) (Bradford, 1976).
To determine the optimal reaction temperature, protease activities were measured at temperatures of 20°C, 30°C, 35°C, 40°C, 45°C, 50°C, 55°C, 60°C and 65°C, respectively, in 2 M NaCl, 50 mM Tris-HCl (pH 8.0). Thermal stability was determined by pre-incubation of the enzyme at 30°C, 40°C, 50°C and 60°C for 0 min, 20 min, 40 min and 60 min, respectively. Then the protease activity was measured at optimum temperature.
The optimal pH was determined by assaying protease activities at the optimum temperature and 2 M NaCl in different buffers of 50 mM K2HPO4/KH2PO4 buffer at pH 6.0, pH 6.5, pH 7.0 and pH 7.5; 50 mM Tris-HCl buffer at pH 7.5, pH 8.0, pH 8.5 and pH 9.0; 50 mM CHES-NaOH buffer at pH 9.0, pH 10.0 and pH 10.5. pH stability was determined by pre-incubation of the enzyme at room temperature at pH 6.0 (50 mM K2HPO4/KH2PO4 buffer), pH 8.0 (50 mM Tris-HCl buffer) and pH 10.0 (50 mM CHES-NaOH buffer) for 0 min, 20 min, 40 min and 60 min, respectively. Then the protease activity was measured at the optimum temperature.
The optimal NaCl concentration was determined by assaying at NaCl concentrations of 0.4 M, 0.5 M, 1.0 M, 1.5 M, 2.0 M, 2.5 M, 3.0 M, 3.5 M and 4.0 M, respectively, at the optimum pH and temperature. Salinity stability was determined by pre-incubation of the enzyme at room temperature with 0 M, 2 M and 4 M NaCl for 0 min, 20 min, 40 min and 60 min, respectively. Then the protease activity was measured at the optimum reaction condition.
The protease activities were measured with additives of 5 mM of metal ions or 5% (v/v) of organic solvents or 10% (v/v) of detergents. The protease inhibitors included 4 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM pepsin inhibitor (Pepstatin A), 1 mM p-chloromercuribenzoate (PCMB), 10 mM dithiothreitol (DTT) and 5 mM ethylene diamine tetraacetic acid (EDTA). The protease activities were measured at their respective optimal conditions (3.5 M NaCl, 50°C, pH 8.5 for HslHlyB and 3 M NaCl, 50°C, pH 7.5 for HslHlyBΔCTE).
The enzyme activity (in terms of tyrosine produced) was determined by reacting with azocasein, casein, gelatin, bovine serum protein, bovine hemoglobin, egg albumin, fish collagen or skim milk at a final concentration of 5 g/L under their respective optimal conditions (3.5 M NaCl, 50°C, pH 8.5 for HslHlyB and 3 M NaCl, 50°C, pH 7.5 for HslHlyBΔCTE).
The kinetic parameters of the purified halolysins were determined against 0.01-9.44 mM azocasein or 0.01-2 mM Suc-AAPF-pNA. The reaction with 0.8 μg protease was performed at the optimal conditions. The Km and Vmax values were calculated from the Michaelis-Menten equation by nonlinear regression (GraphPad Prism 7).
Tris-glycine SDS-PAGE was performed according to the method of King and Laemmli (King and Laemmli, 1971). To prevent the self-degradation of halolysin during the sample pretreatment, TCA-acetone precipitation was used to denature halolysin, referring to Hou’s method (Hou et al., 2021). After that, 20-40 μL of loading buffer containing 8 M urea was added to the protein precipitate with DTT at a final concentration of 100 mM, and the supernatant was used for SDS-PAGE analysis.
After tris-glycine SDS-PAGE analysis, the protein bands were in-gel digested as described by Shevchenko (Shevchenko et al., 1996), and the tryptic peptides were sequenced using Matrix-Assisted Laser Desorption Ionization Time of Flight/Time of Flight Mass Spectrometry (MALDI-TOF/TOF MS) (AB Sciex 5800, USA). The molecular mass of the mature halolysin was determined by gel filtration chromatography as previously described (Hou et al., 2021).
Soybean protein isolate (Sinopharm) was washed three times with sterile distilled water to remove soluble peptides. The reaction with 0.1 mg/mL enzyme and 5% (m/v) soybean protein isolate was carried out under optimum reaction conditions (3.5 M NaCl, 50°C, pH 8.5 for HslHlyB and 3 M NaCl, 50°C, pH 7.5 for HslHlyBΔCTE) for 24 h. The enzyme was inactivated in boiling water bath for 10 min. The protein hydrolysate solution was filtered, then desalted by a C18 column and identified by LC-MS (Agilent 1260/6460 LC/Triple Quadrupole MS).
Hmmer is a sensitive search tool for searching homologs protein by using hidden Markov models. The Pfam hmm model of Peptidase_S8 domain was used to search for potential halolysin genes. Nep, a most intensively-studied halolysin from Nab. magadii (De Castro et al., 2008), was also used to search for potential halolysin by BlastP tool. Three potential halolysins HslHlyA, HslHlyB and HslHlyC were found, with 24.5-56.0% identities to Nep (Table S2). Their amino acid sequences were compared against the SwissProt database. The most similar protein for HslHlyA was found to be SubE, the Bacillus subtilis protease, from Bacillus licheniformis, with 37.8% amino acid identity; the most similar proteins for HslHlyB and HslHlyC were halolysin from Hfx. mediterranei R4, with amino acid identities of 57.4% and 23.9%, respectively. The theoretical molecular masses of HslHlyA (383 aa), HslHlyB (613 aa) and HslHlyC (488 aa) were 39.5, 63.0 and 51.2 kDa and pI values of 4.20, 4.06 and 4.30, respectively. All of them contained Tat signal peptide and S8/S53 peptidase catalytic domain with aspartate-histidine-serine catalytic active sites. HslHlyA and HslHlyC did not contain any CTE while HslHlyB was found to contain two domains, PKD domain and PPC domain, at the C-terminus. SWISS-MODEL homology modeling was used to predict the secondary structure of PKD and PPC domains of HslHlyB, and found that they both have seven β-sheets (Figure S1). The structure of HslHlyB was determined with molecular replacement using a model obtained from Alphafold2 (the predicted local distance difference test score is 87.4). The 3D structure shows that HslHlyB consists of 4 domains, with 2 independent β-sandwich structures at the C-terminus (Figure S1). Some proteases distributed in different families contain CTEs, such as PKD domain, PPC domain and ChBD domain (Huang et al., 2017). These domains are different in amino acid sequences and also structures (Huang et al., 2019). But they are all rich in β-sheets (Huang et al., 2017). There are several studies on CTE function of bacterial proteases (Wang et al., 2010; He et al., 2012), but few studies on CTE of haloarchaeal proteases (Xu et al., 2011; Marem et al., 2018).
The potential halolysin genes were expressed in the non-extracellular protease-producing Hfx. volcanii H1424, a halophilic archaeal host, to investigate whether the encoded protein was active. Only the recombinant strain expressing HslHlyB showed significant hydrolysis zones on milk and gelatin plates, those expressing HslHlyA or HslHlyC could not hydrolyze milk or gelatin (Figures 1A, B). Meanwhile, the extracellular supernatant of the recombinant strain with HslHlyB had the ability to hydrolyze azocasein and Suc-AAPF-pNA, that of HslHlyA could weekly hydrolyze Suc-AAPF-pNA but not azocasein, and that of HslHlyC could not hydrolyze azocasein or Suc-AAPF-pNA (Figure 1C). According to these results, HslHlyB has low sequence similarity to other reported proteases (with the highest identity of 57.4% in SwissProt database) and ability to hydrolyze milk, gelatin, azocasein and tetra-peptide substrate. To our knowledge, no halolysin from the genus Halostella has been characterized. Additionally, this is the first reported halolysin with PKD and PPC domains at the C-terminus.
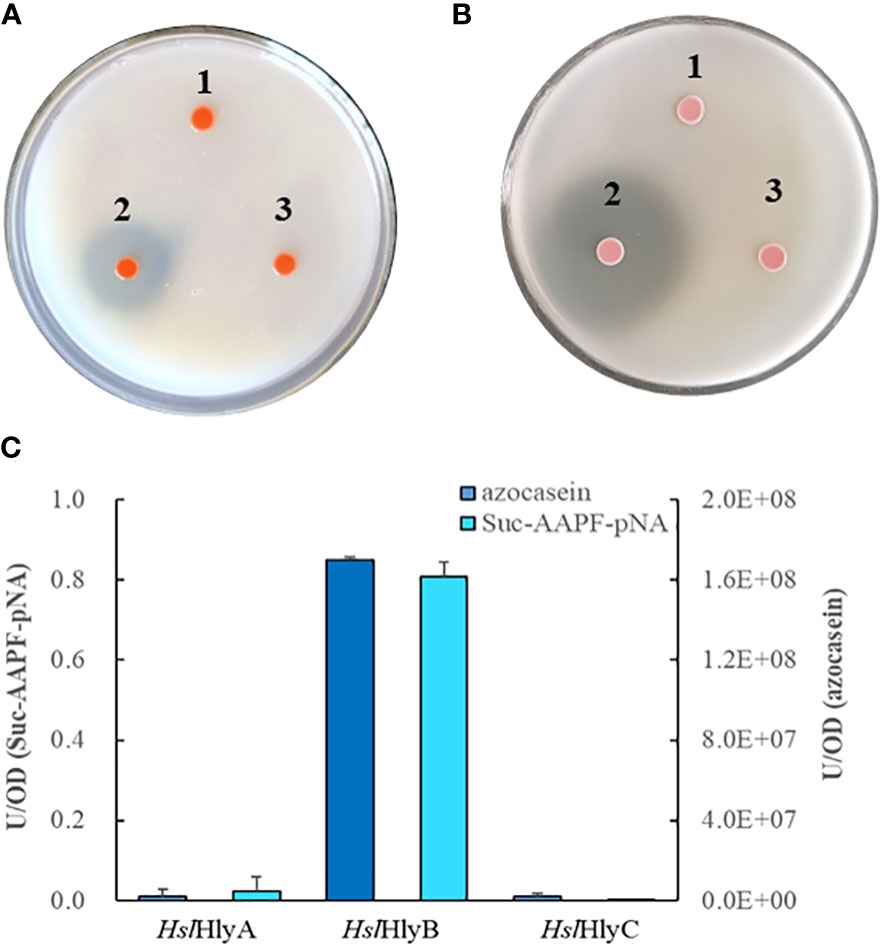
Figure 1 Protein hydrolysis by recombinant Hfx. volcanii cells. Hydrolysis of skim milk (A) and gelatin (B) on YPC agar plates, azocasein and Suc-AAPF-pNA hydrolytic activity (C) of cell-free culture supernatants of recombinant Hfx. volcanii. 1, HslHlyA; 2, HslHlyB; 3, HslHlyC.
CTE is prevalent in halolysins. It has important effects on enzyme activity and enzyme secretion. The novel C-terminal domains of HslHlyB consist of a PKD domain and a PPC domain. To investigate the effect of CTEs on enzymatic activity, HslHlyBΔPKD and HslHlyBΔCTE were constructed, with deletion of HslHlyB PKD domain and the whole CTE domain. In contrast to the strain expressing wild-type HslHlyB, the recombinant strains showed reductions in hydrolysis circle size on milk plates and also decreased activities against azocasein and Suc-AAPF-pNA (Figures 2B, C). The extracellular supernatant activity of the recombinant strain with HslHlyBΔPKD towards azocasein and Suc-AAPF-pNA decreased by 54.5% and 56.5%, respectively. The extracellular supernatant activity of the strain expressing HslHlyBΔCTE showed undetectable protease activity.

Figure 2 Effect of CTE on HslHlyB activity. (A) Domain arrangement of HslHlyB and the chimeras. S: signal peptide, P: propeptide, PKD: polycystic kidney disease domain, PPC: prepeptidase C-terminal domain, HlyA-C:C-terminal extension of HlyA, R4-C: C-terminal extension of R4, Nep-C: C-terminal extension of Nep. (B) Hydrolysis of skim milk by recombinant Hfx. volcanii cells on YPC agar plates. (C) Azocasein and Suc-AAPF-pNA hydrolytic activity of cell-free culture supernatants of recombinant Hfx. volcanii. 1, HslHlyB; 2, HslHlyBΔPKD; 3, HslHlyBΔCTE; 4, HslHlyB-HlyAC; 5, HslHlyB-R4C; 6, HslHlyB-NepC.
The CTE domain is self-stabilizing and can fold independently. Our previous study found that CTE domain can be functionally interchangeable among halolysins (Hou et al., 2020). To investigate the effect of CTE on HslHlyB activity, three chimeras, HslHlyB-HlyAC, HslHlyB-R4C and HslHlyB-NepC, namely, HslHlyB catalytic domain with the CTE of HlyA from Hcc. salifodinae (119 aa), R4 from Hfx. mediterranei (114 aa) and Nep from Nab. magadii (131 aa) (Kamekura et al., 1996; De Castro et al., 2008; Hou et al., 2020) (Figure 2A) were heterologously expressed in Hfx. volcanii H1424. The extracellular proteolytic activities were determined. The strains expressing HslHlyB-NepC cannot hydrolyze milk and had no activity towards azocasein and Suc-AAPF-pNA (Figures 2B, C). The strains expressing HslHlyB-HlyAC can hydrolyze milk. The hydrolysis zone of HslHlyB-R4C expressing strains is larger than those of HslHlyB-HlyAC expressing strains, while both are smaller than that of wildtype HslHlyB expressing strains (Figure 2B). The activity of the culture supernatant of cells expressing HslHlyB-HlyAC towards azocasein and Suc-AAPF-pNA decreased by 89.5% and 87.9%, respectively, and that of HslHlyB-R4C decreased by 93.9% and 89.6%, respectively (Figure 2C). The extracellular activities of recombinant strains expressing HslHlyB-HlyAC and HslHlyB-R4C towards tetrapeptide, a small molecule substrate, decreased less than that of azocasein, indicating that the CTE may affect the hydrolysis ability towards different substrates. The CTE of halolysins R4 from Hfx. mediterranei, HlyA from Hcc. salifodinae, Nep from Nab. magadii significantly reduced or abolished the extracellular enzyme activity of HslHlyB after substitution. The decreased extracellular enzyme activity is possibly because heterologous CTEs affect the correct folding of the HslHlyB catalytic domain. The effect of CTEs on halolysin activity remains to be investigated in depth.
HslHlyB and HslHlyBΔCTE with 6×His tag at the C-terminus were expressed in E. coli BL21 (DE3). Both intracellular lysate and culture supernatant of E. coli were without protease activity after induced expression. The protease precursors were obtained by affinity chromatography and confirmed by MALDI-TOF/TOF MS (Figure 3). The molecular mass of the protease precursor estimated from the HslHlyB protein sequence was 63 kDa, while the apparent molecular masses from SDS-PAGE were 108 kDa and 96 kDa. The presence of two bands of HslHlyB precursor was due to the excision of N-terminal sequences (Figure 3C). As for HslHlyBΔCTE, a single band with an apparent molecular mass of about 68 kDa was obtained. The phenomenon that the apparent molecular mass is much larger than the predicted molecular mass is common in the SDS-PAGE analysis of halophilic proteins (Alves et al., 2004; Karan et al., 2013; Gao et al., 2017), which is caused by the decrease of protein migration rate with the high content of acidic amino acids in halophilic proteins.
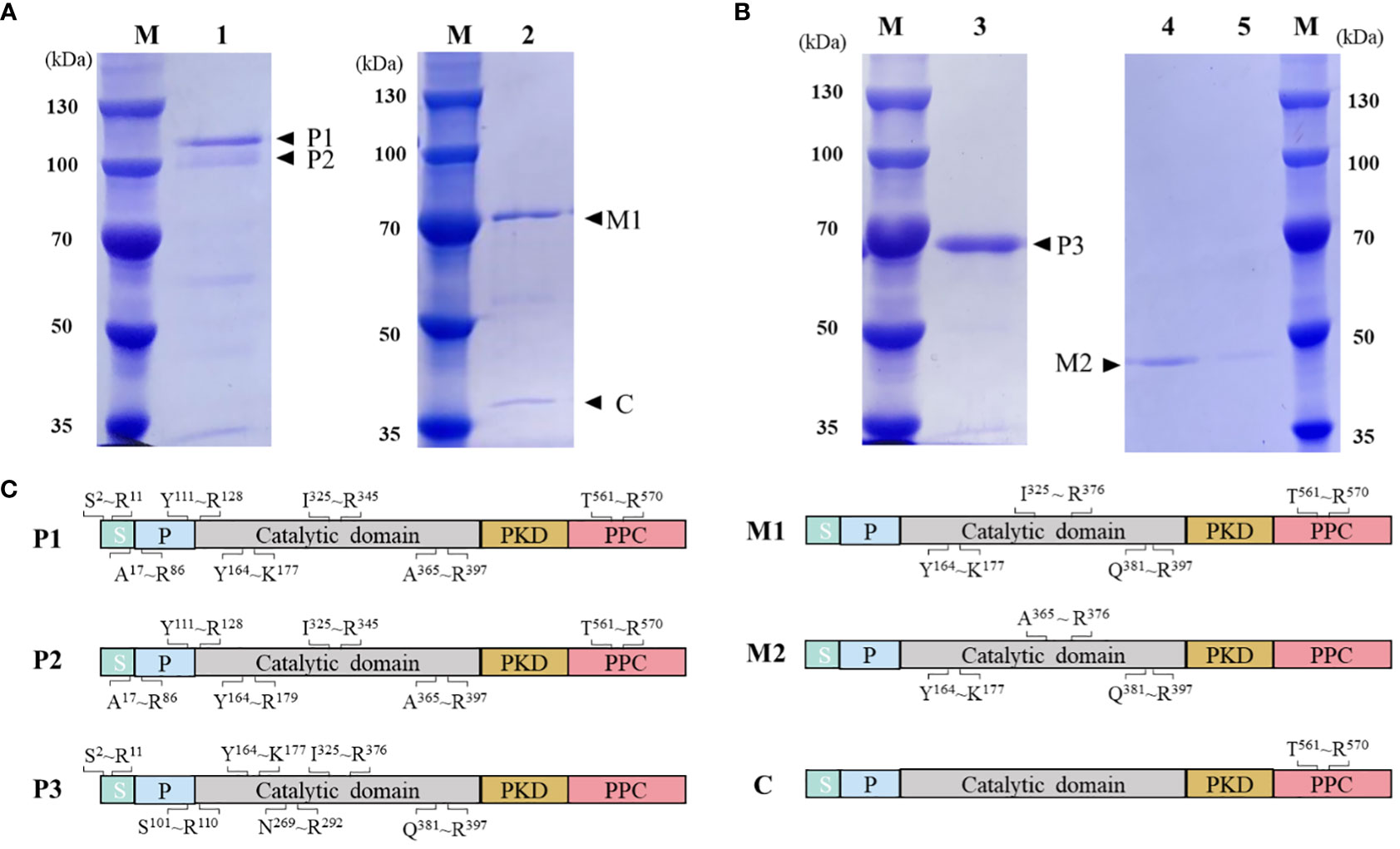
Figure 3 Purification and on-column refolding of HslHlyB and HslHlyBΔCTE. (A) SDS-PAGE analysis of HslHlyB precursor and mature HslHlyB. (B) SDS-PAGE analysis of HslHlyBΔCTE precursor and mature HslHlyBΔCTE. (C) MALDI-TOF/TOF MS analysis of protein bands in SDS-PAGE analysis. Sequenced tryptic peptides are marked. P1 and P2: HslHlyB precursor, P3, HslHlyBΔCTE precursor; C, the cleaved C-terminal fragments; M1, mature HslHlyB; M2, mature HslHlyBΔCTE; M, marker; lane 1, recombinant HslHlyB precursor purified under denaturing conditions; lane 2, refolded product of HslHlyB precursor; lane 3, recombinant HslHlyBΔCTE precursor purified under denaturing conditions; lane 4-5, refolded product of HslHlyBΔCTE precursor; S, signal peptide; P, propeptide; PKD, polycystic kidney disease domain; PPC, prepeptidase C-terminal domain.
On-column refolding was performed to allow the mature enzymes to be obtained by self-processing with its C-terminus bound to Ni-agarose. The refolding product of HslHlyB revealed two bands on SDS-PAGE gel, and that of HslHlyBΔCTE revealed a major band (Figures 3A, B). The HslHlyB refolding product consisted of mature enzyme, and also CTE fraction from autodegradation (Figure 3C). The refolding product of HslHlyBΔCTE corresponded to the catalytic domain identified by MALDI-TOF/TOF MS (Figure 3C). The refolding products were further purified by Superdex 200 GL column (Figures S2A, B). The purified products each revealed a major band on SDS-PAGE gel (Figures S2C, D). Combined with the molecular sieve profiles of protein standards with known molecular mass, the molecular mass of HslHlyB and HslHlyBΔCTE were estimated to be 42 kDa and 20 kDa, respectively. Halolysins, like most subtilases, are autocatalyzed into mature enzymes by cis- and trans-processing (Ruiz et al., 2012; Du et al., 2015). The purified precursors could autocatalyze to mature enzymes in high salt environments. Both CTE truncates and wild-type proteases were able to mature in vitro. However, when HslHlyBΔCTE was expressed in Hfx. volcanii, the recombinant strains showed no extracellular protease activity, similarly to the removal of the CTE from halolysin R4 and SptA (Kamekura et al., 1996; Xu et al., 2011). The immunoblot analysis of CTE truncates of SptA showed that no pro- or mature forms were secreted into the extracellular matrix (Xu et al., 2011). In contrast, the studies on Nep from Nab. magadii revealed that the absence of CTE did not affect the secretion and maturation of the enzyme in the halophilic archaeon (Marem et al., 2018). These results indicated that CTEs may have different effects on the structure of other domains in vivo, thus affecting maturation or secretion for different halolysins. In the case of HslHlyB in this study, as a subtilisin family enzyme with two β-sandwich domains at the C-terminus, its domain distribution is identical to that of fervidolysin (from Fervidobacterium pennivorans) with a sequence identity of 37.69%. The crystal structure of fervidolysin (PDB accession no. 1R6V) suggests that its CTE can create an open hydrophobic pocket that contributes to the fixation of the N-terminal elongated chain of the N-terminal pre-peptide domain (Kim et al., 2004). The study of the metalloprotease PrtV (from Vibrio cholerae) showed that the PKD domain with calcium ions controls the flexibility of the protein domain linker (Edwin et al., 2013). The CTE of HslHlyB probably affects the domain linker of the pre-peptide and plays a regulatory role in proper folding, thus affecting the secretion and autocatalytic maturation of the enzyme in vivo. CTE has also been reported to be involved in maintaining the stability of the catalytic domain. The halolysin SptA from Natrinema sp. J7 without CTE would be more prone to enzyme autolysis (Xu et al., 2011). However, the catalytic domain of HslHlyB remained stable in vitro in the absence of the entire CTE. The structure of halolysins would help dissect the underlying mechanisms.
The optimum reaction temperature for HslHlyB and HslHlyBΔCTE was 50°C. They were able to maintain more than 79% and 88% of the maximum enzyme activity at 40-60°C, respectively (Figures 4A, B). The thermal stability of the enzymes was studied based on the residual enzyme activities of the proteases after incubation at different temperatures (Figures 4C, D). The activity of HslHlyB decreased with increasing incubation time at 60°C, but it could still maintain about 80% of the original enzyme activity after incubation for 60 min. HslHlyB maintained about 91% of the original enzyme activity after incubation at 50°C for 60 min, and the activity of HslHlyB did not change significantly after incubation at 30-40°C. Notably, the activity of HslHlyBΔCTE remained essentially the same after incubation at 30-60°C for 0-60 min. The residual activity of Nep from Nab. magadii was >30% of original activity after incubation at 60°C for 2 h (Giménez et al., 2000), that of HlyHap from Haladaptatus sp. DYF46 was 33.9% after incubation at 60°C for 1 h (Hou et al., 2022), and that of Hly from Hgn. rubrum RO2-11 was <20% after incubation at 60°C for 1 h (Gao et al., 2017). Both HslHlyB and HslHlyBΔCTE have good thermal stability, and HslHlyBΔCTE shows better thermal stability than wild type enzyme.
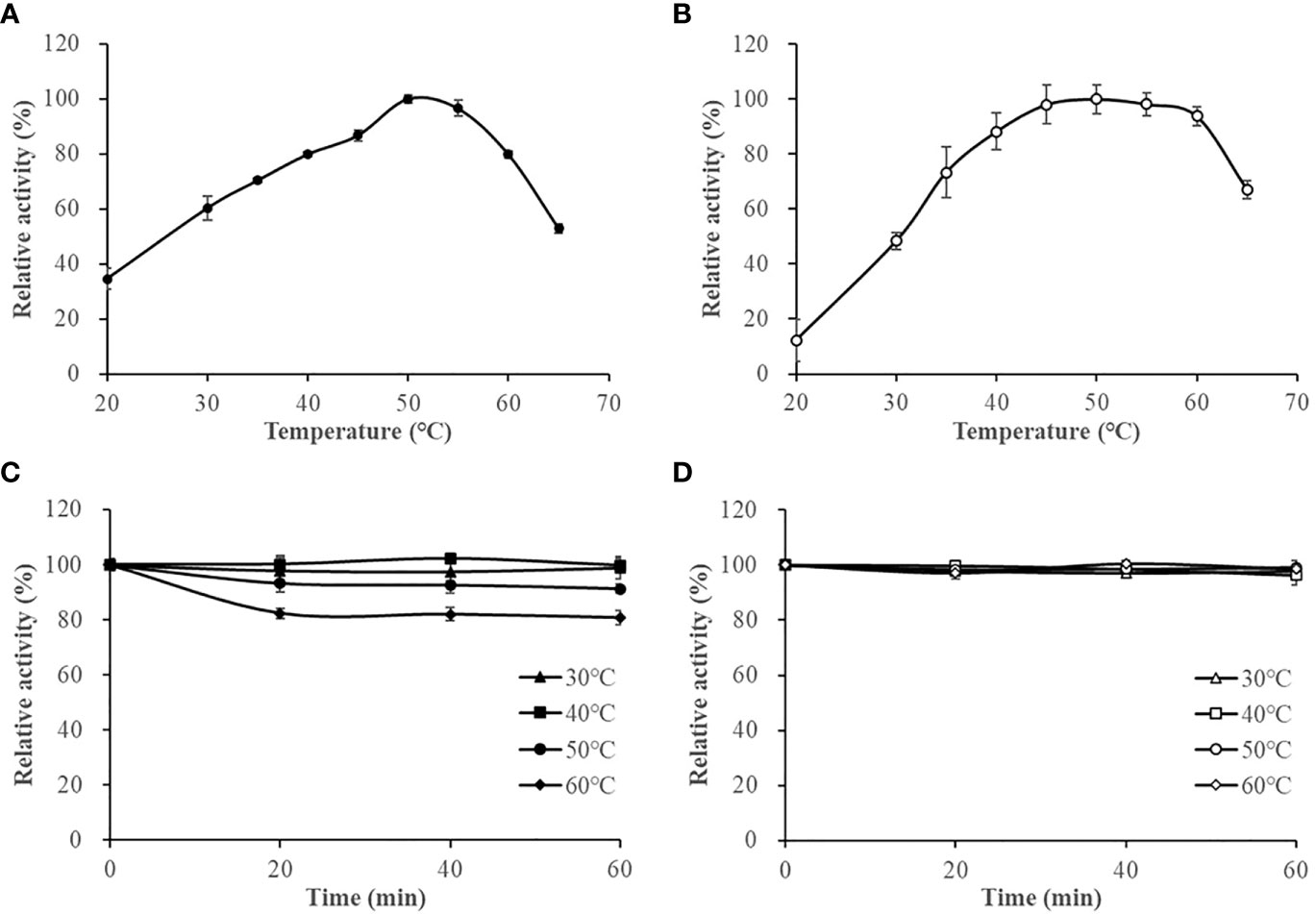
Figure 4 The effect of temperature on HslHlyB (A) and HslHlyBΔCTE activity (B), and the thermal stability of HslHlyB (C) and HslHlyBΔCTE (D). In (A, B), relative activities were calculated with the maximal activity defined as 100%. In (C, D), the initial enzyme activity was set as 100%, upon which the relative activities of enzymes after incubation at different temperatures for 20, 40 or 60 min were calculated. Mean ± SD of three biological replicates.
The activities of HslHlyB and HslHlyBΔCTE in Tris-HCl buffer (pH 7.5-9.0) showed different trends, with HslHlyB activity increasing and then decreasing, and HslHlyBΔCTE activity decreasing within pH rise. Their optimal pH was 8.5 and 7.5, respectively (Figures 5A, B). HslHlyB displayed >83% of the maximal activity within pH 7.5-9.0, and HslHlyBΔCTE displayed >74% of the maximal activity within pH 7.5-8.5. The activities of HslHlyB and HslHlyBΔCTE did not significantly change from 0-60 min when the proteases were incubated at different pH (Figures 5C, D), indicating their good pH stability.
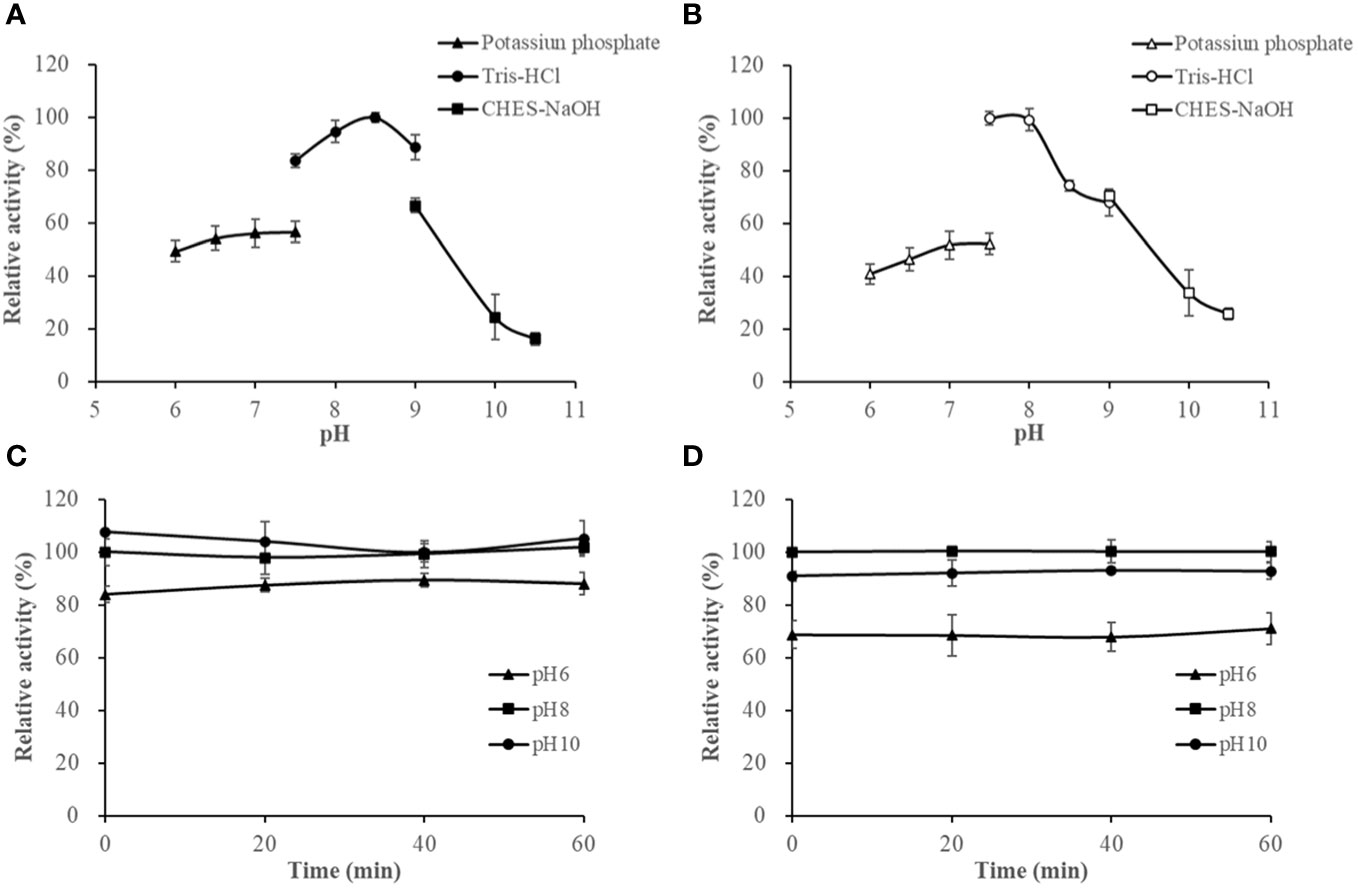
Figure 5 The effect of pH on HslHlyB (A) and HslHlyBΔCTE activity (B), and the pH stability of HslHlyB (C) and HslHlyBΔCTE (D). In (A, B), relative activities were calculated with the maximal activity defined as 100%. In (C, D), the initial enzyme activity at pH 8.0 was set as 100%, upon which the relative activities of enzymes after incubation at different pH for 20, 40 or 60 min were calculated. Mean ± SD of three biological replicates.
The activities of HslHlyB and HslHlyBΔCTE increased with increasing NaCl concentrations at NaCl < 3 M. They maintained more than 93% of the highest catalytic activity in 3-4 M NaCl. Their optimum NaCl concentrations were 3.5 M and 3 M (Figures 6A, B). The salinity stability of HslHlyB and HslHlyBΔCTE was similar (Figures 6C, D). The enzyme activity decreased slightly (<10%) after 60 min incubation at NaCl concentrations of 2-4 M. The activity decreased rapidly after 0-60 min of incubation with a NaCl concentration of 0.8 M. The inactivation rate of HslHlyBΔCTE was faster than that of HslHlyB. HslHlyB and HslHlyBΔCTE were stable at NaCl concentrations >2 M and inactivated at low salt environments. Most halolysin activities increase and then decrease with increasing salt concentration, and the optimal salt concentration for the catalytic reaction is around 2-3 M (Kamekura et al., 1996; Giménez et al., 2000; Shi et al., 2006; Vidyasagar et al., 2006; Elbanna et al., 2015; Dammak et al., 2016; Hou et al., 2021). In this study, the catalytic activities of HslHlyB and HslHlyBΔCTE increased and then remained stable with increasing salt concentration. They reached the highest activity at >3 M salt concentration, thus were more suitable for catalysis in high salt environment.
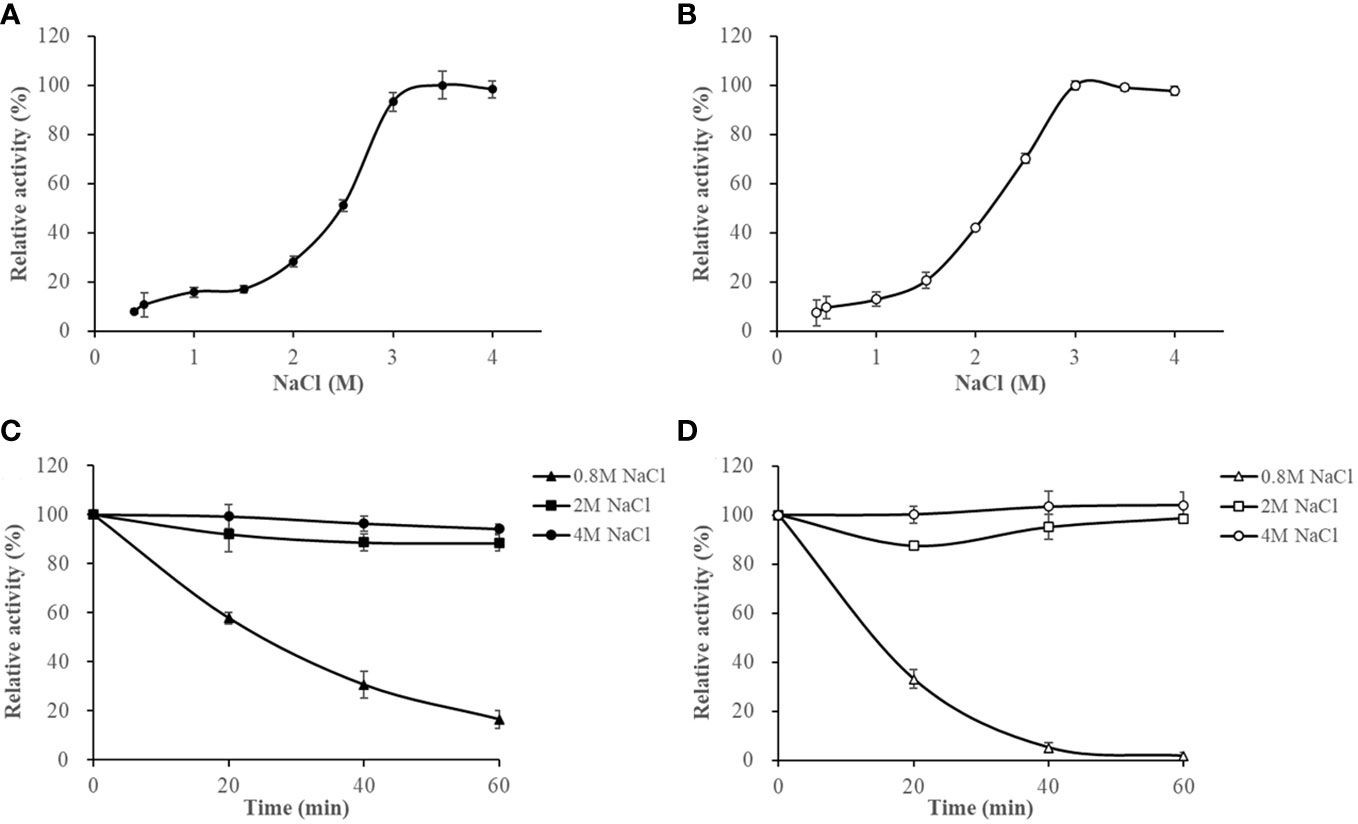
Figure 6 The effect of NaCl on HslHlyB (A) and HslHlyBΔCTE activity (B), and the salinity stability of HslHlyB (C) and HslHlyBΔCTE (D). In (A, B), relative activities were calculated with the maximal activity defined as 100%. In (C, D), the initial enzyme activity was set as 100%, upon which the relative activities of enzymes after incubation at different salinities for 20, 40 or 60 min were calculated. Mean ± SD of three biological replicates.
Metal ions are often used as enzyme cofactors. Different metal ions were reported to have different effects on halolysin activity. For HslHlyB, Fe3+, K+, Mg2+ and Mn2+ had almost no effect on the enzyme activity, Ca2+ and Sr2+ slightly promoted the enzyme activity, and Cu2+ and Zn2+ inhibited 55.1% and 29.6% of the enzyme activity (Figure 7A). For HslHlyBΔCTE, K+, Ca2+, Mg2+, Sr2+ slightly promoted the enzyme activity, Fe3+, Cu2+, Mn2+ and Zn2+ inhibited 25.2%, 44.4%, 56.5% and 22.4% of the enzyme activity (Figure 7A). Cu2+ and Zn2+ significantly inhibited both HslHlyB and HslHlyBΔCTE activities. Fe3+ and Mn2+ had a much higher inhibitory effect on the activity of HslHlyBΔCTE than HslHlyB. The CTE domain of HslHlyB affects the tolerance of halolysin to some metal ions.
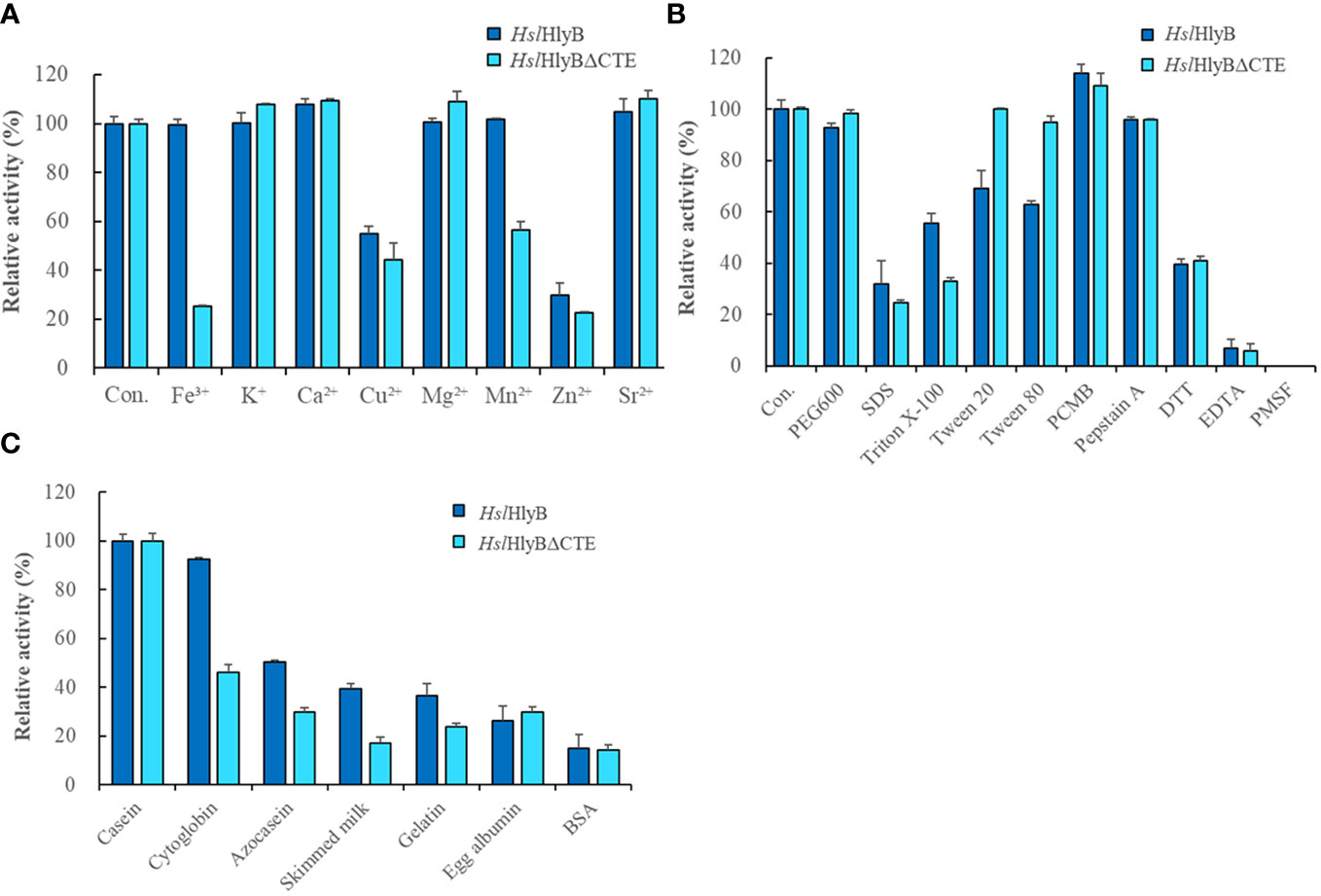
Figure 7 The effect of metal ions (A), detergents, and inhibitors (B) on HslHlyB and HslHlyBΔCTE activity, and proteolytic activity against different substrates of HslHlyB and HslHlyBΔCTE (C). Con.: control. In (A, B), the enzyme assay was carried out with different metal ions at the concentration of 5 mM, with 10% detergents, 1 mM PCMB, 10 mM Pepstatin A, 10 mM DTT, 5 mM EDTA, 4 mM PMSF. In (C), the protein substrate concentration of 5 g/L was used and the enzyme activity was measured by the amount of tyrosine produced. Mean ± SD of three biological replicates.
Different solvents and surfactants decreased the activity of HslHlyB and HslHlyBΔCTE to different degrees (Table 1; Figure 7B). The solvents in which the HslHlyB activity remained above 80% were glycerol, dimethyl sulfoxide (DMSO), and polyethylene glycol (PEG) 600. Isopropyl alcohol and SDS had a significant inhibitory effect on HslHlyB activity (<50%). The solvents in which HslHlyBΔCTE activity maintained more than 80% were methanol, glycerol, acetone, DMSO, dimethylformamide (DMF), Tween 20, Tween 80 and PEG 600. Isopropanol, SDS and TritonX-100 had a significant inhibitory effect on HslHlyBΔCTE activity (<50%). HslHlyBΔCTE had a better tolerance to organic solvents than wild type enzyme.
The effect of protease inhibitors on HslHlyB and HslHlyBΔCTE activity was essentially the same (Figure 7B). 4 mM PMSF completely inhibited the activity of both enzymes, EDTA reduced their activity by 93.0% and 94.3% respectively, Pepstatin A reduced their activity by 4.3%, DTT reduced their activity by 60.3% and 59.1% respectively, and PCMB increased their enzymatic activity by 14.1% and 9.2%. This suggested that HslHlyB and HslHlyBΔCTE may be metal ion-dependent serine proteases and that disulfide bonds are important for their activity.
The data obtained were well described by the Michaelis-Menten equation using azocasein and Suc-AAPF-pNA as reaction substrates (Figure 8). The Km and Vmax of HslHlyB and HslHlyBΔCTE were 0.07 mM, 2814 U/min/mg and 0.16 mM, 4485 U/min/mg against azocasein, 0.36 mM, 8.4×109 μmol/min/mg and 0.27 mM, 2.1×1010 μmol/min/mg against Suc-AAPF-pNA. HslHlyBΔCTE showed higher maximum reaction rates for both substrates, 1.6-fold and 2.5-fold of the wild type, respectively. HslHlyBΔCTE had a higher Km value for azocasein and a lower Km value for Suc-AAPF-pNA than wild type. The above results indicated that HslHlyBΔCTE had better catalytic effect on small molecule substrates, as shown by higher maximum reaction rate and higher substrate affinity. As for azocasein, HslHlyBΔCTE had higher maximum reaction rate but lower substrate affinity than wild-type. Similar phenomenon on the hydrolysis of small peptide substrates by HslHlyB and HslHlyBΔCTE has been reported for Nep from Nab. magadii (De Castro et al., 2008). CTE are involved in protein-protein interactions. PKD domain does not show any proteolysis activity, but can adsorb and swell insoluble proteins (Chen et al., 2007). The PPC domain also plays a key role in binding to the substrate (Xu et al., 2011; He et al., 2012). In the present study, CTE of HslHlyB also affected the affinity of the enzyme against the protein substrate.
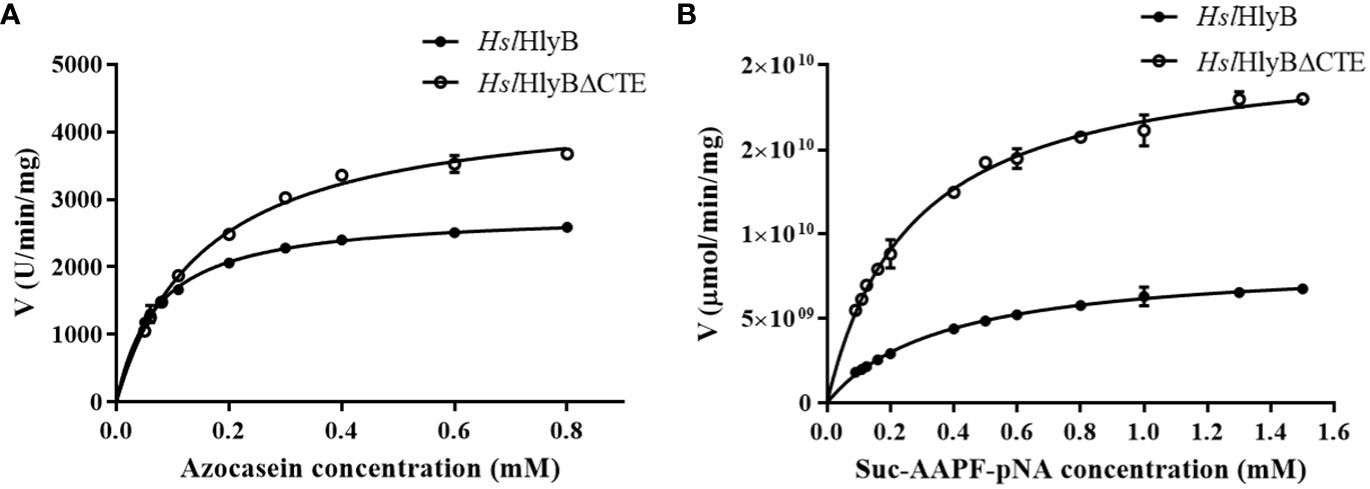
Figure 8 Michaelis-Menten graph of HslHlyB and HslHlyBΔCTE against azocasein (A) and Suc-AAPF-pNA (B). Mean ± SD of three biological replicates.
HslHlyB and HslHlyBΔCTE can hydrolyze casein, bovine hemoglobin, azocasein, skim milk, gelatin, egg albumin and bovine serum albumin, with the highest catalytic activity towards casein (Figure 7C). Their wide substrate spectra make them good candidates for industrial applications.
High-salt fermentation of soybeans is a common production process for soy sauce. The hydrolysis of soybean protein contributes the flavor and raw material utilization of soy sauce. To expand the application of halolysin in high-salt foods, the peptides from hydrolyzed soybean isolate protein by HslHlyB and HslHlyBΔCTE were identified. 128 peptides with an average molecular mass of 1928.87 Da and an average peptide chain length of 16.2 amino acids were identified from soybean protein isolate hydrolysate by HslHlyB (Table S3). 205 peptides with an average molecular mass of 1894.01 Da and an average peptide chain length of 15.6 amino acids were identified from soybean protein isolate hydrolysate by HslHlyBΔCTE (Table S3). The average molecular mass of the peptide profile after hydrolysis by HslHlyBΔCTE was smaller, and the average length of the peptide chains was shorter. Soybean protein isolate hydrolysates by both enzymes showed the highest peptide content in the range of 11-20 amino acids. The amino acid number distribution of peptides generated by HslHlyB was more uniform. The cleavage sites of HslHlyB and HslHlyBΔCTE were counted based on the peptides identified from the soybean protein isolate hydrolysates (Tables S4, 5). Similar to most serine proteases, HslHlyB and HslHlyBΔCTE do not have specific cleavage sites, but they seem to prefer to cleave proteins with acidic amino acids at the P1’ position and proline at the P2 position. Halolysins exert endo-protease activity that hydrolyze protein into peptides. HslHlyB and HslHlyBΔCTE can hydrolyze a wide range of protein substrates, such as soybean protein, which is mainly hydrolyzed to peptides of 11-20 amino acids. The variety of soybean protein hydrolysates were important compounds for soy sauce flavor. There are also several reports on halolysins for accelerating fish sauce fermentation in high-salt or low-salt conditions (Akolkar et al., 2010; Gao et al., 2017; Hou et al., 2020). Halolysins thus revealed potential application in food industry.
In summary, the gene hslhlyB encodes a novel halolysin with PKD and PPC domains at the C-terminus in Hsl. pelagica DL-M4T. HslHlyB and HslHlyBΔCTE, like most halolysins, were highly salt tolerant. They show relatively good tolerance to organic solvents and good thermal stability. The absence of CTE of HslHlyB resulted in the increased activity and thermal stability of the protease while maintaining similar catalytic properties. The CTE affected maturation or secretion of HslHlyB in vivo, while it had no effect on maturation in vitro. Based on the biochemical characterization of halolysin from Hsl. pelagica DL-M4T, HslHlyB and in particular the CTE deletion mutant, are promising salt-tolerant protease candidates for high-salt food processing applications, such as protein hydrolysis in soy sauce.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
DH: conducted experiments, analyzed data and wrote the manuscript, acquired funding. JH: conceived and designed research, wrote and reviewed the manuscript. H-LC: supervised experiments, reviewed and edited the manuscript, acquired funding. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Science and Technology Fundamental Resources Investigation Program of China (No. 2019FY100700), the National Natural Science Foundation of China (No. 32070003, No. 32200012 and No. 31770005), the Natural Science Research of Jiangsu Higher Education Institutions of China (22KJB180016), and the start-up fund from Jiangsu University of Science and Technology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1166287/full#supplementary-material
Akolkar A. V., Deshpande G. M., Raval K. N., Durai D., Nerurkar A. S., Desai A. J. (2008). Organic solvent tolerance of Halobacterium sp. SP1 (1) and its extracellular protease. J. Basic Microb. 48, 421–425. doi: 10.1002/jobm.200800012
Akolkar A. V., Durai D., Desai A. J. (2010). Halobacterium sp. SP1 (1) as a starter culture for accelerating fish sauce fermentation. J. Appl. Microbiol. 109, 44–53. doi: 10.1111/j.1365-2672.2009.04626.x
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Alves V. S., Pimenta D. C., Sattlegger E., Castilho B. A. (2004). Biophysical and functional characterization of Gir2, a highly acidic protein of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 314, 229–234. doi: 10.1016/j.bbrc.2003.12.086
Amoozegar M. A., Siroosi M., Atashgahi S., Smidt H., Ventosa A. (2017). Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 163, 623–645. doi: 10.1099/mic.0.000463
Armenteros J. J. A., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420. doi: 10.1038/s41587-019-0036-z
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Boichem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chaveesuk R., Smith J. P., Simpson B. K. (1994). Production of fish sauce and acceleration of sauce fermentation using proteolytic enzymes. J. Aquat. Food. Prod. T. 2, 59–77. doi: 10.1300/J030v02n03_05
Chen S. X., Sun S. Q., Wang R., Feng H. L., Xiang H. (2021). Haloferax mediterranei halolysin R4 confers antagonistic and defensive capabilities. Appl. Environ. Microb. 87, e02889–e02820. doi: 10.1128/AEM.02889-20
Chen X. L., Xie B. B., Lu J. T., He H. L., Zhang Y. (2007). A novel type of subtilase from the psychrotolerant bacterium Pseudoalteromonas sp. SM9913: catalytic and structural properties of deseasin MCP-01. Microbiology 153, 2116–2125. doi: 10.1099/mic.0.2007/006056-0
Cui H. L., Dyall Smith M. L. (2021). Cultivation of halophilic archaea (class Halobacteria) from thalassohaline and athalassohaline environments. Mar. Life. Sci. Tech. 3, 243–251. doi: 10.1007/s42995-020-00087-3
Cui H. L., Yang X., Mou Y. Z. (2011). Salinarchaeum laminariae gen. nov., sp. nov.: a new member of the family Halobacteriaceae isolated from salted brown alga Laminaria. Extremophiles 15, 625–631. doi: 10.1007/s00792-011-0393-0
Dammak D. F., Smaoui S. M., Ghanmi F., Boujelben I., Maalej S. (2016). Characterization of halo-alkaline and thermostable protease from Halorubrum ezzemoulense strain ETR14 isolated from sfax solar saltern in Tunisia. J. Basic Microb. 56, 337–346. doi: 10.1002/jobm.201500475
De Castro R. E., Ruiz D. M., Giménez M. I., Silveyra M. X., Paggi R. A., Maupin-Furlow J. A. (2008). Gene cloning and heterologous synthesis of a haloalkaliphilic extracellular protease of Natrialba magadii (Nep). Extremophiles 12, 677. doi: 10.1007/s00792-008-0174-6
Du X., Li M. R., Tang W., Zhang Y. X., Zhang L., Wang J., et al. (2015). Secretion of tat-dependent halolysin SptA capable of autocatalytic activation and its relation to haloarchaeal growth. Mol. Microbiol. 96, 548–565. doi: 10.1111/mmi.12955
Edwin A., Rompikuntal P., Bjorn E., Stier G., Wai S. N., Sauer-Eriksson A. E. (2013). Calcium binding by the PKD1 domain regulates interdomain flexibility in Vibrio cholerae metalloprotease PrtV. FEBS Open Bio. 3, 263–270. doi: 10.1016/j.fob.2013.06.003
Elbanna K., Ibrahim I. M., Revol Junelles A. M. (2015). Purification and characterization of halo-alkali-thermophilic protease from Halobacterium sp. strain HP25 isolated from raw salt, lake qarun, fayoum, Egypt. Extremophiles 19, 763–774. doi: 10.1007/s00792-015-0752-3
Elsztein C., Herrera Seitz M. K., Sanchez J. J., De Castro R. E. (2001). Autoproteolytic activation of the haloalkaliphilic archaeon Natronococcus occultus extracellular serine protease. J. Basic Microb. 41, 319–327. doi: 10.1002/1521-4028(200112)41:6<319::AID-JOBM319>3.0.CO;2-8
Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gao R. C., Shi T., Liu X. D., Zhao M. Q., Cui H. L., Yuan L. (2017). Purification and characterisation of a salt-stable protease from the halophilic archaeon Halogranum rubrum. J. Sci. Food Agr. 97, 1412–1419. doi: 10.1002/jsfa.7879
Gao X., Wang J., Yu D. Q., Bian F., Xie B. B., Chen X. L., et al. (2010). Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family. Proc. Natl. Acad. Sci. U.S.A. 107, 17569–17574. doi: 10.1073/pnas.1005681107
Giménez M. I., Studdert C. A., Sánchez J. J., De Castro R. E. (2000). Extracellular protease of Natrialba magadii: purification and biochemical characterization. Extremophiles 4, 181–188. doi: 10.1007/s007920070033
Han D., Cui H. L. (2020). Halostella pelagica sp. nov. and Halostella litorea sp. nov., isolated from salted brown alga Laminaria. Int. J. Syst. Evol. Micr. 70, 1969–1976. doi: 10.1099/ijsem.0.004003
He H. L., Guo J., Chen X. L., Xie B. B., Zhang X. Y., Yu Y., et al. (2012). Structural and functional characterization of mature forms of metalloprotease E495 from arctic sea-ice bacterium Pseudoalteromonas sp. SM495. PloS One 7, e35442. doi: 10.1371/journal.pone.0035442
Hou J., Han D., Zhou Y., Li Y., Cui H. L. (2020). Identification and characterization of the gene encoding an extracellular protease from haloarchaeon Halococcus salifodinae. Microbiol. Res. 236, 126468. doi: 10.1016/j.micres.2020.126468
Hou J., Li S. Y., Zhao Y. J., Cui H. L. (2022). A novel halolysin without c-terminal extension from an extremely halophilic archaeon. Appl. Microbiol. Biot. 106, 3009–3019. doi: 10.1007/s00253-022-11903-4
Hou J., Yin X. M., Li Y., Han D., Lü B., Zhang J. Y., et al. (2021). Biochemical characterization of a low salt-adapted extracellular protease from the extremely halophilic archaeon Halococcus salifodinae. Int. J. Biol. Macromol. 176, 253–259. doi: 10.1016/j.ijbiomac.2021.02.081
Huang J. F., Wu R. B., Liu D., Liao B. Q., Lei M., Wang M., et al. (2019). Mechanistic insight into the binding and swelling functions of prepeptidase c-terminal (PPC) domains from various bacterial proteases. Appl. Environ. Microb. 85, e00611–e00619. doi: 10.1128/AEM.00611-19
Huang J., Wu C., Liu D., Yang X., Wu R., Zhang J., et al. (2017). C-terminal domains of bacterial proteases: structure, function and the biotechnological applications. J. Appl. Microbiol. 122, 12–22. doi: 10.1111/jam.13317
Jaouadi B., Ellouz-Chaabouni S., Rhimi M., Bejar S. (2008). Biochemical and molecular characterization of a detergent-stable serine alkaline protease from Bacillus pumilus CBS with high catalytic efficiency. Biochimie 90, 1291–1305. doi: 10.1016/j.biochi.2008.03.004
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kamekura M., Seno Y., Dyall Smith M. (1996). Halolysin R4, a serine proteinase from the halophilic archaeon Haloferax mediterranei: gene cloning, expression and structural studies. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 1294, 159–167. doi: 10.1016/0167-4838(96)00016-7
Kamekura M., Seno Y., Holmes M. L., Smith M. L. D. (1992). Molecular cloning and sequencing of the gene for a halophilic alkaline serine protease (halolysin) from an unidentified halophilic archaea strain (172P1) and expression of the gene in Haloferax volcanii. J. Bacteriol. 174, 736–742. doi: 10.1128/jb.174.3.736-742.1992
Karan R., Capes M. D., DasSarma P., DasSarma S. (2013). Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnol. 13, 3. doi: 10.1186/1472-6750-13-3
Kim J. S., Kluskens L. D., de Vos W. M., Huber R., van der Oost J. (2004). Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin. J. Mol. Biol. 335, 787–797. doi: 10.1016/j.jmb.2003.11.006
King J., Laemmli U. K. (1971). Polypeptides of the tail fibres of bacteriophage T4. J. Mol. Biol. 62, 465–477. doi: 10.1016/0022-2836(71)90148-3
Kon K., Shimanaga M., Horinouchi M. (2020). “Marine ecology: Intertidal/Littoral zone,” in Japanese Marine life. Eds. Inaba K., Hall-Spencer J. (Singapore: Springer), 241–254. doi: 10.1007/978-981-15-1326-8_20
Lee H. S. (2013). Diversity of halophilic archaea in fermented foods and human intestines and their application. J. Microb. Biotechn. 23, 1645–1653. doi: 10.4014/jmb.1308.08015
Lefebvre O., Moletta R. (2006). Treatment of organic pollution in industrial saline wastewater: a literature review. Water Res. 40, 3671–3682. doi: 10.1016/j.watres.2006.08.027
Liu G. M., Hou J., Cai S. F., Zhao D. H., Cai L., Han J., et al. (2015). A patatin-like protein associated with the polyhydroxyalkanoate (PHA) granules of Haloferax mediterranei acts as an efficient depolymerase in the degradation of native PHA. Appl. Environ. Microb. 81, 3029–3038. doi: 10.1128/AEM.04269-14
Marem A., Okamoto D. N., Oliveira L. C., Ruiz D. M., Paggi R. A., Kondo M. Y., et al. (2018). Functional roles of c-terminal extension (CTE) of salt-dependent peptidase activity of the Natrialba magadii extracellular protease (NEP). Int. J. Biol. Macromol. 113, 1134–1141. doi: 10.1016/j.ijbiomac.2018.03.026
Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Ruiz D. M., Iannuci N. B., Cascone O., De Castro R. E. (2010). Peptide synthesis catalysed by a haloalkaliphilic serine protease from the archaeon Natrialba magadii (Nep). Lett. Appl. Microbiol. 51, 691–696. doi: 10.1111/j.1472-765X.2010.02955.x
Ruiz D. M., Paggi R. A., Giménez M. I., De Castro R. E. (2012). Autocatalytic maturation of the tat-dependent halophilic subtilase nep produced by the archaeon Natrialba magadii. J. Bacteriol. 194, 3700–3707. doi: 10.1128/JB.06792-11
Ryu K., Kim J., Dordick J. S. (1994). Catalytic properties and potential of an extracellular protease from an extreme halophile. Enzyme Microb. Tech. 16, 266–275. doi: 10.1016/0141-0229(94)90165-1
Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. doi: 10.1021/ac950914h
Shi W. L., Tang X. F., Huang Y. P., Gan F., Tang B., Shen P. (2006). An extracellular halophilic protease SptA from a halophilic archaeon Natrinema sp. J7: gene cloning, expression and characterization. Extremophiles 10, 599–606. doi: 10.1007/s00792-006-0003-8
Stroud A., Liddell S., Allers T. (2012). Genetic and biochemical identification of a novel single-stranded DNA-binding complex in Haloferax volcanii. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00224
Studdert C. A., De Castro R. E., Seitz K. H., Sánchez J. J. (1997). Detection and preliminary characterization of extracellular proteolytic activities of the haloalkaliphilic archaeon Natronococcus occultus. Arch. Microbiol. 168, 532–535. doi: 10.1007/s002030050532
Studdert C. A., Herrera Seitz M. K., Plasencia Gil M. I., Sanchez J. J., De Castro R. E. (2001). Purification and biochemical characterization of the haloalkaliphilic archaeon Natronococcus occultus extracellular serine protease. J. Basic Microb. 41, 375–383. doi: 10.1002/1521-4028(200112)41:6<375::AID-JOBM375>3.0.CO;2-0
Sun Y. P., Wang B. B., Wu Z. P., Zheng X. W., Hou J., Cui H. L. (2023). Halorarius litoreus gen. nov., sp. nov., Halorarius halobius sp. nov., Haloglomus halophilum sp. nov., Haloglomus salinum sp. nov., and Natronomonas marina sp. nov., extremely halophilic archaea isolated from tidal flat and marine solar salt. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1105929
Vidyasagar M., Prakash S., Litchfield C., Sreeramulu K. (2006). Purification and characterization of a thermostable, haloalkaliphilic extracellular serine protease from the extreme halophilic archaeon Halogeometricum borinquense strain TSS101. Archaea 2, 51–57. doi: 10.1155/2006/430763
Wang Y. K., Zhao G. Y., Li Y., Chen X. L., Xie B. B., Su H. N., et al. (2010). Mechanistic insight into the function of the c-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913: binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix. J. Biol. Chem. 285, 14285–14291. doi: 10.1074/jbc.M109.087023
Xu Z. S., Du X., Li T.-T., Gan F., Tang B., Tang X. F. (2011). Functional insight into the c-terminal extension of halolysin SptA from haloarchaeon Natrinema sp. J7. PloS One 6, e23562. doi: 10.1371/journal.pone.0023562
Yongsawatdigul J., Rodtong S., Raksakulthai N. (2007). Acceleration of Thai fish sauce fermentation using proteinases and bacterial starter cultures. J. Food Sci. 72, M382–M390. doi: 10.1111/j.1750-3841.2007.00532.x
Keywords: halolysin, C-terminal extension, haloarchaeon, tidal flat, marine solar salt
Citation: Han D, Hou J and Cui H-L (2023) An extracellular protease containing a novel C-terminal extension produced by a marine-originated haloarchaeon. Front. Mar. Sci. 10:1166287. doi: 10.3389/fmars.2023.1166287
Received: 15 February 2023; Accepted: 09 May 2023;
Published: 19 May 2023.
Edited by:
Xue-Wei Xu, Ministry of Natural Resources, ChinaReviewed by:
Ravi Raghavbhai Sonani, University of Virginia, United StatesCopyright © 2023 Han, Hou and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heng-Lin Cui, Y3VpaGVuZ2xpbkB1anMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.