- 1Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
- 2Molecular and Human Genetics Department, Baylor College of Medicine, Houston, TX, United States
The ability for adaptation to track environmental change depends on how efficiently selection can act on heritable genetic variation. Complex life cycles may promote or constrain adaptation depending on the integration or independence of fitness-related traits over development. Reef-building corals exhibit life cycle complexity and are sensitive to increasing temperatures, highlighting the need to understand heritable potential of the thermal stress response and its developmental regulation. We used tag-based RNA-seq to profile holobiont gene expression of inshore and offshore Porites astreoides adults and recruit offspring in response to a 16-day heat stress, and larvae in response to a 4-day heat stress. Host developmental stage affected both broad patterns of host and symbiont expression, and modulated the stress response in both partners, suggesting that symbiotic interactions could vary between host developmental stages and influence the thermal stress response. Populations also exhibited origin-specific treatment responses, but response magnitude differed among life-stages. Inshore parents and recruit offspring exhibited a more robust stress response, exhibiting greater expression profile divergence and differentially expressing more genes compared to offshore-origin corals. This suggests genetic or epigenetic inheritance of regulatory mechanisms giving rise to expression plasticity, although ontogenetic plasticity as a result of the local reef environment during larval development could also explain the origin effect. However, larval populations exhibited the opposite response, with offshore larvae exhibiting a more robust stress response, possibly due to stage-specific effects or exposure duration. Overall, these results show that putatively adaptive regulatory variation persists in thermally naïve life stages, but thermally responsive genes are stage-specific, which could complicate the evolutionary response of corals to climate change.
Introduction
Central to predicting organismal responses to climate change is understanding how fitness landscapes will change over time. If traits are heritable and unconstrained, adaptation may keep pace with environmental change (Bay et al., 2017a). However, organisms with complex life cycles can assume different forms and functions, and even occupy different habitats within their lifetime (Wilbur, 1980). As life stages share a common genome, variable selection over the course of development (Aguirre et al., 2014; Coronado-Zamora et al., 2019) can affect total fitness and, ultimately, adaptation (Albecker et al., 2021). Gene expression is a mechanistic link between genotype and phenotype and can be measured over development to explore the degree of integration or independence between life stages. Though differential expression is common between developmental transitions (Schmid et al., 2005; Reyes-Bermudez et al., 2016; Herrig et al., 2021), it is unclear whether many traits relevant to all life stages, such as the response to stress, also vary over development. As selective pressures intensify in thermally vulnerable species under climate change (Bell and Collins, 2008), understanding how the thermal stress response is modulated over development is key to predicting whether life cycle complexity will promote or constrain adaptation.
Autonomy, or ontogenetic decoupling, is one hypothesis for the evolution of discrete developmental stages, where genes can evolve independently due to stage-specific selective pressures (Moran, 1994). Alternatively, traits can be genetically correlated or expressed across multiple life stages, conferring either a fitness advantage (synergistic pleiotropy) or disadvantage (antagonistic pleiotropy) in subsequent stages (Conner, 2012; Schluter et al., 1991). Synergistic pleiotropy results in consistent selection across all life stages and is thus predicted to result in rapid evolution (Albecker et al., 2021). Genes that are only expressed in discrete life stages, such as in ontogenetic decoupling, are predicted to adapt slower than those with synergistic pleiotropy but faster than those with antagonistic pleiotropy, where expression will differentially affect fitness over developmental time and constrain adaptation (Albecker et al., 2021).
Reef-building corals, the foundation of tropical reef ecosystems, have complex life cycles and are sensitive to rising temperature. Like many marine invertebrates, coral development involves metamorphosis, where free-swimming larvae are restructured into sessile, juvenile polyps (Richmond and Hunter, 1990). Juvenile corals begin to form a calcium carbonate skeleton and invest in growth until they reach the reproductively mature, adult stage. Corals also form a nutritive symbiosis with dinoflagellates in the family Symbiodiniaceae (Muscatine, 1990), which can be acquired directly from maternal colonies during oogenesis (vertical transmission) or from the external environment (horizontal transmission), and mixed mode transmission is also possible (Byler et al., 2013; Quigley et al., 2018). Although this partnership is mutually beneficial under normal conditions, the loss of algal cells or their photosynthetic pigments, known as coral bleaching, can occur when temperatures reach 1°C above local summer maxima for one week (Strong et al., 2011), and potentially lead to starvation and eventual death of the host under prolonged stress. As climate change progresses, increasingly frequent and severe thermal anomalies are resulting in mass bleaching and mortality events on a worldwide scale (Hoegh-Guldberg, 1999; Hughes et al., 2018). Despite the importance of early life stages in the demographic recovery from bleaching (Hughes and Tanner, 2000; Doropoulos et al., 2015), only 2% of all coral heat stress experiments have been performed on larvae and 1% on juveniles, and even fewer studies have included multiple stages (McLachlan et al., 2020). Monitoring of reefs during natural bleaching suggests that adults may differ from earlier life stages in their susceptibility to thermal stress (Álvarez-Noriega et al., 2018), emphasizing the need for comparative, mechanistic studies spanning life stages.
Earlier work examining gene regulation during coral development suggests that life stages express unique genes (Schwarz et al., 2008; Reyes-Bermudez et al., 2016). However, these prior studies focused on identifying differences during major developmental transitions, such as the onset of symbiosis (Schwarz et al., 2008), metamorphosis, and calcification (Reyes-Bermudez et al., 2016), while only one study examined the molecular response to a common stressor, salinity, across multiple life stages (Aguilar et al., 2019a). In a meta-analysis of expression response to stress in Acroporid corals, Dixon et al. (2020) found life stage to be the major factor differentiating transcriptional variation, but most studies were limited to a single life stage, precluding any formal investigation of life-stage differences. Developmental stage has been found to modulate the transcriptional response to stress in other systems, including water and salinity stress in plants (Garg et al., 2016; Liu et al., 2021), viral infection in insects (Schneweis et al., 2017), and pH stress in sea urchins (Devens et al., 2020). It is therefore necessary to explore how the thermal stress response is regulated across developmental stages to inform understanding of coral adaptive capacity.
Contemporary populations that are locally adapted represent promising study systems to understand whether patterns of regulation driving thermal tolerance are heritable and expressed across life stages, thereby increasing total fitness. Coral populations from backreef pools in American Samoa (Bay and Palumbi, 2017b) and nearshore reefs in the Florida Keys (Kenkel et al., 2015a) exhibit local adaptation, with more thermally variable reefs having more tolerant adults (Kenkel et al., 2013a; Palumbi et al., 2014), possibly driven by differences in gene expression plasticity (Barshis et al., 2013; Kenkel and Matz, 2016). Corals from highly variable backreef pools constitutively up or down-regulate stress responsive genes in anticipation of thermal stress, known as front or backloading, and thus have reduced expression plasticity compared to susceptible corals from less variable pools (Barshis et al., 2013). In contrast, locally adapted corals from more variable, inshore reefs in the Florida Keys exhibit greater plasticity of the environmental stress response and reduced bleaching compared to offshore-origin corals (Kenkel et al., 2013a; Kenkel and Matz, 2016). Reciprocal transplants between reefs demonstrate that thermal tolerance and gene expression begin to converge after acclimatization to a common environment, but native corals continue to outperform transplants, indicative of fixed differences between populations (Palumbi et al., 2014; Rose et al., 2015; Kenkel and Matz, 2016). Therefore, despite differences in regulatory responses, persistent population-level variation suggests that expression plasticity is irreversible in adult populations, but it is unclear whether these patterns of regulation are inherited across generations.
Here, we compare the molecular response to heat stress across three life stages of Porites astreoides and their symbionts from inshore and offshore reefs in the Florida Keys. P. astreoides is a brooding coral, meaning gamete fertilization occurs internally and competent planula larvae are released which then settle and metamorphose into juvenile recruits (Richmond and Hunter, 1990). All life stages are symbiotic, with symbionts being vertically inherited from maternal colonies (Thornhill et al., 2006). Although adult populations and their symbionts differ in the magnitude of gene expression plasticity, which could underpin differences in thermal tolerance (Kenkel and Matz, 2016), it is unknown whether larvae and juveniles also exhibit these patterns of regulation. In this study, adult colonies from inshore and offshore populations and their recruit offspring, as well as larvae from separate families, were profiled for gene expression after exposure to controlled thermal stress experiments to ask 1) What are the effects of developmental stage and reef origin on host and symbiont expression? 2) How does host developmental stage affect the response to heat stress? and 3) Are population-level patterns of regulation present in thermally naïve life stages?
Methods
Thermal stress experiments
Porites astreoides adults and recruit offspring were subject to a 16-day common garden heat stress experiment as described in Kenkel et al. (2015a). Briefly, adult colonies were collected from one inshore (Summerland Shoals Patch, 24°36.346 N, 81°25.742 W) and one offshore (Dave’s Ledge, 24°31.887 N, 81°29.013 W) reef site in April 2012 during peak larval release and held at Mote Marine Laboratory’s Tropical Research Lab in Summerland Key, FL under permit #FKNMS-2012-028. Maternal colonies were separated into individual containers with settlement tiles to obtain 50-150 juvenile recruits per tile. As P. astreoides has internal fertilization and maternal colonies can be fertilized by multiple partners, recruits were distinguished based on maternal colony, hereafter referred to as a ‘family’. Parent colonies were then halved using a tile saw after spawning and reared in a common garden environment with their juvenile recruits for 5 weeks. After the acclimation period, paired parent colony halves and recruit tiles were distributed into experimental tanks and exposed to either control (28 +/- 0.4°C) or elevated temperatures (30.9 +/- 1.1°C) for 16 days. Adult and recruit samples from 7 inshore and 5 offshore families per treatment (N=48) were then flash frozen in liquid nitrogen and stored at -80°C.
Larvae from separate families were sampled after exposure to a moderate heat stress for 4 days as described in Zhang et al. (2019). In April 2018 parent colonies were collected from the same inshore and offshore reef sites used in the 2012 experiment and brought to Mote Marine Laboratory’s IC2R3 in Summerland Key, FL under permit #FKNMS-2018-033. Larvae were collected from each parent colony (3 inshore and 4 offshore) and separated by family in floating netwells, with 10 larvae per family per netwell. Temperature was ramped to 32°C over 24 hours in the thermal stress treatment, while the control treatment remained at room temperature (24°C). Pools of ten larvae per family per treatment (N=14) were sampled after 4 days of exposure and frozen at -80°C until processing.
RNA isolation, library preparation, and sequencing
Extraction and library preparation for the adult/recruit samples follow protocols described in Kenkel & Matz (2016). For both sample sets, one microgram of total RNA per sample was used to generate tag-based RNA-seq, or TagSeq, libraries (Meyer et al., 2011; Lohman et al., 2016), with modifications for sequencing on the Illumina platform as described at https://github.com/ckenkel/tag-based_RNAseq
Adult/recruit and larval TagSeq libraries were sequenced independently, on an Illumina HiSeq 2500 in 2013 by the Genomic Sequencing and Analysis Facility at UT Austin and in 2018 by the USC Genome Core, respectively. Adult/recruit libraries were sequenced at an average depth of 6.27 million reads per sample (median = 5.89; range = 0.23-17.4) and larval libraries were sequenced at an average depth of 5.60 million reads per sample (median 5.16; range = 3.06-10.61). Raw sequence data are available at NCBI PRJNA666709.
Data processing
Data cleaning and processing was performed on USC’s Center for Advanced Research Computing following protocols described in https://github.com/ckenkel/tag-based_RNAseq. First, PCR duplicates and reads missing adaptor sequences were removed using custom perl scripts. Adaptor sequences, poly-A tails and sample-specific barcodes were trimmed, and reads were filtered to retain only those exhibiting Q20 over 70% of the read. To generate a host-specific transcriptome, a P. astreoides holobiont assembly (Mansour et al., 2016) (https://www.ncbi.nlm.nih.gov/Traces/wgs/GEHP01) consisting of transcriptomic reads from multiple developmental stages (larvae, recruits, adults) was re-filtered using a hierarchical series of blast searches against potential contaminants following methods described in (Kitchen et al., 2015) (see methods S1 for additional details on assembly quality). SHRiMP (Rumble et al., 2009) was used to competitively map reads to the concatenated host-specific and Symbiodinium spp. (KB8, formerly Clade A) transcriptome (Bayer et al., 2012; Mansour et al., 2016). The number of reads mapping to each isogroup by reference were summed using a custom perl script, separated by isogroup into host and symbiont datasets, and analyzed separately.
Differential gene expression analysis
Outliers were screened using the R package arrayQualityMetrics (Kauffmann et al., 2009) and genes with low abundance transcripts (count <2 in 90% of samples) were removed. One sample outlier was removed from the host dataset (47RH) and four were removed from the symbiont dataset (8AH, 16AH, 31AH, 47RH). 20,185 and 14,124 high-expression genes remained for adult/recruit host and symbiont datasets for 47 and 44 samples respectively. For the host larval expression, 23,806 genes remained. Only ~3,000 genes remained after filtering symbiont expression in the larval dataset so further analyses were not pursued. Variance stabilizing transformation (VST) was applied to count data for principal components analysis (PCA) to visualize the dominant factors driving transcriptional variation.
The package DESeq2 (Love et al., 2014) in R (v3.6.1) was used to statistically evaluate differential expression across life stages, reef origin, and treatment groups (~ stage + origin + treatment) using default settings for the adult and recruit dataset. Due to potential batch effects, the effect of reef origin and treatment on larval differential expression was analyzed independently from the adult and recruit dataset (~ origin + treatment), and the identity of significantly differentially expressed genes by origin and treatment were then compared across datasets. Significance testing was determined using a Wald test after independent filtering using an FDR threshold of 0.1. Multiple test correction was applied to raw p-values following (Benjamini and Hochberg, 1995) and adjusted p-values less than 0.1 were deemed significant. Specific contrasts were used to evaluate the effects of life stage or reef origin on the treatment response. Contrasts were created by using a grouping variable of life stage and treatment (i.e. recruit-control, recruit-heat, adult-control, adult-heat) while controlling for origin effects (~ origin + group). A separate model was created to contrast the treatment response according to reef origin while controlling for life stage (~ stage + group). Scripts and input files for DESeq2 and downstream analyses can be found at https://github.com/mruggeri55/PastGE-lifestage.
Front and backloading analysis
To test whether treatment inducible genes in one population or life stage were not detected in the other due to front/backloading or differences in variance, we generated a correlation plot of the log fold change (LFC) across treatment groups for significant differentially expressed genes following (Barshis et al., 2013). Genes deviating from the one to one line indicate potential candidates for front/backloading whereas those along the one to one line responded similarly in both groups and, therefore, were likely not detected due to differences in variance among groups.
Candidate front/backloaded genes were statistically evaluated as those that were significantly differentially expressed in control conditions (constitutively up/down-regulated) and did not respond significantly to treatment in one group, but were significantly differentially expressed in another group. This analysis was repeated across life stages and reef origin for both the host and symbiont in the adult and recruit dataset only. Front/backloading in larvae could not be determined in relation to adults and recruits due to the potential for batch effects across experiments. Therefore, only front/backloading based on reef origin was explored in larvae.
Discriminant analysis
Discriminant analysis of principal components was performed using the R package adegenet (Jombart, 2008). To explore shifts in expression profiles based on population, a discriminant function was defined by contrasting inshore samples in control and heat conditions including all treatment responsive genes. The number of PCs used to create the function were chosen to capture at least 80% of transcriptional variance. The function was then applied to offshore samples and plotted on the same axis as the inshore response. MCMCglmm (Hadfield, 2010) was used to model the population by treatment interaction to determine whether one population had a significantly greater response to treatment. This analysis was also repeated by contrasting all treatment responsive genes in offshore control and heated conditions and applying the function to inshore samples. In order to determine whether differences in plasticity underlie differences in bleaching phenotypes, a linear model was used to test whether distance along the discriminant function for each family was correlated to changes in previously published bleaching phenotypes, including chlorophyll content of replicate larval pools (Zhang et al., 2019) and bleaching score of the adult individuals sampled in the present study (Kenkel et al., 2015).
Functional enrichment
Gene ontology (GO) annotations were performed by blasting nucleotide sequences to the SwissProt-UniProt database (Boutet et al., 2007) allowing a maximum of 5 alignments and retaining hits with a minimum e-value of 0.0001. Rank-based GO enrichments were performed on signed log p-values for each DESEq2 contrast using the package GO_MWU (Wright et al., 2015) with a FDR threshold of 10%. Protein sequences were further categorized into EuKaryotic Orthologous Groups (KOG) using the online interface of Eggnog-mapper v5.0 (see Methods S1 for details) (Cantalapiedra et al., 2021). The KOGMWU R package (Dixon et al., 2015) was used to calculate delta ranks of KOG classes across life stages based on their treatment response. To test whether KOG delta ranks were correlated across life stages, pairwise Pearson correlations were calculated and visualized using the R package ‘corrplot’.
Results
Life stage drives expression variation in both hosts and symbionts
Life stage was the primary driver of transcriptional variation. Adult and recruit samples separated by stage along the first principal component of variance stabilized counts for both hosts (Figure 1A) and their symbionts (Figure 1B) explaining 31% and 28% of transcriptional variance, respectively. Stage also accounted for the highest proportion of differential expression, with 78.5% (6,944) of all significant host genes (8,851) differentially expressed between adults and recruits (Figure S1A). Adult corals more highly expressed genes involved in proteolysis, protein folding, immune system processes, and ossification compared to recruits (Table S1). In contrast, recruits overexpressed genes involved in signal transduction, ion transport, system processes, and DNA integration (Table S1). Differential expression in the symbiont was also driven by the host developmental stage, with 77% (7,169) of all differentially expressed genes (9,355) regulated based on life stage (Figure S1B). Symbionts in adult corals overexpressed cellular metabolic processes, including photosynthesis, whereas symbionts in recruits overexpressed genes involved in reproduction, cell cycle processes, and DNA metabolic processes (Table S2).
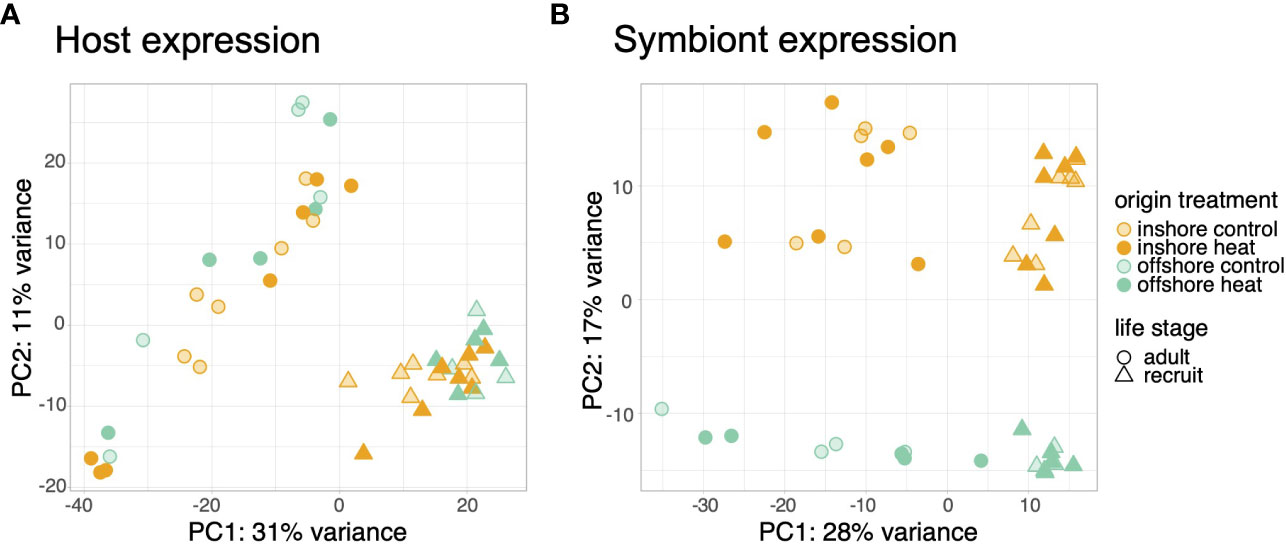
Figure 1 Principal components analysis of variance stabilized count data for the host (A) and symbiont (B) transcriptomes. Samples are colored by population and treatment. Symbols represent different life history stages.
Signatures of reef origin are maintained, even in naive juveniles
Host and symbiont expression was also distinguished by reef origin regardless of life stage or treatment conditions. Host samples separated along the second principal component by origin, which explained ~12.5% of the variance across life stages (Figure S2). Reef origin accounted for the second highest proportion of differentially expressed genes across adult and recruit samples (1,302 genes, 15% of all DEGs; Figure S1A). Larval samples differentially expressed 225 genes based on origin, although treatment had a marginally larger effect (283 genes; Figure S3). Offshore corals exhibited greater expression of genes involved in carbohydrate transport, the TCA cycle, and nervous system development compared to inshore corals and reduced expression of genes involved in cellular component organization, localization, and vesicle-mediated transport (Table S3). Reef origin was also the primary driver of symbiont expression profiles along PC2 (Figure 1; 17% of variance) and accounted for 1,700 (18%) of differentially expressed genes. No GO terms were significantly enriched for origin-associated expression in symbionts.
Moderate duration thermal stress induces moderate transcriptional regulation
Treatment accounted for the smallest number of differentially expressed genes in adults and recruits for both host (605 genes, 7% of all DEGs; Figure S1A) and symbiont (466 genes, 5% of all DEGs; Figure S1B). No discernable pattern was observed due to treatment across the first two principal components in either partner (Figure 1). A significant interaction was detected for 9 genes in which the treatment specific response differed according to host life stage. Five of these were upregulated in adults and downregulated in recruits of which 2 were annotated: mitogen activated protein kinase 6 (MAPK6) and endothelin converting enzyme 2 (ECE2). Whereas three unannotated genes were downregulated in adults and upregulated in recruits in response to thermal stress. One unannotated gene was upregulated in both life stages, but was more strongly regulated in adults compared to recruits (LFC 6.3 vs 1.4). No differential expression was detected for the host origin by treatment interaction, nor for any symbiont interaction terms. Although the majority of differentially expressed genes were regulated by treatment in larvae (283 genes, 56.5% of all DEGs), larval samples did not align along the first or second principal component by treatment (Figure S2). No genes were significantly differentially expressed due to the interaction of treatment based on larval origin.
Treatment response is modulated by life stage
Although few genes were detected for the treatment by life stage interaction, within group analysis revealed that the most treatment responsive genes were largely stage-specific. Recruits mounted the largest detectable expression response (311 DEGs) followed by larvae (283 DEGs) and then adults (194 DEGs). Only 9 genes consistently responded to treatment across all life stages (Figure 2C), including upregulation of the universal stress protein Sll1388, calumenin A, soma ferritin, and malate dehydrogenase. Conversely, expression of cyp1A1 varied, with adults and recruits upregulating and larvae downregulating cyp1A1 under heat stress. The treatment response in symbionts was also modulated by host life stage (Figure 3A). Recruit symbionts had a greater expression response to treatment compared to symbionts in adults (155 DEGs versus 92 DEGs respectively) with only 16 DEGs overlapping between the two life stages (Figure 3B). These 16 core genes included annotations for fatty acid metabolism, RNA processing, metal ion binding, and oxidoreductase activity.
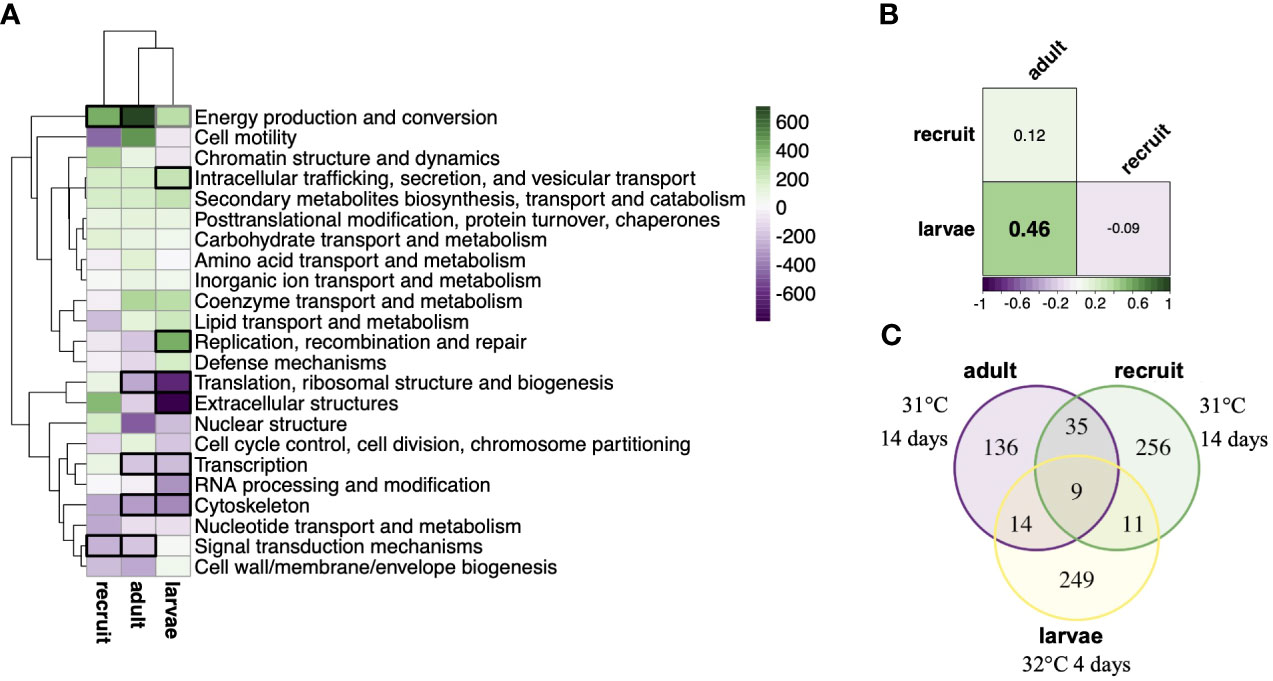
Figure 2 Host transcriptional response to moderate-term thermal stress by life stage. (A) Clustering of KOG enrichments by delta ranks and life stage. Bolded boxes represent KOG classes significantly enriched in response to treatment (black padj < 0.05; gray padj < 0.1). (B) Pairwise Pearson correlation coefficients of KOG delta ranks between life stages. Bolding denotes significant correlation (p < 0.05). (C) Venn diagram of genes significantly differentially expressed in response to treatment (padj < 0.1) in each life stage.
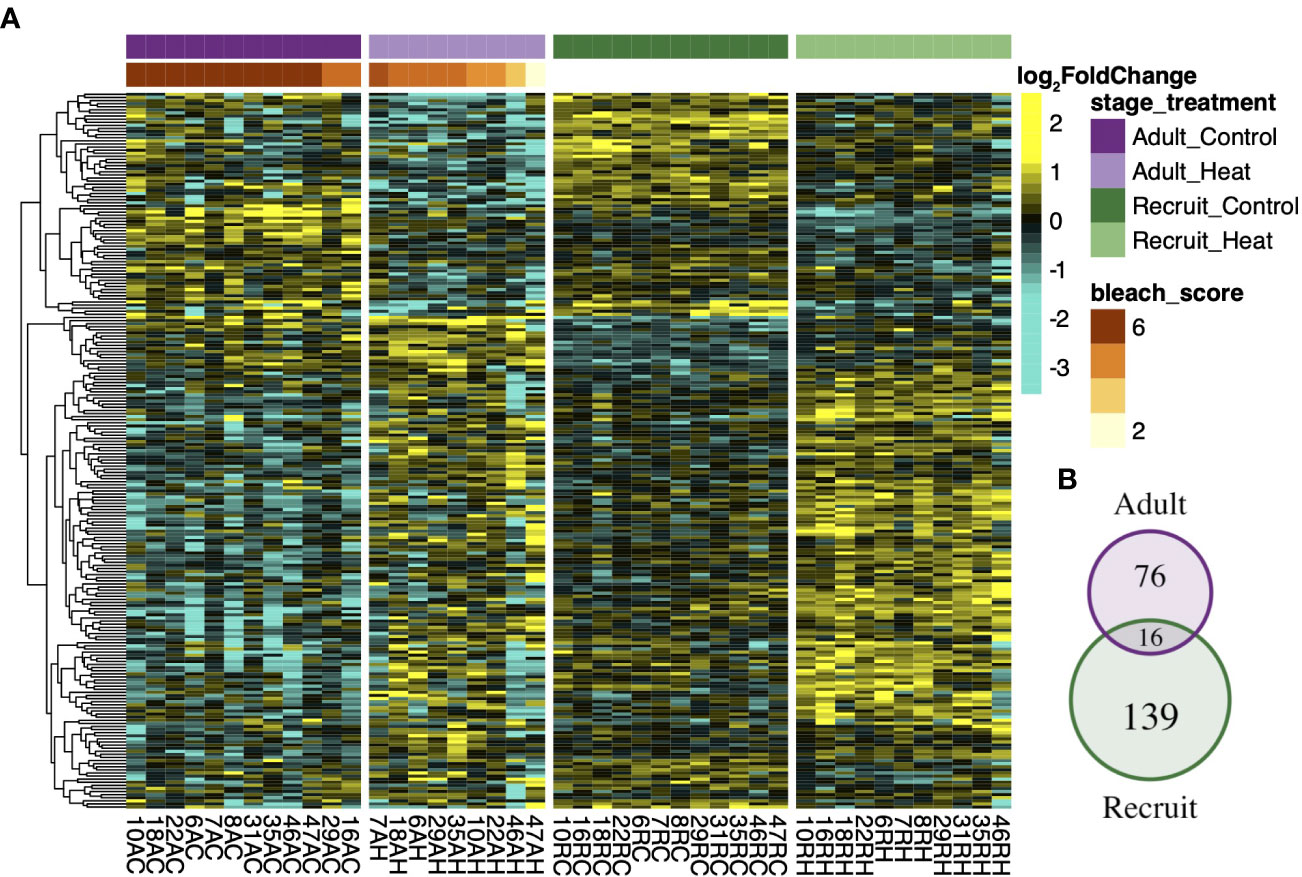
Figure 3 Symbiont transcriptional response to treatment across host developmental stages. Rows of the heatmap (A) represent the log2 fold change of the 231 treatment responsive genes (padj < 0.1) detected in total for symbionts in adult and recruit life stages, as indicated by the venn diagram (B). Each column represents a unique sample where the family number is followed by life stage (R-recruit, A-adult) the treatment condition (C-control, H-heat). Adult corals were ranked by visual bleaching score from 1 (lightest) – 6 (darkest) (Kenkel et al., 2015) which is denoted by the red/orange bar.
The unique differential expression response to treatment between adults and recruits does not appear to be due to differences in variance (detection limits) but rather to differences in transcriptional baselines. Forty-four genes were constitutively expressed in adults, but responsive in recruits (Table S4). Whereas 27 treatment responsive genes unique to the adult life stage were overexpressed in control conditions by the recruit life stage (Table S4). Adults and recruits also exhibited backloading of 54 and 29 genes, respectively. Symbionts exhibited a similar pattern of differing baselines leading to divergent thermal stress responses. Symbionts in adults frontloaded 15 genes and backloaded 28 genes that were responsive in recruit symbionts, while recruit symbionts frontloaded 22 genes and backloaded 17 genes that were responsive in adult symbionts (Table S5).
Functional enrichment analysis also indicates that life stages utilize different biological processes to respond to heat stress. Ontology enrichments in adults included genes involved in upregulation of metabolic processes and downregulation of cellular amide metabolic processes under heat stress (Figure S4A; Table S6), while recruits downregulated genes involved in cell communication, membrane lipid biosynthetic processes, and cellular protein metabolic processes (Figure S4B; Table S7). Both adults and recruits upregulated organic acid catabolic processes and downregulated heterochromatin assembly by small RNA (Figure S4AB). Like adults, larvae also downregulated cellular amide metabolic processes. However, unlike either adults or recruits, larvae upregulated carbohydrate and DNA metabolic processes and downregulated cellular developmental processes in response to heat stress (Figure S4C; Table S8). Among all life stages, energy production and conversion was the only KOG term significantly enriched among upregulated genes (Figure 2A). Although only 23 treatment responsive genes and 2 ontology term enrichments were shared among adults and larvae, a significant correlation in KOG class enrichment in response to treatment was evident (r = 0.46, p<0.05, Figure 2B). However, KOG class enrichment was not significantly correlated in adults and their recruit offspring, in spite of higher relatedness and exposure to identical treatment conditions (r = 0.12; Figure 2B).
Despite the lack of shared regulation at the level of individual genes, some biological processes upregulated by symbionts in response to heat stress were shared across life stages. Both adult and recruit symbionts upregulated genes involved in RNA metabolism and cell cycle processes (Figure S5). However, genes involved in the cellular stress response, photosynthesis, and nitrogen metabolism differentiated thermally responsive symbiont genes across life stages. Recruit symbionts additionally upregulated genes involved in the cellular response to stress and downregulated nitrogen transport, utilization, and metabolism (Table S10), while adult symbionts downregulated photosynthetic processes (Figure S5; Table S9).
Treatment response is also modulated by reef origin, but patterns vary among life-stages
The magnitude of the expression response to heat stress also differed by population. Inshore adults and recruits had a much larger transcriptional response, differentially expressing 316 genes across treatments, whereas only 43 genes were detected as differentially expressed by offshore hosts (Figure 4A). Only 25 treatment responsive genes were differentially expressed in both populations. Universal stress protein 1388, Calumenin-A, and Malate Dehydrogenase were responsive to treatment in both populations, as well as in all life stages, suggesting these may be core genes involved in the cellular stress response. Symbionts also exhibited population-level variation in the magnitude of their treatment response, with inshore symbionts differentially expressing 142 genes compared to 57 genes in offshore symbionts, and only 14 genes showing consistent responses to heat stress between the two populations (Figure S6). Of the annotated genes involved in the core symbiont thermal stress response among life stages, four were also identified as core genes among life stages: At2g29290, ppsD, PP1, and NUDT3. Regulatory protein flaEY, which plays a role in flagellum biogenesis (Purucker et al., 1982), was also upregulated in response to heat stress in both symbiont populations.
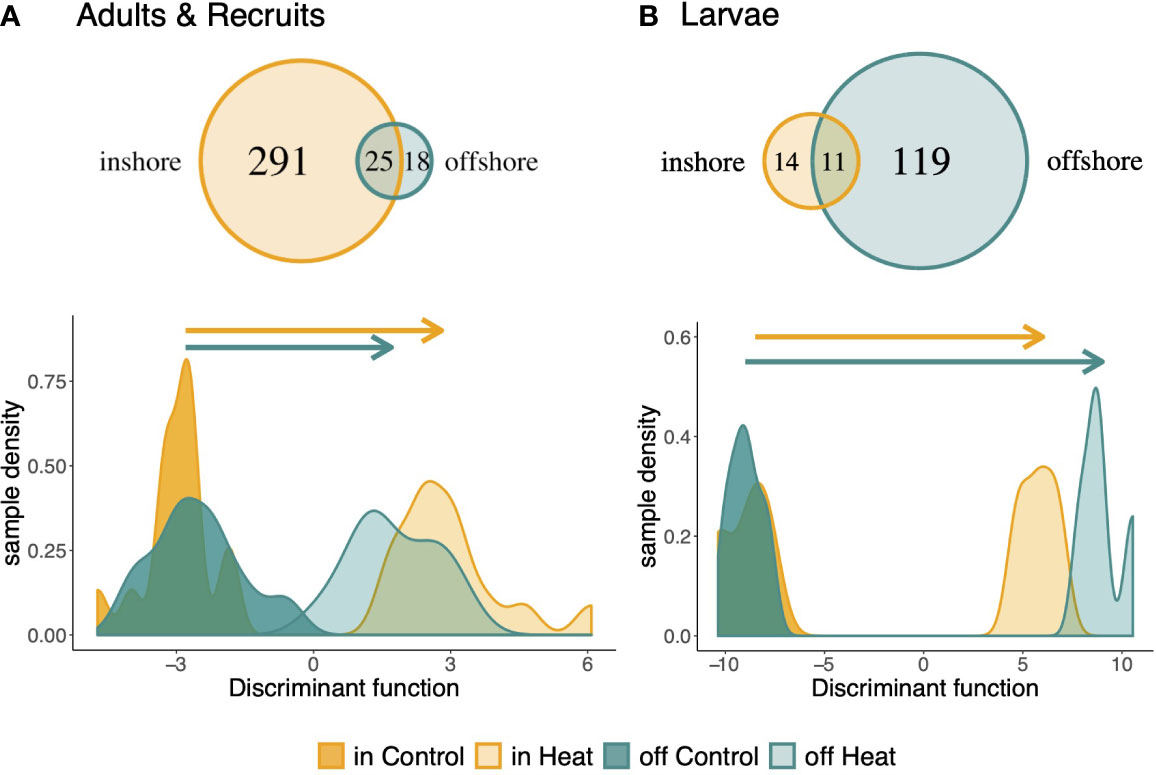
Figure 4 Host transcriptional response to treatment among populations. Venn diagram of the population-specific response to heat stress in adults and recruits (A) and larvae (B). Discriminant analysis of principal components was performed on all treatment responsive genes to explore the magnitude of expression change between populations. The discriminant function was defined by contrasting samples in heat and control from one population and predicting the response of the other.
Unlike life-stage patterns, the low number of treatment responsive genes in offshore adults and recruits does not seem to be due to front or backloading, but rather reduced expression plasticity compared with inshore origin coral. Only 1 inshore responsive gene was frontloaded (LFC > 20 in control; Table S11) in offshore hosts, while 14 were backloaded (LFC -0.3 to -4.7 in control; Table S11). Offshore symbionts may be transcriptionally loading some inshore responsive genes (Table S12), with 11 genes frontloaded (LFC 0.6 to 3.3) and one backloaded (LFC -0.8). However, for both host and symbiont, the total number of front and back-loaded genes in offshore samples is not enough to make up for the greater degree of differential expression in inshore samples.
Discriminant analysis of principal components for all treatment responsive genes supports a significantly less plastic expression response in offshore adults and recruits (PMCMC < 0.01; Figure 4A). The robust inshore host response included upregulation of genes involved in catabolic and cellular metabolism and downregulation of genes involved in actin-filament based processes and heterochromatin assembly. DNA metabolic processes were also upregulated in inshore corals, whereas miRNA and protein metabolic processes were downregulated under heat stress (Table S13). Specifically, protein autoubiquitination was enriched among downregulated genes. Conversely, offshore hosts upregulated two stress response genes (ERP29 and slr1101) and one apoptosis gene (Mzb1). Although offshore-origin adults bleach significantly more than inshore-origin adults (Kenkel et al., 2015b), the distance between adult samples along the discriminant function axis was not correlated with changes in bleaching score (Figure S7A).
Despite a consistent pattern of greater transcriptional response in inshore hosts and symbionts for adult and recruit life stages, larval hosts exhibited the opposite response. Offshore larvae differentially expressed 130 genes in response to treatment, whereas inshore larvae only differentially expressed 25 genes, with 11 genes shared among populations (Figure 4B). Offshore larvae also exhibited greater change in their transcriptional profiles for treatment responsive genes (DAPC, PMCMC<0.01; Figure 4B). Similar to adults, the distance between larval samples along the discriminant function was not correlated with changes in chlorophyll across treatments (Figure S7B). No genes were detected as front or backloaded by inshore larvae.
Discussion
Population origin and developmental stage have major impacts on gene expression and modulate the response of coral hosts and their symbionts to heat stress. A largely stage-specific thermal stress response implies inconsistent selection among life stages that could delay or constrain adaptation. Despite life stage differences, a fixed effect of population regardless of thermal history indicates genetic or irreversible epigenetic variation differentiates population-level expression. Further, the magnitude of the treatment response varied based on population and was consistent across adults and their recruit offspring, suggesting patterns of regulation are heritable. However, differences in response magnitude may be influenced by the duration of heat stress or developmental stage necessitating further exploration. Considerable variation in baseline gene expression and the response to stress was also observed for symbionts among life stages and populations, despite vertical transmission to offspring and similar ITS2 haplotypes between populations (Kenkel et al., 2013a), implying that there is likely more physiological and/or genetic diversity in symbiont communities than has been previously described.
Developmental stage drives differential expression in coral hosts and modulates the thermal stress response
Life stage was the prominent driver of host transcriptional variation (Figure 1A) and modulated the response to thermal stress (Figure 2C). Surprisingly, the thermal stress response was largely stage-specific, with few genes consistently responding to heat stress across adults, recruits, and larvae (Figure 2C). Conversely, the only other study examining transcriptomic responses across life stages to identical treatment conditions reported similar responses to salinity stress between adult and juvenile corals (Aguilar et al., 2019). However, thousands of genes were exclusive to individual life stages with only ~20% DEGs consistently responding to salinity stress (Aguilar et al., 2019), reinforcing a largely stage-specific stress response. Although sparse, there is also evidence for development modulating the stress response in plants (Garg et al., 2016; Liu et al., 2021) and other invertebrates (Schneweis et al., 2017; Devens et al., 2020), suggesting complex life cycles can decouple the molecular response to stress. The lack of consistency in the stress response across life stages has major implications for the evolutionary trajectory of coral. If inducible genes in one life stage affect fitness, but are not expressed in other stages or are co-opted for other functions, it may delay or constrain adaptation (Bay and Palumbi, 2017b).
Stage-specific responses to thermal stress are partly due to differing baselines. Life history stage was the prominent driver of differential expression in the host (Figure S1), suggesting that genome-wide expression varies more over an individual’s lifetime than between locally adapted populations or thermal environments. Developmental modulation of baseline expression could potentially reduce plasticity during environmental perturbations. About one-third of treatment responsive genes in parents or recruits were front/back-loaded by the other stage. Front/backloading of stress responsive genes has been proposed as an adaptive mechanism that increases coral thermal tolerance (Barshis et al., 2013). However, if front/back-loading had a heritable basis, offspring would be expected to exhibit constitutive expression similar to adults. Although constitutive expression could induce a priming effect against heat stress (Barshis et al., 2013), the robust signal of developmental stage, regardless of genetic or environmental factors, suggests these genes are pleiotropic, performing different functions over developmental time.
Pleiotropic genes could have a positive effect on fitness in every developmental stage, and thus increase total fitness, or they could experience divergent selection pressures over time, limiting adaptive potential (Cheverud et al., 1983). Pleiotropy is generally believed to constrain adaptation, however, a meta-analysis on quantitative genetic studies found genetic covariances were just as likely to promote, constrain, or have no effect on the rate of adaptation, though covariances among life stages were not considered (Agrawal and Stinchcombe, 2009). In model organisms, there is evidence suggesting that pleiotropy can constrain evolution through a change in the direction of selection across life stages (Postma and Ågren, 2016; Coronado-Zamora et al., 2019). Although functional genomic evidence for pleiotropy is currently lacking for corals, bleaching resistance in adults can come at the cost of other fitness-related traits (Shore-Maggio et al., 2018; Cornwell et al., 2021), suggesting pleiotropy can constrain thermal adaptation within life stages. Putatively pleiotropic genes identified in the present study include tumor necrosis factor receptor superfamily 19 and a peroxidasin homolog, which have been previously implicated as key players in both the immune (Traylor-Knowles et al., 2021) and environmental stress response (Barshis et al., 2013; Louis et al., 2017) in coral, yet their role in developmental modulation is unknown. The present study provides the first evidence for developmental pleiotropy of thermally responsive genes in corals by controlling for treatment intensity and duration, symbiotic state, and relatedness, emphasizing the need for functional and quantitative genetics to be conducted across multiple life stages to determine the costs and limits of thermal adaptation.
In contrast to one gene having multiple functions, different genes could also perform the same function, but be uniquely expressed during discrete developmental stages, resulting in a stage-specific thermal response. Transcriptome sequencing of five life history stages in two coral species, Acropora palmata and Montastrea cavernosa, found that expressed sequence tags (ESTs) were stage-specific (Schwarz et al., 2008), suggesting that life stages utilize completely different gene sets to achieve the same functions. Similarly, in P. astreoides, the inclusion of sequence data from three life-stages, larvae, newly settled recruits, and adults, increased the size of the transcriptome by 30 Mbp compared to the previously published adult-only reference (Mansour et al., 2016). As developmental stage was the main factor differentiating genome-wide expression in this study (Figure 1A), unique genes may be expressed during each life stage resulting in different treatment responsive genes. Therefore, in addition to differing baselines, ontogenetic decoupling could also explain the lack of consistency in the thermal stress response among developmental stages.
Life stages could also be decoupled physiologically, rather than at the molecular-level (reviewed in (Rivera et al., 2021)), leading to potential differences in thermal susceptibility, and in the identity of treatment responsive genes. However, despite different genes, the response to heat stress in adults and larvae was functionally similar (Figure 2B), consistent with previous research in acroporid species (Dixon et al., 2015), suggesting similar physiological states and a common response to thermal stress regardless of treatment intensity or duration. Conversely, adult and recruit responses were not correlated despite identical treatment conditions and high genetic relatedness, suggesting the recruit stage is fundamentally different from adults and larvae, and may experience stress differently. Physiologically, adults may be investing more energy in sexual reproduction, but differentially expressed genes between these two stages were not enriched for reproductive processes (Table S1), indicating that there are likely other physiological and functional differences. Future work should focus on bleaching phenotypes that can be compared among all stages to establish the resilience/susceptibility of different developmental stages to controlled thermal stress.
Ontogenetic decoupling could delay adaptation because expression of specific genes will only have fitness effects during discrete developmental periods (Albecker et al., 2021). However, decoupling could also break genetic correlations between discrete life stages, reducing antagonistic pleiotropy, and therefore optimizing stage-specific fitness (Moran, 1994). As metamorphosis is often accompanied by changes in habitat, ontogenetic decoupling is predicted to be strongest across metamorphic transitions (Moran, 1994). Interestingly, in addition to differences in the thermal stress response between larvae and post-metamorphic life stages, we also see clear differences in the identity of treatment responsive genes between recruits and adults (Figure 2C), which lack any major morphological or ecological transitions. This suggests that adults and recruits, in addition to larvae, vary in their fitness optima and can evolve somewhat independently. In Drosophila, artificial selection on larval or adult tolerance to extreme temperatures did not confer tolerance in the other life stage (Loeschcke and Krebs, 1996) and the genetic architecture underlying tolerance in each stage was found to be independent (Loeschcke and Krebs, 1996; Freda et al., 2017). A largely decoupled thermal stress response among all three life stages in this study suggests that selection for tolerant coral genotypes in one life stage will likely not translate into greater tolerance in subsequent stages, as the genes under selection will not be expressed. Intervention strategies, such as artificial selection and assisted gene-flow, have been proposed to save vulnerable coral populations from bleaching events (van Oppen et al., 2015), but our results suggest further research is needed to determine the effect of genetic variation on total organismal fitness, including all life stages, before these can be successfully implemented.
Population-level signatures persist in thermally naive life stages
Origin effects on constitutive expression persisted in all life stages despite rearing adults in a common garden for five weeks and larvae and recruits having no exposure to their home environment independent from the maternal colony. Although adult P. astreoides from inshore and offshore reefs in the Florida Keys are known to exhibit population-level divergence (Kenkel et al., 2013b; Kenkel and Matz, 2016), the underlying mechanisms driving expression variation remain unclear. Here, clear differentiation between inshore and offshore expression profiles in juvenile life stages reared in a common environment suggest genetic or irreversible epigenetic variation underlies expression divergence. Population-level differences were also present in the thermal stress response, where inshore adults and recruit offspring exhibited a larger shift in expression of treatment responsive genes and greater differential expression across treatments than offshore-origin adults and recruits (Figure 4A). Inshore adults of P. astreoides also exhibit greater genome-wide expression plasticity in response to transplantation (Kenkel and Matz, 2016) and are generally more thermally tolerant compared to offshore adults (Kenkel et al., 2013a; Kenkel et al., 2015a). Here we show that this expression pattern is consistent in juvenile offspring, with no prior thermal stress experience, indicating that plasticity is potentially heritable.
Though environmental history of juvenile life stages was controlled, origin specific differences in expression could also be due to transgenerational effects induced by the parental environment, including maternal effects, developmental plasticity, and epigenetic inheritance. Some studies have documented an effect of parental environment on offspring physiology in coral (Putnam and Gates, 2015; Bellworthy et al., 2019; Wong et al., 2021), suggesting that underlying expression patterns likely play a role. Further, there is evidence for environmentally induced epigenetic inheritance in coral (Liew et al., 2020), which could drive expression variation even in thermally naïve offspring. It is also important to note that because P. astreoides employ a brooding reproductive strategy, larvae and juveniles were exposed to their home environment during embryonic development. Temperature profiles are similar between inshore and offshore reefs in the fall and spring when embryonic development occurs, yet many other environmental parameters diverge (Kenkel et al., 2015b), which could have induced a plastic response. Though we cannot rule out plastic effects, brooded offspring are commonly retained within reefs (Ayre and Hughes, 2000; Underwood et al., 2007), so persistent population-level differences could facilitate survival of early life stages in their local environment.
Despite consistent population-specific differences in parents and offspring, expression plasticity may not be the primary mechanism driving differences in thermal tolerance. Similar to adults, inshore larvae are more resistant to bleaching than offshore larvae (Zhang et al., 2019). However, unlike population-specific plasticity observed in adults and recruits, offshore larvae demonstrated larger shifts in expression profiles for all treatment responsive genes and differentially expressed more genes across treatments compared to inshore-origin larvae (Figure 4B). Reduced expression plasticity due to constitutive expression of thermally responsive genes, or front/back-loading, has been documented in tolerant coral adults in American-Samoa (Barshis et al., 2013) and the Red Sea (Voolstra et al., 2021). However, we did not detect any front/back-loaded genes in inshore larvae, suggesting this is not the main mechanism driving population variation. Additionally, expression plasticity was not correlated with plasticity in adult bleaching score (Figure S7A) or larval chlorophyll content (Figure S7B), implying that population-level differences in plasticity do not predict the magnitude of bleaching.
Alternatively, the timing of the cellular stress response may vary across populations and underlie differences in thermal tolerance rather than its plasticity (Rivera et al., 2021). Population-specific plasticity in larvae exposed to only 4 days of heat stress was opposite that of adults and recruits exposed to 16 days of heat stress. As offshore-origin larvae had a more robust response to acute stress (Figure 4B), but inshore-origin adults and recruits had a more robust response to moderate heat stress (Figure 4A), the onset of the stress response might occur earlier in offshore corals, artificially leading to differences in population-specific plasticity across life stages. It is also possible that after 16-days of moderate thermal stress, population-specific responses observed in adults and recruits are due to differences in cellular fate, rather than the timing of the cellular stress response. Treatment responsive genes in inshore adults and recruits were mainly involved in metabolism, actin-filament based processes, and heterochromatin organization (Table S13), whereas the offshore-specific response included genes involved in the cellular stress response and apoptosis. This pattern could indicate that offshore adults and recruits may have reached their thermal limit and activated cellular death pathways, while inshore corals are utilizing other pathways to acclimate to warmer conditions. Multiple sampling timepoints during controlled thermal stress are needed to resolve whether differences in response timing or plasticity underlie differences in cell-fate and affect the bleaching response.
Core treatment responsive genes
Despite a largely stage-specific response to treatment, 5 annotated genes consistently responded to heat stress in coral hosts: soma ferritin, cytochrome p450 1a1, calumenin A, universal stress protein, and malate dehydrogenase. Soma ferritin and cytochrome p450 are both involved in oxidative stress and are commonly detected among corals undergoing thermal stress (Edge et al., 2005; Voolstra et al., 2009; Császár et al., 2010), owing to ROS production following damage to the symbiont photosystem. However, cytochrome p450 was upregulated in larvae, and downregulated in adults and recruits. This could be explained due to differences in the treatment duration, as previous research saw upregulation after an acute heat stress (Voolstra et al., 2009), which was more similar to the duration of larval heat exposure in this experiment. Upregulation of calumenin has also previously been implicated in driving greater thermal tolerance in adult corals after thermal preconditioning, while downregulation of calumenin was observed in non-preconditioned, bleaching susceptible corals (Bellantuono et al., 2012). Calumenin is a calcium-binding protein with diverse roles including host-symbiont signaling, calcium homeostasis, and apoptosis (Orrenius et al., 2003; Honoré, 2009; Ganot et al., 2011). In this study, calumenin was upregulated in both inshore and offshore populations (LFC 2.4-2.5) regardless of bleaching susceptibility, as well as all in all life stages (LFC 2.5-3.5), contradicting its role in detecting bleaching variation. Malate dehydrogenase, a key metabolic enzyme, and universal stress protein 1388 were also upregulated, showing a sustained increase in metabolism and stress related expression under high temperatures. Taken together, these genes support a conserved response to thermal stress regardless of life stage, population of origin, or duration of treatment and could represent potential candidates for biomarker development.
Symbiont expression is modulated by host and holobiont condition
Symbiont expression reflected that exhibited by the host, with life stage and population both differentiating expression and modulating the response to heat stress. Stage was the prominent driver of transcriptional variation (Figure 1B), providing the first evidence that symbiont expression is affected by the host’s developmental stage. During development, carbon translocation trades off with symbiont density (Kopp et al., 2016), supported by overexpression of genes involved in somatic reproduction and underexpression of photosynthetic genes in juvenile life stages observed here. This suggests that the metabolic balance between host and symbiont varies over development, even when symbionts are vertically transmitted, and could have implications for how symbiosis is maintained during different developmental stages. Underexpression of photosynthetic genes in juveniles could also reduce oxidative damage and increase bleaching resilience (Lesser, 1996). In support of this, expression of photosynthetic genes in adults responding to heat stress was negatively correlated to bleaching (Figure S8), indicating that juvenile holobionts may be more thermally resilient than adults, but comparable bleaching phenotypes must be measured across life stages to test this hypothesis.
Consistent with host expression, inshore symbionts have a more robust response to heat stress, even in environmentally naive recruits, further supporting heritability of holobiont tolerance. Symbiont community composition is presumed to be similar across life stages and populations, as symbionts are directly inherited by recruits through the process of vertical transmission and the same symbiont haplotypes (ITS2 type A4/A4a) were previously found in adult P. astreoides at these reef sites (Kenkel et al., 2013a). Consequently, high transcriptional variation among similar symbiont communities could be caused by differences in the host environment. However, vertical transmission could also cause local retention of symbiont strains within each population leading to genetic differentiation and expression divergence (Barshis et al., 2014; Parkinson et al., 2016). Though symbiont expression could differ due to long-term acclimatization, symbionts in environmentally naive recruits also exhibit population-level differentiation, indicating genetics of the symbiont and/or host are more likely driving this variation. Restricted gene flow in host populations (Kenkel et al., 2013a) may be driving differences in symbiont expression across reefs, but fine-scale population genetics is needed to test whether genetic differentiation in the symbiont is a possible mechanism for population-level expression divergence observed here.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
CK conceived and designed experiments. CK and YZ performed physiological assays. CK and GA prepared the sequencing libraries. MR performed all analyses and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
CK was supported in part by fellowship funds from the Alfred P. Sloan Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1163552/full#supplementary-material
References
Agrawal A. F., Stinchcombe J. R. (2009). How much do genetic covariances alter the rate of adaptation? Proc. Biol. Sci. 276, 1183–1191. doi: 10.1098/rspb.2008.1671
Aguilar C., Raina J.-B., Fôret S., Hayward D. C., Lapeyre B., Bourne D. G., et al. (2019). Transcriptomic analysis reveals protein homeostasis breakdown in the coral acropora millepora during hypo-saline stress. BMC Genomics 20, 148. doi: 10.1186/s12864-019-5527-2
Aguirre J. D., Blows M. W., Marshall D. J. (2014). The genetic covariance between life cycle stages separated by metamorphosis. Proc. Biol. Sci. 281, 20141091. doi: 10.1098/rspb.2014.1091
Albecker M. A., Wilkins L. G. E., Krueger-Hadfield S. A., Bashevkin S. M., Hahn M. W., Hare M. P., et al. (2021). Does a complex life cycle affect adaptation to environmental change? genome-informed insights for characterizing selection across complex life cycle. Proc. Biol. Sci. 288, 20212122. doi: 10.1098/rspb.2021.2122
Álvarez-Noriega M., Baird A. H., Bridge T. C. L., Dornelas M., Fontoura L., Pizarro O., et al. (2018). Contrasting patterns of changes in abundance following a bleaching event between juvenile and adult scleractinian corals. Coral Reefs 37, 527–532. doi: 10.1007/s00338-018-1677-y
Ayre D. J., Hughes T. P. (2000). Genotypic diversity and gene flow in brooding and spawning corals along the great barrier reef, Australia. Evolution 54, 1590–1605. doi: 10.1111/j.0014-3820.2000.tb00704.x
Barshis D. J., Ladner J. T., Oliver T. A., Palumbi S. R. (2014). Lineage-specific transcriptional profiles of symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 31, 1343–1352. doi: 10.1093/molbev/msu107
Barshis D. J., Ladner J. T., Oliver T. A., Seneca F. O., Traylor-Knowles N., Palumbi S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U. S. A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Bay R. A., Palumbi S. R. (2017b). Transcriptome predictors of coral survival and growth in a highly variable environment. Ecol. Evol. 7, 4794–4803. doi: 10.1002/ece3.2685
Bay R. A., Rose N. H., Logan C. A., Palumbi S. R. (2017a). Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci. Adv. 3, e1701413. doi: 10.1126/sciadv.1701413
Bayer T., Aranda M., Sunagawa S., Yum L. K., Desalvo M. K., Lindquist E., et al. (2012). Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PloS One 7, e35269. doi: 10.1371/journal.pone.0035269
Bell G., Collins S. (2008). Adaptation, extinction and global change. Evol. Appl. 1, 3–16. doi: 10.1111/j.1752-4571.2007.00011.x
Bellantuono A. J., Granados-Cifuentes C., Miller D. J., Hoegh-Guldberg O., Rodriguez-Lanetty M. (2012). Coral thermal tolerance: tuning gene expression to resist thermal stress. PloS One 7, e50685. doi: 10.1371/journal.pone.0050685
Bellworthy J., Spangenberg J. E., Fine M. (2019). Feeding increases the number of offspring but decreases parental investment of red Sea coral. Ecol. Evol. 9, 12245–12258. doi: 10.1002/ece3.5712
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boutet E., Lieberherr D., Tognolli M., Schneider M., Bairoch A. (2007). UniProtKB/Swiss-prot. Methods Mol. Biol. 406, 89–112. doi: 10.1007/978-1-59745-535-0_4
Byler K. A., Carmi-Veal M., Fine M., Goulet T. L. (2013). Multiple symbiont acquisition strategies as an adaptive mechanism in the coral stylophora pistillata. PloS One 8, e59596. doi: 10.1371/journal.pone.0059596
Cantalapiedra C. P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38 (12), 5825–5829. doi: 10.1093/molbev/msab293
Cheverud J. M., Rutledge J. J., Atchley W. R. (1983). Quantitative genetics of development: genetic correlations among age-specific trait values and the evolution of ontogeny. Evolution 37, 895–905. doi: 10.1111/j.1558-5646.1983.tb05619.x
Conner J. K. (2012). Quantitative genetic approaches to evolutionary constraint: how useful?. Evolution 66, 3313–3320.
Cornwell B., Armstrong K., Walker N. S., Lippert M., Nestor V., Golbuu Y., et al. (2021). Widespread variation in heat tolerance and symbiont load are associated with growth tradeoffs in the coral acropora hyacinthus in Palau. Elife 10, e64790. doi: 10.7554/eLife.64790
Coronado-Zamora M., Salvador-Martínez I., Castellano D., Barbadilla A., Salazar-Ciudad I. (2019). Adaptation and conservation throughout the drosophila melanogaster life-cycle. Genome Biol. Evol. 11, 1463–1482. doi: 10.1093/gbe/evz086
Császár N. B. M., Ralph P. J., Frankham R., Berkelmans R., Van Oppen M. J. H. (2010). Estimating the potential for adaptation of corals to climate warming. PloS One 5, e9751. doi: 10.1371/journal.pone.0009751
Devens H. R., Davidson P. L., Deaker D. J., Smith K. E., Wray G. A., Byrne M. (2020). Ocean acidification induces distinct transcriptomic responses across life history stages of the sea urchin heliocidaris erythrogramma. Mol. Ecol. 29, 4618–4636. doi: 10.1111/mec.15664
Dixon G. B., Davies S. W., Aglyamova G. A., Meyer E., Bay L. K., Matz M. V. (2015). CORAL REEFS. genomic determinants of coral heat tolerance across latitudes. Science 348, 1460–1462. doi: 10.1126/science.1261224
Dixon G., Abbott E., Matz M. (2020). Meta-analysis of the coral environmental stress response: Acropora corals show opposing responses depending on stress intensity. Mol. Ecol. 29, 2855–2870.
Doropoulos C., Ward S., Roff G., González-Rivero M., Mumby P. J. (2015). Linking demographic processes of juvenile corals to benthic recovery trajectories in two common reef habitats. PloS One 10, e0128535. doi: 10.1371/journal.pone.0128535
Edge S. E., Morgan M. B., Gleason D. F., Snell T. W. (2005). Development of a coral cDNA array to examine gene expression profiles in montastraea faveolata exposed to environmental stress. Mar. pollut. Bull. 51, 507–523. doi: 10.1016/j.marpolbul.2005.07.007
Freda P. J., Alex J. T., Morgan T. J., Ragland G. J. (2017). Genetic decoupling of thermal hardiness across metamorphosis in drosophila melanogaster. Integr. Comp. Biol. 57, 999–1009. doi: 10.1093/icb/icx102
Ganot P., Moya A., Magnone V., Allemand D., Furla P., Sabourault C. (2011). Adaptations to endosymbiosis in a cnidarian-dinoflagellate association: differential gene expression and specific gene duplications. PloS Genet. 7, e1002187. doi: 10.1371/journal.pgen.1002187
Garg R., Shankar R., Thakkar B., Kudapa H., Krishnamurthy L., Mantri N., et al. (2016). Transcriptome analyses reveal genotype- and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci. Rep. 6, 19228. doi: 10.1038/srep19228
Hadfield J. D. (2010). MCMC methods for multi-response generalized linear mixed models: the MCMCglmm r package. J. Stat. Software 33, 1–22. doi: 10.18637/jss.v033.i02
Herrig D. K., Vertacnik K. L., Kohrs A. R., Linnen C. R. (2021). Support for the adaptive decoupling hypothesis from whole-transcriptome profiles of a hypermetamorphic and sexually dimorphic insect, neodiprion lecontei. Mol. Ecol. 30, 4551–4566. doi: 10.1111/mec.16041
Hoegh-Guldberg O. (1999). Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshw. Res. 50, 839–866. doi: 10.1071/MF99078
Honoré B. (2009). The rapidly expanding CREC protein family: members, localization, function, and role in disease. Bioessays 31, 262–277. doi: 10.1002/bies.200800186
Hughes T. P., Anderson K. D., Connolly S. R., Heron S. F., Kerry J. T., Lough J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Hughes T. P., Tanner J. E. (2000). Recruitment failure, life histories, and long-term decline of Caribbean corals. Ecology 81, 2250–2263. doi: 10.1890/0012-9658(2000)081[2250:RFLHAL]2.0.CO;2
Jombart T. (2008). Adegenet: a r package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Kauffmann A., Gentleman R., Huber W. (2009). arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics 25, 415–416. doi: 10.1093/bioinformatics/btn647
Kenkel C. D., Almanza A. T., Matz M. V. (2015a). Fine-scale environmental specialization of reef-building corals might be limiting reef recovery in the Florida keys. Ecology 96, 3197–3212. doi: 10.1890/14-2297.1
Kenkel C. D., Goodbody-Gringley G., Caillaud D., Davies S. W., Bartels E., Matz M. V. (2013a). Evidence for a host role in thermotolerance divergence between populations of the mustard hill coral (Porites astreoides) from different reef environments. Mol. Ecol. 22, 4335–4348. doi: 10.1111/mec.12391
Kenkel C. D., Matz M. V. (2016). Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1, 14. doi: 10.1038/s41559-016-0014
Kenkel C. D., Meyer E., Matz M. V. (2013b). Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22, 4322–4334. doi: 10.1111/mec.12390
Kenkel C. D., Setta S. P., Matz M. V. (2015b). Heritable differences in fitness-related traits among populations of the mustard hill coral, porites astreoides. Heredity 115, 509–516. doi: 10.1038/hdy.2015.52
Kitchen S. A., Crowder C. M., Poole A. Z., Weis V. M., Meyer E. (2015). De novo assembly and characterization of four anthozoan (Phylum cnidaria) transcriptomes. G3 5, 2441–2452. doi: 10.1534/g3.115.020164
Kopp C., Domart-Coulon I., Barthelemy D., Meibom A. (2016). Nutritional input from dinoflagellate symbionts in reef-building corals is minimal during planula larval life stage. Sci. Adv. 2, e1500681. doi: 10.1126/sciadv.1500681
Lesser M. P. (1996). Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in ymbiotic dinoflagellates. Limnol. Oceanogr. 41, 271–283. doi: 10.4319/lo.1996.41.2.0271
Liew Y. J., Howells E. J., Wang X., Michell C. T., Burt J. A., Idaghdour Y., et al. (2020). Intergenerational epigenetic inheritance in reef-building corals. Nat. Clim. Chang. 10, 254–259.
Liu S., Zenda T., Dong A., Yang Y., Wang N., Duan H. (2021). Global transcriptome and weighted gene co-expression network analyses of growth-stage-specific drought stress responses in maize. Front. Genet. 12, 645443. doi: 10.3389/fgene.2021.645443
Loeschcke V., Krebs R. A. (1996). Selection for heat-shock resistance in larval and in adult drosophila buzzatii: comparing direct and indirect responses. Evolution 50, 2354–2359. doi: 10.2307/2410704
Lohman B. K., Weber J. N., Bolnick D. I. (2016). Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol. Ecol. Resour. 16, 1315–1321. doi: 10.1111/1755-0998.12529
Louis Y. D., Bhagooli R., Kenkel C. D., Baker A. C., Dyall S. D. (2017). Gene expression biomarkers of heat stress in scleractinian corals: promises and limitations. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 191, 63–77. doi: 10.1016/j.cbpc.2016.08.007
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mansour T. A., Rosenthal J. J. C., Brown C. T., Roberson L. M. (2016). Transcriptome of the Caribbean stony coral porites astreoides from three developmental stages. Gigascience 5, 33. doi: 10.1186/s13742-016-0138-1
McLachlan R. H., Price J. T., Solomon S. L., Grottoli A. G. (2020). Thirty years of coral heat-stress experiments: a review of methods. Coral Reefs 39, 885–902. doi: 10.1007/s00338-020-01931-9
Meyer E., Aglyamova G. V., Matz M. V. (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol. Ecol. 20, 3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x
Moran N. A. (1994). ADAPTATION AND CONSTRAINT IN THE COMPLEX LIFE CYCLES OF ANIMALS. Annu. Rev. Ecol. Syst. 25, 573–600. doi: 10.1146/annurev.es.25.110194.003041
Muscatine L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals. ecosystems of the world. In: Coral reefs. Available at: https://ci.nii.ac.jp/naid/10020710664/ (Accessed March 11, 2022).
Orrenius S., Zhivotovsky B., Nicotera P. (2003). Regulation of cell death: the calcium–apoptosis link. Nat Rev Mol Cell Biol 4, 552–565. doi: 10.1038/nrm1150
Palumbi S. R., Barshis D. J., Traylor-Knowles N., Bay R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science 344, 895–898. doi: 10.1126/science.1251336
Parkinson J. E., Baumgarten S., Michell C. T., Baums I. B., LaJeunesse T. C., Voolstra C. R. (2016). Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus symbiodinium. Genome Biol. Evol. 8, 665–680. doi: 10.1093/gbe/evw019
Postma F. M., Ågren J. (2016). Early life stages contribute strongly to local adaptation in arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 113, 7590–7595. doi: 10.1073/pnas.1606303113
Purucker M., Bryan R., Amemiya K., Ely B., Shapiro L. (1982). Isolation of a Caulobacter gene cluster specifying flagellum production by using nonmotile Tn5 insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 79, 6797–6801. doi: 10.1073/pnas.79.22.6797
Putnam H. M., Gates R. D. (2015). Preconditioning in the reef-building coral pocillopora damicornis and the potential for trans-generational acclimatization in coral larvae under future climate change conditions. J. Exp. Biol. 218, 2365–2372. doi: 10.1242/jeb.123018
Quigley K. M., Warner P. A., Bay L. K., Willis B. L. (2018). Unexpected mixed-mode transmission and moderate genetic regulation of symbiodinium communities in a brooding coral. Heredity 121, 524–536. doi: 10.1038/s41437-018-0059-0
Reyes-Bermudez A., Villar-Briones A., Ramirez-Portilla C., Hidaka M., Mikheyev A. S. (2016). Developmental progression in the coral acropora digitifera is controlled by differential expression of distinct regulatory gene networks. Genome Biol. Evol. 8, 851–870. doi: 10.1093/gbe/evw042
Richmond R. H., Hunter C. L. (1990). Reproduction and recruitment of corals: comparisons among the Caribbean, the tropical pacific, and the red Sea. Mar. Ecol. Prog. series. Oldendorf 60, 185–203. doi: 10.3354/meps060185
Rivera H. E., Aichelman H. E., Fifer J. E., Kriefall N. G., Wuitchik D. M., Wuitchik S. J. S., et al. (2021). A framework for understanding gene expression plasticity and its influence on stress tolerance. Mol. Ecol. 30, 1381–1397. doi: 10.1111/mec.15820
Rose N. H., Seneca F. O., Palumbi S. R. (2015). Gene Networks in the Wild: Identifying Transcriptional Modules that Mediate Coral Resistance to Experimental Heat Stress. Genome Biol. Evol. 8, 243–252.
Rumble S. M., Lacroute P., Dalca A. V., Fiume M., Sidow A., Brudno M. (2009). SHRiMP: accurate mapping of short color-space reads. PloS Comput. Biol. 5, e1000386. doi: 10.1371/journal.pcbi.1000386
Schluter D., Price T. D., Rowe L., Grant P. R. (1991). Conflicting selection pressures and life history trade-offs. Proceedings of the Royal Society of London. Series B: Biological Sciences 246, 11–17.
Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., et al. (2005). A gene expression map of arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
Schneweis D. J., Whitfield A. E., Rotenberg D. (2017). Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in frankliniella occidentalis, the primary vector. Virology 500, 226–237. doi: 10.1016/j.virol.2016.10.009
Schwarz J. A., Brokstein P. B., Voolstra C., Terry A. Y., Manohar C. F., Miller D. J., et al. (2008). Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, acropora palmata and montastraea faveolata. BMC Genomics 9, 97. doi: 10.1186/1471-2164-9-97
Shore-Maggio A., Callahan S. M., Aeby G. S. (2018). Trade-offs in disease and bleaching susceptibility among two color morphs of the Hawaiian reef coral, montipora capitata. Coral Reefs 37, 507–517. doi: 10.1007/s00338-018-1675-0
Strong A. E., Liu G., Skirving W., Eakin C. M. (2011). NOAA’s coral reef watch program from satellite observations. Ann. GIS 17, 83–92. doi: 10.1080/19475683.2011.576266
Thornhill D. J., Fitt W. K., Schmidt G. W. (2006). Highly stable symbioses among western Atlantic brooding corals. Coral Reefs 25, 515–519. doi: 10.1007/s00338-006-0157-y
Traylor-Knowles N., Connelly M. T., Young B. D., Eaton K., Muller E. M., Paul V. J., et al. (2021). Gene expression response to stony coral tissue loss disease transmission in m. cavernosa and o. faveolata from Florida. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.681563
Underwood J. N., Smith L. D., Van Oppen M. J. H., Gilmour J. P. (2007). Multiple scales of genetic connectivity in a brooding coral on isolated reefs following catastrophic bleaching. Mol. Ecol. 16, 771–784. doi: 10.1111/j.1365-294X.2006.03187.x
van Oppen M. J. H., Oliver J. K., Putnam H. M., Gates R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U. S. A. 112, 2307–2313. doi: 10.1073/pnas.1422301112
Voolstra C. R., Schnetzer J., Peshkin L., Randall C. J., Szmant A. M., Medina M. (2009). Effects of temperature on gene expression in embryos of the coral montastraea faveolata. BMC Genomics 10, 627. doi: 10.1186/1471-2164-10-627
Voolstra C. R., Valenzuela J. J., Turkarslan S., Cárdenas A., Hume B. C. C., Perna G., et al. (2021). Contrasting heat stress response patterns of coral holobionts across the red Sea suggest distinct mechanisms of thermal tolerance. Mol. Ecol. 30, 4466–4480. doi: 10.1111/mec.16064
Wilbur H. M. (1980). Complex life cycles. Annu. Rev. Ecol. Syst. 11, 67–93. doi: 10.1146/annurev.es.11.110180.000435
Wong K. H., Goodbody-Gringley G., de Putron S. J., Becker D. M., Chequer A., Putnam H. M. (2021). Brooded coral offspring physiology depends on the combined effects of parental press and pulse thermal history. Glob. Change Biol. 27, 3179–3195. doi: 10.1111/gcb.15629
Wright R. M., Aglyamova G. V., Meyer E., Matz M. V. (2015). Gene expression associated with white syndromes in a reef building coral, acropora hyacinthus. BMC Genomics 16, 371. doi: 10.1186/s12864-015-1540-2
Keywords: gene expression, heritability, development, coral, heat stress, adaptation
Citation: Ruggeri M, Zhang Y, Aglyamova GV and Kenkel CD (2023) Divergent transcriptional response to thermal stress among life stages could constrain coral adaptation to climate change. Front. Mar. Sci. 10:1163552. doi: 10.3389/fmars.2023.1163552
Received: 10 February 2023; Accepted: 23 June 2023;
Published: 11 July 2023.
Edited by:
Charles Alan Jacoby, St. Johns River Water Management District, United StatesReviewed by:
Eva Majerová, University of Hawaii at Manoa, United StatesNia Symone Walker, Stanford University, United States
Copyright © 2023 Ruggeri, Zhang, Aglyamova and Kenkel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Ruggeri, bWFyaWFydWdnZXJpNTVAZ21haWwuY29t
 Maria Ruggeri
Maria Ruggeri Yingqi Zhang1
Yingqi Zhang1 Carly D. Kenkel
Carly D. Kenkel